Abstract
In autumn 2020, cultured sea cucumbers (Apostichopus japonicus) in the Xiapu Sea area (Southeast China) exhibited ulcerative skin lesions. To identify the causative pathogen, the Gram-negative strain 10MKXP20 was isolated from the muscle tissue of diseased animals and identified as Photobacterium leiognathi through phenotypic and genetic analysis. The bacterium was sensitive to 14 antibiotics, including tetracycline and enrofloxacin, but resistant to gentamicin, neomycin, kanamycin, rifampicin, amikacin, and amoxicillin. Minimum inhibitory concentration (MIC) assays revealed varying efficacy among the tested antibiotics, with enrofloxacin, doxycycline, and florfenicol showing particularly low MICs, indicating strong inhibitory activity. Challenge experiments via immersion and intramuscular injection confirmed the pathogenicity of P. leiognathi 10MKXP20, with mortality rates being dose-dependent. This study is the first to identify P. leiognathi as the causative agent of ulcerative skin disease in sea cucumbers, providing crucial insights for disease diagnosis and antibiotic treatment.
1 Introduction
The sea cucumber, Apostichopus japonicus, plays an important role in contributing to aquaculture production in the coastal areas of China, Korea, Japan, and Russia (Chang et al., 2009; Lu, 2019). In China, A. japonicus is the most popular species that is mainly cultured in Liaoning, Shandong, Hebei, and Fujian (Fisheries Bureau of the Ministry of Agriculture, 2021). Sea cucumber has been considered a source of high nutritional and medicinal values (Zhao et al., 2021). The rapid development of the sea cucumber aquaculture industry has led to the occurrence of problems that cause huge economic losses. Among them, disease outbreaks are one of the important factors limiting the sustainable development of the sea cucumber aquaculture industry (Li et al., 2012). Skin ulceration syndrome is one of the serious diseases that occurs in sea cucumbers cultured in both hatchery and pond culture systems in China (Yang, 2020; Deng et al., 2008). The disease annually happens in February to March (to animals cultured in hatcheries) and March to April (to animals cultured in ponds) and widely affects sea cucumbers with high mortality (usually 50%) in the adult and mortality can reach up to 100% at the juvenile stage (Deng et al., 2005). Sea cucumbers show syndromes of weak activity, anorexia, swollen mouth, evisceration, skin erosion, and deep ulceration (Li et al., 2012). According to national food safety regulations, sea cucumbers with ulcerated skin are prohibited from being sold in the market. Such sea cucumbers, characterized by ulcerated epidermis, constitute a direct health risk to consumers, as they may harbor pathogenic microorganisms, chemical contaminants, and parasites. These ulcers are often caused by bacterial infections (including Vibrio parahaemolyticus and Salmonella sp.) or fungal infections, can lead to serious food poisoning (Zhang et al., 2018).
Previous studies have reported that bacteria, fungi, and viruses are related to disease outbreaks in cultured sea cucumbers (Wang et al., 2021a, 2021b). Studies have confirmed the existence and pathogenicity of bacterial pathogens in sea cucumbers with skin ulceration syndromes (Becker et al., 2004). For example, the two bacteria, Pseudoalteromonas sp. and Pseudoalteromonas tetraodonis, were isolated from the diseased sea cucumber (A. japonicus) with the symptoms of ulcer spots on the dorsal skin and abdominal parapodia (Liu et al., 2010). Another study reported that Vibrio cyclitrophicus, V. splendidus, V. harveyi, V. tasmaniensis, Photobacterium damselae subsp. damselae, Arthrobacter protophormiae, and Staphylococcus equorum were isolated from A. japonicus suffering from skin ulceration (Deng et al., 2009). The results of the experiment showed that all six major bacterial isolates were pathogenic and exhibited the same symptoms as natural infection. Moreover, V. parahaemolyticus, a common zoonotic bacterium known to cause infections and morbidity in both A. japonicus and humans, has been identified as the primary cause of vibriosis in A. japonicus aquaculture. This condition is particularly prevalent in larvae, which exhibit symptoms such as reduced attachment, body contraction, and collapsing (Ren et al., 2019).
In late October 2020, an outbreak of skin ulcers was detected in A. japonicus cultured in an indoor pond system in Xiapu, China, accompanied by a significant mortality rate. The pathogenicity of Photobacterium damselae subsp. damselae against skin ulcer syndromes in A. japonicus was confirmed in a previous study (Wang et al., 2022). However, during the isolation of bacteria from diseased sea cucumbers, the bacterial isolate, namely 10MKXP20, showed more predominance in the culture plates. In the present study, we isolated and identified the bacterial isolate 10MKXP20 and conducted a challenge test to confirm the pathogenic agents causing skin ulceration in A. japonicus. The findings of this study enhance our understanding of the pathogens causing diseases in sea cucumbers, which facilitates the discovery and development of strategies for preventing and controlling disease outbreaks in the aquaculture industry of sea cucumbers.
2 Materials and methods
2.1 Sea cucumbers
The sea cucumbers were cultured on a farm in Xiapu, China (N26°40’21.99”, E119°57’49.91”). The water temperature in the culture ponds was between 23-25°C, and fed on a commercial formulated diet once a day. In late October 2020, a significant disease outbreak occurred, characterized by body atrophy, anorexia, and extensive ulcerated patches on the body surface. To investigate the cause of the outbreak, 30 diseased (with ulceration in the body surface and external lesions) and 30 healthy sea cucumbers (body weight 32.8 ± 3.7 g) were collected from each group, respectively. All samples were promptly transported in a 200-L tank with aeration to the laboratory within 6 h. Animals that died during transportation were not considered in the subsequent analysis (Deng et al., 2008).
2.2 Histological analysis
Sea cucumber body wall samples were fixed overnight in 10% buffered formalin. For histological analysis, tissues were dehydrated through an ethanol gradient, cleared in xylene, and paraffin-embedded. We sectioned the embedded tissues at 5 µm using a microtome, stained the sections with H&E, and imaged them using a Feica DM5500 light microscope.
2.3 Isolation of bacteria from diseased sea cucumber
For bacterial isolation, tissue samples were aseptically taken from the sea cucumber body wall, respiratory tree, and intestinal tract of a moribund diseased sea cucumber. Tissue samples were streaked onto plates of thiosulphate citrate bile sucrose (TCBS) agar (Huankai Microbial, Hangzhou, China) and tryptone soy agar (BD Difco, Sparks, MD, USA) supplemented with 1% NaCl (TSA-1) plates. The cultures were incubated for 48 h at 25°C. Both media plates showed signs of bacterial development. Re-streaking onto new TSA-1 plates allowed for the purification of five randomly chosen single colonies from the plates. Following an overnight culture in TSB-1, the purified bacterial isolates were kept in 20% glycerol stock at -80°C until further analysis.
2.4 Bacterial identification
The ultrastructural morphology of bacterial isolates was examined via scanning electron microscopy (HT7800, Hitachi, Japan). Specimens underwent chemical fixation in 2.5% glutaraldehyde, followed by graded ethanol dehydration and critical-point drying. Subsequently, the specimens were sputter-coated with a 10 nm gold-palladium alloy. Micrographs were captured at an accelerating voltage of 180 kV, with working distances kept between 8-10 mm and magnifications spanning from ×8,000 (scale bars = 1μm).
Biochemical tests and 16S rRNA gene sequence analysis were used to identify the bacterial isolates that were isolated from diseased sea cucumbers. Biochemical tests of isolates were performed by using a commercial Biolog system (Gen III MicroStation, BIOLOG, USA) according to the manufacturer’s instructions. For 48 h, the growth of bacteria on TCBS agar was examined. The standard soft-agar stabbing method was used to detect motility through a motility test agar (Luria-Bertani (LB) broth supplemented with 0.25% agar). The standard diffusion method was used to identify hemolytic phenotypes using TSA-1 plates supplemented with 5% sheep blood. In all of the tests, the culture temperature was 25°C.
