Abstract
Introduction:
The presence and effect of triclosan (TCS), a non-antibiotic antimicrobial biocide mostly used in personal, household, and healthcare products, on aquatic life is alarming nowadays. Although several studies have addressed TCS toxicity in aquatic organisms, its effects on brain tissues remain poorly explored.
Methods:
In our study, label-free proteomics liquid chromatography- tandem mass spectrometry (LC–MS/MS) was used to analyze the long-term effects of TCS on the brain tissues of Labeo catla. Catla fingerlings (mean weight 12 ± 1.76 g; mean length 12 ± 2.14 cm) were exposed to TCS at 0.073 mg/L, a sublethal concentration corresponding to 1/10th of LC50 and within reported environmental hotspots, for 30 days in 50-L glass tanks with predefined laboratory conditions. After TCS exposure, fish brain tissue samples were collected and used for LC-MS/MS analysis.
Results:
The proteomic analysis suggested that TCS treatment of Catla brain tissues upregulated the proteins related to motor activity, neuron development, and semaphorin complex. In contrast, proteins related to myotube development, meiotic chromosome separation, myosin complex, and plasma membrane were downregulated. Principal component analysis (PCA) revealed significant proteomic alterations. ECT2 and Zcchc11 proteins showed marked upregulation, while EIF4G3B and PBRM1 were significantly downregulated. These results indicate that exposure to triclosan alters critical cellular growth pathways, RNA processing, and translation.
Discussion:
Our findings provide valuable insights into the molecular impact of environmental contaminants on aquatic species. Altered proteins (e.g., actin alpha1a/1b, myosin heavy chain fast skeletal muscle, camsap1b, and plexin B1) were consistently identified as potential candidate biomarkers of TCS neurotoxicity, pending further validation. Overall, our findings highlight the eco-physiological risks of TCS exposure, suggesting that proteomic disruptions in neuronal and muscular processes may translate into impaired fish fitness in contaminated habitats.
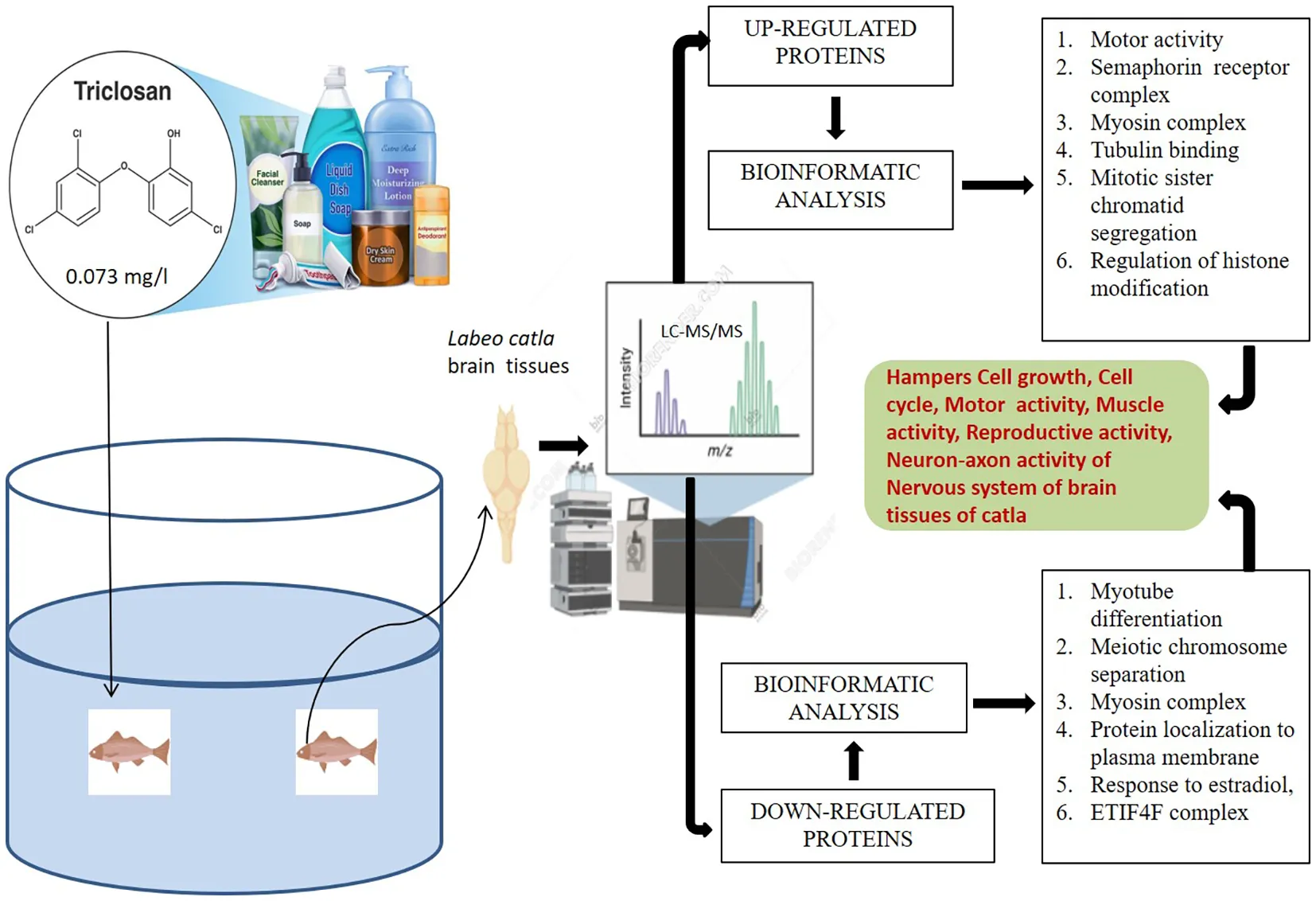
Introduction
Triclosan [TCS; 5-chloro-2-(2,4-dichlorophenoxy)phenol] is a widely used non-antibiotic antimicrobial biocide that is frequently present for many years in personal, household, and healthcare products like soaps, sanitizers, toothpaste, deodorants, mouthwashes, kitchen utensils, textiles, plastics, and medical devices (Yueh and Tukey, 2016; Lu et al., 2018). TCS has a perceptible aromatic odor and low solubility in water but good solubility in organic solvents, including ethanol, methanol, and dimethyl sulfoxide (Montville and Schaffner, 2011). The widespread presence of TCS in aquatic environments worldwide is increasingly alarming. The entry of TCS into aquatic environments and land is due to wastewater and biosolids discharge from different sources (Dann and Hontela, 2011). The extensive use of TCS containing sanitizers and antiseptics during the time of the COVID-19 pandemic has raised concerns about the environment as well as public health (Lu and Guo, 2021). Due to the high bioaccumulation factor, the possibility of the accumulation of TCS in non-target aquatic animals is high (Dhillon et al., 2015; Yao et al., 2018). Recently, it was observed that non-ionic detergents like Tween 20 can inhibit TCS activity in vivo, most probably due to micelle formation (Alfhili and Lee, 2019). Previous studies have suggested that TCS causes toxicity to various aquatic organisms (Weatherly and Gosse, 2017), like fish (Dar et al., 2019), arthropods (Peng et al., 2013), molluscs (Geiß et al., 2016), nematodes (Vingskes and Spann, 2018), algae (Bi et al., 2018), and plants (Prosser et al., 2014). Different studies have demonstrated that fish were very sensitive to TCS toxicity, which has LC50 values ranging from 0.26 to 0.45 mg/L (Orvos et al., 2002). The effects of TCS in the form of biochemical alteration include endocrine disruption and delay in embryonic development in different fish species like Japanese medaka Oryzias latipes (Ishibashi et al., 2004), zebrafish Danio rerio (Oliveira et al., 2009), and major carps like Labeo rohita, Cirrhinus mrigala, Ctenopharyngodon idella, and Cyprinus carpio (Dar et al., 2019; Dar et al., 2020). Loss of equilibrium, fish jaw locked open, erratic swimming, spinal curvature, and quiescence were observed in rainbow trout fish due to the TCS effect (Orvos et al., 2002). Increased lysosomal membrane destabilization, phosphorylation of PKCα and PKCβII isoforms, and phagocytic activity were induced by TCS both in vitro and in vivo (Canesi et al., 2007). Pharmacokinetic studies in humans have suggested that TCS can reach the systemic circulation by the process of rapid absorption through skin and mucous membranes of the oral cavity and gastrointestinal tract and also affect the rate of urinary excretion (Queckenberg et al., 2010; Sandborgh-Englund et al., 2006). The data related to environmental and human toxicology suggest that acute toxicity was seen in aquatic organisms and humans in the form of potential carcinogenicity, endocrine disruption, antimicrobial resistance, and mild genotoxicity (Huang et al., 2014; Li, 2021).
The use of TCS in over-the-counter antiseptics like soap, foam, and liquid wash has been banned by the United States Food and Drug Administration (USFDA) since 2016, although other TCS-containing products like hand sanitizers and wipes are not regulated by the FDA (Food and Drug Administration, 2016). TCS works like a biocide, and food contact products were banned by the European Union, but its use in personal care products is still allowed (EC, 2016). In the modern era, when a pandemic like COVID-19 has shaken mankind, the use of antimicrobial and disinfectant agents like TCS is very relevant (Dhama et al., 2021). However, the excessive and unnecessary use of TCS in daily life products should be restricted to mitigate the health hazards to human beings and negative effects on the environment and ecology (Mukherjee et al., 2021).
Previously, several proteomic studies have been performed to observe the effect of TCS, and they identified affected biological processes and potential proteins or groups of proteins that were altered due to the TCS treatment of different organisms, including aquatic organisms. A proteomic study using Two-Dimensional Difference Gel Electrophoresis (2D DIGE) of TCS-treated 7-day post-fertilization larvae found that the proteins involved in the cytoskeleton, stress response, eye, and neural development were affected. An enzyme study also supported this result, which indicated an impairment of glutathione metabolism and acute neurotoxicity (Falisse and Voisin, 2017). Using the Two-Dimensional Electrophoresis (2DE) system, proteomic analysis of freshwater mussel Dreissena polymorpha treated with TCS for 7 days revealed the altered protein profile of biological processes like calcium binding and stress response (Riva et al., 2012). The proteomic approach is a useful tool for analyzing the upregulated and downregulated proteins in the different body samples of aquatic animals. Proteomic analysis with the help of bioinformatics tools is very useful in experiments like 2D DIGE, Isobaric tag for relative and absolute quantitation (iTRAQ), and label-free proteomics (Fernandez et al., 2024). Label-free proteomic techniques are very rapid, low-cost, and accurate techniques that can produce altered protein profiles of aquatic organisms regardless of sample complexity (Gajahin Gamage et al., 2022).
Labeo catla is the most important and preferred carp among the three Indian major carps. Catla is the most edible major carp, contributing half of the total production from composite fish culture. It is highly demanded due to its faster growth, quality yield, and good size, and it contributes to the livelihood of fish farmers around South Asia (Sahoo et al., 2020). Earlier works were mainly focused on the effect of TCS on mammals and aquarium fish, but the data related to edible fish have not been explored extensively.
To obtain detailed information about the effects of TCS and the mode of operation in the brain tissues of Catla, we used a modern proteomic approach with detailed analysis by advanced bioinformatics tools. Proteomic data related to TCS-treated fish brain tissues are very limited. To obtain elaborate and comprehensive data related to the mode of action of TCS and its toxicity in the brain tissues of Catla, we used label-free proteomics and advanced bioinformatics analysis. To our knowledge, this is the first study applying label-free proteomics to evaluate TCS effects on the brain tissues of the commercially important carp L. catla. Recent studies have further highlighted the ecotoxicological relevance of TCS (Sharma et al., 2022a, Sharma et al., 2022b; Dar et al., 2024, Dar et al., 2025), reinforcing the need to investigate its molecular impacts in food fish species.
