- 1Department of Microbiology, M. D. University, Rohtak, India
- 2Kirori Mal College, University of Delhi, Delhi, India
- 3Centre for Medical Biotechnology, M. D. University, Rohtak, India
Nanoparticles (NPs) possess unique properties due to their higher surface-to-volume ratio and reactivity. Negative environmental impact and high cost of traditional modes of synthesis have driven the shift towards utilization of microbes and plants for synthesising NPs, referred as the biological or ‘Green’ synthesis. This study reported extracellular synthesis of copper NPs (CuNPs) using the supernatant of Bacillus licheniformis CPJN13S. The parameters affecting this process were optimized by OFAT approach and were reported to be 5 mM concentration of copper sulfate (CuSO4), 32 h incubation period, 18 h reaction time, 20:20 filtrate/substrate ratio, 7 pH, and 37 °C temperature. CuNPs produced a characteristic UV-Visible absorption peak between 550–650 nm, Z-average of 305.3 nm and zeta potential value of −24.7 mV. SEM and HR-TEM revealed hexagonal shape of NPs having average size of 12.4 nm. XRD peaks obtained at 2θ positions of 45.52°, 56.52°, and 75.34° matched to diffraction from Cu. Antimicrobial assay conducted using 100 μg/mL CuNPs led to highest inhibition of 20.4% (Bacillus subtilis MTCC No. 441) and 43% (Staphylococcus aureus MTCC No. 737), at 21 h and 27 h, respectively. The results suggest that biological synthesis can serve as the eco-friendly alternative of physical and chemical modes of synthesizing CuNPs and can be used to develop highly effective antibacterial agents.
1 Introduction
Nanoparticles (NPs) are micro particles in the range of 1–100 nm made up of metal, metal oxides or organic matter (Santos et al., 2025). They possess unique physical, chemical and biological properties as well as enhanced reactivity than their bulk forms (Ealia and Saravanakumar, 2017). Metal NPs synthesized by different precursors viz., gold, zinc, copper, iron, and silver, have distinct optoelectrical properties because of their plasma resonance features (Burlec et al., 2023). Various physical (Ultrasonication, laser, pulse laser ablation) and chemical (Microemulsion, chemical vapour deposition, electrochemical) methods are used to synthesise NPs (Khan et al., 2022). However, requirement of extremely high temperature and pressure, high cost of sophisticated instruments, use of toxic chemicals along with difficulties in controlling size, distribution, and shape, have paved the way for biological synthesis (Rani et al., 2022). It includes the synthesis of NPs using bacteria, fungi, plants, etc., and has gained much attention recently owing to environmental safety and convenience (Bokolia et al., 2024). These NPs are formed by the reducing agents present in the supernatant/cell of microorganisms or plant extract. There is no requirement of extreme pressure/temperature and use of hazardous chemicals (Salem and Fouda, 2021).
Bacteria possess the ability of rapid multiplication and can be manipulated easily, therefore, suitable for the production of biological NPs. Conditions such as aeration, temperature, pH, and incubation period, can be adjusted to regulate their growth and produce NPs of desired shape and size (Yusof et al., 2019). They utilize either intracellular or extracellular mechanism to synthesize NPs (Ahir et al., 2025). The metal ions are reduced into their nanoform within the cell in intracellular mode of synthesis whereas extracellular synthesis is mediated by the enzymes present on the plasma membrane or released into the media. Once the metals are reduced from their ionic to atomic form, they undergo nucleation and form NPs. Some extracellular proteins also act as capping agents and stabilize the NPs (Tank et al., 2025).
Cu has long-term stability, resistance against oxidation, and is significant for the normal functioning of organisms. Its NPs exhibit unique properties due to their small size, high surface area-to-volume ratio, and quantum effects leading to more reactions in less time (Crisan et al., 2021). Though numerous studies have reported the antimicrobial properties of CuNPs, its synergistic use with antibiotics has also been reported as an efficient way to enhance the antibiotic efficacy used in clinical practice (Abo-Shama et al., 2020; Agreles et al., 2022). In addition, CuNPs have also been found to exhibit superior antibacterial properties as compared to a number of antibiotics used in present time (Sacoto-Figueroa et al., 2021). For plants, Cu is an essential micronutrient critical to photosynthesis, respiration, antioxidant system, and signal transduction (Raza et al., 2022). These properties have boosted the demand of CuNPs particularly in agriculture and healthcare sector (Rautela et al., 2025; Vigneshwaran et al., 2025).
Bacillus species are known to contain an array of enzymes, polysaccharide, and amino acids, which can reduce metal ions to NPs. However, several physico-chemical factors affect this process, necessitating the optimization of biosynthesis process in order to enhance the quality of NPs. Optimization ensures the production of homogeneous NPs with well-defined properties (Park et al., 2023). Therefore, one factor at a time (OFAT) approach was used in the present study to optimize the following factors; concentration of metal precursor (CuSO4), incubation period, reaction time, filtrate (Bacterial supernatant) to substrate (CuSO4) ratio, pH, and temperature, which affects the synthesis of CuNPs using Bacillus licheniformis CPJN13S. Earlier studies have reported the antibacterial properties of CuNPs against a number of bacteria viz., Klebsiella pneumoniae, Proteus vulgaris, Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Streptococcus mutans (Aytar et al., 2025; Gao et al., 2025; Ibrahim, 2025). However, there are very few reports on the comparative account of antibacterial efficacy of biological (synthesized using bacteria) and chemical CuNPs. Therefore, in the present study, biological CuNPs synthesized using optimized parameters were utilized to check their antibacterial efficacy and compare the same with chemically synthesized CuNPs.
2 Experimental setup
2.1 Optimization of CuNPs biosynthesis using Bacillus licheniformis CPJN13S
Bacillus licheniformis CPJN13S was procured from Lab 312, Department of Microbiology, MDU, Rohtak, Haryana. It is a plant growth promoting endophyte and has already been used for the synthesis of CuNPs in a previous study (Rani et al., 2024). First, CuSO4 concentration was optimized by adding 1 mM, 2 mM, 3 mM, 4 mM, and 5 mM of CuSO4 solution to the equal volume of supernatant obtained by centrifuging 2-days old CPJN13S culture and incubated at 30 °C, 150 rpm for 24 h. The effect of incubation period was determined by deriving the supernatant from 16 h, 24 h, 32 h, 40 h, and 48 h old CPJN13S culture and adding it to the CuSO4 solution with optimized concentration followed by incubation at 30 °C, 150 rpm for 24 h. Next, reaction mixture of CuSO4 and supernatant (optimized CuSO4 concentration and incubation period) was incubated for different time intervals (0 h, 6 h, 12 h, 18 h, & 24 h), at 30 °C and 150 rpm to assess the impact of reaction time. Then, filtrate to substrate ratio was optimized by adding 4 mL, 8 mL, 12 mL, 16 mL, and 20 mL of supernatant derived from CPJN13S culture to 20 mL of CuSO4 solution using optimized factors. Effect of pH (3, 5, 7, 9, and 11) was also determined with optimized factors and incubated at 30 °C and 150 rpm. Finally, the reaction mixture prepared using all the optimized factors were incubated at varying degrees of temperature (23 °C, 30 °C, 37 °C, 44 °C, 51 °C) at 150 rpm for optimized reaction time (Table 1).
