- HIV Biology and Variability Unit, Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid, Spain
Integrase strand transfer inhibitor (INSTI)-containing regimens in HIV-1-infected patients have experienced a global increase. Recently, WHO has emphasized the need to fast-track the transition to dolutegravir (DTG)-based antiretroviral (ARV) treatments. However, continued surveillance of INSTI resistance is recommended. In this study, clinical, epidemiological, and virological features associated with INSTI resistance diagnosed in Spain were analyzed. Samples collected between 2008 and 2021 from HIV-1-infected patients were analyzed in integrase, protease, and reverse transcriptase using Sanger population sequencing. ARV drug resistance was evaluated with the Stanford University HIVdb program. Among 2,696 patients, 174 (6.5%) had INSTI resistance, all of them to first-generation INSTIs, and 71 (2.6%) had also resistance to second-generation INSTIs. Of these, only 5 individuals were exposed to DTG as the only INSTI, in whom resistance development was associated with poor treatment adherence and/or resistance to other ARV classes. Of newly HIV-1-diagnosed individuals, 0.92% harbored INSTI-resistant viruses, with low prevalences maintained along time, and only one had low-level resistance to DTG. Persons who inject drugs, age over 39 years, resistance to other ARV classes, and longer time from diagnosis were associated with INSTI resistance (p < 0.001). Non-subtype B INSTI-resistant viruses lacked the Q148H + G140S resistance pathway and showed lower INSTI resistance levels than subtype B viruses. In conclusion, INSTI resistance is uncommon and associated with long-term infections, older age and additional resistance to other ARV drug classes, and is rare in newly diagnosed HIV-1 infections. Our results also support the preferential use of DTG-containing regimens in first-line treatments, although surveillance of INSTI resistance is encouraged.
Introduction
Integrase strand transfer inhibitors (INSTIs) are a family of antiretrovirals (ARVs) for the treatment of HIV-1 infections. Raltegravir (RAL) and elvitegravir (EVG) are the approved first-generation INSTIs, which are safe and effective for the treatment of ARV-naïve and -experienced patients (DeJesus et al., 2006; Steigbigel et al., 2008; Lennox et al., 2009). While these drugs induce a strong inhibition of HIV-1 replication, they have a modest genetic barrier to resistance, selecting for INSTI resistance mutations that reduce susceptibility to both RAL and EVG (Anstett et al., 2017).
Second-generation INSTIs comprise dolutegravir (DTG) and bictegravir (BIC), which have shown strong suppression of HIV-1 replication, a good safety, high genetic barriers preventing the emergence of drug resistance in large clinical trials (Walmsley et al., 2015; Gallant et al., 2017; Sax et al., 2017; Acosta et al., 2019), and efficacy in treatment-experienced patients (Eron et al., 2013; Castagna et al., 2018).
In the last years, a global increase in the use of INSTI-containing regimens is occurring. In spite of this, very low prevalences of transmitted drug resistance (TDR) to INSTIs have been reported (Inzaule et al., 2018; Casadellá et al., 2020), supporting the use of this drug class in first-line ARV treatments. However, the great increase in the use of second-generation INSTIs and cross-resistance among INSTIs advocate for a continued surveillance of TDR, as well as monitoring their clinical impact.
WHO has proposed DTG as a component of first- and second-line ARV treatments (WHO, 2018) and more recently has emphasized the need to fast-track the transition to DTG-based ARV treatments due to the high prevalence of resistance to non-nucleoside reverse transcriptase inhibitors (NNRTI) in pretreated patients found in some countries (WHO, 2021).
In this study, we analyze the clinical, epidemiological, and virological features of the patients with INSTI resistance included in our database from 2008 to 2021 and the emergence and trends of INSTI resistance. We also focus on DTG to evaluate potential consequences of the widespread inclusion of this drug in first- and second-line ARV drug regimens.
Materials and methods
Patients
Samples from individuals diagnosed of HIV-1 infection in Spain which were sent to the HIV Biology and Variability Unit, at Centro Nacional de Microbiología, Instituto de Salud Carlos III (CNM-ISCIII), for INSTI resistance analysis during 2008–2021 were included in the study.
Nucleic acid extraction, amplification, and sequencing
Nucleic acids were extracted from plasma or whole blood samples. RNA was extracted from 1 ml plasma using NUCLISENS® easyMAG® (BioMérieux, Marcy l’Etoile, France) and DNA was extracted from 200 μl whole blood using QIAamp® DNA DSP blood mini kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. An integrase-coding fragment of pol (HXB2 positions 4,160–5,220) was amplified by RT-PCR followed by nested PCR from RNA, or by nested PCR from DNA. Reagents, PCR thermal profiles, and primers are described in Supplementary Tables 1, 2. The protease-reverse transcriptase (Pr-RT) fragment was amplified as previously described (Gil et al., 2022).
Population sequencing was performed with ABI Prism BigDye Terminator Cycle Sequencing kit and ABI 3730 XL sequencer (Applied Biosystems, Foster City, CA, U.S.A.) at the Genomic Unit of Instituto de Salud Carlos III. Sequences were assembled with SeqMan Pro v.12.2.1 (DNA STAR Lasergene, Madison, WI, USA) and edited with BioEdit v.7.2.5 (Hall, 1999).
Integrase strand transfer inhibitor resistance analysis
The obtained sequences were analyzed with the Stanford University HIV Drug Resistance Database HIVdb program (version 8.9-11) for the detection of drug resistance (Rhee et al., 2003; Shafer, 2006). Sequences predicted to have a potential low-level resistance to INSTIs were considered susceptible for the objectives of this study. Sequences containing drug resistance mutations (DRMs) associated with apolipoprotein B mRNA editing enzyme, catalytic polypeptide (APOBEC) activity, as determined by the HIVdb program, were excluded from further analysis. A single sequence per patient was considered. In the cases where several sequences were available from a patient with INSTI resistance, the one sampled closer to HIV-1 diagnosis showing INSTI resistance was selected.
Codon usage bias in subtype B and non-subtype B HIV-1 infections was analyzed calculating the frequencies of each triplet in our sequence set and the cost of each mutation using the values described previously (Theys et al., 2019).
Phylogenetic analyses and genetic form classification
Sequences were analyzed phylogenetically by an approximately maximum-likelihood method using FastTree2 (Price et al., 2010). In these analyses, the general time reversible model of nucleotide substitution with CAT approximation to account for among-site heterogeneity in substitution rates (GTR + CAT) was used, and the reliability of nodes was assessed with Shimodaira-Hasegawa (SH)-like local support values. Classification of sequences in subtypes and circulating recombinant forms was based on clustering with clade references in these trees. Sequences suspected of intersubtype recombination were subsequently analyzed by bootscanning with SimPlot v3.5 (Lole et al., 1999).
Statistical analysis
Epidemiological factors associated with INSTI resistance were analyzed with chi-squared, Fisher’s exact, and Mann–Whitney tests. The frequencies of DRMs and resistance levels to the different INSTIs were compared between subtype B and non-subtype B viruses with chi-squared and Fisher’s exact tests. The analyses were performed using the STATA statistical software package version 17 (Stata Corporation, College Station, TX, US). Associations were considered statistically significant at a value of p < 0.05.
Ethics statement
Sequences derive from ARV resistance genotypic tests. The use of anonymized, de-identified clinical/demographic and sequence data was reviewed and approved under an exempt protocol by the Bioethics and Animal Well-being Committee of Instituto de Salud Carlos III, with report numbers CEI PI 38_2016-v3 (dated 20 June 2016) and CEI PI 31_2019-v5 (dated 6 November 2019). This study did not require written informed consent by study participants, except for those who donated samples different from the ones obtained in clinical practice.