For genetic identification of isolates, bacterial genomic DNA was first extracted using the TIAamp Bacteria DNA Kit (Beijing Tiangen Biotech Co., Ltd.). The 16S rRNA was amplified by PCR using the primer pair 27F (5′-AGAGTTTGATCCTGGCTCAG-3′)/1492R (5′-ACCTTGTTACGACTT-3′) (Gürtler and Stanisich, 1996). The gyrB gene was amplified by PCR using the primer pair 3F primer (5′−TCCGGCGGTCTGCACGGCGT−3′) and 14R primer (5′−TTGTCCGGGTTGTACTCGTC−3′) (Soler et al., 2004). The PCR reaction was conducted using the Green Taq Mix (Vazyme Biotech Co., Ltd.). The PCR amplification was performed under a three-step protocol: initial denaturation at 98°C for 30 s, followed by 35 cycles of denaturation at 98°C for 30 s and annealing at 60°C for 90 s, with fluorescence data collected during the annealing phase. Finally, the melting curve was analyzed from 65°C to 95°C. The 16S amplicons were then sequenced on a Sanger sequencing platform. Identification of bacterial species using the online BLAST tool available at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) (Altschul et al., 1990). The criterion used for species-level identification was the identification of more than 98.7% of the 16S rRNA gene (Drancourt et al., 2000). Neighbor-Joining method was used to determine the evolutionary history of these bacterial isolates based on the 16S rRNA gene using MEGA7.0 (Kumar et al., 2016, Saitou and Nei, 1987).
2.5 Challenge tests
The bacterial isolate P. leiognathi 10MKXP20 was cultured in TSB supplemented with 1% NaCl overnight at 25°C. To obtain a mid-exponential culture, the overnight culture was diluted 1:10 with fresh TSB-1 medium and incubated with shaking for an additional 3 h at 25°C. Following this, the bacteria were centrifuged, re-suspended in phosphate-buffered saline (PBS), and adjusted to an OD600 of 1.0. The bacterial resuspensions were subsequently used in the experimental infection.
Healthy sea cucumbers (body weight 22.9 ± 2.5 g) without clinical signs were purchased from a local farm (Zhangzhou, Fujian, China). Sea cucumbers were transferred to the laboratory, maintained in 200-L tanks with aeration, and acclimated to the laboratory conditions (water temperature: 22°C, salinity: 30‰) for a week. Prior to the infection, five sea cucumbers were randomly selected for bacterial isolation from the body wall, respiratory tree, and intestinal tract to confirm the absence of P. leiognathi in animals. For the intramuscular injection and immersion infections, the experimental and control groups were randomly divided into four groups, each including three replicates of 20 sea cucumbers. The two groups were housed in 12 tanks, with each tank containing 20 individuals, respectively. In the injection challenge, sea cucumbers in the experimental group were intramuscularly injected with 0.1 mL of bacterial suspension at the final dose of 1.2×104, 1.2×105, 1.2×106, 1.2×107, or 1.2×108 CFU/mL; whereas sea cucumbers in the control group was injected with an equal volume of PBS.
In the immersion infection, sea cucumbers were immersed in a suspension of P. leiognathi 10MKXP20 in 40 L of UV-disinfected seawater containing 1.2×105 or 1.2×107 CFU/mL for 2 h and then moved to 200-L tanks of seawater. The control animals were immersed in the same seawater without the addition of the bacterium. All sea cucumbers in challenge tests were kept at 20 ± 1°C. The presence of clinical signs and mortality of sea cucumbers was observed and recorded daily. For both challenge models, the moribund animals with clinical signs were observed by gross inspection, and the tissues were collected and used for bacterial isolation. Re-isolation and re-identification of bacteria (strain 4CF10XP) were performed.
2.6 Drug sensitivity analysis
The drug sensitivity of pathogenic bacteria was analyzed by the agar plate diffusion method (K-B method) (Wang et al., 2022). Single colonies of strain 10MKXP20 were collected and placed in sterile saline to produce a suspension with a concentration of about 1.2×108 CFU/mL (0.5 MJ). The chemotherapeutic agent discs (Binhe Microbiology Reagent, Hangzhou, China) were placed on the surface of the plates. The plate was incubated at 25°C for 18 h and used for the measurement of the inhibition zone diameter. MIC values of the pathogenic bacteria against eight kinds of antimicrobial agents permitted for use in aquaculture by the Ministry of Agriculture and Rural Affairs (China) were determined by the micro-method (Wang et al., 2022). The suspension of P. leiognathi 10MKXP20 was prepared at a concentration of 2.2×106 CFU/mL for use, and a volume of 200 μL of bacterial suspension was added to the 96-well broth microdilution plate (Nanjing Finn Medical Science, Nanjing, China). The test plate was cultured at 25°C for 18 h, and the results were observed. The sensitivity of the pathogen to the drug was judged by reference to the Clinical and Laboratory Standards Institute (CLSI) (Walker, 1999).
3 Results
3.1 Clinical signs and histology of infected sea cucumbers
Clinical observations of diseased sea cucumbers revealed the presence of small white erosions around their bodies. The body wall of affected individuals exhibited excessive secretion of white mucus, and most exhibited an inability to move in response to mechanical stimuli (Figure 1). Histopathological examination showed that the epidermis was completely lost over a length of 500 μm in affected areas. Collagen bundles originating from the dense connective tissue extended into the underlying loose connective tissue beneath the area of epidermal loss. The upper portion of the “T”-shaped collagen structure formed by these bundles acts to seal the epidermal defect. Additionally, collagen bundles from the dense connective tissue at the center of the lesion formed a plug that extended hundreds of microns outward from the body surface. At the lesion’s periphery, collagen fibers from the dense connective tissue replaced the remaining loose connective tissue beneath the thinned epidermis, which was reduced to less than 100 μm in thickness. In the context of ulceration, the pallid appearance of the lesions is attributable to the exposure of connective tissue that occurs following the destruction of the stratum corneum, the epidermis, and the upper portion of the connective tissue layer (Figure 2).
