- Department of Biology, College of Life Sciences, China Jiliang University, Hangzhou, China
The takeout family genes encode relatively small proteins that are related to olfaction and are regulated by juvenile hormone (JH). The takeout genes modulate various physiological processes, such as behavioral plasticity in the migratory locust Locusta migraloria and feeding and courtship behaviors in Drosophila. Therefore, to understand the regulatory mechanism of these physiological processes, it is important to study the expressions of the takeout genes that are regulated by JH signaling. We used quantitative real-time PCR (qRTPCR) to study the role of JH signaling in the regulation of the takeout family genes in the brown planthopper Nilaparvata lugens (N. lugens) through the application of Juvenile hormone III (JHIII) and the down-regulation of key genes in the JH signaling pathway. The topical application of JHIII induced the expressions of most of the takeout family genes, and their expressions decreased 2 and 3 days after the JHIII application. Down-regulating the brown planthopper JH receptor NlMethoprene-tolerant (NlMet) and its interacting partners, NlTaiman (NlTai) and Nlß-Ftz-F1 (Nlß-Ftz), through RNAi, exhibited distinct effects on the expressions of the takeout family genes. The down-regulation of NlMet and NlKrüppel-homolog 1 (NlKr-h1) increased the expressions of the takeout family genes, while the down-regulation of the Met interacting partners NlTai and Nlß-Ftz decreased the expressions of most of the takeout family genes. This work advanced our understanding of the molecular function and the regulatory mechanism of JH signaling.
Introduction
The takeout family genes encode relatively small proteins that are related to olfaction (Dauwalder et al., 2002; Saito et al., 2006; Hagai et al., 2007). Since the first characterization of the takeout gene in Drosophila melanogaster (Fujikawa et al., 2006), homologs of takeout have been identified from a broad range of insect species, including Phormia regina (Fujikawa et al., 2006), Manduca sexta (Du et al., 2003), Bombyx mori (Saito et al., 2006), Apis mellifera (Hagai et al., 2007), Reticulitermes flavipes (Dauwalder et al., 2002), and Locusta migraloria (Guo et al., 2011). The migratory locust Locusta migraloria takeout modulates behavioral plasticity (Guo et al., 2011), i.e., the switch between attraction and repulsion during the phase transition (Guo et al., 2011). The takeout gene was found to be regulated by the circadian rhythm and affects feeding behavior (So et al., 2000; Meunier et al., 2007), locomotion (Meunier et al., 2007), and male courtship behavior (Dauwalder et al., 2002) in D. melanogaster. Takeout is also involved in the trail-following behavior of the termite Reticulitermes flavipes (Dauwalder et al., 2002). The expressions of the takeout genes are usually male biased (Hagai et al., 2007; Vanaphan et al., 2012) and are regulated by age and nutrition (Du et al., 2003; Hagai et al., 2007). A circadian transcription factor PAR domain protein 1 (Pdp1ε) mediated the regulation of takeout by the circadian rhythm (Dauwalder et al., 2002). The expression of takeout is usually regulated by a crucial hormone in insects, Juvenile hormone (JH; Du et al., 2003; Hagai et al., 2007). However, the regulatory mechanism of takeout expression by JH remained unclear.
JH is secreted by the corpora allata (CA) and belongs to a type of sesquiterpenoid and regulates development, reproduction, polyphenism (a special case of phenotypic plasticity), and behaviors, such as feeding and mating (Jindra et al., 2013). The signal transduction pathway of JH is initiated by the release of the JH ligand, followed by binding to the intracellular receptor Methoprene-tolerant (Met; Bernardo and Dubrovsky, 2012; Jindra et al., 2013) through an interaction between Met and Taiman (Tai), which is an EcR coactivator (Zhu et al., 2006; Li et al., 2011, 2014), possibly also through an interaction between Met and ß-Ftz-F1 (Zhu et al., 2006; Yoo et al., 2011; Bernardo and Dubrovsky, 2012), leading to transcriptional changes of downstream genes and the regulation of developmental and physiological processes (Truman and Riddiford, 2002; Belles et al., 2005; Flatt et al., 2005). JH induced the transcription of Kr-h1 through the binding of the Met-Tai complex to the E-Box at the 5′ of the Kr-h1 gene in the mosquito Aedes aegypti (Zhu et al., 2010; Li et al., 2011, 2014). Works in Tribolium also indicated that the function of Kr-h1 is dependent on the JH receptor Met (Minakuchi et al., 2009). Consistently, we previously showed that the brown planthopper Kr-h1 is induced by JH or its mimics (Jin et al., 2014).
The brown planthopper, Nilaparvata lugens (N. lugens), which is one of the most important insect pests in rice production, exhibits polyphenism, and has the long wing and short wing forms. The long wing form is migratory, and the short wing form is reproductive. Previous studies have shown that the wing form of brown planthopper is regulated by JH and the density and developmental stage of the rice plant (Kisimoto, 1956, 1965; Iwanaga and Tojo, 1986; Ayoade et al., 1999; Bertuso et al., 2002). More recently, it was found that the wing form of the brown planthopper is regulated by two alternative receptors in the insulin signaling pathway and the JNK signaling pathway (Xu et al., 2015; Lin et al., 2016a,b). Interestingly, we found that wounding also affects the wing form through the regulation of the transcription factor Foxo (Lin et al., 2016c).
The regulation of target genes by JH signaling is bidirectional; certain genes are activated by JH, and other genes are repressed or not affected. The activation is mediated by the JH receptor Met (Schwinghammer et al., 2011), and the repression is mediated by Met through the recruitment of the Hairy/Goucho molecular system (Hagai et al., 2007). However, the role of Met and its interacting partners in regulating the expressions of the takeout genes remained unknown, and the role of the takeout genes in wing polyphenism remained unclear due to the lack of knowledge of behavior plasticity. Moreover, the complete identification of the N. lugens genome sequence (Xue et al., 2014) and key biological characteristics, such as migration and behavior plasticity, are important for pest control and predictions of pest outbreaks, making N. lugens an appropriate model for studying the role of gene families, such as the takeout family genes. Here, we use quantitative real-time PCR to study the role of JH signaling in the regulation of N. lugens takeout genes by the topical application of JH or the down-regulation of Met and its interacting partners through RNAi.
