- 1Christian Medical College & Hospital, Vellore, India
- 2Jai Prakash Narayan Apex Trauma Center, All India Institute of Medical Sciences, New Delhi, India
- 3Post Graduate Institute of Medical Education and Research (PGIMER), Chandigarh, India
- 4Microbiology Department, Molecular Genetics and Immunology, University of Kansas, Lawrence, KS, United States
- 5Indian Council of Medical Research (ICMR), New Delhi, National Capital Territory of Delhi, New Delhi, India
With the excessive genome plasticity, Acinetobacter baumannii can acquire and disseminate antimicrobial resistance (AMR) genes often associated with mobile genetic elements (MGEs). Analyzing the genetic environment of resistance genes often provides valuable information on the origin, emergence, evolution, and spread of resistance. Thus, we characterized the genomic features of some clinical isolates of carbapenem-resistant A. baumannii (CRAb) to understand the role of diverse MGEs and their genetic context responsible for disseminating carbapenem resistance genes. For this, 17 clinical isolates of A. baumannii obtained from multiple hospitals in India between 2018 and 2019 were analyzed. AMR determinants, the genetic context of resistance genes, and molecular epidemiology were studied using whole-genome sequencing. This study observed an increased prevalence of blaOXA–23 followed by dual carbapenemases, blaOXA–23, and blaNDM. This study identified three novel Oxford MLST sequence types. The majority of the isolates belonged to the dominant clone, IC2, followed by less prevalent clones such as IC7 and IC8. This study identified variations of AbaR4 and AbGRI belonging to the IC2 lineage. To the best of our knowledge, this is the first study that provides comprehensive profiling of resistance islands, their related MGEs, acquired AMR genes, and the distribution of clonal lineages of CRAb from India.
Introduction
Acinetobacter baumannii is a member of the ESKAPE group of pathogens and is considered to be one of the major global causes of hospital-acquired infections (HAIs) (Lee et al., 2017). A. baumannii is responsible for causing a wide range of infections, with pneumonia being the most commonly observed infection among critically ill patients (Dexter et al., 2015). This pathogen has a propensity to rapidly acquire antibiotic resistance genes and to develop resistance to multiple classes of antimicrobials (Lee et al., 2017). Carbapenems are one of the most commonly used antibiotics for the treatment of Acinetobacter infections. Carbapenem resistance in A. baumannii ranges between 70 and 85% in the Asia-Pacific region (O’Donnell et al., 2021). A study from SENTRY surveillance reported carbapenem resistance rates ranging from 55 to 90% in India (Gales et al., 2019). Both the Center for Disease Control (CDC) and the World Health Organization (WHO) categorized carbapenem-resistant A. baumannii (CRAb) under “Urgent Threat” and as Priority 1: Critical pathogen, respectively. Recently, the WHO Country Office for India developed the Indian Priority Pathogen List (IPPL) and categorized carbapenem-resistant, colistin-resistant A. baumannii under “Critical Priority.”1
With excessive genome plasticity, A. baumannii can acquire and disseminate antimicrobial resistance (AMR) genes that are often associated with various mobile genetic elements (MGEs) (Roca et al., 2012). Carbapenem resistance in A. baumannii is mainly due to genes encoding class D oxacillinases, blaOXA–23–like, blaOXA–51–like, and blaOXA–58–like (Poirel and Nordmann, 2006). The blaOXA–23 gene is the most predominant and is carried on many MGEs, including transposons, plasmids, and resistance islands (RIs) (Pagano et al., 2016). The association of insertion sequence (IS) elements with blaOXA–51-like, blaOXA–23–like, blaNDM–like, and blaOXA–58–like genes was reported earlier (Poirel et al., 2005, 2012; Turton et al., 2006a). Typically, blaOXA–23 is associated with transposons such as Tn2006, Tn2008, and Tn2009, while blaNDM–1 was mobilized by the Tn125-like composite transposon (Pagano et al., 2016). Recent studies have also indicated the role of conjugative plasmids as vehicles for disseminating resistance determinants such as blaOXA–23 in A. baumannii (Salto et al., 2018). Additionally and most importantly, the emergence of A. baumannii RIs carrying clusters of horizontally transferred genes is considered a significant contributor to the multidrug-resistant (MDR) phenotype of A. baumannii (Cameranesi et al., 2020). RIs in A. baumannii are made of transposons and are known to carry genes that confer resistance to multiple antibiotics and heavy metals (Hamidian and Hall, 2018). The AbaR3-type elements are confined to the International Clone 1 (IC1) and represented by ST1, ST19, ST20, and ST81. AbaR3 comprises a Tn6019 backbone and is consistently linked with Tn6018 or its components with multiple antimicrobial resistance regions (MARRs) (Hamidian and Hall, 2011). Similarly, studies have shown that Tn6022 can acquire blaOXA–23 transposon Tn2006 and form AbaR4 islands (Hamidian and Hall, 2017). Table 1 outlines the genomic and epidemiological features of different clones of CRAb.
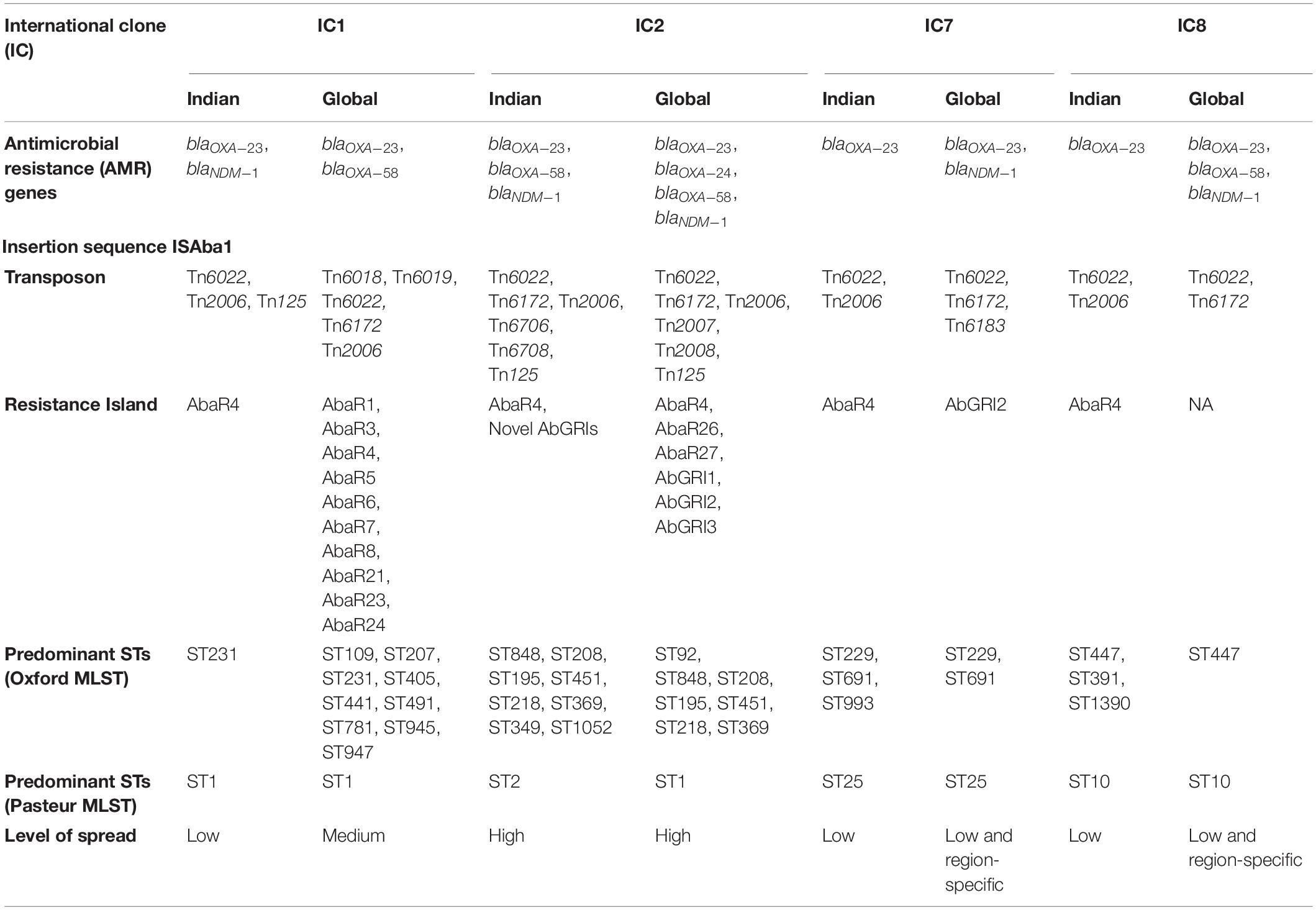
Table 1. Genomic and epidemiological features of different clones of carbapenem-resistant Acinetobacter baumannii–Indian vs. global scenario.
Although the endemic burden of CRAb is a significant public health problem within Indian hospitals, the lack of genomic information makes it difficult to track its persistence (Mancilla-Rojano et al., 2019). Studying the genetic environment of resistance genes often provides valuable information on the origin, emergence, evolution, and spread of resistance in bacterial populations (Hamidian and Nigro, 2019).
We aimed to characterize the prevalent genomic features of clinical isolates of CRAb in India. We also compared the structural configuration of RIs with the complete genetic information and observed structural variations within the genetic environment of resistance genes. We found that the backbone of MGEs and their associated AMR genes among this study isolates were similar to that of the global context.
Materials and Methods
Bacterial Isolates
A total of 17 clinical isolates of A. baumannii collected as a part of a surveillance study were used. Of the 17 isolates included in this study, 13 were from Christian Medical College (CMC), Vellore, three from All India Institute of Medical Sciences (AIIMS) Trauma Center, New Delhi, and one from Post Graduate Institute of Medical Education & Research (PGIMER), Chandigarh (Supplementary Figure 1). Among the isolates, ten isolates were from blood (B; n = 10), six from endotracheal aspirate (ETA; n = 6), and one from pus (P; n = 1). Phenotypic characterization of all the isolates as A. baumannii calcoaceticus (Acb) complex was determined using standard biochemical tests. Confirmation of the Acb complex at the species level was performed by Vitek-MS (Database v2.0, bioMerieux, France) as described earlier and by identifying chromosomally encoded blaOXA–51–like gene by PCR (Turton et al., 2006b).
