- 1Cotton Fiber Bioscience and Utilization Research Unit, U.S. Department of Agriculture, Southern Regional Research Center, Agricultural Research Service, New Orleans, LA, United States
- 2Microbial and Chemical Food Safety Research Unit, U.S. Department of Agriculture, Eastern Regional Research Center, Agricultural Research Service, Wyndmoor, PA, United States
- 3Commodity Utilization Research Unit, U.S. Department of Agriculture, Southern Regional Research Center, Agricultural Research Service, New Orleans, LA, United States
Cotton gin byproduct (CGB), a high-volume byproduct of cotton processing composed of discarded plant materials, is often underutilized despite its abundance and potential value. In this study, CGB was upcycled into a nano-in-nano structured composite with potent antimicrobial properties, offering a sustainable solution for agricultural waste management and pathogen control. Cellulose nanofibers (CNF) were extracted from CGB and used as an active biotemplate for the in situ synthesis of antimicrobial silver nanoparticles (Ag NPs). CNF inherently facilitated the formation of Ag NPs (17.2 ± 4.7 nm, 6.8 wt%) without the need for external reducing or stabilizing agents, resulting in a hybrid nanocomposite (Ag-CNF). This embedded nanostructure demonstrated stability and potent antimicrobial efficacy, achieving >99.99% reduction of S. Typhimurium and Listeria monocytogenes within 10 min, Pseudomonas aeruginosa within 1 h, and Staphylococcus aureus within 24 h at a 0.5 wt% concentration. These findings suggest the potential of Ag-CNF as an eco-friendly antimicrobial material with applications in disease prevention and sustainable waste utilization.
1 Introduction
Harvesting raw cotton involves separating foreign material from the lint and seed during the ginning process. This extraneous material, traditionally referred to as cotton gin trash, is a byproduct consisting of stems, sticks, leaves, and other vegetative matter removed from cotton feedstock bales (Alege et al., 2024). The composition of cotton gin byproduct (CGB) typically includes 24%–37% cellulose, 14%–18% hemicellulose, and 18%–23% lignin (McIntosh et al., 2014). Depending on the process method, cotton harvesting may produce between 30 kg and 450 kg of CGB per bale of cotton. For example, cotton strippers, which produce the majority of CGB in the U.S., generate approximately 170 kg of CGB per bale. During the 2023–2024 growing season, U.S. cotton production reached approximately 15.8 million bales, which corresponds to an annual CGB production of about 2.6 billion kg (Crops-Cotton, 2024).
Current uses for CGB are limited; it may be utilized as soil amendments for erosion control or as a fiber supplement for ruminant animals (Stewart, 2010; Alege et al., 2024). Recent research has explored alternative applications to repurpose this agronomic byproduct. These efforts include developing films (Haque et al., 2020), composites (Ge et al., 2020), particleboard (Holt et al., 2012), and packaging materials (Haque and Naebe, 2021). Other methodologies have been developed including chemical treatments to isolate bio-oils (Subramani et al., 2022) and cellulose nanofibers (Nam et al., 2024), expanding the potential for value-added products derived from CGB.
In this study, we upcycled CGB by transforming it into a nano-in-nano structured composite designed for effective pathogen control. While significant advances have been made in pathogen control technologies, the scale of recent pandemics underscores the ongoing need for more effective and sustainable antimicrobial solutions (Balasubramaniam et al., 2020; Tang et al., 2020). Nanotechnology offers a promising approach, leveraging the exceptionally high surface area of nanoscale materials to enhance reactivity (Singh et al., 2021). Addressing this need, we developed a nanoparticle-embedded nanofiber by extracting cellulose nanofiber (CNF) from CGB through a simple mechanical process and embedding antimicrobial silver nanoparticles (Ag NPs) directly into the CNF matrix. This nested nanostructure is designed to maximize antimicrobial activity through a stable and highly reactive surface.
A further benefit of this embedding technology is its environmentally friendly process. Unlike conventional nanofabrication methods, which often rely on toxic chemical agents (Almatroudi, 2020), the method proposed in this study utilizes CNF’s natural ability to in situ generate Ag NPs, eliminating the need for external agents, such as reducing and capping agents. This green synthesis approach not only reduces environmental impact but also simplifies the fabrication process.
The efficiency of this eco-friendly synthesis was evaluated by measuring the surface plasmon resonance intensity of the CNF embedded with Ag NPs (Ag-CNF) and examining the dispersion of Ag NPs within the nanofiber matrix. The antimicrobial efficacy of Ag-CNF, attributed to its high surface-area nano-in-nano structure, was assessed against opportunistic and foodborne bacterial pathogens. Examination of bacterial cells exposed to Ag-CNF provided insights into the stability of the embedded Ag NPs and their antimicrobial mechanisms.
This study is significant as it demonstrates how CNF can serve as a natural, renewable source for producing NPs and facilitating the release of Ag+ ions that effectively inactivate bacterial pathogens posing public health risks. The unique structure of the Ag-CNF composite makes it a promising candidate for applications requiring potent antimonial effects, such as in medical devices, wound dressings, and food packaging materials.
2 Materials and methods
2.1 Materials
CGB was sourced from the USDA Research Facility in Stoneville, MS, United States. Silver nitrate (AgNO3, 99.9%) was purchased from J. T. Baker (Radnor, PA, United States), and sodium hydroxide (NaOH), methyl methacrylate, butyl methacrylate, methyl ethyl ketone, and benzoyl peroxide were purchased from Sigma-Aldrich (St. Louis, MO, United States). Listeria monocytogenes (ATCC 19116) and Salmonella Typhimurium (ATCC H3380) were obtained from the American Type Culture Collection (Manassas, VA, United States). Trypticase Soy Broth (TSB), modified Oxoid agar (MOX), and Xylose Lactose Tergitol-4 (XLT-4 agar) were purchased from Thermo Fisher Scientific (Waltham, MA, United States). All chemicals were used as supplied, and deionized (DI) water was used as the solvent.
2.2 CNF fabrication
CGB was ground using a Wiley mill (E3300, Eberbach Corp., Belleville, MI, United States) fitted with a 50-mesh sieve. The resulting powder was treated with a 4 wt% NaOH solution at 60°C for 2 h at a sample-to-liquor ratio of 1:20 (w/v) to eliminate extractives. It was then rinsed with DI water until the eluate reached a neutral pH (≈7). The alkali-treated CGB was dispersed in DI water at approximately 0.5 wt% using an Ultra-Turrax mechanical homogenizer (T25, IKA Works, Inc., Wilmington, NC, United States). The obtained slurry underwent high-shear mixing at 8,000 rpm for 10 min with a Silverson L5M-A Laboratory Mixer equipped with a Square Hole High Shear Screen. The cellulose slurry was then processed under intense shearing forces in a high-pressure homogenizer (Microfluidizer M-110P, Microfluidics Corp., Newton, MA, United States). It was passed through a 200 µm ceramic interaction chamber five times, followed by an additional five passes through an 87 µm diamond Z-shaped interaction chamber connected in series with the 200 µm chamber. The operating pressure was maintained at 207 MPa.
