- 1MOE Key Laboratory for Developmental Genes and Human Disease, School of Life Science and Technology, Jiangsu Province High-Tech Key Laboratory for Bio-Medical Research, Southeast University, Nanjing, China
- 2Department of Otolaryngology Head and Neck Surgery, Nanjing Drum Tower Hospital Clinical College of Nanjing Medical University, Jiangsu Provincial Key Medical Discipline (Laboratory), Nanjing, China
- 3Research Institute of Otolaryngology, Nanjing, China
- 4Department of Otolaryngology, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, China
- 5Co-Innovation Center of Neuroregeneration, Nantong University, Nantong, China
- 6Institute for Cardiovascular Science, Department of Cardiovascular Surgery of the First Affiliated Hospital, Medical College, Soochow University, Suzhou, China
- 7Department of Otorhinolaryngology, Affiliated Sixth People’s Hospital of Shanghai Jiao Tong University, Shanghai, China
- 8Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing, China
Exosomes are nanoscale membrane-enclosed vesicles 30–150 nm in diameter that are originated from a number of type cells by the endocytic pathway and consist of proteins, lipids, RNA, and DNA. Although, exosomes were initially considered to be cellular waste, they have gradually been recognized to join in cell-cell communication and cell signal transmission. In addition, exosomal contents can be applied as biomarkers for clinical judgment and exosomes can as potential carriers in a novel drug delivery system. Unfortunately, purification methods of exosomes remain an obstacle. We described some common purification methods and highlight Morpho Menelaus (M. Menelaus) butterfly wings can be developed as efficient methods for exosome isolation. Furthermore, the current research on exosomes mainly focused on their roles in cancer, while related studies on exosomes in the visual and auditory systems are limited. Here we reviewed the biogenesis and contents of exosomes. And more importantly, we summarized the roles of exosomes and provided prospective for exosome research in the visual and auditory systems.
Introduction
Extracellular vesicles (EVs) belong to a class of phospholipid bilayer membrane enclosed vesicles, and they are classified according to their biological origin and size (Shao et al., 2018; van Niel et al., 2018). Over the past decade, EVs have been found adjust and boost intercellular communication (El Andaloussi et al., 2013b), including signal transduction between cells, and thus invovled in cell proliferation, cell migration, immune regulation, etc (Meldolesi, 2018; Seo et al., 2018; Zhang S. et al., 2018). EVs are divided into three main types by their biological processes: exosomes, microvesicles and apoptotic bodies (El Andaloussi et al., 2013b; Shao et al., 2018; Gurunathan et al., 2019). Here, we focus on the exosomes, which were first identified by Johnstone (Johnstone et al., 1987). Exosomes are cell secreted vesicles that can be isolated from various humor, such as blood, plasma, urine, tears, and lymph (Ibrahim and Marban, 2016), and they range in size from 30 to 150 nm.
A new cell-cell communication system has recently been identified, that lie with exosomes’ ability to target recipient cells and to transfer exosomal proteins, lipids, and nucleic acids to these cells (Milane et al., 2015; Zhang et al., 2015; Poe and Knowlton, 2018; Gurunathan et al., 2019). Exosomes have been shown to be closely related to tumorigenesis (Rodrigues et al., 2019), and exosomes have become a new type of clinical targets, for example, circulating exosomes containing glypican-1 have been used for the early judgment of early pancreatic cancer (Melo et al., 2015). Moreover, recent studies that exosomes are a potential system for targeted drug delivery (Kibria et al., 2018; Arrighetti et al., 2019; Sun et al., 2019).
The visual and auditory systems are the two major sensory systems. Many signaling pathways, such as Wnt, FGF pathway, are involved in the development of these two sensory systems (Heavner and Pevny, 2012; Żak et al., 2015). There are many factors that can lead to vision and hearing loss, and aging is a common cause of both. Hereditary factors are also important causes for hearing and vision loss. The Leber’s hereditary optic neuropathy is an inherited disease with mutations mitochondrial DNA that affects both visual and auditory functions (Rance et al., 2012; Bianco et al., 2017; Karanjia et al., 2017). Recent studies showed that exosomes can be secreted by visual and auditory cells (Han et al., 2017; Klingeborn et al., 2018; Breglio et al., 2020). Based on exosomes studies in other fields, exosomes might have similar uses in studying and treating disorders of the visual and auditory systems.
This article reviews the latest research in exosomes, described from exosome biogenesis, detection, and enrichment methods to their functions in visual and auditory systems.
Biogenesis of Exosomes
Exosomes are vesicle-like bodies with a size of 30–150 nm that are produced by a variety of cells, and they have cup-shaped structures when observed under an electron microscope (Shao et al., 2012). Exosomes are originated from a number of type cells by the endosomal membrane pathway. First, the plasma membrane invaginated to produced endocytic vesicles, and multiple endocytic bodies then gather together to form early endosomes. Next, the invagination of early endosomes further transformed into multivesicular bodies (MVBs). Finally, MVBs move and touch to plasma membrane to secrete vesicles into the extracellular space, and these vesicles are defined as exosomes (Figure 1; Shao et al., 2018).
