- 1Department of Pharmacology, Frederick P. Whiddon College of Medicine, University of South Alabama, Mobile, AL, United States
- 2Department of Biology, College of Arts and Sciences, University of South Alabama, Mobile, AL, United States
Y RNAs are a poorly-studied class of small non-coding RNAs (sncRNAs) which have previously been implicated in the pathogenesis of different human diseases, including cardiac and autoimmune conditions, as well as certain cancers. In recent years, however, multiple studies have reported correlations between Y RNA expressions and disease outcomes in viral infections (e.g., IAV, HIV, HPV, and SARS-CoV-2) as well as potential mechanistic roles that Y RNAs may play in host anti-viral defense. These studies suggest that Y RNAs may be associated with upregulation of viral defense proteins as well as altered cell-cell communication during viral infections. In this review, current literature detailing Y RNA effects on human viral infection will be summarized and future directions in the study of these relationships discussed.
Introduction
Y RNAs are a class of small non-coding RNAs (sncRNAs) which were first described in 1981 (Lerner et al., 1981). Their name derives from the observation that they tend to localize in the cytoplasm of cells. Thus, the “Y” in “Y RNA” derives from the “y” in “cytoplasm”, as opposed to the “U” in the word “nucleus”. There are four distinct Y RNAs encoded in the human genome in a “syntenic” cluster on chromosome 7q36: Y RNA1 (hY1), Y RNA3 (hY3), Y RNA4 (hY4), and Y RNA5 (hY5). All four of these genes are transcribed by RNA polymerase III, and the resultant RNAs range from 83 to 112 nucleotides in length. Mature Y RNA primary structures include a triphosphate group at the 5′ end, a polyuridine tail at the 3′ end, and are thought to contain no modified nucleotides (Kowalski and Krude, 2015). After transcription, Y RNAs undergo folding in the nucleus to form distinct secondary structures, which are recognized and bound by the La and Ro60 proteins in a Ro-ribonucleoprotein complex (RoRNP), then exported to the cytoplasm (Figures 1A,B) (Valkov and Das, 2020). Previous studies have revealed roles for Y RNA in DNA replication, RNA quality control, and response to cellular stress (Wang et al., 2014), that said, major gaps in our understanding of their role(s) in human biology remain uncharacterized.
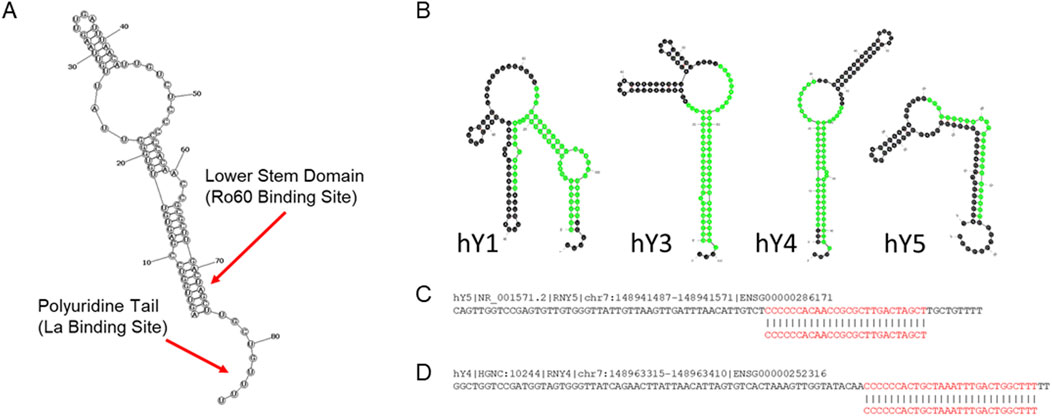
Figure 1. Y RNA biology. (A) Graphical overview of general Y RNA structure. All four human Y RNAs form a complex stem-loop structure. Specific proteins bind to certain Y RNA secondary structure elements. Ro60 binds the lower stem domains of all human Y RNAs to form Ro ribonucleoprotein (RoRNP) complexes. The La protein similarly binds to the 3′ polyuridine tail of all Y RNAs. The large, internal loop structure is thought to associate with multiple other cellular proteins (Teunissen et al., 2000). (B) Human Y RNA structures. YsRNAs are highlighted in green (Nicolas et al., 2012). Structures were generated with mfold (Zuker, 2003). (C,D) Descriptions of hY4 and hY5 ysRNAs correlated with viral severity. Human Y RNAs hY4 and hY5 sequences are shown and the ysRNAs correlated with viral severity excised from each are indicated in red. (C) Plasma levels of the indicated ysRNA excised from hY5 (initially annotated as hsa-miR-1975) is strongly correlated with influenza severity (Liu et al., 2021) and delivery of this ysRNA can inhibit influenza replication (Liu et al., 2019). (D) Plasma levels of the indicated ysRNA excised from hY4 is strongly correlated with SARS-CoV-2 severity (Olliff et al., 2023).