2.3 Ag-CNF fabrication
A 10 mL aqueous solution of AgNO3 at a concentration of 12 mM was introduced into a 50 mL suspension of CNF prepared at a concentration of 0.5 wt%. The mixture was then subjected to thermal treatment at 100°C for 30 min to facilitate the reduction reaction. Following the thermal treatment, the CNF was rinsed multiple times in DI water at a concentration of approximately 0.5 wt% to remove unreacted Ag precursor ions. The washing process was conducted using a centrifuge (Eppendorf 5810, Hamburg, Germany) with an FA-45-6-30 rotor operating at 10,000 × g for 10 min per cycle.
2.4 Characterization
Samples were photographed using a digital camera (RX100, Sony, New York, NY, United States). Optical microscopy images were obtained in reflection mode using a digital microscope (KH-8700, Hirox, Oradell, NJ, United States). Transmission electron microscopy (TEM) images were acquired using a TEM (JEM-2011, Jeol, Peabody, MA, United States) operating at 200 kV. Samples for TEM were prepared following published methods (Thibodeaux and Evans, 1986; Boylston et al., 1991) by embedding the material in a mixture of methyl methacrylate, butyl methacrylate, and benzoyl peroxide, then polymerizing it using a UV cross-linker (UVP CL-1000, Analytik, Jena, Germany). The polymerized sample was cut into approximately 100 nm-thick sections using a microtome (PowerTome Ultramicrotome, Boeckeler Instruments, Tucson, AZ, United States). These thin films were placed on a carbon-film-coated copper grid. The size of Ag NPs was determined by analyzing TEM images with ImageJ software (Schneider et al., 2012). Ultraviolet-visible (UV-vis) spectra were collected using a UV-vis spectrometer (ISR-2600, Shimadzu, Columbia, MD, United States) equipped with an integrating sphere unit. Absorbance spectra were collected in the wavelength range of 200–1,400 nm. The silver content was measured using a graphite furnace atomic absorption spectrometer (240Z AA, Agilent, Santa Clara, CA, United States). Approximately 0.05 g of each sample was digested in 10 mL of 6 M nitric acid (trace metal grade) using a microwave digestion system (MARS 6, CEM Corporation, Matthews, NC, United States). The average of three measurements was determined from an external calibration curve with a silver standard.
2.5 Antimicrobial property tests
The antibacterial properties against opportunistic bacterial pathogens, Pseudomonas aeruginosa (ATCC 9027) and Staphylococcus aureus (ATCC 6538) were tested at the Situ Biosciences Microbial Product Test Laboratory (Wheeling, IL, United States). The minimum inhibitory concentration (MIC) was determined following the ISO 17025 standard. Bacterial cultures were grown in Tryptic Soy Broth (TSB) at 37°C for 24 h and diluted to approximately 1 × 103 colony-forming units (CFU)/mL in phosphate-buffered saline (PBS). Serial 1:1 dilutions (eight-fold) of the test sample were prepared in a 96-well plate, inoculated with 100 µL of the bacterial suspension, and incubated for 48 h. The MIC value was obtained as the average concentration that inhibited bacterial growth across 12 replicates. To evaluate biocidal activity, bacterial suspensions were adjusted to approximately 1 × 108 CFU/mL and 10 µL of the inoculum was added to each of the three test sample replicates. Exposure durations of 1 h and 24 h were used, and the biocidal log reductions were calculated based on the difference in the common logarithms of pathogen populations between the control and test samples.
The antibacterial properties against foodborne bacterial pathogens, Salmonella Typhimurium and L. monocytogenes, were evaluated at the Microbial and Chemical Food Safety Research Unit (Wyndmoor, PA, United States). For determination of biocidal log reductions, bacterial strains were grown in 20 mL of TSB at 37°C for 24 h (172 rpm), centrifuged (10,000 × g, 5°C, 10 min), and the pellets resuspended in PBS. Inoculum levels were adjusted to ∼6 logs CFU/mL by serial dilution. For testing, 1.0 mL of each sample was mixed with 0.5 mL of bacterial suspension and incubated at 37°C for 10 min or 24 h (exposure duration). Aliquots (100 µL) were spread on selective media—XLT-4 agar for S. Typhimurium and MOX for L. monocytogenes—and incubated at 37°C for 24 h. Controls were included to quantify the initial inoculum, and pathogen reductions were calculated by subtracting CFU counts of test samples from controls. A zone of inhibition assay was conducted by pipetting 50 µL of the test sample onto each of three Tryptic Soy Agar replicates inoculated with approximately 1 × 106 CFU/mL. The plates were incubated at 37°C for 24–48 h to evaluate the zones of bacterial growth inhibition.
Scanning electron microscopy (SEM) images of foodborne bacteria were obtained using an FEI Quanta 200F (Hillsboro, OR, United States) following a published procedure (Olanya et al., 2023). Briefly, 100 µL of cells were pipetted onto coverslips, incubated at 25°C for 30 min, and chemically fixed with 2.5% glutaraldehyde. The fixative solution was decanted, and samples were washed twice with 0.1 M imidazole buffer. The samples were then dehydrated using a graded ethanol series (50%, 70%, 90%, and 100%) for 30 min at each concentration and subjected to critical point drying. The samples were sputter-coated with gold for 60 s using an EMS/150R EMS (Hatfield, PA, United States). Control cells were included in the imaging process for comparison.
3 Results and discussion
3.1 From CGB to CNF
Figure 1 shows the fabrication process of CNF using CGB. CGB is a heterogeneous mixture of burs, bracts, sticks, leaf fragments, and gin motes (Figure 1A). Gin motes consist of small, immature, or broken cotton fibers mixed with other plant debris. The composition of CGB varies depending on factors such as the harvesting method, with mechanical harvesting—common in most cotton-growing regions—yielding 30%–48% burs/bracts, 10%–18% sticks, and 34%–60% motes/leaf fragments (Wanjura et al., 2013; Wanjura et al., 2017). These components differ significantly in size, shape, and color.
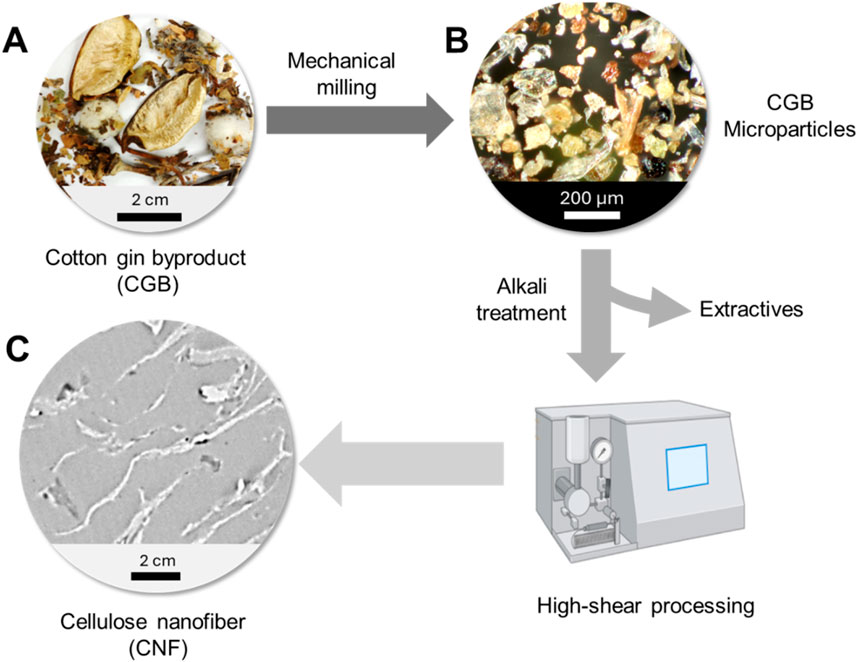
Figure 1. Fabrication of CNF from CGB: (A) photograph of CGB, (B) optical microscopy image of mechanically processed CGB microparticles, and (C) TEM image of CNF.