Antimicrobial Susceptibility Testing
All the isolates were characterized for susceptibility to ceftazidime (30 μg), cefepime (30 μg), piperacillin-tazobactam (100/10 μg), cefoperazone-sulbactam (75/30 μg), imipenem (10 μg), meropenem (10 μg), levofloxacin (5 μg), amikacin (30 μg), netilmicin (30 μg), tobramycin (10 μg), aztreonam (30 μg), tetracycline (30 μg), minocycline (30 μg), and tigecycline (15 μg) using the Kirby Bauer’s disk diffusion (DD) method.
For colistin, broth microdilution (BMD) was performed. Isolates identified as carbapenem-resistant by DD were further subjected to BMD to determine the minimum inhibitory concentration (MIC) for imipenem and meropenem. The susceptibility was interpreted as per the criteria defined by CLSI guidelines (Weinstein et al., 2018, 2019). Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853) were included in every batch for quality control (QC). For colistin susceptibility testing, in addition to QC strains, an mcr-1 positive E. coli isolate and two Klebsiella pneumoniae strains (BA38416 and BA25425) were also included for QC.
Whole-Genome Sequencing and Assembly
The isolates were recovered overnight on blood agar, and genomic DNA was extracted from pure cultures using a QIAamp DNA mini kit (Qiagen, Germany) following the manufacturers’ instructions. DNA was quantified by NanoDrop spectrophotometry (Thermo Fisher Scientific, United States) and Qubit 3.0 fluorometry (Life Technologies, United States) and stored at −20°C until further characterization.
Short read sequencing of the 17 isolates was carried out using the Ion Torrent PGM™ platform using 400-bp chemistry (Life Technologies, United States) or 150-bp paired-end sequencing using HiSeq 2500 platform (Illumina, United States). The PHRED quality score was checked on the sequences, and reads with a score below 20 were discarded. Adapters were trimmed using cutadapt v. 1.8.12 and assessed with FastQC version 0.11.4.3
All genomic DNA was further subjected to Oxford Nanopore MinION sequencing (Oxford Nanopore Technologies, United Kingdom) to obtain long-read sequences. For this, the DNA library was prepared as per the manufacturer’s protocol using the SQK-LSK108 ligation sequencing kit (version R9) and the ONT EXP-NBD103 Native Barcode Expansion Kit (Oxford Nanopore Technologies, United Kingdom). The library was loaded onto the FLO-MIN106 R9 flow cell and run for 48 h using the standard MinKNOW software. The Fast5 files from MinION sequencing were subjected to base calling using Guppy.4
Complete circular genomes for the 17 isolates were obtained using the de novo hybrid assembly of Illumina and Oxford nanopore sequences as described earlier (Wick et al., 2017a). The long reads were error-corrected using the standalone Canu tool (version 1.7) and filtered using Filtlong version 0.2.05 with parameters set at min_length 1000 −90 %. The short reads generated using ion torrent were error-corrected using Ionhammer (Ershov et al., 2019) available in SPAdes, and the default FastA output was converted to FastQ with custom scripts. Additionally, genomes were assembled using the Unicycler hybrid assembly pipeline (version 0.4.6) with the default settings (Wick et al., 2017b). The obtained genome sequence was polished using high-quality Illumina/Ion torrent reads to reduce the base level errors with multiple rounds of Pilon (version 1.22) (Walker et al., 2014). The assembly quality was assessed for completeness, correctness, and contiguity using CheckM version 1.0.5 (Parks et al., 2015). The genome sequences of the chromosomes and plasmids have been deposited in GenBank under the accession numbers–AB01 (CP040080–CP040083), AB02 (CP035672–CP035675), AB03 (CP050388–CP050389), AB06 (CP040050–CP040052), AB10 (CP040053–CP040055), AB11 (CP040056–CP040057), AB13 (CP040087–CP040088), AB14 (CP040259–CP040262), AB15 (CP050385–CP050387), AB16 (CP050523–CP050525), AB18 (CP050390–CP050391), AB19 (CP050410–CP050411), AB20 (CP050412–CP050414), AB23 (CP050432–CP050435), AB26 (CP050401–CP050402), AB27 (CP050421–CP050423), and AB28 (CP050403–CP050409).
Genome Analysis
Further downstream analysis of the 17 complete genome sequences was performed using tools available at the Center for Genomic Epidemiology (CGE).6 AMR genes were identified from the genome sequences using the BLASTn-based ABRicate (version 0.8.10) program7 to query the ResFinder database.8 The Capsular Polysaccharide loci (KL) and the Outer Core Lipooligosaccharide loci (OCL) types were identified using the Kaptive database.9
The presence of ISs was identified using ISFinder.10 Using BLAST analysis, the plasmid repAci types from the complete genomes were identified and characterized. The PHASTER server was used to determine the prophages.11 The prophage regions identified were analyzed for the presence of any AMR genes using the ABRicate (version 0.8.10) program(see text footnote 7). A BLAST similarity search was performed on the individual genomes to identify the comM region that flanks AbaR type genomic islands. Based on the known AbaR sequences collected from published literature, the precise boundary of AbaRs and the respective backbone were curated manually. The sequence types were identified with MLST Finder 2.0 using Oxford MLST and Pasteur MLST schemes.12
Phylogenetic Analysis
The assembled genome sequences were mapped to the reference genome ATCC 17978 (CP012004) using the BWA MEM13 algorithm, and the variants were filtered with FreeBayes available in Snippy (Seemann, 2015). The core SNP genome alignment of all the genomes was generated with Snippy-core. The recombination regions within the core genome alignment were further filtered and removed using the Gubbins (version 2.4.1) algorithm (Croucher et al., 2015). The maximum likelihood (ML) phylogeny was constructed using FastTree version 2.1.8 (Price et al., 2010) using the GTR model with 100 bootstrap replicates. The phylogenetic tree was rooted in the reference genome and labeled using the Interactive Tree of Life software (iTOL v3) (Letunic and Bork, 2021).
Results
Varied Resistance Profile of Acinetobacter baumannii Strains With blaOXA–51 and blaOXA–23 Variants
All 17 isolates were phenotypically identified as Acb-complex and further reconfirmed as A. baumannii using Vitek-MS (Data not shown). Among the 17 isolates, AB01 was pan-susceptible (PSAB), AB23 was multidrug-resistant (MDRAB) but susceptible to carbapenem (CSAB), 12 isolates (AB10, AB11, AB13, AB14, AB15, AB16, AB18, AB19, AB20, AB26, AB27, and AB28) were CRAb, and the remaining three isolates (AB02, AB03, and AB06) were pan-drug resistant (PDRAB). Table 2 outlines the presence of resistance genes among the 17 isolates against different classes of antimicrobials. All the study isolates carried the intrinsic blaOXA–51–like and blaADC–25–like genes. Whole-genome sequencing (WGS) identified seven different variants of blaOXA–51 (blaOXA–66, blaOXA–68, blaOXA–64, blaOXA–144, blaOXA–203, blaOXA–337, and blaOXA–371), a single variant of blaOXA–23 (blaOXA–169), and a single variant of blaNDM–like gene (blaNDM–1). Among the 17 study isolates, ten carried blaOXA–23–like alone, one isolate carried blaNDM–1 alone, and four isolates co-harbored blaOXA–23–like and blaNDM–1. No acquired carbapenemase genes were identified in the remaining two isolates. More than one copy of blaOXA–23 was observed in thirteen isolates (Table 2). As expected, none of our isolates carried blaOXA–24–like or blaOXA–58–like genes. The A. baumannii isolates in this study belonged to diverse sequence types (STs) representing four International clones, ICs (IC1/CC1, IC2/CC2, IC7/CC25, and IC8/CC10), one clonal complex, i.e., CC862, and one singleton (Table 2). Although genes that confer resistance to different antimicrobials were identified across all clonal lineages, the majority of the genes that encode for aminoglycoside modifying enzymes, RMTases, macrolide resistance, sulfonamide resistance, chloramphenicol, and rifampicin resistance were confined to isolates belonging to IC2 (Rodrigues et al., 2021; Hernández-González et al., 2022).
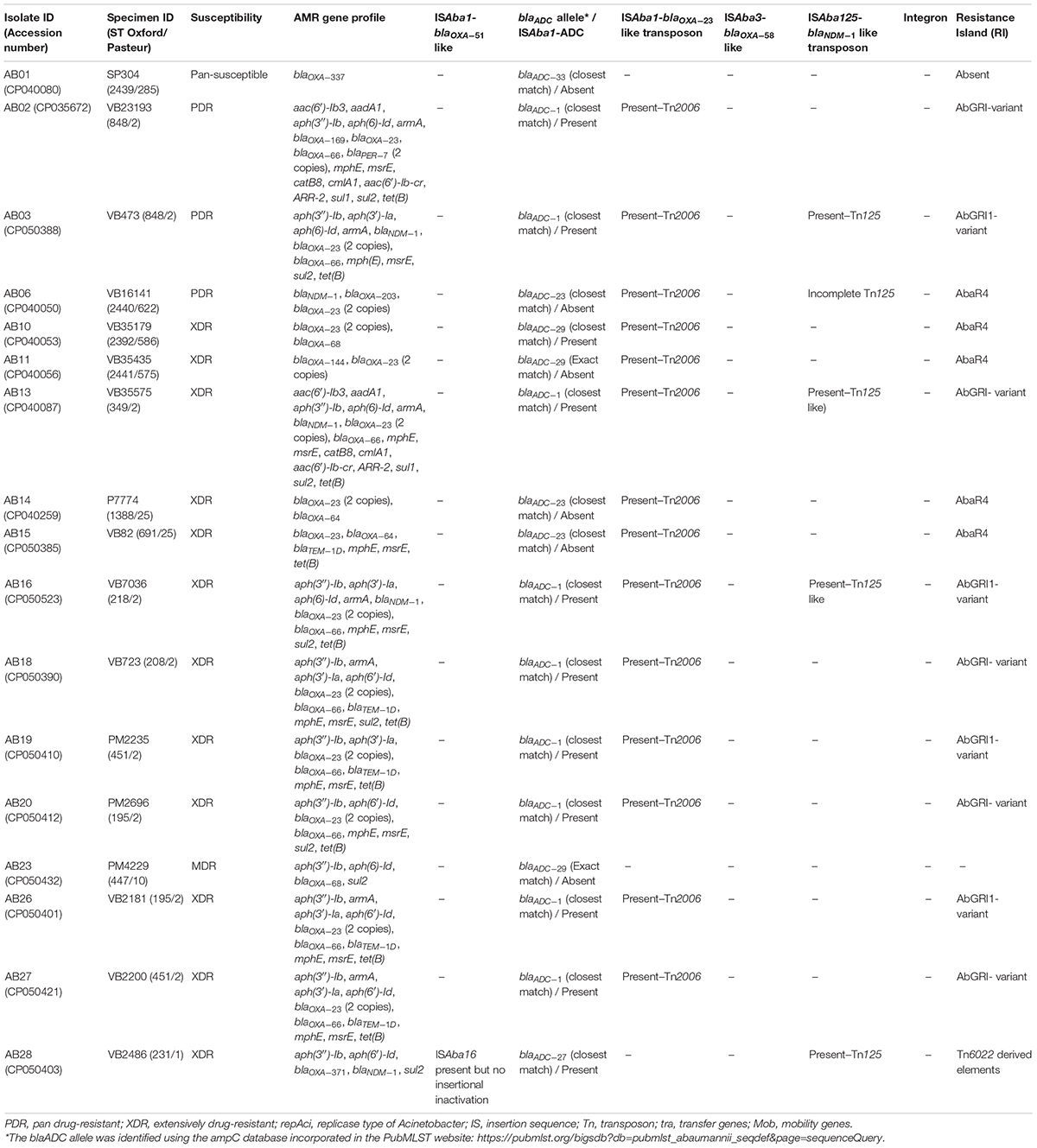
Table 2. Presence of antimicrobial resistance (AMR) genes, mobile genetic elements, and resistance islands among the 17 complete genomes of Acinetobacter baumannii.
