Abstract
Introduction:
The gilthead sea bream (Sparus aurata) is a key species in European aquaculture, but its production is threatened by infectious diseases, including photobacteriosis caused by Photobacterium damselae subsp. piscicida (Phdp). This bacterial pathogen leads to high mortality, particularly in larvae and juveniles, and poses a significant challenge due to rising antibiotic resistance and the limited efficacy of vaccines in early developmental stages. Despite extensive research into Phdp virulence factors, the immune mechanisms in gilthead sea bream remain poorly understood.
Methods:
In this study, we conducted the first RNA-Seq analysis of Phdp-infected gilthead sea bream to characterize the immune response at 24 hours post-infection (hpi). We examined the expression of both coding genes and long non-coding RNAs (lncRNAs) in the head kidney and intestine.
Results:
Our findings revealed a robust immune response, particularly in the head kidney, characterized by significant modulation of genes involved in complement and coagulation cascades, iron metabolism, pathogen recognition, antigen presentation, inflammation and reactive oxygen species (ROS) production and detoxification, among others.
Discussion:
These results provide novel insights into the complex immune response of gilthead sea bream to Phdp and highlight the potential involvement of lncRNAs in modulating immune pathways. This lays the foundation for future research on host-pathogen interactions, which could support the development of preventive and control strategies for disease management, such as selective breeding for disease resistance, vaccine development, or host-derived alternatives to antibiotics.
1 Introduction
Gilthead sea bream (Sparus aurata) is one of the most important aquaculture finfish species in Europe, ranking fourth in European aquaculture production. In 2022, its production reached 344,000 tons, with a market value of 2.014 billion USD (FAO, 2025). However, the production of this Sparidae species can be substantially affected by various infectious diseases (Borrego et al., 2017), which, in turn, can significantly reduce sea bream productivity.
Among the bacterial diseases causing significant mortality in sea bream production, photobacteriosis (also known as fish pasteurellosis or pseudotuberculosis) is one of the most important. This septicemic bacterial disease, caused by the Gram-negative facultative intracellular bacterium Photobacterium damselae subsp. piscicida (thereafter Phdp), affects various marine fish species worldwide, leading to high mortality rates (Romalde, 2002; Andreoni and Magnani, 2014). Larvae and juveniles are particularly susceptible to the disease (Noya et al., 1995). Photobacteriosis often lacks obvious external signs. However, some fish may darken or show mild hemorrhages on the head and gills (Magariños et al., 1996a). Internally, the acute form causes minimal visible changes but leads to multifocal necrosis in the liver, kidney, and spleen, with bacterial accumulations (Magariños et al., 1996a). In the chronic stage, white tubercles (0.5–3.5 mm) may develop in internal organs, though granuloma formation varies by species (Magariños et al., 1996a).
Despite the identification of several virulence factors in this pathogen, its pathobiology remains largely unknown (Abushattal et al., 2020). However, it is now evident that its pathogenicity is linked to multiple virulence factors, including: a polysaccharide capsule that protects the bacterium from immune degradation (Magariños et al., 1996b; Arijo et al., 1998); the 56 kDa apoptotic-inducing protein (AIP56), an exotoxin that selectively triggers apoptosis in fish macrophages and neutrophils (Silva et al., 2010); extracellular products with phospholipase, cytotoxic, and hemolytic activities (Magariños et al., 1992); and the siderophore piscibactin, which enables iron acquisition from the host, ensuring bacterial survival and proliferation (Magariños et al., 1994).
Early diagnosis of Phdp is crucial for implementing appropriate measures to prevent both horizontal and vertical transmission of the disease (Magariños et al., 1995; Romalde et al., 1999). Commercial vaccines against this bacterium are currently available (Miccoli et al., 2019). However, the higher susceptibility of early developmental stages makes vaccination alone insufficient as a control strategy. As a result, antibiotics remain the first-line treatment for managing photobacteriosis outbreaks. Nevertheless, antibiotic resistance has been reported in Phdp isolates from different countries (Thyssen and Ollevier, 2001; Kawanishi et al., 2006; Martínez-Manzanares et al., 2008; Laganà et al., 2011; Zhou et al., 2025), which is a growing concern and further complicates efforts to manage photobacteriosis outbreaks. Additionally, antibiotic residues pose risks to both consumer health (Ljubojević Pelić et al., 2024) and the environment (Kraemer et al., 2019). These challenges highlight the urgent need to develop new, highly effective preventive and curative strategies.
A deeper insight into immune responses at the molecular level is crucial for elucidating host–pathogen interactions and for guiding the development of effective disease control measures in aquaculture. In this context, elucidating the immune mechanisms activated in gilthead sea bream following Phdp infection is highly relevant, as it could enable the development of new antibacterial strategies or selective breeding programs targeting resistance-associated genes. However, information regarding the immune response and resistance markers against this pathogen remains limited. Some studies have shown that exposure to formalin-inactivated Phdp increases antiprotease activity, lysozyme activity, and immunoglobulin production (Hanif et al., 2005). The rapid activation of the nitric oxide response has been proposed as a protective mechanism in juvenile gilthead sea bream against photobacteriosis (Acosta et al., 2004). Vallecillos et al. (2021) also reported that higher serum peroxidase activity under naïve conditions correlates with improved survival following experimental Phdp infection. Both nitric oxide and peroxidase contribute to the oxidative burst, a key antimicrobial defense mechanism in fish, by generating reactive oxygen and nitrogen species that help neutralize pathogens during the early stages of infection. Furthermore, exposure to UV-inactivated Phdp has been shown to increase the expression of pro-inflammatory genes such as interleukin-1 beta, tumor necrosis factor-alpha, and cyclooxygenase-2 in the sea bream liver (Grasso et al., 2015). Short-term immune responses to Phdp in sea bream have also been assessed by Santos et al. (2022), who found that infection induced anemia, neutrophilia, and monocytosis, along with altered expression of pro-inflammatory, anti-inflammatory, and other immune-related genes in the head kidney. Moreover, plasma protease activity, lipid peroxidation, and the activity of glutathione-S-transferase and catalase all increased (Santos et al., 2022). The most comprehensive gene expression analysis of gilthead sea bream infected with Phdp to date was conducted by Pellizzari et al. (2013), who used microarray technology to analyze head kidney samples at 24 and 48 hpi. According to their findings, the authors suggest that Phdp induces an anti-inflammatory response and alternatively activates macrophages, either as a host strategy to minimize tissue damage from excessive inflammation or as a mechanism for Phdp to evade immune defenses.
In this study, we conducted the first RNA-Seq analysis of Phdp-infected gilthead sea bream to gain insights into the immune response of this teleost to Phdp. We examined the expression of both coding genes and long non-coding RNAs (lncRNAs) in the head kidney and intestine at 24 hpi. Our results revealed a strong activation of the immune response, particularly in the head kidney, characterized by a marked overexpression of numerous genes involved in the complement and coagulation cascades, as well as the modulation of several genes encoding proteins associated with iron metabolism (including various hepcidin genes), pathogen recognition, antigen processing and presentation, inflammation, and the synthesis and detoxification of reactive oxygen species (ROS), among others.
Also, numerous lncRNAs were modulated in the head kidney. LncRNAs are a subclass of non-coding RNAs (ncRNAs) that are longer than 200 nucleotides and do not encode proteins. Despite lacking protein-coding potential, lncRNAs play a crucial regulatory role in the immune response (Aune and Spurlock, 2016; Elling et al., 2016). They can modulate gene expression either in cis, by regulating adjacent genes, or in trans, by influencing genes located at distant genomic regions, even on different chromosomes (Ponting et al., 2009). LncRNAs mediate gene activation or repression through various mechanisms, including chromatin remodeling, promoter inactivation, transcriptional interference, recruitment or transport of transcription factors, and epigenetic modifications (Ponting et al., 2009). Since most lncRNAs function primarily in cis, their roles in fish are often inferred based on the functions of neighboring protein-coding genes (Wang et al., 2018). In this study, we observed that several key processes and genes regulated following Phdp infection appear to be tightly controlled by lncRNAs.
2 Materials and methods
2.1 Gilthead sea bream and bacteria
Juvenile specimens of gilthead sea bream (mean length: 8.28 ± 0.83 cm; mean weight: 8.38 ± 2.39 g) were sourced from Grupo Culmarex (Aquicultura Balear, Palma de Mallorca, Spain). The fish were housed in 500 L fiberglass tanks within a recirculating saline water system (salinity ~35 g/L) under a 12-hour light/12-hour dark photoperiod at 25 °C. Before initiating the experiments, they underwent a two-week acclimatization period under laboratory conditions. They were fed daily to satiety with the commercial dry diet Vita 1 (Veronesi, Verona, Italy) at a rate of 2% of their body weight per day. For injection procedures, fish were anesthetized with MS-222 (0.24 mg/mL), while euthanasia for sampling and at the conclusion of survival trials was carried out using an overdose of MS-222 (5 mg/mL). All procedures related to fish handling and challenge experiments were evaluated and approved by the CSIC National Committee on Bioethics (approval number ES360570202001/21/FUN.01/INM06/BNG01).
The pathogenic bacterium Photobacterium damselae subsp. piscicida (thereafter Phdp) strain IA 8.1, generously provided by Dr. Beatriz Magariños (Universidad de Santiago de Compostela, Spain), was cultured overnight on tryptic soy agar (TSA) supplemented with 1% NaCl at 22 °C. Just before injection, a bacterial suspension was prepared in phosphate-buffered saline (PBS), and its final concentration was determined by quantifying colony-forming units (CFU) through 10-fold serial dilutions plated on TSA with 1% NaCl.
2.2 Infection, sampling and survival analysis
A schematic overview of the experimental procedure is presented in Figure 1A. A total of 92 gilthead sea bream were distributed across four tanks: three tanks containing 22 fish each and one tank with 26 fish. Fish in the three tanks with 22 individuals were intraperitoneally (i.p.) injected with 100 µL of a Phdp (2 x 107 CFU/mL). In contrast, the 26 fish in the remaining tank received the same volume of PBS and served as uninfected controls. At 24 hours post-infection (hpi), two fish from each infected tank and six from the control tank were sampled, yielding a total of six individuals per condition. The head kidney and intestine were dissected and stored individually at -80 °C until RNA extraction. The remaining fish (n = 20 per tank; three infected and one control tank) were monitored for mortality over 21 days post-infection. Survival curves were plotted using the Kaplan–Meier method, and differences between groups were assessed using the log-rank (Mantel–Cox) test. The analysis was performed using GraphPad Prism version 9, and fish that survived until the end of the trial were considered censored observations.
Figure 1
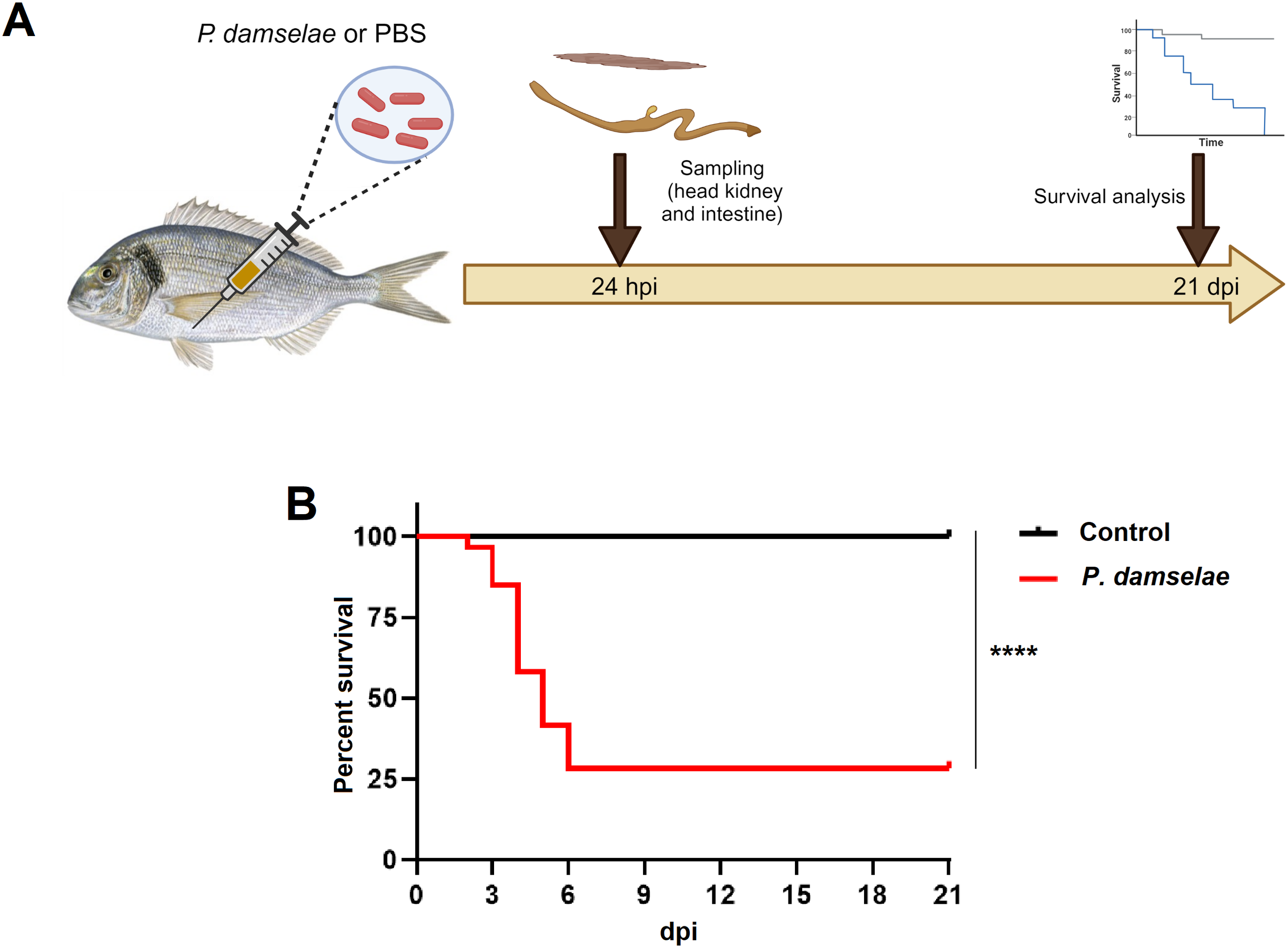
Evaluation of the effect of the intraperitoneal injection of Photobacterium damselae subsp. piscicida in juvenile gilthead seabream. (A) Schematic representation of the experimental design carried out. Head kidney and intestine samples were collected at 24 hpi for RNA-Seq analysis, and survival was monitored for 21 days following the challenge. Figure created with BioRender.com. (B) Kaplan-Meier survival curves of the control and P. damselae-infected fish. (****, p-value < 0.0001).
2.3 RNA isolation and transcriptome sequencing
Total RNA from head kidney and intestine samples obtained from the control and Phdp-infected sea bream was isolated using the Maxwell RSC simplyRNA Tissue kit (Promega) with an automated Maxwell® RSC 48 Instrument in accordance with the instructions provided by the manufacturer. The quantity of RNA was measured with a NanoDrop ND-1000 (NanoDrop Technologies, Inc.); afterwards, RNA integrity was analysed in an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer’s instructions. All samples passed the quality control tests (RIN ≥ 8); however, the four samples per tissue and condition with the highest quality scores were selected for library construction and transcriptome sequencing. This sample size is generally considered sufficient to detect meaningful patterns of differential gene expression when aiming to identify general transcriptional responses of a species to a pathogen challenge.
Double-stranded cDNA libraries were constructed using TruSeq Stranded mRNA LT Sample (Illumina, San Diego, CA, USA). Paired-end 150 bp (PE150) sequencing was performed on an Illumina NovaSeq 6000 sequencer. Both library preparation and sequencing were performed at Macrogen, Inc. (Seoul, Republic of Korea). The raw read sequences obtained were deposited in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under BioProject accession number PRJNA1240102.
