Abstract
Mangrove forests are highly productive coastal ecosystems in which microbial communities drive key biogeochemical processes, such as sulfur and carbon cycling. Lucinid clams and their bacterial symbionts, commonly found in organic-rich habitats such as mangrove fringes, can perform chemosynthesis driven by the oxidation of sulfide and can further modify sediment characteristics through their chemosymbiotic and burrowing activities. However, the impact of these activities on sedimentary microbial communities remains unclear. In this study, we collected sediment samples from areas with varying lucinid densities and conducted amplicon sequencing to examine microbial community structure. Our findings demonstrated that microbial diversity and community composition varied considerably with lucinid density. Among the dominant microbial taxa, sulfur-reducing groups such as Desulfobacterales were significantly more abundant in samples from lucinid-rich regions. Furthermore, the abundance of sulfur-reducing functional genes was higher in lucinid-rich regions. These results indicate that the sulfur cycle is more active in these areas, possibly due to the high organic matter content and the presence of chemosymbiotic lucinids. The accelerated sulfur cycle can enhance carbon fixation by lucinid symbionts, highlighting the ecological importance of chemosymbiotic bivalves in peri-mangrove microbial communities.
1 Introduction
Mangrove forests are unique ecosystems found in the intertidal zones of coastal wetlands. They are characterized by high productivity and a distinct microbial community structure (Liu et al., 2019). Mangroves play crucial ecological roles, including maintaining biodiversity and promoting the circulation of coastal materials and energy flow (Mattone and Sheaves, 2024). Microorganisms function as key components of the mangrove community, playing essential roles in the recycling of biochemical elements, particularly in the carbon sequestration cycle (Zhuang et al., 2020; Sunish, 2024). Furthermore, they drive many of the complex biogeochemical cycles essential for the high productivity of mangrove ecosystems, accelerating processes such as carbon fixation, ammonia oxidation, nitrogen fixation, sulfide oxidation, and Dimethyl sulfoniopropionate (DMSP) catabolism (Lin et al., 2019). For example, in the sulfur cycle, microorganisms link the oxidation of sulfides and the reduction of sulfates to carbon dioxide assimilation (Wu et al., 2021; Zhou et al., 2025). Meanwhile, the construction of microbial communities is important for ecosystem function (Bittleston, 2024), and can greatly affect their ecological functions in biogeochemical cycles. Therefore, understanding the regulatory factors of microbial communities in mangrove ecosystems is crucial due to their ecological significance in coastal areas.
Sulfate reduction is the most important respiratory process in mangrove sediments. Through this process, the microbial community couples the sulfur and carbon cycles (Lin et al., 2019). In one prior study, Liu et al. applied amplicon sequencing to analyze the community structure of bacteria and archaea in mangrove wetland soil, identifying Desulfobacterales and Syntrophobacterales as dominant taxa, further supporting their contributions to sulfur recycling (Liu et al., 2019). However, growing evidence indicates that such activities may also be influenced by sediment physical characteristics, particularly megafauna burrowing (Booth et al., 2023).
Lucinid clams (Lucinidae) are considered dominant benthic organisms in mangrove ecosystems, often inhabiting sediments around peri-mangrove sediments (Rondinelli and Barros, 2010; Lim et al., 2019). Lucinids play a dual role in shaping sediment microbial communities. First, their gills harbor endosymbiotic sulfur-oxidizing bacteria, which oxidize reduced sulfur compounds, potentially influencing the sediment biogeochemistry (Distel and Felbeck, 1987; Taylor et al., 2014; Martin et al., 2020). Second, these microorganisms contribute to the mineralization of organic nitrogen and phosphorus, as well as anaerobic nitrogen fixation (Welsh, 2000). Third, their burrowing activity triggers physical and chemical changes in the sediment (Gadeken and Dorgan, 2022), introducing oxygen, altering nutrient gradients, and creating microhabitats. The chemosynthetic symbiosis involving lucinid clams can significantly impact marine benthic ecosystems by altering sediment nutrients and influencing surrounding biological communities (Newton et al., 2008). These changes can influence microbial diversity, community structure, and functional capacity, including sulfur and nitrogen cycling (Osvatic et al., 2020). Even the interactions between chemosynthetic lucinid bivalves and seagrass have been widely studied (van der Heide et al., 2012; Petersen et al., 2016). However, the impact of these interactions on microbial communities remains a topic of debate.
The distribution of benthic organisms varies across different intertidal zones (Zamprogno et al., 2012), and the distribution patterns of different lucinids also differ (Taylor et al., 2014). Herein, we investigated the role of lucinids in structuring sediment microbiomes and altering biogeochemical processes in mangrove sediments. We hypothesized that this clam’s burrowing activity and endosymbiotic relationships would considerably influence microbial community composition and metabolic functions, thereby highlighting an overlooked ecological interaction in mangrove ecosystems. This study aimed to bridge the knowledge gap in this field by elucidating the role of lucinids in sedimentary microbial composition and potential metabolic processes, which has implications for understanding nutrient cycling and ecosystem functioning in mangrove habitats.
2 Materials and methods
2.1 Sampling area and sample collection
The distribution of Indoaustriella scarlatoi (Zorina, 1978, Lucinidae) in the intertidal mangroves of Huiwen Town, Hainan Island (19˚26′N, 110˚64′E) was assessed through the random placement of sampling quadrats along transects in a mangrove wetland region. These two independent sampling experiments were conducted in January and September, respectively. The quadrat size was 20×20 cm. The vertical distribution of I. scarlatoi in the sediments was estimated using pushcore-sampled columns. Samples were collected from areas with high (the mid-tidal zone) and low (the low-tidal zone) lucinid densities (Figure 1). In each area, three columnar sediment samples were collected using a pushcore sediment sampler. The sampling interval between samples in each area was approximately 10 m. Sediment samples from regions with high lucinid density (the mid-tidal zone) were labeled M1, M2, and M3. Sediment samples from areas with low lucinid density (the low-tidal zone) were labeled L1, L2, and L3. After collection, samples were sliced to a thickness of 10 cm using a sterile scraper, placed in sterile sampling bags, and stored on dry ice. Among them, those collected from a depth range of 0–10 cm were labeled as upper, while those collected from a depth exceeding 10 cm were labeled as lower (i.e. M-upper, L-upper, M-lower, and L-lower). In the laboratory, the samples were stored at −80°C before DNA extraction and microbial community analysis. Meanwhile, the collected sediment samples were acidified in the laboratory, and then the acidified samples were used for the determination of organic matter, including total organic carbon (TOC) and total nitrogen (TN).
