- School of Food and Biological Engineering, Suzhou University, Suzhou, Anhui, China
Introduction: Phytohormone abscisic acid (ABA) plays a pivotal regulatory role in crop responses to abiotic stress. However, the specificities of the coordinated transcriptional and metabolic regulatory network in wheat under ABA signaling remain to be fully elucidated.
Methods: This study systematically investigated the regulatory effects of exogenous ABA on wheat germinating seeds through integrated physiological, transcriptomic, and metabolomic analyses.
Results: Physiological results demonstrated that low-concentration ABA (2 mg·L-1) promoted primary root elongation (12% increase vs. 0 mg·L-1 (CK)), whereas high concentrations (≥4 mg·L-1) significantly inhibited growth (40% root length reduction under 6 mg·L-1 ABA). Concurrently, electrolyte leakage, malondialdehyde (MDA) content, and catalase (CAT) activity markedly increased with ABA concentration (P < 0.05), indicating aggravated oxidative stress. Transcriptomic profiling (CK vs. 6 mg·L-1 ABA) identified 854 differentially expressed genes (DEGs; 470 up-regulated/384 down-regulated). Gene Ontology (GO) enrichment revealed DEGs predominantly involved in “Cellular process”, “Metabolic process”, “Catalytic activity”, and “Transporter activity”. KEGG analysis highlighted activation of “Linoleic acid metabolism”, “Alpha-Linolenic acid metabolism”, “Glycolysis/Gluconeogenesis”, and “Biosynthesis of amino acids” pathways. Metabolomics detected 665 differentially accumulated metabolites (DAMs), with “Lipids”, “Organic acids”, and “Amino acids” exhibiting significant alterations. KEGG enrichment emphasized “benzoxazinoid biosynthesis” and “Nicotinate/nicotinamide metabolism”. Integrative multi-omics analysis uncovered 10 core pathways, such as “Glycolysis/Gluconeogenesis”, “Biosynthesis of amino acids”, and “Cysteine and methionine metabolism”, that orchestrating ABA stress responses. Notably, L-serine and the genes TraesCS3A02G276100 and TraesCS5A02G398300 were recurrently implicated in multiple pathways, indicating their function as key network nodes.
Discussion: This study elucidates the molecular mechanisms by which wheat adapts to ABA stress through dynamic reprogramming of its metabolic and gene expression networks, thereby laying a theoretical foundation for developing future ABA-based seed treatment technologies or stress-resistant breeding strategies.
Introduction
Wheat (Triticum aestivum L.), an allohexaploid species, stands as one of the world’s most vital cereal crops, serving as the primary food source for over 40% of the global population (Mastrangelo and Cattivelli, 2021; Ana et al., 2024). Seed germination in wheat represents a critical physiological process characterized by three distinct phases: rapid initial water uptake, a lag phase (plateau period), and further water absorption (Paparella et al., 2015; Nile et al., 2022). This process involves complex metabolic transitions, progressing from sucrose degradation to the activation of major nutrient reserves (starch, proteins, and lipids), ultimately providing the energy and biomaterials essential for seedling establishment and photosynthesis.
Abscisic acid (ABA), a central phytohormone, extensively regulates multiple aspects of plant physiology, including seed dormancy induction/maintenance, germination suppression, adaptive responses to biotic/abiotic stresses, and developmental modulation under non-stress conditions (Brookbank et al., 2021). ABA exhibits a canonical concentration-dependent biphasic effect: low concentrations typically activate defense mechanisms and moderately promote growth—e.g., rescuing growth defects in ABA-deficient mutants (Cheng et al., 2002; Vishwakarma et al., 2017), while high concentrations inhibit growth or even trigger programmed cell death (Tu et al., 2025). Notably, this biphasic effect is pronounced in root development; for instance, in Arabidopsis, low-dose exogenous ABA promotes primary and lateral root growth, whereas high concentrations suppress elongation (Emenecker and Strader, 2020; Li et al., 2017). ABA maintains root meristem homeostasis by inhibiting quiescent center (QC) cell division and stem cell differentiation (Zhang et al., 2010), and orchestrates resource allocation via long-distance signaling (McAdam et al., 2016).
ABA also modulates central metabolism and nutrient signaling, particularly carbon metabolism and sugar sensing. Basal ABA levels regulate cell cycle gene expression, chloroplast biogenesis, cuticle deposition, epidermal development, plant metabolism (e.g., carbon metabolism/transport), and xylem differentiation (Kishor et al., 2022). However, high ABA concentrations mimicking stress conditions negatively regulate photosynthesis and carbon assimilation, down-regulating photosynthetic genes encoded by the nuclear and chloroplast genomes (Cutler et al., 2010; Fujita et al., 2011; Yamburenko et al., 2015), while promoting soluble sugar accumulation, sucrose transport, and wax biosynthesis regulation (Gibson, 2004; Ma et al., 2017; Li et al., 2021). In crops like wheat, foliar ABA application enhances carbohydrate accumulation and remobilization to grains, thereby increasing yield (Travaglia et al., 2007), highlighting its agronomic potential.
The advent of multi-omics technologies (e.g., transcriptomics, metabolomics) provides powerful tools for deciphering complex stress-response mechanisms. Transcriptomics systematically identifies differentially expressed genes (DEGs) and enriched signaling pathways, while metabolomics captures dynamic reprogramming of small-molecule metabolites. Their integration reveals the “gene-metabolite-phenotype” cascade logic. Plants produce a vast diversity of metabolic compounds (>200,000 reported compounds) (Goodacre et al., 2004), with primary metabolites (e.g., carbohydrates, nucleotides, sulfur-containing compounds) often induced by ABA (Zhu and Assmann, 2017). These metabolites serve as precursors for secondary metabolites (SMs) through core pathways: glycolysis, TCA cycle, pentose phosphate pathway, shikimate pathway, and amino acid metabolism. As a key signal responding to water availability (Yoshida et al., 2019; Dekkers et al., 2015; Munemasa et al., 2015; Nakashima and Yamaguchi-Shinozaki, 2013; LeNoble et al., 2004; Sharp et al., 2000), ABA precisely coordinates metabolic and growth adaptations to environmental pressures (Yoshida et al., 2019). Although significant advances have been made in understanding the ABA signaling pathway in wheat (Walker-Simmons, 1987; Nakamura et al., 2011; Schramm et al., 2012; Chono et al., 2013; Utsugi et al., 2020; Rehal et al., 2022), the specific nature of the ABA-responsive transcriptional-metabolic network in this crucial crop requires further in-depth exploration.
Building upon the well-documented biphasic, concentration-dependent effect of ABA and its central role in metabolic regulation, we propose the overarching hypothesis that exogenous ABA triggers a highly coordinated and dose-dependent reprogramming of the transcriptomic and metabolomic networks in germinating wheat. Specifically, we hypothesize that: at a low concentration, ABA fine-tunes pathways related to carbon metabolism and reserve mobilization, thereby maintaining or moderately promoting adaptive responses such as root growth; whereas at a high concentration, it strongly induces stress-associated genes and metabolites (e.g., antioxidants and specific secondary metabolites) while repressing growth-related pathways, leading to the coordinated suppression of germination progression and enhancement of stress tolerance. To test this, wheat seeds at the germination stage were subjected to a gradient of ABA treatments (0, 2, 4, and 6 mg·L-1), a design intended to mimic basal physiological and stress-response states, respectively. By integrating physiological phenotyping with multi-omics data, this study aims to systematically identify the core regulatory modules and key metabolic hubs underlying the ABA response in wheat. Our findings are expected to provide a molecular framework for understanding wheat adaptation and to inform the development of ABA-based seed treatment technologies for future crop improvement.
Materials and methods
Plant materials and experimental design
Wheat seeds (Triticum aestivum L. cv. Suzhou) were stored at 4 °C. Uniform plump seeds were surface-sterilized with 0.5% (w/v) NaClO for 10min, rinsed 5 times with ultrapure water, and soaked in distilled water for 12h. After blot-drying with sterile filter paper, seeds were germinated on double-layered sterile filter paper in 60mm Petri dishes (30 mL distilled water per dish) under controlled conditions: 25 °C, 12h photoperiod/12h dark, 65% relative humidity.
