Abstract
Neuropathic pain, which results from damage to the somatosensory nervous system, is a global clinical condition that affects many people. Neuropathic pain imposes significant economic and public health burdens and is often difficult to manage because the underlying mechanisms remain unclear. However, mounting evidence indicates a role for neurogenic inflammation and neuroinflammation in pain pattern development. There is increasing evidence that the activation of neurogenic inflammation and neuroinflammation in the nervous system contribute to neuropathic pain. Altered miRNA expression profiles might be involved in the pathogenesis of both inflammatory and neuropathic pain by regulating neuroinflammation, nerve regeneration, and abnormal ion channel expression. However, the lack of knowledge about miRNA target genes prevents a full understanding of the biological functions of miRNAs. At the same time, an extensive study on exosomal miRNA, a newly discovered role, has advanced our understanding of the pathophysiology of neuropathic pain in recent years. This section provides a comprehensive overview of the current understanding of miRNA research and discusses the potential mechanisms of miRNAs in neuropathic pain.
Introduction
Neuropathic pain (NP), which results from damage to the somatosensory nervous system, is a global clinical condition that affects many people worldwide (Scholz et al., 2019). Neuropathic pain is associated with significant economic and public health burdens and is often difficult to manage because the underlying mechanisms remain obscure. However, mounting evidence indicates a role for neurogenic inflammation and neuroinflammation in pain patterns development (Yi et al., 2021). There is increasing evidence that the activation of neurogenic inflammation and neuroinflammation both in the periphery and the central nervous system conduce to the maintenance of neuropathic pain (Walters, 2014; Huh et al., 2017). Our earlier research looked at whether chronic inflammation brought on by an inflammatory reaction in the neurological system brought on by nerve damage might be the source of chronic neuropathic pain (Li et al., 2013).
Recent evidence has suggested that the anti-inflammatory effects of microRNAs are essential for pathogenesis in various contexts, such as inflammation, tumours, respiratory and cardiovascular diseases (Bernardo et al., 2015; Rupaimoole and Slack, 2017; Li Y. et al., 2019; Hill and Tran, 2021). Unfortunately, the pathogenesis of neuropathic pain has not been fully elucidated. Using microarray techniques, recent studies have reported that miRNAs play pivotal roles in the evolution and progression of neuropathic pain by regulating neuronal excitability and plasticity, neuronal ion channels, neuroinflammation, synaptic plasticity and other functions. These results indicate that altered miRNA expression profiles might be involved in the pathogenesis of neuropathic pain. However, the lack of knowledge about miRNA target genes prevents the achievement of a full understanding of the biological functions of miRNAs under these conditions. This review provides insight into miRNA expression profiling studies on neuropathic pain. We also highlight the molecular mechanisms of specific miRNAs that play critical functional roles in neuropathic pain pathogenesis and discuss their potential diagnostic, prognostic, and therapeutic utilization in neuropathic pain clinical management.
The structure and function of miRNAs
Gene structure
MicroRNAs are a varied family of 19–24 nucleotide short noncoding single-stranded RNAs that have fundamental biological activities in posttranscriptional gene silencing. The ribonuclease II enzymes, Drosha and Dicer sequentially cleave precursor RNA transcripts to generate miRNAs (Hutvágner et al., 2001; Gregory et al., 2004; Han et al., 2004). The miRNA-induced silencing complex (miRISC) is then formed by loading miRNAs onto the effector protein Argonaute (Ago) (Schwarz et al., 2003). By using translational repression and/or mRNA instability, miRISC silences mRNA targets by binding to them via sequence complementarity (Schwarz et al., 2003; Macfarlane and Murphy, 2010; Figure 1).
Figure 1

The classical bioformation process of miRNA. miRNAs are typically transcribed as primary miRNAs (pri-miRNAs) by RNA polymerase II (Pol II). The microprocessor complex, composed of the RNase III enzyme Drosha and the double-stranded RNA (dsRNA)-binding protein (dsRBP) DiGeorge critical region 8 (DGCR8), cleaves the pri-miRNA and releases a precursor miRNA (pre-miRNA). The export receptor exportin 5 binds pre-miRNAs and aids their export into the cytoplasm. Then the RNase III endonuclease DICER cleaves the pre-miRNA to release double-stranded miRNA. This miRNA is cleaved by Argonaute (Ago) to form the miRNA-induced silencing complex (miRISC).
The function of miRNAs
MiRNAs, which regulate approximately one-third of human genes, are widely present in mammalian cells. More than 1800 miRNAs, which target 60% of human mRNAs, are found in the human genome (Olive et al., 2015). Numerous human disorders, including neuropathic pain, are linked to mutations in miRNA genes and miRNA pathway genes. Furthermore, the peripheral and central nervous systems, which contain pain-related components, are widely dispersed with microRNAs. Despite not being directly engaged in peptide synthesis, miRNAs are a type of noncoding RNAs that have a significant impact on miRNA stability and protein translation (Saliminejad et al., 2019).
By binding to the 3’UTR of target mRNAs, miRNAs control posttranscriptional gene expression by lowering the majority (84%) of protein products (Lutz et al., 2014). One miRNA can target more than one mRNA transcript, and multiple miRNAs can simultaneously act on one mRNA strand (Lutz et al., 2014; Li et al., 2015). There is a general consensus that miRNAs play pivotal roles in regulating important biological processes, including early development, cell proliferation, cell death and apoptosis, fat metabolism, cell differentiation, and disease progression (Lima et al., 2011). Various disorders, including cardiovascular, cerebrovascular, and neurodegenerative diseases, may be impacted by changed miRNA expression profiles, according to the results of experimental models (Barwari et al., 2016; Wang et al., 2020).
Potential regulatory mechanisms of miRNAs in neuropathic pain
Several studies have reported broad abnormalities in miRNAs in animals following peripheral nerve injury. Several identified miRNAs are involved in neuroinflammation, nerve regeneration, and abnormal ion channel expression (Figure 2), suggesting that deregulation of miRNA expression may be included in the development of neuropathic pain and could be potentially useful diagnostic markers, improving the classification of neuropathic pain. The absence of information on miRNA target genes, however, limits a complete comprehension of the biological roles played by miRNAs. This section gives a thorough overview of what is currently known about miRNAs and explores how they can contribute to neuropathic pain (Table 1).
Figure 2
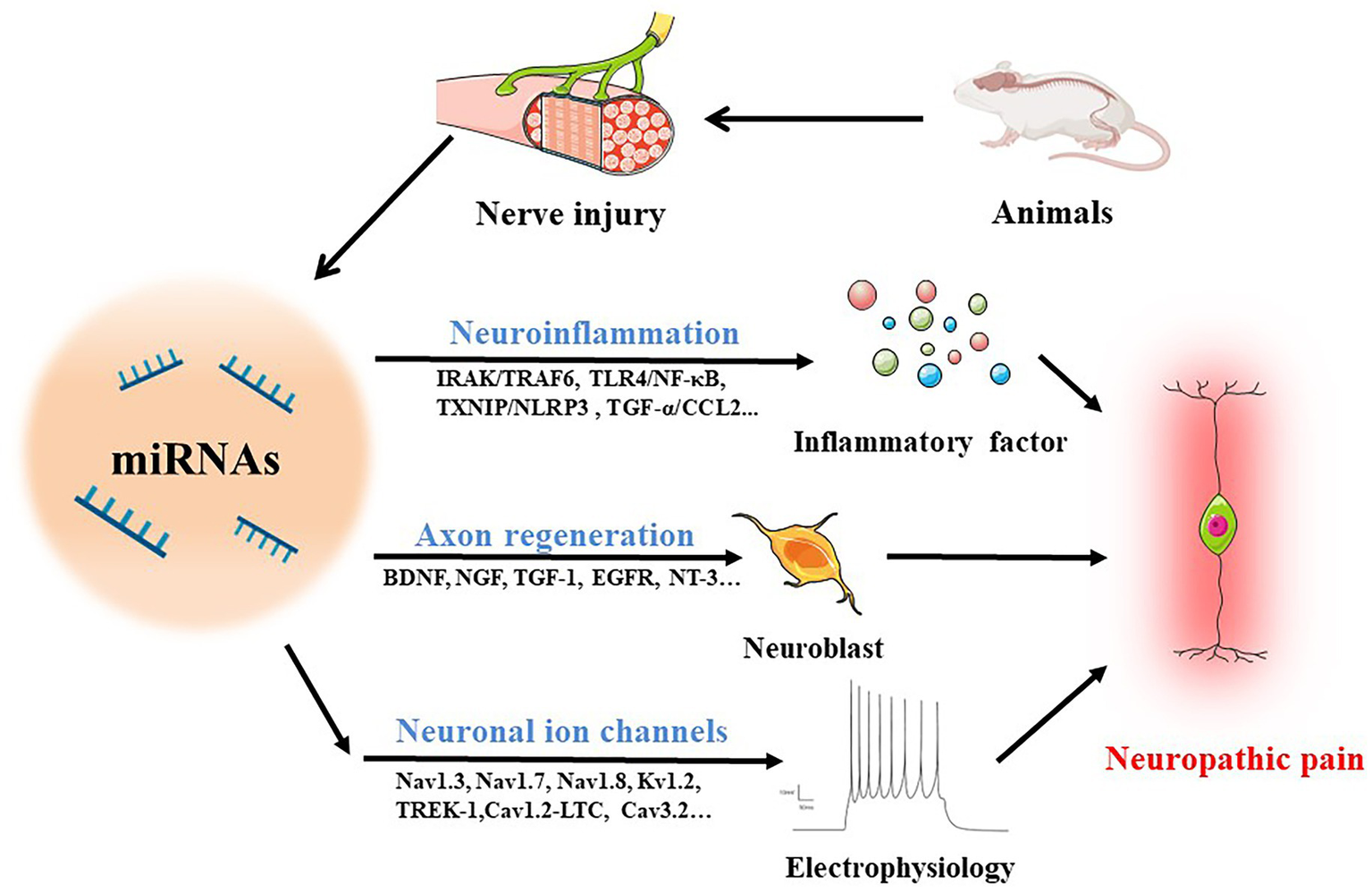
MiRNAs in neuropathic pain. The following routes are used by miRNAs to mediate neuropathic pain in animals with damaged nerves: (1) operating on immune cells to stimulate the production of inflammatory cytokines through IRAK/TRAF6, TLR4/NF-κB, TXNIP/NLRP3, TGF-α/CCL2 and other pathways to regulate neuroinflammation; (2) accelerating axon regeneration by increasing the synthesis of trophic factors such BDNF, NGF, TGF-1, EGFR, and NT-3; (3) mediating neuroelectrophysiological changes by activating ion channels like Nav1.3, Nav1.7, Nav1.8, Kv1.2, TREK-1, Cav1.2-LTC, and Cav3.2.
