- 1Department of Medicinal Chemistry, University of Utah, Salt Lake City, UT, United States
- 2School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, GA, United States
- 3Scripps Institution of Oceanography, University of California, San Diego, San Diego, CA, United States
- 4Parker H. Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA, United States
- 5School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, United States
Halogenated molecules produced by marine algae are thought to be defensive secondary metabolites. The extraordinarily high concentration of bromoform in the seaweed Asparagopsis—up to 8% dry tissue weight—challenges the exclusivity of this paradigm. In this report, we provide evidence that the mbb1 gene which encodes the bromoform producing halogenase is among the most highly transcribed genes in Asparagopsis tissue, with the resulting Mbb1 protein abundance rivaling that of enzymes involved in photosynthesis and carbon fixation. When the seaweed was stressed with light, transcripts for both mbb1 and for proteins involved in photosynthesis were significantly downregulated. Conversely, heat stress modestly upregulated some photosynthesis genes but had no impact on mbb1. Taken together, these findings allow us to posit that bromoform production is not solely a stress-response or self-defense mechanism for A. taxiformis. Instead, we propose that the halogenase Mbb1 likely fulfils a primary metabolic function in this red alga thusly reconceptualizing halogenation biochemistry and pulling it out of the domain of natural product biosynthesis alone.
Introduction
Halogenation of biomolecules is often a bioactivity-defining modification. In addition to numerous fluorinated pharmacophores, aryl chlorination confers antibiotic activity to vancomycin, tyrosine bromination controls Drosophila spermatogenesis, and iodination is critical to the activity of the thyroid hormone (Bianco et al., 2002; Pinchman and Boger, 2013a; Pinchman and Boger, 2013b; Gillis et al., 2015; Su et al., 2024). Halogenating enzymes—halogenases—are considered to be highly specialized secondary metabolic enzymes (Agarwal et al., 2017). The discovery, biochemical characterization, and biotechnological application of halogenases has traditionally involved their study in natural product biosynthetic schema (Agarwal et al., 2017).
As the largest reservoirs of halides, the world’s oceans shape the global halogen cycle. A large fraction of atmospheric halogen flux is derived from halogenated methanes naturally produced by marine algae (Saiz-Lopez and von Glasow, 2012). Just like anthropogenic chlorofluorocarbons, ocean-derived halomethanes degrade the ozone layer. Among these, up to 30% of ozone depletion is attributed to bromoform (CHBr3) that is produced by marine biota (Quack et al., 2004; Salawitch, 2006; Navarro et al., 2015). Seaweeds Asparagopsis taxiformis and Asparagopsis armata are prolific bromoform producers and bromoform has been proposed to protect these seaweeds against biofouling (Figure 1A) (Paul et al., 2006a). Contrary to traditional description of molecules