Abstract
The climate crisis necessitates new and expanded agrochemical options to address the challenges in current agricultural production. The marine flora represents an attractive source of novel bioactives compounds with potential relevance to agriculture (including both crops and livestock applications), human health, and biomaterials. While significant research is currently underway focusing on discovering and characterising bioactives derived directly from algal biomass, an often-overlooked aspect of seaweeds - or marine macro-organisms in general - is their close association with a diverse array of microorganisms, forming what is now referred to as holobiont systems. As such, the marine flora hosts a variety of microbes, including epiphytic and endophytic bacteria and fungi. This reservoir of microbial biodiversity itself offers a promising, yet largely untapped, source of novel bioactives with potential applications in the agriculture and healthcare industries. This mini-review aims to discuss the recent findings in the bioactivities of the Seaweed-Associated Microbiome (SAM) and specifically explore the potential applications of seaweed microbiome-derived bioactives as a novel source of agrochemicals relevant to crop growth, health, and pest management.
1 Introduction
Climate change forecast predicts an increase in overall temperatures and longer wet spells, intensifying (a)biotic pressure on crops as warmer and wetter days stimulate pathogen growth, particularly fungi & moulds (Chaloner et al., 2021). This may in turn necessitate more pesticides application to maintain yields, with known negative impacts on the ecosystem and human health (Sharma et al., 2019). Consequently, the discovery of novel natural compounds that enhance crop yields or resilience to biotic and abiotic pressures is crucial for “climate-proofing” agricultural systems.
Significant research efforts are directed towards identifying alternative microbial sources for sustainable crop protection and biostimulation. Terrestrial microbial sources, such as Bacillus (Fira et al., 2018; Radhakrishnan et al., 2017) and Pseudomonas species (Mehmood et al., 2023; Raio and Puopolo, 2021), are well-established for their biopesticidal and plant growth-promoting properties, while certain marine-derived fungi and bacteria have also shown promise in controlling plant diseases via their secondary metabolites (Nguyen et al., 2022; Qi et al., 2023) and enhancing growth in various agricultural settings, including as biofertilisers (Joshi et al., 2020; Rathod et al., 2023). Among these diverse microbial reservoirs, the marine environment offers a unique and largely underexplored biodiversity that could be leveraged for these critical needs. Specifically, the SAM may produce a plethora of compounds relevant to crop production and health, such as SAM-derived growth regulators, AHLs, defence elicitors or antimicrobials against crop pathogens (Figure 1).
Figure 1
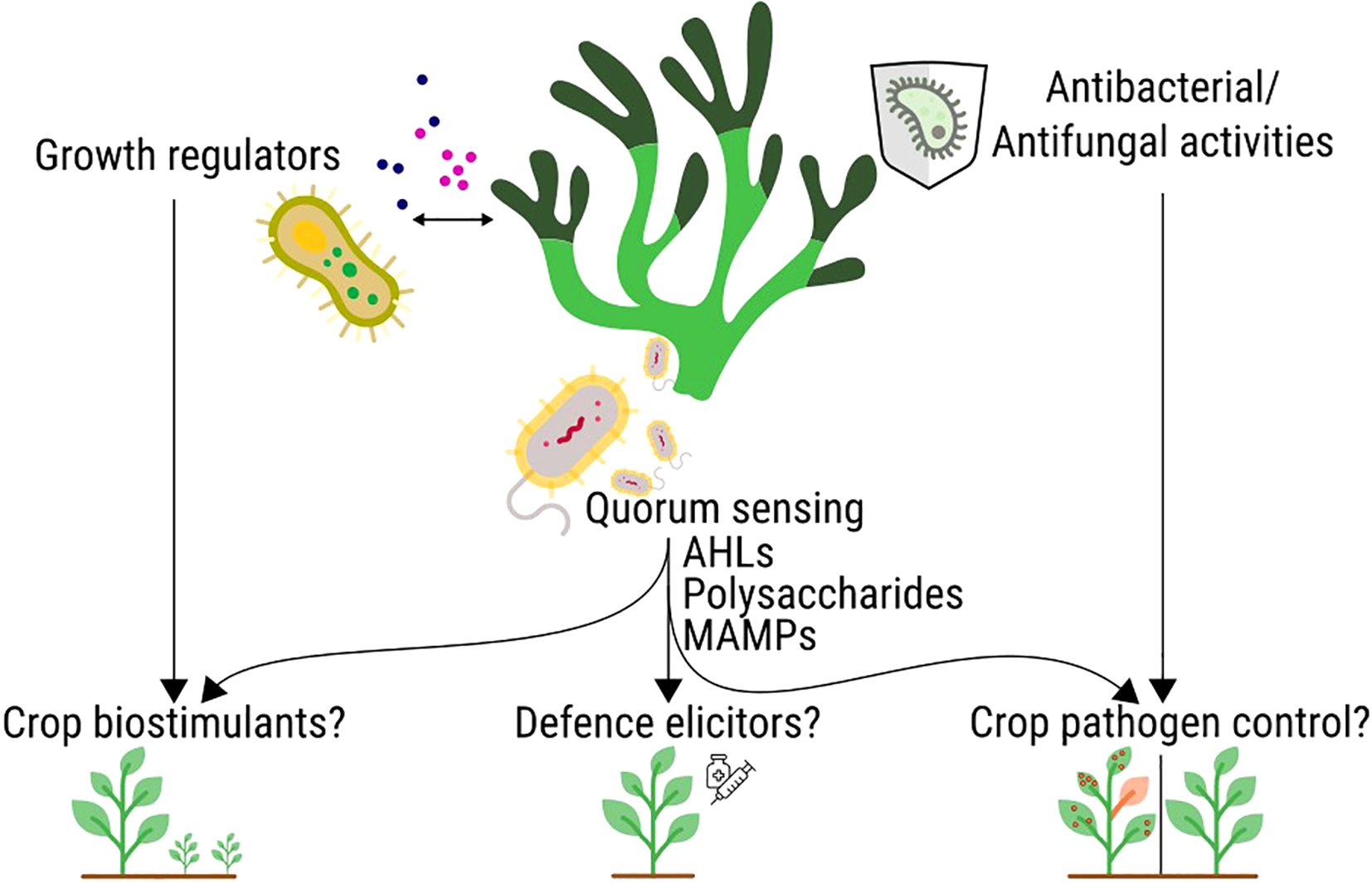
Potential for seaweed-associated microbes to produce bioactive compounds for crop health and as crop biostimulants. Left: Growth regulators produced by the SAM could be used to stimulate crop growth and development. Middle: Quorum sensing molecules, such as AHLs, can display biostimulants, defence elicitor and antimicrobial activities. Right: antimicrobial and antifungal compounds have been described as originating from the SAM, which may be used as novel pesticides for crop pathogen control.
The role and diversity of the seaweed microbiome has garnered significant attention in recent years. Those include non-specific associations, where the seaweed biomass serves as substrate to colonising microbes (Saha and Weinberger, 2019), to symbiotic relationships where seaweed growth and development is directly dependent of the presence of their symbiotic bacteria (Spoerner et al., 2012). Other examples of associations include the protection against pathogens conferred by colonising bacteria – chemically recruited by the seaweed host (Saha and Weinberger, 2019) -, to increased environmental resilience (Ghaderiardakani et al., 2020). Another example showed that Ulva’s microbiome quickly undergoes taxonomic modifications when introduced in a different environment (van der Loos et al., 2024), and similar re-structuring occurs between U. rigida grown in an integrated multi-trophic aquaculture site and the surrounding lagoon area (Califano et al., 2020).
