- 1Human Biology Program, University of Tsukuba, Tsukuba, Japan
- 2Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan
- 3Doctoral Program in Medical Sciences, Faculty of Medicine, University of Tsukuba, Tsukuba, Japan
- 4Department of Natural Science, Graduate School of Technology, Industrial and Social Science, Tokushima University, Tokushima, Japan
- 5Department of Materials Science and Biotechnology, Graduate School of Science and Engineering, Ehime University, Matsuyama, Japan
RNA ligases play important roles in repairing and circularizing RNAs post-transcriptionally. In this study, we generated an allelic knockout of ATP-dependent RNA ligase (Rnl) in the hyperthermophilic archaeon Thermococcus kodakarensis to identify its biological targets. A comparative analysis of circular RNA reveals that the Rnl-knockout strain represses circularization of C/D box sRNAs without affecting the circularization of tRNA and rRNA processing intermediates. Recombinant archaeal Rnl could circularize C/D box sRNAs with a mutation in the conserved C/D box sequence element but not when the terminal stem structures were disrupted, suggesting that proximity of the two ends could be critical for intramolecular ligation. Furthermore, T. kodakarensis accumulates aberrant RNA fragments derived from ribosomal RNA in the absence of Rnl. These results suggest that Rnl is responsible for C/D box sRNA circularization and may also play a role in ribosomal RNA processing.
Introduction
Numerous RNA molecules are involved in controlling gene expression to maintain cellular RNA metabolism. In addition to tRNA, mRNA and rRNA, cells also contain a striking diversity of additional RNA types, such as edited RNAs, circularized RNAs, trans-spliced RNAs, and other non-coding RNAs (Brennicke et al., 1999; Burroughs and Aravind, 2016; Kristensen et al., 2019). We hypothesize that RNA ligase, an enzyme that joins free RNA ends together, is a key player in producing a diverse set of RNAs by altering their structures. The recent finding that RNA ligase is responsible for generating a circular RNA molecule (circRNA) and could selectively modify the ends of the RNA raises the possibility that RNA ligase may also function to regulate cellular RNA metabolism.
ATP-dependent RNA ligase (Rnl) catalyzes the formation of phosphodiester bonds between the 5′-phosphate and 3′-hydroxyl termini of RNA (Uhlenbeck and Gumport, 1982). Rnl can join two single-stranded RNA molecules with or without a complementary bridging polynucleotide. It can also catalyze intramolecular ligation, leading to the formation of a covalently-closed circRNA. The biological functions of Rnl are firmly established in bacterial tRNA restriction/repair (Amitsur, 1987; Omari et al., 2006; Wang et al., 2007; Nandakumar et al., 2008), yeast and plant tRNA splicing (Sidrauski et al., 1996; Abelson et al., 1998; Englert and Beier, 2005), and kinetoplastid mitochondrial RNA editing pathways (McManus et al., 2001; Rusché et al., 2001; Schnaufer et al., 2001).
Many archaea species encode Rnl, and its structure is unique among polynucleotide ligases in that it forms a homodimeric quaternary structure. The crystal structure of Pyrococccus abyssi (PabRnl) and Methanobacterium thermoautotrophicum Rnl (MthRnl) have been solved and were shown to catalyze an intramolecular ligation of single-stranded RNA to form a covalently closed circRNA (Brooks et al., 2008; Gu et al., 2016). MthRnl can also transfer AMP to RNA containing 3′- phosphate termini to form 2′,3′-cyclic phosphate, and can selectively cleave adenosine from the 3′-hydroxyl end of the RNA, to form the 2′,3′-cyclic phosphate (Zhelkovsky and McReynolds, 2014; Yoshinari et al., 2017). Although the biological function of archaea Rnl is not known, RNA immunoprecipitation studies in P. abyssi suggest that it can interact with circular non-coding RNAs, including C/D box guide RNA (Becker et al., 2017).
Here we generated an allelic knockout of the Rnl gene in Thermococcus kodakarensis (TkoRnl; TK1545) and analyzed the change in RNA metabolism using high-throughput RNA-Seq technology. We showed that deletion of TkoRnl selectively dissipates circular C/D box sRNAs and other small RNA species. The conserved C/D box sequence elements were not strictly required for ligation activity of the archaeal Rnl. We also found that deletion of TkoRnl produces aberrant rRNA fragments, suggesting that TkoRnl may also participate in rRNA maturation process.
Materials and Methods
Strains, Media, and Culture Conditions
The T. kodakarensis KUW1 (Sato et al., 2005) and gene disruptant strain were cultivated under anaerobic conditions at 85°C (optimum growth temperature) in a nutrient-rich medium (ASW-YT) or a synthetic medium (ASW-AA). ASW-YT medium (1 L) contains 5 g yeast extract (Y) and 5 g tryptone (T) dissolved in artificial seawater (ASW) (Sato et al., 2003a). ASW-AA medium is a synthetic medium that contains a mixture of vitamins, modified Wolfe’s trace minerals, and the 20 canonical amino acids dissolved in 0.8 × ASW (Sato et al., 2003a; Atomi et al., 2004). Elemental sulfur (2 g) was added into 1 L ASW-YT and ASW-AA media before culturing. For all liquid media, resazurin (0.5 mg/L) was supplemented as an oxygen indicator, and 5.0% Na2S was added until the medium became colorless. For colony isolation, solid ASW-AA medium containing 1 g of Gelrite and 0.4 g of polysulfide per 0.1 L was used. The E. coli Mach1-T1 was used to construct the targeting plasmids and was grown at 37°C in LB medium containing ampicillin (100 mg/L). For RNA isolation and growth analysis, cells were cultured in MA-YT-P medium (0.8 × Marine Art SF1 reagent [Osaka Yakken Co. Ltd., Osaka, Japan], 5 g/L of yeast extract, 5 g/L of trypton, and 5 g/L of sodium pyruvate) which lacks elementary sulfur.
Deletion of TK1545 Gene
A DNA fragment containing the TK1545 gene along with its 5′- and 3′-flanking regions (∼1.0 kbp) was amplified with forward (5′ACTCTCTCCTTTTCTCCAATTTCGG-3′) and reverse (5′-TCAGGATTTTGCAAA GTACTGACTGG-3′) primers using T. kodakarensis KUW1 genomic DNA as a template for PCR and was cloned into the Hinc II site of pUC118 to obtain pUC-TK1545. The RNL coding sequence in pUC-TK1545 was replaced with a DNA fragment containing pyrF gene (TK2276: orotidine 5′-phosphate decarboxylase) and its promoter element as described (Sato et al., 2003b). This was accomplished by amplifying pUC-TK1545 using two outward primers (5′-AGCTGTAAGGGGCCTGTGGACATTTC-3′ and 5′- GATATCACCGAGAAGAGTGGGAGC-3′) complementary to the upstream and downstream sequence of the TkoRNL coding sequence. The amplified plasmid fragment was ligated with a PvuII-PvuII restriction fragment (763 bp) containing the pyrF marker gene derived from pUD2 plasmid (Sato et al., 2003b). All the sequences of the inserted region were verified with DNA sequencing. The resulting targeting plasmid, with pyrF marker gene inserted between the 5′- and 3′-flanking regions of RNL gene, was used to transform T. kodakarensis KUW1 strain. Cells grown in ASW-YT-S0 medium at 85°C for 10 h were harvested and suspended in 200 μL of 0.8 × ASW and kept on ice for 30 min. Then, 3 μg of the plasmid was gently added into the suspended cells and kept on ice for 1 h. Transformants were cultivated in an uracil-free ASW-AA-S0 at 85°C for 40 h. Next, 200 μL of the culture was transferred to a fresh medium and cultivated under the same conditions to enrich transformants displaying uracil prototrophy. The cultures (100 μL) were spread onto ASW-AA-S0 solid medium and incubated at 85°C for 3 days. Only cells that obtained a phenotype exhibiting uracil prototrophy by homologous recombination can grow in the absence of uracil. Single colonies were selected and then cultured in ASW-YT-S0 medium at 85°C for 10 h. Throughout this article, the term wild-type (WT) refers to KUW1 and tk1545 KO refers to TkoRnl deletion in the KUW1 strain. The cells of the tk1545 KO strain were harvested and suspended in distilled water. Genomic DNA was extracted from the cells using phenol-chloroform treatment. The replacement of TK1545 gene with pyrF gene was verified with PCR (Figure 1A).
