Abstract
Indigenous probiotics are widely applied in Penaeus vannamei aquaculture due to their potential to enhance disease resistance and promote growth. However, their biological and environmental safety requires comprehensive evaluation, particularly during the larval stage. This study investigated the effects of shrimp derived Lactobacillus casei HD1 and Bacillus licheniformis WZ1 on larval growth performance, aquatic bacterial communities, potential pathogens, and antibiotic resistance genes (ARGs), by using illumina sequencing and quantitative PCR (qPCR), comparing probiotic-treated and control (CK) groups. Probiotic supplementation significantly improved survival rate, biomass, and individual weight, while reducing concentrations of NH4+-N, NO3−-N, and PO43−-P in rearing water. Compared to the control, α-diversity of the aquatic microbiota increased significantly, accompanied by elevated relative abundances of Bacteroidota, Bacillota, and Balneolota. The results of qPCR showed that no significant changes in abundance of ARGs and Lactobacillus in taxa were observed, whereas Bacillus was significantly enriched within probiotic-adding, compared to CK group. Notably, structural equation modeling (SEM) revealed that probiotics enhanced aquaculture performance through multiple pathways: indirectly by modulating aquatic microbial communities and directly by mitigating waterborne nutrients. These findings support the use of targeted indigenous probiotics as a sustainable strategy to balance productivity with environmental health, reduce reliance on antibiotics, overcome larval survival bottlenecks, and ensure ecological safety. Furthermore, the distinct impacts of different probiotics on microbial structure and host performance highlight the importance of strategic formulation in probiotic combinations for shrimp larval aquaculture.
1 Introduction
Shrimp farming constitutes a cornerstone of the global aquaculture industry, contributing significantly to both economic development and food security. According to the Food and Agriculture Organization (FAO), global shrimp production surpassed 6.8 million tons in 2023, with a market value exceeding 42 billion USD, representing 12.7% of the total value of aquaculture production (FAO, 2024). This sector supports livelihoods for over 10 million individuals, primarily in developing countries across Asia and Latin America. As a high-value aquatic commodity, shrimp holds a pivotal position in international trade with an annual economic value surpassing 100 billion USD (Umamaheswari et al., 2025). Despite its economic significance, the rapid expansion of shrimp aquaculture has been accompanied by substantial challenges, particularly during the larval rearing phase, where frequent disease outbreaks and low survival rates remain critical technical bottlenecks to sustainable industry growth (Thitamadee et al., 2016). Historically, disease management has relied heavily on antibiotics and chemical therapeutics, practices that have contributed to the proliferation of antibiotic resistance genes (ARGs) and pose risks to aquatic ecosystem integrity (Bailey et al., 2014). In light of increasing regulatory restrictions on antibiotic use and the growing demand for cost-effective, environmentally sustainable practices, there is an urgent need to transition toward green and ecologically sound management strategies, such as the application of probiotics and other biocontrol methods, to shift shrimp farming from a quantity-driven to a quality- and sustainability-oriented model (Kronman et al., 2012).
Within shrimp larvae culture systems, the symbiotic microbial communities associated with shrimp play a fundamental regulatory role in host physiology and health (Andriantahina et al., 2013). The establishment and succession of intestinal microbial communities during early development not only influence larval survival and growth performance but also shape microbial colonization patterns that may have lasting effects on shrimp health and productivity in later life stages (Holt et al., 2021; Tamburini et al., 2016; Wang et al., 2020). Concurrently, microbial communities in the rearing water are integral to biogeochemical cycling, particularly in the transformation of nitrogenous compounds. Fluctuations in concentrations of toxic metabolites such as ammonia and nitrite are closely tied to the composition and functional dynamics of the aquatic microbiome (Wang et al., 2020). Critically, the microbial communities in the water and the shrimp gut are interconnected through continuous microbial exchange. The aquatic environment serves as a primary inoculum for the intestinal microbiota, suggesting that manipulation of the water microbiome can be an effective strategy for modulating gut microbial composition and promoting intestinal health (Maynard et al., 2012). Furthermore, the supplementation of probiotics and prebiotics, such as fermentable carbon sources, has been shown to effectively shape the structure and function of the shrimp gut microbiome (Zhou et al., 2009; Thakur et al., 2025).
Lactic acid bacteria (LAB) and Bacillus species are among the most widely studied probiotic candidates in shrimp aquaculture. Lactobacillus in particular, exhibits multiple beneficial traits, including the ability to ferment diverse carbohydrates, thereby enhancing nutrient availability and absorption (El-Saadony et al., 2021). For instance, L. casei functions as a multifunctional probiotic, capable of modulating gut microbiota, improving digestive efficiency, and enhancing immune responses. Dietary supplementation with L. casei has been shown to improve growth performance and reduce the incidence of enteritis in shrimp (Figueras et al., 2022). Similarly, B. licheniformis demonstrates denitrifying activity, contributing to water quality improvement (Seabkongseng et al., 2025), while also producing a suite of extracellular digestive enzymes including proteases, amylases, lipases, and saccharifying enzymes that facilitate nutrient digestion and utilization (Li et al., 2018; Kuebutornye et al., 2019; Ghosh, 2025). These enzymes further assist in the breakdown of otherwise indigestible feed components such as pectin, glucans, cellulose, and hemicellulose (Zhou et al., 2009). Additionally, Bacillus subtilis could secrete phytase in the shrimp gut, enhancing the bioavailability of plant-derived phosphorus and improving lipid metabolism (El-Saadony et al., 2021; Li et al., 2018). However, the majority of research on microbial interventions has focused on grow-out phases, with limited attention devoted to the larval rearing stage, where microbial community assembly and host immune development are particularly vulnerable.
Disruption of the microbial community structure induced by environmental stressors, pathogen invasion, or other biotic and abiotic factors could be frequently associated with the proliferation of opportunistic pathogens and the dissemination of ARGs (Dong et al., 2021a; Dong et al., 2021b; Liu et al., 2019). A stable intestinal microenvironment is maintained through a dynamic equilibrium of interspecies microbial interactions and host-microbe immune crosstalk, which collectively suppress the overgrowth and spread of potential pathogens (Xiong et al., 2018). Perturbations to this equilibrium, whether from external stressors (e.g., environmental fluctuations) or internal challenges (e.g., pathogen incursion), can trigger microbial community shifts and niche reorganization, creating conditions conducive to pathogen dominance and ARG propagation (Su et al., 2017; Luo et al., 2025). For example, in adult shrimp, invasion by Vibrio species disrupts gut microbiota homeostasis, thereby facilitating further colonization and infection. Conversely, supplementation with beneficial microbes such as Rhodobacteraceae taxa has been shown to enhance microbial stability and confer resistance to pathogen invasion (Guo et al., 2020; Dong et al., 2023). Current safety assessments of probiotics primarily focus on the genomic absence of virulence factors and ARGs in the probiotic strains themselves, with insufficient attention paid to the broader ecological impacts—particularly how probiotic application may influence the abundance and dynamics of indigenous pathogens and ARGs within the host-associated and environmental microbiomes (Gadhiya et al., 2025). Moreover, comprehensive safety evaluations of probiotics during the sensitive larval stage remain scarce. Consequently, it is still unclear whether probiotic supplementation during larval rearing could inadvertently promote pathogen expansion or ARG dissemination, thereby limiting the safe and effective deployment of probiotics at this critical developmental phase.
To systematically evaluate the probiotic efficacy and environmental safety of shrimp-derived strains, specifically L. casei HD1 and B. licheniformis WZ1, a mixed probiotic formulation was administered continuously throughout the larval rearing period. Shrimp larval survival, growth performance, and key water quality parameters were monitored and statistically analyzed. Additionally, high-throughput sequencing was employed to characterize the microbial community structure, diversity, and taxonomic composition in both shrimp gut and rearing water, with a specific focus on identifying potential pathogens and ARGs (Dong et al., 2025; Love et al., 2014). This integrated approach enabled a quantitative assessment of the mixed probiotic’s effects on larval development and microbial environmental safety, providing essential data and evaluation criteria to support the safe and sustainable application of probiotics in shrimp aquaculture, particularly during the shrimp larval stage.
2 Materials and methods
2.1 Shrimp source and bacterial isolation from culture system of shrimp
The healthy sub-adult Pacific white shrimp (Penaeus vannamei) with an initial body weight of 8.0 ± 1.2 g were obtained to isolate the potential probiotics strains from the Yongxing Base of the Zhejiang Provincial Marine Aquaculture Research Institute (Zhejiang, China). The specific pathogen-free shrimp larvae (Zoea I) of the brand “Zhehai No.1” from Zhejiang Provincial Marine Aquaculture Research Institute were purchased and cultured in standardized system of shrimp farming, and used for the addition experiments of indigenous probiotics.
To isolate the potential probiotics strains including lactic acid bacteria and Bacillus taxa, serial dilutions of the glycerol stocks obtained from shrimp guts and enrichment culture were plated on marine MRS and R2A liquid media prepared with 75% seawater (salinity 30‰) based on the methods of Guo et al. (2023). Plates were incubated at 28°C for 12 hours, and colonies of bacteria showing different morphological features (e.g., size and color of colonies) were re-streaked on 1/10 marine broth 2216E (MB; Becton–Dickinson) plate to ensure purity. The total DNA genome of the purified strain was extracted by using FastPure Bacteria DNA Isolation Mini Kit (DC103, Vazyme, China), and the full-length sequences of 16S rRNA gene were amplified using the universal primers 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’), and purified for Sanger sequencing (Sangon Biotech, China). These sequences were then submitted to EZBioCloud of the database (https://www.ezbiocloud.net) for taxonomic identification.
After taxonomic annotation, the strains of HD1 and WZ1 were identified as Lacticaseibacillus sp. Bacillus sp. from shrimp gut and rearing water, respectively. Furthermore, those two sequences were mapped to the database of 16S ribosomal RNA gene from curated type bacteria and archaea strain sequences in Genbank with web-blastn suite (http://www.ncbi.nlm.nih.gov) to find the most similar type strains of Lacticaseibacillus sp. and Bacillus sp. Furthermore, to identify the reliable taxonomy of the bacterial isolations of HD1 and WZ1, the full-length sequences of those two strains were aligned against the sequences of L. casei (Type strain NBRC 15883, NR_113333.1), L. casei (Type strain ATCC 393T, NR_041893.1), L. paracasei (strain NBRC 15889, NR_113337.1), L. paracasei (Type strain ATCC 25302T, NR_117987.1), L. mingshuiensis (Type strain 117-1, NR_179363.1), L. hegangensis (Type strain 73-4,NR_180276.1), L. Camelliae (Type strain MCH3-1, NR_041457.1), L. jixianensis (Type strain 159-4, NR_180278.1), L. rhamnosus (Type strain NBRC 3425, NR_113332.1), B. paralicheniformis (Type strain KJ-16, NR_137421.1), B. glycinifermentans (Type strain GO-13, NR_137407.1), B. piscis (Type strain 16MFT21, NR_165685.1), B. haynesii (Type strain NRRL B-41327, NR_157609.1), B. swezeyi (Type strain NRRL B-41294, NR_157608.1), B. sonorensis (Type strain NBRC 101234, NR_113993.1), B. licheniformis (Type strain ATCC 14580T, NR_074923.1), B. licheniformis (strain DSM 13, NR_118996.1), B. licheniformis (strain NBRC 12200, NR_113588.1), B. capparidis (Type strain EGI 6500252, NR_156073.1), B. subtilis (Ehrenberg) Cohn (Type strain ATCC 6051T, NR_102783.2), and Streptomyces rutgersensis (outer group of tree, Type strain DSM 40077, NR_119349.1) by using Muscle (version 5.3, https://drive5.com/muscle/). The evolutionary distances were computed using the Maximum Composite Likelihood method. The evolutionary relationships of taxa were inferred using the UPGMA method and were. Finally, the phylogenetic tree between strains of HD1 and WZ1, and those type strains were conducted in MEGA11 (Tamura et al., 2021).
