Abstract
Viruses have been responsible for many epidemics and pandemics that have impacted human life globally. The COVID-19 pandemic highlighted both our vulnerability to viral outbreaks, as well as the mobilization of the scientific community to come together to combat the unprecedented threat to humanity. Cryo-electron microscopy (cryo-EM) played a central role in our understanding of SARS-CoV-2 during the pandemic and continues to inform about this evolving pathogen. Cryo-EM with its two popular imaging modalities, single particle analysis (SPA) and cryo-electron tomography (cryo-ET), has contributed immensely to understanding the structure of viruses and interactions that define their life cycles and pathogenicity. Here, we review how cryo-EM has informed our understanding of three distinct viruses, of which two - HIV-1 and SARS-CoV-2 infect humans, and the third, bacteriophages, infect bacteria. For HIV-1 and SARS-CoV-2 our focus is on the surface glycoproteins that are responsible for mediating host receptor binding, and host and cell membrane fusion, while for bacteriophages, we review their structure, capsid maturation, attachment to the bacterial cell surface and infection initiation mechanism.
1 Introduction: from blobology to high resolution structural biology
Electron microscopy has a long history of being used as a diagnostic tool to identify new infectious agents, study their structures, and understanding virus-cell interaction (Friedlaender et al., 1955; Roingeard, 2008; Goldsmith and Miller, 2009). Since its invention, the electron microscope has not only been used for viral diagnoses but also in biomedical research to examine the structure of tissues, cells, organelles, and macromolecular complexes, and in pathogenesis studies. Biological samples, during their visualization under an electron microscope, suffered damage due to dehydration and staining. Therefore, rapid freezing of unstained biological samples was developed during the 70s–80s (Dubochet et al., 1988). With the development of sophisticated cryo-electron microscopy (cryo-EM) technique, detailed structures of viruses were determined in their native state. In its early days, cryo-EM structures mostly looked like blobs due to low-resolution imaging, hence the moniker “blobology”. However, the immense potential of cryo-EM was recognized, and with investment of time and resources, cryo-EM has come a long way with steadily improving microscopes, and improved methods for sample purification, specimen preparation, data collection and image analysis (Milne et al., 2013; Thompson et al., 2016; Saibil 2017; Dandey et al., 2018; Noble et al., 2018). Cryo-EM has now emerged as a primary structural biology technique and was recognized as the method of the year in 2015 (2016). The development of direct electron detectors transformed the cryo-EM field, and the world witnessed the resolution revolution that culminated in the Nobel Prize in Chemistry in 2017 being awarded to Jacques Dubochet, Joachim Frank and Richard Henderson for developing freezing method of biomolecules, image-processing software and first 3D model and atomic structure of a protein, respectively (Kühlbrandt, 2014a; Cressey and Callaway, 2017). Thereafter, the number of entries in the Electron Microscopy Data Bank (EMDB), the public archive for three-dimensional EM maps, has rapidly increased (Patwardhan, 2017).
Advances in cryo-EM have improved the quality of imaging and analysis. Moreover, the classification schemes used in cryo-EM image processing have made it possible to separate multiple conformations and compositions of macromolecular complexes within the same dataset. This sets cryo-EM apart from many other structural techniques that rely on homogeneous sample preparations, making it versatile and useful for studying complex samples (Fernández et al., 2013; Kühlbrandt, 2014b; Scheres, 2016). Computational techniques for enhancing local resolution can address issues of lowered resolution due to local sample heterogeneity and conformational flexibility (Kucukelbir et al., 2014; Punjani et al., 2017; Grant et al., 2018). Noticeably, more countries are getting involved in this cutting-edge technique to tackle future threats of infectious diseases. Cryo-EM has been widely used for understanding Zika, Nipah, Ebola, Influenza, SARS, MERS, Langya, Lassa, Dengue, Chikungunya, HIV, and other viruses (Harris et al., 2006; Lyumkis et al., 2013, Sirohi et al., 2016; Yuan et al., 2017; Song et al., 2018, Su et al., 2018; Basore et al., 2019; Dang et al., 2019; Stass et al., 2019, Katz et al., 2022; Guo et al., 2024). With advances in cryo-EM methodology, it is now possible to obtain near-atomic resolutions of viruses and their spike structures, their interaction with host cell receptors, virus entry, assembly and antibody-mediated neutralization (Casasnovas, 2013; Stass et al., 2019; Burton, 2023). All these discoveries can collectively contribute to the development of new vaccines and antiviral therapies.
In this review, we highlight the contribution of two cryo-EM imaging modalities, SPA and cryo-ET, in understanding the protein structures and life cycles of three viruses of relevance to human health, of which two–HIV-1 (Mak and de Marco, 2018; Graham and Zhang, 2023) and SARS-CoV-2 (Zhang et al., 2021a) cause disease in humans, whereas the third, bacteriophages, infects bacteria and has been harnessed for biotechnological applications, and for combatting drug-resistant bacteria (Jo et al., 2023).
2 Contribution of Cryo-EM and its advances in studying virus-mediated infectious diseases
Electron microscopy has been at the forefront for fast detection of any infectious agent and its morphology (Hazelton and Gelderblom, 2003). With the development of cryo-EM, the structures of viruses, their interaction with the host, and disease progression have been successfully studied, with technological advances in the cryo-EM field enabling near-atomic structural insights having transformative impact on molecular and cellular structural biology. (Jiang and Tang, 2017; Creekmore et al., 2021; Graham and Zhang, 2023). Pathogenic viruses are infectious agents that exist in various size and shape in the virosphere. Pathogenic enveloped viruses have in common a membrane-embedded spike glycoprotein responsible for host-receptor binding to begin the infection process. Therefore, the spike protein is an important target to study in detail by cryo-EM (Dokainish et al., 2022). The resolution revolution resulted in an atomic-level understanding of many surface proteins of enveloped pathogenic viruses, thus enabling structure-based drug discovery and vaccine design (Graham et al., 2019; Kim and Jung, 2021). Figure 1 highlights the timeline of developments in cryo-EM technology juxtaposed with breakthroughs in our understanding of the bacteriophage capsid structure, capsid maturation, tail contraction, and surface glycoproteins of HIV-1 and SARS-CoV-2 (Leiman et al., 2004; Jiang et al., 2008; Liu et al., 2008; Lyumkis et al., 2013; Ke et al., 2020; Nakane et al., 2020; Wrapp et al., 2020). Of the two modalities of cryo-EM that have commonly been used to study the structures of viral proteins, single particle analysis (SPA) is the most popular cryo-EM technique for determining atomic resolution structures of “spike” proteins of enveloped viruses (Zhou, 2014; Ward and Wilson, 2017), while cryo-electron tomography (cryo-ET) provides in situ information within a native cellular context, including the distribution of proteins, the presence of any transient conformational changes, viral assembly, and maturation mechanism within the host (Subramaniam et al., 2007; Liu et al., 2010; Ke et al., 2020).
FIGURE 1

Road map of development of cryo-electron microscopy field. Milestones indicated with blue flags starting from development of electron microscope (1931), visualization of first virus under EM (1939), electron diffraction of frozen, hydrated catalase crystals (1974) and development of electron microscopy of frozen biological samples (1983). Then low resolution cryo-EM maps were reconstructed during early 2000. With the development of direct electron detectors around 2011-2013 there was an upsurge of high resolution cryo-EM structures. In 2017 the Nobel Prize in Chemistry was given to the three pioneers (Jacques Dubochet, Joachim Frank, Richard Henderson) for the development of structure determination by cryo-electron microscopy. By the year 2020 atomic resolution (1.22Å) structure was achieved for apoferritin (EMD-11638) by cryo-EM. Green flags indicate the important milestones of the contributions of cryo-EM in bacteriophages (EMD-1086; 5003), HIV Envelope (EMD-5779; 5019) and SARS-CoV2 spike (EMD-21375; 11494) structure determination, the three topics that are the focus of this paper.
3 Structural studies on the HIV-1 Env
3.1 Contribution of cryo-EM/cryo-ET in advancing the knowledge of HIV-1 structural biology
3.1.1 HIV structure
Human immunodeficiency virus-1 (HIV-1) is the causative agent of acquired immunodeficiency syndrome (AIDS). HIV-1 has a lipid envelope consisting of a bilayer membrane and trimeric envelope (Env) glycoprotein are embedded onto the membrane (Blumenthal et al., 2012). HIV-1 Env is composed of a receptor binding gp120 subunit and a fusion mediating gp41 subunit that contains the fusion peptide. Env plays a pivotal role in virus attachment to the host cell surface and in initiating the infection process through the fusion of the viral and host cell membrane (Xiao et al., 2021a). Several Env defense mechanisms, including a rapidly mutating genome that introduces antigenic variation, conformational masking and shielding of antigenic sites by long variable loops, have challenged the development of an effective HIV-1 vaccine (Haynes et al., 2023). However, critical regions that bind to the primary HIV-1 receptor CD4, and co-receptors, remain conserved and are among the primary targets of vaccine and drug development efforts.
Structural determination of Env began with a crystal structure of the gp120 core in complex with the primary HIV-1 receptor CD4 and the fragment antigen binding (Fab) of a coreceptor mimetic antibody 17b that was solved in the ‘90s (Kwong et al., 1998), followed by several other structures of gp120, unliganded or bound to antibodies and receptors. While these structures provided detailed views of gp120 domain organization, its internal architecture and epitopes targeted by antibodies, they were limited their utility in informing Env biology that required the context of the trimer. X-ray crystal structures of soluble HIV-1 Env ectodomains were later determined (Julien et al., 2013; Pancera et al., 2014; Kwon et al., 2015; Stewart-Jones et al., 2016) and while these structures provided information in the Env trimer context, obtaining well diffracting crystals of new complexes remained a bottleneck that hindered throughput.
3.1.2 Cryo-ET of HIV-1 native spikes
Electron microscopy provides a methodological platform for determining structures of biomolecules without the bottleneck of obtaining well diffracting crystals. Cryo-ET structures of SIV/HIV-Env spikes on intact virus membrane determined in early 2000s provided the first views of the HIV-1 Env on the virion surface (Zanetti et al., 2006; Zhu et al., 2006). These were subsequently followed by several cryo-ET structures of Env, either unliganded or in complex with receptors and antibodies, with better resolution enabled by technical advances in microscopic and sample preparation techniques (Figure 2A). These structures shed light on the mechanism of closed-to-open conformational change in the native, membrane-associated Env, (Liu et al., 2008; Tran et al., 2012; Meyerson et al., 2013; Li et al., 2020). Cryo-ET analysis on Env performed in both mature and immature HIV-1 particles provided information on the location and arrangement of structural proteins, e.g., radial arrangement of Gag structural protein along with hexameric capsid (CA) protein stabilization by bundle of six spacer element 1(SP 1) helices, and formation of fullerene cone shaped mature HIV-1 capsid in which capsid-protein hexamers and pentamers form a closed hexagonal surface lattice, tubular assembly of HIV-1 Gag derived protein reveals structural arrangement of C-terminal domain of CA and region which is essential for virus assembly located downstream of CA, tertiary and quaternary structural arrangement for viral assembly (Wright et al., 2007; Zhao et al., 2013; Bharat et al., 2014; Schur et al., 2015).
FIGURE 2

Cryo-ET and single particle cryo-EM of HIV-1 Env. (A) From left to right, CD4-binding site antibody VRC01 and CD4-induced antibody 17b (EMD-5461), CD4-binding site antibody b12 (EMD-5018), sCD4 and CD4-induced antibody m36 (EMD-5554), and V3-glycan antibody 10-1074 and CD4-binding site antibody 3BNC117 (EMD-21413). (B) Cryo-ET study of HIV-1 entry mechanism. From left to right, One (EMD-29292), two (EMD-29293) and three (EMD-29294) CD4 molecules sequentially bound to native HIV-1 Env on membrane brings the host and viral membranes close to each other to initiate the fusion process. (C) Single particle cryo-EM of HIV-1 Env conformational states. From left to right, closed (EMD-8714), open-occluded (EMD-25878, bound antibody not shown), open (EMD-8713, bound CD4 and antibody not shown) conformational states are shown with the Env colored sea green, V1V2 loop colored orange, V3 loop colored pink and CD4 binding site colored medium orchid. Each panel includes a schematic representation.
The HIV-1 assembly and its release in intact human cells suggested that the organization of the Gag protein occurs in the assembly site in immature HIV-1 with no further changes in arrangement. Proteolytic maturation results in two different hexameric lattice rearrangements of the HIV-1 matrix (MA) between immature and mature virions. The membrane-bound MA lattice plays a significant role in mature virions. Env on immature HIV-1 virion structure gives an idea about the Env positioning relative to Gag hexameric arrangement. The structural analysis of Env on mature HIV-1 viral particles revealed substantial glycan shielding, flexibility in the gp41 stalk that exposes the crucial epitopes present in FP, gp120/41 interface, and MPER regions differently and leads to structural variations. (Carlson et al., 2010; Qu et al., 2021; Mangala Prasad et al., 2022). Subsequent cryo-ET studies have increased our knowledge of HIV-1 entry and fusion. These include direct visualization of the hypothesized pre-hairpin intermediate of HIV-1 Env (Ladinsky et al., 2020), visualization of the contact between T-cells and SIV/HIV-1 during viral entry (Sougrat et al., 2007), and studies on HIV-1 and CD4, both on the membrane, showing three CD4 molecules gradually bind to Env through cluster and ring formation ultimately bringing both membranes closer and the transition of Env to an open state (Li et al., 2023) (Figure 2B).
