- 1CRUK Beatson Institute, Glasgow, United Kingdom
- 2School of Infection and Immunity, University of Glasgow, Glasgow, United Kingdom
- 3School of Cancer Sciences, University of Glasgow, Glasgow, United Kingdom
Cell polarity and cell migration both depend on pseudopodia and lamellipodia formation. These are regulated by coordinated signaling acting through G-protein coupled receptors and kinases such as PKB/AKT and SGK, as well as the actin cytoskeletal machinery. Here we show that both Dictyostelium PKB and SGK kinases (encoded by pkbA and pkgB) are dispensable for chemotaxis towards folate. However, both are involved in the regulation of pseudopod formation and thus cell motility. Cells lacking pkbA and pkgB showed a substantial drop in cell speed. Actin polymerization is perturbed in pkbA- and reduced in pkgB- and pkbA-/pkgB- mutants. The Scar/WAVE complex, key catalyst of pseudopod formation, is recruited normally to the fronts of all mutant cells (pkbA-, pkgB- and pkbA-/pkgB-), but is unexpectedly unable to recruit the Arp2/3 complex in cells lacking SGK. Consequently, loss of SGK causes a near-complete loss of normal actin pseudopodia, though this can be rescued by overexpression of PKB. Hence both PKB and SGK are required for correct assembly of F-actin and recruitment of the Arp2/3 complex by the Scar/WAVE complex during pseudopodia formation.
Introduction
Cell migration is a fundamental biological process required for immune cell function, development and differentiation, among other roles. Alterations in migration can play central roles in diseases like cancer (Talmadge and Fidler, 2010; Bravo-Cordero et al., 2012) and infections (Zaki et al., 2006). Cell migration can be random or directional. Directed migration of cells is highly orchestrated and can occur in response to external chemical cues or signaling (Affolter and Weijer, 2005; Wang et al., 2011; Artemenko et al., 2014) called chemotaxis.
Cells move by forming actin rich structures, such as lamellipodia and pseudopodia. These protrusive structures are made through actin polymerization. Actin branching is mediated by the actin related protein complex Arp2/3, whose activation is driven ubiquitously by the recruitment of the Scar/WAVE complex at the front of the cells (Machesky and Insall, 1998; Krause and Gautreau, 2014; Schaks et al., 2019). The Scar/WAVE complex is an approximately 450 kDa hetero-pentameric complex, whose members are Pir121 (Cyfip), Nap1 (NCKAP1), Scar (WAVE), Abi, and HSPC300.
Activation of the Scar/WAVE complex at the leading edge is an essential step for the formation of pseudopodia, that is mediated by GTP bound active Rac1. At the cell front and in pseudopods, active Rac1 binds to the Pir121 subunit of the complex and activates it (Chen et al., 2010; Chen et al., 2017; Schaks et al., 2018). In addition to Rac1, phosphorylation of Scar/WAVE has been described as an additional regulator of the complex (Danson et al., 2007; Lebensohn and Kirschner, 2009; Mendoza et al., 2011). Earlier work flagged ERK2 kinase as important (Danson et al., 2007; Lebensohn and Kirschner, 2009; Mendoza et al., 2011), but we have recently shown that the SepA kinase, and not ERK2, phosphorylates Dictyostelium Scar/WAVE. However, neither controls its activation or inactivation. Still, phosphorylation of Scar/WAVE is useful as an indicator of the active state of the Scar/WAVE complex because it occurs immediately after the complex is activated (Singh et al., 2020; Singh et al., 2021). Phosphorylation of Scar/WAVE tunes cell migration by controlling the Scar/WAVE complex’s stability, lifetime, and recruitment to pseudopodia.
During our screen for kinases that phosphorylate Scar/WAVE, we also identified the Dictyostelium protein kinase B (PKB)/AKT homolog encoded by pkbA and the SGK kinase PkgB (also called PKB-R1) as potential interactors of the Scar/WAVE complex in protein crosslinking and pulldown experiments (Fort et al., 2018; Singh et al., 2020). Both proteins have been implicated in the regulation of actin-dependent processes as endocytosis (Williams et al., 2019) and chemotaxis (Meili et al., 1999; Meili et al., 2000; Kamimura et al., 2008; Kamimura and Devreotes, 2010), but their importance and their crosstalk with the Scar/WAVE complex are not yet understood.
The role of actin in macropinocytosis—in which cells feed by large scale uptake of liquid medium (Lewis, 1931; King and Kay, 2019)—is becoming clearer. Recruitment of the Scar/WAVE complex to macropinocytic cups is associated with F-actin polymerization (Veltman et al., 2016). Cells lacking Scar/WAVE complex members show defects in macropinocytosis. Similarly, cells lacking both PKB and PkgB show substantially reduced levels of macropinocytosis (Williams et al., 2019).
A range of publications describes the importance of PKB and PkgB to Dictyostelium directed cell migration and chemotaxis (Meili et al., 1999; Meili et al., 2000; Kamimura et al., 2008; Kamimura and Devreotes, 2010). Both kinases have been shown to be activated in response to cAMP (Meili et al., 1999; Meili et al., 2000), and PKB and PkgB phosphorylate common substrates after stimulation with cAMP or folate (Liao et al., 2010). These result in changes of the cytoskeleton during cell migration (Srinivasan et al., 2013). Cells lacking pkbA showed cell polarity defects and impaired motility (Meili et al., 1999), but only subtle migration defects were seen in cells lacking pkgB (Meili et al., 2000). In contrast, another report describes that cells lacking pkgB but not pkbA manifest severe defects in gradient sensing and chemotaxis (Kamimura et al., 2008). The chemotactic defects become even more severe in the pkbA-/pkgB- double knockout cells, and result in developmental arrest at the aggregation stage (Meili et al., 2000).
Altogether, this suggests that both kinases have roles in the regulation of cell motility and actin dynamics, though few mechanistic details are understood. In this study, we describe a link between Scar/WAVE phosphorylation, activation and both PKB, and PkgB kinases.
Materials and methods
Cell lines
All the cell lines Ax2, pkbA-, pkgB-, pkbA-/pkgB- used in this study were kind gifts from Dr. Robert R. Kay (Williams et al., 2019).
Dictyostelium cell culture
All cell lines were grown with bacteria Klebsiella aerogenes on SM agar plates. Bacterial lawns were prepared on SM agar plates by spreading bacteria and allowing them to grow overnight at 22°C. Cells were streaked out on bacterial lawns with a sterile inoculation loop for approximately 48 h, by which time cells have cleared areas of bacteria and formed growth zones. Cells from growth zones were collected with a sterile inoculation loop (10 µL), washed 3 times with development buffer (DB) to clean remaining bacteria and used in under agarose chemotaxis or western blotting analyses.
Plasmid constructs
Lifeact-mRFPmars2, mRFPmars2-ArpC4 or Pak-CRIB-mRFPmars2 expression fragments were cloned into pDM1091 using the NgoMIV restriction site to generate co expression constructs with HSPC300-eGFP. To construct extrachromosomal expression vectors of PKB and PkgB, pkbA and pkgB were PCR-amplified using Ax3 genomic DNA. pkbA was amplified using a forward primer (AGATCTAAAATGGGAAAAGGACAAAGTAA) and reverse primer (ACTAGTTAATCCTTTAAGATTGAATCAGC). pkgB was amplified using a forward primer (AGATCTAAAATGTCAACAGCACCAATTAAAC) and reverse primer (ACTAGTTATCTTAAATGTTCAGATTCAGCG). Amplified products were cloned into pDM1033 using BglII and SpeI restriction sites.
Transfection
Cells were grown on SM agar plates in conjunction with Klebsiella aerogenes. Cells were collected from the clearing zones with sterile inoculation loops and transfected as described in Paschke et al., 2019 (Paschke et al., 2019).