2.2 Experimental design and sample collection
After the bacterial isolation was finished, the probiotic-adding experiment was conducted by using the same farm and culture systems of bacterial isolation. Twelve standardized shrimp nursery ponds (cement pool with waterproof paint, 6 m × 5 m × 1.3 m) were randomly divided into two groups with larvae density of 1.33×105 ind./m3 (each group containing six ponds as replicates): the control group (CK) and the probiotics-adding group (Probiotics). All ponds were managed uniformly, including seawater introduction and treatment. The experiment began at 3 dph (days post hatching), and the developmental stage of shrimp larvae had changed from Nauplius to Zoea. In the Probiotics group, the composite probiotics (L. casei HD1 and B. licheniformis WZ1 mixed in a 1:1 ratio) with a composite probiotic solution concentration 1.0 × 109CFU/mL based on our previous studies (Liu et al., 2018; Du et al., 2021) were uniformly applied to the ponds every morning. The amount of feed had been gradually increasing and the residual feed and excreta of shrimp were also accumulating along as the developmental stages of larva. Therefore, the usage of probiotics gradually increases, and the probiotic dosage was set at 3, 5, and 10L per pound (Liu et al., 2018), respectively, during the Zoea I, Mysis I, and Postlarvae I stages. Continuous aeration was maintained throughout the experiment, and the following water parameters were reported: temperature 30-32 °C, pH 8.0-8.3, salinity 28 psu, dissolved oxygen 5–8 mg/L. At the start of the experiment, inorganic nitrogen concentrations were monitored, revealing nitrite nitrogen < 0.5 mg/L and ammonia nitrogen < 0.1 mg/L. Throughout the entire experimental period of 15 days, fundamental water quality parameters, including temperature, pH, salinity, and dissolved oxygen, were monitored daily using a multi-parameter (Jingcheng, China) water quality instrument.
At the end of experiment, water samples and all the shrimps were collected from each pond. The indexes including the survival rate, average body length, weight, total biomass, and weight gain ratios of shrimp larvae were calculated by using the methods of (Wang et al., 2020). Water samples were collected from six randomly selected points within each pond for the analysis of water quality and microbial community. At each point, 50 mL of water was collected and pooled to form a composite sample for each parameter. The water samples were pre-filtered through a 100 μm polycarbonate membrane (Merck KGaA, Germany) and analyzed for NH4+-N, NO2−-N, NO3−-N, and PO43−-P following GB 17378.4-2007. Each measurement was performed in triplicate to ensure analytical precision. Water microbial samples were pre-filtered through a 100 μm membrane, then 100 mL of water was filtered through a 0.22 μm polycarbonate membrane (Merck KGaA, Germany) to capture microbial biomass. All filters were immediately stored at −80 °C until DNA extraction.
2.3 DNA extraction, 16S rRNA gene amplification, and Illumina sequencing
Microbial DNA from aquatic samples was extracted from polycarbonate filter membranes using the PowerSoil® DNA Kit (MOBIO, USA). The V4 region of the bacterial 16S rRNA gene was amplified using primers 515F-Y (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R-B (5′-GGACTACNVGGGTWTCTAAT-3′) with dual barcodes (Wang et al., 2022). To minimize amplification bias, three independent PCR reactions were performed per sample. The total PCR volume was 20 μl, with the following conditions: 95 °C initial denaturation for 3 min; followed by 27 cycles of 95 °C denaturation for 30 s, 55 °C annealing for 30 s, and 72 °C extension for 45 s; with a final extension at 72 °C for 10 min. PCR products from the three reactions per sample were pooled, purified using the TaKaRa PCR Fragment Purification Kit (TaKaRa, Japan), and sequenced on the Illumina NovaSeq platform. All those sequencing data were available in CNCB (China National Center for Bioinformation, https://ngdc.cncb.ac.cn/) with the accession number of PRJCA044428.
2.4 The relative abundance of Lactobacillus sp. Bacillus sp. and ARGs by using qPCR
The copy number of total bacterial 16S rRNA gens and Lactobacillus genus-specific, Bacillus genus-specific and ARGs (including acrB, tetA, ermF, floR, blaTEM) from rearing water were tested by using Quantitative PCR (Bio-Rad, USA). The information of those genes was shown in Supplementary Table S1. The copy numbers of those genes were tested at same time for each sample, and all qPCR experiments were performed in triplicate, with the following process: initial denaturation at 95 °C for 10 minutes; denaturation at 95 °C for 30 seconds; annealing at 60 °C for 30 seconds; and 40 cycles are performed. Finally, the software in qPCR automatically generated a melting curve analysis. The data quality control conditions were: (1) efficiency within the range of (90%-110%); (2) no multiple melting curves in the amplicon; (3) all three replicates are within the detection limit at cycle threshold (Ct) of 31. The construction method of 16S standard curve refers to the method of Lu et al (Lu et al., 2024). The standard curve of the 16S rRNA gene was generated by a ten-fold dilution series of the standard plasmid which was constructed with One step ZTOPO-Blunt/TA (Zomanbio, China) as the vector and containing the 288 bp bacterial targeted fragment amplified from water samples, with R2 higher than 0.999. The copy numbers of the 16S rRNA gene generated according to the standard curves were defined as absolute prokaryotic microbial biomass. The copy numbers of ARGs were calculated according to the following equation: relative gene copy number The relative abundances (copies/16S rRNA gene copies) of ARGs were normalized to 16S rRNA gene copies (Looft et al., 2012; Yu et al., 2025). The formulas with the relationship between Ct values and concentration of gene copies (Copies) for 16S rRNA gene, Bacillus taxa, Lactobacillus taxa and, ARGs were as follows:
2.5 Sequencing data processing process
Raw sequencing data underwent quality control by using trimmomatic (version 0.40, https://github.com/usadellab/Trimmomatic) software (Bolger et al., 2014), and then the paired reads were merged together, and the primer sequences were removed by using USEARCH11 (version 11.0.667, https://drive5.com/usearch) software (Edgar, 2010). The resulting high-quality sequences were denoised and chimeric sequences were using the UNOISE3 algorithm (Edgar, 2016) to call the zero-radius operational taxonomic units (ZOTUs, Supplementary Table S2). Those ZOTU sequences were taxonomically classified using the NCBI RefSeq Targeted Loci Project (Bacteria and Archaea 16S ribosomal RNA Nucleotide sequence record database, release·2025-01-12, https://www.ncbi.nlm.nih.gov/refseq/targetedloci/). Sequences of ZOTUs classified as chloroplasts, mitochondria, unclassified Archaea or Bacteria, and singletons were removed. All those data analysis processes were finished by using the Dix-seq pipeline (version 1.0, https://github.com/jameslz/dix-seq, Dong et al., 2025). Potential pathogens were annotated using the DPiWE database (http://pathogen.umehd.com, Dong et al., 2021a). For sequencing depth normalization, ZOTU tables were rarefied to a minimum depth of 24,900 sequences per sample for downstream analysis. Antibiotic resistance genes (ARGs) were prediction following the method described by Luo et al (Luo et al., 2025).
2.6 Statistical analysis
Alpha- and beta-diversity indices were calculated using QIIME and the ‘vegan’ package in R (v 4.4.1). The coefficient of variation (CV) for alpha-diversity indices was calculated in R. Differences in alpha-diversity indices and beta-diversity between groups were assessed using the Wilcoxon rank-sum test. Differences in the relative abundance of taxonomic units between groups were also analyzed using R. Constrained principal coordinate analysis (CAP) based on Bray-Curtis dissimilarity was performed to visualize microbial community structure differences between CK and probiotics groups samples, and significance was tested using analysis of similarities (ANOSIM). Differential abundance of ZOTUs between the CK and Probiotics groups was identified using the negative binomial generalized linear model in the ‘DESeq2’ R package (Love et al., 2014), with significant ZOTUs selected based on |log2(fold change) | > 2 and adjusted P < 0.05 (-log10 (adjusted P) > 1.3). To investigate correlations between biomarker ZOTUs, host health status, environmental factors, and probiotic supplementation, a partial least squares path model (PLS-PM) was constructed using the ‘plspm’ R package (Dong et al., 2021b). Differences in water physicochemical parameters between groups were analyzed using one-way analysis of variance (ANOVA) with the ‘aov’ function, followed by Duncan’s multiple range test for post-hoc comparisons.
3 Results
3.1 The taxonomy and phylogenetic information of strain HD1 and WZ1
The full-length sequences of 16S rRNA gene in strain HD1 and WZ1 were blast against the database of 16S ribosomal RNA gene from curated type bacteria and archaea strain sequences in Genbank, and top hits results (Supplementary Table S3) showed that For HD1 strain, the top five similar type strains were Lacticaseibacillus casei ATCC 393 (Similarity: 97.05%, Coverage: 98.00%, Accession: NR_041893.1), Lacticaseibacillus casei strain NBRC 15883 (96.66%, 98.00%, NR_113333.1), Lacticaseibacillus casei strain BCRC 10697 (96.49%, 98.00%, NR_115322.1), Lacticaseibacillus zeae strain RIA 482 (96.33%, 98.00%, NR_037122.1), and Lacticaseibacillus chiayiensis strain BCRC 81062 (96.16%, 98.00%, NR_179712.1), and WZ1 were Bacillus licheniformis strain DSM 13 (99.92%, 100.00%, NR_118996.1), Bacillus licheniformis strain BCRC 11702 (99.86%, 100.00%, NR_116023.1), Bacillus paralicheniformis strain KJ-16 (99.80%, 87.00%, NR_137421.1), Bacillus licheniformis strain ATCC 14580 (99.80%, 100.00%, NR_137421.1), Bacillus licheniformis strain NBRC 12200 (99.66%, 100.00%, NR_137421.1). Although the similarity between B. paralicheniformis strain KJ-16 and strain WZ1 was over 99%, the coverage between those two strains was less than 87%, which may not be supported on identifying strain WZ1 as B. paralicheniformis. Therefore, the strain HD1 and WZ1 were preliminarily identified as Lacticaseibacillus casei and Bacillus licheniformis, respectively. Similarly, the phylogenetic relationship between strain HD1 and L. casei strain NBRC 15883 and L. casei strain ATCC 393T was more closed to other Lacticaseibacillus taxa, and B. licheniformis strain NBRC 12200 and B. licheniformis strain DSM 13 were more closed to strain WZ1 (Supplementary Figure S1).
3.2 Effects of probiotic supplementation on water quality and culture performance of shrimp larvae
Following composite probiotic supplementation, analysis of water quality parameters (Supplementary Figure S2) revealed that, compared to the CK group, concentrations of NH4+-N, NO3−-N, and PO43−-P were significantly lower in the Probiotics group (P < 0.05), whereas NO2−-N concentration did not differ significantly. Additionally, probiotic addition significantly improved shrimp larvae growth performance: survival rate, biomass, and individual body weight were significantly higher in the Probiotics group compared to the CK group (P < 0.05, Supplementary Figure S2).
3.3 Effects of probiotic supplementation on microbial diversity in rearing water
Composite probiotic addition significantly increased α-diversity indices (richness, evenness, Shannon, phylogenetic diversity) of water microorganisms in the Probiotics group compared to the CK group (P < 0.05; Figure 1A). For water microorganisms, the coefficient of variation (CV) for richness and phylogenetic diversity was significantly lower in the Probiotics group, whereas CV for evenness and Shannon index was significantly higher (P < 0.05; Figure 1B). Quantitative PCR (qPCR) analysis (Supplementary Table S4; Supplementary Figure S3) revealed that both the absolute and relative abundance (against the total copies of 16S rRNA gene) of Bacillus taxa in the Probiotics group were obviously increased with 7.13 (P < 0.05) and 1.68 (P < 0.05) fold changes, respectively, compared to the control group. However, no significantly different absolute and relative abundances of Lactobacillus taxa were compared between Probiotics and control groups. The correlation analysis between the relative abundance of Lactobacillus and Bacillus and the α-diversity index of aquatic microorganisms revealed that the relative abundance of Lactobacillus significantly affected α-diversity, particularly richness and phylogenetic diversity (P < 0.05; Supplementary Figure S4). However, Bacillus had no significant effect on α-diversity.”
