- Department of Biotechnology, National Institute of Technology, Durgapur, India
Arabidopsis seedling growth is regulated by integration of various environmental and hormonal signals. While the interactions between light and cytokinin signaling pathways have been studied, the molecular mechanism by which their interaction regulate hypocotyl elongation remain unclear. In this study, we demonstrate that MYB4, a positive regulator of photomorphogenesis, physically interacts with HY5 and attenuates HY5-mediated regulation of MYB4. The expression of MYB4 is induced by different wavelengths of light and it genetically interacts with HY5 to regulate hypocotyl length during light-mediated seedling development. The MYB4 and HY5 mediated regulation of hypocotyl length is altered upon cytokinin treatment in a light intensity-dependent manner. Furthermore, on contrary to the light signals, cytokinin suppresses MYB4 expression. MYB4 together with HY5 regulates the expression of the key genes such as ABCG14, ARR4, and CHS involved in cytokinin signaling pathway. Taken together, this study highlights how the HY5-MYB4 module integrates light and cytokinin signals to fine-tune Arabidopsis seedling development.
Introduction
Plants, as photoautotrophs, are highly sensitive to their light environment. Light plays a crucial role not only as an energy source for photosynthesis but also in influencing various developmental and physiological processes throughout the plant’s life cycle. These processes include seed germination, seedling photomorphogenesis, photoperiodic responses, shade avoidance, and flowering (Deng and Quail, 1999; Sullivan and Deng, 2003; Chen et al., 2004; Jiao et al., 2007; Bentsink and Koornneef, 2008; Alvarez-Buylla et al., 2010). Plants have evolved with a sophisticated sensory network that monitors key aspects of their illuminated environment, including light intensity, quality, duration, and direction (Kendrick and Kronenberg, 1994). These various light signals are detected by at least five classes of wavelength-specific photoreceptors, including phytochromes (PHYA-PHYE), cryptochromes (CRY1 and CRY2), phototropins (PHOT1 and PHOT2), F-box-containing flavin-binding proteins (ZTL, FKF1, and LKP2), and UV-B RESISTANCE LOCUS 8 (UVR8) (Paik and Huq, 2019). These photoreceptors are biologically activated by different light signals, leading to widespread transcriptional reprogramming at the genome level (Jing and Lin, 2020).
Extensive genetic and biochemical research has shown that the ELONGATED HYPOCOTYL5 (HY5), a bZIP transcription factor, is a key regulator of light-responsive transcriptional changes. HY5 primarily binds to ACGT-containing cis-elements (such as the G-box and T/G-box) of numerous target genes, thereby modulating various light-regulated physiological and developmental processes in plants (Chattopadhyay et al., 1998, Lee et al., 2007; Zhang et al., 2011). Mutant seedlings lacking HY5 function exhibit significantly elongated hypocotyls under different light conditions (Oyama et al., 1997), indicating that HY5 operates downstream of multiple photoreceptors to promote photomorphogenesis. In addition to inhibiting hypocotyl elongation, HY5 also regulates various other physiological and developmental processes, including root growth, pigment biosynthesis and accumulation, responses to various hormonal signals, and adaptation to low and high temperatures (Marzi et al., 2020; Bhagat et al., 2021; Wang et al., 2021). Similar to HY5, CAM7 (Calmodulin7), also known as Z-box binding factors 3 (ZBF3) (McCormack et al.,2005; Kushwaha et al., 2008) acts as transcription factor and directly interacts with promoters of several light-inducible genes (Kushwaha et al., 2008; Kumar et al., 2016). Previous studies have demonstrated that CAM7 physically interacts with HY5 to enhance the activity of the HY5 promoter and promote photomorphogenesis (Kushwaha et al., 2008; Abbas et al., 2014). More recently, a yeast two-hybrid screen using CAM7 as bait identified MYB4 as one of its interacting partners. CAM7 also interacts genetically with MYB4 to control the hypocotyl length during Arabidopsis seedling development. Additionally, it was reported that both HY5 and CAM7 bind to the promoter of MYB4 and positively regulate its expression (Dutta et al., 2024).
The MYB protein family encompasses a large group of transcription factors that play roles in various plant-specific processes (Dubos et al., 2010). MYB4, a well-studied member of the R2R3-MYB subfamily, is known for regulating the phenylpropanoid metabolic pathway, which contributes to UV-B light resistance (Jin et al., 2000; Zhao et al., 2007; Fornalé et al., 2014; Zhou et al., 2015; Wang et al., 2019). MYB4 functions as a transcriptional repressor by directly targeting the expression of C4H (encodes cinnamate 4-hydroxylase, a crucial enzyme in the biosynthesis of sinapate esters), and AtMYB7, another transcriptional repressor of several flavonoid biosynthesis genes. MYB4 is expressed in various organs of adult plants, including roots, stems, leaves, and flowers (Fornalé et al., 2014). Studies on light-mediated regulation of MYB4 expression in rosette leaves have shown that it is expressed in darkness and is induced under various wavelengths of light, including white, blue, and UV light (Jin et al., 2000). MYB4 has also been found to directly influence flavonoid biosynthesis by repressing the expression of ADT6 (AROGENATE DEHYDRATASE 6) and interfering with the transcriptional activity of MBW (composed of MYB, bHLH, and WD40 proteins) complexes through interaction with bHLH transcription factors (Baudry et al., 2004; Xu et al., 2014; Wang et al., 2019). More recently, it has been shown that the N-terminal MYB domains of MYB4 play a role in its stability and folding under thermal stress in Arabidopsis thaliana (Mitra et al., 2021).
The internal hormonal balance in plants plays a significant role in determining its sensitivity and response to various environmental stimuli. Light signaling components like HY5, PIF3, and PIF4 act as key integrators of light and hormonal pathways by regulating the levels of gibberellin, abscisic acid, auxin, and cytokinin in Arabidopsis (Sibout et al., 2006; Chen et al., 2008; Feng et al., 2008; de Lucas et al., 2008; Lau and Deng, 2010; Rasmussen et al., 2012; Yu et al., 2013; Gangappa and Botto, 2016; van Gelderen, 2018; Yang et al., 2018; Doroshenko et al., 2020; Bhagat et al., 2021; Duan et al., 2021; Dai et al., 2022; Chen et al., 2024; Basu et al., 2025). Among the various hormones, exogenous cytokinin treatment resulted in paler plants that exhibited stunted growth, elevated levels of anthocyanin and diminished apical dominance (Chory et al., 1994). Previously, it was also demonstrated that cytokinins enhance the CHLOROPHYLL A/B BINDING PROTEIN (CAB) and RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN (RBCS) levels (Fierabend and deBoer, 1978; Flores and Tobin, 1986). Additionally, the RESPONSE REGULATOR 4 (ARR4) protein, which mediates cytokinin action, directly interacts with PHYB and stabilizes its active form (Sweere et al., 2001). Despite the significant resistance of hy5 seedlings to systemic cytokinin application in both shoot and root growth inhibition (Cluis et al., 2004), cytokinin induces the accumulation of HY5 regardless of light conditions. The MYB transcription factors in Arabidopsis thaliana also play a crucial role in regulating hormone signaling pathways. For instance, AtMYB59 has been shown to influence cytokinin signaling by modulating the expression of ARR16, a key component of the cytokinin signal transduction pathway (Mu et al., 2009). Since CAM7 genetically interacts with HY5 and MYB4, and that HY5 regulates the MYB4 transcript abundance, we examined the genetic and molecular interrelation between HY5 and MYB4 during Arabidopsis seedling development emphasising on light and cytokinin signaling pathways.
Results
MYB4 physically interacts with HY5
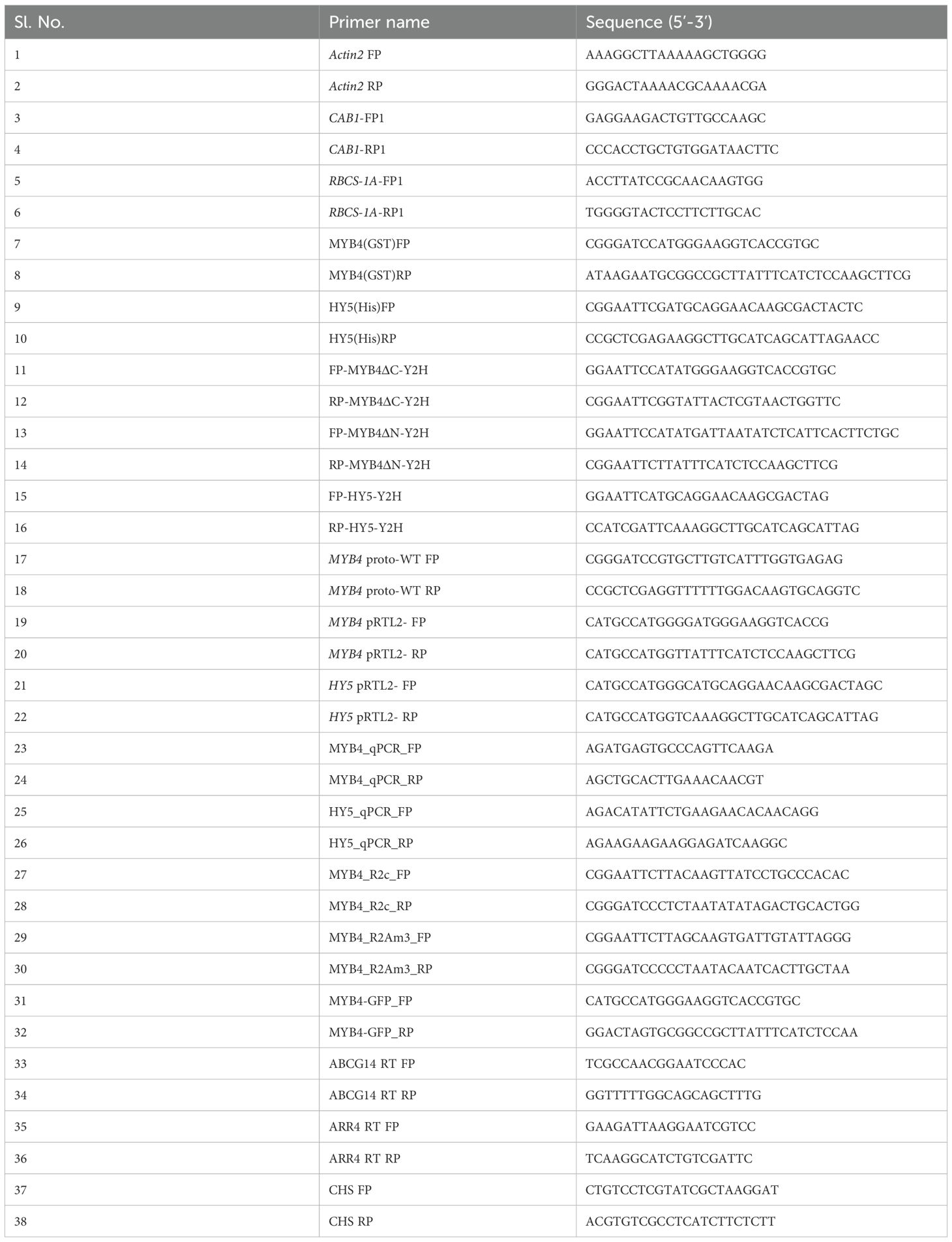
A recent study has demonstrated that MYB4 physically interacts with CAM7, and both CAM7 and HY5 bind to the MYB4 promoter to regulate its transcriptional activation (Dutta et al., 2024). To investigate whether MYB4 physically interacts with HY5, we conducted in vitro pull-down assays using poly-His and GST fusion proteins. GST, GST-CAM7, and GST-MYB4 were incubated with Ni-NTA beads to which HY5-6His was bound. The α-GST immunoblot of the pulled down proteins revealed that GST-MYB4 was selectively pulled down by HY5-6His similar to GST-CAM7, used a positive control (Abbas et al., 2014), however not with GST alone (Figure 1A, Upper Panel). The same membrane was then stripped and re-probed using α-His to examine the equal loading of HY5 (Figure 1A, Lower Panel). These results suggest that MYB4 and HY5 physically interact with each other.
