- 1Biohybrid Systems Group, Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University School of Medicine, Atlanta, GA, United States
- 2Institute of Cell and Tissue Engineering, College of Medicine, The Catholic University of Korea, Seoul, Republic of Korea
Nanobiosensors now allow continuous, nondestructive tracking of stem cell differentiation and organoid maturation. Classical assays such as immunostaining and polymerase chain reaction are invasive snapshots that overlook fast molecular events guiding lineage choice. Nanoscale probes operate inside living constructs, translating genetic, metabolic, and mechanical signals into optical, electrical, or magnetic readouts while leaving viability intact. This review arranges recent progress by cell type. In pluripotent systems CRISPR Cas13a fluorescence resonance energy transfer beacons, single layer molybdenum disulfide nanopores, and dCas9 SunTag reporters reveal minute scale waves of microRNA and transcription factor activity, addressing teratoma risk. Mesenchymal stromal cells use locked nucleic acid beacons, piezoelectric scaffolds, and magnetic tracers to quantify Notch signaling, mechano sensing, and engraftment. Brain, cardiac, and vascular organoids adopt microneedle electrode arrays, stretchable optical membranes, and impedance chips to monitor deep electrophysiology, contractility, and barrier integrity, while quantum dots and metal organic frameworks combine delivery and sensing across other organoid models. Key hurdles remain, including lack of fabrication standards, uncertain probe occupancy limits, and unclear regulatory pathways. Multimodal chips, artificial intelligence driven analytics, and biodegradable sensor substrates offer potential solutions, moving nanobiosensors closer to routine clinical use.
1 Introduction
In regenerative medicine, stem cells and organoids have rapidly become a triple-driving force that powers modeling, therapies, and cell manufacturing all at once (Pazzin et al., 2024). Patient-specific induced pluripotent stem cells (iPSCs) can supply an inexhaustible, genetically matched cell source, while CRISPR-based editing corrects or installs disease mutations with single-nucleotide precision, yielding human-relevant models that outclass traditional animals (Torizal et al., 2024; Yao et al., 2024; Wiley et al., 2015). Nevertheless, the rapid progress in stem cell and organoids has outpaced the tools that are typically used to research them (Kang et al., 2023). Traditional analytical methods such as immunocytochemistry, polymerase chain reaction, and Western blot analysis present significant limitations, being invasive, time-consuming, and destructive to the cellular systems under investigation. These conventional approaches often require cell fixation and staining procedures that preclude real-time monitoring of dynamic biological processes. Consequently, there has been an urgent demand for non-invasive, non-destructive, and label-free sensing techniques capable of providing continuous, real-time monitoring of stem cell differentiation and organoid function.
Nanotechnology has emerged as a paradigm-shifting solution to the challenges of monitoring stem cell differentiation and organoid development, offering highly precise control over cellular fate and enabling ultrasensitive detection of molecular and phenotypic changes in real time (Jarrige et al., 2021). Nanoparticle-based biosensors, due to their physicochemical properties like high surface-area-to-volume ratios, tunable surface chemistry, and the ability to interact with biomolecules at the nanoscale, can be integrated into biological systems to monitor key biomarkers and cellular processes during differentiation. Unlike conventional sensing methods that are often destructive, endpoint-based, or lack spatial and temporal resolution, nanoparticle biosensors can mimic the structural and biochemical features of the extracellular matrix, providing both dynamic guidance cues and real-time feedback to cells (Ferrari, 2023). For example, gold nanorod-based molecular beacons can track lineage-specific miRNA expression in neural stem cells over several days, while carbon quantum dots and MOF nanoparticles can be engineered to deliver differentiation factors and simultaneously report on cellular responses (Hou et al., 2025). Furthermore, these nanostructures can be functionalized to respond to mechanical, electrical, or biochemical stimulus, creating closed-loop systems that not only sense but also actively modulate the stem cell microenvironment (Lin et al., 2024; Jhunjhunwala et al., 2023). This integration of sensing and actuation at the nanoscale is paving the way for advanced, automated, and personalized approaches in regenerative medicine, enabling unprecedented insight into the dynamics of stem cell fate decisions and organoid maturation.
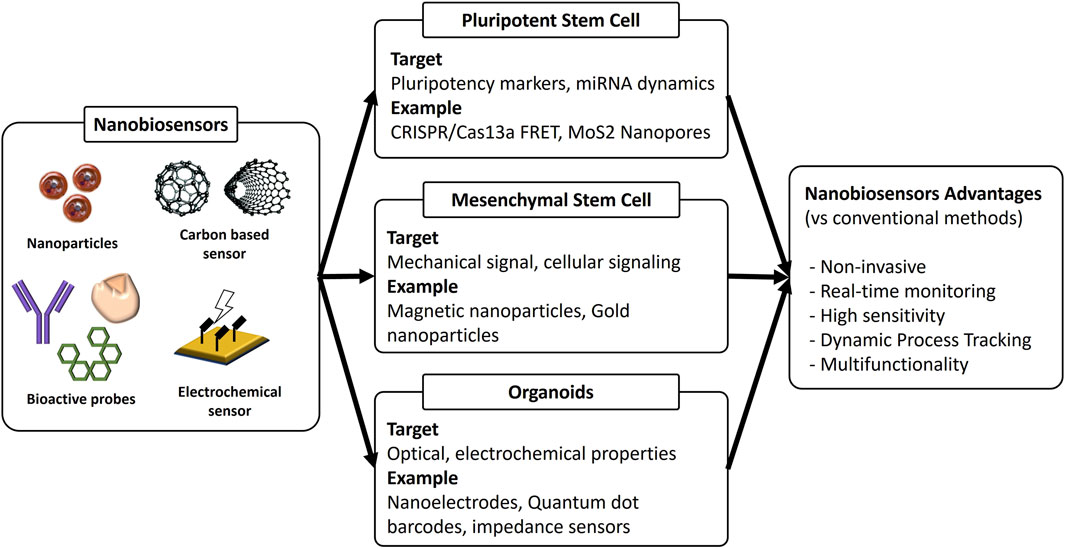
This mini review examines the current state of nanoparticle-based biosensor technologies for monitoring stem cell differentiation and organoid development. Rather than focusing on sensor technology classifications, this review adopts a biologically centered approach, organizing the discussion around specific stem cell types and organoid systems (Figure 1). This perspective provides a more clinically relevant understanding of how nanoparticle biosensors are being applied to advanced regenerative medicine applications.