IC2–the Predominant and Endemic Lineage With Novel Structural Variations
Nine isolates belonged to IC2, and all had either XDR or PDR phenotypes. Six Oxford MLST STs (STOxf) were identified, ST195 (2), ST451 (2), ST848 (2), ST208 (1), ST218 (1), and ST349 (1), but there was a single Pasteur MLST ST2Pas. IC2 isolates predominantly carried either blaOXA–23 alone (7/9) or coproduced blaOXA–23 and blaNDM–1 (2/9). All had blaOXA–23 in Tn2006, an ISAba1-bounded composite transposon in the chromosome (Figure 1). Of the nine isolates, AB03, AB13, and AB16 carried the blaNDM–1 gene on the chromosome with two different structural variations in the genetic context. AB03 and AB16 were associated with the most commonly reported transposon, Tn125 (Figure 2A), while a truncated form of Tn125 (Tn125-like) was identified in AB13, where the genome harbors a single copy of ISAba125 and an incomplete transposase at the left-hand and right-hand extremities of Tn125, respectively (Figure 2B). One to three plasmids were present among the nine genomes (Table 3). The p1AB20 belongs to the plasmid family which encodes repAci6 and was found to carry the aphA6 gene within TnaphA6 that was bounded by two copies of ISAba125 in direct orientation. The most commonly observed prophage elements among the nine IC2 genomes include PHAGE_Acinet_Bphi_B1251_NC_019541, PHAGE_Psychr_Psymv2_NC_023734, and PHAGE_Acinet_ YMC11/11/R3177_NC_041866.
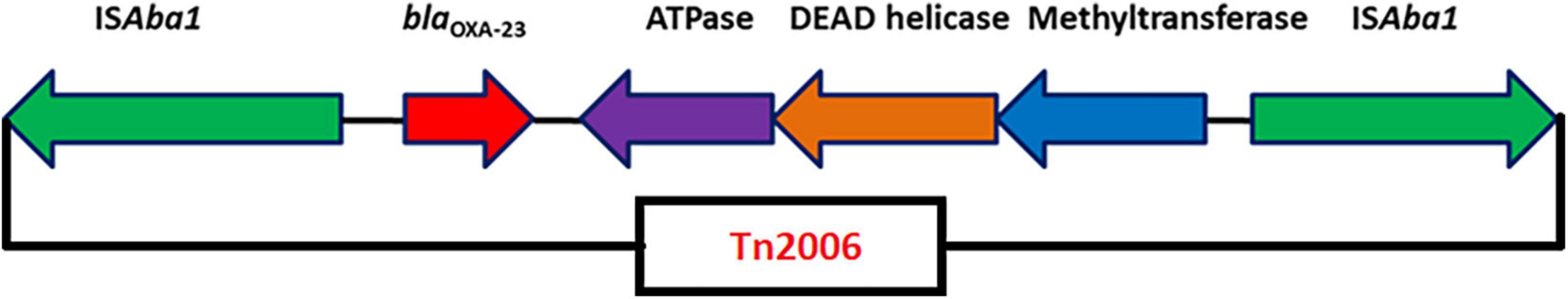
Figure 1. Genetic arrangement of blaOXA–23 identified in this study. The blaOXA–23 gene was flanked by two copies of an insertion sequence, ISAba1 in opposite orientations, forming a composite transposon, Tn2006.
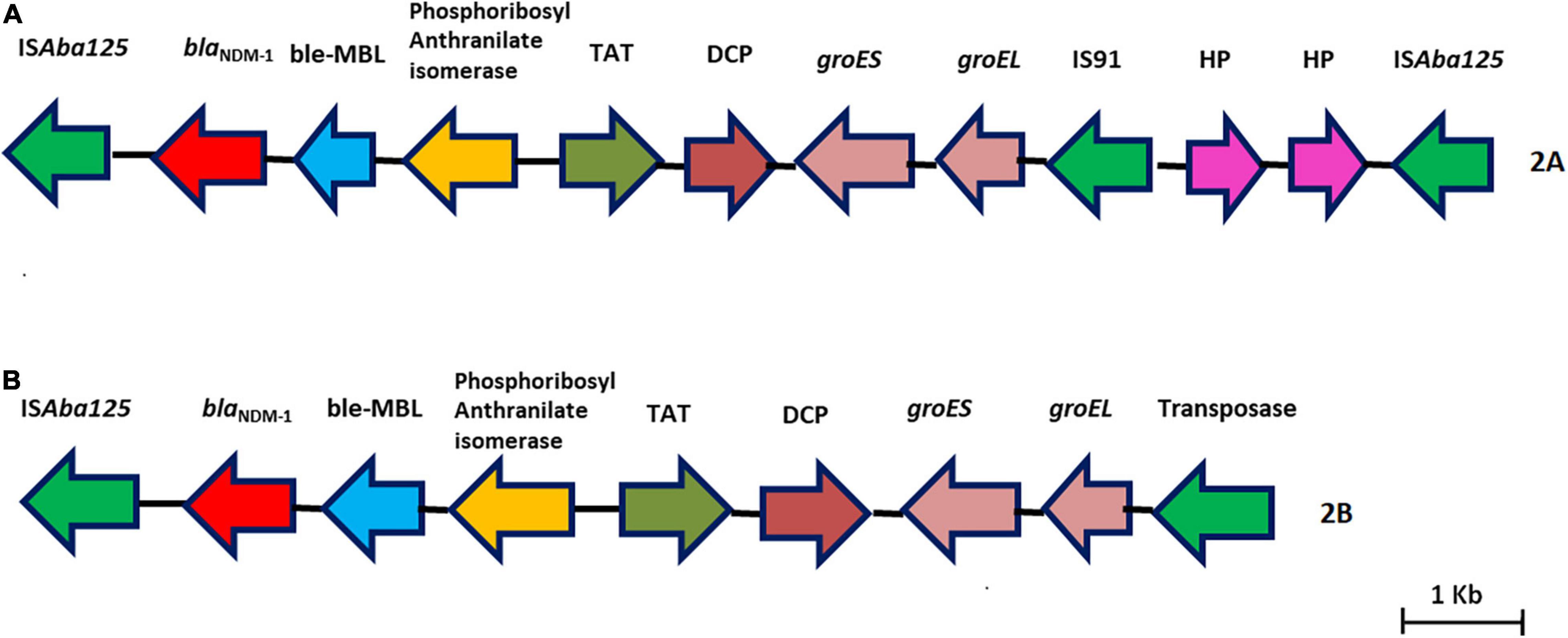
Figure 2. (A,B) Representative genomes showing the genetic environment of the blaNDM–1 gene characterized in this study. (A) Tn125–blaNDM–1 with two copies of ISAba125. (B) Tn125-like–blaNDM–1 with one copy of ISAba125 and a truncated transposase.
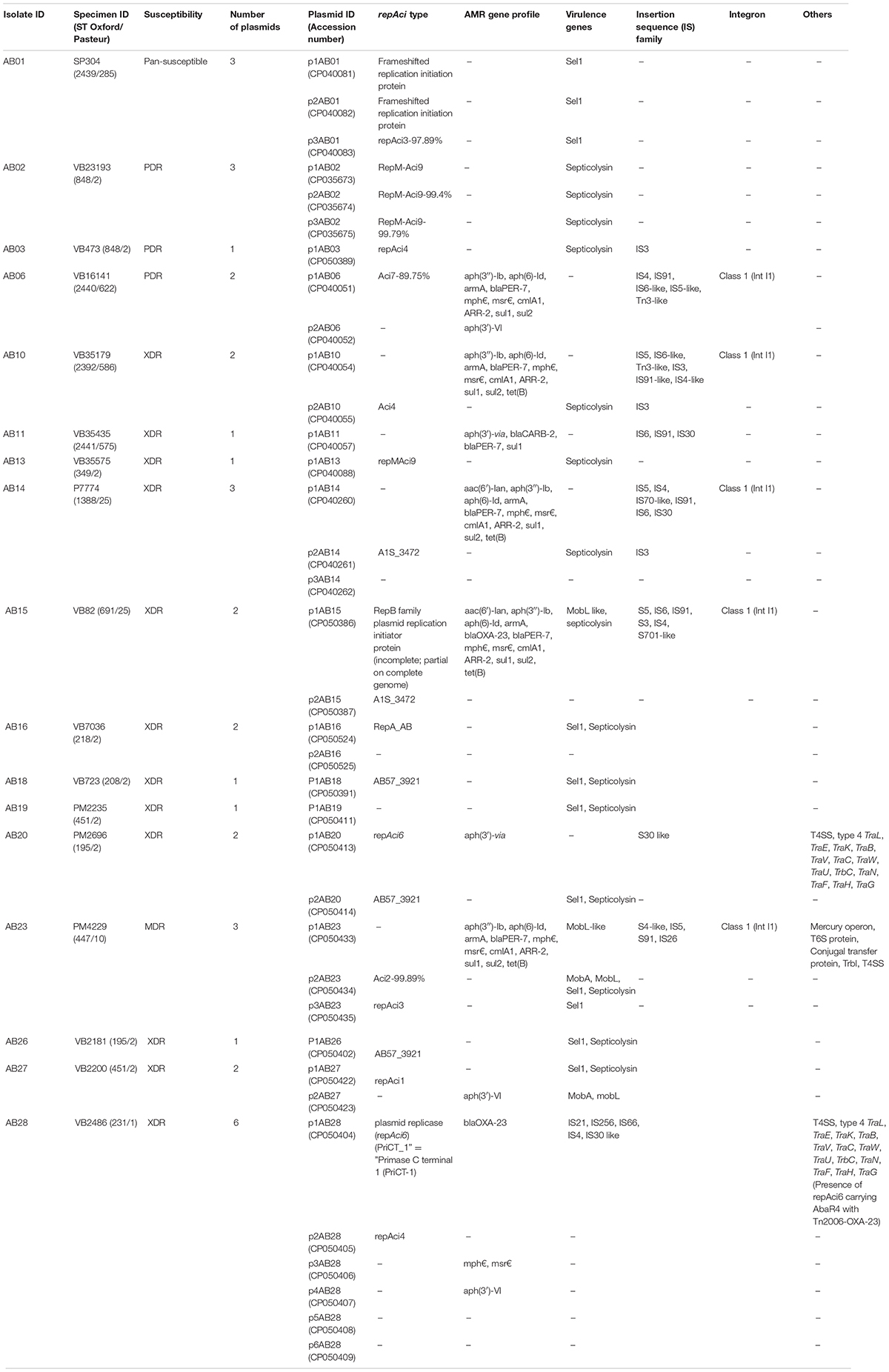
Table 3. Characteristic features of plasmids among the 17 complete genomes of Acinetobacter baumannii.
