Abstract
In the United States, shark meat is sold in grocery stores, seafood markets, and online. The meat is often mislabeled as another species or generically labeled as “shark”. The ambiguity of these generic labels makes it challenging to assess the conservation implications of this practice and for consumers to avoid high trophic-level species that often have high mercury concentrations in their tissues. We purchased and DNA barcoded 29 shark products in the United States to determine their species identity. Our samples consisted of 19 filets sold in grocery stores, seafood markets, and Asian specialty markets (mostly in North Carolina) and 10 ordered online as “jerky”. Ninety three percent of samples (27 of 29) were ambiguously labeled as shark or mako shark but not as a specific species. Of the two samples that were labeled to species, one was mislabeled (e.g., shortfin mako shark labeled as blacktip shark) and the other was correctly labeled. Barcoding indicated that the 29 samples included 11 different species of shark, including three species listed by the IUCN as Critically Endangered: great hammerhead, scalloped hammerhead, and tope. Previous studies have found that the first two species contain very high levels of mercury, illustrating the implications of seafood mislabeling for human health. The availability of shark meat in U.S. grocery stores is surprising given the dramatic decline of shark populations globally. Moreover, the fact that nearly all shark meat is labeled ambiguously or incorrectly amplifies the problem. Accurate, verified product labels for shark meat would benefit consumers and shark conservation efforts, and should be a priority for the seafood industry.
Introduction
Fishing has caused dramatic declines in shark populations worldwide (Baum et al., 2003; Ward-Paige et al., 2012; Valdivia et al., 2017; Roff et al., 2018). As a result, one-third of shark species are threatened with extinction and designated as Critically Endangered, Endangered, or Vulnerable by the International Union for Conservation of Nature (IUCN) (Dulvy et al., 2021). Moreover, a recent study found that despite international efforts to conserve and protect sharks, global shark mortality from fishing is still rapidly increasing (Worm et al., 2024).
The shark fin and meat trade is a global market with the global trade for shark meat steadily expanding since the early 2000s (Dent and Clarke, 2015). Sharks are harvested for food (e.g., shark fin soup and meat) and other commercial products (e.g., squalene which is commonly used in moisturizing skincare products), and in many regions they are targeted specifically for their culturally and economically valuable fins (Clarke, 2004; Man et al., 2014). Although shark finning is generally considered the main threat to shark populations, the market for shark meat (from the bodies of sharks) has been increasing globally (Dent and Clarke, 2015) and has surpassed the fin market in terms of both volume and value (Niedermüller et al., 2021). The troubling growth in the global trade of shark meat is due to several factors, including growing consumer demand for seafood, the overfishing of other stocks, and perversely, regulations designed to reduce shark finning (Dent and Clarke, 2015; Niedermüller et al., 2021; Worm et al., 2024).
Most shark products, especially fins, are exported to countries in East and Southeast Asia such as China, Hong Kong, Taiwan, Singapore, Malaysia, and Vietnam (Vannuccini, 1999; Boon, 2017; Cardeñosa et al., 2020). In contrast, some of the world’s largest consumers of shark meat are in South America and Europe, particularly Italy, Brazil, Uruguay, and Spain (Dent and Clarke, 2015; Niedermüller et al., 2021). In these regions non-threatened shark species are often sold legally; however, the meat is typically labeled incorrectly (e.g., labeling a shortfin mako shark as a blacktip shark) or ambiguously as “shark” (Barbuto et al., 2010; Almerón-Souza et al., 2018; Pazartzi et al., 2019) (Figure 1). Ambiguous labeling is common in the sale of a variety of seafood products, e.g., use of labels such “scallops” or “squid” without species-level identification. Cundy et al. (2023) found in Australia that ambiguously labeled seafood products had higher rates of mislabeling than products labeled to species. For example, samples labeled as “shark” or “flake” were often identified as chimaeras (Callorhinchus spp.) rather than a species of shark. Previous studies have found that shark meat is often mislabeled (Agyeman et al., 2021) or ambiguously labeled as “shark” regardless of species (Bornatowski et al., 2013; Pazartzi et al., 2019; Abdullah et al., 2020). Shark meat is also commonly sold under various ambiguous and colloquial terms that obscure its identity from consumers (Bornatowski et al., 2013). For example, in South Africa shark is sold as “ocean filets’’ or “skomoro”, in Brazil elasmobranchs (i.e., sharks, skates, and rays) are sold as “cação” (Bornatowski et al., 2013), in Australia shark is sold under the term “flake” (Braccini et al., 2020), and in the United Kingdom small sharks are commonly sold as “rock salmon”, “huss”, “rock eel”, and “rigg” (Hobbs et al., 2019). In the United States, the term “dogfish” (referring to sharks in the family Squalidae) is also commonly used, which may mislead consumers into believing they are purchasing a generic fish product rather than a species of shark.
Figure 1

Photos exemplifying how some of the samples were labeled and displayed when purchased. (A) Dusky smooth-hound sold whole, missing the head, labeled as “shark” in English and Chinese at an Asian grocery store in Orlando, FL. (B) Shortfin mako shark sold as jerky and purchased online from the Newport Jerky Company in Rhode Island labeled as “mako shark jerky”. (C) Common thresher shark sold as “fresh shark [steak]” at an Asian grocery store in Duluth, GA. The label also included “wild caught/USA”. (D) Blacktip shark labeled as “wild blacktip shark, fresh never frozen” from a Publix grocery store in Cary, NC. The label also included that it was a “product of Florida” and that it was “responsibly sourced”.
The use of ambiguous, colloquial labeling has been shown to confuse consumers about what they are purchasing. For example, Bornatowski et al. (2015) interviewed customers at fish markets in Southern Brazil and found that 61% of respondents claimed that they ate cação but not shark. Similarly, López de la Lama et al. (2018) found that 77.5% of Peruvians that eat shark meat were unaware that they were consuming shark since it was commonly labeled with the colloquial term “tollo”. Hobbs et al. (2019) reported that shark (primarily spiny dogfish Squalus acanthias) is commonly sold in takeaway fish and chips meals across the United Kingdom. The study also found that the sale of shark is common in UK grocery stores and seafood markets including IUCN Critically Endangered scalloped hammerhead (Sphyrna lewini). Likewise, Munguia-Vega et al. (2022) found that in Mexico, scalloped hammerhead and several other IUCN Endangered and Vulnerable species were being sold generically as “cazon”, a term used in the Mexican seafood trade to mean any shark meat.