that mediate ecological interactions and are often produced in small quantities, the concentration of bromoform in Asparagopsis is exceptionally high with up to 8% Asparagopsis tissue dry weight being bromoform (Gribble, 2000). Asparagopsis is attracting attention as a cattle feed additive; supplementing cattle feed with Asparagopsis reduces emission of the greenhouse gas methane from livestock (Roque et al., 2021; Wasson et al., 2022). A. taxiformis is also a traditional Hawaiian food, known as limu kohu or the superior seaweed (Gribble, 2000).
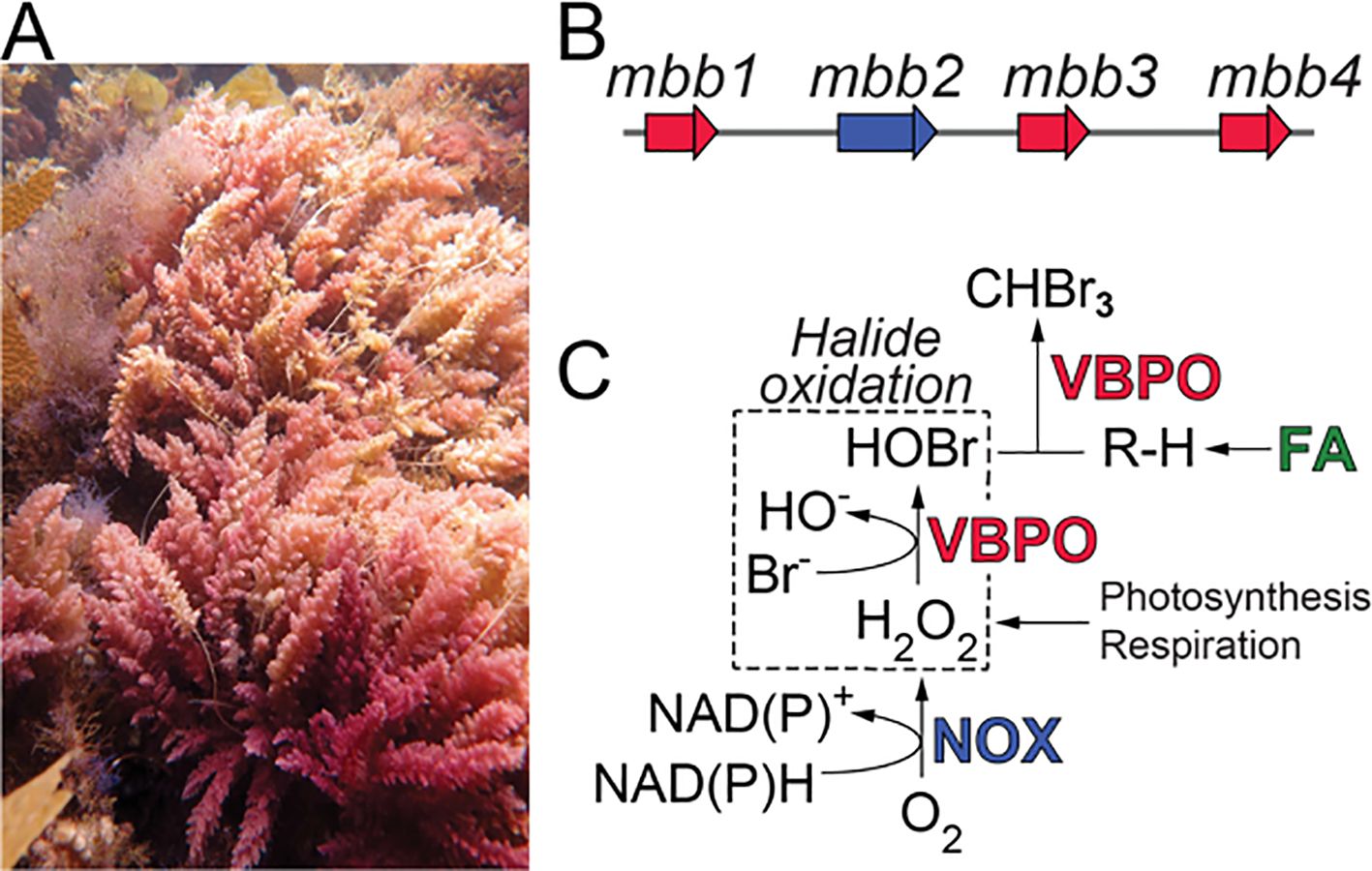
Figure 1. (A) A. taxiformis gametophytes from the Santa Catalina Island, CA. (B) The mbb gene locus. (C) The NOX Mbb2 generates hydrogen peroxide (H2O2) by reduction of molecular oxygen with concomitant oxidation of NAD(P)H. Hydrogen peroxide can also be generated by electron transport during photosynthesis and respiration and is used as an oxidant by VBPOs to catalyze bromide oxidation. A hydrocarbon substrate (R–H) is furnished by the fatty acid (FA) biosynthetic pathway for bromoform biosynthesis.
We have recently reported the genetic and enzymatic route for bromoform biosynthesis in A. taxiformis (Thapa et al., 2020). The marine bromoform biosynthesis (mbb) gene locus in A. taxiformis encodes three vanadium-dependent bromoperoxidases (VBPOs; Mbb1, Mbb3, and Mbb4, Figure 1B) in the vicinity of a hydrogen-peroxide producing NAD(P)H-oxidase (NOX; Mbb2). We demonstrated that among the three VBPOs, Mbb1 and Mbb4 furnished bromoform starting from hydrocarbon substrates derived from fatty acid biosynthesis (R–H in Figure 1C) (Thapa et al., 2020). Isofunctional VBPOs in marine cyanobacteria also produce bromoform (Thapa and Agarwal, 2021). The VBPO Mbb3—while oxidizing bromide—did not catalyze bromoform production (Thapa et al., 2020). Akin to other haloperoxidases, Mbb1 and Mbb4 employed hydrogen peroxide (H2O2) as an electron sink to catalyze bromide oxidation to hypobromous acid (HOBr, Figure 1C). Hydrogen peroxide, in turn, might be produced by the NOX Mbb2, or derived from electron transport processes in photosynthesis and respiration (Thapa et al., 2020). VBPOs and vanadium-dependent chloroperoxidases (VHPOs), in addition to be being involved in natural product biosynthesis schemes, are versatile and broad utility catalysts for C–H activation (Branham et al., 2025; Sharma et al., 2025; Zhao et al., 2025).
While the identity of the primary players in bromoform biosynthesis was thusly established, the rationale for the unusually high bromoform production in Asparagopsis remained unknown; the concentration of bromoform in other seaweeds such as Chondrus crispus (Irish moss) that also encode isofunctional bromoform producing VBPOs is vastly lower (Thapa et al., 2020). To explore the molecular basis of bromoform abundance in Asparagopsis, we undertook a paired multi-omic inquiry into the transcriptomic and proteomic abundance of the mbb gene expression and Mbb enzymes, respectively. Here, we describe observations that are suggestive of diverse roles of VBPOs in Asparagopsis physiology.
