Abstract
Rice (Oryza sativa L.) is a basic staple crop, sustaining nearly half of the global population and underpinning the livelihoods of millions. As climate change exacerbates the frequency of drought, salinity, and nutrient limitations, optimizing rice root system architecture (RSA)—particularly deep root systems—has become essential for ensuring productivity and resilience. Deep RSA, characterized by steeper root growth angles and extensive large lateral roots, enhances access to water and nutrients in deeper soil layers, improving drought tolerance, nutrient use efficiency, and yield stability under environmental stress. This review synthesizes advances in understanding the physiological, genetic, and hormonal regulation of deep root development in rice. Key genes, including DEEPER ROOTING 1 (DRO1), qSOR1, and SOR1, regulate root growth angle and depth, while aquaporins and hormonal pathways (auxin, cytokinin, ethylene, abscisic acid, gibberellin) modulate root dynamics and water transport. The plasticity of RSA allows rice to adapt to diverse environments, with deeper roots conferring resilience to drought and nutrient deficiency, and shallower roots offering advantages in saline soils. Advances in marker-assisted selection, genome editing (CRISPR-Cas9), and RNA-based technologies enable precise manipulation of root traits, accelerating the development of climate-resilient rice varieties. Agronomic practices such as deep fertilizer placement further promote rooting depth and resource use efficiency. Additionally, deep RSA offers potential as a sustainable carbon sink, contributing to climate change mitigation. By leveraging these innovations, deep root systems can enhance rice crop resilience and support sustainable agriculture, ensuring global food security in a changing climate.
1 Introduction: the significance of rice and the importance of deep root systems
Rice (Oryza sativa L.) stands as a cornerstone of global food security, serving as a primary caloric source for nearly half of the world’s population. Its cultivation is particularly vital for the world’s poor, with an estimated 900 million individuals depending on it as either producers or consumers (Verma et al., 2022). This profound reliance underscores the urgent need to safeguard and enhance rice production amidst increasingly challenging environmental conditions.
Historically, the rice breeding programs have predominantly focused on enhancing the above ground characteristics like grain yield, plant height and disease resistance, and little or no attention was paid to the “below ground” characteristics like the roots and the rhizosphere. Significant progress has been made in understanding and manipulating the above-ground traits, such as increasing tiller number and optimizing leaf structure, to improve photosynthesis and yield. However, these improvements alone are insufficient to address environmental stressors like drought and salinity, warranting enhanced resource acquisition from the soil. However, the modern breeding approaches are recognizing the crucial role played by the root system of crop plants in the overall plant performance and resilience (Yoshino et al., 2019). Optimizing plant architecture, including above-ground components like tiller number and leaf area, and below-ground roots, is increasingly recognized as a crucial factor in improving crop productivity and enhancing adaptation to diverse and changing environments (Kitomi et al., 2020).
Root system architecture (RSA) refers to the spatial configuration and developmental dynamics of a plant’s root network within the soil. An efficiently structured RSA is crucial for the effective uptake of vital resources such as water and nutrients, as well as for securing the plant’s physical stability. Beyond resource acquisition, RSA significantly influences a plant’s resilience to environmental stressors (Verma et al., 2022). However, RSA, particularly deep root systems, has historically received less attention from researchers and breeders (Kitomi et al., 2020) owing to the inherent difficulties in observing and quantifying below-ground plant traits. These challenges include labor-intensive excavation, limited imaging technologies, and problems in replicating field conditions. Recent advances in non-invasive imaging technologies and molecular genetics have opened new avenues for the study and manipulation of RSA.
The growing scientific focus on subterranean traits like root architecture reflects an evolving appreciation for the complex interactions between plants and their surroundings. This trend marks a shift toward integrative strategies aimed at boosting crop robustness and yield, recognizing that key physiological adaptations often originate below ground (Karlova et al., 2021). In particular, the role of deep rooting in enhancing access to water and nutrients highlights the direct relationship between root morphology and stress tolerance. Deep root systems in rice offer a key adaptive mechanism that can help stabilize yields under stress, making their study and enhancement essential for ensuring a sustainable food supply for the future. These systems play an indispensable role in augmenting the plant’s capacity to acquire essential resources, including water and nutrients from deeper soil layers - tapping into resources inaccessible to shallow-rooted counterparts, and in improving its overall productivity and tolerance to various environmental stresses (Verma et al., 2022). This foundational link has positioned root architecture as a central target in efforts to improve crop performance in increasingly challenging environments.
This review synthesizes current understanding of the physiological, genetic, and hormonal mechanisms underlying deep root development in rice. It highlights the benefits of deep rooting for resource acquisition and stress resilience, explores recent breakthroughs in genetic and biotechnological approaches, and discusses agronomic strategies for promoting deep root systems. By integrating these insights, we aim to provide a foundation for breeding and management practices that will ensure sustainable rice production in the face of climate change.
2 Deep root architecture in rice
2.1 Rice root system development
The rice root system exhibits a complex and dynamic architecture, fundamentally characterized as a fibrous root system (Wang et al., 2020) (Figure 1A). This system is comprised of several distinct types of roots, each with its own developmental origin and functional role. The primary root, also known as the seminal root, is the first to emerge from the germinating seed, providing initial anchorage and initiating the uptake of water and nutrients. However, it is short-lived in rice and typically degenerates as plant develops, with its function soon overtaken by nodal roots. Nodal roots, also known as crown or adventitious roots, develop post-embryonically from the basal nodes of the stem and form the major component of the mature root system (Wang et al., 2020). These roots are essential for long-term water and nutrient absorption and can be further classified into embryonic crown roots, which emerge early from lower nodes, and post-embryonic crown roots, which develop later from higher stem nodes, expanding the root system as the plant grows. Lateral roots (LR), branching from both primary and nodal roots, significantly increase the surface area of the root system, enhancing water and nutrient uptake. Subsequently, nodal roots, also referred to as crown roots (CR) or adventitious roots, develop post-embryonically from the basal nodes of the stem, forming the bulk of the mature root system (Mai et al., 2014). They are divided into large lateral roots (LLR), which are thicker and support deeper soil exploration, and small lateral roots (SLR), which are finer and more numerous, aiding in precise nutrient and moisture absorption. Finally, brace roots are a specialized type of adventitious root that emerge from above-ground stem nodes later in the plant’s development. They provide additional mechanical support, particularly in tall rice varieties, helping to prevent lodging, and may also contribute to water and nutrient absorption, especially under resource-limited conditions (Verma et al., 2022).
Figure 1
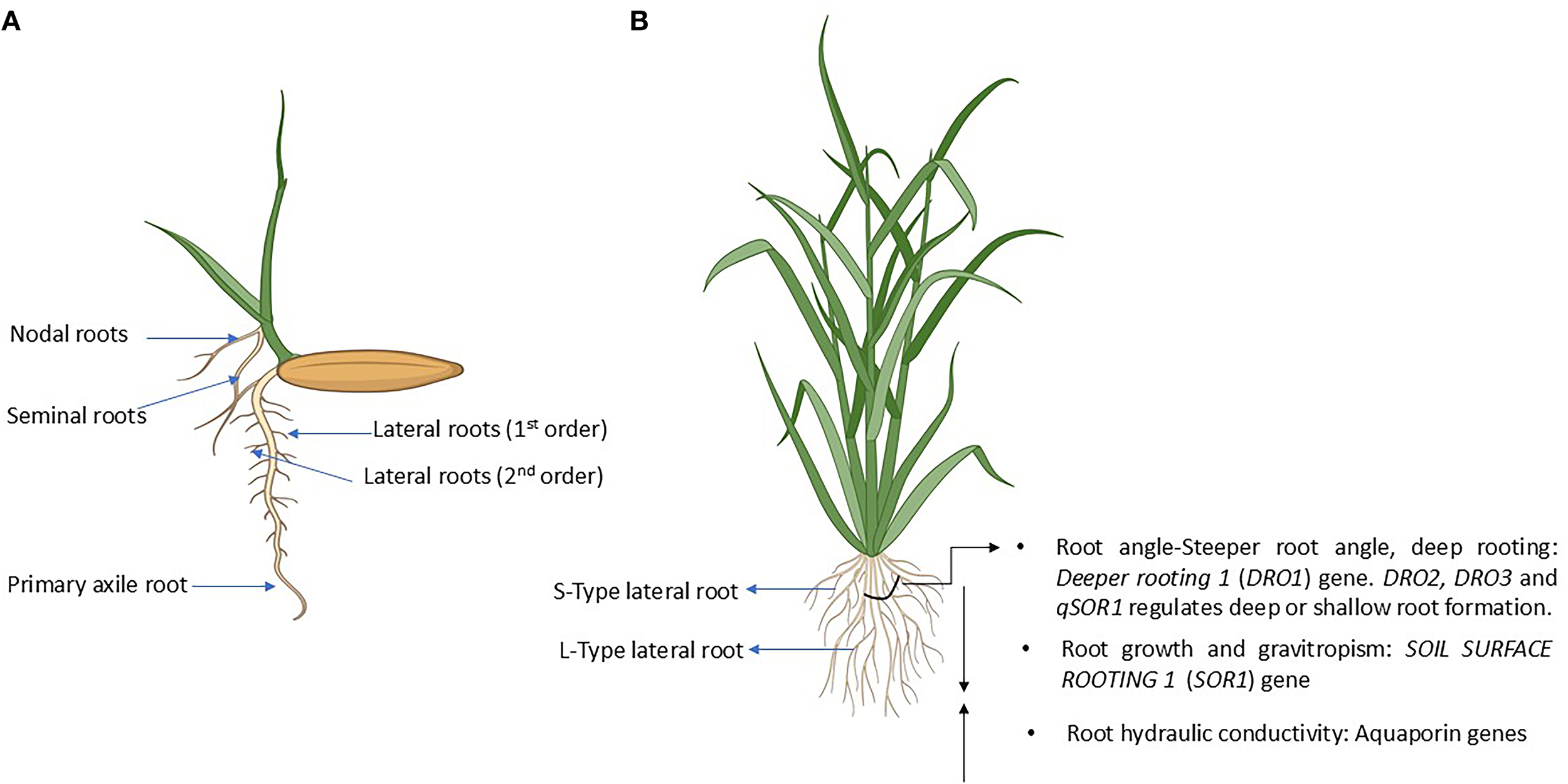
The rice root system and genes influencing deep root development. (A) Structure of rice seedlings showing seminal and nodal roots emerging from the seed. (B) Schematic highlighting beneficial traits conferred by genes promoting deep rooting, including the roles of S-type and L-type lateral roots in nutrient uptake under conditions such as phosphorus deficiency. See text for more details. Rice seedling and plant images created with Biorender (www.bioRender.com).
A key aspect of deep root development is the bifurcation of the radicle and crown roots, which leads to the formation of longer lateral roots. These longer LRs demonstrate positive geotropism, indeterminate growth, emerge intermittently, and harbor expandable long or short LRs till the fifth branching order, allowing them to extend into deeper soil horizons. In contrast, shorter lateral roots are ageotropic, displaying determinate growth, larger in number, and typically remain confined to the upper, shallower layers of the soil. CRs, on the other hand, develop from the stem whilst maintaining similar features evident in root tissues (Mai et al., 2014; Verma et al., 2022) (Figure 1A). The specific timing and patterns of root development are not uniform across all rice cultivars. Different cultivars have evolved diverse root systems that are finely tuned to the particular environmental conditions in which they thrive, including variations in water availability, nutrient levels, and soil type (Pedersen et al., 2021; Verma et al., 2021) For example, in response to water deficit, upland rice cultivars exhibit the remarkable ability to enhance the development of deeper roots, enabling them to tap into water reserves located in the subsoil and thereby combat the effects of drought stress (Lynch and Wojciechowski, 2015; Plett et al., 2020). This inherent adaptability, or plasticity, of the rice root system architecture highlights the potential for selecting and breeding cultivars with root traits that are advantageous for specific environments and stresses. The developmental progression from the initial seminal root to the extensive network of nodal and lateral roots, with some extending to significant depths, reflects a strategic allocation of resources by the plant. This phased development ensures early establishment and then progressively enhances the plant’s capacity to acquire resources from a larger soil volume. Furthermore, the diversity observed in root system development among different rice cultivars suggests a rich genetic reservoir that can be leveraged to breed for improved RSA traits, including the ability to form thicker, branched and deeper root systems that are crucial for adapting to the challenges of climate change.
2.2 Deeper roots and nutrient uptake efficiency
A significant advantage conferred by a deep root architecture is the efficient access and absorption of nutrients from deeper layers of the soil. This efficiency is vital for nitrogen (Wasaya et al., 2018), which is highly mobile within the soil profile and tends to leach downwards with water movement post rainfall or irrigation events (Plett et al., 2020). Rice varieties with well-developed deep root systems are better equipped to exploit these deeper nitrogen reserves, leading to improved nitrogen use efficiency (Verma et al., 2022). Studies have consistently demonstrated that rice varieties characterized by larger root systems, exhibiting increased total root length, root surface area, and root volume, tend to have higher nitrogen uptake efficiency (Liu et al., 2023). The concept of the “Steep, Cheap and Deep” root ideotype, a valuable target for breeding rice varieties with enhanced efficiency in acquiring resources, including both nitrogen and water, from the deeper soil horizons (Galindo-Castañeda et al., 2022; Lynch, 2019). This ideotype emphasizes specific root traits such as a steeper root growth angle, a reduced number of crown roots, longer but fewer lateral roots, and anatomical features that minimize the metabolic cost of root development while maximizing the capture of resources from the subsoil (Lynch, 2019; Galindo-Castañeda et al., 2022). The DEEPER ROOTING 1 (DRO1) gene has been identified as a key genetic factor that regulates root depth and angle of the nodal roots resulting in steeper, deeper roots capable of enhancing the plant’s ability to capture nutrients from deeper soil profiles and potentially circumvent the drought stress (Verma et al., 2022; Galindo-Castañeda et al., 2022; Uga et al., 2011). Thus, breeding for rice varieties suitable for low-input, rain-fed agricultural systems prioritizes the selection of genotypes exhibiting deep and robust root systems (Kim et al., 2020). Conversely, breeding efforts for high-input, irrigated conditions favors rice varieties with shallower root systems, which are more adept at acquiring resources that are typically concentrated in the topsoil layers where fertilizers are applied (Kim et al., 2020). However, with growing concerns regarding environmental sustainability and the imperative to optimize the use of agricultural inputs, current focus is on developing root systems that can efficiently utilize nutrients distributed throughout the entire soil profile.