Materials and Methods
Insects
The brown planthopper (N. lugens) insectary population was provided by Professor Zhu Zeng-Rong, Institute of Insect Sciences, Zhejiang University. The insects were cultured with rice seedling and raised at a temperature = 25°C, relative humidity = 60%, and a photoperiod = 16 L:8 D.
Construction of Phylogenetic Trees and WebLogo Conserved Amino Acid Analysis
A Phylogenetic tree, including 17 brown planthopper Takeout proteins and 65 homologs of other species, was constructed, and the sequences were downloaded from the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/). The phylogenetic tree was constructed by MEGA 6.0 software using the Neighbor-Joining method and a bootstrap value of 1000. The predicted amino acid sequences of 17 N. lugens Takeout proteins were aligned into WebLogo (http://weblogo.berkeley.edu/logo.cgi) and were compared in pairs using the default settings.
JHIII Treatment
The juvenile hormone III (JHIII, Sigma Aldrich, USA) was dissolved in acetone at a concentration of 1 μg/μL, with acetone as a control group, and a volume of 0.2 μL was applied to the back of each brown planthopper at the 5th nymph stage; the brown planthoppers were collected 1 or 3 days after the treatment and ground in TRIzol, and the total RNA was then extracted.
RNA Interference
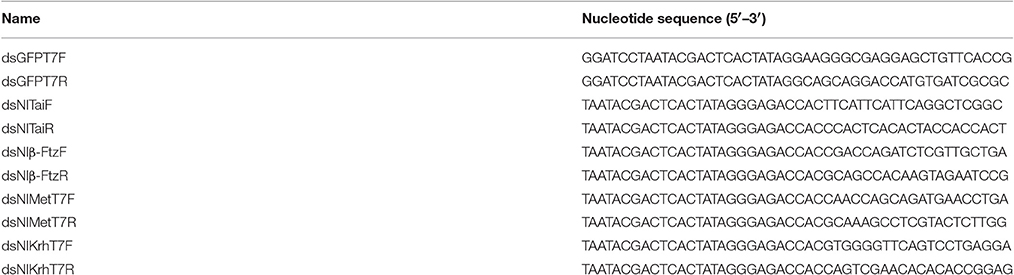
The DNA fragments used for the dsRNA synthesis were amplified through PCR using NlMet, NlKr-h1, NlTai, and Nlβ-Ftz cloned into PMD18-T separately as templates. The primers are listed in Table 1. Double-stranded RNA of NlMet, NlKr-h1, NlTai, and Nlβ-Ftz were synthesized using the RNA Production System-T7 kit (RiboMAX Large Scale, Promega). dsGFP was used as a control. The 5th instar nymphs of N. lugens were injected. The Narishige Injection System (MN-151, Narishige) was used for the dsRNA injection. One or three days after the injection, the insects were collected for RNA extraction.
RNA Extraction and Quantitative Real-Time PCR
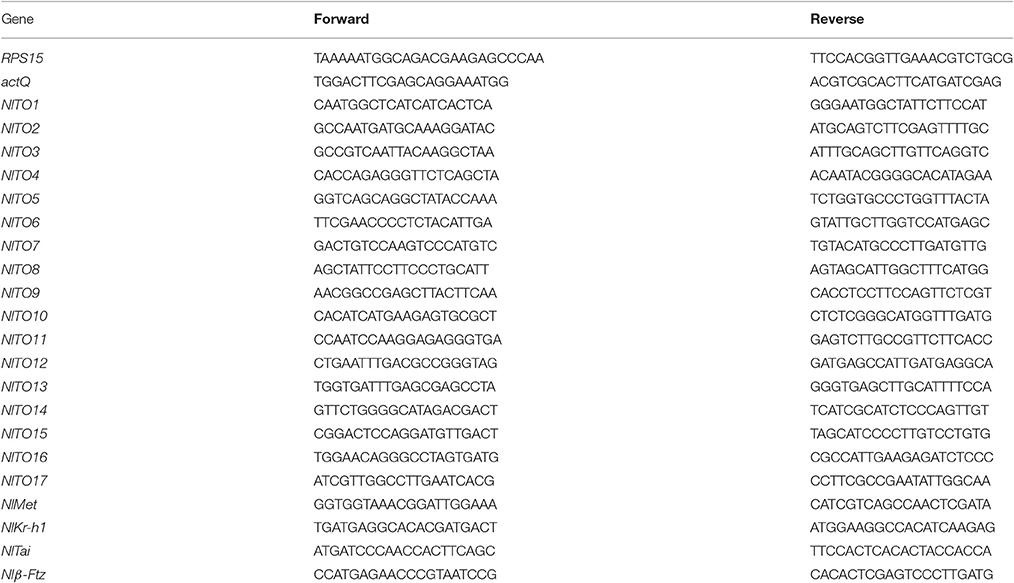
Total RNA was extracted using the TRIzol RNA extraction kit (TaKaRa). Reverse transcription was carried out using the First Strand cDNA Synthesis kit (Roche). The real-time quantitative PCR kit SuperReal PreMix (SYBR Green, Tiagen, Beijing) was used. All primers were synthesized by Sangon (Shanghai). All primers are listed in Table 2. The reference genes were selected based on previous reports (Yuan et al., 2014).
