Abstract
Background:
Neuropathic pain (NP) takes a heavy toll on individual life quality, yet gaps in its molecular characterization persist and effective therapy is lacking. This study aimed to provide comprehensive knowledge by combining transcriptomic and proteomic data of molecular correlates of NP in the anterior cingulate cortex (ACC), a cortical hub responsible for affective pain processing.
Methods:
The NP model was established by spared nerve injury (SNI) in Sprague–Dawley rats. RNA sequencing and proteomic data from the ACC tissue isolated from sham and SNI rats 2 weeks after surgery were integrated to compare their gene and protein expression profiles. Bioinformatic analyses were performed to figure out the functions and signaling pathways of the differentially expressed genes (DEGs) and differentially expressed proteins (DEPs) enriched in.
Results:
Transcriptomic analysis identified a total of 788 DEGs (with 49 genes upregulated) after SNI surgery, while proteomic analysis found 222 DEPs (with 89 proteins upregulated). While Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses of the DEGs suggested that most of the altered genes were involved in synaptic transmission and plasticity, bioinformatics analysis of the DEPs revealed novel critical pathways associated with autophagy, mitophagy, and peroxisome. Notably, we noticed functionally important NP-related changes in the protein that occurred in the absence of corresponding changes at the level of transcription. Venn diagram analysis of the transcriptomic and proteomic data identified 10 overlapping targets, among which only three genes (XK-related protein 4, NIPA-like domain-containing 3, and homeodomain-interacting protein kinase 3) showed concordance in the directions of change and strong correlations between mRNA and protein levels.
Conclusion:
The present study identified novel pathways in the ACC in addition to confirming previously reported mechanisms for NP etiology, and provided novel mechanistic insights for future research on NP treatment. These findings also imply that mRNA profiling alone fails to provide a complete landscape of molecular pain in the ACC. Therefore, explorations of changes at the level of protein are necessary to understand NP processes that are not transcriptionally modulated.
Introduction
Neuropathic pain (NP), affecting approximately 10% of the world’s population, is a common type of pathological pain resulting from a lesion or dysfunction in the somatosensory system (Finnerup et al., 2021). As a debilitating medical condition, NP takes a heavy toll on individual well-being and poses a huge challenge to both pain physicians and neurologists (Scholz et al., 2019). Current therapy predominated by CNS-acting drugs including opioids and anti-depressants fails to meet the clinical need, owing to limited efficacy and safety concerns (Cohen et al., 2021). This dilemma highlights the need for improving our understanding of the molecular basis of NP. Therefore, identifying the gene or protein expression profile in pain-associated areas is essential for understanding NP neurobiology and developing novel therapeutic strategies (Starobova et al., 2018; Gomez-Varela et al., 2019).
The anterior cingulate cortex (ACC) is a pivotal cortical area for affective pain processing (Bliss et al., 2016). Early functional data indicated that the activation of the ACC promotes behavioral sensitization and pain-related aversiveness (Lee et al., 1999; Johansen et al., 2001; Tang et al., 2005; Zhuo, 2006). With the advent of optogenetics for cell-specific manipulations, ACC glutamatergic neurons have been revealed to be effectors of nociceptive responses, while GABAergic interneurons exert opposite effects (Delevich et al., 2015; Kang et al., 2015). Further neurocircuitry studies indicated that the ACC integrates emotional pain signals from the mediodorsal and parafascicular thalamic nuclei (Meda et al., 2019; Zhu et al., 2021), and regulates emotional qualities of pain via distinct pathways differentially from behavioral hypersensitivity, with projections to the spinal dorsal horn (SDH) and striatum for behavioral hyperalgesia (Chen et al., 2018; Zhuang et al., 2021) and mesolimbic dopamine system for generating aversiveness (Gao et al., 2020). Cortical long-term potentiation (LTP) in the ACC has been esteemed as a cellular model of pathological pain (Zhuo, 2008). After nerve injury, cingulate excitatory synapses underwent long-lasting enhancements, which were attributed to enhanced presynaptic release of glutamate and postsynaptic recruitment of AMPARs and NMDARs (Xu et al., 2008). Thus, the genetic deletion of key molecules in the formation of cingulate LTP ameliorated NP in mice models (Li et al., 2010; Wang et al., 2011). Despite this strong evidence focusing on cellular mechanisms, remarkably less is known about the molecular basis underlying the neurobiology of the ACC in NP.
The past decade has witnessed a paradigm shift in the domain of pain research with the advent of multi-omics biology. This approach enables a leap forward from traditional qualitative studies of single targets to systematic investigations of multidimensional cellular networks. The ability to monitor the abundance of pain-related molecules on a genomic scale relies on high-throughput tools including RNA-sequencing and mass spectrometry-based proteomics (Nie et al., 2007; Niederberger and Geisslinger, 2008). In the pain field, analyses of mRNA and protein expression patterns have been frequently reported, but the application was limited at levels of primary sensory neurons and the SDH (Niederberger and Geisslinger, 2008; Starobova et al., 2018). In the last 2 years, emerging data have revealed transcriptomic profiles of the ACC following nerve injury (Su et al., 2021; Dai et al., 2022; Zhang et al., 2022). However, it remains hard to meet the increasing expectations of systematic pain study by investigating changes in biomolecules at a single level.
In the present study, we simultaneously utilized transcriptomic and proteomic approaches to elaborate on the complicated pathological alterations of the ACC in a rat NP model induced by spared nerve injury (SNI). These findings are of value to obtain a comprehensive view of neurobiological mechanisms underlying the role of the ACC in the modulation of NP processing and to suggest novel therapeutic targets for the treatment of NP.
Materials and methods
Animals and experimental design
All experimental procedures were approved by the Ethics Committee of the General Hospital of Norther Theater Command, and performed in line with the guidelines of the National Institute of Health Guide for the Care and Use of Laboratory Animals. In all, 78 adult male Sprague–Dawley rats (200–250 g) were provided by the Department of Experimental Animals of the General Hospital of Northern Theater Command. Among the animals, 36 rats were used for integrative transcriptomic and proteomic analysis, while the others were used for small RNA sequencing. The animals were housed in a reversed 12 h dark/12 h light cycle environment and provided with free access to water and food.
The animals were equally divided into a sham group and an SNI group. NP was elicited by SNI surgery. Two weeks after SNI surgery or sham operation, the rats underwent behavioral tests for confirming the successful establishment of the NP model. One day later, the animals were sacrificed by rapid cervical dislocation. The ACC is located adjacent to the medial frontal cortex, and these two regions have been revealed to exert different roles in pain modulation (Zhuo, 2006; Ong et al., 2019). To guarantee the accuracy of sampling, coronal brain sections (300 μm) containing the ACC were cut with a vibratome in ice-cold artificial cerebrospinal fluid (in mM: 2.6 KCl, 124 NaCl, 1 MgCl2, 0.5 CaCl2, 1.23 NaH2PO4, 26.2 NaHCO3, 5 kynurenic acids, 212.7 sucrose, 10 dextroses, pH 7.4), as described in previous patch-clamp experiments (Bai et al., 2019; Ren et al., 2022). The contralateral ACC was dissected out with a 15G puncher and then snap-frozen on dry ice. Afterward, RNA and protein isolation were performed on six independent pools (for integrative transcriptomic and proteomic analysis) or seven independent pools (for small RNA sequencing) of samples from the ACC, with three animals mixed as one biological pool (Figure 1; Supplementary Figure S1). To minimize the degradation of RNAs, all the materials and instruments used for sampling were treated with 0.01 M phosphate-buffered saline (PBS) containing 0.1% diethylpyrocarbonate overnight before sampling.
Figure 1

Systematic workflow of transcriptome and proteome analysis in the anterior cingulate cortex of a neuropathic pain rat model. (A) Schematic diagram shows the key steps of this study. (B) Transcriptomic workflow. (C) TMT proteomic workflow.
SNI surgery
As described in a previous study (Ma et al., 2022), an incision was made in the hind leg to expose the biceps femoris muscle after deep anesthetization. Then, the sciatic nerve and its terminal branches, including the sural, common peroneal, and tibial nerves, were exposed. The common peroneal and tibial nerves were ligated with a 6.0 silk suture and then sectioned distal to the ligation. The muscle and skin were closed. For sham surgery, the sciatic nerve was exposed only.
Behavioral experiments
The mechanical paw withdrawal threshold (PWT) of bilateral hind paws was calculated via the von Frey filament (VFF) test. Before testing, the animals were handled and habituated in the testing apparatus for 30 min until calming down for three consecutive days. VFFs with increasing forces from 1.4 g to 26 g were applied vertically to the lateral plantar surface of the hind paw five times, each for 5–8 s with a 5-min interval. The minimal force triggering withdrawal responses at least three times in five stimulations was considered as the PWT. Positive signs included vocalization, ipsilateral rear leg vibrating, nibbling, and withdrawal.
An open-field test was conducted to examine the locomotion as well as exploratory behaviors after SNI surgery. After acclimation in the observation room, the rats were placed in the center of the open field (100 cm × 100 cm × 60 cm). The locomotion was recorded for 15 min via a motion-tracking system (Shanghai Mobile Datum Information Technology, Shanghai, China). Anxiety-like behavior was evaluated by the total distance traveled and time spent in the center of the open field.
RNA sequencing
Total RNA was separated with the Trizol reagent (Thermo Fisher, CA, United States). Total RNA purity and quantity were analyzed by Bioanalyzer 2,100 and RNA 6000 Nano LabChip Kit (Agilent, CA, United States). Samples with the parameters of concentration > 50 ng/uL and RIN number > 7.0 were adopted for sequencing library construction. Then, Dynabeads Oligo (dT) (Thermo Fisher) was used for the purification of mRNA with PolyA. Next, the mRNA was fragmented into small fragments with divalent cations via Magnesium RNA Fragmentation Module (NEB, MA, USA). The cleaved fragments were reverse-transcribed to create the cDNA with SuperScript™ II Reverse Transcriptase (Invitrogen, CA, United States), and then were used to synthesize U-labeled second-stranded DNAs with dUTP Solution (Thermo Fisher), RNase H (NEB), and E. coli DNA polymerase I (NEB). An A-base was then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contained a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. Dual-index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. The U-labeled second-stranded DNAs were treated with the heat-labile UDG enzyme (NEB), and then the ligated products were amplified with polymerase chain reaction under the following conditions: initial denaturation at 95°C for 3 min; eight cycles of denaturation at 98°C for 15 s, annealing at 60°C for 15 s, extension at 72°C for 30 s; and final extension at 72°C for 5 min. The final cDNA libraries had a mean insert size of 300 ± 50 bp. Finally, the 2 × 150 bp paired-end sequencing was performed with the aid of an Illumina Novaseq™ 6,000 platform (Illumina, CA, United States).
