- 1Department of Medical Parasitology, Post Graduate Institute of Medical Education and Research, Chandigarh, India
- 2Department of Pharmacology and Therapeutics, All India Institute of Medical Sciences, Gorakhpur, India
- 3Department of Microbiology, Aarupadai Veedu Medical College and Hospital, Puducherry, India
Leishmaniasis remains a significant global health challenge, with over a billion people at risk of infection and limited effective treatment options due to escalating drug resistance. This review explores the underlying mechanisms of drug resistance in Leishmania species, focusing on genomic plasticity as a driving factor for survival and adaptation. Key mechanisms, including genetic mutations, gene amplification, chromosomal rearrangements, and efflux transporters, contribute to the parasite’s ability to evade existing therapies. Advances in genomic and proteomic studies have provided deeper insights into these resistance pathways, enabling the development of novel therapeutic strategies. Additionally, this review highlights current therapeutic approaches, including combination therapies and potential new drug candidates, that address multidrug resistance and explore the vulnerabilities of Leishmania. Understanding these mechanisms and their clinical implications is essential for developing targeted interventions that improve treatment outcomes and combat resistance in leishmaniasis.
1 Introduction
Leishmaniasis is a parasitic disease caused by protozoa of the genus Leishmania and transmitted through the bites of infected female phlebotomine sandflies. More than 90 sandfly species are known to transmit Leishmania parasites. The disease is widespread, affecting approximately89 countries (Torres-Guerrero et al., 2017). It is endemic in Asia, Africa, the Americas, and the Mediterranean region, placing over 1 billion people at risk of infection (World Health Organization, 2025). Leishmaniasis presents in four clinical forms: cutaneous, mucocutaneous, visceral (kala-azar), and post-kala-azar dermal leishmaniasis (Torres-Guerrero et al., 2017). However, many infections remain asymptomatic (Singh et al., 2020). Leishmania can also act as an opportunistic pathogen in immunosuppressed individuals.
Chemotherapy remains the cornerstone of leishmaniasis management and control. Treatment choice depends on multiple factors, including disease type, coexisting conditions, parasite species, and geographic location. Since current drugs cannot fully eliminate the parasite, immunocompetence is crucial to prevent relapse. Pentavalent antimonials, amphotericin B, paromomycin, miltefosine, pentamidine, and sitamaquine are the medications used in current treatments regimen (Kapil et al., 2018). Each drugs have its own mechanism of action, such as antimonials are believed to interfere with the parasite’s energy production, inhibiting glycolysis and fatty acid oxidation. They may also disrupt parasite thiol metabolism, leading to oxidative stress and cell death (Haldar et al., 2011), Amphotericin B binds to ergosterol, forming pores in the membrane and causing parasite death (Stone et al., 2016). Miltefosine disrupts cell membrane integrity and inhibits phospholipid metabolism. It also interferes with mitochondrial function and triggers apoptosis-like cell death in the parasite (Palić et al., 2019). In many areas, antimonials continue to be the major medication used to treat various types of leishmaniasis. However, antimony resistance has made the use of substitute drugs necessary, particularly in the Indian subcontinent. Currently, parenteral paromomycin, amphotericin B (AmB), and the oral miltefosine (MIL) are widely used. The frequency of treatment failure may be significant in patients treated with MIL, which has supplanted antimonials in the kala-azar extermination campaign in nations such as India, even though it has been noted in patients treated with the majority of anti-leishmanials. AmB is highly efficacious but also has associated toxicity such as includes fever, nausea, vomiting, rigors, hypertension or hypotension, and hypoxia when administered in its free deoxycholate form which has been overcome in its liposomal formulation (Laniado-Laborín and Cabrales-Vargas, 2009; Zhang et al., 2025). Unfortunately, resistance to AmB has also been shown to be a concern in laboratory experiments.
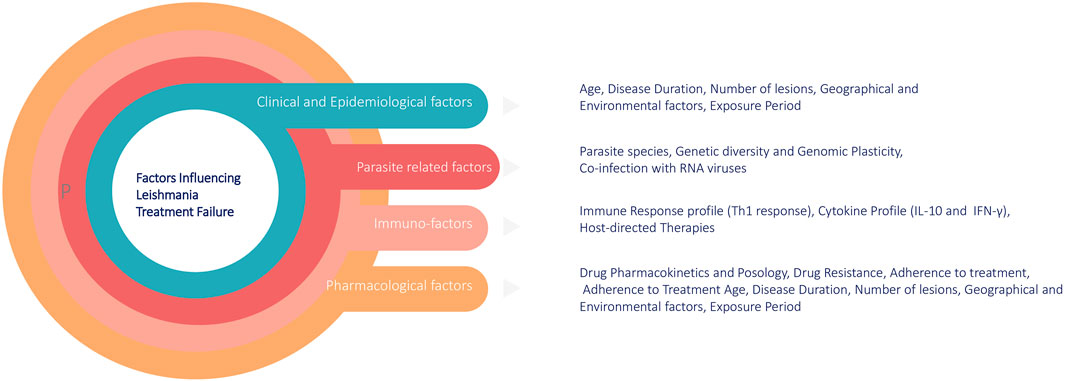
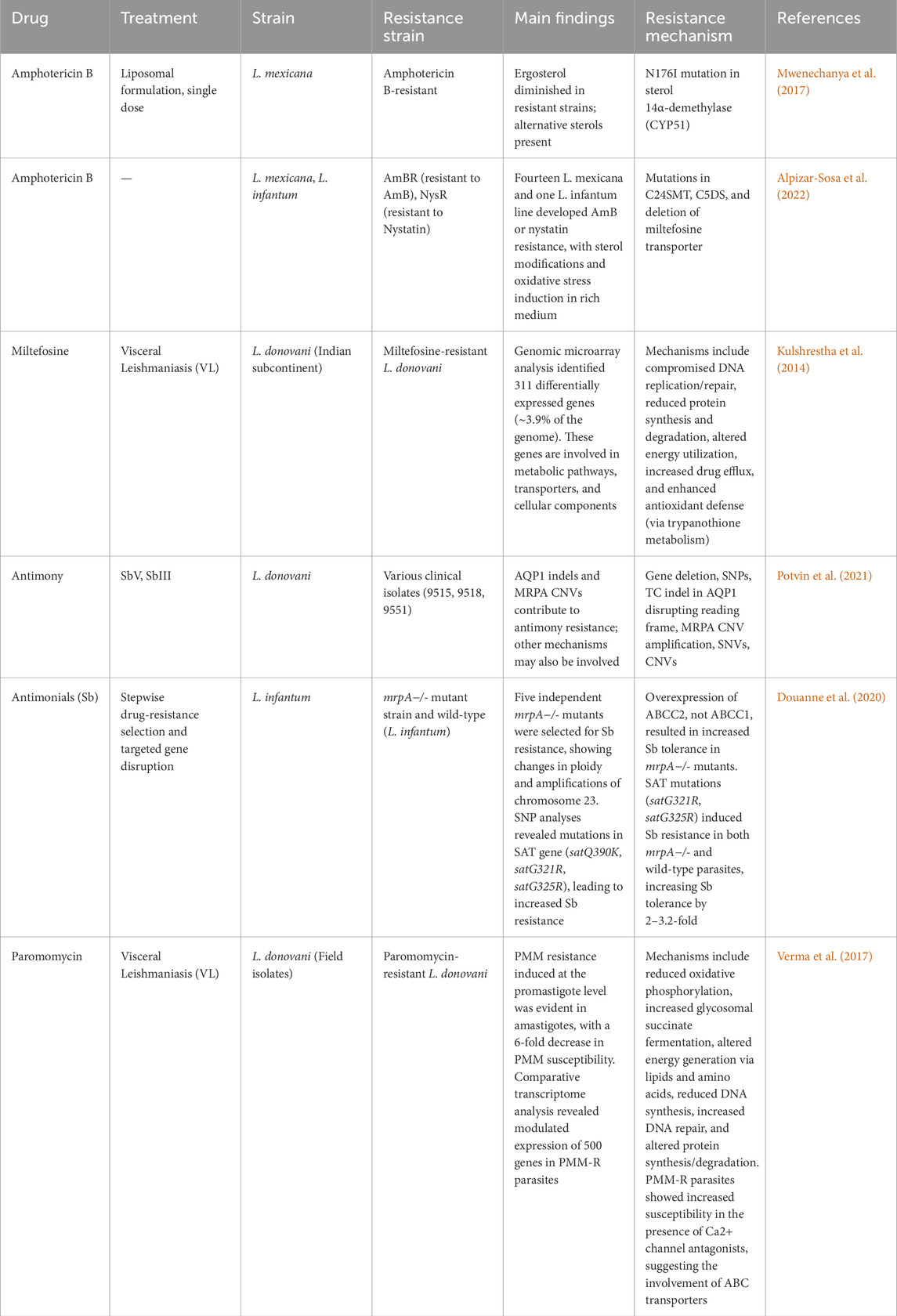
The rise of drug resistance impacts treatment outcomes and is influenced by multiple factors. These include host-related factors (e.g., immune response and cytokine profile), immuno-factors, drug pharmacokinetics (e.g., metabolism and adherence to treatment), and parasite-specific factors, such as genetic plasticity and co-infections (Figure 1). A comprehensive understanding of these factors is crucial for developing targeted interventions to overcome treatment failure. Moreover, the numbers of leishmaniasis cases are increasing worldwide. Some reasons are the lack of vaccines, difficulties in controlling vectors and the increasing number of parasites resistance to chemotherapy. The rise of drug resistance impacts treatment outcome, and understanding its causes, spread, and impact will help us manage the risks it imposes (Moncada-Diaz et al., 2024). Out of various reasons of appearance of drug resistance, genome plasticity is another key factor in the survival of Leishmania parasites and their development of drug resistance in which genetic variations such as mutations, gene amplifications, and chromosomal rearrangements play crucial roles in the appearance of resistance (Kamran et al., 2023). In the following sections we discuss about the Leishmania treatment regimens, mechanism behinds underlying drug resistance to currently available antileishmanial drugs, role of genetic adaptability in drug resistance and currents drugs that are under drug discovery pipelines and potential drug targets.
