Abstract
Introduction:
Polycystic ovary syndrome (PCOS) is a prevalent endocrine disorder affecting 5–18% of reproductive-aged women, characterized by menstrual irregularities, hyperandrogenism, and polycystic ovarian morphology. Beyond its endocrine manifestations, PCOS involves significant metabolic dysfunction, particularly in lipid homeostasis. Elevated triglyceride levels are closely linked to insulin resistance and cardiovascular risk, suggesting a central role for lipid dysregulation in PCOS pathogenesis. However, traditional observational studies struggle to establish causal relationships due to confounding factors and reverse causality.
Methods:
To address these limitations, this study employed a two-sample Mendelian randomization (MR) design to assess the causal effects of lipid-related metabolites—specifically triglyceride-rich lipoprotein subclasses—on PCOS susceptibility. Furthermore, to elucidate potential biological mechanisms, we integrated the MR analysis with in vitro functional experiments, focusing on the role of ketone body metabolism and specifically 3-hydroxybutyrate (3-HB), a major circulating ketone body known to regulate gene expression via epigenetic modifications.
Results:
Our analysis identified a causal contribution of lipid-related metabolites to PCOS. notably, we demonstrated that 3-HB plays a critical role in the development of PCOS. Mechanistic investigations revealed that 3-HB contributes to metabolic and hormonal dysregulation primarily through the modulation of HDAC3 activity, linking ketone body metabolism directly to the disease phenotype.
Conclusion:
This study provides robust causal evidence linking lipid metabolism and ketone bodies to PCOS, moving beyond descriptive associations. by uncovering the specific pathway involving 3-HB and HDAC3, we highlight a novel molecular mechanism underlying PCOS pathogenesis. These findings suggest that targeting the 3-HB/HDAC3 axis could offer new strategies for therapeutic intervention in managing PCOS-related metabolic dysfunction.
1 Introduction
Polycystic Ovary Syndrome (PCOS) is one of the most common endocrine disorders worldwide, affecting approximately 5–18% of women of reproductive age (1). Its clinical features include menstrual irregularities, hyperandrogenism (such as hirsutism and acne), and polycystic ovaries (2, 3). Although the etiology of PCOS is not fully understood, an increasing number of studies have shown that PCOS is not only a manifestation of endocrine disorders but also involves metabolic abnormalities, especially alterations in lipid metabolism. Lipid metabolism disorders, particularly those related to triglycerides, have been confirmed as one of the main phenotypes of PCOS and are closely related to insulin resistance, obesity, and cardiovascular diseases in PCOS (4). Therefore, lipid metabolism may play a core role in the pathogenesis of PCOS.
Traditional observational studies often have difficulty in clearly establishing the relationship between metabolites and PCOS due to confounding factors, reverse causality, and potential biases (5, 6). Mendelian Randomization (MR), as a causal inference method based on genetic variations, can overcome these problems and has shown unique advantages in the study of metabolic diseases, especially PCOS (7). By identifying single nucleotide polymorphisms (SNPs) related to exposure factors (such as metabolites) from genome-wide association studies (GWAS), MR can effectively avoid the confounding factors and reverse causality problems in traditional studies, thereby providing more accurate causal inferences.
Although previous studies have shown a certain association between lipid metabolism and PCOS, most of them have remained at the level of phenotypic association analysis, lacking in-depth causal inference and mechanism exploration (8–10). Therefore, this study aims to reveal the causal relationship between lipid metabolites, especially triglyceride-related lipoprotein particles and PCOS, through two-sample MR analysis, and further explore the potential mechanisms of these metabolites in the occurrence of PCOS. In addition, ketone body metabolism, especially 3-hydroxybutyrate (3-HB), as a key intermediate product in energy metabolism, may also regulate ovarian function through epigenetic mechanisms and be closely related to the occurrence of PCOS. Therefore, the goal of this study is to reveal the causal role of metabolites and ketone body metabolism in the occurrence of PCOS through MR analysis, and further verify the biological effects of 3-HB through in vitro experiments, providing new perspectives and potential targets for the treatment of PCOS.
1.1 Research design
To explore the relationship between metabolites and polycystic ovary syndrome (PCOS), we designed a two-sample Mendelian randomization (MR) study (11). MR studies need to meet the following assumptions (12): (1) a strong and robust correlation between instrumental variables (IVs) and exposure (association assumption); (2) independence of IVs from confounding factors that affect the relationship between exposure and outcome (independence assumption); (3) gene variations only affect the outcome through the exposure factor and not through other means (exclusion restriction assumption).
1.2 Data sources
All data on exposure and outcome in this study were obtained from the IEU website.1 All the data used have been published in public databases and do not require additional ethical approval. The MR analysis we conducted used single nucleotide polymorphisms (SNPs) related to metabolites as exposure factors and PCOS as the outcome. Metabolite data were from the article by Karjalainen et al. (13), and PCOS data were from the EBI database (ID: ebi-a-GCST90044902).
1.3 Selection of instrumental variables
To fulfill the first assumption of MR, SNPs closely related to the exposure factor were selected (p < 5.0 × 10−8, r2 = 0.001, genetic distance = 10,000 kb). To fulfill the second assumption of MR, we queried the Phenoscanner database2 to ensure that the selected SNPs were not related to known confounding factors. Finally, the F-statistic was calculated to assess whether there was weak instrument bias in the selected instrumental variables (14). An F > 10 indicates no weak instrument bias, further supporting the association assumption. The formula for calculating F is: F = [R2/(1 − R2)] × [(N − K −1)/K], where N is the sample size of the exposure factor, K is the number of instrumental variables, and R2 is the proportion of variation in the exposure factor explained by the instrumental variables (15).
1.4 Mendelian randomization analysis
All MR analyses in this study were conducted using the “TwoSampleMR,” “MendelianRandomization,” and “ggplot2” packages in R language (4.3.3). The inverse variance weighted method (IVW) was used as the primary analysis method, and the weighted median, MR-Egger regression, simple model, and weighted model were used as auxiliary analysis methods.
1.5 Sensitivity analysis
The Cochrane Q value and MR-Egger intercept were used to assess heterogeneity and horizontal pleiotropy, respectively (16). Leave-one-out analysis was used to detect whether the association between exposure and outcome was mainly influenced by a single SNP. Sensitivity analysis was visualized using scatter plots, funnel plots, and forest plots to demonstrate the robustness of the MR study. The odds ratio and its 95% confidence interval were used to quantitatively assess the potential causal relationship between the exposure factor and the outcome. All statistical tests were two-tailed, and p < 0.05 was considered statistically significant. The study results were reported in accordance with the STROBE-MR guidelines (17).
1.6 Experimental verification
1.6.1 Materials
KGN human ovarian granulosa cell line (OriCell, Catalog No. H6-1301), fetal bovine serum (FBS), DMEM high glucose medium, 1% penicillin/streptomycin solution. HDAC3-specific inhibitor RGFP966 (MCE, Catalog No. HY-13909), 3-Hydroxybutyrate (MCE, Catalog No. HY-113378). Human Testosterone, T ELISA Kit (cusabio, Catalog No. CSB-E05099h), Human Estradiol, E2 ELISA Kit (cusabio, Catalog No. CSB-E05108h), SYBR qPCR SuperMix Plus (novoprotein, Catalog No. E096-01A), Plus All-in-one 1st Strand cDNA Synthesis SuperMix (novoprotein, Catalog No. E047-01B).
1.6.2 Cell culture
Culture conditions: Cells were cultured in DMEM/F12 medium containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C in a 5% CO₂ incubator. After the cells reached 80–90% confluence, they were divided into different treatment groups for processing.
1.6.3 Experimental grouping
Control group: Only medium. 3-HB treatment group: Given 3-HB. 3-HB inhibition group: Treated with 3-HB -specific inhibitor (RGFP966) and 3-HB simultaneously.
