Abstract
Medical image segmentation faces critical challenges in renal histopathology due to the intricate morphology of glomeruli characterized by small size, fragmented structures, and low contrast against complex tissue backgrounds. While the Segment Anything Model (SAM) excels in natural image segmentation, its direct application to medical imaging underperforms due to (1) insufficient preservation of fine-grained anatomical details, (2) computational inefficiency on gigapixel whole-slide images (WSIs), and (3) poor adaptation to domain-specific features like staining variability and sparse annotations. To address these limitations, we propose V-SAM, a novel framework enhancing SAM's architecture through three key innovations: (1) a V-shaped adapter that preserves spatial hierarchies via multi-scale skip connections, recovering capillary-level details lost in SAM's aggressive downsampling; (2) lightweight adapter layers that fine-tune SAM's frozen encoder with fewer trainable parameters, optimizing it for histopathology textures while avoiding catastrophic forgetting; and (3) a dynamic point-prompt mechanism enabling sub-pixel refinement of glomerular boundaries through gradient aware localization. Evaluated on the HuBMAP Hacking the Human Vasculature and Hacking the Kidney datasets, V-SAM achieves state-of-the-art performance, surpassing 89.31%, 97.65% accuracy, 86.17%, 95.54% F1-score respectively. V-SAM sets a new paradigm for adapting foundation models to clinical workflows, with direct applications in chronic kidney disease diagnosis and biomarker discovery. This work bridges the gap between SAM's generalizability and the precision demands of medical imaging, offering a scalable solution for resource constrained healthcare environments.
1 Introduction
Histopathological analysis of kidney biopsies plays a pivotal role in diagnosing and monitoring chronic kidney diseases, where precise segmentation of functional tissue units (FTUs), such as glomeruli and microvasculature, is critical for quantitative assessment. While convolutional neural networks (CNNs) have advanced medical image analysis enabling breakthroughs in retinal fundus imaging, lesion detection, and MRI interpretation their reliance on large, well-annotated datasets and computational intensity remains a barrier to clinical adoption. The emergence of foundation models like the Segment Anything Model (SAM) offers transformative potential, yet its application to histopathology reveals critical gaps in handling the unique challenges of medical imaging.
Trained on over a billion natural image masks, SAM achieves remarkable zero-shot segmentation by leveraging prompt-guided inference (1). However, its performance on medical images, particularly high-resolution histopathology data, remains inconsistent. Recent benchmarks across 12 medical datasets highlight SAM's limitations in capturing fine-grained anatomical structures, with accuracy variations of up to 40% compared to specialized models (2). In renal histopathology, these shortcomings manifest in three key areas: (1) failure to distinguish texturally similar but biologically distinct regions (e.g., sclerotic vs. healthy glomeruli), (2) oversimplification of irregular microvascular boundaries, and (3) sensitivity to staining artifacts and low-contrast features prevalent in periodic acid-Schiff (PAS)-stained whole-slide images (WSIs).
Prior work in renal tissue analysis underscores these challenges. Hybrid architectures like CNN-TransXNet (3) and DLRS systems (4) have improved glomerular segmentation by combining multi-scale feature extraction with attention mechanisms. Yet, these methods require extensive task-specific tuning and lack the generalizability of foundation models. SAM's prompt driven design could bridge this gap, but its native architecture optimized for natural image semantics struggles with the hierarchical complexity of histology, where a single WSI contains structures spanning four orders of magnitude in scale (from 2μm capillaries to 200μm glomeruli).
To address these limitations, we propose a novel method that integrates SAM with a U-Net adapter to enhance segmentation performance in histological images of kidney biopsy. Our approach involves incorporating a U-Net adapter for downsampling and upsampling, along with a residual block in the image encoder to improve feature extraction. In this framework, SAM guides pixel-level segmentation, while the U-Net adapter extracts multi-scale features and refines the segmentation. The residual block enhances the capture of fine-grained details by learning residuals across layers. The detected regions are transformed into prompts and embedded in the SAM prompt encoder, which guides the segmentation process. This method aims to achieve efficient segmentation with reduced computational requirements by focusing SAM on relevant tissue areas, thereby simplifying the prediction process and eliminating the need for extensive preprocessing. Our approach introduces three key innovations:
The key contribution to this research work is given below:
-
Introduce a U-Net adapter into the SAM framework, enhancing multi-scale feature extraction through skip connections, which improves segmentation accuracy and detail in complex medical images.
-
A Point-Prompt Mechanism enables sub-pixel precision through adaptive refinement of point prompts via spatial gradient backpropagation, achieving precise segmentation of intricate glomerular structures while maintaining computational efficiency.
-
Lightweight modules inserted into SAM's frozen encoder enable domain-specific adaptation through minimal trainable parameters (down/up-projections with ReLU). This preserves SAM's pretrained knowledge while allowing targeted feature refinement for medical textures, reducing overfitting risks on limited histopathology datasets.
-
Experiments using the two datasets demonstrates the effectiveness of V-SAM. The results show significant performance improvements compared to current architectures.
The article is organized into several key sections. Section 2 provides a comprehensive review of related work in kidney segmentation, highlighting current advancements and challenges. Section 3 outlines the methodology employed in the proposed framework, detailing the approach used for improving segmentation accuracy. Section 3.2 presents a description of the datasets used in the experiments, including the data characteristics and their relevance to the study. Section 4 discusses the implementation details, covering the experimental setup, parameters, and hyperparameters that were fine-tuned for optimal performance. Section 5 showcases the results of the experiment, along with an ablation study to analyze the contribution of different components of the framework. Section 6 provides a discussion on the findings, interpreting the results in the context of existing literature. Finally, Section 7 concludes the paper with a summary of key findings and suggests directions for future research in kidney segmentation.
2 Related work
In medical image segmentation, U-Net's architecture has evolved with the integration of residual blocks to enhance gradient flow and feature extraction. Recent advancements like U-Net++ and Attention U-Net further refine segmentation accuracy. The Segment Anything Model (SAM) introduces promotable segmentation, offering interactive and flexible approaches in medical imaging. Table 1 provides an analysis of different studies in kidney segmentation methods. In kidney segmentation research, state-of-the-art models have primarily focused on accurate glomerulus identification, along with the metrics used for their validation. A chord plot (Figure 1) visualizes the contribution of these models across different validation metrics, highlighting which metrics are most commonly employed revealing key trends in evaluation practices.
Table 1
| References | WSIs | Glom. | Approach | Purpose | Performance |
|---|---|---|---|---|---|
| (27) | 47 | 1,245 | UNet SegNet AlexNet |
Using deep learning for semantic segmentation to classify glomeruli for detecting glomerulosclerosis. |
Acc: 0.99 F1: 0.99 CK: 0.99 |
| (28) | 26 | 2,772 | SegNet DeepLabv3+ |
Segmentation and classification of glomeruli for reliable estimation of the Karpinski histological score. |
Rec: 0.47 Pre: 0.97 F1: 0.63 |
| (29) | 400 | 12,418 | LSTM-GCNet 2D V-Net | Locate glomeruli and identify lesions like sclerosis, crescents, or none. |
CK: 0.91 Pre: 0.93 Rec: 0.94 |
| (30) | 61 | 1,334 | Mask-RCNN DeepLabv3 |
Downsampling critical glomeruli features reduces instance segmentation accuracy in high-resolution WSIs. | DSC: 0.95 |
| (31) | 258 | 24,133 | VGG Unet | Developing deep learning models to automate glomeruli quantification in kidney biopsies. |
DSC: 0.83 F1: 0.87 Rec: 0.93 Pre: 0.81 |
| (32) | 348 | 8,665 | Cascade Mask R-CNN |
Developing a robust model for glomeruli segmentation and classification across stains and pathologies. |
Pre: 0.95 Rec: 0.92 F1: 0.94 IoU: 0.87 |
| (33) | 15 | 4,500 | FCN-ResNet DeepLabv3 |
Quantification and classification of glomeruli for histopathologic assessment of renal tissue. |
Acc: 0.97 Pre: 0.92 Rec: 0.90 F1: 0.91 |
| (34) | 146 | 5,459 | Mask R-CNN LSTM |
Examine kidney biopsies to identify glomeruli and differentiate glomerulonephritis types. |
Acc: 0.94 F1: 0.94 ROC: 0.94 |
| (35) | 660 | 5,309 | DS-FNet | Boundary-aware glomerulus segmentation aims to generalize across various staining methods. | DSC: 0.95 |
| (36) | 130 | 2,340 | CNB-MVN-MLS | Evaluating spatial deformation augmentation for glomeruli segmentation in histopathology. | DS: 0.85 |
| (37) | 459 | 1,751 | Omni-Seg | Addressing scale variations in renal WSIs with tissue-optimized multi-network segmentation. | DS: 0.87 |
| (38) | 20 | — | LinkNet | Glomeruli detection through segmentation using neural networks. | Acc: 0.99 DC: 0.94 |
| (39) | — | 500 | HistoStar-GAN UDA-GAN | Enable multi-stain transfer, normalization, and segmentation for unseen stains. |
Pre: 0.85 Rec: 0.90 F1: 0.87 |
| (3) | 20 | — | TransXNet OSRA |
Capture fine details and broader context for precise segmentation. | Acc: 0.85 MIoU: 0.77 |
| (40) | 20 | 21,000 | SegNext MobileNet | Enhancing glomeruli segmentation through cross-species pre-training on diverse kidney tissue data. | MIoU: 0.92 DS: 0.96 |
| (41) | 1,536 | — | ECSA-SUNet | Variable morphology and indistinct boundaries blending with surrounding renal tissue. | DS: 0.92 IoU: 0.92 |
| (42) | 210 | 20,868 | UniMatchsf | The resolve inter-observer variability in analyzing histopathology images. |
Pre: 0.95 Rec: 0.76 DC: 0.81 |
| Ours | 14 | 13,440 | V-SAM | Integration of FTU for segmenting blood vessels and glomeruli. |
Acc: 0.98 Pre: 0.96 Rec: 0.95 F1: 0.95 |
State-of-the-art kidney segmentation methods: purpose, approach, and performance metrics.