In addition, the strategic application of fertilizers in deeper soil layers, a technique known as deep placement (DP), significantly enhanced nutrient uptake in rice by stimulating root elongation and proliferation in the vicinity of the fertilizer (Mumtahina et al., 2023). The expression of specific nitrogen transporter genes within the roots is essential for the efficient uptake of nitrogen, and the development of root traits themselves is often regulated by the availability of nitrogen in the soil, underscoring the complex interplay between root development and nutrient acquisition (Liu et al., 2023). A study by Mumtahina et al. (2023) reported that DP at 15–20 cm depth increased root biomass by 25% and nutrient uptake (e.g., nitrogen, calcium, magnesium) by 18–22% compared to surface application. DP promotes the expression of nitrogen transporter genes (e.g., OsNRT1.1A), which are upregulated by 1.5–2-fold in deeper roots under low-nitrogen conditions, improving NUE (Liu et al., 2023). This inherent plasticity of rice RSA to nutrient availability enhances uptake efficiency and supports climate-smart agriculture by reducing fertilizer runoff (Shrestha et al., 2024).
The potential of deep fertilizer placement in promoting root growth and nutrient uptake accentuates a beneficial interaction between agricultural practices and root system architecture. By strategically managing the availability of nutrients in deeper soil layers, it is possible to encourage rice plants to develop more extensive deep root systems, leading to improved resource utilization and a reduction in environmental impacts associated with fertilizer runoff. The “Steep, Cheap and Deep” root ideotype provides a clear direction for breeding efforts aimed at developing rice varieties that are not only more drought-tolerant but also more efficient in acquiring essential nutrients from deeper soil layers, contributing to a more sustainable approach to rice production.
2.3 Improved water uptake and drought avoidance
A well-developed deep root architecture enhances the efficiency of water uptake in rice plants, useful trait under conditions of water scarcity and drought stress (Verma et al., 2022; Lynch and Wojciechowski, 2015). By extending further into the soil profile, deeper roots enable plants to access water reserves located in the subsoil, which often remain available even when the upper soil layers become dry during prolonged periods without rainfall or irrigation (Han et al., 2016). This ability to tap into deeper water sources is a key mechanism for drought avoidance in many plants (Uga et al., 2013a). Rice plants exhibit this plasticity in their root development, where growth of deeper roots is triggered as a physiological response to water deficit conditions. This adaptive strategy allows them to actively seek out and extract water from the subsoil, thereby mitigating the adverse effects of drought stress on overall plant growth and grain yield (Verma et al., 2022). The angle at which roots grow downwards into the soil, referred to as the root growth angle (RGA), is a critical determinant of the overall rooting depth. Steeper root angles, which result in a more vertical orientation of root growth, improve water uptake in various plant species, including rice, especially when subjected to drought conditions (Uga et al., 2013a). Genes such as DEEPER ROOTING 1 (DRO1), qSOR1 (quantitative trait locus for SOIL SURFACE ROOTING 1) (Kitomi et al., 2020), and SOR1 (SOIL SURFACE ROOTING 1) (Hanzawa et al., 2013) (Figure 1B) play a significant role in regulating RGA in rice, with specific alleles associated with a deeper rooting habit that has been proven to enhance grain yields under drought stress (Uga et al., 2013a). Besides DRO1, the QTLs DRO2 (Uga et al., 2013b) and DRO3 (Uga et al., 2015) along with qSOR1 have also been shown to associate with the formation of deep or shallow rice root systems. In Arabidopsis, the natural variations of the gene EXOCYST70A3 regulates PIN4 distribution that in turn modulates auxin signaling and accumulation (Ogura et al., 2019), however, the homologue of this gene is hardly studied in rice.
Interestingly, while the root hairs are known to improve the accumulation of both water and nutrients in many plant species, their role in water absorption in rice is less prominent compared to their importance in nutrient uptake (Gonzalez et al., 2021). Instead, the expression of aquaporins, which are water channel proteins embedded in plant cell membranes, plays a central role in regulating the water transport capacity of the rice root system (Kim et al., 2020). Changes in aquaporin expression under drought stress directly affect root hydraulic conductivity, a measure of efficient water movement through the root system to meet transpirational demands (Kim et al., 2020). Deeper nodal roots with a low metabolic cost per unit length, achieved through a smaller root diameter and/or increased development of aerenchyma (air-filled spaces within the root cortex), transport water efficiently, potentially through smaller diameter metaxylem vessels. These are particularly adept for water uptake during drought (Fonta et al., 2022). In addition, deeper roots exhibiting high axial hydraulic conductance, coupled with a reduced metabolic cost of root development and maintenance, also contribute to greater shoot biomass accumulation under drought stress (Fonta et al., 2022).
The consistent correlation between deeper rooting, enhanced water uptake, and drought tolerance is a key feature of physiological adaptation in rice. Breeding for increased root depth and optimized root growth angle is therefore a robust strategy for improving rice resilience to water scarcity, a major consequence of climate change. Furthermore, the complex interplay between root architecture, root anatomy, and the expression of aquaporins highlights the multifaceted nature of water uptake in rice roots, suggesting that a holistic approach considering all these interacting factors is necessary for optimizing water acquisition under drought conditions.
3 Deep root architecture in stress mitigation and resilience
The cultivation of rice is increasingly challenged by a wide array of abiotic stresses, including extreme temperatures, drought, heavy metal toxicity and salinity. These environmental pressures are intensifying due to changes in the global climate, resulting in reduced grain production, quality deterioration and substantial crop loss (Hilker and Schmülling, 2019). A well-adapted RSA is intimately associated to a plant’s ability to tolerate abiotic stresses (Table 1).
Table 1
| Abiotic stress | Primary impact on rooting | Key root architectural/morphological changes | Adaptive significance/mechanism | References |
|---|---|---|---|---|
| Drought | Inhibits cell elongation, reduces growth, water deficit | Deeper roots; reduced total length, lateral root growth (severe); increased lateral roots (tolerant genotypes/mild stress); altered root angle, diameter, density | Water uptake, drought avoidance, maintaining stomatal opening, grain filling | Uga et al., 2013a |
| Salinity | Ionic toxicity, osmotic stress, reduced water availability | Reduced root length; deeper, more expansive root systems; altered root hair density; increased root exudation; aerenchyma formation | Ion exclusion/homeostasis, access to less saline water, oxygen transport in waterlogged soils | Sackey et al., 2025 |
| Submergence | Low oxygen availability, inhibits primary root growth | Shallower, more compact root systems; increased adventitious roots (ARs); aerenchyma formation | Oxygen transport, nutrient acquisition, replacement of dysfunctional primary roots | Gong et al., 2025; Wang et al., 2024a |
| Heavy Metal Toxicity | Decreases metabolic activities, morphological abnormalities | Root remodeling; rapid proliferation of lateral roots in stress-free areas | Avoidance of localized stress, detoxification | Wang et al., 2025 |
| Nitrogen Deficiency | Reduces nitrogen absorption, grain yield | Decreased total root length, surface area, number, volume; increased single root length, aerenchyma area, lignin content | Efficient nutrient exploration and uptake | Giehl and von Wirén, 2014; Liu et al., 2023 |
| Phosphorus Deficiency | Lateral root development | Enhanced early root growth, larger root system | Efficient phosphorus uptake and utilization | Liu et al., 2024b; Barrero et al., 2022 |
Overview of major abiotic stresses and their impact on rice root architecture.
3.1 Role in enhancing drought tolerance and avoidance
Drought is a primary factor contributing to significant reductions in yield in rice as the widely bred rice varieties have naturally shallow root systems (Sarma et al., 2023). Under moderate to severe drought conditions, cell elongation and expansion is affected, resulting in the inhibition of lateral root growth, decrease in total root length and reduced biomass (Wang et al., 2025). As the topsoil layers dry out during periods of limited rainfall or irrigation, root system of rice plants extends deeper into the soil profile to access water reserves in the subsoil (Kitomi et al., 2020). This access to a more stable water source allows the plants to maintain their hydration levels, sustain essential physiological processes like photosynthesis, and ultimately minimize the negative impacts of drought on grain yield (Uga et al., 2013a). Some native genotypes are known to exhibit an increase in lateral roots that are crucial for maintaining cellular hydration (Geng et al., 2024). These genotypes exhibit differences in root traits such as root diameter, length, specific area, angle, and density, and therefore, are considered advantageous for improving plant growth under drought conditions (Geng et al., 2024) (Table 1).
At the genetic level, the DEEPER ROOTING1 (DRO1) again plays a central role in regulating root length under drought stress. A functional allele of this gene, specifically the one derived from the deep-rooting cultivar Kinandang Patong (DRO1-KP), promotes a steeper, more downward growth pattern of the roots (Kitomi et al., 2020). This deeper rooting habit, conferred by DRO1, has consistently led to increased grain yields in rice cultivars grown under drought conditions in upland environments (Kitomi et al., 2020). Comparative studies involving IR64 (shallow root in upland conditions), KP (upland cultivar with deep rooting) and near-isogenic lines in IR64 (Dro1-NILs) carrying either the functional DRO1 allele from KP (DRO1-kp) or the non-functional allele from IR64 (DRO1-ir) have demonstrated the direct impact of the DRO1 gene on root architecture. Lines with functional KP allele exhibit significantly greater maximum root depth and a larger proportion of their root system is distributed in the deeper soil layers compared to their counterparts with non-functional allele having shallower roots. These findings provide clear evidence that the functional DRO1 allele directly contributes to deeper rooting and, consequently, to improved drought avoidance in rice (Uga et al., 2013a). Beyond the overall depth of the root system, other root architectural traits also contribute to enhanced drought tolerance. A greater root length density at deeper soil depths, i.e., the total length of roots per unit volume of soil in the lower horizons, allows for an increased exploration of the subsoil and a greater potential for water absorption (Siangliw et al., 2022).
Similarly, a deeper root angle, regulated by DRO1 gene, facilitates rapid penetration of the root system to deeper water reserves (Siangliw et al., 2022). Interestingly, while deep rooting is in itself advantageous for drought avoidance, an increase in root diameter also enhances the root’s ability to penetrate deeper into the soil, especially in soils that are compacted or have a hardpan layer, and further contributes to drought tolerance (Siangliw et al., 2022). Genes like OsABA8ox2 (root biomass), OsERF48, OsERF71, OsNAC10, OsNAC5, OsNAC9, RRS1 have been shown to regulate root diameter, lateral root density, and primary and lateral root length in response to drought stress, thereby regulating the deep root nature of rice plants under different environmental conditions (Zhang et al., 2025) (Figure 2). However, it is important to note that the relationship between root diameter and drought response is complex and may vary depending on the specific rice genotype and the prevailing environmental conditions (Siangliw et al., 2022).
Figure 2
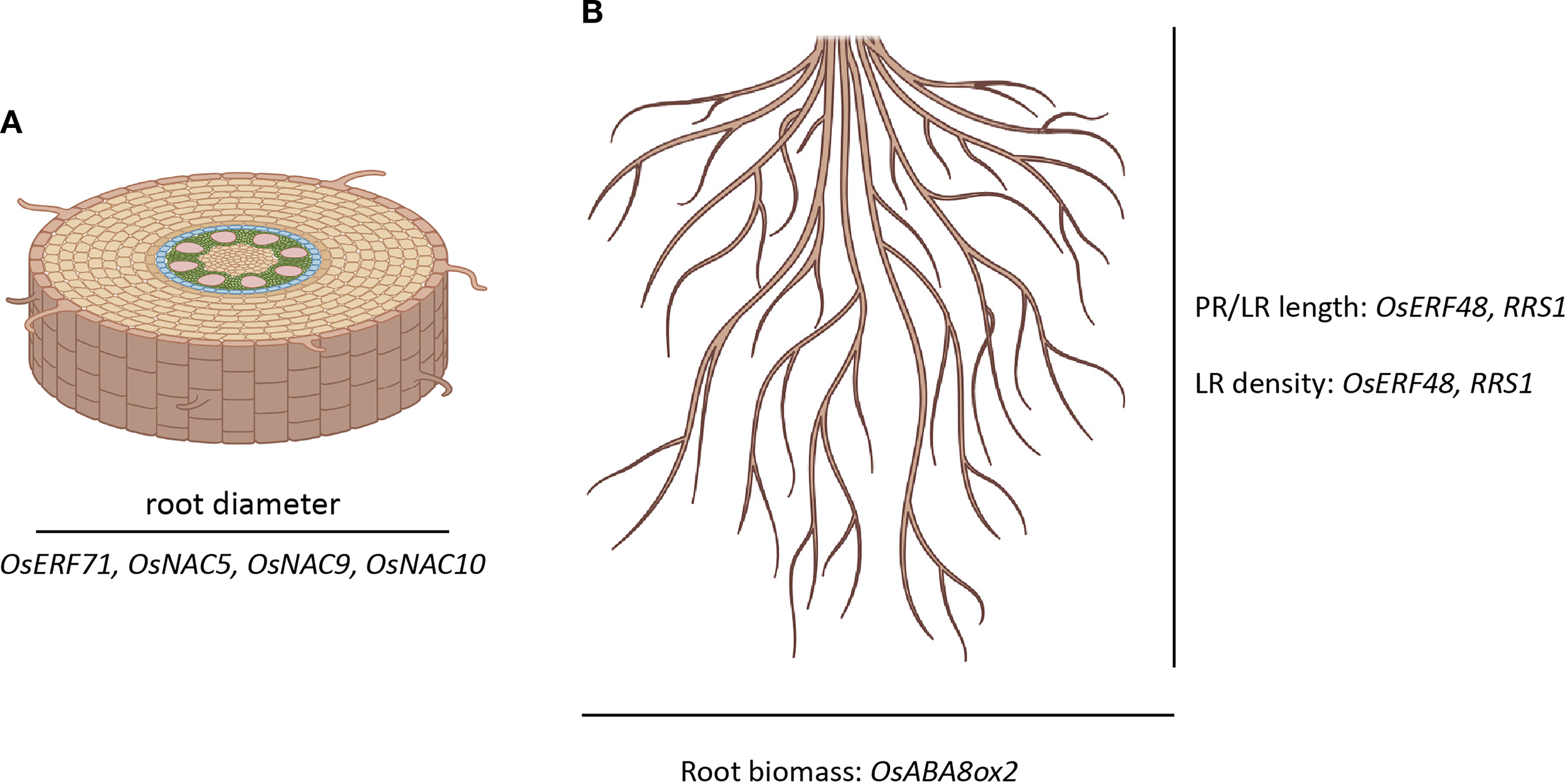
Genes affecting root morphology in drought stress. (A) Genes affecting root diameter. (B) Genes affecting root length and biomass. PR, primary root; LR, Lateral root. The illustration was generated with the help of Biorender (www.bioRender.com) and Adobe Illustrator, Adobe Creative Cloud suite.
