- Lung Health Centre, King Faisal Specialist Hospital and Research Centre, Riyadh, Saudi Arabia
The S100 family of proteins plays a pivotal role in the pathogenesis of lung diseases, including asthma, chronic obstructive pulmonary disease (COPD), cystic fibrosis, pulmonary arterial hypertension (PAH), pulmonary fibrosis, lung cancers, acute lung injury, acute respiratory distress syndrome, COVID-19, and lung transplantation. This review comprehensively examines the contributions of S100 proteins to the progression of these disorders, focusing on their potential as diagnostic and prognostic biomarkers, as well as therapeutic targets. S100A protein-mediated key molecular mechanisms that influence inflammation, airway remodeling, fibrosis, and tumorigenesis in the lungs are discussed. The importance of their normal function is evident from the observation that simultaneous mutations in S100A3 and S100A13 predispose individuals to early-onset pulmonary fibrosis, underscoring their critical role in lung health. Furthermore, sustained S100 protein elevation is explored in the context of long COVID, shedding light on its role in chronic inflammation. These proteins act as damage-associated molecular patterns (DAMPs), activating immune pathways via receptors like TLR4 and RAGE, thereby driving inflammation and immune cell recruitment. Notably, in lung transplantation, elevated levels of S100A8, S100A9, and S100A12 serve as early biomarkers of graft rejection and complications such as graft-vs.-host disease, which indicates their role in mediating immune responses and transplant outcomes. While promising, the clinical application of S100 proteins faces challenges, including disease-specific variability and the need for robust validation across diverse populations. This narrative review underscores the dual potential of S100 proteins as biomarkers and therapeutic targets in respiratory medicine while emphasizing the importance of overcoming current limitations through targeted research and clinical trials.
Introduction
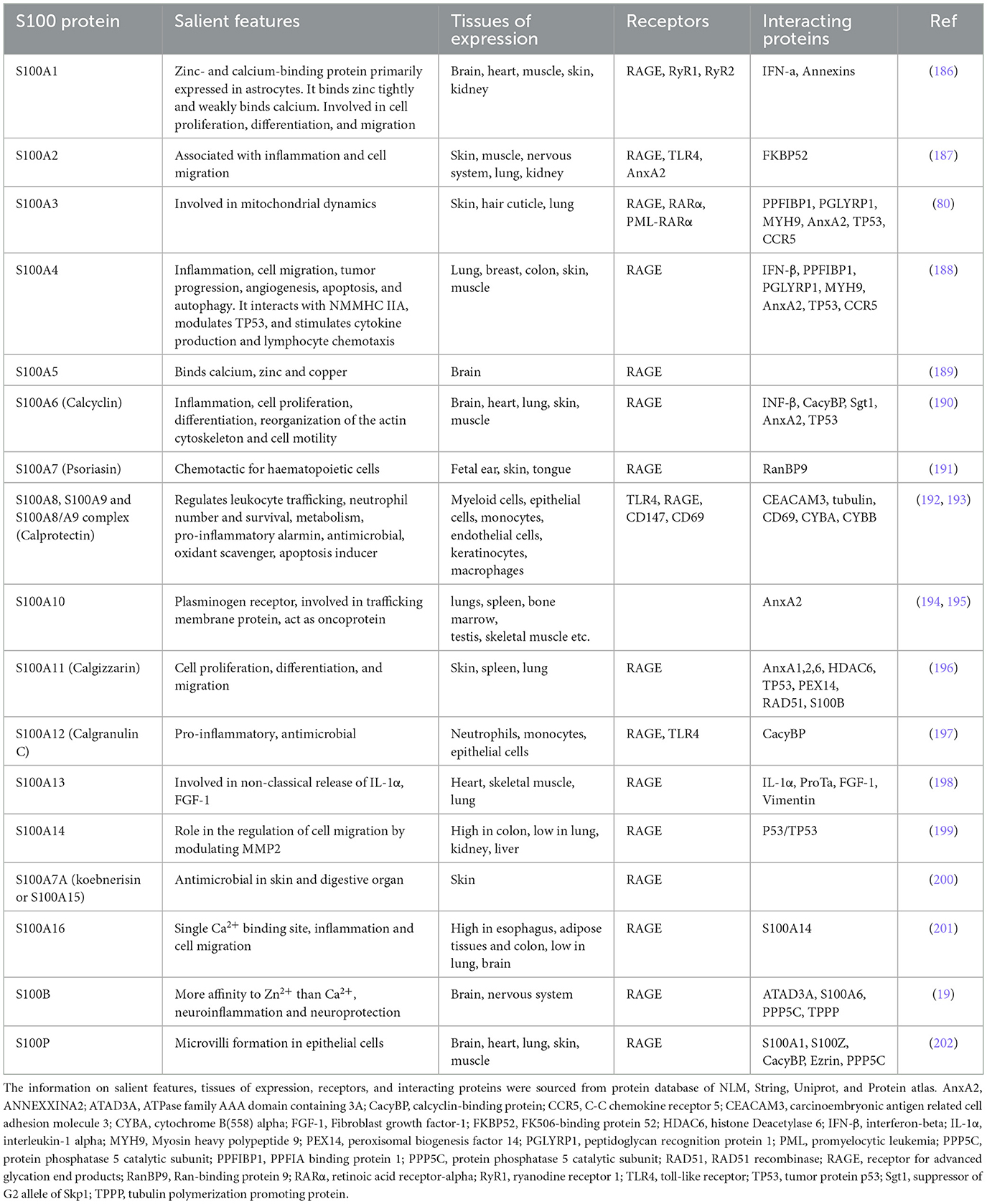
Human diseases have long been associated with the dysregulation of protein expression and functions, which play pivotal roles in maintaining cellular homeostasis (1, 2). Proteins are the driving force of signal transduction, structural maintenance, enzymatic catalysis, and immunological responses, and the perturbations in their expression levels or functional integrity because of genetic alterations, environmental influences, or other factors can result in diseases (1, 3, 4). Many protein families, like S100 family, are evolutionarily conserved to carry out the fundamental processes that maintain the physiological homeostasis of an organism (5, 6). S100 protein family, (Table 1) (7–9) comprises S100A1 to S100A16, S100B, S100G, S100P, and S100Z (10), along with S100-fused-type proteins such as trichohyalin (11), filaggrin (12), filaggrin2 (13), cornulin (14), and repetin (15) (see Table 1 for general details. We are not including S100-fused-type proteins in the table, as the relation of these proteins in lung diseases is almost null).
S100 proteins bind calcium (via EF-hand motifs), as well as zinc and copper ions (6, 16, 17) (Figure 1). Structural analyses show that S100 proteins have at least three active sites on two surfaces, enabling diverse protein interactions for their biological effects, which are often modulated by calcium-induced conformational changes (18). S100 proteins can be categorized into three groups based on their functions: (a) intracellular regulators, (b) dual-function proteins acting intracellularly and extracellularly (19) and (c) primarily extracellular entities (5). Intracellular S100 proteins regulate cell functions like growth, movement, cell cycle, transcription, and differentiation. Extracellularly, they influence inflammation, migration, tissue development, and repair and enhance leukocyte and tumor cell invasiveness (5).

Figure 1. Sequence features of S100 proteins highlighting metal-binding sites and lung cancer-associated mutations. Yellow highlights indicate EF-hand motifs; turquoise highlights, the canonical EF-hand. Brown residues denote Ca2+-binding sites; purple residues, Zn2+-binding sites; and boxed purple residues, Cu2+-binding sites. Brackets represent α-helices. Single-nucleotide mutations associated with lung cancers are indicated.
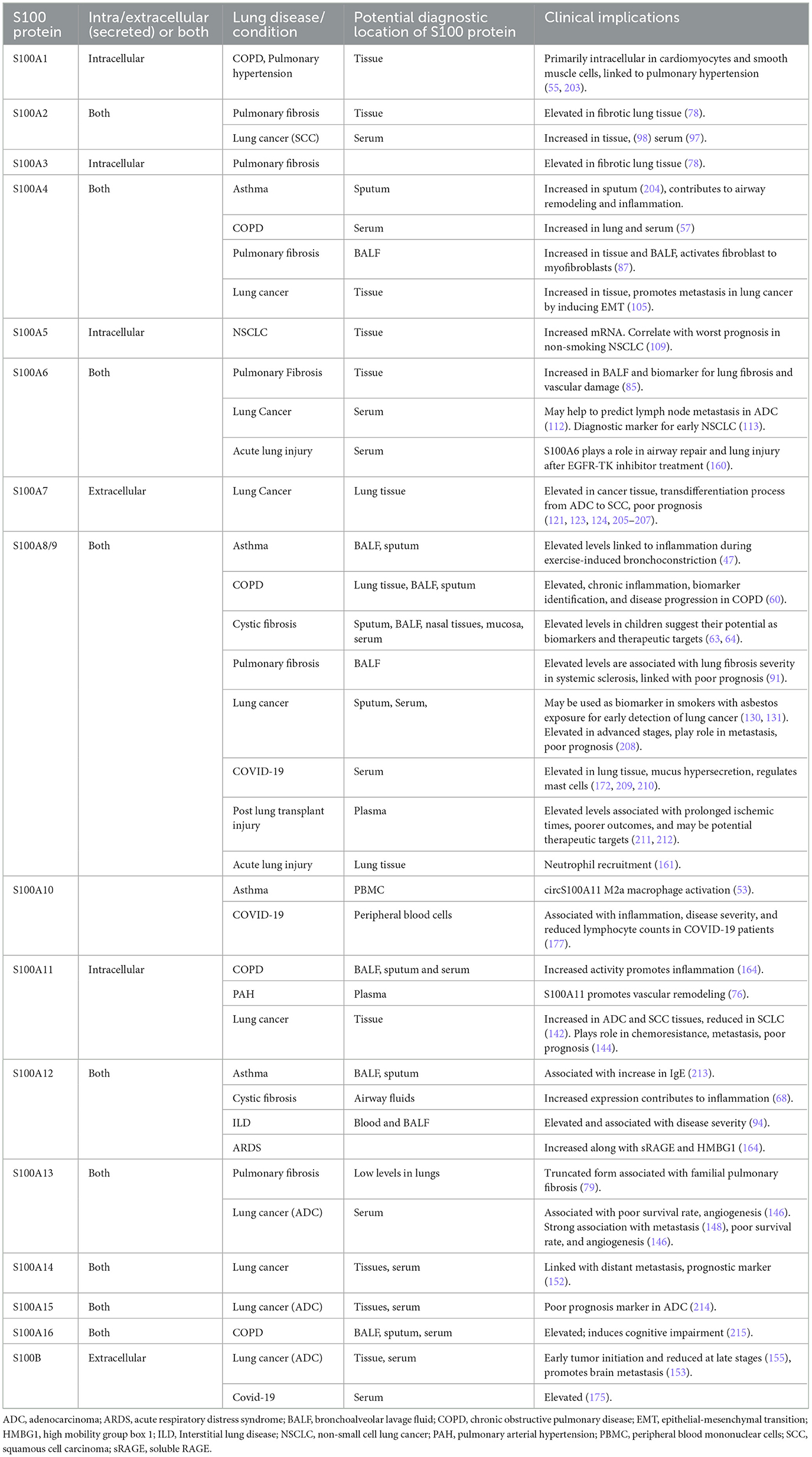
Clinically, dysregulated S100 proteins are valuable diagnostic and prognostic markers in various diseases, including neurodegenerative disorders (20), cardiomyopathy (21), and lung diseases (10) (Figure 2). S100 proteins help in distinguishing between conditions like idiopathic pulmonary fibrosis (IPF) and rheumatoid arthritis-associated interstitial pneumonia (IP) where S100 protein-positive dendritic cells are present only in the latter (22). CD8+ve lymphocytes are more prominent in fibrosing regions surrounding S100-positive dendritic cells than CD4+ve lymphocytes (23). S100A4 and S100B overexpression is associated with poor prognosis and tumor metastasis in lung cancer (10, 20–25) (see Table 2 for roles in lung diseases).
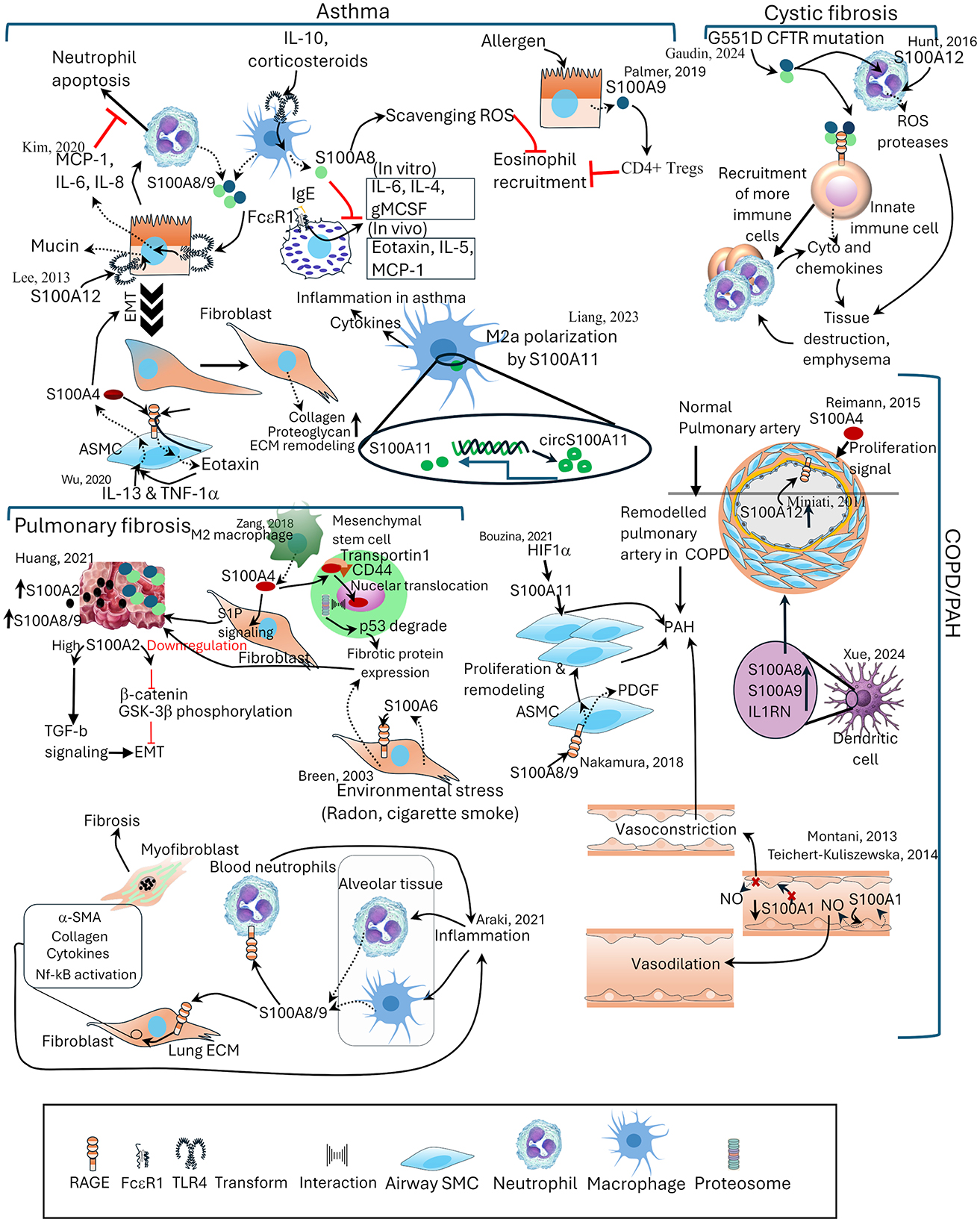
Figure 2. The distinct and shared roles of S100 proteins in lung pathologies. Schematic representation of S100 protein involvement in major lung diseases, including asthma, cystic fibrosis, pulmonary fibrosis, and COPD/pulmonary arterial hypertension (PAH). In asthma, S100A8/A9, S100A4, and S100A11 regulate cytokine production, neutrophil apoptosis, and eosinophil recruitment. In cystic fibrosis, S100A9 and S100A12 contribute to immune cell recruitment, protease release, and emphysematous tissue destruction. In pulmonary fibrosis, S100A2, S100A4, S100A6, S100A8/9, and S100A11 promote fibroblast activation, epithelial–mesenchymal transition, and extracellular matrix remodeling. In COPD/PAH, S100A1, S100A4, S100A8/9, and S100A12 are involved in airway smooth muscle proliferation, vascular remodeling, and vasoregulatory imbalance. Reported interactions with signaling pathways, transcriptional regulators, and environmental stressors are indicated. ECM, extracellular matrix; EMT, epithelial mesenchymal transition; GM-CSF, granulocyte-macrophage colony-stimulating factor; NO, nitric oxide; S1P, sphingosine 1 phosphate; SMC, smooth muscle cell; TGF-β, transforming growth factor- β.