1.6.4 Cell toxicity assay
After the cells reached 80–90% confluence, KGN cells were seeded into 96-well plates at a density of 1 × 104 cells per well. In the 3-HB treatment group, 3-HB was added at predetermined concentrations (0.5, 1, 2, 3, 4, 5 mM), and CCK8 reagent was added after 24 h. In the HDAC3 inhibition group, RGFP966 was added at predetermined concentrations (10, 20, 40, 80, 160 nM), and CCK8 reagent was added after 24 h.
1.6.5 ELISA
After drug treatment, the supernatants of each group were collected, and the contents of testosterone and estradiol were detected according to the kit instructions.
1.6.6 qRT-PCR
Total RNA was extracted from tissue samples using Trizol, and the RNA samples were reverse transcribed into cDNA using the HiFiScript cDNA First Strand Synthesis Kit. qRT-PCR was performed using the CFX ConnectTM Real-Time PCR System (Bio-Rad, USA). The primer sequences of the genes are shown in Table 1. The relative mRNA expression of the target genes in each sample was calculated using the 2−△△CT method.
Table 1
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| GAPDH | CATGAGAAGTATGACAACAGCCT | AGTCCTTCCACGATACCAAAGT |
| FSHR | TCTGTCACTGCTCTAACAGGG | TGCACCTTTTTGGATGACTCG |
| LHCGR | TTCCAAGGGATGAATAACGAGTCT | TGCATGGCTTTGTACTTCTTCAA |
| CYP19A1 | AACAACTCGACCCTTCTTTATG | TTTGAGGGATTCAGCACAG |
| HDAC3 | TGATGACCAGAGTTACAAGCAC | GGGCAACATTTCGGACAG |
| IRS1 | TTGAGAATGTGTGGCTGAGG | TCCTTGACCAAATCCAGGTC |
| StAR | CAGACTTCGGGAACATGCCT | CCCTTGAGGTCGATGCTGAG |
| CYP11A1 | CACTCCTCAAAGCCAGCATCA | ACGAAGCACCAGGTCATTCAC |
| CYP17A1 | GGGCGGCCTCAAATGG | CAGCGAAGGCGAAGGCGATACCCTTA |
| HSD3B2 | TCTCAGATGACACGCCTCAC | GGGCTGAGTAGGAAGCTCAC |
| HSD17B13 | CCTACTTGGAGTCGTTGGTGA | CCAATATGCTCTGTCGTTTTGC |
Primers sequence.
1.6.7 Statistical analysis
Statistical analysis was performed using Graphpad prism 5.0 software. Experimental data are presented as mean ± standard deviation (SD). Comparisons between two groups were made using one-way analysis of variance (One-way ANOVA).
2 Results
2.1 Instrumental variable characteristics
After applying stringent selection criteria—including genome-wide significance screening (p < 1 × 10−5), elimination of variants in linkage disequilibrium (LD pruning), effect allele alignment, MR-PRESSO testing, and F-statistic evaluation—the final set of SNPs used as instrumental variables (IVs) all exhibited F-values exceeding 10. This indicates that each selected IV has a strong and statistically robust association with the respective gut microbiota traits, minimizing the risk of weak instrument bias.
2.2 Genetic evidence for metabolite effects on polycystic ovary syndrome
Mendelian randomization analysis revealed several lipid-related metabolites with significant causal associations with polycystic ovary syndrome (PCOS). Notably, triglyceride measures within low-density lipoprotein (LDL) and very low-density lipoprotein (VLDL) subclasses demonstrated consistent positive effects: higher triglyceride content in LDL particles (IVW OR = 1.167, p = 0.048), increased ratio of triglycerides to total lipids in large LDL (IVW OR = 1.263, p = 0.019), and elevated triglyceride levels in medium-sized LDL (IVW OR = 1.182, p = 0.037) were all linked to greater disease risk. Similarly, both total lipid amount (IVW OR = 1.159, p = 0.046) and particle number (IVW OR = 1.164, p = 0.038) in extremely small VLDL subfractions were positively associated with PCOS. Furthermore, elevated ratios of apolipoprotein B to A1 (IVW OR = 1.153, p = 0.040) and increased levels of 3-hydroxybutyrate (IVW OR = 1.554, p = 0.023) suggested enhanced risk, whereas higher concentrations of cholesterol in very large high-density lipoprotein (HDL) particles were found to be protective (IVW OR = 0.820, p = 0.039). Collectively, these findings highlight the potential role of dysregulated lipid metabolism—particularly involving triglyceride-enriched lipoproteins and ketone body pathways—in the etiology of PCOS. These genetically informed results provide compelling support for the involvement of lipid homeostasis in PCOS pathogenesis, offering novel insights into metabolic mechanisms underlying disease susceptibility. The detailed outcomes are illustrated in Figures 1, 2.
Figure 1
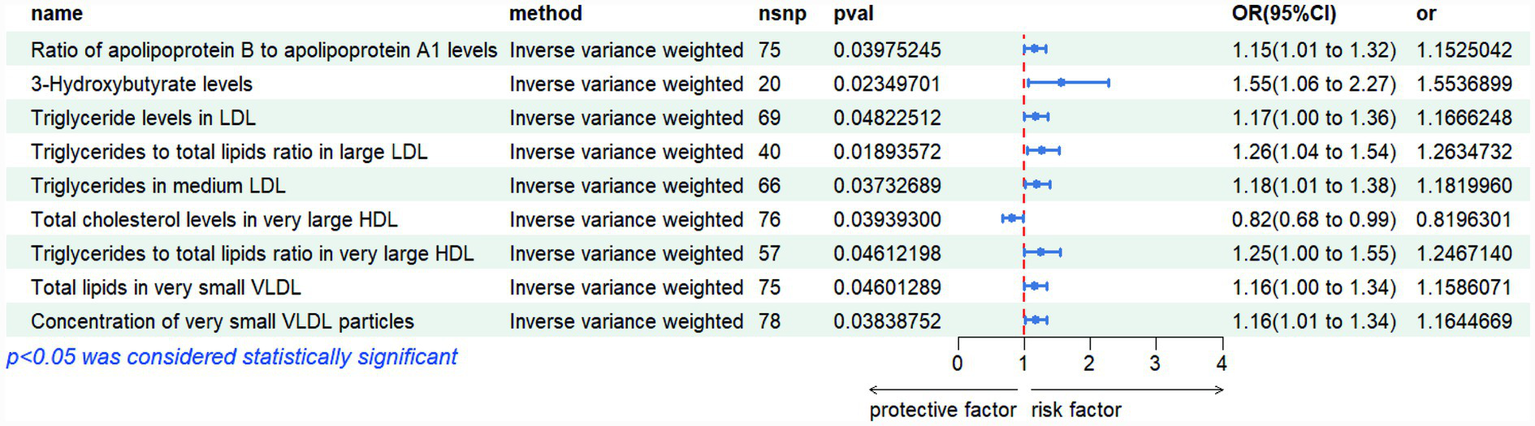
Risk plots of various metabolites IVW results.