2 Leishmaniasis treatment regimens
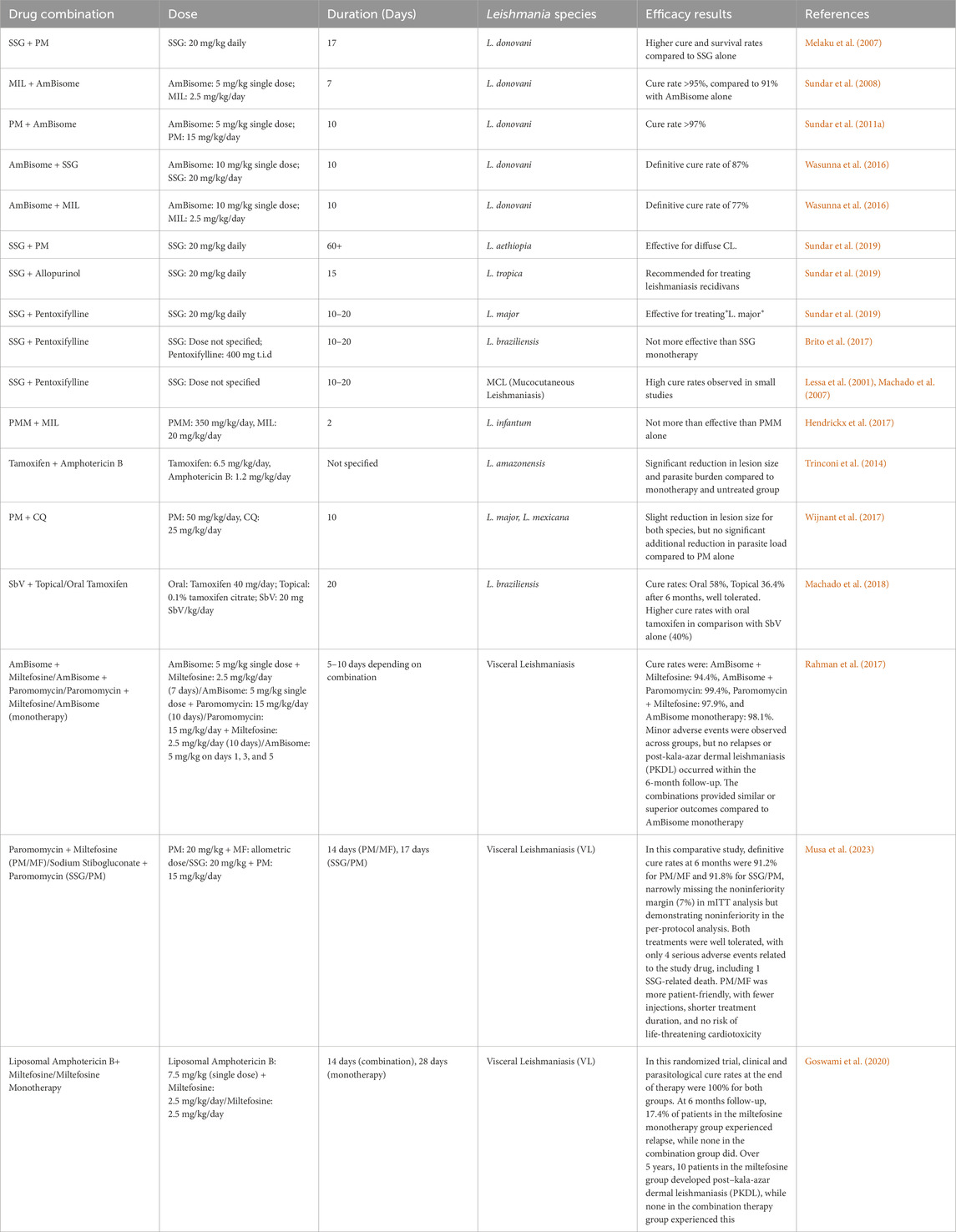
Pentavalent antimonials (SbV) have been the cornerstone in the treatment of leishmaniasis for many decades. The two primary forms used are sodium stibogluconate (SSG) and meglumine antimoniate (MA), each administered as parenteral drugs (IM, IV, or IL) at a standard dose of 20 mg/kg/day for 28–30 days (Haldar et al., 2011). In the 1980s, the World Health Organization (WHO) recommended the use of SbV at an increased dose of 20 mg/kg/day up to a maximum of 850 mg for 20–30 days. Despite their long-standing use and initial success, their effectiveness has gradually declined due to widespread resistance and adverse effects. The main drawback of pentavalent antimonials is their severe toxicity, including cardiotoxicity (e.g., ventricular tachycardia, prolonged QTc interval), pancreatitis, pancytopenia, and nephrotoxicity. Due to these risks, SbV is not recommended for HIV-VL co-infected patients as they experience higher toxicity and reduced efficacy (Haldar et al., 2011). Due to the increasing resistance and toxicity of SbV, alternative treatments such as paromomycin (PM) have gained prominence in managing leishmaniasis. Paromomycin (PM), an aminoglycoside antibiotic, is an affordable and effective option for treating leishmaniasis, administered intramuscularly at 15 mg/kg/day for 21 days. Although PM shows high cure rates (94.6% in a Phase III study in India), its prolonged treatment duration poses a challenge in endemic areas. PM is also potentially nephrotoxic and ototoxic. While it is used alone for visceral leishmaniasis (VL) in some regions, in Africa, it is typically combined with SSG. Topical formulations of PM are used for cutaneous leishmaniasis (CL), though efficacy varies by geographical region, with better outcomes for systemic use in Brazil (Pokharel et al., 2021). While PM remains a viable option, particularly in combination therapies, another potent alternative is Amphotericin B, which is particularly effective in cases resistant to pentavalent antimonials.
Amphotericin B, a polyene antifungal, is highly effective for treating leishmaniasis, particularly in regions with pentavalent antimonial resistance. Administered intravenously at 0.75–1 mg/kg/day for 15–20 days, it has cure rates approaching 100%. However, its use is limited by severe nephrotoxicity and the need for hospitalization during treatment. Liposomal formulations like L-AmB (AmBisome) offer targeted delivery with fewer side effects, making it the preferred treatment for visceral leishmaniasis, especially in HIV-VL co-infections. Despite its high cost and need for a cold chain, L-AmB’s safety profile and variable dosing regimens make it a versatile option across different geographical regions (Frézard et al., 2022). Given the challenges associated with injectable therapies like Amphotericin B, the development of oral treatments such as Miltefosine (MIL) has significantly impacted leishmaniasis management. Miltefosine (MIL) is the first effective oral drug for visceral leishmaniasis, with a 94% cure rate. Initially introduced in 2002 and widely used in India’s kala-azar elimination program, it is now registered in several countries. Administered at 2–2.5 mg/kg for 28 days, MIL works by increasing nitric oxide production in macrophages, disrupting parasite membranes, and damaging mitochondria. However, its long half-life and poor compliance contribute to resistance development. MIL is teratogenic and unsuitable for pregnant women, with common side effects including gastrointestinal issues, renal toxicity, and dehydration (Scarpini et al., 2022) (Table 1).
3 Geographic variations in antileishmanial drug resistance
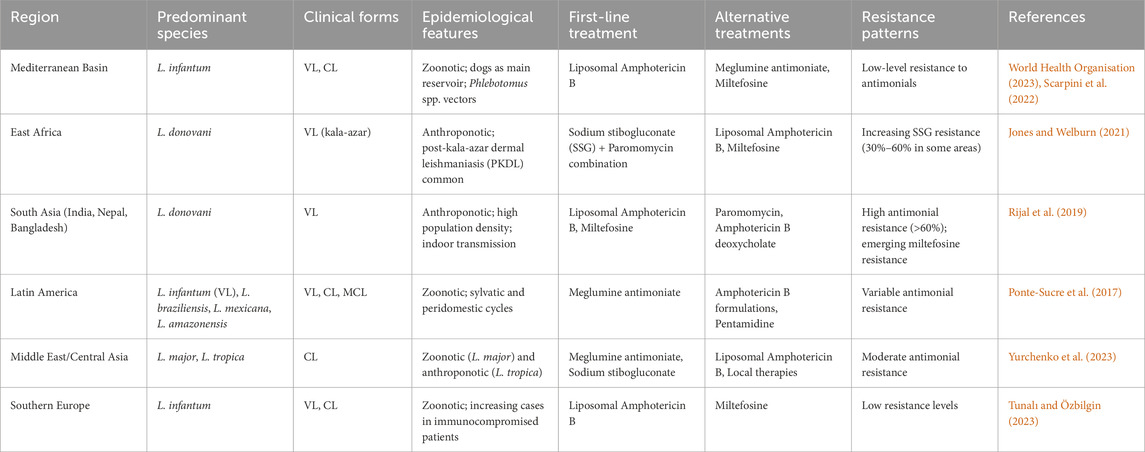
Treatment options for leishmaniasis vary significantly by region due to differences in dominant Leishmania species and emerging drug resistance patterns (Ponte-Sucre et al., 2017). In South Asia, Leishmania donovani shows high resistance to sodium stibogluconate (SSG) and declining susceptibility to miltefosine, necessitating alternatives like liposomal amphotericin B (L-AmB) and combination therapies. In East Africa, SSG remains a primary treatment, though resistance has been reported, prompting the use of SSG-paromomycin combinations. In Latin America, where Leishmania braziliensis and related species prevail, antimonials like meglumine antimoniate are still used, but resistance varies by region, with pentamidine and miltefosine serving as alternatives. The Mediterranean and Middle East face challenges with Leishmania infantum and Leishmania tropica, where antimonial resistance is emerging, leading to increased reliance on L-AmB. Central Asia reports growing SSG resistance in L. tropica, pushing the adoption of thermotherapy and L-AmB (Pigott et al., 2014; Herrera et al., 2020) (Table 2).
4 Molecular mechanisms of drug resistance in Leishmania
4.1 Mechanism underlying antimony resistance in Leishmania
One of the primary mechanisms of antimony resistance in Leishmania is the reduced uptake of the drug by the parasite. Aquaglyceroporin 1 (AQP1) is known to facilitate the uptake of SbIII by the parasite. In drug-resistant parasites, downregulation of AQP1 has been observed, leading to decreased drug uptake and subsequent resistance (de Santana et al., 2025; Hefnawy et al., 2017; Santos et al., 2023). Another significant mechanism involves increased intracellular thiol levels. In drug-sensitive strains, SbIII disrupts thiol homeostasis by inducing the efflux of thiols such as trypanothione (TSH), glutathione (GSH), and cysteine, which maintain thiol redox homeostasis in Leishmania, protecting the parasite from chemical and oxidative stress. The γ-GCS gene encodes an enzyme catalyzing the rate-limiting step of GSH biosynthesis, while the ODC gene encodes an enzyme regulating polyamine biosynthesis. Polyamines are precursor metabolites of trypanothione. Antimony-resistant strains have shown inconsistent upregulation of γ-GCS and overexpression of ODC genes, increasing the intracellular thiol-dependent antioxidant capacity and resulting in resistance to antimony. Additionally, the trypanothione reductase gene is amplified in antimony-resistant isolates, leading to high intracellular trypanothione levels and increased resistance to SbIII (Haldar et al., 2011; Fekrisoofiabadi et al., 2019).
Sequestration and rapid drug efflux also contribute to antimony resistance. ATP-binding cassette (ABC) transporters efflux the drug out of the parasite or sequester it in intracellular vesicles. The two classes of ABC transporters involved in this process are P-glycoprotein (e.g., MRPA) and multi-drug resistance-related protein (e.g., MRP1). Genes encoding these transporters are amplified in antimony-resistant parasites, leading to effective drug efflux and sequestration (Berg et al., 2015). Changes in membrane fluidity have been demonstrated in resistance to antimony combinations, further contributing to the resistance mechanism (Salari et al., 2022). Modulation of cell death through heat shock proteins (e.g., HSP83 and HSP70) has been reported in resistant parasites. Additionally, cell death-related proteins such as protein tyrosine phosphatase (PTP), proliferating cell nuclear antigen (PCNA), and mitogen-activated protein kinase (MAPK) show differential expression, with PTP and PCNA being upregulated and MAPK being downregulated in antimony-resistant strains (Salari et al., 2022; Tandon et al., 2014).
Beyond intrinsic parasite resistance mechanisms, Leishmania also modulates signaling pathways in host macrophages. Drug-resistant parasites have been shown to alter the host-pathogen interaction and the host immune response, contributing to the development of resistance (Mukhopadhyay et al., 2011). Certain proteins are differentially expressed in response to antimony. For example, proteins such as histone 1, H2A, H4, and leucine-rich repeat protein are overexpressed in antimony-resistant parasites, while proteins like the kinetoplastid membrane protein (KMP-11) are under-expressed (Das et al., 2015). Apart from genetic and biochemical adaptations, external factors such as the misuse of antimony drugs has significantly contributed to the development of resistance. Practices such as inadequate dosing, inappropriate treatment regimens, the free availability of drugs, management of patients by unqualified persons, and incomplete treatment courses have led to the development of subtherapeutic levels of antimony in the blood, promoting parasite tolerance and resistance to the drug (Kazemi-Rad et al., 2013).
Further reinforcing these resistance mechanisms, a study on L. donovani compared Sb(V)-sensitive and -resistant strains from kala-azar patients. The resistant strain exhibited cross-resistance to miltefosine and other drugs. Proteomic analysis identified altered programmed cell death (PCD) pathways as central to resistance. Notably, the heat shock protein HSP83 was found to enhance drug resistance by disrupting mitochondrial membrane potential and diminishing drug-induced PCD. Conversely, the protein SKCRP14.1 promoted PCD in response to antimonials but conferred protection against miltefosine-induced PCD (Ennes-Vidal et al., 2017).
