- 1National Facility for Coastal & Marine Research (NFCMR) and Centre for Ocean Research (DST-FIST Sponsored centre), MoES – Earth Science & Technology Cell, Sathyabama Institute of Science and Technology, Chennai, Tamil Nadu, India
- 2Centre for Ocean Research (DST-FIST Sponsored centre), MoES – Earth Science & Technology Cell, Sathyabama Institute of Science and Technology, Chennai, Tamil Nadu, India
- 3Department of Civil and Energy System Engineering, Kyonggi University, Suwon, Republic of Korea
- 4Center for Herbal Pharmacology and Environmental Sustainability, Chettinad Hospital and Research Institute, Chettinad Academy of Research and Education, Kelambakkam, Tamil Nadu, India
- 5Department of Pharmaceutical Biology, Faculty of Pharmaceutical Sciences, UCSI University, Kuala Lumpur, Malaysia
- 6Department of Microbiology, Faculty of Arts Science Commerce and Management, Karpagam Academy of Higher Education, Coimbatore, Tamil Nadu, India
Introduction: Antimicrobial resistance (AMR) is a growing public health concern, and understanding the processes driving its growth is crucial.
Methodology: The study investigates the link between heavy metal/biocide (BMRGs) and antibiotic resistance genes (ARGs) and the presence of mobile genetic elements (MGEs) in the polychaete gut microbiome from four differently polluted estuaries along the Southeast coast of India- Adyar Estuary (S1), Ennore Creek (S2), Penna Sangamam (S3), and Pazhayakayal Estuary (S4), using whole metagenome sequencing.
Results: ICPOES analysis for the tissue, sediment and water revealed the presence of high concentration of Zn, Fe, and Cu in sediment and tissue and high levels of Hg and Zn in water and bioaccumulation of Cu and Fe in polychaete tissues, especially in S2 and S4. A total of 2054 ARGs has been identified in all four samples and mostly belonging to class of Cephalosporin and Carbapenem. MRGs against Cu, Fe and Zn were predominantly found in all four samples. MGEs were extensively present in S4 & S2. The co-occurrence analysis of ARGs, BMRGs & MGEs revealed the presence of high co-occurrence in S2 & S4 samples. Risk score assessment indicated that the S2 and S4 samples have a high potential for spreading AMR across ecosystems.
Discussion: The findings suggest that industrial effluents discharged into rivers and aquatic ecosystems and presence of heavy metals beyond permissible limit could contribute to the spread of AMR through the co-selection of BMRGs.
1 Introduction
Estuarine environments, situated at the interface of terrestrial/freshwater and marine ecosystems, represent critical zones profoundly affected by anthropogenic activities, making them pivotal hotspots for ecological impacts (Lotze et al., 2006; Rodgers et al., 2019). Urbanization is causing surface waters to be under pressure due to the dispersal of chemicals and waste biomass from various environments, including household, clinical, and industrial activities (Pimentel et al., 2007; Ayukekbong et al., 2017). Organic pollutants and microbial contaminants from sewage emissions also contribute to changes in physicochemical properties and microbial composition in river systems (Menon et al., 2020). Landscape diversity, patterns like engineered reservoirs and local waste management, influence the spatial dynamics of aquatic contaminants, making water quality in Low-Middle Income Countries (LMICs) highly variable over time (Liu et al., 2018; Zhang et al., 2020; Ho et al., 2021). Among the array of pollutants infiltrating these ecosystems, heavy metals, pesticides, hydrocarbons, and antibiotics stand out as major concerns, with their ingress posing risks to both human health and ecosystem integrity. While conventional pollutants like organochlorine pesticides (Da et al., 2014) and heavy metals (Wu et al., 2014) have long been recognized, antibiotics and antibiotic resistance genes (ARGs) have emerged as significant pollutants, with their abundance skyrocketing since the widespread manufacture of antibiotics in the mid-20th century (Pruden et al., 2006; Knapp et al., 2010). The overuse and misuse of antibiotics have played a pivotal role in exacerbating this issue, leading to the proliferation of ARGs in various aquatic environments (Drudge et al., 2012; McKinney and Pruden, 2012; Segawa et al., 2013; Zhu et al., 2017).
In India, inland aquatic ecosystems have undergone a steady decline due to the influx of pollutants from domestic, industrial, and agricultural sources (Das et al., 2012, 2023). Recent years have witnessed a noticeable increase in antimicrobial contamination, particularly antibiotics, further compounded by the amalgamation of antimicrobial drug residues with other water pollutants (Mutiyar and Mittal, 2014; Balakrishna et al., 2017; Gothwal and Shashidhar, 2017; Taneja and Sharma, 2019). This amalgamation has facilitated the emergence and dissemination of antimicrobial resistance (AMR) within Indian water bodies, drawing attention to the global research focus on AMR harbored in aquatic environments (EC Com, 2017; WHO et al., 2020). Antimicrobial resistance genes (ARGs) are primarily driven by antibiotic overuse, but recent research has highlighted the role of heavy metals in driving ARGs (Andersson and Hughes, 2014; Lopatkin et al., 2016; Knapp et al., 2017; Shen et al., 2021; Zhou et al., 2021). Co-selection, where heavy metals exacerbate the abundance of ARGs in the environment, is now recognized as a crucial mechanism (Baker-Austin et al., 2006; Seiler and Berendonk, 2012; Yuan et al., 2019; Zhang et al., 2019b, 2019a; Liu et al., 2022). Because of their non-biodegradability, metals can act as a long-term selection pressure, stabilizing the ARG pool in both constructed and natural systems (Stepanauskas et al., 2005). Biocides, such as quaternary ammonium compounds (QACs), biguanides, and bisphenols, have received less attention as a potential co-selection agent (SCENIHR (Scientific Committee on Emerging and Newly Identified Health Risks), 2009; Buffet-Bataillon et al., 2012; Ortega Morente et al., 2013; Tandukar et al., 2013). Metals and biocides can co-select for antibiotic-resistant bacteria through co-resistance, cross resistance, and co-regulation (Chapman, 2003; Yu et al., 2016; Zhao et al., 2019). ARG persistence and proliferation may make the simultaneous transfer of ARGs and BRGs/MRGs via mobile genetic elements a difficult public health issue (Li et al., 2022).
Polychaetes, a diverse taxon in marine ecosystems, play a crucial role in the decay of organic sediment matter, bioturbation of marine sediments, and nutrient transfer to the water column (Hutchings, 1998; López and Richter, 2017; Kauppi et al., 2018; Pamungkas et al., 2019). They are also well-suited proxies for monitoring environmental disturbance and marine pollution due to their continuous exposure to various xenobiotics (Olsgard et al., 2003; Díaz-Castañeda and Reish, 2009; Mdaini et al., 2021). Eco toxicological research has employed polychaetes in both field and lab settings (Bat and Üniversitesi, 2005; Amiard-Triquet et al., 2013; Bouraoui et al., 2014). The study conducted by Dafforn et al. (2013) examines pollution effects on polychaetes in estuarine environments. Heavily polluted estuaries exhibit elevated metal and PAH concentrations. Despite pollution, polychaete diversity thrives in heavily modified estuaries, possibly due to organic enrichment (Dafforn et al., 2013). Against this backdrop, our study aimed to explore the gut microbiota of benthic invertebrates (polychaetes) collected from four polluted estuaries along the Southeast coast of India, namely Ennore Creek (Heavy metal Pollution), Penna Sangamam (Aquaculture wastes), Adyar estuary (Domestic sewage), and Pazhayakayal estuary (Industrial effluents). Model species including Marphysa madrasi (Ennore creek and Penna Sangamam) and Namalycastis jaya (Adyar and Pazhayakayal estuary) were chosen due to their abundance and ecological significance as prey for bottom-feeding fish and crustaceans within each estuary.
Metagenomic sequencing was employed to analyze the resistome in fecal samples of these animals, aiming to elucidate potential correlations between environmentally toxic heavy metals and their ARG counterparts. Additionally, the study hypothesizes that excessive heavy metal and biocide exposure especially in Ennore and Pazhayakayal estuary, exacerbates the evolution of antimicrobial resistance within the gut microbiome of polychaetes. The findings of this study shed light on the complex interplay between heavy metals, biocides and antimicrobial resistance, emphasizing the imperative to regulate the effluent release and polluting the estuaries to mitigate the dissemination of antimicrobial resistance in the environment. The study also highlights the co-occurrences of ARGs, BMRGs and MGEs will lead to transfer of the multi-drug, -metal, -biocide resistance to other water ecosystem. We also hypothesis that both heavy metal and industrial pollutants would lead to spread of AMR to different ecosystem than the other two types of pollutants.
2 Materials and methodology
2.1 Sample site information
We chose four different polluted estuaries along India’s Southeast coast: Adyar Estuary (S1), Ennore Creek (S2), Penna Sangamam (S3), and Pazhayakayal Estuary (S4). The Adyar Estuary lies in the center of Chennai, India. The estuary, where the Adyar River meets the Bay of Bengal, is a historical site. The Adyar River and its estuary are extremely polluted as a result of unmanaged domestic waste and untreated human excreta dumped along its banks (Mukhopadhyay et al., 2020). Additionally, the river gets sewage waste from the city’s nearly 58 drain outlets. Rapid development and urbanization in the 1980s and 1990s contributed to the river’s current contamination (Xiao et al., 2013; Somasundaram and Radhakrishnan, 2023). For the purposes of this study, the coordinates 13.014434°N latitude and 80.27716°E longitude identify the geographical area of interest around the Adyar Estuary.
Ennore Creek is located on the northern suburbs of Chennai, India, where the Kosasthalaiyar River enters the Bay of Bengal. This estuary region is an important component of the region’s hydrological network. According to Raj (2013), Industrial activities, thermal power plants, petroleum-based facilities, automobiles, ports, oil refineries, fertilizer production, leather processing units, and residential areas, including fishing communities, are among the human activities that contribute to estuary pollution. 17 industries are classified as ultra-red, 5 as red, and 4 as orange and green, according to a 2010 TNPCB report that categorized industries based on pollutant and effluent discharge emissions (TNPCB, 2010). The Ennore estuary receives 3.4 million liters of home sewage, fly ash, and 449,000 liters of industrial effluents every day, according to Environmental Information Systems (2017) (Environmental Information Systems, 2017; Sigamani et al., 2024). For the purposes of this study, the coordinates 13.230064°N latitude and 80.321721°E longitude identify the geographical area of interest near Ennore Creek.
Penna Sangamam is located near Nellore, India where the Penna River enters the Bay of Bengal. This estuary region contains a complex balance of marine ecosystems. With its brackish water, it supports a rich range of marine life. The culture of P. vannamei in India has grown to 100,206 ha by 2017-18, accounting for 622,000 t of production (MPEDA, 2018; Salunke et al., 2020). Andhra Pradesh leads in both P. vannamei production and cultured area, with Nellore being an ecologically and economically important location (Sundara Raja Reddy et al., 2016; MPEDA, 2018; Nadella et al., 2021). However, diseases in P. vannamei culture have led to the use of antibiotics, bactericidal and chemical drugs in shrimp aquaculture (Nadella et al., 2021). This excessive use has led to health problems and concerns about microbial resistance, as per the World Health Organization (WHO, 2016; Nadella et al., 2021). For the purpose of this study, the coordinates 14.567599°N latitude and 80.162421°E longitude define the exact geographical area of interest surrounding Penna Sangamam.