Metabarcoding studies regularly find hundreds of bacterial genera from seaweed samples, with variations in composition based on the hosts, abiotic parameters, and geography (Burgunter-Delamare et al., 2023; Burke et al., 2011; Deutsch et al., 2023; Paix et al., 2021; Ramírez-Puebla et al., 2022; van der Loos et al., 2023; Wood et al., 2022). Of particular note is the presence of “functional guilds” within seaweed-associated microbes that specialise in the degradation of seaweed-specific polysaccharides (Khan et al., 2024). The use of metagenome-assembled genomes (MAG) from seaweed holobionts (Weigel Brooke et al., 2022) is likely to yield novel enzymes and pathways that can have biotechnological implications, such as in the degradation of halogenated compounds (Lavecchia et al., 2024), nutrient cycling (Weigel Brooke et al., 2022), or the production of plant growth regulators (Wang et al., 2022). Significant efforts are currently underway to better characterise and understand the role, diversity and dynamics of the seaweed microbiome, a topic extensively reviewed by Saha et al. (2024). A deeper understanding of the seaweed holobiont is expected to lead to higher yields or the creation of tailored biomass through optimising the three-way interaction between seaweed genotype, its environment and microbiome (Li et al., 2023; Simon et al., 2022), or through improved microbiome design (Wichard, 2023).
The reported role(s) of the SAM are likely due, in part, to the microbial community’s production of bioactive compounds. For example, a number of microorganisms such as Maribacter sp. MS6 (Alsufyani et al., 2020), Bacillus pumilus (Singh et al., 2011a) or Azotobacter species (Head and Carpenter, 1975) have been identified as Seaweed Beneficial Microorganisms (SBMs) producing algal growth and morphogenesis-promoting factors (AGMPF), including phytohormones (e.g auxins-like, cytokinins-like), vitamin B12, and providing nutrient fixation (Li et al., 2023). Other beneficial effects of SAMs in disease protection have been uncovered. For example, Phaeobacter sp. BS52 and Pseudoalteromonas sp. PB2-1 can reduce the impact of the macroalgal pathogen Pseudoalteromonas arctica G-MAN6, responsible for bleaching disease in Agarophyton vermiculophyllum and Delisea pulchra (Li et al., 2022). Similarly, the production of pyrenocines by Phaeosphaeria sp. AN596H can inhibit the infection of Ectocarpus siliculosus by several protistan pathogens (Vallet et al., 2018). Finally, the role of SAM in protecting their host against disease may also be modulated by a stimulation of its immune response (Li et al., 2023), although a direct elicitation of algal immune responses by the SAM has yet to be reported.
This mini-review will shift focus from seaweed holobiont systems to explore the potential uses of SAM-derived bioactives in agriculture, specifically as a reservoir of crop biostimulants.
2 Potential for SAM-derived microbes and their bioactives in crop agriculture
2.1 Plant growth promoting regulatory compounds
Among SAM bioactives are phytohormones. For example, Ulva’s microbiome produces cytokinins-like and auxins-like phytohormones, originating from Roseovarius sp. MS2, and Maribacter sp. MS6, respectively (Ghaderiardakani et al., 2017). Those hormones have been shown to induce morphogenesis in Ulva species via promoting cell division and cell differentiation (Wichard, 2023). Other strains and species that phenocopy Roseovarius and Maribacter role have been isolated (i.e Sulfitobacter sp. BPC-C4, and Maribacter sp. BPC-D8) demonstrating the diversity of algae growth-promoting bacteria present within the seaweed holobiont (Ghaderiardakani et al., 2024). In another example, thallusin, a steroid-like compound produced by Maribacter associated with both Monostroma oxyspermum (Matsuo et al., 2005) and Ulva spp (Alsufyani et al., 2020), exert numerous bioactivities, ranging from growth stimulation to morphogenesis and cell wall formation (Alsufyani et al., 2020; Dhiman et al., 2022; Yamamoto et al., 2018). Those bioactivities are structure-dependent as (−)-thallusin and its synthetic derivatives display differential activities in Ulva (Dhiman et al., 2022). Phytohormone production is not limited to Ulva’s microbiome (De Clerck et al., 2018) and has been demonstrated in other phyla including in brown (e.g Ectocarpus) (Burgunter-Delamare et al., 2020), and red algae [e.g Porphyridium purpureum and Pyropia yezoensis (Kim et al., 2024; Matsuda et al., 2018; Mori et al., 2017)]. Therefore, phytohormone/AGMPF production is likely a common feature of SAM. Outside of the SAM, marine bacteria associated with phytoplankton have also been shown to contain biosynthetic genes for plant growth-promoting phytohormones and conversely produce 6 out of the 7 plant growth hormones tested (Khalil et al., 2024), highlighting their potential as a reservoir of plant growth regulators.
The type and structural diversity of plant growth regulators originating from SAM could be leveraged for the biodiscovery of novel crop growth-promoting compounds. Indeed, auxins, cytokinins and steroid compounds are major plant phytohormones controlling a wide range of cellular and developmental processes. For this, plant trials could indicate 1) if those marine-derived growth regulators can be recognised by plant receptors, and 2) whether SAM-derived growth regulators are indeed effective in modulating crop growth, development, and response to environmental stresses. Screening for an impact of SAM-derived growth regulators on plants could be relatively straightforward, using high-throughput phenotyping platforms that measure biomass growth over time (Fort et al., 2019, 2016), following the application of SAM-derived extracts. However, the characterisation of the growth regulators within, and their mode of action in planta will require more extensive research.
Another mechanism by which plant growth & resilience could be modulated by marine bacteria is through the use of plant growth promoting rhizobacteria (PGPR) isolated from the marine environment, as demonstrated by several studies showing improved crop growth and stress responses (notably salt-stress) following inoculation; via a combination of growth-promoting effects or the production of osmoprotectants (Aizaz et al., 2023; Carreiras et al., 2023). These studies underscore the broader potential of marine microbes to act as biofertilizers (Singh et al., 2023).
2.2 Plant defence elicitors
Beyond plant growth regulators are molecules produced by the SAM - such as N-Acyl homoserine lactones (AHLs) - that also hold promise as crop biostimulants and defense elicitors. AHLs represent a class of signalling molecules involved in quorum sensing and biofilm formation in bacteria. AHLs produced by the SAM are involved in seaweed-microbiome interactions. For example, Pseudoalteromonas galatheae isolated from Porphyra haitanensis, was found to produce four types of AHL molecules that stimulate biofilm formation on the seaweed surface (Aslam et al., 2023). In another example, Vibrio anguillarum’s production of three AHLs was reported, with the AHL 3-oxo-C10-HSL involved in the attraction of Ulva zoospores (Joint et al., 2007; Tait et al., 2005). Shewanella algae produces five types of AHLs, with its C4 and C6 AHLs able to induce carpospore liberation in Gracilaria dura (Singh et al., 2015a). Those studies highlight the wide composition and roles of AHLs produced by the SAM.
AHLs can act as strong plant defence elicitors (i.e. priming the plant pathogen defence pathways) (Schenk et al., 2014; Schikora et al., 2016). For example, AHLs can induce resistance against plant pathogens (i.e. Aternaria alternata) when applied to tomatoes (Schuhegger et al., 2006), brassicas (Duan et al., 2023; Shrestha et al., 2020) and barley (Han et al., 2016). AHLs work in plants via priming the induced systemic resistance (ISR) pathway – typically modulated by the plant rhizosphere, and leading to the production of reactive oxygen species, phenolic compounds, callose and lignin accumulation as well as stomatal closure (Zhu et al., 2022). All of which lead to a faster and stronger response when the plant is exposed to pathogens.
In addition to their role in plant pathogen defence, AHLs can also act as crop biostimulants by stimulating root and biomass growth (Nawaz et al., 2020; Moshynets et al., 2019; Ortiz et al., 2024; Shrestha et al., 2020), particularly when applied on plants under salt stress (Zhao et al., 2020). The action of AHLs on plants depend on their structure and given the variety of AHLs produced by the seaweed associated microbiome, an investigation of their potential impact on crop defence and/or growth is warranted.