Upon radicle protrusion (≥2 mm), uniformly germinated seeds were transferred to new Petri- dishes (30 seeds/dish) containing 30 mL of ABA (Hefei BASF Biotechnology Co., Ltd., China; ≥98.5% purity by HPLC) solutions at concentrations of 0 (CK), 2, 4, 6 mg·L-1, prepared from 100 mM stock in 0.01% ethanol. Three biological replicates per treatment were maintained in growth chambers under identical conditions for 7d. To maintain humidity and ABA concentration, 30 mL of corresponding solutions were replenished every 48h. Root length was measured after treatment. Seedlings were flash-frozen in liquid N2 for physiological assays, transcriptomics, and metabolomics.
Physiological parameter analysis
Malondialdehyde (MDA) content was measured following (Nahakpam and Shah, 2011). Samples (0.5g FW) were homogenized in 2 mL 10% (w/v) trichloroacetic acid (TCA) on ice. After centrifugation (10,000 ×g, 10min), 1 mL supernatant was mixed with 2 mL 0.67% (w/v) thiobarbituric acid (TBA). The mixture was incubated at 95 °C for 30min, cooled, and centrifuged (12,000 ×g, 10min). Absorbance at 532, 600, and 450 nm was measured. MDA content = [6.45×(A532 - A600) - 0.56×A450] × V/(W × 1000) (μmol·g-1 FW). Catalase (CAT) activity was measured following (Rao and Sresty, 2000). Reaction mixture: 1.5 mL 0.2 M phosphate buffer (PBS; 50 mM Na2HPO4/NaH2PO4, pH 7.8), 1.0 mL H2O, 0.3 mL 0.1 M H2O2. After pre-incubation at 25 °C for 5min, 0.2 mL enzyme extract was added. CAT activity was calculated from ΔA240·min-1 and expressed as U·g-1 FW·min-1. Relative electrolyte leakage (REL) was measured following (Zhu et al., 2025; Han et al., 2022). Samples (0.5g FW) were rinsed, blotted dry, and incubated in 10 mL ultrapure water at 25 °C for 24h with shaking (100 rpm). Initial conductivity (C1) was measured. After autoclaving (121 °C, 0.1 MPa, 20min), final conductivity (C2) was recorded. REL (%) = (C1/C2) × 100. All assays included three technical replicates.
Transcriptomic analysis, RNA extraction and library construction
Total RNA was extracted from six samples (CK and 6 mg L-1 ABA; 3 biological replicates each) using TRIzol® (Takara). RNA integrity was confirmed by: 1% agarose gel electrophoresis (intact 28S/18S rRNA bands), NanoDrop 2000 (A260/A280 ≥ 1.8; A260/A230 ≥ 2.0) and Qubit® 2.0 Fluorometer, Agilent 2100 Bioanalyzer (RIN ≥ 8.0). Libraries were prepared using NEBNext® Ultra™ RNA Library Prep Kit (Illumina®) with unique indices. Poly(A)+ mRNA was enriched, fragmented, and reverse-transcribed. After end-repair and adapter ligation, libraries were size-selected (200-300 bp) using AMPure XP beads. Final libraries were quantified by qPCR (KAPA Biosystems) and sequenced on Illumina NovaSeq 6000 (150 bp paired-end). Bioinformatic Analysis: Reads were aligned to T. aestivum reference genome (IWGSC RefSeq v2.1) using HISAT2. Gene expression was quantified as FPKM (Trapnell et al., 2010). Differentially expressed genes (DEGs) were identified by DESeq2 (Pertea et al., 2015) (|log2FC| > 1, FDR < 0.05). GO and KEGG enrichment used Blast2GO with Fisher’s exact test (FDR < 0.05).
Metabolomic analysis, metabolite extraction and profiling
Lyophilized samples (100 mg) were extracted with 1.2 mL 80% methanol at 4 °C. After centrifugation (12,000 ×g, 15min), supernatants were analyzed by UPLC (SHIMADZU Nexera X2)-MS/MS (AB Sciex 4500 QTRAP). The liquid chromatography separation was performed on an Agilent SB-C18 column (100mm × 2.1mm, 1.8 μm) maintained at 40 °C. The mobile phase consisted of ultrapure water with 0.1% formic acid (eluent A) and acetonitrile with 0.1% formic acid (eluent B). A gradient elution program was applied as follows: 5% B initially, increased linearly to 95% B over 9 minutes, held for 1 minute, then returned to 5% B within 0.1 minutes and re-equilibrated for 4.9 minutes (total run time: 15 minutes). The flow rate was 0.35 mL/min, and the injection volume was 4 μL (Cao et al., 2015). DAMs were filtered by: VIP > 1.0 (from OPLS-DA model) (Phahlane et al., 2022), |log2FC| ≥ 1, and P<0.05 (Student’s t-test).
qRT-PCR validation of differentially expressed genes
To independently verify the reliability of the transcriptome sequencing data, we randomly selected 12 differentially expressed genes (DEGs) for expression level confirmation by quantitative real-time PCR (qRT-PCR). First-strand cDNA was synthesized from the isolated total RNA using the TaKaRa PrimeScript™ RT Master Mix. All gene-specific primers were designed utilizing the Primer-BLAST tool available on the NCBI website, with their sequences detailed in Supplementary Table 1. The qPCR assays were conducted in a final volume of 20 µL, which contained 10 µL of TaKaRa SYBR® Premix Ex Taq™ II, 0.8 µL of each gene-specific primer (10 µM), 0.4 µL of Rox Reference Dye, 2 µL of cDNA, and 6 µL of ddH2O. Amplification was performed on a Thermo Fisher Quantum Studio 5 real-time PCR system, following the thermal cycling protocol established by Chen et al. (2021). The GADPH gene served as the endogenous control for normalization, and the relative expression of each target gene, based on three biological replicates per condition, was determined using the comparative 2 –ΔΔCT method.
Statistical analysis
Data were processed in Excel 2019 (Microsoft) and analyzed by one-way ANOVA (SPSS 22.0; IBM). Significant differences among treatments were determined by ANOVA followed by Duncan’s multiple range test. Mean values labeled with different letters are significantly different at the 5% level (P<0.05). Values represent mean ± SD of three biological replicates.
Results
Effects of exogenous ABA on morpho-physiological parameters
Increasing ABA concentrations induced a biphasic response in root elongation of wheat germinating seeds (Figure 1A). Specifically, 2 mg·L-1 ABA treatment yielded maximal root length, representing a 1.12-fold increase versus CK (P<0.05). However, concentrations ≥ 4 mg·L-1 significantly suppressed growth, with 6 mg·L-1 ABA reducing root length by 40% relative to CK (Figure 1B).
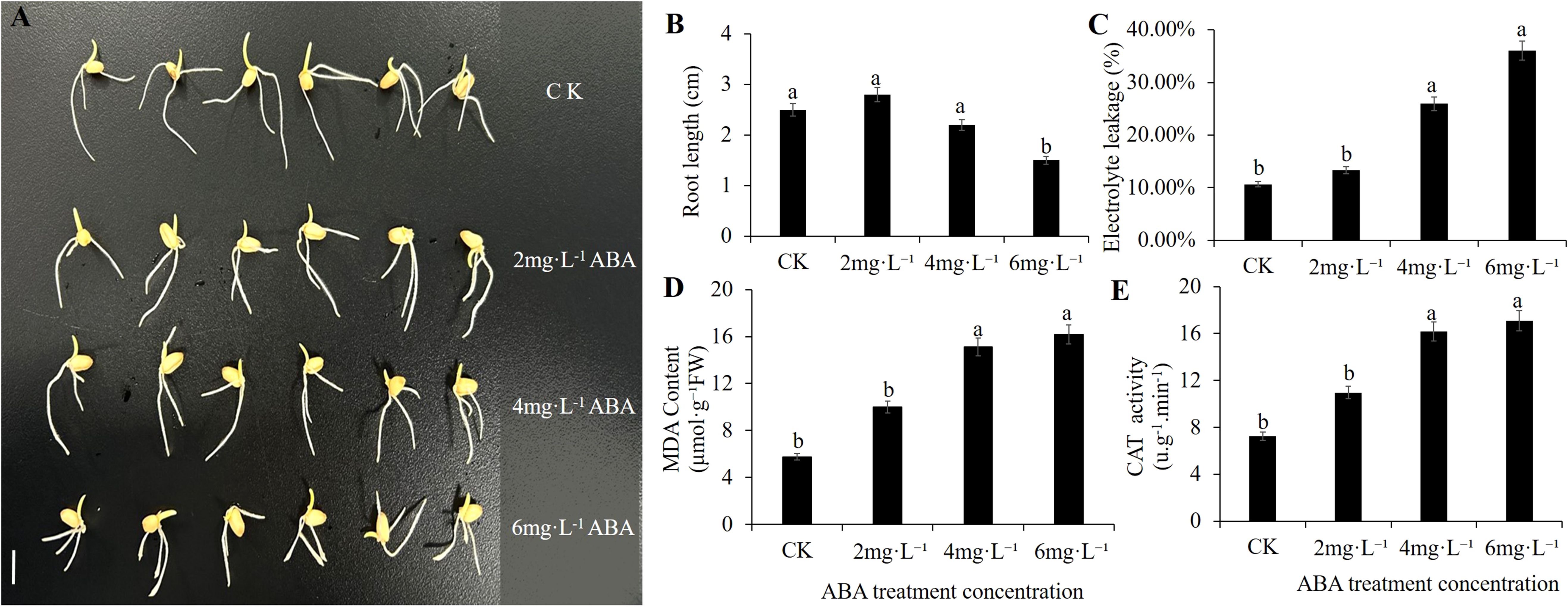
Figure 1. Effect of exogenous ABA treatment on morphological characteristics and physiological traits during wheat seed germination. (A) represent Morphological Characteristics changes under different Exogenous ABA treatments. Bar = 1cm. (B) ABA treatments on wheat root length. (C-E) represent respectively (REL, MDA, and CAT) changes under different Exogenous ABA treatments.