The Pazhayakayal Estuary is located in India’s Tuticorin district, where the Tamirabarani River flows into the Gulf of Mannar. Tuticorin, a heavily industrialized coastal city in India’s Gulf of Mannar region, faces significant metal pollution due to untreated waste, heavy metals from fertilizer and chemical industries, heated effluents from a thermal power station, oil spills from a fishing harbor, and untreated shrimp farm effluents (Rajaram et al., 2021). The city’s harbor activities, including dredging, cargo handling, ship waste disposal, chlorinated hydrocarbons, chemical spills, and metal ores, contribute to the pollution. The intensive energy-generating water source and solid waste with toxic metals pose significant environmental threats through the hydrological cycle (Rajaram et al., 2021). For the purposes of this study, the coordinates 8.658539°N latitude and 78.11311°E longitude define the specific geographical area of interest, Pazhayakayal Estuary. The study areas are showed in Figure 1.
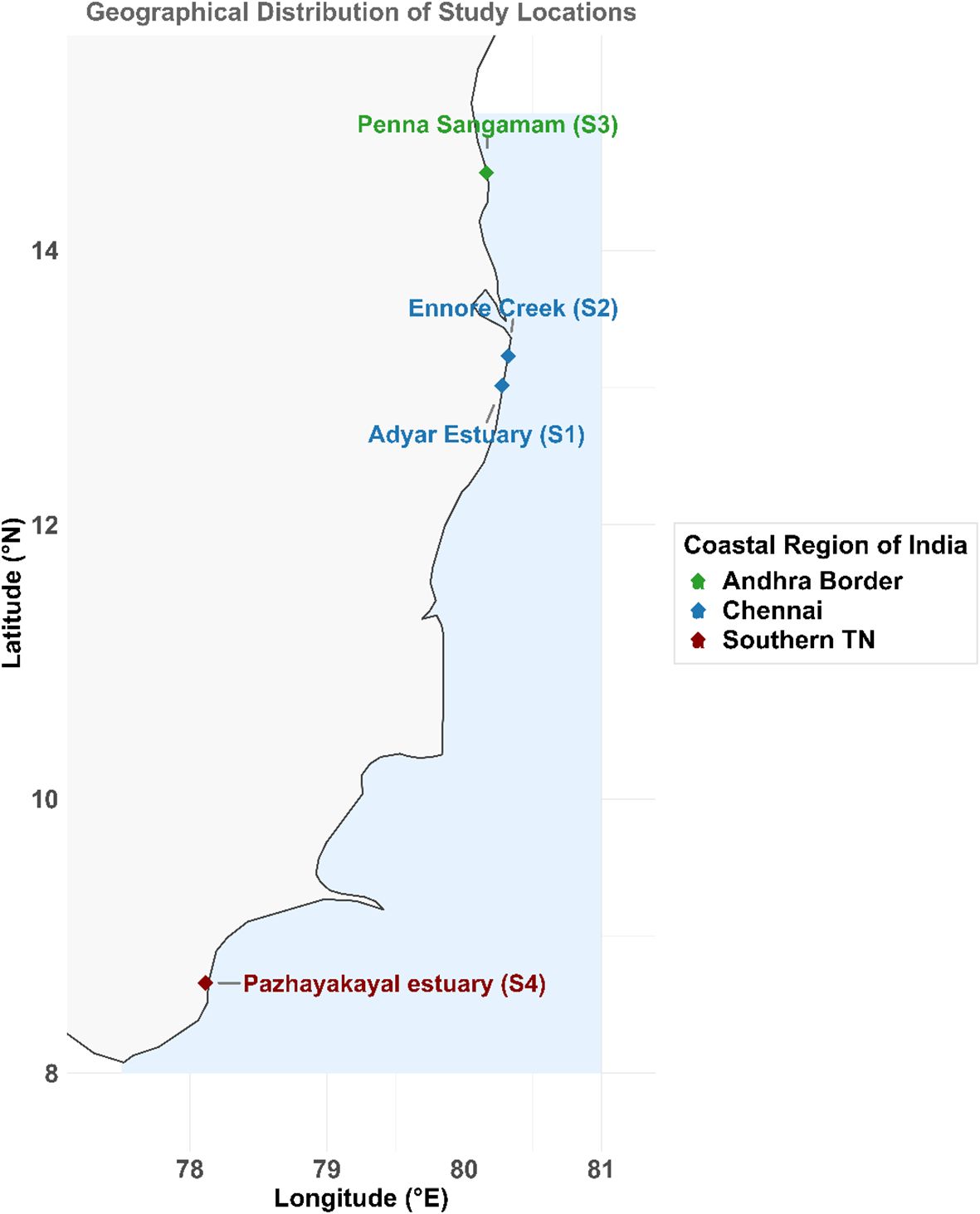
Figure 1. Map of the sample collection sites. The polychaetes were collected from the following four estuaries: Adyar estuary (S1), Ennore Creek (S2), Penna Sangamam (S3) & Pazhayakayal estuary (S4).
2.2 Sample collection and processing
The study collected sediment samples from four estuaries during the low tide phase of the Premonsoon season. The samples were sieved through brass sieves with mesh sizes of 500 mm and 66 mm, and placed in plastic containers. The polychaetes were rinsed five times with sterile water, soaked in 75% ethanol for ~30 seconds to 1 minute for surface sterilization, and then dissectioned to extract the polychaete gut. The surrounding bodily tissues were removed to prevent contamination. Gut samples were taken from beneath the skin and transferred to a 2-ml tube containing phosphate buffer solution. Contaminants were eliminated by washing the samples three times with the phosphate buffer solution. The purified gut contents were stored at -80˚C for DNA extraction. The gut content was isolated using the method described by Song et al., 2022. The detailed isolation protocol is given in Supplementary Section.
2.3 Sample digestion and ICPOES analysis
A 0.5 g of sediment and dried polychaete tissue was placed into Teflon tubes to conduct digestion. The ICPOES grade and concentrated HNO3 was used for digestion in MARS9 Micro digester. The water samples collected were filtered and directly used for ICPOES analysis. The ICPOES analyses were carried out in Sophisticated Analytical Instrument Facility, Indian Institute of Technology, Madras and Environmental Engineering and Environmental Research Laboratory, Thiagarajar College of Engineering, Madurai. The results obtained were corrected with a dilution factor of 5.
2.4 DNA isolation and sequencing
The QIAamp Power fecal DNA Kit was used to extract the DNA from the isolated gut contents. The study used three subsamples from each area. The subsamples extracted DNA was combined and regarded as a single sample that represented a specific location to obtain good quality DNA for Library Preparation and also due to low biomass yield of gut content after washing with PBS. The Qubit dsDNA High sensitivity Assay was used to quantify the sample DNA, and it was then stored at -80˚C until further use. Following the manufacturer’s instructions, the 151 bp fragment sequencing libraries were created and sequenced at MedGenome Labs Ltd. in Bangalore using an Illumina NovaSeq 6000 platform. Whole metagenome sequencing was carried out.
2.5 Sequence pre-processing
The raw paired end reads were checked for initial quality using FASTQC software available in BioBam’s OmicsBox 3.2 platform (https://www.biobam.com/). Following the QC, the adapters were trimmed using cutadapt (V.4.8.) with a Phred quality score 20 and a minimum length filter of 50 bases for the trimmed reads. The alignment to human genome was performed in Omicsbox with database index Homo sapiens (grch38). The denovo assembly and ORF prediction were done using MEGAHIT (version. 1.2.9) and Prodigal (version 2.6.3) respectively.
2.6 Prediction of ARG, BRG, MRG, and MGE
The antibacterial biocide and metal resistance genes database (BacMet, version 2.0) was employed for the predictions of presence of BRG and MRG (Pal et al., 2014). The amino acid sequences of predicted ORFs were used to perform similarity searches against the BacMet database with diamond search in the high sensitive mode (Buchfink et al., 2015). Only BRGs and MRGs with a bit score >50 and a maximum E-value of 10–5 were retained. The Comprehensive Antibiotic Resistance Database (CARD database, version 4.0.2) was employed to detect ARGs in the amino acid sequences of predicted ORFs, which were aligned with the CARD database with diamond as aligner (Jia et al., 2017). Only ARGs with a bit score >50 and a maximum E-value of 10–5 were retained for further analysis. The mobileOG-DB database was utilized to annotate the mobile genetic elements (MGEs), which incorporated 10,776,212 protein sequences from 7 MGE reference databases and provided a core set of MGEs with 6,140 manually validated protein sequences (Brown et al., 2022; Mao et al., 2023). These genes participate in the complete life cycle of MGEs (integration, splicing, replication, recombination, repair, transfer, stability, and defense) (Brown et al., 2022; Mao et al., 2023). The amino acid sequences were assigned using Diamond software, with a maximum E-value of 10–5 and bit score > 50 were used for further analysis.
2.7 Statistical analysis and visualization
All statistical analysis and data sorting in this study were done in R studio (V 4.3.3) and python matplotlib (V 3.11.5). The heavy metal concentrations in water, sediment, and tissue samples were analyzed using one-way ANOVA to assess significant differences among locations. Tukey’s HSD test was performed for post-hoc comparisons, and results were expressed as mean ± SD (letter), where different letters indicate significant differences (p < 0.05). The stacked bar graphs and box plot were plotted using Matplotlib for relative abundance of genes. The PCA analysis was carried using “Vegan” package in R. Venn diagram was plotted using R function “create_venn” in R package “VennDiagram” (Chen and Boutros, 2011). The bar plots with fisher exact test were carried out in R. The resistome risk scores and Pearson correlation coefficient were calculated using Python Programming Language. Weight for each sample and gene categories-ARGs, BMRGs and MGEs were assigned based on their importance derived from a random forest analysis. The resulting weight were used to calculate risk scores with Pearson correlation coefficient. To evaluate resistome risk, we integrated abundance profiles of ARGs, BMRGs, & MGEs across four samples using component-weighted scoring derived from random forest importance metrics. A composite resistome risk score was calculated per sample. Due to the lack of biological replicates and less number of samples, model validation was performed using leave-one-out cross –validation (LOOCV) (Wong, 2015), and bootstrap resampling was used to assess classification robustness. No AUC or MSE metrics were applied. Additionally, pairwise Pearson correlation heat maps were generated for the top 10 co-occurring ARGs, BMRGs, and MGEs per sample, based on shared contig presence. The tools used were Pands library for data manipulation, Scipy and Stats libraries for statistical analysis and Matplotlib and Seaborn libraries for data visualization. Fisher’s exact test was used to test the statistical significance of the number of contigs between groups (R package “stats”). Custom R function “create_co_occurrence” was used to create dataframes for co-occurrence combinations: ARG-BMRG, ARG-MGE, BMRG-MGE, and ARG-BMRG-MGE. The “scipy.stats” library is essential to conduct statistical significance. The Kruskal-Wallis test, which compares multiple groups to determine if they come from specific or same distribution was performed using function “kruskal()” and additionally, the pairwise Mann-Whitney U tests were performed to evaluate the differences between two groups using “mannwhitneyu()” function in Python. Effect sizes were computed using eta-squared (η2) to quantify the proportion of variance explained by group differences. These effect sizes are summarizes in Supplementary Table S1.