Outside of AHLs, other potential plant defence elicitors could be produced by the SAM or its enzymatic activity on seaweed polysaccharides, such as specific polysaccharides and Microbe Associated Molecular Patterns (MAMPs). These include oligosaccharides, chitin fragments, lipopolysaccharides (LPS), and peptidoglycan derivatives, all of which are classes of molecules that have been shown to activate immune responses in plants (Erbs et al., 2010). Alginate oligosaccharides, for example, can induce defence-related gene expression and improve resistance to pathogens when applied exogenously (Peng et al., 2025). LPS from gram negative bacteria are recognised by plant receptors and can trigger an immune responses or act as elicitors (Meena et al., 2022). Some marine bacteria, including SAM-derived ones such as Staphylococcus equorum and Bacillus tropicus, isolated from Gracilaria sp., possess chitinase activity (Ginting et al., 2024) and are able to produce chitin fragments that are well established elicitors that interact with plant lysin motif receptors to activate signalling cascades that bolster plant defence (Saberi Riseh et al., 2024). Marine bacteria, including member of the SAM such as Pseudoalteromonas spp., are known to produce diverse extracellular polysaccharides (EPS) (Daly et al., 2023; Meunier et al., 2024; Xu et al., 2021), some of which may mimic these immune triggering molecules or interfere with host signalling.
Altogether, SAM’s diversity may represent a reservoir of molecules with plant defence elicitor activities, offering a promising, largely unexplored means of natural crop protection and immune modulation. Using reporter gene systems, such as plants carrying a reporter gene (e.g. GFP), under the control of a promoter activated by plant defences pathways such as PATHOGENERIS RELATED 1 (PR1) or NONEXPRESSER OF PR GENES 1 (NPR1) (Halder and Kombrink, 2015), could allow for rapid screening of SAM extracts for plant elicitor bioactivities.
2.3 Antimicrobial compounds against crop pathogens
Finally, while SAM bioactives could be recognised and act on crops, they could also impact crop pathogens themselves. Most research in this area focuses on antimicrobial bioactivities against human-relevant pathogens (Asharaf et al., 2022; Girão et al., 2019; Karthick and Mohanraju, 2018; Manam et al., 2025; Martinez-Delgado and Benitez-Campo, 2025; Vega-Portalatino et al., 2024; Tangestani et al., 2021; Manam et al., 2025). In the case of Manam et al. (2025), the bioactive originates from Bacillus subtilis, an endophyte isolated from Gracilaria edulis. The compound was identified via GC-MS and FT-IR as Pyrrolo[1,2-α] pyrazine-1,4-dione, hexahydro-3-(2-methylpropyl) (PPDHMP), and possess beta-lactamase and cell wall inhibitory activities. Through the use of bioactivity-guided isolation - a systematic approach to purify bioactive compounds from complex mixtures by iteratively separating the mixture into fractions and testing each fraction for bioactivity; followed by mass spectrometry and NMR, decylprodigiosin, a compound with anticancer and antibacterial activity was identified (Girão et al., 2024). The compound was produced by Streptomyces violaceoruber, a bacteria associated with the green seaweed Codium tomentosum. Bioactivities from SAM-derived microbes have also been reported against aquaculture pathogens. A study by Deutsch et al. (2021), found 23 endophytes originating from twenty seaweed species with antimicrobial activities against four aquaculture pathogens. In this example however, the bioactives responsible are not known.
Regarding crop pathogens, compounds like haliangicin, produced by marine bacteria associated with seaweeds [Haliangium luteum (Fudou et al., 2001)] have been found to have strong antibacterial and antifungal effects, which could be useful in protecting plants from harmful pathogens, such as the oomycete Phytophthora capsica (Sun et al., 2016). In addition, the recently identified antibiotic compound kocumarin (4-[(Z)- 2- phenylethyl] benzoic acid), produced by the actinobacterium Kocuria marina CMG S2, isolated from the brown seaweed Pelvetia canaliculata, exhibited significant antimicrobial activities against both fungi and pathogenic bacteria, including crop pathogens such as Aspergillus (Uzair et al., 2018). Other examples of antimicrobials characterised from SAM-derived bacteria include furan derivatives (Karthick and Mohanraju, 2018), bacteriocins (Luz Prieto et al., 2012), alkaloids (Cui et al., 2009; Ravisankar et al., 2013), polyketides (Chakraborty et al., 2018) and massetolides (Desriac et al., 2013). Notably, massetolide A displays antifungal activities against the major crop pathogen Phytophthora infestans (Tran et al., 2007). Marine fungi isolated from seaweed have also been shown to produce interesting compounds like griseofulvin (Petit et al., 2004), known for its antifungal properties, and utilised in crop protection (Aris et al., 2022). Finally, a recent report has shown that Sargassum’s endophyte Bacillus halotolerans is producing antifungal compounds effective against the fungi responsible for chili fruit rot, Fusarium incarnatum (Suji et al., 2024). The above-mentioned studies are of particular importance as they highlight the potential for SAM-derived extracts to contain new crop-relevant compounds with a direct connection between SAM compounds and crop protection. The extensive biodiversity present within the SAM is therefore likely to contain numerous compounds that have not yet been tested specifically against plant pathogens.
3 Challenges in isolation of seaweed-associated microbes
Leveraging the SAM diversity to discover novel compounds with applications in crops first require the isolation of the bacteria and fungi associated with seaweeds. This presents several challenges, including the need for a wide variety of specialised culture media to encompass the SAM metabolic diversity; to separate microbes from different niches (e.g., epiphytes and endophytes), replicating natural growth conditions in the laboratory (Kaur et al., 2023); and account for the “One Strain Many Compounds” (OSMAC) phenomenon, where a single strain can produce different compounds depending on growth conditions (Romano et al., 2018), or when bioactivities -including that of SAM bacteria such as Roseovarius aestuarii or Rathayibacter festucae- change under environmental stress (Hmani et al., 2024).
A fundamental approach involves general isolation and culturing on agar plates. Epiphytes are typically isolated from swabs or streaks of seaweed thalli, while endophytes require surface sterilisation (Abdelrazek et al., 2024; Deutsch et al., 2021). This plating technique is widely used to cultivate a broad range of bacteria and fungi, as demonstrated in studies characterizing bacterial communities associated with green, and brown and red seaweeds often using nutrient-rich media like Zobell Marine Agar or potato dextrose agar to screen for antibacterial activity (Karthick and Mohanraju, 2018), and for fungi focusing on seaweed-associated endophytes (Abeygunawardane et al., 2025; Fan et al., 2020).
Specialised media are required for specific groups like fungi or bacteria with metabolic capabilities difficult to replicate on ex situ cultivation. Using host homogenate as nutrient/carbon source during isolation (e.g adding sterile host biomass to culture media), could significantly improve the diversity of isolated microbial species. While this method was used in plant microbiome research (Armanhi et al., 2018; Sarhan et al., 2019), to our knowledge this has not been employed on seaweed samples and could yield many novel isolates.
4 Conclusion & perspectives
Seaweed-associated microbiomes have been identified as promising sources of bioactive compounds with antimicrobial properties, offering new opportunities for sustainable crop protection strategies (Singh et al., 2015b). Of note, whether some SAM-derived bioactives could act on insects, weeds or nematodes has not been investigated to date. Other potential, more speculative since they have not been tested yet to the authors knowledge, include using SAM extracts as crop biostimulants and defence elicitors. Systematic testing of those SAM-derived compounds on crops/crop pathogens could yield significant impacts on plants given that these compounds might interact differently with land plant receptors or pathways; or offer novel modes of action due to their structural diversity. These represent important avenues for future research. Several SAM-derived compounds that have been characterised to date could already be potential targets for these uses, including the plant growth regulators and AHLs described above, and summarised in Table 1.