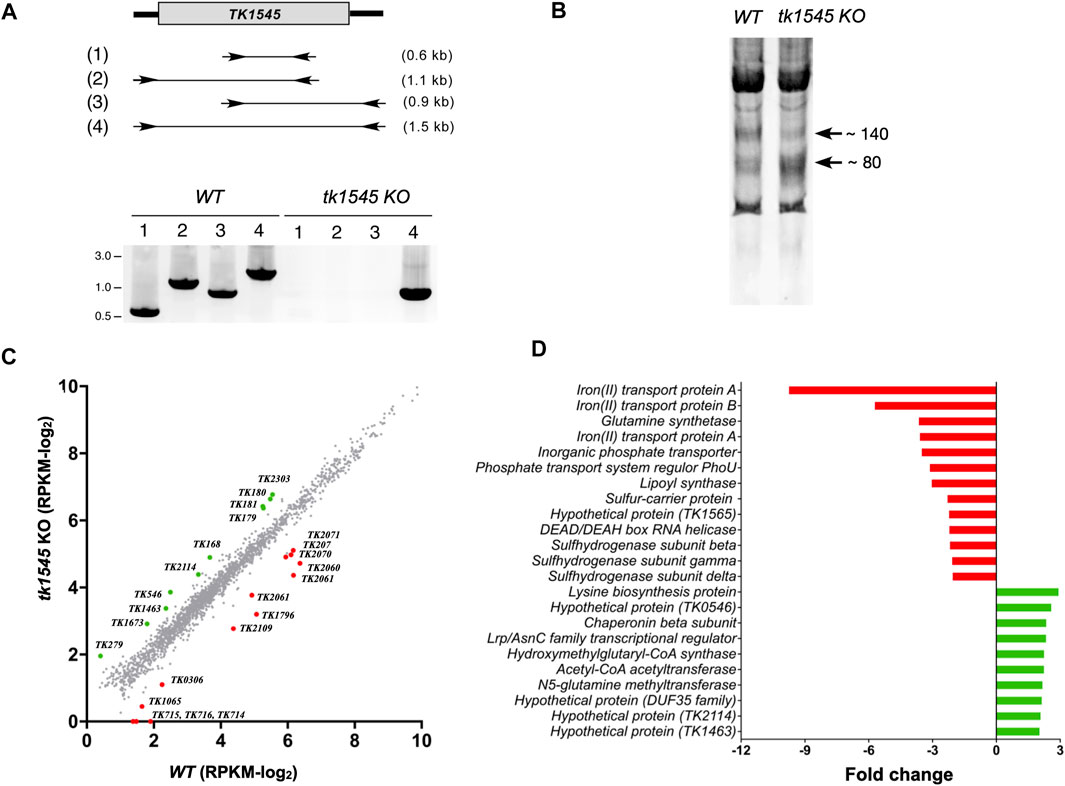
FIGURE 1. Differentially expressed genes by deletion of TkoRnl gene. (A) Generation of RNA ligase knockout in T. kodakarensis. The TK1545 gene was replaced with the pyrF selectable marker (763 bp). The positions of four pairs of PCR primers and their expected product sizes are depicted in (1–4). The lower panel shows an agarose gel electrophoresis of PCR products derived from WT and tk1545 KO genomic DNAs. (B) Total RNA extracted from WT and tk1545 KO strains were separated using 10% denaturing polyacrylamide gel electrophoresis. Ethidium bromide staining of the gel is shown. (C) Transcriptosome analysis of WT and tk1545 KO. Total RNA isolated from WT and tk1545 KO cells were subjected to RNA-Seq analysis. Each dot represents an individual gene and are depicted according to read per million mapped reads (RPKM) values. The RPKM value with less than one RPKM were set to 0. Genes with RPKM of <2.0 in both WT and tk1545 KO were omitted. Differentially expressed genes are depicted as colored dots. Green dots represent the 10 genes that are up-regulated, and red dots represent 13 genes that are down-regulated by more than 2-fold in tk1545 KO (excluding the tk1545 gene). The RPKM for tk1545 from the WT and tk1545 KO were 9.05 and 0.04, respectively. Genes that show greater than two-fold change are listed in (D) and in Supplementary Table S2.
RNA Isolation and RNA-Seq Analysis
T. kodakarensis WT and tk1545 KO (1.6 L) were cultured in MA-YT-P medium at 85°C and harvested when the absorbance at 660 nm reached ∼0.7 (late-log phase). Total RNA was isolated using TRIzol reagent (Invitrogen) following the manufacture’s instruction. The total RNA and small RNA libraries were prepared using standard Illumina protocol by UB Genomics and Bioinformatics Core at The State University of New York (SUNY) at Buffalo, United States. The high-throughput RNA-sequencing was performed using Illumina HighSeq2500 technology (51-base; pair-ended read). Raw reads were trimmed, and FastQC was used to determine the clean reads. For both WT and tk1545 KO datasets, 80 percent of the reads had Phred quality scores >35. The reads were mapped to T. kodakarensis reference genome (NC_006624.1) using bowtie2-2.2.9 with the option preset set to sensitive and alignment type set to local mode. The reads per kilo-base per million mapped reads (RPKM) were calculated using featureCount (Liao et al., 2014). From the WT dataset, 101 million reads were obtained, of which 94.86% mapped to the reference genome, with a median RPKM of 9.8. From the tk1545 KO dataset, 58 million pairs of reads were obtained, of which 94.54% were mapped to the reference genome with a median RPKM of 10.9 (Supplementary Data S1).
For the small RNA-Seq, the 5′-adaptor sequence (5′-GTTCAGAGTTCTACAGTCCGACGATC) and 3′-adaptor sequence (5′-TGGAATTCTCGGGTGCCAAGG) were trimmed using Cutadapt, with 80 percent of the reads having Phred quality scores of >35. The reads aligned to reference genome as described above. We obtained 22 million pairs of reads from the WT sample, of which 79.94% mapped to the reference genome. From the tk1545 KO sample, 16 million reads were obtained, and 81.43% mapped to the genome (Supplementary Data S1).
Analysis of circRNA from RNA-Seq Data
A custom Perl script was used to identify RNA-Seq reads containing circular junction sequences. The screening was done to identify RNA-Seq reads containing two segments of at least 20-nts matching the reference genome; the two matched segments should be encoded in the same direction but inverse order in the reference genome. From total RNA Seq data, 410,073 and 281,828 reads containing circular junctions from the WT and tk1545 KO were identified, respectively (Supplementary Data S2). To reduce redundancy, reads containing similar junction sequences within five nucleotide variations were classified into the same group, which decreased the number of candidate circRNA reads to 12,632 for WT and 9,744 for tk1545 KO. Subsequently, circRNA junction reads with less than 100 independent read counts were eliminated. This criterion identified 113 and 63 reads containing circRNA junction sequences from WT and tk1545 KO, respectively. A similar strategy was used to identify circRNAs from the small RNA-Seq datasets, except that circRNA reads predicted to be longer than 10 kb and read counts of less than 20 were eliminated (Supplementary Data S3).
Detection of circRNA Using RT-PCR
We performed RT-PCR to detect predicted circular RNAs. Primers containing gene-specific sequences were used for reverse transcription reaction. The reaction mixture (40 μL) containing total RNA (200 ng) from either WT or tk1545 KO was incubated with 25 μM gene-specific primer, 0.5 mM dNTP, and 50 U ReverTra Ace-α (Toyobo, Japan) in a supplied reaction buffer at 55°C for 10 min (for primer sequences, see Supplementary Table S1). The reaction was terminated at 95°C for 10 min, and an aliquot (1 μL) was used as a template for PCR. PCR (50 μL) contained 0.2 μM circular junction primer and gene-specific primer, 2.5 U Paq5000 DNA polymerase (Toyobo, Japan) programmed for 25 cycles (95°C for 30 s; 60°C for 30 s; 72°C for 10 s). PCR products were separated on 3% low-range ultra-agarose gel, stained with ethidium bromide, and visualized using UV.
RNA Ligase and RNA Substrates
His-tagged MthRnl and His-tagged T4 RNA Ligase 2 (T4 Rnl2) were produced in E. coli and purified from soluble bacterial extracts using Ni-agarose chromatography as described previously (Ho and Shuman, 2002; Torchia et al., 2008). In vitro transcription was used to synthesize RNAs from PCR amplified linear DNA templates containing a T7 RNA polymerase promoter. RNAs containing 5′ triphosphate were purified by electrophoresis through a non-denaturing 8% polyacrylamide gel. The RNAs were then treated with calf intestinal alkaline phosphatase, extracted by phenol-chloroform, and ethanol precipitated. The in vitro transcribed RNAs and the chemically synthesized 24-mer RNA were labeled at the 5′-end with [γ-32P] ATP using T4 polynucleotide kinase and purified on the non-denaturing polyacrylamide gel.
