Abstract
Coastal marine organisms are often exposed to high levels of biologically harmful ultraviolet radiation (UVR), the most photochemically reactive waveband of sunlight. It is well known that marine organisms at higher trophic levels, such as fish, enhance their UV protection by accumulating UV-protective metabolites from their diet, as primary producers can effectively synthesize these compounds. Among the best studied natural UV-sunscreens in marine organisms are mycosporine-like amino acids (MAAs). They are known for their high molar extinction coefficients in the UVR-region along with pronounced photo- and thermal stability. In the present study we investigated the qualitative and quantitative MAA distribution in organs (eyes, gills, heart, intestine, kidneys, liver, skin, and stomach) of three flatfish species from the Baltic Sea, the benthivorous European flounder (Platichthys flesus) and European plaice (Pleuronectes platessa), and the piscivorous turbot (Scophthalmus maximus), using state-of-the-art analytical methods. Most of the analyzed organ samples contained the MAAs palythine, asterina-330, porphyra-334, usujirene, and palythene at concentrations sufficient for reliable detection and quantification using an established HPLC-UV method. Additionally, in a few samples also shinorine and mycosporine-methylamine-threonine were found. The highest MAA contents (0.04 to 0.25 mg g-1 dry weight) occurred in the eyes of the three fish species, while the other organs exhibited much lower but still detectable concentrations. Our data support the assumed trophic transfer of MAAs from primary producers via the food web to fish. For the first time we show that MAAs are not only found in the eyes but also in internal organs that possibly represent transfer points from the digestive tract to UV-sensitive tissues. The underlying mechanisms are, however, still unknown.
Introduction
High-energy ultraviolet (UV) wavelengths, part of natural sunlight reaching the Earth’s surface, affect many marine organisms (Vincent and Neale, 2000; Karsten, 2008). While solar radiation is a prerequisite for all primary producers, UV light can also be harmful to biomolecules like DNA and proteins, as well as to numerous physiological processes, especially at high doses during summer or at noon, and particularly for organisms living in shallow waters (Pavlov et al., 2019). In terrestrial and aquatic ecosystems, UV radiation (UVR) strongly varies in space and time, influenced by geographic location, time of day, season, clouds, or canopy coverage. In addition, in the coastal-marine realm also waves, tides, currents, and the optical properties of the water column affect UVR strength. For example, suspended particles and chromophoric dissolved organic matter are documented to reduce penetration of UVR into the water column (Pavlov et al., 2019). Consequently, due to the high variability of the underwater radiation climate, marine organisms such as fish experience quite different doses of UVR. Nevertheless, the shallow coastal waters, such as the Baltic Sea, are attractive feeding grounds for consumers because of their generally high productivity, but at the same time require adaptive traits against enhanced UVR.
The Baltic Sea has unique features, as it is enclosed and geologically young, exhibits a horizontal salinity gradient leading to brackish conditions, lacks tidal currents and is geomorphologically diverse due to a dynamic mosaic pattern of eroding cliffs and sand deposition areas (Jurasinski et al., 2018). Besides UVR, anthropogenic pressures and climate change exert additional strong negative effects on biodiversity and ecosystem functioning in the Baltic Sea (Viitasalo and Bonsdorff, 2022). Such stressors include, for example, eutrophication and pollution together with rising water temperatures and sea levels along with decreasing ice covers during winter, thereby impacting food webs and fish stocks (Moll et al., 2024).
At high levels, UVR damages all organisms by causing DNA lesions and generating reactive oxygen species (ROS), which trigger lipid peroxidation, protein degradation, and other disruptions of metabolic pathways (Karsten, 2008; Schuch et al., 2017). Marine organisms sustain fitness and survival through mechanisms such as DNA repair by photolyase and the biosynthesis or accumulation of UV-filtering compounds (Bandaranayake, 1998; Cockell and Knowland, 1999; Dunlap and Shick, 1998; Karsten, 2008). One of the best studied group of natural UV-sunscreens are the so-called mycosporines and mycosporine-like amino acids (MAAs), which consist of about 80 chemically closely related structures (Peng et al., 2023; Robson et al., 2019; Karsten, 2008).
MAAs and mycosporines are colorless, hydrophilic biomolecules with molecular weights usually smaller than 400 Da. They act as photoprotective compounds in many different marine organisms, from cyanobacteria to micro- and macroalgae as well as from many invertebrates to fish (Peng et al., 2023; Bonin et al., 2024). These natural products contain a cyclohexenone (mycosporines) or cyclohexenimine (MAAs) core, which is substituted with an amino acid or its imino alcohol. Such a conjugated structure leads to very high molar extinction coefficients in the UVR-region, ranging from Ɛ = 28,100 to 50,000 M-1 cm-1, with typical absorption maxima between 310 and 360 nm. From the well-studied MAAs with a cyclohexenimine structure like palythine, porphyra-334 or shinorine, it is reported that they are chemically quite stable under heat and irradiation exposure (Karsten, 2008 and references therein). Some MAAs act as antioxidants and are involved in stress response (Shick and Dunlap, 2002; Karsten, 2008; Peng et al., 2023; Wang et al., 2025). For asterina-330, even protective effects against arsenic- and fluoride-induced toxicity in zebrafish hepatocytes have been reported (Mondal et al., 2025).
Karsten (2008) and Peng et al. (2023) described the most abundant MAAs to originate from primary producers, such as planktonic diatoms, dinoflagellates, cyanobacteria, among others, or red macroalgae. Just recently, direct evidence was obtained that also surface scums of cyanobacteria along the southern Baltic Sea coast of Finland contain the MAAs porphyra-334 and shinorine (Vuori et al., 2025). Nevertheless, MAAs also occur in many marine invertebrates like sea urchins, corals or sponges, as well as in fish (Shick and Dunlap, 2002; Bonin et al., 2024). Since marine invertebrates and fish cannot produce MAAs, they must obtain them through their diet, requiring a trophic transfer from primary producers up the food web to higher levels (Bonin et al., 2024 and references therein). Bonin et al. (2024) investigated the eyes of 39, mainly marine fish species from the Northern Hemisphere and reported the presence of aplysiapalythine A, asterina-330, palythene, palythine, porphyra-334, shinorine, and usujirene in most of the samples, although the composition and concentrations were highly variable. Total MAAs covered a wide concentration range from trace amounts up to > 4.2 mg g−1 dry weight, with the highest MAA values measured in zooplanktivorous Sprattus sprattus (Bonin et al., 2024). Previous studies confirmed the presence of MAAs in many warm-temperate to tropical fish species (e.g., Mason et al., 1998; Siebeck and Marshall, 2001), mainly located in the eyes, mucus (Eckes et al., 2008; Zamzow, 2004, 2007), or eggs (Chioccara et al., 1980). In addition, the MAA-related compound gadusol, characterized by an additional hydroxyl substitution at the core and the absence of an amino acid residue, has been reported in the fish roes of three species from the Argentine Sea (Arbeloa et al., 2010). However, the assumed transfer of MAAs from the ingested diet to the eyes is still an unexplored question.
Therefore, the present investigation aims at shedding light on this MAA transfer in various flatfish species by assessing the MAA concentration in different organs of two benthivorous and one piscivorous Baltic flatfish species. The nursery grounds of the three selected species are very shallow waters (Aarnio et al., 1996; Nissling et al., 2007). Adult flounder and plaice extensively use shallow coastal waters during the summer months (Strodtmann, 1906). Adult turbot even spawn in very shallow water in late spring and feed close to the coast during the summer months (Florin and Franzén, 2010). Hence, these species are particularly exposed to UVR both as juveniles and adults. To investigate whether MAAs are present in different organs of flounder, plaice, and turbot, and to assess potential quantitative differences, we employed a combination of targeted and untargeted metabolomics. Metabolites were quantified using a validated HPLC-UV method (Orfanoudaki et al., 2020), while the metabolome was qualitatively analyzed by UHPLC-ESI-HRMS/MS and subsequently visualized and evaluated through feature-based molecular networking (Nothias et al., 2020).
Material and methods
Fish samples and organ dissection
In this study we used adults of three commercially used flatfish species, the European flounder (Platichthys flesus, total length range: 28–31 cm), European plaice (Pleuronectes platessa, total length range: 27–28 cm), and turbot (Scophthalmus maximus, total length range: 32–34 cm), all caught by demersal trawl in October 2023 in the western Baltic Sea and provided by the Thünen-Institute of Baltic Sea Fisheries (Rostock, Germany). The trawls were undertaken with the research vessel “Solea” during cruise 828 at different positions (54°11.7 to 54°12.2 latitude, 11°53.9 to 11°59.8 longitude). Flounder and plaice are benthivorous, feeding on bivalves and other invertebrates while turbot is piscivorous. The western Baltic Sea is a shallow-water region, with most of the Belt Sea under 25 m deep and a maximum depth of 50 m in the Arkona Sea. All three species use shallow waters, especially from spring to autumn.
The freshly caught fish (10 specimens per species) were percussion stunned by a blow to the head and killed by a cut through the spine. Fish were kept on ice prior to dissection of the following organs: eyes, gills, heart, intestine, kidneys, liver, skin (without mucus), and stomach (without content), using binocular, scalpel, and forceps. After dissection, the different fish organs were briefly washed with distilled water to remove external tissue remains, frozen at -18 °C, followed by freeze-drying in a lyophilizer (Alpha 1–4 LSCPlus, Martin Christ GmbH, Osterode am Harz, Germany). The freeze-dried organs were stored at -18 °C prior to MAA extraction.
MAA extraction
Freeze-dried organs that were hard and fibrous, i.e., eyes, gills, intestines, skin, and stomach, were crushed using a Mikro-Dismembrator S 8531722 (Sartorius, Göttingen, Germany). The corresponding samples were placed in a Teflon shake flask with agate beads and ground for 15 min at 2,000 rpm. Softer organs, such as the heart, kidneys or the liver, were disintegrated to the smallest possible size using spatulas and tweezers.