Materials and methods
Test chemical and experimental fish
TCS of analytical grade (purity >98%; CAS Number: 3380-34-5) was purchased from Sigma-Aldrich, Burlington, Massachusetts, USA. The required test solutions for TCS were made by performing a series of dilutions. The first step involved making a 1 g/L stock by dissolving the required quantity of the test substance in methanol. Catla fingerlings (mean weight 12 ± 1.76 g; mean length 12 ± 2.14 cm) were acquired from a privately owned fish farm in Palta, West Bengal, India, and then raised within the hatcheries and breeding facilities division of the institute. The fish were housed in perpetually aerated 50-L glass tanks with predefined laboratory conditions, including dechlorinated water (pH 7.2 ± 0.1) at 24.0 °C ± 1.75 °C and a constant photoperiod with alternate cycles comprising 12 h of light and dark for acclimatization. The fish were fed with commercial feed twice a day, and one-third of the water, together with the leftover feed and the carp excrement, was regularly siphoned off. The specifications set by the Institute Animal Ethics Committee (No. CIFRI/IAEC-23-24/06) were adhered to in all the cases regarding the usage of the experimental carps, collection of biological samples, and experimental procedures.
Experimental exposure
The 96-h LC50 value for TCS in Catla was determined to be 0.73 mg/L in our earlier investigations on acute toxicity (Adhikari et al., 2023). The fish were randomly chosen irrespective of gender and then assigned to distinct exposure groups for a time period of 30 days. The dosage at which the fish were exposed to the aforementioned test had 1/10th (0.073 mg/L) concentrations of the 96-h LC50 value previously derived for the test substance. Each treatment was conducted in triplicate tanks (n = 3), with 15 fish per tank. Tanks, rather than individual fish, were considered biological replicates to avoid pseudoreplication and were maintained in the same conditions as those maintained during the acclimatization period. Following the experimental exposure, the fish were anesthetized using tricaine (150 mg/L) and thereafter dissected with immediate removal of their brains stored in Phosphate Buffered Saline (PBS) for proteomic analysis.
Proteomic analysis via LC–MS/MS
An ACQUITY UPLC system (Waters Corporation, Milford, Massachusetts, USA) was employed to perform liquid chromatography. The trypsinized peptide solutions were separated using a Waters-made ACQUITY UPLC BEH C18 column (150 mm × 2.1 mm × 1.7 µm). To accomplish the chromatographic separation, a gradient elution program was employed with mobile phases A and B (0.1% formic acid in water and acetonitrile, respectively). The flow rate was adjusted to 0.30 mL/min, and 1 µL from each of the individual samples (n = 3) was taken for injection into the system. Mass spectrometry was performed using a SYNAPT G2 QTOF (Waters, USA) outfitted with an electrospray ionization (ESI) source. The sampling cone, extraction cone, capillary energy, and trap collision energy were set at 45.0, 4.50, 3.50 kV, and 6.0 V, respectively. During the increased energy scan, the trap collision energy was scaled up from 20 to 45 V. ProteinLynx Global Server (PLGS) 3.0.2, a program that is utilized for database searches, was used to process the raw mass spectrometry data. The mass spectrometry proteomic data have been deposited in the ProteomeXchange Consortium via PRIDE. Differentially expressed proteins were identified using R/Bioconductor (DEP) packages, followed by principal component analysis (PCA) to visualize proteomic variations.
Enriched Gene Ontology, protein–protein interaction, pathway analysis, and statistical proteomics
To identify and analyze the biological processes and their pathways of upregulated and downregulated proteins induced due to the TCS treatment of Catla and to find their molecular functions and cellular components, the ClueGO v2.5.10 plugin of the Cytoscape 3.9.1 tool was used, and the zebrafish D. rerio protein database was taken as a reference list or background list for this analysis. Protein–protein interactions of upregulated and downregulated proteins of TCS-treated brain tissues of Catla were performed using the Cytoscape 3.9.1 tool, and results were shown in radial network topology format. Kyoto Encyclopedia of Genes and Genomes (KEGG) and WiKiPathways enrichment analyses were performed using the FishEnrichr tool, which used upregulated and downregulated proteomic data. The top observed ratio (control vs. treatment) of each up- and downregulated protein was used for PCA using the R package.
Results
The protein expression patterns of the brain tissues of L. catla exposed to TCS were compared with their control counterpart. After analyzing the LC–MS/MS data of L. catla brain tissue samples, we identified nearly 342 upregulated and 272 downregulated proteins due to TCS treatment. With these upregulated and downregulated proteins, enrichment analysis, protein–protein network analysis, and pathway analysis were performed using Gene Ontology (GO), network analysis, and KEGG and WIKI analyses, respectively and we found that various biological processes, cellular components, and molecular functions were affected by some individual proteins and as well as associated proteins, which will be analyzed and discussed elaborately in this section.
GO analysis
GO enrichment analysis identified 23 biological processes, four cellular components, and seven molecular functions significantly upregulated by TCS treatment (False Discovery Rate (FDR) < 0.05) due to triclosan treatment of L. catla brain tissues (Figure 1). The maximum number of identified upregulated proteins induced by TCS treatment in the biological processes was four. The proteins are related to the fin regeneration process (proteins: chd4a, hspa9, inhbaa, and ttk) and sister chromatid segregation processes (proteins: ncapd3, pds5b, smc3, and top2b). The microtubule cellular components possess the highest number of eight (camsap1b, cep162, ckap5, clasp1a, gas8, kif13bb, kif15, and map4l) upregulated proteins. Out of seven molecular functions, tubulin binding had the maximum number of upregulated proteins, and the 10 upregulated proteins were agtpbp1, camsap1b, ccdc88c, ckap5, clasp1a, gas8, kif13bb, kif15, macf1b, and map4l. Although among the upregulated proteins the maximum proportion of proteins was related to the motor activity (34.29%) term, semaphorin receptor complex (25.71%) term, and myosin complex (11.43%) term (Figure 2), in upregulated proteins, kinesin-like protein (kif13bb), microtubule-associated protein (map41), CLIP-associated protein (clasp1a), sister chromatid cohesion proteins (pds5b), cytoskeleton-associated protein (ckap5), calmodulin-regulated spectrin-associated protein (camsap1b), and plexin proteins (plexinb1 and plexinc1) were well overexpressed in Catla brain due to TCS exposure.
Figure 1
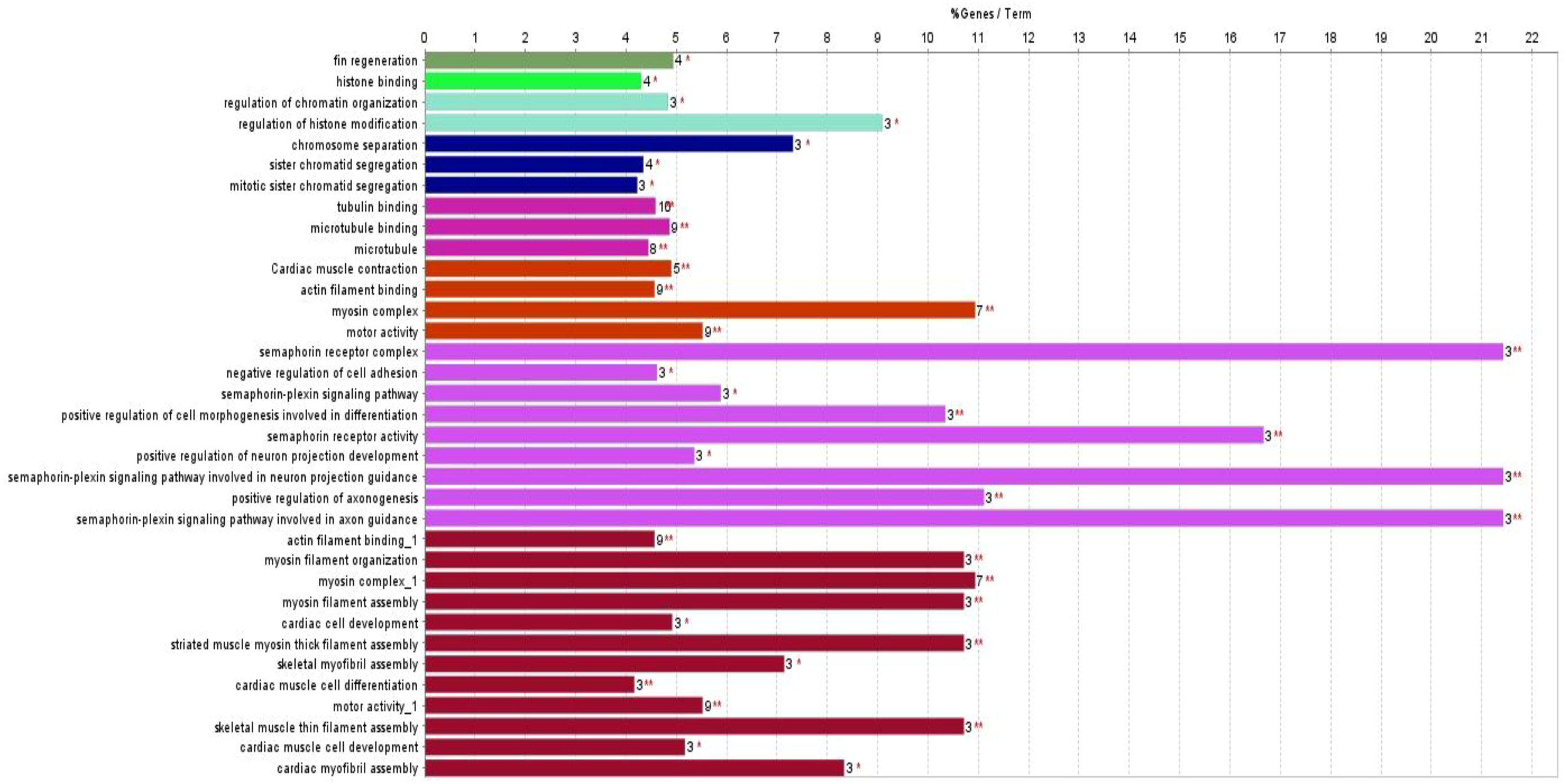
Gene ontology (GO) enrichment analysis of significantly upregulated proteins (adjusted p < 0.05, FDR < 0.05) using ClueGO v2.5.10 plugin of Cytoscape 3.9.1. Terms grouped by biological processes, cellular components, and molecular functions.
Figure 2
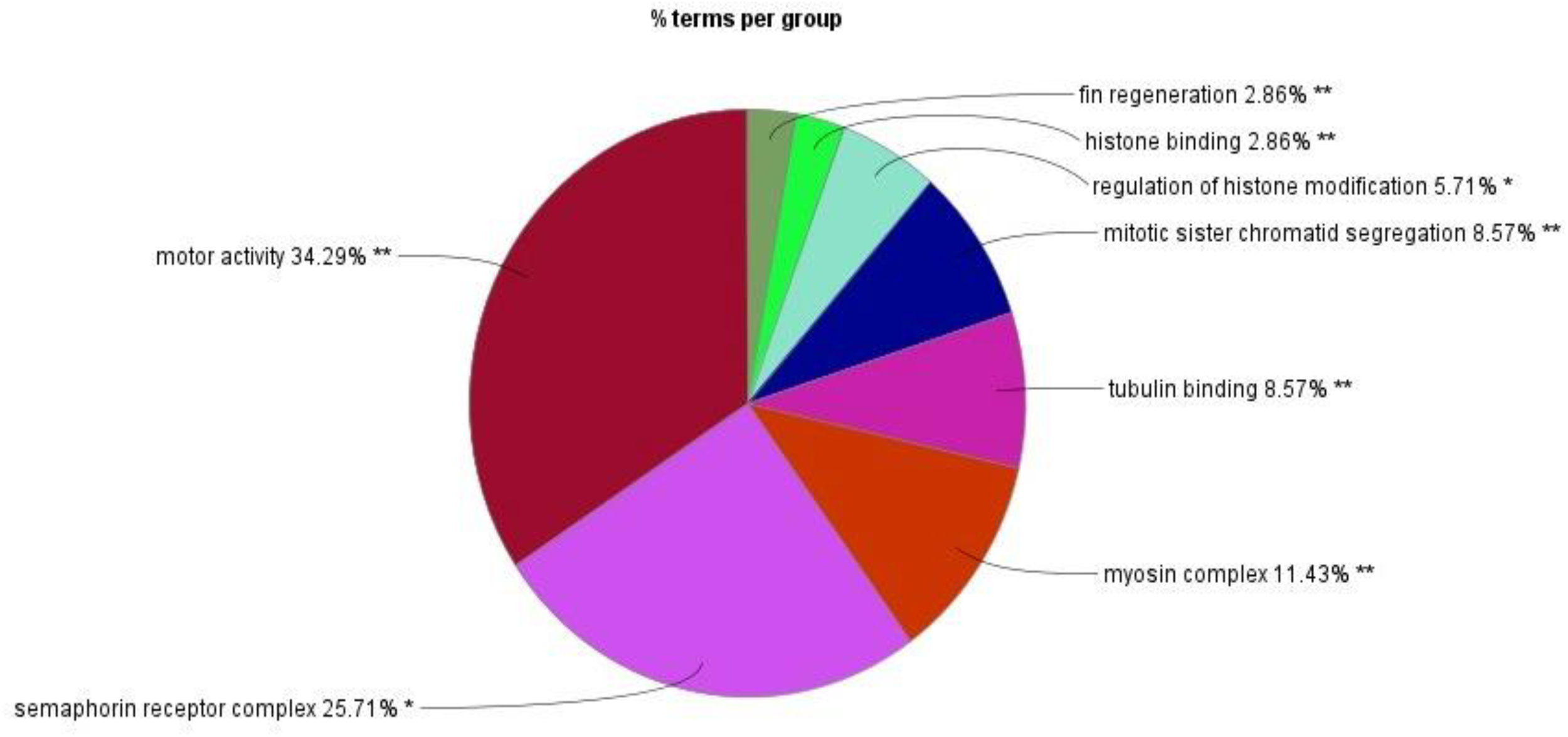
Relative proportion of upregulated proteins across major Gene Ontology (GO) terms identified by ClueGO v2.5.10.
