Abstract
Mycolicibacterium mageritense (M. mageritense) is a rare species among rapidly growing mycobacteria, and M. mageritense pleurisy is very rare. Here, we report for the first time, an immunocompetent patient with pleurisy caused by M. mageritense. The patient had no history of immunodeficiency and no recurrence of lung cancer after surgery. However, 8 months after surgery, he developed a new lung shadow and pleurisy. Although whole-genome analysis of the colony cultured from the patient's pleural fluid revealed M. mageritense, we could not identify it in time, resulting in a poor outcome. M. mageritense pleurisy in this case might have occurred via a bulla rupture of the lung lesion because computed tomography of the patient's chest showed pneumothorax and a lung lesion in contact with thoracic cavity. This case emphasized that nontuberculous mycobacterial pleurisy should be considered in the differential diagnoses of pleural effusion even in immunocompetent patients. Advancement of comprehensive and rapid analyses of genomic data from clinical specimens will lead to better treatment strategies.
Introduction
Nontuberculous mycobacteria (NTM) consist of over 200 species and subspecies. They can cause infectious diseases in humans of all ages at both pulmonary and extrapulmonary sites. In Asia, 31% of NTM-associated infectious diseases are caused by rapidly growing mycobacteria (RGM) (1, 2) and usually require long-term treatment with multidrug antibiotic regimens. They are often refractory to treatment and have a high likelihood of relapse (3).
Mycolicibacterium mageritense (M. mageritense) is a rare RGM related to Mycobacterium fortuitum (M. fortuitum), a low-virulence species that is the most common species among RGMs (4). M. mageritense has never been recognized as a highly pathogenic bacterium that causes serious infectious diseases. Several studies have reported cases of skin and soft tissue infections, pneumonia, and health care-associated infections among patients with or without immunodeficiency (5–7).
NTM pleurisy is very rare, and only one case of pleurisy due to M. mageritense has been reported (8). Here, we report for the first time, a case of severe pleurisy due to M. mageritense in an immunocompetent patient.
Case Description
A 77-year-old man was referred to our hospital for right pleural effusion lasting several months with unknown etiology. During follow-up at our hospital, we observed a nodule in the upper lobe of the right lung, showing high 2-deoxy-2-[18F] fluoro-D-glucose (FDG) uptake [Primary tumor standardized uptake value (SUV)max = 12.4] on positron emission tomography-computed tomography (PET-CT) (Figure 1). After excluding malignant pleural effusion via cytological testing, we diagnosed the patient with stage IA3 squamous cell lung carcinoma and performed combined resection of the right upper lobe and part of the middle and lower lobes via video-assisted thoracic surgery. The patient was a heavy smoker (60 packs/year), with a history of radiation-treated laryngeal cancer. He had complications of chronic obstructive pulmonary disease and angina, which were treated with coronary artery stenting.
Figure 1
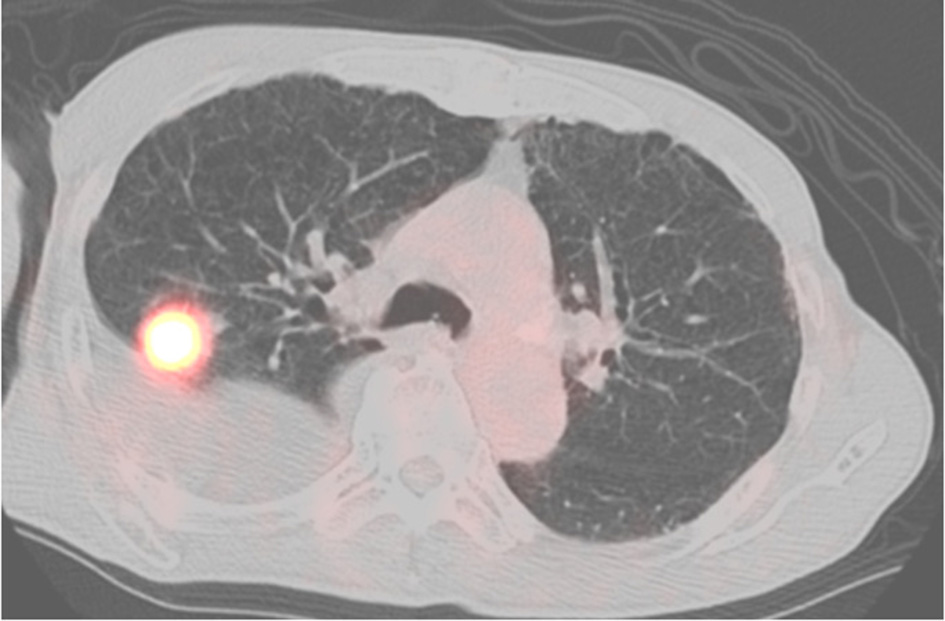
PET-CT scan showing no FDG uptake in the pleura.