Results
Frequency of INSTI resistance and associated factors
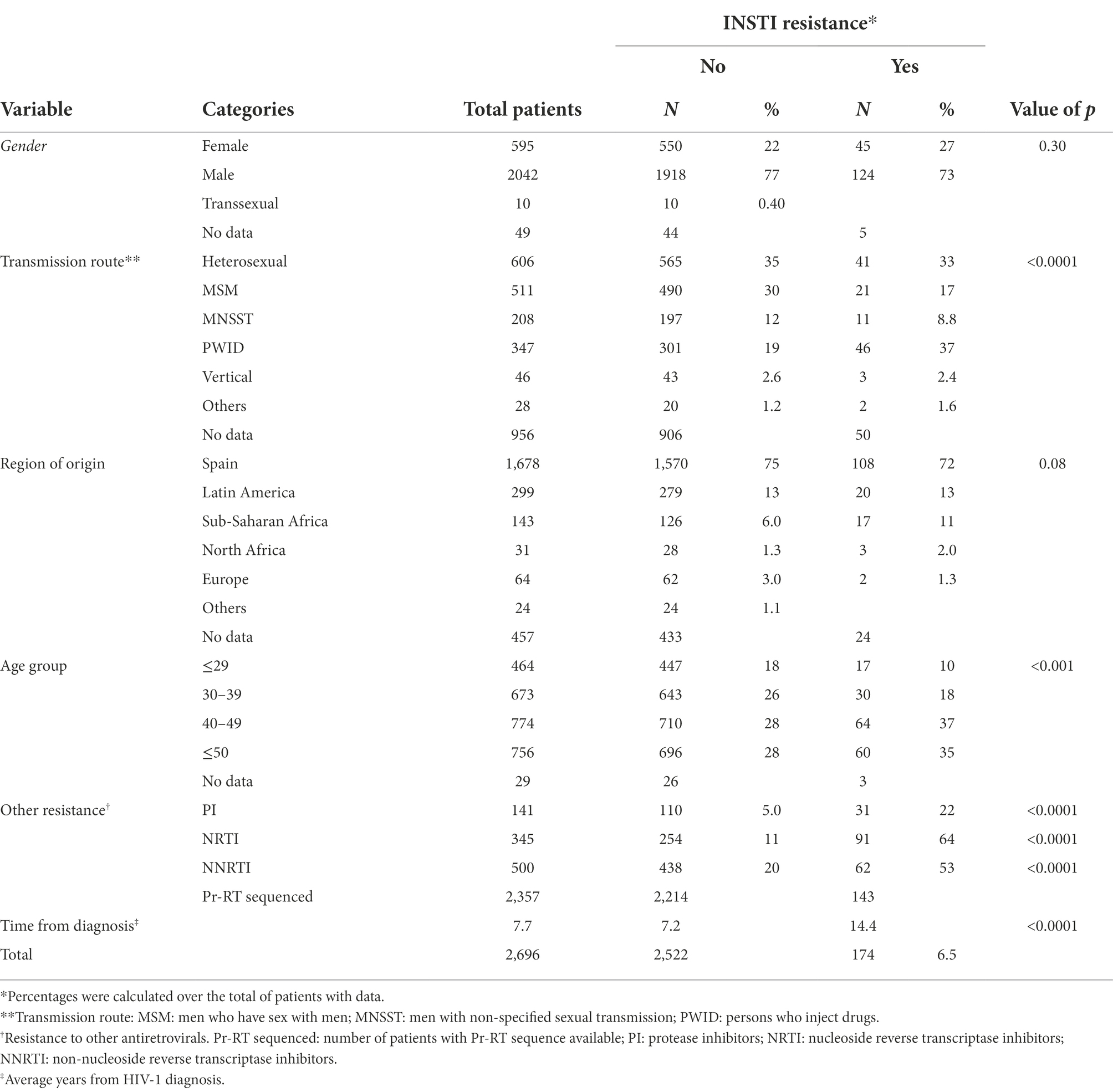
Resistance mutations in integrase conferring, at least, low level of resistance to any INSTI were detected in 174 (6.5%) of 2,696 patients analyzed. A comparison between both populations, with and without INSTI resistance, is shown in Table 1. Persons who inject drugs (PWID) were the main transmission category in the group with INSTI resistance (37% vs. 19% in those without INSTI resistance). The main age groups with INSTI resistance were 40–49 and ≥ 50, with similar percentages in both age groups; joining them together (ages 40 and older), percentages were 73% vs. 56% in the groups with and without INSTI resistance, respectively. The average time from diagnosis was double in patients with INSTI resistance (14.4 years vs. 7.2 years). Differences between groups with and without INSTI resistance with regard to transmission routes, age, and time from HIV-1 diagnosis were statistically significant (p < 0.001; Table 1).
Patients with INSTI resistance also had a higher percentage of DRMs in Pr-RT, compared to the group without INSTI resistance, associated with resistance to protease inhibitors (PI; 22% vs. 5%), to nucleoside reverse transcriptase inhibitors (NRTI; 64% vs. 11%), and to NNRTI (53% vs. 20%), with statistically significant associations for all three drug classes (p < 0.0001; Table 1).
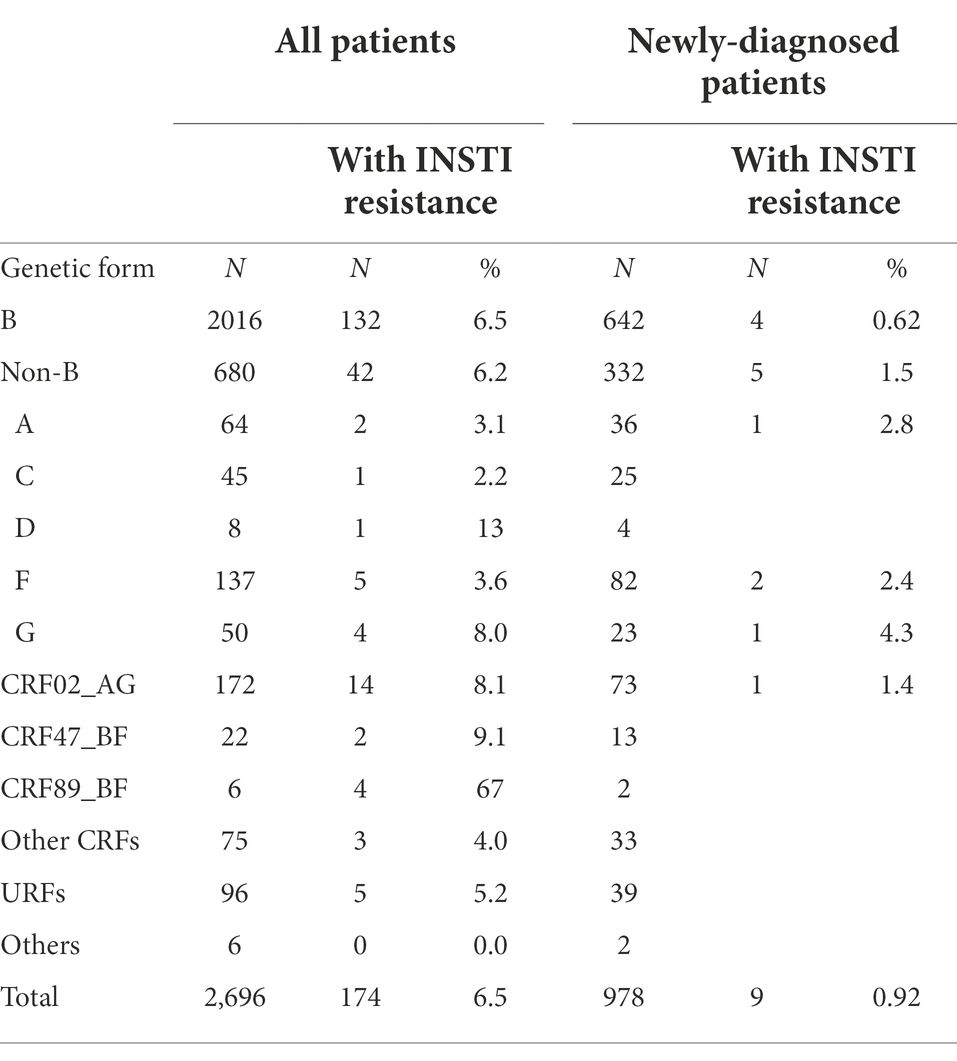
Most (75%) viruses in the studied population were of subtype B. There was no statistically significant difference in the frequency of INSTI resistance between subtype B (6.5%) and non-subtype B (6.2%) viruses. Among non-subtype B viruses, the most frequent genetic form was CRF02_AG, which represented 33% (14/42) of non-B viruses with INSTI resistance, and within which the prevalence of INSTI resistance was 8.1%. The highest prevalence of INSTI resistance was found in CRF89_BF (67%), although only 6 patients were infected with viruses of this genetic form (Table 2).
The most frequent INSTI in the treatment regimens of patients with INSTI resistance was RAL, used in 58%, followed by EVG, used in 7.5%, and DTG, used in 4.6%. The remaining 30% were patients exposed to two different INSTIs or an unspecified INSTI.
INSTI resistance in newly diagnosed and INSTI-naïve patients
Among newly-diagnosed patients (NDs), the prevalence of INSTI resistance was 0.92% (9/978). It was lower in subtype B (0.6%) than in non-subtype B (1.5%) infections (Table 2), but the difference was not statistically significant. The 5 non-subtype B viruses with INSTI resistance belonged to subtypes A, F (two patients), and G, and to CRF02_AG (Table 2). The annual prevalence of INSTI resistance among NDs was in the range of 0.75–1.5%, showing a stable trend in 2015–2020 (Figure 1). In 2021, no patients with INSTI resistance were detected, although only 33 patients were analyzed that year.