The RNA concentration was measured using NanoDrop 1000 (Thermo, USA). The primers were designed in the range of 90–110 bp for the qRT-PCR measurement of the NlTO genes. Three replicates were used for the qRT-PCR reactions of each sample. In total, a 20 μL reaction was used for the qRT-PCR reaction, including 10 μL 2 × SuperReal PreMix, 0.6 μL upstream and downstream primers (10 μmol • L−1), 0.6 μL 50 × ROX Reference Dye, 2 μL cDNA template, and 6.2 μL DEPC-treated water. Using a two-step qRT-PCR amplification procedure, the pre-denaturation was as follows: 95°C 1 min, 1 cycle; The qRT-PCR reactions were as follows: 95°C 3 s, 58°C 30 s, 40 cycles. All data were analyzed using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Statistics and Heatmap
SPSS 20.0 was used for the data analysis. For the analysis of the qRT-PCR experiment, student's t-test was used. A heatmap was constructed using HemI1.03, and the fold changes of relative expressions were logarithmically transformed. The clustering method was a hierarchical average linkage, and the similarity metric was the Pearson distance.
Results
Cloning and Analysis of the Brown Planthopper takeout Family Genes
We searched the brown planthopper N. lugens genome (Xue et al., 2014) and InsectBase (Yin et al., 2016). We identified 17 takeout homologs. We then cloned, sequenced and named all takeout homologs-takeout 1 (TO1) to takeout 17 (TO17). The phylogenetic tree analysis showed that the brown planthopper takeout genes are conserved across the species (Figure 1). NlTO11 clustered with NlTO17, and both clustered with NlTO7 (Figure 1). These three Takeout homologs together clustered with NlTO12 and TcTO22 (Figure 1). NlTO15 clustered with NlTO5, and both clustered together with NlTO14 (Figure 1). Four homologs, including NlTO3, NlTO4, NlTO6, and NlTO13, are relatively distant, and each has close homologs from other species (Figure 1).
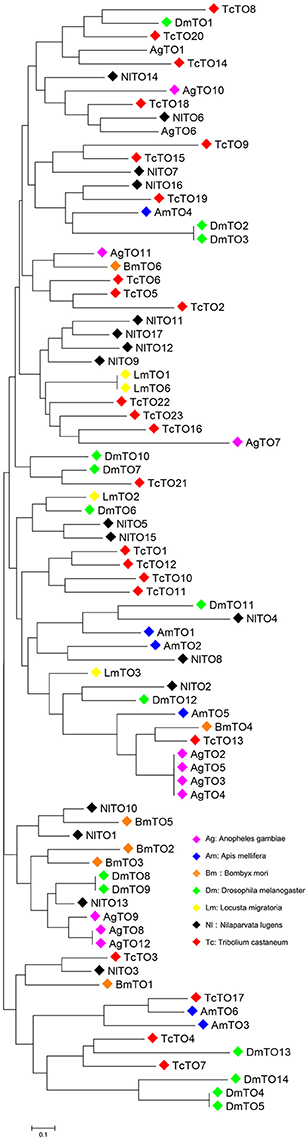
Figure 1. Phylogenetic analysis of Takeout family proteins from N. lugens and other species. AgTO1 (XM_311285), AgTO2 (XM_321081), AgTO3 (XM_003435936), AgTO4 (XM_003435937), AgTO5 (XM_003435938), AgTO6 (XM_321079), AgTO7 (XM_321075), AgTO8 (XM_313180), AgTO9 (XM_313181), AgTO10 (XM_001230706), AgTO11 (XM_309482), AgTO12 (XM_307380), AmTO1 (GB48492-PA), AmTO2 (GB42798-PA), AmTO3 (GB42796-PA), AmTO4 (GB42799-PA), AmTO5 (GB42800-PA), AmTO6 (GB42704-PA), BmTO1 (XP_004927145), BmTO2 (NP_001036949), BmTO3 (NP_001036945), BmTO4 (XP_004923014), BmTO5 (XP_004932669), BmTO6 (XP_012548133), DmTO1 (FBpp0078169), DmTO2 (FBpp0082691), DmTO3 (FBpp0307590), DmTO4 (FBpp0083445), DmTO5 (FBpp0311940), DmTO6 (FBpp0084027), DmTO7 (FBpp0084184), DmTO8 (FBpp0084185), DmTO9 (FBpp0308365), DmTO10 (FBpp0308366), DmTO11 (FBpp0084473), DmTO12 (FBpp0084474), DmTO13 (FBpp0290041), DmTO14 (FBpp0083446), LmTO1 (GU722575), LmTO2 (CO856064), LmTO3 (CO825835), LmTO6 (KM503135), TcTO1 (XP_967109), TcTO2 (EFA05096), TcTO3 (EFA05095), TcTO4 (XP_966559), TcTO5 (XP_974592), TcTO6 (XP_974610), TcTO7 (XP_966559), TcTO8 (XP_008190426), TcTO9 (EFA05633), TcTO10 (XP_970866), TcTO11 (XP_970866), TcTO12 (EFA05635), TcTO13 (KYB27715), TcTO14 (XP_973361), TcTO15 (EFA03576), TcTO16 (EFA03557), TcTO17 (XP_015836023), TcTO18 (XP_972960), TcTO19 (XP_972997), TcTO20 (XP_001812695), TcTO21 (XP_015840904), TcTO22 (EEZ98654), TcTO23 (XP_974890). Ag, Anopheles gambia; Am, Apis mellifera; Bm, Bombyx mori; Dm, Drosophila melanogaster; Lm, Locusta migraloria; Tc, Tribolium castaneum.
We aligned the predicted Takeout protein sequences and graphical presentation of the sequence conservation by the overall height (Figure 2). The conserved amino acids were distributed throughout the entire Takeout protein sequence. A comparison of these Takeout proteins revealed two highly conserved cysteine residues (C) at the N terminal, four highly conserved glycine residues (G) and two highly conserved proline residues (F, Figure 2) in the middle of the protein.
