- 1Crop Biology Resource Centre, South African Sugarcane Research Institute, Mount Edgecombe, South Africa
- 2Department of Biology, School of Life Sciences, University of KwaZulu-Natal, Westville, South Africa
- 3Unit for Environmental Sciences and Management, North-West University, Potchefstroom, South Africa
- 4BeauSci Ltd., Waterbeach, Cambridge, United Kingdom
Pre-commercialization studies on environmental biosafety of genetically modified (GM) crops are necessary to evaluate the potential for sexual hybridization with related plant species that occur in the release area. The aim of the study was a preliminary assessment of factors that may contribute to gene flow from sugarcane (Saccharum hybrids) to indigenous relatives in the sugarcane production regions of Mpumalanga and KwaZulu-Natal provinces, South Africa. In the first instance, an assessment of Saccharum wild relatives was conducted based on existing phylogenies and literature surveys. The prevalence, spatial overlap, proximity, distribution potential, and flowering times of wild relatives in sugarcane production regions based on the above, and on herbaria records and field surveys were conducted for Imperata, Sorghum, Cleistachne, and Miscanthidium species. Eleven species were selected for spatial analyses based on their presence within the sugarcane cultivation region: four species in the Saccharinae and seven in the Sorghinae. Secondly, fragments of the nuclear internal transcribed spacer (ITS) regions of the 5.8s ribosomal gene and two chloroplast genes, ribulose-bisphosphate carboxylase (rbcL), and maturase K (matK) were sequenced or assembled from short read data to confirm relatedness between Saccharum hybrids and its wild relatives. Phylogenetic analyses of the ITS cassette showed that the closest wild relative species to commercial sugarcane were Miscanthidium capense, Miscanthidium junceum, and Narenga porphyrocoma. Sorghum was found to be more distantly related to Saccharum than previously described. Based on the phylogeny described in our study, the only species to highlight in terms of evolutionary divergence times from Saccharum are those within the genus Miscanthidium, most especially M. capense, and M. junceum which are only 3 million years divergent from Saccharum. Field assessment of pollen viability of 13 commercial sugarcane cultivars using two stains, iodine potassium iodide (IKI) and triphenyl tetrazolium chloride, showed decreasing pollen viability (from 85 to 0%) from the north to the south eastern regions of the study area. Future work will include other aspects influencing gene flow such as cytological compatibility and introgression between sugarcane and Miscanthidium species.
Introduction
Commercial sugarcane (Saccharum hybrids) was thought to have arisen from an interspecific hybridization event between S. spontaneum and S. officinarum in Java in the late 1800's (Paterson et al., 2013). Recent literature, though, suggests that the heritage is more complicated, especially when considering the nuclear phyologenetic relationships (Lloyd Evans and Joshi, 2016a). The complex ancestry, the polyploid and aneuploid nature of modern sugarcane makes conventional breeding challenging (Butterfield et al., 2001). Notwithstanding these issues, in excess of 60 “N” sugarcane cultivars have been released in the South African industry since 1955, but environmental constraints affect sexual hybridization because floral induction, flowering synchronicity between selected parental germplasm and pollen fertility are problematic at sub-tropical latitudes (Brett, 1950; Horsley and Zhou, 2013). Attempts to increase genetic diversity by intergeneric crossing of commercial hybrids and members of the “Saccharum complex” have met with either limited or no success, even under controlled conditions with human intervention, and there are no reports of such hybridization in the wild (Bonnett et al., 2008; Cheavegatti-Gianotto et al., 2011; Organisation for Economic Cooperation and Development, 2013).
Cultivar improvement using genetic modification (GM) technology is being explored and a range of traits have been introduced to sugarcane (reviews by Lakshmanan et al., 2005; Brumbley et al., 2008; Meyer and Snyman, 2013). Commercial cultivation of GM sugarcane has only been approved in Indonesia (Xue et al., 2014) and more recently, Brazil1, but research of this nature is underway in most sugarcane-producing countries.
In South Africa, legislation governs the use and cultivation of GM crops [namely the Genetically Modified Organisms Act (Act 15 of 1997) and the National Environmental Management Act (Act 107 of 1998)]. One aspect of GM crop cultivation that requires assessment prior to commercial release is establishing the likelihood of lateral gene flow between related plant species. Hybridization is only possible between a crop plant and a wild relative if a number of barriers to gene flow are traversed (McGeoch et al., 2009). According to den Nijs et al. (2004), successful gene transfer (barrier crossing) requires plant populations to: (a) overlap spatially; (b) overlap temporally (flowering periods); and (c) be sufficiently close biologically that the resulting hybrids are fertile, facilitating introgression of genetic material into a new population. The probability of and extent of gene flow varies according to these limiting factors (Légère, 2005).
Gene flow from transgenic crops to wild relatives may have negative environmental effects if the hybrid plants inherit an increased capacity for invasiveness and weediness of a species (e.g., by conferring a trait such as herbicide tolerance to a specific/related active ingredient would be problematic if that was the only mechanism of eradication) (Andow and Zwahlen, 2006). Furthermore, gene flow from GM plants may be difficult to contain, demonstrated by transgene movement in rice (traits such as high protein content, disease and insect resistance and herbicide and salt tolerance), creeping bentgrass (herbicide tolerance), and oilseed rape (herbicide tolerance) (Rieger et al., 2002; Warwick et al., 2003; Chen et al., 2004; Watrud et al., 2004; Zapiola et al., 2008). This could lead to the evolution of highly competitive weeds and the degeneration of the genetic diversity in indigenous grasses.
This study was conducted to assess the likelihood of gene flow from commercial sugarcane to wild relatives in the sugar production regions of South Africa. Factors such as spatial overlap, proximity, flowering synchrony and pollen viability are prerequisites for hybridization to occur. Therefore, if close relatives occur in areas where sugarcane is cultivated, then transgenic sugarcane presents a likelihood for gene flow to these species. To assess this possibility, the objectives are as follows: (i) review the literature to identify the wild relatives of Saccharum, collate what is known about gene flow between cultivated Saccharum hybrids and wild relatives in South Africa, determine overlapping flowering times and assess pollen viability of commercial sugarcane; (ii) quantify the distribution of wild Saccharum relatives and assess the spatial overlap of their distributions with commercial sugarcane plantations; (iii) determine phylogenetic relationships within the Saccharum complex to confirm which species are most closely related to cultivated sugarcane; (iv) make an assessment of the likelihood of gene flow potential between related species and cultivated sugarcane.
Materials and Methods
Phytogeography of Saccharum Wild Relatives in South Africa
Wild relatives which diverged from Saccharum <7.3 million years ago (based on chloroplast sequence chronograms) were identified from a global phylogeny based on chloroplast genomes/regions for the Poaceae (Skendzic et al., 2007; Soreng et al., 2015; Lloyd Evans and Joshi, 2016a). Eleven species of the Sorghinae and Saccharinae subtribes of the Andropogoneae were selected for spatial analyses based on their presence within the sugarcane cultivation region of South Africa: four species that belong to Saccharinae and seven to Sorghinae (Organisation for Economic Cooperation and Development, 2013; Fish et al., 2015; Soreng et al., 2015). Grass nomenclature is in accordance with The Plant List (2013).
Herbarium specimens were sourced from 11 South African herbaria. All specimen data were captured and a gap analysis conducted for the study area to identify where insufficient information was available regarding the occurrence of wild relatives. At these sites, sugarcane field margins were examined for the target species, especially at the preferred habitats of sugarcane relatives such as disturbed and waterlogged areas. Collections were made during flowering periods, May to July, of 2016 and 2017. Field data of collected species were recorded and specimens accessioned in the A. P. Goossens Herbarium (PUC) and National Herbarium (PRE). Herbarium distribution records of the new collections were added to the master database to construct a distribution map per species with ArcGIS (student edition version 10.3, Esri, USA) to confirm their presence in sugarcane cultivation areas (Supplementary Figure 1).
Plant Material
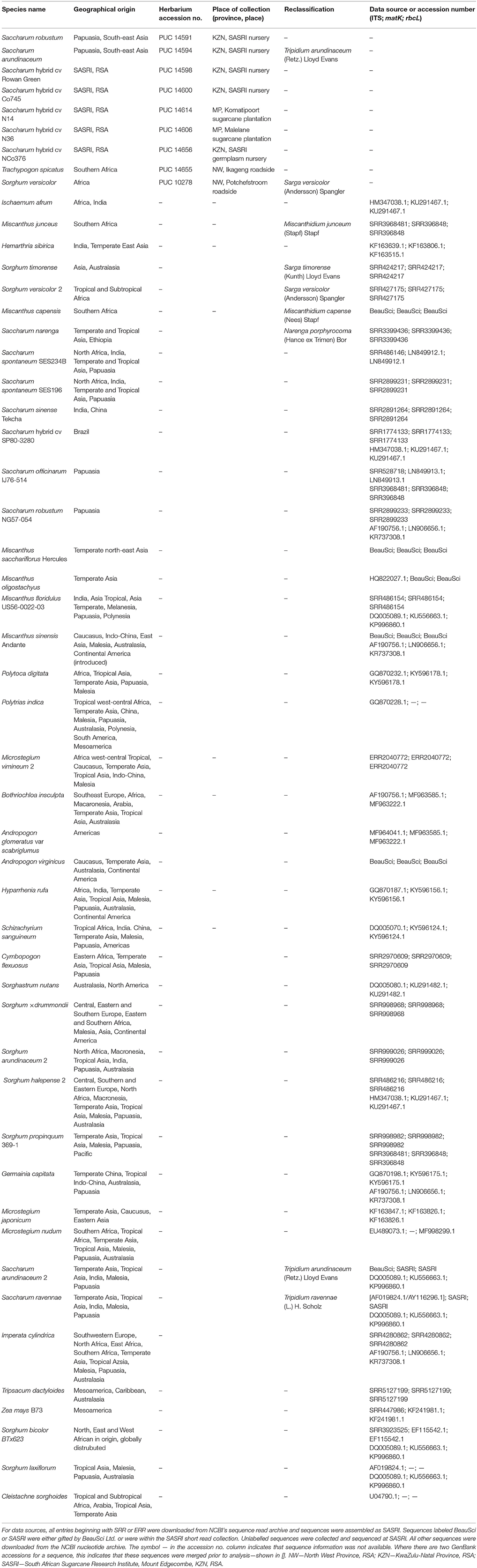
Leaf samples from Saccharum hybrid parental breeding lines were collected at SASRI, Mount Edgecombe (23 May 2016). Leaf samples from commercial sugarcane cultivars were collected from grower plantations (4–7 July 2016). Herbarium records and iSpot2 were used to pinpoint localities and habitat types where selected wild relatives of Saccharum have been collected in the past and are known to occur. Samples of plant leaf material were collected from these locations, for which plant specimens are deposited in the A.P. Goossens Herbarium (PUC) (Table 1). The leaf material was decontaminated with 70% (v/v) ethanol and stored in 50 ml plastic tubes (Thermo Scientific Group) filled with 15 g silica gel. Related species and outgroups that could not be collected in the field were sourced from GenBank genetic sequence database (Table 2).
Table 1. Herbarium accession numbers of the different flowering sugarcane cultivars tested for pollen viability and sampled for genomic DNA extraction in 2016 and 2017.
DNA Extraction, Amplification, and Sequencing
Between 0.10–0.15 g of dry plant leaf material per species was homogenized in liquid nitrogen and genomic DNA was isolated (GeneJET Plant Genomic DNA Purification kit; Thermo Fisher Scientific, USA) according to the manufacturer's protocol. The purity and concentration of the DNA was assessed (NanoDrop ND-1000 spectrophotometer; NanoDrop Technologies, Inc., Thermo Scientific Group).
DNA sequences of the internal transcribed spacer (ITS) regions of the 5.8s ribosomal gene as well as that of two chloroplast genes, ribulose-bisphosphate carboxylase (rbcL) and maturase K (matK) were used to design primers (Table 3). Amplification of the above three regions was done via Polymerase Chain Reaction (PCR) on a C1000 Thermal Cycler (BioRad, USA). The reaction mixture included 2X KAPA Taq readyMix PCR kit (1x PCR buffer, 2 U Taq DNA polymerase, 0.2 mM of each DNTP, 1.5 mM MgCl2 and stabilizers), 0.5 μM forward and reverse specific primers, 5–50 ng DNA template and nuclease-free water. For each primer set (Table 3) the initial denaturation step was at 94°C for 3 min, followed by denaturation at 94°C for 60 s. Annealing temperatures varied depending on the primer set: 50°C for 30 s (ITS and rbcL) and 48°C for 40 s for matK.; the extension step was at 72°C for 30 s (ITS and rbcL) and 60 s for matK. There were 35 thermocycles for ITS and rbcL and 40 for matK. The final extension step was at 72°C for 10 min. PCR products were visualized on a 1% (w/v) agarose gel and cleaned-up (GeneJET PCR purification kit; Thermo Fisher Scientific, USA).