Table 1
| Mechanisms | miRNA | Target | Objectives | Models | Results | References |
|---|---|---|---|---|---|---|
| Neuroinflammation | ||||||
| Infiltration of immune cells | miR-590-3p ↑ | RAP1A ↓ | C57BL/6 mice | DPNP | T cell infiltration was reduced, which in turn hinders DPNP progression. | Wu et al. (2020) |
| miR-214-3p ↑ | CSF1 ↓ | SD rats | SNL | Attenuated the neuroinflammation and pain behavior. | Liu L. et al. (2020) | |
| miR-146a-5p ↑ | IRAK1 / TRAF6 ↓ | SD rats | CCI | Inhibited the development of CCI-induced neuropathic pain. | Wang et al. (2018) | |
| Neuroinflammation | miR-381 ↑ | HMGB1 and CXCR4 ↓ | SD rats | CCI | Inhibited neuropathic pain development. | Zhan et al. (2018) |
| miR-362-3p ↑ | BAMBI ↓ | C57BL/6 mice | CCI | NP progression was suppressed. | Zhang et al. (2022) | |
| miR-183 ↑ | TGF-α/CCL2/CCR2 ↓ | C57BL/6 mice | OA | Osteoarthrotic pain was ameliorated. | Tao et al. (2021) | |
| miR-28-5p ↑ | ZEB1 ↓ | SD rats | CCI | Reduced neuropathic pain. | Bao et al. (2018) | |
| miR-128-3p ↑ | ZEB1 ↓ | SD rats | CCI | Alleviated the progression of neuropathic pain | Zhang et al. (2020) | |
| miR-136 ↑ | ZEB1 ↓ | SD rats | CCI | Inhibited neuropathic pain development. | Shen et al. (2019) | |
| miR-150 ↑ | ZEB1 ↓ | SD rats | CCI | Inhibited neuropathic pain in vivo. | Yan et al. (2018a,b) | |
| miR-200b/ | ZEB1 ↓ | SD rats | CCI | Reduced neuropathic pain development in vivo. | Yan et al. (2018a,b) | |
| miR-429 ↑ | ||||||
| miR-23a ↑ | CXCR4/TXNIP/NLRP3 ↓ | C57BL/6 mice | pSNL | Inhibited neuropathic pain development. | Pan et al. (2018) | |
| miR-140 ↑ | S1PR1 ↓ | SD rats | CCI | Suppressed CCI-stimulated neuropathic pain. | Li et al. (2021) | |
| miR-144 ↑ | RASA1 ↓ | C57BL/6 mice | CCI | Facilitated the inhibition of neuropathic pain development. | Zhang et al. (2020) | |
| miR-216a-5p ↑ | KDM3A ↓ | SD rats | CCI | Alleviated neuropathic pain in rats. | Wang and Li W. et al. (2021) | |
| miR-93 ↑ | STAT3 ↓ | SD rats | CCI | Inhibited neuropathic pain development of CCI rats. | Yan et al. (2017) | |
| miR-155 ↓ | SOCS1 ↑ | SD rats | CCI | Attenuated neuropathic pain. | Tan et al. (2015) | |
| miR-155 ↓ | TRPA1 ↓ | SD rats | Chemotherapeutic Oxaliplatin | Supprressed the OXL-induced neuropathic pain. | Miao et al. (2019) | |
| miR-221 ↓ | SOCS1 ↑ | SD rats | CCI | Alleviateed neuropathic pain and neuroinflammation. | Xia et al. (2016) | |
| miR-221 ↓ | SOCS3 ↑ | SD rats | DPN | Reduced pain and decreased expression of inflammatory factors. | Wu et al. (2021) | |
| miR-22 ↑ | Mtf1 ↑ | Kunming mice | CFA | Promoted the development and maintenance of inflammatory pain. | Hao et al. (2022) | |
| Nerve regeneration | miR-192-5p ↑ | XIAP ↑ | SD rats | SNI | Decreased the apoptosis of nerve cells, and promote the repair and regeneration of peripheral nerve injury. | Liu X. et al. (2020) |
| miR-210 ↑ | EFNA3 ↓ | CF-1 mice | SNC | Promoted sensory axon regeneration and inhibit apoptsosis. | Hu et al. (2016) | |
| miR-135s ↑ | KLF4 ↓ | C57BL/6 mice | ONI | Facilitated RGC axon regeneration after optic nerve injury in adult mice. | van Battum et al. (2018) | |
| miR-125b ↑ | JAK/STAT ↓ | mice | SCI | Promoted the repair and regeneration following spinal cord injury. | Dai et al. (2018) | |
| miR-155 ↓ | SPRR1A ↑ | C57BL/6 mice | SCI | Enhanced neuron survival and axon growth. | Gaudet et al. (2016) | |
| miR-19a ↑ | PTEN ↑ | SD rats and C57BL/6 mice | ONC | Promoted axon regeneration after optic nerve crush in adult mice. | Mak et al. (2020) | |
| miR-21 ↑ | PTEN ↓ | SD rats | SNI | Promoted neurite growth. | Kar et al. (2021) | |
| miR-21 ↑ | TGFβI/TIMP3/EPHA4 ↓ | SD rats | Nerve injury | Promoted SC proliferation and axon regeneration. | Ning et al. (2020) | |
| miR-21 ↑ | EGFR ↑ | SD rats | ONC | Enhanced the axon regeneration after ONC. | Li et al. (2018) | |
| miR-199a-3p ↑ | mTOR ↓ | SD rats | SNI | Attenuated neurite growth. | Kar et al. (2021) | |
| miR-26a ↑ | GSK3β/Smad1 ↓ | CF-1 mice | SNC | Supported mammalian axon regeneration in vivo. | Jiang et al. (2015) | |
| miR-455-5p ↓ | GSK3β/Tau ↑ | Rats | SNI | Promoted axonal growth and regeneration. | Su et al. (2020) | |
| miR let-7 ↑ | Ntn1 ↓ | SD rats | SNI | Reduced axon outgrowth. | Wang et al. (2019) | |
| miR-9 ↑ | Dcc ↓ | |||||
| miR-9 ↑ | FoxP1 ↓ | CF-1 mice | SNC | Inhibited axon regeneration in vitro and in vivo. | Jiang et al. (2017) | |
| miR-138 ↓ | SIRT1 ↑ | CF-1 mice | SNC | Promoted mammalian axon regeneration. | Liu et al. (2013) | |
| Neuronal ion channels | ||||||
| Voltage-gated sodium channel | miR-7a ↑ | β2 subunit ↓ | SD rats | SNL and CCI | Suppressed neuropathic pain. | Sakai et al. (2013) |
| miR-96 ↑ | Nav1.3 ↓ | SD rats | CCI | Alleviated neuropathic pain. | Chen et al. (2014) | |
| miR-384-5p ↑ | SCN3A ↓ | SD rats | CCI | Significantly repressed mechanical allodynia and heat hyperalgesia in CCI rats. | Ye et al. (2020) | |
| miR-182 ↑ | Nav1.7 ↓ | SD rats | SNI | Alleviated SNI-induced neuropathic pain. | Cai et al. (2018) | |
| miR-30b ↑ | Nav1.3 ↓ | SD rats | SNL | Suppressed neuropathic pain. | Su et al. (2017) | |
| miR-30b-5p ↑ | Nav1.6 ↓ | SD rats | Chemotherapeutic Oxaliplatin | Attenuated pain. | Li L. et al. (2019) | |
| miR-30b ↑ | Nav1.7 ↓ | SD rats | SNI | Alleviated neuropathic pain. | Shao et al. (2016) | |
| miR-183 ↑ | Nav1.3/Nav1.7/Nav1.8 ↓ | C57BL/6 mice | DMM | Inhibited the expression of pain-related factors and ameliorated OA pain. | Tao et al. (2021) | |
| Voltage-gated potassium channels | miR-17-92 ↓ | multiple voltage-gated potassium channels ↑ | SD rats | SNL | Alleviated mechanical allodynia induced by nerve injury. | Sakai et al. (2013) |
| miR-137 ↓ | Kv1.2 ↑ | SD rats | CCI | Alleviated mechanical allodynia and thermal hyperalgesia. | Zhang et al. (2021) | |
| miR-183-5P ↑ | TREK-1 ↓ | SD rats | CCI | Efficiently ameliorated neuropathic pain. | Shi et al. (2018) | |
| Voltage-gated calcium channels | miR-219 ↑ | CaMKIIγ ↓ | Kunming mice | CFA and CCI | Prevented and reversed neuropathic pain and spinal neuronal sensitization induced by CFA. | Pan et al. (2014) |
| miR-124a ↑ | MeCP2 ↓ | C57BL/6 mice | Formalin | Decreased inflammatory nociception. | Kynast et al. (2013) | |
| miR-103 ↑ | Cav1.2-LTC ↓ | Wistar rats | SNL | Successfully relieve pain. | Favereaux et al. (2011) | |
| miR-32-5p ↑ | Cav3.2 ↓ | SD rats | CCI | Reversed mechanical allodynia. | Qi et al. (2022) | |
| miR-183 ↑ | α2δ-1 and α2δ-2 | C57BL/6 mice | SNI | Prevented elevation of basal mechanical sensitivity in nociceptors | Peng et al. (2017) | |
miRNAs and neuropathic pain.
RAP1A, Ras-related protein 1A; DPNP, diabetic peripheral neuropathic pain; CSF1, Colony-stimulating factor 1; SD, Sprague–Dawley; SNL, sciatic nerve injury; IRAK, IL-1R-associated kinase; TRAF6, Tumor necrosis factor receptor-associated factor 6; CCI, chronic constriction nerve injury; HMGB1, high mobility group box 1; CXCR4, Chemokine CXC receptor 4; BAMBI, bone morphogenetic protein and activin membrane-bound inhibitor; TGF-α, Tumor necrosis factor α; CCL2, CC chemokine ligand 2; CCR2, CC chemokine receptor type-2; ZEB1, Zinc finger E-box-binding homeobox 1; TXNIP, Thioredoxin interacting protein; NLRP3, NOD-like receptor protein 3; S1PR1, Sphingosine-1-phosphate receptor 1; RASA1,RAS P21 protein activator 1; KDM3A, lysine-specific demethylase 3A; STAT, signal transducer and activator of transcription; SOCS, Suppressor of cytokine signaling; TRPA1, transient receptor potential cation channel subfamily A member 1; Mtf1, Mitochondrial Transcription Factor 1; OXL, Oxaliplatin; DPN, diabetic peripheral neuropathy; CFA, Complete Freund’s adjuvant; OA, osteoarthritic; XIAP, X-linked inhibitor of apoptosis protein; SNI, sciatic nerve injury; EFNA3, ephrin-A3; SNC, sciatic nerve crush; KLF4, kruppel-like factor 4; ONI, optic nerve injury; RGC, retinal ganglion cell; JAK, Janus kinase gene; STAT, signal transducer and activator of transcription; SCI, spinal cord injury; SPRR1A, small proline-rich repeat protein 1A; PTEN, phosphatase and tensin homolog; ONC, Optic nerve crush; TGFβI, transforming growth factor-beta-induced protein; TIMP3, tissue inhibitor of metalloproteinases 3; EPHA4, erythropoietin-producing human hepatocellular receptor A4; EGFR, epidermal growth factor receptor; mTOR, mechanistic target of rapamycin; GSK3β, glycogen synthase kinase 3β; Ntn1, netrin-1; Dcc, deleted in colorectal cancer; FoxP1, forkhead box protein P1; SIRT1, Sirtuin 1; Nav, voltage-gated sodium channels; DMM, destabilization of the medial meniscus; Kv, voltage-gated potassium channels; TREK-1, TWIK-related K channel 1; MeCP2, Methyl-CpG binding protein 2; Cav1.2-LTC, Cav1.2-comprising L-type calcium channel; CaMKII, calmodulin (CaM)-activated kinase II.
The regulation of neuroinflammation in neuropathic pain development
MiRNAs have been demonstrated to be involved in almost all known biological processes and many pathophysiological conditions, including neuropathic pain. The activation of thermoreceptors and mechanoreceptors in sensory neurons, which takes place in chronic inflammatory pain conditions, leads to the pathophysiology of neuropathic pain, which includes polyneuropathies, fibromyalgia syndrome, complex regional pain syndrome (CRPS), and postherpetic neuralgia (PHN) (Sayed and Abdellatif, 2011). Numerous miRNAs are becoming more widely recognized as master switches in the pathophysiology condition and as regulators of various neuroinflammation and neuronal gene expression (Figure 2). Differential expression of various miRNAs has been directly reported in the dorsal root ganglion (DRG) after the induction of inflammatory and neuropathic pain (Fang et al., 2022). Therefore, it is necessary to discuss how miRNAs regulate the infiltration of immune cells and neuroinflammation after nerve injury.
Immune cell infiltration
It is well known that neuropathic pain is associated with immediate immune cell infiltration following peripheral nerve injury. Recent studies suggest that miRNAs secreted from immune and nonimmune cells exert a pivotal effect on immune regulation (Figure 2). miR-590-3p regulates the infiltration of immune cells into neural tissues in diabetic peripheral neuropathic pain (Wu et al., 2020). Similar to this, after spinal nerve ligation (SNL), there was a decrease in miR-214-3p in the spinal astrocytes of rats, which caused them to become overactive by upregulating CSF1 (Liu L. et al., 2020). Additionally, alterations in the microglial cells’ miRNA profiles imply that these cells have distinct functions depending on the tissue and/or the stage of the pathology.
Neuroinflammation
Early research shown that miRNAs can either activate or suppress the immune system, suggesting that they may play a part in the onset and progression of inflammatory and autoimmune illnesses, including neuropathic pain. During the early-to-late phases of the illnesses, there have also been reports of changes in the expression of certain miRNAs (Figure 2). By signalling through the IRAK/TRAF6, TLR4/NF-κB, TXNIP/NLRP3 inflammasome, MAP kinase, and TNF-α and TLR5 signalling pathways, miRNAs mediate their effects in neuropathic pain (Gada et al., 2021).