WSI, Whole Slide Image; Glom, Number of Glomeruli; Pre, Precision; Rec, Recall; Acc, Accuracy; F1, F1-Score; CK, Cohen Kappa; DSC, Dice-Similarity Coefficient; DS, Dice Score; IoU, Intersection over Union; MIoU, Mean Intersection over Union; ROC, Receiver Operating Characteristic; Sens, Sensitivity; Spec, Specificity.
Figure 1

Chord plot to visualize the distributions of different models contribution for kidney glomerulus identification.
2.1 UNet
An early approach to semantic segmentation using CNNs was the Fully Convolutional Network (FCN), which struggles to recover fine details during upsampling due to spatial reduction in the downsampling process, even with transpose convolutions, resulting in coarse outputs. To address this, Ronneberger et al. (5) introduced skip connections in U-Net's encoder-decoder framework, improving segmentation accuracy. However, U-Net still faces challenges in capturing multi-scale features due to the fixed receptive field of its convolutional kernels. To improve this, Kaur et al. (6) proposed a modified U-Net model for the detection of glomeruli in images of whole-slide kidney tissue. The model improves feature extraction by adjusting filter numbers, feature map dimensions, and adding extra convolution blocks to the encoder and decoder. MLP-UNet (7) introduces a novel design avoiding traditional convolutions and self-attention mechanisms, and compares various approaches, including U-Net. For the first time, it trains the TransUNet model on the kidney WSI dataset. The TransXNet block (3), designed for glomerular segmentation, captures both fine details and broader context through a two-phase process: down-sampling with CNNs for detailed features and up-sampling with deconvolution to restore spatial resolution and enhance feature representation. UNet++ (8), an enhanced version of U-Net with a nested layer network, improves kidney biomarker predictions. DeepLabV3 (9) further enhances segmentation accuracy with atrous convolutions and atrous spatial pyramid pooling (ASPP), making it ideal for identifying four histopathological structures in acute renal tubular injury: glomeruli, necrotic tubules, healthy tubules, and tubules with casts.
2.2 Segment Anything Model (SAM)
The Segment Anything Model (SAM) has shown promise in medical image segmentation, leveraging its ability to generalize across diverse datasets with zero-shot learning. Its flexible prompt-based design allows for efficient segmentation of complex structures, reducing reliance on large annotated datasets. SAM's adaptability makes it a valuable tool for automated and accurate medical image analysis. SAM, originally designed for natural images, requires improvements for optimal performance on medical images (10). Leveraging SAM's ability to generate pixel-level annotations from box annotations, the authors use these SAM-generated labels to train a segmentation model. The SAM-assisted molecular-empowered learning (SAM-L) approach reduces the efforts to label the annotations of lay annotators by requiring only weak box annotations (11). A novel detector method is used to perform automatic prompted segmentation, offering a low-cost alternative to other SAM customization processes for specific tasks. det-SAM (12) features a detection head in its architecture, which provides additional domain-specific information to SAM through prompt engineering, enhancing its performance in medical segmentation tasks. DeSAM (13) introduces a decoupled SAM architecture with domain-specific prompt tuning, enhancing generalization across diverse medical imaging tasks. The framework adapts SAM's segmentation capability through anatomical-aware prompt engineering while preserving its foundational strengths. SAM2-UNet (14) integrates SAM's vision transformer encoder into a U-Net structure, demonstrating robust feature transferability for both natural and medical image segmentation. This hybrid approach achieves strong performance by combining SAM's pretrained representations with U-Net's hierarchical decoding.
Skip connections also play a vital role in preserving spatial details within encoder-decoder architectures. SAttisUNet (15) replaces direct skip pathways with an attentive module that fuses Swin Transformer features using cross-covariance attention, enhancing multi-scale context representation. The 3D framework in (16) employs convolutional blocks (CIRU, CIRC) to aggregate encoder outputs, prompt embeddings, and raw image features for improved prediction quality. Meanwhile, Yang et al. (17) propose a Deformable Squeeze-and-Attention (DSA) block that leverages deformable convolutions to adaptively refine segmentation details by learning flexible receptive fields. In contrast, our V-SAM incorporates structurally-guided skip connections that deliver multi-resolution encoder features through attention-based pathways aligned with U-shaped topology. Tailored for histological precision, these connections emphasize glomerular boundary fidelity using class-aware fusion. Unlike existing approaches, V-SAM addresses the unique challenges of glomerulus segmentation–namely, the need for fine-grained boundary preservation and prompt-driven contextual adaptation.
2.3 Adapter layer
The use of adapter layers in medical image segmentation has recently gained significant attention due to their ability to enhance model efficiency without the need for extensive retraining. Zhu et al. (18) introduced AdaptFormer, a transformer-based model that incorporates adapter layers, showing improved performance in medical image segmentation tasks, particularly for CT and MRI scans. Li et al. (19) also explored this concept with their dual adapter networks, which optimized segmentation accuracy for abdominal organ segmentation in CT images by combining a general-purpose backbone with task-specific adapters. Wang et al. (20) demonstrated the effectiveness of transformer-adapter networks for brain tumor segmentation in MRI scans, highlighting their capacity to extract key features with fewer parameters. Finally, Yuan et al. (21) developed dynamic adapters for multi-class segmentation tasks in lung CT scans, achieving better accuracy and efficiency in complex cases. These advancements emphasize the power of adapter layers to improve the performance of medical image segmentation models, especially when data are limited.
3 Methodology
Applying SAM to medical image segmentation has garnered increasing attention, with many approaches focusing on transferring knowledge from natural image domains. While SAM has demonstrated promising results in tasks involving organs or regions with large and well-defined structures, such as liver or lung segmentation, its performance declines when dealing with finer anatomical details. Tasks like blood vessel or glomerulus segmentation, which involve low-contrast boundaries, intricate morphologies, and small-scale structures, still present significant challenges highlighting the need for further architectural adaptation. In this study, we propose V-SAM, a novel architecture designed to overcome these limitations and enhance segmentation performance in the medical domain, specifically for glomerulus segmentation in kidney histology images. V-SAM integrates a promptable paradigm to improve lesion localization and more effectively capture the complex and subtle structural variations inherent in glomerular regions. It incorporates three key components: promptable information (points) for precise target area localization, a V-shaped structure to capture low-level glomerulus details, and skip connections to preserve and recover spatial information throughout the encoding-decoding process, as illustrated in Figure 2.