To extract MAAs, 500 µL of 100% methanol was added to approximately 10 mg of freeze-dried, powdered fish organ sample in an Eppendorf reaction tube. The samples were vortexed (Vortex-Genie 2, Scientific Industries, Inc., Bohemia, New York) and extracted for 15 min in an ultrasonic bath (Sonorex TK 52, Bandelin electronic GmbH & Co. KG, Berlin, Germany) at ambient temperature. After centrifugation (Centrifuge 5804R, Eppendorf, Hamburg, Germany) for 10 min at 12,500 rpm and 25 °C, the supernatant was carefully removed using a Pasteur pipette and transferred to a reaction tube. This extraction procedure was repeated two more times, and the resulting extracts were combined. The pooled extracts were dried overnight under a stream of compressed air, and the resulting residue was re-dissolved in 300 µL of ultra-pure water. After vortexing, the solution was subjected to 5 min of ultrasonication followed by 5 min centrifugation at 12,500 rpm. The final supernatant was filtered through cotton wool directly into an HPLC vial and stored at -18 °C until being analyzed.
MAA analysis by HPLC-UV
Analyses were performed on a Merck–Hitachi Elite La Chrom instrument (Tokyo, Japan) equipped with an organizer (SYSTEM O/B), a quaternary pump (L-2100), a degasser (Degasys DG-2410; Uniflows Co.,Ltd., Tokyo, Japan), an autosampler (L-2200), a column oven (L-2300), and a diode-array detector (L-2455). Separation conditions were according to the method described by Orfanoudaki et al. (2020). A YMC-Pack ODS column (250 mm × 4.60 mm, 5 μm; YMC Europe, Dinslaken, Germany) in combination with an appropriate guard column served as stationary phase. The mobile phase consisted of 20 mM ammonium formate and 0.25% (v/v) formic acid in water (A) and methanol (B). The applied gradient was as follows: 100% A from 0 to 20 min, 80% A at 30 min, 2% A at 35 min, and held at this composition for 5 min (total runtime of 40 min); followed by 15 min of re-equilibration under the initial conditions. Flow rate, temperature, and injection volume were adjusted to 0.65 mL min-1, 9 °C and 5.0 μL, respectively. Detection wavelengths were set to 310, 330, and 350 nm. The area under the curve of target peaks was determined with EZChrom Elite software using the chromatograms recorded at 330 nm. Chromatograms were constructed with OriginPro 2020 software (OriginLab Corporation, Northampton, Massachusetts, USA).
As in our former study (Bonin et al., 2024), calibration was performed using stock solutions of shinorine, palythine, asterina-330, porphyra-334, and aplysiapalythine A, which came from a previous study (Orfanoudaki et al., 2019). For each MAA, at least six calibration levels were prepared by diluting standards in water, with each of them being analyzed in triplicate. Calibration curves were established by plotting peak area against concentration and analyzed via linear regression using Microsoft Excel. The resulting regression equations and coefficients of determination (R2) were as follows: shinorine (y = 262406x – 11866, R2 = 0.9998), palythine (y = 139440x + 16461, R2 = 0.9997), asterina-330 (y = 196159x – 4343.6, R2 = 0.9992), porphyra-334 (y = 256858x + 979.62, R2 = 0.9995), and aplysiapalythine A (y = 149303x + 1628.5, R2 = 0.9997). Limits of detection (LOD) and quantification (LOQ) were calculated from the regression models as 3.3 and 10 times the residual standard deviation divided by the slope, respectively. The LODs ranged from 0.05 to 0.15 µg mL-1, and the LOQs from 0.16 to 0.46 µg mL-1. Usujirene and palythene were quantified using the calibration curve of aplysiapalythine A. Data visualization of the absolute quantification results was performed in RStudio, utilizing the packages readxl for data import, tidyverse for data manipulation, ggpubr and FSA for statistical plotting, patchwork for image generation, and system fonts as well as extra font to import additional fonts.
Untargeted metabolomics analysis by UHPLC-ESI-HRMS/MS
Analyses of fish organ extracts, blanks (ultra-pure water), and standard mixtures (calibrators and a sample containing mycosporine-methylamine-threonine from Orfanoudaki et al., 2020) were performed according to the method described by Zwerger et al. (2023), with minor modifications. The experiments were conducted on a Vanquish system (Thermo Scientific, Waltham, USA) consisting of a binary pump (VC-P10-A), an autosampler (VC-A12-A), a column oven (VC-C10-A), and a variable wavelength detector (VC-D40-A) connected to a Thermo Scientific Exploris 120 Orbitrap HRMS unit. Separation was carried out on a Luna Omega C18 100 Å column (100 mm × 2.1 mm; particle size 1.6 µm; Phenomenex, Torrance, USA) protected by a Security Guard ULTRA guard C18 pre-column. The mobile phase comprised water with 0.25% formic acid and 20 mM ammonium formate (A) and acetonitrile (B). The applied gradient was as follows: 0 min, 0% B; 10 min, 5% B; 11 min, 90% B; 13 min, 90% B. Finally, the column was re-equilibrated with the original solvent composition (i.e., 0% B) for 20 min, which corresponds to a total run time of 33 min. The flow rate, column oven temperature, auto-sampler temperature, and injection volume were adjusted to 0.3 mL min-1, 17 °C, 20 °C, and 1 µL, respectively. The detection wavelengths were set to 330 and 350 nm, the data collection rate to 2.0 Hz, the response time to 2.0 s, and the peak width to 0.2 min.
The system was controlled by Thermo Scientific Xcalibur 4.4 software. Calibration of the mass analyzer was done via the Thermo Scientific proprietary calibration mix and the respective automatic calibration function. The mass spectrometric parameters were as follows: heated-ESI ionization source, static spray voltage (positive: 3500 V), sheath gas (N2): 30 arbitrary units, auxiliary gas (N2): 17 arbitrary units, sweep gas (N2): 0 arbitrary units. Temperature of the ion transfer tube and vaporizer was adjusted to 370 and 420 °C, respectively. MS data (range 70–1000 m/z) were recorded from 0 to 20 min with a resolution of 60,000 FWHM for MS1. The RF lens parameter was set to 70%. Data-dependent experiments were conducted with stepped collision energy mode and normalized collision energy type using HCD collision energies of 15, 30, and 45% at a resolution of 15,000 FWHM. The number of dependent scans was set to 3. The following selection of filters was employed: intensity threshold filter (1.0E5), dynamic exclusion (auto), isotope exclusion (assigned), charge state (perform dependent scans on singly charged precursors only), and apex filter (desired apex window: 75%). In addition, a specific exclusion list was created for the measurement using HPLC-grade water as a background extract with an IODA Mass Spec notebook (Zuo et al., 2021).
Raw data were converted using MSConvert (Chambers et al., 2012) and subsequently processed in mzmine 4.5.37 (mzio GmbH, Bremen, Germany) (Schmid et al., 2023) before submission to the feature-based molecular networking (FBMN) workflow on GNPS (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp) for molecular network creation (Nothias et al., 2020). The molecular network was visualized using Cytoscape software (Shannon et al., 2003). Metabolite annotation was performed with SIRIUS 6.1.1 (Dührkop et al., 2015, 2019, 2021). Detailed parameters for data processing and annotation are provided in the Supplementary Material.
Calculation of sun protection factors in fish eyes
Extracts of fish eyes, differentiated in right and left eye, were prepared as described above and diluted at a 1:10 ratio using distilled water prior to analysis. The Sun Protection Factor (SPF) was determined following the method of Fonseca and Rafaela (2013). Sample absorbance was measured across the UV-B wavelength range (290–320 nm) at 5 nm intervals on a nanodrop ONEc (Thermo Scientific, Waltham, USA), with each measurement repeated three times. The SPF was then calculated using the Mansur equation, as follows:
CF = correction factor (10),
EE = erythemogenic effect of radiation with wavelength,
I = solar intensity spectrum,
Abs (λ) = spectrophotometric absorbance values at wavelength.
The values of EE (λ) x I(λ) are constant according to Sayre et al. (1979). To assess the relationship between the SPF factor and the total concentration of MAAs in the eye extracts, Pearson’s correlation coefficients were calculated using Microsoft Excel.
Results
A comprehensive approach was chosen to investigate the MAA distribution across various flatfish organs, combining both targeted and untargeted metabolomics analyses (Figure 1). From each of ten individuals per species, up to eight different organs were dissected, freeze-dried, and extracted with methanol. The resulting extracts were analyzed using HPLC-UV with a validated method originally developed for the phytochemical profiling of MAAs in algae (Orfanoudaki et al., 2020). Holistic compound annotation was subsequently performed using untargeted metabolomics. UHPLC-HRMS/MS experiments in combination with dedicated data processing and visualization via feature-based molecular networking (Nothias et al., 2020), enabled a qualitative exploration of the metabolite profiles. The identities of known MAAs could be confirmed with high confidence, while structural suggestions were proposed for unknown metabolites.
Figure 1

Study workflow: Fish were dissected and methanolic extracts prepared from eight different fish organs (i.e., left and right eyes, gills, heart, intestine, kidneys, liver, skin, and stomach). MAA concentrations were quantified based on HPLC-UV analyses. Additionally, untargeted metabolomics using UHPLC-HRMS/MS followed by feature-based molecular networking enabled qualitative analysis of the samples.