Generic umbrella terms also allow for species that are subject to international trade restrictions under the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) to enter the market undetected. Moreover, ambiguous labeling and mislabeling hinder efforts to monitor and manage species-specific population trends, undermining stock assessments, sustainable catch quotas, and international reporting obligations and potentially allowing declines in vulnerable species to go unnoticed.
Although shark products are widely traded in the global food system, substantial information gaps remain regarding their trade frequency, geographic extent, species identity, and numerous other aspects of the shark trade (Dent and Clarke, 2015; Van Houtan et al., 2020; Niedermüller et al., 2021). Limited information on the shark trade constrains our ability to quantitatively evaluate the implications of shark consumption for human health (e.g., exposure to heavy metals) and marine biodiversity conservation. Improved species-specific market labels and trade data are essential to more accurately evaluate these risks and inform policy decisions. Between 2010 and 2023, 17.1% of commercial seafood samples (i.e., bony fish and shellfish) analyzed in the United States were found to have unacceptable market names (Ahles et al., 2025). Despite the potential conservation and public health implications, only four studies have used molecular techniques (e.g., DNA barcoding) to quantify the accuracy of shark product labeling in the United States (Wallace et al., 2012; Cardeñosa, 2019; Hellberg et al., 2019; Eppley and Coote, 2025). DNA barcoding (i.e., used for species identification based on short, standardized DNA sequences) is an essential tool when investigating shark product mislabeling because in most cases when shark products are sold it is extremely difficult to visually identify the species because distinguishing morphological characteristics are removed.
The purpose of this study was to quantify the identity of “shark” meat sold in the United States. We purchased 30 samples from grocery stores and online jerky vendors. Using standard DNA barcoding techniques, we identified 29 of the 30 samples to the species level. We found that Critically Endangered sharks, including great hammerhead (Sphyrna mokarran) and scalloped hammerhead, are available in grocery stores. Moreover, of the 29 samples that were successfully identified to the species level, 93% were labeled ambiguously and one of the two products labeled at the species level was mislabeled.
Methods
Sample collection
This study was conducted by students and instructors (including undergraduate teaching assistants) in the Seafood Forensics course (BIOL 221) at The University of North Carolina at Chapel Hill. A total of 30 shark products were collected from September 2021 to September 2022, including 19 raw shark steaks and 11 packages of shark jerky from seafood markets, grocery stores, Asian markets, and online vendors in the United States. Jerky is a preserved meat product that can be made from various animal species, in which the meat is salted, seasoned, and dehydrated. All raw shark steaks were purchased on the east coast of the United States (i.e., North Carolina (n = 13), Florida (n = 3), Georgia (n = 2), and Washington, D. C. (n = 1)). For each sample, we recorded how the seller had labeled the item (e.g., “shark steak”, “Mako shark”, “jerky”, etc.) and the price (Supplementary Table S1). Steak samples were kept frozen at -20 °C and jerky samples were kept at room temperature in their unopened original packaging until DNA extraction.
DNA laboratory procedures
DNA extractions were completed using the Qiagen DNeasy Blood and Tissue Kit, following the manufacturer’s protocol but using a one-hour digestion at 55°C and eluted with 25 μl of molecular biology grade water. Polymerase Chain Reaction (PCR) was used to amplify three mitochondrial DNA gene fragments, specifically: 610 base pairs (bp) of cytochrome c oxidase subunit I (COI; (Ivanova et al., 2007)), 464 bp of cytochrome B (CytB; (Wolf et al., 2000)), and 990 bp of NADH dehydrogenase subunit 2 (ND2; (Vella et al., 2017)). Separate PCR reactions were performed for each primer set across all 30 extracted samples (Supplementary Table S2), resulting in a total of 90 PCR reactions. We used three sets of primers (i.e., COI, ND2, and CytB) on each sample because the COI region lacked the resolution to distinguish between closely related species in some cases (e.g., blacktip and spinner sharks). We utilized a modified version of Spencer and Bruno (2019) PCR protocol with two additional sets of primers (i.e., ND2 and CytB). Separate PCR reactions were run for each primer set by adding 1.3 μl of each primer at a concentration of 10 uM to individual 0.2 ml illustra puReTaq Ready-To-Go PCR bead tubes. We then added 20 -100 ng of DNA to each tube. Molecular biology grade water was added to each tube to bring the final volume to 25 μl: 18.8 μl with the COI primer and 21.4 μl for the tubes with either the CytB or ND2 primers. To ensure that there was no DNA contamination, control tubes that contained only primer and molecular biology grade water were used for each sample and primer. Once the PCR bead was dissolved, we placed the tubes into a Bio-Rad T100 Thermal Cycler and used the following thermal cycling protocol for all three primer sets (Willette et al., 2017; Spencer and Bruno, 2019).
-
Initial denaturing: 95°C for 5 min
-
Denature: 94°C for 30 s
-
Annealing: 50°C for 45 s
-
Final extension: 72°C for 10 min
(steps 2 – 4 were repeated for 35 cycles)
To assess the results of the PCR amplification we used gel electrophoresis following the same protocol as Spencer and Bruno (2019). The PCR products were sent to Eton Bioscience in Durham, North Carolina for purification and Sanger sequencing in the forward direction. We followed the protocol from Spencer and Bruno (2019) for the COI amplicon and used a newly designed tagged primer consisting of M13F-21 (5’ TGTAAAACGACGGCCAGT 3’) annealed to a gene-specific primer for sequencing. The ND2 and CytB amplicons were sequenced using their respective forward primers (Supplementary Table S2).
Sequencing analysis
Primers and bases with more than a 1% per base error rate were trimmed from the forward sequences of each amplicon in Geneious Prime (v.2024.0.3) leaving zero ambiguous or low-quality bases. Primer sequences were conservatively trimmed, allowing up to 3 mismatches in the primer-binding regions. Sequences with greater than 100 bp remaining were retained for further analysis to maintain enough sequence length for reliable identification. Sequences were then assigned to a species or genus with a 98% minimum sequence similarity threshold, using the megablast algorithm of the Basic Local Alignment Search Tool (BLAST) in Geneious Prime. The top 100 BLAST hits for each sequence are available at (https://doi.org/10.6084/m9.figshare.22943474.v1). In the case of disagreement between CytB and ND2 (i.e., sample 1), we preferred the ND2 assignment because this gene was able to achieve better overall identification success than CytB for our samples.