Cloning of Small RNA Fragments
Total RNA (15 µg) from WT and tk1545 KO were separated using 10% denaturing polyacrylamide gel, and RNA ranging from 70 to 120 nt was isolated by elution from the gel with Tris-EDTA (TE) buffer. RNA was ethanol-precipitated and resuspended to 50 µL with TE buffer. The isolated small RNA was used for the adapter-mediated RNA cloning with SOLiD Small RNA Expression Kit (Ambion). Ligation and reverse-transcription were performed according to the manufacturers’ instructions. cDNA was amplified using Taq DNA polymerase, inserted into TOPO cloning vector (Invitrogen), and transformed into E. coli DH5α. The region of cDNA insertion within the TOPO vector was PCR amplified directly from the bacterial colonies, and product size was analyzed on 1.8% agarose gel. The majority (90%) of the plasmid recovered contained primer-dimer insert. Therefore, PCR product corresponding to >40 bp cDNA insert was regarded as a “positive” clone and was sequenced to reveal the identity of cloned RNA species. PCR product length corresponding to <40 bp insert was omitted, as it likely represented a fragment derived from a primer dimer.
Results
Deletion of T. kodakarensis Rnl Gene
TK1545 encoding for T. kodakarensis ATP-dependent RNA ligase (TkoRnl) was removed via homologous recombination using a non-replicating targeting vector carrying orotidine-5′-monophosphate decarboxylase gene (pyrF) flanked by ∼700 bp of upstream and downstream TK1545 DNA sequence. The plasmid was transformed into T. kodakarensis strain KUW1 (∆pryF) and the Ura+ transformants were recovered. Deletion of TK1545 locus was confirmed using PCR analysis (Figure 1A). Expression of TK1545 was verified using RNA-Seq analysis, as described below. No significant difference in growth phenotype was observed between the WT and tk1545 KO strains in nutrient-rich medium, at optimal (85°C) and elevated temperatures (93°C) (Supplementary Figure S1), which implies that TkoRnl is not essential for viability. Polyacrylamide gel electrophoresis analysis of the small RNA population suggested that the relative distribution of short RNA species, ranging from 80 to 140 nt, differed significantly between the two strains (Figure 1B). Compared to the parental strain, the level of ∼140-nt species was reduced while that of the ∼80-nt species was increased in the tk1545 KO strains. Therefore, both total and small RNA-Seq analyses were performed to evaluate the physiological consequences of TkoRnl deletion.
Transcriptome Analysis
Total and small RNAs isolated from the wild-type and tk1545 KO strains were sequenced on an Illumina HighSeq platform sized at 51-bp and mapped uniquely to the annotated genome. The gene expression abundance was normalized using RPKM (Supplementary Data S2), and scatterplots were used to assess the expression variation of the genes between the WT and tk1545 KO from the total RNA-Seq dataset (Figure 1C). Finally, 23 genes were identified that showed altered expression changes of >2-fold, of which 13 were up-regulated, and 10 were down-regulated in tk1545 KO compared to the WT (Figure 1D; Supplementary Table S2). Many of these genes that exhibit differential gene expression are encoded on the same polycistronic transcription unit; iron transport proteins (TK0714, TK0715, and TK0716), phosphate transporter proteins (TK2060 and TK2061), sulfur reductase subunits (TK2071 and TK2072), and Acetyl-CoA acetyltransferase pathway (TK0179, TK0180, and TK0181). The genes encoded within each operon were either enhanced or reduced to a similar extent in tk1545 KO. Transcriptome analysis from the small RNA-Seq data is shown in Supplementary Figure S2.
Computational Predictions of Circular RNAs
It has been widely reported that non-coding RNAs, including tRNAs, C/D box sRNA, and rRNA processing intermediates are circularized in archaea (Tang et al., 2002; Starostina et al., 2004; Danan et al., 2012; Randau, 2012; Su et al., 2013; Becker et al., 2017). We hypothesize that if Rnl is responsible for generating circRNAs, we would identify its RNA target by comparing the circRNA reads obtained from WT and tk1545 KO. Our criteria for detecting circular reads from the RNA-Seq data were as follows: 1) the 51-nts RNA-Seq reads containing two segments, and each segment has a minimum 20-nts match to the reference genome sequence; 2) the two matched segments within the read are encoded in the same transcriptional direction, but are positioned in inverse order in the reference genome (Danan et al., 2012); and 3) the two matched segments are fused to form a unique circular junction sequence. Variation in the circular junction within five nucleotides in the locus was classified into the same group to reduce redundancy (Supplementary Data S2). The predicted length of individual circRNA was deduced from the distance between the two homologous segments in the genome reference. For the total RNA-Seq data, we selected reads that support more than 100 counts (Supplementary Data S2). For the small RNA-Seq data, we selected read supported more than 20 counts and did not include reads that were predicted to be longer than 10 kb (Supplementary Data S3).
In the WT T. kodakarensis, 31 circRNA reads were detected from the small RNA Seq data set, many of which were derived from C/D box sRNAs (Supplementary Table S2). The C/D box sRNA molecule has four sequence elements: the C box and C’ box motifs with the consensus sequence RUGAUGA, and the D box and D’ box motifs with the consensus sequence CUGA. Of the 61 putative T. kodakarensis C/D box sRNAs we identified, 26 C/D Box sRNAs had circRNA reads (Supplementary Table S3). Analysis of P. abyssi RNA-Seq dataset detected 24 circRNA reads (Toffano-Nioche et al., 2013), many of which were shown to be circularized (Becker et al., 2017). Other circRNA reads detected from T. kodakarensis include protein coding genes (TK0058 [HAD superfamily hydrolase], TK2034 [Universal stress protein], TK2109 [lipoyl synthase], TK0894 [hypothetical protein], TK1980 [ferredoxin oxidoreductase, alpha subunit], TK0135 [ferredoxin oxidoreductase, beta subunit]) and non-coding RNAs designated here as ncRNA01, ncRNA02, and ncRNA03 (Table 1; Supplementary Data S2). Some of these RNAs were previously reported as C/D box sRNA (Jäger et al., 2014). CircRNAs were also detected in abundance from rRNA operon and tRNATrp as previously reported (Danan et al., 2012).
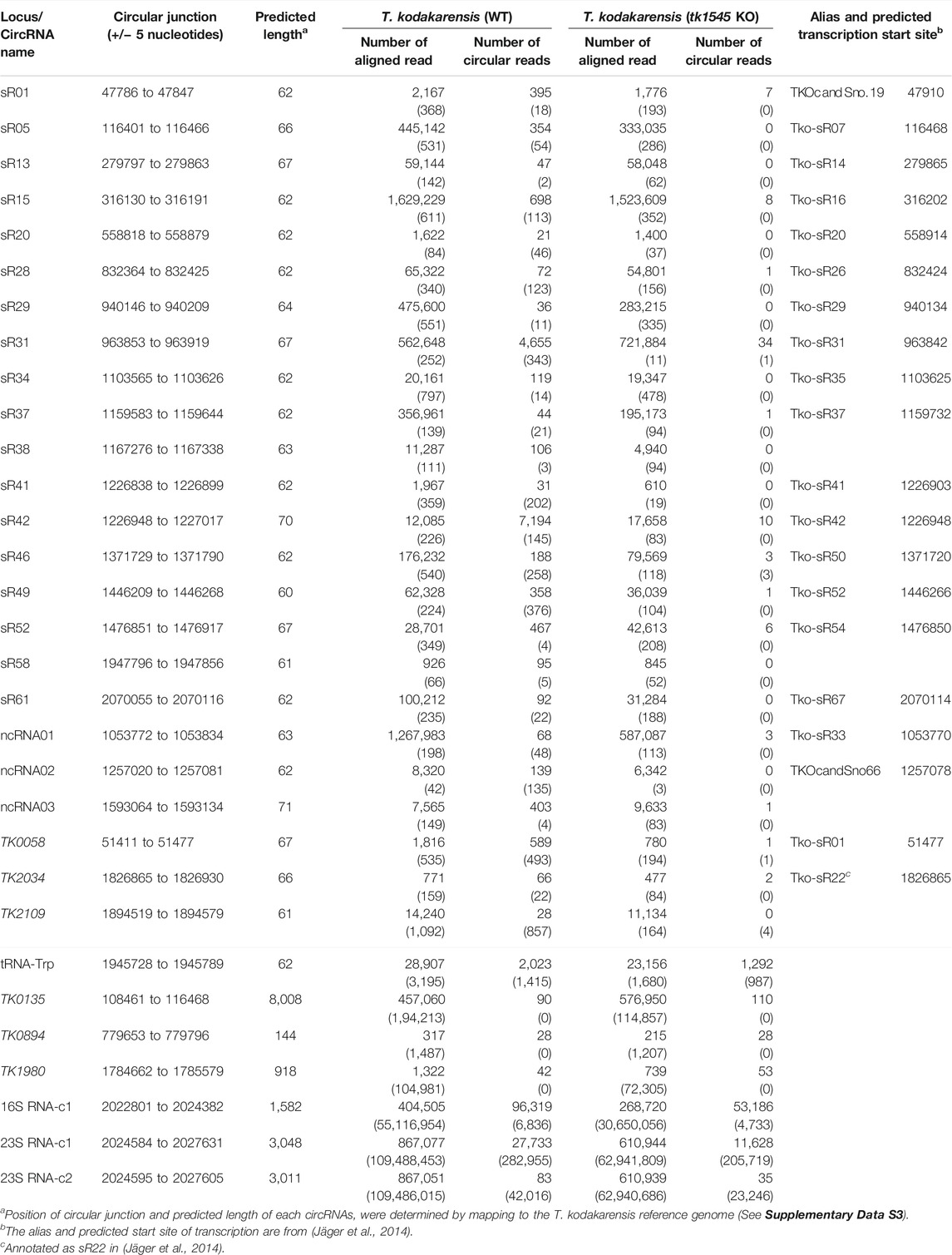
TABLE 1. List of circRNAs in T. kodakarensis. The top part of the table shows list of circRNAs that were detected in WT but were either absent or significantly reduced in tk1545 KO small RNA-Seq dataset. The bottom part of the table shows a list of circRNAs that were detected in both WT and tk1545 KO. The value in parenthesis shows the number of reads detected in total RNA-Seq dataset.