Table 3. The primers used for the amplification and sequencing of the internal transcribed spacer (ITS), ribulose-bisphosphate carboxylase (rbcL), and maturase K (matK) gene fragments used as the basis for the phylogenetic analyses.
Sequencing reactions were performed with the same primers as those used for PCR using the BigDye Terminator V1.3 cycle sequencing kit (Applied Biosystems, USA). This was followed by fluorescence-based DNA analysis using capillary electrophoresis technology on the Applied Biosystems 3500 Genetic Analyser. Sequences were analyzed and trimmed using Sequencing Analysis V5.3.1 (Applied Biosystems).
Sequence Assembly
The 5.8s genomic ITS cassette along with the chloroplastic matK and rbcL genes were chosen for phylogenetic analysis. In those cases where no ITS, matK, or rbcL sequences could be found in GenBank, sequences were assembled from short read data (either mined from NCBI's SRA archive3 or made available through on-going collaborations) (Table 2) using a bait-and-assemble assembly method described previously (Lloyd Evans and Joshi, 2016b). Third party data assembled for this study are noted in Table 2 and the assemblies are provided as Supplementary File 1.
Sequence Alignments
The ITS cassette (18s rRNA partial, ITS1 complete, 5.8s rRNA, ITS2 complete, 28s rRNA partial) region was aligned as described previously (Martin et al., 2017). Briefly, DNA sequences (Table 2) were aligned with SATÉ (Liu et al., 2009) using MAFFT (Katoh and Standley, 2013) as the aligner, MUSCLE (Edgar, 2004) as the sub-alignment joiner and RAxML as the tree estimator. The final RAxML tree was used as input for PRANK (Löytynoja et al., 2012) an indel-aware alignment optimizer. PRANK was run for 5 generations, using RAxML (identifying the most likely tree from 100 samples) for Maximum Likelihood (ML) tree estimation until both the alignment and the tree topology stabilized. The chloroplastic matK and rbcL sequences were aligned with SATÉ.
Long-branch attraction and incomplete sampling (Philippe et al., 2017) can be major confounding effects in phylogenetic inference. In an attempt to minimize these effects, at least two exemplars for each sequence were included in the initial alignment and as many species and genera were sampled as possible. To test for long-branch attraction a custom PERL script was written. This script removed one sequence at a time from the final alignment. The reduced alignment was analyzed with RAxML where the most likely tree was identified from 100 random replicates. After the analysis, all trees were compared and where the initial reference tree and the resampled tree differed significantly the deleted sequence was labeled as responsible for long-branch effects and was removed from all subsequent analyses. The sequences remaining after this test were re-aligned using SATÉ and PRANK, as described above. These sequences yielded the final alignment. The final ITS alignment and phylogeny along with the matK alignment and phylogeny and the rbcL alignment and phylogeny were deposited in TreeBase4.
Wherever possible, the entire ITS cassette was used. However, where no alternate data was available, the shorter assemblies from existing sequence data were integrated into the alignment and padded with Ns.
Partition Analyses
The ITS cassette was divided into 18s rRNA, ITS1, 5.8s rRNA, ITS2, and 28s rRNA regions, whilst the entire matK and rbcL genes were analyzed as a single partition. Best-fit evolutionary models were determined using jModelTest2 (Darriba et al., 2012) under the AICc criterion. The best fit models were found to be: 18s RNA: TVM + G; ITS1: TPM3uf + G; 5.8s rRNA: JC + G; ITS2: GTR + G; 28s rRNA: GTR + G; matK: TVM + G; and rbcL: HKY + I.
Phylogenetic Analyses
Phylogenetic analyses were run for the ITS cassette along with separate analyses for matK and rbcL. Non-parametric bootstrap tests (using the above partitioning schema) and SH-aLRT analyses were run with IQ-Tree (Nguyen et al., 2015). Neighbor-Joining analyses were run with APE (Paradis et al., 2004). Bayesian Inference (BI; again using the above partitioning schema) was run with MrBayes (Ronquist and Huelsenbeck, 2003). IQ-Tree analyses were run for 2,000 replicates. MrBayes analyses were run with 50,000,000 generations with sampling every 100th tree. Two independent MrBayes analyses, each of two independent runs, were conducted. To avoid any potential over-partitioning of the data, the posterior distributions and associated parameter variables were monitored for each partition using Tracer v 1.6 (Rambaut et al., 2017). High variance and low effective sample sizes were used as signatures of over-sampling. Burn-in was determined by topological convergence and was judged to be sufficient when the average standard deviation of split frequencies was <0.002 along with the use of the Cumulative and Compare functions of AWTY (Nylander et al., 2008). The first 30% of sampled trees were discarded as burn-in.
Phylogenetic analyses (ML and BI) were summarized with Sumtrees (Sukumaran and Holder, 2010) prior to drawing with FigTree (2017) and finishing with Adobe Illustrator to generate publication-quality figures. The ITS only, matK, only and rbcL only tree topologies were deposited in TreeBase4.
Chronogram Generation With r8s
The application r8s (Sanderson, 2003) was employed for chronogram generation. An optimal tree topology was generated and was used for analysis. Parameters were adjusted for ML branch lengths on all trees and divergence timings were estimated with a smoothing factor of 100, the Penalized Likelihood method using the Truncated Newton optimization framework with analytical gradients generated by r8s. To generate 95% confidence intervals on branch times, the non-parametric bootstrap trees generated by IQ-Tree were used as input to r8s. All trees were concatenated into a single nexus file using a custom PERL script and an r8s block was appended so that r8s could be executed over all trees with parameters as defined above. The profile command of r8s was employed to individually summarize the distribution of ages at all given nodes of the tree (employing a custom PERL wrapper). Priors for the main nodes were defined as follows: root, fixed age of 13.8 million years ago, Tripsacum–Germainia node, fixed age of 9.2 million years ago (Estep et al., 2014), Sarga–Miscanthidium node, minimum age of 7.4 million years ago, Miscanthus–Miscanthidium node fixed age 3.4 million years ago, S. spontaneum–S. sinense node, minimum age of 1.4 million years ago (Lloyd Evans and Joshi, 2016a). All other nodes were unconstrained.
Pollen Viability Testing
Pollen samples from commercial sugarcane cultivars were collected during the flowering season (July 2016 and 2017) from nine different sites in South Africa, two in Mpumalanga and seven in KwaZulu-Natal (Figure 1). Sites 1–5 are situated in the irrigated region while sites 6–9 are rain-fed. Fresh pollen was collected from anthers in dehiscence, from three separate inflorescences per cultivar per site. Inflorescence collection was between 6.00 and 8.30 h and viability tests conducted in the field immediately thereafter (Amaral et al., 2013).
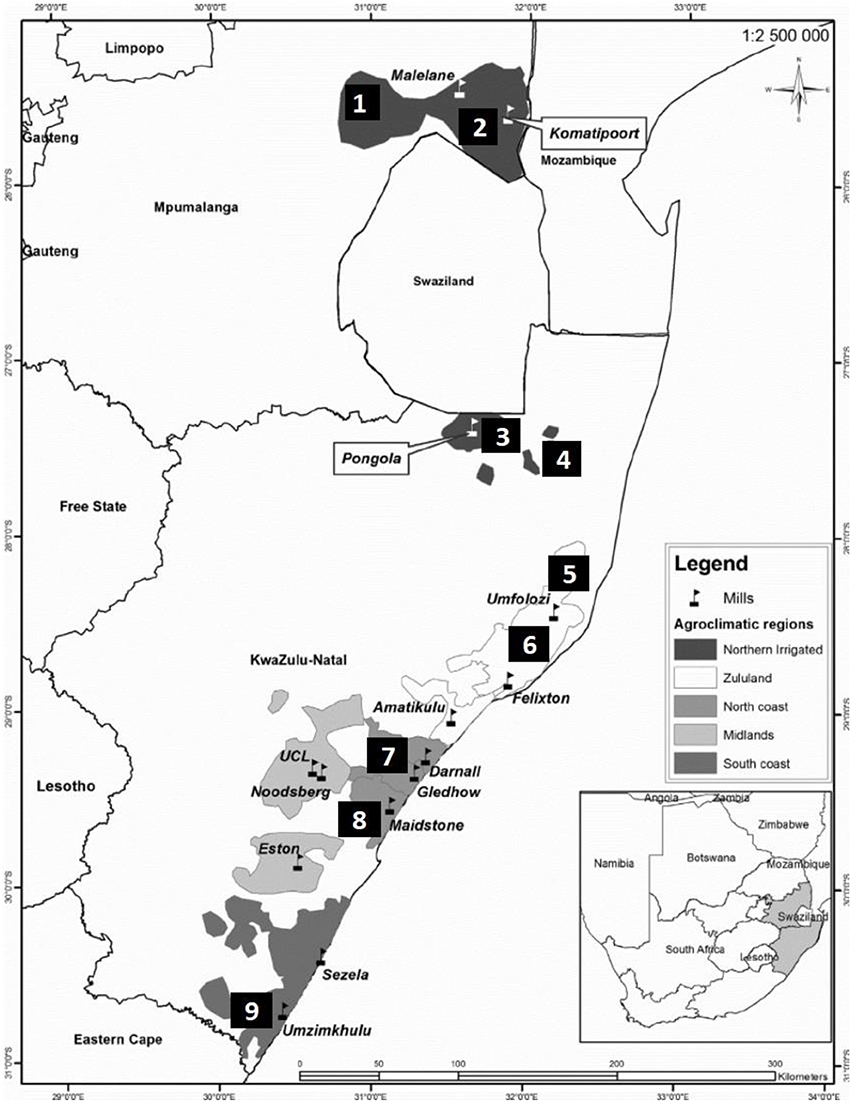
Figure 1. Sugarcane production regions and locations of sugar mills in the Mpumalanga and KwaZulu-Natal provinces of South Africa. Sites for pollen collection were as follows: 1: Malelane; 2: Komatipoort (Mpumalanga); 3: Pongola; 4: Jozini; 5: Mtubatuba; 6: Empangeni; 7: Umhlali; 8: Mount Edgecombe; and 9: Port Shepstone (KwaZulu-Natal).
Two stains were used to estimate pollen viability: 2,3,5-triphenyl tetrazolium chloride (TTC) (Soares et al., 2013) and iodine potassium iodide (IKI) (Huang et al., 2004). Pollen grains were stained with IKI [1% (w/v) iodine and 2% (w/v) potassium iodide in distilled water] for 5 min, while those stained with TTC [1% (w/v) TTC and 5% (w/v) sucrose in distilled water] were examined after 15 min of incubation in direct sunlight. Viewing was under a compound microscope (Model 11, Wild, Heerbrugg Switzerland) at 100 × magnification and counting was aided using a grid stuck to the underside of each glass slide. A random count of a minimum of 100–150 pollen grains was performed for each cultivar replicate, and the percentage viability was determined as the ratio of viable pollen grains (intense dark color for IKI and deep pink for TTC) divided by the total number of grains.
An average from three pollen counts per cultivar per locality was used for calculating percentage pollen viability. All statistical analyses were carried out using Statistica (version 13; Dell Inc., USA). The Kolmogorov-Smirnoff and Lilliefors tests for normality showed that the data did not meet the assumptions of normality in the distribution of all variables. Therefore the Kruskal-Wallis analysis of variance (ANOVA; non-parametric statistics) for comparing multiple independent groups was used to determine differences between determinants measured.
Environmental data including relative humidity, soil water content at 100 mm depth, minimum and maximum temperatures were extracted from the SASRI weather web5. Automatic weather stations were situated at each of the sampling sites. Data was extracted from the first of May 2016 and 2017 up to the day at which sampling took place for each of the sites. Mean values were used for each environmental variable at each site. Day length data with the same time resolution and period was obtained online6. The non-parametric Spearman rank correlation coefficient was calculated as a measure of correlation between all possible pairs of variables and significance was tested at the 0.05 level.
Desk-Top Study of Hybridization
Prominent literature was consulted to assess gene flow potential. Printed evidence of reproductive compatibility and the formation of hybrids between commercial sugarcane with target related species were used to assess the likelihood of hybridization. The numbers of publications which reported hybridization were recorded. Successes were scored if the publications reported formation of hybrid progeny (FitzJohn et al., 2007; McGeoch et al., 2009; Organisation for Economic Cooperation and Development, 2013) and ranked accordingly. In cases where literature recorded hybridization evidence between Saccharum hybrids and wild relatives, the following approaches were undertaken: (i) if target species were reported to hybridize with Saccharum hybrids, the number of publications and successes were recorded and scored 1 per event; (ii) if species not found in South Africa hybridized with Saccharum hybrids, and the genus is present in the sugar production area, the species from such genera were treated as reproductively compatible with commercial sugarcane and the number of publications and successes recorded and scored 0.5 per event. The wild relative-Saccharum crosses with most hybrids ranked the highest and species with fewer hybrids were ranked lower.