Pleural fluid analysis at surgery showed the following: total protein, 4.8 g/dL; glucose, 85.6 mg/dL; carcinoembryonic antigen, 2.6 ng/mL; neutrophils, 2.0%; lymphocytes, 78.0%; monocytes, 12.0%; eosinophils, 0.0%; adenosine deaminase, 21.0 U/L; and lactate dehydrogenase (LDH), 140 U/L, with negative bacterial and acid-fast bacterial cultures.
Eight months postsurgery, he lost his appetite. A CT scan showed increased pleural effusion with pneumothorax and new centrilobular nodular shadows in contact with the pleura (Figure 2). This time, the acid-fast bacterial smear test was positive. We diagnosed him with NTM pleurisy after excluding tuberculosis via PCR. However, we could not correctly identify the species using conventional methods such as AccuProbe (Gen-Probe Inc., San Diego, CA, USA), COBAS AMPLICOR (Roche Diagnostic, Tokyo, Japan), and a DNA-DNA hybridization assay (Kyokuto Pharmaceutical Industrial, Tokyo, Japan), although the growth and appearance of the colonies suggested M. fortuitum, a low-virulence organism. Table 1 shows the susceptibility results for this strain. We tried treating the pleurisy by drainage and single antibiotics, levofloxacin, and imipenem/cilastatin at the time of exacerbation. However, we could not control the effusion, and the patient died of aspiration pneumonia and CO2 narcosis.
Figure 2
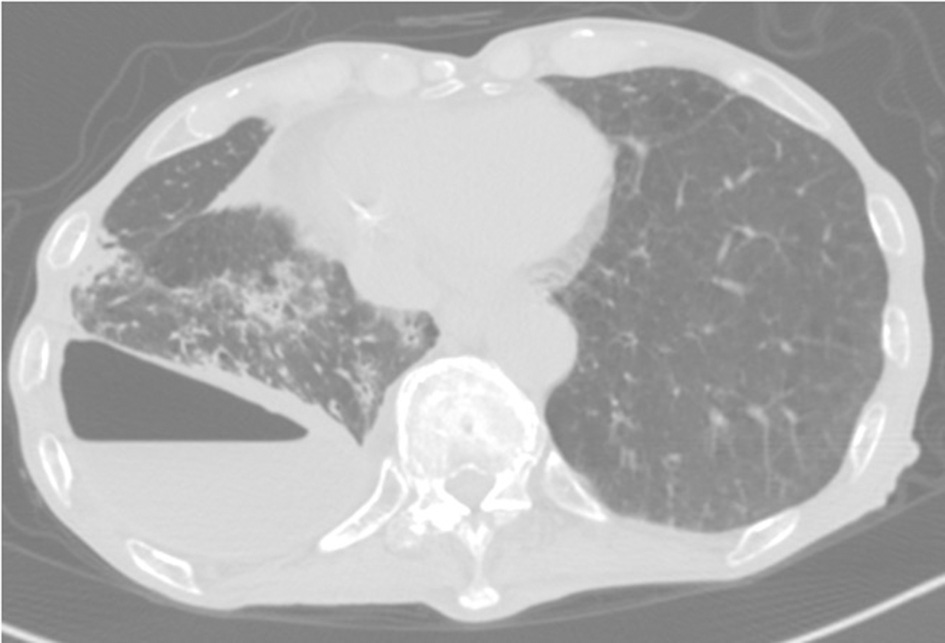
CT scan showing pneumothorax and centrilobular nodular shadows with ipsilateral increasing pleural effusion.