Methods
Detailed materials and methods are available in the Supplementary Material.
Results
Field collected A. taxiformis tetrasporophyte samples were cultivated under conditions that allowed for robust growth such that a weekly biomass increase of up to 70% was observed for tissue samples used in this study (Supplementary Figure S1) (Dishon et al., 2023; Hargrave et al., 2024; Resetarits et al., 2024). The proteomic abundance of stress response proteins, such as superoxide dismutase (SOD), heat shock protein 40 (Hsp40), and the molecular chaperone DnaK were much lower than that of primary metabolic proteins such as phycobilisome component CpeB, RuBisCO small subunit RbcS, and ATP synthase alpha subunit AtpA (Supplementary Figure S2) (Kültz, 2005). Taken together, these physiological and molecular data suggest that the algal samples used in this study were experiencing low stress conditions. Sanger sequencing of the mitochondrial cox2–3 spacer PCR amplicons confirmed that the specimens used in this study belonged to the A. taxiformis lineage 2 haplotypes (Nahor et al., 2022).
Replicate cDNA libraries were prepared and RNA-Seq was used to query global gene expression profiles (Supplementary Table S1; Supplementary Figure S3). A high degree of transcript overlap was observed among three biological replicates (Supplementary Figure S4). The bimodal distribution of transcript abundances, denoted as mean values for transcripts per million (TPM), was consistent across all three biological replicates (Figure 2A). Expectedly for a phototrophic organism, some of the highest abundance transcripts belonged to components of the photosynthetic complexes (Supplementary Data Sheet 1). The bromoform producing VBPO encoding gene mbb1 was among the most abundant transcripts based on the average TPM values in the A. taxiformis transcriptomes (ranked 75, Supplementary Data Sheet 1). Other genes in the mbb locus, mbb2–4, were found at much lower abundance levels (mbb2 ranked 20,361; mbb3 ranked 4,002; mbb4 ranked 1,679). These RNA-Seq-derived observations agreed with quantitative reverse transcription PCR (qRT-PCR) data wherein the abundance of the mbb1 transcript, relative to the housekeeping actin gene, were much higher than that of mbb2–4 (Figure 2B; Supplementary Table S2).
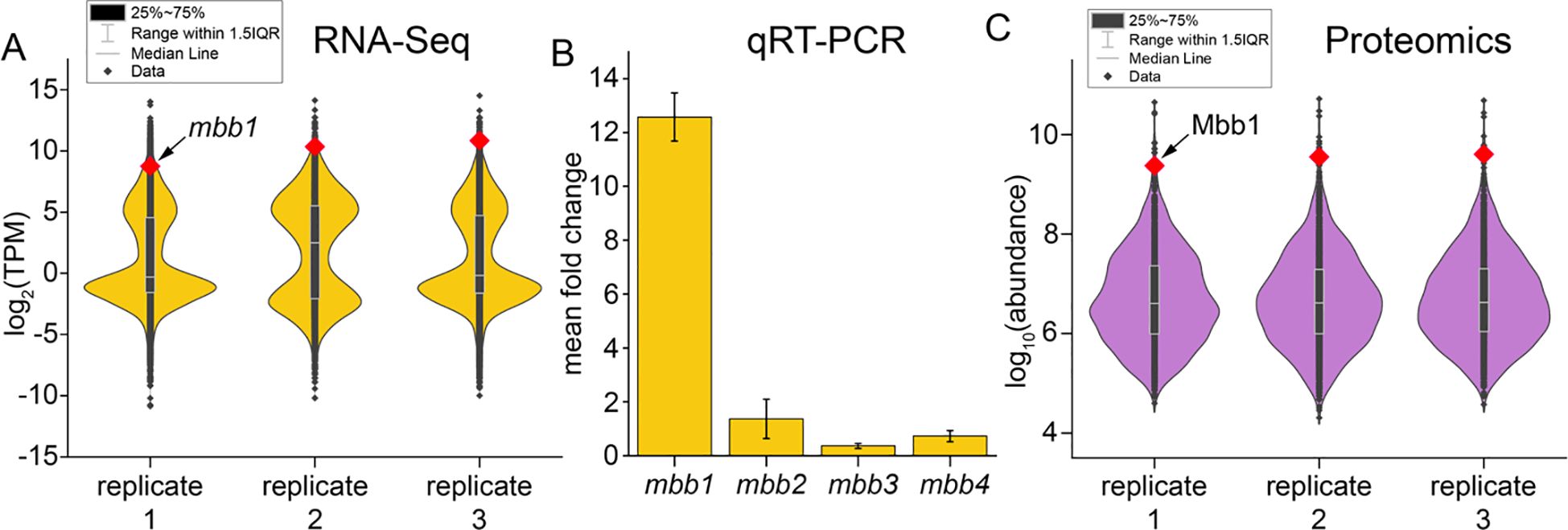
Figure 2. (A) Violin plots denoting normalized transcript abundance for the three biological replicates. Each transcript is represented as a data point (black diamond) with the mbb1 transcript highlighted in red. Median line represents middle value of each dataset when all values are sorted from smallest to largest. The “25–75%” refers to the middle 50% of the dataset, specifically the range between the 25th percentile (lower quartile) and the 75th percentile (upper quartile). Range within 1.5 interquartile range (IQR) refers to the data range between 25th and 75th percentiles. Data points that fall below the lower limit (25th value – 1.5×IQR) or above the upper limit (75th value + 1.5×IQR) are considered outliers. (B) Steady-state mRNA abundance levels for mbb1–4 genes as determined experimentally using RT-qPCR with the same total RNA sample that was used for RNA-Seq. Mean fold change represents mean ± standard deviation of fold changes for three replicates; fold change calculated relative to housekeeping actin encoding gene. (C) Violin plots depicting normalized protein abundances for the three replicate tissue samples. Abundance of each protein is represented as a data point with the Mbb1 protein abundance highlighted in red. Statistical representation is identical to that in panel (A).