Table 1
| Bioactive functions | Type of molecule | Examples of SAM origin(s) | Seaweed host | Role |
|---|---|---|---|---|
| Crop growth promoting compounds | Auxin-like | Maribacter sp. MS6 (Spoerner et al., 2012), Neptunomonas spp (Matsuda et al., 2018) | Ulva spp., Pyropia yezoensis | Proposed impact on crop growth and development |
| Cytokinin-like | Roseovarius sp. MS2 (Spoerner et al., 2012); Halomonas sp. MS1 (Morales-Reyes et al, 2022) | Ulva spp. | Proposed impact on crop growth and development | |
| Thallusin | Maribacter sp. BPC-D8 & Sulfitobacter sp. BPC-C4 (Ghaderiardakani et al., 2024); Maribacter sp. MS6 (Spoerner et al., 2012) | Ulva spp. | Proposed impact on crop growth and development | |
| N-Acyl Homoserine Lactones (AHLs) | Pseudoalteromonas galatheae (Aslam et al., 2023); Vibrio anguillarum (Joint et al., 2007); Shewanella algae (Singh et al., 2015a) | Porphyra haitanensis; Ulva spp.; Gracilaria dura | Proposed as crop biostimulants | |
| Osmoprotectants promotion | Marine microbes consorsium (Carreiras et al., 2023), possible SAM-derived | n/a | Proposed as stimulating plant stress tolerance (Carreiras et al., 2023) | |
| Plant Growth Promoting Rhizobacteria (PGPR) | Marine microbes (Bacillus subtilis, Nitratireductor aquimarinus, Halopseudomonas pachastrellae (Aizaz et al., 2023)), possible SAM-derived | n/a | Proposed as biofertilizers (Aizaz et al., 2023; Carreiras et al., 2023) | |
| Antimicrobials | PPDHMP | Bacillus subtilis (Manam et al., 2025) | Gracilaria edulis | Antimicrobial. Role against crop pathogens to be determined |
| Decylprodigiosin | Streptomyces violaceoruber (Girão et al., 2024) | Codium tomentosum | Antimicrobial. Role against crop pathogens to be determined | |
| Haliangicin | Haliangium luteum (Fudou et al., 2001) | n.d | Antimicrobial. Role against Phytophtora capsica (Sun et al., 2016) | |
| Kocumarin | Kocuria marina (Uzair et al., 2018) | Pelvetia canaliculata | Antimicrobial, role against Aspergillus spp (Uzair et al., 2018) | |
| Furan derivatives | Pseudomonas stutzeri, Alcanivorax dieselolei, Exiguobacterium profundum, Vibrio sp (Karthick and Mohanraju, 2018) | Gracilaria corticata; Ulva lactuca; Turbinaria ornata; Mastophora rosea | Antimicrobials, including crop pathogens | |
| Bacteriocins | Bacillus spp (Luz Prieto et al., 2012) | Ulva spp. | Antibacterials, including crop pathogens | |
| Alkaloids | Pseudomonas sp (Ravisankar et al., 2013); Aspergillus ochraceus (Cui et al, 2009) | Padina tetrastromatica; Sargassum kjellmanianum | Antimicrobials, including crop pathogens | |
| Polyketides | Bacillus amyloliquefaciens (Chakraborty et al, 2018) | Kappaphycus alvarezii | Antimicrobials, griseofulvin effective against crop fungal pathogens (Aris et al., 2022) | |
| Massetolides | Pseudomonas sp. (Gerard et al, 1997) | n.d | Antifungal, massetolide A effective against Phytophtora infestans (Tran et al., 2007) | |
| Other Antifungal compounds (unspecified chemical nature) | Bacillus halotolerans (Suji et al., 2024) | Sargassum wightii | Antifungal (Fusarium incarnatum, Suji et al., 2024). Other B. halotolerans strains induced resistance against Botrytis cinerea (Tsalgatidou et al., 2023) | |
| Plant defence elicitors | N-Acyl Homoserine Lactones (AHLs) | Pseudoalteromonas galatheae (Aslam et al., 2023); Vibrio anguillarum (Joint et al., 2007); Shewanella algae (Singh et al., 2015a) | Gracilaria corticata; Ulva lactuca; Turbinaria ornata; Mastophora rosea | Priming of plant defence system, polyphenols & ROS production (Zhu et al., 2022) |
| Alginate derived oligosaccharides | General class of molecules (from seaweed, processed by SAM) | n/a | Priming of plant defence system & polyphenols production (Peng et al., 2025) | |
| Chitin fragments | Staphylococcus equorum and Bacillus tropicus (Ginting et al., 2024) | Gracilaria sp. | Priming of plant defence system & phytoalexin production (Saberi Riseh et al., 2024) | |
| Lipopolysaccharides (LPS) | General class of molecules (gram-negative bacteria) | n/a | Priming of plant defence system & hypersensitive response (Erbs et al., 2010) | |
| Peptidoglycan derivatives | General class of molecules | n/a | Priming of plant defence system, chitinase activity & polyphenols production (Erbs and Newman, 2012) | |
| Extracellular Polysaccharides (EPS) |
Bacillus licheniformis (Singh et al., 2011b) Pseudoalteromonas spp (Xu et al., 2021) |
Gracilaria. dura; Fucus evanescens | Priming of plant defence system, hypersensitive response & ROS production (Drira et al., 2021) |
Type of molecules, origins and roles of SAM-derived classes of bioactives with a potential on crops.
n/a, Not applicable; nd, Not determined.
However, while potential is significant, practical application of SAM-derived bioactives in agriculture will likely face hurdles. Focused and systematic research is needed to bridge this gap, particularly in i) isolating and characterising individual potential compounds, ii) understanding their mode of action in crops/soils; and iii) assess their effectiveness and environmental impact(s) compared to existing phytochemicals. These, particularly the characterisation of the compounds (e.g via bioactivity-guided fractionation), the use of specialised instrumentation, and cost in both time and expertise needed, represent major challenges. The industrial production and purification of those compounds similarly require extensive research, as large batch cultivation of the target marine microorganism could be difficult. An attractive option could be to first decipher the metabolic pathways leading to bioactive accumulation in the desirable microbe itself via genomics and metabolomics (Castro-Falcón et al., 2025; Molina et al., 2025; Tsalgatidou et al., 2022), and then transfer the genes responsible via synthetic biology to microbial factories for heterologous production (Chaudhary et al., 2024). Finally, matrix/synergistic effects between compounds within the SAM should also be considered, and creating rhizosphere SAM-derived communities will require extensive testing.
In conclusion, while the exploration of seaweed-associated microbiomes as sources of crop protective bioactives & biostimulants is still in its early stages, the diversity of SAM-derived metabolites offers a compelling case for further investigation.