2.2 Biosynthesis and characterization of CuNPs using optimized parameters
CuNPs obtained using CPJN13S with optimized parameters were characterized for UV-Visible absorption spectrum (Shimadzu) in the range of 400–800 nm. Polydispersity index (PDI), particle size analysis, and zeta potential, were measured on the Zetasizer (Malvern). Scanning electron microscopy (SEM) images were obtained at an accelerating voltage of 15 kV (Zeiss Geminisem). The aqueous suspension of CuNPs was transferred onto an amorphous carbon-coated copper grid, dried, and analyzed for high resolution transmission electron microscopy (HR-TEM) (Hitachi). Samples were scanned from 30° to 80° of 2θ in increments of 0.02° with a 4-s counting time for XRD (X-ray diffraction) analysis (Rigaku).
2.3 Antibacterial assay
The antibacterial activity of CuNPs was checked against Bacillus subtilis MTCC No. 441 and Staphylococcus aureus MTCC No. 737. The solution containing 100 μg/mL CuNPs (1.5 mL) was added to 50 mL of tryptic soy broth (TSB) inoculated with 1 × 108 CFU mL-1 of B. subtilis and S. aureus culture and incubated at 30 °C, 150 rpm. Autoclaved water served as control. Bacterial growth studies were conducted in triplicates by measuring the absorbance at 600 nm within every 3 h for 30 h. The percent inhibition was calculated using:
Where A0 is the absorbance of the control and A is the absorbance of test sample (Wu et al., 2019).
3 Results and discussion
3.1 Optimization factors
3.1.1 Concentration of CuSO4 solution
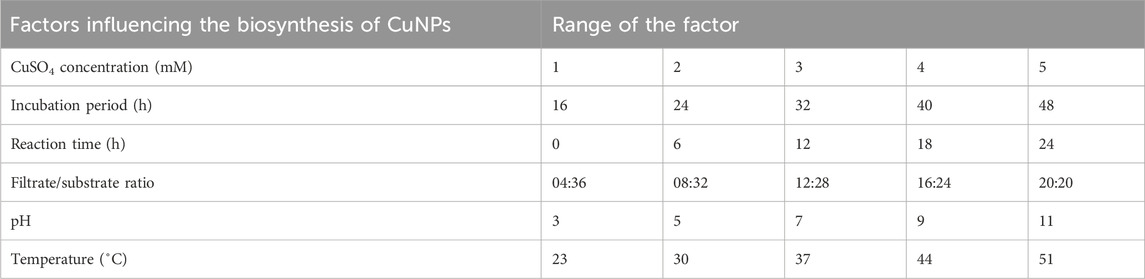
Five concentrations of CuSO4 solution (1–5 mM) were used to analyze their effect on the biosynthesis of CuNPs. The colour of CuSO4 solution changed from blue to green upon the addition of an equal volume of CPJN13S supernatant. The optical properties of CuSO4 solution change with the formation of nanoparticles, causing the solution to turn green and act as the preliminary confirmation of nanoparticle synthesis (Amaliyah et al., 2020; Rani et al., 2024). Since color change depends on the excitation on surface plasmon vibration, the formation of CuNPs was confirmed by UV-Visible spectrophotometer in the range of 400–800 nm.
Generally, CuNPs exhibit a characteristic UV-Visible absorption peak between 550–650 nm (Gopalakrishnan and Muniraj, 2021; Aziz et al., 2024). In the present study, lower concentrations of CuSO4 (1–3 mM) did not produce a characteristic absorption peak of CuNPs. Only higher concentrations led to the formation of CuNPs absorption peak with 5 mM concentration showing highest absorbance (λmax615 nm = 0.239) (Figure 1). Similar study done on the synthesis of CuONPs using Streptomyces sp. Reported increase in the rate of NPs formation with increase in the concentration of CuSO4 from 1 mM to 8 mM and achieved maximum absorbance at 5 mM concentration (Bukhari et al., 2021). Noor et al. (2020) also observed the characteristic peaks of CuNPs synthesized from Aspergillus niger STA9 at/above 5 mM CuSO4 concentration while concentrations lower than 5 mM did not produce the absorption peak.
3.1.2 Incubation period
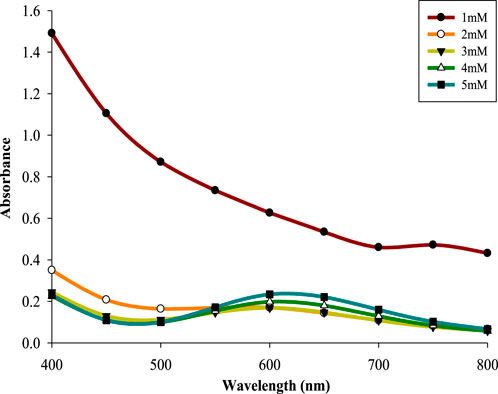
CPJN13S was incubated in TSB for different time intervals (16–48 h) to determine the effect of incubation period on biosynthesis of CuNPs, Supernatant derived from each time interval was added to 5 mM concentration of CuSO4 solution. Incubation period above 24 h resulted into the formation of a characteristic absorption peak with maximum value of absorbance (λmax614 nm = 0.281) obtained at 32 h (Figure 2). This can be attributed to the formation of secondary metabolites at late-exponential and early stationary phase leading to the reduction of metal salt into NPs (Kaur et al., 2016; Seyedsayamdost, 2019). A further increase in the incubation time caused a slight reduction in absorbance. An earlier study done in our laboratory have also reported 32 h as the optimized incubation period (Rani et al., 2025).