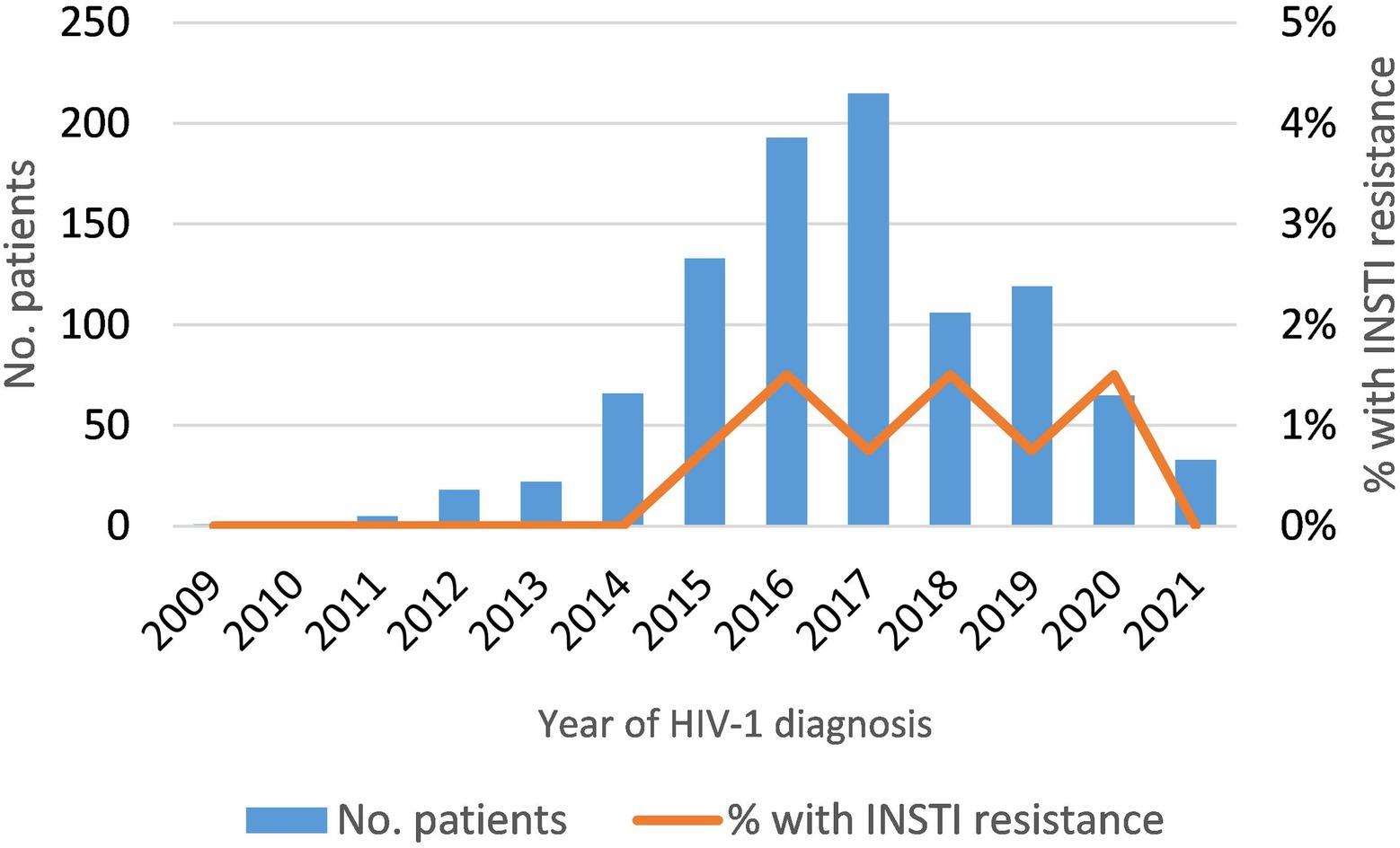
Figure 1. Temporal trend of INSTI resistance in newly-diagnosed HIV-1 patients analyzed in this study (2009–2021).
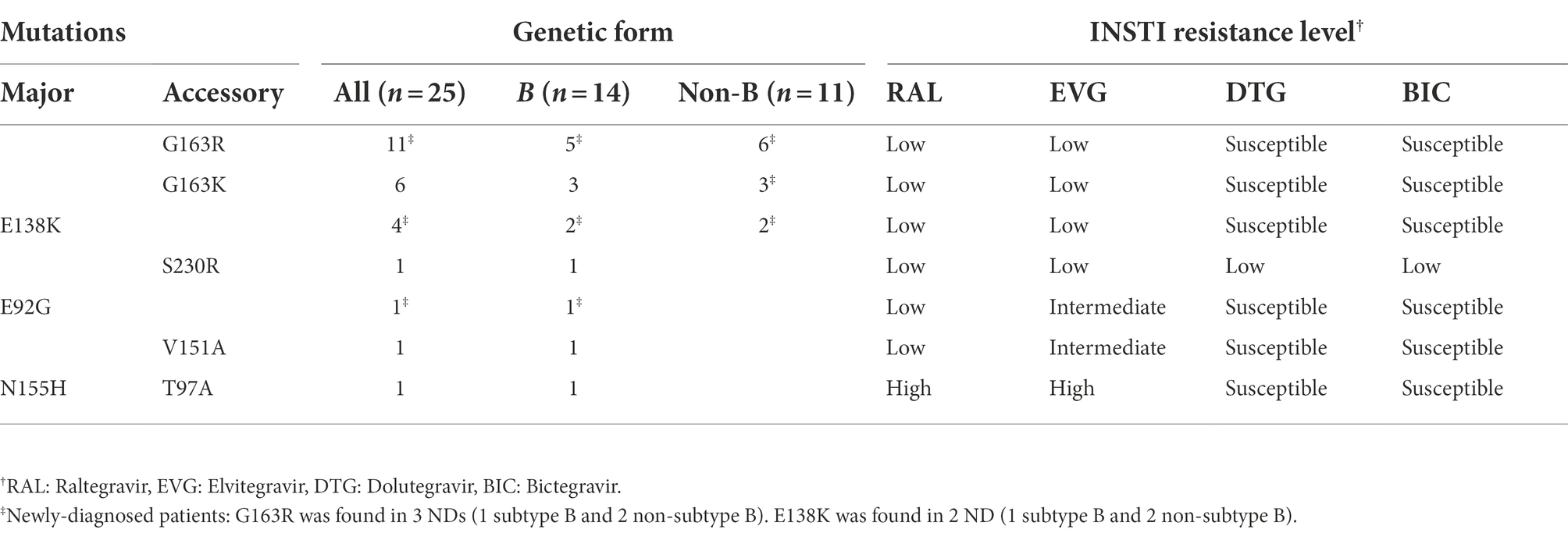
Among 174 patients with INSTI resistance, only 25 (14%) were INSTI-naïve, 11 of whom were infected with non-subtype B and 14 with subtype B strains (Table 3). Among INSTI-naïve patients with INSTI resistance, resistance levels were low in most (88%) of them, with the most common mutation being G163K/R (INSTI accessory mutations), detected in 17 (68%), followed by E138K, detected in 4 (16%). In these patients, resistance to DTG or BIC was found in only one infection, carrying the S230R accessory mutation, which is associated with low-level resistance to these INSTIs (Table 3).
Resistance levels to INSTIs
All INSTI-resistant viruses exhibited, at least, a predicted low-level resistance to RAL, with 70, 66, and 12% showing high-level resistance to RAL, EVG, and both DTG and BIC, respectively (Table 4). We also compared resistance levels to different INSTIs. Regarding EVG, resistance levels to this INSTI were similar to those observed to RAL, although 3.5% (6/174) RAL-resistant viruses were susceptible to EVG and 15% (8/52) viruses with low or intermediate resistance to RAL had high-level resistance to EVG, which was associated with T66A/I and E92Q mutations. Interestingly, 59 and 61% of RAL-resistant viruses were susceptible to DTG and BIC, respectively, and only 14% (6/43) viruses with low-level RAL resistance showed intermediate resistance to these second-generation INSTIs (Table 4), due to R263K mutation.
INSTI resistance levels in subtype B and non-subtype B viruses
Despite the lack of statistical differences in the frequency of INSTI-resistant viruses between patients infected with B and non-B strains, non-subtype B strains exhibited lower resistance levels than subtype B strains (Figure 2). High-level resistance to RAL was found in 79% (104/132) and 43% (18/42) of subtype B and non-subtype B infections, respectively. Similarly, high-level resistance to EVG was found in 73% (97/132) and 43% (18/42) of the respective categories. Interestingly, of non-B strains resistant to first-generation INSTIs, 81% (34/42) were susceptible to both second-generation INSTIs, a proportion that was higher than in subtype B strains, in which 52% (69/132) and 55% (72/132) viruses resistant to first-generation INSTIs where susceptible to DTG and BIC, respectively. Differences in INSTI resistance levels between B and non-B strains were statistically significant, ranging from p = 0.0001 for RAL to p = 0.013 for BIC (Figure 2).