For the preparation of extracts, the protocol by Bonin et al. (2024) was slightly modified. Preliminary tests to determine the optimal method for tissue homogenization and MAA extraction from fish organs were carried out using herring eyes (Clupea harengus), which were available from a previous study (Bonin et al., 2024). A fine, homogeneous powder could be obtained using a micro-dismembrator, in which freeze-dried sample material was placed in a Teflon capsule filled with agate beads and shaken at high frequency for several minutes. However, this method proved unsuitable for organs such as liver, heart, and kidney, as it resulted in a sticky mass rather than a free-flowing powder. Such matrices were therefore manually minced into the smallest possible pieces using a spatula and tweezers. Pulverizing the dried samples under additional cooling with liquid nitrogen did not bring any advantages either, but rather led to an accumulation of humidity, resulting in a moist mass instead of a powder.
Various methanol-water mixtures (i.e., 0%, 25%, 50%, 75%, and 100% methanol) were tested for extraction. Although the 75:25 (v/v) water-methanol mixture showed the highest extraction efficiency in a single step, removing the solvent would have required freeze-drying, a rather time-consuming step given the large number of samples to be analyzed. In contrast, pure methanol allowed for easier and faster solvent evaporation under an air stream, while still providing nearly exhaustive extraction efficiency. Therefore, 100% methanol was chosen for practical reasons.
To determine the optimal number of extraction cycles, one sample (herring eyes as mentioned above) was extracted five times, and each extract was analyzed individually by HPLC. Already after three extraction steps, 97-98% of the total MAA content were recovered, making additional extractions unnecessary. Thus, triple extraction was considered both efficient and time-saving for processing large sample numbers.
In the analyzed organ samples, the MAAs palythine, asterina-330, porphyra-334, usujirene, and palythene (list in order of increasing retention time) were present most of the time at concentrations sufficient for reliable detection or quantification using the available HPLC-UV method. Additionally, in a few specimens, shinorine (e.g., stomach of flounder #7) and mycosporine-methylamine-threonine (e.g., stomach of flounder #3, #4, #5, and #8) were also detected.
The average total MAA concentration in the eyes of flounder (n = 10 per organ) was approximately 0.15 mg g-1 dry weight (DW), around 0.05 mg g-1 DW in plaice, and 0.10 mg g-1 DW in turbot (Figure 2). In samples of flounder, palythine was the most abundant MAA across all organs. Concentrations in the eyes ranged from 0.04 to 0.25 mg g-1 DW lyophilized tissue and were significantly higher than in any other organ (Figure 3, Supplementary Figure S2). Asterina-330 followed a similar trend but at lower levels, ranging from 0.003 to 0.04 mg g-1 DW. Notably, one flounder (#1) exhibited unusually high MAA levels across all organs, including for instance 0.07 mg g-1 DW palythine in the kidneys (Figure 3, Supplementary Figure S2).
Figure 2
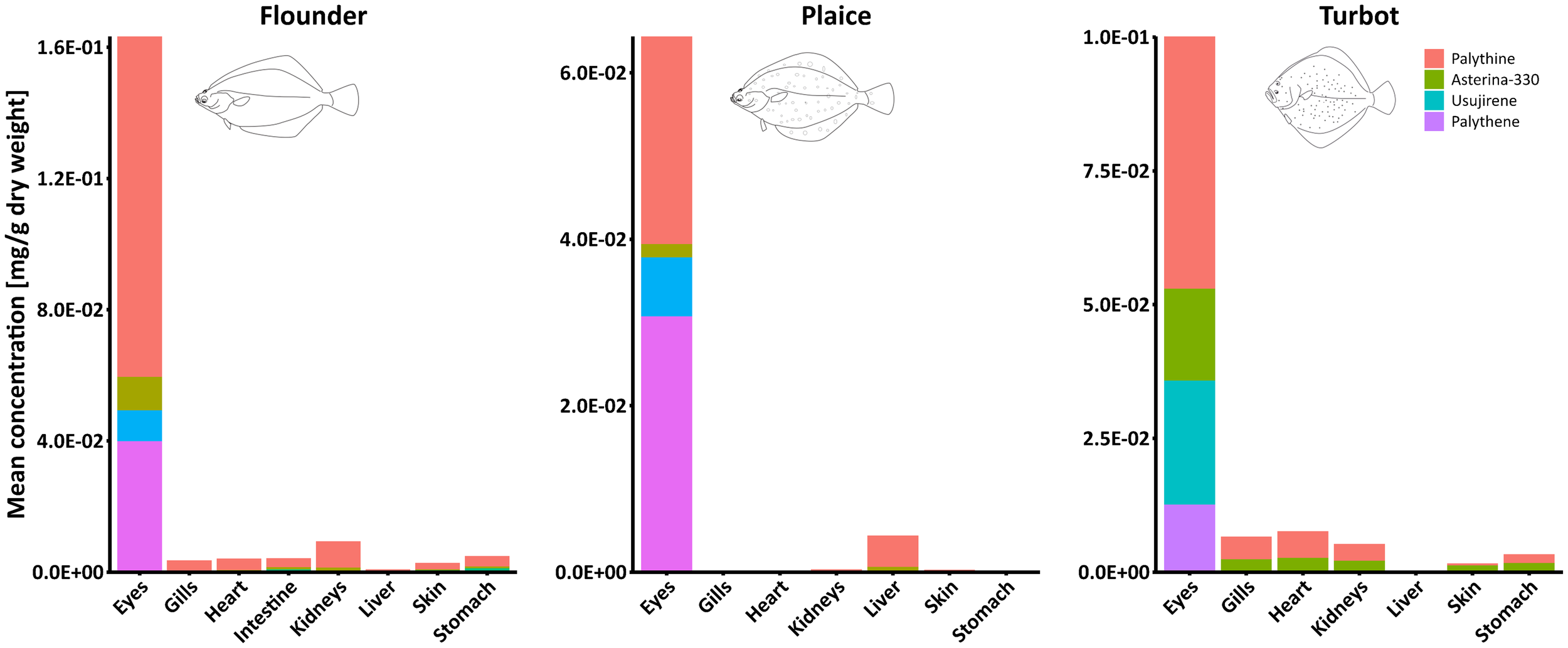
Stacked bar plots showing the mean concentrations of five MAAs in various organs of the flounder, plaice, and turbot (left to right). Each bar represents the average concentration of MAAs per organ, with segments corresponding to individual compounds. Note different y axis scales.
Figure 3
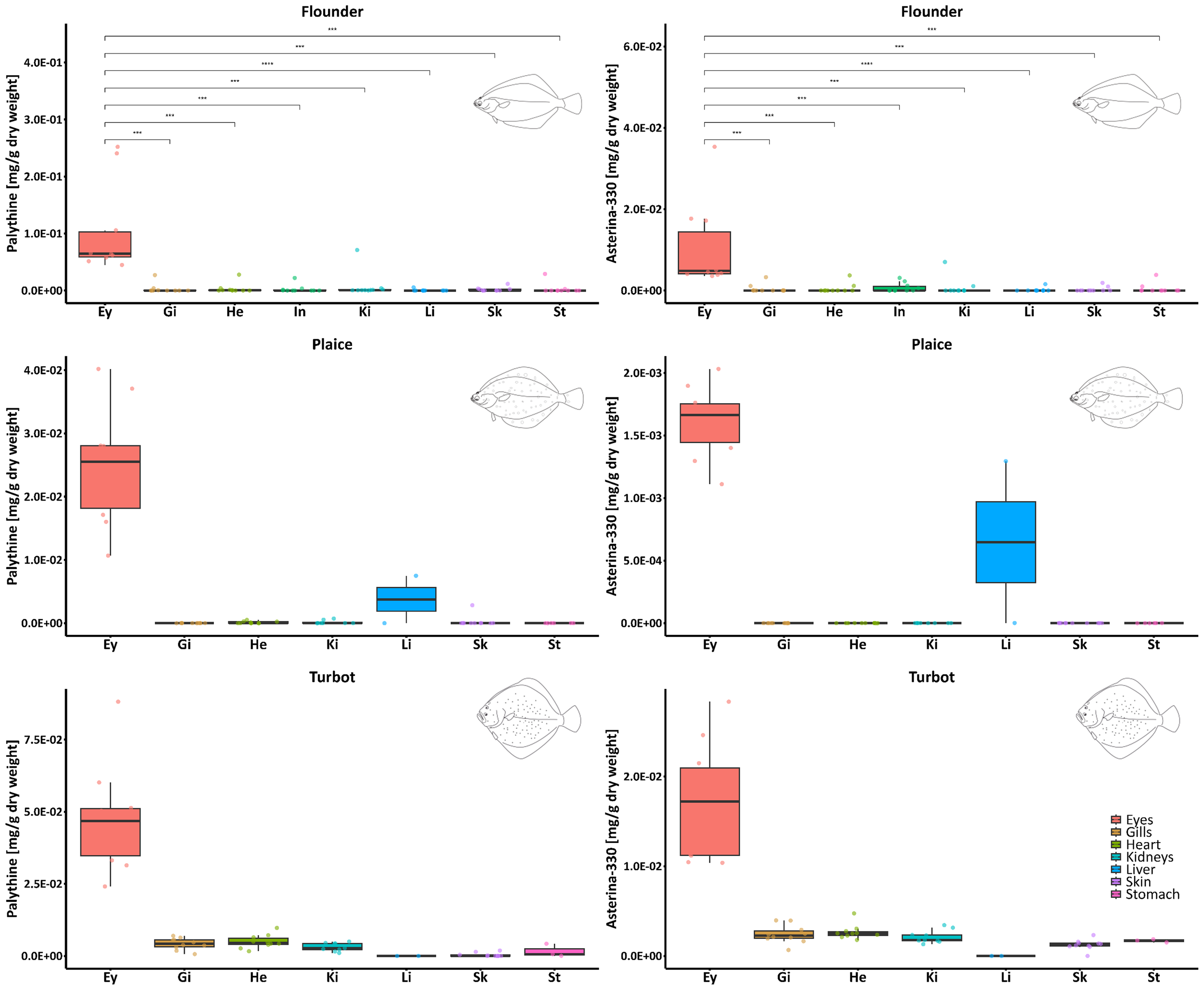
Boxplots showing the distribution of palythine (left side) and asterina-330 (right side) concentrations (mg g-1 DW) across different fish organs and species (top to bottom). Each box represents the interquartile range (IQR), with the median marked as a horizontal line. Individual data points are overlaid as jittered dots. Statistical comparisons between eyes and all other organs of the flounder were performed using the Wilcoxon rank-sum test. Significant differences are indicated by asterisks (***p ≤ 0.001, ****p ≤ 0.0001). Non-significant comparisons (p > 0.05) are not shown. Since complete organ data were available only for the flounder set, statistical comparisons could be performed exclusively for this species. Concentrations reported for the eyes represent the average of values obtained from the left and right eye. Note different y axis scales.