The first step in extracting CNF from CGB involves mechanical milling, converting the waste into a powdered form. The optical microscopy image of the obtained particles shows that their size was reduced to below 300 μm (Figure 1B). The resulting microparticles underwent an alkali treatment to remove impurities (∼19%), such as sugars, inorganic salts, fertilizers, and waxes (Sluiter et al., 2005; Jordan et al., 2019; Jordan et al., 2021). High-shear processing was then applied to fibrillate the particles into CNF. The high-shear approach enables efficient fibrillation while preserving the structural integrity of the cellulose (Abolore et al., 2024). Furthermore, this method is advantageous for minimizing the use of chemical additives. The final product consisted of fibers measuring several hundred nanometers in thickness and several microns in length (Figure 1C).
3.2 From CNF to Ag-CNF
CGB contains naturally occurring reducing agents, such as lignin and hemicellulose, approximately 20% and 10%, respectively, based on its dry weight (Jordan et al., 2019). Hemicellulose, a branched polysaccharide, contains a variety of sugar units, including galactose. Its vicinal diols—two hydroxyl groups on adjacent carbon atoms—can undergo oxidation, forming dialdehyde compounds. Lignin, a complex, heterogeneous polymer composed of phenolic units linked by various aliphatic and ether bonds, exhibits redox activity due to its multiple functional groups, such as methoxy groups, hydroxyl groups, and aldehyde groups. The phenolic hydroxyl groups play a key role in electron donation, enhancing lignin’s reducing capability. As demonstrated in our previous study (Nam et al., 2023a), these inherent redox properties of hemicellulose and lignin enabled CGB microparticles to effectively reduce Ag+ ions for the in situ formation of Ag NPs.
The natural reducing agents inherent in CNF were utilized to reduce Ag+ ions for the in situ synthesizing Ag NPs. This approach offers two key advantages. First, it eliminates the need for external reducing agents, such as hydrazine or sodium borohydride, which are often toxic and environmentally hazardous. Second, it enables the direct embedding of Ag NPs within the CNF matrix. This embedding is highly advantageous as it creates a hybrid nanocomposite fiber, in which the Ag NPs are stabilized, ensuring their long-term functionality.
The heat treatment of CNF in an aqueous solution facilitated the in situ synthesis of Ag NPs, indicated by a noticeable color change in the CNF from yellow to brown (Figures 2A,B, respectively). This color change is attributed to the unique optical properties of Ag NPs, which arise from their surface plasmon resonance. When embedded within a cellulose matrix, Ag NPs significantly alter the material’s color, ranging from yellow to dark brown depending on their concentration (Nam and Condon, 2014; Nam et al., 2021b; Nam et al., 2022; Nam et al., 2023b). The nanoscopic dimensions of CNF contributed to the stability of its aqueous suspensions. At a concentration of 0.01 wt%, the CNF suspension appeared nearly colorless (Figure 2C), indicating its high dispersion and transparency. In contrast, the Ag-CNF suspension exhibited a vibrant yellow hue (Figure 2D), signifying the successful formation of Ag NPs.
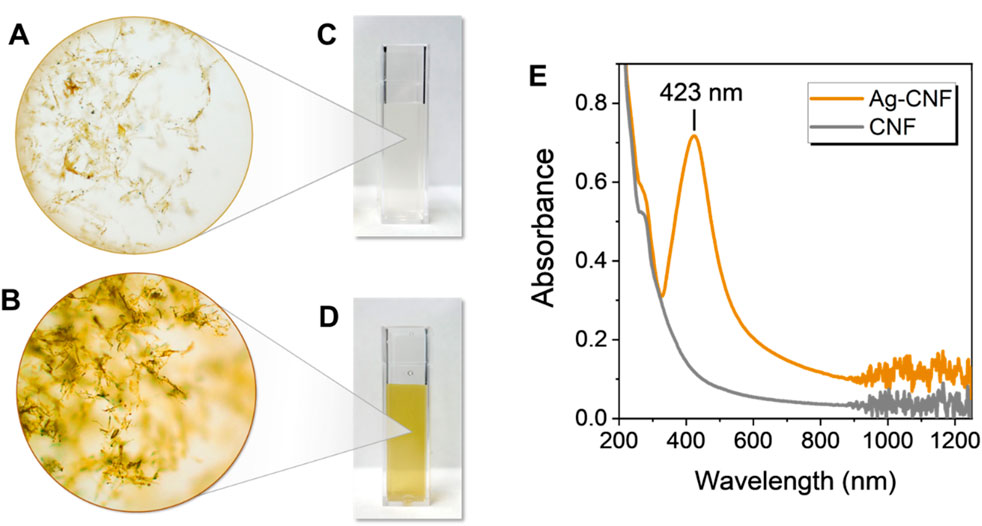
Figure 2. Optical microscopy images of (A) CNF and (B) Ag-CNF. Photographs of (C) CNF and (D) Ag-CNF suspensions (0.01 wt%). (E) UV-vis spectra of Ag-CNF and CNF suspensions.
Surface plasmon resonance is a unique optical phenomenon that occurs when conduction electrons on the surface of metallic nanoparticles resonate with incident light of a specific wavelength. This resonance results in distinctive absorption peaks, providing an indicator of NP formation. To confirm the formation of Ag NPs, UV-vis spectra of CNF and Ag-CNF were collected (Figure 2E). CNF showed no distinctive peaks in its spectrum, while Ag-CNF exhibited a strong surface plasmon resonance peak centered at 423 nm, characteristic of Ag NPs. This result suggests that the components in CGB, primarily hemicellulose and lignin, created a robust natural reducing system capable of facilitating Ag NP synthesis. It should be noted that no external stabilizing or capping agents were used during the synthesis. Stabilizing agents are typically necessary to control particle growth and prevent aggregation, as NPs tend to cluster into large particles. The CNF matrix not only acted as an effective reducing agent but also served as an active biotemplate to generate and stabilize NPs.
To examine the morphology and spatial distribution of Ag NPs formed by CNF, TEM images of CNF were obtained before (Figures 3A,B) and after treatment (Figures 3C,D). Prior to treatment, the TEM image did not show distinct structural details of CNF at both low and high magnifications (Figures 3A,B, respectively) because of the low electron densities of its carbon-based organic components. In addition to hemicellulose and lignin, cellulose (31 wt%) is another major component of CGB (Jordan et al., 2019). After treatment, the TEM image at high magnification clearly shows that CNF was filled with numerous spherical NPs (Figure 3D). These NPs, with higher electron density than the CNF matrix, appeared darker and more discernible. The NPs were mostly well-separated from each other and relatively uniformly distributed throughout the matrix. The average diameter of NPs determined from the TEM images was 10.2 ± 3.4 nm. Previous studies have shown that NPs embedded within the fiber matrix exhibit greater resistance to leaching compared to those placed on the surface of fibers (Nam et al., 2020; Nam et al., 2021a; Nam et al., 2021b; Nam et al., 2022; Nam et al., 2023b). The concentration of embedded Ag NPs was determined to be 6.8 wt%, indicating CNF’s effectiveness as an NP producer and scaffold.