Figure 1
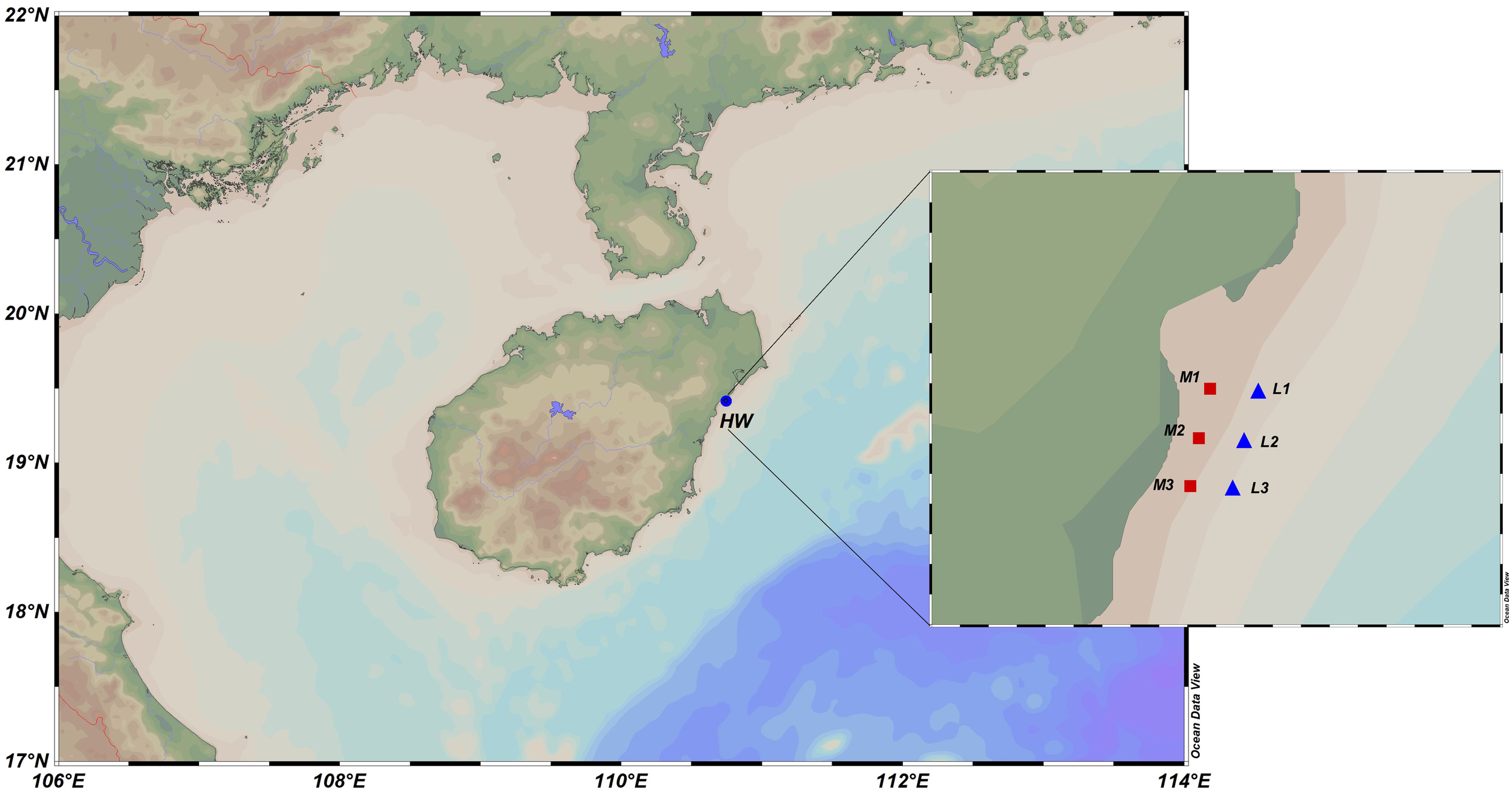
Schematic diagram of the sampling sites. The red square area labeled M represents the sampling points with a relatively high density of Lucinidae in the mid-tidal zone, while the blue triangular area L indicates the sampling points with a relatively low density of Lucinidae in the low-tidal zone. The panel in the upper left corner shows the main target species, I. scarlatoi, collected in this study. M1, M2, M3 represent three sampling points collected in the M site. For each sampling point, one sediment sample column was collected, and each sediment sample column was divided into two samples: upper and lower. Similarly, L1, L2, L3 represent the corresponding meaning.
2.2 DNA extraction and amplicon sequencing
DNA was extracted from all sediment samples using the DNeasy® PowerSoil Pro Kit (QIAGEN GmbH, Germany). DNA concentrations were subsequently measured with a Qubit 2.0 fluorometer (Invitrogen, Singapore). The V3-V4 region of the 16S rRNA gene was amplified with primers 341F (5’-CCTAYGGGRBGCASCAG-3’) and 806R (5’-GGACTACNNGGGTATCTAAT-3’) (Youssef et al., 2009; Caporaso et al., 2011). Each PCR reaction contained 15 µL of Phusion High-Fidelity PCR Master Mix (New England Biolabs), 0.2 µM of each primer, and 10 ng of genomic DNA template. The PCR conditions included an initial denaturation step at 98°C for 1 minute, followed by 30 cycles of 98°C for 10 seconds, 50°C for 30 seconds, and 72 °C for 30 seconds, with a final extension step of 72°C for 5 minutes. The 16S rRNA gene sequencing library was prepared using the NEBNext DNA Sample Preparation Kit, quantified with a Qubit fluorometer and qPCR, and sequenced on the Illumina NovaSeq platform with 250 bp paired-end reads.
2.3 Amplicon sequencing data processing
Cutadapt and fastp v0.23.1 software (Bokulich et al., 2013) were employed to splice and filter the raw data. FLASH v1.2.11 (Magoč and Salzberg, 2011) was applied to concatenate the reads of each sample to obtain effective tags. To minimize the effect of sequencing depth on subsequent analyses of alpha and beta diversity, the number of sequences for all samples was reduced to 20,000, while the average sequencing coverage for each sample still reached 99.09% following this reduction. Based on the filtered data, the DADA2 module in QIIME2 software (Li et al., 2020) was applied for noise reduction to obtain the final amplicon sequence variants (ASVs) and feature list (Wang et al., 2021).
2.4 Bioinformatic analysis
The classify-sklearn algorithm of QIIME2 was applied for each representative sequence of ASVs (Bokulich et al., 2018; Bolyen et al., 2019) and the Silva138.1 database (Quast et al., 2013) was applied for taxonomic annotation. Species-based abundances were calculated based on the taxonomic annotation. Alpha diversity was calculated using phyloseq in R (Xia et al., 2018), and shared and sample-unique taxa were determined using the abundance data. The Ade4 and ggplot2 software packages, along with R, were applied to reduce the dimensionality of the original variables for principal component analysis (PCA). To further explore the differences in community compositions among grouped samples, statistical analysis methods such as STAMP v2.1.3 (Parks et al., 2014) and Welch’s t-tests were chosen, with a significance level threshold of 0.05. PICRUSt2 software was applied to perform functional prediction analysis of microbial communities in the samples (Douglas et al., 2020).
3 Results
3.1 Microbial community divergence in lucinid-abundant and non-abundant regions
The water temperatures at the two sampling sites are both approximately 20°C ± 2°C, and the salinities are both approximately 32‰, but their respective TOC contents differ. It can be observed that the organic matter content of the Jan-M-upper (0.01369% ± 0.00151%) is slightly higher than that of the Jan-L-upper (0.01181% ± 0.00416%), although no significant difference was detected (Figure 2).
Figure 2

The relative content of TOC and TN in sediments in January. The M-upper represents the microbial community collected from the upper layer (0–10 cm) of the M sediments, and M-lower represents those collected from the deeper layer (> 10 cm in depth). Similarly, L-upper and L-lower represent the microbial community samples collected from the upper and deeper layers of the L site sediments, respectively.
The density of I. scarlatoi in the M and L sites was 437.5 ± 62.5 individuals/m2 and 37.5 ± 12.5 individuals/m2 in January, respectively. While the clams were distributed within the top 10 cm of the sediments, primarily at depths of 4–8 cm. In September, we also found that the density of lucinids at the M site was higher than that at the L site, and their distribution depth was also 4-8 cm.
PCA analysis was conducted based on the relative abundance of ASVs in January (Figure 3a). Overall, the samples were predominantly separated along PC1 based on their sampling depths. At depths where the clams were predominantly located, microbial communities from the two sites were distinctly separated along PC2 (Figure 3a). These results indicate that the higher abundance of lucinids in the M site played a major role in shaping the microbial communities, contributing to the observed differentiation. We also performed non-metric multidimensional scaling (nMDS) analysis on the sediment samples and found that the results were consistent with the patterns revealed by the PCA analysis (Figure 3b). We also performed PCA and nMDS analyses on another independent experiment and found the same pattern of conclusions (Figures 3c, d).