Interestingly, recent research has shown that Y RNAs can also be cleaved to produce yet another type of sncRNA, termed Y RNA-derived small RNAs (ysRNAs) typically upregulated during cellular stress (Verhagen and Pruijn, 2011). It was previously thought that these ysRNAs are a subclass of microRNAs (miRNAs), but research has now shown that the biogenesis of ysRNAs is independent of the cellular factors driving the biogenesis of miRNAs (e.g., Argonaute and Dicer) (Nicolas et al., 2012). While the cellular mechanism of ysRNA biogenesis is not yet fully understood, recent research indicates that the mechanism of cleavage is dependent on an intact binding site for Ro60 in the Y RNA structure and is potentially mediated by RNAse L (Billmeier et al., 2022; Donovan et al., 2017).
Notably, potential links between Y RNAs, their associated proteins, and various pathologies, including autoimmune conditions, cardiovascular and neural disease, and specific cancers, have been suggested (Cambier et al., 2017; Repetto et al., 2015; Scheckel et al., 2016; Tebaldi et al., 2018; Dr et al., 2019; Ma et al., 2022; Lunavat et al., 2015; Campo et al., 2025; Boccitto and Wolin, 2019). In addition, a number of studies have now reported significant correlations between Y RNA and ysRNA expressions and activities and viral disease (Table 1), and in light of this, this review summarizes recently reported associations of, and putative roles for, Y RNAs during viral infection.
Human papilloma virus (HPV)
In 2020, Guglas et al. reported a potential relationship between Y RNA expression and head and neck squamous cell carcinoma (HNSCC). They observed an overall decrease in hY1 expression in HNSCC tumor cells compared to non-malignant dysplastic keratinocytes, but noted increased hY1 expression in tumors from HPV-positive patients versus HPV-negative ones (Guglas et al., 2020a). In a follow up study in 2023, Guglas et al. similarly found hY3 expression was significantly higher in advanced-stage HPV-positive tumors, and more notably, that elevated hY1 levels were linked to better survival in HPV-positive patients. The study also explored Y RNA interactions with host cellular defense, finding that experimental overexpression of hY1 in HNSCC cell lines led to an upregulation of immune-related pathways, including Fc-gamma receptor signaling and phagocytosis (Guglas et al., 2020b; Guglas et al., 2023). Collectively, the observed correlations between hY1 expression and (i) distinct outcomes in HSNCC patients with and without HPV infection, and (ii) host cell immune response pathway activations, strongly support a role for Y RNAs, particularly hY1, in HPV-related oncogenesis and HNSCC pathology.
Respiratory syncytial virus (RSV)
In 2022 Pålsson et al. (2022) performed an experiment aimed at identifying host small non-coding RNAs (sncRNAs) present in bronchoalveolar lavage fluid (BALF) capable of inhibiting respiratory syncytial virus (RSV) infection. Notably, they found that two of the sncRNAs they identified corresponded to specific ∼30 nt ysRNA sequences apparently excised from full length hY4 and hY5. To assess their antiviral potential, mimics of these ysRNAs were transfected into A549 lung epithelial cells prior to infection with GFP-tagged RSV virions. Subsequent GFP quantification confirmed significant reductions in RSV infectivity in cells transfected with either ysRNA as compared to controls. Further, cells transfected with either of these ysRNAs also exhibited significantly reduced nucleolin antibody binding compared to controls, suggesting that these ysRNAs interfere with the ability of RSV to bind nucleolin, a known RSV co-receptor required for viral entry. As such, this study represents one of the most definitive characterizations of specific roles for ysRNAs in innate viral defense yet reported.
Kaposi’s sarcoma herpesvirus (KSHV)
KSHV is the etiological agent of Kaposi’s sarcoma (KS). In 2018 Ikoma et al. characterized KSHV miRNAs in plasma taken from KSHV infected individuals in Uganda and confirmed that their expressions correlated with KSHV plasma viremia. They also found exosomal small RNA profiles were highly atypical with strikingly high levels of Y RNAs (as much as 93% of the exosomal small RNAs detected) (Ikoma et al., 2018). Notably, high levels of exosomal Y RNAs was also previously observed in chronic lymphocytic leukemia (CCL) with hY4 constituting the most abundant small RNA in CCL patient plasma exosomes, and interestingly, addition of these exosomes trigger monocyte activation through Toll-like receptor (TLR) 7/8 signaling leading to enhanced tumor growth (Farahani et al., 2015). That said, Ikoma et al. similarly found hY4 to be the most abundant small RNA in exosomes isolated from plasma taken from individuals with asymptomatic malaria infection, and as TLR7/8 signaling can induce KSHV reactivation from latency, their observations support a model in which malaria infection results in increased KSHV reactivation rates in co-infected individuals and so, account for the observed association between KSHV sero-positivity and malaria in Ugandan people (Ikoma et al., 2018).
