- 1School of Food and Bioengineering, Chengdu University, Chengdu, Sichuan, China
- 2School of Environment and Energy, Peking University Shenzhen Graduate School, Shenzhen, China
- 3Department of Pharmaceutical Microbiology, Faculty of Pharmacy Medical University of Gdańsk, Gdańsk, Poland
Thermophiles from extreme thermal environments have shown tremendous potential regarding ecological and biotechnological applications. Nevertheless, thermophilic cyanobacteria remain largely untapped and are rarely characterized. Herein, a polyphasic approach was used to characterize a thermophilic strain, PKUAC-SCTB231 (hereafter B231), isolated from a hot spring (pH 6.62, 55.5°C) in Zhonggu village, China. The analyses of 16S rRNA phylogeny, secondary structures of 16S-23S ITS and morphology strongly supported strain B231 as a novel genus within Trichocoleusaceae. Phylogenomic inference and three genome-based indices further verified the genus delineation. Based on the botanical code, the isolate is herein delineated as Trichothermofontia sichuanensis gen. et sp. nov., a genus closely related to a validly described genus Trichocoleus. In addition, our results suggest that Pinocchia currently classified to belong to the family Leptolyngbyaceae may require revision and assignment to the family Trichocoleusaceae. Furthermore, the complete genome of Trichothermofontia B231 facilitated the elucidation of the genetic basis regarding genes related to its carbon-concentrating mechanism (CCM). The strain belongs to β-cyanobacteria according to its β-carboxysome shell protein and 1B form of Ribulose bisphosphate Carboxylase-Oxygenase (RubisCO). Compared to other thermophilic strains, strain B231contains a relatively low diversity of bicarbonate transporters (only BicA for HCO3− transport) but a higher abundance of different types of carbonic anhydrase (CA), β-CA (ccaA) and γ-CA (ccmM). The BCT1 transporter consistently possessed by freshwater cyanobacteria was absent in strain B231. Similar situation was occasionally observed in freshwater thermal Thermoleptolyngbya and Thermosynechococcus strains. Moreover, strain B231 shows a similar composition of carboxysome shell proteins (ccmK1-4, ccmL, -M, -N, -O, and -P) to mesophilic cyanobacteria, the diversity of which was higher than many thermophilic strains lacking at least one of the four ccmK genes. The genomic distribution of CCM-related genes suggests that the expression of some components is regulated as an operon and others in an independently controlled satellite locus. The current study also offers fundamental information for future taxogenomics, ecogenomics and geogenomic studies on distribution and significance of thermophilic cyanobacteria in the global ecosystem.
Introduction
Thermophilic cyanobacteria are widely distributed in hot spring ecological niches (Miller et al., 2007; Alcorta et al., 2020). Moreover, many studies have demonstrated the importance of thermophilic cyanobacteria as primary photosynthetic producers of geothermal ecosystems, accounting for a large part of those ecosystems’ biomass and productivity (Esteves-Ferreira et al., 2018; Chen et al., 2021). Besides, high-value-added products can be harvested from thermophilic cyanobacteria and have been applied to numerous industries, e.g., agricultural, pharmaceutical and nutraceutical (Patel et al., 2019).
Isolation of thermophilic cyanobacteria from diverse ecosystems is critical for multidisciplinary studies regarding their morphology, genetics, physiology, biochemistry, and for providing potential strains for biotechnology and industrial applications (Cordeiro et al., 2020). It is, however, challenging to assign the taxonomy of thermophilic cyanobacteria due to their simple morphology. As a result, the diversity of thermophilic cyanobacterial genera might be severely underestimated, and community efforts are being made to rectify it. Synechococcus-like strains (Tang et al., 2022c) and Leptolyngbya-like strains (Mai et al., 2018; Yao et al., 2021) have undergone an extensive reevaluation based on multi-locus sequence analysis and genomic data, providing new insights into genetic diversity of the two genera. Furthermore, polyphasic taxonomic classification approaches have been widely utilized for cyanobacterial identification, particularly for understudied or unresolved polyphyletic families/genera/species and identification of novel families and genera (Raabova et al., 2019; Shalygin et al., 2020). Consequently, these reassignments will better taxonomically resolve cyanobacterial genera and families. Especially, the increased number of genomic sequences will facilitate the development of taxogenomics and complement the traditional 16S rRNA-based taxa identification.
The aquatic environments where thermophilic cyanobacteria live are characterized by low availability of CO2, primarily due to external factors, e.g., temperature, pH and gas exchange (Durall and Lindblad, 2015). Meanwhile, the availability of dissolved inorganic carbon in form of carbonates is highly variable. Therefore, to survive in the hostile aquatic habitat, the thermophilic cyanobacteria utilize CO2-concentrating mechanisms (CCM) to ensure that the Ribulose bisphosphate Carboxylase-Oxygenase (RubisCO) with low affinity for CO2 is surrounded by high CO2 levels and functions regardless of thermal stress (Galmés et al., 2015, 2016). Therefore, investigating the molecular component at the genomic level is a prerequisite for understanding the thermophilic cyanobacterial CCM and its relationship with their niches. In addition, under the current scenario of global warming, the studies on the molecular components of cyanobacterial CCM in relation to their specific habitats may shed light on the evolution of hot spring genomes as an example of selective pressure in warmer environments.
In our previous study, we isolated a Leptolyngbya-like strain, B231, from a green microbial mat of a hot spring in Zhonggu village, Sichuan, China, which can grow at 47°C and/or at the concentration of 0.1 M NaHCO3 (Tang et al., 2018a). Herein, thorough polyphasic characterization for strain B231, including 16S rRNA phylogeny, the secondary structure of 16S-23S ITS, and morphology description has been achieved. According to the botanical code, a new genus name Trichothermofontia sichuanensis gen. et sp. nov. has been proposed for strain B231 as the first representative of the genus Trichothermofontia. Moreover, based on our research interests, the molecular basis of CCM has been investigated for strain B231 by computational identification, and the CCM component has been further related to its adaptation. The current study lays a foundation for future taxogenomic, ecogenomic and geogenomic studies on the distribution and significance of thermophilic cyanobacteria.
Materials and methods
Origins, cultivation, and basic physiological assessment of Trichothermofontia sichuanensis B231
The strain B231, capable of forming mats, used in the present study was initially isolated from a hot spring around Zhonggu village in Ganzi Prefecture of Sichuan Province, China. Sample collection was done on 12 May 2016, with the humidity close to 71%. The ambient temperature at the time of collection was 15°C, and the light intensity was around 1,000 lux. The temperature of the hot spring, its pH, and the concentration of total dissolved solids were 55.5°C, 6.62, and 492 mmol L−1, respectively. Information about the sampling site and preliminary taxonomic assignment of the strain was detailed in our previous studies (Tang et al., 2018a,b). A unicyanobacterial culture of B231, recovered from 10% DMSO stocks maintained at −80°C for over 2 years, was used to establish experimental cultures as described previously (Tang et al., 2018a). Briefly, the recovered strain was cultivated at 45°C in 150 mL BG-11 medium in 500 mL Erlenmeyer flasks agitated at 100 rpm under 16 l:8D photoperiod at 45 μmol m−2 s−1 provided by fluorescent tubes unless stated otherwise. The strain initially denoted and maintained in Peking University Algae Collection as PKUAC-SCTB231 has also been deposited in the Freshwater Algae Culture Collection at the Institute of Hydrobiology (FACHB-collection) with accession number FACHB-3573. The strain was assessed for the capacity to utilize nitrite and nitrate using the modifications of BG-11, essentially as described earlier (Liang et al., 2019). Briefly, BG-11 medium without nitrogen was supplemented with 0, 0.075, 0.5, 1.5, 5, 5.7 g L−1 NaNO3; 1.218 g L−1 NaNO2. The effect of bicarbonate has been tested in regular BG-11 medium supplemented with 0, 0.1, 0.3, 0.5, 0.7, 1.0 M sodium bicarbonate. The growth of the strain has been assessed qualitatively due to the mat-forming character of the strain.
Genome sequencing, assembly, and annotation
Integrated sequencing strategies employing MGISEQ-2000 PE150 and Oxford Nanopore Technologies PromethION sequencing platforms were applied for whole-genome sequencing of the strain B231. The sequencing was performed by a commercial provider BGI-TECH (Wuhan, China). The genomic DNA was isolated using FastDNA™ SPIN Kit for Soil (MP Biomedicals, Irvine, CA, United States), and its integrity verified with agarose electrophoresis. After sequencing, a total of 806,280,450 bp of short read data and 1,717,621,070 bp of the long read data of an average length of 14,717 bp were used for the assembly of the genome yielding a single circular chromosome and no plasmids. The assembly has been performed from long-read data using Flye v2.7 (Kolmogorov et al., 2019) module integrated into the commercial Geneious Prime 2022.2 package (Kearse et al., 2012) and subsequently refined with short-read data using Geneious mapper on default settings. The genome of strain B231 was annotated using a modified pipeline previously established by Tang et al. (2019). Briefly, gene prediction and annotation were automatically performed using the NCBI prokaryotic genome annotation pipeline (O'Leary et al., 2016), and further using the RAST annotation system (Brettin et al., 2015) to minimize poor calls. The genome annotation of strain B231 was summarized in Supplementary Table 1. The complete genome has been deposited in GenBank under accession number CP110848.
Phylogenetic inference of 16S rRNA
The full-length 16S rRNA gene sequence of strain B231 was extracted from its genome sequence. Additional 97 16S rRNA gene sequences of cyanobacterial references were retrieved from NCBI through BLASTN search. Muscle complemented in Mega7 (Kumar et al., 2016) was employed to generate multiple sequence alignments, and manual editing comprising trimming to the same length and adjusting poorly aligned regions were carried out where necessary. Sequences of the alignment were trimmed to the same length (1,013 bp). The alignment of 16S rRNA gene sequences was subjected to Bayesian Inference using MrBayes v3.2.7 (Ronquist et al., 2012). The following parameters were applied: NST = 6, Rates = equal, MCMC Ngen = 10,000,000. Default settings were used for all the other parameters. The Bayesian analysis had a mean estimated sample size (ESS) exceeding 4,700 for all parameters, far above the average of 200 typically accepted as sufficient by phylogeneticists (Drummond et al., 2006). The final average standard deviation of split frequencies was 0.002624. The potential scale reduction factor (PSRF) value for all the estimated parameters in the Bayesian analysis was 1.00, indicating that convergence of the MCMC chains was statistically achieved (Gelman and Rubin, 1992).
Analysis of 16S-23S ITS
The conserved domains of the 16S-23S ITS region: D1-D1’, D2, D3, boxA, and D4; and its variable regions (V2, boxB, and V3) were identified as previously described (Iteman et al., 2000). The tRNAs presented in the spacer were identified by tRNAscan-SE v1.3.1 (Lowe and Eddy, 1997). The secondary structures of the identified fragments were individually determined by Mfold web server (Zuker, 2003). Except for using the structure, draw mode untangle with loop fix, default conditions in Mfold were used in all cases.
Whole-genome comparisons
A genome dataset was compiled for whole-genome comparisons, including genomes of strain B231 and 17 representative focus taxa (one representative from the family Trichocoleusaceae, eight from the family Oculatellaceae and nine from the family Leptolyngbyaceae as references). The quality of each genome was assessed using CheckM (Parks et al., 2015) to ensure a high-quality dataset with near completeness (≥95%) and low contamination (<5%). Moreover, the corresponding protein sequences of each genome were downloaded from the NCBI database.
Three indices useful for genus delineation were calculated to summarize the similarity or distance between genomes. The whole genome average nucleotide identity (ANI) and average amino acid identity (AAI) between genomes were calculated using the ANI/AAI calculator with default settings.1 The percentages of conserved proteins (POCP) between genomes were determined according to the method described previously (Qin et al., 2014).
Phylogenomic reconstruction
The phylogenomic relationship between strain B231 and focal taxa was elucidated by the concatenated protein sequences from 647 single-copy genes shared by all the genomes. The homologous gene clusters identified by OrthoMCL (Li et al., 2003) were used to refine single-copy genes shared by all the genomes, which were concatenated employing a custom Perl script. Multisequence alignment was performed using MAFFT v7.453 (Standley, 2013). The supergene alignment was subjected to phylogenomic inference using IQ-TREE v2.1.3 (Minh et al., 2020). A total of 546 protein models were used to select the optimal substitution model for phylogenomic analysis using ModelFinder implemented in IQ-TREE. Bootstrap tests (1,000 replicates) were carried out for the assessment of tree topologies using UltraFast Bootstrap (Hoang et al., 2018).
Investigation on CCM in strain B231
Orthologous proteins involved in CCM of strain B231 were identified as previously described (Tang et al., 2022d). Briefly, amino acid sequences of 28 proteins involved in CCM of Synechocystis sp. PCC 6803 were retrieved as a reference protein set. Orthologous genes in strain B231 were identified with the bidirectional best hit (BBH) criterion using BLASTP with the following thresholds: E-value cut-off of 1E-6, ≥30% identity and 70% coverage, and manually curated. Amino acid sequences of RubisCO large subunit (rbcL) were collected for strain B231 and reference cyanobacteria to infer the protein function and classification. Protein sequences of genes ccmK, -L, -M, -N, -O, and -P encoding carboxysome shell proteins were also collected for phylogenetic reconstruction. All the phylogenetic analyses were performed using Maximum-Likelihood (ML) algorithm as previously described (Tang et al., 2022c).
Morphology investigation
All microscopic operations were performed essentially as described by Tang et al. (2021). In short, strain B231 was inspected at 400× magnification using light microscopy (LM, DP72, OLYMPUS, Japan), equipped with an image acquisition system (U-TV0.63XC, OLYMPUS, Japan). Microscopic investigations were also conducted using scanning electron microscopy (SEM; SU8100, HITACHI, Japan), and using transmission electron microscopy (TEM; HT7800, HITACHI, Japan). Cell measurements have been performed on 100 representative cells selected from six independent micrographs and presented as a range.