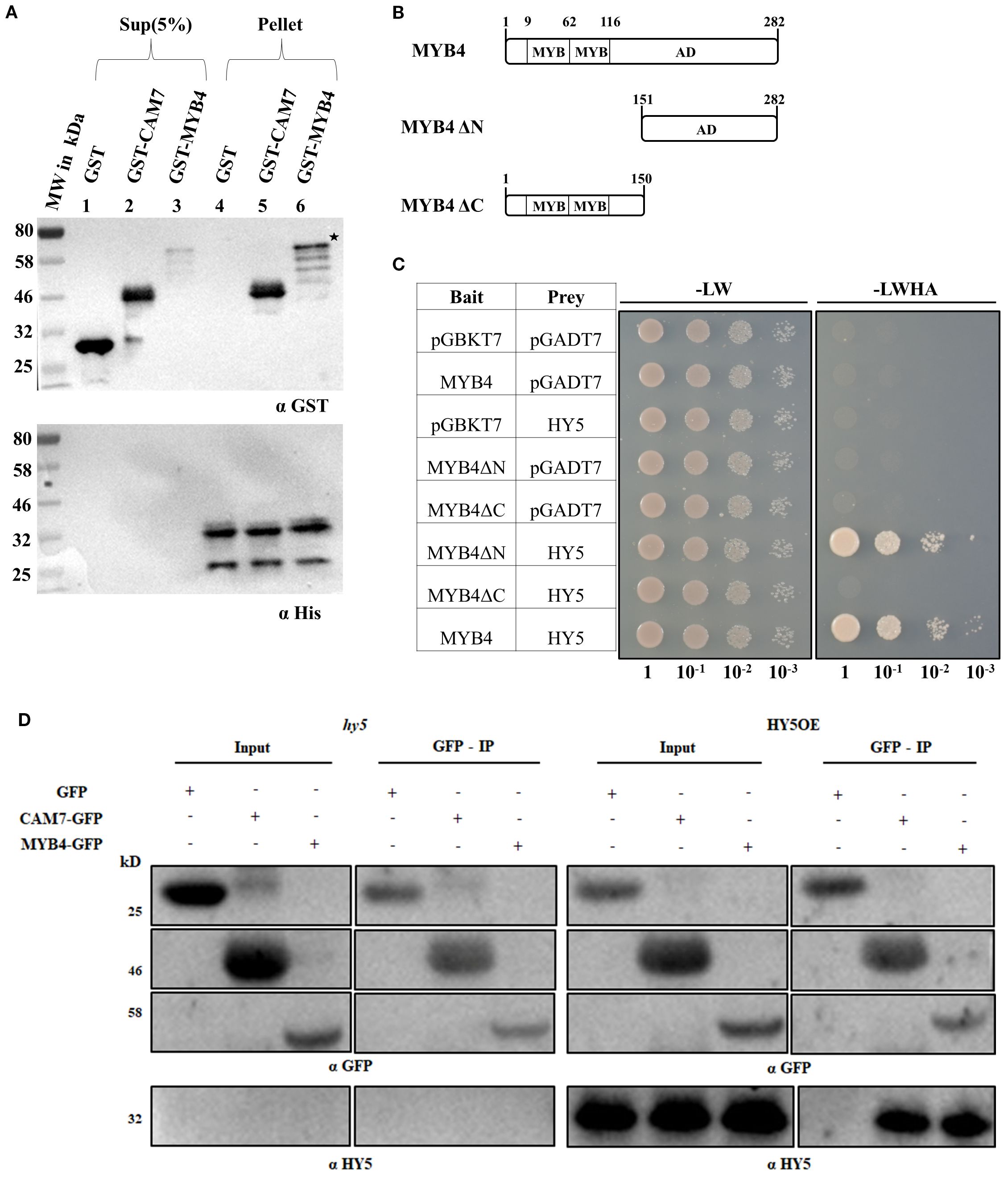
Figure 1. MYB4 physically interacts with HY5. (A) (Upper Panel). In vitro binding of MYB4 and HY5. Approximately 2 µg of HY5-6His was bound to Ni-NTA beads. GST, GST-CAM7 and GST-MYB4 protein was added in equimolar ratio. Supernatant (Sup5%) and pellet fractions were fractionated by SDS-PAGE, blotted, and probed with anti-GST antibodies. Lanes 2 and 5 show HY5-6His with GST-CAM7 (positive control), lanes 3 and 6 show HY5-6His with GST-MYB4, and lanes 1 and 4 show HY5-6His with GST (negative control). (Lower Panel). The membrane was stripped and re-probed with anti-His antibody (for loading control). The position of the full-length GST-MYB4 protein in elute fractions are indicated by asterisk. MW (molecular weight) is in kDa (Kilo Dalton). (B) Diagram illustrating different segments of MYB4, including the full-length protein and various fragments, used in yeast two-hybrid experiments. Each segment is labelled with corresponding amino acid numbers. (C) Co-transformed yeast cells were grown in 2D [lacking Leucine (L) and Tryptophan (W) amino acids] and 4D [lacking Leucine (L), Tryptophan (W), Adenine (A) and Histidine (H) nutrients] selective media to test the protein-protein interactions. The empty vectors (pGBKT7 and pGADT7) and their combinations with MYB4 (pGBKT7 + pGADT7-MYB4; pGBKT7 + pGADT7-MYB4ΔN; pGBKT7 + pGADT7-MYB4ΔC) and HY5 (pGBKT7-HY5 + pGADT7) were used as negative controls. Numbers below the panel indicate dilutions of the starting culture spotted on dropout media. (D) MYB4 interacts with HY5 in vivo. CAM7-GFP and MYB4-GFP were transiently co-expressed with empty vector as negative control in rosette leaves of 30-d old Arabidopsis hy5 and HY5OE (Lee et al., 2007; Abbas et al., 2014) lines. The protein samples were immunoprecipitated with anti-GFP antibody. Both input (5%) and IP fractions were immunoblotted with anti-GFP antibody (upper panel) and the co-immunoprecipitated proteins were detected with anti-HY5 antibody (lower panel). The interaction between CAM7 and HY5 served as the positive control. The experiment was repeated three times and similar results were obtained.
MYB4 has two distinct domains; the N-terminal domain (9-116 aa) comprises of two MYB domains and the C-terminal domain (117-282 aa) comprises of transcriptional activation domain (Mitra et al., 2019) (Figure 1B). To determine the HY5 interacting domain of MYB4, we conducted yeast two-hybrid assays to examine the domain wise protein-protein interaction. As shown in Figure 1C, the C terminal domain of MYB4 interacted with HY5, however not the N-terminal domain. The negative controls involving empty vectors (AD and BD) and their respective combinations with HY5 and MYB4 (full length and truncated) did not exhibit any physical interaction. These results suggest that MYB4 physically interacts with HY5 through the C terminal domain.
To validate this physical interaction in vivo, we performed co-immunoprecipitation assay. The rosette leaves of 30-d old hy5 mutant (Oyama et al., 1997) and HY5 OE line (Lee et al., 2007) were infiltrated with Agrobacterium harbouring either GFP, GFP-CAM7 (positive control, Abbas et al., 2014), or GFP-MYB4 constructs, resulting in transient expression of these proteins. As shown in Figure 1D, HY5 was co-immunoprecipitated with GFP-CAM7 and GFP-MYB4 but not with GFP alone in HY5 OE background. However, in hy5 mutant background, used as negative control, HY5 wasn’t coimmunoprecipitated with any of the transiently expressed proteins. These results further confirm that MYB4 physically interacts with HY5.
MYB4 genetically interacts with HY5 to regulate photomorphogenic growth
MYB4 and HY5 work as positive regulators of photomorphogenesis at various wavelengths of light (Koornneef et al., 1980; Oyama et al., 1997; Ang et al., 1998; Chattopadhyay et al., 1998; Brown and Jenkins, 2008; Dutta et al., 2024). To further investigate the role of MYB4 in photomorphogenesis, we first examined whether MYB4 expression is induced during dark-to-light transition. The semi-quantitative and quantitative PCR analyses revealed that MYB4 expression was induced at various wavelengths of light during dark to light transition. As shown in Figures 2A–D and Supplementary Figure 2, the level of induction varied with context to time and wavelength of light tested. We then determined the physiological significance of the physical interaction between MYB4 and HY5 through genetic studies. For that, the myb4 was genetically crossed with hy5 mutant, and the homozygous myb4 hy5 double mutant lines were generated Supplementary Figure 3). To determine the genetic relationship between MYB4 and HY5, 6-day-old wild-type (segregated WT of F2 population), myb4, hy5, and myb4 hy5 seedlings were grown in dark and at various wavelengths of light. Dark-grown seedlings exhibited hypocotyl length similar to the WT background (Figures 3A, B). The myb4 mutant seedlings displayed significantly increased hypocotyl length as compared to WT under white light (WL), blue light (BL), and far-red light (FRL) (Figures 3C–H). No such effect was observed under red light (RL) condition (Figures 3I, J). Consistent with the previous findings, hy5 mutant exhibited a significantly elongated hypocotyl compared to WT under all light conditions tested (Ang et al., 1998). The hypocotyl of myb4 hy5 double mutant was found to be significantly more elongated than each of the single mutants in WL, BL, and FRL, however not in RL (Figures 3C–J). These results suggest that MYB4 and HY5 work in an additive manner to control the hypocotyl length under WL, BL and FRL.