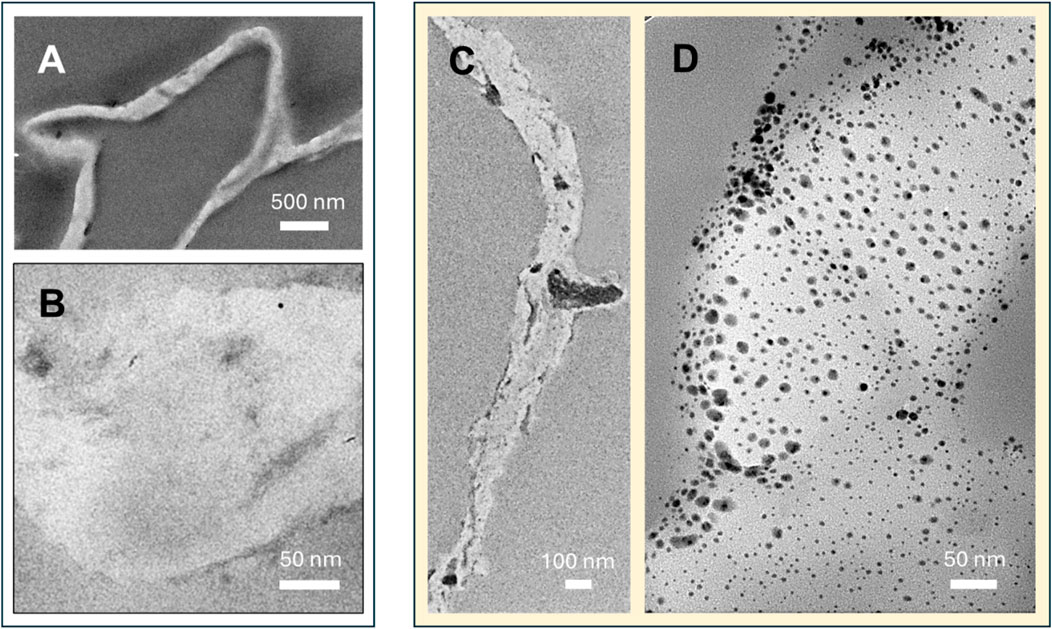
Figure 3. TEM images of CNF at (A) low and (B) high magnifications. TEM images of Ag-CNF at (C) low and (D) high magnifications.
3.3 Pathogen control of Ag-CNF
Figure 4 shows the antimicrobial activity against the opportunistic bacterial pathogens P. aeruginosa and S. aureus. These bacterial strains are clinically significant due to their association with severe infections (Hendricks et al., 2001; Pastar et al., 2013) and notable antibiotic resistance (Ramakant et al., 2011). P. aeruginosa is a leading cause of hospital-acquired infections and is known for its intrinsic resistance to many antibiotics. S. aureus is responsible for a range of conditions from minor skin infections to life-threatening diseases with increasing resistance to multiple antibiotics. The MICs of Ag-CNF were determined to be 0.07% w/v for P. aeruginosa and 0.11% w/v for S. aureus (Figure 4A). Their corresponding concentrations of Ag were 48.9 mg/L and 74.8 mg/L, respectively. These Ag concentrations represent the total Ag, including Ag NPs embedded within the CNF matrix. The concentration of Ag+ ions released from the Ag NPs, which are primarily responsible for antimicrobial activity, is lower than the measured MIC values. As will be discussed later, the primary mechanism of antimicrobial activity of Ag-CNF is the release of Ag+ ions rather than the direct disruption of Ag NPs. The enhanced efficacy at these low concentrations is attributed to the unique nano-in-nano structure of the Ag-CNF composite, which maximizes the surface exposure of Ag NPs. This configuration facilitates the efficient release of Ag+ ions, promoting greater interaction with bacterial cells. In contrast, CNF alone, at a concentration of 0.15% w/v, did not exhibit inhibitory effects on bacterial growth.
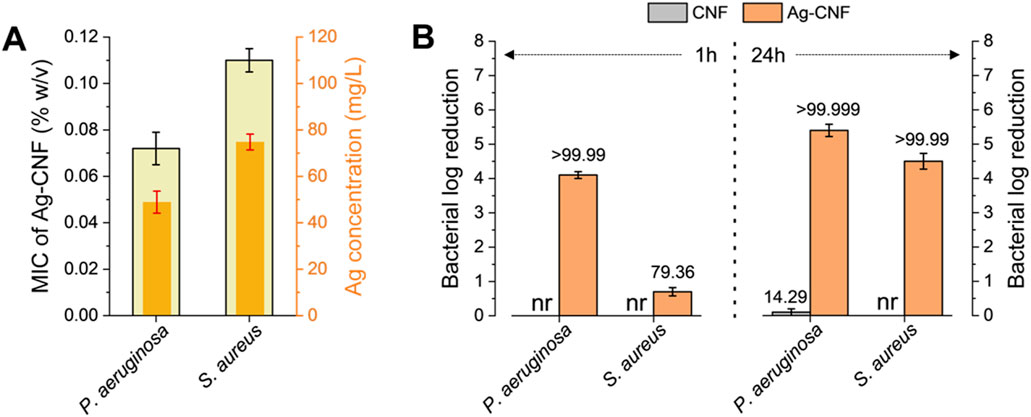
Figure 4. (A) MICs of Ag-CNF with corresponding Ag concentrations, against P. aeruginosa and S. aureus. (B) Log reductions in P. aeruginosa and S. aureus populations following exposure to CNF and Ag-CNF for 1 h and 24 h. nr denotes no reduction. Numbers atop each bar represent percent reductions.
Figure 4B shows the bactericidal efficacy of CNF and Ag-CNF at a concentration of 0.15% w/v against P. aeruginosa and S. aureus. Ag-CNF exhibited substantial bacterial reductions, achieving >5.4 Log CFU/mL for P. aeruginosa and >4.6 Log CFU/mL for S. aureus within 24 h. When the exposure duration was reduced to 1 h, the reductions were 4.1 Log CFU/mL for P. aeruginosa and 0.7 CFU/mL for S. aureus, corresponding to percent reductions of over 99.99% and 79.36%, respectively. These results suggest that Ag-CNF is more effective against P. aeruginosa than against S. aureus, and that the duration of exposure plays an important role in antimicrobial efficacy. The superior efficacy of Ag-CNF against Gram-negative P. aeruginosa may be attributed to its cell wall structure, whose outer membrane interacts more readily with Ag+ ions (More et al., 2023). On the other hand, S. aureus, a Gram-positive bacterium with a thicker peptidoglycan layer in its cell wall, may be less susceptible (More et al., 2023). CNF without embedded Ag NPs exhibited negligible antibacterial effects; after 1 h of exposure, no reduction in bacterial count was observed for either P. aeruginosa or S. aureus. Even after 24 h, CNF alone resulted in only a 14.3% reduction for P. aeruginosa and no measurable reduction for S. aureus.