Transcriptomic analysis
A total of 544.78 million 2 × 150 bp paired-end reads were acquired after the RNA-seq approach. The reads were further filtered by Cutadapt1 (version: cutadapt-1.9). Then, the sequence quality was verified using FastQC2 (version: 0.11.9). All raw sequence data have been submitted to the NGDC Genome Sequence Archive (GSA) with accession number <CRA009767>. After that, a total of 79.19 Gbps of cleaned, paired-end reads were produced. The reads of all samples were aligned to the rat reference genome using the HISAT2 package3 (version: hisat2-2.0.4). The mapped reads of each specimen were assembled via StringTie (version: string tie-1.3.4d) with default parameters. Then, the transcriptomes from all specimens were merged to reconstruct a comprehensive transcriptome through gffcompare software (version: gffcompare-0.9.8). Then, ballgown and StringTie were utilized to perform expression abundance for mRNAs by calculating fragment per kilobase of transcript per million mapped reads (FPKM) value. Principal component analysis (PCA) was performed with the princomp function of R. The heatmaps were created via OmicShare (1GENE DENOVO). The analysis of differential gene expression was performed by DESeq24. Those with absolute fold change (FC) ≥ 2 and false discovery rate (FDR) of less than 0.05 were considered differentially expressed genes (DEGs), which underwent subsequent enrichment analysis of Gene Ontology (GO) functions5 and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways6 with the aid of the clusterProfilter R software package.
Small RNA sequencing and analysis
Total RNA was used as input material for RNA sample preparation. Sequencing libraries were generated using NEB Next® Multiplex Small RNA Library Prep Set for Illumina® (NEB E7300L). Briefly, 3′ and 5′ adaptors were ligated to the 3′ and 5′ end of small RNA, respectively. Then the first strand of cDNA was synthesized after hybridization with reverse transcription primer. The double-stranded cDNA library was generated through PCR enrichment. After purification and size selection, libraries with insertions between18 ~ 40 bp were ready for sequencing on Illumina sequencing with SE50. Subsequently, library quality was assessed on the Agilent 5400 system and quantified by qPCR (1.5 nM). The Qualified libraries were pooled and sequenced on Illumina platforms with SE50 strategy in Novogene Bioinformatics Technology Co., Ltd. (Beijing, China), according to effective library concentration and the data amount required.
Cleaning data were obtained by cleaning low-quality tags, removing adapter sequences, and filtering adaptor-ligated contaminants. The final reads were mapped to the reference sequence by Bowtie (version: 1.0.1) (Langmead et al., 2009). Clean reads were compared against small RNAs using the Rfadm database to annotate small RNS sequences. The miRBase20.0 was used to look for known miRNAs. The hairpin structures were used to predict novel miRNAs using miRDeep2 software (version: mirdeep2_0_0_5). miRNA expression levels were estimated by TPM (transcript per million) through the following criteria (Zhou et al., 2010): Normalization formula: Normalized expression = mapped read count/Total reads*1000000. Differential expression analysis of the two groups was performed using the DESeq R package (1.8.3). The p-values were adjusted using the Benjamini & Hochberg method. The corrected p-value of 0.05 was set as the threshold for significantly differential expression by default. p-value < 0.01 and |log2(fold change)| > 1 were set as the threshold for significantly differential expression by default.
Predicting the target gene of miRNA was performed by miRanda (version: miRanda-3.3a) (Enright et al., 2003) and RNAhybrid (version: RNAhybrid v2.0) (Krüger and Rehmsmeier, 2006). The targeted genes were subsequently performed for GO enrichment analysis and KEGG analyses. GOseq-based Wallenius non-central hyper-geometric distribution was implemented for GO enrichment analysis (Young et al., 2010), while KOBAS software was used for the analysis of KEGG pathways (Mao et al., 2005).
Proteomics
After three washes in 0.01 M PBS, the tissues were ultrasonically treated for 5 min with a RIPA working solution (Beyotime, Shanghai, China). Then, the tissues were centrifuged (12,000 rpm, 4°C, 15 min) to procure the supernatant. Next, the protein solution was incubated with 5 mM dithiothreitol (Sigma-Aldrich, MO, United States) at 55°C for 20 min for reduction, followed by incubation in 15 mM iodoacetamide (Sigma-Aldrich) at room temperature (RT) for 30 min for alkylation. After digestion with filter-aided sample preparation, the solution was transferred to an Amicon filter (Millipore, MA, United States), followed by centrifugation (13,800 rpm, 25°C, 60 min) twice and the addition of 100 mM TEAB and 20% acetonitrile (Thermo Fisher). Then, the protein solution was centrifuged (13,800 rpm, 25°C, 60 min) after the addition of enzymatic buffer (100 mM TEAB, pH 7.5). At last, mass spectrometry (MS)-grade trypsin (Promega, WI, United States) was added for the first 12 h digestion, and sequencing-grade Lys-C (Wako, Japan) was added for the second 4 h digestion. After digestion, the filtrate was collected following centrifugation (13,800 rpm, 25°C, 60 min). Then concentrated to 1 mg/mL TMT reagent (Thermo Fisher) was added to 41 uL acetonitrile for re-dissolution, and this mixture was then added to the corresponding peptide segment solution and incubated (RT, 2 h) for labeling. The reaction was terminated by the addition of 5% hydroxylamine (8 uL, RT, 30 min, Sigma-Aldrich).
For each sample, 2 μg of total peptides were procured and analyzed with a nanoUPLC (EASY-nLC1200, Thermo Fisher) coupled to a Q Exactive HFX Orbitrap instrument (Thermo Fisher) with a nano-electrospray ion source. Separation was performed using a reversed-phase column (100 um ID × 15 cm, Reprosil-Pur 120C18AQ, 1.9 um, Germany). H2O with 0.1% formic acid (Sigma-Aldrich) and 2% acetonitrile (ANPEL Laborotory Technologies, Shanghai, China), and H2O with 80% ACN and 0.1% formic acid were used as mobile phases in phase A and B, respectively. Sample separation was executed at 300 nL/min flow rate with a 90 min gradient. Solvent B was set as 2–5% for 2 min, 5–22% for 68 min, 22–45% for 16 min, 45–95% for 2 min, and 95% for 2 min.
The data-dependent acquisition was processed in profile and positive mode via Orbitrap analyzer at a resolution of 120,000 (@200 m/z) and m/z range of 350–1,600 for MS1, and at a resolution of 45 k with a fixed first mass of 110 m/z for MS2. The automatic gain control target for MS1 was set to 3E6 with a max IT of 30 ms, and 1E5 for MS2 with a max IT of 96 ms. The top 20 most intense ions were fragmented by higher-energy collisional dissociation with a normalized collision energy of 32% and isolation window of 0.7 m/z. The dynamic exclusion time window was set at 45 s, and single-charged peaks and peaks with a charge exceeding six were excluded from the data-dependent acquisition.
Proteomic analysis
The vendor’s raw MS files were processed with Proteome Discoverer (version: 2.4, Thermo Fisher) and the built-in Sequest HT search engine. MS spectra lists were searched against their species-level UniProt FASTA databases (UniProt-Rattus norvegicus-10,116-2021-8.fasta), with Carbamidomethyl [C], TMTpro (K), TMT pro (N-term) as the fixed modification and Oxidation (M) and Acetyl (Protein N-term) as variable modifications. The FDR was set to 0.01 for both peptide and peptide-spectrum match levels. Peptide identification was conducted with a fragment mass deviation of 0.02 Da and an initial precursor mass deviation of up to 10 ppm. The total peptide amount was used for normalization, and the razor peptide and the unique peptide were used for the quantification of proteins. The other parameters were reserved as default.
Differential protein screening followed threshold conditions of a p-value of less than 0.05 and 1.2-fold FC. GO and KEGG enrichment analyses were conducted on the differentially expressed proteins (DEPs) for protein function probing. The STRING database (version: 11.0)7 was utilized to analyze the relationships and construct the interaction network of DEPs via R language (version: 3.6.1, drawing package ggplot2).
Data analysis
All of the normally distributed continuous variables were expressed as the mean ± standard deviation. Means were compared by unpaired t-tests for normally distributed data or by the Wilcoxon rank-sum (Mann–Whitney) test for non-normal data. Correlation coefficients were calculated using Pearson’s correlation test. Statistical analysis was conducted using the SPSS statistical software (IBM Corp., version: 26.0, NY, United States). A two-tailed p-value < 0.05 was considered significant.
Results
Behavioral evaluation of the rats after SNI
Behavioral assays were conducted 2 weeks after surgery to confirm the establishment of NP models. Compared with the sham group, SNI-treated rats displayed a significant decrease in PWT in the ipsilateral (Figures 2A,B) and contralateral hind paw (Figure 2C), indicating the existence of mirror pain. The open field test showed that SNI rats traveled less distance (Figures 2D,E) and spent less time in the central area (Figure 2F), implicating the existence of hypolocomotion and anxiety-like emotions. The behavioral performance of these rats was consistent with SNI models described in previous studies (Liu and Chen, 2014; Li Q. Y. et al., 2022). These behavioral data provided evidence of nociceptive hypersensitivity and behavioral anxiety in SNI-treated rats.
Figure 2

SNI-induced mechanical hyperalgesia and anxiety-like behaviors in rats. (A) Diagram showing the von Frey test in rats. (B,C) SNI-treated rats showed decreased mechanical withdrawal threshold in both ipsilateral (B) and contralateral (C) hind paws. (D) Representative motion trails of sham (left) and SNI (right) rats in the open field. (E,F) SNI-treated rats traveled less distance in the open field (E) and spent less time in the center of the open field (F) than sham rats. Unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, SNI vs. sham.
Transcriptomic signatures in the ACC of rats following SNI
The transcriptome data were acquired using the RNA-seq method from rat ACC tissue 2 weeks after SNI or sham surgery. A total of 44,000,392 clean reads were averagely generated after filtering for low-quality from 45,398,261 raw sequencing reads. The number of identified genes and their proportion and distribution to the total gene number in the database of each sample were counted and calculated (Supplementary Table S1). The distribution and density of gene expression levels are shown in Supplementary Figures S2A,B. PCA showed that the two groups of samples distinctively clustered into different categories, implying distinct gene expression patterns of NP rats (Figure 3A). The volcano plot revealed 40 upregulated DEGs and 739 downregulated DEGs in the ACC of rats 14 days after SNI (Figures 3B,C). The heatmap data showed that the top 100 DEGs according to the q-value could significantly distinguish these two groups (Figure 3D). Most of the DEGs (89.7%) were mRNAs (Supplementary Figure S2C). In addition, a few differentially expressed long intergenic non-coding RNAs (lincRNA, N = 27), microRNAs (miRNA, N = 5), small nucleolar RNAs (snoRNA, N = 7), ribosomal RNAs (rRNA, N = 5), and small nuclear RNA (snRNA, N = 3), were also identified. The top 20 upregulated and downregulated differentially expressed protein-coding RNAs are shown in Table 1, while the significant non-coding RNAs (ncRNA) are listed in Supplementary Table S2.
Figure 3

General transcriptomic outcomes. (A) Principal component analysis indicated a near-complete separation of genes in the sham (blue) and SNI (green) groups. (B) In all, 40 genes were upregulated, and 739 genes were downregulated. (C) Volcano plot shows differentially expressed genes. The red and blue dots indicate significantly upregulated and downregulated genes, respectively. (D) Hierarchical cluster analysis. Red indicates upregulation, and blue indicates downregulation. The dendrograms represent the classification of genes. The number in the color scale indicates the z-score.