One of the most crucial receptors for innate immunity, TLR stimulates the generation of pro-inflammatory cytokines, initiates the synthesis of inflammatory mediators that cause fever, pain, and other inflammation, and exerts some regulatory control over the inflammatory response (Tang et al., 2023). Tumour necrosis factor receptor-associated factor 6 (TRAF6), a critical mediator of TLR signalling, NF-κB activation, and proinflammatory cytokine and interferon expression, was upregulated after exposure to TNF-α or IL-1β in cultured astrocytes from mice following SNL. The protein known as IL-1 receptor-associated kinase (IRAK) is involved in the signaling of MyD88. The link between IRAK and TRAF6 encourages additional nuclear factor κ-activated B-cell light chain enhancer (NF-κB) and Janus kinase N-terminal region activation (JAK) (Zarezadeh Mehrabadi et al., 2022). By inhibiting IRAK1 and TRAF6 mRNA 3’UTR sections in TLR signaling pathways and lowering their protein production, targeting miRNA-146a-5p has a detrimental impact on NF-κB activation, the NLRP3 inflammasome signaling pathway, and NP levels (Wang et al., 2018; Hou et al., 2021). miR-381 overexpression alleviated neuropathic pain behaviours in chronic constriction nerve injury (CCI) rats by inhibiting the expression of HMGB1. Moreover, this effect was reversed by miR-381 inhibitors (Xia et al., 2018; Zhan et al., 2018). Similarly, overexpression of miR-362-3p significantly suppressed the elevation of the levels of proinflammatory cytokines by regulating the expression level of BAMBI, which in turn hindered the neuroinflammatory process and NP in CCI mice (Zhang et al., 2022).
Members of the transforming growth factor β (TGF-β) family are secreted cytokines that control a range of biological processes, including as cell division, migration, survival, and differentiation (Nickel et al., 2018). The TGF-family includes transforming growth factor α (TNF-α), which is closely related to NP. A member of the C-C chemokine family, C-C motif chemokine ligand 2 (CCL2) has a strong affinity for C-C chemokine receptor type 2 (CCR2). According to a recent study, overexpression of miR-183 inhibits the TGF-α/CCL2/CCR2 signalling axis, which in turn inhibits the expression of proinflammatory cytokines (IL-6, IL-1β, TNF-α), as well as pain-related markers (TRPV1, Nav1.3, Nav1.7, Nav1.8), in the DRG (Tao et al., 2021). Moreover, zinc finger E-box-binding homeobox 1 (ZEB1) is a transcription factor that is involved in various diseases by inhibiting ZEB1 expression. miR-28-5p, miR-128-3p, miR-136, miR-150, miR-200b and miR-429 can coordinate the progression of neuroinflammation and neuropathic pain by inhibiting ZEB1 expression (Bao et al., 2018; Yan et al., 2018a,b; Shen et al., 2019).
The G-protein-coupled receptor family, which includes the chemokine CXC receptor 4 (CXCR4), controls the development of glia cells and neurons in the central nervous system (Bianchi and Mezzapelle, 2020). The involvement of CXCR4 in various nociceptive stimulus response mechanisms is becoming more and more clear. A multifunctional protein called thioredoxin-interacting protein (TXNIP) is necessary for numerous cellular functions including metabolic processes, growth, division, and cell death (Wondafrash et al., 2020). And inflammation is brought on by the intracellular complex known as the NOD-like receptor family pyrin domain-containing protein 3 (NLRP3) inflammasome, which promotes the development and release of the pro-inflammatory cytokines interleukin-1 (IL-1) and IL-18 (He et al., 2016). They all contribute significantly to the onset of inflammation. MiR-23a controls NP in spinal glial cells by specially targeting CXCR4 and the TXNIP/NLRP3 inflammasome axis. To reduce hyperalgesia, intrathecal injection of miR-23a mimicked spinal CXCR4 downregulation by a lentivirus and blocked TXNIP or NLRP3 overexpression (Pan et al., 2018).
When rats were used as a model for NP, the expression of miR-140 and miR-144 was downregulated in the DRG. Furthermore, by targeting sphingosine-1-phosphate receptor 1 (S1PR1) and RASP21 protein activator 1(RASA1), respectively, intrathecal injection of miR-140 and miR-144 agomir resulted in decreased inflammatory factor secretion and ameliorated hyperalgesia (Zhang et al., 2020; Li et al., 2021). Additionally, miR-216a-5p reduced the neuropathic pain that rats experienced after CCI by targeting KDM3A and deactivating the Wnt/β-catenin signalling pathway (Wang and Li, 2021). Signal transducer and activator of transcription 3 (STAT3)’ 3’UTR is the direct target of miR-93, which prevents the disease from developing in CCI rats (Yan et al., 2017).
NF-κB is a key mediator in the inflammatory process. The M1 and M2 macrophages are activated by the activation of the NF-κB pathway, which causes them to release pro-inflammatory cytokines and speed up the inflammatory response process (Poladian et al., 2023). Meanwhile, MAPK is a mitogen activated kinase that controls a number of physiological and pathological processes, including NP, by phosphorylating serine/threonine and tyrosine (Zhang et al., 2023). However, the balance of Th1-Th2 cells, the control of cytokine signaling negative feedback, and the reduction of Th2-induced inflammation are all regulated by a unique family of proteins known as cytokine signaling inhibitors (SOCS). Several inflammatory and anti-inflammatory cytokines stimulate SOCS1 and SOCS3, which then block cytokine function (Sobah et al., 2021). Many researchers noted that miR-155 or miR-221 inhibition alleviated neuropathic pain and neuroinflammation by enhancing suppressor of cytokine signalling 1 (SOCS1) expression via NF-κB and p38-MAPK inhibition (Tan et al., 2015; Xia et al., 2016; Liu Y. et al., 2021). Additionally, inhibition of miR-221 reduced pain and decreased the expression of inflammatory factors (PEG2, BK, IL-6, IL-1β, and TNF-α) by targeting SOCS3 in diabetic peripheral neuropathy (DPN) (Wu et al., 2021). Additionally, spinal cord miR-155 expression was increased in oxaliplatin-induced peripheral neuropathic pain, and the intrathecal injection of a miR-155 inhibitor reduced hyperalgesia in rats, potentially through blocking oxidative stress-TRPA1 pathways (Miao et al., 2019). Complete Freund’s adjuvant (CFA)-induced mechanical allodynia and heat hyperalgesia were reduced by knocking down or blocking miRNA-22, but overexpressing miRNA-22 resulted in pain-like behaviours. In order to activate RNA polymerase II and elevate Mtf1 expression, the enhanced miRNA-22 physically bonded to the Mtf1 promoter. Increased expression of p-ERK1/2, GFAP, and c-Fos in the dorsal horn is proof that the increased Mtf1 expression faciliated spinal central sensitization (Hao et al., 2022).
These results suggest that epigenetic interventions against miRNAs to alleviate neuroinflammation may potentially provide novel therapeutic avenues in treating peripheral nerve injury-induced nociceptive hypersensitivity and neuropathic pain.
Regulation of nerve regeneration
After peripheral nerve injury, the survival of neurons is an essential prerequisite for neural regeneration and functional recovery. According to earlier research, prolonged pain causes spinal cord and peripheral nerve cell body damage or possibly cell death (Cohen et al., 2021).
Recent studies have identified that injured peripheral neurons can activate the secretion of intrinsic neurotrophic factors that promote neuronal survival and axon regeneration, such as nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), and neurotrophin-3 (NT-3) (Keefe et al., 2017). In this regard, miRNAs can also play a role in nerve regeneration (Figure 2). Coincidentally, it was discovered that the DRG has aberrant expression of a number of miRNAs that target genes involved in nerve regeneration (Bai et al., 2007). Moreover, it was suggested that, by increasing the expression of the X-linked inhibitor of apoptosis protein (XIAP), downregulation of miR-192-5p can decrease the apoptosis of nerve cells and aid in the regeneration process following sciatic nerve injury (SNI) (Liu X. et al., 2020). Additionally, the overexpression of miR-210 contributed to neuronal survival by preventing apoptosis through targeting ephrin-A3 (EFNA3), thereby promoting the regeneration of axons (Hu et al., 2016). Kruppel-like factor 4 (KLF4), a really well intrinsic inhibitor of axonal outgrowth and regeneration, can be efficiently silenced by miR-135a and miR-135b to stimulate axonal outgrowth and cortical neuron migration (van Battum et al., 2018).
In spinal cord injury (SCI) rats, direct 3D fiber hydrogel scaffold implantation combined with continuous supply of a cocktail of axon miRNAs (miR-132, miR-222, and miR-431) dramatically improved axon regeneration (Zhang et al., 2021). In earlier research, miR-125b overexpression supported axon regeneration after spinal cord injury through controlling the JAK/STAT pathway. Furthermore, through lowering neuronal apoptosis and the inflammatory reaction, miR-125b demonstrated a neuronal protective effect (Dai et al., 2018). In vivo, miR-155 deletion enhanced injury-induced expression of SPRR1A, a regeneration-related gene, in neurons and reduced inflammatory signalling in macrophages, thereby enhancing axon regeneration (Gaudet et al., 2016). Mice with an optic nerve compression greatly increased their ability to regenerate their axons in vivo by enhancing the levels of miR-19a in their retinal ganglion cells (Mak et al., 2020). Additionally, injury-induced changes in the expression of miR-21 and miR-199a-3p changed the ability of axons to develop by altering both systemic and intra-axonal protein synthesis through control of the PTEN/mTOR pathway (Kar et al., 2021). The PTEN/mTOR pathway is a major factor in determining axonal regeneration. The tumor suppressor phosphatase and tensin homologue (PTEN) is a PIP3 phosphatase, and an inhibitor of mTOR signaling. The serine/threonine protein kinase known as the mechanistic target of rapamycin (mTOR), whose activation improved protein synthesis and mRNA translation (Tang, 2018). They synergistically regulate axon growth. MiRNA-21 controls the expression of TGF-I, TIMP3, and EPHA4 target genes, which is crucial for increasing Schwann cell (SC) proliferation and axon regeneration during the healing of damaged peripheral nerves (Ning et al., 2020). Moreover, endogenous miR-26a in sensory neurons enhanced the regeneration of sensory axons after spinal cord injury (SNI) by promoting the induced activation of Smad1 and inhibiting the expression of glycogen synthase kinase 3β (GSK3β) (Jiang et al., 2015).
In contrast, many miRNAs can also negatively regulate neuronal regeneration, thereby supporting neuropathic progression. By controlling the epidermal growth factor receptor (EGFR) pathway, miR-21, for instance, promotes the hyperactivation of astrocytes and the development of glial scar tissue, preventing the regeneration of axons (Li et al., 2018). Additionally, miR-455-5p inhibition suppressed axonal growth and regeneration and downregulated activation of the GSK3β/Tau protein pathway in murine sensory neurons (Su et al., 2020). miR let-7 and miR-9 hindered axonal regeneration through inhibition of the protein levels of Ntn1 and Dcc (Wang et al., 2019), respectively. Likewise, researchers found that axon regeneration is also driven by miR-9 through regulation of FoxP1 triggered by injury. However, sensory neurons with high endogenous miR-9 levels were unable to regenerate their axons (Jiang et al., 2017). Additionally, a novel mechanism for regulating the capacity for intrinsic axon regeneration is provided by the mutual negative response regulatory loop formed by miR-138 and SIRT1 (Liu et al., 2013).
These findings present a novel idea for the future study of axon regeneration in neuropathic pain. There are still many obstacles to be overcome in order to create therapies that achieve complete regeneration and functional recovery of neurons, even though substantial advances have been made in comprehending the fundamental mechanisms of peripheral nerve regeneration and how these pathways can be effectively utilized to promote regeneration after peripheral nerve injury (PNI).
Regulation of neuronal ion channels
After the nerve fiber is injured, the structure and function of ion channels in the nerve endings and DRG of the spinal cord may change, leading to the ectopic discharge of neurons, and neuropathic pain (Jiang et al., 2022). Thus, ion channels play a key role in neuronal excitability and may be targets of miRNAs under pain conditions (Figure 2). Notably, voltage-gated channels involved in the pain pathway have become the main targets of neuropathic pain treatment interventions.
Voltage-gated sodium channels
Voltage-gated sodium channel Nav1.3, an isoform that is sensitive to tetrodotoxin and is encoded by SCN3A, can produce sodium ion currents with quick repriming dynamics. These currents can promote repetitive firing patterns and ectopic discharge in damaged neurons, which can enhance neuronal hyperexcitability and are strongly linked to neuropathic pain (Lindia et al., 2005). Similar sodium-ion channels also have Nav1.6, encoded by SCN8A, and Nav 1.7, encoded by SCN9A (Bennett et al., 2019).
MiR-96 administered intrathecally inhibited the expression of Nav1.3 brought on by CCI. Further investigation indicated that miR-96 decreased the in vitro expression of Nav1.3 mRNA in embryonic DRG neurons (Chen et al., 2014). By controlling SCN3A, miR-384-5p inhibits the emergence of neuropathic pain (Ye et al., 2020). By controlling Nav1.7 in rats, miR-182 reduced the neuropathic pain brought on by SNI (Cai et al., 2018). Surprisingly, overexpression of miR-30b lowered the expression of Nav1.3, Nav1.6, and Nav1.7 both in DRG neurons and the spinal cord, which greatly reduced neuropathic pain brought on by SNL or oxaliplatin (Shao et al., 2016; Su et al., 2017; Li L. et al., 2019). Consistently, miR-183 overexpression attenuated osteoarthritic pain by inhibiting the production of Nav1.3, Nav1.7, and Nav1.8 (Tao et al., 2021).
miR-7a overexpression in primary sensory neurons of injured DRGs suppressed the increase in the β2 subunit of the voltage-gated sodium channel and normalized the long-lasting hyperexcitability of nociceptive neurons (Sakai et al., 2013).