Flowering Times
Flowering times were assessed using literature, herbarium specimens and collections made during field surveys. Saccharum hybrids flower from March to August in South Africa (Sithole and Singels, 2013; Zhou, 2013). Plant specimens with inflorescences, dates of collections and occurrence in the study area were used to analyse flowering times in addition to collections sampled during the study. The overlapping percentages between the flowering time of Saccharum hybrids and each wild relative was calculated by dividing the number of overlapping months with the total number of months of sugarcane flowering. The wild relatives with more overlapping months were ranked the highest and species with less overlap were ranked lower.
Spatial Assessment
The qualitative assessment to determine the likelihood of wild relatives co-occurring with cultivated sugarcane, which may enhance gene flow potential, was based on the following factors: prevalence, spatial overlap, proximity, distribution potential, gene flow potential, and flowering times (Ellstrand et al., 1999; Chapman and Burke, 2006; Schmidt and Bothma, 2006; Tesso et al., 2008; McGeoch et al., 2009; Andriessen, 2015). All target species were assessed and ranked per factor, whereby species with highest rank was scored 11 and species with lowest rank was scored 1. In the cases where no information was available for a species, the species could not be ranked and was scored 0 (no evidence equates to no ranking). It would be inaccurate to rank species without data, as it would inflate the likelihood scores for the areas where these species were found.
Sugarcane production areas for Limpopo, Mpumalanga and KwaZulu-Natal were obtained from the 2015 National Land Cover dataset. These areas were then overlaid with a grid of quarter-degree squares (QDS) using ArcGIS to provide 113 mapping units for the spatial assessment (Robertson and Barker, 2006). Some of these QDS overlap with Mozambique and Swaziland, but no data was available for these areas. It should be noted that wild relatives may be present in those jurisdictions and did not form part of this study.
The presence of wild relatives in QDS of sugarcane cultivation areas were used to calculate their prevalence, i.e., how common these species are in the study area. The number of individuals per species per QDS within the sugarcane cultivation area was determined. The proportion of individuals per species within QDS was calculated. The same procedure was followed for QDS bordering sugarcane cultivation areas. These proportions were summed to determine the proportional prevalence of each species in the study area. These prevalence values were then sorted from highest to lowest proportion of individuals per species within and bordering sugarcane QDS and scored.
Spatial overlap is the notion of similarity in distribution patterns (or shared occurrences). It was calculated for each species by dividing the number of QDS that overlap with sugarcane cultivation areas with the total number of QDS for sugarcane cultivation areas. This derived a percentage of overlap per species. Species were ranked from highest to lowest based on overlap percentage, with the highest rank scoring 11 and lowest rank scoring 1.
Pollen of graminoids can travel up to 700 m from the donor plant (Schmidt and Bothma, 2006). This was set as the cut-off for proximity measures both during field work and extracting data from herbarium specimens. The herbarium record database was used to construct a table of habitat notes per species and the presence or absence of wild relatives in the vicinity of sugarcane fields were noted. These records were combined with confirmations from the literature and field surveys. Species with more occurrences within the 700 m zone (high proximity) were ranked higher than species with few or no records in sugarcane fields and margins.
Weedy grasses are often spread by different modes of transport (Milton, 2004). Transport networks therefore gives an indication of the potential for weedy relatives of sugarcane to spread, with denser networks implying higher chances for migrations. Road and railway networks were used to calculate the spatial distribution potential of wild relatives across the study area. For each species the number of railway lines and roads per QDS were counted respectively. Totals of QDS containing railways and roads per species were summed. Higher totals were considered indicative of a wild relative's ability to disperse and ranked as highest likelihood for the species to spread to sugarcane fields (Knispel et al., 2008).
Likelihood Scores
Likelihood scores were calculated per species to determine which Saccharum relatives might present a higher likelihood for gene flow with sugarcane based on relatedness, flowering time and spatial assessment. Factors were weighted equally for relatedness and spatial assessments (Butler et al., 2007). Relatedness was calculated from the phylogenetic classification and hybridization events, and spatial assessment involved prevalence, spatial overlap, proximity, and distribution potential. Thereafter, spatial, temporal (flowering time) and relatedness assessments were weighted 1:1:2 to come up with a final likelihood score. This weighting was based on the assumption that gene flow and relatedness are not correlated due to reproductive barriers such as flowering time (Panova et al., 2006), and that gene flow likelihood is evenly dependent on temporal and spatial assessment factors. Relatedness is weighted more as it becomes the determining factor for gene flow when prevalence, spatial overlap, proximity, distribution potential or flowering time provide the required compatibility for pollen from one species to reach the stigma of another species.
Likelihood maps indicating various levels of potential for gene flow to occur between Saccharum hybrids and wild relatives within sugarcane production areas of eastern South Africa was constructed based on the factor scores per species and summed per grid. The following classes were used for assessing the likelihood for gene flow: Sorghastrum nudipes scored 6 and there was no sugarcane QDS containing only this wild relative species. QDS with sugarcane plantations without wild relatives (0–12); sugarcane QDS plantations with wild relatives: very low (13–43); low (44–86); high (87–129); very high (130–172).
Results
Assessing Hybridization Potential From the Literature
A literature review of hybridization events between cultivated sugarcane and its relatives, revealed 39 hybridization incidents were reported in 23 different studies dating from 1935 to 2014 (reviews by Bourne, 1935; Gao et al., 2014). From these, there were only three claims of spontaneous hybridization (Parthasarathy, 1948; Ellstrand et al., 1999), with the remaining crosses requiring human intervention in artificially controlled conditions using experimental procedures that maximized flowering, pollination and seedling survival. Crosses were performed to integrate the beneficial traits of one species to another to enhance agronomic traits such as growth, ratoonability and biomass accumulation (Brett, 1950; Piperidis et al., 2000; Aitken et al., 2007; Gao et al., 2014).
The genus previously known as Erianthus (now divided into Tripidium and Saccharum) was utilized in 18 of the artificial man-made crosses, predominantly with Saccharum arundinaceum (synonym Erianthus arundinaceus, Tripidium arundinaceum). Similarly, the number of crosses made with cultivated sugarcane was mainly with the Saccharum genus (10 crosses) and with S. arundinaceum (4 crosses). Other genera which have been crossed with sugarcane include Bambusa, Imperata, Miscanthidium, Sorghum, and Zea. Of the 18 species that have been involved in hydridization with sugarcane, seven occur in South Africa and comprise 30.77% of the total hybridization events. The highest number of seedling survival in cultivation was 1,371, resulting from Saccharum hybrids × Sorghum bicolor (L.) Moench, representing a 9.7% recovery rate from 14,141 total seedlings produced from the crosses (Hodnett et al., 2010). The lowest seedling survival was from a cross involving Zea mays L., where only one from more than 1,000 seedlings survived (Bonnett et al., 2008). One of the reported crosses involving S. bicolor failed with no true seedlings obtained (Bourne, 1935). With the exclusion of the former attempt, 48.72% studies used molecular markers to verify the presence of the maternal and paternal alleles from putative hybrids, whereas the remaining crosses (51.38%) relied on visual inspection of inherited morphological characteristics against those of parent lines as well as chromosome counts (Khanyi, 2018).
Imperata cylindrica, Sorghum arundinaceum, S. × drummondii, and S. halepense were the only species that were found to be reproductively compatible with Saccharum species based on assessed literature (Table 4). Miscanthidium capense and Miscanthidium junceum were not part of any species-specific hybridization studies, but were scored as compatible reproductive species based on the literature reporting on other species of the genus hybridizing with Saccharum species (Table 4). Miscanthidium hybridization is especially documented in the literature (17 publications) of which six reported successes. Hybridization potential between Miscanthidium and Saccharum ranked highest, I. cylindrica was reported in five publications with one success and S. halepense was recorded in two publications with one success (Table 4). There were considerably more publications on other Sorghum species hybridizing with Saccharum species, which was not included in the analyses due to uncertainty regarding the generic divisions within the Sorghum complex.
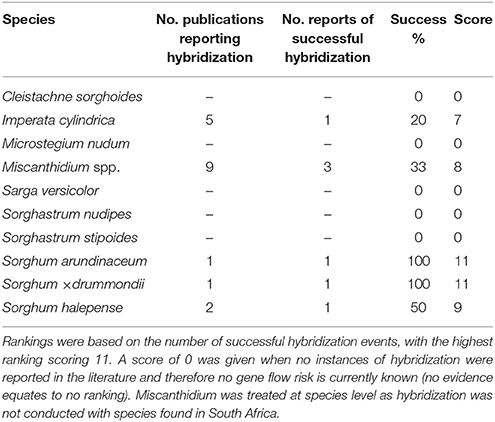
Table 4. Summary of gene flow reports between Saccharum hybrids and wild relatives from the literature for genera present in the sugarcane cultivation areas.
Occurrence of Andropogoneae in Sugarcane Cultivation Areas
A total of 815 herbarium specimens of 11 Saccharum wild relative species were sourced from 11 herbaria. These records were supplemented by 34 observations of Saccharum wild relatives during field visits to sugarcane cultivation areas in South Africa. All 11 wild relatives of the Andropogoneae have been recorded from sugarcane cultivation areas. Six species occurred throughout the sugar cultivation region, but M. capense (previously Miscanthus capensis), Sorghum × drummondii, and Sorghastrum stipoides were restricted to the southern parts, and Cleistachne sorghoides, and S. nudipes to the northern parts of the cultivation area.
Pollen Viability of Commercial Sugarcane Cultivars
A total of 11 sugarcane cultivars were tested for pollen viability during 2016 from six sites in the study area. Pollen viability tests during 2017 included two additional cultivars, N39 and N58, from site 9. No significant difference in pollen viability using two stains, IKI (40.5%) and TCC (38.1%), was observed when comparing 42 individual counts (Kruskal-Wallis ANOVA; p = 0.622), therefore results presented are those obtained using the TTC stain for 2016 and 2017 (Figures 2A,B, respectively).
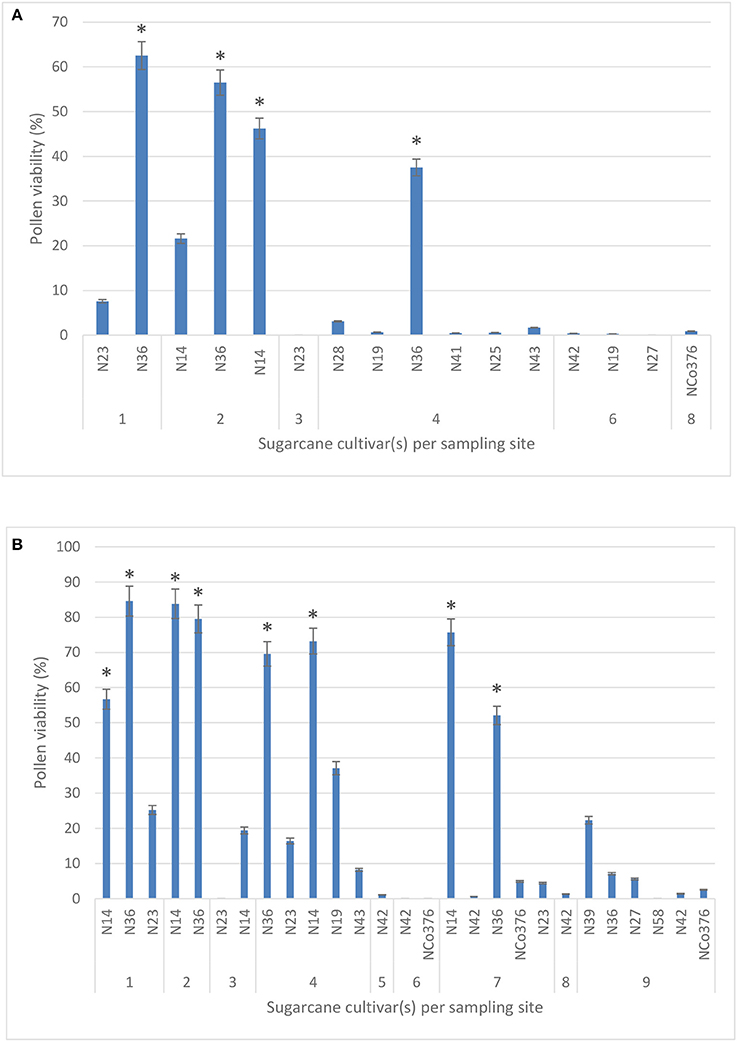
Figure 2. Pollen viability (%) of 11 sugarcane cultivars measured across six study sites using the 2,3,5-triphenyl tetrazolium chloride stain in 2016 (A) and 2017 (B). Sites −1: Malelane; 2: Komatipoort (Mpumalanga); 3: Pongola; 4: Jozini; 5: Mtubatuba; 6: Empangeni; 7: Umhlali; 8: Mount Edgecombe; and 9: Port Shepstone (KwaZulu-Natal). Values are means ± SE (n = 3). Significant differences are indicated by a * (ANOVA).