Physiological assessments revealed no significant changes in electrolyte leakage, malondialdehyde (MDA), or catalase (CAT) activity content at 2 mg·L-1 ABA versus CK (Figures 1C–E). Elevated ABA concentrations (4 – 6 mg·L-1) triggered significant increases in these markers (P<0.01), indicating progressive oxidative damage. Collectively, wheat exhibited biphasic adaptation to ABA stress: initial ROS scavenging enhancement followed by physiological dysfunction upon exceeding tolerance thresholds.
Transcriptomic profiling and DEG functional annotation
Given the pronounced phenotypic divergence between CK and 6 mg·L-1 ABA groups, transcriptome sequencing was performed to decipher molecular mechanisms. High-quality data were obtained (Q30: 92.93–93.50%; GC content: 53.50–55.38%; Supplementary Table 2). Pairwise comparison identified 854 differentially expressed genes (DEGs; |log2FC| > 1, adj. P<0.05), comprising 470 up-regulated and 384 down-regulated genes (Figure 2A, Supplementary Table 4). Hierarchical clustering analysis revealed a clear separation between the CK and 6 mg/L ABA-treated samples based on the 854 differentially expressed genes (DEGs) (Figure 2B). This result indicates that exogenous ABA stress induces significant alterations in the transcriptome of germinating wheat seeds. Gene Ontology (GO) enrichment categorized DEGs into three domains (Figure 2C). First, Cellular components including “Intracellular”, “Protein-containing complex”, and “Cellular anatomical entity” were significantly enriched. Second, Biological processes including “Cellular process” and “Metabolic process”, reflecting dynamic stress adaptation. Third, Molecular functions including “Binding”, “Catalytic activity”, and “Transporter activity” indicated roles in biochemical reactions and transmembrane transport. KEGG pathway analysis highlighted ABA-responsive pathways: “Linoleic acid metabolism”, “Alpha-Linolenic acid metabolism”, “Glycolysis/Gluconeogenesis”, “Biosynthesis of amino acids”, and “Cysteine and methionine metabolism” (Figure 2D), underscoring their roles in ROS scavenging, energy provision, and protein homeostasis.
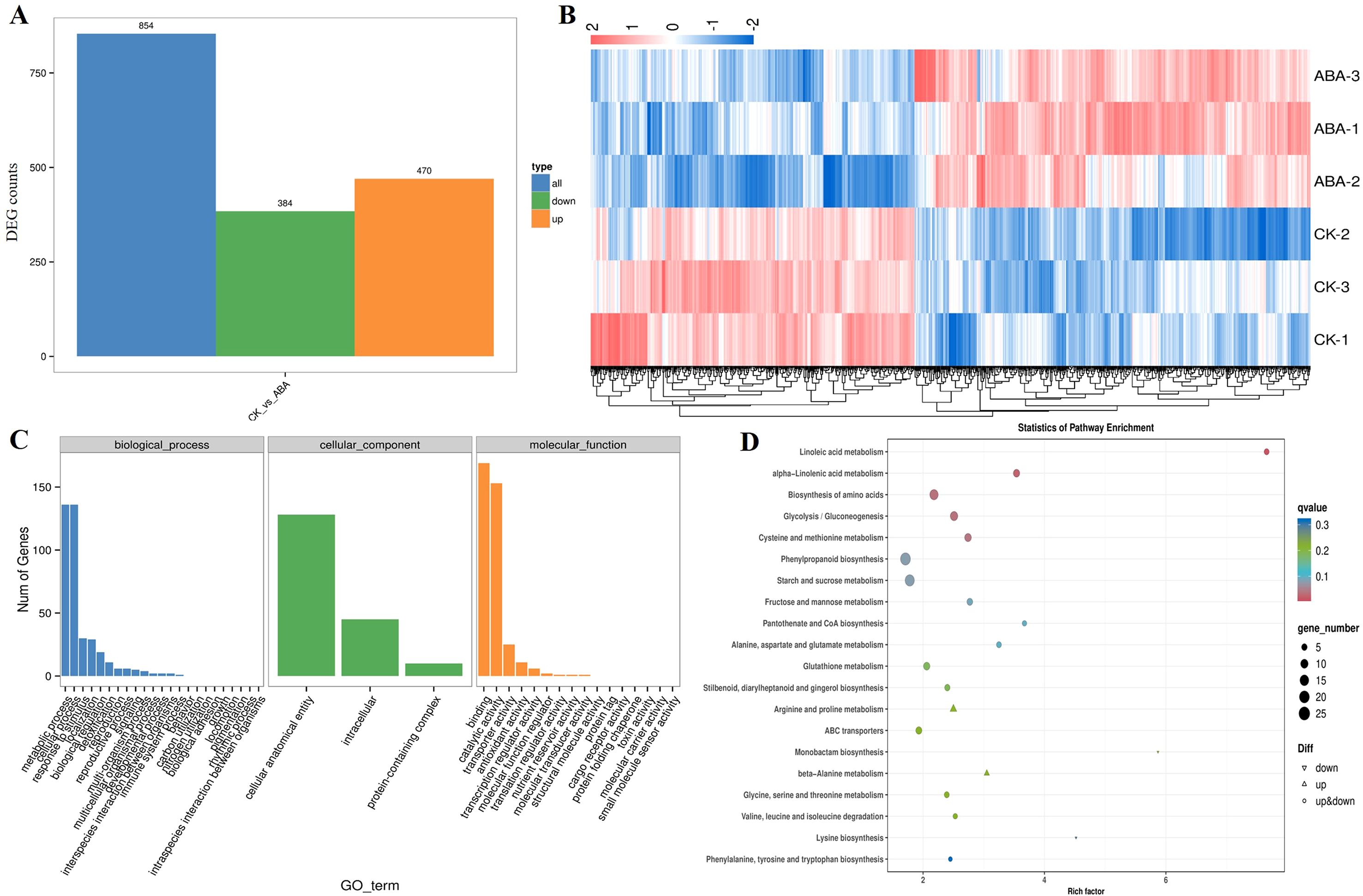
Figure 2. Transcriptomic analysis of germinating wheat seeds under exogenous ABA treatment: CK vs. 6 mg·L-1 ABA. (A) Identification of differentially expressed genes (DEGs) between CK and 6 mg·L-1 ABA treatment groups in germinating wheat seeds under exogenous ABA application. (B) Hierarchical clustering heatmap of DEG expression profiles between CK and 6 mg·L-1 ABA treatments. (C) Gene Ontology (GO) enrichment analysis of DEGs categorized in cellular components, biological processes and molecular functions. (D) KEGG pathway enrichment analysis of DEGs from CK vs 6 mg·L-1 ABA comparison.