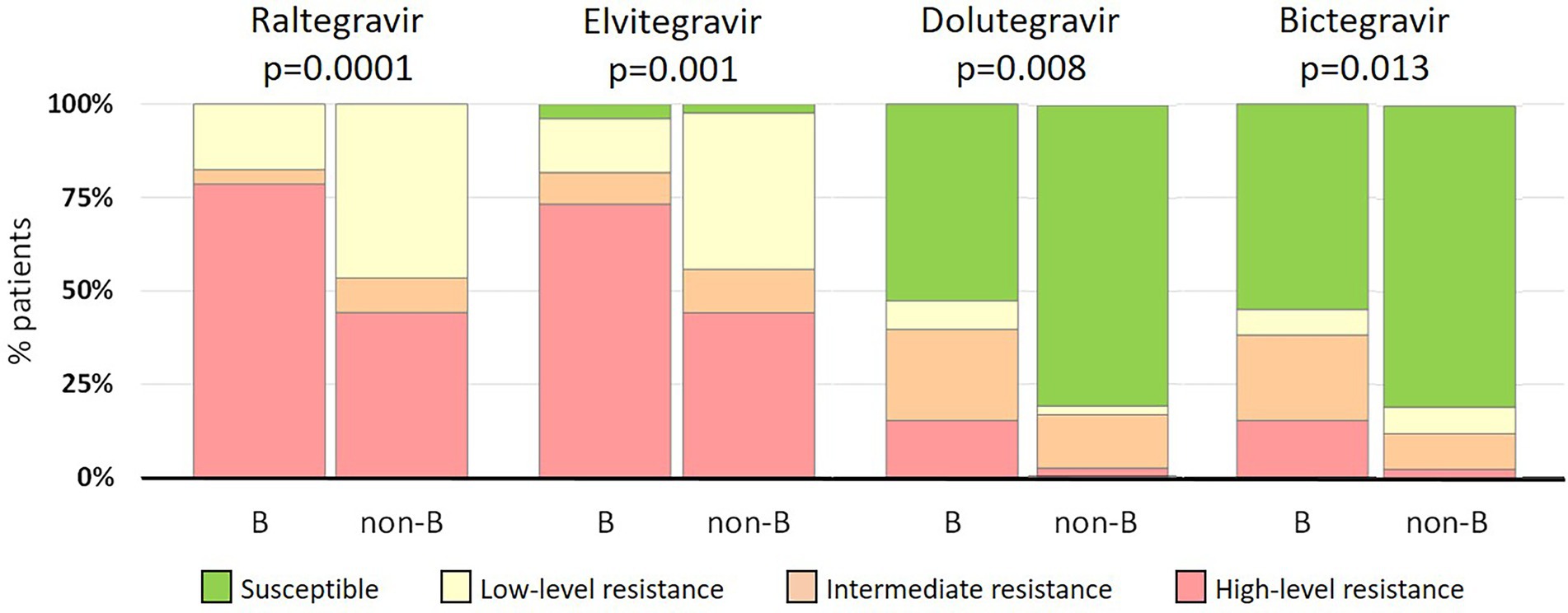
Figure 2. Comparison of INSTI resistance levels between subtype B (n = 132) and non-subtype B (n = 42) infections.
INSTI resistance mutations
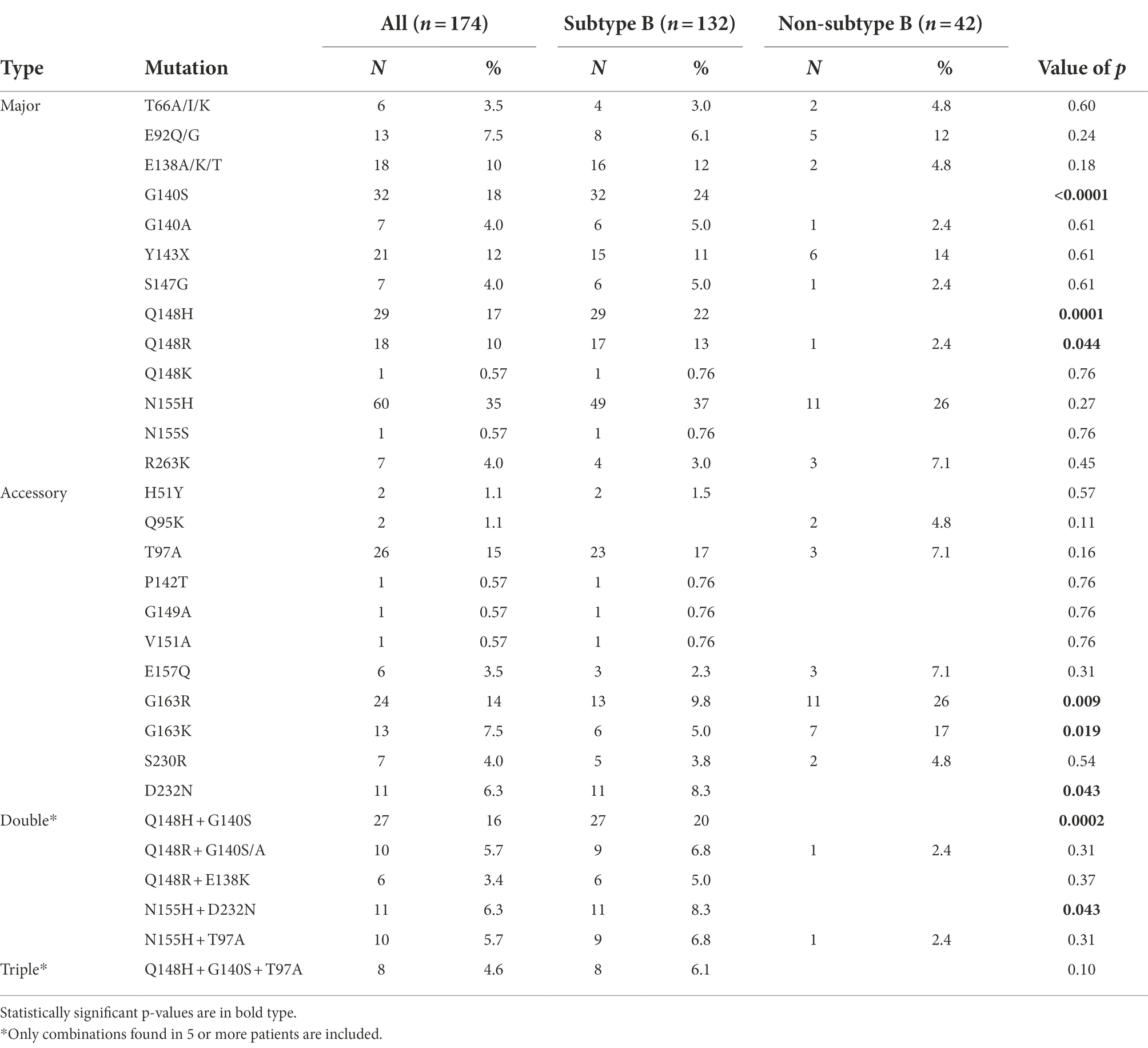
The most frequent major INSTI DRM was N155H, which was found in 35% INSTI-resistant viruses, followed by G140S, Q148H, and Y143X, found in 18, 17, and 12%, respectively. Regarding the accessory DRMs, T97A was detected in 15% of the patients, followed by G163R, G163K, and D232N, detected in 14, 7.5, and 6.3%, respectively. The most frequent INSTI DRM combination was Q148H + G140S, which was observed in 16% of INSTI-resistant viruses, with the additional presence of T97A in 8 of them. G118R mutation, associated with DTG resistance, was not found in any patient. The frequencies of all INSTI DRMs are shown in Table 5.
Different DRM patterns were observed in subtype B and non-subtype B viruses. Among major DRMs, G140S (p < 0.0001), Q148H (p = 0.0001), and Q148R (p = 0.044) were found mainly or exclusively in subtype B-infected patients (Table 5). Similarly, the accessory DRM D232N (p = 0.043) was associated with subtype B infections, while G163R (p = 0.009) and G163K (p = 0.019) were detected mainly in non-subtype B infections (Table 5; Supplementary Figure 1). G163K/R was frequent among F subtype and BF recombinant viruses, as 67% of non-B viruses with these mutations were of subtype F in integrase, and 4 of 6 CRF89_BF INSTI-resistant viruses carried G163R.
Codon usage among subtype B and non-subtype B HIV-1 strains
Large differences in the frequency of codon usage were observed between subtype B and non-subtype B strains in amino acids E92 (GAG, 64% vs. 5.7%; and GAA, 29% vs. 92%), G140 (GGC, 72% vs. 7.8%; and GGA, 3.6% vs. 74%), G163 (GGA, 80% vs. 47%; and GGG, 8% vs. 41%), and D232 (GAT, 78% vs. 22%; and GAC, 11% vs. 73%) (Supplementary Table 3). However, only for G140 there was a difference in the substitution cost of change from the most frequent codon to the resistance-associated amino acid (G140S), which was higher for the non-subtype B strains (GGA, score 7.4) than for subtype B viruses (GGC, score 1) (Supplementary Table 3).
Regarding the R263K and G118R mutations, which are associated with DTG resistance, there were no differences in codon usage, although the substitution cost was higher for G118R (score 5.8) than for R263K (score 1; Supplementary Table 3).
