- 1College of Animal Science and Technology, Yunnan Agricultural University, Kunming, China
- 2College of Biology and Agriculture, Zunyi Normal University, Zunyi, China
Bile acids play a dual role by aiding lipid absorption and acting as signaling molecules by interacting with various receptors. Bile acids are perpetually recycled via enterohepatic circulation and are biotransformation by gut microbiota, making bile acid metabolism a critical regulator of intestinal homeostasis. The intestinal epithelium prominently expresses two key bile acid receptors - the farnesoid X receptor (FXR) and G protein-coupled bile acid receptor 1 (TGR5) - which play indispensable roles in maintaining bile acid homeostasis and intestinal barrier function. Due to the abundant expression of bile acid receptors and the importance of the intestine in preventing pathogen invasion, researchers are increasingly focused on the function of bile acids in this system. This article focuses on the effect of bile acids and their receptors, FXR and the TGR5, in modulating intestinal barrier function.
1 Introduction
The core structure of bile acids (BAs) consists of 17 carbon atoms arranged in three hexane rings and one pentane ring (1). BAs possess both hydrophilic and hydrophobic regions, enabling them to form mixed micelles with lipids and their digestive products. This character allows cholesterol and other lipophilic substances to dissolve in bile, promoting the emulsification and absorption of fats and fat-soluble vitamins, while also regulating cholesterol stability (2, 3). Furthermore, BAs are recognized as important signaling molecules that play a role in regulating lipid metabolism, glucose metabolism, and energy metabolism, exhibiting hormone-like functions (4). Moreover, bile acids play a crucial role in the intestinal barrier. Different bile acids have varying effects on intestinal epithelial cells. Lithocholic acid (LCA) and deoxycholic acid (DCA) may promote epithelial renewal by inducing programmed cell death, while ursodeoxycholic acid (UDCA) may protect epithelial cells (5–7). The role of BAs is also crucial in inflammatory bowel diseases (IBDs). Research has shown that patients with Crohn’s disease have a smaller bile acid pool, and the ratio of glycine to taurine complexes significantly increases. The decrease in bile acid pool size is related to disease activity (8). The deficiency of secondary BAs produced by gut microbiota can promote intestinal inflammation, while DCA and LCA can alleviate inflammation in mouse colitis models (9).
Given the high expression levels of bile acid receptors and the gut’s critical role in preventing pathogen invasion, the significance of BAs in the gut garners increasing attention from researchers (10). This review aims to elucidate the functions of bile acid metabolism and its intricate relationship with the intestinal barrier, detailing the regulatory mechanisms by which BAs and their primary receptors, FXR and TGR5, influence the mechanical, mucosal, microbial, and immune barriers of the intestine.
2 BAs metabolism
In the liver, BAs are synthesized from cholesterol through the action of at least 17 enzymes (11, 12). There are two primary pathways for BA synthesis: the classical (neutral) pathway and the alternative (acidic) pathway. The classical pathway is facilitated by cholesterol 7 α-hydroxylase (CYP7A1), resulting in the production of cholic acid (CA) and chenodeoxycholic acid (CDCA). Conversely, the alternative pathway is mediated by sterol 27-hydroxylase (CYP27A1), which exclusively produces CDCA. The BAs generated through these pathways are referred to as primary BAs (13). In the liver, primary BAs can conjugate with glycine or taurine at the C-24 position to form conjugated BAs (14).
Bile acids synthesized in the liver are secreted into the bile duct and stored in the gallbladder, facilitated by the bile salt export pump (BSEP) and the phosphatidylcholine transporter (ABCB4). Upon food intake, cholecystokinin released from the duodenum triggers gallbladder contraction, leading to the release of bile into the intestine (15–17). As BAs traverse the intestine, a small fraction of unbound BAs is reabsorbed through passive diffusion, while the majority is actively absorbed via the apical sodium-dependent bile acid transporter (ASBT) in the distal ileum, subsequently entering the liver through the portal vein (13, 18). The reabsorbed and primary BAs undergo further processing in the liver before being secreted back into the gallbladder and re-entering the intestine, thereby establishing the enterohepatic circulation of BAs.
A portion of the BAs that are not absorbed is converted into secondary BAs through microbial action in the distal ileum and colon. For instance, microbe with bile salt hydrolase (BSH) activity deconjugate CA and CDCA, yielding unconjugated primary BAs, which are then transformed into secondary BAs via microbial 7-dehydroxylation (19, 20). Deoxycholic acid (DCA), derived from CA, is reabsorbed and returned to the liver from the colon, while lithocholic acid (LCA), produced from CDCA, poses a health risk; only a small amount of LCA is transported to the liver for detoxification through sulfation before entering the bile, with the majority excreted in feces (21).
The bile acid pool contains approximately 3 g of BAs, which circulate 6–15 times daily, with about 0.2–0.5 g excreted in feces (22). This loss of BAs is compensated by the de novo synthesis, ensuring the stability of the bile acid pool (23). Furthermore, the excretion of BAs in feces serves as a primary mechanism for the body to eliminate cholesterol (24).
3 BA receptors
Bile acid receptors can be classified into two main types: nuclear receptors and membrane receptors. The nuclear receptors encompass FXR (25, 26), vitamin D receptor (VDR) (27), pregnane X receptor (PXR) (28), and constitutive androstane receptor (CAR) (29). In contrast, the membrane receptors include the G protein-coupled receptor 1 (TGR5) (7) and sphingosine 1-phosphate receptor 2 (S1PR2) (30). Among these, the nuclear receptor FXR and the membrane receptor TGR5 are the most thoroughly investigated receptors to date.
3.1 The nuclear receptor FXR
The nuclear receptor FXR was the first bile acid receptor to be identified (25) and is predominantly expressed in tissues that participate in the hepatic-intestinal circulation of BAs (12). FXR can be activated by BAs to exert its biological effects, although its activation potential varies among different BAs. Hydrophilic BAs, such as UDCA does not activate FXR and Muricholic acids (MCA) are known to antagonize FXR, whereas hydrophobic BAs activate FXR in the following order: CDCA > LCA = DCA > CA (31, 32). Upon activation, FXR regulates the expression of various genes either as monomers or in heterodimeric complexes by binding to the retinoic acid X receptor (RXR) on DNA (33, 34). FXR serves as a sensor for intracellular bile acid levels and plays a crucial role in maintaining bile acid homeostasis (Figure 1).