A similar trend was observed in the samples of plaice (Supplementary Figure S3), where higher concentrations of both palythine and asterina-330 were found in the eyes compared to other organs. Interestingly, in plaice #4, slightly higher amounts of palythine (0.007 mg g-1 DW) and asterina-330 (0.001 mg g-1 DW) were detected in the liver. In turbot, these two MAAs were again most abundant in the eyes, while concentrations were comparatively uniform and low in all other organs analyzed (Supplementary Figure S4). Apart from the accumulation in fish eyes, no specific distribution patterns of MAAs in the inspected organs of the three fish species were observed.
Since shinorine, aplysiapalythine A, mycosporine-methylamine-threonine, as well as usujirene and palythene, were found at relatively low concentrations in some samples, their presence was further confirmed using UHPLC-VWD-HRMS/MS analysis (Figure 4). For this purpose, an MAA standard mix from previous investigations was employed to compare both chromatographic retention times and mass spectrometric data. Extracted ion chromatograms (XICs) demonstrated that in total eight MAAs were detectable within a ±5 ppm window around the exact masses of quantified (or annotated) MAAs in the organ extracts of all three flatfish species, even in samples with low concentrations (Figure 4, Supplementary Figure S1).
Figure 4
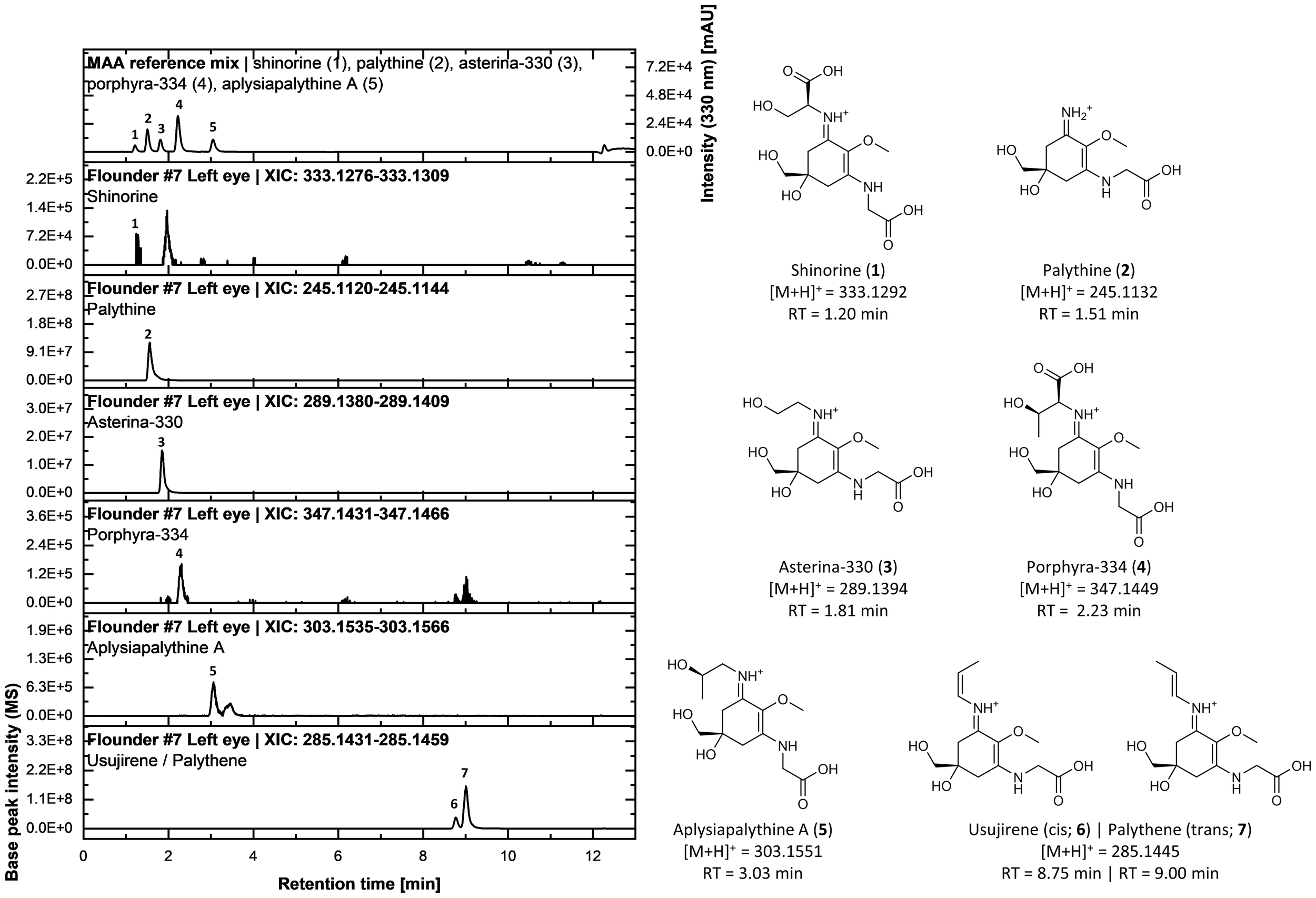
UHPLC analysis of a standard mixture of five mycosporine-like amino acids (MAAs) [i.e., shinorine (1), palythine (2), asterina-330 (3), porphyra-334 (4), and aplysiapalythine A (5)] alongside extracted ion chromatograms (XICs) from the methanolic extract of the left eye of flounder specimen #7. The upper chromatogram displays the UV absorbance trace at 330 nm. Below, XICs were generated with a mass tolerance of 5 ppm based on the calculated exact masses of the protonated molecular species ([M+H]+) for each MAA. The right panel shows the chemical structures of the MAAs, including their calculated exact masses and chromatographic retention times (RT). The identity of usujirene (6) and palythene (7) in the sample was confirmed via evaluation of fragmentation spectra and in-house data.
To further explore the chemical space of fish organ extracts, feature-based molecular networking (FBMN) (Nothias et al., 2020) was employed. Using the bioinformatic tool SIRIUS (Dührkop et al., 2019), all features were annotated with putative compound classes. Applying the NPClassifier chemotaxonomy (class level) (Kim et al., 2021), the most frequent annotations were amino acids (21.1%), followed by dipeptides (18.9%), and tripeptides (11.9%). Among these, 13 features (0.2%) were annotated as mycosporines or mycosporine-like amino acids. Spectral matching via the GNPS community library revealed putative hits for palythine, palythinol, porphyra-334, and usujirene (or palythene, because both are structural isomers). This high number of putative MAA annotations can be partly attributed to previously submitted MS² spectra from our lab that are now publicly available (Zwerger et al., 2023).
Figure 5 shows the complete FBMN consisting of 7,610 nodes and 8,942 edges, organized into many clusters. Large MAA clusters were not formed with many MAA-related features remaining singletons due to cosine scores below 0.6. These include shinorine, poprhyra-334, and mycosporine-methylamine-threonine. Palythine was represented as a small cluster containing its protonated species [M+H]+, a demethylated derivative [M-CH3+H]+, and a sodium adduct [M+Na]+. Asterina-330 and aplysiapalythine A, differing only by a methyl group, formed a small cluster (Figure 5A).