To validate the reliability of the transcriptome data, we performed qRT-PCR analysis on 12 randomly selected differentially expressed genes (DEGs) identified from the comparison between the CK and 6 mg·L-1 ABA treatment. The qRT-PCR results correlated well with the RNA-seq data (Supplementary Figure 1), confirming the high reproducibility of our transcriptomic dataset.
Metabolomic profiling and DAM functional annotation
Untargeted LC/MS metabolomics detected 21,651 metabolic features, annotated as 6,080 compounds. Principal component analysis (PCA) showed clear separation between CK and 6 mg/L ABA groups (Figure 3A). Volcano plot analysis identified 665 differentially accumulated metabolites (DAMs; VIP ≥1, |FC|≥2), with 345 up- regulated and 320 down-regulated (Figure 3B, Supplementary Table 5). These DAMs were systematically classified into 16 distinct groups. Ketones, aldehydes, and esters (104, 15.6%) constituted the largest group, followed by lipids (103, 15.5%), saccharides (40, 6.0%), organic acids (45, 6.8%), terpenoids (53, 8.0%), amino acids (30, 4.5%), alkaloids (24, 3.6%), flavonoids (22, 3.3%), polyphenols (14, 2.1%), alcohols (13, 2.0%), steroids (11, 1.7%), nucleotides (10, 1.5%), quinones (7, 1.0%), coumarins (5, 0.8%), and phenylpropanoids (1, 0.2%). Additionally, other compounds (94, 14.1%) and unannotated compounds (89, 13.3%) comprised the classification (Supplementary Figure 2). Cluster analysis revealed distinct metabolic profiles between treatments (Figure 3C). KEGG enrichment identified “Benzoxazinoid biosynthesis” and “Nicotinate and nicotinamide metabolism” as pathways with the largest changes (Figure 3D).
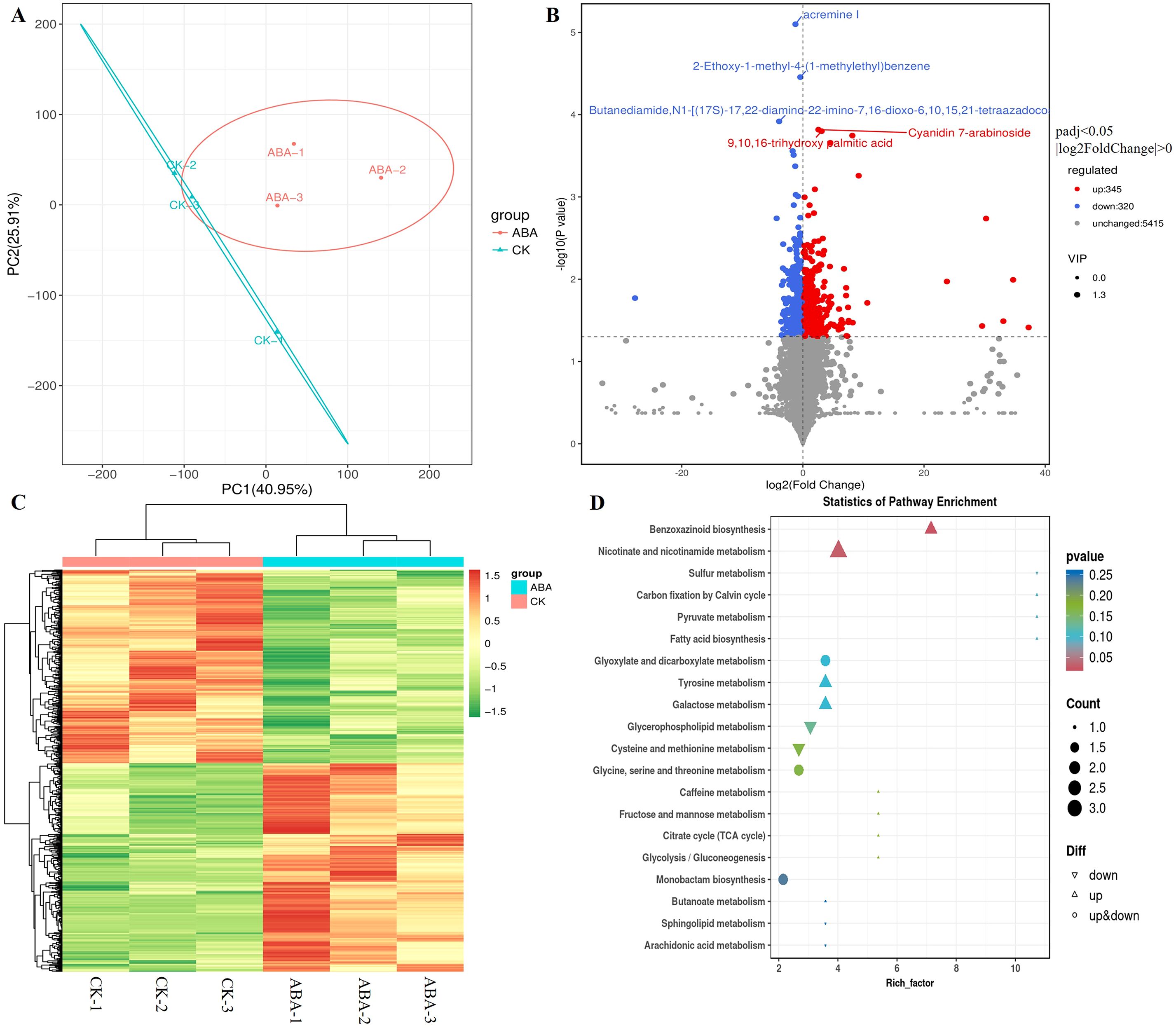
Figure 3. Metabolomic analysis of germinating wheat seeds under exogenous ABA treatment: CK vs. 6 mg·L-1 ABA. (A) Principal component analysis (PCA) of metabolomes from CK and 6 mg·L-1 ABA-treated germinating wheat seeds under exogenous ABA application. (B) Volcano plot of differentially accumulated metabolites (DAMs) between CK and 6 mg·L-1 ABA groups. (C) Hierarchical clustering heatmap of differentially accumulated metabolites (DAMs). (D) KEGG pathway enrichment analysis of DAMs.
Integrative transcriptomic-metabolomic analysis
Co-expression network analysis (Pearson |r| > 0.8, P<0.01) revealed strong positive correlations between DEGs and DAMs (Figure 4A). O2PLS integration highlighted 15 key gene-metabolite pairs (Figure 4B). Joint KEGG enrichment identified 10 core pathways including: Glycolysis/Gluconeogenesis, Biosynthesis of amino acids, Cysteine and methionine metabolism、Starch and sucrose metabolism, ABC transporters, and others (Figure 4C). Crucially, the metabolite L-serine was enriched in five pathways (Biosynthesis of amino acids, Cysteine and methionine metabolism, Glycine/serine/threonine metabolism, ABC transporters, Fructose and mannose metabolism). Key genes demonstrated cross-pathway regulation: TraesCS3A02G276100, TraesCS3B02G309700, TraesCS3D02G276000 and TraesCS5B02G078300 co-regulated Starch and sucrose metabolism, Fructose and mannose metabolism, and Glycolysis/Gluconeogenesis. TraesCS5A02G398300, TraesCS5D02G407800, TraesCS7A02G102700 modulated Biosynthesis of amino acids and Cysteine and methionine metabolism. NewGene_15595, TraesCS1D02G062800, TraesCS7A02G015900, TraesCS7D02G012500 participated in Biosynthesis of amino acids and Glycolysis/Gluconeogenesis. These multi-pathway hubs (L-serine and highlighted genes) constitute critical key network nodes in ABA response networks (Figure 5, Supplementary Table 3).