Table 1
| Gene name | Description | Average | p-value | FC | |
|---|---|---|---|---|---|
| Sham | SNI | ||||
| Downregulated genes | |||||
| Vom1r88 | Vomeronasal 1 receptor 88 | 0.10 | 0.00 | 0.00599 | 0.00 |
| Chrm5 | Cholinergic receptor, muscarinic 5 | 0.04 | 0.00 | 0.00153 | 0.00 |
| Drd3 | Dopamine receptor D3 | 0.22 | 0.01 | 0.00142 | 0.02 |
| AC128967 | Zinc finger protein 42-like | 0.16 | 0.01 | 0.00791 | 0.07 |
| Ccdc3 | Coiled-coil domain containing 3 | 0.32 | 0.03 | 0.00009 | 0.09 |
| Matn3 | Matrilin 3 | 0.09 | 0.01 | 0.00426 | 0.10 |
| Fshb | Follicle-stimulating hormone subunit beta | 0.15 | 0.02 | 0.00060 | 0.13 |
| Trappc3l | Trafficking protein particle complex 3-like | 0.53 | 0.07 | 0.00088 | 0.13 |
| Npy2r | Neuropeptide Y receptor Y2 | 0.43 | 0.06 | 0.00019 | 0.13 |
| Cd300le | Cd300 molecule-like family member E | 0.45 | 0.06 | 0.00073 | 0.14 |
| Pag1 | Phosphoprotein membrane anchor with glycosphingolipid microdomains 1 | 0.70 | 0.10 | 0.00004 | 0.15 |
| Pcdha4 | Protocadherin alpha 4 | 0.43 | 0.07 | 0.00001 | 0.16 |
| Il13ra1 | Interleukin 13 receptor, alpha 1-like | 0.60 | 0.10 | 0.00140 | 0.17 |
| Syt9 | Synaptotagmin 9 | 0.95 | 0.19 | 0.00000 | 0.20 |
| LOC100909830 | m7GpppN-mRNA hydrolase-like | 0.28 | 0.06 | 0.00019 | 0.22 |
| Nos1 | Nitric oxide synthase 1 | 1.16 | 0.25 | 0.00000 | 0.22 |
| Pcdhga5 | Protocadherin gamma subfamily A, 5 | 0.36 | 0.08 | 0.00000 | 0.22 |
| Cxcl10 | C-X-C motif chemokine ligand 10 | 0.45 | 0.10 | 0.00245 | 0.23 |
| Pcdhga8 | Protocadherin gamma subfamily A, 8 | 0.69 | 0.17 | 0.00015 | 0.23 |
| Acvr1c | Activin A receptor type 1C | 0.57 | 0.14 | 0.00001 | 0.23 |
| Upregulated genes | |||||
| LOC687560 | Hypothetical protein | 7.4 | 20.5 | 0.00422 | 1384.90 |
| Olr1104 | Olfactory receptor 1104 | 70.0 | 169.8 | 0.00487 | 650.96 |
| Trim29 | Tripartite motif-containing 29 | 635.9 | 1144.1 | 0.00727 | 255.43 |
| AABR07000740 | Uncharacterized LOC103690177 | 62.0 | 108.4 | 0.00093 | 18.75 |
| Tfap2a | Transcription factor AP-2 alpha | 19.8 | 31.8 | 0.00334 | 9.70 |
| Gp6 | Glycoprotein 6 (platelet) | 145.3 | 229.4 | 0.00027 | 5.29 |
| S100a9 | S100 calcium binding protein A9 | 891.1 | 1387.1 | 0.00004 | 4.71 |
| Pcp2 | PURKINJE cell protein 2 | 611.2 | 932.0 | 0.00587 | 4.55 |
| Pou5f1 | POU class 5 homeobox 1 | 2428.6 | 3697.5 | 0.00128 | 3.08 |
| LOC100362054 | mCG114696-like | 91.5 | 134.9 | 0.00007 | 3.00 |
| Sox15 | SRY box 15 | 83.2 | 121.5 | 0.00000 | 2.55 |
| Scp2d1 | SCP2 sterol-binding domain containing 1 | 251.0 | 362.6 | 0.00026 | 2.30 |
| LOC108348155 | Paired immunoglobulin-like type 2 receptor beta-2 | 26.1 | 37.3 | 0.00026 | 2.30 |
| Prr32 | Proline rich 32 | 342.7 | 484.2 | 0.00415 | 2.25 |
| Prss22 | Serine protease 22 | 16.6 | 23.4 | 0.00005 | 2.21 |
| Mia | MIA SH3 domain containing | 222.2 | 313.5 | 0.00011 | 2.19 |
| Ahsp | Alpha hemoglobin stabilizing protein | 36.8 | 51.6 | 0.00490 | 2.17 |
| Mpp4 | Membrane palmitoylated protein 4 | 202.9 | 282.9 | 0.00000 | 2.13 |
| Creb3l4 | cAMP responsive element binding protein 3-like 4 | 162.4 | 225.8 | 0.00013 | 2.06 |
| Tnni3 | Troponin I3, cardiac type | 270.6 | 373.9 | 0.00448 | 2.03 |
Differentially expressed protein-coding genes in the anterior cingulate cortex after nerve injury by RNA-Seq.
Twenty downregulated and upregulated differentially expressed protein-coding genes with the lowest p-values in the rat anterior cingulate cortex after SNI. The genes are ranked on the average of respective absolute fold change.
FC, fold change; SNI, spared nerve injury.
To understand the functions of the DEGs identified, GO analysis was performed for enrichment analysis and classifications, with the top 10 enriched items in each classification shown in Figures 4A–C. GO analysis identified biological processes (BP) associated with adhesion molecules, ion transport, homophilic cell adhesion via plasma membrane, regulation of DNA-templated transcription, regulation of ion transmembrane transport, potassium ion transport, and chemical synaptic transmission. Enriched cellular component (CC) terms associated with membrane (including caveola, presynaptic, and postsynaptic membrane) and synapse (including glutamatergic and GABAergic synapses). Moreover, identified enriched molecular functions (MF) associated with protein binding, calcium ion binding, ion channel activity, protein serine/threonine kinase activity, steroid hormone receptor activity, and potassium channel activity. We then used the DEGs for KEGG pathway enrichment (Figure 4D). The DEGs were significantly enriched in the classifications of the calcium signaling pathway, cholinergic synapse, axon guidance, cAMP signaling pathway, signaling pathways in charge of regulation of stem cell pluripotency, longevity regulating pathway, estrogen signaling pathway, parathyroid hormone synthesis, secretion and action, insulin resistance, and AMPK signaling pathway. All these suggest that peripheral nerve injury-induced synaptic activity changes in the ACC.
Figure 4

Bioinformatic analysis for differentially expressed genes in sham and NP rats. (A) Biological process. (B) Cell components. (C) Molecular function. Darker colors indicate higher statistical significance. (D) Bubble chart shows KEGG analysis of differential genes. The horizontal axis represents a rich factor (ratio of the sum of differential genes enriched in a pathway to the number of genes annotated by the pathway). Bubble size indicates the number of genes included in each pathway, and different colors indicate different p-values.
MiRNAomic signatures in the ACC of rats following SNI
Differing from mRNAs, small non-coding RNAs exert functions by silencing the targeted mRNAs to regulate post-transcriptional protein accumulation and play a pivotal role in ontogenesis, tumorigenesis, and the development of various human disorders (Esteller, 2011). Interestingly, the domain of pain study is experiencing rapid growth in the research of relationships between small RNAs and chronic pain (Bali and Kuner, 2014). Our RNA-sequencing data displayed above implied the change of small RNAs in the ACC under the condition of NP. To gain a complete picture of miRNAomic signatures in the ACC, small RNA sequencing was further performed in another batch of rats receiving SNI or sham surgery (Supplementary Figure S1).
A total of 11,040,407 clean reads were averagely generated after filtering for low quality from 11,355,792 raw sequencing reads. The number of identified genes and their proportion and distribution to the total gene number in the database of each sample are displayed in Supplementary Table S3. The distribution and density of gene expression levels are shown in Supplementary Figure S3. Most small RNAs were 21-23 nt in length in all the libraries, with 22 nt being the most frequent length, and more than 40.29% were miRNA in the catalog of small RNA in all 14 libraries. A total of 662 mature miRNAs and 165 novel miRNAs were identified in these small RNA libraries.
Concerning miRNA sequencing data analysis, a total of 33 DEmiRNAs were identified, with 31 known and 2 novel miRNAs. Also, 21 and 12 miRNAs were upregulated and downregulated, respectively (Supplementary Figures S4A,B), and a detailed literature review concerning the role of these miRNAs in pain modulation is presented in Supplementary Table S4. Cluster analysis of the DEmiRNAs is shown in Supplementary Figure S4C. Next, we predicted gene targets of 33 DEmiRNAs through the online miRNA targets prediction database to discover the potential miRNA-regulated pathway in NP. A total of 470,667 and 651,166 targeted genes were predicted from the databases RNAhybrid and miRanda, respectively. By intersecting the genes that were predicted in the two databases, we obtained 73,258 predicted genes. GO enrichment and KEGG analyses were finally performed on these genes, and the top 10 terms in BP, CC, and MF and KEGG pathways are shown separately (Supplementary Figure S5). Intriguingly, synapse-related terms and pathways were highly enriched, which was in accordance with the transcriptomic data mentioned above.
Proteomic signatures in the ACC of rats following SNI
To quantify the proteomic changes in the ACC of SNI rats, LC-MS/MS was adopted to analyze protein extracts. In total, 6,967 credible proteins were identified, most of which were distributed within the range of 0–200 Da molecular weights (Supplementary Figure S6A) and 0–20 peptide numbers (Supplementary Figure S6B). The results of PCA analysis (Figure 5A) showed that the same group of samples were more concentrated in spatial distribution. Within our identified proteins, we found 89 significantly upregulated and 133 downregulated proteins (Figures 5B,C). The top 20 upregulated/downregulated proteins are shown in Table 2. A heat map further shows the differential expression changes between groups and within each sample of the significantly altered proteins (Figure 5D).