For both years, 2016 and 2017, the highest mean percentage viability was observed in cultivar N36 (62.5 and 84.6%, respectively), followed by N14 (46.2 and 83.8%, respectively) in the northern irrigated regions of Mpumalanga. Pollen from all the other cultivars (N19, N23, N25, N27, N28, N41, N42, N43, and NCo376) during the same year had lower mean percentages of viability ranging from 0 to 7.6%, while pollen from N23, N42, N58, and NCo376 was not viable in 2017. In 2017, pollen viability decreased from 84.6% in the northern irrigated regions (site 1) to 0% in the southern rain-fed coastal regions of the study area (site 9) (Figure 2), likely due to less favorable environmental conditions. None of the sites had optimal conditions required for flowering (reviewed by Cheavegatti-Gianotto et al., 2011; Organisation for Economic Cooperation and Development, 2013), but percentage pollen viability had a significant positive correlation with both mean maximum temperature (r = 0.6) and day length (r = 0.5), and a significant negative correlation with soil water content (r = −0.4) (results not shown). It must be noted that different cultivars were planted at the sampling sites.
Flowering Times
Information sourced from herbarium labels and field surveys highlighted that I. cylindrica and S. arundinaceum flower throughout the year, suggesting a 100% flowering synchrony with Saccharum hybrids (Table 5). Miscanthidium capense has an 83% overlap in flowering time with Saccharum hybrids. More than 66% of flowering synchrony was further depicted for Microstegium nudum, M. junceum, S. × drummondii, and S. halepense (Table 5).
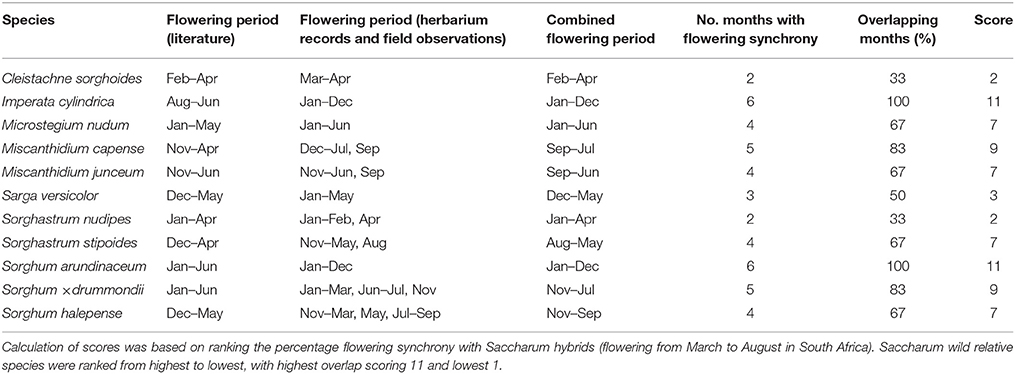
Table 5. Flowering times of Saccharum wild relatives (based on literature, herbarium specimens, and field observations) in sugarcane cultivation areas.
Determining Genetic Relatedness Using Phylogenetic Analyses
The initial experimental design was based on chloroplast phylogenies. However, during the course of the study, the paper of Folk et al. (2017) highlighted the importance of ancient reticulate evolution and parallel organellar capture in plant evolution. As a result of that paper, we performed an ITS-based phylogeny to check for reticulate evolution in the Andropogoneae. The overall ITS cassette phylogeny (Figure 3) is consistent with previous genomic studies of the Andropogoneae (Estep et al., 2014; Welker et al., 2015). However, we have increased resolution of the core Saccharinae and from our analyses, Saccharum sensu stricto (Saccharum spontaneum and its sister group) is sister to Miscanthidium and Narenga with good support. This crown group is in turn sister to Miscanthus (with moderate support). The entire grouping is, in turn, sister to Sarga (with moderate support).
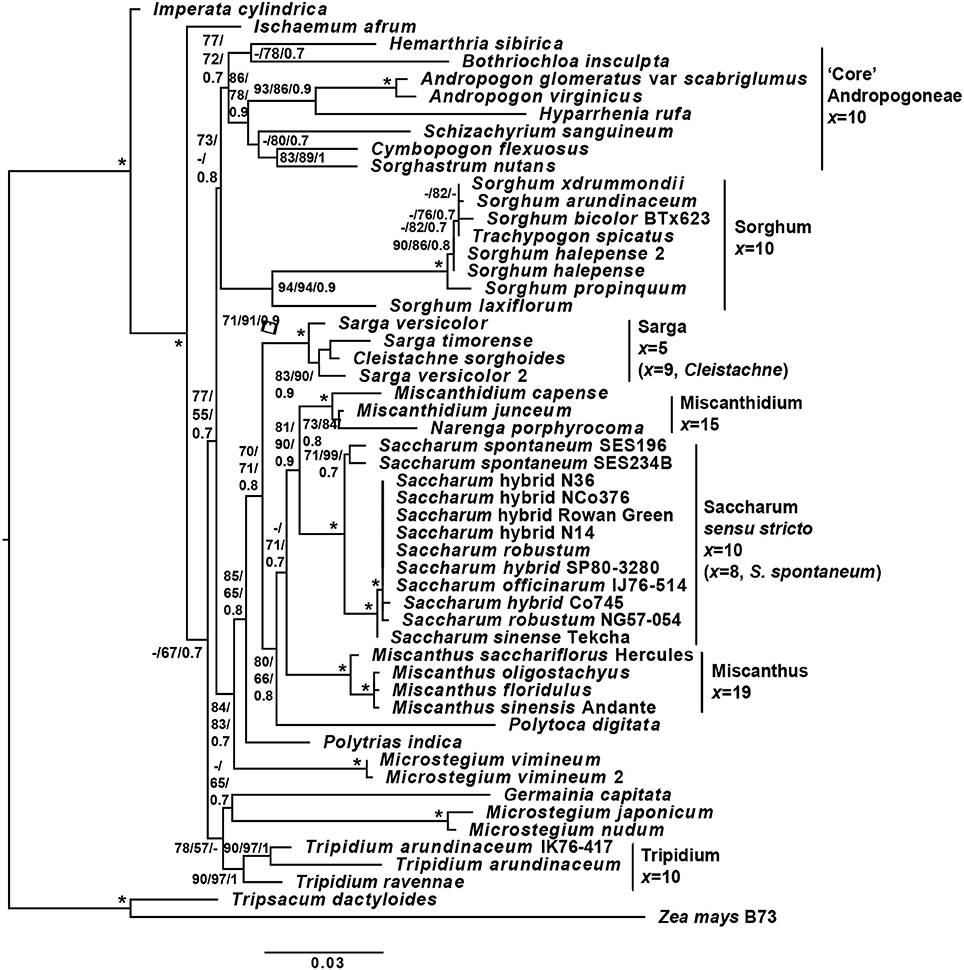
Figure 3. Phylogeny of sugarcane and related genera, based on the ITS cassette. A phylogeny of Saccharum, Sorghum and related genera based on the ITS (18s rRNA partial, ITS1 complete, 5.8s rRNA complete, ITS2 complete and 28s rRNA partial) genomic cassette. Tree terminals are the species name and cultivar or accession, where appropriate. Numbers at nodes represent SH-aLRT/non-parametric bootstrap/Bayesian inference support values. Bars to the right of the tree represent major clades, with associated base or monoploid (x) chromosome numbers. Branch lengths (scale on the bottom) correspond to the expected numbers of substitutions per sides. Monoploid chromosome numbers are derived from: Sorghum and Sarga —Gu et al. (1984); Miscanthus—Adati (1958); Miscanthidium —Strydom et al. (2000); Saccharum spontaneum —Ha et al. (1999); Saccharum officinarum —Li et al. (1959); Tripidium—Jagathesan and Devi (1969); and Cleistachne—Celarier (1958). The code *represents complete support for a node (100% SH-aLRT, 100% non-parametric boostrap and Bayesian inference of 1), whilst—represents support that is below the threshold (65% for SH-aLRT, 50% for non-parametric bootstrap and 0.7 for Bayesian inference). Within Saccharum sensu stricto, between the sister relationship of Saccharum robustum NG57-054, Saccharum hybrid cv Co745 and Saccharum officinarum IJ76-514 with the remaining species there was insufficient sequence divergence within the ITS cassette to yield any meaningful branch supports between the species. The Tripsacinae (Tripsacum dactyoides and Zea mays) were employed as an outgroup.
In common with the findings of Hodkinson et al. (2002) we also see Polytoca digitata within this grouping. Microstegium is clearly not monophyletic and we place Microstegium vimineum (with good support) as an outgroup to the entire clade that might be described as the “Saccharinae.” The core Andropogoneae is sister to the Saccharinae and Sorghum is placed as sister to the core Andropogoneae, although with only moderate support (73% SH-aLRT and 0.8 BI). Though the support for the placement of Sorghum is not strong, all independent tree topologies (SH-aLRT, maximum likelihood and Bayesian inference) agree on the topology and our placement of Sorghum as sister to the core Andropogoneae is consistent with the work of Hawkins et al. (2015) who analyzed multiple genes. This confirms the presence of reticulate evolution in the origins of Andropogoneae and casts doubt on many conclusions determined from chloroplast only datasets.
Of the two chloroplastic genes chosen for this study, matK provided only a relatively weak phylogenetic signal with over 50% of sequences undetermined and rbcL provided no phylogenetic signal (data submitted to TreeBase). Both chloroplastic genes failed IQ-Tree statistical testing for phylogenetic signal. Moreover, as the chloroplastic signal for many of the genera (particularly Imperata and Sorghum) differ (compare: Estep et al., 2014; Hawkins et al., 2015 and Burke et al., 2016) combining genomic (ITS) and chloroplastic (matK and rbcL) data would be detrimental to the overall topology of the phylogeny, particularly as genomic data is currently considered to present the “true” evolutionary signal (Estep et al., 2014).
The Maximum Likelihood phylogeny was converted into a chronogram (Figure 4) using r8s (Sanderson, 2003) with 95% branch confidence values determined by re-analyzing the non-parametric bootstrap tree set generated by IQ-Tree. Broadly, timings are consistent with previous work (Estep et al., 2014; Lloyd Evans and Joshi, 2016a) with only the genera Miscanthus and Miscanthidium lying within the 3.4 million year window where wild hybridization is possible as determined by Lloyd Evans and Joshi (2016a) when analyzing wild (i.e., not human mediated) hybridization within the Andropogoneae, specifically the Saccharinae. As it is placed within Sarga, C. sorghoides is the only other South African genus (apart from Miscanthidium) that lies within the 7.4 million year window chosen as a divergence cut-off for this project.
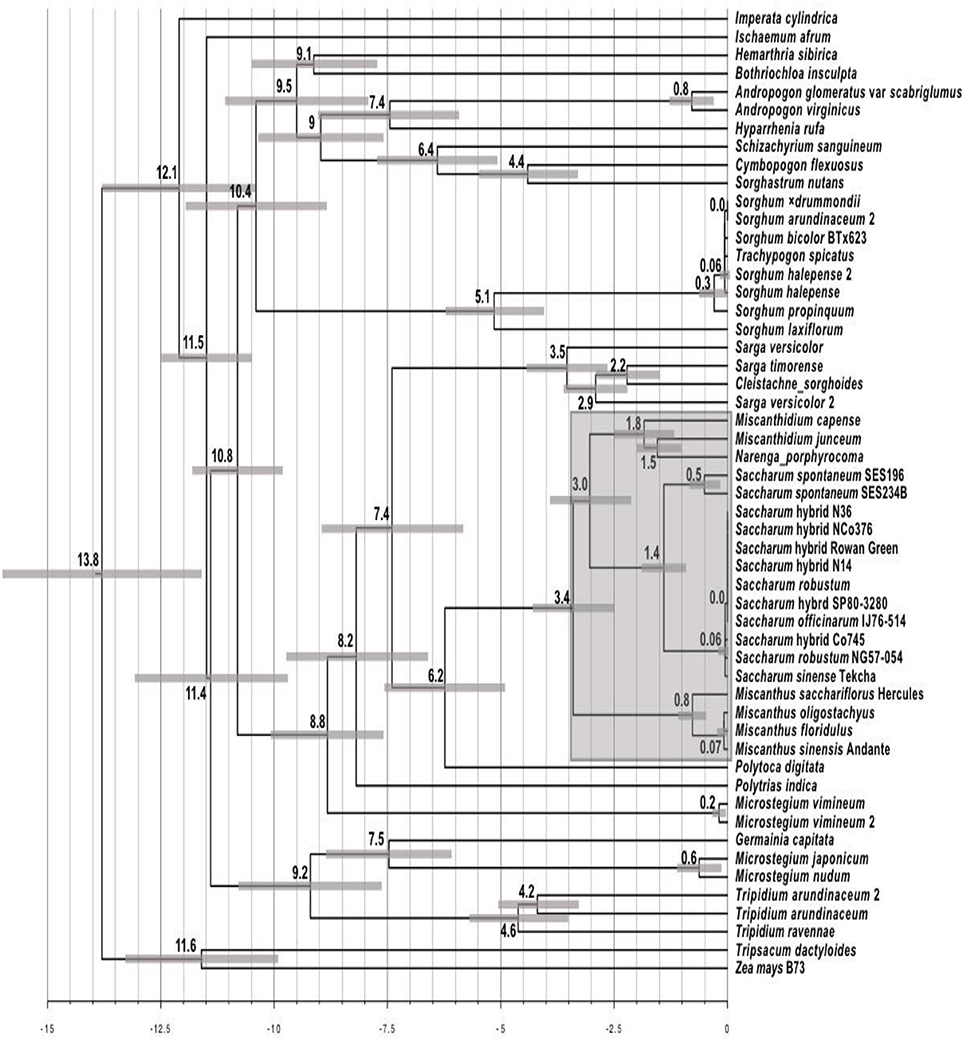
Figure 4. Chronogram derived from the alignment of Andropogoneae ITS cassette sequences. The chronogram was generated with r8s from the Maximum Likelihood ITS phylogeny from Figure 3. The scale at the bottom represents millions of years before present. Numbers at nodes represent the age of that node as millions of years before present. Scale bars at nodes represent the central 95% of the age distribution (i.e., 95% confidence interval) as determined by bootstrap resampling. The shaded region centered on Saccharum represents the 3.4 million year window in which wild hybridizations between Saccharum and other genera is possible.
