- 1Laboratory of Parasitology, Department of Veterinary Medicine, S. Seifullin Kazakh Agrotechnical Research University, Astana, Kazakhstan
- 2Laboratory of Biodiversity and Genetic Resources, National Center for Biotechnology, Astana, Kazakhstan
- 3Institute of Parasitology, Justus Liebig University Giessen, Giessen, Germany
Introduction: Cryptosporidium spp. and Cystoisospora spp. are significant unicellular parasites that cause gastrointestinal infections in both humans and animals globally. Among these, Cryptosporidium felis and Cystoisospora felis are particularly important for feline health and pose potential zoonotic risks, especially for individuals with compromised immune systems. Kazakhstan, characterized by its diverse climate zones and an increasing population of pets, provides an excellent context for studying the epidemiology and genetic diversity of these parasites. In Kazakhstan, the mandatory registration of pets offers a valuable opportunity to explore the distribution and molecular characteristics of these parasites. This study focuses on the prevalence, genetic diversity, and zoonotic potential of Cryptosporidium and Cystoisospora from companion and shelter cats across five major cities in Kazakhstan.
Methods: Overall, from five cities, 1301 fecal samples were collected and studied. Samples were study by direct modified Sheather’s flotation technique was applied using a sugar solution. Samples were screened using the 18S rRNA gene for Cryptosporidium and the ITS-1 gene for Cystoisospora. Nucleotide sequences were aligned with the MUSCLE multiple sequence alignment program. Phylograms were constructed with the MEGA11 software using the Maximum Likelihood (ML) method.
Results and discussion: In total, we examined 1,301 fecal samples and found that 31 (2.4%) contained Cryptosporidium spp., including 10 identified as Cryptosporidium felis. Additionally, 121 samples (9.3%) tested positive for Cystoisospora felis. The studied Cryptosporidium parvum isolates obtained in this study belong to subtype IIdA15G1, which is dominant and clusters well with previously reported sequences from different countries on the gp60 gene. Shelter cats are more susceptible to these parasites, with a prevalence of 3.1% for Cryptosporidium and a notably higher rate of 19.0% for Cystoisospora. In contrast, companion cats showed lower rates, at 1.6% for Cryptosporidium and 5.1% for Cystoisospora. Our findings identified the species Cystoisospora felis, Cryptosporidium parvum, and Cryptosporidium felis, with a determined subtype of XIXa.
1 Introduction
Cryptosporidium spp. and Cystoisospora spp. are unicellular parasites of significant medical and veterinary importance, causing gastrointestinal infections in a wide range of hosts, including humans, domestic animals, and wildlife (Lappin, 2005; Morelli et al., 2021; Guo et al., 2022). These pathogens are particularly concerning due to their ability to induce severe diarrheal disease, their environmental resilience, and, in some cases, their zoonotic potential (Santin, 2020; Baptista et al., 2021). Among the affected species, domestic cats (Felis catus) serve as important hosts for Cryptosporidium felis (C. felis) and Cystoisospora felis, which can lead to clinical illness in feline populations and pose potential risks to human health, especially in immunocompromised individuals (Santin, 2020).
Enteric unicellular parasites are found worldwide and are primarily maintained in nature through fecal-oral transmission, which means that more cases tend to arise in crowded and unsanitary environments (Certad et al., 2017; Relat and O’Connor, 2020). Cryptosporidium spp. oocysts are immediately infectious upon being excreted by the host, whereas Cystoisospora spp. must first sporulate outside the host before they can spread (Matsubayashi et al., 2011; Dubey, 2018). Cryptosporidium is transmitted through the fecal-oral route via both direct and indirect means. Direct transmission occurs when an individual ingests oocysts from the feces of an infected host, while indirect transmission happens through the consumption of water or food contaminated with these oocysts. Over 20 different Cryptosporidium species have been linked to human cryptosporidiosis, with the most prevalent being C. hominis, C. parvum, C. meleagridis, C. felis, and C. canis (Checkley et al., 2015). Among these, C. felis primarily infects cats, making it a species adapted specifically to its host (Feng et al., 2018; Zahedi and Ryan, 2020). However, infections in humans from C. felis are frequently observed in developing nations (Caccio et al., 2002; Xiao, 2010; Cieloszyk et al., 2012; Beser et al., 2015; de Lucio et al., 2016), and there have been documented cases of zoonotic transmission of C. felis between pet cats and their owners (Power et al., 2011).
Similarly, Cystoisospora follows a comparable transmission pattern, but its oocysts must sporulate in the environment to become infectious (Matsubayashi et al., 2011; Dubey, 2018). Once ingested, its oocysts release sporozoites that invade intestinal cells, resulting in mucosal inflammation and malabsorptive diarrhea. Cystoisospora felis and Cystoisospora rivolta are obligate intracellular coccidian parasites that primarily infect domestic and wild felids (Shafiei et al., 2016). Current evidence suggests that Cystoisospora felis and C. rivolta are not transmissible to humans, with human cystoisosporiasis exclusively caused by the morphologically similar but genetically distinct C. belli (Lindsay, 2019; Guzmán-Lara et al., 2020). Although zoonotic risks appear negligible, the morphological similarity between Cystoisospora species warrants continued differentiation from truly zoonotic coccidian-like Cryptosporidium (Li et al., 2020).
In Kazakhstan, the epidemiology of these parasites in feline populations remains understudied, despite the country’s diverse climatic conditions and the increasing number of companion animals. Since September 2023, Kazakhstan has implemented a mandatory companion animal registration system, which reported approximately 88,514 registered cats by March 2025 (TAÑBA in digits, 2025). The aim of this study is to genetically characterize Cryptosporidium spp. and Cystoisospora felis infections in domestic and shelter cats across major cities in Kazakhstan, using 18S rRNA, gp60, and ITS-1 (Power et al., 2011; Shafiei et al., 2016; Rojas-Lopez et al., 2020; Koseoglu et al., 2022) gene markers. This study aims to determine the prevalence and distribution of these protozoan infections in feline populations across five major cities (Astana, Almaty, Shymkent, Oral, and Kostanay).