3.1.3 High-resolution cryo-EM structural studies of HIV-1 SOSIP
The development of stabilized HIV-1 Env ectodomain trimers (Sanders et al., 2002; Sanders et al., 2013a; Sliepen et al., 2019) synergized with the cryo-EM resolution revolution to turbo charge HIV-1 Env structural biology, making it faster and easier to obtain high resolution structures of the HIV-1 Env trimer (Lyumkis et al., 2013; Lee et al., 2016; Ozorowski et al., 2017). Overall features of Env trimer embedded in virions surface showed remarkable similarities with the structures of the engineered soluble trimers (Harris et al., 2011; Torrents de la Peña et al., 2019; Li et al., 2020). Cryo-EM has enabled high resolution visualization of Env conformational states that are relevant for viral entry and for antibody access to sites of vulnerability. The HIV-1 Env has a prefusion, closed state in which the gp41 helices that are anchored to the viral membrane act as a support to rearrange other parts of the spike when it changes to the receptor-bound open conformation (Figure 2C). HIV Env binding to CD4 induces extensive conformational changes in Env that eventually opens the Env and exposes the co-receptor binding sites (Ozorowski et al., 2017). Other partially open intermediates have been observed (Figure 2C) that open new avenues for the development of therapeutics to prevent viral infection (Scharf et al., 2015; Yang et al., 2022; Dam et al., 2023).
The development of stabilized Env ectodomain constructs also synergized with development of improved methods for broadly neutralizing antibody (bNAb) isolation through high throughput single-cell B-cell receptor (BCR) amplification and novel soluble Env selection tools together with the culture of memory B cells that allowed the identification and isolation of more potent and broader bNAbs (Walker et al., 2009; Walker et al., 2011; Huang et al., 2012) Single particle cryo-EM analysis uses recombinant soluble stabilized trimeric Env ectodomain constructs that mimic the native spike structure. The clade A BG505 SOSIP.664 has been used as the gold standard HIV native-like engineered Env trimer immunogen allowing the high-resolution structure determination of the Env trimer (Sanders et al., 2013b) in complex with various naturally-elicited antibodies (Figure 3A) and vaccine-elicited antibodies (Figure 3B) to reveal how antibodies can access Env sites of vulnerability by overcoming the barriers posed by the glycan shield, variable loops, and conformational masking (Ananthaswamy et al., 2019; Saunders et al., 2019; Seabright et al., 2020; Yang et al., 2022; Banach et al., 2023; Henderson et al., 2023; Holt et al., 2023; Wang et al., 2023; Saunders et al., 2024; Wiehe et al., 2024; Williams et al., 2024). Another clade B virus improved version of Env with MPER region lacking the cytoplasmic tail (Env ΔCT) was reported that contributed insights into a more native-like Env structure (Lee et al., 2016).
FIGURE 3

High resolution cryo-EM structures of HIV SOSIP Env with antibodies. (A) Naturally-elicited antibodies in complex with HIV SOSIP Env spike: From left to right (first row), CD4 binding site Ab VRC01 (EMD-40281), glycan cluster Ab 2G12 (EMD-20224), V1/V2-apex directed Ab PGT145 (EMD-29288). From left to right (second row), V3-glycan targeted Ab DH270.6 (EMD-20818), gp120-gp41 interface Ab 8ANC195 (EMD-0485), fusion peptide-targeting Ab VRC34 (EMD-28617), membrane proximal external region targeted Ab 4E10 (EMD-25045). (B) Vaccine-elicited antibodies in complex with HIV SOSIP Env spike: From left to right (first row), V3-glycan mouse Ab MU89 (EMD-27706), CD4 binding site macaque Ab DH1285 (EMD-41823), FP-targeting human Ab 2C06 (EMD-29725), MPER targeted human Ab DH1317.4 (EMD-44246). (C) Cryo-EM based polyclonal epitope mapping (cryo-EMPEM): From left to right (first row), polyclonal antibodies were selected from rhesus macaques Rh.32034 (Grp1): pAbC-1(C3/V5 targeted, EMD-23223) and pAbC-2(N241/N289 glycan hole targeted, EMD-23224); From left to right (second row) Rh.33104 (Grp1): pAbC-1(N241/N289 glycan hole targeted, EMD-23227) and pAbC-2(N241/N289 glycan hole targeted, EMD-23228); From left to right (third row) Rh.33311 (Grp2): pAbC-1(FP targeted, EMD-23236), pAbC-2(N611-glycan targeted, EMD-23237), and pAbC-5(C3/V5 targeted, EMD-23240). Antibody heavy chain in medium purple and light chain in pink, Env gp120 in dark gray and gp41 in light steel blue throughout Figure 3. Each panel includes a schematic representation (created with BioRender.com).
Cryo-EM has impacted the ongoing HIV-1 vaccine development on multiple fronts, that include the facilitation of the design of native-like functional Env immunogens to elicit broadly neutralizing antibodies (bNAbs), high resolution definition of antibody epitopes to establish conserved sites of vulnerability, visualization of antibody virus co-evolution and evaluation of vaccine-induced antibodies for their precision of targeting the desired sites (Lyumkis et al., 2013; Saunders et al., 2019; LaBranche et al., 2019; Torrents de la Peña et al., 2019; Antanasijevic et al., 2020; Cottrell et al., 2021; Roark et al., 2021; Henderson et al., 2023; Saunders et al., 2024; Wiehe et al., 2024). Structural determination by cryo-EM facilitated the discovery of a novel category of Fab-dimerized glycan reactive antibodies that target a Env glycan cluster (Williams et al., 2021). A rapid EM-based polyclonal epitope mapping (EMPEM) method has facilitated direct high resolution structural determination of antibody epitopes from vaccine sera (Figure 3C) (Bianchi et al., 2018; Antanasijevic et al., 2021).
3.2 Advantages of cryo-ET over SPA to study HIV-1 viral infection and assembly
Cryo-electron tomography (cryo-ET) and subtomogram averaging (STA) are techniques to study dynamic biological processes and provide unprecedented structural details at moderately high resolution while conserving spatial arrangement in native environments. Cryo-ET has also broadened its applicability by combining with other methods such as correlative light and electron microscopy (CLEM), cryo-focused ion beam (FIB) milling, ion-abrasion scanning electron microscopy (SEM), or focused ion beam-scanning electron microscopy (FIB-SEM) which reveal a wealth of information with new insights (Felts et al., 2010; Jun et al., 2011; Earl and Subramaniam 2013; Blanco-Rodriguez et al., 2020; Zila et al., 2021). Cryo-ET is a rapidly growing field, with method developments being simultaneously carried out to achieve quicker, better, and more detailed outcomes, namely, denoising (Narasimha et al., 2008) and segmentation to reveal intermediate virus assembly stages (Bartesaghi et al., 2005; Narasimha et al., 2008; Woodward et al., 2015; Ladinsky et al., 2020). A newly developed scalable, end-to-end framework for single particle cryo-ET data analysis enables resolution of conformational heterogeneity in situ during on-the-fly preprocessing of tilt series without subtomogram averaging followed by high-resolution refinement and classification (Liu et al., 2023). Despite numerous solved structures of HIV-1 and receptor complexes, several aspects of the HIV-1 entry mechanism remain unresolved and continued advances in structural determination will enable discoveries to further elucidate the HIV-1 entry and fusion mechanism.
4 Structural studies on the SARS-CoV-2 S protein
4.1 Cryo-EM structure of SARS-CoV-2 spikes of viral variants
The outbreak of severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 had already shown that coronaviruses can cross the species barrier to emerge as highly pathogenic viruses (Lu et al., 2015). More recently, SARS-CoV-2 caused a global pandemic that started in December 2019 and was subsequently named COVID-19 (Zhu et al., 2020). The highly infectious disease is characterized by fever, acute respiratory illness, and pneumonia. The SARS-CoV-2 surface spike (S) glycoprotein has a trimeric structure similar to SARS-CoV and many other enveloped pathogenic viruses. This spike consists of three S1-S2 heterodimers that bind the cellular receptor angiotensin-converting enzyme 2 (ACE2) and mediate fusion of the viral and cellular membranes through extensive conformational change (Song et al., 2018; Lan et al., 2020; Wrapp et al., 2020). The pre-fusion S protein consists of S1 subunit that includes N-terminal domain (NTD), receptor-binding domain (RBD) and S2 subunit that includes fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), transmembrane (TM) and cytoplasmic tail (CT) (Wang et al., 2020; Wrapp et al., 2020).
The first SARS-CoV-2 trimeric spike high-resolution structures were determined soon after the pandemic begun (Walls et al., 2020; Wrapp et al., 2020). Previous research on the MERS-CoV and SARS-CoV S proteins facilitated by cryo-EM were instrumental in the discovery of the pre-fusion S protein stabilizing two proline (2P) mutations used in several vaccines that were rapidly developed to combat the COVID-19 pandemic (Kirchdoerfer et al., 2016; Pallesen et al., 2017; Walls et al., 2020). Single particle cryo-EM analysis was pivotal to characterizing the structures and conformations of the SARS-CoV-2 S protein and in the structure-based design of engineered S protein constructs (Cai et al., 2020; Henderson et al., 2020; Hsieh et al., 2020; Lu et al., 2022; Shi et al., 2023). A 6P version of pre-fusion SARS-CoV-2 S protein that exhibited better immunogenicity than the S-2P is a component of next-generation stabilized spike vaccines (Carreño et al., 2023; Zhu et al., 2024). While studies of the pre-fusion S protein dominated the cryo-EM bandwidth, innovative studies on the postfusion conformation of the S protein enabled a comprehensive understanding of the fusion mechanism and the impact of mutations on the process (Yang S. et al., 2022; Shi et al., 2023). Lipid nanodiscs have been shown to play a crucial role in visualizing the membrane interacting regions in postfusion SARS-CoV-2 spike proteins using cryo-EM. The full-length postfusion spike in the lipid bilayer allows for the visualization of structural details in the membrane-interacting regions FP and TM during the final stage of membrane fusion. Additionally, mutations in the fusion peptide proximal region (FPPR) completely inhibit its interaction with FP, subsequently blocking membrane fusion activity (Shi et al., 2023). Another study focused on the SARS-CoV-2 membrane (M) protein, an essential component of the viral assembly when placed on a lipid nanodisc revealed structural details and ion channel activity of the M protein (Dolan et al., 2022).
Early in the COVID-19 pandemic, a D614G substitution was identified in the SD2 subdomain of the SARS-CoV-2 S protein that was retained in all subsequent variants (Korber et al., 2020). Cryo-EM analysis of D614G substitution revealed an altered spike conformation with higher proportion of RBD up states, that was accompanied by enhanced sensitivity to neutralization (Yurkovetskiy et al., 2020; Zhang et al., 2021b; Gobeil et al., 2021; Weissman et al., 2021). Subsequently several variants of SARS-CoV2 emerged and swept local and global populations, with some major lineages designated as variants of concern (VOCs) due to increased transmissibility, disease severity, resistance to neutralizing antibodies elicited by vaccines, or reduced efficacy of treatments (Carabelli et al., 2023; Aleem, Akbar Samad and Vaqar, 2024). Figure 4 shows the evolution of SARS-CoV-2 through the course of the COVID-19 pandemic and highlights the role played by cryo-EM in our rapid understanding of evolving SARS-CoV-2 variants (Wrapp et al., 2020; Zhang et al., 2021a; Gobeil et al., 2021; Cerutti et al., 2022; Mannar et al., 2022; Stalls et al., 2022; Wrobel et al., 2022; Mannar et al., 2024; Zhang et al., 2024). The Omicron variant is currently circulating worldwide and has evolved into several sublineages that have successively swept global populations (Parsons and Acharya, 2023). Cryo-EM studies were instrumental in defining the altered structure of the Omicron spikes relative to previous variants and in elucidating how these changes impacted SARS-CoV-2 biology (Cerutti et al., 2022; Gobeil et al., 2022; Mannar et al., 2022; Stalls et al., 2022).