Under agarose chemotaxis
Cellular morphology, pseudopod dynamics and cell migration were measured by an under-agarose folate chemotaxis assay as described previously (Singh et al., 2020; Singh and Insall, 2022). In brief, 0.4% SeamKem GTG agarose in Lo-Flo medium (Formedium) was boiled. After cooling, folic acid to a final concentration of 10 μM was added. 5 ml of agarose-folate mix was poured into the 1% BSA-coated 50 mm glass bottom dishes (MatTek). A 5 mm wide well was cut with a sterile scalpel and filled with 200 μL of 2 × 106 cells/ml. Cell migration was imaged after 4–6 h with 10x and 60x differential interference contrast (DIC) microscopy. To examine the localization of labelled proteins in the pseudopods, cells were also imaged by AiryScan confocal microscopy.
Western blotting
Cells were lysed by directly adding NuPAGE LDS sample buffer (Invitrogen) containing 20 mM DTT, HALT protease and phosphatase inhibitors (Thermo Fisher Scientific) onto the cells. Samples were boiled at 100°C for 5 min. Proteins were separated on 10% Bis-Tris NuPAGE gels (Invitrogen) or on hand-poured low-bis acrylamide (0.06% bis acrylamide and 10% acrylamide) gels, then separated at 150 V, 90 min. Proteins were transferred onto 0.45 μM nitrocellulose membrane. Membranes were blocked in TBS+5% non-fat milk. Primary antibodies, rabbit anti-Scar (Blagg et al., 2003), Abi (Pollitt and Insall, 2008), Nap1 and sheep anti-Pir121 (Ibarra et al., 2006), rat anti-GFP(3H9, Chromotek, RRID; AB_10773374) were used at 1:1000 dilution. Goat anti rabbit IgG (H + L) Dylight 800(Invitrogen; SA5-35571; RRID:AB_2556775) Rabbit anti Sheep IgG (H + L) Dylight 800 (Invitrogen, SA5-10058; RRID:AB_2556638) or Goat anti-Rat IgG (H + L) Dylight 800 (Invitrogen, SA5-10024; RRID:AB_2556604) fluorescently conjugated secondary antibodies at 1:10,000 dilution (were used to detect the protein bands using an Odyssey CLx Imaging system (LI-COR Biosciences). Mccc1 was used as a loading control (Davidson et al., 2013a). Streptavidin Alexa 680 conjugate was used to probe Mccc1 at 1:1000 dilution (Invitrogen, S32358).
Phosphatase treatment
Cells grown in a 35 mm Petri dish were lysed in 100 μL TN/T buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.1% Triton X-100), kept on ice for 5 min and cleared by centrifugation (13,000 rpm, 4°C, 5 min). Proteins were dephosphorylated using 1 μL Lambda phosphatase at 30°C (NEB; P0753S) for 1 h. Protein dephosphorylation was assessed by western blotting.
Microscopy
To determine the morphology, chemotactic speed and directionality of cells, phase contrast time lapse microscopy was performed at 10x/0.3NA on a Nikon ECLIPSE TE-2000-R inverted microscope equipped with a Retiga EXI CCD monochromatic camera. Images of cells migrating under agarose in a folate gradient were captured per minute for 45 min. DIC images were taken every 3 s for 3 min with 60x/1.4 NA to observe the pseudopod formation. HSPC300-eGFP expressing cells were used to determine the activation of the Scar/WAVE complex. Localization of the Scar/WAVE complex, Arp2/3 complex, and F- actin was examined by a 63x/1.4 NA objective on an AiryScan Zeiss 880 inverted confocal microscope. The brightness and contrast of the images were adjusted to visualize the cell protrusions and defects using ImageJ.
Quantification of data and statistics
Each experiment was performed at least thrice. Speed of cells, chemotaxis efficiency index, directedness, circularity, cell area were calculated from the movies of cells migrating under agarose up a folate gradient using homemade plugins in ImageJ (17). Pseudopodia dynamics was quantified from DIC movies of cells migrating under agarose up a folate gradient by counting the number of frames during a pseudopodia extension manually using ImageJ. The Scar/WAVE complex dynamics; patch lifetime and patch frequency were calculated from the movies of cells migrating under agarose up a folate gradient using homemade plugins in ImageJ (Singh et al., 2021).
Graphs and statistical analyses were performed using Prism. To quantify the proportion of Scar/WAVE phosphorylation, the total intensity of all bands and the lowest band were determined using the Odyssey CLx Imaging system (LI-COR Biosciences, Lincoln, NE, United States). The percentage of phosphorylation was calculated by using the formula: % phosphorylation = (Total intensity−intensity of lowest band)*100/Total intensity. Statistical significance analyses were performed by non-parametric statistics, such as one-way ANOVA and Dunn’s multiple comparison tests, as described for each result. The experimental groups were tested for normal distribution using the Shapiro-walk test of Prism 7 software (Graphpad, Graphpad, San Diego, CA, United States). Sample sizes are provided in the figure legends. n refers to; independent experiments in western blotting, total number of cells from 3 independent experiments for cell speed, pseudopod dynamics, circularity, solidity and cell area.
Results
Scar phosphorylation is diminished in pkbA-/pkgB- cells
We recently showed that Scar/WAVE phosphorylation occurs after the activation of the complex and is to a large extent mediated by SepA kinase (Singh et al., 2020; Singh and Insall, 2020). However, in sepA- cells phosphorylation of the Scar/WAVE was not entirely eliminated, meaning that other kinases are also active. We therefore examined a range of mutants in known kinases implicated in cell migration.
We recently established that the most effective way to measure the different phosphorylation states of Scar/WAVE uses western blots from low-bis acylamide gels. Phosphorylated proteins are visualised as a ladder of slower-migrating bands that are abolished by phosphatase treatment (Singh et al., 2020; Singh et al., 2021). We tested how far PKB and SGK contribute to Scar/WAVE phosphorylation in wild type (Ax2), pkbA-, pkgB- and pkbA-/pkgB- cells. In migrating Dictyostelium cells, multiple phosphorylated Scar bands can be resolved (Figure 1A), giving distinct peaks on the intensity plot (asterisks, Figure 1B). Lambda phosphatase treatment resolved all of the Scar phosphorylation bands into one single band and intensity peak (Figures 1A,B), confirming that the banding pattern is exclusively driven by different patterns of phosphorylation. The number of phosphorylated bands was unaltered in pkbA- and pkgB- cells, but interestingly, Scar phosphorylation was greatly diminished in the pkbA-/pkgB- double knockout cells (Figures 1C–E). We analyzed Scar band intensities by drawing a line across bands in ImageJ and plotted the values in graphs. Each peak on the graph represents one band of Scar (asterisks, Figure 1D). The intensity profile shows distinct multiple sharp peaks in Ax2, pkbA- cells and pkgB-, but only one in pkbA-/pkgB- cells (asterisks, Figure 1D). Quantification of the fraction of Scar in the upper bands shows a trend of reduction, which is 50% less in pkbA-/pkgB- cells (Figure 1E). This graph underestimates the total change in Scar phosphorylation, because the higher bands that are lost from pkbA-/pkgB- cells contain several phosphates (Singh et al., 2020). These results suggest that Scar/WAVE activation is altered in pkbA-/pkgB- cells but not in pkbA- or pkgB-.