Based on the genetic configurations, three different variants of RIs were identified among the nine genomes. Of which, AB03, AB16, AB19, and AB26 carried variants of AbGR1, which included the presence of a partial region of Tn6172 with the aminoglycoside resistance genes; aph(6) and aph(3)-I, mobilization gene; mobL, transposable element; CR2, phosphoglucosamine mutase; pgm, arsenic resistance encoding gene; arsR, tetracycline efflux protein; tet(B), and tetracycline resistance transcriptional repressor gene; tetR(B) along with △Tn6022 and Tn2006 (Figure 3). AB18, AB20, and AB27 carried AbGRI variants with complex, diverse structures (Figure 4). AB18 and AB20 had a single copy of an ISAba1 element, sul2, rcr2, and a hypothetical protein inserted at the tniCA element on the Tn6172 backbone. Two copies of Tn2006 were observed in both the genomes but differed at the insertion site. In AB18, one copy of Tn2006 was inserted at orf4 while the second copy was inserted between orfBA and the tetracycline resistance transcriptional repressor gene, tetR(B). In AB20, one copy of Tn2006 was observed between the Tn6022 element and the plasmid linker, whereas another Tn2006 was inserted near orfBA. AB27 had two copies of ISAba1, one copy inserted at the tniE on the Tn6022 backbone and the second copy inserted at tniA of Tn6172. Two copies of Tn2006 were seen, one present on Tn6022 at orf4 with the second adjacent to orfBA on Tn6172. Insertion of sul2, rcr2, and hypothetical protein at the left inverted repeat of the Tn6172 element was also observed. Additionally, insertion of the arsenic resistance encoding gene, arsR, and the mobilization gene, mobL, was present on the Tn6172 element of all three genomes (Figure 4).
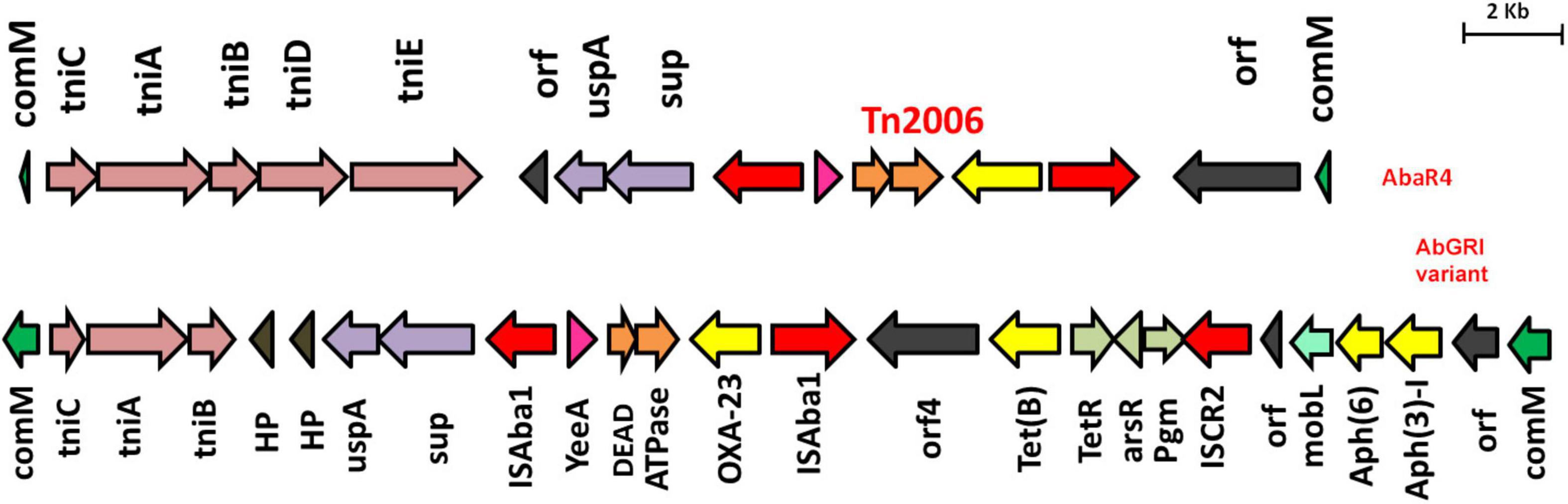
Figure 3. Structures of AbaR4 and variants of AbGRI1 identified in this study. The top figure depicts the typical AbaR4 island, while the bottom figure indicates the AbGRI1 variant due to additional mobL (light green arrow) and arsenic resistance gene, arsR (light gray arrow). The yellow arrow indicates antimicrobial resistance genes, and the red arrow depicts insertion elements.
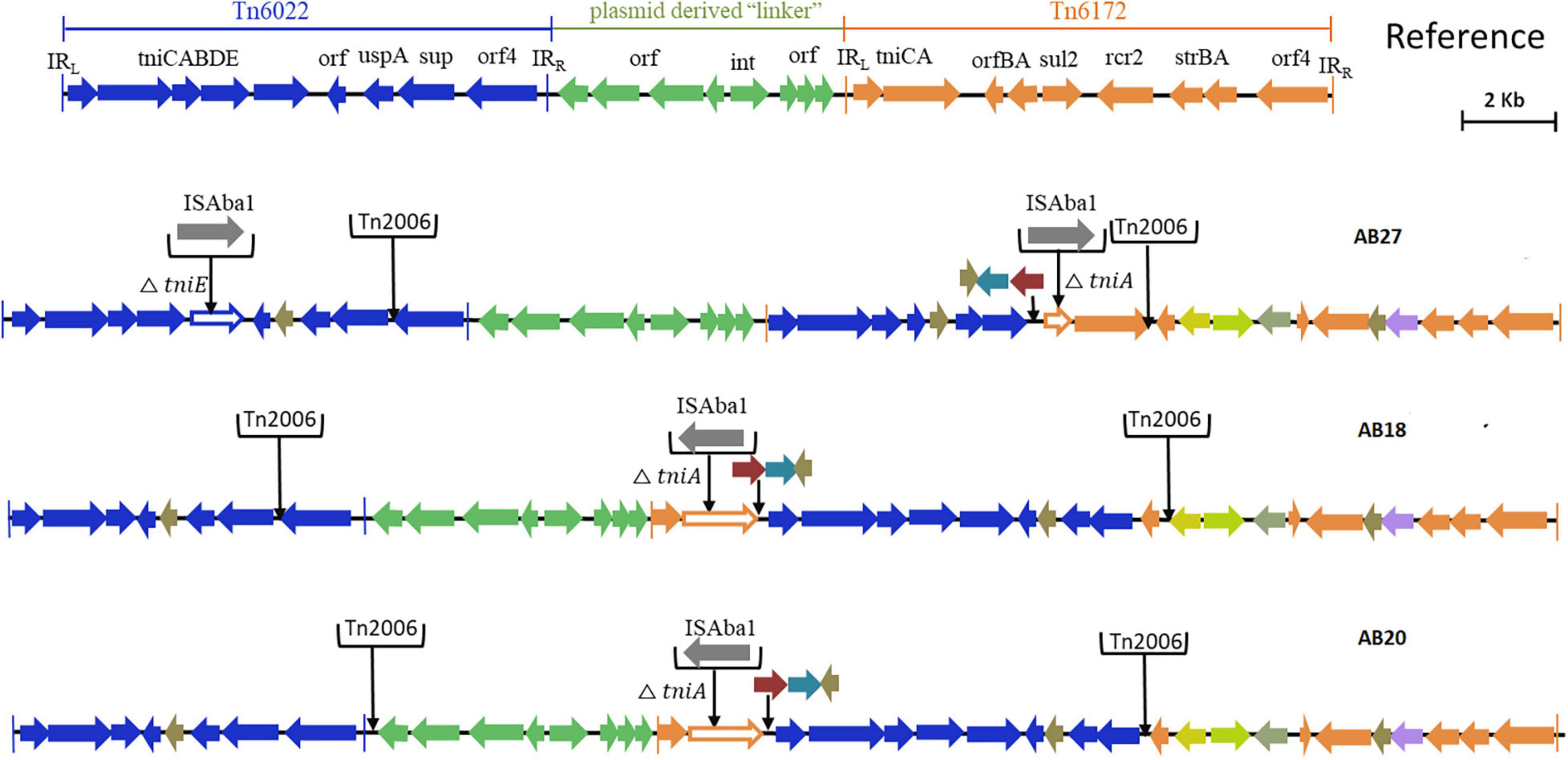
Figure 4. Structures of variants of AbGRI resistance island identified in this study. The typical AbGRI1 structure with an intact “Tn6022-linker-Tn6172” backbone is shown as a reference. The Tn6022 or Tn6022-derived part is shown in blue, the Tn6172 part or its partial segments are shown in orange, and the linker region is shown in green. The black arrows shown downward indicate the insertion of insertion sequence (IS) element or transposon or additional genes.
Interestingly, AB02 and AB13 carried the novel Tn6022-derived, plasmid linker, and Tn6172-derived elements. Insertion of a single copy of Tn2006 and △CR2-△Tn10-MARR-like region in Tn6172 was observed in both the genomes (Figures 5A,B). However, one minor difference was identified between the genomes, where AB02 carried blaPER–7 within the class 1 integron of the Tn6172-derived element (Figure 5A), while AB13 was found to carry a class 1 integron but was devoid of blaPER–7 (Figure 5B).