Figure 2
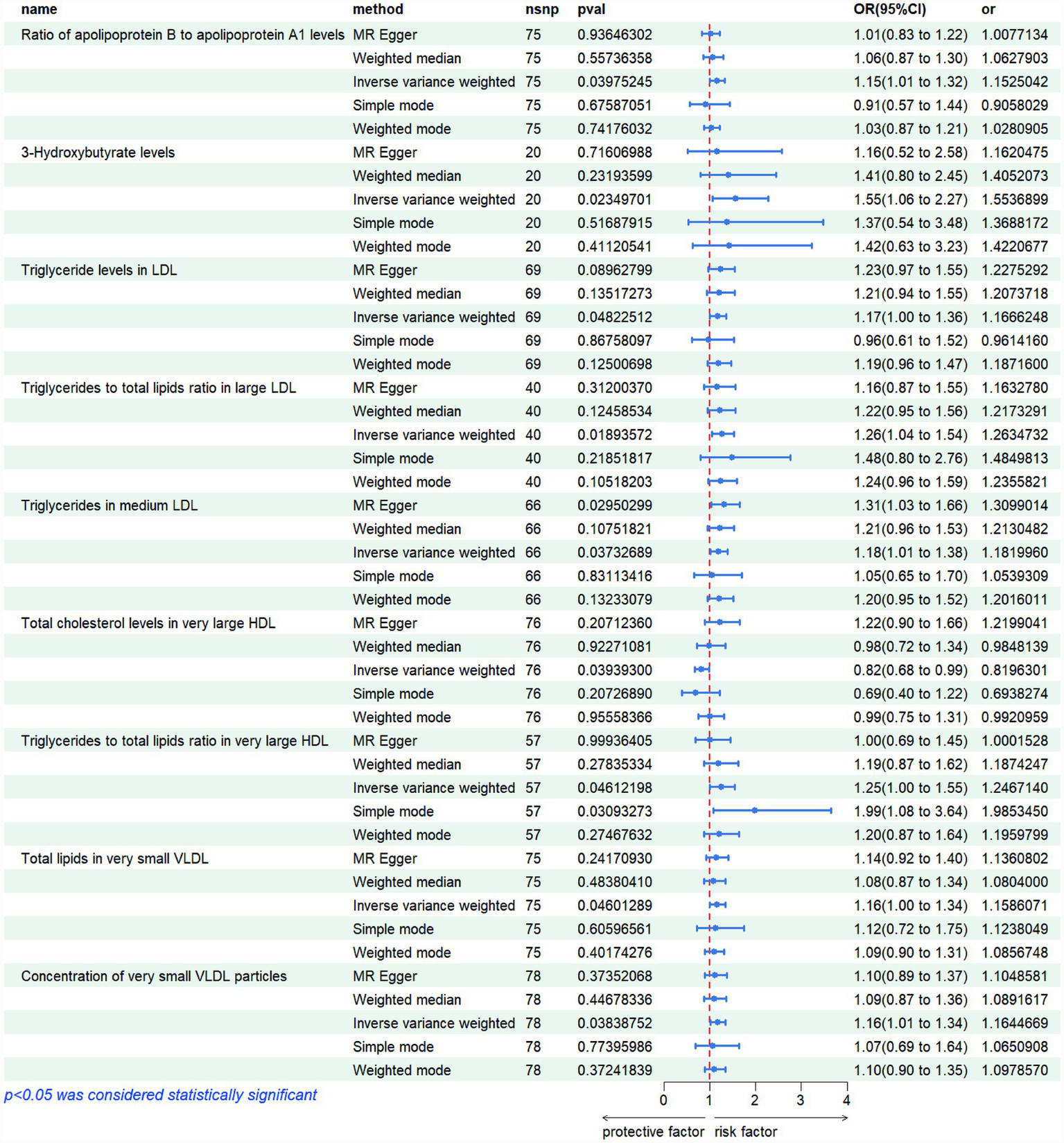
Risk graphs of all results for various metabolites.
2.3 Sensitivity analysis
The sensitivity analyses provided further validation of the robustness of the primary findings. Results obtained from multiple Mendelian randomization approaches—such as the weighted median and MR-Egger methods—were highly aligned with those derived from the inverse variance weighted method, showing no major discrepancies. The MR-Egger regression intercept test revealed no evidence of significant horizontal pleiotropy (p > 0.05), suggesting that the genetic variants primarily affected the outcome via the exposure of interest, thereby minimizing concerns about pleiotropy-induced bias. Although Cochran’s Q test indicated the presence of heterogeneity for certain exposures, the observed associations remained statistically significant under the random-effects model, implying that heterogeneity did not produce spurious results. Furthermore, the leave-one-out analysis demonstrated that no individual SNP disproportionately influenced the overall causal estimate, confirming that the findings were not driven by a single outlying genetic variant. Overall, these sensitivity assessments collectively reinforce the credibility and stability of the main analytical outcomes, lending strong support to the study’s core conclusions. The corresponding results are presented in Figures 3–5.
Figure 3
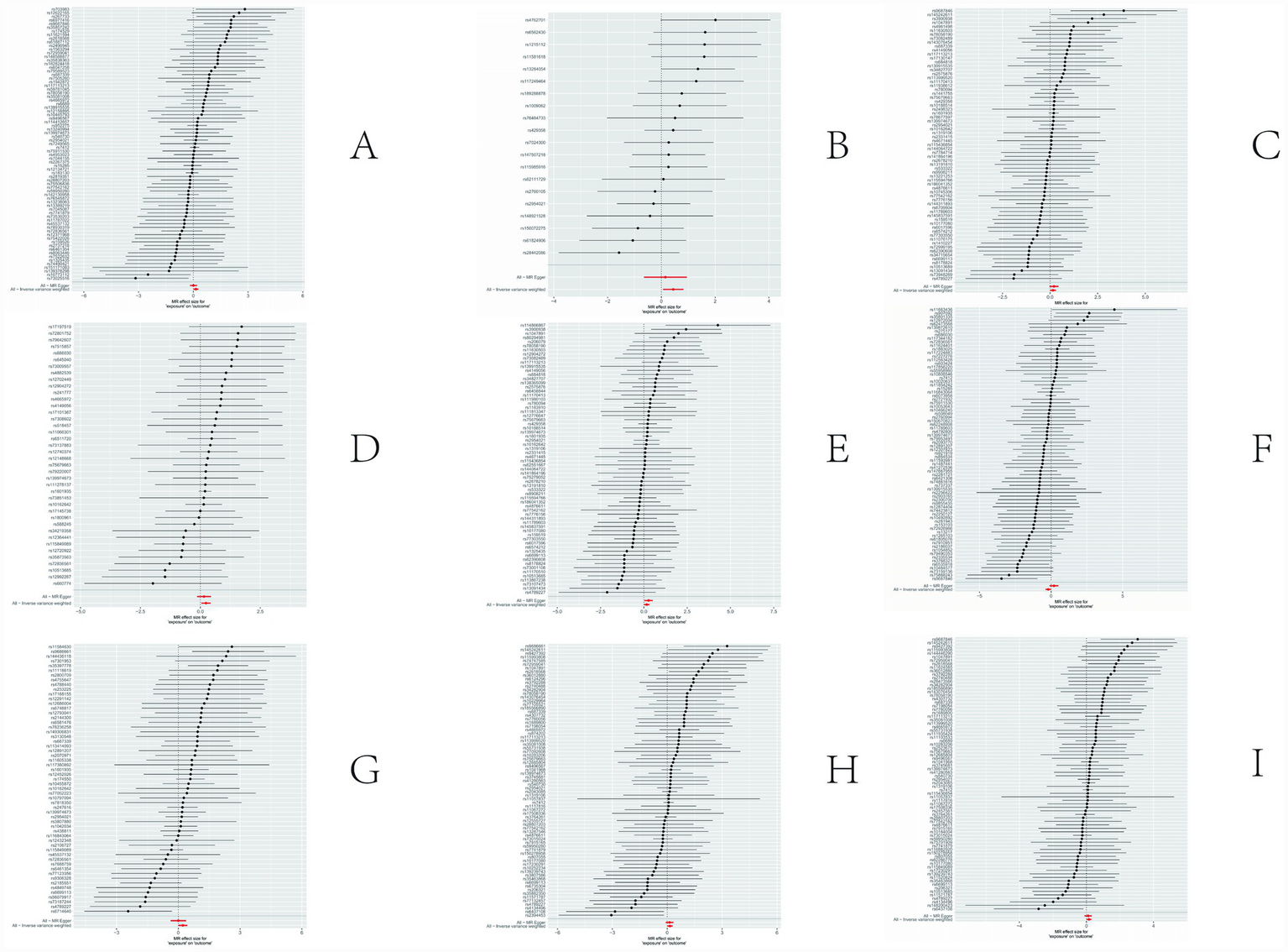
Results of sensitivity analysis-forest. (A) Ratio of apolipoprotien B to apolipoprotien A1 levels; (B) 3-hydroxybutyrate levels; (C) triglyceride levels in LDL; (D) triglycerides to total lipids ratio in large LDL; (E) triglyceride in medium LDL; (F) total cholesterol levels in very large HDL; (G) triglycerides to total lipids ratio in very large HDL; (H) total lipids in very small VLDL; (I) concentration of very small VLDL particles.