Following sequencing similarity analysis with BLAST, we assessed the phylogenetic relationships between each query sample sequence and its top five BLAST matches based on the BLAST scores. We added up to four additional accessions when available for species which were represented by only one accession in each alignment. For each sample, we downloaded the top five BLAST hit accessions and supplemented the dataset with additional accessions for species represented by only a single accession in each alignment. For each best BLAST match, two congeners were added to the tree, represented by three accessions each. Sample sequences and reference sequences for each marker gene were aligned to the mitochondrial genome of Mobula alfredi (coastal manta ray; NC068734) using the Geneious assembler algorithm in Geneious Prime to standardize read directions. Using the assembled reads, a multiple sequence alignment was performed with Clustal Omega (v.1.2.3), and the region of the target gene with maximum query sequence coverage was extracted for each target gene region (COI = 161bp, ND2 = 100bp, CytB = 310bp). A new multiple sequence alignment of the trimmed extracted sequences was made with Clustal Omega. A RAXML tree was then created from the alignments using a GTR Gamma model with an algorithm incorporating rapid bootstrapping and search for the best scoring maximum likelihood tree (Figure 2). The tree was rooted using Mobula alfredi (NC068734) as the outgroup. We selected this ray species because its complete mitogenome allowed it to serve as an outgroup across all three genes.
Figure 2
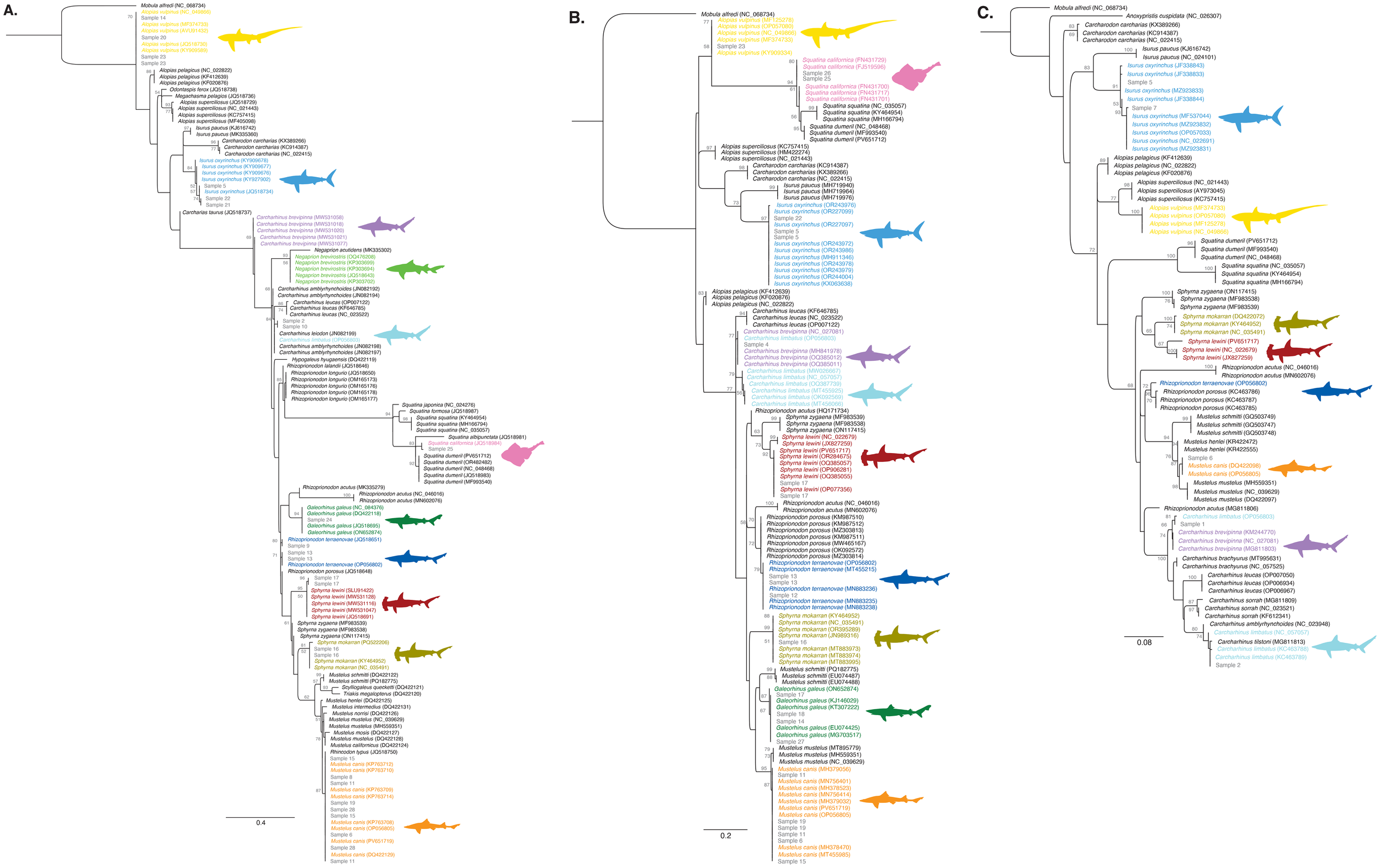
Phylogenetic species identification based on maximum likelihood analyses of (A) NADH dehydrogenase subunit 2 (ND2), (B) cytochrome c oxidase subunit I (COI), and (C) cytochrome B (CytB). Branches are labeled with support values ≥50% from four bootstrap replicates. Reference sequences include GenBank accession numbers in parentheses, and colors correspond to the identified shark species.
All samples were categorized as either ambiguously labeled (i.e., as “shark” without species-level identification), mislabeled (labeled as a specific species but genetically identified as a different species), or correctly labeled (the labeled species matched the genetic identification). Samples labeled as “mako shark” or “thresher shark” were also considered ambiguously labeled, as these common names refer to multiple, distinct species — there are two species of mako shark (Isurus oxyrinchus and Isurus paucus) and three species of thresher shark (Alopias pelagicus, A. superciliosus, and A. vulpinus). The conservation status of each species was determined using the IUCN Red List of Threatened Species (http://www.iucnredlist.org).