Voltage-gated potassium channels
Voltage-gated potassium channels play a crucial role in controlling the excitability of neurons by altering the production of action potentials, the pace at which neurons fire, or the release of neurotransmitters (Kim and Nimigean, 2016). miR-17-92, a miRNA cluster that includes six different members, downregulated the expression of potassium channels and reduced outward potassium currents, especially type A currents, resulting in the generation of mechanical allodynia (Sakai et al., 2017). Additionally, blocking miR-137 reduced mechanical allodynia and thermal hyperalgesia, recovered aberrant Kv currents and the overactivity of DRG neurons, and restored the expression of the potassium channel Kv1.2 (Zhang et al., 2021). Additionally, miR-183-5p contributed to the control of CCI-induced NP by suppressing TREK-1 expression (a Kv channel) (Shi et al., 2018).
Calcium channels
In addition to voltage-gated sodium channels and potassium channels, calcium channels also play an indispensable role in the process of pain sensitization after nerve injury due to their involvement in neurotransmitter release and the regulation of neuronal excitability and intracellular changes, including gene induction (Rahman and Dickenson, 2013). MiR-219 and miR-124a, which negatively influenced the expression of spinal CaMKIIγand the proinflammatory marker MeCP2, were dramatically downregulated in murine spinal neurons following the development of inflammatory pain by either CFA or formalin injection (Kynast et al., 2013; Pan et al., 2014). Additionally, miR-103 expression was shown to be downregulated in spinal neurons of SNL rats, which appeared to simultaneously control the translational levels of the three components that make up the Cav1.2-comprising L-type calcium channel (Cav1.2-LTC), a calcium ion channel associated with pain sensitization (Favereaux et al., 2011). Additionally, by targeting Cav3.2 channels, histone methylation-mediated miR-32-5p decreased expression in trigeminal ganglion (TG) neurons controls trigeminal NP (Qi et al., 2022). By controlling the auxiliary voltage-gated calcium channel subunits α2δ-1 and α2δ-2, the miR-183 cluster in mice regulated more than 80% of NP-regulated genes and attributed to scaling basal mechanical sensitivity and mechanical allodynia (Peng et al., 2017).
These findings suggest that miRNA-mediated channel dysfunction is a significant contributor to the pathogenesis of nerve injury-induced NP, highlight the significance of abnormal afferent input in the persistence of neuropathic pain and the promise of targeted chemogenetic silencing as a potential neuropathic pain therapy.
Role of exosomal miRNAs in neuropathic pain
Exosomes (Exos), a class of nanosized EVs with sizes from 40 to 200 nm, are released from all cell types and participate in paracrine interactions between various cells, including neurons, glial cells, mesenchymal stem cells, endothelial cells, and leukocytes (Mathivanan et al., 2010). As another type of secreted factor, these biological nanocarriers, which are rich in a variety of genetic materials, including miRNAs, long noncoding RNA, proteins, and lipids, can be easily distributed in biofluids and modulate biochemical responses and cell viability during physiological and pathological conditions in neurodegenerative or inflammatory diseases (Cata et al., 2022). Blood, saliva, breast milk, urine, and other bodily fluids all contain significant amounts of exosomal miRNAs (Zhang et al., 2015; Ding et al., 2019).
Pain is frequently caused by inflammation. A variety of cytokines, chemokines, and additional elements contribute to the emergence of acute inflammatory pain. Chronic inflammation can start the processes that lead to cerebral and peripheral sensitization (Jiang et al., 2020). Exosomal miRNAs can be transported to different sites after autocrine production, acting on macrophages, microglia, neurons or other tissue cells, and regulate the process of neuropathic pain by participating in the secretion of inflammatory factors, and oxidative stress, and regulating neural remodelling or nerve regeneration (Figure 3). Exosomes have the ability to regulate the release from cells of nociceptive mediators, which are involved in neuroinflammation and are recognized to sensitize sensory terminals (Groot and Lee, 2020). For instance, immunological cells such as T lymphocytes and antigen-presenting dendritic cells (DCs) can release and absorb exosomal miRNAs, indicating that exosomal transfer of miRNAs may constitute a novel method of intercellular communication (Torralba et al., 2018). As a result, it is believed that exosomal miRNA transmission is significant for a number of systems and processes, such as the immune reaction and neuron–glia communication (Kalani et al., 2014).
Figure 3

Roles of exosomal miRNAs in neuropathic pain. Exosomal miRNAs can be transported to different sites after autocrine production, acting on macrophages, microglia, neurons or other tissue cells, and regulate the process of neuropathic pain by participating in the secretion of inflammatory factors, and oxidative stress, and regulating neural remodelling or nerve regeneration.
Interestingly, these exosomal miRNAs can have both proinflammatory and anti-inflammatory effects. As mentioned above, some exosomal miRNAs can release cytokines or other proinflammatory mediators that directly act on target organs (Zhang et al., 2014; Console et al., 2019). For example, exosomes from chondrocytes, neutrophils, and synovial fibroblasts encouraged macrophages to produce IL-1 and metalloproteinases. Furthermore, the regulation of the synthesis of these inflammatory chemicals has been linked to the transfer of miR-206 and miR-449a-5p in the cargo of exosomes (Liu et al., 2018; Ni et al., 2019). MiR-449a-5p in particular mediates ATG4B inhibition, which in turn controls macrophage autophagy, encourages inflammosome activation, and exacerbates the inflammatory response.
Evidence suggests that miR-21 was increased in serum exosomes from neuropathic mice. Further research revealed that miR-21-5p-containing macrophage-derived exosomes promoted pyroptosis via A20, encouraging a proinflammatory phenotype and exacerbating podocyte damage in diabetic nephropathy mice (Ding et al., 2021). Notably, the DRG’s hyperalgesia and macrophage recruitment were reduced by either intrathecal miR-21-5p antagomir addition or miR-21 conditional deletion in sensory neurons. A20 is an inhibitor of the NF-κB signaling pathway. Similar research showed that after nerve damage, DRG sensory neurons released EVs that were miR-23a-enriched and were then ingested by macrophages to improve M1 polarization in vitro. Additionally, by blocking A20 to promote NF-κB signaling, an EV-miR-23a antagomir delivered intrathecally reduced M1 macrophages and improved neuropathic hyperalgesia (Zhang et al., 2021).
Conversely, other exosomal miRNAs exert anti-inflammatory and analgesic effects in chronic pain models via the transfer of therapeutic factors to injured neurons in the central nervous system (CNS) and peripheral nervous system (PNS) (Ren J. et al., 2019). The promotion of inflammation is thought to be the leading cause of pain. Exosomal miRNAs can also suppress the production of proinflammatory cytokines such as IL-1β, IL-6, TNF-α, and PGE2 in injured areas and stimulate the release of IL-10, leading to antinociceptive effects (Cata et al., 2022). These miRNAs have the ability to modify nociception, and intrathecal administration of miR-124, miR-103, miR-23b, miR-25, and miR-544, for example, reduced inflammatory and neuropathic pain by modifying intracellular neuronal, astrocytic, and microglial activities (Favereaux et al., 2011; Willemen et al., 2012; Wang et al., 2018; Zhao et al., 2019; Li et al., 2020). MiR-124 inhibits GRK2 expression, thereby regulating the M1/M2 phenotypic balance of the spinal cord.
Similar to the effect of some neurotrophic factors, including GDNF, IGF-1, BDNF, NGF, and FGF-1, exosomal miRNAs can enhance axonal growth and neuronal viability and intensify therapeutic effects (Lv et al., 2021). To the best of our knowledge, the targeted and modular EV loading (TAMEL) method has not been implemented in experimental pain studies. Despite the fact that many studies have tried to provide new and more effective mechanistic insights into the function of exosomal miRNAs in NP, the cellular and molecular functions of exosomal miRNAs and their downstream targets remain to be elucidated.
Emerging role of exosomal miRNAs in neuropathic pain management
Standard analgesics, such as acetaminophen, nonsteroidal anti-inflammatory drugs, local anesthetics, and, to a lesser extent, opioids, may be helpful in controlling acute pain. Unfortunately, due to ineffectiveness or undesirable side effects, there are few clinically useful analgesics for the treatment of neuropathic pain (Amaechi et al., 2021). Therefore, present research investigations should give top priority to the identification and development of mechanism-based therapies for the amelioration of neuropathic pain.
Exosomes have the capacity to control NP and can be secreted by a variety of cell types, including stem cells, according to laboratory research (Ren J. et al., 2019). Further evidence suggests that stem cell-derived exosomes can largely mimic the functional effects of parental stem cells and have been identified as key players in the stem cell repair of damaged tissues (Keshtkar et al., 2018). Stem cell-derived exosomes can not only play a role in nerve repair but also avoid the risk of immunosuppression, genetic modification and malignant transformation caused by stem cell transplantation due to their paracrine effect, providing a new therapeutic strategy and research target for neuropathic pain (Liu W. Z. et al., 2021). It is believed that stem cell-derived exosomes can transfer neurotrophic factors, such as GDNF, IGF-1, BDNF, NGF, and FGF-1, to injured neurons. Additionally, intrathecal infusion of mesenchymal stem cell exosomes reduces neuropathic pain in spinal cord injured rats by causing microglia to become polarized from M1 to M2 and preventing the release of inflammatory cytokines such TNF-α, IL-1, IL-6, and NF-κB (Harrell et al., 2019; Arabpour et al., 2021; Liu W. Z. et al., 2021). Among them, the regulatory effect of the miR-216a-5p/TLR4 axis on microglial polarization has been demonstrated (Liu W. et al., 2020).
Mesenchymal stem cells (MSCs) are one of the most promising stem cell types for the treatment of various ischaemic diseases and tissue damage due to their multidirectional differentiation potential and extensive immune regulatory functions (Uccelli et al., 2008). A recent laboratory study indicated that MSCs can migrate to the injured nerve tissue and stimulate the regeneration of injured neurons (Norte-Muñoz et al., 2021). MSC-derived exosomes regulate neurite growth by controlling the number and total length of neurites through the transfer of miR-133b to nerve cells (Xin et al., 2012, 2013; Ren Z. W. et al., 2019). Exosomes enriched in miR-17-92 clusters may increase neuroplasticity and functional recovery by targeting PTEN to activate the PI3K/Akt/mTOR/GSK-3β signalling pathway (Xin et al., 2017). Similarly, SC-derived small exosomes containing miR-21-5p negatively regulate PTEN to improve sensory neuron growth and survival (Cong et al., 2021). In order to increase the capacity for neurite outgrowth in vitro and nerve regeneration in vivo, SCs-extracellular vehicles (EVs) transported miR-23b-3p from mechanically activated SCs to neurons and decreased neuronal neuropilin 1(Nrp1) expression (Xia et al., 2020). Umbilical cord mesenchymal stem cell-derived exosomes boost axon regrowth and spinal cord functional improvement via miR-199a-3p/145-5p targeting of Cblb/Cbl-mediated NGF/TrkA signaling in rats (Wang et al., 2021). Following spinal cord damage, exosomes released from neural stem cells exposed to IGF-1 decreased apoptosis and promoted nerve regeneration, at least in part through a miR-219a-2-3p/YY1 mechanism (Ma et al., 2019). Similarly, miR-181c-5p negatively regulates Bcl-2-interacting cell death mediators (BIM), which can effectively inhibit neuronal apoptosis and regulate the cell vitality of cortical neurons to promote axon regeneration (Zhang et al., 2019; Figure 4).