3.1.3 Reaction time
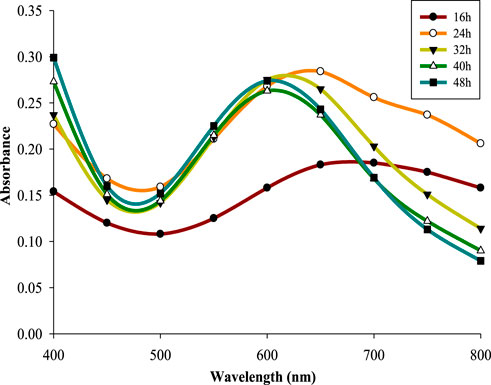
Reaction time has significant effect on the stability and production of NPs (Rajesh et al., 2016). 5 mM CuSO4 solution mixed with supernatant retrieved from 32 h old CPJN13S culture, was incubated for different reaction times (0–24 h). All the reaction times led to the formation of characteristic absorption peak of CuNPs with absorbance showing a consistent increase upon increasing the reaction time up to 18 h (λmax602 nm = 0.342). Previous study has also reported increase in absorbance with increasing reaction times from 24 h to 96 h during synthesis of CuNPs using Aspergillus niger STA9 (Noor et al., 2020). However, reaction time above 18 h resulted in lower absorbance indicating its negative influence on biosynthesis (Figure 3). CuNPs synthesized using cell-free culture extract of Pseudomonas fluorescens MAL2 underwent increase in absorbance upon increasing the reaction time up to 90 min above which it reduced indicating the role of optimized reaction time in sustaining the stability and aggregation of CuNPs (El-Saadony et al., 2020).
3.1.4 Filtrate to substrate ratio
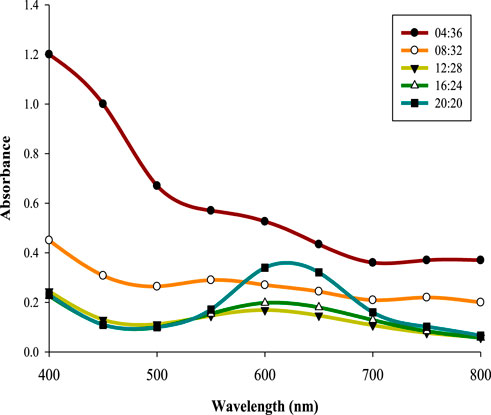
Five varying volumes of filtrate derived from 32 h old CPJN13S culture were mixed with five different volumes of 5 mM substrate in the ratio of 04:36–20:20 (mL), and incubated for 18 h at 30 °C, 150 rpm. Lower volumes of supernatant (04:36, 08:32, and 12:28) did not produce absorption peak as observed in case of higher volumes with maximum absorbance obtained with an equal ratio of the supernatant and CuSO4, i.e., 20:20 (λmax600 nm = 0.34). An earlier study has also reported equal ratio (1:1) of filtrate and substrate to result in highest absorbance of CuONPs at 550 nm as compared to 1:0.5 and 1:2 ratio (Bukhari et al., 2021). Similar observation was made during the green synthesis of CuNP from the polysaccharides of Bacillus sp. (Banerjee et al., 2024). Thus an equal ratio of supernatant and substrate is optimum for biosynthesis of CuNPs (Figure 4).
3.1.5 pH
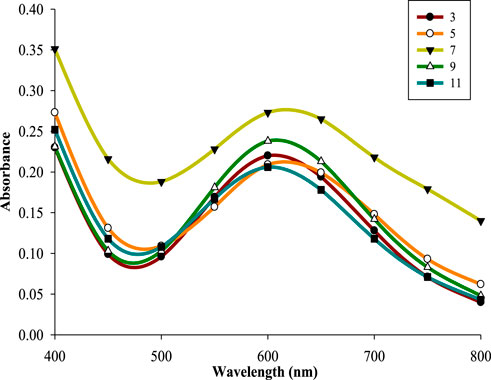
Variations in pH can affect shape as well as size of the NPs (Rajesh et al., 2016; Alhalili, 2022). Therefore, effect of different pH (3–11) was analyzed using optimized CuSO4 concentration, incubation period, reaction time, and filtrate to substrate ratio, at 30 °C and 150 rpm. Neutral pH (7) led to highest absorbance (λmax610 nm = 0.277) while lower and higher pH reduced the same by decreasing the rate of reduction of copper ions into atoms (Figure 5). This is in accordance with previous studies reporting maximum synthesis of NPs under neutral pH condition (Tiwari et al., 2014; Lv et al., 2018). However, slightly acidic and basic conditions have also been reported as optimal for green synthesis of CuNPs rather than neutral pH depending upon the specific mode of synthesis and type of reducing agent/s (Shende et al., 2015; Amjad et al., 2021).
3.1.6 Temperature
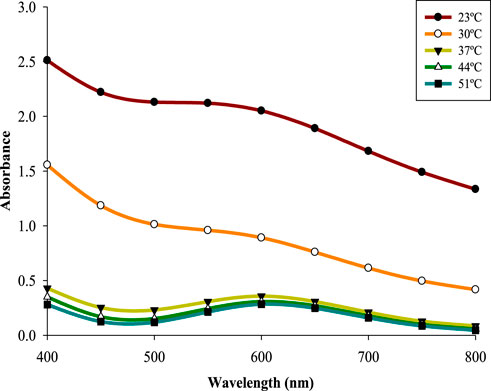
Five different temperatures 23 °C–51 °C were used to study the effect of temperature on the biosynthesis of CuNPs. Characteristic peaks were observed only above 30 °C with 37 °C being the optimum one (λmax600 nm = 0.358) (Figure 6). Reduced absorbance at lower temperatures is attributed to lowering of microbial metabolism as noticed in case of CuNPs synthesized using Shewanella loihica (Lv et al., 2018). The conversion rate of Cu2+ ions increased when the temperature went up due to their enhanced availability to the reducing agents present in supernatant and reduced risk of secondary processes (Gopalakrishnan and Muniraj, 2021; Amjad et al., 2021). However, a temperature above 37 °C decreased the conversion rate of copper ions probably due to the enhanced nuclei surface activity promoting the agglomeration of particles or enzyme denaturation (Lv et al., 2018; Nagar and Devra, 2018).