2.4 Trimming, RNA-Seq and differential expression analysis
RNA-Seq data were processed using CLC Genomics Workbench v. 22.0 (CLC Bio, Aarhus, Denmark) for read filtering, trimming, and mapping to the gilthead sea bream genome (Version fSpaAur1.1; BioProject number PRJEB31901). Raw reads underwent trimming to remove adaptor sequences and low-quality reads, with a quality score threshold set at 0.05. Mapping was performed using the gilthead sea bream genome with the following parameters: length fraction = 0.8, similarity fraction = 0.8, mismatch cost = 2, insertion cost = 3, and deletion cost = 3. Gene expression was quantified as transcripts per million (TPM). Differential expression analysis was conducted to identify differentially expressed genes (DEGs), with genes retained for further analysis if they exhibited fold changes (FC) > |2| and False Discovery Rate (FDR)-adjusted p-values < 0.01.
2.5 Identification of long noncoding RNAs and their neighbouring coding genes
To identify the lncRNAs modulated in the head kidney and the intestine at 24 hpi with Phdp, we followed a pipeline previously carried out in our group for other fish species infected with different pathogens (Valenzuela-Muñoz et al., 2019; Pereiro et al., 2020a; Romero et al., 2024). For this, a sea bream reference transcriptome was constructed by de novo assembly of the trimmed reads using all the samples and with an overlap criterion of 70% and a similarity of 0.9 to exclude paralogous sequence variants. The assembly settings were set as follows: mismatch cost = 2, deletion cost = 3, insert cost = 3, and a minimum contig length = 200 base pairs. The resulting contigs were annotated using BLASTx (e-value < 1 × 10−5) against the gilthead sea bream genome peptide database, as well as the protein databases for all bony fish species available in NCBI GenBank and UniProt/Swiss-Prot. Contigs with a significant match were subsequently removed. Non-annotated contigs longer than 200 bp were retrieved from the de novo assembly, and reads were mapped to those contigs to retain only those with an average coverage >50. For the final filtering of putative lncRNAs, contigs were selected after discarding all potential coding sequences using the “Find ORF” tool in CLC Genomics Workbench v. 22.0, as well as those contigs exhibiting significant coding potential as estimated by the Coding Potential Assessment Tool (CPAT; http://lilab.research.bcm.edu/; Wang et al., 2013). The contigs that passed all the filters were considered putative lncRNAs and retained for further analyses. RNA-Seq and differential expression analysis of the potential lncRNAs were conducted using the methodology described above for the coding transcripts.
To determine the position of the differentially expressed (DE) lncRNAs on the gilthead sea bream genome, they were mapped to the gilthead sea bream using the following parameters: length fraction = 0.8, similarity fraction = 0.8, mismatch cost = 2, insertion cost = 3 and deletion cost = 3. The flanking coding genes of the differentially expressed lncRNAs were retrieved 10,000 bp up and downstream of each lncRNA. To analyze the correlation between lncRNAs and their flanking genes, a Shapiro–Wilk normality test was performed to select the most appropriate correlation coefficient. Neither of the 2 variables followed a normal distribution. Therefore, we performed a Spearman’s correlation analysis to evaluate the adjustment of the TPM values of the DE lncRNAs and their neighbouring DEGs.
2.6 Bioinformatic analyses
Venn diagrams were constructed with the Venny 2.1 tool (http://bioinfogp.cnb.csic.es/tools/venny/. Using the TPM values of the selected DEGs or DE lncRNAs, heatmaps were constructed using the average linkage method with Euclidean distance in the Clustvis web tool (Metsalu and Vilo, 2015; https://biit.cs.ut.ee/clustvis/). PCA plots were also constructed with Clustvis using the TPM values of all the coding transcripts and lncRNAs predicted for gilthead sea bream.
Gene Ontology (GO) enrichment analyses of the DEGs were performed in OmicsBox v1.3.11 (https://www.biobam.com/omicsbox) using Fisher’s exact test enrichment analysis and a false discovery rate (FDR) ≤ 0.05. Only the 30 biological process terms most significantly enriched were represented. The KEGG Mapper tool (Kanehisa and Sato, 2020) was used to identify DEGs encoding proteins involved in pathways cataloged in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
2.7 cDNA synthesis and RNA-Seq validation by quantitative PCR
RNA from the same samples used in the RNA-Seq experiment was reverse-transcribed into cDNA using the NZY First-Strand cDNA Synthesis Kit (NZYtech), following the manufacturer’s protocol and employing 0.5 μg of input RNA per reaction. To validate the RNA-Seq findings, we performed qPCR on selected immune-related genes (complement component c7 –c7–, fibrinogen gamma chain –fgg–, serotransferrin –tf–, melanotransferrin –meltf–, toll-like receptor 5 –tlr5–, D-amino acid oxidase –dao–, interleukin-1 beta –il1b-) that showed significant regulation in both the head kidney and intestine. Primer specificity and amplification efficiency were assessed following the approach described by Pfaffl (2001). Each qPCR reaction was carried out in a final volume of 25 μL, comprising 12.5 μL of SYBR GREEN PCR Master Mix (Applied Biosystems), 10.5 μL of ultrapure water (Sigma–Aldrich), 0.5 μL of each primer (10 μM), and 1 μL of the synthesized cDNA. Reactions were performed in technical triplicates using a 7300 Real-Time PCR System (Applied Biosystems) under the following cycling conditions: initial denaturation at 95 °C for 10 minutes, followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute for annealing and extension. Gene expression levels were normalized to the 18s rRNA gene using the Pfaffl method (Pfaffl, 2001). Fold changes were determined by comparing the normalized expression values in infected samples to those in the respective control groups. The sequences of all primers used in this study are provided in Supplementary Table S1.
The fold-changes obtained in the RNA-Seq analysis for those seven genes in the head kidney and the intestines were compared to those obtained through qPCR. A Pearson’s correlation analysis was carried out with the GraphPad Prism version 9 to estimate the degree of concordance between both methods.
3 Results
3.1 Survival and overall transcriptome modulation in gilthead sea bream after P. damselae subsp. piscicida infection
The infection of juvenile sea bream with the pathogenic bacterium Phdp resulted in a survival rate of 28.33% after 21 days of mortality monitoring. In contrast, the uninfected control group maintained a 100% survival rate (Figure 1B). Therefore, these results indicate that the infection was highly lethal for the fish.
To elucidate the impact of the infection at the transcriptome level, RNA-Seq analysis of head kidney and intestine samples was conducted at 24 hpi. A summary of the number of raw reads, high-quality reads after trimming and mapping results is shown in Supplementary Table S2. The PCA of both tissues revealed a clear distinction between samples from infected fish and those from control individuals (Figure 2A), highlighting significant transcriptional differences between the groups.
Figure 2
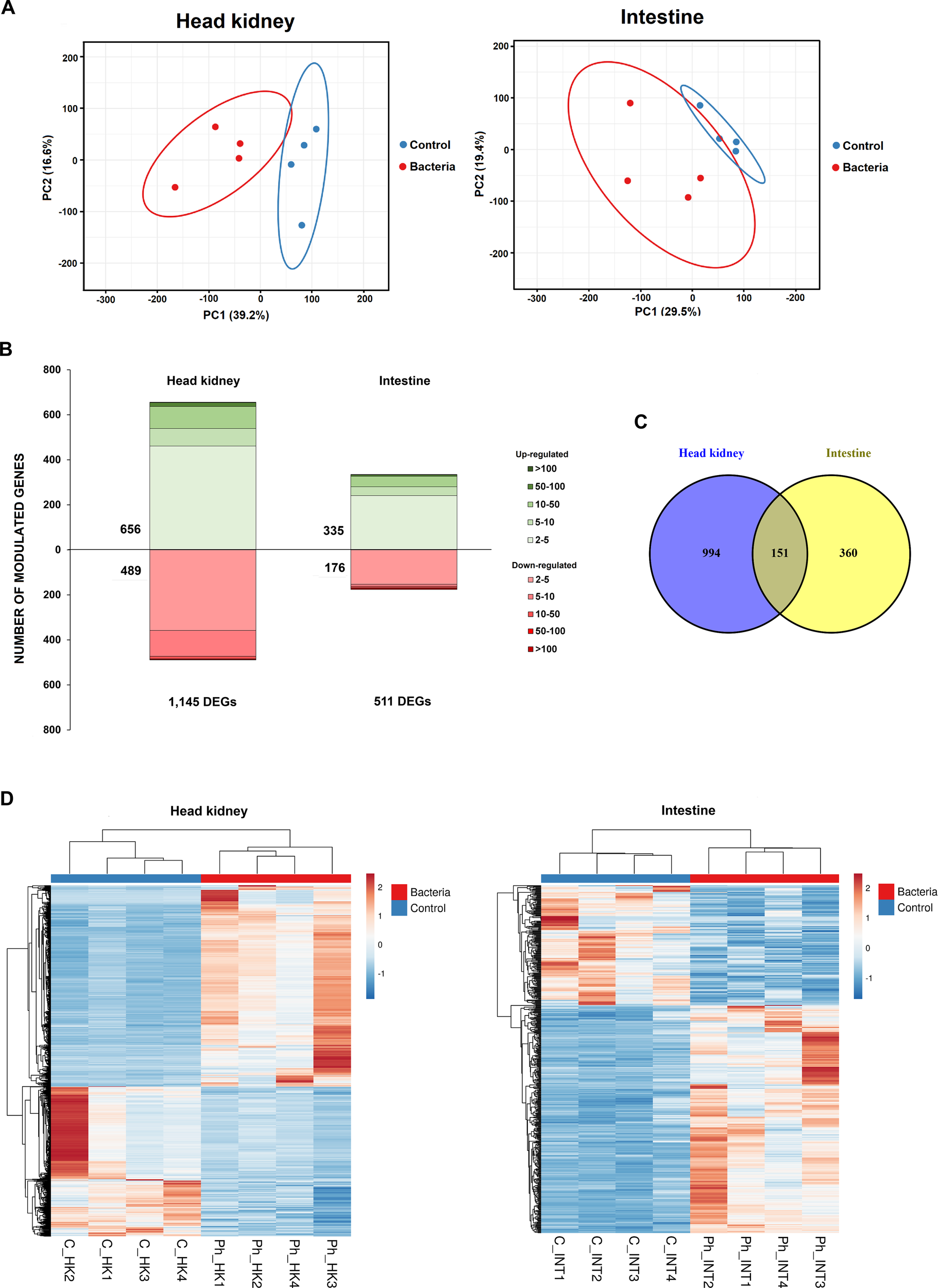
Overall transcriptome response of juvenile sea bream at 24 hpi with Phdp. (A) Principal component analysis (PCA) of head kidney and intestine samples from Phdp-infected and control individuals. (B) Stacked column charts representing the number and intensity (in fold-change value) of the DEGs between the infected and the control samples. (C) Venn diagram showing the number of shared and exclusive DEGs in response to infection with Phdp (24 hpi) between both tissues. (D) Heatmaps showing the expression (TPM Values) of DEGs in the head kidney and intestine.
The differential expression analysis identified 1,145 DEGs in the head kidney (656 up-regulated and 489 down-regulated) and 511 DEGs in the intestine (225 up-regulated and 176 down-regulated), based on a fold change (FC) > |2| and FDR-adjusted p-values < 0.01 (Figure 2B; Supplementary File S1). As shown in the corresponding heatmaps constructed with TPM values, the DEGs exhibited a consistent expression pattern across the biological replicates (Figure 2D). Among the DEGs, 151 were commonly affected in both the head kidney and intestine (Figure 2C), with most of these genes involved in immune response. The complete list of DEGs is provided in Supplementary File S1, while Supplementary File S2 contains the 151 DEGs that are commonly modulated in both tissues.
Validation of the RNA-Seq results by qPCR revealed a strong positive Pearson’s correlation (r = 0.884), underscoring the robustness and reliability of the RNA-Seq data (Supplementary Figure S1).
3.2 GO enrichment analyses of the DEGs
In order to investigate the biological processes primarily affected by the DEGs after infection with Phdp, GO enrichment analyses were conducted (Figure 3). In the head kidney, a strong representation of immune terms was found to be highly enriched, such as those related to the antigen processing and presentation (e.g. ‘peptide antigen transport’, ‘positive regulation of antigen processing and presentation of peptide antigen via MHC class I’ or ‘antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent’), complement system (‘regulation of complement activation, alternative pathway’ or ‘complement-dependent cytotoxicity’), among other immune terms (‘positive regulation of type IIa hypersensitivity’, ‘positive regulation of apoptotic process in another organism’, ‘defense response to protozoan’, ‘cellular response to interleukin-1’ or ‘antimicrobial humoral response’) (Figure 3A). It is also worth mentioning the high representation of GO terms related to metabolism, particularly amino acid metabolism and lipid metabolism or iron homeostasis, among others. On the other hand, immune terms were also enriched in the intestine, such as ‘opsonization’, ‘antimicrobial humoral immune response mediated by antimicrobial peptide’, ‘positive regulation of chemokine production’, ‘positive regulation of humoral immune response’, ‘cellular response to interleukin-1’, ‘positive regulation of inflammatory response’ or ‘cellular response to lipopolysaccharide’. In this tissue, terms involved in different metabolic processes were also observed, including the term also enriched in the head kidney ‘intracellular iron homeostasis’ (Figure 3A). Indeed, the GO enrichment analysis of the 151 genes commonly affected in both tissues also showed a strong enrichment of terms with a role in iron homeostasis (‘iron ion export across plasma membrane’, ‘intracellular iron homeostasis’), but also in several immune terms (‘modulation of apoptotic process in another organism’, ‘antimicrobial humoral response’, ‘cellular response to interleukin-1’, ‘regulation of humoral immune response’, ‘defense response to Gram-negative bacterium’, ‘defense response to Gram-positive bacterium’, or ‘cellular response to lipopolysaccharide’ (Figure 3B).
Figure 3
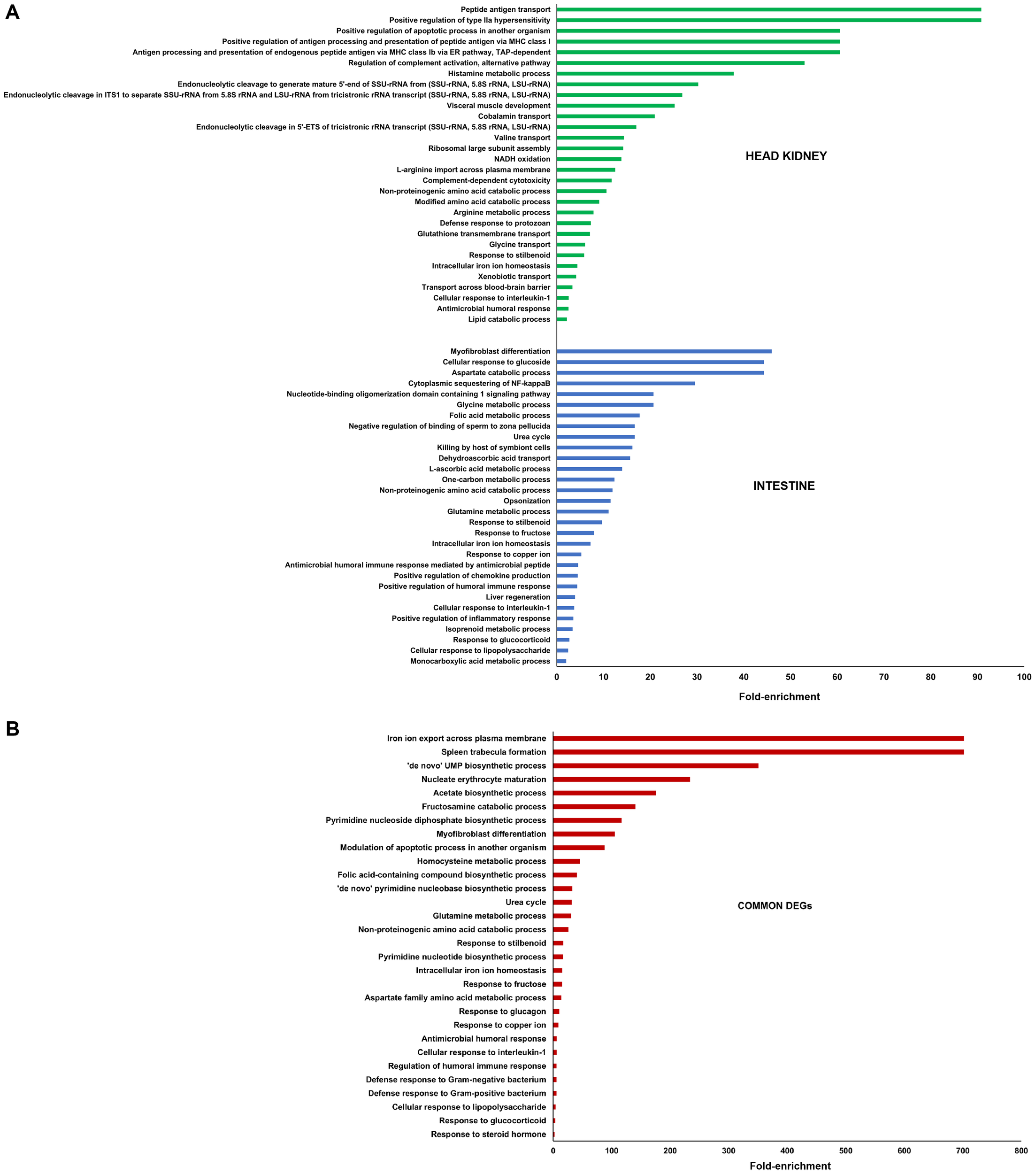
GO enrichment analysis (biological processes) of (A) the DEGs in head kidney and intestine samples between Phdp-infected and control sea bream, and (B) the DEGs commonly modulated in both tissues. Only the 30 most significantly enriched terms are represented.