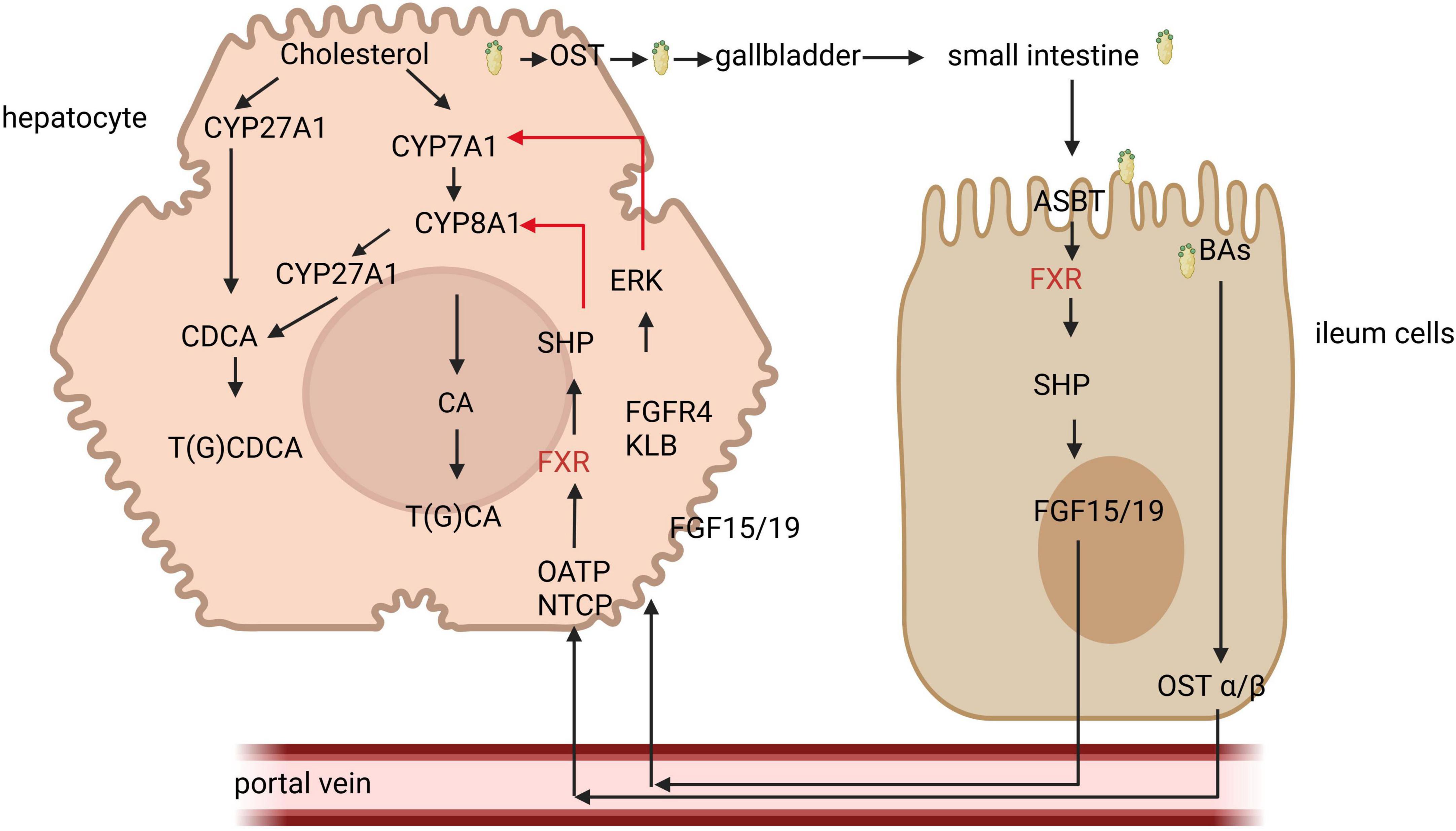
Figure 1. The role of farnesoid X receptor (FXR) signal in bile acid regulation. Bile acids (Bas) are synthesized from cholesterol via both classical and alternative pathways. The classical pathway is regulated by the key enzyme CYP7A1, whereas the alternative pathway is facilitated by CYP27A1. The primary BAs produced are stored in the gallbladder. Upon ingestion of food, the gallbladder contracts, releasing BAs into the intestine. These BAs are then absorbed in the distal ileum and transported back to the liver through the portal vein. Within liver cells, BAs activate FXR receptors, which in turn promote the expression of small hetero-dimer partner (SHP), resulting in the inhibition of CYP8A1. Additionally, BAs that enter intestinal cells activate FXR, leading to the secretion of FGF15/19, which subsequently travels to the liver via the portal vein to inhibit CYP7A1, thereby reducing bile acid synthesis. NTCP, sodium taurocholate co-transporting polypeptide; FGF, fibroblast growth factor; ERK; extracellular signal-regulated kinases.
In the intestine, BAs enter intestinal cells through the action of the apical sodium-dependent bile acid transporter (ASBT) and subsequently bind to the ileal bile acid binding protein (IBABP), facilitating their transport from the apical surface to the basolateral membrane. With the assistance of the organic solute transporter (OST) and organic anion transporting polypeptides (OATP), BAs then enter the portal vein and are transported to the liver. In the liver, they bind to the fibroblast growth factor receptor 4 (FGFR4) and β Klotho (KLB), leading to the downregulation of the rate-limiting enzyme CYP7A1 in the classical pathway of the extracellular signal regulated kinase (ERK), thereby inhibiting bile acid synthesis (35).
In liver cells, BAs stimulate FXR, which induces the expression of the small hetero-dimer partner (SHP) and inhibits the expression of the key enzyme CYP8B1 (sterol 12α-hydroxylase) involved in bile acid synthesis, thus regulating bile acid metabolism by modulating genes associated with bile acid secretion (36). The activation of FXR can also improve liver disease. In patients with non-alcoholic fatty liver disease (NAFLD) or in mouse models, treatment with FXR agonist obeticholic acid (OCA) produced a series of related liver effects, reducing triglycerides (TAGs) and inflammation, and alleviating steatohepatitis and liver fibrosis (37). FXR also influences molecules related to bile acid transport, with IBABP exhibiting a high affinity for BAs. In intestinal epithelial cells, FXR can regulate bile acid homeostasis by modulating the IBABP gene (38). Additionally, FXR is implicated in intestinal immune regulation and the maintenance of intestinal barrier function (39, 40). Activation of FXR has been shown to mitigate inflammation in animal models of inflammatory bowel disease (IBD), alleviate colitis symptoms, protect the intestinal epithelial barrier, and reduce the loss of goblet cells (41).
3.2 The membrane receptor TGR5
G protein-coupled bile acid receptor 1 is a significant bile acid membrane receptor that is predominantly expressed in the gallbladder, ileum, and colon (42–44). In the liver, TGR5 regulates microcirculation, inflammation, regeneration, bile secretion and proliferation, as well as gallbladder filling (45). TGR5 has also been identified as a negative regulator of liver inflammation. Mice lacking TGR5 are more susceptible to liver injury after intraperitoneal injection of lipopolysaccharide, leading to increased levels of inflammatory cytokines and enhanced liver cell apoptosis (46). The potency of BAs in activating TGR5 follows the hierarchy: LCA > DCA > CDCA > CA (47). Upon binding with BAs, TGR5 triggers the release of a complex comprising G proteins—αs, β, and γ. This interaction facilitates the exchange of GDP for GTP within the G protein complex, resulting in the dissociation of the complex and the formation of G protein - αs and β - γ dimers. The G protein - αs subunit activates adenylate cyclase, which in turn promotes the synthesis of cyclic adenosine monophosphate (cAMP) and the activation of protein kinase A (PKA). This cascade initiates downstream signaling pathways, while cAMP production also influences energy and glucose metabolism (48). BAs regulate metabolic processes differently across various tissue types (Figure 2). For instance, the elevation of cAMP levels in brown adipocytes, induced by bile acid treatment, enhances the activity and oxygen consumption of type 2 iodothyronine deiodinase (D2), thereby contributing to the regulation of energy homeostasis (49). Additionally, glucagon-like peptide-1 (GLP-1) can lower blood glucose levels, and BAs in the intestine stimulate GLP-1 secretion from intestinal endo-crine cells via TGR5, thus impacting glucose metabolism (50). Furthermore, TGR5 activation in pancreatic beta cells promotes insulin secretion through the G-αs/cAMP/Ca2+ signaling pathway (51). TGR5 also plays a crucial role in maintaining intestinal barrier function; in intestinal cells, TGR5 activation by BAs stimulates myosin light chain kinase (MLCK) signaling, thereby enhancing intestinal barrier protection (52) (Figure 2). Moreover, TGR5 is involved in the regulation of bile acid metabolism, as it modulates the expression of cholesterol 12α-hydroxylase (CYP8B1), a key enzyme in bile acid synthesis, leading to a reduction in the proportion of 12α-hydroxy BAs in the bile acid pool (53).