Figure 2
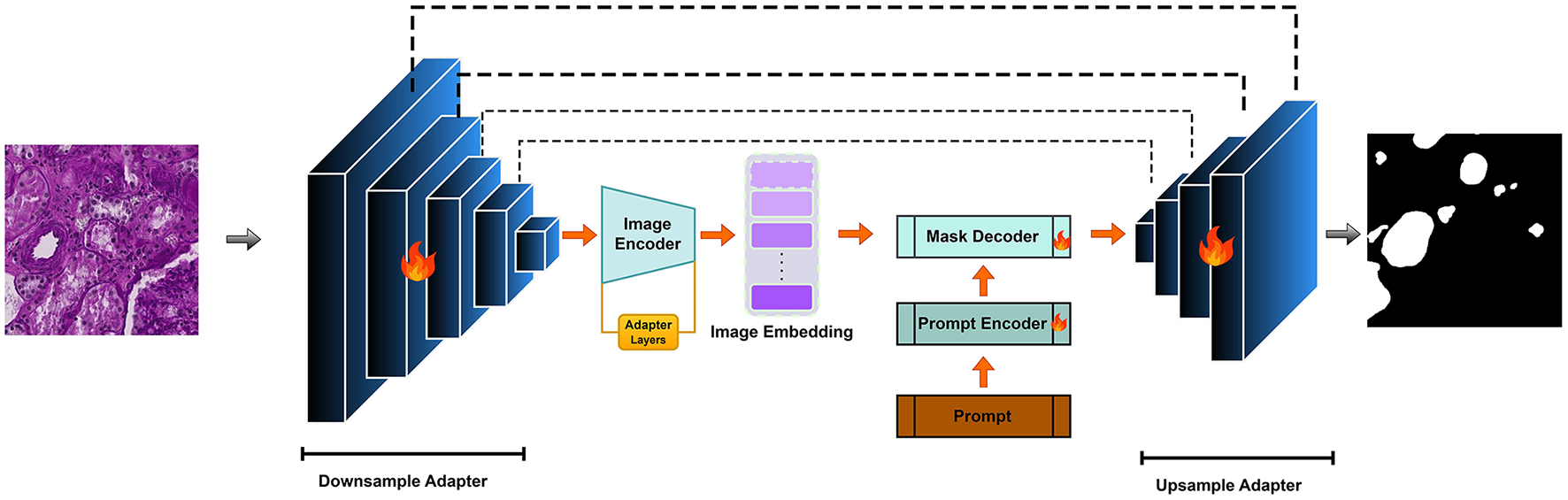
V-SAM Model: integrating a U-Net adapter into the SAM framework for enhanced multi-scale feature extraction.
Segmenting FTU in kidney images presents a unique challenge due to the small size and complex shape of the glomerulus. The kidney's anatomical complexity makes it difficult for conventional segmentation models [e.g., U-Net (5), DeepLabv3+ (22), TransUNet (23), Swin-Unet (24), UNet++ (23)]. Inspired by the success of the Segment Anything Model (SAM) and its promptable segmentation paradigm (e.g., bounding boxes, points, and text), we propose to adopt this paradigm to improve the localization of glomeruli and address the unique challenges presented by kidney images. However, SAM alone struggles to achieve precise segmentation in regions with low contrast or intricate structures, as seen in kidney tissue. This challenge is particularly critical in glomerulus segmentation.
3.1 V-SAM framework
Following the principles of SAM, our V-SAM model uses a two-step upsampling scheme to recover the image resolution. Unlike the long-stride upsampling strategy in the original SAM, V-SAM introduces a pair of V-shaped adapters to improve segmentation accuracy. The pipeline is divided into two major phases: the downsampling encoder and the upsampling decoder. In the following sections, the image encoder is treated as part of the downsampling process, while the mask decoder is responsible for the upsampling.
3.1.1 Downsampling adapter
The downsampling encoder in V-SAM serves as the hierarchical feature extractor tailored for glomerulus segmentation in gigapixel WSIs. Designed as a U-Net-inspired contraction path with V-shaped adapters, it progressively condenses spatial resolution while expanding channel capacity to capture multi-scale glomerular features from capillary-level textures to global structural contexts.
Each downsampling block implements a resolution halving operation governed by:
where fi and fi + 1 denote input/output feature maps. The MaxPool (kernel=3, stride=2) first suppresses spatial redundancy while preserving dominant activations critical for detecting glomerular capsules. The subsequent Conv layer (kernel=3, padding=1) doubles channel depth (except at f4) to expand receptive fields without sacrificing feature density – a deliberate trade-off for WSI processing where excessive downsampling erodes subtle tuft boundaries.
The five-stage feature pyramid is structured as:
With C = 256 matching the Channel Attention Module (CAM)'s latent dimension, this scaling ensures compatibility with SAM's pretrained weights while preventing channel explosion. The exponential channel growth (C → 2C at f4) compensates for spatial information loss, preserving texture gradients vital for distinguishing sclerotic glomeruli (collapsed capillaries) from healthy ones.
V-SAM integration: The deepest features f4 feed directly into SAM's image decoder as positional priors, while f0-f3 are routed via V-shaped skip connections to the upsampling decoder (Figure 2). This dual-path strategy merges SAM's semantic understanding (via f4) with U-Net's boundary precision (via skip features), crucial for resolving overlapping glomeruli in crowded WSI regions. The adapter layers (Figure 3) refine these features through lightweight down/up-projections, enabling domain adaptation without compromising SAM's generalization. For better understanding the architecture pseudo-code for V-SAM is given in Algorithm 1.
Figure 3

Left shows the ViT block capturing context, while the right depicts the adapter layer for enhanced multi-scale feature extraction and segmentation accuracy.
Algorithm 1
| 1: Input: Image I of size 224 × 224 × 3 |
| 2: Output: Segmentation mask S of size 224 × 224 × N |
| 3: let f0, f1, f2, f3, f4 be the feature maps at each downsampling stage |
| 4: let SAM_output be the output from SAM decoder |
| 5: let S_refined be the final refined segmentation output |
| 6: Step 1: Pre-process Input Image |
| 7: I ← normalize(I) |
| 8: Step 2: Downsampling Encoder |
| 9: fori = 0 to 3 do |
| 10: fi ← Encoder_Block(I, in_channels, out_channels) |
| 11: I ← MaxPool(I, kernel = 3, stride = 2) |
| 12: I ← Conv(I, kernel = 3, padding = 1) |
| 13: end for |
| 14: Step 3: Adapter Layer |
| 15: f4 ← Adapter_Layer(f4) |
| 16: Step 4: SAM Image Encoder Processing |
| 17: SAM_encoded ← SAM_Encoder(f4) |
| 18: Step 5: SAM Decoder Processing |
| 19: SAM_output ← SAM_Decoder(SAM_encoded) |
| 20: Step 6: Upsampling Decoder |
| 21: fori = 3 down to 0 do |
| 22: ri ← Upsample_Block(SAM_output, fi) |
| 23: SAM_output ← Upsample(SAM_output, scale_factor = 2) |
| 24: end for |
| 25: Step 7: Final Refinement |
| 26: S_refined ← Final_Refinement(SAM_output, f1, f0) |
| 27: Step 8: Final Segmentation Mask Output |
| 28: S ← Final_Convolution(S_refined) |
| 29: Return:S |
V-SAM Algorithm
3.1.2 Adapter layer in image encoder
We integrate adapter layers into the image encoder of the Segment Anything Model (SAM) to adapt the pre-trained encoder for medical image segmentation tasks. Adapter layers serve as lightweight modules inserted into the encoder, introducing task-specific transformations without modifying the majority of the model's pre-trained parameters. This allows for computational efficiency by reducing the number of trainable parameters, while maintaining the encoder's generalization capability. The adapter layer is composed of two main components: a down-projection and an up-projection. The adapter layer and ViT block can be visualize in Figure 3 is The down-projection reduces the dimensionality of the feature map hl, and the up-projection restores the original dimension. The transformation of the input feature map hl through the adapter layer is mathematically defined as:
Where hl represents the output from the previous layer, Wdown and Wup are the weight matrices for the down-projection and up-projection layers, respectively. The activation function is denoted by σ, and bdown and bup correspond to the bias terms for the down-projection and up-projection layers.