Despite these findings, the collective literature on S100 proteins in lung diseases remains limited, including their roles in COVID-19 and lung transplantation. This review aims to provide a comprehensive exploration of the diagnostic, prognostic, and therapeutic potential of S100 proteins in these contexts, offering a detailed analysis to bridge existing knowledge gaps.
Metal ion binding and conformational changes of S100 proteins for intra- and extracellular functions
The EF-hand motif of many S100 proteins have Zn2+/Cu2+ binding sites in addition to Ca2+ metal ions(26–30). This unique feature provides them the versatility of performing both intracellular and extracellular functions (26, 31, 32).
The Ca+ switch for intracellular functions
The EF-hand motif of S100 proteins binds Ca+ ions, triggering a conformational change often described as the “S100 Ca+-switch.” This structural rearrangement exposes previously buried hydrophobic surfaces, creating docking sites for a wide array of intracellular targets such as enzymes, cytoskeletal proteins, and transcriptional regulators (33, 34). Through these interactions, S100 proteins regulate fundamental cellular processes, including proliferation, differentiation, apoptosis, and motility. Thus, Ca+-dependent conformational dynamics are central to the intracellular signaling roles of S100 proteins (8, 19).
Transition metal-dependent structural rearrangements for extracellular functions
In addition to Ca+ binding, S100 proteins possess unique transition metal-binding sites at their dimer interface, particularly for Zn+ and Cu+ (27–29, 33). Binding of these metals induces structural changes distinct from those caused by Ca+ (35, 36). These rearrangements enable S100 proteins to interact with cell surface receptors, most notably the receptor for advanced glycation end products (RAGE) and toll-like receptor 4 (TLR4). These interactions mediate extracellular signaling through both autocrine and paracrine pathways, which connects them to regulation of the immune system, inflammation, and many diseases.
Distinction of S100 proteins from other EF-hand proteins
While classical EF-hand proteins like calmodulin also undergo Ca+-induced conformational changes, S100 proteins stand out due to their dual/triple metal-binding capability and the resulting distinct conformational responses (37, 38). Ca+ binding exposes hydrophobic pockets for intracellular interactions, whereas Zn+/Cu+ binding at the dimer interface enables extracellular receptor engagement (5, 26, 39). This adaptability allows S100 proteins to serve as both intracellular regulators and extracellular signaling molecules—an evolutionary specialization not shared by simpler Ca+ sensors (26, 39).
The structural plasticity of S100 proteins, governed by their ability to bind multiple metal ions, underpins their dual roles. By coupling Ca+-induced conformational changes to intracellular signaling and Zn+/Cu+-induced rearrangements to extracellular receptor interactions, S100 proteins uniquely bridge intracellular regulation with extracellular communication (39, 40). This property sets them apart from other EF-hand proteins and explains their prominent involvement in processes ranging from cytoskeletal dynamics to cancer metastasis and inflammation (39).
S100 proteins induce inflammation and airway remodeling in asthma
Asthma is a chronic inflammatory disease of the airways characterized by bronchoconstriction, elevated levels of allergen-specific IgE, airway hyperresponsiveness and remodeling (41). Until now, S100A4, S100A8/S100A9 (calprotectin), S100A11, and S100A12 have been implicated in the pathophysiology of asthma, exhibiting both similarities and differences in their mechanisms of action.
S100A4, also known as fibroblast-specific protein 1 (FSP1), contributes to asthma by promoting inflammation and epithelial-mesenchymal transition (EMT) in the airway (42). Similarly, in pleural fibrosis, S100A4 has been demonstrated to stimulate the production of transforming growth factor-β (TGF-β) and facilitate epithelial-mesenchymal transition (EMT) in pleural mesothelial cells (43). While this specific mechanism has not been investigated in asthma, it is plausible that S100A4 plays a similar role in the airway remodeling observed in asthmatic patients. Notably, during episodes of exacerbated inflammation, cytokines such as IL-13 and TNF-α trigger the release of S100A4 from airway smooth muscle cells. The secreted S100A4 subsequently engages the RAGE, thereby activating the Akt/NF-κB signaling pathway (44). This activation results in the synthesis of eotaxin and further production of S100A4, consequently establishing a positive feedback loop that could perpetuate inflammation in individuals with asthma. Diagnostically, elevated levels of S100A4, like calprotectin, in the sputum of asthmatic patients correlate with airway hyperresponsiveness, providing evidence of its role in disease exacerbation. Thus, S100A4 neutralizing antibodies have shown promising results of reducing airway hyperresponsiveness and inflammation and preventing fibrosis in animal models (42).
The S100A8/A9 heterodimer plays a dual role in asthma pathogenesis, depending on the inflammatory milieu and asthma subtype. During infection and inflammation, extracellular S100A8/9 levels rise and engage TLR4 on bronchial epithelial cells, activating MAPK and NF-κB pathways to induce neutrophil survival cytokines such as MCP-1, IL-6, and IL-8 (45, 46), thereby intensifying airway inflammation. Aligned to that, elevated S100A8/A9 levels are observed in the serum and sputum of asthmatic patients, particularly during episodes of exercise-induced bronchoconstriction (47), without any difference between the subgroups of asthma or compared to COPD (48, 49). In addition, elevated expression of S100A8/9 was observed in lungs of mouse model of asthma, a finding that aligns with observations in human asthma patients. In these patients, calprotectin levels were associated with several clinical parameters, including the ratio of forced expiratory volume in one second to forced vital capacity, smoking history, body mass index, and the percentage of neutrophils in the blood (49). In contrast, in allergic, Th2-driven asthma, S100A8/A9 exerts a regulatory function. In wild-type mice, Alternaria alternata challenge augmented S100A8/A9 release into the alveolar space and elevated its expression in the epithelium. Compared to wild-type, S100A9-deficient mouse model displayed severe airway inflammation, marked by elevated IL-13, CCL11, CCL24, serum IgE, eosinophil recruitment, and increased airway resistance and elastance. The study suggests S100A9-mediated protection occurs via regulation of CD4+ T CD25low regulatory T (Treg) cells (50). However, S100A9 levels in sputum are seen higher in neutrophilic uncontrolled asthma patients compared to controlled asthma cases (51). A therapeutic potential for S100A9 was demonstrated in rats by significantly reducing isometric tension of isolated tracheal spirals (52). This dual functionality underscores its context-specific nature, acting as an inflammatory amplifier in innate immune settings and a modulator in adaptive, allergic responses, with its net impact depending on the prevailing immunological profile of the disease.
S100A11 has an immunomodulatory effect in asthma. S100A11-gene derived circular RNA (circS100A11) is significantly higher in monocytes of pediatric asthma patients. circS100A11 enhances S100A11 expression that promotes STAT6-mediated M2a macrophage activation and exacerbates lung inflammation in mouse model (53). However, an airway smooth muscle cell (ASMC) relaxing effect by S100A11 is also reported in an allergen-induced asthma model (54). Recombinant S100A11 treatment in OVA-challenged rat results in a reduced airway hyperresponsiveness (AHR), and it reduces acetylcholine-induced myosin light chain phosphorylation in ASMC, in a calcium-independent manner. It denotes there may be cell-type specificity existing in response to S100A11 (54). Whether S100A11 has any impact on mast cells, histamine release or any other broncho-constrictive pathways still need to be addressed. The role of S100A11 in promoting inflammation to ward off infections/allergens while also providing a compensatory relaxation effect in ASM cells underscores the complexity of S100 proteins in asthma and their potential as targets for nuanced therapeutic strategies (53, 54).
S100A12, as well as S100A8 and S100A9, was shown to activate TLR4 and RAGE in normal bronchial epithelial cells and lung carcinoma cells in vitro to produce MUC5AC, a predominant protein in mucin (51). Since mucin production is a common feature in severe asthma, this observation underscores the importance of these S100 proteins in airway congestion, and their regulation could be of therapeutic value.
It is evident that S100 proteins contribute to inflammation and remodeling in asthma, often via RAGE and TLR4, yet vary in cellular targets and mechanisms. Diagnostically, they may serve as markers of severity and phenotype; prognostically, they could predict progression in severe asthma.
S100 proteins increase chronic inflammation in COPD
COPD is a progressive disease marked by persistent airflow limitation due to neutrophilic airway inflammation, emphysema, and vascular remodeling. S100 proteins play critical roles in both the inflammatory and structural components of COPD. Serum levels of S100A1 distinguish cachectic COPD patients from non-cachectic ones, establishing it as a biomarker for COPD progression, particularly in the context of cachexia (55).
Increased S100A4 levels in the remodeled intrapulmonary arteries may be an indication of this protein's involvement in vascular remodeling of COPD patients (56). Likewise, elevated S100A4 levels in the serum in conjunction with sphingosine 1 phosphate (S1P) correlate with reduction in lung function (57).
The predominant role of S100A8/9-mediated RAGE activation in COPD is evident from the observation that lower levels of S100A8/9 in RAGE-deficient mice result in decreased cigarette smoke-induced inflammation (58). Chronic inflammation, reduced lung function (59), and IL-17-related signaling in COPD are linked to upregulated S100A8 and S100A9 or their heterodimer in dendritic cells (60). Additionally, increased S100A8/A9 levels in smokers with COPD indicate their potential as biomarkers for diagnosis and tracking disease progression (61).
Elevated S100A12 levels in the airways and blood are associated with poor prognosis in COPD, making it a potential biomarker for disease progression (62). S100A12 effect is mediated through RAGE, while its soluble form, sRAGE, functions as a decoy receptor that limits the inflammation. Low sRAGE levels are linked to severe emphysema and chronic cor pulmonale, promoting the activation of neutrophils and macrophages and contributing to tissue damage.
S100 proteins regulate neutrophil-mediated inflammation in cystic fibrosis
Cystic fibrosis (CF) is characterized by chronic neutrophilic inflammation and progressive lung damage due to mutations in the CFTR gene. S100 proteins, particularly calprotectin and S100A12, play critical roles in sustaining this inflammation. A marked increase in exocytosis of S100A8/A9 in the airways of CF patients contributes to the perpetuation of neutrophilic inflammation (63, 64). The G551D CFTR mutation leads to dysregulated calcium signaling, which in turn activates S100A8/A9 and promotes the release of pro-inflammatory cytokines. These proteins drive neutrophil degranulation, resulting in the release of proteases and reactive oxygen species (ROS), which cause damage to the airway epithelium and exacerbate lung injury. Elevated levels of S100A8 associated with hyperactive immune response have been observed in experimental models of CF (65, 66).
Coupled with a deficiency of sRAGE (67), increased levels of S100A12 in the airways interact with RAGE, followed by activation of the p38 MAPK pathway in neutrophils leading to the continuous release of pro-inflammatory mediators, contributing to chronic inflammation, worsening CF progression, and impaired lung function (11, 67, 68).
S100 proteins regulate vascular remodeling in pulmonary arterial hypertension
Pulmonary arterial hypertension (PAH) is characterized by increased pulmonary artery pressure due to vascular remodeling, which results in right heart failure (69). S100 proteins have been implicated in the regulation of vascular homeostasis and remodeling in PAH.
Vascular endothelium-derived S100A1 regulates vascular effects by influencing nitric oxide (NO) production (70, 71). Reduced lung endothelial S100A1 levels may diminish NO expression, which leads to pulmonary vasoconstriction and potentially to PAH (72). The therapeutic potential of S100A1 in PAH was demonstrated by the administration of exogenous S100A1 to S100A1 knockout (KO) mice, leading to improvements in pulmonary artery pressure, vascular resistance, and endothelial cell survival (73).
S100A8/A9 also contributes to vascular remodeling in PAH by promoting smooth muscle cell proliferation and migration. Through RAGE signaling, S100A8/A9 enhances the expression of pro-inflammatory cytokines and growth factors, including PDGF, which accelerates the pathogenesis of pulmonary vascular remodeling (74).
Elevated levels of S100A11 are observed in the plasma of PAH patients (75). Under hypoxic conditions, hypoxia-inducible factor 1-α (HIF-1-α) induces upregulation of S100A11 mRNA in rat lungs, along with increased taurine levels. Administration of taurine attenuates HIF-1-α-induced transcriptional activation of S100A11, suppressing vascular remodeling. This suggests that S100A11 is a potential therapeutic target for vascular remodeling in pulmonary diseases and that taurine could be a treatment to inhibit hypoxia-induced vascular remodeling (76).
S100 proteins mediate EMT in pulmonary fibrosis
Pulmonary fibrosis (PF) is characterized by the excessive deposition of extracellular matrix (ECM) components and progressive scarring of lung tissue (77). Several S100 proteins, notably S100A2, S100A3, S100A4, S100A6, S100A8/A9 and S100A13 are deeply implicated in the mechanisms underlying fibrotic progression.
Elevated levels of S100A2 are found in lung tissues of PF patients. Its downregulation inhibits TGF-β1-induced EMT by blocking β-catenin expression and GSK-3β phosphorylation in A549 cells. Lithium chloride, a Wnt/β-catenin pathway activator, reverses EMT inhibition caused by S100A2 silencing, suggesting a potential treatment for PF by the inhibition of S100A2 (78).
S100A3 and S100A13 mutations are particularly relevant in the context of familial early-onset pulmonary fibrosis (PF), with our research showing that these mutations disrupt key cellular processes that contribute to fibrosis (Figure 3). S100A3 mutations impair calcium signaling, disrupting cellular homeostasis, while S100A13 mutations affect mitochondrial function and cytoskeletal dynamics via vimentin, driving early fibrotic changes. These dual disruptions in S100A3 and S100A13 affect both intracellular and extracellular processes essential for fibrosis. Our findings suggest that targeting these proteins, or their downstream effects, could help reverse the defective signaling pathways and provide therapeutic benefit in familial PF cases, potentially preventing excessive fibrotic remodeling (79–82).