Results
Twenty nine of the 30 samples were successfully identified to the species level with at least one of the three genes. Twenty-seven were labeled ambiguously as “shark”: all 10 of the jerky samples and 17 of the 19 filets. One of the two samples labeled at the species level was mislabeled (i.e., sold as blacktip shark but was shortfin mako) and only one sample was correctly labeled, and it was a blacktip shark. Twenty nine of the 30 samples were identified as shark and came from 11 species, including three species listed by the IUCN as Critically Endangered: great hammerhead, scalloped hammerhead, and tope (Galeorhinus galeus) (Figure 3, Supplementary Table S1). The average price in USD of the fresh shark meat was $6.30/lb ± 2.88 ($13.86/kg ± 6.51) with prices ranging from $2.99/lb ($6.59/kg) to $11.99/lb ($26.41/kg). The average price of the shark jerky was $5.08/oz ± 0.99 ($207.37/kg ± 37.21).
Figure 3
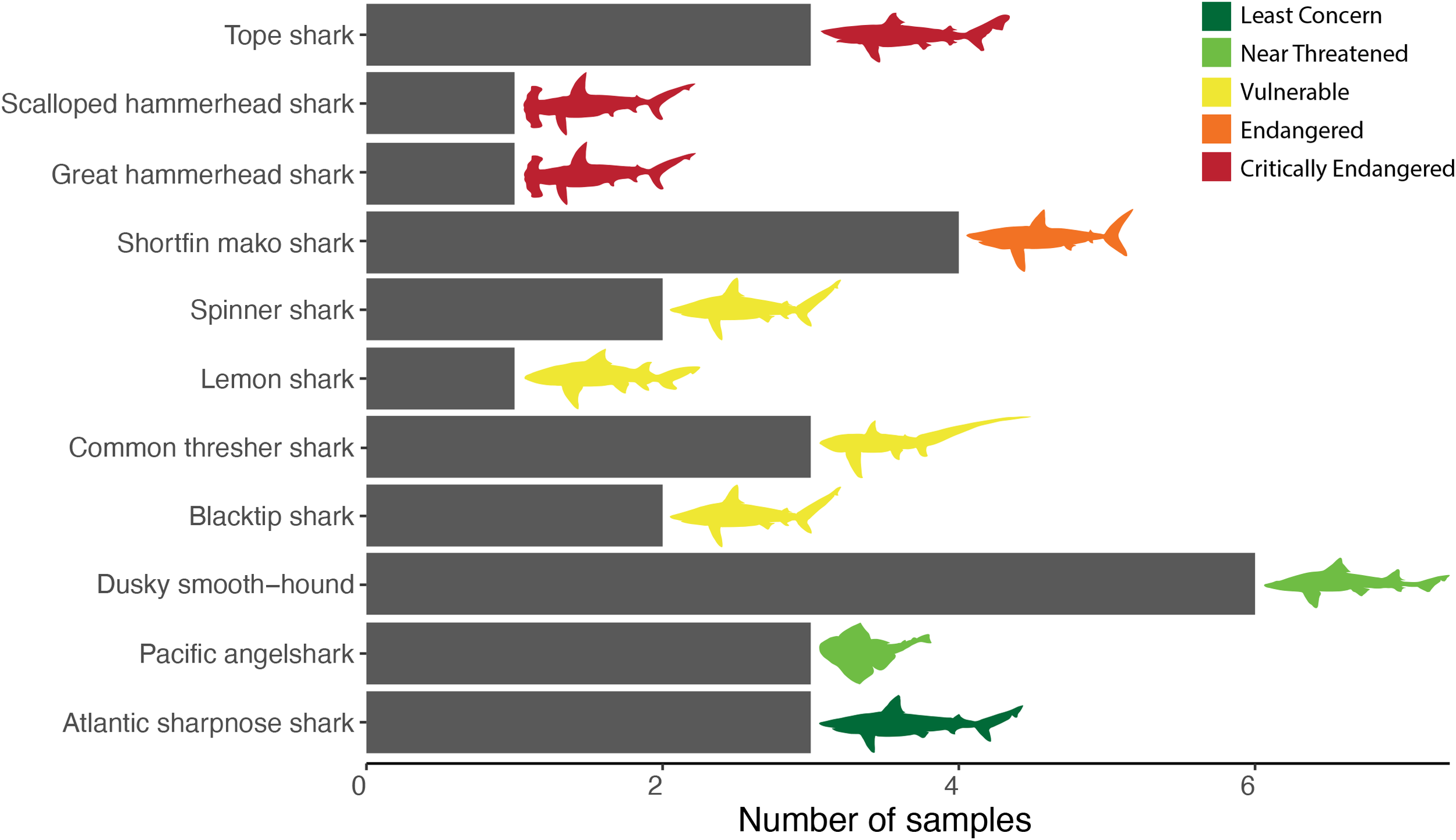
The number of each species found in our analysis using DNA barcoding to identify products labeled as shark. Species are color-coded to depict IUCN status. Figure includes all identified samples (n = 29) and only the identity of each sample from the DNA barcoding results.
Discussion
Overall, we found a high level of ambiguous and incorrect shark product labeling and evidence of IUCN listed endangered species in the U.S. food supply. Thirty one percent of our samples were classified by the IUCN as Endangered or Critically Endangered species (Figure 3). All nine of these samples were also either ambiguously or incorrectly labeled. Out of the 29 samples, only one was correctly labeled and it was from a blacktip shark. Although our sample size is relatively small (n = 29), we still detected multiple Critically Endangered and CITES Appendix II species being sold in U.S. markets. These results are concordant with those of similar studies that were conducted in the United States and Canada (Wallace et al., 2012; Hellberg et al., 2019). For example, all 35 shark products (i.e., jerky, shark fin soup, cartilage pills, and fresh or grilled fillets) collected from Orange and Los Angeles Counties, CA, USA and online by Hellberg et al. (2019) were labeled ambiguously.