Figure 1
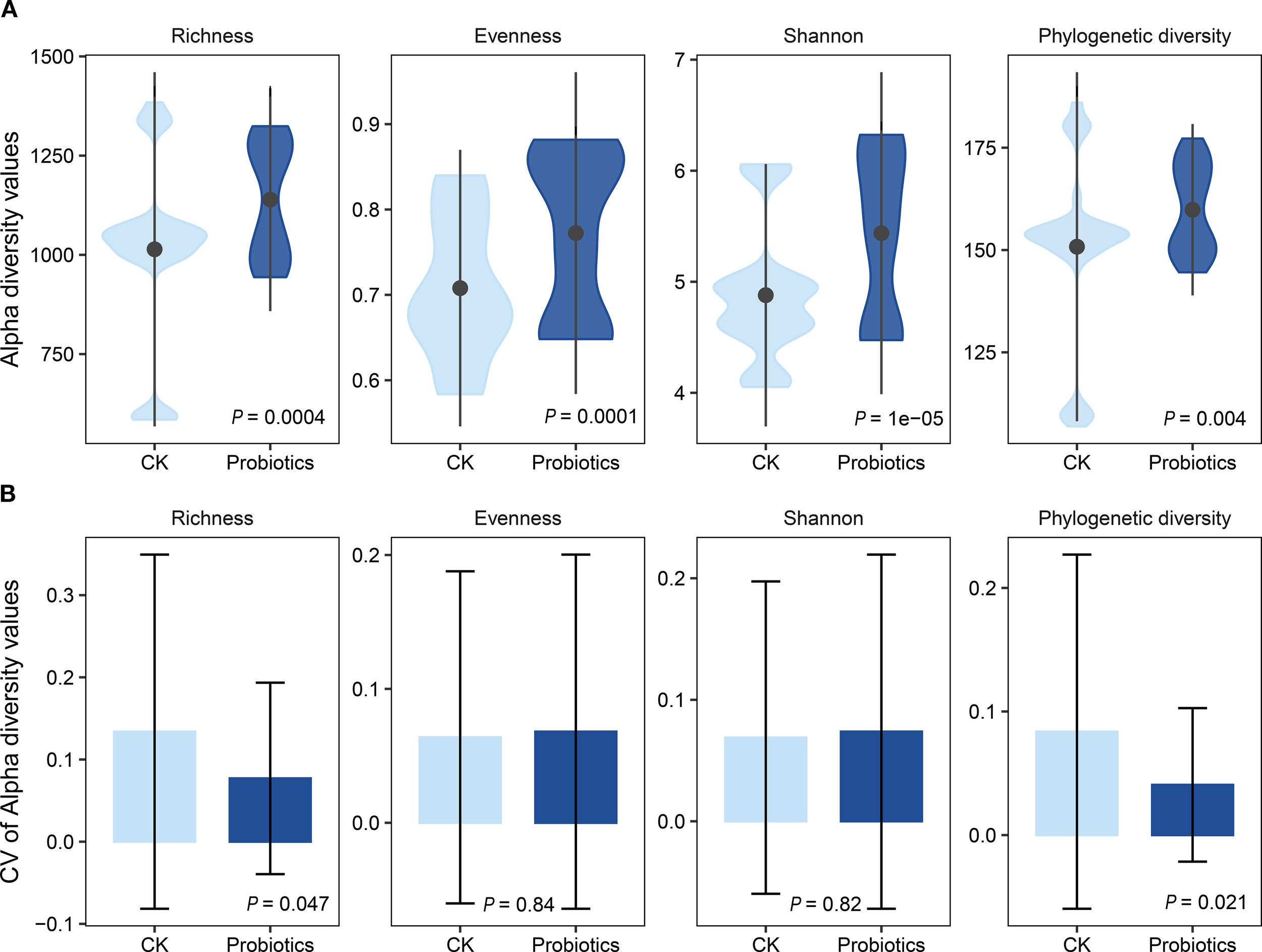
The effects of probiotics addition on alpha diversity indices of the bacterial community in shrimp larvae rearing water. (A) Comparison of alpha diversity indices including richness, evenness, Shannon, and phylogenetic diversity indexes between Probiotics and control groups in rearing water bacterial community. Violin plots show the frequency distributions of the diversity indices, and the black points and lines inside the violin plots display the median, and 95% confidence interval within each data set, respectively; (B) Coefficient of variation (CV) for the diversity indices between Probiotics and control groups in shrimp larvae rearing water. Asterisks denote levels of significance (*P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis test).
Integrated analysis of water quality parameters, growth performance, and probiotic supplementation was performed using redundancy analysis (RDA). The results revealed significant separation between the microbial communities in the CK and probiotics compartments (Figure 2A). The CAP model explained 15.49% (R2 = 0.92, P = 0.042) of the compositional variation in bacterial communities between the control and probiotics-added groups in rearing water. Bray-Curtis and weighted Unifrac distances between samples within the Probiotics group were significantly lower than those within the CK group. Similarly, the coefficient of variation (CV) calculated for both distance metrics was significantly lower in the Probiotics group compared to the CK group (P < 0.05; Figure 2B).
Figure 2
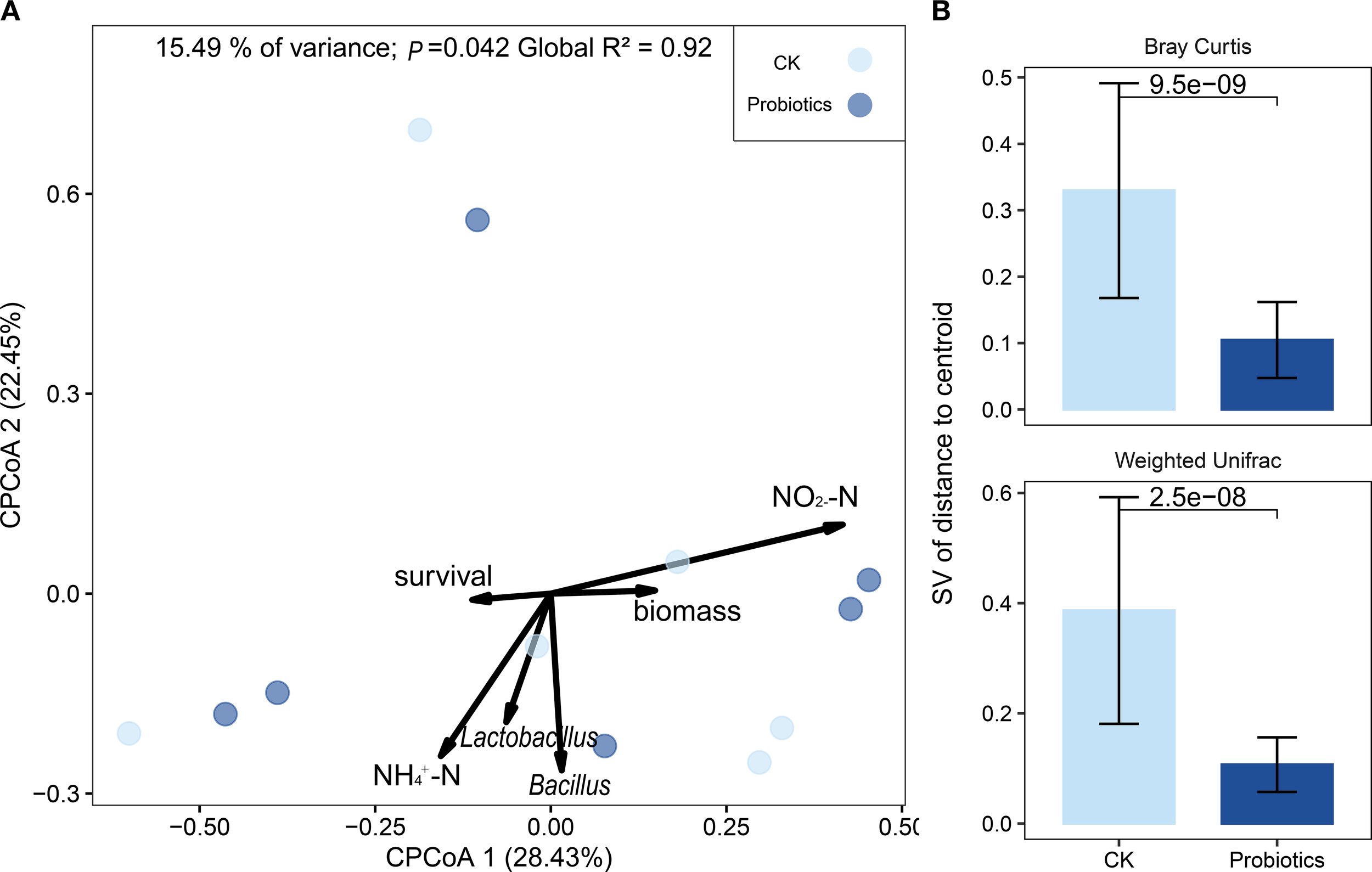
Effects of probiotics addition on the structure of the bacterial community in rearing water. (A) Constrained principal coordinate analysis (CAP) based on Bray-Curtis dissimilarity constrained by NO2−-N and NH4+-N in rearing water, survival and biomass of shrimp larvae, and relative abundance of Bacillus and Lactobacillus in total bacterial community from larvae rearing system of shrimp larvae. The non-redundant indicators including hydrochemistry, culture performance of shrimp, and relative abundance of probiotics were selected by using multiple collinear tests with variance inflation factor (VIF < 10). The model of CAP explaining 15.49% variation in composition of bacterial community between the control and probiotics-added groups in rearing water, and permutational multivariate analysis of variance (PERMANOVA, global R2 = 0.92, P = 0.042) was used to test the difference across sample types and treatments. (B) Compare the differences in bacterial communities between the control group and the probiotic group in the aquaculture water and the coefficient of variation (CV) of bacterial community differences. Asterisks denote levels of significance (*P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis test).
3.4 Effects of probiotic supplementation on the microbial composition of water
The impact of composite probiotic supplementation on bacterial composition was analyzed for the top 10 bacterial groups at the phylum and family levels based on relative abundance. At the phylum level (Figure 3A), Alphaproteobacteria exhibited the highest relative abundance in the water, followed by Bacteroidota and Actinobacteriota. Probiotic addition significantly increased the relative abundances of Bacteroidota, Bacillota, and Balneolota in the Probiotics group compared to the CK group (P < 0.05). At the family level (Figure 3B), Roseobacteraceae displayed the highest relative abundance in the water, followed by Flavobacteriaceae. Probiotic addition significantly enhanced the relative abundance of Saprospiraceae and Balneolaceae in the Probiotics group compared to the CK group (P < 0.05). Furthermore, alterations in the relative abundance of other uncultured Roseobacteraceae bacteria at the genus level were dominant taxa both in the Probiotics group compared to the CK group, and observed more enrichment in the Probiotics group compared to the CK group (P < 0.05, Supplementary Table S5).
Figure 3
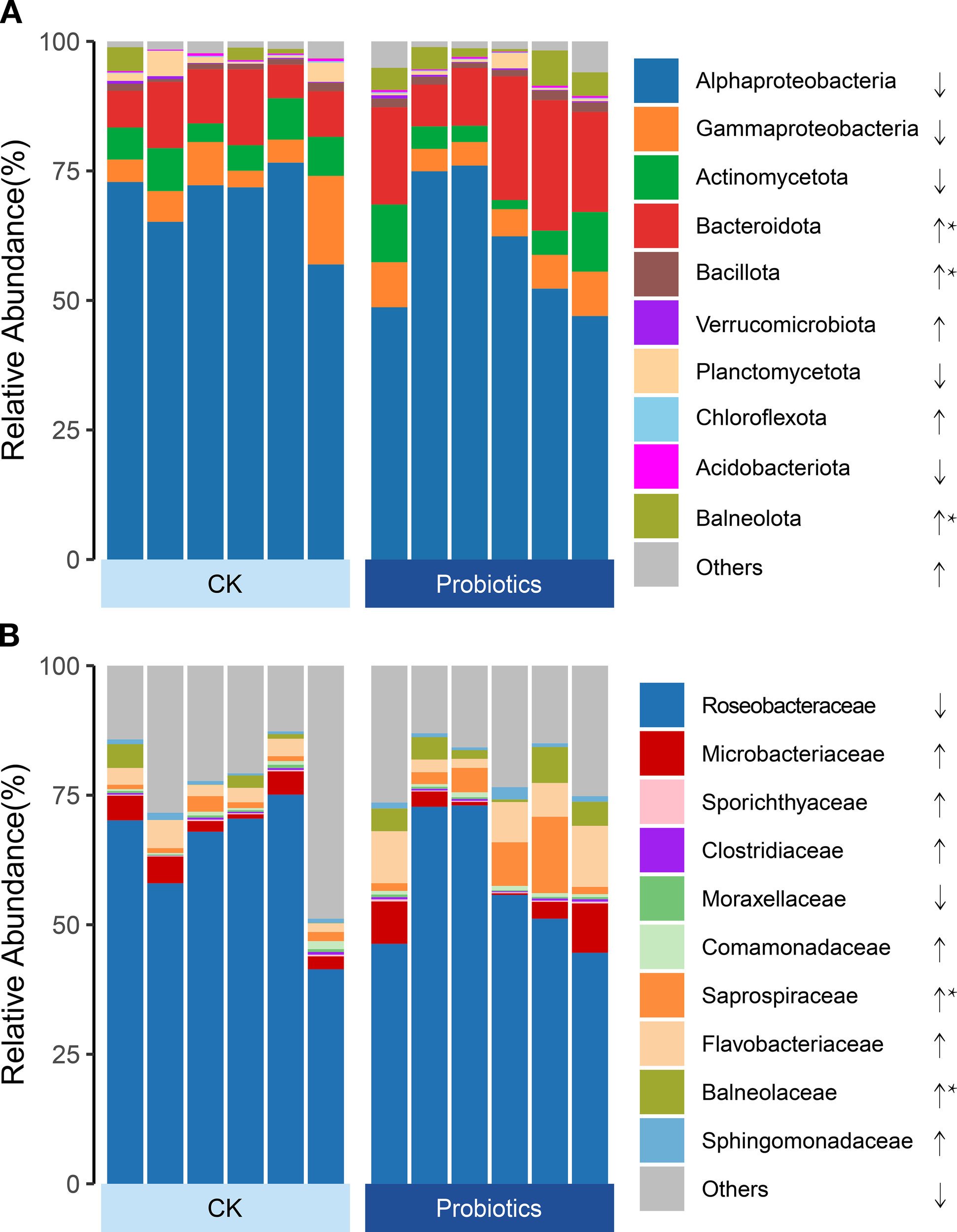
The different relative abundances of the dominant taxa (> 0.001%) between control and Probiotics groups in rearing water bacterial communities. Relative abundances of the dominant bacterial phylum classes (A), and families (B) between control and Probiotics groups in rearing water. Data were statistically compared between control and Probiotics groups by using Wilcoxon Rank Sum test. Significant differences across entire samples compartments are indicated with * (P < 0.05), and ** (P < 0.01). The decreasing or increasing average relative abundance of taxa in Probiotics groups compared to control are indicated with “↓”, “↑”.