2 Materials and methods
2.1 Sample collection and analysis
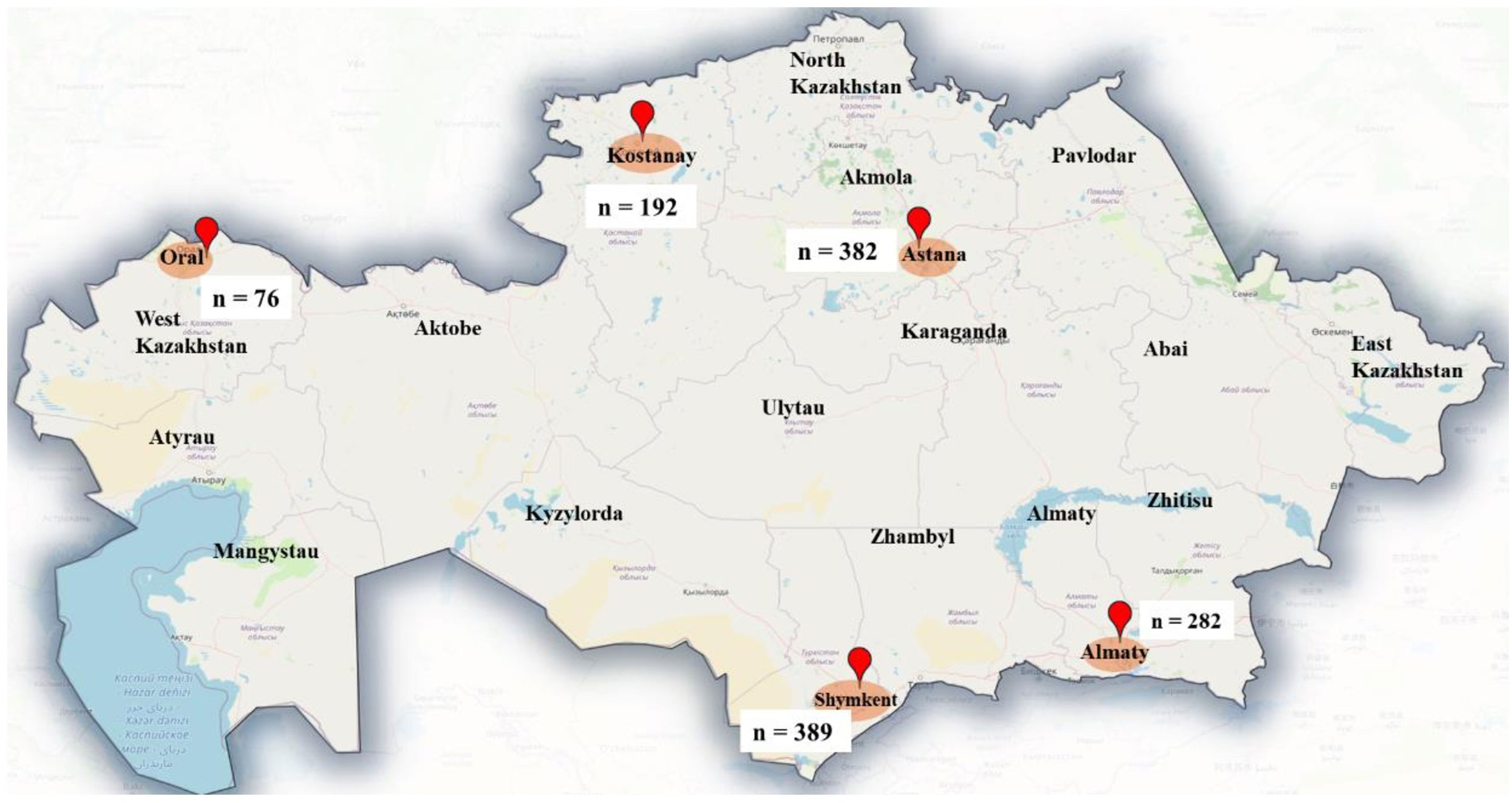
The distribution of sampling sites suggests a broad geographic coverage, spanning the western, northern, central, southern, and southeastern regions of Kazakhstan. This ensures a representative dataset for studying the prevalence of parasitic infections in cats across diverse climatic and ecological zones (Figure 1).
Overall, from five cities, 1301 samples were collected and studied. The samples were collected either during visits to one of the local veterinary clinics or upon the rescue of strays at shelters involved in the study. The fecal samples were sent to the Parasitology Laboratory of S. Seifullin Kazakh Agro Technical Research University in Astana for analysis within a span of 1 to 3 days, using coolers for transport. Sample collection occurred during the following intervals: from September 2023 to January 2025.
All fecal samples were visually inspected for the presence of cestode proglottids, after which the direct modified Sheather’s flotation technique was applied using a sugar solution (specific gravity: 1.3) as the flotation medium for worm eggs and coccidian oocysts (Kassai et al., 2013). The identification of the parasitic fecal stages relied on their morphological features (Kassai et al., 2013).
2.2 DNA extraction
Genomic DNA was isolated from approximately 150-200 mg of fecal matter using the QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany) following the instructions provided by the manufacturer; samples mixed with InhibitEX buffer were incubated for 10 minutes at 95°C. The extracted and purified DNA samples (50 µl) were stored at −20°C until further molecular analyses were performed.
2.3 Molecular identification
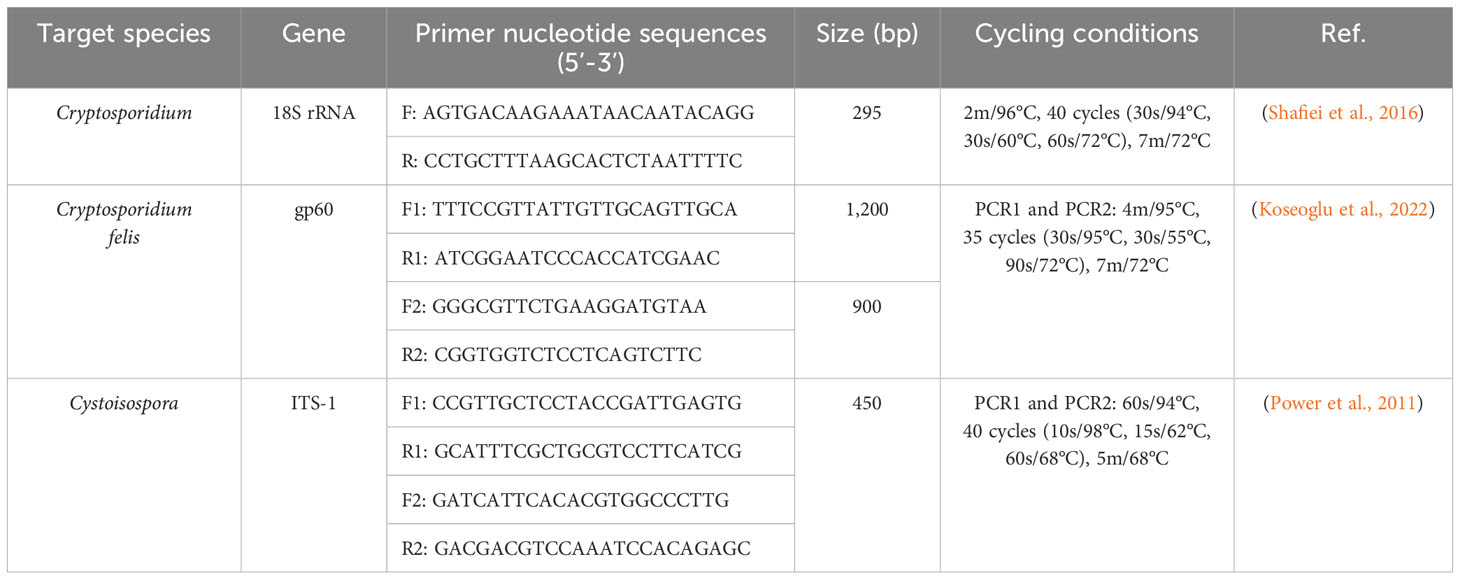
The molecular identification of Cryptosporidium and Cystoisospora was conducted by extracting DNA and amplifying with specific target genes from fecal samples using a thermocycler (Eppendorf Mastercycler, Hamburg Germany) (Table 1). Samples were screened using the 18S rRNA gene for Cryptosporidium and the ITS-1 gene for Cystoisospora (Power et al., 2011; Shafiei et al., 2016). The nested PCR was performed for the gp60 gene for C. felis (Rojas-Lopez et al., 2020; Koseoglu et al., 2022; Yun et al., 2023) to identify their subtypes. A negative template control sample (nuclease-free water) was included in each PCR to verify the absence of contamination with the PCR reaction mixture. Agarose gels (1.5%) were prepared in 1× TAE solution with 8 ng/µL ethidium bromide (Sigma, E1510). Electrophoresis was performed using 10 μL PCR products with a DirectLoad 100 bp Low ladder ready-to-use (Sigma, D3687-1VL) for 50 min at 120 V. The PCR-amplified target gene fragment was purified using a QIAquick PCR Purification Kit, (QIAGEN, Germany, Cat.: 28106), following the manufacturer’s protocols. Sequencing was performed according to the manual for Seq Studio Genetic Analyzer (Thermo Fisher Scientific Applied Biosystems). The resulting nucleotide sequences were visually checked by the Bioedit program version 7.0. The nucleotide sequences of the studied species were compared with other sequences in the NCBI gene bank database by using the BLAST options (http://blast.ncbi.nlm.nih.gov). The nucleotide sequences of the studied species were deposited in NCBI GenBank database.
2.4 Phylogenetic analysis
Nucleotide sequences were aligned with the MUSCLE multiple sequence alignment program for 18s rRNA, gp60 for Cryptosporidium species and ITS-1 genes for Cystoisospora isolates. Phylograms were constructed with the MEGA11 software (Tamura et al., 2021) using the Maximum Likelihood (ML) method.