The investigation was extended to include foodborne pathogens, specifically S. Typhimurium and L. monocytogenes, as these bacteria are significant public health concerns. S. Typhimurium is known for causing gastroenteritis through contaminated food products, and L. monocytogenes is among the most virulent foodborne pathogens, causing a serious infection of listeriosis (Vázquez-Boland et al., 2001). Figure 5 shows the results of the zone of inhibition assay conducted to evaluate the antimicrobial activity of CNF and Ag-CNF at a concentration of 0.15% w/v against these pathogens. The CNF exhibited no inhibitory effect on either S. Typhimurium or L. monocytogenes, as evidenced by the absence of inhibition zones. However, Ag-CNF demonstrated clear zones of inhibition, measuring 2.9 ± 0.2 mm for L. monocytogenes and 2.3 ± 0.3 mm for S. Typhimurium (p < 0.01 compared to CNF). The presence of inhibition zones indicates effective suppression of bacterial growth with a slightly greater efficacy observed against Gram-positive bacterium L. monocytogenes.
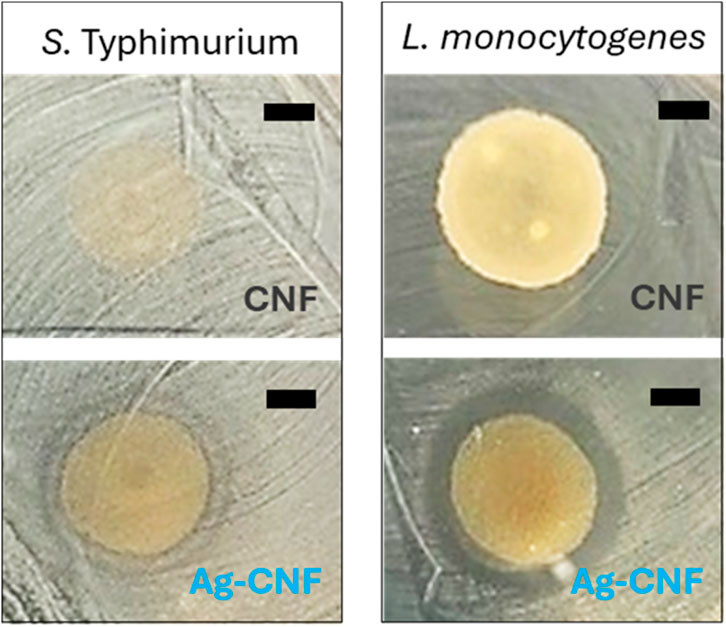
Figure 5. Photographs of inhibition zones for CNF (top) and Ag-CNF (bottom) for S. Typhimurium (left) and L. monocytogenes (right). Scale bars represent 5 mm.
Motivated by the results of the zone of inhibition test, the log reductions of S. Typhimurium and L. monocytogenes were measured after exposure to Ag-CNF and CNF for 10 min and 24 h (Figure 6). Notably, Ag-CNF exhibited greater potency against these foodborne pathogens compared to the previously tested opportunistic pathogens. Within 24 h, Ag-CNF achieved inactivation levels exceeding 6.6 Log CFU/mL for both S. Typhimurium and L. monocytogenes (p < 0.001 compared to CNF). Remarkably, within just a 10-min exposure, Ag-CNF resulted in reductions of >4.9 Log CFU/mL for S. Typhimurium and >5.5 Log CFU/mL for L. monocytogenes (p < 0.01). CNF alone exhibited slight reductions in bacterial counts: with a 10 min-exposure, the log reductions were 0.69 Log CFU/mL for S. Typhimurium and 1.03 Log CFU/mL for L. monocytogenes (p > 0.05). Increasing the exposure time to 24 h did not significantly influence the log reductions (p > 0.05; Table 1). These reductions can be attributed to the mild antimicrobial properties of flavonoids present in CGB (Ma et al., 2013; Nam et al., 2021b).
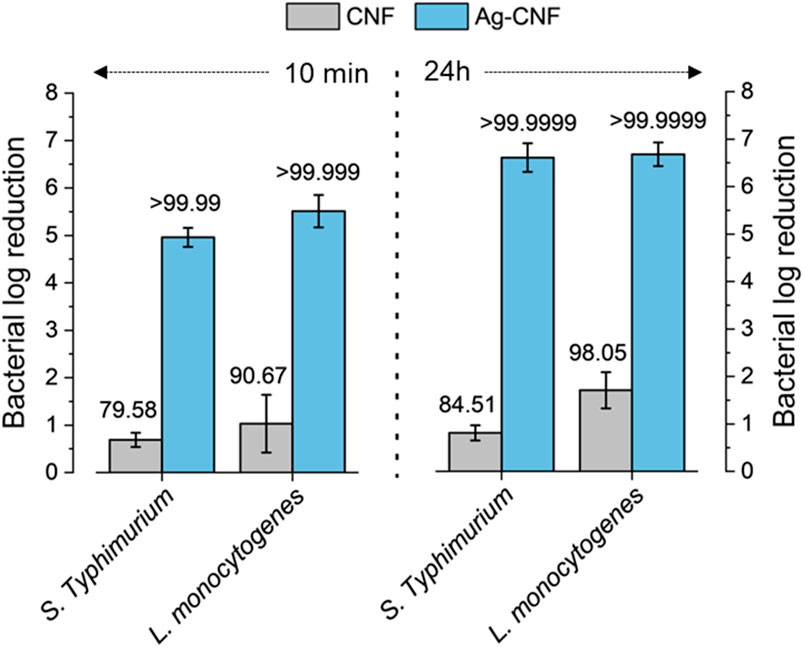
Figure 6. Log reductions in S. Typhimurium and L. monocytogenes populations following exposure to CNF and Ag-CNF for 10 min and 24 h. Numbers atop each bar represent percent reductions.
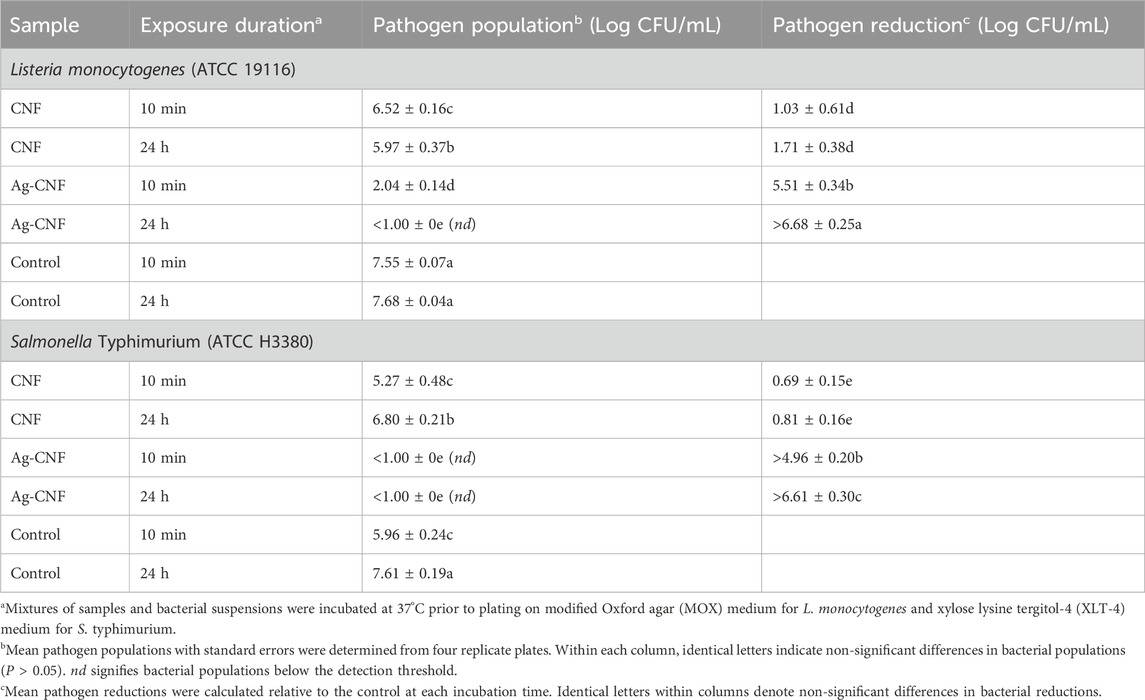
Table 1. In vitro assessment of L. monocytogenes and S. Typhimurium populations and their reductions following exposure to CNF and Ag-CNF for 10 min and 24 h.