Figure 4
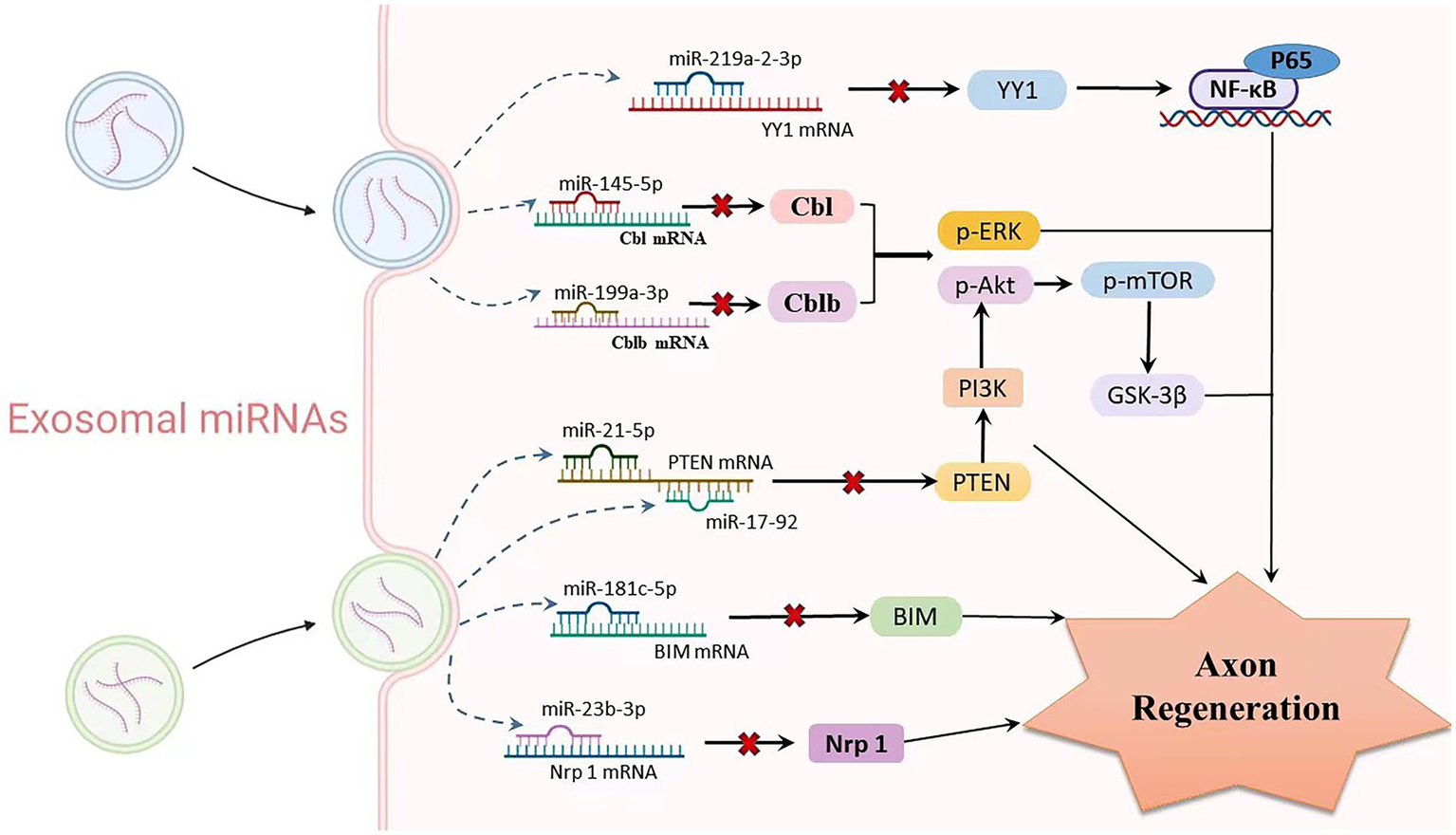
Relationship between exosomal miRNAs and axon regeneration. As one of the important cargos of exosomes, miRNAs are involved in mediating the process of axon regeneration. Exosomes produced by various stem cells enter neurons or glial cells through paracrine routes, where multiple miRNAs are released. Upregulation of exo-miR-219a-2-3p inhibits YY1 expression, thereby suppressing the NF-κB-p65 pathway and exerting neuroprotective effects. miR-199a-3p/145-5p suppresses Cblb and Cbl expression and causes upregulation of p-Erk and p-Akt, subsequently promoting neurite outgrowth. The miR-17-92 cluster and miR-21-5p carrying exosomes downregulate PTEN levels and subsequently activate the PI3K/Akt/mTOR signalling pathway, thereby increasing neuronal dendritic plasticity. MiR-181c-5p negatively regulates BIM mRNA, inhibits BIM expression level, reduces neuronal apoptosis, and promotes axon regeneration. Targeted inhibition of Nrf1 mRNA levels by miR-23b-39 subsequently promotes axonal regeneration.
A recent study suggested that miR-29-enriched exosomes originate from stem cells and alleviate proinflammatory responses in an osteoarthritis in a rat model (Le et al., 2016). Similarly, miR-199-3p overexpression attenuated TRX-induced PHN by targeting MECP2 in mice (Wang et al., 2022). ExomiR-181c-5p was incorporated by microglial cells and prevented inflammatory substances from being released. In addition, intrathecal exomiR-181c-5p treatment reduced neuroinflammatory symptoms and neuropathic pain in CCI rats (Zhang et al., 2022). In order to reduce neuroinflammation, human umbilical cord MSC-derived exosomes upregulated the expression of proteins associated with autophagy (LC3-II and beclin1) and blocked the activation of NLRP3 inflammasomes through miR-146a-5p/TRAF6 in the spinal cord dorsal horn (Hua et al., 2022). Furthermore, cyclooxygenase-2 (PTGS2) was downregulated in rat synovial fibroblasts after xenogenic injection of human MSC exosomes enriched with miR-26a-5p to lessen pathogenic alterations (Jin et al., 2020).
MSC-derived exosomes can exert analgesic effects in chronic pain models via the transfer of certain miRNAs in the CNS and PNS (Hmadcha et al., 2020). Exosomal miRNAs can be important biomarkers, and compared with free-floating miRNAs, they have the following advantages: (Scholz et al., 2019) exosomes contain a range of miRNAs, making them reliable carriers for the study of miRNAs; (Yi et al., 2021) the bilayer membrane shape of exosomes can improve miRNA stability, susceptibility to miRNA amplification, and the likelihood of unfavorable outcomes; and (Huh et al., 2017) exosomes can cross the blood–brain or blood-spinal cord barriers (Ding et al., 2019). Despite the potential advantages of MSC-derived exosomes, the low therapeutic effect due to poor survival of transplanted cells in damaged tissues is still the largest obstacle in stem cell therapy.
These data suggest that stem cell-derived exosomal miRNAs can manage pain by reducing proinflammatory cytokines and promoting neuronal regeneration and differentiation, which presents a novel therapeutic strategy for the treatment of nerve injury.
Perspectives on this review
In conclusion, neuropathic pain poses a substantial threat to patients’ lives, health, and quality of life, and the absence of safe and effective treatment options continues to be a significant therapeutic challenge. In recent years, the in-depth study of miRNA in the development of neuropathic pain has provided great opportunities for its clinical transformation, especially its outstanding contribution in neuroinflammation, nerve regeneration and other aspects has brought another bright prospect for the clinical treatment of neuropathic pain. Even though it has been demonstrated that miRNA is a viable candidate for NP therapy, obstacles such its bioactivity, stability, safety, and tissue specificity still need to be overcome. On the journey to the target, miRNA’s transmembrane efficiency and enzymatic reaction are additional factors that must be taken into account. The study of delivery molecules, including exosomes, liposomes, viral vectors, as well as miRNA mimics and inhibitors, is therefore currently given significant emphasis. Last but not least, patient safety has to be the top priority in clinical application, making it difficult for us to effectively assess the immunological response that exogenous miRNA treatment may trigger. Further preclinical research and clinical trials are crucial measures to support clinical transformation because current research is still confined to cell and animal investigations. With further research on the mechanism of action of miRNAs and the use of the latest miRNA gene chips and other high-throughput technologies, miRNAs may become a new biological marker for disease diagnosis and will provide new targets and methods for the pathogenesis and intervention strategies of neuropathic pain in the future.
Funding
This work was supported by grants from the Tianjin Natural Science Foundation Project (grant no. 20JCYBJC00370), Tianjin, China, and the Tianjin Key Medical Discipline (Specialty) Construction Project (grant no. TJYXZDXK-017A).
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Author contributions
Y-YZ wrote the manuscript and made illustrations. Z-JW, L-JZ, and T-XN provided advice for the manuscript. BL and JL provided the supervision and comments on the manuscript. All authors have read and approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
| miRISC | miRNA-induced silencing complex |
| DRG | Dorsal root ganglion |
| SNL | Spinal nerve ligation model |
| CSF1 | Colony-stimulating factor 1 |
| IRAK | IL-1R-associated kinase |
| TRAF6 | Tumour necrosis factor receptor-associated factor 6 |
| TLR | Toll-like receptors |
| NF-κB | Nuclear factor κB |
| TXIP | Thioredoxin-interacting protein |
| NLRP3 | NOD-like receptor protein 3 |
| TNF-α | Tumour necrosis factor α |
| IL-1β | Interleukin-1β |
| 3’UTR | 3′ untranslated region |
| CCI | Chronic constriction nerve injury |
| HMGB1 | High mobility group box 1 |
| BAMBI | Bone morphogenetic protein and activin membrane-bound inhibitor |
| NP | Neuropathic pain |
| TRPV1 | Transient receptor potential (TRP) vanilloid 1 |
| TGF-α | Transforming growth factor-α |
| CCL2 | CC chemokine ligand 2 |
| CCR2 | CC chemokine receptor type-2 |
| ZEB1 | Zinc finger E-box-binding homeobox 1 |
| CXCR4 | Chemokine CXC receptor 4 |
| TXNIP | Thioredoxin interacting protein |
| S1PR1 | Sphingosine-1-phosphate receptor 1 |
| RASA1 | RAS P21 protein activator 1 |
| KDM3A | Lysine-specific demethylase 3A |
| STAT3 | Activator of transcription 3 |
| SOCS1 | Suppressor of cytokine signalling 1 |
| MAPK | Mitogen-activated protein kinase |
| p38 | p38 mitogen-active protein kinase |
| PEG2 | Prostaglandin E2 |
| SOCS3 | Suppressor of cytokine signalling 3 |
| DPN | Diabetic peripheral neuropathy |
| CFA | Complete Freund’s adjuvant |
| Mtf1 | Mitochondrial Transcription Factor 1 |
| p-ERK1/2 | Phosphorylation of ERK1/2 |
| GFAP | Glial fibrillary acidic protein |
| NGF | Nerve growth factor |
| BDNF | Brain-derived neurotrophic factor |
| NT-3 | Neurotrophin-3 |
| FGF | Fibroblast growth factor |
| XIAP | X-linked inhibitor of apoptosis protein |
| EFNA3 | Ephrin-A3 |
| KLF4 | Kruppel-like factor 4 |
| SCI | Spinal cord injury |
| JAK | Janus kinase gene |
| STAT | Signal transducer and activator of transcription |
| SPRR1A | Small proline-rich repeat protein 1A |
| PTEN | Phosphatase and tensin homologue |
| mTOR | Mechanistic target of rapamycin |
| SC | Schwann cells |
| GFβI | Transforming growth factor-beta-induced protein |
| TIMP3 | Tissue inhibitor of metalloproteinases 3 |
| EPHA4 | Erythropoietin-producing human hepatocellular receptor A4 |
| GSK3β | Glycogen synthase kinase 3β |
| EGFR | Epidermal growth factor receptor |
| Ntn1 | Netrin-1 |
| Dcc | Deleted in colorectal cancer |
| FoxP1 | Forkhead box protein P1 |
| SIRT1 | Sirtuin 1 |
| Nav | Voltage-gated sodium channels |
| SNL | Spinal nerve ligation model |
| Kv | Voltage-gated potassium |
| TREK-1 | TWIK-related K channel 1 |
| MeCP2 | Methyl-CpG binding protein 2 |
| Cav1.2-LTC | Cav1.2-comprising L-type calcium channel |
| CaMKII | Calmodulin (CaM)-activated kinase II |
| TG | Trigeminal ganglion |
| EVs | Extracellular vesicles |
| DCs | Dendritic cells |
| CNS | Central nervous system |
| PNS | Peripheral nervous system |
| MS | Mechanical stimulation |
| Nrp | Neuropilin |
| PHN | Postherpetic neuralgia |
| TRX | Thioredoxin |
| MECP2 | Methyl-CpG-binding protein 2 |
| IGF-1 | Insulin-like growth factor I |
| MSCs | Mesenchymal stem cells |
| PTGS2 | Prostaglandin-endoperoxide synthase 2. |
References
1
Amaechi O. Huffman M. M. Featherstone K. (2021). Pharmacologic therapy for acute pain. Am. Fam. Physician104, 63–72. (Accessed July 1, 2021)
2
Arabpour M. Saghazadeh A. Rezaei N. (2021). Anti-inflammatory and M2 macrophage polarization-promoting effect of mesenchymal stem cell-derived exosomes. Int. Immunopharmacol.97:107823. doi: 10.1016/j.intimp.2021.107823
3
Bai G. Ambalavanar R. Wei D. Dessem D. (2007). Downregulation of selective microRNAs in trigeminal ganglion neurons following inflammatory muscle pain. Mol. Pain3:15. doi: 10.1186/1744-8069-3-15
4
Bao Y. Wang S. Xie Y. Jin K. Bai Y. Shan S. (2018). MiR-28-5p relieves neuropathic pain by targeting Zeb1 in CCI rat models. J. Cell. Biochem.119, 8555–8563. doi: 10.1002/jcb.27096
5
Barwari T. Joshi A. Mayr M. (2016). MicroRNAs in cardiovascular disease. J. Am. Coll. Cardiol.68, 2577–2584. doi: 10.1016/j.jacc.2016.09.945
6
Bennett D. L. Clark A. J. Huang J. Waxman S. G. Dib-Hajj S. D. (2019). The role of voltage-gated sodium channels in pain signaling. Physiol. Rev.99, 1079–1151. doi: 10.1152/physrev.00052.2017
7