Statements
Author contributions
SM: Writing – original draft, Writing – review & editing. ED: Writing – original draft, Writing – review & editing. AF: Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors acknowledge funding from Research Ireland (AMicrobioM project, grant #22/FFP-P/11555). The article is also based upon work from COST Action CA20106 “Tomorrow’s wheat of the sea’: Ulva, a model for an innovative mariculture”, supported by COST (European Cooperation in Science and Technology).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abdelrazek H. M. Shams El-Din N. G. Ghozlan H. A. Sabry S. A. Abouelkheir S. S. (2024). Distribution and functional perspective analysis of epiphytic and endophytic bacterial communities associated with marine seaweeds, Alexandria shores, Egypt. BMC Microbiol.24, 293. doi: 10.1186/s12866-024-03426-x
2
Abeygunawardane S. Thambugala K. M. Kumara W. Daranagama D. Abeygunawardane S. Thambugala K. M. et al . (2025). Seaweed-associated fungal endophytes from southern Sri Lanka and their biocontrol potential against selected fungal phytopathogens. Stud. Fungi10. doi: 10.48130/sif-0025-0002
3
Aizaz M. Lubna Ahmad W. Khan I. Asaf S. Bilal S. et al . (2023). Exploring the potential of halotolerant bacteria from coastal regions to mitigate salinity stress in wheat: physiological, molecular, and biochemical insights. Front. Plant Sci.14. doi: 10.3389/fpls.2023.1224731
4
Alsufyani T. Califano G. Deicke M. Grueneberg J. Weiss A. Engelen A. H. et al . (2020). Macroalgal–bacterial interactions: identification and role of thallusin in morphogenesis of the seaweed Ulva (Chlorophyta). J. Exp. Bot.71, 3340–3349. doi: 10.1093/jxb/eraa066
5
Aris P. Wei Y. Mohamadzadeh M. Xia X. (2022). Griseofulvin: an updated overview of old and current knowledge. Molecules27 (20), 7034. doi: 10.3390/molecules27207034
6
Armanhi J. S. L. de Souza R. S. C. Damasceno N. de B. de Araújo L. M. Imperial J. et al . (2018). A community-based culture collection for targeting novel plant growth-promoting bacteria from the sugarcane microbiome. Front. Plant Sci.8. doi: 10.3389/fpls.2017.02191
7
Asharaf S. Chakraborty K. Chakraborty R. D. (2022). Seaweed-associated heterotrophic bacteria: are they future novel sources of antimicrobial agents against drug-resistant pathogens? Arch. Microbiol.204, 232. doi: 10.1007/s00203-022-02835-8
8
Aslam M. Pei P. Ye P. Li T. Liang H. Zhang Z. et al . (2023). Unraveling the Diverse Profile of N-Acyl Homoserine Lactone Signals and Their Role in the Regulation of Biofilm Formation in Porphyra haitanensis-Associated Pseudoalteromonas galatheae. Microorganisms11 (9), 2228. doi: 10.3390/microorganisms11092228
9
Burgunter-Delamare B. KleinJan H. Frioux C. Fremy E. Wagner M. Corre E. et al . (2020). Metabolic complementarity between a brown alga and associated cultivable bacteria provide indications of beneficial interactions. Front. Mar. Sci.7. doi: 10.3389/fmars.2020.00085
10
Burgunter-Delamare B. Rousvoal S. Legeay E. Tanguy G. Fredriksen S. Boyen C. et al . (2023). The Saccharina latissima microbiome: Effects of region, season, and physiology. Front. Microbiol.13. doi: 10.3389/fmicb.2022.1050939
11
Burke C. Thomas T. Lewis M. Steinberg P. Kjelleberg S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J.5, 590–600. doi: 10.1038/ismej.2010.164
12
Califano G. Kwantes M. Abreu M. H. Costa R. Wichard T. (2020). Cultivating the macroalgal holobiont: effects of integrated multi-trophic aquaculture on the microbiome of Ulva rigida (Chlorophyta). Front. Mar. Sci.7. doi: 10.3389/fmars.2020.00052
13
Carreiras J. Cruz-Silva A. Fonseca B. Carvalho R. C. Cunha J. P. Proença Pereira J. et al . (2023). Improving grapevine heat stress resilience with marine plant growth-promoting Rhizobacteria consortia. Microorganisms11, 856. doi: 10.3390/microorganisms11040856
14
Castro-Falcón G. Guillén-Matus D. G. Silva E. B. D. Guo W. Ross A. Sá Magalhães Serafim M. et al . (2025). Structure elucidation, biosynthetic gene cluster distribution, and biological activities of ketomemicin analogs in salinispora. Mar. Drugs23, 126. doi: 10.3390/md23030126
15
Chakraborty K. Thilakan B. Kizhakkekalam V. K. (2018). Antibacterial aryl-crowned polyketide from Bacillus subtilis associated with seaweed Anthophycus longifolius. J. Appl. Microbiol.124, 108–125. doi: 10.1111/jam.13627
16
Chaloner T. M. Gurr S. J. Bebber D. P. (2021). Plant pathogen infection risk tracks global crop yields under climate change. Nat. Clim. Change11, 710–715. doi: 10.1038/s41558-021-01104-8
17
Chaudhary R. Nawaz A. Fouillaud M. Dufossé L. Haq I. Mukhtar H. (2024). Microbial cell factories: biodiversity, pathway construction, robustness, and industrial applicability. Microbiol. Res.15, 247–272. doi: 10.3390/microbiolres15010018
18
Cui C.-M. Li X.-M. Li C.-S. Sun H.-F. Gao S.-S. Wang B.-G. (2009). Benzodiazepine alkaloids from marine-derived endophytic fungus Aspergillus ochraceus. Helv. Chim. Acta92, 1366–1370. doi: 10.1002/hlca.200900084
19
Daly G. Decorosi F. Viti C. Adessi A. (2023). Shaping the phycosphere: Analysis of the EPS in diatom-bacterial co-cultures. J. Phycol.59, 791–797. doi: 10.1111/jpy.13361
20
De Clerck O. Kao S.-M. Bogaert K. A. Blomme J. Foflonker F. Kwantes M. et al . (2018). Insights into the evolution of multicellularity from the sea lettuce genome. Curr. Biol.28, 2921–2933.e5. doi: 10.1016/j.cub.2018.08.015
21
Desriac F. Jégou C. Balnois E. Brillet B. Le Chevalier P. Fleury Y. (2013). Antimicrobial peptides from marine proteobacteria. Mar. Drugs11, 3632–3660. doi: 10.3390/md11103632
22
Deutsch Y. Gur L. Berman Frank I. Ezra D. (2021). Endophytes from algae, a potential source for new biologically active metabolites for disease management in aquaculture. Front. Mar. Sci.8. doi: 10.3389/fmars.2021.636636
23
Deutsch Y. Ofek-Lalzar M. Borenstein M. Berman-Frank I. Ezra D. (2023). Re-introduction of a bioactive bacterial endophyte back to its seaweed (Ulva sp.) host, influences the host’s microbiome. Front. Mar. Sci.10. doi: 10.3389/fmars.2023.1099478
24
Dhiman S. Ulrich J. F. Wienecke P. Wichard T. Arndt H.-D. (2022). Stereoselective total synthesis of (–)-Thallusin for bioactivity profiling. Angew. Chem. Int. Ed.61, e202206746. doi: 10.1002/anie.202206746
25
Drira M. Elleuch J. Ben Hlima H. Hentati F. Gardarin C. Rihouey C. et al . (2021). Optimization of Exopolysaccharides Production by Porphyridium sordidum and Their Potential to Induce Defense Responses in Arabidopsis thaliana against Fusarium oxysporum. Biomolecules11, 282. doi: 10.3390/biom11020282
26
Duan Y. Han M. Grimm M. Ponath J. Reichelt M. Mithöfer A. et al . (2023). Combination of bacterial N-acyl homoserine lactones primes Arabidopsis defenses via jasmonate metabolism. Plant Physiol.191, 2027–2044. doi: 10.1093/plphys/kiad017
27
Erbs G. Molinaro A. Dow J. M. Newman M.-A. (2010). “Lipopolysaccharides and plant innate immunity,” in Endotoxins: Structure, Function and Recognition. Eds. WangX.QuinnP. J. (Springer Netherlands, Dordrecht), 387–403. doi: 10.1007/978-90-481-9078-2_17
28
Erbs G. Newman M.-A. (2012). The role of lipopolysaccharide and peptidoglycan, two glycosylated bacterial microbe-associated molecular patterns (MAMPs), in plant innate immunity. Mol. Plant Pathol.13, 95–104. doi: 10.1111/j.1364-3703.2011.00730.x
29
Fan B. Dewapriya P. Li F. Blümel M. Tasdemir D. (2020). Pyrenosetins A–C, new decalinoylspirotetramic acid derivatives isolated by bioactivity-based molecular networking from the seaweed-derived fungus Pyrenochaetopsis sp. FVE-001. Mar. Drugs18, 47. doi: 10.3390/md18010047
30
Fira D. Dimkić I. Berić T. Lozo J. Stanković S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol.285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
31
Fort A. Lebrault M. Allaire M. Esteves-Ferreira A. A. McHale M. Lopez F. et al . (2019). Extensive variations in diurnal growth patterns and metabolism among Ulva spp. Strains. Plant Physiol.180, 109–123. doi: 10.1104/pp.18.01513
32
Fort A. Ryder P. McKeown P. C. Wijnen C. Aarts M. G. Sulpice R. et al . (2016). Disaggregating polyploidy, parental genome dosage and hybridity contributions to heterosis in Arabidopsis thaliana. New Phytol.209, 590–599. doi: 10.1111/nph.13650
33
Fudou R. Iizuka T. Sato S. Ando T. Shimba N. Yamanaka S. (2001). Haliangicin, a novel antifungal metabolite produced by a marine myxobacterium 2. Isolation and structural elucidation. J. Antibiot. (Tokyo)54, 153–156. doi: 10.7164/antibiotics.54.153
34
Gerard J. Lloyd R. Barsby T. Haden P. Kelly M. T. Andersen R. J . (1997). Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 60 (3), 223–229. doi: 10.1021/np9606456
35
Ghaderiardakani F. Coates J. C. Wichard T. (2017). Bacteria-induced morphogenesis of Ulva intestinalis and Ulva mutabilis (Chlorophyta): a contribution to the lottery theory. FEMS Microbiol. Ecol.93, fix094. doi: 10.1093/femsec/fix094
36
Ghaderiardakani F. Quartino M. L. Wichard T. (2020). Microbiome-dependent adaptation of seaweeds under environmental stresses: A perspective. Front. Mar. Sci.7. doi: 10.3389/fmars.2020.575228
37
Ghaderiardakani F. Ulrich J. F. Barth E. Quartino M. L. Wichard T. (2024). Algal growth and morphogenesis-promoting factors released by cold-adapted bacteria contribute to the resilience and morphogenesis of the seaweed Ulva (Chlorophyta) in Antarctica (Potter cove). J. Plant Growth Regul. 1–18. doi: 10.1007/s00344-024-11507-4
38
Ginting E. L. Sumilat D. A. Rumampuk N. D. C. Kabense R. Moko E. M. Siby M. S. (2024). Identification of seaweed-associated chitinolytic bacteria capable in forming chitosan from Tongkaina waters, North Sulawesi. AACL Bioflux17 (3), 997–1007.