3.2 Characterization of biosynthesized CuNPs
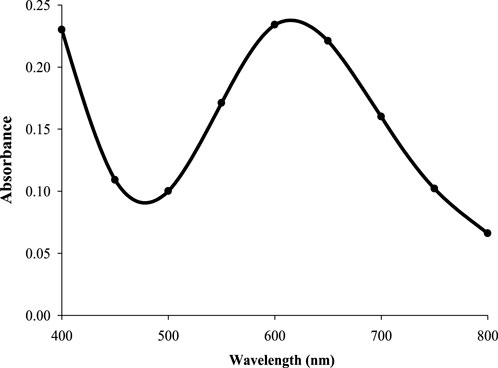
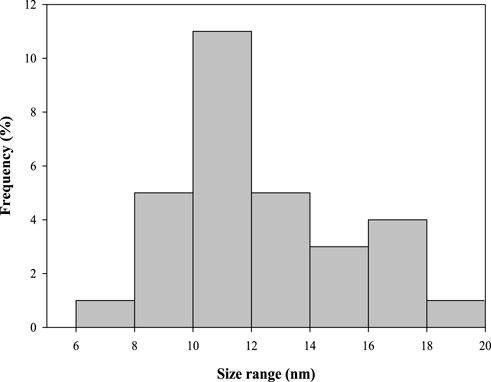
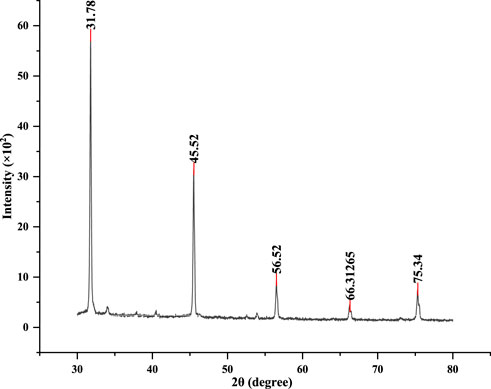
CuNPs were biosynthesised using the optimized parameters and characterized. The color of CuSO4 solution changed from blue to dark green upon addition of CPJN13S supernatant and exhibited an absorbance peak between 550 and 650 nm (Figure 7) as observed earlier (El-Saadony et al., 2020; Bukhari et al., 2021). PDI, Particle size, and zeta potential, were measured on the zetasizer. PDI is a measure of particle size distribution and its value below 0.4 (0.386 in present study) indicates the monodispersity of sample formulation (Danaei et al., 2018). Z-average depicting hydrodynamic size and zeta potential of NPs were reported to be 305.3 nm and −24.7 mV, respectively. Highly negative zeta potential prevents the aggregation and ensures long-term stability of biogenic CuNPs as reported earlier (Singh et al., 2023; Habibah et al., 2024) (Figures 8a,b). In a previous study, Cu/CuO NPs synthesised from Stenotrophomonas sp. and Priestia megaterium, also had highly negative zeta potential (Talebian et al., 2023; Mohamed et al., 2024). SEM and TEM revealed polygonal shape of the CuNPs (Figures 9a,b). Analysis of TEM images (ImageJ software) showed the NPs lying in range of 7.7–18.4 nm with an average size of 12 nm (Figure 10). Earlier studies have reported the synthesis of spherical CuNPs with an average size of 30 nm using bacterial strains isolated from Antarctica and polygonal CuNPs with an average size of 13.025 nm using Pantoea agglomerans (John et al., 2021; Rani et al., 2024). The XRD spectrum revealed three sharp peaks at 2θ positions of 45.52°, 56.52°, and 75.34° (JCPDS No. 040836) that matched to diffraction from Cu (Figure 11). According to JCPDS data (80–0,076), the exhibited diffraction peaks at 2θ = 31.78° and 66.31° corresponds to different planes of monoclinic phase of CuONPs, indicating partial oxidation of the CuNPs. The results are in accordance with previous investigations that reported similar diffraction peaks for Cu/CuONPs (Phul et al., 2018; Thakar et al., 2022; Manyasree et al., 2017). A comparative study of biosynthesized Cu/CuO NPs based on previous studies has been given in Table 2.
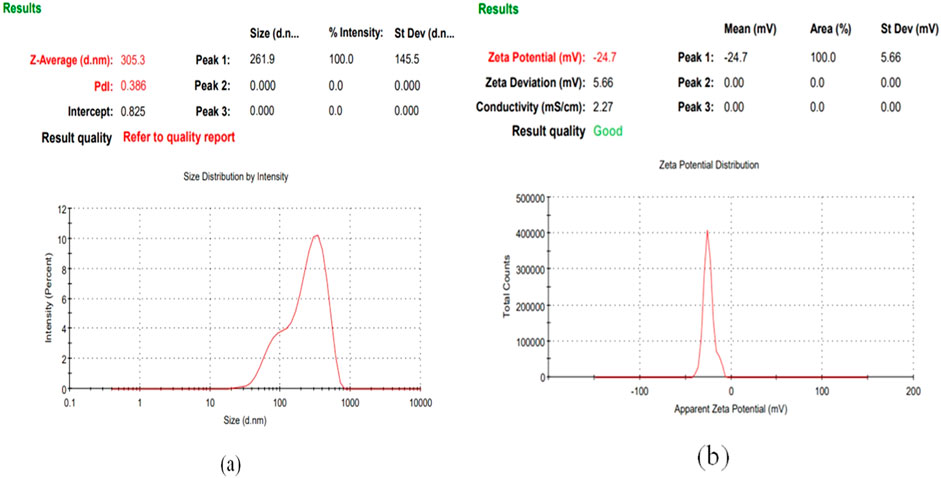
Figure 8. Size distribution (a) and Zeta potential analysis of CuNPs biosynthesized using optimized parameters (b).
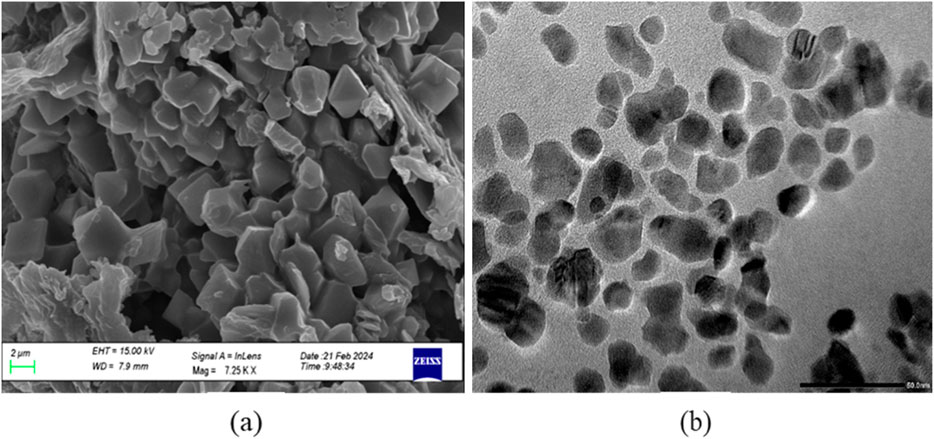
Figure 9. SEM micrograph (Scale 2 µm) (a) and TEM electron micrograph of CuNPs biosynthesized using optimized parameters (Scale 50 nm) (b).