Bernardo B. C. Ooi J. Y. Y. Lin R. C. Y. McMullen J. R. (2015). miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart. Future Med. Chem.7, 1771–1792. doi: 10.4155/fmc.15.107
8
Bianchi M. E. Mezzapelle R. (2020). The chemokine receptor CXCR4 in cell proliferation and tissue regeneration. Front. Immunol.11:2109. doi: 10.3389/fimmu.2020.02109
9
Cai W. Zhao Q. Shao J. Zhang J. Li L. Ren X. et al . (2018). MicroRNA-182 alleviates neuropathic pain by regulating Nav1.7 following spared nerve injury in rats. Sci. Rep.8:16750. doi: 10.1038/s41598-018-34755-3
10
Cata J. P. Uhelski M. L. Gorur A. Dougherty P. M. (2022). Nociception and pain: new roles for Exosomes. Neuroscientist28, 349–363. doi: 10.1177/10738584211027105
11
Chen H.-P. Zhou W. Kang L.-M. Yan H. Zhang L. Xu B.-H. et al . (2014). Intrathecal miR-96 inhibits Nav1.3 expression and alleviates neuropathic pain in rat following chronic construction injury. Neurochem. Res.39, 76–83. doi: 10.1007/s11064-013-1192-z
12
Cohen S. P. Vase L. Hooten W. M. (2021). Chronic pain: an update on burden, best practices, and new advances. Lancet397, 2082–2097. doi: 10.1016/S0140-6736(21)00393-7
13
Cong M. Shen M. Wu X. Li Y. Wang L. He Q. et al . (2021). Improvement of sensory neuron growth and survival via negatively regulating PTEN by miR-21-5p-contained small extracellular vesicles from skin precursor-derived Schwann cells. Stem Cell Res Ther12:80. doi: 10.1186/s13287-020-02125-4
14
Console L. Scalise M. Indiveri C. (2019). Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta488, 165–171. doi: 10.1016/j.cca.2018.11.009
15
Dai J. Xu L. J. Han G. D. Sun H. L. Zhu G. T. Jiang H. T. et al . (2018). MicroRNA-125b promotes the regeneration and repair of spinal cord injury through regulation of JAK/STAT pathway. Eur. Rev. Med. Pharmacol. Sci.22, 582–589. doi: 10.26355/eurrev_201802_14271
16
Ding S.-Q. Chen J. Wang S.-N. Duan F.-X. Chen Y.-Q. Shi Y.-J. et al . (2019). Identification of serum exosomal microRNAs in acute spinal cord injured rats. Exp. Biol. Med.244, 1149–1161. doi: 10.1177/1535370219872759
17
Ding X. Jing N. Shen A. Guo F. Song Y. Pan M. et al . (2021). MiR-21-5p in macrophage-derived extracellular vesicles affects podocyte pyroptosis in diabetic nephropathy by regulating A20. J. Endocrinol. Investig.44, 1175–1184. doi: 10.1007/s40618-020-01401-7
18
Fang Z.-H. Liao H.-L. Tang Q.-F. Liu Y.-J. Zhang Y.-Y. Lin J. et al . (2022). Interactions among non-coding RNAs and mRNAs in the trigeminal ganglion associated with neuropathic pain. J. Pain Res.15, 2967–2988. doi: 10.2147/JPR.S382692
19
Favereaux A. Thoumine O. Bouali-Benazzouz R. Roques V. Papon M.-A. Salam S. A. et al . (2011). Bidirectional integrative regulation of Cav1.2 calcium channel by microRNA miR-103: role in pain. EMBO J.3018, 3830–3841. doi: 10.1038/emboj.2011.249
20
Gada Y. Pandey A. Jadhav N. Ajgaonkar S. Mehta D. Nair S. (2021). New vistas in microRNA regulatory interactome in neuropathic pain. Front. Pharmacol.12:778014. doi: 10.3389/fphar.2021.778014
21
Gaudet A. D. Mandrekar-Colucci S. Hall J. C. E. Sweet D. R. Schmitt P. J. Xu X. et al . (2016). miR-155 deletion in mice overcomes neuron-intrinsic and neuron-extrinsic barriers to spinal cord repair. J. Neurosci.36, 8516–8532. doi: 10.1523/JNEUROSCI.0735-16.2016
22
Gregory R. I. Yan K.-P. Amuthan G. Chendrimada T. Doratotaj B. Cooch N. et al . (2004). The microprocessor complex mediates the genesis of microRNAs. Nature432, 235–240. doi: 10.1038/nature03120
23
Groot M. Lee H. (2020). Sorting mechanisms for microRNAs into extracellular vesicles and their associated diseases. Cells9:1044. doi: 10.3390/cells9041044
24
Han J. Lee Y. Yeom K.-H. Kim Y.-K. Jin H. Kim V. N. (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev.18, 3016–3027. doi: 10.1101/gad.1262504
25
Hao L.-Y. Zhang M. Tao Y. Xu H. Liu Q. Yang K. et al . (2022). miRNA-22 upregulates in dorsal horn neurons and is essential for inflammatory pain. Oxidative Med. Cell. Longev.2022:8622388. doi: 10.1155/2022/8622388
26
Harrell C. R. Jovicic N. Djonov V. Arsenijevic N. Volarevic V. (2019). Mesenchymal stem cell-derived Exosomes and other extracellular vesicles as new remedies in the therapy of inflammatory diseases. Cells8:1605. doi: 10.3390/cells8121605
27
He Y. Hara H. Núñez G. (2016). Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci.41, 1012–1021. doi: 10.1016/j.tibs.2016.09.002
28
Hill M. Tran N. (2021). miRNA interplay: mechanisms and consequences in cancer. Dis. Model. Mech.14:dmm047662. doi: 10.1242/dmm.047662
29
Hmadcha A. Martin-Montalvo A. Gauthier B. R. Soria B. Capilla-Gonzalez V. (2020). Therapeutic potential of Mesenchymal stem cells for cancer therapy. Front. Bioeng. Biotechnol.8:43. doi: 10.3389/fbioe.2020.00043
30
Hou J. Deng Q. Deng X. Zhong W. Liu S. Zhong Z. (2021). MicroRNA-146a-5p alleviates lipopolysaccharide-induced NLRP3 inflammasome injury and pro-inflammatory cytokine production via the regulation of TRAF6 and IRAK1 in human umbilical vein endothelial cells (HUVECs). Ann. Transl. Med.9:1433. doi: 10.21037/atm-21-3903
31
Hu Y.-W. Jiang J.-J. Yan G. Wang R.-Y. Tu G.-J. (2016). MicroRNA-210 promotes sensory axon regeneration of adult mice in vivo and in vitro. Neurosci. Lett.622, 61–66. doi: 10.1016/j.neulet.2016.04.034
32
Hua T. Yang M. Song H. Kong E. Deng M. Li Y. et al . (2022). Huc-MSCs-derived exosomes attenuate inflammatory pain by regulating microglia pyroptosis and autophagy via the miR-146a-5p/TRAF6 axis. J. Nanobiotechnol.20:324. doi: 10.1186/s12951-022-01522-6
33
Huh Y. Ji R.-R. Chen G. (2017). Neuroinflammation, bone marrow stem cells, and chronic pain. Front. Immunol.8:1014. doi: 10.3389/fimmu.2017.01014
34
Hutvágner G. McLachlan J. Pasquinelli A. E. Bálint E. Tuschl T. Zamore P. D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science293, 834–838. doi: 10.1126/science.1062961
35
Jiang J. Hu Y. Zhang B. Shi Y. Zhang J. Wu X. et al . (2017). MicroRNA-9 regulates mammalian axon regeneration in peripheral nerve injury. Mol. Pain13:1744806917711612. doi: 10.1177/1744806917711612
36
Jiang B.-C. Liu T. Gao Y.-J. (2020). Chemokines in chronic pain: cellular and molecular mechanisms and therapeutic potential. Pharmacol. Ther.212:107581. doi: 10.1016/j.pharmthera.2020.107581
37
Jiang J. J. Liu C. M. Zhang B. Y. Wang X. W. Zhang M. Saijilafu Z. S. R. et al . (2015). MicroRNA-26a supports mammalian axon regeneration in vivo by suppressing GSK3β expression. Cell Death Dis.6:e1865. doi: 10.1038/cddis.2015.239
38
Jiang M. Wang Y. Wang J. Feng S. Wang X. (2022). The etiological roles of miRNAs, lncRNAs, and circRNAs in neuropathic pain: a narrative review. J. Clin. Lab. Anal.36:e24592. doi: 10.1002/jcla.24592
39
Jin Z. Ren J. Qi S. (2020). Human bone mesenchymal stem cells-derived exosomes overexpressing microRNA-26a-5p alleviate osteoarthritis via down-regulation of PTGS2. Int. Immunopharmacol.78:105946. doi: 10.1016/j.intimp.2019.105946
40
Kalani A. Tyagi A. Tyagi N. (2014). Exosomes: mediators of neurodegeneration, neuroprotection and therapeutics. Mol. Neurobiol.49, 590–600. doi: 10.1007/s12035-013-8544-1
41
Kar A. N. Lee S.-J. Sahoo P. K. Thames E. Yoo S. Houle J. D. et al . (2021). MicroRNAs 21 and 199a-3p regulate axon growth potential through modulation of Pten and mTor mRNAs. eNeuro8. doi: 10.1523/ENEURO.0155-21.2021
42
Keefe K. M. Sheikh I. S. Smith G. M. (2017). Targeting neurotrophins to specific populations of neurons: NGF, BDNF, and NT-3 and their relevance for treatment of spinal cord injury. Int. J. Mol. Sci.18:548. doi: 10.3390/ijms18030548
43
Keshtkar S. Azarpira N. Ghahremani M. H. (2018). Mesenchymal stem cell-derived extracellular vesicles: novel frontiers in regenerative medicine. Stem Cell Res Ther9:63. doi: 10.1186/s13287-018-0791-7
44
Kim D. M. Nimigean C. M. (2016). Voltage-gated potassium channels: a structural examination of selectivity and gating. Cold Spring Harb. Perspect. Biol.8:a029231. doi: 10.1101/cshperspect.a029231
45
Kynast K. L. Russe O. Q. Möser C. V. Geisslinger G. Niederberger E. (2013). Modulation of central nervous system-specific microRNA-124a alters the inflammatory response in the formalin test in mice. Pain154, 368–376. doi: 10.1016/j.pain.2012.11.010
46
Le L. T. T. Swingler T. E. Crowe N. Vincent T. L. Barter M. J. Donell S. T. et al . (2016). The microRNA-29 family in cartilage homeostasis and osteoarthritis. J. Mol. Med.94, 583–596. doi: 10.1007/s00109-015-1374-z
47
Li Y. Huang J. Guo M. Zuo X. (2015). MicroRNAs regulating signaling pathways: potential biomarkers in systemic sclerosis. Genomics Proteomics Bioinformatics13, 234–241. doi: 10.1016/j.gpb.2015.07.001
48
Li C. Li X. Zhao B. Wang C. (2020). Exosomes derived from miR-544-modified mesenchymal stem cells promote recovery after spinal cord injury. Arch. Physiol. Biochem.126, 369–375. doi: 10.1080/13813455.2019.1691601
49
Li H.-J. Pan Y.-B. Sun Z.-L. Sun Y.-Y. Yang X.-T. Feng D.-F. (2018). Inhibition of miR-21 ameliorates excessive astrocyte activation and promotes axon regeneration following optic nerve crush. Neuropharmacology137, 33–49. doi: 10.1016/j.neuropharm.2018.04.028
50
Li L. Shao J. Wang J. Liu Y. Zhang Y. Zhang M. et al . (2019). MiR-30b-5p attenuates oxaliplatin-induced peripheral neuropathic pain through the voltage-gated sodium channel Na1.6 in rats. Neuropharmacology153, 111–120. doi: 10.1016/j.neuropharm.2019.04.024
51
Li J. Wei G.-H. Huang H. Lan Y.-P. Liu B. Liu H. et al . (2013). Nerve injury-related autoimmunity activation leads to chronic inflammation and chronic neuropathic pain. Anesthesiology118, 416–429. doi: 10.1097/ALN.0b013e31827d4b82
52
Li Y. Yin Z. Fan J. Zhang S. Yang W. (2019). The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct. Target. Ther.4:47. doi: 10.1038/s41392-019-0080-7
53
Li J. Zhu Y. Ma Z. Liu Y. Sun Z. Wu Y. (2021). miR-140 ameliorates neuropathic pain in CCI rats by targeting S1PR1. J. Recept. Signal Transduct. Res.41, 401–407. doi: 10.1080/10799893.2020.1818091