To assess the stability of Ag NPs embedded with CNF, foodborne bacterial cells exposed to Ag-CNF were examined. Figure 7 shows SEM images comparing S. Typhimurium cells that were unexposed, those exposed to CNF, and those exposed to Ag-CNF for 24 h. Previous studies have demonstrated that Ag NPs can attach to bacterial membranes, causing physical disruption (Morones et al., 2005; El Badawy et al., 2011). However, in this study, SEM images revealed no NPs on the bacterial surfaces, and the integrity of the bacterial membranes appeared consistent across all samples, whether exposed to Ag-CNF, CNF, or unexposed control. Similar observations were made for L. monocytogenes (Figure 8). These findings suggest that the Ag NPs remained embedded and stable within the CNF matrix, serving as a reservoir for the release of Ag+ ions. Ag+ ions are well known for their potent antimicrobial properties, which arise from several mechanisms: 1) the generation of reactive oxygen species (ROS), leading to oxidative stress and damage to DNA, proteins, and lipids; 2) membrane disruption through binding to membrane proteins and phospholipids, which results in leakage of cellular contents; and 3) protein inactivation due to their high affinity for thiol groups in proteins, inhibiting enzymatic activities and disrupting essential metabolic processes (Yamanaka et al., 2005; Park et al., 2009; Quinteros et al., 2016). These combined actions enable Ag+ ions to effectively inactivate a broad spectrum of bacterial pathogens.
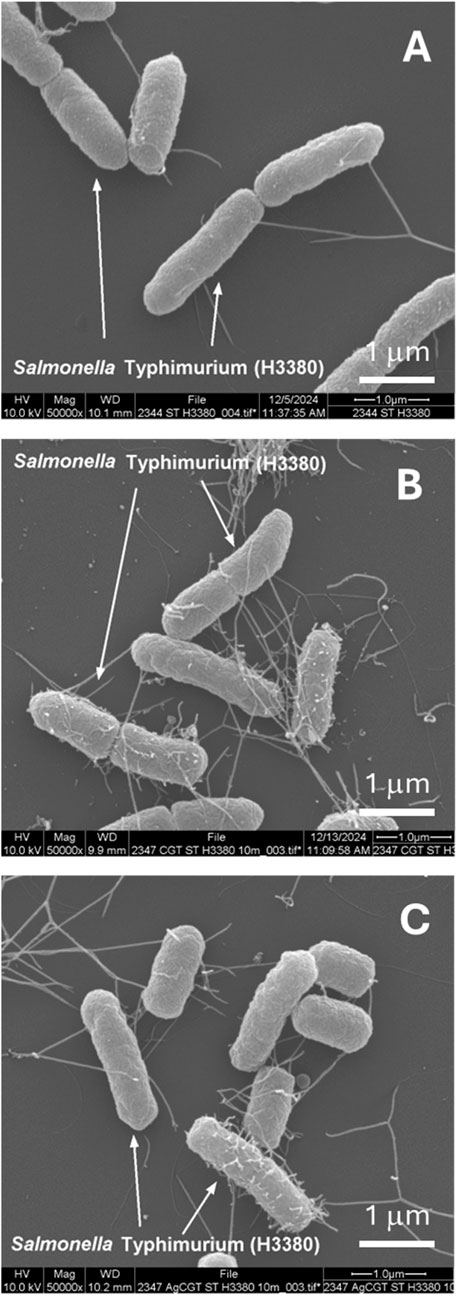
Figure 7. SEM images of S. Typhimurium cells: (A) unexposed (control), (B) exposed to CNF for 24 h, and (C) exposed to Ag-CNF for 24 h.
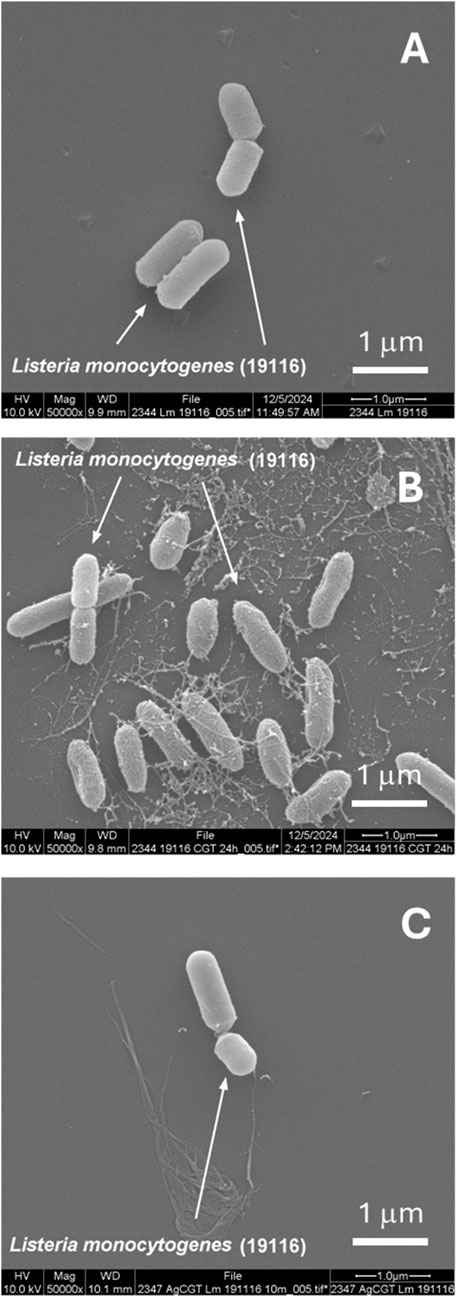
Figure 8. SEM images of L. monocytogenes cells: (A) unexposed (control), (B) exposed to CNF for 24 h, and (C) exposed to Ag-CNF for 24 h.
Like other NPs, Ag NPs tend to aggregate into larger particles, posing a challenge during synthesis and storage. However, the Ag NPs generated within the CNF matrix are expected to exhibit enhanced stability, because the CNF network prevents particle aggregation through electrostatic interactions. Additionally, the CNF matrix serves as a protective barrier, shielding Ag NPs from physical degradation and surface contamination over time.