4.2 Complex mechanisms of miltefosine resistance in Leishmania
Recent studies have identified multiple mechanisms underlying miltefosine (MF) resistance in Leishmania species, posing significant challenges to effective treatment. This resistance involves genetic, biochemical, and immunological factors that reduce the drug’s efficacy. A study conducted by Caroline R. Espada et al. (2019) on Leishmania (V.) braziliensis clinical isolates found that decreased MF susceptibility was linked to reduced drug accumulation. This reduction was attributed to diminished Ros3 mRNA expression rather than polymorphisms in MT-Ros3 complex genes, which are crucial for drug uptake. This finding underscores the need for new molecules or modifications to MF to overcome MT-Ros3 dependence and enhance treatment efficacy (Espada et al., 2019). Beyond reduced drug accumulation, MF resistance in Leishmania exhibits remarkable stability, as indicated by a consistent EC50 value, even after prolonged culture without drug exposure. This suggests a specific and persistent mechanism likely involving changes in transporter expression or translocation machinery. Notably, this resistance is largely exclusive to MF, with some exceptions showing reduced susceptibility to SbIII, hinting at a complex interplay between different resistance pathways (Vacchina et al., 2016).
The overexpression of multidrug resistance proteins, such as MRPA, is another critical factor in MF resistance. MRPA, an ATP-binding cassette (ABC) transporter, actively pumps MF out of the parasite’s cells, thereby reducing its intracellular concentration and effectiveness (Khanra et al., 2017). Additionally, other ABC transporters like ABCB4, ABCG4, and ABCG6 contribute to this resistance by enhancing drug efflux (Pérez-Victoria et al., 2011; Da Costa et al., 2018). MF resistance is also influenced by immune evasion strategies, as seen in L. donovani. This impairment weakens the host’s ability to mount an effective immune response, characterized by a lack of Th1-type immune responses and reduced production of essential cytokines and antibodies. Consequently, the host struggles to control and eliminate the parasite, even with MF treatment, complicating treatment outcomes and resistance management (Khanra et al., 2017).
In Leishmania infantum, resistance has been linked to the deletion of the MSL locus, affecting key enzymes (NUC1 and NUC2) involved in MF susceptibility. This deletion also leads to increased baseline lipid content, including ergosterol, which may act as a reservoir for MF, contributing to resistance. Moreover, isolates from relapsed patients demonstrated better control of lipid perturbations and nitric oxide accumulation in macrophages, suggesting a role in modulating host immune responses (Carnielli et al., 2022). Expanding on these species-specific variations, whole genome sequencing of L. donovani identified a significant mutation in the LdMT gene, leading to transporter inactivation. This mutation, along with changes in membrane fluidity and gene expression—such as the upregulation of genes related to surface proteins and phosphoglycan biosynthesis, and the downregulation of stress response and folate transport genes—highlights specific genetic and metabolic adaptations rather than a general resistance profile. Despite reduced metacyclogenesis, these resistant parasites maintain their ability to invade and replicate in host cells (Vacchina et al., 2016). Together, these findings underscore the multifaceted nature of MF resistance in Leishmania, integrating genetic mutations, metabolic alterations, and immune evasion strategies, significantly impacting treatment efficacy. Understanding these mechanisms is crucial for addressing the growing challenge of drug resistance in leishmaniasis.
4.3 Sterol mutations and amphotericin B resistance in Leishmania
Resistance to amphotericin B (AmB) in Leishmania primarily involves mutations in the sterol biosynthesis pathway, resulting in altered membrane sterol composition. Key mutations in genes such as C24SMT, C5DS, and C14DM lead to the loss of ergosterol and the accumulation of other sterol precursors, which reduce the binding affinity of AmB to the parasite membrane and decrease drug sensitivity (Mukherjee et al., 2020). This resistance mechanism has been observed in both laboratory-generated resistant strains and clinical isolates, underscoring its relevance in both experimental and field settings (Pountain et al., 2019).The altered sterol profile, often featuring ergosta-7,22-dienol or cholesta-5,7,22-trienol, compromises the drug’s efficacy, indicating that mutations in sterol metabolism are central to AmB resistance in Leishmania (Yao and Wilson, 2016; Alpizar-Sosa et al., 2022). Further studies have shown that resistance is associated with an increased conversion of β-sitosterol into stigmasterol, significantly raising the IC50 by four times compared to wild-type strains. This sterol alteration, observed in both promastigotes and axenic amastigotes, highlights stigmasterol’s role in AmB resistance, despite the reduced infectivity of the resistant strain in vitro (Bansal et al., 2020).
Additionally, in L. martiniquensis, AmB resistance is linked to increased metacyclogenesis, growth, and infectivity, with resistant strains persisting longer in mice without causing clinical disease. These asymptomatic hosts could act as reservoirs, enhancing transmission, which underscores the need for vigilant monitoring of AmB-resistant Leishmania martiniquensis, particularly in relapsing and HIV-coinfected patients (Mano et al., 2023). The sterol biosynthesis pathway depicted in Figure 2 highlights how alterations at key enzymatic steps can lead to the production of alternative sterols that diminish AmB efficacy. Moreover, other studies have found that Leishmania can develop resistance to AmB through the loss of ergosterol and its replacement with cholestane-type sterols. This process, involving mutations in the enzyme sterol 14α-demethylase, disrupts sterol synthesis and affects AmB binding. Additionally, these resistant strains exhibit increased sensitivity to oxidative stress (Mwenechanya et al., 2017). Other studies have identified mutations in sterol biosynthesis enzymes, such as SMT, as significant contributors to AmB resistance. These mutations can alter sterol composition and membrane properties, affecting AmB binding and efficacy. Additionally, resistance mechanisms have been linked to changes in the miltefosine transporter and increased membrane fluidity (Pountain et al., 2019).
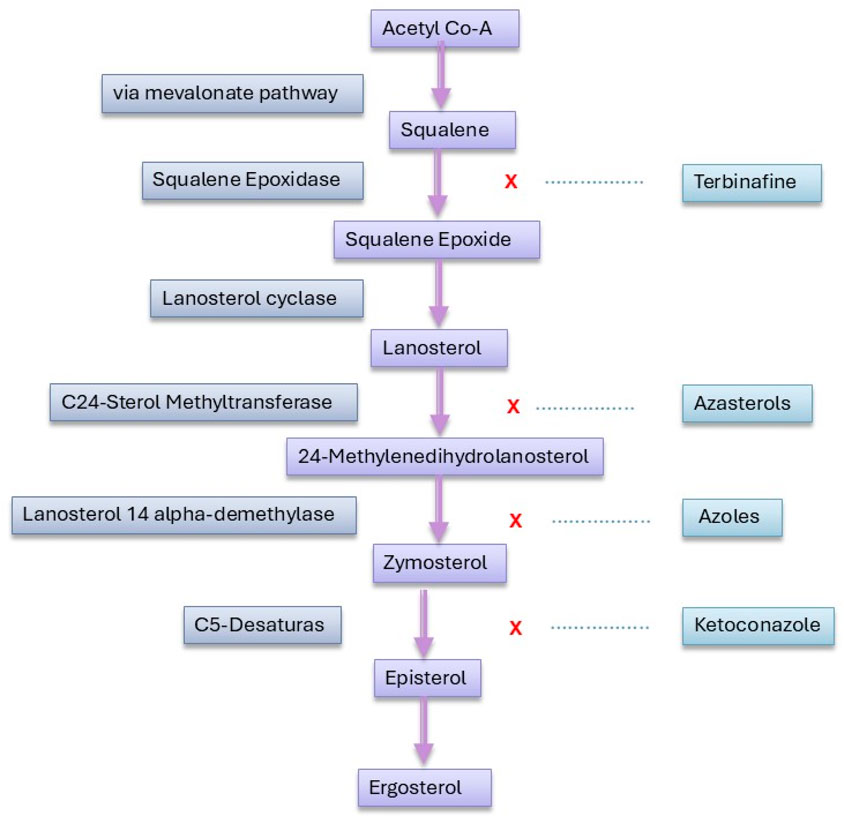
Figure 2. Enzymatic drug targets and inhibition sites in the sterol biosynthesis pathway of Leishmania.
Further research suggests that resistance mechanisms to AmB in Leishmania involve complex interactions between genetic mutations and metabolic changes. For instance, AmB-resistant Leishmania lines show decreased levels of oligohexoses, which may influence virulence, and increased levels of protective thiols such as trypanothione and glutathione. These findings indicate that metabolic adaptations, beyond primary genetic mutations, play a crucial role in shaping the resistance profile of Leishmania parasites (Pountain and Barrett, 2019). Additionally, it has been found that the enzyme L-asparaginase (LdAI) is crucial for L. donovani’s resistance to AmB, with its overexpression enhancing survival under treatment (Singh et al., 2017). Elevated levels of the protein Sir2 in resistant parasites lead to increased MDR1 expression, enhanced drug efflux, reduced ROS levels, and decreased apoptosis, contributing to higher resistance to AmB. Conversely, inhibiting or deleting Sir2 increases drug susceptibility, making Sir2 a potential resistance marker for visceral leishmaniasis (Purkait et al., 2015). Lastly, some studies suggest that Leishmania resists AmB by protecting against membrane ion leakage and oxidative damage, despite normal ergosterol levels. This resistance mechanism involves altered cell signalling due to AmB’s membrane-thinning effects (Cohen, 2016).
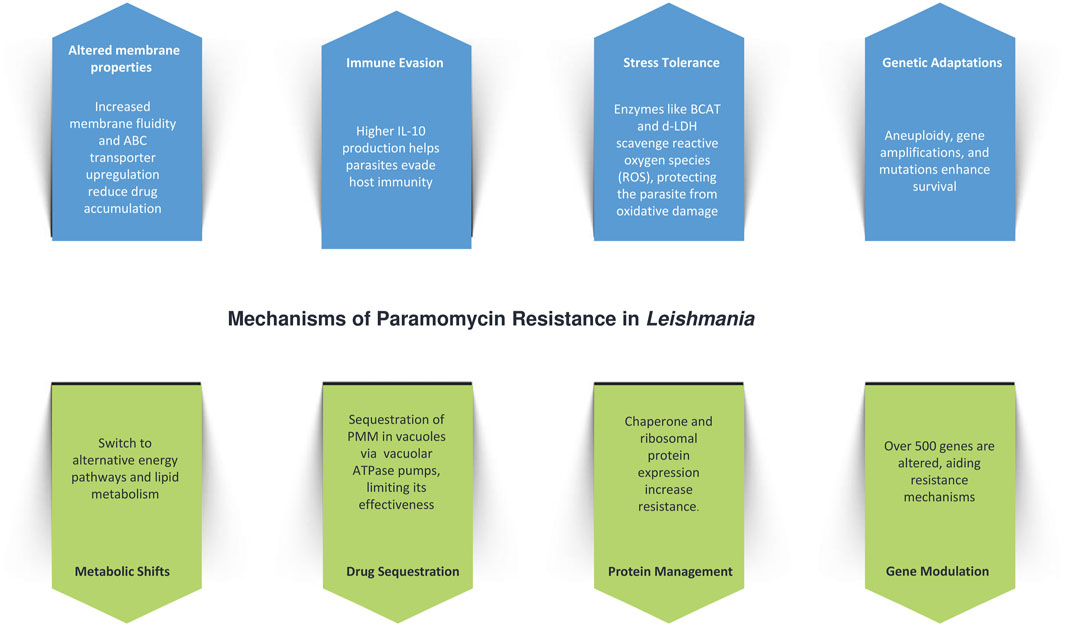
4.4 Physiological and genetic adaptations to paromomycin resistance in Leishmania
Paromomycin (PMM) resistance in Leishmania is driven by a combination of physiological, genetic, and metabolic adaptations (Figure 3). One of the key mechanisms is increased membrane fluidity, which impairs drug penetration and decreases intracellular accumulation, thereby reducing the drug’s effectiveness. This is accompanied by the upregulation of ATP-binding cassette (ABC) transporters, such as MDR1 and MRPA, which enhance drug efflux and further contribute to resistance. The resistant parasites also exhibit improved tolerance to host defense mechanisms, such as nitrosative stress, and evade immune responses through increased interleukin-10 (IL-10) production, which facilitates immune evasion (Shaw et al., 2019).