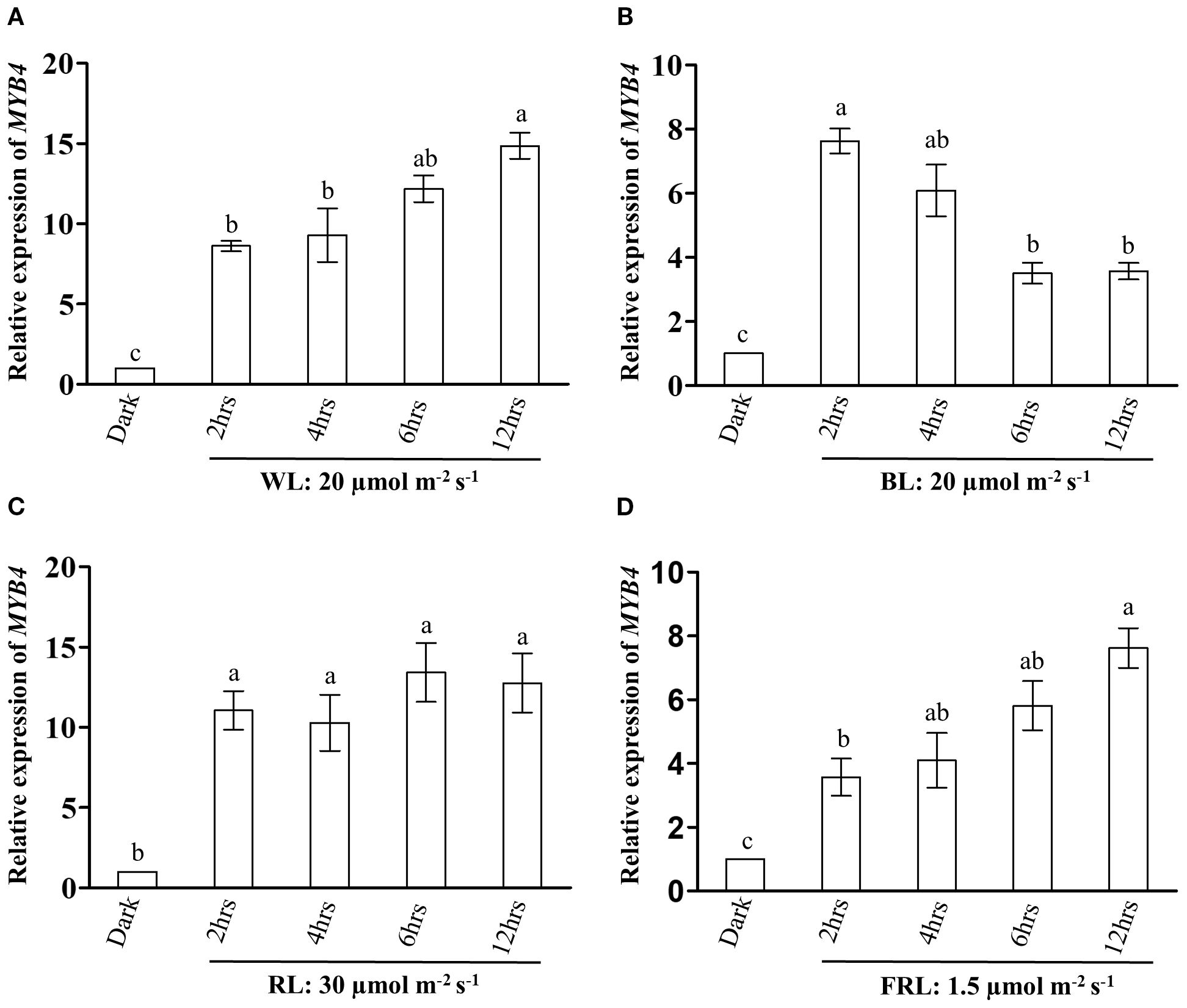
Figure 2. The expression of MYB4 is induced during dark to light transitions. (A, D) Transcript abundance of MYB4 in 5-day-old dark grown WT (ecotype Landsberg erecta) seedlings transferred either to white light (WL: 20 µmolm-2s-1) (A) blue light (BL: 20 µmolm-2s-1) (B) red light (RL:30 µmolm-2s-1) (C) or far-red light (FR: 1.5 µmolm-2s-1) (D) for various time points. ACTIN2 was used as the internal control. Error bars represent ± SE of three biological replicates. Different alphabets denote statistically significant differences (p < 0.05) of MYB4 transcript level. The data were compared by using one-way ANOVA factorial analysis followed by Tukey’s HSD test.
Figure 3. MYB4 works additively with HY5 to regulate the hypocotyl elongation. (A, B) Visible Phenotype (A) and Quantification of hypocotyl length (B) of 6-day-old wild type (Segregated WT, Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant dark. Scale bar = 5 mm. The error bars indicate ± SD (n=20). (C, D) Visible Phenotype (C) of 6-day-old wild type (Segregated WT, Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant WL (15 µmolm-2s-1). Scale bar = 5 mm. Quantification (D) of hypocotyl length of WT (Segregated WT Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown in various fluence of WL. The error bars indicate ± SD (n=20). (E, F) Visible Phenotype (E) and Quantification of hypocotyl length (F) of 6-day-old wild type (Segregated WT, Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant BL (20 µmolm-2s-1). Scale bar = 5 mm. The error bars indicate ± SD (n=20). (G, H) Visible Phenotype (G) and Quantification of hypocotyl length (H) of 6-day-old wild type (Segregated WT, Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant FRL (1.5 µmolm-2s-1). Scale bar = 5 mm. The error bars indicate ± SD (n=20). (I, J) Visible Phenotype (I) and Quantification of hypocotyl length (J) of 6-day-old wild type (Segregated WT, Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant RL (30 µmolm-2s-1). Scale bar = 5 mm. The error bars indicate ± SD (n=20). (K) Real-time PCR analyses of RBCS-1A and CAB1 in 6-day-old WT (Segregated WT Ws-Ler), myb4, hy5 and myb4 hy5 seedlings grown under constant white light (30 µmolm-2s-1). Error bars represent ± SE of the mean of three biological replicates. Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. ACTIN2 was used as endogenous control. (L) Quantification of accumulation of chlorophyll in 6-day-old WT (Segregated WT Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant white light (30 µmolm-2s-1). Error bars represent ± SE of the mean of three biological replicates. Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. (M) Quantification of accumulation of anthocyanin in 6-day-old WT (Segregated WT Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant white light (30 µmolm-2s-1). Error bars represent ± SE of the mean of three biological replicates. Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test.
MYB4 modulates HY5-mediated physiological responses
We then examined whether the genetic interaction between MYB4 and HY5 could influence the expression of light-inducible genes. To determine this, we assessed the transcript levels of RBCS-1A and CAB1 in 6-day-old WT, myb4, hy5, and myb4 hy5 seedlings grown under constant WL (30 μmol m-2 s-1). As previously reported, the expression of these genes was significantly decreased in hy5 background as compared to the WT (Ang et al., 1998; Chattopadhyay et al., 1998). However, as shown in Figure 3K, the additional mutation of MYB4 in hy5 mutant, did not alter the expression of RBCS-1A and CAB1, resulting in the expression pattern similar to that of hy5 in myb4 hy5 double mutant. These results collectively suggest that additional mutation in MYB4 in hy5 background does not affect the expression of light inducible genes.The accumulation of chlorophyll and anthocyanin is an important physiological response controlled by light signaling pathways (Lifschitz et al., 1990; Neff and Chory, 1998; Fankhauser and Casal, 2004). It has been reported earlier that hy5 mutant seedlings show a reduced level of chlorophyll and anthocyanin contents (Oyama et al., 1997; Shin et al., 2007). To examine the significance of the genetic interplay between MYB4 and HY5 in the accumulation of chlorophyll and anthocyanin, we assessed the chlorophyll and anthocyanin levels in 6-day-old WT, myb4, hy5, and myb4 hy5 double mutant seedlings grown in WL (30 µmolm-2s-1). As shown in Figures 3L, M, while chlorophyll content between myb4 and WT seedlings was similar (Figure 3L), an increase in anthocyanin accumulation was observed in myb4 mutants when compared to the WT seedlings (Figure 3M). The additional mutation of MYB4 in hy5 mutant seedlings did not alter the level of chlorophyll accumulation (Figure 3L), suggesting that HY5 regulates the chlorophyll accumulation independently of MYB4. However, as shown in Figure 3M, the anthocyanin content of myb4 hy5 double mutant was similar to WT background, suggesting that MYB4 and HY5 work antagonistically to regulate the anthocyanin accumulation.
MYB4 regulates HY5 mediated activation of its own promoter
Since HY5 binds to the MYB4 promoter and positively regulates its expression, and given that HY5 and MYB4 physically interact, we aimed to investigate the physiological relevance of this physical interaction in mediating the transcriptional regulation of MYB4. To investigate this, we analysed the DNA-protein interaction between the MYB4 promoter and HY5 in the absence or presence of MYB4. Previously it has been reported that the MYB4 promoter contains three E-Boxes, and HY5 specifically binds to the third E-Box (Dutta et al., 2024). We conducted electrophoretic mobility shift assays (EMSA) using purified GST-HY5 and GST-MYB4 fusion proteins, along with both wild-type and mutated versions of the MYB4 promoter fragments (Figure 4A). As shown in Figure 4B, HY5 exhibited strong binding to the wild-type MYB4 promoter fragment, however not to the mutated one. The binding affinity of HY5 to wild-type MYB4 promoter fragment is decreased in the presence of MYB4 (Figure 4B, lane nos. 6 and 7). The reduction in HY5 binding affinity became more pronounced with increasing concentrations of MYB4. However, the DNA-protein complex of HY5 with the wild-type promoter fragment remained unchanged in the presence of GST alone (Figure 4B, lane no. 4). Additionally, no DNA-protein complex was formed between MYB4 and the wild-type promoter fragment (Figure 4B, lane no. 3).
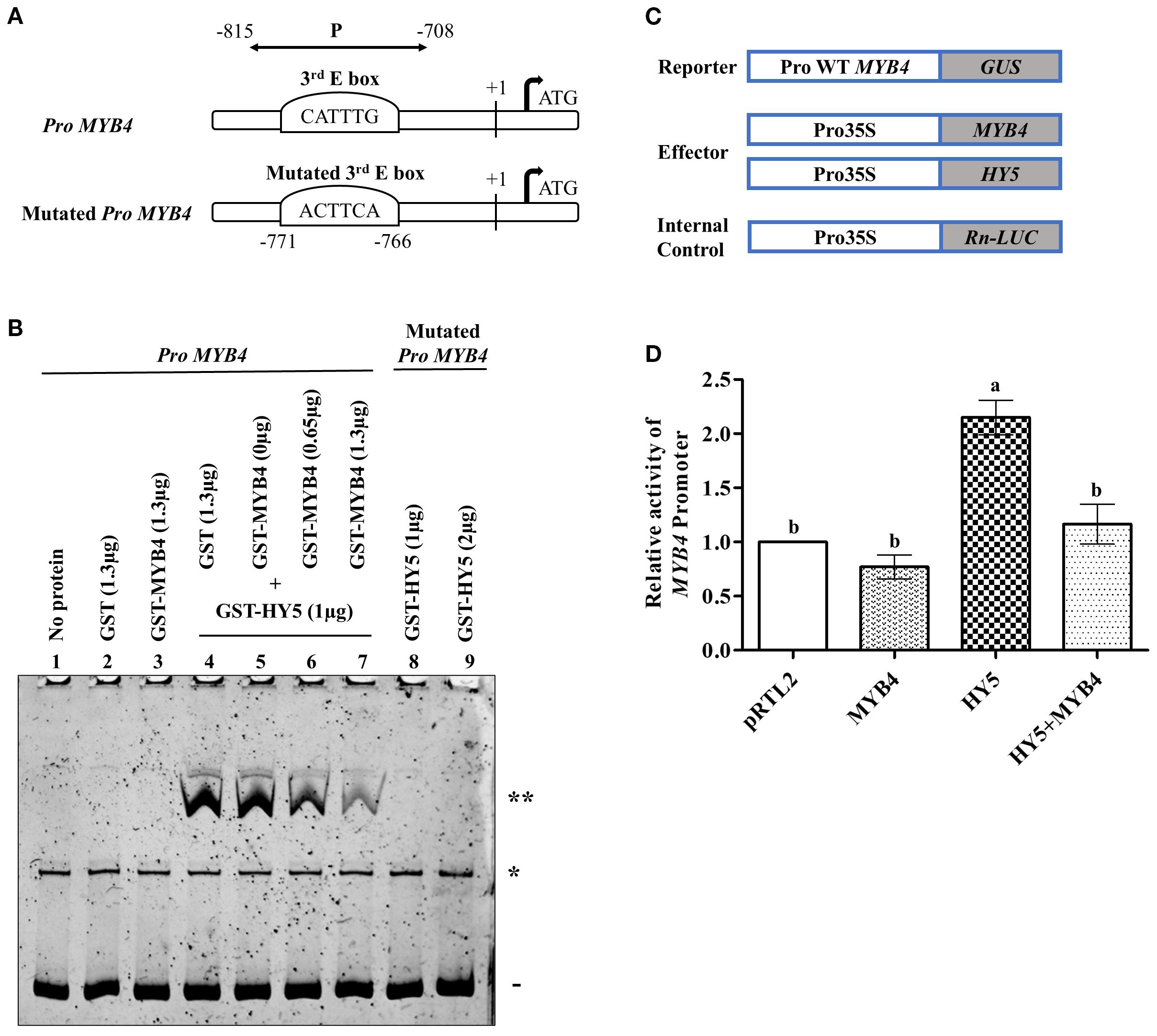
Figure 4. MYB4 autoregulates the HY5 mediated regulation of its own promoter. (A) A visual depiction of the MYB4 promoter showing the wild type and mutated sequence of the 3rd E-Box along with their positions. The (+1) notation denotes the transcription initiation site. The double-headed line labeled as “P” denotes the location of the DNA fragment (from -708 to -815) used in DNA-protein interaction analyses during the competitive EMSA experiment. (B) Electrophoretic mobility shift assay (EMSA) analysis of HY5 protein binding to MYB4 promoter in presence or absence of MYB4 protein. Lane 1 had no protein added, lane 2 contained 1.3 µg of GST and lane 3 had 1.3 µg of GST-MYB4 as control. In lane 4-7, 1 µg of GST-HY5 was added with 100 ng of wild type MYB4 promoter fragments (107 bp long: -708 to -815 bp) along with GST or GST-MYB4 in increasing concentrations. In lane 8-9, 1 µg and 2 µg of GST-HY5 were respectively added with 100 ng of mutated MYB4 promoter fragments as control. The DNA-protein complexes were separated using an 8% native polyacrylamide gel and visualized using SYBR® Green EMSA staining. The presence of a double asterisk (**) signifies the DNA-protein complex, a solid line (-) indicates the free probe, and a single asterisk (*) denotes a spurious band consistently observed across all lanes. (C) Constructs used in the protoplast experiment in different combination to measure the MYB4 promoter activity. (D) The relative MYB4 promoter activity values in Arabidopsis protoplasts transiently transformed with the indicated effectors and reporters constructs. The experiments were repeated three times with similar results, and a representative result has been shown. The error bars indicate ± SD. Different alphabets denote statistically significant differences (p < 0.05) of MYB4 promoter activity. The data were compared by using one-way ANOVA factorial analysis followed by Tukey’s HSD test.