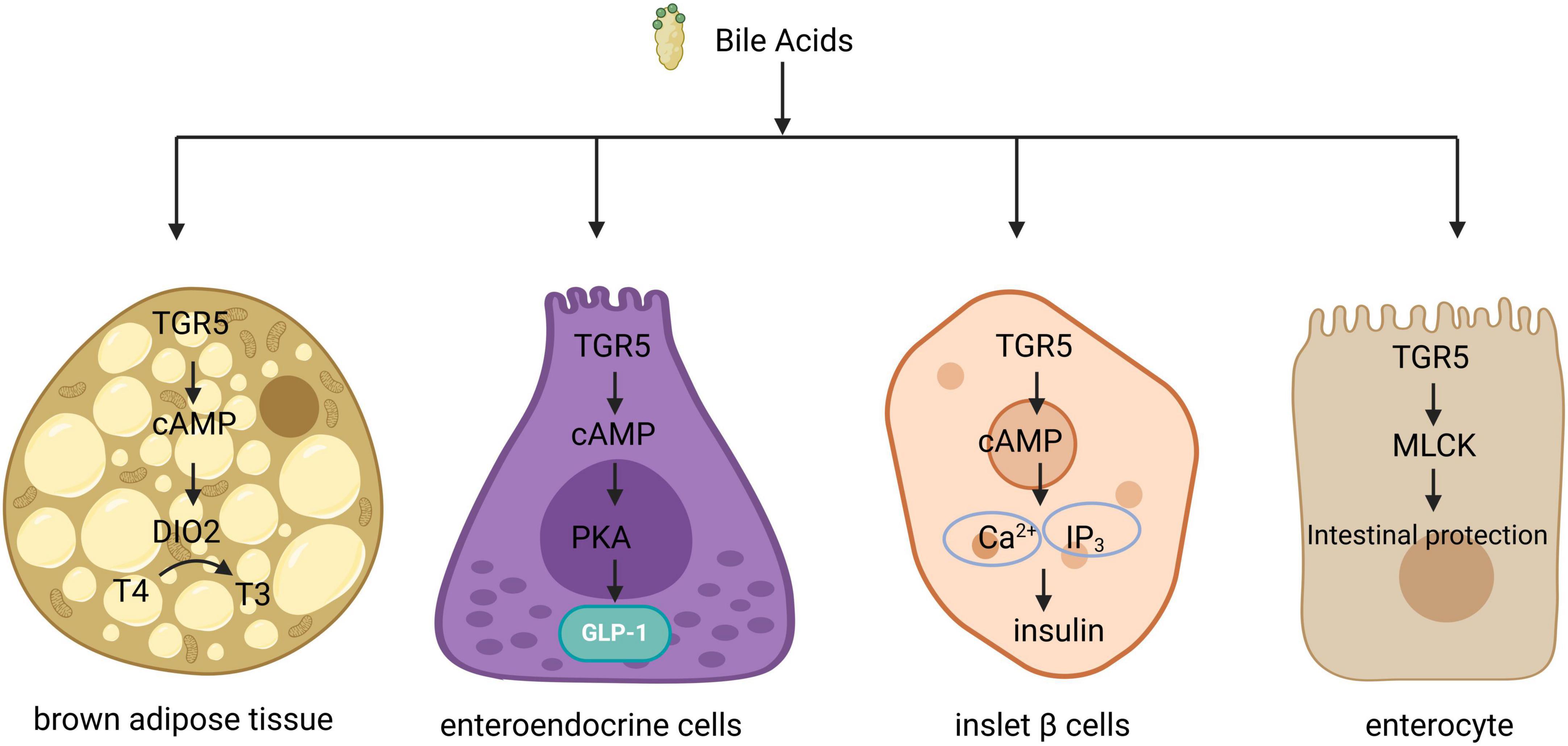
Figure 2. Signal transduction of TGR5 in different organizations. In brown adipose tissue, TGR5 activation facilitates the conversion of thyroid hormone T4 to T3 by inducing type 2 deiodinase, thereby enhancing energy metabolism. In intestinal endocrine cells, TGR5 activation triggers the activation of protein kinase A (PKA), which subsequently promotes the secretion of glucagon-like peptide-1 (GLP-1). In pancreatic beta cells, TGR5 activation results in elevated levels of intracellular cyclic adenosine monophosphate (cAMP) and Ca2+, leading to an increase in insulin secretion. Additionally, in intestinal epithelial cells, TGR5 activation stimulates the MLCK signaling pathway, contributing to epithelial protection. TGR5, G protein-coupled bile acid receptor 1.
4 The role of BAs in the intestinal barrier
4.1 Mechanical barriers
The intestinal epithelium consists of a single layer of columnar cells approximately 20 μm thick, and the epithelial layer not only comprises intestinal epithelial cells but also includes secretory cells such as goblet cells, Paneth cells, and intestinal endocrine cells (54). Within the intact epithelial layer, tight junctions (TJs) and adherens junctions (AJs) serve as critical cellular connections that contribute to the mechanical barrier of the intestine (55, 56). TJs are intercellular connections located at the apical region of cell contact, playing a vital role in determining epithelial permeability and regulating paracellular transport pathways (57). They are essential connections between epithelial cells, formed by proteins such as claudins, occludin, and junctional adhesion molecules (JAMs), which effectively prevent the invasion of intestinal bacteria and toxins (54). Alterations in the expression, post-translational modifications, localization, or activity of tight junction proteins or their regulatory factors can influence the permeability to larger molecules (58). Damage to epithelial cells disrupts the intestinal barrier, resulting in a loss of barrier function and allowing substances from the intestinal lumen to penetrate the submucosal layer (54). AJs, located on the lateral membranes of epithelial cells beneath the TJs, primarily function to maintain intercellular contact, cell polarity, motility, and proliferation (56). The structure of adherens junctions mainly consists of transmembrane glycoproteins from the classical cadherin superfamily (such as E-cadherin) and members of the catenin family (including p120 catenin, β-catenin, and α-catenin), which collectively regulate the formation, maintenance, and functionality of these connections (59). Additionally, adherens junctions provide mechanical strength to adjacent epithelial cells (60).
Bile acids and their receptors are intricately linked to the mechanical barrier function of the intestine (Figure 3). Research indicates that BAs can enhance epithelial regeneration by activating the membrane receptor TGR5 in intestinal stem cells (ISCs). It has been demonstrated that the release of endogenous BAs in the intestinal lumen is sufficient to coordinate the renewal and proliferation of ISCs (7). Chenodeoxycholic acid (CDCA) has been shown to promote the proliferation of piglet jejunal epithelial cells (IPEC-J2), accelerate cell cycle progression in the S and G2/M phases, improve mitochondrial function, reduce intracellular reactive oxygen species (ROS) production in IPEC-J2 cells (61), thereby exerting a beneficial effect on these cells and protecting intestinal epithelial barrier function from lipopolysaccharide-induced damage via the FXR-MLCK pathway (62). In vitro, studies have revealed that CDCA, deoxycholic acid (DCA), and cholic acid (CA) can induce a transient reduction in transepithelial resistance in Caco-2 cells and modulate intestinal permeability by promoting the self-phosphorylation of epidermal growth factor (EGF) receptors, dephosphorylation of occludin, and rearrangement of tight junctions (63). Lithocholic acid (LCA), on the other hand, can downregulate the expression of tight junction proteins and genes in IPEC-J2 cells (64) and induce apoptosis in IPEC-J2 cells by activating CD95 clusters and caspase 8 (65), suggesting a detrimental effect of LCA on the intestinal barrier (66). The receptors TGR5 and FXR for BAs have also been identified as crucial in the regulation of intestinal barrier function. Tauroursodeoxycholic acid (TUDCA) has been shown to ameliorate epithelial barrier damage induced by Escherichia coli in IPEC-J2 cells through TGR5 activation (52) and to reverse the decrease in mRNA expression of ZO-1 (zonula occludens 1), JAM, occludin, and claudin-4 in the mouse intestine (66). FXR in the ileum regulates Angiopoietins 1, Inducible Nitric Oxide Synthase, and Interleukin-18, ensuring adequate intestinal protection during periods of heightened microbial exposure while preventing excessive protein production that could lead to inflammation and intestinal diseases (40).