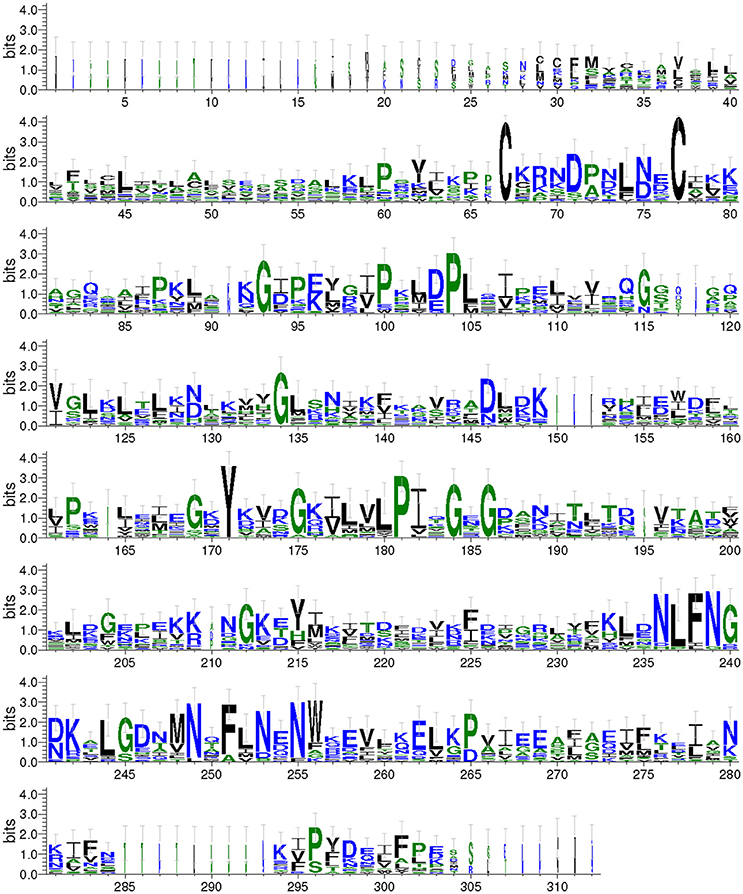
Figure 2. WebLogo of the Takeout protein sequences. The data are the alignment of 17 Takeout proteins from N. lugens. The X-axis represents the amino acid position. The Y-axis (bits) represents the relative proportion of the amino acids at one position. The height of the logo varied inversely with the variability at the position.
Male Biased Expressions of Brown Planthopper takeout Family Genes
To study whether the expressions of the takeout genes in brown planthopper are male biased, we measured the expressions of the takeout family genes using qRT-PCR and compared the expressions in males to those in females in the two wing forms. The results showed that the expressions of 16 of the 17 takeout genes are male biased (Figure 3), which is consistent with previous studies by Dauwalder et al. in D. melanogaster (Dauwalder et al., 2002). However, in contrast, we found one takeout gene, NlTO16, that was more highly expressed in females than in males (Figure 3), i.e., the fold change is 7.5 times in the long-wing form and 6 times in the short-wing form. The expression of NlTO16 in the long wing and short wing forms was not significantly different.
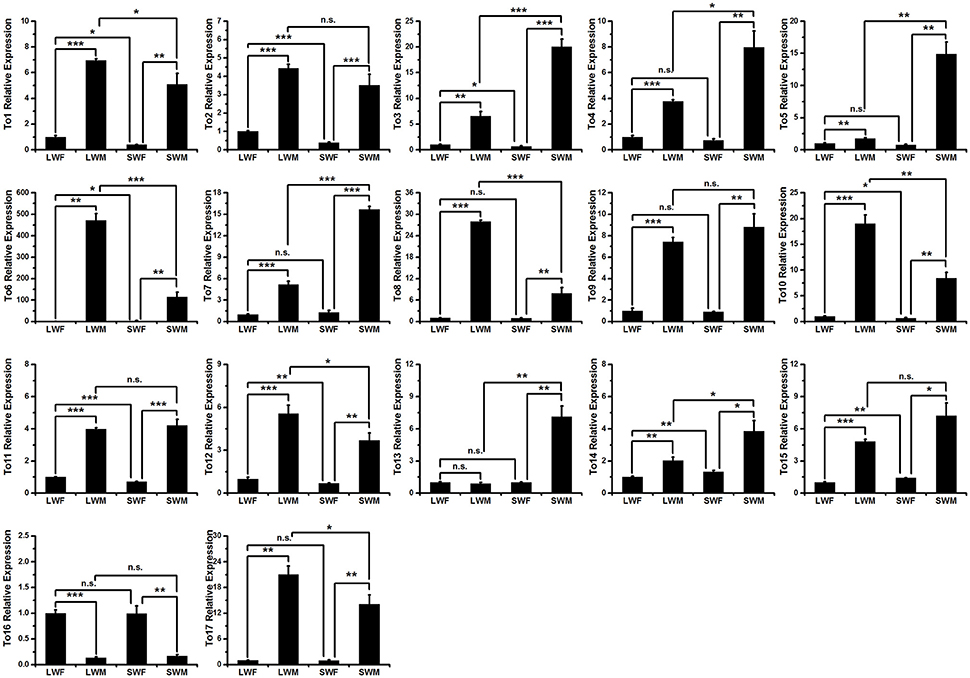
Figure 3. Expressions of the takeout family genes in four types of brown planthopper adults. LWF, long-wing female; LWM, long-wing male; SWF, short-wing female; SWM, short-wing male. Student's t-test was used, *P < 0.05, **P < 0.01, ***P < 0.001.