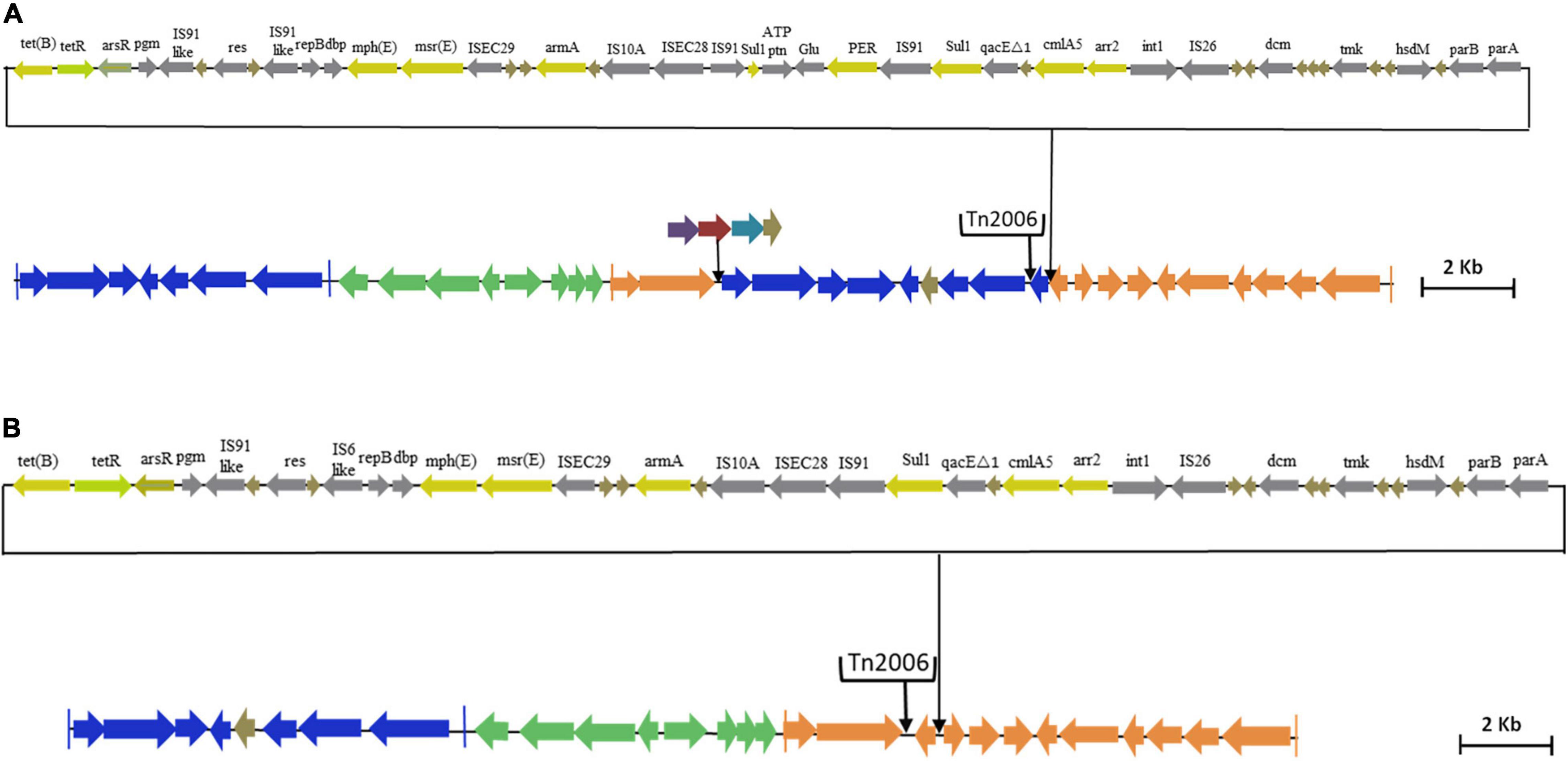
Figure 5. (A) Genetic backbone of AB13 carrying AbGRI variant identified in this study. The Tn6022 or Tn6022-derived part is shown in blue, the Tn6172 part or its partial segments are shown in orange, and the linker region is shown in green. The black arrows in the downward direction indicate the insertion of IS element, transposon, or additional genes. The pale yellow arrow indicates AMR genes, the light green arrow represents the tetracycline repressor gene, the gray arrow represents insertion elements, and the brown arrow indicates hypothetical protein. (B) Genetic backbone of AB02 carrying AbGRI variant identified in this study. The Tn6022 or Tn6022-derived part is shown in blue, the Tn6172 part or its partial segments are shown in orange, and the linker region is shown in green. The black arrows shown downward indicate the insertion of IS element or transposon or additional genes. The pale yellow arrow indicates AMR genes, the light green arrow represents the tetracycline repressor gene, the gray arrow represents insertion elements, and the brown arrow indicates hypothetical protein.
IC7 and IC8–the Emerging Lineage of CRAb Isolates in India
Two genomes, namely, AB14 and AB15, belonged to IC7 and were represented by ST1388 Oxf/ST25Pas and ST691 Oxf/ST25Pas, and both were XDR. AB14 and AB15 carried three and two plasmids, respectively. AB14 harbored blaOXA–23 in Tn2006 on the chromosome alone, whereas AB15 had it in both the chromosome (blaOXA–23 in Tn2006) and on an incomplete RepB family plasmid (blaOXA–23 in △Tn2006). Both AB14 and AB15 showed the presence of two prophage regions, namely, PHAGE_Mannhe_vB_MhM_3927AP2_NC_028766 and PHAGE_Acinet_YMC11/11/R3177_NC_041866. AbaR4 that were mapped to Tn6022 backbone and Tn2006 linked blaOXA–23 locus was present in both the genomes (Figure 3).
AB10, AB11, and AB23 belonged to IC8. Of the three, AB10 and AB11 were XDR and corresponded to novel STs: ST2392 Oxf/ST586Pas (AB10) and ST2441 Oxf/ST575Pas (AB11), whereas AB23 corresponded to ST447Oxf/ST10Pas and had an MDR phenotype. AB10 and AB11 were blaOXA–23 producers, while AB23 was found to be a non-carbapenemase producer. Two, one, and three plasmids were identified in AB10, AB11, and AB23, respectively. Three intact prophages such as PHAGE_Pelagi_HTVC010P_NC_020481, PHAGE_Acinet_Bphi_B1251_NC_019541, and PHAGE_Acinet_ vB_AbaS_TRS1_NC_031098 were seen in AB10. In AB11, PHAGE_Acinet_YMC11/11/R3177_NC_041866, PHAGE_ Acinet_Bphi_B1251_NC_019541, and PHAGE_Mannhe_ vB_MhM_3927AP2_NC_028766 were observed. A single prophage, PHAGE_Acinet_Bphi_B1251_NC_019541, was present in AB23. Similar to IC7, both XDR isolates carried AbaR4 on the chromosome but were absent in the MDR isolate, AB23 (Figure 3).
IC1 Lineage With Tn6022-Derived Elements
AB28 had an XDR phenotype that corresponded to ST231 Oxf/ST1Pas and belonged to IC1. Interestingly, in AB28, which carried a variant of blaOXA–51 (blaOXA–371), an insertion of ISAba16, TnpB, and an IS66 transposase, there was no upstream presence or insertional inactivation (Figure 6). AB28 carried Tn125 linked blaNDM–1 on the chromosome (Figure 2A). AB28 harbored six plasmids. Of which, p1AB28 carried blaOXA–23 on repAci6 family plasmid and several plasmid transfer (tra) genes. When we analyzed and compared the p1AB28 plasmid sequence with the reference plasmid, pA85-3 (accession number–KJ493819), we found the presence of a complete blaOXA–23 gene with one complete and an incomplete copy of an ISAba1 locus. However, some transposon-related genes such as uspA and sulP were intact. IS66 family transposase with its accessory protein, tnpB, was also encoded within the p1AB28 plasmid but was absent in the pA85-3 reference plasmid. The plasmid, p1AB28, also carried putative tra genes that are required for mating pair formation and trwC and trwB genes that are needed for plasmid mobilization (Figure 7). AB28 carried five different prophages as follows: PHAGE_Stx2_c_Stx2a_F451_NC_049924, PHAGE_Acinet_Bphi_B1251_NC_019541, PHAGE_Psychr_ pOW20_A_NC_020841, PHAGE_Escher_SH2026Stx1_NC_ 049919, and PHAGE_Acinet_vB_AbaS_TRS1_NC_031098. Notably, AB28 encoded Tn6022-derived elements in which the insertion of an IS256 family transposase, ISAba42, was observed between tniE and orf [Tn6022 (tniE-orf)::ISAba42] (Figure 8).
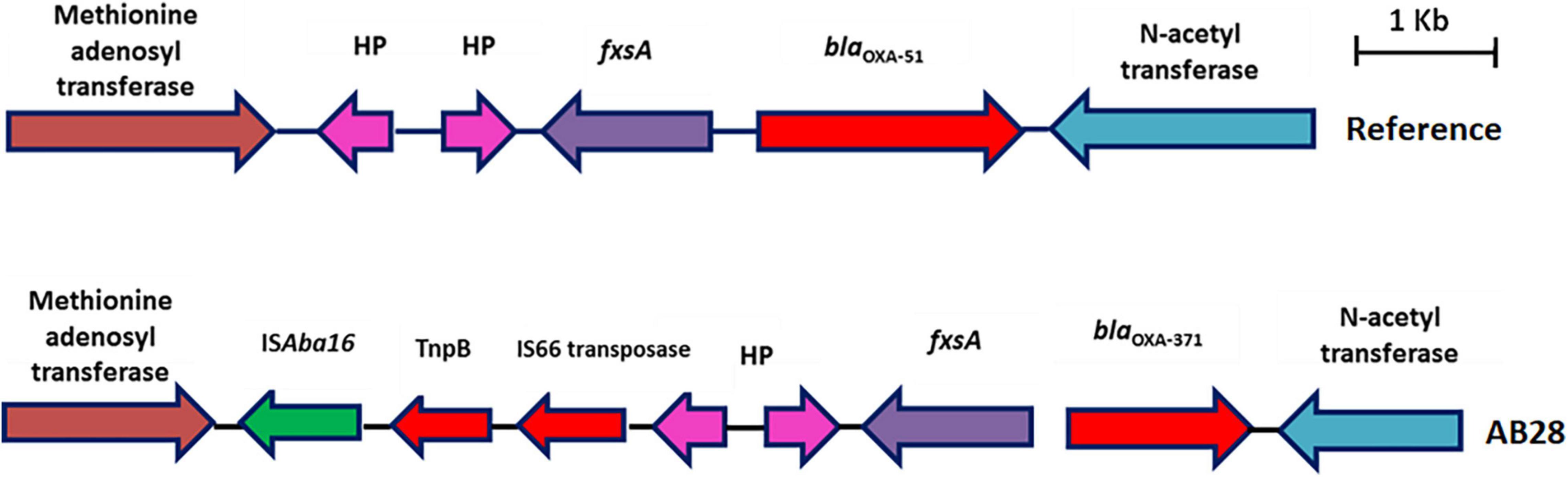
Figure 6. Genetic backbone of blaOXA–51. Two types of genetic structures were identified in this study. Sixteen isolates were identified with typical backbone, whereas one isolate with blaOXA–371 was identified with insertion sequence, ISAba16, TnpB, and IS66 family transposase.
