- 1Department of Plant Biotechnology, Institute of Agricultural Sciences, Banaras Hindu University, Varanasi, India
- 2Department of Veterinary Physiology and Biochemistry, Faculty of Veterinary and Animal Sciences, Institute of Agricultural Sciences, Banaras Hindu University, Varanasi, India
The rapid advancement and integration of engineered nanomaterials (ENMs) into consumer products, industrial processes, biomedical applications, and environmental technologies have revolutionized multiple sectors. However, their increased production and environmental release raise critical concerns about unintended interactions with microbial ecosystems. ENMs, including metal-based nanoparticles (silver, titanium dioxide, zinc oxide) and carbon nanomaterials (graphene, carbon nanotubes), possess unique physicochemical properties such as high surface area-to-volume ratios, tunable reactivity, and antimicrobial potential that allow them to interact directly with microbial cells or indirectly influence their habitats. This review critically examines the emerging evidence on ENM–microbiome interactions across human, aquatic, terrestrial, and agricultural systems. In human-associated microbiomes, especially the gut, ENMs can induce dysbiosis by disrupting microbial diversity, altering metabolite production (e.g., short-chain fatty acids), and impairing gut barrier integrity, contributing to inflammation and metabolic disorders. In environmental settings, ENMs influence key microbial functions like nitrogen fixation, organic matter decomposition, and biogeochemical cycling, potentially undermining ecosystem stability and agricultural productivity. Moreover, ENMs are increasingly implicated in accelerating antimicrobial resistance by promoting horizontal gene transfer and enriching resistance genes in microbial communities. The review highlights methodological advances such as high-throughput sequencing, meta-omics approaches, in vitro colon simulators, and in vivo models that have enhanced the assessment of ENM-induced microbiome alterations. Despite these advances, significant gaps remain in understanding long-term and low-dose effects, dose–response relationships, and ecological thresholds. Addressing these gaps through multidisciplinary research and regulatory frameworks is essential for ensuring the safe and sustainable deployment of nanotechnologies in a microbiome-sensitive world.
1 Introduction
The emergence and widespread integration of engineered nanomaterials (ENMs) into consumer products, industrial processes, environmental technologies, and biomedical applications has revolutionized multiple sectors of modern life (Bora et al., 2022; Rocco, 2025). These nanoscale materials, typically ranging from 1 to 100 nm in size, exhibit novel physicochemical properties such as high surface area-to-volume ratio, tunable reactivity, and enhanced mechanical or optical behavior (Kumar A. Y. N. et al., 2024). Common ENMs include metal-based nanoparticles (NPs), such as silver nanoparticles (AgNPs), titanium dioxide nanoparticles (TiO2 NPs), zinc oxide nanoparticles (ZnO NPs), carbon-based nanomaterials (CNMs) like graphene, carbon nanotubes (CNTs), and composite nanostructures (Fritea et al., 2021). While these innovations have brought significant technological and economic benefits, their increasing production and environmental release have raised growing concerns about potential biological and ecological risks, especially concerning their interactions with microbial communities (Rajpal et al., 2025). Microbial ecosystems spanning human-associated microbiomes to those found in aquatic, terrestrial, and agricultural environments play fundamental roles in maintaining physiological and ecological equilibrium (Ma L. C. et al., 2023). In the human body, particularly within the gastrointestinal tract, the gut microbiota contributes to digestion, immune regulation, synthesis of essential vitamins and short-chain fatty acids (SCFAs), and defense against pathogens (See et al., 2025). Similarly, in natural environments, microbial communities regulate biogeochemical cycles, decompose organic matter, support plant growth through symbiosis, and influence soil and water quality. Any disturbance in microbial diversity, abundance, or metabolic function, termed as dysbiosis, can have cascading effects on host health and ecosystem resilience (Prasad et al., 2024; Zaman et al., 2025).
ENMs are increasingly recognized as potential disruptors of microbial homeostasis. When released into the environment through waste streams, or ingested or inhaled by humans and animals, ENMs can interact directly with microbial cells or indirectly influence their surrounding microenvironments (Ray et al., 2009; Kumar et al., 2022). Their mechanisms of action include generation or induction of reactive oxygen species (ROS), disruption of membrane integrity, interference with DNA and protein function, and modulation of microbial metabolic and signaling pathways (Kumar R. et al., 2024). These effects can lead to reduced microbial diversity, shifts in community composition, alterations in metabolite production, and perturbation of key functional processes such as nutrient cycling, energy metabolism, and host–microbe communication (Wang et al., 2017). In human systems, such alterations have been linked to inflammatory bowel diseases, metabolic disorders, immune dysregulation, and neurobehavioral conditions (Xuan et al., 2023). In environmental settings, ENMs can affect microbial-driven processes like nitrogen fixation, denitrification, and organic matter decomposition, ultimately threatening soil fertility, water quality, and ecosystem services (Hochella et al., 2019; Huang et al., 2024).
Furthermore, ENMs may play a role in accelerating antimicrobial resistance (AMR) by enhancing horizontal gene transfer, altering microbial stress responses, and promoting the persistence of resistance genes in various environmental matrices (Wang X. et al., 2024; Piergiacomo et al., 2022). This poses a dual risk: while ENMs are often employed for their antimicrobial properties, their indiscriminate or chronic use could inadvertently select for resistant strains, complicating both clinical and ecological health management (Alav and Buckner, 2024).
Given the breadth of these interactions, the current review aims to provide a comprehensive overview of the current understanding of ENM–microbiome dynamics, the effects of various NMs on the human gut microbiota, aquatic and soil microbial ecosystems, and the potential for ENMs to influence microbial resistance development. It also evaluates the methodologies used to study these effects, including high-throughput sequencing, omics technologies, and in vitro/in vivo models, while highlighting regulatory and scientific challenges in assessing long-term risks. Finally, key knowledge gaps are identified and future directions are proposed for ensuring that the deployment of nanotechnologies is conducted safely and sustainably in a microbiome-sensitive world.
2 ENMs as anthropogenic sources of environmental and human microbiome perturbation
ENMs, especially AgNPs, ZnO NPs, TiO2 NPs, etc., are now ubiquitous across consumer, medical, agricultural, and industrial applications—ranging from antimicrobial coatings and textiles to water treatment, pesticides, cosmetics, and electronics (Figure 1). AgNPs are incorporated into food packaging materials for their antimicrobial properties, helping to extend shelf life and reduce spoilage (Martirosyan and Schneider, 2014). Titanium dioxide and silica (e.g., TiO2, SiO2 NPs) are common food additives or colourants (for example, TiO2 as food colourant E171) or anticaking agents in powdered products (Villamayor et al., 2023). ENMs are also used in nano-encapsulation of bioactive compounds, improving bioavailability of vitamins or nutraceuticals, and in smart packaging systems (with sensors, diffusion barriers, or controlled antimicrobial release) (Martirosyan and Schneider, 2014). ENMs are widely used in cosmetic formulations to improve efficacy, texture, appearance, and UV protection. Common nanomaterials include nano-TiO2 and nano-zinc oxide, which serve as UV filters in sunscreens because, at the nanoscale, they offer strong UV protection while remaining transparent on skin (Pastrana et al., 2018; Catalano et al., 2021). Other ENMs include nano-silica, used to improve spreadability, reduce greasiness, act as anti-caking agents in powders and lipsticks, and enhance pigment dispersion (Ferreira et al., 2023). ENMs are increasingly integrated into drugs and medical products due to their unique physicochemical properties, such as high surface area, tunable size, and surface functionality. In drug and vaccine delivery, ENMs like liposomes, polymeric nanoparticles, and metal-based nanoparticles enhance bioavailability, facilitate targeted drug delivery and sustained release to specific tissues, and reduce systemic side effects. Applications of ENMs in medical devices, diagnostic imaging, wound dressings, antimicrobial textiles/implants, etc., are also increasing exponentially (Cai et al., 2023; Kurul et al., 2025); ENMs are increasingly applied in agriculture to enhance nutrient efficiency, promote plant growth, and control pests. Nano-fertilizers using metal or oxide NPs (e.g., ZnO, Fe3O4) and carrier platforms (e.g., nanoclay, chitosan) help minimize nutrient leaching and increase uptake (N, P, K) in crops (Yin et al., 2018). Nanopesticides formulated with ENMs offer targeted delivery, lower chemical doses, and longer persistence against pathogens (Adisa et al., 2019). Beyond these applications, ENMs find widespread use in general industrial sectors such as coatings, electronics, energy, and construction. TiO2 NPs are used in self-cleaning and UV-resistant coatings, paints, and solar panel coatings to improve durability and reduce maintenance (Moloi et al., 2021; Hussein, 2023). Graphene, CNTs, and hybrid nanocomposites enhance electrical and thermal properties in devices, lightweight materials, and structural composites (Cataldi et al., 2020). Silver, zinc oxide, and silicon dioxide ENMs are used in textiles, antimicrobial surfaces, filtration systems, and sensors, due to their unique optical, catalytic or antimicrobial functionalities (Moloi et al., 2021; Singh et al., 2023).

Figure 1. Applications of engineered nanomaterials (ENMs), exposure pathways, and associated microbiome perturbations. The schematic tree illustrates the diverse applications of ENMs across major sectors including medicine and healthcare, water purification, food and packaging, agriculture, electronics and energy, industry, textiles, and coatings and paints. Exposure through ingestion, inhalation, dermal absorption, or soil/water contamination can disrupt human and environmental microbiomes, causing functional, compositional, and resistance-related shifts.
The ongoing and widespread use of ENMs brings them into frequent contact with environmental and human microbial communities, representing emerging anthropogenic sources of microbial disruption. ENMs in food (additives, packaging, nano-encapsulation), drugs or cosmetics are ingested or absorbed, while agricultural nano-fertilizers and nanopesticides enter soil and water, exposing soil and gut microbiota (Kah, 2015; Khan et al., 2021; Yadav et al., 2023). Improperly managed industrial nano-effluents serve as significant sources of toxicants to the environment and the food chain causing microbiome perturbations (Naz et al., 2024). These perturbations are dose-dependent, often manifest first in metabolic or enzymatic function and substrate utilization before major shifts in diversity or taxonomy (Avila-Arias et al., 2023). Given the environmental persistence and bioaccumulation potential of many ENMs, understanding realistic exposure levels (in water, soil, diet), their transformation (coating, aggregation, ion release), and their functional effects on microbial communities is critical for risk assessment and regulatory oversight.
3 ENMs and human microbiome
The human gut microbiota is a complex community of trillions of microorganisms, primarily bacteria that live in the gastrointestinal tract, especially the colon. These microbes play a crucial role in maintaining human health by aiding in digestion, synthesizing essential vitamins, regulating the immune system, and protecting against harmful pathogens. The composition of the gut microbiota is influenced by various factors, including diet, antibiotics, age, and lifestyle. A balanced microbiota supports overall health, while disruptions, known as dysbiosis, have been linked to numerous conditions such as inflammatory bowel disease, obesity, diabetes, and mental health disorders (Thursby and Juge, 2017; Hou et al., 2022). ENMs, such as Ag NPs, TiO2 NPs, ZnO NPs, and CNMs are increasingly used in food additives, packaging, pharmaceuticals, and cosmetics. When ingested, either directly through food or indirectly via environmental exposure, these NMs can interact with the gut microbiota and potentially disrupt gut health. Studies have shown that certain ENMs can alter the composition and diversity of the microbial community, leading to dysbiosis, inflammation, and impaired gut barrier function (Utembe et al., 2022). For example, Ag NPs and TiO2 NPs have been associated with reduced beneficial bacteria (like Lactobacillus and Bifidobacterium) and increased pro-inflammatory species (Utembe et al., 2022; Ma Y. et al., 2023). Additionally, ENMs can affect microbial metabolism and the production of SCFAs, which are vital for maintaining intestinal health (Figure 2). While the long-term impacts on human health are still being investigated, emerging evidence suggests that chronic exposure to ENMs may contribute to gastrointestinal disorders, immune dysregulation, and metabolic disturbances by disrupting the delicate balance of the gut ecosystem (Table 1) (Tang et al., 2021; Wojciechowska et al., 2023).

Figure 2. Effects of engineered nanomaterials (ENMs) on human gut microbiota and environmental microbial communities. This schematic illustrates the multifaceted impacts of ENMs, including metal-based nanoparticles and carbon nanomaterials, on microbial ecosystems. In the human gut, ENMs disrupt microbial diversity and composition, reducing beneficial taxa, altering short-chain fatty acid (SCFA) production, and compromising gut barrier integrity, which may lead to dysbiosis, inflammation, and metabolic disorders. In environmental systems (aquatic, soil, and agricultural), ENMs interfere with microbial-mediated biogeochemical processes such as nitrogen fixation, denitrification, and organic matter decomposition, potentially impairing nutrient cycling and ecosystem resilience.
3.1 Impact of titanium dioxide nanoparticles on gut microbiota and their mechanism of interaction
TiO2 NPs appear to exert a limited effect on gut microbiota diversity; however, they tend to influence the overall abundance of gut bacteria more noticeably (Table 1). Notably, TiO2 NPs have been found to affect specific bacterial groups, including Lactobacillus, as well as members of the Firmicutes and Proteobacteria phyla, while leaving overall microbial diversity at higher taxonomic levels relatively unaffected (Lin et al., 2014; Sohm et al., 2015; Burke et al., 2015; Zhang et al., 2022; Ma Y. et al., 2023). For instance, a total of 42 bacterial species were reported to undergo significant changes upon TiO2 NPs exposure, suggesting a risk of dysbiosis (Zhang et al., 2022). These compositional shifts are accompanied by disruptions in key metabolic pathways, particularly those associated with oxidative phosphorylation and energy metabolism, which are critical for maintaining gut health (Figure 2). In vitro studies also indicated that TiO2 NPs can inhibit the growth of beneficial bacteria like Lactobacillus and Bifidobacterium, through an exposure of diets containing 0.1% TiO2 NPs for 90 days to 1 mg/kg TiO2 NP for 7 days respectively, further implicating them in adverse gut effects (Wu et al., 2023). Such microbial disturbances have been associated with health risks, including colitis and obesity (Gangadoo et al., 2021). Although some evidence suggests that TiO2 NPs might enhance probiotic diversity under certain conditions, the prevailing data highlight their potential to negatively impact gut microbial communities (Kumar A. et al., 2024). Notably, these effects appear to be dose- and duration-dependent, underscoring the need for more comprehensive research to clarify the complex interactions between TiO2 NPs and the gut microbiome.
TiO2 NPs interact with gut microbiota primarily through mechanisms that induce dysbiosis linked to various health disorders, inflammation, immune modulation, and metabolic pathway disruption (Figure 2). Studies have shown that oral exposure to TiO2 NPs leads to significant shifts in microbial composition; for instance, decreasing beneficial bacteria like Veillonella while increasing genera such as Lactobacillus gasseri, Turicibacter, Lachnospiraceae NK4A136 group, etc., thereby impairing normal gut function and metabolism (Chen et al., 2019; Yan et al., 2020; Rinninella et al., 2021; Bianchi et al., 2024). These NPs also induce ROS within the gut, causing oxidative stress and inflammation that facilitate lipopolysaccharide (LPS) leakage from Gram-negative bacteria, further exacerbating gut inflammation-a process implicated in chronic metabolic diseases like obesity and diabetes. Additionally, TiO2 NPs may directly engage with intestinal immune cells, altering the microbiota–immune system axis and perpetuating inflammatory responses (Lamas et al., 2023). Metabolically, TiO2 NP exposure disrupts critical bacterial pathways including energy metabolism, detoxification, amino acid metabolism and SCFA production, which are vital for maintaining gut barrier integrity and regulating host energy homeostasis (Utembe et al., 2022; Wu et al., 2023; Pinget et al., 2019; Yang et al., 2022). Liquid chromatography-mass spectrometry (LC-MS/MS)-based untargeted metabolomics analysis demonstrated that TiO2 NPs disrupt critical metabolic pathways in gut bacteria, particularly tryptophan and arginine metabolism, which are essential for maintaining gut and host health. In vivo studies showed that mice fed a diet containing TiO2 NPs (0.1 wt% for 8 weeks) exhibited significant changes in urinary metabolites, with notable alterations in the tryptophan metabolism pathway. Furthermore, different neuroprotective metabolites such as tryptamine, N-Methyltryptamine, 6-Hydroxymelatonin, and N-Acetylserotonin, were significantly decreased in both bacterial cultures and the urine of treated mice (Wu et al., 2023). These interlinked mechanisms, i.e., microbial imbalance, oxidative-inflammation signaling, immune dysregulation, and metabolic impairment highlight how TiO2 NPs can detrimentally reshape the gut ecosystem and contribute to metabolic health disturbances.
The interactions between NPs and gastrointestinal microbiota are governed by multiple parameters, including the surface charge and physicochemical properties of both NPs and bacterial cells, the electrostatic interactions with digested food matrices, the chemical composition and bioactivity of dietary components, as well as dynamic physicochemical conditions within the gastrointestinal tract, such as pH gradients, enzymatic activity, ionic strength, and the presence of bile salts and other metabolites (Siemer et al., 2018; Baranowska-Wójcik, 2021). Analysis of the impact of engineered NPs on both commensal and pathogenic microorganisms in a gastrointestinal study showed that food-grade and model NPs readily form stable complexes with both probiotic and pathogenic bacteria under simulated gastrointestinal conditions. NP size emerged as the key determinant of NP–bacteria interactions, with smaller negatively charged NPs binding more efficiently to bacterial surfaces than larger positively charged ones, regardless of charge similarity. Additionally, low gastric pH enhanced complexation, while polymer coatings on NPs inhibited binding via steric repulsion, highlighting factors influencing NP–microbe interactions in the GI tract (Siemer et al., 2018). Toxicity analysis of five TiO2 NPs (10–50 nm) with different crystal phases using Escherichia coli revealed that antibacterial activity decreased with larger particle size and higher rutile content. Smaller anatase TiO2 NPs induced greater ROS production, membrane damage, and internalization, while higher pH and ionic strength reduced their antibacterial efficacy (Lin et al., 2014). Further, it has been reported that oral TiO2 NPs aggravated acute colitis by activating the NLRP3 inflammasome, leading to IL-1β and IL-18 release, ROS generation, and increased intestinal permeability. TiO2 crystals accumulated in the spleen of treated mice, and elevated titanium levels were detected in UC patients with active disease (Ruiz et al., 2017). These findings suggest potential harm of TiO2 NPs in individuals with compromised gut barrier and pre-existing inflammation, like IBD.
3.2 AgNPs and gut microbiota: exploring their impact and interaction mechanisms
The impacts of AgNPs on the human gut microbiota are complex and multifaceted, involving direct antimicrobial activity, disruption of microbial community structure, modulation of host metabolism, and impairment of gut barrier integrity (Table 1). AgNPs exert direct antimicrobial effects by interacting with bacterial cells, where they damage cell membranes, interfere with metabolic pathways, and inhibit DNA replication, ultimately leading to bacterial death (Bruna et al., 2021; Rodrigues et al., 2024). A key mechanism underlying these effects is the induction of oxidative stress within gut bacteria, characterized by increased nitric oxide production, lipid peroxidation, and DNA damage, all of which can destabilize the gut microbial ecosystem and promote dysbiosis (Li et al., 2019; Adeyemi et al., 2020; Manuja et al., 2021).
Such antimicrobial actions lead to significant alterations in the structural composition of the gut microbiota, with notable shifts in community diversity and abundance (Figure 2). AgNPs have been shown to selectively inhibit the growth of certain Gram-negative bacteria, reducing the prevalence of beneficial taxa like Bacteroides while favoring opportunistic and potentially pathogenic groups such as Enterococcus (More et al., 2023; Xie et al., 2023). These compositional changes result in a measurable loss of microbial diversity, particularly after short-term AgNP exposure (Bi et al., 2020; Wang X. et al., 2023). This loss of diversity has been directly associated with dysbiosis and adverse health outcomes, including gastrointestinal disorders (Ghebretatios et al., 2021). Interestingly, some studies suggest that microbial diversity may partially recover over extended exposure periods; however, metabolic perturbations often persist, indicating enduring effects on host physiology (Wang X. et al., 2023). Structural alterations are particularly prominent among microorganisms involved in energy, amino acid, and lipid metabolism (Wang, X. et al., 2022). Notably, in studies where mice received oral gavage of AgNPs once daily for 120 consecutive days, there were significant alterations in gut microbial functions associated with amino acid, purine, pyrimidine, lipid, and energy metabolism, as well as downstream effects on liver metabolic pathways (Wang, X. et al., 2022). Further, AgNPs significantly impact bacterial adhesion to the intestinal epithelium, a critical step in colonization and biofilm development. AgNPs exert a dual role in adhesion dynamics—primarily inhibiting bacterial attachment and biofilm formation, yet potentially triggering compensatory virulent behaviors under certain conditions. These adhesion-disrupting effects are crucial for their application in gut environments, where they may influence both pathogen colonization and commensal biofilm stability (Saeki et al., 2021; Afrasiabi and Partoazar, 2024; González-Fernández et al., 2025).
Moreover, AgNPs can compromise the integrity of the intestinal barrier, which serves as a critical defense against the translocation of bacteria and toxins from the gut lumen into the systemic circulation (Figure 2). Disruption of this barrier has been linked to increased gut permeability, low-grade inflammation, and the potential development of systemic health issues (Shayo et al., 2024; Li W. et al., 2024). While AgNPs hold promise in controlling pathogenic bacteria, their ability to perturb microbial balance and host metabolic processes raises significant concerns regarding their widespread use in consumer products and medical applications. The inherent resilience of the gut microbiome may offer some degree of recovery from AgNP-induced dysbiosis; however, the persistence of metabolic and barrier-related disruptions highlights the need for cautious evaluation of AgNP exposure and its long-term implications for gut and overall health.