Spatial Assessment Within the Sugarcane Cultivation Region
Imperata cylindrica, S. arundinaceum, and M. capense showed the highest prevalence within sugarcane cultivation areas (Table 6). Three species from Sorghinae, namely C. sorghoides, S. nudipes, and Sorghum × drummondii showed low prevalence within sugarcane QDS (Table 6). The highest spatial overlap of wild relatives with QDS containing sugarcane plantations revealed a similar outcome to the prevalence rankings (Table 7). In both cases, i.e., prevalence and spatial overlap, the highest and lowest score values differed substantially. I. cylindrica showed the highest likelihood for spatial congruence with sugarcane and S. nudipes the least.
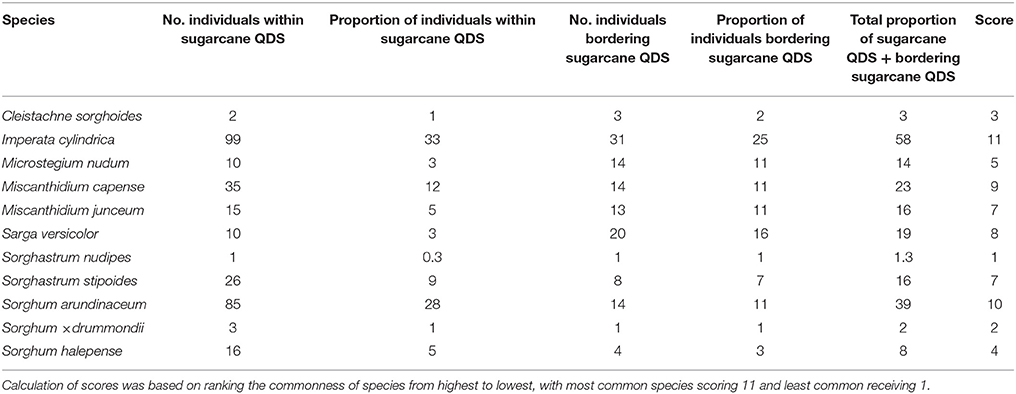
Table 6. Prevalence or commonness of individuals (based on herbarium specimens) of Saccharum wild relatives in sugarcane cultivation areas.
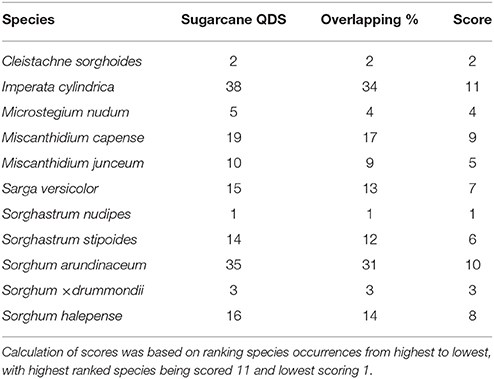
Table 7. Spatial overlap (shared occurrence) of Saccharum wild relatives (based on herbarium specimens) with sugarcane cultivation areas (113 QDS).
No collections or observations were made of five wild relatives within sugarcane fields within 700 m of the field margin (Table 8). These species can therefore not be considered as common weeds of sugarcane plantations besides the prevalence and spatial overlap with some sugarcane QDS. In general, members of Sorghum scored higher rankings for proximity to sugarcane plantations, except for Sarga versicolor (Table 8), and this is ascribed to preferences for habitat associated with sugarcane fields. Imperata cylindrica also ranked high, indicating its ability to colozise sugarcane fields. Miscanthidium species were moderately associated with sugarcane fields (Table 8). Both I. cylindrica and M. capense were found to be weeds in sugarcane plantations during field surveys although these species were not documented in South African literature as such.
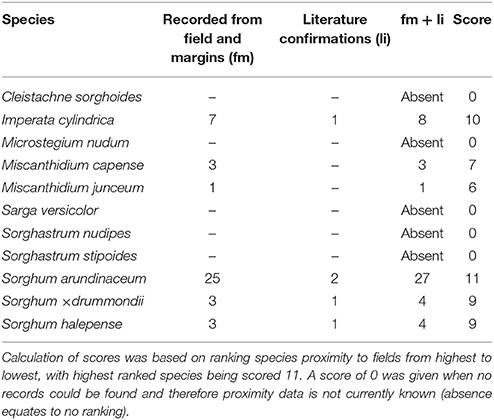
Table 8. Proximity or closeness of Saccharum wild relatives (based on herbarium specimens, field observations and literature) to sugarcane fields in cultivation areas.
Imperata cylindrica, M. junceum, and S. arundinaceum were ranked highest in terms of having extensive road and railway networks associated with their QDS of occurrence (Table 9). These networks present a higher likelihood for these species to spread into and within sugar cultivation areas compared with species that have fewer distribution networks. Species that are in isolated QDS and that are normally restricted to certain locations will also lack these distribution networks.
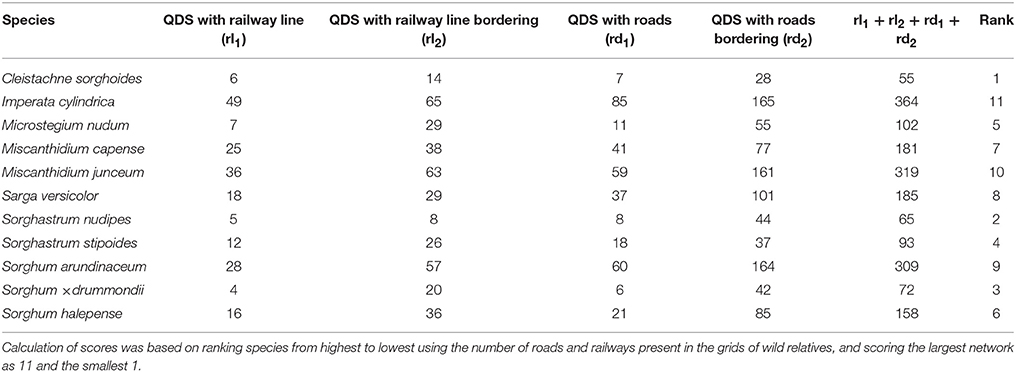
Table 9. Distribution potential of Saccharum wild relatives (based on road and railway networks) in sugarcane cultivation areas.
Gene Flow Likelihood
Imperata cylindrica scored the highest during the spatial and temporal assessment, followed by S. arundinaceum and M. capense (Table 10). M. junceum, Sorghum × drummondii, and S. halepense are further species with high scores. However, based on the relatedness assessment, I. cylindrica and the above Sorghum species are not closely related with commercial sugarcane (Figure 2) and are therefore not candidates to consider for gene flow. A likelihood score based on spatial, temporal and relatedness assessments (Figure 5) highlighted the two Miscanthidium species. Although S. arundinaceum had the highest overall score its distance from Saccharum in the phylogeny generated in our study makes it low risk for out crossing. Species with low scores are not considered to present any likelihood for gene flow, especially if these species have diverged from Saccharum at more than 7.3 million years (e.g., Sorghum).
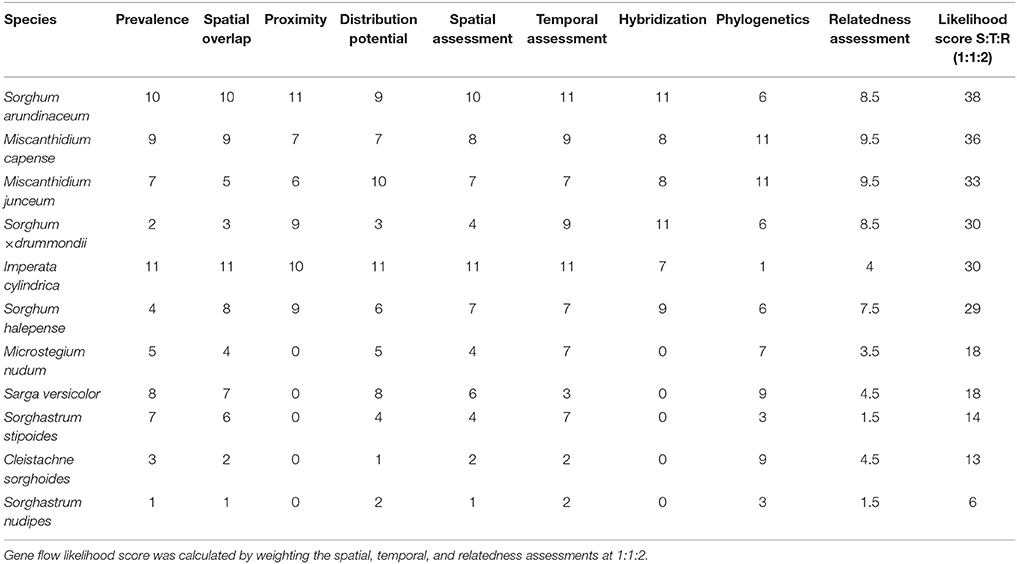
Table 10. Score per species calculated by equal weighting of factors obtained for each of spatial (prevalence, spatial overlap, proximity, and distribution potential), temporal (flowering time), and relatedness [hybridization and phylogenetics (Figure 3)] assessments.
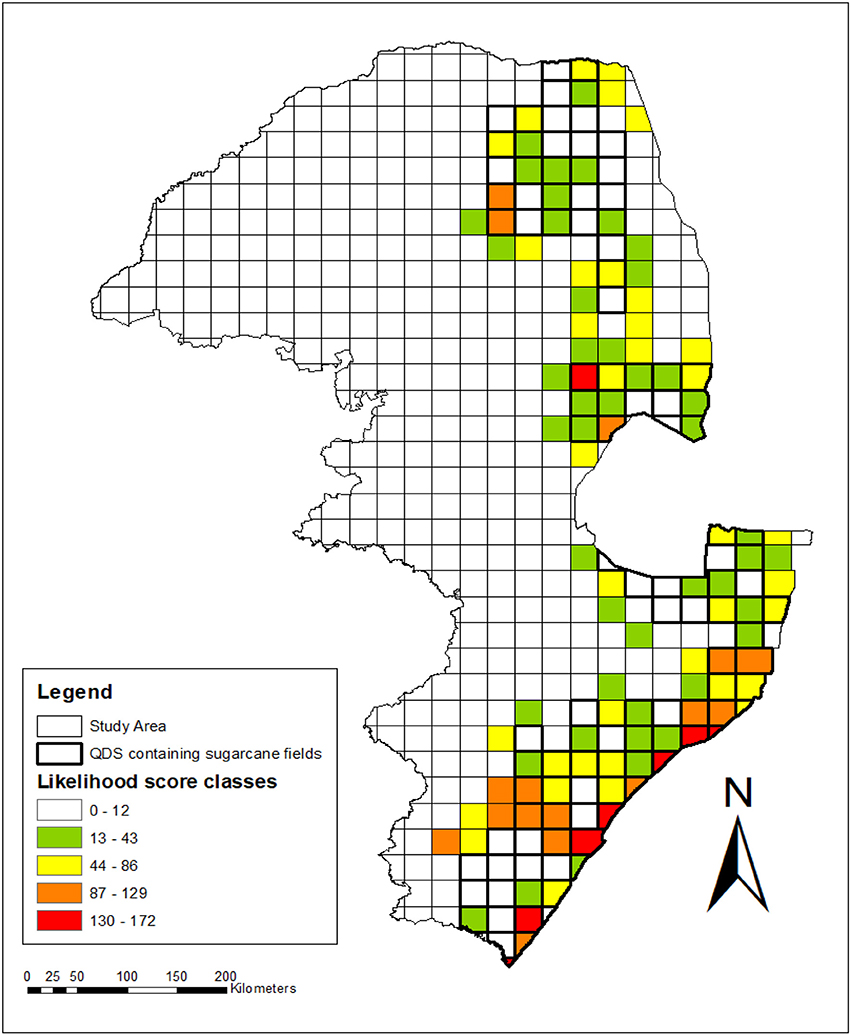
Figure 5. Spatial, temporal and relatedness assessment indicating the levels of likelihood for gene flow to occur between sugarcane and wild relatives in the sugar production region of South Africa. Grid values were calculated by summing the likelihood scores allocated per species (from Table 10) for all the species recorded per grid. QDS with sugarcane plantations are indicated with bold lines, whereas other QDS of the study area without sugarcane plantations are not shown with bold lines. Likelihood for gene flow: Sorghastrum nudipes scored 6 and there was no sugarcane QDS containing only this wild relative species. QDS with sugarcane plantations without wild relatives (0–12); sugarcane QDS plantations with wild relatives: very low (13–43); low (44-86); high (87–129); very high (130–172).