2.5 Statistical analysis
The frequency of gastrointestinal unicellular parasite isolates detected from fecal samples was statistically compared using the chi-square test, with a 95% confidence interval. The P-value was calculated, with statistical significance established at P < 0.05.
3 Results
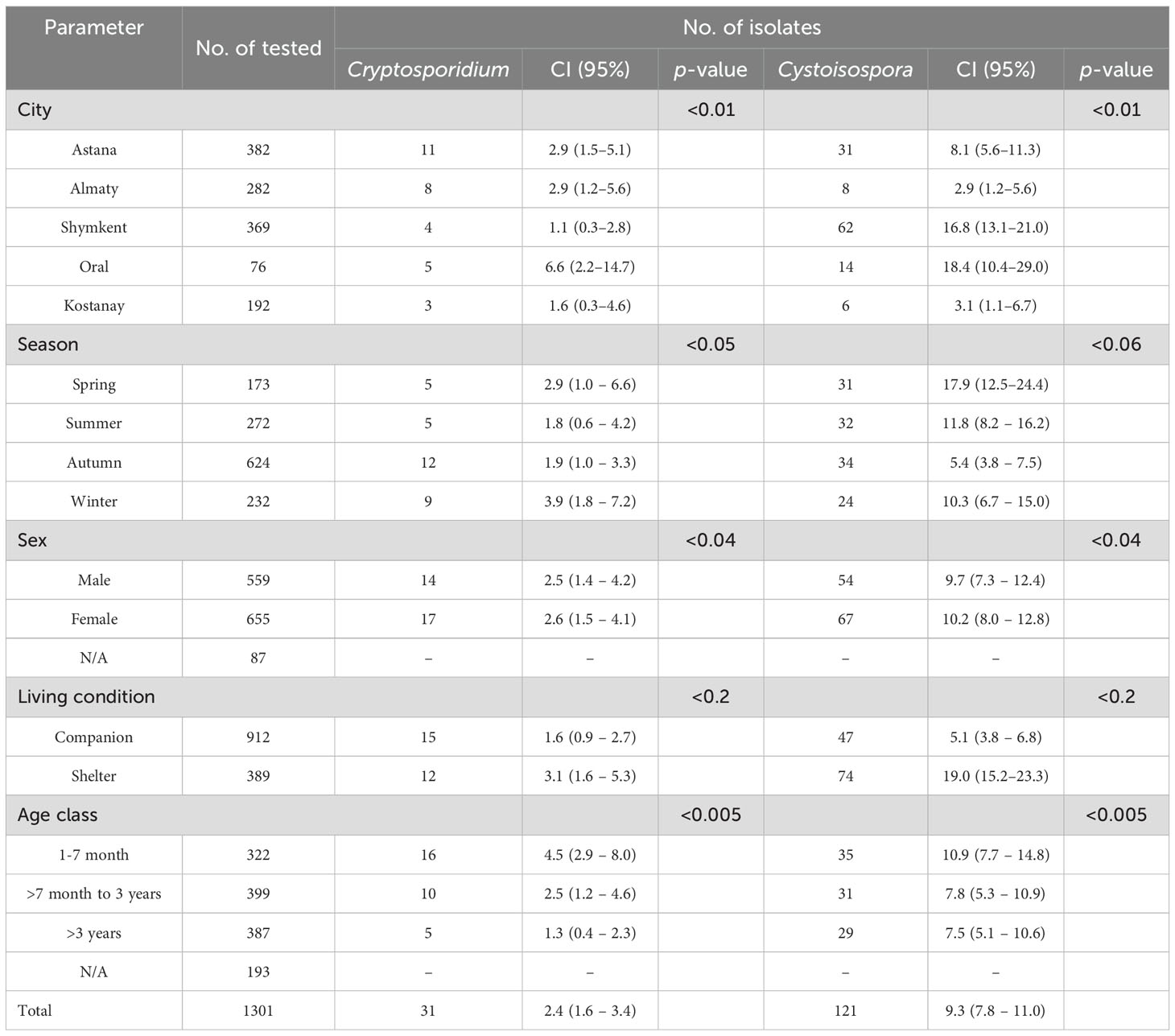
Among the 1301 cats examined, 912 (70.1%) were classified as companion cats, whereas 389 (29.9%) originated from shelters. The distribution of cats sampled across various cities included Shymkent (389), Astana (382), Almaty (282), Kostanay (192), and Oral (76) (Table 2).
When examining the gender ratio, the results revealed 655 males (54.0%) and 559 females (46.0%). Approximately a quarter of the cats (N = 387; 38.9%) were older than three years, 322 (32.1%) were aged between 1 and 7 months, and 399 (40.1%) fell within the range of 7 months to three years. Overall, the findings indicated that gastrointestinal protozoan parasites were detected in 152 out of 1301 (14.44%; 95% CI 1.0–13.55) fecal samples from the cats examined. Multiple protozoa parasites were detected in nine fecal samples, they were co-infected with Cystoisospora and Cryptosporidium (data not shown).
The coproscopic analysis revealed the presence of Cystoisospora felis in 94 out of 1301 samples and C. rivolta in 26 out of 1301 samples. Additionally, Cryptosporidium spp. was detected in 31 out of 1301 samples. As shown in Figure 2, observed C. felis large ovoid oocysts measuring 32-53 × 26-43 µm, light yellow or light brown. C. rivolta oval or ovoid, size 23-29 × 20-26 µm, light brown, may be colorless.
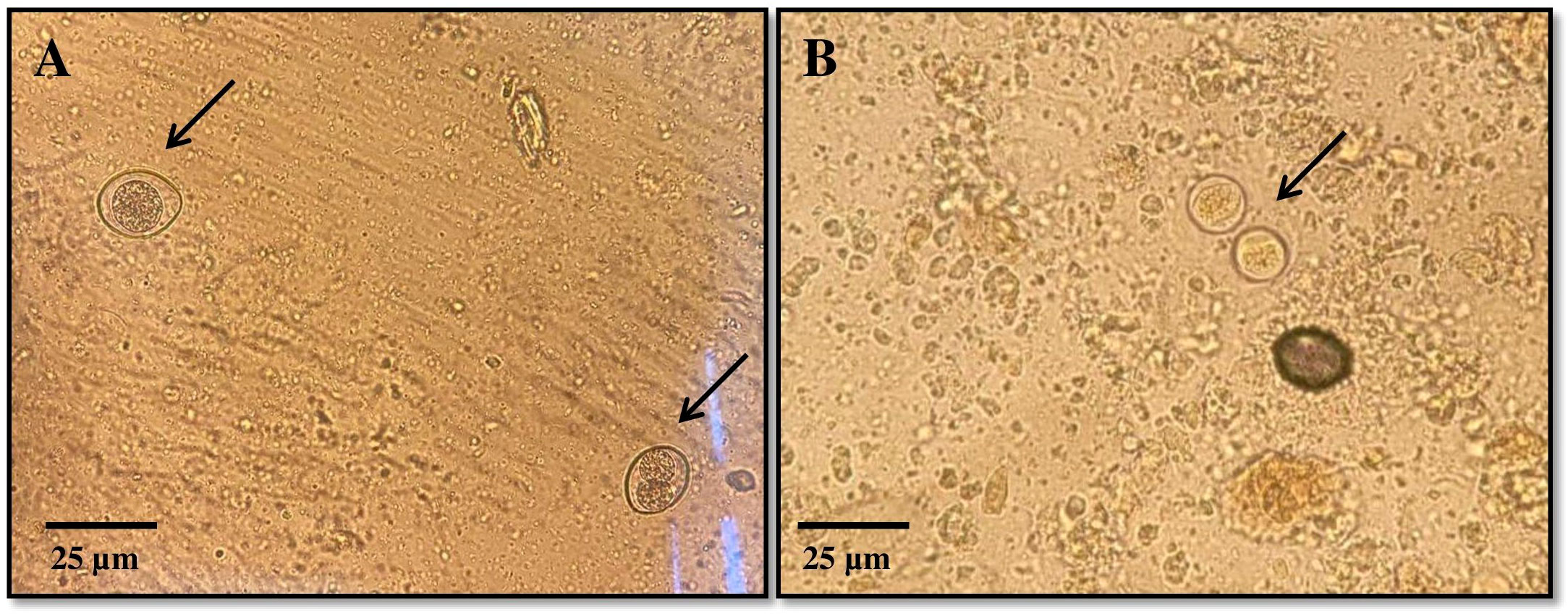
Figure 2. Representative images of Cystoisospora spp. oocysts detected during coprological analysis: (A) – Cystoisospora felis, (B) – Cystoisospora rivolta.