To further gain insight into this regulation, we conducted transient expression assays in Arabidopsis protoplasts using a construct in which the wild-type MYB4 promoter (DNA fragment containing HY5 but not MYB4 binding sites) was fused to the β-glucuronidase reporter gene (Pro AtMYB4-GUS) (Figure 4C). As shown in Figure 4D, while HY5 increased the MYB4 promoter activity by ~2.2-fold, MYB4 alone did not alter the promoter activity. However, when both MYB4 and HY5 were introduced into the protoplast, MYB4 could drastically reduce the HY5 mediated increase in MYB4 promoter activity (Figure 4D). Since we omitted the MYB4 binding site in the transactivation assay, the negative regulation is probably not due to direct binding to the promoter which supports our EMSA results. These results collectively suggest that the physical interaction between MYB4 and HY5 modulates HY5-mediated transcriptional activation of MYB4.
MYB4 and HY5 mediate the integration of light and cytokinin signaling
Previous studies have shown that HY5 protein is stabilized by light (Ang et al., 1998; Wang et al., 2021; Fang et al., 2024) and cytokinin (Vandenbussche et al., 2007). As this study revealed the genetic interplay between HY5 and MYB4 during photomorphogenic growth, we wanted to explore the genetic interaction between MYB4 and HY5 upon cytokinin treatment. To examine this, we first analyzed whether cytokinin alters MYB4 expression. For this, we monitored MYB4 transcript levels in WT seedlings grown on MS plates, with or without 2 µM and 4 µM trans-zeatin. It was observed that at lower light intensity (15 µmol m-2s-1) MYB4 expression was significantly reduced at 4 µM trans-zeatin, however not at 2 µM, suggesting that higher concentrations of cytokinin represses MYB4 expression (Supplementary Figure 4). Next, we examined the hypocotyl length of 6-day-old WT, myb4, hy5, myb4 hy5 mutants in both cytokinin-treated and untreated seedlings grown in the darkness. It was observed that the dark-grown seedlings of each genotype studied exhibited similar hypocotyl length in absence or presence of different concentrations of cytokinin. (Supplementary Figure 5). These results suggest that mutation in MYB4 doesn’t alter the cytokinin mediated inhibition of hypocotyl elongation in dark.
To determine the effect of light and cytokinin on hypocotyl growth, we examined hypocotyl length of 6-day-old WT, myb4, hy5, and myb4 hy5 double mutant seedlings grown under lower and higher WL fluences on MS plates, with or without 1 µM, 2 µM, and 4 µM trans-zeatin. As shown in Figures 5A, C, under lower intensity of WL (15 μmol m−2 s−1), among the various cytokinin concentrations tested, only the highest concentration (4 µM) was able to rescue the etiolated growth of the myb4 mutant. However, at higher WL intensity (60 μmol m−2 s−1), both 2 µM and 4 µM cytokinin were able to suppress hypocotyl elongation, resulting in hypocotyl length in the myb4 mutant similar to that of the WT (Figures 5B, D). Consistent with earlier reports (Vandenbussche et al., 2007), we observed that similar to the control condition, hy5 mutants exhibited elongated hypocotyl as compared to the WT upon cytokinin treatment. Notably, the hypocotyl length of the myb4 hy5 double mutant was significantly higher than that of hy5 mutant under all the conditions tested (Figures 5A–D). These results suggest that the additional loss of MYB4 function alters the hy5 phenotype upon cytokinin treatment. At lower intensity of WL and lower cytokinin concentration, MYB4 and HY5 function additively to regulate hypocotyl length which is similar to the effect of WL alone. However, at the same light intensity but higher cytokinin concentration this genetic interaction is altered and MYB4 and HY5 exhibit synergistic mode of interaction. At higher light intensity even lower cytokinin concentration is sufficient for the cytokinin mediated synergistic interaction between MYB4 and HY5. Thus, the genetic interaction between MYB4 and HY5 is modulated by both light intensity and cytokinin levels.
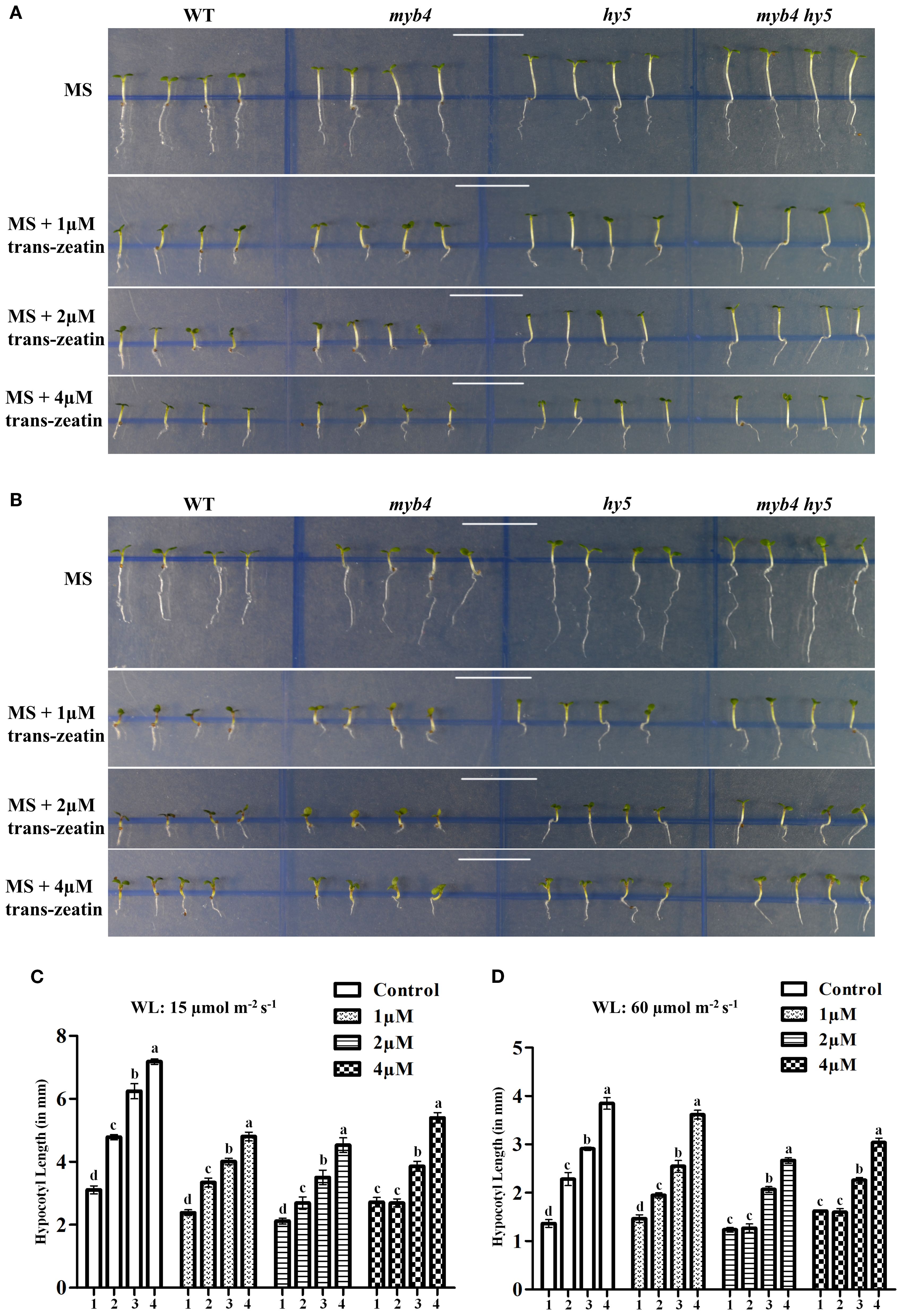
Figure 5. MYB4 and HY5 mediates the integration of light and cytokinin signaling. (A, B) Visible phenotype of 6-day-old WT (Segregated WT Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant WL (15 µmolm-2s-1) (A) or in constant WL (60 µmolm-2s-1) (B) in 0 µM, 1 µM, 2 µM or 4 µM (Top to bottom) trans-Zeatin concentration. Error bars represent ± SD (n=10). Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. 1, 2, 3, 4 under the bar represents WT, myb4, hy5 and myb4 hy5 double mutant respectively. Scale bar = 10 mm. (C, D) Quantification of hypocotyl length of 6-day-old WT (Segregated WT Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant WL (15 µmolm-2s-1) (C) or in constant WL (60 µmolm-2s-1) (D) in 0 µM, 1 µM, 2 µM or 4 µM (Top to bottom) trans-Zeatin concentration. Error bars represent ± SD (n=10). Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. 1, 2, 3, 4 under the bar represents WT, myb4, hy5 and myb4 hy5 double mutant respectively. Scale bar = 10 mm.