39
Girão M. Freitas S. Martins T. P. Urbatzka R. Carvalho M. F. Leão P. N. (2024). Decylprodigiosin: a new member of the prodigiosin family isolated from a seaweed-associated Streptomyces. Front. Pharmacol.15. doi: 10.3389/fphar.2024.1347485
40
Girão M. Ribeiro I. Ribeiro T. Azevedo I. C. Pereira F. Urbatzka R. et al . (2019). Actinobacteria isolated from Laminaria ochroleuca: A source of new bioactive compounds. Front. Microbiol.10. doi: 10.3389/fmicb.2019.00683
41
Halder V. Kombrink E. (2015). Facile high-throughput forward chemical genetic screening by in situ monitoring of glucuronidase-based reporter gene expression in Arabidopsis thaliana. Front. Plant Sci.6. doi: 10.3389/fpls.2015.00013
42
Han S. Li D. Trost E. Mayer K. F. Vlot A. C. Heller W. et al . (2016). Systemic responses of barley to the 3-hydroxy-decanoyl-homoserine lactone producing plant beneficial endophyte Acidovorax radicis N35. Front. Plant Sci.7. doi: 10.3389/fpls.2016.01868
43
Head W. D. Carpenter E. J. (1975). Nitrogen fixation associated with the marine macroalga Codium fragile. Limnol. Oceanogr.20, 815–823. doi: 10.4319/lo.1975.20.5.0815
44
Hmani I. Ghaderiardakani F. Ktari L. Bour M. E. Wichard T. (2024). High-temperature stress induces bacteria-specific adverse and reversible effects on Ulva (Chlorophyta) growth and its chemosphere in a reductionist model system. Bot. Mar.67, 131–138. doi: 10.1515/bot-2023-0053
45
Joint I. Tait K. Wheeler G. (2007). Cross-kingdom signalling: exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Philos. Trans. R. Soc B Biol. Sci.362, 1223–1233. doi: 10.1098/rstb.2007.2047
46
Joshi H. Shourie A. Singh A. (2020). “Chapter 25 - Cyanobacteria as a source of biofertilizers for sustainable agriculture,” in Advances in Cyanobacterial Biology. Eds. SinghP. K.KumarA.SinghV. K.ShrivastavaA. K. (Cambridge, MA: Academic Press), 385–396. doi: 10.1016/B978-0-12-819311-2.00025-5
47
Karthick P. Mohanraju R. (2018). Antimicrobial potential of epiphytic bacteria associated with seaweeds of little Andaman, India. Front. Microbiol.9. doi: 10.3389/fmicb.2018.00611
48
Kaur M. Saini K. C. Mallick A. Bast F . (2023). Seaweed-associated epiphytic bacteria: Diversity, ecological and economic implications. Aquat. Bot. 189, 103698.
49
Khalil A. Bramucci A. R. Focardi A. Le Reun N. Willams N. L. R. Kuzhiumparambil U. et al . (2024). Widespread production of plant growth-promoting hormones among marine bacteria and their impacts on the growth of a marine diatom. Microbiome12, 205. doi: 10.1186/s40168-024-01899-6
50
Khan T. Song W. Nappi J. Marzinelli E. M. Egan S. Thomas T. (2024). Functional guilds and drivers of diversity in seaweed-associated bacteria. FEMS Microbes5, xtad023. doi: 10.1093/femsmc/xtad023
51
Kim K. H. Kim J. M. Baek J. H. Jeong S. E. Kim H. Yoon H. S. et al . (2024). Metabolic relationships between marine red algae and algae-associated bacteria. Mar. Life Sci. Technol.6, 298–314. doi: 10.1007/s42995-024-00227-z
52
Lavecchia A. Fosso B. Engelen A. H. Borin S. Manzari C. Picardi E. et al . (2024). Macroalgal microbiomes unveil a valuable genetic resource for halogen metabolism. Microbiome12, 47. doi: 10.1186/s40168-023-01740-6
53
Li J. Weinberger F. de Nys R. Thomas T. Egan S. (2023). A pathway to improve seaweed aquaculture through microbiota manipulation. Trends Biotechnol.41, 545–556. doi: 10.1016/j.tibtech.2022.08.003
54
Li J. Weinberger F. Saha M. Majzoub M. E. Egan S. (2022). Cross-host protection of marine bacteria against macroalgal disease. Microb. Ecol.84, 1288–1293. doi: 10.1007/s00248-021-01909-2
55
Luz Prieto M. O’Sullivan L. Tan S. P. McLoughlin P. Hughes H. O’Connor P. M. et al . (2012). Assessment of the bacteriocinogenic potential of marine bacteria reveals lichenicidin production by seaweed-derived Bacillus spp. Mar. Drugs10, 2280–2299. doi: 10.3390/md10102280
56
Manam M. Srivatsa S. Osborne W. J. (2025). Endophytic bacteria of Gracilaria edulis in combating human bacterial pathogens by PPDHMP – A crude to single molecule product development approach. Microb. Pathog.202, 107431. doi: 10.1016/j.micpath.2025.107431
57
Martinez-Delgado J. Benitez-Campo N. (2025). Probiotic potential of bacteria associated with the mangrove epiphytic algae Bostrychia calliptera and Rhizoclonium riparium. PeerJ13, e19073. doi: 10.7717/peerj.19073
58
Matsuda R. Handayani M. L. Sasaki H. Takechi K. Takano H. Takio S. (2018). Production of indoleacetic acid by strains of the epiphytic bacteria Neptunomonas spp. isolated from the red alga Pyropia yezoensis and the seagrass Zostera marina. Arch. Microbiol.200, 255–265. doi: 10.1007/s00203-017-1439-1
59
Matsuo Y. Imagawa H. Nishizawa M. Shizuri Y. (2005). Isolation of an algal morphogenesis inducer from a marine bacterium. Science307, 1598–1598. doi: 10.1126/science.1105486
60
Meena M. Yadav G. Sonigra P. Nagda A. Mehta T. Swapnil P. et al . (2022). Role of elicitors to initiate the induction of systemic resistance in plants to biotic stress. Plant Stress5, 100103. doi: 10.1016/j.stress.2022.100103
61
Mehmood N. Saeed M. Zafarullah S. Hyder S. Rizvi Z. F. Gondal A. S. et al . (2023). Multifaceted impacts of plant-beneficial pseudomonas spp. in managing various plant diseases and crop yield improvement. ACS Omega8, 22296–22315. doi: 10.1021/acsomega.3c00870
62
Meunier L. Costa R. Keller-Costa T. Cannella D. Dechamps E. George I. F. (2024). Selection of marine bacterial consortia efficient at degrading chitin leads to the discovery of new potential chitin degraders. Microbiol. Spectr.12, e00886–e00824. doi: 10.1128/spectrum.00886-24
63
Molina D. Angamarca E. Marinescu G. C. Popescu R. G. Tenea G. N. (2025). Integrating metabolomics and genomics to uncover antimicrobial compounds in Lactiplantibacillus plantarum UTNGt2, a cacao-originating probiotic from Ecuador. Antibiotics14, 123. doi: 10.3390/antibiotics14020123
64
Morales-Reyes C. F. Ghaderiardakani F. Wichard T . (2022). Genome sequence of Halomonas sp. strain MS1, a metallophore-producing, algal growth-promoting marine bacterium isolated from the green seaweed Ulva mutabilis (Chlorophyta). Microbiol. Resour. Announc. 11, e00685–22. doi: 10.1128/mra.00685-22
65
Mori I. C. Ikeda Y. Matsuura T. Hirayama T. Mikami K. (2017). Phytohormones in red seaweeds: a technical review of methods for analysis and a consideration of genomic data. Bot. Mar.60, 153–170. doi: 10.1515/bot-2016-0056
66