3.5 Correlation analysis among key ZOTUs, water quality parameters, and growth performance in rearing water
The effects of composite probiotic treatment on bacterial community structure and composition were analyzed by using DESeq2 (Supplementary Figure S5). The results revealed that probiotic addition significantly increased the relative abundance of 12 ZOTUs (belonging to families including Cryomorphaceae, Flavobacteriaceae), while significantly decreasing the relative abundance of 7 ZOTUs (belonging to families such as Maritalea, Nitrospinota) in rearing water (P < 0.05).Correlations between key differential ZOTUs and water quality parameters/growth performance were calculated using Spearman correlation analysis with Z-score standardization (Figure 4). Differential ZOTUs exhibited stronger negative correlations with survival rate and the relative abundances of Bacillus and Lactobacillus, but stronger positive correlations with NO2−-N, NO3−-N, PO43−-P, and the weight.
Figure 4
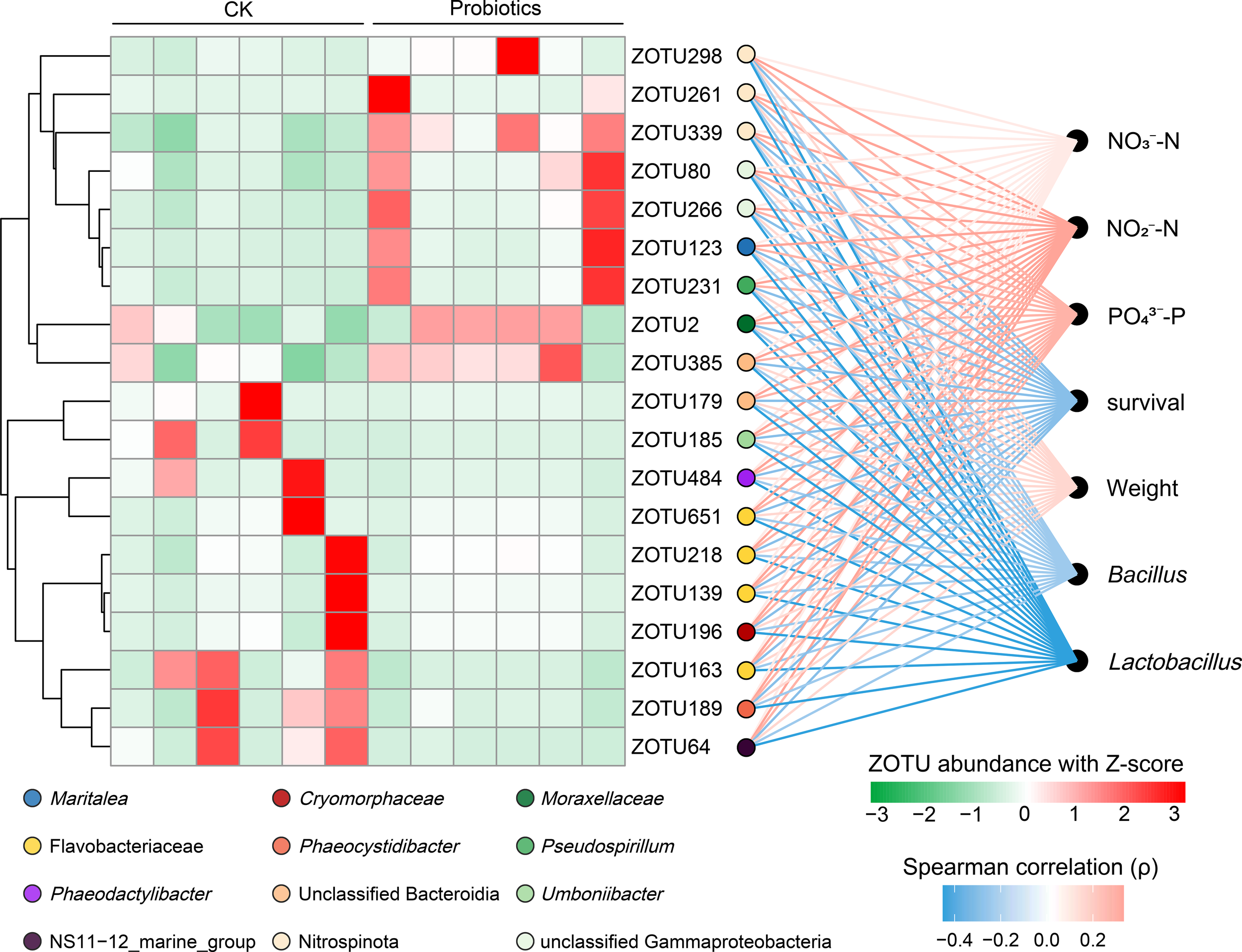
Differences in ZOTUs between control and probiotics-added groups associated with biotic and abiotic factors in the shrimp larval rearing system. The correlation between the relative abundance of top 19 key differential ZOTUs and the non-redundant indicators in cultivation system of shrimp larvae. The heatmap shows the relative abundances of the 19 ZOTUs normalized by Z-score scales in the control and probiotics-added groups. The significant positive correlations (ρ > 0.3, P < 0.05) between relative abundances of ZOTUs and Ln transformation of non-redundant indicators (including culture performance, environmental factors, and the relative abundance of probiotics) were plot as red lines, whereas those with significant negative correlations (ρ < −0.3, P < 0.05) as blue. The colors of dots present different genera. All the P values were adjusted using the Benjamini-Hochberg method.
3.6 Effects of probiotic supplementation on the composition of potential pathogenic bacteria and antibiotic resistance genes in aquatic environments of shrimp larvae
The distribution of potential pathogenic bacteria, antibiotic resistance genes (ARGs), and microbial composition were annotated to assess the risk of probiotic supplementation. The results for potential pathogenic bacteria (Figure 5A) showed that in aquatic microbial communities, the relative abundance of most potential pathogenic bacteria increased in the probiotic group compared to the control group; however, these changes were not statistically significant. Next, we used the PICRUSt2 software package to predict ARG distribution. The results of qPCR and PICRUSt2 predictions (Figure 5B; Supplementary Table S4) indicated decreases in most antibiotic resistance genes (e.g., multidrug, beta-lactam, and tetracycline-resistance genes), while increases occurred in a few genes (e.g., vancomycin and quinolone-resistance genes). Nevertheless, none of the ARG changes was statistically significant.
Figure 5
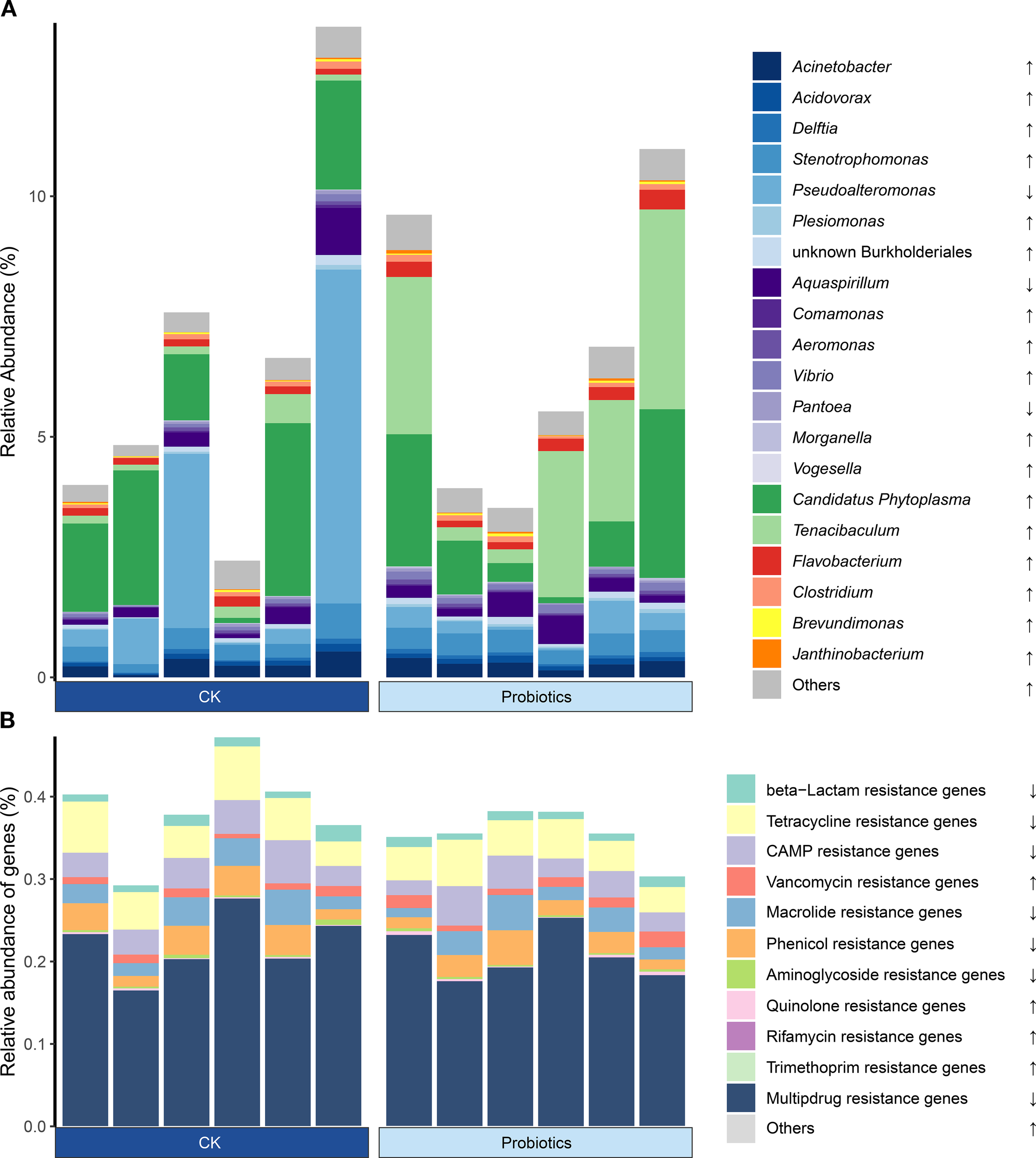
The different relative abundances of potential pathogenic bacteria (A) and predicted antibiotic resistance genes (B) between the control and probiotics-added groups in the bacterial communities of rearing water. Data were statistically compared between control and Probiotics groups by using Wilcoxon Rank Sum test. Significant differences across entire samples compartments are indicated with * (P < 0.05), and ** (P < 0.01). The decreasing or increasing average relative abundance of taxa in Probiotics groups compared to control are indicated with “↓”, “↑”.