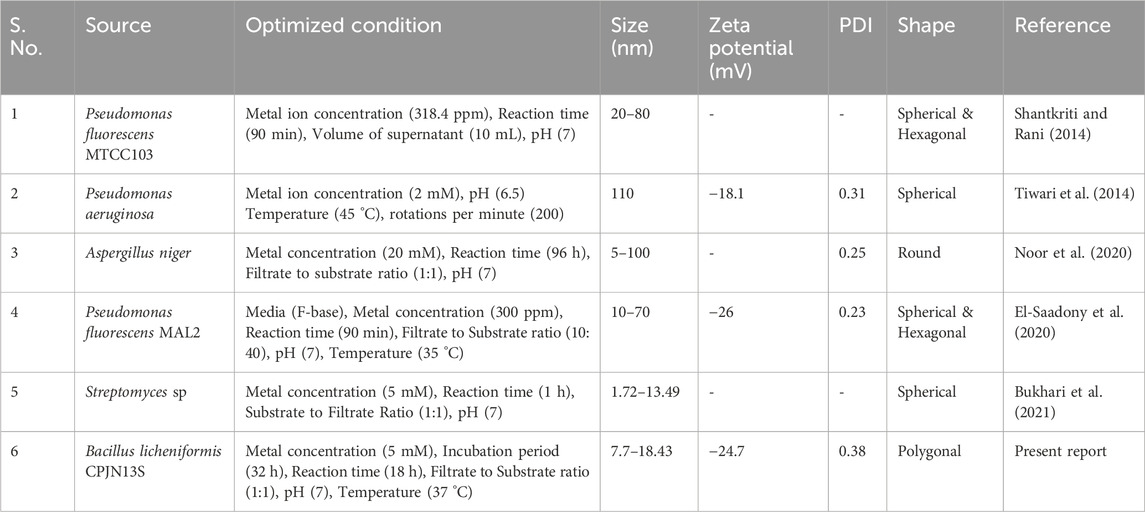
Table 2. A comparative analysis of the present study of biosynthesis, optimization and characterization of CuNPs with previous reports.
3.3 Antimicrobial assays of CuNPs
The antimicrobial activity of CuNPs was studied against B. subtilis and S. aureus by analyzing their growth curves in the presence of CuNPs. It was observed that 100 μg/mL of biosynthesized CuNPs lowered the growth rate of both the bacteria. However, maximum inhibition was noticed at 21 h and 27 h wherein a significant decrease of 20.4% and 43% was observed in the growth of B. subtilis and S. aureus, respectively, as compared to their respective control sample (Figures 12a,b). Moreover, S. aureus showed more sensitivity towards CuNPs as compared to B. subtilis and was more negatively affected. Several studies have demonstrated the bactericidal effect of biogenic CuNPs synthesized from bacteria (John et al., 2021; Talebian et al., 2023; Rani et al., 2025), fungi (Murthy et al., 2020; Nassar et al., 2023; Noor et al., 2025), and plants (Rambau et al., 2024; Kasthuri et al., 2024; Ibrahim, 2025), against a variety of bacterial strains. In addition, bimetallic nanoparticles of copper and silver synthesised biologically have also displayed significant antibacterial activity (Merugu et al., 2021; Al-Haddad et al., 2020).
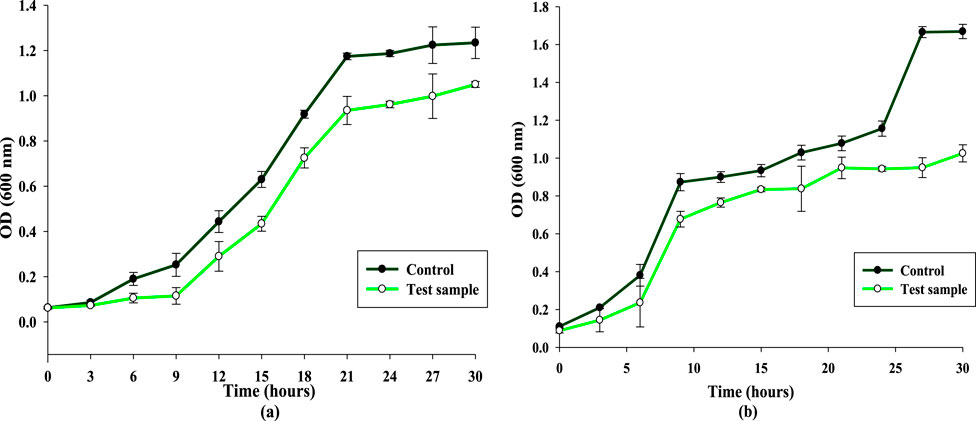
Figure 12. Effect of biosynthesized CuNPs on the growth curve dynamics (a) Bacillus subtilis. (b) Staphylococcus aureus.