Closely related species with high spatial congruity pose the highest likelihood for gene flow and certain areas can be flagged where this is the case. No sugarcane QDS with very high likelihood for gene flow was found in Limpopo but there were two of high likelihood in Modjadjiskloof and Tzaneen (Figure 5). There was one QDS with very high likelihood in Nelspruit in addition to one QDS with high likelihood in Mpumalanga province. Thirteen QDS with high and 7 with very high likelihood were identified for KwaZulu-Natal, namely Durban, Felixton, Gingindlovu, Port Edward, Port Shepstone, Richards Bay, and Verulam. Overall it appears as if coastal and southern-inland KwaZulu-Natal have the highest likelihood for gene flow to occur based on relatedness, temporal and spatial congruity (Figure 5).
Discussion
Several studies have assessed the potential hybridization between plants and their closest relatives in GM scenarios (Ellstrand et al., 1999; FitzJohn et al., 2007; McGeoch et al., 2009) and similar evaluations have been made in sugarcane (Bonnett et al., 2008; Cheavegatti-Gianotto et al., 2011; Organisation for Economic Cooperation and Development, 2013). Our study was designed to consider these factors in a South African context. A review by Ellstrand et al. (1999) listed sugarcane amongst the world's important crop species which hybridize with wild relatives in agricultural systems. Commercial sugarcane cultivars have not been reported to spontaneously hybridize with any related genera and in the two published reviews that assessed the likelihood of GM sugarcane outcrossing with wild species there was no evidence of natural hybridization (Bonnett et al., 2008; Cheavegatti-Gianotto et al., 2011).
Imperata, Sorghum, Narenga, and Zea are genera found in South Africa that have been artificially crossed with sugarcane, and evidence of introgression has been confirmed on a molecular level (except in Imperata) (Bonnett et al., 2008; Hodnett et al., 2010). It was evident that sugarcane has a considerably low success of producing hybrids compared with its progenitors (i.e., Saccharum officinarum) (Piperidis et al., 2000; Aitken et al., 2007). Cheavegatti-Gianotto et al. (2011) noted that even when the barriers to hybridization were eliminated in artificial crosses (i.e., where flowering was synchronized, male pollen viability was increased and numerous florets were hand pollinated), there was poor growth and low survival in seedlings of the progeny. Even though Saccharum has previously crossed with Sorghum and Miscanthidium (Bourne, 1935; Brett, 1954; Gupta et al., 1978), Bonnett et al. (2008) concluded that these genera are unlikely to interbreed either spontaneously or without intervention from breeders due to the low survival rate of the seedlings.
Although the spatial assessment, both prevalence and spatial overlap, confirmed that I. cylindrica, S. arundinaceum, and M. capense had the highest spatial congruence within sugarcane cultivation areas (Tables 4, 6–8), and synchronous flowering times could facilitate gene flow (Table 5), evidence gathered in the present study using phylogenetic analyses of the ITS cassette demonstrated that commercial sugarcane cultivars were sister to Miscanthidium species and Narenga, but were only distantly related to S. arundinaceum and I. cylindrica (Figure 3).
It is generally accepted (Kellogg, 2013) that the “core” Andropogoneae (Figure 3) defines the dividing line between species that could be part of the Saccharinae and those that are not. Our phylogeny (Figure 3) clearly places I. cylindrica and Ischaemum afrum outside the Saccharinae. The same is true for genus Tripidium (Asiatic species). We also place Sorghum as sister to the core Andropogoneae (as has also been reported by Hawkins et al., 2015). This means that Sorghum is over 11 million years distant from Saccharum; well outside the natural hybridization window. Polytrias indica and M. vimineum form outgroups to the core Saccharinae. Sarga is sister to the core Saccharinae, but this is essentially an Asiatic genus; the one exception being C. sorghoides, which is native to Eastern Africa from Mpumalanga to Ethiopia (Clayton et al., 2006). However, with a base chromosomal number of 9 (Celarier, 1958), Cleistachne is unlikely to be karyotypically compatible with sugarcane.
Miscanthus and Polytoca, which are sister to Saccharum are Asiatic species as well. The next grouping, which is directly sister to Saccharum sensu stricto includes the African Miscanthidium species as well as Narenga porphyrocoma, which is mainly Asiatic, but has a rump population in Ethiopia. In an African context, at least in terms of evolutionary distance, these are the species most likely to hybridize with Saccharum. Narenga–Saccharum hybrids have been generated in breeding programmes, but they tend to be male sterile and suffer chromosomal loss in the F2 generation (Price, 1957). Chloroplast data (D Lloyd Evans, personal communication) indicates that Narenga hybridized with Saccharum more recently than Miscanthidium, and thus may contain more compatible chromosomes.
Miscanthidium species have a base chromosome number of 15 and show no recent hybridization with sugarcane (the two genera have been isolated for at least 2.5 million years). Thus it is likely that Miscanthidium and Saccharum are not chromosomally compatible. As an Asiatic and Ethiopian species, S. narenga poses no threat to gene flow with South African sugarcane, but could be a bridge species in a broader African context. It should be noted however, that of all the genera presented in the phylogeny (Figure 3) only the Asiatic and Polynesian species, Miscanthus floridulus has categorically been demonstrated to have hybridized with Saccharum in the wild (Lloyd Evans and Joshi, 2016a).
As sugarcane hybrids are based on a small number of inter-related parental lines, it is hardly surprising that these cultivars could not be resolved in the ITS phylogeny and the ITS cassette itself does not possess sufficient characters to resolve recently diverged species or cultivars. However, we see that the two S. spontaneum accessions are clearly divergent from the other Saccharum species or cultivars. S. sinense cv Tekcha emerges as ancestral to the remaining Saccharum species with 100% support. This is not unexpected as S. sinense accessions are ancient hybrids of S. officinarum and S. spontaneum (Irvine, 1999). As a grouping, S. robustum NG57-054, Saccharum hybrid cv Co745 and S. officinarum IJ76-514 were also resolved from the sugarcane hybrids with 100% support, though resolution within the monophyletic grouping was not possible.
The chronogram (Figure 4) provides timings for the radiation events undergone by species analyzed in this study. Few genera lie within the 3.4 million year window where wild hybridization is possible between Saccharum and other genera. Even if this window is extended to 7.4 million years, this only adds an additional two genera. All members of Sorghum (including Trachypogon spicatus) can be excluded as they are 10.4 million years divergent from Saccharum. The same applies to I. cylindrica, which is 12.1 million years divergent. Interestingly, the chronogram places Tripidium species (which sugarcane breeders have been attempting to introgress into Saccharum hybrid cultivars for over 50 years with poor success) as 11.4 million years divergent from Saccharum. The Southern African species, C. sorghoides lies within the genus Sarga which is 7.4 million years divergent from sugarcane. However, this species poses low risk of hybridization as it lies outside the wild hybridization window. The only species of high concern in terms of divergence times from Saccharum are those within the genus Miscanthidium, most especially M. capense, and M. junceum which are estimated to be approximately 3 million years divergent from Saccharum (Figure 4).
An unexpected finding was that commercial sugarcane cultivars N36 and N14 had pollen viability of up to 80% in some regions of South Africa (Figure 2). Even though no similar studies conducted field assessments across the sugarcane cultivation regions in South Africa, sugarcane seldom produces viable pollen under natural conditions at Mount Edgecombe (site 8) (Brett, 1950; Horsley and Zhou, 2013). Pollen viability gradually decreased from the northern inland (85%) to the south coastal regions (0%) of the study. Within certain study sites (e.g., site 5), some cultivars showed pollen viability of 70%, while others had <10%. A similar study in Brazil reported 100% viable pollen in some cultivars while others showed pollen viability of <9%, under the same environmental conditions (Melloni et al., 2015). Pollen viability has also been closely associated with genotype (Nair, 1975; Pagliarini, 2000; Melloni et al., 2015).
There is a higher likelihood for gene flow when potential pollen recipients flower at the same time as donor crop species when they are in close proximity (Ellstrand et al., 1999; Chapman and Burke, 2006; Schmidt and Bothma, 2006; FitzJohn et al., 2007; Bonnett et al., 2008; Tesso et al., 2008; Nieh et al., 2014). In the current study, there is only one related species with flower synchrony and shared habitat, M. capense, which presents the highest potential for gene flow (Table 10 and Figure 5). Although, as discussed previously, all verified hybrids between sugarcane and numerous species within the Andropogoneae have been created through human mediation. Moreover, in all cases hybrids are typically male sterile (Bremer, 1961; Kandasami, 1961; Aitken et al., 2007; Sobhakumari and Nair, 2014) and in F2 and subsequent generations there is considerable chromosomal loss. Thus no sugarcane hybrid reported thus far is a true hybrid, they are always intergeneric (partial) hybrids. Primarily this is due to chromosome number incompatibility (Figures 3, 4) and reflects the divergent evolutionary history of the major lineages within the Andropogoneae. Whilst there are reports of possible hybridizations between Saccharum species and related species in the wild, there have been no reports of wild hybridizations with modern hybrid sugarcane cultivars (Cheavegatti-Gianotto et al., 2011). Again this is an issue of chromosomal compatibility. Wild type Saccharum officinarum has a base chromosome count of 60 or 80 (typically the latter), but modern hybrids have a chromosome count of about 136 chromosomes—this is variable in different hybrids, but there are typically 10% S. spontaneum chromosomes and 90% S. officinarum chromosomes (Bremer, 1961). As a consequence, chromosomal incompatibility is far more likely between modern commercial sugarcane hybrids and wild species than between sugarcane's ancestors and wild species. Indeed, even back crosses of commercial hybrids with their immediate ancestors (S. spontaneum and S. officinarum) often lead to problems of male sterility (Babu, 1990). For crosses between sugarcane hybrid and wild species of low ploidy, not only is there an issue of chromosome incompatibility due to evolutionary distance, there is the added problem of lack of meiotic pairing due to differential chromosome numbers.
In our study, I. cylindrica, M. capense, M. junceum, S. arundinaceum, S. × drummondii, and S. halepense were found in relatively close proximity to sugarcane fields (Supplementary Figure 1). The latest review of invasive grasses of South Africa (Visser et al., 2017) reported Sorghum × drummondii and S. halepense amongst 256 weedy grasses that were introduced to agricultural systems. Weedy relatives may be considered as higher risk for gene flow potential when they are geographically associated with GM crops (Bonnett et al., 2008; Organisation for Economic Cooperation and Development, 2013). In general, most problematic weeds of sugarcane are in the Andropogoneae (Cheavegatti-Gianotto et al., 2011; Organisation for Economic Cooperation and Development, 2013). Imperata cylindrica and members of Sorghum have been documented as aggressive weeds of agricultural fields including sugarcane plantations in many countries (Van Oudtshoorn, 1999; Firehun and Tamado, 2006; Bonnett et al., 2008; Organisation for Economic Cooperation and Development, 2013; Takim et al., 2014). Sorghum arundinaceum and S. × drummondii are considered as weeds of sugarcane in South Africa (Van Oudtshoorn, 1999; Milton, 2004; Fish et al., 2015). Studies from Nigeria reported I. cylindrica amongst problem weeds of sugarcane (Takim et al., 2014), and both S. arundinaceum and S. × drummondii are regarded as major weeds of sugarcane in Ethiopia (Firehun and Tamado, 2006). For South African situations assessed in this study, although M. capense and M. junceus may be considered to be weeds in sugarcane fields, they are not considered to be “weedy”7.
Vehicles are amongst the main factors associated with the spread of weedy grasses in South Africa (Milton, 2004). The transport network therefore gives an indication of the potential for weedy relatives of sugarcane to spread, with denser networks implying higher chances for migrations. Furthermore, sugarcane relatives are often associated with roadsides as a preferred habitat (Retief and Herman, 1997; Van Oudtshoorn, 1999; Fish et al., 2015). Potential distribution networks of related species in our study show that most would be able to spread from the areas in which they are found, for example, M. capense is associated with vast road and rail networks (Table 9), which suggests that anthropogenic activities can enhance seed dispersal and increase gene flow potential (Andow and Zwahlen, 2006) in weedy species.