Here, hl is the input feature map from the encoder layer, The term σ is a non-linear activation function ReLU. The output represents the adapted feature map, which is used for downstream segmentation tasks. For fine-tuning, the parameters of the adapter layers, i.e., Wdown, Wup, bdown, bup, are trained, while the other encoder parameters are kept frozen. The fine-tuning objective is to minimize the segmentation loss , which measures the difference between the model's predicted segmentation and the ground truth ygt:
Where is the segmentation output generated from the adapted feature map, and ygt is the ground truth label. The parameter efficiency of the adapter layer is given by:
This results in significantly fewer trainable parameters compared to fine-tuning the entire encoder. The computational complexity of the adapter layer is:
Since r ≪ d, the computational cost is minimal. Furthermore, by training only the adapter layer parameters, we reduce the risk of overfitting, which is particularly beneficial when working with smaller medical datasets. This efficiency enables faster convergence and makes the approach suitable for resource-constrained environments.
3.1.3 Prompt (point-based)
Unlike the default static point prompt strategy in SAM, our method introduces an enhanced point-based prompt mechanism tailored for multi-class medical segmentation and improved spatial precision. The proposed approach provides sparse but highly informative guidance by integrating three key innovations:
Here, generates positional embeddings using sinusoidal encoding, while maps discrete class identifiers (e.g., glomeruli, vessels, uncertain regions) to learned embeddings. The concatenation ⊕ allows the prompt to encode both spatial and semantic (class-level) information. This formulation supports multi-class segmentation directly within the prompting mechanism, a functional extension beyond SAM's binary (foreground/background) prompt treatment.
To integrate the point prompts with the model's features, we introduce a constrained cross-attention mechanism between encoded prompts and encoder outputs fi:
where σ denotes row-wise softmax, and W{q, k, v} are linear projection matrices. This constrained attention formulation allows the model to selectively focus on relevant spatial regions while significantly reducing memory usage compared to dense attention used in SAM. Finally, we introduce a gradient-based refinement step during training, which adapts the prompt coordinates by backpropagating segmentation loss gradients with respect to spatial positions:
where η is the learning rate. This approach allows the prompt positions to be optimized during training for finer localization accuracy, resulting in sub-pixel alignment without any additional inference cost.
3.1.4 Upsampling adapter
The V-SAM decoder extends SAM's mask generation through a hierarchical refinement process that progressively recovers spatial resolution using multi-scale encoder features. Given the initial mask source and transformed mask tokens from SAM's core processing, our architecture introduces three sequential upsampling stages:
where ⊕ denotes channel-wise concatenation with skip connections from the downsampling encoder's feature pyramid. Each Upsamplei operation consists of 2 × bilinear upscaling, 3 × 3 convolution with stride 1, ReLU activation and feature fusion with encoder skip connection fi + 1.
The final resolution enhancement to is achieved through an additional refinement layer:
where provides high-frequency spatial details from the initial encoder stage. This contrasts with SAM's original approach that directly upscales low-resolution logits () through a single 4 × bilinear interpolation, resulting in lost boundary details critical for glomerulus segmentation.
The complete upsampling chain reduces effective stride from 16 × to 2 × through:
preserving morphological details through four gradual refinement stages rather than SAM's single coarse upsampling. This proves essential for maintaining capsule boundary continuity and resolving sub-glomerular structures in high-magnification WSIs.
3.2 Datasets
In this following section we will briefly explain the characteristics of two datasets used in our experiments.
3.2.1 HuBMAP - Hacking the Human Vasculature (HubMAP-1)
The HubMAP-1 dataset, originally part of the https://www.kaggle.com/competitions/hubmap-hacking-the-human-vasculature challenge on Kaggle, is now publicly available for open use. This dataset aims to support mapping the human vascular system, focusing on functional tissue units like glomeruli and blood vessels. It includes 2D periodic acid-Schiff (PAS)-stained tiles (512 × 512 pixels) from 14 whole slide images (WSIs) of human kidney histology, selected specifically for microvascular structure segmentation. The demographic data from the data set are shown in Figure 4, with expert-verified annotations for accuracy. Additional tile information provided by dataset distributor is provided in Table 2, and training set examples are displayed in Figure 5.
Figure 4
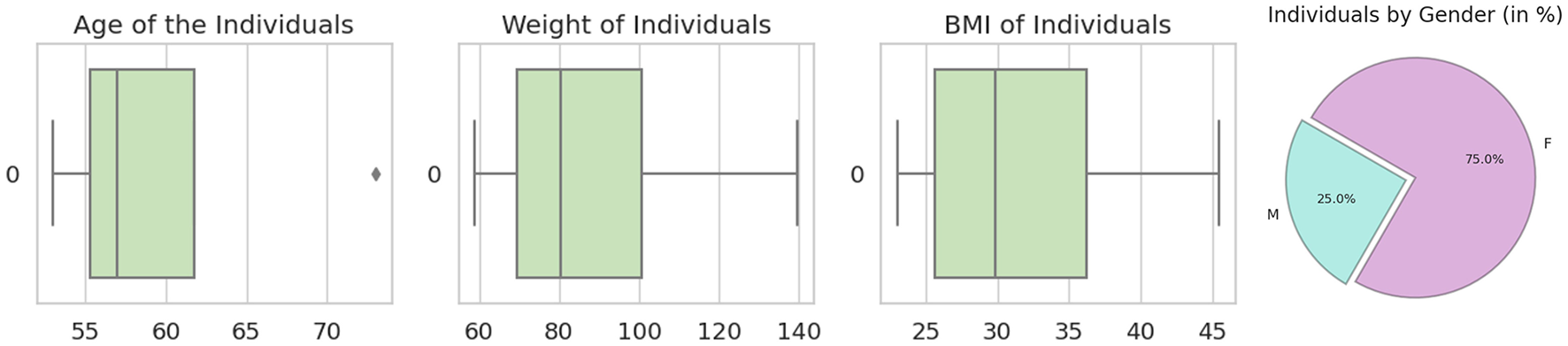
From left to right: the first image shows the age distribution among patients, the second displays weight distribution, the third illustrates body mass index (BMI) distribution, and the last image presents the dataset's gender distribution (male vs. female) in percentages.
Table 2
| Attributes | Description |
|---|---|
| source_wsi | The tile is extracted from which WSI |
| ID | ID number of the tile |
| i | j | The location of the upper corner from which the WSI is cropped |
| Age, sex, race, height, weight, BMI | Demographics of the tissue donors |
Attributes and metadata of the HubMAP-1 dataset.
Figure 5

Some examples of image tiles taken from HubMAP-1.
3.2.2 HuBMAP—hacking the kidney (HubMAP-2)
The National Institutes of Health (NIH) funded the HubMAP-2 project, which is now publicly available on Kaggle. https://www.kaggle.com/competitions/hubmap-kidney-segmentation, project aimed to create a detailed map of the human vascular system, with a particular focus on functional tissue units known as glomeruli. The dataset includes 11 fresh frozen and 9 Formalin Fixed Paraffin Embedded (FFPE) PAS kidney images. The TIFF images, with dimensions exceeding 19,780 × 26,840 (ranging from 182.65 MB to 4.87 GB), feature annotations of glomeruli in two formats: RLE-encoded and unencoded (JSON). Additional metadata is provided in Table 3, and sample image tiles from the training set are shown in Figure 6. Demographics for HubMAP-2 is given in Figure 7.
Table 3
| Attributes | Description |
|---|---|
| width/height pixels | Image resolution details |
| patient number | Tissue donor patient identifier |
| Race, Ethnicity, Sex, Age | Demographics of the tissue donors |
Attributes and metadata of the HubMAP-2 dataset.
Figure 6

Some examples of image tiles taken from HubMAP-2.
Figure 7

Demographic distribution by tissue composition. Left: age, sex, and race distribution in relation to medulla percentage. Right: age, sex, and race distribution in relation to cortex percentage for HubMAP-2 dataset.