FIGURE 4

Structural determination of SARS-CoV-2 variants of concern (VoC) (built with nextstrain/ncov). 2019–2024 evolution of SARS-CoV-2 spikes of key emerging variants of concern (VoC): 2019-nCoV (EMD-21375), D614G (EMD-22825), Alpha (B.1.1.7) (EMD-14229), Beta (B.1.351) (EMD-27505), Gamma (P.1) (EMD-24987), Delta (B.1.617.2) (EMD-24981), Omicron BA.1 (B.1.1.529) (EMD-25896), Omicron BA.2 (EMD-26433), Omicron (XBB.1.5) (EMD-43320), Omicron (XBB.1.16) (EMD-42860), Omicron (EG.5.1) (EMD-44000). All cryo-EM structures (side and top view) of spike show the three monomers in dark gray, light steel blue and thistle colors and mutations in spike are shown in red color.
Overall structure of the SARS-CoV-2 spike protein is schematically illustrated in Figure 5A. Recognition and binding to the host cell receptor is the initial step of virus entry into the host cell. Figure 5B depicts the cryo-EM structure of the SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (Mannar et al., 2024). Cryo-EM structural studies revealed that ACE2 can only interact with the RBD (receptor-binding domain) of the soluble S glycoprotein trimer when the soluble trimer is in RBD-up conformation. The structures also show that that the N-terminal domain (NTD) slightly shifts outward after RBD-ACE2 binding, but no substantial changes occur in the S2 subunit (Xiao et al., 2021b; Jackson et al., 2022). Furthermore, the cryo-EM structure of the soluble S trimer in complex with ACE2 at serological and endosomal pH levels of 7.4 and 5.5, respectively, showed no difference in stoichiometries. Visualization of the soluble S trimer without a ligand at different low pH levels, ranging from pH 5.5 to pH 4.0, revealed a reduction in conformational heterogeneity, ultimately resulting in all RBDs adopting a down conformation (Zhou et al., 2020). Most studies with the ACE2 receptor have used the ACE2 ectodomain, although a cryo-EM structure of the full-length ACE2 protein bound to the receptor-binding domain (RBD) of the SARS-CoV-2 S-glycoprotein was reported (Yan et al., 2020).
FIGURE 5
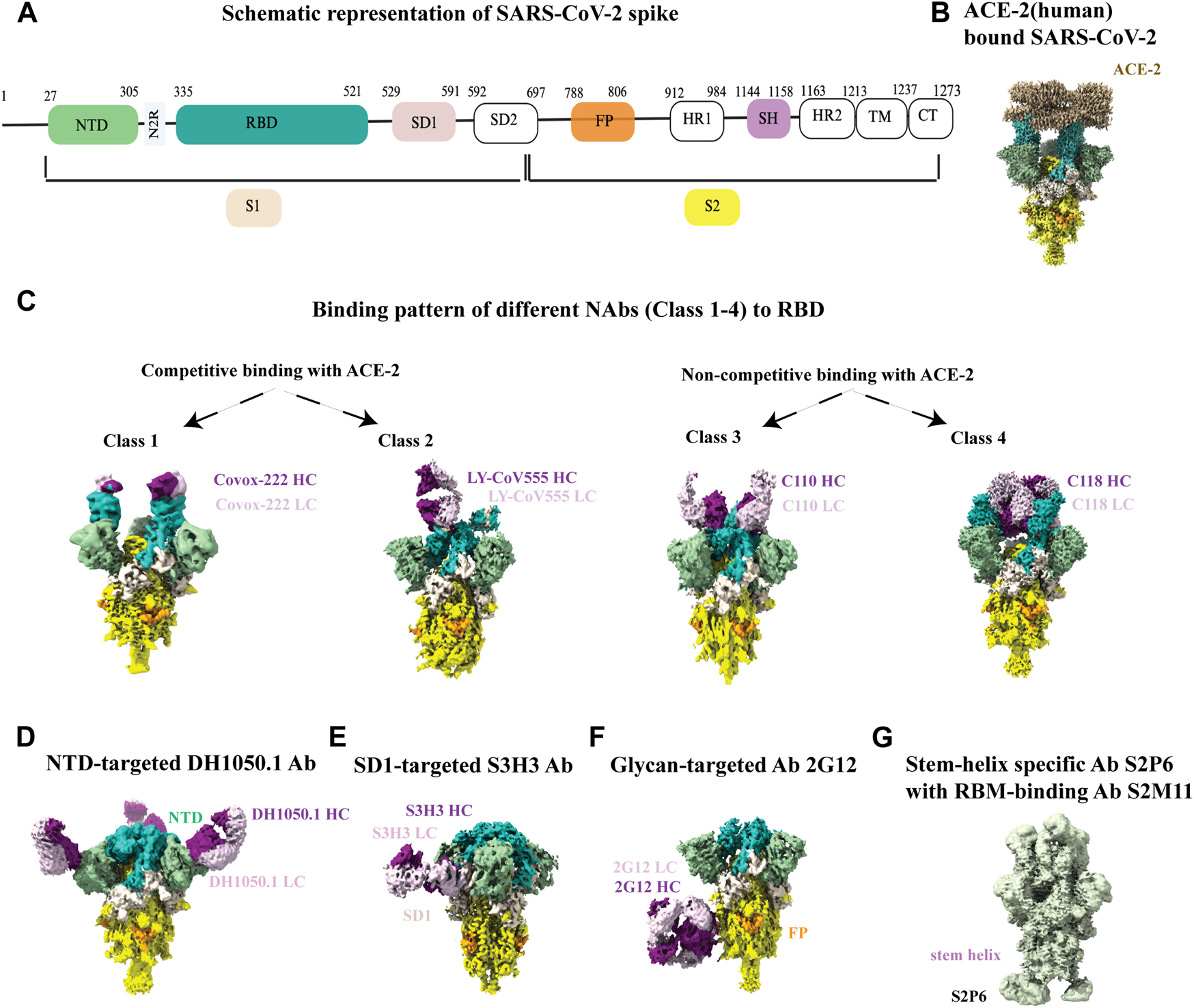
Cryo-EM studies of SARS-CoV-2 spikes and its interaction with antibodies. (A) Schematic representation of SARS-CoV2 spike with its different parts color labeled with S1 (antique white), S2 (yellow), N-terminal domain (NTD, dark sea green), receptor binding domain (RBD, sea green), SD1 (misty rose), fusion peptide (FP, orange) and stem helix region (pink violet).The same color scheme was maintained throughout Figure 5 except 5G. Antibody heavy chain is shown in purple and light chain is in thistle color. (B) ACE-2 bound (human) SARS-CoV-2 XBB.1.5 spike (EMD-43324). (C) Binding patterns of different NAbs (class 1, EMD-13869), (class 2, EMD-23156), (class 3, EMD-22732), (class 4, EMD-24504) to RBD. (D) Binding of NTD-targeted Ab DH1050.1 (EMD-23277). (E) Binding of SD1-targeted Ab S3H3 (EMD-32564). (F) Binding of glycan-targeted Ab 2G12 (EMD-23094). (G) Binding of stem-helix specific Ab S2P6 along with RBM-bound S2M11 (EMD-24533).
For SARS-CoV-2, although the dominant cellular receptor is ACE2, there exists an ACE2-independent pathway for cell entry. Accessory proteins on the membrane or alternative receptors, such as CD147 (extracellular matrix metalloproteinase), NRP1 (Neuropilin-1, a transmembrane protein), CD26 (also known as DPP4, dipeptidyl peptidase IV), AGTR2 (Angiotensin II receptor type 2), C-type lectins, LDLRAD3 and few others, have been implicated in mediating virus attachment and entry in the absence of ACE2 (Alipoor and Mirsaeidi, 2022; Lim et al., 2022). The spike protein is the target of almost all neutralizing antibodies found in the sera of a recovering person or elicited by vaccines. Throughout the pandemic, cryo-EM was also instrumental in visualizing antibody binding to the SARS-CoV-2 spike, enabling our understanding of the immune response in atomic level detail and informing the development of vaccines and therapeutics (Barnes et al., 2020; Jette et al., 2021; Jones et al., 2021; Li et al., 2021; Pinto et al., 2021; Williams et al., 2021; Hong et al., 2022; Liu et al., 2022) RBD-directed neutralizing antibodies are grouped in four different classes (1-4) based on the cryo-EM structural analysis of ACE-2 competitive or non-competitive binding site (Figure 5C). There are other neutralizing antibodies available that binds to the N-terminal domain (NTD) (Figure 5D), highly conserved epitopes in SD1 region (Figure 5E), high mannose glycan cluster of S2 subunit (Figure 5F), and stem-helix region in S2 domain (Figure 5G). Non-competing antibody cocktails could be a potential solution to prevent mutational escape from antibody regimens. While escape mutants arose in the presence of a single antibody, no resistance mutants were detected in the presence of non-competing or partially competing RBD binding site antibodies, (Baum et al., 2020). Aside from antibody-bound structures, the cryo-EM structure of the SARS-CoV-2 spike protein with linoleic acid (LA) bound to the hydrophobic pocket of the RBD to lock the spike in a stable conformation and occlude its most immunogenic epitopes, highlights the ability of cryo-EM to determine structures of small molecules bound to spike at high resolution and yield important mechanistic insights (Toelzer et al., 2023).
4.2 Cryo-ET of SARS-CoV-2 virus reveals intact virion, the mechanisms of receptor binding and antibody neutralization
Single-particle cryo-EM analysis has accelerated the process of determining structures of soluble SARS-CoV-2 S protein constructs. However, to visualize the spike structure and distribution on mature viruses in their natural environment, cryo-ET and subtomogram averaging (STA) have been used for in situ analysis (Figure 6A, left panel) (Yao et al., 2020). Cryo-ET studies of infected Vero E6 cells have revealed structural insights into the viral replication compartment, including the budding and assembly mechanism of virions, and the knowledge of packaging of extra-large genomes inside virion (Figure 6A, middle panel) (Klein et al., 2020). Furthermore, cryo-ET studies of SARS-CoV-2 provide information on the spatial arrangements of RNA filaments. Additionally, subtomogram averaging of vRNP complexes has shown a preferred stacking orientation of RNPs (Figure 6A, right panel) (Klein et al., 2020; Yao et al., 2020).
FIGURE 6

Cryo-electron tomography of membrane bound SARS-CoV-2 spikes. (A) From left to right, SARS-CoV-2 virus with spikes from subtomogram averaging (12Å) put back on membrane surface and glimpse of inside, EMD-30430, a representative tomogram slice of SARS-CoV-2 infected and cryo-FIB milled VeroE6 cells (black arrows indicate virions), EMD-11865, and in situ assembly of the ribonucleoprotein inside SARS-CoV-2 virus, EMD-30429. (B) From left to right, In situ spike structures reveal flexibility mediated by three hinges, EMD-11223, prefusion spike viewed at a 40° angle from the normal axis on membrane in a 3 RBD-down conformation, EMD-30426, and post-fusion conformation, EMD-30428.
The S protein on the surface of intact SARS-CoV-2 shows significant conformational diversity, including at its receptor binding site and in its spatial arrangement relative to the viral membrane. This provides a fundamental understanding of how the S protein interacts with the ACE2 receptor and neutralizing antibodies (Ke et al., 2020). The spike protein has shown some flexibility due to the presence of three hinges (Figure 6B, left panel) that are shielded by glycans and few such spikes together can interact with the host cell (Turoňová et al., 2020). Hinge glycans are mostly conserved and are held responsible for the bending of membrane-anchored spikes (Figure 6B, middle panel) (Yao et al., 2020). Additional insights on glycan shielding of the SARS-CoV-2 spike have been provided by molecular dynamics (MD) simulation studies (Casalino et al., 2020). A study on human coronavirus NL63 (HCoV-NL63) spikes reported greater oligomannose glycan shielding compared to SARS-CoV-2 spikes indicating better evasion from the host immune response for the former. Furthermore, molecular MD simulation of the HCoV-NL63 full length spikes show the function of hinge glycans in regulating spike flexibility (Chmielewski et al., 2023). The prefusion spike has intrinsic instability and therefore convert to postfusion state (Figure 6B, right panel) with spontaneous S1 dissociation (Cai et al., 2020; Yao et al., 2020).
The pre-fusion and post-fusion states of SARS-CoV-2 spike structures have been successfully resolved using cryo-EM SPA and cryo-ET techniques. However, the transient intermediate states of the fusion process remain to be detected. Cryo-ET is a powerful technique that can help capture these transient states. For example, a study reported the results of capturing extended and partially folded intermediate states of S2 by using an antiviral lipopeptide entry inhibitor (Marcink et al., 2022). Additionally, time-resolved cryo-EM and other complementary structural techniques hold the promise of enabling visualization of transient intermediates that occur along the viral entry pathways at rapid timescales (Harder et al., 2023; Bennett et al., 2024).