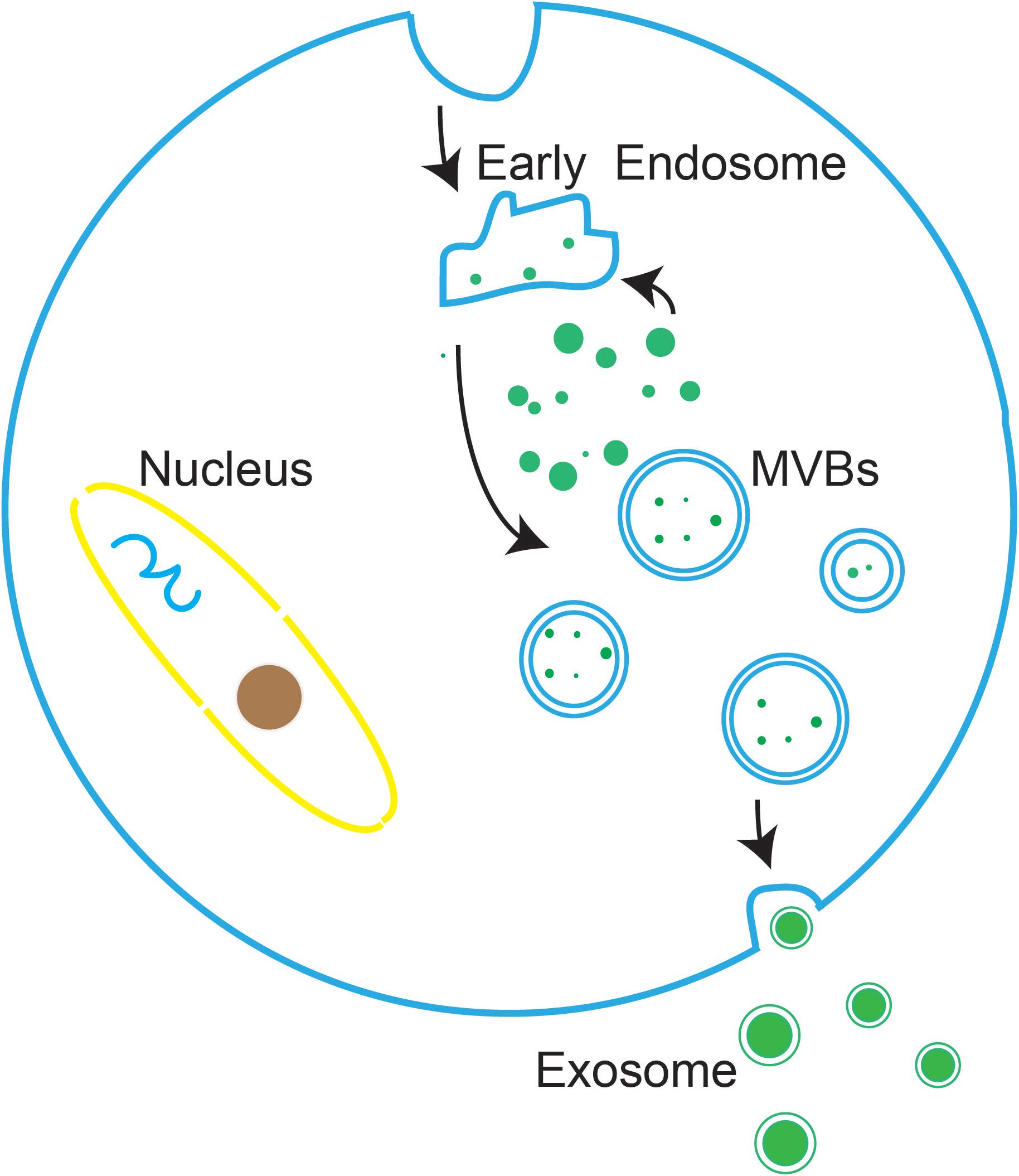
Figure 1. The biogenesis of exosomes. the plasma membrane invaginated to produced endocytic vesicles, which are further transformed into MVBs. MVBs then move and touch to the plasma membrane to secrete vesicles into the extracellular environment, at which point the vesicles are defined as exosomes.
Exosome formation involves content sorting and release, and the precise regulation of this process requires the coordinated functions of many different proteins. Subunits of endosomal sorting complex required for transport (ESCRT) are necessary for exosome biogenesis, including ESCRT complexs from 0 to III (Farooqi et al., 2018). There are two mechanisms for exosome secretion, ESCRT-independent and ESCRT-dependent.
In the ESCRT-dependent mechanism, the ESCRT-0 complex recruit proteins by ubiquitin or clathrin. ESCRT-I and ESCRT-III control the budding process, the formation of ILVs, and the subsequent formation of exosomes. The ESCRT-independent mechanism involves syntenin, ALIX and ESCRT-III. Syntenin recruits proteins to form cargo cluster, and then ALIX, and ESCRT-III regulate the budding process, the formation of ILVs, and the subsequent formation of exosomes. The proteins recruitment and cargo clustering can be achieved by either the ESCRT-dependent or the ESCRT-independent mechanisms (van Niel et al., 2018).
Compositions of Exosomes
Exosomes contain a variety of bioactive substances, mainly proteins, nucleotides, and lipids, and they are encapsulated by a phospholipid bilayer membrane that their internal bioactive substances avoid proteases degradation in the extracellular fluid (Tao et al., 2017). The phospholipid bilayer of exosomal membrane is composed of glycerophospholipids, sphingolipids, cholesterol, diglycerides, phospholipids and polyglycerophospholipids (Subra et al., 2010). Therefore, these exosomal proteins and nucleotides have potential use as reference index for disease judgment.
Membrane transport and fusion proteins are the protein with the largest proportion on the surface of exosomes (Ibrahim and Marban, 2016; Farooqi et al., 2018), and these include tetraspanins (e.g., CD9, CD63, and CD81), heat shock proteins (e.g., Hsp70, Hsp90), cytoskeletal proteins (e.g., actin, fibronectin), and viral proteins (e.g., LAMP1, Tsg101). Other proteins also present in exosomes including signal transduction proteins, vesicle biogenesis proteins, and enzymes. Moreover, there are a variety of RNAs in exosomes, such as mRNAs, miRNAs, ncRNAs, and LncRNAs (Sexton et al., 2019; Figure 2).