Figure 3
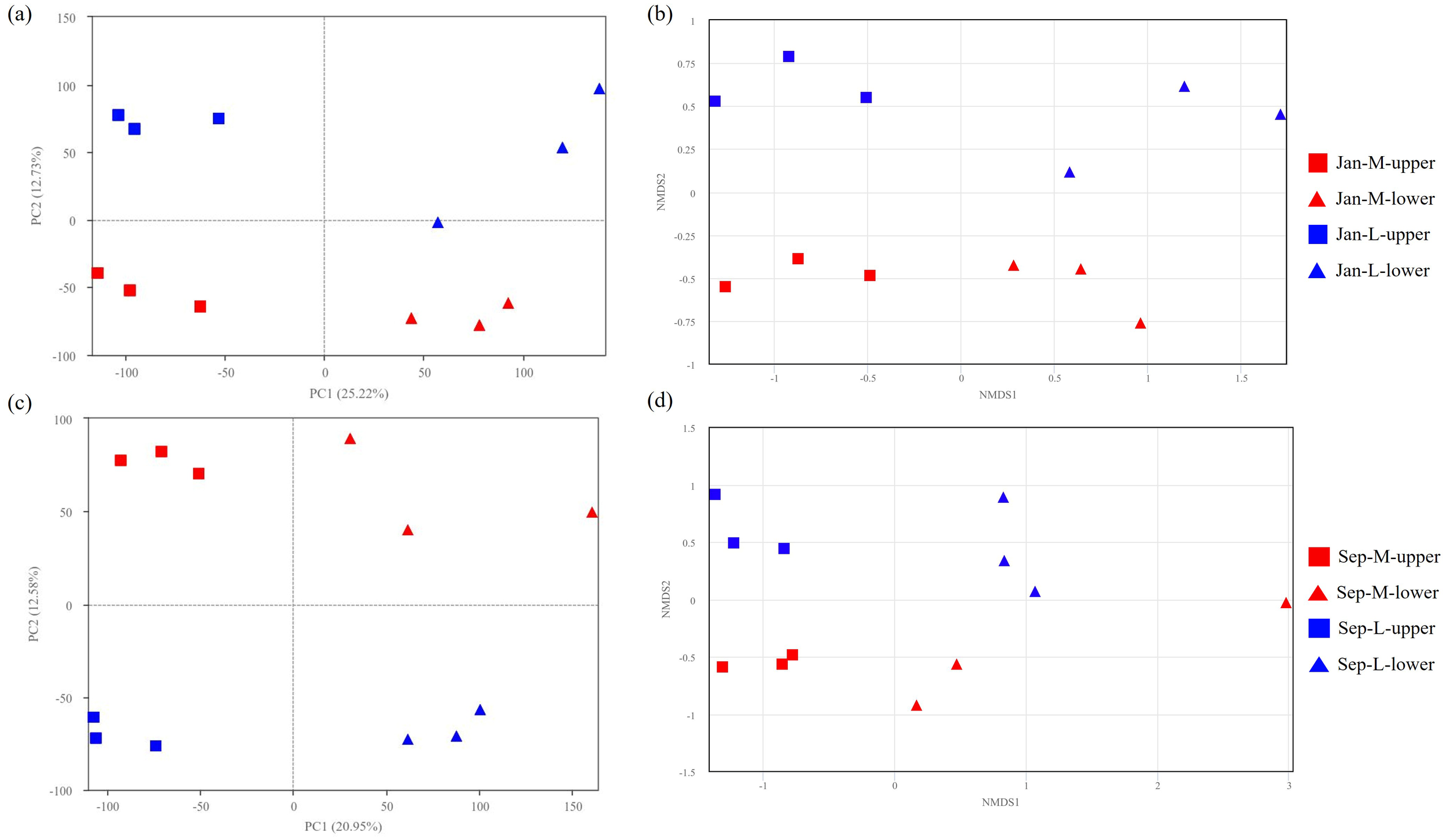
PCA plots (a) and nMDS analyses (b) of samples from M and L sites in January. PCA plots (c) and nMDS analyses (d) of samples from M and L sites in September.
The α-diversity and richness of microbial communities with abundant lucinid clams (M sites) are significantly higher than those with fewer lucinids (L sites, Figure 4a) in both upper and lower layers. Taken together, these results indicate that the upper and lower layers showed significantly higher diversities in the lucinid-abundant zone (that is, the M site), indicating that the presence of symbiotic lucinids may contribute to the diversity of the microbial community. Similarly, in another independent experiment, the microbial community diversity and richness in the M site with abundant lucinids were significantly higher than those in the L site with fewer lucinids (Figure 4b).
Figure 4
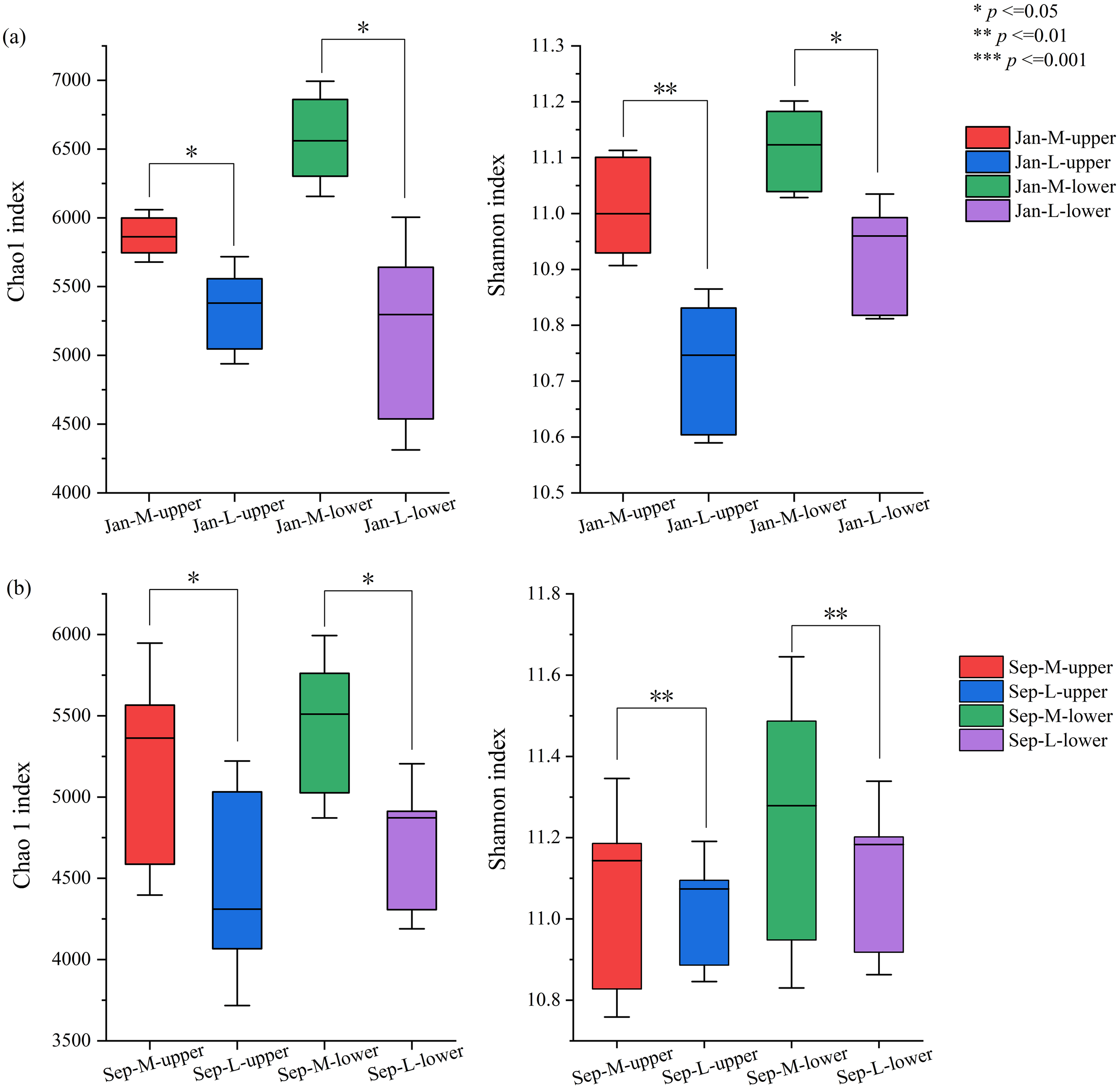
Comparisons of α-diversities in January (a) or September (b). Statistical analyses of microbial community indices in different regions (*p < 0.05, **p < 0.01) show that the species richness in the M site is higher than that in L site, and the species diversity in the M site is relatively superior.