Human immunodeficiency virus type 1 (HIV-1), dengue (DENV), and measles virus
At times, dying cells, especially through necrosis, release cellular RNA complexed with riboproteins capable of binding TLRs and stimulating innate immune response via increased interferon signaling (Boccitto and Wolin, 2019). Relatedly, in 2022, (Vabret et al., 2022) observed an interaction between Y RNAs and RIG-1-like receptors (RLRs) suggesting a potential role for Y RNAs in host cell innate viral immunity. RLRs primarily function as sensors for viral RNA that trigger interferon (IFN-I and IFN-III) responses when viral RNA is detected. While RLRs are known to bind foreign RNA, their interaction with endogenous RNA remains less understood. That said, after infecting HEK293 cells with HIV-1, or with the positive single stranded dengue or measles viruses, Vabret et al. identified hY4 as significantly associating with RIG-1 in response to all three infections. Interestingly, in HIV-1-infected cells, only endogenous RNAs were found to be in complex with RIG-1, distinguishing it from dengue and measles infections. Further experiments in HIV-1-infected Jurkat T cells, found hY4 underwent increased triphosphorylation, quantified using 5′-PPPseq sequencing, and that this increased triphosphorylation was required for viral downregulation of DUSP11, a phosphatase normally charged with removing these phosphate groups. Importantly, HIV-1 infection significantly reduced DUSP11 levels in Jurkat T cells and CD4+ T cells, and DUSP11-knockout cells showed increased hY4 binding to RIG-1 compared to controls. Further, the study confirmed that this effect is dependent on the HIV vpr gene, as strains lacking vpr failed to suppress DUSP11, induce hY4 triphosphorylation, or enhance IFN-I signaling. Taken together, these findings strongly suggest that HIV-1 alters Y RNA modifications to manipulate host immune activation.
Moloney murine leukemia virus (MMLV)
Relatedly, a number of retroviruses (e.g., Rous sarcoma virus, MMLV, and HIV-1) package SRP RNA, the core RNA of host signal recognition particles, into their viral particles before release. While MLV and HIV virions contain similar numbers of SRP RNAs (∼7 and 12 respectively), MLV virions contain significantly higher levels of Y RNAs. Like MLV, HIV-1 does package at least some Y RNA, although at much lower levels than SRP RNA. In contrast, Y RNAs represent the most prevalent ncRNAs in MLV virions with mouse Y RNAs mY1 and mY3 enriched in MLV particles similar to that of SRP RNA in HIV-1 (Garcia et al., 2009).
Influenza a virus (IAV) and enterovirus 71 (EV71)
The most comprehensive characterization of the ability of a ysRNA to inhibit viral infection performed to date was conducted by Liu et al., (2019). In this study, (Liu et al., 2019) focused on hsa-miR-1975, a ysRNA excised from hY5 initially misclassified as a microRNA (Figure 1C), after finding that hsa-miR-1975 was the most upregulated small RNA in human lung epithelial cells following IAV infection. Importantly, they showed that transfecting A549 cells with an hsa-miR-1975 mimic significantly lowered viral nucleoprotein levels and titers, while transfection with an hsa-miR-1975 sponge conversely increased viral replication. Notably, the antiviral effects associated with increased levels of hsa-miR-1975 was shown to be IFN-β-dependent, as inhibition of IFN-β expression nullified the protective impact of hsa-miR-1975. This effect was absent in Vero cells, which lack IFN-β signaling, further confirming the role of hsa-miR-1975 in modulating the immune response to IAV infection. Since IAV infection induces apoptosis and Y RNAs are cleaved in a caspase-dependent manner, the researchers also examined hsa-miR-1975 levels in IAV-infected cells treated with a pan-caspase inhibitor. They found that blocking apoptosis reduced hsa-miR-1975 upregulation (and concurrent hY5 downregulation) linking its production to apoptosis initiation and also showed that hsa-miR-1975 inhibits IAV replication through directing increased interferon-β (IFN-β) expression.
A follow-up study by this group in 2021 further investigated the clinical relevance of hsa-miR-1975 in influenza patients. In this work, Liu et al. (2021) discovered that hsa-miR-1975 is packaged into exosomes by infected cells and transferred to neighboring cells, enhancing their antiviral response. They found delivery of exosomes containing hsa-miR-1975 to recipient cells significantly increased IFN-β expression and impeded IAV replication. Furthermore, they also found serum samples from influenza-infected patients were characterized by significantly higher levels of hsa-miR-1975, and additionally, hsa-miR-1975 levels, along with other clinical factors, effectively predicted respiratory failure requiring mechanical ventilation.