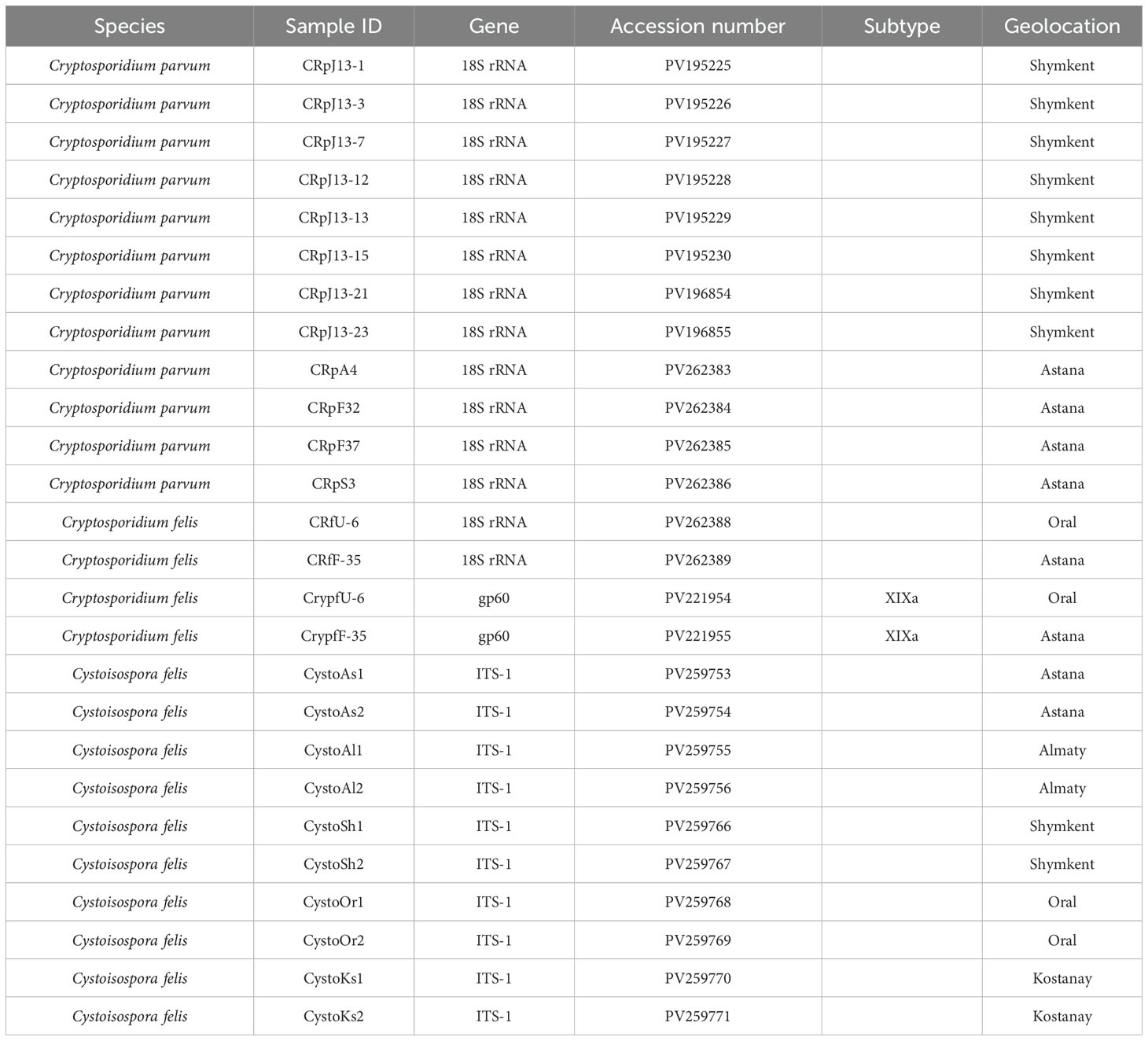
The isolates of C. felis identified through the 18S rRNA gene were sequenced using the gp60 gene to determine the subtype of C. felis. Sequence analysis of the gp60 gene revealed that 2 out of the 31 isolates were successfully identified as C. felis and revealed that isolates in the present study relate to subtype XIXa. Meanwhile, 12 out of 31 isolates were confirmed as C. parvum using the 18S rRNA gene. The 121 samples tested positive for Cystoisospora sp., with 10 of them identified as C. felis (Table 3).
Cystoisospora felis has been successfully identified in five cities across Kazakhstan. Notably, the cities of Almaty and Kostanay showed no positive samples for either C. felis or C. parvum, highlighting a significant absence of these parasites in those regions. The Figure 3 presents a phylogenetic tree illustrating the relationships among C. felis XIXa subtypes based on the gp60 gene sequence. The evolutionary history was inferred by using the Maximum Likelihood method and Tamura-Nei model (Tamura et al., 2021). This analysis involved 21 nucleotide sequences. There were a total of 1332 positions in the final dataset.
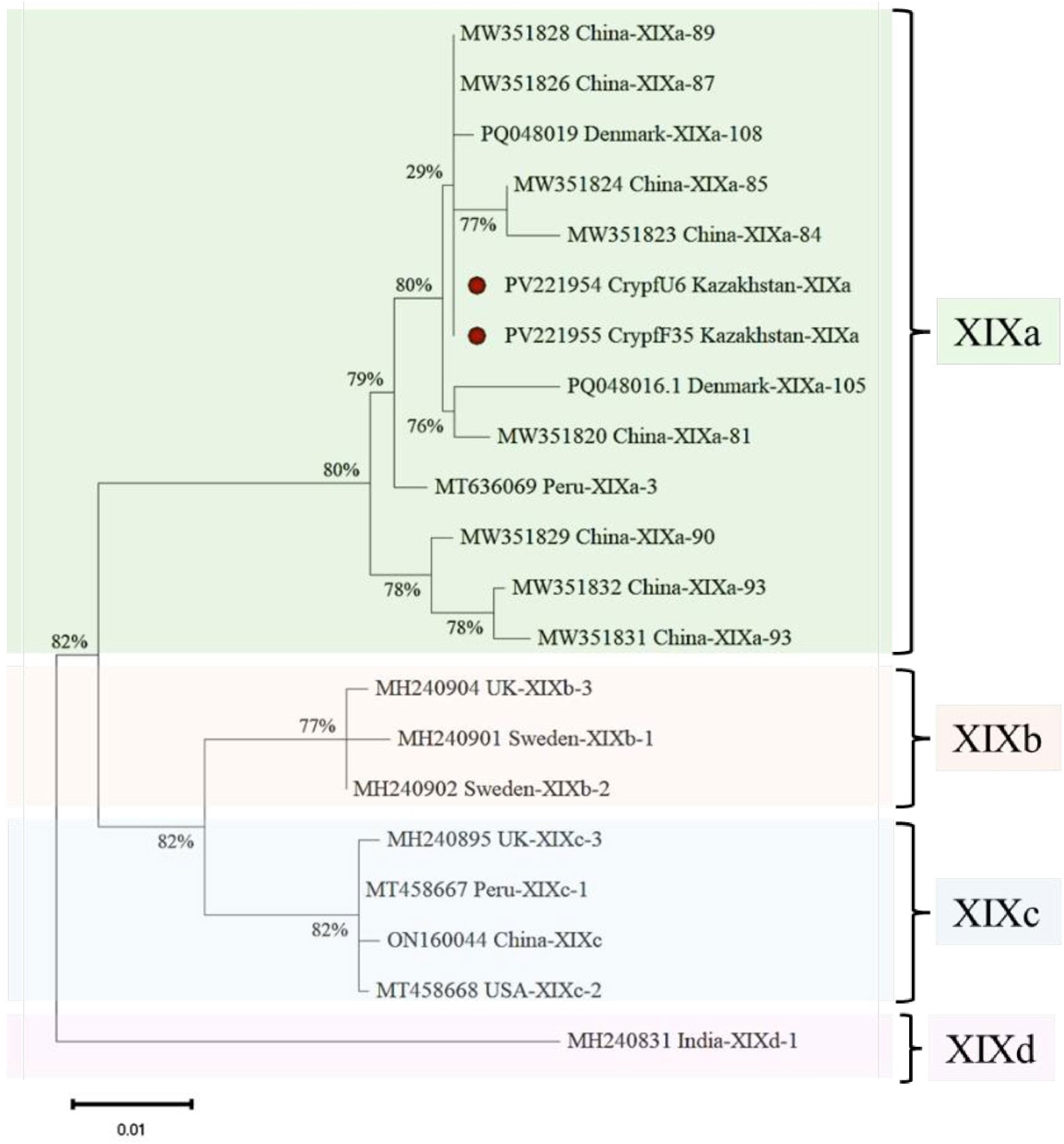
Figure 3. Phylogenetic analysis of XIXa subtypes of C. felis based on the gp60 gene sequence. The nucleotide sequences obtained in this study were compared with those of C. felis retrieved from GenBank.