2 Pluripotent stem cells
The clinical translation of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) faces a critical safety bottleneck: teratoma formation risk from residual undifferentiated cells. Even minute subpopulations of pluripotent cells can initiate tumor formation, as demonstrated by studies showing that cells retaining pluripotency characteristics remain present following differentiation protocols and can recover their tumorigenic potential under appropriate culture conditions (Ramirez et al., 2007). Traditional pluripotency markers including OCT4, NANOG, and SOX2 provide only static, endpoint snapshots of cellular state, fundamentally failing to capture the dynamic micro-RNA oscillations and transcription factor fluctuations that preceded and control germ layer commitment (Wang W. et al., 2020). This limitation creates a dangerous gap in safety monitoring, as classical markers miss the rapid molecular transitions occurring at sub-hour timescales during differentiation initiation (Fu et al., 2017). Recent evidence demonstrates that pluripotency maintenance involves complex regulatory networks where factors like Gas5 long non-coding RNA form positive feedback loops with Sox2, Oct4, and Nanog, creating dynamic expression patterns that conventional assays cannot detect (Gonzales et al., 2021).
2.1 CRISPR/Cas13a FRET sensors
The development of CRISPR/Cas13a-based fluorescence resonance energy transfer (FRET) beacons represents a paradigm shift in real-time microRNA detection within differentiating stem cells (Kutsche et al., 2018; Papadimitriou et al., 2023). This groundbreaking proof-of-principle study employed cytoplasmically expressed Cas13a coupled with exogenous guide RNA, though the initial signal-to-background ratio remained modest and required further optimization for clinical applications (Wang Y. et al., 2020). Recently, two significant advancements were made that substantially improved the sensitivity and temporal resolution of CRISPR-based biosensors (Wang Y. et al., 2020; Montagud-Martínez et al., 2023). First, the guide RNA (gRNA) was strategically pre-assembled with a quencher-labeled single-stranded DNA (ssDNA) reporter, creating a sophisticated detection mechanism where target binding unleashed Cas13a′s collateral cleavage activity, thereby releasing the fluorophore and significantly magnifying the detection signal (Montagud-Martínez et al., 2023; Iwasaki and Batey, 2020). Second, microfluidic integration technology confined reagents to picolitre volumes, compressing reaction time to under 15 min while maintaining high sensitivity and specificity (Degliangeli et al., 2014; Xiao et al., 2018).
The integration of CRISPR/Cas13a systems with stem cell monitoring has revolutionized understanding of microRNA dynamics during differentiation processes (Ishikawa et al., 2020). These systems enable unprecedented single-cell resolution tracking of lineage commitment, capturing the rapid molecular transitions that occur at sub-hour timescales during differentiation initiation (Lu and Yoo, 2018). The ability to detect miRNA-124, a critical neuronal differentiation marker, with such high temporal resolution has opened new avenues for studying neural stem cell fate decisions and optimizing differentiation protocols (Papadimitriou et al., 2023; Ishikawa et al., 2020).
2.2 Monolayer nanopores
While CRISPR beacons excel at detecting relative microRNA ratios, solid-state nanopores address the critical need for measuring absolute transcription dynamics with single-molecule precision (Li et al., 2021; Ren et al., 2025). Transition-metal dichalcogenides such as molybdenum disulfide (MoS2) uniquely combine atomic thickness, exceptional mechanical robustness, and highly tunable electronic properties that make them ideal for nanopore applications (Xiao et al., 2018; Li et al., 2021). The sophisticated ability to temporal-stamp each individual blockade event enabled these researchers to reconstruct detailed bursty transcription kinetics in differentiating embryoid bodies with unprecedented resolution (Wheat et al., 2020). Solid-state nanopores, especially MoS2 nanopores, allow single-molecule discrimination of transcript isoforms and real-time measurement of transcriptional bursting. Although large arrays remain in development, proof-of-concept studies demonstrate detection of individual mRNA molecules from live cell extracts, offering direct observation of stochastic gene expression that underlies iPSC fate decisions (Zhong et al., 2023). This capability reveals burst frequency and amplitude for key pluripotency genes (e.g., OCT4, SOX2) at the single-cell level.
2.3 Optical nanobiosensors
A complementary non-destructive approach utilizes dCas9-SunTag scaffolds strategically fused to promoters of interest, providing an alternative method for monitoring transcriptional activity (Kumar et al., 2023; Papikian et al., 2019; Pflueger et al., 2018). The fluorescence signal is dramatically amplified through a sophisticated tandem array of antibody-binding peptides, with each peptide recruiting ten dye-conjugated single-chain variable fragment (scFv) components (Pflueger et al., 2018). Another research reported dCas9-CRISPR system, functioned as programmable transcriptional biosensors in ESCs, enabling real-time functional validation of microRNA promoter activity by targeting genomic regions with sgRNAs and detecting resultant changes in miRNA expression. In mouse ESCs, these tools confirmed that miR-335 expression is exclusively regulated by the host gene Mest’s promoter, demonstrating their utility for mapping lineage-specific regulatory elements in pluripotent systems (Kumar et al., 2023).