Relatedly, in 2010 Cui et al. identified marked upregulation of ysRNA hsa-miR-1975 in human epidermoid carcinoma (Hep2) cells infected with another single-stranded RNA virus, Enterovirus 71, and common cause of hand, foot, and mouth disease (HFMD) in infants and young children (Cui et al., 2010). Additionally of note and further supporting the potential utility of ysRNAs as biomarkers for influenza severity, a 2024 study by Ko et al. (2024) similarly identified significant ysRNA upregulations in mice infected with IAV.
SARS-CoV-2
Due to the global impact of SARS-CoV-2 in recent years, and its continued impact on human health, our laboratory recently performed a pilot study to investigate how the serum levels of various sncRNAs correlate with disease outcome in patients infected with SARS-CoV-2 (Olliff et al., 2023). Samples were stratified according to patients who were SARS-CoV-2 negative, patients who recovered from mild disease, patients with severe disease as defined by the requirement of admission into an intensive care unit, and those who died of SARS-CoV-2 infection. Small RNAs were isolated from samples taken during active infection and also following resolution of symptoms (when possible) then sequenced. Five sncRNAs were identified as significantly differential expressed between the outcome cohorts, but only one, a ysRNA excised from human hY4 (Figure 1D), was consistently expressed at reproducibly detectable levels (>100 reads per million). Notably, this ysRNA exhibited significant differential expression when sequencing data from patients who recovered from mild SARS-CoV-2 infection (12,068 average RPM) were compared to those who suffered from severe disease (1,303 RPM) (p = 0.019), those who died from SARS-CoV-2 infection (2,496 RPM) (p = 0.032), and the combined average RPM of those who suffered from severe infection/suffered fatalities (1,932 RPM) (p = 0.025). Furthermore, there was significant differential expression between the combined data of patients who were negative for SARS-CoV-2 and those who recovered from mild disease (10,666 RPM) compared to those who suffered from severe disease or who died from COVID-19 (p = 0.005) (Olliff et al., 2023).
Further supporting a potential role for ysRNAs during SARS-CoV-2 infection, a study by Nersisyan et al. (2023), published later in 2023, similarly identified significantly lower ysRNA levels in the platelets of COVID-19 patients as compared to healthy controls, studies by Driedonks et al. (2024a) and Driedonks et al. (2024b) reported significant ysRNA upregulations in cultured lung cells infected with SARS-CoV-2, and a study by Ranches et al., in 2025 found various Y RNA species as the most abundant differentially upregulated ncRNAs in SARS-CoV-2 Delta-infected normal human bronchial epithelial (NHBE) cells compared to noninfected controls (Ranches et al., 2025).
Discussion
Emerging research continues to reveal the multifaceted roles of Y RNAs and their derivatives in human pathophysiology. These sncRNAs have now been implicated in a remarkably diverse array of disease processes through distinct yet interconnected molecular mechanisms, positioning them as crucial regulators of cellular homeostasis and intercellular communication. As an example, a seminal study by Cambier et al. (2017) found that cardiosphere-derived extracellular vesicles containing Y RNA fragments exert significant cardioprotective effects following myocardial infarction. These protective mechanisms appear to operate through sophisticated modulation of fibroblast activation states and extracellular matrix remodeling processes, potentially offering novel avenues for post-infarction therapeutic intervention. Complementary work by Repetto et al. (2015) further solidified these findings by identifying distinct ysRNA signatures in patients with coronary artery disease, suggesting the potential utility of these molecules as diagnostic biomarkers or therapeutic targets in cardiovascular pathologies. Similarly, pioneering research by Scheckel et al. (2016) demonstrated that Y RNA dysregulation contributes substantially to the aberrant RNA splicing patterns characteristic of Alzheimer’s disease neuropathology. These effects appear mediated through specific interactions with heterogeneous nuclear ribonucleoproteins (hnRNPs), key regulators of neuronal RNA processing. Expanding on these observations, Tebaldi et al. (2018) found that Y RNA expression dynamics significantly influence neuronal progenitor cell differentiation pathways, implying potential roles in both neurodevelopmental disorders and neurodegenerative processes. These findings collectively suggest that Y RNAs may serve as critical nodes in the complex regulatory networks governing neural development and maintenance.