Figure 4 demonstrates that the majority of the studied C. parvum isolates obtained in this study belong to subtype IIdA15G1, which is dominant and clusters well with previously reported sequences from different countries on the gp60 gene. This analysis involved 22 nucleotide sequences, the final dataset comprised a total of 1,035 positions.
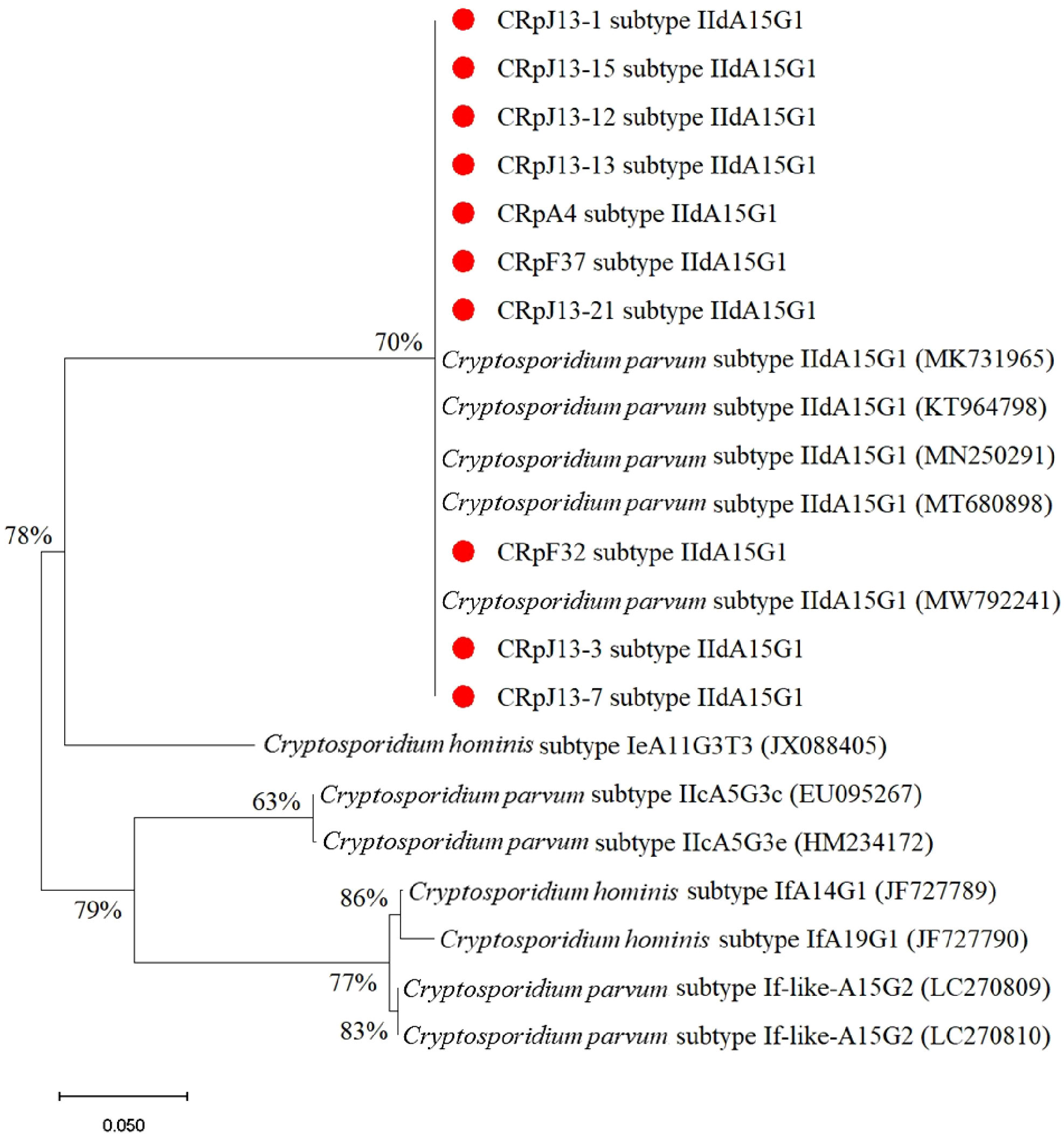
Figure 4. Phylogenetic analysis of subtypes of C. parvum based on the gp60 gene sequence. The nucleotide sequences obtained in this study were compared with other subtypes retrieved from GenBank. Red circles indicate new isolates obtained in this study.
C. parvum sequences form well-supported clades. The highest support (94%) is observed at one of the internal nodes within the C. parvum cluster. Cryptosporidium viatorum (JN846708) is a sister group to C. parvum with moderate bootstrap support (59%). Cryptosporidium andersoni (HQ259590) is more distantly related. C. felis sequences (PV262388 CRfU-6 and PV262389 CRfF-35) show 61% bootstrap support (Figure 5).