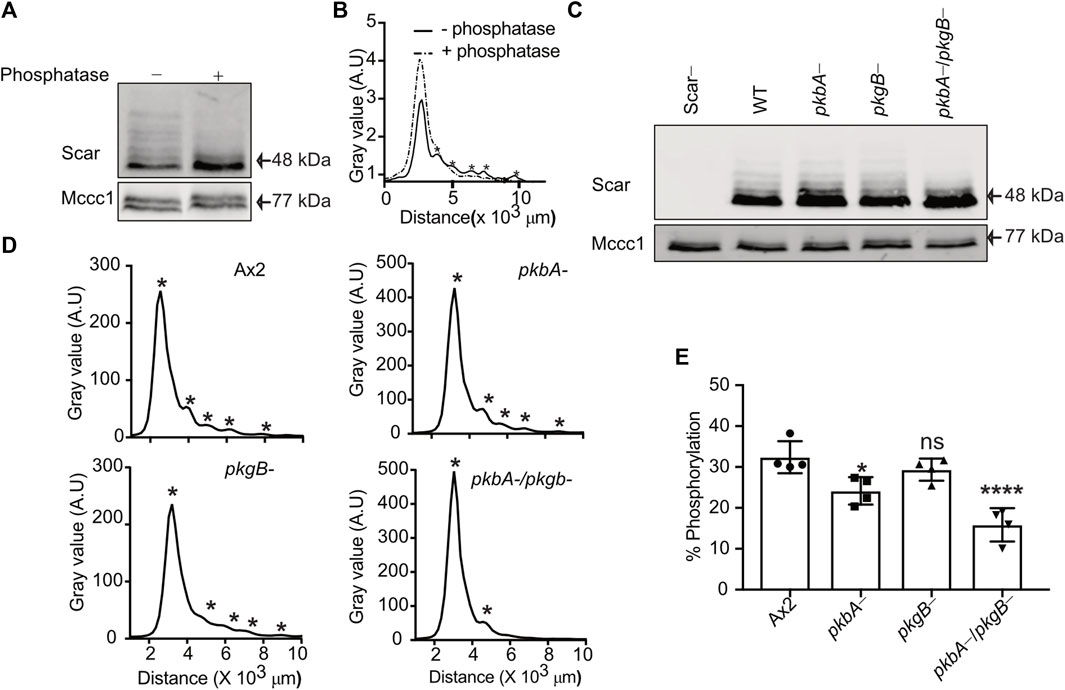
FIGURE 1. Phosphorylation of Scar is diminished in Dictyostelium pkbA-/pkgB-. (A) Assay of Scar phosphorylation. Ax2 cell lysates treated with phosphatase were analyzed by western blotting. (B) Density quantification shows multiple peaks (solid line) resolved into one (dotted line) after phosphatase treatment. Asterisks at the peaks represents Scar bands. (C) Assay of Scar phosphorylation in Ax2, pkbA-, pkgB- and pkbA-/pkgB- cells. Whole cell lysates were analysed by western blotting. (D) Density quantification shows flattening of multiple peaks (asterisks) in pkbA-, pkgB-, and loss in pkbA-/pkgB-. (E) Shows ratio of upper (more-phosphorylated) band to aggregate intensity of total Scar (bars show mean ± SD, n = 4 independent experiment, *p ≤ 0.05, ****p ≤ 0.0001, One-way Annova, Dunnett’s multiple comparison test). Mccc1 was used as loading control.
A role for PKB and PkgB in cell motility
Phosphorylation of Scar/WAVE impacts cell motility and protrusion dynamics (Singh et al., 2020). To assess the role of PKB and PkgB in cell migration, we assayed the ability of Ax2, pkbA-, pkgB- and pkbA-/pkgB- cells to move up a folate gradient under agarose chemotaxis and examined their migratory phenotypes (Figure 2; Supplementary Videos S1–S3). Like Ax2, pkbA- cells formed pseudopodia and migrated efficiently. However, pkgB- and pkgB-/pkbA- cells do not form proper pseudopodia and migrated with the aid of thin, short-lived protrusions (arrows, Figure 2A; Supplementary Video S1). The formation of pseudopods in pkbA- cells was more frequent compared to Ax2, but once formed they lasted for a similar time (Figures 2B,C). Since pkgB- and pkbA-/pkgB- cells do not form normal pseudopods, their lifetime and frequency could not be quantified. Instead, we quantified cell spreading (cell area) and polarity (circularity and solidity). In both cases pkgB-/pkbA- mutants were substantially compromised, while pkgB- cells showed slight defects and pkbA- cells seemed unaffected (Figures 2D,E; Supplementary Figure S1). All kinase mutants appear to migrate slower than the Ax2 parental cells (Figure 2F), though the double mutant differs hugely. Directedness and chemotactic efficiency index are unaffected in all mutants (Figures 2G,H). The same is not true of shape, which is of course profoundly affected by the loss of normal pseudopod formation. The pkbA -/pkgB- double knockout has ∼20% smaller projected area and a substantially higher circularity and solidity (Figures 2D,E,G,H; Supplementary Figure S1). All together these results show that loss of both pkbA and pkgB impairs formation of pseudopodia, which results in defective cell motility, but has no obvious effect on guidance.
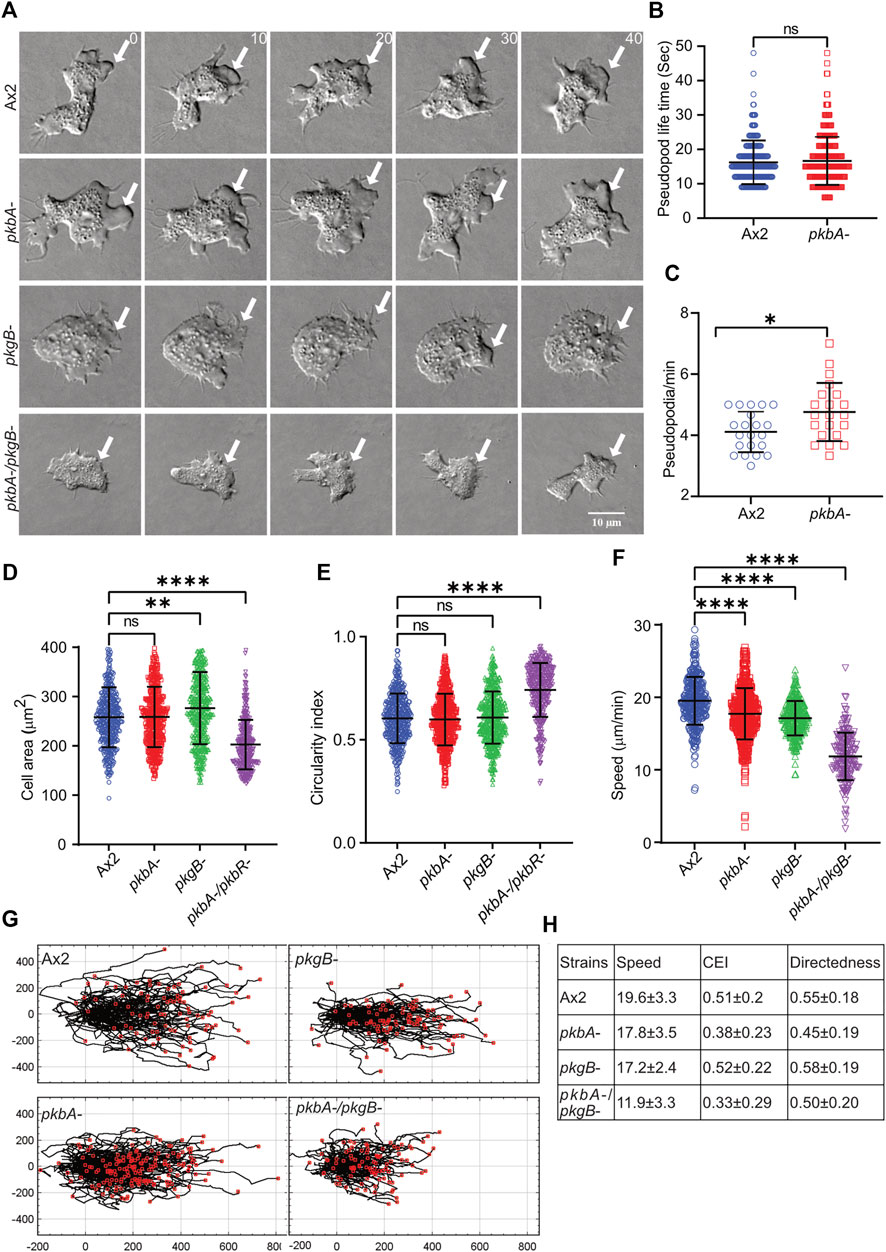
FIGURE 2. pkgB is required for efficient pseudopodia formation and cell migration. (A) Representative images of >20 cells showing pseudopodia formation in Ax2 and mutant cells. Ax2, pkbA-, pkgB- and pkbA-/pkgB- cells were allowed to migrate under agarose sensing a folate gradient and were imaged by DIC microscopy at a frame interval of 2 s (1f/2 s). Pseudopods are formed in Ax2 and single mutant pkbA-, but not in pkgB- and double mutant pkbA-/pkgB- (arrows). (B–C) Effect of pkbA- on pseudopods dynamics. Pseudopod lifetime and generation rate was measured from DIC videos using ImageJ from single frames, and generation rate calculated from the number of pseudopods lasting at least 2 frames. Lifetime of pseudopodia is equal in both Ax2 and pkbA- cells, but pkbA- generates pseudopods more frequently. (Mean ± SD, n > 20 cells, n > 25 cells from 3 experiments; *p ≤ 0.05, 1-way ANOVA, Mann-Whitney test). (D,E) Shows quantification of cell area and circularity index. Cell area is unchanged in Ax2 and pkbA-, increased in pkgB-, but decreased in pkbA-/pkgB-. Circularity index is only increased in pkbA-/pkgB-. (mean ± SD; n = 416Ax2, 474pkbA-, 458pkgB- and 366pkbA-/pkgB- over 3 independent experiments, **p ≤ 0.01, ****p ≤ 0.0001, 1-way ANOVA, Dunn’s multiple comparison test). (F) Shows quantification of cell speed. Mutants migrate slower than Ax2 cells and (mean ± SD; n = 361Ax2, 496pkbA-, 298pkgB- and 200pkbA-/pkgB- over 3 independent experiments, ****p ≤ 0.0001, One-way ANOVA, Dunn’s multiple comparison test). (G) Shows cell tracks of migrating cells. (H) Shows quantified values of speed, chemotactic efficiency index (CEI) and directedness of cells.