5 Bacteriophages, the bacterial viruses against infectious diseases including multidrug-resistant (MDR) strain-mediated diseases
Bacteriophages infect bacteria and are an emerging therapeutic tool to combat MDR bacteria (Viertel et al., 2014; Taati et al., 2020; Hibstu et al., 2022). Bacteriophages are the most abundant entity in any ecosystem where their host bacteria are present (Clokie et al., 2011; Romero-Calle et al., 2019). There are two types of bacteriophages: lytic or virulent, and lysogenic or temperate. Lytic phages, which have a short replication cycle and large burst size, are often preferred for therapeutic purposes (Fernández et al., 2019). Cocktails of phages can also be effective in treating bacterial infections and minimizing phage resistance (Chan et al., 2013; Abedon et l. 2021; Duc et al., 2023). Better and broader application of phages requires a deeper understanding of their structural components in different conformations and how the components function (Helen and Elena, 2019). In 1959 the first image of negatively stained T2 phage was produced (Brenner et al., 1959). EM has contributed to the detection of bacteriophage, their morphology and classification, and structure determination. Advances in cryo-EM technology have allowed researchers to study phage structures by single particle analysis and cryo-electron tomography in unprecedented detail (Helen and Elena, 2019; Belnap et al., 2021). Phages usually have a capsid head, head-tail interface (neck) and tail part, and the latter part participates in the recognition of host receptors, determining host range, and initiating infection (Chaban et al., 2015). A schematic representation of a long-tailed phage is shown in Figure 7A with different parts labelled with representative cryo-EM structures (Fokine et al., 2013; Yap et al., 2016; Ayala et al., 2023; Yang et al., 2023; Zhu et al., 2023; Mukherjee et al., 2024; Sonani et al., 2024; Wang et al., 2024). The maturation process of bacteriophage capsid head from immature procapsid takes place through a series of transient intermediates that have been extensively studied by cryo-EM (Ionel et al., 2011; Veesler et al., 2012; Huet et al., 2019; Wang et al., 2022). The capsid assembly and maturation process is closely connected to genome packaging inside phage capsid and cryo-EM has made substantial contributions in understanding this process (Rao and Black, 2010; Bayfield et al., 2019; Orlov et al., 2022; Hawkins et al., 2023). The major unit of head-tail interface is a dodecameric portal protein located in the vertex connected to the tail. It is a well-studied component of tailed dsDNA bacteriophages showing their diversity by cryo-EM with continuous improvement in the resolution (Orlova et al., 1999; Cerritelli et al., 2003; Dedeo et al., 2019; Javed et al., 2021). Phage tail recognizes the host cell through its receptor-binding protein and sends a signal to the phage head that are followed by a cascade of conformational change throughout phage tail and the formation of a channel to transfer the genome from phage capsid to the host cytoplasm (Figure 7B) (Cuervo et al., 2013; Arnaud et al., 2017; Wang et al., 2024). The visualization of infection and genome delivery mechanism of phages started in early 2000 using cryo-ET (Böhm et al., 2001; Liu et al., 2011). This is still a poorly understood process that involves sequential conformational changes in phage structural components mainly the tail components which are now a heavily studied area enabled by current advances of cryo-ET (Fu et al., 2011; Guerrero-Ferreira et al., 2019). With the high-resolution snapshots of the entire event of phage-host interactions, it was possible to capture pre and post infection/injection structures of short and long-tailed phages (Figure 7C) (Hu et al., 2015; Chen et al., 2021), high-resolution well studied structures of intermediate stages of long-tail contraction (Figure 7D) (Leiman et al., 2004), formation of transmembrane channel during short-tailed phage infection (Figure 7E) (Wang et al., 2019). All these have helped to propose a general mechanism of phage infection to destroy host bacteria (Hu et al., 2015; Nováček et al., 2016; Wang et al., 2019).
FIGURE 7

Structural studies on bacteriophages using cryo-EM. (A) Schematic diagram of a long-tailed bacteriophage with its parts labeled. Structure of different parts of bacteriophages were solved either individually or partially together and a representative cryo-EM map is shown for each part: capsid (EMD-40228), portal protein (EMD-43145), collar and whiskers (EMD-5528), tail tube (EMD-29354), tail sheath (EMD-36127), base plate (EMD-8064), tail fiber (EMD-34968), tail pin (EMD-35824) in figure A (created with BioRender.com). (B) Phage infection initiation mechanism is shown with a schematic model. (C) DNA pre injection and post injection states are shown in short tailed (left, EMD-31315, 31318) and long-tailed phages (right, EMD-2774, 6082). (D) Close view of detailed structural changes during intermediate stages of long tail contraction (EMD-1089, 1086). (E) Cryo-EM map of trans envelope channel formation during short-tailed phage infection (EMD-9006). (F) Schematic diagram of various application of phages (created with BioRender.com).
Phage therapy, which is over a century old, has been revived to combat the current crisis of antimicrobial resistance (AMR). Thanks to advanced technologies, such as high-throughput sequencing, synthetic biology, and genetic engineering, phages are now being used for other applications apart from antibacterial activities (Chen et al., 2019; Olsen et al., 2020; Lenneman et al., 2021). Once the high-resolution structural details and sequencing data for a new phage are available, genetical modification or phage structural engineering can be done to create a better version for therapeutic or industrial purposes (Gibb et al., 2021; Guo et al., 2021). By understanding the structure of phages, they can be applied in potential capsid-based delivery systems (e.g., drug, antigen), bio-nanotechnology, and can combat infectious diseases more effectively. For example, a study demonstrated that the icosahedral bacteriophage capsid can be used as a system to carry host cell hemagglutinin in an arrangement that blocks the entire influenza viral surface by binding to it in a defined multivalent mode, which inhibits the infection process (Lauster et al., 2020). Additionally, a phage display-based model was used to study the diversity within the receptor-binding domain (RBD) of SARS-CoV-2 Env proteins (Pérez-Massón et al., 2024). There are many other applications of phages beyond phage therapy or drug delivery. In Figure 7F, the multitude of applications of phage is described in a schematic presentation.
6 Discussion
Cryo-EM has had a significant impact on the field of structural virology and helped understand infection process of bacterial viruses to animal/human viruses. With the structural knowledge in hand, virus entry and infection mechanism models were proposed. The detailed understanding of the mechanism of HIV-1 virus attachment and entry into host cells using advanced cryo-EM technology has revealed previously unknown target sites for therapeutic intervention. There still remains a constant need for more effective and safe drugs and new targets to overcome the growing resistance of a highly mutating virus HIV-1 (Bean, 2005; Adamson and Freed, 2009). Identification of host-related targets can be a potential solution as the virus may find it difficult to develop resistance against the drug (Puhl et al., 2019). There are small molecule compounds identified through computer-aided molecular docking tools or high throughput screening to block the virus-CD4 receptor interaction (early phase), coreceptor entrance (advanced phase), and membrane fusion (final phase) (Briz et al., 2006; Yu et al., 2014; Xiao et al., 2020). APOBEC3G (A3G) is a key component of the human innate immune system to defend against invading viruses, whereas the viral infectivity factor (Vif) of HIV-1 virus helps them to neutralize cellular defenses. A recent report provides valuable information on APOBEC3G-Vif interaction, a prime event of viral immune suppression to overcome host defenses (Li et al., 2023). As a result, this has become a potential target for new therapeutics against HIV-1. Single Particle cryo-EM analysis was used to study the interaction of the HIV-1 capsid with the first-in-class capsid-targeting antiretroviral compound GS-6207 (lenacapavir) (Highland et al., 2023).
The COVID-19 pandemic caused by SARS-CoV-2 and its variants caught much scientific attention. While whole genome analysis and structural studies were the forefront at identifying and characterizing new variants, antibody isolation techniques coupled with cryo-EM structural determination accelerated our knowledge of how the immune system responds to SARS-CoV-2 and led to the development of antibody-based therapeutics. While therapeutic antibodies rapidly became ineffective against new variants, new computational tools coupled with structural analysis were employed to successfully redesign antibodies to bind the evolved variants (Desautels et al., 2024).
Antibiotic-resistant bacterial infection is a growing public threat that needs immediate attention. Phage therapy is a powerful alternative strategy to address the increasing number of drug-resistant bacterial infection and biofilm-mediated infection where lytic phages are deployed to infect and destroy the drug-resistant bacterial cells. Bacteriophages are plentiful in all ecosystems and play a role in controlling bacterial populations and evolution. The use of phages to address antibiotic resistance is gaining attention within the “One Health” framework, which encompasses not only humans, but also other animals (Kittler et al., 2017; Garvey, 2020). The most promising side of phage therapy is that there are no harmful effects so far reported. Still phage therapy is uninviting for many reasons such as immune response, selection of effective phages, lack of knowledge to setup well-developed clinical trials, and occurrence of phage resistance (Fernández et al., 2019; Hitchcock et al., 2023; Noor et al., 2024). In this review we mostly focused our discussion on structural studies of phages by cryo-EM and how this technique contributed to further understanding phage structures and proposing model for infection, assembly and egress (Guerrero-Ferreira and Wright, 2013; Linares et al., 2020; Conners et al., 2023). Current maps and models derived from cryo-EM data also showed the interior of the phage structure to reveal the highly ordered hexagonal packaging arrangement of DNA (Bayfield et al., 2019; Orlov et al., 2022).
7 Future epidemics and other emerging infectious disease threats: potential of cryo-EM
The COVID-19 pandemic has shown the aftermath of a pandemic causing severe damage to the health system, death toll, and socioeconomic condition of any country. At the same time, it showed us that extensive international collaboration in health science sectors can have a substantial impact on controlling the healthcare crisis and other collateral damages caused by a pandemic. Along with the advances in cryo-EM, other innovative approaches are being developed and integrated with cryo-EM for a comprehensive understanding of various events of physiological processes. The integrative approaches include CLEM (correlative light electron microscopy), FIB-SEM(focused ion beam scanning electron microscopy), Mass spectrometry, and time-resolved TEM (Tuijtel et al., 2019; Amann et al., 2023; de Beer et al., 2023; Lorenz 2024). These integrated cryo-EM approaches will contribute to overcoming the challenges of infectious disease studies especially where viral protein purification is difficult or for visualizing rapid time-scale events. With the current knowledge of vaccine technology and programs such as Defense Advances Research Program Agency (DARPA), Coalition for Epidemic Preparedness Innovations (CEPI) and Advanced Research Projects Agency for Health (ARPA-H), precautionary measures and infrastructures built to handle future pandemics and any new emerging disease threats are needed for pandemic preparedness (Sempowski et al., 2020). Modern state-of-the-art cryo-EM modalities combined with advanced image processing and other modern technologies, including AI-based structure prediction tools, have the potential to accelerate and enable structure-based design of interventions to combat infectious diseases (Jumper et al., 2021; Schoehn et al., 2023).
Statements
Author contributions
MD: Conceptualization, Investigation, Methodology, Validation, Visualization, Writing–original draft, Writing–review and editing. PA: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Funded by NIH grants R01 AI145687, R01 AI165947, R01 AI165147, U54 AI170752, U01 AI169587, P01 AI131251, and UM1 AI144371.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abedon S. T. Danis-Wlodarczyk K. M. Wozniak D. J. (2021). Phage cocktail development for bacteriophage therapy: toward improving spectrum of activity breadth and depth. Pharm. (Basel)14 (10), 1019. 10.3390/ph14101019
2
Adamson C. S. Freed E. O. (2009). Anti-HIV-1 therapeutics: from FDA-approved drugs to hypothetical future targets. Mol. Interv.9 (2), 70–74. 10.1124/mi.9.2.5
3
Aleem A. Akbar Samad A. B. Vaqar S. (2024). Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19). StatPearls copyright © 2024. Treasure Island (FL): StatPearls Publishing.
4
Alipoor S. D. Mirsaeidi M. (2022). SARS-CoV-2 cell entry beyond the ACE2 receptor. Mol. Biol. Rep.49 (11), 10715–10727. 10.1007/s11033-022-07700-x
5
Amann S. J. Keihsler D. Bodrug T. Brown N. G. Haselbach D. (2023). Frozen in time: analyzing molecular dynamics with time-resolved cryo-EM. Structure31 (1), 4–9.
6
Ananthaswamy N. Fang Q. AlSalmi W. Jain S. Chen Z. Klose T. et al (2019). A sequestered fusion peptide in the structure of an HIV-1 transmitted founder envelope trimer. Nat. Commun.10 (1), 873. 10.1038/s41467-019-08825-7
7
Antanasijevic A. Sewall L. M. Cottrell C. A. Carnathan D. G. Jimenez L. E. Ngo J. T. et al (2021). Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM. Nat. Commun.12 (1), 4817. 10.1038/s41467-021-25087-4
8
Antanasijevic A. Ueda G. Brouwer P. J. M. Copps J. Huang D. Allen J. D. et al (2020). Structural and functional evaluation of de novo-designed, two-component nanoparticle carriers for HIV Env trimer immunogens. PLoS Pathog.16 (8), e1008665. 10.1371/journal.ppat.1008665
9
Arnaud C. A. Effantin G. Vivès C. Engilberge S. Bacia M. Boulanger P. et al (2017). Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection. Nat. Commun.8 (1), 1953. 10.1038/s41467-017-02049-3
10
Ayala R. Moiseenko A. V. Chen T.-H. Kulikov E. E. Golomidova A. K. Orekhov P. S. et al (2023). Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers. Nat. Commun.14 (1), 8205. 10.1038/s41467-023-43824-9
11
Banach B. B. Pletnev S. Olia A. S. Xu K. Zhang B. Rawi R. et al (2023). Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site. Nat. Commun.14 (1), 7593. 10.1038/s41467-023-42098-5
12
Barnes C. O. Jette C. A. Abernathy M. E. Dam K.-M. A. Esswein S. R. Gristick H. B. et al (2020). SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature588 (7839), 682–687. 10.1038/s41586-020-2852-1
13
Basore K. Kim A. S. Nelson C. A. Zhang R. Smith B. K. Uranga C. et al (2019). Cryo-EM structure of chikungunya virus in complex with the Mxra8 receptor. Cell177 (7), 1725–1737. e1716.