3.7 Pathways of probiotics affecting the physical and chemical properties of water and the growth performance of shrimp larvae
A fitness-well structural equation modeling (SEM, goodness of fits index, GoFs > 0.57) was employed to analyze the interrelationships among variables in the system at three levels (Figure 6). In the shrimp larvae intestinal model: at the first level, composite probiotics exerted a significant direct positive effect on shrimp farming performance, irrespective of the pathway (waterborne microorganisms or water quality parameters). At the second level, composite probiotics indirectly influenced shrimp growth performance by regulating the dynamic balance of intestinal and water microbial communities. Regulation of dynamic changes in the intestinal microbial community also significantly enhanced shrimp farming performance; however, the water microbial pathway did not yield a significant improvement. Additionally, composite probiotics regulated potential pathogenic bacteria and ARGs. Potential pathogenic bacteria and ARGs exerted a significant negative impact on shrimp growth performance. The intestinal microbial community had a significant positive influence on potential pathogenic bacteria and ARGs. At the third level, composite probiotics improved shrimp farming performance by regulating environmental factors.
Figure 6
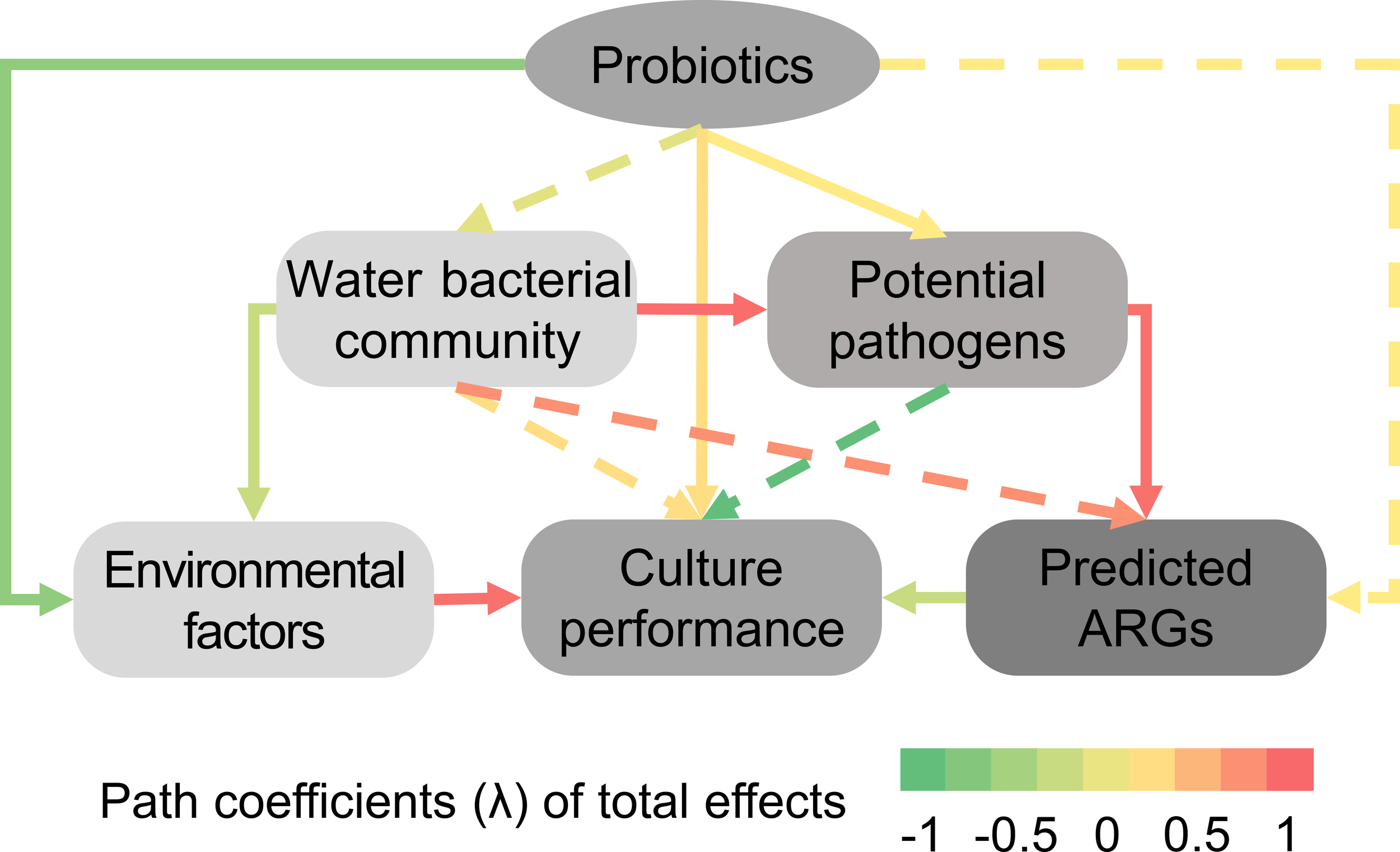
Effects of probiotics addition on environmental factors, culture performance of shrimp larvae, bacterial communities, potential pathogenic bacteria, and predicted antibiotic resistance genes (ARGs) by partial least squares path models (PLS-PM). Directed graph illustrating differences in rearing water microbiota response to probiotic addition, based on total effects derived from PLS-PM analysis. Path coefficients (λ) of total effects are reflected with red and green indicating positive and negative effects, respectively. Dashed arrows show coefficients that were insignificant (P > 0.1). The environmental factors included NO2−-N, and NH4+-N, the culture performance included survival rate and biomass of shrimps, and relative abundance of probiotics. The goodness of fits index (GoFs) for this PLS-PMs are 0.574.
4 Discussion
4.1 Adding indigenous probiotics from shrimp could improve the culture performance of larvae and water quality in shrimp culture system
The aquatic environment serves as the fundamental medium for the survival and development of cultured aquatic species. As fish and shrimp reside in water throughout their life cycle, their health and growth performance are intrinsically linked to water quality. Thus, maintaining optimal water conditions is essential for sustainable aquaculture. However, intensive farming systems often face challenges such as heightened environmental stress, frequent disease outbreaks, and effluent nutrients due to high stocking densities and excessive feeding (Dubey et al., 2024). Accumulation of organic waste and subsequent water quality deterioration are key drivers of pathogenic microbial proliferation. In this context, probiotics have been extensively adopted in aquaculture as feed additives or water amendments. This practice aims to enhance the living environment of aquatic organisms and modulate the microbiota associated with them (Butt et al., 2021). Over the past decade, numerous bacterial strains have been screened and evaluated as potential probiotics. Probiotic formulations may consist of single strains or, more commonly, multiple strains, with composite formulations typically exhibiting enhanced efficacy (Mathan Muthu et al., 2024; Wang et al., 2019). Probiotics exert multiple beneficial effects on the host by modulating host or environmental microbial communities, including improved feed utilization, enhanced growth, strengthened immunity, and increased survival rates (El-Saadony et al., 2022; Thakur et al., 2025). Among the most studied genera are Lactobacillus and Bacillus, which are valued for their antagonistic activity against pathogens, ability to regulate intestinal microbiota, and secretion of extracellular digestive enzymes that aid nutrient digestion and growth in aquatic animals (Kewcharoen and Srisapoome, 2019; Ringø et al., 2020; Wang et al., 2019).
In the present study, probiotic supplementation significantly improved shrimp larval growth performance and water quality. Specifically, the addition of probiotics significantly increased larval survival rate biomass, and individual body weight (Supplementary Figure S1). These results are consistent with numerous studies demonstrating that probiotics enhance feed utilization and nutrient absorption (Ringø et al., 2020; Rohani et al., 2022). The growth-promoting effects may stem from improved nutrient digestion, absorption, and assimilation facilitated by probiotics (Abarike et al., 2018; Mathan Muthu et al., 2024). Evidence indicates that probiotics enhance shrimp growth through multiple mechanisms, including increased digestive enzyme activity, provision of essential nutrients, and improved water quality. For instance, Xie et al. (2019) reported that shrimp fed a diet supplemented with a composite probiotic (B. subtilis, B. licheniformis, and Lactobacillus spp) for 8 weeks exhibited significantly higher final body weight (FBW), weight gain (WG), and specific growth rate (SGR) compared to controls, along with elevated lipase and amylase activities. Similarly, Lee et al. (2022) demonstrated in a 12-week trial that supplementation with the commercial probiotic SYNSEA (B. subtilis and Lactobacillus) significantly increased final body weight, average pond yield, and weight gain percentage in shrimp. Wang et al. (Wang et al., 2020) also observed improved growth performance in Micropterus salmoides following dietary supplementation with L. casei K17, which was attributed to significantly enhanced intestinal amylase and lipase activities.
Notably, the benefits of probiotics extend beyond gut health, contributing to overall host resilience through pathogen inhibition and water quality improvement (Butt et al., 2021; Rohani et al., 2022). Seabkongseng et al. (2025) reported that supplementation with B. velezensis S141 significantly improved the survival rates of shrimp and concurrently reduced the pathogen loads, when infected with white spot syndrome virus (WSSV), acute hepatopancreatic necrosis disease (AHPND), and Enterocytozoon hepatopenaei (EHP). Kewcharoen and Srisapoome (2019) isolated B. subtilis AQAHBS001 from shrimp intestine, which not only enhanced growth performance and intestinal morphology (increased microvillus height and intestinal wall thickness) but also effectively suppressed Vibrio growth and improved resistance to AHPND. Hamza et al. (2018) found that the cell-free supernatant of B. licheniformis inhibited the growth of pathogenic V. alginolyticus and P. gessardii, exhibiting anti-adhesion and antibacterial properties. Kong et al. (2019) demonstrated that recombinant L. casei expressing viral and bacterial antigens enhanced innate immunity in Cyprinus carpio and conferred protection against Aeromonas veronii infection.
In the current study, probiotic supplementation not only improved shrimp growth performance but also significantly reduced concentrations of NH4+-N, NO3−-N, and PO43−-P in the culture water (P < 0.05). The role of probiotics in water quality improvement is well established. Khademzade et al. (2020) reported that supplementation with Bacillus cereus and Lactobacillus acidophilus significantly reduced NH4+-N and NO3−-N levels in shrimp culture systems. A study on the probiotic of tilapia was observed that application of the probiotic AquaStar® Pond (Bacillus sp., Pediococcus sp., Enterococcus sp.) resulted in significantly lower total TAN and NH3 concentrations compared to the control (Redhwan et al., 2024). Zhang et al. (2020) found that co-amendment of sucrose and B. licheniformis in Penaeus vannamei larval rearing water significantly reduced NH4+-N, NO2−-N, and NO3−-N after 20 days. Additionally, probiotics may indirectly improve water quality by promoting beneficial algal growth and suppressing harmful algal blooms (Raza et al., 2025).
4.2 The different processes of indigenous probiotic mixture between regulating the growth performance of larvae shrimp and water environment
Intensive aquaculture systems often lead to elevated stress responses in farmed organisms, increased disease incidence, and pollution of water, sediment, and surrounding environments. Under such conditions, probiotic application is recognized as an effective strategy for modulating microbial communities in pond ecosystems and improving water and sediment quality (Thakur et al., 2025). Probiotics enhance host health by stimulating immune responses and competitively excluding pathogens; they may also reduce the emergence frequency of antibiotic resistance genes (ARGs), offering a viable alternative to antibiotics (Rohani et al., 2022). Probiotics improve feed utilization, water quality, and stress resilience in diverse aquaculture species through mechanisms like competitive exclusion, immune modulation, and antimicrobial compound production (Kewcharoen and Srisapoome, 2022; Sha et al., 2016; Zhang et al., 2020). Structural equation modeling (SEM) analysis in this study suggests that the composite probiotic enhances shrimp farming performance by regulating the complex interactions among the host microbiome, host immune and metabolic states, and environmental factors (Figure 6).
The applied probiotics exhibited distinct spatial distribution patterns: Bacillus significantly increased in relative abundance within aquatic microbial communities, whereas Lactobacillus did not show a significant change in the water microbiota. This differential distribution may reflect the specific colonization capabilities of probiotic strains in distinct microenvironments, potentially explained by niche differentiation theory (Bydalek et al., 2024; Zhao et al., 2024). Through spatial niche partitioning and functional complementarity, the two probiotics may efficiently utilize aquaculture environmental resources, potentially leading to synergistic enhancement of shrimp larval growth performance.