In addition to drug efflux and membrane adaptations, L. donovani reinforces its resistance by withstanding nitric oxide (NO) stress, particularly in the amastigote stage, a critical phase for survival within host macrophages (Hendrickx et al., 2014). This is supported by metabolic shifts, including the upregulation of enzymes like branched-chain aminotransferase (BCAT) and d-lactate dehydrogenase (d-LDH), which help scavenge reactive oxygen species (ROS) and protect the parasite from oxidative damage (Rastrojo et al., 2018). Genetic adaptations, such as aneuploidy and ribosomal RNA gene amplifications, enhance these defense mechanisms. Whole-genome sequencing of resistant clones has revealed no single mutation consistently linked to resistance, but multiple genetic variations—such as single nucleotide variants (SNVs) and copy number variations (CNVs)—have been observed. These variations are concentrated in genes related to protein synthesis, mitochondrial function, and virulence factors like HSP78 and sterol 24-C-methyltransferase, along with CNVs affecting mitochondrial transport, vesicular trafficking, and protein turnover (Shaw et al., 2019; Hendrickx et al., 2021).
Metabolic adaptations are also central to PMM resistance, with resistant parasites exhibiting a shift away from oxidative phosphorylation towards glycosomal succinate fermentation, and increased reliance on lipid and amino acid metabolism for energy. Reduced DNA synthesis paired with enhanced DNA repair, alongside decreased protein synthesis and degradation, further support survival under drug pressure. Transcriptomic analysis has shown modulation of over 500 genes, while calcium channel antagonists, such as verapamil and amlodipine, increase PMM susceptibility, implicating ABC transporters in the resistance pathway. Interestingly, while PMM-resistant parasites modulate NO levels in infected macrophages, ROS levels remain unaffected (Verma et al., 2017).
Another significant aspect of PMM resistance involves drug sequestration. PMM is internalized via endocytosis and sequestered in vacuoles, where vacuolar ATPase pumps are upregulated to isolate the drug, reducing its efficacy. The stress induced by PMM leads to the increased expression of chaperone proteins, which aid in protein folding and turnover, helping the parasite cope with the drug’s effects. Additionally, the upregulation of ribosomal proteins enhances protein synthesis, and glycolytic enzyme overexpression boosts energy production, collectively contributing to the parasite’s survival and resistance (Chawla et al., 2011).
A comparative overview of resistance mechanisms across major antileishmanial drugs is provided in Table 3, highlighting recurrent themes such as transporter upregulation, metabolic adaptations, and genetic mutations.
5 Genomic plasticity and drug resistance in Leishmania
5.1 Atypical genome of Leishmania
The Leishmania genome exhibits unique characteristics compared to other eukaryotes, with variations in chromosome numbers and gene sets (Iv. Gerasimov et al., 2023). Recent genomic assemblies, such as that of Leishmania major, revealed a 32.8 Mb genome containing 11,238 genes distributed across 36 chromosomes (Camacho et al., 2021). Initially thought to be strictly diploid, Leishmania populations display mosaic aneuploidy, where chromosomal copy numbers vary between strains and species (Bussotti et al., 2018; Zackay et al., 2018).
Unlike typical eukaryotes, Leishmania genes lack introns and are organized into unidirectional polycistronic transcription units without functional clustering (Bartholomeu et al., 2021). Transcription is constitutive, mediated by RNA polymerase II, but lacks canonical promoters (Saha, 2020). Epigenetic mechanisms, including histone modifications and DNA accessibility, regulate transcription initiation (Chandra et al., 2017), while termination is determined by base J (Reynolds et al., 2016). Since transcriptional regulation is minimal, gene expression is primarily controlled post-transcriptionally via mRNA stability, translation efficiency, and protein degradation (Grünebast and Clos, 2020).
5.2 Genetic diversity and genomic plasticity
Large-scale genomic studies have revealed extensive genetic diversity in Leishmania, influencing its geographical distribution and clinical manifestations (Llanes et al., 2022; Franssen et al., 2020). Single-cell sequencing has identified multiple karyotypes within a single clone (Imamura et al., 2020; Negreira et al., 2022), and mixed-genotype infections are common even within the same host tissue (Cupolillo et al., 2020).
5.2.1 Mechanisms of genomic plasticity
Large-scale genomic studies have highlighted the extensive genetic diversity in Leishmania, which influences its geographical distribution and clinical manifestations (Ruang-Areerate et al., 2023; Hadermann et al., 2023). Single-cell sequencing has revealed multiple karyotypes within a single clone (Imamura et al., 2020; Negreira et al., 2023), and mixed-genotype infections are frequently observed even within the same host tissue (Bharati, 2022). Genomic plasticity in Leishmania is driven by several mechanisms, including mosaic aneuploidy—a common feature that enables rapid adaptation under stress, with non-random, strain-specific patterns indicating selective pressure (Sterkers et al., 2014; Dujardin et al., 2014; Bussotti et al., 2018). Additionally, gene copy number variations (CNVs) such as tandem amplifications, deletions, and extrachromosomal circular or linear DNA contribute to genomic diversity (Black et al., 2023). Homologous recombination, facilitated by repeated sequences near DNA double-strand breaks, further promotes gene rearrangements (da Silva, 2021), while telomeric instability due to replicative stress in subtelomeric regions enhances genomic variability (Damasceno et al., 2016). Despite predominantly clonal expansion, evidence suggests genetic exchange between parasites, possibly through sexual recombination, which may enhance long-term survival (Figure 4) (Van den Broeck et al., 2020).
5.3 Genomic adaptations and drug resistance
Leishmania parasites exhibit remarkable genomic plasticity that underpins their adaptability to drug pressure. The parasite genome demonstrates significant instability characterized by aneuploidy, copy number variations (CNVs), and single nucleotide polymorphisms that collectively facilitate rapid adaptation to therapeutic interventions (Laffitte et al., 2016). These genomic alterations enable Leishmania to develop resistance through various mechanisms including altered drug transport, target modification, and enhanced metabolic detoxification pathways (Ponte-Sucre et al., 2017). The genomic instability of Leishmania serves as an evolutionary advantage, allowing rapid selection of resistant populations under drug pressure. Whole genome sequencing studies have revealed extensive chromosomal amplifications and deletions occurring in response to drug exposure showing more frequent copy number alterations (Leprohon et al., 2014). These changes often correlate with altered expression of genes involved in stress response, metabolism, and drug transport, establishing a genetic foundation for resistance development (Ubeda et al., 2008).
5.4 Gaps in current knowledge and future directions
While genomic plasticity contributes to resistance, the precise evolutionary trajectory of resistant Leishmania strains under drug pressure remains unclear, necessitating longitudinal studies to track genomic changes in response to treatment (Santi and Murta, 2022). Additionally, the role of host immune modulation in drug resistance requires further exploration, as studies suggest that resistant parasites alter host immune responses, though the molecular mechanisms remain poorly characterized (Costa-da-Silva et al., 2022). Beyond genetic mutations, epigenetic modifications such as histone modifications, DNA methylation, and non-coding RNAs may regulate resistance-related genes, warranting further investigation (Afrin et al., 2019). Furthermore, cross-resistance to different drug classes, including miltefosine and amphotericin B, has been observed, yet the underlying mechanisms remain inadequately understood (Zhang et al., 2025). Lastly, while genomic and transcriptomic analyses have identified numerous resistance-associated genes, their functional roles remain speculative, highlighting the need for gene knockout or overexpression studies to validate their contributions (Bharadava et al., 2024).
6 Drug pipeline advances in leishmaniasis
Significant global efforts are driving advancements in the leishmaniasis drug pipeline, with clinical trials aiming to refine treatments, enhance prevention strategies, strengthen immune responses, and improve diagnostics These efforts span various forms of the leishmaniasis addressing challenges such as drug resistance, treatment adherence, and accessibility. Recent studies have also emphasized the role of host-directed therapies and novel drug delivery systems to enhance treatment efficacy. In visceral leishmaniasis (VL), treatment trials have emphasized refining the use of Amphotericin B, a highly effective but costly drug. Liposomal Amphotericin B has been tested in single- and multiple-dose regimens, showing strong efficacy, especially in Indian patients; however, its high cost limits its accessibility (Lee et al., 2024). To improve outcomes, combination therapies including Amphotericin B, Miltefosine, and Paromomycin are being evaluated, with trials like NCT01122771 exploring shorter regimens to increase treatment adherence and reduce toxicity (Rahman et al., 2017). For patients co-infected with HIV, Miltefosine has demonstrated potential in Ethiopian trials, though the rise of drug resistance in endemic regions remains a concern (Ritmeijer et al., 2006).
Beyond treatment strategies, significant prevention efforts have been focused heavily on vector control, with long-lasting insecticide-treated nets (LLINs) proving effective in reducing transmission rates in endemic areas (Garlapati et al., 2021). Additionally, trials are assessing immune response modulation with agents like N-Acetylcysteine, often paired with Sodium Stibogluconate, to boost host resistance to infection, offering promise for high-risk individuals (Magalhães et al., 2022). In diagnostics, rapid diagnostic tests (RDTs) and cost-effective assays such as LAMP are being developed to improve early detection and timely treatment, crucial for reducing disease severity and transmission (Erber et al., 2022). New treatment options, like Sitamaquine and lipid-based Amphotericin B formulations, are being tested for patients resistant to traditional drugs (Sundar et al., 2011b). For relapse prevention in HIV co-infected patients, combination therapies with drugs like Pentamidine are also under study (Diro et al., 2015). Vaccination is an emerging focus, with trials for the LEISH-F3 + SLA-SE vaccine indicating favorable safety and immunogenicity in healthy adults, marking an important step toward an immune-based prophylactic for VL (Coler et al., 2015; Lacey et al., 2022). While these advances mark significant progress, several challenges persist, including resistance, high relapse rates, and limited diagnostic resources. Ongoing trials assessing predictive biomarkers and less invasive monitoring methods seek to address these limitations and enhance patient outcomes, though high costs and limited access continue to restrict broader application of effective treatments like liposomal formulations.
While visceral leishmaniasis has been the focus of extensive clinical trials, parallel efforts are advancing treatment strategies for cutaneous leishmaniasis (CL), where pentavalent antimonials remain a frontline choice despite concerns around toxicity and variable efficacy across different regions. These drugs show limited efficacy against certain species, like L. major and Leishmania tropica, prompting dose optimization trials and alternative delivery methods to reduce side effects. Miltefosine remains a key option, particularly in areas with limited injectable access, though issues of teratogenicity and resistance drive interest in more accessible oral formulations (van Henten et al., 2021). Combination therapies, such as Miltefosine with Paromomycin or liposomal Amphotericin B, are also being investigated to improve effectiveness and reduce treatment duration (Intakhan et al., 2024). Fexinidazole has shown promise as a short-course oral therapy for various forms of leishmaniasis (de Morais-Teixeira et al., 2019).
Nanoparticle formulations of Amphotericin B, including topical options, are being trialed for safer, localized treatment of CL lesions (Fairuz et al., 2022). Adjunct therapies with azole antifungals like Itraconazole are also under evaluation for their immunomodulatory effects, potentially benefiting resistant cases when combined with first-line treatments (Fischer et al., 2024). Immunotherapy is an expanding field, with trials assessing cytokine modulators such as GM-CSF and interferon-gamma alongside conventional treatments to enhance immune response and healing, particularly in immunocompromised patients (Akbari et al., 2021). Advances in diagnostics for CL, focusing on point-of-care tests and molecular tools like qPCR and LAMP, support accurate, early detection, enabling species-specific treatment in resource-limited settings (Erber et al., 2022).