3 Mesenchymal stem cells
Mesenchymal stromal cells (MSCs) function as the mechanosensitive engines of regenerative orthopedics, supporting modern approaches to bone grafting, cartilage resurfacing and tendon reconstruction (Lee et al., 2014; Kang et al., 2024). Late in the lineage-commitment timeline researchers can confirm differentiation by measuring RUNX2 for osteoblasts, PPARγ for adipocytes and COL2A1 for chondrocytes, but these transcripts emerge only after the decisive cues have already nudged the cell toward a specific fate. In fact, the earliest choices unfold over milliseconds to hours and are dictated by biophysical stimuli such as plasma-membrane stretch, cytoskeletal tension, integrin clustering, transient calcium influx and shifts in membrane potential (An et al., 2015). Substrate stiffness, nanoscale surface texture and fluid shear convert into these intracellular events, activating mechanosensitive ion channels together with RhoA and ROCK signaling well before any osteogenic or chondrogenic gene is detectable. For that reason, a lineage-centric biosensor suited to MSC culture must capture mechanical or electro-mechanical parameters directly and preferably do so without external wiring so that scaffold designers and bioreactor operators can intervene while cell identity is still flexible rather than after the transcriptional trajectory has become locked in (Bagnaninchi and Drummond, 2011; Ding et al., 2024; Lee et al., 2024).
3.1 Osteogenic differentiation tracking
Mesenchymal stem cell osteogenic differentiation represents a cornerstone process in tissue engineering and regenerative medicine applications. Double-stranded locked nucleic acid (LNA)/DNA nanosensors enable real-time monitoring of Notch1-Dll4 signaling pathway roles at the single mesenchymal stem cell level (Zhao et al., 2022). These sensors track Dll4 mRNA expression dynamics during osteogenic differentiation, allowing observation of differentiation process heterogeneity (Zhao et al., 2022). Barium titanate nanoparticles (BT NPs) combined with alginate polymers provide a novel biocompatible three-dimensional scaffold approach for inducing osteogenic stem cell differentiation (Amaral et al., 2019). BT NP/alginate 3D scaffolds demonstrate osteogenic differentiation induction potential without requiring osteogenic supplements, with BMP-2 and ALP mRNA significantly upregulated after 21 days. The scaffolds exhibit highly interconnected pores and surface nano topography favorable for mesenchymal stem cell differentiation, with mineralization nodules formation and morphological changes from spindle to cuboid shape. Another study reported a dual-color nano sensor in which PLGA nanoparticles co-encapsulate molecular beacons for GAPDH (housekeeping control) and ALP (early osteogenic marker), enabling live, non-destructive read-out of MSC differentiation (Wiraja et al., 2016). Because the particles slowly degrade intracellularly, ALP mRNA dynamics could be followed continuously for 18 days, and the ratio metric fluorescence closely matched conventional RT-qPCR data, validating both accuracy and viability preservation. Applied in standard 2-D culture and on tricalcium-phosphate-loaded PCL films, the sensor revealed an earlier ALP peak on the osteo-inductive scaffold, demonstrating its utility for rapid screening of next-generation bone-graft materials.
3.2 Magnetic nanoparticle-enhanced delivery systems
Magnetic nanoparticle-enhanced extracellular vesicles (GMNPE-EVs) derived from bone marrow mesenchymal stem cells provide an effective therapeutic strategy for diabetic osteoporosis through miR-15b-5p delivery. These systems transfer miR-15b-5p to osteoclasts, downregulating GFAP expression and inhibiting osteoclast differentiation (Xu et al., 2024). In vitro experiments demonstrate that GMNPE-EVs effectively deliver miR-15b-5p to osteoclasts, while in vivo tests confirm therapeutic potential in alleviating diabetic osteoporosis. Gold nanoparticles have proven effective for mesenchymal stem cell tracking in tumor-targeted delivery systems (Xu et al., 2023). Mesenchymal stem cell-mediated gold nanoparticle delivery showed 2.4- to 9.3-fold enhanced performance in tumor accumulation and penetration compared to conventional enhanced permeability and retention (EPR) effect-based delivery.
3.3 Superparamagnetic iron oxide nanoparticle tracking
Superparamagnetic iron oxide nanoparticles (SPIOs) serve as powerful tools for stem cell tracking applications. Cubic iron oxide nanoparticles with 22 nm edge length (CIONs-22) are optimized for magnetic particle imaging (MPI), showing superior MPI performance compared to commercialized tracers like Vivotrax (Wang Q. et al., 2020). These magnetic properties ensure high sensitivity and resolution for MPI application-ns, with efficient cellular uptake enabling real-time and prolonged monitoring of stem cells transplanted into hindlimb ischemia mice.
4 Organoids
Organoids are self-organizing, three-dimensional mini tissues that recapitulate key aspects of native architecture, multicellular heterogeneity, and dynamic physiology far better than two-dimensional cultures (Septiana and Pawitan, 2024; Thangam et al., 2024). Their complex luminal structures, steep oxygen and nutrient gradients, and millimeter-scale thickness make conventional end-point assays, fixation, sectioning, bulk PCR, both invasive and poorly representative of spatiotemporal variation. Nanobiosensors meet these challenges by embedding ultrasensitive optical, electrical or magnetic probes directly within the organoid matrix, where they can track local gene expression, metabolic flux and mechanical cues without disrupting growth. Because nanoparticles can be functionalized to respond to specific biomolecules or physical forces, they translate otherwise invisible events, such as hypoxic shifts in a liver organoid core or calcium sparks in cerebral organoids, into real-time signals. This continuous, non-destructive feedback is essential for validating developmental fidelity, optimizing culture conditions and, ultimately, qualifying organoids for personalized drug testing and regenerative-medicine applications.