The biological impact of AgNPs on gut microbiota is strongly shaped by particle size, shape, surface charge, concentration, surface coating, and the existing microbial community. Smaller AgNPs (<10 nm) exhibit heightened toxicity due to their larger surface-area-to-volume ratios, accelerated release of Ag+ ions, and greater ROS generation, which promotes oxidative stress in microbial cells (Zhang L. et al., 2018; Menichetti et al., 2023; Dinç, 2025; Sati et al., 2025). NP shape also affects efficacy; spherical, triangular, hexagonal, cubic or rod-shaped forms of AgNPs displayed variable antimicrobial activity. However, the anti-microbial potential based upon morphological attributes varied contrastingly according to the variable experimental conditions (Van Dong et al., 2012; Alshareef et al., 2017; Hanan et al., 2018; Vanlalveni et al., 2024). Therefore, a generalization may draw false narrative. Surface charge modulates interactions with bacteria: positively charged AgNPs strongly adhere to negatively charged bacterial membranes via electrostatic attraction, increasing membrane disruption and antimicrobial potency, whereas negatively charged or neutral coatings can reduce this effect (El Badawy et al., 2011; Alshareef et al., 2017). The concentration of AgNPs is critical—higher doses induce more pronounced disturbances in microbial diversity, metabolic function, and intestinal inflammation, while low or sub-threshold levels may have minimal or reversible effects; for instance, a dose-dependent alteration in gut microbiota α- and β-diversity has been reported in mice orally exposed for 28 days to food pellets supplemented with increasing doses of AgNPs (0, 46, 460, or 4,600 ppb). They also observed that higher doses shifted the phyla balance (Firmicutes vs. Bacteroidetes), but low doses had lesser or no overt toxicity (van Den Brule et al., 2015). Similarly, oral exposure to AgNPs (0, 1, 5, 10, 25, 50 mg/kg bw/day for 4 weeks) induced dose-dependent toxicity in mice, evidenced by body weight suppression and reduced intestinal dendritic cell numbers, confirming that both systemic and local immune effects scale with dose (Ren et al., 2023). However, contrasting results have also been reported documenting no changes in the Firmicutes/Bacteroidetes ratio in gut microbiota of rats and mice exposed to AgNPs at dose levels of 9 mg/kg bw/day and 10 mg/kg bw/day, respectively (Wilding et al., 2016; Hadrup et al., 2012). The duration of NP exposure may also have significant effects. In mice, administration of AgNPs and Ag nanowires (0.5 and 2.5 mg/kg) for 14 days reduced gut microbial diversity and altered community structure. After 28 days, partial recovery of the microbiota was observed, although metabolic disturbances, particularly in gut metabolites, persisted (Wang X. et al., 2023). Surface coatings, such as PEG, PVP, or chitosan, are routinely used to reduce toxicity by limiting dissolution and aggregation; PEG-coated AgNPs, for instance, exhibit reduced cytotoxicity and attenuated gut microbiota disruption (Caballero-Díazet al., 2013; Das et al., 2017; Menichetti et al., 2023; Vanlalveni et al., 2024). Finally, the baseline gut microbiota composition and host factors shape the response to AgNPs exposure—individual differences in microbial communities, and host-specific parameters determine susceptibility to dysbiosis and ecological shifts (Ren et al., 2023). Even, AgNPs exposure during critical developmental periods leads to enduring gut dysbiosis, neurobehavioral, and metabolic alterations as evidenced in mice (Lyu et al., 2021). Together, these factors underscore that the effects of AgNPs on gut health are not solely dependent on dosage but are the result of complex nanoparticle–microbiome interactions that must be addressed in designing safe, targeted applications.
3.3 ZnO NPs and gut microbiota dynamics
ZnO NPs exert multifaceted effects on the human gut microbiota, influencing microbial diversity, community composition, and metabolic activity (Table 1) (Figure 2). Research shows that these effects are highly dependent on factors such as concentration and the individual’s health status. For example, ZnO NPs [0 mg/kg (control), 25 mg/kg, 50 mg/kg, and 100 mg/kg] were fed to poultry birds for 9 weeks; at the highest concentration (100 mg/kg), ZnO NPs tended to reduce microbial richness and diversity, particularly impacting beneficial bacteria such as Lactobacillus (Feng et al., 2017). Exposure of the human intestinal microbiome to ZnO NPs at 0.1, 2.5, and 50 mg/L revealed clear dose-dependent effects. While low concentrations caused minimal changes, higher doses (especially 50 mg/L) significantly altered microbial composition, reduced diversity, and disrupted metabolic functions (Zhang et al., 2021). However, the response is not uniform across all populations. In children with Autism Spectrum Disorder, ZnO NPs actually increased gut bacterial diversity, whereas a decrease was observed in healthy children, suggesting that pre-existing health conditions may modulate the microbial response to ZnO NP exposure (Yu et al., 2021). Metabolically, ZnO NPs have been associated with a reduction in the production of SCFAs, which play essential roles in gut health and host metabolism (Zhang et al., 2021). Additionally, alterations in gut microbiota composition due to ZnO NPs have been linked to changes in plasma metabolites, indicating a complex and systemic metabolic impact (Feng et al., 2017). The effects of ZnO NPs also extend to microbial resistance traits. While medium concentrations were shown to reduce antibiotic resistance genes, low concentrations paradoxically enriched tetracycline resistance genes, revealing a nuanced influence on the gut resistome (Zhang et al., 2021). Interestingly, not all effects of ZnO NPs are negative. In certain contexts, such as in broiler chickens, ZnO NPs have been found to support the growth of beneficial bacteria, enhance gut health, and boost immune function (Qu et al., 2023). This duality highlights the complexity of ZnO NP interactions with the gut microbiome and underscores the need for further targeted studies to fully elucidate their health implications.
ZnO NPs influence gut microbiota through several mechanisms of action, with their effects varying depending on individual health status, such as in children with Attention-Deficit Hyperactivity Disorder or Autism Spectrum Disorder, as well as in various animal models. One of the primary mechanisms is their antibacterial activity. ZnO NPs have been shown to inhibit the growth of gut bacteria, particularly in healthy children, where they significantly reduced the number of live bacterial cells (Zhou et al., 2021; Yu et al., 2021). However, this response appears to differ in children with ASD, where ZnO NP exposure was associated with an increase in gut bacterial diversity, suggesting a more complex interaction with pre-existing microbial communities (Yu et al., 2021). Additionally, ZnO NPs are known to modulate microbial diversity by altering the overall community structure of gut bacteria. Higher concentrations, particularly 100 mg/kg, orally, for 9 days have been linked to a notable decline in beneficial bacteria such as Lactobacillus in poultry birds, with bacterial richness showing a negative correlation with ZnO NPs concentration indicating that excessive exposure may contribute to gut dysbiosis (Feng et al., 2017). Beyond their antimicrobial properties, ZnO NPs also possess anti-inflammatory effects. In conditions such as ulcerative colitis, they have been reported to help restore gut homeostasis by activating the Nrf2 signaling pathway and reducing levels of pro-inflammatory cytokines (Li et al., 2017). These findings suggest that while ZnO NPs can beneficially modulate gut microbiota and reduce inflammation under certain conditions, their overuse or inappropriate application may lead to harmful effects. Therefore, careful consideration of dosage and individual health status is essential when evaluating the therapeutic or dietary use of ZnO nanoparticles.
3.4 Carbon nanomaterials-gut microbiota interaction
CNMs, including single-walled carbon nanotubes (SWCNTs) and graphene oxide (GO), CNM-based nanozymes, quantum dots have a significant and multifaceted impact on the human gut microbiota, influencing both microbial composition and metabolic processes (Table 1) (Bantun et al., 2022; Zhao et al., 2024). These materials become integrated into the gut’s carbon flow, where they affect microbial fermentation and alter the production of key metabolites such as butyrate (Cui et al., 2023). This interaction has the potential to influence the proliferation and differentiation of intestinal stem cells, raising concerns about the possible health risks associated with CNM exposure. One major impact of CNMs is on the microbial composition within the gut. Studies in rodents show that single-walled CNTs and GO, when ingested, alter microbial composition shifting the Firmicutes/Bacteroidetes balance and increasing pro-inflammatory taxa like Alistipes and Lachnospiraceae (Chen et al., 2018; Utembe et al., 2022). CNM exposure can inhibit the growth of beneficial probiotic bacteria while promoting the growth of opportunistic pathogens, leading to dysbiosis (Figure 2) (Ma Y. et al., 2023; Wojciechowska et al., 2023; Chen et al., 2025). Interestingly, CNMs may also lead to an increase in butyrate-producing bacteria, as some of these microbes can utilize CNMs as a carbon source. This shift in population dynamics can have downstream effects on gut health and homeostasis (Cui et al., 2023). Beyond compositional changes, CNMs also influence gut microbial metabolism. Through fermentation, CNMs are converted into organic metabolites such as butyrate, which plays a critical role in gut physiology. However, the excessive production of butyrate due to CNM metabolism can disrupt normal intestinal processes by affecting the proliferation and differentiation of intestinal stem cells (Cui et al., 2023). These metabolic shifts could have long-term implications for intestinal function and overall health. Although the adverse effects of CNMs on gut microbiota are becoming increasingly evident, the broader implications for human health are still being explored. Responses to CNM exposure appear to vary among individuals, likely due to differences in baseline microbiota composition and genetic predispositions. Therefore, more comprehensive research is necessary to understand the full scope of health risks and to identify potential strategies for mitigating the negative effects of CNMs on the gut microbiome (Wojciechowska et al., 2023; Zickgraf et al., 2023; Chen et al., 2025).
The mechanisms through which CNMs affect the human gut microbiome are complex, involving both direct interactions with microbial populations and indirect influences via metabolic pathways. Recent research has shown that CNMs, such as single-walled carbon nanotubes and GO, can be utilized by gut microbiota as a novel carbon source. This fermentation process results in the production of beneficial metabolites such as butyrate, a SCFA, essential for gut health and the regulation of intestinal cellular functions (Cui et al., 2023). CNMs enhance microbial fermentation by selectively supporting the growth of specific butyrate-producing bacteria, thereby increasing butyrate levels in the gut. This has important implications for maintaining intestinal health and promoting epithelial integrity (Cui et al., 2023). However, the impact of CNMs on microbial metabolism is concentration-dependent. While moderate levels may support SCFA production, high concentrations of CNMs can suppress key metabolic pathways, disrupting SCFA synthesis and potentially leading to adverse physiological outcomes (Figure 2) (Cui et al., 2023; Wojciechowska et al., 2023). In addition to their metabolic effects, CNMs possess antimicrobial properties. They can compromise microbial cell integrity through oxidative stress, membrane disruption, and other toxic effects. The antimicrobial efficacy of CNMs including GO, CNTs, fullerenes, and carbon quantum dots is strongly governed by their physical and chemical attributes: size, shape, and surface functionalization. (Maksimova, 2019). Smaller GO sheets, for instance, exhibit higher oxidative stress–mediated killing when used as surface coatings, while larger sheets in suspension are more effective at entrapping and isolating bacteria, with smaller ones (∼0.01 µm2) showing a four-fold increase in antibacterial activity relative to larger sheets (∼0.65 µm2) in coatings (Perreault et al., 2015). The shape is equally decisive: sharp, high-aspect-ratio edges such as those in GO nanowalls or CNT tips mechanically disrupt bacterial membranes, significantly reducing survival rates of E. coli and S. aureus (Mohammed et al., 2020). Surface functionalization further modulates activity: oxygenated groups (–COOH, –OH), reduced forms (rGO), or conjugation with Cu2+ or Ag nanoparticles enhance dispersion, surface charge, ROS production, and targeted adhesion, with rGO–Cu hybrids achieving two orders of magnitude greater antibacterial effect compared to rGO alone (Tu et al., 2021). Thus, tuning CNM size, tailoring sharp morphologies, and engineering surface chemistries—especially via metal decoration or functional groups—synergistically optimize antimicrobial performance (Cobos et al., 2020; Khairol Anuar et al., 2021). Although CNMs hold promise for modulating gut microbial activity and enhancing certain health-related functions, their long-term impact on microbial diversity and gut health remains uncertain. Prolonged or high-level exposure may disrupt normal microbial communities and even promote the development of antibiotic resistance (Figure 2) (Xie et al., 2016). Therefore, while the therapeutic potential of CNMs is notable, careful evaluation of their safety and biological interactions is essential.
The interactions of ENMs with the human microbiome cannot be fully understood without considering their broader ecological footprint. ENMs released into soil, water, and air do not remain confined to the environment but can re-enter the food chain and drinking water, ultimately influencing human exposure. For instance, agricultural ENMs that alter soil or rhizosphere microbial communities may indirectly affect nutritional and microbial inputs to the human gut via crops (Mgadi et al., 2024; Dixit et al., 2024); similarly, ENM contamination of aquatic systems can influence water-borne microbiota that serve as reservoirs of antimicrobial resistance genes (Cui and Smith, 2022). Studies show that exposure to polluted or microbe-rich environments correlates with shifts in gut microbiome composition and diversity, suggesting a soil-plant-gut axis of microbial transmission and those environmental pollutants select for microbiome functionality in the gut (Ma et al., 2025; De Filippis et al., 2024). Thus, environmental and human microbiomes are interconnected through shared exposure pathways, bioaccumulation, and resistome exchange, highlighting the need to view ENM–microbiome interactions not as isolated domains but as part of a continuum linking ecosystem health and human health.
4 ENMs and environmental microbial communities
ENMs, such as nano-TiO2 and various metal NPs, have a profound impact on water-borne microbial communities, influencing key ecological functions like nutrient cycling and overall ecosystem health (Table 2). Chronic exposure to ENMs can disrupt the structure and function of microbial communities, leading to significant changes in biogeochemical processes essential for ecosystem sustainability. For instance, long-term exposure to nano-TiO2 has been found to reduce biofilm biomass and alter microbial community composition by decreasing the presence of taxa involved in nitrogen fixation and denitrification, the processes critical to nutrient cycling (Binh et al., 2016; Passarelli et al., 2020). Similarly, metal NPs such as silver and copper exhibit toxicity toward microbial populations, impairing enzymatic activities necessary for processes like nitrogen fixation and organic matter decomposition (Peyrot et al., 2014; Chhipa, 2021). ENMs also affect microbial activity by suppressing fundamental metabolic processes including respiration and photosynthesis, which are essential for maintaining ecosystem functions (Binh et al., 2016). Additionally, the presence of ENMs can lead to a shift in microbial community composition, favoring resistant taxa while reducing those crucial for nutrient cycling, thereby impairing key ecosystem services (Binh et al., 2016). These disruptions can have cascading effects on ecosystem health, such as reduced soil fertility and compromised ecosystem functionality, ultimately threatening agricultural productivity and environmental stability (Yadav and Yadav, 2024). Despite these concerns, some studies suggest that under certain conditions, ENMs may also enhance specific microbial functions or increase microbial resilience, highlighting a complex and context-dependent interaction between ENMs and microbial communities. As such, a comprehensive understanding of the dual roles of ENMs, both beneficial and detrimental is essential for evaluating their environmental impact and guiding their responsible use (Yadav and Yadav, 2024).
4.1 Impact of ENMs on aquatic ecosystems
ENMs, such as nano-TiO2 and various metal NPs, profoundly affect aquatic microbial communities, disrupting ecological processes like nutrient cycling and ecosystem stability (Table 2) (Figure 2). In freshwater systems, chronic exposure to nano-TiO2 can drastically reduce biofilm biomass and alter community composition, particularly diminishing nitrogen-fixing bacteria like Azotobacter and denitrifiers such as Pseudomonas, which play essential roles in nitrogen cycling (Binh et al., 2014). Short-term exposure further highlights species-specific effects: nano-TiO2 has been shown to depress Bacillus subtilis and Aeromonas hydrophila, while paradoxically enhancing growth of Arthrobacter and Klebsiella, likely by photo-oxidizing organic matter into more bioavailable forms (Binh et al., 2014). Simultaneously, metal NPs like copper oxide (CuO NPs) disrupt critical microbial function such as nitrate reduction by damaging cell membranes, down-regulating enzymes like NADH dehydrogenase, cytochromes, nitrate and nitrite reductases in model denitrifier Paracoccus denitrificans, ultimately reducing denitrification efficiency by roughly 36% (from ∼98% to ∼62%) at concentrations between 0.05–0.25 mg/L (Su et al., 2015).
ENMs also impair microbial metabolic pathways integral to ecosystem function. Nano-TiO2 and ZnO NPs suppress respiration and photosynthesis in primary producers; for instance, ZnO exposure lowers photosynthetic activity in cyanobacteria like Microcystis aeruginosa. Nano-TiO2 inhibits metabolic activity in algae and bacteria, reshaping taxa distribution by reducing sensitive algal species and allowing more resistant cyanobacteria to thrive, raising concerns around harmful algal blooms (Chen B. et al., 2022). These disruptions carry serious ecological consequences (Figure 2). In agricultural runoff zones, ENMs can reduce microbial diversity and soil nutrient availability, decreasing fertility and crop yield. In wetlands, altered biofilm and microbial enzyme activity impairs organic matter decomposition, affecting carbon sequestration and water purification. However, not all effects are negative: low concentrations of iron oxide NPs have been observed to stimulate growth and denitrification in P. denitrificans and other denitrifiers, illustrating the nuanced, context-dependent nature of ENM–microbe interactions. Therefore, ENMs exert complex dual roles in aquatic ecosystems: they can both disrupt and potentially enhance microbial community structure and function. A deeper, context-sensitive understanding of these interactions is essential to assess environmental risks and guide responsible applications of nanotechnology.
The influence of ENMs on bioaccumulation and ecotoxicological effects within keystone microbial species is profound and multifaceted. Silver and copper NPs, for instance, readily accumulate in microbial cells, disrupting physiological functions and reshaping community structures based on concentration and particle type (He et al., 2014; Von Moos and Slaveykova, 2014). In freshwater sediments, exposure to citrate- and PVP-coated AgNPs at 0, 25, 50, 75, 100, and 125 mg/L caused dose-dependent inhibition of microbial functional diversity. At the highest concentration (125 mg/L), citrate-AgNP significantly reduced microbial catabolic activity by up to 80% and diminished both substrate richness and diversity, impairing organic matter degradation and broader nutrient cycling (Kusi et al., 2020). This bioaccumulation at lower trophic levels also poses a risk of trophic transfer, potentially leading to biomagnification in higher organisms.
Ecotoxicologically, ENMs can both generate ROS at their surfaces (e.g., via catalytic activity or metal ion release) and induce ROS production within organisms (e.g., via mitochondrial dysfunction or activation of NADPH oxidases) (Mendoza and Brown, 2019; Ge et al., 2019). This oxidative stress disrupts essential enzymatic and metabolic functions such as nitrogen fixation, respiration, and organic matter decomposition, all critical for ecosystem sustainability (Von Moos and Slaveykova, 2014; Zhai et al., 2018; Gambardella and Pinsino, 2022). In microbial and algal models (e.g., Chlorella vulgaris), exposure to CNTs and metal-oxide NPs triggered elevated antioxidant enzyme activity (e.g., superoxide dismutase) alongside ROS-induced damage and reduced cell viability (Pereira et al., 2014). In environmental contexts, even low levels of AgNPs reduced metabolic diversity in organic-matter-associated microbial communities, which in turn indirectly stunted invertebrate growth in aquatic food webs (Zhai et al., 2018; Guo et al., 2019; Bao et al., 2016; Kusi et al., 2020). The toxicity of these materials is closely tied to their physicochemical characteristics such as size, shape, surface chemistry, ion dissolution rates, all of which modulate interactions with microbial cells (Triboulet et al., 2013; He et al., 2014; Pereira et al., 2014; Von Moos and Slaveykova, 2014; Kusi et al., 2020). For instance, the ecotoxicity of silver varies significantly between metallic AgNPs and their sulfide-transformed forms (Ag2S), with reduced bioavailability and toxicity in soils (Courtois et al., 2019). Copper NPs similarly trigger oxidative stress and metabolic shifts in bacteria, affecting glutathione levels and antioxidant enzyme pathways (Figure 2) (Rana and Kalaichelvan, 2013; Triboulet et al., 2013; Javurek et al., 2017).
Despite these adverse outcomes, there is emerging potential for ENMs in bioremediation applications, where targeted use of certain NPs can enhance microbial degradation of pollutants. This highlights a complex dual role: while ENMs may pose significant ecological hazards, they also offer novel opportunities for environmental management. However, addressing this duality requires nuanced and context-aware research to assess long-term impacts and guide responsible use of nanotechnologies.
4.2 Impact of ENMs on soil microbiome
ENMs significantly influence soil microbial diversity and enzymatic activities, thereby affecting soil health and agricultural productivity (Figure 2). Exposure to metal ENMs such as AgNPs, ZnO NPs and TiO2 NPs can significantly alter microbial metabolic diversity and key enzyme functions in soils (Table 2) (Rajput et al., 2023; Islam, 2025). In one study, metal ENMs like AgNPs and ZnO NPs altered community-level physiological profiles of soil bacteria and significantly decreased the diversity indices, such as Shannon’s diversity index, Evenness diversity index, and Simpson’s diversity index while TiO2 NPs had no detectable impact (Asadishad et al., 2018; Chavan and Nadanathangam, 2020; Yadav and Yadav, 2024). Further, soil amendments with AgNPs, ZnO NPs, and CuO NPs showed dose-dependent effects on enzymatic activity: ZnO and CuO either enhanced or had no effect at moderate doses (1–10 mg/kg), while AgNPs at higher concentrations (100 mg/kg) inhibited enzymes such as dehydrogenase, phosphatase, and urease (Tripathi et al., 2023). High levels of ZnO, TiO2, and CeO2 (around 1,000 mg/kg) also reduced populations of Azotobacter and other nutrient-solubilizing bacteria, along with suppressed enzyme activities. AgNPs are reported to suppress populations of essential nitrogen-fixing and nitrifying microbes, including Rhizobium and Nitrosomonas, which disrupts nitrogen cycling processes in soil ecosystems (Shah and Belozerova, 2009). Likewise, ZnO NPs have been found to impair microbial respiration and inhibit key enzymatic activities, such as dehydrogenase and urease, both of which are vital for organic matter breakdown and nutrient recycling (Parada et al., 2019). In another study, acute exposure to CuO NPs (10–500 mg kg-1) through nano-pesticide markedly inhibited soil denitrification, with the highest dose (500 mg kg-1) causing an 11-fold increase in nitrate accumulation and a 10.2%–24.1% reduction in N2O emissions. This suppression was linked to decreased activities of nitrate reductase and nitric oxide reductase, along with inhibited electron transport system activity, altered expression of denitrifying functional genes, and shifts in bacterial community composition (Zhao et al., 2020). Similarly, only 90-day exposure of agricultural soils to TiO2 NPs (1 and 500 mg kg-1) significantly inhibited nitrification enzyme activities and reduced the abundance of ammonia-oxidizing microorganisms, as indicated by decreased amoA gene copies. This suppression cascaded to reduce denitrification enzyme activity, with declines in nirK and nirS gene abundances, alongside marked shifts in bacterial community structure, even at the lowest realistic NP concentration (Simonin et al., 2016). These disruptions can impair soil nutrient cycling. Specifically, enzymes critical for organic matter decomposition and nutrient release such as urease, phosphatase, and dehydrogenase show decreased activity upon ENM exposure, threatening soil fertility (Figure 2) (Asadishad et al., 2018; Chavan and Nadanathangam, 2019). Moreover, ZnO and CuO NPs may reprogram microbial metabolic pathways by upregulating nitrogen fixation genes (nifH, amoA) while downregulating denitrification genes (norB, nosZ), leading to imbalances in nitrogen cycling (Luche et al., 2016; Sun et al., 2022; Tripathi et al., 2023). Despite these negative impacts, low to moderate ENM levels could stimulate microbial resilience. Biogenic NMs like nanozeolite and nanochitosan, combined with plant probiotics, were found to enhance dehydrogenase, alkaline phosphatase, and fluorescein diacetate hydrolase activities—doubling or tripling enzyme performance under agricultural conditions (Upadhayay et al., 2023), a promising application in sustainable agriculture.