Figure 1
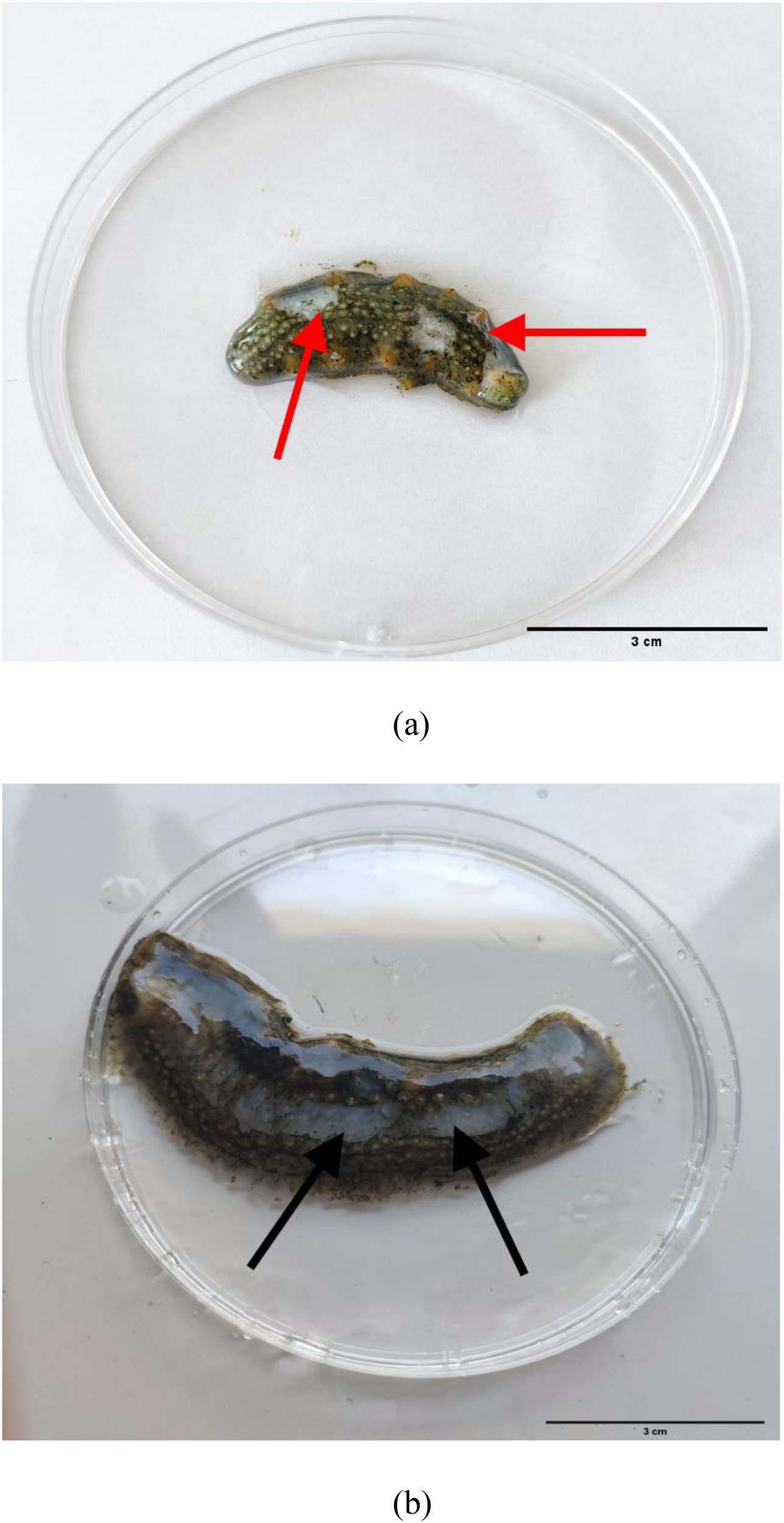
Clinical signs of sea cucumber infected with P. leiognathi 10MKXP20 under natural (A), and artificial infections (B). Red arrows represent pathological symptoms under natural infection, while black arrows represent pathological symptoms under bacterium P. leiognathi 10MKXP20 infection.
Figure 2
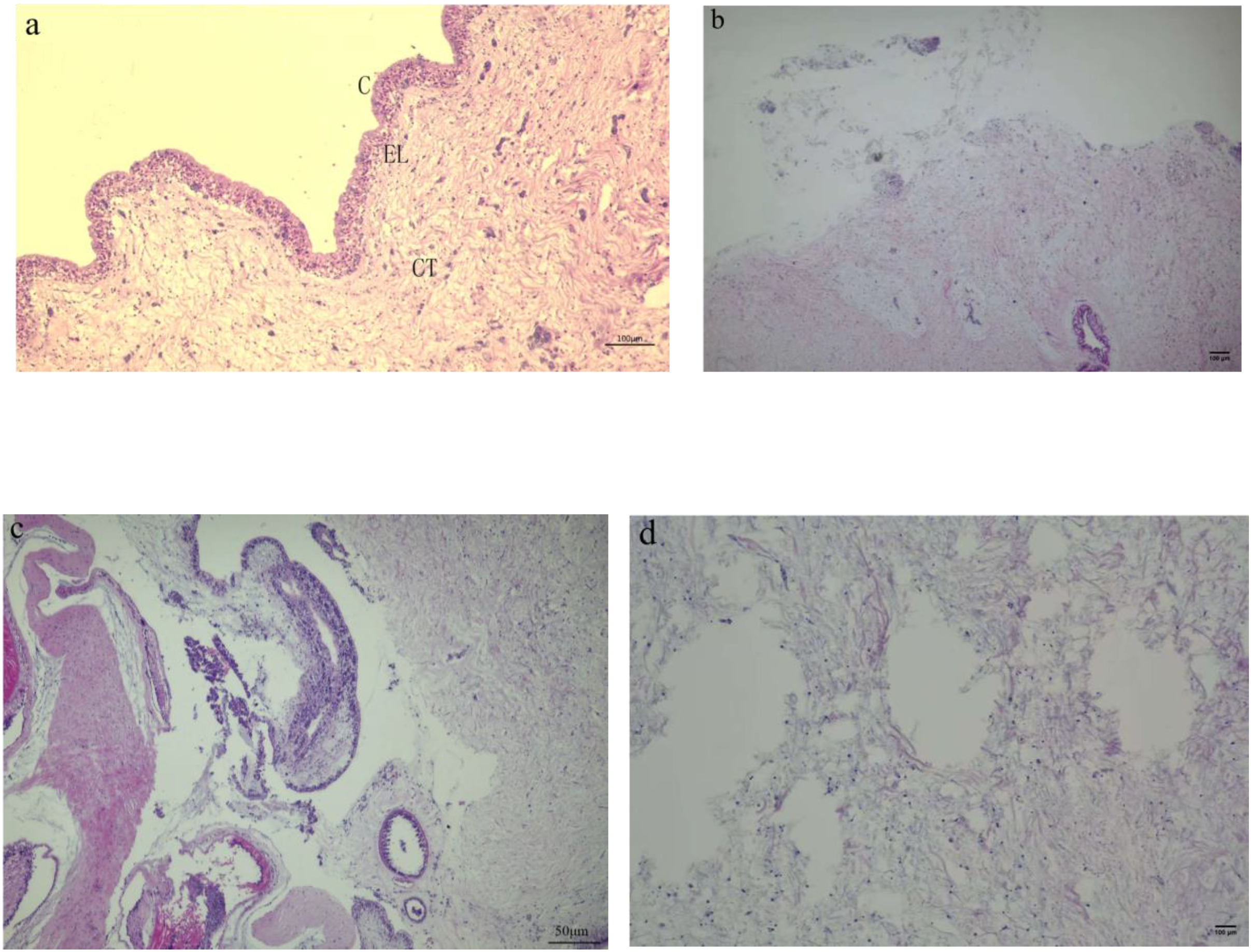
Histopathological change of the body wall of sea cucumbers infected with P. leiognathi 10MKXP20. (A) body wall of healthy control and (B) infected animals with the blur of the boundary between cells and the dissolution and disappearance of the epidermis. Scale bars: 100 μm (A, B, D), 50 μm (C). (C) Stratum corneum; EL, Epithelium layer; CT, Connective tissue. (A, C) are transverse sections, while (B, D) are longitudinal sections.
3.2 Identification of bacteria from diseased sea cucumbers
A total of five bacterial colonies with similar appearance were isolated from the ulcerative lesions of diseased sea cucumbers. One isolate, 10MKXP20, was used for further investigation. On TCBS agar, 10MKXP20 formed rapidly growing, raised colonies that were round with neat and round in the edge, reaching 1-2 mm in diameter after 24 h of incubation at 28°C. The colonies appeared green and the bacterium was identified to be Gram-negative. The transmission electron microscopic observation showed that the cell of 10MKXP20 was rod-shaped, and arranged in a single dispersed arrangement, with a size of 3.7-0.9 μm×1.2-1.5 μm. The morphological characteristics of the bacterial strain are shown in Figure 3. The bacterial isolate 10MKXP20 was identified as Photobacterium leiognathid based on the homology of its 16S rRNA and gyrB gene sequences. The sequences shared 99.79% and 99.83% homology, respectively, with those of P. leiognathi. The sequence for 10MKXP20 was deposited in GenBank under accession number OR835662. Phylogenetic tree analysis also revealed that P. leiognathi 10MKXP20 formed a distinct cluster with P. leiognathi (accession numbers NR_029253.1 and NR_15541.1) (Figure 4).
