- 1Yazhou Bay Innovation Institute, Hainan Tropical Ocean University, Sanya, China
- 2Key Laboratory of Utilization and Conservation for Tropical Marine Bioresources, Ministry of Education, Hainan Tropical Ocean University, Sanya, China
- 3Hainan Key Laboratory for Conservation and Utilization of Tropical Marine Fishery Resources, Hainan Tropical Ocean University, Sanya, China
A species of sea slug discovered in the South China Sea was characterized based on its morphology, mitochondrial genome, and phylogenetic position. The slug exhibits brown coloration, resembling seaweed. It features detachable, feather-like cerata capable of independent movement. The mitochondrial genome (14,452 bp; GenBank accession: PV637185) contains 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs, and a non-coding control region. Nucleotide composition showed a strong AT bias (65.55%), with AT-skew and GC-skew values of -0.1421 and 0.0607, respectively. Phylogenetic based on the cytochrome c oxidase subunit I (cox1) gene sequence analysis revealed a close relationship with Polybranchia orientalis, while BLAST analysis demonstrated high similarity between the cox1 gene of this sea slug and that of P. orientalis. Since the complete genome sequence of P. orientalis is currently unavailable, an additional phylogenetic analysis was performed using the 13 protein-coding genes (PCGs) of other sea slugs, which showed that this species clustered within a clade comprising several other species of the family Plakobranchidae. Integrating cox1 sequence data and morphological evidence, we assign this slug to the species Polybranchia orientalis sp. nov. Interestingly, the cerata exhibit green pigmentation, and algal chloroplast genes were detected within them, suggesting kleptoplasty.
1 Introduction
The mitochondrial genome (mtDNA), as the genetic material of organelles, is characterized by a small molecular weight, relatively conserved structure, moderate evolutionary rate, and matrilineal inheritance (usually without recombination). It serves as one of the core markers for studying species phylogeny, population genetic structure, comparative genomics, and molecular evolutionary mechanisms (Kumar et al., 2007; Nohara et al., 2009). Mitochondria contain their own genome, typically a circular DNA molecule. Mitochondrial genomes are highly diverse, with significant variations in size and morphology across different species (Ma et al., 2019).
Recent years have witnessed significant breakthroughs in sacoglossan research, particularly in systematics and functional mechanisms. Integrative taxonomic studies driven by molecular phylogenetics have revealed extensive cryptic diversity: taxonomic research was conducted on the molecular biological characteristics and ecological traits of the marine shellfish genus Oxynoe (Heterobranchia: Sacoglossa), successfully describing seven new species (Krug et al., 2018). Meanwhile, investigations of Plakobranchus and Elysia Risso uncovered pseudocryptic species and novel population distributions across the Caribbean, Mediterranean, and West Pacific regions. Notably, next-generation sequencing (NGS) of Hawaiian Plakobranchus populations demonstrated that their kleptoplasts originate from previously unrecognized bryopsidalean algal diversity (Chlorophyta) (Krug et al., 2016; Wade and Sherwood, 2017). Concurrently, transcontinental lineage analyses of Placida have exposed the evolutionary complexity of multi-species complexes (Cetra et al., 2021), fundamentally reshaping traditional morphology-based classification frameworks.
Parallel functional ecological research has elucidated environmental adaptation mechanisms in kleptoplasty. Experimental work confirmed that Elysia viridis (Montagu) achieves cellular-level carbon and nitrogen fixation and metabolic integration via functional kleptoplasts, with plastid recognition potentially mediated by scavenger receptors and thrombospondin-type-1 repeat proteins (TSP-1) (Clavijo et al., 2020). Thermal stress studies demonstrated significantly reduced amylose production and photosynthetic efficiency in the sister species E. timida Risso and E. cornigera Nuttall under elevated temperatures, indicating climate change threats to photo-symbiotic stability (Laetz and Waegele, 2019). Furthermore, the broad-spectrum algal host utilization strategy of Plakobranchus cf. ianthobapsus offers new perspectives for invasive species management (Wade and Sherwood, 2018), while 3D reconstruction of the renopericardial system in E. viridis revealed evolutionary innovations in circulatory-excretory functions (Neusser et al., 2018). Collectively, these advances establish sacoglossa as a model system for studying animal-plastid symbiotic interactions.
However, due to their low population density and effective mimicry defense mechanisms that make them difficult to detect, obtaining sea slug samples remains a challenge. Numerous sea slug species remain unidentified. During our field investigations, a species with external morphological characteristics strikingly similar to Polybranchia orientalis was discovered among sea grapes, here it was named Polybranchia orientalis sp. nov. There has been no previous report of this species in the southeastern coastal area of China. Moreover, a search of publicly available databases revealed no existing genetic information on this species. Therefore, in this study, we conducted a comprehensive identification and phylogenetic analysis of this sea slug species. This was achieved through in-depth mitochondrial genome sequence analysis, combined with a detailed examination of its external morphological features. Our aim is to provide a robust theoretical foundation for the classification and identification of sea slugs, contributing to a better understanding of their biodiversity and evolutionary relationships.
2 Materials and methods
2.1 Sample collection, genomic DNA extraction, and quality assessment
Slugs were collected during low tide in June 2024 from the coastal area of Sanya, Hainan Province, China (Longitude: 109.496745, Latitude: 18.2069). Their leaf-like cerata were sampled and stored in 95% ethanol. Genomic DNA was extracted from tissue samples using the Rapid Tissue Genomic DNA Extraction Kit (Beijing Aidlab Biotechnologies Co., Ltd.). Following extraction, the quality of the genomic DNA was assessed by agarose gel electrophoresis. Only DNA samples that met quality standards were used for subsequent experiments.
2.2 Primer design, PCR amplification, and sequencing assembly
Primers designed to match generally conserved regions of target mtDNA were used to amplify short fragments from cox1, 16S, nd1, cytb, 12S and nd4. Specific primers were designed based on these conserved regions sequences and used to amplify the remained mtDNA sequence in several PCR reactions (Supplementary Table S1). Primers were designed to produce amplicons with overlaps of about 100 bp. The PCR reaction was performed using the LA Taq polymerase. The PCR conditions were as follows: initial denaturation 94°C for 2 min, then 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min/kb, followed by the final extension at 72°C for 10 min. The total volume for PCR and LA-PCR was 50 μl, of which Takara LATaq (5 U/μl) was 0.5 μl, 10×LATaq Buffer II (Mg2+) was 5 μl, dNTP mixture (2.5 mM) was 8 μl, template was 60 ng, and the total volume was then made up with distilled water. The final concentration of the forward and reverse primers was 0.2~1.0 μM, and that of MgCl2 was 2.0 mM. The PCR products were sequenced directly, or if needed first cloned into a pMD18-T vector (Takara, JAP) and then sequenced, by the dideoxynucleotide procedure, using an ABI 3730 automatic sequencer (Sanger sequencing) using the same set of primers. All obtained fragments were quality-proofed (electropherogram) and BLASTed to confirm that the amplicon is the actual target sequence. Whenever the quality was sub-optimal, sequencing was repeated. All obtained fragments were BLASTed to confirm that the amplicon is the target sequence. Mitogenome was assembled stepwise with the help of DNAstar v7.1 (Burland, 2000) program, making sure that the overlaps were identical, and that no NUMTS were incorporated into the sequence.
2.3 Sequence annotation
After quality-proofing of the obtained fragments, the complete mt genome sequence was assembled manually using DNAstar v7.1 softwar (Burland, 2000). Mt genome was annotated roughly following the procedure described before (Zhang et al., 2018; Zou et al., 2017). First, raw mitogenomic sequences were imported into MITOS web servers to determine the approximate boundaries of genes. Exact positions of protein-coding genes (PCGs) were found by searching for ORFs (employing genetic code 5, the invertebrate mitochondrion). All tRNAs were identified using ARWEN, DOGMA and MITOS. The precise boundaries of rrnL and rrnS were determined via a comparison with homologs.