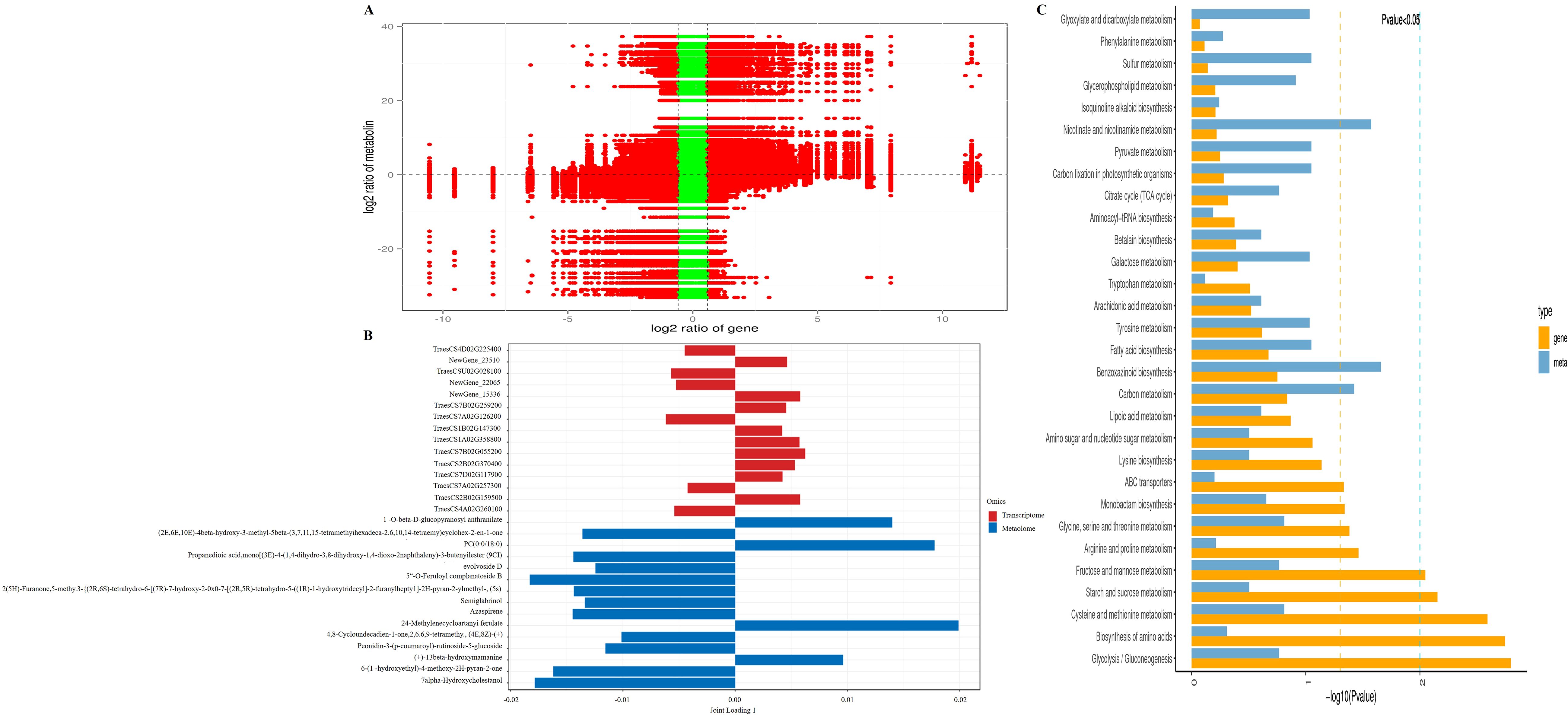
Figure 4. Integrated transcriptomic and metabolomic analysis of germinating wheat seeds under exogenous ABA treatment: CK vs. 6 mg·L-1ABA. (A) Co-expression network analysis of differentially expressed genes (DEGs) and differentially accumulated metabolites (DAMs) in germinating wheat seeds under exogenous ABA application. (B) Two-way orthogonal partial least squares-discriminant analysis (O2PLS-DA) of transcriptomic and metabolomic datasets. (C) Joint KEGG pathway enrichment analysis of DEGs and DAMs.
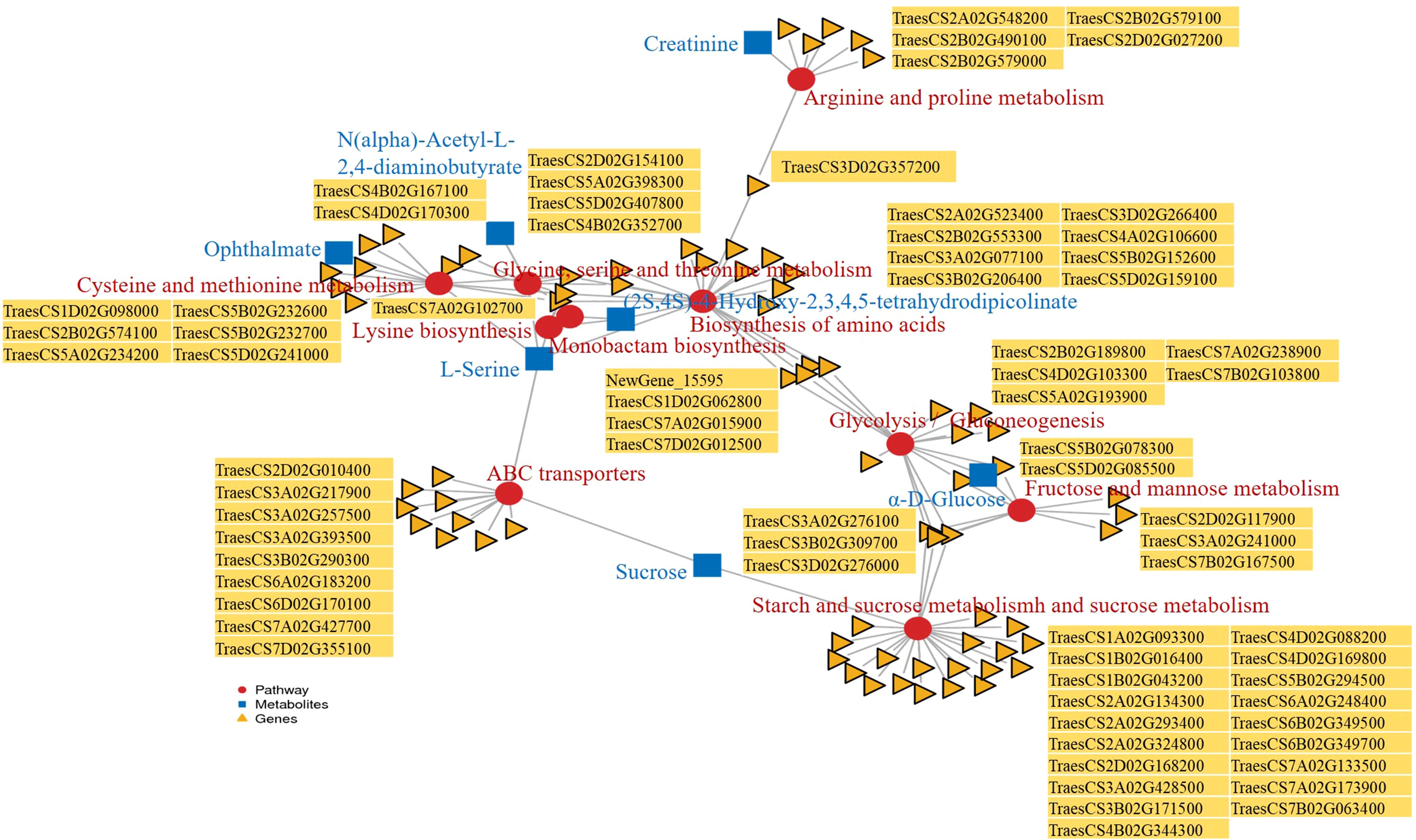
Figure 5. Pathway enrichment analysis of differentially expressed genes (DEGs) and differentially accumulated metabolites (DAMs) from germinating wheat seeds under exogenous ABA treatment: CK vs. 6 mg·L-1 ABA. Red circles represent pathways, blue squares represent metabolites, yellow triangles represent genes.
Discussion
Biphasic physiological response to exogenous ABA and critical threshold
As a key phytohormone, ABA regulates plant growth and stress adaptation through multifaceted signaling (Chen et al., 2020). Basal ABA levels are essential for growth modulation across tissues (Jiang et al., 2025), while its accumulation underpins adaptive responses to environmental challenges (Chen et al., 2006). This study delineated a concentration-dependent biphasic response in wheat roots: 2 mg·L-1 ABA increased root length (1.12-fold vs. CK), whereas 6 mg·L-1 caused 40% reduction. This biphasicity correlated with ROS dynamics: low ABA mildly induced ROS, activating CAT (initially unchanged), but concentrations ≥4 mg·L-1 triggered ROS burst (2.1-fold MDA increase), causing membrane peroxidation (electrolyte leakage surge) and homeostasis collapse. These findings align with ABA’s biphasic model, identifying 4 mg·L-1 as the oxidative damage threshold.