Figure 3
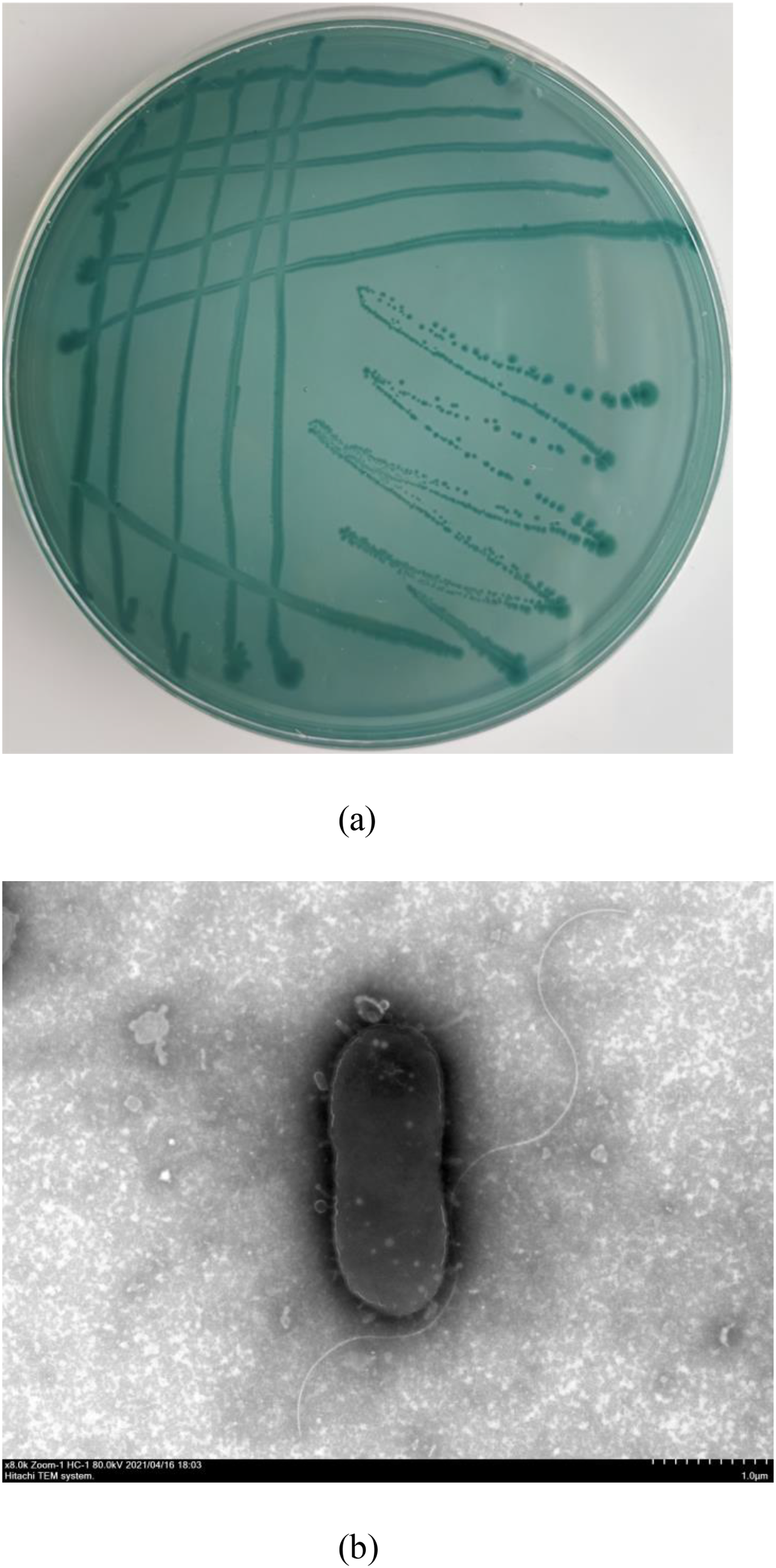
Characteristics of colony morphology (A) and cell appearance observed by scanning electron microscopy (B) of P. leiognathi 10MKXP20.
Figure 4
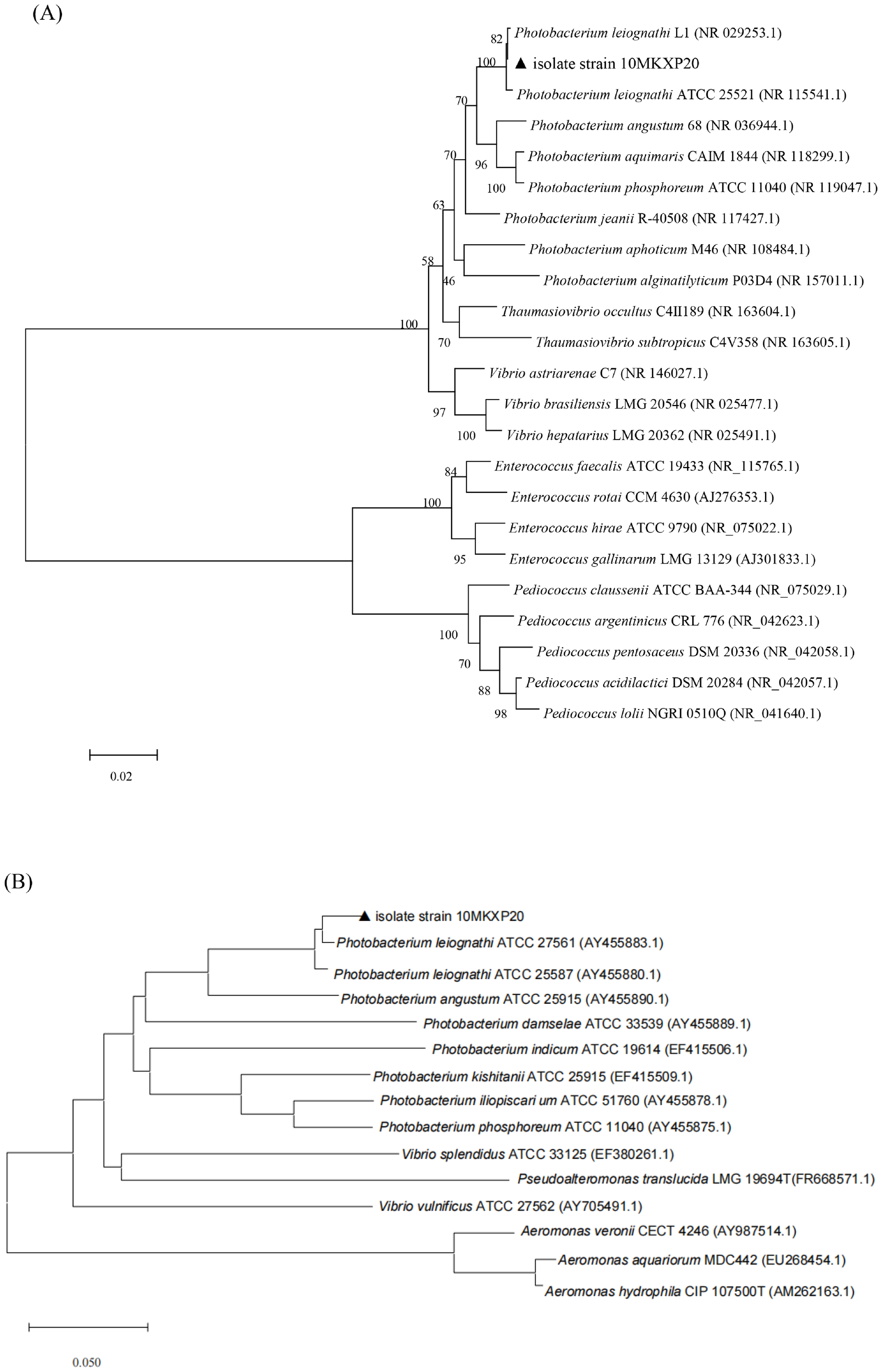
Neighbor-Joining tree shows the phylogenetic positions of P. leiognathi 10MKXP20 as compared with P. leiognathi strains isolated from other hosts obtained from GenBank. The evolutionary distances were computed using the Tamura-Nei method and are in units of the number of base substitutions per site. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The scale bar corresponds to 1 substitution per 100 nucleotide positions. (A) 16S rRNA phylogenetic tree, (B) gyrB phylogenetic tree.