4 Conclusion
In the present study, OFAT-mediated optimization of the biosynthesis of CuNPs using Bacillus licheniformis CPJN13S resulted into 5 mM concentration of CuSO4, 32 h incubation period, 18 h reaction time, equal filtrate to substrate ratio, 7 pH, and 37 °C temperature, to be the optimum values. These CuNPs were found to be effective against B. subtilis and S. aureus. The optimization studies are immensely crucial for the largescale application of CuNPs in the field of biomedicine to assist in replacing the widely consumed antibiotics and several related areas. Nevertheless, there is a need to keep screening more non-pathogenic microbes for their ability to synthesize NPs. Endophytes are a reliable source of metabolites which can be harnessed to synthesize various metallic NPs as utilized in the present study. In addition, more studies targeted on identifying and characterizing the reducing and capping agents involved in the reduction and stabilization of NPs, respectively, are required to be conducted.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
SR: Data curation, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. SD: Methodology, Writing – review and editing. PK: Formal Analysis, Writing – review and editing. PD: Formal Analysis, Writing – review and editing. KA: Formal Analysis, Writing – review and editing. AD: Conceptualization, Funding acquisition, Supervision, Validation, Writing – review and editing. PS: Conceptualization, Funding acquisition, Supervision, Validation, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. SR would like to acknowledge the Council of Scientific & Industrial Research, New Delhi, India, for CSIR fellowship. PK and PD acknowledge UGC for the fellowship support. The authors would like to acknowledge the Department of Science and Technology, Government of India, for providing FIST grant (Grant No. 1196 SR/FST/LS-I/2017/4) to the Department of Microbiology, MDU, Rohtak. The authors acknowledge the support from CIL (central instrumentation laboratory), MDU, Rohtak for carrying out the characterization of CuNPs. The authors are thankful to Department of Science and Technology, Govt. of Haryana, India (Grant No. HSCSIT/R&D/2022/2968) for providing infrastructural and financial support to undertake this research work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnano.2025.1663115/full#supplementary-material
References
Abo-Shama, U. H., El-Gendy, H., Mousa, W. S., Hamouda, R. A., Yousuf, W. E., Hetta, H. F., et al. (2020). Synergistic and antagonistic effects of metal nanoparticles in combination with antibiotics against some reference strains of pathogenic microorganisms. Infect. drug resist 13, 351–362. doi:10.2147/idr.s234425
Agreles, M. A. A., Cavalcanti, I. D. L., and Cavalcanti, I. M. F. (2022). Synergism between metallic nanoparticles and antibiotics. Appl. Microbiol. Biotechnol. 106, 3973–3984. doi:10.1007/s00253-022-12001-1
Ahir, S., Mehta, V. N., Raval, J. B., Kagdi, J. H., Kailasa, S. K., and Jha, S. (2025). Microbe-assisted synthesis of nanomaterials: insights and applications. ChemistrySelect 10, e02966. doi:10.1002/slct.202502966
Al-Haddad, J., Alzaabi, F., Pal, P., Rambabu, K., and Banat, F. (2020). Green synthesis of bimetallic copper–silver nanoparticles and their application in catalytic and antibacterial activities. Clean. Technol. Envir. 22, 269–277. doi:10.1007/s10098-019-01765-2
Alhalili, Z. (2022). Green synthesis of copper oxide nanoparticles CuO NPs from Eucalyptus Globoulus leaf extract: adsorption and design of experiments. Arab. J. Chem. 15, 103739. doi:10.1016/j.arabjc.2022.103739
Amaliyah, S., Pangesti, D. P., Masruri, M., Sabarudin, A., and Sumitro, S. B. (2020). Green synthesis and characterization of copper nanoparticles using Piper retrofractum Vahl extract as bioreductor and capping agent. Heliyon 6, e04636. doi:10.1016/j.heliyon.2020.e04636
Amjad, R., Mubeen, B., Ali, S. S., Imam, S. S., Alshehri, S., Ghoneim, M. M., et al. (2021). Green synthesis and characterization of copper nanoparticles using Fortunella margarita leaves. Polymers 13, 4364. doi:10.3390/polym13244364
Aytar, E. C., Basılı, T., Durmaz, A., Aydın, B., Seyfeli, R. C., Kahyaoğlu, İ. M., et al. (2025). Zero-waste production of copper nanoparticles and activated carbon from Erigeron canadensis L.: a sustainable approach for environmental and health applications. ChemistrySelect 10, e202405073. doi:10.1002/slct.202405073
Aziz, N. M. A., Goda, D. A., Abdel-Meguid, D. I., El-Sharouny, E. E., and Soliman, N. A. (2024). A comparative study of the biosynthesis of CuNPs by Niallia circulans G9 and Paenibacillus sp. S4c strains: characterization and application as antimicrobial agents. Microbe Cell Fact. 23, 156. doi:10.1186/s12934-024-02422-0
Banerjee, A., Roy, R. K., Sarkar, S., López, J. L., Vuree, S., and Bandopadhyay, R. (2024). Synthesis of hot spring origin bacterial cell wall polysaccharide-based copper nanoparticles with antibacterial property. Electron. J. Biotechnol. 68, 11–19. doi:10.1016/j.ejbt.2023.11.005
Bokolia, M., Baliyan, D., Kumar, A., Das, R., Kumar, R., and Singh, B. (2024). Biogenic synthesis of nanoparticles using microbes and plants: mechanisms and multifaceted applications. J. Environ. Anal. Chem. 105, 2069–2097. doi:10.1080/03067319.2024.2305238
Bukhari, S. I., Hamed, M. M., Al-Agamy, M. H., Gazwi, H. S., Radwan, H. H., and Youssif, A. M. (2021). Biosynthesis of copper oxide nanoparticles using Streptomyces MHM38 and its biological applications. J. Nanomater. 2021, 1–16. doi:10.1155/2021/6693302
Burlec, A. F., Corciova, A., Boev, M., Batir-Marin, D., Mircea, C., Cioanca, O., et al. (2023). Current overview of metal nanoparticles’ synthesis, characterization, and biomedical applications, with a focus on silver and gold nanoparticles. Pharmaceuticals 16, 1410. doi:10.3390/ph16101410
Crisan, M. C., Teodora, M., and Lucian, M. (2021). Copper nanoparticles: synthesis and characterization, physiology, toxicity and antimicrobial applications. Appl. Sci. 12, 141. doi:10.3390/app12010141