4 Implementation
The experiments were carried out on a single NVIDIA RTX 3090 GPU using the PyTorch framework. To prevent overfitting, we applied data augmentations and other image slide pre-processing techniques, detailed in Table 4. Pre-trained weights from SAM on natural images were utilized to accelerate convergence and enhance training stability. During training, we froze the parameters of SAM's prompt encoder for the same purpose. While SAM was originally designed for larger input sizes, such as 1024x1024, we adapt it to handle smaller inputs like 224x224, similar to how TransUNet adapts Transformer-based architectures for reduced image sizes. In TransUNet (25), the model effectively processes smaller input sizes by leveraging a Transformer encoder-decoder structure that captures long-range dependencies while preserving fine-grained details through skip connections. For our implementation, the input resolution is set to 224x224, with a batch size of 8, enabling efficient processing while maintaining segmentation accuracy. The model was trained end-to-end using the Adam optimizer (26), with an initial learning rate of 0.0001 to speed up convergence. The parameters setting is listed in Table 5. The masking for HubMAP-1 and HubMAP-2 is given in Figures 8, 9 respectively.
Table 4
| Attributes | Description |
|---|---|
| CLAHE | True |
| Diagonal shift | True |
| Horizontal shift | True |
| Hue saturation | True |
| IAAPiecewiseAffine | 0.3 |
| Optical distortion | 0.3 |
| Random brightness | True |
| Random contrast | True |
| Vertical shift | True |
Augmentation parameters for the experiment.
Table 5
| Parameters | Setting |
|---|---|
| Batch size | 8 |
| Epoch | 100 |
| Initial learning rate | 1 × 10−4 |
| Loss function | L BCE + LDice |
| Optimizer | Adam |
| Patch size | 224 × 224 |
Parameters settings for the experiment.
Figure 8

From left to right: in first image, display the dataset image. In the second image, depict polygons drawn on the corresponding image. In the third image, exhibit the binary mask of that image.
Figure 9

From left to right: in first image, display the dataset image. In the second image, depict polygons drawn on the corresponding image.
The loss function used is a combination of binary cross-entropy and the Dice coefficient. It is formulated as:
Where Ŷb and Yb denote the predicted probabilities and the ground truth values (both flattened) for the b-th image in the batch, respectively, and N represents the batch size.
5 Results
5.1 Results on HubMAP-1
The results demonstrate that V-SAM achieves exceptional performance on the HubMAP-1 dataset, with an Accuracy of 89.31%, a Recall of 87.29%, and F1-Score of 86.17%. These scores consistently surpass a range of existing models, including both convolution-based and transformer-based architectures. Compared to UNet++ (ResNet-50), V-SAM delivers a ↑3.69% gain in Accuracy, ↑3.77% in Recall, and ↑3.26% in F1-Score. Against nnUNet, which uses a standard UNet backbone, V-SAM improves Accuracy by ↑3.12%, Recall by ↑3.02%, and F1-Score by ↑2.18%. When compared with DRA-Net (ResNet-34), V-SAM achieves ↑2.05% higher Accuracy, ↑1.66% higher Recall, and ↑1.48% higher F1-Score. Notably, although DET-SAM employs the same ViT-B backbone, V-SAM significantly outperforms it by ↑4.52% in Accuracy, ↑3.54% in Recall, and ↑5.03% in F1-Score.
These consistent improvements highlight key limitations in convolution-based models like ResNet, which struggle with modeling long-range dependencies and preserving fine-grained spatial details due to their localized receptive fields. While DET-SAM shares the ViT-B transformer backbone with V-SAM, its performance is hindered by suboptimal prompting and lack of targeted skip connection design. In contrast, V-SAM incorporates a structurally guided attention mechanism, topology-aware prompting, and class-sensitive skip connections, which collectively contribute to robust segmentation of fine structures such as glomeruli. Full quantitative results are summarized in Table 6, and the corresponding performance trend is visualized in Figure 10.
Table 6
| Model | Backbone | Accuracy (%) | Recall (%) | F1-score (%) |
|---|---|---|---|---|
| UNet++ (43) | ResNet-50 | 85.62 | 83.52 | 82.91 |
| nnUNet (44) | UNet | 86.19 | 84.27 | 83.99 |
| DRA-Net (45) | ResNet-34 | 87.26 | 85.63 | 84.69 |
| DET-SAM (12) | ViT-B | 84.79 | 83.75 | 81.14 |
| V-SAM | ViT-B | 89.31 | 87.29 | 86.17 |
Comparison of V-SAM with different models for HubMAP-1.
The bold values showing best results.
Figure 10
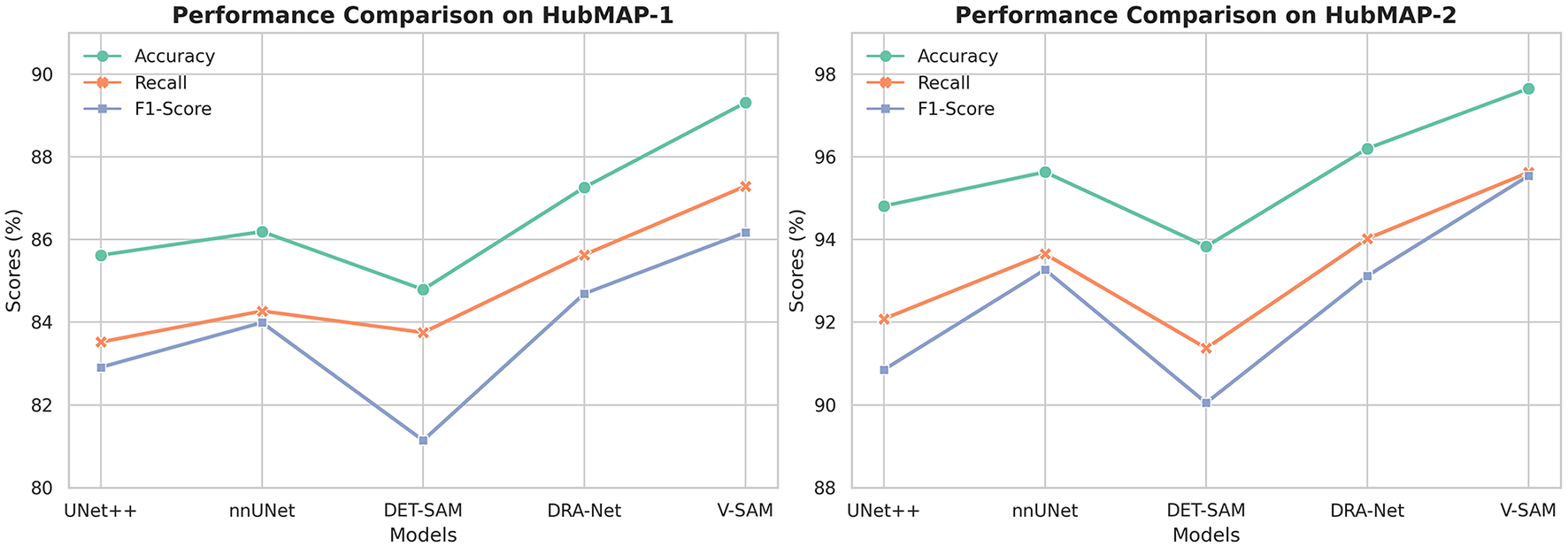
The trend of performance improvement across different models on both datasets. From left to right, the figure shows the incremental growth in Accuracy, Recall, and F1-Score for HubMAP-1 and HubMAP-2, respectively.
Furthermore, V-SAM's performance is compared with recent state-of-the-art (SOTA) models on the HubMAP-1 dataset. V-SAM achieves an Accuracy of 89.31% and an F1-Score of 86.17%, surpassing UniMatch_sf (MiT-B1 backbone) by 4.77% and Omni-Seg (ReS-UNet backbone) by 3.87% in F1-Score, while achieving a remarkable 28.17% improvement over FastAI U-Net (ResNet-50 backbone), which only achieves an F1-Score of 58.00%. This performance gap underscores the effectiveness of V-SAM's transformer-based architecture, which excels at capturing global context and long-range dependencies, unlike convolutional backbones such as ResNet or ReS-UNet. Although UniMatch_sf and Omni-Seg demonstrate strong performance, their reliance on convolutional architectures limits their ability to model complex structures and global context effectively. In contrast, V-SAM's advanced fine-tuning techniques and adaptability to complex structures in medical images contribute to its superior performance, making it a robust and reliable model for medical image segmentation tasks. The comparison with SOTA models is provided in Table 7.