We also measured the primary root length in WT, myb4, hy5, and myb4 hy5 double mutant seedlings grown under cytokinin treatment in darkness and under various intensities of white light. While cytokinin suppresses primary root length in WT in both dark and WL (Liu et al., 2022), our results showed no significant differences in inhibition ratios between WT and myb4 mutant seedlings across all conditions (Supplementary Figures 6A–C). Consistent with previous findings by Cluis et al. (2004), the hy5 mutant exhibited resistance to cytokinin treatment under both the intensities of the white light. The myb4 hy5 double mutant displayed primary root inhibition patterns similar to the hy5 mutant in white light, indicating that HY5 works independent of MYB4 in regulating primary root length upon cytokinin treatment (Supplementary Figures 6B, C).
MYB4 and HY5 differentially regulate the expression of cytokinin responsive genes
Our study shows that while cytokinin rescues the etiolated growth of myb4 seedlings, the primary root length of myb4 remains similar to WT in both presence and absence of cytokinin. The ATP-binding cassette (ABC) transporter subfamily G14 (ABCG14) in Arabidopsis is primarily active in roots and is crucial for transporting cytokinin to the shoots. When ABCG14 expression is disrupted, it leads to significant stunted growth in the shoots, which can be reversed by applying trans-zeatin externally (Ko et al., 2014; Zhang et al., 2014). To explore whether altered ABCG14 levels is responsible for the suppression of the hypocotyl length of myb4, we assessed the transcript levels of ABCG14 in 6-day-old WT, myb4, hy5, and myb4 hy5 seedlings grown under low (15 μmol m-2 s-1) and high (60 μmol m-2 s-1) white light in MS or in MS with 2 μM and 4 μM trans-zeatin (Figures 6A, B). Under control conditions (without cytokinin), at both low (15 μmol m-2 s-1) and high intensities (60 μmol m-2 s-1) of WL, the expression of ABCG14 in the myb4 mutant is similar to that in the WT, whereas the hy5 mutants exhibit a significant upregulation (~2.2-fold increase) of ABCG14 expression compared to WT (Figures 6A, B). An additional mutation in MYB4 could suppress the elevated expression of ABCG14 in the hy5 mutant, resulting in expression levels similar to WT in the myb4 hy5 double mutant. Earlier it was reported that the expression of ABCG14 was induced by cytokinin treatment (Ko et al., 2014). When we assessed the transcript levels of ABCG14 in 6-day-old WT, myb4, hy5, and myb4 hy5 seedlings grown under constant low white light (15 μmol m-2 s-1) and under constant high white light (60 μmol m-2 s-1) in MS with 2 μM and 4 μM trans-zeatin, we observed that in low light intensity and low cytokinin concentrations, the expression of ABCG14 is significantly reduced in myb4 mutant than that of WT (Figures 6A, B). The expression level pattern of ABCG14 remains same like the untreated for both myb4 and hy5 mutant in all other conditions. Interestingly, unlike the untreated conditions, in myb4 hy5 double mutant, the expression of ABCG14 is similar to that of hy5 mutants. This suggests that upon 2 μM cytokinin treatment at low intensity of WL, HY5 works downstream to MYB4 to regulate the expression of ABCG14 (Figure 6A).
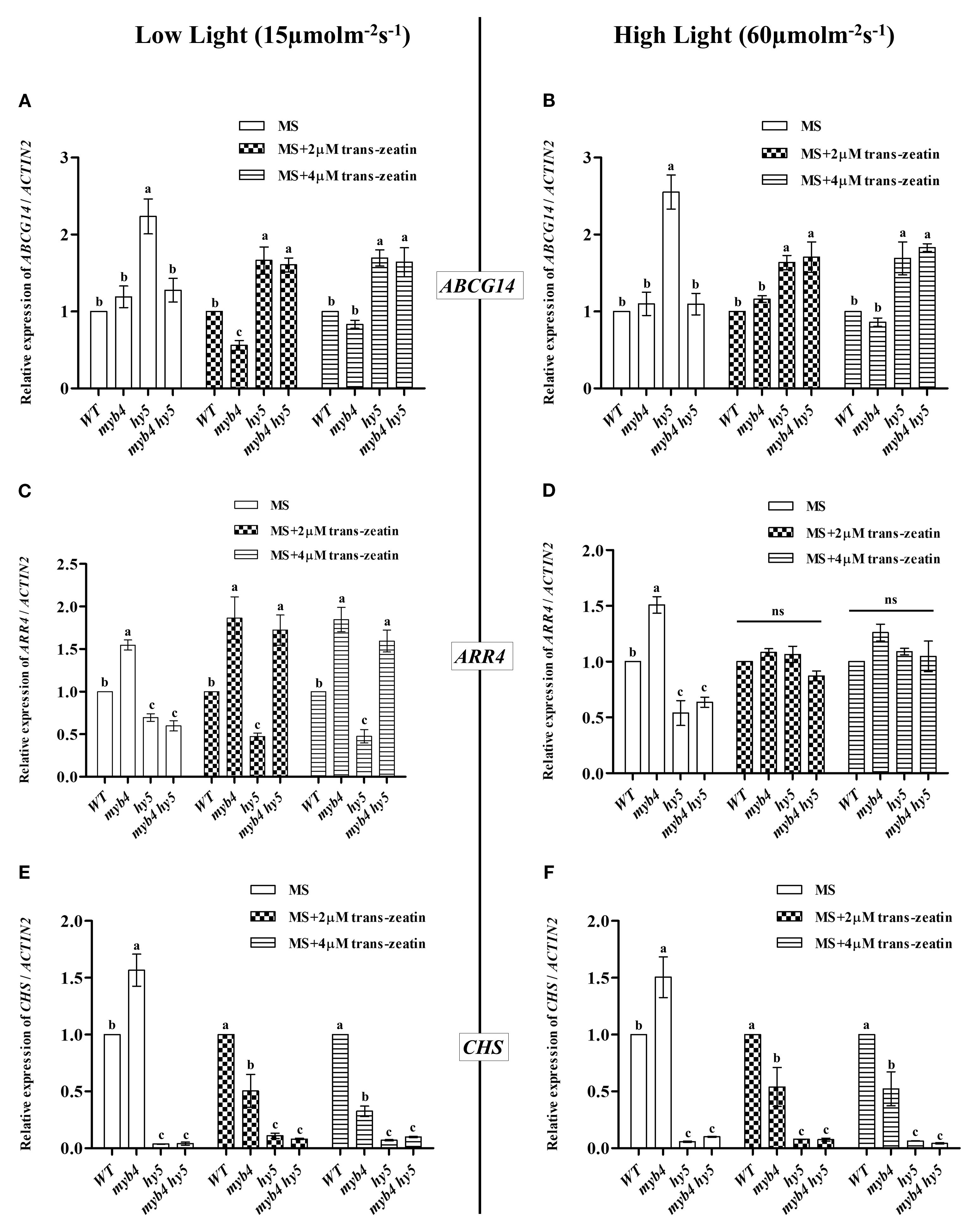
Figure 6. Differential regulation of cytokinin responsive genes by MYB4 and HY5. (A, B) Real-time PCR analyses of ABCG14 in 6-day-old wild type (Segregated wild type Ws-Ler), myb4, hy5 and myb4 hy5 seedlings grown under constant white light (15 µmolm-2s-1) (A) and constant white light (60 µmolm-2s-1) (A) in normal MS media (Control) or in MS media supplemented with 2 µM and 4 µM trans-zeatin (Treated). Error bars represent ± SE of the mean of three biological replicates. Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. ACTIN2 was used as endogenous control. (C, D) Real-time PCR analyses of ARR4 in 6-day-old wild type (Segregated wild type Ws-Ler), myb4, hy5 and myb4 hy5 seedlings grown under constant white light (15 µmolm-2s-1) (C) and constant white light (60 µmolm-2s-1) (D) in normal MS media (Control) or in MS media supplemented with 2 µM and 4 µM trans-zeatin (Treated). Error bars represent ± SE of the mean of three biological replicates. Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. ACTIN2 was used as endogenous control. (E, F) Real-time PCR analyses of CHS in 6-day-old wild type (Segregated wild type Ws-Ler), myb4, hy5 and myb4 hy5 seedlings grown under constant white light (15 µmolm-2s-1) (E) and constant white light (60 µmolm-2s-1) (F) in normal MS media (Control) or in MS media supplemented with 2 µM and 4 µM trans-zeatin (Treated). Error bars represent ± SE of the mean of three biological replicates. Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. ACTIN2 was used as endogenous control.
Sweere et al. (2001) suggested a potential mechanism through which cytokinin and light signaling pathways interact to control hypocotyl elongation through RESPONSE REGULATOR 4 (ARR4). To determine if ARR4 plays a role in the altered response of myb4 mutant seedlings, we measured ARR4 transcript levels in 6-day-old WT, myb4, hy5, and myb4 hy5 seedlings grown under continuous low white light (15 μmol m−2 s−1) and high white light (60 μmol m−2 s−1) conditions, both in untreated MS and in MS supplemented with 2 μM and 4 μM trans-zeatin (Figures 6C, D). The results show that ARR4 expression was significantly increased in the myb4 mutant but decreased in the hy5 mutant. In the myb4 hy5 double mutant, ARR4 expression was similar to that in the hy5 mutant, indicating that HY5 functions downstream of MYB4 in regulating ARR4 expression. As it was previously reported that the ARR4 gene expression is also induced by cytokinin (Sweere et al., 2001), we checked the transcript levels of ARR4 in 6-day-old WT, myb4, hy5, and myb4 hy5 seedlings grown under constant low white light (15 μmol m-2 s-1) and under constant high white light (60 μmol m-2 s-1) in MS with 2 μM and 4 μM trans-zeatin. In low light, the pattern of expression of ARR4 is similar to that of the untreated for both myb4 and hy5 mutants. Interestingly, the expression of ARR4 in myb4 hy5 double mutant is similar to that of myb4 mutants, suggesting that under cytokinin treatment in low light, MYB4 works downstream to HY5 to regulate the expression of ARR4. However, in high light, there were no significant differences in the ARR4 transcript in any cytokinin concentrations tested (Figure 6D). This suggests that the role of MYB4 and HY5 in the regulation of cytokinin induced ARR4 expression is specific to low intensity of WL.
Seedlings treated with exogenous cytokinins accumulate anthocyanin pigments, with increased transcript levels of CHALCONE SYNTHASE (CHS) gene, a key player in the anthocyanin biosynthesis pathway (Deikman et al., 1995). HY5 enhances anthocyanin biosynthesis by directly binding to CHS promoter resulting in its transcriptional activation (Ang et al., 1998; Chattopadhyay et al., 1998). Conversely, MYB4, a known negative regulator of anthocyanin biosynthesis, indirectly repress CHS expression (Wang et al., 2020). Given these observations, we asked how loss of MYB4 and HY5 functions influence CHS regulation under cytokinin treatment. To answer this, we grew 6-day-old WT, myb4, hy5, and myb4 hy5 seedlings under constant low white light (15 μmol m−2 s−1) and high white light (60 μmol m−2 s−1) on MS media without trans-zeatin and with 2 µM and 4 µM trans-zeatin (Figures 6E, F). Consistent with previous findings, we observed that in untreated conditions, CHS transcript level was significantly elevated in the myb4 mutant whereas it was significantly reduced in the hy5 mutant. In the myb4 hy5 double mutant, CHS expression was similar to the hy5 mutant, indicating that HY5 acts downstream of MYB4 in regulating CHS expression under control condition (Figures 6E, F). However, under both low and high cytokinin treatment across both light intensities, CHS expression was significantly reduced (~2-3 fold) in the myb4 mutant. In contrast, CHS expression patterns were similar in hy5 and myb4 hy5 double mutants across both treated and untreated conditions. These findings suggest that MYB4 plays opposing roles in the regulation of CHS expression depending on the presence or absence of cytokinin and HY5 works downstream to MYB4 in this regulation.
