- 1Department of Convergence Study on the Ocean Science and Technology, Korea Maritime and Ocean University, Busan, Republic of Korea
- 2College of Pharmacy, Yeungnam University, Gyeongsan, Gyeong-buk, Republic of Korea
- 3Research Institute of Cell Culture, Yeungnam University, Gyeongsan, Gyeong-buk, Republic of Korea
The Antarctic marine environment is a promising region for the discovery of extraordinary microbial taxa and the exploration of their applications. This study aimed to investigate the phylogenetic diversity of culturable actinomycetes from deep sea bottom sediment of the Amundsen Sea, Antarctica, and their potential to produce novel metabolites. A total of 24 actinomycete strains were isolated and assigned to the following genera based on the 16S rRNA gene sequence analysis: Blastococcus (3 strains), Corynebacterium (2 strains), Microbacterium (3 strains), Micrococcus (1 strain), Modestobacter (3 strains), Nocardioides (3 strains), Rhodococcus (6 stains), and Streptomyces (3 strains). The 15 strains were selected and chemically investigated. A molecular networking analysis was performed using tandem mass spectrometry (MS/MS) to analyze the metabolites produced by 15 strains. This analysis was used to annotate known compounds and identify potential novel compounds. The molecular network that resulted from this analysis contained 3,702 parent ions. Of these, 1,414 parent ions (38.20%) were identified as genus-specific compounds. Feature-based molecular networking analysis revealed the presence of 269 annotated compounds within the GNPS library. This study demonstrates the diversity of the marine actinomycetes isolated from Amundsen Sea sediment. Additionally, the molecular networking analysis suggests the capacity for the production of biologically active compounds.
1 Introduction
Antarctica is proving to be a promising region for the discovery of extraordinary microbial taxa, attributable to its unique extreme environment. Antarctic microorganisms have evolved unique adaptations in response to extreme environmental conditions, such as low temperatures and relatively scarce nutrients. These conditions contribute to a richer biodiversity and a higher potential for discovering bioactive compounds (Avila, 2016; Medeiros et al., 2025). Despite its potential as a promising region, the Antarctic marine environment has been less studied than the other marine regions due to accessibility challenges. However, some exemplary research has been accomplished in the field of benthic ecosystems through sediment profiles obtained by the box, gravity, and multicore sediment samples. These sediments represent an opportunity to explore the sources of novel bacterial diversity and chemical potential (Millán-Aguiñaga et al., 2019).
Actinomycetes are phylogenetically defined as Gram-positive bacteria with a high guanine-cytosine (G+C) content in their genomic DNA (Gao and Gupta, 2012). Actinomycetes have been widely used as sources of structurally diverse and biologically active secondary metabolites (Genilloud, 2017). Nevertheless, given the recurrent genetic exchange between species from similar ecosystems, the discovery of novel antibiotic molecules from bacteria has decreased in recent years (Núñez-Montero and Barrientos, 2018). Consequently, recent studies have focused on the investigation of actinomycetes from unconventional and isolated environments, such as polar regions, to discover novel bioactive metabolites (Qin et al., 2016). For instance, an Actinomycetes strain Streptomyces sp. INACH3013 was isolated from the terrestrial soil of the Antarctic continent and has been shown to possess the capacity to inhibit the growth of seven Gram-negative and eight Gram-positive foodborne pathogens (Lavin et al., 2016). In a separate study, a total of more than a dozen Antarctic strains were isolated from soil samples collected from Barrientos Island, which showed antimicrobial activity against Candida albicans, Staphylococcus aureus, methicillin-resistant S. aureus, and Pseudomonas aeruginosa (Lee et al., 2012). A similar antimicrobial result was reported for other Nocardioides sp. strains isolated from Antarctic soils (Gesheva and Vasileva-Tonkova, 2012). While these studies demonstrated the pharmacological potential of Antarctic microorganisms, they did not clarify the chemical information on the responsible molecules for the bioactivities.
Tandem mass (MS/MS) data-based molecular networking is widely used, approaching the Global Natural Products Social Molecular Networking (GNPS; https://gnps.ucsd.edu) platform for visualization and annotation of secondary metabolites from crude extracts that have undergone no purification process (Nothias et al., 2017). It has been demonstrated rapid dereplication of compounds in crude extracts. Furthermore, it has been shown to promote the targeted isolation of new structures within the expanding chemical spaces (Purves et al., 2016). Here, we report the phylogenetic tree of the cultured actinomycete strains from deep sea bottom sediment of the Amundsen Sea. In addition, the chemical potential of the microbial strains was investigated using molecular networking, which revealed the metabolite profile.
2 Materials and methods
2.1 Sediment sample collection
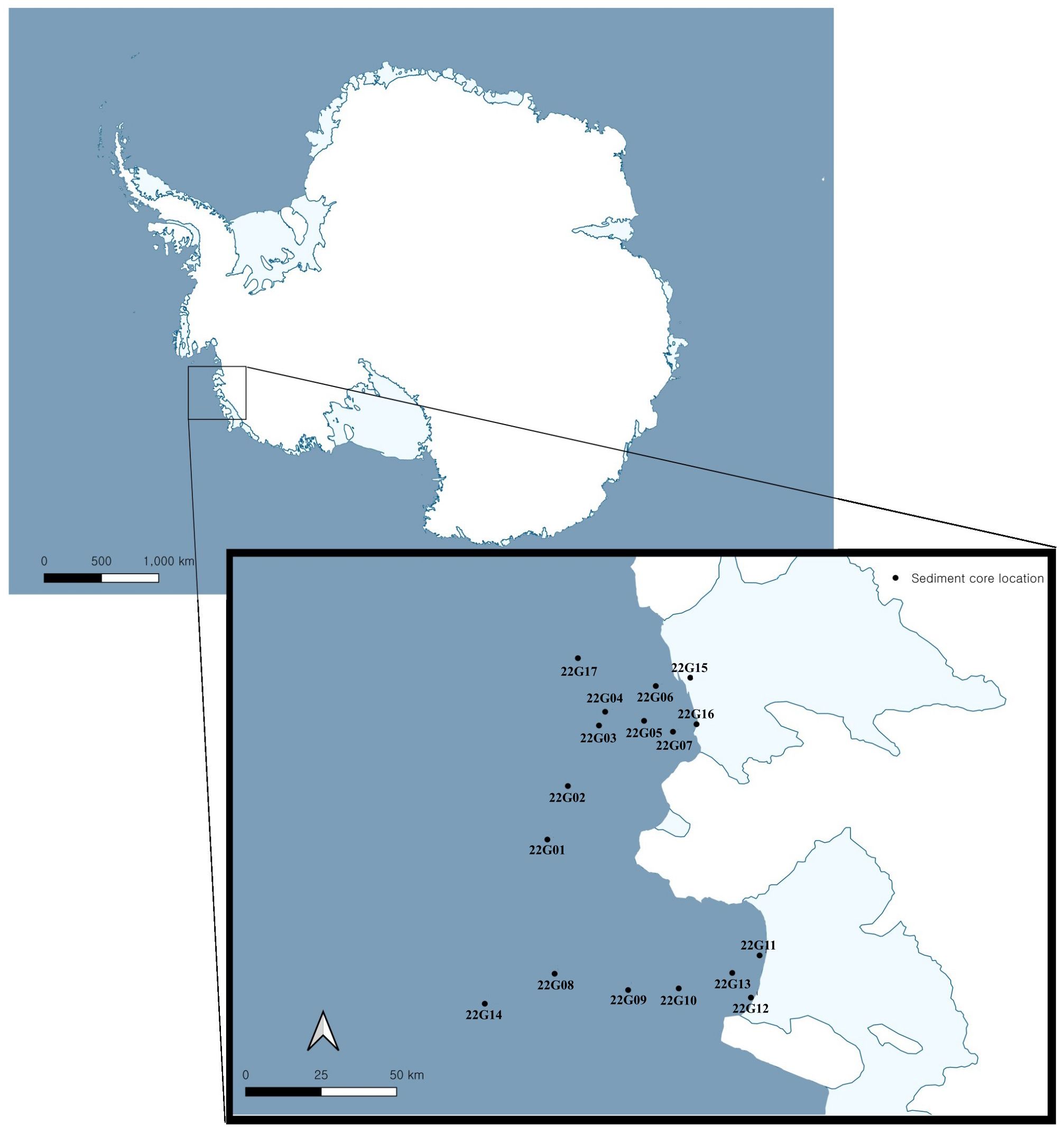
Marine sediment samples were collected from the Amundsen Sea, Antarctica, at a depth range of 694–1249 m using a box corer during the RV ARAON research cruise between January and February of 2022 (ANA12C). At each location, approximately 50 g of muddy sediments were collected aseptically into sterile conical tubes. Some samples were treated on-board for actinomycete isolation, while others were maintained at a temperature of 4°C during a one-month transportation back to the laboratory in Korea.