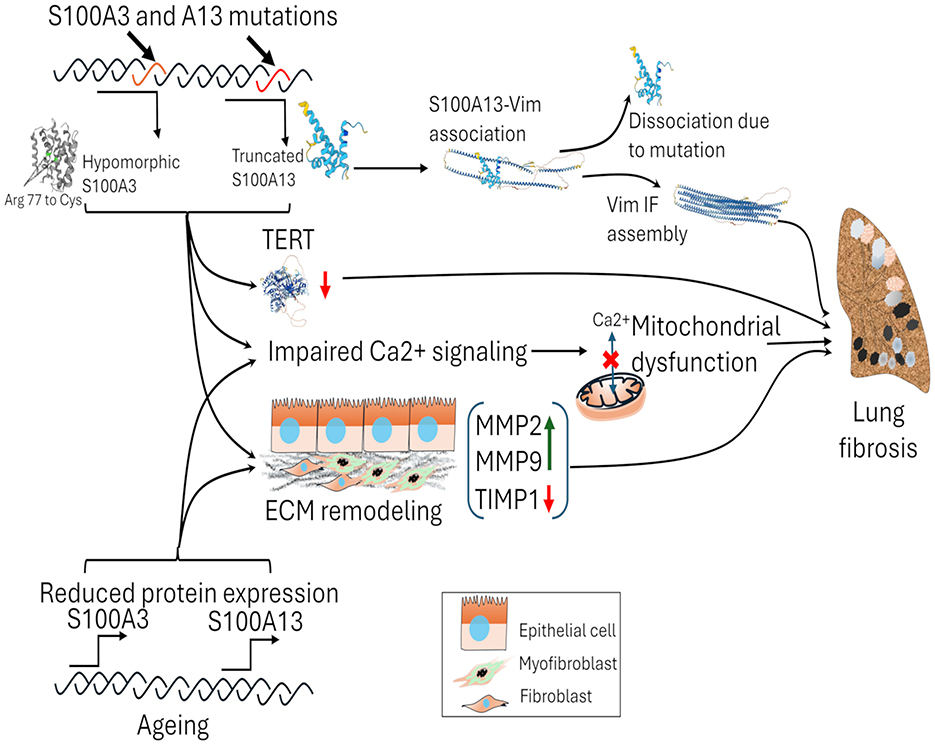
Figure 3. The impact of S100A3 and S100A13 mutations and the reduction of wild-type S100A3 and S100A13 while aging on lung fibrosis. Mutations in S100A3 (c.229C > T; Arg 77 to Cys) and S100A13 (c.238–241delATTG) proteins lead to functional alterations, affecting calcium signaling and telomerase reverse transcriptase (TERT) expression. The mutant variants of S100A3 and S100A13 affect Ca2+ signaling, mitochondrial dysfunction, and ECM remodeling by the increased expression of MMPs and decrease in TIMP1. S100A13 interacts with vimentin intermediate filaments (IF), but mutations cause dissociation, leading to defects in vimentin IF assembly. These genetic changes contribute to mitochondrial dysfunction and tissue damage. In sporadic cases of pulmonary fibrosis, age-related declines in S100A3 and S100A13 protein expression can contribute to susceptibility to developing pulmonary fibrosis. TERT, telomerase reverse transcriptase; Vim IF, vimentin intermediate filament.
M2 macrophage-released S100A4 activates lung fibroblasts through sphingosine 1 phosphate (S1P) signaling pathway to drive fibrosis (83–85). Nuclear translocation of S100A4 by making a complex with CD44 and transportin1 enhances the fibrogenic potential of mesenchymal progenitor cells. The nuclear S100A4 interacts with the proteasome to degrade p53 is crucial in fibrogenesis (86). In vivo studies have demonstrated that S100A4 deficiency protects against pulmonary fibrosis, consistent with its abnormal increase in human IPF (87).
S100A6 plays a major role in maintaining lung integrity by involving itself in tissue repair and fibroblast proliferation in response to mechanical stress (88, 89). S100A6 is elevated in BALF samples from PF-systemic sclerosis patients compared to smoker and non-smoker controls (85). The interaction between S100A6 and RAGE plays a vital role in mediating inflammatory and oxidative damage from prolonged cigarette smoke or radon exposure. This underscores S100A6 as a potential biomarker and therapeutic target against environmental-induced lung damage.
Elevated S100A8/A9 expression in lung, BALF and blood is correlated with the severity of PF-systemic sclerosis patients as well as sarcoidosis (90, 91). The main sources of S100A8/9 in the lung are macrophages and neutrophils. Upon an inflammatory signal, they release S100A8/9, which is released into the lung ECM and blood. The fibroblasts in the ECM get activated via RAGE and transdifferentiate into myofibroblasts. The expression of pro-inflammatory cytokines, collagen, and α-SMA are all found elevated and associated with myofibroblast formation (92). Moreover, during acute exacerbations of IPF, increased serum S100A8/A9 concentrations are linked to poor prognostic outcomes and reduced survival, proposing their use as prognostic markers. Exposure to zinc oxide nanoparticles can elevate respiratory S100A8 and S100A9 levels, potentially increasing lung inflammation and exacerbating fibrotic and cancerous conditions (93).
Elevated S100A12 levels in blood and BALF of patients with idiopathic interstitial pneumonias (IIP) and IPF are associated with disease severity and can be used as prognostic markers, particularly in IPF, where higher levels indicate a poorer prognosis (94). S100A12 inhibits physiological fibroblast migration for tissue repair through RAGE-p38 MAPK signaling. Targeting the S100A12-RAGE-p38 MAPK pathway could be beneficial for pulmonary disorders with abnormal tissue remodeling (95).
In pulmonary fibrosis, S100 proteins collectively drive inflammation, fibroblast activation, and ECM deposition, often via RAGE-mediated pathways. However, there are notably divergent roles among them; for example, S100A4 and S100A6 directly promote fibroblast activity and remodeling, while S100A8/9 and S100A12 amplify inflammation and serve as prognostic markers. S100A2 uniquely regulates EMT. However, normal function of S100A3 and S10013 appears to be important for normal physiology of lungs, and certain mutations in S100A3 and S100A13 contribute to familial PF. On the other hand, S100A6 responds to environmental triggers and leads to its abnormal expression leads to fibrogenesis. These contrasting functions underscore the complexity of S100 proteins in PF and their promise as tailored diagnostic and treatment targets.
S100 proteins in lung cancer
Lung cancer, particularly non-small cell lung carcinoma (NSCLC), is a heterogeneous disease encompassing various subtypes, each characterized by distinct molecular and clinical features (96). The S100 proteins, present primarily in NSCLC and its early-stage expression significantly influence tumor progression and therapy resistance, emerging as potential biomarkers and therapeutic targets in disease management. Their specific role in small cell lung carcinoma (SCLC) is limited and, in some cases, yields negative results. A comprehensive figure capturing the roles of S100 proteins in lung cancer is provided in Figure 4.
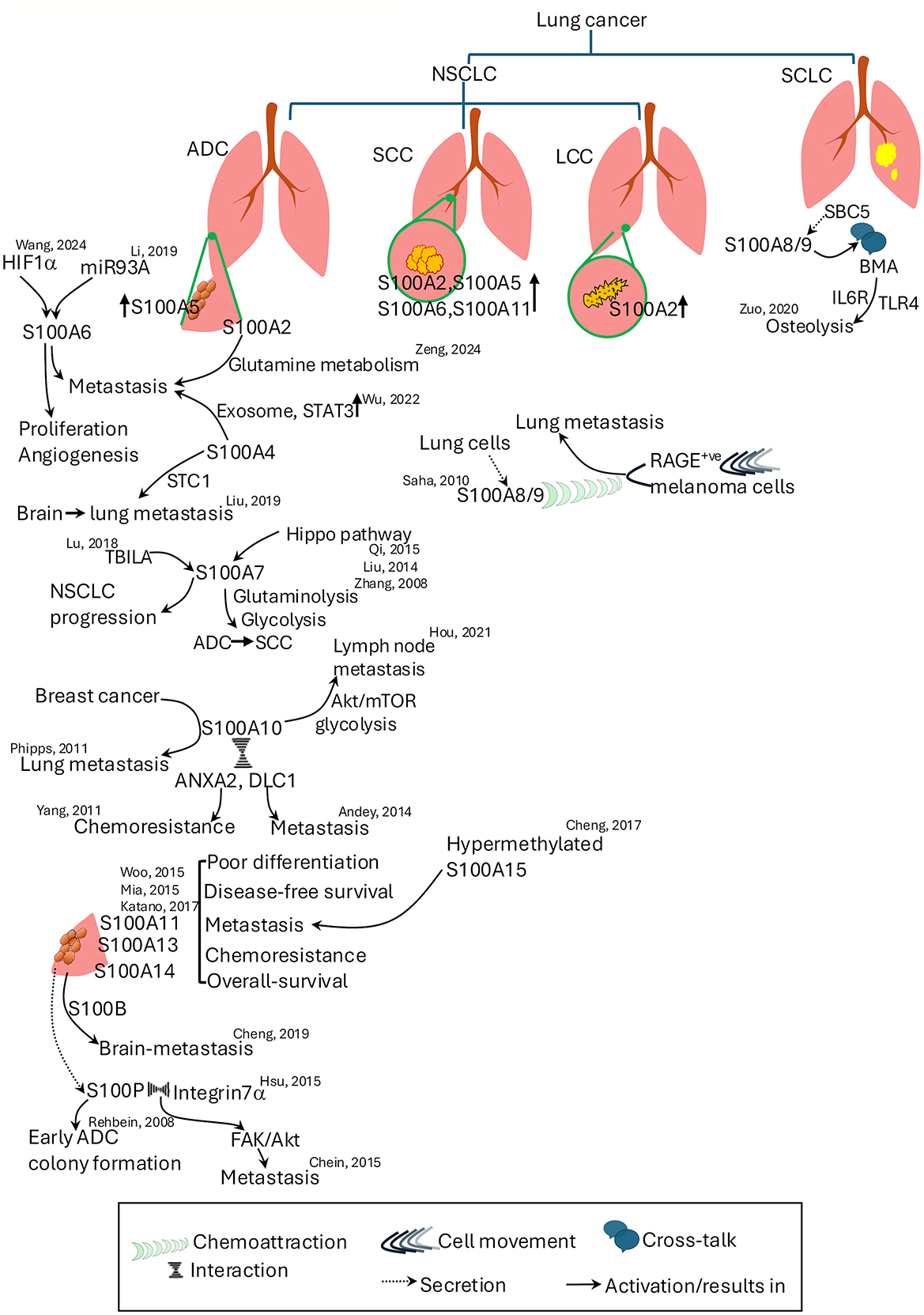
Figure 4. Roles of S100 proteins in lung cancer subtypes and metastasis. S100 proteins contribute to various aspects of lung cancer progression, metastasis, and chemoresistance. The figure illustrates the subtype-specific expression and functions of S100 proteins in non-small cell lung cancer Key roles include regulation of proliferation, angiogenesis, metabolic reprogramming, and metastatic dissemination. Cross-talk between S100 proteins and other signaling pathways, including Hippo, Akt/mTOR, STAT3, and RAGE-mediated mechanisms, is shown. Specific S100 proteins associated with metastasis to brain, lymph nodes, and lungs are highlighted. References denote supporting studies. ADC, adenocarcinoma; ANXA2, annexin A2; Akt, Ak strain transforming; BMA, bone marrow adipocytes; DLC1, deleted in liver cancer 1; FAK, focal adhesion kinase; HIF1-α, hypoxia inducing factor 1–α; LCC, large cell carcinoma; mTOR, mammalian target of rapamycin; NSCLC, non-small cell lung carcinoma; SBC5, small cell carcinoma-5; SCC, squamous cell carcinoma; SCLC, small cell lung cancer; STC1, stanniocalcin 1; TBILA, TGFβ-induced lncRNA; TLR4, toll-like receptor 4.
Elevated levels of S100A2 in the serum of NSCLC patients serve as a potential diagnostic and prognostic biomarker, especially in early-stage disease and development of metastasis (97–99). Lung adenocarcinoma (ADC), squamous cell carcinoma (SCC), large cell carcinoma, and atypical carcinoids show high S100A2 expression, while small cell lung carcinoma (SCLC) lacks S100A2 expression (100). Studies reveal that TFAP2A, a transcriptional regulator, increases S100A2 expression, a distinct molecular marker for pre-invasive stages of ADC (101) and this elevation contributes to ADC metastasis by regulating glutamine metabolism (102). Although S100A2 mutations that can be attributed to NSCLC are rare, alterations in the gene have been identified in lung SCC samples (103). Even though S100A3 does not have a direct effect on pathogenesis of lung cancer, it alters the response of lung cancer cells to all-trans retinoic acid (ATRA) treatment by interacting with retinoic acid receptor-alpha (RARα) transcription factor, which results in the degradation of RARα and promyelocytic leukemia (PML)-RARα receptor (104).
High expression of S100A4 facilitates NSCLC metastasis and immunosuppression via exosomes and the STAT3 pathway, which results in poor tumor differentiation, inhibition of autophagy, and worse prognosis (105, 106). S100A4 enhances breast-to-lung metastasis through stanniocalcin 1 (STC1). Inhibiting S100A4 reduces STC1-induced metastatic colonization, indicating its promise as a therapeutic target (107). S100A4 influences lung cancer cell metabolism by regulating mitochondrial function and oxygen consumption, with reduced levels promoting a shift to glycolysis and less aggressive behavior (108).
Increased expression of S100A5 mRNA has been noted in NSCLC, and it is correlated with worse prognosis in non-smoking NSCLC patients (109). Bioinformatic analysis of TCGA-derived lung SCC data identified S100A5 as a key immune-related differentially expressed gene (DEG) for constructing a prognostic model. Integration of S100A5 with ten other genes enables effective prognosis assessment, and this model offers insights for personalized immunotherapy and improved diagnostic strategies for SCC (110).
S100A6 signaling through RAGE may be involved in lung cancer pathogenesis (111), and it is a promising diganostic marker, like S100A2, for early stage NSCLC detection. Its differential expression distinguishes NSCLC from SCLC, correlating with advanced stages and metastasis in lung ADC (112) and worse outcomes in older SCC patients and poorly differentiated tumors (113–115). Hypermethylation of S100A6 promotor confers radiation resistance in NSCLC cell line H1299 (116). Overexpression of S100A6, driven by miR-193a (117) or by HIF-1-α-induced hypermethylation (118) of the S100A6 promoter region, has been linked to the promotion of lung cancer cell proliferation, invasion, migration, and angiogenesis. However, a study suggests that S100A6 expression and its post-translational modifications correlate with improved outcomes in stage 1 NSCLC patients, especially in tumors without p53 expression, suggesting a pro-apoptotic role and potential interactions with p53 (119).
S100A7 act as metabolic regulator in lung ADC (120), driving glycolytic and glutaminolytic pathways, and Hippo pathway-mediated overexpression of it accelerates trans-differentiation from lung ADC to SCC and is associated with poor prognosis (121–123). Silencing S100A7 reduces proliferation, NF-κB activity, and proliferation in lung cancer cells (122, 124). TGFβ-induced lncRNA (TBILA) activates the S100A7-JAB1 signaling pathway, which plays a critical role in regulating the cell cycle and contributes to the progression of NSCLC (125).
S100A8/A9 plays a role in metastasis, as shown in SBC5 (small cell lung carcinoma cell line) invasion via the S100A8/A9-IL6R-TLR4 pathway, a key mechanism facilitating osteolytic activity in bone metastases (126). RAGE-expressing melanoma cells are chemotactically attracted by S100A8/A9 to lung (127). In NSCLC, S100A8, S100A9, and S100A12 proteins serve as potential biomarkers and assist in monitoring therapeutic responses (128, 129). Elevated S100A8 and/or S100A9 levels in male NSCLC and subtype patients, smokers, and those with advanced disease correlate with survival outcomes, suggesting their potential as prognostic markers (130–132). Increased plasma S100A8 levels in NSCLC patients with venous thromboembolism (VTE) suggest its use as a biomarker for VTE diagnosis (133).