INSTI resistance in patients exposed only to DTG
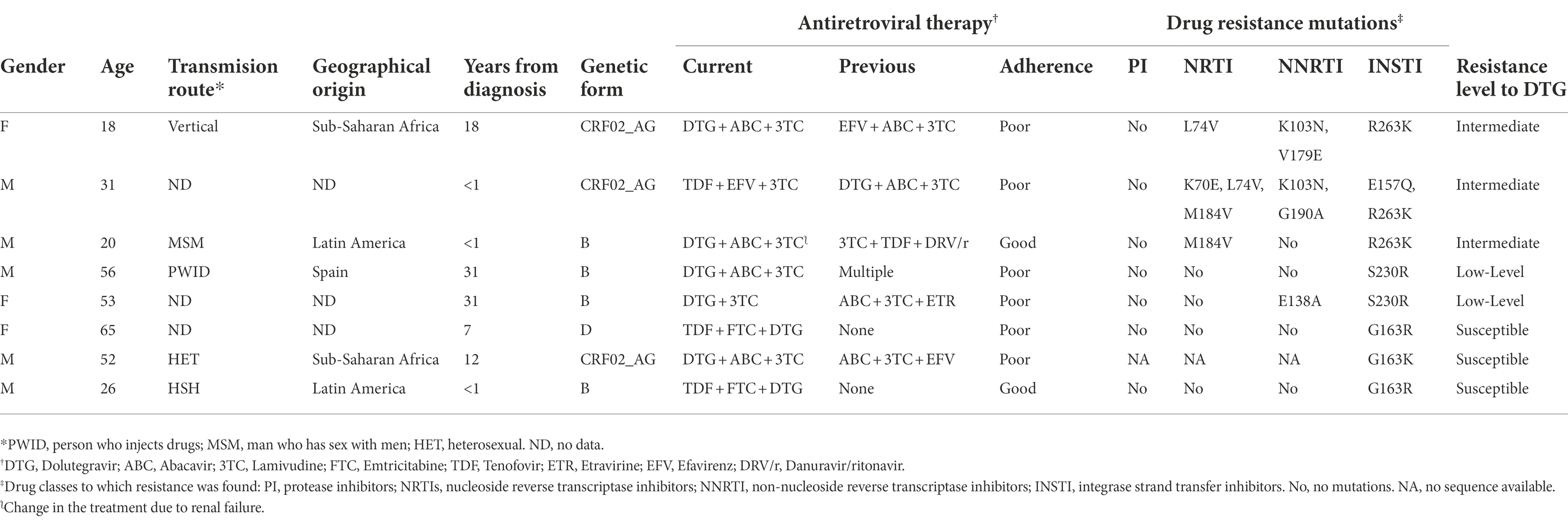
In 8 (4.6%) of the 174 patients with INSTI resistance, DTG was the only INSTI used in the treatment regimen. Three of them only had G163R/K, which keep viruses susceptible to DTG, while 5 showed intermediate (n = 3) or low (n = 2) level resistance to DTG, associated with R263K and S230R mutations, respectively (Table 6). All patients with intermediate-level resistance to DTG had DRMs to NNRTI and NRTI, which were M184V and K103N in two of them (Table 6). Four of 5 patients with DTG resistance had poor therapeutic adherence (as assessed by their physicians when the sample was sent for resistance testing) and in the fifth, the treatment regimen had been changed after renal failure (Table 6).
Discussion
In this study we have analyzed epidemiological and virological features associated with INSTI resistance in our cohort of 2,696 patients, including 25% infected with non-subtype B strains. INSTI resistance was associated with PWID, patients 40 years of age or older, and long-term infections. PWID frequently have difficulties to fit antiretroviral treatment in their daily routine, increasing their risk for reduced adherence (Lert and Kazatchkine, 2007; Vervoort et al., 2007). Other factors, like focusing in acquisition of illegal drugs, interactions of these drugs with treatments that induce side effects, homelessness, or difficulties in obtaining medication, have been also associated with reduced adherence in PWID (Witteveen and van Ameijden, 2002; Vervoort et al., 2007). Close follow-up and new strategies to improve adherence in HIV-1-infected PWID are necessary to prevent emergence of INSTI resistance in this population.
Accumulation of DRMs to PI, NRTI, and NNRTI was also associated with INSTI resistance. These DRMs can reduce the activity of ARVs used in combination treatments with INSTI, increasing the risk of a functional INSTI monotherapy and explaining the emergence of INSTI resistance (Cahn et al., 2013; Naeger et al., 2016; Modica et al., 2019; Underwood et al., 2022). Also, the presence of DRMs in Pr-RT could indicate a reduced adherence of the patient to the treatment, which can enhance the appearance of INSTI resistance mutations (Nachega et al., 2011; von Wyl et al., 2013; Acosta et al., 2022).
Transmitted INSTI resistance is currently a rare event, which we found only in 0.92% of the 978 NDs, with a stable trend in the last years (2015–2020). Moreover, the most frequent INSTI resistance mutation found among these patients was the polymorphic G163K/R mutation (Tzou et al., 2020), which is only associated with a low-level resistance to RAL and EVG. The low frequency of transmitted INSTIs resistance mutations has been reported in previous surveillance studies and meta-analyses (Doyle et al., 2015; Hauser et al., 2018; Inzaule et al., 2018; Raffaelli et al., 2018; Alvarez et al., 2019; Liu et al., 2019; Casadellá et al., 2020; Mbisa et al., 2020; Tzou et al., 2020; Semengue et al., 2021). This scenario supports the use of INSTIs in first-line therapeutic regimens. However, continued surveillance of ARV resistance is strongly recommended (WHO, 2014), as increase in the prevalence of resistance to INSTI could occur with the growing use of these drugs. Therefore, updated surveillance data will allow choosing the most appropriate treatment strategy at the global and national levels.
The three main RAL resistance pathways, N155H, Q148H/R/K, and Y143C/R (Cooper et al., 2008; Anstett et al., 2017), the latter at low frequency, were found among INSTI-resistant viruses in our study. N155H and Q148X affect virus fitness, to a lesser extent in the case of N155H. Thus, under RAL treatment, N155H emerged earlier than Q148X (Hu and Kuritzkes, 2010). Such difference in fitness could explain the high frequency of N155H found in our study, where INSTI-resistant viruses were mainly exposed to RAL. Fitness loss can be compensated by secondary mutations, which appear sequentially, inducing higher levels of resistance to RAL (Hu and Kuritzkes, 2010; Anstett et al., 2017). This can explain the frequent finding of Q148H/R + G140S/A, Q148R + E138K, and N155H + T97A combinations in our study. Moreover, viruses with Q148H + G140S, which was the most frequent combination in our cohort, are fitter than viruses with single mutations or double mutants with N155H, and therefore can be preferably selected along time under RAL pressure (Hu and Kuritzkes, 2010). Later, along the evolution of drug resistance, other mutations can emerge, such as T97A, which we have found frequently with the Q148H + G140S combination.
The most common INSTI resistance mutations in persons with virological failure under DTG-containing regimens were R263K, G118R, N155H, and Q148H/R (Eron et al., 2013; Castagna et al., 2018; Rhee et al., 2019), with R263K and G118R being predominant in INSTI-naïve patients (Cahn et al., 2013, 2022; Underwood et al., 2022; Vavro et al., 2022). However, G118R was not found in our cohort, probably due to the preferential codon usage in this position, GGC/GGT, whose change to arginine has a high substitution cost compared to the rare GGA/GGG codons. This bias in codon usage is found in almost all HIV-1 subtypes (Brenner et al., 2016; Theys et al., 2019) and represents an additional genetic barrier to the development of DTG resistance. Indeed, the presence of these rare triplets has been postulated to be a requirement for the development of G118R mutation (Brenner et al., 2016).
The frequency of INSTI resistance in subtype B and non-subtype B infections was similar, although the latter group showed lower resistance levels to all INSTIs. This is mainly due to the different INSTI mutation patterns observed, with the Q148H + G140S combination, frequent in subtype B, being absent among non-subtype B infections, similarly to other observational studies (Brenner et al., 2011; Doyle et al., 2015; Fourati et al., 2015; Hauser et al., 2018; Raffaelli et al., 2018; Modica et al., 2019; Sánchez et al., 2020; Scutari et al., 2020; Tzou et al., 2020). This bias is related to the different codon usage in G140, which has a higher substitution cost for acquiring the G140S resistance mutation in non-subtype B strains (Theys et al., 2019). In addition, the G163K/R polymorphic mutations were associated to subtype F and BF recombinant viruses, and D232N in combination with N155H was associated to subtype B, similarly to the findings in other studies (Sánchez et al., 2020; Tzou et al., 2020).
DTG has a high genetic barrier (Kobayashi et al., 2011; Quashie et al., 2012; Rhee et al., 2019), which seems to be higher for non-subtype B strains, according to the absence in them of the Q148H + G140S resistance pathway found in this and previous studies. (Brenner et al., 2011; Doyle et al., 2015; Fourati et al., 2015; Modica et al., 2019; Sánchez et al., 2020; Scutari et al., 2020; Tzou et al., 2020). Non-subtype B strains are predominant in most low- and middle-income countries (Hemelaar et al., 2019), which is an additional advantage for the use of DTG in these countries, where genotyping testing capacity for identification of DRMs that can jeopardize DTG-based therapies is frequently less available.