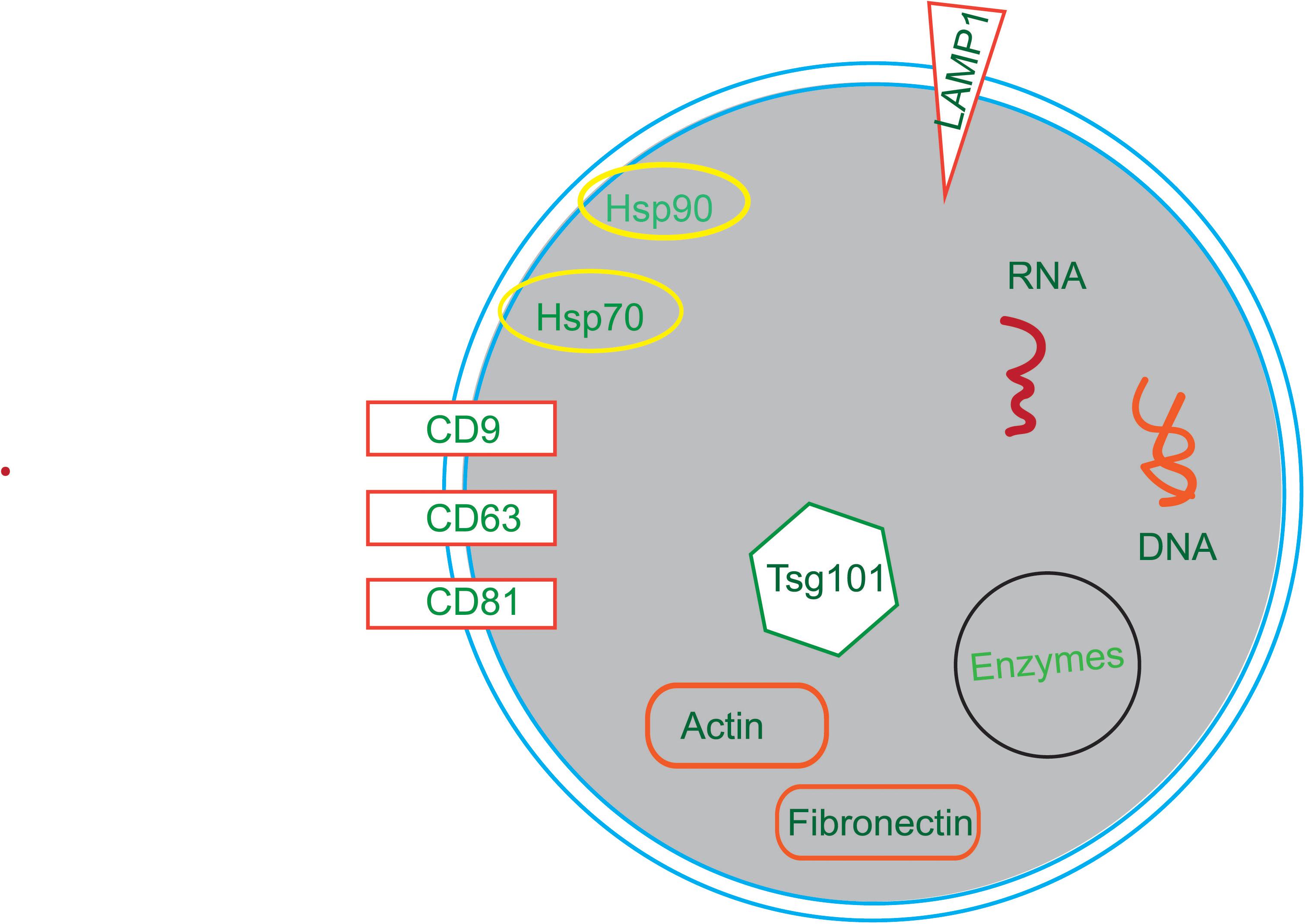
Figure 2. The compositions of exosomes. Exosomes are composed of in a variety of bioactive substances, mainly proteins, nucleotides and lipids. Proteins include tetraspanins (e.g., CD9, CD63, and CD81), heat shock proteins (e.g., Hsp70, Hsp90), cytoskeletal proteins (e.g., Actin, Fibronectin), viral proteins (e.g., Tsg101) and enzymes. Nucleotides include RNA and DNA.
Isolation and Purification of Exosomes
The isolation and purification of exosomes shows significant promise for biomedical research (Witwer et al., 2013; Gardiner et al., 2016), but there are still difficulties to be solve in using exosomes in research and clinical applications. The main challenge is that the size and physicochemical properties of exosomes overlap with those of lipoprotein and protein complexes. Based on the size and biological characteristics of exosomes, many enrichment methods have been used to purify them, including differential ultracentrifugation, gradient ultracentrifugation, size-exclusion chromatography (Fonseca et al.), immunoaffinity enrichment, and co-precipitation. More speed and efficient methods are being exploited to purified exosomes.
Ultracentrifugation
In cell culture medium, blood, urine, and breast milk, exosomes are often mixed with other impurities that have similar physical and chemical properties, including lipoproteins, cell fragments, etc. According to their specific size and density distribution range, exosomes can be effectively separated from these particles by ultracentrifugation and density gradient centrifugation. Ultracentrifugation is considered the most common traditional strategy for exosome enrichment (Gardiner et al., 2016). Combinations of different speeds and times are used in ultracentrifugation to remove other impurities in the sample step-by-step, and finally the separation and enrichment of exosomes are achieved. First, whole cells and large apoptotic bodies are depleted for 15–30 min by low-speed centrifugation (600–2,000 × g) at 4°C (Crewe et al., 2018; Langevin et al., 2019). Next, the supernatant centrifuged for 45–60 min by 12,000 × g centrifugation at 4°C to remove MVBs (Langevin et al., 2019). Lastly, the supernatant is moved to a new ultracentrifuge tube and centrifuged for 1–2 h by 100,000 × g-120,000 × g centrifugation at 4°C (Witwer et al., 2013; Langevin et al., 2019). The supernatant is deleted and the particles are resuspended in 100 μL phosphate-buffered saline. Exosomes cannot be separated completely by particle size using this protocol, because sedimentation is based on the density and other non-exosome vesicles can also be enriched (Zhang M. D. et al., 2018). EVs have a unique lipid membrane structure that encapsulates a certain amount of nucleic acids and proteins, resulting in a density range of 1.13–1.19 mg/ml (Thery et al., 2006). Density-gradient centrifugation involves centrifuging through an iodixanol or sucrose density gradient and different particles settle at different points in the gradient based on their different densities (Shao et al., 2018). Compared to differential centrifugation, the density-gradient centrifugation approach results in purer exosomes, but it requires a longer time to reach equilibrium and thus leads to greater damage to the instrument.
Size-Exclusion Chromatography (SEC)
SEC is a chromatography technique that differentiate molecules in a solution depend on their size and molecular weight (Boing et al., 2014; Burgess, 2018). The purification column is made up of spherical beads with a specific aperture pores, and examples of commonly used materials are Sephadex, Sepharose, and Sephacryl. When the sample flows into column, large molecules block out the pores, while small molecules can diffuse into the pores. Therefore, larger molecules pass through the column faster than small molecules, and exosomes are separated due to their size. Purified exosomes can be isolated from complex biological media such as milk, urine, and plasma using SEC (Lozano-Ramos et al., 2015; Blans et al., 2017; Kreimer and Ivanov, 2017; Shao et al., 2018).
Immunoaffinity Enrichment
Immunoaffinity enrichment is based on antibodies to specific exosome marker proteins. Proteins such as CD9, CD63, and CD81 are located on the exosome surface, and tumor-associated markers (HER2, EpCAM) are also present on tumor-associated exosomes (Taylor and Gercel-Taylor, 2008; Woo et al., 2016; Barok et al., 2018). Antibodies against these proteins linked with beads or other substrates by covalent or high-affinity interactions, and these antibodies bind to exosomes using low-speed centrifugation or magnetic techniques (Witwer et al., 2013). Taylor and Gercel-Taylor have successfully isolated circulating exosomes secreted from tumors using EpCAM magnetic beads (Taylor and Gercel-Taylor, 2008). This method has the potential for high specificity and efficiency (Tauro et al., 2012) and is usually performed using commercially available kits.