3 Results and discussion
3.1 Analysis of metals
The ICPOES analysis was carried out for sediment, water and polychaete tissues. The Table 1 shows the concentration of 8 heavy metals- Cadmium (Cd), Chromium (Cr), Copper (Cu), Iron (Fe), Mercury (Hg), Nickel (Ni), Lead (Pb) and Zinc (Zn). In S1, Zn (35.06 ± 9.269 mg/L) and Cu (10.195 ± 1.791 mg/L) showed higher accumulation in tissue samples, while Fe (4.942 ± 4.9 mg/L) was majorly present in sediment. S2 had significantly higher Hg (5.472 ± 1.051 mg/L) and Ni (0.0038 ± 0.006 mg/L) in water, and Cr (4.157 ± 4.884 mg/L) in sediment. S3 showed comparatively lower metal concentrations across all the matrices, except Fe (23.428 ± 29.327 mg/L) and Zn (2.5 ± 2.5 mg/L) in sediment. S4 showed the highest Hg (13.325 ± 0.837 mg/L), Zn (0.906 ± 0.342 mg/L), and Fe (24.671 ± 22.558 mg/L) in water and sediment. Bioaccumulation of Cu and Zn in the polychaete tissue was notable in S1, while Hg in water from S4 represented higher contamination. ICPOES analysis revealed some interesting facts about the accumulation of 8 heavy metals in sediment, water and polychaete tissues from the four estuaries. The sediment samples possessed high concentration of Zn, Fe, Cr whereas most of the heavy metals are very negligible in water samples. The tissue samples showed high concentration of Zn, Fe and Cr. Metals in sediments can be taken up by deposit-feeding animals, entering the food chain and eventually exerting an adverse effect on human health (Pan and Wang, 2012). The bioavailability of metals associated with contaminated sediment has been a longstanding topic in ecotoxicological study (Bryan and Langston, 1992; Fan et al., 2014). Marphysa madrasi and Namalycastis jaya being deposit feeder’s bio accumulated the heavy metals from the sediment.
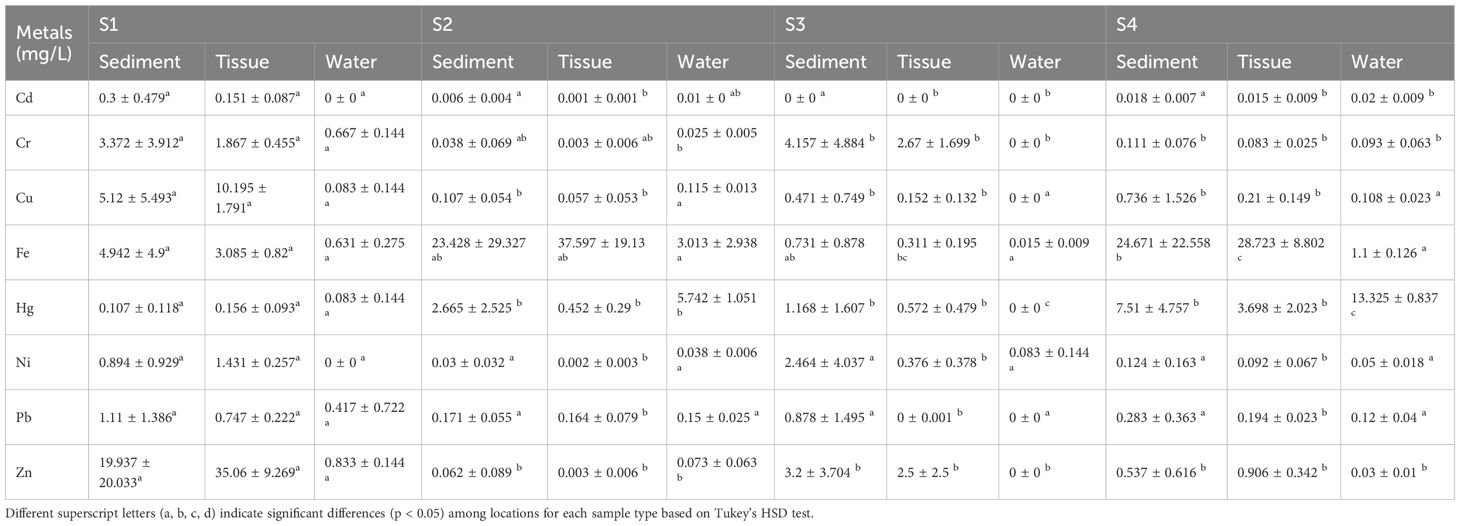
Table 1. Mean ± standard deviation (SD) of heavy metal concentrations (mg/L) in sediment, tissue, and water samples from four sampling sites (S1, S2, S3, S4).
3.2 Profiles of ARGs, BMRGs and MobileOGs/MGEs
Wastewater from an industrial park, which often contained tremendous chemicals such as antimicrobials, heavy metals, surfactants, and disinfectants, might be the “best-case scenario” for AMR development in the environment (Bengtsson-Palme et al., 2019; Ma et al., 2020). The metals and antibiotic residues are transported from their source to estuaries and marine ecosystems through natural and anthropogenic routes. Their accumulation inflicts significant stress on the growth and survival of aquatic organisms and emergence of multidrug and metal resistance in microorganisms. The tolerance threshold for metals and their nanoparticles varies among organisms, hinging on each organism’s oxidative stress response mechanisms and their microbiome (Anas et al., 2008; Biju et al., 2008; Sheeba et al., 2017). Microorganisms, particularly those inhabiting metal-polluted ecosystems, are renowned for developing resistance against metals, with some even demonstrating resilience against extremely high metal concentrations (Jose et al., 2011; Sheeba et al., 2020; Chekidhenkuzhiyil et al., 2024). The current study investigated the profiles of ARGs, BMRGs and MGEs present in the polychaete gut microbiome from four differently polluted estuaries across the Southeast coast of India.
A total of 2054 antibiotic resistant genes belonging to 27 classes of antibiotics were detected in the polychaete gut microbiome S1, S2, S3 and S4 (Figure 2A). 714 genes are detected in S1, 1018 genes in S2, 261 genes in S3 and 2640 genes in S4. The genes belonging to class Cephalosporin were present in higher percentage making upto more than 30% of the total antibiotic classes detected. The second most abundant class of antibiotics resistant genes was Carbapenem in all four samples, followed by Tetracycline antibiotic. Fluoroquinolone antibiotic was the third most abundant class of ARGs present in S1, followed by Peptide antibiotic and Macrolide antibiotic. In S2, the ARGs belonging to the classes like Glycopeptide antibiotic, Cephamycin and Diaminopyrimidine antibiotic were also detected in significant percentage. In S3, Cephamycin, Lincoasamide antibiotic and Peptide antibiotic classes were also detected. In S4, Glycopeptide antibiotic and Cephamycin were the third and fourth most abundance antibiotic classes. The predominance of certain classes of ARGs, such as cephalosporins and carbapenems, in all samples suggests a widespread adaptation of microbial communities to these antibiotics. The identification of these resistance genes indicates that antibiotic resistance is prevalent in the studied estuarine ecosystems. Interestingly, cephalosporin-resistant genes accounted for a significant proportion of the ARGs, underscoring the widespread presence of resistance to beta-lactam antibiotics, which are commonly used in both human and veterinary medicine.
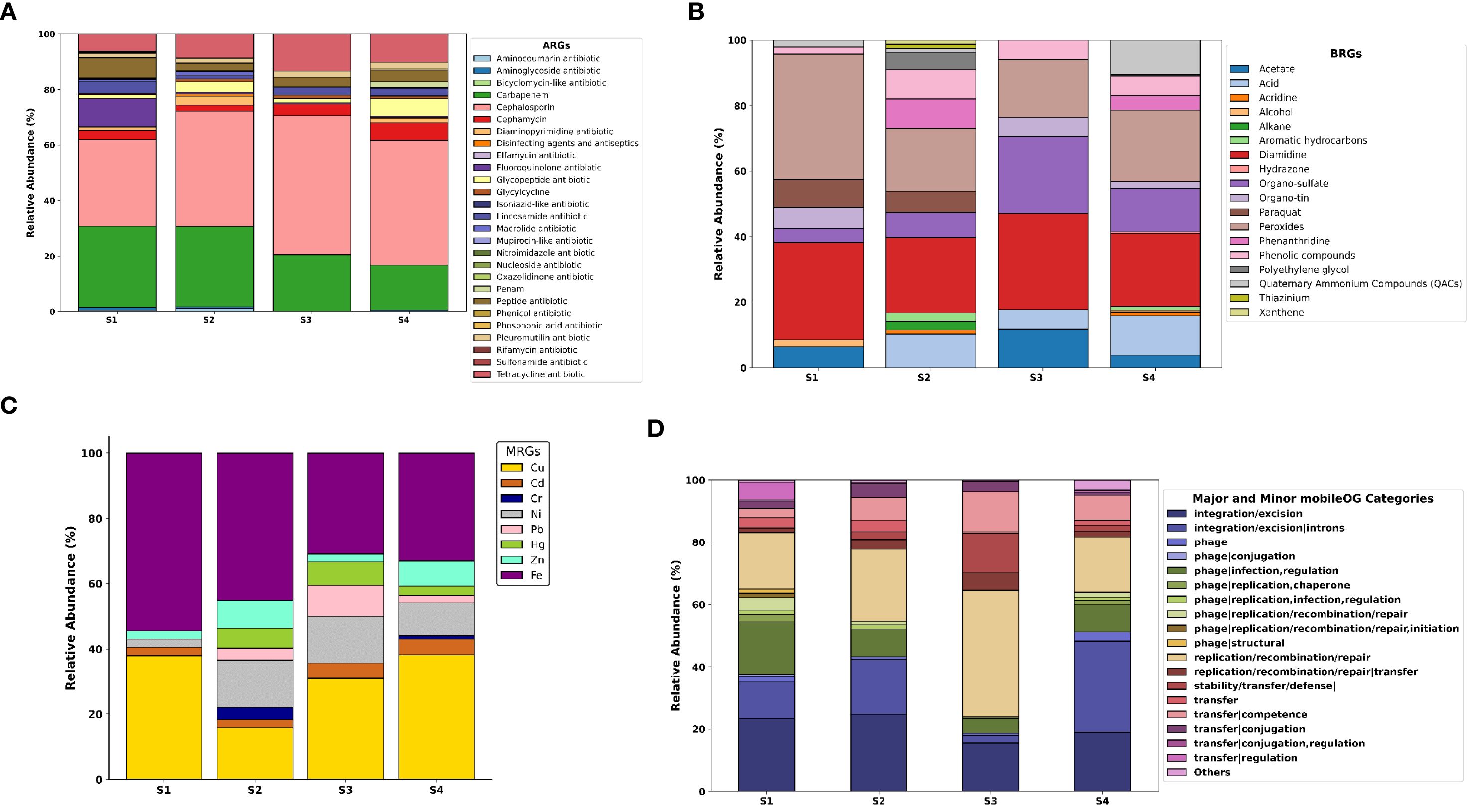
Figure 2. Profiles of MRG and ARG abundances and types in polychaete gut microbiome. (A) types of ARGs annotated and their relative abundance levels in all four gut microbiomes, (B) types of MRGs annotated and their relative abundance levels in all four gut microbiomes, (C) types of MRGs annotated and their relative abundance levels in all four gut microbiomes, (D) types of mobileOGs/MGEs annotated and their relative abundance.