Conclusions
Phylogenetic analyses of the ITS cassette showed that the closest wild relative species to commercial sugarcane were M. capense, M. junceum, and N. porphyrocoma. Sorghum was found to be more distantly related to Saccharum than previously described. Similarly, Imperata is so distant from Saccharum that it poses no risk of gene flow. In the wild, no hybrids between modern sugarcane hybrid cultivars and any species have been reported. All documented wild hybrids are between sugarcane's ancestors (S. officinarum, S. robustum, and S. spontaneum) and a small number of closely related species. The phytogeography assessment indicated that the only wild relatives likely to be recipients of gene flow in the study area are Miscanthidium species—M. capense was observed to be a weed in cultivated sugarcane plantations but it does not have characteristics that make it “weedy.” Consequently, even although some commercial sugarcane cultivars do produce fertile pollen—especially in northern irrigated areas of KZN, there is a low likelihood of hybrids occurring in the natural environment. Therefore in a future scenario where GM sugarcane is cultivated in South Africa, the risk of gene flow to wild relatives is low.
Author Contributions
SaS conceived the idea and acquired funding for this research. SB and StS designed the experiments. HK and DK conducted the experimental work, analyzed data and interpreted results for their MSc degrees at North-West University, South Africa. Supervision was provided by SB and StS and co-supervision by JvdB, DC, and SaS. SaS co-ordinated the paper writing and submission. DLE performed assemblies from short read data, generated the alignments, performed and interpreted the phylogenies. All authors reviewed, revised, and approved the final version of the manuscript.
Conflict of Interest Statement
Author DLE acts as an unpaid Senior Informatics Specialist for BeauSci Ltd. The relationship is based on data sharing only.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding was provided by Biosafety SA via the Technology Innovation Agency and the South African Department of Science and Technology. We thank BeauSci Ltd., Cambridge, UK for providing additional short read sequence data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2018.00072/full#supplementary-material
Footnotes
2. ^https://www.ispotnature.org
3. ^https://ncbi.nlm.nih.gov/sra
4. ^http://purl.org/phylo/treebase/phylows/study/TB2:S22812
5. ^http://www.sasa.org.za/sasri
References
Adati, S. (1958). Studies on the genus Miscanthus with special reference to the Japanese species suitable for breeding purposes as fodder crops. Bull. Fac. Agric. Mie Univ. 17, 1–112.
Aitken, K., Li, J., Wang, L., Qing, C., Fan, Y. H., and Jackson, P. (2007). Introgression of Erianthus rockii into sugarcane and verification of intergeneric hybrids using molecular markers. Genet. Resour. Crop Evol. 54, 1395–1405. doi: 10.1007/s10722-006-9124-2
Amaral, A. L., Santos, J. M., Câmara, T. M. M., Filho, L. S. C., and Barbosa, G. V. S. (2013). Pollen storage methodology in sugarcane. Comun. Técn. 129, 1–11. Available online at: https://www.embrapa.br/busca-de-publicacoes/-/publicacao/953861/metodologia-de-conservacao-de-polen-de-cana-de-acucar
Andow, D. A., and Zwahlen, C. (2006). Assessing environmental risks of transgenic plants. Ecol. Lett. 9, 196–214. doi: 10.1111/j.1461-0248.2005.00846.x
Andriessen, M. (2015). Assessment of Brassica Napus Gene Flow Potential to Wild Relatives in Northern South Africa. Dissertation/master's thesis, North West University, Potchefstroom.
Bonnett, G. D., Nowak, E., Olivares-Villegas, J. J., Berding, N., Morgan, T., and Aitken, K. S. (2008). Identifying the risks of transgene escape from sugarcane crops to related species, with particular reference to Saccharum spontaneum in Australia. Trop. Plant Biol. 1, 58–71. doi: 10.1007/s12042-007-9002-x
Bourne, B. A. (1935). A comparative study of certain morphological characters of sugarcane x sorgo hybrids. J. Agric. Res. 5, 539–552.
Bremer, G. (1961). Problems in breeding and cytology of sugar cane. Euphytica 10, 59–78. doi: 10.1007/BF00037206
Brett, P. G. C. (1950). Flowering and pollen fertility in relation to sugarcane breeding in Natal. Proc. Int. Soc. Sug. Cane Technol. 7, 43–56.
Brett, P. G. C. (1954). Saccharum-Miscanthidium hybrids. J. Genet. 52, 542–546. doi: 10.1007/BF02985077
Brumbley, S. M., Snyman, S. J., Gnanasambandam, A., Joyce, P., Hermann, S. R., da Silva, J. A. G., et al. (2008). “Sugarcane,” in A Compendium of Transgenic Crop Plants, Vol. 7: Sugar, Tuber and Fiber Crops. eds C. Kole and T. C. Hall (Oxford: Blackwell), 1–58.
Burke, S. V., Wysocki, W. P., Zuloaga, F. O., Craine, J. M., Pires, J. C., Edger, P. P., et al. (2016). Evolutionary relationships in Panicoid grasses based on plastome phylogenomics (Panicoideae; Poaceae). BMC Plant Biol. 16:140. doi: 10.1186/s12870-016-0823-3
Butler, S. J., Vickery, J. A., and Norris, K. (2007). Farmland biodiversity and the footprint of agriculture. Science 315, 381–384. doi: 10.1126/science.1136607
Butterfield, M. K., D'Hont, A., and Berding, N. (2001). The sugarcane genome: a synthesis of current understanding, and lessons for breeding and biotechnology. Proc. S. Afr. Sug. Technol. Assoc. 75, 1–5. Available online at: http://www.sasta.co.za/wp-content/uploads/Proceedings/2000s/2001_butterfield_THE%20SUGARCANE%20GENOME.pdf
Celarier, R. P. (1958). Cytotaxonomy of the Andropogoneae: III. Subtribe Sorgheae, genus Sorghum. Cytologia 23, 395–418. doi: 10.1508/cytologia.23.395
Chapman, M. A., and Burke, J. M. (2006). Letting the gene out of the bottle: the population genetics of genetically modified crops. New Phytol. 170, 429–443. doi: 10.1111/j.1469-8137.2006.01710.x
Cheavegatti-Gianotto, A., de Abreu, H. M. C., Arruda, P., Bespalhok Filho, J. C., Burnquist, W. L., Creste, S., et al. (2011). Sugarcane (Saccharum x officinarum): a reference study for the regulation of genetically modified cultivars in Brazil. Trop. Plant Biol. 4, 62–89. doi: 10.1007/s12042-011-9068-3
Chen, L. J., Lee, D. S., Song, Z. P., Suh, H. S., and Lu, B.-R. (2004). Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Ann. Bot. 93, 67–73. doi: 10.1093/aob/mch006
Clayton, W. D., Vorontsova, M. S., Harman, K. T., and Williamson, H. (2006) onwards. GrassBase — The Online World Grass Flora. Available online at: http://www.kew.org/data/grasses-db.html (Accessed November 8, 2006).
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
den Nijs, H. C. M., Bartsch, D., and Sweet, J. (2004). “Introgression from genetically modified plants into wild relatives,” ed H. C. M. den Nijs (Wallingford; Oxfordshire: CAB International), 422.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ellstrand, N. C., Prentice, H. C., and Hancock, J. F. (1999). Gene flow and introgression from domesticated plants into their wild relatives. Annu. Rev. Ecol. Syst. 30, 539–563. doi: 10.1146/annurev.ecolsys.30.1.539
Estep, M., McKain, M., Vela Diaz, D., Zhong, J., Hodge, J., Hodkinson, T., et al. (2014). Allopolyploidy, diversification, and the Miocene grassland expansion. Proc. Natl. Acad. Sci. U.S.A. 111, 15149–15154. doi: 10.1073/pnas.1404177111
FigTree, (2017). FigTree. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (Accessed November 10, 2017).
Firehun, Y., and Tamado, T. (2006). Weed flora in the Rift Valley sugarcane plantations of Ethiopia as influenced by soil types and agronomic practices. Weed Biol. Manag. 6, 139–150. doi: 10.1111/j.1445-6664.2006.00207.x
Fish, L., Mashau, A. C., Moeaha, M. J., and Nembudani, M. T. (2015). Identification Guide to Southern African Grasses. An Identification Manual Keys, Descriptions and Distributions. Strelitzia 36. Pretoria: South African National Biodiversity Institute.
FitzJohn, R. G., Armstrong, T. T., Newstrom-Lloyd, L. E., Wilton, A. D., and Cochrane, M. (2007). Hybridisation within Brassica and allied genera: evaluation of potential for transgene escape. Euphytica 158, 209–230. doi: 10.1007/s10681-007-9444-0
Folk, R. A., Mandel, J. R., and Freudenstein, J. V. (2017). Ancestral gene flow and parallel organellar genome capture result in extreme phylogenomic discord in a lineage of angiosperms. Syst. Biol. 66, 320–337. doi: 10.1093/sysbio/syw083
Gao, Y.-J., Liu, X.-H., Zhang, R.-H., Zhou, H., Liao, J.-X., Duan, W.-X., et al. (2014). Verification of progeny from crosses between sugarcane (Saccharum spp.) and an intergeneric hybrid (Erianthus arundinaceus × Saccharum spontaneum) with molecular markers. Sugar Tech. 17, 31–35. doi: 10.1007/s12355-014-0310-9
Gu, M. H., Ma, H. T., and Liang, G. H. (1984). Karyotype analysis of seven species in the genus Sorghum. J. Hered. 75, 196–202. doi: 10.1093/oxfordjournals.jhered.a109911
Gupta, S. C., de Wet, J. M. J., and Harlan, J. R. (1978). Morphology of Saccharum-Sorghum hybrid derivatives. Am. J. Bot. 65, 936–942. doi: 10.1002/j.1537-2197.1978.tb06158.x
Ha, S., Moore, P. H., Heinz, D., Kato, S., Ohmido, N., and Fukui, K. (1999). Quantitative chromosome map of the polyploid Saccharum spontaneum by multicolor fluorescence in situ hybridization and imaging methods. Plant Mol. Biol. 39, 1165–1173. doi: 10.1023/A:1006133804170
Hawkins, J. S., Ramachandran, D., Henderson, A., Freeman, J., Carlise, M., Harris, A., et al. (2015). Phylogenetic reconstruction using four low-copy nuclear loci strongly supports a polyphyletic origin of the genus Sorghum. Ann. Bot. 116, 291–299. doi: 10.1093/aob/mcv097
Hodkinson, T. R., Chase, M. W., Lledo, M. D., Salamin, N., and Renvoize, S. A. (2002). Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnLintron and trnL-F intergenic spacers. J. Plant Res. 115, 381–392. doi: 10.1007/s10265-002-0049-3
Hodnett, G. L., Hale, A. L., Packer, D. J., Stelly, D. M., da Silva, J., and Rooney, W. L. (2010). Elimination of a reproductive barrier facilitates intergeneric hybridization of Sorghum bicolor and Saccharum. Crop Sci. 50, 1188–1195. doi: 10.2135/cropsci2009.09.0486
Horsley, T., and Zhou, M. (2013). Effect of photoperiod treatments on pollen viability and flowering at the South African Sugarcane Research Institute. Proc. S. Afr. Sug. Technol. Assoc. 86, 286–290. Available online at: https://www.sasta.co.za/wp-content/uploads/Proceedings/2010s/2013-45Horsley.pdf
Huang, Z., Zhu, J., Mu, X., and Lin, J. (2004). Pollen dispersion, pollen viability and pistil receptivity in Leymus chinensis. Ann. Bot. 93, 295–301. doi: 10.1093/aob/mch044
Irvine, J. E. (1999). Saccharum species as horticultural classes. Theor. Appl. Genet. 98, 186–194. doi: 10.1007/s001220051057
Jagathesan, D., and Devi, K. R. (1969). Cytogenetical studies in Erianthus, I. Chromosome morphology in 2 N = 20 forms. Caryologia 22, 359–368.
Kandasami, P. A. (1961). Interspecific and intergeneric hybrids of Saccharum spontaneum L. Cytologia 26, 117–123. doi: 10.1508/cytologia.26.117
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kellogg, E. A. (2013). “Pylogenetic relationships of Saccharinae and Sorghinae,” in: Genomics of the Saccharinae, ed A. H. Paterson (New York, NY: Springer), 3–21.
Khanyi, H. (2018). Relatedness of Saccharum Species Hybrids and Wild Relatives in Eastern South Africa. Dissertation/master's thesis, North West University, Potchefstroom.