4.1 Brain organoids
Electrophysiological and metabolic read-out of brain organoids has advanced far beyond occasional calcium-imaging snapshots (Yousuf et al., 2025; Park et al., 2024). Zips et al. used aerosol-jet and ink-jet additive printing to fabricate high-aspect-ratio microneedle electrode arrays composed of the conductive polymer poly (3,4-ethylenedioxythiophene):polystyrene sulfonate blended with multi-walled carbon nanotubes, creating nano-electrodes able to penetrate 400–600 µm brain spheroids and cerebral organoids (Zips et al., 2023). The microneedles had 20 µm tips, centimeter scale mechanical flexibility, and week-long electrochemical stability transform them into a practical biosensing platform that captures deep extracellular signals unreachable by conventional planar microelectrode arrays. By delivering reliable recordings from both engineered neutrosphere cultures and self-assembled organoids, the work closes a critical depth-sensing gap and illustrates how additive-manufactured nano-electrodes can advance three-dimensional neural-tissue electrophysiology. Multimodal “Phase-Zero” platforms have been reported to go a step further by threading a broadband vis/NIR fiber through the same chamber, turning the setup into a combined spectrophotometer and electrophysiology rig (Dutta et al., 2020). Local-field-potential spectral exponents in the 30–50 Hz band are logged in parallel with the optical redox read-out of cytochrome-c oxidase; together, these metrics chart excitation–inhibition balance and mitochondrial health during neurodevelopment or drug exposure in a way that single-modality systems cannot.
4.2 Cardiac organoids
Mechanical, biochemical, and electrical cues in cardiac organoids require similarly integrated monitoring (Son et al., 2025). Stretchable PDMS wave-guide membranes patterned with micro-optical structures record sub-percent changes in organoid length as shifts in transmitted light intensity, enabling real-time quantification of systolic and diastolic strain under physiologically relevant after-load (Sannino et al., 2023). When human PSC-derived constructs on these membranes receive low-dose doxorubicin, contractility drops are detected hours before visible beating irregularities emerge, providing an early-warning screen for cardiotoxic compounds. On-chip electrochemical sensors add biochemical depth: aptamer-coated gold microelectrodes incorporated into the same heart-on-a-chip quantify femtomolar surges of creatine-kinase-MB and troponin-T, correlating biomarker release with beat-rate slow-down and viability loss (Devarasetty et al., 2017). At the sub-cellular scale, genetically encoded biosensors that dock a Förster-pair to the ryanodine receptor let investigators visualize cAMP transients exactly where excitation–contraction coupling begins. In β-adrenergic stimulation experiments, these reporters reveal sub-second cAMP spikes that foreshadow maladaptive remodeling, showing how optical genetics, electrochemistry, and mechanics can converge on a single micro-tissue platform.
4.3 Vascular organoids
Adding vasculature introduces new sensing demands that nanotechnology is beginning to meet (Kim J-E. et al., 2025). Electric cell–substrate impedance sensing, miniaturized onto transparent indium-tin-oxide grids, now follows tight-junction maturation in endothelial sheets derived from vascular organoids, delivering second-by-second barrier-resistance curves while leaving the epithelium available for fluorescence or trans-endothelial electric-potential imaging (Ouahoud et al., 2024; Eshetie et al., 2023; Schoon et al., 2025). Perfusable organoid-on-chip devices extend this principle: parallel microchannels lined with HUVECs are fluidically coupled to developing kidney or liver organoids, whose endogenous endothelial buds anastomose with the channel to form macro vessels (Kim Y. et al., 2025). A vascularized tumor-organoid chip grows its own micro-vessels and lets nanosensors watch endothelial sprouting, cancer cell escapes and Notch signaling live (Du et al., 2025). The open channels accept quantum-dot barcodes or plasmonic gold probes, so oxygen use, and cytokine bursts can be measured during drug tests without harming the tissue. A companion smart vascular graft places a laser-written graphene strain lattice inside flexible silicone, sensing blood-flow strain down to three ten-thousandths of a percent through more than thirty-two thousand cycles (Ma et al., 2025). The porous graphene acts both as a strain gauge and a high-area surface ready for electrochemical coatings, streaming wireless data that can flag clots or early wall thickening. Together these platforms show how nanosensors can give continuous, patient-specific feedback from both in-vitro disease models and implanted vascular devices.
4.4 Other organoids
Beyond brain, cardiac, and vascular organoids, nanobiosensors are increasingly being applied to other organoid types, most notably kidney, liver, and intestinal organoids, to advance disease modeling, drug screening, and tissue engineering. In kidney organoids, biosensors such as ATP/ADP ratio sensors and impedance-based devices have been integrated to enable real-time, non-invasive assessment of nephrotoxicity, transporter function, and barrier integrity, providing a sensitive platform for evaluating drug-induced injury and organoid maturation in vitro (Susa et al., 2023; Tabibzadeh and Morizane, 2024; Tabibzadeh et al., 2023). High-throughput, automated imaging systems combined with label-free biosensor algorithms further facilitate large-scale toxicity screening and functional analysis of personalized kidney organoids, achieving accuracy comparable to conventional biological assays. Organ-on-a-chip technologies have also been successfully merged with kidney and liver organoids, incorporating microfluidic flow and multisensor arrays to monitor biochemical, mechanical, and electrophysiological parameters continuously, thereby enhancing organoid maturation and physiological relevance for regenerative medicine applications (Ferrari et al., 2020; Zhang et al., 2017). Additionally, nanomaterial-based biosensors are being explored across diverse organoid models, including intestinal and tumor organoids, to monitor cell differentiation, tissue barrier function, and responses to mechanical or chemical stimuli, underscoring the broad utility of nanobiosensors in advancing 3D organoid research and translational biomedicine (Yousafzai and Hammer, 2023; Shen et al., 2023).