Does the mRNA abundance of the mbb1 transcript translate to the abundance of the Mbb1 VBPO protein in A. taxiformis tissue? To investigate this question, proteomic analyses were conducted using the same A. taxiformis tissue samples that were used to generate the transcriptomic profiles (Supplementary Data Sheet 2). High confidence datasets were generated wherein two or more unique peptides were mapped to each protein sequence (Supplementary Data Sheet 3). Among these, 80.3% peptides were common across all three biological replicates signifying fidelity of the proteomic inventory (Supplementary Figure S5). A search for posttranslational modifications revealed that 66 proteins were phosphorylated, 110 were methylated, and 250 were acetylated (Supplementary Figure S6). Across the three biological replicates, Mbb1 bore two modifications—Lys530 acetylation and Arg539 methylation (Supplementary Figure S7). Gene ontology (GO) functional enrichment analysis was conducted to classify genes and proteins (Supplementary Data Sheets 4, 5). Similar categories were found to be shared across the transcriptome and proteome datasets, suggesting consistent expression of gene and protein functional groups (Supplementary Figure S8).
Between the three biological replicates, the mean abundance of Mbb1 denoted it to be the eleventh most abundant protein in the entire A. taxiformis proteome (Figure 2C). The Mbb1 protein abundance rivals that of the photosynthesis-related RuBisCO small and large subunits (RbcS and RbcL, respectively) and phycobilisome complex members CpeA, CpeB, ApcA, and ApcB—in phototrophs, these are expectedly the most abundant proteins. Mbb4 was detected at far less abundance (rank ~900) while Mbb2 and Mbb3 were not detected, at all.
The high expression of the mbb1 gene and the extraordinarily high abundance of the Mbb1 protein are not tied to molecular response to stress—the laboratory cultivated algae samples used here do not bear the molecular hallmarks of stress response (vide supra). To further investigate whether mbb1 expression is responsive to stress experienced by the algae, A. taxiformis tissues were exposed to light and to heat stress using well established growth protocols (Dishon et al., 2023; Hargrave et al., 2024; Resetarits et al., 2024). Under these stress conditions, differential gene expression was queried by transcriptomic profiling, in addition to measurement of photosynthetic efficiencies and hydrogen peroxide concentrations. Under elevated photosynthetically active radiation (PAR) exposure, photosynthetic efficiency significantly decreased in 10 and 60 min under 700 or 1,000 µmol photons m−² s−¹, respectively (Supplementary Figure S9A). Hydrogen peroxide production increased, but the increase was only significant under the 1,000 µmol photons m−² s−¹treatment (Supplementary Figure S9B). Heat stress was assessed by exposing A. taxiformis to 21°C (normal) and at elevated temperatures of 26°C and 31°C for 30 min. Photosynthetic efficiency modestly decreased at the highest temperature (Supplementary Figure S9C), but a clear trend of significantly increased hydrogen peroxide concentration was observed at both temperatures (Supplementary Figure S9D).
Unexpected correlations of mbb1 gene expression alterations were observed with other primary metabolism genes in response to light and heat stress (Figure 3). Under light stress conditions—which was accompanied by decrease in photosynthetic efficiency—we observed a decrease in mbb1 gene expression together with a general decrease in the expression of other genes annotated to be involved in photosynthesis (Figures 3A, B). In contrast, under heat stress which resulted in minimal changes in photosynthetic efficiency, the mbb1 gene expression was not significantly altered. Here, photosynthesis gene expression was also not significantly altered or was slightly increased (Figures 3C, D; Supplementary Figure S9C).