Table 7
| Model | Publication/ Year | Backbone | Accuracy (%) | F1-Score (%) |
|---|---|---|---|---|
| FastAI U-Net (46) | EESS-23 | ResNet-50 | — | 58.00 |
| UniMatch_sf (42) | MIDL-24 | MiT-B1 | — | 81.40 |
| Omni-Seg (47) | SPIE.MI-23 | ReS-UNet | — | 82.30 |
| V-SAM | — | ViT-B | 89.31 | 86.17 |
Comparison of V-SAM with SOTA for HubMAP-1.
The bold values showing best results.
The segmentation results reveal a clear distinction in the performance of different models. UNet++ captures most major vascular structures but struggles with fine details and boundary precision, often displaying over-segmentation in certain regions, as highlighted in the red boxes. DRA-Net demonstrates improved continuity and connected component detection but still faces challenges with boundary refinement and misses smaller structural elements. Det-SAM further enhances boundary precision and reduces over-segmentation; however, it under-segments certain areas, leading to the loss of finer vessel branches. In contrast, the V-SAM achieves the most accurate segmentation, closely replicating the ground truth. It effectively captures both large and small structures with clean, precise boundaries and maintains detail even in complex regions. This superior performance is likely attributed to advanced spatial, channel, and temporal attention mechanisms that enhance feature extraction and boundary refinement. Additionally, V-SAM appears to generalize better, handling variations in noise and texture more effectively than the other models. Overall, V-SAM strikes the optimal balance between accuracy and detail preservation, making it the best-performing model in this evaluation.
5.2 Results on HubMAP-2
The performance of V-SAM on the HubMAP-2 dataset is thoroughly evaluated against several competitive models in Table 8, using Accuracy, Recall, and F1-Score as key metrics. V-SAM achieves the highest results across all three: 97.65% Accuracy, 95.62% Recall, and 95.54% F1-Score. Compared to UNet++ (ResNet-50), V-SAM shows significant gains of ↑2.84% in Accuracy, ↑3.54% in Recall, and ↑4.69% in F1-Score, indicating its superior ability to capture fine-grained features and global structure. Against nnUNet, which uses a self-configuring UNet-based pipeline, V-SAM improves performance by ↑2.12% in Accuracy, ↑1.97% in Recall, and ↑2.27% in F1-Score. Although nnUNet performs strongly due to its robust configuration heuristics, it still relies on standard convolutional designs, which limits its ability to model long-range dependencies.
Table 8
| Model | Backbone | Accuracy (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| UNet++ (43) | ResNet-50 | 94.81 | 92.08 | 90.85 |
| nnUNet (44) | UNet | 95.63 | 93.65 | 93.27 |
| DRA-Net (45) | ResNet-34 | 96.20 | 94.02 | 93.12 |
| DET-SAM (12) | ViT-B | 93.83 | 91.37 | 90.05 |
| V-SAM | ViT-B | 97.65 | 95.62 | 95.54 |
Comparison of V-SAM with different models for HubMAP-2.
The bold values showing best results.
In comparison to DRA-Net (ResNet-34), which introduces attention mechanisms and dynamic refinement, V-SAM achieves ↑1.45% higher Accuracy, ↑1.60% better Recall, and ↑2.42% improved F1-Score, showcasing its more effective context modeling and class-sensitive segmentation. While DET-SAM uses the same ViT-B backbone as V-SAM, its performance is notably lower across all metrics, with V-SAM achieving ↑3.82% better Accuracy, ↑4.25% higher Recall, and ↑5.49% stronger F1-Score. This gap underscores the impact of V-SAM's tailored optimization, prompting, and skip-connection design, which DET-SAM lacks due to limited adaptation for dense medical tasks. A detailed comparison of all models is summarized in Table 8, and the corresponding performance trends are visualized on the right side of Figure 10.
Table 9 further compares V-SAM with state-of-the-art (SOTA) models on the HubMAP-2 dataset. V-SAM achieves an Accuracy of 97.65% and an F1-Score of 95.54%, outperforming LinkNet (EfficientNet-B3 backbone) in F1-Score by 1.21%, DS-FNet (AU-Net backbone) by 0.36%, and SegNeXt (MSCAN-S backbone) by 0.08%. While LinkNet achieves the highest Accuracy (99.70%), its reliance on EfficientNet limits its ability to balance precision and recall, resulting in a lower F1-Score. This limitation stems from the inherent constraints of convolutional architectures, which struggle to model long-range dependencies and global context effectively. Similarly, DS-FNet and SegNeXt, despite their strong performance, are constrained by their convolutional architectures, which are less effective at capturing fine-grained details and global structures compared to transformer-based models like V-SAM. The F1-Score comparison among both datasets are given in Figure 11.
Table 9
| Model | Publication / Year | Backbone | Accuracy (%) | F1- Score (%) |
|---|---|---|---|---|
| EnsembleDLNet (33) | AJSE-22 | ResNet-50 | 97.50 | 91.50 |
| DS-FNet (35) | CMIG-22 | AU-Net | — | 95.05 |
| UNet (48) | Soft Computing-23 | EfficientNet-B4 | 99.68 | 90.60 |
| LinkNet (38) | Digital Imaging-23 | EfficientNet-B3 | 99.70 | 94.20 |
| SegNeXt (40) | Neurocomputing-24 | MSCAN-S | — | 95.33 |
| CNN-TransXNet (3) | IJCIS-24 | TransXNet | 85.13 | 82.80 |
| V-SAM | — | ViT-B | 97.65 | 95.54 |
Comparison of InFeNet with SOTA for HubMAP-2.
The bold values showing best results.
Figure 11

The variations in F1-Score per epoch by different model on both datasets from (left to right) for HubMAP-1 and HubMAP-2 respectively.
In contrast, CNN-TransXNet (TransXNet backbone) performs poorly, with V-SAM achieving 12.52% higher Accuracy and 12.61% better F1-Score. This significant performance gap is likely due to inefficiencies in CNN-TransXNet's hybrid convolutional-transformer design, which fails to fully leverage the strengths of either architecture. V-SAM, on the other hand, excels due to its pure transformer-based architecture (ViT-B backbone), which is specifically designed to model global dependencies and intricate structures. Additionally, V-SAM's advanced fine-tuning techniques and optimization strategies ensure that it adapts effectively to the dataset, delivering precise and balanced results. These features solidify V-SAM's position as a leading model for medical image segmentation tasks, as demonstrated by its superior performance across multiple metrics. The comparison with SOTA models is provided in Table 9.
The segmentation results provide further insight into the performance of each model when applied to kidney histology images (Figure 12). UNet++ captures the primary structures but struggles with accurately segmenting the finer regions. Over-segmentation is observed in areas such as the top-left and top-right corners, where boundary details are not preserved, and gaps in segmentation appear in some areas, as highlighted in the red boxes. DRA-Net improves upon these issues by showing better overall segmentation continuity, though boundary artifacts persist, and small structures remain inadequately segmented. Det-SAM performs moderately well, showing better refinement of major structures, but there is a tendency to under-segment, particularly in regions containing finer details or smaller clusters. In contrast, the V-SAM model excels by closely matching the ground truth, accurately capturing both large and small features with minimal errors. It achieves clean and precise boundaries, successfully preserving structural integrity even in complex regions. This superior performance is likely due to the model's ability to utilize spatial, channel, and temporal attention mechanisms that enhance feature extraction and refinement of segmentation masks. Additionally, V-SAM demonstrates better generalization, effectively handling variability in texture and shape across different regions of the images. Therefore, V-SAM maintains the optimal balance between segmentation accuracy and detail preservation, making it the best-performing model for this task.
Figure 12

Visual comparison of the proposed V-SAM model with SOTA models on the HubMAP-1 and HubMAP-2. From left to right, the first four images represent HubMAP-1, while the following four images depict HubMAP-2.
6 Discussion
The experimental results demonstrate that the proposed V-SAM model achieves superior performance compared to several state-of-the-art (SOTA) segmentation models, including UNet++, nnUNet, DRA-Net, and DET-SAM, on both the HubMAP-1 and HubMAP-2 datasets. These improvements are largely attributed to V-SAM's transformer-based architecture, specifically the ViT-B backbone, which effectively captures both global contextual relationships and fine-grained structural details. Robust data augmentation and careful fine-tuning of the attention mechanism further enhance the model's ability to handle complex textures and morphological variations common in medical images. V-SAM achieved an accuracy of 89.31%, a Recall of 87.29%, and an F1-score of 86.17% on HubMAP-1, and an accuracy of 97.65%, Recall of 95.62%, and F1-score of 95.54% on the more challenging HubMAP-2 dataset. These results indicate consistent improvements across both coarse and fine structures, as further validated by visual comparisons showing V-SAM's ability to delineate both large anatomical regions and smaller entities like glomeruli critical for accurate diagnosis and treatment planning.