Physiological and biochemical characteristics of P. leiognathi 10MKXP20 were determined, and the results revealed that the bacterium can utilize N-acetyl-D-glucosamine, N-Acetyl-β-D-mannosamine, α-D-glucose, D-mannose, D-gluconic acid, inosine, D-arabinol, glycerol, D-glucose-6-phosphate, and L-aspartic acid (Table 1).
Table 1
| Blood biochemical item | 10MKXP20 | 4CF10XP | Blood biochemical item | 10MKXP20 | 4CF10XP |
|---|---|---|---|---|---|
| Dextrin | – | – | Mucic acid | – | – |
| D-Maltose | – | – | Quinic acid | – | – |
| D-Trehalose | – | – | D-Saccharic acid | – | – |
| Gentiobiose | – | – | p-Hydroxy-Phenylacetic acid | – | – |
| Sucrose | – | – | Methyl pyruvate | – | – |
| Stachyose | – | – | Citric acid | – | – |
| D-Raffinose | – | – | α-Keto-Glutaric acid | – | – |
| α-D-Lactose | – | – | D-Malic acid | – | – |
| D-Melibiose | – | – | L-Malic acid | – | – |
| β-Methyl-D-Glucoside | – | – | Bromo-Succinic acid | – | – |
| D-Salicin | – | – | γ-Amino-Butryric acid | – | – |
| N-Acetyl-D-Glucosamine | + | + | α-Hydroxy-Butyric acid | – | – |
| N-Acetyl-β-Dmannosamine | + | + | β-Hydroxy-D, L-Butyric acid | – | + |
| N-AcetyNeuraminic acid | – | – | c-Propionic acid | – | – |
| α-D-Glucose | + | + | Acetic acid | – | – |
| D-Mannose | + | – | Formic acid | – | – |
| D-Fructose | – | – | pH 6 | + | + |
| D-Gluconic acid | + | + | pH 5 | – | – |
| L-Fructose | – | – | 1% NaCl | + | + |
| Inosine | + | + | 4% NaCl | + | + |
| D-Sorbitol | – | – | 1% Sodium lactate | + | + |
| D-Mannitol | – | – | Fusidic acid | + | + |
| D-Arabitol | + | + | D-Serine | + | + |
| Glycerol | + | + | Troleandomycin | + | + |
| D-Glucose-6-PO4 | + | + | SVRifamycin SV | + | + |
| D-Fructose-6-PO4 | – | – | Minocycline | – | – |
| D-Aspartic acid | – | – | Lincomycin | – | – |
| D-Serine | – | – | Guanidine HCl | – | – |
| Gelatin | – | – | Niaproof 4 | + | + |
| Glycyl-L-Prolin | – | – | Vancomycin | + | + |
| L-Aspartic acid | + | + | Tetrazolium violet | – | – |
| L-Glutamic acid | – | – | Nalidixic acid | – | – |
| L-Histidine | – | – | Lithium chloride | + | + |
| L-Pyroglutamic acid | – | – | Potassium tellurite | – | – |
| L-Serine | + | + | Aztreonam | – | – |
| D-Galacturonic acid | – | – | Sodium butyrate | – | – |
| L-Galactonic acid lactone | – | – | Sodium bromate | – | – |
Physiological and biochemical characteristics of P. leiognathi 10MKXP20 and 4CF10XP.
“+” denotes positive; “-” denotes negative; “ND” denotes no data; “d” denotes 26%-75% positive.
The antibiotic sensitivity results displayed that P. leiognathi 10MKXP20 was sensitive to 14 kinds of antibiotics (such as tetracycline, enrofloxacin, and ciprofloxacin), resistant to 6 antibiotics (including gentamicin, neomycin, kanamycin, rifampicin, amikacin, and amoxicillin), intermediate to 6 antibiotics (streptomycin, polymyxin B, erythromycin, azithromycin, cefoperazone, and ampicillin), and noneffective to neomycin sulfate and thiamphenicol (Table 2). The MIC value of P. leiognathi 10MKXP20 for the tested antibiotics were 0.008 μg/mL (enrofloxacin), 1 μg/mL (neomycin sulfate), 0.25 μg/mL (thiamphenicol), 1 μg/mL (florfenicol), 0.06 μg/mL (doxycycline hyclate), 0.125 μg/mL (flumequine), 2 μg/mL (sodium sulfamonomethoxine), and 1.2/0.06 μg/mL (mixture of sulfamethoxazole and rimethoprim).
Table 2
| Antibiotics | Pharmic content (μg) | Inhibition zone diameter (mm) | MIC value (μg/mL) | Sensitivity |
|---|---|---|---|---|
| Gentamicin | 10±2.5 | 15 | – | R |
| Tetracycline | 30 | 32 | – | S |
| Streptomycin | 10 | 18 | – | I |
| Polymyxin B | 300 | 16 | – | I |
| Erythromycin | 15 | 20 | – | I |
| Zithromycin | 15 | 22 | – | I |
| Enrofloxacin | 5 | 37 | – | S |
| Neomycin | 30 | 18 | – | R |
| Kanamycin | 30 | 19 | – | R |
| Ciprofloxacin | 5 | 44 | – | S |
| Rifampicin | 5 | 18 | – | R |
| Ceftriaxone | 30 | 42 | – | S |
| Ofloxacin | 5 | 36 | – | S |
| Cefoperazone | 75 | 26 | – | I |
| Norfloxacin | 10 | 38 | – | S |
| Cefradine | 30 | 26 | – | S |
| Ampicillin | 10 | 22 | – | I |
| Amicacin | 30±7.5 | 16 | – | R |
| Amoxicillin | 10 | 18 | – | R |
| Cotrimoxazole | 23.75ugSMZ/1.25ugTMP | 25 | – | S |
| Enrofloxacin | 5 | 36 | 0.008 | S |
| Neomycin sulfate | – | – | 1 | ND |
| Thiamphenicol | – | – | 0.25 | ND |
| Florfenicol | – | – | 1 | S |
| Doxycycline hyclate | – | – | 0.06 | S |
| Flumequine | – | – | 0.125 | S |
| Sodium sulfamonomethoxine | – | – | 2 | S |
| A mixture of sulfamethoxazole and rimethoprim | – | – | 1.2/0.06 | S |
Results of antibiotic sensitivity test of P. leiognathi 10MKXP20.