Figure 5
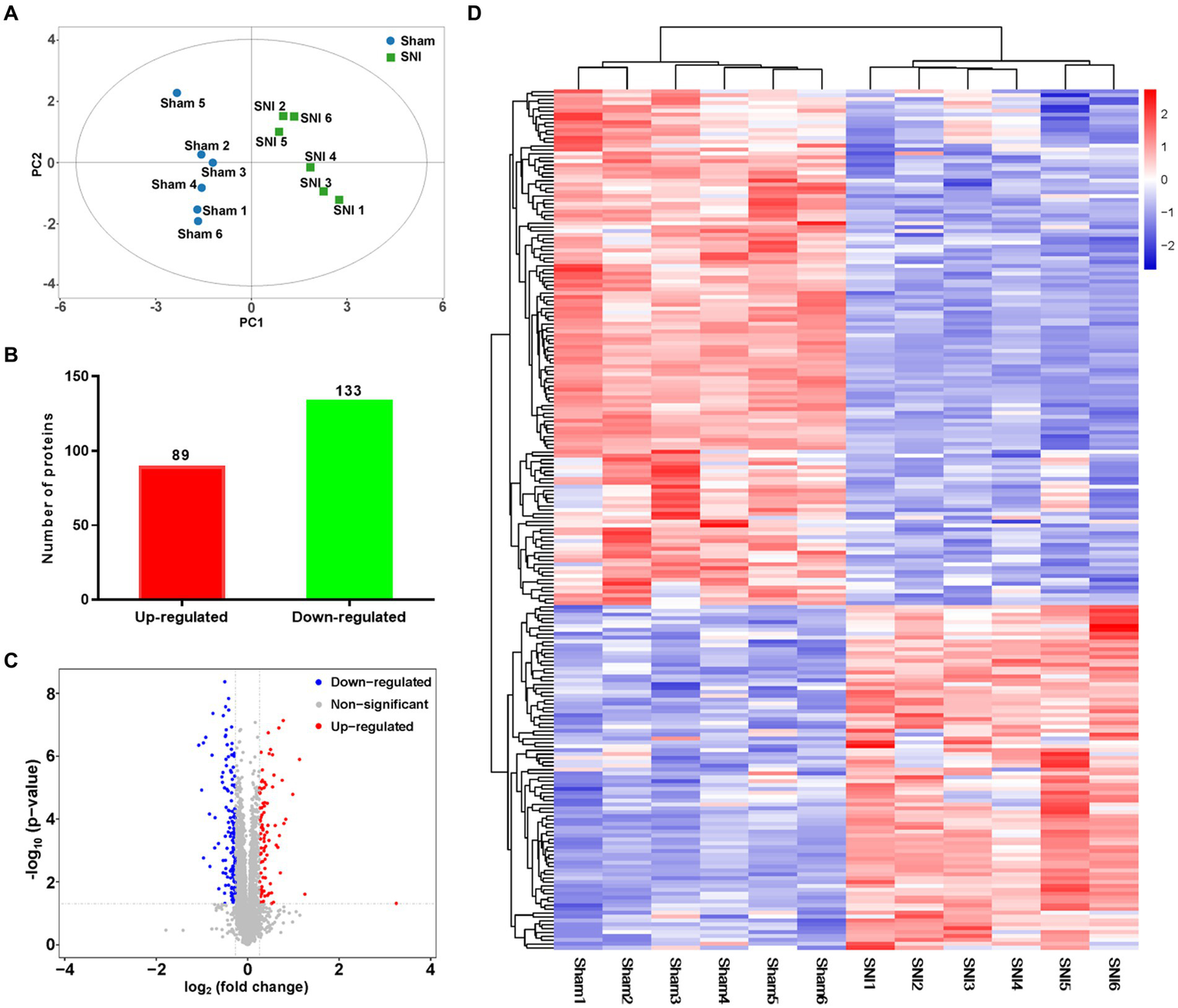
General proteomic outcomes. (A) Principal component analysis indicated a near-complete separation of proteins in the sham (blue) and SNI (green) groups. (B) In all, 89 proteins were upregulated, and 133 proteins were downregulated. (C) Volcano plot shows differentially expressed proteins. The red and blue dots indicate significantly upregulated and downregulated proteins, respectively. (D) Hierarchical cluster analysis. Red indicates upregulation, and blue indicates downregulation. The dendrograms represent the classification of proteins. The number in the color scale indicates the z-score.
Table 2
| Accession | Description | UP | Average | p-value | FC | |
|---|---|---|---|---|---|---|
| Sham | SNI | |||||
| Downregulated proteins | ||||||
| D3ZF21 | GPRIN family member 3 (Gprin3) | 12 | 1189.9 | 978.3 | 0.00015 | 0.82 |
| D3ZCF8 | ATP-binding cassette, subfamily A (ABC1), member 8a (Abca8a) | 12 | 1189.0 | 972.4 | 0.00528 | 0.82 |
| P60203 | Myelin proteolipid protein (Plp1) | 11 | 9661.4 | 8010.0 | 0.04107 | 0.83 |
| P63036 | DnaJ homolog subfamily A member 1 (Dnaja1) | 10 | 1146.8 | 924.0 | 0.02215 | 0.81 |
| B0BND0 | Glycerophosphocholine cholinephosphodiesterase ENPP6 (Enpp6) | 9 | 1223.4 | 910.8 | 0.00028 | 0.74 |
| P57113 | Maleylacetoacetate isomerase (Gstz1) | 8 | 1146.7 | 943.2 | 0.04171 | 0.82 |
| Q9JKA9 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Hcn2) | 8 | 636.5 | 519.8 | 0.00639 | 0.82 |
| D3ZDU5 | Profilin (Pfn2) | 7 | 3492.1 | 2814.2 | 0.01542 | 0.81 |
| Q63553 | SNF-related serine/threonine-protein kinase (Snrk) | 6 | 260.9 | 214.6 | 0.01073 | 0.82 |
| Q63327 | Myelin-associated oligodendrocyte basic protein (Mobp) | 6 | 928.1 | 733.7 | 0.01040 | 0.79 |
| Q811T3 | Kv3.3c voltage gated potassium channel subunit splice variant C (Kcnc3) | 5 | 285.1 | 229.3 | 0.00033 | 0.80 |
| Q5XIU4 | B-cell receptor-associated protein (Bcap29) | 5 | 465.9 | 369.8 | 0.00182 | 0.79 |
| Q7TP52 | Carboxymethylenebutenolidase homolog (Cmbl) | 4 | 278.1 | 230.0 | 0.01243 | 0.83 |
| G3V6B9 | Tyrosine-protein phosphatase non-receptor type (Ptpn4) | 4 | 240.5 | 193.7 | 0.00501 | 0.81 |
| M0R4S2 | Apolipoprotein D (Apod) | 4 | 273.5 | 215.8 | 0.00492 | 0.79 |
| P07722 | Myelin-associated glycoprotein (Mag) | 4 | 1484.1 | 1103.0 | 0.00215 | 0.74 |
| A0A0G2JUM8 | Cytochrome c oxidase assembly factor 6 (Coa6) | 3 | 243.0 | 199.3 | 0.02272 | 0.82 |
| G3V8G6 | Hyaluronan and proteoglycan link protein 2 (Hapln2) | 3 | 178.2 | 146.0 | 0.03783 | 0.82 |
| D3ZSV7 | Similar to THUMP domain containing 3, isoform CRAa (Thumpd3) | 3 | 210.4 | 169.1 | 0.00196 | 0.80 |
| D4A2K5 | Nuclear receptor interacting protein 3, isoform CRAa (Nrip3) | 3 | 99.4 | 76.1 | 0.01244 | 0.77 |
| Upregulated proteins | ||||||
| A0A0G2K6S9 | Myosin-11 (Myh11) | 39 | 7.4 | 20.5 | 0.03966 | 1.52 |
| C0JPT7 | Filamin A (Flna) | 39 | 70.0 | 169.8 | 0.02728 | 1.37 |
| A0A0G2JWK7 | Transgelin (Tagln) | 11 | 635.9 | 1144.1 | 0.01146 | 1.56 |
| Q62720 | Zinc transporter 1 (Slc30a1) | 9 | 62.0 | 108.4 | 0.00035 | 1.21 |
| Q5FVG5 | Similar to tropomyosin 1, embryonic fibroblast-rat, isoform CRAc (Tpm2) | 7 | 19.8 | 31.8 | 0.03293 | 1.80 |
| P07150 | Annexin A1 (Anxa1) | 6 | 145.3 | 229.4 | 0.03493 | 1.21 |
| D3ZC96 | Tetratricopeptide repeat protein 39B (Ttc39b) | 5 | 891.1 | 1387.1 | 0.00768 | 1.52 |
| A9CMA6 | Transmembrane protein 163 (Tmem163) | 5 | 611.2 | 932.0 | 0.00054 | 1.33 |
| A0A0G2K1G8 | Solute carrier family 7, member 14 (Slc7a14) | 4 | 2428.6 | 3697.5 | 0.01619 | 1.27 |
| P09330 | Ribose-phosphate pyrophosphokinase 2 (Prps2) | 4 | 91.5 | 134.9 | 0.00253 | 1.26 |
| Q64361 | Latexin (Lxn) | 4 | 83.2 | 121.5 | 0.00746 | 1.21 |
| D3ZQL1 | ER membrane protein complex subunit 7 (Emc7) | 4 | 251.0 | 362.6 | 0.04083 | 1.20 |
| Q923Z2 | Tropomyosin 1, alpha, isoform CRA_a (Tpm1) | 3 | 26.1 | 37.3 | 0.04930 | 1.31 |
| Q5U204 | Ragulator complex protein LAMTOR3 (Lamtor3) | 3 | 342.7 | 484.2 | 0.01251 | 1.30 |
| Q5BJT2 | Ubiquitin-like protein 3 (Ubl3) | 3 | 16.6 | 23.4 | 0.00637 | 1.25 |
| P63255 | Cysteine-rich protein 1 (Crip1) | 3 | 222.2 | 313.5 | 0.03531 | 1.24 |
| M0R4L3 | PTS1-BP (Pex5) | 3 | 36.8 | 51.6 | 0.01220 | 1.23 |
| Q27W01 | RNA-binding protein 8A (Rbm8a) | 3 | 202.9 | 282.9 | 0.00407 | 1.23 |
| D4A4F9 | RCG20461 (Stum) | 3 | 162.4 | 225.8 | 0.00186 | 1.23 |
| F1LYA6 | Abl-interactor 2 (Abi2) | 3 | 270.6 | 373.9 | 0.00209 | 1.22 |
Differentially expressed proteins in the anterior cingulate cortex after nerve injury by TMT.
Twenty downregulated and upregulated differentially expressed proteins with the lowest p-values in the rat anterior cingulate cortex after SNI. The proteins are ranked on the average of respective absolute fold change.
FC, fold change; SNI, spared nerve injury.
To obtain an overview of the effects of SNI on the ACC, GO functional annotation of these 123 DEPs was performed, and the top 10 items according to the p-value in each classification are listed in Figures 6A–C. The enrichment results showed that the DEPs were significantly enriched in BPs such as mitochondrion organization, alternative mRNA splicing via spliceosome, plasma membrane bounded cell projection assembly, lipid oxidation, cellular response to leptin stimulus, postsynaptic cytoskeleton organization, and regulation of biological quality. For MF, the DEPs were mainly associated with the structural constituent of the myelin sheath, protein binding, as well as the activity of cyclic nucleotide (cNMP)-gated ion channels, molecular adaptors, metalloendopeptidase inhibitors, electron transfer, and calcium channel regulators. The top CC categories mainly included cytoplasm, intracellular organelle part, and main axon. Through the KEGG database, a total of 10 statistically significant pathways were obtained (Figure 6D). The DEPs were mainly involved in mitophagy, spliceosome, autophagy, and peroxisome. In addition, the KEGG analysis indicated that NP etiology in the ACC shared similar pathways with other neurological diseases, such as amyotrophic lateral sclerosis (ALS), spinocerebellar ataxia (SCA), and Huntington’s disease (HD). These results demonstrated that autophagic and mitochondrial mechanisms are involved in cingulate neurons after peripheral nerve injury.
Figure 6

Bioinformatic analysis for differentially expressed proteins in sham and NP rats. (A) Biological process. (B) Cell components. (C) Molecular function. Darker colors indicate higher statistical significance. (D) Bubble chart shows KEGG analysis of differential proteins. The horizontal axis represents a rich factor (ratio of the sum of differential proteins enriched in a pathway to the number of proteins annotated by the pathway). Bubble size indicates the number of proteins included in each pathway, and different colors indicate different p-values.