As the field continues to evolve, Y RNAs are increasingly becoming recognized not merely as cellular housekeeping elements, but as central players in the complex interplay between RNA biology and human pathophysiology. The remarkable diversity of Y RNA disease involvements likely stems from several unique structural and biochemical properties of Y RNAs. First, their ability to serve as molecular scaffolds for ribonucleoprotein complexes enables participation in diverse cellular processes. Second, functioning as competing endogenous RNAs (ceRNAs) may allow them to influence miRNA availability and activity (Ma et al., 2022). Third, their capacity to generate bioactive small RNA fragments through regulated processing creates additional layers of regulatory potential. Finally, their involvement in both intracellular signaling and intercellular communication via extracellular vesicles positions them as multimodal signaling molecules (Lunavat et al., 2015; Ikoma et al., 2018; Farahani et al., 2015; Liu et al., 2019; Olliff et al., 2023; Nersisyan et al., 2023; Dhahbi et al., 2013; van Balkom et al., 2015; Chakrabortty et al., 2015; Lovisa et al., 2020; Tong et al., 2023; Driedonks et al.). That said, despite significant recent advances, critical knowledge gaps persist in our understanding of Y RNA biology. The field currently lacks comprehensive characterization of tissue-specific Y RNA functions and detailed mechanistic explanations for their disease associations. Standardization of detection methodologies and functional assays remains an urgent priority, as highlighted by Guglas et al. (Shaw et al., 2025), given the methodological variability across current studies. Three key areas warrant focused investigation in future research: (i) the development of sophisticated Y RNA-specific knockout models to enable precise functional characterization, (ii) high-resolution structural studies to elucidate the molecular details of Y RNA-protein interactions, and (iii) comprehensive longitudinal clinical studies to correlate Y RNA expression dynamics with disease progression and treatment responses. These investigations will be essential for translating our growing understanding of Y RNA biology into clinically relevant applications, potentially yielding novel diagnostic tools and therapeutic strategies for a wide range of human diseases (Gulìa et al., 2020).
Particularly relevant to this review, despite being relatively understudied, Y RNAs and their cleavage products (ysRNAs) are rapidly emerging as important regulators of host-pathogen interactions, with growing evidence suggesting their involvement in antiviral defense mechanisms and disease prognosis (Table 1). Recent studies have begun to uncover fascinating roles for these small non-coding RNAs in modulating immune responses to viral infections, though much remains to be learned about their precise mechanisms of action. The current body of research, while limited, provides compelling justification for deeper investigation into how Y RNAs participate in cellular defense pathways and how their expression patterns might serve as clinical biomarkers. Several key findings have particularly highlighted their potential significance: (i) in influenza infection, ysRNA fragments have been shown to enhance interferon responses and correlate with disease severity (Liu et al., 2019; Liu et al., 2021; Ko et al., 2024), (ii) in HIV infections specific Y RNAs interact with innate immune sensors to amplify antiviral signaling (Vabret et al., 2022), and similarly, (iii) our group recently showed that a ysRNA excised from hY4 is significantly less abundant in the serum of patients with severe COVID-19 compared to the serum of patients with mild disease or healthy controls. Lower ysRNA levels in severe COVID-19 cases raises important questions about its role in modulating immune responses and whether its restoration could mitigate disease progression (Olliff et al., 2023). While further research is clearly needed to elucidate whether dysregulation of this hY4-excised ysRNA is a cause or consequence of severe COVID-19, our recent work suggests that higher plasma ysRNA levels may be directly involved with priming cells for an enhanced interferon response (Olliff et al., 2023). And, finally, (iv) alterations in Y RNA and ysRNA expression profiles and/or associations with antiviral immunity pathways have also been observed in DENV (Vabret et al., 2022), Measles Virus (Vabret et al., 2022), EV71 (Cui et al., 2010), KSHV (Ikoma et al., 2018), HPV (Guglas et al., 2020a; Guglas et al., 2020b), and RSV (Pålsson et al., 2022) infections, suggesting these molecules may play broad roles in host defense across diverse viral pathogens.
Also of note, Y RNAs are remarkably conserved, maintaining more than 90% sequence similarity across vertebrate species, and Y RNA orthologs have also been identified in several bacterial species as well as in Caenorhabditis elegans. That said, two of the most fundamental and evolutionarily preserved roles of Y RNAs in vertebrates is (i) participation in the initiation of chromosomal DNA replication and (ii) serving as integral components of Ro ribonucleoprotein (Ro RNP) complexes which also contain the highly conserved Ro60 protein—a chaperone that binds misfolded non-coding RNAs (Valkov and Das, 2020). Research in Deinococcus radiodurans, the first bacterium found to possess a Ro60 ortholog, demonstrated that Ro60 and Y RNAs cooperate with 3′–5′ exoribonucleases to modulate transcriptome RNA profiles during stress. In this well-characterized example, Y RNAs act as a scaffold that links Ro60 to the exoribonuclease polynucleotide phosphorylase, thereby enhancing the enzyme’s ability to degrade structured RNAs (Sim and Wolin, 2018). The highly conserved nature of both Y RNAs and Ro60, together with their functional partnership, underscores an ancient evolutionary relationship in which Y RNAs regulate Ro60’s activity in RNA quality control. As such, although the roles of Y RNAs and their processed fragments (ysRNAs) in antiviral defense remain incompletely understood, as most reported Y RNA viral associations involve RNA viruses (Table 1), it is tempting to speculate that Y RNA association with a ribonuclease complex targeting highly structured RNAs might be involved in innate antiviral immunity.