2.4 Data processing and analysis
The mitochondrial genome map was generated online using the CGView server (http://cgview.ca/). The base composition, amino acid usage, and relative synonymous codon usage (RSCU) of the mitochondrial genome of P. orientalis sp. nov. were analyzed using MEGA version 12. The AT and GC skew values were calculated using the formulas: AT-skew = (A - T)/(A + T) and GC-skew = (G - C)/(G + C).
2.5 Phylogenetic analysis
We downloaded the complete mitochondrial genome sequences of 23 species with high similarity to P. orientalis sp. nov. from the NCBI database (https://www.ncbi.nlm.nih.gov/genbank/) following a BLAST search (Supplementary Table S2). The cox1 genes of the above 23 species were extracted using Extract tool integrated in Phylosuite v1.2.3 (Xiang et al., 2023; Zhang et al., 2019). The cox1 gene sequence of P. orientalis sp. nov. was used as the query for BLAST search on NCBI, and subsequently, cox1 sequences of 7 species with high similarity were selected and downloaded. However, these seven cox1 sequences were incomplete. Multiple sequence alignment was performed using MAFFT (Katoh and Standley, 2013) implemented in Phylosuite v1.2.3, and each sequence was aligned with MAFFT using ‘–auto’ strategy and normal alignment mode. Subsequently, the optimal evolutionary model was selected via ModelFinder v2.2.0 (Kalyaanamoorthy et al., 2017) based on the Bayesian Information Criterion (BIC) (Supplementary Table S3), with GTR+F+I+G4 identified as the best-fit model. Bayesian inference (BI) analyses were conducted in MrBayes 3.2.6 (Ronquist et al., 2012) using the best-fit models, with Markov chain Monte Carlo (MCMC) simulations run for 1,000,000 generations. Two parallel runs (each containing 4 chains) were performed, with the first 25% of sampled trees discarded as burn-in.
Using Phylosuite v1.2.3, we extracted the 13 mitochondrial protein-coding genes (PCGs) from these 23 species and aligned them using the integrated MAFFT software. The aligned PCG sequences were trimmed with Gblocks and subsequently concatenated. The dataset comprising 13 PCGs was initially partitioned into separate groups according to their gene names. Subsequently, we used ModelFinder v2.2.0 to partition the concatenated dataset into 14 partitions and select the best-fit partition model for each partition using BIC criterion. Bayesian inference phylogenies were inferred using a partition model (two parallel runs, each containing 4 chains, 1,000,000 generations) in which the initial 25% of the sampled data were discarded as burn-in. Average standard deviation of split frequencies (ASDSF) = 0.000428. The phylogenetic trees constructed based on the concatenated PCGs and single cox1 gene were visualized and manually adjusted using the online platform iTOL (https://itol.embl.de/).
3 Results
3.1 Morphological characteristics of P. orientalis sp. nov.
This sea slug resembles a clump of brown seaweed with a somewhat flattened dorsal surface. On the head, there is a pair of tentacles conical and placed at the anterior extremity of the dorsal surface. Like other sap-sucking sea slugs, it feeds on algae and changes color depending on the algae it consumes—appearing brown at times and green at others. The body is adorned with numerous feather-like cerata (Figures 1A, B). Cerata flat, numerous, fan-shaped, with toothed margins, uneven with long footstalk. These cerata exhibit a fine branching pattern resembling the veins of a leaf. When gently manipulated, the cerata readily detach; upon detachmentappe, they tend to adhere to human skin and retain the ability to undergo movement in seawater (Figure 1C). These branches are composed of multiple granular structures with distinct pigmentation (Figure 1D). Foot elongate and ample, broader anteriorly and narrower at the posterior end.

Figure 1. The morphological characteristics of the sample. (A) Dorsal view; (B) Ventral view; (C) Feather-like structure; (D) Microscopic structure of the internal branching structure.
In most cases, this type of animal is hermaphroditic and reproduces through cross-fertilization. Egg laying occurs either during copulation or several hours afterward. The eggs are bound together by a gelatinous substance secreted by the albumen gland, forming long, spiral-shaped ribbon-like structures (Figure 2A). Microscopic observation revealed numerous developing embryos, as shown in Figure 2B.
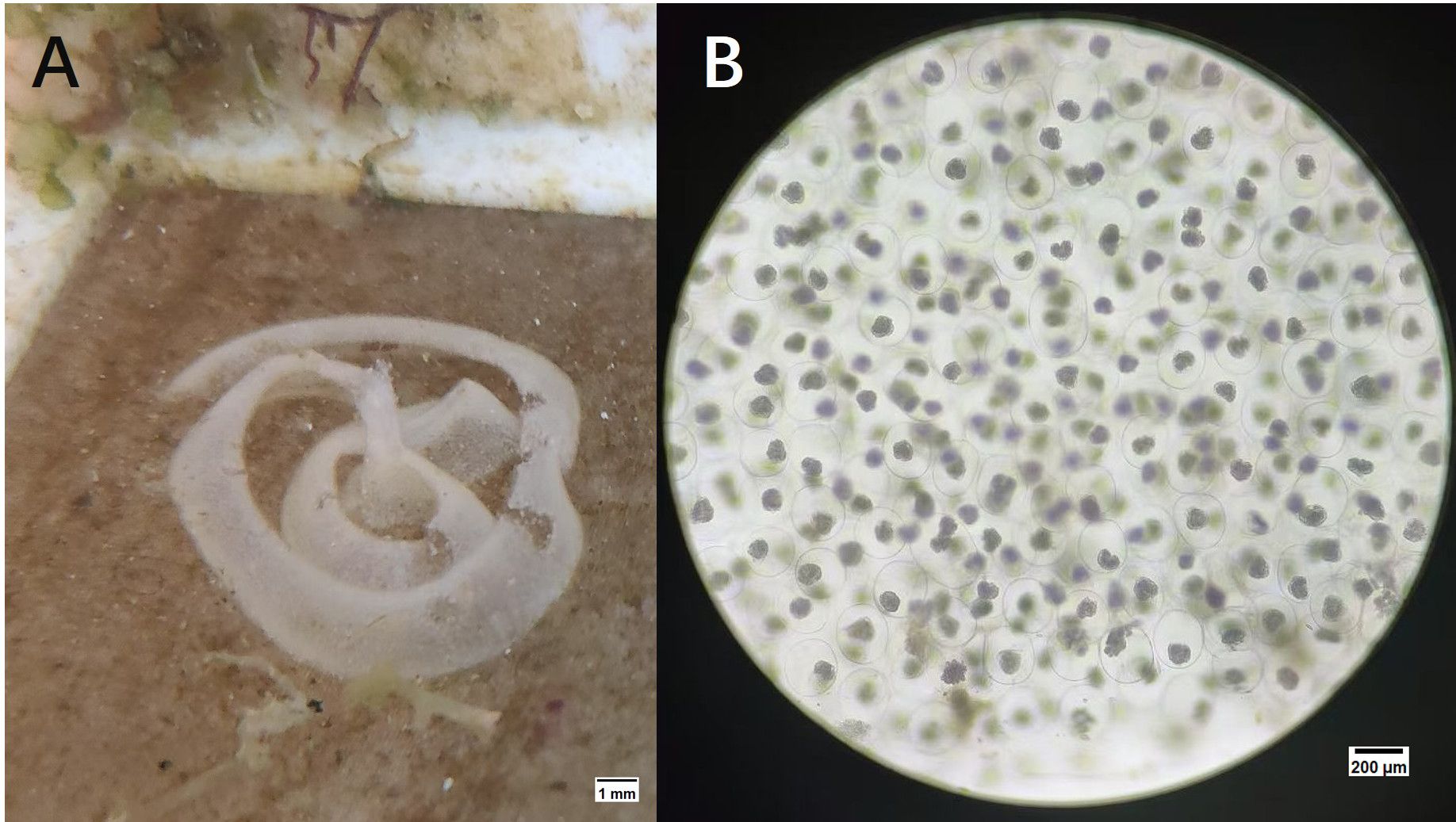
Figure 2. The eggs of the P. orientalis sp. nov. (A) The egg band of P. orientalis sp. nov.; (B) The developing embryo of P. orientalis sp. nov.