5 Discussion
Broad adoption of nanobiosensors for stem cell and organoid research is slowed by three interconnected challenges. First, little standardization exists; particle size, surface chemistry, and readout protocols vary from laboratory to laboratory, making results hard to reproduce and to compare. Second, safety guidance is vague. Investigators have yet to agree on a probe occupancy limit that defines how much RNA or protein a sensor can bind before it perturbs differentiation, and the long-term fate of many nanomaterials in living tissue remains uncertain. Third, the regulatory path is unclear; moving a prototype into a fully compliant Good Manufacturing Practice workflow demands costly retooling and exhaustive documentation that most academic centers cannot support.
Next-generation sensor platforms now merge optical, electrochemical, magnetic, and mechanical readouts on a single chip to capture the full complexity of lineage choice. Artificial intelligence tools sift these high dimensional data streams, uncovering faint molecular patterns that predict fate decisions and automatically compensating for sensor drift. Emerging systems even pair real time sensing with on board actuators such as microfluidic pumps or drug releasing nanoparticles, allowing the culture environment to adjust itself in response to live feedback and thereby reducing batch variability while enabling truly personalized cell products.
The coming years are likely to bring biodegradable sensor substrates, gene circuit smart beacons, and wireless implantable networks that extend monitoring from the dish into the patient. Success will depend on community driven benchmarks covering device specifications, data formats, and analysis pipelines. International working groups must align quality metrics, while explainable artificial intelligence frameworks will help regulators trust machine guided judgements. Should these pieces align, nanobiosensors will finally offer regenerative medicine precise and continuous control over cell fate and tissue function from the bioreactor all the way to clinical application.
Author contributions
YS: Conceptualization, Writing – original draft, Writing – review and editing. G-JJ: Conceptualization, Funding acquisition, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant No. RS-2023-00237493) and the Ministry of Trade, Industry and Energy (MOTIE), Republic of Korea (Grant No. RS-2024-00424600 and RS-2024-00433255).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amaral, D. L., Zanette, R. S., Almeida, C. G., Almeida, L. B., de Oliveira, L. F., Marcomini, R. F., et al. (2019). In vitro evaluation of barium titanate nanoparticle/alginate 3D scaffold for osteogenic human stem cell differentiation. Biomed. Mater. 14 (3), 035011. doi:10.1088/1748-605x/ab0a52
An, J. H., Kim, S. U., Park, M.-K., and Choi, J. W. (2015). Electrochemical detection of human mesenchymal stem cell differentiation on fabricated gold nano-dot cell chips. J. Nanosci. Nanotechnol. 15 (10), 7929–7934. doi:10.1166/jnn.2015.11225
Bagnaninchi, P. O., and Drummond, N. (2011). Real-time label-free monitoring of adipose-derived stem cell differentiation with electric cell-substrate impedance sensing. Proc. Natl. Acad. Sci. 108 (16), 6462–6467. doi:10.1073/pnas.1018260108
Degliangeli, F., Kshirsagar, P., Brunetti, V., Pompa, P. P., and Fiammengo, R. (2014). Absolute and direct microRNA quantification using DNA–gold nanoparticle probes. J. Am. Chem. Soc. 136 (6), 2264–2267. doi:10.1021/ja412152x
Devarasetty, M., Forsythe, S., Shupe, T. D., Soker, S., Bishop, C. E., Atala, A., et al. (2017). Optical tracking and digital quantification of beating behavior in bioengineered human cardiac organoids. Biosensors 7 (3), 24. doi:10.3390/bios7030024
Ding, Y., Tous, C., Choi, J., Chen, J., and Wong, W. W. (2024). Orthogonal inducible control of Cas13 circuits enables programmable RNA regulation in mammalian cells. Nat. Commun. 15 (1), 1572. doi:10.1038/s41467-024-45795-x
Du, Y., Wang, Y. R., Bao, Q. Y., Xu, X. X., Xu, C., Wang, S., et al. (2025). Personalized vascularized tumor organoid-on-a-chip for tumor metastasis and therapeutic targeting assessment. Adv. Mater. 37 (6), 2412815. doi:10.1002/adma.202412815
Dutta, A., Karanth, S. S., Bhattacharya, M., Liput, M., Augustyniak, J., Cheung, M., et al. (2020). A proof of concept ‘phase zero’ study of neurodevelopment using brain organoid models with Vis/near-infrared spectroscopy and electrophysiology. Sci. Rep. 10 (1), 20987. doi:10.1038/s41598-020-77929-8
Eshetie, K., Najafova, N., Graefen, B., Aliyeva, L., Tauseef, M., and Fazal, N. (2023). SPHK1 maintains endothelial barrier function and reanneals adherens junctions, thus hastens the recovery process. J. Immunol. 210 (Suppl. ment_1), 70. doi:10.4049/jimmunol.210.supp.70.03