DTG remains a therapeutic option in most infections with INSTI resistance in our cohort, among which 59% are still susceptible to DTG. In fact, DTG 50 mg twice daily, instead of the usual once daily regimen, with an optimized background therapy has been shown to be efficient in the VIKING and PRESTIGIO trials, in which patients with failing regimens and different INSTI resistance levels were enrolled (Eron et al., 2013; Castagna et al., 2018), although patients with Q148X mutations showed a reduced response to the therapy (Castagna et al., 2014).
Virological failure in INSTI-naïve patients who are prescribed DTG-containing regimens is rare and has been associated with low adherence, DTG monotherapy, previous presence of resistance mutations in Pr-RT, and the interaction of INSTI with other drugs, which reduce the effective DTG plasma concentration (Cahn et al., 2013; Cevik et al., 2020; Cahn et al., 2022; Revollo et al., 2022; Underwood et al., 2022; Vavro et al., 2022). These patients were also rare in our cohort, where only 5 individuals exposed to DTG as the only INSTI developed mutations associated to intermediate- or low-level resistance to DTG. The emergence of such mutations could be related to low therapeutic adherence and the presence of previous drug resistance mutations in Pr-RT. Indeed, the presence of M184V and K103N could have contributed to the development of DTG resistance in two of them.
As a limitation, our cohort includes a greater representation of patients with INSTI resistance compared to the HIV-1-infected population, due to a higher representation of patients with therapeutic failure in the samples received in our laboratory for ARV resistance testing. However, this bias allows us to have a higher number of samples for analysis of INSTI resistance mutations and their associated features.
INSTI resistance levels determined in our study were based on the Stanford HIVdb algorithm, which uses phenotypic data to assign a value to each mutation to predict the resistance level of the virus. However, since there are fewer phenotypic studies with non-subtype B strains than with subtype B viruses, the contributions of some mutations to the resistance level of non-subtype B strains may not be well characterized and, therefore, the level of resistance could be underestimated. Additional phenotypic analyses with non-subtype B strains are needed to improve the resistance level predictions with the available algorithms.
In conclusion, in our study, INSTI resistance was uncommon and was associated with long-term infections, additional resistance mutations to other ARV classes, and probably with low therapeutic adherence. The few cases of INSTI resistance mutations in regimens containing DTG and the low level of transmitted resistance to DTG in our cohort supports the WHO recommendation for the use of DTG in first- and second-line treatments. However, the expected increase in the use of DTG in the next years and its use in settings with high prevalence of NRTI and NNRTI resistance, such as low- and middle-income countries, encourage the implementation of surveillance systems to detect the potential emergence and spread of DTG-resistant strains.
Members of the Spanish Group for the Study of Antirretroviral Drug Resistance
Aragón: Hospital Universitario Miguel Servet: Ana Mª Martínez-Sapiña, Piedad Arazo; Hospital Clínico Universitario Lozano Blesa: Sonia Algarate; Hospital de Barbastro: Marta Lalana Garcés. Basque Country: Hospital Universitario Basurto: Josefa Muñoz, Mª Carmen Nieto, Sofía Ibarra, Estibaliz Ugalde; Hospital Universitario de Cruces: Luis Elorduy, Elena Berciartua, Josune Goikoetxea, Laura Guío, Mª José Blanco; Hospital de Galdakao: Mª José López de Goicoechea, José Mayo; Hospital Universitario Donostia: Carlos Gustavo Cilla, José Antonio Iribarren, Mª Yolanda Salicio, Maitane Aranzamendi, Maialen Ibarguren. Hospital Universitario de Álava: Juan Carlos Gainzarain, Zuriñe Ortiz de Zárate, Miguel Ángel Morán, Ester Sáez de Arana, José Joaquín Portu, Carmen Gómez-González. Cantabria: Hospital Universitario Marqués de Valdecilla: M. Eliecer Cano. Castilla y León: Hospital El Bierzo: Sonia Belén Paredes. Hospital Clínico Universitario de Valladolid: Carmen Hinojosa, Begoña Monteagudo; Hospital Universitario Río Hortega: Belén Lorenzo, Jessica Abadía; Hospital Virgen de la Concha: Teresa Martín-Domínguez, Rosa Martínez-González. Castilla-La Mancha: Hospital Virgen de la Salud: César Gómez-Hernando, José Largo-Pau; Hospital Universitario de Guadalajara: Alejandro González-Praetorius. Hospital Virgen de la Luz: Enrique Prada, Paloma Geijo; Hospital Santa Bárbara: Ana Cosmen; Comunidad Valenciana: Hospital Universitari Sant Joan d’Alacant: Fernando Buñuel, Ana Infante. Extremadura: Hospital de Mérida: Julián Sánchez. Complejo Hospitalario Universitario de Cáceres: Guadalupe Rodríguez. Galicia: Complejo Hospitalario Universitario de Ferrol: Ana Mariño, Patricia Ordóñez, Hortensia Álvarez, Nieves Valcarce, Sabela Sánchez; Complejo Hospitalario Universitario de A Coruña: Ángeles Cañizares, Mª Ángeles Castro, Luz Moldes Suárez. Hospital Universitario Lucus Augusti: Ramón Rabuñal, María José García País, Mª José Gude González, Pilar Alonso, Eva María Romay, Antonio Moreno; Complejo Hospitalario Universitario de Ourense: Juan García Costa, Ricardo Fernández-Rodríguez, Raúl Rodríguez-Pérez, Luis Canoura, María Dolores Díaz-López, Mª Genoveva Naval-Calviño; Complejo Hospitalario Universitario de Vigo: Celia Miralles, Antonio Ocampo, Sonia Pérez-Castro, Jorge Julio Cabrera; Sandra Cortizo Vidal, Guillermo Pousada, Luis Morano; Complejo Hospitalario de Pontevedra: Julio Díz-Arén, Matilde Trigo, Mª Ángeles Pallarés. La Rioja: Hospital San Pedro: José Ramón Blanco, Miriam Blasco. Madrid: Hospital de Fuenlabrada: Julio García Díez, Laura María Molina Esteban, Santiago Prieto-Menchero, Mª Isabel García-Arata; Hospital Clínico San Carlos: Esther Culebras, Iciar Rodríguez-Avial; Hospital Universitario Fundación Jiménez Díaz: Raquel Téllez, Miguel Górgolas, Ángel Luis Castaño, Olalla Calabia, Alfonso Cabello. Hospital Universitario de la Princesa: Laura Cardeñoso. Hospital Universitario Príncipe de Asturias: Juan Cuadros. Fundación Hospital de Alcorcón: Mª José Goyanes, Carolina Campelo. Murcia: Hospital J.M. Morales Meseguer: Pedro Antequera. Navarra: Hospital Universitario de Navarra: Carmen Martín, Carmen Ezpeleta-Baquedano, Aitziber Aguinaga, Irati Arregui García, Ana Miqueleiz, Mª Gracia Ruiz de Alda. Hospital Reina Sofía de Tuleda: Leticia Raquel Armendáriz López, Marta Adelantado Lacasa.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by Bioethics and Animal Well-being Committee of Instituto de Salud Carlos III. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
HG, ED, and MT conceived the study and supervised the experimental work. ED and HG performed drug resistance analyses and data curation. HG performed statistical analyses and wrote the manuscript draft, with contributions to the text by MT and ED. SB and MM-L performed experimental work. The members of the Spanish Group for the Study of Antiretroviral Drug Resistance recruited patients and obtained epidemiological and clinical data. All authors read and approved the text.
Funding
This work was funded through Acción Estratégica en Salud Intramural (AESI), Instituto de Salud Carlos III, projects “Estudios sobre vigilancia epidemiológica molecular del VIH-1 en España,” PI16CIII/00033 and “Epidemiología molecular del VIH-1 en España y su utilidad para investigaciones biológicas y en vacunas”, PI19CIII/0042; and scientific agreements with Consellería de Sanidade, Government of Galicia (MVI 1004/16) and Osakidetza-Servicio Vasco de Salud, Government of Basque Country (MVI 1001/16).