3.3 Main immune mechanisms affected by the infection
3.3.1 Complement and coagulation cascades
A glance at the 60 most overexpressed DEGs in the head kidney following Phdp infection reveals a significant number of genes associated with the complement and coagulation pathways, two closely interconnected processes (Supplementary File S1). The KEGG mapper tool further indicated that ‘Complement and coagulation cascades’ was the immune KEGG pathway most impacted by the infection in the head kidney, with the highest number of modulated genes (Figure 4A). This suggests that it is likely the immune mechanism most affected by the infection in this tissue, both in terms of gene number and intensity. The expression levels of all DEGs involved in these pathways were visualized in a heatmap, which highlighted a strong and widespread induction at 24 hpi in the head kidney (Figure 4B). While to a lesser extent, some genes in these pathways were also modulated in the intestine (Figure 4C). Notably, seven genes encoding different isoforms of the central complement component C3 were induced by the bacterial challenge in the head kidney, whereas only two were affected in the intestine (Figure 4D). However, in the intestine, a gene encoding a C3 component that was overexpressed in the head kidney (FC = 21.58) exhibited the opposite response, being inhibited after infection (FC = -2.93) (Figure 4D).
Figure 4
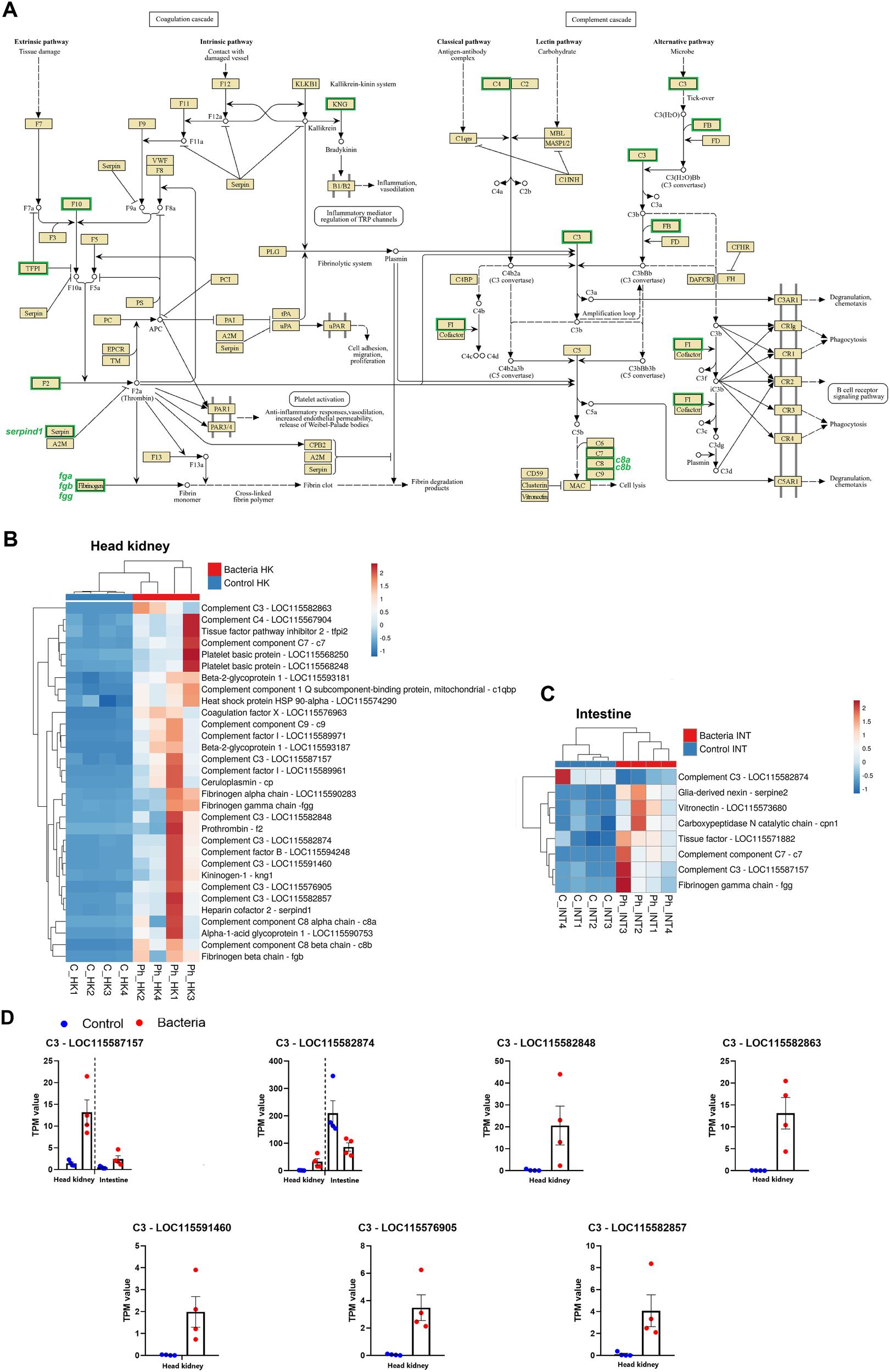
Impact of Phdp infection on the complement and coagulation cascades in juvenile gilthead sea bream. (A) Schematic representation of complement- and coagulation-related genes modulated in the head kidney at 24 hpi. (B) Heatmap showing the expression (TPM values) of complement- and coagulation-related genes in the head kidney. (C) Heatmap showing the expression (TPM values) of these genes in the intestine. (D) Expression patterns of differentially expressed C3 complement component genes in both tissues following bacterial challenge.
3.3.2 Hepcidins and other iron homeostasis-related genes
Iron homeostasis is crucial during bacterial infections, as it regulates iron availability—a key nutrient for bacterial pathogens. Meanwhile, hepcidins not only play a vital role in iron homeostasis but also exhibit direct antimicrobial properties, as they are classified as antimicrobial peptides (AMPs). The expression of several genes related to iron homeostasis was altered following infection, both in the head kidney (Figure 5A) and in the intestine (Figure 5B). In the head kidney, a significant induction was observed for the genes ferritin middle subunit, metalloreductase STEAP4 (steap4), hephaestin-like protein 1 (hephl1), natural resistance-associated macrophage protein 2 (= solute carrier family 11 member 2, slc11a2), haptoglobin (hp), transferrin receptor protein 1 (tfrc), ferric-chelate reductase 1 (frrs1), serotransferrin (tf), and ceruloplasmin (cp), all encoding for proteins involved in iron transport and regulation (Figure 5A). In contrast, two genes encoding for solute carrier family 40 member 1 (slc40a1), melanotransferrin (meltf), heme transporter HRG1 (= solute carrier family 48 member 1, slc48a1) and three different hemoglobin subunits were inhibited (Figure 5A). These proteins play key roles in iron transport, which may be downregulated to limit iron availability to pathogens. In the intestine, although a lower number of genes was affected, an overexpression of steap4, tfrc, tf and hp was also observed, together with the overexpression of heme oxygenase (hmox1), whereas meltf was also inhibited in this tissue, together with sideroflexin-1 (sfxn1) (Figure 5B). Regarding hepcidins, 9 isoforms were affected by the challenge in the head kidney (Figures 5A, C), and six in the intestine (Figures 5B, C) with some being upregulated and others downregulated after infection.
Figure 5
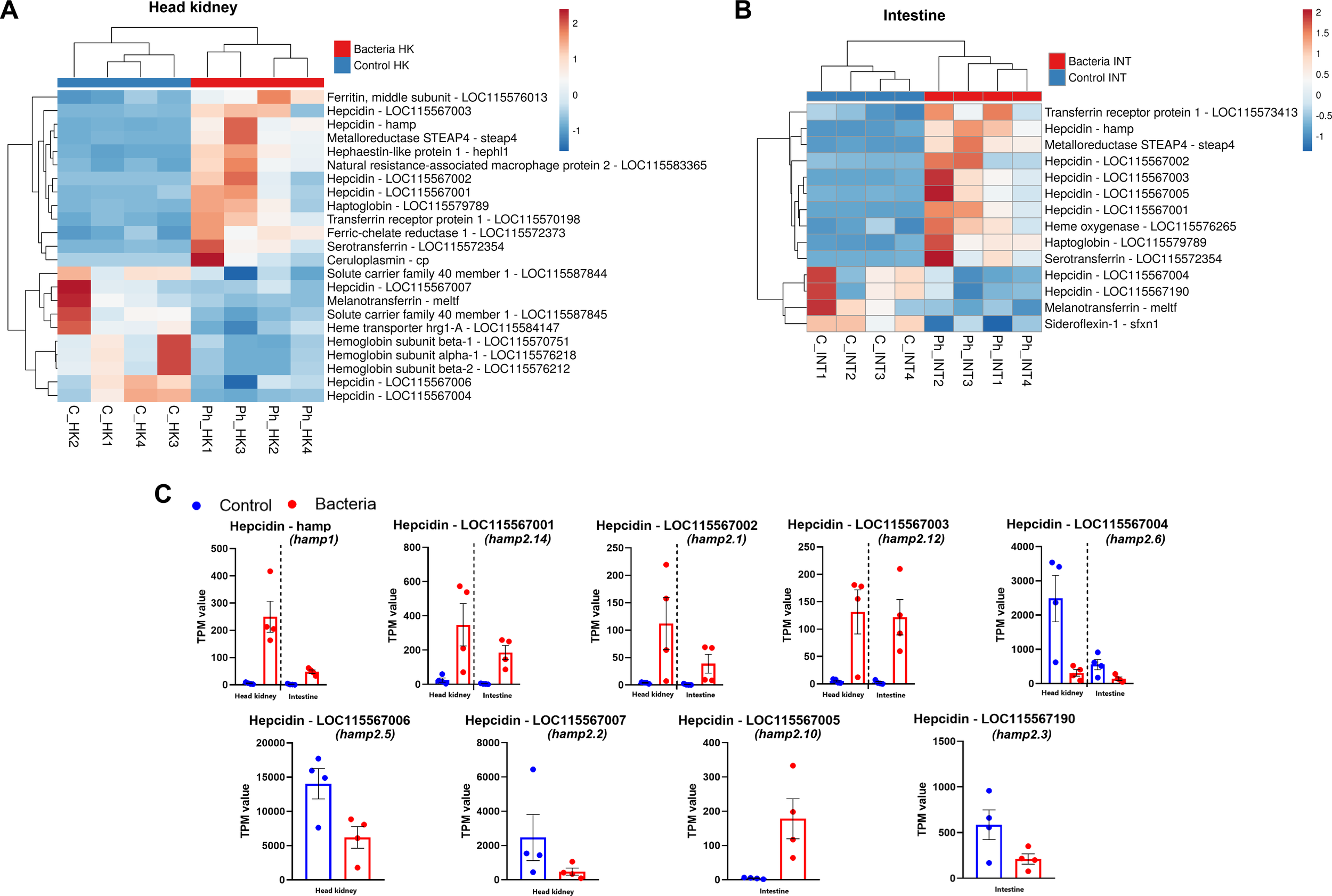
Phdp infection alters the expression of numerous genes involved in iron homeostasis. (A) Heatmap showing the expression (TPM values) of iron homeostasis-related DEGs in the head kidney. (B) Heatmap showing the expression (TPM values) of these genes in the intestine. (C) Expression patterns of differentially expressed hepcidin-like genes in both tissues following bacterial challenge. The nomenclature established by Serna-Duque et al. (2022a) is included in parentheses.
3.3.3 Pathogen recognition
A wide variety of genes encoding pathogen recognition receptors (PRRs) were found to be modulated at 24 hpi with Phdp, especially in the head kidney (Figure 6A). Among these, three genes encoding Toll-like receptors (TLRs) were overexpressed: tlr1, tlr5, and tlr13, whereas only tlr5 was significantly induced in the intestine (Figure 6B). Undoubtedly, the most widely affected type of PRRs by bacterial infection were lectins. Various genes encoding C-type lectins, F-type lectins, galectins, rhamnose-binding lectins, and sialic acid-binding lectins were affected by the challenge in both tissues (Figures 6A, B). Interestingly, while some were strongly overexpressed, others were downregulated. For example, the genes annotated as C-type lectin domain family 4 member D and C-type lectin domain family 4 member G were induced in the head kidney. In contrast, the infection inhibited those annotated as C-type lectin domain family 4 member M, C-type lectin domain family 10 member A, and C-type lectin domain family 12 member B (Figure 6A). Another example is the overexpression of galectin-3 and the downregulation of galectin-4 genes in the intestine (Figure 6B). Among other regulations, two genes encoding the macrophage receptor MARCO, a scavenger receptor involved in the recognition and uptake of pathogens, were found to be inhibited in the head kidney (Figure 6A).