TK1545 is Required for C/D Box sRNA Circularization
Analysis of tk1545 KO RNA-Seq data revealed that 24 out of 31 circRNAs that were detected in WT were either absent or significantly reduced in tk1545 KO (Table 1). These included 18 C/D box sRNAs, three non-coding RNAs, and three protein-coding genes (TK0058, TK2109, and TK 2034). Similar results were obtained when RNA-Seq analysis of WT and tk1545 KO were analyzed on a SOLiD sequencing platform (Supplementary Table S4) (Liu, 2022). Figure 2A shows a sequence alignment of circRNAs that were affected in tk1545 KO. All these circRNAs have a similar length (61–71 nts) and homology to the C/D box sRNA. The terminal ends are generally GC-rich, and sequences at the termini can hybridize to form a stem, a structural characteristic found in C/D box sRNA. Secondary structure analysis suggests that hybridization between the two terminal ends could be critical for RNA circularization (Supplementary Figure S3). The majority of the RNAs that were circularized (21 out of 24 shown in Figure 2A) could potentially form three or more base pairings to form a terminal stem (Supplementary Figure S3A). In contrast, 16 out of 17 non-circular C/D box sRNAs are less likely to form a terminal stem with two or less base pairings (Supplementary Figure S3B). We note that relative abundance of six C/D box sRNAs (sR14, sR35, sR38, sR41, sR54, and sR61) out of sixty-one C/D box sRNAs that we identified, were reduced by 2-fold or more in tk1545 KO compared to the WT (Figure 2B).
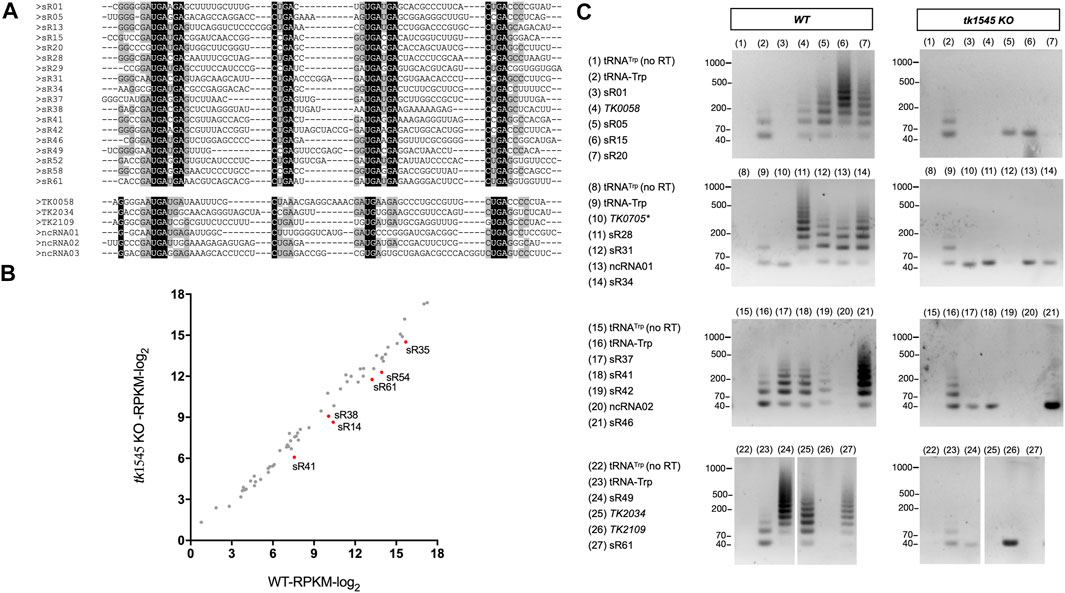
FIGURE 2. Effect on RNA circularization by TK1545 knockout. (A) Sequence alignment of circRNAs which were reduced or absent in tk1545 KO (see Table 1). Putative circular C/D box RNAs are listed on top and other circRNAs are listed on the bottom. The positions of conserved motifs (C box, C′ box, D box, and D′ box) are indicated. Identical nucleotides are highlighted in black with white text and conserved nucleotides are highlighted in gray. (B) Expression of C/D box RNA. Expression analysis of sixty-one T. kodakarensis C/D box RNA listed on Supplementary Table S3. Red dots represent the six C/D box RNAs reduced by more than 2-fold in tk1545 KO. (C) RT-PCR analysis was used to detect circRNAs from total RNA isolated from WT and tk1545 KO. The primer set used to detect circular RNA species is indicated on the right of the gel. A circular tRNATrp intron primer was used as a positive control (tRNA-Trp). As a negative control, assay was performed without reverse transcriptase treatment using tRNATrp intron primers [tRNATrp (no RT)], or with the TK0705 primers (Gene ID 3234407: NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, which is not circularized in both WT and tk1545 KO). The size of the molecular weight markers is indicated on the left. PCR product that migrates ∼40 nts most likely represented a product derived from a primer dimer.
There were no significant changes in the level of circRNA reads derived from tRNATrp intron and 16S and 23S rRNAs. We note that circularization of TK0135, TK0894 and TK1980, all of which have predicted circRNA size of >100 nts, were not significantly affected by TkoRnl deletion. Circular TK0894, TK1980 and TK0135 were not detected in the whole RNA-seq data (Table 1), suggesting that these RNAs may likely circularize after the degradation or processing of the transcript into small RNA.
RT-PCR analysis was performed to verify whether the circular RNA species are present in T. kodakarensis (Figure 2C). In this procedure, reverse transcription primer was designed to hybridize the gene specific sequence. If this primer hybridizes to circular RNA, reverse transcription will generate a long “rolling-circle” single-stranded cDNA. Subsequent PCR with circular junction and gene specific primers generate a ladder of DNA fragments, which can be visualized on the gel electrophoresis (Starostina et al., 2004). A tRNATrp primer was used as a positive control because tRNATrp introns accumulate circRNA reads in both WT and tk1545 KO, generating a ladder of DNA fragments from WT and tk1545 KO RNAs (Figure 2C, tRNA-Trp) but not when reverse transcriptase was omitted in the reaction (no RT). Out of the 18 candidate circRNAs, 16 were detected circularized in the WT, but not in tk1545 KO. A circular form of C/D box sR01 and TK2109 could not be detected in WT or tk1545 KO (Figure 2C; lanes 3 and 26, respectively), possibly due to a heterogeneous mixture of circular junction sequences in these RNAs. While most of the circular junction sequences represent ligation between the predicted 5′-end and 3′-end, some of the circRNA reads had a few nucleotides missing at the circular junction, which may have affected the PCR amplification step.
RNA Ligation Activity on C/D Box sRNA
To determine whether Rnl preferentially recognizes C/D box sRNA sequence elements, we assayed for the ligation activity in vitro using a 5′-monophosphate terminated C/D box sR42 and sR31 sRNAs as substrates. These two C/D box sRNAs were selected because both are highly enriched and circularized in the WT sample (Table 1; Figure 2C). We previously showed that TkoRnl is capable of circularizing 24-mer single-stranded RNA, but the circularization activity was weak compared to the M. thermoautotrophicus enzyme (MthRnl) (Yoshinari et al., 2017; Zhang and Tripathi, 2017); thus, MthRnl was used for the ligation assay. MthRnl is a homolog of TkoRnl (NCBI BLAST E-value of 6 × 10−60). The biochemical activity of MthRnl has been extensively characterized and all amino acid residues found to be essential for the MthRnl ligation activity are conserved in TkoRnl (Zhelkovsky and McReynolds, 2014; Torchia et al., 2008; Gu et al., 2016; Yoshinari et al., 2017).