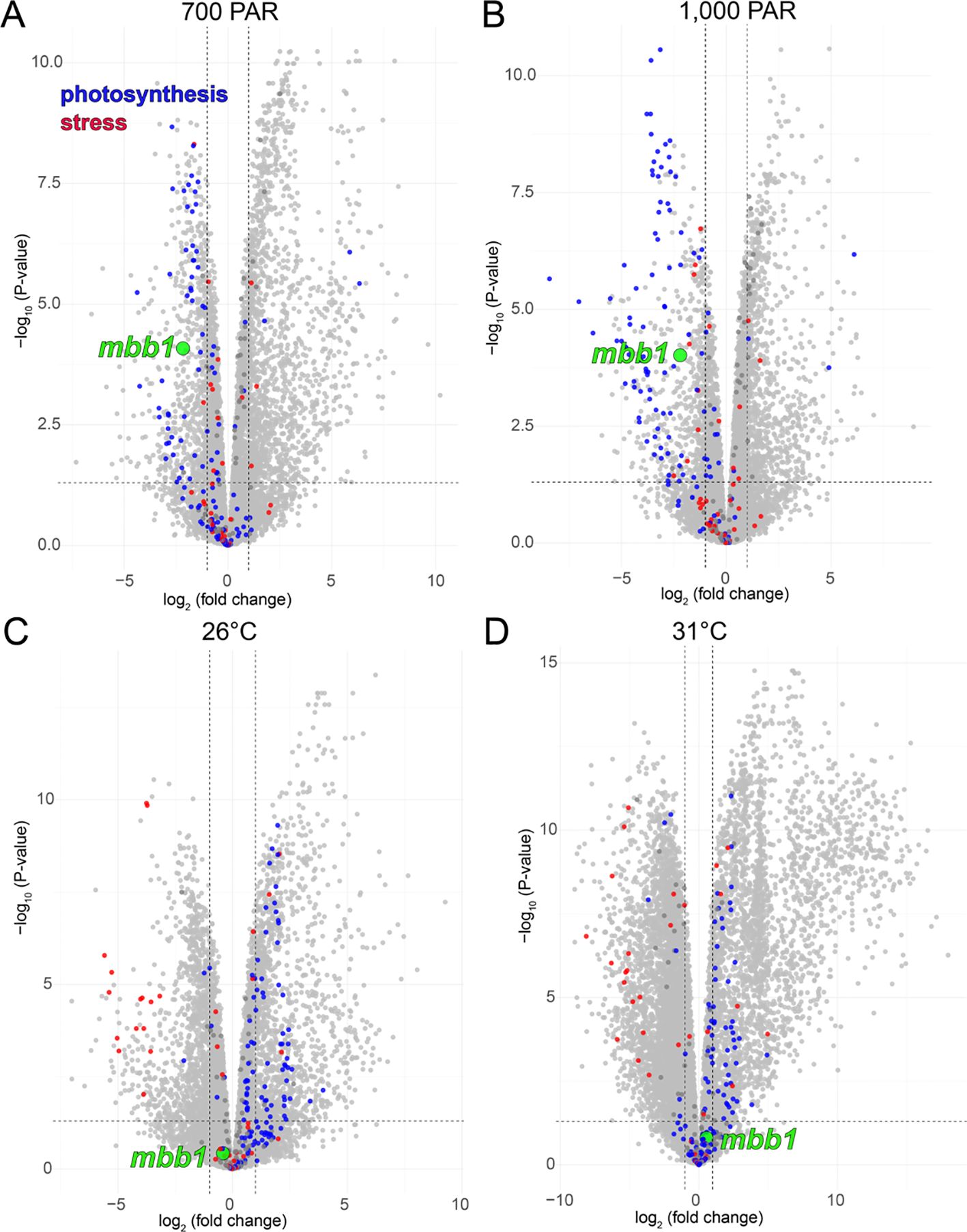
Figure 3. Volcano plots demonstrating differential gene expression in A. taxiformis under (A) 700 PAR light stress, (B) 1000 PAR light stress, (C) 26°C heat stress, and (D) 31°C heat stress as compared to physiological growth conditions of 25 PAR and 21°C. The node corresponding to mbb1 is colored green, and nodes corresponding to genes annotated to be involved in photosynthesis and stress response are colored blue and red, respectively.
Discussion
Taken together, these data allow us to posit that under stressful conditions, differences in mbb1 gene expression tracks with those of genes encoding proteins that are involved in photosynthesis. High light stress led to a general decrease in photosynthetic gene expression along with a concomitant depression in mbb1 gene expression (Figures 3A, B). Taken together, 48 photosynthetic genes were downregulated at both 700 and 1000 µmol photons m−² s−¹, while only 2 were upregulated in both conditions (Supplementary Data Sheet 6; Supplementary Figure S10). Of these 48 light-responsive downregulated photosynthetic genes, 41 encoded proteins related to those found in the light harvesting complexes (LHCs) (Engelken et al., 2012). In red algae, LHC proteins are generally pigment-binding, intrinsic membrane proteins that are part of the photosystem I light-harvesting antenna (You et al., 2023). The 41 LHC-constituting A. taxiformis proteins were aligned with structurally characterized orthologs from Rhodophyta and Cryptophyta photosystems and used to construct a phylogenetic tree (Supplementary Table S3; Supplementary Figure S11) (Pi et al., 2018; You et al., 2023; Si et al., 2024). The A. taxiformis proteins were highly sequence diverse, forming clades with different LHC families and reflecting the diverse functions of LHC proteins in light harvesting. While the structures of the orthologs are characterized, their functions are not. LHC proteins are thought to be important in improving photosynthetic efficiency and their sequence variability in different organisms reflects different ecological and physiological requirements (You et al., 2023). Thus, a downregulation of many LHC proteins under light stress is consistent with an overall decrease in photosynthetic efficiency that would help to prevent cellular damage. Simultaneously, expression of mbb1 was decreased, as described above, which could point towards the involvement of Mbb1 in a primary physiological process related to photosynthesis which would align with its unusually high transcriptomic and proteomic abundance.