Therefore, the ability of rice plants to modify their root architecture in response to changing environmental conditions is crucial for adaptation to the dynamic nature of drought stress (Sandhu et al., 2016). Under conditions of progressive drought, rice plants exhibiting plasticity in various root traits, such as increasing the root length density in deeper soil layers to enhance water uptake, promoting lateral root branching to maximize the surface area for absorption, or adjusting the root growth angle to facilitate quicker penetration into deeper, moister soil (Kano et al., 2011; Tran et al., 2015; Sandhu et al., 2016), leads to better yield and crop resilience. This dynamic ability to alter root architecture is essential for the plant to optimize its water uptake strategy as the soil moisture profile rapidly changes during a drought event (Sandhu et al., 2016).
3.2 Contribution to nutrient deficiency resilience
Nutrient deficiencies, such as those of nitrogen and phosphorus, also compel plants to alter their RSA to efficiently explore soil zones with limited nutrient availability (Giehl and von Wirén, 2014). Under low nitrogen conditions (LN) there is a significant reduction in nitrogen absorption and grain yield and is often accompanied by a decrease in total root length, surface area, root number and root volume (Giehl and von Wirén, 2014; Liu et al., 2023). On the other hand, under LN conditions, upland rice genotypes exhibit adaptive responses, such as promoting the growth of the primary root and developing a deeper overall root system, a key strategy for maximizing the capture of nitrogen from a larger soil profile (Kato et al., 2006; Galindo-Castañeda et al., 2022; Liu et al., 2023). Although, shallow root systems are considered more advantageous for the uptake of relatively immobile nutrients like phosphorus, deep roots still contribute to the plant’s phosphorus nutrition indirectly (Uga et al., 2013a). For instance, the small S-type lateral roots in rice, which are very fine and densely covered with root hairs, play a significant role in phosphorus uptake by increasing overall surface area of the roots. These roots can develop on both the main root axes (including nodal roots that contribute to depth) and the larger L-type lateral roots (Gonzalez et al., 2021) (Figure 1B). Paradoxically, single root length, root aerenchyma area, and root lignin content may increase under LN (Liu et al., 2023) (Table 1).
In addition, the plasticity of root architecture in response to water availability indirectly affects the phosphorus uptake by causing modifications in root structure (Liu et al., 2024b). For example, reduced water availability leads to a decrease in nodal root thickness and an increase in secondary root branching in rice (De Bauw et al., 2019). Both these changes enhance the efficiency of phosphorus uptake, even in situations where the root-to-shoot ratio is decreased due to water stress (De Bauw et al., 2019). At the molecular level, OsAMT genes are associated with efficient nitrogen accumulation under LN, while PSTOL1 gene is known to be crucial for phosphorous acquisition efficiency (PAE) and root biomass (Mai et al., 2014; Barrero et al., 2022) (Table 2).
Table 2
| Gene/QTL | Chromosome location | Primary function/impact on rooting | Associated abiotic stress(es) | References |
|---|---|---|---|---|
| DRO1 (DEEPER ROOTING 1) | Chr 9 | Controls root growth angle, promotes deeper rooting, influences gravitropism | Drought | Uga et al., 2011, 2013a |
| DRO2 (DEEPER ROOTING 2) | Chr 4 | Contributes to root growth angle and gravitropic curvature | Drought | Uga et al., 2013b |
| SUB1A (SUBMERGENCE 1A) | Chr 9 | Confers tolerance to complete submergence; regulates anaerobic metabolism, ROS, senescence | Submergence, Drought | Xu et al., 2006 |
| PSTOL1 (PHOSPHORUS STARVATION TOLERANCE 1) | Not specified | Enhances phosphorus acquisition efficiency (PAE) and root biomass; promotes early root growth | Phosphorus Deficiency | Gamuyao et al., 2012 |
| SPDT (SULTR-like Phosphorus Distribution Transporter/SULTR3;4) | Not specified | Influences phosphorus use efficiency (PUE) and grain quality (less P to grain) | Phosphorus Deficiency | Yamaji et al., 2017 |
| OsHKT1;5 (High-Affinity Potassium Transporter) | Saltol locus, Chr 1 | Selectively extracts Na+ from xylem, maintains K+ levels; preferentially expressed in roots | Salinity | Ren et al., 2005; Kobayashi et al., 2017; Ismail and Horie, 2017 |
| SOS pathway (SOS1, SOS2, SOS3) | Not specified | Modulates Na+ efflux, facilitates K+ retention, maintains ion homeostasis | Salinity | Ji et al., 2013 |
Key genes and QTLs influencing deep rooting and abiotic stress tolerance in rice.
Another significant abiotic stressor is heavy metal toxicity, which can decrease or completely halt metabolic activities, induce morphological abnormalities and lead to reduced crop yield (Sarma et al., 2023). To avoid heavy metal toxicity, rice remodels its root architecture by rapidly proliferating lateral roots in stress-free areas, a process mediated by reactive oxygen species (ROS)-directed auxin signaling (Wang et al., 2025).
Breeding for deeper rooting in rice could therefore improve the efficiency of nutrient use and potentially reduce the overall requirement for chemical fertilizer inputs, leading to more sustainable agricultural practices. This dependence of root responses to water and phosphorus availability suggests that breeding strategies aimed at improving resilience to one type of stress may also have indirect effects on the plant’s ability to cope with other limitations, favoring the need for a comprehensive approach to developing broadly adapted rice varieties.
3.3 Impact on salinity and submergence stress tolerance
Salinity significantly impacts rice plants, particularly during their seedling and reproductive stages; where high concentrations of sodium and chloride ions induce ionic toxicity, osmotic imbalance and nutritional imbalance, thereby inhibiting seed germination, stunting root and shoot growth, and ultimately reducing overall yield and quality (Sarma et al., 2023; Haque et al., 2021). Studies on salt-tolerance have revealed that tolerant rice varieties usually demonstrate deeper and more expansive root systems, which allows them to reach less saline water sources that are located at greater depths (Sackey et al., 2025). While deep root architecture is beneficial for enhancing drought and, to some extent, nutrient deficiency resilience in rice, its role in the plant’s tolerance to salinity stress is more complex and can even be disadvantageous in certain contexts (Verma et al., 2022). In saline paddy soils, where high concentrations of salt can create a reducing environment and other associated stresses, a shallower root growth angle (RGA) is more beneficial for rice plants (Uga et al., 2011, 2013a). The qSOR1 (quantitative trait locus for SOIL SURFACE ROOTING 1) gene (Kitomi et al., 2020) in rice, which is a homolog of the DRO1 gene (known for promoting deeper rooting), actually promotes shallower root growth. This allows the roots to develop on the surface of the soil, thereby avoiding the more stressful subsurface conditions that are prevalent in saline paddies. Studies focusing on shallower rooting habit, facilitated by qSOR1, have reported increased rice yields in saline environments compared to cultivars that develop deeper root systems (Uga et al., 2013a) (Table 2).
Soil salinity itself has a critical impact on the root system architecture of rice plants, typically inhibiting the growth of the primary root and altering the overall RSA (Shelden and Munns, 2023). Plants subjected to salt stress often respond by redistributing their root mass between the main roots and the lateral roots as a strategy to cope with the osmotic and ionic imbalances caused by the high salt concentrations in the soil solution (Seo et al., 2020). By altering the density of root hairs and increased root exudation, rice plants strive to enhance ionic balance (Oburger et al., 2014; Holz et al., 2018). In rice, research posits that salinity stress often leads to a reduction in the overall length of the roots, a decrease in the depth of the rooting system, and a lower total root weight. However, the magnitude of these effects can vary considerably depending on the specific genotype of the rice plant (Julkowska et al., 2017) (Table 1).
In submergence, conditions of low oxygen availability severely inhibit primary root growth in rice, impeding photosynthesis, depleting energy reserves, and ultimately resulting in plant mortality (Wang et al., 2024a) (Table 1). As a wetland plant, rice possesses several adaptive traits to cope with submergence such as the formation of well-developed aerenchyma in the leaves, sheaths, roots, and internodes; thereby creating interconnected gas spaces, facilitating oxygen transport from the parts of the plant above water to the submerged roots (Sarma et al., 2023). A specific type of aerenchyma known as lysigenous aerenchyma can form constitutively in roots and is further induced under waterlogging conditions (Wang et al., 2024a). Another critical adaptation is the formation of adventitious roots (ARs) as the primary root system often becomes dysfunctional under submerged conditions. As a result, rice root systems tend to become shallower and more compact with an accompanied increase in ARs. Both pre-existing and newly formed ARs emerge to replace the compromised primary roots, enabling efficient oxygen and nutrient acquisition (Phukan et al., 2023). Conversely, aerenchyma can increase under low nitrogen conditions (Liu et al., 2023).
In addition, the ability of rice plants to regenerate their roots under conditions of submergence further enhances their resilience to nutrient deficiencies (Ijaz et al., 2019). Unlike other cereal crops such as corn and wheat, which have a limited capacity for root regeneration, rice can develop new adventitious roots from submerged nodes on the stem. This ability allows rice to recover from root damage caused by flooding or nutrient stress, helping to maintain nutrient uptake even when parts of the original root system are compromised.
To summarize the effects on stress mitigation resilience, deep and shallow rooting have contrasting benefits in drought and salinity conditions that emphasize the importance of considering the particular environmental challenges when breeding for improved root system architecture in rice (Uga et al., 2013a). This concept of “soil-surface roots” (SOR) stands in direct contrast to the usual emphasis on deep rooting for enhancing drought tolerance Kitomi et al. (2020) reported that the homologs of DRO1 have different roles in regulating RGA in rice, indicating that the optimal root system architecture is not a universal characteristic but rather depends on the specific environmental context. Thus, genotypes of rice that can modulate their RSA in response to different stresses effectively are more likely to exhibit greater tolerance to salinity or drought stresses (Shelden and Munns, 2023; Vázquez-Glaría et al., 2021). These alterations could be either in root length, branching patterns or in anatomical features that might help the plant to exclude or tolerate the uptake of high concentrations of salt ions (Vázquez-Glaría et al., 2021; Shelden and Munns, 2023; Geng et al., 2024). Further, a root system optimized for water uptake in drought (e.g., dense, highly lignified) might be less efficient for nutrient acquisition in well-watered conditions or for oxygen transport in flooded conditions. This complex interplay highlights that engineering a multi-stress tolerant rice variety requires a sophisticated understanding of these trade-offs and the capacity to fine-tune specific adaptations based on the predominant stress conditions.
4 Deep root architecture and carbon sequestration in agricultural soils
4.1 Potential of rice deep roots for increased soil organic carbon storage
Soil is recognized as a substantial reservoir of carbon, estimated to be around 2,300 Gt carbon situated at a 3m depth, and constitutes about three times the current pool of atmospheric CO2 (Schlesinger and Bernhardt, 2020). The source of soil organic carbon stocks (SOC) primarily comes from the aboveground plant biomass, roots, and root exudates. This carbon, which is ‘fixed’ by photosynthesis, enters the soil through the decomposition of root biomass and the release of various organic compounds as root exudates (Kell, 2012, 2011). Therefore, cropland and grazing land soils, that is estimated to be around 5 billion hectares globally, have the tremendous potential to sequester and store atmospheric carbon (Sanderman et al., 2017). Breeding crop plants, including rice, with root systems that are not only deeper but also “bushier” has emerged as a promising strategy for enhancing the amount of organic carbon stored in agricultural soils over the long term (Pett-Ridge et al., 2018). By encouraging the growth of roots into deeper soil profiles, the concentration of carbon sequestered in these deeper layers will be more stable than the roots in the upper soil layer, where the stored carbon is released back to the atmosphere as CO2 after the decomposition of the roots (Pett-Ridge et al., 2018). Carbon that is sequestered in the deeper horizons of the soil has the potential to remain stable for extended periods, ranging from centuries to even millennia, making it an effective approach for mitigating the increase in atmospheric carbon dioxide concentrations (Kell, 2011, 2012).
The global cultivated rice area is estimated to be around 116 million hectares annually, which provides for an excellent concept of developing RSAs that are capable of sequestering carbon in agricultural soils and have a substantial impact on increasing SOC worldwide, thereby reducing carbon emissions significantly (Kell, 2011). Furthermore, the development of deep-rooted rice varieties capable of sequestering more carbon would offer a compelling dual benefit: enhancing plant performance in terms of stress tolerance and nutrient uptake, coupled with a positive impact on environmental sustainability by achieving ‘negative carbon emission’.
4.2 Mechanisms of carbon input and stabilization in deeper soil profiles
Roots are estimated to contribute 30% to 40%, of the total organic carbon inputs into the soil, that achieves long-term stabilization within the soil matrix (Kell, 2011). Stabilization of carbon in the soil is crucial as it prevents the rapid decomposition of carbon by soil microorganisms and its subsequent release back into the atmosphere as carbon dioxide. There are two principal mechanisms by which roots release carbon back into the soil: the deposition of root biomass as roots grow and die, and the release of a diverse array of carbon-containing root exudates into the surrounding soil (Kell, 2011, 2012). These root exudates, can include sugars, amino acids, organic acids, and other metabolites that serve as a vital source of energy and nutrients for the complex community of microorganisms that inhabit the rhizosphere (Pett-Ridge et al., 2018). While it is well-established that deep root systems can increase the input of carbon into the deeper layers of the soil profile, the specific factors that govern the long-term fate and stability of this below-ground carbon in its various molecular forms are not yet fully understood (Kell, 2011). In recent years, scientists have focused on root biochemistry and found that the stable lipophilic complex polyester, suberin, is a good source for stable SOC (Lorenz et al., 2007; Eckardt et al., 2023) (Figure 3). Suberin is biochemically very stable and can interact with soil minerals and occlusion in topsoil microaggregates (Kell, 2012; Lin and Simpson, 2016). Microaggregates are essentially small clusters of soil particles bound together by organic matter and microbial by-products and can physically protect root-derived carbon from microbial decomposition. Thus, deeper soil horizons may offer a more favorable environment for the stabilization of newly added root-derived carbon as compared to the topsoil by the formation of microaggregates (Kell, 2011).