Elevated levels of S100A10 are associated with advanced cancer progression, lymph node metastasis, and poor prognosis in lung cancer types, particularly in ADC and SCC (134, 135) attributed to its role in enhanced cell proliferation, invasion via the Akt-mTOR pathway, and increased glycolysis (136). In breast cancer, elevated S100A10 corresponds to lung metastasis, especially the aggressive triple-negative subtype, as supported by both human data and S100A10-deficient mouse models (137, 138). Mechanistically, the interaction of S100A10 with tumor suppressor DLC1 facilitates metastasis, while its binding with AnxA2 contributes to chemotherapy resistance (139, 140). Additionally, co-elevated levels of S100A10, fibronectin, and tenascin-C in lung tumor ECM highlight their potential as a combined biomarker for predicting patient survival (141).
In ADC and SCC, elevated S100A11 expression in patient lung tissues and serum is associated with poor differentiation, KRAS mutations, shorter disease-free survival (142), advanced tumor stages and metastasis (143), and chemoresistance, as reducing its expression sensitizes cancer cells to chemotherapy like cisplatin (144). In contrast to NSCLC, the expression of S100A11 is low in SCLC (145).
Elevated expression of S100A13 in early-stage NSCLC is associated with poorer overall survival and disease-free survival rates. It contributes to enhanced angiogenesis within tumors, promotes invasive behavior of lung cancer cells, and serves as a potential prognostic marker, with higher levels observed in more aggressive cancers (146, 147).
Analyses of lung ADC cases have shown frequent upregulation of S100A14 in tumor tissues and serum correlating strongly with poor differentiation, metastasis, advanced disease stage, smoking history, EGFR mutations, and unfavorable patient outcomes (148, 149). Murine studies have also confirmed that S100A14 is involved in lung metastasis, and in vivo knockdown reaffirms its metastasis-promoting effects (150).
S100A15 has gained attention as an important biomarker in lung cancer progression and prognosis, particularly in lung ADC. Analysis of 178 lung cancer specimens revealed that increased nuclear S100A15 expression is associated with distant metastasis and reduced survival in patients on first-line therapy and predicting three-year mortality (151). Hypomethylation of the S100A15 promoter at sites −423 and −248 correlates with disease progression and decreased one-year survival (151). S100A15 also modulates immune response in NSCLC. Upregulation of S100A15 alongside DOK2 in patients pre- and post-chemotherapy identifies it as a potential biomarker for tumor staging and prognosis (152).
High serum S100B levels are proposed as a sensitive biomarker for early detection of brain metastasis in lung ADC (153, 154), promoting proliferation, migration and invasion inhibiting apoptosis as seen in the PC14/B cell line.
S100P plays a stage-dependent and context-dependent role in lung cancer as observed from two different studies. Rehbein et al. (155) report lung ADC expresses S100P in early/T1 stage, but not in advanced/T2 stage, suggesting early tumor initiation rather than aggressive growth in advanced stages. Overexpression of S100P in H358 cell lines promoted colony formation but paradoxically reduced proliferation and migration. Moreover, S100P expression was found to regulate itself by transcriptional feedback (155). In contrast, Hsu et al. (156) report S100P as a pro-metastatic oncogenic driver in lung cancer. S100P promotes migration, invasion, EMT, and metastasis via integrin α7 and downstream FAK/AKT/Src/ZEB1 signaling. Chein et al. (157) also suggest metastatic potential of S100P as Keap1 mediated reduction in S100P levels and decreases metastasis of NSCLC cells. It was also noted that knocking down S100P expression by shRNA in NSCLC animal models reduced angiogenesis and metastasis (158). S100P along with GATA3 and napsin A expression help to distinguish lung-derived bladder adenocarcinoma from primary bladder adenocarcinoma (159).
S100 proteins in acute lung injury (ALI) and acute respiratory distress syndrome (ARDS)
ALI and ARDS are conditions characterized by the rapid onset of inflammation and damage to lung tissue, leading to impaired gas exchange and respiratory failure. S100 proteins play critical roles in modulating release of proinflammatory cytokines, inflammatory pathways and neutrophils and macrophages responses during these lung injuries.
In ALI, S100A6 is involved in airway epithelial recovery and may affect inflammation and lung damage following EGFR-tyrosine kinase inhibitor treatment (160). Upregulation of S100A6, S100A8, and StefinA3 during airway epithelial repair with gefitinib treatment can increase neutrophil retention, worsening ALI (160).
ALI highlights the role of S100A8/A9 in neutrophil recruitment via TLR4 pathways in alveolar epithelial cells (161). While both proteins can influence neutrophil influx and inflammation, the heterodimer S100A8/A9 exhibits distinct effects. S100A9 promotes mild inflammation through mast cell degranulation and chemokine upregulation, but unlike S100A8, does not induce proinflammatory mediators. Both S100A8 and S100A9 can reduce neutrophil influx in LPS-induced lung injury, potentially through shared mechanisms like sirtuin-1 activation and STAT3 signaling. These findings highlight the distinct roles of S100A8, S100A9, and their heterodimer in lung homeostasis (162).
Elevated levels of S100A12 in BALF and pulmonary tissue suggest its association with neutrophil activation and inflammation. Proinflammatory effects of S100A12 are likely mediated through its interaction with the RAGE receptor, contributing to endothelial activation and further exacerbating lung injury (163). In ARDS, patients exhibit elevated sRAGE, HMGB1, and S100A12 levels, with decreased esRAGE and AGEs. These changes in RAGE isoforms and ligands, including S100A12, differentiate ARDS patients, suggesting a potential role of the RAGE/S100A12 axis in the disease process (164). S100A12 levels in BALF offer promise in distinguishing ARDS from conditions like CF and COPD (165).
Role of S100 proteins as biomarkers in COVID-19 and long COVID
Elevated mRNA expression of S100A6, S100A8, S100A9, and S100P, have been identified in the nasal swabs of COVID-19 patients. They also identified thioredoxin significantly upregulated in those patients. Thioredoxin inhibitor Auranofin has been found effective to mitigate SARS-CoV-2 replication in hamster model. However, a relationship between S100 proteins and thioredoxin was not elucidated in this study (166). S100A8/A9 is most predictive of severe disease and long COVID, driving cytokine storms and chronic inflammation via TLR4/RAGE (167). In severe COVID-19, elevated S100A8/9 levels drive emergency myelopoiesis, leading to the generation of immature neutrophil subsets and resulting in dysfunctional innate immune responses (168, 169). S100A8/9 can activate these immature neutrophils, and macrophages via TLR4 to induce the production of IL-6, TNF-1α, and S100A8 itself in a positive feedback loop to sustain this cycle of events (169). It has been shown that S100A8/A9 induces IL-8 release from bronchial cells and triggers pro-inflammatory responses in endothelial cells (170). High serum levels of S100A8/A9 in patients at hospital admission correlate with poor outcomes and predict severe disease (171). Transcriptomic analyses have shown overexpression of S100A8, S100A9, S100P and S100A12 in lung tissue from fatal COVID-19 patients (172, 173). S100B levels are also found significantly higher in 38% of ICU admitted COVID-19 patients without any clinical evidence of brain injury. It was also higher in patients succumbed to death compared to those who survived. S100B levels in those patients were correlated with IL-6 levels, illness severity and lymphocyte count. However, the exact cellular source of S100B in these patients remains elusive (174, 175). Tissue hypoxia, critical illness and systemic inflammation may be activating/injuring glial cells to secrete S100B (176). Additionally, the levels of S100A4, S100A9, and S100A10 have been shown to influence inflammation and disease severity, associating them with ALI and reduced lymphocyte counts in COVID-19 patients (177).
In the context of long COVID, sustained elevation of S100A8/A9 and inflammatory cytokines like IL-1β, IL-6, and TNFα indicate a chronic pro-inflammatory state, driven by a TLR4/RAGE feedback loop (178). This ongoing inflammation contributes to multi-organ symptoms such as fatigue, brain fog, and persistent inflammation, even after the virus is cleared (179). The continuous expression of proinflammatory cytokines is key to maintain long COVID symptoms (180). Targeting S100 proteins and their pathways offers a potential therapeutic strategy in this condition. Early treatments using inhibitors like ezrin peptides (181) and tocilizumab show promise in disrupting this inflammatory cycle (182). Additionally, inhibition of the binding of S100A8/A9 to TLR4 by paquinimod has shown it can reverse abnormal neutrophil activity and reduce mortality in coronavirus-infected mice (183). Additionally, long-term longitudinal studies have revealed specific perturbations in the immune system, including upregulated expression of S100A8/A9 and associated markers, even 6 months after acute SARS-CoV-2 infection (40). This persistent immune activation underscores the potential for S100 proteins to serve as both biomarkers and therapeutic targets in the management of COVID-19 and its long-term sequelae (Figure 5).
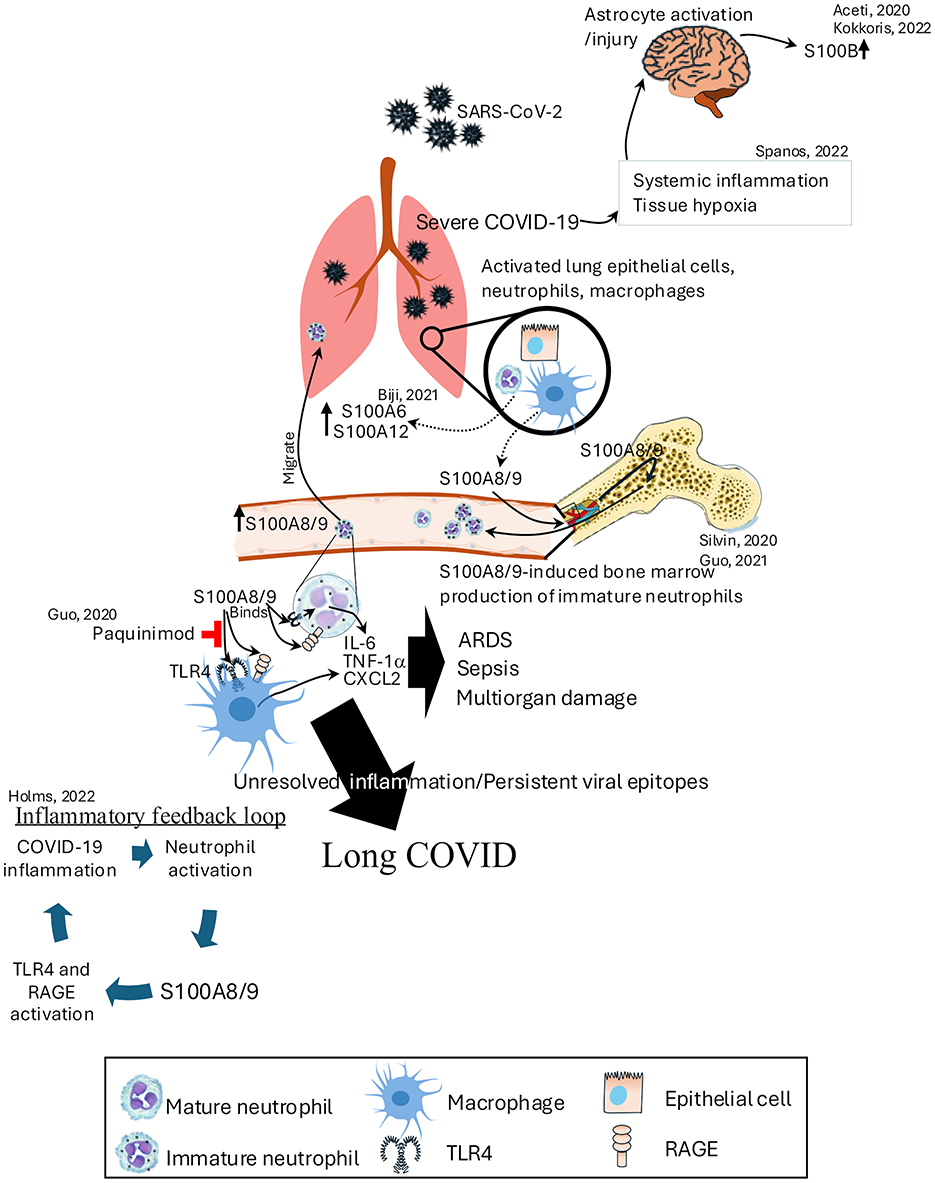
Figure 5. Role of S100 proteins in the pathogenesis of COVID-19 and long COVID. SARS-CoV-2 infection activates lung epithelial cells, neutrophils, and macrophages, leading to the release of S100A8/9, which binds to TLR4 and RAGE receptors. This interaction triggers an inflammatory feedback loop, promoting neutrophil activation and the production of immature neutrophils in the bone marrow. Elevated S100A8/9 levels contribute to severe COVID-19 by inducing ARDS, sepsis, and multiorgan damage through the release of IL-6, TNF-α, and CXCL2. On the other hand, severe COVID-19 increases the systemic inflammation and tissue hypoxia that leads to increased expression of S100B levels. Persistent viral epitopes and unresolved inflammation perpetuate long COVID, with S100A8/9 continuing to drive TLR4 and RAGE activation. ARDS, acute respiratory distress syndrome; CXCL2, chemokine (C-X-C Motif) ligand 2; TNF-α, tumor necrosis factor-alpha.
S100 proteins as early biomarkers and therapeutic targets of graft rejection in lung transplantation
In the context of lung transplantation, elevated levels of S100 proteins such as S100A8, S100A9, and S100A12 can serve as early biomarkers of graft rejection or complications like graft-vs.-host disease. Higher plasma S100A8/A9 levels are associated with prolonged ischemic times and poorer outcomes post-lung transplantation. Treatment with an anti-S100A8/A9 antibody in bronchiolitis obliterans syndrome post-lung transplantation reduces myofibroblast infiltration and inflammation. Because of the damage-associated molecular patterns (DAMPs), they interact with receptors like TLR4 and RAGE, leading to the recruitment and activation of immune cells and the secretion of pro-inflammatory cytokines (184, 185). This inflammatory response can be indicative of transplant rejection or other immune-mediated events, making S100 proteins valuable for monitoring and managing post-transplant inflammation and immune responses in lung transplant patients.
Clinical relevance and biomarker potential
S100 proteins mediates its effect through signaling pathways like RAGE and TLR4, influencing inflammatory mechanisms common to many lung diseases. Their functions vary by context, for example, S100A4 is involved in both tissue remodeling and metastasis, while S100A11 affects inflammation and chemotherapy resistance depending on the microenvironment. These insights suggest S100 proteins could serve as biomarkers for disease severity, prognosis, and therapeutic response; for instance, high levels of S100A8/A9 may indicate severe COVID-19 or pulmonary fibrosis, and S100A12 and S100A8/A9 can help monitor graft rejection in lung transplant patients. The main challenge lies in validating these proteins as reliable biomarkers and integrating them into clinical practice.
Conclusion
In recent years, there has been significant progress in unraveling the roles of S100 proteins in pulmonary diseases, offering potential therapeutic avenues. Despite advancements in understanding S100 protein biology, gaps persist in comprehending the mechanism of many S100 proteins in the etiology of many diseases. Interestingly, the ongoing COVID-19 pandemic has brought to light the potential implication of S100 proteins in tissue damage, highlighting the imperative for further exploration in this field. Continued research on the intricate interactions and signaling mechanisms of S100 proteins is crucial for devising diagnostic biomarkers and innovative therapeutic targets to tackle lung diseases effectively. The ongoing research on S100 proteins may promise future development of tailored therapies in the domain of respiratory medicine.
Points for clinical practice and future research
S100 proteins are emerging as promising biomarkers and therapeutic targets, with significant potential in lung diseases, such as elevated levels of S100A8/A9 (calprotectin) correlating with severe COVID-19 and cytokine storms, which suggests their utility as predictive markers. Monitoring these proteins may also help identify patients at risk for long COVID. Given their role in inflammation, airway remodeling, and tumor progression, S100 proteins are valuable for therapeutic development in pulmonary diseases. However, further research is needed to understand their molecular mechanisms in inflammation, protein-protein interaction, and synergy with other S100 proteins in disease progression and tumor metastasis, as well as their broader potential as cross-disease biomarkers, to enhance clinical applications.