Bacillus primarily exerts indirect effects by improving water quality, thereby promoting shrimp growth. This genus is characterized by aerobic metabolism, robust extracellular enzyme secretion, and high metabolic activity, enabling strong adaptability to aquatic environments (Kuebutornye et al., 2019). By efficiently degrading organic detritus, such as uneaten feed, feces, and biological remains, Bacillus could be significantly reduced concentrations of harmful nitrogenous compounds (e.g., NH4+-N, NO2−-N, NO3−-N, TAN), and also helpful to improve the rearing environment of shrimp. Our previous research also indicates that exogenous additives (e.g., carbon sources, probiotics) could shape the gut microbiota of shrimp by modulating the surrounding microbial community (Guo et al., 2022; Liu et al., 2018). Numerous studies confirm the efficacy of Bacillus in water quality management, with Bacillus strains commonly dominating biofloc systems, where they contribute to sustained water quality improvement (Ghosh, 2025; Nayak, 2021; Qiu et al., 2023). Moreover, supplementation of Bacillus, such as B. subtilis) and B. licheniformis (Monier et al., 2023) not only could improve water quality with reducing NH3, NO2−-N, NO3−-N (Mohamed et al., 2024, and Huang TY. et al., 2025) in shrimp culture system, but also enhance digestive enzyme activities (e.g., protease, amylase), immunity and stress resistance in shrimp (Zokaeifar et al., 2014; Sadat Hoseini Madani et al., 2018) and modify flavor quality of shrimp meat by increasing polyunsaturated fatty acid content (Liu et al., 2025).
In contrast, Lactobacillus may act more directly on the host. Studies show that heat-inactivated Lactobacillus can adhere to intestinal mucus and utilize steric hindrance to block pathogen colonization in fish (Bernardeau and Vernoux, 2024). Du et al. (2022) demonstrated that L. plantarum HC-2 colonizes the host intestine via secretion of adhesion proteins, primarily in the foregut, followed by the midgut. Lactobacillus can competitively occupy host cell surface receptors through adhesion protein expression, effectively inhibiting pathogen adhesion and colonization (Evangelista et al., 2025; Lau and Chye, 2018). In this study, the relative abundance of lactic acid bacteria in the aquatic microbiota was not obviously difference between probiotics and CK groups. In this study, we observed no statistically significant variation in the relative abundance of lactic acid bacteria within the aquatic microbiota between the probiotic group and the CK group. According to extant literature, exogenous lactobacilli demonstrate a higher propensity for intestinal colonization. It is reasonable to hypothesize that exogenous lactobacilli strains do not inhabit the water column freely, but rather colonize the intestine of shrimp. However, the present experiment did not reveal alterations in the abundance of lactic acid bacteria within the shrimp intestine. Subsequent experiments are planned to provide further evidence to support or refute this hypothesis.
4.3 The addition of indigenous probiotics did not significantly change the distribution of the potential pathogens and ARGs in shrimp aquaculture water microbial community
The rapid expansion of the aquaculture industry has been accompanied by significant challenges, including frequent disease outbreaks, low survival rates, and environmental nutrients from wastewater discharge. Historically, antibiotics have been widely used to manage disease (Gadhiya et al., 2025). Although effective in the short term, excessive antibiotic use leads to several adverse consequences: disruption of host microbiota, particularly depletion of beneficial microbes, proliferation of antibiotic-resistant bacteria (ARBs), accelerated horizontal transfer of antibiotic ARGs to human-associated microbiomes, and potential environmental contamination (Cheruvari and Kammara, 2025). Consequently, identifying safe and effective alternatives to antibiotics has become a research priority, with biosafety as a critical concern. Recent studies confirm that probiotics, prebiotics, and synbiotics can reduce pathogen loads and enhance host disease resistance without promoting antimicrobial resistance (Butt et al., 2021; Rohani et al., 2022).
However, administered probiotics are typically exogenous to the host’s indigenous microbial community. Research indicates that microbial community diversity strongly influences the success of exogenous microbial colonization, with less diverse communities being more susceptible to invasion (van Elsas et al., 2012; Van Nevel et al., 2013). The introduction of exogenous microbes may therefore alter microbial composition and diversity, potentially facilitating the persistence of potential pathogens or the spread of ARGs. Such disruptive effects have been documented. For instance, Liu et al. (2018) reported that adding commercial probiotics (Bacillus, Lactobacillus, photosynthetic bacteria) to the shrimp larval rearing medium caused a significant change in the planktonic microbial community by day 2, which diminished by days 4 and 7. Probiotics substantially reshaped the planktonic bacterial community within one week. Zhang et al. (2023) further demonstrated that inoculation with Escherichia coli correlated positively with the abundance of potential pathogens such as Prevotella, Aeromonas, and Selenomonas. Moreover, Huang X. et al. (2025) showed that genetically engineered, chlorine-resistant E. coli MG1655 was able to transfer exogenous DNA to indigenous bacteria, thereby conferring enhanced chlorine resistance and environmental fitness and highlighting potential ecological risks. Conversely, exogenous additives such as carbon sources and probiotics can also exert beneficial effects by reshaping aquatic microbial communities, increasing similarity between water and host intestinal microbiota, and enhancing microbial community stability (Feng et al., 2018; Thakur et al., 2025). Supporting this, Dong et al. (2023) and Guo et al. (2022) demonstrated that carbon source supplementation (e.g., sucrose, glucose) in shrimp aquaculture increased microbial community similarity between shrimp gut and rearing water, and strengthened bacterial interaction network stability. The findings of the present study align with these positive regulatory effects: the probiotic treatment significantly reduced the beta diversity coefficient and enhanced structural stability of microbial communities compared to the control (Figure 2C). This suggests that the applied probiotics not only improve shrimp farming performance but also promote microbial community resilience.
Most commercial aquaculture probiotics, isolated from host intestines, have proven effective in controlling pathogenic diseases, serving as safe antibiotic alternatives (Gadhiya et al., 2025). For example, Sekar et al. (2019) screened three Bacillus strains (B. subtilis KA1, B. licheniformis KA2, B. subtilis KA3), among which B. subtilis KA1 exhibited strong antagonism against Vibrio parahaemolyticus and white spot syndrome virus (WSSV). Dietary supplementation with this strain increased beneficial gut bacteria and effectively suppressed specific pathogens (Ralstonia pickettii, Aeromonas veronii, Pseudochrobactrum brenneri). Han et al. (2015) found that supplementation with B. licheniformis improved growth performance (FBW, WG, SGR) in Oreochromis niloticus and significantly enhanced serum lysozyme activity, thereby boosting disease resistance. In contrast, in the present experiment, the addition of shrimp-derived probiotics improved larval growth performance, yet did not elicit significant changes in the abundance of potential pathogenic bacteria in rearing water. Regarding ARGs, the abundance of certain types (e.g., β-lactam, vancomycin, aminoglycoside resistance genes) showed a decreasing trend, while others (e.g., tetracycline, CAMP resistance genes) increased slightly; however, none of these changes were statistically significant (P > 0.05). During Zoea I, water microbiota plays a major role in colonization; external sources dominate in Zoea II, and gut microbiota succession intensifies thereafter. Approximately 66.7% of adult shrimp intestinal prokaryotes originate from larval stages, underscoring the long-term impact of early microbial community assembly (Heyse et al., 2021; Wang et al., 2020; Zhang et al., 2025). This emphasizes the importance of microbial safety when applying probiotics during early development.
In summary, the shrimp-derived composite probiotics used in this experiment improved environmental biosafety and public health. Supplementing probiotics significantly improved the survival rate, biomass, and water quality of shrimp without causing significant changes in microbial community diversity, structural stability, potential pathogen abundance, or the risk of ARGs transmission in aquaculture water. However, the experiment has some limitations, including only testing in shrimp larvae period and no specific pathogen challenge. In future research, we will extend the experimental period and increase the challenge of specific pathogen attack, such as Vibrio to verify the environmental biosafety of shrimp-derived composite probiotics.
5 Conclusion
In conclusion, supplementation with a mixed shrimp-derived probiotic (L. casei HD1 and B. licheniformis WZ1) during the larval rearing stage effectively lowered nitrogen and phosphorus nutrients (e.g., ammonia nitrogen, reactive phosphorus), improves larval survival rates, and protects the natural microbial community structure without promoting pathogen proliferation or antibiotic resistance gene (ARGs) transmission, thereby improving the environment and biosafety of larval shrimp farming. However, different probiotic strains exhibit diverse mechanisms in modulating microbial community dynamics in both aquatic and intestinal environments, thereby eliciting variable impacts on larval growth performance. Water-based probiotics primarily modulate the physicochemical properties of the aquatic environment and indirectly influence shrimp performance through mediation by the aquatic microbial community. These findings indicate that shrimp derived probiotics can improve the water quality and aquaculture performance of shrimp, providing some ideas or strategies for enhancing the environmental biosafety and public health of larvae shrimp farming.
Statements
Data availability statement
All core datasets generated or analyzed during the study are fully disclosed within the published article. Additional supporting data are available in the supplementary materials. All the amplicon sequencing data were available in CNCB (China National Center for Bioinformation, https://ngdc.cncb.ac.cn/) with the accession number of PRJCA044428.
Ethics statement
All animal procedures in this study strictly adhered to the institutional animal care standards of Zhejiang Mariculture Research Institute, which operates under standing approval from its Animal Ethics Committee for aquaculture research complying with the Chinese standard Guidelines for Ethical Review of Laboratory Animal Welfare (GB/T 35892-2018). As per institutional policy, individual project identifiers are not issued for studies involving established protocols meeting predefined ethical criteria.
Author contributions
JY: Visualization, Conceptualization, Investigation, Writing – original draft, Methodology, Data curation, Formal Analysis. CW: Writing – original draft, Formal Analysis, Visualization, Methodology, Validation, Investigation, Conceptualization. PD: Data curation, Project administration, Visualization, Investigation, Validation, Funding acquisition, Writing – review & editing, Software, Methodology, Supervision. CC: Formal Analysis, Project administration, Resources, Investigation, Writing – review & editing. HC: Project administration, Resources, Formal Analysis, Writing – review & editing, Investigation. JC: Resources, Investigation, Project administration, Writing – review & editing. XL: Resources, Methodology, Writing – review & editing, Investigation. KEW: Resources, Investigation, Writing – review & editing. KAW: Resources, Writing – review & editing, Project administration, Conceptualization, Methodology. DZ: Project administration, Funding acquisition, Supervision, Writing – review & editing, Data curation, Formal Analysis, Methodology, Conceptualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by the Wenzhou Science and Technology Project (KN20210008), Zhejiang Key Laboratory of Coastal Biological Germplasm Resources Conservation and Utilization (2024E10128), China Agriculture Research System of MOF and MARA (CARS-48), Municipal Key R&D Program of Ningbo, China (2022Z177), High-Level Science & Technology Project Cultivation Program of Ningbo University (GJPY2025039), the National Natural Science Foundation of China (32303039), State Key Laboratory for Managing Biotic and Chemical Threats to the Quality, Safety of Agro-products (2021DG700024-KF202425), Special Fund for Henan Agriculture Research System (HARS-22-16-T), and Henan Provincial Department of Science and Technology Research Project (242102111013, 252102111033)..
Acknowledgments
We are grateful to Ming Li, Guoxi Li, Yanting Wang and Bianzhi Liu from Henan Agricultural University for their technical support. We are also grateful to the personnel from Ningning Su, Guangqing Yu, Zhenjiang Yang, and Xiaocheng Hang in the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1683189/full#supplementary-material
Supplementary Table 1The information of primers in this research.
Supplementary Table 2The summary of illumina sequence characteristics across all samples.
Supplementary Table 3The results of strain WZ1 and HD1 sequences blastn top hits against the 16S rRNA gene from curated type bacteria and archaea strain sequences in Genbank.
Supplementary Table 4qPCR analysis for the gene copies of 16S rRNA gene, Bacillus taxa, Lactobacillus taxa and, ARGs in probiotic-adding and control groups.
Supplementary Table 5Average (mean ± SD) relative abundance of dominant taxa (genus level) in the control and treatment groups.
Supplementary Figure 1The phylogenetic tree of the taxonomic relationship between strain HD1, WZ1, and other 21 type strains. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches. This tree involved 23 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option).
Supplementary Figure 2Effects of probiotics on the environmental factors and culture performance in aquaculture system of shrimp larvae. Data were statistically compared between control and Probiotics groups by using Wilcoxon Rank Sum test. Significant differences across entire samples compartments are indicated with * (P < 0.05), and ** (P < 0.01).
Supplementary Figure 3The relative abundances of Bacillus and Lactobacillus in rearing water bacterial communities differed significantly between the control and probiotics-added groups. Data were statistically compared between control and Probiotics groups by using Wilcoxon Rank Sum test.
Supplementary Figure 4The relationship between relative abundances of genus Bacillus and Lactobacillus and the values of Alpha diversity in rearing water bacterial communities. The Pearson correlation analysis was used to statistically test the relationship of data, and correlation coefficient (r) and P value were also showed in the figures.
Supplementary Figure 5The difference of ZOTUs between rearing water bacterial communities in the control and probiotics-added groups by the “DESeq2” R package. The data are log2 fold-changes, and the dot size represents the Ln-transformation of average abundance. Significantly differential ZOTUs with adjusted P < 0.05 are denoted as color circles, and not significant as grey.