3.2 Mitochondrial genome structure of P. orientalis sp. nov.
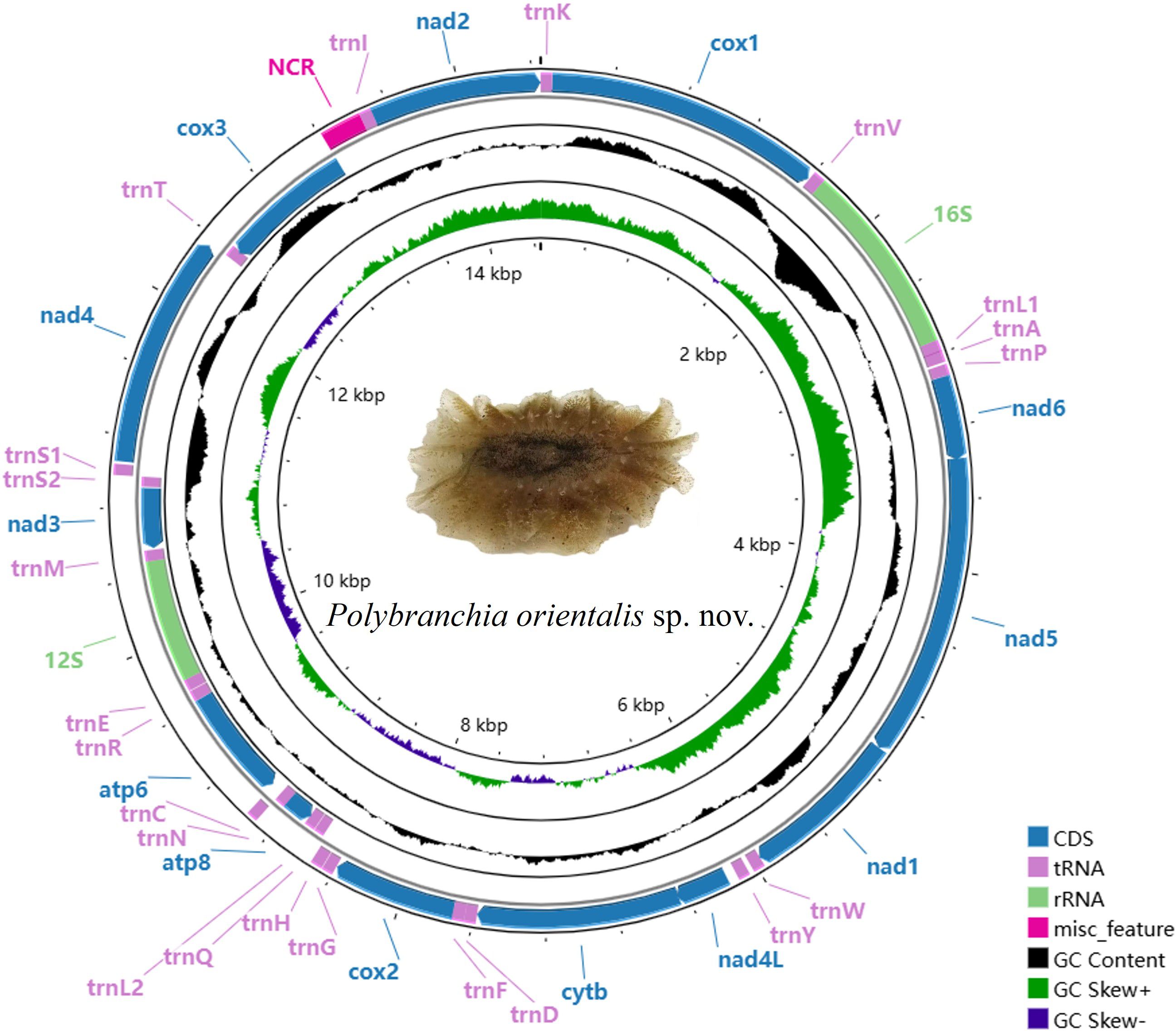
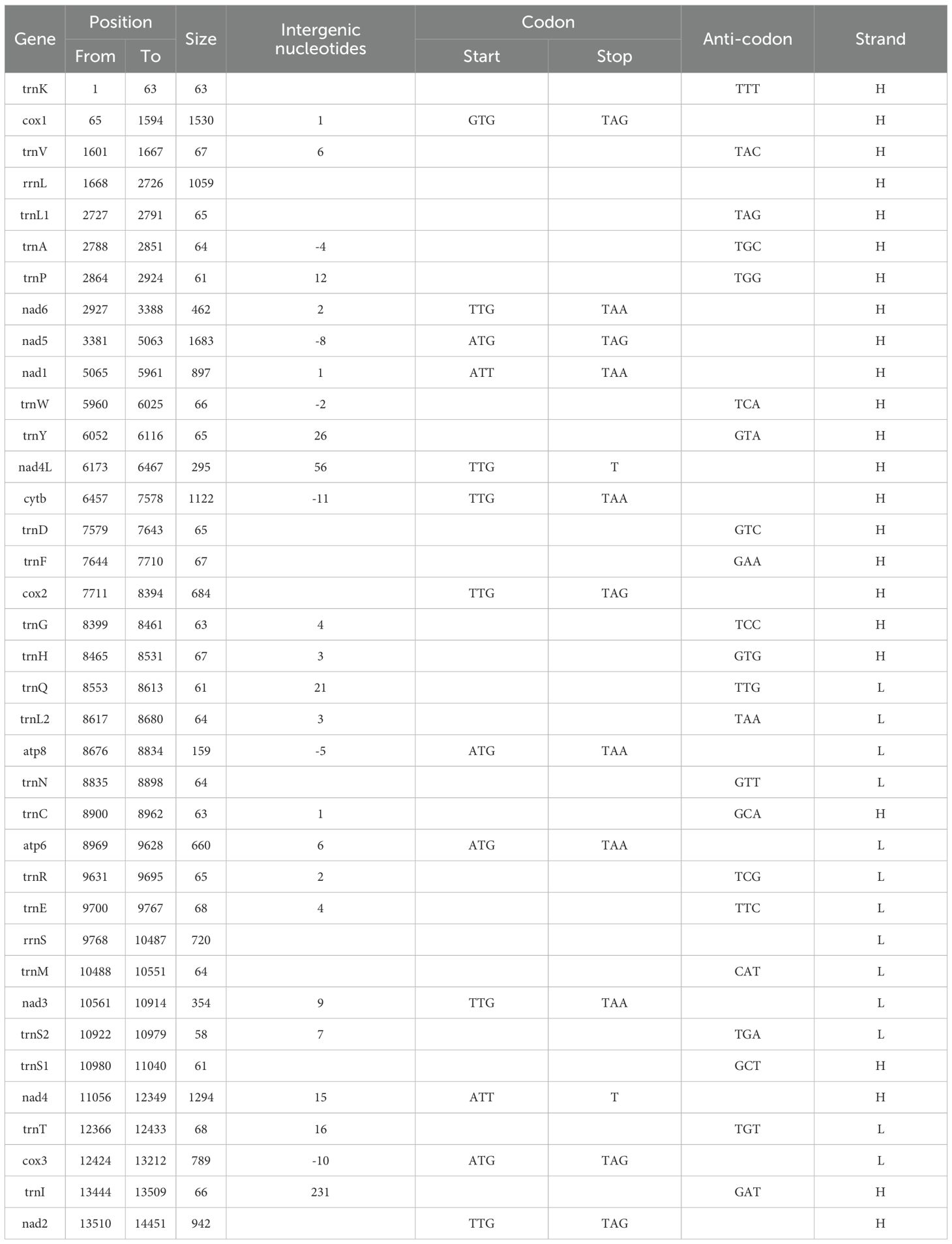
The mitochondrial genome of P. orientalis sp. nov., after sequencing and assembly, was found to be 14,452 bp in length (GenBank accession number: PV637185). It contains 13 PCGs, 22 tRNA genes, 2 rRNA genes, and one putative non-coding control region (Figure 3). The total length of the PCGs is 10,871 bp, tRNA genes account for 1,415 bp, rRNA genes 1,779 bp, and the control region is 231 bp, corresponding to 75.22%, 9.79%, 12.31%, and 1.60% of the entire mitochondrial genome, respectively.