ENMs profoundly affect plant–microbe interactions, especially those involving mycorrhizal fungi and nitrogen-fixing bacteria, with outcomes that vary depending on material type, concentration, and microbial partners (Table 2). Arbuscular mycorrhizal fungi, essential for enhancing plant nutrient uptake, can be adversely impacted by ENMs, which inhibit fungal growth and disrupt nutrient exchange between plants and soil (Yu M. et al., 2020; Vera-Reyes et al., 2023). Additionally, nitrogen-fixing bacteria such as Bradyrhizobium diazoefficiens are sensitive to ENM exposure: cerium oxide nanoparticles (CeO2 NPs) and multi-walled carbon nanotubes (MWCNTs) can suppress their growth, reduce nodulation competitiveness, and interfere with plant–bacteria signaling critical for effective symbiosis (Mortimer et al., 2020). ENMs further reshape the soil microbiome, altering processes such as mineralization and nitrogen fixation (Figure 2). Their antimicrobial properties can decrease beneficial microbial populations, further disrupting plant–microbe symbioses. This disruption undermines soil functions that are key to plant health. However, context matters: in certain situations, NMs have been shown to enhance plant growth and mitigate abiotic stress by supporting beneficial microbial interactions and functioning as “smart fertilizers” (Ma et al., 2022; Berrios et al., 2023; Sodhi et al., 2025). At low concentrations (∼5 mg/kg Ag and 50 mg/kg Zn and Ti), ENMs in biosolids did not adversely affect Medicago truncatula growth or metal accumulation in shoots. Instead, they enhanced symbiotic interactions with rhizobia (Sinorhizobium meliloti), as reflected by a higher nodule number, and significantly increased total soil microbial biomass. Furthermore, ENM exposure at low concentrations altered microbial community composition, increasing Gram-negative and anaerobic bacteria while reducing eukaryotic abundance. These findings suggest potential stimulatory effects of transformed ENMs on soil microbial activity and plant–rhizobia symbiosis (Chen et al., 2017). Exposure to ZnO NPs as nanofertilizer at 10 mg/kg and 100 mg/kg significantly reshaped the rhizospheric bacterial community structure, particularly altering the abundance of Cyanobacteria and key plant growth-promoting taxa, while alpha diversity remained stable. These compositional shifts were more pronounced at the higher dose (100 mg/kg), suggesting dose-dependent effects of ZnO NPs on soil microbial ecology. While 10 mg/kg ZnO NPs also coincided with enhanced lettuce biomass and photosynthetic rate, no additional plant growth benefits were observed at 100 mg/kg, despite the intensified microbial community changes (Xu et al., 2018). Therefore, the effects of ENMs on soil microbial communities are complex. High concentrations of metal-based NPs tend to inhibit diversity and enzyme function compromising nutrient cycling and soil fertility, while the use of certain nanobiofertilizers at controlled doses shows potential for improving microbial function and crop productivity (Zhang et al., 2024). Therefore, ENMs offer potential agricultural benefits such as improved nutrient delivery and stress resistance, while their adverse effects on crucial microbial partnerships raise concerns about their long-term ecological impact. This underscores the need for targeted research to balance ENM applications benefits against ecological risks.
5 Cross-talk between ENMs and microbial resistance
ENMs are increasingly implicated in exacerbating antimicrobial resistance among microbial communities by facilitating the persistence and dissemination of antimicrobial resistance genes (ARGs). Notably, metal oxide NPs such as ZnO NPs have been shown to promote horizontal gene transfer among bacteria. In laboratory and soil studies, ZnO NPs increased transformation frequency in E. coli by approximately 1.8-fold and enhanced the copy number of metal resistance–linked genes, indicating co-selection of resistance mechanisms (Markowicz et al., 2023; Otinov et al., 2020; Wang X. et al., 2024; Alav and Buckner, 2024). Furthermore, AgNPs and CuNPs stimulate the conjugative transfer of ARG-laden plasmids across bacterial genera; for instance, silver ions and AgNPs have been shown to facilitate plasmid-mediated resistance gene transfer, and CuNPs promoted multi-antibiotic resistance gene spread in environmental bacteria (Yu K. et al., 2020). The mechanisms underlying these effects include enhanced horizontal gene transfer among bacterial communities, particularly fostering the spread of ARGs through oxidative stress, increased membrane permeability, induction of the bacterial SOS response, and upregulation of conjugation-related genes (Figure 2) (Yu K. et al., 2020). Sub-lethal exposure to ENMs like nano-alumina (Al2O3) has been shown to induce ROS, which damage cell membranes and create pores that facilitate plasmid uptake; this also triggers the SOS DNA damage response that further promotes plasmid transformation and conjugation (e.g., pBR322 into E. coli and S. aureus)—a striking increase in horizontal gene transfer efficiency linked directly to NP presence (Ding et al., 2016). Similarly, nanofullerene (nC60) exposure increases ROS generation and membrane disruption, upregulating genes crucial for DNA transfer and conjugative machinery (e.g., trbBp, korA/B) in exposed bacteria (Ji et al., 2020; Amaro et al., 2021). ENMs also act as environmental ARG reservoirs. For example, certain NPs can adsorb plasmids or ARG-containing DNA, protecting them from degradation and increasing their environmental persistence. CeO2 NPs, in particular, have been investigated both for their facilitation and inhibition of antimicrobial resistance genes propagation; while some studies identified increased conjugation, others found that CeO2 can suppress antimicrobial resistance genes transfer by reducing ROS and downregulating horizontal gene transferring genes (Yu K. et al., 2020; Sharma et al., 2025). Additionally, ENMs impose selective pressure on microbial communities: resistant strains, better able to cope with metal-induced stress, proliferate while susceptible strains decline leading to enrichment of antimicrobial resistance traits (Neethu et al., 2022).
Comprehensive reviews reinforce the notion that ENMs in environments from various sources such as wastewater treatment plants exert selective pressure on microbial communities and enhance horizontal gene transfer by increasing membrane permeability through both direct interaction and ROS-mediated damage, concurrently inducing genetic changes linked to conjugation (Figure 2) (Cui and Smith, 2022; Li et al., 2025). Additionally, ENMs contribute to these effects by adsorbing extracellular ARGs and plasmids, protecting them from degradation and facilitating persistence and uptake in microbial populations (Ding et al., 2016; Cui and Smith, 2022; Xu et al., 2023; Kalli et al., 2023; Kaushik et al., 2023; Mosaka et al., 2023; Li et al., 2025). These processes lead to accelerated spread of resistance genes in microbial communities, with ENMs acting as vectors and catalysts for antimicrobial resistant gene propagation in wastewater and natural environments, a trend confirmed by increased antimicrobial resistant gene and mobile genetic element abundance following NP exposure (Ding et al., 2016; Cui and Smith, 2022; Li et al., 2025). However, research also suggests a potential dual role: emerging NP designs with antioxidant or electrochemical properties may attenuate ARG transmission, indicating a path toward mitigating antimicrobial resistance spread via ENMs. The ENMs may also hold beneficial potential in antimicrobial resistance mitigation by their use in targeted drug-delivery systems for antibiotics and/or having synergistic effects reducing the overall required dosage, lowering selective pressure and slowing resistance development (Ribeiro et al., 2022; AlQurashi et al., 2025; Sharma et al., 2025). For instance, encapsulation of enrofloxacin in PLGA and lignin NPs mitigated its disruptive effects on the gut microbiome compared to free enrofloxacin, with lignin-encapsulated Enro showing minimal impact on microbial diversity. Notably, NP delivery delayed the rise in ARG expression between 24 and 72 h, suggesting a potential to reduce antibiotic-induced resistome shifts in the gut (Herrera et al., 2024). Nano-ZnO exposures have been reported to cause dose-dependent alterations in gut microbiota composition and diversity, suppressed SCFA production, and shifted key microbial functional pathways. While medium doses (2.5 mg/L) reduced several ARGs by inhibiting host bacteria, low doses (0.1 mg/L) unexpectedly enriched tetracycline resistance genes, highlighting potential risks to gut health and resistome stability (Zhang et al., 2021). Another study reported that during cultivation of leachate microbiota, ARG diversity and abundance dropped significantly (1.4–3.2 log), with NPs—especially metal oxides (CuO, ZnO)—enhancing this attenuation in an ARG-specific manner. The attenuation was driven by metal-induced bacterial growth inhibition, dissolved ion stress, and internalized NPs inducing oxidative stress (ROS), which together reduced horizontal ARG transfer and damaged resistance genes (Su et al., 2019). Altogether, ENMs may promote antimicrobial resistance—yet, with mindful design, they may also serve as tools to counteract microbial resistance dissemination in environmental as well as in vivo settings.
Another crucial aspect is ENMs in combination with existing pollutants, such as heavy metals, can synergistically drive microbial resistance, posing a complex environmental hazard (Balta et al., 2025). ENMs like TiO2 NPs and ZnO NPs interact with heavy metals to alter their bioavailability, which may enhance metal uptake by microbes and foster resistance via genetic mutations or horizontal gene transfer. This synergy not only increases toxicity to microbial populations but also pressures communities to develop resistance mechanisms such as efflux pumps and biofilm formation (Dickinson et al., 2019; Wu et al., 2021). The combined presence of ENMs and pollutants alters microbial community structures, often favoring resistant strains over susceptible ones and intensifying resistance traits through continuous stress. Moreover, co-contamination with ENMs and organic pollutants can result in synergistic toxicity effects that are typically underestimated in conventional environmental risk assessments (Tufail et al., 2022; Zhu et al., 2024; Olawade et al., 2024). This increases the overall health risks posed by resistant pathogens by creating niches that sustain and spread antimicrobial resistance (Balta et al., 2025). Despite these challenges, the interaction between ENMs and pollutants also presents opportunities for novel antimicrobial strategies. By understanding these synergistic effects, researchers hope to develop targeted nanoparticle-based interventions that can disrupt resistant microbial communities or enhance contaminant removal, offering a balanced approach to combating antimicrobial resistance.
6 Tools and techniques for assessing ENM-Microbiome interactions
Assessing the impact of ENMs on microbiome interactions requires a comprehensive, multi-tiered approach that integrates high-throughput sequencing (16S rRNA, metagenomics, metatranscriptomics), functional assays (enzyme activity, SCFA profiling, metaproteomics, metabolomics), resistome analysis (qPCR, metagenomics), microscopy techniques (TEM, confocal), and in vitro/in vivo models (organoids and organ-on-a-chip technologies, gnotobiotic mice, “Humanized” murine models) to evaluate compositional, functional, and genetic changes. Advanced tools like meta-analyses and machine learning further enhance the interpretation of complex omics data and support a holistic understanding of ENM–microbiome dynamics (Galloway-Peña and Hanson, 2020; Moreno-Indias et al., 2021; Mortimer et al., 2021). A global meta-analysis involving over 2,100 observations demonstrated clear negative effects of ENMs on soil microbial diversity, biomass, and functional enzyme activity, with machine learning models effectively predicting key determinants of impact, highlighting the power of these computational tools for ecological risk assessment. Beyond statistical modeling, mass spectrometry-based multi-omics approaches, including genomics, proteomics, lipidomics, and metabolomics provide essential systems-level insights into how ENMs alter microbial physiology and cellular pathways, revealing widespread adaptations and stress responses at multiple biological layers (Day et al., 2023). Community structure analysis further shows that ENMs, especially metal NPs, can profoundly disrupt microbial populations, diminishing critical functions like nutrient cycling and pollutant degradation by suppressing enzymatic activities.
Despite the robustness of these methodological frameworks, significant variability across studies underscores the need for standardized protocols. Differences in ENM types, doses, exposure durations, microbial communities, and omics platforms often limit comparability and reproducibility. Addressing this heterogeneity through harmonized experimental designs and cross-laboratory validation will be crucial to ensure consistent and reliable assessments of ENM-induced ecological risks across diverse environments.
6.1 Marker gene sequencing for microbiome analysis
Marker gene sequencing is a widely used and cost-effective approach in microbiome analysis that targets conserved genetic regions (e.g., 16S rRNA for bacteria and archaea, ITS for fungi, 18S rRNA for eukaryotes) to profile microbial community composition and diversity (Figure 3). This technique enables identification and relative quantification of taxa within complex microbial ecosystems by amplifying and sequencing these phylogenetic markers. It provides insights into taxonomic structure but has limited resolution for functional and strain-level analysis compared to whole metagenome sequencing (De la Cuesta-Zuluaga and Escobar, 2016; Douglas et al., 2018; Johnson et al., 2019; Galloway-Peña and Hanson, 2020; Bharti and Grimm, 2021).

Figure 3. General workflow for marker gene sequencing in microbiome analysis. The schematic outlines the traditional metagenomics workflow using amplicon sequencing of marker genes (16S rRNA for bacteria/archaea, 18S rRNA for eukaryotes, and ITS for fungi). Key steps include DNA extraction, amplification of target regions, high-throughput sequencing, and bioinformatics analysis for taxonomic profiling and diversity assessment.
6.1.1 16S ribosomal RNA (rRNA) gene sequencing
16S rRNA gene sequencing is a cornerstone method for profiling bacterial and archaeal communities by targeting conserved and hypervariable regions (e.g., V1–V2, V3–V4, V4–V5) of the ∼1,500 bp 16S gene. Primer choice such as V3–V4 for broad bacterial coverage or V1–V2 for higher species resolution is critical to capture the desired taxonomic groups accurately (Na et al., 2023; Combrink et al., 2023; Lee et al., 2023). Post-sequencing, raw reads undergo quality filtering, chimera removal, and demultiplexing using pipelines like QIIME 2 or Mothur (Galloway-Peña and Hanson, 2020; Combrink et al., 2023; Lewis et al., 2021). Feature tables can be generated as Operational Taxonomic Units (OTUs) by clustering at ∼97% similarity which is effective for legacy comparisons and mitigating sequencing noise, or as Amplicon Sequence Variants (ASVs) via denoising tools like DADA2 for single-nucleotide resolution, improved reproducibility, and finer ecological insights (Galloway-Peña and Hanson, 2020; Lewis et al., 2021; Jeske and Gallert, 2022; Chiarello et al., 2022; Daly et al., 2024). Finally, taxonomic assignment is performed against curated databases (e.g., SILVA, Greengenes) and downstream analyses include diversity metrics and phylogenetic comparisons to reveal community structure and dynamics (Figure 3) (Johnson et al., 2019; Lewis et al., 2021; Combrink et al., 2023).
Using 16S rRNA gene sequencing targeting the V3–V4 region, a study revealed that long-term dietary exposure to Ag, SiO2, and TiO2 NPs significantly altered the gut microbiota composition and β-diversity in mice. ASV analysis using DADA2 method showed a dose-dependent increase in Cyanobacteria and a marked reduction in Tenericutes and Turicibacter with TiO2 NPs, while SiO2 and TiO2 also suppressed SCFA production. These shifts in microbial composition and metabolic function were largely reversible after an 8-week recovery period without NP exposure, indicating transient but notable perturbations of the gut microbiome by dietary NMs (Perez et al., 2021). Another research employed 16S rRNA gene sequencing (targeting V1–V3 regions with 27f/534r primers) to investigate the influence of plant-derived nanoparticles on the gut microbiome of germ-free mice colonized with human fecal bacteria. Sequencing on the 454 Jr. platform and QIIME 2-based analysis revealed significant shifts in microbiota composition upon NP exposure, with notable enrichment of Lachnospiraceae, Bacteroidaceae, Coriobacteriaceae, and Ruminococcaceae families. OTUs were clustered at 97% similarity, and hierarchical clustering highlighted differences between in vitro and in vivo bacterial uptake of plant-derived nanoparticles, suggesting these NPs selectively modulate gut microbial communities and may alter functional pathways (Teng et al., 2025). Further, 16S rRNA gene sequencing revealed that oral exposure to SiO2NPs in young mice significantly altered gut microbiota composition and diversity, with increased abundances of Firmicutes and Patescibacteria and distinct shifts in β-diversity profiles. OTU-based analysis (97% similarity) and LEfSe identified 41 bacterial clades with differential abundance, suggesting SiO2NP-induced microbiome dysbiosis, which was associated with neurobehavioral impairments via disruption of the gut–brain axis (Diao et al., 2021). Oral exposure to lead-based (CsPbBr3) perovskite nanoparticles (CPB-PNPs) induced significant, dose-dependent alterations in gut microbiota composition as revealed by 16S rRNA gene sequencing. ASV-based analysis showed reduced alpha diversity indices (Chao1, Shannon, Simpson) and a marked shift in β-diversity, as evidenced by principal coordinate analysis (PCoA). High-dose CPB-PNPs increased pro-inflammatory taxa such as Clostridia and decreased beneficial Muribaculaceae, disrupting the Firmicutes/Bacteroidetes ratio. Differential abundance analysis further identified enrichment of microbial taxa linked to intestinal inflammation. These microbiota perturbations were associated with compromised gut barrier integrity and colitis-like phenotypes in exposed mice (Mei et al., 2023).
6.1.2 18S rRNA and internal transcribed spacer (ITS) sequencing
18S rRNA and ITS amplicon sequencing are pivotal techniques for profiling fungal and other eukaryotic communities in microbiome studies (Figure 3). The 18S rRNA gene, with its conserved and hypervariable regions (V1–V9), enables broad phylogenetic placement across diverse eukaryotes, though it typically resolves taxa only down to genus level. In contrast, the ITS regions (ITS1 and ITS2), located between 18S, 5.8S, and 28S genes, exhibit high variability and are therefore standard markers for species- and strain-level identification of fungi (Banos et al., 2018; Gao et al., 2021; Olivier et al., 2023). These amplicon data are processed through pipelines like QIIME 2, LotuS2, or ITSx, which include quality filtering, chimera removal, OTU/ASV clustering, and taxonomic assignment using curated databases such as SILVA for 18S and UNITE for ITS (Gao et al., 2021; Özkurt et al., 2022). This dual-marker approach offers comprehensive insights into eukaryotic microbiome structure and diversity. Using long-read sequencing of the nearly complete rRNA operon (16S-ITS-23S), a clear enhancement in species-level resolution of Lactobacillaceae has been reported compared to shorter amplicons. RibDif2 analysis revealed substantial allele overlap in V3–V4 (n = 43 overlaps) and whole 16S (∼11), while complete 16S-ITS-23S showed zero predicted overlaps. Empirical MinION™ data confirmed these predictions: full-length operon sequencing identified 100% of target species with fewer misclassifications, outperforming both V3–V4 (80% correct) and single 16S (∼95%), thus highlighting the combined power of 18S rRNA/ITS region (rRNA operon) for accurate microbiome profiling (Olivier et al., 2023).
Most of the available reports focused on 16S rRNA gene sequencing to track bacterial community changes, and responses of eukaryotic microbes (fungi, protozoa) via 18S/ITS remains largely unexplored in NP exposure contexts. Using 16S and 18S rRNA gene sequencing with PNA clamps and ITS qPCR, a study demonstrated that nanoscale sulfur (pristine and stearic acid-coated) modulated both bacterial and fungal communities in the tomato rhizosphere. While bacterial ASV diversity increased under nano-sulfur treatments, eukaryotic communities, particularly fungi and ciliates, showed resilience with minimal diversity shifts. ITS qPCR revealed no significant changes in total fungal abundance, but differential analysis indicated reduced relative abundance of Fusarium oxysporum in nano-sulfur treatments compared to controls. Enrichment of sulfur-oxidizing bacteria (Thiobacillus) and subtle shifts in fungal taxa suggest nano-sulfur may suppress soil-borne pathogens indirectly by altering microbiome composition and functional interactions (Steven et al., 2024).
6.2 Whole-genome shotgun (WGS) metagenomics
Whole-Genome Shotgun (WGS) metagenomics is an untargeted sequencing method that captures the entire genetic content of all microorganisms in a sample that include bacteria, archaea, fungi, viruses, and eukaryotes, providing comprehensive taxonomic and functional insights. Compared to 16S rRNA amplicon sequencing, WGS delivers higher species- and strain-level resolution, detects greater microbial diversity, and identifies functional genes such as antibiotic resistance and metabolic pathways (Ranjan et al., 2016; Jovel et al., 2016; Keepers et al., 2019; Brumfield et al., 2020). However, it is more expensive, requires deeper sequencing coverage, and demands advanced computational infrastructure due to large data volumes and complexity (Jovel et al., 2016; Fox et al., 2024). WGS metagenomics offers a powerful, holistic view of microbial communities and their functional potential, making it indispensable for studies that extend beyond taxonomic profiling into functional and ecological questions.
WGS metagenomics begins with sample collection and total DNA extraction, followed by random fragmentation and high-throughput sequencing (e.g., Illumina, PacBio, Nanopore). After quality control and contaminant removal (using tools like FastQC and Trimmomatic), reads are assembled de novo into contigs and binned into putative genomes (MAGs) using assemblers (e.g., MEGAHIT, metaSPAdes) and binning tools (e.g., MetaBAT, CONCOCT). These assemblies and unassembled reads are then annotated taxonomically (with tools like Kraken2, GTDB-Tk for MAGs, or Kraken2, MetaPhlAn for reads) and functionally profiled against databases such as KEGG, COG, eggNOG, and CARD to reveal metabolic pathways and resistance genes (Pérez-Cobas et al., 2020; Bharti and Grimm, 2021; Saenz et al., 2022). The final step involves comparing taxonomic and functional profiles across samples to assess microbial diversity, community structure, and functional potential (Figure 4).