“R”, resistant; “I”, intermediateb; “S”, sensitive; “ND”, no critical value of drug resistance.
3.3 Cumulative mortality
The cumulative mortality of sea cucumbers challenged with P. leiognathi 10MKXP20 by intramuscular injection and immersion was recorded for 14 days (Figure 5). Clinical signs of infected sea cucumbers were similar to those of naturally infected individuals, with the presence of white lesions that enlarged and progressed to ulcerations throughout the body. However, no signs were observed in the control group. The time to first observation of mortality in the sea cucumbers in the two challenge models was dose-dependent. In the intramuscular injection challenge, the highest mortality (90%) was found at the highest bacterial dose (1.2×108 CFU/mL), whereas no mortality was observed in the control group (Figure 5A). Similarly, in the immersion challenge, the results showed the highest mortality (60%) occurred at 1.2×107 CFU/mL, followed by 35% at 1.2×106 CFU/mL, 25% at 1.2×105 CFU/mL, and 0% in the control group.
Figure 5
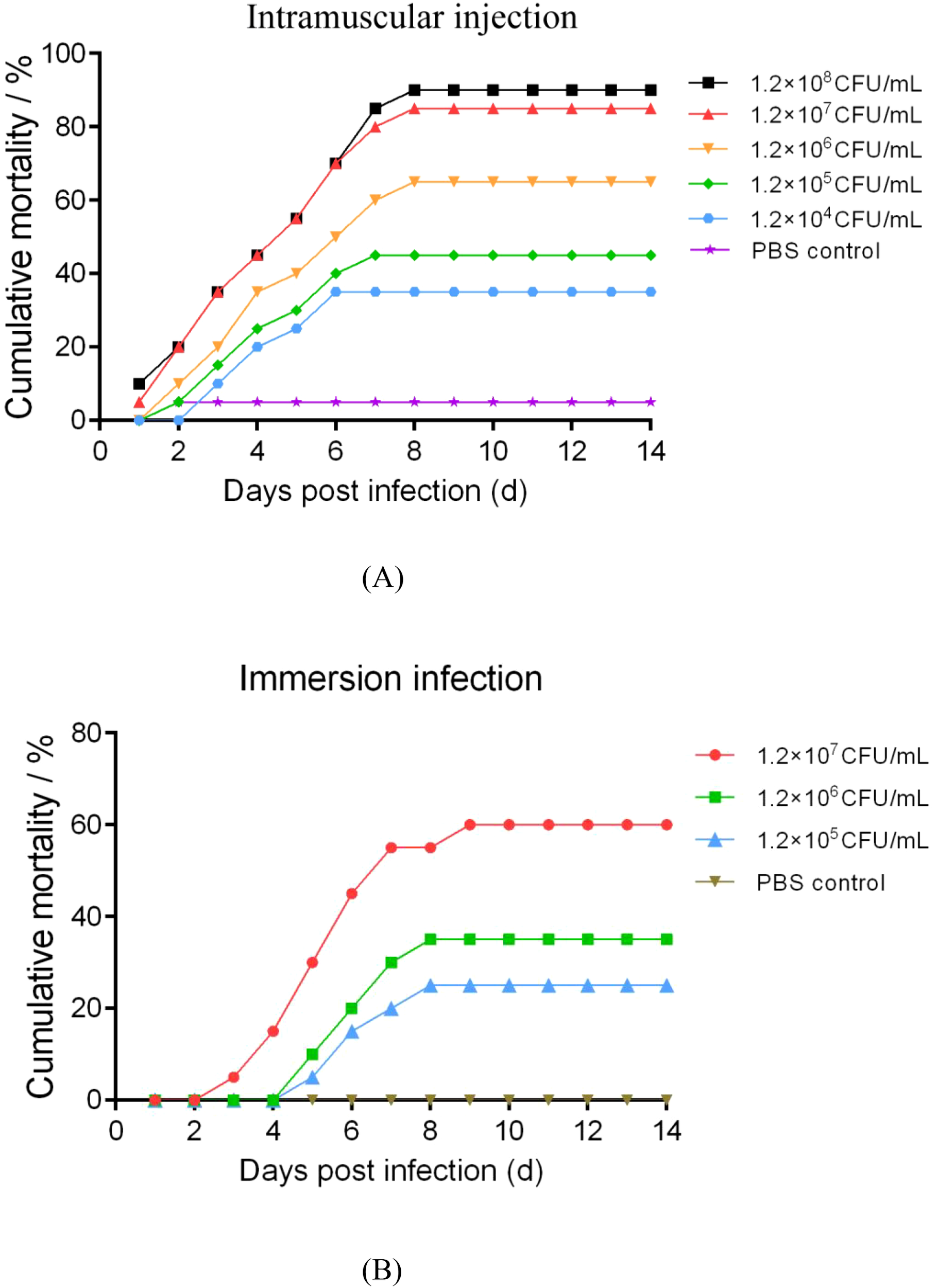
Cumulative mortality of sea cucumbers artificially infected with Photobacterium leiognathi 10MKXP20 by intramuscular injection (A) and immersion (B) infections.
4 Discussion
The skin ulceration syndrome has been known to occur worldwide in sea cucumber species, including A. japonicus (in China), Isostichopus fuscus (in Equator), and Holothuria scabra (in Madagascar, Australia, and New Caledonia). The disease was described by the first appearance of a white lesion, followed by the extension over the whole body, degradation of the cuticle, epidermis, connective tissue, and ossicles of the sea cucumbers. Similar results were observed in our current study, with the presence of white lesions and whitish mucus on the body surface of the sea cucumbers, and a body lying motionless on the bottom of the culture tank. Previous studies have reported that bacterial agents are the main pathogens causing skin ulceration disease in sea cucumbers (Tangestani and Kunzmann, 2019). This is evidenced by the discovery of P. damselae subsp. damselae in Cynoglossus semilaevis by Shao et al. (2019). In this study, the bacterium P. leiognathi 10MKXP20 was isolated and identified from the diseased sea cucumbers for the first time. Herein, the strain 10MKXP20 was cultured and purified on TCBS agar and the colony showed a green color on the surface of culture agar, which is similar to the description of P. leiognathi isolated from pony fish (Secutor ruconius) (Thirukumar et al., 2022). Also, the observations found that P. leiognathi is a Gram-negative and rod-shaped bacterium. Similar results have been reported with the characteristics of other strains of P. leiognathi (Dunlap, 1984). The results obtained from the phenotypic and biochemical characteristics of the bacterial strain 10MKXP20 isolated from sea cucumbers indicate the characteristic features of P. leiognathi. This identification was confirmed by sequencing the 16S rRNA and gyrB genes. The 16S rRNA sequencing is commonly applied as a molecular tool for bacterial identification (Drancourt et al., 2000), while the gyrB is considered suitable for bacterial phylogenetic analysis, being present in all bacterial taxa and possessing important features, including limitation in horizontal transmission (Watanabe et al., 2001). The combined use of these analyses has been successfully applied to identify bacterial species, such as V. harveyi, V. campbellii, and P. leiognathi (Chiu et al., 2007). Therefore, these results suggest the reliability of identifying 10MKXP20 as P. leiognathi 10MKXP20.