Figure 4
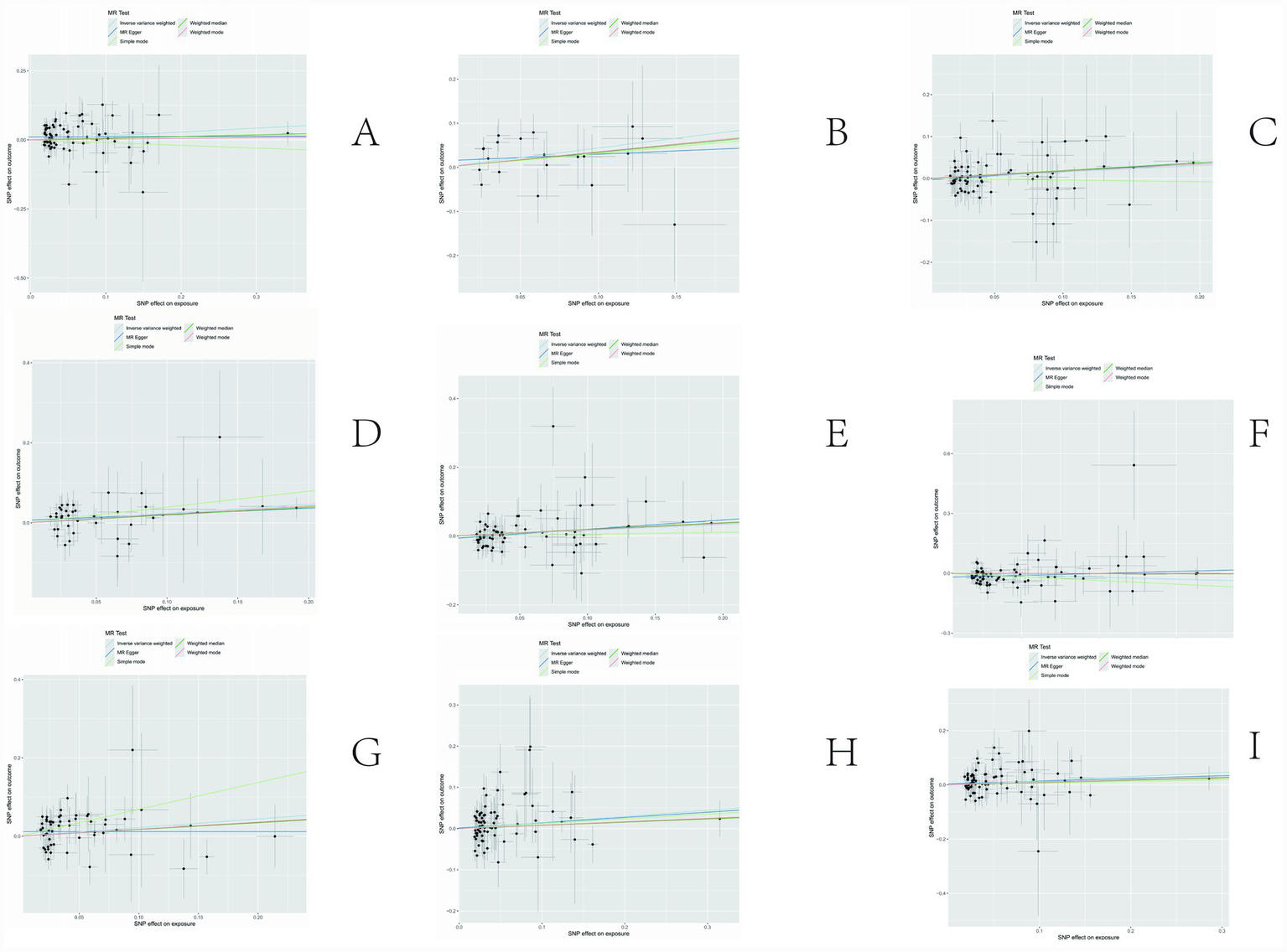
Results of sensitivity analysis-scatter. (A) Ratio of apolipoprotien B to apolipoprotien A1 levels; (B) 3-hydroxybutyrate levels; (C) triglyceride levels in LDL; (D) triglycerides to total lipids ratio in large LDL; (E) triglyceride in medium LDL; (F) total cholesterol levels in very large HDL; (G) triglycerides to total lipids ratio in very large HDL; (H) total lipids in very small VLDL; (I) concentration of very small VLDL particles.
Figure 5
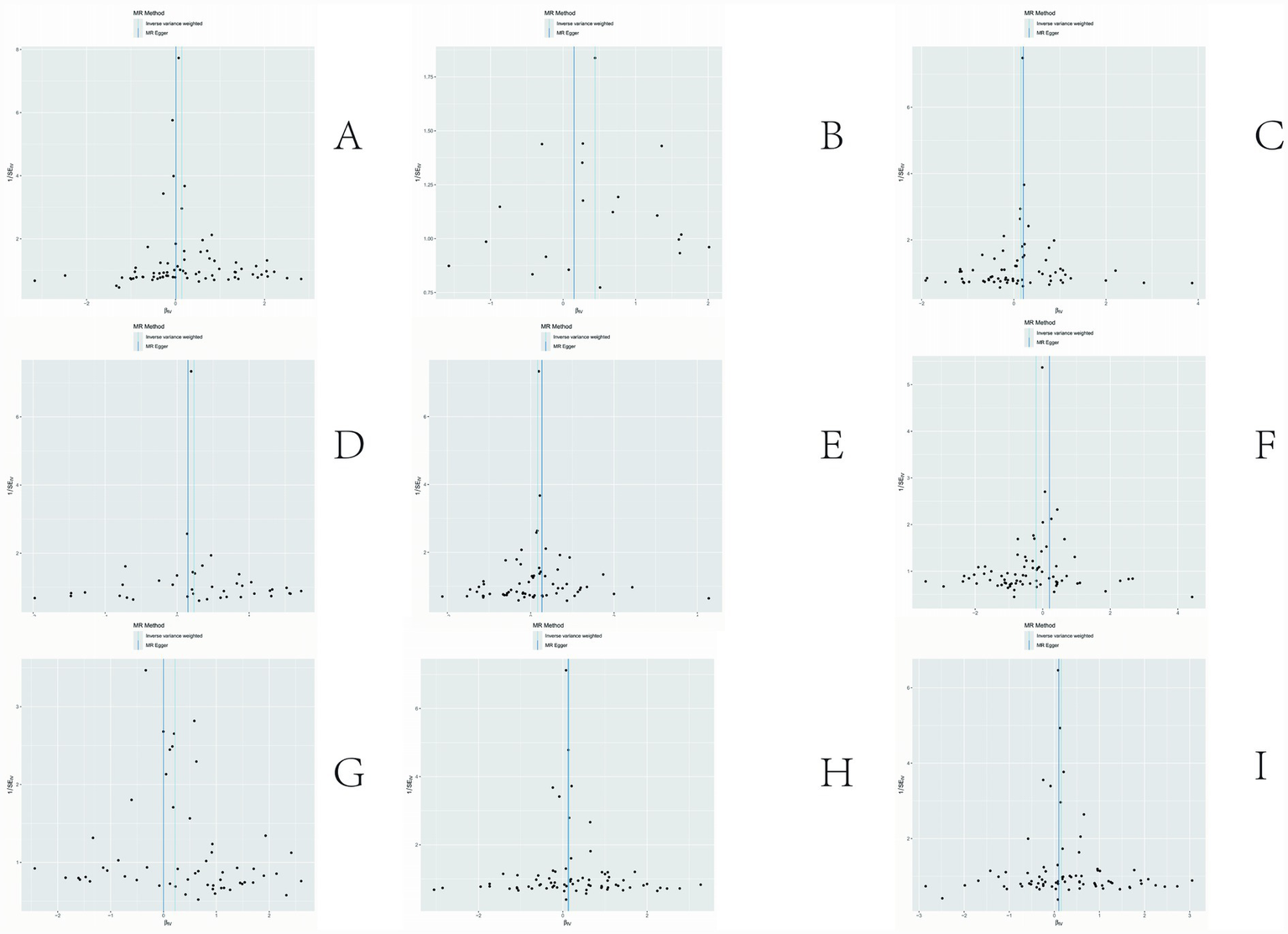
Results of sensitivity analysis-funnelplot. (A) Ratio of apolipoprotien B to apolipoprotien A1 levels; (B) 3-hydroxybutyrate levels; (C) triglyceride levels in LDL; (D) triglycerides to total lipids ratio in large LDL; (E) triglyceride in medium LDL; (F) total cholesterol levels in very large HDL; (G) triglycerides to total lipids ratio in very large HDL; (H) total lipids in very small VLDL; (I) concentration of very small VLDL particles.