Taxonomic evaluation
The classification system in this study was applied according to Komárek et al. (2014). Briefly, taxonomy assignment was determined based on phylogenetic position of the corresponding entity, as well as its morphological and ecological characters. The taxon description follows the prescriptions of the Botanical Code, International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen code; Turland et al., 2018).
Results and discussion
General genomic characteristics of strain B231
The complete genome of strain B231 has been achieved by integrating Oxford Nanopore and DNBSEQ sequencing systems yielding genome coverage of 387× and 181×, respectively. The B231 genome comprises a single circular chromosome with a size of 4,436,989 bp and a GC content of 53.9%. Bioinformatic annotation indicates that two ribosomal RNA (rrn) operons, 45 tRNA genes and 4,352 protein-coding sequences (CDS), were present in the B231 genome (Supplementary Table 1). The two ribosomal RNA operons differed in length by an 11 bp insertion in the ITS region, two single bp insertions in the 23S rRNA gene region and 2 bp difference between the two variants of 16S rRNA gene, indicating their 99.98% identity. None of the above-mentioned differences fundamentally impacted the phylograms or predicted secondary structures of the ITS region, as the 11 bp insertion was outside the regions of interest. Approximately 48.2% of protein-coding genes (2,098 out of 4,352) protein-coding genes were predicted to be hypothetical proteins. It was not surprising to identify such a high percentage of hypothetical protein in the B231 genome since similar observations are typical in the genomes of other thermophilic cyanobacteria (Cheng et al., 2020; Kono et al., 2022; Tang et al., 2022a,b).
Phylogeny reconstruction of 16S rRNA gene
The Bayesian phylograms (Figure 1; Supplementary Figure 1) inferred by 16S rRNA gene sequences categorize the cyanobacterial strains into three well-defined families. Genera were also taxonomically resolved within families with the support of high posterior probabilities. The focal strain B231 is closely grouped with five thermophilic strains isolated from various hot springs in Zhonggu village (Table 1). Moreover, five other uncultured cyanobacteria were also closely related to strain B231. These 11 strains formed a well-supported clade distinct from the two described genera within the family Trichocoleusaceae. This result suggests the phylogenetically novel clade and a new genus within the family Trichocoleusaceae. In fact, the family Trichocoleusaceae is a monophyletic family that has been recently proposed by dividing the Leptolyngbyaceae into family-level clades based on molecularly-supported data (Mai et al., 2018). To date, the genus Trichocoleus (Mühlsteinova et al., 2014) was the only described member of the family Trichocoleusaceae. The phylogram generated in this study suggests the existence of two more genera in this family. The genus Pinocchia (Dvorak et al., 2015) is still classified within the family Leptolyngbyaceae, which contradicts the 16S-based phylogeny obtained in this study. Therefore, we propose that the genus Pinocchia should be reclassified into the family Trichocoleusaceae.
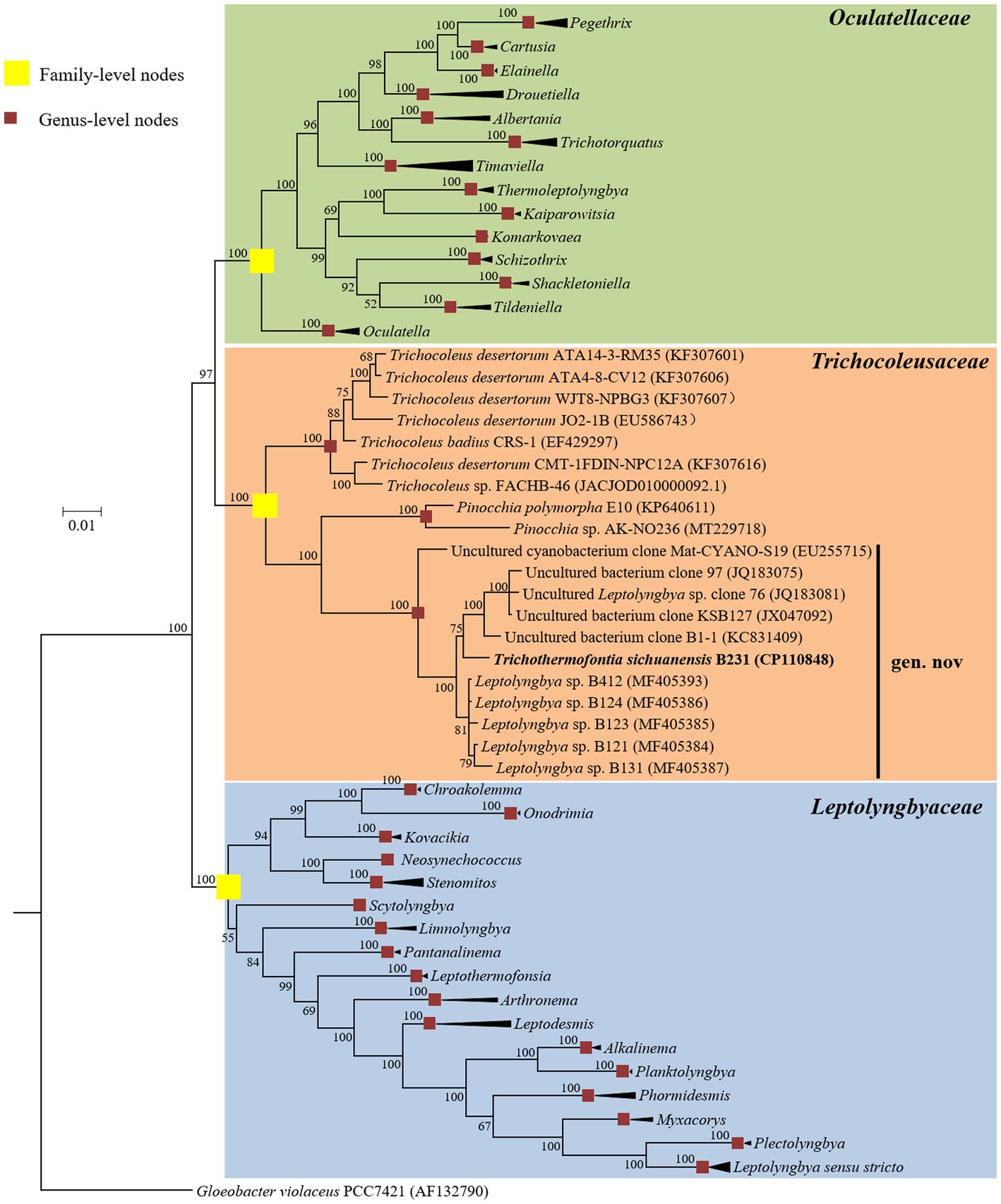
Figure 1. Bayesian inference of 16S rRNA gene sequences representing 98 cyanobacterial strains. Collapsed genera are indicated by black polygons, with a length corresponding to the distance from the most basal sequence to the most diverged sequence of the genus. The complete phylogram refers to Supplementary Figure 1. Posterior probabilities (%) are given above the nodes.
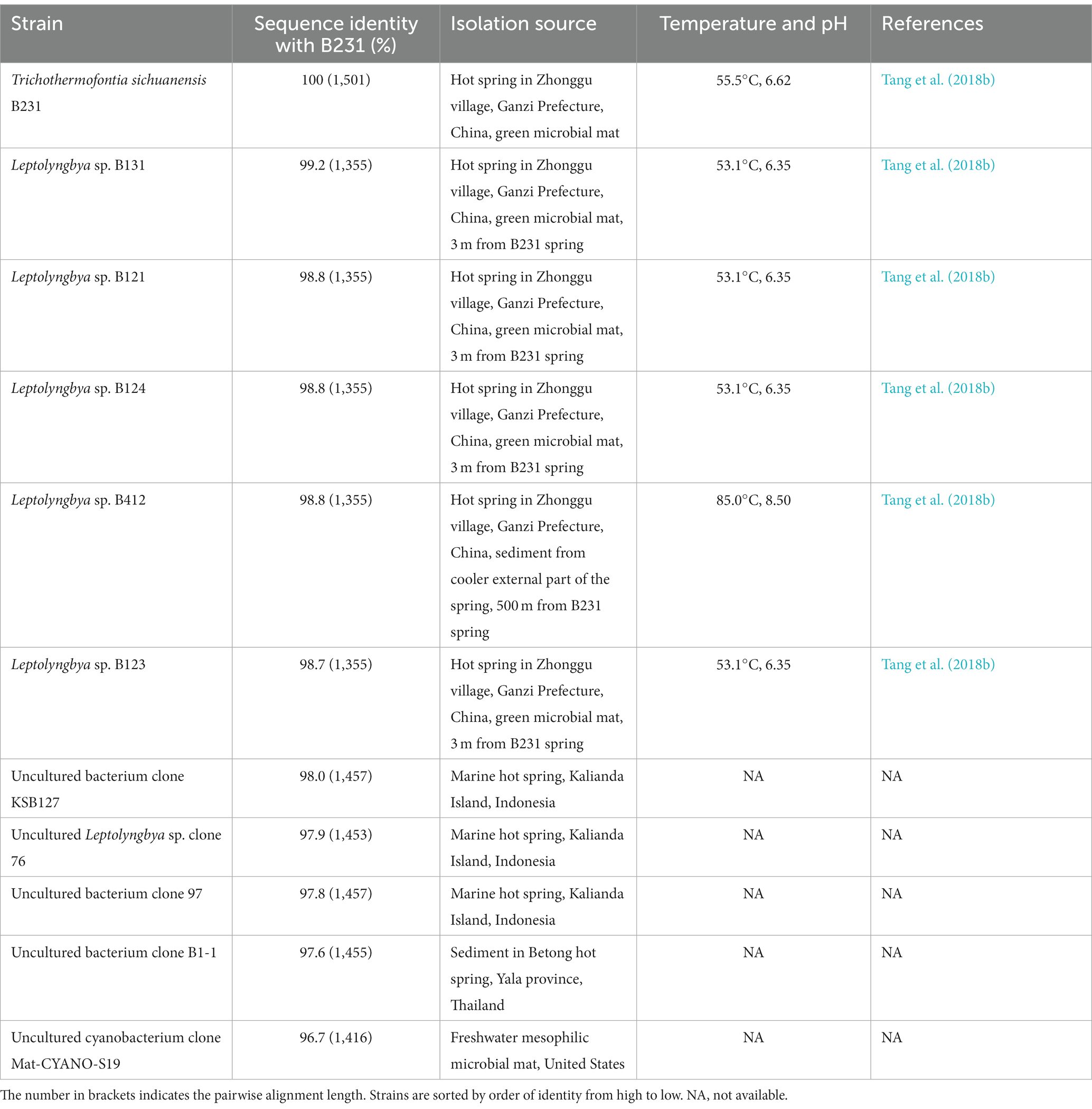
Table 1. The sequence identities of 16S rRNA gene between strain B231 and other closely related strains.
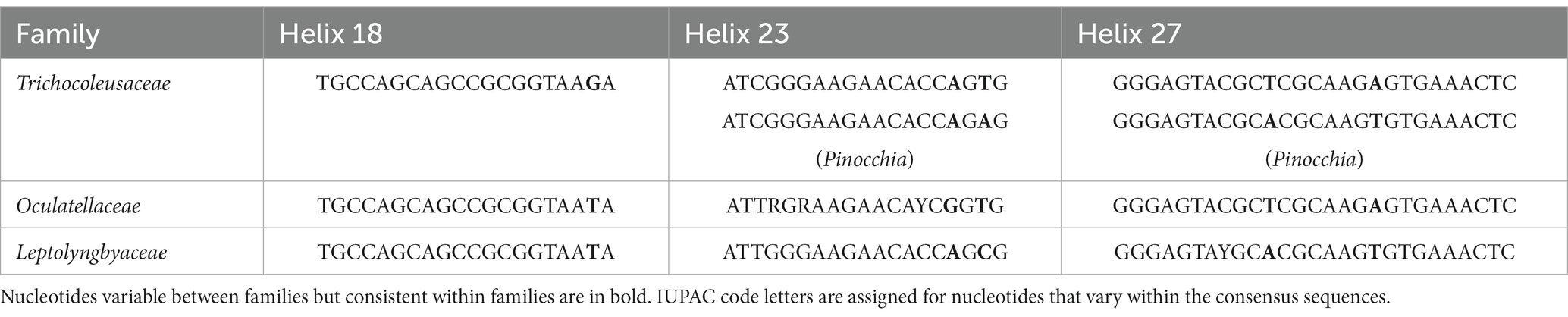
Furthermore, the helices within 16S rRNA have been studied. Only helices 18, 23 and 27 were investigated as they were considered the most informative for family distinction (Mai et al., 2018). As shown in Table 2, one distinctive nucleotide was found in helix 18 between Trichocoleusaceae and Oculatellaceae/Leptolyngbyaceae. Within helix 23, two types of distinctive nucleotides were present in Trichocoleusaceae, both capable of differentiating them from Oculatellaceae and Leptolyngbyaceae. However, the distinctive nucleotides in helix 27 were shared among families. Taken together, several nucleotide positions in different helices were considered informative indicators of family-level classification. Again, these results confirm the recognition of the novel clade and Pinocchia to join Trichocoleus as members of the family Trichocoleusaceae.
Within the novel clade, all the strains show high 16S rRNA sequence identities (97.6%–99.2%) to the strain B231 except for clone Mat-CYANO-S19 (Table 1). Therefore, according to the recommended threshold of 16S rRNA gene identity for bacterial species (98%–99%) or genera (94.5%–95%) demarcation (Yarza et al., 2014), clone Mat-CYANO-S19 can be proposed to be new species and the remaining 10 strains to be another new species. Such delineation was also supported by the 16S rRNA phylogeny (Figure 1). Besides, the novel clade shows 92.8%–93.5% of 16S rRNA sequence identities to Trichocoleus strains and 91.3%–92.9% to Pinocchia strains. This result was consistent with the taxonomic delineation of this novel clade as a new genus member within the family Trichocoleusaceae.