Moshynets O. V. Babenko L. M. Rogalsky S. P. Iungin O. S. Foster J. Kosakivska I. V. et al . (2019). Priming winter wheat seeds with the bacterial quorum sensing signal N-hexanoyl-L-homoserine lactone (C6-HSL) shows potential to improve plant growth and seed yield. PloS One14, e0209460. doi: 10.1371/journal.pone.0209460
67
Nawaz M. S. Arshad A. Rajput L. Fatima K. Ullah S. Ahmad M. et al . (2020). Growth-stimulatory effect of quorum sensing signal molecule N-acyl-homoserine lactone-producing multi-trait Aeromonas spp. on wheat genotypes under salt stress. Front. Microbiol.11, 553621. doi: 10.3389/fmicb.2020.553621
68
Nguyen M. V. Han J. W. Kim H. Choi G. J. (2022). Phenyl ethers from the marine-derived fungus Aspergillus tabacinus and their antimicrobial activity against plant pathogenic fungi and bacteria. ACS Omega7, 33273–33279. doi: 10.1021/acsomega.2c03859
69
Ortiz J. Dias N. Alvarado R. Soto J. Sanhueza T. Rabert C. et al . (2024). N- acyl homoserine lactones (AHLs) type signal molecules produced by rhizobacteria associated with plants that growing in a metal(oids) contaminated soil: A catalyst for plant growth. Microbiol. Res.281, 127606. doi: 10.1016/j.micres.2024.127606
70
Paix B. Layglon N. Le Poupon C. D’Onofrio S. Misson B. Garnier C. et al . (2021). Integration of spatio-temporal variations of surface metabolomes and epibacterial communities highlights the importance of copper stress as a major factor shaping host-microbiota interactions within a Mediterranean seaweed holobiont. Microbiome9, 201. doi: 10.1186/s40168-021-01124-8
71
Peng C. Xu W. Wang X. Meng F. Zhao Y. Wang Q. et al . (2025). Alginate oligosaccharides trigger multiple defense responses in tobacco and induce resistance to Phytophthora infestans. Front. Plant Sci.16. doi: 10.3389/fpls.2025.1506873
72
Petit K. E. Mondeguer F. Roquebert M. F. Biard J. F. Pouchus Y. F. (2004). Detection of griseofulvin in a marine strain of Penicillium waksmanii by ion trap mass spectrometry. J. Microbiol. Methods58, 59–65. doi: 10.1016/j.mimet.2004.03.004
73
Qi L. Du H.-F. Sun T.-T. Li L. Zhang Y.-H. Liu Y.-F. et al . (2023). Natural products from marine fungi as a source against agricultural pathogenic fungi. Appl. Microbiol. Biotechnol.107, 5003–5017. doi: 10.1007/s00253-023-12657-3
74
Radhakrishnan R. Hashem A. Abd_Allah E. F. (2017). Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol.8. doi: 10.3389/fphys.2017.00667
75
Raio A. Puopolo G. (2021). Pseudomonas chlororaphis metabolites as biocontrol promoters of plant health and improved crop yield. World J. Microbiol. Biotechnol.37, 99. doi: 10.1007/s11274-021-03063-w
76
Ramírez-Puebla S. T. Weigel B. L. Jack L. Schlundt C. Pfister C. A. Mark Welch J. L. (2022). Spatial organization of the kelp microbiome at micron scales. Microbiome10, 52. doi: 10.1186/s40168-022-01235-w
77
Rathod K. Rana S. Dhandukia P. Thakker J. N. (2023). Investigating marine Bacillus as an effective growth promoter for chickpea. J. Genet. Eng. Biotechnol.21, 137. doi: 10.1186/s43141-023-00608-4
78
Ravisankar A. Gnanambal M. E. K. Sundaram L. R. (2013). A newly isolated Pseudomonas sp., epibiotic on the seaweed, Padina tetrastromatica, off Southeastern Coast of India, reveals antibacterial action. Appl. Biochem. Biotechnol.171, 1968–1985. doi: 10.1007/s12010-013-0473-y
79
Romano S. Jackson S. A. Patry S. Dobson A. D. W. (2018). Extending the “One strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs16, 244. doi: 10.3390/md16070244
80
Saberi Riseh R. Gholizadeh Vazvani M. Vatankhah M. Kennedy J. F. (2024). Chitin-induced disease resistance in plants: A review. Int. J. Biol. Macromol.266, 131105. doi: 10.1016/j.ijbiomac.2024.131105
81
Saha M. Dittami S. M. Chan C. X. Raina J.-B. Stock W. Ghaderiardakani F. et al . (2024). Progress and future directions for seaweed holobiont research. New Phytol.244, 364–376. doi: 10.1111/nph.20018
82
Saha M. Weinberger F. (2019). Microbial “gardening” by a seaweed holobiont: Surface metabolites attract protective and deter pathogenic epibacterial settlement. J. Ecol.107, 2255–2265. doi: 10.1111/1365-2745.13193
83
Sarhan M. S. Hamza M. A. Youssef H. H. Patz S. Becker M. ElSawey H. et al . (2019). Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media – A review. J. Adv. Res.19, 15–27. doi: 10.1016/j.jare.2019.04.002
84
Schenk S. T. Hernández-Reyes C. Samans B. Stein E. Neumann C. Schikora M. et al . (2014). N-Acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell26, 2708–2723. doi: 10.1105/tpc.114.126763
85
Schikora A. Schenk S. T. Hartmann A. (2016). Beneficial effects of bacteria-plant communication based on quorum sensing molecules of theN-acyl homoserine lactone group. Plant Mol. Biol.90, 605–612. doi: 10.1007/s11103-016-0457-8
86
Schuhegger R. Ihring A. Gantner S. Bahnweg G. Knappe C. Vogg G. et al . (2006). Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ.29, 909–918. doi: 10.1111/j.1365-3040.2005.01471.x
87
Sharma A. Kumar V. Shahzad B. Tanveer M. Sidhu G. P. S. Handa N. et al . (2019). Worldwide pesticide usage and its impacts on ecosystem. SN Appl. Sci.1, 1446. doi: 10.1007/s42452-019-1485-1
88
Shrestha A. Grimm M. Ojiro I. Krumwiede J. Schikora A. (2020). Impact of quorum sensing molecules on plant growth and immune system. Front. Microbiol.11. doi: 10.3389/fmicb.2020.01545
89
Simon C. McHale M. Sulpice R. (2022). Applications of Ulva biomass and strategies to improve its yield and composition: A perspective for Ulva aquaculture. Biology11, 1593. doi: 10.3390/biology11111593
90
Singh R. P. Baghel R. S. Reddy C. R. K. Jha B. (2015a). Effect of quorum sensing signals produced by seaweed-associated bacteria on carpospore liberation from Gracilaria dura. Front. Plant Sci.6. doi: 10.3389/fpls.2015.00117
91
Singh R. P. Bijo A. J. Baghel R. S. Reddy C. R. K. Jha B. (2011a). Role of bacterial isolates in enhancing the bud induction in the industrially important red alga Gracilaria dura. FEMS Microbiol. Ecol.76, 381–392. doi: 10.1111/j.1574-6941.2011.01057.x
92
Singh P. FNU K. Encarnação T. (2023). “Marine bacteria for biofertilizers,” in Marine Organisms: A Solution to Environmental Pollution? Uses in Bioremediation and in Biorefinery. Eds. EncarnaçãoT.Canelas PaisA. (Springer International Publishing, Cham), 189–203. doi: 10.1007/978-3-031-17226-7_9
93
Singh R. P. Kumari P. Reddy C. R. K. (2015b). Antimicrobial compounds from seaweeds-associated bacteria and fungi. Appl. Microbiol. Biotechnol.99, 1571–1586. doi: 10.1007/s00253-014-6334-y