7 Emerging drug targets in Leishmania
Recent advances in molecular and cellular biology have identified several promising drug targets that could pave the way for novel therapeutic interventions (Jain et al., 2022). One notable target is cyclin-dependent kinase 12 (CDK12), whose inhibition has demonstrated efficacy against Leishmania parasites, suggesting its potential as a therapeutic target for visceral leishmaniasis (Wyllie et al., 2018). Another promising target is the cytochrome bc1 complex, with inhibitors disrupting mitochondrial function in Leishmania species, leading to parasite death (Saldivia et al., 2024). Additionally, the proteasome has been identified as a viable target, with inhibitors like GNF6702 exhibiting broad-spectrum antiprotozoal activity against Leishmania species by selectively targeting the parasite’s proteasome without affecting host cells (Khare et al., 2016). Enzymes in the purine salvage pathway, such as adenine phosphoribosyltransferase (APRT) and hypoxanthine-guanine phosphoribosyltransferase (HGPRT), are essential for Leishmania survival and have been explored as potential targets (Boitz et al., 2012). Another critical pathway is the trypanothione system, which is unique to trypanosomatids and replaces the glutathione system in these parasites. Inhibitors of trypanothione reductase (TR) and trypanothione synthetase (TryS) have shown potent antileishmanial activity in preclinical studies (Beniwal et al., 2025; González-Montero et al., 2024). Additionally, sterol biosynthesis in Leishmania has emerged as a promising target. The enzyme sterol 14α-demethylase (CYP51), which is involved in ergosterol biosynthesis, has been successfully targeted by azole compounds, such as posaconazole and ketoconazole (Bhusal et al., 2024; Emami et al., 2017). Furthermore, protein kinases, particularly mitogen-activated protein kinases (MAPKs) and cyclin-dependent kinases (CDKs), play crucial roles in Leishmania proliferation and differentiation, making them attractive targets for kinase inhibitors (Naula et al., 2005). Other emerging targets include leishmanial proteases, such as cysteine proteases (e.g., CPA, CPB) and metalloproteases, which are involved in parasite virulence and immune evasion (de Oliveira et al., 2025). Polyamine biosynthesis in Leishmania represents another promising drug target, as enzymes like ornithine decarboxylase (ODC) and spermidine synthase are critical for parasite survival. Inhibiting key steps in polyamine metabolism could disrupt growth and redox balance in the parasite, offering potential therapeutic strategies for leishmaniasis (Carter et al., 2022). Finally, host-directed therapies that modulate immune responses, such as targeting host cytokines (e.g., IL-10, TGF-β) or enhancing macrophage leishmanicidal activity, represent a promising complementary approach (Kumar et al., 2017). Despite these advancements, significant research gaps remain. These include a lack of understanding of drug resistance mechanisms, the need for better in vitro and in vivo models, and the absence of effective vaccines. Addressing these gaps will require interdisciplinary collaboration, increased funding, and the integration of omics technologies to identify novel biomarkers and therapeutic targets.
8 Conclusion
The emergence of multi-drug resistant Leishmania strains presents a major challenge to disease management, significantly compromising clinical outcomes. The parasite’s unique genomic dynamics—manifested through mechanisms like gene amplifications, chromosomal rearrangements, and mosaic aneuploidy—facilitates rapid adaptation to pharmacological stress and drives phenotypic diversity. This genomic flexibility, coupled with specific mutations in drug targets, overexpression of efflux transporters, and alterations in sterol biosynthesis pathways, enables Leishmania to withstand various therapeutic interventions and develop resistance to multiple drugs.
Recent advances in understanding the molecular underpinnings of resistance highlight the critical role of genomic plasticity in the rapid emergence of resistant phenotypes. This knowledge underscores the urgent need for innovative therapeutic strategies that go beyond traditional approaches. By integrating genomic insights with advanced drug discovery techniques, it is possible to design targeted interventions, such as combination therapies and inhibitors that specifically disrupt resistance pathways or exploit the parasite’s genomic vulnerabilities.
Despite significant progress, the effective translation of these findings into clinical practice remains a challenge. Leishmania’s inherent genomic complexity and adaptability necessitate a multifaceted approach that combines genetic, biochemical, and pharmacological perspectives. Future research should aim to deepen our understanding of drug resistance mechanisms, with a focus on developing precision medicines tailored to specific resistance profiles. Additionally, exploring combination therapies or repurposing existing drugs to counteract known resistance mechanisms may offer promising avenues for managing resistant Leishmania cases. Enhanced diagnostic tools for early detection of resistant strains, coupled with therapeutic strategies that disrupt key resistance pathways, will be essential to ensuring the long-term success of anti-leishmanial therapies and improving patient outcomes. Expanding our understanding of molecular adaptations across different Leishmania species and strains will be key to designing next-generation therapies and sustainable treatment regimens.
Author contributions
CB: Conceptualization, Investigation, Methodology, Writing – original draft. SS: Conceptualization, Data curation, Validation, Writing – original draft, Writing – review and editing. DD: Data curation, Investigation, Writing – review and editing. RS: Conceptualization, Formal Analysis, Supervision, Visualization, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
VL, Visceral Leishmaniasis; CL, Cutaneous Leishmaniasis; SSG, Sodium Stibogluconate; AmB, Amphotericin B; L-AmB, Liposomal Amphotericin B; MIL, Miltefosine; PM, Paromomycin; MRPA, Multidrug Resistance Protein A; AQP1, Aquaglyceroporin 1; PKDL, Post-Kala-Azar Dermal Leishmaniasis; HSP, Heat Shock Protein; ABC, ATP-Binding Cassette; MDR1, Multidrug Resistance 1; MSL, Miltefosine Sensitivity Locus; SMT, Sterol Methyltransferase; SC5D, Sterol C5-Desaturase; RDT, Rapid Diagnostic Test; LAMP, Loop-Mediated Isothermal Amplification; CNV, Copy Number Variation; SNP, Single Nucleotide Polymorphism; LLINs, Long-Lasting Insecticide-Treated Nets.
References
Afrin, F., Khan, I., and Hemeg, H. A. (2019). Leishmania-host interactions-an epigenetic paradigm. Front. Immunol. 10, 492. doi:10.3389/fimmu.2019.00492
Akbari, M., Oryan, A., and Hatam, G. (2021). Immunotherapy in treatment of leishmaniasis. Immunol. Lett. 233, 80–86. doi:10.1016/j.imlet.2021.03.011
Alpizar-Sosa, E. A., Ithnin, N. R., Wei, W., Pountain, A. W., Weidt, S. K., Donachie, A. M., et al. (2022). Amphotericin B resistance in Leishmania mexicana: alterations to sterol metabolism and oxidative stress response. PLoS Neglected Trop. Dis. 16 (9), e0010779. doi:10.1371/journal.pntd.0010779
Bansal, R., Sen, S. S., Muthuswami, R., and Madhubala, R. (2020). Stigmasterol as a potential biomarker for amphotericin B resistance in Leishmania donovani. J. Antimicrob. Chemother. 75 (4), 942–950. doi:10.1093/jac/dkz515
Bartholomeu, D. C., Teixeira, S. M. R., and Cruz, A. K. (2021). Genomics and functional genomics in Leishmania and Trypanosoma cruzi: statuses, challenges and perspectives. Mem. Inst. Oswaldo Cruz 116, e200634. doi:10.1590/0074-02760200634
Beniwal, P., Bhusal, C. K., Choudhary, G., Sehgal, R., Medhi, B., Prakash, A., et al. (2025). In silico screening and Molecular Dynamic simulations of FDA-Approved drugs as an inhibitor of Trypanothione Reductase of Leishmania donovani. Exp. Parasitol. 26, 108942. doi:10.1016/j.exppara.2025.108942
Berg, M., García-Hernández, R., Cuypers, B., Vanaerschot, M., Manzano, J. I., Poveda, J. A., et al. (2015). Experimental resistance to drug combinations in Leishmania donovani: metabolic and phenotypic adaptations. Antimicrob. agents Chemother. 59 (4), 2242–2255. doi:10.1128/AAC.04231-14
Bharadava, K., Upadhyay, T. K., Kaushal, R. S., Ahmad, I., Alraey, Y., Siddiqui, S., et al. (2024). Genomic insight of leishmania parasite: in-depth review of drug resistance mechanisms and genetic mutations. ACS Omega 9 (11), 12500–12514. doi:10.1021/acsomega.3c09400
Bharati, K. (2022). Human genetic polymorphism and Leishmaniasis. Infect. Genet. Evol. 98, 105203. doi:10.1016/j.meegid.2021.105203
Bhusal, C. K., Beniwal, P., Singh, S., Kaur, D., Kaur, U., Kaur, S., et al. (2024). Possibility of re-purposing antifungal drugs posaconazole and isavuconazole against promastigote form of Leishmania major. Indian J. Med. Res. 160 (5), 466–478. doi:10.25259/IJMR_569_2024
Black, J. A., Reis-Cunha, J. L., Cruz, A. K., and Tosi, L. R. O. (2023). Life in plastic, it's fantastic! How Leishmania exploit genome instability to shape gene expression. Front. Cell Infect. Microbiol. 13, 1102462. doi:10.3389/fcimb.2023.1102462
Boitz, J. M., Strasser, R., Hartman, C. U., Jardim, A., and Ullman, B. (2012). Adenine aminohydrolase from Leishmania donovani: unique enzyme in parasite purine metabolism. J. Biol. Chem. 287 (10), 7626–7639. doi:10.1074/jbc.M111.307884
Brito, G., Dourado, M., Guimarães, L. H., Meireles, E., Schriefer, A., de Carvalho, E. M., et al. (2017). Oral pentoxifylline associated with pentavalent antimony: a randomized trial for cutaneous leishmaniasis. Am. J. Trop. Med. Hyg. 96 (5), 1155–1159. doi:10.4269/ajtmh.16-0435
Bussotti, G., Gouzelou, E., Côrtes Boité, M., Kherachi, I., Harrat, Z., Eddaikra, N., et al. (2018). Leishmania genome dynamics during environmental adaptation reveal strain-specific differences in gene copy number variation, karyotype instability, and telomeric amplification. MBio 9 (6), 013999-18–e2128. doi:10.1128/mBio.01399-18
Camacho, E., González-De la Fuente, S., Solana, J. C., Rastrojo, A., Carrasco-Ramiro, F., Requena, J. M., et al. (2021). Gene annotation and transcriptome delineation on a de novo genome assembly for the reference Leishmania major Friedlin strain. Genes 12 (9), 1359. doi:10.3390/genes12091359