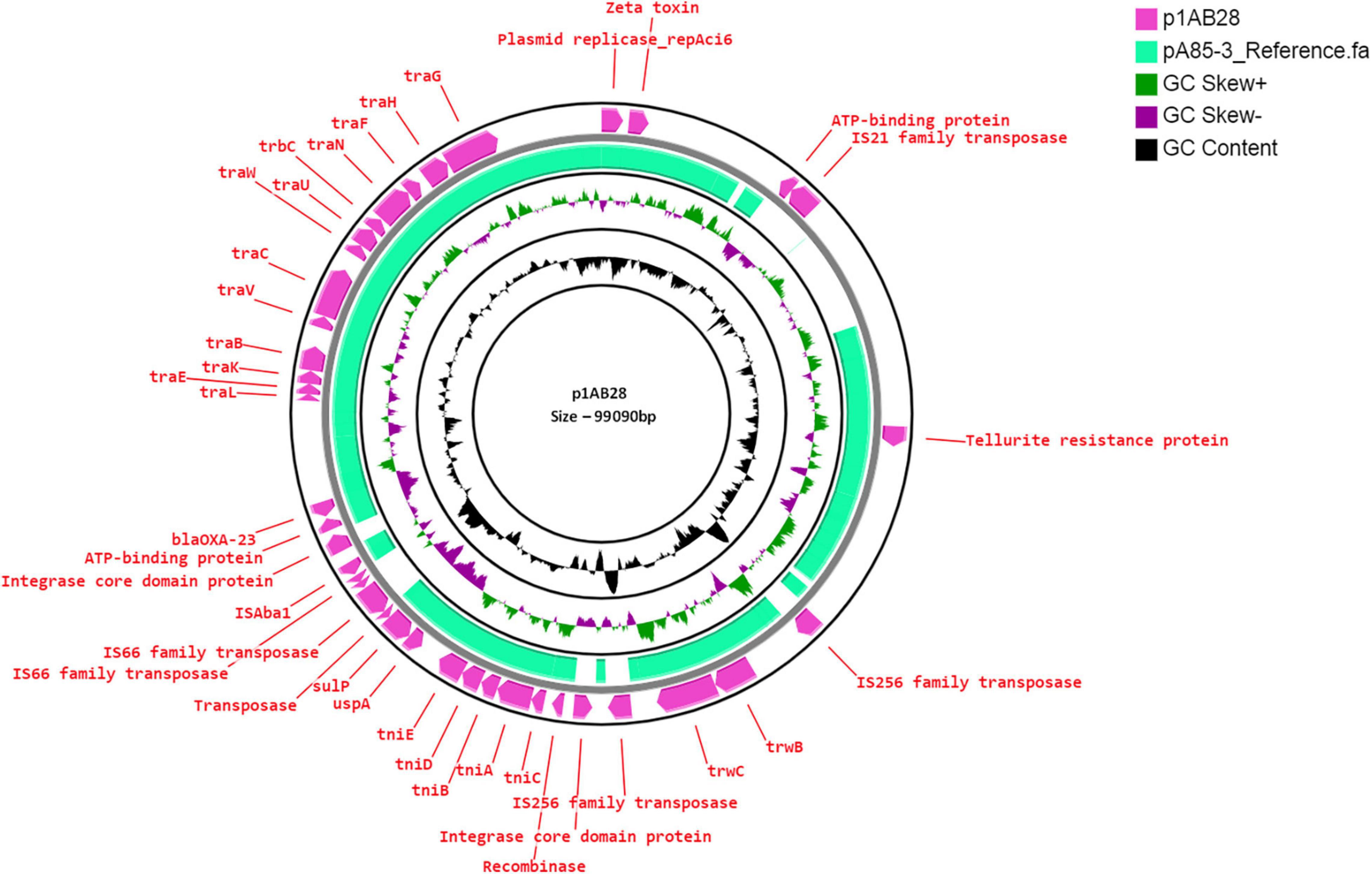
Figure 7. Circular representation of repAci6 plasmid (pink arrow), p1AB28, of Acinetobacter baumannii displayed using CG view server with the reference plasmid pA85-3 (accession number–KJ493819) (green-colored region). The two inner circles represent GC content and GC skew. The pink-colored arrow represents the presence of the OXA-23 gene along with the plasmid replication gene, repAci6, tra genes, and plasmid mobilization genes in p1AB28.
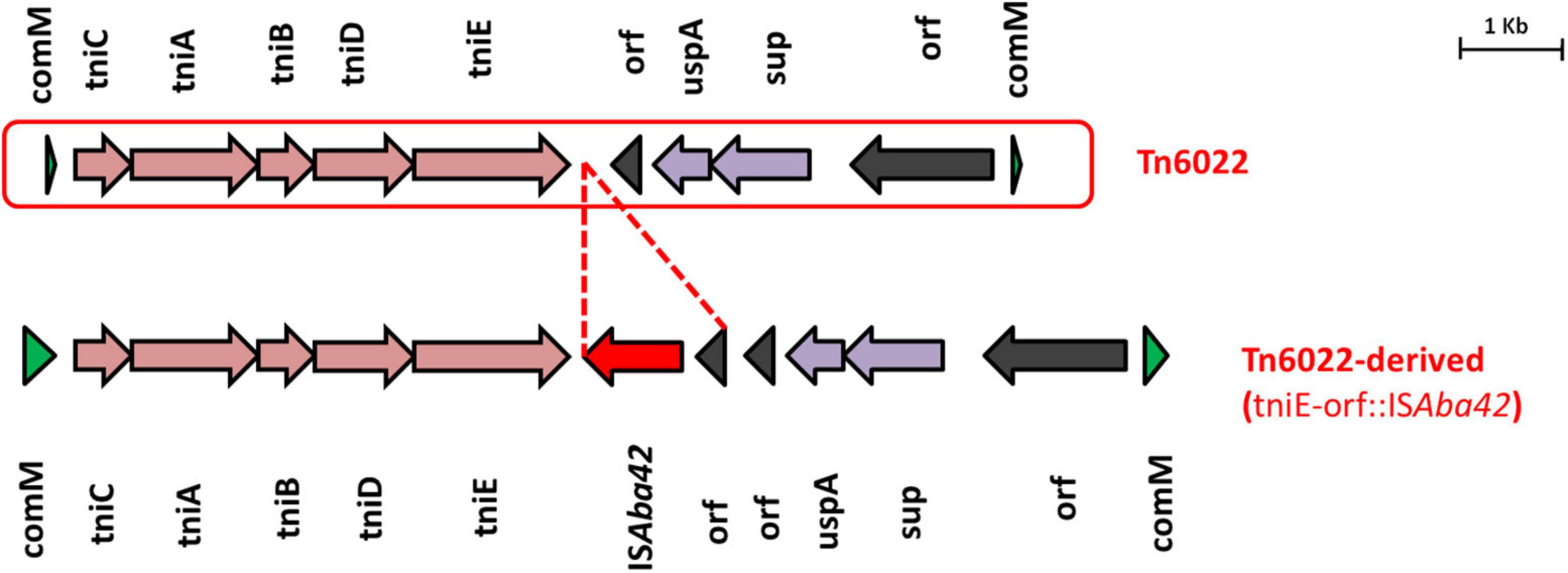
Figure 8. Structures of Tn6022 and Tn6022-like elements. The typical Tn6022 backbone is shown at the top of the figure. Tn6022-derived element observed in this study is displayed at the bottom of the figure and showed the insertion of ISAba42 (red arrow) with an additional orf. Appropriate names of the elements found within the genetic configurations are given. Dotted lines in red are used to depict the insertion of genetic elements.
Pan-Susceptible Singleton and Pan-Drug Resistant CC862
The PSAB, AB01 represented as a singleton and belonged to the novel ST, ST2439 Oxf/ST285Pas, while the PDRAB, AB06 belonged to CC862 and was represented by another novel ST, ST2440 Oxf/ST622Pas. As expected, AB01 did not harbor any of the AMR determinants except blaADC–25–like, blaOXA–337. Three plasmids were present with no AMR genes. No intact prophage and RI were present.
AB06 carried dual carbapenemases, blaOXA–23 and blaNDM–1, on the chromosome, and they were found to be carried on transposon, Tn2006, and Tn125, respectively. Two plasmids were observed with genes encoding resistance to β-lactamases, aminoglycosides, macrolides, and sulfonamides. Three intact prophages, PHAGE_Pseudo_phiCTX_NC_003278, PHAGE_Acinet_Bphi_B1251_NC_019541, and PHAGE_Acinet_YMC11/11/R3177_NC_041866, were present. In addition, AB06 possesses the commonly reported AbaR4 RI (Figure 3).
Phylogenetic Analysis of Core Genomes of CRAb
Analysis of core genomes of CRAb revealed the presence of multiple AMR genes among the IC2 isolates. Clone-specific OCL types such as OCL1 to IC2, OCL5 to IC7, and OCL2 to IC8 were observed. Diverse KL types were identified among the study isolates, and the tree showed the presence of ST-specific KL types within a specific clonal lineage. AbaR4 was present among the IC1, IC7, and IC8 isolates, while AbaR4 and AbGRI variants were observed only among the IC2 isolates (Figure 9).