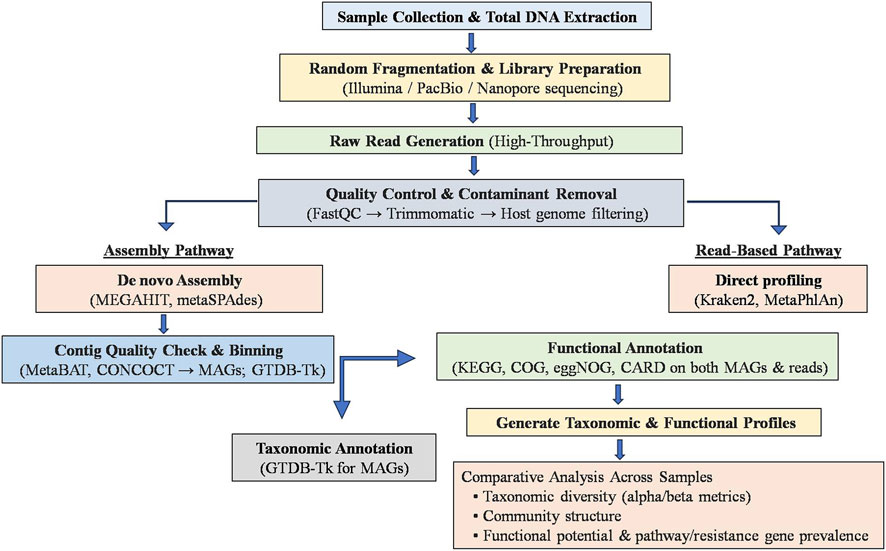
Figure 4. General workflow for whole genome shotgun (WGS) metagenomics in microbiome analysis. The schematic illustrates the WGS metagenomics workflow starting from sample collection and total DNA extraction, followed by random fragmentation, library preparation, and high-throughput sequencing (e.g., Illumina, PacBio, Nanopore). Quality control and contaminant removal (using tools such as FastQC and Trimmomatic) are performed before downstream analysis. Two pathways are depicted: the assembly-based pathway involves de novo assembly (MEGAHIT, metaSPAdes), binning (MetaBAT, CONCOCT), and taxonomic annotation (GTDB-Tk); the read-based pathway uses direct profiling tools (Kraken2, MetaPhlAn). Both approaches undergo functional annotation (KEGG, COG, eggNOG, CARD) to generate taxonomic and functional profiles, supporting comparative analyses of microbial diversity, community structure, and functional potentials across samples.
WGS metagenomics and traditional metagenomics (e.g., amplicon sequencing of marker genes like 16S, 18S, or ITS) both aim to profile microbial communities, but they differ fundamentally in scope and resolution. WGS metagenomics sequences all DNA fragments randomly, enabling species- and strain-level identification, comprehensive detection of bacteria, archaea, fungi, viruses, and functional gene content including antibiotic resistance and metabolic pathways. In contrast, marker gene-based metagenomics targets specific loci (like 16S, 18S, ITS) to characterize community composition more cost-effectively and with simpler bioinformatics, though typically limited to genus-level resolution and offering limited insight into functional potential. While WGS metagenomics demands deeper sequencing, greater computational resources, and more complex analysis pipelines, it reveals richer diversity including rare taxa and accurate functional profiling, making it ideal for comprehensive ecological or clinical microbiome studies (Ranjan et al., 2016; Rausch et al., 2019; Brumfield et al., 2020; Pérez-Cobas et al., 2020; Wang Z. et al., 2023).
Whole-genome shotgun metagenomics revealed that ZnO NPs significantly altered soil microbial taxonomic and functional diversity, reducing gene abundance related to carbon degradation and nitrogen cycling while increasing genes for CO2 fixation and sulfur metabolism. ZnO NPs also disrupted microbial network complexity by decreasing connectivity and modularity among taxa, with archaeal, fungal, and viral communities showing more pronounced responses than bacteria (Sun et al., 2025). Additionally, WGS metagenomic profiling of nano-ZnO–polluted soils revealed a dose-dependent enrichment of biofilm-related genes, particularly those involved in exopolysaccharide biosynthesis and cell attachment, with stronger effects seen for nanoparticles than bulk ZnO at moderate concentrations (50–500 mg/kg) (Dinesh et al., 2023b). Further, multidrug resistance genes and mobile genetic elements (MGEs) co-occurred with biofilm genes, indicating enhanced potential for horizontal gene transfer under nano-ZnO exposure (Dinesh et al., 2023a). At higher ZnO levels (≥500 mg/kg), nano-ZnO suppressed microbial biomass, respiration, and enzyme activity, reflecting microbial stress and community destabilization (Dinesh et al., 2023b). These findings underscore that nano-ZnO can reshape soil microbiome function and resistance dynamics via gene-level shifts in stress response and community interaction mechanisms. WGS metagenomic analysis of wastewater microbial communities exposed to gold NPs revealed that NP morphology (nanospheres vs. nanorods) strongly influenced both taxonomic composition and functional gene profiles. CTAB-coated nanospheres caused significant shifts in microbial structure and enriched antibiotic resistance genes, metal resistance genes, and plasmid-associated genes, whereas nanorods had subtler effects. These results highlight how NP design parameters can modulate microbiome diversity and functional potential in engineered ecosystems (Metch et al., 2018).
Therefore, both WGS metagenomic analysis and traditional marker-based metagenomics using next-generation sequencing provide culture-independent approaches to comprehensively characterize the human gut microbiome and environmental microbial communities. These methods enable the detection of dysbiosis, detailed taxonomic profiling, and the discovery of novel functional genes and metabolic pathways, including those associated with health, disease, and the resistome. Despite these advances, challenges remain. High-throughput sequencing (HTS) data quality can be compromised by sequencing errors, sampling bias, and variable 16S copy number across taxa—factors that skew abundance and diversity estimates (Kembel et al., 2012; Di Bella et al., 2013; Khachatryan et al., 2020; Beaudry et al., 2021; Fox et al., 2024). Moreover, the rapid growth of sequencing data demands continuous development of scalable, reproducible, and integrative bioinformatics frameworks to keep pace with analytical needs. Continued innovation in HTS technologies and computational tools will be critical to unlocking the full potential of microbiome science in both environmental and biomedical contexts.
6.3 Metaproteomics for microbiome analysis
Metaproteomics is a powerful, culture-independent approach for microbiome analysis that investigates the entire protein complement expressed by complex microbial communities in their native environment. Starting from biomass-enriched samples (e.g., feces, soil), proteins are extracted, digested into peptides (commonly via phenol extraction and FASP protocols), and then analyzed using LC–MS/MS (Tanca et al., 2014; Zhang et al., 2016; Heyer et al., 2019; Do et al., 2024). This workflow enables identification of active microbial species and their expressed enzymes, revealing metabolic pathways and biomarkers linked to community function such as over 600 microbial species and 250 protein families in gut samples (Tanca et al., 2014; Heyer et al., 2019; 2025). Bioinformatic tools like MetaProteomeAnalyzer, MaxQuant, and Unipept facilitate peptide-spectrum matching, taxonomic and functional annotation, and robust protein quantification (Starr et al., 2018; Kruk et al., 2024; Do et al., 2024; Nebauer et al., 2024). When combined with metagenomics or metatranscriptomics, metaproteomics offers a holistic view of microbial activity, host–microbe interactions, and ecosystem dynamics, advancing both research and diagnostic applications (Heyer et al., 2025). Metaproteomics, which analyzes the expressed proteins of microbial communities, provides a direct snapshot of active biological processes; improvements in mass spectrometry and bioinformatics have enhanced the ability to profile functional shifts within microbiomes (Jagtap et al., 2015; Young et al., 2015; Zhang X. et al., 2018; Do et al., 2024; Valdés-Mas et al., 2025).
While significant progress has been made in understanding NM–microbiome interactions using approaches such as 16S/18S rRNA amplicon sequencing for taxonomic profiling, WGS metagenomics for functional gene prediction, and culture-based assays for targeted microbial analyses, these methods offer only partial insights into microbial function. Notably, metaproteomics, a powerful tool capable of directly assessing protein-level responses, microbial activity, stress adaptations, and metabolic pathway alterations in complex communities, remains largely absent in this field. This represents a critical research gap, as integrating metaproteomics could provide a more comprehensive understanding of how NMs influence microbiome functionality beyond genetic potential, capturing real-time microbial responses to NP exposure. By integrating metagenomic sequencing with fecal proteomics, Valdés-Mas et al. (2025) applied metagenome-informed metaproteomics (MIM) to both mouse models and human IBD patients, enabling species-level resolution of host–microbiome–diet interactions. In IBD cases, they uncovered a dual signature of “compositional dysbiosis” i.e., shifts in microbial taxa and “functional dysbiosis” i.e., reduced protein activity from beneficial commensals in response to inflammatory signals. MIM also accurately reconstructed dietary exposure profiles and in vivo nutritional compliance using dietary-specific protein markers. Utilizing microbiome transfer experiments, the study revealed early-onset, species-specific microbiome and host proteomic responses, and identified candidate fecal host-microbiome protein biomarkers that outperformed traditional calprotectin (S100A8/S100A9) in predicting IBD (Valdés-Mas et al., 2025). These findings demonstrate that a combined dietary–microbial–host proteomic analysis can functionally dissect trans-kingdom interactions and offers advancements toward personalized diagnostics and therapeutics in gut-related diseases.
6.4 Metabolomics in microbiome analysis
Metabolomics in microbiome analysis employs techniques like LC–MS, GC–MS, and NMR to profile small molecules (metabolites) produced or transformed by microbial communities, offering a direct snapshot of microbial activity, biochemical interactions, and signaling. By illuminating changes in metabolic pathways such as SCFA production, amino acid metabolism, energy metabolism or xenobiotic transformations, metabolomics complements genomic and transcriptomic data to reveal functional and ecological impacts of microbiota under different conditions. Metabolomics begins with strategic experimental design followed by sample collection and rapid quenching to preserve metabolic states, then extraction of metabolites using solvents (e.g., methanol, acetone, chloroform) optimized for polarity range. Extracted metabolites are analyzed via LC–MS, GC–MS, or NMR, allowing separation and detection of wide-ranging compounds with sensitivity and structural resolution. Raw data undergo preprocessing including noise reduction, peak detection, retention time alignment, and normalization using tools like XCMS, MZmine, and MS-DIAL. Metabolite identification is achieved through spectral database matching (e.g., HMDB, METLIN), followed by statistical analysis (PCA, PLS-DA) and pathway enrichment (KEGG, MetaboAnalyst) to interpret biological significance (Kumar et al., 2020; Han et al., 2021; Chen Y. et al., 2022; Muhamadali et al., 2023).
Untargeted metabolomics revealed that TiO2 NPs inhibited growth of key beneficial bacteria (L. reuteri, L. gasseri, B. animalis, B. longum) through membrane damage and disrupted metabolic pathways, including tryptophan and arginine metabolism in vitro. In vivo, mice fed with TiO2 NPs showed altered urinary metabolite profiles, notably reduced neuroprotective metabolites and perturbed tryptophan metabolism, all indicative of gut microbiome–mediated host metabolic dysregulation (Wu et al., 2023). Studies integrating microbiome profiling with metabolomic analysis have shown that ZnO NPs (25–100 mg/kg in poultry) significantly disrupt gut microbial community structure and metabolism. Metabolomic shifts included altered levels of glucose, choline, lactate, methionine, indole derivatives, and pro-inflammatory arachidonic acid metabolites correlating with declines in beneficial taxa like Lactobacillus and Bifidobacterium, and increased E. coli and other opportunists. These multi-omics approaches reveal that NM exposure can rewire host–microbe metabolic interactions, with implications for inflammatory and metabolic health (Feng et al., 2017). In an in vitro digestion and fermentation model, ZnO, TiO2, and Ag NPs induced significant shifts in gut microbial genera, particularly Bifidobacterium, Sutterella, Escherichia, and Bacteroides as determined by 16S rRNA sequencing. Metabolomic profiling revealed NP-specific modulation of metabolites, including indole derivatives, peptides, and protein metabolism intermediates, with TiO2 notably increasing pro-inflammatory arachidonic acid pathway metabolites like prostaglandin E2 and leukotriene B4. These findings suggest that metallic NPs exposure can disrupt both microbial composition and metabolic by-products linked to gut inflammation and disease pathways (Vaccari et al., 2023). Oral exposure to Ag NPs and silver nanowires (Ag NWs) at 0.5–2.5 mg/kg in mice significantly disrupted gut microbiota structure reducing diversity and suppressing Gram-negative bacteria within 14 days, while the community largely recovered by 28 days. Despite this recovery, fecal and systemic metabolomics revealed persistent increases in gut-derived 1H-indole-3-carboxylic acid and elevated gut and blood serotonin levels, linking NPs exposure to microbial metabolic shifts and neurochemical alterations (Wang X. et al., 2023). Therefore, for microbiome studies, metabolomics insights alone or preferably integrated with metagenomics, metatranscriptomics, or metaproteomics data can be useful to uncover mechanistic links between microbial composition, metabolic activity, and environmental or host interactions.
Together, these meta-omics technologies transcend traditional culture-based methods, enabling comprehensive analysis of microbial species, functional capacity, and metabolic activity in relation to health and disease. However, despite their power, challenges remain in data integration and interpretation. The sheer complexity of meta-omics datasets necessitates robust bioinformatics frameworks and harmonized analytical standards to fully leverage these approaches in microbiome-targeted therapies and precision medicine (Wang et al., 2015; Puig-Castellvi et al., 2023; Do et al., 2024).
6.5 Use of in vitro models and in vivo animal studies
In Vitro Colon Simulators (e.g., SHIME®, TIM-2, CoMiniGut, SIMGI) are dynamic, multi-stage fermentation systems that faithfully reproduce human colonic conditions and microbial ecosystems. For ENM–microbiome interaction studies, they offer unparalleled ability to simulate realistic dosing, track microbiota and metabolite alterations, and integrate with host-response models making them an essential tool in assessing NMs safety and biological impact in the gut. Using a dynamic, multi-stage in vitro colon simulator inoculated with human fecal microbiota, Zhang et al. (2021) demonstrated that exposure to nano-ZnO caused dose-dependent declines in SCFA production and significant perturbations in microbial composition and diversity. Following cessation of exposure, microbial diversity largely rebounded; however, SCFA levels remained suppressed in relation to ZnO concentration. Moreover, the study revealed contrasting effects on the gut resistome: a medium concentration (2.5 mg/L) reduced the abundance of many antibiotic-resistance genes, whereas a low concentration (0.1 mg/L) notably enriched tetracycline resistance genes (Zhang et al., 2021).
Intestinal organoids and gut-on-chip systems as in vitro models and in vivo animal models together provide a complementary and powerful framework for evaluating how ENMs interact with and impact the microbiome, with valuable insights for human health risk assessment. Human intestinal organoids, derived from stem cells and often cultured within biomimetic hydrogels or perfusable microfluidic platforms, offer improved physiological relevance over 2D cultures for investigating NM–microbiome interactions in the gut. These 3D structures faithfully recapitulate crypt–villus architecture and cellular diversity including enterocytes, goblet cells, Paneth cells, and M cells—enabling physiologically relevant assessments of nanotoxicity and uptake dynamics (Prasad et al., 2021; Bantun et al., 2022; Yuan and Liu, 2023; Shi et al., 2024). When co-cultured with microbiota, they reveal critical insights into cross-kingdom metabolism: for instance, gut microbes have been shown to enzymatically degrade CNMs (e.g., GO, SWCNTs), fermenting them into butyrate, which then modulates epithelial stem cell proliferation and barrier integrity (Cui et al., 2023). Moreover, exposure to nanoplastics or engineered inorganic nanoparticles triggers inflammatory signaling in organoids, particularly through M cell-mediated pathways and disrupts commensal composition by altering the Firmicutes/Bacteroidetes ratio, with downstream effects on mucus secretion and mucosal immunity (Hao et al., 2022; Qiao et al., 2024). A study aimed to elucidate how nano-sized food additives interact with commensal and pathogenic bacteria within the gastrointestinal tract utilized human gastric organoids to demonstrate that silica NP–H. pylori complexes retained bacterial adherence but significantly attenuated bacterial internalization and pathogenic signaling, including CagA phosphorylation and IL-8 secretion. These findings underscore the potential of ENMs to modulate host–microbiome interactions within the gastric niche (Siemer et al., 2018).
Gut-on-chip platforms are microfluidic devices that reconstitute key physiological and structural aspects of the human intestine such as epithelial villi, mucus layers, and peristaltic flow, while enabling co-culture with microbial communities under controlled, real-time conditions (Trujillo-de Santiago et al., 2018; Xiang et al., 2020; Thomas et al., 2023). These systems allow precise dosing of ENMs and measurement of outcomes like barrier integrity, cytokine production, microbial colonization, and metabolite exchange. Gut-on-chip models are faster and free from ethical issues than animal studies, making them ideal for high-throughput screening of nanoparticles and their acute effects on gut–microbe interactions.
In vivo animal models, particularly zebrafish and rodents, offer critical physiological and systemic context for assessing long-term effects of ENM exposure capturing microbiome perturbations, immune modulation, metabolic shifts, and organ-level responses. Zebrafish are especially valuable as high-throughput vertebrate models due to their conserved intestinal physiology, genetic tractability, and amenability to germ-free and fluorescent-transgenic approaches, enabling real-time visualization of host–microbe–ENM interactions (Xia et al., 2022). Rodent studies, while resource-intensive and ethically demanding, complement these findings by providing detailed insights into ENM-induced shifts in microbial communities, metabolite profiles, inflammatory markers, and histopathology across tissues. However, these in vivo approaches require careful design to balance throughput, ethical considerations, and translational relevance (e.g., dosing regimen, exposure duration), underscoring the need for integrated strategies that leverage both zebrafish and murine models (Ashammakhi et al., 2020; Xia et al., 2022; Zhong et al., 2022; Liu et al., 2024).
However, both models have limitations. Gut-on-chip systems lack full immune and multi-organ integration and are best suited for mechanistic, short-term investigations. Animal studies provide whole-body responses but are less amenable to mechanistic dissection and suffer from inter-species variability. Thus, a hybrid strategy using gut-on-chip platforms for mechanistic hypothesis testing followed by targeted validation in animal models offers a robust pathway to translate findings to human-relevant insights. By standardizing ENM dosing, dynamic microenvironment conditions, co-culture complexity, and integrated omics readouts, the synergy between these approaches will enable safer ENM development and improved prediction of their microbiome-related effects.
7 Risk assessment and long-term implications
Assessing the risks and long-term implications of ENMs on microbiome interactions requires a robust, integrative approach combining cutting-edge tools, environmental and regulatory expertise. A global meta-analysis of over 2,100 observations has shown that ENMs negatively impact soil microbial diversity, biomass, and functional enzyme activities, with artificial intelligence models like random forests effectively predicting these outcomes based on ENM characteristics (Pietroiusti et al., 2016). To further elucidate these effects, multi-omics technologies such as metagenomics, metaproteomics, and metabolomics are increasingly being recommended by regulatory bodies, including the European Food Safety Authority (EFSA), to standardize microbiome assessments and identify biomarkers of disturbance under controlled exposure scenarios. For instance, mass spectrometry-based omics workflows enable comprehensive profiling of microbial functional responses to ENMs, revealing shifts in pathways linked to nutrient cycling and immune signaling (Mortimer et al., 2021).
Environmental and gut microbiome changes following ENM exposure evaluated through both acute (e.g., 90-day rodent studies) and long-term trials indicate persistent effects on gut barrier integrity, immune modulation, and SCFA production (Pietroiusti et al., 2016; Javurek et al., 2017; Tang et al., 2021; Utembe et al., 2022). Developmental exposure to AgNPs in mice resulted in persistent gut dysbiosis, characterized by reduced levels of beneficial taxa (Bifidobacterium, Mucispirillum) and enrichment of inflammatory-associated bacteria (Prevotella, Enterococcus). These microbiome alterations correlated with disruptions in metabolic pathways, reduced microglial counts in the brain, and behavioral and metabolic changes in offspring, highlighting the long-term impacts of early-life NMs exposure on the gut–microbiome–brain axis (Lyu et al., 2021). Moreover, emerging evidence indicates that CNMs can be fermented by gut bacteria into bioactive metabolites like butyrate, which then modulate host physiology, such as intestinal stem cell differentiation highlighting not only toxicological but also transformative microbial–host interactions (Cui et al., 2023). Animal models are essential for capturing chronic, systemic effects, but high-throughput in vitro systems such as gut-on-chip platforms are increasingly integrated into tiered risk frameworks to enhance hazard identification without excessive animal use. These tiered strategies align with emerging regulatory recommendations that prioritize microbiome endpoints and make use of NAMs (new approach methodologies), standardized sampling, and shared microbiome databases to improve cross-study comparability and regulatory confidence (OP EUROPA, 2027).
While the impacts of ENMs on microbiome include dysbiosis, impaired function of key microbial taxa, altered host-microbe interactions, chronic gut inflammation, immune dysfunction, metabolic and neurological disorders, their roles as environmental disruptors remain under-evaluated (Tang et al., 2021; Lamas et al., 2020; Li P. et al., 2024). EFSA’s roadmap emphasizes establishing core microbiome definitions, bioindicator panels, harmonized analytical protocols, and cross-lab networks to resolve this gap (Debode et al., 2024). Overall, advancing ENM risk assessment necessitates a combined strategy linking global meta-analyses, multi-omics, sophisticated modeling, and tiered experimental designs supported by regulatory-driven standardization and data sharing. This holistic approach promises more reliable prediction, monitoring, and mitigation of ENM effects on microbial ecosystems and public health.
8 Regulatory and mitigation strategies
Regulation of ENMs currently leverages existing legislative frameworks rather than relying on bespoke NM laws. In the United States, the Toxic Substances Control Act (TSCA) empowers the Environmental Protection Agency (EPA) to require pre-manufacture notices and implement information-gathering mandates for new nanoforms, while the Food and Drug Administration (FDA), the Occupational Safety and Health Administration (OSHA), and Consumer Product Safety Commission regulate ENMs in food, drugs, cosmetics, and workplaces under broader statutes and hazard communication standards (Lin, 2001; Tang et al., 2024; El-Kalliny et al., 2023; Ghosh and Kumar, 2024). The European Union (EU) employs Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH) legislation, Classification, Labelling and Packaging (CLP) Regulation, and the Biocidal Products Regulation to enforce registration, hazard evaluation, and labeling of materials at or above one ton annually, and maintains transparency via national NM registries (e.g., France, Denmark, Norway, Belgium) and the EU Observatory for Nanomaterials (Environment EC, 2024b; Environment EC, 2024a; ECHA, 2025; Pavlicek et al., 2021). Standardization efforts led by international organizations like Organisation for Economic Co-operation and Development (OECD), International Organization for Standardization (ISO) Technical Committee (TC) 229 (ISO/TC 229), and World Health Organization (WHO) further support harmonized definitions, testing protocols, and “precautionary approach” workplace guidelines. Despite this multilayered oversight, challenges include fragmented global definitions, evolving physicochemical properties, detection limitations, and often a disconnect between innovation pace and regulation leading experts to call for adaptive, coordinated, and evidence-driven governance that leverages “Safe-by-Design” approaches and precautionary risk frameworks (Gottardo et al., 2021; Kraegeloh et al., 2018; Resnik, 2019; Salieri et al., 2021).