Co-precipitation
Recently, polymer co-precipitation strategies have been exploited to enrich exosomes. The common methods are protamine precipitation, acetate precipitation, protein organic solvent precipitation, and hydrophilic polymers precipitation (Brownlee et al., 2014; Deregibus et al., 2016; Gallart-Palau et al., 2016). These reagents precipitate EVs by reducing the hydration and thus the solubility of EVs (Shao et al., 2018). Therefore, it is possible to isolate exosomes using low centrifugal forces. Based on this method, many exosome extraction kits have been developed, for instance Total Exosome Isolation (Invitrogen, United States).
New Enrichment Methods
In the past few years, many exosome enrichment methods have been developed, here we summarize some of the recent progress.
Microfluidic filtering is a novel technology that extracts exosomes from a small amount of liquid (10–9 to 10–18 liters) through channels ranging from tens to hundreds of micrometers (Whitesides, 2006). With recent developments in nanomaterials, some emerging nanomaterials have been used for the capture of exosomes. For example, Lim et al. (2019) produced nanowires with CD63, CD9, and CD81 antibodies attached to their surfaces for capturing exosomes.
Recently, Chai et al. developed a noval microvortex chips method using Morpho Menelaus (M. Menelaus) butterfly wings modified by lipid nanoprobe, which, when integrated into microfluidic chips, greatly improved the efficiency of EV enrichment by over 70%. M. Menelaus wings have an original three-dimensional (3D) microgroove structure linked with many intersection points, and these microgrooves are distributed on wing surface parallelly. Due to this structure of M. Menelaus wings and the lipid bilayer structure of EVs, the lipid nanoprobe modified M. Menelaus wings can be applied to isolate and purified EVs (Figure 3) (Han et al., 2020). These results demonstrated that the efficiency is greatly improved by using new microvortex chips. Based on this method, enrichment exosomes by M. Menelaus wings is possible. There are many exosomal marker proteins, which are present on the surface of exosome (e.g., CD9, CD63, and CD81). Based on M. Menelaus wings structure, antibodies of these marker proteins can be modified on M. Menelaus wings. When samples enter M. Menelaus wings, antibodies of these marker proteins will bind to marker proteins on exosome surface and thus capture exosomes effectively.

Figure 3. Scheme of isolation and enrichment of EVs by M. Menelaus wings. M. Menelaus wings were modified by lipid nanoprobes. When samples through M. Menelaus wings, the lipid nanoprobes can insert into the EV membranes to capture EVs.
Characterization and Detection of Exosomes
Exosomes not only have extremely small particle size, but also have a high heterogeneity and diversity among individual exosomes due to different cell sources, cell states, and secretory pathways (Tkach and Thery, 2016). Thus it is a challenge to accurately detect and characterize exosomes. Here we review some methods for the characterization and detection of exosomes.
Scanning Electron Microscopy (SEM)
SEM is a common method to observe the morphology of exosomes (van der Pol et al., 2010; Sokolova et al., 2011). The surface of the exosome sample is scanned by an electron beam to produce the images. Information about the three-dimensional surface morphology and the elemental composition of the exosomes is obtained through various signals that result from the interaction between the electron beam and atoms in the sample (Shao et al., 2018). Under SEM, most exosomes show a cup-shaped morphology (Thery et al., 2006).
Transmission Electron Microscopy (TEM)
TEM is another method commonly used for exosome characterization (Thery et al., 2006). Compared to SEM, TEM has superior resolution and can resolve objects less than 1nm in size. Using short wavelength electrons, TEM can also detect the cup-shaped morphology of exosomes. Exosome samples need to be stained with special chemical reagents such as osmium tetroxide and phosphotungstic acid in order to be observed by TEM (Shao et al., 2012).
Nanoparticle Tracking Analysis (NTA)
NTA was first applied for exosome detection and characterization in 2011 (Sokolova et al., 2011). NTA is a technique using optical particle to track nanoparticles for measuring concentration and size distribution of the nanoparticles (Dragovic et al., 2011; Sokolova et al., 2011; Gardiner et al., 2013). The principle of NTA is that the random Brownian motion of nanoparticles in liquids can be recorded with a high-speed camera (Bachurski et al., 2019), and this information can be used in the Stokes-Einstein equation to determine the size and concentration of the tracked particles (Dragovic et al., 2011; Kestens et al., 2017). Compared with other methods, NTA is now apply for exosome analysis at the single particle level in many studies.
Dynamic Light Scattering (DLS)
DLS is also lied with the Brownian motion of the particles, and this method can detect particle sizes ranging from 5 to 10 nm to 6 μm through fluctuations in the scattered light intensity (Erdbrugger and Lannigan, 2016). When particles do Brownian motion, the scattered light waves emitted by all particles interferes with each other, and their intensities change over time (Shao et al., 2018). The hydrodynamic radius of the particles is calculated by using the Siegert relationship to convert the intensity autocorrelation function into the scattering electric field correlation function (Sitar et al., 2015).
Tunable Resistive Pulse Sensing (TRPS)
TRPS is an approach to quickly characterizing the particle size distribution and concentration of nanoparticles at the single particle level (Coumans et al., 2014; Vogel et al., 2016). The TRPS device has a membrane with a pore, and a current through the pore is produced by applying a voltage to the membrane (Blundell et al., 2015). The exosome sample is placed on one side of the membrane and only one particle is allowed to pass through the pore at a time under the drive force of the pressure and voltage difference (Koritzinsky et al., 2017). The particle concentration and size are determined by particle passing through the pore frequency and the drop in current, respectively.
Western Blotting and Enzyme-Linked Immunosorbent Assay (ELISA)
Proteins are one of the main components of exosomes, and the quantification and identification of exosomal proteins is essential not only for explaining exosome biogenesis and transport, but also for identifying disease markers (Shao et al., 2018). Western blotting and ELISA are conventional protein analysis techniques. Western blotting, also called immunoblotting, is the most common protein assay approach and is widely used in molecular biology research. This process relies on antibody-protein interactions and generally apply to detect the existence of target proteins in exosomes.
ELISA is another available approach for protein quantification. ELISA also relies on antibody-protein interactions, but it uses a specific antibody attached to an enzyme. The antigen and antibody are directly proportional to the antibody, and the absorbance of the enzyme is measured to quantify the protein (Engvall and Perlmann, 1971; Aydin, 2015; Knight et al., 2018). Compared to western blotting, ELISA can be faster and scaled up for high throughput measurements.
Mass Spectrometry (MS)
MS has high specificity and sensitivity, can not only recognize and characterize the molecular components of vesicles, but can also be used for high-throughput peptide profiling (Kreimer et al., 2015; Pocsfalvi et al., 2016). Sample-preparation for proteomics analysis normally follows three steps: (1) SDS-PAGE separation (Lin et al., 2004; Pisitkun et al., 2004), (2) isoelectric focusing-based fractionation (Choi et al., 2012), and (3) two-dimensional liquid chromatography (Gonzalez-Begne et al., 2009). Over the past decade, the integration of qualitative MS applications and quantitative protein analysis has been greatly improved in terms of sensitivity, resolution, and speed. There are two main technical methodologies for MS quantitation: label-based and label-free quantitation (Choi et al., 2015). Label-free quantitation is more widely applied than label-based.