Incubation of MthRnl with either 5′-monophosphate terminated sR42 or sR31 RNAs generates a circRNA molecule that migrates slower than the linear pRNA on a denaturing PAGE (Figure 3A). The circularity of the slower migrating RNA product was verified by its resistance to alkaline phosphatase and RNase R treatment (data not shown). Under identical conditions, MthRnl could not ligate a non-structured 67-mer RNA, suggesting that a structure on the C/D box sRNAs is necessary for RNA circularization. Ligation in the absence of added ATP reflects the presence of pre-adenylated ligase intermediate in the enzyme preparation (Torchia et al., 2008; Gu et al., 2016). We also note that inclusion of ATP in the reaction did not affect circularization of sR42 and sR31 RNAs. This contrasts with ligation of a 24-mer pRNA substrate, which accumulates AppRNA intermediate and suppress the circularization in the presence of ATP, as shown previously for MthRnl (Torchia et al., 2008; Gu et al., 2016), and T4 Rnl2 (Ho and Shuman, 2002; Yin et al., 2003; Nandakumar et al., 2004).
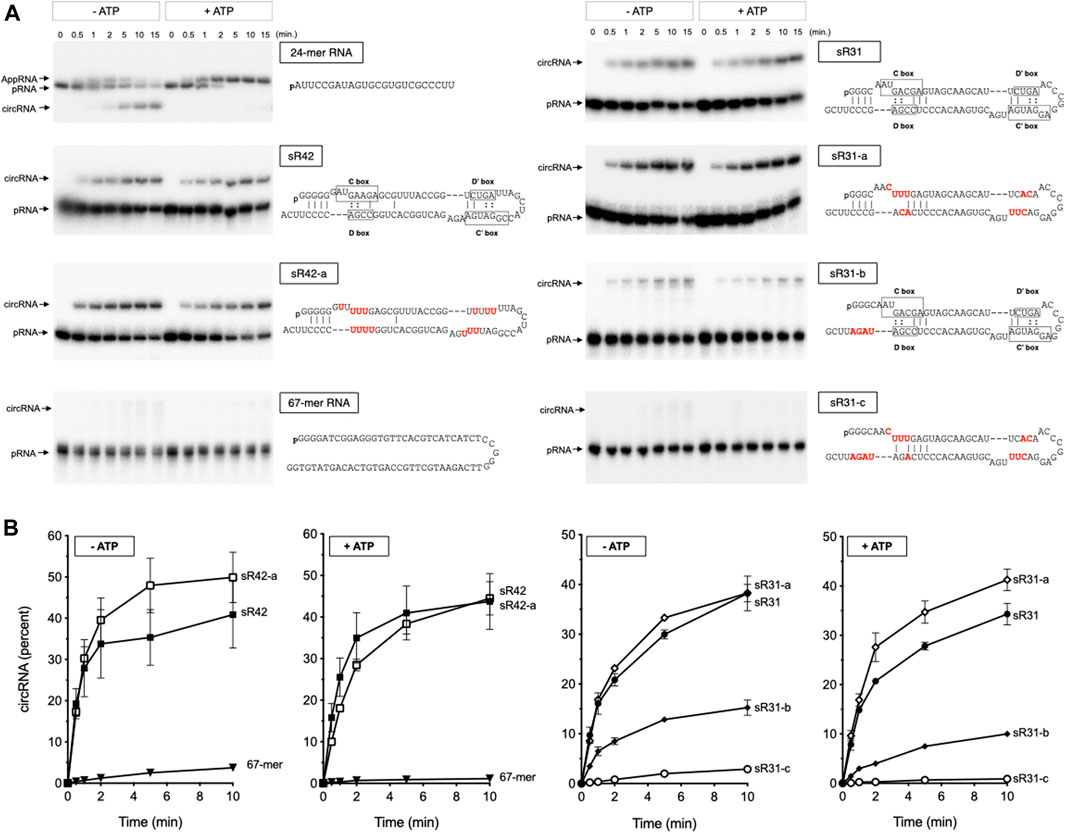
FIGURE 3. Characterization of RNA circularization activity of MthRnl on C/D box sRNA. (A) MthRnl (360 ng) was incubated with 2 pmol of indicated pRNA at 70°C in a reaction mixture (40 μL) that contained 50 mM Tris-HCl (pH 6.5), 1 mM MgCl2, in the presence or absence of 1 mM ATP. Aliquots (3 μL) were withdrawn at the times indicated and the products were separated by denaturing polyacrylamide gel. Positions of pRNA, AppRNA, and circRNA are indicated on the left. The sequences of pRNA substrates are illustrated on the right. The conserved C/D box sequence elements in sR42 and sR31 are highlighted, and the variant nucleotides in a mutant form of sR42 and sR31 RNAs are colored in red. (B) The yield of circular RNA products by MthRnl in the presence (+ATP) and absence (−ATP) of ATP is plotted as a function of time. The data shown represent the average of three separate experiments with SE bars.
The circularization activity was not significantly affected when the conserved C/D box sequence was substituted with different bases (Figure 3A; sR42-a and sR31-a). However, a mutant form of sR31 RNA that can alleviate hybridization between the terminus was a poor substrate for ligation (Figures 3A,B; sR31-b). Furthermore, MthRnl was inert for circularizing the RNA when both the C/D box and the 3′-terminal sequences were altered (Figures 3A,B; sR31-c). As a control reaction, we showed that bacteriophage T4 Rnl2 could efficiently circularize both the linear 67-mer and sR31-c RNAs (Supplementary Figure S4). We conclude that conserved C/D box sequence elements are not strictly required for circularization by MthRnl. The sequence surrounding the termini could be important for guiding the two termini in close proximity to be recognized by archaea Rnl to allow for intramolecular ligation.
Other circRNAs in T. kodakarensis
It has been widely reported that tRNA introns and rRNA processing intermediates are circularized in various archaea species (Danan et al., 2012; Su et al., 2013; Becker et al., 2017; Jüttner et al., 2019; Qi et al., 2020; Breuer et al., 2021). In both WT and tk1545 KO, high-levels of circRNA reads were detected from the 16S-23S rRNA operon and tRNATrp intron (Supplementary Data S2). Inspection of circular junction sequences in total RNA-Seq data reveal that tRNATrp intron (1,415 reads), 16S circRNA-1 (6,835 reads) and 23S circRNA-1 (282,955 reads) are likely cleaved at the Bulge-Helix-Bulge (BHB) motifs by tRNA splicing endonuclease and joined by tRNA ligase RtcB to form a circular rRNA processing intermediate, as reported (Trotta et al., 1997; Englert et al., 2011; Popow et al., 2011). The circularized tRNATrp intron was also detected in our RT-PCR analysis in abundance, both in WT and tk1545 KO (Figure 2C), implying that deletion of TkoRnl does not have an impact on the RtcB ligation pathway.
Effect on rRNA Processing by Rnl Knock-Out
In addition to the circular rRNA processing intermediate, we also detected a high level of circular junction reads (42,016 reads) near the predicted 5′-and 3′-ends of 23S rRNA (Figure 4A; 23S circRNA-2). Recent studies in Pyrococcus furiosus suggest that 3′-end of 23S rRNA could be fused to the 5′-end by an RNA rearrangement as a consequence of excision of 40-nts helix 98 (H98) located ∼100 nucleotides upstream of the mature 3′-end (Birkedal et al., 2020). Similar to P. furiosus rRNA, the H98 could be excised in T. kodakarensis, evinced by low coverage of RNA-Seq reads within the equivalent segment (Figure 4B).