References
1
Abarike E. D. Cai J. Lu Y. Yu H. Chen L. Jian J. et al . (2018). Effects of a commercial probiotic BS containing Bacillus subtilis and Bacillus licheniformis on growth, immune response and disease resistance in Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol.82, 229–238. doi: 10.1016/j.fsi.2018.08.037
2
Andriantahina F. Liu X. Feng T. Xiang J. (2013). Current status of genetics and genomics of reared penaeid shrimp: information relevant to access and benefit sharing. Mar. Biotechnol. (New York N.Y.)15 (4), 399-412. doi: 10.1007/s10126-013-9500-9
3
Bailey L. C. Forrest C. B. Zhang P. Richards T. M. Livshits A. DeRusso P. A. (2014). Association of antibiotics in infancy with early childhood obesity. JAMA Pediatr.168, 1063–1069. doi: 10.1001/jamapediatrics.2014.1539
4
Bernardeau M. Vernoux J.-P. (2024), 658–688).
5
Bolger A. M. Lohse M. Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
6
Butt U. D. Lin N. Akhter N. Siddiqui T. Li S. Wu B. (2021). Overview of the latest developments in the role of probiotics, prebiotics and synbiotics in shrimp aquaculture. Fish Shellfish Immunol.114, 263–281. doi: 10.1016/j.fsi.2021.05.003
7
Bydalek F. Webster G. Barden R. Weightman A. J. Kasprzyk-Hordern B. Wenk J. (2024). Microbial community and antimicrobial resistance niche differentiation in a multistage, surface flow constructed wetland. Water Res.254, 121408. doi: 10.1016/j.watres.2024.121408
8
Cheruvari A. Kammara R. (2025). Bacteriocins future perspectives: Substitutes to antibiotics. Food Control168, 110834. doi: 10.1016/j.foodcont.2024.110834
9
Dong P. Chen Y. Wei Y. Zhao X. Wang T. Jiang S. et al . (2025). Dix-seq: An integrated pipeline for fast amplicon data analysis. Innovation Life3, 100120. doi: 10.59717/j.xinn-life.2024.100120
10
Dong P. Guo H. Huang L. Zhang D. Wang K. (2023). Glucose addition improves the culture performance of Pacific white shrimp by regulating the assembly of Rhodobacteraceae taxa in gut bacterial community. Aquaculture567, 739254. doi: 10.1016/j.aquaculture.2023.739254
11
Dong P. Guo H. Wang Y. Cheng H. Wang K. Hong M. et al . (2021a). DPiWE: a curated database for pathogenic bacteria involved in water environment. J. Fish China45 (11), 1921–1933. doi: jfc.20210612935
12
Dong P. Guo H. Wang Y. Wang R. Chen H. Zhao Y. et al . (2021b). Gastrointestinal microbiota imbalance is triggered by the enrichment of Vibrio in subadult Penaeus vannamei with acute hepatopancreatic necrosis disease. Aquaculture533, 736199. doi: 10.1016/j.aquaculture.2020.736199
13
Du S. Chen W. Yao Z. Huang X. Chen C. Guo H. et al . (2021). Enterococcus faecium are associated with the modification of gut microbiota and shrimp post-larvae survival. Anim. microbiome3, 88. doi: 10.1186/s42523-021-00152-x
14
Du Y. Li H. Shao J. Wu T. Xu W. Hu X. et al . (2022). Adhesion and colonization of the probiotic lactobacillus plantarum HC-2 in the intestine of litopenaeus vannamei are associated with bacterial surface proteins. Front. Microbiol.13, 878874. doi: 10.3389/fmicb.2022.878874
15
Dubey D. Toppo K. Kumar S. Dutta V. (2024). Intensive aquaculture affects lake’s trophic status and aquatic floral diversity. Environ. Science: Adv.3, 1628–1642. doi: 10.1039/D4VA00038B
16
Edgar R. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics26, 2460–2461. doi: 10.1093/bioinformatics/btq461
17
Edgar R. (2016). UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv, 081257. doi: 10.1101/081257
18
El-Saadony M. T. Alagawany M. Patra A. K. Kar I. Tiwari R. Dawood M. A. O. et al . (2021). The functionality of probiotics in aquaculture: An overview. Fish Shellfish Immunol.117, 36–52. doi: 10.1016/j.fsi.2021.07.007
19
El-Saadony M. T. Swelum A. A. Abo Ghanima M. M. Shukry M. Omar A. A. Taha A. E. et al . (2022). Shrimp production, the most important diseases that threaten it, and the role of probiotics in confronting these diseases: A review. Res. Vet. Sci.144, 126–140. doi: 10.1016/j.rvsc.2022.01.009
20
Evangelista A. G. Janotto L. d. S. d. A. C. M. de Macedo R. E. F. Luciano F. B. (2025). Cellular effect of lactic acid bacteria and their metabolites against animal production bacteria. Total Environ. Microbiol.1, 100013. doi: 10.1016/j.temicr.2025.100013
21
FAO . (2024). The State of World Fisheries and Aquaculture 2024 – Blue Transformation in action. Rome. doi: 10.4060/cd0683en
22
Feng P. Ye Z. Kakade A. Virk A. K. Li X. Liu P. (2018). A review on gut remediation of selected environmental contaminants: possible roles of probiotics and gut microbiota. Nutrients11 (1), 22. doi: 10.3390/nu11010022
23
Figueras A. Chizhayeva A. Amangeldi A. Oleinikova Y. Alybaeva A. Sadanov A. (2022). Lactic acid bacteria as probiotics in sustainable development of aquaculture. Aquat. Living Resour.35, 2022011. doi: 10.1051/alr/2022011
24
Gadhiya A. Katariya S. Khapandi K. Chhatrodiya D. (2025). Probiotics as a sustainable alternative to antibiotics in aquaculture: a review of the current state of knowledge. Microbe8, 100426. doi: 10.1016/j.microb.2025.100426
25
Ghosh T. (2025). Recent advances in the probiotic application of the Bacillus as a potential candidate in the sustainable development of aquaculture. Aquaculture594, 741432. doi: 10.1016/j.aquaculture.2024.741432
26
Guo H. Huang L. Hu S. Chen C. Huang X. Liu W. et al . (2020). Effects of carbon/nitrogen ratio on growth, intestinal microbiota and metabolome of shrimp (Litopenaeus vannamei). Front. Microbiol., 11, 652. doi: 10.3389/fmicb.2020.00652
27
Guo H. Dong P. Gao F. Huang L. Wang S. Wang R. et al . (2022). Sucrose addition directionally enhances bacterial community convergence and network stability of the shrimp culture system. NPJ Biofilms Microbiomes8, 22. doi: 10.1038/s41522-022-00288-x
28
Guo H. Fu X. He J. Wang R. Yan M. Wang J. et al . (2023). Gut bacterial consortium enriched in a biofloc system protects shrimp against Vibrio parahaemolyticus infection. Microbiome11, 230. doi: 10.1186/s40168-023-01663-2
29
Hamza F. Kumar A. R. Zinjarde S. (2018). Efficacy of cell free supernatant from Bacillus licheniformis in protecting Artemia salina against Vibrio alginolyticus and Pseudomonas gessardii. Microb. Pathog.116, 335–344. doi: 10.1016/j.micpath.2018.02.003
30
Han B. Long W. Q. He J. Y. Liu Y. J. Si Y. Q. Tian L. X. (2015). Effects of dietary Bacillus licheniformis on growth performance, immunological parameters, intestinal morphology and resistance of juvenile Nile tilapia (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol.46, 225–231. doi: 10.1016/j.fsi.2015.06.018
31
Heyse J. Props R. Kongnuan P. De Schryver P. Rombaut G. Defoirdt T. et al . (2021). Rearing water microbiomes in white leg shrimp (Litopenaeus vannamei) larviculture assemble stochastically and are influenced by the microbiomes of live feed products. Environ. Microbiol.23, 281–298. doi: 10.1111/1462-2920.15310
32
Holt C. Bass D. Stentiford D. van der Giezen M. (2021). Understanding the role of the shrimp gut microbiome in health and disease. J. Invertebrate Pathol.186, 107387. doi: 10.1016/j.jip.2020.107387
33
Huang X. Tan Z. Wei J. Bai X. (2025). Super-robust synthetic microorganism can get chlorine resistance in advance and transfer their inserted DNA sequence in genome to indigenous bacteria in water. Water Res.281, 123594. doi: 10.1016/j.watres.2025.123594
34
Huang T.-Y. Ju H.-J. Huang M.-Y. Kuo Q.-M. Su W.-T. (2025). Optimal nitrite degradation by isolated Bacillus subtilis sp. N4 and applied for intensive aquaculture water quality management with immobilized strains. J. Environ. Manage., 374, 123896. doi: 10.1016/j.jenvman.2024.123896
35
Kewcharoen W. Srisapoome P. (2019). Probiotic effects of Bacillus spp. from Pacific white shrimp (Litopenaeus vannamei) on water quality and shrimp growth, immune responses, and resistance to Vibrio parahaemolyticus (AHPND strains). Fish Shellfish Immunol.94, 175–189. doi: 10.1016/j.fsi.2019.09.013
36
Kewcharoen W. Srisapoome P. (2022). Potential synbiotic effects of a Bacillus mixture and chitosan on growth, immune responses and VP((AHPND)) resistance in Pacific white shrimp (Litopenaeus vannamei, Boone 1931). Fish Shellfish Immunol.127, 715–729. doi: 10.1016/j.fsi.2022.07.017
37
Khademzade O. Zakeri M. Haghi M. Mousavi S. M. (2020). The effects of water additive Bacillus cereus and Pediococcus acidilactici on water quality, growth performances, economic benefits, immunohematology and bacterial flora of whiteleg shrimp (Penaeus vannamei Boone 1931) reared in earthen ponds. Aquaculture Res.51, 1759–1770. doi: 10.1111/are.14525
38
Kong Y. D. Kang Y. H. Tian J. X. Zhang D. X. Zhang L. Tao L. T. et al . (2019). Oral immunization with recombinant Lactobacillus casei expressing flaB confers protection against Aeromonas veronii challenge in common carp, Cyprinus carpio. Fish Shellfish Immunol.87, 627–637. doi: 10.1016/j.fsi.2019.01.032
39
Kronman M. P. Zaoutis T. E. Haynes K. Feng R. Coffin S. E. (2012). Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics130, e794–e803. doi: 10.1542/peds.2011-3886
40
Kuebutornye F. K. A. Abarike E. D. Lu Y. (2019). A review on the application of Bacillus as probiotics in aquaculture. Fish Shellfish Immunol.87, 820–828. doi: 10.1016/j.fsi.2019.02.010
41
Lau L. Y. J. Chye F. Y. (2018). Antagonistic effects of Lactobacillus plantarum 0612 on the adhesion of selected foodborne enteropathogens in various colonic environments. Food Control91, 237–247. doi: 10.1016/j.foodcont.2018.04.001
42
Lee J. W. Chiu S. T. Wang S. T. Liao Y. C. Chang H. T. Ballantyne R. et al . (2022). Dietary SYNSEA probiotic improves the growth of white shrimp, Litopenaeus vannamei and reduces the risk of Vibrio infection via improving immunity and intestinal microbiota of shrimp. Fish Shellfish Immunol.127, 482–491. doi: 10.1016/j.fsi.2022.06.071
43
Li E.-C. Xu C. Wang X. Shifeng W. Zhao Q. Zhang M. et al . (2018). Gut Microbiota and its Modulation for Healthy Farming of Pacific White Shrimp Litopenaeus vannamei. Rev. Fisheries Sci. Aquaculture26, 1–19. doi: 10.1080/23308249.2018.1440530
44
Liu S. Luo G. Wu G. Fan B. Liu W. Tan H. et al . (2025). Impact of commercial probiotics (Bacillus subtilis) addition during biofloc start-up: water quality, nitrogen and microbial community. Aquaculture, 609, 742845. doi: 10.1016/j.aquaculture.2025.742845
45
Liu Z. Qiuqian L. Yao Z. Wang X. Huang L. Zheng J. et al . (2018). Effects of a commercial microbial agent on the bacterial communities in shrimp culture system. Front. Microbiol.9. doi: 10.3389/fmicb.2018.02430
46
Liu J. Wang K. Wang Y. Chen W. Jin Z. Yao Z. et al . (2019). Strain-specific changes in the gut microbiota profiles of the white shrimp Litopenaeus vannamei in response to cold stress. Aquaculture503, 357–366. doi: 10.1016/j.aquaculture.2019.01.026
47
Looft T. Johnson T. A. Allen H. K. Bayles D. O. Alt D. P. Stedtfeld R. D. et al . (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. U.S.A.109, 1691–1696. doi: 10.1073/pnas.1120238109
48