New Detection Methods
As the development of exosome detection technologies have developed in recent years, more and more studies have focused on the detection of single vesicles. Total internal reflection fluorescence microscopy, based on aptamer fluorescent DNA nanodevice on target exosome surfaces, is an ultra-sensitive method that has been developed to directly quantify and visualize tumor exosomes in plasma samples at the single-vesicle level (He et al., 2019). Localized surface plasmon resonance imaging platform detects single exosome by using a nano-microarray with gold sensing element atop quartz nanopillars binding anti-CD63 (Raghu et al., 2018). Single particle interferometric reflectance imaging sensor, which is based on interference reflectance, and requires LED lights and a COMS camera, is used to count exosomes (Daaboul et al., 2016).
Applications of Exosomes
Exosomes mediate a number of biological processes, including physiological and pathological processes. Exosomes contain various important biomolecules (e.g., proteins and RNAs) that regulate intercellular communications, and clinical applications of exosomes have also achieved breakthroughs (Thery, 2015; Meldolesi, 2018; Huang and Deng, 2019). Currently, most of the exosome research is focused on the tumors, and it has been identified that exosomes encourage cell polarity and control cell motility by altering extracellular matrix (ECM) components (Sung et al., 2015). Exosomes can also stimulate angiogenic activities (Zhang et al., 2016), and tumor-derived exosomes can activate the immune response (Greening et al., 2015). In contrast to research in tumors, few studies have focused on exosomes in diseases of the eye and the inner ear. Based on the roles of tumor-derived exosomes, eye cells-derived exosomes might play similar roles. Consequently, we summary the applications and exosomes’ functions reported in the visual and auditory systems.
Exosomes in the Visual System
The eye is a vital sensory organ, and eye diseases can have significant negative impacts on daily life. Age-related macular degeneration, cataract, diabetic retinopathy, and glaucoma are the four most widespread eye diseases (Saldanha et al., 2017). Aging is the main cause of eye disease, and age-related visual impairment involves corneal endothelial cells’ death, a decline in the number and sensitivity of neurons in the retina, and changes in the tissue structure of eye (Lei et al., 2011; Gambato et al., 2015; Nadal-Nicolás et al., 2018; Wang et al., 2018).
Exosomes have been identified promote cells and the ECM communication and to participate in ECM assembly and adhesion (Hoshino et al., 2013; Mead et al., 2013; Wang et al., 2017). Glaucoma is a common eye disease, and its pathogenesis involves the ECM of the trabecular meshwork (TM), oxidative stress, the TGFβ signaling pathway, and apoptosis (Suri et al., 2018). Human TM explants exposure in dxamethasone induces fibronectin production, and reduces the interaction of exosomes for the fibronectin surface, and this might explain the abnormal amassing of ECM substances in steroid-induced glaucoma patients (Dismuke et al., 2016). Studies have shown that alters in the ECM in the lamina cribrosa and TM are behind the irreparable vision loss in glaucoma (Klingeborn et al., 2017), and illumination the exosome-related defects in the ECM in both tissues will likely lead to a better explanation of the pathogenesis of glaucoma.
Exosomes have been shown effect immune regulation in many diseases, including tumors (Seo et al., 2018), inflammatory diseases (Cypryk et al., 2018), autoimmune diseases (Anel et al., 2019), and neurodegenerative disorders (Tofaris, 2017). However, there have been few studies showing the roles of exosomes in immune regulation in eye diseases. Corneal transplantation is an effective treatment for blindness, but immune rejection is still a significant problem, and studies have indicated that exosomes are involved in recovery process after corneal transplantation (Coulson-Thomas et al., 2013; Harrell et al., 2018). Short collagen-like peptides (CLPs) such as CLP-PGE promote the regeneration of stable corneal tissues and nerves by stimulating the production of exosomes by corneal epithelial cells (Jangamreddy et al., 2018). In addition, mouse-derived exosomes have been identified that as a medium, involved in signal transmission between cells during corneal wound healing (Han et al., 2017).
Mesenchymal stem cell (MSC) derived exosomes show therapeutic potential in various diseases. MSC-derived exosomes can relieve the symptoms of type two diabetes mellitus (Sun et al., 2018), and hypoxia-conditioned bone marrow MSC-derived exosomes can promote heart repair after ischemic injury by reducing myocardial apoptosis (Zhu et al., 2018). Similarly, MSC-derived exosomes have been identified make significant contributions in the visual system. The degree of in vitro wound healing by corneal MSCs (cMSCs) of human corneal epithelial cells is higher than controls, and corneas treated in vivo with cMSC-derived exosomes also have a higher degree of wound healing (Samaeekia et al., 2018). In the rat retinal detachment model, MSC-derived exosomes can inhibit apoptosis of photoreceptor cells and maintain normal retinal structure (Ma et al., 2020). MSC-derived exosomes inhibit the migration of inflammatory cells and decline the infiltration of white blood cells to ease the progress of experimental autoimmune uveitis (Bai et al., 2017). MSC-derived exosomes also improve visual function by downregulating the vascular endothelial growth factor-A produced (He et al., 2018).
Exosomes not only participate in pathological and physiological processes, but also have therapeutic and diagnostic potential. Adeno-associated virus (AAV), a small, non-enveloped virus (Samulski and Muzyczka, 2014), is often used as a gene therapy carrier with its high efficiency and safety (Wassmer et al., 2017). Exosomes (exo) can be combined with AVV to produce exo-AAVs that have higher gene transfer efficiency compared to AVVs alone (Hudry et al., 2016). Wassmer et al. packaged the green fluorescent protein gene into AAV2 and exo-AAV2 vectors and injected them into mouse eyes through intravitreal (IVT). The data indicated that the exo-AAV2 vector was superior compare to AAV in terms of delivery and retinal transduction and that exo-AAV2 could enhance gene transduction in the ganglion cell layer and in the deep retina (Wassmer et al., 2017). Owing to exosomes can pass across the blood-brain barrier, and the endothelial barrier (El Andaloussi et al., 2013a; Hudry et al., 2016), they might provide a chance to treat of eye diseases.