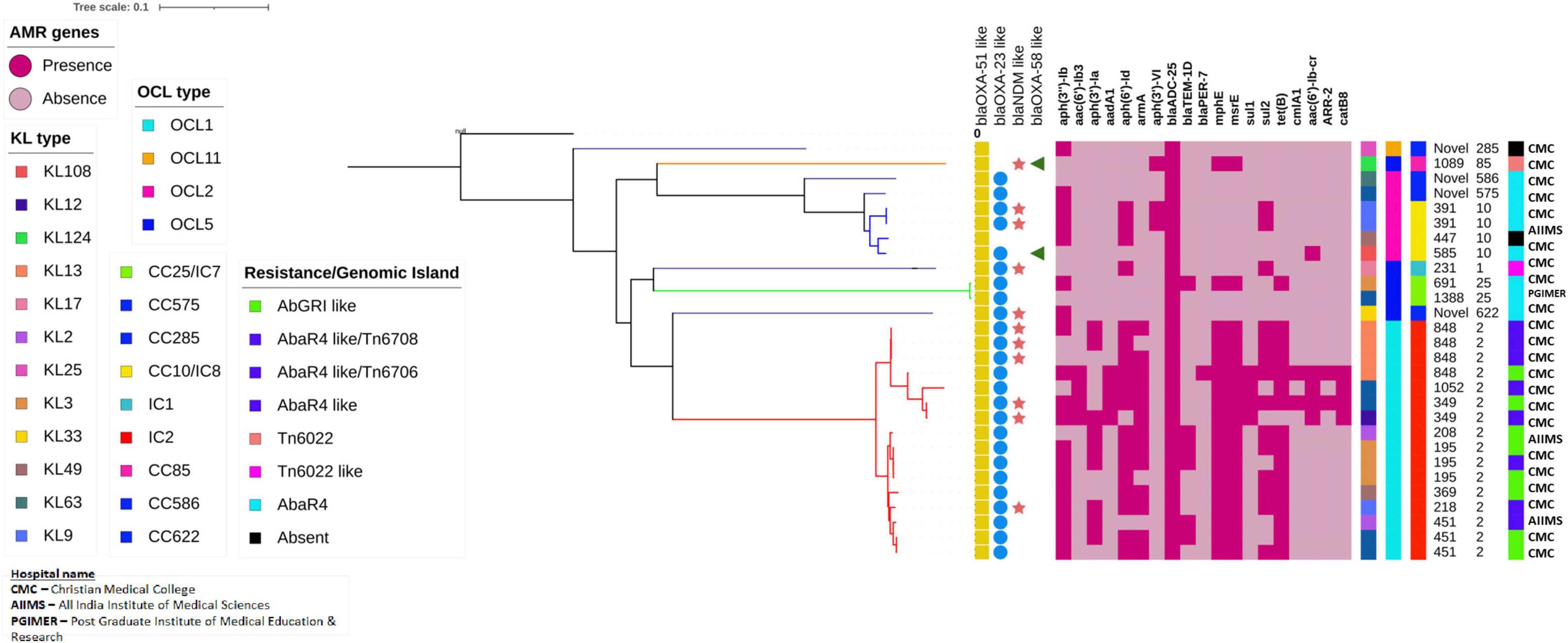
Figure 9. Single nucleotide polymorphism (SNP)-based phylogenetic tree of carbapenem-resistant Acinetobacter baumannii sequenced in this study. The color-filled shape denoted presence, while the empty shape denoted the absence of the respective traits. The heat map represents the presence or absence of AMR genes; Dark red indicates the presence of the respective gene, while light red indicates the lack of the respective gene. The capsular types (KL), outer core lipopolysaccharide types (OCL), International clones/clonal complexes, and resistance/genomic islands of the CRAb isolates were represented by color-coded boxes as given in the legend. The Oxford and Pasteur scheme sequence types (STs) were given as text labels.
Discussion
Acinetobacter baumannii has become an important hospital-acquired pathogen and is of major concern due to the rapid emergence of MDR, XDR, and PDR strains (Agoba et al., 2018; Havenga et al., 2019). Carbapenem resistance rates of more than 85% in A. baumannii have been reported from previous studies in India and are typically associated with isolates carrying either blaOXA–23 alone or both blaOXA–23 and blaNDM–1, which concurs with this study (Vijayakumar et al., 2016; Vijayakumar et al., 2019; Vijayakumar et al., 2020). The majority of the isolates (13/17) in this study encoded more than one copy of the blaOXA–23 gene. However, we could not find any high-level carbapenem resistance genes in these isolates. Earlier, Hua et al. (2016) reported the presence of multiple copies of blaOXA–23 among CRAb as a common phenomenon without an increase in carbapenem resistance.
This study showed the endemicity of IC2 along with the emergence of sporadic clones, such as IC7 and IC8. Although previous studies from India reported the predominance of IC2, the presence of isolates that belongs to IC7 and IC8 indicates the dissemination of CRAb and reinforces the fact that the International clones of CRAb isolates are widespread among hospitals in India.
Several studies have reported that the blaOXA–23 gene has relocated to chromosomes and plasmids with the help of transposons (Hamidian and Nigro, 2019; Graña-Miraglia et al., 2020). Fourteen CRAb isolates were identified with Tn2006-linked blaOXA–23 in this study. Although experimental observations were not performed, carbapenem resistance in these isolates could be due to the ISAba1-mediated overexpression of the blaOXA–23 gene in Tn2006. Occasionally, carbapenem resistance in A. baumannii could happen due to the overexpression of blaOXA–51 variants by insertion of ISAba1 (Wong et al., 2019). In this study, the presence of ISAba16 was observed in one genome; however, insertional inactivation of blaOXA–51–like was not seen.
In A. baumannii, the blaNDM–1 gene can be encoded by either chromosomes or plasmids (Bonnin et al., 2012). However, this study observed A. baumannii isolates harboring blaNDM–1 only in chromosomes. Unlike Enterobacteriaceae, in which blaNDM–1 is often observed with a single copy of truncated ISAba125 on plasmids, the dissemination of blaNDM–1 in A. baumannii is always associated with a complete Tn125 (Poirel et al., 2012; Dortet et al., 2014). In contrast with the above statement, one genome in this study was identified with Tn125-like linked blaNDM–1, suggesting that it could have acquired blaNDM–1 from other species.
The presence of repAci6 harboring blaOXA–23 and belonging to IC1 was identified in the p1AB28 plasmid. Comparative analysis revealed that AB28 carries a plasmid closely related to the reference, as it harbors the blaOXA–23 gene in a different context (Hamidian et al., 2016). The p1AB28 plasmid is conjugative and can spread carbapenem resistance by disseminating the blaOXA–23 gene into diverse clones. However, further studies are warranted to confirm the same. Another genome, AB20, belonged to IC2 and carried a repAci6 conjugative plasmid. This plasmid harbors the aphA6 gene on the TnaphA6 transposon which encodes an aminoglycoside (3′) phosphotransferase and confers resistance to amikacin. Previous studies from European and Asian countries have reported isolates of A. baumannii with large conjugative plasmid such as repAci6, carrying both the blaOXA–23 and aphA6 genes, which contribute to the dissemination of resistance to carbapenems and amikacin, respectively (Towner et al., 2011; Nigro and Hall, 2016). Earlier studies by Costa et al. (2018) reported the presence of AMR and virulence genes within the prophage regions of A. baumannii genomes. This study showed at least one prophage region in all the genomes except the PSAB. However, no prophages with AMR genes were detected.
Genomic analysis of AbaRs in this study unveiled novel genetic configurations specific to backbones, which involve either insertion of MGEs or structural modifications driven by known MGEs. For example, insertion of ISAba42 within the Tn6022 backbone leads to a truncated form of the tniE transposition gene, thereby forming the Tn6022 derived element. Furthermore, in this study, we identified an isolate (AB28) that belonged to IC1 but lacked an AbaR3 type island. Instead, it carried an IC2-specific Tn6022-derived backbone, which indicates the possibility of independent acquisition. Tn6022-derived elements and AbaR4 and AbGRI variants are typically confined to IC2. In this study, we also found that none of the IC2 isolates carried AbaR4; instead, it was present among isolates belonging to other ICs such as IC7 and IC8. All the study isolates belonging to IC2 possessed either the AbGRI1 variant or the AbGRI variant with complex chimeric structures. Although the genetic events behind this process are unclear, such complex structural variation in the AbaR backbones might have resulted either due to the target sequences favorable for MGE insertion or due to the exposure of AbaRs with different MGEs in different clones. These findings indicate that AbaRs with diverse backbones might have evolved separately.
Conclusion
Overall, to the best of our knowledge, this study is the first that provides comprehensive profiling of RIs together with the MGEs, acquired AMR genes, and the distribution of clonal lineages among CRAb from India. Although this study provides a clear picture of the Indian scenario, further comparative analysis with an extensive collection of global isolates is required to understand the structural diversity and the evolution of these MGEs that drive the genome plasticity of A. baumannii.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SV: laboratory methods, data analysis and interpretation, and manuscript writing. JJ: data analysis, interpretation, and manuscript writing. KV: hybrid genome assembly and other bioinformatics methods. PM, PR, SA, IB, and KW: manuscript correction. AN and AB: data analysis. BV: study design and supervising, manuscript writing, and manuscript correction. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge Ms. Catherine Truman, Clinical Pharmacist, Christian Medical College and Hospital, India, for her valuable input in language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.869653/full#supplementary-material
Supplementary Figure 1 | A map of India showing the location of three hospitals from where the samples were collected as a part of the study. The red color represents PGIMER, Chandigarh, the green color represents AIIMS-Trauma, New Delhi, and the light red color represents CMC, Vellore. Map outline was created using mapchart.net. Republished from mapchart.net under a CC BY license, with permission from MapChart, original copyright 2021.
Footnotes
- ^ https://dbtindia.gov.in
- ^ https://github.com/marcelm/cutadapt
- ^ http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- ^ https://github.com/gnetsanet/ONT-GUPPY
- ^ https://github.com/rrwick/Filtlong
- ^ http://www.genomicepidemiology.org/services/
- ^ https://github.com/tseemann/abricate
- ^ https://cge.food.dtu.dk/services/ResFinder/
- ^ https://kaptive-web.erc.monash.edu/
- ^ https://www-is.biotoul.fr/blast.php
- ^ https://phaster.ca/
- ^ https://cge.food.dtu.dk/services/MLST/
- ^ https://github.com/lh3/bwa
References
Agoba, E. E., Govinden, U., Peer, A. K., Osei Sekyere, J., and Essack, S. Y. (2018). IS Aba1 regulated OXA-23 carbapenem resistance in Acinetobacter baumannii strains in Durban, South Africa. Microb. Drug Resist. 24, 1289–1295. doi: 10.1089/mdr.2017.0172
Bonnin, R. A., Poirel, L., Naas, T., Pirs, M., Seme, K., Schrenzel, J., et al. (2012). Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 18, E362–E365.