Mitigation strategies for ENM-associated risks follow a dual pathway: reducing hazard through material design and minimizing exposure across the life cycle. “Safe-by-Design” approaches actively tailor physicochemical properties such as size, shape, surface charge, encapsulation to improve stability and biocompatibility while limiting harmful byproducts (Kraegeloh et al., 2018). These principles are implemented in EU projects (e.g., NANoREG, NanoReg2, SUN, caLIBRAte) that integrate risk pre-assessment, modeling (QSAR, AOP pathways), iterative stakeholder engagement, and green synthesis to align early-stage innovation with safety and regulatory preparedness (Kraegeloh et al., 2018; Isigonis et al., 2019; Schmutz et al., 2020). On the operational front, occupational controls adhere to the hierarchy of hazard mitigation: elimination/substitution, engineering controls (e.g., HEPA-filtered hoods, gloveboxes, sealed balances), administrative best practices, and personal protective equipment as per WHO and OSHA guidance (OSHA, 2025; McLean et al., 2024). Environmental surveillance and risk assessment rely on life-cycle analyses, environmental monitoring, and computational evaluation to forecast fate, transport, and ecological impacts, while AI-augmented characterization and risk modeling promise faster assessments and reduced animal testing. Together, these approaches couple proactive material design with rigorous exposure controls to manage ENM risks to environmental and human microbiomes.
9 Future directions and research gaps
Future research on the impact of ENMs on microbiome interactions points toward several critical directions. Firstly, the field must advance standardized life-cycle exposure models that track ENMs from synthesis through environmental release, transformation, and biological uptake—an area having significant research gap (e.g., bioaccumulation of transformation products) (Pietroiusti et al., 2016). Secondly, integrating in vitro and in vivo systems with multi-omics technologies (genomics, proteomics, metabolomics) and predictive machine learning that offers the promise of mechanistic insight and reliable ecological risk forecasting. For instance, recent meta-analyses and machine learning applications have effectively predicted ENM impacts on soil microbial communities and identified functional gene disruptions. Additionally, a One-Health research framework that encompasses soil, plant, animal, and human microbiomes is urgently needed to explore systemic ENM effects across ecological and host-associated networks. Furthermore, the identification of microbiome-based biomarkers such as shifts in SCFA profiles, immune-metabolite signatures, or specific community compositions could serve as early warnings of ENM-induced dysbiosis. Finally, establishing standardized protocols and NM research modules, shared data repositories, and inter-laboratory collaborations will be essential to ensure reproducibility, regulatory trust, and comprehensive understanding of long-term ENM–microbiome interactions.
10 Conclusion
ENMs are transforming numerous industries due to their unique physicochemical properties, yet their interactions with microbial ecosystems raise significant ecological and health concerns. This review highlights the profound effects of ENMs on human-associated microbiota and environmental microbial communities, revealing their capacity to alter microbial diversity, metabolic activity, and functional roles in nutrient cycling and host health. Metal-based NPs, such as Ag, TiO2, ZnO, and CNMs, can induce dysbiosis, impair enzymatic activities critical for biogeochemical processes, and potentially accelerate the spread of antimicrobial resistance genes. While advanced methodologies like high-throughput sequencing, meta-omics approaches, and in vitro/in vivo models have deepened our understanding of ENM–microbiome interactions, challenges persist in standardizing protocols and assessing long-term risks. Given the dual nature of ENMs, offering both technological advantages and potential ecological hazards, it is imperative to adopt a precautionary approach toward their deployment. Future research should focus on elucidating dose- and context-dependent effects, exploring safer-by-design NMs, and developing regulatory frameworks that account for microbiome sensitivity. Only through such multidisciplinary efforts we can ensure the sustainable use of nanotechnologies while preserving microbial diversity and ecosystem resilience.
Author contributions
AC: Writing – original draft, Writing – review and editing. MG: Conceptualization, Writing – original draft, Writing – review and editing.
Funding
The authors declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adeyemi, O. S., Shittu, E. O., Akpor, O. B., Rotimi, D., and Batiha, G. E. S. (2020). Silver nanoparticles restrict microbial growth by promoting oxidative stress and DNA damage. EXCLI Journal 19, 492–500. doi:10.17179/EXCLI2020-1244
Adisa, I. O., Pullagurala, V. L. R., Peralta-Videa, J. R., Dimkpa, C. O., Elmer, W. H., Gardea-Torresdey, J. L., et al. (2019). Recent advances in nano-enabled fertilizers and pesticides: a critical review of mechanisms of action. Environ. Sci. Nano 6 (7), 2002–2030. doi:10.1039/C9EN00265K
Afrasiabi, S., and Partoazar, A. (2024). Targeting bacterial biofilm-related genes with nanoparticle-based strategies. Front. Microbiol. 15, 1387114. doi:10.3389/fmicb.2024.1387114
Alav, I., and Buckner, M. M. C. (2024). Non-antibiotic compounds associated with humans and the environment can promote horizontal transfer of antimicrobial resistance genes. Crit. Reviews Microbiology 50 (6), 993–1010. doi:10.1080/1040841X.2023.2233603
AlQurashi, D. M., AlQurashi, T. F., Alam, R. I., Shaikh, S., and Tarkistani, M. A. M. (2025). Advanced nanoparticles in combating antibiotic resistance: current innovations and future directions. J. Nanotheranostics 6 (2), 9. doi:10.3390/jnt6020009
Alshareef, A., Laird, K., and Cross, R. B. M. (2017). Shape-dependent antibacterial activity of silver nanoparticles on Escherichia coli and Enterococcus faecium bacterium. Appl. Surf. Sci. 424, 310–315. doi:10.1016/j.apsusc.2017.03.176
Amaro, F., Morón, Á., Díaz, S., Martín-González, A., and Gutiérrez, J. C. (2021). Metallic nanoparticles—friends or foes in the battle against antibiotic-resistant bacteria? Microorganisms 9 (2), 364. doi:10.3390/microorganisms9020364
Ansari, M. A., Khan, H. M., Khan, A. A., Cameotra, S. S., Saquib, Q., and Musarrat, J. (2014). Interaction of Al2O3 nanoparticles with Escherichia coli and their cell envelope biomolecules. J. Applied Microbiology 116 (4), 772–783. doi:10.1111/jam.12423
Asadishad, B., Chahal, S., Akbari, A., Cianciarelli, V., Azodi, M., Ghoshal, S., et al. (2018). Amendment of agricultural soil with metal nanoparticles: effects on soil enzyme activity and microbial community composition. Environ. Sci. and Technol. 52 (4), 1908–1918. doi:10.1021/acs.est.7b05389
Ashammakhi, N., Nasiri, R., De Barros, N. R., Tebon, P., Thakor, J., Goudie, M., et al. (2020). Gut-on-a-chip: current progress and future opportunities. Biomaterials 255, 120196. doi:10.1016/j.biomaterials.2020.120196
Avila-Arias, H., Nies, L. F., Gray, M. B., Barreto-Hernández, E., and Turco, R. F. (2023). Soil activity and microbial community response to nanometal oxides were not due exclusively to a particle size effect. Environ. Sci. Nano 10 (1), 129–144. doi:10.1039/D2EN00762B
Balta, I., Lemon, J., Gadaj, A., Cretescu, I., Stef, D., Pet, I., et al. (2025). The interplay between antimicrobial resistance, heavy metal pollution, and the role of microplastics. Front. Microbiol. 16, 1550587. doi:10.3389/fmicb.2025.1550587
Banos, S., Lentendu, G., Kopf, A., Wubet, T., Glöckner, F. O., and Reich, M. (2018). A comprehensive fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms. BMC Microbiology 18 (1), 190. doi:10.1186/s12866-018-1331-4
Bantun, F., Singh, R., Alkhanani, M. F., Almalki, A. H., Alshammary, F., Khan, S., et al. (2022). Gut microbiome interactions with graphene based nanomaterials: challenges and opportunities. Sci. Total Environ. 830, 154789. doi:10.1016/j.scitotenv.2022.154789
Bao, S., Wang, H., Zhang, W., Xie, Z., and Fang, T. (2016). An investigation into the effects of silver nanoparticles on natural microbial communities in two freshwater sediments. Environ. Pollut. 219, 696–704. doi:10.1016/j.envpol.2016.06.071
Baranowska-Wójcik, E. (2021). Factors conditioning the potential effects TiO2 NPs exposure on human microbiota: a mini-review. Biol. Trace Elem. Res. 199 (12), 4458–4465. doi:10.1007/s12011-021-02578-5
Beaudry, M. S., Wang, J., Kieran, T. J., Thomas, J., Bayona-Vásquez, N. J., Gao, B., et al. (2021). Improved microbial community characterization of 16S rRNA via metagenome hybridization capture enrichment. Front. Microbiol. 12, 644662. doi:10.3389/fmicb.2021.644662
Berrios, L., Yeam, J., Holm, L., Robinson, W., Pellitier, P. T., Chin, M. L., et al. (2023). Positive interactions between mycorrhizal fungi and bacteria are widespread and benefit plant growth. Curr. Biol. 33 (14), 2878–2887.e4. doi:10.1016/j.cub.2023.06.010
Bharti, R., and Grimm, D. G. (2021). Current challenges and best-practice protocols for microbiome analysis. Briefings Bioinformatics 22 (1), 178–193. doi:10.1093/bib/bbz155
Bi, Y., Marcus, A. K., Robert, H., Krajmalnik-Brown, R., Rittmann, B. E., Westerhoff, P., et al. (2020). The complex puzzle of dietary silver nanoparticles, mucus and microbiota in the gut. J. Toxicol. Environ. Health, Part B 23 (2), 69–89. doi:10.1080/10937404.2019.1710914
Bianchi, M. G., Chiu, M., Taurino, G., Bergamaschi, E., Turroni, F., Mancabelli, L., et al. (2024). Amorphous silica nanoparticles and the human gut microbiota: a relationship with multiple implications. J. Nanobiotechnology 22 (1), 45. doi:10.1186/s12951-024-02305-x
Binh, C. T. T., Tong, T., Gaillard, J. F., Gray, K. A., and Kelly, J. J. (2014). Common freshwater bacteria vary in their responses to short-term exposure to nano-TiO2. Environ. Toxicology Chemistry 33 (2), 317–327. doi:10.1002/etc.2442
Binh, C. T. T., Adams, E., Vigen, E., Tong, T., Alsina, M. A., Gaillard, J. F., et al. (2016). Chronic addition of a common engineered nanomaterial alters biomass, activity and composition of stream biofilm communities. Environ. Sci. Nano 3 (3), 619–630. doi:10.1039/C5EN00274E
Bora, K. A., Hashmi, S., Zulfiqar, F., Abideen, Z., Ali, H., Siddiqui, Z. S., et al. (2022). Recent progress in bio-mediated synthesis and applications of engineered nanomaterials for sustainable agriculture. Front. Plant Sci. 13, 999505. doi:10.3389/fpls.2022.999505
Brumfield, K. D., Huq, A., Colwell, R. R., Olds, J. L., and Leddy, M. B. (2020). Microbial resolution of whole genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data. PLoS One 15 (2), e0228899. doi:10.1371/journal.pone.0228899
Bruna, T., Maldonado-Bravo, F., Jara, P., and Caro, N. (2021). Silver nanoparticles and their antibacterial applications. Int. Journal Molecular Sciences 22 (13), 7202. doi:10.3390/ijms22137202
Burke, D. J., Pietrasiak, N., Situ, S. F., Abenojar, E. C., Porche, M., Kraj, P., et al. (2015). Iron oxide and titanium dioxide nanoparticle effects on plant performance and root associated microbes. Int. J. Mol. Sci. 16 (10), 23630–23650. doi:10.3390/ijms161023630
Caballero-Díaz, E., Pfeiffer, C., Kastl, L., Rivera-Gil, P., Simonet, B., Valcárcel, M., et al. (2013). The toxicity of silver nanoparticles depends on their uptake by cells and thus on their surface chemistry. Part. and Part. Syst. Charact. 30 (12), 1079–1085. doi:10.1002/ppsc.201300215
Cai, L., Zhang, L., Yang, J., Zhu, X., Wei, W., Ji, M., et al. (2023). Encapsulating antibiotic and protein-stabilized nanosilver into sandwich-structured electrospun nanofibrous scaffolds for MRSA-infected wound treatment. ACS Appl. Mater. and Interfaces 15 (42), 48978–48995. doi:10.1021/acsami.3c10994
Caixeta, M. B., Araújo, P. S., Rodrigues, C. C., Gonçalves, B. B., Araújo, O. A., Bevilaqua, G. B., et al. (2021). Risk assessment of iron oxide nanoparticles in an aquatic ecosystem: a case study on Biomphalaria glabrata. J. Hazard. Mater. 401, 123398. doi:10.1016/j.jhazmat.2020.123398
Catalano, R., Slomberg, D. L., Picard, C., Hucher, N., Vidal, V., Saint-Antonin, F., et al. (2021). In situ determination of engineered nanomaterial aggregation state in a cosmetic emulsion–toward safer-by-design products. Environ. Sci. Nano 8 (12), 3546–3559. doi:10.1039/D1EN00345C
Cataldi, P., Steiner, P., Raine, T., Lin, K., Kocabas, C., Young, R. J., et al. (2020). Multifunctional biocomposites based on polyhydroxyalkanoate and graphene/carbon nanofiber hybrids for electrical and thermal applications. ACS Appl. Polym. Mater. 2 (8), 3525–3534. doi:10.1021/acsapm.0c00539
Chavan, S., and Nadanathangam, V. (2019). Effects of nanoparticles on plant growth-promoting bacteria in Indian agricultural soil. Agronomy 9 (3), 140. doi:10.3390/agronomy9030140
Chavan, S., and Nadanathangam, V. (2020). Shifts in metabolic patterns of soil bacterial communities on exposure to metal engineered nanomaterials. Ecotoxicol. Environ. Saf. 189, 110012. doi:10.1016/j.ecoenv.2019.110012
Chen, C., Tsyusko, O. V., McNear Jr, D. H., Judy, J., Lewis, R. W., and Unrine, J. M. (2017). Effects of biosolids from a wastewater treatment plant receiving manufactured nanomaterials on Medicago truncatula and associated soil microbial communities at low nanomaterial concentrations. Sci. Total Environ. 609, 799–806. doi:10.1016/j.scitotenv.2017.07.188
Chen, H., Zhao, R., Wang, B., Zheng, L., Ouyang, H., Wang, H., et al. (2018). Acute oral administration of single-walled carbon nanotubes increases intestinal permeability and inflammatory responses: association with the changes in gut microbiota in mice. Adv. Healthcare Materials 7 (13), 1701313. doi:10.1002/adhm.201701313
Chen, Z., Han, S., Zhou, D., Zhou, S., and Jia, G. (2019). Effects of oral exposure to titanium dioxide nanoparticles on gut microbiota and gut-associated metabolism in vivo. Nanoscale 11 (46), 22398–22412. doi:10.1039/C9NR07580A
Chen, B., Pan, Y., Chen, Y., Zhang, Z., Yang, Z., Zheng, M., et al. (2022). TiO2 nanoparticles exert an adverse effect on aquatic microbial communities. Sci. Total Environ. 831, 154942. doi:10.1016/j.scitotenv.2022.154942
Chen, A., Gong, Y., Wu, S., Du, Y., Liu, Z., Jiang, Y., et al. (2025). Navigating a challenging path: precision disease treatment with tailored oral nano-armor-probiotics. J. Nanobiotechnology 23 (1), 72. doi:10.1186/s12951-025-03141-3
Chen, Y., Li, E. M., and Xu, L. Y. (2022). Guide to metabolomics analysis: a bioinformatics workflow. Metabolites 12 (4), 357. doi:10.3390/metabo12040357
Cherchi, C., and Gu, A. Z. (2010). Impact of titanium dioxide nanomaterials on nitrogen fixation rate and intracellular nitrogen storage in Anabaena variabilis. Environ. Science and Technology 44 (21), 8302–8307. doi:10.1021/es101658p
Chhipa, H. (2021). Nano-toxicity to microbes: potential implications of nanomaterials on microbial activity. Nanotoxicology Nanoecotoxicology 1, 99–123. doi:10.1007/978-3-030-63241-0_4
Chiarello, M., McCauley, M., Villéger, S., and Jackson, C. R. (2022). Ranking the biases: the choice of OTUs vs. ASVs in 16S rRNA amplicon data analysis has stronger effects on diversity measures than rarefaction and OTU identity threshold. PloS One 17 (2), e0264443. doi:10.1371/journal.pone.0264443
Choi, O. K., and Hu, Z. Q. (2009). Nitrification inhibition by silver nanoparticles. Water Science Technology 59 (9), 1699–1702. doi:10.2166/wst.2009.205
Cobos, M., De-La-Pinta, I., Quindós, G., Fernández, M. J., and Fernández, M. D. (2020). Graphene oxide–silver nanoparticle nanohybrids: synthesis, characterization, and antimicrobial properties. Nanomaterials 10 (2), 376. doi:10.3390/nano10020376
Combrink, L., Humphreys, I. R., Washburn, Q., Arnold, H. K., Stagaman, K., Kasschau, K. D., et al. (2023). Best practice for wildlife gut microbiome research: a comprehensive review of methodology for 16S rRNA gene investigations. Front. Microbiol. 14, 1092216. doi:10.3389/fmicb.2023.1092216
Courtois, P., Rorat, A., Lemiere, S., Guyoneaud, R., Attard, E., Levard, C., et al. (2019). Ecotoxicology of silver nanoparticles and their derivatives introduced in soil with or without sewage sludge: a review of effects on microorganisms, plants and animals. Environ. Pollution 253, 578–598. doi:10.1016/j.envpol.2019.07.053
Cui, H., and Smith, A. L. (2022). Impact of engineered nanoparticles on the fate of antibiotic resistance genes in wastewater and receiving environments: a comprehensive review. Environ. Res. 204, 112373. doi:10.1016/j.envres.2021.112373
Cui, X., Wang, X., Chang, X., Bao, L., Wu, J., Tan, Z., et al. (2023). A new capacity of gut microbiota: fermentation of engineered inorganic carbon nanomaterials into endogenous organic metabolites. Proc. Natl. Acad. Sci. 120 (20), e2218739120. doi:10.1073/pnas.2218739120
Daly, S. E., Feng, J., Daeschel, D., Kovac, J., and Snyder, A. B. (2024). The choice of 16S rRNA gene sequence analysis impacted characterization of highly variable surface microbiota in dairy processing environments. Msystems 9 (11), e00620–24. doi:10.1128/msystems.00620-24
Das, P., Williams, C. J., Fulthorpe, R. R., Hoque, M. E., Metcalfe, C. D., and Xenopoulos, M. A. (2012). Changes in bacterial community structure after exposure to silver nanoparticles in natural waters. Environ. Science and Technology 46 (16), 9120–9128. doi:10.1021/es3019918
Das, B., Tripathy, S., Adhikary, J., Chattopadhyay, S., Mandal, D., Dash, S. K., et al. (2017). Surface modification minimizes the toxicity of silver nanoparticles: an in vitro and in vivo study. JBIC J. Biol. Inorg. Chem. 22 (6), 893–918. doi:10.1007/s00775-017-1468-x
Day, N., Zhang, T., Gaffrey, M. J., Thrall, B. D., and Qian, W. J. (2023). Assessment of the biological impact of engineered nanomaterials using mass spectrometry-based MultiOmics approaches. Impact Eng. Nanomater. Genomics Epigenomics, 331–354. doi:10.1002/9781119896258.ch15
De Filippis, F., Valentino, V., Sequino, G., Borriello, G., Riccardi, M. G., Pierri, B., et al. (2024). Exposure to environmental pollutants selects for xenobiotic-degrading functions in the human gut microbiome. Nat. Commun. 15 (1), 4482. doi:10.1038/s41467-024-48739-7
De la Cuesta-Zuluaga, J., and Escobar, J. S. (2016). Considerations for optimizing microbiome analysis using a marker gene. Front. Nutrition 3, 26. doi:10.3389/fnut.2016.00026
Debode, F., Caulier, S., Demeter, S., Dubois, B., Gelhay, V., Hulin, J., et al. (2024). Roadmap for the integration of environmental microbiomes in risk assessments under EFSA's remit. EFSA Support. Publ. 21 (2), 8602E. doi:10.2903/sp.efsa.2024.EN-8602
Di Bella, J. M., Bao, Y., Gloor, G. B., Burton, J. P., and Reid, G. (2013). High throughput sequencing methods and analysis for microbiome research. J. Microbiological Methods 95 (3), 401–414. doi:10.1016/j.mimet.2013.08.011
Diao, J., Xia, Y., Jiang, X., Qiu, J., Cheng, S., Su, J., et al. (2021). Silicon dioxide nanoparticles induced neurobehavioral impairments by disrupting microbiota–gut–brain axis. J. Nanobiotechnology 19 (1), 174. doi:10.1186/s12951-021-00916-2
Dickinson, A. W., Power, A., Hansen, M. G., Brandt, K. K., Piliposian, G., Appleby, P., et al. (2019). Heavy metal pollution and co-selection for antibiotic resistance: a microbial palaeontology approach. Environ. International 132, 105117. doi:10.1016/j.envint.2019.105117
Dinç, B. (2025). Comprehensive toxicity assessment of silver nanoparticles on bacteria, human vein endothelial cells, and Caenorhabditis elegans. Results Chem. 14, 102092. doi:10.1016/j.rechem.2025.102092
Dinesh, R., Sreena, C. P., Sheeja, T. E., Charles, S., Srinivasan, V., Sajith, V., et al. (2023a). Metagenomics indicates abundance of biofilm related genes and horizontal transfer of multidrug resistant genes among bacterial communities in nano zinc oxide polluted soil. Sci. Total Environ. 859, 160032. doi:10.1016/j.scitotenv.2022.160032
Dinesh, R., Sreena, C. P., Sheeja, T. E., Kumar, I. V., Praveena, R., Charles, S., et al. (2023b). Soil polluted with nano ZnO reveals unstable bacterial communities and decoupling of taxonomic and functional diversities. Sci. Total Environ. 889, 164285. doi:10.1016/j.scitotenv.2023.164285
Ding, C., Pan, J., Jin, M., Yang, D., Shen, Z., Wang, J., et al. (2016). Enhanced uptake of antibiotic resistance genes in the presence of nanoalumina. Nanotoxicology 10 (8), 1051–1060. doi:10.3109/17435390.2016.1161856
Dixit, M., Ghoshal, D., Meena, A. L., Ghasal, P. C., Rai, A. K., Choudhary, J., et al. (2024). Changes in soil microbial diversity under present land degradation scenario. Total Environ. Adv. 10, 200104. doi:10.1016/j.teadva.2024.200104
Do, K., Mehta, S., Wagner, R., Bhuming, D., Rajczewski, A. T., Skubitz, A. P., et al. (2024). A novel clinical metaproteomics workflow enables bioinformatic analysis of host-microbe dynamics in disease. Msphere 9 (6), e00793–23. doi:10.1128/msphere.00793-23
Douglas, G. M., Beiko, R. G., and Langille, M. G. (2018). “Predicting the functional potential of the microbiome from marker genes using PICRUSt,” in Microbiome analysis: methods and protocols (New York, NY: Springer New York), 169–177. doi:10.1007/978-1-4939-8728-3_11
ECHA (2025). Biocidal products regulation (BPR). Available online at: https://echa.europa.eu/regulations/biocidal-products-regulation/understanding-bpr (Accessed on July 12, 2025).