Figure 6
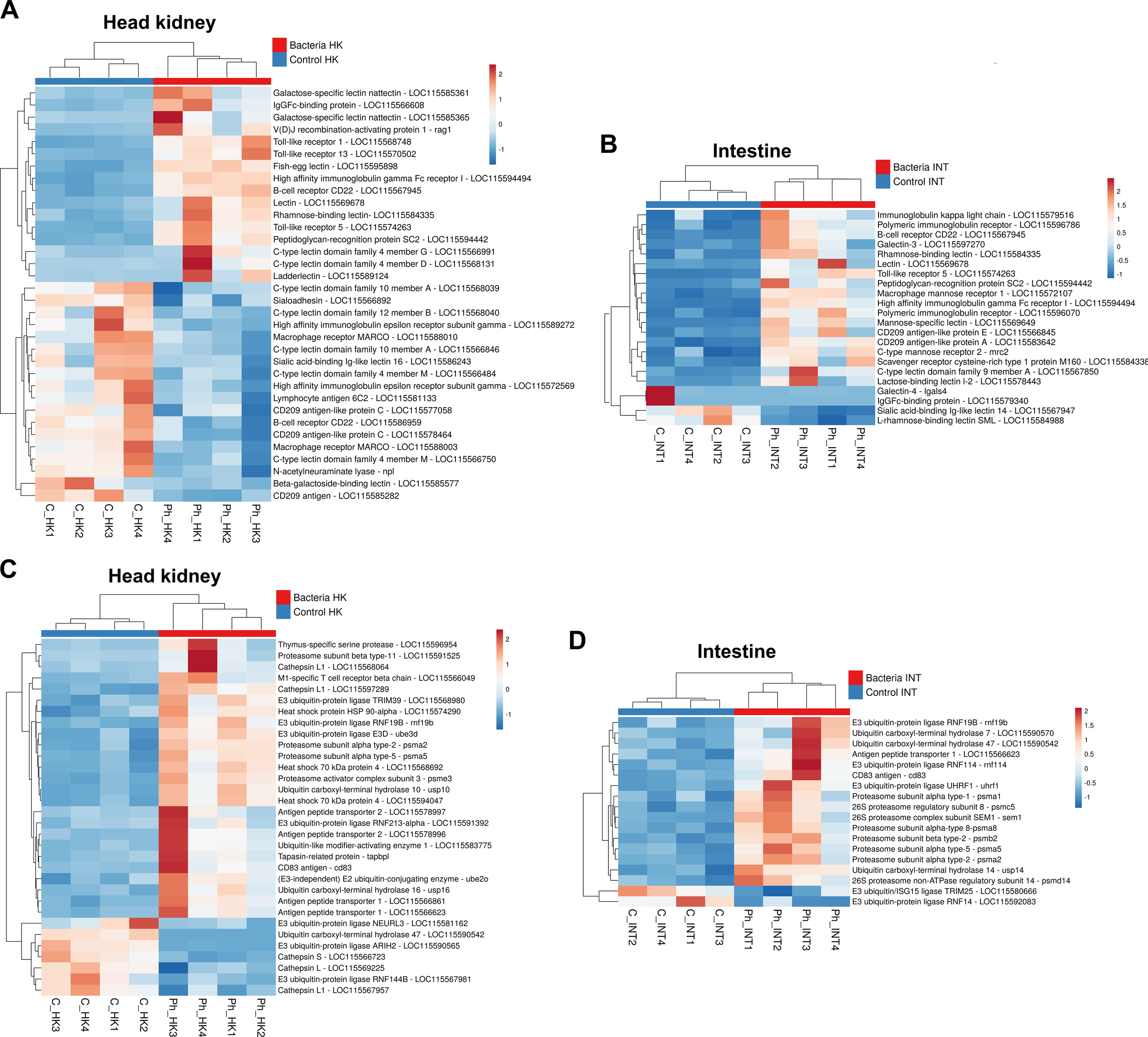
Genes involved in pathogen recognition and antigen presentation are broadly modulated by Phdp. (A) Heatmaps showing the expression (TPM values) of various pattern recognition receptors (PRRs) and other molecules involved in pathogen recognition and degradation in the head kidney, and (B) in the intestine. (C) Heatmaps illustrating the expression (TPM values) of DEGs involved in antigen presentation in the head kidney and (D) in the intestine.
Certain immunoglobulins and immunoglobulin receptors were also modulated in both tissues (Figures 6A, B), as was the gene V(D)J recombination-activating protein 1 (rag1), which is responsible for creating the diversity of antibodies and T-cell receptors, which was induced in the head kidney (Figure 6A).
3.3.4 Antigen presentation
In line with the GO enrichment of multiple terms related to antigen processing and presentation in the head kidney, a broad repertoire of DEGs associated with this process was identified in this tissue (Figure 6C), with a more limited presence in the intestine (Figure 6D). Among these DEGs, several ubiquitination-related proteins, primarily E3 ubiquitin-protein ligases, ubiquitin carboxyl-terminal hydrolases, and various 26S proteasome subunits, were observed. Additionally, genes involved in the peptide-loading complex, such as antigen peptide transporter 1 (tap1), antigen peptide transporter 2 (tap2), and tapasin-related protein (tapbpl), were differentially expressed. Notably, tap1 was significantly overexpressed in both tissues, whereas tap2 and tapbpl were induced exclusively in the head kidney (Figures 6C, D).
Furthermore, genes encoding lysosomal proteases, specifically cathepsins, which are involved in the degradation of extracellular antigens in endolysosomes, were also modulated in the head kidney at 24 hpi (Figure 6C). Interestingly, different cathepsin L1 isoforms exhibited opposite expression patterns, while cathepsin S was notably downregulated, emphasizing the functional diversification of these proteases.
Heat shock proteins (HSPs), particularly HSP70 and HSP90, play crucial roles in antigen processing and presentation within the major histocompatibility complex (MHC) class I and II pathways. The genes encoding these HSPs were significantly upregulated in the head kidney at 24 hpi (Figure 6C). Additionally, other upregulated genes involved in antigen presentation in the head kidney included those encoding CD83 antigen (cd83), thymus-specific serine protease (prss16), and M1-specific T cell receptor beta chain (trb), further underscoring the activation of immune-related pathways in response to infection.
3.3.5 Cytokines and cytokine receptors
As expected, numerous cytokines and their associated receptors were modulated following the challenge with Phdp in both the head kidney (Figure 7A) and intestine (Figure 7B). Among these, several CXC chemokines and CC chemokines were induced by the bacteria, with the exception of a gene annotated as C-C motif chemokine 3, which was downregulated in the head kidney. The pro-inflammatory cytokine interleukin-1β (il1b) was significantly induced in both tissues, as was the interleukin-27 subunit beta (ebi3) gene, which encodes a cytokine involved in both pro-inflammatory and anti-inflammatory responses (Figures 7A, B). Additionally, the genes interleukin-6 (il6) and interferon-gamma (ifng) were overexpressed in the head kidney.
Figure 7
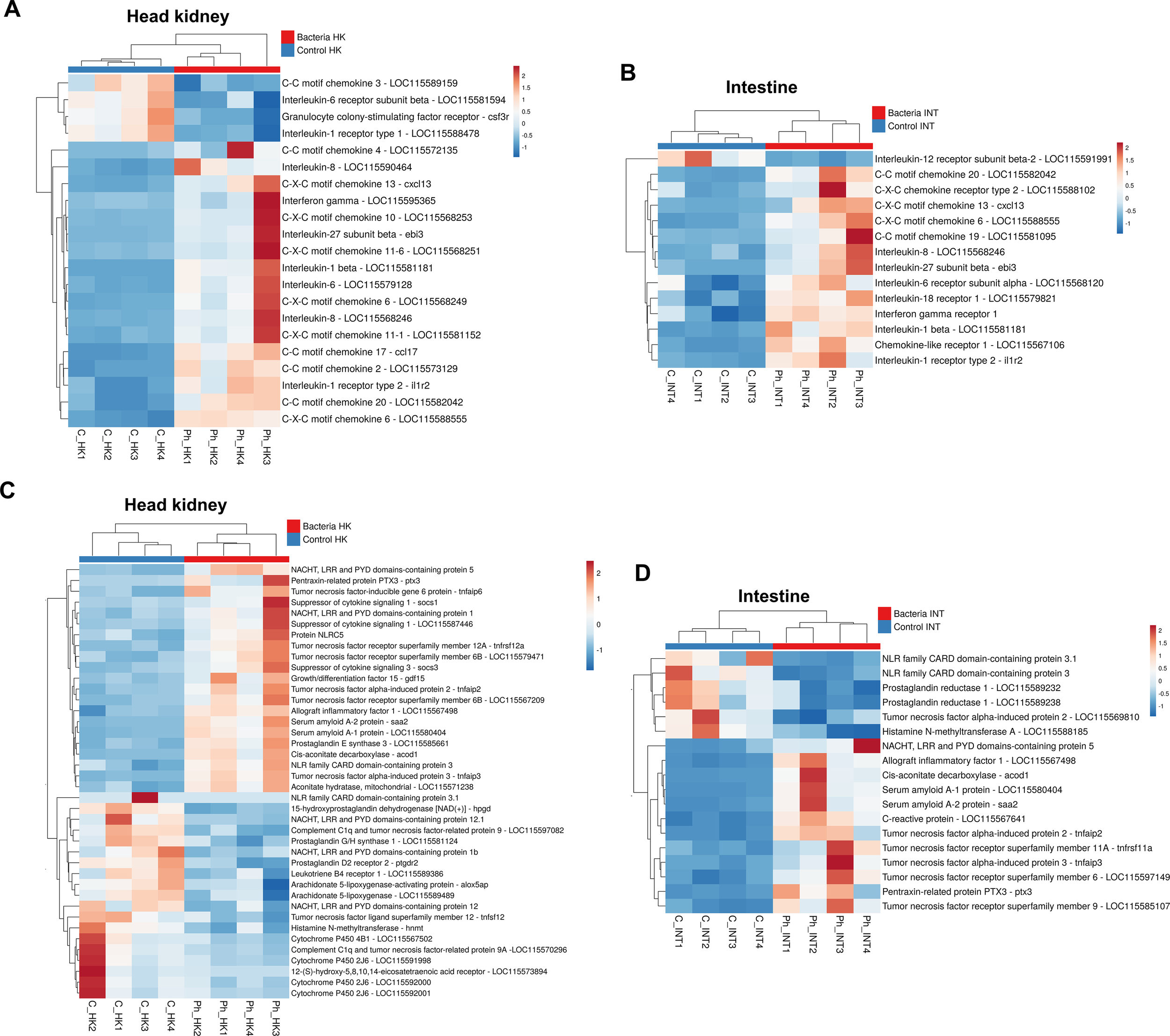
Expression patterns of genes encoding various cytokines and cytokine receptors, acute-phase proteins, and genes involved in inflammation. (A) Heatmaps showing the expression (TPM values) of cytokines and their associated receptors in the head kidney, and (B) in the intestine. (C) Heatmaps illustrating the expression (TPM values) of DEGs involved in the acute-phase response and inflammation in the head kidney and (D) in the intestine.
Several cytokine receptors were also modulated. Among these, the interleukin-1 receptor type 1 (il1r1) was downregulated in the head kidney, whereas interleukin-1 receptor type 2 (il1r2), acting as a decoy receptor, was overexpressed in both tissues (Figures 7A, B). This, along with the downregulation of the interleukin-12 receptor subunit beta-2 (il12rb2) in the intestine, may indicate a regulatory mechanism to control the hyperactivation of the inflammatory response.
3.3.6 Other acute phase and inflammation-related genes
Genes encoding key acute phase response proteins, such as serum amyloid and pentraxins/C-reactive proteins, were modulated by Phdp infection. The genes for serum amyloid A-1 (saa1), serum amyloid A-2 (saa2), and pentraxin-3 (ptx3) were upregulated in both the head kidney (Figure 7C) and the intestine (Figure 7D), while a gene annotated as C-reactive protein was also induced in the intestine. The pro-inflammatory gene allograft inflammatory factor 1 (aif1) was significantly overexpressed in both tissues. Despite the broad induction of pro-inflammatory genes, anti-inflammatory genes were also upregulated after bacterial infection, including suppressor of cytokine signaling 1 (socs1), suppressor of cytokine signaling 3 (socs3), and growth differentiation factor 15 (gdf15) in the head kidney (Figure 7C).
Cis-aconitate decarboxylase (acod1) and aconitate hydratase, mitochondrial (aco2), both involved in the synthesis of the anti-inflammatory and antimicrobial metabolite itaconate during the Krebs cycle, were significantly upregulated in the head kidney (Figure 7C), with acod1 also induced in the intestine (Figure 7D); this suggests an increase in the synthesis of this metabolite following infection with Phdp.
Moreover, different members of the NACHT, LRR, and PYD domains-containing (NLRP) family, as well as the NLR family CARD domain-containing (NLRC) family -both of which play critical roles in the inflammatory process- were modulated following Phdp infection in both tissues. Some of these genes were overexpressed, while others were inhibited. In addition, members of the tumor necrosis factor superfamily were also affected, with the majority being overexpressed after the challenge (Figures 7C, D). Lastly, the infection predominantly downregulated genes involved in the metabolism of prostaglandins, leukotrienes, and histamine.
3.3.7 Reactive oxygen species production and antioxidant defense mechanisms
Different genes involved in the synthesis of ROS and ROS detoxification were affected by the infection in both tissues (Figures 8A, B). Interestingly, a gene encoding eosinophil peroxidase (epx), which produces highly oxidizing agents, was inhibited after infection in the head kidney (Figure 8A). Among the DEGs with a role in ROS production, genes encoding members of the amino acid oxidase family, involved in the synthesis of hydrogen peroxide (H2O2) as a byproduct, were modulated in both organs. Curiously, a gene encoding D-amino acid oxidase (dao) was downregulated in both tissues, whereas genes encoding L-amino acid oxidases were overexpressed: three in the head kidney and two in the intestine (Figure 8C). Finally, the gene cytochrome b-245 light chain (cyba), involved in the NADPH oxidase complex, was overexpressed in the intestine (Figure 8B).
Figure 8
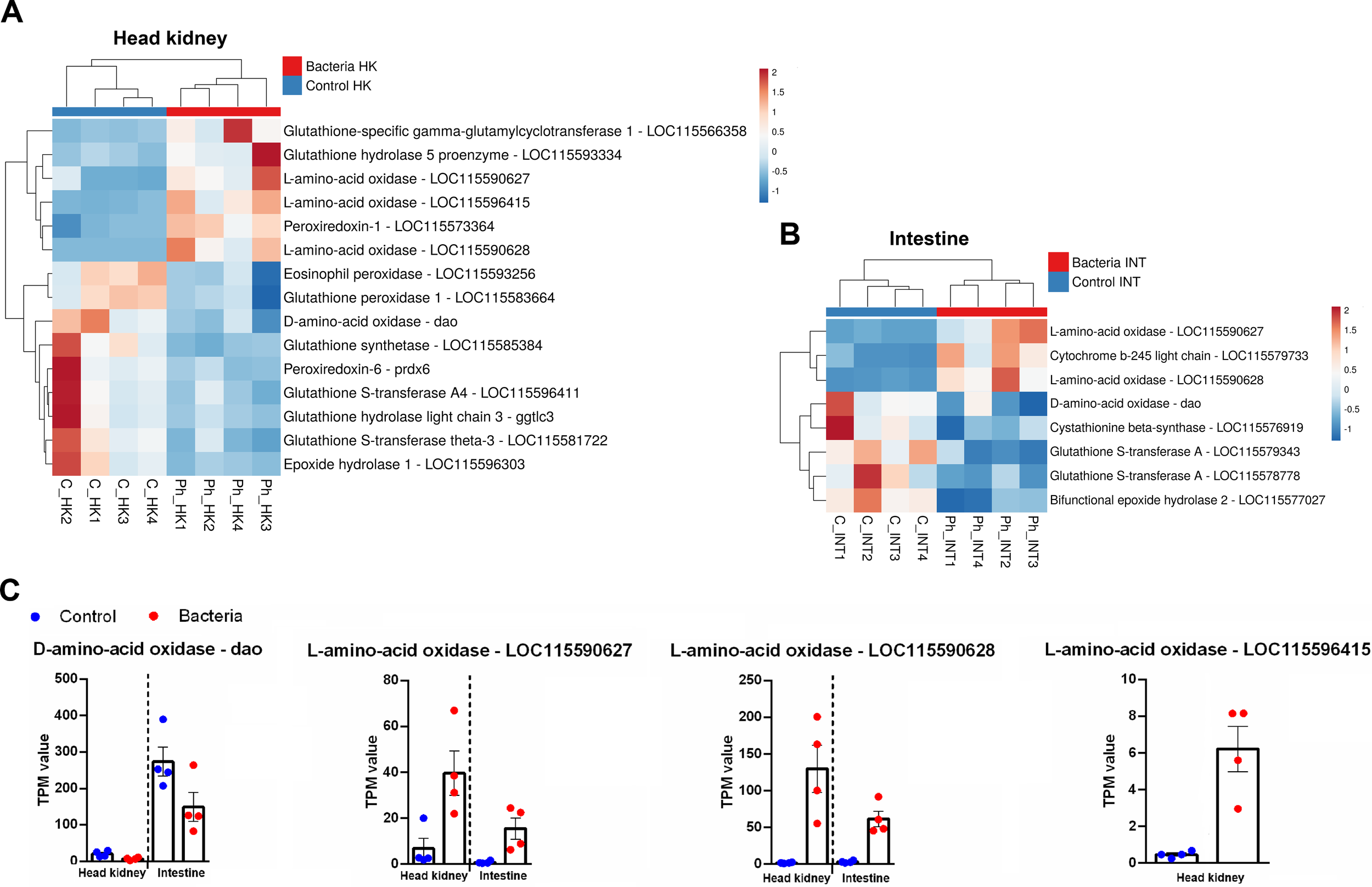
ROS synthesis and detoxification-related genes were modulated by Phdp. (A) Heatmaps showing the expression (TPM values) of these genes in the head kidney and (B) in the intestine. (C) Expression patterns of DEGs belonging to the amino acid oxidase family in both tissues following bacterial challenge.