Although V-SAM leverages point-based prompts to guide segmentation, the inclusion of sparse point prompts is deliberate. This approach addresses limitations observed in purely automated models, particularly for tasks involving complex, low-contrast structures such as kidney glomeruli, where fully automated methods often struggle to achieve the necessary precision. These prompts enable precise localization and targeted correction, offering enhanced control and reliability in edge cases where automated models tend to fail. The integration of gradient-based refinement further allows sub-pixel accuracy with minimal computational cost, making the approach viable for semi-supervised clinical workflows. This trade-off balances automation with interpretability and expert oversight, which is often essential in real-world medical applications.
Despite its strong performance, V-SAM does come with certain limitations. The generalizability to other imaging modalities and anatomical regions remains an open area for exploration. Future work will focus on reducing dependence on manual prompts while retaining performance gains, potentially through active learning or self-supervised refinement mechanisms. Overall, V-SAM presents a promising advancement in medical image segmentation, offering a compelling balance between automation and precision, and setting the stage for more robust and adaptable diagnostic tools in computational pathology.
7 Conclusion
In this study, we introduced V-SAM, a novel and efficient architecture tailored for medical image segmentation, with a specific focus on glomerulus segmentation in kidney images. V-SAM integrates a promptable paradigm, a V-shaped structure, and skip connections to effectively capture fine-grained details and preserve critical spatial information. The addition of adapter layers facilitates efficient fine-tuning of the pre-trained SAM, while the point-based prompt mechanism enhances the model's ability to localize low-contrast and fragmented structures with greater accuracy. Moreover, the upsampling adapter improves segmentation results by recovering high-resolution details. Experimental evaluations on the HubMAP-1 and HubMAP-2 datasets highlight V-SAM's superior performance. On HubMAP-1, V-SAM outperforms DET-SAM with improvements of approximately 5.3%, 4.2%, and 6.2% in accuracy, Recall, and F1-score, respectively. On the more challenging HubMAP-2 dataset, V-SAM surpasses DET-SAM by about 4.0%, 4.6%, and 6.1% in accuracy, Recall, and F1-score, respectively. These results demonstrate V-SAM's ability to capture global context and long-range dependencies, offering a clear advantage over traditional convolutional backbones.
In conclusion, V-SAM demonstrates its potential as a robust and efficient solution for medical image segmentation, particularly for complex tasks such as segmentation of the glomerulus in kidney images. With its adaptability, computational efficiency, and superior performance, V-SAM is well suited for clinical applications. Future work will focus on expanding V-SAM to other medical imaging tasks and further refining its architecture to optimize both performance and real-time usability in clinical settings.
Statements
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.kaggle.com/competitions/hubmap-kidney-segmentation; https://www.kaggle.com/competitions/hubmap-hacking-the-human-vasculature.
Author contributions
MW: Conceptualization, Data curation, Methodology, Writing – original draft. CW: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing. MA: Software, Visualization, Writing – review & editing. JS: Formal analysis, Investigation, Writing – review & editing. SA: Funding acquisition, Resources, Validation, Writing – review & editing. FA: Formal analysis, Investigation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The Researchers would like to thank the Deanship of Graduate Studies and Scientific Research at Qassim University for financial support (QU-APC-2025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. During the preparation of this work, the author(s) utilized ChatGPT for LaTeX formatting and QuillBot for sentence restructuring. Following the use of these tools, the author(s) thoroughly reviewed and edited the content, taking full responsibility for the final version of the publication.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
It holds that authors are fully responsible for the content of their manuscript, even those parts produced by an AI tool, and are thus liable for any breach of publication ethics (in the writing process).
References
1.
Ke L Ye M Danelljan M Tai YW Tang CK Yu F et al . Segment anything in high quality. In: 37th Conference on Neural Information Processing Systems (NeurIPS 2023). (2024). p. 36. Available online at: https://arxiv.org/abs/2306.01567
2.
He S Bao R Li J Stout J Bjornerud A Grant PE et al . Computer-Vision Benchmark Segment-Anything Model (SAM) in Medical Images: Accuracy in 12 Datasets. (2023). Available online at: https://arxiv.org/abs/2304.09324
3.
Liu Y A . Hybrid CNN-TransXNet Approach for advanced glomerular segmentation in renal histology imaging. Int J Comp Intellig Syst. (2024) 17:126. 10.1007/s44196-024-00523-7
4.
Suzuki N Kojima K Malvica S Yamasaki K Chikamatsu Y Oe Y et al . Deep learning-based histopathological assessment of tubulo-interstitial injury in chronic kidney diseases. Commun Med. (2025) 5:3. 10.1038/s43856-024-00708-3
5.
Ronneberger O Fischer P Brox T . U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015: 18th International Conference.Munich: Springer (2015). p. 234–241.
6.
Kaur G Garg M Gupta S Juneja S Rashid J Gupta D et al . Automatic identification of glomerular in whole-slide images using a modified UNet model. Diagnostics. (2023) 13:3152. 10.3390/diagnostics13193152
7.
Saikia FN Iwahori Y Suzuki T Bhuyan MK Wang A Kijsirikul B . MLP-UNet: Glomerulus segmentation. IEEE Access. (2023) 11:53034–47. 10.1109/ACCESS.2023.3280831
8.
Oghli MG Bagheri SM Shabanzadeh A Mehrjardi MZ Akhavan A Shiri I et al . Fully automated kidney image biomarker prediction in ultrasound scans using Fast-Unet++. Sci Rep. (2024) 14:4782. 10.1038/s41598-024-55106-5
9.
Nguyen TTU Nguyen AT Kim H Jung YJ Park W Kim KM et al . Deep-learning model for evaluating histopathology of acute renal tubular injury. Sci Rep. (2024) 14:9010. 10.1038/s41598-024-58506-9
10.
Chen C Miao J Wu D Zhong A Yan Z Kim S et al . Ma-sam: Modality-agnostic sam adaptation for 3D medical image segmentation. Med Image Anal. (2024) 98:103310. 10.1016/j.media.2024.103310
11.
Li X Deng R Tang Y Bao S Yang H Huo Y . Leverage weekly annotation to pixel-wise annotation via zero-shot segment anything model for molecular-empowered learning. In: Medical Imaging 2024: Digital and Computational Pathology. New York: SPIE (2024). p. 133–139.
12.
Ning X Ni Y Liang K Wang D Purwanto E Man KL . Customizing SAM for histologic image segmentation of kidney biopsy by a detector approach: det-SAM. In: 2023 International Conference on Cyber-Enabled Distributed Computing and Knowledge Discovery (CyberC). Jiangsu: IEEE (2023). p. 225–233.
13.
Gao Y Xia W Hu D Wang W Gao X . DeSAM: Decoupled Segment Anything Model for Generalizable Medical Image Segmentation. In:LinguraruMGDouQFeragenAGiannarouSGlockerBLekadirKetal., editors. Medical Image Computing and Computer Assisted Intervention-MICCAI 2024. Cham: Springer Nature Switzerland (2024). p. 509–19.
14.
Xiong X Wu Z Tan S Li W Tang F Chen Y et al . SAM2-UNet: Segment Anything 2 Makes Strong Encoder for Natural and Medical Image Segmentation. (2024). Available online at: https://arxiv.org/abs/2408.08870
15.
Elahi MT Lee W Phan P . SAttisUNet: UNet-like swin transformer with attentive skip connections for enhanced medical image segmentation. In: 2023 International Conference on Machine Learning and Applications (ICMLA). (2023). p. 1994–1999.
16.
Kuang Y Ma X Zhao J Wang G Zeng Y Liu S . A novel 3D medical image segmentation model using improved SAM. In: 2024 IEEE International Conference on Systems, Man, and Cybernetics (SMC)Kuching: IEEE. (2024). p. 2528–2534.
17.