Intriguingly, the habitat niches were quite distinct among strains from the novel clade (Table 1). The six strains isolated from China were originally recovered from three separate freshwater hot springs located at a high altitude (3,200 m; Tang et al., 2018a,b). The other five strains, for which only their molecular signatures were available, also mostly show thermal origin. Three were from marine hot springs in Indonesia, one from a mesophilic microbial mat in the United States and one from a hot spring sediment in Thailand. The distinct habitat niches suggested that these strains might be classified into at least three ecotypes. The ecotype determination of uncultured bacterium clone B1-1 appears impractical in light of the minimal ecological information. In addition, the acclimation of these strains to diverse habitats strongly suggests the underlying genetic diversity and a wide distribution among representatives of this genus. Verifying this speculation will be an interesting topic that could be explored using phylogenomic and ecogenomic approaches providing that more isolates and genome sequences are available. Unfortunately, the data on uncultured strains were restricted to molecular signatures isolated from environments, hindering further detailed comparisons (e.g., physiological, ecological and genomic studies) that would require isolates.
Secondary structures of 16S-23S ITS
In the present study, the phylogeny of 16S-23S ITS has not been reconstructed mainly for two reasons. First, the 16S-23S ITS of important reference cyanobacteria, Pinocchia strains, only contained conserved tRNAlle, while another conserved tRNAAla was missing (Dvorak et al., 2015). Second, the 16S-23S ITS of strain B231 lacks the V2 region (Table 3); indicating significant operon heterogeneity in these strains. Taken together, the current dataset of 16S-23S ITS sequences was extraordinarily divergent and would most likely result in a misleading taxonomic assignment (Johansen et al., 2011). However, sequence comparisons of 16S-23S ITS were still performed among representatives from the three families. Excluding two highly conserved tRNAs from ITS sequences, the length of the remaining ITS sequences tremendously varied from 230 to 535 bp (Table 3). Such an immense discrepancy was primarily attributed to the length differences in D1-D1’ (51–141 bp), V2 (0–218 bp), boxB (33–70 bp), and V3 (21–161 bp). Within family Trichocoleusaceae, enormous variations were also observed among genera (Table 3). As a result, the nucleotide differences of these domains result in divergences of the secondary structures among representative strains from family Trichocoleusaceae (Supplementary Figure 2).
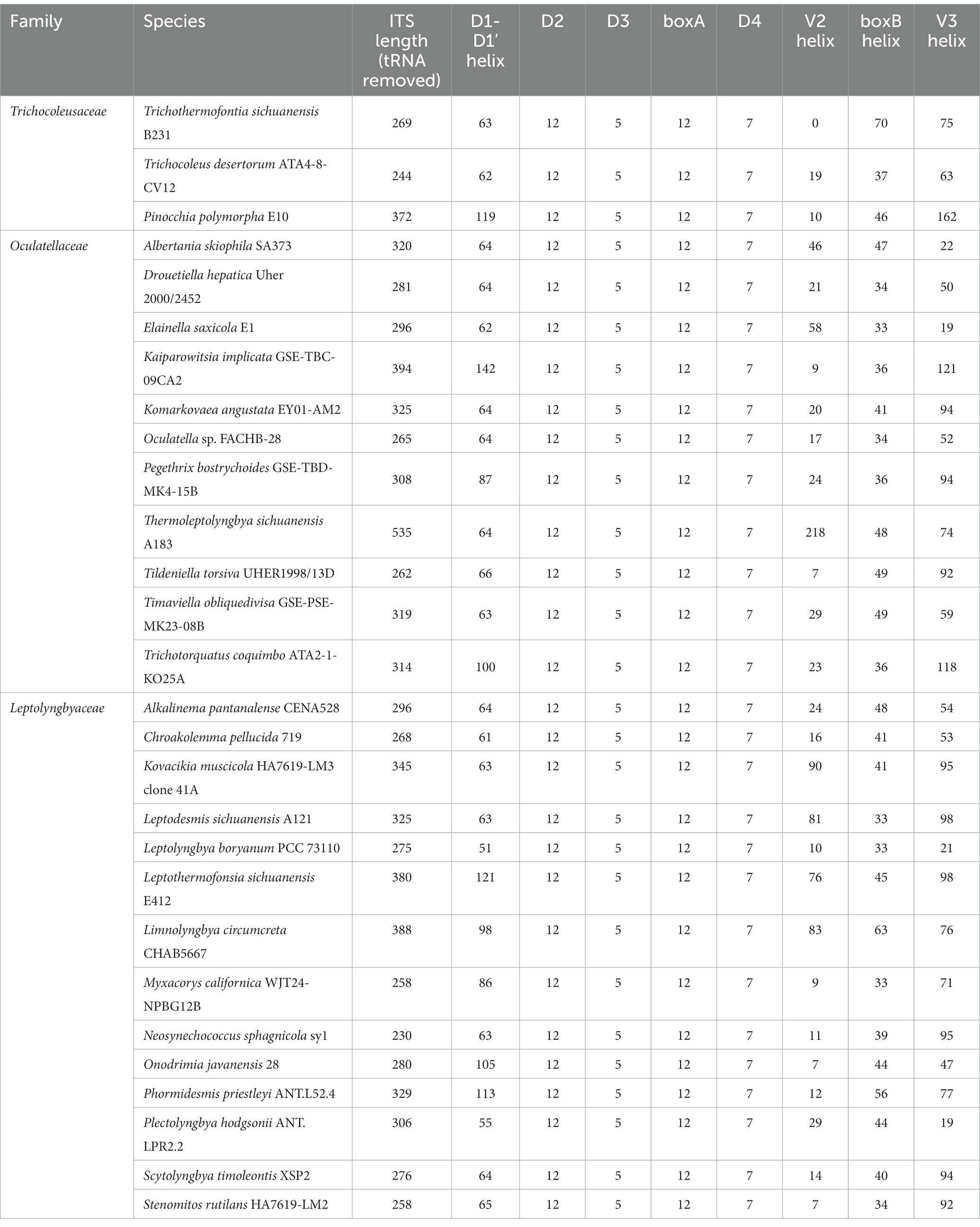
Table 3. The Length summary (bp) of corresponding regions within 16S-23S ITS of strain B231 and focal taxa.
The inferred D1-D1’ helix of strain B231 differs from the other two inferred structures (Supplementary Figure 2A). The most similar structure to D1-D1’ helix of strain B231 was that of T. desertorum. Both D1-D1’ helices vary in the overall length and topology while retaining the basal stem structures of five base pairs followed by a right asymmetrical loop.
V2 helix was absent in the 16S-23S ITS of strain B231. A similar structure of the V2 helix was shared by the other two representative strains from the family Trichocoleusaceae, comprising one stem and a hairpin loop (Supplementary Figure 2B). However, the V2 helices of the two strains differ in the stem length and residues of hairpins.
All three strains share a basal stem structure (AGCA-TGCT) in boxB helices (Supplementary Figure 2C). Strain B231 shows a much longer residue length than the other two strains (Table 3), resulting in a divergent boxB helix structure.
The V3 helix of strain B231 consists of an asymmetrical loop and a 4-residue hairpin loop, and two stems (Supplementary Figure 2D). The V3 helix of strain B231 was different from those of the other two strains, while a basal stem structure (TGTC-GACA) was shared by all the strains.
Conclusively, the phylogeny of 16S rRNA and the result of 16S-23S ITS secondary structure analysis supports the delineation of strain B231 as a novel genus within this family.
Genome comparisons
To our best knowledge, there are no genomic-level comparisons within the family Trichocoleusaceae or between members of the family Trichocoleusaceae and genera from other different families. Therefore, it was crucial to elucidate a snapshot of genomic divergences between Strain B231 and focal taxa. Herein, three indices of whole genome comparisons between strain B231 and representative species from Trichocoleusaceae, Oculatellaceae and Leptolyngbyaceae were presented (Table 4).
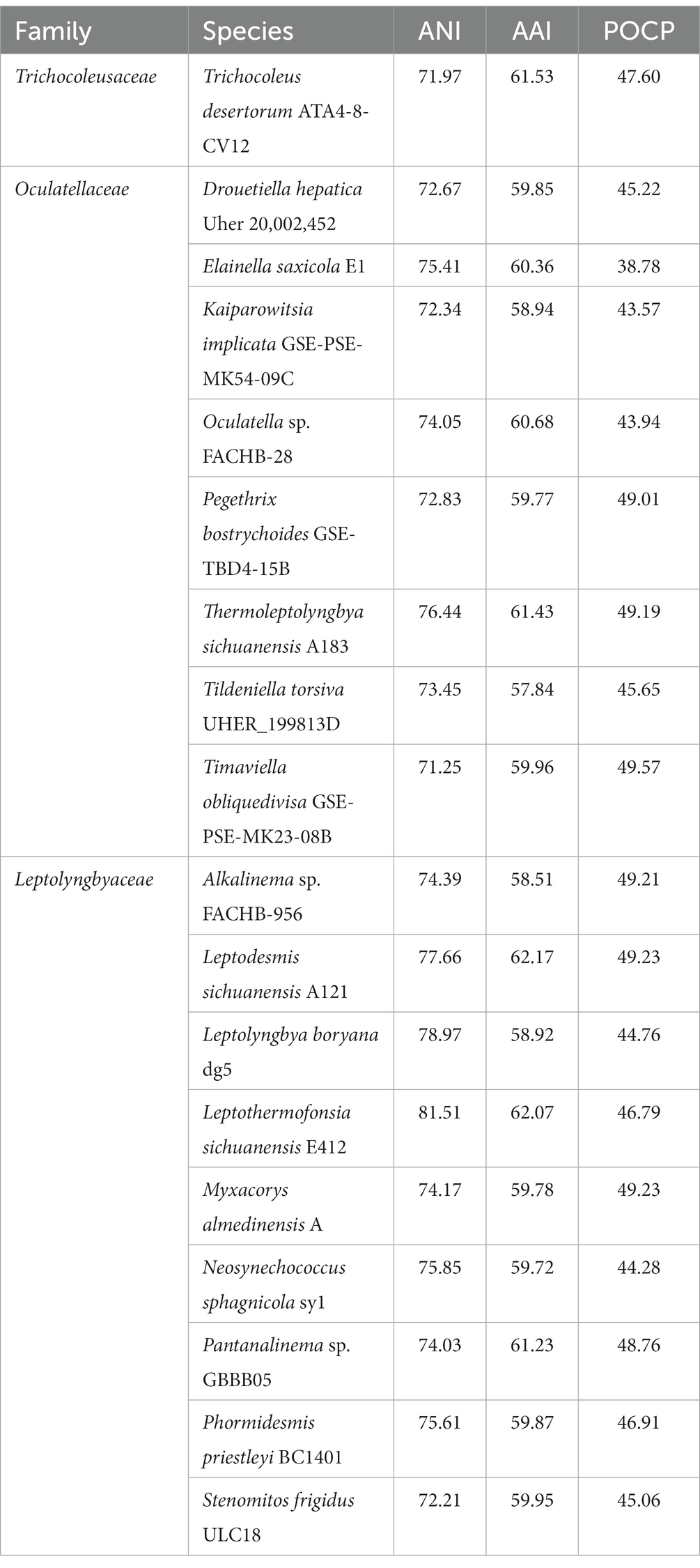
Table 4. Indices values (%) of whole genome comparisons between strain B231 and representative species from Trichocoleusaceae, Oculatellaceae and Leptolyngbyaceae.
Considerable divergences in genomes were observed between strain B231 and the other 18 strains, as revealed by the ANI and AAI values (Table 4). The ANI and AAI values were less than 82 and 63%, respectively. This result conforms to the suggested values for genus (ANI < 83%, AAI ≤ 70%) delimitation (Walter et al., 2017; Jain and Rodriguez, 2018). However, in some cases, misleading results might be achieved for the classification of the prokaryotic genus using ANI or AAI (Pannekoek et al., 2016). Therefore, the POCP specific for genus delineation was also calculated between strain B231 and the other focal taxa. The POCP values range from 38.78% to 49.23% (Table 4), all within the threshold (<50%) for the definition of a prokaryotic genus (Qin et al., 2014). Taken together, all the results verify the genus demarcation of strain B231 as a novel genus within the family Trichocoleusaceae. This conclusion was in accordance with the results of molecular phylogeny (Figure 1).
Phylogenomic analysis
Analysis of homologous gene clusters generated 647 single-copy genes shared by the genomes studied. The concatenated alignments of these genes possess 216,534 aligned amino acid sites. Using the optimal substitution model (LG + F + R5), the ML inference of the supergene alignment provides a phylogeny with strong bootstrap support for all branches (Figure 2), defining representative species from each described genus. Within the Trichocoleusaceae clade, strain B231 is quite divergent from the described genus Trichocoleus and Neosynechococcus. Strain B231 is well-separated by the long branches from other described genera in the family, suggesting the taxon as a novel genus.