Author contributions
VR: Writing – original draft, Writing – review & editing, Conceptualization. SA: Writing – review & editing. EA: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. We acknowledge that Google Gemini and Microsoft Copilot were used in refining, correcting, and editing the main text.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ADC, adenocarcinoma; ALI, acute lung injury; ARDS, acute respiratory distress syndrome; ATRA, all-trans retinoic acid; BALF, broncho alveolar lavage fluid; COPD, chronic obstructive pulmonary disease; CXCR4, C-X-C chemokine receptor type 4; DAMP, damage-associated molecular pattern; DLC1, deleted in liver cancer 1; DNp63, DeltaNp63; EMT, epithelial-mesenchymal transition; FAK, focal adhesion kinase; FDG, F-fluorodeoxyglucose; GSK-3b, glycogen synthase kinase-3beta; HMGB1, high mobility group box 1; IFN-gR, interferon gamma; IL6R, interleukin 6 receptor; ILD, interstitial lung disease; IP, interstitial pneumonia; IPF, idiopathic pulmonary fibrosis; JAB1, c-Jun activation domain binding protein-1; Keap1, Kelch-like ECH-associated protein 1; lncRNA, long non-coding RNA; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; mTOR, the mammalian target of rapamycin; NDUFS2, NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial; NO, nitric oxide; Nrf2, nuclear factor erythroid 2–related factor 2; NSCLC, non-small cell lung cancer; NuRD, nucleosome remodeling and deacetylase; PAH, pulmonary arterial hypertension; PBMC, peripheral blood mononuclear cells; PF, pulmonary fibrosis; PH, pulmonary hypertension; PML, promyelocytic leukemia; RAGE, receptor for advanced glycation endproducts; RAR-a, retinoic acid receptor alpha; SCC, squamous cell carcinoma; SCLC, small cell lung cancer; sRAGE, solubleRAGE; STAT3, signal transducer and activator of transcription 3; TAZ, transcriptional coactivator with PDZ-binding motif; TCGA, the cancer genome atlas; TEAD, TEA domain family member 1; TERT, telomerase reverse transcriptase; TFAP2A, transcription factor AP-2 alpha; TGF-b1, transforming growth factor-beta1; TLR, toll-like receptor; TTF1, thyroid transcription factor 1; VTE, venous thromboembolism; YAP, yes-associated protein 1.
References
1. Poluri KM, Gulati K, Tripathi DK, Nagar N. Protein networks in human disease. In: Protein-Protein Interactions: Pathophysiological and Therapeutic Aspects: Volume II. Singapore: Springer Nature Singapore (2023). p. 1–41. doi: 10.1007/978-981-99-2423-3_1
2. Uversky VN. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front Mol Biosci. (2014) 1:6. doi: 10.3389/fmolb.2014.00006
3. Neagu, M, Constantin C. Signal transduction in immune cells and protein kinases. In:Engin AB, Engin A, , editors. Protein Kinase-Mediated Decisions Between Life and Death. Cham: Springer International Publishing (2021). p. 133–49. doi: 10.1007/978-3-030-49844-3_5
4. Poluri KM, Gulati K, Sarkar S, editors. Structural and functional properties of proteins. In: Protein-Protein Interactions: Principles and Techniques: Volume I. Singapore: Springer Singapore (2021). p. 1–60. doi: 10.1007/978-981-16-1594-8_1
5. Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, et al. Functions of S100 proteins. Curr Mol Med. (2013) 13:24–57. doi: 10.2174/156652413804486214
6. Donato R, Geczy CL, Weber DJ. S100 proteins. In:Kretsinger RH, Uversky VN, Permyakov EA, , editors. Encyclopedia of Metalloproteins. New York, NY: Springer New York (2013). p. 1863–74. doi: 10.1007/978-1-4614-1533-6_48
7. Moore BW. A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun. (1965) 19:739–44. doi: 10.1016/0006-291X(65)90320-7
8. Donato R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim Biophys Acta. (1999) 1450:191–231. doi: 10.1016/S0167-4889(99)00058-0
9. Eckert RL, Broome A-M, Ruse M, Robinson N, Ryan D, Lee K. S100 proteins in the epidermis. J Invest Dermatol. (2004) 123:23–33. doi: 10.1111/j.0022-202X.2004.22719.x
10. Sattar Z, Lora A, Jundi B, Railwah C, Geraghty P. The S100 protein family as players and therapeutic targets in pulmonary diseases. Pulm Med. (2021) 2021:5488591. doi: 10.1155/2021/5488591
11. Hunt WR, Helfman BR, McCarty NA, Hansen JM. Advanced glycation end products are elevated in cystic fibrosis-related diabetes and correlate with worse lung function. J Cyst Fibros. (2016) 15:681–8. doi: 10.1016/j.jcf.2015.12.011
12. Presland RB, Haydock PV, Fleckman P, Nirunsuksiri W, Dale BA. Characterization of the human epidermal profilaggrin gene. Genomic organization and identification of an S-100-like calcium binding domain at the amino terminus. J Biol Chem. (1992) 267:23772–81. doi: 10.1016/S0021-9258(18)35905-2
13. Wu Z, Hansmann B, Meyer-Hoffert U, Gläser R, Schröder J-M. Molecular identification and expression analysis of filaggrin-2, a member of the S100 fused-type protein family. PLoS ONE. (2009) 4:e5227. doi: 10.1371/journal.pone.0005227
14. Contzler R, Favre B, Huber M, Hohl D. Cornulin, a new member of the “fused gene” family, is expressed during epidermal differentiation. J Invest Dermatol. (2005) 124:990–7. doi: 10.1111/j.0022-202X.2005.23694.x
15. Krieg P, Schuppler M, Koesters R, Mincheva A, Lichter P, Marks F. Repetin (Rptn), a new member of the “fused gene” subgroup within the S100 gene family encoding a murine epidermal differentiation protein. Genomics. (1997) 43:339–48. doi: 10.1006/geno.1997.4818
16. Sedaghat F, Notopoulos A. S100 protein family and its application in clinical practice. Hippokratia. (2008) 12:198–204.
17. Weisz J, Uversky VN. Zooming into the dark side of human annexin-S100 complexes: dynamic alliance of flexible partners. Int J Mol Sci. (2020) 21:5879. doi: 10.3390/ijms21165879
18. Zimmer DB, Wright Sadosky P, Weber DJ. Molecular mechanisms of S100-target protein interactions. Microsc Res Tech. (2003) 60:552–9. doi: 10.1002/jemt.10297
19. Donato R, Sorci G, Riuzzi F, Arcuri C, Bianchi R, Brozzi F, et al. S100B's double life: intracellular regulator and extracellular signal. Biochim Biophys Acta. (2009) 1793:1008–22. doi: 10.1016/j.bbamcr.2008.11.009
20. Li Y, Wang J, Sheng JG, Liu L, Barger SW, Jones RA, et al. S100β increases levels of β-amyloid precursor protein and its encoding mRNA in rat neuronal cultures. J Neurochem. (1998) 71:1421–8. doi: 10.1046/j.1471-4159.1998.71041421.x
21. Imbalzano E, Mandraffino G, Casciaro M, Quartuccio S, Saitta A, Gangemi S. Pathophysiological mechanism and therapeutic role of S100 proteins in cardiac failure: a systematic review. Heart Fail Rev. (2016) 21:463–73. doi: 10.1007/s10741-016-9529-8
22. Yoshinouchi T, Ohtsuki Y, Ueda R, Sato S, Ueda N. Myofibroblasts and S-100 protein positive cells in idiopathic pulmonary fibrosis and rheumatoid arthritis-associated interstitial pneumonia. Eur Respir J. (1999) 14:579–84. doi: 10.1034/j.1399-3003.1999.14c16.x
23. Shimizu S, Yoshinouchi T, Ohtsuki Y, Fujita J, Sugiura Y, Banno S, et al. The appearance of S-100 protein-positive dendritic cells and the distribution of lymphocyte subsets in idiopathic nonspecific interstitial pneumonia. Respir Med. (2002) 96:770–6. doi: 10.1053/rmed.2002.1345
24. Kimura K, Endo Y, Yonemura Y, Heizmann CW, Schafer BW, Watanabe Y, et al. Clinical significance of S100A4 and E-cadherin-related adhesion molecules in non-small cell lung cancer. Int J Oncol. (2000) 16:1125–56. doi: 10.3892/ijo.16.6.1125
25. Jiang W, Jia Q, Liu L, Zhao X, Tan A, Ma N, et al. S100B promotes the proliferation, migration and invasion of specific brain metastatic lung adenocarcinoma cell line. Cell Biochem Funct. (2011) 29:582–8. doi: 10.1002/cbf.1791
26. Gilston BA, Skaar EP, Chazin WJ. Binding of transition metals to S100 proteins. Sci China Life Sci. (2016) 59:792–801. doi: 10.1007/s11427-016-5088-4
27. Baudier J, Holtzscherer C, Gerard D. Zinc-dependent affinity chromatography of the S100b protein on phenyl-Sepharose. A rapid purification method. FEBS Lett. (1982) 148:231–4. doi: 10.1016/0014-5793(82)80813-2
28. Baudier J, Glasser N, Haglid K. Gerard D. Purification, characterization and ion binding properties of human brain S100b protein. Biochim Biophys Acta. (1984) 790:164–73. doi: 10.1016/0167-4838(84)90220-6
29. Nishikawa T, Lee IS, Shiraishi N, Ishikawa T, Ohta Y, Nishikimi M. Identification of S100b protein as copper-binding protein and its suppression of copper-induced cell damage. J Biol Chem. (1997) 272:23037–41. doi: 10.1074/jbc.272.37.23037
30. Wheeler LC, Donor MT, Prell JS, Harms MJ. Multiple evolutionary origins of ubiquitous Cu2+ and Zn2+ binding in the S100 protein family. PLoS ONE. (2016) 11:e0164740. doi: 10.1371/journal.pone.0164740
31. Sivaraja V, Kumar TK, Rajalingam D, Graziani I, Prudovsky I, Yu C. Copper binding affinity of S100A13, a key component of the FGF-1 nonclassical copper-dependent release complex. Biophysical J. (2006) 91:1832–43. doi: 10.1529/biophysj.105.079988
32. Moroz OV, Burkitt W, Wittkowski H, He W, Ianoul A, Novitskaya V, et al. Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Biochem. (2009) 10:11. doi: 10.1186/1471-2091-10-11
33. Mäler L, Sastry M, Chazin WJ. A structural basis for S100 protein specificity derived from comparative analysis of apo and Ca(2+)-calcyclin. J Mol Biol. (2002) 317:279–90. doi: 10.1006/jmbi.2002.5421
34. Bhattacharya S, Bunick CG, Chazin WJ. Target selectivity in EF-hand calcium binding proteins. Biochim Biophys Acta. (2004) 1742:69–79. doi: 10.1016/j.bbamcr.2004.09.002
35. Ite K, Yonezawa K, Kitanishi K, Shimizu N, Unno M. Optimal mutant model of human S100A3 protein citrullinated at Arg51 by peptidylarginine deiminase type III and its solution structural properties. ACS Omega. (2020) 5:4032–42. doi: 10.1021/acsomega.9b03618
36. Moroz O, Wilson K, Bronstein I. The role of zinc in the S100 proteins: insights from the X-ray structures. Amino Acids. (2010) 41:761–72. doi: 10.1007/s00726-010-0540-4
37. Chazin WJ. Relating form and function of EF-hand calcium binding proteins. Acc Chem Res. (2011) 44:171–9. doi: 10.1021/ar100110d
38. Zuo W, Tian M, Qi J, Zhang G, Hu J, Wang S, et al. The functions of EF-hand proteins from host and zoonotic pathogens. Microbes Infect. (2025) 27:105276. doi: 10.1016/j.micinf.2023.105276
39. Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. (2003) 60:540–51. doi: 10.1002/jemt.10296
40. Singh P, Ali SA. Multifunctional role of S100 protein family in the immune system: an update. Cells. (2022) 11:2274. doi: 10.3390/cells11152274
41. Hammad H, Lambrecht BN. The basic immunology of asthma. Cell. (2021) 184:1469–85. doi: 10.1016/j.cell.2021.02.016
42. Liu S, Liu M, Zhong J, Chen S, Wang Z, Gao X, et al. Anti-S100A4 antibody administration alleviates bronchial epithelial-mesenchymal transition in asthmatic mice. Open Med. (2023) 18:20220622. doi: 10.1515/med-2022-0622
43. Ning Q, Li F, Wang L, Li H, Yao Y, Hu T, et al. S100A4 amplifies TGF-β-induced epithelial-mesenchymal transition in a pleural mesothelial cell line. J Investig Med. (2018) 66:334–9. doi: 10.1136/jim-2017-000542
44. Wu Y, Zhang W, Gunst SJ. S100A4 is secreted by airway smooth muscle tissues and activates inflammatory signaling pathways via receptors for advanced glycation end products. Am J Physiol Lung Cell Mol Physiol. (2020) 319:L185–l195. doi: 10.1152/ajplung.00347.2019
45. Kim DH, Gu A, Lee JS, Yang EJ, Kashif A, Hong MH, et al. Suppressive effects of S100A8 and S100A9 on neutrophil apoptosis by cytokine release of human bronchial epithelial cells in asthma. Int J Med Sci. (2020) 17:498–509. doi: 10.7150/ijms.37833
46. Chatziparasidis G, Kantar A. Calprotectin: an ignored biomarker of neutrophilia in pediatric respiratory diseases. Children. (2021) 8:428. doi: 10.3390/children8060428
47. Gharib SA, Nguyen EV, Lai Y, Plampin JD, Goodlett DR, Hallstrand TS. Induced sputum proteome in healthy subjects and asthmatic patients. J Allergy Clin Immunol. (2011) 128:1176–84.e6. doi: 10.1016/j.jaci.2011.07.053
48. Decaesteker T, Bos S, Lorent N, Everaerts S, Vanoirbeek J, Bullens D, et al. Elevated serum calprotectin (S100A8/A9) in patients with severe asthma. J Asthma. (2022) 59:1110–5. doi: 10.1080/02770903.2021.1914649
49. Lee YG, Hong J, Lee PH, Lee J, Park SW, Kim D, et al. Serum calprotectin is a potential marker in patients with asthma. J Korean Med Sci. (2020) 35:e362. doi: 10.3346/jkms.2020.35.e362
50. Palmer LD, Maloney KN, Boyd KL, Goleniewska AK, Toki S, Maxwell CN, et al. The innate immune protein S100A9 protects from T-helper cell type 2-mediated allergic airway inflammation. Am J Respir Cell Mol Biol. (2019) 61:459–68. doi: 10.1165/rcmb.2018-0217OC
51. Lee TH, Jang AS, Park JS, Kim TH, Choi YS, Shin HR, et al. Elevation of S100 calcium binding protein A9 in sputum of neutrophilic inflammation in severe uncontrolled asthma. Ann Allergy Asthma Immunol. (2013) 111:268–75.e1. doi: 10.1016/j.anai.2013.06.028
52. Yin LM, Li HY, Zhang QH, Xu YD, Wang Y, Jiang YL, et al. Effects of S100A9 in a rat model of asthma and in isolated tracheal spirals. Biochem Biophys Res Commun. (2010) 398:547–52. doi: 10.1016/j.bbrc.2010.06.116
53. Liang Q, Fu J, Wang X, Liu L, Xiao W, Gao Y, et al. circS100A11 enhances M2a macrophage activation and lung inflammation in children with asthma. Allergy. (2023) 78:1459–72. doi: 10.1111/all.15515
54. Cheng M, Shi YL, Shang PP, Chen YJ, Xu YD. Inhibitory effect of S100A11 on airway smooth muscle contraction and airway hyperresponsiveness. Curr Med Sci. (2022) 42:333–40. doi: 10.1007/s11596-022-2559-7
55. Mokari-Yamchi A, Sharifi A. Kheirouri S. Increased serum levels of S100A1, ZAG, and adiponectin in cachectic patients with COPD. Int J Chron Obstruct Pulmon Dis. (2018) 13:3157–63. doi: 10.2147/COPD.S172996
56. Reimann S, Fink L, Wilhelm J, Hoffmann J, Bednorz M, Seimetz M, et al. Increased S100A4 expression in the vasculature of human COPD lungs and murine model of smoke-induced emphysema. Respir Res. (2015) 16:127. doi: 10.1186/s12931-015-0284-5
57. Qin HY Li MD, Xie GF, Cao W, Xu DX, Zhao H, et al. Associations among S100A4, sphingosine-1-phosphate, and pulmonary function in patients with chronic obstructive pulmonary disease. Oxid Med Cell Longev. (2022) 2022:6041471. doi: 10.1155/2022/6041471
58. Chen M, Wang T, Shen Y, Xu D, Li X, An J, et al. Knockout of RAGE ameliorates mainstream cigarette smoke-induced airway inflammation in mice. Int Immunopharmacol. (2017) 50:230–5. doi: 10.1016/j.intimp.2017.06.018
59. Huang SJ, Ding ZN, Xiang HX, Fu L, Fei J. Association between serum S100A8/S100A9 heterodimer and pulmonary function in patients with acute exacerbation of chronic obstructive pulmonary disease. Lung. (2020) 198:645–52. doi: 10.1007/s00408-020-00376-9
60. Xue T, Zheng L, Dong F, Zhou G, Zhong X. [Single-cell RNA sequencing combined experimental verifies the core genes of dendritic cells in chronic obstructive pulmonary disease]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. (2024) 40:97–105.