2.4 Analysis of metabolite target sites
Based on the above analysis, we found that an elevated level of 3-Hydroxybutyrate increases the risk of polycystic ovary syndrome, and its main target site is Histone deacetylase 3. Therefore, we speculate that Histone deacetylase 3 may be related to the pathogenesis of polycystic ovary syndrome. To further verify the biological effect of 3-Hydroxybutyrate, we used the KGN human ovarian granulosa cell line in vitro.
2.5 Cytotoxicity assay results
The CCK8 assay results indicated that 1 mM 3-HB had a minor effect on the viability of KGN cells within 24 h; similarly, 40 nM RGFP966 (HDAC3 inhibition) had a minor impact on the viability of KGN cells within 24 h. Subsequently, 1 mM 3-HB and 40 nM RGFP966 were selected for the experiment (Figure 6).
Figure 6
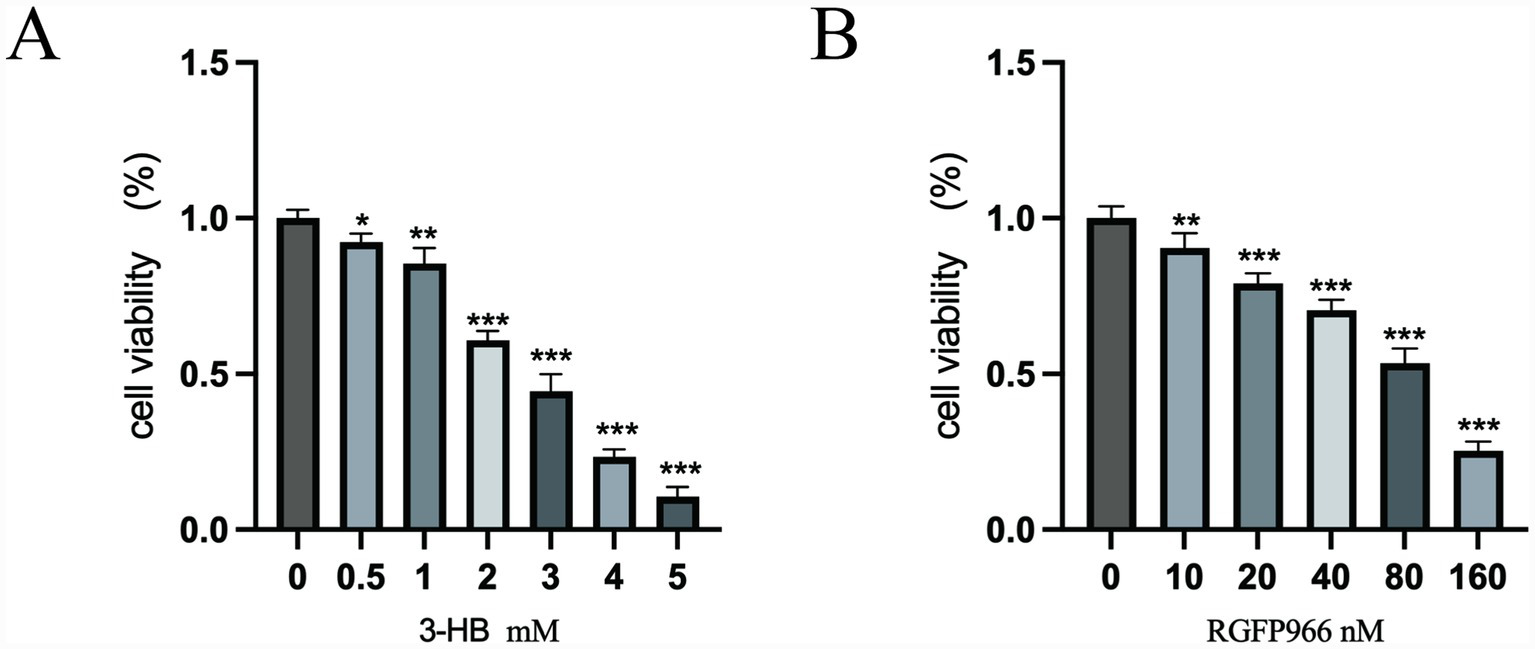
(A) 3-HB; (B) RGFP966. *p < 0.05, **p < 0.01, ***p < 0.001.
2.6 Measurement of testosterone and estradiol levels in cell culture supernatants
The primary pathological of PCOS involves hyperandrogenism and disrupted estrogen homeostasis. ELISA analyses of cell culture supernatants revealed that 3-HB induces dysregulation of testosterone and estradiol levels. Furthermore, pharmacological blockade of 3-HB alleviates this hormonal imbalance between testosterone and estradiol (Figure 7).
Figure 7
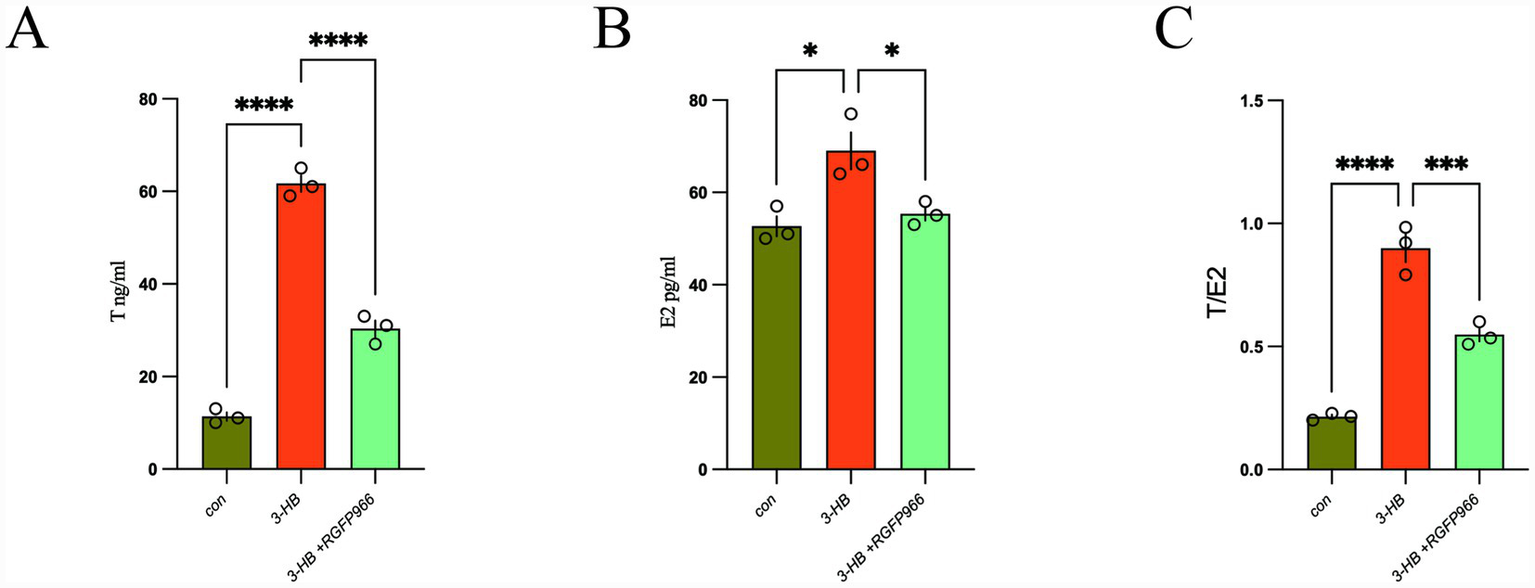
(A) Testosterone; (B) estrogen; (C) testosterone/estrogen ratio. *p < 0.05, ***p < 0.001, ****p < 0.0001.