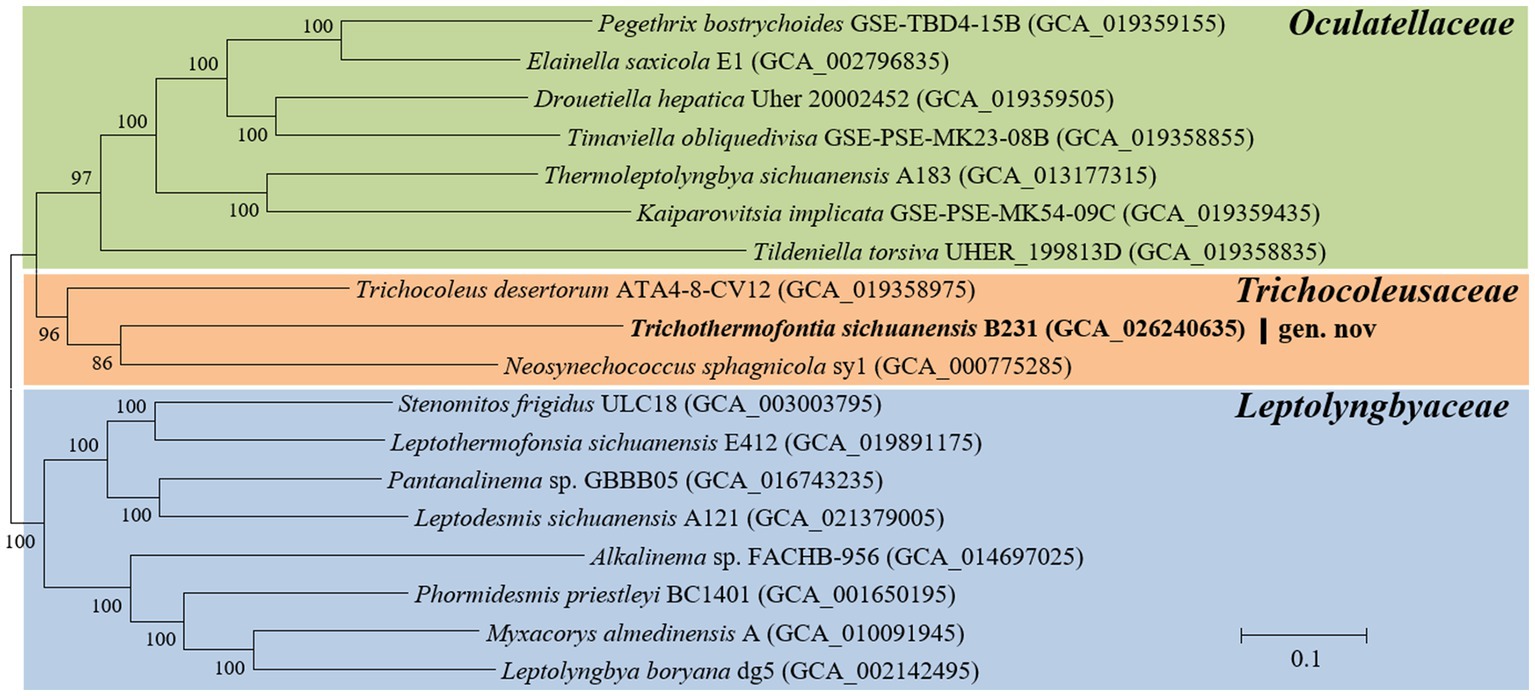
Figure 2. ML phylogenomic inference of concatenated protein alignment of 647 single-copy genes shared by all genomes. Strain no. in bold represent the strain identified in this study. Bootstrap values are indicated at nodes. Scale bar = 10% substitutions per site.
The phylogenomic topology was almost identical to that of the 16S rRNA gene (Figure 1). However, one exception was present in the phylogram. The phylogenetic analysis of the 16S rRNA gene suggests the clear affiliation of Neosynechococcus sphagnicola sy1 to the family Leptolyngbyaceae (Figure 1; Dvorak et al., 2014), whereas sy1 was located in a position within the genome-scale phylogram, forming a clade with two genera from family Trichocoleusaceae (Figure 2). This position of the strain was consistent with the phylogenomic results reported in previous studies (Tang et al., 2022a,b). Taken together, the discordant two trees suggest that currently used approaches cannot determine whether the taxonomic position of sy1 is accurate. The inconsistent phylogenies imply that an extensive study of this organism encompassing phenotypical, chemotaxonomical, physiological and genotypical studies should be carried out.
Classification of CCM in strain B231
The aquatic environments where most thermophilic cyanobacteria live usually suffer from low availability of CO2, primarily due to external factors, e.g., temperature, pH and gas exchange. On the other hand the availability of soluble carbonates is highly variable depending on the geochemistry of the habitat. Therefore, it was crucial to investigate the molecular component and organization of CCM of thermophilic strains in relation to their habitats.
Phylogenetic analysis of the molecular marker, rbcL, was frequently used to indicate the carboxysome type of cyanobacteria (Klanchui et al., 2017). Herein, the ML phylogram of rbcL positions strain B231 within the category of β-cyanobacteria (Figure 3), indicating the presence of RubisCO 1B form in this thermophile. This result was in accordance with a previous study that surveyed 17 thermophilic cyanobacteria and allocated them to the β-cyanobacteria (Tang et al., 2022d). Nevertheless, strain B231 clusters with none of these thermophilic cyanobacteria but is uniquely positioned among mesophilic cyanobacteria (Figure 3), suggesting considerable genetic diversity of rbcL amino acid sequences among these thermophiles. Furthermore, the evolutionary relationship based on habitats or morphology cannot be elucidated from the present rbcL phylogram, suggesting that the phylogenetic inference of rbcL may be unsuitable for elaborating the relationship between evolution and cyanobacterial habitat and environments (Komárek, 2016).
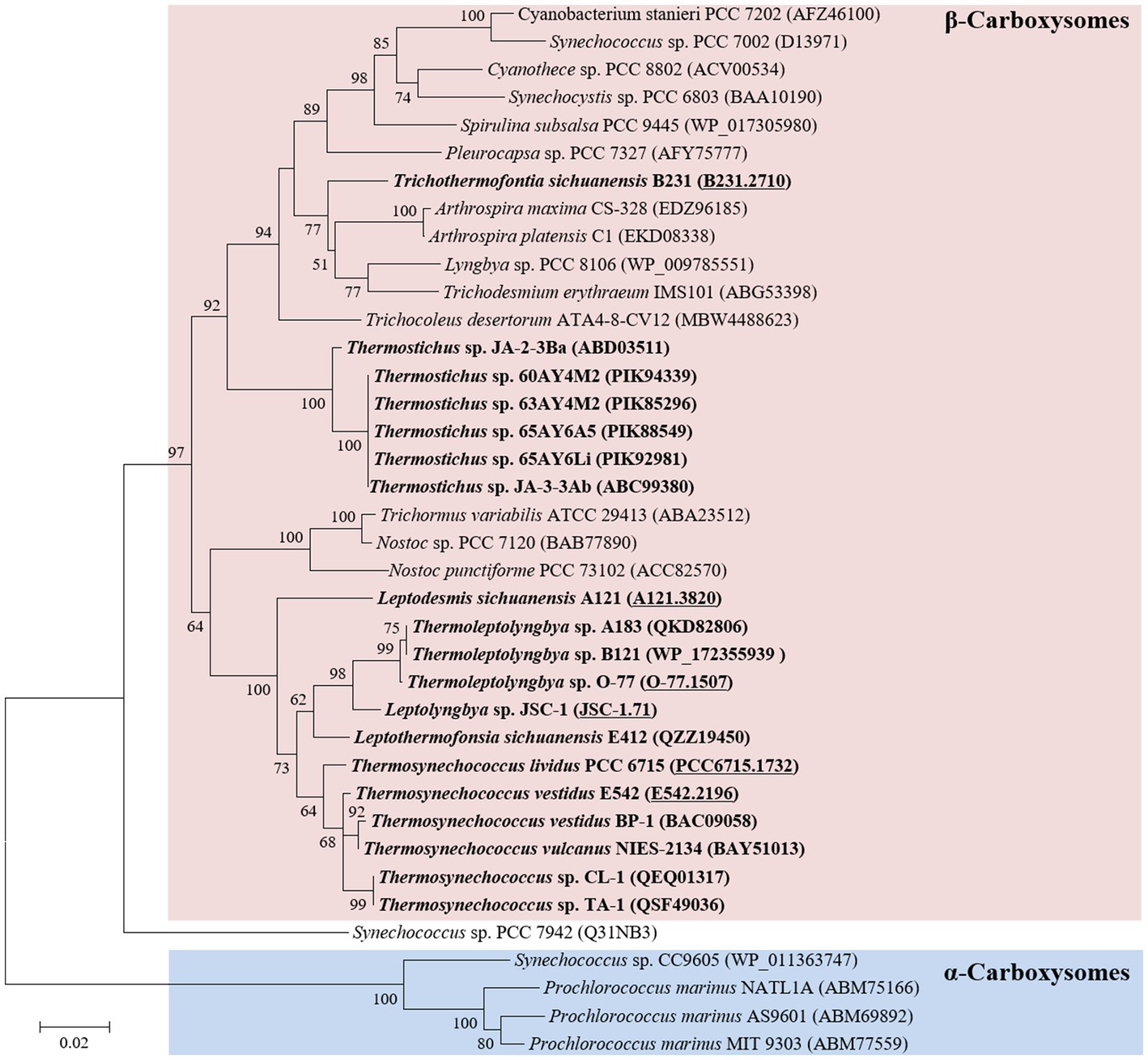
Figure 3. ML phylogenetic inference of RubisCO large subunit protein sequences. The thermophilic cyanobacterium investigated in this study and previously reported thermophiles are indicated in bold. The accession numbers underscored refer to the gene IDs in Supplementary Table 1 or the reference (Tang et al., 2022d).
Genes encoding Ci uptake systems in strain B231
In general, the cyanobacterial CCM comprises two primary components: Ci uptake systems and carboxysomes. Cyanobacteria have been reported to have up to five different systems to actively acquire and transport Ci into the cells. Two for the uptake of CO2 and three for the transport of HCO3− (Pronina et al., 2017). Herein, three different transport systems (Figure 4) have been identified in strain B231, including two CO2 uptake systems, NDH-13 and NDH-14 complex, and one HCO3− transport system, BicA. Existence of both CO2 uptake systems in strain B231 aligns with the previous studies indicating that these CO2 transporters were present in β-cyanobacteria living in freshwater, brackish or eutrophic lakes (Durall and Lindblad, 2015). This is in sharp contrast to oceanic α-cyanobacteria (e.g., Prochlorococcus species) and marine β-cyanobacteria (e.g., Trichodesmium species) that contain only one or even lack them entirely (Pronina et al., 2017). Taken together, the presence of NDH-13 and NDH-14 might be relevant to the environments where these cyanobacteria inhabit. Indeed, a low-CO2 inducible high-affinity CO2 uptake system (NDH-13 complex) and a constitutive low-affinity CO2 uptake system (NDH-14 complex), may allow the thermophilic cyanobacteria more alternative strategies to survive in environments with significant CO2 fluctuation, particularly in hot springs. In addition, the protein sequences of five genes (ndhD4, ndhF4, cupB, ndhD3, ndhF3) encoding the two CO2 uptake systems exhibit different identities with the sequences of non-thermophilic reference cyanobacteria (Synechocystis PCC 6803), ranging from 60.4 to 68.6%, but a high degree of homology (84.4% identity) to cupA gene of NDH-13 complex.
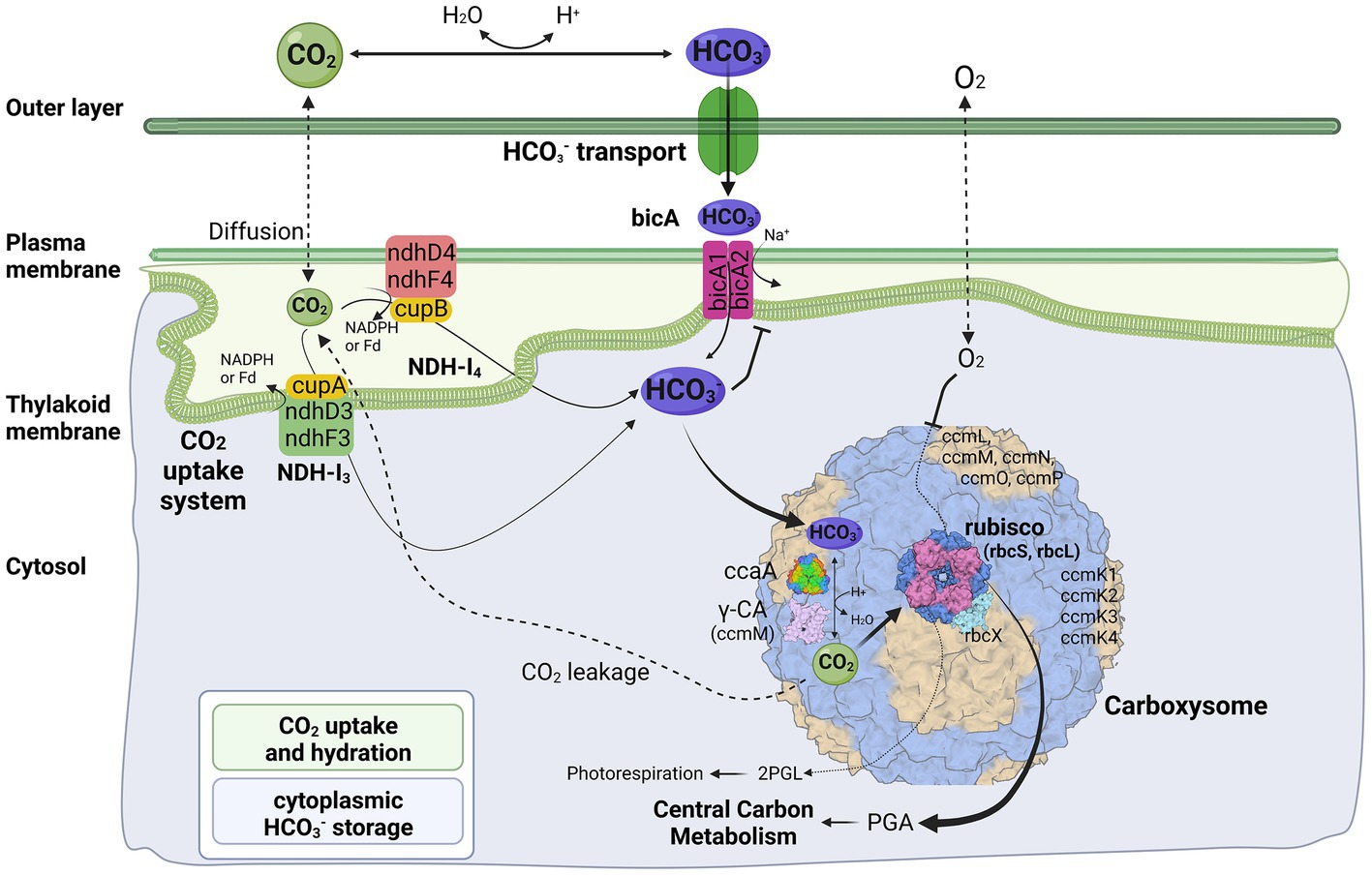
Figure 4. Molecular components of CCM for Trichothermofontia sichuanensis B231. 2PGL, 2-phosphoglycolate; PGA, phosphoglyceric acid. Three-dimensional structures of proteins visualized using related structures of 2YBV, 6OWF, 5SWC, 5BS1, 6KI1. Figure created with BioRender.com.