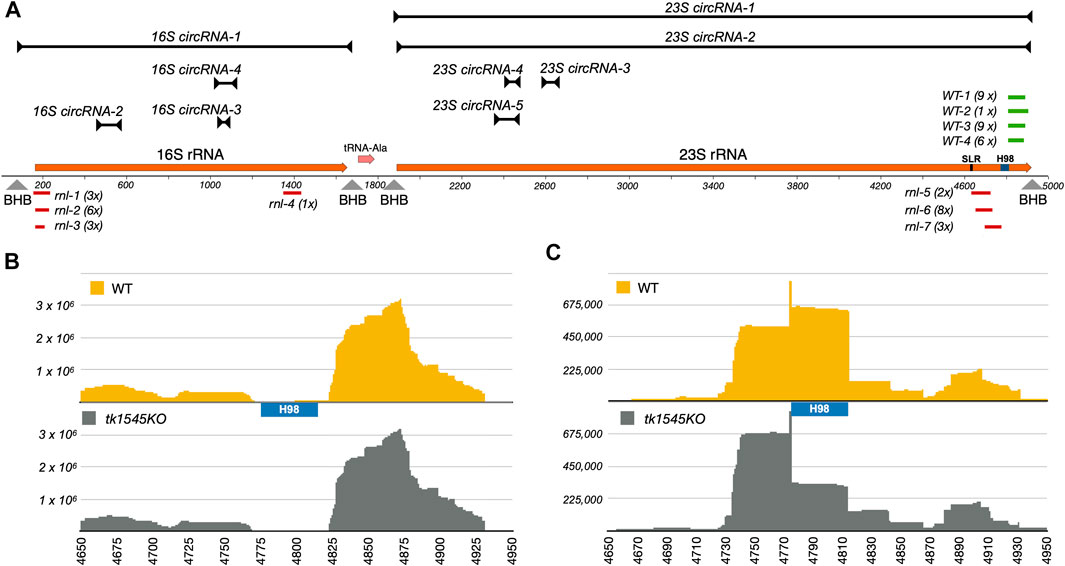
FIGURE 4. Effect of TK1545 knockout on rRNA circularization and processing. (A) Circular RNAs derived from T. kodakarensis 16S-tRNAAla-23S rRNA operon. A diagram on the bottom represents a 5 kb segment of T. kodakarensis 16S-tRNAAla-23S rRNA operon, corresponding to 2022701–2027700 of NC_006624.1 reference sequence. The 16S rRNA, tRNAAla, and 23S rRNA genes are colored in orange. Positions of SRL and H98 are annotated. Arrowheads indicate predicted positions of bulge-helix-bulge motifs (BHB) on the primary transcript. The black lines above the diagram represent the expected size and position of highly represented circRNAs (Supplementary Data S2). Predicted length and position were determined from total RNA-Seq reads containing the junction sequence. Small RNAs fragments derived from WT and T. kodakarensis were cloned into TOPO vector via adaptor-mediate RNA ligation. The cloned fragments derived from WT (WT: green) or tk1545 KO (rnl: red) were aligned to the reference genome. The number on the right represents number of identical RNA fragments recovered in the RNA cloning experiment (i.e., 2 x indicates twice). See Supplementary Figure S5 for circular junction and alignment of small RNA fragments with the 16S-tRNAAla-23S rRNA operon. (B) Read-coverage in 23S rRNA at the 3′-end from total RNA-Seq dataset from WT (top) and tk1545 KO (bottom). (C) Read-coverage in 23S rRNA at the 3′-end from small RNA-Seq dataset from WT (top) and tk1545 KO (bottom). The read counts (y-axis) for the tk1545 KO were normalized to the WT read counts. The position of H98 is marked below the WT read-coverage.
We noted earlier that the relative distribution of small RNA species was altered in tk1545 KO (Figure 1B). While this change is partially attributable to the reduced level of circular C/D box sRNAs in tk1545 KO, it cannot solely account for the observed differences because a fraction of C/D box sRNAs are likely circularized. We therefore cloned and sequenced the small RNA fragments accumulated in WT and tk1545 KO. RNA populations in the range of 70–120 nts were isolated from WT and tk1545 KO cells by gel electrophoresis, annealed to an adapter oligonucleotide containing six degenerate nucleotides at the 3′ end, ligated, and then converted into cDNAs by reverse transcription. The cDNA fragment was cloned into a plasmid using TA cloning and transformed into bacteria. Plasmids isolated from the individual colonies were sequenced using the Sanger method. Note that the adaptor sequence used for RNA cloning has a degenerate sequence at the ends, which allowed us to verify that each obtained clone was derived from an independent RNA and that no single clone was over-represented during PCR amplification.
Alignment of sequences retrieved from small RNA cloning shows that all the fragments obtained were derived from 16S and 23S rRNAs. There were no overlaps between the fragments recovered from WT and tk1545 KO, indicating that this difference is a consequence of Rnl deletion. Fragments isolated from WT were all derived from 23S rRNA, between the excision site of H98 to the 3′-end of the predicted 23S rRNA (WT-1 through WT-4 colored as green in Figure 4A; Supplementary Figure S5). In the tk1545 KO, nearly half of the cDNA fragments matched the 23S rRNA. Notably, all the fragments mapped upstream of the H98 near the sarcin-ricin loop (SRL) (rnl-5 through rnl-7; colored in red and Supplementary Figure S5). The remaining half of the fragments were derived from 16S rRNA that mapped at the 5′-end of 16S rRNA (rnl-1 through rnl-3; colored in red). Consistent with this finding, analysis of small RNA-Seq data reveals that the reads-coverage upstream of H98 are more abundant in the TkoRnl deletion strain than the WT. (Figure 4C). Taken together, our results suggest that Rnl may participate in rRNA processing, either directly by joining the breaks near the SRL or indirectly through the formation of circular C/D box sRNA.
Discussion
Here we generated an allelic knock-out of ATP-dependent RNA ligase in T. kodakarensis to determine the biological targets of archaeal Rnl. Whereas TkoRnl was not essential for the growth of T. kodakarensis under standard laboratory conditions, we showed that its absence abolishes circularization of C/D box sRNAs. The conserved C/D box sequence element, however, was not sufficient for circularization because not all C/D box sRNAs were circularized in T. kodakarensis. Furthermore, we demonstrated that recombinant Rnl was capable of forming circular C/D box sRNA with a mutation in the conserved sequence element. The archaeal Rnl could not circularize unstructured RNA of a similar length or C/D box sRNAs that have disrupted terminal stem structures. We conclude that archaeal Rnl may preferentially recognize the terminal stem, and the proximity of the two ends could be critical for intramolecular ligation.
We also detected numerous circular RNA-Seq reads derived from 16S and 23S rRNAs in T. kodakarensis. As expected, circularization of tRNA intron or rRNA processing intermediates was not affected in the absence of Rnl, which implies that Rnl does not affect the RtcB ligation pathway. Our finding that C/D box sRNAs are prime substrates for archaeal Rnl is consistent with the previous findings that circular C/D box sRNA-Rnl complexes were detected in P. abyssi (Becker et al., 2017).
The functional significance of circular C/D Box sRNA is unclear. C/D box sRNAs have been reported to function as a guide RNA for methylating tRNAs and rRNAs. Circular C/D Box sRNA may alter the specificity to guide RNA to regulate rRNA methylation. However, we did not detect significant differences in the expression, or the overall read coverage, of rRNA genes from the whole transcriptome RNA-seq analysis, between the WT and TkoRnl deletion strains. Comparative transcriptomics analysis revealed that TkoRnl may alter that abundance of subset of C/D Box sRNAs (Figure 2B). TkoRnl could also be involved by regulating the expression of genes involved in sulfur or iron metabolism (Figure 1; Supplementary Table S2). We note that only one biological replicate was analyzed in this study. While it is clear that TkoRnl is responsible for C/D box sRNA circularization, further analysis is necessary to evaluate the biological function of Rnl in archaea.
Similar to P. furiosus, T. kodakarensis appears to excise H98 from 23S rRNA (Figure 4B), consistent with the finding that the H98 is not present in the cryo-EM structure of T. kodakarensis 70S rRNA (Birkedal et al., 2020; Sas-Chen et al., 2020). We found that T. kodakarensis accumulates ∼90 nts fragments consisting of a sequence that matches the H98 3′-cleavage site to the predicted 3′-end of 23S rRNA. We speculate that the excision of H98 releases the 3′-end fragment and may have accumulated in T. kodakarensis. Intriguingly, we did not retrieve the same fragments from the TkoRnl deletion strain. Instead, we recovered fragments that mapped upstream of H98 near the SRL. SRL interacts with the translational elongation factors that hydrolyze GTP during translocation (Wool et al., 1992; Szewczak et al., 1993; Schmeing et al., 2009), and the cleavage or modification by ribotoxins could block ribosome translocation (Wool et al., 1992; Szewczak et al., 1993; Schmeing et al., 2009).
While it is tempting to speculate that Rnl directly participates in joining the breakage upon excision of H98, we fail to detect any RNA-Seq reads suggesting such “cis-splicing” events near the 3′-end of T. kodakarensis 23S rRNA. Furthermore, TkoRnl is not likely involved in rearranging the 3′- and 5′-ends of the 23S rRNA as observed in P. furious (Birkedal et al., 2020) because permuted reads containing junction sequence between the 5′- and 3′- ends of 23S rRNA were detected in both WT and TkoRnl deletion strain (Table 1; Supplementary Data S2, S3). Therefore, Rnl ligation activity may not act on rRNA directly. It is plausible that Rnl may act indirectly through the formation of circular C/D box sRNA, which in turn could regulate rRNA processing.