2.2 Isolation of actinomycetes
The sediment samples were treated immediately after collection. They were pretreated using the stamping technique as follows: sediments were aseptically placed into a sterile petri dish, air-dried in a laminar flow hood, and then heated for 15 min at 57°C. The treated samples were pressed into a sterile form plug, and inoculated onto 1/10 MA (10% Marine broth (2216 Difco, US), 1.8% agar), 1/10 SYPA (0.1% soluble starch, 0.04% yeast extract, 0.02% peptone, and 1.8% agar) and SWA (1.8% agar) media plates by circular stamping, thereby inducing a serial dilution effect (Hameş-Kocabaş and Uzel, 2012). All media were prepared using 75% artificial seawater, which was prepared using commercial sea salt (Blue Treasure SPS Sea Salt, Qingdao Sea-Salt Aquarium Technology Co., Ltd., China). Two isolation methods were employed for the remaining samples moved to the laboratory: serial dilution and the stamping technique. The serial dilution process was carried out as follows: 1 g of sediment sample was added to a sterile 50 mL conical tube containing 9 mL of 1/2 MB (50% Marine broth (2216 Difco, US), 1% NaCl) and shaken at 180 rpm using an orbital shaker at 4°C for 7 days. From these tubes, 1 mL of an aliquot was transferred and mixed with another 9 mL of 1/2 MB to make a 10-2 dilution factor. Following the dilution process, 0.1 mL of each sample was plated onto 1/10 MA, SYPA, and SWA media plates. Some media were supplemented with filter-sterilized cycloheximide (25 μg/mL) after autoclaving. The agar plates were incubated at room temperature (20–25°C) and observed for at least six months. Colonies with suspected actinomycetes morphology were subcultured on SYPA media and incubated at 28°C for 7 days. The pure cultures were preserved with 20% (v/v) glycerol and stored at -80°C.
2.3 16S rRNA gene amplification and phylogenetic analysis
The 16S rRNA gene was amplified by PCR for all strains using universal primers: 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’) and sequenced by BioFact (Daejeon, Korea). PCR was performed in a Hushrun PCR cycler (BioFact, Deajeon, Korea) using an initial denaturation at 95°C for 15 min, followed by 30 cycles at 95°C for 20 sec, 56°C for 30 sec, and 72°C for 5 min. The BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al., 1990) and the web-based tool EZTaxon (https://www.ezbiocloud.net) (Chun et al., 2007) were used to compare the partial 16S rRNA gene sequences with the GenBank database. Sequences were aligned with closely related species, and the phylogenetic tree was constructed using maximum likelihood (ML) methods with a bootstrap analysis of 1,000 replicates using Molecular Evolutionary Genetics Analysis software version 12 (MEGA12) (Kumar et al., 2024).
2.4 Fermentation and extraction
All strains were precultured (28°C, 180 rpm for 7 days) in 15 mL SYP broth (1% soluble starch, 0.4% yeast extract, 0.2% peptone in 75% artificial seawater). The fermentation broth was inoculated into 1 L of SYP broth in a 2.5-L Erlenmeyer flask (1.5% v/v inoculation ratio). Following inoculation at 28°C with shaking at 180 rpm for 7 days, the culture was extracted with 1 L of ethyl acetate (EtOAc). The organic layer was then separated using a separation funnel. The organic phase was concentrated under a vacuum, yielding the crude extract. The crude extract was subjected to open column chromatography on silica gel 60 (Merck, Germany) and eluted with a step gradient of CH2Cl2-MeOH (99:1, 90:10, and 0:100) to yield three fractions. The extracts of most strains were tested for their antibacterial activity against methicillin-resistant S. aureus and Acinetobacter baumannii by determining their growth kinetics (Lee et al., 2025).