Among these, atp8, atp6, nad3, cox3, 12S-rRNA, and 8 tRNA genes (trnQ, trnL2, trnN, trnR, trnE, trnM, trnS2, trnT) are encoded by the L-strand, while the remaining genes are encoded by the H-strand. Among the 13 PCGs, four different start codons were identified: GTG, TTG, ATG, and ATT. Stop codons include TAA, TAG, and an incomplete stop codon represented as T– (Table 1).
3.3 Nucleotide composition and skewness of the mitochondrial genome
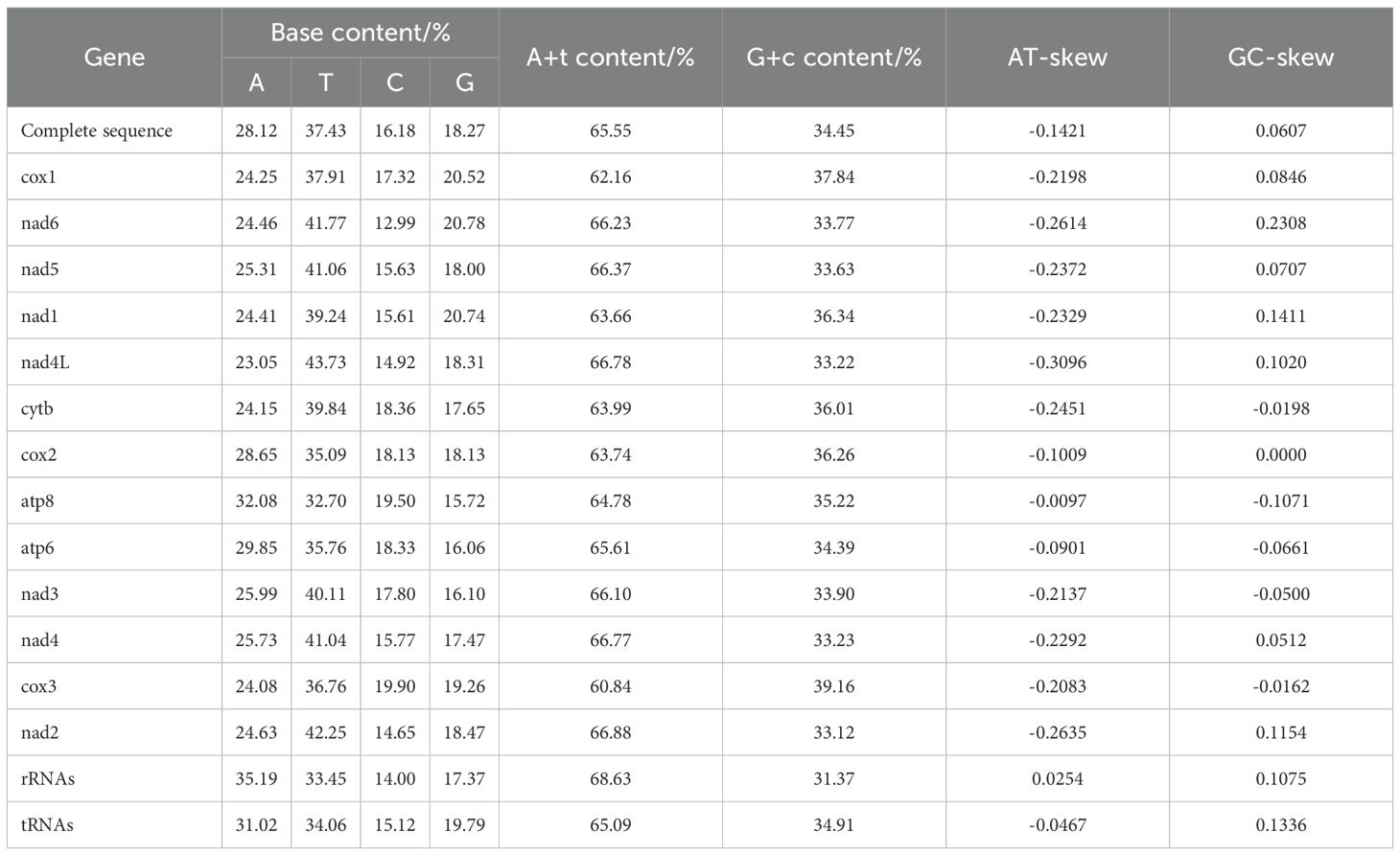
The nucleotide composition of the P. orientalis sp. nov. mitochondrial genome, from highest to lowest, is T (37.43%), A (28.12%), G (18.27%), and C (16.18%). The A+T content accounts for 65.55% of the entire mitochondrial genome, while the G+C content is 34.45%. The AT-skew and GC-skew values are -0.1421 and 0.0607, respectively, indicating a clear AT bias in nucleotide composition. In other regions of the mitochondrial genome, the A+T content similarly exceeds the G+C content.
Except for the rRNAs, the AT-skew values for the entire mitochondrial genome, tRNAs, and all 13 PCGs are negative. Among the 13 PCGs, the GC-skew values of cytb, atp8, atp6, nad3, and cox3 are negative, while the remaining PCGs, tRNAs, and rRNAs exhibit positive GC-skew values. Additionally, relatively high proportions of thymine (T) and adenine (A) were observed across PCGs, tRNAs, and rRNAs (Table 2).
3.4 Mitochondrial protein-coding genes
The mitochondrial genome of P. orientalis sp. nov. contains 13 protein-coding genes (PCGs) with a total length of 10,871 bp. Among them, nad5 is the longest gene at 1,683 bp, while atp8 is the shortest at 159 bp. The genes atp8, atp6, nad3, and cox3 are encoded on the L-strand, whereas the remaining genes are located on the H-strand. Several overlapping regions exist between adjacent genes, including a 2 bp overlap between nad6 and nad5, an 8 bp overlap between nad5 and nad1, and a 56 bp overlap between nad4L and cytb.
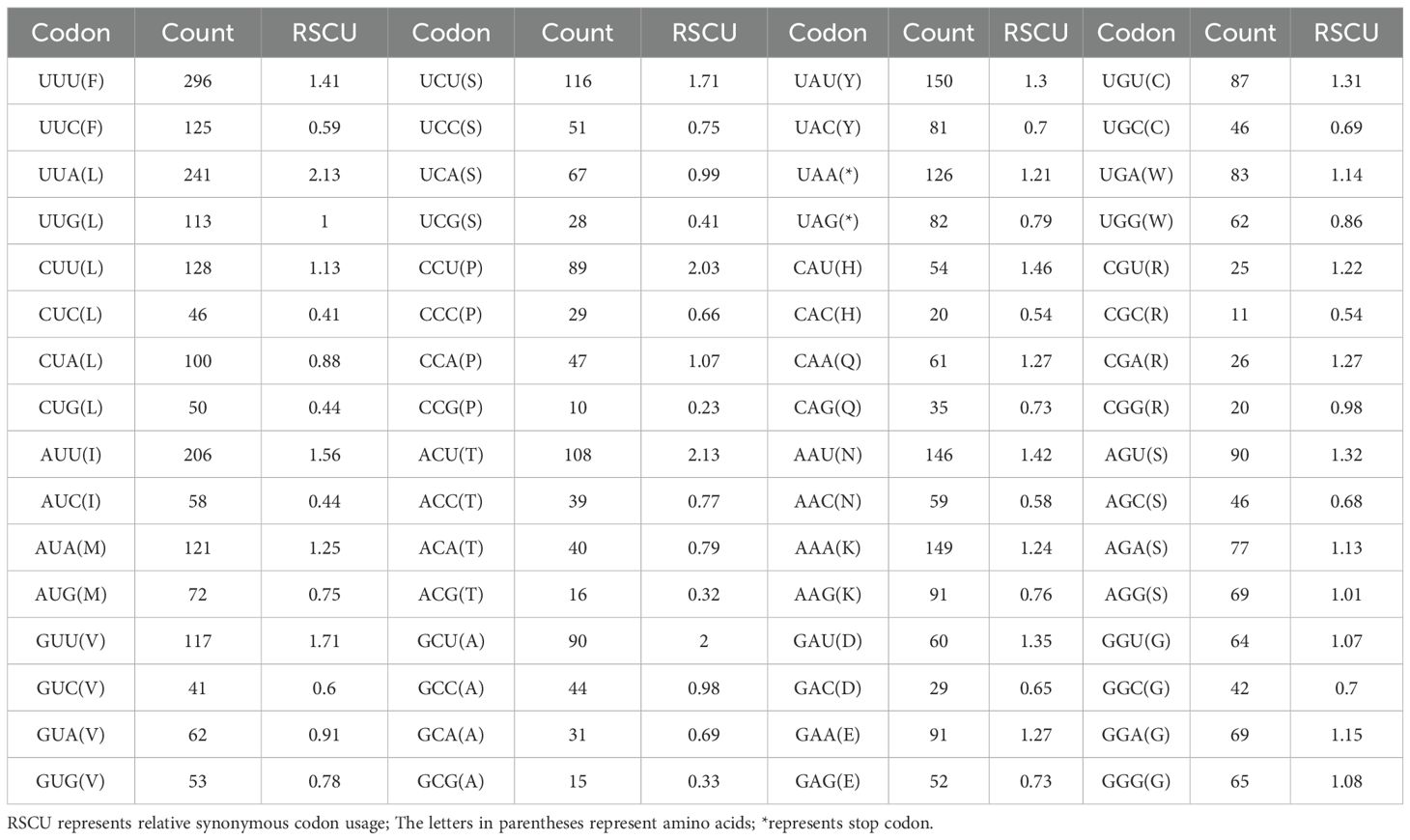
Analysis of relative synonymous codon usage (RSCU) revealed a total of 29 preferred codons. The highest RSCU value was 2.13, while the lowest was 0.23. Four codons had RSCU values equal to or greater than 2: UUA (Leu), CCU (Pro), ACU (Thr), and GCU (Ala), all of which end with either A or U (Table 3).