2.7 The results of the qRT-PCR experiment
Using qRT-PCR, we demonstrated that 3-HB significantly up-regulates the transcriptional levels of 10 genes—HDAC3, CYP19A1, IRS1, FSHR, LHCGR, StAR, CYP11A1, CYP17A, HSD3B2, and HSD17B13. Furthermore, pharmacological inhibition of 3-HB leads to a partial but consistent down-regulation in the expression of these genes (Figure 8).
Figure 8
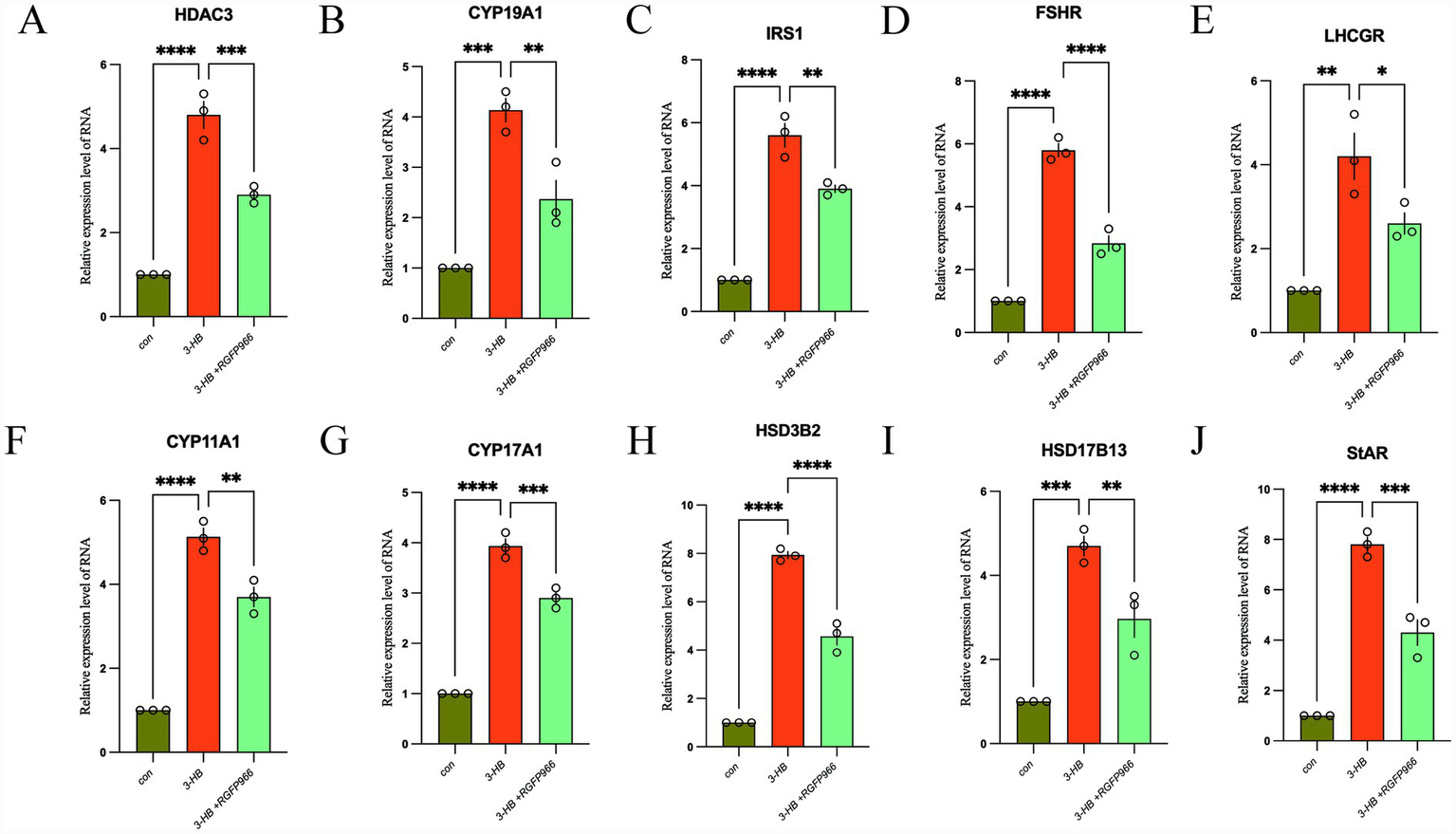
The expression changes of HDAC3 (A), CYP19A1 (B), IRS1 (C), FSHR (D), LHCGR (E), CYP11A1 (F), CYP17A1 (G), HSD3B2 (H), HSD17B13 (I), and StAR (J) genes. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3 Discussion
This study explored the potential role of 3-Hydroxybutyrate (3-HB) in the pathophysiology of Polycystic Ovary Syndrome (PCOS), combining Mendelian Randomization (MR) approaches with in vitro cellular assays. A central focus was placed on the involvement of Histone deacetylase 3 (HDAC3) as a mediating pathway. The results reveal a causal association between elevated 3-HB levels and increased PCOS susceptibility, highlighting HDAC3 as a key player in this molecular interplay.
3.1 Insights from Mendelian randomization analysis
The MR analysis demonstrated a statistically significant link between higher circulating 3-HB concentrations and an elevated risk of developing PCOS. Notably, 3-HB showed consistent positive associations with lipid-related metabolites, particularly triglycerides present in low-density lipoprotein (LDL) and very-low-density lipoprotein (VLDL) particles. These observations align with existing evidence suggesting that disturbances in lipid metabolism—especially those involving triglyceride-rich lipoproteins—and altered ketone body homeostasis may contribute to PCOS etiology (18, 19). Additionally, we identified a correlation between 3-HB and the apolipoprotein B/A1 ratio, indicating a multifaceted influence of 3-HB on metabolic and cardiovascular risk factors associated with PCOS. Sensitivity analyses confirmed the robustness of these findings, minimizing concerns about horizontal pleiotropy and reinforcing the validity of our conclusions (20).
3.2 Impact of 3-HB on gene expression in PCOS-related pathways
To further elucidate the biological mechanisms, we conducted in vitro experiments assessing how 3-HB modulates the expression of genes implicated in PCOS. Our data indicate that 3-HB exerts substantial regulatory effects on several critical genes involved in endocrine function, insulin signaling, and metabolic regulation.
A marked increase in HDAC3 mRNA levels was observed following 3-HB exposure. As a core epigenetic enzyme, HDAC3 regulates gene transcription by removing acetyl groups from histones and other proteins, thereby influencing diverse cellular processes (21). In the context of PCOS, HDAC3 may serve as a mediator of ovarian dysfunction and insulin resistance through epigenetic reprogramming. This positions HDAC3 as a pivotal factor in 3-HB-driven pathogenic pathways, warranting deeper investigation (22).