3.2 Differences of microbial compositions
We subsequently compared the microbial community compositions of the sediments in M and L site. The results showed that 221 orders were commonly detected in the samples from Jan-M-upper and Jan-L-upper. In contrast, the number of common microbial orders in the samples from Jan-M-lower and Jan-L-lower was 193 (Figures 5a, b). There were 48 unique orders in the Jan-M-upper samples, accounting for 0.42% of the total abundance of the microbial community (Figure 5a). These unique microbial orders included Thermodesulfovibrionia which was regarded as functional group of sulfate-reducing bacteria (SRB). In contrast, 36 unique orders were observed in the Jan-L-upper samples, with Desulfuromonadia being a typical representative which may be involved in SRB. These microbial orders were not found in the Jan-M-upper samples, and they accounted for 0.26% of the total abundance of the microbial community (Figure 5a). For the Jan-M-lower, 20 exclusive orders were detected, representing 0.12%, while 57 orders, or 0.58%, were only found in the Jan-L-lower (Figure 5b). The unique microbial order in Jan-M-lower includes Syntrophomonadales which was regarded as a functional group of strict anaerobic bacteria. In contrast, among the unique microbial orders observed in Jan-L-lower, Nitrosopumilales was a typical representative, and it may also be involved in the processes related to aerobic ammonium oxidation. As the abundance of these unique microbial communities was at a relatively low level, their contribution to the differences in the entire microbial community was not obvious.
Figure 5
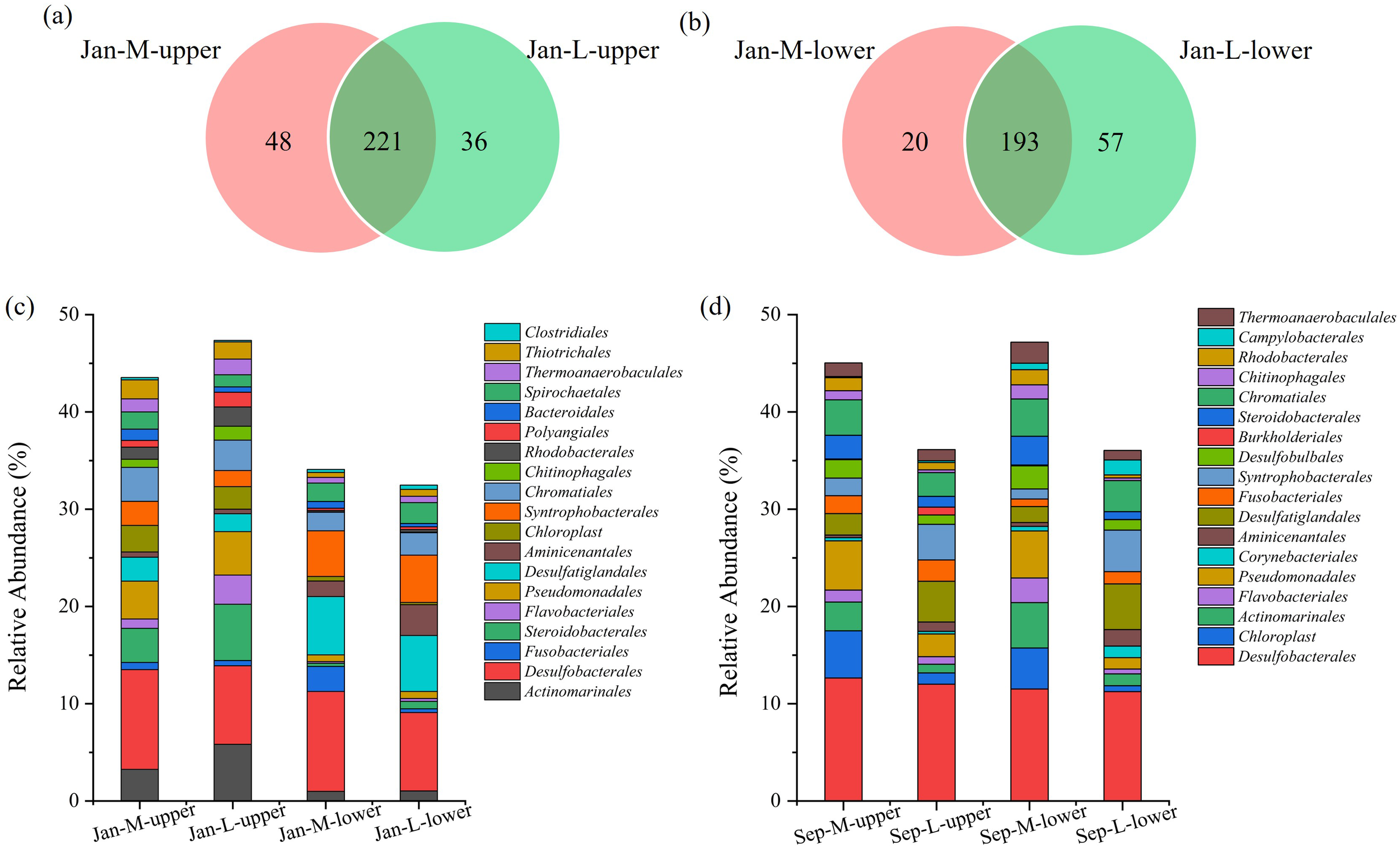
Comparisons of microbial communities at the order level and abundances of the top 20 orders. (a) Venn diagram at the order level of the sediments in Jan-M-upper and Jan-L-upper, (b) Venn diagram at the order level of the sediments of Jan-M-lower and Jan-L-lower. Bar chart of the relative abundances of the top 20 taxa in the samples from the M and L sites at the order level in January (c) or September (d).
We subsequently compared the abundance of dominant taxa, represented by the top 20 abundant taxa in these samples at the order level, between the M and L sites (Figures 5c, 6a). Microbes belonging to these taxa were present in both regions, but showed differences in abundance. Regarding the most abundant taxa, Desulfobacterales accounted for a significantly higher proportion in the Jan-M-upper samples compared with those from Jan-L-upper (Figure 6c), and this significance was maintained in comparisons of deeper-layer samples from Jan-M-lower and Jan-L-lower (Figure 6d). A sulfur-reducing-related group, Desulfatiglandales, was more abundant in the Jan-M-upper than in the Jan-L-upper, although without reaching significance (Figure 6c). Additionally, Bacteroidales exhibited a significantly higher abundance in the Jan-M-upper compared with the Jan-L-upper (Figure 6c). Conversely, Actinomarinales, Flavobacteriales, and Polyangiales were significantly more abundant in the Jan-L-upper site (Figure 6c). Furthermore, Desulfatiglandales and Syntrophobacterales, both containing anaerobic species (Chen et al., 2020; Zehnle et al., 2023), exhibited higher abundances in the Jan-M-lower and Jan-L-lower sites. These results underscore the significant compositional differences in microbial communities between the M and L sites, and highlight the role of lucinids in shaping the microbial structure in each area. Many of these differentially distributed bacteria were associated with sulfur metabolism or organic matter degradation, which are closely linked to the sulfide-cleaning role of the lucinid clams. Meanwhile, we also found that Chromatiales shows a slightly higher abundance in Jan-M-upper than in Jan-L-upper samples. However, the abundance difference of Chromatiales between the two sample types is not statistically significant (Figure 6a). We also observed similar conclusions in another independent experiment. For example, the abundance of Desulfobacterales was higher at the M site, Chromatiales showed a slightly higher abundance in the Sep-M-upper samples than in the Sep-L-upper samples, and Desulfatiglandales also exhibited higher abundances at the Sep-M-lower and Sep-L-lower sites (Figure 5d).