Two homologs of BicA were present in strain B231 (Figure 4). However, the protein sequences of the two homologs show distinct identities to that of Synechocystis PCC 6803, namely 66.5% and 36.7%. Such discrepancy in CDS of bicA genes in strain B231 requires future studies to elucidate the possible evolutionary processes and functional differences. In addition, the HCO3− transport systems, sbt regulator and BCT1, were not present in strain B231. The absence of sbt regulator in strain B231 was consistent with the previous finding that the thermophilic cyanobacteria typically have bicA rather than sbt as suggested by their dominant presence. At the same time, a lack of BCT1 was observed in several thermophilic cyanobacteria, e.g., Thermoleptolyngbya and Thermosynechococcus strains (Tang et al., 2022d). Meanwhile, thermophiles without both sbt regulator and BCT1 were rare (Tang et al., 2022d). This result suggests that the BicA in strain B231 may be sufficient to manage the transport of dissolved inorganic carbon in its habitat.
Genes encoding carboxysomes in strain B231
The cyanobacterial carboxysomes comprise protein shells and two encapsulated enzymes, RubisCO and carbonic anhydrase (CA; Kerfeld and Melnicki, 2016). For β-cyanobacteria, shell proteins are normally encoded by ccmKLMNO operon and ccmP (Melnicki et al., 2021). Strain B231 contains ccmK1-4 (Figure 4), a typical gene set in the β-cyanobacterial genome (MacCready et al., 2020). It is known that ccmK1 and ccmK2 are the main structural proteins of the carboxysome shell and share high sequence conservation (Cai et al., 2016). Phylogenetic analysis suggests that the two homologs of ccmK1/2 in strain B231 form a separate cluster, and both group into the clade of ccmK2 (Figure 5A). Although 10 amino acid-long C-terminal extension of ccmK1 can distinguish ccmK1 and ccmK2 (Kerfeld et al., 2005), the remaining part of ccmK1/2 protein sequences share a similarity as high as 92.2%, leading to their assembly into one cluster. The structural and functional specificity of the two ccmK genes requires future careful investigation. The ccmK3 and ccmK4 of strain B231 were relatively divergent from other cyanobacteria, as suggested by long branches and substitution rates (Figures 5B,C). The ccmK3 and ccmK4 of strain B231 show a moderate degree of homology to that of Synechocystis PCC 6803, 61.5% and 74.5%, respectively. The ccmK3/K4 present in the strain B231 may function as adjusting the properties of carboxysome for rapid adaptation to environmental changes in thermal regions through, e.g., expanding the range of permeability properties of metabolite channels in carboxysome shells (Sommer et al., 2019).
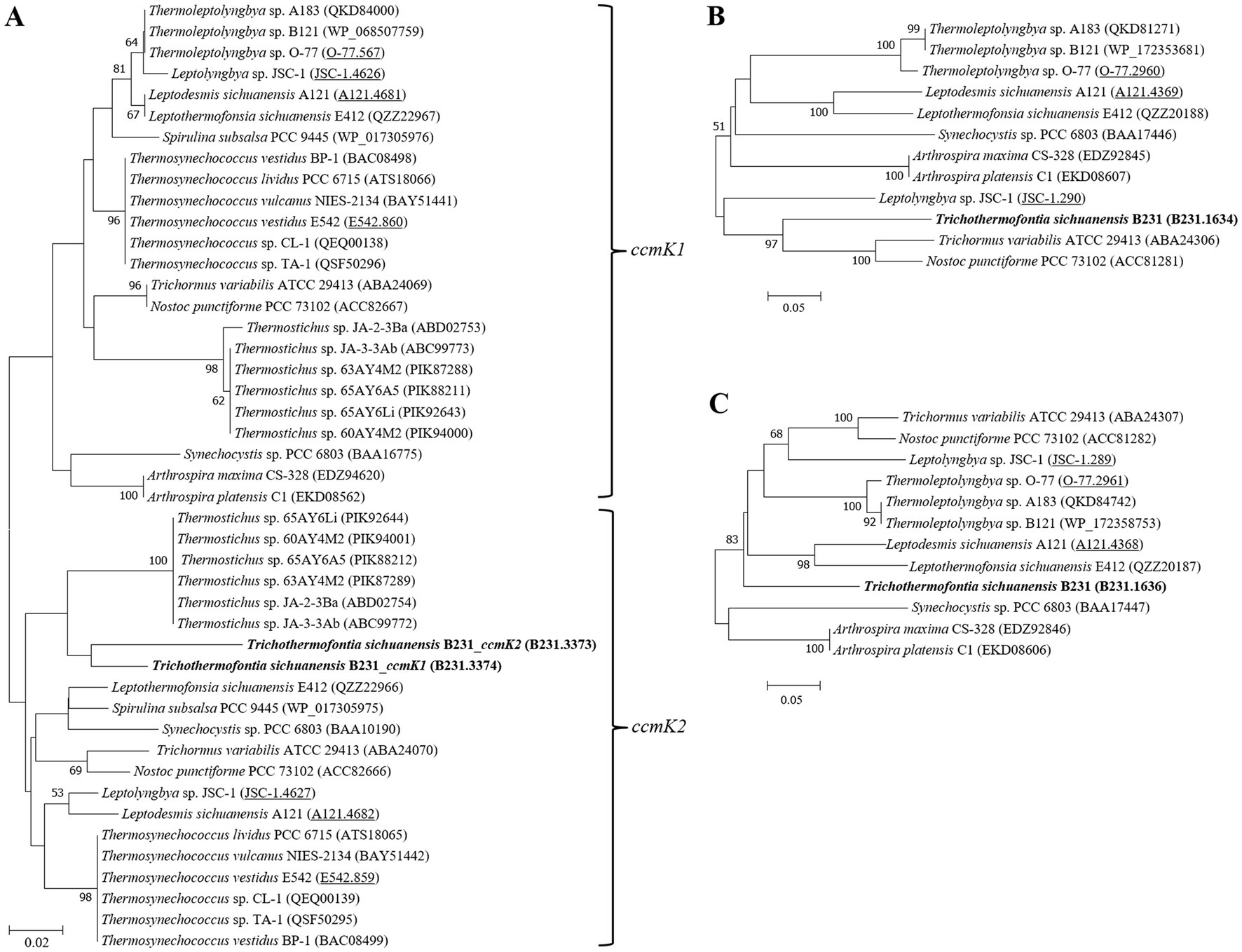
Figure 5. ML phylogenetic inference of protein sequences of ccmK1/2 (A), ccmK3 (B), and ccmK4 (C). The thermophilic cyanobacterium investigated in this study is indicated in bold. The accession numbers underscored refer to the gene IDs in Supplementary Table 1 or the reference (Tang et al., 2022d).
Apart from ccmK, other genes encoding carboxysome shell proteins, ccmL, -M, -N, -O, and -P, were also present in strain B231 (Figure 4). Only ccmM and ccmN show weak homologs (46.2, 32.1%) to that of Synechocystis PCC 6803, while the other three genes exhibit an identity of 62.2%, 61.4%, and 72.0%, respectively. Phylogenetic analysis of these genes suggests extensive genetic divergence between strain B231 and reference cyanobacteria, as suggested by the assignments of strain B231 into separate branches (Supplementary Figure 3).
The carboxysomal CA catalyzes the conversion of HCO3− into CO2, a substrate for RubisCO. The subunits of RubisCO, rbcL and rbcS, and RubisCO assembly chaperone, rbcX, were present in strain B231 (Figure 4). The rbcL of strain B231 shows high sequence conservation to Synechocystis PCC 6803 (89.7% identity), whereas moderate conservation (66.4% identity) was observed in rbcS. The protein sequence of rbcX was more divergent, showing 58% identity with the reference protein. As for CA, only one type of β-CA, carboxysomal ccaA (Figure 4), was present in strain B231, with a similarity of 56.4% to the reference protein. Moreover, Strain B231 shows a similar primary structure (Supplementary Figure 4) of amino acid residues in γ-CA-like domain of ccmM to that with CA activity in Thermosynechococcus BP-1 (Peña et al., 2010) and Nostoc PCC 7120 (de Araujo et al., 2014), indicating that the ccmM protein of strain B231 may possess CA activity. Thus, the CA activities of ccmM and ccaA might confer strain B231 with more alternative strategies for regulating carboxysome function. Intriguingly, previous studies (Peña et al., 2010; de Araujo et al., 2014; Tang et al., 2022d) suggest that the CA activity of ccmM (γ-CA) was usually present in cyanobacteria lacking ccaA, while cyanobacteria comprising both ccmM and ccaA are likely to have non-functional γ-CA domain and only ccaA function as CA activity. Future experimental studies should be performed to elucidate the function and relationship of the two identified proteins with potential CA activity in strain B231.
Genomic organization of CCM-related genes in strain B231
Investigation of the genomic organization of CCM-related genes in strain B231 may provide insights into the function and evolution of these genes. As shown in Figure 6, genes encoding each NDH-1 complex separately cluster together. The two homologs of bicA genes are distantly located on the genome (Figure 6). Regarding the gene organization of the carboxysomal shell proteins, a typical main carboxysome locus (MCL) was present. It contained ccmK1/2 genes, followed by ccmL, -M, -N, and -O (Figure 6). The sequential arrangement of ccmK2 and ccmK1 in the MCL may facilitate protein complex assembly or balancing of the shell protein stoichiometry during the translation of MCL genes (Cai et al., 2016), while ccmK, -L, -M, -N, and -O may be co-regulated in an MCL as an operon (Rae et al., 2013; Billis et al., 2014; Sommer et al., 2017). In addition to the MCL, the other three shell proteins form two satellite loci, ccmP and an operon with ccmK3/4 genes. The separated organization of K1/K2 and K3/K4 paralogs may be associated with the structural segregation of the two groups (Sommer et al., 2019). Moreover, the expression of ccmK3/4 from a satellite locus may increase the flexibility of carboxysome shell assembly and permeability (Sommer et al., 2019), and may provide differing metabolite selectivities (Sommer et al., 2017). The localization of genes encoding rbcL, rbcS and rbcX for RubisCO was consistent with the previous findings that the three genes always cluster in an operon in β-cyanobacteria (Badger et al., 2002). In addition, the RubisCO gene cluster was remote from the MCL, suggesting that this gene cluster was a satellite locus to MCL and may conduct independent expression regulation. The role of rbcX in thermophilic cyanobacteria is still uncertain and requires future investigations.
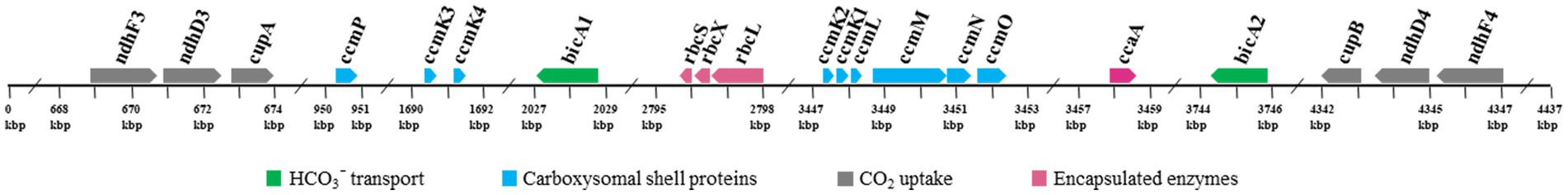
Figure 6. Genomic organization of CCM-related genes for Trichothermofontia sichuanensis B231. Solid arrow boxes refer to genes and the direction of transcription.