2.5 LC-HR-MS/MS analysis of microbial fractions
LC-HR-MS/MS was conducted using a Thermo Fisher Scientific Vanquish UHPLC System integrated with a Thermo Scientific Q Exactive Plus mass spectrometer at the Core Research Support Center for Natural Products and Medicinal Materials (CRCNM). The fractions were prepared at a concentration of 1 mg/mL in methanol for analysis. The chromatographic separation was performed on an Acquity UPLC BEH C18 column (2.1 mm × 100 mm, 130Å, 1.7 μm). The column temperature was set to 25°C. The injection volume was set to 4 μL with a sample concentration of 1 mg/mL, corresponding to an injected amount of 4 μg. The flow rate was maintained at 0.3 mL/min throughout the analysis. The mobile phase consisted of two solvents: solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). A gradient elution was performed starting with 5% solvent B, which was held constant for the first 2 min. The percentage of solvent B was then increased to 20% over the next 6 min and further increased to 100% over the following 6 min, reaching 100% at 14 min. This condition was maintained for 5 min. The system operated in positive ionization mode under the following acquisition parameters: an MS range of m/z 100–1500, an MS2 range of m/z 50–1500. The raw data of LC-HR-MS/MS were processed using MZmine version 4.3.0. The mass detection was performed by setting the noise levels at 2.0E2 for MS and 1.0E1 for MS2. Chromatograms were constructed with a minimum time span of 0.01 min, a minimum height of 3000, and an m/z tolerance of 0.0 (or 20.0 ppm). The chromatographic deconvolution was carried out using a baseline cutoff algorithm with the following parameters: a minimum peak height of 1500, a peak duration range of 0.01–3.00 min, a baseline level of 1000, an m/z range for MS2 scan pairing of 0.02 Da, and a retention time (RT) range for MS2 scan pairing of 0.1 min. Deisotoping was performed with an isotope peak grouper algorithm, using an m/z tolerance of 0.0 (or 20.0 ppm) and an RT tolerance of 0.1 min. Alignment was conducted using the join aligner module, with an m/z tolerance of 0.0 (or 20.0 ppm), an absolute RT tolerance of 0.1 min, a weight for m/z of 70, and a weight for RT of 25. Finally, peaks were gap-filled with a peak finder module, applying an intensity tolerance of 10.0%, an m/z tolerance of 0.0 (or 20.0 ppm), and an absolute RT tolerance of 0.2 min.
2.6 Molecular networking
Feature-based molecular networking (FBMN) was conducted using the GNPS platform (https://gnps.ucsd.edu). The precursor ion and fragment ion tolerances were set to 0.02 Da. The molecular network was generated with a minimum cosine score of 0.7 and at least six matched fragment ions and peaks. MS/MS spectra were filtered to retain the top six peaks within a 50 Da window across each spectrum. Spectral data in the network were matched against the GNPS spectral library. The visualization of the molecular network was performed using Cytoscape version 3.10.2 (https://www.cytoscape.org/). The full FBMN workflow and results are accessible via the GNPS repository under Task ID eed2645813084a80a05f3936bbc76e69.
3 Results and discussion
3.1 Culturable actinomycete diversity and phylogeny
A total of 17 sediment samples were collected from the Amundsen Sea in Antarctica for the purpose of investigating the diversity of actinomycetes. This investigation employed a culture-dependent analytical approach. The latitude, longitude, and sediment depth were recorded (Figure 1; Supplementary Table S1). A total of 24 actinomycete strains were isolated based on actinomycete-like morphology from the sediment samples. Strains were considered distinct if they differed in at least one of the following criteria: location, date of isolation, culture medium, isolation method, or morphological characteristics.
The BLAST analysis of the 16S rRNA gene sequences (1378–1471 bp) from the strains showed 98.57% or higher similarity value with sequences of previously described species. Based on the phylogenetic tree in Figure 2, the actinomycete strains isolated from the Amundsen Sea were classified into five taxonomic orders: Micrococcales (four isolates), Kitasatosporales (three isolates), Propionibacteriales (three isolates), Mycobacteriales (eight isolates), and Geodermatophilales (six isolates). In the order Micrococcales, three isolates (LMA519, LMA520, and LMA521) and one isolate (LMA743) respectively formed different phyletic lineages within the genera Microbacterium and Micrococcus. Three isolates (LMA499, LMA744, and LMA746) formed phyletic lines encompassed by the genus Streptomyces. Three isolates (LMA649, LMA688, and LMA915) were identified as Nocardioides salarius J112 by BLAST analysis with 99.06 to 99.14% similarity. Eight isolates (LMA660, LMA661, LMA648, LMA686, LMA741, LMA742, LMA745, and LMA916) formed diverse phylogenetic lines within the evolutionary radiation encompassed by the two families Corynebacteriaceae and Nocardiaceae. The isolates LMA660 and LMA661 showed a low level of similarity to the known species Corynebacterium pilbarense IMMIB WACC-658 with similarity percentages of 98.63% and 98.70%, respectively. In the order Geodermatophilales, six isolates (LMA501, LMA503, LMA687, LMA689, LMA690, and LMA917) formed different phyletic lineages within the genera Blastococcus and Modestobacter. The strains LMA503, LMA687, and LMA917 were assigned to the genus Blastococcus. LMA503 showed a low level of similarity to the known species Blastococcus deserti SYSU D8006, with a similarity percentage of 98.57%. LMA687 and LMA917 exhibited a 98.77% and 98.71% similarity, respectively, to the known species Blastococcus litoris GP-S2-8 and Blastococcus aggregatus DSM 4725. The isolates LMA501, LMA689, and LMA690 were identified as belonging to the genus Modestobacter (Modestobacter marinus 42H12-1). All partial 16S rRNA gene sequences were submitted to GenBank. The accession numbers obtained are listed in Table 1.
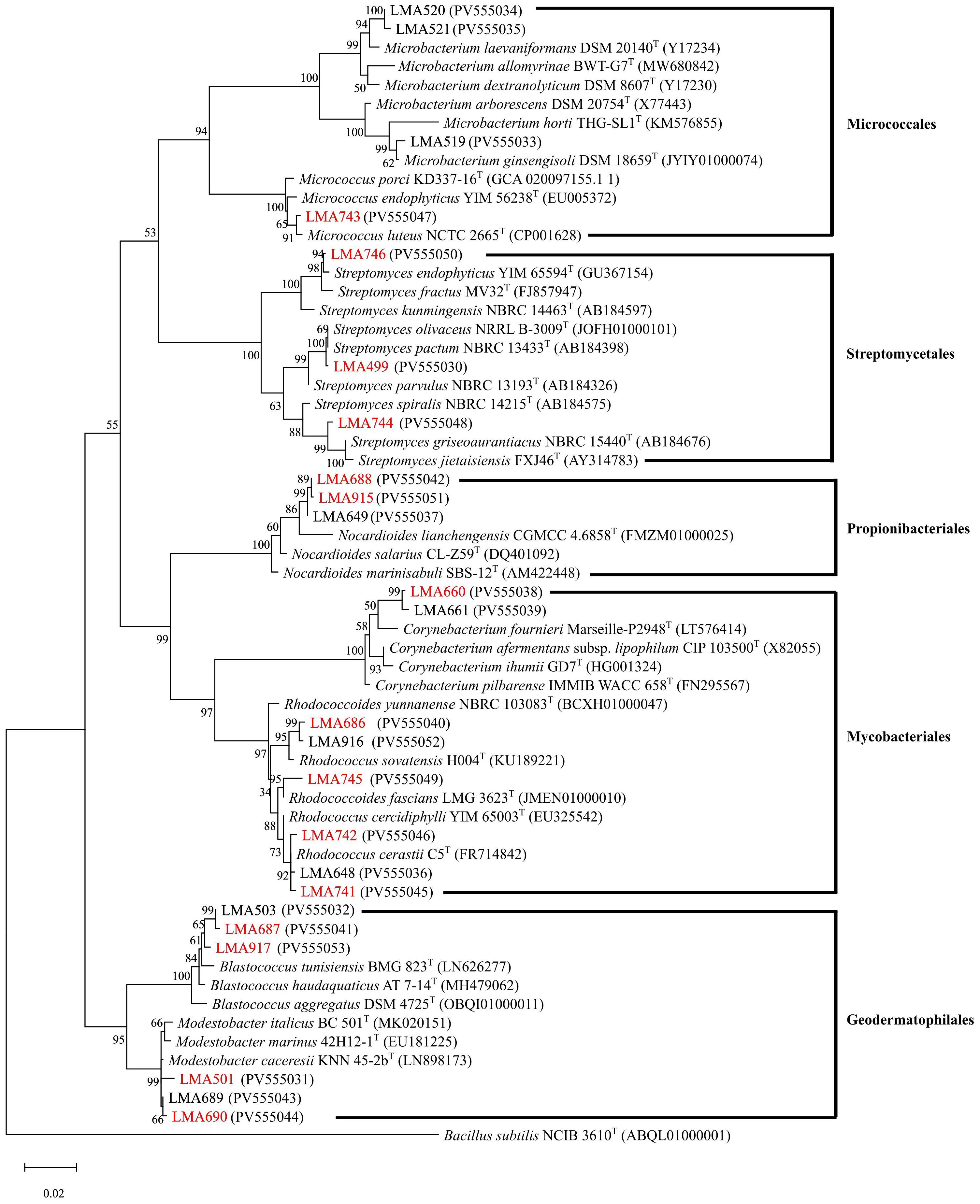
Figure 2. Maximum likelihood phylogenetic tree based on the alignment of partial 16S rRNA sequences showing relationships among the isolates. The phylogeny was inferred using the Maximum Likelihood method and the Tamura-Nei model (Tamura and Nei, 1993). The percentage of replicate trees on which the associated taxa clustered together (1,000 replicates) is shown next to the branches. GenBank accession numbers are shown in parentheses. The tree was rooted to the outgroup Bacillus subtilis NCIB 3610T. The scale bar represents 0.02 substitutions per nucleotide position.