14
Bartesaghi A. Sapiro G. Subramaniam S. (2005). An energy-based three-dimensional segmentation approach for the quantitative interpretation of electron tomograms. IEEE Trans Image Proc.14 (9), 1314–1323.
15
Baum A. Fulton B. O. Wloga E. Copin R. Pascal K. E. Russo V. et al (2020). Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science369 (6506), 1014–1018. 10.1126/science.abd0831
16
Bayfield O. W. Klimuk E. Winkler D. C. Hesketh E. L. Chechik M. Cheng N. et al (2019). Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Proc. Natl. Acad. Sci. U. S. A.116 (9), 3556–3561. 10.1073/pnas.1813204116
17
Bean P. (2005). New drug targets for HIV. Clin. Infect. Dis.41 (Suppl. 1), S96–S100. 10.1086/429504
18
Blanco-Rodriguez G. Gazi A. Monel B. Frabetti S. Scoca V. Mueller F. et al (2020). Remodeling of the core leads HIV-1 preintegration complex into the nucleus of human lymphocytes. J. Virol.9 (11). 10.1128/jvi.00135-00120
19
Belnap D. M. (2021). “Detection of bacteriophages: electron microscopy and visualization,” in Bacteriophages: biology, technology, therapy. Editors HarperD. R.AbedonS. T.BurrowesB. H.McConvilleM. L. (Cham: Springer International Publishing), 561–620.
20
Bennett A. L. Edwards R. Kosheleva I. Saunders C. Bililign Y. Williams A. et al (2024). Microsecond dynamics control the HIV-1 Envelope conformation. Sci. Adv.10 (5), eadj0396. 10.1126/sciadv.adj0396
21
Bharat T. A. Castillo Menendez L. R. Hagen W. J. Lux V. Igonet S. Schorb M. et al (2014). Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc. Natl. Acad. Sci.111 (22), 8233–8238. 10.1073/pnas.1401455111
22
Bianchi M. Turner H. L. Nogal B. Cottrell C. A. Oyen D. Pauthner M. et al (2018). Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity49 (2), 288–300. 10.1016/j.immuni.2018.07.009
23
Blumenthal R. Durell S. Viard M. (2012). HIV entry and envelope glycoprotein-mediated fusion. J. Biol. Chem.287 (49), 40841–40849. 10.1074/jbc.R112.406272
24
Böhm J. Lambert O. Frangakis A. S. Letellier L. Baumeister W. Rigaud J. L. (2001). FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr. Biol.11 (15), 1168–1175. 10.1016/s0960-9822(01)00349-9
25
Brenner S. Streisinger G. Horne R. W. Champe S. P. Barnett L. Benzer S. et al (1959). Structural components of bacteriophage. J. Mol. Biol.1 (3), 281–IN21. 10.1016/s0022-2836(59)80035-8
26
Briz V. Poveda E. Soriano V. (2006). HIV entry inhibitors: mechanisms of action and resistance pathways. J. Antimicrob. Chemother.57 (4), 619–627. 10.1093/jac/dkl027
27
Burton D. R. (2023). Antiviral neutralizing antibodies: from in vitro to in vivo activity. Nat. Rev. Immunol.23 (11), 720–734. 10.1038/s41577-023-00858-w
28
Cai Y. Zhang J. Xiao T. Peng H. Sterling S. M. Walsh R. M. Jr. et al (2020). Distinct conformational states of SARS-CoV-2 spike protein. Science369 (6511), 2020.05.16.099317–1592. 10.1101/2020.05.16.099317
29
Carabelli A. M. Peacock T. P. Thorne L. G. Harvey W. T. Hughes J. Peacock S. J. et al (2023). SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol.21 (3), 162–177. 10.1038/s41579-022-00841-7
30
Carlson L. A. de Marco A. Oberwinkler H. Habermann A. Briggs J. A. Kräusslich H. G. et al (2010). Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog.6 (11), e1001173.
31
Carreño J. M. Raskin A. Singh G. Tcheou J. Kawabata H. Gleason C. et al (2023). An inactivated NDV-HXP-S COVID-19 vaccine elicits a higher proportion of neutralizing antibodies in humans than mRNA vaccination. Sci. Transl. Med.15 (683), eabo2847. 10.1126/scitranslmed.abo2847
32
Casalino L. Gaieb Z. Goldsmith J. A. Hjorth C. K. Dommer A. C. Harbison A. M. et al (2020). Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci.6 (10), 1722–1734. 10.1021/acscentsci.0c01056
33
Casasnovas J. M. (2013). Virus-receptor interactions and receptor-mediated virus entry into host cells. Struct. Phys. Viruses Integr. Textb.68, 441–466. 10.1007/978-94-007-6552-8_15
34
Cerritelli M. E. Trus B. L. Smith C. S. Cheng N. Conway J. F. Steven A. C. (2003). A second symmetry mismatch at the portal vertex of bacteriophage T7: 8-fold symmetry in the procapsid core. J. Mol. Biol.327 (1), 1–6. 10.1016/s0022-2836(03)00117-7
35
Cerutti G. Guo Y. Liu L. Liu L. Zhang Z. Luo Y. et al (2022). Cryo-EM structure of the SARS-CoV-2 Omicron spike. Cell Rep.38 (9), 110428. 10.1016/j.celrep.2022.110428
36
Chaban Y. Lurz R. Brasilès S. Cornilleau C. Karreman M. Zinn-Justin S. et al (2015). Structural rearrangements in the phage head-to-tail interface during assembly and infection. Proc. Natl. Acad. Sci. U. S. A.112 (22), 7009–7014. 10.1073/pnas.1504039112
37
Chan B. K. Abedon S. T. Loc-Carrillo C. (2013). Phage cocktails and the future of phage therapy. Future Microbiol.8 (6), 769–783. 10.2217/fmb.13.47
38
Chen W. Xiao H. Wang L. Wang X. Tan Z. Han Z. et al (2021). Structural changes in bacteriophage T7 upon receptor-induced genome ejection. Proc. Natl. Acad. Sci.118 (37), e2102003118. 10.1073/pnas.2102003118
39
Chen Y. Batra H. Dong J. Chen C. Rao V. B. Tao P. (2019). Genetic engineering of bacteriophages against infectious diseases. Front. Microbiol.10, 954. 10.3389/fmicb.2019.00954
40
Chmielewski D. Wilson E. A. Pintilie G. Zhao P. Chen M. Schmid M. F. et al (2023). Structural insights into the modulation of coronavirus spike tilting and infectivity by hinge glycans. Nat. Commun.14 (1), 7175. 10.1038/s41467-023-42836-9
41
Clokie M. R. Millard A. D. Letarov A. V. Heaphy S. (2011). Phages in nature. Bacteriophage1 (1), 31–45. 10.4161/bact.1.1.14942
42
Conners R. León-Quezada R. I. McLaren M. Bennett N. J. Daum B. Rakonjac J. et al (2023). Cryo-electron microscopy of the f1 filamentous phage reveals insights into viral infection and assembly. Nat. Commun.14 (1), 2724. 10.1038/s41467-023-37915-w
43
Cottrell C. A. Manne K. Kong R. Wang S. Zhou T. Chuang G. Y. et al (2021). Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies. Cell Rep.37 (5), 109922. 10.1016/j.celrep.2021.109922
44
Creekmore B. C. Chang Y.-W. Lee E. B. (2021). The Cryo-EM effect: structural biology of neurodegenerative disease aggregates. J. Neuropathology Exp. Neurology80 (6), 514–529. 10.1093/jnen/nlab039
45
Cressey D. Callaway E. (2017). Cryo-electron microscopy wins chemistry Nobel. Nature550 (7675).
46
Cuervo A. Chagoyen M. Pulido-Cid M. Camacho A. Carrascosa J. L. (2013). Structural characterization of T7 tail machinery reveals a conserved tubular structure among other Podoviridae family members and suggests a common mechanism for DNA delivery. Bacteriophage3 (4), e27011. 10.4161/bact.27011
47
Dandey V. P. et al (2018). Spotiton: New features and applications. J. Struct. Biol.202 (2), 161–169.
48
Dam K.-M. A. Fan C. Yang Z. Bjorkman P. J. (2023). Intermediate conformations of CD4-bound HIV-1 Env heterotrimers. Nature623 (7989), 1017–1025. 10.1038/s41586-023-06639-8
49
Dang H. V. Chan Y. Park Y. Broder C. Veesler D. (2019). Cryo-EM Structure of Nipah Virus Fusion Glycoprotein in Complex with a Monoclonal Antibody Reveals Mechanism of Neutralization. Microsc. Microanal.25 (S2), 1328–1329.
50
Dedeo C. L. Cingolani G. Teschke C. M. (2019). Portal protein: the orchestrator of capsid assembly for the dsDNA tailed bacteriophages and herpesviruses. Annu. Rev. Virol.6 (1), 141–160. 10.1146/annurev-virology-092818-015819
51
Desautels T. A. Arrildt K. T. Zemla A. T. Lau E. Y. Zhu F. Ricci D. et al (2024). Computationally restoring the potency of a clinical antibody against Omicron. Nature629 (8013), 878–885. 10.1038/s41586-024-07385-1
52
Dokainish H. M. Re S. Mori T. Kobayashi C. Jung J. Sugita Y. (2022). The inherent flexibility of receptor binding domains in SARS-CoV-2 spike protein. Elife11, e75720. 10.7554/eLife.75720
53
Dolan K. A. Dutta M. Kern D. M. Kotecha A. Voth G. A. Brohawn S. G. (2022). Structure of SARS-CoV-2 M protein in lipid nanodiscs. eLife11, e81702. 10.7554/eLife.81702
54
de Beer M. Daviran D. Roverts R. Rutten L. Macías-Sánchez E. Metz J. R. et al (2023). Precise targeting for 3D cryo-correlative light and electron microscopy volume imaging of tissues using a FinderTOP. Communi. Bio.6 (1), 510
55
Dubochet J. Adrian M. Chang J. J. Homo J. C. Lepault J. McDowall A. W. et al (1988). Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys.21 (2), 129–228.
56
Duc H. M. Zhang Y. Hoang S. M. Masuda Y. Honjoh K.-I. Miyamoto T. (2023). The use of phage cocktail and various antibacterial agents in combination to prevent the emergence of phage resistance. Antibiotics12 (6), 1077. 10.3390/antibiotics12061077
57
Earl L. A. Lifson J. D. Subramaniam S. (2013). Catching HIV ‘in the act’with 3D electron microscopy. Trends Microbiol.21 (8), 397–404.
58
Fernández I. S. Bai X. C. Hussain T. Kelley A. C. Lorsch J. R. Ramakrishnan V. (2013). Molecular architecture of a eukaryotic translational initiation complex. Science342 (6160), 1240585.
59
Felts R. L. Narayan K. Estes J. D. Shi D. Trubey C. M. Fu J. et al (2010). 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc Natl Acad Sci U S A107 (30), 13336–13341.