Figure 5
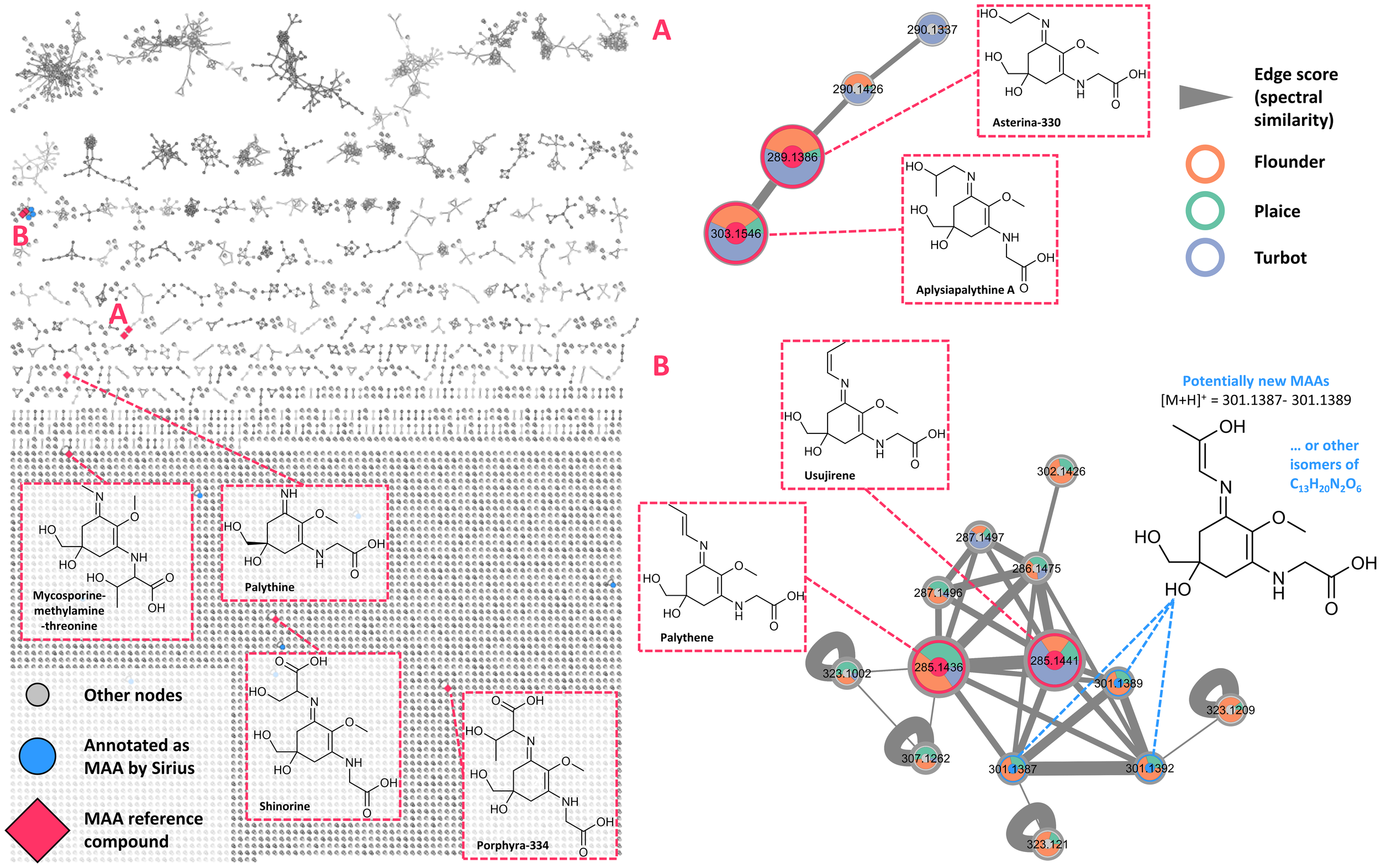
Feature-based molecular network generated from all organ extracts analyzed in this study. Each node represents a feature, with unannotated nodes shown in grey. Nodes annotated as potential mycosporine-like amino acids (MAAs) via SIRIUS are highlighted in blue. Nodes corresponding to confirmed MAA reference compounds (i.e., shinorine, palythine, asterina-330, porphyra-334, aplysiapalythine A, mycosporine-methylamine-threonine, usujirene, and palythene) are marked with red diamonds. (A) Cluster comprising asterina-330 and aplysispalythine A, as well as several nodes (m/z 301.1387–301.1389) potentially representing novel MAAs or isomeric variants of the formula C13H20N2O6. (B) Cluster containing nodes annotated as the cis/trans pair usujirene and palythene. Species-specific occurrence of each node is indicated by colored rings: orange … flounder, light green … plaice, blue … turbot.
Usujirene and palythene were found in a moderately sized cluster (Figure 5B), largely composed of various adducts ([M+Na]+: 307.1262, [M+K]+: 323.1002) and unannotated nodes. For three of these nodes (m/z 301.1387, 301.1389, 301.1392), SIRIUS suggested a potential classification as MAAs. These ions were observed in multiple samples, most prominently in the left eye of flounder #4 (Figure 6A). The molecular formula predicted for these unknown chemical structures (C13H20N2O6) does not correspond to any reported MAAs. Upon closer inspection of the eye extracts from flounder #4 and #7 as well as plaice #5 and #6, poorly resolved yet UV-active peaks at 330 nm were observed around a retention time of 6 min during UHPLC analysis. Extracted ion chromatograms (XICs) within a ±5 ppm mass window around the theoretical exact mass corresponding to the molecular formula C13H21N2O6 (quasimolecular ion, [M+H]+) showed the same retention time, suggesting that the signals represent the same compounds. Cosine scores between these features and known usujirene/palythene nodes ranged from 0.63 to 0.80, indicating strong structural similarity. One possibility is a hydroxylated derivative of usujirene or palythene, as shown in Figure 5B. This hypothetical MAA structure (Figure 6B) is also consistent with typical MAA fragmentation behavior, as characteristic losses, such as the methyl group, water, or CO2, can be observed in the MS² spectrum of the corresponding precursor ion.
Figure 6
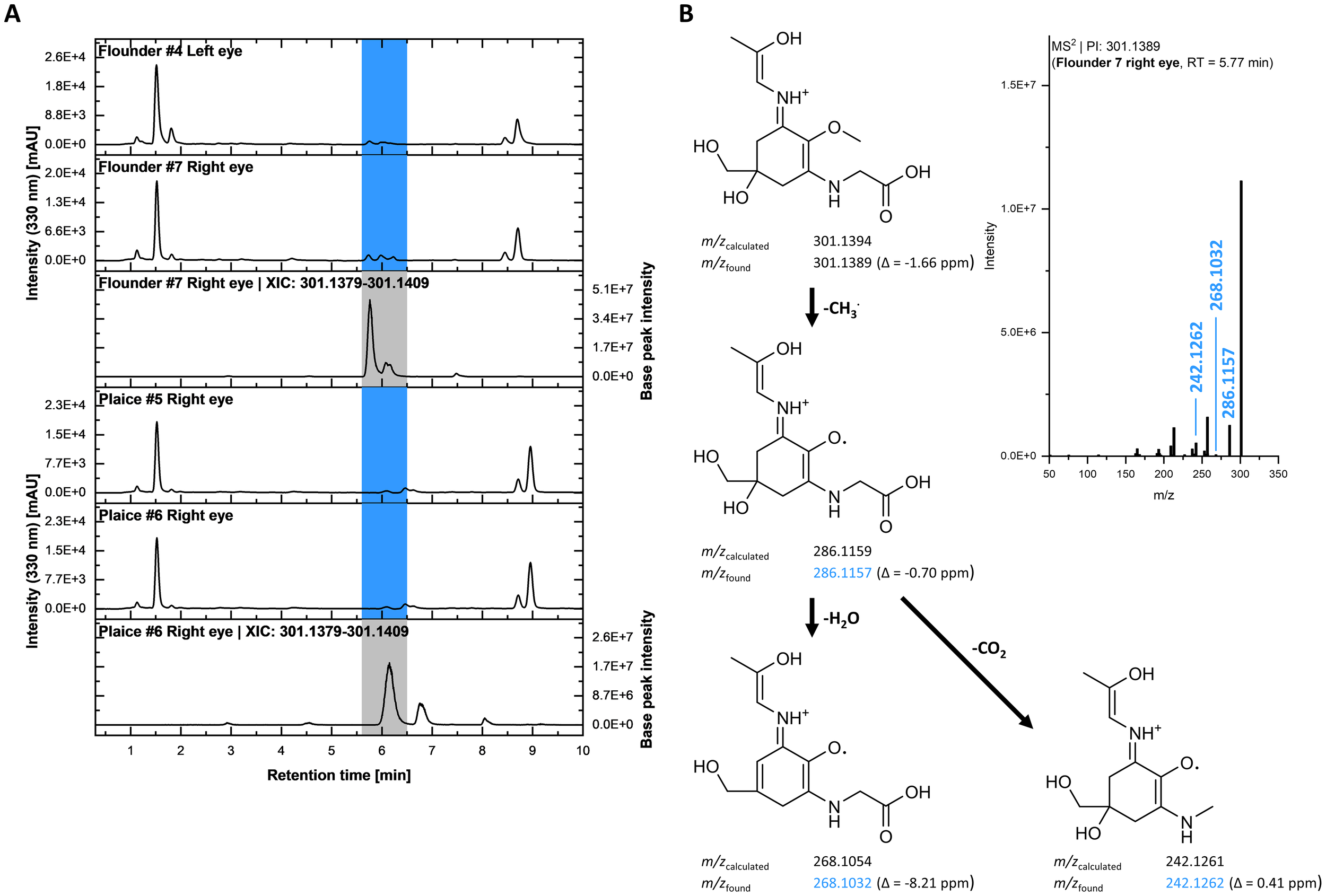
(A) UHPLC chromatograms of eye extracts from flounder (#4 left eye, #7 right eye) and plaice specimens (#5 right eye, #6 right eye). Peaks highlighted in blue indicate UV-absorbing compounds (detection wavelength = 330 nm), potentially corresponding to yet unidentified MAAs. The respective bottom traces show extracted ion chromatograms (XICs) using a 5 ppm mass window around the theoretical m/z of a compound with the molecular formula C13H21N2O6 (protonated species). (B) Top right: MS² spectrum of the ion at m/z 301.1389, eluting at 5.77 min in the extract of the right eye of flounder #7. Left: Fragmentation tree of a putative MAA candidate (shown as the cis isomer, one of two possible isomeric forms) matching the UV signal and MS² spectrum, with characteristic MAA fragmentation reactions. Experimentally observed fragment m/z values are shown along with their corresponding mass errors.