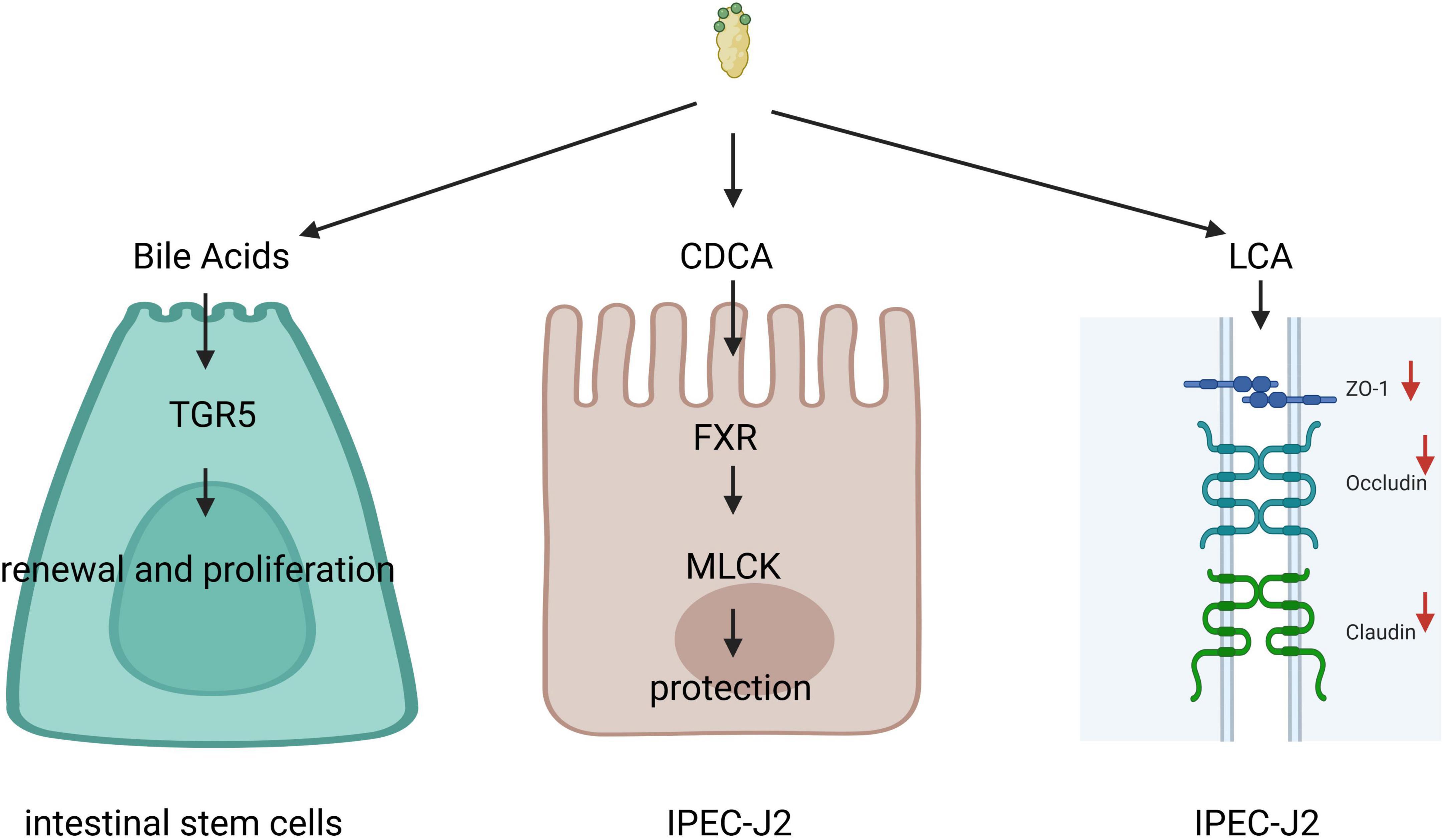
Figure 3. Bile acids (Bas) and their receptors are intricately linked to the mechanical barrier function of the intestine. Bile acids can promote epithelial regeneration by activating the membrane receptor G protein-coupled bile acid receptor 1 (TGR5) in intestinal stem cells (ISCs), chenodeoxycholic acid (CDCA) can protect intestinal epithelial barrier function through the farnesoid X receptor (FXR)-myosin light chain kinase (MLCK) pathway. Lithocholic acid (LCA) can downregulate the expression of tight junction proteins and genes in IPEC-J2.
4.2 Mucosal barrier
The intestinal epithelium is covered by a mucus layer secreted by goblet cells, which comprises water, phospholipids, mucins (MUC), secreted immunoglobulin A (IgA), antimicrobial peptides, and various defense factors (54). This mucus layer plays a crucial role in preventing the adhesion and invasion of microorganisms and harmful substances, shielding epithelial cells from physical abrasion, and facilitating epithelial renewal and differentiation (67, 68). The mucus layer is structured into two distinct layers: an inner mucus layer that is tightly bound to the epithelial cells, creating a barrier that prevents bacterial penetration and maintains a sterile environment at the epithelial surface, and an outer mucus layer that is more loosely associated and provides a habitat for gut microbiota (69, 70). Notably, the inner mucus layer in the small intestine is thinner than that in the large intestine, likely to optimize nutrient absorption (71). Mucin, the primary component of the mucus layer, is essential for protecting the intestine from microbial infections (72). The human mucin family consists of 24 members (MUC1 to MUC24), categorized into secreted mucins and transmembrane mucins. Transmembrane mucins include MUC1, MUC3A/B, MUC4, MUC13, MUC15-17, MUC20, and MUC21, while secreted mucins encompass MUC2, MUC5AC/B, MUC6, MUC7, and MUC19 (73). Impairments in mucin production can compromise intestinal barrier function and contribute to the development of intestinal-related diseases, such as inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), and cancer (56).
The mucus produced by goblet cells, particularly the mucin component, is crucial for protecting the intestinal mucosa (74, 75). The secretion of mucus is regulated by endoplasmic reticulum stress and autophagy. TUDCA has been shown to alleviate endoplasmic reticulum stress, stimulate the secretion of mucus from goblet cells, increase the thickness of mucus layers, and bolster the protective function of chemical barriers (76). Chenodeoxycholic acid (CDCA) activates FXR in normal rat gastric epithelial cells in a dose-dependent manner, leading to an upregulation of MUC2 protein expression and an enhancement of the chemical barrier (77). Research indicates that BAs can activate the FXR/nuclear transcription factor-κB (NF-κB) signaling pathway, which contributes to the promotion of MUC2 expression (78). Conversely, BAs can also compromise chemical barriers; for instance, deoxycholic acid (DCA) induces endoplasmic reticulum stress in the intestinal mucosa in vitro and exerts toxic effects on goblet cells, resulting in damage and a diminished protective capacity of chemical barriers (79). Additionally, bile acid receptors are significant in maintaining chemical barriers. Evidence suggests that the knockout or inhibition of the bile acid re-ceptor FXR leads to a reduction in MUC2 levels in mouse intestinal organoids, but the application of the FXR agonist GW4064 to activate FXR in mice has been found to mitigate radiation Ninduced intestinal damage (80). However, the specific knockout of FXR in the mouse liver appears to enhance colonic mucosal barrier function (81).
4.3 Microbial barriers
The gut microbial community comprises bacteria, fungi, viruses, archaea, and protozoa, with bacteria constituting over 70% of this population (82, 83). The majority of gut bacteria belong to two primary phyla: Bacteroidetes and Firmicutes (84). These intestinal bacteria play a crucial role in enhancing the host’s digestive efficiency by breaking down dietary polysaccharides into absorbable compounds, such as short-chain fatty acids (85). Additionally, they provide protection against pathogenic infections. Firstly, pathogenic bacteria face competition for nutrients from the gut microbiota, which restricts their ability to colonize the gut (86, 87). Secondly, the gut microbiota can stimulate the host’s immune response, thereby preventing pathogen invasion. For instance, the immune response triggered by symbiotic bacteria activating epithelial Toll-like receptors (TLRs) limits the proliferation of Salmonella typhimurium serotypes (88). In sterile mice, a reduced proliferation rate of intestinal epithelial cells, shorter crypt-villus length, and diminished angiogenesis have been observed; however, short-term supplementation with specific bacteria has demonstrated a proliferative effect in these mice, suggesting that a normal microbiota can stimulate cryptosystem cell activity, thereby influencing the proliferation rates in the colon and small intestine (89). Furthermore, the microbiota contributes to the integrity of the epithelial barrier. For example, butyrate produced by intestinal bacteria promotes wound healing by enhancing oxygen utilization, boosting anaerobic conditions, and activating compensatory hypoxia inducible factors (HIF), ultimately improving barrier function (90). Additionally, deoxycholic acid (DCA) generated by bacteria can facilitate mucosal wound healing by modulating local Prostaglandin E2 levels (91).