The Effect of JH on the Expressions of takeout Family Genes
Our previous study showed that the expression of brown planthopper NlKr-h1 is induced by JH or its mimics (Jin et al., 2014). The expression of NlKr-h1 was significantly high (≈5 times) 1 day after the JH treatment (Jin et al., 2014). Therefore, we measured the expressions of the takeout family genes 1 day after the JHIII treatment (Figure 4). The result showed that 14 of the 17 takeout genes are up-regulated, and the expression of 13 takeout genes was significantly high 1 day after the JHIII treatment (Figure 4B). However, the expression of 12 takeout genes was significantly high 1 h after the JHIII treatment (Figure 4A), and the fold changes are lower than those following a 1 day treatment. The expressions of NlTO9, 10, 13, 14, and 17 increased more than 6-fold 1 day after the JHIII treatment. Only three genes, NlTO 3, 6, and 7, are down-regulated after the JHIII treatment and decreased by <6 times 1 day after the JHIII treatment (Figure 4B). This finding is consistent with previous studies in the honey bee A. mellifera (Hagai et al., 2007) and the tobacco hornworm Manduca sexta (Du et al., 2003) in which the expression of the takeout gene is regulated by JH. However, when we measured the expressions of the takeout genes 2 and 3 days after the JHIII treatment, the expressions of 9/15 of the 17 genes were significantly reduced (Figures 4C,D).
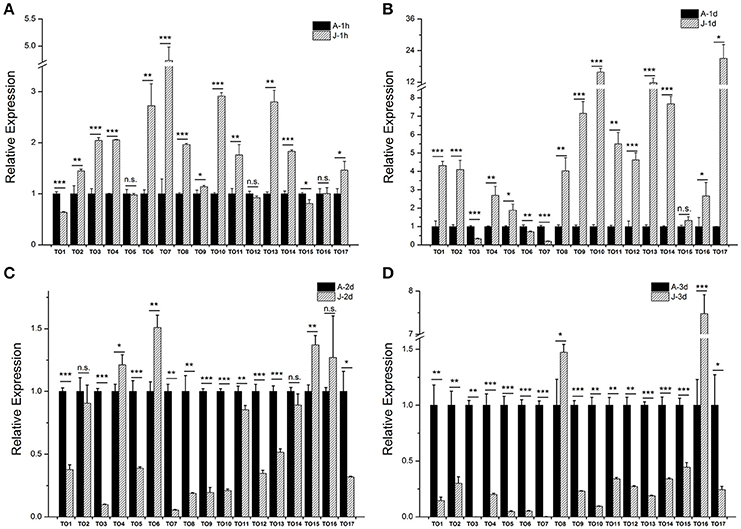
Figure 4. The effect of juvenile hormone III (JHIII) on the expressions of the takeout family genes. The measurements of the takeout family genes were carried out 1 h (A), 1 day (B), 2 days (C), and 3 days (D) after the JHIII treatment. The expressions of the takeout genes TO1-TO17 after the JHIII treatment were compared with the control nymphs that were treated with acetone. Student's t-test was used, *P < 0.05, **P < 0.01, ***P < 0.001.
The Expressions of the takeout Family Genes in Met and Interacting Proteins Down-Regulated Brown Planthoppers
To further understand the regulatory role of JH signaling in the expressions of the takeout family genes, we used RNAi to down-regulate the expressions of the JH receptor or its interacting proteins and then measured the fold changes of the takeout family genes 1 and 3 days after the dsRNA injection. The expressions of half of the takeout family genes are not changed significantly 1 day after the NlMet dsRNA and NlKr-h1 dsRNA injections. One day after the NlMet dsRNA injection, the expressions of 10 NlTO genes are not changed significantly. The fold changes of five genes are <2, and the fold changes of the remaining genes are <4 (Figure 5, Table 3). However, 1 day after the NlKr-h1 dsRNA injection, the expressions of 7 NlTO genes are not changed significantly. The fold changes of the eight genes are <2, and fold changes of the remaining genes are <4 (Figure 5, Table 3). Three days after the dsRNA injection, the majority of the takeout genes are up-regulated, and only four and two takeout genes are down-regulated 1 and 3 days after the injection, respectively (Figure 5, Table 3). In addition, only a few genes showed no significant changes (NlTO12 for NlMet and NlTO8, 12, and 16 for NlKr-h1 dsRNA; Figure 5, Table 3). In summary, the takeout family genes showed relatively stable expressions 1 day after the NlMet and NlKr-h1 dsRNA injections, while after 3 days, the expressions of the majority of the takeout family genes changed significantly (Figure 5, Table 3).
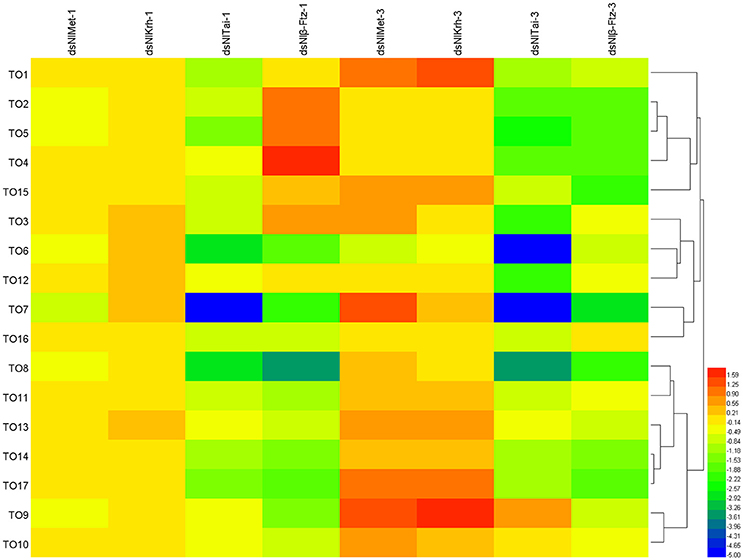
Figure 5. The expressions of the takeout family genes in NlMet, NlKr-h1, NlTai, and β-Ftz down-regulated brown planthopper. The measurements of the takeout family genes were carried out 1 and 3 days after the dsRNA injection. The expressions of the takeout genes TO1–TO17 after the NlMet, NlKr-h1, NlTai, or β-Ftz dsRNA injections were compared with the control, which was injected with GFP dsRNA. Heatmap was used for the visualization of the expression changes. NlMet-1, NlKr-h1-1, NlTai-1, and β-Ftz-1 indicate 1 day after the dsRNA injection, and NlMet-3, NlKr-h1-3, NlTai-3, and β-Ftz-3 indicate 3 days after the dsRNA injection.