54
Lima R. T. Busacca S. Almeida G. M. Gaudino G. Fennell D. A. Vasconcelos M. H. (2011). MicroRNA regulation of core apoptosis pathways in cancer. Eur. J. Cancer47, 163–174. doi: 10.1016/j.ejca.2010.11.005
55
Lindia J. A. Köhler M. G. Martin W. J. Abbadie C. (2005). Relationship between sodium channel NaV1.3 expression and neuropathic pain behavior in rats. Pain117, 145–153. doi: 10.1016/j.pain.2005.05.027
56
Liu X. Cui X. Guan G. Dong Y. Zhang Z. (2020). microRNA-192-5p is involved in nerve repair in rats with peripheral nerve injury by regulating XIAP. Cell Cycle19, 326–338. doi: 10.1080/15384101.2019.1710916
57
Liu Y. Lin L. Zou R. Wen C. Wang Z. Lin F. (2018). MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle17, 2411–2422. doi: 10.1080/15384101.2018.1526603
58
Liu W.-Z. Ma Z.-J. Li J.-R. Kang X.-W. (2021). Mesenchymal stem cell-derived exosomes: therapeutic opportunities and challenges for spinal cord injury. Stem Cell Res Ther12:102. doi: 10.1186/s13287-021-02153-8
59
Liu W. Rong Y. Wang J. Zhou Z. Ge X. Ji C. et al . (2020). Exosome-shuttled miR-216a-5p from hypoxic preconditioned mesenchymal stem cells repair traumatic spinal cord injury by shifting microglial M1/M2 polarization. J. Neuroinflammation17:47. doi: 10.1186/s12974-020-1726-7
60
Liu Y. Wan X. Yuan Y. Huang J. Jiang Y. Zhao K. et al . (2021). Opposite effects of miR-155 in the initial and later stages of lipopolysaccharide (LPS)-induced inflammatory response. J. Zhejiang Univ. Sci. B22, 590–598. doi: 10.1631/jzus.B2000826
61
Liu C.-M. Wang R.-Y. Saijilafu J. Z.-X. Zhang B.-Y. Zhou F.-Q. (2013). MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev.27, 1473–1483. doi: 10.1101/gad.209619.112
62
Liu L. Xu D. Wang T. Zhang Y. Yang X. Wang X. et al . (2020). Epigenetic reduction of miR-214-3p upregulates astrocytic colony-stimulating factor-1 and contributes to neuropathic pain induced by nerve injury. Pain161, 96–108. doi: 10.1097/j.pain.0000000000001681
63
Lutz B. M. Bekker A. Tao Y.-X. (2014). Noncoding RNAs: new players in chronic pain. Anesthesiology121, 409–417. doi: 10.1097/ALN.0000000000000265
64
Lv B. Zhang X. Yuan J. Chen Y. Ding H. Cao X. et al . (2021). Biomaterial-supported MSC transplantation enhances cell-cell communication for spinal cord injury. Stem Cell Res. Ther.12:36. doi: 10.1186/s13287-020-02090-y
65
Ma K. Xu H. Zhang J. Zhao F. Liang H. Sun H. et al . (2019). Insulin-like growth factor-1 enhances neuroprotective effects of neural stem cell exosomes after spinal cord injury via an miR-219a-2-3p/YY1 mechanism. Aging11, 12278–12294. doi: 10.18632/aging.102568
66
Macfarlane L.-A. Murphy P. R. (2010). MicroRNA: biogenesis, function and role in cancer. Curr. Genomics11, 537–561. doi: 10.2174/138920210793175895
67
Mak H. K. Yung J. S. Y. Weinreb R. N. Ng S. H. Cao X. Ho T. Y. C. et al . (2020). MicroRNA-19a-PTEN axis is involved in the developmental decline of axon regenerative capacity in retinal ganglion cells. Mol. Ther. Nucleic Acids21, 251–263. doi: 10.1016/j.omtn.2020.05.031
68
Mathivanan S. Ji H. Simpson R. J. (2010). Exosomes: extracellular organelles important in intercellular communication. J. Proteome73, 1907–1920. doi: 10.1016/j.jprot.2010.06.006
69
Miao F. Wang R. Cui G. Li X. Wang T. Li X. (2019). Engagement of MicroRNA-155 in exaggerated oxidative stress signal and TRPA1 in the dorsal horn of the spinal cord and neuropathic pain during chemotherapeutic oxaliplatin. Neurotox. Res.36, 712–723. doi: 10.1007/s12640-019-00039-5
70
Ni Z. Kuang L. Chen H. Xie Y. Zhang B. Ouyang J. et al . (2019). The exosome-like vesicles from osteoarthritic chondrocyte enhanced mature IL-1β production of macrophages and aggravated synovitis in osteoarthritis. Cell Death Dis.10:522. doi: 10.1038/s41419-019-1739-2
71
Nickel J. Ten Dijke P. Mueller T. D. (2018). TGF-β family co-receptor function and signaling. Acta Biochim. Biophys. Sin. Shanghai50, 12–36. doi: 10.1093/abbs/gmx126
72
Ning X.-J. Lu X.-H. Luo J.-C. Chen C. Gao Q. Li Z.-Y. et al . (2020). Molecular mechanism of microRNA-21 promoting Schwann cell proliferation and axon regeneration during injured nerve repair. RNA Biol.17, 1508–1519. doi: 10.1080/15476286.2020.1777767
73
Norte-Muñoz M. Lucas-Ruiz F. Gallego-Ortega A. García-Bernal D. Valiente-Soriano F. J. de la Villa P. et al . (2021). Neuroprotection and axonal regeneration induced by bone marrow mesenchymal stromal cells depend on the type of transplant. Front. Cell Dev. Biol.9:772223. doi: 10.3389/fcell.2021.772223
74
Olive V. Minella A. C. He L. (2015). Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci. Signal.8:re2. doi: 10.1126/scisignal.2005813
75
Pan Z. Shan Q. Gu P. Wang X. M. Tai L. W. Sun M. et al . (2018). miRNA-23a/CXCR4 regulates neuropathic pain via directly targeting TXNIP/NLRP3 inflammasome axis. J. Neuroinflammation15:29. doi: 10.1186/s12974-018-1073-0
76
Pan Z. Zhu L.-J. Li Y.-Q. Hao L.-Y. Yin C. Yang J.-X. et al . (2014). Epigenetic modification of spinal miR-219 expression regulates chronic inflammation pain by targeting CaMKIIγ. J. Neurosci.34, 9476–9483. doi: 10.1523/JNEUROSCI.5346-13.2014
77
Peng C. Li L. Zhang M.-D. Bengtsson G. C. Parisien M. Belfer I. et al . (2017). miR-183 cluster scales mechanical pain sensitivity by regulating basal and neuropathic pain genes. Science356, 1168–1171. doi: 10.1126/science.aam7671
78
Poladian N. Orujyan D. Narinyan W. Oganyan A. K. Navasardyan I. Velpuri P. et al . (2023). Role of NF-κB during Mycobacterium tuberculosis infection. Int. J. Mol. Sci.24:1772. doi: 10.3390/ijms24021772
79
Qi R. Cao J. Sun Y. Li Y. Huang Z. Jiang D. et al . (2022). Histone methylation-mediated microRNA-32-5p down-regulation in sensory neurons regulates pain behaviors via targeting Cav3.2 channels. Proc. Natl. Acad. Sci. U. S. A.119:e2117209119. doi: 10.1073/pnas.2117209119
80
Rahman W. Dickenson A. H. (2013). Voltage gated sodium and calcium channel blockers for the treatment of chronic inflammatory pain. Neurosci. Lett.557, 19–26. doi: 10.1016/j.neulet.2013.08.004
81
Ren J. Liu N. Sun N. Zhang K. Yu L. (2019). Mesenchymal stem cells and their Exosomes: promising therapeutics for chronic pain. Curr. Stem Cell Res. Ther.148, 644–653. doi: 10.2174/1574888X14666190912162504
82
Ren Z. W. Zhou J. G. Xiong Z. K. Zhu F. Z. Guo X. D. (2019). Effect of exosomes derived from MiR-133b-modified ADSCs on the recovery of neurological function after SCI. Eur. Rev. Med. Pharmacol. Sci.23, 52–60. doi: 10.26355/eurrev_201901_16747
83
Rupaimoole R. Slack F. J. (2017). MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov.163, 203–222. doi: 10.1038/nrd.2016.246
84
Sakai A. Saitow F. Maruyama M. Miyake N. Miyake K. Shimada T. et al . (2017). MicroRNA cluster miR-17-92 regulates multiple functionally related voltage-gated potassium channels in chronic neuropathic pain. Nat. Commun.8:16079. doi: 10.1038/ncomms16079
85
Sakai A. Saitow F. Miyake N. Miyake K. Shimada T. Suzuki H. (2013). miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain136, 2738–2750. doi: 10.1093/brain/awt191
86
Saliminejad K. Khorram K. H. R. Soleymani Fard S. Ghaffari S. H. (2019). An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell. Physiol.234, 5451–5465. doi: 10.1002/jcp.27486
87
Sayed D. Abdellatif M. (2011). MicroRNAs in development and disease. Physiol. Rev.91, 827–887. doi: 10.1152/physrev.00006.2010
88
Scholz J. Finnerup N. B. Attal N. Aziz Q. Baron R. Bennett M. I. et al . (2019). The IASP classification of chronic pain for ICD-11: chronic neuropathic pain. Pain1601, 53–59. doi: 10.1097/j.pain.0000000000001365
89
Schwarz D. S. Hutvágner G. Du T. Xu Z. Aronin N. Zamore P. D. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cells115, 199–208. doi: 10.1016/s0092-8674(03)00759-1
90
Shao J. Cao J. Wang J. Ren X. Su S. Li M. et al . (2016). MicroRNA-30b regulates expression of the sodium channel Nav1.7 in nerve injury-induced neuropathic pain in the rat. Mol. Pain12:174480691667152. doi: 10.1177/1744806916671523
91
Shen F. Zheng H. Zhou L. Li W. Zhang Y. Xu X. (2019). LINC00657 expedites neuropathic pain development by modulating miR-136/ZEB1 axis in a rat model. J. Cell. Biochem.120, 1000–1010. doi: 10.1002/jcb.27466
92
Shi D.-N. Yuan Y.-T. Ye D. Kang L.-M. Wen J. Chen H.-P. (2018). MiR-183-5p alleviates chronic constriction injury-induced neuropathic pain through inhibition of TREK-1. Neurochem. Res.43, 1143–1149. doi: 10.1007/s11064-018-2529-4
93
Sobah M. L. Liongue C. Ward A. C. (2021). SOCS proteins in immunity, inflammatory diseases, and immune-related cancer. Front. Med.8:727987. doi: 10.3389/fmed.2021.727987
94
Su S. Shao J. Zhao Q. Ren X. Cai W. Li L. et al . (2017). MiR-30b attenuates neuropathic pain by regulating voltage-gated Sodium Channel Nav1.3 in rats. Front. Mol. Neurosci.10:126. doi: 10.3389/fnmol.2017.00126
95
Su H. Xiaohui X. He X. Liu C. Wang G. Zhou C. (2020). The miR-455-5p/ERα36 axis regulates mammalian neuronal viability and axonal regeneration. Neurosci. Lett.735:135159. doi: 10.1016/j.neulet.2020.135159
96
Tan Y. Yang J. Xiang K. Tan Q. Guo Q. (2015). Suppression of microRNA-155 attenuates neuropathic pain by regulating SOCS1 signalling pathway. Neurochem. Res.40, 550–560. doi: 10.1007/s11064-014-1500-2
97
Tang B. L. (2018). Promoting axonal regeneration through exosomes: an update of recent findings on exosomal PTEN and mTOR modifiers. Brain Res. Bull.143, 123–131. doi: 10.1016/j.brainresbull.2018.10.008
98
Tang X. Xu Q. Yang S. Huang X. Wang L. Huang F. et al . (2023). Toll-like receptors and thrombopoiesis. Int. J. Mol. Sci.24:1010. doi: 10.3390/ijms24021010
99
Tao Z. Zhou Y. Zeng B. Yang X. Su M. (2021). MicroRNA-183 attenuates osteoarthritic pain by inhibiting the-mediated/signalling axis. Bone Joint Res.10, 548–557. doi: 10.1302/2046-3758.108.BJR-2019-0308.R2
100