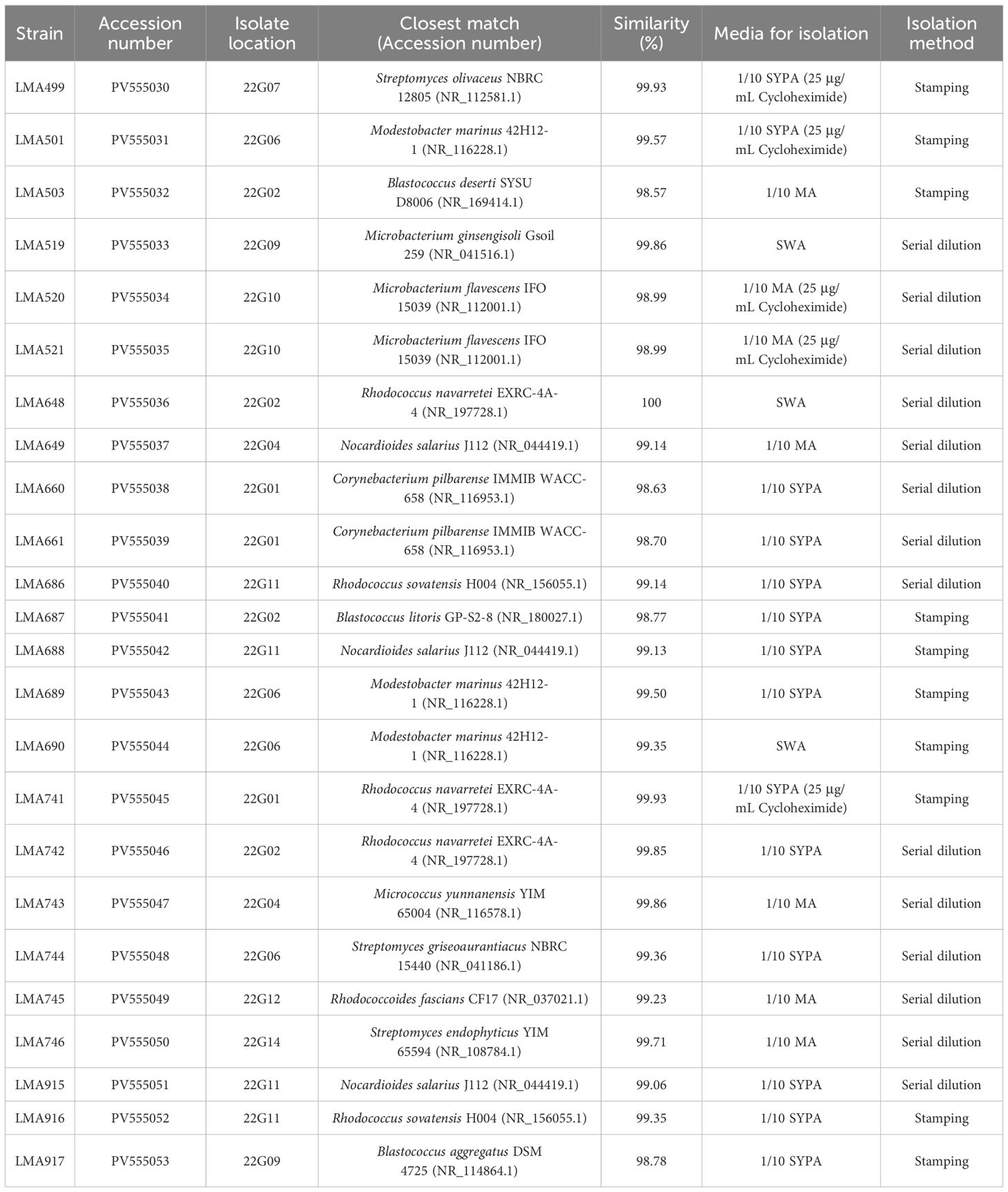
Table 1. Identification of total isolates from the Amundsen Sea, Antarctica, based on 16S rRNA gene sequence similarity and summary of isolation methods.
3.2 Specialized metabolite potential
Fifteen strains (designated with red names in Figure 2) were selected for metabolite analysis based on their phylogenetic relationships. The selected strains belonged to the genera Rhodococcus, Corynebacterium, Nocardioides, Blastococcus, Modestobacter, Micrococcus, and Streptomyces. The extraction yields from 1 L cultures varied significantly among different genera, ranging from 31.2 mg to 73.9 mg. Approximately 70 mg of extract was obtained from strains belonging to the genus Streptomyces, 50 mg from Rhodococcus and Blastococcus strains, and around 30 mg from Modestobacter, Corynebacterium, Nocardioides, and Micrococcus strains following EtOAc extraction. The fermentative extracts were tested for bioactivity against both Gram-positive and Gram-negative bacterial pathogens. Nevertheless, the results obtained did not reveal any significant antibiotic activity (data not shown).