60
Fernández L. Gutiérrez D. García P. Rodríguez A. (2019). The perfect bacteriophage for therapeutic applications-A quick guide. Antibiot. (Basel)8 (3), 126. 10.3390/antibiotics8030126
61
Fokine A. Zhang Z. Kanamaru S. Bowman V. D. Aksyuk A. A. Arisaka F. et al (2013). The molecular architecture of the bacteriophage T4 neck. J. Mol. Biol.425 (10), 1731–1744. 10.1016/j.jmb.2013.02.012
62
Friedlaender M. Moore D. H. Koprowski H. (1955). Studies with the electron microscope of virus-host relationships in ehrlich ascites tumor cells: ii. The localization and possible development of anopheles a virus within the endoplasmic reticulum of the host cell. J. Exp. Med.102 (4), 371–378. 10.1084/jem.102.4.371
63
Fu X. Walter M. H. Paredes A. Morais M. C. Liu J. (2011). The mechanism of DNA ejection in the Bacillus anthracis spore-binding phage 8a revealed by cryo-electron tomography. Virology421 (2), 141–148. 10.1016/j.virol.2011.08.028
64
Garvey M. (2020). Bacteriophages and the One health approach to combat multidrug resistance: is this the way?Antibiot. (Basel)9 (7), 414. 10.3390/antibiotics9070414
65
Gibb B. Hyman P. Schneider C. L. (2021). The many applications of engineered bacteriophages-an overview. Pharm. (Basel)14 (7), 634. 10.3390/ph14070634
66
Gobeil S. M. Henderson R. Stalls V. Janowska K. Huang X. May A. et al (2022). Structural diversity of the SARS-CoV-2 Omicron spike. Mol. Cell82 (11), 2050–2068.e6. 10.1016/j.molcel.2022.03.028
67
Gobeil S. M. Janowska K. McDowell S. Mansouri K. Parks R. Manne K. et al (2021). D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep.34 (2), 108630. 10.1016/j.celrep.2020.108630
68
Goldsmith C. S. Miller S. E. (2009). Modern uses of electron microscopy for detection of viruses. Clin. Microbiol. Rev.22 (4), 552–563. 10.1128/CMR.00027-09
69
Graham B. S. Gilman M. S. McLellan J. S. (2019). Structure-based vaccine antigen design. Annu. Rev. Med.70, 91–104. 10.1146/annurev-med-121217-094234
70
Graham M. Zhang P. (2023). Cryo-electron tomography to study viral infection. Biochem. Soc. Trans.51 (4), 1701–1711. 10.1042/BST20230103
71
Grant T. Rohou A. Grigorieff N. (2018). cis TEM, user-friendly software for single-particle image processing. Elife7, e35383
72
Guerrero-Ferreira R. C. Hupfeld M. Nazarov S. Taylor N. M. Shneider M. M. Obbineni J. M. et al (2019). Structure and transformation of bacteriophage A511 baseplate and tail upon infection of Listeria cells. Embo J.38 (3), e99455. 10.15252/embj.201899455
73
Guerrero-Ferreira R. C. Wright E. R. (2013). Cryo-electron tomography of bacterial viruses. Virology435 (1), 179–186. 10.1016/j.virol.2012.08.022
74
Guo D. Chen J. Zhao X. Luo Y. Jin M. Fan F. et al (2021). Genetic and chemical engineering of phages for controlling multidrug-resistant bacteria. Antibiot. (Basel)10 (2), 202. 10.3390/antibiotics10020202
75
Guo Y. Wun S. Li W. Yang H. Shi T. Ju B. et al (2024). The cryo-EM structure of homotetrameric attachment glycoprotein from langya henipavirus. Nat. Commun.1 (1), 812
76
Harder O. F. Barrass S. V. Drabbels M. Lorenz U. J. (2023). Fast viral dynamics revealed by microsecond time-resolved cryo-EM. Nat. Commun.14 (1), 5649. 10.1038/s41467-023-41444-x
77
Harris A. Cardone G. Winkler D. C. Heymann J. B. Brecher M. White J. M. et al (2006). Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci.103 (50), 19123–19127.
78
Harris A. Borgnia M. J. Shi D. Bartesaghi A. He H. Pejchal R. et al (2011). Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci.108 (28), 11440–11445.
79
Hawkins D. Bayfield O. W. Fung H. K. H. Grba D. N. Huet A. Conway J. F. et al (2023). Insights into a viral motor: the structure of the HK97 packaging termination assembly. Nucleic Acids Res.51 (13), 7025–7035. 10.1093/nar/gkad480
80
Haynes B. F. Wiehe K. Borrow P. Saunders K. O. Korber B. Wagh K. et al (2023). Strategies for HIV-1 vaccines that induce broadly neutralizing antibodies. Nat. Rev. Immunol.23 (3), 142–158. 10.1038/s41577-022-00753-w
81
Hazelton P. R. Gelderblom H. R. (2003). Electron microscopy for rapid diagnosis of infectious agents in emergent situations. Emerg. Infect. Dis.9 (3), 294–303. 10.3201/eid0903.020327
82
Helen E. W. Elena V. O. (2019). Bacteriophages: their structural organisation and function. Bacteriophages. S. Renos. Rijeka, IntechOpen: Ch. 2.
83
Henderson R. Edwards R. J. Mansouri K. Janowska K. Stalls V. Gobeil S. M. C. et al (2020). Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol.27 (10), 925–933. 10.1038/s41594-020-0479-4
84
Henderson R. Zhou Y. Stalls V. Wiehe K. Saunders K. O. Wagh K. et al (2023). Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage. Nat. Commun.14 (1), 2782. 10.1038/s41467-023-38108-1
85
Highland C. M. A. Tan A. Ricaña C. L. Briggs J. A. G. Dick R. A. (2023). Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM. Proc. Nat. Acad. Sci.120 (18), e2220545120.
86
Hibstu Z. Belew H. Akelew Y. Mengist H. M. (2022). Phage therapy: a different approach to fight bacterial infections. Biologics16, 173–186. 10.2147/BTT.S381237
87
Hitchcock N. M. Devequi Gomes Nunes D. Shiach J. Valeria Saraiva Hodel K. Dantas Viana Barbosa J. Alencar Pereira Rodrigues L. et al (2023). Current clinical landscape and global potential of bacteriophage therapy. Viruses15 (4), 1020. 10.3390/v15041020
88
Holt G. T. Gorman J. Wang S. Lowegard A. U. Zhang B. Liu T. et al (2023). Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design. Cell Rep.42 (7), 112711. 10.1016/j.celrep.2023.112711
89
Hong Q. Han W. Li J. Xu S. Wang Y. Xu C. et al (2022). Molecular basis of receptor binding and antibody neutralization of Omicron. Nature604 (7906), 546–552. 10.1038/s41586-022-04581-9
90
Hsieh C.-L. Goldsmith J. A. Schaub J. M. DiVenere A. M. Kuo H.-C. Javanmardi K. et al (2020). Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science369 (6510), 1501–1505. 10.1126/science.abd0826
91
Hu B. Margolin W. Molineux I. J. Liu J. (2015). Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc. Natl. Acad. Sci. U. S. A.112 (35), E4919–E4928. 10.1073/pnas.1501064112
92
Huang J. Ofek G. Laub L. Louder M. K. Doria-Rose N. A. Longo N. S. et al (2012). Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature491 (7424), 406–412. 10.1038/nature11544
93
Huet A. Duda R. L. Boulanger P. Conway J. F. (2019). Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. U. S. A.116 (42), 21037–21046. 10.1073/pnas.1909645116
94
Ionel A. Velázquez-Muriel J. A. Luque D. Cuervo A. Castón J. R. Valpuesta J. M. et al (2011). Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. J. Biol. Chem.286 (1), 234–242. 10.1074/jbc.M110.187211
95
Jackson C. B. Farzan M. Chen B. Choe H. (2022). Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol.23 (1), 3–20. 10.1038/s41580-021-00418-x
96
Javed A. Villanueva H. Shataer S. Vasciaveo S. Savva R. Orlova E. V. (2021). Cryo-EM structures of two bacteriophage portal proteins provide insights for antimicrobial phage engineering. Viruses13 (12), 2532. 10.3390/v13122532
97
Jette C. A. Cohen A. A. Gnanapragasam P. N. P. Muecksch F. Lee Y. E. Huey-Tubman K. E. et al (2021). Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. Cell Rep.36 (13), 109760. 10.1016/j.celrep.2021.109760
98
Jiang W. Baker M. L. Jakana J. Weigele P. R. King J. Chiu W. (2008). Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature451 (7182), 1130–1134. 10.1038/nature06665
99
Jiang W. Tang L. (2017). Atomic cryo-EM structures of viruses. Curr. Opin. Struct. Biol.46, 122–129. 10.1016/j.sbi.2017.07.002
100
Jo S. J. Kwon J. Kim S. G. Lee S. J. (2023). The biotechnological application of bacteriophages: what to do and where to go in the middle of the post-antibiotic era. Microorganisms11 (9), 2311. 10.3390/microorganisms11092311
101
Jones B. E. Brown-Augsburger P. L. Corbett K. S. Westendorf K. Davies J. Cujec T. P. et al (2021). The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med.13 (593), eabf1906. 10.1126/scitranslmed.abf1906
102
Julien J.-P. Cupo A. Sok D. Stanfield R. L. Lyumkis D. Deller M. C. et al (2013). Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science342 (6165), 1477–1483. 10.1126/science.1245625
103
Jumper J. Evans R. Pritzel A. Green T. Figurnov M. Ronneberger O. et al (2021). Highly accurate protein structure prediction with AlphaFold. Nature596 (7873), 583–589. 10.1038/s41586-021-03819-2
104
Ke Z. Oton J. Qu K. Cortese M. Zila V. McKeane L. et al (2020). Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature588 (7838), 498–502. 10.1038/s41586-020-2665-2
105
Kim H.-u. Jung H. S. (2021). Cryo-EM as a powerful tool for drug discovery: recent structural based studies of SARS-CoV-2. Appl. Microsc.51 (1), 13. 10.1186/s42649-021-00062-x
106
Kirchdoerfer R. N. Cottrell C. A. Wang N. Pallesen J. Yassine H. M. Turner H. L. et al (2016). Pre-fusion structure of a human coronavirus spike protein. Nature531 (7592), 118–121. 10.1038/nature17200
107
Kittler S. Wittmann J. Mengden R. A. L. P. Klein G. Rohde C. Lehnherr H. (2017). The use of bacteriophages as One-Health approach to reduce multidrug-resistant bacteria. Sustain. Chem. Pharm.5, 80–83. 10.1016/j.scp.2016.06.001
108
Klein S. Cortese M. Winter S. L. Wachsmuth-Melm M. Neufeldt C. J. Cerikan B. et al (2020). SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun.11 (1), 5885. 10.1038/s41467-020-19619-7
109
Korber B. Fischer W. M. Gnanakaran S. Yoon H. Theiler J. Abfalterer W. et al (2020). Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell182 (4), 812–827.e19. 10.1016/j.cell.2020.06.043
110
Kucukelbir A. Sigworth F. J. Tagare H. D. (2014). Quantifying the local resolution of cryo-EM density maps. Nat. Methods11 (1), 63–65.
111
Kühlbrandt W. (2014a). The resolution revolution. Science343 (6178), 1443–1444.
112
Kühlbrandt W. (2014b). Cryo-EM enters a new era. Elife3, e03678
113
Kwon Y. D. Pancera M. Acharya P. Georgiev I. S. Crooks E. T. Gorman J. et al (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol.22 (7), 522–531. 10.1038/nsmb.3051
114
Kwong P. D. Wyatt R. Robinson J. Sweet R. W. Sodroski J. Hendrickson W. A. (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature393 (6686), 648–659. 10.1038/31405
115
LaBranche C. C. Henderson R. Hsu A. Behrens S. Chen X. Zhou T. et al (2019). Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancestor of CH235 lineage CD4bs broadly neutralizing antibodies. PLoS Pathog.15 (9), e1008026. 10.1371/journal.ppat.1008026
116
Ladinsky M. S. Gnanapragasam P. N. Yang Z. Behrens S. West A. P. Kay M. S. et al (2020). Electron tomography visualization of HIV-1 fusion with target cells using fusion inhibitors to trap the pre-hairpin intermediate. Elife9, e58411
117
Lan J. Ge J. Yu J. Shan S. Zhou H. Fan S. et al (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature581 (7807), 215–220. 10.1038/s41586-020-2180-5
118
Lauster D. Klenk S. Ludwig K. Nojoumi S. Behren S. Adam L. et al (2020). Phage capsid nanoparticles with defined ligand arrangement block influenza virus entry. Nat. Nanotechnol.15 (5), 373–379. 10.1038/s41565-020-0660-2
119
Lee J. H. Ozorowski G. Ward A. B. (2016). Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science351 (6277), 1043–1048. 10.1126/science.aad2450
120
Leiman P. G. Chipman P. R. Kostyuchenko V. A. Mesyanzhinov V. V. Rossmann M. G. (2004). Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell118 (4), 419–429. 10.1016/j.cell.2004.07.022
121
Lenneman B. R. Fernbach J. Loessner M. J. Lu T. K. Kilcher S. (2021). Enhancing phage therapy through synthetic biology and genome engineering. Curr. Opin. Biotechnol.68, 151–159. 10.1016/j.copbio.2020.11.003
122
Li D. Edwards R. J. Manne K. Martinez D. R. Schäfer A. Alam S. M. et al (2021). In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell184 (16), 4203–4219.e32. 10.1016/j.cell.2021.06.021
123
Li Y. L. Langley C. A. Azumaya C. M. Echeverria I. Chesarino N. M. Emerman M. et al (2023). The structural basis for HIV-1 Vif antagonism of human APOBEC3G. Nature615 (7953), 728–733. 10.1038/s41586-023-05779-1
124
Li Z. Li W. Lu M. Bess J. Jr Chao C. W. Gorman J. et al (2020). Subnanometer structures of HIV-1 envelope trimers on aldrithiol-2-inactivated virus particles. Nat. Struct. Mol. Biol.27 (8), 726–734. 10.1038/s41594-020-0452-2
125
Lim S. Zhang M. Chang T. L. (2022). ACE2-Independent alternative receptors for SARS-CoV-2. Viruses14 (11), 2535. 10.3390/v14112535
126
Linares R. Arnaud C.-A. Degroux S. Schoehn G. Breyton C. (2020). Structure, function and assembly of the long, flexible tail of siphophages. Curr. Opin. Virology45, 34–42. 10.1016/j.coviro.2020.06.010
127
Liu C. Zhou D. Nutalai R. Duyvesteyn H. M. E. Tuekprakhon A. Ginn H. M. et al (2022). The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants. Cell Host Microbe30 (1), 53–68.e12. 10.1016/j.chom.2021.11.013
128
Liu J. Bartesaghi A. Borgnia M. J. Sapiro G. Subramaniam S. (2008). Molecular architecture of native HIV-1 gp120 trimers. Nature455 (7209), 109–113. 10.1038/nature07159
129
Liu J. Chen C.-Y. Shiomi D. Niki H. Margolin W. (2011). Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology417 (2), 304–311. 10.1016/j.virol.2011.06.005
130
Liu J. Wright E. R. Winkler H. (2010). 3D visualization of HIV virions by cryoelectron tomography. Methods Enzymol.483, 267–290. 10.1016/S0076-6879(10)83014-9
131
Liu H. F. Zhou Y. Huang Q. Piland J. Jin W. Mandel J. et al (2023). nextPYP: a comprehensive and scalable platform for characterizing protein variability in situ using single-particle cryo-electron tomography. Nat. Metho.20 (12), 1909–1919.