Table 1
| Antibiotics | MIC (μg/mL) | Susceptibility |
|---|---|---|
| Clarithromycin | 16 | R |
| Azithromycin | >32 | R |
| Cefoxitin | 16 | S |
| Imipenem | <0.5 | S |
| Meropenem | 2 | S |
| Faropenem | 4 | |
| Amikacin | 4 | S |
| Tobramycin | >16 | R |
| Minomycin | 2 | I |
| Doxycycline | 4 | I |
| Linezolid | <4 | S |
| Moxifloxacin | <0.25 | S |
| Ciprofloxacin | <0.5 | S |
| Levofloxacin | <0.5 | S |
| Sulfamethoxazole–Trimethoprim | <2 | S |
Susceptibility of the isolate.
MIC, minimal inhibitory concentration; S, susceptible; I, intermediate; R, resistant.
Later, whole-genome sequencing revealed that his pleural effusion culture isolate was M. mageritense. The sequencing was performed following the method described before using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA) (9). We used mlstverse software (9) for accessory-genome multilocus sequence type analysis and found only one profile matching M. mageritense with a score of 0.998 (Table 2).
Table 2
| Species | Strain | Score |
|---|---|---|
| Mycolicibacterium mageritense | CIP 104973 | 0.998 |
MLST score of the isolate.
Discussion
M. mageritense was originally discovered from human sputum in 1997 (10) and has been detected from human samples collected from surgical wounds, blood, sinuses, and joint fluid (11). As with other NTM species, M. mageritense rarely develops into pleurisy, and only one case of pleurisy caused by M. mageritense, which occurred in an immunocompromised host, has been reported (8). Our case is the first to have occurred in an immunocompetent host.
In the NTM pleurisy pathogenesis, two possible mechanisms are considered. One is the direct extension of lung lesions into the pleura. The other is a hematogenous route (12). Because our patient's chest CT showed pneumothorax and centrilobular shadows contacting the thoracic cavity, which were previously unseen, M. mageritense pleurisy in our patient might have occurred via bulla rupture of a lung lesion. We also investigated the possibility that thoracic surgery might have caused the infection. However, his pleural effusion did not increase over several months postsurgery; therefore, we think this possibility was very low. Although we could not rule out disseminated NTM, the patient had no abnormal findings other than lung disease.
In treating rare mycobacterial diseases such as those caused by M. mageritense, we usually determine treatment regimens by referring to previous case reports about the organism or established treatment regimens for related organisms, then modify the regimen individually as per drug-susceptibility tests. Precisely identifying the pathogen is the first step. However, due to the lack of a clinically available identification technique, we could not identify M. mageritense in time in this case. Advancement of comprehensive and rapid analysis of genomic data from clinical specimens will lead to clinical sequencing in NTM and thus will help clinicians evaluate the pathogenicity and choose the proper treatment timing and regimen.
In conclusion, this report describes an immunocompetent patient with both early-stage lung cancer and pleural effusion caused by M. mageritense. In patients with pleural effusion, RGM pleurisy should be considered as a differential diagnosis, even in patients who are not immunocompromised. Accurately identifying rare organisms using genomic data may enable establishing proper treatment strategies.
Funding
This work was supported in part by AMED (grant numbers JP20fk0108129, JP21fk0108129h0702, JP21lm0203007), a GSK Research grant (grant number A-32), JSPS KAKENHI (grant numbers JP21K16118, JP21K08194), the Smoking Research Foundation, Takeda Science Foundation, and the Japan Intractable Diseases (Nanbyo) Research Foundation (grant number 2020B02).
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The datasets supporting the conclusions of this article are included within the article. Whole genome sequence analysis was deposited to BioProject (PRJDB12517), BioSample (SAMD00414014), Nucleotide (BPWM01000001-BPWM01000192).
Author contributions
TNii and TK drafted the manuscript. KF supervised the writing of the manuscript and was responsible for the clinical data. YA and MO contributed to critically reviewing the manuscript. HK organized and contributed to managing the case report. KH, HS, TNit, and AKa performed the mycobacterial culturing and analyzed the culture isolates. YM, DM, and SN performed the whole-genome analysis. All authors contributed to writing the final manuscript.
Acknowledgments
We thank Traci Raley, MS, ELS, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
Shinnick TM Good RC . Mycobacterial taxonomy. Eur J Clin Microbiol Infect Dis. (1994) 13:884–901. 10.1007/BF02111489
2.