Nonetheless, the phylogenetic analysis suggests a possible link between the archaea Rnl, C/D box sRNA circularization, and H98 processing. A high abundance of circular C/D box sRNA molecules was detected in T. kodakarensis, P. furiosus, P. abyssi, and Methanopyrus kandleri (Starostina et al., 2004; Danan et al., 2012; Su et al., 2013; Toffano-Nioche et al., 2013). They all encode a homodimeric type-3 Rnl (Gu et al., 2016) and possess H98 or an equivalent structural element in their large subunit of rRNA. In T. kodakarensis and P. furiosus, H98 is excised evinced by discontinuous RNA-Seq map coverage ((Birkedal et al., 2020) and this study). While many species from Methanomicrobiales and Archaeoglobales encode Rnl, the helix equivalent of H98 is replaced with a short linker sequence (Birkedal et al., 2020). In contrast, Haloferax volcanii, Nanoarchaeum equitans, Sulfolobus solfataricus, Sulfolobus acidocaldarius, and Pyrobaculum aerophilum, do not encode homolog of type-3 Rnl. Circular tRNA intron and rRNA intermediates are present in abundance, but only a modest number of circular C/D box RNAs were reported in H. volcanii, S. solfataricu, S. acidocaldarius, and N. equitans (Danan et al., 2012; Randau, 2012; Becker et al., 2019). The large subunits of S. acidocaldarius and P. aerophilum rRNAs were shown to retain H98 evinced by a continuous read coverage at the 3′-end (Birkedal et al., 2020). Because many RNA-Seq data are depleted for rRNA, it is difficult to evaluate its read coverage. Availability of complete RNA-Seq data from other archaea species could provide further insight into the role of Rnl and its relationship to small RNA circularization and rRNA processing.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE186817.
Author Contributions
CKH designed research. YL, YT, AH, and CKH performed research. YL, YT, and CKH analyzed data. KM and MS data curation. HH and CKH provide resources. CKH wrote the paper.
Funding
This material is based upon work supported by National Science Foundation 1050984 (to CKH) and Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research KAKENHI 21K06984 (to CKH). Open Access publication charge was provided by National Institute of Advanced Industrial Science and Technology.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Katsuhiko Murakami (Penn State University) for valuable discussion and Thomas Mayers (University of Tsukuba) for editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.811548/full#supplementary-material
References
Abelson, J., Trotta, C. R., and Li, H. (1998). tRNA Splicing. J. Biol. Chem. 273, 12685–12688. doi:10.1074/jbc.273.21.12685
Amitsur, M., Levitz, R., and Kaufmann, G. (1987). Bacteriophage T4 Anticodon Nuclease, Polynucleotide Kinase and RNA Ligase Reprocess the Host Lysine tRNA. EMBO J. 6, 2499–2503. doi:10.1002/j.1460-2075.1987.tb02532.x
Atomi, H., Fukui, T., Kanai, T., Morikawa, M., and Imanaka, T. (2004). Description ofThermococcus Kodakaraensissp. nov., a Well Studied Hyperthermophilic Archaeon Previously Reported asPyrococcussp. KOD1. Archaea 1, 263–267. doi:10.1155/2004/204953
Becker, H. F., Héliou, A., Djaout, K., Lestini, R., Regnier, M., and Myllykallio, H. (2017). High-throughput Sequencing Reveals Circular Substrates for an Archaeal RNA Ligase. RNA Biol. 14, 1075–1085. doi:10.1080/15476286.2017.1302640
Becker, H. F., L'Hermitte-Stead, C., and Myllykallio, H. (2019). Diversity of Circular RNAs and RNA Ligases in Archaeal Cells. Biochimie 164, 37–44. doi:10.1016/j.biochi.2019.06.011
Birkedal, U., Beckert, B., Wilson, D. N., and Nielsen, H. (2020). The 23S Ribosomal RNA from Pyrococcus Furiosus Is Circularly Permuted. Front. Microbiol. 11, 582022. doi:10.3389/fmicb.2020.582022
Brennicke, A., Marchfelder, A., and Binder, S. (1999). RNA Editing. Fems Microbiol. Rev. 23, 297–316. doi:10.1111/j.1574-6976.1999.tb00401.x
Breuer, R., Gomes-Filho, J.-V., and Randau, L. (2021). Conservation of Archaeal C/D Box sRNA-Guided RNA Modifications. Front. Microbiol. 12, 654029. doi:10.3389/fmicb.2021.654029
Brooks, M. A., Meslet-Cladiére, L., Graille, M., Kuhn, J., Blondeau, K., Myllykallio, H., et al. (2008). The Structure of an Archaeal Homodimeric Ligase Which Has RNA Circularization Activity. Protein Sci. 17, 1336–1345. doi:10.1110/ps.035493.108
Burroughs, A. M., and Aravind, L. (2016). RNA Damage in Biological Conflicts and the Diversity of Responding RNA Repair Systems. Nucleic Acids Res. 44, 8525–8555. doi:10.1093/nar/gkw722
Danan, M., Schwartz, S., Edelheit, S., and Sorek, R. (2012). Transcriptome-wide Discovery of Circular RNAs in Archaea. Nucleic Acids Res. 40, 3131–3142. doi:10.1093/nar/gkr1009
Englert, M., and Beier, H. (2005). Plant tRNA Ligases Are Multifunctional Enzymes that Have Diverged in Sequence and Substrate Specificity from RNA Ligases of Other Phylogenetic Origins. Nucleic Acids Res. 33, 388–399. doi:10.1093/nar/gki174
Englert, M., Sheppard, K., Aslanian, A., Yates, J. R., and Söll, D. (2011). Archaeal 3'-phosphate RNA Splicing Ligase Characterization Identifies the Missing Component in tRNA Maturation. Proc. Natl. Acad. Sci. 108, 1290–1295. doi:10.1073/pnas.1018307108
Gu, H., Yoshinari, S., Ghosh, R., Ignatochkina, A. V., Gollnick, P. D., Murakami, K. S., et al. (2016). Structural and Mutational Analysis of Archaeal ATP-dependent RNA Ligase Identifies Amino Acids Required for RNA Binding and Catalysis. Nucleic Acids Res. 44, 2337–2347. doi:10.1093/nar/gkw094
Ho, C. K., and Shuman, S. (2002). Bacteriophage T4 RNA Ligase 2 (gp24.1) Exemplifies a Family of RNA Ligases Found in All Phylogenetic Domains. Proc. Natl. Acad. Sci. 99, 12709–12714. doi:10.1073/pnas.192184699
Jäger, D., Förstner, K. U., Sharma, C. M., Santangelo, T. J., and Reeve, J. N. (2014). Primary Transcriptome Map of the Hyperthermophilic Archaeon Thermococcus Kodakarensis. BMC Genomics 15, 684–699. doi:10.1186/1471-2164-15-684
Jüttner, M., Weiß, M., Ostheimer, N., Reglin, C., Kern, M., Knüppel, R., et al. (2019). A Versatile Cis-Acting Element Reporter System to Study the Function, Maturation and Stability of Ribosomal RNA Mutants in Archaea. Nucleic Acids Res. 48, 2073–2090. doi:10.1093/nar/gkz1156
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Liao, Y., Smyth, G. K., and Shi, W. (2014). Featurecounts: An Efficient General Purpose Program For Assigning Sequence Reads To Genomic Features. Bioinformat. 30, 923–930. doi:10.1093/bioinformatics/btt656
Liu, Y. (2022). Genetic And Functional Analyses Of Archaeal ATP-Dependent RNA Ligase. Ph.D. thesis Tsukuba (Japan): University of Tsukuba.