Moreover, treatment with 3-HB led to increased expression of FSHR (follicle-stimulating hormone receptor), LHCGR (luteinizing hormone receptor), and CYP19A1 (aromatase)—genes essential for gonadal hormone production and follicular development. Dysregulation of these receptors is commonly linked to hyperandrogenism, disrupted ovulation, and impaired folliculogenesis in PCOS (22, 23). Given that FSHR and LHCGR govern follicular maturation and steroidogenesis, while CYP19A1 controls estrogen biosynthesis, their altered expression under 3-HB influence may amplify hormonal imbalances characteristic of PCOS.
Additionally, 3-HB induced upregulation of IRS1 (insulin receptor substrate 1), a crucial component of the insulin signaling cascade closely tied to insulin resistance (24). Although enhanced IRS1 expression might initially appear beneficial for insulin sensitivity, in the context of PCOS, such changes could paradoxically disrupt downstream signaling, contributing to metabolic dysregulation. This suggests that 3-HB may indirectly worsen insulin resistance despite apparent activation of early insulin pathway elements.
Collectively, these findings suggest that 3-HB influences multiple genetic pathways relevant to PCOS pathogenesis via HDAC3-mediated regulation. By altering the expression of genes involved in steroidogenesis, insulin action, and ovarian physiology, 3-HB may drive key clinical features of PCOS, including menstrual irregularities, hyperandrogenemia, and metabolic disturbances (25, 26).
3.3 Role of HDAC3 in epigenetic regulation
HDAC3 plays a fundamental role in chromatin remodeling and transcriptional control through histone deacetylation. In this study, its upregulation was directly correlated with 3-HB treatment. Importantly, pharmacological inhibition of HDAC3 significantly reduced the effects of 3-HB on target gene expression, indicating that HDAC3 activity is functionally required for 3-HB’s regulatory actions. This implies that HDAC3 acts as a central node in the network connecting 3-HB to PCOS-related molecular alterations, particularly those affecting hormonal balance and metabolic function (27, 28).
These results position HDAC3 not only as a potential contributor to PCOS progression but also as a critical effector in the 3-HB signaling axis. Through modulation of gene networks governing insulin response, steroid hormone synthesis, and cellular energy metabolism, HDAC3 may accelerate disease development in individuals with elevated 3-HB levels (29, 30).
3.4 Proposed mechanistic framework linking 3-HB and PCOS
Integrating the genetic and functional evidence, we propose that 3-HB contributes to PCOS development primarily through HDAC3-dependent epigenetic regulation. By altering the expression of genes involved in sex hormone biosynthesis and insulin signal transduction, 3-HB may promote hyperandrogenism and estrogen deficiency. Concurrently, its influence on insulin-related genes may exacerbate metabolic impairments commonly seen in PCOS. Together, these pathways highlight 3-HB as a biologically active metabolite with significant implications for both reproductive and metabolic health in PCOS (31, 32). Targeting the 3-HB–HDAC3 axis may therefore represent a novel therapeutic strategy for managing or even preventing PCOS progression (33, 34).
3.5 Limitations and directions for future research
While this study provides compelling evidence linking 3-HB to PCOS through MR and experimental models, certain limitations must be acknowledged. First, the findings are based solely on cell culture systems without validation in human tissue samples or animal models. Future studies should incorporate clinical cohorts and in vivo experiments to confirm these mechanisms and assess translational potential. Second, although HDAC3 was identified as a major mediator, it is likely that 3-HB affects PCOS through additional epigenetic regulators or alternative signaling cascades. Further exploration into other possible pathways—such as DNA methylation, non-coding RNAs, or mitochondrial function—could provide a more comprehensive understanding of 3-HB’s role in metabolic-endocrine crosstalk.
In summary, this work uncovers a novel mechanism whereby 3-HB modulates PCOS-associated gene networks via HDAC3 activation, offering new insights into the metabolic underpinnings of PCOS. HDAC3 emerges as a promising therapeutic target, paving the way for innovative interventions aimed at disrupting the pathological loop between ketone body metabolism and reproductive dysfunction in PCOS.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://gwas.mrcieu.ac.uk/.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
JX: Writing – original draft, Formal analysis, Data curation. LL: Validation, Formal analysis, Writing – original draft. SY: Writing – review & editing, Methodology. PL: Data curation, Writing – original draft, Conceptualization. BH: Funding acquisition, Writing – review & editing. J-lK: Validation, Supervision, Writing – review & editing.
Funding
The author(s) declared that financial support was received for this work and/or its publication. This work was supported by the Health Research Project of Hunan Provincial Health Commission (No. W20243071 to Jia Xu, No. 20258047 to Bing He), Joint Project of Hunan Provincial Natural Science Foundation (No. 2025JJ80906 to Jia Xu, No. 2024JJ9448 Bing He, No. 2024JJ9446 to Lan Li), Youth Project of Hunan Provincial Department of Educationt (No. 24B0354 to Jia Xu), Research Fund Project of Hunan University of Traditional Chinese Medicine (No. 2022XYLH027 to Jia Xu), Construction Project of National Famous Traditional Chinese Medicine Expert Inheritance Studio in 2022, National Medical Education Letter (2022) No. 75 to Bing He, Hunan Provincial Traditional Chinese Medicine Backbone Talent Project (2024) No. 3 to Bing He, Key Project of Hunan Provincial Department of Educationt (No. 24A0257 to Shuo Yang), and Changsha Natural Science Foundation (No. KQ2402176 to Shuo Yang).
Acknowledgments
Thank Minna K, Karjalainen for the data from her previous research.
Conflict of interest
The author(s) declared that this work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declared that Generative AI was not used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1.
Helvaci N Yildiz BO . Polycystic ovary syndrome as a metabolic disease. Nat Rev Endocrinol. (2025) 21:230–44. doi: 10.1038/s41574-024-01057-w
2.
Escobar-Morreale HF . Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol. (2018) 14:270–84. doi: 10.1038/nrendo.2018.24,
3.
Dilliyappan S Kumar AS Venkatesalu S Palaniyandi T Baskar G Sivaji A et al . Polycystic ovary syndrome: recent research and therapeutic advancements. Life Sci. (2024) 359:123221. doi: 10.1016/j.lfs.2024.123221
4.
Han W Zhang M Wang H Yang Y Wang L . Lipid accumulation product is an effective predictor of metabolic syndrome in non-obese women with polycystic ovary syndrome. Front Endocrinol. (2023) 14:1279978. doi: 10.3389/fendo.2023.1279978
5.
Wang X Han H Shi X Nie X Zhu R Jin J et al . Genetic insights of blood lipid metabolites on polycystic ovary syndrome risk: a bidirectional two-sample Mendelian randomization study. Front Endocrinol. (2024) 15:1391826. doi: 10.3389/fendo.2024.1391826
6.
Jia MJ Zhou L Liu X-N Li H-L . Genetically predicted serum metabolites mediate the association between inflammatory proteins and polycystic ovary syndrome: a Mendelian randomization study. Front Med. (2024) 11:1433612. doi: 10.3389/fmed.2024.1433612
7.
Ma Y Cai J Liu L-W Wen T Huang W Hou W et al . Causal relationships exist between polycystic ovary syndrome and adverse pregnancy and perinatal outcomes: a Mendelian randomization study. Front Endocrinol. (2024) 15:1327849. doi: 10.3389/fendo.2024.1327849
8.
Zhao H Xing C Zhang J He B . Comparative efficacy of oral insulin sensitizers metformin, thiazolidinediones, inositol, and berberine in improving endocrine and metabolic profiles in women with PCOS: a network meta-analysis. Reprod Health. (2021) 18:171. doi: 10.1186/s12978-021-01207-7,
9.
Ferrer MJ Silva AF Abruzzese GA Velázquez ME Motta AB . Lipid metabolism and relevant disorders to female reproductive health. Curr Med Chem. (2021) 28:5625–47. doi: 10.2174/0929867328666210106142912,
10.