Discussion
A signal transduction pathway relies on its interconnected transcriptional regulatory network for proper functional execution. In this study, we show that MYB4 is functionally connected with HY5 to integrate light and cytokinin responses. Furthermore, our results suggest that MYB4, through its physical interaction, attenuates HY5 from binding to the MYB4 promoter. Multiple lines of experimental evidences, including in vitro and in vivo assays, demonstrate a physical interaction between MYB4 and HY5 proteins. Domain-specific interaction studies further reveal that HY5 interacts with the C-terminal domain (transcriptional activation domain, Mitra et al., 2019) of MYB4. Depending on the growth conditions, two or more substrates can compete for binding to the same domain of a protein. For example, upon blue light exposure, photoactivated CRY2 competes with PAP2 for binding to the WD40 domain of COP1, thereby stabilizing PAP2 and resulting in anthocyanin biosynthesis (Ponnu et al., 2019). Previous studies have shown that CAM7 interacts with HY5 (Abbas et al., 2014), and recent findings indicate that CAM7 interacts with the C-terminal domain of MYB4 as well (Dutta et al., 2024). This raises intriguing questions about whether CAM7, HY5 and MYB4 compete for binding with each other or form a transient or obligatory multiprotein complex to regulate photomorphogenesis. One plausible functional significance of MYB4-HY5 physical interaction could be that MYB4 modulates HY5-mediated regulation of MYB4 expression in order to maintain the balance between light and cytokinin mediated hypocotyl elongation. The EMSA results show that the presence of MYB4 significantly reduces HY5-binding to the MYB4 promoter which is further supported by trans-activation experiments in Arabidopsis protoplasts.
Genetic analysis of the myb4 hy5 double mutant shows that long hypocotyl phenotype of hy5 is further enhanced in myb4 hy5 double mutant, resulting in an exaggerated tall phenotype under WL, BL, and FRL, but not in RL. Thus, MYB4 and HY5 function additively under those wavelengths of light. While light-inducible genes like RBCS-1A and CAB1 display expression levels similar to WT in myb4 mutants, their expression in myb4 hy5 mutant is similar to hy5 mutant. Additionally, while HY5 regulate chlorophyll accumulation independent of MYB4, they work antagonistically to regulate anthocyanin levels. Therefore, MYB4 and HY5 coordinate both independently and dependently to regulate photomorphogenic growth.
Integration of light and hormone signaling pathway in Arabidopsis is well studied (Feierabend and de Boer, 1978; Tian and Reed, 1999; Moore et al., 2003; Xiong et al., 2023). Among the various phytohormones, cytokinin mimics the effect of light signals, resulting in the de-etiolation of dark-grown seedlings and the inhibition of hypocotyl elongation at lower intensities of light (Chory et al., 1994; Deikman and Hammer, 1995; Vandenbussche et al., 2007). Our study demonstrates that upon cytokinin treatment, the elongated hypocotyl phenotype of myb4 in WL, is suppressed. At higher light intensities, where the effect of cytokinin is otherwise diminished, exogenous cytokinin could still suppress the hypocotyl elongation of myb4 mutants. One possible explanation for this observation is that, due to impaired light responses in the myb4 mutant, the inhibitory effect of light on cytokinin-mediated suppression of hypocotyl elongation is rescued. Additionally, cytokinin application downregulates MYB4 expression. The genetic interaction between HY5 and MYB4, typically additive, becomes synergistic under cytokinin treatment in WL. These altered morphologies are probably due to changes in the dynamics of physical interactions and transcriptional regulation under different growth conditions. The shift in genetic interaction following treatment with exogenous signals or environmental stimuli, which alters the regulatory dynamics between genes, is not unprecedented. Sandhu et al. (2012) explores the intricate interactions between photoreceptors and brassinosteroid-inactivating P450 enzymes, demonstrating how light treatments influence flowering and gene expression through various genetic pathways. Additionally, the genetic interaction between HY5 and GBF1 also varies in a light intensity- and wavelength-dependent manner (Singh et al., 2012). Apart from regulating light-responsive genes, MYB4 and HY5 regulate the expression of genes involved in cytokinin signaling pathway. While the expression of ABCG14, which is involved in the transport of cytokinin from root to shoot, is significantly downregulated in the myb4 mutant, the expression of ARR4, an A-type response regulator, is significantly upregulated as compared to the corresponding cytokinin-treated WT seedlings. Furthermore, HY5 and MYB4 work in the same pathway to regulate the expression of ABCG14 and ARR4. The reduced expression of ABCG14 in myb4 might be responsible for altered cytokinin export to shoot resulting in the observed stunted growth. Whereas in light signaling MYB4 negatively regulates CHS expression, it positively regulates CHS expression upon cytokinin treatment. Since plants are continuously exposed to changing environmental cues, understanding how they integrate and respond to fluctuating light signals and internal hormone levels provides insights into their intricate mechanisms of adaptability and survival.
In summary, HY5 binds to the MYB4 promoter, and positively regulates its expression (Dutta et al., 2024). Conversely, MYB4 physically interacts with HY5, inhibiting HY5 to bind to its promoter, thus autoregulating its own expression (Figure 7). Furthermore, under higher light intensity, myb4 mutants show hypersensitivity to lower cytokinin concentrations, whereas under lower light intensity, higher cytokinin levels are required to observe this effect. This stoichiometric balances between HY5 and MYB4 provide maintain homeostasis and accordingly control hypocotyl elongation in response to light and cytokinin.

Figure 7. Simplified Model depicting the role of MYB4 and HY5 in light and cytokinin signaling pathway. Light stimulates both HY5 and MYB4 expression, whereas cytokinin stimulates HY5 but suppresses MYB4. HY5 interacts with the 3rd E box on the MYB4 promoter (Dutta et al., 2024) to enhance MYB4 expression. As a feedback response, MYB4 interacts with HY5, preventing its binding to MYB4 promoter, thus finely regulating its expression to balance light and cytokinin signals in controlling hypocotyl growth during Arabidopsis seedling development.
Methods
Plant materials and growth conditions
Arabidopsis thaliana plants used in this study were homozygous myb4 mutant (CS26404), a Ds transposon tagged mutant in Ler background (Dutta et al., 2024) and homozygous hy5-ks50 mutant, a T-DNA insertion mutant in Ws background (Oyama et al., 1997). The myb4 hy5 double mutants were created through genetic crosses using standard methods outlined by Yadav et al. (2005) by crossing homozygous myb4 mutant with hy5 homozygous mutant lines. The homozygosity of both the mutation in T2 generation were confirmed by the inability of gene-specific primer sets, which spanned the full length of MYB4 (MYB4FP and MYB4RP primers) and HY5 (HY5FP and HY5RP) to amplify the MYB4 and HY5 gene respectively from genomic DNA isolated from the plants. One homozygous myb4 hy5 double mutant plant, used for different experiments was additionally confirmed by assessing the transcript levels of both MYB4 and HY5 using RT-PCR analyses. To analyse double mutants from different ecotype backgrounds, a segregated WT line of Ws-Ler from the T2 generation was marked as WT and used as a control to compare phenotypic and molecular differences.
Arabidopsis seeds were surface-sterilized and sown on Murashige and Skoog (MS) plates, stratified in the dark at 4°C to break dormancy before being transferred to growth chambers. They were then grown under specific wavelengths of light at designated light intensities at 22 °C. For different phenotypic studies, seedlings were photographed, and their hypocotyl lengths were measured using ImageJ software. For cytokinin responsive experiments, seeds were sown on MS plates with different concentration of trans-Zeatin.
In vitro pull-down assay
The full-length coding sequences (CDS) of CAM7 and MYB4 were cloned into the pGEX‐4T2 vector to produce fusion proteins with Glutathione S‐transferase (GST). The GST (negative control), GST-CAM7 (positive control) and GST-MYB4 were overexpressed and purified from E. coli BL21 (DE3) using Glutathione Sepharose 4B beads (Amersham Biosciences). The full-length CDS of HY5 was cloned into the pET‐20b (+) vector, adding a 6× Histidine tag to the C-terminus. The HY5-His protein was overexpressed in E. coli BL21 (DE3) and purified with Ni‐NTA Agarose beads (Qiagen). The in vitro binding assay was performed following the protocol from Senapati et al. (2019). For the assay, 2μg of HY5-His was added to microcentrifuge tubes containing Ni-NTA Agarose beads and incubated with in vitro binding buffer (50 mM Tris-Cl pH 7.5, 100 mM NaCl, 0.2% glycerol, 0.1% Triton-X100, 1 mM EDTA, 1 mM PMSF, 0.1% NP-40, and protease inhibitor cocktail; Sigma, Calcutta, India) for 2 hours at 4 °C. After washing off the unbounded proteins, GST, GST-CAM7 or GST-MYB4 was added in equimolar amounts and incubated overnight at 4 °C. Following incubation, the supernatant was collected by centrifugation, and the beads were washed three times with binding buffer. The beads were then resuspended in 5X sodium dodecyl sulfate (SDS) loading buffer and boiled for 10 minutes. Both the supernatant (5%) and pellet fractions were analyzed using 12% SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotted with anti-GST antibodies to detect protein interactions. The same membrane was then stripped and probed using α-His to examine the equal loading of HY5-His.
Yeast two-hybrid assay
For the domain-wise interaction study of MYB4 with HY5, full-length CDS and truncated version of MYB4 was combined with the GAL4 DNA-binding domain (BD-MYB4- bait) and the full-length CDS of HY5 was combined with the GAL4 activation domain (AD-HY5- prey). The fusion constructs were then introduced into yeast AH109 strain using polyethylene glycol/lithium acetate transformation (Clontech), and the growth were assessed of the co-transformed yeast cells on 2D plates (lacking leucine and tryptophan) and 4D plates (lacking leucine, tryptophan, adenine, and histidine). As previously demonstrated by Ang et al., 1998, COP1 and HY5 were used as a positive control for this study.
Co‐immunoprecipitation assays
Co-IP assay was performed as described previously (Senapati et al., 2019) with slight modifications. Full length CDS of MYB4 was cloned into pCAMBIA 1303 to yield a GFP fusion protein. Total protein was extracted from the leaves of 30-d old hy5 and HY5OE plants transiently expressing GFP, CAM7-GFP (Senapati et al., 2019) or MYB4-GFP using Co‐IP buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% NP-40, 10% glycerol and protease inhibitor cocktail). 500 μg of total protein for each sample was incubated with 15 μl of anti-GFP monoclonal antibody (Sigma) for 6h at 4 °C. To this 30 μl of pre blocked protein A‐agarose beads (Sigma) was added and incubated for 2 hr at 4 °C. Then the beads were washed three times with Co‐IP buffer, and after the last wash the beads were boiled in 5X protein loading dye for 10 min to elute the protein and along with that 5% input was run in SDS‐PAGE. Immunoblots were developed using anti-GFP antibody (Sigma, dilution- 1:2500) and co-immunoprecipitated protein was detected by stripping and re-probing with anti-HY5 antibody (Srivastava et al., 2015; Sigma, dilution- 1:5000).