Exosomal RNA is significantly increased in both myopic and normal aqueous humor (AH) samples, and there are several specific miRNAs that might be useful biomarkers of myopia (Chen et al., 2019). Exosomes are widely found in all kinds of fluids, including tears, so it might be possible to find exosomal markers of in eye diseases from tears. Some studies have identified the appearance of exosomal marker proteins in tears, such as CD9, CD63 (Aass et al., 2015; Matheis et al., 2015; Grigor’eva et al., 2016).
Exosomes in Hearing System
Deafness is a common sensory disease and sensing hair cells (HCs) in the inner ear are sensors for recognizing sound. In mammals, HC loss due to noise, aging, and ototoxic drugs is irreparable and is thus the major cause of permanent hearing loss (Brigande and Heller, 2009; Furness, 2015). Recent studies showed that exosomes are present in the inner ear, and the release of inner ear exosomes is decreased and the protein profile of exosomes is significantly changed after treatment with ototoxic drugs such as neomycin and cisplatin (Wong et al., 2018). Exo-AAV has been successfully used as a non-toxic and noval gene vector for gene therapy of retinal diseases in mouse models, and György et al. have demonstrated that exo-AAV is also an effective gene vector for inner ear HCs (György et al., 2017). The addition of the exo-AAV vector to the in vitro culture medium showed that about 95% of the vector was transduced into inner and outer HCs (IHCs and OHCs). The vector was also injected into the mouse cochlea in vivo through round window (RW), and the transduction efficiency in IHCs and OHCs was 88 and 25%, respectively. LHFPL5, also called Tmhs, is an essential component of the mechanical transduction mechanism of OHCs and IHCs, and its deletion can cause hearing loss (Xiong et al., 2012). Exo-AAV- HA-Lhfpl5 injection through RW could partically rescue hearing in Lhfpl5-/- mice (György et al., 2017), thus exo-AAV vectors might provide a new method in gene therapy for treating clinical deafness.
Exosomes can mediate intercellular communication between different cells by delivery of exosomal proteins, nucleic acid and lipids to recipient cells (Colombo et al., 2014). Heat-shock 70-kDa protein (HSP70) is widely found in exosomes (Fonseca et al., 2016), and previous study by Lisa L Cunningham et al. showed that supporting cells (SCs) require HSP70 to protect HCs (May et al., 2013). Recently, they found that SCs derived exosomes are significantly increased by heat shock, and these exosomes can improve promote HCs survival under neomycin exposure. Furthermore, they found that HSP70 in SCs-derived exosomes interact with TLR4 receptors on the cell membrane of HCs, which can protect HCs from damage (Breglio et al., 2020).
Exosomes are likely associated with the pathological processes of inner ear diseases, for instance sensorineural hearing loss and genetic deafness. At present, although there are only a few reports on exosomes in the auditory system, we believe that exosomes will make significant contributions in future clinical treatment of inner ear diseases and prediction of deafness.
Conclusion and Perspectives
Exosomes contain abundant proteins and nucleic acids, and they are considered to be a medium for transmitting information between cells. Exosomes have significant functions not only in the metastasis and growth of tumor cells, but also in normal physiological processes. The phospholipid bilayer structure and rich proteins and nucleic acids of exosomes also make them useful as biomarkers for medical diagnose. Due to their heterogeneity and not restricted by the blood-brain barrier, exosomes can design as targeted drug-delivery vehicles, and will likely be important components of novel therapeutic strategies.
Many new methods for analyzing exosomes have been developed in recent years, but the isolation and analysis of exosomes remains a challenge because the molecular interactions and the precise functions of exosomes are still difficult to analyze. New technologies for single exosome analysis are required to reveal the unique molecular functions and diversity of exosomes. Exosomes exist not only in various body fluids, but also in tissues, and the requirement for extraction specified steps are different in different samples. Therefore, further studying the functions of exosomes in disease processes, it is vital to establish standard protocols based on different exosomal sample types to be extracted to avoid influence from human normal physiological factors.
Research on exosomes in the visual and auditory systems is still a relatively young field, especially the field of hearing, and the related research is summarized in Figure 4. To further study exosomes in the field of vision and hearing, it is necessary to develop animal models for exosom research, such as mouse, zebrafish or Drosophila models. With the help of animal models, we can more clearly explain the function of exosomes. It was reported that exosome reporter mice has been used to show exosomes involved in central nervous system communication (Men et al., 2019).
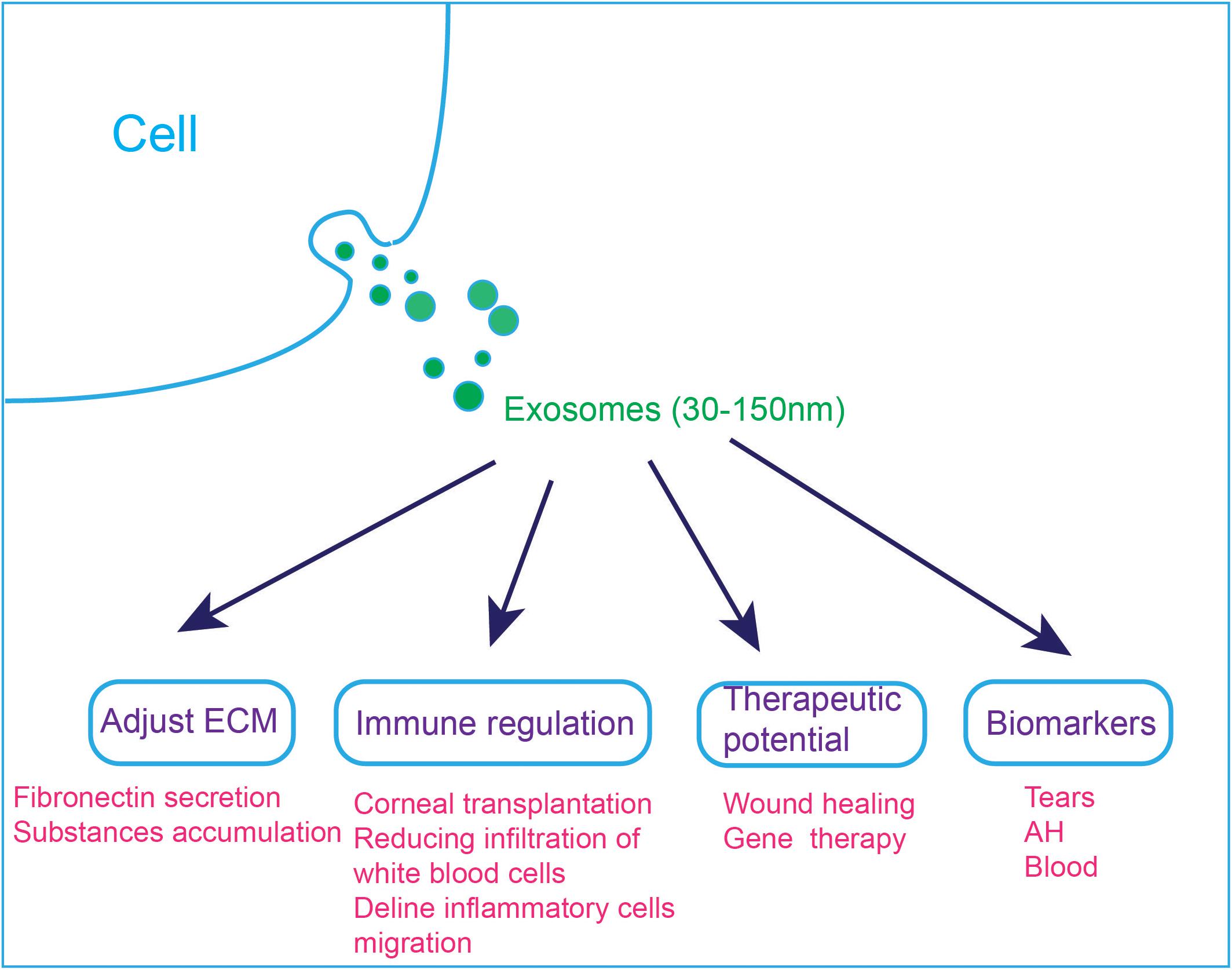