Primary BAs are transformed into secondary BAs by intestinal microorganisms. While these microorganisms metabolize BAs, the BAs simultaneously influence the gut microbiota (92). CA can inhibit bacterial growth by causing membrane damage. As the concentration of CA increases, the internal pH level of the bacteria gradually decreases, ultimately leading to the complete dissipation of both pH and transmembrane potential. Furthermore, potassium ion leakage was observed when the CA concentration exceeded 2 mM, and an increase in the leakage of other cellular components was noted when the CA concentration surpassed 4 mM (93). Additionally, bile acid salts that penetrate bacterial cells may inflict damage on bacterial nucleic acids, triggering SOS responses and causing oxidative damage to DNA, thereby hindering bacterial reproduction (94, 95). The antibacterial efficacy of various BAs differs, with bound BAs exhibiting lower antibacterial activity compared to their unbound counter-parts (96). This discrepancy may arise from the ability of unbound BAs to passively diffuse across membranes, resulting in intracellular toxicity, whereas bound BAs are fully ionized at physiological pH and only exert toxicity when specific transport proteins facilitate their entry into the cell (97). Furthermore, bacterial tolerance to BAs varies among species. The distinct structural characteristics of cell walls in Gram-negative bacteria allow them to exclude antibiotics and other antibacterial agents, potentially resulting in greater resistance to BAs (98). In contrast, Gram-positive bacteria tend to be more susceptible to bile action (97). For instance, Gram-negative bacteria such as Salmonella and Escherichia coli can thrive in the gallbladder, where bile concentrations are extremely high (99), while the growth of Lactobacillus acidophilus, a Gram-positive bacterium, is significantly inhibited in the presence of BAs (100).
4.4 Immune barrier
The mucosa associated lymphoid tissue (MALT) located in the intestinal lamina propria represents the largest immune organ in the human body, playing a pivotal role in the intestinal immune response (101). MALT is primarily divided into two components: the induction site and the effector site. The induction site comprises Peyer’s patches (PP), mesenteric lymph nodes (MLN), and isolated lymphoid follicles (ILF), which are crucial for the initial activation and differentiation of immune cells (102). The effector sites include the intestinal lamina propria and epithelium, which are essential for maintaining the integrity of immune cells and barriers (103). Immune cells are categorized into innate immune cells (such as innate lymphocytes, dendritic cells (DCs), macrophages, and natural killer cells) and adaptive immune cells (including B and T cells) (104). DCs and macrophages play a key role in recognizing antigens and presenting them to lymph nodes, thereby facilitating the generation of specific T and B cells. These specific T and B cells subsequently migrate back to the mucosal lamina propria to act as effector cells or to persist as memory cells (105). T cells are involved in cellular immunity, while immunoglobulin A (IgA) secreted by B cells serves as the primary antibody in the intestine, predominantly in the form of Secretory Immunoglobulin A (SIgA). SIgA functions by obstructing the entry of antigens, microorganisms, and exogenous proteins at the intestinal surface (102). Typically, SIgA is produced in response to microbial stimulation (103) and is present throughout the entire intestine (106). Upon re-infection, the titer of IgA antibodies rises significantly more rapidly and remains elevated for an extended duration, thereby enhancing the body’s ability to combat infections (102).
Bile acids play a crucial role in regulating intestinal mucosal homeostasis and the inflammatory response through various receptors, including FXR and TGR5, along with their associated signaling pathways (107). Studies have demonstrated that treatment with INT-747, an FXR agonist, in mice with colitis results in a reduction of pro-inflammatory cytokines such as Interleukin-1 beta (IL-1β) and Interleukin-6 (IL-6), as well as chemo-kines like C-C motif ligand 2 (41). Activation of FXR inhibits NF-κB activity by preventing the clearance of nuclear helper receptors at the binding sites for tumor necrosis factor (TNF) and IL-1β (108). In vitro studies indicate that FXR activation can enhance the release of proinflammatory cytokines, while Caco-2 cells treated with FXR antagonists show a significant decrease in the secretion of IL-6 and TNF (109). In vivo, FXR activation has been associated with reduced levels of TNF in models of dextran sulfate sodium (DSS) colitis (110). Furthermore, the knockout of the FXR receptor alleviates intestinal barrier dysfunction induced by lipopolysaccharide (LPS) damage and mitigates inflammatory injury. This protective effect is attributed to the decreased production of inflammatory cytokines, which aids in maintaining the integrity of tight junctions (111). Additionally, BAs can promote the differentiation of monocytes into DCs that secrete low levels of IL-12 and TNF-α via the TGR5-cAMP pathway (112), while also inhibiting the activation of the NOD-like receptor thermal protein domain associated protein 3 (NLRP3) inflammasome through the TGR5-cAMP-PKA axis (113), underscoring the significant role of BAs in modulating inflammatory responses. Research has found that the levels of pro-inflammatory cytokines (IL-1 β and TNF α) in the colon of low birth weight (LBW) animals are elevated and UDCA is significantly reduced. However, after supplementing UDCA, UDCA can induce M2 polarization of macrophages, inhibit NF - κ B, and exert anti-inflammatory effects in the intestine (114). BAs can also regulate the differentiation of T lymphocytes, which to some extent affects the homeostasis of the intestinal immune barrier. T helper cells expressing interleukin-17A (Th17 cells) help resist extracellular pathogens, while secondary BAs isodeoxycholic acid (isoDCA) can inhibit Th17 cell differentiation by suppressing ROR γ t (retinal acid receptor related nuclear receptor γ t), which may be closely related to IBD (115).
In general, the intestine serves not only as a crucial digestive organ but also functions as a barrier that separates the luminal contents from the body’s internal environment through the intestinal mucosa. Acting as a physical, biochemical, and immune barrier, it engages with the external environment to safeguard the internal milieu from harmful substances. While certain BAs may exert detrimental effects on the gut, they are vital for preserving the integrity of the gut barrier. BAs play a significant role in cell proliferation and apoptosis, regulate both mucosal and mechanical barriers, inhibit the growth of specific harmful bacteria, and are involved in the immune response within the gut.
5 Conclusion
Since the identification of the bile acid receptor FXR, there has been a growing interest in the role of BAs as signaling molecules that influence cell growth and immune responses. The interaction between BAs and their key receptors, FXR and TGR5, along with the intestinal barrier, is crucial for maintaining intestinal barrier integrity. This article reviews the functions of bile acid metabolism and the intestinal barrier, detailing the regulatory mechanisms by which BAs and their primary receptors, FXR and TGR5, influence the mechanical, mucosal, microbial, and immune barriers of the intestine. This underscores the significant role of BAs and their receptors in preserving intestinal barrier homeostasis. Nonetheless, further investigation is needed to enhance our understanding of the intricate relationship between BAs and the intestinal barrier, clarify their potential mechanisms of action, and identify associated risks, ultimately aiming to develop more effective treatment strategies for intestinal diseases.