The significant increase in MDA content observed in our study is consistent with the findings of Rehal et al. (2022) in wheat seedlings, reinforcing that membrane lipid peroxidation is a key consequence of ABA-induced oxidative stress. However, the observed physiological response may exhibit species-specific variations. For instance, Xie et al. (2021) found less root elongation at any ABA concentration above 0 mg/L in Arabidopsis, which is inconsistent with the present findings. This discrepancy could be attributed to intrinsic differences between the plant species investigated, such as variations in ABA receptor sensitivity, antioxidant capacity, and the efficiency of downstream signaling pathways, which collectively determine the physiological response to phytohormone-induced stress. However, to further refine the dose-response relationship and precisely determine the toxicity threshold, future studies could be strengthened by incorporating a wider concentration gradient (e.g., up to 8 and 10 mg·L-1) and calculating the half-maximal inhibitory concentration (IC50). This approach would provide a more quantitative assessment of the inhibitory effects and enhance the predictive power of the model for agricultural applications.
Metabolic adaptation to stress and ABA-mediated linolenic acid regulation
Plants deploy metabolic reprogramming to cope with stress (Cramer et al., 2011), where metabolomics predicts adaptive capacity (Sweetlove et al., 2008; Fiers et al., 2005). Core strategies include osmotic adjustment (Krasensky and Jonak, 2012) and protective secondary metabolite accumulation—e.g., phenolics, proline, and soluble sugars maintain water potential, stabilize proteins/membranes, and scavenge ROS (Dixon and Paiva, 1995; Takahashi et al., 2020).
Our transcriptomics under 6 mg·L-1ABA identified 854 DEGs. GO enrichment highlighted “metabolic process” and “transporter activity”, indicating metabolic and transport network remodeling. KEGG analysis pinpointed three core pathways: Linolenic acid metabolism, Gene TraesCS5B02G078300 enrichment suggests jasmonate (JA) precursor synthesis for defense signaling. Cysteine-methionine metabolism: Supplies glutathione precursors for ROS scavenging and alleviating oxidative damage induced by ABA. The glycolysis/luconeogenesis pathway, highlighted by the recurrent enrichment of gene TraesCS3A02G276100, is implicated in sustaining energy homeostasis and generating carbon precursors for the potential synthesis of compatible solutes under ABA-induced stress.
Metabolomics corroborated these findings: lipids (15.5% of DAMs) and organic acids (6.8%) dominated the profile. Pathway analysis further revealed benzoxazinoid (stress resistance) and nicotinate (NAD+ synthesis) activation. Integratively, we propose an ABA-induced adaptation axis: ABA triggers lipid oxidation for signaling (e.g., JA precursors) while mobilizing secondary metabolites (e.g., benzoxazinoids) to mitigate oxidative damage. Crucially, linolenic acid metabolism holds strategic importance, as exogenous ABA specifically induces its genes and promotes endodermal deposition (Barberon et al., 2016; Cottle and Kolattukudy, 1982; Boher et al., 2013; Kosma et al., 2014, 2014; Yadav et al., 2014; Verdaguer et al., 2016; Shukla et al., 2021).
Gene-metabolite network drives stress adaptation
Co-expression networks (|r| > 0.8, P<0.01) identified two hub types. First, a metabolite hub is represented by the accumulation of L-Serine which is involved in five pathways and showed a strong correlation with TraesCS7A02G015900 expression (r = 0.92, P<0.001). As a carbon-nitrogen flux node, it balances resource allocation. Second, a gene hub represented TraesCS3A02G276100 (glycolytic enzyme) co-regulated starch/sucrose, fructose/mannose, and glycolysis, coordinating carbon partitioning for energy homeostasis.
Of particular note, our integrated analysis revealed that the expression changes of amino acid biosynthesis genes (e.g., TraesCS5A02G398300) were highly correlated with the abundance of the lysine precursor (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate. We speculate that this co-variation may suggest a potential mechanism for regulating reactive oxygen species (ROS) homeostasis through nitrogen metabolism. Based on these correlations, we propose a working model wherein under high ABA stress, energy supply (glycolysis), the ROS scavenging system, and nitrogen metabolism (amino acid balance) form a functionally interconnected network. Our data indicate that ABA treatment was accompanied by attenuated glycolysis, decreased ATP levels, altered amino acid metabolic profiles, and accumulated ROS, collectively pointing to a state of overall metabolic dysfunction (Figure 6). It must be emphasized that this model is proposed based on correlative data obtained under specific experimental conditions, and the underlying causal relationships and molecular mechanisms require future validation through functional gain-/loss-of-function experiments and metabolic flux analysis.
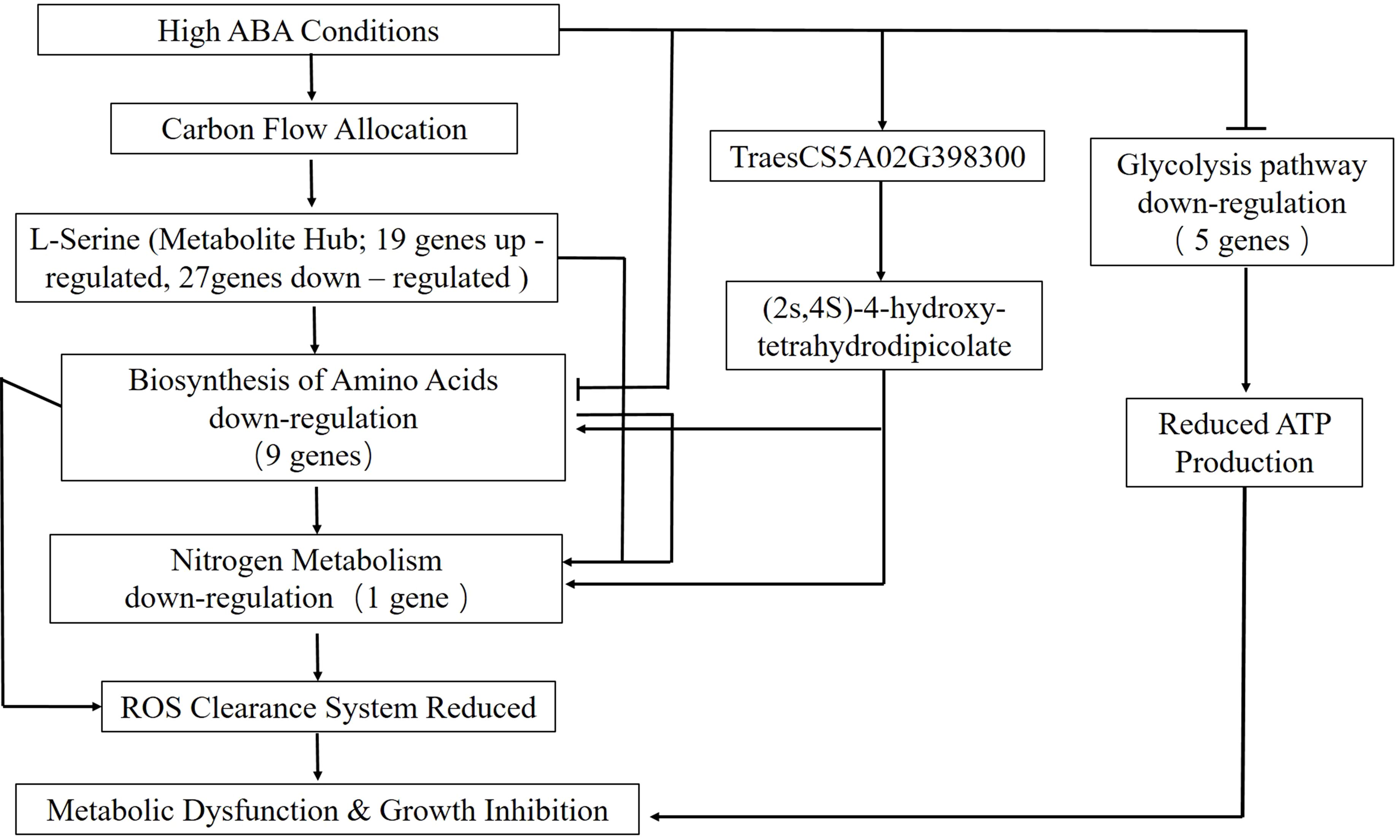
Figure 6. Regulatory mechanism of wheat seed germination under high ABA concentration. arrows for induction, T-bars for inhibition.