Gene expression analysis
Total plant RNA was isolated from the 6-day-old Arabidopsis thaliana seedlings grown under required light conditions using RNeasy plant mini kit (Qiagen). cDNA was synthesized from 1µg of total RNA using RevertAid H Minus First Strand cDNA synthesis Kit (Thermo Scientific). qPCR was then performed using Power SYBR® Green PCR Master Mix (Applied 509 Biosystems) in StepOnePlusTM Real-Time PCR Systems using respective qPCR gene specific primers. ACTIN2, a housekeeping gene was used as reference gene to normalize the transcript level of the qPCR values. To analyze the relative changes in gene expression, the common 2-ΔΔCT algorithm was used. First, ΔCt value is calculated by normalizing samples Ct to ACTIN2 Ct values. This value for different samples was then normalized to the Ct values of the experimental control and the ΔΔCt values were obtained. Fold expression was calculated by the formula, 2-ΔΔCT, which was then plotted on the graph. The primers used for qPCR analysis has been mentioned in Table 1.
Analysis of pigment accumulation
The levels of chlorophyll and anthocyanin were determined following the protocol described by Holm et al. (2002). Specifically, for chlorophyll content estimation, 6-day-old seedlings grown under continuous white light (WL) were weighed and quickly frozen in liquid nitrogen. The frozen tissues were grounded in a 1.5 ml microcentrifuge tube, and chlorophyll was extracted in the dark using 80% acetone until the pellet became colourless. Chlorophyll a and b levels were calculated using MacKinney’s specific absorption coefficients: chlorophyll a = 12.7(A663) - 2.69(A645), and chlorophyll b = 22.9(A645) - 4.48(A663). The total chlorophyll content was expressed as micrograms of chlorophyll a and b per gram of fresh tissue weight.
For anthocyanin content estimation, 6-day-old seedlings grown under continuous WL were weighed, rapidly frozen in liquid nitrogen, and ground. Total plant pigments were extracted overnight in 0.3 mL of 1% HCl in methanol. After adding 0.2 mL of water, chlorophyll was separated from anthocyanin by adding an equal volume of chloroform. The anthocyanin level was assessed by spectrophotometric measurements of the aqueous phase (A530 - A657) and normalized to the total fresh weight of the tissue. Flowering times were determined as described by Yadav et al. (2005).
Electrophoretic mobility shift assay
GST, GST-MYB4, and GST-HY5 proteins were overexpressed in E. coli BL21 (DE3) and affinity purified using Glutathione Sepharose 4B beads (Amersham Biosciences). For the DNA binding assays, wild type MYB4 promoter spanning the 3rd E box and mutated 3rd E box generated through primer-based site-directed mutagenesis, were used as probes. All of these fragments were cloned into the pBluescript SK+ vector, followed by PCR amplification and purification to produce the probes. Approximately 100 ng of the DNA fragment was incubated with the purified protein in a reaction mixture containing 1X binding buffer (15 mM HEPES, pH 7.5, 35 mM KCl, 1 mM MgCl2, 1 mM EDTA, pH 8.0, 2% glycerol, and 1 mM DTT) in a total volume of 20 µl. The incubation was carried out at room temperature for 20 minutes. Following incubation, the reactions were resolved on a 7% native polyacrylamide gel, stained with SYBR® Green EMSA nucleic acid gel stain (Molecular Probe, Invitrogen), and visualized using the iBright 750 imaging system (Invitrogen) for image documentation.
Transient expression assay in Arabidopsis protoplast
Arabidopsis protoplasts were isolated and transformed following the protocol outlined by Yoo et al. (2007). Wild-type MYB4 promoter fragments containing the three E-boxes were PCR amplified and cloned into the pGAL-UAS-GUS vector. The full-length coding sequences (CDS) of HY5 and MYB4 were individually cloned into the pRTL2 vector to create the effector constructs. Both the reporter and effector constructs were introduced into Col-0 protoplasts. Following transfection, the protoplasts were incubated under 20 µmol/m²/s white light for 10 hours before harvesting. The GUS activity measurement was carried out as mentioned by Yoo et al. (2007) and Chakraborty et al. (2019).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
AP: Investigation, Conceptualization, Writing – review & editing, Data curation, Writing – original draft, Validation, Methodology. RB: Writing – original draft, Investigation, Writing – review & editing. SC: Writing – review & editing, Funding acquisition, Writing – original draft, Supervision.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work is supported by a research grant of Science and Engineering Research Board (SERB), Government of India (CRG/2021/000063) to SC. SC also acknowledges Sir J.C. Bose National Fellowship Award Grant from SERB (2011-21), Government of India. A.P. and RB are recipients of CSIR-SRF from Council of Scientific and Industrial Research (CSIR), Government of India. SC acknowledges the (DST-FIST) instrumentation facility of the department (provided by Department of Science and Technology, Govt. of India).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphgy.2025.1657264/full#supplementary-material
Supplementary Figure 1 | Full Blot of co-immunoprecipitation assay showing interaction between MYB4 and HY5.
Supplementary Figure 2 | The expression of MYB4 is stimulated during dark to light transitions. (A–D) Semiquantitative PCR to show the transcript abundance of MYB4 in 5-day-old dark grown WT (ecotype Landsberg erecta) seedlings transferred either to white light (WL: 20 µmolm-2s-1) (A), blue light (BL: 20 µmolm-2s-1) (B), red light (RL:30 µmolm-2s-1) (C), or far-red light (FR: 1.5 µmolm-2s-1) (D), at various time points. Each PCR reaction was sampled at 25 cycles to monitor the amplification process before saturation occurred. The semiquantitative PCR amplification of ACTIN2 for the same reaction setup was used as endogenous control.
Supplementary Figure 3 | Confirmation of myb4 hy5 double mutant. Semiquantitative PCR to confirm myb4 hy5 double mutant plant by using gene specific primers. ACTIN2 was used as endogenous control.
Supplementary Figure 4 | Expression of MYB4 is downregulated upon cytokinin treatment. Real-time PCR analyses of MYB4 in 6-day-old wild type seedlings grown under constant white light (15 µmolm-2s-1) in normal MS media (Control) or in MS media supplemented with 2 µM and 4 µM trans-zeatin (Treated). Error bars represent ± SE of the mean of three biological replicates. Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. ACTIN2 was used as endogenous control.
Supplementary Figure 5 | Response of myb4 hy5 double mutants to external cytokinin treatment grown under dark condition. (A, B). Visible phenotype of 6-day-old WT (Segregated wild type Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant dark (A); Quantification of hypocotyl length (B) of 6-day-old WT (Segregated wild type Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under constant dark. Error bars represent ± SD (n=10). Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. 1, 2, 3, 4 under the bar represents WT, myb4, hy5 and myb4 hy5 double mutant respectively.
Supplementary Figure 6 | HY5 works independently of MYB4 to regulate the root growth inhibition under cytokinin treatment. (A–C) Quantification of primary root length of 6-day-old WT (Segregated wild type Ws-Ler), myb4, hy5 and myb4 hy5 double mutant seedlings grown under dark (A), constant WL (15 µmolm-2s-1) (B) and WL (60 µmolm-2s-1) (C) in absence or in presence of different concentrations of trans-zeatin. Error bars represent ± SD (n=8). Different alphabets denote statistically significant differences (p < 0.05) by One-way ANOVA followed by a Tukey-Kramer post-hoc test. 1, 2, 3, 4 under the bar represents WT, myb4, hy5 and myb4 hy5 double mutant respectively.
References
Abbas N., Maurya J. P., Senapati D., Gangappa S. N., and Chattopadhyay S. (2014). Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell. 26, 1036–1052. doi: 10.1105/tpc.113.122515
Alvarez-Buylla E. R., Benítez M., Corvera-Poiré A., Chaos Cador A., de Folter S., Gamboa de Buen A., et al. (2010). Flower development. Arabidopsis Book. 8 e0127. doi: 10.1199/tab.0127
Ang L. H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., et al. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. doi: 10.1016/S1097-2765(00)80022-2
Basu R., Pal A., and Chattopadhyay S. (2025). Arabidopsis ARA4 modulates HY5-mediated seedling growth and ABA responsiveness. Plant J.: Cell Mol. Biol. 122, e70260. doi: 10.1111/tpj.70260
Baudry A., Heim M. A., Dubreucq B., Caboche M., Weisshaar B., and Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bentsink L. and Koornneef M. (2008). Seed dormancy and germination. Arabidopsis Book. 6, e0119. doi: 10.1199/tab.0119
Bhagat P. K., Verma D., Sharma D., and Sinha A. K. (2021). HY5 and ABI5 transcription factors physically interact to fine tune light and ABA signaling in Arabidopsis. Plant Mol. Biol. 107, 117–127. doi: 10.1007/s11103-021-01187-z
Brown B. A. and Jenkins G. I. (2008). UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 146, 576–588. doi: 10.1104/pp.107.108456
Chakraborty M., Gangappa S., Jay J. P., Sethi V., Srivastava A., Singh A., et al. (2019). Functional interrelation of MYC 2 and HY 5 plays an important role in arabidopsis seedling development. Plant J. 99, 1080–1097. doi: 10.1111/tpj.14381
Chattopadhyay S., Ang L. H., Puente P., Deng X. W., and Wei N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 10, 673–683. doi: 10.1105/tpc.10.5.673
Chen M., Chory J., and Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87–117. doi: 10.1146/annurev.genet.38.072902.092259
Chen Y., Ince Y.Ç., Kawamura A., Favero D. S., Suzuki T., and Sugimoto K. (2024). ELONGATED HYPOCOTYL5-mediated light signaling promotes shoot regeneration in Arabidopsis thaliana. Plant Physiol. 196, 2549–2564. doi: 10.1093/plphys/kiae474
Chen H., Zhang J., Neff M. M., Hong S. W., Zhang H., Deng X. W., et al. (2008). Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. U. S. A. 105, 4495–4500. doi: 10.1073/pnas.0710778105
Chory J., Reinecke D., Sim S., Washburn T., and Brenner M. (1994). A role for cytokinins in de-etiolation in Arabidopsis. Plant Physiol. 104, 339–347. doi: 10.1104/pp.104.2.339
Cluis C. P., Mouchel C. F., and Hardtke C. S. (2004). The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 38, 332–347. doi: 10.1111/j.1365-313X.2004.02052.x
Dai X., Wang J., Wang L., Liu Z., Li Q., Cai Y., et al. (2022). HY5 inhibits in vitro shoot stem cell niches initiation via directly repressing pluripotency and cytokinin pathways. Plant J.: Cell Mol. Biol. 110, 781–801. doi: 10.1111/tpj.15703
Deikman J. and Hammer P. E. (1995). Induction of anthocyanin accumulation by cytokinins in arabidopsis thaliana. Plant Physiol. 108, 47–57. doi: 10.1104/pp.108.1.47
de Lucas M., Davière J. M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J. M., Lorrain S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
Deng X. W. and Quail P. H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10, 121–129. doi: 10.1006/scdb.1999.0287
Doroshenko A. S., Danilova M. N., Andreeva A. A., Kudryakova N. V., Kuznetsov V. V., and Kusnetsov V. V. (2020). The Transcription Factor HY5 Is Involved in the Cytokinin-Dependent Regulation of the Expression of Genes Encoding Proteins Associated with Bacterial Plastid RNA-Polymerase during De-Etiolation of Arabidopsis thaliana. Doklady. Biochem. Biophys 492, 124–129. doi: 10.1134/S1607672920020076
Duan X., Xu S., Xie Y., Li L., Qi W., Parizot B., et al. (2021). Periodic root branching is influenced by light through an HY1-HY5-auxin pathway. Curr. Biol.: CB 31, 3834–3847.e5. doi: 10.1016/j.cub.2021.06.055
Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., and Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Dutta S., Basu R., Pal A., Kunalika M. H., and Chattopadhyay S. (2024). The homeostasis of AtMYB4 is maintained by ARA4, HY5, and CAM7 during Arabidopsis seedling development. Plant J 120 (6), 2515–2535. doi: 10.1111/tpj.17126
Fang K., Yao X., Tian Y., He Y., Lin Y., Lei W., et al. (2024). Ubiquitin-specific protease UBP14 stabilizes HY5 by deubiquitination to promote photomorphogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 121, e2404883121. doi: 10.1073/pnas.2404883121
Fankhauser C. and Casal J. J. (2004). Phenotypic characterization of a photomorphogenic mutant. Plant J. 39, 747–760. doi: 10.1111/j.1365-313X.2004.02148.x
Feierabend J. and de Boer J. (1978). Comparative analysis of the action of cytokinin and light on the formation of ribulosebisphosphate carboxylase and plastid biogenesis. Planta 142, 75–82. doi: 10.1007/BF00385123
Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Flores S. and Tobin E. M. (1986). Benzyladenine modulation of the expression of two genes for nuclear-encoded chloroplast proteins in Lemna gibba: apparent post-transcriptional regulation. Planta 168, 340–349. doi: 10.1007/BF00392359
Fornalé S., Lopez E., Salazar-Henao J. E., Fernandez-Nohales P., Rigau J., and Caparros-Ruiz D. (2014). AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 55, 507–516. doi: 10.1093/pcp/pct187
Gangappa S. N. and Botto J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Holm M., Ma L. G., Qu L. J., and Deng X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. doi: 10.1101/gad.969702
Jiao Y., Lau O. S., and Deng X. W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230. doi: 10.1038/nrg2049
Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jing Y. and Lin R. (2020). Transcriptional regulatory network of the light signaling pathways. N. Phytol. 227, 683–697. doi: 10.1111/nph.16602
Kendrick R. E. and Kronenberg G. H. M. (1994). Photomorphogenesis in plants (Dordrecht, The Netherlands: Kluwer Academic Publishers).