Figure 6
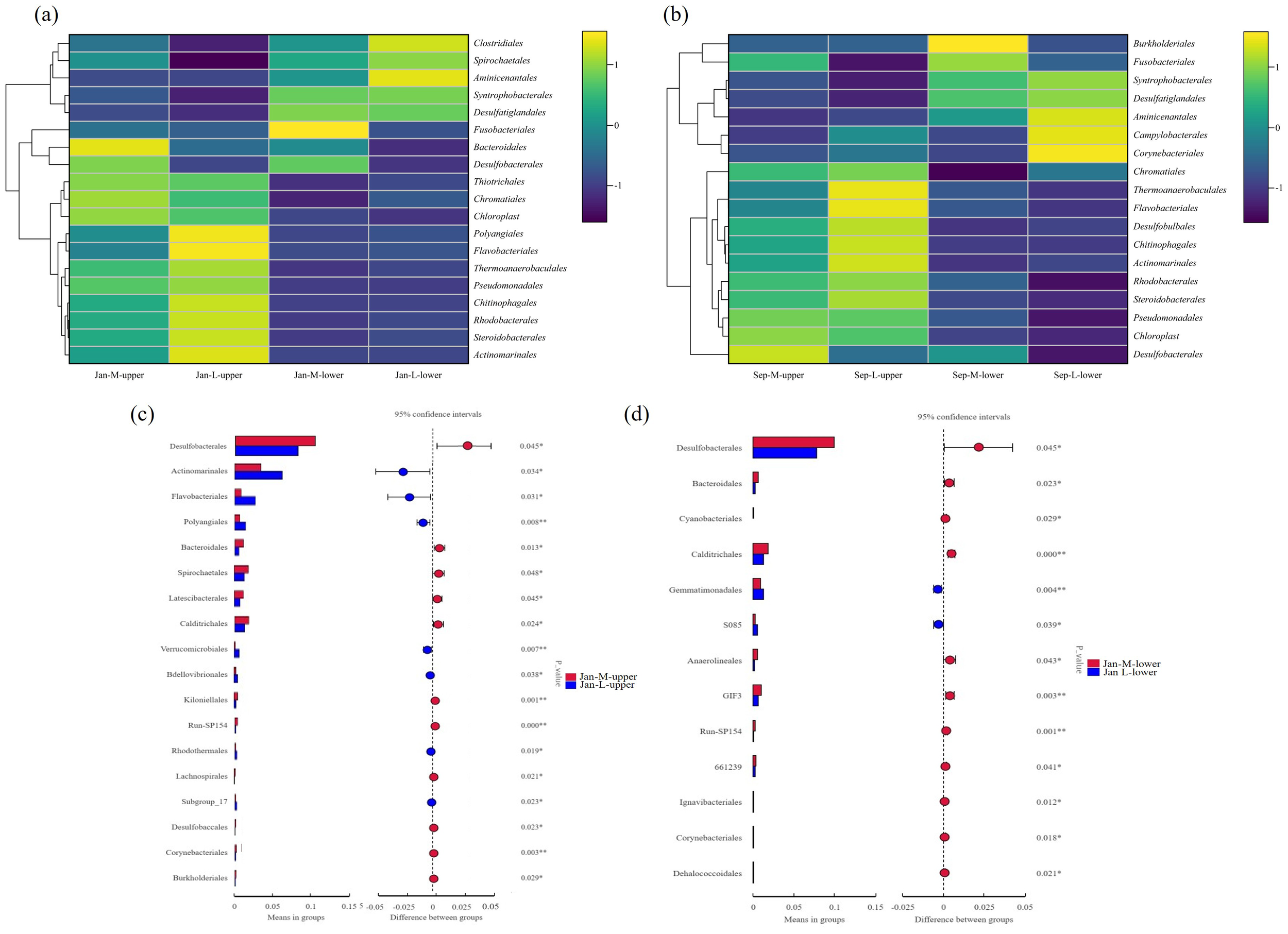
Statistical analyses of differentially distributed microbes. Clustering heat maps of dominant groups in the M and L sites at the order level in January (a) or September (b). Statistical analyses of differentially distributed microbes in Jan-M-upper and Jan-L-upper (c). Differences were calculated using T-tests. Significant differences in the abundances of microbes in the deeper-layer (Jan-M-lower and Jan-L-lower) (d). Significances were calculated using T-test.
We also performed phylum- and genus-level taxonomic characterizations of microbial communities across different areas and observed the same pattern in January. The phylum Desulfobacterota, which encompasses Desulfobacterales, exhibited higher abundances at the M site than at the L site (Figure 7a). Meanwhile, the genera Sva0081_sediment_group and SEEP-SRB1, which are classified under Desulfobacterales, also showed consistent trends (Figure 7b). Similarly, both the phylum Desulfobacterota to which Desulfatiglandales belong and the genus Desulfatiglans within Desulfatiglandales displayed higher abundances in M site compared to L site (Figure 7). These findings are consistent with those observed at the order level.
Figure 7

Comparisons of microbial communities at the different levels and abundances of the top 20 orders in January. The level of phylum (a) and genus (b).
3.3 Relative abundance and functional gene prediction of dominant populations
We next predicted the relative abundance of functional genes in microbial communities (Figure 8), identifying significant differences in functional genes associated with nitrogen and sulfur cycles between the M and L sites. Among the various nitrogen cycle-associated genes, those involved in the process of dissimilatory nitrate reduction to ammonia (nirBD and narGHI) exhibited higher abundance in the Jan-M-upper site.
Figure 8
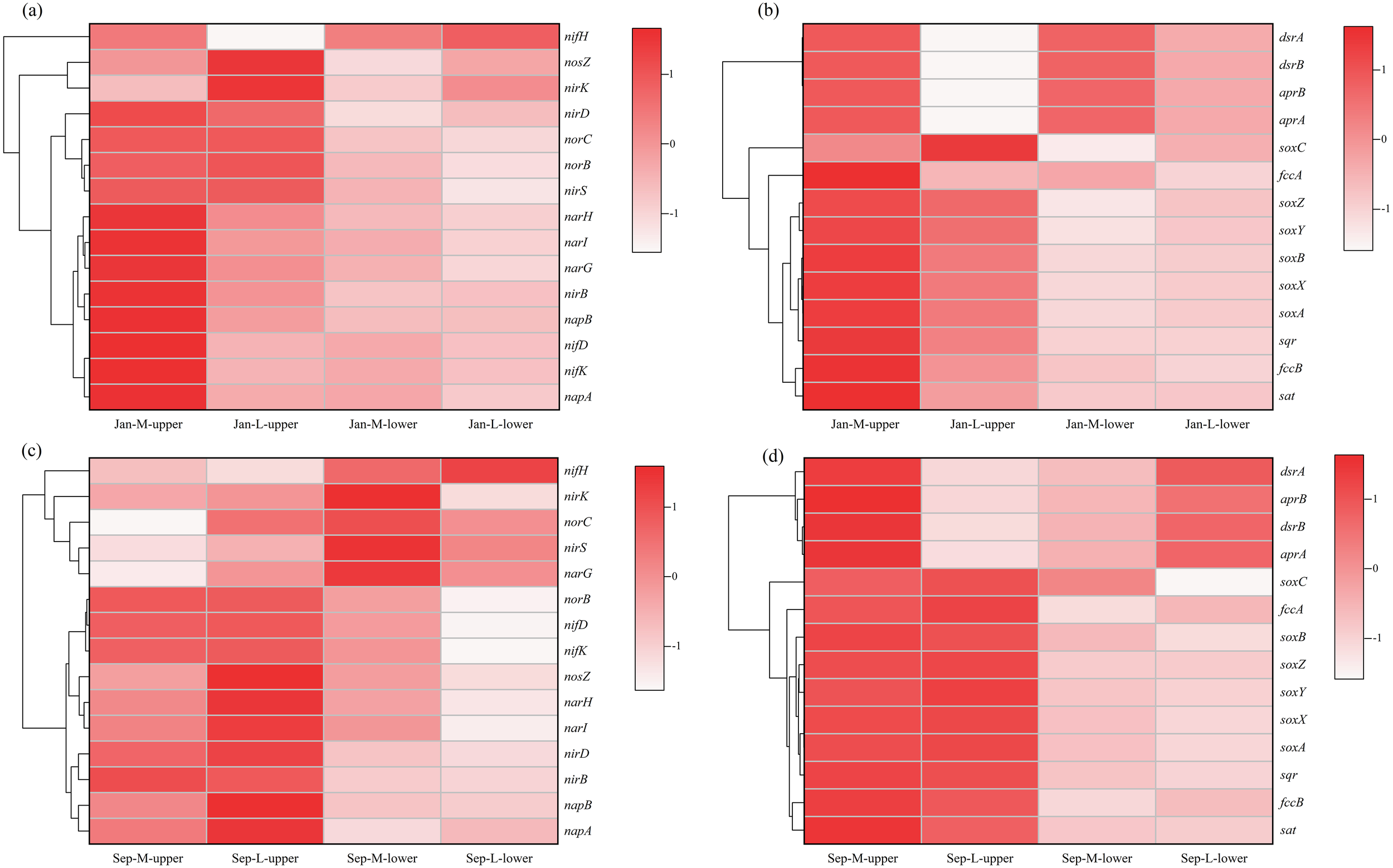
Clustering heat maps of functional gene prediction in M site and L site. Functional genes associated with nitrogen (a) and sulfur (b) in January. Functional genes associated with nitrogen (c) and sulfur (d) in September.