Various genes involved in ROS detoxification were modulated; some were overexpressed, and others were inhibited after the bacteria challenge. As an example, a gene encoding for peroxiredoxin-1 (prdx1) was overexpressed in the head kidney, whereas a gene encoding for peroxiredoxin-6 (prdx6) was downregulated (Figure 8A). Notably, genes involved in detoxification through the use of glutathione, either by synthesizing it, conjugating it with reactive compounds, or hydrolyzing its derivatives to neutralize harmful substances, were mainly inhibited after the infection in both tissues (Figures 8A, B) with some exceptions in the head kidney. Epoxide hydrolase 1 (ephx1) and epoxide hydrolase 2 (ephx2), which play critical roles in the detoxification of reactive epoxide compounds, were inhibited in the head kidney (Figure 8A) and the intestine (Figure 8B), respectively.
3.3.8 Other immune genes
In addition to the genes mentioned above, several other immune-related genes were affected by the infection in both tissues (Supplementary Figures S2, S3). For instance, multiple GTPases of immunity-associated proteins (GIMAPs), which regulate various lymphoid populations, were upregulated in both the head kidney (Supplementary Figure S2) and the intestine (Supplementary Figure S3).
Additionally, several genes associated with the type I interferon system were modulated in the head kidney, including interferon regulatory factor 1 (irf1), interferon regulatory factor 4 (irf4), stimulator of interferon genes protein STING (tmem173), interferon-induced very large GTPase 1 (gvin1), and four genes annotated as interferon-induced protein 44 (ifi44), three of which were upregulated, while one was downregulated following infection (Supplementary Figure S2). In the intestine, an ifi44 gene that remained unchanged in the head kidney was downregulated (Supplementary Figure S3).
Some genes with a role in the degradation of pathogens and not mentioned in the other categories (e.g. complement, hepcidins) were also affected by the challenge, such as granzyme B (induced in the head kidney), lysozyme G (inhibited in the head kidney) o bactericidal permeability-increasing protein (induced in the intestine).
Finally, among the repertoire of immune-related genes, it is also noteworthy that arginase-1 (arg1) was upregulated in the intestine. Beyond its role in arginine metabolism, this gene is highly expressed in M2 macrophages, contributing to an anti-inflammatory profile (Supplementary Figure S3).
3.4 Identification of potential lncRNAs in gilthead sea bream
The de novo assembly of the gilthead sea bream transcriptome using all the samples yielded 325,850 contigs with a minimum length of 200 bp. After annotation against a custom teleost database, annotated contigs were removed, retaining 294,695 non-annotated contigs. These were mapped to the de novo assembly, selecting 14,553 contigs with a coverage >50. Using the ‘Find ORF’ option in CLC Genomics Workbench, 11,451 contigs were retained. A subsequent BLASTx search against the gilthead sea bream genome peptide database further reduced the number to 10,366 contigs. Finally, the CPAT tool was used to assess coding potential, filtering out additional contigs and identifying 10,150 as putative lncRNAs.
3.5 The infection of juvenile sea bream with P. damselae induces the modulation of lncRNAs potentially involved in the immune response
A principal component analysis (PCA) of the RNA-Seq results for potential lncRNAs revealed distinct patterns between tissues. While control and infected samples did not form separate clusters in the intestine, two well-defined clusters were observed in the head kidney, indicating greater differentiation in lncRNA expression within this hematopoietic tissue (Figure 9A). These findings were further supported by differential expression analyses, which identified 366 differentially expressed (DE) lncRNAs in the head kidney (200 upregulated, 166 downregulated) and only 23 in the intestine (14 upregulated, 9 downregulated) (Figure 9B). Notably, none of the 23 DE lncRNAs modulated after the Phdp challenge in the intestine were affected in the head kidney (Figure 9C). Heatmaps displaying TPM expression values showed consistent expression patterns across samples (Figure 9D).
Figure 9
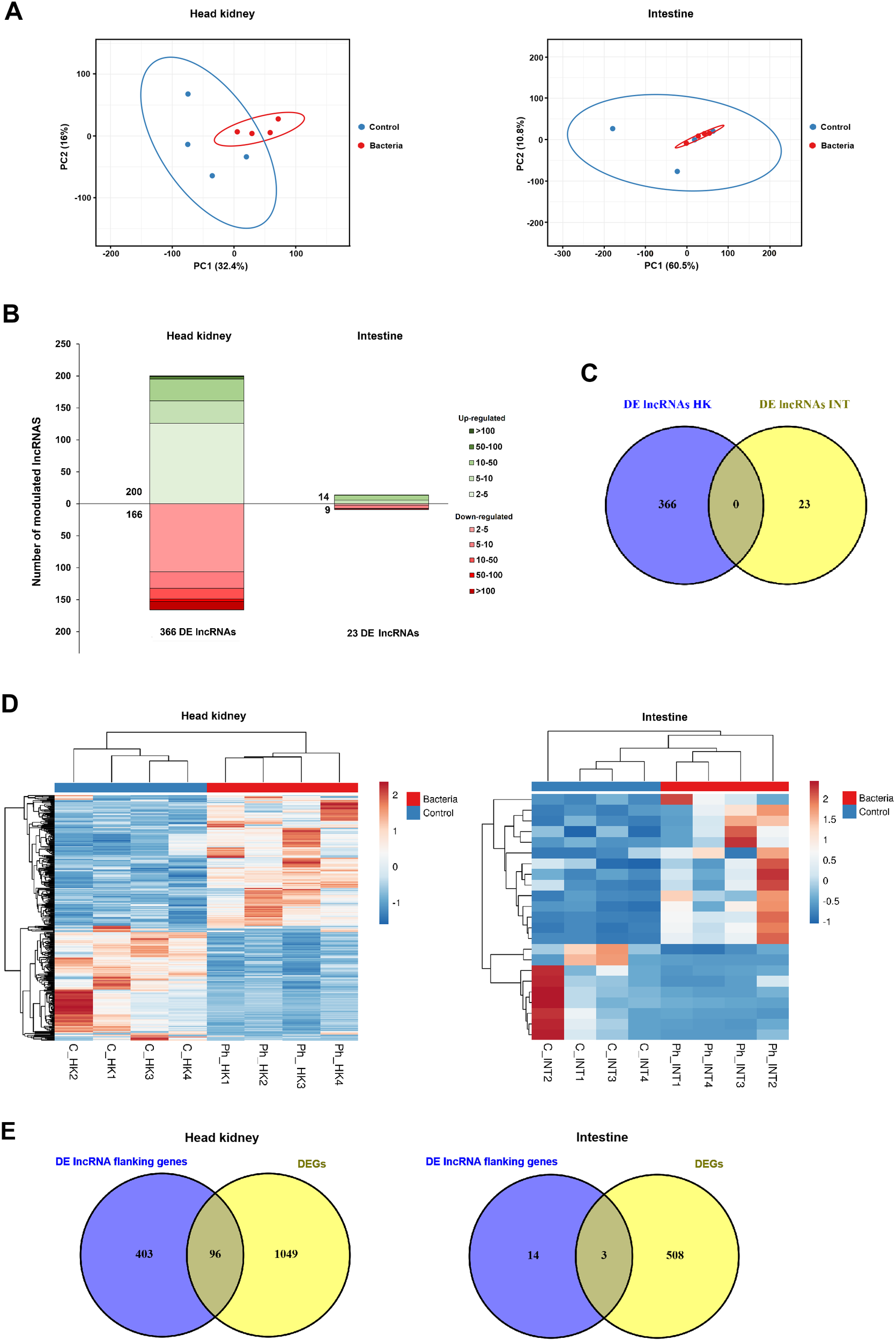
Overall overview of the modulation of lncRNAS in the head kidney and the intestine of juvenile gilthead sea bream at 24 hpi with Phdp. (A) PCA of the expression of the lncRNAs identified in this work in both tissues. (B) Stacked column charts representing the number and intensity (in fold-change value) of the DE lncRNAs between the infected and the control samples. (C) Venn diagram showing the number of shared and exclusive DE lncRNAs in response to infection with Phdp (24 hpi) between both tissues. (D) Heatmaps showing the expression (TPM Values) of DE lncRNAs in the head kidney, and intestine. (E) Venn diagrams representing the number of DEGs located within the 10 kb regions of DE lncRNAs in each tissue.
In order to elucidate the potential target modulations of the DE lncRNAs, the genes surrounding the DE lncRNAs within a 10 kb window were identified by mapping these lncRNAs to the gilthead sea bream genome. In the head kidney, 96 of the 1,145 DEGs were located near DE lncRNAs, whereas in the intestine, only 3 of the 511 DEGs were in proximity to DE lncRNAs, which is likely due to the low number of DE lncRNAs in this tissue (Figure 9E). Indeed, GO enrichment analysis of the coding genes neighboring DE lncRNAs in the intestine yielded no significantly enriched terms. In contrast, numerous GO biological processes were significantly enriched for coding genes near DE lncRNAs in the head kidney (Figure 10). Among these, several terms related to the immune response were identified, particularly those associated with cytotoxic T cells and the adaptive immune response. Meanwhile, the enrichment analysis of the 96 DEGs flanking DE lncRNAs revealed an even stronger enrichment in immune-related terms, primarily those linked to antigen processing and presentation, as well as the adaptive immune response, among others (Figure 10). This enrichment result is due to the high representation of immune genes among those 96 DEGs flanking DE lncRNAs (Supplementary File S3).
Figure 10
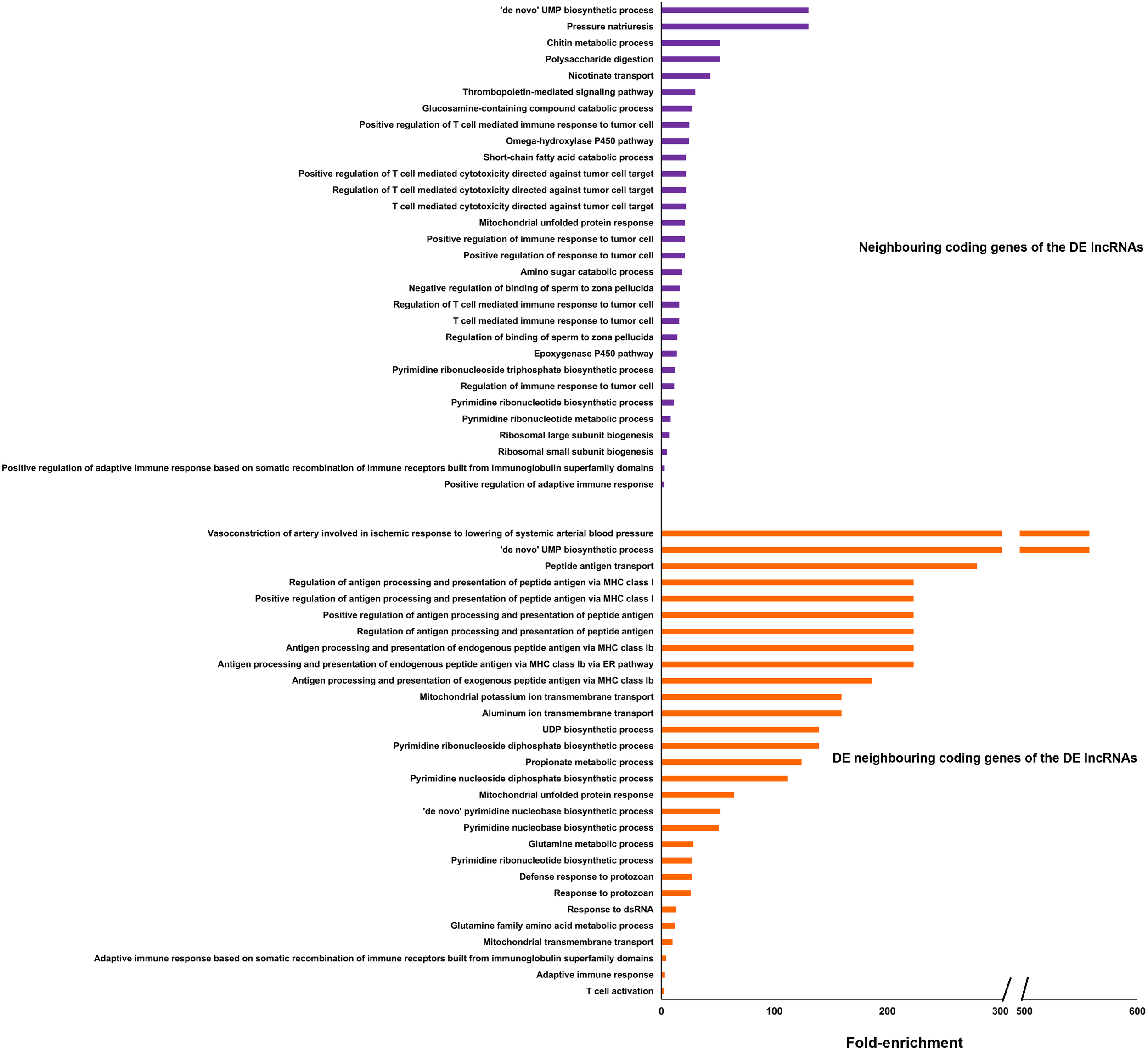
GO enrichment analysis (biological processes) of all coding genes located in the vicinity of the DE lncRNAs in the head kidney and those neighboring genes differentially expressed after the challenge. Only the 30 most significantly enriched terms are shown.
Since lncRNAs typically regulate the expression of nearby genes by acting as local modulators (cis-acting regulation), their expression often shows a positive or inverse correlation with that of adjacent coding genes. We selected a subset of immune-related genes from the 96 DEGs located near DE lncRNAs in the head kidney and compared their expression (TPM values) with that of their neighboring DE lncRNAs. (Figure 11A). Additionally, a Spearman’s correlation analysis was performed to assess the relationship between the TPM values of DE lncRNAs and their adjacent DEGs (Figure 11B). For instance, two hepcidin genes, differentially expressed in the head kidney after Phdp infection, were located near a DE lncRNA. The expression of these coding genes showed a positive and significant correlation with that of the lncRNA. On the other hand, certain DEGs, such as transferrin receptor protein-1 (tfrc), toll-like receptor 13 (tlr13), C-C motif chemokine 19 (ccl19), C-C motif chemokine 20 (ccl20), C-C chemokine receptor type 3 (ccr3), and one gene encoding antigen peptide transporter 1 (tap1), were located within the 10 kb window of two different DE lncRNAs. Most correlations between coding genes and lncRNAs were positive and statistically significant (Figure 11B). However, negative correlations, such as the one between the complement component C9 (c9) and its neighboring DE lncRNA, were also observed.