Vague and inaccurate labeling can greatly impact the conservation of marine predators by concealing the sale of endangered species. Although consumers are indeed being sold “shark”, there are at least 536 species of sharks (Dulvy et al., 2021) ranging in size, diet, habitat, and conservation status. For example, scalloped hammerhead sharks are Critically Endangered with a population that continues to decline globally (Rigby et al., 2019). Several aspects of the life history of hammerhead sharks make them especially vulnerable to fishing (Gallagher et al., 2014). Despite their status, the scalloped hammerhead was identified as one of the species labeled ambiguously as “shark” in our study. Ensuring transparency and traceability is essential for maintaining legal and sustainable trade in sharks and for preventing the sale of protected species (Niedermüller et al., 2021). In the United States, the Food and Drug Administration (FDA) has issued a Seafood List specifying acceptable common names, market names, and scientific names required for commercial trade (The United States Food and Drug Administration, 2025). However, as of 2025, the acceptable market name for all listed shark species is “shark” (The United States Food and Drug Administration, 2025), diminishing the regulatory effectiveness of species-specific labeling. In contrast, the European Union has implemented a more comprehensive labeling requirement under Regulation (EU) 1379/2013 which mandates that seafood labels include the scientific name, method of harvest or production, catch area, country of origin, fishing gear used, and other relevant information (Council of the European Union & European Parliament, 2013). Sellers in the United States should be required to provide species-specific names, and when shark meat is not a food security necessity, consumers should avoid purchasing products that lack species-level labeling or traceable sourcing (Niedermüller et al., 2021).
Shark fins are a highly sought out product throughout Asia (Eriksson and Clarke, 2015) specifically for shark fin soup. The lower caudal fin of hammerheads (Sphyrna spp.) and the shortfin mako (Isurus oxyrinchus) are considered to have the best quality fin needles for consumption (Clarke et al., 2007), resulting in a high demand for these species. Cardeñosa et al. (2022) found that scalloped hammerhead was the third most common species sold in a Hong Kong fish market as fins. The shortfin mako was the sixth most common species (Cardeñosa et al., 2022). Additionally, Almerón-Souza et al. (2018) found scalloped hammerhead to be the most abundant species when testing shark meat sold in Brazil. These and other shark species threatened with extinction are present-to-common in the food supply in numerous countries around the world.
Eight of our 29 samples were labeled as “mako shark” (n = 7) or “shortfin mako” (n = 1). A higher demand for mako shark fins and meat could be why most mislabeled meat products were labeled as mako. Makos (i.e., Isurus oxyrinchus and Isurus paucus), threshers (i.e., Alopias spp.), larger hammerheads (i.e., Sphyrna lewini, Sphyrna mokarran, and Sphyrna zygaena), and all requiem sharks (e.g., Carcharhinus brevipinna, Negaprion brevirostris, Carcharhinus limbatus, and Rhizoprionodon terraenovae) are all classified under CITES Appendix II which require export permits due to their extreme vulnerability if trade is not strictly controlled. Fifty eight percent of our samples were listed under CITES Appendix II. Catch location was only available for four out of the 30 samples, three being labeled as “caught in the USA” and one labeled as “caught in Hawaii”. Due to this we were unable to determine if the shark meat had been locally sourced in the United States for most of the samples. All of the shark species that we identified are commonly caught in the United States and internationally (IUCN, 2022) which could create potential for CITES violations. Mislabeling is sometimes used to disguise CITES restricted or endangered species (e.g., Sphyrna lewini, Sphyrna mokarran, and Isurus oxyrinchus) being harvested for the fin trade since different species of shark meat can look identical when fileted or dried as jerky.
The Shark Conservation Act of 2010 (H.R. 81. Shark conservation act of 2010, Public Law 111-348., Pub. L. No. 111–348 (2010) requires that all sharks in the United States, with one exception (Mustelus canis), be brought to shore with their fins naturally attached. This law has forced fishermen to land the entire shark rather than only taking the fins and disposing the body at sea, thereby potentially increasing the amount of shark meat being sold as a byproduct. Shark meat is oftentimes considered to be “junk meat” due to its unpleasant smell and taste which is a result of high amounts of urea in the tissue (Suryaningsih et al., 2020). In Europe and North America, few countries restrict the sale of species categorized as Critically Endangered by the IUCN (Frank and Wilcove, 2019). The market cost for most of our samples was very low compared to the other species of fish, especially in grocery stores. For example, a scalloped hammerhead filet was only $4.99/lb ($10.99/kg) while red snapper (Lutjanus campechanus) can commonly be sold as much as $27/lb ($59.4/kg) (Spencer et al., 2020). Given the massive ecological cost of harvesting such an important, rare, and long-lived predator (e.g., scalloped hammerhead) it is remarkable how low their market price is.
Shark products can contain heavy metals including arsenic and mercury (Garcia Barcia et al., 2020) with the highest concentrations in muscle tissue (Tiktak et al., 2020). Consumption of these heavy metals can have negative effects on human health (Tiktak et al., 2020; Souza-Araujo et al., 2021). For example, García Barcia et al. (2022) found high levels of mercury in meat and fin products from large hammerhead species, Sphyrna mokarran, S. zygaena, and S. lewini, and advised consumers to avoid these species. They also recommended that “species specific advisories to be issued for meat and fin products from oceanic whitetip (Carcharhinus longimanus) and dusky smooth-hound sharks (Mustelus canis), which should be avoided by women of childbearing age again due to high mercury levels” (García Barcia et al., 2022). We were sold three of these five species (i.e., scalloped hammerhead, great hammerhead, and dusky smooth-hound) all of which were labeled generically as “shark”. Consumption of mercury can cause damage to the brain and central nervous system while arsenic can lead to skin, bladder, and lung cancer (Ratcliffe et al., 1996; Honda et al., 2006; Ravenscroft et al., 2011; Skalny et al., 2022). Consumption of these metals has been related to fetal cognitive development complications and infant death (Honda et al., 2006; Farzan et al., 2013; Tolins et al., 2014; Quansah et al., 2015). The ambiguous labeling of shark meat can be detrimental to human health. When consumers are purchasing ambiguously labeled or mislabeled shark meat, they have no way to know what species they are consuming and what the associated health risks might be.
Our sampling and inferences about shark steak mislabeling are biased towards North Carolina. However, the jerky samples were purchased from national vendors (all based in the United States) therefore the inferences made from those samples are representative of the geographic region. Due to our limited sample size, future work should include a larger, more geographically widespread sampling to better characterize the sale of shark products. The species composition of shark meat being sold in the United States and the rest of the world is still largely unknown. Increased monitoring and accurate, verified product labels for shark meat are necessary given the high proportion of species threatened with extinction found within our small sample size. Fields et al. (2020) recently used genetic stock identification to identify source populations of the scalloped hammerheads found in the Hong Kong fin trade. Genetic stock identification could also be used to decipher what specific populations or regions the shark meat is coming from (e.g., scalloped hammerheads from the Eastern Pacific). With a comprehensive genetic database of shark stocks in the United States, and for vulnerable populations, this method could provide a better understanding of whether these CITES listed species are being imported illegally and if these sharks are being fished from overexploited regions.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because all shark meat used in the study was purchased from grocery stores where the shark was already deceased and butchered.