94
Singh R. P. Shukla M. K. Mishra A. Kumari P. Reddy C. R. K. Jha B. (2011b). Isolation and characterization of exopolysaccharides from seaweed associated bacteria Bacillus licheniformis. Carbohydr. Polym.84, 1019–1026. doi: 10.1016/j.carbpol.2010.12.061
95
Spoerner M. Wichard T. Bachhuber T. Stratmann J. Oertel W. (2012). Growth and Thallus morphogenesis of Ulva mutabilis (Chlorophyta) depends on A combination of two bacterial species excreting regulatory factors. J. Phycol.48, 1433–1447. doi: 10.1111/j.1529-8817.2012.01231.x
96
Suji H. A. Manikandan K. Sudha A. Muthukumar A. Jeyalakshmi C. Charumathi M. et al . (2024). Whole genome sequence of seaweed endophyte Bacillus halotolerans strain AUPP for antagonistic activity against Fusarium incarnatum causing chilli fruit rot. Sci. Rep.14, 31881. doi: 10.1038/s41598-024-83317-3
97
Sun Y. Feng Z. Tomura T. Suzuki A. Miyano S. Tsuge T. et al . (2016). Heterologous production of the marine myxobacterial antibiotic haliangicin and its unnatural analogues generated by engineering of the biochemical pathway. Sci. Rep.6, 22091. doi: 10.1038/srep22091
98
Tait K. Joint I. Daykin M. Milton D. L. Williams P. Cámara M. (2005). Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol.7, 229–240. doi: 10.1111/j.1462-2920.2004.00706.x
99
Tangestani M. Broady P. Varsani A. (2021). An investigation of antibacterial activity of New Zealand seaweed-associated marine bacteria. Future Microbiol.16, 1167–1179. doi: 10.2217/fmb-2021-0023
100
Tran H. Ficke A. Asiimwe T. Höfte M. Raaijmakers J. M. (2007). Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol.175, 731–742. doi: 10.1111/j.1469-8137.2007.02138.x
101
Tsalgatidou P. C. Thomloudi E.-E. Baira E. Papadimitriou K. Skagia A. Venieraki A. et al . (2022). Integrated genomic and metabolomic analysis illuminates key secreted metabolites produced by the novel endophyte bacillus halotolerans cal.l.30 involved in diverse biological control activities. Microorganisms10 (2), 399. doi: 10.3390/microorganisms10020399
102
Tsalgatidou P. C. Thomloudi E. E. Delis C. Nifakos K. Zambounis A. Venieraki A. et al . (2023). Compatible consortium of endophytic Bacillus halotolerans strains Cal.l.30 and Cal.f.4 promotes plant growth and induces systemic resistance against Botrytis cinerea. Biology12 (6), 779. doi: 10.3390/biology12060779
103
Uzair B. Menaa F. Khan B. A. Mohammad F. V. Ahmad V. U. Djeribi R. et al . (2018). Isolation, purification, structural elucidation and antimicrobial activities of kocumarin, a novel antibiotic isolated from actinobacterium Kocuria marina CMG S2 associated with the brown seaweed Pelvetia canaliculata. Microbiol. Res.206, 186–197. doi: 10.1016/j.micres.2017.10.007
104
Vallet M. Strittmatter M. Murúa P. Lacoste S. Dupont J. Hubas C. et al . (2018). Chemically-mediated interactions between macroalgae, their fungal endophytes, and protistan pathogens. Front. Microbiol.9. doi: 10.3389/fmicb.2018.03161
105
van der Loos L. M. D’hondt S. Engelen A. H. Pavia H. Toth G. B. Willems A. et al . (2023). Salinity and host drive Ulva-associated bacterial communities across the Atlantic–Baltic Sea gradient. Mol. Ecol.32, 6260–6277. doi: 10.1111/mec.16462
106
van der Loos L. M. De Wilde C. Willems A. De Clerck O. Steinhagen S. (2024). The cultivated sea lettuce (Ulva) microbiome: Successional and seasonal dynamics. Aquaculture585, 740692. doi: 10.1016/j.aquaculture.2024.740692
107
Vega-Portalatino E. J. Rosales-Cuentas M. M. Tamariz-Angeles C. Olivera-Gonzales P. Espinoza-Espinoza L. A. Moreno-Quispe L. A. et al . (2024). Diversity of endophytic bacteria with antimicrobial potential isolated from marine macroalgae from Yacila and Cangrejos beaches, Piura-Peru. Arch. Microbiol.206, 372. doi: 10.1007/s00203-024-04098-x
108
Wang J. Tang X. Mo Z. Mao Y. (2022). Metagenome-assembled genomes from pyropia haitanensis microbiome provide insights into the potential metabolic functions to the seaweed. Front. Microbiol.13. doi: 10.3389/fmicb.2022.857901
109
Weigel Brooke L. Miranda Khashiff K. Fogarty Emily C. Watson Andrea R. Pfister Catherine A. (2022). Functional insights into the kelp microbiome from metagenome-assembled genomes. mSystems7, e01422–e01421. doi: 10.1128/msystems.01422-21
110
Wichard T. (2023). From model organism to application: Bacteria-induced growth and development of the green seaweed Ulva and the potential of microbe leveraging in algal aquaculture. Semin. Cell Dev. Biol.134, 69–78. doi: 10.1016/j.semcdb.2022.04.007
111
Wood G. Steinberg P. D. Campbell A. H. Vergés A. Coleman M. A. Marzinelli E. M. (2022). Host genetics, phenotype and geography structure the microbiome of a foundational seaweed. Mol. Ecol.31, 2189–2206. doi: 10.1111/mec.16378
112
Xu F. Cha Q.-Q. Zhang Y.-Z. Chen X.-L. (2021). Degradation and utilization of alginate by marine pseudoalteromonas: a review. Appl. Environ. Microbiol.87, e00368–e00321. doi: 10.1128/AEM.00368-21
113
Yamamoto H. Takagi Y. Yamasaki N. Mitsuyama T. Kasai Y. Imagawa H. et al . (2018). Syntheses of thallusin analogues and their algal morphogenesis-inducing activities. Tetrahedron74, 7173–7178. doi: 10.1016/j.tet.2018.10.048
114
Zhao Q. Yang X.-Y. Li Y. Liu F. Cao X.-Y. Jia Z.-H. et al . (2020). N-3-oxo-hexanoyl-homoserine lactone, a bacterial quorum sensing signal, enhances salt tolerance in Arabidopsis and wheat. Bot. Stud.61, 1–12. doi: 10.1186/s40529-020-00283-5
115
Zhu L. Huang J. Lu X. Zhou C. (2022). Development of plant systemic resistance by beneficial rhizobacteria: Recognition, initiation, elicitation and regulation. Front. Plant Sci.13. doi: 10.3389/fpls.2022.952397
Summary
Keywords
seaweed associated microbiome, plant growth promoting (PGP) activities, phytohomones, defence elicitors, antimicrobials
Citation
McKenna S, Da Silva Pereira EH and Fort A (2025) Seaweed-associated microbes as a novel source of crop agrochemicals. Front. Mar. Sci. 12:1629196. doi: 10.3389/fmars.2025.1629196
Received
15 May 2025
Accepted
14 July 2025
Published
31 July 2025
Volume
12 - 2025
Edited by
Thomas Wichard, Friedrich Schiller University Jena, Germany
Reviewed by
Puja Kumari, Scottish Association for Marine Science, United Kingdom
Dilek Ünal, Bilecik Şeyh Edebali University, Türkiye
Updates
Copyright
© 2025 McKenna, Da Silva Pereira and Fort.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antoine Fort, Antoine.fort@tus.ie
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.