Figure 11
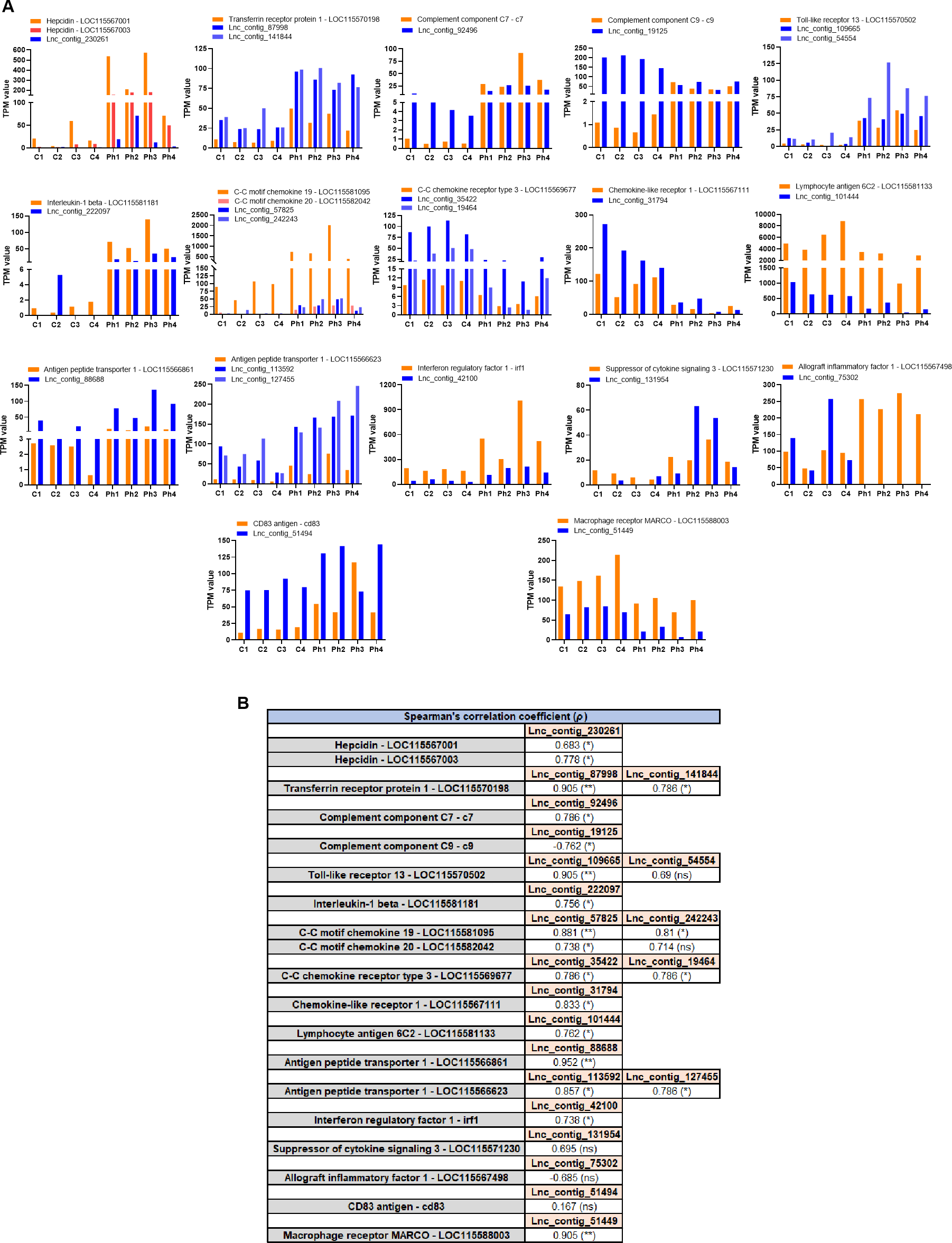
Certain immune genes modulated after Phdp challenge appear to be tightly regulated in cis by lncRNAs. Among the 96 DEGs located near DE lncRNAs, several immune genes were selected to visualize the potential cis-regulation of these genes by lncRNAs. (A) Representation of the TPM values of the selected differentially expressed immune genes and their neighboring DE lncRNAs. (B) Spearman’s correlation analysis of the expression of the lncRNAs and their flanking immune genes. (*p<0.05; **p<0.01; ns, not significant).
4 Discussion
Currently, commercial vaccines are available for Phdp in gilthead sea bream. However, the high incidence of the disease during early developmental stages limits the effectiveness of these preventive strategies in fully mitigating the economic impact of photobacteriosis. Additionally, the excessive use of antibiotics has contributed to the emergence of multiple Phdp strains resistant to commonly used antimicrobial agents in aquaculture, such as erythromycin, kanamycin, sulfamonomethoxine, oxytetracycline, flumequine, chloramphenicol, and oxolinic acid, among others (Thyssen and Ollevier, 2001; Kawanishi et al., 2006; Martínez-Manzanares et al., 2008; Laganà et al., 2011; Zhou et al., 2025). Understanding the gilthead sea bream’s response to Phdp infection is crucial for developing new preventive and therapeutic strategies, with RNA-Seq analysis serving as a valuable tool for examining the transcriptomic response to pathogens. Indeed, the whole transcriptomic response of gilthead sea bream to non-bacterial pathogens, such as viruses and parasites, has been previously analyzed (Piazzon et al., 2019; Pereiro et al., 2023). To the best of our knowledge, this is the first study to comprehensively investigate the full spectrum of gene modulations following Phdp infection in gilthead sea bream. The head kidney, the primary hematopoietic tissue in fish, and the intestine, a key mucosal organ with a high Phdp binding capacity (Magariños et al., 1996c), were selected for analysis due to their crucial roles in the immune response to bacterial pathogens. Noya et al. (1995) found that larvae and juveniles are particularly susceptible to Phdp, which is reflected in our results, where juvenile gilthead sea bream of approximately 8 grams showed a survival rate of only 28.33% following infection. The early immune response is crucial due to the rapid mortality observed after infection with Phdp, which has also been reported in previous studies for this species (Santos et al., 2022), as well as for European sea bass (Dicentrarchus labrax) (Mosca et al., 2014) and Senegalese sole (Solea senegalensis) (Núñez-Díaz et al., 2016). For this reason, samples in our study were collected at 24 hpi, a time point selected to capture the onset of host immune activation before the onset of significant mortality. Also, this study represents the first analysis of lncRNA expression following infection with Phdp in a teleost fish species.
Among the immune systems most affected in the head kidney, and to a lesser extent in the intestine, are the complement system and coagulation, two closely linked defense systems that exhibit significant cross-talk between their respective cascades (Amara et al., 2008). Acosta et al. (2006) reported that the complement system plays a key role in immune resistance to Phdp in various fish species, including sea bream, either through direct bacterial killing or by opsonizing the bacteria to facilitate subsequent phagocytosis. We found that numerous genes involved in both pathways were highly induced at 24 hpi, which is consistent with the overexpression of complement genes observed in other fish species following Phdp infection (Núñez-Díaz et al., 2016; Yu et al., 2022). However, Pellizzari et al. (2013) did not observe any modulation of complement-related genes in the sea bream head kidney at either 24 or 48 hpi with Phdp, despite the microarray used containing probes for several genes involved in this pathway. Interestingly, an RNA-Seq analysis of the liver and spleen from cobia (Rachycentron canadum) infected with Phdp revealed an extensive inhibition of complement and coagulation-related genes in both tissues (Tran et al., 2018). It cannot be ruled out that, in the specific case of cobia, the inhibition of these pathways could be contributing to its susceptibility to this bacterium. Even so, it is important to note that excessive early activation of the complement and coagulation systems has also been linked to increased susceptibility of turbot (Scophthalmus maximus) to Aeromonas salmonicida subsp. salmonicida, as observed when comparing full-sibling families with different susceptibility levels to the bacterium (Pereiro et al., 2024). Although both mechanisms, coagulation and complement, are pivotal components of the early defense response against bacteria, excessive activation of the complement system can exacerbate sepsis-induced coagulation and may even lead to septic coagulopathy (Wei et al., 2024).
Regarding the complement gene c3, expansion of this gene has occurred in teleosts through a process of tandem gene duplication, resulting in the identification of nine c3 genes in gilthead sea bream to date (Najafpour et al., 2020). Variations in the sequence and protein domains of the nine predicted C3 proteins in sea bream, along with the presence of unique cysteine and N-glycosylation residues in each isoform, suggest functional diversity linked to their structure (Najafpour et al., 2020). Indeed, in zebrafish (Danio rerio), it has been shown that c3 genes possess differential roles in the inflammatory response (Forn-Cuní et al., 2014). In this study, we observed the modulation of seven distinct c3-like genes. Five of these were significantly induced exclusively in the head kidney following Phdp infection, while the remaining two were affected in both tissues. However, whereas one of these genes was upregulated in response to the infection, the other was downregulated in both the head kidney and the intestine. Our findings may indicate the functional diversification of c3 genes in response to Phdp infection.
Many pathogenic bacteria affecting mammals and other vertebrates have developed siderophilic tendencies, meaning they rely on iron acquisition for survival and virulence (Ganz, 2018). In 1994, it was demonstrated that iron availability in host fluids correlates with the virulence of Phdp and that this bacterium can utilize heme, hemoglobin, and ferric ammonium citrate as sole iron sources in vitro (Fouz et al., 1994; Magariños et al., 1994). At that time, chemical tests and cross-feeding assays confirmed that Phdp produced a siderophore (Magariños et al., 1994), although its specific siderophore, later named piscibactin, was not formally characterized until 2012 (Souto et al., 2012). To counteract the effects of bacterial siderophores, the host employs iron sequestration as a strategy to restrict pathogen access to this essential nutrient (Ganz, 2018). Our RNA-Seq results showed the modulation of several genes involved in iron homeostasis in both the head kidney and the intestine. For example, serotransferrin (tf) and transferrin receptor protein 1 (tfrc) were found to be overexpressed in both tissues after infection, consistent with the overexpression of tf reported by Santos et al. (2022) following Phdp infection in sea bream. This overexpression likely contributes to reduced iron availability for bacteria, as these proteins transport iron from the blood into cells (Ullah and Lang, 2023). Once inside the cell, ferritin stores intracellular Fe³+ and limits the growth of intracellular pathogens (Ullah and Lang, 2023). Notably, a gene annotated as ferritin was also overexpressed in the head kidney. In addition, ceruloplasmin and hephaestin-like protein 1, both ferroxidases that catalyze the oxidation of Fe²+ to Fe³+ (Knutson, 2017), were overexpressed in our results. This suggests that their upregulation may facilitate iron import into cells through the transferrin-transferrin receptor system. The metalloreductase STEAP4 also increases iron import into the cell (Scarl et al., 2017), and consequently, the steap4 gene was induced following the Phdp challenge. This iron sequestration may help explain the reduced expression of hemoglobin genes in the head kidney, and could also be associated with the anemic condition observed in gilthead sea bream (Santos et al., 2022), hybrid striped bass (Morone chrysops × Morone saxatilis) (Acerete et al., 2009), and meagre (Argyrosomus regius) (Peixoto et al., 2017) infected with Phdp. However, this reduction could also be attributed to the hemolytic activity of Phdp (Rivas et al., 2011), or possibly to a combination of both factors. iron metabolism dysregulation and the hemolytic activity of the pathogen.
Regarding hepcidin, this peptide, primarily produced in the liver but also in other tissues, plays a crucial role in iron metabolism by regulating iron transport across the intestinal epithelium and preventing its release from macrophages and hepatocytes. It achieves this by promoting the degradation of the iron exporter ferroportin, thereby reducing iron availability during inflammation and infection (Ullah and Lang, 2023). However, hepcidin exhibits dual functions and, in addition to its role in iron homeostasis, it is also a cysteine-rich antimicrobial peptide (AMP) (Verga Falzacappa and Muckenthaler, 2005). Unlike mammals, which possess only a single hepcidin gene, most fish species have at least two: one hepcidin-1 and one or more members of the hepcidin-2 family (Pereiro et al., 2012; Neves et al., 2015; Serna-Duque et al., 2022a). In gilthead sea bream, fifteen hepcidin genes have been identified, including one hepcidin-1 and fourteen hepcidin-2 genes (Serna-Duque et al., 2022a). While hepcidin-1 primarily regulates iron homeostasis, hepcidin-2 functions exclusively as an antimicrobial peptide (AMP) (Pereiro et al., 2012; Neves et al., 2015; Serna-Duque et al., 2022a, b). Our findings show that hepcidin-1 (hamp1) was significantly upregulated following Phdp infection in both tissues, indicating the activation of iron-regulatory hepcidin. Meanwhile, eight hepcidin-2 genes were modulated in response to the Phdp challenge, exhibiting different expression patterns, with some genes being overexpressed and others inhibited after the challenge. A differential expression pattern was also previously reported by Serna-Duque et al. (2022a), who observed that while most hepcidin genes were upregulated in the sea bream head kidney at 4 hpi following Vibrio anguillarum infection, two were downregulated. This variable expression of the different hepcidin-2 members is likely due to the fact that, in addition to their roles in iron metabolism and as AMPs, fish hepcidins also possess immunomodulatory properties (Álvarez et al., 2022; Li et al., 2023; Cervera et al., 2024). Given the extensive expansion of hepcidin-2 genes in gilthead sea bream, it is possible that different hepcidin-2 genes fulfill distinct immunomodulatory functions.
The results also showed a broad modulation of PRRs, particularly different types of lectins. While several lectins were overexpressed at 24 hpi, others were downregulated. This was also reported in the microarray study by Pellizzari et al. (2013), who found that certain lectins were induced in the gilthead sea bream head kidney after infection, while others were repressed. Additionally, we observed the overexpression of TLRs in the head kidney (tlr1, tlr5, and tlr13), whereas in the intestine, only tlr5 was induced. Functionally, TLR1 is mainly involved in the recognition of bacterial lipoproteins, TLR5 detects flagellin, and TLR13 recognizes bacterial RNA (Kawai and Akira, 2010; Hidmark et al., 2012). Interestingly, and similarly to what was observed for complement- and coagulation-related genes, Tran et al. (2018) reported a downregulation of tlr2 (TLR2 forms heterodimers with TLR1) and tlr5 in the liver of cobia at 24 hpi following Phdp infection. Also, we observed that two genes annotated as macrophage receptor MARCO, which is primarily expressed in macrophages and known for its ability to bind unopsonized pathogens and facilitate their ingestion by macrophages (Xing et al., 2021), were inhibited in the head kidney. This could indicate two possibilities: a reduction in macrophage numbers, potentially due to apoptosis induced by the Phdp exotoxin AIP56, as previously described (do Vale et al., 2005), or changes in macrophage activity, such as shifts in M1 or M2 polarization (Murthy et al., 2015). Santos et al. (2022) previously observed that circulating monocytes decreased in number at 9 hpi but then increased beyond the levels observed in uninfected sea bream. In contrast, Pellizzari et al. (2013), based on their microarray results (including the overexpression of arginase-1 and arginase-2 genes), suggested that Phdp infection could induce M2 macrophage polarization. Indeed, we also observed the overexpression of the arginase-1 gene in the intestine at 24 hpi with Phdp. However, further investigation is required to fully elucidate this question.
But, in addition to the activation of innate immune receptors, specific pathogen recognition appears to be triggered following infection with Phdp. For instance, rag1, which encodes a protein essential for the recombination of immunoglobulin genes during B cell development and TCR gene rearrangement in T cells (Bassing et al., 2002), was induced in the head kidney. This increase in rag1 expression was also observed in the head kidney of silver pomfret (Pampus argenteus) following Phdp infection (Nawaz et al., 2022).
In agreement with Pellizzari et al. (2013), genes involved in antigen presentation were predominantly induced in sea bream following infection with Phdp, particularly those associated with antigen degradation via the proteasome. This pathway is responsible for processing endogenous peptides for presentation through the MHC class I pathway (Rivett and Hearn, 2004). Notably, genes encoding different cathepsins exhibited both upregulation and downregulation in the head kidney after infection, suggesting potential functional specialization even within the same cathepsin type (e.g., cathepsin L). While two cathepsin L genes were induced post-infection, two others were downregulated, a pattern also observed for cathepsin S. In mammals, both cathepsin L and S contribute to the degradation of extracellular antigens within endolysosomes for antigen presentation via MHC class II (Perišić Nanut et al., 2021). However, as previously noted for other gene families expanded in teleosts, these duplications may have led to functional diversification, potentially explaining the opposing expression patterns observed. Interestingly, while tap1 and tap2 were induced, we did not observe significant modulation in MHC-I or MHC-II gene expression. A previous study reported that mhc1 was slightly induced in the sea bream head kidney following Phdp infection, whereas mhc2 showed no significant modulation (Santos et al., 2022). However, in a gilthead sea bream-specific microarray study, a gene annotated as mhc2 was significantly induced at 48 hpi in the head kidney following Phdp infection (Pellizzari et al., 2013).