Author contributions
SR: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. TY: Conceptualization, Investigation, Writing – review & editing. KJO: Conceptualization, Investigation, Writing – review & editing. EW: Formal analysis, Writing – review & editing. MA: Conceptualization, Investigation, Writing – review & editing. EH: Investigation, Writing – review & editing. PL: Investigation, Writing – review & editing. SG: Investigation, Writing – review & editing. WB: Investigation, Writing – review & editing. JB: Investigation, Writing – review & editing. RC: Investigation, Writing – review & editing. IF: Investigation, Writing – review & editing. AG: Investigation, Writing – review & editing. KH: Investigation, Writing – review & editing. DH: Investigation, Writing – review & editing. TH: Investigation, Writing – review & editing. BL: Investigation, Writing – review & editing. JL: Investigation, Writing – review & editing. GM: Investigation, Writing – review & editing. KM: Investigation, Writing – review & editing. AP-S: Investigation, Writing – review & editing. AG: Investigation, Writing – review & editing. AR: Investigation, Writing – review & editing. AT: Investigation, Writing – review & editing. RW: Investigation, Writing – review & editing. AZ: Investigation, Writing – review & editing. JFB: Conceptualization, Funding acquisition, Investigation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was funded in part by the Department of Biology at The University of North Carolina at Chapel Hill. Samples were collected and analyzed as part of the Seafood Forensics CURE class at UNC-CH (BIOL 221). The project was also partially funded by the National Science Foundation (OCE #2128592 to JFB).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1604454/full#supplementary-material
References
1
Abdullah A. Nurilmala M. Muttaqin E. Yulianto I. (2020). DNA-based analysis of shark products sold on the Indonesian market towards seafood labelling accuracy program. Biodiversitas21, 1385–1390. doi: 10.13057/biodiv/d210416
2
Agyeman N. A. Blanco-Fernandez C. Steinhaussen S. L. Garcia-Vazquez E. MaChado-Schiaffino G. (2021). Illegal, unreported, and unregulated fisheries threatening shark conservation in african waters revealed from high levels of shark mislabelling in Ghana. Genes12, 1002. doi: 10.3390/genes12071002
3
Ahles S. DeWitt C. A. M. Hellberg R. S. (2025). A meta-analysis of seafood species mislabeling in the United States. Food Control171, 111110. doi: 10.1016/j.foodcont.2024.111110
4
Almerón-Souza F. Sperb C. Castilho C. L. Figueiredo P. I. C. C. Gonçalves L. T. MaChado R. et al . (2018). Molecular identification of shark meat from local markets in southern Brazil based on DNA barcoding: evidence for mislabeling and trade of endangered species. Front. Genet.9. doi: 10.3389/fgene.2018.00138
5
Barbuto M. Galimberti A. Ferri E. Labra M. Malandra R. Galli P. et al . (2010). DNA barcoding reveals fraudulent substitutions in shark seafood products: The Italian case of “palombo” (Mustelus spp.). Food Res. Int.43, 376–381. doi: 10.1016/j.foodres.2009.10.009
6
Baum J. K. Myers R. A. Kehler D. G. Worm B. Harley S. J. Doherty P. A. (2003). Collapse and conservation of shark populations in the northwest atlantic. Science299, 389–392. doi: 10.1126/science.1079777
7
Boon P. Y. (2017). The shark and ray trade in Singapore (Selangor, Malaysia: TRAFFIC Southeast Asia Regional Office, Petaling Jaya).
8
Bornatowski H. Braga R. R. Kalinowski C. Vitule J. R. S. (2015). Buying a pig in a poke”: the problem of elasmobranch meat consumption in southern Brazil. EBL6, 196–202. doi: 10.14237/ebl.6.1.2015.451
9
Bornatowski H. Braga R. R. Vitule J. R. S. (2013). Shark mislabeling threatens biodiversity. Science340, 923–923. doi: 10.1126/science.340.6135.923-a
10
Braccini M. Blay N. Harry A. Newman S. J. (2020). Would ending shark meat consumption in Australia contribute to the conservation of white sharks in South Africa? Mar. Policy120, 104144. doi: 10.1016/j.marpol.2020.104144
11
Cardeñosa D. (2019). Genetic identification of threatened shark species in pet food and beauty care products. Conserv. Genet.20, 1383–1387. doi: 10.1007/s10592-019-01221-0
12
Cardeñosa D. Fields A. T. Babcock E. A. Shea S. K. Feldheim K. A. Chapman D. D. (2020). Species composition of the largest shark fin retail-market in mainland China. Sci. Rep.10, 12914. doi: 10.1038/s41598-020-69555-1
13
Cardeñosa D. Shea S. K. Zhang H. Fischer G. A. Simpfendorfer C. A. Chapman D. D. (2022). Two thirds of species in a global shark fin trade hub are threatened with extinction: Conservation potential of international trade regulations for coastal sharks. Conserv. Letters15, e12910. doi: 10.1111/conl.12910
14
Clarke S. (2004). Understanding pressures on ®shery resources through trade statistics: a pilot study of four products in the Chinese dried seafood market. Fish and Fisheries5, 53–74. doi: 10.1111/j.1467-2960.2004.00137.x
15
Clarke S. Milner-Gulland E. J. Bjørndal T. (2007). Social, economic, and regulatory drivers of the shark fin trade. Mar. Resource Economics22, 305–327. doi: 10.1086/mre.22.3.42629561
16
Council of the European Union & European Parliament (2013). Regulation (EU) No 1379/2013 of the European Parliament and of the Council of 11 December 2013 on the common organisation of the markets in fishery and aquaculture products, amending Council Regulations (EC) No 1184/2006 and (EC) No 1224/2009 and repealing Council Regulation (EC) No 104/2000. Off. J. Eur. Union.
17
Cundy M. E. Santana-Garcon J. McLennan A. G. Ayad M. E. Bayer P. E. Cooper M. et al . (2023). Seafood label quality and mislabelling rates hamper consumer choices for sustainability in Australia. Sci. Rep.13, 10146.