The GO process enrichment analysis identified 30 biological processes, four cellular components, and two molecular functions in downregulated proteins caused by triclosan treatment of L. catla (Figure 3). Three biological processes that had the maximum number of downregulated proteins were myotube differentiation (six proteins: acta1a, acta1b, dock5, mybpc1, myo18ab, and ripor2), skeletal muscle organ development (six proteins: acta1a, acta1b, lamb2, mybpc1, myo18ab, and ripor2), and muscle fiber development (six proteins: acta1a, acta1b, mybpc1, mybpha, myo18ab, and ripor2). The actin cytoskeleton cellular components possess the highest number of fourteen (LOC100329748, LOC100329813, inhbaa, mybpc1, mybpha, myh14, myh6, myh7ba, myh9a, myha, myhc4, myo18ab, myo5ab, and si:ch 211-94l19.4) downregulated proteins after TCS treatment. Out of the two molecular functions in motor activity, one has the maximum number of downregulated proteins. Approximately fourteen proteins (LOC100329748, LOC100329813, kif13ba, kif13, kif4, myh14, myh6, myh7ba, myh9a, myha, myhc4, myo18ab, myo5ab, and si:ch211-94l19.4) were downregulated in motor activity. However, the maximum proportion of proteins that were downregulated by TCS exposure was as follows: myotube differentiation (36.11%), meiotic chromosome separation (22.22%), myosin complex (11.11%), and protein localization to the plasma membrane (11.11%) (Figure 4). In our proteomic analysis, we found that myosin heavy chain fast skeletal muscle protein (myha), myosin binding protein c (mybpc1), myosin heavy chain 7B (myh7ba), actin alpha 1 (acta1a and acta1b), lamanin subunit beta 2 (lamb2), and rho family interacting cell polarization regulator 2 (ripo2) were well downregulated in Catla brain tissues as a result of TCS treatment.
Figure 3
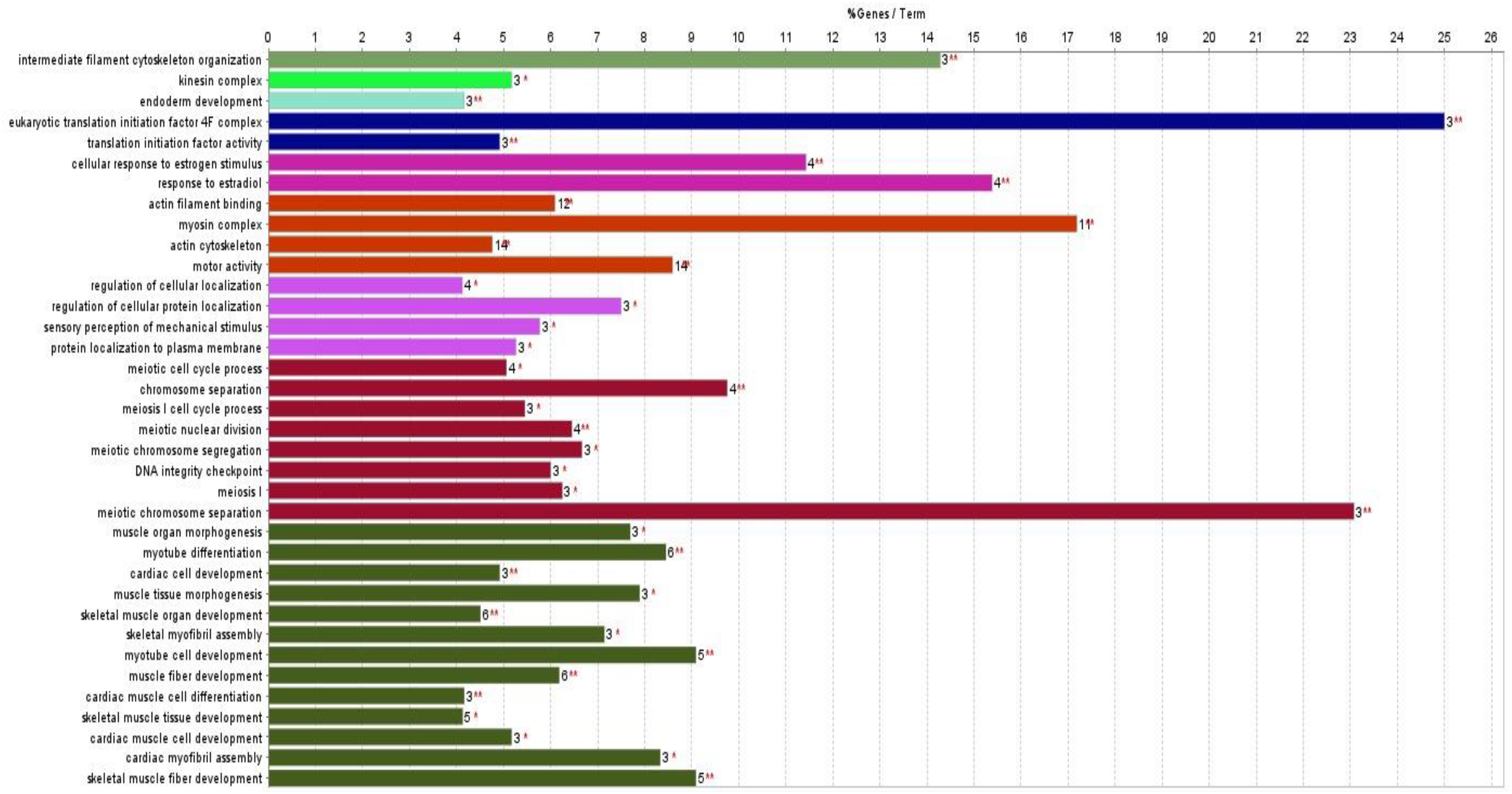
Gene Ontology (GO) enrichment analysis of significantly downregulated proteins (adjusted p < 0.05, FDR < 0.05) using ClueGO v2.5.10. Terms grouped by biological processes, cellular components, and molecular functions.
Figure 4
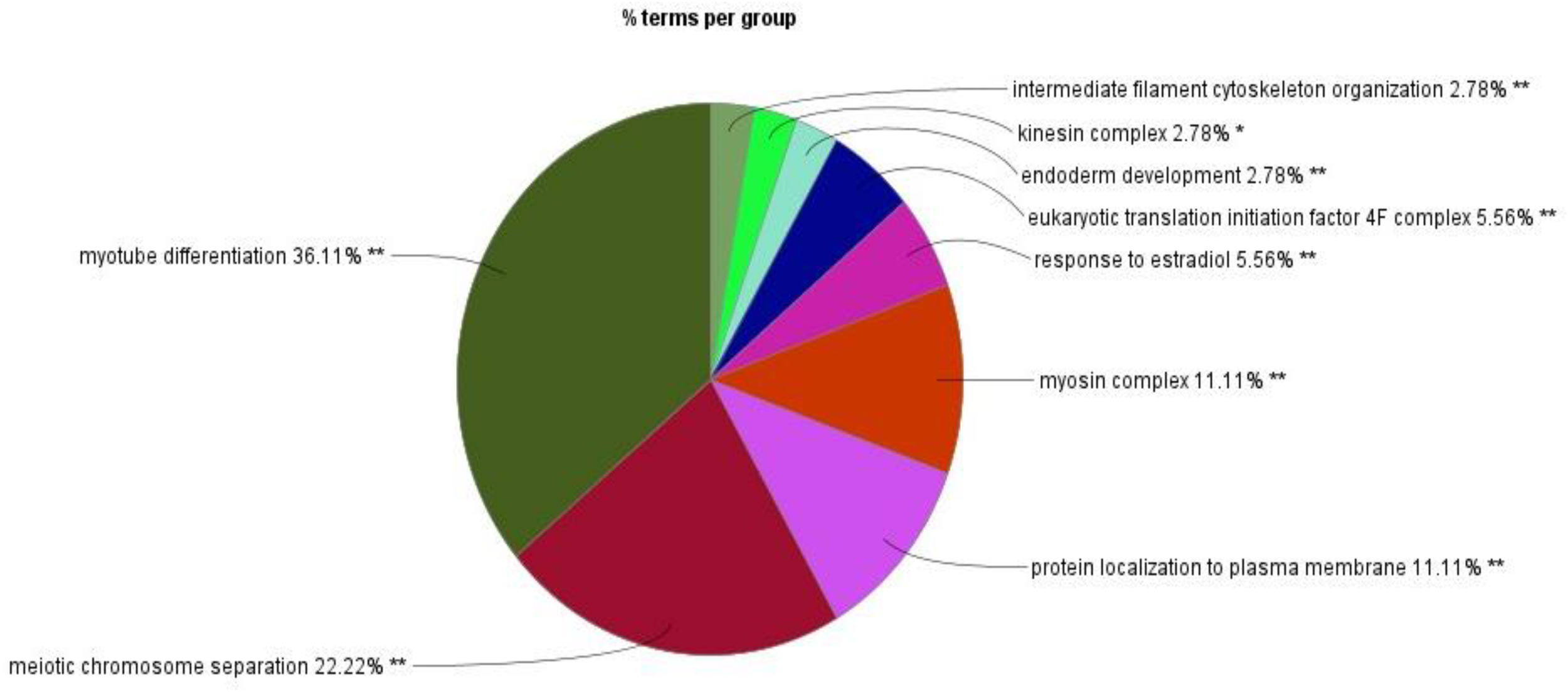
Relative proportion of downregulated proteins across major Gene Ontology (GO) terms identified by ClueGO.
Network analysis
The protein–protein interaction network analysis showed that most upregulated proteins clustered in two main GO categories, “actin filament binding” and “positive regulation of axonogenesis”, which were interconnected to a maximum number of other related biological processes (Figure 5). The actin filament binding term was associated with twelve other related terms or processes, and the positive regulation of axonogenesis was associated with eight other terms or processes. The proteins upregulated in the kinesin complex did not show association with other terms or processes.