Comparative analysis indicates that strain B231 shows a distinct molecular component to the phylogenetically closely related neighbor, Trichocoleus ATA4-8-CV12 (Supplementary Table 2). For the CO2 uptake systems, strain B231 has one set of genes encoding NDH-14 complex, while Trichocoleus ATA4-8-CV12 has two sets of homologs. The constitutive low-affinity CO2 uptake system (NDH-14 complex) with an extra set of genes may be indispensable for the strain to survive in its terrestrial origin (desert) where gaseous CO2 is the primary carbon source (Mühlsteinova et al., 2014) and bicarbonate is less available, or simply the aftermath of strain evolutionary history. More studies are needed on the topic of multiple paralogs of carbon uptake systems in cyanobacteria. Both strains have only one type of HCO3− transport system, namely BicA in strain B231 and BCT1 in strain ATA4-8-CV12. A different type of HCO3− transport system could be attributed to the distinct traits of the two transporters and the habitat of the two strains. First, BicA exhibits low affinity and high flux rate to HCO3− and the genes encoding BicA were found to be primarily constitutively expressed (Price et al., 2004). Therefore, the BicA transport system might be necessary for strain B231 to acclimatize to the exogenous HCO3− in its habitat (pH 6.62) where HCO3− remains dominant dissolved inorganic specie between pH 6 and 10. Second, BCT1 encoded by cmpABCD operon shows a high affinity for HCO3− and was induced under low levels of Ci and enhanced by high light conditions (Omata et al., 2002). Thus, strain ATA4-8-CV12 may use BCT1 to compensate for the functions of constitutive genes (e.g., BicA) for adaptation to high-light conditions in deserts. In addition, strain B231 has one type of β-CA (ccaA) and ccmM (γ-CA) with CA activity, whereas Trichocoleus ATA4-8-CV12 contains two types of β-CA (ccaA and ecaB) and its ccmM appears to have non-functional γ-CA domain (Supplementary Figure 4). The presences of other genes related to CCM are consistent with the two strains. However, the phylogenetic analysis suggests that the protein sequences are divergent (Figures 3, 5; Supplementary Figure 3). Though the genome of Trichocoleus ATA4-8-CV12 was only assembled as a draft, preliminary analysis suggests that its clustering pattern of CCM-related genes in the genome was in accordance with that of strain B231.
Overall, the molecular component and organization of strain B231 were similar to that of other reported thermophilic cyanobacteria (Supplementary Table 2; Tang et al., 2022d). However, distinct characteristics were evident in strain B231. A relatively low diversity of bicarbonate transporters distinguishes strain B231 from other thermophilic strains, each possessing at least two HCO3− transport systems (Tang et al., 2022d). The low diversity may be related to the habitat of strain B231 where a higher variety of transporters with similar functionalities is not required. In addition, strain B231 contains a higher abundance of different types of carboxysomal CAs, β-CA (ccaA) and γ-CA (ccmM), compared to other thermophilic strains (β-CA or γ-CA), but still below those of other freshwater cyanobacteria that possess the abundance of CAs, α-/β-/γ-CAs (Tang et al., 2022d). The CA activities of ccmM and ccaA may provide strain B231 with alternative strategies to ensure that the RubisCO with low affinity for CO2 was surrounded by high CO2 levels and regularly function regardless of thermal stress in this habitat. Strain B231 with freshwater origin lacks the BCT1 transporter, which is near-ubiquitous in freshwater cyanobacteria. Moreover, strain B231 shows a similar composition of carboxysome shell proteins to mesophilic cyanobacteria, the diversity of which was higher than many thermophilic strains lacking at least one of the four ccmK genes (Tang et al., 2022d).
Morphological and physiological features of strain B231
The primary physiological characteristics of the B231 strain were assessed by monitoring its growth at the temperature of 47°C in derivatives of BG-11 medium; the unmodified medium, supplemented with bicarbonate up to the concentration of 1.0 M, and nitrogen-free. The strain could grow in all tested nitrogen conditions, showing optimal growth with moderate (a third of the regular BG-11 medium nitrate content) to high nitrogen availability (five times the regular BG-11 medium nitrate content) and in the presence of 17.65 mM nitrite. Interestingly, despite the lack of heterocytes the strain could grow in a nitrogen-free medium, which is consistent with the presence of the nifHDK gene cluster and associated genes required for molecular nitrogen fixation. The strain grew optimally from ambient up to the 0.1 M bicarbonate concentration. Higher loads of dissolved inorganic carbon had a deleterious effect on the strain, concurrent with its highly restricted bicarbonate transport system revealed earlier. The results are summarized in Table 5.
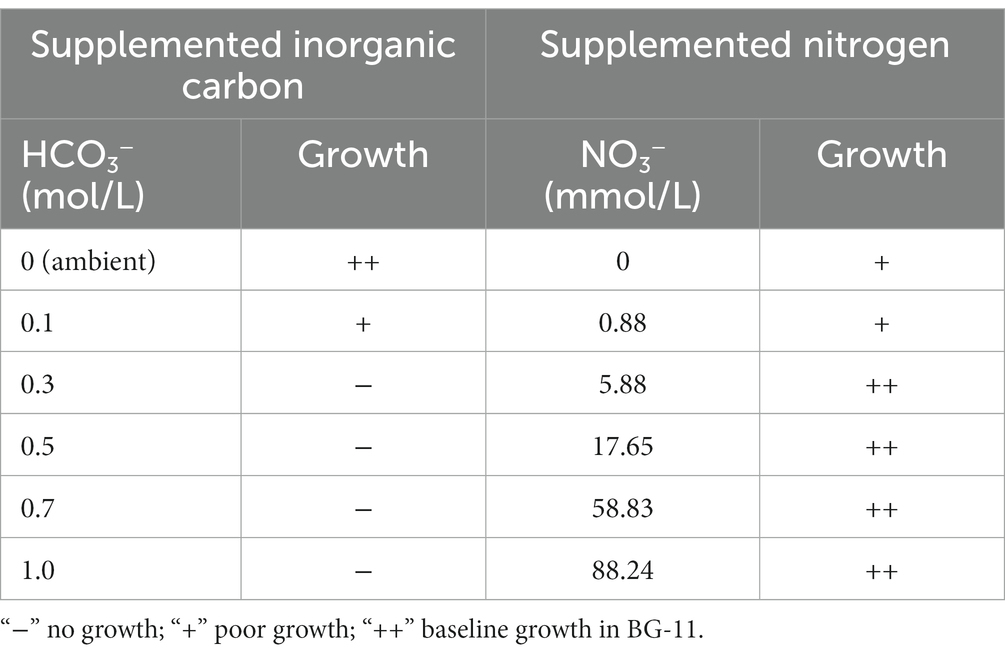
Table 5. Physiological characteristics of Trichothermofontia B231 grown in 47°C, 150 μmol m−2 s−1 under varying carbonate and nitrate concentrations.
Light, scanning, and transmission microscopy were utilized for the morphological description and delineation of strain B231 as the representative of a new genus and to illustrate its key characteristics. The morphology of the strain is presented in Figure 7. The unicyanobacterial culture consists of blue-green filaments (Figures 7A,B), sometimes forming clumps or flat mats when grown in stationary liquid cultures. Trichomes of the cells grown in the liquid medium are solitary or in colony-like mats, straight or bent, moderately entangled or curved, and contain elongated cylindrical barrel-shaped cells terminated with mostly sharply pointed, conical apical cells (Figures 7A,C). Cells grown on a solid medium had multiple trichomes in one sheath (Figure 7B).
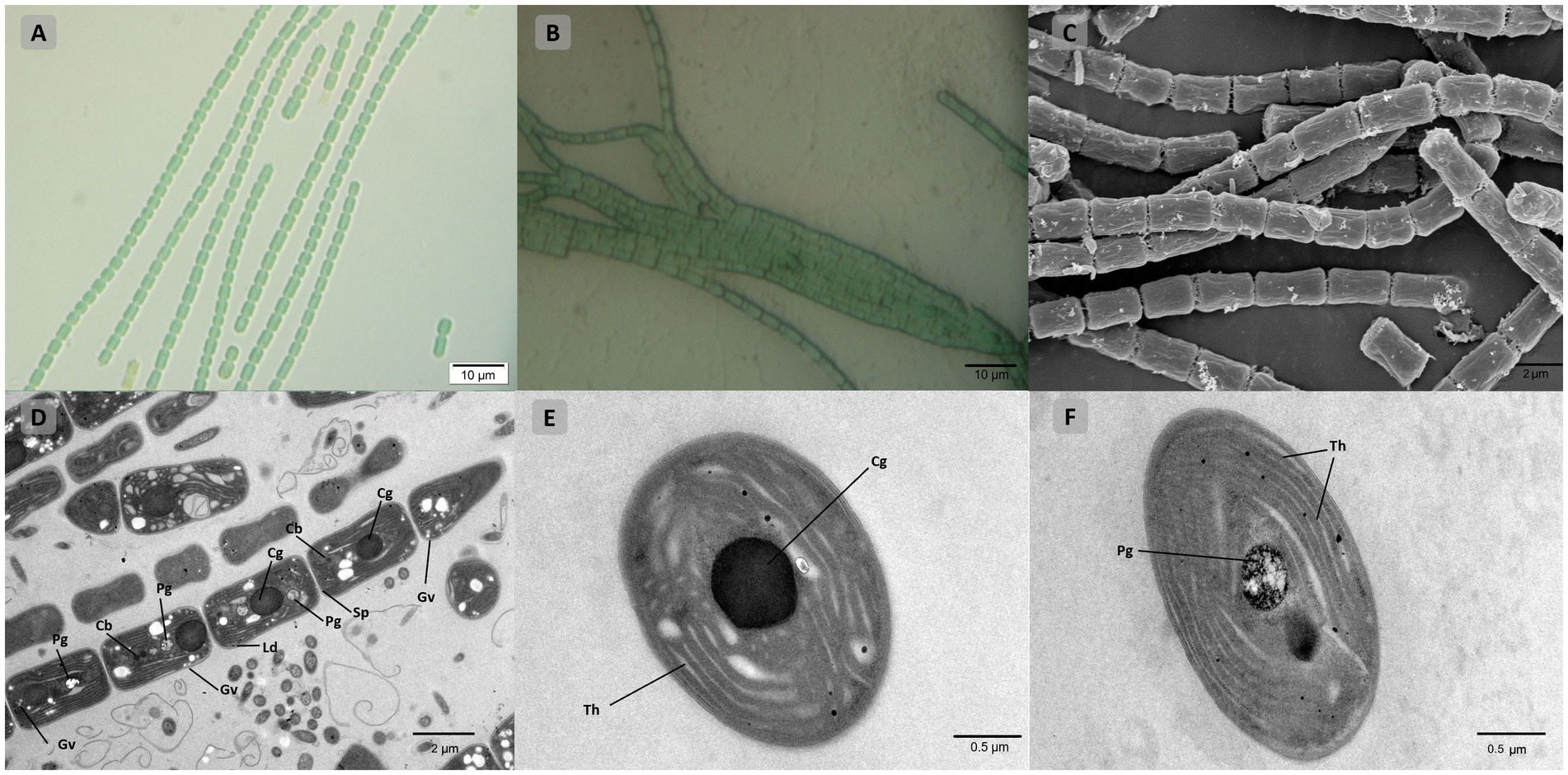
Figure 7. Microscopic morphology of Trichothermofontia B231. (A) Light microscopy image of liquid-medium grown cultures; (B) light microscopy image of solid-medium grown cultures; (C) SEM image; (D–F) TEM images. Cb, carboxysome; Cg, cyanophycin granule; Ld, lipid droplets; Pg, polyphosphate granule; Sp, septum; Th, thylakoid membrane, Gv, gas vesicles. Magnifications were 1,000× (A,B); 5,000× (C,D); 20,000× (E,F).
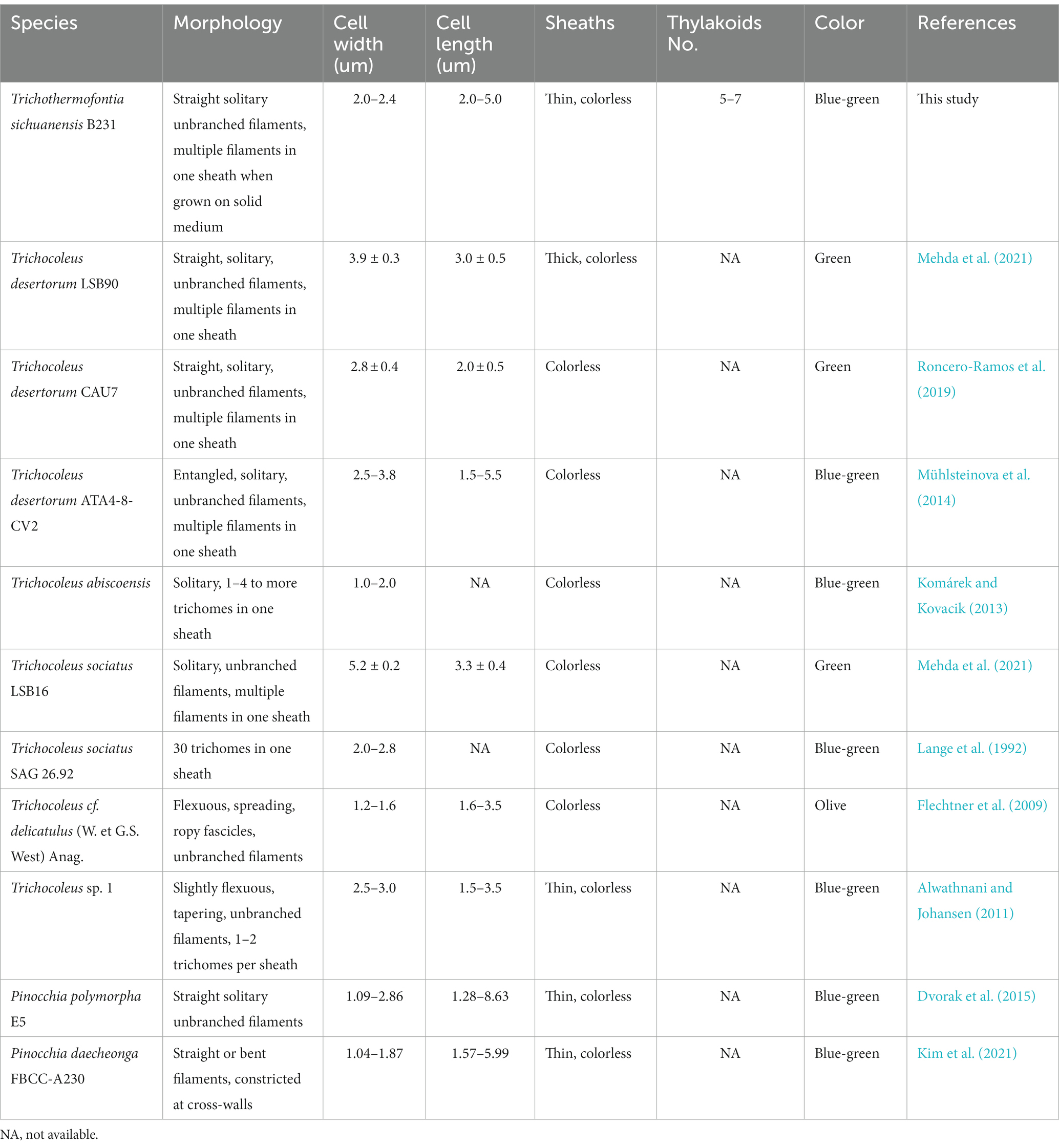
Cells of the strain were longer than they were wide (2.0–2.4 μm wide, 3.4–5.0 μm long), with a length-to-width ratio ranging from 1.4–2.0, and with an irregular ratio in apical cells. The trichomes of this strain were thinner than those of other members of Trichocoleus (Table 6), which typically have length-to-width below 1.0 and Pinocchia with very diverse measurements for both cell lengths and widths. The sheath was not evident in liquid cultures but more apparent in solidified medium, cells were connected with hyaline bridges, slightly constricted at poorly visible cross-walls, with the content mostly homogeneous (Figures 7A,B). The number of thylakoids (Figures 7E,F) varies from 5 to 8 layers, and peripheral thylakoids were arrayed layer by layer, parallel to the long axis of the filament, occasionally with an irregular arrangement. The presence of large cyanophycin granules, often in the center of the cell, characterized the cytoplasm (Figures 7D,E). The relevant biosynthetic genes encoding cyanophycin synthase (cphA) and cyanophycinase (cphB) were identified in the genome. The carboxysomes were observed in the cytoplasm in small numbers, similar to related Pinocchia strains (Dvorak et al., 2015; Kim et al., 2021). Gas vesicles and polyphosphate granules were also observed in the cytoplasm (Figure 7D). Gas vesicles were composed of cylindrical structures, typically two proteins GvpA forming the vesicle core and GvpC providing structural support and were responsible for the shape of gas vesicles (Miklaszewska et al., 2012). Analysis of the B231 genome confirms the presence of the gvpA, gvpC genes. The other genes involved in gas vesicle formation were also identified.