Asparagopsis, and other Rhodophytes possess dedicated gland cells—corps en cerise—to sequester the highly abundant halogenated metabolites (Paul et al., 2006b; Salgado et al., 2008; Paul and Pohnert, 2011). These physiological adaptations, in addition to the omics-driven observations described in this study allow us to propose that bromide oxidation and bromoform biosynthesis is likely not a specialized secondary metabolite production process alone—akin to how we conceptualize the biosynthesis of other halogenated natural products—but may have primary roles in Asparagopsis biology and physiology. Getting firm molecular evidence to support these hypotheses will require the development of forward and reverse genetic tools to manipulate Asparagopsis and pairing them with imaging-based techniques to query subcellular localization of bromoform biosynthetic enzymes such as Mbb1. With due acknowledgement that these tools are not yet in hand, findings described herein set the stage to query the broader ecological interconnections between seaweed biology, atmospheric chemistry, and contemporary directions in marine aquaculture.
Natural products and other specialized secondary metabolites produced by seaweeds have important roles in mediating mutualistic and antagonistic interspecies interactions in the marine environment (Paul VJ. et al., 2006; Rasher et al., 2011; Andras et al., 2012). Bromoform has been postulated to serve a similar role; it was shown to inhibit the growth of epiphytic marine bacteria that may colonize seaweed surfaces (Paul et al., 2006a). Typically, specialized allelopathic molecules have potent bioactivities and are produced in small quantities using tightly regulated molecular processes. Bromoform production in A. taxiformis defies this characterization; it is constitutively produced and in remarkably high quantities using a VBPO that accumulates at very high abundance in the macroalgal tissue. The integrative multi-omics data presented herein now allows us to posit that rather than a specialized metabolite, bromoform could be a byproduct of a primary metabolic activity such as reactive oxygen species (ROS) detoxification by the VBPO Mbb1 that consumes hydrogen peroxide as a substrate. Halide oxidation is inextricably tied to ROS chemistry; akin to heme- and vanadium-dependent peroxidases, flavin-dependent halogenases also produce hypohalous acid upon halide oxidation while involving peroxide intermediates (Phintha et al., 2021). It is likely that the bromoform producing A. taxiformis VBPOs transcend halogenation biochemistry from secondary metabolite natural product biosynthetic pathways to primary metabolism.
Data availability statement
The transcriptomics data have been deposited to the NCBI with the BioProject accession number PRJNA1108735.
Author contributions
ZL: Conceptualization, Formal Analysis, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing, Data curation, Software. SI: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. MH: Investigation, Methodology, Writing – review & editing, Resources. RS: Investigation, Writing – review & editing, Data curation, Formal Analysis, Visualization. LZ: Writing – review & editing, Resources. RX: Resources, Writing – review & editing, Formal Analysis. JS: Resources, Writing – review & editing, Conceptualization, Funding acquisition, Supervision. ES: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft. VA: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing, Validation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The authors are grateful for financial support from the National Institutes of Health (GM142882 to VA) and the National Science Foundation (NSF; MCB-2129491 to ES and MCB-2129492 to JS). SI was supported by NSF grant MCB-2129490. We also thank the Builders Initiative for providing funding to the Smith Lab which helped to support all living cultures and experimental work presented here.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1664275/full#supplementary-material
Supplementary Data Sheet 6 | Changes in gene expression upon Asparagopsis stress treatments.