Cameranesi, M. M., Paganini, J., Limansky, A. S., Moran-Barrio, J., Salcedo, S. P., Viale, A. M., et al. (2020). Acquisition of plasmids conferring carbapenem and aminoglycoside resistance and loss of surface-exposed macromolecule structures as strategies for the adaptation of Acinetobacter baumannii CC104O/CC15P strains to the clinical setting. Microbial Genom. 6:mgen000360. doi: 10.1099/mgen.0.000360
Costa, A. R., Monteiro, R., and Azeredo, J. (2018). Genomic analysis of Acinetobacter baumannii prophages reveals remarkable diversity and suggests profound impact on bacterial virulence and fitness. Sci. Rep. 8:15346.
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Dexter, C., Murray, G. L., Paulsen, I. T., and Peleg, A. Y. (2015). Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology, and pathogenesis. Expert Rev. Anti-infective Therapy 13, 567–573. doi: 10.1586/14787210.2015.1025055
Dortet, L., Poirel, L., and Nordmann, P. (2014). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed Res. Int. 2014:249856. doi: 10.1155/2014/249856
Ershov, V., Tarasov, A., Lapidus, A., and Korobeynikov, A. (2019). IonHammer: homopolymer-space hamming clustering for IonTorrent read error correction. J. Comput. Biol. 26, 124–127. doi: 10.1089/cmb.2018.0152
Gales, A. C., Seifert, H., Gur, D., Castanheira, M., Jones, R. N., and Sader, H. S. (2019). Antimicrobial susceptibility of Acinetobacter calcoaceticus–Acinetobacter baumannii complex and Stenotrophomonas maltophilia clinical isolates: results from the SENTRY antimicrobial surveillance program (1997–2016). Open Forum Infectious Dis. 6, S34–S46.
Graña-Miraglia, L., Evans, B. A., López-Jácome, L. E., Hernández-Durán, M., Colín-Castro, C. A., Volkow-Fernández, P., et al. (2020). Origin of OXA-23 variant OXA-239 from a recently emerged lineage of Acinetobacter baumannii international clone V. mSphere 5:e00801-19. doi: 10.1128/mSphere.00801-19
Hamidian, M., Ambrose, S. J., and Hall, R. M. (2016). A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid 87, 43–50. doi: 10.1016/j.plasmid.2016.09.001
Hamidian, M., and Hall, R. M. (2017). Origin of the AbGRI1 antibiotic resistance island found in the comM gene of Acinetobacter baumannii GC2 isolates. J. Antimicrobial Chemotherapy 72, 2944–2947. doi: 10.1093/jac/dkx206
Hamidian, M., and Hall, R. M. (2011). AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J. Antimicrob. Chemother. 1, 2484–2491. doi: 10.1093/jac/dkr356
Hamidian, M., and Hall, R. M. (2018). The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1–structure, origin and evolution. Drug Resist. Updat 41, 26–39.
Hamidian, M., and Nigro, S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microbial Genom. 5:e000306. doi: 10.1099/mgen.0.000306
Havenga, B., Ndlovu, T., Clements, T., Reyneke, B., Waso, M., and Khan, W. (2019). Exploring the antimicrobial resistance profiles of WHO critical priority list bacterial strains. BMC Microbiol. 19:303. doi: 10.1186/s12866-019-1687-0
Hernández-González, I. L., Mateo-Estrada, V., and Castillo-Ramirez, S. (2022). The promiscuous and highly mobile resistome of Acinetobacter baumannii. Microbial Genom. 8:000762. doi: 10.1099/mgen.0.000762
Hua, X., Shu, J., Ruan, Z., Yu, Y., and Feng, Y. (2016). Multiplication of blaOXA-23 is common in clinical Acinetobacter baumannii, but does not enhance carbapenem resistance. J. Antimicrobial Chemotherapy 71, 3381–3385. doi: 10.1093/jac/dkw310
Lee, C. R., Lee, J. H., Park, M., Park, K. S., Bae, I. K., Kim, Y. B., et al. (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7:55. doi: 10.3389/fcimb.2017.00055
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Mancilla-Rojano, J., Castro-Jaimes, S., Ochoa, S. A., Bobadilla del Valle, M., Luna-Pineda, V. M., et al. (2019). Whole-genome sequences of five Acinetobacter baumannii strains from a child with leukemia M2. Front. Microbiol. 10:132. doi: 10.3389/fmicb.2019.00132
Nigro, S. J., and Hall, R. M. (2016). Structure and context of Acinetobacter transposons carrying the OXA-23 carbapenemase gene. J. Antimicrobial Chemotherapy 71, 1135–1147.
O’Donnell, J. N., Putra, V., and Lodise, T. P. (2021). Treatment of patients with serious infections due to carbapenem-resistant Acinetobacter baumannii: how viable are the current options? Pharmacotherapy: J. Hum. Pharmacol. Drug Therapy 41, 762–780. doi: 10.1002/phar.2607
Pagano, M., Martins, A. F., and Barth, A. L. (2016). Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 47, 785–792. doi: 10.1016/j.bjm.2016.06.005
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Poirel, L., and Nordmann, P. (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12, 826–836.
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., and Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089. doi: 10.1128/AAC.05620-11
Poirel, L., Marqué, S., Héritier, C., Segonds, C., Chabanon, G., and Nordmann, P. (2005). OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 202–208.
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Roca, J., Subirà, I., Espinal, P., Vila-Farrés, X., and Vila Estapé, J. (2012). The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. doi: 10.3389/fmicb.2012.00148
Rodrigues, D. L., Morais-Rodrigues, F., Hurtado, R., Dos Santos, R. G., Costa, D. C., Barh, D., et al. (2021). Pan-resistome insights into the multidrug resistance of Acinetobacter baumannii. Antibiotics 10:596. doi: 10.3390/antibiotics10050596
Salto, I. P., Tejerizo, G. T., Wibberg, D., Pühler, A., Schlüter, A., and Pistorio, M. (2018). Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci. Rep. 8:7783. doi: 10.1038/s41598-018-26180-3
Seemann, T. (2015). Snippy: Fast Bacterial Variant Calling from NGS Reads. San Francisco, CA: GitHub.
Towner, K. J., Evans, B., Villa, L., Levi, K., Hamouda, A., Amyes, S. G., et al. (2011). Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D β-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 2154–2159. doi: 10.1128/AAC.01661-10
Turton, J. F., Ward, M. E., Woodford, N., Kaufmann, M. E., Pike, R., Livermore, D. M., et al. (2006a). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258, 72–77. doi: 10.1111/j.1574-6968.2006.00195.x
Turton, J. F., Woodford, N., Glover, J., Yarde, S., Kaufmann, M. E., and Pitt, T. L. (2006b). Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44, 2974–2976. doi: 10.1128/JCM.01021-06
Vijayakumar, S., Anandan, S., Prabaa, D., Kanthan, K., Vijayabaskar, S., Kapil, A., et al. (2019). Insertion sequences and sequence types profile of clinical isolates of carbapenem-resistant A. baumannii collected across India over four year period. J. Infect. Public Health 23, 1022–1028. doi: 10.1016/j.jiph.2019.11.018
Vijayakumar, S., Gopi, R., Gunasekaran, P., Bharathy, M., Walia, K., Anandan, S., et al. (2016). Molecular characterization of invasive carbapenem-resistant Acinetobacter baumannii from a tertiary care hospital in South India. Infect. Dis. Therapy 5, 379–387. doi: 10.1007/s40121-016-0125-y
Vijayakumar, S., Wattal, C., Oberoi, J. K., Bhattacharya, S., Vasudevan, K., Anandan, S., et al. (2020). Insights into the complete genomes of carbapenem-resistant Acinetobacter baumannii harbouring blaOXA-23, blaOXA-420 and blaNDM-1 genes using a hybrid-assembly approach. Access Microbiol. 2:acmi000140. doi: 10.1099/acmi.0.000140
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Weinstein, M. P., Patel, J. B., and Bobenchik, A. M. (2018). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical, and Laboratory Standards Institute.
Weinstein, M. P., Patel, J. B., Bobenchik, A. M., Campeau, S., Cullen, S. K., and Gallas, M. F. (2019). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and laboratory standards institute.
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017a). Completing bacterial genome assemblies with multiplex MinION sequencing. Microbial Genom. 3:e000132. doi: 10.1099/mgen.0.000132
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017b). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comp. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wong, M. H., Chan, B. K., Chan, E. W., and Chen, S. (2019). Over-expression of ISAba1-linked intrinsic and exogenously acquired OXA type carbapenem-hydrolyzing-class D-ß-lactamase-encoding genes is key mechanism underlying carbapenem resistance in Acinetobacter baumannii. Front. Microbiol. 10:2809. doi: 10.3389/fmicb.2019.02809
Keywords: CRAb, OXA–23, Tn2006, IC2, AbGRI1 variant, AbaR4
Citation: Vijayakumar S, Jacob JJ, Vasudevan K, Mathur P, Ray P, Neeravi A, Baskaran A, Kirubananthan A, Anandan S, Biswas I, Walia K and Veeraraghavan B (2022) Genomic Characterization of Mobile Genetic Elements Associated With Carbapenem Resistance of Acinetobacter baumannii From India. Front. Microbiol. 13:869653. doi: 10.3389/fmicb.2022.869653
Received: 04 February 2022; Accepted: 20 May 2022;
Published: 15 June 2022.
Edited by:
Raffaele Zarrilli, University of Naples Federico II, ItalyReviewed by:
Santiago Castillo Ramírez, National Autonomous University of Mexico, MexicoMehrad Hamidian, University of Technology Sydney, Australia
Copyright © 2022 Vijayakumar, Jacob, Vasudevan, Mathur, Ray, Neeravi, Baskaran, Kirubananthan, Anandan, Biswas, Walia and Veeraraghavan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Balaji Veeraraghavan, vbalaji@cmcvellore.ac.in
 Saranya Vijayakumar
Saranya Vijayakumar Jobin John Jacob
Jobin John Jacob Karthick Vasudevan
Karthick Vasudevan Purva Mathur
Purva Mathur Pallab Ray
Pallab Ray Ayyanraj Neeravi
Ayyanraj Neeravi Ashtawarthani Baskaran
Ashtawarthani Baskaran Agilandeeswari Kirubananthan
Agilandeeswari Kirubananthan Shalini Anandan
Shalini Anandan Indranil Biswas
Indranil Biswas Kamini Walia
Kamini Walia Balaji Veeraraghavan
Balaji Veeraraghavan