Figure 3
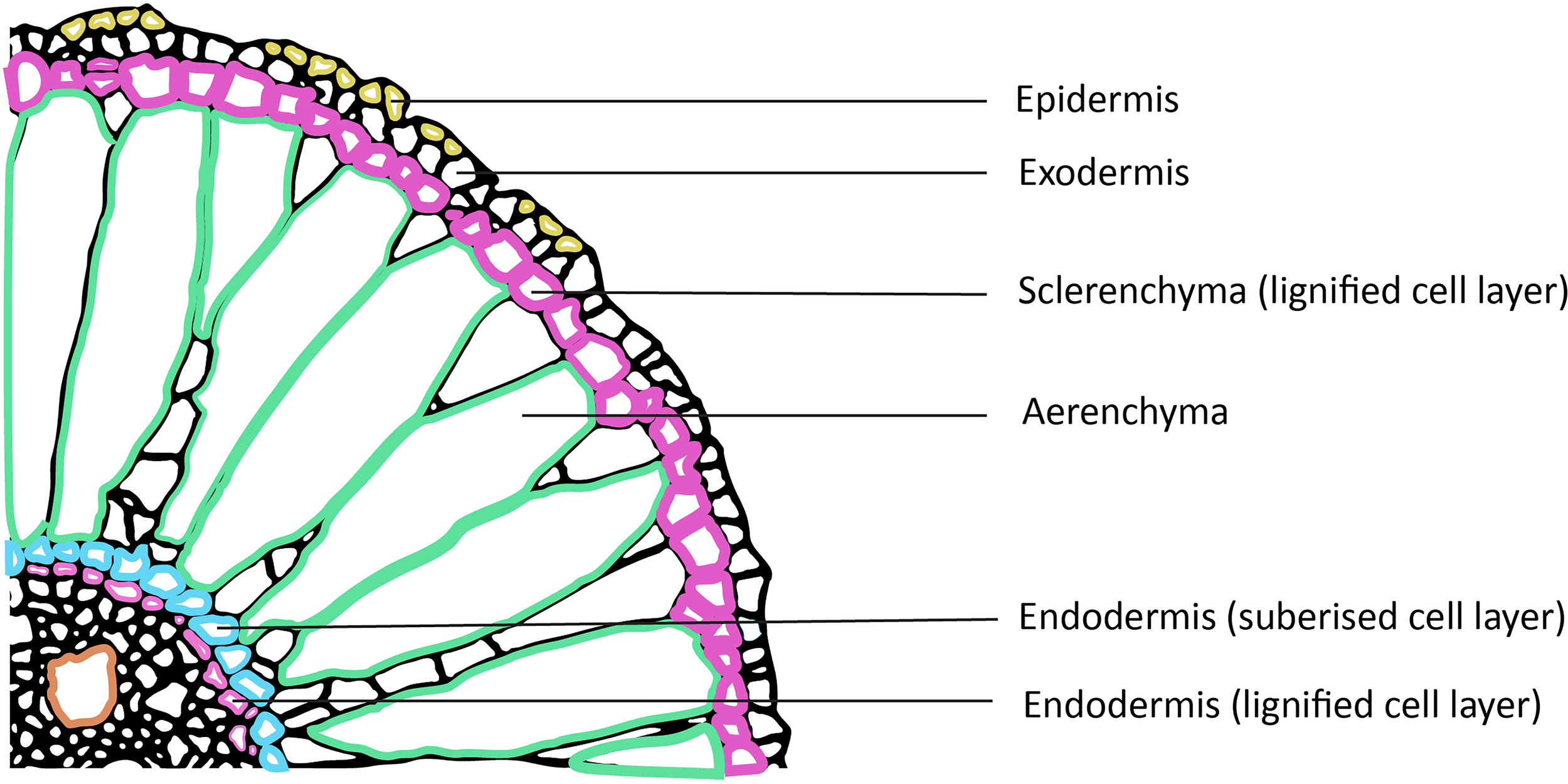
Cross section of a mature rice root focusing on the secondary cell wall thickenings. The sclerenchyma and endodermis along with the inner cortex are lignified to provide mechanical strength. Suberin deposition in the Casparian strip of the endodermis takes place at a later stage and surrounds the entire cell. Suberin is an excellent source of soil organic carbon stocks (SOC) and is recently the focus of attention for carbon sequestration. See text for more details. Figure of root section adapted from Rebouillat et al. (2009).
Interestingly, recent research suggests that mineral weathering processes occurring in the soil, particularly the uptake of silicon by rice plants, can indirectly promote the storage of carbon in paddy fields (Edwards et al., 2015). Kusa et al. (2021)) reported that aluminum released by mineral weathering can bind with organic matter, including carbon derived from rice roots, leading to the formation of more persistent forms of soil carbon. While it was initially hypothesized that organic acids secreted by rice roots played a major role in this mineral weathering effect, the findings by (Kusa et al., 2021) indicate that other mechanisms, possibly involving specific chelating sites located on the cell walls of rice roots, may be more significant in facilitating the dissolution of minerals, the release of aluminum and the subsequent production of persistent soil carbon.
Thus, breeding programs that prioritize traits that lead to increased root biomass, deposition of suberin, and promote the release of root exudates that contribute to the formation of stable carbon pools in the soil could be another promising avenue for increasing the carbon sequestration potential of deep-rooted rice varieties.
5 Ecological perspective of deep root architecture in rice cultivation
5.1 Influence on soil microbial communities
Plant species identity significantly influences the bacterial community structure in the rhizosphere. Plant roots selectively shape the microbiome across different root zones (rhizosphere, rhizoplane, and endosphere) through traits like root exudation, architecture, and morphology, which affect bacterial colonization and function (Schmid et al., 2019). Soil harbors a wide range of microbial communities, such as bacteria, fungi, archaea, protozoans, and nematodes, which are vital for sustainable agriculture. These microbes drive key processes that support soil fertility, plant health, and ecosystem resilience by facilitating nitrogen fixation, phosphorus solubilization, producing growth-promoting substances, enhancing nutrient uptake, improving soil structure, and detoxifying harmful substances (Kiprotich et al., 2025).
The composition and dynamics of root-associated microbiomes in rice are shaped by various factors, including the geographical location of cultivation, soil type, genetic characteristics of the rice cultivar, and the agricultural practices used (Edwards et al., 2015). Root exudation is currently the well-studied in the development of rhizosphere microbiome. Root exudates like sugars, amino acids and organic acids, secreted by the plant are responsible for attracting bacteria from the soil to the root system. The root exudation profile of a plant presents a unique fingerprint which contributes heavily to the recruitment, colonization and function of specific rhizobacteria (Sasse et al., 2018). Soil microbes convert essential nutrients into forms that plants can readily absorb. Additionally, beneficial microbes enhance pathogen resistance, improve water retention, and produce growth-promoting hormones (Edwards et al., 2015).
Specific root exudates can even be linked to the recruitment of certain bacterial taxa (Vieira et al., 2020). The production of bacterial secondary metabolites such as siderophores, cyclic lipopeptides, and antibiotics is an important indicator of overall rhizosphere microbiome functioning and can also be dictated by plant root exudates (Figure 4).
Figure 4
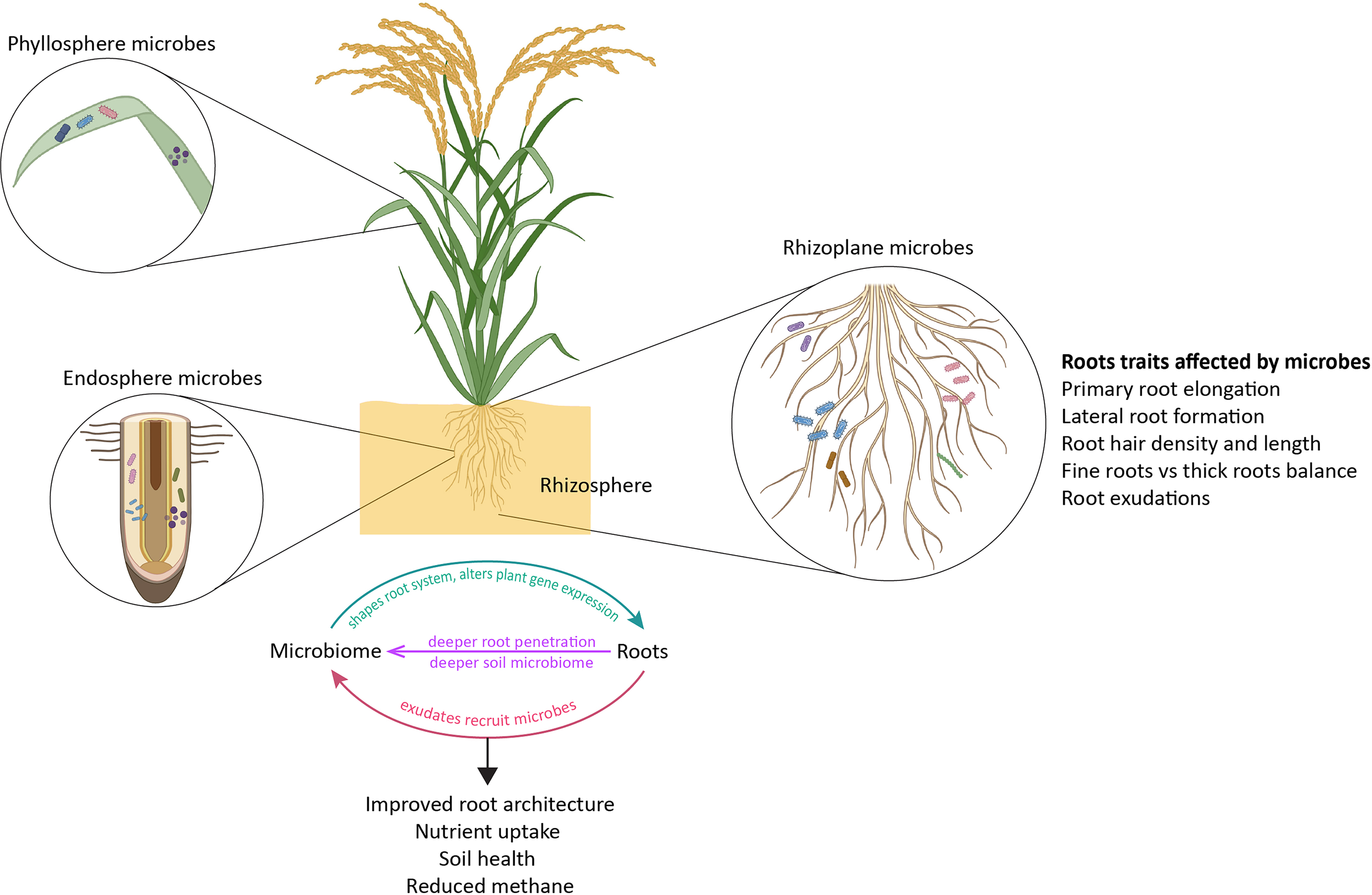
The interrelationship between microbes and root growth. The microbial communities in the rhizosphere are extensively shaped by the plant species. At the same time, the microbiome also influences the root architecture of the plant species like rice. See text for more details. The illustration was generated with the help of Biorender (www.bioRender.com) and Adobe Illustrator, Adobe Creative Cloud suite.
Generalist microbial taxa, characterized by greater abundance and diversity, are adept at exploiting a wide range of root phenotypic traits and environmental conditions to utilize available metabolites and nutrients, thereby promoting plant growth (Zhang et al., 2017). In contrast, specialist taxa are more prevalent in fine roots, likely due to the larger surface area and higher nutrient availability of fine roots, which also exhibit greater bacterial OTU richness (Gaiero et al., 2013). This suggests that plants with more fine roots or root hairs may recruit diverse microbial communities for mutualistic benefits. Specialist taxa may colonize fine roots passively through stochastic processes, such as root hair wounding, and their lower diversity and abundance compared to generalists may result from limited cellular mechanisms for colonizing woody root traits. However, some specialist OTUs demonstrate the potential to colonize woody roots, as indicated by their increased abundance from root hairs to primary roots (Saleem et al., 2018). A study of 10 indica rice varieties demonstrated that host genotype significantly shapes the root-associated microbiome. Analysis of α and β-diversity indices revealed genotype-specific microbiome structures. Machine learning identified key taxa, including Anaerolineae, α-Proteobacteria, and genera such as Desulfobacteria and Ca. Entotheonella, as vital for enhancing plant fitness. Rice rhizobiomes were enriched in genes supporting sulfur oxidation/reduction, biofilm formation, nitrogen fixation, denitrification, and phosphorus metabolism (Singh et al., 2022).
A study investigated the root-associated microbiomes of rice using high-throughput sequencing, examining plants grown under controlled greenhouse conditions and in field settings across multiple sites. Three root compartments, endosphere (root interior), rhizoplane (root surface), and rhizosphere (soil near roots) was found to host distinct microbial communities. In greenhouse conditions, microbiome composition varied with soil source and rice genotype, while in field conditions, geographical location and cultivation practices (organic vs. conventional) influenced microbial diversity. Microorganisms associated with the roots of plants have an important function in plant growth and in soil carbon sequestration. Rice cultivation is the second largest anthropogenic source of atmospheric CH4, which is a significant greenhouse gas (Hernández et al., 2015). Methanogenic archaea, linked to methane emissions, were present in all compartments of field-grown rice (Edwards et al., 2015). Depending on the developmental stage of the rice plant and the prevalent environmental conditions during the cultivation period, the exudates might have different compositions, that leads to temporal shifts in the structure and function of the rhizosphere over the plant’s life cycle (Hernández et al., 2015).