Figure 5
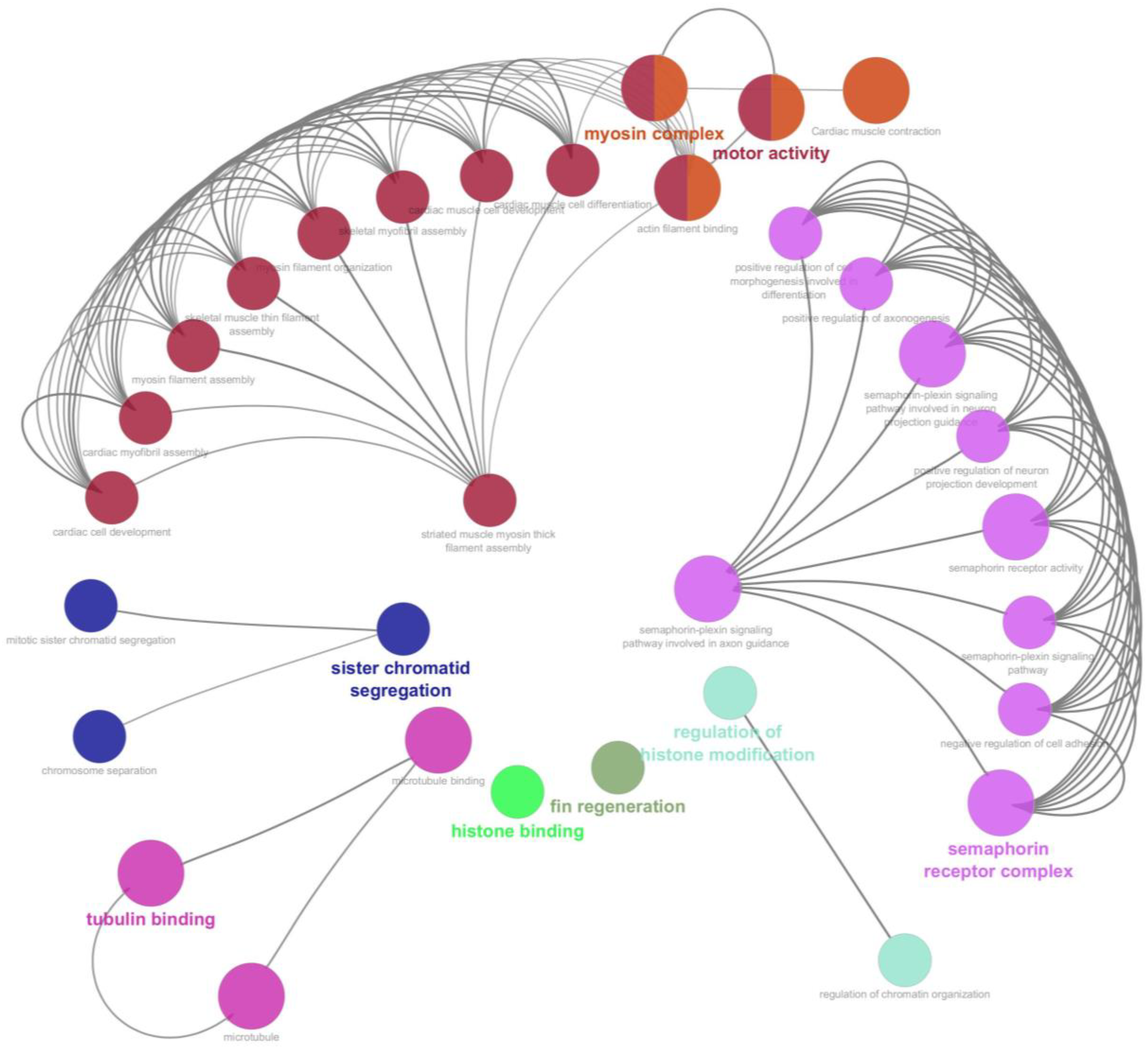
Protein–protein interaction (PPI) network of significantly upregulated proteins (FDR < 0.05) constructed using Cytoscape 3.9.1. Node colors represent Gene Ontology (GO) clusters, and edges represent significant associations.
In protein–protein interaction network analysis, we found that most of the proteins were associated with two main GO categories, “muscle organ morphogenesis” and “meiotic chromosome separation” terms, which were interconnected to a maximum number of other related biological processes (Figure 6). The muscle organ morphogenesis was associated with twelve other terms related to biological processes or terms, and meiotic chromosome separation was associated with seven other terms related to biological processes or terms. The downregulated proteins in kinesin complex, intermediate filament cytoskeleton organizations, and endoderm development terms did not show any association with any other group of proteins or terms.
Figure 6
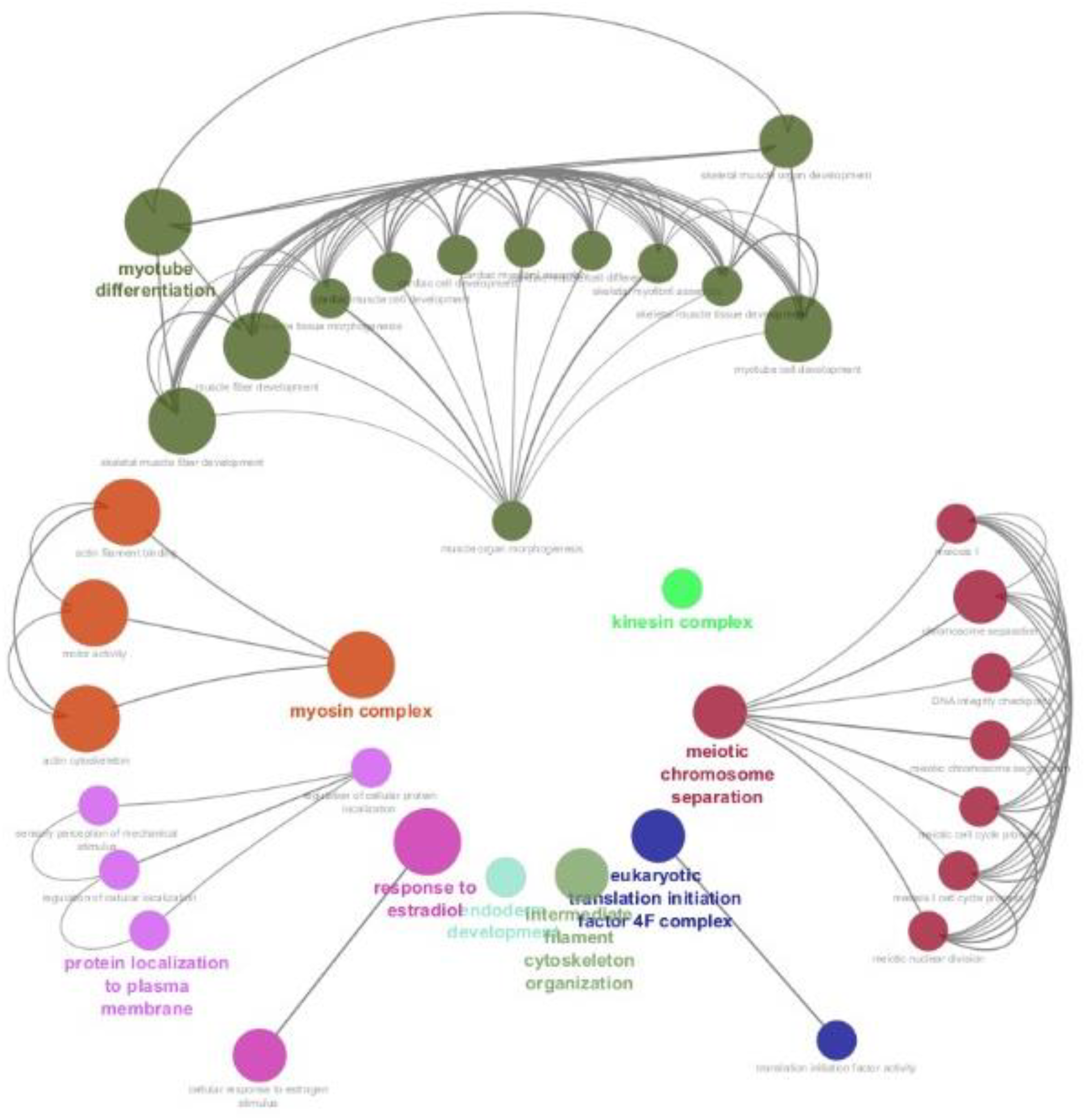
Protein–protein interaction (PPI) network of significantly downregulated proteins (FDR < 0.05). Node clusters represent enriched biological processes. Edge weights indicate significance of associations.
KEGG pathway and WiKiPathways analysis
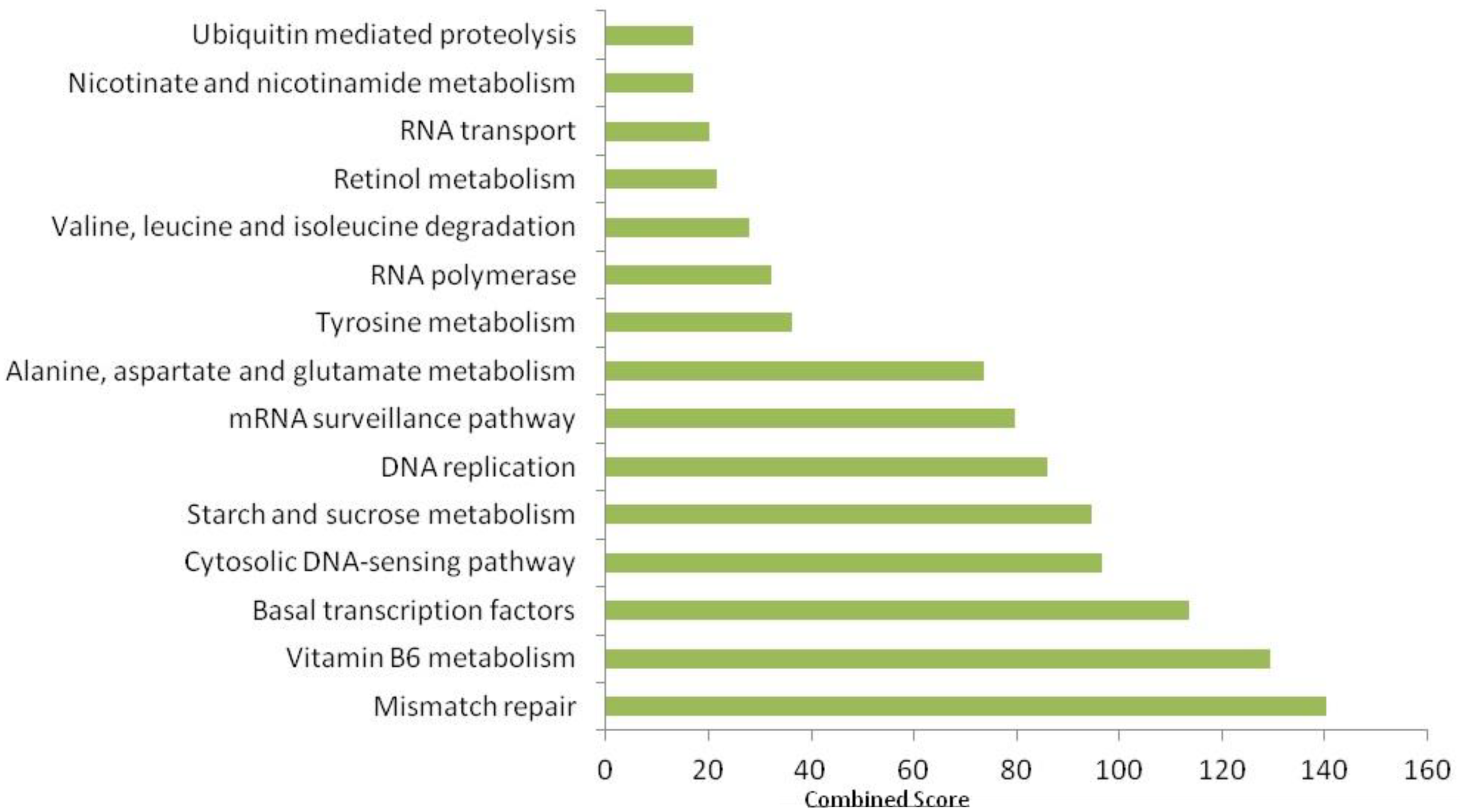
KEGG pathway enrichment analysis using FishEnrichr identified 41 pathways for upregulated proteins and 29 pathways for downregulated proteins. According to KEGG pathway enrichment analysis, it was clear that the maximum number of upregulated proteins (Figure 7) was mainly associated with endocytosis (fam21c, gbf1, asap2b, and eea1), RNA transport (casc3, nup160, and srrm1), adrenergic signaling in cardiomyocytes (myh7l, atp2b1a, and atp1a3b), the regulation of actin cytoskeleton (nckap1, itga3b, and spata13) pathways; downregulated proteins (Figure 8) were associated with tight junction (magi1b, msna, arhgef1a, myh9a, and tj), RNA transport (gemin5, eif4g3b, and eif4g1a), cell cycle (espl1 and anapc1), and cell adhesion molecule (nrxn1a and nrxn3a). WiKiPathways enrichment analysis using FishEnrichr identified 14 pathways for upregulated proteins and 10 pathways for downregulated proteins. According to WiKiPathways analysis, the maximum number of proteins was downregulated (Figure 9) in the glycolysis and gluconeogenesis pathway (eno3 and eno2) and upregulated (Figure 10) in the mRNA processing (prpf6, dhx15, and srrm1) and striated muscle contraction (mybpc2b and mybpc1) pathways.