The antibacterial activity test was conducted at the preliminary screening stage using unpurified crude extracts and only two representative strains. To overcome these limitations and further explore the metabolic potential of the isolates, a molecular networking analysis was performed which yielded 3,702 parent ions (nodes) when the MS/MS data were analyzed by bacterial genus, as shown in Figure 3. The production of parent ions, with the exclusion of media-derived components, showed a mix of genus-specific and shared patterns (Figure 4A). This specificity highlights the potential for discovering bioactive metabolites, as each strain contributes a unique chemical diversity. Within the molecular network, 1,414 parent ions (38.20%) were identified as unique to an individual genus. Remarkably, 203 metabolites (5.48% of the total ions) were specific to Rhodococcus, 104 metabolites (3.81%) to Corynebacterium, 133 metabolites (3.59%) to Nocardioides, 55 metabolites (1.49%) to Blastococcus, 52 metabolites (1.40%) to Modestobacter, 56 metabolites (1.51%) to Micrococcus, and 811 metabolites (21.91%) to Streptomyces (Figure 4A). Interestingly, the majority of detected ions (2,288 ions, 61.80%) were shared across isolates from multiple genera, indicating the presence of broadly conserved or widely distributed metabolic features.

Figure 3. The resulting molecular network consisted of 3,702 parent ions. Feature-based molecular networking (FBMN) analysis revealed that 269 compounds were annotated in the GNPS spectral library. Node colors indicate annotation status: black nodes represent features with a match to a known compound in the GNPS library (putatively annotated), while blue nodes represent features with no match (unknown or potentially novel compounds).
In the FBMN analysis, a total of 3,702 nodes were detected, among which 269 compounds were successfully annotated in the GNPS library (Figures 4B–D). The remaining 3,433 nodes (92.73%) were unannotated, highlighting the extensive presence of yet-to-be-identified metabolites. The analysis revealed that several bacterial genera share common classes of compounds, highlighting conserved biosynthetic pathways across diverse species. For instance, cyclic dipeptides (diketopiperazines) such as cyclo(l-Val-l-Pro) and cyclo(l-Leu-l-Pro) were found in Rhodococcus, Corynebacterium, Nocardioides, Blastococcus, Modestobacter, Micrococcus, and Streptomyces. These compounds are known for their bioactive properties, such as antifungal effects against Fusarium sp. and Aspergillus sp (Soldatou et al., 2021), antibacterial activity against S. aureus and Bacillus subtilis (El-Gendy and Rateb, 2015), and vancomycin-resistant Enterococcus (Rhee, 2002), as well as quorum-sensing modulation (Holden et al., 1999). Their occurrence across the genus suggests a fundamental role in bacterial metabolism. Similarly, fatty acid amides, including palmitamide and 9-octadecenamide, were detected in Rhodococcus, Corynebacterium, Nocardioides, Blastococcus, Modestobacter, Micrococcus, and Streptomyces. Indole derivatives (particularly indole-3-acetic acid and indoxyl sulfate) and adenine derivatives were found to be prevalent in genera such as Rhodococcus, Corynebacterium, Nocardioides, and Blastococcus. Fatty acids and their esters, including linoleic acid and sorbitane monopalmitate, were also commonly detected, indicating the role of lipid metabolism across these bacterial genera.
In addition to the common compound classes, we identified several genus-specific compounds that further differentiate the metabolic profiles of each bacterial genus. For example, Rhodococcus exhibited a wide range of diketopiperazines, indole derivatives, and polyphenols, along with the unique presence of β-caryophyllene oxide and monactin. The detection of β-caryophyllene oxide suggests its potential role in biotechnological applications, highlighting its importance in microbial secondary metabolism. The monactins, known family of macrocyclic antibiotics, were found in a Figure 4B cluster, which suggests that Rhodococcus species may possess antimicrobial properties (Phillies et al., 1975).
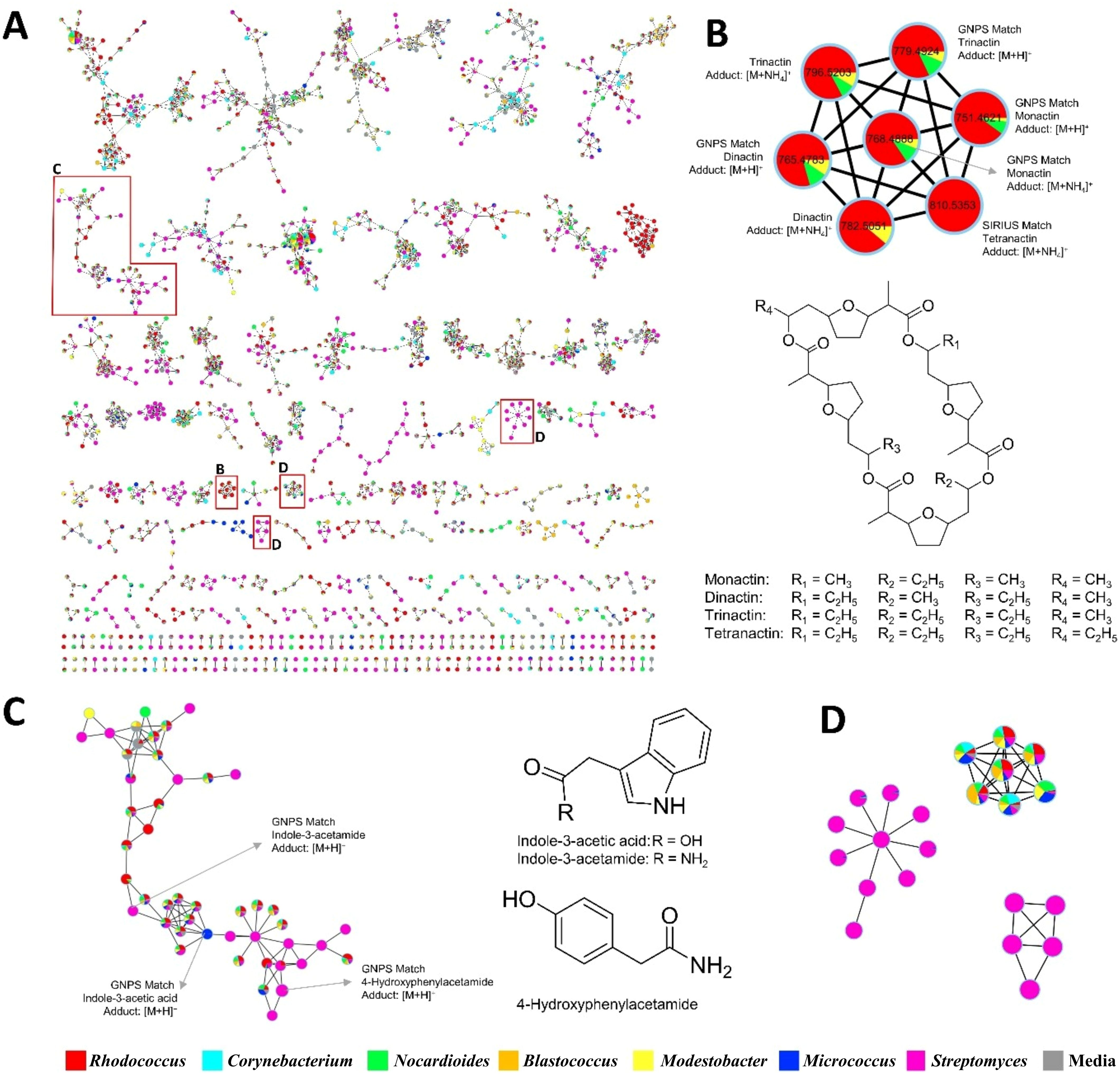
Figure 4. Several important clusters are zoomed in to highlight: (A) Genus-based visualization of the molecular network from Antarctic actinomycete strains, (B) clusters with newly found compounds in the genus, (C) clusters with both GNPS library matches and unannotated features, (D) clusters with no spectral matches, potentially representing new type of compounds. These zoomed views emphasize the diversity and taxonomic specificity revealed by the FBMN analysis.