132
Lorenz U. J. (2024). Microsecond time-resolved cryo-electron microscopy. Curr. Opin. Struc. Virology87, 102840
133
Lu G. Wang Q. Gao G. F. (2015). Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol.23 (8), 468–478. 10.1016/j.tim.2015.06.003
134
Lu M. Chamblee M. Zhang Y. Ye C. Dravid P. Park J.-G. et al (2022). SARS-CoV-2 prefusion spike protein stabilized by six rather than two prolines is more potent for inducing antibodies that neutralize viral variants of concern. Proc. Natl. Acad. Sci.119 (35), e2110105119. 10.1073/pnas.2110105119
135
Lyumkis D. Julien J.-P. De Val N. Cupo A. Potter C. S. Klasse P.-J. et al (2013). Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science342 (6165), 1484–1490. 10.1126/science.1245627
136
Mak J. de Marco A. (2018). Recent advances in retroviruses via cryo-electron microscopy. Retrovirology15 (1), 23. 10.1186/s12977-018-0405-6
137
Mangala Prasad V. Leaman D. P. Lovendahl K. N. Croft J. T. Benhaim M. A. Hodge E. A. et al (2022). Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the Gag lattice. Cell185 (4), 641–653. e617.
138
Mannar D. Saville J. W. Poloni C. Zhu X. Bezeruk A. Tidey K. et al (2024). Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein. Nat. Commun.15 (1), 1854. 10.1038/s41467-024-46104-2
139
Mannar D. Saville J. W. Zhu X. Srivastava S. S. Berezuk A. M. Tuttle K. S. et al (2022). SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science375 (6582), 760–764. 10.1126/science.abn7760
140
Marcink T. C. Kicmal T. Armbruster E. Zhang Z. Zipursky G. Golub K. L. et al (2022). Intermediates in SARS-CoV-2 spike-mediated cell entry. Sci. Adv.8 (33), eabo3153. 10.1126/sciadv.abo3153
141
Meyerson J. R. Tran E. E. Kuybeda O. Chen W. Dimitrov D. S. Gorlani A. et al (2013). Molecular structures of trimeric HIV-1 Env in complex with small antibody derivatives. Proc. Natl. Acad. Sci.110 (2), 513–518. 10.1073/pnas.1214810110
142
Milne J. L. Borgnia M. J. Bartesaghi A. Tran E. E. Earl L. A. Schauder D. M. et al (2013). Cryo‐electron microscopy–a primer for the non‐microscopist. FEBS J.280 (1), 28–45. 10.1111/febs.12078
143
Mukherjee A. Kizziah J. L. Hawkins N. T. C. Nasef M. O. Parker L. K. Dokland T. (2024). Structure of the portal complex from Staphylococcus aureus pathogenicity island 1 transducing particles in situ and in isolation. J. Mol. Biol.436 (4), 168415. 10.1016/j.jmb.2023.168415
144
Nakane T. Kotecha A. Sente A. McMullan G. Masiulis S. Brown P. M. G. E. et al (2020). Single-particle cryo-EM at atomic resolution. Nature587 (7832), 152–156. 10.1038/s41586-020-2829-0
145
Narasimha R. Aganj I. Bennett A. E. Borgnia M. J. Zabransky D. Sapiro G. et al (2008). Evaluation of denoising algorithms for biological electron tomography. J. Struct. Biol.164 (1), 7–17.
146
Noor I. Nasir M. H. Ur Rehman A. Javed N. Waheed W. Waheed A. et al (2024). Medicinal and immunological aspects of bacteriophage therapy to combat antibiotic resistance. Explor. Med.5 (2), 215–231. 10.37349/emed.2024.00217
147
Noble A. J. et al (2018). Routine single particle CryoEM sample and grid characterization by tomography. Elife7.
148
Nováček J. Šiborová M. Benešík M. Pantůček R. Doškař J. Plevka P. (2016). Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate. Proc. Natl. Acad. Sci. U. S. A.113 (33), 9351–9356. 10.1073/pnas.1605883113
149
Olsen N. S. Hendriksen N. B. Hansen L. H. Kot W. (2020). A new high-throughput screening method for phages: enabling crude isolation and fast identification of diverse phages with therapeutic potential. Phage New. Rochelle1 (3), 137–148. 10.1089/phage.2020.0016
150
Orlov I. Roche S. Brasilès S. Lukoyanova N. Vaney M. C. Tavares P. et al (2022). CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper. Nat. Commun.13 (1), 7283. 10.1038/s41467-022-34999-8
151
Orlova E. V. Dube P. Beckmann E. Zemlin F. Lurz R. Trautner T. A. et al (1999). Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat. Struct. Biol.6 (9), 842–846. 10.1038/12303
152
Ozorowski G. Pallesen J. de Val N. Lyumkis D. Cottrell C. A. Torres J. L. et al (2017). Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature547 (7663), 360–363. 10.1038/nature23010
153
Pallesen J. Wang N. Corbett K. S. Wrapp D. Kirchdoerfer R. N. Turner H. L. et al (2017). Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci.114 (35), E7348–E7357. 10.1073/pnas.1707304114
154
Pancera M. Zhou T. Druz A. Georgiev I. S. Soto C. Gorman J. et al (2014). Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature514 (7523), 455–461. 10.1038/nature13808
155
Patwardhan A. (2017). Trends in the electron microscopy data bank (EMDB). Acta Crystallogr., Sect. D: Struct. Biol.73 (6), 503–508.
156
Parsons R. J. Acharya P. (2023). Evolution of the SARS-CoV-2 Omicron spike. Cell Rep.42 (12), 113444. 10.1016/j.celrep.2023.113444
157
Pérez-Massón B. Quintana-Pérez Y. Tundidor Y. Pérez-Martínez D. Castro-Martínez C. Pupo-Meriño M. et al (2024). Studying SARS-CoV-2 interactions using phage-displayed receptor binding domain as a model protein. Sci. Rep.14 (1), 712. 10.1038/s41598-023-50450-4
158
Pinto D. Sauer M. M. Czudnochowski N. Low J. S. Tortorici M. A. Housley M. P. et al (2021). Broad betacoronavirus neutralization by a stem helix–specific human antibody. Science373 (6559), 1109–1116. 10.1126/science.abj3321
159
Puhl A. C. Garzino Demo A. Makarov V. A. Ekins S. (2019). New targets for HIV drug discovery. Drug Discov. Today24 (5), 1139–1147. 10.1016/j.drudis.2019.03.013
160
Punjani A. Rubinstein J. L. Fleet D. J. Brubaker M. A. (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14 (3), 290–296.
161
Qu K. Ke Z. Zila V. Anders-Össwein M. Glass B. Mücksch F. et al (2021). Maturation of the matrix and viral membrane of HIV-1. Science373 (6555), 700–704.
162
Rao V. B. Black L. W. (2010). Structure and assembly of bacteriophage T4 head. Virol. J.7, 356. 10.1186/1743-422X-7-356
163
Roark R. S. Li H. Williams W. B. Chug H. Mason R. D. Gorman J. et al (2021). Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth. Science371 (6525), eabd2638. 10.1126/science.abd2638
164
Roingeard P. (2008). Viral detection by electron microscopy: past, present and future. Biol. Cell100 (8), 491–501. 10.1042/BC20070173
165
Romero-Calle D. Guimarães Benevides R. Góes-Neto A. Billington C. (2019). Bacteriophages as alternatives to antibiotics in clinical care. Antibiot. (Basel)8 (3), 138. 10.3390/antibiotics8030138
166
Saibil H. R. (2017). Blob-ology and biology of cryo-EM: an interview with Helen Saibil. BMC biol.15, 1–7.
167
Sanders R. W. Derking R. Cupo A. Julien J.-P. Yasmeen A. de Val N. et al (2013a). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP. 664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog.9 (9), e1003618. 10.1371/journal.ppat.1003618
168
Sanders R. W. Derking R. Cupo A. Julien J. P. Yasmeen A. de Val N. et al (2013b). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog.9 (9), e1003618. 10.1371/journal.ppat.1003618
169
Sanders R. W. Vesanen M. Schuelke N. Master A. Schiffner L. Kalyanaraman R. et al (2002). Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virology76 (17), 8875–8889. 10.1128/jvi.76.17.8875-8889.2002
170
Saunders K. O. Counts J. Thakur B. Stalls V. Edwards R. Manne K. et al (2024). Vaccine induction of CD4-mimicking HIV-1 broadly neutralizing antibody precursors in macaques. Cell187 (1), 79–94.e24. 10.1016/j.cell.2023.12.002
171
Saunders K. O. Lee E. Parks R. Martinez D. R. Li D. Chen H. et al (2021). Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature594 (7864), 553–559. 10.1038/s41586-021-03594-0
172
Saunders K. O. Wiehe K. Tian M. Acharya P. Bradley T. Alam S. M. et al (2019). Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science366 (6470), eaay7199. 10.1126/science.aay7199
173
Scharf L. Wang H. Gao H. Chen S. McDowall A. W. Bjorkman P. J. (2015). Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell162 (6), 1379–1390. 10.1016/j.cell.2015.08.035
174
Scheres S. H. (2016). Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol.579, 125–157.
175
Schoehn G. Chenavier F. Crépin T. (2023). Advances in structural virology via cryo-EM in 2022. Viruses15 (6), 1315. 10.3390/v15061315
176
Schur F. K. Hagen W. J. Rumlová M. Ruml T. Müller B. Kräusslich H.-G. et al (2015). Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature517 (7535), 505–508. 10.1038/nature13838
177
Seabright G. E. Cottrell C. A. van Gils M. J. D'Addabbo A. Harvey D. J. Behrens A. J. et al (2020). Networks of HIV-1 envelope glycans maintain antibody epitopes in the face of glycan additions and deletions. Structure28 (8), 897–909. 10.1016/j.str.2020.04.022
178
Sempowski G. D. Saunders K. O. Acharya P. Wiehe K. J. Haynes B. F. (2020). Pandemic preparedness: developing vaccines and therapeutic antibodies for COVID-19. Cell181 (7), 1458–1463. 10.1016/j.cell.2020.05.041
179
Shi W. Cai Y. Zhu H. Peng H. Voyer J. Rits-Volloch S. et al (2023). Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane. Nature619 (7969), 403–409. 10.1038/s41586-023-06273-4
180
Sirohi D. Chen Z. Sun L. Klose T. Pierson T. C. Rossmann M. G. et al (2016). The 3.8 Å resolution cryo-EM structure of Zika virus. Science352 (6284), 467–470.
181
Sliepen K. Han B. W. Bontjer I. Mooij P. Garces F. Behrens A.-J. et al (2019). Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun.10 (1), 2355. 10.1038/s41467-019-10262-5
182
Sonani R. R. Palmer L. K. Esteves N. C. Horton A. A. Sebastian A. L. Kelly R. J. et al (2024). An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano. Nat. Commun.15 (1), 756. 10.1038/s41467-024-44959-z
183
Song W. Gui M. Wang X. Xiang Y. (2018). Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog.14 (8), e1007236. 10.1371/journal.ppat.1007236
184
Sougrat R. Sougrat A. Lifson J. D. Bennett A. E. Bess J. W. Zabransky D. J. et al (2007). Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog.3 (5), e63
185
Stalls V. Lindenberger J. Gobeil S. M. Henderson R. Parks R. Barr M. et al (2022). Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike. Cell Rep.39 (13), 111009. 10.1016/j.celrep.2022.111009
186
Stass R. Ng W. M. Kim Y. C. Huiskonen J. T. (2019). Structures of enveloped virions determined by cryogenic electron microscopy and tomography. Adv. virus Res.105, 35–71. 10.1016/bs.aivir.2019.07.009
187
Stewart-Jones G. B. Soto C. Lemmin T. Chuang G. Y. Druz A. Kong R. et al (2016). Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell165 (4), 813–826. 10.1016/j.cell.2016.04.010
188
Su Z. Wu C. Shi L. Luthra P. Pintilie G. D. Johnson B. et al (2018). Electron cryo-microscopy structure of Ebola virus nucleoprotein reveals a mechanism for nucleocapsid-like assembly. Cell172 (5), 966–978. e912.