Torralba D. Baixauli F. Villarroya-Beltri C. Fernández-Delgado I. Latorre-Pellicer A. Acín-Pérez R. et al . (2018). Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun.9:2658. doi: 10.1038/s41467-018-05077-9
101
Uccelli A. Moretta L. Pistoia V. (2008). Mesenchymal stem cells in health and disease. Nat. Rev. Immunol.8, 726–736. doi: 10.1038/nri2395
102
van Battum E. Y. Verhagen M. G. Vangoor V. R. Fujita Y. Derijck A. A. H. A. O'Duibhir E. et al . (2018). An image-based miRNA screen identifies miRNA-135s as regulators of CNS axon growth and regeneration by targeting Krüppel-like factor 4. J. Neurosci.38, 613–630. doi: 10.1523/JNEUROSCI.0662-17.2017
103
Walters E. T. (2014). Neuroinflammatory contributions to pain after SCI: roles for central glial mechanisms and nociceptor-mediated host defense. Exp. Neurol.258, 48–61. doi: 10.1016/j.expneurol.2014.02.001
104
Wang X. Chen Q. Yi S. Liu Q. Zhang R. Wang P. et al . (2019). The microRNAs and down-regulate the axon-guidance genes and during peripheral nerve regeneration. J. Biol. Chem.294, 3489–3500. doi: 10.1074/jbc.RA119.007389
105
Wang Y. Lai X. Wu D. Liu B. Wang N. Rong L. (2021). Umbilical mesenchymal stem cell-derived exosomes facilitate spinal cord functional recovery through the miR-199a-3p/145-5p-mediated NGF/TrkA signaling pathway in rats. Stem Cell Res. Ther.12:117. doi: 10.1186/s13287-021-02148-5
106
Wang W. Li R. (2021). MiR-216a-5p alleviates chronic constriction injury-induced neuropathic pain in rats by targeting KDM3A and inactivating Wnt/β-catenin signaling pathway. Neurosci. Res.170, 255–264. doi: 10.1016/j.neures.2020.08.001
107
Wang Z. Liu F. Wei M. Qiu Y. Ma C. Shen L. et al . (2018). Chronic constriction injury-induced microRNA-146a-5p alleviates neuropathic pain through suppression of IRAK1/TRAF6 signaling pathway. J. Neuroinflammation15:179. doi: 10.1186/s12974-018-1215-4
108
Wang Z. Shen W. Zhu M. Xu M. Qiu M. Zhang D. et al . (2022). MiR-199-3p suppressed inflammatory response by targeting MECP2 to alleviate TRX-induced PHN in mice. Cell Transplant.31:9636897221108192. doi: 10.1177/09636897221108192
109
Wang X. Zhou Y. Gao Q. Ping D. Wang Y. Wu W. et al . (2020). The role of exosomal microRNAs and oxidative stress in neurodegenerative diseases. Oxidative Med. Cell. Longev.2020:3232869. doi: 10.1155/2020/3232869
110
Willemen H. L. D. M. Huo X.-J. Mao-Ying Q.-L. Zijlstra J. Heijnen C. J. Kavelaars A. (2012). MicroRNA-124 as a novel treatment for persistent hyperalgesia. J. Neuroinflammation9:143. doi: 10.1186/1742-2094-9-143
111
Wondafrash D. Z. Nire'a A. T. Tafere G. G. Desta D. M. Berhe D. A. Zewdie K. A. (2020). Thioredoxin-interacting protein as a novel potential therapeutic target in diabetes mellitus and its underlying complications. Diabetes Metab. Syndr. Obes.13, 43–51. doi: 10.2147/DMSO.S232221
112
Wu Y. Gu Y. Shi B. (2020). miR-590-3p alleviates diabetic peripheral neuropathic pain by targeting RAP1A and suppressing infiltration by the T cells. Acta Biochim. Pol.67, 587–593. doi: 10.18388/abp.2020_5451
113
Wu X. Wang X. Yin Y. Zhu L. Zhang F. Yang J. (2021). Investigation of the role of miR-221 in diabetic peripheral neuropathy and related molecular mechanisms. Adv. Clin. Exp. Med.30, 623–632. doi: 10.17219/acem/131217
114
Xia B. Gao J. Li S. Huang L. Zhu L. Ma T. et al . (2020). Mechanical stimulation of Schwann cells promote peripheral nerve regeneration via extracellular vesicle-mediated transfer of microRNA 23b-3p. Theranostics10, 8974–8995. doi: 10.7150/thno.44912
115
Xia L.-X. Ke C. Lu J.-M. (2018). NEAT1 contributes to neuropathic pain development through targeting miR-381/HMGB1 axis in CCI rat models. J. Cell. Physiol.233, 7103–7111. doi: 10.1002/jcp.26526
116
Xia L. Zhang Y. Dong T. (2016). Inhibition of MicroRNA-221 alleviates neuropathic pain through targeting suppressor of cytokine signaling 1. J. Mol. Neurosci.59, 411–420. doi: 10.1007/s12031-016-0748-1
117
Xin H. Katakowski M. Wang F. Qian J.-Y. Liu X. S. Ali M. M. et al . (2017). MicroRNA cluster miR-17-92 cluster in Exosomes enhance neuroplasticity and functional recovery after stroke in rats. Stroke48, 747–753. doi: 10.1161/STROKEAHA.116.015204
118
Xin H. Li Y. Buller B. Katakowski M. Zhang Y. Wang X. et al . (2012). Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells30, 1556–1564. doi: 10.1002/stem.1129
119
Xin H. Li Y. Liu Z. Wang X. Shang X. Cui Y. et al . (2013). MiR-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cells31, 2737–2746. doi: 10.1002/stem.1409
120
Yan X.-T. Ji L.-J. Wang Z. Wu X. Wang Q. Sun S. et al . (2017). MicroRNA-93 alleviates neuropathic pain through targeting signal transducer and activator of transcription 3. Int. Immunopharmacol.46, 156–162. doi: 10.1016/j.intimp.2017.01.027
121
Yan X.-T. Lu J.-M. Wang Y. Cheng X.-L. He X.-H. Zheng W.-Z. et al . (2018a). XIST accelerates neuropathic pain progression through regulation of miR-150 and ZEB1 in CCI rat models. J. Cell. Physiol.233, 6098–6106. doi: 10.1002/jcp.26453
122
Yan X.-T. Zhao Y. Cheng X.-L. He X.-H. Wang Y. Zheng W.-Z. et al . (2018b). Inhibition of miR-200b/miR-429 contributes to neuropathic pain development through targeting zinc finger E box binding protein-1. J. Cell. Physiol.233, 4815–4824. doi: 10.1002/jcp.26284
123
Ye G. Zhang Y. Zhao J. Chen Y. Kong L. Sheng C. et al . (2020). miR-384-5p ameliorates neuropathic pain by targeting SCN3A in a rat model of chronic constriction injury. Neurol. Res.42, 299–307. doi: 10.1080/01616412.2020.1723313
124
Yi M.-H. Liu Y. U. Liu K. Chen T. Bosco D. B. Zheng J. et al . (2021). Chemogenetic manipulation of microglia inhibits neuroinflammation and neuropathic pain in mice. Brain Behav. Immun.92, 78–89. doi: 10.1016/j.bbi.2020.11.030
125
Zarezadeh Mehrabadi A. Aghamohamadi N. Khoshmirsafa M. Aghamajidi A. Pilehforoshha M. Massoumi R. et al . (2022). The roles of interleukin-1 receptor accessory protein in certain inflammatory conditions. Immunology166, 38–46. doi: 10.1111/imm.13462
126
Zhan L.-Y. Lei S.-Q. Zhang B.-H. Li W.-L. Wang H.-X. Zhao B. et al . (2018). Overexpression of miR-381 relieves neuropathic pain development via targeting HMGB1 and CXCR4. Biomed. Pharmacother.107, 818–823. doi: 10.1016/j.biopha.2018.08.053
127
Zhang X. Guo H. Xie A. Liao O. Ju F. Zhou Y. (2020). MicroRNA-144 relieves chronic constriction injury-induced neuropathic pain via targeting RASA1. Biotechnol. Appl. Biochem.67, 294–302. doi: 10.1002/bab.1854
128
Zhang J. Li S. Li L. Li M. Guo C. Yao J. et al . (2015). Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics13, 17–24. doi: 10.1016/j.gpb.2015.02.001
129
Zhang N. Lin J. Lin V. P. H. Milbreta U. Chin J. S. Chew E. G. Y. et al . (2021). A 3D fiber-hydrogel based non-viral gene delivery platform reveals that microRNAs promote axon regeneration and enhance functional recovery following spinal cord injury. Adv. Sci.8:e2100805. doi: 10.1002/advs.202100805
130
Zhang X. A. Liu Z. Shu Q. Yuan S. Xing Z. Song J. (2019). LncRNA SNHG6 functions as a ceRNA to regulate neuronal cell apoptosis by modulating miR-181c-5p/BIM signalling in ischaemic stroke. J. Cell. Mol. Med.23, 6120–6130. doi: 10.1111/jcmm.14480
131
Zhang Y. Liu J. Wang X. Zhang J. Xie C. (2021). Extracellular vesicle-encapsulated microRNA-23a from dorsal root ganglia neurons binds to A20 and promotes inflammatory macrophage polarization following peripheral nerve injury. Aging13, 6752–6764. doi: 10.18632/aging.202532
132
Zhang J. Rong L. Shao J. Zhang Y. Liu Y. Zhao S. et al . (2021). Epigenetic restoration of voltage-gated potassium channel Kv1.2 alleviates nerve injury-induced neuropathic pain. J. Neurochem.156, 367–378. doi: 10.1111/jnc.15117
133
Zhang Y. U. Ye G. Zhao J. Chen Y. Kong L. Sheng C. et al . (2022). Exosomes carried miR-181c-5p alleviates neuropathic pain in CCI rat models. An. Acad. Bras. Cienc.94:e20210564. doi: 10.1590/0001-3765202220210564
134
Zhang B. Yin Y. Lai R. C. Tan S. S. Choo A. B. H. Lim S. K. (2014). Mesenchymal stem cells secrete immunologically active exosomes. Stem Cells Dev.23, 1233–1244. doi: 10.1089/scd.2013.0479
135
Zhang X. Zhang Y. Cai W. Liu Y. Liu H. Zhang Z. et al . (2020). MicroRNA-128-3p alleviates neuropathic pain through targeting ZEB1. Neurosci. Lett.729:134946. doi: 10.1016/j.neulet.2020.134946
136
Zhang H. Zhou Y. Wen D. Wang J. (2023). Noncoding RNAs: master regulator of fibroblast to myofibroblast transition in fibrosis. Int. J. Mol. Sci.24:1801. doi: 10.3390/ijms24021801
137
Zhang W. Zhou L. Zhang C. (2022). LncRNA Miat promotes neuropathic pain through miR-362-3p/BAMBI signaling axis. Exp. Cell Res.420:113359. doi: 10.1016/j.yexcr.2022.113359
138
Zhao L. Jiang X. Shi J. Gao S. Zhu Y. Gu T. et al . (2019). Exosomes derived from bone marrow mesenchymal stem cells overexpressing microRNA-25 protect spinal cords against transient ischemia. J. Thorac. Cardiovasc. Surg.157, 508–517. doi: 10.1016/j.jtcvs.2018.07.095
Summary
Keywords
neuropathic pain, miRNAs, exosomes, neuroinflammation, review
Citation
Zhao Y-Y, Wu Z-J, Zhu L-J, Niu T-X, Liu B and Li J (2023) Emerging roles of miRNAs in neuropathic pain: From new findings to novel mechanisms. Front. Mol. Neurosci. 16:1110975. doi: 10.3389/fnmol.2023.1110975
Received
29 November 2022
Accepted
30 January 2023
Published
17 February 2023
Volume
16 - 2023
Edited by
Silke Neumann, University of Otago, New Zealand
Reviewed by
Stanislava Jergova, University of Miami, United States; Michaela B. Kirschner, University Hospital Zürich, Switzerland; Xin Luo, Guangdong-Hong Kong-Macao Greater Bay Area Center for Brain Science and Brain-Inspired Intelligence, China
Updates
Copyright
© 2023 Zhao, Wu, Zhu, Niu, Liu and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Li, xtjing@163.comBin Liu, liubin78@163.com
This article was submitted to Pain Mechanisms and Modulators, a section of the journal Frontiers in Molecular Neuroscience
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.