Carnielli, J. B., Dave, A., Romano, A., Forrester, S., de Faria, P. R., Monti-Rocha, R., et al. (2022). 3′ Nucleotidase/nuclease is required for Leishmania infantum clinical isolate susceptibility to miltefosine. EBioMedicine 86, 104378. doi:10.1016/j.ebiom.2022.104378
Carter, N. S., Kawasaki, Y., Nahata, S. S., Elikaee, S., Rajab, S., Salam, L., et al. (2022). Polyamine metabolism in leishmania parasites: a promising therapeutic target. Med. Sci. (Basel) 10 (2), 24. doi:10.3390/medsci10020024
Chandra, U., Yadav, A., Kumar, D., and Saha, S. (2017). Cell cycle stage-specific transcriptional activation of cyclins mediated by HAT2-dependent H4K10 acetylation of promoters in Leishmania donovani. PLoS Pathog. 13 (9), e1006615. doi:10.1371/journal.ppat.1006615
Chawla, B., Jhingran, A., Panigrahi, A., Stuart, K. D., and Madhubala, R. (2011). Paromomycin affects translation and vesicle-mediated trafficking as revealed by proteomics of paromomycin–susceptible–resistant Leishmania donovani. PloS one 6 (10), e26660. doi:10.1371/journal.pone.0026660
Cohen, B. E. (2016). The role of signaling via aqueous pore formation in resistance responses to amphotericin B. Antimicrob. agents Chemother. 60 (9), 5122–5129. doi:10.1128/AAC.00878-16
Coler, R. N., Duthie, M. S., Hofmeyer, K. A., Guderian, J., Jayashankar, L., Vergara, J., et al. (2015). From mouse to man: safety, immunogenicity and efficacy of a candidate leishmaniasis vaccine LEISH-F3+ GLA-SE. Clin. and Transl. Immunol. 4 (4), e35. doi:10.1038/cti.2015.6
Costa-da-Silva, A. C., Nascimento, D. O., Ferreira, J. R. M., Guimarães-Pinto, K., Freire-de-Lima, L., Morrot, A., et al. (2022). Immune responses in leishmaniasis: an overview. Trop. Med. Infect. Dis. 7 (4), 54. doi:10.3390/tropicalmed7040054
Cupolillo, E., Cavalcanti, A. S., Ferreira, G. E. M., Boité, M. C., Morgado, F. N., and Porrozzi, R. (2020). Occurrence of multiple genotype infection caused by Leishmania infantum in naturally infected dogs. PLoS Negl. Trop. Dis. 14 (7), e0007986. doi:10.1371/journal.pntd.0007986
Da Costa, K. M., Valente, R. C., Salustiano, E. J., Gentile, L. B., Freire-de-Lima, L., Mendonça-Previato, L., et al. (2018). Functional characterization of ABCC proteins from Trypanosoma cruzi and their involvement with thiol transport. Front. Microbiol. 9, 205. doi:10.3389/fmicb.2018.00205
Damasceno, J. D., Obonaga, R., Santos, E. V., Scott, A., McCulloch, R., and Tosi, L. R. (2016). Functional compartmentalization of Rad9 and Hus1 reveals diverse assembly of the 9-1-1 complex components during the DNA damage response in Leishmania. Mol. Microbiol. 101 (6), 1054–1068. doi:10.1111/mmi.13441
Das, S., Shah, P., Tandon, R., Yadav, N. K., Sahasrabuddhe, A. A., Sundar, S., et al. (2015). Over-expression of cysteine leucine rich protein is related to SAG resistance in clinical isolates of Leishmania donovani. PLoS Neglected Trop. Dis. 9 (8), e0003992. doi:10.1371/journal.pntd.0003992
da Silva, M. S. (2021). DNA double-strand breaks: a double-edged sword for trypanosomatids. Front. Cell Dev. Biol. 9, 669041. doi:10.3389/fcell.2021.669041
de Morais-Teixeira, E., Rabello, A., and Aguiar, M. M. (2019). In vitro activity and in vivo efficacy of fexinidazole against New World Leishmania species. J. Antimicrob. Chemother. 74 (8), 2318–2325. doi:10.1093/jac/dkz172
de Oliveira, R. É., Albino, S. L., Olimpio de Moura, R., and Nascimento, IJDS (2025). Targeting cysteine protease B to discover antileishmanial drugs: directions and advances. Eur. J. Med. Chem. 289, 117500. doi:10.1016/j.ejmech.2025.117500
de Santana, M. B. R., Miranda, G. O., and Carvalho, L. P. (2025). ATP-binding cassette transporters and drug resistance in cutaneous leishmaniasis. Int. J. Infect. Dis. 151, 107315. doi:10.1016/j.ijid.2024.107315
Diro, E., Ritmeijer, K., Boelaert, M., Alves, F., Mohammed, R., Abongomera, C., et al. (2015). Use of pentamidine as secondary prophylaxis to prevent visceral leishmaniasis relapse in HIV infected patients, the first twelve months of a prospective cohort study. PLoS neglected Trop. Dis. 9 (10), e0004087. doi:10.1371/journal.pntd.0004087
Douanne, N., Wagner, V., Roy, G., Leprohon, P., Ouellette, M., and Fernandez-Prada, C. (2020). MRPA-independent mechanisms of antimony resistance in Leishmania infantum. Int. J. Parasitol. Drugs Drug Resist. 13, 28–37. doi:10.1016/j.ijpddr.2020.03.003
Dujardin, J. C., Mannaert, A., Durrant, C., and Cotton, J. A. (2014). Mosaic aneuploidy in Leishmania: the perspective of whole genome sequencing. Trends Parasitol. 30 (12), 554–555. doi:10.1016/j.pt.2014.09.004
Emami, S., Tavangar, P., and Keighobadi, M. (2017). An overview of azoles targeting sterol 14α-demethylase for antileishmanial therapy. Eur. J. Med. Chem. 135, 241–259. doi:10.1016/j.ejmech.2017.04.044
Ennes-Vidal, V., Menna-Barreto, R. F., Branquinha, M. H., Dos Santos, A. L., and D'Avila-Levy, C. M. (2017). Why calpain inhibitors are interesting leading compounds to search for new therapeutic options to treat leishmaniasis? Parasitology 144 (2), 117–123. doi:10.1017/S003118201600189X
Erber, A. C., Sandler, P. J., de Avelar, D. M., Swoboda, I., Cota, G., and Walochnik, J. (2022). Diagnosis of visceral and cutaneous leishmaniasis using loop-mediated isothermal amplification (LAMP) protocols: a systematic review and meta-analysis. Parasites and vectors 15 (1), 34. doi:10.1186/s13071-021-05133-2
Espada, C. R., Magalhães, R. M., Cruz, M. C., Machado, P. R., Schriefer, A., Carvalho, E. M., et al. (2019). Investigation of the pathways related to intrinsic miltefosine tolerance in Leishmania (Viannia) braziliensis clinical isolates reveals differences in drug uptake. Int. J. Parasitol. Drugs Drug Resist. 11, 139–147. doi:10.1016/j.ijpddr.2019.02.005
Fairuz, S., Nair, R. S., and Billa, N. (2022). Orally administered amphotericin B nanoformulations: physical properties of nanoparticle carriers on bioavailability and clinical relevance. Pharmaceutics 14 (9), 1823. doi:10.3390/pharmaceutics14091823
Fekrisoofiabadi, M., Fekri, M., Moradabadi, A., Vahidi, R., Khaleghi, M., Ram, M., et al. (2019). Evaluation of MDR1 and MRPA genes expression in different types of dry cutaneous leishmaniasis. BMC Res. Notes 12, 803–804. doi:10.1186/s13104-019-4784-0
Fischer, T., Fischer, M., Schliemann, S., and Elsner, P. (2024). Treatment of mucocutaneous leishmaniasis–A systematic review. JDDG J. der Deutschen Dermatologischen Gesellschaft 22, 763–773. doi:10.1111/ddg.15424
Franssen, S. U., Durrant, C., Stark, O., Moser, B., Downing, T., Imamura, H., et al. (2020). Global genome diversity of the Leishmania donovani complex. Elife 9, e51243. doi:10.7554/eLife.51243
Frézard, F., Aguiar, M. M. G., Ferreira, L. A. M., Ramos, G. S., Santos, T. T., Borges, G. S. M., et al. (2022). Liposomal amphotericin B for treatment of leishmaniasis: from the identification of critical physicochemical attributes to the design of effective topical and oral formulations. Pharmaceutics 15 (1), 99. doi:10.3390/pharmaceutics15010099
Garlapati, R., Iniguez, E., Serafim, T. D., Mishra, P. K., Rooj, B., Sinha, B., et al. (2021). Towards a sustainable vector-control strategy in the post kala-azar elimination era. Front. Cell. Infect. Microbiol. 11, 641632. doi:10.3389/fcimb.2021.641632
González-Montero, M. C., Andrés-Rodríguez, J., García-Fernández, N., Pérez-Pertejo, Y., Reguera, R. M., Balaña-Fouce, R., et al. (2024). Targeting trypanothione metabolism in trypanosomatids. Molecules 29 (10), 2214. doi:10.3390/molecules29102214
Goswami, R. P., Rahman, M., Das, S., Tripathi, S. K., and Goswami, R. P. (2020). Combination therapy against Indian visceral Leishmaniasis with Liposomal Amphotericin B (FungisomeTM) and short-course miltefosine in comparison to miltefosine monotherapy. Am. J. Trop. Med. Hyg. 103 (1), 308–314. doi:10.4269/ajtmh.19-0931
Grünebast, J., and Clos, J. (2020). Leishmania: responding to environmental signals and challenges without regulated transcription. Comput. Struct. Biotechnol. J. 18, 4016–4023. doi:10.1016/j.csbj.2020.11.058
Hadermann, A., Heeren, S., Maes, I., Dujardin, J. C., Domagalska, M. A., and Van den Broeck, F. (2023). Genome diversity of Leishmania aethiopica. Front. Cell Infect. Microbiol. 13, 1147998. doi:10.3389/fcimb.2023.1147998
Haldar, A. K., Sen, P., and Roy, S. (2011). Use of antimony in the treatment of leishmaniasis: current status and future directions. Mol. Biol. Int. 2011 (1), 571242. doi:10.4061/2011/571242
Hefnawy, A., Berg, M., Dujardin, J. C., and De Muylder, G. (2017). Exploiting knowledge on Leishmania drug resistance to support the quest for new drugs. Trends Parasitol. 33 (3), 162–174. doi:10.1016/j.pt.2016.11.003
Hendrickx, S., Boulet, G., Mondelaers, A., Dujardin, J. C., Rijal, S., Lachaud, L., et al. (2014). Experimental selection of paromomycin and miltefosine resistance in intracellular amastigotes of Leishmania donovani and L. infantum. Parasitol. Res. 113, 1875–1881. doi:10.1007/s00436-014-3835-7
Hendrickx, S., Reis-Cunha, J. L., Forrester, S., Jeffares, D. C., and Caljon, G. (2021). Experimental selection of paromomycin resistance in Leishmania donovani amastigotes induces variable genomic polymorphisms. Microorganisms 9 (8), 1546. doi:10.3390/microorganisms9081546
Hendrickx, S., Van den Kerkhof, M., Mabille, D., Cos, P., Delputte, P., Maes, L., et al. (2017). Combined treatment of miltefosine and paromomycin delays the onset of experimental drug resistance in Leishmania infantum. PLoS neglected Trop. Dis. 11 (5), e0005620. doi:10.1371/journal.pntd.0005620
Herrera, G., Barragán, N., Luna, N., Martínez, D., De Martino, F., Medina, J., et al. (2020). An interactive database of Leishmania species distribution in the Americas. Sci. Data 7 (1), 110. doi:10.1038/s41597-020-0451-5
Imamura, H., Monsieurs, P., Jara, M., Sanders, M., Maes, I., Vanaerschot, M., et al. (2020). Evaluation of whole genome amplification and bioinformatic methods for the characterization of Leishmania genomes at a single cell level. Sci. Rep. 10 (1), 15043. doi:10.1038/s41598-020-71882-2
Intakhan, N., Saeung, A., Rodrigues Oliveira, S. M., Pereira, M. D., and Chanmol, W. (2024). Synergistic effects of artesunate in combination with amphotericin B and miltefosine against leishmania infantum: potential for dose reduction and enhanced therapeutic strategies. Antibiotics 13 (9), 806. doi:10.3390/antibiotics13090806