A total of 73 biocidal resistance genes (BRGs) were detected (Figure 2B). Among these, 11 genes were identified in the S1 sample, 44 in S2, 9 in S3, and 48 in S4. BRGs associated with peroxides comprised nearly 30% in S1, followed by diamidines. Additionally, genes related to paraquat, acetate, alcohol, organo-sulfates, organo-tin, phenolic compounds, and quaternary ammonium compounds (QACs) were present in S1. In S2, diamidine- and peroxide-related BRGs were found at higher percentages than others. The sample also contained genes linked to acids, acridine, alkanes, aromatic hydrocarbons, organo-sulfates, paraquat, phenanthridine, phenolic compounds, polyethylene glycol, QACs, thiazinium, and xanthene. In S3, the major BRGs were related to diamidines, organo-sulfates, and peroxides. Additionally, genes associated with acids, organo-tin, and phenolic compounds were detected. In S4, the most prevalent BRGs were linked to diamidines, peroxides, and organo-sulfates. The second most common BRGs were associated with acids, QACs, phenanthridine, and phenolic compounds. The bar chart (Figure 2C) represents the metal resistant genes detected in polychaete gut microbiome related to the 8 heavy metals for which ICPOES analysis was done. The total number of MRGs detected were 78–24 in S1, 33 in S2, 24 in S3 and 70 in S4. In S1, Iron MRGs are the most abundant followed by Copper. Other resistant genes related to metals Zn, Ni and Cd were also detected. In S2, a diverse profile of genes resistant metal Fe was dominant followed by Cu. S2 had a significant amount of MRGs related to Ni and Zn. Other resistant genes for Cd, Cr, Pb, and Hg were also present in S2. In S3, the predominant MRGs were Cu and Fe, followed by Ni and Pb. In S4, Cu and Fe represent the higher percentage of MRGs. Zn and Ni also showed significantly higher percentage of resistant genes. BMRGs, particularly those related to peroxides and diamidines, were also detected in varying concentrations across the samples. These genes are associated with environmental stressors such as biocides used in agriculture and industry, which further contribute to microbial resistance. The detection of these genes highlights the potential for these estuarine ecosystems to harbor microbial communities capable of resisting a wide range of environmental contaminants, including both metals and biocides. The detection of a large number of MGEs, which are involved in the horizontal transfer of genetic material, particularly ARGs and BMRGs, is critical in understanding how resistance traits can spread between microorganisms in the ecosystem.
The results and subsequent analysis confirm our hypothesis that polychaete gut microbiome from Ennore Creek and Pazhayakayal Estuary, exposed to heavy metal and industrial effluent pollution respectively, had elevated presence of ARGs, BMRGs, and MGEs in these two sites. The bar chart (Figure 2D) represents the major and minor mobileOG (MGE) categories detected in the polychaete gut microbiomes. In S1, the genes associated with Integration/Excision are found predominantly, followed by genes associated with phage- infection, regulation and replication/recombination/repair. Other categories of mobileOGs were also detected. Similarly, in S2, the genes associated with Integration/Excision and replication/recombination/repair were present in higher percentages. The other major mobileOG categories present were phage- infection, regulation, transfer- Competence and Conjugation. Other categories were also detected. In S3, about 40% of genes were associated with replication/recombination/repair, followed by integration/excision, stability/transfer/defense, and transfer- competence. In S4, integration/excision-introns were the predominant associated genes present, followed by integration/excision, replication/recombination/repair, phage-infection, regulation and transfer-competence. The prevalence of MGEs, such as those associated with integration/excision, replication/recombination, and transfer-competence, suggests that these elements play a pivotal role in the mobilization of resistance genes across the polychaete gut microbiome. The environment significantly influences the emergence and transmission of Antimicrobial Resistance (AMR) determinants and pathogenic bacteria (Kotwani et al., 2021). The effluent is released into the environment through soil or water, creating reservoirs for antibiotic residues and vectors for resistance genes (Kotwani et al., 2021). Resistance to antibiotics can be disseminated through vertical gene transfer (VGT) and horizontal gene transfer (HGT), which occur between live bacterial cells. Both HGT and VGT coexist in the natural environment (Li et al., 2019). The presence of antimicrobials, heavy metals, biocides, natural chemicals, and xenobiotics, such as solvents like toluene, aids in the selection and propagation of antibiotic-resistant genes (ARGs) (Kotwani et al., 2021). Cross-resistance and co-resistance to these hazards lead to co-selection of genes and the sharing of antibiotic-resistant genes, mobile genetic elements (MGEs), plasmids, and virulence factors with other bacteria (Singer et al., 2016). The potential for horizontal gene transfer through MGEs could facilitate the rapid dissemination of resistance traits, further complicating efforts to control resistance in environmental contexts. These wild polychaetes are widely collected from these estuaries especially from Ennore creek for fishing and exported for shrimp industries across the country and international farms (Babu et al., 2021). Being involved in the food chain, this resistance against Cephalosporins and Carbapenem is more likely carried to the humans. Filter feeders, periphyton grazers, and detritivores ingest ARG-bearing particles and bacteria; experimental work demonstrated bioaccumulation from primary consumers (Daphnia) to fish guts and regular detection of specific ARGs (eg., tetM, tetX, qnrS and sul2) across trophic steps (Wang et al., 2024). Chironomidae larvae enriched tet(A), sul2, and kan from sediments and transferred ARGs to fish gut microbiota via feeding interactions, with conjugated plasmids implicated in spread (Ding et al., 2021). Benthic flatfish and intensive aquaculture species concentrate indicator genes such as sul1, intI1, tet(A), blaTEM, and serve as vectors to human consumers via the seafood supply chain (Lin et al., 2023; Bourdonnais et al., 2024). Although quantitative transfer efficiency across trophic steps (e.g., percent of an ARG pool moving from primary feeder to predator per feeding event) is not consistently reported. The future studies should focus on prevalence, relative abundances, copies per 16S rRNA gene and spatial decay rate of antibiotic genes.
The co-selection of metal and antibiotic resistance in environmental samples may result from anthropogenic influences or from different ecosystems polluted by different metals found in their source mineral ores. Co-selection is not limited to the model of ecology, metals, or the genetic inheritance of microorganisms through resistivity. It might possibly just be the result of innate genetic ability (Sherpa et al., 2020). The co-selection studies provide a clear example of perfect co-selection for both antibiotic and heavy metal resistivity. When particular resistant genes are located at the same loci on transferable genetic elements, such a plasmid, co-resistance happens (Chapman, 2003). They coexist as a result of their genetic connection. Based on their ability to change other non-resistant microbes or give them the capacity to tolerate and resist, ten years’ worth of theories and evidence have demonstrated that metal and antibiotic-resistance genes are phylogenetically related, particularly within mobile genetic plasmids. This has been demonstrated through functional genomics and gene sequencing studies (Novick and Roth, 1968; Foster, 1983). Biofilms are potentially advantageous micro-environments for the selection, co-selection, (Matviichuk et al., 2022) transmission, and spread of resistant genes (RGs) or resistant bacteria due to the persistent coexistence of antibiotics (and most likely other co-selectors), microorganisms (including pathogens), and antibiotic resistance determinants (ARGs, MGEs, etc.).
3.3 Kruskal-Wallis and PCA based statistical evaluation of ARGs, BMRGs and MGEs in polychaete gut microbiomes
The comparison of abundance of the three types of genetic elements (ARGs, BMRGs and MobileOGs/MGEs) were performed and the result is represented as a bar plot in Figure 3. Kruskal-Wallis test was performed to determine the significant differences among the four polychaete gut microbiomes for each genetic element type. A kruskal-wallis test revealed significant intergroup differences for all these gene categories, with overall effect sizes (η2) ranging from small to medium (ARGs: η2 = 0.068, BMRGs: η2 = 0.084, MGEs: η2 = 0.121). Additionally, pairwise Mann-Whitney U test was conducted indicated by asterisks in the plot to determine the specific differences between each gut microbiome pairs. The results showed that S1 and S2 showed highest median abundances in case of ARGs. Whereas S3 and S4 have low median abundances. Meanwhile S4 showed high variability, indicated by the spread of data points. In case of BMRGs, S2 and S4 showed the highest median abundances. Whereas S1 and S3 showed lower median abundance of BMRGs presence. S2 showed the highest variability of BMRGs. For MGEs, S1 showed the highest median abundance, followed by S3. S2 and S4 showed significantly lower median abundances but the high outliers. S1 showed the highest variability. The effect sizes pattern indicates that while differences between some sites are statistically significant, their biological magnitude varies, with medium-strength effects primarily observed in contrast involving S4 and S1. The Supplementary Table S1 gives a detailed explanation of effect sizes.
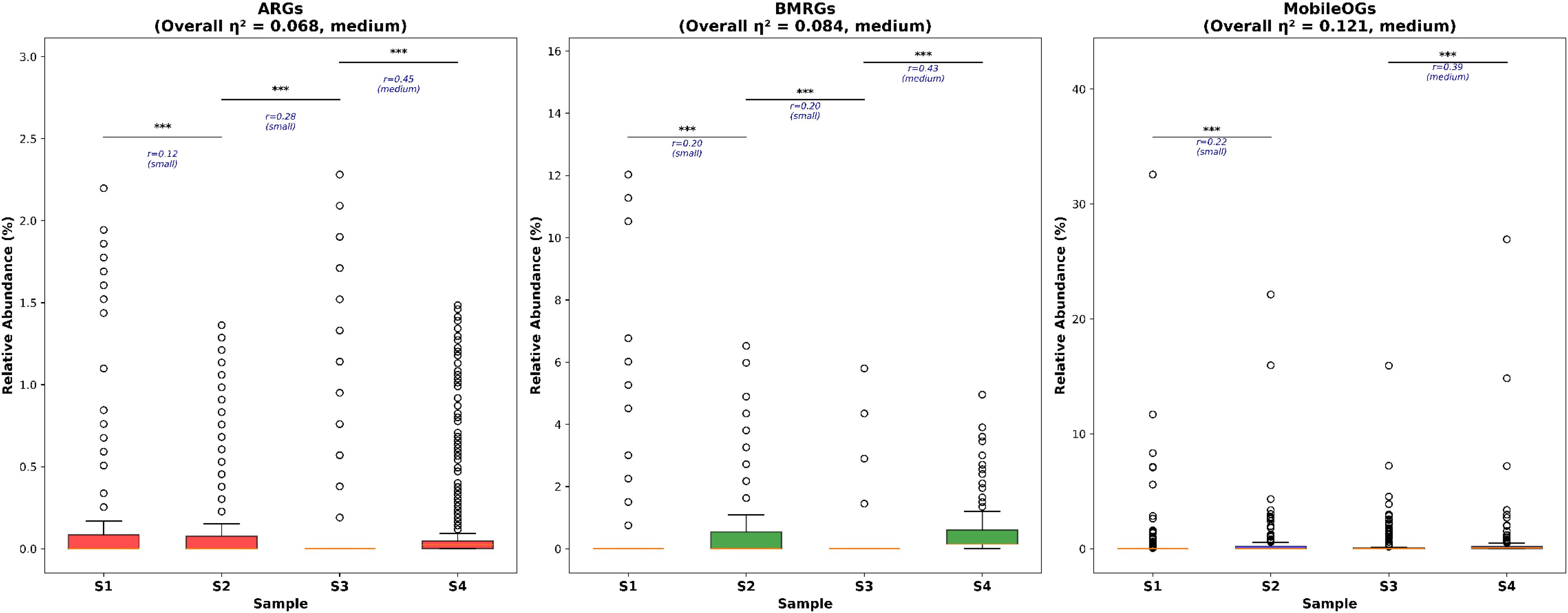
Figure 3. Comparison of total detected numbers of ARGs, BMRGs and MobileOGs in polychaete gut microbiome among different estuaries (Kruskal Wallis test followed by Pairwise Mann-Whitney U test, P < 0.05*, P < 0.01**, P <0.001***, P<0.0001***) Effect sizes (rank-biserial correlation, r) are displayed below significance markers, with interpretations: negligible (r <0.1), small (0.1 ≤ r <0.3), medium (0.3 ≤ r < 0.5), and large (r ≥ 0.5).
The principal component analysis was performed to analyze the patterns of gene abundance in the polychaete gut microbiomes. The resulting plots (Figures 4A–C) display the distinct patterns across ARGs, BMRGs and MGEs. For ARGs, PC1 explains 68.3% of variance. For BMRGs, it was 44.7% variance and for MGEs, PC1 accounted for 62.1%. In ARGs, S1 and S3 were separated along PC1. S2 and S4 were more closely grouped. For BMRGs, S1 and S3 are again separated, S2 and S4 showed more separation compared to ARGs. For MGEs, S1 and S4 were widely separated along PC1 where S2 and S3 were more closely grouped. These findings suggest that the abundance and distribution of these genes are greatly affected by the environmental condition.