Chemotherapeutic agents play an important role in controlling bacterial infections in aquatic animals. In this study, the antibiotic sensitivity patterns of the isolate P. leiognathi 10MKXP20 were evaluated. The results found that P. leiognathi 10MKXP20 was sensitive to 14 kinds of antibiotics, including tetracycline, enrofloxacin, and ciprofloxacin. This result is similar to the observation in the previous study that the bacterial isolates, V. cyclitrophicus, V. splendidus, V. harveyi, and Photobacterium sp. isolated from A. japonicus were sensitive to most tested antibiotics (Deng et al., 2009). Our findings suggested that these antibiotics are suitable therapeutical medicines for controlling infection in sea cucumbers. This agrees with the previous findings in the case of Aeromonas hydrophila isolated from blunt snout bream (Megalobrama amblycephala) with clinical signs of hemorrhagic septicemia (Tran et al., 2015). However, the bacterium shows resistance to six kinds of antibiotics (including gentamicin, neomycin, kanamycin, rifampicin, amikacin, and amoxicillin), which implies that it may carry antibiotic resistance genes and has a risk of horizontal transfer of these genes to other bacteria (Jian et al., 2021; Li et al., 2023). Indeed, it is known that the bacteria can transfer and receive genes resistant to antibiotics from other Gram-negative bacteria (Marchandin et al., 2003). Thus, in this situation, the correct strategies used for controlling the infection by P. leiognathi 10MKXP20 in sea cucumbers are considered to decrease the horizontal transmission of antibiotic-resistant genes among bacteria.
The previous study has demonstrated that P. damselae is a pathogen causing skin ulcers on A. japonicus (Wang et al., 2022). In this current study, we isolated P. leiognathi 10MKXP20 from infected sea cucumbers. The observed clinical signs with white lesions and whitish mucus around the body surface of A. japonicus in the present study resembled those reported in a previous study (Martinez-Manzanares et al., 2008). The results demonstrated that the time-to-first-mortality in sea cucumbers exhibited a dose-dependent relationship with bacterial concentration, consistent with prior findings in snakehead fish (Channa striata) infected with the Gram-negative bacterium, A. hydrophila (Duc et al., 2013). Cumulative mortality in sea cucumbers was highest at the maximum dose levels of 1.2×107 CFU/mL (immersion challenge model) and 1.2×108 CFU/mL (injection challenge model), aligning with mortality patterns observed in silver pomfret (Pampus argenteus) infected with P. damselae subsp. damselae 69TBY1 (Tao et al., 2018). Experimental infection studies confirmed that P. leiognathi 10MKXP20 is the causative agent of skin ulcers in sea cucumbers. Data from the immersion challenge model suggest that the bacterium may infect hosts via waterborne transmission and skin surface penetration. Additionally, the presence of this pathogen in aquaculture systems could increase disease susceptibility among cultured organisms, as stressors such as high culture density and low quality of environmental conditions may promote disease propagation (Fouz et al., 2000). This agrees with the previous reports in turbot, eel, and silver pomfret infected with P. damselae subsp. damselae. Here, we provide the first demonstration that P. leiognathi 10MKXP20 exerts pathogenicity against the sea cucumber and independently elicits typical skin ulceration lesions that are indistinguishable from the skin ulcer syndromes. Our findings indicate that the members of the genus Photobacterium should be incorporated into the aetiological spectrum of bacterial pathogens affecting A. japonicus, offering a novel candidate to explain mass-mortality events in cultured sea cucumber populations. While previous studies have predominantly implicated Vibrio spp. as the primary causative agents of sea cucumber bacterial diseases, the identification of P. leiognathi 10MKXP20 expands the taxonomic breadth of recognized pathogens and provides a new perspective for disease prevention and control. Furthermore, the antimicrobial susceptibility profile generated in this study furnishes essential data for diversifying antibiotic options available to aquaculturists managing P. leiognathi 10MKXP20 infections.
In conclusion, our current study isolated and identified the Gram-negative bacterium P. leiognathi 10MKXP20 from the skin ulceration of sea cucumber (A. japonicus). The bacterium was demonstrated to be a potential pathogen of sea cucumbers, causing white lesions and whitish mucus around the body surface and high mortality upon challenge test. Our findings provide basic information on the bacterial pathogens causing diseases in sea cucumbers, subsequently leading to the exploration of effective methods that prevent and control diseases in the aquaculture industry of sea cucumbers.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal study was approved by science and technology (Research involving animals) of Fisheries Research Institute of Fujian. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QY: Supervision, Writing – review & editing, Writing – original draft. LW: Investigation, Methodology, Writing – original draft. YZ: Formal Analysis, Validation, Writing – original draft. MZ: Data curation, Software, Writing – review & editing. HL: Resources, Investigation, Writing – review & editing. YW: Investigation, Methodology, Writing – review & editing. YF: Project administration, Visualization, Writing – review & editing. WX: Formal Analysis, Investigation, Writing – review & editing. JW: Data curation, Project administration, Writing – review & editing. SL: Funding acquisition, Resources, Writing – review & editing. NT: Validation, Writing – review & editing. QL: Methodology, Project administration, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Acknowledgments
This work was supported by the fund of Fujian Aquatic Seed Industry Innovation and Industrialization Project under Grant No. 2021FJSCZY03; the Specialized Research Foundation of Fujian Provincial Department of Science & Technology of China under Grant No. 2022R10130010 and 2023J011325.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Altschul S. F. Gish W. Miller W. Myers E. W. Lipman D. J. (1990). Basic local alignment search tool. Journal of molecular biology215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
2
Becker P. Gillan D. Lanterbecq D. Jangoux M. Rasolofonirina R. Rakotovao J. et al . (2004). The skin ulceration disease in cultivated juveniles of Holothuria scabra (Holothuroidea, Echinodermata). Aquaculture.242, 13–30. doi: 10.1016/j.aquaculture.2003.11.018
3
Chang Y. Feng Z. Yu J. Ding J. (2009). Genetic variability analysis in five populations of the sea cucumber Stichopus (Apostichopus) japonicus from China, Russia, South Korea and Japan as revealed by microsatellite markers. Mar. Ecol.30, 455–461. doi: 10.1111/j.1439-0485.2009.00292.x
4
Chiu H. H. Chou H. H. Jean W. D. Shieh W. Y. (2007). Isolation and characterization of marine luminous bacteria from shallow coastal waters of Taiwan. J. Microbiol. Immunol. Infect.40, 14–23.
5
Deng H. He C. Zhou Z. Liu C. Tan K. Wang N. et al . (2009). Isolation and pathogenicity of pathogens from skin ulceration disease and viscera ejection syndrome of the sea cucumber Apostichopus japonicus. Aquaculture.287, 18–27. doi: 10.1016/j.aquaculture.2008.10.015
6
Deng H. Zhou Z.-C. Wang N.-B. Liu C. (2008). The syndrome of sea cucumber (Apostichopus japonicus) infected by virus and bacteria. Virol. Sin.23, 63–67. doi: 10.1007/s12250-008-2863-9
7
Deng H. Sui X.L. Chen Q. et al . (2005). A New disease Found in Apostichopus japonicus in Dalian Area During wintering. J Fish Sci. 24(2), 40–41. doi: 10.16378/j.cnki.1003-1111.2005.02.014
8
Drancourt M. Bollet C. Carlioz A. Martelin R. Gayral J.-P. Raoult D. (2000). 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol.38, 3623–3630. doi: 10.1128/JCM.38.10.3623-3630.2000
9
Duc P. M. Tuan T. N. Hatai K. (2013). Aeromonas hydrophila infection in fingerlings of snakehead Channa striata in Viet Nam. Fish Pathol.48, 48–51. doi: 10.3147/jsfp.48.48
10
Dunlap P. V. (1984). Physiological and morphological state of the symbiotic bacteria from light organs of ponyfish. Biol. Bull.167, 410–425. doi: 10.2307/1541286
11
Fisheries Bureau of the Ministry of Agriculture (2021). China fishery statistical yearbook (Beijing, China: Chinese Agricultural Press).