Another advantage of the developed nano-in-nano structure is its ability to address concerns related to cost, environmental impact, potential toxicity, and recycling in Ag NP applications. Unlike conventional Ag NP-based approaches that often require high Ag loading, this unique nanostructure minimizes overall Ag usage while maintaining effectiveness, thereby reducing environmental impact and toxicity concerns. The embedded Ag NPs within the CNF prevent rapid depletion of Ag NPs, enabling a controlled release mechanism that ensures long-term antimicrobial efficacy. This sustained release not only lowers the required Ag concentration, reducing costs and potential safety issues but also decreases Ag waste.
4 Conclusion
Nanotechnology provides innovative solutions for pathogen control by enhancing the efficiency of antimicrobial materials. In this study, a nano-in-nano structured hybrid composite was developed using CGB as a renewable resource for CNF and leveraging the unique capability of CNF as a reagent-free, active bioplatform for the in situ synthesis of Ag NPs. This approach enabled the uniform dispersion of Ag NPs within the CNF matrix, enhancing surface reactivity for antimicrobial performance. The resulting Ag NP-embedded CNF exhibited strong antibacterial activity against opportunistic and foodborne pathogens, achieving reductions of >6.8 Log CFU/mL within as little as 10 min at a concentration of 0.5 wt%. Electron microscopy analysis confirmed the stability of embedded NPs and indicated that their antimicrobial action occurs primarily through ionization. This hybrid composite can be formulated into dispersions, coatings, films, and composites. As a water-dispersible nanomaterial, it can be easily integrated into biomedical dressings, filtration membranes, and antimicrobial surface coatings. Additionally, its incorporation into flexible packaging films offers potential applications in food safety. By repurposing agricultural waste into high-performance functional materials, this approach not only enhances antimicrobial effectiveness but also contributes to environmental sustainability.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SN: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review and editing. OO: Data curation, Formal Analysis, Investigation, Methodology, Writing – review and editing. JJ: Investigation, Methodology, Writing – review and editing. JU: Investigation, Methodology, Writing – review and editing. ZH: Data curation, Writing – review and editing. MK: Investigation, Writing – review and editing. DF: Project administration, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the Agricultural Research Service Innovation Fund.
Acknowledgments
The authors would like to thank the Louisiana State University Shared Instrument Facility for their assistance in data collection. Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by USDA. USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abolore, R. S., Jaiswal, S., and Jaiswal, A. K. (2024). Green and sustainable pretreatment methods for cellulose extraction from lignocellulosic biomass and its applications: a review. Carbohydr. Polym. Technol. Appl. 7, 100396. doi:10.1016/j.carpta.2023.100396
Alege, F. P., Tumuluru, J. S., Holt, G. A., Donohoe, S. P., Delhom, C. D., Wanjura, J. D., et al. (2024). Cotton gin by-products utilization: past, present, and future. J. Cotton Sci. 28, 79–107. doi:10.56454/sfrm7188
Almatroudi, A. (2020). Silver nanoparticles: synthesis, characterisation and biomedical applications. Open Life Sci. 15, 819–839. doi:10.1515/biol-2020-0094
Balasubramaniam, B., Prateek Ranjan, S., Saraf, M., Kar, P., Singh, S. P., Thakur, V. K., et al. (2020). Antibacterial and antiviral functional materials: chemistry and biological activity toward tackling COVID-19-like pandemics. ACS Pharmacol. and Transl. Sci. 4, 8–54. doi:10.1021/acsptsci.0c00174
Boylston, E. K., Hinojosa, O., and Hebert, J. J. (1991). A quick embedding method for light and electron microscopy of textile fibers. Biotech. Histochem. 66, 122–124. doi:10.3109/10520299109110564
Crops-Cotton, S. B. (2024). USDA national agricultural statistics Service. Available online at: https://www.nass.usda.gov/Data_and_Statistics/index.php.
El Badawy, A. M., Silva, R. G., Morris, B., Scheckel, K. G., Suidan, M. T., and Tolaymat, T. M. (2011). Surface charge-dependent toxicity of silver nanoparticles. Environ. Sci. and Technol. 45, 283–287. doi:10.1021/es1034188
Ge, C., Cheng, H. N., Miri, M. J., Hailstone, R. K., Francis, J. B., Demyttenaere, S. M., et al. (2020). Preparation and evaluation of composites containing polypropylene and cotton gin trash. J. Appl. Polym. Sci. 137, 49151. doi:10.1002/app.49151
Haque, A. N., and Naebe, M. (2021). Flexible water-resistant semi-transparent cotton gin trash/poly (vinyl alcohol) bio-plastic for packaging application: effect of plasticisers on physicochemical properties. J. Clean. Prod. 303, 126983. doi:10.1016/j.jclepro.2021.126983
Haque, A. N., Remadevi, R., Wang, X., and Naebe, M. (2020). Mechanically milled powder from cotton gin trash for diverse applications. Powder Technol. 361, 679–686. doi:10.1016/j.powtec.2019.11.056
Hendricks, K. J., Burd, T. A., Anglen, J. O., Simpson, A. W., Christensen, G. D., and Gainor, B. J. (2001). Synergy between Staphylococcus aureus and Pseudomonas aeruginosa in a rat model of complex orthopaedic wounds. J. Bone Jt. Surg. Am. 83-A, 855–861. doi:10.2106/00004623-200106000-00006
Holt, G. A., Chow, P., Wanjura, J. D., Pelletier, M. G., Coffelt, T. A., and Nakayama, F. S. (2012). Termite resistance of biobased composition boards made from cotton byproducts and guayule bagasse. Ind. Crop. Prod. 36, 508–512. doi:10.1016/j.indcrop.2011.10.005
Jordan, J. H., Easson, M. W., Dien, B., Thompson, S., and Condon, B. D. (2019). Extraction and characterization of nanocellulose crystals from cotton gin motes and cotton gin waste. Cellulose 26, 5959–5979. doi:10.1007/s10570-019-02533-7
Jordan, J. H., Easson, M. W., Thompson, S., Wu, Q. L., and Condon, B. D. (2021). Lignin-containing cellulose nanofibers with gradient lignin content obtained from cotton gin motes and cotton gin trash. Cellulose 28, 757–773. doi:10.1007/s10570-020-03549-0
Ma, M., Li, R., Du, Y., Tang, Z., and Zhou, W. (2013). Analysis of antibacterial properties of naturally colored cottons. Text. Res. J. 83, 462–470. doi:10.1177/0040517512447585