Contrary to the suggestion by Pellizzari et al. (2013), who proposed that the high mortality of juvenile gilthead sea bream caused by photobacteriosis is associated with an anti-inflammatory response, our findings suggest the opposite: a strong pro-inflammatory response. We observed the overexpression of various pro-inflammatory cytokines in the head kidney and the intestine, including IL-1β (il1b), the prototypical pro-inflammatory cytokine, and other genes involved in inflammation and the acute phase response. Notably, the gdf15 gene, which encodes a member of the TGF-β superfamily, was induced in the head kidney, suggesting a sepsis-like state. This is significant, as GDF15, a protective anti-inflammatory cytokine, is upregulated during sepsis in mammals (Luan et al., 2019) and fish (Pereiro et al., 2020b). However, a balance between proinflammatory and anti-inflammatory responses is typically observed following infection to prevent excessive inflammation (Cicchese et al., 2018), as seen in our results.
Similar to inflammation-related genes, those involved in ROS production and detoxification exhibited a balanced expression pattern. However, genes associated with ROS detoxification were generally downregulated, which may reflect a host strategy to reduce its antioxidant capacity in response to the oxidative defenses of Phdp. This bacterium can counteract host superoxide radicals through the production of superoxide dismutase (SOD) and catalase, both of which are important virulence factors in many pathogenic bacteria (Barnes et al., 1999; Díaz-Rosales et al., 2006). Vallecillos et al. (2021) found that higher plasma peroxidase activity under naïve conditions shows moderate heritability and is positively correlated with resistance to Phdp in gilthead sea bream, suggesting its potential as a humoral marker for selective breeding. However, total peroxidase activity in the plasma of sea bream following a Phdp challenge did not show significant changes, although lipid peroxidation and the activities of glutathione S-transferase and catalase were significantly increased in the liver (Santos et al., 2022). The peroxidase family consists of enzymes capable of reducing H2O2 and other hydroperoxides to water, with several substrates being oxidized during this process. While some of these enzymes act as antioxidants (e.g., glutathione peroxidase and peroxiredoxins), others use H2O2 to generate even more reactive oxidants as part of the immune response (e.g., eosinophil peroxidase and myeloperoxidase) (Kurutas, 2016). Given the dual nature of peroxidases, it remains unclear whether the sea bream exhibiting higher resistance display an increased prooxidant or antioxidant state (Vallecillos et al., 2021).
Amino acid oxidases (AAOs) are flavoenzymes that catalyze the oxidative deamination of L-amino acids (L-amino acid oxidases, LAOs) or D-amino acids (D-amino acid oxidases, DAOs) into α-keto acids, generating ammonia and H2O2 as byproducts (Zhang et al., 2004). While the antibacterial role of LAOs mediated by H2O2 production has been extensively documented, including in fish (Kitani and Nagashima, 2020), the antibacterial function of vertebrate DAOs is poorly understood. Although some studies have reported a potential role in antibacterial defense, DAOs are primarily associated with regulatory functions, particularly in the brain, where they help control D-serine levels (Sasabe et al., 2014). In our study, we found that a DAO-encoding gene was significantly downregulated in both the head kidney and intestine at 24 hpi. In contrast, and as expected given their known antibacterial role, two LAO-encoding genes were significantly upregulated in both tissues, while a third LAO showed significant induction only in the head kidney. These findings underscore the importance of oxidative deamination of amino acids during bacterial infection.
The role of lncRNAs in regulating various aspects of the immune response has been well documented in mammals in recent years (Ghahramani Almanghadim et al., 2024). However, functional studies of lncRNAs in fish remain scarce and are primarily based on the inferred functions of their flanking coding genes (Wang et al., 2018). In this study, we analyzed the potential modulatory role of lncRNAs in the transcriptomic response to Phdp. Our findings suggest that lncRNAs play a more prominent role in the head kidney during photobacteriosis, as only a small subset of lncRNAs was modulated in the intestine, and these were not directly linked to the immune response. Given that lncRNAs primarily regulate the expression of neighboring genes through cis-acting mechanisms, we inferred their potential regulatory functions by examining the roles of their flanking coding genes. Consistent with the major pathways affected at the coding gene level in our RNA-Seq results, our analysis revealed that lncRNAs appear to tightly regulate processes such as complement activation, iron homeostasis, pathogen recognition, antigen presentation, and inflammation. A significant correlation was observed between DEGs involved in these processes and DE lncRNAs located in their vicinity. However, further functional studies will be necessary to confirm these regulatory interactions.
5 Conclusions
This study presents the first comprehensive RNA-Seq analysis of gilthead sea bream infected with Phdp, as well as the first investigation of long non-coding RNA (lncRNA) expression associated with photobacteriosis in a teleost fish. We found that Phdp infection triggered a broad modulation of immune-related processes, with the complement and coagulation pathways, iron homeostasis, antigen recognition and presentation, and inflammation-related genes being strongly regulated after the challenge. Notably, this immune response appears to be fine-tuned by lncRNAs. Moreover, certain gene families expanded through duplication events in gilthead sea bream displayed contrasting expression patterns following infection (e.g., complement component c3, hepcidin, and cathepsin L genes), providing insights into the functional diversification of these gene families. Overall, our findings offer valuable insights into the immune response of juvenile gilthead sea bream to Phdp infection and lay the foundation for future research aimed at deepening our understanding of host-pathogen interactions. This is an essential step toward developing new preventive and therapeutic strategies.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by CSIC National Committee on Bioethics (approval number ES360570202001/21/FUN.01/INM06/BNG01). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
PP: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. AF: Data curation, Methodology, Writing – review & editing. BN: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was financially supported by project PID2023-148810OB-C21 from the Spanish Ministerio de Ciencia, Innovación y Universidades (MICIU), the Agencia Estatal de Investigación (AEI), and the European Regional Development Fund (ERDF) (MICIU/AEI/10.13039/501100011033/FEDER, EU), as well as by the project Controlling Microbiomes Circulations for Better Food Systems (CIRCLES) from the European Union Horizon 2020 Program (Grant Agreement No. 818290). We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative (PROA) through its Unit of Information Resources for Research (URICI).
Acknowledgments
We wish to thank the IIM-CSIC aquarium staff for their technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1604207/full#supplementary-material
Supplementary Table 1Primer pairs used for qPCR validation of RNA-Seq results.
Supplementary Table 2Summary of the number of raw reads, high-quality reads after trimming and mapping results.
Supplementary Figure 1Validation of RNA-Seq data by qPCR. Fold-change values for the selected genes obtained by RNA-Seq and qPCR in (A) the head kidney and (B) the intestine. (C) Pearson’s correlation between RNA-Seq and qPCR fold-change values resulted in a r = 0.884, confirming the consistency between both methods.
Supplementary Figure 2Heatmap representing the expression (TPM values) of other immune genes differentially expressed in the head kidney that were not included in the main categories described in this work.
Supplementary Figure 3Heatmap representing the expression (TPM values) of other immune genes differentially expressed in the intestine that were not included in the main categories described in this work.
Supplementary File S1Complete repertoire of DEGs in the head kidney and intestine at 24 hpi with Phdp.
Supplementary File S2List of the 151 DEGs commonly affected in the head kidney and intestine at 24 hpi with Phdp.
Supplementary File S3List of the 96 DEGs located near DE lncRNAs in the head kidney.
References
1
Abushattal S. Vences A. Osorio C. R. (2020). A virulence gene typing scheme for Photobacterium damselae subsp. piscicida, the causative agent of fish photobacteriosis, reveals a high prevalence of plasmid-encoded virulence factors and of type III secretion system genes. Aquaculture521, 735057. doi: 10.1016/j.aquaculture.2020.735057
2
Acerete L. Espinosa E. Josa A. Tort L. (2009). Physiological response of hybrid striped bass subjected to Photobacterium damselae subsp. piscicida. Aquaculture298, 16–23. doi: 10.1016/j.aquaculture.2009.09.023
3
Acosta F. Ellis A. E. Vivas J. Padilla D. Acosta B. Déniz S. et al . (2006). Complement consumption by Photobacterium damselae subsp. piscicida in seabream, red porgy and seabass normal and immune serum. Effect of the capsule on the bactericidal effect. Fish Shellfish Immunol.20, 709–717. doi: 10.1016/j.fsi.2005.08.011
4
Acosta F. Galarreta C. M. Ellis A. E. Díaz R. Gómez V. Padilla D. et al . (2004). Activation of the nitric oxide response in gilthead seabream after experimental infection with Photobacterium damselae subsp. piscicida. Fish Shellfish Immunol.16, 581–588. doi: 10.1016/j.fsi.2003.09.010
5
Álvarez C. A. Santana P. A. Salinas-Parra N. Beltrán D. Guzmán F. Vega B. et al . (2022). Immune modulation ability of hepcidin from teleost fish. Animals12, 1586. doi: 10.3390/ani12121586
6
Amara U. Rittirsch D. Flierl M. Bruckner U. Klos A. Gebhard F. et al . (2008). Interaction between the coagulation and complement system. Adv. Exp. Med. Biol.632, 71–79. doi: 10.1007/978-0-387-78952-1_6
7
Andreoni F. Magnani M. (2014). Photobacteriosis: prevention and diagnosis. J. Immunol. Res.2014, 793817. doi: 10.1155/2014/793817
8
Arijo S. Borrego J. J. Zorrilla I. Balebona M. C. Moriñigo M. A. (1998). Role of the capsule of Photobacterium damsela subsp. piscicida in protection against phagocytosis and killing by gilt-head seabream (Sparus aurata, L) macrophages. Fish Shellfish Immunol.8, 63–72. doi: 10.1006/fsim.1997.0122
9
Aune T. M. Spurlock C. F. 3rd (2016). Long non-coding RNAs in innate and adaptive immunity. Virus Res.212, 146–160. doi: 10.1016/j.virusres.2015.07.003
10
Barnes A. C. Balebona M. C. Horne M. T. Ellis A. E. (1999). Superoxide dismutase and catalase in Photobacterium damselae subsp. piscicida and their roles in resistance to reactive oxygen species. Microbiology145, 483–494. doi: 10.1099/13500872-145-2-483
11
Bassing C. H. Swat W. Alt F. W. (2002). The mechanism and regulation of chromosomal V(D)J recombination. Cell109, S45–S55. doi: 10.1016/s0092-8674(02)00675-x
12
Borrego J. J. Labella A. M. Castro D. Ortiz-Delgado J. B. Sarasquete C. (2017). Updated of the pathologies affecting cultured gilthead seabream, Sparus aurata. Ann. Aquac. Res.4, 1033. doi: 10.47739/2379-0881/1033
13
Cervera L. Arizcun M. Mercado L. Chaves-Pozo E. Cuesta A. (2024). Hepcidin and dicentracin peptides show preventive antiviral applications against NNV infection in European sea bass through immunomodulatory roles. Aquaculture583, 740592. doi: 10.1016/j.aquaculture.2024.740592
14
Cicchese J. M. Evans S. Hult C. Joslyn L. R. Wessler T. Millar J. A. et al . (2018). Dynamic balance of pro- and anti-inflammatory signals controls disease and limits pathology. Immunol. Rev.285, 147–167. doi: 10.1111/imr.12671
15
Díaz-Rosales P. Chabrillón M. Arijo S. Martinez-Manzanares E. Moriñigo M. A. Balebona M. C. (2006). Superoxide dismutase and catalase activities in Photobacterium damselae ssp. piscicida. J. Fish. Dis.29, 355–364. doi: 10.1111/j.1365-2761.2006.00726.x
16
do Vale A. Silva M. T. dos Santos N. M. Nascimento D. S. Reis-Rodrigues P. Costa-Ramos C. et al . (2005). AIP56, a novel plasmid-encoded virulence factor of Photobacterium damselae subsp. piscicida with apoptogenic activity against sea bass macrophages and neutrophils. Mol. Microbiol.58, 1025–1038. doi: 10.1111/j.1365-2958.2005.04893.x
17
Elling R. Chan J. Fitzgerald K. A. (2016). Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur. J. Immunol.46, 504–512. doi: 10.1002/eji.201444558
18
FAO (2025). Fishery and Aquaculture Statistics – Yearbook 2022. FAO Yearbook of Fishery and Aquaculture Statistics. Rome.
19
Forn-Cuní G. Reis E. S. Dios S. Posada D. Lambris J. D. Figueras A. et al . (2014). The evolution and appearance of C3 duplications in fish originate an exclusive teleost c3 gene form with anti-inflammatory activity. PloS One9, e99673. doi: 10.1371/journal.pone.0099673
20
Fouz B. Toranzo A. E. Biosca E. G. Mazoy R. Amaro C. (1994). Role of iron in the pathogenicity of Vibrio damsela for fish and mammals. FEMS Microbiol. Lett.121, 181–188. doi: 10.1111/j.1574-6968.1994.tb07097.x
21
Ganz T. (2018). Iron and infection. Int. J. Hematol.107, 7–15. doi: 10.1007/s12185-017-2366-2
22
Ghahramani Almanghadim H. Karimi B. Valizadeh S. Ghaedi K. (2024). Biological functions and affected signaling pathways by Long Non-Coding RNAs in the immune system. Noncoding. RNA Res.10, 70–90. doi: 10.1016/j.ncrna.2024.09.001
23
Grasso V. Padilla D. Bravo J. Román L. Rosario I. Acosta B. et al . (2015). Immunization of sea bream (Sparus aurata) juveniles against Photobacterium damselae subsp. piscicida by short bath: Effect on some pro-inflammatory molecules and the Mx gene expression. Fish Shellfish Immunol.46, 292–296. doi: 10.1016/j.fsi.2015.06.030
24
Hanif A. Bakopoulos V. Leonardos I. Dimitriadis G. J. (2005). The effect of sea bream (Sparus aurata) broodstock and larval vaccination on the susceptibility by Photobacterium damsela subsp. piscicida and on the humoral immune parameters. Fish Shellfish Immunol.19, 45–61. doi: 10.1016/j.fsi.2004.12.009
25
Hidmark A. von Saint Paul A. Dalpke A. H. (2012). Cutting edge: TLR13 is a receptor for bacterial RNA. J. Immunol.189, 2717–2721. doi: 10.4049/jimmunol.1200898
26
Kanehisa M. Sato Y. (2020). KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci.29, 28–35. doi: 10.1002/pro.3711
27
Kawai T. Akira S. (2010). The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol.11, 373–384. doi: 10.1038/ni.1863
28
Kawanishi M. Kijima M. Kojima A. Ishihara K. Esaki H. Yagyu K. et al . (2006). Drug resistance and random amplified polymorphic DNA analysis of Photobacterium damselae ssp. piscicida isolates from cultured Seriola (yellowtail, amberjack and kingfish) in Japan. Lett. Appl. Microbiol.42, 648–653. doi: 10.1111/j.1472-765X.2005.01820.x
29
Kitani Y. Nagashima Y. (2020). L-Amino acid oxidase as a fish host-defense molecule. Fish Shellfish Immunol.106, 685–690. doi: 10.1016/j.fsi.2020.08.028
30
Knutson M. D. (2017). Iron transport proteins: Gateways of cellular and systemic iron homeostasis. J. Biol. Chem.292, 12735–12743. doi: 10.1074/jbc.R117.786632
31
Kraemer S. A. Ramachandran A. Perron G. G. (2019). Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms7, 180. doi: 10.3390/microorganisms7060180
32
Kurutas E. B. (2016). The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: current state. Nutr. J.15, 71. doi: 10.1186/s12937-016-0186-5
33
Laganà P. Caruso G. Minutoli E. Zaccone R. Santi D. (2011). Susceptibility to antibiotics of Vibrio spp. and Photobacterium damsela ssp. piscicida strains isolated from Italian aquaculture farms. New Microbiol.34, 53–63.