Ko D., Kang J., Kiba T., Park J., Kojima M., Do J., et al. (2014). Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. U. S. A. 111, 7150–7155. doi: 10.1073/pnas.1321519111
Koornneef M., Rolff E., and Spruit C. J. P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.). Heynh. Z. Pflanzenphysiol. 100S, 147–160. doi: 10.1016/S0044-328X(80)80208-X
Kumar S., Mazumder M., Gupta N., Chattopadhyay S., and Gourinath S. (2016). Crystal structure of Arabidopsis thaliana calmodulin7 and insight into its mode of DNA binding. FEBS Lett. 590, 3029–3039. doi: 10.1002/1873-3468.12349
Kushwaha R., Singh A., and Chattopadhyay S. (2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell. 20, 1747–1759. doi: 10.1105/tpc.107.057612
Lau O. S. and Deng X. W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13, 571–577. doi: 10.1016/j.pbi.2010.07.001
Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 (3), 731–749. doi: 10.1105/tpc.106.047688
Lifschitz S., Gepstein S., and Horwitz B. A. (1990). Phytochrome regulation of greening in wild type and long-hypocotyl mutants ofArabidopsis thaliana. Planta 181, 234–238. doi: 10.1007/BF02411544
Liu S., Strauss S., Adibi M., Mosca G., Yoshida S., Dello Ioio R., et al. (2022). Cytokinin promotes growth cessation in the Arabidopsis root. Curr. Biol.: CB 32, 1974–1985.e3. doi: 10.1016/j.cub.2022.03.019
Marzi D., Brunetti P., Mele G., Napoli N., Calo` L., Spaziani E., et al. (2020). Light controls stamen elongation via cryptochromes, phytochromes and COP1 through HY5 and HYH. Plant J. 103, 379–394. doi: 10.1111/tpj.14736
McCormack E., Tsai Y. C., and Braam J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Mitra M., Agarwal P., Kundu A., Banerjee V., and Roy S. (2019). Investigation of the effect of UV-B light on Arabidopsis MYB4 (AtMYB4) transcription factor stability and detection of a putative MYB4-binding motif in the promoter proximal region of AtMYB4. PloS One 14 (8), e0220123. doi: 10.1371/journal.pone.0220123
Mitra M., Agarwal P., and Roy S. (2021). The N-terminal MYB domains affect the stability and folding aspects of Arabidopsis thaliana MYB4 transcription factor under thermal stress. Protoplasma 258, 633–650. doi: 10.1007/s00709-020-01590-1
Moore B., Zhou L., Rolland F., Hall Q., Cheng W. H., Liu Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science (New York N.Y.) 300, 332–336. doi: 10.1126/science.1080585
Mu R. L., Cao Y. R., Liu Y. F., Lei G., Zou H. F., Liao Y., et al. (2009). An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 19, 1291–1304. doi: 10.1038/cr.2009.83
Neff M. M. and Chory J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. doi: 10.1104/pp.118.1.27
Oyama T., Shimura Y., and Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. doi: 10.1101/gad.11.22.2983
Paik I. and Huq E. (2019). Rapid examination of phytochrome-phytochrome interacting factor (PIF) interaction by in vitro coimmunoprecipitation assay. Methods Mol. Biol. (Clifton N.J.) 2026, 21–28. doi: 10.1007/978-1-4939-9612-4_2
Ponnu J., Riedel T., Penner E., Schrader A., and Hoecker U. (2019). Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. Proc. Natl. Acad. Sci. U. S. A. 116, 27133–27141. doi: 10.1073/pnas.1909181116
Rasmussen A., Mason M. G., De Cuyper C., Brewer P. B., Herold S., Agusti J., et al. (2012). Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 158, 1976–1987. doi: 10.1104/pp.111.187104
Sandhu K. S., Hagely K., and Neff M. N. (2012). Genetic interactions between brassinosteroid-inactivating P450s and photomorphogenic photoreceptors in arabidopsis thaliana. G3 Genes|Genomes|Genetics 2, 1585–1593. doi: 10.1534/g3.112.004580
Senapati D., Kushwaha R., Dutta S., Maurya J. P., Biswas S., Gangappa S., et al. (2019). COP1 regulates the stability of CAM7 to promote photomorphogenic growth. Plant Direct 3, e00144.
Shin J., Park E., and Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J.: Cell Mol. Biol. 49, 981–994. doi: 10.1111/j.1365-313X.2006.03021.x
Sibout R., Sukumar P., Hettiarachchi C., Holm M., Muday G. K., and Hardtke C. S. (2006). Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2, e202. doi: 10.1371/journal.pgen.0020202
Singh A., Ram H., Abbas N., and Chattopadhyay S. (2012). Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J. Biol. Chem. 287, 25995–26009. doi: 10.1074/jbc.M111.333906
Srivastava A. K., Senapati D., Srivastava A., Chakraborty M., Gangappa S. N., and Chattopadhyay S. (2015). Short Hypocotyl in White Light1 Interacts with Elongated Hypocotyl5 (HY5) and Constitutive Photomorphogenic1 (COP1) and Promotes COP1-Mediated Degradation of HY5 during Arabidopsis Seedling Development. Plant Physiol. 169 (4), 2922–2934. doi: 10.1104/pp.15.01184
Sullivan J. A. and Deng X. W. (2003). From seed to seed: the role of photoreceptors in Arabidopsis development. Dev. Biol. 260, 289–297. doi: 10.1016/s0012-1606(03)00212-4
Sweere U., Eichenberg K., Lohrmann J., Mira-Rodado V., Baurle I., et al. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science (New York, N.Y.) 294 (5544), 1108–1111. doi: 10.1126/science.1065022
Tian Q. and Reed J. W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development (Cambridge England) 126, 711–721. doi: 10.1242/dev.126.4.711
Vandenbussche F., Habricot Y., Condiff A. S., Maldiney R., Van der Straeten D., and Ahmad M.. (2007). HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. : Cell Mol. Biol. 49 (3), 428–441. doi: 10.1111/j.1365-313X.2006.02973.x
van Gelderen K., Kang C., Paalman R., Keuskamp D., Hayes S., and Pierik R. (2018). Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 30, 101–116. doi: 10.1105/tpc.17.00771
Wang W., Paik I., Kim J., Hou X., Sung S., and Huq E. (2021). Direct phosphorylation of HY5 by SPA kinases to regulate photomorphogenesis in Arabidopsis. N. Phytol. 230, 2311–2326. doi: 10.1111/nph.17332
Wang X. C., Wu J., Guan M. L., Zhao C. H., Geng P., and Zhao Q. (2020). Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J.: Cell Mol. Biol. 101, 637–652. doi: 10.1111/tpj.14570
Xiong H., Lu D., Li Z., Wu J., Ning X., Lin W., et al. (2023). The DELLA-ABI4-HY5 module integrates light and gibberellin signals to regulate hypocotyl elongation. Plant Commun. 4, 100597. doi: 10.1016/j.xplc.2023.100597
Xu W., Grain D., Bobet S., Le Gourrierec J., Thevenin J., Kelemen Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. N. Phytol. 202, 132–144. doi: 10.1111/nph.12620
Yadav V., Mallappa C., Gangappa S. N., Bhatia S., and Chattopadhyay S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 17, 1953–1966. doi: 10.1105/tpc.105.032060
Yang B., Song Z., Li C., Jiang J., Zhou Y., Wang R., et al. (2018). RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 14, e1007839. doi: 10.1371/journal.pgen.1007839
Yoo S. D., Cho Y. H., and Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yu Y., Wang J., Zhang Z., Quan R., Zhang H., Deng X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet. 9, e1004025. doi: 10.1371/journal.pgen.1004025
Zhang H., He H., Wang X., Wang X., Yang X., Li L., et al. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. : Cell Mol. Biol. 65 (3), 346–358. doi: 10.1111/j.1365-313X.2010.04426.x
Zhang K., Novak O., Wei Z., Gou M., Zhang X., Yu Y., et al. (2014). Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 5, 3274. doi: 10.1038/ncomms4274
Zhao J., Zhang W., Zhao Y., Gong X., Guo L., Zhu G., et al. (2007). SAD2, an importin-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19, 3805–3818. doi: 10.1105/tpc.106.048900
Keywords: MYB4, HY5, photomorphogenesis, cytokinin, seedling development
Citation: Pal A, Basu R and Chattopadhyay S (2025) MYB4 and HY5 integrate light and cytokinin signaling pathways during Arabidopsis seedling development. Front. Plant Physiol. 3:1657264. doi: 10.3389/fphgy.2025.1657264
Received: 01 July 2025; Accepted: 01 September 2025;
Published: 19 September 2025.
Edited by:
Alexander Heyl, Adelphi University, United StatesReviewed by:
Vikas Garhwal, Indian Institute of Science Education and Research Kolkata, IndiaHan Dong, Henan Agricultural University, China
Copyright © 2025 Pal, Basu and Chattopadhyay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sudip Chattopadhyay, c3VkaXBjaGF0dG9AZ21haWwuY29t; c3VkaXAuY2hhdHRvcGFkaHlheUBidC5uaXRkZ3AuYWMuaW4=
 Abhideep Pal
Abhideep Pal Riya Basu
Riya Basu Sudip Chattopadhyay
Sudip Chattopadhyay