18
Dent F. Clarke S. (2015). State of the global market for shark products. FAO Fisheries and Aquaculture technical paper, (590), I. Available online at: http://libproxy.lib.unc.edu/login?url=https://www.proquest.com/scholarly-journals/state-global-market-shark-products/docview/1708482071/se-2?accountid=14244.
19
Dulvy N. K. Pacoureau N. Rigby C. L. Pollom R. A. Jabado R. W. Ebert D. A. et al . (2021). Overfishing drives over one-third of all sharks and rays toward a global extinction crisis. Curr. Biol.31, 4773–4787.e8. doi: 10.1016/j.cub.2021.08.062
20
Eppley M. G. Coote T. (2025). DNA barcoding reveals mislabeling of endangered sharks sold as swordfish in New England fish markets. Conserv. Genet.26:381–390. doi: 10.1007/s10592-025-01675-5
21
Eriksson H. Clarke S. (2015). Chinese market responses to overexploitation of sharks and sea cucumbers. Biol. Conserv.184, 163–173. doi: 10.1016/j.biocon.2015.01.018
22
Farzan S. F. Karagas M. R. Chen Y. (2013). In utero and early life arsenic exposure in relation to long-term health and disease. Toxicol. Appl. Pharmacol.272, 384–390. doi: 10.1016/j.taap.2013.06.030
23
Fields A. T. Fischer G. A. Shea S. K. H. Zhang H. Feldheim K. A. Chapman D. D. (2020). DNA Zip-coding: identifying the source populations supplying the international trade of a critically endangered coastal shark. Anim. Conserv.23, 670–678. doi: 10.1111/acv.12585
24
Frank E. G. Wilcove D. S. (2019). Long delays in banning trade in threatened species. Science363, 686–688. doi: 10.1126/science.aav4013
25
Gallagher A. Serafy J. Cooke S. Hammerschlag N. (2014). Physiological stress response, reflex impairment, and survival of five sympatric shark species following experimental capture and release. Mar. Ecol. Prog. Ser.496, 207–218. doi: 10.3354/meps10490
26
Garcia Barcia L. Argiro J. Babcock E. A. Cai Y. Shea S. K. H. Chapman D. D. (2020). Mercury and arsenic in processed fins from nine of the most traded shark species in the Hong Kong and China dried seafood markets: The potential health risks of shark fin soup. Mar. pollut. Bull.157, 111281. doi: 10.1016/j.marpolbul.2020.111281
27
García Barcia L. Valdes A. E. Wothke A. Fanovich L. Mohammed R. S. Shea S. Chapman D. (2023). Health risk assessment of globally consumed shark-derived products. Exposure and Health15 (2), 409–423.
28
Hellberg R. S. Isaacs R. B. Hernandez E. L. (2019). Identification of shark species in commercial products using DNA barcoding. Fisheries Res.210, 81–88. doi: 10.1016/j.fishres.2018.10.010
29
Hobbs C. A. D. Potts R. W. A. Bjerregaard Walsh M. Usher J. Griffiths A. M. (2019). Using DNA barcoding to investigate patterns of species utilisation in UK shark products reveals threatened species on sale. Sci. Rep.9, 1028. doi: 10.1038/s41598-018-38270-3
30
Honda S. Hylander L. Sakamoto M. (2006). Recent advances in evaluation of health effects on mercury with special reference to methylmercury – A minireview. Environ. Health Prev. Med.11, 171–176. doi: 10.1265/ehpm.11.171
31
H.R. 81. Shark conservation act of 2010, Public Law 111-348., Pub. L. No. 111–348 (2010).
32
IUCN (2022). “The IUCN Red List of Threatened Species,” in IUCN Red List of Threatened Species. IUCN. Available online at: https://www.iucnredlist.org/en.
33
Ivanova N. V. Zemlak T. S. Hanner R. H. Hebert P. D. N. (2007). Universal primer cocktails for fish DNA barcoding: BARCODING. Mol. Ecol. Notes7, 544–548. doi: 10.1111/j.1471-8286.2007.01748.x
34
López de la Lama R. de la Puente S. Riveros J. C. (2018). Attitudes and misconceptions towards sharks and shark meat consumption along the Peruvian coast. PloS One13, e0202971. doi: 10.1371/journal.pone.0202971
35
Man Y. B. Wu S. C. Wong M. H. (2014). Shark fin, a symbol of wealth and good fortune may pose health risks: the case of mercury. Environ. Geochem Health36, 1015–1027. doi: 10.1007/s10653-014-9598-3
36
Munguia-Vega A. Terrazas-Tapia R. Dominguez-Contreras J. F. Reyna-Fabian M. Zapata-Morales P. (2022). DNA barcoding reveals global and local influences on patterns of mislabeling and substitution in the trade of fish in Mexico. PloS One17, e0265960. doi: 10.1371/journal.pone.0265960
37
Niedermüller S. Ainsworth G. de Juan S. Garcia R. Ospina-Alvarez A. Pita P. et al . (2021). The shark and ray meat network a deep dive into a global affair. WWF MMI Rome. Available online at: https://wwfeu.awsassets.panda.org/downloads/a4_shark_2021_low.pdf.
38
Pazartzi T. Siaperopoulou S. Gubili C. Maradidou S. Loukovitis D. Chatzispyrou A. et al . (2019). High levels of mislabeling in shark meat – Investigating patterns of species utilization with DNA barcoding in Greek retailers. Food Control98, 179–186. doi: 10.1016/j.foodcont.2018.11.019
39
Quansah R. Armah F. A. Essumang D. K. Luginaah I. Clarke E. Marfoh K. et al . (2015). Association of arsenic with adverse pregnancy outcomes/infant mortality: A systematic review and meta-analysis. Environ. Health Perspect.123, 412–421. doi: 10.1289/ehp.1307894
40
Ratcliffe H. E. Swanson G. M. Fischer L. J. (1996). Human exposure to mercury: A critical assessment of the evidence of adverse health effects. J. Toxicol. Environ. Health49, 221–270. doi: 10.1080/00984108.1996.11667600
41
Ravenscroft P. Brammer H. Richards K. (2011). Arsenic pollution: A global synthesis. (West Sussex, United Kingdom: John Wiley & Sons).