compared with the Jan-L-upper site. However, genes associated with the denitrification from nitrite to nitrogen (nirK and nosZ) were less abundant in the M site. This tendency may be associated with differences in microbial structure and sediment redox conditions. The results suggest the nitrate respiration are important in mangrove sediments, but were implemented by different taxa through various pathways. For sulfur cycle-related genes, sulfur reduction-associated functional genes (sat, aprAB and dsrAB) demonstrated a significantly higher abundance in the M site. Moreover, sulfur oxidation genes (sqr, fccAB and sox) also showed significantly higher abundance in the Jan-M-upper site. These oxidation genes catalyze the conversion of sulfides to sulfate or other oxidized sulfur compounds, suggesting the presence of an active sulfur cycle in the M site.
4 Discussion
In the PCA analysis, samples from various sampling zones and depths exhibited a distinct distribution pattern, possibly associated with the varied distribution densities of lucinids across different sites and depths. Meanwhile, the distribution of macrofauna such as lucinids and the differences in sediment redox conditions influence each other (Taylor et al., 2014; van der Geest et al., 2020). The M site demonstrated higher species richness and microbial diversity, which could be attributed to the fact that the presence of lucinids altered the geochemical characteristics of the surrounding sediments (Distel and Felbeck, 1987; Meyer et al., 2008; Osvatic et al., 2020), thereby providing more favorable environmental conditions for microbial development growth.
Desulfobacterales, which are SRB capable of reducing sulfate to H2S (Zhang et al., 2022), exhibited a significantly higher abundance in the M site, regardless of whether they were located in the surface or deeper layers. This suggests that sulfate reduction may be more active in the M site. Similarly, another group involved in sulfate reduction, Thermodesulfovibrionia (Umezawa et al., 2021), was only detected in the M site. Although the L site also contained some specific SRB, such as Desulfuromonadia (Waite et al., 2020; Langwig et al., 2022), the estimation of the abundances of sulfur-metabolizing genes was significantly higher in the M site than the L site, with this difference being particularly pronounced in the surface layer. The possible reason for this could be that L-upper is situated in the low-tide zone, where it undergoes relatively longer immersion by seawater. The prolonged seawater immersion leads to a more sustained anoxic state in the sediment environment of this area, providing a more stable reducing environment for sulfate-reducing bacteria. The Desulfuromonadia group may possess greater competitive advantages in such an environment characterized by long-term immersion and stable anoxia (Aullo et al., 2013; Xie et al., 2021), resulting in a higher relative abundance in L-upper. In contrast, M-upper is located in the mid-tide zone, where frequent alternations between wet and dry conditions occur. The growth of Thermodesulfovibrionia still relies on the supply of organic carbon in the surrounding environment (Frank et al., 2016). These results indicate that the sulfate-reduction and organic matter-degrading bacteria, such as Desulfobacterales and Thermodesulfovibrionia, are more abundant in the M site, where lucinid densities were higher. Through the determination of TOC in sediments, it was found that the organic matter concentration in Jan-M-upper was higher than that in Jan-L-upper. High concentrations of organic matter can drive the sulfur cycle process. Desulfobacterales can use organic matter as both a carbon source and an energy source (Higashioka et al., 2013), so an increase in organic matter concentration could promote the growth and reproduction of this group. In turn, the sulfide produced by these bacteria through sulfate reduction could provide an important energy source for lucinid clams (Higashioka et al., 2013).
The symbiotic bacteria of lucinids are primarily sulfur-oxidizing gamma-proteobacteria (Zauner et al., 2022; Alcaraz et al., 2024). The same taxonomic groups of the symbiotic bacteria were also existed in the environment. As a symbiotic bacterium of lucinids, Chromatiales exhibits a slightly higher abundance in the Jan-M-upper sample compared with that in the Jan-L-upper sample. However, the difference in abundance is not statistically significant. Symbiotic bacteria can utilize physiological processes such as chemosynthesis to supply essential nutrients to the host organisms, playing a crucial role in the survival and metabolism of the hosts (Ratinskaia et al., 2024). These endosymbiotic bacteria oxidize H2S to fix CO2 and synthesize organic matter (Powell and Somero, 1986), thereby providing energy and nutrients to hosts. The interaction between SRB and the symbiotic bacteria of lucinids promotes sulfur cycling in the environment. When SRB abundance is high and lucinid density is large, sulfur cycling efficiency is significantly enhanced, likely due to an increase in organic matter input. The increased organic matter stimulates active sulfate reduction, producing substantial amounts of H2S (Zhou et al., 2025). This H2S provides abundant substrates for the symbiotic bacteria of lucinids, thereby sustaining the high lucinid density. Furthermore, the highly activated sulfur cycle may accelerate the carbon fixation driven by the sulfide-oxidizing process of the lucinid symbiont (Osvatic et al., 2021). Meanwhile, the results of functional genes also showed that high-abundance sulfur reduction genes (sat, aprAB and dsrAB) can facilitate the reduction of sulfate to sulfides, which may lead to an increase in sulfide concentration in sediments (Yu et al., 2021) and provide energy for the chemosynthesis of symbionts in lucinid.
The abundance of genes associated with denitrification of nitrite to nitrogen gas (nirK and nosZ) were lower in the M site, a trend that may be related to differences in microbial community structure and sediment redox conditions. In the environment, the presence of lucinid clams, such as through processes like bio-irrigation, alters the redox environment of sediments, which in turn influences denitrification.
Specifically, the feeding and excretion activities of lucinid may introduce organic matter into sediments, which can then serve as a carbon source for SRB (Lebata, 2001). Research has also shown that the respiration of lucinids may deplete dissolved oxygen in the sediments, creating locally anoxic conditions in their activity zones. These conditions are favorable for the growth and metabolic activities of SRB (Zhang et al., 2023). Therefore, the consumption of sulfur by lucinid symbiotic bacteria could be regarded as an ecological disturbance factor with the potential to trigger changes in microbial community structure (Breusing et al., 2020). Meanwhile, behaviors such as bioturbation by lucinids can cause bioturbation in sediments (Mermillod-Blondin et al., 2004; Volkenborn et al., 2007), enhancing sediment aeration and redox potential, reducing the accumulation of harmful substances like H2S in sediments, and improving the substrate environment. Over time, these changes may drive the succession of microbial communities and are beneficial for the growth of mangrove plant roots and the survival of microorganisms.
Although this study observed similar phenomena in two different sampling sites, which include the high abundance of Desulfobacterales in areas with more lucinid clams, it would be more robust to incorporate additional manipulative experiments to control and investigate the effects of lucinids on environmental microbes. Additionally, as numerous environmental factors influence microbial communities under field conditions, and this study cannot comprehensively cover all environmental variables and factors, manipulative experiments can further elaborate on the regulatory relationship between lucinid clams and microbial communities based on the patterns discovered in this study.
5 Conclusion
In this study, we demonstrated that the diversity and structure of microbial communities vary with varied lucinid density. Specifically, microbial groups involved in sulfur and nitrogen cycling, such as Desulfobacterales, are more abundant in sediments rich in lucinids, highlighting the role of lucinid clams in reshaping microbial communities through bioturbation and chemosymbiotic activities. The coexistence of SRB and lucinid holobiont, likely driven by higher organic matter input, actively drives the sulfur cycle and enhances carbon fixation in these mangrove ecosystems.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1237586.