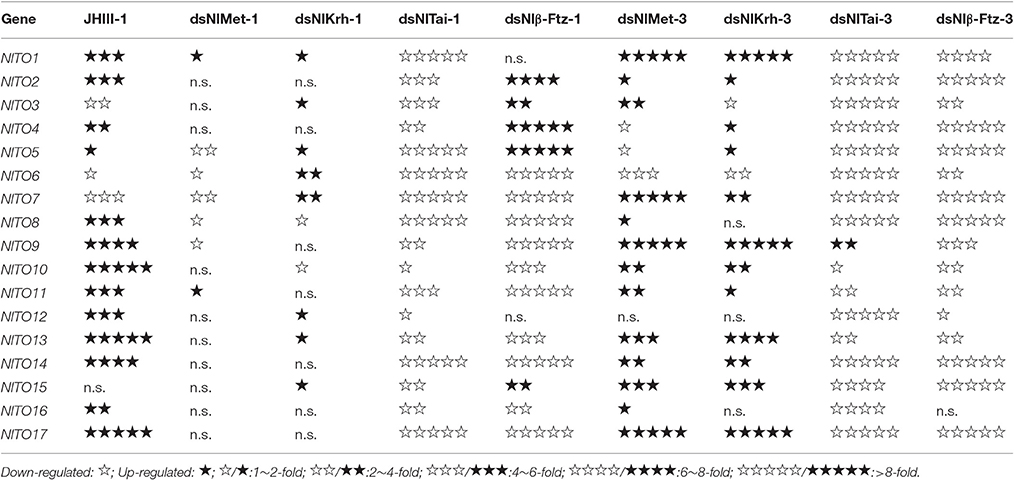
Table 3. Expression changes of the takeout genes by juvenile hormone III (JHIII) treatment and RNAi.
However, after the injections of NlTai and Nlβ-Ftz dsRNA, the expressions of the takeout family genes are mainly down-regulated, and the majority of them are significantly different from the control, which was injected with dsGFP (Figure 5, Table 3). The expressions of the takeout genes, except for NlTO9, are all down-regulated and significantly different after the NlTai dsRNA injection (Figure 5, Table 3). One day after the injection, five genes are up-regulated, two genes are not significantly changed, and the remaining genes are down-regulated significantly (Figure 5, Table 3). All genes were down-regulated 3 days after the Nlβ-Ftz dsRNA injection (Figure 5, Table 3). These results indicated that NlTai and Nlβ-Ftz are probably more important for maintaining or inducing the expressions of the takeout family genes, and NlMet and NlKr-h1 are more important for down-regulating the takeout family genes.
Discussion
Our analysis showed that the brown planthopper takeout family genes are conserved across species (Figure 1). However, the functions of these proteins in N. lugens are unknown. It is well-documented that JH is involved in the wing polyphenism of the brown planthopper (Iwanaga and Tojo, 1986; Bertuso et al., 2002). The role of Locusta migraloria takeout in the behavioral phase change is reminiscent of the role of takeout in N. lugens because the brown planthopper is polyphenism. Due to the limited knowledge regarding the behavioral phase change in the brown planthopper, determining whether Takeout proteins play a role in this process remains to be explored in the future.
Our experiments showed that the brown planthopper takeout family genes are induced 1 h or 1 day after the topical application of JHIII (Figures 4A,B), while the expression levels of most of the takeout genes are reduced 2 and 3 days after the JHIII treatment (Figures 4C,D). When we down-regulated the expressions of the JH receptor NlMet and its downstream target NlKr-h1, as well as the NlMet interacting proteins NlTai and Nlβ-Ftz, the expression patterns of the takeout family genes are distinct. When NlMet and NlKr-h1 are down-regulated, i.e., 1 day after the dsRNA injection, the expressions of the majority of the takeout genes are either not significantly changed or only have slightly changed (Figure 5, Table 3). While after 3 days, the expressions of most of the takeout family genes are increased significantly (Figure 5, Table 3). Overall, the effects of the down-regulation of NlKr-h1 on the expressions of the takeout family genes are similar to those of the down-regulation of NlMet. However, the down-regulation of the NlMet interacting proteins NlTai and Nlβ-Ftz through RNAi led to a down-regulation of most of the takeout family genes 1 and 3 days after the dsRNA injection. This finding indicates distinct roles of NlMet and its interacting proteins in regulating the takeout family genes. NlMet and its interacting proteins NlTai and Nlβ-Ftz might act through different mechanisms in regulating the expressions of the takeout family genes. As mentioned above, JH could either up-regulate or down-regulate gene expression. In this study, we found that in addition to the crucial role of the JH receptor Met, its interacting proteins NlTai and Nlβ-Ftz also play important roles in regulating the expressions of the takeout family genes. However, the roles of NlTai and Nlβ-Ftz are distinct from those of NlMet in regulating the expressions of the takeout genes. This result is consistent with the direct activation of target genes by Met and the repression of target genes with the cooperation of the Hairy/Grouche molecular system (Hagai et al., 2007).
The interaction of Met and Tai in the mosquito Aedes aegypti is dependent on JH (Li et al., 2011, 2014). Here, we show that Met and its interacting proteins play distinct roles in regulating the expressions of the takeout family genes. Although the expressions of most of the takeout family genes significantly increased 3 days after the down-regulation of NlMet and its downstream transcription factor NlKr-h1, there is only a slight effect 1 day after the dsRNA injection, i.e., the expressions of most of the takeout family genes are not significantly changed or only slightly changed.