Iv. Gerasimov, E. S., Novozhilova, T. S., Zimmer, S. L., and Yurchenko, V. (2023). Kinetoplast genome of leishmania spp. is under strong purifying selection. Trop. Med. Infect. Dis. 8 (8), 384. doi:10.3390/tropicalmed8080384
Jain, S., Sahu, U., Kumar, A., and Khare, P. (2022). Metabolic pathways of leishmania parasite: source of pertinent drug targets and potent drug candidates. Pharmaceutics 14 (8), 1590. doi:10.3390/pharmaceutics14081590
Jones, C. M., and Welburn, S. C. (2021). Leishmaniasis beyond East Africa. Front. Vet. Sci. 8, 618766. doi:10.3389/fvets.2021.618766
Kamran, M., Bhattacharjee, R., Das, S., Mukherjee, S., and Ali, N. (2023). The paradigm of intracellular parasite survival and drug resistance in leishmanial parasite through genome plasticity and epigenetics: perception and future perspective. Front. Cell. Infect. Microbiol. 13, 1001973. doi:10.3389/fcimb.2023.1001973
Kapil, S., Singh, P. K., and Silakari, O. M. (2018). An update on small molecule strategies targeting leishmaniasis. Eur. J. Med. Chem. 157, 339–367. doi:10.1016/j.ejmech.2018.08.012
Kazemi-Rad, E., Mohebali, M., Khadem-Erfan, M. B., Saffari, M., Raoofian, R., Hajjaran, H., et al. (2013). Identification of antimony resistance markers in Leishmania tropica field isolates through a cDNA-AFLP approach. Exp. Parasitol. 135 (2), 344–349. doi:10.1016/j.exppara.2013.07.018
Khanra, S., Sarraf, N. R., Das, A. K., Roy, S., and Manna, M. (2017). Miltefosine resistant field isolate from Indian kala-azar patient shows similar phenotype in experimental infection. Sci. Rep. 7 (1), 10330. doi:10.1038/s41598-017-09720-1
Khare, S., Nagle, A. S., Biggart, A., Lai, Y. H., Liang, F., Davis, L. C., et al. (2016). Proteasome inhibition for treatment of leishmaniasis, Chagas disease and sleeping sickness. Nature 537 (7619), 229–233. doi:10.1038/nature19339
Kulshrestha, A., Sharma, V., Singh, R., and Salotra, P. (2014). Comparative transcript expression analysis of miltefosine-sensitive and miltefosine-resistant Leishmania donovani. Parasitol. Res. 113, 1171–1184. doi:10.1007/s00436-014-3755-6
Kumar, R., Chauhan, S. B., Ng, S. S., Sundar, S., and Engwerda, C. R. (2017). Immune checkpoint targets for host-directed therapy to prevent and treat leishmaniasis. Front. Immunol. 8, 1492. doi:10.3389/fimmu.2017.01492
Lacey, C., Musa, A., Younis, B., Osman, M., Wiggins, R., Keding, A., et al. (2022). LEISH2b-A phase 2b study to assess the safety, efficacy, and immunogenicity of the Leishmania vaccine Chad63-KH in post-kala azar dermal leishmaniasis. Wellcome Open Res. 7, 200. doi:10.12688/wellcomeopenres.17951.1
Laffitte, M. N., Leprohon, P., Papadopoulou, B., and Ouellette, M. (2016). Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Res 5, 2350. doi:10.12688/f1000research.9218.1
Laniado-Laborín, R., and Cabrales-Vargas, M. N. (2009). Amphotericin B: side effects and toxicity. Rev. Iberoam. Micol. 26 (4), 223–227. doi:10.1016/j.riam.2009.06.003
Lee, J. S., Cohen, R. M., Khan, R. A., Burry, J., Casas, E. C., Chung, H. Y., et al. (2024). Paving the way for affordable and equitable liposomal amphotericin B access worldwide. Lancet Glob. Health 12 (9), e1552–e1559. doi:10.1016/S2214-109X(24)00225-0
Leprohon, P., Fernandez-Prada, C., Gazanion, É., Monte-Neto, R., and Ouellette, M. (2014). Drug resistance analysis by next generation sequencing in Leishmania. Int. J. Parasitol. Drugs Drug Resist 5 (1), 26–35. doi:10.1016/j.ijpddr.2014.09.005
Lessa, H. A., Machado, P., Lima, F., Cruz, A. A., Bacellar, O., Guerreiro, J., et al. (2001). Successful treatment of refractory mucosal leishmaniasis with pentoxifylline plus antimony. Am. J. Trop. Med. Hyg. 65 (2), 87–89. doi:10.4269/ajtmh.2001.65.87
Llanes, A., Cruz, G., Morán, M., Vega, C., Pineda, V. J., Ríos, M., et al. (2022). Genomic diversity and genetic variation of Leishmania panamensis within its endemic range. Infect. Genet. Evol. 103, 105342. doi:10.1016/j.meegid.2022.105342
Machado, P. R., Lessa, H., Lessa, M., Guimaraes, L. H., Bang, H., Ho, J. L., et al. (2007). Oral pentoxifylline combined with pentavalent antimony: a randomized trial for mucosal leishmaniasis. Clin. Infect. Dis. 44 (6), 788–793. doi:10.1086/511643
Machado, P. R., Ribeiro, C. S., França-Costa, J., Dourado, M. E., Trinconi, C. T., Yokoyama-Yasunaka, J. K., et al. (2018). Tamoxifen and meglumine antimoniate combined therapy in cutaneous leishmaniasis patients: a randomised trial. Trop. Med. and Int. Health 23 (9), 936–942. doi:10.1111/tmi.13119
Magalhães, L. S., Melo, E. V., Damascena, N. P., Albuquerque, A. C., Santos, C. N., Rebouças, M. C., et al. (2022). Use of N-acetylcysteine as treatment adjuvant regulates immune response in visceral leishmaniasis: pilot clinical trial and in vitro experiments. Front. Cell. Infect. Microbiol. 12, 1045668. doi:10.3389/fcimb.2022.1045668
Mano, C., Kongkaew, A., Tippawangkosol, P., Somboon, P., Roytrakul, S., Pescher, P., et al. (2023). Amphotericin B resistance correlates with increased fitness in vitro and in vivo in Leishmania (Mundinia) martiniquensis. Front. Microbiol. 14, 1156061. doi:10.3389/fmicb.2023.1156061
Melaku, Y., Collin, S. M., Keus, K., Gatluak, F., Ritmeijer, K., and Davidson, R. N. (2007). Treatment of kala-azar in southern Sudan using a 17-day regimen of sodium stibogluconate combined with paromomycin: a retrospective comparison with 30-day sodium stibogluconate monotherapy. Am. J. Trop. Med. Hyg. 77 (1), 89–94. doi:10.4269/ajtmh.2007.77.89
Moncada-Diaz, M. J., Rodríguez-Almonacid, C. C., Quiceno-Giraldo, E., Khuong, F. T. H., Muskus, C., and Karamysheva, Z. N. (2024). Molecular mechanisms of drug resistance in leishmania spp. Pathogens 13 (10), 835. doi:10.3390/pathogens13100835
Mukherjee, S., Moitra, S., Xu, W., Hernandez, V., and Zhang, K. (2020). Sterol 14-α-demethylase is vital for mitochondrial functions and stress tolerance in Leishmania major. PLoS Pathog. 16 (8), e1008810. doi:10.1371/journal.ppat.1008810
Mukhopadhyay, R., Mukherjee, S., Mukherjee, B., Naskar, K., Mondal, D., Decuypere, S., et al. (2011). Characterisation of antimony-resistant Leishmania donovani isolates: biochemical and biophysical studies and interaction with host cells. Int. J. Parasitol. 41 (13-14), 1311–1321. doi:10.1016/j.ijpara.2011.07.013
Musa, A. M., Mbui, J., Mohammed, R., Olobo, J., Ritmeijer, K., Alcoba, G., et al. (2023). Paromomycin and miltefosine combination as an alternative to treat patients with visceral leishmaniasis in eastern Africa: a randomized, controlled, multicountry trial. Clin. Infect. Dis. 76 (3), e1177–e1185. doi:10.1093/cid/ciac643
Mwenechanya, R., Kovářová, J., Dickens, N. J., Mudaliar, M., Herzyk, P., Vincent, I. M., et al. (2017). Sterol 14α-demethylase mutation leads to amphotericin B resistance in Leishmania mexicana. PLoS neglected Trop. Dis. 11 (6), e0005649. doi:10.1371/journal.pntd.0005649
Naula, C., Parsons, M., and Mottram, J. C. (2005). Protein kinases as drug targets in trypanosomes and Leishmania. Biochim. Biophys. Acta 1754 (1-2), 151–159. doi:10.1016/j.bbapap.2005.08.018
Negreira, G. H., de Groote, R., Van Giel, D., Monsieurs, P., Maes, I., de Muylder, G., et al. (2023). The adaptive roles of aneuploidy and polyclonality in Leishmania in response to environmental stress. EMBO Rep. 24 (9), e57413. doi:10.15252/embr.202357413
Negreira, G. H., Monsieurs, P., Imamura, H., Maes, I., Kuk, N., Yagoubat, A., et al. (2022). High throughput single-cell genome sequencing gives insights into the generation and evolution of mosaic aneuploidy in Leishmania donovani. Nucleic Acids Res. 50 (1), 293–305. doi:10.1093/nar/gkab1203
Palić, S., Bhairosing, P., Beijnen, J. H., and Dorlo, T. P. C. (2019). Systematic review of host-mediated activity of miltefosine in leishmaniasis through immunomodulation. Antimicrob. Agents Chemother. 63 (7), e02507-18–e02518. doi:10.1128/AAC.02507-18
Pérez-Victoria, J. M., Bavchvarov, B. I., Torrecillas, I. R., Martínez-García, M., López-Martín, C., Campillo, M., et al. (2011). Sitamaquine overcomes ABC-mediated resistance to miltefosine and antimony in Leishmania. Antimicrob. agents Chemother. 55 (8), 3838–3844. doi:10.1128/AAC.00065-11
Pigott, D. M., Bhatt, S., Golding, N., Duda, K. A., Battle, K. E., Brady, O. J., et al. (2014). Global distribution maps of the leishmaniases. Elife 3, e02851. doi:10.7554/eLife.02851
Pokharel, P., Ghimire, R., and Lamichhane, P. (2021). Efficacy and safety of paromomycin for visceral leishmaniasis: a systematic review. J. Trop. Med. 24, 8629039. doi:10.1155/2021/8629039
Ponte-Sucre, A., Gamarro, F., Dujardin, J. C., Barrett, M. P., López-Vélez, R., García-Hernández, R., et al. (2017). Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS neglected Trop. Dis. 11 (12), e0006052. doi:10.1371/journal.pntd.0006052
Potvin, J. E., Leprohon, P., Queffeulou, M., Sundar, S., and Ouellette, M. (2021). Mutations in an aquaglyceroporin as a proven marker of antimony clinical resistance in the parasite Leishmania donovani. Clin. Infect. Dis. 72 (10), e526–e532. doi:10.1093/cid/ciaa1236
Pountain, A. W., and Barrett, M. P. (2019). Untargeted metabolomics to understand the basis of phenotypic differences in amphotericin B-resistant Leishmania parasites. Wellcome Open Res.;4, 176. doi:10.12688/wellcomeopenres.15452.1
Pountain, A. W., Weidt, S. K., Regnault, C., Bates, P. A., Donachie, A. M., Dickens, N. J., et al. (2019). Genomic instability at the locus of sterol C24-methyltransferase promotes amphotericin B resistance in Leishmania parasites. PLoS Neglected Trop. Dis. 13 (2), e0007052. doi:10.1371/journal.pntd.0007052
Purkait, B., Singh, R., Wasnik, K., Das, S., Kumar, A., Paine, M., et al. (2015). Up-regulation of silent information regulator 2 (Sir2) is associated with amphotericin B resistance in clinical isolates of Leishmania donovani. J. Antimicrob. Chemother. 70 (5), 1343–1356. doi:10.1093/jac/dku534
Rahman, R., Goyal, V., Haque, R., Jamil, K., Faiz, A., Samad, R., et al. (2017). Safety and efficacy of short course combination regimens with AmBisome, miltefosine and paromomycin for the treatment of visceral leishmaniasis (VL) in Bangladesh. PLoS neglected Trop. Dis. 11 (5), e0005635. doi:10.1371/journal.pntd.0005635