Figure 7
Top 15 KEGG pathways enriched in significantly upregulated proteins (FDR < 0.05). Enrichment analysis performed using FishEnrichr.
Figure 8
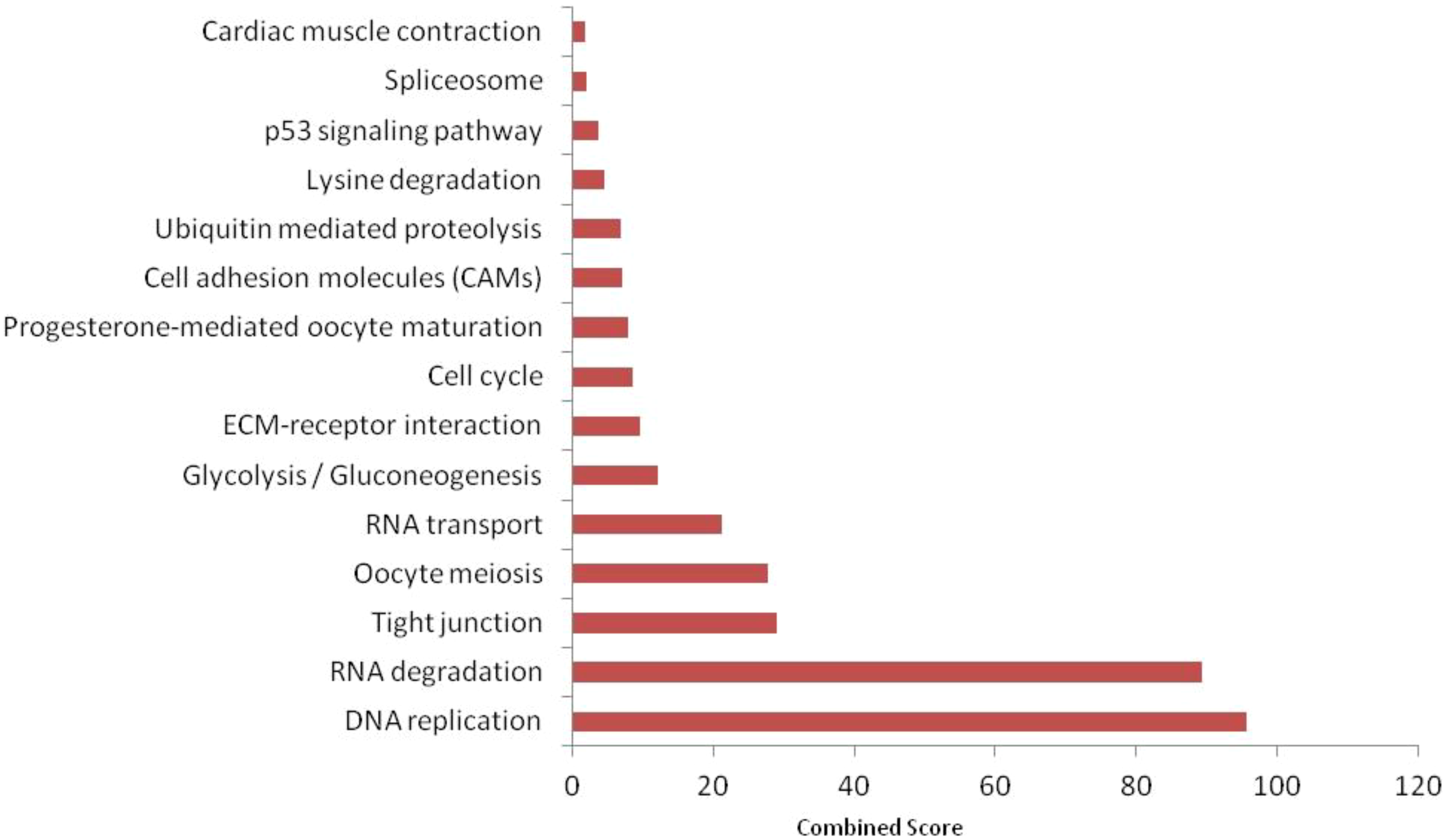
Top 15 KEGG pathways enriched in significantly downregulated proteins (FDR < 0.05) identified by FishEnrichr. Combined score integrates enrichment significance and fold-change impact.
Figure 9
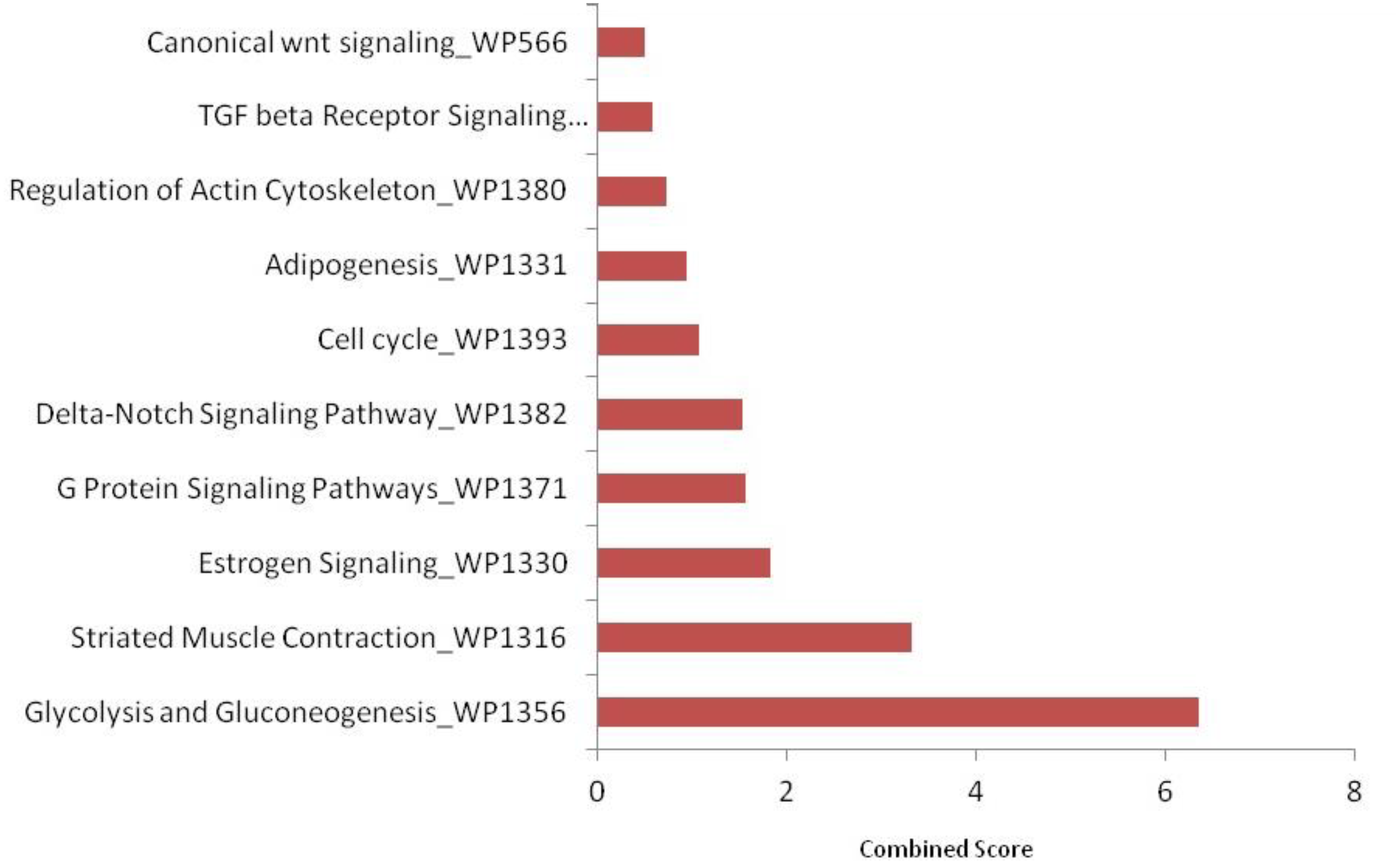
Top 10 WikiPathways enriched in significantly downregulated proteins (FDR < 0.05). Identified using FishEnrichr analysis.
Figure 10
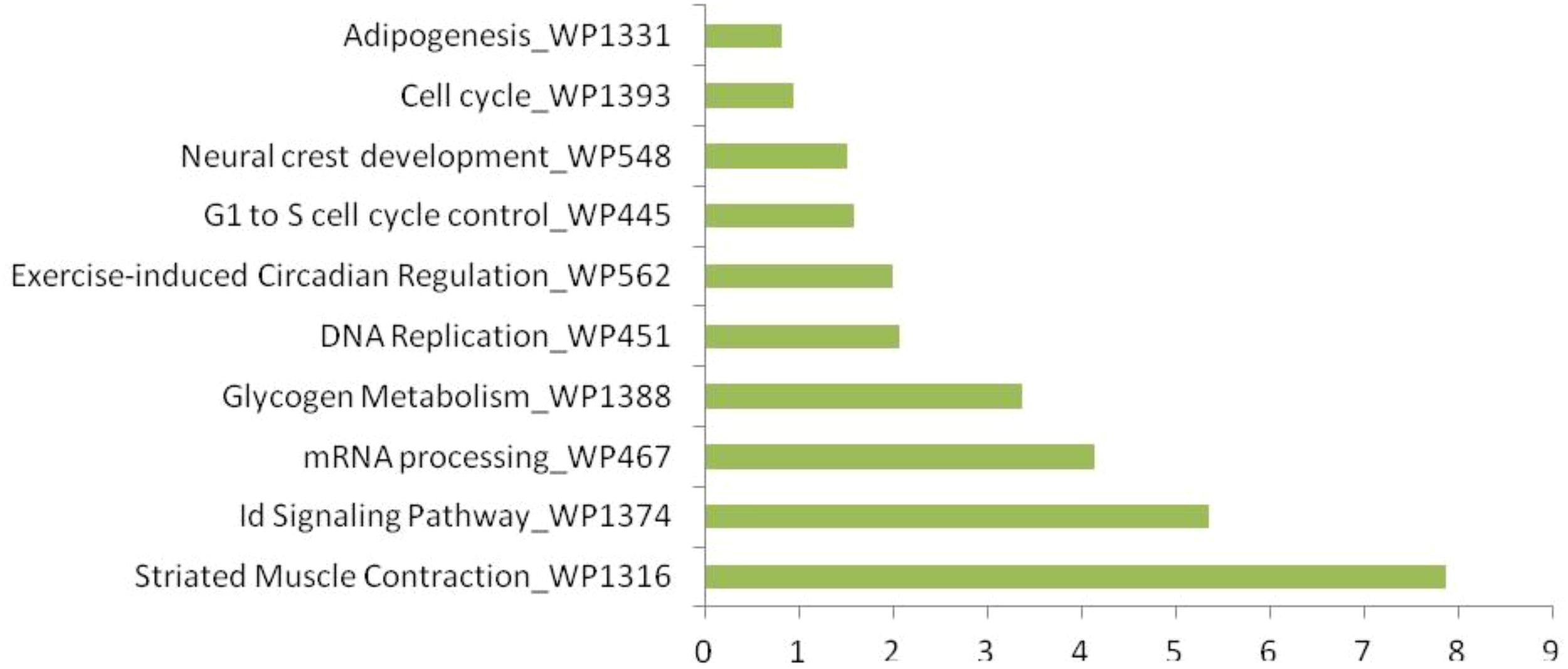
Top 10 WikiPathways enriched in significantly upregulated proteins (FDR < 0.05). Pathways related to mRNA processing, muscle contraction, and cell cycle were prominent.
Statistical proteomics
PCA revealed a distinct separation between the control and triclosan-treated groups, highlighting significant proteomic shifts (Figure 11). ECT2 and Zcchc11 were highly upregulated in the treated group, suggesting their involvement in cellular growth and division. Conversely, EIF4G3B and PBRM1 were downregulated, indicating a suppression of mRNA translation initiation and chromatin remodeling. These expression patterns suggest a disruption of critical cellular processes.