El Badawy, A. M., Silva, R. G., Morris, B., Scheckel, K. G., Suidan, M. T., and Tolaymat, T. M. (2011). Surface charge-dependent toxicity of silver nanoparticles. Environ. Science and Technology 45 (1), 283–287. doi:10.1021/es1034188
El-Kalliny, A. S., Abdel-Wahed, M. S., El-Zahhar, A. A., Hamza, I. A., and Gad-Allah, T. A. (2023). Nanomaterials: a review of emerging contaminants with potential health or environmental impact. Discov. Nano 18 (1), 68. doi:10.1186/s11671-023-03787-8
Environment EC (2024a). CLP regulation. Available online at: https://environment.ec.europa.eu/topics/chemicals/classification-labelling-and-packaging-chemicals_en (Accessed on July 12, 2025).
Environment EC (2024b). REACH regulation. Available online at: https://environment.ec.europa.eu/topics/chemicals/reach-regulation (Accessed on July 12, 2025).
Evariste, L., Braylé, P., Mouchet, F., Silvestre, J., Gauthier, L., Flahaut, E., et al. (2021). Graphene-based nanomaterials modulate internal biofilm interactions and microbial diversity. Front. Microbiol. 12, 623853. doi:10.3389/fmicb.2021.623853
Fajardo, C., Saccà, M. L., Costa, G., Nande, M., and Martin, M. (2014). Impact of Ag and Al2O3 nanoparticles on soil organisms: in vitro and soil experiments. Sci. Total Environ. 473, 254–261. doi:10.1016/j.scitotenv.2013.12.043
Feng, Y., Min, L., Zhang, W., Liu, J., Hou, Z., Chu, M., et al. (2017). Zinc oxide nanoparticles influence microflora in ileal digesta and correlate well with blood metabolites. Front. Microbiology 8, 992. doi:10.3389/FMICB.2017.00992
Ferreira, L., Pires, P. C., Fonseca, M., Costa, G., Giram, P. S., Mazzola, P. G., et al. (2023). Nanomaterials in cosmetics: an outlook for European regulatory requirements and a step forward in sustainability. Cosmetics 10 (2), 53. doi:10.3390/cosmetics10020053
Fox, J. D., Sims, A., Ross, M., Bettag, J., Wilder, A., Natrop, D., et al. (2024). Bioinformatic methodologies in assessing gut microbiota. Microbiol. Res. 15 (4), 2554–2574. doi:10.3390/microbiolres15040170
Fritea, L., Banica, F., Costea, T. O., Moldovan, L., Dobjanschi, L., Muresan, M., et al. (2021). Metal nanoparticles and carbon-based nanomaterials for improved performances of electrochemical (bio) sensors with biomedical applications. Materials 14 (21), 6319. doi:10.3390/ma14216319
Gabrielyan, L., Hovhannisyan, A., Gevorgyan, V., Ananyan, M., and Trchounian, A. (2019). Antibacterial effects of iron oxide (Fe3O4) nanoparticles: distinguishing concentration-dependent effects with different bacterial cells growth and membrane-associated mechanisms. Appl. Microbiology Biotechnology 103 (6), 2773–2782. doi:10.1007/s00253-019-09653-x
Galloway-Peña, J., and Hanson, B. (2020). Tools for analysis of the microbiome. Dig. Diseases Sciences 65 (3), 674–685. doi:10.1007/s10620-020-06091-y
Gambardella, C., and Pinsino, A. (2022). Nanomaterial ecotoxicology in the terrestrial and aquatic environment: a systematic review. Toxics 10 (7), 393. doi:10.3390/toxics10070393
Gangadoo, S., Nguyen, H., Rajapaksha, P., Zreiqat, H., Latham, K., Cozzolino, D., et al. (2021). Inorganic nanoparticles as food additives and their influence on the human gut microbiota. Environ. Sci. Nano 8 (6), 1500–1518. doi:10.1039/D1EN00025J
Gao, B., Chi, L., Zhu, Y., Shi, X., Tu, P., Li, B., et al. (2021). An introduction to next generation sequencing bioinformatic analysis in gut microbiome studies. Biomolecules 11 (4), 530. doi:10.3390/biom11040530
García, A., Delgado, L., Torà, J. A., Casals, E., González, E., Puntes, V., et al. (2012). Effect of cerium dioxide, titanium dioxide, silver, and gold nanoparticles on the activity of microbial communities intended in wastewater treatment. J. Hazardous Materials 199, 64–72. doi:10.1016/j.jhazmat.2011.10.057
Ge, Y., Schimel, J. P., and Holden, P. A. (2012). Identification of soil bacteria susceptible to TiO2 and ZnO nanoparticles. Appl. Environmental Microbiology 78 (18), 6749–6758. doi:10.1128/AEM.00941-12
Ge, D., Du, Q., Ran, B., Liu, X., Wang, X., Ma, X., et al. (2019). The neurotoxicity induced by engineered nanomaterials. Int. Journal Nanomedicine 14, 4167–4186. doi:10.2147/IJN.S203352
Ghebretatios, M., Schaly, S., and Prakash, S. (2021). Nanoparticles in the food industry and their impact on human gut microbiome and diseases. Int. J. Mol. Sci. 22 (4), 1942. doi:10.3390/IJMS22041942
Ghosh, M., and Kumar, R. (2024). “Regulatory issues in nanotechnology,” in Nanotechnology theranostics in livestock diseases and management (Singapore: Springer Nature Singapore), 765–788. doi:10.1007/978-981-16-1610-5_31
González-Fernández, S., Blanco-Agudín, N., Rodríguez, D., Fernández-Vega, I., Merayo-Lloves, J., and Quirós, L. M. (2025). Silver nanoparticles: a versatile tool against infectious and non-infectious diseases. Antibiotics 14 (3), 289. doi:10.3390/antibiotics14030289
Gottardo, S., Mech, A., Drbohlavová, J., Małyska, A., Bøwadt, S., Sintes, J. R., et al. (2021). Towards safe and sustainable innovation in nanotechnology: state-of-play for smart nanomaterials. NanoImpact 21, 100297. doi:10.1016/j.impact.2021.100297
Guilloteau, E., Djouina, M., Caboche, S., Waxin, C., Deboudt, K., Beury, D., et al. (2022). Exposure to atmospheric Ag, TiO2, Ti and SiO2 engineered nanoparticles modulates gut inflammatory response and microbiota in mice. Ecotoxicol. Environ. Saf. 236, 113442. doi:10.1016/j.ecoenv.2022.113442
Guo, Y., Cichocki, N., Schattenberg, F., Geffers, R., Harms, H., and Müller, S. (2019). AgNPs change microbial community structures of wastewater. Front. Microbiology 9, 3211. doi:10.3389/fmicb.2018.03211
Hadrup, N., Loeschner, K., Bergström, A., Wilcks, A., Gao, X., Vogel, U., et al. (2012). Subacute oral toxicity investigation of nanoparticulate and ionic silver in rats. Archives Toxicology 86 (4), 543–551. doi:10.1007/s00204-011-0759-1
Han, S., Van Treuren, W., Fischer, C. R., Merrill, B. D., DeFelice, B. C., Sanchez, J. M., et al. (2021). A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 595 (7867), 415–420. doi:10.1038/s41586-021-03707-9
Hanan, N. A., Chiu, H. I., Ramachandran, M. R., Tung, W. H., Mohamad Zain, N. N., Yahaya, N., et al. (2018). Cytotoxicity of plant-mediated synthesis of metallic nanoparticles: a systematic review. Int. J. Mol. Sci. 19 (6), 1725. doi:10.3390/ijms19061725
Hao, W., Cha, R., Wang, M., Zhang, P., and Jiang, X. (2022). Impact of nanomaterials on the intestinal mucosal barrier and its application in treating intestinal diseases. Nanoscale Horizons 7 (1), 6–30. doi:10.1039/D1NH00315A
He, X., Aker, W. G., Leszczynski, J., and Hwang, H. M. (2014). Using a holistic approach to assess the impact of engineered nanomaterials inducing toxicity in aquatic systems. J. Food Drug Analysis 22 (1), 128–146. doi:10.1016/j.jfda.2014.01.011
Herrera, G., Paudel, S., Lupini, S., Astete, C., Sabliov, C., and Rodrigues, D. (2024). Biodegradable nanoparticles aid the gut microbial community in delaying antibiotic resistance emergence. Environ. Sci. Nano 11 (11), 4501–4512. doi:10.1039/D4EN00382A
Heyer, R., Schallert, K., Büdel, A., Zoun, R., Dorl, S., Behne, A., et al. (2019). A robust and universal metaproteomics workflow for research studies and routine diagnostics within 24 h using phenol extraction, FASP digest, and the MetaProteomeAnalyzer. Front. Microbiol. 10, 1883. doi:10.3389/fmicb.2019.01883
Heyer, R., Wolf, M., Benndorf, D., Uzzau, S., Seifert, J., Grenga, L., et al. (2025). Metaproteomics in the one health framework for unraveling microbial effectors in microbiomes. Microbiome 13 (1), 134. doi:10.1186/s40168-025-02119-5
Hochella Jr, M. F., Mogk, D. W., Ranville, J., Allen, I. C., Luther, G. W., Marr, L. C., et al. (2019). Natural, incidental, and engineered nanomaterials and their impacts on the Earth system. Science 363 (6434), eaau8299. doi:10.1126/science.aau8299
Hoecke, K. V., Quik, J. T., Mankiewicz-Boczek, J., Schamphelaere, K. A. D., Elsaesser, A., Meeren, P. V. D., et al. (2009). Fate and effects of CeO2 nanoparticles in aquatic ecotoxicity tests. Environ. Science and Technology 43 (12), 4537–4546. doi:10.1021/es9002444
Hou, K., Wu, Z. X., Chen, X. Y., Wang, J. Q., Zhang, D., Xiao, C., et al. (2022). Microbiota in health and diseases. Signal Transduction Targeted Therapy 7 (1), 135. doi:10.1038/s41392-022-00974-4
Huang, X., Auffan, M., Eckelman, M. J., Elimelech, M., Kim, J. H., Rose, J., et al. (2024). Trends, risks and opportunities in environmental nanotechnology. Nat. Rev. Earth and Environ. 5 (8), 572–587. doi:10.1038/s43017-024-00567-5
Hussein, H. S. (2023). The state of the art of nanomaterials and its applications in energy saving. Bull. Natl. Res. Centre 47 (1), 7. doi:10.1186/s42269-023-00984-4
Isigonis, P., Hristozov, D., Benighaus, C., Giubilato, E., Grieger, K., Pizzol, L., et al. (2019). Risk governance of nanomaterials: review of criteria and tools for risk communication, evaluation, and mitigation. Nanomaterials 9 (5), 696. doi:10.3390/nano9050696
Islam, S. (2025). Toxicity and transport of nanoparticles in agriculture: effects of size, coating, and aging. Front. Nanotechnol. 7, 1622228. doi:10.3389/fnano.2025.1622228
Jagtap, P. D., Blakely, A., Murray, K., Stewart, S., Kooren, J., Johnson, J. E., et al. (2015). Metaproteomic analysis using the galaxy framework. Proteomics 15 (20), 3553–3565. doi:10.1002/pmic.201500074
Javurek, A. B., Suresh, D., Spollen, W. G., Hart, M. L., Hansen, S. A., Ellersieck, M. R., et al. (2017). Gut dysbiosis and neurobehavioral alterations in rats exposed to silver nanoparticles. Sci. Reports 7 (1), 2822. doi:10.1038/s41598-017-02880-0
Jeske, J. T., and Gallert, C. (2022). Microbiome analysis via OTU and ASV-based pipelines—A comparative interpretation of ecological data in WWTP systems. Bioengineering 9 (4), 146. doi:10.3390/bioengineering9040146
Ji, Q., Zhang, C., and Li, D. (2020). Influences and mechanisms of nanofullerene on the horizontal transfer of plasmid-encoded antibiotic resistance genes between E. coli strains. Front. Environ. Sci. and Eng. 14 (6), 108. doi:10.1007/s11783-020-1287-0
Johnson, J. S., Spakowicz, D. J., Hong, B. Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Communications 10 (1), 5029. doi:10.1038/s41467-019-13036-1
Jovel, J., Patterson, J., Wang, W., Hotte, N., O'Keefe, S., Mitchel, T., et al. (2016). Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiology 7, 459. doi:10.3389/fmicb.2016.00459
Kah, M. (2015). Nanopesticides and nanofertilizers: emerging contaminants or opportunities for risk mitigation? Front. Chemistry 3, 64. doi:10.3389/fchem.2015.00064
Kalli, M., Noutsopoulos, C., and Mamais, D. (2023). The fate and occurrence of antibiotic-resistant bacteria and antibiotic resistance genes during advanced wastewater treatment and disinfection: a review. Water 15 (11), 2084. doi:10.3390/w15112084
Kang, S., Herzberg, M., Rodrigues, D. F., and Elimelech, M. (2008). Antibacterial effects of carbon nanotubes: size does matter. Langmuir 24 (13), 6409–6413. doi:10.1021/la800951v
Kaushik, M., Sarkar, N., Singh, A., and Kumar, P. (2023). Nanomaterials to address the genesis of antibiotic resistance in Escherichia coli. Front. Cell. Infect. Microbiol. 12, 946184. doi:10.3389/fcimb.2022.946184
Keepers, K. G., Pogoda, C. S., White, K. H., Anderson Stewart, C. R., Hoffman, J. R., Ruiz, A. M., et al. (2019). Whole genome shotgun sequencing detects greater lichen fungal diversity than amplicon-based methods in environmental samples. Front. Ecol. Evol. 7, 484. doi:10.3389/fevo.2019.00484
Kembel, S. W., Wu, M., Eisen, J. A., and Green, J. L. (2012). Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Computational Biology 8 (10), e1002743. doi:10.1371/journal.pcbi.1002743
Khachatryan, L., de Leeuw, R. H., Kraakman, M. E., Pappas, N., Te Raa, M., Mei, H., et al. (2020). Taxonomic classification and abundance estimation using 16S and WGS—A comparison using controlled reference samples. Forensic Sci. Int. Genet. 46, 102257. doi:10.1016/j.fsigen.2020.102257
Khairol Anuar, N. K., Tan, H. L., Lim, Y. P., So’aib, M. S., and Abu Bakar, N. F. (2021). A review on multifunctional carbon-dots synthesized from biomass waste: design/fabrication, characterization and applications. Front. Energy Research 9, 626549. doi:10.3389/fenrg.2021.626549
Khan, S. T., Adil, S. F., Shaik, M. R., Alkhathlan, H. Z., Khan, M., and Khan, M. (2021). Engineered nanomaterials in soil: their impact on soil microbiome and plant health. Plants 11 (1), 109. doi:10.3390/plants11010109
Kraegeloh, A., Suarez-Merino, B., Sluijters, T., and Micheletti, C. (2018). Implementation of safe-by-design for nanomaterial development and safe innovation: why we need a comprehensive approach. Nanomaterials 8 (4), 239. doi:10.3390/nano8040239
Kruk, M. E., Mehta, S., Murray, K., Higgins, L., Do, K., Johnson, J. E., et al. (2024). An integrated metaproteomics workflow for studying host-microbe dynamics in bronchoalveolar lavage samples applied to cystic fibrosis disease. mSystems 9 (7), e00929–23. doi:10.1128/msystems.00929-23
Kumar, A. Y. N., Kurian, A., Yadav, P. K., Prasad, R., and Ghosh, M. (2024). “General methods for generation, characterization, and functionalization of different types of nanomaterials,” in Nanotechnology theranostics in livestock diseases and management. Editors M. Prasad, R. Kumar, M. Ghosh, S. M. Syed, and S. Chakravarti (Singapore: Livestock Diseases and Management. Springer). doi:10.1007/978-981-16-1610-5_2
Kumar, R., Ghosh, M., Kumar, S., and Prasad, M. (2020). Single cell metabolomics: a future tool to unmask cellular heterogeneity and virus-host interaction in context of emerging viral diseases. Front. Microbiol. 11, 1152. doi:10.3389/fmicb.2020.01152
Kumar, C. V., Karthick, V., Kumar, V. G., Inbakandan, D., Rene, E. R., Suganya, K. U., et al. (2022). The impact of engineered nanomaterials on the environment: release mechanism, toxicity, transformation, and remediation. Environ. Research 212, 113202. doi:10.1016/j.envres.2022.113202
Kumar, A., Pramanik, J., Batta, K., Bamal, P., Gaur, M., Rustagi, S., et al. (2024). Impact of metallic nanoparticles on gut microbiota modulation in colorectal cancer: a review. Cancer Innov. 3 (6), e150. doi:10.1002/cai2.150
Kumar, R., Lakhani, P., Kumar, A. Y. N., and Ghosh, M. (2024). “Nanotoxicity and mechanisms,” in Nanotechnology theranostics in livestock diseases and management. Editors M. Prasad, R. Kumar, M. Ghosh, S. M. Syed, and S. Chakravarti (Singapore: Livestock Diseases and Management. Springer). doi:10.1007/978-981-16-1610-5_29
Kurul, F., Turkmen, H., Cetin, A. E., and Topkaya, S. N. (2025). Nanomedicine: how nanomaterials are transforming drug delivery, bio-imaging, and diagnosis. Next Nanotechnol. 7, 100129. doi:10.1016/j.nxnano.2024.100129
Kusi, J., Scheuerman, P. R., and Maier, K. J. (2020). Emerging environmental contaminants (silver nanoparticles) altered the catabolic capability and metabolic fingerprinting of microbial communities. Aquat. Toxicol. 228, 105633. doi:10.1016/j.aquatox.2020.105633
Lamas, B., Martins Breyner, N., and Houdeau, E. (2020). Impacts of foodborne inorganic nanoparticles on the gut microbiota-immune axis: potential consequences for host health. Part. Fibre Toxicol. 17 (1), 19. doi:10.1186/s12989-020-00349-z
Lamas, B., Evariste, L., and Houdeau, E. (2023). Dysregulation along the gut microbiota-immune system axis after oral exposure to titanium dioxide nanoparticles: a possible environmental factor promoting obesity-related metabolic disorders. Environ. Pollut. 330, 121795. doi:10.1016/j.envpol.2023.121795
Lee, H. B., Jeong, D. H., Cho, B. C., and Park, J. S. (2023). Comparative analyses of eight primer sets commonly used to target the bacterial 16S rRNA gene for marine metabarcoding-based studies. Front. Mar. Sci. 10, 1199116. doi:10.3389/fmars.2023.1199116
Lewis, S., Nash, A., Li, Q., and Ahn, T. H. (2021). Comparison of 16S and whole genome dog microbiomes using machine learning. BioData Min. 14 (1), 41. doi:10.1186/s13040-021-00270-x
Li, J., Chen, H., Wang, B., Cai, C., Yang, X., Chai, Z., et al. (2017). ZnO nanoparticles act as supportive therapy in DSS-induced ulcerative colitis in mice by maintaining gut homeostasis and activating Nrf2 signaling. Sci. Rep. 7 (1), 43126. doi:10.1038/SREP43126
Li, J., Tang, M., and Xue, Y. (2019). Review of the effects of silver nanoparticle exposure on gut bacteria. J. Appl. Toxicol. 39 (1), 27–37. doi:10.1002/JAT.3729
Li, C., He, Y., Shi, Y., Zhou, M., and Ma, J. (2020). Title: toxicity-effects of copper oxide nanoparticles to soil microbial community: responses of microbial biomass, enzymatic activity, and community structure. Environ. Pollut. 265, 113847. doi:10.1016/j.envpol.2020.113847
Li, W., Zhan, M., Wen, Y., Chen, Y., Zhang, Z., Wang, S., et al. (2024). Recent progress of oral functional nanomaterials for intestinal microbiota regulation. Pharmaceutics 16 (7), 921. doi:10.3390/pharmaceutics16070921
Li, X., Lin, L., Liu, Q., Wei, L., Li, L., Liao, J., et al. (2025). Fate of extracellular antibiotic resistant genes in wastewater treatment plants: characteristics, persistence, transformation, removal and potential risk. Energy and Environ. Sustain. 1 (2), 100022. doi:10.1016/j.eesus.2025.100022
Li, P., Qu, R., Li, M., Sheng, P., Jin, L., Huang, X., et al. (2024). Impacts of food additives on gut microbiota and host health. Food Res. Int. 196, 114998. doi:10.1016/j.foodres.2024.114998
Lin, H. (2001). Engineered nanomaterials. Patty's Toxicol., 1–16. doi:10.1002/0471125474.tox118.pub2
Lin, X., Li, J., Ma, S., Liu, G., Yang, K., Tong, M., et al. (2014). Toxicity of TiO2 nanoparticles to Escherichia coli: effects of particle size, crystal phase and water chemistry. PloS One 9 (10), e110247. doi:10.1371/journal.pone.0110247
Liu, C., Huang, D., Sheng, X., Zhu, J., Dong, S., Chen, S., et al. (2024). Integrated physiological, intestinal microbiota, and metabolomic responses of adult zebrafish (Danio rerio) to subacute exposure to antimony at environmentally relevant concentrations. Ecotoxicol. Environ. Saf. 277, 116326. doi:10.1016/j.ecoenv.2024.116326
Luche, S., Eymard-Vernain, E., Diemer, H., Van Dorsselaer, A., Rabilloud, T., and Lelong, C. (2016). Zinc oxide induces the stringent response and major reorientations in the central metabolism of Bacillus subtilis. J. Proteomics 135, 170–180. doi:10.1016/j.jprot.2015.07.018
Lyu, Z., Ghoshdastidar, S., Rekha, K. R., Suresh, D., Mao, J., Bivens, N., et al. (2021). Developmental exposure to silver nanoparticles leads to long term gut dysbiosis and neurobehavioral alterations. Sci. Rep. 11 (1), 6558. doi:10.1038/s41598-021-85919-7
Ma, Y., Tiwari, J., and Bauddh, K. (2022). Plant-mycorrhizal fungi interactions in phytoremediation of geogenic contaminated soils. Front. Microbiol. 13, 843415. doi:10.3389/fmicb.2022.843415