To confirm that the defects in motility and pseudopod generation are due to PKB and PkgB depletion, we expressed individual kinases in pkgB- and pkbA-/pkgB- cells and observed the rescue of pseudopods in migrating cells in an under-agarose folate chemotaxis assay (Supplementary Figures S2A,B, Supplementary Videos S2, S3). Rescued pkgB-/pkgB cells, somewhat unexpectedly, migrated with a slower speed than pkgB- with unaffected cell polarity (circularity and solidity) (Supplementary Figures S2C,D,E,F). This effect is possibly due to a dominant effect of overexpression of pkgB (Kamimura et al., 2008). Expression of either pkbA or pkgB in pkbA-/pkgB- cells substantially restores pseudopod formation, increases cell speed and polarity (Supplementary Figures S2B,C,D,E; Supplementary Video S3). This may be due to the overlap of substrates between the two kinases, meaning that overexpression of PKB may compensate for the lack of PkgB (Kamimura et al., 2008; Williams et al., 2019).
Both PKB and PkgB are required for proper dynamics and spatiotemporal distribution of actin
Pseudopods are actin rich structures, which drive the movement of cells during migration. Cells deficient in pkgB- and pkbA-/pkgB- cells do not form proper pseudopodia (Figure 2A; Supplementary Video S1). This may be attributed to poor actin polymerization and distribution during cell migration (Kamimura et al., 2008; Liao et al., 2010; Srinivasan et al., 2013). To examine actin polymerization and dynamics in Ax2 and mutant cells, we expressed Lifeact-mRFPmars2 (F-actin marker). F-actin efficiently labels the pseudopods in migrating Ax2 and pkbA- cells (asterisk, Figure 3A). Unlike Ax2, pkbA- cells had a strong F-actin enrichment in pseudopods and in the uropod (arrows, Figure 3A; Supplementary Video S4). Both Ax2 and pkbA- cells had approximately similar F-actin intensities in their pseudopods (Figure 3B). However, a striking feature of polymerized actin in the pseudopods of pkbA- was the static nature of its actin structures, when pseudopodia were growing due to polymerization of new actin (arrows in pkbA- panel; Figure 3A; Supplementary Figure S3A; Supplementary Video S4). Approximately 32% of Ax2, 88% of pkbA-, 100% of pkgB- and 32% of pkbA-/pkgB- cells have F-actin accumulation in the rear or cell body (Supplementary Figures S3A,B). This may be due to poor pseudopodia detachment from the substratum or enhanced adhesion, which could be the reason for the slightly slower migration of pkbA- cells, and lack of actin reaching pseudopodia formation in pkgB- cells.
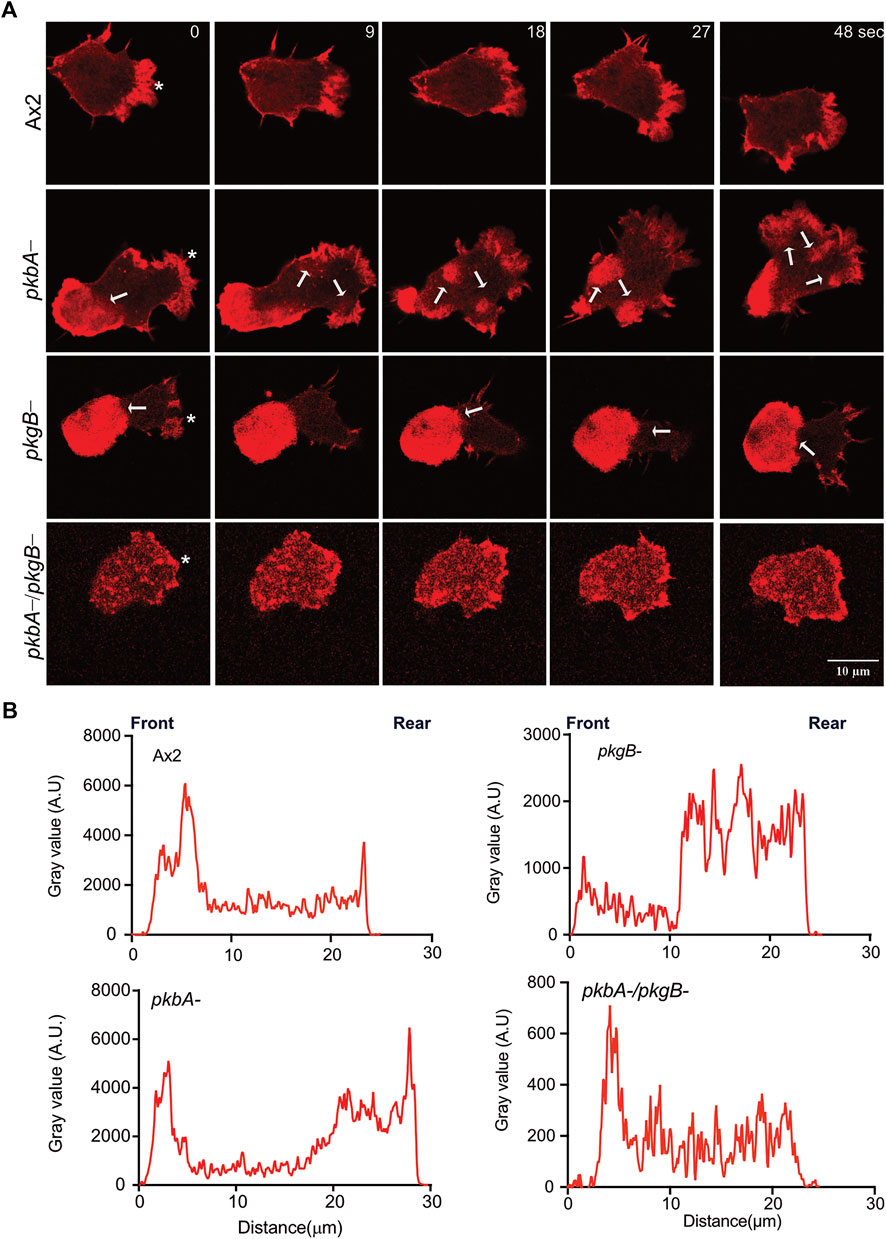
FIGURE 3. Actin localizes to Ax2 and pkbA- protrusions, but not in pkgB- and pkbA-/pkgB-. (A) Subcellular localization of F-actin (Red). Life-act-mRFPmars2 was expressed in Ax2, and mutant cells. Cells were allowed to migrate under agarose mediated by a folate gradient and imaged by AiryScan confocal microscopy at a frame interval of 3 s (1f/3 s). Ax2 and pkbA- cells show F-actin in pseudopodia (asterisk). F-actin accumulation is absent in pkgB- protrusions, and highly reduced in pkbA-/pkgB-. Static F-actin in the cell body is present in both pkbA- and pkgB-. (B) Quantification of the florescence intensities in cells. Using ImageJ, intensity profile plots were generated by drawing a straight line across the cell. The amount of F-actin is similar in Ax2 and pkbA-, but reduced in pkgB- and pkbA-/pkgB- cells, where F-actin is mostly localized in the cell body. Images are representative of ≥10 cells from 3 independent experiments.