In summary, recent Y RNA discoveries not only reveal new layers of complexity in the host-virus arms race but also open promising avenues for diagnostic and therapeutic development. However, critical gaps remain in our understanding of the molecular pathways through which Y RNAs and ysRNAs exert their effects, their tissue-specific functions during infection, and how different viruses may have evolved to counteract or exploit these RNA-mediated defenses. Addressing these questions through systematic research will likely yield important insights into fundamental virological processes while potentially identifying novel targets for antiviral interventions and prognostic tools for viral diseases. The preliminary nature of current findings underscores both the challenges and opportunities in this nascent field of study, where each new discovery may significantly reshape our understanding of RNA biology in the context of infection and immunity.
Author contributions
NO: Writing – original draft. AM: Writing – review and editing. GC: Writing – review and editing. GB: Conceptualization, Funding acquisition, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Funding was provided in part by NSF 231178 (GB) and NSF 2030080 (GB) awarded by the Division of Molecular and Cellular Biosciences and also by NSF grant 2243532 (GB) awarded by the Division of Integrative Organismal Systems.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Billmeier, M., Green, D., Hall, A. E., Turnbull, C., Singh, A., Xu, P., et al. (2022). Mechanistic insights into non-coding Y RNA processing. RNA Biol. 19 (1), 468–480. doi:10.1080/15476286.2022.2057725
Boccitto, M., and Wolin, S. L. (2019). Ro60 and Y RNAs: structure, functions, and roles in autoimmunity. Crit. Rev. Biochem. Mol. Biol. 54 (2), 133–152. doi:10.1080/10409238.2019.1608902
Cambier, L., de Couto, G., Ibrahim, A., Echavez, A. K., Valle, J., Liu, W., et al. (2017). Y RNA fragment in extracellular vesicles confers cardioprotection via modulation of IL-10 expression and secretion. EMBO Mol. Med. 9 (3), 337–352. doi:10.15252/emmm.201606924
Campo, A., Aliquò, F., Velletri, T., and Campo, S. (2025). YRNAs: biosynthesis, structure, functions and involvment in cancer development. Discov. Oncol. 16 (1), 176. doi:10.1007/s12672-025-01957-x
Chakrabortty, S. K., Prakash, A., Nechooshtan, G., Hearn, S., and Gingeras, T. R. (2015). Extracellular vesicle-mediated transfer of processed and functional RNY5 RNA. RNA 21 (11), 1966–1979. doi:10.1261/rna.053629.115
Cui, L., Guo, X., Qi, Y., Qi, X., Ge, Y., Shi, Z., et al. (2010). Identification of microRNAs involved in the host response to enterovirus 71 infection by a deep sequencing approach. J. Biomed. Biotechnol. 2010, 1–8. doi:10.1155/2010/425939
Dhahbi, J. M., Spindler, S. R., Atamna, H., Boffelli, D., Mote, P., and Martin, D. I. (2013). 5'-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol. Genomics 45 (21), 990–998. doi:10.1152/physiolgenomics.00129.2013
Donovan, J., Rath, S., Kolet-Mandrikov, D., and Korennykh, A. (2017). Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23 (11), 1660–1671. doi:10.1261/rna.062000.117
Driedonks, T. A. P., and Nolte-'t Hoen, E. N. M. (2019). Circulating Y-RNAs in extracellular vesicles and ribonucleoprotein complexes; implications for the immune System. Front. Immunol. 9, 3164. doi:10.3389/fimmu.2018.03164
Driedonks, T. A. P., Ressel, S., Tran Ngoc Minh, T., Buck, A. H., and Nolte-'t Hoen, E. N. M. (2024a). Intracellular localisation and extracellular release of Y RNA and Y RNA binding proteins. J. Extracell. Biol. 3 (1), e123. doi:10.1002/jex2.123
Driedonks, T. A. P., Nyberg, L. H., Conte, A., Ma, Z., Pekosz, A., Duban, E., et al. (2024b). Viral and host small RNA transcriptome analysis of SARS-CoV-1 and SARS-CoV-2-infected human cells reveals novel viral short RNAs. Heliyon 10 (3), e24570. doi:10.1016/j.heliyon.2024.e24570