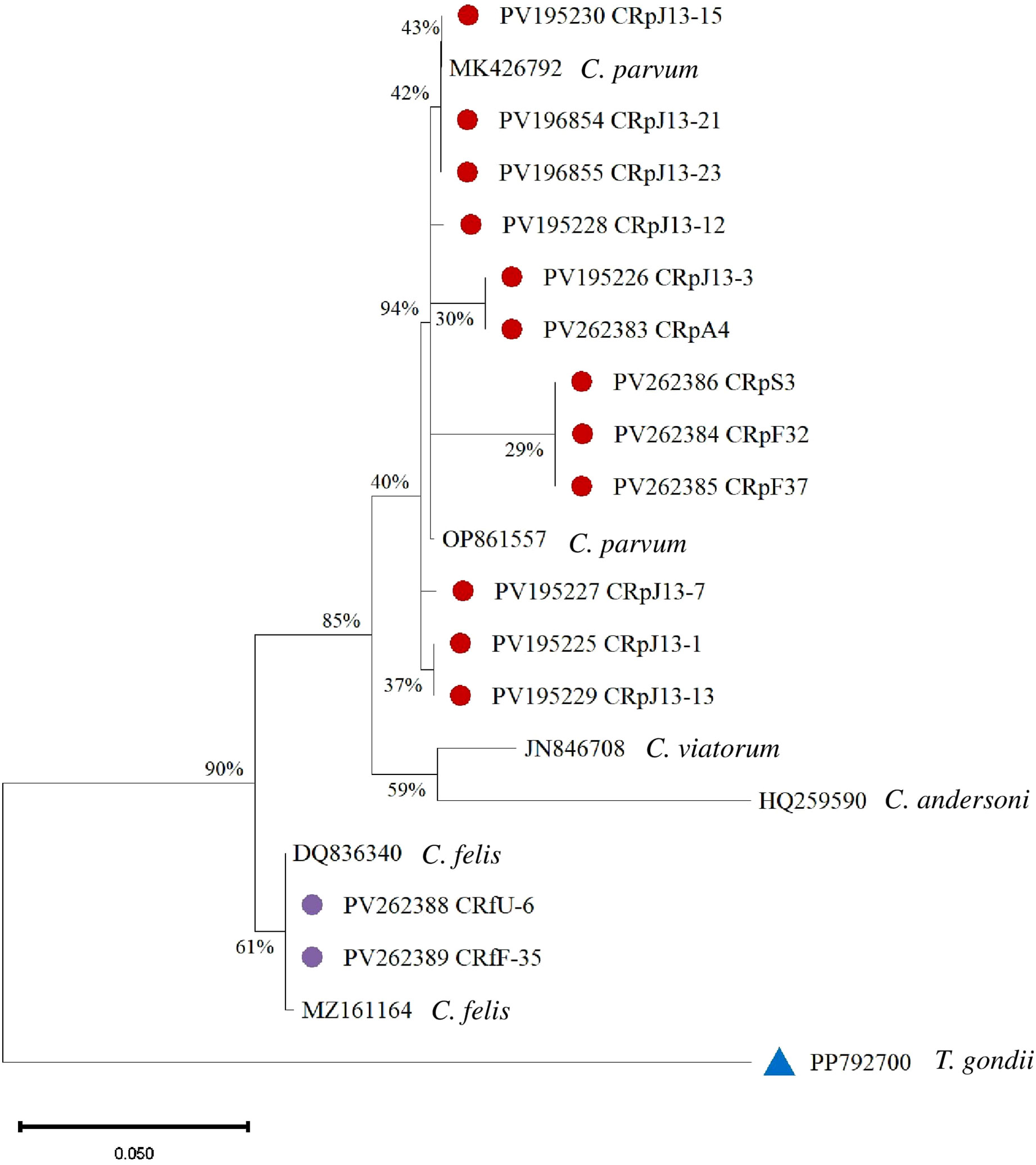
Figure 5. Phylogenetic analysis of Cryptosporidium based on the 18S rRNA gene. Isolates from this study are marked with red and purple circles, and a blue triangle represents the outgroup.
Figure 6 illustrates the sequencing and phylogenetic analyses of the amplification products of the ITS-1 region in Cystoisospora. A comparative analysis of 10 Cystoisospora felis samples was conducted against sequences available in GenBank, revealing a homology range of 86% to 96% compared to the reference isolates. Notably, other species within the Cystoisospora genus were distinctly categorized based on these findings. The phylogenetic tree was rooted using Cryptosporidium baileyi as an outgroup.
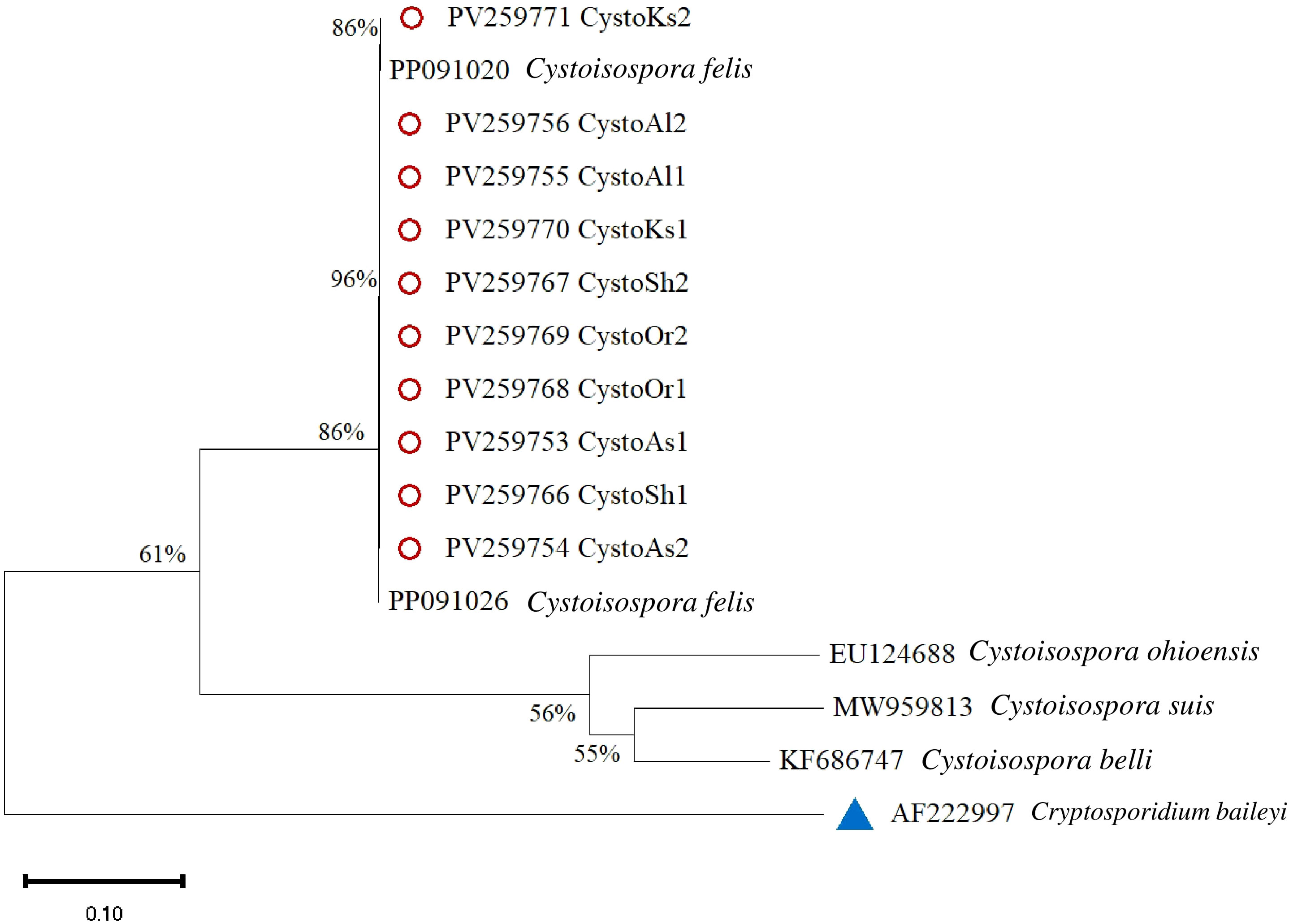
Figure 6. Phylogenetic analysis of Cystoisospora based on the ITS-1 gene. The phylogenetic tree was constructed using the maximum likelihood method with 1,000 bootstrap replicates. Divergence = 0.10. Red circles mark isolates from this study, and a blue triangle represents the outgroup.
4 Discussion
The present study provides significant insights into the molecular epidemiology of Cryptosporidium and Cystoisospora infections in domestic and shelter cats across five major cities in Kazakhstan (Figure 1). The identification of zoonotic species and conserved subtypes suggests both public health relevance and stable transmission patterns within local feline populations.
A total of 1301 fecal samples were examined, with 31 (2.4%) testing positive for Cryptosporidium spp. and 121 (9.3%) for Cystoisospora sp., and 10 samples were successfully sequenced and confirmed as Cystoisospora felis at the species level. Cryptosporidium parvum was detected in 12 cases, underscoring its zoonotic potential and public health significance. This pathogen poses a risk not only to immunocompromised individuals but also to immunocompetent populations, particularly children, the elderly, and those exposed to contaminated water or animal contact (Appelbee et al., 2005; Meng et al., 2021; Taghipour et al., 2021).
Meanwhile, the phylogenetic analysis of Cystoisospora felis confirmed its close relation to isolates previously reported in different geographical regions (Figure 5). The ITS-1 sequencing of Cystoisospora felis demonstrated substantial genetic similarity to isolates from each city, indicating a conserved genetic lineage within this species.