In contrast, in pkgB- cells, F-actin is mainly accumulated in the cell body, but not in protrusions (Figure 3A; Supplementary Video S4). Intensity plots confirmed that the protrusions of pkgB- have much reduced F-actin (Figure 3B). Impaired actin polymerization in the protrusions, and accumulation of F-actin in the cell body could explain the slight migratory defect in those cells (Figure 2D). Interestingly, little actin localization with much reduced intensity compared to Ax2 was observed at the cell periphery and front of pkbA-/pkgB- cells (Figures 3A,B; Supplementary Video S4). These results suggest that PkgB is crucial, but both PKB and PkgB are important for regulation of actin polymerization in pseudopods.
The Scar/WAVE and Arp2/3 complexes activation is altered in pkgB- and pkbA-/pkgB-
Stability of the Scar/WAVE complex is important for its activity modulation (Ura et al., 2012; Davidson et al., 2013b). Hence, we investigated the influence of PKB and PkgB loss on the stability of the Scar/WAVE complex in Ax2 and mutant strains. We expressed HSPC300-eGFP in Ax2 and mutant cells and purified the complex using a GFP-TRAP pulldown. Western blot analysis for Pir121, Nap1, Scar, Abi and GFP showed stable complex formation in all cell lines (Supplementary Figure S4A).
The recruitment for Scar/WAVE complex, and the Arp2/3 complex activation it mediates, are pre-requisites for the polymerization of actin in pseudopods or lamellipods (Insall and Machesky, 2009; Krause and Gautreau, 2014). To measure the effects of pkbA and pkgB mutation on the recruitment of the Scar/WAVE and activation of the Arp2/3 complex, we co-expressed HSPC300-eGFP and ArpC4-mRFPmars2 (Veltman et al., 2012) in mutant strains and examined recruitment in migrating cells. The Scar/WAVE complex localizes in the protrusions of all strains with marked differences in the intensities, dynamics, and activity. The Scar/WAVE and Arp2/3 complexes colocalised in the protrusions of Ax2 (Figure 4A; Supplementary Video S5). The recruitment of both Scar and Arp2/3 complexes was compromised in the protrusion of pkbA-, pkgB- and pkbA-/pkgB- cells (Figures 4A–C; Supplementary Figures S4B,C; Supplementary Video S5). Both complexes accumulated substantially in the cell body of pkgB- cells (see pkgB- panel in Figures 4A–C; Supplementary Figure S4B; Supplementary Video S5). Intensity plots of the Scar/WAVE and Arp2/3 complexes indicate similar intensity in protrusions of all cells, but the mutants have more cytosolic accumulation (Figures 4A,B; Supplementary Video S5).
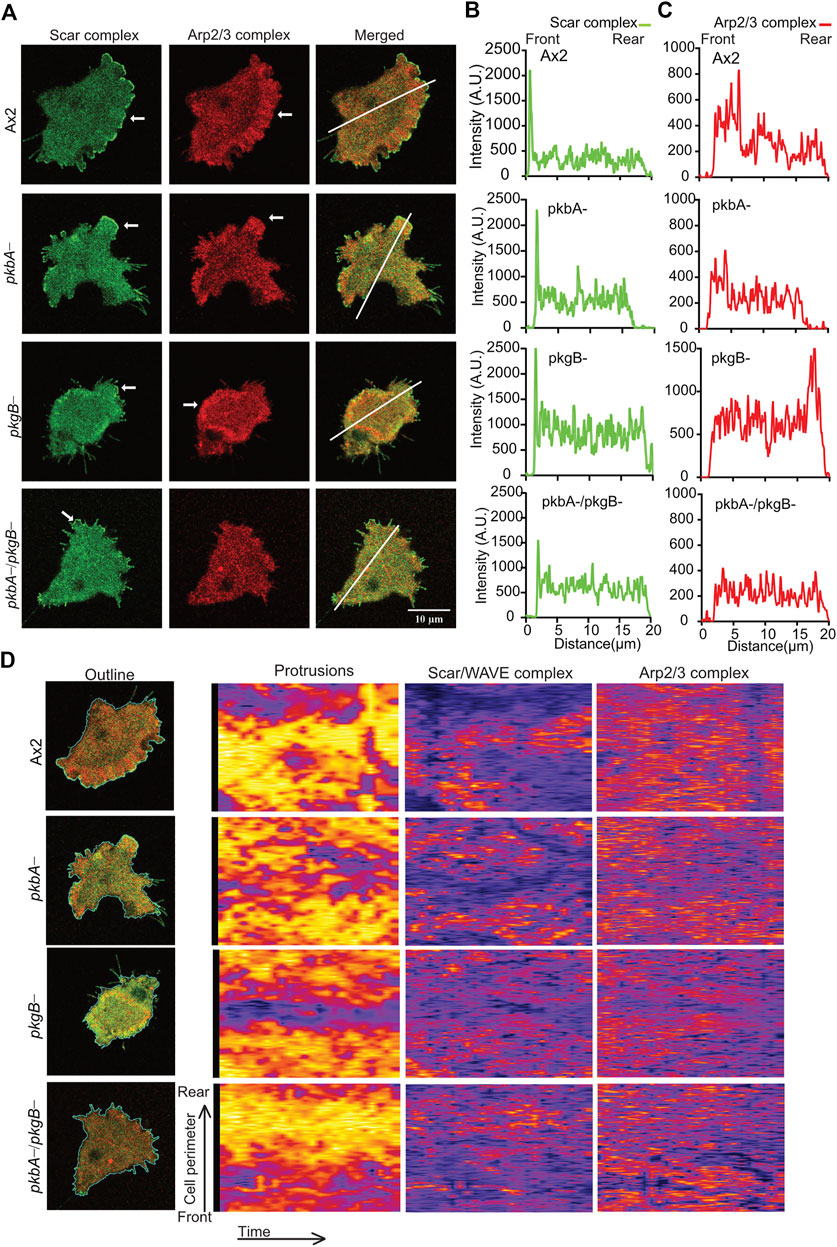
FIGURE 4. The recruitment of Scar/WAVE and Arp2/3 complexes in Ax2 and mutant cells. (A) Subcellular localization of the Scar/WAVE and the Arp2/3 complexes in pseudopods. Ax2, pkbA-, pkgB- and pkbA-/pkgB- cells were labelled with HSPC300-eGFP (Scar/WAVE complex) and mRFPmars2-ArpC4 (Arp2/3 complex) and allowed to migrate under agarose following a folate gradient and imaged by AiryScan confocal microscopy at a frame interval of 3 s (1f/3 s). The Scar/WAVE complex is localized to protrusions of pseudopodia of all cell types (arrows, panel 1). The Arp2/3 complex is enriched in the pseudopodia of Ax2 and pkbA- cells, in the cell body of pkgB-, but is lacking in the pkbA-/pkgB- cells (arrows, panel 2). (B,C) Quantification of the Scar/WAVE and Arp2/3 complex florescence intensities along the indicated line across the cell (panel 3 of A). (D) Quantitative assessment of the Scar/WAVE and Arp2/3 complex recruitment in cells. Panel 1; shows the outline of cells used for the generation of kymographs using ImageJ. Panel 2 represents protrusions, panel 3; the Scar/WAVE complex and panel 4; the Arp2/3 complex. Images are representative of ≥10 cells from 3 independent experiments.