Lastly, the Sun Protection Factor (SPF) values were determined in the fish eye extracts of flounder, plaice, and turbot and compared with the total amount of MAAs in mg g-1 DW (Figure 7). Spectrophotometric analysis was performed across the UV range of 290–320 nm, and SPF values were calculated using the Mansur equation. Overall, a general trend was observed in which higher MAA concentrations were associated with increased UV-protective capacity as measured by SPF, with the fish species exhibiting the highest MAA levels also showing the highest SPF values (e.g., flounder). Eye extracts of flounder had significantly higher SPF values (range: 3.3–8.6; average: 5.7 ± 1.7) than extracts of plaice (range: 1.2–6.6; average: 3.2 ± 1.1) or turbot (range: 1.6–4.4; average: 3.2 ± 0.9). Combining data across all species revealed a modest but statistically significant positive correlation (r = 0.28, p = 0.03) between MAA content and SPF values. However, correlations within individual species were weaker, with only turbot showing a positive, though not statistically significant, relationship (r = 0.39, p = 0.09). Notably, differences in MAA content between the left and right eyes of individual fish were also reflected in corresponding SPF variations, further supporting a relationship between MAA concentration and UV-protective capacity. While some individuals (e.g., flounder #1) exhibited conspicuously different total MAA values of 0.17 and 0.33 mg g-1 DW in the right and left eye, respectively, others (e.g., flounder #4) showed almost identical contents (Figure 7). Nevertheless, while in flounder 6 out of 10 right eyes contained higher total MAA levels compared to the corresponding left eyes, in plaice and turbot 8 out of 10 left eyes exhibited a higher total content (Figure 7). Hence, at least in plaice and turbot a trend for preferential MAA enrichment in the left eyes was visible.
Figure 7
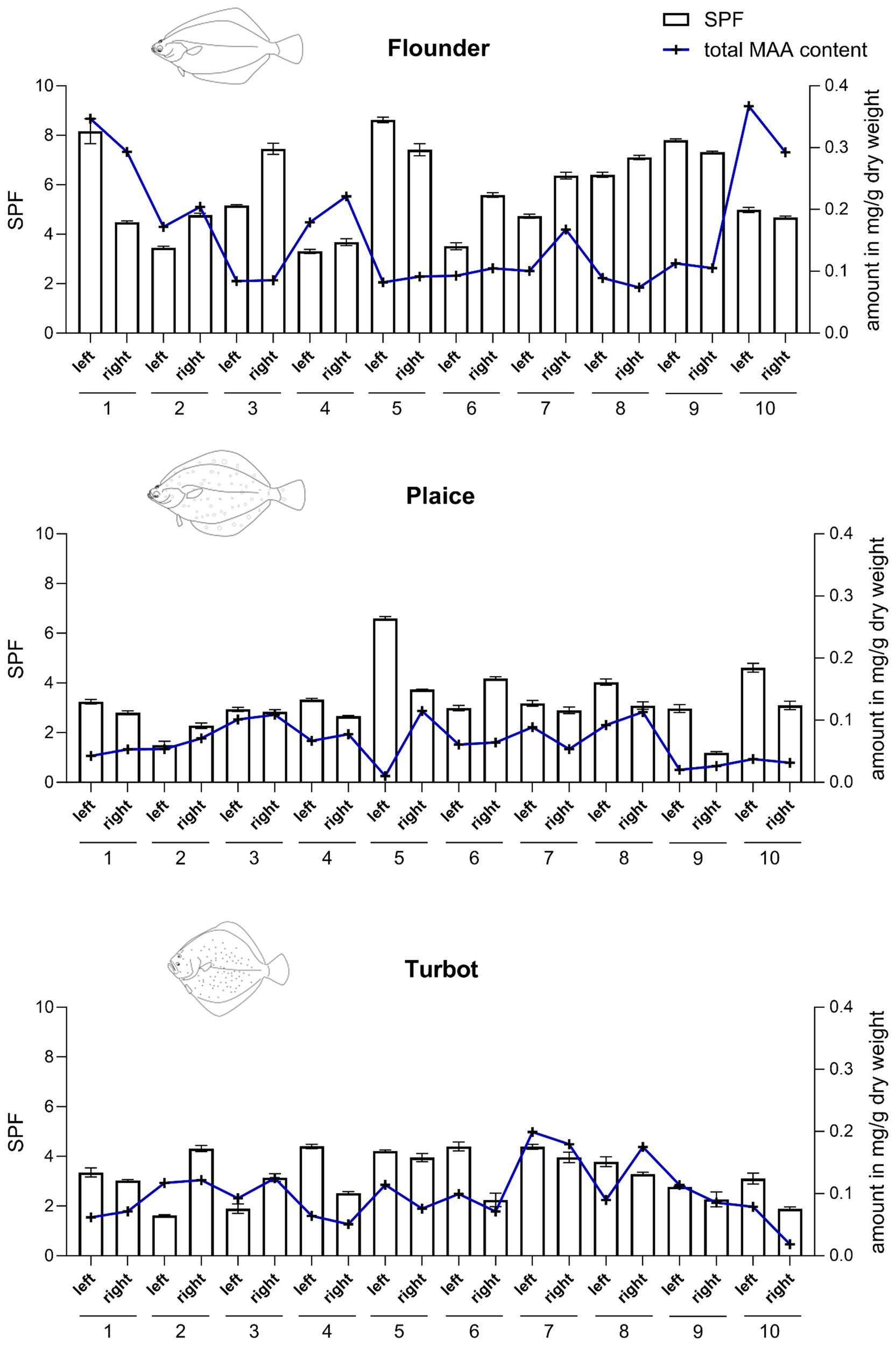
Sun Protection Factor (SPF) values and total mycosporine-like amino acid (MAA) content in eye extracts of European flounder, European plaice, and turbot (top to bottom). SPF determination followed the method described by Fonseca and Rafaela (2013). Spectrophotometric measurements were conducted from 290 to 320 nm at 5 nm intervals, with all readings taken in triplicate. SPF values were calculated using the Mansur equation. Results displayed as bar chart represent mean ± SD of three measurements.
Discussion
Results of the HPLC-UV analysis indicated the presence of eight MAAs, including shinorine, palythine, asterina-330, porphyra-334, and usujirene/palythene. These compounds are well-documented in algae, supporting a dietary origin (Bonin et al., 2024, and references therein). Across all three flatfish species, palythine was the dominant MAA in the eyes, followed by palythene, usujirene, and asterina-330. Results from turbot were slightly different, with higher levels of usujirene relative to palythene. Concentrations in the eyes were significantly higher than in all other organs inspected, particularly in flounder, where palythine reached up to 0.25 mg g-1 DW. European flounder often use very shallow waters (<1m to only a few meters water depth) outside the spawning season (e.g., Dando, 2011, U. Krumme, unpublished data), which is linked to exposure to elevated UVR.
With average total MAA concentrations in eye extracts ranging from 0.05 to 0.15 mg g-1 DW across the three investigated species, our results are consistent with previously reported values in marine fish. Comparable levels have been documented in the eyes of species such as Blicca bjoerkna, Coryphaenoides rupestris, Gadus morhua, and Scomber scombrus. In our earlier study, however, flounder eyes exhibited even higher concentrations exceeding 0.5 mg g-1 DW, suggesting that factors like season, diet or inter-individual differences also influence MAA accumulation (Bonin et al., 2024). An additional interesting observation is that left and right eyes of the same individual fish can have up to 2-fold different total MAA concentrations, which is true for all three flatfish species investigated. Since similar data are completely missing in the literature, we can only speculate on the underlying mechanisms. In plaice and turbot, a trend for preferential enrichment of total MAA concentration in the left eyes was visible. Flatfish undergo a metamorphosis where one eye migrates to the other side of their head during larval development (Bao, 2022). In plaice both eyes are located on the right side of the body, i.e., the left eye migrated to the right side. In turbot it is exactly the opposite, i.e., both eyes are located on the left side of the body with the right eye migrating to the left side (Bao, 2022). This unique adaptation allows flatfish to lie on the seafloor with both eyes facing upwards, camouflaged against the bottom. The migration of the eyes involves an array of complex processes including changes in bone structure (e.g., bone resorption), tissue growth and gene expression, but enables flatfish to adopt a bottom-dwelling lifestyle (Bao, 2022). Our sample size was low and hence the effect could be completely random, but an uneven distribution pattern between the left and right eye of individual flatfish might also be related to differences in the acquisition of MAAs from the diet during the eye migration process or differences in the supply of vasculature of the eyes or the distribution of receptors in the eyes.
Previous studies mainly investigated fish eyes (Bonin et al., 2024 and references therein), mucus (Zamzow, 2004), and roes (Chioccara et al., 1980). In contrast, we report here for the first time the presence of MAAs in the skin and, notably, also in several internal organs of three fish species from the Baltic Sea, although at lower and more variable levels. Palythine and asterina-330 were occasionally detected, but often near or below quantification limits. No consistent organ-specific pattern was observed. The relatively high inter-individual variability, as for example observed in flounder, likely reflects seasonal or dietary influences. Our data are in line with earlier studies, which confirm the preferential occurrence of MAAs in the eyes of numerous fish species from different biogeographic regions (Bonin et al., 2024; Mason et al., 1998; Siebeck and Marshall, 2001). In addition, it was previously reported that tropical marine fish also possess UV-absorbing compounds in their mucus (Zamzow, 2004), and our own preliminary HPLC-based data confirm the presence of palythine in the mucus of Western Baltic spring spawning herring (Clupea harengus) (Karsten, unpublished results). Zamzow (2004) exposed the tropical wrasse Thalassoma duperrey to controlled UVR in combination with sunscreen-enriched and -poor diet, and demonstrated that fish treated with UVR raise the UV absorbance of their mucus only if UV-sunscreens are provided in their diet. Consequently, the quality of the diet is essential for fish to enrich MAAs in the eyes or the epidermal mucus, as both structures are predominantly exposed to UVR if present. In addition, flatfish species avoid too much sun by regular burrowing into the sediment.
However, in the present study we did not investigate any reproductive tissue such as gonads. Chioccara et al. (1980) reported MAAs in ripe eggs of various fish species, and also in many marine invertebrates reproductive tissues are known to preferentially accumulate these UV-sunscreen compounds (for review see: Shick and Dunlap, 2002). The reproductive tissue in many fish species undergoes strong changes in biomass during development and reproduction. Non-mature fish typically exhibit small gonads, and ripe fish exactly the opposite. Our sampling was undertaken in October 2023, and hence outside the spawning season. Therefore, we did not expect to find MAAs in non-mature gonads. In future studies it would be important to address the seasonal dynamics of MAA patterns in gonadal tissue.