Author contributions
GS: Conceptualization, Writing – original draft, Writing – review and editing. YX: Conceptualization, Writing – original draft, Writing – review and editing. LY: Writing – review and editing. WC: Writing – review and editing. HJ: Writing – review and editing. WS: Visualization, Writing – review and editing. QL: Visualization, Writing – review and editing. LF: Supervision, Writing – review and editing. SX: Project administration, Writing – review and editing. DL: Project administration, Writing – review and editing. JZ: Writing – review and editing, Writing – original draft. SZ: Funding acquisition, Supervision, Writing – review and editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. National Natural Science Foundation of China (32360808; 31760645; 31260592; 31060331), Key Science and Technology Project of Yunnan Province (202202AE090032), Sci-Tech Service Station of Farmer Academician in Fuyuan County, and the National Key R&D Program of China (2024YFD1800404).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Fiorucci S, Distrutti E, Carino A, Zampella A, Biagioli M. Bile acids and their receptors in metabolic disorders. Prog Lipid Res. (2021) 82:101094. doi: 10.1016/j.plipres.2021.101094
2. Hofmann A, Mysels K. Bile salts as biological surfactants. Colloids Surf. (1987) 30:145–73. doi: 10.1016/0166-6622(87)80207-x
3. Monte M, Marin J, Antelo A, Vazquez-Tato J. Bile acids: Chemistry, physiology, and pathophysiology. WJG. (2009) 15:804–804. doi: 10.3748/wjg.15.804
4. Kuipers F, Bloks V, Groen A. Beyond intestinal soap–bile acids in metabolic control. Nat Rev Endocrinol. (2014) 10:488–98. doi: 10.1038/nrendo.2014.60
5. Zhang H, Xu H, Zhang C, Tang Q, Bi F. Ursodeoxycholic acid suppresses the malignant progression of colorectal cancer through TGR5-YAP axis. Cell Death Discov. (2021) 7:207. doi: 10.1038/s41420-021-00589-8
6. Barrasa J, Olmo N, Lizarbe M, Turnay J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol In Vitro. (2012) 27:964–77. doi: 10.1016/j.tiv.2012.12.020
7. Sorrentino G, Perino A, Yildiz E, El Alam G, Bou Sleiman M, Gioiello A, et al. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology. (2020) 159:956–968.e8. doi: 10.1053/j.gastro.2020.05.067
8. Vantrappen G, Ghoos Y, Rutgeerts P, Janssens J. Bile acid studies in uncomplicated Crohn’s disease. Gut. (1977) 18:730–5. doi: 10.1136/gut.18.9.730
9. Sinha S, Haileselassie Y, Nguyen L, Tropini C, Wang M, Becker L, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. (2020) 27:659–670.e5. doi: 10.1016/j.chom.2020.01.021
10. Shi L, Jin L, Huang W. Bile acids, intestinal barrier dysfunction, and related diseases. Cells. (2023) 12:1888–1888. doi: 10.3390/cells12141888
11. Hylemon P, Harder J. Biotransformation of monoterpenes, bile acids, and other isoprenoids in anaerobic ecosystems. FEMS Microbiol Rev. (1998) 22:475–88. doi: 10.1111/j.1574-6976.1998.tb00382.x
12. Hofmann A, Hagey L. Bile acids: Chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol Life Sci. (2008) 65:2461–83. doi: 10.1007/s00018-008-7568-6
13. Wahlström A, Sayin S, Marschall H, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. (2016) 24:41–50. doi: 10.1016/j.cmet.2016.05.005
14. Boland C. When bile acids don’t get amidated. Gastroenterology. (2013) 144:868–70. doi: 10.1053/j.gastro.2013.03.014
15. Hofmann A. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med. (1999) 159:2647. doi: 10.1001/archinte.159.22.2647
16. Kullak-ublick G, Stieger B, Meier P. Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology. (2003) 126:322–42. doi: 10.1053/j.gastro.2003.06.005
17. Trauner M, Boyer J. Bile salt transporters: Molecular characterization, function, and regulation. Physiol Rev. (2003) 83:633–71. doi: 10.1152/physrev.00027.2002
18. Chiang J. Bile acid metabolism and signaling. Comp Physiol. (2013) 3:1191–212. doi: 10.1002/cphy.c120023
19. Jones B, Begley M, Hill C, Gahan C, Marchesi J. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. (2008) 105:13580–5. doi: 10.1073/pnas.0804437105
20. Ridlon J, Kang D, Hylemon P. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. (2005) 47:241–59. doi: 10.1194/jlr.r500013-jlr200
21. Hofmann A. Detoxification of lithocholic acid. A toxic bile acid: Relevance to drug hepatotoxicity. Drug Metab Rev. (2004) 36:703–22. doi: 10.1081/dmr-200033475
22. Dawson P, Karpen S. Intestinal transport and metabolism of bile acids. J Lipid Res. (2014) 56:1085–99. doi: 10.1194/jlr.r054114
23. Bustos A, Font de Valdez G, Fadda S, Taranto MP. New insights into bacterial bile resistance mechanisms: The role of bile salt hydrolase and its impact on human health. Food Res Int. (2018) 112:250–62. doi: 10.1016/j.foodres.2018.06.035
24. Dietschy J, Turley S. Control of cholesterol turnover in the mouse. J Biol Chem. (2002) 277:3801–4. doi: 10.1074/jbc.r100057200
25. Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, et al. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun. (2002) 298:714–9. doi: 10.1016/s0006-291x(02)02550-0
26. Makishima M, Okamoto A, Repa J, Tu H, Learned R, Luk A, et al. Identification of a nuclear receptor for bile acids. Science. (1999) 284:1362–5. doi: 10.1126/science.284.5418.1362
27. Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. (2002) 296:1313–6. doi: 10.1126/science.1070477
28. Goodwin B, Gauthier K, Umetani M, Watson M, Lochansky M, Collins J, et al. Identification of bile acid precursors as endogenous ligands for the nuclear xenobiotic pregnane X receptor. Proc Natl Acad Sci USA. (2002) 100:223–8. doi: 10.1073/pnas.0237082100
29. Zhang J, Huang W, Qatanani M, Evans R, Moore D. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem. (2004) 279:49517–22. doi: 10.1074/jbc.m409041200
30. Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. (2011) 55:267–76. doi: 10.1002/hep.24681
31. Jiang C, Xie C, Lv Y, Li J, Krausz K, Shi J, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. (2015) 6:10166. doi: 10.1038/ncomms10166
32. Bookout A, Jeong Y, Downes M, Yu R, Evans R, Mangelsdorf D. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. (2006) 126:789–99. doi: 10.1016/j.cell.2006.06.049
33. Zhang Y, Edwards PA. FXR signaling in metabolic disease. FEBS Lett. (2007) 582:10–8. doi: 10.1016/j.febslet.2007.11.015
34. Parks D, Blanchard S, Bledsoe R, Chandra G. Bile acids: Natural ligands for an orphan nuclear receptor. Science. (1999) 284:1365–8. doi: 10.1126/science.284.5418.1365
35. Inagaki T, Choi M, Moschetta A, Peng L, Cummins C, McDonald J, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. (2005) 2:217–25. doi: 10.1016/j.cmet.2005.09.001
36. Lin S, Wang S, Wang P, Tang C, Wang Z, Chen L, et al. Bile acids and their receptors in regulation of gut health and diseases. Prog Lipid Res. (2023) 89:101210. doi: 10.1016/j.plipres.2022.101210
37. Huang S, Wu Y, Zhao Z, Wu B, Sun K, Wang H, et al. A new mechanism of obeticholic acid on NASH treatment by inhibiting NLRP3 inflammasome activation in macrophage. Metabolism. (2021) 120:154797. doi: 10.1016/j.metabol.2021.154797
38. Coppola C, Gosche J, Arrese M, Ancowitz B, Madsen J, Vanderhoof J, et al. Molecular analysis of the adaptive response of intestinal bile acid transport after ileal resection in the rat. Gastroenterology. (1998) 115:1172–8. doi: 10.1016/s0016-5085(98)70088-5
39. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. (2009) 183:6251–61. doi: 10.4049/jimmunol.0803978
40. Inagaki T, Moschetta A, Lee Y, Peng L, Zhao G, Downes M, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA. (2006) 103:3920–5. doi: 10.1073/pnas.0509592103