As shown in Table 6, the overall morphology of strain B231 was more similar to Pinocchia spp. (Dvorak et al., 2015; Kim et al., 2021) than to other members of the Trichocoleusaceae family (Lange et al., 1992; Flechtner et al., 2009; Alwathnani and Johansen, 2011; Komárek and Kovacik, 2013; Roncero-Ramos et al., 2019; Mehda et al., 2021). This difference can be linked to the habitat. The family appears to have two morphologies, probably driven by environmental conditions. The primarily terrestrial strains, such as T. desertorum, have a very thick sheath capable of maintaining multiple trichomes. Meanwhile, freshwater strains, such as Pinocchia spp. and the B231, exhibit phenotypic plasticity showing marginal sheath when grown in liquid cultures and thick sheath when grown on a solid medium.
Conclusion
In this manuscript, the results of 16S rRNA phylogeny, secondary structures of 16S-23S ITS and strain’s morphology strongly supported the B231 as a novel genus within Trichocoleusaceae and its delineation as a representative of this new taxon. Phylogenomic inference and three genome-based indices (ANI, AAI, and POCP) also support the delineation at the genus level. Consequently, we have proposed a new genus Trichothermofontia sichuanensis gen. et sp. nov. Moreover, based on the results, we suggest family-level revision of Pinocchia from Leptolyngbyaceae to Trichocoleusaceae. The delineation was strongly supported at the molecular level, and its morphological distinction from Trichocoleus sp. can be attributed to the different habitats (terrestrial vs. freshwater). In addition, the obtained complete genome of the newly delineated Trichothermofontia B231 facilitates the elucidation of genetic basis regarding genes related to CCM. The strain belongs to β-cyanobacteria according to its β-carboxysome shell protein with 1B form of RubisCO. Compared to other thermophilic strains, strain B231 contains a relatively low diversity of bicarbonate transporters (only BicA for HCO3− transport) but a higher abundance of different types of carbonic anhydrase (CA), β-CA (ccaA) and γ-CA (ccmM). Strain B231 with freshwater origin lacks the BCT1 transporter, which is consistently possessed by freshwater cyanobacteria. Furthermore, strain B231 shows a similar composition of carboxysome shell proteins (ccmK1-4, ccmL, -M, -N, -O, and -P) to mesophilic cyanobacteria, the diversity of which was higher than many thermophilic strains lacking at least one of the four ccmK genes. The genomic distribution of CCM-related genes suggests various regulations of expression. Overall, the first complete genome of a new genus representative obtained in this study provides insight into the genomic features, CCM components, and the fundamental information for future global taxogenomic, ecogenomic and geogenomic studies.
Taxonomic treatment and description of Trichothermofontia sichuanensis Daroch, Tang, and Zhou et al. gen. et sp. nov.
Phylum: Cyanobacteria
Order: Synechococcales
Family: Trichocoleusaceae
Genus: Trichothermofontia, gen. nov.
Description: Filaments solitary, entangled, lacking false branching, forming small or short fragments in a nutritional deficiency state. Sheath colorless, thin. Trichomes straight or curved, terminated with apical, rounded cells, with invisible crosswalls that separate each cell. Cells mostly longer than wide, with multilayer peripheral thylakoids.
Etymology: Gr. fem. n. thrix (gen. trichos), hair; Gr. masc. Adj. thermos, hot; L. masc. n. fons (gen. fontis). “Tricho” referring to Greek word for hair, representing exhibiting typical thin morphology of trichome, “thermo” referring to Greek word for heat and representing thermophilic (high temperature tolerant) character of the strain, “fontia”—a genus epithet derived from the Latin word fons meaning spring, since all representative of the genus to date were hot spring isolates.
Type species: Trichothermofontia sichuanensis Daroch, Tang, and Zhou et al. sp. nov.
Trichothermofontia sichuanensis Daroch, Tang, and Zhou et al. gen. et. sp. nov.
Diagnosis: Differing from other species of the genus based on the 16S rRNA sequence identity.
Description Colony bright green, flat and tightly packed in a recognizable interleaved or tangled reticular morphology on agar plates. Filaments long or short, blue-green, no branching, typically solitary straight or bent, moderately entangled or curved (Figures 7A,B). Trichomes contained elongated cylindrical barrel-shaped cells terminated with conical, sharply pointed apical cells, mat-forming in stationary liquid culture, yielding solitary filaments when shaken. Cells rectangular in cross-section, connected with hyaline bridges. Intracellular connections between vegetative cells not present. Cells typically longer (3.4–4.0 μm), than wide (2.0–2.4 μm), with a length-to-width ratio ranging from 1.4 to 2.0 (Figures 7A–D), with peripheral thylakoids arrayed in 5 to 8 layers (Figures 7E,F). The sheath minimal in liquid cultures (Figure 7A), and visible in solid-medium grown cultures (Figure 7B). Carboxysomes observed in small numbers. Large centrally positioned cyanophycin granule (Figures 7D,E), and membrane-delimited vesicles (polyphosphate granules and gas vesicles) in the cytoplasm. Development of heterocytes not observed, genetic toolkit for molecular nitrogen fixation detected.
Etymology: “sichuanensis” species epithet derives from the name of the collection province is B231 (=FACHB-3573).
Type locality: Thermal spring, Zhonggu village in Ganzi Prefecture of Sichuan Province, China.
Ecology of type locality: the sample occurred as a macroscopic dark green mat attached to the rock.
Habitat: Thermal springs in Ganzi Prefecture of Sichuan Province, China (30°36′39″ N, 101°41′9″ E; Supplementary Figure 5).
Holotype here designated: The dried inactive holotype was deposited in the Herbarium of North Minzu University with the voucher number: NMU00231 (contact: Lei Zhang, zhangsanshi-0319@163.com).
Reference strain: The culture of Trichothermofontia sichuanensis Daroch, Tang, and Zhou et al. gen. et sp. nov. was initially denoted and deposited in Peking University Algae Collection as PKUAC-SCTB231 has also been deposited in the Freshwater Algae Culture Collection at the Institute of Hydrobiology (FACHB-collection) with accession number FACHB-3573 as Trichocoleusaceae sp. species after identification and authentication based on the full-length sequencing of the 16S rRNA gene along with folding of the secondary structures of the 16S–23S ITS region. After proper identification and authentication, the culture is maintained in the FACHB under the accession number FACHB-3573.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JT: conceptualization, methodology, validation, formal analysis, investigation, data curation, writing-original draft, writing-review and editing, visualization, supervision, project administration, and funding acquisition. HZ: formal analysis, investigation, data curation, and writing-original draft. YJ: formal analysis, investigation, data curation, and writing-review and editing. DY: formal analysis, investigation, and data curation. KW: methodology, validation, data curation, and writing-review and editing. L-MD: methodology, software, and data curation. MD: conceptualization, methodology, resources, data curation, writing-original draft, writing-review and editing, supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (31970092, 32071480, and 3221101094) and Tenure-Track Fund to MD. Funding bodies had no influence over the design and execution of this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1111809/full#supplementary-material
Footnotes
References
Alcorta, J., Alarcón-Schumacher, T., Salgado, O., and Díez, B. (2020). Taxonomic novelty and distinctive genomic features of hot spring cyanobacteria. Front. Genet. 11:568223. doi: 10.3389/fgene.2020.568223
Alwathnani, H., and Johansen, J. R. (2011). Cyanobacteria in soils from a mojave desert ecosystem. Monogr. West. N. Am. Nat. 5:19. doi: 10.3398/042.005.0103
Badger, M. R., Hanson, D., and Price, G. D. (2002). Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 29, 161–173. doi: 10.1071/PP01213
Billis, K., Billini, M., Tripp, H. J., Kyrpides, N. C., and Mavromatis, K. (2014). Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation. PLoS One 9:e109738. doi: 10.1371/journal.pone.0109738
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Cai, F., Bernstein, S. L., and Wilson, S. C. (2016). Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol. 170, 1868–1877. doi: 10.1104/pp.15.01822
Chen, M.-Y., Teng, W.-K., Zhao, L., Hu, C.-X., Zhou, Y.-K., Han, B.-P., et al. (2021). Comparative genomics reveals insights into cyanobacterial evolution and habitat adaptation. ISME J. 15, 211–227. doi: 10.1038/s41396-020-00775-z
Cheng, Y. I., Lin, C., Chiu, Y. F., Hsueh, H. T., and Chu, H. A. (2020). Comparative genomic analysis of a novel strain of Taiwan hot-spring cyanobacterium Thermosynechococcus sp. CL-1. Front. Microbiol. 11:82. doi: 10.3389/fmicb.2020.00082
Cordeiro, R., Luz, R., Vasconcelos, V., Gonçalves, V., and Fonseca, A. (2020). Cyanobacteria phylogenetic studies reveal evidence for polyphyletic genera from thermal and freshwater habitats. Diversity 12:298. doi: 10.3390/d12080298
de Araujo, C., Arefeen, D., Tadesse, Y., Long, B. M., Price, G. D., Rowlett, R. S., et al. (2014). Identification and characterization of a carboxysomal γ-carbonic anhydrase from the cyanobacterium Nostoc sp. PCC 7120. Photosynth. Res. 121, 135–150. doi: 10.1007/s11120-014-0018-4
Drummond, A. J., Ho, S. Y., Phillips, M. J., and Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. doi: 10.1371/journal.pbio.0040088
Durall, C., and Lindblad, P. (2015). Mechanisms of carbon fixation and engineering for increased carbon fixation in cyanobacteria. Algal Res. 11, 263–270. doi: 10.1016/j.algal.2015.07.002
Dvorak, P., Hindak, F., Hasler, P., Hindakova, A., and Poulickova, A. (2014). Morphological and molecular studies of Neosynechococcus sphagnicola, gen. Et sp. nov. (cyanobacteria, Synechococcales). Phytotaxa 170, 024–034. doi: 10.11646/phytotaxa.170.1.3
Dvorak, P., Jahodářová, E., Hasler, P., Gusev, E., and Poulíčková, A. (2015). A new tropical cyanobacterium Pinocchia polymorpha gen. Et sp. nov. derived from the genus Pseudanabaena. Fottea 15, 113–120. doi: 10.5507/fot.2015.010
Esteves-Ferreira, A. A., Inaba, M., Fort, A., Araújo, W. L., and Sulpice, R. (2018). Nitrogen metabolism in cyanobacteria: metabolic and molecular control, growth consequences and biotechnological applications. Crit. Rev. Microbiol. 44, 541–560. doi: 10.1080/1040841X.2018.1446902
Flechtner, V., Johansen, J., and Belnap, J. (2009). The biological soil crusts of the san nicolas island: enigmatic algae from a geographically isolated ecosystem. West. N. Am. Nat. 68, 405–436. doi: 10.3398/1527-0904-68.4.405
Galmés, J., Hermida-Carrera, C., Laanisto, L., and Niinemets, Ü. (2016). A compendium of temperature responses of Rubisco kinetic traits: variability among and within photosynthetic groups and impacts on photosynthesis modeling. J. Exp. Bot. 67, 5067–5091. doi: 10.1093/jxb/erw267
Galmés, J., Kapralov, M. V., Copolovici, L. O., Hermida-Carrera, C., and Niinemets, Ü. (2015). Temperature responses of the Rubisco maximum carboxylase activity across domains of life: phylogenetic signals, trade-offs, and importance for carbon gain. Photosynth. Res. 123, 183–201. doi: 10.1007/s11120-014-0067-8
Gelman, A., and Rubin, D. B. (1992). Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 457–472.