References
Agarwal V., Miles Z. D., Winter J. M., Eustáquio A. S., El Gamal A. A., and Moore B. S. (2017). Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem. Rev. 117, 5619–5674. doi: 10.1021/acs.chemrev.6b00571
Andras T. D., Alexander T. S., Gahlena A., Mitchell Parry R., Fernandez F., Jubanek J, et al. (2012). Seaweed allelopathy against coral: surface distribution of a seaweed secondary metabolite by imaging mass spectrometry. J. Chem. Ecol. 38, 1203–1214. doi: 10.1007/s10886-012-0204-9
Bianco A. C., Salvatore D., Gereben B., Berry M. j. J., and Larsen P. R. (2002). Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr. Rev. 23, 38–89. doi: 10.1210/edrv.23.1.0455
Branham P. J., Saha N., Oyelere S. E., and Agarwal V. (2025). Halogenase-assisted biocatalytic derivatization of aminothiazoles and cephalosporin antibiotics. J. Org. Chem. 90, 3507–3511. doi: 10.1021/acs.joc.4c03043
Dishon G., Resetarits H. M., Tsai B., Jones A. L., Agarwal V., and Smith J. E. (2023). The effect of light intensity, spectrum, and photoperiod on the physiological performance of Asparagopsis taxiformis tetrasporophytes. Algal Res. 76, 103304. doi: 10.1016/j.algal.2023.103304
Engelken J., Funk C., and Adamska I. (2012). “The extended light-harvesting complex (LHC) protein superfamily: classification and evolutionary dynamics,” in Functional Genomics and Evolution of Photosynthetic Systems. Eds. Burnap R. and Vermaas W. (Springer Netherlands, Dordrecht), 265–284.
Gillis E. P., Eastma K. J., Hill M. D., Donnelly D. J., and Meanwell N. A. (2015). Applications of fluorine in medicinal chemistry. J. Med. Chem. 58, 8315–8359. doi: 10.1021/acs.jmedchem.5b00258
Gribble G. W. (2000). The natural production of organobromine compounds. Environ. Sci. pollut. Res. Int. 7, 37–47. doi: 10.1065/espr199910.002
Hargrave M. S., Islam S., Agarwal V., and Smith J. E. (2024). The influence of light stress on bromoform synthesis and concentration in the red seaweed Asparagopsis taxiformis. J. Appl. Phycol. 36, 321–329. doi: 10.1007/s10811-023-03129-2
Kültz D. (2005). Molecular and evolutionary basis of the cellular stress response. Ann. Rev. Physiol. 67, 225–257. doi: 10.1146/annurev.physiol.67.040403.103635
Nahor O., Luzzatto-Knaan T., and Israel Á. (2022). A new genetic lineage of Asparagopsis taxiformis (Rhodophyta) in the Mediterranean Sea: As the DNA barcoding indicates a recent Lessepsian introduction. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.873817
Navarro M. A., Atlas E. L., Saiz-Lopez A., Rodriguez-Lloveras X., Kinnison D. E., Lamarque J. F, et al. (2015). Airborne measurements of organic bromine compounds in the Pacific tropical tropopause layer. Proc. Natl. Acad. Sci. U.S.A. s 112, 13789. doi: 10.1073/pnas.1511463112
Paul C. and Pohnert G. (2011). Production and role of volatile halogenated compounds from marine algae. Nat. Prod. Rep. 28, 186–195. doi: 10.1039/C0NP00043D
Paul N. A., Cole L., De Nys R., and Steinberg P. D. (2006b). Ultrastructure of the gland cells of the red alga asparagopsis armata (Bonnemaisoniaceae)1. J. Phycol. 42, 637–645. doi: 10.1111/j.1529-8817.2006.00226.x
Paul N. A., de Nys R., and Steinberg P. D. (2006a). Chemical defence against bacteria in the red alga Asparagopsis armata: linking structure with function. Mar. Ecol. Prog. Ser. 306, 87–101. doi: 10.3354/meps306087
Paul V. J., Puglisi M. P., and Ritson-Williams R. (2006). Marine chemical ecology. Nat. Prod. Rep. 23, 153–180. doi: 10.1039/b404735b
Phintha A., Prakinee K., Jaruwat A., Lawan N., Visitsatthawong S., Kantiwiriyawanitch C, et al. (2021). Dissecting the low catalytic capability of flavin-dependent halogenases. J. Biol. Chem. 296, 100068. doi: 10.1074/jbc.RA120.016004
Pi X., Tian L., Dai H. E., Qin X., Cheng L., Kuang T, et al. (2018). Unique organization of photosystem I–light-harvesting supercomplex revealed by cryo-EM from a red alga. Proc. Natl. Acad. Sci. U.S.A. 115, 4423–4428. doi: 10.1073/pnas.1722482115
Pinchman J. R. and Boger D. L. (2013a). Investigation into the functional impact of the vancomycin C-ring aryl chloride. Bioorg. Med. Chem. Lett. 23, 4817–4819. doi: 10.1016/j.bmcl.2013.06.080