Ferrari, E. (2023). Gold nanoparticle-based plasmonic biosensors. Biosensors 13 (3), 411. doi:10.3390/bios13030411
Ferrari, E., Palma, C., Vesentini, S., Occhetta, P., and Rasponi, M. (2020). Integrating biosensors in organs-on-chip devices: a perspective on current strategies to monitor microphysiological systems. Biosensors 10 (9), 110. doi:10.3390/bios10090110
Fu, J., Wiraja, C., Chong, R., Xu, C., and Wang, D.-A. (2017). Real-time and non-invasive monitoring of embryonic stem cell survival during the development of embryoid bodies with smart nanosensor. Acta Biomater. 49, 358–367. doi:10.1016/j.actbio.2016.11.027
Gonzales, K. A. U., Polak, L., Matos, I., Tierney, M. T., Gola, A., Wong, E., et al. (2021). Stem cells expand potency and alter tissue fitness by accumulating diverse epigenetic memories. Science. 374 (6571), eabh2444. doi:10.1126/science.abh2444
Hou, Y., Chen, M., Yang, L., and Lee, K.-B. (2025). Real-time intracellular monitoring of miRNA dynamics during induced pluripotent stem cell neuronal differentiation via plasmon-enhanced nanobiosensing. Nano Lett. 25, 10402–10411. doi:10.1021/acs.nanolett.5c01840
Ishikawa, M., Aoyama, T., Shibata, S., Sone, T., Miyoshi, H., Watanabe, H., et al. (2020). miRNA-based rapid differentiation of purified neurons from hPSCs advancestowards quick screening for neuronal disease phenotypes in vitro. Cells 9 (3), 532. doi:10.3390/cells9030532
Iwasaki, R. S., and Batey, R. T. (2020). SPRINT: a Cas13a-based platform for detection of small molecules. Nucleic Acids Res. 48 (17), e101–e. doi:10.1093/nar/gkaa673
Jarrige, M., Frank, E., Herardot, E., Martineau, S., Darle, A., Benabides, M., et al. (2021). The future of regenerative medicine: cell therapy using pluripotent stem cells and acellular therapies based on extracellular vesicles. Cells 10 (2), 240. doi:10.3390/cells10020240
Jhunjhunwala, A., Kim, J., Kubelick, K. P., Ethier, C. R., and Emelianov, S. Y. (2023). In vivo photoacoustic monitoring of stem cell location and apoptosis with caspase-3-responsive nanosensors. ACS Nano 17 (18), 17931–17945. doi:10.1021/acsnano.3c04161
Kang, M.-J., Cho, Y.-W., and Kim, T.-H. (2023). Progress in nano-biosensors for non-invasive monitoring of stem cell differentiation. Biosensors 13 (5), 501. doi:10.3390/bios13050501
Kang, Y., Na, J., Karima, G., Amirthalingam, S., Hwang, N. S., and Kim, H. D. (2024). Mesenchymal stem cell spheroids: a promising tool for vascularized tissue regeneration. Tissue Eng. Regen. Med. 21 (5), 673–693. doi:10.1007/s13770-024-00636-2
Kim, J.-E., Jeong, G.-J., Yoo, Y. M., Bhang, S. H., Kim, J. H., Shin, Y. M., et al. (2025a). 3D bioprinting technology for modeling vascular diseases and its application. Biofabrication 17 (2), 022014. doi:10.1088/1758-5090/adc03a
Kim, Y., Goswami, I., Gill, E., Mahmoodi, S. R., Consiglio, A. N., Velazquez, J., et al. (2025b). Vascular microphysiological system for investigating endothelial barrier function during organ preservation and reperfusion. Small 21 (11), 2410168. doi:10.1002/smll.202410168
Kumar, P., Courtes, M., Lemmers, C., Le Digarcher, A., Coku, I., Monteil, A., et al. (2023). Functional mapping of microRNA promoters with dCas9 fused to transcriptional regulators. Front. Genet. 14, 1147222. doi:10.3389/fgene.2023.1147222
Kutsche, L. K., Gysi, D. M., Fallmann, J., Lenk, K., Petri, R., Swiersy, A., et al. (2018). Combined experimental and system-level analyses reveal the complex regulatory network of miR-124 during human neurogenesis. Cell Syst. 7 (4), 438–52.e8. doi:10.1016/j.cels.2018.08.011
Lee, J., Kim, H., Lim, H.-R., Kim, Y. S., Hoang, T. T. T., Choi, J., et al. (2024). Large-scale smart bioreactor with fully integrated wireless multivariate sensors and electronics for long-term in situ monitoring of stem cell culture. Sci. Adv. 10 (7), eadk6714. doi:10.1126/sciadv.adk6714
Lee, T.-J., Jang, J., Kang, S., Bhang, S. H., Jeong, G.-J., Shin, H., et al. (2014). Mesenchymal stem cell-conditioned medium enhances osteogenic and chondrogenic differentiation of human embryonic stem cells and human induced pluripotent stem cells by mesodermal lineage induction. Tissue Eng. Part A 20 (7-8), 1306–1313. doi:10.1089/ten.tea.2013.0265
Li, X., Song, G., Dou, L., Yan, S., Zhang, M., Yuan, W., et al. (2021). The structure and unzipping behavior of dumbbell and hairpin DNA revealed by real-time nanopore sensing. Nanoscale 13 (27), 11827–11835. doi:10.1039/d0nr08729g
Lin, X., Song, D., Shao, T., Xue, T., Hu, W., Jiang, W., et al. (2024). A multifunctional biosensor via MXene assisted by conductive metal–organic framework for healthcare monitoring. Adv. Funct. Mater. 34 (11), 2311637. doi:10.1002/adfm.202311637