FBMN provided deeper insights into MAA-related chemical space. Several known MAAs formed small clusters with adducts or derivatives. Notably, three unknown features (m/z 301.1387–301.1392) clustered with usujirene/palythene and revealed characteristic MAA fragmentation, suggesting structural similarity. Their identity remains unknown, but they may represent hydroxylated derivatives or other biosynthetically/structurally related MAAs. The presence of these unknown structures, predominantly in eye extracts, points to eyes as potential hotspots for MAA diversity. Given that the eyes of fish are usually not used for human consumption and are rather considered as a waste product, cooperation with fishers, anglers or fisheries research institutes can provide a suitable source of novel MAAs for future functional or photoprotective applications. In addition, further studies involving compound isolation and structural elucidation (e.g., via NMR) are required.
For decades, MAAs have been studied natural UV protective compounds in marine ecosystems (Cockell and Knowland, 1999; Robson et al., 2019; Peng et al., 2023). Their main presence is among different marine micro- and macroalgal groups and marine invertebrates (Karsten, 2008; Dunlap and Shick, 1998). While primary producers are considered as the key organisms synthesizing MAAs, the consumers typically acquire these UV-sunscreens from their diet (Karsten, 2008). The first reports on MAAs in higher trophic levels in the marine realm date back to Grant et al. (1980) and Planck et al. (1981), who reported the occurrence of palythine and palythene in the ovaries of Atlantic cod (Gadus morhua). Bonin et al. (2024), and references therein, systematically screened the MAA distribution patterns in the eyes of several fish species from various biogeographic regions, and almost all samples contained these UV-sunscreens in different composition and quantity. Our data indicate that based on strongly improved analytical techniques, the MAA composition in fish eyes can be more complex than reported previously (Bonin et al., 2024, and references therein).
Previous studies support the obvious trophic transfer of MAAs in the food web from the primary producers via the primary consumers (e.g., zooplankton) to higher trophic levels such as fish (Bonin et al., 2024, and references therein). In addition, our data indicate that the assumed dietary MAA transfer might also happen from fish to fish, as turbot is a piscivorous species with a relatively narrow prey spectrum. It primarily feeds on other bottom-dwelling fish, such as sand-eels and gobies, but also consumes crustaceans and bivalves (Van der Hammen et al., 2013). However, the underlying mechanisms for the internal MAA transfer from the fish diet to the fish eye are still unknown. Our results on the qualitative and quantitative MAA patterns in different fish organs clearly indicate that the UV-sunscreens can be taken up from the diet into the fish without being degraded during digestion or by digestive fluids. This supports the assumption that distinct carriers in the digestive tract are involved which enable the selective transfer of MAAs across membranes into the blood system followed by directed transport through different organs to the eyes. Alternatively, the binding of MAAs from the blood at certain receptors could be especially strong in organs like the eyes, where the presence of such receptors and the retention of MAAs would obviously be most beneficial. However, the underlying biochemical and molecular mechanisms still remain to be elucidated.
The eyes of all vertebrates are functionally and structurally quite similar, and can be considered as highly vulnerable organs against biologically harmful UVR (Bonin et al., 2024). Excessive exposure to ultraviolet radiation (UVR) can cause cellular damage and inflammation in ocular tissues, potentially leading to permanent impairment or loss of visual function. From a chemical standpoint, MAAs represent ideal UV-sunscreen compounds because of conjugated double bonds which result in extraordinarily high molar extinction coefficients in the UVR-waveband along with a pronounced photo-stability (Whittock et al., 2022). Consequently, the high amounts of MAAs in fish eyes as shown in our study and previous publications (Bonin et al., 2024, and references therein) indicate a photoprotective role, although experimental evidence is still missing. Further, it is not yet clear whether MAAs accumulate in eyes of all vertebrates and whether MAAs in vertebrate eyes accumulate uniformly or are compartmentalized within specific structures such as the cornea, the lens, or retinal layers. However, numerous investigations on marine invertebrates such as corals, sea urchins and on marine primary producers unambiguously proved the UV-protective role of MAAs (Karsten, 2008; Peng et al., 2023, and references therein). The general picture is that UVR stimulates MAA biosynthesis and accumulation in primary producers or the uptake of MAA-rich diet in consumers (Karsten, 2008). The higher the MAA levels in an organism, the better protected are physiological and biochemical processes under UVR exposure.
The quantification of SPF values in eye extracts from flounder, plaice, and turbot revealed a generally positive correlation with total MAA content, i.e., the more MAAs were measured the higher the SPF values. Intra-individual differences between left and right eyes confirmed the positive relationship between SFP values and total MAA content. These findings point towards the functional significance of MAAs as naturally occurring photo-protective compounds in fish eyes. Some deviations from the overall trend, however, suggest that other components or structural factors may also influence the level of photoprotection, necessitating further systematic studies of UV protection mechanisms in higher marine organisms.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because the investigated fish was a by-product of commercial fishery. Thus no ethical approval was required.
Author contributions
FH: Data curation, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. RS: Data curation, Formal Analysis, Methodology, Writing – original draft. CK: Data curation, Formal Analysis, Methodology, Writing – original draft, Writing – review & editing. UwK: Resources, Writing – review & editing. JG: Conceptualization, Funding acquisition, Writing – review & editing. UlK: Conceptualization, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. MG: Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Fish sampling by the Thünen Institute of Baltic Sea Fisheries was co-funded by the European Commission’s Data Collection Framework. This research was funded in part by the FWF-DFG DACH project UVision under grant numbers KA899/45-1 (German Research Foundation, DFG) and Austrian Science Fund (FWF) [Grant-DOI 10.55776/I6122]. For open access purposes, the author has applied a CC BY public copyright license to any author accepted manuscript version arising from this submission.
Acknowledgments
The authors thank Susann Vogler, University of Rostock for dissecting and freeze-drying the fish organs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1688685/full#supplementary-material
References
1
Aarnio K. Bonsdorff E. Rosenback N. (1996). Food and feeding habits of juvenile flounder Platichthys flesus (L.), and turbot Scophthalmus maximus L. in the åland archipelago, northern Baltic Sea. J. Sea Res.36, 311–320. doi: 10.1016/S1385-1101(96)90798-4
2
Arbeloa E. M. Uez M. J. Bertolotti S. G. Churio M. S. (2010). Antioxidant activity of gadusol and occurrence in fish roes from Argentine Sea. Food Chem.119, 586–591. doi: 10.1016/j.foodchem.2009.06.061
3
Bandaranayake W. M. (1998). Mycosporines: are they nature’s sunscreens? Nat. Prod. Rep.15, 159–172. doi: 10.1039/A815159Y
4
Bao B. (2022). Flatfish Metamorphosis (Singapore: Springer Nature Singapore Pte Ltd). doi: 10.1007/978-981-19-7859-3
5
Bonin J. Hammerle F. J. Ganzera M. Krumme U. Karsten U. (2024). UV-absorbing mycosporine-like amino acids in the eyes of temperate marine and freshwater fish species. Front. Mar. Sci.11. doi: 10.3389/fmars.2024.1426861
6
Chambers M. C. Maclean B. Burke R. Amodei D. Rudermann D. L. Neumann S. et al . (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol.30, 918–920. doi: 10.1038/nbt.2377
7
Chioccara F. Delia Gala A. De Rosa M. (1980). Mycosporine aminoacids and related compounds from the eggs of fishes. Bull. Soc Chim. Belg.89, 1101–1106. doi: 10.1002/bscb.19800891212
8
Cockell C. S. Knowland J. (1999). Ultraviolet radiation screening compounds. Biol. Rev. Cambridge Philos. Soc74, 311–345. doi: 10.1017/s0006323199005356
9
Dando P. R. (2011). Site fidelity, homing and spawning migrations of flounder Platichthys flesus in the Tamar estuary, South West England. Mar. Ecol. Prog. Ser.430, 183–196. doi: 10.3354/meps09116
10
Dührkop K. Fleischauer M. Ludwig M. Aksenov A. A. Melnik A. V. Meusel M. et al . (2019). SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods16, 299–302. doi: 10.1038/s41592-019-0344-8
11