Figure 4. Applications of exosomes in the visual and auditory systems. (1) Exosomes are involved in cell ECM changes. Dexamethasone treatment decreases the affinity of exosomes to fibronectin, which can explain the abnormal accumulation of ECM substances in steroid-induced glaucoma patients. (2) Exosomes are involved in immune regulation, and they can reduce immune rejection after corneal transplantation. MSC-derived exosomes ameliorate autoimmune uveoretinitis by inhibit the migration of inflammatory cells and decline the infiltration of white blood cells into the eye. (3) Exosomes can accelerate wound healing and exo-AAVs are considered to be a novel gene therapy tool. (4) Exosomes in tears, AH and blood have the potential to be used as diagnostic markers.
The discovery of more and more exosomal disease markers and the chance as a novel drug delivery vehicle are expected to apply in disease judgment, and thus improve patients’ outcomes.
Author Contributions
PJ and SZ wrote the manuscript. CC, SG, and MT collated the resource. LL, GY, and RC wrote and reviewed the manuscript.
Funding
This work was supported by grants from National Key R&D Program of China (No. 2017YFA0103903), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010303), National Natural Science Foundation of China (Nos. 81970882 and 81970892), Natural Science Foundation from Jiangsu Province (BE2019711 and BK20190062), Boehringer Ingelheim Pharma GmbH, K. C. Wong Education Foundation, Jiangsu Provincial Medical Youth Talent of the Project of Invigorating Health Care through Science, Technology and Education (QNRC2016002), National Science Foundation for Young Scientists of China (81700913), and the Fundamental Research Funds for the Central Universities (2242020R40137).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aass, C., Norheim, I., Eriksen, E. F., Thorsby, P. M., and Pepaj, M. (2015). Single unit filter-aided method for fast proteomic analysis of tear fluid. Anal. Biochem. 480, 1–5. doi: 10.1016/j.ab.2015.04.002
Anel, A., Gallego-Lleyda, A., de Miguel, D., Naval, J., and Martínez-Lostao, L. (2019). Role of exosomes in the regulation of t-cell mediated immune responses and in autoimmune disease. Cells 8:154. doi: 10.3390/cells8020154
Arrighetti, N., Corbo, C., Evangelopoulos, M., Pastò, A., Zuco, V., and Tasciotti, E. (2019). Exosome-like nanovectors for drug delivery in cancer. Curr. Med. Chem. 26, 6132–6148. doi: 10.2174/0929867325666180831150259
Aydin, S. (2015). A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 72, 4–15. doi: 10.1016/j.peptides.2015.04.012
Bachurski, D., Schuldner, M., Nguyen, P. H., Malz, A., Reiners, K. S., Grenzi, P. C., et al. (2019). Extracellular vesicle measurements with nanoparticle tracking analysis - An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 8:1596016. doi: 10.1080/20013078.2019.1596016
Bai, L., Shao, H., Wang, H., Zhang, Z., Su, C., Dong, L., et al. (2017). Effects of mesenchymal stem cell-derived exosomes on experimental autoimmune uveitis. Sci. Rep. 7:4323. doi: 10.1038/s41598-017-04559-y
Barok, M., Puhka, M., Vereb, G., Szollosi, J., Isola, J., and Joensuu, H. (2018). Cancer-derived exosomes from HER2-positive cancer cells carry trastuzumab-emtansine into cancer cells leading to growth inhibition and caspase activation. BMC Cancer 18:504. doi: 10.1186/s12885-018-4418-2
Bianco, A., Bisceglia, L., Russo, L., Palese, L. L., D’Agruma, L., Emperador, S., et al. (2017). High mitochondrial DNA copy number is a protective factor from vision loss in Heteroplasmic Leber’s Hereditary Optic Neuropathy (LHON). Invest. Ophthalmol. Vis. Sci. 58, 2193–2197. doi: 10.1167/iovs.16-20389
Blans, K., Hansen, M. S., Sorensen, L. V., Hvam, M. L., Howard, K. A., Moller, A., et al. (2017). Pellet-free isolation of human and bovine milk extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 6:1294340. doi: 10.1080/20013078.2017.1294340
Blundell, E., Mayne, L. J., Billinge, E. R., and Platt, M. (2015). Emergence of tunable resistive pulse sensing as a biosensor. Anal. Methods 7, 7055–7066. doi: 10.1039/c4ay03023k
Boing, A. N., van der Pol, E., Grootemaat, A. E., Coumans, F. A. W., Sturk, A., and Nieuwland, R. (2014). Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 3:23430. doi: 10.3402/jev.v3.23430
Breglio, A. M., May, L. A., Barzik, M., Welsh, N. C., Francis, S. P., Costain, T. Q., et al. (2020). Exosomes mediate sensory hair cell protection in the inner ear. J. Clin. Investig. 130, 2657–2672. doi: 10.1172/JCI128867
Brigande, J. V., and Heller, S. (2009). Quo vadis, hair cell regeneration? Nat. Neurosci. 12, 679–685. doi: 10.1038/nn.2311
Brownlee, Z., Lynn, K. D., Thorpe, P. E., and Schroit, A. J. (2014). A novel “salting-out” procedure for the isolation of tumor-derived exosomes. J. Immunol. Methods 407, 120–126. doi: 10.1016/j.jim.2014.04.003
Burgess, R. R. (2018). A brief practical review of size exclusion chromatography: rules of thumb, limitations, and troubleshooting. Protein Expr. Purif. 150, 81–85. doi: 10.1016/j.pep.2018.05.007
Chen, C.-F., Hua, K., Woung, L.-C., Lin, C.-H., Chen, C.-T., Hsu, C.-H., et al. (2019). Expression profiling of exosomal miRNAs derived from the aqueous humor of myopia patients. Tohoku J. Exp. Med. 249, 213–221. doi: 10.1620/tjem.249.213