Lu, Y.-L., and Yoo, A. S. (2018). Mechanistic insights into microRNA-induced neuronal reprogramming of human adult fibroblasts. Front. Neurosci. 12, 522. doi:10.3389/fnins.2018.00522
Ma, Z., Zhang, J., Zou, S., Huang, K., Li, W., Elhousseini Hilal, M., et al. (2025). Smart vascular grafts with integrated flow biosensors for hemodynamic real-time monitoring and vascular healthcare. ACS Nano 19, 7661–7676. doi:10.1021/acsnano.4c09980
Montagud-Martínez, R., Márquez-Costa, R., and Rodrigo, G. (2023). Programmable regulation of translation by harnessing the CRISPR-Cas13 system. Chem. Commun. 59 (18), 2616–2619. doi:10.1039/d3cc00058c
Ouahoud, S., Giugliano, F. P., and Muncan, V. (2024). Monitoring intestinal organoid–derived monolayer barrier functions with electric cell–substrate impedance sensing (ECIS). Bio-protocol 14 (5), e4947. doi:10.21769/bioprotoc.4947
Papadimitriou, E., Koutsoudaki, P. N., Thanou, I., Karagkouni, D., Karamitros, T., Chroni-Tzartou, D., et al. (2023). A miR-124-mediated post-transcriptional mechanism controlling the cell fate switch of astrocytes to induced neurons. Stem Cell Rep. 18 (4), 915–935. doi:10.1016/j.stemcr.2023.02.009
Papikian, A., Liu, W., Gallego-Bartolomé, J., and Jacobsen, S. E. (2019). Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 10 (1), 729. doi:10.1038/s41467-019-08736-7
Park, S. B., Lim, B., Kim, K. Y., and Koh, B. (2024). Long and short-term effect of mTOR regulation on cerebral organoid growth and differentiations. Tissue Eng. Regen. Med. 21 (1), 159–169. doi:10.1007/s13770-023-00611-3
Pazzin, D. B., Previato, T. T. R., Budelon Gonçalves, J. I., Zanirati, G., Xavier, F. A. C., da Costa, J. C., et al. (2024). Induced pluripotent stem cells and organoids in advancing neuropathology research and therapies. Cells 13 (9), 745. doi:10.3390/cells13090745
Pflueger, C., Tan, D., Swain, T., Nguyen, T., Pflueger, J., Nefzger, C., et al. (2018). A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. 28 (8), 1193–1206. doi:10.1101/gr.233049.117
Ramirez, M. A., Pericuesta, E., Fernandez-Gonzalez, R., Pintado, B., and Gutierrez-Adan, A. (2007). Inadvertent presence of pluripotent cells in monolayers derived from differentiated embryoid bodies. Int. J. Dev. Biol. 51 (5), 397–408. doi:10.1387/ijdb.062255mr
Ren, X., Weng, Y., Zhang, Z., Pan, G., Li, C., and Cao, Y. (2025). Nanopore-based electroporation enables high-efficiency, rapid RNA-mediated reprogramming of primary fibroblasts into human iPSCs. Nano Lett. 25, 10310–10320. doi:10.1021/acs.nanolett.5c01219
Sannino, A., Velarte, A., Otín, A., Artigas, J. I., and Oliván-Viguera, A. (2023). A flexible PDMS-based optical biosensor for stretch monitoring in cardiac tissue samples. Sensors 23 (23), 9454. doi:10.3390/s23239454
Schoon, R. M., van der Meer, W. J., van Stalborch, A.-M. D., van Buul, J. D., and Huveneers, S. (2025). VE-cadherin RGD motifs are dispensable for cell–cell junctions, endothelial barrier function and monocyte extravasation. Tissue Barriers, 2478349. doi:10.1080/21688370.2025.2478349
Septiana, W. L., and Pawitan, J. A. (2024). Potential use of organoids in regenerative medicine. Tissue Eng. Regen. Med. 21 (8), 1125–1139. doi:10.1007/s13770-024-00672-y
Shen, C., Zhang, Z.-j., Li, X.-x., Huang, Y.-p., Wang, Y.-x., Zhou, H., et al. (2023). Intersection of nanomaterials and organoids technology in biomedicine. Front. Immunol. 14, 1172262. doi:10.3389/fimmu.2023.1172262
Son, Y. H., Won, J., Park, Y. I., Park, S.-J., and Jeong, G. J. (2025). Mitochondrial dysfunction and fibrosis in atrial fibrillation: molecular signaling in fast-pacing organoid models. J. Industrial Eng. Chem. doi:10.1016/j.jiec.2025.06.038
Susa, K., Kobayashi, K., Galichon, P., Matsumoto, T., Tamura, A., Hiratsuka, K., et al. (2023). ATP/ADP biosensor organoids for drug nephrotoxicity assessment. Front. Cell Dev. Biol. 11, 1138504. doi:10.3389/fcell.2023.1138504
Tabibzadeh, N., and Morizane, R. (2024). Advancements in therapeutic development: kidney organoids and organs on a chip. Kidney Int. 105 (4), 702–708. doi:10.1016/j.kint.2023.11.035
Tabibzadeh, N., Satlin, L. M., Jain, S., and Morizane, R. (2023). Navigating the kidney organoid: insights into assessment and enhancement of nephron function. Am. J. Physiology-Renal Physiology 325 (6), F695–F706. doi:10.1152/ajprenal.00166.2023
Thangam, T., Parthasarathy, K., Supraja, K., Haribalaji, V., Sounderrajan, V., Rao, S. S., et al. (2024). Lung organoids: systematic review of recent advancements and its future perspectives. Tissue Eng. Regen. Med. 21 (5), 653–671. doi:10.1007/s13770-024-00628-2