Love M. I. Huber W. Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15 (12), 550. doi: 10.1186/s13059-014-0550-8
49
Lu Z. Lin W. Li Q. Wu Q. Ren Z. Mu C. et al . (2024). Recirculating aquaculture system as microbial community and water quality management strategy in the larviculture of Scylla paramamosain. Water Res.252, 121218. doi: 10.1016/j.watres.2024.121218
50
Luo H. Xie K. Dong P. Zhang Y. Ren T. Sui C. et al . (2025). Assessing the risks of potential pathogens and antibiotic resistance genes among heterogeneous habitats in a temperate estuary wetland: a meta-analysis. Microb. Ecol.87, 172. doi: 10.1007/s00248-024-02484-y
51
Mathan Muthu C. M. Vickram A. S. Bhavani Sowndharya B. Saravanan A. Kamalesh R. Dinakarkumar Y. (2024). A comprehensive review on the utilization of probiotics in aquaculture towards sustainable shrimp farming. Fish Shellfish Immunol.147, 109459. doi: 10.1016/j.fsi.2024.109459
52
Maynard C. L. Elson C. O. Hatton R. D. Weaver C. T. (2012). Reciprocal interactions of the intestinal microbiota and immune system. Nature489, 231–241. doi: 10.1038/nature11551
53
Mohamed H. A. Ayyat M. S. Mahgoub S. A. Mahmoud H. K. Alkhedaide A. Q. (2024). Does the use of two probiotic bacteria (Latiplantibacillus plantarum and Bacillus toyonensis) as water additives enhance growth performance, the immune responses, antioxidative maintenance, water quality and intestinal bacterial counts of Nile tilapia? Aquaculture Rep.39, 102471. doi: 10.1016/j.aqrep.2024.102471
54
Monier M. N. Kabary H. Elfeky A. Saadony S. El-Hamed N. N. B. A. Eissa M. E. H. et al . (2023). The effects of Bacillus species probiotics (Bacillus subtilis and B. licheniformis) on the water quality, immune responses, and resistance of whiteleg shrimp (Litopenaeus vannamei) against Fusarium solani infection. Aquaculture Int.31, 3437–3455. doi: 10.1007/s10499-023-01136-1
55
Nayak S. K. (2021). Multifaceted applications of probiotic Bacillus species in aquaculture with special reference to Bacillus subtilis. Rev. Aquaculture13, 862–906. doi: 10.1111/raq.12503
56
Qiu Z. Xu Q. Li S. Zheng D. Zhang R. Zhao J. et al . (2023). Effects of probiotics on the water quality, growth performance, immunity, digestion, and intestinal flora of giant freshwater prawn (Macrobrachium rosenbergii) in the biofloc culture system. Water15 (6), 1211. doi: 10.3390/w15061211
57
Raza B. Ramzan M. N. Yang W. (2025). A review: Improving aquaculture rearing water quality by removal of nutrients using microalgae, challenges and future prospects. Aquaculture598, 741959. doi: 10.1016/j.aquaculture.2024.741959
58
Redhwan A. Eissa E.-S. H. Ezzo O. H. Abdelgeliel A. S. Munir M. B. Chowdhury A. J. K. et al . (2024). Effects of water additive mixed probiotics on water quality, growth performance, feed utilization, biochemical analyses and disease resistance against Aeromonas sobria of Nile tilapia. Desalination Water Treat319, 100480. doi: 10.1016/j.dwt.2024.100480
59
Ringø E. Van Doan H. Lee S. H. Soltani M. Hoseinifar S. H. Harikrishnan R. et al . (2020). Probiotics, lactic acid bacteria and bacilli: interesting supplementation for aquaculture. J. Appl. Microbiol.129, 116–136. doi: 10.1111/jam.14628
60
Rohani M. F. Islam S. M. Hossain M. K. Ferdous Z. Siddik M. A. Nuruzzaman M. et al . (2022). Probiotics, prebiotics and synbiotics improved the functionality of aquafeed: Upgrading growth, reproduction, immunity and disease resistance in fish. Fish Shellfish Immunol.120, 569–589. doi: 10.1016/j.fsi.2021.12.037
61
Sadat Hoseini Madani N. Adorian T. J. Ghafari Farsani H. Hoseinifar S. H. (2018). The effects of dietary probiotic Bacilli (Bacillus subtilis and Bacillus licheniformis) on growth performance, feed efficiency, body composition and immune parameters of whiteleg shrimp (Litopenaeus vannamei) Postlarvae. Aquaculture Res.49, 1926–1933. doi: 10.1111/are.13648
62
Seabkongseng T. Limkul S. Sriphuttha C. Phiwthong T. Aunkam P. Suwannathit R. et al . (2025). Supplementation of Bacillus velezensis S141 in feed as a probiotic enhances growth performance, pathogenic tolerances, and immune system in shrimp. Aquaculture604, 742448. doi: 10.1016/j.aquaculture.2025.742448
63
Sekar A. Kim M. Jeon H. Kim K. (2019). Screening and selection of bacteria inhibiting white spot syndrome virus infection to Litopenaeus vannamei. Biochem. Biophys. Rep.19, 100663. doi: 10.1016/j.bbrep.2019.100663
64
Sha Y. Wang L. Liu M. Jiang K. Xin F. Wang B. (2016). Effects of lactic acid bacteria and the corresponding supernatant on the survival, growth performance, immune response and disease resistance of Litopenaeus vannamei. Aquaculture452, 28–36. doi: 10.1016/j.aquaculture.2015.10.014
65
Su H. Liu S. Hu X. Xu X. Xu W. Xu Y. et al . (2017). Occurrence and temporal variation of antibiotic resistance genes (ARGs) in shrimp aquaculture: ARGs dissemination from farming source to reared organisms. Sci. Total Environ.607-608, 357–366. doi: 10.1016/j.scitotenv.2017.07.040
66
Tamburini S. Shen N. Wu H. C. Clemente J. C. (2016). The microbiome in early life: implications for health outcomes. Nat. Med.22, 713–722. doi: 10.1038/nm.4142
67
Tamura K. Stecher G. Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol.38, 3022–3027. doi: 10.1093/molbev/msab120
68
Thakur K. Singh B. Kumar S. Sharma D. Sharma A. K. Jindal R. et al . (2025). Potential of probiotics and postbiotics in aquaculture: Connecting current research gaps and future perspectives. Microbe8, 100431. doi: 10.1016/j.microb.2025.100431
69
Thitamadee S. Prachumwat A. Srisala J. Jaroenlak P. Salachan P. V. Sritunyalucksana K. et al . (2016). Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture452, 69–87. doi: 10.1016/j.aquaculture.2015.10.028
70
Umamaheswari T. Sugumar G. Rajakumar M. Felix N. Athithan S. Veerabhadran K. (2025). Value chain analysis of Penaeus vannamei in Tamil Nadu, India. Aquaculture International, 2025, 33 (3), 229. doi: 10.1007/s10499-025-01910-3
71
van Elsas J. D. Chiurazzi M. Mallon C. A. Elhottova D. Kristufek V. Salles J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A.109, 1159–1164. doi: 10.1073/pnas.1109326109
72
Van Nevel S. De Roy K. Boon N. (2013). Bacterial invasion potential in water is determined by nutrient availability and the indigenous community. FEMS Microbiol. Ecol.85, 593–603. doi: 10.1111/1574-6941.12145
73
Wang Y. Wang K. Huang L. Dong P. Wang S. Chen H. et al . (2020). Fine-scale succession patterns and assembly mechanisms of bacterial community of Litopenaeus vannamei larvae across the developmental cycle. Microbiome, 8 (1), 106. doi: 10.1186/s40168-020-00879-w
74
Wang R. Chen H. Zhu Y. Al-Masqari Z. A. Yan M. Wang G. et al . (2022). Survival status of Penaeus vannamei is associated with the homeostasis and assembly process of the intestinal bacterial community. Aquaculture558, 738398. doi: 10.1016/j.aquaculture.2022.738398
75
Wang Y. C. Hu S. Y Chui C. S. Lui C. H. . (2019). Multiple-strain probiotics appear to be more effective in improving the growth performance and health status of white shrimp, Litopenaeus vannamei, than single probiotic strains. Fish Shellfish Immunol84, 1050–1058. doi: 10.1016/j.fsi.2018.11.017
76
Wang Y. Wang K. Huang L. Dong P. Wang S. Chen H. et al . (2020). Fine-scale succession patterns and assembly mechanisms of bacterial community of Litopenaeus vannamei larvae across the developmental cycle. Microbiome8, 106. doi: 10.1186/s40168-020-00879-w
77
Xie J. J. Liu Q. Q. Liao S. Fang H. H. Yin P. Xie S. W. et al . (2019). Effects of dietary mixed probiotics on growth, non-specific immunity, intestinal morphology and microbiota of juvenile pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol.90, 456–465. doi: 10.1016/j.fsi.2019.04.301
78
Xiong J. Dai W. Qiu Q. Zhu J. Yang W. Li C. (2018). Response of host-bacterial colonization in shrimp to developmental stage, environment and disease. Mol. Ecol.27, 3686–3699. doi: 10.1111/mec.14822
79
Yu D. Wang T. Zhang L. Gao N. Huang Y. Zhang J. et al . (2025). Identification of body fluid sources based on microbiome antibiotic resistance genes using high-throughput qPCR. Forensic Sci. International: Genet.77, 103241. doi: 10.1016/j.fsigen.2025.103241
80
Zhang F. Chen H. Yu J. Chen R. Zhang D. Chen C. et al . (2025). Source tracking of larval bacterial community of Pacific white shrimp across the developmental cycle: Ecological insights for microbial management and pathogen prevention in larviculture. Aquaculture609, 742800. doi: 10.1016/j.aquaculture.2025.742800
81
Zhang N. Liang C. Kan P. Yangyao J. Lu D. Yao Z. et al . (2023). Indigenous microbial community governs the survival of Escherichia coli O157:H7 in constructed wetlands. J. Environ. Manage.334, 117524. doi: 10.1016/j.jenvman.2023.117524
82
Zhang Z. Yang Z. Zheng G. Lin Q. Zhuo X. Zhang G. (2020). Effects of addition of sucrose and probiotics on whiteleg shrimp Litopenaeus vannamei postlarvae performance, water quality, and microbial community. North Am. J. Aquac., 82 (1), 43-53. doi: 10.1002/naaq.10116
83
Zhao Z. Wang Y. Wei Y. Peng G. Wei T. He J. et al . (2024). Distinctive patterns of bacterial community succession in the riverine micro-plastisphere in view of biofilm development and ecological niches. J. Hazardous Materials480, 135974. doi: 10.1016/j.jhazmat.2024.135974
84
Zhou X.-X. Wang Y.-B. Li W.-F. (2009). Effect of probiotic on larvae shrimp (Penaeus vannamei) based on water quality, survival rate and digestive enzyme activities. Aquaculture287, 349–353. doi: 10.1016/j.aquaculture.2008.10.046
85
Zokaeifar H. Babaei N. Saad C. R. Kamarudin M. S. Sijam K. Balcazar J. L. (2014). Administration of Bacillus subtilis strains in the rearing water enhances the water quality, growth performance, immune response, and resistance against Vibrio harveyi infection in juvenile white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol.36, 68–74. doi: 10.1016/j.fsi.2013.10.007
Summary
Keywords
Penaeus vannamei , indigenous probiotics, microbial community dynamics, potential pathogens, antibiotic resistance genes, environmental safety
Citation
Yu J, Wu C, Dong P, Chen C, Chen H, Chen J, Liu X, Wang K, Wang K and Zhang D (2025) Risk assessment of shrimp-derived probiotics on culture performance and environmental biosafety in shrimp larvae rearing system. Front. Mar. Sci. 12:1683189. doi: 10.3389/fmars.2025.1683189
Received
10 August 2025
Accepted
15 September 2025
Published
09 October 2025
Volume
12 - 2025
Edited by
Seyyed Morteza Hoseini, Iranian Fisheries Science Research Institute (IFSRI), Iran
Reviewed by
Chittaranjan Baruah, Darrang College, India; Melika Ghelichpour, University of Tehran, Iran; Maryam Mirbakhsh, Agricultural Research, Education and Extension Organization (AREEO), Iran
Updates
Copyright
© 2025 Yu, Wu, Dong, Chen, Chen, Chen, Liu, Wang, Wang and Zhang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengsheng Dong, dpsh@henau.edu.cn; Demin Zhang, zhangdemin@nbu.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.