Author contributions
JS: Conceptualization, Investigation, Writing – original draft. YG: Conceptualization, Writing – review & editing. SJ: Investigation, Writing – original draft. KZ: Funding acquisition, Resources, Writing – review & editing. FL: Resources, Writing – original draft. ZZ: Resources, Writing – original draft. HZ: Funding acquisition, Resources, Writing – original draft. JZ: Conceptualization, Writing – original draft. WM: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work were supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2022QNLM030004-3), the National Key Research and Development Program of China (2023YFC2811501), the Key Laboratory of Ocean Observation and Information of Hainan Province (Open Fund Project No. HKLOOI-OF-2023-05), and the Oceanographic Data Center, IOCAS.
Conflict of interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Alcaraz C. M. Séneca J. Kunert M. Pree C. Sudo M. Petersen J. M. (2024). Sulfur-oxidizing symbionts colonize the digestive tract of their lucinid hosts. ISME J.18, wrae200. doi: 10.1093/ismejo/wrae200
2
Aullo T. Ranchou-Peyruse A. Ollivier B. Magot M. (2013). Desulfotomaculum spp. and related gram-positive sulfate-reducing bacteria in deep subsurface environments. Front. Microbiol.4, 362. doi: 10.3389/fmicb.2013.00362
3
Bittleston L. S. (2024). Connecting microbial community assembly and function. Curr. Opin. Microbiol.80, 102512. doi: 10.1016/j.mib.2024.102512
4
Bokulich N. A. Kaehler B. D. Rideout J. R. Dillon M. Bolyen E. Knight R. et al . (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome6, 90. doi: 10.1186/s40168-018-0470-z
5
Bokulich N. A. Subramanian S. Faith J. J. Gevers D. Gordon J. I. Knight R. et al . (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods10, 57–59. doi: 10.1038/nmeth.2276
6
Bolyen E. Rideout J. R. Dillon M. R. Bokulich N. A. Abnet C. C. Al-Ghalith G. A. et al . (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol.37, 852–857. doi: 10.1038/s41587-019-0209-9
7
Booth J. M. Fusi M. Marasco R. Daffonchio D. (2023). The microbial landscape in bioturbated mangrove sediment: A resource for promoting nature-based solutions for mangroves. Microbial. Biotechnol.16, 1584–1602. doi: 10.1111/1751-7915.14273
8
Breusing C. Perez M. Beinart R. A. Young C. R. (2020). Differences in cofactor, oxygen and sulfur requirements influence niche adaptation in deep-sea vesicomyid clam symbioses. bioRxiv2020, 2010.2019.345819. doi: 10.1101/2020.10.19.345819
9
Caporaso J. G. Lauber C. L. Walters W. A. Berg-Lyons D. Lozupone C. A. Turnbaugh P. J. et al . (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci.108, 4516–4522. doi: 10.1073/pnas.1000080107
10
Chen Y.-T. Zeng Y. Li J. Zhao X.-Y. Yi Y. Gou M. et al . (2020). Novel syntrophic isovalerate-degrading bacteria and their energetic cooperation with methanogens in methanogenic chemostats. Environ. Sci. Technol.54, 9618–9628. doi: 10.1021/acs.est.0c01840
11
Distel D. L. Felbeck H. (1987). Endosymbiosis in the lucinid clams Lucinoma aequizonata, Lucinoma annulata and Lucina floridana: a reexamination of the functional morphology of the gills as bacteria-bearing organs. Mar. Biol.96, 79–86. doi: 10.1007/BF00394840
12
Douglas G. M. Maffei V. J. Zaneveld J. R. Yurgel S. N. Brown J. R. Taylor C. M. et al . (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol.38, 685–688. doi: 10.1038/s41587-020-0548-6
13
Frank Y. A. Kadnikov V. V. Lukina A. P. Banks D. Beletsky A. V. Mardanov A. V. et al . (2016). Characterization and genome analysis of the first facultatively alkaliphilic thermodesulfovibrio isolated from the deep terrestrial subsurface. Frontiers in Microbiology7, 2000. doi: 10.3389/fmicb.2016.02000
14
Gadeken K. Dorgan K. (2022). Sediment macrofaunal response to the diel oxygen cycle. Mar. Ecol. Prog. Ser.703, 67–80. doi: 10.3354/meps14217
15
Higashioka Y. Kojima H. Watanabe M. Fukui M. (2013). Desulfatitalea tepidiphila gen. nov., sp. nov., a sulfate-reducing bacterium isolated from tidal flat sediment. Int. J. Syst. Evol. Micr.63, 761–765. doi: 10.1099/ijs.0.043356-0
16
Langwig M. V. De Anda V. Dombrowski N. Seitz K. W. Rambo I. M. Greening C. et al . (2022). Large-scale protein level comparison of Deltaproteobacteria reveals cohesive metabolic groups. ISME J.16, 307–320. doi: 10.1038/s41396-021-01057-y
17
Lebata M. J. (2001). Oxygen, sulphide and nutrient uptake of the mangrove mud clam Anodontia edentula (Family: Lucinidae). Mar. pollut. Bull.42, 1133–1138. doi: 10.1016/s0025-326x(01)00113-8
18
Li M. Shao D. Zhou J. Gu J. Qin J. Chen W. et al . (2020). Signatures within esophageal microbiota with progression of esophageal squamous cell carcinoma. Chin. J. Cancer Res.32, 755–767. doi: 10.21147/j.issn.1000-9604.2020.06.09
19
Lim S. J. Davis B. G. Gill D. E. Walton J. Nachman E. Engel A. S. et al . (2019). Taxonomic and functional heterogeneity of the gill microbiome in a symbiotic coastal mangrove lucinid species. ISME J.13, 902–920. doi: 10.1038/s41396-018-0318-3
20
Lin X. Hetharua B. Lin L. Xu H. Zheng T. He Z. et al . (2019). Mangrove sediment microbiome: adaptive microbial assemblages and their routed biogeochemical processes in Yunxiao mangrove national nature reserve, China. Microbial. Ecol.78, 57–69. doi: 10.1007/s00248-018-1261-6
21
Liu M. Huang H. Bao S. Tong Y. (2019). Microbial community structure of soils in Bamenwan mangrove wetland. Sci. Rep.9, 8406. doi: 10.1038/s41598-019-44788-x
22
Magoč T. Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics27, 2957–2963. doi: 10.1093/bioinformatics/btr507
23
Martin B. C. Middleton J. A. Fraser M. W. Marshall I. P. G. Scholz V. V. Hausl B. et al . (2020). Cutting out the middle clam: lucinid endosymbiotic bacteria are also associated with seagrass roots worldwide. Isme J.14, 2901–2905. doi: 10.1038/s41396-020-00771-3
24
Mattone C. Sheaves M. (2024). Mangrove forest ecological function is influenced by the environmental settings and the benthic fauna composition. Estuarine Coast. Shelf Sci.309, 108959. doi: 10.1016/j.ecss.2024.108959
25
Mermillod-Blondin F. Rosenberg R. François-Carcaillet F. Norling K. Mauclaire L. (2004). Influence of bioturbation by three benthic infaunal species on microbial communities and biogeochemical processes in marine sediment. Aquat. Microbial. Ecol.36, 271–284. doi: 10.3354/ame036271
26
Meyer E. Nilkerd B. Glover E. Taylor J. (2008). Ecological Importance of chemoautotrophic lucinid bivalves in a peri – mangrove community in Eastern Thailand. Raffles Museum Bull. Zool. Supplement18, 41–55.