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Iteman, I., Rippka, R., Tandeau, D. M. N., and Herdman, M. (2000). Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 146, 1275–1286. doi: 10.1099/00221287-146-6-1275
Jain, C., and Rodriguez, R. L. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Johansen, J. R., Kovacik, L., Casamatta, D. A., Iková, K. F., and Kaštovský, J. (2011). Utility of 16S-23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: Leptolyngbya corticola sp. nov. (Pseudanabaenaceae, cyanobacteria). Nova Hedwigia 92, 283–302. doi: 10.1127/0029-5035/2011/0092-0283
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kerfeld, C. A., and Melnicki, M. R. (2016). Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31, 66–75. doi: 10.1016/j.pbi.2016.03.009
Kerfeld, C. A., Sawaya, M. R., Tanaka, S., Nguyen, C. V., Phillips, M., Beeby, M., et al. (2005). Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938. doi: 10.1126/science.1113397
Kim, D.-H., Choi, H. J., Ki, J.-S., and Lee, O.-M. (2021). Pinocchia daecheonga sp. nov. (Synechococcales, cyanobacteria) isolated from a Daecheong Lake in Geum River, Republic of Korea. Phytotaxa 510, 135–147. doi: 10.11646/phytotaxa.510.2.2
Klanchui, A., Cheevadhanarak, S., Prommeenate, P., and Meechai, A. (2017). Exploring components of the CO2-concentrating mechanism in alkaliphilic cyanobacteria through genome-based analysis. Comput. Struct. Biotechnol. J. 15, 340–350. doi: 10.1016/j.csbj.2017.05.001
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Komárek, J. (2016). A polyphasic approach for the taxonomy of cyanobacteria: principles and applications. Eur. J. Phycol. 51, 346–353. doi: 10.1080/09670262.2016.1163738
Komárek, J., Kaštovský, J., Mares, J., and Johansen, J. (2014). Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 86, 295–335.
Komárek, J., and Kovacik, L. (2013). Schizotrichacean cyanobacteria from Central Spitsbergen (Svalbard). Polar Biol. 36, 1811–1822. doi: 10.1007/s00300-013-1402-9
Kono, M., Martinez, J., Sato, T., and Haruta, S. (2022). Draft genome sequence of the thermophilic unicellular cyanobacterium Synechococcus sp. strain C9. Microbiol. Resour. Announc. 11:e0029422. doi: 10.1128/mra.00294-22
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lange, O. L., Kidron, G. J., Budel, B., Meyer, A., and Abeliovich, E. K. (1992). Taxonomic composition and photosynthetic characteristics of the biological soil crusts' covering sand dunes in the western Negev Desert. Funct. Ecol. 6, 519–527. doi: 10.2307/2390048
Li, L., Stoeckert, C. J. Jr., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Liang, Y., Tang, J., Luo, Y., Kaczmarek, M. B., Li, X., and Daroch, M. (2019). Thermosynechococcus as a thermophilic photosynthetic microbial cell factory for CO2 utilisation. Bioresour. Technol. 278, 255–265.
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
MacCready, J. S., Basalla, J. L., and Vecchiarelli, A. G. (2020). Origin and evolution of carboxysome positioning systems in cyanobacteria. Mol. Biol. Evol. 37, 1434–1451. doi: 10.1093/molbev/msz308
Mai, T., Johansen, J. R., Pietrasiak, N., Bohunicka, M., and Martin, M. P. (2018). Revision of the Synechococcales (cyanobacteria) through recognition of four families including Oculatellaceae fam. Nov. and Trichocoleaceae fam nov. and seven new genera containing 14 species. Phytotaxa 365, 1–59. doi: 10.11646/phytotaxa.365.1.1
Mehda, S., Muñoz-Martín, M., Oustani, M., Hamdi-Aïssa, B., Perona, E., and Mateo, P. (2021). Microenvironmental conditions drive the differential cyanobacterial community composition of biocrusts from the sahara desert. Microorganisms 9:487. doi: 10.3390/microorganisms9030487
Melnicki, M. R., Sutter, M., and Kerfeld, C. A. (2021). Evolutionary relationships among shell proteins of carboxysomes and metabolosomes. Curr. Opin. Microbiol. 63, 1–9. doi: 10.1016/j.mib.2021.05.011
Miklaszewska, M., Waleron, M., Morin, N., Calusinska, M., Wilmotte, A., Tandeau De Marsac, N., et al. (2012). Elucidation of the gas vesicle gene clusters in cyanobacteria of the genus Arthrospira (Oscillatoriales, Cyanophyta) and correlation with ITS phylogeny. Eur. J. Phycol. 47, 233–244. doi: 10.1080/09670262.2012.692817
Miller, S. R., Castenholz, R. W., and Pedersen, D. (2007). Phylogeography of the thermophilic cyanobacterium Mastigocladus laminosus. Appl. Environ. Microbiol. 73, 4751–4759. doi: 10.1128/AEM.02945-06
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mühlsteinova, R., Johansen, J. R., Pietrasiak, N., Martin, M. P., Osorio-Santos, K., and Warren, S. D. (2014). Polyphasic characterization of Trichocoleus desertorum sp. nov. (Pseudanabaenales, cyanobacteria) from desert soils and phylogenetic placement of the genus Trichocoleus. Phytotaxa 163, 241–261. doi: 10.11646/phytotaxa.163.5.1
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, 733–745. doi: 10.1093/nar/gkv1189
Omata, T., Takahashi, Y., Yamaguchi, O., and Nishimura, T. (2002). Structure, function and regulation of the cyanobacterial high-affinity bicarbonate transporter, BCT1. Funct. Plant Biol. 29, 151–159. doi: 10.1071/PP01215
Pannekoek, Y., Qi-Long, Q., Zhang, Y.-Z., and van der Ende, A. (2016). Genus delineation of Chlamydiales by analysis of the percentage of conserved proteins justifies the reunifying of the genera chlamydia and Chlamydophila into one single genus chlamydia. Pathogens Dis 74:ftw071. doi: 10.1093/femspd/ftw071
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Patel, A., Matsakas, L., Rova, U., and Christakopoulos, P. (2019). A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Bioresour. Technol. 278, 424–434. doi: 10.1016/j.biortech.2019.01.063
Peña, K. L., Castel, S. E., de Araujo, C., Espie, G. S., and Kimber, M. S. (2010). Structural basis of the oxidative activation of the carboxysomal γ-carbonic anhydrase, CcmM. Proc. Natl. Acad. Sci. 107, 2455–2460. doi: 10.1073/pnas.0910866107
Price, G. D., Woodger, F. J., Badger, M. R., Howitt, S. M., and Tucker, L. (2004). Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. 101, 18228–18233. doi: 10.1073/pnas.0405211101
Pronina, N. A., Kupriyanova, E. V., and Igamberdiev, A. U. (2017). “Photosynthetic carbon metabolism and CO2-concentrating mechanism of cyanobacteria” in Modern topics in the phototrophic prokaryotes: Metabolism, bioenergetics, and omics. ed. P. C. Hallenbeck (Cham: Springer International Publishing), 271–303.
Qin, Q., Xie, B., Zhang, X., Chen, X., Zhou, B., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Raabova, L., Kovacik, L., Elster, J., and Strunecky, O. (2019). Review of the genus Phormidesmis (cyanobacteria) based on environmental, morphological, and molecular data with description of a new genus Leptodesmis. Phytotaxa 395, 1–16. doi: 10.11646/phytotaxa.395.1.1
Rae, B. D., Long, B. M., Badger, M. R., and Price, G. D. (2013). Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379. doi: 10.1128/MMBR.00061-12
Roncero-Ramos, B., Muñoz-Martín, M., Chamizo, S., Fernández-Valbuena, L., Mendoza, D., Perona, E., et al. (2019). Polyphasic evaluation of key cyanobacteria in biocrusts from the most arid region in Europe. PeerJ 7:e6169. doi: 10.7717/peerj.6169
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shalygin, S., Shalygina, R., Redkina, V., Gargas, C., and Johansen, J. (2020). Description of Stenomitos kolaenensis and S. hiloensis sp. nov. (Leptolyngbyaceae, cyanobacteria) with an emendation of the genus. Phytotaxa 440, 108–128. doi: 10.11646/phytotaxa.440.2.3
Sommer, M., Cai, F., Melnicki, M., and Kerfeld, C. A. (2017). β-Carboxysome bioinformatics: identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints. J. Exp. Bot. 68, 3841–3855. doi: 10.1093/jxb/erx115
Sommer, M., Sutter, M., Gupta, S., Kirst, H., Turmo, A., Lechno-Yossef, S., et al. (2019). Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of Beta carboxysome shells. Plant Physiol. 179, 156–167. doi: 10.1104/pp.18.01190
Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Tang, J., Du, L., Li, M., Yao, D., Waleron, M., Waleron, K. F., et al. (2022a). Characterization of a novel hot-spring cyanobacterium Leptodesmis sichuanensis sp. nov. and genomic insights of molecular adaptations into its habitat. Front. Microbiol. 12:739625. doi: 10.3389/fmicb.2021.739625
Tang, J., Du, L.-M., Liang, Y.-M., and Daroch, M. (2019). Complete genome sequence and comparative analysis of Synechococcus sp. CS-601 (SynAce01), a cold-adapted cyanobacterium from an oligotrophic Antarctic habitat. Int. J. Mol. Sci. 20:152. doi: 10.3390/ijms20010152
Tang, J., Jiang, D., Luo, Y., Liang, Y., Li, L., Shah, M. M. R., et al. (2018a). Potential new genera of cyanobacterial strains isolated from thermal springs of western Sichuan, China. Algal Res. 31, 14–20. doi: 10.1016/j.algal.2018.01.008
Tang, J., Li, L., Li, M., Du, L., Shah, M. R., Waleron, M., et al. (2021). Description, taxonomy, and comparative genomics of a novel species, Thermoleptolyngbya sichuanensis sp. nov., isolated from Hot Springs of Ganzi, Sichuan, China. Front. Microbiol. 12:696102. doi: 10.3389/fmicb.2021.696102
Tang, J., Liang, Y., Jiang, D., Li, L., Luo, Y., Shah, M. M. R., et al. (2018b). Temperature-controlled thermophilic bacterial communities in hot springs of western Sichuan, China. BMC Microbiol. 18:134. doi: 10.1186/s12866-018-1271-z
Tang, J., Shah, M. R., Yao, D., Du, L., Zhao, K., Li, L., et al. (2022b). Polyphasic identification and genomic insights of Leptothermofonsia sichuanensis gen. Sp. nov., a novel thermophilic cyanobacteria within Leptolyngbyaceae. Front. Microbiol. 13:765105. doi: 10.3389/fmicb.2022.765105
Tang, J., Yao, D., Zhou, H., Du, L., and Daroch, M. (2022c). Reevaluation of Parasynechococcus-like strains and genomic analysis of their microsatellites and compound microsatellites. Plan. Theory 11:1060. doi: 10.3390/plants11081060
Tang, J., Zhou, H., Yao, D., Riaz, S., You, D., Klepacz-Smółka, A., et al. (2022d). Comparative genomic analysis revealed distinct molecular components and organization of CO2-concentrating mechanism in thermophilic cyanobacteria. Front. Microbiol. 13:876272. doi: 10.3389/fmicb.2022.876272
Turland, N., Wiersema, J., Barrie, F. R., Greuter, W., and Smith, G. F. (2018). International code of nomenclature for algae, fungi, and plants (Shenzhen code) adopted by the nineteenth international botanical congress Shenzhen, China, July 2017. In: International code of nomenclature for algae, fungi, and plants (Shenzhen code) adopted by the nineteenth international botanical congress Shenzhen, China, July 2017.
Walter, J. M., Coutinho, F. H., Dutilh, B. E., Swings, J., Thompson, F. L., and Thompson, C. C. (2017). Ecogenomics and taxonomy of cyanobacteria phylum. Front. Microbiol. 8:2132. doi: 10.3389/fmicb.2017.02132
Yao, D., Cheng, L., Du, L., Li, M., Daroch, M., and Tang, J. (2021). Genome-wide investigation and analysis of microsatellites and compound microsatellites in Leptolyngbya-like species, cyanobacteria. Life 11:1258. doi: 10.3390/life11111258
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F. O., Ludwig, W., Schleifer, K.-H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Keywords: 16S rRNA, 16S-23S ITS, CO2-concentrating mechanism, thermophilic cyanobacterium, Trichocoleusaceae, Leptolyngbyaceae, Pinocchia, Trichothermofontia
Citation: Tang J, Zhou H, Jiang Y, Yao D, Waleron KF, Du L-M and Daroch M (2023) Characterization of a novel thermophilic cyanobacterium within Trichocoleusaceae, Trichothermofontia sichuanensis gen. et sp. nov., and its CO2-concentrating mechanism. Front. Microbiol. 14:1111809. doi: 10.3389/fmicb.2023.1111809
Edited by:
Brian P. Hedlund, University of Nevada, Las Vegas, United StatesReviewed by:
Paulina Corral, Sevilla University, SpainNicole Pietrasiak, New Mexico State University, United States
Peerada Prommeenate, National Science and Technology Development Agency (NSTDA), Thailand
Copyright © 2023 Tang, Zhou, Jiang, Yao, Waleron, Du and Daroch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurycy Daroch, m.daroch@pkusz.edu.cn
 Jie Tang
Jie Tang Huizhen Zhou
Huizhen Zhou Ying Jiang
Ying Jiang Dan Yao
Dan Yao Krzysztof F. Waleron
Krzysztof F. Waleron Lian-Ming Du
Lian-Ming Du Maurycy Daroch
Maurycy Daroch