The study also revealed key epidemiological patterns. Shelter cats exhibited a higher prevalence of both Cryptosporidium (3.1%) and Cystoisospora (19.0%) compared to companion cats (1.6% and 5.1%, respectively), likely due to increased exposure to contaminated environments and stress-related immunosuppression. Additionally, younger cats (1-7 months) had a significantly higher infection rate for Cryptosporidium (4.5%) and Cystoisospora (10.9%) than older age groups, underscoring the susceptibility of juvenile felines. Seasonal variations also played a role, with winter and spring showing the highest infection rates, potentially due to the increased environmental persistence of oocysts in colder temperatures. Environmental studies confirm that oocyst survival is significantly longer in soils and waters kept at lower temperatures (Peng et al., 2008; Li et al., 2010). Epidemiological data in cats show a higher prevalence in winter, reinforcing how seasonal conditions influence transmission (Armon et al., 2016).
A global meta-analysis by Taghipour et al. (2021) reported an overall prevalence of Cryptosporidium in cats at 4.0%, with higher rates in shelter and stray populations compared to household pets, aligning with the elevated rates seen in shelter cats in Kazakhstan (Taghipour et al., 2021). In South Korea, Yun et al. (2023) found Cryptosporidium spp. in 3.6% of cats, with Cystoisospora spp. detected in 7.6%, similar to the Kazakhstan findings for Cystoisospora felis. Additionally, the prevalence of Cryptosporidium is reported to be 6.5% in shelter cats versus 1.8% in pet cats, with higher rates observed in animals under one year of age (Yun et al., 2023), which confirms our findings in this work. Mendoza and Otranto (2023) also emphasized that younger animals are more vulnerable due to immature immune systems and shelter environments pose a greater risk for protozoan infections due to increased exposure and stress-immunosuppression (Mendoza and Otranto, 2023).
In this study, 12 case of C. parvum infection was detected in stray and companion cats. The presence of C. parvum in domestic cats poses potential zoonotic risks, especially for individuals with compromised immune systems (Jiang et al., 2020). Molecular characterization of studied C. felis isolates (2/14) provided critical insights into the genetic structure of the detected parasites. Notably, the C. felis isolates identified in this study belonged to the XIXa subtype, which has been reported globally, reinforcing the genetic homogeneity of this subtype (Lucio-Forster et al., 2010; Xiao and Feng, 2017; Rojas-Lopez et al., 2020; Mendoza and Otranto, 2023). The gp60 subtyping of C. felis revealed sequence variations that align with global reports, contributing to the growing database of Cryptosporidium genetic diversity (de Oliveira et al., 2021). Of the 12 Cryptosporidium isolates analyzed in this study, 10 were successfully sequenced, and all were identified as belonging to the IIdA15G1 subtype. As illustrated in Figure 4, these isolates cluster within the C. parvum IIdA15G1 clade, which also includes reference sequences retrieved from GenBank (accession numbers: MK731965, KT964798, MN250291, MT680898, MW792241). The bootstrap support for this clade ranges from 70% to 78%, reflecting a robust phylogenetic relationship among isolates of this subtype. In contrast, reference isolates of the C. hominis lineage comprising subtypes IeA11G3T3, IfA14G1, and IfA19G1 form a clearly distinct and well-supported branch (bootstrap up to 86%), demonstrating clear genetic separation from C. parvum. Additionally, the IIcA5G3c subtype (EU095267, HM234172) and the If-like subtypes (LC270809, LC270810) each form discrete phylogenetic lineages, further underscoring the genetic heterogeneity within C. parvum.
These findings underscore the importance of continuous surveillance and molecular monitoring of Cryptosporidium and Cystoisospora infections in felines, particularly in regions with close human-animal interactions. Given the zoonotic potential of C. parvum, targeted public health interventions, including improved hygiene practices and regular veterinary screenings, are essential to mitigate transmission risks. Future studies should explore the role of additional host factors and environmental conditions influencing infection dynamics, as well as assess the broader public health implications of C. felis in humans.
Overall, this study contributes to the understanding of unicellular parasite infections in cats in Kazakhstan, emphasizing the need for integrated One Health approaches to monitor and control these infections in both animal and human populations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal studies were approved by Animal Ethics Committee of the S. Seifullin Kazakh Agrotechnical University (extract from protocol No. 2 dated November 03, 2022). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
LL: Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Formal analysis, Methodology, Writing – original draft, Resources, Data curation. RU: Writing – original draft, Software, Visualization, Data curation, Validation, Methodology, Formal analysis. NM: Data curation, Visualization, Formal analysis, Validation, Writing – original draft. VY: Formal analysis, Visualization, Data curation, Software, Writing – original draft. AA: Formal analysis, Data curation, Writing – original draft, Visualization, Software. AS: Visualization, Software, Formal Analysis, Writing – original draft, Data curation, Methodology, Validation. CH: Data curation, Methodology, Conceptualization, Writing – review & editing. VK: Data curation, Supervision, Writing – review & editing, Methodology, Conceptualization, Resources, Writing – original draft, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan AR19679420 (The study of the genetic diversity of zoonotic parasites of cats circulating in Kazakhstan).
Acknowledgments
The authors would like to express their sincere gratitude to the staff of veterinary clinics and shelters for their assistance in collecting the samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Appelbee A. J., Thompson R. C., and Olson M. E. (2005). Giardia and Cryptosporidium in mammalian wildlife–current status and future needs. Trends Parasitol. 21, 370–376. doi: 10.1016/j.pt.2005.06.004
Armon R., Gold D., Zuckerman U., and Kurzbaum E. (2016). Environmental aspects of Cryptosporidium. J. Vet. Med. Res. 3, 1048.