Corynebacterium strains shared many compound classes with Rhodococcus, but they were distinguished by the presence of biotin methyl ester, phenacetin, and deoxycholate, which may serve specific functions in metabolism or stress responses. Additionally, fatty acid derivatives such as palmitamide, 9Z-octadecenamide, 13Z-docosenamide, and linoleic acid further corroborate the unique fatty acid synthase system in Corynebacterium. This system produces coenzyme A (CoA)-linked fatty acids instead of acyl carrier protein (ACP)-linked intermediates (Takeno et al., 2013). The presence of biotin methyl ester and lumichrome indicates the potential for vitamin biosynthesis, consistent with findings from metabolic engineering studies (Feierabend et al., 2021).
Conversely, Nocardiodes strains were enriched in aromatic amines such as anthranilate and 4-hydroxyphenylacetamide. These compounds are often associated with secondary metabolic processes and may play a role in bacterial survival under certain environmental conditions. A range of fatty acid derivatives including linoleic acid, palmitamide, palmitoyl ethanolamide, 13Z-docosenamide, and 9Z-octadecenamide, have been identified, aligning with the known fatty acid composition of this genus (Yoon et al., 2005; Mitzscherling et al., 2022). The detection of dipeptides (Val-Leu, Phe-Pro, His-Pro, Phe-Glu) and cyclic dipeptides [e.g., cyclo(l-Val-l-Pro), cyclo(l-Leu-l-Pro)] supports the presence of peptidoglycan-derived amino acids, which are integral to the cell walls of Nocardioides species. Additionally, the presence of adenosine and 5’-deoxyadenosine aligns with the high G+C content characteristic of Nocardioides genomes (Yoon et al., 2005).
In Blastococcus strains, they showed a distinct profile dominated by histamine-based lipids and bioactive compounds such as Denatonium and Nerolidol, which suggests potential antimicrobial and bioactive properties (Hezbri et al., 2024; Urzì et al., 2004). The detection of fatty acids such as linoleic acid, oleic acid, and 12(13)-epoxy-9Z-octadecenoic acid further aligns with the fatty acid profile of Blastococcus, which predominantly includes isopalmitic acid (iso-C16:0), cis-palmitoleic acid (iso-C16:1 H), and cis-10-heptadecenoic acid (C17:1 ω8c) (Hezbri et al., 2024; Urzì et al., 2004). Furthermore, palmitamide and 9Z-octadecenamide are amide derivatives that are structurally related to the polar lipids in Blastococcus, such as phosphatidylcholine and phosphatidylethanolamine (Montero-Calasanz et al., 2022; Yang et al., 2019).
Modestobacter strains shared some similarities with Blastococcus, because of the production of indigo red and deoxycholate. Metabolite profiling of Modestobacter strains identified palmitamide and octadecanamide, both fatty acid amides, consistent with the presence of iso-C16:0 and cis-9-octadecenoic acid (C18:1 ω9c) fatty acids in Modestobacter species. Additionally, phospholipid-related metabolites such as lauryldiethanolamine and N,N-dimethyldodecylamine N-oxide were observed, which may correspond to the diverse polar lipids reported in Modestobacter species, including phosphatidylethanolamine and phosphatidylinositol (Jiang et al., 2021). The identification of adenosine and N6-(Δ2-isopentenyl)-adenine also supports the nucleotide-derived metabolic profile of Modestobacter, which has been implicated in menaquinone biosynthesis (Jiang et al., 2021).
Micrococcus strains produced unique compounds such as carbazole, a nitrogen-containing polycyclic aromatic compound, and tris(2-butoxyethyl) phosphate. These compounds may offer insights into the ecological adaptability of Micrococcus strains and their potential industrial applications. Metabolites such as cyclo(l-Val-l-Pro), Phe-Pro, cyclo(l-Leu-l-Pro), N-acetyl-tyramine, lumichrome, xanthine, and indole-3-acetate were identified. These compounds support the production of bioactive peptides and alkaloids in the genus Micrococcus, including thiazolyl peptide antibiotics (e.g., micrococcin and kocurin) and carotenoid-derived pigments with antimicrobial properties (Tizabi and Hill, 2023). The identification of pyrrolo[1,2-a]pyrazine-1,4-dione, an alkaloid-related compound, aligns with the genus’s ability to produce bioactive nitrogen-containing metabolites (Tizabi and Hill, 2023).
Finally, Streptomyces were found to produce a diverse array of secondary metabolites, including cyclo(l-Val-l-Pro), Phe-Pro, His-Pro, Val-Leu, and Leu-Gly, which are commonly associated with antimicrobial and signaling activities in actinomycetes. Furthermore, the presence of nerolidol, a sesquiterpene alcohol, suggests the potential for further antimicrobial activity in these strains (Krist et al., 2015). The detection of linoleic acid and 9,12-octadecadiynoic acid, both with cytotoxic and antimicrobial properties, further underscores the adaptability of Streptomyces in extreme environments (Pacios-Michelena et al., 2021).
4 Conclusion
The actinomycete strains, which were isolated from the Amundsen Sea in Antarctica, were clustered into 5 different groups based on the phylogenetic analysis of the 16S rRNA gene sequences. They showed 98.57%−100% similarity with sequences of previously described species, including the genera Microbacterium, Micrococcus, Streptomyces, Nocardioides, Corynebacterium, Rhodococcus, Blastococcus, and Modestobacter. A total of 87.5% of the isolated strains were taxonomically assigned to rare actinomycete genera, thereby representing phylogenetic diversity. Culturable rare actinomycetes are generally less isolated by conventional methods and less studied than the genus Streptomyces (Subramani and Aalbersberg, 2013). This is due to their slow growth in laboratory environments and the lack of selective isolation methods (Parra et al., 2023). This study employed six types of isolation medium with two different loading methods. Additionally, prepared agar media plates were observed for a long time period, close to a year. The impact of these variations on the process of microbial isolation remains to be elucidated. However, these various attempts have resulted in the emergence of diversity among the isolated strains, indicating the necessity for additional trials. Additionally, unexplored environments in extreme conditions, including polar ecosystems, are promising sources of rare actinomycetes (Millán-Aguiñaga et al., 2019; Soldatou et al., 2021). The Amundsen Sea is a previously untapped region that has largely remained unexplored in terms of its microbial diversity. Due to its extreme environment and the lack of research, it is expected to be inhabited by unexplored actinomycetes.