41. Gadaleta R, van Erpecum K, Oldenburg B, Willemsen E, Renooij W, Murzilli S, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. (2011) 60:463–72. doi: 10.1136/gut.2010.212159
42. Keitel V, Cupisti K, Ullmer C, Knoefel W, Kubitz R, Häussinger D. The membrane-bound bile acid receptor TGR5 is localized in the epithelium of human gallbladders. Hepatology. (2009) 50:861–70. doi: 10.1002/hep.23032
43. Pols T, Noriega L, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol. (2010) 54:1263–72. doi: 10.1016/j.jhep.2010.12.004
44. Schaap F, Trauner M, Jansen P. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. (2013) 11:55–67. doi: 10.1038/nrgastro.2013.151
45. Häussinger D, Keitel V. Role of TGR5 (GPBAR1) in liver disease. Semin Liver Dis. (2018) 38:333–9. doi: 10.1055/s-0038-1669940
46. Wang Y, Chen W, Yu D, Forman B, Huang W. The G-Protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology. (2011) 54:1421–32. doi: 10.1002/hep.24525
47. Kuhre R, Wewer Albrechtsen N, Larsen O, Jepsen S, Balk-Møller E, Andersen D, et al. Bile acids are important direct and indirect regulators of the secretion of appetite– and metabolism-regulating hormones from the gut and pancreas. Mol Metab. (2018) 11:84–95. doi: 10.1016/j.molmet.2018.03.007
48. Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. (2003) 278:9435–40. doi: 10.1074/jbc.m209706200
49. Watanabe M, Houten S, Mataki C, Christoffolete M, Kim B, Sato H, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. (2006) 439:484–9. doi: 10.1038/nature04330
50. Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun. (2005) 329:386–90. doi: 10.1016/j.bbrc.2005.01.139
51. Kumar D, Rajagopal S, Mahavadi S, Mirshahi F, Grider J, Murthy K, et al. Activation of transmembrane bile acid receptor TGR5 stimulates insulin secretion in pancreatic β cells. Biochem Biophys Res Commun. (2012) 427:600–5. doi: 10.1016/j.bbrc.2012.09.104
52. Song M, Zhang F, Fu Y, Yi X, Feng S, Liu Z, et al. Tauroursodeoxycholic acid (TUDCA) improves intestinal barrier function associated with TGR5-MLCK pathway and the alteration of serum metabolites and gut bacteria in weaned piglets. J Animal Sci Biotechnol. (2022) 13:73. doi: 10.1186/s40104-022-00713-3
53. Versteeg B, Himschoot M, van den Broek I, Bom R, Speksnijder A, Schim van der Loeff MF, et al. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Sex Transm Infect. (2015) 91:415–22. doi: 10.1136/sextrans-2014-051790
54. Pan P, Song Y, Du X, Bai L, Hua X, Xiao Y, et al. Intestinal barrier dysfunction following traumatic brain injury. Neurol Sci. (2019) 40:1105–10. doi: 10.1007/s10072-019-03739-0
55. Turner J. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. (2009) 9:799–809. doi: 10.1038/nri2653
56. Ghosh S, Nukavarapu S, Jala V. Effects of heavy metals on gut barrier integrity and gut microbiota. Microb Host. (2023) 2:e230015. doi: 10.1530/mah-23-0015
57. Otani T, Furuse M. Tight junction structure and function revisited. Trends Cell Biol. (2020) 30:805–17. doi: 10.1016/j.tcb.2020.08.004
58. Katzenberger R, Ganetzky B, Wassarman D. The gut reaction to traumatic brain injury. Fly. (2015) 9:68–74. doi: 10.1080/19336934.2015.1085623
59. Hartsock A, Nelson W. Adherens and tight junctions: Structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. (2007) 1778:660–9. doi: 10.1016/j.bbamem.2007.07.012
60. Ivanov A, Naydenov N. Dynamics and regulation of epithelial adherens junctions: Recent discoveries and controversies. Int Rev Cell Mol Biol. (2013) 303:27–99. doi: 10.1016/B978-0-12-407697-6.00002-7
61. Xu L, Li Y, Wei Z, Bai R, Gao G, Sun W, et al. Chenodeoxycholic acid (CDCA) promoted intestinal epithelial cell proliferation by regulating cell cycle progression and mitochondrial biogenesis in IPEC-J2 cells. Antioxidants. (2022) 11:2285. doi: 10.3390/antiox11112285
62. Song M, Ye J, Zhang F, Su H, Yang X, He H, et al. Chenodeoxycholic acid (CDCA) protects against the lipopolysaccharide-induced impairment of the intestinal epithelial barrier function via the FXR-MLCK pathway. J Agric Food Chem. (2019) 67:8868–74. doi: 10.1021/acs.jafc.9b03173
63. Raimondi F, Santoro P, Barone M, Pappacoda S, Barretta M, Nanayakkara M, et al. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am J Physiol Liver Physiol. (2008) 294:G906–13. doi: 10.1152/ajpgi.00043.2007
64. Lin S, Yang X, Yuan P, Yang J, Wang P, Zhong H, et al. Undernutrition shapes the gut microbiota and bile acid profile in association with altered gut-liver FXR signaling in weaning pigs. J Agric Food Chem. (2019) 67:3691–701. doi: 10.1021/acs.jafc.9b01332
65. Katona B, Anant S, Covey D, Stenson W. Characterization of enantiomeric bile acid-induced apoptosis in colon cancer cell lines. J Biol Chem. (2009) 284:3354–64. doi: 10.1074/jbc.m805804200
66. Wang W, Zhao J, Gui W, Sun D, Dai H, Xiao L, et al. Tauroursodeoxycholic acid inhibits intestinal inflammation and barrier disruption in mice with non-alcoholic fatty liver disease. Br J Pharmacol. (2018) 175:469–84. doi: 10.1111/bph.14095
67. Farhadi A, Banan A, Fields J, Keshavarzian A. Intestinal barrier: An interface between health and disease. J Gastroenterol Hepatol. (2003) 18:479–97. doi: 10.1046/j.1440-1746.2003.03032.x
68. Alonso C, Vicario M, Pigrau M, Lobo B, Santos J. Intestinal barrier function and the brain-gut axis. Adv Exp Med Biol. (2014) 817:73–113. doi: 10.1007/978-1-4939-0897-4_4
69. Johansson M, Larsson J, Hansson G. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proc Natl Acad Sci USA. (2010) 108:4659–65. doi: 10.1073/pnas.1006451107
70. Johansson M, Phillipson M, Petersson J, Velcich A, Holm L, Hansson G. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. (2008) 105:15064–9. doi: 10.1073/pnas.0803124105
71. Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: Thickness and physical state in vivo. Am J Physiol Liver Physiol. (2001) 280:G922–9. doi: 10.1152/ajpgi.2001.280.5.g922
72. McGuckin M, Lindén S, Sutton P, Florin T. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. (2011) 9:265–78. doi: 10.1038/nrmicro2538
73. Cox K, Liu S, Lwin T, Hoffman R, Batra S, Bouvet M. The mucin family of proteins: Candidates as potential biomarkers for colon cancer. Cancers. (2023) 15:1491–1491. doi: 10.3390/cancers15051491
74. Modilevsky S, Naama M, Bel S. Goblet and paneth cells: Producers of the intestinal barrier. Encyclop Cell Biol. (2023) 6:66–71. doi: 10.1016/b978-0-12-821618-7.00140-1
75. Shekels L, Lyftogt C, Ho S. Bile acid-induced alterations of mucin production in differentiated human colon cancer cell lines. Int J Biochem Cell Biol. (1996) 28:193–201. doi: 10.1016/1357-2725(95)00125-5
76. Naama M, Telpaz S, Awad A, Ben-Simon S, Harshuk-Shabso S, Modilevsky S, et al. Autophagy controls mucus secretion from intestinal goblet cells by alleviating ER stress. Cell Host Microbe. (2023) 31:433–446.e4. doi: 10.1016/j.chom.2023.01.006
77. Xu Y, Watanabe T, Tanigawa T, Machida H, Okazaki H, Yamagami H, et al. Bile acids induce Cdx2 expression through the farnesoid X receptor in gastric epithelial cells. J Clin Biochem Nutr. (2009) 46:81–6. doi: 10.3164/jcbn.09-71
78. Yu J, Zheng J, Qi J, Yang K, Wu Y, Wang K, et al. Bile acids promote gastric intestinal metaplasia by upregulating CDX2 and MUC2 expression via the FXR/NF-κB signalling pathway. Int J Oncol. (2019) 54:879–92. doi: 10.3892/ijo.2019.4692
79. Huang D, Xiong M, Xu X, Wu X, Xu J, Cai X, et al. Bile acids elevated by high-fat feeding induce endoplasmic reticulum stress in intestinal stem cells and contribute to mucosal barrier damage. Biochem Biophys Res Commun. (2020) 529:289–95. doi: 10.1016/j.bbrc.2020.05.226