In the mosquito Aedes aegypti, JH activated the phospholipase C (PLC) pathway and protein kinase C (PKC) and immediately increased the levels of inositol 1,4,5-trisphosphate (IP3), diacylglycerol (DAG), and intracellular calcium, thereby activating calcium/calmodulin-dependent protein kinase II (CaMKII; Liu et al., 2015; Ojani et al., 2016). Met protein is phosphorylated upon JH binding (Liu et al., 2015). The increased expressions of the takeout genes by the down-regulation of NlMet and NlKr-h1 indicates a possibly distinct mechanism in the regulation of the takeout genes by Met and its interacting partners or regulation at different levels, i.e., at the transcriptional, translational or post-translational levels. It is possible that the initial regulation of JH signaling upon ligand binding was affected by the phosphorylation of the Met protein, which leads to the initial unresponsiveness of the takeout family genes even though Met transcription was down-regulated, i.e., down-regulating NlMet resulted in a change in the phosphorylation of the Met proteins and its down-stream signaling components. Based on our previous study on Kr-h1, the genes downstream of JH action are prone to be induced 1 day after the JH application (Jin et al., 2014). In this study, we found that the takeout genes are induced 1 h and 1 day after the JHIII application and are reduced 2 and 3 days after the treatment. This result indicates a possible feedback mechanism in regulating the expressions of the takeout genes after the induction by JHIII. Additionally, Met and its interacting proteins may act at different developmental stages; in this study, we only tested the expression changes of the takeout family genes in brown planthoppers treated at the 5th instar nymph stage. In the future, studies that measure the gene expression levels in other stages and different tissues are to be carried out.
The takeout family genes are regulated by JH signaling in N. lugens. Although previous studies have shown that takeout is involved in feeding and migration, this work advanced our understanding of the molecular function and the regulatory mechanism of JH signaling. Furthermore, this work could help in the development of potential small molecules or the identification of target genes for regulating the expressions of the takeout genes behaviors of N. lugens, such as feeding and migration, which could be an efficient and environment friendly approach for the control of this pest in the future. The functions of the takeout family genes, including its role in polymorphism, remain unclear. Additional experiments are required for the understanding of the mechanisms regulating the takeout family genes by JH signaling.
Author Contributions
XL designed the study; LZ and YJ performed the experiment; XL and LZ analyzed the data and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The National Natural Science Foundation of China (No. 31471771, No. 31672023), the Natural Science Foundation of Zhejiang Province (No. Y14C140007), and the National Basic Research Program (973) of China (No. 2010CB126200) supported this work.
References
Ayoade, O., Morooka, S., and Tojo, S. (1999). Enhancement of short wing formation and ovarian growth in the genetically defined macropterous strain of the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 45, 93–100. doi: 10.1016/S0022-1910(98)00103-6
Belles, X., Martin, D., and Piulachs, M. D. (2005). The mevalonate pathway and the synthesis of juvenile hormone in insects. Annu. Rev. Entomol. 50, 181–199. doi: 10.1146/annurev.ento.50.071803.130356
Bernardo, T. J., and Dubrovsky, E. B. (2012). The Drosophila juvenile hormone receptor candidates methoprene-tolerant (MET) and germ cell-expressed (GCE) utilize a conserved LIXXL motif to bind the FTZ-F1 nuclear receptor. J. Biol. Chem. 287, 7821–7833. doi: 10.1074/jbc.M111.327254
Bertuso, A. G., Morooka, S., and Tojo, S. (2002). Sensitive periods for wing development and precocious metamorphosis after precocene treatment of the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 48, 221–229. doi: 10.1016/S0022-1910(01)00167-6
Dauwalder, B., Tsujimoto, S., Moss, J., and Mattox, W. (2002). The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 16, 2879–2892. doi: 10.1101/gad.1010302
Du, J., Hiruma, K., and Riddiford, L. M. (2003). A novel gene in the takeout gene family is regulated by hormones and nutrients in Manduca larval epidermis. Insect Biochem. Mol. Biol. 33, 803–814. doi: 10.1016/S0965-1748(03)00079-1
Flatt, T., Tu, M. P., and Tatar, M. (2005). Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays 27, 999–1010. doi: 10.1002/bies.20290
Fujikawa, K., Seno, K., and Ozaki, M. (2006). A novel takeout-like protein expressed in the taste and olfactory organs of the blowfly, Phormia regina. FEBS J. 273, 4311–4321. doi: 10.1111/j.1742-4658.2006.05422.x
Guo, W., Wang, X., Ma, Z., Xue, L., Han, J., Yu, D., et al. (2011). CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genet. 7:e1001291. doi: 10.1371/journal.pgen.1001291
Hagai, T., Cohen, M., and Bloch, G. (2007). Genes encoding putative takeout/juvenile hormone binding proteins in the honeybee (Apis mellifera) and modulation by age and juvenile hormone of the takeout-like gene GB19811. Insect Biochem. Mol. Biol. 37, 689–701. doi: 10.1016/j.ibmb.2007.04.002
Iwanaga, K., and Tojo, S. (1986). Effects of juvenile hormone and rearing density on wing dimorphism and oöcyte development in the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 32, 585–590. doi: 10.1016/0022-1910(86)90076-4
Jin, M. N., Xue, J., Yao, Y., and Lin, X. D. (2014). Molecular characterization and functional analysis of Krüppel-homolog 1 (Kr-h1) in the brown planthopper, Nilaparvata lugens (Stål). J. Integr. Agric. 13, 1972–1981. doi: 10.1016/S2095-3119(13)60654-1
Jindra, M., Palli, S. R., and Riddiford, L. M. (2013). The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 58, 181–204. doi: 10.1146/annurev-ento-120811-153700
Kisimoto, R. (1956). Effect of crowding during the larval period on the determination of the wing-form of an adult plant-hopper. Nature 178, 641–642. doi: 10.1038/178641a0
Kisimoto, R. (1965). Studies on polymorphism and its role playing in the population growth of brown planthopper, Nilaparvata lugens (Stal). Bull. Shikoku Agric. Exp. Stn. 13, 101–106.