In addition, several isolates exhibited 16S rRNA gene sequence similarity lower than 98.7% with the reported strain of their respective genera. Sequence analysis identified LMA503 with 98.57% 16S rRNA gene similarity with Blastococcus deserti. The other isolated strains, LMA660 and LMA661, were identified as Corynebacterium pilbarense with 98.63% and 98.70% similarity, respectively, with the reported strain. Recently, the threshold for bacterial species classification has been set at 16S rRNA gene sequence similarity values of less than 98.7% (Chun et al., 2018).
Molecular networking enables the timely, efficient chemical dereplication and the discovery of new metabolites when dealing with complex mixtures, such as bacterial culture extracts. In this study, a total of 15 strains were selected for chemical investigation. The GNPS platform was used to build a molecular network of 3,702 parent ions based on bacterial genus. In addition to the common classes of compounds, we identified several genus-specific compounds that further distinguish the metabolic profiles of each genus. This specificity, with each strain contributing a proportion of unique chemical diversity, highlights the potential for the discovery of bioactive metabolites. Furthermore, among the 3,702 ions, 269 compounds were successfully annotated in the GNPS library. The unannotated metabolites support the idea that Antarctic actinomycetes have potential for chemical diversity and the discovery of new specialized metabolites (Figures 4C, D).
In conclusion, we have demonstrated much untapped potential in Antarctic marine environments as rich sources of both novel species and rare genera of actinomycetes. Further research is necessary to characterize the genomic features of these strains and confirm their exact taxonomic position. In addition, molecular networking has been demonstrated to be a valuable tool for matching identical and related molecules, allowing the identification of metabolite patterns for each genus. The findings of this study have provided a prioritized list of strains for future natural product discovery based on their metabolic uniqueness and taxonomic novelty.
Data availability statement
The datasets generated for this study can be found in online repositories. The names of the repository and accession number can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PV555030–PV555053; https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=eed2645813084a80a05f3936bbc76e69.
Author contributions
JL: Formal Analysis, Data curation, Investigation, Writing – original draft, Writing – review & editing, Methodology. VS: Visualization, Methodology, Investigation, Formal Analysis, Writing – review & editing, Writing – original draft, Data curation. SK: Writing – original draft, Formal Analysis, Data curation. HC: Conceptualization, Writing – review & editing, Funding acquisition, Supervision, Resources, Project administration. IY: Supervision, Conceptualization, Writing – review & editing, Resources, Funding acquisition, Project administration.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by a grant from the National Research Foundation of Korea (RS-2022-NR071935 and 2021-KS211525 for I.H) funded by the Korean government. On-board research opportunity was supported by Korea Polar Research Institute (KOPRI) grant funded by the Ministry of Oceans and Fisheries (KOPRI PE21900). Also, this study was supported by 2022 Yeungnam University Research Grant funded by Yeungnam University (for H.C).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript to check grammatical errors and polish the sentences.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1650636/full#supplementary-material
References
Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Avila C. (2016). Biological and chemical diversity in Antarctica: from new species to new natural products. Biodiversity 17, 5–11. doi: 10.1080/14888386.2016.1176957
Chun J., Lee J. H., Jung Y., Kim M., Kim S., Kim B. K., et al. (2007). EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261. doi: 10.1099/ijs.0.64915-0
Chun J., Oren A., Ventosa A., Christensen H., Arahal D. R., da Costa M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
El-Gendy B. E. D. M. and Rateb M. E. (2015). Antibacterial activity of diketopiperazines isolated from a marine fungus using t-butoxycarbonyl group as a simple tool for purification. Bioorganic Med. Chem. Lett. 25, 3125–3128. doi: 10.1016/j.bmcl.2015.06.010
Feierabend M., Renz A., Zelle E., Nöh K., Wiechert W., and Dräger A. (2021). High-quality genome-scale reconstruction of corynebacterium glutamicum ATCC 13032. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.750206
Gao B. and Gupta R. S. (2012). Phylogenetic framework and molecular signatures for the main clades of the phylum actinobacteria. Microbiol. Mol. Biol. Rev. 76, 66–112. doi: 10.1128/mmbr.05011-11
Genilloud O. (2017). Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 34, 1203–1232. doi: 10.1039/c7np00026j
Gesheva V. and Vasileva-Tonkova E. (2012). Production of enzymes and antimicrobial compounds by halophilic Antarctic Nocardioides sp. grown on different carbon sources. World J. Microbiol. Biotechnol. 28, 2069–2076. doi: 10.1007/s11274-012-1009-2
Hameş-Kocabaş E. E. and Uzel A. (2012). Isolation strategies of marine-derived actinomycetes from sponge and sediment samples. J. Microbiol. Methods 88, 342–347. doi: 10.1016/j.mimet.2012.01.010
Hezbri K., Kammoun I., Sbissi I., Klenk H. P., Montero-Calasanz M., del C., et al. (2024). Blastococcus brunescens sp. nov., a member of the Geodermatophilaceae isolated from sandstone collected from the Sahara Desert in Tunisia. Int. J. Syst. Evol. Microbiol. 74, 1–9. doi: 10.1099/ijsem.0.006317
Holden M. T. G., Chhabra S. R., De Nys R., Stead P., Bainton N. J., Hill P. J., et al. (1999). Quorum-sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 33, 1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x
Jiang Z. M., Zhang B. H., Sun H. M., Zhang T., Yu L. Y., and Zhang Y. Q. (2021). Properties of Modestobacter deserti sp. nov., a Kind of Novel Phosphate-Solubilizing Actinobacteria Inhabited in the Desert Biological Soil Crusts. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.742798