Finally, the PPI network was constructed, with relevant hub molecules identified (Figure 7A). According to the degree rank, the top 10 molecules were apolipoprotein B-100 (Apob, degree = 7), dynactin subunit 6 (Dctn6, degree = 4), electron transfer flavoprotein subunit beta (Etfb, degree = 4), heat shock 70 kDa protein 4 (Hspa4, degree = 6), mitochondrial ribosomal protein 63 (Mrpl57, degree = 6), peptidyl-prolyl cis-trans isomerase F (Ppif, degree = 6), RNA-binding protein 8A (Rbm8a, degree = 5), superoxide dismutase, mitochondrial (Sod2, degree = 6), serine/arginine-rich splicing factor 2 (Srsf2, degree = 6), and transferrin receptor protein 1 (Tfrc, degree = 5) (Figure 7B).
Figure 7

Interaction networks analyzed for differentially expressed proteins. (A) Protein-protein interaction. Node color represents the regulation type. Pink nodes indicate upregulated genes and blue nodes indicate downregulated genes. Node size represents the degree value. A larger node size indicates a higher degree value. Different line styles denote different sources of protein-protein interaction. (B) Ten proteins with maximum interaction counts are listed.
Comparison of NP-related changes in mRNA and protein
We combined the proteomic and transcriptomic data, and then subjected them to Venn diagram analysis, with 10 overlapping targets finally found (Figure 8A). We looked at concordance in the directions of change between mRNA and protein and identified four genes for mRNA and protein with trends that were concordantly decreasing, including glycerol-3-phosphate acyltransferase 1, mitochondrial (Gpam), XK-related protein 4 (Xkr4), NIPA-like domain-containing 3 (Nipal3), and homeodomain-interacting protein kinase 3 (Hipk3). In addition, we identified six genes with decreasing mRNA and increasing protein levels with NP, including interactor protein for cytohesin exchange factors 1 (Ipcef1), abl-interactor 2 (Abi2), lysosome membrane protein 2 (Scarb2), solute carrier family 7, member 14 (Slc7a14), aquaporin-1 (Aqp1), and neurexin-1-beta (Nrxn1). We further tested the intensity of the association between the proteomic and transcriptomic data of the four targets with consistent trends. Spearman correlation analysis showed strong correlations between mRNA and protein levels in Nipal3 (r = 0.761), Hipk3 (r = 0.720), and Xkr4 (r = 0.641), but no significant correlation in Gpam (p = 0.075) (Figures 8B–E).
Figure 8

Integrated analysis of transcriptomic and proteomic data. (A) Venn diagram showing 10 overlapping targets between transcriptomic and proteomic data. (B–E) There is a significant correlation between transcriptomic and proteomic data of Nipal2 (B), Hipk3 (C), and Xkr4 (D) instead of Gpam (E). Pearson’s correlation test.
Discussion
Although molecular alterations within the ACC after NP have been investigated in prior studies, these data have been limited to RNA-seq of mRNA expression. Thus, the present study utilized a combination of quantitative proteomic and transcriptomic approaches to examine molecular changes within the ACC as a consequence of prolonged exposure to peripheral nerve injury. We were able to confirm previously reported molecular alterations underlying synaptic transmission and plasticity via RNA-seq. However, herein we provided another perspective of understanding through proteomics, indicating the mitochondrial involvement at the cortical level.
Transcriptomic profiling of the ACC in NP model rats
During the pathological process of NP, noxious signals are transmitted from the periphery to the ACC via the thalamus, which consequently leads to enhancements of cingulate synaptic plasticity and neuronal excitability. LTP of glutamatergic transmission in several pain-related sensory central synapses including the ACC is considered the classic experimental model for studying synaptic mechanisms of chronic pain (Bliss et al., 2016). Our transcriptomic data showed differentially expressed mRNAs enriched in synaptic functions and pathways in the ACC between NP and sham rats, which was subsequently supported by our miRNAome data. Next, we will summarize breakthroughs and insights of these GO terms and KEGG pathways relevant to cingulate synaptic mechanisms under the condition of NP.
Nerve injury triggers excessive glutamate release, which induces Ca2+ influx into postsynaptic neurons via NMDAR opening. The expression of NP-related cingulate LTP depends on a complex cascade of Ca2+-calmodulin (CaM) dependent signaling pathways. As a pivotal hub of the cascade reactions, adenylyl cyclase 1 (AC1) catalyzes ATP to cyclic AMP (cAMP), which subsequently activates cAMP-dependent protein kinase (PKA). PKA drives the synthesis of several downstream plasticity proteins via phosphorylation of cAMP response element-binding protein (CREB) in the nucleus. These biochemical events ultimately lead to enhanced phosphorylation and membrane insertion of NMDAR and Ca2+-permeable AMPAR, thus increasing postsynaptic sensitivity (Xu et al., 2008; Bliss et al., 2016). In addition, AC1 is pivotal for the generation of presynaptic LTP via downstream PKA-dependent signaling (Zhuo, 2016). Inhibiting AC1 activity by selective inhibitors blocks behavioral sensitization and injury-related anxiety in chronic pain models, which is a hopeful strategy for the management of NP (Li et al., 2020). Apart from glutamatergic mechanisms, ACC pathophysiology is featured with cortical disinhibition. Nerve injury facilitates GABA removal from the synaptic cleft, decreases GABA release, and suppresses inhibitory postsynaptic currents in cingulate pyramidal neurons (Narita et al., 2011; Blom et al., 2014). The loss of GABAergic control predisposes hyperactivity of cingulate pyramidal neurons and contributes to pain-related aversion. Thus, upregulating GABAergic inhibitory tone within the ACC provides long-lasting pain relief (Juarez-Salinas et al., 2019).
Our KEGG analysis indicated important roles of cholinergic synapse and estrogen signaling pathways in the ACC under the condition of NP. Accumulating evidence indicated that estrogen in the ACC drove emotional pain via estrogen receptor-β/PKA and G protein-coupled estrogen receptor-1/protein kinase B pathways to promote NMDAR-mediated synaptic plasticity (Xiao et al., 2013; Zang et al., 2020), while cholinergic systems alleviated NP through the activation of GABAAR-mediated inhibitory transmission via muscarinic M1 receptors (Koga et al., 2019). In addition to calcium signaling, our GO analysis emphasized the activity of voltage-gated potassium channels (Kv). In 2017, Gao et al. revealed that NP-related cingulate pyramidal neuron hyperactivity was associated with a reduction in Kv2-mediated currents, and functional restoration of Kv2 could reduce neural hyperexcitability and produce analgesia (Gao et al., 2017).
Interestingly, we also noticed the enrichment of adhesion molecules and caveola in the GO terms. Synaptic adhesion molecules play key roles in the modulation of synapse development, neural circuits, and behaviors (Kurshan and Shen, 2019). In accordance with these functions, synaptic adhesion molecules in the ACC have been implicated in a series of brain diseases and dysfunctions (Thomas et al., 2002, 2004). Under the condition of NP, cingulate activation alternates the content of specific adhesion molecules. Among them, neural cell adhesion molecule 1 contributes to behavioral sensitization via spine organization and NMDAR-dependent LTP (Ko et al., 2018; Pan et al., 2018). Caveola, a special type of membrane raft, is implicated in normal physiological processes of the central nervous system (CNS) including endocytosis, cell metabolism, signaling transduction (Allen et al., 2007), spine morphology (Hering et al., 2003), axon guidance (Guirland and Zheng, 2007), and receptor trafficking (Francesconi et al., 2009). As a main scaffolding protein in the caveola, caveolin-1 (Cav-1) plays a key role in synapse formation and plasticity. The dysregulation of Cav-1 is implicated in abnormal neuronal signaling in CNS pathological processes (Gaudreault et al., 2004). Yang et al. first demonstrated that Cav-1 in ACC neurons contributes to NP via promoting membrane insertion of NMDAR (Yang et al., 2015).
Proteomic profiling of the ACC in NP model rats
Autophagy, an evolutionarily-conserved intracellular process, delivers aged proteins and damaged organelles into the lysosome for degradation. It is pivotal for neuronal survival and synaptic maintenance/plasticity in the CNS and for receptor turnover, synaptic integrity, and myelination in the peripheral nervous system (PNS) (Liu et al., 2019). Pain bears common features with neurodegenerative diseases, such as HD, SCA, and ALS, in which autophagy plays a critical role in the pathology (Deneubourg et al., 2022). As an early event in the origin of NP, elevated autophagic activity in injured nerves is esteemed as an adaptation to stress and to help diminish myelin debris (Marinelli et al., 2014). Although autophagy is differentially regulated in spinal neurons and glial cells following peripheral nerve injury, disrupted autophagic activity is considered to play a key role in the induction and development of NP (Zhang et al., 2013; Liao et al., 2022). In general, autophagic impairment contributes to NP, while autophagic upregulation suppresses pain behavior, possibly by suppressing inflammatory responses (Liao et al., 2022). Correspondingly, drugs aiming at upregulating autophagic levels have been screened out for alleviating NP in preclinical models (Shi et al., 2013; Yin et al., 2018). Emerging data indicated that activating cingulate microglial autophagy ameliorated NP via neuroinflammation suppression (Meng et al., 2020). In accordance with this report, our protein profiling analysis identified the enrichment of significant proteins in autophagic pathways. Thus, autophagy in the cortical pain matrix remains an interesting topic for future studies.
The “mitotoxicity theory” of NP has its origin in chemotherapy-related neuropathy and later proved to be true of peripheral nerve injury. These factors induce mitochondrial dysfunction in primary sensory neurons by disturbing mitochondrial functions (including bioenergetics, transport, fusion, and mitophagy) and enhancing nitro-oxidative stress, which ultimately contributes to axonal growth defects and subsequent sensory neuron sensitization (Bennett et al., 2014; Doyle and Salvemini, 2021). Mitophagy, selective autophagic degradation of damaged mitochondria, constitutes a major mitochondrial quality control pathway. The dysregulation of mitophagy in the PNS leads to aggregation of impaired mitochondria, reactive oxygen species (ROS) production, microglial activation, and myelinoclasis, and as a result, the development of NP (Dai et al., 2020; Huang et al., 2022). At the level of the SDH, impaired mitophagy in microglial cells facilitates NLRP3 inflammasome-mediated neuroinflammation via mitochondrial ROS production, finally promoting pain-related plastic changes in the CNS (Shao et al., 2021). Correspondingly, strategies aiming at improving mitophagy have shown curative effects in experimental NP models (Doyle and Salvemini, 2021; Shao et al., 2021). However, whether mitophagy impairment in supraspinal structures underlines NP remains elusive. Herein, our analysis highlights the key role of mitochondrial dysfunction in the ACC in NP pathophysiology.
Apart from mitochondria, peroxisomes are another type of intracellular organelles maintaining the redox cellular state. Genetic deficit of peroxisome could induce severe demyelination, axonal degeneration, and neuroinflammation (Powers and Moser, 1998; Trompier et al., 2014). Peroxisome involvement has been reported in the development of specific degenerative diseases (Trompier et al., 2014), as well as NP. Chemotherapy-related neuropathy involves peroxisome alterations at the levels of both dorsal root ganglion and SDH (Zanardelli et al., 2014). Correspondingly, peroxisome proliferator-activated receptor (PPAR) agonists emerged as a potential therapeutic strategy (Lan et al., 2007) for NP, owing to their anti-inflammatory and anti-oxidative effects (Freitag and Miller, 2014; Zanardelli et al., 2014). Emerging evidence also hints at the potential role of PPAR signaling in NP suppression via microglial modulation at the level of the ACC (Georgieva et al., 2019). Interestingly, our KEGG analysis further supports the role of cingulate peroxisome in NP pathophysiology.