61. Merkel D, Rist W, Seither P, Weith A, Lenter MC. Proteomic study of human bronchoalveolar lavage fluids from smokers with chronic obstructive pulmonary disease by combining surface-enhanced laser desorption/ionization-mass spectrometry profiling with mass spectrometric protein identification. Proteomics. (2005) 5:2972–80. doi: 10.1002/pmic.200401180
62. Miniati M, Monti S, Basta G, Cocci F, Fornai E, Bottai M. Soluble receptor for advanced glycation end products in COPD: relationship with emphysema and chronic cor pulmonale: a case-control study. Respir Res. (2011) 12:37. doi: 10.1186/1465-9921-12-37
63. Gaudin C, Ghinnagow R, Lemaire F, Villeret B, Sermet-Gaudelus I, Sallenave JM. Abnormal functional lymphoid tolerance and enhanced myeloid exocytosis are characteristics of resting and stimulated PBMCs in cystic fibrosis patients. Front Immunol. (2024) 15:1360716. doi: 10.3389/fimmu.2024.1360716
64. McMorran BJ, Patat SA, Carlin JB, Grimwood K, Jones A, Armstrong DS, et al. Novel neutrophil-derived proteins in bronchoalveolar lavage fluid indicate an exaggerated inflammatory response in pediatric cystic fibrosis patients. Clin Chem. (2007) 53:1782–91. doi: 10.1373/clinchem.2007.087650
65. Thomas GR, Costelloe EA, Lunn DP, Stacey KJ, Delaney SJ, Passey R, et al. G551D cystic fibrosis mice exhibit abnormal regulation of inflammation in lungs and macrophages. J Immunol. (2000) 164:3870–7. doi: 10.4049/jimmunol.164.7.3870
66. Tirkos S, Newbigging S, Nguyen V, Keet M, Ackerley C, Kent G, et al. Expression of S100A8 correlates with inflammatory lung disease in congenic mice deficient of the cystic fibrosis transmembrane conductance regulator. Respir Res. (2006) 7:51. doi: 10.1186/1465-9921-7-51
67. Makam M, Diaz D, Laval J, Gernez Y, Conrad CK, Dunn CE, et al. Activation of critical, host-induced, metabolic and stress pathways marks neutrophil entry into cystic fibrosis lungs. Proc Natl Acad Sci U S A. (2009) 106:5779–83. doi: 10.1073/pnas.0813410106
68. Foell D, Seeliger S, Vogl T, Koch HG, Maschek H, Harms E, et al. Expression of S100A12 (EN-RAGE) in cystic fibrosis. Thorax. (2003) 58:613–7. doi: 10.1136/thorax.58.7.613
69. Lai YC, Potoka KC, Champion HC, Mora AL, Gladwin MT. Pulmonary arterial hypertension: the clinical syndrome. Circ Res. (2014) 115:115–30. doi: 10.1161/CIRCRESAHA.115.301146
70. Pleger ST, Harris DM, Shan C, Vinge LE, Chuprun JK, Berzins B, et al. Endothelial S100A1 modulates vascular function via nitric oxide. Circ Res. (2008) 102:786–94. doi: 10.1161/CIRCRESAHA.108.172031
71. Weinberger B, Heck DE, Laskin DL, Laskin JD. Nitric oxide in the lung: therapeutic and cellular mechanisms of action. Pharmacol Ther. (1999) 84:401–11. doi: 10.1016/S0163-7258(99)00044-3
72. Montani D, Günther S, Dorfmüller P, Perros F, Girerd B, Garcia G, et al. Pulmonary arterial hypertension. Orphanet J Rare Dis. (2013) 8:97. doi: 10.1186/1750-1172-8-97
73. Teichert-Kuliszewska K, Tsoporis JN, Desjardins JF, Yin J, Wang L, Kuebler WM, et al. Absence of the calcium-binding protein, S100A1, confers pulmonary hypertension in mice associated with endothelial dysfunction and apoptosis. Cardiovasc Res. (2014) 105:8–19. doi: 10.1093/cvr/cvu241
74. Nakamura K, Sakaguchi M, Matsubara H, Akagi S, Sarashina T, Ejiri K, et al. Crucial role of RAGE in inappropriate increase of smooth muscle cells from patients with pulmonary arterial hypertension. PLoS ONE. (2018) 13:e0203046. doi: 10.1371/journal.pone.0203046
75. Bouzina H, Hesselstrand R, Rådegran G. Plasma insulin-like growth factor binding protein 1 in pulmonary arterial hypertension. Scand Cardiovasc J. (2021) 55:35–42. doi: 10.1080/14017431.2020.1782977
76. Amano H, Maruyama K, Naka M, Tanaka T. Target validation in hypoxia-induced vascular remodeling using transcriptome/metabolome analysis. Pharmacogenomics J. (2003) 3:183–8. doi: 10.1038/sj.tpj.6500177
77. Jiang M, Bu W, Wang X, Ruan J, Shi W, Yu S, et al. Pulmonary fibrosis: from mechanisms to therapies. J Transl Med. (2025) 23:515. doi: 10.1186/s12967-025-06514-2
78. Huang G, Zhang J, Qing G, Liu D, Wang X, Chen Y, et al. S100A2 silencing relieves epithelial-mesenchymal transition in pulmonary fibrosis by inhibiting the Wnt/β-catenin signaling pathway. DNA Cell Biol. (2021) 40:18–25. doi: 10.1089/dna.2020.6030
79. Al-Mutairy EA, Imtiaz FA, Khalid M, Al Qattan S, Saleh S, Mahmoud LM, et al. An atypical pulmonary fibrosis is associated with co-inheritance of mutations in the calcium binding protein genes S100A3 and S100A13. Eur Respir J. (2019) 54:1802041. doi: 10.1183/13993003.02041-2018
80. Al-Mutairy EA, Al Qattan S, Khalid M, Al-Enazi AA, Al-Saif MM, Imtiaz F, et al. Wild-type S100A3 and S100A13 restore calcium homeostasis and mitigate mitochondrial dysregulation in pulmonary fibrosis patient-derived cells. Front Cell Dev Biol. (2023) 11:1282868. doi: 10.3389/fcell.2023.1282868
81. Qattan SA, Al-Mohanna F, Raveendran VV, Abdulqawi R, Saleh RA, Almutairy EA. S100A3 regulates the expression of telomerase reverse transcriptase (TERT), in abstract. Am Thoracic Soc. (2023) 207:A2231. doi: 10.1164/ajrccm-conference.2023.207.1_MeetingAbstracts.A2231
82. Raveendran VV, AlQattan S, Alaiya A, Abdulqawi R, Saleh RA, Mohammed SF, et al. Interaction of S100A13 With vimentin in pulmonary fibrosis, in abstract. Am Thoracic Soc. (2023) 207:A2232. doi: 10.1164/ajrccm-conference.2023.207.1_MeetingAbstracts.A2232
83. Zhang W, Ohno S, Steer B, Klee S, Staab-Weijnitz CA, Wagner D, et al. S100a4 is secreted by alternatively activated alveolar macrophages and promotes activation of lung fibroblasts in pulmonary fibrosis. Front Immunol. (2018) 9:1216. doi: 10.3389/fimmu.2018.01216
84. Li Y, Bao J, Bian Y, Erben U, Wang P, Song K, et al. S100A4(+) Macrophages Are Necessary for Pulmonary Fibrosis by Activating Lung Fibroblasts. Front Immunol. (2018) 9:1776. doi: 10.3389/fimmu.2018.01776
85. Landi C, Bargagli E, Carleo A, Refini RM, Bennett D, Bianchi L, et al. Bronchoalveolar lavage proteomic analysis in pulmonary fibrosis associated with systemic sclerosis: S100A6 and 14-3-3epsilon as potential biomarkers. Rheumatology. (2019) 58:165–78. doi: 10.1093/rheumatology/key223
86. Xia H, Herrera J, Smith K, Yang L, Gilbertsen A, Benyumov A, et al. Hyaluronan/CD44 axis regulates S100A4-mediated mesenchymal progenitor cell fibrogenicity in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. (2021) 320:L926–41. doi: 10.1152/ajplung.00456.2020
87. Southern BD Li H, Mao H, Crish JF, Grove LM, Scheraga RG, et al. A novel mechanoeffector role of fibroblast S100A4 in myofibroblast transdifferentiation and fibrosis. J Biol Chem. (2024) 300:105530. doi: 10.1016/j.jbc.2023.105530
88. Breen EC, Tang K. Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. J Cell Biochem. (2003) 88:848–54. doi: 10.1002/jcb.10398
89. Breen EC, Fu Z, Normand H. Calcyclin gene expression is increased by mechanical strain in fibroblasts and lung. Am J Respir Cell Mol Biol. (1999) 21:746–52. doi: 10.1165/ajrcmb.21.6.3312
90. Lin L, Zhao Y, Li Z, Li Y, Wang W, Kang J, et al. Expression of S100A9 and KL-6 in common interstitial lung diseases. Medicine. (2022) 101:e29198. doi: 10.1097/MD.0000000000029198
91. Hesselstrand R, Wildt M, Bozovic G, Andersson-Sjöland A, Andréasson K, Scheja A, et al. Biomarkers from bronchoalveolar lavage fluid in systemic sclerosis patients with interstitial lung disease relate to severity of lung fibrosis. Respir Med. (2013) 107:1079–86. doi: 10.1016/j.rmed.2013.03.015
92. Araki K, Kinoshita R, Tomonobu N, Gohara Y, Tomida S, Takahashi Y, et al. The heterodimer S100A8/A9 is a potent therapeutic target for idiopathic pulmonary fibrosis. J Mol Med. (2021) 99:131–45. doi: 10.1007/s00109-020-02001-x
93. Juang YM, Lai BH, Chien HJ, Ho M, Cheng TJ, Lai CC. Changes in protein expression in rat bronchoalveolar lavage fluid after exposure to zinc oxide nanoparticles: an iTRAQ proteomic approach. Rapid Commun Mass Spectrom. (2014) 28:974–80. doi: 10.1002/rcm.6866
94. Li Y, He Y, Chen S, Wang Q, Yang Y, Shen D, et al. S100A12 as biomarker of disease severity and prognosis in patients with idiopathic pulmonary fibrosis. Front Immunol. (2022) 13:810338. doi: 10.3389/fimmu.2022.810338
95. Tanaka N, Ikari J, Anazawa R, Suzuki M, Katsumata Y, Shimada A, et al. S100A12 inhibits fibroblast migration via the receptor for advanced glycation end products and p38 MAPK signaling. In Vitro Cell Dev Biol Anim. (2019) 55:656–64. doi: 10.1007/s11626-019-00384-x
96. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. (2014) 14:535–46. doi: 10.1038/nrc3775
97. Wang T, Liang Y, Thakur A, Zhang S, Yang T, Chen T, et al. Diagnostic significance of S100A2 and S100A6 levels in sera of patients with non-small cell lung cancer. Tumour Biol. (2016) 37:2299–304. doi: 10.1007/s13277-015-4057-z
98. Diederichs S, Bulk E, Steffen B, Ji P, Tickenbrock L, Lang K, et al. S100 family members and trypsinogens are predictors of distant metastasis and survival in early-stage non-small cell lung cancer. Cancer Res. (2004) 64:5564–9. doi: 10.1158/0008-5472.CAN-04-2004
99. Bargagli E, Olivieri C, Prasse A, Bianchi N, Magi B, Cianti R, et al. Calgranulin B (S100A9) levels in bronchoalveolar lavage fluid of patients with interstitial lung diseases. Inflammation. (2008) 31:351–4. doi: 10.1007/s10753-008-9085-z
100. Wang T, Liang Y, Thakur A, Zhang S, Liu F, Khan H, et al. Expression and clinicopathological significance of S100 calcium binding protein A2 in lung cancer patients of Chinese Han ethnicity. Clin Chim Acta. (2017) 464:118–22. doi: 10.1016/j.cca.2016.11.027
101. Shimada A, Kano J, Ishiyama T, Okubo C, Iijima T, Morishita Y, et al. Establishment of an immortalized cell line from a precancerous lesion of lung adenocarcinoma, and genes highly expressed in the early stages of lung adenocarcinoma development. Cancer Sci. (2005) 96:668–75. doi: 10.1111/j.1349-7006.2005.00100.x
102. Zeng T, Ren W, Zeng H, Wang D, Wu X, Xu G. TFAP2A activates S100A2 to mediate glutamine metabolism and promote lung adenocarcinoma metastasis. Clin Respir J. (2024) 18:e13825. doi: 10.1111/crj.13825
103. Strazisar M, Rott T, Glavac D. Frequent polymorphic variations but rare tumour specific mutations of the S100A2 on 1q21 in non-small cell lung cancer. Lung Cancer. (2009) 63:354–9. doi: 10.1016/j.lungcan.2008.06.005
104. Gianni M, Terao M, Kurosaki M, Paroni G, Brunelli L, Pastorelli R, et al. Correction: S100A3 a partner protein regulating the stability/activity of RARalpha and PML-RARalpha in cellular models of breast/lung cancer and acute myeloid leukemia. Oncogene. (2023) 42:254–8. doi: 10.1038/s41388-022-02564-8
105. Wu X, Zhang H, Jiang G, Peng M, Li C, Lu J, et al. Exosome-transmitted S100A4 induces immunosuppression and non-small cell lung cancer development by activating STAT3. Clin Exp Immunol. (2022) 210:309–20. doi: 10.1093/cei/uxac102
106. Stewart RL, Carpenter BL, West DS, Knifley T, Liu L, Wang C, et al. S100A4 drives non-small cell lung cancer invasion, associates with poor prognosis, and is effectively targeted by the FDA-approved anti-helminthic agent niclosamide. Oncotarget. (2016) 7:34630–42. doi: 10.18632/oncotarget.8969
107. Liu A, Li Y, Lu S, Cai C, Zou F, Meng X. Stanniocalcin 1 promotes lung metastasis of breast cancer by enhancing EGFR-ERK-S100A4 signaling. Cell Death Dis. (2023) 14:395. doi: 10.1038/s41419-023-05911-z
108. Liu L, Qi L, Knifley T, Piecoro DW, Rychahou P, Liu J, et al. S100A4 alters metabolism and promotes invasion of lung cancer cells by up-regulating mitochondrial complex I protein NDUFS2. J Biol Chem. (2019) 294:7516–27. doi: 10.1074/jbc.RA118.004365
109. Liu Y, Cui J, Tang YL, Huang L, Zhou CY, Xu JX. Prognostic roles of mRNA expression of S100 in non-small-cell lung cancer. Biomed Res Int. (2018) 2018:9815806. doi: 10.1155/2018/9815806
110. Pu J, Teng Z, Yang W, Zhu P, Zhang T, Zhang D, et al. Construction of a prognostic model for lung squamous cell carcinoma based on immune-related genes. Carcinogenesis. (2023) 44:143–52. doi: 10.1093/carcin/bgac098
111. Hsieh HL, Schäfer BW, Sasaki N, Heizmann CW. Expression analysis of S100 proteins and RAGE in human tumors using tissue microarrays. Biochem Biophys Res Commun. (2003) 307:375–81. doi: 10.1016/S0006-291X(03)01190-2
112. Jia R, Sui Z, Zhang H, Yu Z. Identification and validation of immune-related gene signature for predicting lymph node metastasis and prognosis in lung adenocarcinoma. Front Mol Biosci. (2021) 8:679031. doi: 10.3389/fmolb.2021.679031
113. Lee HS, Park JW, Chertov O, Colantonio S, Simpson JT, Fivash MJ, et al. Matrix-assisted laser desorption/ionization mass spectrometry reveals decreased calcylcin expression in small cell lung cancer. Pathol Int. (2012) 62:28–35. doi: 10.1111/j.1440-1827.2011.02783.x
114. Ishii A, Suzuki M, Satomi K, Kobayashi H, Sakashita S, Kano J, et al. Increased cytoplasmic S100A6 expression is associated with pulmonary adenocarcinoma progression. Pathol Int. (2009) 59:623–30. doi: 10.1111/j.1440-1827.2009.02417.x
115. He X, Xu X, Khan AQ, Ling W. High expression of S100A6 predicts unfavorable prognosis of lung squamous cell cancer. Med Sci Monit. (2017) 23:5011–7. doi: 10.12659/MSM.904279
116. Kim EH, Park AK, Dong SM, Ahn JH, Park WY. Global analysis of CpG methylation reveals epigenetic control of the radiosensitivity in lung cancer cell lines. Oncogene. (2010) 29:4725–31. doi: 10.1038/onc.2010.223
117. Li P, Lv X, Zhang Z, Xie S. S100A6/miR193a regulates the proliferation, invasion, migration and angiogenesis of lung cancer cells through the P53 acetylation. Am J Transl Res. (2019) 11:4634–49.