Krist S., Banovac D., Tabanca N., Wedge D. E., Gochev V. K., Wanner J., et al. (2015). Antimicrobial activity of nerolidol and its derivatives against airborne microbes and further biological activities. Nat. Prod. Commun. 10, 143–148. doi: 10.1177/1934578x1501000133
Kumar S., Stecher G., Suleski M., Sanderford M., Sharma S., and Tamura K. (2024). MEGA12: Molecular Evolutionary Genetic Analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 41, 1–9. doi: 10.1093/molbev/msae263
Lavin P. L., Yong S. T., Wong C. M. V. L., and De Stefano M. (2016). Isolation and characterization of Antarctic psychrotroph Streptomyces sp. strain INACH3013. Antarct. Sci. 28, 433–442. doi: 10.1017/S0954102016000250
Lee J., Shin H., Kim H. S., Park J. W., Akinniyi G., Kang S., et al. (2025). Diversity and bioactivities of ulleung island-derived marine actinomycetes. Ocean Sci. J. 60, 1–12. doi: 10.1007/s12601-024-00192-9
Lee L. H., Cheah Y. K., Sidik S. M., Mutalib N. S. A., Tang Y. L., Lin H. P., et al. (2012). Molecular characterization of Antarctic actinobacteria and screening for antimicrobial metabolite production. World J. Microbiol. Biotechnol. 28, 2125–2137. doi: 10.1007/s11274-012-1018-1
Medeiros W., Kralova S., Oliveira V., Ziemert N., and Sehnal L. (2025). Antarctic bacterial natural products: from genomic insights to drug discovery. Nat. Prod. Rep. 42, 774–787. doi: 10.1039/d4np00045e
Millán-Aguiñaga N., Soldatou S., Brozio S., Munnoch J. T., Howe J., Hoskisson P. A., et al. (2019). Awakening ancient polar actinobacteria: Diversity, evolution and specialized metabolite potential. Microbiol. (United Kingdom) 165, 1169–1180. doi: 10.1099/mic.0.000845
Mitzscherling J., Maclean J., Lipus D., Bartholomäus A., Mangelsdorf K., Lipski A., et al. (2022). Nocardioides alcanivorans sp. nov., a novel hexadecane-degrading species isolated from plastic waste. Int. J. Syst. Evol. Microbiol. 72, 1–11. doi: 10.1099/ijsem.0.005319
Montero-Calasanz M., del C., Yaramis A., Rohde M., Schumann P., Klenk H. P., et al. (2022). Genotype–phenotype correlations within the Geodermatophilaceae. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.975365
Nothias L. F., Petras D., Schmid R., Dührkop K., Rainer J., Sarvepalli A., et al. (2017). Feature-based molecular networking in the GNPS analysis environment. Physiol. Behav. 176, 100–106. doi: 10.1177/0022146515594631.Marriage
Núñez-Montero K. and Barrientos L. (2018). Advances in antarctic research for antimicrobial discovery: A comprehensive narrative review of bacteria from antarctic environments as potential sources of novel antibiotic compounds against human pathogens and microorganisms of industrial importance. Antibiotics 7 (4), 90. doi: 10.3390/antibiotics7040090
Pacios-Michelena S., Aguilar González C. N., Alvarez-Perez O. B., Rodriguez-Herrera R., Chávez-González M., Arredondo Valdés R., et al. (2021). Application of streptomyces antimicrobial compounds for the control of phytopathogens. Front. Sustain. Food Syst. 5. doi: 10.3389/fsufs.2021.696518
Parra J., Beaton A., Seipke R. F., Wilkinson B., Hutchings M. I., and Duncan K. R. (2023). Antibiotics from rare actinomycetes, beyond the genus Streptomyces. Curr. Opin. Microbiol. 76, 102385. doi: 10.1016/j.mib.2023.102385
Phillies G. D. J., Asher I. M., and Eugene H. (1975). Nonactin, monactin, dinactin, trinactin, and tetranactin. BIOPOLY MERS 14, 2311–2327. doi: 10.1002/bip.1975.360141107
Purves K., Macintyre L., Brennan D., Hreggviðsson G., Kuttner E., Ásgeirsdóttir M. E., et al. (2016). Using molecular networking for microbial secondary metabolite bioprospecting. Metabolites 6 (1), 2. doi: 10.3390/metabo6010002
Qin S., Li W. J., Dastager S. G., and Hozzein W. N. (2016). Editorial: Actinobacteria in special and extreme habitats: Diversity, function roles, and environmental adaptations. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01415
Rhee K. H. (2002). Isolation and characterization of Streptomyces sp. KH-614 producing anti-VRE (vancomycin-resistant enterococci) antibiotics. J. Gen. Appl. Microbiol. 48, 321–327. doi: 10.2323/jgam.48.321
Soldatou S., Eldjárn G. H., Ramsay A., van der Hooft J. J. J., Hughes A. H., Rogers S., et al. (2021). Comparative metabologenomics analysis of polar actinomycetes. Mar. Drugs 19, 1–21. doi: 10.3390/MD19020103
Subramani R. and Aalbersberg W. (2013). Culturable rare Actinomycetes: Diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 97, 9291–9321. doi: 10.1007/s00253-013-5229-7
Takeno S., Takasaki M., Urabayashi A., Mimura A., Muramatsu T., Mitsuhashi S., et al. (2013). Development of fatty acid-producing Corynebacterium glutamicum strains. Appl. Environ. Microbiol. 79, 6775–6783. doi: 10.1128/AEM.02003-13
Tamura K. and Nei M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Tizabi D. and Hill R. T. (2023). Micrococcus spp. as a promising source for drug discovery: A review. J. Ind. Microbiol. Biotechnol. 50 (1), kuad017. doi: 10.1093/jimb/kuad017
Urzì C., Salamone P., Schumann P., Rohde M., and Stackebrandt E. (2004). Blastococcus saxobsidens sp. nov., and emended descriptions of the genus Blastococcus Ahrens and Moll 1970 and Blastococcus aggregatus Ahrens and Moll 1970. Int. J. Syst. Evol. Microbiol. 54, 253–259. doi: 10.1099/ijs.0.02745-0
Yang Z. W., Asem M. D., Li X., Li L. Y., Salam N., Alkhalifah D. H. M., et al. (2019). Blastococcus deserti sp. nov., isolated from a desert sample. Arch. Microbiol. 201, 193–198. doi: 10.1007/s00203-018-1604-1
Keywords: Antarctica, marine actinomycetes, phylogenetic diversity, molecular networking, secondary metabolites
Citation: Lee J, Silviani V, Kang S, Choi H and Yang I (2025) Diversity and metabolic potential of culturable marine actinomycetes from the sediment of Amundsen Sea, Antarctica. Front. Mar. Sci. 12:1650636. doi: 10.3389/fmars.2025.1650636
Received: 20 June 2025; Accepted: 30 July 2025;
Published: 18 August 2025.
Edited by:
Santhiyagu Prakash, Tamil Nadu Fisheries University, IndiaReviewed by:
Manikandan Gurusamy, Tshwane University of Technology, South AfricaAna Catarina Fonseca, University of Porto, Portugal
Copyright © 2025 Lee, Silviani, Kang, Choi and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyukjae Choi, aDVjaG9pQHl1LmFjLmty; Inho Yang, aWh5YW5nQGttb3UuYWMua3I=
†These authors have contributed equally to this work
 Jeonghee Lee
Jeonghee Lee Velina Silviani
Velina Silviani Soojin Kang
Soojin Kang Hyukjae Choi
Hyukjae Choi Inho Yang
Inho Yang