Acknowledgments
We would like to thank José Antonio Taboada, from Consellería de Sanidade, Xunta de Galicia, and Daniel Zulaika, from Osakidetza-Servicio Vasco de Salud, for their support of this study, and the personnel at the Genomic Unit, Instituto de Salud Carlos III, for technical assistance in sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1051096/full#supplementary-material
Footnotes
References
Acosta, R. K., D'Antoni, M. L., Mulato, A., Yant, S. R., Cihlar, T., and White, K. L. (2022). Forgiveness of INSTI-containing regimens at drug concentrations simulating variable adherence in vitro. Antimicrob. Agents Chemother. 66:e0203821. doi: 10.1128/aac.02038-21
Acosta, R. K., Willkom, M., Martin, R., Chang, S., Wei, X., Garner, W., et al. (2019). Resistance analysis of bictegravir-emtricitabine-tenofovir alafenamide in HIV-1 treatment-naive patients through 48 weeks. Antimicrob. Agents Chemother. 63:e02533-18. doi: 10.1128/AAC.02533-18
Alvarez, M., Casas, P., de Salazar, A., Chueca, N., Guerrero-Beltran, C., Rodríguez, C., et al. (2019). Surveillance of transmitted drug resistance to integrase inhibitors in Spain: implications for clinical practice. J. Antimicrob. Chemother. 74, 1693–1700. doi: 10.1093/jac/dkz067
Anstett, K., Brenner, B., Mesplede, T., and Wainberg, M. A. (2017). HIV drug resistance against strand transfer integrase inhibitors. Retrovirology 14:36. doi: 10.1186/s12977-017-0360-7
Brenner, B. G., Lowe, M., Moisi, D., Hardy, I., Gagnon, S., Charest, H., et al. (2011). Subtype diversity associated with the development of HIV-1 resistance to integrase inhibitors. J. Med. Virol. 83, 751–759. doi: 10.1002/jmv.22047
Brenner, B. G., Thomas, R., Blanco, J. L., Ibanescu, R. I., Oliveira, M., Mesplède, T., et al. (2016). Development of a G118R mutation in HIV-1 integrase following a switch to dolutegravir monotherapy leading to cross-resistance to integrase inhibitors. J. Antimicrob. Chemother. 71, 1948–1953. doi: 10.1093/jac/dkw071
Cahn, P., Pozniak, A. L., Mingrone, H., Shuldyakov, A., Brites, C., Andrade-Villanueva, J. F., et al. (2013). Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 382, 700–708. doi: 10.1016/S0140-6736(13)61221-0
Cahn, P., Sierra, M. J., Arribas, J. R., Antinori, A., Ortiz, R., Clarke, A. E., et al. (2022). Three-year durable efficacy of dolutegravir plus lamivudine in antiretroviral therapy - naive adults with HIV-1 infection. AIDS 36, 39–48. doi: 10.1097/QAD.0000000000003070
Casadellá, M., Santos, J. R., Noguera-Julian, M., Micán-Rivera, R., Domingo, P., Antela, A., et al. (2020). Primary resistance to integrase strand transfer inhibitors in Spain using ultrasensitive HIV-1 genotyping. J. Antimicrob. Chemother. 75, 3517–3524. doi: 10.1093/jac/dkaa349
Castagna, A., Ferrara, M., Galli, L., Comi, L., Sterrantino, G., Cenderello, G., et al. (2018). Long-term efficacy of dolutegravir in treatment-experienced subjects failing therapy with HIV-1 integrase strand inhibitor-resistant virus. J. Antimicrob. Chemother. 73, 177–182. doi: 10.1093/jac/dkx371
Castagna, A., Maggiolo, F., Penco, G., Wright, D., Mills, A., Grossberg, R., et al. (2014). Dolutegravir in antiretroviral-experienced patients with raltegravir- and/or elvitegravir-resistant HIV-1: 24-week results of the phase III VIKING-3 study. J. Infect. Dis. 210, 354–362. doi: 10.1093/infdis/jiu051
Cevik, M., Orkin, C., and Sax, P. E. (2020). Emergent resistance to dolutegravir among INSTI-Naïve patients on first-line or second-line antiretroviral therapy: a review of published cases. Open Forum Infect. Dis. 7:ofaa202. doi: 10.1093/ofid/ofaa202
Cooper, D. A., Steigbigel, R. T., Gatell, J. M., Rockstroh, J. K., Katlama, C., Yeni, P., et al. (2008). Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359, 355–365. doi: 10.1056/NEJMoa0708978
DeJesus, E., Berger, D., Markowitz, M., Cohen, C., Hawkins, T., Ruane, P., et al. (2006). Antiviral activity, pharmacokinetics, and dose response of the HIV-1 integrase inhibitor GS-9137 (JTK-303) in treatment-naive and treatment-experienced patients. J. Acquir. Immune Defic. Syndr. 43, 1–5. doi: 10.1097/01.qai.0000233308.82860.2f
Doyle, T., Dunn, D. T., Ceccherini-Silberstein, F., De Mendoza, C., Garcia, F., Smit, E., et al. (2015). Integrase inhibitor (INI) genotypic resistance in treatment-naive and raltegravir-experienced patients infected with diverse HIV-1 clades. J. Antimicrob. Chemother. 70, 3080–3086. doi: 10.1093/jac/dkv243
Eron, J. J., Clotet, B., Durant, J., Katlama, C., Kumar, P., Lazzarin, A., et al. (2013). Safety and efficacy of dolutegravir in treatment-experienced subjects with raltegravir-resistant HIV type 1 infection: 24-week results of the VIKING study. J. Infect. Dis. 207, 740–748. doi: 10.1093/infdis/jis750
Fourati, S., Charpentier, C., Amiel, C., Morand-Joubert, L., Reigadas, S., Trabaud, M. A., et al. (2015). Cross-resistance to elvitegravir and dolutegravir in 502 patients failing on raltegravir: a French national study of raltegravir-experienced HIV-1-infected patients. J. Antimicrob. Chemother. 70, 1507–1512. doi: 10.1093/jac/dku535
Gallant, J., Lazzarin, A., Mills, A., Orkin, C., Podzamczer, D., Tebas, P., et al. (2017). Bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir, abacavir, and lamivudine for initial treatment of HIV-1 infection (GS-US-380-1489): a double-blind, multicentre, phase 3, randomised controlled non-inferiority trial. Lancet 390, 2063–2072. doi: 10.1016/S0140-6736(17)32299-7
Gil, H., Delgado, E., Benito, S., Georgalis, L., Montero, V., Sánchez, M., et al. (2022). Transmission clusters, predominantly associated with men who have sex with men, play a main role in the propagation of HIV-1 in northern Spain (2013–2018). Front. Microbiol. 13:782609. doi: 10.3389/fmicb.2022.782609
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41, 95–98.
Hauser, A., Hofmann, A., Meixenberger, K., Altmann, B., Hanke, K., Bremer, V., et al. (2018). Increasing proportions of HIV-1 non-B subtypes and of NNRTI resistance between 2013 and 2016 in Germany: results from the national molecular surveillance of new HIV-diagnoses. PLoS One 13:e0206234. doi: 10.1371/journal.pone.0206234
Hemelaar, J., Elangovan, R., Yun, J., Dickson-Tetteh, L., Fleminger, I., Kirtley, S., et al. (2019). Global and regional molecular epidemiology of HIV-1, 1990-2015: a systematic review, global survey, and trend analysis. Lancet Infect. Dis. 19, 143–155. doi: 10.1016/S1473-3099(18)30647-9
Hu, Z., and Kuritzkes, D. R. (2010). Effect of raltegravir resistance mutations in HIV-1 integrase on viral fitness. J. Acquir. Immune Defic. Syndr. 55, 148–155. doi: 10.1097/QAI.0b013e3181e9a87a
Inzaule, S. C., Hamers, R. L., Noguera-Julian, M., Casadellá, M., Parera, M., Rinke de Wit, T. F., et al. (2018). Primary resistance to integrase strand transfer inhibitors in patients infected with diverse HIV-1 subtypes in sub-Saharan Africa. J. Antimicrob. Chemother. 73, 1167–1172. doi: 10.1093/jac/dky005
Kobayashi, M., Yoshinaga, T., Seki, T., Wakasa-Morimoto, C., Brown, K. W., Ferris, R., et al. (2011). In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob. Agents Chemother. 55, 813–821. doi: 10.1128/AAC.01209-10
Lennox, J. L., DeJesus, E., Lazzarin, A., Pollard, R. B., Madruga, J. V., Berger, D. S., et al. (2009). Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet 374, 796–806. doi: 10.1016/S0140-6736(09)60918-1
Lert, F., and Kazatchkine, M. D. (2007). Antiretroviral HIV treatment and care for injecting drug users: an evidence-based overview. Int. J. Drug Policy 18, 255–261. doi: 10.1016/j.drugpo.2007.05.002
Liu, L., Dai, L., Yao, J., Pan, P., Li, L., Liu, Z., et al. (2019). Lack of HIV-1 integrase inhibitor resistance among 392 antiretroviral-naïve individuals in a tertiary care hospital in Beijing, China. AIDS 33, 1945–1947. doi: 10.1097/QAD.0000000000002282
Lole, K. S., Bollinger, R. C., Paranjape, R. S., Gadkari, D., Kulkarni, S. S., Novak, N. G., et al. (1999). Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73, 152–160. doi: 10.1128/JVI.73.1.152-160.1999