118. Wang T, Zhu G, Wang B, Hu M, Gong C, Tan K, et al. Activation of hypoxia inducible factor-1 alpha-mediated DNA methylation enzymes (DNMT3a and TET2) under hypoxic conditions regulates S100A6 Transcription to promote lung cancer cell growth and metastasis. Antioxid Redox Signal. 2024. doi: 10.1089/ars.2023.0397
119. De Petris L, Orre LM, Kanter L, Pernemalm M, Koyi H, Lewensohn R, et al. Tumor expression of S100A6 correlates with survival of patients with stage I non-small-cell lung cancer. Lung Cancer. (2009) 63:410–7. doi: 10.1016/j.lungcan.2008.06.003
120. Zhang J, Wang X, Song C, Li Q. Identification of four metabolic subtypes and key prognostic markers in lung adenocarcinoma based on glycolytic and glutaminolytic pathways. BMC Cancer. (2023) 23:152. doi: 10.1186/s12885-023-10622-x
121. Qi Z, Li T, Kong F, Li Y, Wang R, Wang J, et al. The characteristics and function of s100a7 induction in squamous cell carcinoma: heterogeneity, promotion of cell proliferation and suppression of differentiation. PLoS ONE. (2015) 10:e0128887. doi: 10.1371/journal.pone.0128887
122. Liu G, Wu Q, Liu G, Song X, Zhang J. Knockdown of S100A7 reduces lung squamous cell carcinoma cell growth in vitro and in vivo. Int J Clin Exp Pathol. (2014) 7:8279–89.
123. Zhang H, Zhao Q, Chen Y, Wang Y, Gao S, Mao Y, et al. Selective expression of S100A7 in lung squamous cell carcinomas and large cell carcinomas but not in adenocarcinomas and small cell carcinomas. Thorax. (2008) 63:352–9. doi: 10.1136/thx.2007.087015
124. Hu M, Ye L, Ruge F, Zhi X, Zhang L, Jiang WG. The clinical significance of Psoriasin for non-small cell lung cancer patients and its biological impact on lung cancer cell functions. BMC Cancer. (2012) 12:588. doi: 10.1186/1471-2407-12-588
125. Lu Z, Li Y, Che Y, Huang J, Sun S, Mao S, et al. The TGFβ-induced lncRNA TBILA promotes non-small cell lung cancer progression in vitro and in vivo via cis-regulating HGAL and activating S100A7/JAB1 signaling. Cancer Lett. (2018) 432:156–68. doi: 10.1016/j.canlet.2018.06.013
126. Luo G, Tang M, Zhao Q, Lu L, Xie Y, Li Y, et al. Bone marrow adipocytes enhance osteolytic bone destruction by activating 1q213(S100A7/8/9-IL6R)-TLR4 pathway in lung cancer. J Cancer Res Clin Oncol. (2020) 146:2241–53. doi: 10.1007/s00432-020-03277-9
127. Saha A, Lee YC, Zhang Z, Chandra G, Su SB, Mukherjee AB. Lack of an endogenous anti-inflammatory protein in mice enhances colonization of B16F10 melanoma cells in the lungs. J Biol Chem. (2010) 285:10822–31. doi: 10.1074/jbc.M109.083550
128. Lim MY, Thomas PS. Biomarkers in exhaled breath condensate and serum of chronic obstructive pulmonary disease and non-small-cell lung cancer. Int J Chronic Dis. (2013) 2013:578613. doi: 10.1155/2013/578613
129. Rostila AM, Anttila SL, Lalowski MM, Vuopala KS, Toljamo TI, Lindström I, et al. Reactive oxygen species-regulating proteins peroxiredoxin 2 and thioredoxin, and glyceraldehyde-3-phosphate dehydrogenase are differentially abundant in induced sputum from smokers with lung cancer or asbestos exposure. Eur J Cancer Prev. (2020) 29:238–47. doi: 10.1097/CEJ.0000000000000537
130. Koh HM, An HJ, Ko GH, Lee JH, Lee JS, Kim DC, et al. Prognostic role of S100A8 and S100A9 protein expressions in non-small cell carcinoma of the lung. J Pathol Transl Med. (2019) 53:13–22. doi: 10.4132/jptm.2018.11.12
131. Huang H, Huang Q, Tang T, Gu L, Du J, Li Z, et al. Clinical significance of calcium-binding protein S100A8 and S100A9 expression in non-small cell lung cancer. Thorac Cancer. (2018) 9:800–4. doi: 10.1111/1759-7714.12649
132. Su YJ, Xu F, Yu JP, Yue DS, Ren XB, Wang CL. Up-regulation of the expression of S100A8 and S100A9 in lung adenocarcinoma and its correlation with inflammation and other clinical features. Chin Med J. (2010) 123:2215–20.
133. Liu Y, Gao L, Fan Y, Ma R, An Y, Chen G, et al. Discovery of protein biomarkers for venous thromboembolism in non-small cell lung cancer patients through data-independent acquisition mass spectrometry. Front Oncol. (2023) 13:1079719. doi: 10.3389/fonc.2023.1079719
134. Hou YL, Zhang JH, Guo JB, Chen H. Clinical significance of serum S100A10 in lung cancer. J Int Med Res. (2021) 49:3000605211049653. doi: 10.1177/03000605211049653
135. Sato K, Saiki Y, Arai K, Ishizawa K, Fukushige S, Aoki K, et al. S100A10 upregulation associates with poor prognosis in lung squamous cell carcinoma. Biochem Biophys Res Commun. (2018) 505:466–70. doi: 10.1016/j.bbrc.2018.09.118
136. Wang H, Sun Z, Zhao W, Geng B. [S100A10 promotes proliferation and invasion of lung adenocarcinoma cells by activating the Akt-mTOR signaling pathway]. Nan Fang Yi Ke Da Xue Xue Bao. (2023) 43:733–40.
137. Phipps KD, Surette AP, O'Connell PA, Waisman DM. Plasminogen receptor S100A10 is essential for the migration of tumor-promoting macrophages into tumor sites. Cancer Res. (2011) 71:6676–83. doi: 10.1158/0008-5472.CAN-11-1748
138. Bharadwaj AG, Dahn ML, Liu R-Z, Colp P, Thomas LN, Holloway RW, et al. S100A10 has a critical regulatory function in mammary tumor growth and metastasis: insights using MMTV-PyMT oncomice and clinical patient sample analysis. Cancers. (2020) 12:3673. doi: 10.3390/cancers12123673
139. Yang X, Popescu NC, Zimonjic DB. DLC1 interaction with S100A10 mediates inhibition of in vitro cell invasion and tumorigenicity of lung cancer cells through a RhoGAP-independent mechanism. Cancer Res. (2011) 71:2916–25. doi: 10.1158/0008-5472.CAN-10-2158
140. Andey T, Marepally S, Patel A, Jackson T, Sarkar S, O'Connell M, et al. Cationic lipid guided short-hairpin RNA interference of annexin A2 attenuates tumor growth and metastasis in a mouse lung cancer stem cell model. J Control Release. (2014) 184:67–78. doi: 10.1016/j.jconrel.2014.03.049
141. Gocheva V, Naba A, Bhutkar A, Guardia T, Miller KM, Li CM, et al. Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proc Natl Acad Sci U S A. (2017) 114:E5625–34. doi: 10.1073/pnas.1707054114
142. Woo T, Okudela K, Mitsui H, Tajiri M, Rino Y, Ohashi K, et al. Up-Regulation of S100A11 in Lung Adenocarcinoma - Its Potential Relationship with Cancer Progression. PLoS ONE. (2015) 10:e0142642. doi: 10.1371/journal.pone.0142642
143. Tian T, Hao J, Xu A, Hao J, Luo C, Liu C, et al. Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis. Cancer Sci. (2007) 98:1265–74. doi: 10.1111/j.1349-7006.2007.00514.x
144. Zagryazhskaya A, Surova O, Akbar NS, Allavena G, Gyuraszova K, Zborovskaya IB, et al. Tudor staphylococcal nuclease drives chemoresistance of non-small cell lung carcinoma cells by regulating S100A11. Oncotarget. (2015) 6:12156–73. doi: 10.18632/oncotarget.3495
145. Hao J, Wang K, Yue Y, Tian T, Xu A, Hao J, et al. Selective expression of S100A11 in lung cancer and its role in regulating proliferation of adenocarcinomas cells. Mol Cell Biochem. (2012) 359:323–32. doi: 10.1007/s11010-011-1026-8
146. Miao S, Qiu T, Zhao Y, Wang H, Sun X, Wang Y, et al. Overexpression of S100A13 protein is associated with tumor angiogenesis and poor survival in patients with early-stage non-small cell lung cancer. Thorac Cancer. (2018) 9:1136–44. doi: 10.1111/1759-7714.12797
147. Pierce A, Barron N, Linehan R, Ryan E, O'Driscoll L, Daly C, et al. Identification of a novel, functional role for S100A13 in invasive lung cancer cell lines. Eur J Cancer. (2008) 44:151–9. doi: 10.1016/j.ejca.2007.10.017
148. Katono K, Sato Y, Kobayashi M, Saito K, Nagashio R, Ryuge S, et al. Clinicopathological Significance of S100A14 Expression in Lung Adenocarcinoma. Oncol Res Treat. (2017) 40:594–602. doi: 10.1159/000478100
149. Ding F, Wang D, Li XK, Yang L, Liu HY, Cui W, et al. Overexpression of S100A14 contributes to malignant progression and predicts poor prognosis of lung adenocarcinoma. Thorac Cancer. (2018) 9:827–35. doi: 10.1111/1759-7714.12654
150. Sugino T, Ichikawa-Tomikawa N, Tanaka M, Shishito N, Miura T, Abe M, et al. Identification of S100A14 as a metastasis-promoting molecule in a murine organotropic metastasis model. Clin Exp Metastasis. (2019) 36:411–22. doi: 10.1007/s10585-019-09979-w
151. Chen YC, Lin MC, Hsiao CC, Zheng YX, Chen KD, Sung MT, et al. Increased S100A15 expression and decreased DNA methylation of its gene promoter are involved in high metastasis potential and poor outcome of lung adenocarcinoma. Oncotarget. (2017) 8:45710–24. doi: 10.18632/oncotarget.17391
152. Chen YC, Hsiao CC, Chen KD, Hung YC, Wu CY, Lie CH, et al. Peripheral immune cell gene expression changes in advanced non-small cell lung cancer patients treated with first line combination chemotherapy. PLoS ONE. (2013) 8:e57053. doi: 10.1371/journal.pone.0057053
153. Chen L, Hu X, Wu H, Jia Y, Liu J, Mu X, et al. Over-expression of S100B protein as a serum marker of brain metastasis in non-small cell lung cancer and its prognostic value. Pathol Res Pract. (2019) 215:427–32. doi: 10.1016/j.prp.2018.11.011
154. Pang X, Min J, Liu L, Liu Y, Ma N, Zhang H. S100B protein as a possible participant in the brain metastasis of NSCLC. Med Oncol. (2012) 29:2626–32. doi: 10.1007/s12032-012-0169-0
155. Rehbein G, Simm A, Hofmann HS, Silber RE, Bartling B. Molecular regulation of S100P in human lung adenocarcinomas. Int J Mol Med. (2008) 22:69–77. doi: 10.3892/ijmm.22.1.69
156. Hsu YL, Hung JY, Liang YY, Lin YS, Tsai MJ, Chou SH, et al. S100P interacts with integrin alpha7 and increases cancer cell migration and invasion in lung cancer. Oncotarget. (2015) 6:29585–98. doi: 10.18632/oncotarget.4987
157. Chien MH, Lee WJ, Hsieh FK, Li CF, Cheng TY, Wang MY, et al. Keap1-Nrf2 interaction suppresses cell motility in lung adenocarcinomas by targeting the S100P protein. Clin Cancer Res. (2015) 21:4719–32. doi: 10.1158/1078-0432.CCR-14-2880
158. Bulk E, Hascher A, Liersch R, Mesters RM, Diederichs S, Sargin B, et al. Adjuvant therapy with small hairpin RNA interference prevents non-small cell lung cancer metastasis development in mice. Cancer Res. (2008) 68:1896–904. doi: 10.1158/0008-5472.CAN-07-2390
159. Raspollini MR, Comin CE, Crisci A, Chilosi M, et al. The use of placental S100 (S100P), GATA3 and napsin A in the differential diagnosis of primary adenocarcinoma of the bladder and bladder metastasis from adenocarcinoma of the lung. Pathologica. (2010) 102:33–5.