Root architecture determines the location of microbial associations within root systems, which, when integrated with soil vertical gradients, determines the functions and the metabolic capability of rhizosphere microbial communities (Galindo-Castañeda et al., 2024). Conversely, the establishment of a diverse root microbiome significantly alters the transcriptional profiles of rice plants. The presence of complex microbial communities triggered a major shift, downregulating one-third to one-fourth of leucine-rich repeat receptor-like kinases and nucleotide-binding leucine-rich repeat receptor gene families in rice roots. While some gene expression changes were consistent across all soil types, a significant portion of the transcriptional response varied by soil source, underscoring the role of root microbiomes in shaping plant transcriptomes and suggesting the involvement of key immune receptor families in recruiting and maintaining rhizosphere microbiomes (Santos-Medellín et al., 2022).
It is imperative to understand how root systems influence the microbial communities inhabiting deeper soil layers for predicting the long-term ecological sustainability of cultivating rice varieties with altered root architecture. Changes in the subsoil microbiome could have implications for nutrient availability not only for the rice crop itself but also for subsequent crops grown in rotation, as well as impacting broader soil health parameters.
5.2 Effects on nutrient cycling processes in the rhizosphere and bulk soil
Rice roots are associated with diverse microbial communities that are the primary drivers of essential nutrient cycling processes occurring in both the immediate vicinity of the roots (the rhizosphere) and in the bulk soil (Verma et al., 2022). These processes include atmospheric nitrogen fixation into forms that can be utilized by plants, the release of essential nutrients by breakdown of complex organic matter, the solubilization of mineral nutrients such as phosphorus, and the various transformations of nitrogen compounds, including nitrification and denitrification (Liu et al., 2023). Rice plants with deep root systems access and influence different pools of nutrients that are present at varying depths within the soil profile, such as the reserves of mobile nutrients like nitrate that may have leached down from the upper soil layers. Furthermore, the presence of roots at greater depths can directly influence the activity of microbial communities that are involved in nutrient transformations within those specific soil zones. Submerged rice roots develop aerenchyma that facilitates the transport of oxygen from the shoots to the roots and then to the surrounding soil. This released oxygen is known as radial oxygen loss (ROL), that creates an oxidized environment around the roots in an erstwhile anoxic flooded soil (Lai et al., 2012; Duyen et al., 2022). This ROL, by oxidation of the rhizosphere, helps in the reduction of phyto-toxins like sulfides and ferrous iron (Smith and Luna, 2013). The presence of high ROL is also known to reduce the concentration of methane by significantly reducing the activity of methanogens, slowing down the process of methanogenesis, and behavior of the entire microbial community; although the sensitivity to oxygen is remains variable amongst the species (Kiener and Leisinger, 1983; Tholen et al., 2007; Zheng et al., 2018). However, the specific mechanisms by which deep root systems influence the dynamics of the associated microbial community are not fully understood and requires further investigation.
The ability to influence nutrient cycling by the deep root systems at different soil depths could be the focus of breeding programs for specific root architectures as a strategy to manipulate the availability of nutrients in the soil to the advantage of the rice plant. This could result in more efficient nutrient uptake and a reduced need for excessive external inputs in the form of fertilizers. However, any changes in root architecture and the associated microbial communities, particularly in flooded rice paddies, can have implications for the production and emission of greenhouse gases like methane and nitrous oxide, which are themselves key components of nutrient cycling processes. Therefore, a comprehensive ecological perspective on deep-rooted rice cultivation must also include an evaluation of its impact on these broader biogeochemical cycles.
6 Strategies for better root systems - the future for climate change?
6.1 Genetic engineering and genome editing for enhanced deep root traits
Genetically improving root system architecture (RSA), particularly by enhancing deep root traits, is increasingly recognized as an effective strategy for developing climate-resilient crops such as rice. The advent of genetic engineering and advanced genome editing technologies, especially CRISPR-Cas9, has opened unprecedented opportunities to precisely manipulate the rice genome and tailor root systems for improved performance under challenging environmental conditions (Uga et al., 2013a). Key genes (Table 3) that regulate root growth angle and depth, such as DEEPER ROOTING 1 (DRO1) and its homologs like qSOR1 (quantitative trait locus for SOIL SURFACE ROOTING 1), have emerged as promising targets for genetic manipulation (Uga et al., 2011; Uga et al., 2013b). Overexpression of the functional DRO1 allele has been consistently associated with a steeper root growth angle, resulting in deeper rooting and significantly enhancing the plant’s capacity to avoid drought stress and maintain yield under water-limited conditions (Uga et al., 2013a). Conversely, manipulation of qSOR1 can promote a shallower root growth angle, which is advantageous for rice cultivated in saline paddy soils (Uga et al., 2013b).
Table 3
| Factor | Response | References |
|---|---|---|
|
DRO1
(DEEPER ROOTING 1) |
Controls root growth angles by negatively regulating auxin and induces cell elongation in the root tip that causes asymmetric root growth and downward bending of the root in response to gravity. | Uga et al., 2013a |
|
DRO2
(DEEPER ROOTING 2) |
Responsible for higher root length density and deeper rooting | Uga et al., 2013b |
|
DRO3
(DEEPER ROOTING 3) |
It affects root growth angle (RGA) in plants with a functional DRO1 allele. | Uga et al., 2015 |
|
qSOR1
(QTL for SOIL SURFACE ROOTING 1) |
It is a DRO1 homolog involved in root gravitropic responses. | Kitomi et al., 2020 |
|
DROT1
(DROUGHT1) |
Specifically expressed in vascular bundles of root, shoot and leaves. It induces drought resistance by increasing cellulose content in the cell wall structure which leads to improved root function and water uptake under drought conditions. | Sun et al., 2022 |
|
OsFBX257
(F-BOX PROTEIN CODING GENE 257) |
Overexpression of this gene increases root length, root depth and crown root allowing for better water absorption. | Sharma et al., 2023 |
|
OsNMCP1
(NUCLEAR MATRIX CONSTITUENT PROTEINS 1) |
Regulates drought resistance and root growth through chromatin accessibility modulation by interacting with a chromatin remodeler OsSWI3C in rice | Yang et al., 2020; Siangliw et al., 2022 |
|
OsRSL3
(ROOT HAIR DEFECTIVE-SIX LIKE 3) |
Four SNPs in OsRSL3 were found that caused amino acid changes and significantly associated with increase in median metaxylem diameter. | Siangliw et al., 2022 |
|
OsRSL3
(ROOT HAIR DEFECTIVE-SIX LIKE 3) |
Four SNPs in OsRSL3 were found that caused amino acid changes and significantly associated with increase in median metaxylem diameter. | Siangliw et al., 2022 |
|
OsERF71
(ETHYLENE RESPONSE FACTOR 71) |
Overexpression of this gene in roots elevated the expression levels of genes enabling cell wall loosening and lignin biosynthetic genes leading to changes in root structure, the formation of enlarged aerenchyma, and high lignification levels. | Lee et al., 2016 |
|
OsPIN1b
(PIN- FORMED 1b) |
Knockout of this gene alters auxin distribution and root growth patterns. | Sun et al., 2018 |
|
OsPIN3t
(PIN PROTEIN 3T) |
Involved in control of polar auxin transport and knockdown of this gene is related to crown root abnormalities in the seedling stage. | Zhang et al., 2012 |
|
OsERA1
(ENHANCED RESPONSE TO ABA 1) |
Negative regulator of ABA signaling, transgenic rice lines have increased ABA sensitivity, enhanced primary root growth in non-stress conditions. | Ogata et al., 2020 |
|
OsSAUR11
(SMALL AUXIN-UP RNA 11) |
Positively regulates deep rooting in rice. | Xu et al., 2023 |
Genes associated with root architecture in rice (Oryza sativa).
In addition to these regulators of root angle, recent CRISPR-Cas9 studies have targeted other genes involved in root development and stress responses. For instance, knockout of OsPIN1b—a gene crucial for the polar transport of auxin—has demonstrated its essential role in root gravitropism and overall root system architecture, as disruption of OsPIN1b alters auxin distribution and root growth patterns (Yu et al., 2021). Similarly, CRISPR-Cas9-mediated mutagenesis of OsERA1, a negative regulator of abscisic acid (ABA) signaling, has produced rice lines with heightened ABA sensitivity. These edited lines exhibit enhanced primary root growth even under non-stressed conditions and display stronger physiological responses to ABA, a key hormone in drought stress signaling (Ogata et al., 2020). Collectively, these advances illustrate how precise genome editing can be leveraged to unravel the genetic basis of RSA and to develop rice varieties with optimized root traits for improved stress resilience and yield stability.
In addition to direct modification of DNA, RNA-based technologies are also being explored for their potential to enhance root development and stress resilience in rice. For example, Yu et al. (2021) demonstrated that overexpressing the human RNA demethylase FTO protein in rice modulates the plant RNA N6-methlyadenosine (m6A) modifications, resulting in ~50% increases in yield and biomass, by stimulating root meristem cell proliferation, deeper roots, enhanced bud formation, increased photosynthetic efficiency and drought tolerance. These results have shown RNA methylation pathways to be a promising avenue for improving root development and the ability of rice to withstand abiotic stresses.
Identifying candidate genes that are associated with desirable root traits through comprehensive genome-wide expression analyses conducted under various stress conditions is another crucial step in guiding genetic engineering efforts aimed at enhancing deep root architecture in rice (Fujino et al., 2019). By comparing the transcriptome responses of rice varieties that exhibit contrasting root architectures and levels of stress tolerance, researchers can pinpoint specific genes that play key roles in these processes, making them potential targets for genetic modification to improve deep rooting and overall stress adaptation (Fujino et al., 2019).
The precision and efficiency of CRISPR-Cas9 technology have transformed the landscape of plant breeding, enabling rapid and targeted improvements in root traits of rice that were previously difficult to achieve through conventional breeding methods. Accelerating the rate of genetic gain in this way is essential for developing climate-resilient rice varieties capable of meeting the demands of a changing climate. As our understanding of the key regulatory genes and pathways involved in root development continues to grow, the integration of genome editing tools with advanced genetic knowledge offers unprecedented potential for designing rice varieties with root systems optimized for specific environmental challenges such as drought and salinity.
6.2 Marker-assisted selection for breeding deep-rooted rice varieties
Marker-assisted selection (MAS) utilizes DNA markers tightly linked to specific genes or quantitative trait loci (QTLs) and is a valuable biotechnological approach that allows plant breeders to efficiently select for desirable traits. MAS offers several advantages over traditional phenotypic selection, particularly for complex traits like root architecture that can be challenging, time-consuming, or labor-intensive to measure directly in the field (Uga et al., 2013a). Extensive QTL mapping studies conducted in rice have successfully identified specific genomic regions and associated DNA markers that are linked to various root morphological traits that are relevant to the development of deep root systems. These traits include maximum root length, total root number, root volume, root dry weight, root penetration ability, root thickness, and root growth angle (Liu et al., 2024a). For example, the DRO1 QTL, which plays a significant role in determining the depth of the root system in rice, has been precisely mapped to a specific region on chromosome 9. DNA markers associated with Dro1 are now routinely used in breeding programs to select for deeper rooting (Liu et al., 2024a). Similarly, QTLs related to increased root length and root thickness have been identified across different rice populations, allowing breeders to target and introgress these beneficial traits into elite rice varieties (Uga et al., 2011).
A key advantage of MAS is the ability to select for desired root traits at the seedling stage, significantly accelerating the breeding process. For example (Zheng et al., 2000) utilized QTL mapping to identify genetic regions controlling root penetration ability and thickness, enabling early selection and reducing the time required to develop improved rice lines. Furthermore, MAS facilitates the pyramiding of multiple beneficial QTLs—combining traits such as deep rooting, disease resistance, and enhanced yield—into a single variety, resulting in cultivars with superior resilience and productivity (Uga et al., 2011). The effectiveness of MAS in rice breeding programs has been well-documented for a wide range of traits, including resistance to various biotic and abiotic stresses, as well as characteristics related to grain yield and quality (Ogata et al., 2020). The advent of high-throughput genotyping technologies and the expanding knowledge of the rice genome further increase the efficiency and applicability of MAS, enabling rapid and cost-effective analysis of DNA markers in large populations.
By leveraging DNA markers linked to key root traits, MAS allows breeders to overcome the challenges of directly selecting for below-ground characteristics. This approach enables selection based on the plant’s genetic potential, leading to more efficient and precise breeding for deep rooting and other desirable root system attributes. Ultimately, MAS accelerates the development of rice varieties with improved root systems, better adapted to the environmental challenges posed by climate change.
6.3 Application of plant growth regulators to promote deep root development
Plant growth regulators (PGRs), or phytohormones, are naturally synthesized compounds that play essential roles in regulating plant growth and development. As central integrators, phytohormones coordinate external environmental cues with internal genetic programs to shape the spatial distribution of the root system within the soil. Auxin acts as a positive regulator of crown root development by promoting cell growth and division (Wang et al., 2018), while cytokinins serve as negative regulators of this process. The antagonistic yet coordinated interaction between auxin and cytokinin is mediated by genes such as OsERF3 and Wuschel-related homeobox (WOX) OsWOX11, which are crucial for crown root initiation (Zhao et al., 2015). Overexpression of ORR3, a B-type cytokinin response regulator, suppresses cell division and influences auxin synthesis and transport, thereby negatively regulating young rice root growth (Wei et al., 2025). Recently, it was shown that the overexpression of OsSAUR11, a novel small auxin-up RNA gene was found to enhance deep rootedness in rice (Xu et al., 2023). Additionally, studies in Arabidopsis and Prunus have shown that auxin can negatively regulate DRO1, indicating a complex feedback loop where auxin levels modulate root angles for optimal soil penetration (Dardick et al., 2019). The gaseous hormone ethylene also plays a key role in determining rice root angle by modulating auxin-mediated root gravitropism (Kong et al., 2024). Unlike Arabidopsis, in rice, the ABA pathway mediates ethylene-induced inhibition of root elongation( (Yang et al., 2007). Furthermore, high ethylene concentrations reduce bioactive gibberellin (GA) content, affecting cell proliferation in the root meristem and primary root elongation (Qin et al., 2022) (Figure 5).