3.5 tRNA and rRNA genes of P. orientalis sp. nov.
The mitochondrial genome of P. orientalis sp. nov. contains 22 tRNA genes, ranging in length from 58 to 68 bp. The longest tRNAs are trnE and trnT, while the shortest is trnS2, with a total tRNA length of 1,415 bp. Eight of these tRNAs (trnQ, trnL2, trnN, trnR, trnE, trnM, trnS2, trnT) are encoded on the L-strand, while the remaining tRNAs are located on the H-strand.
The total length of the rRNA genes is 1,779 bp, comprising two genes: 12S rRNA and 16S rRNA, located on the L- and N-strands, respectively. The lengths of 16S rRNA and 12S rRNA are 1,059 bp and 720 bp, respectively. Their overall A+T content is 65.09%, with nucleotide composition as follows: A = 35.19%, T = 33.45%, G = 17.38%, and C = 14.00%. The 12S rRNA gene is situated between trnM and trnE, while the 16S rRNA gene is located between trnV and trnL1. A 6 bp overlap is observed between 16S rRNA and trnV, and a 4 bp overlap between 12S rRNA and trnE. The AT skew (0.0254) of the two rRNA genes is slightly lower than the GC skew (0.0175), indicating a bias toward G and C nucleotides.
3.6 Phylogenetic analysis
To determine the molecular taxonomic position of this sea slug, a Bayesian tree was constructed using Phylosuite software based on the 13 protein-coding genes (PCGs) and cox1 sequences of sea slug species retrieved from GenBank. BLAST searches revealed no existing information on this species; however, its cox1 sequence showed a high similarity of 99.26% to that of P. orientalis (AB501309.1). Phylogenetic analysis based on cox1 sequences further indicated that this species first clustered with P. orientalis, and then together with congeneric sequences (e.g., P. mexicana, P. schmekelae, P. jannae) formed a distinct clade within the family Caliphyllidae (Figure 4). Since the complete genome sequence of P. orientalis is currently unavailable, an additional phylogenetic analysis was performed using the 13 PCGs of other sea slugs, which showed that this species clustered within a clade comprising species of the family Plakobranchidae (Figure 5).
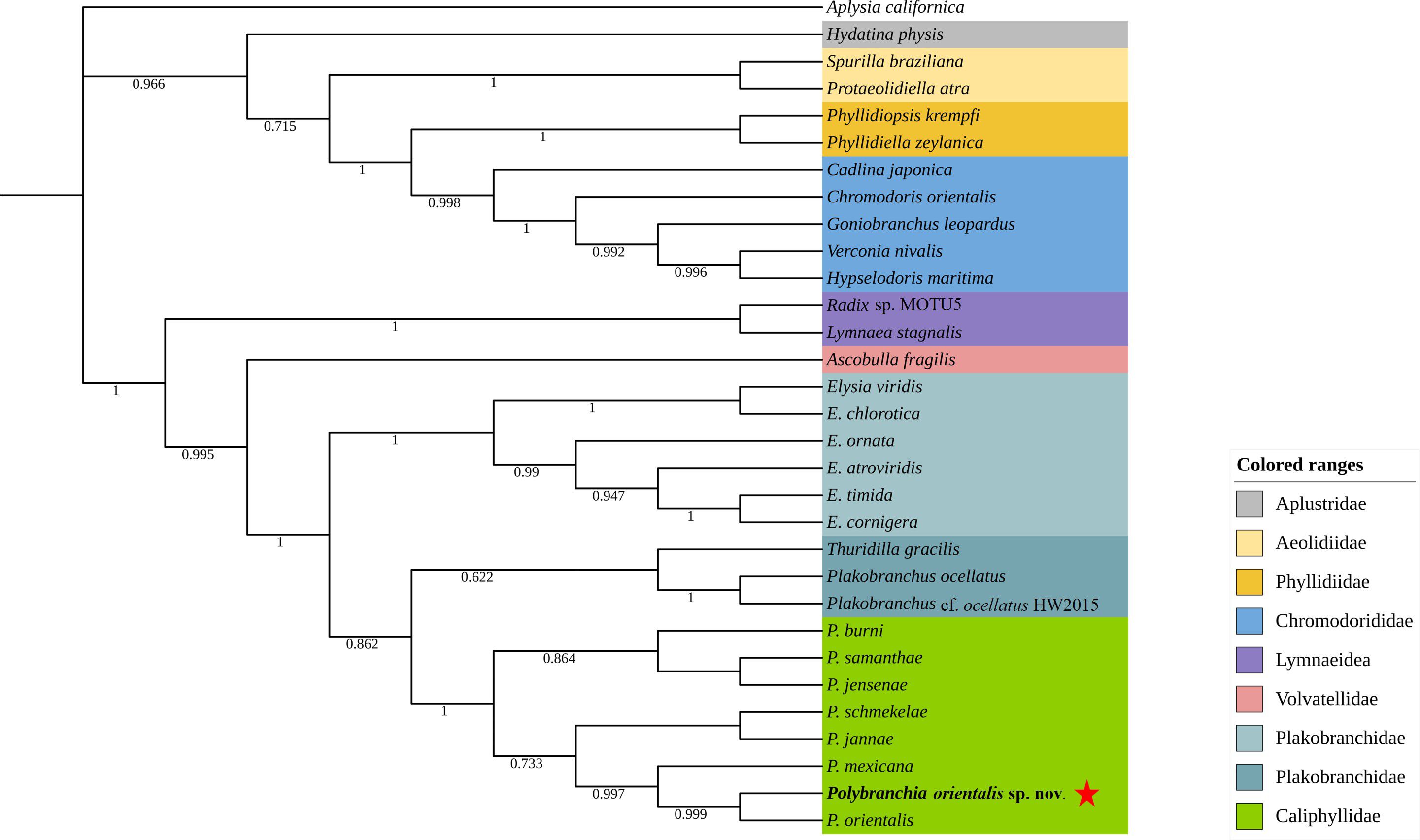
Figure 4. Phylogenetic tree constructed based on the sequences of cox1 of the mitochondrial genome. Star stands for P. orientalis sp. nov.