Dührkop K. Nothias L. F. Fleischauer M. Reher R. Ludwig M. Hoffmann M. A. et al . (2021). Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol.39, 462–471. doi: 10.1038/s41587-020-0740-8
12
Dührkop K. Shen H. Meusel M. Rousu J. Böcker S. (2015). Searching molecular structure databases with tandem mass spectra using CSI: FingerID. Proc. Natl. Acad. Sci. U.S.A.112, 12580–12585. doi: 10.1073/pnas.1509788112
13
Dunlap W. C. Shick J. M. (1998). Ultraviolet radiation-absorbing mycosporine-like amino acids in coral reef organisms: a biochemical and environmental perspective. J. Phycol.34, 418–430. doi: 10.1046/j.1529-8817.1998.340418.x
14
Eckes M. J. Siebeck U. E. Dove S. Grutter A. S. (2008). Ultraviolet sunscreens in reef fish mucus. Mar. Ecol. Prog. Ser.353, 203–211. doi: 10.3354/meps07210
15
Florin A. B. Franzén F. (2010). Spawning site fidelity in Baltic Sea turbot (Psetta maxima). Fish. Res.102, 207–213. doi: 10.1016/j.fishres.2009.12.002
16
Fonseca A. P. Rafaela N. (2013). Determination of sun protection factor by UV-Vis spectrophotometry. Health Care: Curr. Rev.1, 1. doi: 10.4172/hccr.1000108
17
Grant P. T. Plack P. A. Thomson R. H. (1980). Gadusol, a metabolite from fish eggs. Tetrahedron Lett.21, 4043–4044. doi: 10.1016/S0040-4039(00)92866-1
18
Jurasinski G. Janssen M. Voss M. Böttcher M. Brede M. Burchard H. et al . (2018). Understanding the coastal ecocline: Assessing sea–land interactions at non-tidal, low-lying coasts through interdisciplinary research. Front. Mar. Sci.26. doi: 10.3389/fmars.2018.00342
19
Karsten U. (2008). “ Defense strategies of algae and cyanobacteria against solar ultraviolet radiation,” in Algal chemical ecology. Ed. AmslerC. D. ( Springer-Verlag, Berlin), 273–296. doi: 10.1007/978-3-540-74181-7_13
20
Kim H. W. Wang M. Leber C. A. Nothias L. F. Reher R. Kang K. B. et al . (2021). NPClassifier: A deep neural network-based structural classification tool for natural products. J. Nat. Prod.84, 2795–2807. doi: 10.1021/acs.jnatprod.1c00399
21
Mason D. S. Schafer F. Shick J. M. Dunlap W. C. (1998). Ultraviolet radiation-absorbing mycosporine-like amino acids (MAAs) are acquired from their diet by medaka fish (Oryzias latipes) but not by SKH-1 hairless mice. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol.120, 587–598. doi: 10.1016/S1095-6433(98)10069-7
22
Moll D. Asmus H. Blöcker A. Böttcher U. Conradt J. Färber L. et al . (2024). A climate vulnerability assessment of the fish community in the Western Baltic Sea. Sci. Rep.14, 16184. doi: 10.1038/s41598-024-67029-2
23
Mondal P. Sen A. Shaw P. Dey Bhowmik A. Mandal S. Chattopadhyay A. et al . (2025). UV-absorbing compound asterina-330 mitigates the cytotoxic effect of arsenic and fluoride in zebrafish hepatocytes. Nucleus. doi: 10.1007/s13237-025-00573-y
24
Nissling A. Jacobsson M. Hallberg N. (2007). Feeding ecology of juvenile turbot Scophthalmus maximus and flounder Pleuronectes flesus at Gotland, Central Baltic Sea. J. Fish Biol.70, 1877–1897. doi: 10.1111/j.1095-8649.2007.01463.x
25
Nothias L. F. Petras D. Schmid R. Dührkop K. Rainer J. Sarvepalli A. et al . (2020). Feature-based molecular networking in the GNPS analysis environment. Nat. Methods17, 905–908. doi: 10.1038/s41592-020-0933-6
26
Orfanoudaki M. Hartmann A. Kamiya M. West J. A. Ganzera M. (2020). Chemotaxonomic study of bostrychia spp. (Ceramiales, rhodophyta) based on their mycosporine-like amino acid content. Molecules25, 3273. doi: 10.3390/molecules25143273
27
Orfanoudaki M. Hartmann A. Karsten U. Ganzera M. (2019). Chemical profiling of mycosporine-like amino acids in twenty-three red algal species. J. Phycol.55, 393–403. doi: 10.1111/jpy.12827
28
Pavlov A. K. Leu E. Hanelt D. Bartsch I. Karsten U. Hudson S. R. et al . (2019). “ The underwater light climate in kongsfjorden and its ecological implications,” in The Ecosystem of Kongsfjorden, Svalbard. Eds. HopH.WienckeC. ( Springer, Berlin, Germany), 137–170. doi: 10.1007/978-3-319-46425-1_5
29
Peng J. Guo F. Liu S. Fang H. Xu Z. Wang T. (2023). Recent advances and future prospects of mycosporine-like amino acids. Molecules28, 5588. doi: 10.3390/molecules28145588
30
Planck P. A. Fraser N. W. Grant P. T. Middleton C. Mitchell A. I. Thomson R. H. (1981). Gadusol, an enolic derivative of cyclohexane-1,3-dione present in the roes of cod and other marine fish. Biochem. J.199, 741–747. doi: 10.1042/bj1990741
31
Robson T. M. Aphalo P. J. Banaś A. K. Barnes P. W. Brelsford C. C. Jenkins G. I. et al . (2019). A perspective on ecologically relevant plant-UV research and its practical application. Photochem. Photobiol. Sci.18, 970–988. doi: 10.1039/c8pp00526e
32
Sayre R. M. Agin P. P. Levee G. J. Marlowe E. (1979). Comparison of in vivo and in vitro testing of sun screening formulas. Photochem. Photobiol.29, 559–566. doi: 10.1111/j.1751-1097.1979.tb07090.x
33
Schmid R. Heuckeroth S. Korf A. Smirnov A. Myers O. Dyrlund T. S. et al . (2023). Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol.41, 447–449. doi: 10.1038/s41587-023-01690-2
34
Schuch A. P. Moreno N. C. Schuch N. J. Menck C. F. M. Garcia C. C. M. (2017). Sunlight damage to cellular DNA: Focus on oxidatively generated lesions. Free Rad. Biol. Med.107, 110–124. doi: 10.1016/j.freeradbiomed.2017.01.029
35
Shannon P. Markiel A. Ozier O. Baliga N. Wang J. T. Ramage D. et al . (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504. doi: 10.1101/gr.1239303
36
Shick J. M. Dunlap W. C. (2002). Mycosporine-like amino acids and related gadusols: Biosynthesis, accumulation, and UV-protective functions in aquatic organisms. Ann. Rev. Physiol.64, 223–262. doi: 10.1146/annurev.physiol.64.081501.155802
37
Siebeck U. E. Marshall N. J. (2001). Ocular media transmission of coral reef fish - Can coral reef fish see ultraviolet light? Vision Res.41, 133–149. doi: 10.1016/S0042-6989(00)00240-6
38
Strodtmann S. (1906). “ Laichen und Wandern der Ostseefische, 2. Bericht, Helgoland”, Wiss. Meeresunt. N. F. Abt.7, 132–216.
39
Van der Hammen T. Poos J. J. van Overzee H. M. J. Heessen H. J. L. Magnusson A. Rijnsdorp A. D. (2013). Population ecology of turbot and brill: What can we learn from two rare flatfish species? J. Sea Res.84, 96–108. doi: 10.1016/j.seares.2013.07.001
40
Viitasalo M. Bonsdorff E. (2022). Global climate change and the Baltic Sea ecosystem: direct and indirect effects on species, communities and ecosystem functioning. Earth Syst. Dynam.13, 711–747. doi: 10.5194/esd-13-711-2022
41
Vincent W. F. Neale P. J. (2000). “ Mechanisms of UV damage to aquatic organisms,” in The Effects of UV Radiation in the Marine Environment. Eds. de MoraS. J.DemersS.VernetS. M. V. ( Cambridge University Press, Cambridge, UK), 149–176. doi: 10.1017/CBO9780511535444.007
42
Vuori I. Gaiani G. Arsın S. Delbaje E. Järn J. Snårbacka R. et al . (2025). Direct evidence of microbial sunscreen production by scum-forming cyanobacteria in the baltic sea. Environ. Microbiol. Rep.17, e70056. doi: 10.1111/1758-2229.70056
43
Wang K. Qin L. Lin H. Yao M. Cao J. Zhang Q. et al . (2025). Pharmacological effects of antioxidant mycosporine-glycine in alleviating ultraviolet B-induced skin photodamage: insights from metabolomic and transcriptomic analyses. Antioxidants14, 30. doi: 10.3390/antiox14010030
44
Whittock A. L. Abiola T. T. Stavros V. G. (2022). A perspective on femtosecond pump-probe spectroscopy in the development of future sunscreens. J. Phys. Chem. A126, 2299–2308. doi: 10.1021/acs.jpca.2c01000
45
Zamzow J. P. (2004). Effects of diet, ultraviolet exposure, and gender on the ultraviolet absorbance of fish mucus and ocular structures. Mar. Biol.144, 1057–1064. doi: 10.1007/s00227-003-1286-2
46
Zamzow J. P. (2007). Ultraviolet-absorbing compounds in the mucus of shallow-dwelling tropical reef fishes correlate with environmental water clarity. Mar. Ecol. Prog. Ser.343, 263–271. doi: 10.3354/meps06890
47
Zuo Z. Cao L. Nothia L. F. Mohimani H. (2021). MS2Planner: improved fragmentation spectra coverage in untargeted mass spectrometry by iterative optimized data acquisition. Bioinformatics37, i231–i236. doi: 10.1093/bioinformatics/btab279
48
Zwerger M. J. Hammerle F. Siewert B. Ganzera M. (2023). Application of feature-based molecular networking in the field of algal research with special focus on mycosporine-like amino acids. J. Appl. Phycol.35, 1377–1392. doi: 10.1007/s10811-023-02906-3
Summary
Keywords
fish eyes, fish organs, metabolomics, ultraviolet radiation, UV-sunscreens
Citation
Hammerle FJ, Schöpf R, Karg CA, Krumme U, Gostner JM, Karsten U and Ganzera M (2025) Photoprotective mycosporine-like amino acids in different organs of Baltic flatfish species revealed by targeted and untargeted metabolomics analyses. Front. Mar. Sci. 12:1688685. doi: 10.3389/fmars.2025.1688685
Received
19 August 2025
Accepted
10 October 2025
Published
22 October 2025
Volume
12 - 2025
Edited by
Daniela Giordano, National Research Council (CNR), Italy
Reviewed by
Susan Murch, University of British Columbia, Canada; Rocío Isla Naveira, Instituto Nacional de Investigación y Desarrollo Pesquero, Argentina
Updates
Copyright
© 2025 Hammerle, Schöpf, Karg, Krumme, Gostner, Karsten and Ganzera.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Markus Ganzera, Markus.Ganzera@uibk.ac.at
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.