80. Yang J, Liu M, Lv L, Guo J, He K, Zhang H, et al. Metformin alleviates irradiation-induced intestinal injury by activation of FXR in intestinal epithelia. Front Microbiol. (2022) 13:932294. doi: 10.3389/fmicb.2022.932294
81. Ijssennagger N, van Rooijen K, Magnúsdóttir S, Ramos Pittol J, Willemsen E, de Zoete M, et al. Ablation of liver FXR results in an increased colonic mucus barrier in mice. JHEP Rep. (2021) 3:100344. doi: 10.1016/j.jhepr.2021.100344
82. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. (2020) 19:55–71. doi: 10.1038/s41579-020-0433-9
83. Yu M, Yu B, Chen D. The effects of gut microbiota on appetite regulation and the underlying mechanisms. Gut Microbes. (2024) 16:2414796. doi: 10.1080/19490976.2024.2414796
84. Ley R, Hamady M, Lozupone C, Turnbaugh P, Ramey R, Bircher J, et al. Evolution of mammals and their gut microbes. Science. (2008) 320:1647–51. doi: 10.1126/science.1155725
85. Martens E, Chiang H, Gordon J. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. (2008) 4:447–57. doi: 10.1016/j.chom.2008.09.007
86. Deriu E, Liu J, Pezeshki M, Edwards R, Ochoa R, Contreras H, et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. (2013) 14:26–37. doi: 10.1016/j.chom.2013.06.007
87. Stecher B, Macpherson A, Hapfelmeier S, Kremer M, Stallmach T, Hardt W. Comparison of Salmonella enterica serovar typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect Immun. (2005) 73:3228–41. doi: 10.1128/iai.73.6.3228-3241.2005
88. Vaishnava S, Behrendt C, Ismail A, Eckmann L, Hooper L. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. (2008) 105:20858–63. doi: 10.1073/pnas.0808723105
89. Jones R, Luo L, Ardita C, Richardson A, Kwon Y, Mercante J, et al. Symbiotic lactobacilli stimulate gut epithelial proliferationviaNox-mediated generation of reactive oxygen species. EMBO J. (2013) 32:3017–28. doi: 10.1038/emboj.2013.224
90. Kelly C, Zheng L, Campbell E, Saeedi B, Scholz C, Bayless A, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. (2015) 17:662–71. doi: 10.1016/j.chom.2015.03.005
91. Jain U, Lai C, Xiong S, Goodwin V, Lu Q, Muegge B, et al. Temporal regulation of the bacterial metabolite deoxycholate during colonic repair is critical for crypt regeneration. Cell Host Microbe. (2018) 24:353–363.e5. doi: 10.1016/j.chom.2018.07.019
92. Cai J, Rimal B, Jiang C, Chiang J, Patterson A. Bile acid metabolism and signaling, the microbiota, and metabolic disease. Pharmacol Ther. (2022) 237:108238. doi: 10.1016/j.pharmthera.2022.108238
93. Kurdi P, Kawanishi K, Mizutani K, Yokota A. Mechanism of growth inhibition by free bile acids in Lactobacilli and Bifidobacteria. J Bacteriol. (2006) 188:1979–86. doi: 10.1128/jb.188.5.1979-1986.2006
94. Kandell R, Bernstein C. Bile salt/acid induction of DNA damage in bacterial and mammalian cells: Implications for colon cancer. Nutr Cancer. (1991) 16:227–38. doi: 10.1080/01635589109514161
95. Bernstein C, Bernstein H, Payne C, Beard S, Schneider J. Bile salt activation of stress response promoters in Escherichia coli. Curr Microbiol. (1999) 39:68–72. doi: 10.1007/s002849900420
96. Sannasiddappa T, Lund P, Clarke S. In vitro antibacterial activity of unconjugated and conjugated bile salts on Staphylococcus aureus. Front Microbiol. (2017) 8:1581. doi: 10.3389/fmicb.2017.01581
97. Tian Y, Gui W, Koo I, Smith P, Allman E, Nichols R, et al. The microbiome modulating activity of bile acids. Gut Microbes. (2020) 11:979–96. doi: 10.1080/19490976.2020.1732268
98. Miller S. Antibiotic resistance and regulation of the gram-negative bacterial outer membrane barrier by host innate immune molecules. MBio. (2016) 7:e1541–1516. doi: 10.1128/mbio.01541-16
99. Begley M, Gahan C, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. (2004) 29:625–51. doi: 10.1016/j.femsre.2004.09.003
100. Noh D, Gilliland S. Influence of bile on cellular integrity and β-galactosidase activity of Lactobacillus acidophilus. J Dairy Sci. (1993) 76:1253–9. doi: 10.3168/jds.s0022-0302(93)77454-8
101. Wershil B, Furuta G. Gastrointestinal mucosal immunity. J Allergy Clin Immunol. (2008) 121:S380–3. doi: 10.1016/j.jaci.2007.10.023
102. Velikova T, Kaouri I, Bakopoulou K, Gulinac M, Naydenova K, Dimitrov M, et al. Mucosal immunity and trained innate immunity of the gut. Gastroenterol Insights. (2024) 15:661–75. doi: 10.3390/gastroent15030048
103. Mowat A, Agace W. Regional specialization within the intestinal immune system. Nat Rev Immunol. (2014) 14:667–85. doi: 10.1038/nri3738
104. Di Sabatino A, Santacroce G, Rossi C, Broglio G, Lenti M. Role of mucosal immunity and epithelial–vascular barrier in modulating gut homeostasis. Intern Emerg Med. (2023) 18:1635–46. doi: 10.1007/s11739-023-03329-1
105. Murphy B. Mucosal immunity to viruses. Mucosal Immunol. (2005) 71:799–813. doi: 10.1016/b978-012491543-5/50047-4
106. Mu Q, Kirby J, Reilly C, Luo X. Leaky gut as a danger signal for autoimmune diseases. Front Immuno. (2017) 8:598. doi: 10.3389/fimmu.2017.00598
107. Sun R, Xu C, Feng B, Gao X, Liu Z. Critical roles of bile acids in regulating intestinal mucosal immune responses. Therap Adv Gastroenterol. (2021) 14:17562848211018098. doi: 10.1177/17562848211018098
108. Gadaleta R, Oldenburg B, Willemsen E, Spit M, Murzilli S, Salvatore L, et al. Activation of bile salt nuclear receptor FXR is repressed by pro-inflammatory cytokines activating NF-κB signaling in the intestine. Biochim Biophys Acta. (2011) 1812:851–8. doi: 10.1016/j.bbadis.2011.04.005
109. Horikawa T, Oshima T, Li M, Kitayama Y, Eda H, Nakamura K, et al. Chenodeoxycholic acid releases proinflammatory cytokines from small intestinal epithelial cells through the farnesoid X receptor. Digestion. (2019) 100:286–94. doi: 10.1159/000496687
110. Tang K, Kong D, Peng Y, Guo J, Zhong Y, Yu H, et al. Ginsenoside RC attenuates DSS-induced ulcerative colitis, intestinal inflammatory, and barrier function by activating the farnesoid X receptor. Front Pharmacol. (2022) 13:1000444. doi: 10.3389/fphar.2022.1000444
111. O’Guinn M, Handler D, Hsieh J, Mallicote M, Feliciano K, Gayer CP. FXR deletion attenuates intestinal barrier dysfunction in murine acute intestinal inflammation. Am J Physiol Liver Physiol. (2024) 327:G175–87. doi: 10.1152/ajpgi.00063.2024
112. Ichikawa R, Takayama T, Yoneno K, Kamada N, Kitazume M, Higuchi H, et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology. (2012) 136:153–62. doi: 10.1111/j.1365-2567.2012.03554.x
113. Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. (2016) 45:802–16. doi: 10.1016/j.immuni.2016.09.008
114. Pi Y, Wu Y, Zhang X, Lu D, Han D, Zhao J, et al. Gut microbiota-derived ursodeoxycholic acid alleviates low birth weight-induced colonic inflammation by enhancing M2 macrophage polarization. Microbiome. (2023) 11:19. doi: 10.1186/s40168-022-01458-x
Keywords: bile acids, bile acid receptors, FXR, TGR5, intestinal barrier
Citation: Song G, Xie Y, Yi L, Cheng W, Jia H, Shi W, Liu Q, Fang L, Xue S, Liu D, Zhu J and Zhao S (2025) Bile acids affect intestinal barrier function through FXR and TGR5. Front. Med. 12:1607899. doi: 10.3389/fmed.2025.1607899
Received: 08 April 2025; Accepted: 28 May 2025;
Published: 07 July 2025.
Edited by:
Rui Albuquerque Carvalho, University of Coimbra, PortugalReviewed by:
Leticia Muñoz, University of Alcalá, SpainSei Higuchi, St. John’s University, United States
Copyright © 2025 Song, Xie, Yi, Cheng, Jia, Shi, Liu, Fang, Xue, Liu, Zhu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junhong Zhu, MjAyNDAyMkB5bmF1LmVkdS5jbg==; Sumei Zhao, MjAwMDAyNUB5bmF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Guangyao Song
Guangyao Song Yuxiao Xie1,2†
Yuxiao Xie1,2† Lanlan Yi
Lanlan Yi Wenjie Cheng
Wenjie Cheng