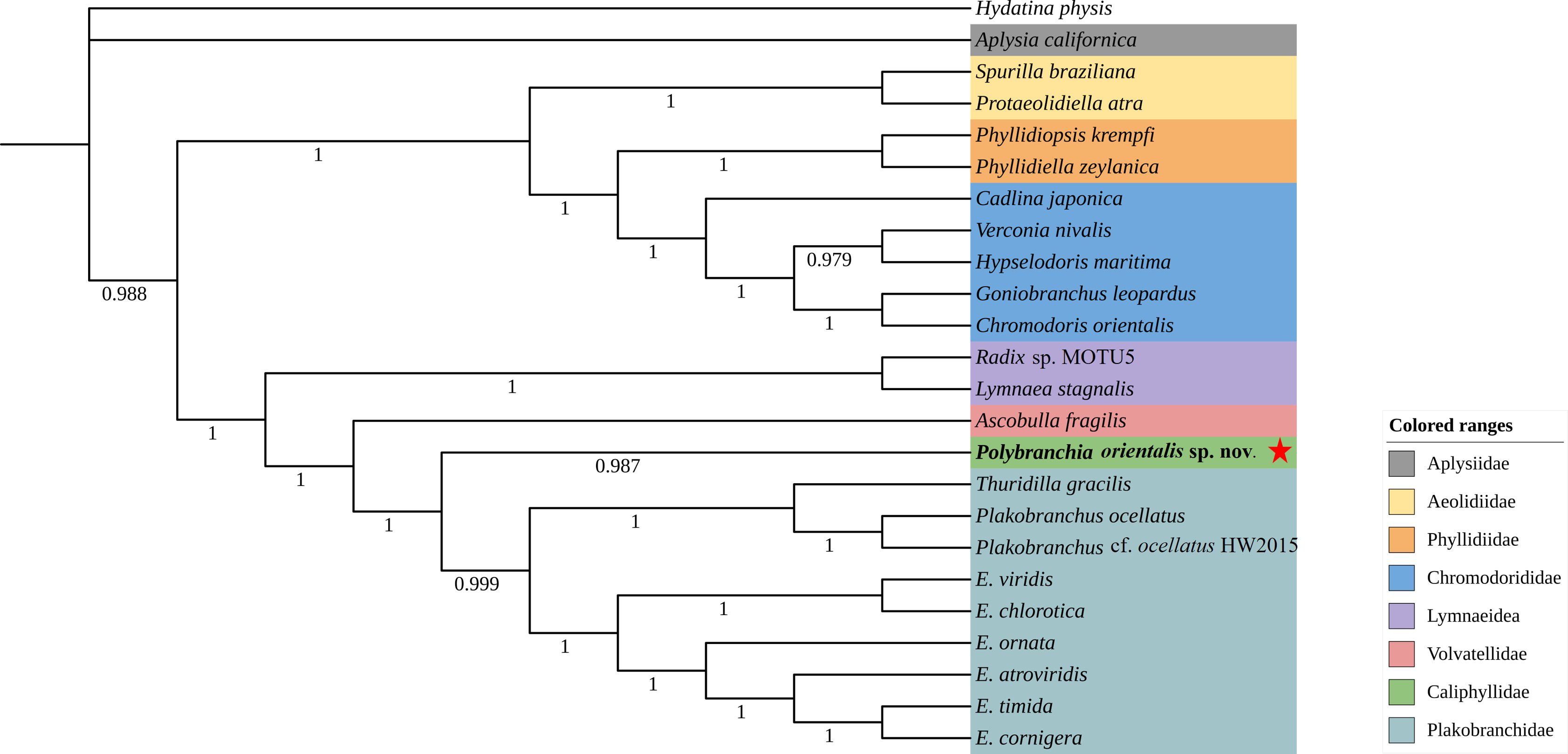
Figure 5. Phylogenetic tree constructed based on the sequences of 13 PCGs of the mitochondrial genome. Note: Star stands for P. orientalis sp. nov.
Morphologically, this sea slug closely resembles P. orientalis, with both species displaying a clump-like appearance similar to brown seaweed and bearing feather-like cerata on their dorsum. A key distinction lies in the cerata: the feathery cerata of P. orientalis are nearly transparent and speckled with cinnamon brown throughout, with two dorso-medial yellowish patches (Medrano et al., 2019), while the cerata of P. orientalis sp. nov. are accompanied by granular structures with green pigmentation (Figure 1). In contrast, sea slugs of the family Plakobranchidae, such as Thuridilla gracilis, Elysia ornata, and Plakobranchus ocellatus, exhibit striking morphological differences from P. orientalis sp. nov. For example, Plakobranchus ocellatus exhibits a dorso-ventrally flattened body with broad parapodia meeting centrally. Its translucent, gray-cream body displays distinctive ocelli with yellow and blue centers (Angela et al., 2022).
Collectively, these phylogenetic and morphological findings support the interpretation of this sea slug within the genus Polybranchia, here referred to as P. orientalis sp. nov.
3.7 Kleptoplasty in P. orientalis sp. nov.
DNA was extracted from cerata (not whole body) and sequence of chloroplast related gene were detected, as shown in the Appendix. BLASTn analysis of chloroplast gene sequences identified in P. orientalis sp. nov. revealed high similarity (>95%) with chloroplast genomes from various species. Among the top matches, Kryptoperidinium foliaceum (96.53%), Navicula arenaria (96.53%) and Pseudonitzschia brasiliana (96.95%) demonstrated the highest sequence similarity (Table 4).The results preliminarily demonstrate that this species exhibits one of the main characteristics of sacoglossa, namely kleptoplasty, literally meaning “stolen plastids” (Vries et al., 2014). This indicates that P. orientalis sp. nov. can utilize algal chloroplasts to carry out photosynthesis and provide energy for itself, which holds significant importance in the process of adaptive evolution in animals.
4 Discussion
This study reports the first identification of P. orientalis sp. nov. in the South China Sea region. We characterize its mitochondrial genome structure and elucidate its evolutionary relationships. Based on the following lines of evidence, we suggest this sea slug belongs to the genus Polybranchia. (1) Phylogenetic analysis based on cox1 revealed that this species occupies a distinct taxonomic position within the genus Polybranchia. (2) In terms of mitochondrial genome sequences, this species shows complete divergence from other species. (3) While morphologically similar to its congener P. orientalis, it exhibits distinct morphological differences in key diagnostic features.
P. orientalis sp. nov. resembles a clump of brown seaweed. Its body bears numerous feather-like cerata characterized by fine, leaf-vein-like branching patterns composed of multiple granular structures exhibiting distinct pigmentation. (Figure 1). The P. orientalis found on the south-west coast of India differs from P. orientalis sp. nov. in two key morphological traits: the cerata of this P. orientalis are dark green in color, and its dorsum bears a white spot with a slight swelling, which indicates the position of the heart (Ravinesh et al., 2014). In contrast, the P. orientalis from the Philippines can be distinguished from P. orientalis sp. nov. by the following characteristics: its pericardium is white and typically very conspicuous; the ceratal peduncle has snowy white pigment in the depression; the cerata are translucent and speckled with cinnamon brown throughout, accompanied by two dorso-medial yellowish patches (some of which have a single black dot) (Medrano et al., 2019).
In addition, we elucidated the structural characteristics and evolutionary relationships of the mitochondrial genome of P. orientalis sp. nov. Similar to the mitochondrial genomes of other reported sea slug species in terms of size and composition, the mitochondrial genome of P. orientalis sp. nov. is circular, with a total length of 14,452 base pairs. It comprises 37 genes, including 22 tRNAs, 13 PCGs, and 12S and 16S rRNAs. Both the entire mitochondrial genome and its individual components exhibit a high AT content. Moreover, the whole mitochondrial genome and PCGs display AT bias, consistent with findings in species such as Thuridilla gracilis, E. ornata, and E. cornigera (Medina et al., 2011; Rauch et al., 2017). This AT bias may be associated with evolutionary pressures during mitochondrial DNA replication and transcription (Zhong et al., 2002).