12
Fouz B. Toranzo A. E. Milán M. Amaro C. (2000). Evidence that water transmits the disease caused by the fish pathogen Photobacterium damselae subsp. damselae. J. Appl. Microbiol.88, 531–535. doi: 10.1046/j.1365-2672.2000.00992.x
13
Gürtler V. Stanisich V. A. (1996). New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology.142, 3–16. doi: 10.1099/13500872-142-1-3
14
Jian Z. Zeng L. Xu T. Sun S. Yan S. Yang L. et al . (2021). Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol.61, 1049–1070. doi: 10.1002/jobm.202100201
15
Kumar S. Stecher G. Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol.33, 1870–1874. doi: 10.1093/molbev/msw054
16
Li C. Feng W. Qiu L. Xia C. Su X. Jin C. et al . (2012). Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol.33, 436–441. doi: 10.1016/j.fsi.2012.04.013
17
Li C. Xu R. Zhang X. Chen D. Qiu Y. Chen W. et al . (2023). Genome analysis of pathogenic Vibrio harveyi strain TS-628. J. Fisheries Res.45, 415–426. doi: 10.3390/ijms242216202
18
Liu H. Zheng F. Sun X. Hong X. Dong S. Wang B. et al . (2010). Identification of the pathogens associated with skin ulceration and peristome tumescence in cultured sea cucumbers Apostichopus japonicus (Selenka). J. Invertebr. Pathol.105, 236–242. doi: 10.1016/j.jip.2010.05.016
19
Lu Z. (2019). Bacterial component analysis of the gut of Apostichopus japonicus cultured in Dalian, Yantai and Fujian. J. Fisheries Res.41, 187–194. doi: 10.14012/j.cnki.fjsc.2019.03.002
20
Marchandin H. Godreuil S. Darbas H. Jean-Pierre H. Jumas-Bilak E. Chanal C. et al . (2003). Extended-spectrum β-lactamase TEM-24 in an Aeromonas clinical strain: acquisition from the prevalent Enterobacter aerogenes clone in France. Antimicrob. Agents Chemother.47, 3994. doi: 10.1128/AAC.47.12.3994-3995.2003
21
Martinez-Manzanares E. Tapia-Paniagua S. T. Diaz-Rosales P. Chabrillon M. Morinigo M. A. (2008). Susceptibility of Photobacterium damselae subsp. piscicida strains isolated from Senegalese sole, Solea Senegalensis Kaup, and gild head seabream, Sparus aurata L., to several antibacterial agents. J. Fish Disease.31, 73–76. doi: 10.1111/j.1365-2761.2007.00872.x
22
Ren Y. Li Q. Wang Y. N. et al . (2019). Research progress on coelomocytes of echinoderms. Journal of Agricultural Science and Technology, 21(02), 91–97. doi: 10.13304/j.nykjdb.2018.0065
23
Saitou N. Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol.4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
24
Shao P. Yong P. Zhou W. Sun J. Wang Y. (2019). First isolation of Photobacterium damselae subsp. damselae from half-smooth tongue sole suffering from skin-ulceration disease. Aquaculture.511, 734208. doi: 10.1016/j.aquaculture.2019.734208
25
Tangestani M. Kunzmann A. (2019). Isolation and characterization of bacteria from the lesion of juvenile sea cucumber Holothuria scabra (Jaeger 1938) with symptom of skin ulceration disease. Iran. J. Fish. Sci.18, 915–923. doi: 10.22092/ijfs.2019.118391
26
Tao Z. Shen C. Zhou S.-M. Yang N. Wang G.-L. Wang Y.-J. et al . (2018). An outbreak of Photobacterium damselae subsp. damselae infection in cultured silver pomfret Pampus argenteus in Eastern China. Aquaculture.492, 201–205. doi: 10.1016/j.aquaculture.2018.04.013
27
Thirukumar S. Selvaraj U. Rath S. Shankar K. Muthuvel A. (2022). Evaluation of bioactive secondary metabolites from ponyfish associated bacteria (Photobacterium leiognathi). J. Trop. Biodivers. Biotechnol.7, 71758. doi: 10.22146/jtbb.71758
28
Tran N. T. Gao Z.-X. Milton J. Lin L. Zhou Y. Wang W.-M. (2015). Pathogenicity of Aeromonas hydrophila to blunt snout bream Megalobrama amblycephala. IJSRP.5, 1–7. doi: 10.5114/ceji.2015.52825
29
Walker R. D. (1999). Standards for antimicrobial susceptibility testing. (Chicago, IL, USA: American journal of veterinary research). 60 (9), 1034.
30
Wang C. Wang W. Yao L. Sang S. Li C. Zhang Q. (2021a). Histopathological study of covert mortality nodavirus infection in sea cucumber (Apostichopus japonicus). Aquaculture.545, 737161. doi: 10.1016/j.aquaculture.2021.737161
31
Wang L. Yang Q. Li H. Yang F. Wu J. Jiang X. et al . (2022). Isolation and identification of Photobacterium damselae causing disease in cultured Apostichopus japonicus in southern China. J. Fishery Sci. China.29, 1800–1811. doi: 10.12264/JFSC2022-0193
32
Wang C. Yao L. Wang W. Sang S. Hao J. Li C. et al . (2021b). First report on natural infection of Nodavirus in an Echinodermata, sea cucumber (Apostichopus japonicas). Viruses.13, 636. doi: 10.3390/v13040636
33
Watanabe K. Nelson J. Harayama S. Kasai H. (2001). ICB database: the gyrB database for identification and classification of bacteria. Nucleic Acids Res.29, 344–345. doi: 10.1093/nar/29.1.344
34
Yang Q. (2020). Optimization of culture medium and fermentation conditions of Bacillus amyloliquefaciens CQN-2 isolated from aquatic animal. J. Fisheries Res.42, 339–347. doi: 10.14012/j.cnki.fjsc.2020.04.005
35
Zhao Z. Zhou Z. Dong Y. Pan Y. Jiang J. Jiang Y. et al . (2021). Association of intestinal fungal communities with the body vesicular syndrome: An emerging disease of sea cucumber (Apostichopus japonicus). Aquaculture.530, 735758. doi: 10.1016/j.aquaculture.2020.735758
36
Zhang Z. Xing R. Lv Z Shao Y. Zhang W. Zhao X. et al . (2018). Analysis of gut microbiota revealed Lactococcus garviaeae could be an indicative of skin ulceration syndrome in farmed sea cucumber Apostichopus japonicus. Fish & shellfish immunolog80, 148–154. doi: 10.1016/j.fsi.2018.06.001
Summary
Keywords
Photobacterium leiognathi 10MKXP20, sea cucumber, pathogen, ulceration, drug sensitivity, minimum inhibitory concentrations
Citation
Yang Q, Wang L, Zhong Y, Zhang M, Li H, Wang Y, Fan Y, Xing W, Wu J, Li S, Tran NT and Lin Q (2025) Isolation, identification and characterization of Photobacterium leiognathi 10MKXP20 from sea cucumber (Apostichopus japonicus) cultured in Southeast China. Front. Mar. Sci. 12:1679287. doi: 10.3389/fmars.2025.1679287
Received
04 August 2025
Accepted
15 October 2025
Published
04 November 2025
Volume
12 - 2025
Edited by
Ming Guo, Ningbo University, China
Reviewed by
Xulu Chang, Henan Normal University, China
Chunming Tan, China Agricultural University, China
Updates
Copyright
© 2025 Yang, Wang, Zhong, Zhang, Li, Wang, Fan, Xing, Wu, Li, Tran and Lin.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiuhua Yang, qhyang1314@163.com; Qi Lin, xmqlin@sina.com; Ngoc Tuan Tran, tranntts@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.