Danaei, M. R. M. M., Dehghankhold, M., Ataei, S., Hasanzadeh Davarani, F., Javanmard, R., Dokhani, A., et al. (2018). Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 10, 57. doi:10.3390/pharmaceutics10020057
Ealia, S. A. M., and Saravanakumar, M. P. (2017). A review on the classification, characterisation, synthesis of nanoparticles and their application. Mater. Sci. Eng. 263, 032019. doi:10.1088/1757-899X/263/3/032019
El-Saadony, M. T., Abd El-Hack, M. E., Taha, A. E., Fouda, M. M., Ajarem, J. S., N. Maodaa, S., et al. (2020). Ecofriendly synthesis and insecticidal application of copper nanoparticles against the storage pest Tribolium castaneum. Nanomaterials 10, 587. doi:10.3390/nano10030587
Gao, J., Lu, H., Yin, H., Zou, G., Chen, X., Jiang, T., et al. (2025). Enhanced antibacterial activity through photothermal therapy of copper nanoparticles on titanium-based implants. Materialia 43, 102514. doi:10.1016/j.mtla.2025.102514
Gopalakrishnan, V., and Muniraj, S. J. M. T. P. (2021). Neem flower extract assisted green synthesis of copper nanoparticles–optimisation, characterisation and anti-bacterial study. Mater. Today Proc. 36, 832–836. doi:10.1016/j.matpr.2020.07.013
Habibah, F. F., Rizki, W. O. S., Ivansyah, A. L., Astuti, D. I., and Hertadi, R. (2024). Green synthesis of copper ions nanoparticles functionalized with rhamnolipid as potential antibacterial agent for pathogenic bacteria. Heliyon 10, e24242. doi:10.1016/j.heliyon.2024.e24242
Ibrahim, A. A. (2025). Microwave-Assisted biogenic synthesis of pure copper oxide nanoparticles by Lepidium Sativum L. leaves extract: characterization and evaluation of antibacterial activity. J. Biotechnol. 406, 82–90. doi:10.1016/j.jbiotec.2025.07.003
John, M. S., Nagoth, J. A., Zannotti, M., Giovannetti, R., Mancini, A., Ramasamy, K. P., et al. (2021). Biogenic synthesis of copper nanoparticles using bacterial strains isolated from an antarctic consortium associated to a psychrophilic marine ciliate: characterization and potential application as antimicrobial agents. Mar. Drugs. 19, 263. doi:10.3390/md19050263
Kasthuri, K., Kumar, J. K., Rajkumar, P., Kalpana, S., and Balasubramani, V. (2024). Feasible antibacterial nanomaterials: copper oxide nanoparticles synthesized using dandelion leaf extract. Inorg. Chem. Commune. 167, 112804. doi:10.1016/j.inoche.2024.112804
Kaur, P., Thakur, R., and Chaudhury, A. (2016). Biogenesis of copper nanoparticles using peel extract of Punica granatum and their antimicrobial activity against opportunistic pathogens. GCLR 9, 33–38. doi:10.1080/17518253.2016.1141238
Khan, Y., Sadia, H., Ali Shah, S. Z., Khan, M. N., Shah, A. A., Ullah, N., et al. (2022). Classification, synthetic, and characterization approaches to nanoparticles, and their applications in various fields of nanotechnology: a review. Catalysts 12, 1386. doi:10.3390/catal12111386
Lv, Q., Zhang, B., Xing, X., Zhao, Y., Cai, R., Wang, W., et al. (2018). Biosynthesis of copper nanoparticles using Shewanella loihica PV-4 with antibacterial activity: novel approach and mechanisms investigation. J. Hazard. Mater. 347, 141–149. doi:10.1016/j.jhazmat.2017.12.070
Manyasree, D., Peddi, K. M., and Ravikumar, R. (2017). CuO nanoparticles: synthesis, characterization and their bactericidal efficacy. Int. J. Appl. Pharm. 9, 71–74. doi:10.22159/ijap.2017v9i6.71757
Merugu, R., Garimella, S., Velamakanni, R., Vuppugalla, P., Chitturi, K. L., and Jyothi, M. (2021). Synthesis, characterization and antimicrobial activity of bimetallic silver and copper nanoparticles using fruit pulp aqueous extracts of Moringa oleifera. Mater. Today Proc. 44, 153–156. doi:10.1016/j.matpr.2020.08.549
Mohamed, S. H., Othman, B. A., Abd-Elhalim, B. T., and Seada, M. N. A. (2024). Copper nanoparticles biosynthesis by Priestia megaterium and its application as antibacterial and antitumor agents. Sci. Rep. 14, 23615. doi:10.1038/s41598-024-72598-3
Murthy, H. A., Desalegn, T., Kassa, M., Abebe, B., and Assefa, T. (2020). Synthesis of green copper nanoparticles using medicinal plant Hagenia abyssinica (Brace) JF. Gmel. leaf extract: antimicrobial properties. J. Nanomater. 2020, 3924081–12. doi:10.1155/2020/3924081
Nagar, N., and Devra, V. (2018). Green synthesis and characterization of copper nanoparticles using Azadirachta indica leaves. Mater. Chem. Phys. 213, 44–51. doi:10.1016/j.matchemphys.2018.04.007
Nassar, A. R. A., Atta, H. M., Abdel-Rahman, M. A., El Naghy, W. S., and Fouda, A. (2023). Myco-synthesized copper oxide nanoparticles using harnessing metabolites of endophytic fungal strain Aspergillus terreus: an insight into antibacterial, anti-Candida, biocompatibility, anticancer, and antioxidant activities. BMC Complement. Med. Ther. 23, 261. doi:10.1186/s12906-023-04056-y
Noor, S., Shah, Z., Javed, A., Ali, A., Hussain, S. B., Zafar, S., et al. (2020). A fungal based synthesis method for copper nanoparticles with the determination of anticancer, antidiabetic and antibacterial activities. J. Microbiol. Methods 174, 105966. doi:10.1016/j.mimet.2020.105966
Noor, S., Hassan, M. A., Aslam, M. A., Muhammad, S. A., Quosain Naqvi, T., Batool Alvi, G., et al. (2025). Fungal biosystems for the fabrication of copper nanoparticles: an effective approach for the treatment of bacterial biofilms. ACS Appl. Mater. Interfaces. 17, 42706–42716. doi:10.1021/acsami.5c07764
Park, J., Kim, Y. M., Hong, S., Han, B., Nam, K. T., and Jung, Y. (2023). Closed-loop optimization of nanoparticle synthesis enabled by robotics and machine learning. Matter 6, 677–690. doi:10.1016/j.matt.2023.01.018
Phul, R., Kaur, C., Farooq, U., and Ahmad, T. (2018). Ascorbic acid assisted synthesis, characterization and catalytic application of copper nanoparticles. Mater. Sci. Eng. Int. J. 2, 90–94. doi:10.15406/mseij.2018.02.00040