The absence of P. mexicana, P. orientalis, P. jannae and other species of the genus Polybranchia (family Caliphyllidae) from the 13 PCGs-based phylogenetic tree is attributed to the unavailability of their complete mitochondrial genome sequences in public databases. Currently, published information on the mitochondrial genomes of sea slugs remains limited. Therefore, additional mitochondrial genome data are needed in future studies to provide robust support for elucidating their evolutionary history and genetic divergence.
This species belongs to sacoglossan sea slugs and therefore possesses some characteristics of this group. Sacoglossans feed by ingesting the cellular contents of algae. Notably, these slugs sequester living chloroplasts from the algae they consume and utilize them within their own tissues—a highly unusual phenomenon known as kleptoplasty. Based on Table 3 and Figure 5, P. orientalis sp. nov. is likely to possess a kleptoplastic structure, similar to that of Plakobranchus ocellatus, E. viridis, and E. timida and other species in the family Plakobranchidae. These species are typical examples of photosynthetic sea slugs (Angela et al., 2022; Cruz et al., 2020; Laetz and Waegele, 2017). Among them, Plakobranchus ocellatus is a “solar-powered” sea slug renowned for its ability to retain chloroplasts for extended periods (Greve et al., 2017). We collected the cerata from the dorsal region of P. orientalis sp. nov. and detected chloroplast genes through specific PCR and high-throughput sequencing. These genes showed a high degree of similarity (96.53%) with those of Kryptoperidinium foliaceum and Navicula arenaria. Therefore, it is reasonable to hypothesize that P. orientalis sp. nov. may possess kleptoplastic capabilities.
Possessing kleptoplastic ability implies a redefinition of energy acquisition and survival for sacoglossans (Laetz and Waegele, 2017). Functionally, it enables mixed trophic strategies that buffer against resource scarcity and abiotic stress (Angela et al., 2022); evolutionarily, it drives trait divergence, favors host genomic adaptations over horizontal gene transfer (HGT), and demonstrates that complex traits can be acquired via organelle sequestration (Maeda et al., 2021). From P. ocellatus’ months-long photosynthesis to Placida sp.’s transient energy boost, kleptoplasty exemplifies how symbiotic “theft” evolves into a powerful adaptive strategy (Fan et al., 2014)—reshaping ecological niches and evolutionary trajectories within Sacoglossa (Cruz et al., 2015; Kano et al., 2015). The P. orientalis sp. nov. may possess kleptoplastic capabilities. Future research should focus on experimentally verifying the presence of this remarkable feature.
5 Conclusion
Collectively, the evidence presented herein demonstrates the first discovery of P. orientalis sp. nov., in the South China Sea. This species is characterized by its distinctive feather-like cerata. Phylogenetic analyses consistently place it within the genus Polybranchia of the family Caliphyllidae. Our findings further suggest that P. orientalis sp. nov. may possess kleptoplastic capabilities. These discoveries not only expand our understanding of the biodiversity of sacoglossa sea slugs but also provide a valuable foundation for further research on kleptoplasty.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. PV637185.
Ethics statement
The animal study was approved by The Animal Care Committee of Hainan Tropical Ocean University, China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MM: Resources, Writing – original draft, Supervision, Project administration, Writing – review & editing, Methodology, Funding acquisition. WY: Data curation, Formal Analysis, Writing – original draft, Software, Writing – review & editing, Investigation. JJ: Supervision, Funding acquisition, Writing – review & editing, Resources, Project administration, Writing – original draft. TX: Formal Analysis, Data curation, Writing – original draft, Software, Investigation. GX: Software, Investigation, Writing – original draft, Data curation, Formal Analysis.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National key research and development program of China (2024YFD2401803), the National Natural Science Foundation of China (Grant No. 32360923), the Hainan Provincial Natural Science Foundation of China (322RC716), the Hainan Provincial Natural Science Foundation of China (422MS083), the Scientific Research Foundation of Hainan Tropical Ocean University (RHDRC202206). Any field work in this study complied with the current laws of China, where it was performed.
Acknowledgments
We thank Dr. Haishan Wang from Hainan Tropical Ocean University for help with classification of mollusks. We also acknowledge the reviewers for their constructive feedback.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1650005/full#supplementary-material
References
Angela R. D., Jussi E., and Geir J. (2022). The role of parapodia and lack of photoacclimation in kleptoplasts of the sacoglossan sea slug Plakobranchus ocellatus. Coral. Reefs. 41, 319–332. doi: 10.1007/s00338-022-02224-z
Burland T. G. (2000). DNASTAR’s lasergene sequence analysis software. Methods Mol. Biol. 132, 71–91. doi: 10.1385/1-59259-192-2:71
Cetra N., Gregoric D. E. G., and Roche A. (2021). A new species of placida (Gastropoda: sacoglossa) from southern South America. Malacologia 64, 109–120. doi: 10.4002/040.064.0106
Clavijo J. M., Frankenbach S., Fidalgo C., Serodio J., Donath A., Preisfeld A., et al. (2020). Identification of scavenger receptors and thrombospondin-type-1 repeat proteins potentially relevant for plastid recognition in Sacoglossa. Ecol. Evol. 10, 12348–12363. doi: 10.1002/ece3.6865
Cruz S., Cartaxana P., Newcomer R., Dionísio G., Calado R., Serôdio J., et al. (2015). Photoprotection in sequestered plastids of sea slugs and respective algal sources. Sci. Rep. 5, 7904. doi: 10.1038/srep07904
Cruz S., LeKieffre C., Cartaxana P., Hubas C., Thiney N., Jakobsen S., et al. (2020). Functional kleptoplasts intermediate incorporation of carbon and nitrogen in cells of the sacoglossa sea slug Elysia viridis. Sci. Rep. 10, 10548. doi: 10.1038/s41598-020-66909-7
Fan X., Qiao H., Xu D., Cao S., Zhang X., Mou S., et al. (2014). Short-term retention of kleptoplasty from a green alga (Bryopsis) in the sea slug Placida sp. Ys001. Biologia 69, 635–643. doi: 10.2478/s11756-014-0355-y
Greve C., Lui M. R. T., Sivalingam S., Ludwig K. U., Waegele H., and Donath A. (2017). The complete mitochondrial genome of the ‘solar-powered’ sea slug Plakobranchus cf. Ocellatus (Heterobranchia: Panpulmonata: Sacoglossa). Mitochondrial. DNA B. 2, 130–131. doi: 10.1080/23802359.2016.1247667
Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., Haeseler >A. V., and Jermiin L. S. (2017). Modelfinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kano Y., Neusser T. P., Fukumori H., Jorger K. M., and Schrodl M. (2015). Sea-slug invasion of the land. Biol. J. Linn. Soc 116, 253–259. doi: 10.1111/bij.12578