The frequency and lifetime of Scar/WAVE is in general highly dynamic. In the mutants, recruitment seemed transient and oscillatory. To quantify the differences in the lifetime of the Scar/WAVE and Arp2/3 complexes in our cells, we generated whole-boundary kymographs and intensity graphs using a custom-made ImageJ plugin. In all cell lines, protrusion (Figure 4D, protrusions column) is highly correlated with Scar/WAVE recruitment (compare Figure 4D, Scar/WAVE column), with protrusions centered on areas rich in Scar/WAVE. WT Ax2 cells have large, long-lasting pseudopods centred on longer-lasting Scar patches (dotted line; Figures 5A–D). The lifetimes of these patches are reduced in pkbA- and reduced further still in both pkgB- and double mutant cells (Figures 5B–E). In pkgB- cells, Scar mostly accumulated in the periphery of the cell body, and in the pkbA-/pkgB-, Scar accumulation was observed mostly as puncta in protrusions (dotted line; Figure 5A).
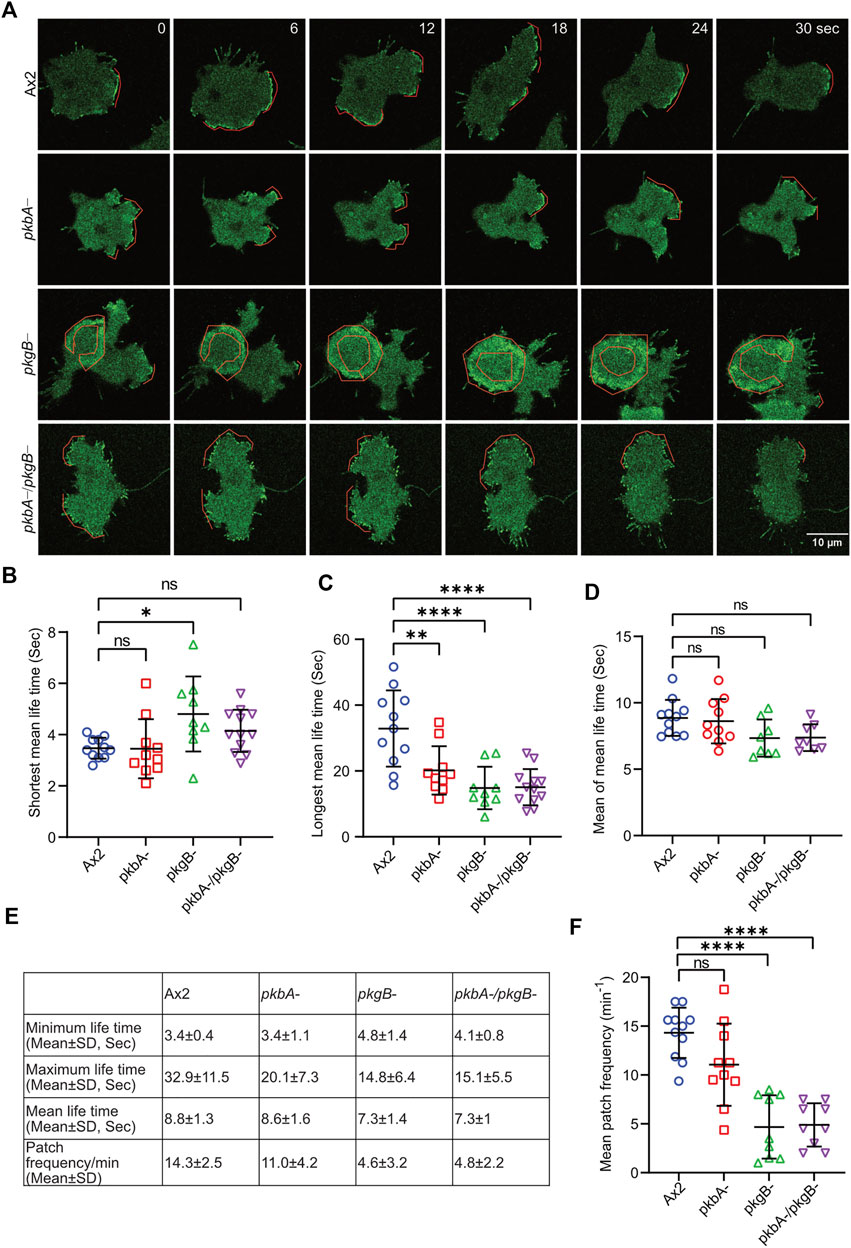
FIGURE 5. The recruitment of the Scar/WAVE complex in Ax2 and mutant cells. Subcellular localization of the Scar/WAVE complex in protrusions. (A) Ax2, pkbA-, pkgB- and pkbA-/pkgB- cells were labelled with HSPC300-eGFP (Scar/WAVE complex) and allowed to migrate under agarose following a folate gradient and imaged by AiryScan confocal microscopy at a frame interval of 3 s (1f/3 s). The Scar/WAVE complex is localized to protrusions of pseudopodia of all cell types (dotted line). (B–D) Quantification of lifetime to Scar/WAVE recruitment. The lifetime of the Scar/WAVE complex accumulation was quantified using homemade ImageJ plugin. Mean lifetimes of shortest (B), longest (C) and mean (D) Scar/WAVE duration were plotted. (n > 9 cells over three experiments, mean ± SD, ns = non-significant *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, One-way ANOVA, Dunn’s multiple comparison test). (E) Scar/WAVE lifetimes in protrusions of cell lines. (F) Scar/WAVE recruitment frequency in the protrusions of cell-lines.
The same correlation holds true in pkbA- cells, though they have a broader spread of shorter-lived Scar/WAVE flashes (Figures 4D, 5B–E–5E). In both pkgB- and pkbA-/pkgB- cells, Scar/WAVE and Arp2/3 complex flashes were transient (compare Arp2/3 column of Figure 4D with Supplementary Figure S4B). Moreover, Scar/WAVE recruitment frequency was highly reduced in mutant cell-lines, compared to Ax2 (Figures 5E,F). This suggests that both PKB and PkgB are required for the proper activation of the Scar/WAVE complex and coupling from active Scar/WAVE to the Arp2/3 complex.
Discussion
We report that PKB and PkgB regulate Scar/WAVE phosphorylation and pseudopod formation in Dictyostelium. We have shown that Scar/WAVE phosphorylation is strongly reduced in pkbA-/pkgB- cells. Single mutants were largely unaffected, and PKB and PkgB, both share a conserved kinase motif, suggesting redundancy in their substrates. The classical Akt/Sgk motif RXRXXS∗/T∗ is not found within the serine residues that have been found to be important for Scar phosphorylation (Singh et al., 2020), nor is any member of the Scar/WAVE complex among the known targets of these enzymes. It therefore seems likely that PKB and PkgB control Scar phosphorylation through other, intermediate kinases. This assumption is supported by phospho-proteomics data published for pkbA-, pkgB- and double mutant which include known kinases as KrsB and Pats1 (Williams et al., 2019). However, there is a strong possibility that both PKB and PkgB are in some way involved in the regulation of SepA which has been previously reported to be essential for Scar/WAVE phosphorylation. The sepA knockout showed a similar level of reduction in phosphorylation, making a connection between the kinases likely (Singh et al., 2020). How this crosstalk between the different kinases is mediated, will need to be further investigated in the future.
We used folate chemotaxis to assay the morphological and cytoskeletal changes caused by loss of pkbA and pkgB. These changes appear to be driven by changes in the phosphorylation state of Scar/WAVE. This is likely to be true for other chemoattractants - cAMP and folate induce an identical set of substrate phosphorylation and operate through common pathways (Liao et al., 2010; Srinivasan et al., 2013), but we have focussed on folate because loss of PKB affects development. While common signaling networks are involved in the response to both chemoattractants, differences have been observed before when comparing folate to cAMP chemotaxis in the same mutant cell line (Nichols et al., 2019). cAMP chemotaxis does not only depend on the actin cytoskeleton but also on cell differentiation and the correct expression of developmental markers to make cells able to respond efficiently to cAMP.
Previous studies have reported that both kinases are important for cAMP chemotaxis (Meili et al., 1999; Meili et al., 2000; Kamimura et al., 2008; Kamimura and Devreotes, 2010). In contrast, this work reveals that both PKB and PkgB are dispensable for chemotaxis to folate but important for pseudopod formation and cell speed. This is less likely to represent changes in mechanism, and more likely a complication of developmental changes in pkbA and pkgB mutants. There are also assay-specific differences in the way cells experience the gradient of chemoattractant. We view cells in a self-generated gradient when performing chemotaxis under agarose, because this gives optimal conditions for visualising Scar/WAVE complex recruitment during pseudopod formation. The behavior of cells is different when they respond to a chemoattractant that is released from micropipettes.