Figure 11
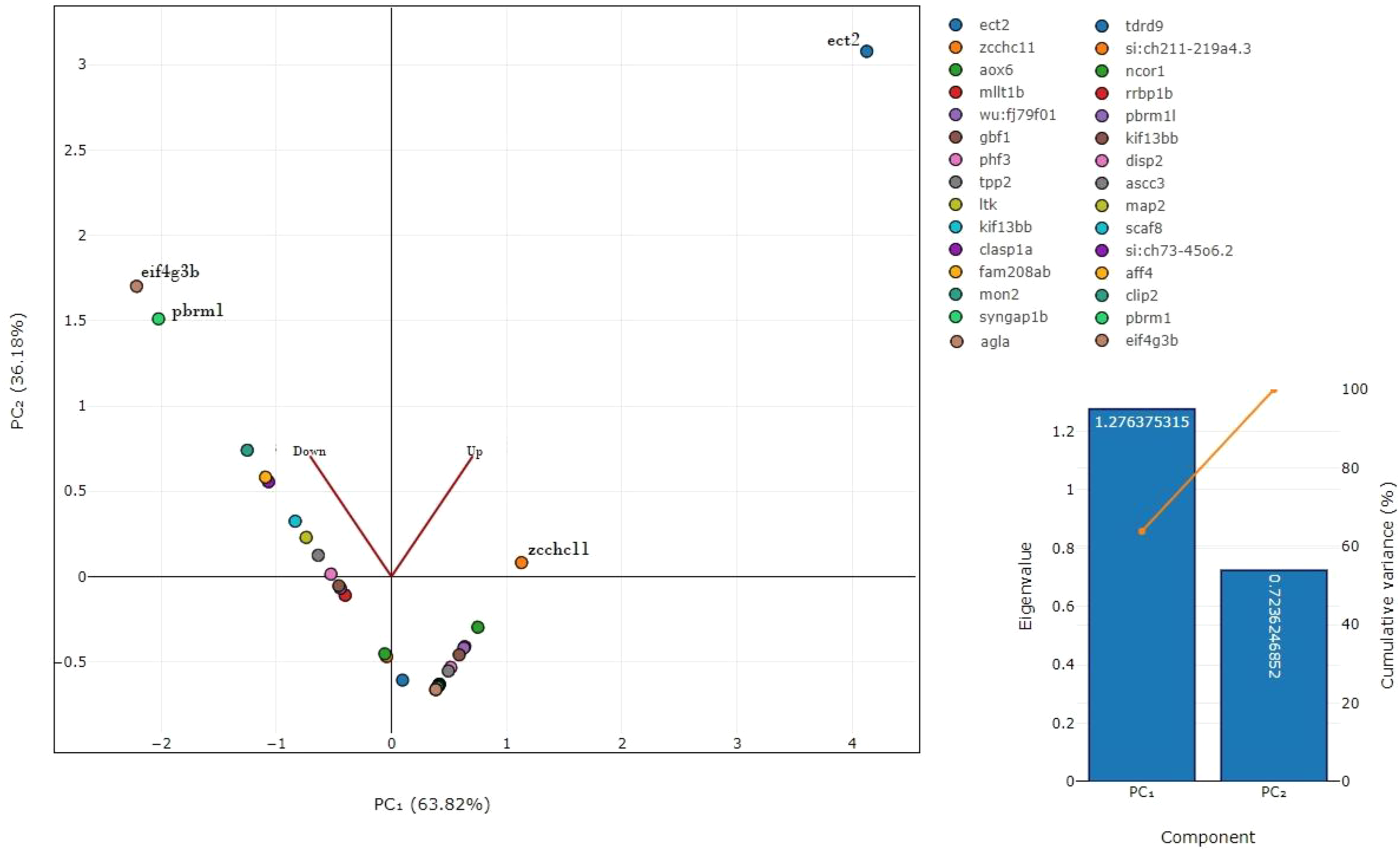
Principal component analysis (PCA) of significantly altered proteins. Clear separation observed between control and triclosan (TCS)-treated groups, highlighting major proteomic shifts. Scree plot shows variance explained by principal components (PCs).
Discussions
One of the challenges in designing experiments with ecotoxicological subjects was selecting the optimum TCS concentration for treatment, which can induce biological processes without causing mortality. The TCS concentration we used for treatment to obtain proteomic data was 0.073 mg/L, which was 1/10 of the 96-h LC50 value for Catla (Adhikari et al., 2023). While lower concentrations (50–100 µg/L) also caused neurodevelopmental changes in zebrafish (Falisse et al., 2017), our chosen sublethal concentration ensured measurable proteomic alterations without mortality.
Several in vitro and in vivo antibiotic exposure studies have been conducted in aquatic animals, and the effects were identified at various cellular and molecular levels, with varying concentrations of antibiotics and exposure time (Fernandez et al., 2021). However, the information related to the effect of TCS on brain tissue is still very scarce. Our main focus of this study was to detail the investigation of the effects of TCS on the brain tissues of L. catla at the cellular and molecular levels using a proteomic approach with the help of bioinformatics analysis. TCS is lipophilic in nature, accumulating in fat-rich tissues like the brain, liver, and muscle tissues (Dar et al., 2022).
The LC–MS/MS data of TCS-treated brain samples suggested that various proteins were upregulated and some proteins were downregulated with respect to their control counterparts. The GO analysis suggested that the maximum number of upregulated proteins involved in fin regeneration and sister chromatid, microtubule cellular component, and tubulin-binding molecular function, through the maximum proportion of upregulated proteins, was involved in motor activities (34.29%), semaphorin receptor complex (25.71%), and myosin complex (11.43%). Among upregulated proteins, kinesin-like protein (kif13bb), microtubule-associated protein (map41), CLIP-associated protein (clasp1a), sister chromatid cohesion proteins (pds5b), cytoskeleton-associated protein (ckap5), calmodulin-regulated spectrin-associated protein (camsap1b), and plexin proteins (plexinb1 and plexinc1) were well overexpressed in Catla brain tissues due to TCS treatment.
The GO analysis of the LC–MS/MS data of TCS-exposed Catla brain tissue samples identified downregulated proteins, which were mainly associated with myotube differentiation (36.11%), meiotic chromosome separation (22.22%), myosin complex (11.11%), and protein localization to the plasma membrane (11.11%) processes. We found some well downregulated proteins due to TCS exposure in Catla in the aforementioned biological processes, and these proteins were myosin heavy chain fast skeletal muscle protein (myha), myosin binding protein c (mybpc1), myosin heavy chain 7B (myh7ba), actin alpha 1 (acta1a and acta1b), lamanin subunit beta 2 (lamb2), and rho family interacting cell polarization regulator 2 (ripo2).
In our study, some proteins were highly downregulated, and some proteins were highly upregulated due to TCS exposure to Catla brain tissues. Highly downregulated proteins were nuclear receptor corepressor 1 isoform (ncor1), poly(ADP ribose) polymerase, ATP-dependent RNA helicase (tdrd9), microtubule-associated proteins (map2), CAP-Gly domain-containing linker protein 2 isoform (clip2), and polybromo 1-like protein (pbrm1l). Nuclear receptor corepressor 1 (ncor1) regulates the activity of thyroid hormone in liver tissues and the hypothalamic pituitary–thyroid axis both in vitro and in vivo (Ritter et al., 2021). ncor1 also hampered lipid oxidation by suppressing PPARα activity. Previous studies have explained that the thyroid axis was disturbed and that oxidative stress was induced due to TCS exposure, which will hamper growth and development as well as behavior and reproduction (Oliveira et al., 2009). The CAP-Gly domain impacts the initial interaction of motor microtubules as well as growing mobility along microtubules (Fan and McKenney, 2023). Two important microtubule-associated proteins, like tan and map2, in vertebrates were closely associated with the nervous system. They mainly promote the assembly and stability of microtubules and may be associated with neural polarity (Goedert et al., 1991). Chronic TCS treatment caused critical impairment of motor neurons in zebrafish (Muth-Kothnen et al., 2012). Similarly, we found that highly downregulated proteins due to TCS exposure in the brain tissues of Catla were related to cellular processes associated with brain functions.
Highly upregulated proteins due to TCS exposure to brain tissues of Catla were protein ECT2 isoform, aldehyde oxidase 6 (aox6), PHD finger protein isoform (phf3), tripeptidyl-peptidase 2 (tpp2), RNA uridylyl transferase (zcchc11), and CLIP-associated protein 1 isoform (clasp1a). Epithelial cell transforming 2 (ECT2) is an oncoprotein that functions as a signaling protein for cell division, cell growth, and other cellular processes (Chen et al., 2015). Many cell division, cell cycle, and apoptosis-related proteins were affected due to TCS exposure (Kim et al., 2015). Enhanced glutathione peroxidase 1 (gpx1) and aldehyde oxidase 1 (aox1) activities were seen as a result of TCS exposure of C57BL/6 mice (Alfhili and Lee, 2019). CLIP-associating proteins (CLASPs) form an evolutionarily conserved protein family that regulates microtubule dynamics and the organizational networks of microtubules (Lawrence et al., 2020). Highly overexpressed proteins in brain tissues due to TCS treatment are mainly cell growth, cell cycle, and microtubule-related cellular processes. The effects of other downregulated and upregulated proteins, including those proteins induced by TCS exposure on brain tissues of Catla, were discussed elaborately with the help of different bioinformatics analyses in the latter part of the discussion.
In our study, slow myosin heavy chain proteins were upregulated, and fast myosin heavy chain proteins were downregulated. The slow myosin protein is associated with long-term movement of muscle with longer interaction, while fast myosin is associated with fast, rapid, and powerful movements with shorter interaction with actin (Shchepkin et al., 2020). The proteomic analysis of zebrafish (D. rerio) and freshwater mussel D. polymorpha suggested the alteration of cytoskeletal myosin protein levels due to TCS treatment (Falisse and Voisin, 2017; Riva et al., 2012). Our study observed that tubulin-binding proteins were upregulated after the TCS treatment of Catla brain tissues. Previously, Riva et al. (2012) showed that Tubulin alpha-2/alpha-4 chain, Tubulin beta-4 chain, and calcium-binding proteins were upregulated due to the TCS treatment of freshwater mussel D. polymorpha. Our data are positively correlated with previous data related to TCS treatment.
Earlier studies have suggested that the TCS treatment of zebrafish upregulated some neurodevelopment and neural maturation genes (Kim et al., 2018; Liu et al., 2018). Chronic TCS exposure led to the severe impairment of motor neurons in zebrafish (Muth-Kothnen et al., 2012). In our study, we also identified some upregulated proteins related to axonogenesis, semaphorin activity, and semaphorin-plexin pathways involved in axon guidance, which was supported by previous studies performed by Kim et al. (2018).
In a previous study, it was observed that actin α1a and cytoplasmic 2 protein were downregulated by TCS exposure in zebrafish (Alfhili and Lee, 2019). In our study, we also found that actin α1 protein was downregulated by the TCS treatment of Catla. Chromosomal stickiness decreased mitotic activity, and effects in the ana-telophase bridge were seen in the bulb of Allium cepa after TCS treatment (Herrero et al., 2012). In our experiment, we also found proteins related to meiotic chromosome separation, which were downregulated after TCS treatment. We found that actin alpha1a and actin alpha 2b proteins were downregulated in brain samples due to the TCS treatment of Catla. Our data were supported by the previous experiments where Falisse and Voisin (2017) and Riva et al. (2012) showed that actin alpha 1, skeletal muscle, and actin cytoplasmic 2 protein were under-expressed due to TCS exposure.
Previous studies have shown that the overexpression or knockdown of ANXA13 can effectively inhibit the proliferation, invasion, migration, and apoptosis of lung adenocarcinoma and colorectal cancer cells (Jiang et al., 2017; You et al., 2019; Ni et al., 2019; Xue et al., 2020). In several studies, it was found that TCS can downregulate some genes, which will regulate apoptosis in different animal cell lines. In our study, we also found that the ANXA13 protein was downregulated, which can induce apoptosis in fish. N-Ethylmaleimide-sensitive factor (NSF), also known as vesicle-fusing ATPase, is an enzyme of homohexameric AAA ATPase that plays an important role in membrane fusion, transferring membrane vesicles from one compartment to another inside eukaryotic cells. Vesicle trafficking malfunction may cause diseases in the nervous system, like dysregulation of synaptic vesicle fusion in the pre-synapse, which leads to neurodegenerative disease (Cui et al., 2022). We found that the vesicle-fusing ATPase protein was downregulated due to TCS treatment, which can lead to neurodegenerative diseases in Catla. Earlier studies have also depicted that TCS can induce several neurodegenerative diseases in animal systems (Kim et al., 2018; Liu et al., 2018).