Mbisa, J. L., Ledesma, J., Kirwan, P., Bibby, D. F., Manso, C., Skingsley, A., et al. (2020). Surveillance of HIV-1 transmitted integrase strand transfer inhibitor resistance in the UK. J. Antimicrob. Chemother. 75, 3311–3318. doi: 10.1093/jac/dkaa309
Modica, S., Rossetti, B., Lombardi, F., Lagi, F., Maffeo, M., D'Autilia, R., et al. (2019). Prevalence and determinants of resistance mutations in HIV-1-infected patients exposed to integrase inhibitors in a large Italian cohort. HIV Med. 20, 137–146. doi: 10.1111/hiv.12692
Nachega, J. B., Marconi, V. C., van Zyl, G. U., Gardner, E. M., Preiser, W., Hong, S. Y., et al. (2011). HIV treatment adherence, drug resistance, virologic failure: evolving concepts. Infect. Disord. Drug Targets 11, 167–174. doi: 10.2174/187152611795589663
Naeger, L. K., Harrington, P., Komatsu, T., and Deming, D. (2016). Effect of dolutegravir functional monotherapy on HIV-1 virological response in integrase strand transfer inhibitor resistant patients. Antivir. Ther. 21, 481–488. doi: 10.3851/IMP3033
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Quashie, P. K., Mesplède, T., Han, Y. S., Oliveira, M., Singhroy, D. N., Fujiwara, T., et al. (2012). Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. J. Virol. 86, 2696–2705. doi: 10.1128/JVI.06591-11
Raffaelli, C. S., Rossetti, B., Paglicci, L., Colafigli, M., Punzi, G., Borghi, V., et al. (2018). Impact of transmitted HIV-1 drug resistance on the efficacy of first-line antiretroviral therapy with two nucleos(t)ide reverse transcriptase inhibitors plus an integrase inhibitor or a protease inhibitor. J. Antimicrob. Chemother. 73, 2480–2484. doi: 10.1093/jac/dky211
Revollo, B., Viñuela, L., de la Mora, L., García, F., Noguera-Julián, M., Parera, M., et al. (2022). Integrase resistance emergence with dolutegravir/lamivudine with prior HIV-1 suppression. J. Antimicrob. Chemother. 77, 1738–1740. doi: 10.1093/jac/dkac082
Rhee, S. Y., Gonzales, M. J., Kantor, R., Betts, B. J., Ravela, J., and Shafer, R. W. (2003). Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31, 298–303. doi: 10.1093/nar/gkg100
Rhee, S. Y., Grant, P. M., Tzou, P. L., Barrow, G., Harrigan, P. R., Ioannidis, J. P. A., et al. (2019). A systematic review of the genetic mechanisms of dolutegravir resistance. J. Antimicrob. Chemother. 74, 3135–3149. doi: 10.1093/jac/dkz256
Sánchez, D., Arazi-Caillaud, S., Zapiola, I., Fernández-Giuliano, S., Bologna, R., Mangano, A., et al. (2020). Impact of genotypic diversity on selection of subtype-specific drug resistance profiles during raltegravir-based therapy in individuals infected with B and BF recombinant HIV-1 strains. J. Antimicrob. Chemother. 75, 1567–1574. doi: 10.1093/jac/dkaa042
Sax, P. E., Pozniak, A., Montes, M. L., Koenig, E., DeJesus, E., Stellbrink, H. J., et al. (2017). Coformulated bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir with emtricitabine and tenofovir alafenamide, for initial treatment of HIV-1 infection (GS-US-380-1490): a randomised, double-blind, multicentre, phase 3, non-inferiority trial. Lancet 390, 2073–2082. doi: 10.1016/S0140-6736(17)32340-1
Scutari, R., Alteri, C., Vicenti, I., Di, C. D., Zuccaro, V., Incardona, F., et al. (2020). Evaluation of HIV-1 integrase resistance emergence and evolution in patients treated with integrase inhibitors. J. Glob. Antimicrob. Resist. 20, 163–169. doi: 10.1016/j.jgar.2019.07.015
Semengue, E. N. J., Armenia, D., Inzaule, S., Santoro, M. M., Dambaya, B., Takou, D., et al. (2021). Baseline integrase drug resistance mutations and conserved regions across HIV-1 clades in Cameroon: implications for transition to dolutegravir in resource-limited settings. J. Antimicrob. Chemother. 76, 1277–1285. doi: 10.1093/jac/dkab004
Shafer, R. W. (2006). Rationale and uses of a public HIV drug-resistance database. J. Infect. Dis. 194, S51–S58. doi: 10.1086/505356
Steigbigel, R. T., Cooper, D. A., Kumar, P. N., Eron, J. E., Schechter, M., Markowitz, M., et al. (2008). Raltegravir with optimized background therapy for resistant HIV-1 infection. N. Engl. J. Med. 359, 339–354. doi: 10.1056/NEJMoa0708975
Theys, K., Libin, P. J. K., Van, L. K., and Abecasis, A. B. (2019). An evolutionary model-based approach to quantify the genetic barrier to drug resistance in fast-evolving viruses and its application to HIV-1 subtypes and Integrase inhibitors. Antimicrob. Agents Chemother. 63:e00539-19. doi: 10.1128/AAC.00539-19
Tzou, P. L., Rhee, S. Y., Descamps, D., Clutter, D. S., Hare, B., Mor, O., et al. (2020). Integrase strand transfer inhibitor (INSTI)-resistance mutations for the surveillance of transmitted HIV-1 drug resistance. J. Antimicrob. Chemother. 75, 170–182. doi: 10.1093/jac/dkz417
Underwood, M., Horton, J., Nangle, K., Hopking, J., Smith, K., Aboud, M., et al. (2022). Integrase inhibitor resistance mechanisms and structural characteristics in antiretroviral therapy-experienced, Integrase inhibitor-naive adults with HIV-1 infection treated with Dolutegravir plus two nucleoside reverse transcriptase inhibitors in the DAWNING study. Antimicrob. Agents Chemother. 66:e0164321. doi: 10.1128/AAC.01643-21
Vavro, C., Ruel, T., Wiznia, A., Montañez, N., Nangle, K., Horton, J., et al. (2022). Emergence of resistance in HIV-1 Integrase with Dolutegravir treatment in a pediatric population from the IMPAACT P1093 study. Antimicrob. Agents Chemother. 66:e0164521. doi: 10.1128/AAC.01645-21
Vervoort, S. C., Borleffs, J. C., Hoepelman, A. I., and Grypdonck, M. H. (2007). Adherence in antiretroviral therapy: a review of qualitative studies. AIDS 21, 271–281. doi: 10.1097/QAD.0b013e328011cb20
von Wyl, V., Klimkait, T., Yerly, S., Nicca, D., Furrer, H., Cavassini, M., et al. (2013). Adherence as a predictor of the development of class-specific resistance mutations: the Swiss HIV cohort study. PLoS One 8:e77691. doi: 10.1371/journal.pone.0077691
Walmsley, S., Baumgarten, A., Berenguer, J., Felizarta, F., Florence, E., Khuong-Josses, M. A., et al. (2015). Brief report: dolutegravir plus Abacavir/lamivudine for the treatment of HIV-1 infection in antiretroviral therapy-naive patients: week 96 and week 144 results from the SINGLE randomized clinical trial. J. Acquir. Immune Defic. Syndr. 70, 515–519. doi: 10.1097/QAI.0000000000000790
WHO (2014). Surveillance of HIV drug resistance in adults initiating antoretroviral therapy. Available at: https://www.who.int/teams/global-hiv-hepatitis-and-stis-programmes/hiv/treatment/hiv-drug-resistance/hiv-drug-resistance-surveillance/pretreatment-hiv-drug-resistance-adult (Accessed September 16, 2014).
WHO (2018). Updated recommnedations on first-line and second-line antirretroviral regimens and post-exposure prophylaxis and recommendations on early infant diagnosis of HIV. Available at: https://www.who.int/publications/i/item/WHO-CDS-HIV-18.51 (Accessed December 21, 2018).
WHO (2021). HIV drug resistance report 2021. Available at: https://www.who.int/publications/i/item/9789240038608 (Accessed November 22, 2021).
Keywords: HIV-1, Spain, integrase strand transfer inhibitors, resistance mutation, antirretroviral resistance
Citation: Gil H, Delgado E, Benito S, Moreno-Lorenzo M, Thomson MM and the Spanish Group for the Study of Antiretroviral Drug Resistance (2022) Factors associated with HIV-1 resistance to integrase strand transfer inhibitors in Spain: Implications for dolutegravir-containing regimens. Front. Microbiol. 13:1051096. doi: 10.3389/fmicb.2022.1051096
Edited by:
Bin Su, Capital Medical University, ChinaReviewed by:
Peter Kojo Quashie, University of Ghana, GhanaSunee Sirivichayakul, Chulalongkorn University, Thailand
Copyright © 2022 Gil, Delgado, Benito, Moreno-Lorenzo, Thomson and the Spanish Group for the Study of Antiretroviral Drug Resistance. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Horacio Gil, hgil@isciii.es; Michael M. Thomson, mthomson@isciii.es
†These authors have contributed equally to this work and share first authorship
 Horacio Gil
Horacio Gil Elena Delgado
Elena Delgado Sonia Benito
Sonia Benito María Moreno-Lorenzo
María Moreno-Lorenzo Michael M. Thomson
Michael M. Thomson the Spanish Group for the Study of Antiretroviral Drug Resistance
the Spanish Group for the Study of Antiretroviral Drug Resistance