Pinchman J. R. and Boger D. L. (2013b). Probing the role of the vancomycin e-ring aryl chloride: selective divergent synthesis and evaluation of alternatively substituted E-ring analogues. J. Med. Chem. 56, 4116–4124. doi: 10.1021/jm4004494
Quack B., Atlas E., Stroud P. V., Schauffler S., and Wallace D. W. R. (2004). Oceanic bromoform sources for the tropical atmosphere. Geophys. Res. Lett. 31. doi: 10.1029/2004GL020597
Rasher D. B., Stout E. P., Engel S., Kubanek J., and Hay M. E. (2011). Macroalgal terpenes function as allelopathic agents against reef corals. Proc. Natl. Acad. Sci. U.S.A. 108, 17726. doi: 10.1073/pnas.1108628108
Resetarits H. M., Dishon G., Agarwal V., and Smith J. E. (2024). The effects of temperature and CO2 enrichment on the red seaweed Asparagopsis taxiformis from Southern California with implications for aquaculture. J. Phycol. 60, 1567–1584. doi: 10.1111/jpy.13526
Roque B. M., Venegas M., Kinley R. D., de Nys R., Duarte T. L., Yang X, et al. (2021). Red seaweed (Asparagopsis taxiformis) supplementation reduces enteric methane by over 80 percent in beef steers. PloS One 16, e0247820. doi: 10.1371/journal.pone.0247820
Saiz-Lopez A. and von Glasow R. (2012). Reactive halogen chemistry in the troposphere. Chem. Soc. Rev. 41, 6448–6472. doi: 10.1039/c2cs35208g
Salawitch R. J. (2006). Atmospheric chemistry: biogenic bromine. Nature 439, 275–277. doi: 10.1038/439275a
Salgado L. T., Vianna N. B., Andrade L. R., Leal R. N., da Gama B. A. P., Attias M, et al. (2008). Intra-cellular storage, transport and exocytosis of halogenated compounds in marine red alga Laurencia obtusa. J. Struct. Biol. 162, 345–355. doi: 10.1016/j.jsb.2008.01.015
Sharma M., et al. (2025). Intermolecular 1,2,4-thiadiazole synthesis enabled by enzymatic halide recycling with vanadium-dependent haloperoxidases. J. Am. Chem. Soc. 147, 10698–10705. doi: 10.1021/jacs.5c01175
Si L., Pascoe C. A., Jones S. K., Barthel S. G., Davis K. M., and Biegasiewicz K. F. (2024). Structural basis for the distinct core-antenna assembly of cryptophyte photosystem II. Nat. Commun. 15, 6812. doi: 10.1038/s41467-024-51206-y
Su Q., Xu B., Chen X., and Rokita S. E. (2024). Misregulation of bromotyrosine compromises fertility in male Drosophila. Proc. Natl. Acad. Sci. U.S.A. 121, e2322501121. doi: 10.1073/pnas.2322501121
Thapa H. R. and Agarwal V. (2021). Obligate brominating enzymes underlie bromoform production by marine cyanobacteria. J. Phycol. 57, 1131–1139. doi: 10.1111/jpy.13142
Thapa H. R., Lin Z., Yi D., Smith J. E., Schmidt E. W., and Agarwal V. (2020). Genetic and biochemical reconstitution of bromoform biosynthesis in Asparagopsis lends insights into seaweed reactive oxygen species enzymology. ACS Chem. Biol. 15, 1662–1670. doi: 10.1021/acschembio.0c00299
Wasson D. E., Yarish C., and Hristov A. N. (2022). Enteric methane mitigation through Asparagopsis taxiformis supplementation and potential algal alternatives. Front. Anim. Sci. 3. doi: 10.3389/fanim.2022.999338
You X., Zhang X., Cheng J., Xiao Y., Ma J., Sun S, et al. (2023). In situ structure of the red algal phycobilisome–PSII–PSI–LHC megacomplex. Nature 616, 199–206. doi: 10.1038/s41586-023-05831-0
Keywords: Asparagopsis, bromoform, halogenase, proteomics, transcriptomics, methane
Citation: Lin Z, Islam S, Hargrave MS, Singh R, Zhou L, Xie R, Smith JE, Schmidt EW and Agarwal V (2025) Detection of unusually high transcriptomic and proteomic abundance of bromoform-synthesizing halogenase in marine macroalgae Asparagopsis taxiformis. Front. Mar. Sci. 12:1664275. doi: 10.3389/fmars.2025.1664275
Received: 11 July 2025; Accepted: 25 August 2025;
Published: 07 October 2025.
Edited by:
Rafael R. Robaina, University of Las Palmas de Gran Canaria, SpainReviewed by:
José Ángel Huerta Ocampo, National Council of Science and Technology (CONACYT), MexicoZubaida Parveen Patwary, University of the Sunshine Coast, Australia
Copyright © 2025 Lin, Islam, Hargrave, Singh, Zhou, Xie, Smith, Schmidt and Agarwal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer E. Smith, amVzMDEzQHVjc2QuZWR1; Eric W. Schmidt, ZXdzMUB1dGFoLmVkdQ==; Vinayak Agarwal, dmFnYXJ3YWxAZ2F0ZWNoLmVkdQ==
†These authors have contributed equally to this work
 Zhenjian Lin
Zhenjian Lin Shahima Islam2†
Shahima Islam2† Lingjie Zhou
Lingjie Zhou Eric W. Schmidt
Eric W. Schmidt Vinayak Agarwal
Vinayak Agarwal