Chang H Shi B Ge H Liu C Wang L Ma C et al . Acupuncture improves the emotion domain and lipid profiles in women with polycystic ovarian syndrome: a secondary analysis of a randomized clinical trial. Front Endocrinol. (2023) 14:1237260. doi: 10.3389/fendo.2023.1237260
11.
Sun S Jiao M Han C Zhang Q Shi W Shi J et al . Causal effects of genetically determined metabolites on risk of polycystic ovary syndrome: a Mendelian randomization study. Front Endocrinol (Lausanne). (2020) 11:621. doi: 10.3389/fendo.2020.00621
12.
Huang X Guo X Gao W Xiong Y Chen C Zheng H et al . Causal association between years of schooling and the risk of traumatic brain injury: a two-sample mendelian randomization analysis. J Affect Disord. (2024) 354:483–90. doi: 10.1016/j.jad.2024.03.045,
13.
Karjalainen MK Karthikeyan S Oliver-Williams C Sliz E Allara E Fung WT et al . Genome-wide characterization of circulating metabolic biomarkers. Nature. (2024) 628:130–8. doi: 10.1038/s41586-024-07148-y,
14.
Guan J Liu T Yang K Chen H . Dried fruit intake and lower risk of type 2 diabetes: a two-sample mendelian randomization study. Nutr Metab (Lond). (2024) 21:46. doi: 10.1186/s12986-024-00813-z
15.
Li YT Li BH Meng FL . Evaluating the relationship between standing height, body mass index, body fat percentage with risk of inguinal hernia: a Mendelian randomization study. Sci Rep. (2024) 14:26402. doi: 10.1038/s41598-024-78122-x
16.
Jiang H Zhang K Zhang X . Mendelian randomization analysis of the association between childhood overweight or obesity and gestational diabetes mellitus. Diabetes Obes Metab. (2024) 26:6016–22. doi: 10.1111/dom.15975,
17.
Skrivankova VW Richmond RC Woolf BAR Yarmolinsky J Davies NM Swanson SA et al . Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. (2021) 326:1614–21. doi: 10.1001/jama.2021.18236
18.
Newman JC Verdin E . Ketone bodies as signaling metabolites. Trends Endocrinol Metab. (2014) 25:42–52. doi: 10.1016/j.tem.2013.09.002,
19.
Hartman AL Rho JM . The new ketone alphabet soup: BHB, HCA, and HDAC. Epilepsy Curr. (2014) 14:355–7. doi: 10.5698/1535-7597-14.6.355,
20.
Guo X Puttabyatappa M Thompson RC Padmanabhan V . Developmental programming: contribution of epigenetic enzymes to antral follicular defects in the sheep model of PCOS. Endocrinology. (2019) 160:2471–84. doi: 10.1210/en.2019-00389,
21.
Li B Yu Y Liu K Zhang Y Geng Q Zhang F et al . Β-Hydroxybutyrate inhibits histone deacetylase 3 to promote claudin-5 generation and attenuate cardiac microvascular hyperpermeability in diabetes. Diabetologia. (2021) 64:226–39. doi: 10.1007/s00125-020-05305-2
22.
Wang H Cai H Wang X Zhang M Liu B Chen Z et al . HDAC3 maintains oocyte meiosis arrest by repressing amphiregulin expression before the LH surge. Nat Commun. (2019) 10:5719. doi: 10.1038/s41467-019-13671-8,
23.
Jansen E Laven JS Dommerholt HB Polman J van Rijt C van den Hurk C et al . Abnormal gene expression profiles in human ovaries from polycystic ovary syndrome patients. Mol Endocrinol. (2004) 18:3050–63. doi: 10.1210/me.2004-0074,
24.
Salinas I Sinha N Sen A . Androgen-induced epigenetic modulations in the ovary. J Endocrinol. (2021) 249:R53–64. doi: 10.1530/JOE-20-0578,
25.
Liu HJ Liu H Jin L Wang X Shi J He Y et al . Reactive oxygen species in polycystic ovary syndrome: mechanistic insights into pathogenesis and therapeutic opportunities. Redox Biol. (2025) 85:103776. doi: 10.1016/j.redox.2025.103776,
26.
Stener-Victorin E Deng Q . Epigenetic inheritance of polycystic ovary syndrome - challenges and opportunities for treatment. Nat Rev Endocrinol. (2021) 17:521–33. doi: 10.1038/s41574-021-00517-x,
27.
Kaymaz N Kara Ö Şirin H Kasap T Uzun ME . Role of hyperandrogenism on disordered eating behaviors in adolescents with PCOS and interplay with insulin resistance. J Pediatr Endocrinol Metab. (2025) 39:37–45. doi: 10.1515/jpem-2025-0151
28.
Xia XY Chen Y Zhang XJ Wang J Xu HY Zhao R . Gut microbiota dysbiosis and short-chain fatty acid depletion in phlegm-dampness polycystic ovary syndrome: a cross-sectional 16S rRNA sequencing analysis. BMC Endocr Disord. (2025) 25:255. doi: 10.1186/s12902-025-02076-y,
29.
Tao P Yan X Wang Z . Mechanistic role of the KRTAP5-AS1/miR-199b-5p/CYP19A1 axis in polycystic ovary syndrome pathogenesis. J Ovarian Res. (2025) 18:176. doi: 10.1186/s13048-025-01746-8,
30.
Azumah R Hummitzsch K Anderson RA Rodgers RJ . Genes in loci genetically associated with polycystic ovary syndrome are dynamically expressed in human fetal gonadal, metabolic and brain tissues. Front Endocrinol. (2023) 14:1149473. doi: 10.3389/fendo.2023.1149473,
31.
Chen W Pang Y . Metabolic syndrome and PCOS: pathogenesis and the role of metabolites. Meta. (2021) 11:37–45. doi: 10.3390/metabo11120869,
32.
Bulow NS Wissing ML Macklon N Pinborg A Løssl K . Reproductive outcomes after letrozole stimulated versus artificial frozen-thawed embryo transfer cycles in women with PCOS and/or oligo-anovulation: a systematic review and meta-analysis. Hum Reprod Update. (2025) 31:445–63. doi: 10.1093/humupd/dmaf011,
33.
Heidarzadehpilehrood R Pirhoushiaran M Abdollahzadeh R Binti Osman M Sakinah M Nordin N et al . A review on CYP11A1, CYP17A1, and CYP19A1 polymorphism studies: candidate susceptibility genes for polycystic ovary syndrome (PCOS) and infertility. Genes. (2022) 13:302. doi: 10.3390/genes13020302,
34.
Parker J Briden L Gersh FL . Recognizing the role of insulin resistance in polycystic ovary syndrome: a paradigm shift from a glucose-centric approach to an insulin-centric model. J Clin Med. (2025) 14:4021. doi: 10.3390/jcm14124021,
Summary
Keywords
3-hydroxybutyrate, gene expression, HDAC3, lipid metabolism, Mendelian randomization, metabolic dysfunction, polycystic ovary syndrome
Citation
Xu J, Li L, Yang S, Li P, He B and Kuang J-l (2026) Investigating the causal relationship of lipid metabolism in polycystic ovary syndrome: a Mendelian randomization study on the regulatory role of 3-Hydroxybutyrate in gene expression. Front. Med. 13:1747593. doi: 10.3389/fmed.2026.1747593
Received
10 January 2026
Revised
13 January 2026
Accepted
14 January 2026
Published
04 February 2026
Volume
13 - 2026
Edited by
Giulia Rastrelli, University of Florence, Italy
Reviewed by
Radha Chaube, Banaras Hindu University, India
Yoshitaka Imamichi, Fukui Prefectural University, Japan
Updates
Copyright
© 2026 Xu, Li, Yang, Li, He and Kuang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing He, 320026@hnucm.edu.cn; Ji-lin Kuang, kuangjlabc@sina.com
†These authors share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.