Ma, H., Cornadó, D., and Raaijmakers, J. M. (2025). The soil-plant-human gut microbiome axis into perspective. Nat. Commun. 16 (1), 7748. doi:10.1038/s41467-025-62989-z
Ma, L. C., Zhao, H. Q., Wu, L. B., Cheng, Z. L., and Liu, C. (2023). Impact of the microbiome on human, animal, and environmental health from a one health perspective. Sci. One Health 2, 100037. doi:10.1016/j.soh.2023.100037
Ma, Y., Zhang, J., Yu, N., Shi, J., Zhang, Y., Chen, Z., et al. (2023). Effect of nanomaterials on gut microbiota. Toxics 11 (4), 384. doi:10.3390/toxics11040384
Maksimova, Y. G. (2019). Microorganisms and carbon nanotubes: interaction and applications (review). Appl. Biochemistry Microbiology 55 (1), 1–12. doi:10.1134/S0003683819010101
Manuja, A., Kumar, B., Kumar, R., Chhabra, D., Ghosh, M., Manuja, M., et al. (2021). Metal/Metal oxide nanoparticles: toxicity concerns associated with their physical state and remediation for biomedical applications. Toxicol. Rep. 8, 1970–1978. doi:10.1016/j.toxrep.2021.11.020
Markowicz, A., Borymski, S., Adamek, A., and &Sułowicz, S. (2023). The influence of ZnO nanoparticles on horizontal transfer of resistance genes in lab and soil conditions. Environ. Res. 223, 115420. doi:10.1016/j.envres.2023.115420
Martirosyan, A., and Schneider, Y. J. (2014). Engineered nanomaterials in food: implications for food safety and consumer health. Int. Journal Environmental Research Public Health 11 (6), 5720–5750. doi:10.3390/ijerph110605720
McLean, P., Hanlon, J., Salmatonidis, A., Galea, K. S., Brooker, F., Citterio, C., et al. (2024). Safe (r)-by-design principles in the thermoplastics industry: guidance on release assessment during manufacture of nano-enabled products. Front. Public Health 12, 1398104. doi:10.3389/fpubh.2024.1398104
Mei, L., Guo, J., He, R., Ding, X., Yin, W., and Gu, Z. (2023). CsPbBr3 perovskite nanoparticles causes colitis-like symptom via promoting intestinal barrier damage and gut microbiota dysbiosis. Small 19 (32), 2301129. doi:10.1002/smll.202301129
Mendoza, R. P., and Brown, J. M. (2019). Engineered nanomaterials and oxidative stress: current understanding and future challenges. Curr. Opinion Toxicology 13, 74–80. doi:10.1016/j.cotox.2018.09.001
Menichetti, A., Mavridi-Printezi, A., Mordini, D., and Montalti, M. (2023). Effect of size, shape and surface functionalization on the antibacterial activity of silver nanoparticles. J. Funct. Biomaterials 14 (5), 244. doi:10.3390/jfb14050244
Metch, J. W., Burrows, N. D., Murphy, C. J., Pruden, A., and Vikesland, P. J. (2018). Metagenomic analysis of microbial communities yields insight into impacts of nanoparticle design. Nat. Nanotechnology 13 (3), 253–259. doi:10.1038/s41565-017-0029-3
Mgadi, K., Ndaba, B., Roopnarain, A., Rama, H., and Adeleke, R. (2024). Nanoparticle applications in agriculture: overview and response of plant-associated microorganisms. Front. Microbiology 15, 1354440. doi:10.3389/fmicb.2024.1354440
Mohammed, H., Kumar, A., Bekyarova, E., Al-Hadeethi, Y., Zhang, X., Chen, M., et al. (2020). Antimicrobial mechanisms and effectiveness of graphene and graphene-functionalized biomaterials. A scope review. Front. Bioengineering Biotechnology 8, 465. doi:10.3389/fbioe.2020.00465
Moloi, M. S., Lehutso, R. F., Erasmus, M., Oberholster, P. J., and Thwala, M. (2021). Aquatic environment exposure and toxicity of engineered nanomaterials released from nano-enabled products: current status and data needs. Nanomaterials 11 (11), 2868. doi:10.3390/nano11112868
More, P. R., Pandit, S., Filippis, A. D., Franci, G., Mijakovic, I., and &Galdiero, M. (2023). Silver nanoparticles: bactericidal and mechanistic approach against drug resistant pathogens. Microorganisms 11 (2), 369. doi:10.3390/microorganisms11020369
Moreno-Indias, I., Lahti, L., Nedyalkova, M., Elbere, I., Roshchupkin, G., Adilovic, M., et al. (2021). Statistical and machine learning techniques in human microbiome studies: contemporary challenges and solutions. Front. Microbiology 12, 635781. doi:10.3389/fmicb.2021.635781
Mortimer, M., Li, D., Wang, Y., and Holden, P. A. (2020). Physical properties of carbon nanomaterials and nanoceria affect pathways important to the nodulation competitiveness of the symbiotic N2-Fixing bacterium Bradyrhizobium diazoefficiens. Small 16 (21), 1906055. doi:10.1002/smll.201906055
Mortimer, M., Wang, Y., and Holden, P. A. (2021). Molecular mechanisms of nanomaterial-bacterial interactions revealed by omics—the role of nanomaterial effect level. Front. Bioeng. Biotechnol. 9, 683520. doi:10.3389/fbioe.2021.683520
Mosaka, T. B., Unuofin, J. O., Daramola, M. O., Tizaoui, C., and Iwarere, S. A. (2023). Inactivation of antibiotic-resistant bacteria and antibiotic-resistance genes in wastewater streams: current challenges and future perspectives. Front. Microbiology 13, 1100102. doi:10.3389/fmicb.2022.1100102
Muhamadali, H., Winder, C. L., Dunn, W. B., and Goodacre, R. (2023). Unlocking the secrets of the microbiome: exploring the dynamic microbial interplay with humans through metabolomics and their manipulation for synthetic biology applications. Biochem. J. 480 (12), 891–908. doi:10.1042/BCJ20210534
Na, H. S., Song, Y., Yu, Y., and Chung, J. (2023). Comparative analysis of primers used for 16S rRNA gene sequencing in oral microbiome studies. Methods Protoc. 6 (4), 71. doi:10.3390/mps6040071
Naz, S., Chatha, A. M. M., Khan, N. A., Ullah, Q., Zaman, F., Qadeer, A., et al. (2024). Unraveling the ecotoxicological effects of micro and nano-plastics on aquatic organisms and human health. Front. Environ. Sci. 12, 1390510. doi:10.3389/fenvs.2024.1390510
Nebauer, D. J., Pearson, L. A., and Neilan, B. A. (2024). Critical steps in an environmental metaproteomics workflow. Environ. Microbiol. 26 (5), e16637. doi:10.1111/1462-2920.16637
Neethu, C. S., Saravanakumar, C., Purvaja, R., Robin, R. S., and Ramesh, R. (2022). Arsenic resistance and horizontal gene transfer are associated with carbon and nitrogen enrichment in bacteria. Environ. Pollut. 311, 119937. doi:10.1016/j.envpol.2022.119937
Olawade, D. B., Wada, O. Z., Egbewole, B. I., Fapohunda, O., Ige, A. O., Usman, S. O., et al. (2024). Metal and metal oxide nanomaterials for heavy metal remediation: novel approaches for selective, regenerative, and scalable water treatment. Front. Nanotechnol. 6, 1466721. doi:10.3389/fnano.2024.1466721
Olivier, S. A., Bull, M. K., Strube, M. L., Murphy, R., Ross, T., Bowman, J. P., et al. (2023). Long-read MinION™ sequencing of 16S and 16S-ITS-23S rRNA genes provides species-level resolution of lactobacillaceae in mixed communities. Front. Microbiol. 14, 1290756. doi:10.3389/fmicb.2023.1290756
OP EUROPA (2027). EFSA strategy. Available online at: https://op.europa.eu/webpub/efsa/strategy-2027 (Accessed July 12, 2025).
OSHA (2025). Identifying hazard control options: the hierarchy of controls. Available online at: https://www.osha.gov/sites/default/files/Hierarchy_of_Controls_02.01.23_form_508_2.pdf.
Otinov, G. D., Lokteva, A. V., Petrova, A. D., Zinchenko, I. V., Isaeva, M. V., Kovtunov, E. A., et al. (2020). Positive and negative effects of metal oxide nanoparticles on antibiotic resistance genes transfer. Antibiotics 9 (11), 742. doi:10.3390/antibiotics9110742
Özkurt, E., Fritscher, J., Soranzo, N., Ng, D. Y., Davey, R. P., Bahram, M., et al. (2022). LotuS2: an ultrafast and highly accurate tool for amplicon sequencing analysis. Microbiome 10 (1), 176. doi:10.1186/s40168-022-01365-1
Parada, J., Rubilar, O., Fernández-Baldo, M. A., Bertolino, F. A., Durán, N., Seabra, A. B., et al. (2019). The nanotechnology among US: are metal and metal oxides nanoparticles a nano or mega risk for soil microbial communities? Crit. Reviews Biotechnology 39 (2), 157–172. doi:10.1080/07388551.2018.1523865
Passarelli, C., Cui, X., Valsami-Jones, E., and Underwood, G. J. (2020). Environmental context determines the impact of titanium oxide and silver nanoparticles on the functioning of intertidal microalgal biofilms. Environ. Sci. Nano 7 (10), 3020–3035. doi:10.1039/D0EN00440E
Pastrana, H., Avila, A., and Tsai, C. S. (2018). Nanomaterials in cosmetic products: the challenges with regard to current legal frameworks and consumer exposure. Nanoethics 12 (2), 123–137. doi:10.1007/s11569-018-0317-x
Pavlicek, A., Part, F., Rose, G., Praetorius, A., Miernicki, M., Gazsó, A., et al. (2021). A European nano-registry as a reliable database for quantitative risk assessment of nanomaterials? A comparison of national approaches. NanoImpact 21, 100276. doi:10.1016/j.impact.2020.100276
Pereira, M. M., Mouton, L., Yéprémian, C., Couté, A., Lo, J., Marconcini, J. M., et al. (2014). Ecotoxicological effects of carbon nanotubes and cellulose nanofibers in Chlorella vulgaris. J. Nanobiotechnology 12 (1), 15. doi:10.1186/1477-3155-12-15
Perez, L., Scarcello, E., Ibouraadaten, S., Yakoub, Y., Leinardi, R., Ambroise, J., et al. (2021). Dietary nanoparticles alter the composition and function of the gut microbiota in mice at dose levels relevant for human exposure. Food Chem. Toxicol. 154, 112352. doi:10.1016/j.fct.2021.112352
Pérez-Cobas, A. E., Gomez-Valero, L., and Buchrieser, C. (2020). Metagenomic approaches in microbial ecology: an update on whole-genome and marker gene sequencing analyses. Microb. Genomics 6 (8), e000409. doi:10.1099/mgen.0.000409
Perreault, F., De Faria, A. F., Nejati, S., and Elimelech, M. (2015). Antimicrobial properties of graphene oxide nanosheets: why size matters. ACS Nano 9 (7), 7226–7236. doi:10.1021/acsnano.5b02067
Peyrot, C., Wilkinson, K. J., Desrosiers, M., and Sauvé, S. (2014). Effects of silver nanoparticles on soil enzyme activities with and without added organic matter. Environ. Toxicol. Chem. 33 (1), 115–125. doi:10.1002/etc.2398
Piergiacomo, F., Brusetti, L., and Pagani, L. (2022). Understanding the interplay between antimicrobial resistance, microplastics and xenobiotic contaminants: a leap towards one health? Int. J. Environ. Res. Public Health 20 (1), 42. doi:10.3390/ijerph20010042
Pietroiusti, A., Magrini, A., and Campagnolo, L. (2016). New frontiers in nanotoxicology: gut microbiota/microbiome-mediated effects of engineered nanomaterials. Toxicol. Applied Pharmacology 299, 90–95. doi:10.1016/j.taap.2015.12.017
Pinget, G., Tan, J., Janac, B., Kaakoush, N. O., Angelatos, A. S., O'Sullivan, J., et al. (2019). Impact of the food additive titanium dioxide (E171) on gut microbiota-host interaction. Front. Nutrition 6, 57. doi:10.3389/fnut.2019.00057
Prasad, M., Kumar, R., Buragohain, L., Kumari, A., and Ghosh, M. (2021). Organoid technology: a reliable developmental biology tool for organ-specific nanotoxicity evaluation. Front. Cell Developmental Biology 9, 696668. doi:10.3389/fcell.2021.696668
Prasad, N. K., Kaur, L., and Modi, D. R. (2024). Impact of soil bacteria on biochemical cycles. Acta fytotechnica zootechnica 27 (4), 351–365. doi:10.15414/afz.2024.27.04.351-365
Puig-Castellvi, F., Pacheco-Tapia, R., Deslande, M., Jia, M., Andrikopoulos, P., Chechi, K., et al. (2023). Advances in the integration of metabolomics and metagenomics for human gut microbiome and their clinical applications. TrAC Trends Anal. Chem. 167, 117248. doi:10.1016/j.trac.2023.117248
Qiao, X., Bao, L., Liu, G., and Cui, X. (2024). Nanomaterial journey in the gut: from intestinal mucosal interaction to systemic transport. Nanoscale 16, 19207–19220. doi:10.1039/D4NR02480J
Qu, J., Zuo, X., Xu, Q., Li, M., Zou, L., Tao, R., et al. (2023). Effect of two particle sizes of nano zinc oxide on growth performance, immune function, digestive tract morphology, and intestinal microbiota composition in broilers. Animals 13 (9), 1454. doi:10.3390/ani13091454
Rajpal, V. R., Nongthongbam, B., Bhatia, M., Singh, A., Raina, S. N., Minkina, T., et al. (2025). The nano-paradox: addressing nanotoxicity for sustainable agriculture, circular economy and SDGs. J. Nanobiotechnology 23 (1), 314. doi:10.1186/s12951-025-03371-5
Rajput, V. D., Kumari, A., Upadhyay, S. K., Minkina, T., Mandzhieva, S., Ranjan, A., et al. (2023). Can nanomaterials improve the soil microbiome and crop productivity? Agriculture 13 (2), 231. doi:10.3390/agriculture13020231
Rana, S., and Kalaichelvan, P. T. (2013). Ecotoxicity of nanoparticles. Int. Sch. Res. Notices 2013 (1), 1–11. doi:10.1155/2013/574648
Ranjan, R., Rani, A., Metwally, A., McGee, H. S., and Perkins, D. L. (2016). Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophysical Research Communications 469 (4), 967–977. doi:10.1016/j.bbrc.2015.12.083
Rausch, P., Rühlemann, M., Hermes, B. M., Doms, S., Dagan, T., Dierking, K., et al. (2019). Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Microbiome 7 (1), 133. doi:10.1186/s40168-019-0743-1
Ray, P. C., Yu, H., and Fu, P. P. (2009). Toxicity and environmental risks of nanomaterials: challenges and future needs. J. Environmental Science Health. Part C, Environ. Carcinogenesis and Ecotoxicology Reviews 27 (1), 1–35. doi:10.1080/10590500802708267
Ren, Q., Ma, J., Li, X., Meng, Q., Wu, S., Xie, Y., et al. (2023). Intestinal toxicity of metal nanoparticles: silver nanoparticles disorder the intestinal immune microenvironment. ACS Appl. Mater. and Interfaces 15 (23), 27774–27788. doi:10.1021/acsami.3c05692
Resnik, D. B. (2019). How should engineered nanomaterials be regulated for public and environmental health? AMA J. Ethics 21 (4), 363–369. doi:10.1001/amajethics.2019.363
Ribeiro, A. I., Dias, A. M., and Zille, A. (2022). Synergistic effects between metal nanoparticles and commercial antimicrobial agents: a review. ACS Appl. Nano Mater. 5 (3), 3030–3064. doi:10.1021/acsanm.1c03891
Rinninella, E., Cintoni, M., Raoul, P., Mora, V., Gasbarrini, A., and Mele, M. C. (2021). Impact of food additive titanium dioxide on gut microbiota composition, microbiota-associated functions, and gut barrier: a systematic review of in vivo animal studies. Int. J. Environ. Res. Public Health 18 (4), 2008. doi:10.3390/ijerph18042008
Rocco, L. (2025). Engineered nanomaterials for environmental and health applications. Nanomaterials 15 (7), 516. doi:10.3390/nano15070516
Rodrigues, D. F., Jaisi, D. P., and Elimelech, M. (2013). Toxicity of functionalized single-walled carbon nanotubes on soil microbial communities: implications for nutrient cycling in soil. Environ. Science and Technology 47 (1), 625–633. doi:10.1021/es304002q
Rodrigues, A. S., Batista, J. G., Rodrigues, M. Á., Thipe, V. C., Minarini, L. A., Lopes, P. S., et al. (2024). Advances in silver nanoparticles: a comprehensive review on their potential as antimicrobial agents and their mechanisms of action elucidated by proteomics. Front. Microbiol. 15, 1440065. doi:10.3389/fmicb.2024.1440065
Rui, M., Ma, C., Hao, Y., Guo, J., Rui, Y., Tang, X., et al. (2016). Iron oxide nanoparticles as a potential iron fertilizer for peanut (Arachis hypogaea). Front. Plant Science 7, 815. doi:10.3389/fpls.2016.00815
Ruiz, P. A., Morón, B., Becker, H. M., Lang, S., Atrott, K., Spalinger, M. R., et al. (2017). Titanium dioxide nanoparticles exacerbate DSS-induced colitis: role of the NLRP3 inflammasome. Gut 66 (7), 1216–1224. doi:10.1136/gutjnl-2015-310297
Sadiq, I. M., Chowdhury, B., Chandrasekaran, N., and Mukherjee, A. (2009). Antimicrobial sensitivity of Escherichia coli to alumina nanoparticles. Nanomedicine Nanotechnol. Biol. Med. 5 (3), 282–286. doi:10.1016/j.nano.2009.01.002
Saeki, E. K., Yamada, A. Y., De Araujo, L. A., Anversa, L., Garcia, D. D. O., De Souza, R. L. B., et al. (2021). Subinhibitory concentrations of biogenic silver nanoparticles affect motility and biofilm formation in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 11, 656984. doi:10.3389/fcimb.2021.656984
Saenz, C., Nigro, E., Gunalan, V., and Arumugam, M. (2022). MIntO: a modular and scalable pipeline for microbiome metagenomic and metatranscriptomic data integration. Front. Bioinforma. 2, 846922. doi:10.3389/fbinf.2022.846922
Salieri, B., Barruetabeña, L., Rodríguez-Llopis, I., Jacobsen, N. R., Manier, N., Trouiller, B., et al. (2021). Integrative approach in a safe by design context combining risk, life cycle and socio-economic assessment for safer and sustainable nanomaterials. NanoImpact 23, 100335. doi:10.1016/j.impact.2021.100335
Sati, A., Ranade, T. N., Mali, S. N., Ahmad Yasin, H. K., and Pratap, A. (2025). Silver nanoparticles (AgNPs): comprehensive insights into bio/synthesis, key influencing factors, multifaceted applications, and toxicity─ a 2024 update. ACS Omega 10 (8), 7549–7582. doi:10.1021/acsomega.4c11045
Schmutz, M., Borges, O., Jesus, S., Borchard, G., Perale, G., Zinn, M., et al. (2020). A methodological safe-by-design approach for the development of nanomedicines. Front. Bioeng. Biotechnol. 8, 258. doi:10.3389/fbioe.2020.00258
See, M. S., Ching, X. L., Khoo, S. C., Abidin, S. Z., Sonne, C., and Ma, N. L. (2025). Aquatic microbiomes under stress: the role of gut microbiota in detoxification and adaptation to environmental exposures. J. Hazard. Mater. Adv. 17, 100612. doi:10.1016/j.hazadv.2025.100612
Shah, V., and Belozerova, I. (2009). Influence of metal nanoparticles on the soil microbial community and germination of lettuce seeds. Water, Air, Soil Pollution 197 (1), 143–148. doi:10.1007/s11270-008-9797-6
Sharma, D., Gautam, S., Singh, S., Srivastava, N., Khan, A. M., and Bisht, D. (2025). Unveiling the nanoworld of antimicrobial resistance: integrating nature and nanotechnology. Front. Microbiol. 15, 1391345. doi:10.3389/fmicb.2024.1391345
Shayo, G. M., Elimbinzi, E., and Shao, G. N. (2024). Preparation methods, applications, toxicity and mechanisms of silver nanoparticles as bactericidal agent and superiority of green synthesis method. Heliyon 10 (17), e36539. doi:10.1016/j.heliyon.2024.e36539
Shi, Y., Han, X., Zou, S., and Liu, G. (2024). Nanomaterials in organoids: from interactions to personalized medicine. ACS Nano 18 (49), 33276–33292. doi:10.1021/acsnano.4c13330
Sielska, A., and Skuza, L. (2025). Copper nanoparticles in aquatic environment: release routes and oxidative stress-mediated mechanisms of toxicity to fish in various life stages and future risks. Curr. Issues Mol. Biol. 47 (6), 472. doi:10.3390/cimb47060472
Siemer, S., Hahlbrock, A., Vallet, C., McClements, D. J., Balszuweit, J., Voskuhl, J., et al. (2018). Nanosized food additives impact beneficial and pathogenic bacteria in the human gut: a simulated gastrointestinal study. Npj Sci. Food 2 (1), 22. doi:10.1038/s41538-018-0030-8
Simonin, M., and Richaume, A. (2015). Impact of engineered nanoparticles on the activity, abundance, and diversity of soil microbial communities: a review. Environ. Sci. Pollut. Res. 22 (18), 13710–13723. doi:10.1007/s11356-015-4171-x
Simonin, M., Richaume, A., Guyonnet, J. P., Dubost, A., Martins, J. M., and Pommier, T. (2016). Titanium dioxide nanoparticles strongly impact soil microbial function by affecting archaeal nitrifiers. Sci. Rep. 6 (1), 33643. doi:10.1038/srep33643