Earlier studies have observed that TCS could alter biochemical pathways to disrupt endocrine and delay embryonic development in several fish species, including Japanese medaka O. latipes (Ishibashi et al., 2004), zebrafish D. rerio (Oliveira et al., 2009), and major carps like L. rohita, C. mrigala, and C. carpio (Dar et al., 2019, Dar et al., 2020). In this study, it was observed that the proteins related to endoderm development were downregulated (grna and msna) after exposure to TCS.
In a recent study, it was observed that lipid metabolism was disturbed in female zebrafish, while amino acid metabolism, especially involving phenylalanine, alanine, aspartate, and glutamate metabolism, was hindered in male zebrafish (Li et al., 2024). In our study, we also found that proteins related to alanine, aspartate, and glutamate metabolism were upregulated. The amino acids like phenylalanine, alanine, aspartate, and glutamate metabolism were correlated with apoptosis (Feng et al., 2017). Due to TCS exposure, phenylalanine and tyrosine metabolism were significantly altered in zebrafish. Phenylalanine and tyrosine act as precursor molecules for dopamine (Costa and Schoenbaum, 2022), a neurotransmitter associated with reward signaling and important for social and motor behaviors (Dai et al., 2022). Our KEGG analysis found that tyrosine metabolism was hampered in brain tissues due to TCS exposure. TCS exposure can hinder cell cycle and proliferation by downregulating cell cycle-related genes in the brain tissues of zebrafish, potentially disrupting reproductive endocrine function. In KEGG analysis, we observed that cell cycle-related proteins were downregulated in the brain tissues of Catla treated with TCS. In KEGG pathway analysis, it was evident that endocytosis pathways are primarily affected by TCS exposure, as indicated by upregulated proteins. A previous experiment performed by Li et al. (2020) showed that TCS can induce endocytosis in human hepatoma HepG2 cells. In the pathway analysis of upregulated proteins, we found that some proteins were involved in the regulation of the actin cytoskeleton pathway, which was supported by earlier experiments performed by Falisse and Voisin (2017). In our KEGG studies, we found that proteins related to the tight junction pathway were downregulated. Myosin-9 (myh9a), a tight junction-related protein, was associated with thyroid-related pathways, which were downregulated due to TCS treatment. In earlier studies, it has been established that TCS had a negative association with thyroxin (Homburg et al., 2022).
In a recent study, it was observed that TCS treatment impaired lipid and energy metabolism, including glycolipid metabolism, the citrate cycle, and glycolysis (Li et al., 2024). In our study, according to WiKiPathways analysis, we similarly found that the glycolysis, gluconeogenesis, glycogen metabolism, and adipogenesis pathways were affected due to TCS exposure. Adipogenesis plays a crucial role in lipid metabolism by storing excess energy in lipid droplets. Previously, Li et al. (2024) found that the MAPK signaling pathway, calcium signaling pathway, and Wnt signaling pathways were hindered due to TCS exposure in zebrafish. According to WiKiPathways analysis, we also found that some cell signaling pathways, like the G protein signaling pathway, the delta notch signaling pathway, and the Wnt signaling pathway, were hampered by the effect of TCS treatment.
In upregulated and downregulated protein–protein interaction networks, nodes of the same color and interconnectedness collectively represent major biological processes affected by TCS exposure in Catla brain tissues. In our radial network topology model of the protein–protein interaction network, downregulated proteins were interconnected and clustered into three main biological processes: muscle organ morphogenesis, meiotic chromosome separation, and myosin complex. In the upregulated protein–protein interaction network, upregulated proteins were interconnected and clustered into two major biological processes, including motor activity, especially actin filament binding and axonogenesis. The upregulation of ECT2 and Zcchc11 suggests that triclosan may induce a compensatory mechanism related to cell cycle regulation and RNA stabilization in the brain tissue of L. catla. ECT2 (known for its role in cytokinesis) and Zcchc11 (implicated in RNA uridylation) could contribute to stress response mechanisms. However, the downregulation of EIF4G3B and PBRM1 points to an inhibition of protein synthesis and chromatin structure modulation. These alterations may disrupt neuronal function and gene expression regulation, reflecting the neurotoxic effects of triclosan. Our proteomic data analysis of TCS-treated Catla brain tissues showed a high degree of resemblance with previous data related to TCS exposure experiments on several organisms (Falisse and Voisin, 2017; Riva et al., 2012). However, there are a few limitations of the present study. First, brains from five fish per tank were pooled to generate sufficient protein yield, which may have masked inter-individual variability. Second, protein annotation was performed using D. rerio databases due to the absence of a complete L. catla proteome reference, introducing potential cross-species annotation bias. Third, while proteomics provides valuable insights into pathways and candidate biomarkers, functional validation (e.g., behavioral assays, histology, or transcriptomic confirmation) is required to establish causality. These limitations highlight the need for integrative approaches in future studies to confirm the ecological and physiological consequences of triclosan exposure. Detailed cellular, molecular, genetic, histological, and long-term behavioral studies in the near future will give a clear insight into the effect of TCS exposure on aquatic life.
Conclusion
Our study generated the whole proteome profile of the brain tissues of Catla using label-free proteomics (LC–MS/MS). Exposure to sublethal, environmentally relevant concentrations of TCS significantly altered neuronal and muscular pathways. From an eco-physiological perspective, these proteomic alterations suggest potential impairments in locomotion, feeding behavior, and overall fitness, which could negatively affect aquaculture productivity and ecological balance in contaminated habitats. Altered proteins (actin alpha1a/1b, myosin heavy chain fast skeletal muscle, camsap1b, and plexin B1) were consistently identified as potential candidate biomarkers of TCS exposure, pending validation. Future research integrating proteomics with behavioral and histological endpoints will provide a more complete understanding of TCS neurotoxicity in fish.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
All experiments were performed according to the animal utilization protocol approved by the Institutional Animal Ethics Committee, ICAR-CIFRI, Kolkata, India (CIFRI-IAEC/17/2023-24) for the experimental setup. All procedures were made with maximal efforts to minimize fish suffering. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BD: Conceptualization, Data curation, Funding acquisition, Resources, Supervision, Visualization, Writing – original draft. HC: Conceptualization, Data curation, Formal analysis, Methodology, Writing – review & editing. SGh: Data curation, Formal analysis, Investigation, Writing – review & editing. SGa: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. AA: Data curation, Investigation, Methodology, Writing – review & editing. VK: Data curation, Investigation, Methodology, Writing – review & editing. SN: Data curation, Investigation, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work is financially supported by grants-in-aid from the National Mission for Clean Ganga (Ad-35012/1/2023-NMCG-NMCG).
Acknowledgments
The authors acknowledged the director of ICAR-CIFRI for providing the facility to carry out the research work, and also the National Mission for Clean Ganga for providing funds for research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1672496/full#supplementary-material
References
1
Adhikari A. Das B. K. Ganguly S. Nag S. K. Sadhukhan D. Raut S. S. (2023). Emerging contaminant triclosan incites endocrine disruption, reproductive impairments and oxidative stress in the commercially important carp, Catla (Labeo catla): An insight through molecular, histopathological and bioinformatic approach. Comp. Biochem. Physiol. Part - C: Toxicol. Pharmacol.268, 109605. doi: 10.1016/j.cbpc.2023.109605
2
Alfhili M. A. Lee M. (2019). Triclosan: An update on biochemical and molecular mechanisms. Oxid. Med. Cell. Longev., 1607304. doi: 10.1155/2019/1607304
3
Bi R. Zeng X. Mu L. Hou L. Liu W. Li P. et al . (2018). Sensitivities of seven algal species to triclosan, fluoxetine and their mixtures. Sci. Rep.8, 1–10. doi: 10.1038/s41598-018-33785-1
4
Canesi L. Ciacci C. Lorusso L. C. Betti M. Gallo G. Pojana G. et al . (2007). Effects of triclosan on Mytilus galloprovincialis hemocyte function and digestive gland enzyme activities: possible modes of action on non target organisms. Comp. Biochem. Physiol. C Toxicol. Pharmacol.145, 464–472. doi: 10.1016/j.cbpc.2007.02.002
5
Chen J. Xia H. Zhang X. Karthik S. Pratap S. V. Ooi L. L. et al . (2015). ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J. Hepatol.62, 1287–1295. doi: 10.1016/j.jhep.2015.01.014
6
Costa K. M. Schoenbaum G. (2022). Dopamine. Curr. Biol.32, R817–R824. doi: 10.1016/j.cub.2022.06.060
7
Cui L. Li H. Xi Y. Hu Q. Liu H. Fan J. et al . (2022). Vesicle trafficking and vesicle fusion: mechanisms, biological functions, and their implications for potential disease therapy. Mol. Biomed.21, 3(1):29. doi: 10.1186/s43556-022-00090-3
8
Dai B. Sun F. Tong X. Ding Y. Kuang A. Osakada T. et al . (2022). Responses and functions of dopamine in nucleus accumbens core during social behaviors. Cell Rep.40, 111246. doi: 10.1016/j.celrep.2022.111246
9
Dann A. B. Hontela A. (2011). Triclosan: Environmental exposure, toxicity and mechanisms of action. J. Appl. Toxicol.31, 285–311. doi: 10.1002/jat.1660
10
Dar O. I. Aslam R. Pan D. Sharma S. Andotra M. Kaur A. et al . (2022). Source, bioaccumulation, degradability and toxicity of triclosan in aquatic environments: A review. Environ. Technol. Innov.25, 102122. doi: 10.1016/j.eti.2021.102122
11
Dar O. I. Sharma S. Singh K. Kaur A. (2019). Teratogenicity and accumulation of triclosan in the early life stages of four food fish during the bioassay. Ecotoxicol. Environ. Saf.176, 346–354. doi: 10.1016/j.ecoenv.2019.03.102
12
Dar O. I. Sharma S. Singh K. Sharma A. Bhardwaj R. Kaur A. (2020). Biomarkers for the toxicity of sublethal concentrations of triclosan to the early life stages of carps Sci. Rep.10, 1–16. doi: 10.1038/s41598-020-73042-y
13
Dar O. I. Vinothkanna A. Aslam B. Furkh A. Sharma S. Kaur A. et al . (2024). Dynamic alterations in physiological and biochemical indicators of Cirrhinus mrigala hatchlings: A sublethal exposure of triclosan. Sci. Tot Environ.924, 171701. doi: 10.1016/j.scitotenv.2024.171701
14
Dar O. I. Vinothkanna A. Ke X. Chen L. Gao Y. Wang P. et al . (2025). Triclosan-mediated metabolic oxidative stress-triggered cytoskeletal alterations in zebrafish gills and intestine: An integrated biomolecular and NMR-based metabolomics study. J. Hazard Mater492, 138251. doi: 10.1016/j.jhazmat.2025.138251
15
Dhama K. Patel S. K. Kumar R. Masand R. Rana J. Yatoo M. I. et al . (2021). The role of disinfectantsand sanitizers during COVID-19 pandemic: advantagesand deleterious effects on humans and the environment. Environ. Sci. pollut. Res. Int.28, 34211–34228. doi: 10.1007/s11356-021-14429-w
16
Dhillon G. S. Kaur S. Pulicharla R. Brar S. K. Cledón M. Verma M. (2015). Triclosan: Current status, occurrence, environmental risks and bioaccumulation potential. Int. J. Environ. Res. Pub. Heal.12, 5657–5684. doi: 10.3390/ijerph120505657
17
EC (European Commission) . (2016). Commission Implementing Decision (EU) 2016/110 of 27 January 2016 Not Approving Triclosan as an Existing Active Substance for Use in Biocidal Products for Product-Type 1. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32016D0110&from=EN (Accessed July 20, 2025).