Conclusion
This study elucidates a biphasic regulation of root growth by exogenous ABA in wheat germinating seeds: low concentrations (2 mg·L-1) mildly promote elongation, whereas high levels (≥4 mg·L-1) suppress growth via oxidative stress (MDA accumulation, electrolyte leakage). Integrated transcriptomics and metabolomics identified 10 core pathways—notably Glycolysis/Gluconeogenesis, Biosynthesis of amino acids, and Cysteine and methionine metabolism—as response hubs. Critical regulators include the metabolite L-serine and multi-pathway genes (TraesCS3A02G276100, TraesCS5A02G398300), which orchestrate energy provision, ROS clearance, and osmoprotection to maintain homeostasis. Collectively, wheat dynamically balances ABA stress through transcriptional and metabolic reprogramming, providing novel insights for stress-resilient crop breeding.
Data availability statement
The data supporting the findings of this study are openly available in EBI Metagenomics under accession number PRJNA1311629, and the direct URL is: https://www.ebi.ac.uk/ena/browser/view/PRJNA1311629.
Author contributions
XW: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. XS: Data curation, Investigation, Writing – original draft. CC: Formal Analysis, Software, Writing – original draft. CW: Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by Suzhou University Doctoral Research Start-up Fund (Grant No. 2021BSK049), College Students’ Innovation and Entrepreneurship Training Program Project of Suzhou University (Grant No. ZCXM24-209), Research and Reform Practice Project on the “Four Emerging Fields” of Suzhou University (Grant No. szxy2024xnjy01), Anhui Provincial College Student Innovation Training Program (Grant No. S202510379126).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphgy.2025.1684534/full#supplementary-material
Supplementary Figure 1 | qRT-PCR verification of RNA-seq sequencing results.
Supplementary Figure 2 | Classification Diagram of 665 Differentially Accumulated Metabolites (DAMs) of CK vs. 6 mg·L-1 ABA.
Supplementary TABLE 4 | Differentially Expressed Genes (DEGs) Identified from the Pairwise Comparison between CK vs. 6 mg·L-1 ABA.
Supplementary TABLE 5 | Differentially Accumulated Metabolites (DAMs) Identified between CK vs. 6 mg·L-1 ABA.
References
Ana G. G., Manuel A. G. R., and Pilar P. (2024). The highly dynamic satellitomes of cultivated wheat species. Ann. Bot. 134, 975–992. doi: 10.1093/aob/mcae132
Barberon M., Vermeer J. E. M., Bellis D. D., Wang P., Naseer S., Andersen T. G., et al. (2016). Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164, 447–459. doi: 10.1016/j.cell.2015.12.021
Boher P., Serra O., Soler M., Molinas M., and Figueras M. (2013). The potato suberin feruloyl transferase FHT which accumulates in the phellogen is induced by wounding and regulated by abscisic and salicylic acids. J. Exp. Bot. 64, 3225–3236. doi: 10.1093/jxb/ert163
Brookbank B. P., Patel J., Gazzarrini S., and Nambara E. (2021). Role of basal ABA in plant growth and development. Genes (Basel) 12, 1936. doi: 10.3390/genes12121936
Cao M., Fraser K., Huege J., Featonby T., Rasmussen S., and Jones C. (2015). Predicting retention time in hydrophilic interaction liquid chromatography mass spectrometry and its use for peak annotation in metabolomics. Metabolomics 11, 696–706. doi: 10.1007/s11306-014-0727-x
Chen F. Q., Ji X. Z., Bai M. X., Zhuang Z. L., and Peng Y. L. (2021). Network analysis of different exogenous hormones on the regulation of deep sowing tolerance in maize seedlings. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.739101
Chen K., Li G. J., Bressan R. A., Song C. P., Zhu J. K., and Zhao Y. (2020). Abscisic acid dynamics, signaling, and functions in plants. J. Integr.Plant Biol. 62, 25–54. doi: 10.1111/jipb.12899
Chen C. W., Yang Y. W., Lur H. S., Tsai Y. G., and Chang M. C. (2006). novel function of abscisic acid in the regulation of rice (Oryza sativa L.) root growth and development. Plant Cell Physiol. 47, 1–13. doi: 10.1093/pcp/pci216
Cheng W. H., Endo A., Zhou L., Penney J., Chen H. C., Arroyo A., et al. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 14, 2723–2743. doi: 10.1105/tpc.006494
Chono M., Matsunaka H., Seki M., Fujita M., Kiribuchi-Otobe C., Oda S., et al. (2013). Isolation of a wheat (Triticum aestivum L.) mutant in ABA 8’-hydroxylase gene: effect of reduced ABA catabolism on germination inhibition under field condition. Breed Sci. 63, 104–115. doi: 10.1270/jsbbs.63.104
Cottle W. and Kolattukudy P. E. (1982). Abscisic acid stimulation of suberization: Induction of enzymes and deposition of polymeric components and associated waxes in tissue cultures of potato tuber. Plant Physiol. 70, 775–780. doi: 10.1104/pp.70.3.775
Cramer G. R., Urano K., Delrot S., Pezzotti M., and Shinozaki K. (2011). Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 11, 163. doi: 10.1186/1471-2229-11-163
Cutler S. R., Rodriguez P. L., Finkelstein R. R., and Abrams S. R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Dekkers B. J. W., Costa M. C. D., Maia J., Bentsink L., Ligterink W., and Hilhorst H. W. M. (2015). Acquisition and loss of desiccation tolerance in seeds: From experimental model to biological relevance. Planta 241, 563–577. doi: 10.1007/s00425-014-2240-x
Dixon R. A. and Paiva N. L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell. 7, 1085–1097. doi: 10.1105/tpc.7.7.1085
Emenecker R. J. and Strader L. C. (2020). Auxin-abscisic acid interactions in plant growth and development. Biomolecules 10, 281. doi: 10.3390/biom10020281
Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., Stiekema W., et al. (2005). The 14–amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 17, 2542–2553. doi: 10.1105/tpc.105.034009
Fujita Y., Fujita M., Shinozaki K., and Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Gibson S. I. (2004). Sugar and phytohormone response pathways: Navigating a signaling network. J. Exp. Bot. 55, 253–264. doi: 10.1093/jxb/erh048
Goodacre R., Vaidyanathan S., Dunn W. B., Harrigan G. G., and Kell D. B. (2004). Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 22, 245–252. doi: 10.1016/j.tibtech.2004.03.007
Han G., Qiao Z., Li Y., Yang Z., Zhang Z., Zhang Y., et al. (2022). bMYB48 positively regulates salt gland development of Limonium bicolor and salt tolerance of plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1039984
Jiang J. M., Zhu Q. H., Li Y., Wu J. Y., Zou C. H., Li H. B., et al. (2025). Abscisic acid enhances DNA damage response through the nuclear shuttling of clathrin light chain 2 in plant cells. Sci. Adv. 11, eadt2842. doi: 10.1126/sciadv.adt2842
Kishor P. B. K., Tiozon R. N., Fernie A. R., and Sreenivasulu N. (2022). Abscisic acid and its role in the modulation of plant growth, development, and yield stability. Trends Plant Sci. 27, 1283–1295. doi: 10.1016/j.tplants.2022.08.013
Kosma D. K., Murmu J., Razeq F. M., Santos P., Bourgault R., Molina I., et al. (2014). At MYB 41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J. 80, 216–229. doi: 10.1111/tpj.12624
Krasensky J. and Jonak C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. doi: 10.1093/jxb/err460
LeNoble M. E., Spollen W. G., and Sharp R. E. (2004). Maintenance of shoot growth by endogenous ABA: Genetic assessment of the involvement of ethylene suppression. J. Exp. Bot. 55, 237–245. doi: 10.1093/jxb/erh031
Li X., Chen L., Forde B. G., and Davies W. J. (2017). The biphasic root growth response to abscisic acid in Arabidopsis involves interaction with ethylene and auxin signalling pathways. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01493