Figure 5
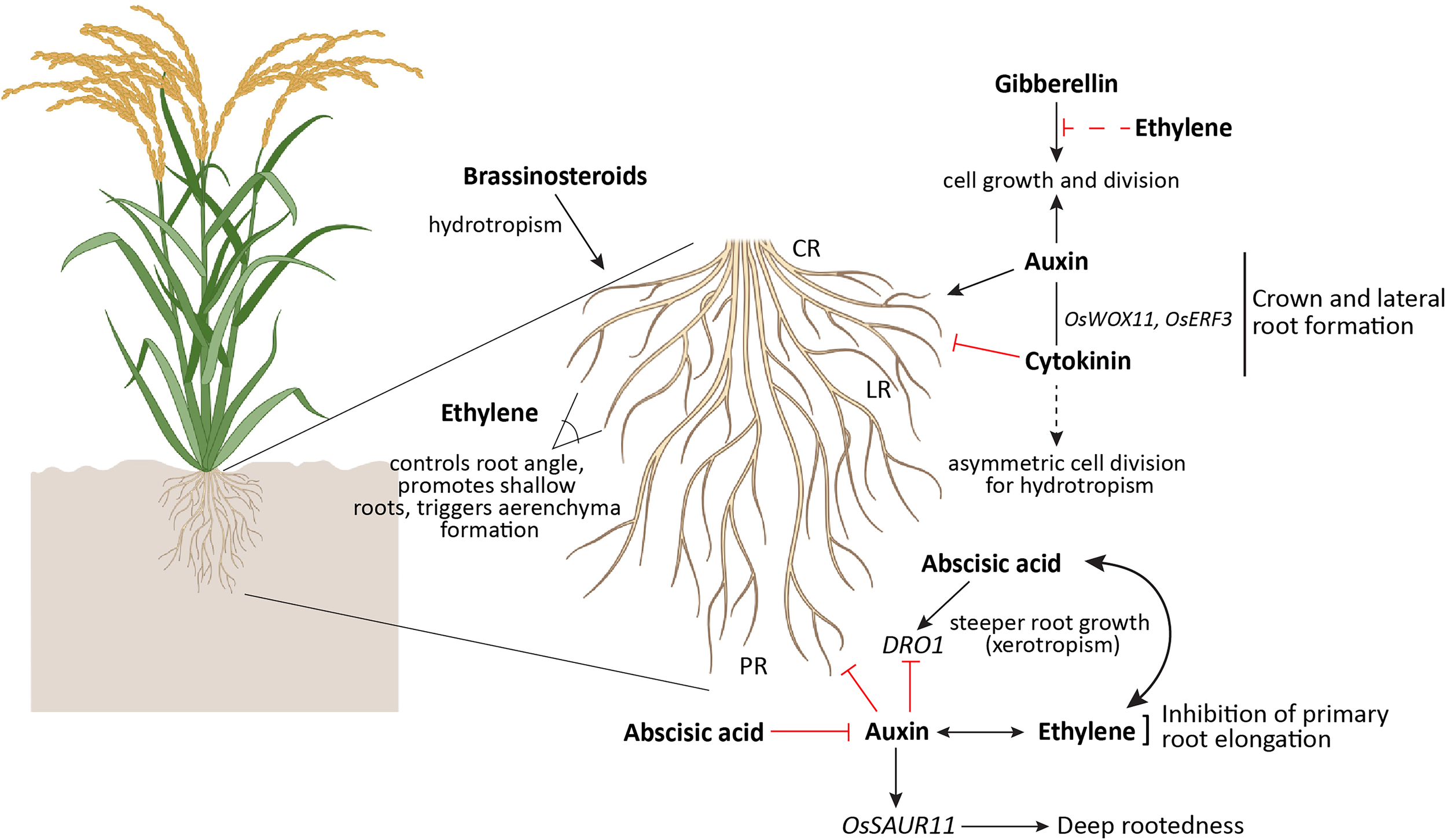
Hormonal regulation of deep root architecture in rice. Auxin promotes crown root (CR) formation and lateral root (LR) initiation by stimulating cell growth and division but inhibits primary root elongation and negatively regulates DRO1 at high concentrations, affecting downward bending. Cytokinin generally inhibits root growth and crown root development but induces asymmetric cell division, influencing hydrotropism. Ethylene controls root angle to promote shallower rooting and inhibits primary root growth while regulating auxin-mediated gravitropism. Abscisic acid (ABA) enhances steeper root growth via DRO1 and drives asymmetric cortical cell expansion for hydrotropism, also blocking auxin movement to pause xerobranching. Gibberellin (GA) supports cell division and elongation necessary for root extension but is negatively influenced by high ethylene levels. Brassinosteroids are essential for root hydrotropism, and jasmonic acid (JA) modulates root architecture and drought resilience. Ethylene is a key phytohormone that triggers aerenchyma formation. The rice plant and roots were generated with the help of Biorender (www.bioRender.com) and the illustration was prepared in Adobe Illustrator, Adobe Creative Cloud suite.
In addition to these hormones, calcium signaling plays a pivotal role in regulating root architecture in rice. Biotic and abiotic stresses trigger physical changes at the root cell’s plasma membrane leading to its depolarization and rapid opening of specialized calcium permeable channels on the plasma membrane and internal cellular compartments like endoplasmic reticulum and vacuoles (Wilkins et al., 2016). A rapid and transient increase in the concentration of free calcium ions (Ca+) occurs within the cytoplasm of the root cells, developing a unique calcium signature that encodes the type and severity of the stress, and key Ca+ sensors get activated to act on downstream targets (Tong et al., 2021; Wilkins et al., 2016). In rice, calcium signaling participates in the root architecture via the OsCBL3-OsCIPK31 module, where OsCBL3 (a calcineurin B-like (CBL) protein) acting as a plasma membrane localized calcium sensor, interacts with the protein kinase OsCIPK31 (Yu et al., 2025). Upon their interaction, the autoinhibitory domains of OsCIPK31 are relieved and this activates the downstream signaling cascade. Further downstream, OsCIPK31 phosphorylates OsCYP2, a cyclophilin involved in auxin-mediated lateral root initiation (Yu et al., 2025). The OsCBL3-OsCIPK31 module regulates root growth by integrating ABA and auxin signaling pathways. Loss of OsCBL3 or OsCIPK31 function leads to elevated ABA levels and suppressed IAA signaling, resulting in inhibited root elongation and lateral root formation. Conversely, treatment with ABA inhibitors or low concentrations of IAA partially restores normal root development in the mutants. The absence of OsCYP2 disrupts lateral root formation even in OsCIPK31 overexpression lines, confirming its role as a critical effector in this calcium-dependent pathway. In summary, calcium signaling via the OsCBL3–OsCIPK31–OsCYP2 axis orchestrates rice root development by fine-tuning hormonal responses, offering insights into genetic strategies for improving root traits under stress conditions (Yu et al., 2025) (Figure 6).
Figure 6
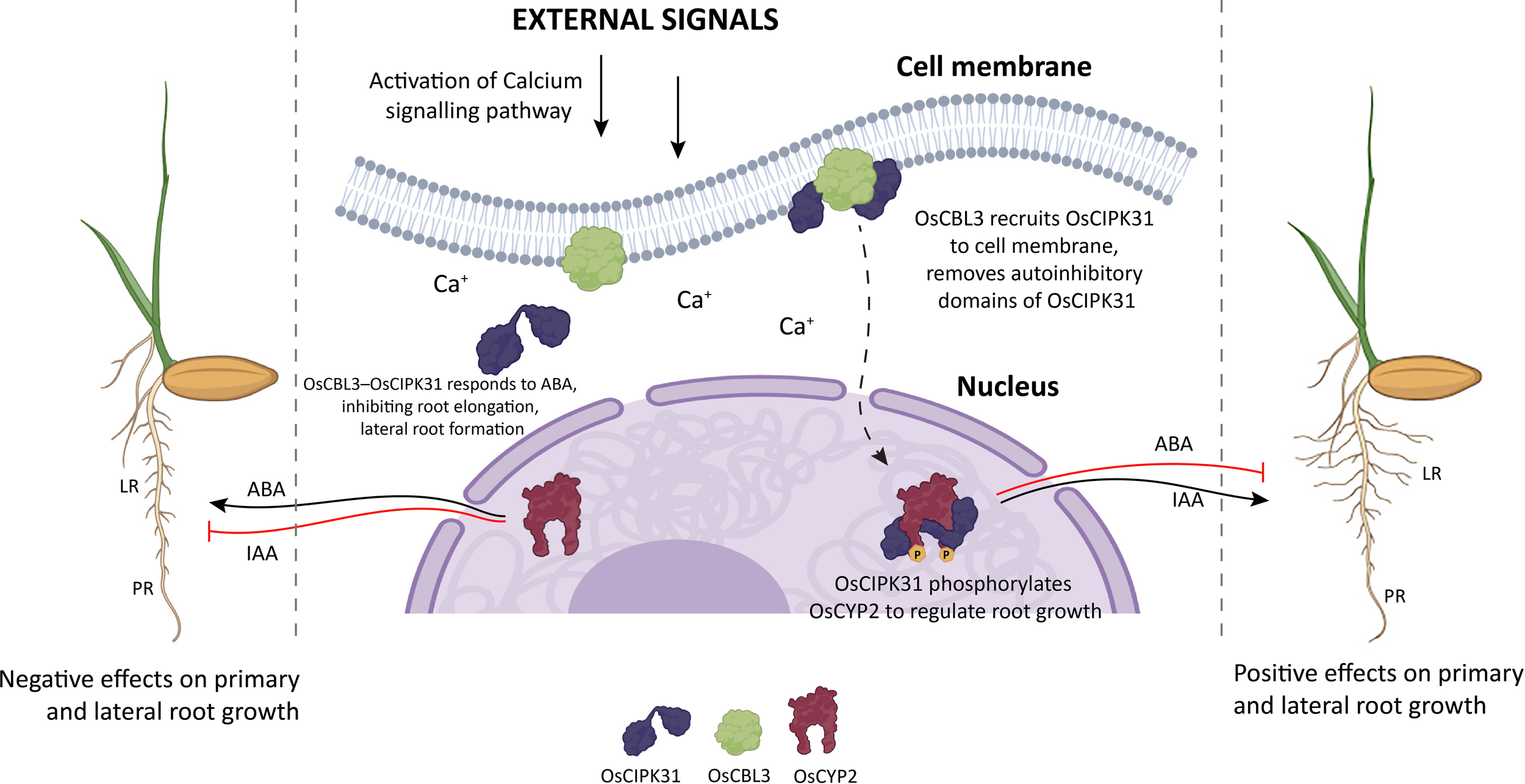
Model depicting the OsCBL3–OsCIPK31–OsCYP2 axis of calcium signaling regulating root architecture as proposed by Yu et al. (2025). OsCBL3 (calcineurin B-like calcium sensor) interacts with the protein kinase OsCIPK31 only at the plasma membrane. OsCIPK31 phosphorylates OsCYP2 in the nucleus to regulate root growth via the auxin and ABA pathways. PR, primary root; LR, lateral root. See text for more details. Figure generated with the help of Biorender (www.bioRender.com) and Adobe Illustrator, Adobe Creative Cloud suite.
The exogenous application of synthetic or naturally derived PGRs presents a practical strategy to promote deep root development and enhance rice resilience to environmental stresses. For instance, beta-cyclocitral, a molecule derived from beta-carotene, has been identified as a conserved regulator of root growth across plant species, including rice. Application of beta-cyclocitral stimulates root stem cell division, leading to enhanced root growth, increased branching, and improved performance under salt stress (Dickinson et al., 2019). Other PGRs, such as 2-diethylaminoethyl hexanoate (DA-6) and combinations of potassium 3-indole-butyrate, potassium 1-naphthylacetate, and either 6-benzylaminopurine or 1-triacontanol, have been shown to improve root growth and increase yield in mechanically transplanted rice seedlings (Sabagh et al., 2021). Foliar sprays of PGRs like 5-aminolevulinic acid (5-ALA) and diethyl toluidine acetate (DTA-6) have also been reported to mitigate oxidative damage to rice roots caused by salt stress (Wang et al., 2024b). Overall, manipulating root growth and development through PGR application offers a flexible and less genetically invasive alternative to genetic engineering. This approach can enhance deep rooting in existing rice varieties and may complement genetic modifications, providing a multifaceted strategy for improving root architecture and stress resilience in rice.
7 Conclusion & future perspectives
Deep root architecture in rice offers a promising avenue for enhancing climate resilience and sustainability in agricultural systems. By improving access to subsoil water and nutrients, deep-rooted genotypes confer greater tolerance to drought, salinity, and nutrient limitations—crucial traits that enhance survival under intensifying climate stress. The contribution of deep rootedness of crop plants like rice to carbon sequestration underscores their relevance to climate mitigation strategies and has recently been the focus of global research to develop climate smart crops. To harness these benefits, a mechanistic understanding of the physiological, genetic, and environmental factors governing root development is essential; as root architecture is shaped by complex interactions among hormonal signaling, gene regulation, and soil conditions. Elucidating these processes will enable targeted manipulation of root traits through advanced breeding and biotechnological approaches, including genome editing with CRISPR, marker-assisted selection, and high-throughput phenotyping to accelerate the development of rice cultivars with optimized root systems. When integrated with sustainable agronomic practices and precision application of plant growth regulators, these technologies can enhance root functionality across diverse agroecological contexts. On the other hand, the ecological implications of cultivating deep-rooted rice—particularly their influence on soil microbiome and nutrient cycling—must be considered to ensure long-term soil health and ecosystem stability. Investigating these belowground interactions will inform best management practices and support the environmental sustainability of improved cultivars.
In summary, the integration of deep root architecture into rice breeding programs is anticipated to transform climate-resilient agriculture. With advancements in techniques for genomic selection and high-throughput phenotyping, root traits can be increasingly tailored to specific edaphic and climatic conditions, accelerating the development of stress-tolerant cultivars. Future research thereby, would need to focus on elucidating the molecular networks governing root plasticity, enabling the dynamic adaptation of crops to an ever-changing environmental stimuli. Concurrently, innovations in rhizosphere microbiome profiling and soil health analytics will enhance our understanding of how deep-rooted systems influence nutrient cycling, carbon sequestration, and ecosystem functionality. By coupling these insights with scalable agronomic practices, next-generation rice varieties are expected to not only withstand abiotic stressors but also contribute to regenerative agricultural landscapes.