Yang W Dong Z Xu M Xu L Geng D Li Y et al . Optimizing transformer-based network via advanced decoder design for medical image segmentation. Biomed Phys & Eng Express. (2025). 10.1088/2057-1976/adaec7
18.
Zhu X Li Y Wu Z Zhang T . AdaptFormer: adapter-based transformers for medical image segmentation. IEEE Trans Med Imaging. (2023) 42:1234–45.
19.
Li J Chen R Liu X Huang Y . Dual adapter networks for segmentation of abdominal organs in CT scans. Med Image Anal. (2023) 80:101568.
20.
Lin H Zou J Deng S Wong KP Aviles-Rivero AI Fan Y et al . Volumetric medical image segmentation via fully 3d adaptation of segment anything model. Biocybern Biomed Eng. (2025) 45:1–10. 10.1016/j.bbe.2024.11.001
21.
Yuan Y Zhang W Wei Z Xu C . Dynamic adapters for multi-class medical image segmentation. IEEE J Biomed Health Inform. (2023) 27:3456–68.
22.
Li X Davis RC Xu Y Wang Z Souma N Sotolongo G et al . Deep learning segmentation of glomeruli on kidney donor frozen sections. J Med Imaging. (2021) 8:067501. 10.1117/1.JMI.8.6.067501
23.
Zhou Z Siddiquee MM Tajbakhsh N Liang J Chang CH . UNet++: a nested u-net architecture for medical image segmentation. Med Image Anal. (2021) 67:101255.
24.
Hea C . Swin-Unet: unifying convolutional networks and transformers for medical image segmentation. Med Image Analy. (2021) 79:102406.
25.
Chen J Lu Y Yu Q Luo X Adeli E Wang Y et al . TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. (2021). Available online at: https://arxiv.org/abs/2102.04306
26.
Kingma DP . Adam: A method for stochastic optimization. arXiv [preprint] arXiv:14126980. (2014). 10.48550/arXiv.1412.6980
27.
Bueno G Fernandez-Carrobles MM Gonzalez-Lopez L Deniz O . Glomerulosclerosis identification in whole slide images using semantic segmentation. Comput Methods Programs Biomed. (2020) 184:105273. 10.1016/j.cmpb.2019.105273
28.
Altini N Cascarano GD Brunetti A Marino F Rocchetti MT Matino S et al . Semantic segmentation framework for glomeruli detection and classification in kidney histological sections. Electronics. (2020) 9:503. 10.3390/electronics9030503
29.
Zeng C Nan Y Xu F Lei Q Li F Chen T et al . Identification of glomerular lesions and intrinsic glomerular cell types in kidney diseases via deep learning. J Pathol. (2020) 252:53–64. 10.1002/path.5491
30.
Jha A Yang H Deng R Kapp ME Fogo AB Huo Y . Instance segmentation for whole slide imaging: end-to-end or detect-then-segment. J Med Imag. (2021) 8:014001. 10.1117/1.JMI.8.1.014001
31.
Li X Davis RC Xu Y Wang Z Souma N Sotolongo G et al . Deep learning segmentation of glomeruli on kidney donor frozen sections. J Med Imaging. (2021) 8:067501. 10.1117/1.JMI.8.6.067501
32.
Jiang L Chen W Dong B Mei K Zhu C Liu J et al . A deep learning-based approach for glomeruli instance segmentation from multistained renal biopsy pathologic images. Am J Pathol. (2021) 191:1431–41. 10.1016/j.ajpath.2021.05.004
33.
Gu Y Ruan R Yan Y Zhao J Sheng W Liang L et al . Glomerulus semantic segmentation using ensemble of deep learning models. Arab J Sci Eng. (2022) 47:14013–24. 10.1007/s13369-022-06608-9
34.
Yang CK Lee CY Wang HS Huang SC Liang PI Chen JS et al . Glomerular disease classification and lesion identification by machine learning. Biomed J. (2022) 45:675–85. 10.1016/j.bj.2021.08.011
35.
Silva J Souza L Chagas P Calumby R Souza B Pontes I et al . Boundary-aware glomerulus segmentation: toward one-to-many stain generalization. Comp Med Imag Graph. (2022) 100:102104. 10.1016/j.compmedimag.2022.102104
36.
Allender F Allègre R Wemmert C Dischler JM . Data augmentation based on spatial deformations for histopathology: an evaluation in the context of glomeruli segmentation. Comp Meth Prog Biomed. (2022) 221:106919. 10.1016/j.cmpb.2022.106919
37.
Deng R Liu Q Cui C Yao T Long J Asad Z et al . Omni-seg: a scale-aware dynamic network for renal pathological image segmentation. IEEE Trans Biomed Eng. (2023) 70:2636–44. 10.1109/TBME.2023.3260739
38.
Singh Samant S Chauhan A Dn J Singh V . Glomerulus detection using segmentation neural networks. J Digit Imaging. (2023) 36:1633–42. 10.1007/s10278-022-00764-y
39.
Vasiljević J Feuerhake F Wemmert C Lampert T . HistoStarGAN: A unified approach to stain normalisation, stain transfer and stain invariant segmentation in renal histopathology. Knowl-Based Syst. (2023) 277:110780. 10.1016/j.knosys.2023.110780
40.
Andreini P Bonechi S Dimitri GM . Enhancing glomeruli segmentation through cross-species pre-training. Neurocomputing. (2024) 563:126947. 10.1016/j.neucom.2023.126947
41.
Zhou C Liu X Liang S Lai Y Che M Li M et al . Swin Transformer based detection and segmentation networks for measurement and quantification analysis of arteriolar vessels from renal whole slide images. Biomed Signal Process Control. (2024) 96:106619. 10.1016/j.bspc.2024.106619
42.
Zhang I Denholm J Hamidinekoo A Ålund O Bagnall C Huix JP et al . Utilizing Weak-to-Strong Consistency for Semi-Supervised Glomeruli Segmentation. In: Medical Imaging with Deep Learning. (2024).
43.
Zhou Z Siddiquee MMR Tajbakhsh N Liang J . UNet++: a nested U-Net architecture for medical image segmentation. arXiv [preprint] arXiv:1807.10165. 10.48550/arXiv.1807.10165
44.
Isensee F Petersen J Klein A Zimmerer D Jaeger PF Kohl S et al . nnU-Net: self-adapting framework for u-net-based medical image segmentation. arXiv [preprint] arXiv:1809.10486. (2018). 10.48550/arXiv.1809.10486
45.
Huang Z Wang L Xu L . DRA-Net: Medical image segmentation based on adaptive feature extraction and region-level information fusion. Sci Rep. (2024) 14:9714. 10.1038/s41598-024-60475-y
46.
Sultan Y Wang Y Scanlon J D'lima L . Microvasculature segmentation in human biomolecular atlas program (HuBMAP). arXiv [preprint] arXiv:230803203. (2023). 10.48550/arXiv.2308.03203
47.
Hu F Deng R Bao S Yang H Huo Y . Multi-scale multi-site renal microvascular structures segmentation for whole slide imaging in renal pathology. In: Medical Imaging 2024: Digital and Computational Pathology. New York: SPIE (2024). p. 310–316.
48.
Shubham S Jain N Gupta V Mohan S Ariffin MM Ahmadian A . Identify glomeruli in human kidney tissue images using a deep learning approach. Soft Computing. (2023) 27:2705–16. 10.1007/s00500-021-06143-z
Summary
Keywords
V-SAM, deep learning, adapter layer, Segment Anything Model, point-based prompt
Citation
Wajeeh Us Sima M, Wang C, Arshad M, Shaikh JA, Alkhalaf S and Alturise F (2025) Leveraging advanced feature extraction for improved kidney biopsy segmentation. Front. Med. 12:1591999. doi: 10.3389/fmed.2025.1591999
Received
11 March 2025
Accepted
13 May 2025
Published
18 June 2025
Volume
12 - 2025
Edited by
Shuoyu Xu, Bio-totem Pte Ltd, China
Reviewed by
Hanxue Gu, Duke University, United States
Yifan Gao, University of Science and Technology of China, China
Updates
Copyright
© 2025 Wajeeh Us Sima, Wang, Arshad, Shaikh, Alkhalaf and Alturise.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengliang Wang wangcl@cqu.edu.cnSalem Alkhalaf s.alkhalaf@qu.edu.sa
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.