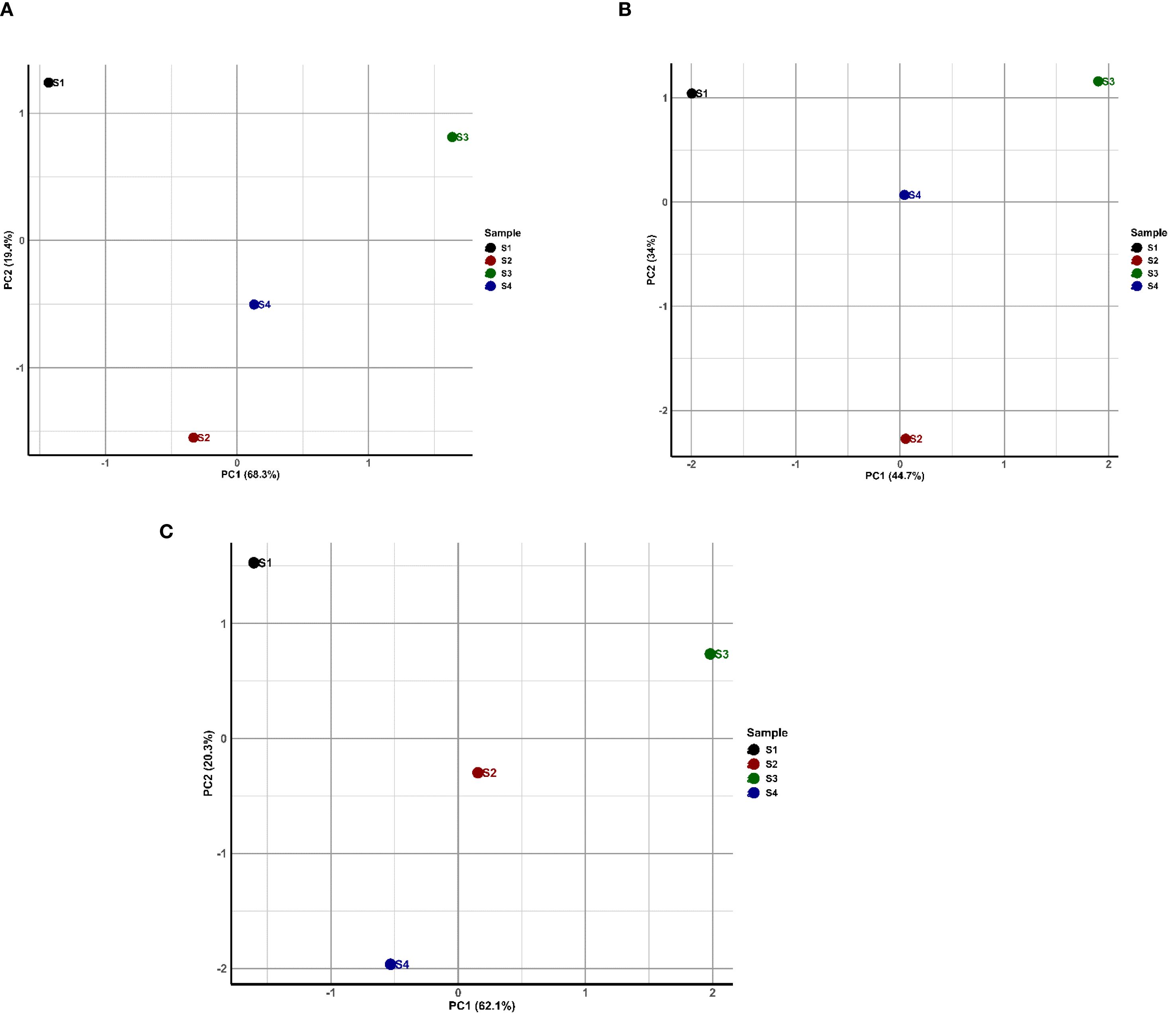
Figure 4. Principal component analysis (PCA) of (A) Antibiotic Resistance Genes (ARGs), (B) Biocide and Metal Resistance Genes (BMRGs), and (C) Mobile Genetic Elements (MGEs) abundance of polychaete gut microbiome. Each plot shows the first two principal components, with the percentage of variance explained by each component indicated on the axes. Data were log-transformed prior to analysis. Points represent individual samples, with their spatial relationships reflecting similarities in gene abundance profiles.
3.4 Co-occurrences of ARGs, BMRGs and MGEs in the assembled contigs
The presence of co-occurrence of resistance genes on contigs were investigated, in the categories of ARGs, BRGs/MRGs, and MGEs to evaluate the potential for co-selection. Figures 5A, B provide an overview of co-occurring ARGs, BMRGs and MGEs. Four combination of co-occurring genes were checked- ARGs+ BMRGs, ARGs+ MGEs, BMRGs+ MGEs and ARGs+ BMRGs+ MGEs. From the Venn diagrams in Figure 5A, we identified a total of 714 ARGs, 133 BMRGs, and 1,754 MGEs in sample S1. Among these, 19 contigs contained co-occurring ARGs and BMRGs, 9 contigs contained BMRGs and MGEs, 13 contigs contained ARGs and MGEs, and 9 contigs contained all three gene types. In sample S2, a total of 1,018 ARGs, 184 BMRGs, and 4,074 MGEs were detected. The co-occurring genes were identified in 19 contigs for ARGs and BMRGs, 16 contigs for BMRGs and MGEs, 18 contigs for ARGs and MGEs, and 11 contigs for all three gene types. For sample S3, we detected 261 ARGs, 69 BMRGs, and 1,676 MGEs. The number of co-occurring contigs was 8 for ARGs and BMRGs, 8 for BMRGs and MGEs, and 8 for ARGs and MGEs. Additionally, 7 contigs contained all three gene types. For S4, 37 contigs have both ARGs and MGEs, 54 contigs have ARGs and BMRGs, 38 contigs have both MGEs and BMRGs and 32 contigs have all three gene types. Figure 5B showed the contigs proportion with the co-occurrence for all four samples. The asterisks represent the P- value of Fisher’s exact test results. Sample S4 had the highest proportion of contigs carrying multiple co-occurrences (Fisher’s exact test, P < 0.0001). The second most predominant co-occurrences were detected in S2, followed by S1 and S3. S3 had the least co-occurrences. In S1, S2 and S4, the co-occurrences of ARGs and BMRGs were high, followed by ARGS and MGEs. The co-occurrences of BMRGs and MGEs are comparatively low compared to other two combinations. The analysis of gene co-occurrence revealed interesting patterns of overlap between ARGs, BMRGs, and MGEs. The co-occurrence of these genes on the same contigs, particularly in S4, suggests the possibility of gene clustering, where resistance traits for antibiotics, biocides, and metals are found together. This phenomenon is a clear example of co-selection, where exposure to one stressor (e.g., heavy metals) may lead to the selection of microorganisms resistant to multiple environmental pressures, including antibiotics and biocides. The high co-occurrence of ARGs and BMRGs in the samples from S1, S2, and S4 further supports the idea of co-selection, where the presence of both types of resistance genes on the same genetic elements might enhance their persistence and spread in microbial communities. Correlation based heat maps detailing the top 10 co-occurring gene pairs for each sample, highlighting their Pearson correlation strengths, are provided in Supplementary Figures S1–S16. These findings highlight the interconnection between metal resistance, antibiotic resistance, and the presence of MGEs. The environmental selection pressure created by heavy metals and antibiotics may lead to the co-selection of resistance genes, making it more difficult to manage microbial resistance in such ecosystems.
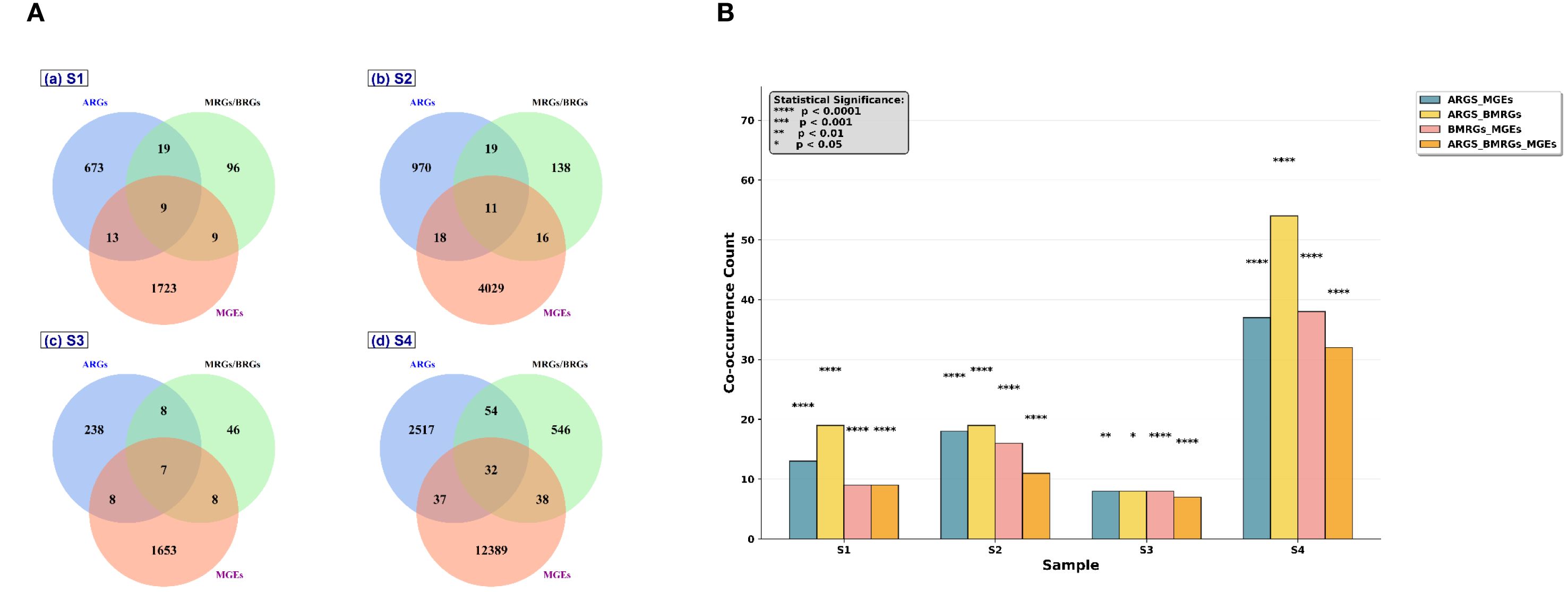
Figure 5. Overview of co-occurrences of ARGs, BRGs, MRGs and MGEs in assembled contigs. (A) Venn diagram showing the number of contigs carrying ARGs, BRGs/MRGs, MGEs and their combinations in polychaete gut microbiome. (B) The proportion of contigs with co-occurrences of ARGs and BRGs/MRGs, ARGs and MGEs, BRGs/MRGs and MGEs, BRGs/MRGs, ARGs and MGEs in polychaete gut microbiome. Asterisks stand for significant statistical difference between groups (Fisher’s exact test; P < 0.05*, P < 0.01**, P <0.001***, P<0.0001****).