Torizal, F. G., Noorintan, S. T., and Gania, Z. (2024). Bioengineering tooth and periodontal organoids from stem and progenitor cells. Organoids 3 (4), 247–265. doi:10.3390/organoids3040015
Wang, Q., Ma, X., Liao, H., Liang, Z., Li, F., Tian, J., et al. (2020c). Artificially engineered cubic iron oxide nanoparticle as a high-performance magnetic particle imaging tracer for stem cell tracking. ACS Nano 14 (2), 2053–2062. doi:10.1021/acsnano.9b08660
Wang, W., Lu, G., Su, X., Tang, C., Li, H., Xiong, Z., et al. (2020a). Pten-mediated Gsk3β modulates the naïve pluripotency maintenance in embryonic stem cells. Cell Death and Dis. 11 (2), 107. doi:10.1038/s41419-020-2271-0
Wang, Y., Yang, L.-Z., and Chen, L.-L. (2020b). Protocol for dynamic imaging of RNA in living cells by CRISPR-Cas13 system. Star. Protoc. 1 (1), 100037. doi:10.1016/j.xpro.2020.100037
Wheat, J. C., Sella, Y., Willcockson, M., Skoultchi, A. I., Bergman, A., Singer, R. H., et al. (2020). Single-molecule imaging of transcription dynamics in somatic stem cells. Nature 583 (7816), 431–436. doi:10.1038/s41586-020-2432-4
Wiley, L. A., Burnight, E. R., Songstad, A. E., Drack, A. V., Mullins, R. F., Stone, E. M., et al. (2015). Patient-specific induced pluripotent stem cells (iPSCs) for the study and treatment of retinal degenerative diseases. Prog. Retin. Eye Res. 44, 15–35. doi:10.1016/j.preteyeres.2014.10.002
Wiraja, C., Yeo, D. C., Chong, M. S., and Xu, C. (2016). Nanosensors for continuous and noninvasive monitoring of mesenchymal stem cell osteogenic differentiation. Small 12 (10), 1342–1350. doi:10.1002/smll.201502047
Xiao, M., Man, T., Zhu, C., Pei, H., Shi, J., Li, L., et al. (2018). MoS2 nanoprobe for microRNA quantification based on duplex-specific nuclease signal amplification. ACS Appl. Mater. Interfaces 10 (9), 7852–7858. doi:10.1021/acsami.7b18984
Xu, C., Wang, Z., Liu, Y., Duan, K., and Guan, J. (2024). Delivery of miR-15b-5p via magnetic nanoparticle-enhanced bone marrow mesenchymal stem cell-derived extracellular vesicles mitigates diabetic osteoporosis by targeting GFAP. Cell Biol. Toxicol. 40 (1), 52. doi:10.1007/s10565-024-09877-2
Xu, L., Xu, M., Sun, X., Feliu, N., Feng, L., Parak, W. J., et al. (2023). Quantitative comparison of gold nanoparticle delivery via the enhanced permeation and retention (EPR) effect and mesenchymal stem cell (MSC)-based targeting. ACS Nano 17 (3), 2039–2052. doi:10.1021/acsnano.2c07295
Yao, B., Lei, Z., Gonçalves, M. A., and Sluijter, J. P. (2024). Integrating prime editing and cellular reprogramming as novel strategies for genetic cardiac disease modeling and treatment. Curr. Cardiol. Rep. 26 (11), 1197–1208. doi:10.1007/s11886-024-02118-2
Yousafzai, M. S., and Hammer, J. A. (2023). Using biosensors to study organoids, spheroids and organs-on-a-chip: a mechanobiology perspective. Biosensors 13 (10), 905. doi:10.3390/bios13100905
Yousuf, M., Rochet, J. C., Singh, P., and Hussain, M. M. (2025). Advancing brain organoid electrophysiology: minimally invasive technologies for comprehensive characterization. Adv. Mater. Technol. 10 (7), 2401585. doi:10.1002/admt.202401585
Zhang, Y. S., Aleman, J., Shin, S. R., Kilic, T., Kim, D., Mousavi Shaegh, S. A., et al. (2017). Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. 114 (12), E2293–E2302. doi:10.1073/pnas.1612906114
Zhao, Y., Yang, R., Bousraou, Z., Richardson, K., and Wang, S. (2022). Probing notch1-dll4 signaling in regulating osteogenic differentiation of human mesenchymal stem cells using single cell nanobiosensor. Sci. Rep. 12 (1), 10315. doi:10.1038/s41598-022-14437-x
Zhong, Z.-D., Xie, Y.-Y., Chen, H.-X., Lan, Y.-L., Liu, X.-H., Ji, J.-Y., et al. (2023). Systematic comparison of tools used for m6A mapping from nanopore direct RNA sequencing. Nat. Commun. 14 (1), 1906. doi:10.1038/s41467-023-37596-5
Zips, S., Huang, B., Hotte, S., Hiendlmeier, L., Wang, C., Rajamani, K., et al. (2023). Aerosol jet-printed high-aspect ratio micro-needle electrode arrays applied for human cerebral organoids and 3D neurospheroid networks. ACS Appl. Mater. and Interfaces 15 (30), 35950–35961. doi:10.1021/acsami.3c06210
Keywords: nano biosensor, stem cell, organoid, differentiation, sensor
Citation: Son YH and Jeong G-J (2025) Nanobiosensors for monitoring of stem-cell differentiation and organoids. Front. Nanotechnol. 7:1652480. doi: 10.3389/fnano.2025.1652480
Received: 23 June 2025; Accepted: 18 July 2025;
Published: 24 July 2025.
Edited by:
Hyo-Ryoung Lim, Pukyong National University, Republic of KoreaReviewed by:
Xiangwei Zhao, Southeast University, ChinaCopyright © 2025 Son and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gun-Jae Jeong, amdqODE0QGNhdGhvbGljLmFjLmty
 Young Hoon Son
Young Hoon Son Gun-Jae Jeong
Gun-Jae Jeong