Katoh K. and Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Krug P. J., Berriman J. S., and Valdes A. (2018). Phylogenetic systematics of the shelled sea slug genus Oxynoe Rafinesque 1814 (Heterobranchia: Sacoglossa), with integrative descriptions of seven new species. Invertebr. Syst. 32, 950–1003. doi: 10.1071/is17080
Krug P. J., Vendetti J. E., and Valdes A. (2016). Molecular and morphological systematics of Elysia Risso 1818 (Heterobranchia: sacoglossa) from the caribbean region. Zootaxa 4148, 1–137. doi: 10.11646/zootaxa.4148.1.1
Kumar N., Lakra W. S., Majumdar K. C., Goswami M., and Ravinder K. (2007). Genetic diversity in the Indian population of Penaeus monodon (Fabricius 1798) as revealed by mtDNA sequence analysis. Aquac. Res. 38, 862–869. doi: 10.1111/j.1365-2109.2007.01740.x
Laetz E. M. J. and Waegele H. (2017). Chloroplast digestion and the development of functional kleptoplasty in juvenile Elysia timida (Risso 1818) as compared to short-term and non-chloroplast-retaining sacoglossan slugs. PloS One 12, e0182910. doi: 10.1371/journal.pone.0182910
Laetz E. M. J. and Waegele H. (2019). Comparing amylose production in two solar-powered sea slugs: the sister taxa Elysia timida and E. cornigera(Heterobranchia: Sacoglossa). J. Mollus. Stud. 85, 166–171. doi: 10.1093/mollus/eyy047
Ma M. J. L., Zhang H., Jiang P., Sin S. T. K., Lam W. K. J., Cheng S. H., et al. (2019). Topologic analysis of plasma mitochondrial DNA reveals the coexistence of both linear and circular molecules. Clin. Chem. 65, 1161–1170. doi: 10.1373/clinchem.2019.308122
Maeda T., Takahashi S., Yoshida T., Shimamura S., Takaki Y., Nagai Y., et al. (2021). Chloroplast acquisition without the gene transfer in kleptoplastic sea slugs, Plakobranchus ocellatus. eLife 10, e60176. doi: 10.7554/elife.60176
Medina M., Lal S., Valles Y., Takaoka T. L., Dayrat B. A., Boore J. L., et al. (2011). Crawling through time: Transition of snails to slugs dating back to the Paleozoic, based on mitochondrial phylogenomics. Mar. Genom. 4, 51–59. doi: 10.1016/j.margen.2010.12.006
Medrano S., Krug P. J., Gosliner T. M., Kumar A. B., and Valdes A. (2019). Systematics of polybranchia pease 1860 (Mollusca: Gastropoda: Sacoglossa) based on molecular and morphological data. Zool. J. Linn. Soc 186, 76–115. doi: 10.1093/zoolinnean/zly050
Neusser T. P., Hanke F., Haszprunar G., and Joerger K. M. (2018). ‘Dorsal vessels’? 3D-reconstruction and ultrastructure of the renopericardial system of Elysia viridis (Montagu 1804) (Gastropoda: Sacoglossa), with a discussion of function and homology. J. Mollus. Stud. 85, 79–91. doi: 10.1093/mollus/eyy049
Nohara K., Takeuchi H., Tsuzaki T., Suzuki N., Tominaga O., and Seikai T. (2009). Genetic variability and stock structure of red tilefish Branchiostegus japonicus inferred from mtDNA sequence analysis. Fish. Sci. 76, 75–81. doi: 10.1007/s12562-009-0188-8
Rauch C., Christa G., Vries J., Woehle C., and Gould S. B. (2017). Mitochondrial Genome Assemblies of Elysia timida and Elysia cornigera and the response of Mitochondrion-Associated Metabolism during Starvation. Genome Biol. Evol. 9, 1873–1879. doi: 10.1093/gbe/evx129
Ravinesh R., Jabir T., Bijukumar A., and Hatha A. A. M. (2014). First record of sea slug Polybranchia orientalis (Sacoglossa: Caliphyllidae) from the south-west coast of India. Mar. Biodivers. Rec. 7, e100. doi: 10.1017/s1755267214000761
Ronquist F., Teslenko M., Mark P., Ayres D. L., Darling A., Hohna S., et al. (2012). Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Vries J. D., Christa G., and Gould S. B. (2014). Plastid survival in the cytosol of animal cells. Trends Plant Sci. 19, 347–350. doi: 10.1016/j.tplants.2014.03.010
Wade R. M. and Sherwood A. R. (2017). Molecular determination of kleptoplast origins from the sea slug Plakobranchus ocellatus (Sacoglossa, Gastropoda) reveals cryptic bryopsidalean (Chlorophyta) diversity in the Hawaiian Islands. J. Phycol. 53, 467–475. doi: 10.1111/jpy.12503
Wade R. M. and Sherwood A. R. (2018). Updating Plakobranchus cf. Ianthobapsus (Gastropoda, Sacoglossa) host use: Diverse algal-animal interactions revealed by NGS with implications for invasive species management. Mol. Phylogenet. Evol. 128, 172–181. doi: 10.1016/j.ympev.2018.07.010
Xiang C. Y., Gao F., Jakovlic I., Lei H. P., Hu Y., Zhang H., et al. (2023). Using Phylosuite for molecular phylogeny and tree-based analyses. IMeta 2, e87. doi: 10.1002/imt2.87
Zhang D., Gao F., Jakovlic I., Zou H., Zhang J., Li W. X., et al. (2019). Phylosuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Notes 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang D., Li W. X., Zou H., Wu S. G., Li M., Jakovlić I., et al. (2018). Mitochondrial genomes of two diplectanids (Platyhelminthes: Monogenea) expose paraphyly of the order Dactylogyridea and extensive tRNA gene rearrangements. Parasite. Vector. 11, 601. doi: 10.1186/s13071-018-3144-6
Zhong D., Zhao G. J., Zhang Z. S., and Xun A. L. (2002). Advance in the entire balance and local unbalance of base distribution in genome. Hereditas 24, 351–355. doi: 10.3321/j.issn:0253-9772.2002.03.033
Zou H., Jakovlić I., Chen R., Zhang D., Zhang J., Li W. X., et al. (2017). The complete mitochondrial genome of parasitic nematode Camallanus cotti: Extreme discontinuity in the rate of mitogenomic architecture evolution within the Chromadorea class. BMC Genomics 18, 840. doi: 10.1186/s12864-017-4237-x
Appendix
Partial chloroplast sequence:
1 TGCTTTGGTA TTTTAATGTT CCCTACATTA TTAACAGCTA CTTCTTGCTT CATTATCGGT
61 TTCATCGCAG CTCCTCCAGT AGATATCGAT GGTATCCGTG AGCCAGTAGC AGGTTCATTA
121 TTATACGGTA ACAACATCAT CACAGGTTCA ATCATCCCTA GTTCAAACGC AATTGGTATG
181 CACTTCTACC CAATCTGGGA AGCAGCTTCT ATCGATGAAT GGTTATACAA CGGTGGTCCT
241 TACCAATTAA TCGTATTACA CTTCTTATTA GGTGTAGCTT CTTACATGGG TCGTGAATGG
301 GAATTATCAT ACCGTTTAGG TATGCGTCCT TGGATCTTCG TAGCATTCTC TGCTCCAGTA
361 GCAGCAGCTT CTGCAGTATT CTTAGTATAC CCAATCGGTC AAGGTTCATT CTCTGATGGT
421 ATGCCTTTAG GTATTTCTGG TACTTTCAAC TTCATGTTAG TATTCCAAGC TGAGCACAAC
481 ATCTTAATGC ACCCATTCCA CATGGCTGGT GTTGCTGGTG TATTCGGTGG TTCTTTATTC
541 TCTGCTATGC ACGGTTCATT AGTAACTTCT TCTTTAATTC GTGAAACAAC TGAAAACGAA
601 TCTACTAACT ACGGTTACAA ATTCGGTCAA GAAGAAGAAA CTTACAACAT CGTTGCAGCT
661 CACGGTTACT TCGGTCGTTT AATCTTCCAA TACGCTTCAT TCAACAACTC TCGTGCTTTA
721 CACTTCTTCT TAGCTTTATG GCCAGTTTTA GGTATCTGGT TAACTGCTAT GGGTATTAGT
781 ACAATGGCGT TCAACTTAAA CGGTTTCAAC TTCAACCAAT CAGTTGTTGA TTCTCAAGG
Keywords: Polybranchia orientalis sp. nov., mitochondrial genome, kleptoplasty, sacoglossa, South China Sea
Citation: Mao M, Wang Y, Jiang J, Tang X and Geng X (2025) Complete mitochondrial genome of Polybranchia orientalis sp. nov. from the South China Sea. Front. Mar. Sci. 12:1650005. doi: 10.3389/fmars.2025.1650005
Received: 21 June 2025; Accepted: 29 September 2025;
Published: 13 October 2025.
Edited by:
Haoya Tong, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Daniel Garcia-Souto, University of Vigo, SpainYundong Li, South China Sea Fisheries Research Institute (CAFS), China
Hee-seung Hwang, Seoul National University, Republic of Korea
Copyright © 2025 Mao, Wang, Jiang, Tang and Geng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jielan Jiang, amllbGFuMDZAZm94bWFpbC5jb20=
 Mingguang Mao1,2,3
Mingguang Mao1,2,3 Yaru Wang
Yaru Wang Jielan Jiang
Jielan Jiang