Integration of the transcriptomic and proteomic data
Integrative multi-omic analysis revealed several targets that may be involved in NP pathophysiology. Non-imprinted in Prader-Willi/Angelman Syndrome (NIPA)-like domain-containing (NPAL) proteins shares high similarity at the levels of amino acid and domain structure with NIPA proteins that function as transporters or receptors. However, knowledge concerning the function of NPAL is still lacking. So far, there has only been one report showing that the disruption of the function of the mouse Nipal3 gene induced increased IgE Levels, lung dysfunction, as well as behavior deficits (Grzmil et al., 2009). HIPK3 belongs to a family of highly-conserved serine/threonine kinases that mediate developmental processes through the regulation of corresponding transcription factors. It has been reported to participate in apoptosis, inflammation and oxidative stress modulation, steroidogenic gene expression, and glucose metabolism (Rochat-Steiner et al., 2000; Lan et al., 2007; Shojima et al., 2012; Liu et al., 2020). As a phospholipid-scrambling protein in the process of efferocytosis, Xkr4 is specifically expressed in the brain where it, upon activation by caspase 6, induces the exposure of PtdSer on dendrites, axons, and synapses to induce microglial responses for neural network remodeling (Suzuki et al., 2014; Maruoka et al., 2021). Gpam, a key member of the mitochondrial enzyme complex, is involved in the biosynthesis process of triacylglycerol and glycerolphospholipids. Loss-of-function mutation of Gpam has been reported to result in corticospinal tract hypomyelination in patients with cerebral palsy (Li N. et al., 2022).
Although molecular changes in the ACC in the case of NP have been explored in prior studies, these have been limited to RNAseq for mRNA expression (Su et al., 2021; Dai et al., 2022; Zhang et al., 2022). One predisposed assumption of these studies was that changes in mRNA determine changes in gene function (protein levels). However, it should be noted that the transcriptome, especially in the brain, does not often precisely indicate protein abundance (Sharma et al., 2015; Liu et al., 2016). A scientific merit of the present study is that through the combination of proteomic profiling and RNA sequencing, we confirmed priorly reported transcriptional changes in the ACC of NP rats, such as synaptic transmission and plasticity, and also provided another layer of understanding concerning the mitochondrial involvement in NP at the cortical level. Surprisingly, in this study, we observed a low correlation between proteomic and RNA-seq data, with only 10 overlapping targets identified. Even in the overlapping targets, the direction of alteration of a protein under the condition of NP cannot be effectively predicted based on alterations in its coding mRNA. One technical reason for this low correlation is the different sensitivity of these approaches based on the current technical level, with RNA-seq profiles reaching tens of million RNA counts in a regular project, compared to tens of thousands in protein mass spectroscopy tests. Hence, comparing these two sources would unavoidably look at the top fractions of very different collections of reads. Importantly, this phenomenon also suggests that processes driving NP-related alterations at the level of protein may involve changes in rates of mRNA or protein turnover or be post-translation dominated (Vogel and Marcotte, 2012).
Post-translational modifications, in particular fast phosphorylation-based control mechanisms that have not been evaluated in our study, have been linked to advanced brain functions. Synaptic signaling and plasticity have been highlighted as pivotal for the phosphorylation of cytoskeletal, scaffolding, and other synaptic proteins, as well as neuroreceptors (Woolfrey and Dell'Acqua, 2015; Dieterich and Kreutz, 2016). Several excellent reviews have discussed the key role of phosphorylation of synaptic proteins in the ACC under the condition of NP (Qiu et al., 2011; Bliss et al., 2016). Given the above, integration of quantitative phosphoproteomic analysis in the multi-omics is imperative to gain a deeper understanding of molecular substrates of NP. Apart from expression levels, neural activities also change the synthesis and degradation rate of synaptic proteins. Therefore, the interrogation of protein turnover rather than merely expression at the level of synapses has gained increasing interest in the domain of pain research. Recently, Ko et al. identified a series of proteins in the ACC exhibiting changed protein turnover rates but not total levels that contributed to the development of NP via synaptosomal proteome (Ko et al., 2018, 2020). Thus, changes in protein turnover partially account for the low correlation between proteomic and transcriptomic findings identified herein.
Limitations
First, this study was performed only at a one-time point after SNI. The transition toward a persistent pain condition could be reflected by time-dependent alterations in cerebral protein composition in NP models (Ujcikova et al., 2022). Thus, the dynamics of molecular events after SNI surgery in the ACC remains a topic of future discussion. Second, the study was performed 2 weeks after SNI surgery, when negative pain affections including anxiety have been stably elicited together with behavioral hyperalgesia. Thus, current multi-omic data failed to clarify molecular changes specific to the sensory or emotional aspects of NP. Third, as mentioned above, integrative multi-omics involving phospho-proteomics, metabolomics, and even lipidomics might outline more detailed differences in the molecular landscape for NP. Last but not least, although several potential pathways have been identified herein, functional verification is urgently needed to be performed in the future.
Conclusion
Overall, these findings revealed that synapse- and mitochondria-related targets in the ACC are potentially involved in NP. Our data provide evidence for understanding the etiology of NP and the theoretical basis for new therapeutic approaches.
Funding
This research was supported by the National Natural Science Foundation of China (Nos. 82101318 to YB, 81971133 to G-BL, 82130034 and 82221001 to Y-QL), Li Yunqing Expert Workstation of Yunnan Province (No. 202005AF150014 to Y-QL), the Innovation Capability Support Program of Shaanxi (No. 2021TD-57 to Y-QL), and the National Science and Technology Innovation 2030 Major Program (No. 2021ZD0204403 to Y-QL).
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was reviewed and approved by the Ethics Committee of the General Hospital of Norther Theater Command.
Author contributions
YB, G-BL, and Y-QL designed the study. X-TQ, CG, and L-TM performed the experiments. YB and CG wrote the manuscript. YB, CG, X-NL, F-SH, M-MZ, and Q-YZ performed the data analysis. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2023.1164426/full#supplementary-material
SUPPLEMENTARY FIGURE 1Systematic workflow of miRNAome analysis in the anterior cingulate cortex of neuropathic pain rat model. (A) Schematic diagram shows the key steps of this study. (B) miRNAomic workflow.
SUPPLEMENTARY FIGURE 2The quality of RNA-seq data from each ACC sample. (A) Boxplot showing the distribution of gene expression levels in each ACC sample. (B) Distribution diagram showing the density of gene expression levels in each ACC sample. (C) The constituent ratio of the identified DEGs. RPKM, reads per kilobase million.
SUPPLEMENTARY FIGURE 3The quality of small RNA sequencing data from each ACC sample. (A) Boxplot showing the distribution of gene expression levels in each ACC sample. (B) Distribution diagram showing the density of gene expression levels in each ACC sample. TPM, transcript per million.
SUPPLEMENTARY FIGURE 4Small RNA sequencing outcomes. (A) 21 miRNAs were up-regulated, and 12 miRNAs were down-regulated. (B) Volcano plot showed differentially expressed miRNAs. The red and green dots indicate significantly up-regulated and down-regulated miRNAs, respectively. (C) Hierarchical cluster analysis. Red indicates up-regulation, and blue indicates down-regulation. The dendrograms represent the classification of miRNAs. The number in the color scale indicates the z-score.
SUPPLEMENTARY FIGURE 5Bioinformatic analysis for the target genes of differentially expressed miRNAs in sham and NP rats. (A) Biological process. (B) Cell components. (C) Molecular function. Darker colors indicate higher statistical significance. (D) Bubble chart shows KEGG analysis of the target genes. The horizontal axis represents a rich factor (ratio of the sum of differential genes enriched in a pathway to the number of genes annotated by the pathway). Bubble size indicates the number of genes included in each pathway, and different colors indicate different p-values.
SUPPLEMENTARY FIGURE 6Information on the identified proteins in the rats. (A) Distribution of the molecular weight corresponding to each qualitative protein. (B) Distribution of the peptide number corresponding to each qualitative protein..