McManus, M. T., Shimamura, M., Grams, J., and Hajduk, S. L. (2001). Identification of Candidate Mitochondrial RNA Editing Ligases from Trypanosoma Brucei. Rna-A Publ. Rna Soc. 7, 167–175. doi:10.1017/s1355838201002072
Nandakumar, J., Ho, C. K., Lima, C. D., and Shuman, S. (2004). RNA Substrate Specificity and Structure-Guided Mutational Analysis of Bacteriophage T4 RNA Ligase 2. J. Biol. Chem. 279, 31337–31347. doi:10.1074/jbc.m402394200
Nandakumar, J., Schwer, B., Schaffrath, R., and Shuman, S. (2008). RNA Repair: an Antidote to Cytotoxic Eukaryal RNA Damage. Mol. Cel. 31, 278–286. doi:10.1016/j.molcel.2008.05.019
Omari, K. E., Ren, J., Bird, L. E., Bona, M. K., Klarmann, G., LeGrice, S. F. J., et al. (2006). Molecular Architecture and Ligand Recognition Determinants for T4 RNA Ligase. J. Biol. Chem. 281, 1573–1579. doi:10.1074/jbc.m509658200
Popow, J., Englert, M., Weitzer, S., Schleiffer, A., Mierzwa, B., Mechtler, K., et al. (2011). HSPC117 Is the Essential Subunit of a Human tRNA Splicing Ligase Complex. Science 331, 760–764. doi:10.1126/science.1197847
Qi, L., Li, J., Jia, J., Yue, L., and Dong, X. (2020). Comprehensive Analysis of the Pre-ribosomal RNA Maturation Pathway in a Methanoarchaeon Exposes the Conserved Circularization and Linearization Mode in Archaea. RNA Biol. 17, 1427–1441. doi:10.1080/15476286.2020.1771946
Randau, L. (2012). RNA Processing in the Minimal Organism Nanoarchaeum Equitans. Genome Biol. 13, R63. doi:10.1186/gb-2012-13-7-r63
Rusché, L. N., Huang, C. E., Piller, K. J., Hemann, M., Wirtz, E., and Sollner-Webb, B. (2001). The Two RNA Ligases of the Trypanosoma Brucei RNA Editing Complex: Cloning the Essential Band IV Gene and Identifying the Band V Gene. Mol. Cel. Biol. 21, 979–989. doi:10.1128/mcb.21.4.979-989.2001
Sas-Chen, A., Thomas, J. M., Matzov, D., Taoka, M., Nance, K. D., Nir, R., et al. (2020). Dynamic RNA Acetylation Revealed by Quantitative Cross-Evolutionary Mapping. Nature 583, 638–643. doi:10.1038/s41586-020-2418-2
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. (2005). Improved and Versatile Transformation System Allowing Multiple Genetic Manipulations of the Hyperthermophilic Archaeon Thermococcus Kodakaraensis. Appl. Environ. Microbiol. 71, 3889–3899. doi:10.1128/aem.71.7.3889-3899.2005
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. (2003a). Targeted Gene Disruption by Homologous Recombination in the Hyperthermophilic Archaeon Thermococcus Kodakaraensis KOD1. J. Bacteriol. 185, 210–220. doi:10.1128/jb.185.1.210-220.2003
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. (2003b). Targeted Gene Disruption by Homologous Recombination in the Hyperthermophilic Archaeon Thermococcus Kodakaraensis KOD1. J. Bacteriol. 185, 210–220. doi:10.1128/jb.185.1.210-220.2003
Schmeing, T. M., Voorhees, R. M., Kelley, A. C., Gao, Y.-G., Murphy, F. V., Weir, J. R., et al. (2009). The Crystal Structure of the Ribosome Bound to EF-Tu and Aminoacyl-tRNA. Science 326, 688–694. doi:10.1126/science.1179700
Schnaufer, A., Panigrahi, A. K., Panicucci, B., Igo, R. P., Salavati, R., Stuart, K., et al. (2001). An RNA Ligase Essential for RNA Editing and Survival of the Bloodstream Form of Trypanosoma Brucei. Science 291, 2159–2162. doi:10.1126/science.1058955
Sidrauski, C., Cox, J. S., and Walter, P. (1996). tRNA Ligase Is Required for Regulated mRNA Splicing in the Unfolded Protein Response. Cell 87, 405–413. doi:10.1016/s0092-8674(00)81361-6
Starostina, N. G., Marshburn, S., Johnson, L. S., Eddy, S. R., Terns, R. M., and Terns, M. P. (2004). Circular Box C/D RNAs in Pyrococcus Furiosus. Proc. Natl. Acad. Sci. 101, 14097–14101. doi:10.1073/pnas.0403520101
Su, A. A. H., Tripp, V., and Randau, L. (2013). RNA-seq Analyses Reveal the Order of tRNA Processing Events and the Maturation of C/D Box and CRISPR RNAs in the Hyperthermophile Methanopyrus Kandleri. Nucleic Acids Res. 41, 6250–6258. doi:10.1093/nar/gkt317
Szewczak, A. A., Moore, P. B., Chang, Y. L., and Wool, I. G. (1993). The Conformation of the Sarcin/ricin Loop from 28S Ribosomal RNA. Proc. Natl. Acad. Sci. 90, 9581–9585. doi:10.1073/pnas.90.20.9581
Tang, T. H., Rozhdestvensky, T. S., d’Orval, B. C., Bortolin, M.-L., Huber, H., Charpentier, B., et al. (2002). RNomics in Archaea Reveals a Further Link between Splicing of Archaeal Introns and rRNA Processing. Nucleic Acids Res. 30, 921–930. doi:10.1093/nar/30.4.921
Toffano-Nioche, C., Ott, A., Crozat, E., Nguyen, A. N., Zytnicki, M., Leclerc, F., et al. (2013). RNA at 92°C. RNA Biol. 10, 1211–1220. doi:10.4161/rna.25567
Torchia, C., Takagi, Y., and Ho, C. K. (2008). Archaeal RNA Ligase Is a Homodimeric Protein that Catalyzes Intramolecular Ligation of Single-Stranded RNA and DNA. Nucleic Acids Res. 36, 6218–6227. doi:10.1093/nar/gkn602
Trotta, C. R., Miao, F., Arn, E. A., Stevens, S. W., Ho, C. K., Rauhut, R., et al. (1997). The Yeast tRNA Splicing Endonuclease: A Tetrameric Enzyme with Two Active Site Subunits Homologous to the Archaeal tRNA Endonucleases. Cell 89, 849–858. doi:10.1016/s0092-8674(00)80270-6
Uhlenbeck, O. C., and Gumport, R. I. (1982). 2 T4 RNA Ligase. The Enzymes 15, 31–58. doi:10.1016/s1874-6047(08)60274-7
Wang, L. K., Nandakumar, J., Schwer, B., and Shuman, S. (2007). The C-Terminal Domain of T4 RNA Ligase 1 Confers Specificity for tRNA Repair. Rna 13, 1235–1244. doi:10.1261/rna.591807
Wool, I. G., Glück, A., and Endo, Y. (1992). Ribotoxin Recognition of Ribosomal RNA and a Proposal for the Mechanism of Translocation. Trends Biochem. Sci. 17, 266–269. doi:10.1016/0968-0004(92)90407-z
Yin, S., Ho, C. K., and Shuman, S. (2003). Structure-function Analysis of T4 RNA Ligase 2. J. Biol. Chem. 278, 17601–17608. doi:10.1074/jbc.m300817200
Yoshinari, S., Liu, Y., Gollnick, P., and Ho, C. K. (2017). Cleavage of 3′-terminal Adenosine by Archaeal ATP-dependent RNA Ligase. Sci. Rep. 7, 11662. doi:10.1038/s41598-017-11693-0
Zhang, L., and Tripathi, A. (2017). Archaeal RNA Ligase from Thermoccocus Kodakarensis for Template Dependent Ligation. RNA Biol. 14, 36–44. doi:10.1080/15476286.2016.1239688
Keywords: circular RNA, RNA ligase, thermococcus kodakarensis KOD1, rRNA processing, C/D box sRNAs
Citation: Liu Y, Takagi Y, Sugijanto M, Nguyen KDM, Hirata A, Hori H and Ho CK (2022) Genetic and Functional Analyses of Archaeal ATP-Dependent RNA Ligase in C/D Box sRNA Circularization and Ribosomal RNA Processing. Front. Mol. Biosci. 9:811548. doi: 10.3389/fmolb.2022.811548
Received: 09 November 2021; Accepted: 08 February 2022;
Published: 25 March 2022.
Edited by:
Teng Ma, Capital Medical University, ChinaReviewed by:
Jinwei Zhang, National Institutes of Health (NIH), United StatesJörg Soppa, Goethe University Frankfurt, Germany
Copyright © 2022 Liu, Takagi, Sugijanto, Nguyen, Hirata, Hori and Ho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: C. Kiong Ho, a2lvbmdob0BtZC50c3VrdWJhLmFjLmpw
†These authors have contributed equally to this work and share first authorship
 Yancheng Liu
Yancheng Liu Yuko Takagi
Yuko Takagi Milyadi Sugijanto
Milyadi Sugijanto Kieu Duong My Nguyen
Kieu Duong My Nguyen Akira Hirata
Akira Hirata Hiroyuki Hori
Hiroyuki Hori C. Kiong Ho
C. Kiong Ho