Farahani, M., Rubbi, C., Liu, L., Slupsky, J. R., and Kalakonda, N. (2015). CLL exosomes modulate the transcriptome and behaviour of recipient stromal cells and are selectively enriched in miR-202-3p. PLoS One 10 (10), e0141429. doi:10.1371/journal.pone.0141429
Garcia, E. L., Onafuwa-Nuga, A., Sim, S., King, S. R., Wolin, S. L., and Telesnitsky, A. (2009). Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J. Virol. 83, 12526–12534. doi:10.1128/JVI.01219-09
Guglas, K., Kolenda, T., Stasiak, M., Kopczyńska, M., Teresiak, A., Ibbs, M., et al. (2020a). YRNAs: new insights and potential novel approach in head and neck squamous cell carcinoma. Cells 9 (5), 1281. doi:10.3390/cells9051281
Guglas, K., Kołodziejczak, I., Kolenda, T., Kopczyńska, M., Teresiak, A., Sobocińska, J., et al. (2020b). YRNAs and YRNA-Derived fragments as new players in cancer research and their potential role in diagnostics. Int. J. Mol. Sci. 21 (16), 5682. doi:10.3390/ijms21165682
Guglas, K., Kolenda, T., Kozłowska-Masłoń, J., Severino, P., Teresiak, A., Bliźniak, R., et al. (2023). The impact of YRNAs on HNSCC and HPV infection. Biomedicines 11 (3), 681. doi:10.3390/biomedicines11030681
Gulìa, C., Signore, F., Gaffi, M., Gigli, S., Votino, R., Nucciotti, R., et al. (2020). Y RNA: an overview of their role as potential biomarkers and molecular targets in human cancers. Cancers (Basel) 12 (5), 1238. doi:10.3390/cancers12051238
Ikoma, M., Gantt, S., Casper, C., Ogata, Y., Zhang, Q., Basom, R., et al. (2018). KSHV oral shedding and plasma viremia result in significant changes in the extracellular tumorigenic miRNA expression profile in individuals infected with the malaria parasite. PLoS One 13 (2), e0192659. doi:10.1371/journal.pone.0192659
Ko, E. A., Zhou, T., and Ko, J. H. (2024). Insight into noncanonical small noncoding RNAs in Influenza A virus infection. Virus Res. 350, 199474. doi:10.1016/j.virusres.2024.199474
Kowalski, M. P., and Krude, T. (2015). Functional roles of non-coding Y RNAs. Int. J. Biochem. and Cell Biol. 66, 20–29. doi:10.1016/j.biocel.2015.07.003
Lerner, M. R., Boyle, J. A., Hardin, J. A., and Steitz, J. A. (1981). Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 211 (4480), 400–402. doi:10.1126/science.6164096
Liu, Y. M., Tseng, C. H., Chen, Y. C., Yu, W. Y., Ho, M. Y., Ho, C. Y., et al. (2019). Exosome-delivered and Y RNA-derived small RNA suppresses influenza virus replication. J. Biomed. Sci. 26 (1), 58. doi:10.1186/s12929-019-0553-6
Liu, Y. M., Chen, H. C., Chen, Y. C., Yu, W. Y., Ho, M. Y., Ho, C. Y., et al. (2021). miR-1975 serves as an indicator of clinical severity upon influenza infection. Eur. J. Clin. Microbiol. Infect. Dis. 40 (1), 141–149. doi:10.1007/s10096-020-04008-1
Lovisa, F., Di Battista, P., Gaffo, E., Damanti, C. C., Garbin, A., Gallingani, I., et al. (2020). RNY4 in circulating exosomes of patients with pediatric Anaplastic large cell lymphoma: an active player? Front. Oncol. 10, 238. doi:10.3389/fonc.2020.00238
Lunavat, T. R., Cheng, L., Kim, D. K., Bhadury, J., Jang, S. C., Lässer, C., et al. (2015). Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells--Evidence of unique microRNA cargos. RNA Biol. 12 (8), 810–823. doi:10.1080/15476286.2015.1056975
Marques, T. M., and Gama-Carvalho, M. (2022). Network approaches to Study endogenous RNA competition and its impact on tissue-specific microRNA functions. Biomolecules 12, 332. doi:10.3390/biom12020332
Nersisyan, S., Montenont, E., Loher, P., Middleton, E. A., Campbell, R., Bray, P., et al. (2023). Characterization of all small RNAs in and comparisons across cultured megakaryocytes and platelets of healthy individuals and COVID-19 patients. J. Thrombosis Haemostasis 21 (11), 3252–3267. doi:10.1016/j.jtha.2023.07.028
Nicolas, F. E., Hall, A. E., Csorba, T., Turnbull, C., and Dalmay, T. (2012). Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett. 586 (8), 1226–1230. doi:10.1016/j.febslet.2012.03.026