3.5 Resistance risk assessment
The resistome risk assessment was done by finding weights of importance using machine learning technique called random forest analysis. The results were shown in Figure 6. The sample S4 showed the highest risk score, followed by S2 and then by S1 and S3. The Figures 6B–D show the comparison of individual gene contributions to the overall resistome risk score in each sample. Pearson correlation coefficients and p-value revealed the contribution of each gene types to risk score. ARGs vs Resistome Risk Score (RSS) (Figure 6B) revealed that ARGs showed very weak correlation (R = 0.02). Meanwhile BMRGs (Figure 6C) showed weak to moderate positive correlation (R = 0.27). MGEs vs RSS showed very strong positive correlation (R = 0.96). The resistance risk score and MGEs showed a strong positive relation, according to correlation analysis, indicating that MGEs were vital in promoting environmental resistome risk. The resistome risk assessment using random forest analysis provided an important understanding of the relative risk posed by each estuary. Sample S4 exhibited the highest risk score, followed by S2, indicating that these sites harbor microbiomes with a greater potential to disseminate resistance. The positive correlation between the abundance of MGEs and the resistome risk score (R = 0.96) suggests that MGEs are a major driver in the spread of resistance traits across microbial populations in these ecosystems. The model validation performed to account for the small dataset and absence of biological replicates. Leave-one-out cross-validation (LOOCV) and bootstrap resampling confirmed the robustness of the model achieving perfect classification accuracy (100%) and a bootstrap mean of 1.0 (Figure 6E, G). Additionally, the Explained Variance of Risk (EVR) separation score (1.44) indicated strong differentiation between high- and low-risk sites, ensuring that the observed risk rankings were not artefacts of overfitting. Overall, the integration of random forest-derived weights, gene abundance data, and robust validation demonstrates that industrial effluent-impacted estuaries (S4) post the highest risk resistome risk, followed by heavy metal-polluted estuaries (S2), with domestic and aquaculture-impacted estuaries (S1 and S3) showing comparatively lower risk. A similar study on Waste Water Treatment Plants (WWTPs) around Hangzhou Bay (Su et al., 2023) reported that Industrial effluents pose a substantial risk to marine environments and public health. The study calls for improved wastewater treatment processes and ecological considerations in industrial effluent management.
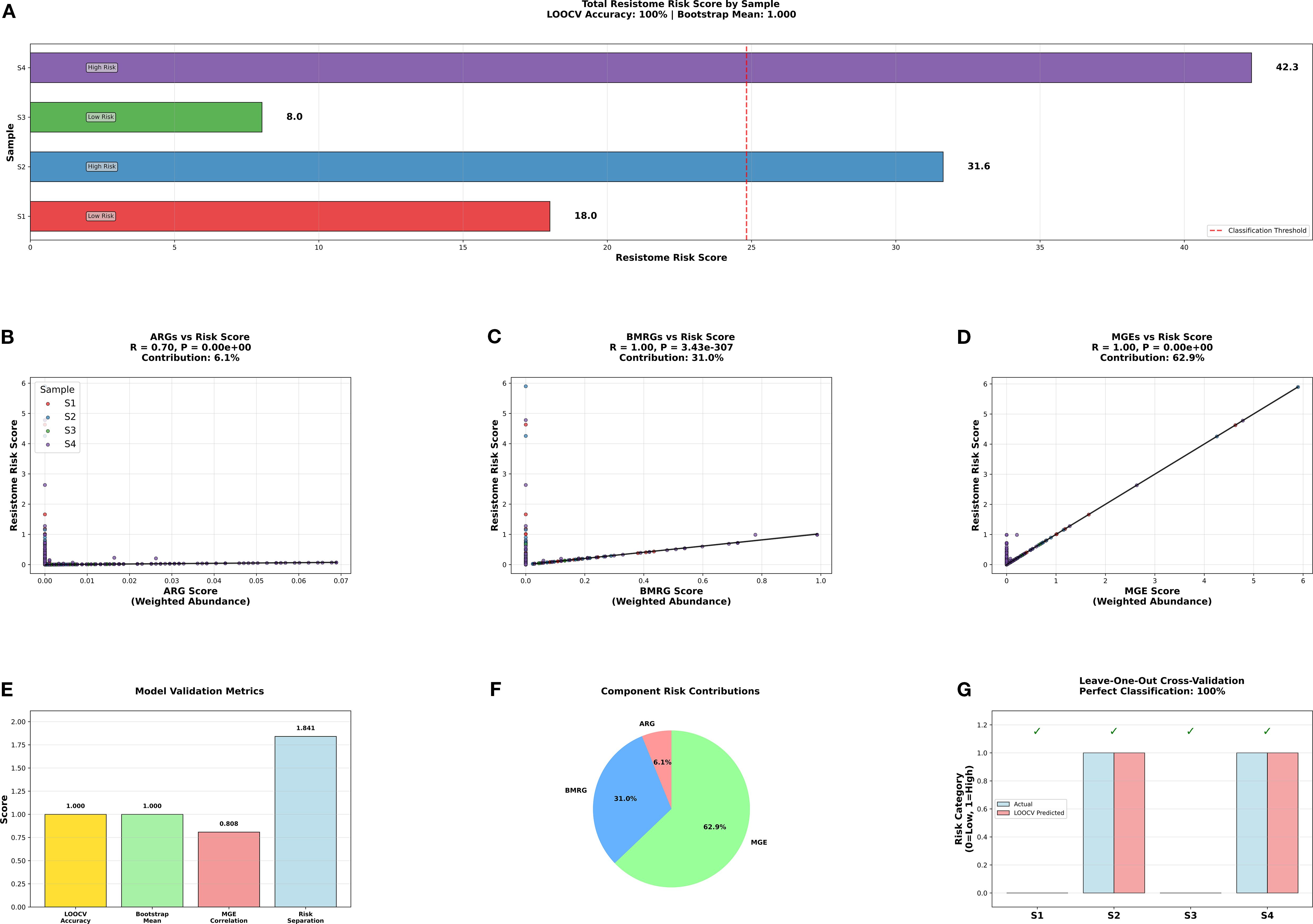
Figure 6. Resistance risk assessment and its association with ARGs, BMRGs, and MGEs. The resistome risk score in each sample (A–D) shows the comparison of individual gene contributions to the overall resistome risk score in each sample. (E) shows the Model Validation Metrics- LOOCV accuracy 91.0), boostrap Value (1.0), MGEs correlation, and EVR separation score (1.44). (F) shows the contributions of each egen categories to rresistome risk. (G) shows LOOCV classification. (LOOCV- Leave-one-out cross-validation, EVR- Explained Variance of Risk Separation).
This also reinforces the role of MGEs in facilitating the horizontal transfer of resistance genes, which could contribute to the persistence and spread of antibiotic and metal resistance in estuarine environments. Compared to the domestic and aquaculture polluted estuaries, the risk score is high in industrial effluents polluted estuary, followed by heavy metal polluted estuary. This calls for a stringent effluent discharge management.
4 Conclusion
Our comprehensive study on polychaete gut microbiomes from four Southeast coast Indian estuaries representing four different types of pollution namely- Domestic Pollution, Heavy Metal Pollution, Aquaculture waste and Industrial Effluents, reveals an alarming connection between heavy metal and industrial pollution and spread of AMR. Over 2,000 ARGs, particularly against cephalosporins and carbapenems which are the most widely used antibiotics in human and veterinary health management is a serious concern. The small number of samples (n=4) and absence of biological replicates limit statistical power and the generalizability of the findings, despite validation with LOOCV and Bootstrap resampling. The study lacks investigation into the presence of ARGs, BMRGs and MGEs across different seasons. The strong co-occurrence patterns between antibiotic, metal, and biocide resistance (R = 0.96), demonstrates how industrially polluted estuaries play a significant role in antimicrobial resistance evolution and the presence of high number of MGEs leads to transfer of these resistant genes. Formation of biofilms among bacteria can lead to enhanced co-occurrence and cross-resistance of Resistant Genes. Comparatively the estuaries with domestic and aquaculture pollution are not a major concern in AMR development. The effluent release into these estuaries should be managed strictly and effectively to stop the further development of AMR in the future. The future study should focus on trophic transfer of ARGs with these wild polychaetes used as primary consumer in food chain.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1164039.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
MG: Formal analysis, Writing – original draft, Writing – review & editing, Software, Methodology, Data curation, Validation, Conceptualization, Resources. RM: Methodology, Visualization, Project administration, Validation, Conceptualization, Writing – original draft, Supervision, Funding acquisition, Writing – review & editing, Investigation, Resources. SS: Writing – review & editing, Data curation, Software, Writing – original draft, Resources. SC: Writing – review & editing, Writing – original draft. KR: Writing – review & editing, Writing – original draft. RRM: Writing – review & editing, Writing – original draft, Methodology, Funding acquisition, Investigation, Conceptualization, Validation. BR: Validation, Conceptualization, Methodology, Supervision, Writing – review & editing, Funding acquisition, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The authors are grateful to the Management of Sathyabama Institute of Science and Technology for providing the necessary facilities to carry out the research work. We are thankful for the support provided by UCSI University for the support provided by the Center of Excellence for Research, Value Innovation and Entrepreneurship (CERVIE) and Research Excellence and Innovation Grant (REIG) with code REIG-FPS-2025/038.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1637421/full#supplementary-material
References
Amiard-Triquet C., Mouneyrac C., and Berthet B. (2013). “Polychaetes in ecotoxicology,” in Encyclopedia of Aquatic Ecotoxicology (Springer Netherlands, Dordrecht), 893–908. doi: 10.1007/978-94-007-5704-2_82
Anas A., Akita H., Harashima H., Itoh T., Ishikawa M., and Biju V. (2008). Photosensitized breakage and damage of DNA by cdSe–ZnS quantum dots. J. Phys. Chem. B 112, 10005–10011. doi: 10.1021/jp8018606
Andersson D. I. and Hughes D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478. doi: 10.1038/nrmicro3270
Ayukekbong J. A., Ntemgwa M., and Atabe A. N. (2017). The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob. Resist. Infect. Control 6, 47. doi: 10.1186/s13756-017-0208-x
Babu B., Sathiyaraj G., Mandal A., Kandan S., Biju N., Palanisamy S., et al. (2021). Surveillance of disease incidence in shrimp farms located in the east coastal region of India and in vitro antibacterial efficacy of probiotics against Vibrio parahaemolyticus. J. Invertebr Pathol. 179, 107536. doi: 10.1016/j.jip.2021.107536
Baker-Austin C., Wright M. S., Stepanauskas R., and McArthur J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182. doi: 10.1016/j.tim.2006.02.006
Balakrishna K., Rath A., Praveenkumarreddy Y., Guruge K. S., and Subedi B. (2017). A review of the occurrence of pharmaceuticals and personal care products in Indian water bodies. Ecotoxicol Environ. Saf. 137, 113–120. doi: 10.1016/j.ecoenv.2016.11.014
Bat L. and Üniversitesi S. (2005). A review of sediment toxicity bioassays using the amphipods and polychaetes. Available online at: https://www.researchgate.net/publication/284107581 (Accessed February 16, 2024).