Rajesh, K. M., Ajith, B., Reddy, Y. A. K., Suneetha, Y., and Reddy, P. S. (2016). Synthesis of copper nanoparticles and role of pH on particle size control. Mater. Today Proc. 3, 1985–1991. doi:10.1016/j.matpr.2016.04.100
Rambau, U., Masevhe, N. A., and Samie, A. (2024). Green synthesis of gold and copper nanoparticles by Lannea discolor: characterization and antibacterial activity. Inorganics 12, 36. doi:10.3390/inorganics12020036
Rani, S., Kumar, P., Dahiya, P., Dang, A. S., and Suneja, P. (2022). Biogenic synthesis of zinc nanoparticles, their applications, and toxicity prospects. Front. Microbiol. 13, 824427. doi:10.3389/fmicb.2022.824427
Rani, S., Kumar, P., Dahiya, P., Gupta, A., Arora, K., Dang, A. S., et al. (2024). Effect of biopriming and nanopriming on physio-biochemical characteristics of Cicer arietinum L. under drought stress. Plant Stress 12, 100466. doi:10.1016/j.stress.2024.100466
Rani, S., Kumar, P., Dahiya, P., Mehta, A., Dang, A. S., and Suneja, P. (2025). Optimization, characterization and antibacterial activity of copper nanoparticles biosynthesized using Pantoea agglomerans CPHN2. Indian J. Microbiol. 65, 1345–1356. doi:10.1007/s12088-025-01455-2
Rautela, G., Rizvi, R., and Ansari, S. (2025). Role of biosynthesized copper nanoparticles in agriculture and other fields. JNST, 990–994. doi:10.30799/jnst.351.25100102
Raza, A., Ahmad, S., Mateen, A., Arshad, A., Rehman, A., and Oliveira, H. A. (2022). Effect of copper nanoparticles on growth parameters of maize seedlings. Int. J. Nanotechnol. 19, 1143–1157. doi:10.1504/IJNT.2022.129766
Sacoto-Figueroa, F. K., Bello-Toledo, H. M., González-Rocha, G. E., Luengo Machuca, L., Lima, C. A., Meléndrez-Castro, M., et al. (2021). Molecular characterization and antibacterial activity of oral antibiotics and copper nanoparticles against endodontic pathogens commonly related to health care-associated infections. Clin. Oral Investig. 25, 6729–6741. doi:10.1007/s00784-021-03959-9
Salem, S. S., and Fouda, A. (2021). Green synthesis of metallic nanoparticles and their prospective biotechnological applications: an overview. Biol. Trace Elem. Res. 199, 344–370. doi:10.1007/s12011-020-02138-3
Santos, P. A., Biraku, X., Nielsen, E., Ozketen, A. C., Ozketen, A. A., and Hakki, E. E. (2025). Agricultural nanotechnology for a safe and sustainable future: current status, challenges, and beyond. J. Sci. Food Agric. 105, 3159–3169. doi:10.1002/jsfa.13922
Seyedsayamdost, M. R. (2019). Toward a global picture of bacterial secondary metabolism. J. Ind. Microbiol. Biotechnol. 46, 301–311. doi:10.1007/s10295-019-02136-y
Shantkriti, S., and Rani, P. (2014). Biological synthesis of copper nanoparticles using Pseudomonas fluorescens. Int. J. Curr. Microbiol. App. Sci. 3, 374–383.
Shende, S., Ingle, A. P., Gade, A., and Rai, M. (2015). Green synthesis of copper nanoparticles by Citrus medica Linn. (Idilimbu) juice and its antimicrobial activity. World J. Microbiol. Biotechnol. 31, 865–873. doi:10.1007/s11274-015-1840-3
Singh, D., Jain, D., Rajpurohit, D., Jat, G., Kushwaha, H. S., Singh, A., et al. (2023). Bacteria assisted green synthesis of copper oxide nanoparticles and their potential applications as antimicrobial agents and plant growth stimulants. Front. Chem. 11, 1154128. doi:10.3389/fchem.2023.1154128
Talebian, S., Shahnavaz, B., Nejabat, M., Abolhassani, Y., and Rassouli, F. B. (2023). Bacterial-mediated synthesis and characterization of copper oxide nanoparticles with antibacterial, antioxidant, and anticancer potentials. Front. Bioeng. Biotechnol. 11, 1140010. doi:10.3389/fbioe.2023.1140010
Tank, D., Bishnoi, A., Goswami, S., Ambegaonkar, N. J., Patel, P., Chahar, M., et al. (2025). Recent advances in phyto-and microorganisms-mediated synthesis of copper nanoparticles and their emerging applications in healthcare, environment, agriculture and food industry. Bioprocess Biosyst. Eng. 1–34. doi:10.1007/s00449-025-03196-4
Thakar, M. A., Jha, S. S., Phasinam, K., Manne, R., Qureshi, Y., and Babu, V. H. (2022). X ray diffraction (XRD) analysis and evaluation of antioxidant activity of copper oxide nanoparticles synthesized from leaf extract of Cissus vitiginea. Mater. Today Proc. 51, 319–324. doi:10.1016/j.matpr.2021.05.410
Tiwari, M., Narayanan, K., Thakar, M. B., Jagani, H. V., and Venkata Rao, J. (2014). Biosynthesis and wound healing activity of copper nanoparticles. IET Nanobiotechnol 8, 230–237. doi:10.1049/iet-nbt.2013.0052
Vigneshwaran, C., Suganya, C., Vasantharaj, K., Kirubakaran, D., and Sarathi, C. (2025). Biogenic synthesis of copper nanoparticles from Lantana camara flower extract: Antioxidant and anticancer activities. Biomed. Mater. Devices, 1–12. doi:10.1007/s44174-025-00371-9
Wu, Y. P., Liu, X. Y., Bai, J. R., Xie, H. C., Ye, S. L., Zhong, K., et al. (2019). Inhibitory effect of a natural phenolic compound, 3-p-trans-coumaroyl-2-hydroxyquinic acid against the attachment phase of biofilm formation of Staphylococcus aureus through targeting sortase A. RSC Adv. 9, 32453–32461. doi:10.1039/c9ra05883d
Keywords: nanoparticles, copper, microorganisms, green synthesis, OFAT, antimicrobial activity
Citation: Rani S, Dhankher S, Kumar P, Dahiya P, Arora K, Dang AS and Suneja P (2025) Biosynthesis, optimization, and characterization of CuNPs using Bacillus licheniformis CPJN13S and their antibacterial activity. Front. Nanotechnol. 7:1663115. doi: 10.3389/fnano.2025.1663115
Received: 10 July 2025; Accepted: 09 October 2025;
Published: 20 October 2025.
Edited by:
Marcelo Maraschin, Federal University of Santa Catarina, BrazilReviewed by:
Aparna Banerjee, Universidad Autonoma de Chile Sede Talca, ChileSalma Hesham, Ain Shams University, Egypt
Copyright © 2025 Rani, Dhankher, Kumar, Dahiya, Arora, Dang and Suneja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pooja Suneja, cG9vamFwYXZpdEBnbWFpbC5jb20=
 Simran Rani
Simran Rani Shikha Dhankher1
Shikha Dhankher1 Pradeep Kumar
Pradeep Kumar Priyanka Dahiya
Priyanka Dahiya Amita Suneja Dang
Amita Suneja Dang Pooja Suneja
Pooja Suneja