Footnotes
1.^ https://cutadapt.readthedocs.io/en/stable/
2.^ http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
3.^ https://daehwankimlab.github.io/hisat2/
4.^ http://bioconductor.org/packages/release/bioc/html/DESeq2.html
References
1
Allen J. A. Halverson-Tamboli R. A. Rasenick M. M. (2007). Lipid raft microdomains and neurotransmitter signalling. Nat. Rev. Neurosci.8, 128–140. doi: 10.1038/nrn2059
2
Bai Y. Ma L. T. Chen Y. B. Ren D. Chen Y. B. Li Y. Q. et al . (2019). Anterior insular cortex mediates hyperalgesia induced by chronic pancreatitis in rats. Mol. Brain12:76. doi: 10.1186/s13041-019-0497-5
3
Bali K. K. Kuner R. (2014). Noncoding RNAs: key molecules in understanding and treating pain. Trends Mol. Med.20, 437–448. doi: 10.1016/j.molmed.2014.05.006
4
Bennett G. J. Doyle T. Salvemini D. (2014). Mitotoxicity in distal symmetrical sensory peripheral neuropathies. Nat. Rev. Neurol.10, 326–336. doi: 10.1038/nrneurol.2014.77
5
Bliss T. V. Collingridge G. L. Kaang B. K. Zhuo M. (2016). Synaptic plasticity in the anterior cingulate cortex in acute and chronic pain. Nat. Rev. Neurosci.17, 485–496. doi: 10.1038/nrn.2016.68
6
Blom S. M. Pfister J. P. Santello M. Senn W. Nevian T. (2014). Nerve injury-induced neuropathic pain causes disinhibition of the anterior cingulate cortex. J. Neurosci.34, 5754–5764. doi: 10.1523/jneurosci.3667-13.2014
7
Chen T. Taniguchi W. Chen Q. Y. Tozaki-Saitoh H. Song Q. Liu R. H. et al . (2018). Top-down descending facilitation of spinal sensory excitatory transmission from the anterior cingulate cortex. Nat. Commun.9:1886. doi: 10.1038/s41467-018-04309-2
8
Cohen S. P. Vase L. Hooten W. M. (2021). Chronic pain: an update on burden, best practices, and new advances. Lancet397, 2082–2097. doi: 10.1016/s0140-6736(21)00393-7
9
Dai C. Q. Guo Y. Chu X. Y. (2020). Neuropathic pain: the dysfunction of Drp1, mitochondria, and ROS homeostasis. Neurotox. Res.38, 553–563. doi: 10.1007/s12640-020-00257-2
10
Dai W. Huang S. Luo Y. Cheng X. Xia P. Yang M. et al . (2022). Sex-specific transcriptomic signatures in brain regions critical for neuropathic pain-induced depression. Front. Mol. Neurosci.15:886916. doi: 10.3389/fnmol.2022.886916
11
Delevich K. Tucciarone J. Huang Z. J. Li B. (2015). The mediodorsal thalamus drives feedforward inhibition in the anterior cingulate cortex via parvalbumin interneurons. J. Neurosci.35, 5743–5753. doi: 10.1523/jneurosci.4565-14.2015
12
Deneubourg C. Ramm M. Smith L. J. Baron O. Singh K. Byrne S. C. et al . (2022). The spectrum of neurodevelopmental, neuromuscular and neurodegenerative disorders due to defective autophagy. Autophagy18, 496–517. doi: 10.1080/15548627.2021.1943177
13
Dieterich D. C. Kreutz M. R. (2016). Proteomics of the synapse--a quantitative approach to neuronal plasticity. Mol. Cell. Proteomics15, 368–381. doi: 10.1074/mcp.R115.051482
14
Doyle T. M. Salvemini D. (2021). Mini-review: mitochondrial dysfunction and chemotherapy-induced neuropathic pain. Neurosci. Lett.760:136087. doi: 10.1016/j.neulet.2021.136087
15
Enright A. J. John B. Gaul U. Tuschl T. Sander C. Marks D. S. (2003). MicroRNA targets in Drosophila. Genome Biol.5:R1. doi: 10.1186/gb-2003-5-1-r1
16
Esteller M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet.12, 861–874. doi: 10.1038/nrg3074
17
Finnerup N. B. Kuner R. Jensen T. S. (2021). Neuropathic pain: from mechanisms to treatment. Physiol. Rev.101, 259–301. doi: 10.1152/physrev.00045.2019
18
Francesconi A. Kumari R. Zukin R. S. (2009). Regulation of group I metabotropic glutamate receptor trafficking and signaling by the caveolar/lipid raft pathway. J. Neurosci.29, 3590–3602. doi: 10.1523/jneurosci.5824-08.2009
19
Freitag C. M. Miller R. J. (2014). Peroxisome proliferator-activated receptor agonists modulate neuropathic pain: a link to chemokines?Front. Cell. Neurosci.8:238. doi: 10.3389/fncel.2014.00238
20
Gao S. H. Shen L. L. Wen H. Z. Zhao Y. D. Chen P. H. Ruan H. Z. (2020). The projections from the anterior cingulate cortex to the nucleus accumbens and ventral tegmental area contribute to neuropathic pain-evoked aversion in rats. Neurobiol. Dis.140:104862. doi: 10.1016/j.nbd.2020.104862
21
Gao S. H. Shen L. L. Wen H. Z. Zhao Y. D. Ruan H. Z. (2017). Inhibition of metabotropic glutamate receptor subtype 1 alters the excitability of the commissural pyramidal neuron in the rat anterior cingulate cortex after chronic constriction injury to the sciatic nerve. Anesthesiology127, 515–533. doi: 10.1097/aln.0000000000001654
22
Gaudreault S. B. Dea D. Poirier J. (2004). Increased caveolin-1 expression in Alzheimer's disease brain. Neurobiol. Aging25, 753–759. doi: 10.1016/j.neurobiolaging.2003.07.004
23
Georgieva M. Wei Y. Dumitrascuta M. Pertwee R. Finnerup N. B. Huang W. (2019). Fatty acid suppression of glial activation prevents central neuropathic pain after spinal cord injury. Pain160, 2724–2742. doi: 10.1097/j.pain.0000000000001670
24
Gomez-Varela D. Barry A. M. Schmidt M. (2019). Proteome-based systems biology in chronic pain. J. Proteome190, 1–11. doi: 10.1016/j.jprot.2018.04.004
25
Grzmil P. Konietzko J. Boehm D. Hoelter S. M. Aguilar A. Javaheri A. et al . (2009). Targeted disruption of the mouse Npal3 gene leads to deficits in behavior, increased IgE levels, and impaired lung function. Cytogenet. Genome Res.125, 186–200. doi: 10.1159/000230003
26
Guirland C. Zheng J. Q. (2007). Membrane lipid rafts and their role in axon guidance. Adv. Exp. Med. Biol.621, 144–155. doi: 10.1007/978-0-387-76715-4_11
27
Hering H. Lin C. C. Sheng M. (2003). Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J. Neurosci.23, 3262–3271. doi: 10.1523/jneurosci.23-08-03262.2003
28
Huang Z. Xiao P. Y. Chen J. Y. Zeng Q. Huang B. X. Yu J. et al . (2022). Mammalian sterile 20-like kinase 1 mediates neuropathic pain associated with its effects on regulating Mitophagy in Schwann cells. Oxidative Med. Cell. Longev.2022, 3458283–3458217. doi: 10.1155/2022/3458283
29
Johansen J. P. Fields H. L. Manning B. H. (2001). The affective component of pain in rodents: direct evidence for a contribution of the anterior cingulate cortex. Proc. Natl. Acad. Sci. U. S. A.98, 8077–8082. doi: 10.1073/pnas.141218998
30
Juarez-Salinas D. L. Braz J. M. Etlin A. Gee S. Sohal V. Basbaum A. I. (2019). GABAergic cell transplants in the anterior cingulate cortex reduce neuropathic pain aversiveness. Brain142, 2655–2669. doi: 10.1093/brain/awz203
31
Kang S. J. Kwak C. Lee J. Sim S. E. Shim J. Choi T. et al . (2015). Bidirectional modulation of hyperalgesia via the specific control of excitatory and inhibitory neuronal activity in the ACC. Mol. Brain8:81. doi: 10.1186/s13041-015-0170-6
32
Ko H. G. Choi J. H. Park D. I. Kang S. J. Lim C. S. Sim S. E. et al . (2018). Rapid turnover of cortical NCAM1 regulates synaptic reorganization after peripheral nerve injury. Cell Rep.22, 748–759. doi: 10.1016/j.celrep.2017.12.059
33
Ko H. G. Park D. I. Lee J. H. Turck C. W. Kaang B. K. (2020). Proteomic analysis of synaptic protein turnover in the anterior cingulate cortex after nerve injury. Mol. Brain13:19. doi: 10.1186/s13041-020-0564-y
34
Koga K. Matsuzaki Y. Migita K. Shimoyama S. Eto F. Nakagawa T. et al . (2019). Stimulating muscarinic M(1) receptors in the anterior cingulate cortex reduces mechanical hypersensitivity via GABAergic transmission in nerve injury rats. Brain Res.1704, 187–195. doi: 10.1016/j.brainres.2018.10.013
35
Krüger J. Rehmsmeier M. (2006). RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res.34, W451–W454. doi: 10.1093/nar/gkl243
36
Kurshan P. T. Shen K. (2019). Synaptogenic pathways. Curr. Opin. Neurobiol.57, 156–162. doi: 10.1016/j.conb.2019.03.005
37
Lan H. C. Li H. J. Lin G. Lai P. Y. Chung B. C. (2007). Cyclic AMP stimulates SF-1-dependent CYP11A1 expression through homeodomain-interacting protein kinase 3-mediated Jun N-terminal kinase and c-Jun phosphorylation. Mol. Cell. Biol.27, 2027–2036. doi: 10.1128/mcb.02253-06
38
Langmead B. Trapnell C. Pop M. Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol.10:R25. doi: 10.1186/gb-2009-10-3-r25
39
Lee D. E. Kim S. J. Zhuo M. (1999). Comparison of behavioral responses to noxious cold and heat in mice. Brain Res.845, 117–121. doi: 10.1016/s0006-8993(99)01956-3
40
Li Q. Y. Chen S. X. Liu J. Y. Yao P. W. Duan Y. W. Li Y. Y. et al . (2022). Neuroinflammation in the anterior cingulate cortex: the potential supraspinal mechanism underlying the mirror-image pain following motor fiber injury. J. Neuroinflammation19:162. doi: 10.1186/s12974-022-02525-8
41
Li X. H. Chen Q. Y. Zhuo M. (2020). Neuronal adenylyl cyclase targeting central plasticity for the treatment of chronic pain. Neurotherapeutics17, 861–873. doi: 10.1007/s13311-020-00927-1
42
Li X. Y. Ko H. G. Chen T. Descalzi G. Koga K. Wang H. et al . (2010). Alleviating neuropathic pain hypersensitivity by inhibiting PKMzeta in the anterior cingulate cortex. Science330, 1400–1404. doi: 10.1126/science.1191792
43
Li N. Zhou P. Tang H. He L. Fang X. Zhao J. et al . (2022). In-depth analysis reveals complex molecular aetiology in a cohort of idiopathic cerebral palsy. Brain145, 119–141. doi: 10.1093/brain/awab209
44
Liao M. F. Lu K. T. Hsu J. L. Lee C. H. Cheng M. Y. Ro L. S. (2022). The role of autophagy and apoptosis in neuropathic pain formation. Int. J. Mol. Sci.23:2685. doi: 10.3390/ijms23052685
45
Liu Y. Beyer A. Aebersold R. (2016). On the dependency of cellular protein levels on mRNA abundance. Cells165, 535–550. doi: 10.1016/j.cell.2016.03.014
46
Liu M. G. Chen J. (2014). Preclinical research on pain comorbidity with affective disorders and cognitive deficits: challenges and perspectives. Prog. Neurobiol.116, 13–32. doi: 10.1016/j.pneurobio.2014.01.003
47
Liu B. Hou Q. Ma Y. Han X. (2020). HIPK3 mediates inflammatory cytokines and oxidative stress markers in monocytes in a rat model of Sepsis through the JNK/c-Jun signaling pathway. Inflammation43, 1127–1142. doi: 10.1007/s10753-020-01200-5
48
Liu X. Zhu M. Ju Y. Li A. Sun X. (2019). Autophagy dysfunction in neuropathic pain. Neuropeptides75, 41–48. doi: 10.1016/j.npep.2019.03.005
49
Ma L. T. Bai Y. Cao P. Ren K. X. Chen J. Zhang T. et al . (2022). The analgesic effects of β-elemene in rats with neuropathic pain by inhibition of spinal astrocytic ERK activation. Mol. Pain18:17448069221121562. doi: 10.1177/17448069221121562
50