Bengtsson-Palme J., Milakovic M., Švecová H., Ganjto M., Jonsson V., Grabic R., et al. (2019). Industrial wastewater treatment plant enriches antibiotic resistance genes and alters the structure of microbial communities. Water Res. 162, 437–445. doi: 10.1016/j.watres.2019.06.073
Biju V., Itoh T., Anas A., Sujith A., and Ishikawa M. (2008). Semiconductor quantum dots and metal nanoparticles: syntheses, optical properties, and biological applications. Anal. Bioanal Chem. 391, 2469–2495. doi: 10.1007/s00216-008-2185-7
Bouraoui Z., Ghedira J., Capri F., Chouba L., and Boussetta H. (2014). Cytochemical responses of Hediste diversicolor (Nereidae, Polychaete) sampled from polluted sites along the Tunisian coast. Rev. Gestão Costeira Integrada 14, 119–127. doi: 10.5894/rgci480
Bourdonnais E., Le Bris C., Brauge T., and Midelet G. (2024). Tracking antimicrobial resistance indicator genes in wild flatfish from the English Channel and the North Sea area: A one health concern. Environ. pollut. 343, 123274. doi: 10.1016/j.envpol.2023.123274
Brown C. L., Mullet J., Hindi F., Stoll J. E., Gupta S., Choi M., et al. (2022). mobileOG-db: a manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl. Environ. Microbiol. 88 (18), e0099122. doi: 10.1128/aem.00991-22
Bryan G. W. and Langston W. J. (1992). Bioavailability, accumulation and effects of heavy metals in sediments with special reference to United Kingdom estuaries: a review. Environ. pollut. 76, 89–131. doi: 10.1016/0269-7491(92)90099-V
Buchfink B., Xie C., and Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Buffet-Bataillon S., Tattevin P., Bonnaure-Mallet M., and Jolivet-Gougeon A. (2012). Emergence of resistance to antibacterial agents: the role of quaternary ammonium compounds—a critical review. Int. J. Antimicrob. Agents 39, 381–389. doi: 10.1016/j.ijantimicag.2012.01.011
Chapman J. S. (2003). Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior Biodegradation 51, 271–276. doi: 10.1016/S0964-8305(03)00044-1
Chekidhenkuzhiyil J., Chandran S., Kaliyath D. R., Sukumaran V., Raju G. K. T., and Abdulaziz A. (2024). Influence of cadmium and zinc contamination on the sediment microbiome of estuarine and coastal ecosystems in the Southwest Coast of India. Environ. Sci. pollut. Res. 31, 54684–54694. doi: 10.1007/s11356-024-34851-0
Chen H. and Boutros P. C. (2011). VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf. 12, 35. doi: 10.1186/1471-2105-12-35
Da C., Liu G., and Yuan Z. (2014). Analysis of HCHs and DDTs in a sediment core from the Old Yellow River Estuary, China. Ecotoxicol Environ. Saf. 100, 171–177. doi: 10.1016/j.ecoenv.2013.10.034
Dafforn K. A., Kelaher B. P., Simpson S. L., Coleman M. A., Hutchings P. A., Clark G. F., et al. (2013). Polychaete richness and abundance enhanced in anthropogenically modified estuaries despite high concentrations of toxic contaminants. PloS One 8 (9), e77018. doi: 10.1371/journal.pone.0077018
Das M. K., Das S., and Srivastava P. K. (2023). An overview on the prevalence and potential impact of antimicrobials and antimicrobial resistance in the aquatic environment of India. Environ. Monit Assess. 195. doi: 10.1007/s10661-023-11569-z
Das M. K., Naskar M., Mondal M. L., Srivastava P. K., Dey S., and Rej A. (2012). Influence of ecological factors on the patterns of fish species richness in tropical Indian rivers. Acta Ichthyol Piscat 42, 47–58. doi: 10.3750/AIP2011.42.1.06
Díaz-Castañeda V. and Reish D. J. (2009). “Polychaetes in environmental studies,” in Shain D. H. (Ed.), Annelids in Modern Biology (Hoboken: Wiley-Blackwell), 203–227. doi: 10.1002/9780470455203.ch11
Ding C., Ma J., Jiang W., Zhao H., Shi M., Cui G., et al. (2021). Chironomidae larvae: A neglected enricher of antibiotic resistance genes in the food chain of freshwater environments. Environ. pollut. (Barking, Essex: 1987) 285, 117486. doi: 10.1016/j.envpol.2021.117486
Drudge C. N., Elliott A. V. C., Plach J. M., Ejim L. J., Wright G. D., Droppo I. G., et al. (2012). Diversity of integron- and culture-associated antibiotic resistance genes in freshwater floc. Appl. Environ. Microbiol. 78, 4367–4372. doi: 10.1128/AEM.00405-12
EC Com (2017). A European One Health Action Plan against Antimicrobial Resistance (AMR) CONTENTS. Available online at: https://health.ec.europa.eu/system/files/2020- 01/amr_ 2017_ actionplan_0.pdf (Accessed February 16, 2024).
Fan W., Xu Z., and Wang W.-X. (2014). Metal pollution in a contaminated bay: Relationship between metal geochemical fractionation in sediments and accumulation in a polychaete. Environ. pollut. 191, 50–57. doi: 10.1016/j.envpol.2014.04.014
Foster T. J. (1983). Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol. Rev. 47, 361–409. doi: 10.1128/mr.47.3.361-409.1983
Gothwal R. and Shashidhar (2017). Occurrence of high levels of fluoroquinolones in aquatic environment due to effluent discharges from bulk drug manufacturers. J. Hazard Toxic Radioact Waste 21. doi: 10.1061/(ASCE)HZ.2153-5515.0000346
Ho J. Y., Jong M.-C., Acharya K., Liew S. S. X., Smith D. R., Noor Z. Z., et al. (2021). Multidrug-resistant bacteria and microbial communities in a river estuary with fragmented suburban waste management. J. Hazard Mater 405, 124687. doi: 10.1016/j.jhazmat.2020.124687
Hutchings P. (1998). Biodiversity and functioning of polychaetes in benthic sediments. Biodivers Conserv. 7, 1133–1145. doi: 10.1023/A:1008871430178
Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., Tsang K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jose J., Giridhar R., Anas A., Loka Bharathi P. A., and Nair S. (2011). Heavy metal pollution exerts reduction/adaptation in the diversity and enzyme expression profile of heterotrophic bacteria in Cochin estuary, India. Environ. pollut. 159, 2775–2780. doi: 10.1016/j.envpol.2011.05.009
Kauppi L., Bernard G., Bastrop R., Norkko A., and Norkko J. (2018). Increasing densities of an invasive polychaete enhance bioturbation with variable effects on solute fluxes. Sci. Rep. 8, 7619. doi: 10.1038/s41598-018-25989-2
Knapp C. W., Callan A. C., Aitken B., Shearn R., Koenders A., and Hinwood A. (2017). Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environ. Sci. pollut. Res. 24, 2484–2494. doi: 10.1007/s11356-016-7997-y
Knapp C. W., Dolfing J., Ehlert P. A. I., and Graham D. W. (2010). Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 44, 580–587. doi: 10.1021/es901221x
Kotwani A., Joshi J., and Kaloni D. (2021). Pharmaceutical effluent: a critical link in the interconnected ecosystem promoting antimicrobial resistance. Environmental science and pollution research international. 28 (25), 32111–32124. doi: 10.1007/s11356-021-14178-w
Li B., Qiu Y., Song Y., Lin H., and Yin H. (2019). Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ. Int. 131, 105007. doi: 10.1016/j.envint.2019.105007
Li X., Rensing C., Vestergaard G., Arumugam M., Nesme J., Gupta S., et al. (2022). Metagenomic evidence for co-occurrence of antibiotic, biocide and metal resistance genes in pigs. Environ. Int. 158. doi: 10.1016/j.envint.2021.106899
Lin X., Tan A., Deng Y., Liu W., Zhao F., and Huang Z. (2023). High occurrence of antibiotic resistance genes in intensive aquaculture of hybrid snakehead fish. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.1088176
Liu C., Li G., Qin X., Xu Y., Wang J., Wu G., et al. (2022). Profiles of antibiotic- and heavy metal-related resistance genes in animal manure revealed using a metagenomic analysis. Ecotoxicol Environ. Saf. 239. doi: 10.1016/j.ecoenv.2022.113655
Liu J., Shen Z., and Chen L. (2018). Assessing how spatial variations of land use pattern affect water quality across a typical urbanized watershed in Beijing, China. Landsc Urban Plan 176, 51–63. doi: 10.1016/j.landurbplan.2018.04.006
Lopatkin A. J., Huang S., Smith R. P., Srimani J. K., Sysoeva T. A., Bewick S., et al. (2016). Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 1, 16044. doi: 10.1038/nmicrobiol.2016.44
López E. and Richter A. (2017). Non-indigenous species (NIS) of polychaetes (Annelida: Polychaeta) from the Atlantic and Mediterranean coasts of the Iberian Peninsula: an annotated checklist. Helgol Mar. Res. 71, 19. doi: 10.1186/s10152-017-0499-6
Lotze H. K., Lenihan H. S., Bourque B. J., Bradbury R. H., Cooke R. G., Kay M. C., et al. (2006). Depletion, degradation, and recovery potential of estuaries and coastal seas. Sci. (1979) 312, 1806–1809. doi: 10.1126/science.1128035
Ma B., Tian G., and Kong L. (2020). Spatial-temporal characteristics of China’s industrial wastewater discharge at different scales. Environ. Sci. pollut. Res. 27, 8103–8118. doi: 10.1007/s11356-019-07488-7
Mao C., Li Q., Komijani M., Huang J., and Li T. (2023). Metagenomic analysis reveals the dissemination mechanisms and risks of resistance genes in plateau lakes. iScience 26. doi: 10.1016/j.isci.2023.107508
Matviichuk O., Mondamert L., Geffroy C., Gaschet M., Dagot C., and Labanowski J. (2022). River biofilms microbiome and resistome responses to wastewater treatment plant effluents containing antibiotics. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.795206
McKinney C. W. and Pruden A. (2012). Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ. Sci. Technol. 46, 13393–13400. doi: 10.1021/es303652q
Mdaini Z., Telahigue K., Hajji T., Rabeh I., El Cafsi M., Tremblay R., et al. (2021). Comparative biomarker responses to urban pollution in three polychaete species: Perinereis cultrifera, Diopatra neapolitana, and Marphysa sanGuinea from the lagoon of Tunis. Environ. Monit Assess. 193. doi: 10.1007/s10661-021-08906-5
Menon N. G., Mohapatra S., Padhye L. P., Tatiparti S. S. V., and Mukherji S. (2020). Review on occurrence and toxicity of pharmaceutical contamination in Southeast Asia 63–91. doi: 10.1007/978-981-32-9771-5_4
MPEDA (2018). Annual Report 2017-18 (Cochin, India). Available online at: https://mpeda.gov.in/wp-content/uploads/2020/11/1550120514MPEDAAR201718.pdf.
Mukhopadhyay M., Sampath S., Muñoz-Arnanz J., Jiménez B., and Chakraborty P. (2020). Plasticizers and bisphenol A in Adyar and Cooum riverine sediments, India: occurrences, sources and risk assessment. Environ. Geochem Health 42, 2789–2802. doi: 10.1007/s10653-020-00516-3
Mutiyar P. K. and Mittal A. K. (2014). Occurrences and fate of selected human antibiotics in influents and effluents of sewage treatment plant and effluent-receiving river Yamuna in Delhi (India). Environ. Monit Assess. 186, 541–557. doi: 10.1007/s10661-013-3398-6
Nadella R. K., Panda S. K., MadhuSudana Rao B., Pani Prasad K., Raman R. P., and Mothadaka M. P. (2021). Antibiotic resistance of culturable heterotrophic bacteria isolated from shrimp (Penaeus vannamei) aquaculture ponds. Mar. pollut. Bull. 172. doi: 10.1016/j.marpolbul.2021.112887
Novick R. P. and Roth C. (1968). Plasmid-linked resistance to inorganic salts in staphylococcus aureus. J. Bacteriol 95, 1335–1342. doi: 10.1128/jb.95.4.1335-1342.1968
Olsgard F., Brattegard T., and Holthe T. (2003). Polychaetes as surrogates for marine biodiversity: Lower taxonomic resolution and indicator groups. Biodivers Conserv. 12, 1033–1049. doi: 10.1023/A:1022800405253
Ortega Morente E., Fernández-Fuentes M. A., Grande Burgos M. J., Abriouel H., Pérez Pulido R., and Gálvez A. (2013). Biocide tolerance in bacteria. Int. J. Food Microbiol. 162, 13–25. doi: 10.1016/j.ijfoodmicro.2012.12.028
Pal C., Bengtsson-Palme J., Rensing C., Kristiansson E., and Larsson D. G. J. (2014). BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 42, D737–D743. doi: 10.1093/nar/gkt1252
Pamungkas J., Glasby C. J., Read G. B., Wilson S. P., and Costello M. J. (2019). Progress and perspectives in the discovery of polychaete worms (Annelida) of the world. Helgol Mar. Res. 73, 4. doi: 10.1186/s10152-019-0524-z
Pan K. and Wang W.-X. (2012). Trace metal contamination in estuarine and coastal environments in China. Sci. Total Environ. 421–422, 3–16. doi: 10.1016/j.scitotenv.2011.03.013
Pimentel D., Cooperstein S., Randell H., Filiberto D., Sorrentino S., Kaye B., et al. (2007). Ecology of increasing diseases: population growth and environmental degradation. Hum. Ecol. 35, 653–668. doi: 10.1007/s10745-007-9128-3
Pruden A., Pei R., Storteboom H., and Carlson K. H. (2006). Antibiotic resistance genes as emerging contaminants: studies in Northern Colorado. Environ. Sci. Technol. 40, 7445–7450. doi: 10.1021/es060413l
Raj M. V. (2013).Water quality Parameters and it influences in the Ennore estuary and near Coastal Environment with respect to Industrial and Domestic sewage. Available online at: www.isca.in (Accessed February 16, 2024).