18
Falisse E. Voisin A. S. (2017). Impacts of triclosan exposure on zebrafish early-life stage: toxicity and acclimation Mechanisms. Aquat. Toxicol.189, 97–107. doi: 10.1016/j.aquatox.2017.06.003
19
Fan X. McKenney R. J. (2023). Control of motor landing and processivity by the CAP-Gly domain in the KIF13B tail. Nat. Commun.5:14, 4715. doi: 10.1038/s41467-023-40425-4
20
Feng L. Li W. Liu Y. Jiang W. D. Kuang S. Y. Wu P. et al . (2017). Protective role of phenylalanine on the ROS-induced oxidative damage,apoptosis and tight junction damage via Nrf2, TOR and NF-κB signalling molecules in the gill of fish. Fish Shellfish Immunol.60, 185–196. doi: 10.1016/j.fsi.2016.11.048
21
Fernandez R. Colas-Ruiz N. R. Lara-Martín P. A. Fernández-Cisnal R. Hampel M. (2024). Proteomic analysis in the brain and liver of sea bream (Sparus aurata) exposed to the antibiotics ciprofloxacin, sulfadiazine, and trimethoprim. Environ. Pollut. 356, 124308. doi: 10.1016/j.envpol.2024.124308
22
Food and Drug Administration (FDA) . (2016). Safety and effectiveness of consumer antiseptics; topical antimicrobial drug products for over-the-counter human use. Available online: https://www.federalregister.gov/documents/2016/09/06/2016-21337/safety-and-effectiveness-of-consumer-antiseptics-topical-antimicrobial-drug-products-for (Accessed October 5, 2025).
23
Gajahin Gamage N. T. Miyashita R. Takahashi K. Asakawa S. Senevirathna J. D. M. (2022). Proteomic applications in aquatic environment studies. Proteomes.10, 32. doi: 10.3390/proteomes10030032
24
Geiß C. Ruppert K. Heidelbach T. Oehlmann J. (2016). The antimicrobial agents triclocarban and triclosan as potent modulators of reproduction in Potamopyrgus antipodarum (Mollusca: Hydrobiidae). J. Environ. Sci. Heal. A51, 1173–1179. doi: 10.1080/10934529.2016.1206388
25
Goedert M. Goedert M. Crowther R. A. Crowther R. A. Garner C. C. Garner C. C. (1991). Molecular characterization of microtubule-associated proteins tau and map2. Trends Neuroscien.14, 193–199. doi: 10.1016/0166-2236(91)90105-4
26
Herrero O. Perez Martin J. M. Fernandez Freire P. Carvajal Lopez L. Peropadre A. Hazen M. J. (2012). Toxicological evaluation of three contaminants of emerging concern by use of the Allium cepa test. Mutat. Res. 7431-2, 20–24. doi: 10.1016/j.mrgentox.2011.12.028
27
Homburg M. Rasmussen Å. K. Ramhøj L. Feldt-Rasmussen U. (2022). The influence of triclosan on the thyroid hormone system in humans - A systematic review. Front. Endocrinol. (Lausanne).13, 883827. doi: 10.3389/fendo.2022.883827
28
Huang C. L. Ma H. W. Yu C. P. (2014). Substance flow analysis and assessment of environmental exposure potential for triclosan in mainland China. Sci. Total Environ.499, 265–275. doi: 10.1016/j.scitotenv.2014.08.032
29
Ishibashi H. Matsumura N. Hirano M. Matsuoka M. Shiratsuchi H. Ishibashi Y. et al . (2004). Effects of triclosan on the early lifestages and reproduction of medaka Oryzias latipes and induction of hepatic vitellogenin. Aquat. Toxicol.67, 167–179. doi: 10.1016/j.aquatox.2003.12.005
30
Jiang G. Wang P. Wang W. Li W. Dai L. Chen K. (2017). Annexin A13 promotes tumor cell invasion in vitro and is associated with metastasis in human colorectal cancer. Oncotarget.8, 21663–21673. doi: 10.18632/oncotarget.15523
31
Kim S. H. Hwang K. A. Shim S. M. Choi K. C. (2015). Growth and migration of LNCaP prostate cancer cells are promoted by triclosan and benzophenone-1 via an androgen receptor signaling pathway. Environ. Toxicol. Pharmacol.39:2, 568–576. doi: 10.1016/j.etap.2015.01.003
32
Kim J. Oh H B. Ryu B. (2018). Triclosan affects axon formation in the neural development stages of zebrafish embryos (Danio rerio) Environ. Pollut.236, 304–312. doi: 10.1016/j.envpol.2017.12.110
33
Lawrence E. J. Zanic M. Rice L. M. (2020). CLASPs at a glance. J. Cell Sci.133, 243097. doi: 10.1242/jcs.243097
34
Li L. (2021). Toxicity evaluation and by-products identification of triclosan ozonation and chlorination. Chemosphere263, 128223. doi: 10.1016/j.chemosphere.2020.128223
35
Li X. Shang Y. Yao W. Li Y. Tang N. An J. et al . (2020). Comparison of transcriptomics changes induced by TCS and MTCS exposure in human hepatoma hepG2 cells. ACS Omega5, 10715–10724. doi: 10.1021/acsomega.0c00075
36
Li Z. Xian H. Ren X. Ye R. Zhong Y. Huang Y. et al . (2024). Insight into Triclosan-induced endocrine disruption: evidence from the National Health and Nutrition examination survey and zebrafish model. EnvHealth2:7, 424–440. doi: 10.1021/envhealth.4c00045
37
Liu J. Sun L. Zhang H. (2018). Response mechanisms to joint exposure of triclosan and its chlorinated derivatives on zebrafish (Danio rerio) behaviour. Chemosphere193, 820–832. doi: 10.1016/j.chemosphere.2017.11.106
38
Lu J. Guo J. (2021). Disinfection spreads antimicrobial resistance. Science371, 474. doi: 10.1126/science.abg4380
39
Lu J. Wang Y. Li J. Mao L. Hoang S. Duarte T. (2018). Triclosan at environmentally relevant concentrations promotes horizontal transfer of multidrug resistance genes within and across bacterial genera. Environ. Int.121, 1217–1226. doi: 10.1016/j.envint.2018.10.040
40
Montville R. Schaffner D. W. (2011). A meta-analysis of the published literature on the effectiveness of antimicrobial soaps. J. @ Food Protec.74, 1875–1882. doi: 10.4315/0362-028X.JFP-11-122
41
Mukherjee S. Boral S. Siddiqi H. Mishra A. Meikap B. C. (2021). Present cum future of SARS-CoV-2 virus and its associated control of virus-laden air pollutants leading to potential environmental threat—a global review. J. Environ. Chem. Eng.9, 104973. doi: 10.1016/j.jece.2020.104973
42
Muth-Kothnen E. Wichmann A. Delov V. Fenske M. (2012). The classification of motor neuron defects in the zebrafish embryo toxicity test (ZFET) as an animal alternative approach to assess developmental neurotoxicity Neurotoxicol. Teratol.34, 413–424. doi: 10.1016/j.ntt.2012.04.006
43
Ni J. S. Zheng H. Huang Z. P. Hong Y. G. Ou Y. L. Tao Y. P. et al . (2019). MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol. Lett.17, 2317–2327. doi: 10.3892/ol.2018.9848
44
Oliveira R. Domingues I. Grisolia C. K. Soares A. M. (2009). Effects of triclosan on zebrafish early-life stages and adults. Environ. Sci. pollut.16, 679–688. doi: 10.1007/s11356-009-0119-3
45
Orvos D. R. Versteeg D. J. Inauen J. Capdevielle M. Rothenstein A. Cunningham V. (2002). Aquatic toxicity of triclosan. Environ. Toxicol. Chem.21, 1338–1349. doi: 10.1002/etc.5620210703
46
Peng Y. Luo Y. Nie X. P. Liao W. Yang Y. F. Ying G. G. (2013). Toxic effects of triclosan on the detoxification system and breeding of Daphnia magna. .Ecotoxicology22, 1384–1394. doi: 10.1007/s10646-013-1124-3
47
Prosser R. S. Lissemore L. Topp E. Sibley P. K. (2014). Bioaccumulation of triclosan and triclocarban in plants grown in soils amended with municipal dewatered biosolids. Environ. Toxicol. Chem.33, 975–984. doi: 10.1002/etc.2505
48
Queckenberg C. Meins J. Wachall B. (2010). Absorption, pharmacokinetics, and safety of triclosan after dermal administration. Antimicrobial Agents Chemotherapy.54, 570–572. doi: 10.1128/AAC.00615-09
49
Ritter M. J. Amano I. Imai N. Soares De Oliveira L. Vella K. R. Hollenberg A. N. (2021). Nuclear Receptor CoRepressors, NCOR1 and SMRT, are required for maintaining systemic metabolic homeostasis. Mol. Metab.53, 101315. doi: 10.1016/j.molmet.2021.101315
50
Riva C. Cristoni S. Binelli A. (2012). Effects of triclosan in the freshwater mussel Dreissena polymorpha: a proteomic investigation,” Aquat. Toxicol.118-119, 62–71. doi: 10.1016/j.aquatox.2012.03.013
51
Sahoo L. Das P. Sahoo B. Das G. Meher P. K. Udit U. K. et al . (2020). The draft genome of Labeo catla. BMC Res. Notes13, 411. doi: 10.1186/s13104-020-05240-w
52
Sandborgh-Englund G. Adolfsson-Erici M. Odham G. Ekstrand J. (2006). Pharmacokinetics of triclosan following oral ingestion in humans. J. Toxicol. Env. Heal. Part A.69, 1861–1873. doi: 10.1080/15287390600631706
53
Sharma S. Dar O. I. Andotra M. Sharma S. Bhagat A. Thakur S. et al . (2022a). Cellular, molecular and genomic alterations in the hatchlings of Labeo rohita after exposure to Triclosan. Front. Environ. Sci.10, 992435. doi: 10.3389/fenvs.2022.992435
54
Sharma S. Dar O. I. Thakur S. Kesavan A. K. Kaur A. (2022b). Environmentally relevant concentrations of Triclosan cause transcriptomic and biomolecular alterations in the hatchlings of Labeo rohita. Environ. Toxicol. Pharmacol.96, 104004. doi: 10.1016/j.etap.2022.104004
55
Shchepkin D. V. Nabiev S. R. Koubassova N. A. (2020). Comparison of functional characteristics of myosin in fast and slow skeletal muscles. Bull. Exp. Biol. Med.169, 338–341. doi: 10.1007/s10517-020-04882-x
56
Vingskes A. K. Spann N. (2018). The toxicity of a mixture of two antiseptics, triclosan and triclocarban, on reproduction and growth of the nematode Caenorhabditis elegans. Ecotoxicology27, 420–429. doi: 10.1007/s10646-018-1905-9
57
Weatherly L. M. Gosse J. A. (2017). Triclosan exposure, transformation, and human health effects. J. Toxicol. Env. Heal. B20, 447–469. doi: 10.1080/10937404.2017.1399306
58
Xue G. L. Zhang C. Zheng G. L. Zhang L. J. Bi J. W. (2020). Annexin A13 predicts poor prognosis for lung adenocarcinoma patients and accelerates the proliferation and migration of lung adenocarcinoma cells by modulating epithelial-mesenchymal transition. Fundam. Clin. Pharmacol.34, 687–696. doi: 10.1111/fcp.12555
59
Yao L. Zhao J. L. Liu Y. S. Zhang Q. Q. Jiang Y. X. Liu S. (2018). Personal care products in wild fish in two main Chinese rivers: Bioaccumulation potential and human health risks. Sci. Total Environ.621, 1093–1102. doi: 10.1016/j.scitotenv.2017.10.117
60
You C. Jin L. Xu Q. Shen B. Jiao X. Huang X. (2019). Expression of miR-21 and miR-138 in colon cancer and its effect on cell proliferation and prognosis. Oncol. Lett.17, 2271–2277. doi: 10.3892/ol.2018.9864
61
Yueh M. F. Tukey R. H. (2016). Triclosan: A widespread environmental toxicant with many biological effects. Annu. Rev. Pharmacol. Toxicol.56, 251–272. doi: 10.1146/annurev-pharmtox-010715-103417
Summary
Keywords
triclosan, bioinformatics, proteomics, Labeo catla , brain
Citation
Das BK, Chakraborty HJ, Ghosh S, Ganguly S, Adhikari A, Kumar V and Kumar Nag S (2025) Environmentally relevant concentrations of triclosan modulate the brain proteome profile of Labeo catla. Front. Mar. Sci. 12:1672496. doi: 10.3389/fmars.2025.1672496
Received
25 July 2025
Accepted
10 October 2025
Published
23 October 2025
Volume
12 - 2025
Edited by
Yafei Duan, South China Sea Fisheries Research Institute, China
Reviewed by
Rodolfo Rondon, Instituto Antártico Chileno (INACH), Chile; Owias Iqbal Dar, Hainan University, China
Updates
Copyright
© 2025 Das, Chakraborty, Ghosh, Ganguly, Adhikari, Kumar and Kumar Nag.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Basanta Kumar Das, basantakumard@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.