Mao X. Cai T. Olyarchuk J. G. Wei L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics21, 3787–3793. doi: 10.1093/bioinformatics/bti430
51
Marinelli S. Nazio F. Tinari A. Ciarlo L. D'Amelio M. Pieroni L. et al . (2014). Schwann cell autophagy counteracts the onset and chronification of neuropathic pain. Pain155, 93–107. doi: 10.1016/j.pain.2013.09.013
52
Maruoka M. Zhang P. Mori H. Imanishi E. Packwood D. M. Harada H. et al . (2021). Caspase cleavage releases a nuclear protein fragment that stimulates phospholipid scrambling at the plasma membrane. Mol. Cell81, 1397–1410.e9. doi: 10.1016/j.molcel.2021.02.025
53
Meda K. S. Patel T. Braz J. M. Malik R. Turner M. L. Seifikar H. et al . (2019). Microcircuit mechanisms through which Mediodorsal thalamic input to anterior cingulate cortex exacerbates pain-related aversion. Neuron102, 944–959.e3. doi: 10.1016/j.neuron.2019.03.042
54
Meng X. L. Fu P. Wang L. Yang X. Hong G. Zhao X. et al . (2020). Increased EZH2 levels in anterior cingulate cortex microglia aggravate neuropathic pain by inhibiting autophagy following brachial plexus avulsion in rats. Neurosci. Bull.36, 793–805. doi: 10.1007/s12264-020-00502-w
55
Narita M. Niikura K. Nanjo-Niikura K. Narita M. Furuya M. Yamashita A. et al . (2011). Sleep disturbances in a neuropathic pain-like condition in the mouse are associated with altered GABAergic transmission in the cingulate cortex. Pain152, 1358–1372. doi: 10.1016/j.pain.2011.02.016
56
Nie L. Wu G. Culley D. E. Scholten J. C. Zhang W. (2007). Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit. Rev. Biotechnol.27, 63–75. doi: 10.1080/07388550701334212
57
Niederberger E. Geisslinger G. (2008). Proteomics in neuropathic pain research. Anesthesiology108, 314–323. doi: 10.1097/01.anes.0000299838.13368.6e
58
Ong W. Y. Stohler C. S. Herr D. R. (2019). Role of the prefrontal cortex in pain processing. Mol. Neurobiol.56, 1137–1166. doi: 10.1007/s12035-018-1130-9
59
Pan W. Zhang G. F. Li H. H. Ji M. H. Zhou Z. Q. Li K. Y. et al . (2018). Ketamine differentially restores diverse alterations of neuroligins in brain regions in a rat model of neuropathic pain-induced depression. Neuroreport29, 863–869. doi: 10.1097/wnr.0000000000001045
60
Powers J. M. Moser H. W. (1998). Peroxisomal disorders: genotype, phenotype, major neuropathologic lesions, and pathogenesis. Brain Pathol.8, 101–120. doi: 10.1111/j.1750-3639.1998.tb00139.x
61
Qiu S. Li X. Y. Zhuo M. (2011). Post-translational modification of NMDA receptor GluN2B subunit and its roles in chronic pain and memory. Semin. Cell Dev. Biol.22, 521–529. doi: 10.1016/j.semcdb.2011.06.003
62
Ren D. Li J. N. Qiu X. T. Wan F. P. Wu Z. Y. Fan B. Y. et al . (2022). Anterior cingulate cortex mediates hyperalgesia and anxiety induced by chronic pancreatitis in rats. Neurosci. Bull.38, 342–358. doi: 10.1007/s12264-021-00800-x
63
Rochat-Steiner V. Becker K. Micheau O. Schneider P. Burns K. Tschopp J. (2000). FIST/HIPK3: a Fas/FADD-interacting serine/threonine kinase that induces FADD phosphorylation and inhibits fas-mediated Jun NH(2)-terminal kinase activation. J. Exp. Med.192, 1165–1174. doi: 10.1084/jem.192.8.1165
64
Scholz J. Finnerup N. B. Attal N. Aziz Q. Baron R. Bennett M. I. et al . (2019). The IASP classification of chronic pain for ICD-11: chronic neuropathic pain. Pain160, 53–59. doi: 10.1097/j.pain.0000000000001365
65
Shao S. Xu C. B. Chen C. J. Shi G. N. Guo Q. L. Zhou Y. et al . (2021). Divanillyl sulfone suppresses NLRP3 inflammasome activation via inducing mitophagy to ameliorate chronic neuropathic pain in mice. J. Neuroinflammation18:142. doi: 10.1186/s12974-021-02178-z
66
Sharma K. Schmitt S. Bergner C. G. Tyanova S. Kannaiyan N. Manrique-Hoyos N. et al . (2015). Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci.18, 1819–1831. doi: 10.1038/nn.4160
67
Shi G. Shi J. Liu K. Liu N. Wang Y. Fu Z. et al . (2013). Increased miR-195 aggravates neuropathic pain by inhibiting autophagy following peripheral nerve injury. Glia61, 504–512. doi: 10.1002/glia.22451
68
Shojima N. Hara K. Fujita H. Horikoshi M. Takahashi N. Takamoto I. et al . (2012). Depletion of homeodomain-interacting protein kinase 3 impairs insulin secretion and glucose tolerance in mice. Diabetologia55, 3318–3330. doi: 10.1007/s00125-012-2711-1
69
Starobova H. S. W. A. H. Lewis R. J. Vetter I. (2018). Transcriptomics in pain research: insights from new and old technologies. Mol. Omics14, 389–404. doi: 10.1039/c8mo00181b
70
Su S. Li M. Wu D. Cao J. Ren X. Tao Y. X. et al . (2021). Gene transcript alterations in the spinal cord, anterior cingulate cortex, and amygdala in mice following peripheral nerve injury. Front. Cell Dev. Biol.9:634810. doi: 10.3389/fcell.2021.634810
71
Suzuki J. Imanishi E. Nagata S. (2014). Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem.289, 30257–30267. doi: 10.1074/jbc.M114.583419
72
Tang J. Ko S. Ding H. K. Qiu C. S. Calejesan A. A. Zhuo M. (2005). Pavlovian fear memory induced by activation in the anterior cingulate cortex. Mol. Pain1:6. doi: 10.1186/1744-8069-1-6
73
Thomas A. J. Davis S. Ferrier I. N. Kalaria R. N. O'Brien J. T. (2004). Elevation of cell adhesion molecule immunoreactivity in the anterior cingulate cortex in bipolar disorder. Biol. Psychiatry55, 652–655. doi: 10.1016/j.biopsych.2003.10.015
74
Thomas A. J. Ferrier I. N. Kalaria R. N. Davis S. O'Brien J. T. (2002). Cell adhesion molecule expression in the dorsolateral prefrontal cortex and anterior cingulate cortex in major depression in the elderly. Br. J. Psychiatry181, 129–134. doi: 10.1017/s0007125000161847
75
Trompier D. Vejux A. Zarrouk A. Gondcaille C. Geillon F. Nury T. et al . (2014). Brain peroxisomes. Biochimie98, 102–110. doi: 10.1016/j.biochi.2013.09.009
76
Ujcikova H. Robles D. Yue X. Svoboda P. Lee Y. S. Navratilova E. (2022). Time-dependent changes in protein composition of medial prefrontal cortex in rats with neuropathic pain. Int. J. Mol. Sci.23:955. doi: 10.3390/ijms23020955
77
Vogel C. Marcotte E. M. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet.13, 227–232. doi: 10.1038/nrg3185
78
Wang H. Xu H. Wu L. J. Kim S. S. Chen T. Koga K. et al . (2011). Identification of an adenylyl cyclase inhibitor for treating neuropathic and inflammatory pain. Sci. Transl. Med.3:65ra3. doi: 10.1126/scitranslmed.3001269
79
Woolfrey K. M. Dell'Acqua M. L. (2015). Coordination of protein phosphorylation and Dephosphorylation in synaptic plasticity. J. Biol. Chem.290, 28604–28612. doi: 10.1074/jbc.R115.657262
80
Xiao X. Yang Y. Zhang Y. Zhang X. M. Zhao Z. Q. Zhang Y. Q. (2013). Estrogen in the anterior cingulate cortex contributes to pain-related aversion. Cereb. Cortex23, 2190–2203. doi: 10.1093/cercor/bhs201
81
Xu H. Wu L. J. Wang H. Zhang X. Vadakkan K. I. Kim S. S. et al . (2008). Presynaptic and postsynaptic amplifications of neuropathic pain in the anterior cingulate cortex. J. Neurosci.28, 7445–7453. doi: 10.1523/jneurosci.1812-08.2008
82
Yang J. X. Hua L. Li Y. Q. Jiang Y. Y. Han D. Liu H. et al . (2015). Caveolin-1 in the anterior cingulate cortex modulates chronic neuropathic pain via regulation of NMDA receptor 2B subunit. J. Neurosci. Res.35, 36–52. doi: 10.1523/jneurosci.1161-14.2015
83
Yin Y. Yi M. H. Kim D. W. (2018). Impaired autophagy of GABAergic interneurons in neuropathic pain. Pain Res. Manag.2018:9185368. doi: 10.1155/2018/9185368
84
Young M. D. Wakefield M. J. Smyth G. K. Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol.11:R14. doi: 10.1186/gb-2010-11-2-r14
85
Zanardelli M. Micheli L. Cinci L. Failli P. Ghelardini C. Di Cesare Mannelli L. (2014). Oxaliplatin neurotoxicity involves peroxisome alterations. PPARγ agonism as preventive pharmacological approach. PLoS One9:e102758. doi: 10.1371/journal.pone.0102758
86
Zang K. K. Xiao X. Chen L. Q. Yang Y. Cao Q. L. Tang Y. L. et al . (2020). Distinct function of estrogen receptors in the rodent anterior cingulate cortex in pain-related aversion. Anesthesiology133, 165–184. doi: 10.1097/aln.0000000000003324
87
Zhang Y. Jiang S. Liao F. Huang Z. Yang X. Zou Y. et al . (2022). A transcriptomic analysis of neuropathic pain in the anterior cingulate cortex after nerve injury. Bioengineered13, 2058–2075. doi: 10.1080/21655979.2021.2021710
88
Zhang E. Yi M. H. Ko Y. Kim H. W. Seo J. H. Lee Y. H. et al . (2013). Expression of LC3 and Beclin 1 in the spinal dorsal horn following spinal nerve ligation-induced neuropathic pain. Brain Res.1519, 31–39. doi: 10.1016/j.brainres.2013.04.055
89
Zhou L. Chen J. Li Z. Li X. Hu X. Huang Y. et al . (2010). Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One5:e15224. doi: 10.1371/journal.pone.0015224
90
Zhu X. Tang H. D. Dong W. Y. Kang F. Liu A. Mao Y. et al . (2021). Distinct thalamocortical circuits underlie allodynia induced by tissue injury and by depression-like states. Nat. Neurosci.24, 542–553. doi: 10.1038/s41593-021-00811-x
91
Zhuang X. Huang L. Gu Y. Wang L. Zhang R. Zhang M. et al . (2021). The anterior cingulate cortex projection to the dorsomedial striatum modulates hyperalgesia in a chronic constriction injury mouse model. Arch. Med. Sci.17, 1388–1399. doi: 10.5114/aoms.2019.85202
92
Zhuo M. (2006). Molecular mechanisms of pain in the anterior cingulate cortex. J. Neurosci. Res.84, 927–933. doi: 10.1002/jnr.21003
93
Zhuo M. (2008). Cortical excitation and chronic pain. Trends Neurosci.31, 199–207. doi: 10.1016/j.tins.2008.01.003
94
Zhuo M. (2016). Neural mechanisms underlying anxiety-chronic pain interactions. Trends Neurosci.39, 136–145. doi: 10.1016/j.tins.2016.01.006
Summary
Keywords
spared nerve injury, neuropathic pain (NP), anterior cingulate cortex (ACC), transcriptomics, proteomics
Citation
Qiu X-T, Guo C, Ma L-T, Li X-N, Zhang Q-Y, Huang F-S, Zhang M-M, Bai Y, Liang G-B and Li Y-Q (2023) Transcriptomic and proteomic profiling of the anterior cingulate cortex in neuropathic pain model rats. Front. Mol. Neurosci. 16:1164426. doi: 10.3389/fnmol.2023.1164426
Received
12 February 2023
Accepted
25 May 2023
Published
15 June 2023
Volume
16 - 2023
Edited by
Jie Zhang, Xiamen University, China
Reviewed by
Bin Pan, Medical College of Wisconsin, United States; Hermona Soreq, Hebrew University of Jerusalem, Israel
Updates
Copyright
© 2023 Qiu, Guo, Ma, Li, Zhang, Huang, Zhang, Bai, Liang and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Bai, sydbaiyang@163.com; Guo-Biao Liang, liangguobiao6708@vip.163.com; Yun-Qing Li, deptanat@fmmu.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.