160. Harada C, Kawaguchi T, Ogata-Suetsugu S, Yamada M, Hamada N, Maeyama T, et al. EGFR tyrosine kinase inhibition worsens acute lung injury in mice with repairing airway epithelium. Am J Respir Crit Care Med. (2011) 183:743–51. doi: 10.1164/rccm.201002-0188OC
161. Chakraborty D, Zenker S, Rossaint J, Hölscher A, Pohlen M, Zarbock A, et al. Alarmin S100A8 activates alveolar epithelial cells in the context of acute lung injury in a TLR4-dependent manner. Front Immunol. (2017) 8:1493. doi: 10.3389/fimmu.2017.01493
162. Hiroshima Y, Hsu K, Tedla N, Wong SW, Chow S, Kawaguchi N, et al. S100A8/A9 and S100A9 reduce acute lung injury. Immunol Cell Biol. (2017) 95:461–72. doi: 10.1038/icb.2017.2
163. Wittkowski H, Sturrock A, van Zoelen MA, Viemann D, van der Poll T, Hoidal JR, et al. Neutrophil-derived S100A12 in acute lung injury and respiratory distress syndrome. Crit Care Med. (2007) 35:1369–75. doi: 10.1097/01.CCM.0000262386.32287.29
164. Jabaudon M, Blondonnet R, Roszyk L, Pereira B, Guérin R, Perbet S, et al. Soluble forms and ligands of the receptor for advanced glycation end-products in patients with acute respiratory distress syndrome: an observational prospective study. PLoS ONE. (2015) 10:e0135857. doi: 10.1371/journal.pone.0135857
165. Lorenz E, Muhlebach MS, Tessier PA, Alexis NE, Hite DR, Seeds MC, et al. Different expression ratio of S100A8/A9 and S100A12 in acute and chronic lung diseases. Respir Med. (2008) 102:567–73. doi: 10.1016/j.rmed.2007.11.011
166. Biji A, Khatun O, Swaraj S, Narayan R, Rajmani RS, Sardar R, et al. Identification of COVID-19 prognostic markers and therapeutic targets through meta-analysis and validation of Omics data from nasopharyngeal samples. EBioMedicine. (2021) 70:103525. doi: 10.1016/j.ebiom.2021.103525
167. Mellett L, Khader SA. S100A8/A9 in COVID-19 pathogenesis: impact on clinical outcomes. Cytokine Growth Factor Rev. (2022) 63:90–7. doi: 10.1016/j.cytogfr.2021.10.004
168. Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020 182:1401–18.e18. doi: 10.1016/j.cell.2020.08.002
169. Guo Q, Zhao Y, Li J, Liu J, Yang X, Guo X, et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe. 2021 29:222–35.e4. doi: 10.1016/j.chom.2020.12.016
170. Ahmad A, Li C, Wu L, Wu C, Chen J, Jiang C, et al. Myeloid related protein-8/14 stimulates interleukin-8 production in airway epithelial cells. Am J Respir Cell Mol Biol. (2003) 29:523–30. doi: 10.1165/rcmb.2002-0286OC
171. Chen L, Long X, Xu Q, Tan J, Wang G, Cao Y, et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell Mol Immunol. (2020) 17:992–4. doi: 10.1038/s41423-020-0492-x
172. Wu M, Chen Y, Xia H, Wang C, Tan CY, Cai X, et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc Nat Acad Sci. (2020) 117:28336–43. doi: 10.1073/pnas.2018030117
173. Lei H. A single transcript for the prognosis of disease severity in COVID-19 patients. Sci Rep. (2021) 11:12174. doi: 10.1038/s41598-021-91754-7
174. Kokkoris S, Stamataki E, Emmanouil G, Psachoulia C, Ntaidou T, Maragouti A, et al. Serum inflammatory and brain injury biomarkers in COVID-19 patients admitted to intensive care unit: a pilot study. eNeurologicalSci. (2022) 29:100434. doi: 10.1016/j.ensci.2022.100434
175. Aceti A, Margarucci LM, Scaramucci E, Orsini M, Salerno G, Di Sante G, et al. Serum S100B protein as a marker of severity in Covid-19 patients. Sci Rep. (2020) 10:18665. doi: 10.1038/s41598-020-75618-0
176. Spanos M, Shachar S, Sweeney T, Lehmann HI, Gokulnath P, Li G, et al. Elevation of neural injury markers in patients with neurologic sequelae after hospitalization for SARS-CoV-2 infection. iScience. (2022) 25:104833. doi: 10.1016/j.isci.2022.104833
177. Bagheri-Hosseinabadi Z, Abbasi M, Kahnooji M, Ghorbani Z, Abbasifard M, et al. The prognostic value of S100A calcium binding protein family members in predicting severe forms of COVID-19. Inflamm Res. (2022) 71:369–76. doi: 10.1007/s00011-022-01545-7
178. Holms RD. Long COVID (PASC) is maintained by a self-sustaining pro-inflammatory TLR4/RAGE-loop of S100A8/A9 > TLR4/RAGE signalling, inducing chronic expression of IL-1b, IL-6 and TNFa: anti-inflammatory ezrin peptides as potential therapy. Immuno. (2022) 2:512–33. doi: 10.3390/immuno2030033
179. Tandon P, Abrams ND, Avula LR, Carrick DM, Chander P, Divi RL, et al. Unraveling links between chronic inflammation and long COVID: workshop report. J Immunol. (2024) 212:505–12. doi: 10.4049/jimmunol.2300804
180. Abonia JP, Austen KF, Rollins BJ, Joshi SK, Flavell RA, Kuziel WA, et al. Constitutive homing of mast cell progenitors to the intestine depends on autologous expression of the chemokine receptor CXCR2. Blood. (2005) 105:4308–13. doi: 10.1182/blood-2004-09-3578
181. Holms RD, Ataullakhanov RI. Ezrin peptide therapy from HIV to COVID: inhibition of inflammation and amplification of adaptive anti-viral immunity. Int J Mol Sci. (2021) 22:11688. doi: 10.3390/ijms222111688
182. Menzella F, Fontana M, Salvarani C, Massari M, Ruggiero P, Scelfo C, et al. Efficacy of tocilizumab in patients with COVID-19 ARDS undergoing noninvasive ventilation. Critical Care. (2020) 24:589. doi: 10.1186/s13054-020-03306-6
183. Guo Q, Zhao Y, Li J, Liu J, Guo X, Zhang Z, et al. Small molecules inhibit SARS-CoV-2 induced aberrant inflammation and viral replication in mice by targeting S100A8/A9-TLR4 axis. BioRxiv. (2020). doi: 10.1101/2020.09.09.288704
184. Roh JS, Sohn DH. Damage-associated molecular patterns in inflammatory diseases. Immune Netw. (2018) 18:e27. doi: 10.4110/in.2018.18.e27
185. Prantner D, Nallar S, Vogel SN. The role of RAGE in host pathology and crosstalk between RAGE and TLR4 in innate immune signal transduction pathways. FASEB J. (2020) 34:15659. doi: 10.1096/fj.202002136R
186. Wright NT, Cannon BR, Zimmer DB, Weber DJ, et al. S100A1: structure, function, and therapeutic potential. Curr Chem Biol. (2009) 3:138–45. doi: 10.2174/2212796810903020138
187. Shimamoto S, Kubota Y, Tokumitsu H, Kobayashi R, et al. S100 proteins regulate the interaction of Hsp90 with Cyclophilin 40 and FKBP52 through their tetratricopeptide repeats. FEBS Lett. (2010) 584:1119–25. doi: 10.1016/j.febslet.2010.02.055
188. Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. (2006) 66:5173–80. doi: 10.1158/0008-5472.CAN-05-3087
189. Schäfer BW, Fritschy JM, Murmann P, Troxler H, Durussel I, Heizmann CW, et al. Brain S100A5 is a novel calcium-, zinc-, and copper ion-binding protein of the EF-hand superfamily. J Biol Chem. (2000) 275:30623–30. doi: 10.1074/jbc.M002260200
190. Robaszkiewicz K, Jurewicz E, Moraczewska J, Filipek A, et al. Ca(2+)-dependent binding of S100A6 to cofilin-1 regulates actin filament polymerization-depolymerization dynamics. Cell Calcium. (2021) 99:102457. doi: 10.1016/j.ceca.2021.102457
191. Watson PH, Leygue ER, Murphy LC. Psoriasin (S100A7). Int J Biochem Cell Biol. (1998) 30:567–71. doi: 10.1016/S1357-2725(97)00066-6
192. Scott NR, Swanson RV, Al-Hammadi N, Domingo-Gonzalez R, Rangel-Moreno J, Kriel BA, et al. S100A8/A9 regulates CD11b expression and neutrophil recruitment during chronic tuberculosis. J Clin Invest. (2020) 130:3098–112. doi: 10.1172/JCI130546
193. Raquil MA, Anceriz N, Rouleau P, Tessierc PA, et al. Blockade of antimicrobial proteins S100A8 and S100A9 inhibits phagocyte migration to the alveoli in streptococcal pneumonia. J Immunol. (2008) 180:3366–74. doi: 10.4049/jimmunol.180.5.3366
194. Madureira PA, O'Connell PA, Surette AP, Miller VA, Waisman DM. The biochemistry and regulation of S100A10: a multifunctional plasminogen receptor involved in oncogenesis. J Biomed Biotechnol. (2012) 2012:353687. doi: 10.1155/2012/353687
195. Zhou X, Shi M, Cao J, Yuan T, Yu G, Chen Y. S100 calcium binding protein A10, a novel oncogene, promotes the proliferation, invasion, and migration of hepatocellular carcinoma. Front Genet. (2021) 12:695036. doi: 10.3389/fgene.2021.695036
196. Zhang L, Zhu T, Miao H, Liang B, et al. The calcium binding protein S100A11 and its roles in diseases. Front Cell Dev Biol. (2021) 9:693262. doi: 10.3389/fcell.2021.693262
197. Jackson E, Little S, Franklin DS, Gaddy JA, Damo SM. Expression, purification, and antimicrobial activity of S100A12. J Vis Exp. (2017) 123:55557. doi: 10.3791/55557-v
198. Prudovsky I, Mandinova A, Soldi R, Bagala C, Graziani I, Landriscina M, et al. The non-classical export routes: FGF1 and IL-1α point the way. J Cell Sci. (2003) 116:4871–81. doi: 10.1242/jcs.00872
199. Zhu M, Wang H, Cui J, Li W, An G, Pan Y, et al. Calcium-binding protein S100A14 induces differentiation and suppresses metastasis in gastric cancer. Cell Death Dis. (2017) 8:e2938. doi: 10.1038/cddis.2017.297
200. Wolf R, Ruzicka T, Yuspa SH. Novel S100A7 (psoriasin)/S100A15 (koebnerisin) subfamily: highly homologous but distinct in regulation and function. Amino Acids. (2011) 41:789–96. doi: 10.1007/s00726-010-0666-4
201. Basnet S, Vallenari EM, Maharjan U, Sharma S, Schreurs O, Sapkota D, et al. An update on S100A16 in human cancer. Biomolecules. (2023) 13:1070. doi: 10.3390/biom13071070
202. Austermann J, Nazmi AR, Heil A, Fritz G, Kolinski M, Filipek S, et al. Generation and characterization of a novel, permanently active S100P mutant. Biochim Biophys Acta. (2009) 1793:1078–85. doi: 10.1016/j.bbamcr.2008.11.012
203. Most P, Bernotat J, Ehlermann P, Pleger ST, Reppel M, Börries M, et al. S100A1: a regulator of myocardial contractility. Proc Nat Acad Sci. (2001) 98:13889–94. doi: 10.1073/pnas.241393598
204. Huang X, Qu D, Liang Y, Huang Q, Li M, Hou C, et al. Elevated S100A4 in asthmatics and an allergen-induced mouse asthma model. J Cell Biochem. (2019) 120:9667–76. doi: 10.1002/jcb.28245
205. Liu C-C, Yeh H-I. Nicotine: a double-edged sword in atherosclerotic disease. Acta Cardiologica Sinica. (2014) 30:108–13.
206. Tsuta K, Tanabe Y, Yoshida A, Takahashi F, Maeshima AM, Asamura H. Utility of 10 immunohistochemical markers including novel markers (desmocollin-3, glypican 3, S100A2, S100A7, and Sox-2) for differential diagnosis of squamous cell carcinoma from adenocarcinoma of the Lung. J Thorac Oncol. (2011) 6:1190–9. doi: 10.1097/JTO.0b013e318219ac78
207. Wang R, Li Y, Hu E, Kong F, Wang J, Liu J, et al. S100A7 promotes lung adenocarcinoma to squamous carcinoma transdifferentiation, and its expression is differentially regulated by the Hippo-YAP pathway in lung cancer cells. Oncotarget. (2017) 8:24804–14. doi: 10.18632/oncotarget.15063
208. Katono K, Sato Y, Jiang SX, Kobayashi M, Saito K, Nagashio R, et al. Clinicopathological significance of S100A10 expression in lung adenocarcinomas. Asian Pac J Cancer Prev. (2016) 17:289–94. doi: 10.7314/APJCP.2016.17.1.289
209. Shi H, Zuo Y, Yalavarthi S, Gockman K, Zuo M, Madison JA, et al. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. J Leukoc Biol. (2021) 109:67–72. doi: 10.1002/JLB.3COVCRA0720-359R
210. Norman GL, Navaz SA, Kanthi Y, Albesa R, Mahler M, Knight JS, et al. Circulating calprotectin as a predictive and severity biomarker in patients with COVID-19. Diagnostics. (2022) 12:1324. doi: 10.3390/diagnostics12061324
211. Nakata K, Okazaki M, Kawana S, Kubo Y, Shimizu D, Tanaka S, et al. S100A8/A9 as a prognostic biomarker in lung transplantation. Clin Transplant. (2023) 37:e15006. doi: 10.1111/ctr.15006
212. Shimizu D, Okazaki M, Sugimoto S, Kinoshita R, Nakata K, Tanaka S, et al. Inhibiting S100A8/A9 attenuates airway obstruction in a mouse model of heterotopic tracheal transplantation. Biochem Biophys Res Commun. (2022) 629:86–94. doi: 10.1016/j.bbrc.2022.08.087
213. Recto KA, Huan T, Lee DH, Lee GY, Gereige J, Yao C, et al. Transcriptome-wide association study of circulating IgE levels identifies novel targets for asthma and allergic diseases. Front Immunol. (2023) 14:1080071. doi: 10.3389/fimmu.2023.1080071
214. Kobayashi M, Nagashio R, Saito K, Aguilar-Bonavides C, Ryuge S, Katono K, et al. Prognostic significance of S100A16 subcellular localization in lung adenocarcinoma. Hum Pathol. (2018) 74:148–55. doi: 10.1016/j.humpath.2018.01.001
Keywords: S100A proteins, lung diseases, pulmonary fibrosis, lung transplantation, COVID-19
Citation: Raveendran VV, AlQattan S and AlMutairy E (2025) A review on clinical implications of S100 proteins in lung diseases. Front. Med. 12:1618772. doi: 10.3389/fmed.2025.1618772
Received: 26 April 2025; Accepted: 18 September 2025;
Published: 14 October 2025.
Edited by:
Yun-Ju Lai, University of Massachusetts Lowell, United StatesReviewed by:
Yuhao Xie, St. John's University, United StatesHsiang-Chi Huang, National Cheng Kung University, Taiwan
Copyright © 2025 Raveendran, AlQattan and AlMutairy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vineesh V. Raveendran, dnJhdmVlbmRyYW5Aa2ZzaHJjLmVkdS5zYQ==; Eid AlMutairy, ZWFsbXV0YWlyeTQ1QGtmc2hyYy5lZHUuc2E=
 Vineesh V. Raveendran
Vineesh V. Raveendran Somaya AlQattan
Somaya AlQattan