We know that mechanism of chemotaxis does not principally work through initiation of new protrusions, but rather through changes to pseudopod dynamics (Andrew and Insall, 2007; Bosgraaf and Van Haastert, 2009). Pseudopods control chemotactic steering (Andrew and Insall, 2007; Bosgraaf and Van Haastert, 2009), persistence, polarity, cell shape and motility (Fort et al., 2018). However, in our assays pkgB- and pkbA-/pkgB- cells move towards folate sources without forming proper pseudopods, which raises the possibility of pseudopodia independent chemotaxis, using other transient protrusions. The defects in pseudopod formation in pkgB- is more severe than in pkbA-. This might be explained by a higher level of substrate phosphorylation by PkgB compared to PKB (Kamimura et al., 2008).
As the loss of pseudopod formation in pkbA-/pkgB- cells can be rescued by extrachromosomal expression of either pkbA or pkgB. This is because both kinase share substrates and over-expression of either kinase has been shown to rescue phenotypes of pkbA-/pkgB- cells (Meili et al., 2000; Williams et al., 2019).
To generate and extend pseudopodia, cell substrate adhesion is essential (Ladoux and Nicolas, 2012). The defects observed in pkgB- and pkbA-/pkgB- may be due to the inability of pseudopods to attach with substratum. Talin integrates, adhesion molecules like integrins (homolog of SibA&B in Dictyostelium) to the actin cytoskeleton (Niewohner et al., 1997; Simson et al., 1998). In Dictyostelium Talin B has been identified as one of the substrates of PKB and PkgB (Chen et al., 2017) and cells lacking TalinB have severe adhesion and motility defects (Tsujioka et al., 1999; Tsujioka et al., 2004). We have also reported that adhesion stimulates activation and phosphorylation of Scar/WAVE in pseudopods of migrating cells (Singh et al., 2020; Singh and Insall, 2020).
Additionally, we observed impaired activation of the Scar/WAVE and Arp2/3 complexes in the protrusions of pkgB- and pkbA-/pkgB- cells. This leads to a strongly reduced level of F-actin at the leading edge of the cell, due to the disrupted interaction between both complexes (Scar/WAVE at the front, Arp2/3 at rear in pkgB- or barely expressed in the double mutant). The hyperaccumulation of the Scar/WAVE, Arp2/3 complexes and F-actin in cell body indicates that PkgB is involved in the recruitment of the Scar/WAVE complex in the pseudopodia. The eventual destination of the Scar/WAVE complex is exceptionally complicated, being affected by positive feedback, retrograde actin flow, and a poorly-understood, activation-dependent degradation mechanism. We note that in pkgB- cells, Scar mostly accumulated in the periphery of the cell body, whereas in the pkbA-/pkgB-, it was observed mostly as puncta in protrusions (Figure 5A). It is not yet clear what causes this difference.
The undermined Scar/WAVE and Arp2/3 complexes recruitment in pkbA-, pkgB- and pkbA-/pkgB- hints that both PKB and PkgB seem important for Arp2/3 complex localization and activation.
Overall, this study finds that PkgB independently or together with PKB regulates pseudopodia formation and cell migration via the Scar/WAVE and Arp2/3 complexes and raises multiple questions regarding regulation of cell motility.
Data availability statement
All the raw data for the figures and videos used in this study can be requested from RI (Robert.Insall@glasgow.ac.uk) and SS (Shashi.Singh@glasgow.ac.uk).
Author contributions
SS; conceptualization, investigation, methodology, data curation, analysis, writing original draft. PP; resources, review and editing, LT; Software, review and editing, RI; Supervision, administration funding acquisition, review and editing the manuscript.
Funding
This work was supported by Cancer Research United Kingdom core Grant Number A17196, A31287 and Multidisciplinary Award A20017 to RHI.
Acknowledgments
We thank Robert R. Kay for providing cells.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.965921/full#supplementary-material
References
Affolter, M., and Weijer, C. J. (2005). Signaling to cytoskeletal dynamics during chemotaxis. Dev. Cell. 9 (1), 19–34. doi:10.1016/j.devcel.2005.06.003
Andrew, N., and Insall, R. H. (2007). Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat. Cell. Biol. 9 (2), 193–200. doi:10.1038/ncb1536
Artemenko, Y., Lampert, T. J., and Devreotes, P. N. (2014). Moving towards a paradigm: Common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell. Mol. Life Sci. 71 (19), 3711–3747. doi:10.1007/s00018-014-1638-8
Blagg, S. L., Stewart, M., Sambles, C., and Insall, R. H. (2003). PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13 (17), 1480–1487. doi:10.1016/s0960-9822(03)00580-3
Bosgraaf, L., and Van Haastert, P. J. (2009). Navigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PLoS One 4 (8), e6842. doi:10.1371/journal.pone.0006842
Bravo-Cordero, J. J., Hodgson, L., and Condeelis, J. (2012). Directed cell invasion and migration during metastasis. Curr. Opin. Cell. Biol. 24 (2), 277–283. doi:10.1016/j.ceb.2011.12.004
Chen, B., Chou, H. T., Brautigam, C. A., Xing, W., Yang, S., Henry, L., et al. (2017). Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. Elife 6, e29795. doi:10.7554/eLife.29795
Chen, Z., Borek, D., Padrick, S. B., Gomez, T. S., Metlagel, Z., Ismail, A. M., et al. (2010). Structure and control of the actin regulatory WAVE complex. Nature 468 (7323), 533–538. doi:10.1038/nature09623
Danson, C. M., Pocha, S. M., Bloomberg, G. B., and Cory, G. O. (2007). Phosphorylation of WAVE2 by MAP kinases regulates persistent cell migration and polarity. J. Cell. Sci. 120 (23), 4144–4154. doi:10.1242/jcs.013714
Davidson, A. J., King, J. S., and Insall, R. H. (2013). The use of streptavidin conjugates as immunoblot loading controls and mitochondrial markers for use with Dictyostelium discoideum. Biotechniques 55 (1), 39–41. doi:10.2144/000114054
Davidson, A. J., Ura, S., Thomason, P. A., Kalna, G., and Insall, R. H. (2013). Abi is required for modulation and stability but not localization or activation of the SCAR/WAVE complex. Eukaryot. Cell. 12 (11), 1509–1516. doi:10.1128/EC.00116-13
Fort, L., Batista, J. M., Thomason, P. A., Spence, H. J., Whitelaw, J. A., Tweedy, L., et al. (2018). Fam49/CYRI interacts with Rac1 and locally suppresses protrusions. Nat. Cell. Biol. 20 (10), 1159–1171. doi:10.1038/s41556-018-0198-9
Ibarra, N., Blagg, S. L., Vazquez, F., and Insall, R. H. (2006). Nap1 regulates Dictyostelium cell motility and adhesion through SCAR-dependent and -independent pathways. Curr. Biol. 16 (7), 717–722. doi:10.1016/j.cub.2006.02.068
Insall, R. H., and Machesky, L. M. (2009). Actin dynamics at the leading edge: From simple machinery to complex networks. Dev. Cell. 17 (3), 310–322. doi:10.1016/j.devcel.2009.08.012
Kamimura, Y., and Devreotes, P. N. (2010). Phosphoinositide-dependent protein kinase (PDK) activity regulates phosphatidylinositol 3, 4, 5-trisphosphate-dependent and -independent protein kinase B activation and chemotaxis. J. Biol. Chem. 285 (11), 7938–7946. doi:10.1074/jbc.M109.089235