Rastrojo, A., García-Hernández, R., Vargas, P., Camacho, E., Corvo, L., Imamura, H., et al. (2018). Genomic and transcriptomic alterations in Leishmania donovani lines experimentally resistant to antileishmanial drugs. Int. J. Parasitol. Drugs Drug Resist. 8 (2), 246–264. doi:10.1016/j.ijpddr.2018.04.002
Reynolds, D. L., Hofmeister, B. T., Cliffe, L., Siegel, T. N., Anderson, B. A., Beverley, S. M., et al. (2016). Base J represses genes at the end of polycistronic gene clusters in Leishmania major by promoting RNAP II termination. Mol. Microbiol. 101 (4), 559–574. doi:10.1111/mmi.13408
Rijal, S., Sundar, S., Mondal, D., Das, P., Alvar, J., and Boelaert, M. (2019). Eliminating visceral leishmaniasis in South Asia: the road ahead. BMJ 364, k5224. doi:10.1136/bmj.k5224
Ritmeijer, K., Dejenie, A., Assefa, Y., Hundie, T. B., Mesure, J., Boots, G., et al. (2006). A comparison of miltefosine and sodium stibogluconate for treatment of visceral leishmaniasis in an Ethiopian population with high prevalence of HIV infection. Clin. Infect. Dis. 43 (3), 357–364. doi:10.1086/505217
Ruang-Areerate, T., Ruang-Areerate, P., Manomat, J., Naaglor, T., Piyaraj, P., Mungthin, M., et al. (2023). Genetic variation and geographic distribution of Leishmania orientalis and Leishmania martiniquensis among Leishmania/HIV co-infection in Thailand. Sci. Rep. 13 (1), 23094. doi:10.1038/s41598-023-50604-4
Saha, S. (2020). Histone modifications and other facets of epigenetic regulation in trypanosomatids: leaving their mark. mBio 11 (5), 010799-20–e1120. doi:10.1128/mBio.01079-20
Salari, S., Bamorovat, M., Sharifi, I., and Almani, P. G. N. (2022). Global distribution of treatment resistance gene markers for leishmaniasis. J. Clin. Lab. Anal. 36 (8), e24599. doi:10.1002/jcla.24599
Saldivia, M., Lima, APCA, and Mottram, J. C. (2024). A promising pipeline of preclinical drug candidates for leishmaniasis and chronic Chagas' disease. Trends Parasitol. 40 (3), 211–213. doi:10.1016/j.pt.2024.02.002
Santi, A. M. M., and Murta, S. M. F. (2022). Impact of genetic diversity and genome plasticity of leishmania spp. in treatment and the search for novel chemotherapeutic targets. Front. Cell Infect. Microbiol. 12, 826287. doi:10.3389/fcimb.2022.826287
Santos, G. A., Sousa, J. M., Aguiar, AHBM, Torres, K. C. S., Coelho, A. J. S., Ferreira, A. L., et al. (2023). Systematic review of treatment failure and clinical relapses in leishmaniasis from a multifactorial perspective: clinical aspects, factors associated with the parasite and host. Trop. Med. Infect. Dis. 8 (9), 430. doi:10.3390/tropicalmed8090430
Scarpini, S., Dondi, A., Totaro, C., Biagi, C., Melchionda, F., Zama, D., et al. (2022). Visceral leishmaniasis: epidemiology, diagnosis, and treatment regimens in different geographical areas with a focus on pediatrics. Microorganisms 10 (10), 1887. doi:10.3390/microorganisms10101887
Shaw, C. D., Imamura, H., Downing, T., Blackburn, G., Westrop, G. D., Cotton, J. A., et al. (2019). Genomic and metabolomic polymorphism among experimentally selected paromomycin-resistant leishmania donovani strains. Antimicrob. Agents Chemother. 64 (1), e00904-19–e00919. doi:10.1128/AAC.00904-19
Singh, J., Khan, M. I., Yadav, S. P., Srivastava, A., Sinha, K. K., Das, P., et al. (2017). L-Asparaginase of Leishmania donovani: metabolic target and its role in Amphotericin B resistance. Int. J. Parasitol. Drugs Drug Resist. 7 (3), 337–349. doi:10.1016/j.ijpddr.2017.09.003
Singh, O. P., Hasker, E., Boelaert, M., Sacks, D., and Sundar, S. (2020). Xenodiagnosis to address key questions in visceral leishmaniasis control and elimination. PLOS Neglected Trop. Dis. 14 (8), e0008363. doi:10.1371/journal.pntd.0008363
Sterkers, Y., Crobu, L., Lachaud, L., Pagès, M., and Bastien, P. (2014). Parasexuality and mosaic aneuploidy in Leishmania: alternative genetics. Trends Parasitol. 30 (9), 429–435. doi:10.1016/j.pt.2014.07.002
Stone, N. R., Bicanic, T., Salim, R., and Hope, W. (2016). Liposomal amphotericin B (AmBisome®): a review of the pharmacokinetics, pharmacodynamics, clinical experience and future directions. Drugs 76, 485–500. doi:10.1007/s40265-016-0538-7
Sundar, S., Chakravarty, J., and Meena, L. P. (2019). Leishmaniasis: treatment, drug resistance and emerging therapies. Expert Opin. Orphan Drugs 7 (1), 1–10. doi:10.1080/21678707.2019.1552853
Sundar, S., Rai, M., Chakravarty, J., Agarwal, D., Agrawal, N., Vaillant, M., et al. (2008). New treatment approach in Indian visceral leishmaniasis: single-dose liposomal amphotericin B followed by short-course oral miltefosine. Clin. Infect. Dis. 47 (8), 1000–1006. doi:10.1086/591972
Sundar, S., Sinha, P. K., Dixon, S. A., Buckley, R., Miller, A. K., Mohamed, K., et al. (2011b). Pharmacokinetics of oral sitamaquine taken with or without food and safety and efficacy for treatment of visceral leishmaniais: a randomized study in Bihar, India. Am. J. Trop. Med. Hyg. 84 (6), 892–900. doi:10.4269/ajtmh.2011.10-0409
Sundar, S., Sinha, P. K., Rai, M., Verma, D. K., Nawin, K., Alam, S., et al. (2011a). Comparison of short-course multidrug treatment with standard therapy for visceral leishmaniasis in India: an open-label, non-inferiority, randomised controlled trial. Lancet 377 (9764), 477–486. doi:10.1016/S0140-6736(10)62050-8
Tandon, R., Chandra, S., Baharia, R. K., Das, S., Misra, P., Kumar, A., et al. (2014). Characterization of the proliferating cell nuclear antigen of Leishmania donovani clinical isolates and its association with antimony resistance. Antimicrob. agents Chemother. 58 (6), 2997–3007. doi:10.1128/AAC.01847-13
Torres-Guerrero, E., Quintanilla-Cedillo, M. R., Ruiz-Esmenjaud, J., and Arenas, R. (2017). Leishmaniasis: a review, F1000Res., 6, 750, doi:10.12688/f1000research.11120.1
Trinconi, C. T., Reimão, J. Q., Yokoyama-Yasunaka, J. K., Miguel, D. C., and Uliana, S. R. (2014). Combination therapy with tamoxifen and amphotericin B in experimental cutaneous leishmaniasis. Antimicrob. agents Chemother. 58 (5), 2608–2613. doi:10.1128/AAC.01315-13
Tunalı, V., and Özbilgin, A. (2023). Knock, knock, knocking on Europe's door: threat of leishmaniasis in Europe with a focus on Turkey. Curr. Res. Parasitol. Vector Borne Dis. 4, 100150. doi:10.1016/j.crpvbd.2023.100150
Ubeda, J. M., Légaré, D., Raymond, F., Ouameur, A. A., Boisvert, S., Rigault, P., et al. (2008). Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 9 (7), R115. doi:10.1186/gb-2008-9-7-r115
Vacchina, P., Norris-Mullins, B., Abengozar, M. A., Viamontes, C. G., Sarro, J., Stephens, M. T., et al. (2016). Genomic appraisal of the multifactorial basis for in vitro acquisition of miltefosine resistance in Leishmania donovani. Antimicrob. Agents Chemother. 60 (7), 4089–4100. doi:10.1128/AAC.00478-16
Van den Broeck, F., Savill, N. J., Imamura, H., Sanders, M., Maes, I., Cooper, S., et al. (2020). Ecological divergence and hybridization of Neotropical Leishmania parasites. Proc. Natl. Acad. Sci. U. S. A. 117 (40), 25159–25168. doi:10.1073/pnas.1920136117
van Henten, S., Tesfaye, A. B., Abdela, S. G., Tilahun, F., Fikre, H., Buyze, J., et al. (2021). Miltefosine for the treatment of cutaneous leishmaniasis—a pilot study from Ethiopia. PLoS neglected Trop. Dis. 15 (5), e0009460. doi:10.1371/journal.pntd.0009460
Verma, A., Bhandari, V., Deep, D. K., Sundar, S., Dujardin, J. C., Singh, R., et al. (2017). Transcriptome profiling identifies genes/pathways associated with experimental resistance to paromomycin in Leishmania donovani. Int. J. Parasitol. Drugs Drug Resist. 7 (3), 370–377. doi:10.1016/j.ijpddr.2017.10.004
Wasunna, M., Njenga, S., Balasegaram, M., Alexander, N., Omollo, R., Edwards, T., et al. (2016). Efficacy and safety of AmBisome in combination with sodium stibogluconate or miltefosine and miltefosine monotherapy for African visceral leishmaniasis: phase II randomized trial. PLoS neglected Trop. Dis. 10 (9), e0004880. doi:10.1371/journal.pntd.0004880
Wijnant, G. J., Van Bocxlaer, K., Yardley, V., Murdan, S., and Croft, S. L. (2017). Efficacy of paromomycin-chloroquine combination therapy in experimental cutaneous leishmaniasis. Antimicrob. agents Chemother. 61 (8), 003588-17–e1128. doi:10.1128/AAC.00358-17
World Health Organisation (2023). Leishmaniasis. Available online at: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis.
World Health Organization (2025). Leishmaniasis. Geneva: World Health Organization. Available online at: https://www.who.int/health-topics/leishmaniasis#tab=tab_1.
Wyllie, S., Thomas, M., Patterson, S., Crouch, S., De Rycker, M., Lowe, R., et al. (2018). Cyclin-dependent kinase 12 is a drug target for visceral leishmaniasis. Nature 560 (7717), 192–197. doi:10.1038/s41586-018-0356-z
Yao, C., and Wilson, M. E. (2016). Dynamics of sterol synthesis during development of Leishmania spp. parasites to their virulent form. Parasites and vectors 9, 200–202. doi:10.1186/s13071-016-1470-0
Yurchenko, V., Chistyakov, D. S., Akhmadishina, L. V., Lukashev, A. N., Sádlová, J., and Strelkova, M. V. (2023). Revisiting epidemiology of leishmaniasis in central Asia: lessons learnt. Parasitology 150 (2), 129–136. doi:10.1017/S0031182022001640
Zackay, A., Cotton, J. A., Sanders, M., Hailu, A., Nasereddin, A., Warburg, A., et al. (2018). Genome wide comparison of Ethiopian Leishmania donovani strains reveals differences potentially related to parasite survival. PLoS Genet. 14 (1), e1007133. doi:10.1371/journal.pgen.1007133
Keywords: leishmaniasis, drug resistance, genomic plasticity, antileishmanial therapy, drug targets
Citation: Bhusal CK, Sinha S, Kaur D and Sehgal R (2025) Unravelling drug resistance in leishmaniasis: genomic adaptations and emerging therapies. Front. Mol. Biosci. 12:1573618. doi: 10.3389/fmolb.2025.1573618
Received: 09 February 2025; Accepted: 12 May 2025;
Published: 26 May 2025.
Edited by:
Surendra Kumar Prajapati, Henry M Jackson Foundation for the Advancement of Military Medicine (HJF), United StatesReviewed by:
Debabrata Mandal, National Institute of Pharmaceutical Education and Research, IndiaRakhee Rathnam Kalari Kandy, University of Texas MD Anderson Cancer Center, United States
Copyright © 2025 Bhusal, Sinha, Kaur and Sehgal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rakesh Sehgal, c2VoZ2FscGdpQGdtYWlsLmNvbQ==
 Chandra Kanta Bhusal
Chandra Kanta Bhusal Shweta Sinha
Shweta Sinha Davinder Kaur
Davinder Kaur Rakesh Sehgal
Rakesh Sehgal