Statements
Author contributions
RT: Conceptualization, Writing – original draft, Writing – review & editing. UC: Writing – original draft. CH: Writing – original draft. PB: Conceptualization, Funding acquisition, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. PB would like to acknowledge The Assam Royal Global University, Guwahati for providing partial support in the form of a Seed Grant (RGU/Ch(Acad)/(07)/Botany (29)).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Biorender (www.bioRender.com) Adobe Illustrator Creative Cloud Suite was used to prepare the illustrations.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Barrero L. S. Willmann M. R. Craft E. J. Akther K. M. Harrington S. E. Garzon-Martinez G. A. et al . (2022). 'Identifying genes associated with abiotic stress tolerance suitable for CRISPR/Cas9 editing in upland rice cultivars adapted to acid soils. Plant Direct6, e469. doi: 10.1002/pld3.469
2
Dardick C. D. Guseman J. M. Hollender C. A. (2019). DEEPER ROOTING genes and methods of use thereof, US Patent 10,513,708 B2. Available online at: https://patents.google.com/patent/US10513708B2/en.
3
De Bauw P. Vandamme E. Lupembe A. Mwakasege L. Senthilkumar K. Merckx R. (2019). Architectural root responses of rice to reduced water availability can overcome phosphorus stress. Agronomy9, 11. doi: 10.3390/agronomy9010011
4
Dickinson A. J. Lehner K. Mi J. Jia K. P. Mijar M. Dinneny J. et al . (2019). β-Cyclocitral is a conserved root growth regulator. Proc. Natl. Acad. Sci. U.S.A.116, 10563–10567. doi: 10.1073/pnas.1821445116
5
Duyen D. V. Kwon Y. Kabange N. R. Lee J. Y. Lee S. M. Kang J. W. et al . (2022). Novel QTL associated with aerenchyma-mediated radial oxygen loss (ROL) in rice. Plants (Basel)11 (6), 788. doi: 10.3390/plants11060788
6
Eckardt N. A. Ainsworth E. A. Bahuguna R. N. Broadley M. R. Busch W. Carpita N. C. et al . (2023). Climate change challenges, plant science solutions. Plant Cell35, 24–66. doi: 10.1093/plcell/koac303
7
Edwards J. Johnson C. Santos-Medellín C. Lurie E. Podishetty N. K. Bhatnagar S. et al . (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A.112, E911–E920. doi: 10.1073/pnas.1414592112
8
Fonta J. E. Vejchasarn P. Henry A. Lynch J. P. Brown K. M. (2022). Many paths to one goal: Identifying integrated rice root phenotypes for diverse drought environments. Front. Plant Sci.13, 959629. doi: 10.3389/fpls.2022.959629
9
Fujino K. Hirayama Y. Kaji R. (2019). Marker-assisted selection in rice breeding programs in Hokkaido. Breed Sci.69, 383–392. doi: 10.1270/jsbbs.19062
10
Gaiero J. R. McCall C. A. Thompson K. A. Day N. J. Best A. S. Dunfield K. E. (2013). Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am. J. Bot.100, 1738–1750. doi: 10.3732/ajb.1200572
11
Galindo-Castañeda T. Hartmann M. Lynch J. P. (2024). Location: root architecture structures rhizosphere microbial associations. J. Exp. Bot.75, 594–604. doi: 10.1093/jxb/erad421
12
Galindo-Castañeda T. Lynch J. P. Six J. Hartmann M. (2022). Improving soil resource uptake by plants through capitalizing on synergies between root architecture and anatomy and root-associated microorganisms. Front. Plant Sci.13, 827369. doi: 10.3389/fpls.2022.827369
13
Gamuyao R. Chin J. H. Pariasca-Tanaka J. Pesaresi P. Catausan S. Dalid C. et al . (2012). The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature488, 535–539. doi: 10.1038/nature11346
14
Geng A. Lian W. Wang Y. Liu M. Zhang Y. Wang X. et al . (2024). Molecular mechanisms and regulatory pathways underlying drought stress response in rice. Int. J. Mol. Sci.25 (2), 1185. doi: 10.3390/ijms25021185
15
Giehl R. F. von Wirén N. (2014). Root nutrient foraging. Plant Physiol.166, 509–517. doi: 10.1104/pp.114.245225
16
Gong W. Vinarao R. Proud C. Wood S. Snell P. Fukai S. et al . (2025). Genomic regions and molecular markers associated with deeper rooting to improve grain yield in aerobic rice (Oryza sativa L.) production systems. Rice (N Y)18, 24. doi: 10.1186/s12284-025-00784-6
17
Gonzalez D. Postma J. Wissuwa M. (2021). Cost-benefit analysis of the upland-rice root architecture in relation to phosphate: 3D simulations highlight the importance of S-type lateral roots for reducing the pay-off time. Front. Plant Sci.12, 641835. doi: 10.3389/fpls.2021.641835
18
Han J.-H. Shin N.-H. Moon J.-H. Chin J. H. Yoo S.-C. (2016). A simple phenotyping method for deep-rooting rice grown in pots. J. Plant Biotechnol.43, 444–449. doi: 10.5010/JPB.2016.43.4.444
19
Hanzawa E. Sasaki K. Nagai S. Obara M. Fukuta Y. Uga Y. et al . (2013). Isolation of a novel mutant gene for soil-surface rooting in rice (Oryza sativa L.). Rice (N Y)6, 30. doi: 10.1186/1939-8433-6-30
20
Haque M. A. Rafii M. Y. Yusoff M. M. Ali N. S. Yusuff O. Datta D. R. et al . (2021). Advanced breeding strategies and future perspectives of salinity tolerance in rice. Agronomy11, 1631. doi: 10.3390/agronomy11081631
21
Hernández M. Dumont M. G. Yuan Q. Conrad R. (2015). Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl. Environ. Microbiol.81, 2244–2253. doi: 10.1128/AEM.03209-14
22
Hilker M. Schmülling T. (2019). Stress priming, memory, and signalling in plants. Plant Cell Environ.42, 753–761. doi: 10.1111/pce.13526
23
Holz M. Zarebanadkouki M. Kuzyakov Y. Pausch J. Carminati A. (2018). Root hairs increase rhizosphere extension and carbon input to soil. Ann. Bot.121, 61–69. doi: 10.1093/aob/mcx127
24
Ijaz B. Sudiro C. Jabir R. Schiavo F. Hyder M. Yasmin T. (2019). Adaptive behaviour of roots under salt stress correlates with morpho-physiological changes and salinity tolerance in rice. Int. J. Agric. Biol.21, 667–674. doi: 10.17957/IJAB/15.0943
25
Ismail A. M. Horie T. (2017). Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol.68, 405–434. doi: 10.1146/annurev-arplant-042916-040936
26
Ji H. Pardo J. M. Batelli G. Van Oosten M. J. Bressan R. A. Li X. (2013). The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol. Plant6, 275–286. doi: 10.1093/mp/sst017
27
Julkowska M. M. Koevoets I. T. Mol S. Hoefsloot H. Feron R. Tester M. A. et al . (2017). Genetic components of root architecture remodeling in response to salt stress. Plant Cell29, 3198–3213. doi: 10.1105/tpc.16.00680
28
Kano M. Inukai Y. Kitano H. Yamauchi A. (2011). Root plasticity as the key root strait for adaptation to various intensities of drought stress in rice. Plant Soil342, 117–128. doi: 10.1007/s11104-010-0675-9
29
Karlova R. Boer D. Hayes S. Testerink C. (2021). Root plasticity under abiotic stress. Plant Physiol.187, 1057–1070. doi: 10.1093/plphys/kiab392
30
Kato Y. Abe J. Kamoshita A. Yamagishi J. (2006). Genotypic Variation in Root Growth Angle in Rice (Oryza sativa L.) and its Association with Deep Root Development in Upland Fields with Different Water Regimes. Plant Soil287, 117–129. doi: 10.1007/s11104-006-9008-4
31
Kell D. B. (2011). Breeding crop plants with deep roots: their role in sustainable carbon, nutrient and water sequestration. Ann. Bot.108, 407–418. doi: 10.1093/aob/mcr175
32
Kell D. B. (2012). Large-scale sequestration of atmospheric carbon via plant roots in natural and agricultural ecosystems: why and how. Philos. Trans. R Soc. Lond. B Biol. Sci.367, 1589–1597. doi: 10.1098/rstb.2011.0244
33
Kiener A. Leisinger T. (1983). Oxygen sensitivity of methanogenic bacteria. Syst. Appl. Microbiol.4, 305–312. doi: 10.1016/S0723-2020(83)80017-4
34
Kim Y. Chung Y. S. Lee E. Tripathi P. Heo S. Kim K. H. (2020). Root response to drought stress in rice. Int. J. Mol. Sci.21 (4), 1513. doi: 10.3390/ijms21041513
35
Kiprotich K. Muema E. Wekesa C. Ndombi T. Muoma J. Omayio D. et al . (2025). Unveiling the roles, mechanisms and prospects of soil microbial communities in sustainable agriculture. Discov. Soil2, 10. doi: 10.1007/s44378-025-00037-4
36
Kitomi Y. Hanzawa E. Kuya N. Inoue H. Hara N. Kawai S. et al . (2020). Root angle modifications by the. Proc. Natl. Acad. Sci. U.S.A.117, 21242–21250. doi: 10.1073/pnas.2005911117
37
Kobayashi N. I. Yamaji N. Yamamoto H. Okubo K. Ueno H. Costa A. et al . (2017). OsHKT1;5 mediates Na. Plant J.91, 657–670. doi: 10.1111/tpj.13595
38
Kong X. Xiong Y. Song X. Wadey S. Yu S. Rao J. et al . (2024). Ethylene regulates auxin-mediated root gravitropic machinery and controls root angle in cereal crops. Plant Physiol.195, 1969–1980. doi: 10.1093/plphys/kiae134
39
Kusa K. Moriizumi M. Hobara S. Kaneko M. Matsumoto S. Kasuga J. et al . (2021). Mineral weathering and silicon uptake by rice plants promote carbon storage in paddy fields. Soil Sci. Plant Nutr.67, 162–170. doi: 10.1080/00380768.2021.1878471
40
Lai W.-L. Zhang Y. Chen Z.-H. (2012). Radial oxygen loss, photosynthesis, and nutrient removal of 35 wetland plants. Ecol. Eng.39, 24–30. doi: 10.1016/j.ecoleng.2011.11.010
41
Lee D. K. Jung H. Jang G. Jeong J. S. Kim Y. S. Ha S. H. et al . (2016). Overexpression of the osERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol.172, 575–588. doi: 10.1104/pp.16.00379
42
Lin L. H. Simpson M. J. (2016). Enhanced extractability of cutin- and suberin-derived organic matter with demineralization implies physical protection over chemical recalcitrance in soil. Organic Geochem.97, 111–121. doi: 10.1016/j.orggeochem.2016.04.012
43
Liu L. Cui K. Qi X. Wu Y. Huang J. Peng S. (2023). Varietal responses of root characteristics to low nitrogen application explain the differing nitrogen uptake and grain yield in two rice varieties. Front. Plant Sci.14, 1244281. doi: 10.3389/fpls.2023.1244281
44
Liu Y. Gao J. Zhao Y. Fu Y. Yan B. Wan X. et al . (2024b). Effects of different phosphorus and potassium supply on the root architecture, phosphorus and potassium uptake, and utilization efficiency of hydroponic rice. Sci. Rep.14, 21178. doi: 10.1038/s41598-024-72287-1
45
Liu G. Peiwen Z. Yi L. Deyan K. Jiahong W. Lijun L. et al . (2024a). Molecular marker-assisted selection of a new water-saving and drought-resistant rice (WDR) restoration line, hanhui 8200, for enhanced resistance to rice blast. Agronomy14, 1504. doi: 10.3390/agronomy14071504
46
Lorenz K. Lal R. Preston C. M. Nierop K. G. J. (2007). Strengthening the soil organic carbon pool by increasing contributions from recalcitrant aliphatic bio(macro)molecules. Geoderma142, 1–10. doi: 10.1016/j.geoderma.2007.07.013
47
Lynch J. P. (2019). Root phenotypes for improved nutrient capture: an underexploited opportunity for global agriculture. New Phytol.223, 548–564. doi: 10.1111/nph.15738
48
Lynch J. P. Wojciechowski T. (2015). Opportunities and challenges in the subsoil: pathways to deeper rooted crops. J. Exp. Bot.66, 2199–2210. doi: 10.1093/jxb/eru508
49
Mai C. D. Phung N. T. To H. T. Gonin M. Hoang G. T. Nguyen K. L. et al . (2014). Genes controlling root development in rice. Rice (N Y)7, 30. doi: 10.1186/s12284-014-0030-5
50
Mumtahina N. Matsuoka A. Yoshinaga K. Moriwaki A. Uemura M. Shimono H. et al . (2023). Deep placement of fertilizer enhances mineral uptake through changes in the root system architecture in rice. Plant Soil490, 189–200. doi: 10.1007/s11104-023-06066-8
51
Oburger E. Gruber B. Schindlegger Y. Schenkeveld W. D. C. Hann S. Kraemer S. M. et al . (2014). Root exudation of phytosiderophores from soil-grown wheat. New Phytol.203, 1161–1174. doi: 10.1111/nph.12868
52
Ogata T. Ishizaki T. Fujita M. Fujita Y. (2020). CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PloS One15, e0243376. doi: 10.1371/journal.pone.0243376
53
Ogura T. Goeschl C. Filiault D. Mirea M. Slovak R. Wolhrab B. et al . (2019). Root system depth in arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell178, 400–412.e16. doi: 10.1016/j.cell.2019.06.021
54
Pedersen O. Sauter M. Colmer T. D. Nakazono M. (2021). Regulation of root adaptive anatomical and morphological traits during low soil oxygen. New Phytol.229, 42–49. doi: 10.1111/nph.16375
55
Pett-Ridge J. Nuccio E. Mcfarlane K. (2018). Deeply rooted: Evaluating plant rooting depth as a means for enhanced soil carbon sequestration (No. LLNL-CONF-750320). Livermore, CA (United States): Lawrence Livermore National Lab.