Hoefsloot W van Ingen J Andrejak C Angeby K Bauriaud R Bemer P et al . The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J. (2013) 42:1604–13. 10.1183/09031936.00149212
3.
Park IK Olivier KN . Nontuberculous mycobacteria in cystic fibrosis and non–cystic fibrosis bronchiectasis. Semin Respir Crit Care Med. (2015) 36:217–24. 10.1055/s-0035-1546751
4.
Brown-Elliott BA Wallace RJ Jr . Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev. (2002) 15:716–46. 10.1128/CMR.15.4.716-746.2002
5.
Lipika Singhal L Bhagat A Gupta M Gulati N Dalal AK Chander J . Multiple recalcitrant draining sinuses caused by mycobacterium mageritense following laparoscopic cholecystectomy: a case report and brief review of literature. Jpn J Infect Dis. (2020) 73:256–8. 10.7883/yoken.JJID.2019.343
6.
Huth RG Brown-Elliott BA Wallace RJ Jr . Mycobacterium mageritense pulmonary disease in patient with compromised immune system. Emerg Infect Dis. (2011) 17:556–8. 10.3201/eid1703.101279
7.
Ali S Khan FA Fisher F . Catheter-related bloodstream infection caused by Mycobacterium mageritense. J Clin Microbiol. (2007) 45:273. 10.1128/JCM.01224-06
8.
Hirabayashi R Nakagawa A Takegawa H Tomii K . A case of pleural effusion caused by Mycobacterium fortuitum and Mycobacterium mageritense coinfection. BMC Infectious Dis. (2019) 19:720. 10.1186/s12879-019-4366-8
9.
Matsumoto Y Kinjo T Motooka D Nabeya D Jung N Uechi K et al . Comprehensive subspecies identification of 175 nontuberculous mycobacteria species based on 7547 genomic profiles. Emerg Microbes Infect. (2019) 8:1043–53. 10.1080/22221751.2019.1637702
10.
Domenech P Jimenez MS Menendez MC Bull TJ Samper S Manrique A et al . Mycobacterium mageritense sp. Int J Syst Bacteriol. (1997) 47:535–40. 10.1099/00207713-47-2-535
11.
Wallace RJ Jr Brown-Elliott BA Hall L Roberts G Wilson RW et al . Clinical and laboratory features of Mycobacterium mageritense. J Clin Microbiol. (2002) 40:2930–35. 10.1128/JCM.40.8.2930-2935.2002
12.
Park S Jo KW Lee SD Kim WS Shim TS . Clinical characteristics and treatment outcomes of pleural effusions in patients with nontuberculous mycobacterial disease. Respir Med. (2017) 133:36–41. 10.1016/j.rmed.2017.11.005
Summary
Keywords
pleural effusion, rapidly growing mycobacteria, nontuberculous mycobacteria, Mycolicibacterium mageritense , immunocompetent
Citation
Niitsu T, Kuge T, Fukushima K, Matsumoto Y, Abe Y, Okamoto M, Haduki K, Saito H, Nitta T, Kawano A, Matsuki T, Motooka D, Tsujino K, Miki K, Nakamura S, Kida H and Kumanogoh A (2021) Pleural Effusion Caused by Mycolicibacterium mageritense in an Immunocompetent Host: A Case Report. Front. Med. 8:797171. doi: 10.3389/fmed.2021.797171
Received
18 October 2021
Accepted
01 November 2021
Published
24 November 2021
Volume
8 - 2021
Edited by
Yuko Ishida, Wakayama Medical University, Japan
Reviewed by
Toshikazu Kondo, Wakayama Medical University, Japan; Fukumi Furukawa, Takatsuki Red Cross Hospital, Japan; Koichi Tsuneyama, Tokushima University, Japan
Updates
Copyright
© 2021 Niitsu, Kuge, Fukushima, Matsumoto, Abe, Okamoto, Haduki, Saito, Nitta, Kawano, Matsuki, Motooka, Tsujino, Miki, Nakamura, Kida and Kumanogoh.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kiyoharu Fukushima fukushima@imed3.med.osaka-u.ac.jp
†These authors have contributed equally to this work
This article was submitted to Pulmonary Medicine, a section of the journal Frontiers in Medicine
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.