Knispel, A. L., McLachlan, S. M., Van Acker, R. C., and Friesen, L. F. (2008). Gene flow and multiple herbicide resistance in escaped canola populations. Weed Sci. 56, 72–80. doi: 10.1614/WS-07-097.1
Kress, W. J., Erickson, D. L., Jones, F. A., Swenson, N. G., Perez, R., Sanjur, O., et al. (2009). Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl. Acad. Sci. U.S.A. 106, 18621–18626. doi: 10.1073/pnas.0909820106
Lakshmanan, P., Geijskes, R. J., Aitken, K. S., Grof, C. L. P., Bonnett, G. D., and Smith, G. R. (2005). Invited review: sugarcane biotechnology: the challenges and opportunities. In Vitro Cell Dev. Biol. Plant 41, 345–363. doi: 10.1079/IVP2005643
Légère, A. (2005). Risks and consequences of gene flow from herbicide-resistant crops: canola (Brassica napus L.) as a case study. Pest Manag. Sci. 61, 292–300. doi: 10.1002/ps.975
Li, H. W., Shang, K. C., Hsiao, Y. Y., and Yang, P. C. (1959). Cytological studies of sugarcane and its relatives. Cytologia 24, 220–236. doi: 10.1508/cytologia.24.220
Liu, K., Raghavan, S., Nelesen, S., Linder, C. R., and Warnow, T. (2009). Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science 324, 1561–1564. doi: 10.1126/science.1171243
Lloyd Evans, D., and Joshi, S. V. (2016a). Complete chloroplast genomes of Saccharum spontaneum, Saccharum officinarum and Miscanthus floridulus (Panicoideae: Andropogoneae) reveal the plastid view on sugarcane origins. Syst. Biodivers. 14, 548–571. doi: 10.1080/14772000.2016.1197336
Lloyd Evans, D., and Joshi, S. V. (2016b). Elucidating modes of activation and herbicide resistance by sequence assembly and molecular modelling of the acetolactate synthase complex in sugarcane. J. Theor. Biol. 407, 184–197. doi: 10.1016/j.jtbi.2016.07.025
Löytynoja, A., Vilella, A. J., and Goldman, N. (2012). Accurate extension of multiple sequence alignments using a phylogeny-aware graph algorithm. Bioinformatics 28, 1684–1691. doi: 10.1093/bioinformatics/bts198
Martin, L. A., Lloyd Evans, D., Castlebury, L. A., Sifundza, J. T., Comstock, J. C., Rutherford, R. S., et al. (2017). Macruropyxis fulva sp. nov., a new rust (Pucciniales) infecting sugarcane in southern Africa. Australas. Plant Pathol. 46, 63–74. doi: 10.1007/s13313-016-0460-1
McGeoch, M. A., Kalwij, J. M., and Rhodes, J. I. (2009). A spatial assessment of Brassica napus gene flow potential to wild and weedy relatives in the Fynbos biome. S. Afr. J. Sci. 105, 109–115. doi: 10.4102/sajs.v105i3/4.57
Melloni, M. L. G., Melloni, M. N. G., Scarpari, M. S., Garcia, J. C., Landell, M. G. A., and Pinto, L. R. (2015). Flowering of sugarcane genotypes under different artificial photoperiod conditions. Am. J. Plant Sci. 6, 456–463. doi: 10.4236/ajps.2015.63051
Meyer, G. M., and Snyman, S. J. (2013). Progress in research on genetically modified sugarcane in South Africa and associated regulatory requirements. Acta Hortic. 974, 43–50. doi: 10.17660/ActaHortic.2013.974.4
Milton, S. J. (2004). Grasses as invasive alien plants in South Africa. S. Afr. J. Sci. 100, 69–75. Available online at: http://hdl.handle.net/10520/EJC96211
Nair, M. K. (1975). Cytogenetics of Saccharum officinarum L. and S. spontaneum L. IV. Chromosome number and meiosis in S. officinarum x S. spontaneum hybrids. Caryologia 28, 1–14.
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, Q. A. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nieh, S. C., Lin, W. S., Hsu, Y. H., Shieh, G. J., and Kuo, B. J. (2014). The effect of flowering time and distance between pollen source and recipient on maize. GM Crops Food 5, 287–295. doi: 10.4161/21645698.2014.947805
Nylander, J. A., Wilgenbusch, J. C., Warren, D. L., and Swofford, D. L. (2008). AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583. doi: 10.1093/bioinformatics/btm388
Organisation for Economic Cooperation and Development (OECD) (2013). Consensus Document on the Biology Of Sugarcane (Saccharum spp.), Series on the Harmonisation of Regulatory Oversight in Biotechnology No. 56, OECD Environment Directorate (Paris).
Pagliarini, M. S. (2000). Meiotic behavior of economically important plant species: the relationship between fertility and male sterility. Genet. Mol. Biol. 23, 997–1002. doi: 10.1590/S1415-47572000000400045
Panova, M., Hollander, J., and Johannesson, K. (2006). Site-specific genetic divergence in parallel hybrid zones suggests nonallopatric evolution of reproductive barriers. Mol. Ecol. 15, 4021–4031. doi: 10.1111/j.1365-294X.2006.03067.x
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Parthasarathy, N. (1948). Origin of noble sugar-canes (Saccharum officinarum L.). Nature 161:608. doi: 10.1038/161608a0
Paterson, A. H., Moore, P. H., and Tew, T. L. (2013). “The gene pool of Saccharum species and their improvement,” in Genomics of the Saccharinae. ed A. H. Paterson (New York, NY: Springer), 43–71.
Philippe, H., Vienne, D. M. D., Ranwez, V., Roure, B., Baurain, D., and Delsuc, F. (2017). Pitfalls in supermatrix phylogenomics. Eur. J. Taxon 283, 1–25. doi: 10.5852/ejt.2017.283
Piperidis, G., Christopher, M. J., Carroll, B. J., Berding, N., and D'Hont, A. (2000). Molecular contribution to selection of intergeneric hybrids between sugarcane and the wild species Erianthus arundinaceus. Genome 43, 1033–1037. doi: 10.1139/g00-059
Price, S. (1957). Cytological studies in Saccharum and allied genera. III. Chromosome numbers in interspecific hybrids. Bot. Gaz. 118, 146–159.
Rambaut, A., Suchard, M. A., Xie, D., and Drummond, A. J. (2017). Tracer, Version 1.5. Available online at: http://tree.bio.ed.ac.uk/software/tracer/ (Accessed December 19, 2017).
Retief, E., and Herman, P. P. J. (1997). Plants of The Northern Provinces of South Africa: Keys and Diagnostic Characters. Strelitzia 6. Pretoria: South African National Biodiversity Institute.
Rieger, M. A., Lamond, M., Preston, C., Powles, S. B., and Roush, R. T. (2002). Pollen-mediated movement of herbicide resistance between commercial canola fields. Science 296, 2386–2388. doi: 10.1126/science.1071682
Robertson, M. P., and Barker, N. P. (2006). A technique for evaluating species richness maps generated from collections data. S. Afr. J. Sci. 102, 77–84. Available online at: http://hdl.handle.net/10520/EJC96492
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sanderson, M. J. (2003). r8s: Divergence Estimates on Phylogenies. Available online at: https://sourceforge.net/projects/r8s/files/?source=navbar (Accessed January 10, 2018).
Schmidt, M., and Bothma, G. (2006). Risk assessment for transgenic sorghum in Africa: crop-to-crop gene flow in Sorghum bicolor (L.) Moench. Crop Sci. 46, 790–798. doi: 10.2135/cropsci2005.06-0117
Sithole, P., and Singels, A. (2013). Predicting Sugarcane Flowering. Available online at: http://www.sasa.org.za/Libraries/TheLinkMay2013/LinkMay2013P6_7.sflb.ashx (Accessed September 1, 2017).
Skendzic, E. M., Columbus, J. T., and Cerros-Tlatilpa, R. (2007). Phylogenetics of Andropogoneae (Poaceae: Panicoideae) based on nuclear ribosomal internal transcribed spacer and chloroplast trnL-F sequences. Aliso 23, 530–544. doi: 10.5642/aliso.20072301.40
Soares, T. L., Jesus, O. N., Souza, E. H., Santos-Serejo, J. A., and Oliveira, E. J. (2013). Morphology and viability of pollen grains from passion fruit species (Passiflora spp.). Acta Bot. Brasilica 27, 779–787. doi: 10.1590/S0102-33062013000400018
Sobhakumari, V. P., and Nair, N. V. (2014). The cytological mechanism of male sterility in intergeneric hybrids of Sorghum × Saccharum. Cytologia 79, 79–83. doi: 10.1508/cytologia.79.79
Soreng, R. J., Peterson, P. M., Romaschenko, K., Davidse, G., Zuloaga, F. O., Judziewicz, E. J., et al. (2015). A worldwide phylogenetic classification of the Poaceae (Gramineae). J. Syst. Evol. 53, 117–137. doi: 10.1111/jse.12150
Stanford, A., Harden, R., and Parks, C. (2000). Phylogeny and biogeography of Juglans (Juglandaceae) based on MATK and ITS sequence data. Am. J. Bot. 87, 872–882. doi: 10.2307/2656895
Strydom, A., Spies, J. J., and Van Wyk, S. M. C. (2000). Poaceae: chromosome studies on african plants. 14. Panicoideae. Bothalia 39, 201–214. doi: 10.4102/abc.v30i2.559
Sukumaran, J., and Holder, M. T. (2010). DendroPy: a Python library for phylogenetic computing. Bioinformatics 26, 1569–1571. doi: 10.1093/bioinformatics/btq228
Takim, F. O., Fadayomi, O., Alabi, M. A., and Olawuyi, O. J. (2014). Impact of natural weed infestation on the performance of selected sugarcane varieties in the southern Guinea savanna of Nigeria. Ethiopian J. Environ. Stud. Manage. 7, 279–288. doi: 10.4314/ejesm.v7i3.7
Tesso, T., Kapran, I., Grenier, C., Snow, A., Sweeney, P., Pedersen, J., et al. (2008). The potential for crop-to-wild gene flow in sorghum in Ethiopia and Niger: a geographic survey. Crop Sci. 48, 1425–1431.
The Plant List (2013). Version 1.1. Available online at: http://www.theplantlist.org/ (Accessed May 3, 2017).
Visser, V., Wilson, J. R. U., Canavan, K., Canavan, S., Fish, L., Le Maitre, D., et al. (2017). Grasses as invasive plants in South Africa revisited: patterns, pathways and management. Bothalia 47, 1–29. doi: 10.4102/abc.v47i2.2169
Warwick, S. I., Simard, M.-J., Légère, A., Beckie, H. J., Braun, L., Zhu, B., et al. (2003). Hybridization between transgenic Brassica napus L. and its wild relatives: Brassica rapus L., Raphanus raphanistrum L., Sinapis arvensis L., and Erucastrum gallicum (Willd.) O.E. Schulz. Theor. Appl. Genet. 107, 528–539. doi: 10.1007/s00122-003-1278-0
Watrud, L. S., Lee, E. H., Fairbrother, A., Burdick, C., Reichman, J. R., Bollman, M., et al. (2004). Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass CP4 EPSPS as a marker. Proc. Natl. Acad. Sci. U.S.A. 101, 14533–14538. doi: 10.1073/pnas.0405154101
Welker, C. A. D., Souza-Chies, T. T., Longhi-Wagner, H. M., Peichoto, M. C., McKain, M. R., and Kellogg, E. A. (2015). Phylogenetic analysis of Saccharum s.l. (Poaceae; Andropogoneae), with emphasis on the circumscription of the South American species. Am. J. Bot. 102, 248–263. doi: 10.3732/ajb.1400397
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. Innis, D. Gelfand, J. Sninsky, and T. White (San Diego, CA: Academic Press), 315–322.
Xue, B., Guo, J., Que, Y., Fu, Z., Wu, L., and Xu, L. (2014). Selection of suitable endogenous reference genes for relative copy number detection in sugarcane. Int. J. Mol. Sci. 15, 8846–8862. doi: 10.3390/ijms15058846
Yu, J., Xue, J.-H., and Zhou, S.-L. (2011). New universal matK primers for DNA barcoding angiosperms. J. Syst. Evol. 49, 176–181. doi: 10.1111/j.1759-6831.2011.00134.x
Zapiola, M. L., Campbell, C. K., Butler, M. D., and Mallory-Smith, C. A. (2008). Escape and establishment of transgenic glyphosate-resistant creeping bentgrass Agrostis stolonifera in Oregon, USA: a 4-year study. J. Appl. Ecol. 45, 486–494. doi: 10.1111/j.1365-2664.2007.01430.x
Keywords: gene flow, hybridization, pollen viability, phytogeography, spatial assessment, phylogeny, Miscanthidium
Citation: Snyman SJ, Komape DM, Khanyi H, van den Berg J, Cilliers D, Lloyd Evans D, Barnard S and Siebert SJ (2018) Assessing the Likelihood of Gene Flow From Sugarcane (Saccharum Hybrids) to Wild Relatives in South Africa. Front. Bioeng. Biotechnol. 6:72. doi: 10.3389/fbioe.2018.00072
Received: 23 January 2018; Accepted: 17 May 2018;
Published: 07 June 2018.
Edited by:
Andrew F. Roberts, International Life Sciences Institute, United StatesReviewed by:
Graham Bonnett, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaAlan John Gray, Centre for Ecology & Hydrology, Edinburgh, United Kingdom
Copyright © 2018 Snyman, Komape, Khanyi, van den Berg, Cilliers, Lloyd Evans, Barnard and Siebert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandy J. Snyman, c2FuZHkuc255bWFuQHN1Z2FyLm9yZy56YQ==
 Sandy J. Snyman
Sandy J. Snyman Dennis M. Komape3
Dennis M. Komape3 Hlobisile Khanyi
Hlobisile Khanyi Johnnie van den Berg
Johnnie van den Berg Dyfed Lloyd Evans
Dyfed Lloyd Evans Sandra Barnard
Sandra Barnard Stefan J. Siebert
Stefan J. Siebert