Baptista R. P., Cooper G. W., and Kissinger J. C. (2021). Challenges for Cryptosporidium population studies. Genes (Basel) 12, 894. doi: 10.3390/genes12060894
Beser J., Toresson L., Eitrem R., Troell K., Winiecka-Krusnell J., and Lebbad M. (2015). Possible zoonotic transmission of Cryptosporidium felis in a household. Infect. Ecol. Epidemiol 5, 28463. doi: 10.3402/iee.v5.28463
Caccio S., Pinter E., Fantini R., Mezzaroma I., and Pozio E. (2002). Human infection with Cryptosporidium felis: Case report and literature review. Emerg. Infect. Dis. 8, 85–86. doi: 10.3201/eid0801.010269
Certad G., Viscogliosi E., Chabé M., and Cacciò S. M. (2017). Pathogenic mechanisms of Cryptosporidium and Giardia. Trends Parasitol. 33, 561–576. doi: 10.1016/j.pt.2017.02.006
Checkley W., White A. C., Jaganath D., Arrowood M. J., Chalmers R. M., Chen X. M., et al. (2015). A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 15, 85–94. doi: 10.1016/S1473-3099(14)70772-8
Cieloszyk J., Goni P., Garcia A., Remacha M. A., Sanchez E., and Clavel A. (2012). Two cases of zoonotic cryptosporidiosis in Spain by the unusual species Cryptosporidium ubiquitum and Cryptosporidium felis. Enferm Infecc Microbiol. Clin. 30, 549–551. doi: 10.1016/j.eimc.2012.04.011
de Lucio A., Merino F. J., Martinez-Ruiz R., Bailo B., Aguilera M., Fuentes I., et al. (2016). Molecular genotyping and sub-genotyping of Cryptosporidium spp. isolates from symptomatic individuals attending two major public hospitals in Madrid, Spain. Infect. Genet. Evol. 37, 49–56. doi: 10.1016/j.meegid.2015.10.026
de Oliveira A. G. L., Sudré A. P., Bergamo do Bomfim T. C., and Santos H. L. C. (2021). Molecular characterization of Cryptosporidium spp. in dogs and cats in the city of Rio de Janeiro, Brazil, reveals potentially zoonotic species and genotype. PLoS One 16, e0255087. doi: 10.1371/journal.pone.0255087
Dubey J. P. (2018). A review of Cystoisospora felis and C. rivolta-induced coccidiosis in cats. Vet. Parasitol. 263, 34–48. doi: 10.1016/j.vetpar.2018.09.016
Feng Y., Ryan U. M., and Xiao L. (2018). Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 34, 997–1011. doi: 10.1016/j.pt.2018.07.009
Guo Y., Ryan U., Feng Y., and Xiao L. (2022). Emergence of zoonotic Cryptosporidium parvum in China. Trends Parasitol. 38, 335–343. doi: 10.1016/j.pt.2021.12.002
Guzmán-Lara M. D., Kruth P. S., Rangel-Díaz J., Juárez-Estrada M. A., Soriano-Vargas E., and Barta J. R. (2020). Cystoisospora felis infection in a captive jaguar cub (Panthera onca) in Michoacán, México. Vet. Parasitol. Reg. Stud. Rep. 19, 100371. doi: 10.1016/j.vprsr.2020.100371
Jiang W., Roellig D. M., Lebbad M., Beser J., Troell K., Guo Y., et al. (2020). Subtype distribution of zoonotic pathogen Cryptosporidium felis in humans and animals in several countries. Emerg. Microbes Infect. 9, 2446–2454. doi: 10.1080/22221751.2020.1840312
Kassai T., Deplazes P., Eckert J., von Samson-Himmelstjerna G., and Zahner H. (2013). Lehrbuch der Parasitologie für die Tiermedizin [Textbook of Parasitology for Veterinary Medicine]. Third Edition. Acta Vet. Hung 61, 147–148. doi: 10.1556/AVet.2012.062
Koseoglu A. E., Can H., Karakavuk M., Güvendi M., Değirmenci Döşkaya A., Manyatsi P. B., et al. (2022). The study investigates the molecular prevalence and subtyping of Cryptosporidium spp. in fecal samples from stray cats in İzmir, Turkey. BMC Vet. Res. 18, 89. doi: 10.1186/s12917-022-03190-y
Lappin M. R. (2005). Enteric protozoal diseases. The Veterinary clinics of North America. Small Anim. Pract. 35, 81–88. doi: 10.1016/j.cvsm.2004.08.004
Li X., Atwill E. R., Dunbar L. A., and Tate K. W. (2010). Effect of daily temperature fluctuation during the cool season on the infectivity of Cryptosporidium parvum. Appl. Environ. Microbiol. 76, 989–993. doi: 10.1128/AEM.02103-09
Li J., Wang Z., Karim M. R., and Zhang L. (2020). Detection of human intestinal protozoan parasites in vegetables and fruits: a review. Parasit Vectors 13, 380. doi: 10.1186/s13071-020-04255-3
Lindsay D. S. (2019). Cystoisospora species insights from development in vitro. Front. Vet. Sci. 5. doi: 10.3389/fvets.2018.00335
Lucio-Forster A., Griffiths J. K., Cama V. A., Xiao L., and Bowman D. D. (2010). Minimal zoonotic risk of cryptosporidiosis from pet dogs and cats. Trends Parasitol. 26, 174–179. doi: 10.1016/j.pt.2010.01.004
Matsubayashi M., Carreno R. A., Tani H., Yoshiuchi R., Kanai T., Kimata I., et al. (2011). Phylogenetic identification of Cystoisospora spp. from dogs, cats, and raccoon dogs in Japan. Vet. Parasitol. 176, 270–274. doi: 10.1016/j.vetpar.2010.11.008
Mendoza R. and Otranto D. (2023). Zoonotic parasites associated with predation by dogs and cats. Parasit Vectors 16, 55. doi: 10.1186/s13071-023-05670-y
Meng X. Z., Li M. Y., Lyu C., Qin Y. F., Zhao Z. Y., Yang X. B., et al. (2021). The global prevalence and risk factors of Cryptosporidium infection among cats during 1988-2021: A systematic review and meta-analysis. Microb. Pathog. 158, 105096. doi: 10.1016/j.micpath.2021.105096
Morelli S., Diakou A., Di Cesare A., Colombo M., and Traversa D. (2021). Canine and feline parasitology: analogies, differences, and relevance for human health. Clin. Microbiol. Rev. 34, e0026620. doi: 10.1128/CMR.00266-20
Peng X., Murphy T., and Holden N. M. (2008). Evaluation of the effect of temperature on the die-off rate for Cryptosporidium parvum oocysts in water, soils, and feces. Appl. Environ. Microbiol. 74, 7101–7107. doi: 10.1128/AEM.01442-08
Power M. L., Holley M., Ryan U. M., Worden P., and Gillings M. R. (2011). Identification and differentiation of Cryptosporidium species by capillary electrophoresis single-strand conformation polymorphism. FEMS Microbiol. Lett. 314, 34–41. doi: 10.1111/j.1574-6968.2010.02134.x
Relat R. M. B. and O’Connor R. M. (2020). Cryptosporidium: host and parasite transcriptome in infection. Curr. Opin. Microbiol. 58, 138–145. doi: 10.1016/j.mib.2020.09.012
Rojas-Lopez L., Elwin K., Chalmers R. M., Enemark H. L., Beser J., and Troell K. (2020). Development of a gp60-subtyping method for Cryptosporidium felis. Parasit Vectors 13, 39. doi: 10.1186/s13071-020-3906-9
Santin M. (2020). Cryptosporidium and Giardia in ruminants. Vet. Clin. North Am. Food Anim. Pract. 36, 223–238. doi: 10.1016/j.cvfa.2019.11.005
Shafiei R., Najjari M., Kargar Kheirabad A. K., and Hatam G. (2016). Severe diarrhea due to Cystoisospora belli infection in an HTLV-1 woman. Iran J. Parasitol. 11, 121–125.
Taghipour A., Khazaei S., Ghodsian S., Shajarizadeh M., Olfatifar M., Foroutan M., et al. (2021). Global prevalence of Cryptosporidium spp. in cats: A systematic review and meta-analysis. Res. Vet. Sci. 137, 77–85. doi: 10.1016/j.rvsc.2021.04.015
Tamura K., Stecher G., and Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
TAÑBA in digits (2025).Animal accounting information system. Available online at: https://tanba.kezekte.kz/en/tanba-public/stat (Accessed April 03, 2025).
Xiao L. (2010). Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 124, 80–89. doi: 10.1016/j.exppara.2009.03.018
Xiao L. and Feng Y. (2017). Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 8-9, 14–32. doi: 10.1016/j.fawpar.2017.09.002
Yun C. S., Moon B.-Y., Lee K., Kang S. M., Ku B.-K., and Hwang M.-H. (2023). The detection and phylogenetic characterization of Cryptosporidium, Cystoisospora, and Giardia duodenalis of cats in South Korea. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1296118
Keywords: Cryptosporidium parvum, gp60 subtyping, molecular epidemiology, zoonotic parasites, Kazakhstan
Citation: Lider L, Uakhit R, Manapov N, Yerzhanova V, Andreyev A, Smagulova A, Hermosilla C and Kiyan V (2025) Molecular genetic characterization of Cryptosporidium and Cystoisospora protozoan infections in cats from large cities of Kazakhstan. Front. Parasitol. 4:1608542. doi: 10.3389/fpara.2025.1608542
Received: 16 April 2025; Accepted: 30 June 2025;
Published: 14 July 2025.
Edited by:
Andrea Dellarupe, National Scientific and Technical Research Council (CONICET), ArgentinaReviewed by:
Aleksandra Uzelac, University of Belgrade, SerbiaMousa Tavassoli, Urmia University, Iran
Beatriz Cancino-Faure, Universidad Católica del Maule, Chile
Copyright © 2025 Lider, Uakhit, Manapov, Yerzhanova, Andreyev, Smagulova, Hermosilla and Kiyan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vladimir Kiyan, dnNraXlhbkBnbWFpbC5jb20=
 Lyudmila Lider1
Lyudmila Lider1 Rabiga Uakhit
Rabiga Uakhit Alexandr Andreyev
Alexandr Andreyev Ainura Smagulova
Ainura Smagulova Carlos Hermosilla
Carlos Hermosilla Vladimir Kiyan
Vladimir Kiyan