Li J. J., Liu B. Y., Li X. Y., Li D. M., Han J. Y., Zhang Y., et al. (2021). Exogenous abscisic acid mediates berry quality improvement by altered endogenous plant hormones level in “Ruiduhongyu. Grapevine. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.739964
Ma Q. J., Sun M. H., Lu J., Liu Y. J., Hu D. G., and Hao Y. J. (2017). Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 174, 2348–2362. doi: 10.1104/pp.17.00502
Mastrangelo A. M. and Cattivelli L. (2021). What makes bread and durum wheat different? Trends Plant Sci. 26, 677–684. doi: 10.1016/j.tplants.2021.01.004
McAdam S. A. M., Brodribb T. J., and Ross J. J. (2016). Shoot-derived abscisic acid promotes root growth. Plant Cell Environ. 39, 652–659. doi: 10.1111/pce.12669
Munemasa S., Hauser F., Park J., Waadt R., Brandt B., and Schroeder J. I. (2015). Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 28, 154–162. doi: 10.1016/j.pbi.2015.10.010
Nahakpam S. and Shah K. (2011). Expression of key antioxidant enzymes under combined effect of heat and cadmium toxicity in growing rice seedlings. Plant Growth Regul. 63, 23–35. doi: 10.1007/s10725-010-9508-3
Nakamura S., Abe F., Kawahigashi H., Nakazono K., Tagiri A., Matsumoto T., et al. (2011). Wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell. 23, 3215–3229. doi: 10.1105/tpc.111.088492
Nakashima K. and Yamaguchi-Shinozaki K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nile S. H., Thiruvengadam M., Wang Y., Samynathan R., Shariati M. A., Rebezov M., et al. (2022). Nano-priming as emerging seed priming technology for sustainable agriculture—recent developments and future perspectives. J. Nanobiotechnol. 20, 254. doi: 10.1186/s12951-022-01423-28
Paparella S., Araújo S. S., Rossi G., Wijayasinghe M., Carbonera D., and Balestrazzi A. (2015). Seed priming: state of the art and new perspectives. Plant Cell Rep. 34, 1281–1293. doi: 10.1007/s00299-015-1784-y
Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., and Salzberg S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Phahlane C. J., Laurie S. M., Shoko T., Manhivi V. E., and Sivakumar D. (2022). Comparison of caffeoylquinic acids and functional properties of domestic sweet potato (Ipomoea batatas (L.) Lam.) storage roots with established overseas varieties. Foods 11, 1329. doi: 10.3390/foods11091329
Rao K. M. and Sresty T. (2000). Antioxidative parameters in the seedlings of pigeonpea (Cajanus cajan (L.) Millspaugh) in response to Zn and Ni stresses. Plant Sci. 157, 113–128. doi: 10.1016/s0168-9452(00)00273-9
Rehal P. K., Tuan P. A., Nguyen T. N., Cattani D. J., Humphreys D. G., and Ayele B. T. (2022). Genetic variation of seed dormancy in wheat (Triticum aestivum L.) is mediated by transcriptional regulation of abscisic acid metabolism and signaling. Plant Sci. 324, 111432. doi: 10.1016/j.plantsci.2022.111432
Schramm E. C., Nelson S. K., and Steber C. M. (2012). Wheat ABA-insensitive mutants result in reduced grain dormancy. Euphytica 188, 35–49. doi: 10.1007/s10681-012-0669-1
Sharp R. E., LeNoble M. E., Else M. A., Thorne E. T., and Gherardi F. (2000). Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: Evidence for an interaction with ethylene. J. Exp. Bot. 51, 1575–1584. doi: 10.1093/jexbot/51.350.1575
Shukla V., Han J. P., Cléard F., Lefebvre-Legendre L., Gully K., Flis P., et al. (2021). Suberin plasticity to developmental and exogenous cues is regulated by a set of MYB transcription factors. Proc. Natl. Acad. Sci. U.S.A. 118, e2101730118. doi: 10.1073/pnas.2101730118
Sweetlove L. J., Fell D., and Fernie A. R. (2008). Getting to grips with the plant metabolic network. Biochem. J. 409, 27–41. doi: 10.1042/BJ20071115
Takahashi F., Kuromori T., Urano K., Yamaguchi-Shinozaki K., and Shinozaki K. (2020). Drought stress responses and resistance in plants: from cellular responses to long-distance intercellular communication. Front. Plant Sci. 11. doi: 10.3389/fpls
Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., Baren M. J., et al. (2010). Transcript assembly and quantification by RNA Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Travaglia C., Cohen A. C., Reinoso H., Castillo C., and Bottini R. (2007). Exogenous abscisic acid increases carbohydrate accumulation and redistribution to the grains in wheat grown under field conditions of soil water restriction. J. Plant Growth Regul. 26, 285–289. doi: 10.1007/s00344-007-9018-3
Tu D. H., Tian X., Liang C. J., Zhou P., Wang Y. P., Xing D., et al. (2025). Physiological and biochemical responses of pepper to cadmium stress: The mitigating effects of exogenous abscisic acid. Ecotoxicol Environ. Saf. 299, 118370. doi: 10.1016/j.ecoenv.2025.118370
Utsugi S., Ashikawa I., Nakamura S., and Shibasaka M. (2020). TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J. Plant Res. 133, 245–256. doi: 10.1007/s10265-020-01166-3
Verdaguer R., Soler M., Serra O., Garrote A., Fernandez S., Company-Arumi D., et al. (2016). Silencing of the potato StNAC103 gene enhances the accumulation of suberin polyester and associated wax in tuber skin. J. Exp. Bot. 67, 5415–5427. doi: 10.1093/jxb/erw305
Vishwakarma K., Upadhyay N., Kumar N., Yadav G., Singh J., Mishra R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00161
Walker-Simmons M. (1987). ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 84, 61–67. doi: 10.1104/pp.84.1.61
Xie Q. J., Essemine J., Pang X. C., Chen H. Y., Jin J., and Cai W. M. (2021). Abscisic acid regulates the root growth trajectory by reducing auxin transporter PIN2 protein levels in Arabidopsis thaliana. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.632676
Yadav V., Molina I., Ranathunge K., Castillo I. Q., Rothstein S. J., and Reed J. W. (2014). ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell. 26, 3569–3588. doi: 10.1105/tpc.114.129049
Yamburenko M. V., Zubo Y. O., and Borner T. (2015). Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C dependent activation of nuclear genes: Repression by guanosine-3-5-bisdiphosphate and activation by sigma factor 5. Plant J. 82, 1030–1041. doi: 10.1111/tpj.12876
Yoshida T., Obata T., Feil R., Lunn J. E., Fujita Y., Yamaguchi-Shinozaki K., et al. (2019). The role of abscisic acid signaling in maintaining the metabolic balance required for Arabidopsis growth under nonstress conditions. Plant Cell. 31, 84–105. doi: 10.1105/tpc.18.00766
Zhang H., Han W., De S. I., Talboys P., Loya R., Hassan A., et al. (2010). ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 64, 764–774. doi: 10.1111/j.1365-313X.2010.04367.x
Zhu M. and Assmann S. M. (2017). Metabolic signatures in response to abscisic acid (ABA) treatment in Brassica napus guard cells revealed by metabolomics. Sci. Rep. 7, 12875. doi: 10.1038/s41598-017-13166-w
Keywords: wheat, abscisic acid (ABA), stress response, multi-omics analysis, metabolic reprogramming
Citation: Wang X, Chen C, Sun X and Wang C (2025) Integrated transcriptomic and metabolomic analysis reveals the molecular mechanisms underlying wheat germinating seed response to exogenous abscisic acid stress. Front. Plant Physiol. 3:1684534. doi: 10.3389/fphgy.2025.1684534
Received: 12 August 2025; Accepted: 22 October 2025;
Published: 06 November 2025.
Edited by:
Raoudha Abdellaoui, Institut des Régions Arides, TunisiaReviewed by:
Wu Xu, University of Louisiana at Lafayette, United StatesMai Duy Luu Trinh, University of Copenhagen, Denmark
Jacob Sebesta, National Renewable Energy Laboratory (DOE), United States
Copyright © 2025 Wang, Chen, Sun and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolei Wang, d3hsMHZpcEAxNjMuY29t
 Xiaolei Wang
Xiaolei Wang Chuchu Chen
Chuchu Chen