Mcintosh, S., Vancov, T., Palmer, J., and Morris, S. (2014). Ethanol production from cotton gin trash using optimised dilute acid pretreatment and whole slurry fermentation processes. Bioresour. Technol. 173, 42–51. doi:10.1016/j.biortech.2014.09.063
More, P. R., Pandit, S., Filippis, A., Franci, G., Mijakovic, I., and Galdiero, M. (2023). Silver nanoparticles: bactericidal and mechanistic approach against drug resistant pathogens. Microorganisms 11, 369. doi:10.3390/microorganisms11020369
Morones, J. R., Elechiguerra, J. L., Camacho, A., Holt, K., Kouri, J. B., Ramirez, J. T., et al. (2005). The bactericidal effect of silver nanoparticles. Nanotechnology 16, 2346–2353. doi:10.1088/0957-4484/16/10/059
Nam, S., Chavez, S. E., Hillyer, M. B., Condon, B. D., Shen, H., and Sun, L. (2021a). Interior vs. exterior incorporation of silver nanoparticles in cotton fiber and washing durability. AATCC J. Res. 8, 1–12. doi:10.14504/ajr.8.6.1
Nam, S., and Condon, B. (2014). Internally dispersed synthesis of uniform silver nanoparticles via in situ reduction of [Ag(NH3)2]+ along natural microfibrillar substructures of cotton fiber. Cellulose 21, 2963–2972. doi:10.1007/s10570-014-0270-y
Nam, S., Easson, M., Jordan, J. H., He, Z., Zhang, H., Santiago Cintron, M., et al. (2023a). Unveiling the hidden value of cotton gin waste: natural synthesis and hosting of silver nanoparticles. ACS Omega 8, 31281–31292. doi:10.1021/acsomega.3c03653
Nam, S., Easson, M. W., Jordan, J. H., He, Z., Lima, I. M., Hillyer, M. B., et al. (2024). Repurposing cotton gin trash for cellulose nanofibril-silver hybrid and ultralight silver-infused aerogel. ACS Omega 9, 38195–38204. doi:10.1021/acsomega.4c05728
Nam, S., Hillyer, M. B., Condon, B. D., Lum, J. S., Richards, M. N., and Zhang, Q. (2020). Silver nanoparticle-infused cotton fiber: durability and aqueous release of silver in laundry water. J. Agric. Food Chem. 68, 13231–13240. doi:10.1021/acs.jafc.9b07531
Nam, S., Hillyer, M. B., He, Z., Chang, S., and Edwards, J. V. (2022). Self-induced transformation of raw cotton to a nanostructured primary cell wall for a renewable antimicrobial surface. Nanoscale Adv. 4, 5404–5416. doi:10.1039/d2na00665k
Nam, S., Hinchliffe, D. J., Hillyer, M. B., Gary, L., and He, Z. (2023b). Washable antimicrobial wipes fabricated from a blend of nanocomposite raw cotton fiber. Molecules 28, 1051. doi:10.3390/molecules28031051
Nam, S., Selling, G. W., Hillyer, M. B., Condon, B. D., Rahman, M. S., and Chang, S. (2021b). Brown cotton fibers self-produce Ag nanoparticles for regenerating their antimicrobial surfaces. ACS Appl. Nano Mater. 12, 13112–13122. doi:10.1021/acsanm.1c02640
Olanya, O. M., Yosief, H. O., Ashby, R. D., Niemira, B. A., Sarker, M. I., Ukuku, D. O., et al. (2023). Inactivation of foodborne and other pathogenic bacteria with pyrrolidine based fatty acid amide derivatives. J. Food Saf. 43, e13079. doi:10.1111/jfs.13079
Park, H. J., Kim, J. Y., Kim, J., Lee, J. H., Hahn, J. S., Gu, M. B., et al. (2009). Silver-ion-mediated reactive oxygen species generation affecting bactericidal activity. Water Res. 43, 1027–1032. doi:10.1016/j.watres.2008.12.002
Pastar, I., Nusbaum, A. G., Gil, J., Patel, S. B., Chen, J., Valdes, J., et al. (2013). Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8, e56846. doi:10.1371/journal.pone.0056846
Quinteros, M. A., Cano Aristizábal, V., Dalmasso, P. R., Paraje, M. G., and Páez, P. L. (2016). Oxidative stress generation of silver nanoparticles in three bacterial genera and its relationship with the antimicrobial activity. Toxicol. Vitro 36, 216–223. doi:10.1016/j.tiv.2016.08.007
Ramakant, P., Verma, A. K., Misra, R., Prasad, K. N., Chand, G., Mishra, A., et al. (2011). Changing microbiological profile of pathogenic bacteria in diabetic foot infections: time for a rethink on which empirical therapy to choose? Diabetologia 54, 58–64. doi:10.1007/s00125-010-1893-7
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi:10.1038/nmeth.2089
Singh, Y. D., Ningthoujam, R., Panda, M. K., Jena D, B., Babu, P. J., and Mishra, A. K. (2021). Insight from nanomaterials and nanotechnology towards COVID-19. Sensors Int. 2, 100099. doi:10.1016/j.sintl.2021.100099
Sluiter, A., Ruiz, R., Scarlata, C., Sluiter, J., and Templeton, D. (2005). Determination of extractives in biomass. Denver, CO: National Renewable Energy Laboratory, 1–12.
Subramani, V. B., Shi, C., Moghaddam, L., Atanda, L., Ramirez, J., Del Río, J. C., et al. (2022). Cotton farming sustainability: formation of trans-isoeugenol/bio-aromatics, 5-chloromethylfurfural, C13–C17 liquid hydrocarbons and fertilizer from cotton gin trash. J. Clean. Prod. 363, 132404. doi:10.1016/j.jclepro.2022.132404
Tang, Z., Kong, N., Zhang, X., Liu, Y., Hu, P., Mou, S., et al. (2020). A materials-science perspective on tackling COVID-19. Nat. Rev. Mater. 5, 847–860. doi:10.1038/s41578-020-00247-y
Thibodeaux, D. P., and Evans, J. P. (1986). Cotton fiber maturity by image analysis. Text. Res. J. 56, 130–139. doi:10.1177/004051758605600211
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640. doi:10.1128/cmr.14.3.584-640.2001
Wanjura, J. D., Baker, K., and Barnes, E. (2017). Cotton ginners handbook (2016 revised edition): harvesting. J. Cotton Sci. 21, 70–80. doi:10.56454/axfa4722
Wanjura, J. D., Boman, R. K., Kelley, M. S., Ashbrook, C. W., Faulkner, W. B., Holt, G. A., et al. (2013). Evaluation of commercial cotton harvesting systems in the southern high plains. Appl. Eng. Agric. 29, 321–332. doi:10.13031/aea.29.9884
Keywords: cotton gin byproduct, cellulose nanofiber, nanocomposite, green synthesis, pathogen control
Citation: Nam S, Olanya OM, Jordan JH, Uknalis J, He Z, Kashem MNH and Fang D (2025) From cotton gin byproduct to nano-in-nano structured hybrid composite for effective pathogen control. Front. Nanotechnol. 7:1567693. doi: 10.3389/fnano.2025.1567693
Received: 28 January 2025; Accepted: 23 April 2025;
Published: 06 May 2025.
Edited by:
Hayrunnisa Nadaroglu, Atatürk University, TürkiyeReviewed by:
Amit K. Roy, Northeastern University, United StatesPradeep Kumar, University of the Witwatersrand, South Africa
Copyright © 2025 Nam, Olanya, Jordan, Uknalis, He, Kashem and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunghyun Nam, c3VuZ2h5dW4ubmFtQHVzZGEuZ292
†ORCID: Md Nayeem Hasan Kashem, https://orcid.org/0000-0003-0179-7050
 Sunghyun Nam
Sunghyun Nam Ocen Modesto Olanya
Ocen Modesto Olanya Jacobs H. Jordan
Jacobs H. Jordan Joseph Uknalis2
Joseph Uknalis2 Zhongqi He
Zhongqi He