Li, M., Liu, P., Wiley, J. D., Ojani, R., Bevan, D. R., Li, J., et al. (2014). A steroid receptor coactivator acts as the DNA-binding partner of the methoprene-tolerant protein in regulating juvenile hormone response genes. Mol. Cell. Endocrinol. 394, 47–58. doi: 10.1016/j.mce.2014.06.021
Li, M., Mead, E. A., and Zhu, J. (2011). Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc. Natl. Acad. Sci. U.S.A. 108, 638–643. doi: 10.1073/pnas.1013914108
Lin, X., Xu, Y., Yao, Y., Wang, B., Lavine, M. D., and Lavine, L. C. (2016a). JNK signaling mediates wing form polymorphism in brown planthoppers (Nilaparvata lugens). Insect Biochem. Mol. Biol. 73, 55–61. doi: 10.1016/j.ibmb.2016.04.005
Lin, X., Yao, Y., Wang, B., Emlen, D. J., and Lavine, L. C. (2016b). Ecological Trade-offs between migration and reproduction are mediated by the nutrition-sensitive insulin-signaling pathway. Int. J. Biol. Sci. 12, 607–616. doi: 10.7150/ijbs.14802
Lin, X., Yao, Y., Wang, B., Lavine, M. D., and Lavine, L. C. (2016c). FOXO links wing form polyphenism and wound healing in the brown planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 70, 24–31. doi: 10.1016/j.ibmb.2015.12.002
Liu, P., Peng, H. J., and Zhu, J. (2015). Juvenile hormone-activated phospholipase C pathway enhances transcriptional activation by the methoprene-tolerant protein. Proc. Natl. Acad. Sci. U.S.A. 112, E1871–E1879. doi: 10.1073/pnas.1423204112
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Meunier, N., Belgacem, Y. H., and Martin, J. R. (2007). Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J. Exp. Biol. 210, 1424–1434. doi: 10.1242/jeb.02755
Minakuchi, C., Namiki, T., and Shinoda, T. (2009). Kruppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 325, 341–350. doi: 10.1016/j.ydbio.2008.10.016
Ojani, R., Liu, P., Fu, X., and Zhu, J. (2016). Protein kinase C modulates transcriptional activation by the juvenile hormone receptor methoprene-tolerant. Insect Biochem. Mol. Biol. 70, 44–52. doi: 10.1016/j.ibmb.2015.12.001
Saito, K., Su, Z. H., Emi, A., Mita, K., Takeda, M., and Fujiwara, Y. (2006). Cloning and expression analysis of takeout/JHBP family genes of silkworm, Bombyx mori. Insect Mol. Biol. 15, 245–251. doi: 10.1111/j.1365-2583.2006.00612.x
Schwinghammer, M. A., Zhou, X., Kambhampati, S., Bennett, G. W., and Scharf, M. E. (2011). A novel gene from the takeout family involved in termite trail-following behavior. Gene 474, 12–21. doi: 10.1016/j.gene.2010.11.012
So, W. V., Sarov-Blat, L., Kotarski, C. K., McDonald, M. J., Allada, R., and Rosbash, M. (2000). takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol. Cell. Biol. 20, 6935–6944. doi: 10.1128/MCB.20.18.6935-6944.2000
Truman, J. W., and Riddiford, L. M. (2002). Endocrine insights into the evolution of metamorphosis in insects. Annu. Rev. Entomol. 47, 467–500. doi: 10.1146/annurev.ento.47.091201.145230
Vanaphan, N., Dauwalder, B., and Zufall, R. A. (2012). Diversification of takeout, a male-biased gene family in Drosophila. Gene 491, 142–148. doi: 10.1016/j.gene.2011.10.003
Xu, H. J., Xue, J., Lu, B., Zhang, X. C., Zhuo, J. C., He, S. F., et al. (2015). Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467. doi: 10.1038/nature14286
Xue, J., Zhou, X., Zhang, C. X., Yu, L. L., Fan, H. W., Wang, Z., et al. (2014). Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15, 521. doi: 10.1186/s13059-014-0521-0
Yin, C., Shen, G., Guo, D., Wang, S., Ma, X., Xiao, H., et al. (2016). InsectBase: a resource for insect genomes and transcriptomes. Nucleic Acids Res. 44, D801–D807. doi: 10.1093/nar/gkv1204
Yoo, J., Ko, S., Kim, H., Sampson, H., Yun, J. H., Choe, K. M., et al. (2011). Crystal structure of Fushi tarazu factor 1 ligand binding domain/Fushi tarazu peptide complex identifies new class of nuclear receptors. J. Biol. Chem. 286, 31225–31231. doi: 10.1074/jbc.M111.252916
Yuan, M., Lu, Y., Zhu, X., Wan, H., Shakeel, M., Zhan, S., et al. (2014). Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS ONE 9:e86503. doi: 10.1371/journal.pone.0086503
Zhu, J., Busche, J. M., and Zhang, X. (2010). Identification of juvenile hormone target genes in the adult female mosquitoes. Insect Biochem. Mol. Biol. 40, 23–29. doi: 10.1016/j.ibmb.2009.12.004
Keywords: brown planthopper, juvenile hormone, Met, Taiman, ß-Ftz-F1, takeout
Citation: Lin X, Zhang L and Jiang Y (2017) Distinct Roles of Met and Interacting Proteins on the Expressions of takeout Family Genes in Brown Planthopper. Front. Physiol. 8:100. doi: 10.3389/fphys.2017.00100
Received: 01 September 2016; Accepted: 07 February 2017;
Published: 21 February 2017.
Edited by:
Arash Zibaee, University of Gilan, IranReviewed by:
Takashi Koyama, Instituto Gulbenkian de Ciência, PortugalMauro Mandrioli, University of Modena and Reggio Emilia, Italy
Copyright © 2017 Lin, Zhang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinda Lin, bGlueGluZGFAY2psdS5lZHUuY24=
 Xinda Lin
Xinda Lin Ling Zhang
Ling Zhang Yanyun Jiang
Yanyun Jiang