56
Phukan U. J. Jindal S. Laldinsangi C. Singh P. K. Longchar B. (2023). A microscopic scenario on recovery mechanisms under waterlogging and submergence stress in rice. Planta259, 9. doi: 10.1007/s00425-023-04285-y
57
Plett D. C. Ranathunge K. Melino V. J. Kuya N. Uga Y. Kronzucker H. J. (2020). The intersection of nitrogen nutrition and water use in plants: new paths toward improved crop productivity. J. Exp. Bot.71, 4452–4468. doi: 10.1093/jxb/eraa049
58
Qin H. Pandey B. K. Li Y. Huang G. Wang J. Quan R. et al . (2022). Orchestration of ethylene and gibberellin signals determines primary root elongation in rice. Plant Cell34, 1273–1288. doi: 10.1093/plcell/koac008
59
Rebouillat J. Dievart A. Verdeil J. L. Escoute J. Giese G. Breitler J. C. et al . (2009). Molecular genetics of rice root development. Rice2, 15–34. doi: 10.1007/s12284-008-9016-5
60
Ren Z. H. Gao J. P. Li L. G. Cai X. L. Huang W. Chao D. Y. et al . (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet.37, 1141–1146. doi: 10.1038/ng1643
61
Sabagh A. E. L. Mbarki S. Hossain A. Iqbal M. A. Islam M. S. Raza A. et al . (2021). Potential role of plant growth regulators in administering crucial processes against abiotic stresses. Front. Agron.3. doi: 10.3389/fagro.2021.648694
62
Sackey O. K. Feng N. Mohammed Y. Z. Dzou C. F. Zheng D. Zhao L. et al . (2025). A comprehensive review on rice responses and tolerance to salt stress. Front. Plant Sci.16, 1561280. doi: 10.3389/fpls.2025.1561280
63
Saleem M. Law A. D. Sahib M. R. Pervaiz Z. H. Zhang Q. (2018). Impact of root system architecture on rhizosphere and root microbiome. Rhizosphere6, 47–51. doi: 10.1016/j.rhisph.2018.02.003
64
Sanderman J. Hengl T. Fiske G. J. (2017). Soil carbon debt of 12,000 years of human land use. Proc. Natl. Acad. Sci. U.S.A.114, 9575–9580. doi: 10.1073/pnas.1706103114
65
Sandhu N. Raman K. A. Torres R. O. Audebert A. Dardou A. Kumar A. et al . (2016). Rice root architectural plasticity traits and genetic regions for adaptability to variable cultivation and stress conditions. Plant Physiol.171, 2562–2576. doi: 10.1104/pp.16.00705
66
Santos-Medellín C. Edwards J. Nguyen B. Sundaresan V. (2022). Acquisition of a complex root microbiome reshapes the transcriptomes of rice plants. New Phytol.235, 2008–2021. doi: 10.1111/nph.18261
67
Sarma B. Kashtoh H. Lama Tamang T. Bhattacharyya P. N. Mohanta Y. K. Baek K. H. (2023). Abiotic stress in rice: visiting the physiological response and its tolerance mechanisms. Plants (Basel)12 (23), 3948. doi: 10.3390/plants12233948
68
Sasse J. Martinoia E. Northen T. (2018). Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci.23 (1), 25–41.
69
Schlesinger W. Bernhardt E. (2020). Biogeochemistry: An Analysis of Global Change, Fourth Edition. Amsterdam: Elsevier Science.
70
Schmid M. W. Hahl T. van Moorsel S. J. Wagg C. De Deyn G. B. Schmid B. (2019). Feedbacks of plant identity and diversity on the diversity and community composition of rhizosphere microbiomes from a long-term biodiversity experiment. Mol. Ecol.28, 863–878. doi: 10.1111/mec.14987
71
Seo D. H. Seomun S. Choi Y. D. Jang G. (2020). Root Development and Stress Tolerance in rice: The Key to Improving Stress Tolerance without Yield Penalties. Int. J. Mol. Sci.21 (5), 1807. doi: 10.3390/ijms21051807
72
Sharma E. Bhatnagar A. Bhaskar A. Majee S. M. Kieffer M. Kepinski S. et al . (2023). Stress-induced F-Box protein-coding gene OsFBX257 modulates drought stress adaptations and ABA responses in rice. Plant Cell Environ.46, 1207–1231. doi: 10.1111/pce.14496
73
Shelden M. C. Munns R. (2023). Crop root system plasticity for improved yields in saline soils. Front. Plant Sci.14, 1120583. doi: 10.3389/fpls.2023.1120583
74
Shrestha K. Miyazaki A. Yoshii D. Sakata M. Moritsuka N. (2024). Effect of localized fertilizer application on deep root development during early growth in upland rice under upland condition. Plant Product. Sci.27, 240–252. doi: 10.1080/1343943X.2024.2376296
75
Siangliw J. L. Thunnom B. Natividad M. A. Quintana M. R. Chebotarov D. McNally K. L. et al . (2022). Response of Southeast Asian rice root architecture and anatomy phenotypes to drought stress. Front. Plant Sci.13, 1008954. doi: 10.3389/fpls.2022.1008954
76
Singh A. Kumar M. Chakdar H. Pandiyan K. Kumar S. C. Zeyad M. T. et al . (2022). Influence of host genotype in establishing root associated microbiome of. Front. Microbiol.13, 1033158. doi: 10.3389/fmicb.2022.1033158
77
Smith K. E. Luna T. O. (2013). Radial oxygen loss in wetland plants: Potential impacts on remediation of contaminated sediments. J. Environ. Eng.139, 496–501. doi: 10.1061/(ASCE)EE.1943-7870.0000631
78
Sun H. Tao J. Bi Y. Hou M. Lou J. Chen X. et al . (2018). OsPIN1b is involved in rice seminal root elongation by regulating root apical meristem activity in response to low nitrogen and phosphate. Sci. Rep.8, 13014. doi: 10.1038/s41598-018-29784-x
79
Sun X. Xiong H. Jiang C. Zhang D. Yang Z. Huang Y. et al . (2022). Natural variation of DROT1 confers drought adaptation in upland rice. Nat. Commun.13, 4265. doi: 10.1038/s41467-022-31844-w
80
Tholen A. Pester M. Brune A. (2007). Simultaneous methanogenesis and oxygen reduction by Methanobrevibacter cuticularis at low oxygen fluxes. FEMS Microbiol. Ecol.62, 303–312. doi: 10.1111/j.1574-6941.2007.00390.x
81
Tong T. Li Q. Jiang W. Chen G. Xue D. Deng F. et al . (2021). Molecular evolution of calcium signaling and transport in plant adaptation to abiotic stress. Int. J. Mol. Sci.22 (22), 12308. doi: 10.3390/ijms222212308
82
Tran T. Kano-Nakata M. Suralta R. Menge D. Mitsuya S. Inukai Y. et al . (2015). Root plasticity and its functional roles were triggered by water deficit but not by the resulting changes in the forms of soil N in rice. Plant Soil386, 65–76. doi: 10.1007/s11104-014-2240-4
83
Uga Y. Kitomi Y. Yamamoto E. Kanno N. Kawai S. Mizubayashi T. et al . (2015). A QTL for root growth angle on rice chromosome 7 is involved in the genetic pathway of DEEPER ROOTING 1. Rice (N Y)8, 8. doi: 10.1186/s12284-015-0044-7
84
Uga Y. Okuno K. Yano M. (2011). Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot.62, 2485–2494. doi: 10.1093/jxb/erq429
85
Uga Y. Sugimoto K. Ogawa S. Rane J. Ishitani M. Hara N. et al . (2013a). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet.45, 1097–1102. doi: 10.1038/ng.2725
86
Uga Y. Yamamoto E. Kanno N. Kawai S. Mizubayashi T. Fukuoka S. (2013b). A major QTL controlling deep rooting on rice chromosome 4. Sci. Rep.3, 3040. doi: 10.1038/srep03040
87
Vázquez-Glaría A. Eichler-Löbermann B. Loiret F. G. Ortega E. Kavka M. (2021). Root-system architectures of two Cuban rice cultivars with salt stress at early development stages. Plants (Basel)10 (6), 1194. doi: 10.3390/plants10061194
88
Verma P. K. Verma S. Pandey N. (2022). Root system architecture in rice: impacts of genes, phytohormones and root microbiota. 3 Biotech.12, 239. doi: 10.1007/s13205-022-03299-9
89
Verma P. K. Verma S. Tripathi R. D. Pandey N. Chakrabarty D. (2021). CC-type glutaredoxin, OsGrx_C7 plays a crucial role in enhancing protection against salt stress in rice. J. Biotechnol.329, 192–203. doi: 10.1016/j.jbiotec.2021.02.008
90
Vieira S. Sikorski J. Dietz S. Herz K. Schrumpf M. Bruelheide H. et al . (2020). Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J.14, 463–475. doi: 10.1038/s41396-019-0543-4
91
Wang J. Han M. Huang Y. Zhao J. Liu C. Ma Y. (2024a). Flooding tolerance of rice: regulatory pathways and adaptive mechanisms. Plants (Basel)13 (9), 1178. doi: 10.3390/plants13091178
92
Wang C. Li X. Caragea D. Bheemanahallia R. Jagadish S. V. K. (2020). Root anatomy based on root cross-section image analysis with deep learning. Comput. Electron. Agric.175, 105549. doi: 10.1016/j.compag.2020.105549
93
Wang X. Liu X. Su Y. Shen H. (2025). Rice responses to abiotic stress: key proteins and molecular mechanisms. Int. J. Mol. Sci.26 (3), 896. doi: 10.3390/ijms26030896
94
Wang Y. Zhang T. Wang R. Zhao Y. (2018). Recent advances in auxin research in rice and their implications for crop improvement. J. Exp. Bot.69, 255–263. doi: 10.1093/jxb/erx228
95
Wang Y. Zhao L. M. Feng N. Zheng D. Shen X. F. Zhou H. et al . (2024b). Plant growth regulators mitigate oxidative damage to rice seedling roots by NaCl stress. PeerJ12, e17068. doi: 10.7717/peerj.17068
96
Wasaya A. Zhang X. Fang Q. Yan Z. (2018). Root phenotyping for drought tolerance: A review. Agronomy8, 241. doi: 10.3390/agronomy8110241
97
Wei G. Yu W. Chen X. Yun H. Wang T. Wang N. et al . (2025). The overexpression of ORR3 negatively regulates the growth of young rice roots by reducing the cell size and the number in the root meristematic zone. Plants (Basel)14 (11), 1627. doi: 10.3390/plants14111627
98
Wilkins K. A. Matthus E. Swarbreck S. M. Davies J. M. (2016). Calcium-mediated abiotic stress signaling in roots. Front. Plant Sci.7, 1296. doi: 10.3389/fpls.2016.01296
99
Xu K. Lou Q. Wang D. Li T. Chen S. Li T. et al . (2023). Overexpression of a novel small auxin-up RNA gene, OsSAUR11, enhances rice deep rootedness. BMC Plant Biol.23, 319. doi: 10.1186/s12870-023-04320-w
100
Xu K. Xu X. Fukao T. Canlas P. Maghirang-Rodriguez R. Heuer S. et al . (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature442, 705–708. doi: 10.1038/nature04920
101
Yamaji N. Takemoto Y. Miyaji T. Mitani-Ueno N. Yoshida K. T. Ma J. F. (2017). Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature541, 92–95. doi: 10.1038/nature20610
102
Yang J. Chang Y. Qin Y. Chen D. Zhu T. Peng K. et al . (2020). A lamin-like protein OsNMCP1 regulates drought resistance and root growth through chromatin accessibility modulation by interacting with a chromatin remodeller OsSWI3C in rice. New Phytol.227, 65–83. doi: 10.1111/nph.16518
103
Yang C. Xu Z. Song J. Conner K. Vizcay Barrena G. Wilson Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell19, 534–548. doi: 10.1105/tpc.106.046391
104
Yoshino K. Numajiri Y. Teramoto S. Kawachi N. Tanabata T. Tanaka T. et al . (2019). Towards a deeper integrated multi-omics approach in the root system to develop climate-resilient rice. Mol. Breed.39, 165. doi: 10.1007/s11032-019-1058-4
105
Yu Q. Liu S. Yu L. Xiao Y. Zhang S. Wang X. et al . (2021). RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat. Biotechnol.39, 1581–1588. doi: 10.1038/s41587-021-00982-9
106
Yu S. Zheng S. Ning J. Shi Y. Guo D. Luo R. et al . (2025). The rice OsCBL3-OsCIPK31 module regulates root development via abscisic acid and auxin signaling pathways. Crop J.13, 694–704. doi: 10.1016/j.cj.2025.02.006
107
Zhang Q. Li J. Zhang W. Yan S. Wang R. Zhao J. et al . (2012). The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J.72, 805–816. doi: 10.1111/j.1365-313X.2012.05121.x
108
Zhang Q. Saleem M. Wang C. (2017). Probiotic strain Stenotrophomonas acidaminiphila BJ1 degrades and reduces chlorothalonil toxicity to soil enzymes, microbial communities and plant roots. AMB Express7, 227. doi: 10.1186/s13568-017-0530-y
109
Zhang Y. Wu X. Wang X. Dai M. Peng Y. (2025). Crop root system architecture in drought response. J. Genet. Genomics52, 4–13. doi: 10.1016/j.jgg.2024.05.001
110
Zhao Y. Cheng S. Song Y. Huang Y. Zhou S. Liu X. et al . (2015). The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell27, 2469–2483. doi: 10.1105/tpc.15.00227
111
Zheng H. G. Babu R. C. Pathan M. S. Ali L. Huang N. Courtois B. et al . (2000). Quantitative trait loci for root-penetration ability and root thickness in rice: comparison of genetic backgrounds. Genome43, 53–61. doi: 10.1139/g99-065
112
Zheng H. Fu Z. Zhong J. Long W. (2018). Low methane emission in rice cultivars with high radial oxygen loss. Plant Soil431, 1–10. doi: 10.1007/s11104-018-3747-x
Summary
Keywords
deep root architecture, drought tolerance, nutrient use efficiency, root growth angle, DEEPER ROOTING1 (DRO1) , carbon sequestration, marker assisted selection, CRISPR
Citation
Tiwary R, Chettri U, Hasnu C and Borah P (2025) Dig deeper – insights into the genetic, physiological and climatic implications of deep root architecture of rice plants. Front. Plant Physiol. 3:1665196. doi: 10.3389/fphgy.2025.1665196
Received
13 July 2025
Accepted
31 August 2025
Published
16 September 2025
Volume
3 - 2025
Edited by
Shashank Sagar Saini, Institute for Agricultural and Forestry Systems of the Mediterranean (ISAFOM, CNR), Italy
Reviewed by
Swarnendra Banerjee, Southern Federal University, Russia
Ashwani Kumar, Govt Post Graduate College Berinag, India
Updates
Copyright
© 2025 Tiwary, Chettri, Hasnu and Borah.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pratikshya Borah, pborah3@rgu.ac; pratikshyaborah@south.du.ac.in
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.