Kamimura, Y., Xiong, Y., Iglesias, P. A., Hoeller, O., Bolourani, P., and Devreotes, P. N. (2008). PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr. Biol. 18 (14), 1034–1043. doi:10.1016/j.cub.2008.06.068
King, J. S., and Kay, R. R. (2019). The origins and evolution of macropinocytosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374 (1765), 20180158. doi:10.1098/rstb.2018.0158
Krause, M., and Gautreau, A. (2014). Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell. Biol. 15 (9), 577–590. doi:10.1038/nrm3861
Ladoux, B., and Nicolas, A. (2012). Physically based principles of cell adhesion mechanosensitivity in tissues. Rep. Prog. Phys. 75 (11), 116601. doi:10.1088/0034-4885/75/11/116601
Lebensohn, A. M., and Kirschner, M. W. (2009). Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell. 36 (3), 512–524. doi:10.1016/j.molcel.2009.10.024
Liao, X. H., Buggey, J., and Kimmel, A. R. (2010). Chemotactic activation of Dictyostelium AGC-family kinases AKT and PKBR1 requires separate but coordinated functions of PDK1 and TORC2. J. Cell. Sci. 123 (6), 983–992. doi:10.1242/jcs.064022
Machesky, L. M., and Insall, R. H. (1998). Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8 (25), 1347–1356. doi:10.1016/s0960-9822(98)00015-3
Meili, R., Ellsworth, C., and Firtel, R. A. (2000). A novel Akt/PKB-related kinase is essential for morphogenesis in Dictyostelium. Curr. Biol. 10 (12), 708–717. doi:10.1016/s0960-9822(00)00536-4
Meili, R., Ellsworth, C., Lee, S., Reddy, T. B., Ma, H., and Firtel, R. A. (1999). Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 18 (8), 2092–2105. doi:10.1093/emboj/18.8.2092
Mendoza, M. C., Er, E. E., Zhang, W., Ballif, B. A., Elliott, H. L., Danuser, G., et al. (2011). ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell. 41 (6), 661–671. doi:10.1016/j.molcel.2011.02.031
Nichols, J. M. E., Paschke, P., Peak-Chew, S., Williams, T. D., Tweedy, L., Skehel, M., et al. (2019). The atypical MAP kinase ErkB transmits distinct chemotactic signals through a core signaling module. Dev. Cell. 48 (4), 491–505 e9. doi:10.1016/j.devcel.2018.12.001
Niewohner, J., Weber, I., Maniak, M., Muller-Taubenberger, A., and Gerisch, G. (1997). Talin-null cells of Dictyostelium are strongly defective in adhesion to particle and substrate surfaces and slightly impaired in cytokinesis. J. Cell. Biol. 138 (2), 349–361. doi:10.1083/jcb.138.2.349
Paschke, P., Knecht, D. A., Williams, T. D., Thomason, P. A., Insall, R. H., Chubb, J. R., et al. (2019). Genetic engineering of Dictyostelium discoideum cells based on selection and growth on bacteria. J. Vis. Exp. 143. doi:10.3791/58981
Pollitt, A. Y., and Insall, R. H. (2008). Abi mutants in Dictyostelium reveal specific roles for the SCAR/WAVE complex in cytokinesis. Curr. Biol. 18 (3), 203–210. doi:10.1016/j.cub.2008.01.026
Schaks, M., Giannone, G., and Rottner, K. (2019). Actin dynamics in cell migration. Essays Biochem. 63 (5), 483–495. doi:10.1042/EBC20190015
Schaks, M., Singh, S. P., Kage, F., Thomason, P., Klünemann, T., Steffen, A., et al. (2018). Distinct interaction sites of rac GTPase with WAVE regulatory complex have non-redundant functions in vivo. Curr. Biol. 28 (22), 3674–3684.e6. e6. doi:10.1016/j.cub.2018.10.002
Simson, R., Wallraff, E., Faix, J., Niewohner, J., Gerisch, G., and Sackmann, E. (1998). Membrane bending modulus and adhesion energy of wild-type and mutant cells of Dictyostelium lacking talin or cortexillins. Biophys. J. 74 (1), 514–522. doi:10.1016/S0006-3495(98)77808-7
Singh, S. P., and Insall, R. H. (2020). Adhesion stimulates Scar/WAVE phosphorylation in mammalian cells. Commun. Integr. Biol. 14 (1), 1–4. doi:10.1080/19420889.2020.1855854
Singh, S. P., and Insall, R. H. (2022). Under-agarose chemotaxis and migration assays for Dictyostelium. Methods Mol. Biol. 2438, 467–482. doi:10.1007/978-1-0716-2035-9_27
Singh, S. P., Thomason, P. A., and Insall, R. H. (2021). Extracellular signalling modulates scar/WAVE complex activity through Abi phosphorylation. Cells 10 (12), 3485. doi:10.3390/cells10123485
Singh, S. P., Thomason, P. A., Lilla, S., Schaks, M., Tang, Q., Goode, B. L., et al. (2020). Cell-substrate adhesion drives Scar/WAVE activation and phosphorylation by a Ste20-family kinase, which controls pseudopod lifetime. PLoS Biol. 18 (8), e3000774. doi:10.1371/journal.pbio.3000774
Srinivasan, K., Wright, G. A., Hames, N., Housman, M., Roberts, A., Aufderheide, K. J., et al. (2013). Delineating the core regulatory elements crucial for directed cell migration by examining folic-acid-mediated responses. J. Cell. Sci. 126 (1), 221–233. doi:10.1242/jcs.113415
Talmadge, J. E., and Fidler, I. J. (2010). AACR centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res. 70 (14), 5649–5669. doi:10.1158/0008-5472.CAN-10-1040
Tsujioka, M., Machesky, L. M., Cole, S. L., Yahata, K., and Inouye, K. (1999). A unique talin homologue with a villin headpiece-like domain is required for multicellular morphogenesis in Dictyostelium. Curr. Biol. 9 (7), 389–392. doi:10.1016/s0960-9822(99)80169-9
Tsujioka, M., Yoshida, K., and Inouye, K. (2004). Talin B is required for force transmission in morphogenesis of Dictyostelium. EMBO J. 23 (11), 2216–2225. doi:10.1038/sj.emboj.7600238
Ura, S., Pollitt, A. Y., Veltman, D. M., Morrice, N. A., Machesky, L. M., and Insall, R. H. (2012). Pseudopod growth and evolution during cell movement is controlled through SCAR/WAVE dephosphorylation. Curr. Biol. 22 (7), 553–561. doi:10.1016/j.cub.2012.02.020
Veltman, D. M., King, J. S., Machesky, L. M., and Insall, R. H. (2012). SCAR knockouts in Dictyostelium: WASP assumes SCAR's position and upstream regulators in pseudopods. J. Cell. Biol. 198 (4), 501–508. doi:10.1083/jcb.201205058
Veltman, D. M., Williams, T. D., Bloomfield, G., Chen, B. C., Betzig, E., Insall, R. H., et al. (2016). A plasma membrane template for macropinocytic cups. Elife 5, e20085. doi:10.7554/eLife.20085
Wang, Y., Chen, C. L., and Iijima, M. (2011). Signaling mechanisms for chemotaxis. Dev. Growth Differ. 53 (4), 495–502. doi:10.1111/j.1440-169x.2011.01265.x
Williams, T. D., Peak-Chew, S. Y., Paschke, P., and Kay, R. R. (2019). Akt and SGK protein kinases are required for efficient feeding by macropinocytosis. J. Cell. Sci. 132 (2), jcs224998. doi:10.1242/jcs.224998
Keywords: cell motility, pkbA, pkgB, actin, Scar/WAVE complex, Arp2/3 complex
Citation: Singh SP, Paschke P, Tweedy L and Insall RH (2022) AKT and SGK kinases regulate cell migration by altering Scar/WAVE complex activation and Arp2/3 complex recruitment. Front. Mol. Biosci. 9:965921. doi: 10.3389/fmolb.2022.965921
Received: 10 June 2022; Accepted: 28 July 2022;
Published: 29 August 2022.
Edited by:
Victoria Sanz-Moreno, Queen Mary University of London, United KingdomReviewed by:
Kyra J. E. Borgman, Institut Curie, FranceArnon Henn, Technion Israel Institute of Technology, Israel
Copyright © 2022 Singh, Paschke, Tweedy and Insall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shashi Prakash Singh, U2hhc2hpLlNpbmdoQGdsYXNnb3cuYWMudWs=
 Shashi Prakash Singh
Shashi Prakash Singh Peggy Paschke
Peggy Paschke Luke Tweedy
Luke Tweedy Robert H. Insall
Robert H. Insall