189
Subramaniam S. Bartesaghi A. Liu J. Bennett A. E. Sougrat R. (2007). Electron tomography of viruses. Curr. Opin. Struct. Biol.17 (5), 596–602. 10.1016/j.sbi.2007.09.009
190
Taati M. Khoshbayan A. Chegini Z. Farahani I. Shariati A. (2020). Bacteriophages, a new therapeutic solution for inhibiting multidrug-resistant bacteria causing wound infection: lesson from animal models and clinical trials. Drug Des. Devel Ther.14, 1867–1883. 10.2147/DDDT.S251171
191
Thompson R. F. Walker M. Siebert C. A. Muench S. P. Ranson N. A. (2016). An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods100, 3–15.
192
Tuijtel M. W. Koster A. J. Jakobs S. Faas F. G. A. Sharp T. H. (2019). Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins. Sci. Rep.9 (1), 1369
193
Toelzer C. Gupta K. Berger I. Schaffitzel C. (2023). Cryo-EM reveals binding of linoleic acid to SARS-CoV-2 spike glycoprotein, suggesting an antiviral treatment strategy. Acta Crystallogr. D. Struct. Biol.79 (Pt 2), 111–121. 10.1107/S2059798323000049
194
Torrents de la Peña A. Rantalainen K. Cottrell C. A. Allen J. D. van Gils M. J. Torres J. L. et al (2019). Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLoS Pathog.15 (7), e1007920. 10.1371/journal.ppat.1007920
195
Tran E. E. Borgnia M. J. Kuybeda O. Schauder D. M. Bartesaghi A. Frank G. A. et al (2012). Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog.8 (7), e1002797. 10.1371/journal.ppat.1002797
196
Turoňová B. Sikora M. Schürmann C. Hagen W. J. H. Welsch S. Blanc F. E. C. et al (2020). In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science370 (6513), 203–208. 10.1126/science.abd5223
197
Veesler D. Quispe J. Grigorieff N. Potter C. S. Carragher B. Johnson J. E. (2012). Maturation in action: CryoEM study of a viral capsid caught during expansion. Structure20 (8), 1384–1390. 10.1016/j.str.2012.05.011
198
Viertel T. M. Ritter K. Horz H.-P. (2014). Viruses versus bacteria—novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J. Antimicrob. Chemother.69 (9), 2326–2336. 10.1093/jac/dku173
199
Walker L. M. Huber M. Doores K. J. Falkowska E. Pejchal R. Julien J. P. et al (2011). Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature477 (7365), 466–470. 10.1038/nature10373
200
Walker L. M. Phogat S. K. Chan-Hui P. Y. Wagner D. Phung P. Goss J. L. et al (2009). Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science326 (5950), 285–289. 10.1126/science.1178746
201
Walls A. C. Park Y. J. Tortorici M. A. Wall A. McGuire A. T. Veesler D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell181 (2), 281–292. 10.1016/j.cell.2020.02.058
202
Wang C. Duan J. Gu Z. Ge X. Zeng J. Wang J. (2024). Architecture of the bacteriophage lambda tail. Structure32 (1), 35–46.e3. 10.1016/j.str.2023.10.006
203
Wang C. Tu J. Liu J. Molineux I. J. (2019). Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat. Microbiol.4 (6), 1049–1056. 10.1038/s41564-019-0403-z
204
Wang C. Zeng J. Wang J. (2022). Structural basis of bacteriophage lambda capsid maturation. Structure30 (4), 637–645.e3. 10.1016/j.str.2021.12.009
205
Wang M. Y. Zhao R. Gao L. J. Gao X. F. Wang D. P. Cao J. M. (2020). SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front. Cell Infect. Microbiol.10, 587269. 10.3389/fcimb.2020.587269
206
Wang S. Matassoli F. Zhang B. Liu T. Shen C. H. Bylund T. et al (2023). HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide. Cell Rep.42 (7), 112755. 10.1016/j.celrep.2023.112755
207
Ward A. B. Wilson I. A. (2017). The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol. Rev.275 (1), 21–32. 10.1111/imr.12507
208
Weissman D. Alameh M. G. de Silva T. Collini P. Hornsby H. Brown R. et al (2021). D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe29 (1), 23–31.e4. 10.1016/j.chom.2020.11.012
209
Wiehe K. Saunders K. O. Stalls V. Cain D. W. Venkatayogi S. Martin Beem J. S. et al (2024). Mutation-guided vaccine design: a process for developing boosting immunogens for HIV broadly neutralizing antibody induction. Cell Host Microbe32, 693–709.e7. 10.1016/j.chom.2024.04.006
210
Williams W. B. Alam S. M. Ofek G. Erdmann N. Montefiori D. C. Seaman M. S. et al (2024). Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans. Cell.187, 2919–2934.e20. 10.1016/j.cell.2024.04.033
211
Williams W. B. Meyerhoff R. R. Edwards R. J. Li H. Manne K. Nicely N. I. et al (2021). Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies. Cell184 (11), 2955–2972.e25. 10.1016/j.cell.2021.04.042
212
Woodward C. L. Cheng S. N. Jensen G. J. (2015). Electron cryotomography studies of maturing HIV-1 particles reveal the assembly pathway of the viral core. J. Virol.89 (3), 1267–1277.
213
Wrapp D. Wang N. Corbett K. S. Goldsmith J. A. Hsieh C.-L. Abiona O. et al (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science367 (6483), 1260–1263. 10.1126/science.abb2507
214
Wright E. R. Schooler J. B. Ding H. J. Kieffer C. Fillmore C. Sundquist W. I. et al (2007). Electron cryotomography of immature HIV‐1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J.26 (8), 2218–2226. 10.1038/sj.emboj.7601664
215
Wrobel A. G. Benton D. J. Roustan C. Borg A. Hussain S. Martin S. R. et al (2022). Evolution of the SARS-CoV-2 spike protein in the human host. Nat. Commun.13 (1), 1178. 10.1038/s41467-022-28768-w
216
Xiao T. Cai Y. Chen B. (2021a). HIV-1 entry and membrane fusion inhibitors. Viruses13 (5), 735. 10.3390/v13050735
217
Xiao T. Frey G. Fu Q. Lavine C. L. Scott D. A. Seaman M. S. et al (2020). HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes. Nat. Chem. Biol.16 (5), 529–537. 10.1038/s41589-020-0496-y
218
Xiao T. Lu J. Zhang J. Johnson R. I. McKay L. G. A. Storm N. et al (2021b). A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent. Nat. Struct. Mol. Biol.28 (2), 202–209. 10.1038/s41594-020-00549-3
219
Yan R. Zhang Y. Li Y. Xia L. Guo Y. Zhou Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science367 (6485), 1444–1448. 10.1126/science.abb2762
220
Yang F. Wang L. Zhou J. Xiao H. Liu H. (2023). In situ structures of the ultra-long extended and contracted tail of myoviridae phage P1. Viruses15 (6), 1267. 10.3390/v15061267
221
Yang K. Wang C. White K. I. Pfuetzner R. A. Esquivies L. Brunger A. T. (2022a). Structural conservation among variants of the SARS-CoV-2 spike postfusion bundle. Proc. Natl. Acad. Sci.119 (16), e2119467119. 10.1073/pnas.2119467119
222
Yang S. Hiotis G. Wang Y. Chen J. Wang J.-H. Kim M. et al (2022b). Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack. Nat. Commun.13, 6393. 10.1038/s41467-022-34008-y
223
Yang Z. Dam K.-M. A. Bridges M. D. Hoffmann M. A. DeLaitsch A. T. Gristick H. B. et al (2022c). Neutralizing antibodies induced in immunized macaques recognize the CD4-binding site on an occluded-open HIV-1 envelope trimer. Nat. Commun.13 (1), 732. 10.1038/s41467-022-28424-3
224
Yao H. Song Y. Chen Y. Wu N. Xu J. Sun C. et al (2020). Molecular architecture of the SARS-CoV-2 virus. Cell183 (3), 730–738. 10.1016/j.cell.2020.09.018
225
Yap M. L. Klose T. Arisaka F. Speir J. A. Veesler D. Fokine A. et al (2016). Role of bacteriophage T4 baseplate in regulating assembly and infection. Proc. Natl. Acad. Sci.113 (10), 2654–2659. 10.1073/pnas.1601654113
226
Yu F. Lu L. Liu Q. Yu X. Wang L. He E. et al (2014). ADS-J1 inhibits HIV-1 infection and membrane fusion by targeting the highly conserved pocket in the gp41 NHR-trimer. Biochim. Biophys. Acta1838 (5), 1296–1305. 10.1016/j.bbamem.2013.12.022
227
Yuan Y. Cao D. Zhang Y. Ma J. Qi J. Wang Q. et al (2017). Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun.8 (1), 15092
228
Yurkovetskiy L. Wang X. Pascal K. E. Tomkins-Tinch C. Nyalile T. P. Wang Y. et al (2020). Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell183 (3), 739–751. 10.1016/j.cell.2020.09.032
229
Zanetti G. Briggs J. A. Grünewald K. Sattentau Q. J. Fuller S. D. (2006). Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog.2 (8), e83.
230
Zhang J. Cai Y. Xiao T. Lu J. Peng H. Sterling S. M. et al (2021a). Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science372 (6541), 525–530. 10.1126/science.abf2303
231
Zhang J. Xiao T. Cai Y. Lavine C. L. Peng H. Zhu H. et al (2021b). Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science374 (6573), 1353–1360. 10.1126/science.abl9463
232
Zhang Q. E. Lindenberger J. Parsons R. J. Thakur B. Parks R. Park C. S. et al (2024). SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition. bioRxiv. 10.1101/2024.02.12.580004
233
Zhao G. Perilla J. R. Yufenyuy E. L. Meng X. Chen B. Ning J. et al (2013). Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature497 (7451), 643–646. 10.1038/nature12162
234
Zhou T. Tsybovsky Y. Gorman J. Rapp M. Cerutti G. Chuang G. Y. et al (2020). Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe28 (6), 867–879. 10.1016/j.chom.2020.11.004
235
Zhou Z. H. (2014). Structures of viral membrane proteins by high-resolution cryoEM. Curr. Opin. Virology5, 111–119. 10.1016/j.coviro.2014.04.001
236
Zhu J. Batra H. Ananthaswamy N. Mahalingam M. Tao P. Wu X. et al (2023). Design of bacteriophage T4-based artificial viral vectors for human genome remodeling. Nat. Commun.14 (1), 2928. 10.1038/s41467-023-38364-1
237
Zhu N. Zhang D. Wang W. Li X. Yang B. Song J. et al (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med.382 (8), 727–733. 10.1056/NEJMoa2001017
238
Zhu P. Liu J. Bess J. Chertova, Jr. E. Lifson J. D. Grisé H. et al (2006). Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature441 (7095), 847–852.
239
Zhu Z. Zhang L. Li S. Gao Y. Wang Y. Ma X. et al (2024). S–6P exhibits better immunogenicity than S–2P at lower doses of COVID-19 mRNA vaccines. Decoding Infect. Transm.2, 100017. 10.1016/j.dcit.2024.100017
240
Zila V. Margiotta E. Turoňová B. Müller T. G. Zimmerli C. E. Mattei S. et al (2021). Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell184 (2), 1032–1046.e1018.
Summary
Keywords
cryo-EM, cryo-ET, HIV-1, SARS-CoV-2, bacteriophage, 3D reconstructions, structure-based design, vaccine development
Citation
Dutta M and Acharya P (2024) Cryo-electron microscopy in the study of virus entry and infection. Front. Mol. Biosci. 11:1429180. doi: 10.3389/fmolb.2024.1429180
Received
07 May 2024
Accepted
12 June 2024
Published
24 July 2024
Volume
11 - 2024
Edited by
Edward T. Eng, New York Structural Biology Center, United States
Reviewed by
Kiefer Ramberg, Trinity College Dublin, Ireland
Ruby Sharma, Albert Einstein College of Medicine, United States
Updates
Copyright
© 2024 Dutta and Acharya.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moumita Dutta, moumita.dutta@duke.edu; Priyamvada Acharya, priyamvada.acharya@duke.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.