42
Rigby C. L. Dulvy N. K. Barreto R. Carlson J. Fernando D. Fordham S. et al . (2019). “Sphyrna lewini,” in The IUCN Red List of Threatened Species. IUCN.
43
Roff G. Brown C. J. Priest M. A. Mumby P. J. (2018). Decline of coastal apex shark populations over the past half century. Commun. Biol.1, 223. doi: 10.1038/s42003-018-0233-1
44
Skalny A. V. Aschner M. Sekacheva M. I. Santamaria A. Barbosa F. Ferrer B. et al . (2022). Mercury and cancer: Where are we now after two decades of research? Food Chem. Toxicol.164, 113001. doi: 10.1016/j.fct.2022.113001
45
Souza-Araujo J. Souza-Junior O. G. Guimarães-Costa A. Hussey N. E. Lima M. O. Giarrizzo T. (2021). The consumption of shark meat in the Amazon region and its implications for human health and the marine ecosystem. Chemosphere265, 129132. doi: 10.1016/j.chemosphere.2020.129132
46
Spencer E. T. Bruno J. F. (2019). Fishy business: red snapper mislabeling along the coastline of the southeastern United States. Front. Mar. Sci.6. doi: 10.3389/fmars.2019.00513
47
Spencer E. T. Richards E. Steinwand B. Clemons J. Dahringer J. Desai P. et al . (2020). A high proportion of red snapper sold in North Carolina is mislabeled. PeerJ8, e9218. doi: 10.7717/peerj.9218
48
Suryaningsih W. Supriyono Hariono B. Kurnianto M. (2020). Improving the quality of smoked shark meat with ozone water technique. IOP Conf. Ser.: Earth Environ. Sci.411, 12048. doi: 10.1088/1755-1315/411/1/012048
49
The United States Food and Drug Administration (2025). The Seafood List. Available online at: https://www.hfpappexternal.fda.gov/scripts/fdcc/index.cfm?set=SeafoodList&sort=MARKET_NAME&order=ASC&startrow=1&type=basic&search=shark (Accessed April 25, 2025).
50
Tiktak G. P. Butcher D. Lawrence P. J. Norrey J. Bradley L. Shaw K. et al . (2020). Are concentrations of pollutants in sharks, rays and skates (Elasmobranchii) a cause for concern? A systematic review. Mar. pollut. Bull.160, 111701. doi: 10.1016/j.marpolbul.2020.111701
51
Tolins M. Ruchirawat M. Landrigan P. (2014). The developmental neurotoxicity of arsenic: cognitive and behavioral consequences of early life exposure. Ann. Global Health80, 303. doi: 10.1016/j.aogh.2014.09.005
52
Valdivia A. Cox C. E. Bruno J. F. (2017). Predatory fish depletion and recovery potential on Caribbean reefs. Sci. Adv.3, e1601303. doi: 10.1126/sciadv.1601303
53
Van Houtan K. S. Gagné T. O. Reygondeau G. Tanaka K. R. Palumbi S. R. Jorgensen S. J. (2020). Coastal sharks supply the global shark fin trade. Biol. Lett.16, 20200609. doi: 10.1098/rsbl.2020.0609
54
Vannuccini S. (1999). Shark utilization, marketing and trade (Rome: Food and Agriculture Organization of the United Nations).
55
Vella A. Vella N. Schembri S. (2017). A molecular approach towards taxonomic identification of elasmobranch species from Maltese fisheries landings. Mar. Genomics36, 17–23. doi: 10.1016/j.margen.2017.08.008
56
Wallace L. J. Boilard S. M. A. L. Eagle S. H. C. Spall J. L. Shokralla S. Hajibabaei M. (2012). DNA barcodes for everyday life: Routine authentication of Natural Health Products. Food Res. Int.49, 446–452. doi: 10.1016/j.foodres.2012.07.048
57
Ward-Paige C. A. Keith D. M. Worm B. Lotze H. K. (2012). Recovery potential and conservation options for elasmobranchs. J. Fish Biol.80, 1844–1869. doi: 10.1111/j.1095-8649.2012.03246.x
58
Willette D. A. Simmonds S. E. Cheng S. H. Esteves S. Kane T. L. Nuetzel H. et al . (2017). Using DNA barcoding to track seafood mislabeling in Los Angeles restaurants. Conserv. Biol.31, 1076–1085. doi: 10.1111/cobi.12888
59
Wolf C. Burgener M. Hübner P. Lüthy J. (2000). PCR-RFLP analysis of mitochondrial DNA: differentiation of fish species. LWT - Food Sci. Technol.33, 144–150. doi: 10.1006/fstl.2000.0630
60
Worm B. Orofino S. Burns E. S. D’Costa N. G. Manir Feitosa L. Palomares M. L. D. et al . (2024). Global shark fishing mortality still rising despite widespread regulatory change. Science383, 225–230. doi: 10.1126/science.adf8984
Summary
Keywords
shark meat, mislabeling, DNA barcoding, shark conservation, wildlife trade
Citation
Ryburn SJ, Yu T, Ong KJ, Wisely E, Alston MA, Howie E, LeRoy P, Giang SE, Ball W, Benton J, Calhoun R, Favreau I, Gutierrez A, Hallac K, Hanson D, Hibbard T, Loflin B, Lopez J, Mock G, Myers K, Pinos-Sánchez A, Garcia AMS, Romero AR, Thomas A, Williams R, Zaldivar A and Bruno JF (2025) Sale of critically endangered sharks in the United States. Front. Mar. Sci. 12:1604454. doi: 10.3389/fmars.2025.1604454
Received
01 April 2025
Accepted
05 August 2025
Published
10 September 2025
Volume
12 - 2025
Edited by
Xiong Xiong, Nanjing Tech University, China
Reviewed by
Chuanjiang Zhou, Henan Normal University, China
Marcela Alvarenga, Centro de Investigacao em Biodiversidade e Recursos Geneticos (CIBIO-InBIO), Portugal
Updates
Copyright
© 2025 Ryburn, Yu, Ong, Wisely, Alston, Howie, LeRoy, Giang, Ball, Benton, Calhoun, Favreau, Gutierrez, Hallac, Hanson, Hibbard, Loflin, Lopez, Mock, Myers, Pinos-Sánchez, Garcia, Romero, Thomas, Williams, Zaldivar and Bruno.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Savannah J. Ryburn, savannahjryburn@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.