27
Newton I. L. G. Girguis P. R. Cavanaugh C. M. (2008). Comparative genomics of vesicomyid clam (Bivalvia: Mollusca) chemosynthetic symbionts. BMC Genomics9, 585. doi: 10.1186/1471-2164-9-585
28
Osvatic J. T. Wilkins L. G. E. Leibrecht L. Leray M. Zauner S. Polzin J. et al . (2021). Global biogeography of chemosynthetic symbionts reveals both localized and globally distributed symbiont groups. Proc. Natl. Acad. Sci.118 (29), e2104378118. doi: 10.1073/pnas.2104378118
29
Osvatic J. Windisch J. Yuen B. Hausl B. Polzin J. Petersen J. (2020). Chemosymbiotic lucinid clams modify the physical, chemical and biological characteristics of marine sediments globally (EGU General Assembly 2020), Online, EGU2020–19150. doi: 10.5194/egusphere-egu2020-19150
30
Parks D. H. Tyson G. W. Hugenholtz P. Beiko R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics30, 3123–3124. doi: 10.1093/bioinformatics/btu494
31
Petersen J. M. Kemper A. Gruber-Vodicka H. Cardini U. van der Geest M. Kleiner M. et al . (2016). Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat. Microbiol.2, 16195. doi: 10.1038/nmicrobiol.2016.195
32
Powell M. A. Somero G. N. (1986). Hydrogen sulfide oxidation is coupled to oxidative phosphorylation in mitochondria of solemya reidi. Science233, 563–566. doi: 10.1126/science.233.4763.563
33
Quast C. Pruesse E. Yilmaz P. Gerken J. Schweer T. Yarza P. et al . (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res.41, D590–D596. doi: 10.1093/nar/gks1219
34
Ratinskaia L. Malavin S. Zvi-Kedem T. Vintila S. Kleiner M. Rubin-Blum M. et al . (2024). Metabolically-versatile Ca. Thiodiazotropha symbionts of the deep-sea lucinid clam Lucinoma kazani have the genetic potential to fix nitrogen. ISME Commun.4, ycae076. doi: 10.1093/ismeco/ycae076
35
Rondinelli S. F. Barros F. (2010). Evaluating shellfish gathering (Lucina pectinata) in a tropical mangrove system. J. Sea Res.64, 401–407. doi: 10.1016/j.seares.2010.06.002
36
Sunish K. S. (2024). “Mangrove Microbiome Dynamics: Exploring Diversity and Ecological Interactions,” in Mangroves in a Changing World: Adaptation and Resilience (Springer Nature Switzerland, Cham), 87–110.
37
Taylor J. D. Glover E. A. Williams S. T. (2014). Diversification of chemosymbiotic bivalves: origins and relationships of deeper water Lucinidae. Biol. J. Linn. Soc.111, 401–420. doi: 10.1111/bij.12208
38
Umezawa K. Kojima H. Kato Y. Fukui M. (2021). Dissulfurispira thermophila gen. nov., sp. nov., a thermophilic chemolithoautotroph growing by sulfur disproportionation, and proposal of novel taxa in the phylum Nitrospirota to reclassify the genus Thermodesulfovibrio. Systematic Appl. Microbiol.44, 126184. doi: 10.1016/j.syapm.2021.126184
39
van der Geest M. van der Heide T. Holmer M. de Wit R. (2020). First field-based evidence that the seagrass-lucinid mutualism can mitigate sulfide stress in seagrasses. Front. Mar. Sci.7, 11. doi: 10.3389/fmars.2020.00011
40
van der Heide T. Govers L. L. de Fouw J. Olff H. van der Geest M. van Katwijk M. M. et al . (2012). A three-stage symbiosis forms the foundation of seagrass ecosystems. Science336, 1432–1434. doi: 10.1126/science.1219973
41
Volkenborn N. Polerecky L. Hedtkamp S. I. C. van Beusekom J. E. E. de Beer D. (2007). Bioturbation and bioirrigation extend the open exchange regions in permeable sediments. Limnology Oceanogr.52, 1898–1909. doi: 10.4319/lo.2007.52.5.1898
42
Waite D. W. ChuvoChina M. Pelikan C. Parks D. H. Yilmaz P. Wagner M. et al . (2020). Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Systematic Evolutionary Microbiol.70, 5972–6016. doi: 10.1099/ijsem.0.004213
43
Wang Y. Guo H. Gao X. Wang J. (2021). The intratumor microbiota signatures associate with subtype, tumor stage, and survival status of esophageal carcinoma. Front. Oncol.11. doi: 10.3389/fonc.2021.754788
44
Welsh D. T. (2000). Nitrogen fixation in seagrass meadows: Regulation, plant–bacteria interactions and significance to primary productivity. Ecol. Lett.3, 58–71. doi: 10.1046/j.1461-0248.2000.00111.x
45
Wu B. Liu F. Fang W. Yang T. Chen G.-H. He Z. et al . (2021). Microbial sulfur metabolism and environmental implications. Sci. Total Environ.778, 146085. doi: 10.1016/j.scitotenv.2021.146085
46
Xia Y. Sun J. Chen D.-G. (2018). “Community Diversity Measures and Calculations,” in Statistical Analysis of Microbiome Data with R (Springer Singapore, Singapore), 167–190.
47
Xie L. Yoshida N. Ishii S.i. Meng L. (2021). Isolation and Polyphasic Characterization of Desulfuromonas versatilis sp. Nov., an Electrogenic Bacteria Capable of Versatile Metabolism Isolated from a Graphene Oxide-Reducing Enrichment Culture. Microorganisms9 (9), 1953. doi: 10.3390/microorganisms9091953
48
Youssef N. Sheik Cody S. Krumholz Lee R. Najar Fares Z. Roe Bruce A. Elshahed Mostafa S. (2009). Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl. Environ. Microbiol.75, 5227–5236. doi: 10.1128/AEM.00592-09
49
Yu X. Zhou J. Song W. Xu M. He Q. Peng Y. et al . (2021). SCycDB: A curated functional gene database for metagenomic profiling of sulphur cycling pathways. Mol. Ecol. Resour.21, 924–940. doi: 10.1111/1755-0998.13306
50
Zamprogno G. C. Fernandes F. C. Fernandes L. L. (2012). Temporal and spatial variation of rocky shores intertidal benthic communities in Southeast Brazil. Iheringia Série Zoologia102 (4), 375–383. doi: 10.1590/S0073-47212012000400003
51
Zauner S. Vogel M. Polzin J. Yuen B. Mußmann M. El-Hacen E.-H. M. et al . (2022). Microbial communities in developmental stages of lucinid bivalves. ISME Commun.2, 56. doi: 10.1038/s43705-022-00133-4
52
Zehnle H. Otersen C. Benito Merino D. Wegener G. (2023). Potential for the anaerobic oxidation of benzene and naphthalene in thermophilic microorganisms from the Guaymas Basin. Front. Microbiol.14. doi: 10.3389/fmicb.2023.1279865
53
Zhang W. Han S. Zhang D. Shan B. Wei D. (2023). Variations in dissolved oxygen and aquatic biological responses in China’s coastal seas. Environ. Res.223, 115418. doi: 10.1016/j.envres.2023.115418
54
Zhang Z. Zhang C. Yang Y. Zhang Z. Tang Y. Su P. et al . (2022). A review of sulfate-reducing bacteria: Metabolism, influencing factors and application in wastewater treatment. J. Cleaner Production376, 134109. doi: 10.1016/j.jclepro.2022.134109
55
Zhou Z. Tran P. Q. Cowley E. S. Trembath-Reichert E. Anantharaman K. (2025). Diversity and ecology of microbial sulfur metabolism. Nat. Rev. Microbiol.23, 122–140. doi: 10.1038/s41579-024-01104-3
56
Zhuang W. Yu X. Hu R. Luo Z. Liu X. Zheng X. et al . (2020). Diversity, function and assembly of mangrove root-associated microbial communities at a continuous fine-scale. NPJ Biofilms Microbiomes6, 52. doi: 10.1038/s41522-020-00164-6
Summary
Keywords
mangrove ecosystem, lucinids, microbial community, sulfur cycle, amplicon sequencing
Citation
Shi J, Guo Y, Jin S, Zhao K, Li F, Zhong Z, Zhang H, Zhang J and Wang M (2025) Lucinid-associated microbial community divergence in peri-mangrove ecosystems. Front. Mar. Sci. 12:1606783. doi: 10.3389/fmars.2025.1606783
Received
06 April 2025
Accepted
07 July 2025
Published
24 July 2025
Volume
12 - 2025
Edited by
Luca Rindi, University of Pisa, Italy
Reviewed by
Li Jing Jiang, State Oceanic Administration, China
Ludovica Pedicini, University of Pisa, Italy
Updates
Copyright
© 2025 Shi, Guo, Jin, Zhao, Li, Zhong, Zhang, Zhang and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhua Zhang, zhangjianhua@qut.edu.cn; Minxiao Wang, wangminxiao@qdio.ac.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.