34
Li X. Chi H. Dalmo R. A. Tang X. Xing J. Sheng X. et al . (2023). Anti-microbial activity and immunomodulation of recombinant hepcidin 2 and NK-lysin from flounder (Paralichthys olivaceus). Int. J. Biol. Macromol.253, 127590. doi: 10.1016/j.ijbiomac.2023.127590
35
Ljubojević Pelić D. Radosavljević V. Pelić M. Živkov Baloš M. Puvača N. Jug-Dujaković J. et al . (2024). Antibiotic residues in cultured fish: implications for food safety and regulatory concerns. Fishes9, 484. doi: 10.3390/fishes9120484
36
Luan H. H. Wang A. Hilliard B. K. Carvalho F. Rosen C. E. Ahasic A. M. et al . (2019). GDF15 is an inflammation-induced central mediator of tissue tolerance. Cell178, 1231–1244. doi: 10.1016/j.cell.2019.07.033
37
Magariños B. Bonet R. Romalde J. L. Martínez M. J. Congregado F. Toranzo A. E. (1996b). Influence of the capsular layer on the virulence of Pasteurella piscicida for fish. Microb. Pathog.21, 289–297. doi: 10.1006/mpat.1996.0062
38
Magariños B. Romalde J. L. Lemos M. L. Barja J. L. Toranzo A. E. (1994). Iron uptake by Pasteurella piscicida and its role in pathogenicity for fish. Appl. Environ. Microbiol.60, 2990–2998. doi: 10.1128/aem.60.8.2990-2998.1994
39
Magariños B. Romalde J. L. Noya M. Barja J. L. Toranzo A. E. (1996c). Adherence and invasive capacities of the fish pathogen Pasteurella piscicida. FEMS Microbiol. Lett.138, 29–34. doi: 10.1111/j.1574-6968.1996.tb08130.x
40
Magariños B. Santos Y. Romalde J. L. Rivas C. Barja J. L. Toranzo A. E. (1992). Pathogenic activities of live cells and extracellular products of the fish pathogen Pasteurella piscicida. J. Gen. Microbiol.138, 2491–2498. doi: 10.1099/00221287-138-12-2491
41
Magariños B. Toranzo A. E. Romalde J. L. (1995). Different susceptibility of gilthead sea bream and turbot to Pasteurella piscicida infection by the water route. Bull. Eur. Assoc. Fish. Pathol.15, 88–90.
42
Magariños B. Toranzo A. E. Romalde J. L. (1996a). Phenotypic and pathobiological characteristics of Pasteurella piscicida. Annu. Rev. Fish. Dis.6, 41–64. doi: 10.1016/S0959-8030(96)90005-8
43
Martínez-Manzanares E. Tapia-Paniagua S. T. Díaz-Rosales P. Chabrillón M. Moriñigo M. A. (2008). Susceptibility of Photobacterium damselae subsp. piscicida strains isolated from Senegalese sole, Solea Senegalensis Kaup, and gilthead seabream, Sparus aurata L., to several antibacterial agents. J. Fish. Dis.31, 73–76. doi: 10.1111/j.1365-2761.2007.00872.x
44
Metsalu T. Vilo J. (2015). ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res.43, W566–W570. doi: 10.1093/nar/gkv468
45
Miccoli A. Saraceni P. R. Scapigliati G. (2019). Vaccines and immune protection of principal Mediterranean marine fish species. Fish Shellfish Immunol.94, 800–809. doi: 10.1016/j.fsi.2019.09.065
46
Mosca F. Ciulli S. Volpatti D. Romano N. Volpe E. Bulfon C. et al . (2014). Defensive response of European sea bass (Dicentrarchus labrax) against Listonella Anguillarum or Photobacterium damselae subsp. piscicida experimental infection. Vet. Immunol. Immunopathol.162, 83–95. doi: 10.1016/j.vetimm.2014.10.002
47
Murthy S. Larson-Casey J. L. Ryan A. J. He C. Kobzik L. Carter A. B. (2015). Alternative activation of macrophages and pulmonary fibrosis are modulated by scavenger receptor, macrophage receptor with collagenous structure. FASEB J.29, 3527–3536. doi: 10.1096/fj.15-271304
48
Najafpour B. Cardoso J. C. R. Canário A. V. M. Power D. M. (2020). Specific evolution and gene family expansion of complement 3 and regulatory factor H in fish. Front. Immunol.11. doi: 10.3389/fimmu.2020.568631
49
Nawaz M. Li X. Yue X. Gouife M. Huang K. Chen S. et al . (2022). Transcriptome profiling and differential expression analysis of the immune-related genes during the acute phase of infection with Photobacterium damselae subsp. damselae in silver pomfret (Pampus argenteus). Fish Shellfish Immunol.131, 342–348. doi: 10.1016/j.fsi.2022.10.020
50
Neves J. V. Caldas C. Vieira I. Ramos M. F. Rodrigues P. N. (2015). Multiple hepcidins in a teleost fish, Dicentrarchus labrax: different hepcidins for different roles. J. Immunol.195, 2696–2709. doi: 10.4049/jimmunol.1501153
51
Noya M. Magariños B. Lamas J. (1995). Interactions between peritoneal exudate cells (PECs) of gilthead seabream (Sparus aurata) and Pasteurella piscicida. A morphological study. Aquaculture131, 11–21. doi: 10.1016/0044-8486(94)00353-P
52
Núñez-Díaz J. A. Fumanal M. Mancera J. M. Moriñigo M. A. Balebona M. C. (2016). Two routes of infection with Photobacterium damselae subsp. piscicida are effective in the modulation of the transcription of immune related genes in Solea Senegalensis. Vet. Immunol. Immunopathol.179, 8–17. doi: 10.1016/j.vetimm.2016.07.009
53
Peixoto M. J. Salas-Leitón E. Brito F. Pereira L. F. Svendsen J. C. Baptista T. et al . (2017). Effects of dietary Gracilaria sp. and Alaria sp. supplementation on growth performance, metabolic rates and health in meagre (Argyrosomus regius) subjected to pathogen infection. J. Appl. Phycol.29, 433–447. doi: 10.1007/s10811-016-0917-1
54
Pellizzari C. Krasnov A. Afanasyev S. Vitulo N. Franch R. Pegolo S. et al . (2013). High mortality of juvenile gilthead sea bream (Sparus aurata) from photobacteriosis is associated with alternative macrophage activation and anti-inflammatory response: results of gene expression profiling of early responses in the head kidney. Fish Shellfish Immunol. 34, 1269–1278. doi: 10.1016/j.fsi.2013.02.007
55
Pereiro P. Figueras A. Novoa B. (2012). A novel hepcidin-like in turbot (Scophthalmus maximus L.) highly expressed after pathogen challenge but not after iron overload. Fish Shellfish Immunol.32, 879–889. doi: 10.1016/j.fsi.2012.02.016
56
Pereiro P. Figueras A. Novoa B. (2023). RNA-Seq analysis of juvenile gilthead sea bream (Sparus aurata) provides some clues regarding their resistance to the nodavirus RGNNV genotype. Fish Shellfish Immunol.134, 108588. doi: 10.1016/j.fsi.2023.108588
57
Pereiro P. Lama R. Moreira R. Valenzuela-Muñoz V. Gallardo-Escárate C. Novoa B. et al . (2020a). Potential involvement of lncRNAs in the modulation of the transcriptome response to nodavirus challenge in European sea bass (Dicentrarchus labrax L.). Biology9, 165. doi: 10.3390/biology9070165
58
Pereiro P. Librán-Pérez M. Figueras A. Novoa B. (2020b). Conserved function of zebrafish (Danio rerio) Gdf15 as a sepsis tolerance mediator. Dev. Comp. Immunol.109, 103698. doi: 10.1016/j.dci.2020.103698
59
Pereiro P. Tur R. García M. Figueras A. Novoa B. (2024). Unravelling turbot (Scophthalmus maximus) resistance to Aeromonas salmonicida: transcriptomic insights from two full-sibling families with divergent susceptibility. Front. Immunol.15. doi: 10.3389/fimmu.2024.1522666
60
Perišić Nanut M. Pečar Fonović U. Jakoš T. Kos J. (2021). The role of cysteine peptidases in hematopoietic stem cell differentiation and modulation of immune system function. Front. Immunol.12. doi: 10.3389/fimmu.2021.680279
61
Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res.29, e45. doi: 10.1093/nar/29.9.e45
62
Piazzon M. C. Mladineo I. Naya-Català F. Dirks R. P. Jong-Raadsen S. Vrbatović A. et al . (2019). Acting locally - affecting globally: RNA sequencing of gilthead sea bream with a mild Sparicotyle chrysophrii infection reveals effects on apoptosis, immune and hypoxia related genes. BMC Genomics20, 200. doi: 10.1186/s12864-019-5581-9
63
Ponting C. P. Oliver P. L. Reik W. (2009). Evolution and functions of long noncoding RNAs. Cell136, 629–641. doi: 10.1016/j.cell.2009.02.006
64
Rivas A. J. Balado M. Lemos M. L. Osorio C. R. (2011). The Photobacterium damselae subsp. damselae hemolysins damselysin and HlyA are encoded within a new virulence plasmid. Infect. Immun.79, 4617–4627. doi: 10.1128/IAI.05436-11
65
Rivett A. J. Hearn A. R. (2004). Proteasome function in antigen presentation: immunoproteasome complexes, peptide production, and interactions with viral proteins. Curr. Protein Pept. Sci.5, 153–561. doi: 10.2174/1389203043379774
66
Romalde J. L. (2002). Photobacterium damselae subsp. piscicida: an integrated view of a bacterial fish pathogen. Int. Microbiol.5, 3–9. doi: 10.1007/s10123-002-0051-6
67
Romalde J. L. Magariños B. Lores F. Osorio C. R. Toranzo E. (1999). Assessment of a magnetic bead-EIA based kit for rapid diagnosis of fish pasteurellosis. J. Microbiol. Methods382, 147–154. doi: 10.1016/s0167-7012(99)00088-3
68
Romero A. Rey-Campos M. Pereiro P. Librán-Pérez M. Figueras A. Novoa B. (2024). Transcriptomic analysis of turbot (Scophthalmus maximus) treated with zymosan a reveals that lncRNAs and inflammation-related genes mediate the protection conferred against Aeromonas salmonicida. Fish Shellfish Immunol.147, 109456. doi: 10.1016/j.fsi.2024.109456
69
Santos P. Peixoto D. Ferreira I. Passos R. Pires P. Simões M. et al . (2022). Short-term immune responses of gilthead seabream (Sparus aurata) juveniles against Photobacterium damselae subsp. piscicida. Int. J. Mol. Sci.23, 1561. doi: 10.3390/ijms23031561
70
Sasabe J. Suzuki M. Imanishi N. Aiso S. (2014). Activity of D-amino acid oxidase is widespread in the human central nervous system. Front. Synaptic. Neurosci.6. doi: 10.3389/fnsyn.2014.00014
71
Scarl R. T. Lawrence C. M. Gordon H. M. Nunemaker C. S. (2017). STEAP4: its emerging role in metabolism and homeostasis of cellular iron and copper. J. Endocrinol.234, R123–R134. doi: 10.1530/JOE-16-0594
72
Serna-Duque J. A. Cuesta A. Esteban M. Á. (2022a). Massive gene expansion of hepcidin, a host defense peptide, in gilthead seabream (Sparus aurata). Fish Shellfish Immunol.124, 563–571. doi: 10.1016/j.fsi.2022.04.032
73
Serna-Duque J. A. Espinosa Ruiz C. Martínez Lopez S. Sánchez-Ferrer Á. Esteban M. Á. (2022b). Immunometabolic involvement of hepcidin genes in iron homeostasis, storage, and regulation in gilthead seabream (Sparus aurata). Front. Mar. Sci.9. doi: 10.3389/fmars.2022.1073060
74
Silva M. T. Dos Santos N. M. do Vale A. (2010). AIP56: a novel bacterial apoptogenic toxin. Toxins2, 905–918. doi: 10.3390/toxins2040905
75
Souto A. Montaos M. A. Rivas A. J. Balado M. Osorio C. R. Rodríguez J. et al . (2012). Structure and biosynthetic assembly of piscibactin, a siderophore from Photobacterium damselae subsp. piscicida, predicted from genome analysis. Eur. J. Org. Chem.2012, 5693–5700. doi: 10.1002/ejoc.201200818
76
Thyssen A. Ollevier F. (2001). In vitro antimicrobial susceptibility of Photobacterium damselae subsp. piscicida to 15 different antimicrobial agents. Aquaculture200, 259–269. doi: 10.1016/S0044-8486(01)00517-8
77
Tran H. B. Lee Y. H. Guo J. J. Cheng T. C. (2018). De novo transcriptome analysis of immune response on cobia (Rachycentron canadum) infected with Photobacterium damselae subsp. piscicida revealed inhibition of complement components and involvement of MyD88-independent pathway. Fish Shellfish Immunol.77, 120–130. doi: 10.1016/j.fsi.2018.03.041
78
Ullah I. Lang M. (2023). Key players in the regulation of iron homeostasis at the host-pathogen interface. Front. Immunol.14. doi: 10.3389/fimmu.2023.1279826
79
Valenzuela-Muñoz V. Pereiro P. Álvarez-Rodríguez M. Gallardo-Escárate C. Figueras A. Novoa B. (2019). Comparative modulation of lncRNAs in wild-type and rag1-heterozygous mutant zebrafish exposed to immune challenge with spring viraemia of carp virus (SVCV). Sci. Rep.9, 14174. doi: 10.1038/s41598-019-50766-0
80
Vallecillos A. Chaves-Pozo E. Arizcun M. Perez R. Afonso J. M. Berbel C. et al . (2021). Genetic parameters for Photobacterium damselae subsp. piscicida resistance, immunological markers and body weight in gilthead seabream (Sparus aurata). Aquaculture543, 736892. doi: 10.1016/j.aquaculture.2021.736892
81
Verga Falzacappa M. V. Muckenthaler M. U. (2005). Hepcidin: iron-hormone and anti-microbial peptide. Gene364, 37–44. doi: 10.1016/j.gene.2005.07.020
82
Wang M. Jiang S. Wu W. Yu F. Chang W. Li P. et al . (2018). Non-coding RNAs function as immune regulators in teleost fish. Front. Immunol.9. doi: 10.3389/fimmu.2018.02801
83
Wang L. Park H. J. Dasari S. Wang S. Kocher J. P. Li W. (2013). CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res.41, e74. doi: 10.1093/nar/gkt006
84
Wei X. Tu Y. Bu S. Guo G. Wang H. Wang Z. (2024). Unraveling the intricate web: complement activation shapes the pathogenesis of sepsis-induced coagulopathy. J. Innate. Immun.16, 337–353. doi: 10.1159/000539502
85
Xing Q. Feng Y. Sun H. Yang S. Sun T. Guo X. et al . (2021). Scavenger receptor MARCO contributes to macrophage phagocytosis and clearance of tumor cells. Exp. Cell Res.408, 112862. doi: 10.1016/j.yexcr.2021.112862
86
Yu Y. Wang Y. Liao M. Shi L. Li Y. Li B. et al . (2022). Comparative analysis of spleen transcriptome of immune response in Sebastes schlegeli against Photobacterium damselae subsp. damselae infection. Aquac. Res.53, 232–242. doi: 10.1111/are.15569
87
Zhang H. Yang Q. Sun M. Teng M. Niu L. (2004). Hydrogen peroxide produced by two amino acid oxidases mediates antibacterial actions. J. Microbiol.42, 336–339.
88
Zhou D. Zhang B. Qiu Y. Li X. Zhang J. (2025). First report and pathogenicity analysis of Photobacterium damselae subsp. piscicida in cage-cultured black rockfish (Sebastes schlegelii) associated with skin ulcers. Microorganisms13, 441. doi: 10.3390/microorganisms13020441
Summary
Keywords
transcriptome, lncRNAs, gilthead sea bream, Photobacterium damselae subsp. piscicida, immune response
Citation
Pereiro P, Figueras A and Novoa B (2025) Deciphering the transcriptomic and lncRNA landscape of gilthead sea bream (Sparus aurata) in response to Photobacterium damselae subsp. piscicida infection. Front. Mar. Sci. 12:1604207. doi: 10.3389/fmars.2025.1604207
Received
01 April 2025
Accepted
20 August 2025
Published
09 September 2025
Volume
12 - 2025
Edited by
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), India
Reviewed by
Sofia Priyadarsani Das, National Taiwan Ocean University, Taiwan
Feng-Jie Su, National Taiwan University, Taiwan
Updates
Copyright
© 2025 Pereiro, Figueras and Novoa.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patricia Pereiro, patriciapereiro@iim.csic.es
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.