Rajaram R., Ganeshkumar A., Vinothkumar S., and Arun G. (2021). Ecological risk assessment of toxic metals contamination in Tuticorin coast of Gulf of Mannar, Southern India. Chem. Ecol. 37, 132–148. doi: 10.1080/02757540.2020.1819986
Rodgers K., McLellan I., Peshkur T., Williams R., Tonner R., Hursthouse A. S., et al. (2019). Can the legacy of industrial pollution influence antimicrobial resistance in estuarine sediments? Environ. Chem. Lett. 17, 595–607. doi: 10.1007/s10311-018-0791-y
Salunke M., Kalyankar A., Khedkar C. D., Shingare M., and Khedkar G. D. (2020). A review on shrimp aquaculture in India: historical perspective, constraints, status and future implications for impacts on aquatic ecosystem and biodiversity. Rev. Fisheries Sci. Aquaculture 28, 283–302. doi: 10.1080/23308249.2020.1723058
SCENIHR (Scientific Committee on Emerging and Newly Identified Health Risks) (2009). Assessment of the Antibiotic Resistance Effects of Biocides. Available online at: https://ec.europa.eu/health/ph_risk/committees/04_scenihr/docs/scenihr_o_021.pdf (Accessed February 17, 2024).
Segawa T., Takeuchi N., Rivera A., Yamada A., Yoshimura Y., Barcaza G., et al. (2013). Distribution of antibiotic resistance genes in glacier environments. Environ. Microbiol. Rep. 5, 127–134. doi: 10.1111/1758-2229.12011
Seiler C. and Berendonk T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00399
Sheeba V. A., Abdulaziz A., Gireeshkumar T. R., Ram A., Rakesh P. S., Jasmin C., et al. (2017). Role of heavy metals in structuring the microbial community associated with particulate matter in a tropical estuary. Environ. pollut. 231, 589–600. doi: 10.1016/j.envpol.2017.08.053
Sheeba V. A., Anas A., Jasmin C., Vincent M., and Parameswaran P. S. (2020). Response of particle-associated bacteria to long-term heavy metal contamination in a tropical estuary. World J. Microbiol. Biotechnol. 36, 65. doi: 10.1007/s11274-020-02842-1
Shen Q., Tang J., Wang X., Li Y., Yao X., Sun H., et al. (2021). Fate of antibiotic resistance genes and metal resistance genes during the thermophilic fermentation of solid and liquid swine manures in an ectopic fermentation system. Ecotoxicol Environ. Saf. 213, 111981. doi: 10.1016/j.ecoenv.2021.111981
Sherpa M. T., Najar I. N., Das S., and Thakur N. (2020). Distribution of antibiotic and metal resistance genes in two glaciers of North Sikkim, India. Ecotoxicol Environ. Saf. 203, 111037. doi: 10.1016/j.ecoenv.2020.111037
Sigamani S., J.A. D., Dony D. M., S. S., U. B., Kolandhasamy P., et al. (2024). Bioaccumulation and health risk of metal contamination from different tiers of food chain in Ennore estuary, Southeast coast of India. Mar. pollut. Bull. 200. doi: 10.1016/j.marpolbul.2024.116154
Singer A. C., Shaw H., Rhodes V., and Hart A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01728
Somasundaram R. and Radhakrishnan N. (2023). Establishment the relationship between water quality parameter and micro plastic concentration for Adyar and Cooum estuary. Environ. Qual. Manage. 33, 121–133. doi: 10.1002/tqem.22028
Song J., Li T., Zheng Z., Fu W., Long Z., Shi N., et al. (2022). Carbendazim shapes microbiome and enhances resistome in the earthworm gut. Microbiome 10. doi: 10.1186/s40168-022-01261-8
Stepanauskas R., Glenn T. C., Jagoe C. H., Tuckfield R. C., Lindell A. H., and McArthur J. V. (2005). Elevated microbial tolerance to metals and antibiotics in metal-contaminated industrial environments. Environ. Sci. Technol. 39, 3671–3678. doi: 10.1021/es048468f
Su Z., Wen D., Gu A. Z., Zheng Y., Tang Y., and Chen L. (2023). Industrial effluents boosted antibiotic resistome risk in coastal environments. Environ. Int. 171, 107714. doi: 10.1016/j.envint.2022.107714
Sundara Raja Reddy B. C., Jayaraju N., Sreenivasulu G., Suresh U., and Reddy A. N. (2016). Heavy metal pollution monitoring with foraminifera in the estuaries of Nellore coast, East coast of India. Mar. pollut. Bull. 113, 542–551. doi: 10.1016/j.marpolbul.2016.08.051
Tandukar M., Oh S., Tezel U., Konstantinidis K. T., and Pavlostathis S. G. (2013). Long-term exposure to benzalkonium chloride disinfectants results in change of microbial community structure and increased antimicrobial resistance. Environ. Sci. Technol. 47, 9730–9738. doi: 10.1021/es401507k
Taneja N. and Sharma M. (2019). Antimicrobial resistance in the environment: The Indian scenario. Indian J. Med. Res. 149, 119. doi: 10.4103/ijmr.IJMR_331_18
TNPCB (2010). “Action plan for critically polluted area,” in Manal. Available online at: https://tnpcb.gov.in/pdf/Action_plan_Manali12092016.pdf.
Wang C., Song Y., Liang J., Wang Y., Zhang D., and Zhao Z. (2024). Antibiotic resistance genes are transferred from manure-contaminated water bodies to the gut microbiota of animals through the food chain. Environ. pollut. 363. doi: 10.1016/j.envpol.2024.125087
WHO (2016). WHO GLOBAL PRINCIPLES FOR THE CONTAINMENT OF ANTIMICROBIAL RESISTANCE IN ANIMALS INTENDED FOR FOOD Report of a WHO Consultation with the participation of the Food and Agriculture Organization of the United Nations and the Office International des Epizooties (Geneva: World Health Organization). Available online at: http://www.who.int/foodsafety/publications/containment-amr/en/.
WHO, FAO, and WOAH (2020). Technical Brief on Water, Sanitation, Hygiene and Wastewater Management to Prevent Infections and Reduce the Spread of Antimicrobial Resistance (Rome, Italy: World Health Organization). Edition(1), Pp 32. Available online at: https://www.who.int/publi catio ns/i/item/9789240006416.
Wong T.-T. (2015). Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recognit 48, 2839–2846. doi: 10.1016/j.patcog.2015.03.009
Wu G., Shang J., Pan L., and Wang Z. (2014). Heavy metals in surface sediments from nine estuaries along the coast of Bohai Bay, Northern China. Mar. pollut. Bull. 82, 194–200. doi: 10.1016/j.marpolbul.2014.02.033
Xiao R., Bai J., Huang L., Zhang H., Cui B., and Liu X. (2013). Distribution and pollution, toxicity and risk assessment of heavy metals in sediments from urban and rural rivers of the Pearl River delta in southern China. Ecotoxicology 22, 1564–1575. doi: 10.1007/s10646-013-1142-1
Yu Z., He P., Shao L., Zhang H., and Lü F. (2016). Co-occurrence of mobile genetic elements and antibiotic resistance genes in municipal solid waste landfill leachates: A preliminary insight into the role of landfill age. Water Res. 106, 583–592. doi: 10.1016/j.watres.2016.10.042
Yuan L., Li Z.-H., Zhang M.-Q., Shao W., Fan Y.-Y., and Sheng G.-P. (2019). Mercury/silver resistance genes and their association with antibiotic resistance genes and microbial community in a municipal wastewater treatment plant. Sci. Total Environ. 657, 1014–1022. doi: 10.1016/j.scitotenv.2018.12.088
Zhang J., Lu T., Chai Y., Sui Q., Shen P., and Wei Y. (2019a). Which animal type contributes the most to the emission of antibiotic resistance genes in large-scale swine farms in China? Sci. Total Environ. 658, 152–159. doi: 10.1016/j.scitotenv.2018.12.175
Zhang S., Wang Y., Song H., Lu J., Yuan Z., and Guo J. (2019b). Copper nanoparticles and copper ions promote horizontal transfer of plasmid-mediated multi-antibiotic resistance genes across bacterial genera. Environ. Int. 129, 478–487. doi: 10.1016/j.envint.2019.05.054
Zhang X., Zhi X., Chen L., and Shen Z. (2020). Spatiotemporal variability and key influencing factors of river fecal coliform within a typical complex watershed. Water Res. 178, 115835. doi: 10.1016/j.watres.2020.115835
Zhao Y., Cocerva T., Cox S., Tardif S., Su J.-Q., Zhu Y.-G., et al. (2019). Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci. Total Environ. 656, 512–520. doi: 10.1016/j.scitotenv.2018.11.372
Zhou Q., Zhou T., Feng F., Huang S., and Sun Y. (2021). The response of copper resistance genes, antibiotic resistance genes, and intl1/2 to copper addition during anaerobic digestion in laboratory. Ecotoxicol Environ. Saf. 210, 111822. doi: 10.1016/j.ecoenv.2020.111822
Keywords: antimicrobial resistance, polychaete, gut microbiome, metagenome, metal resistance, mobile genetic elements
Citation: Ganesan M, Mani R, Saikumar S, Chang SW, Ravi K, Mani RR and Ravindran B (2025) Studies on environmental resistomes in polychaete gut microbiome from polluted estuaries. Front. Mar. Sci. 12:1637421. doi: 10.3389/fmars.2025.1637421
Received: 29 May 2025; Accepted: 02 September 2025;
Published: 29 September 2025.
Edited by:
Lingshi Yin, Hunan Agricultural University, ChinaReviewed by:
Naga Radha Srinivas Tanuku, Council of Scientific and Industrial Research (CSIR), IndiaYan Fang, Ludong University, China
Copyright © 2025 Ganesan, Mani, Saikumar, Chang, Ravi, Mani and Ravindran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ravi Mani, cmF2aW1pY3JvMjAxOEBnbWFpbC5jb20=; Ravishankar Ram Mani, UmF2aXNoYW5rYXJAdWNzaXVuaXZlcnNpdHkuZWR1Lm15; Balasubramani Ravindran, S2FsYW1yYXZpQGdtYWlsLmNvbQ==
 Mirunalini Ganesan
Mirunalini Ganesan Ravi Mani
Ravi Mani Sakthinarenderan Saikumar2
Sakthinarenderan Saikumar2 Soon Woong Chang
Soon Woong Chang Ravishankar Ram Mani
Ravishankar Ram Mani Balasubramani Ravindran
Balasubramani Ravindran