Olliff, N. S., Hunt, M. A., Paudel, S. S., Nguyen, K. N., Delcher, H. A., DeMeis, J. D., et al. (2023). Human YRNA 4 (hY4) plasma levels are a prognostic indicator of SARS-CoV-2 infection clinical severity. Micropubl. Biol. 2023. doi:10.17912/micropub.biology.000925
Pålsson, S. A., Sekar, V., Kutter, C., Friedländer, M. R., and Spetz, A. L. (2022). Inhibition of respiratory syncytial virus infection by small non-coding RNA fragments. Int. J. Mol. Sci. 23 (11), 5990. doi:10.3390/ijms23115990
Ranches, G., Hackl, H., Zaderer, V., Ploner, M., Posch, W., Wilflingseder, D., et al. (2025). Differentially expressed ncRNAs as key regulators in infection of human bronchial epithelial cells by the SARS-CoV-2 Delta variant. Mol. Ther. Nucleic Acids 36 (2), 102559. doi:10.1016/j.omtn.2025.102559
Repetto, E., Lichtenstein, L., Hizir, Z., Tekaya, N., Benahmed, M., Ruidavets, J. B., et al. (2015). RNY-derived small RNAs as a signature of coronary artery disease. BMC Med. 13, 259. doi:10.1186/s12916-015-0489-y
Scheckel, C., Drapeau, E., Frias, M. A., Park, C. Y., Fak, J., Zucker-Scharff, I., et al. (2016). Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. Elife 5, e10421. doi:10.7554/eLife.10421
Shaw, A., Ajit, K., Chataignier, M., and Gullerova, M. (2025). Non-coding Y RNA fragments in a complex with YBX1 modulate PARP1 residency at DNA double strand breaks. Nucleic Acids Res. 53 (11), gkaf517. doi:10.1093/nar/gkaf517
Sim, S., and Wolin, S. L. (2018). Bacterial Y RNAs: gates, tethers, and tRNA mimics. Microbiol. Spectr. 6 (4), 6.4.04. doi:10.1128/microbiolspec.RWR-0023-2018
Tebaldi, T., Zuccotti, P., Peroni, D., Köhn, M., Gasperini, L., Potrich, V., et al. (2018). HuD is a neural translation enhancer acting on mTORC1-Responsive genes and counteracted by the Y3 small non-coding RNA. Mol. Cell 71 (2), 256–270.e10. doi:10.1016/j.molcel.2018.06.032
Teunissen, S. W., Kruithof, M. J., Farris, A. D., Harley, J. B., Venrooij, W. J., and Pruijn, G. J. (2000). Conserved features of Y RNAs: a comparison of experimentally derived secondary structures. Nucleic acids Res. 28 (2), 610–619. doi:10.1093/nar/28.2.610
Tong, F., Tang, G., and Wang, X. (2023). Characteristics of human and microbiome RNA profiles in saliva. RNA Biol. 20 (1), 398–408. doi:10.1080/15476286.2023.2229596
Vabret, N., Najburg, V., Solovyov, A., Gopal, R., McClain, C., Šulc, P., et al. (2022). Y RNAs are conserved endogenous RIG-I ligands across RNA virus infection and are targeted by HIV-1. iScience 25 (7), 104599. doi:10.1016/j.isci.2022.104599
Valkov, N., and Das, S. (2020). Y RNAs: biogenesis, function and implications for the cardiovascular System. Adv. Exp. Med. Biol. 1229, 327–342. doi:10.1007/978-981-15-1671-9_20
van Balkom, B. W., Eisele, A. S., Pegtel, D. M., Bervoets, S., and Verhaar, M. C. (2015). Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 4, 26760. doi:10.3402/jev.v4.26760
Verhagen, A. P., and Pruijn, G. J. (2011). Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. BioEssays 33 (9), 674–682. doi:10.1002/bies.201100048
Wang, I., Kowalski, M. P., Langley, A. R., Rodriguez, R., Balasubramanian, S., Hsu, S. T., et al. (2014). Nucleotide contributions to the structural integrity and DNA replication initiation activity of noncoding y RNA. Biochemistry 53 (37), 5848–5863. doi:10.1021/bi500470b
Keywords: Y RNA, ysRNA, microRNA, virus, SARS-CoV-2, HIV, influenza A virus, innate immunity
Citation: Olliff NS, Mabien AA, Cole GM and Borchert GM (2025) Y RNA and Y RNA-derived ysRNA associations with viral pathogens. Front. RNA Res. 3:1679653. doi: 10.3389/frnar.2025.1679653
Received: 04 August 2025; Accepted: 08 September 2025;
Published: 19 September 2025.
Edited by:
Nikolay Shirokikh, Australian National University, AustraliaReviewed by:
Anindhya S. Das, Brown University, United StatesCopyright © 2025 Olliff, Mabien, Cole and Borchert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Glen M. Borchert, Ym9yY2hlcnRAc291dGhhbGFiYW1hLmVkdQ==
 Nathaniel S. Olliff1
Nathaniel S. Olliff1 Glen M. Borchert
Glen M. Borchert