Singh, P., Ali, S. W., and Kale, R. D. (2023). Antimicrobial nanomaterials as advanced coatings for self-sanitizing of textile clothing and personal protective equipment. ACS Omega 8 (9), 8159–8171. doi:10.1021/acsomega.2c06343
Sodhi, G. K., Wijesekara, T., Kumawat, K. C., Adhikari, P., Joshi, K., Singh, S., et al. (2025). Nanomaterials–plants–microbes interaction: plant growth promotion and stress mitigation. Front. Microbiol. 15, 1516794. doi:10.3389/fmicb.2024.1516794
Sohm, B., Immel, F., Bauda, P., and &Pagnout, C. (2015). Insight into the primary mode of action of TiO2 nanoparticles on Escherichia coli in the dark. Proteomics 15 (1), 98–113. doi:10.1002/pmic.201400101
Starr, A. E., Deeke, S. A., Li, L., Zhang, X., Daoud, R., Ryan, J., et al. (2018). Proteomic and metaproteomic approaches to understand host–microbe interactions. Anal. Chemistry 90 (1), 86–109. doi:10.1021/acs.analchem.7b04340
Steven, B., Hassani, M. A., LaReau, J. C., Wang, Y., and White, J. C. (2024). Nanoscale sulfur alters the bacterial and eukaryotic communities of the tomato rhizosphere and their interactions with a fungal pathogen. NanoImpact 33, 100495. doi:10.1016/j.impact.2024.100495
Su, Y., Zheng, X., Chen, Y., Li, M., and Liu, K. (2015). Alteration of intracellular protein expressions as a key mechanism of the deterioration of bacterial denitrification caused by copper oxide nanoparticles. Sci. Reports 5 (1), 15824. doi:10.1038/srep15824
Su, Y., Wu, D., Xia, H., Zhang, C., Shi, J., Wilkinson, K. J., et al. (2019). Metallic nanoparticles induced antibiotic resistance genes attenuation of leachate culturable microbiota: the combined roles of growth inhibition, ion dissolution and oxidative stress. Environ. International 128, 407–416. doi:10.1016/j.envint.2019.05.007
Sun, C., Hu, K., Mu, D., Wang, Z., and Yu, X. (2022). The widespread use of nanomaterials: the effects on the function and diversity of environmental microbial communities. Microorganisms 10 (10), 2080. doi:10.3390/microorganisms10102080
Sun, J., Yang, W., Li, M., Zhang, S., Sun, Y., and Wang, F. (2025). Metagenomic analysis reveals soil microbiome responses to microplastics and ZnO nanoparticles in an agricultural soil. J. Hazard. Mater. 492, 138164. doi:10.1016/j.jhazmat.2025.138164
Tanca, A., Palomba, A., Pisanu, S., Deligios, M., Fraumene, C., Manghina, V., et al. (2014). A straightforward and efficient analytical pipeline for metaproteome characterization. Microbiome 2 (1), 49. doi:10.1186/s40168-014-0049-2
Tang, M., Li, S., Wei, L., Hou, Z., Qu, J., and Li, L. (2021). Do engineered nanomaterials affect immune responses by interacting with gut microbiota? Front. Immunology 12, 684605. doi:10.3389/fimmu.2021.684605
Tang, W., Zhang, X., Hong, H., Chen, J., Zhao, Q., and Wu, F. (2024). Computational nanotoxicology models for environmental risk assessment of engineered nanomaterials. Nanomaterials 14 (2), 155. doi:10.3390/nano14020155
Teng, Y., Luo, C., Qiu, X., Mu, J., Sriwastva, M. K., Xu, Q., et al. (2025). Plant-nanoparticles enhance anti-PD-L1 efficacy by shaping human commensal microbiota metabolites. Nat. Communications 16 (1), 1295. doi:10.1038/s41467-025-56498-2
Thomas, D. P., Zhang, J., Nguyen, N. T., and Ta, H. T. (2023). Microfluidic gut-on-a-chip: fundamentals and challenges. Biosensors 13 (1), 136. doi:10.3390/bios13010136
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. Journal 474 (11), 1823–1836. doi:10.1042/BCJ20160510
Triboulet, S., Aude-Garcia, C., Carrière, M., Diemer, H., Proamer, F., Habert, A., et al. (2013). Molecular responses of mouse macrophages to copper and copper oxide nanoparticles inferred from proteomic analyses. Mol. and Cell. Proteomics 12 (11), 3108–3122. doi:10.1074/mcp.M113.030742
Tripathi, S., Mahra, S., Tiwari, K., Rana, S., Tripathi, D. K., Sharma, S., et al. (2023). Recent advances and perspectives of nanomaterials in agricultural management and associated environmental risk: a review. Nanomaterials 13 (10), 1604. doi:10.3390/nano13101604
Trujillo-de Santiago, G., Lobo-Zegers, M. J., Montes-Fonseca, S. L., Zhang, Y. S., and Alvarez, M. M. (2018). Gut-microbiota-on-a-chip: an enabling field for physiological research. Microphysiological Systems 2, 7. doi:10.21037/mps.2018.09.01
Tu, Y., Li, P., Sun, J., Jiang, J., Dai, F., Li, C., et al. (2021). Remarkable antibacterial activity of reduced graphene oxide functionalized by copper ions. Adv. Funct. Mater. 31 (13), 2008018. doi:10.1002/adfm.202008018
Tufail, M. A., Iltaf, J., Zaheer, T., Tariq, L., Amir, M. B., Fatima, R., et al. (2022). Recent advances in bioremediation of heavy metals and persistent organic pollutants: a review. Sci. Total Environ. 850, 157961. doi:10.1016/j.scitotenv.2022.157961
Upadhayay, V. K., Chitara, M. K., Mishra, D., Jha, M. N., Jaiswal, A., Kumari, G., et al. (2023). Synergistic impact of nanomaterials and plant probiotics in agriculture: a tale of two-way strategy for long-term sustainability. Front. Microbiol. 14, 1133968. doi:10.3389/fmicb.2023.1133968
Utembe, W., Tlotleng, N., and Kamng'ona, A. W. (2022). A systematic review on the effects of nanomaterials on gut microbiota. Curr. Res. Microb. Sci. 3, 100118. doi:10.1016/j.crmicr.2022.100118
Vaccari, F., Zhang, L., Giuberti, G., Grasso, A., Bandini, F., Garcia-Perez, P., et al. (2023). The impact of metallic nanoparticles on gut fermentation processes: an integrated metabolomics and metagenomics approach following an in vitro digestion and fecal fermentation model. J. Hazard. Mater. 453, 131331. doi:10.1016/j.jhazmat.2023.131331
Valdés-Mas, R., Leshem, A., Zheng, D., Cohen, Y., Kern, L., Zmora, N., et al. (2025). Metagenome-informed metaproteomics of the human gut microbiome, host, and dietary exposome uncovers signatures of health and inflammatory bowel disease. Cell 188 (4), 1062–1083.e36. doi:10.1016/j.cell.2024.12.016
van Den Brule, S., Ambroise, J., Lecloux, H., Levard, C., Soulas, R., De Temmerman, P. J., et al. (2015). Dietary silver nanoparticles can disturb the gut microbiota in mice. Part. Fibre Toxicology 13 (1), 38. doi:10.1186/s12989-016-0149-1
Van Dong, P., Ha, C. H., Binh, L. T., and Kasbohm, J. (2012). Chemical synthesis and antibacterial activity of novel-shaped silver nanoparticles. Int. Nano Lett. 2 (1), 9. doi:10.1186/2228-5326-2-9
Vanlalveni, C., Ralte, V., Zohmingliana, H., Das, S., Anal, J. M. H., Lallianrawna, S., et al. (2024). A review of microbes mediated biosynthesis of silver nanoparticles and their enhanced antimicrobial activities. Heliyon 10 (11), e32333. doi:10.1016/j.heliyon.2024.e32333
Vera-Reyes, I., Vázquez-Núñez, E., Castellano, L. E., Bautista, D. I. A., Soto, J. H. V., and Valle-García, J. D. (2023). Biointeractions of plants–microbes–engineered nanomaterials. Physicochem. Interactions Engineered Nanoparticles Plants, 201–231. doi:10.1016/B978-0-323-90558-9.00001-2
Villamayor, N., Villaseñor, M. J., and Ríos, Á. (2023). Monitoring nanomaterials in food: a critical overview, perspectives, and challenges. Explor. Foods Foodomics 1 (1), 43–61. doi:10.37349/eff.2023.00005
Von Moos, N., and Slaveykova, V. I. (2014). Oxidative stress induced by inorganic nanoparticles in bacteria and aquatic microalgae–state of the art and knowledge gaps. Nanotoxicology 8 (6), 605–630. doi:10.3109/17435390.2013.809810
Wang, L., Hu, C., and Shao, L. (2017). The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. Journal Nanomedicine 12, 1227–1249. doi:10.2147/IJN.S121956
Wang, S., Alenius, H., El-Nezami, H., and Karisola, P. (2022). A new look at the effects of engineered ZnO and TiO2 nanoparticles: evidence from transcriptomics studies. Nanomaterials, 12 (8), 1247. doi:10.3390/nano12081247
Wang, W. L., Xu, S. Y., Ren, Z. G., Tao, L., Jiang, J. W., and Zheng, S. S. (2015). Application of metagenomics in the human gut microbiome. World Journal Gastroenterology WJG 21 (3), 803. doi:10.3748/wjg.v21.i3.803
Wang, X. L., Yu, N., Wang, C., Zhou, H. R., Wu, C., Yang, L., et al. (2022). Changes in gut microbiota structure: a potential pathway for silver nanoparticles to affect the host metabolism. ACS Nano 16 (11), 19002–19012. doi:10.1021/acsnano.2c07924
Wang, X., Cui, X., Wu, J., Bao, L., and Chen, C. (2023). Oral administration of silver nanomaterials affects the gut microbiota and metabolic profile altering the secretion of 5-HT in mice. J. Mater. Chem. B 11 (9), 1904–1915. doi:10.1039/d2tb02756a
Wang, X., Li, J., and Pan, X. (2024). How micro-/nano-plastics influence the horizontal transfer of antibiotic resistance genes-a review. Sci. Total Environ. 944, 173881. doi:10.1016/j.scitotenv.2024.173881
Wang, Z., Bergemann, C. M., Simonin, M., Avellan, A., Kiburi, P., and Hunt, D. E. (2024). Interactions shape aquatic microbiome responses to Cu and Au nanoparticle treatments in wetland manipulation experiments. Environ. Res. 252, 118603. doi:10.1016/j.envres.2024.118603
Wang, Z., Dalton, K. R., Lee, M., Parks, C. G., Beane Freeman, L. E., Zhu, Q., et al. (2023). Metagenomics reveals novel microbial signatures of farm exposures in house dust. Front. Microbiology 14, 1202194. doi:10.3389/fmicb.2023.1202194
Wilding, L. A., Bassis, C. M., Walacavage, K., Hashway, S., Leroueil, P. R., Morishita, M., et al. (2016). Repeated dose (28-day) administration of silver nanoparticles of varied size and coating does not significantly alter the indigenous murine gut microbiome. Nanotoxicology 10 (5), 513–520. doi:10.3109/17435390.2015.1078854
Wojciechowska, O., Costabile, A., and Kujawska, M. (2023). The gut microbiome meets nanomaterials: exposure and interplay with graphene nanoparticles. Nanoscale Adv. 5 (23), 6349–6364. doi:10.1039/D3NA00696D
Wu, S. Y., and Duncan, L. W. (2020). Recruitment of an insect and its nematode natural enemy by olfactory cues from a saprophytic fungus. Soil Biol. Biochem. 144, 107781. doi:10.1016/j.soilbio.2020.107781
Wu, S., Gaillard, J. F., and Gray, K. A. (2021). The impacts of metal-based engineered nanomaterial mixtures on microbial systems: a review. Sci. Total Environ. 780, 146496. doi:10.1016/j.scitotenv.2021.146496
Wu, Y., Cao, X., Du, H., Guo, X., Han, Y., McClements, D. J., et al. (2023). Adverse effects of titanium dioxide nanoparticles on beneficial gut bacteria and host health based on untargeted metabolomics analysis. Environ. Res. 228, 115921. doi:10.1016/j.envres.2023.115921
Xia, H., Chen, H., Cheng, X., Yin, M., Yao, X., Ma, J., et al. (2022). Zebrafish: an efficient vertebrate model for understanding role of gut microbiota. Mol. Med. 28 (1), 161. doi:10.1186/s10020-022-00579-1
Xiang, Y., Wen, H., Yu, Y., Li, M., Fu, X., and Huang, S. (2020). Gut-on-chip: recreating human intestine in vitro. J. Tissue Engineering 11, 2041731420965318. doi:10.1177/2041731420965318
Xie, Y., Wu, B., Zhang, X. X., Yin, J., Mao, L., and Hu, M. (2016). Influences of graphene on microbial community and antibiotic resistance genes in mouse gut as determined by high-throughput sequencing. Chemosphere 144, 1306–1312. doi:10.1016/j.chemosphere.2015.09.076
Xie, W., Liu, L., Xiong, Z., Cui, H., Cao, L., and Tang, Y. (2023). Exploring the effects of silver nanoparticles on the bacterial microbiota composition of normal human skin. Next Nanotechnol. 1, 100009. doi:10.1016/j.nxnano.2023.100009
Xiong, T., Yuan, X., Wang, H., Leng, L., Li, H., Wu, Z., et al. (2018). Implication of graphene oxide in Cd-contaminated soil: a case study of bacterial communities. J. Environmental Management 205, 99–106. doi:10.1016/j.jenvman.2017.09.067
Xu, J., Luo, X., Wang, Y., and Feng, Y. (2018). Evaluation of zinc oxide nanoparticles on lettuce (Lactuca sativa L.) growth and soil bacterial community. Environ. Sci. Pollut. Res. 25 (6), 6026–6035. doi:10.1007/s11356-017-0953-7
Xu, Y., Li, H., Li, X., and Liu, W. (2023). What happens when nanoparticles encounter bacterial antibiotic resistance? Sci. Total Environ. 876, 162856. doi:10.1016/j.scitotenv.2023.162856
Xuan, L., Ju, Z., Skonieczna, M., Zhou, P. K., and Huang, R. (2023). Nanoparticles-induced potential toxicity on human health: applications, toxicity mechanisms, and evaluation models. MedComm 4 (4), e327. doi:10.1002/mco2.327
Yadav, A., and Yadav, K. (2024). Exploring the effect of engineered nanomaterials on soil microbial diversity and functions: a review. J. Environ. Nanotechnol. 13 (1), 48–62. doi:10.13074/jent.2024.03.241503
Yadav, N., Garg, V. K., Chhillar, A. K., and Rana, J. S. (2023). Recent advances in nanotechnology for the improvement of conventional agricultural systems: a review. Plant Nano Biol. 4, 100032. doi:10.1016/j.plana.2023.100032
Yamini, V., Shanmugam, V., Rameshpathy, M., Venkatraman, G., Ramanathan, G., Garalleh, H. A., et al. (2023). Environmental effects and interaction of nanoparticles on beneficial soil and aquatic microorganisms. Environ. Res. 236, 116776.
Yan, J., Wang, D., Li, K., Chen, Q., Lai, W., Tian, L., et al. (2020). Toxic effects of the food additives titanium dioxide and silica on the murine intestinal tract: mechanisms related to intestinal barrier dysfunction involved by gut microbiota. Environ. Toxicol. Pharmacol. 80, 103485. doi:10.1016/j.etap.2020.103485
Yang, C., Tan, Y., Li, F., Wang, H., Lin, Y., Lu, F., et al. (2022). Intestinal microecology of mice exposed to TiO2 nanoparticles and bisphenol A. Foods 11 (12), 1696. doi:10.3390/foods11121696
Yin, J., Wang, Y., and Gilbertson, L. M. (2018). Opportunities to advance sustainable design of nano-enabled agriculture identified through a literature review. Environ. Sci. Nano 5 (1), 11–26. doi:10.1039/C7EN00766C
Young, J. C., Pan, C., Adams, R. M., Brooks, B., Banfield, J. F., Morowitz, M. J., et al. (2015). Metaproteomics reveals functional shifts in microbial and human proteins during a preterm infant gut colonization case. Proteomics 15 (20), 3463–3473. doi:10.1002/pmic.201400563
Yu, R., Ahmed, T., Jiang, H., Zhou, G., Zhang, M., Lv, L., et al. (2021). Impact of zinc oxide nanoparticles on the composition of gut microbiota in healthy and autism spectrum disorder children. Materials 14 (19), 5488. doi:10.3390/MA14195488
Yu, K., Chen, F., Yue, L., Luo, Y., Wang, Z., and Xing, B. (2020). CeO2 nanoparticles regulate the propagation of antibiotic resistance genes by altering cellular contact and plasmid transfer. Environ. Science and Technology 54 (16), 10012–10021. doi:10.1021/acs.est.0c01870
Yu, M., Wang, Q., Tao, W., Liu, G., Liu, W., Wang, L., et al. (2020). Interactions between arbuscular mycorrhizal fungi and soil properties jointly influence plant C, N, and P stoichiometry in west Lake, hangzhou. RSC Advances 10 (65), 39943–39953. doi:10.1039/D0RA08185J
Yuan, Q., and Liu, Y. (2023). Utilization of intestinal organoid models for assessment of micro/nano plastic-induced toxicity. Front. Environ. Sci. 11, 1285536. doi:10.3389/fenvs.2023.1285536
Zaman, W., Ayaz, A., and Puppe, D. (2025). Biogeochemical cycles in plant–soil systems: significance for agriculture, interconnections, and anthropogenic disruptions. Biology 14 (4), 433. doi:10.3390/biology14040433
Zhai, Y., Brun, N. R., Bundschuh, M., Schrama, M., Hin, E., Vijver, M. G., et al. (2018). Microbially-mediated indirect effects of silver nanoparticles on aquatic invertebrates. Aquat. Sci. 80 (4), 44. doi:10.1007/s00027-018-0594-z
Zhang, X., Ning, Z., Mayne, J., Moore, J. I., Li, J., Butcher, J., et al. (2016). MetaPro-IQ: a universal metaproteomic approach to studying human and mouse gut microbiota. Microbiome 4 (1), 31. doi:10.1186/s40168-016-0176-z
Zhang, X., Deeke, S. A., Ning, Z., Starr, A. E., Butcher, J., Li, J., et al. (2018). Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat. Communications 9 (1), 2873. doi:10.1038/s41467-018-05357-4
Zhang, T., Zhu, X., Guo, J., Gu, A. Z., Li, D., and Chen, J. (2021). Toxicity assessment of nano-ZnO exposure on the human intestinal microbiome, metabolic functions, and resistome using an in vitro colon simulator. Environ. Science and Technology 55 (10), 6884–6896. doi:10.1021/ACS.EST.1C00573
Zhang, J. H., Shi, J. Q., Chen, Z. J., and Jia, G. (2022). Effects of nano titanium dioxide on gut microbiota based on human digestive tract microecology simulation system in vitro. Beijing da xuexue bao. Yi xue ban= J. Peking Univ. Health Sci. 54 (3), 468–476. doi:10.19723/j.issn.1671-167X.2022.03.011
Zhang, H., Zheng, T., Wang, Y., Li, T., and Chi, Q. (2024). Multifaceted impacts of nanoparticles on plant nutrient absorption and soil microbial communities. Front. Plant Science 15, 1497006. doi:10.3389/fpls.2024.1497006
Zhang, L., Wu, L., Si, Y., and Shu, K. (2018). Size-dependent cytotoxicity of silver nanoparticles to azotobacter vinelandii: growth inhibition, cell injury, oxidative stress and internalization. PloS One 13 (12), e0209020. doi:10.1371/journal.pone.0209020
Zhao, S., Su, X., Wang, Y., Yang, X., Bi, M., He, Q., et al. (2020). Copper oxide nanoparticles inhibited denitrifying enzymes and electron transport system activities to influence soil denitrification and N2O emission. Chemosphere 245, 125394. doi:10.1016/j.chemosphere.2019.125394
Zhao, K., Zhao, Y., Wang, Y., Han, B., and Lian, M. (2024). Progress in antibacterial applications of nanozymes. Front. Chem. 12, 1478273. doi:10.3389/fchem.2024.1478273
Zhong, X., Li, J., Lu, F., Zhang, J., and Guo, L. (2022). Application of zebrafish in the study of the gut microbiome. Animal Models Exp. Med. 5 (4), 323–336. doi:10.1002/ame2.12227
Zhou, G., Yu, R., Ahmed, T., Jiang, H., Zhang, M., Lv, L., et al. (2021). Biosynthesis and characterization of zinc oxide nanoparticles and their impact on the composition of gut microbiota in healthy and attention-deficit hyperactivity disorder children. Front. Microbiology 12, 700707. doi:10.3389/FMICB.2021.700707
Zhu, K., Li, X., Chen, Y., Huang, Y., Yang, Z., Guan, G., et al. (2024). Recent advances on the spherical metal oxides for sustainable degradation of antibiotics. Coord. Chem. Rev. 510, 215813. doi:10.48550/arXiv.2403.17316
Keywords: engineered nanomaterials, microbiome, dysbiosis, antimicrobial resistance, ecosystem health, meta-omics technologies
Citation: Chowdhury A and Ghosh M (2025) Engineered nanomaterials and the microbiome: assessing disruptions in environmental and human microbial communities. Front. Nanotechnol. 7:1666431. doi: 10.3389/fnano.2025.1666431
Received: 15 July 2025; Accepted: 24 November 2025;
Published: 10 December 2025.
Edited by:
Sarmistha Saha, GLA University, IndiaReviewed by:
Minghui Li, Army Medical University, ChinaPradeep Mankodi, Maharaja Sayajirao University of Baroda, India
Copyright © 2025 Chowdhury and Ghosh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mayukh Ghosh, Z2hvc2gubWF5dWtoODdAZ21haWwuY29t
 Alonkrita Chowdhury
Alonkrita Chowdhury Mayukh Ghosh
Mayukh Ghosh