Choi, D.-S., Choi, D.-Y., Hong, B. S., Jang, S. C., Kim, D.-K., Lee, J., et al. (2012). Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J. Extracell. Vesicles 1:10.3402/jev.v1i0.18704. doi: 10.3402/jev.v1i0.18704
Choi, D. S., Kim, D. K., Kim, Y. K., and Gho, Y. S. (2015). Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom. Rev. 34, 474–490. doi: 10.1002/mas.21420
Colombo, M., Raposo, G., and Théry, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289. doi: 10.1146/annurev-cellbio-101512-122326
Coulson-Thomas, V. J., Caterson, B., and Kao, W. W. (2013). Transplantation of human umbilical mesenchymal stem cells cures the corneal defects of mucopolysaccharidosis VII mice. Stem Cells 31, 2116–2126. doi: 10.1002/stem.1481
Coumans, F. A. W., van der Pol, E., Boing, A. N., Hajji, N., Sturk, G., van Leeuwen, T. G., et al. (2014). Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J. Extracell. Vesicles 3:25922. doi: 10.3402/jev.v3.25922
Crewe, C., Joffin, N., Rutkowski, J. M., Kim, M., Zhang, F., Towler, D. A., et al. (2018). An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell 175, 695–708.e13. doi: 10.1016/j.cell.2018.09.005
Cypryk, W., Nyman, T. A., and Matikainen, S. (2018). From inflammasome to exosome-does extracellular vesicle secretion constitute an inflammasome-dependent immune response? Front. Immunol. 9:2188. doi: 10.3389/fimmu.2018.02188
Daaboul, G. G., Gagni, P., Benussi, L., Bettotti, P., Ciani, M., Cretich, M., et al. (2016). Digital detection of exosomes by interferometric imaging. Sci. Rep. 6, 37246–37246. doi: 10.1038/srep37246
Deregibus, M. C., Figliolini, F., D’Antico, S., Manzini, P. M., Pasquino, C., De Lena, M., et al. (2016). Charge-based precipitation of extracellular vesicles. Int. J. Mol. Med. 38, 1359–1366. doi: 10.3892/ijmm.2016.2759
Dismuke, W. M., Klingeborn, M., and Stamer, W. D. (2016). Mechanism of fibronectin binding to human trabecular meshwork exosomes and its modulation by dexamethasone. PLoS One 11:e0165326. doi: 10.1371/journal.pone.0165326
Dragovic, R. A., Gardiner, C., Brooks, A. S., Tannetta, D. S., Ferguson, D. J. P., Hole, P., et al. (2011). Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 7, 780–788. doi: 10.1016/j.nano.2011.04.003
El Andaloussi, S., Lakhal, S., Mäger, I., and Wood, M. J. A. (2013a). Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 65, 391–397. doi: 10.1016/j.addr.2012.08.008
El Andaloussi, S., Maeger, I., Breakefield, X. O., and Wood, M. J. A. (2013b). Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 348–358. doi: 10.1038/nrd3978
Engvall, E., and Perlmann, P. (1971). “Enzyme-linked immunosorbent assay (ELISA),” in Proceedings of the Nineteenth Colloquium, Protides of the Biological Fluids, (Brugge: Pergamon Press), 553–556.
Erdbrugger, U., and Lannigan, J. (2016). Analytical challenges of extracellular vesicle detection: a comparison of different techniques. Cytometry Part A 89A, 123–134. doi: 10.1002/cyto.a.22795
Farooqi, A. A., Desai, N. N., Qureshi, M. Z., Librelotto, D. R. N., Gasparri, M. L., Bishayee, A., et al. (2018). Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 36, 328–334. doi: 10.1016/j.biotechadv.2017.12.010
Fonseca, P., Vardaki, I., Occhionero, A., and Panaretakis, T. (2016). Metabolic and signaling functions of cancer cell-derived extracellular vesicles. Int. Rev. Cell Mol. Biol. 326, 175–199. doi: 10.1016/bs.ircmb.2016.04.004
Furness, D. N. (2015). Molecular basis of hair cell loss. Cell Tissue Res. 361, 387–399. doi: 10.1007/s00441-015-2113-z
Gallart-Palau, X., Serra, A., and Sze, S. K. (2016). Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol. Neurodegener. 11:13. doi: 10.1186/s13024-016-0108-1
Gambato, C., Longhin, E., Catania, A. G., Lazzarini, D., Parrozzani, R., and Midena, E. (2015). Aging and corneal layers: an in vivo corneal confocal microscopy study. Graefes Arch. Clin. Exp. Ophthalmol. 253, 267–275. doi: 10.1007/s00417-014-2812-2
Gardiner, C., Di Vizio, D., Sahoo, S., Thery, C., Witwer, K. W., Wauben, M., et al. (2016). Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J. Extracell. Vesicles 5:32945. doi: 10.3402/jev.v5.32945
Gardiner, C., Ferreira, Y. J., Dragovic, R. A., Redman, C. W. G., and Sargent, I. L. (2013). Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2:19671. doi: 10.3402/jev.v2i0.19671
Gonzalez-Begne, M., Lu, B. W., Han, X. M., Hagen, F. K., Hand, A. R., Melvin, J. E., et al. (2009). Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J. Proteome Res. 8, 1304–1314. doi: 10.1021/pr800658c
Greening, D. W., Gopal, S. K., Xu, R., Simpson, R. J., and Chen, W. S. (2015). Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 40, 72–81. doi: 10.1016/j.semcdb.2015.02.009
Grigor’eva, A. E., Tamkovich, S. N., Eremina, A. V., Tupikin, A. E., Kabilov, M. R., Chernykh, V. V., et al. (2016). Characteristics of exosomes andmicroparticles discovered in human tears. Biomed. Khim. 62, 99–106. doi: 10.18097/PBMC20166201099
Gurunathan, S., Kang, M.-H., Jeyaraj, M., Qasim, M., and Kim, J.-H. (2019). Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 8:307. doi: 10.3390/cells8040307
György, B., Sage, C., Indzhykulian, A. A., Scheffer, D. I., Brisson, A. R., Tan, S., et al. (2017). Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol. Ther. 25, 379–391. doi: 10.1016/j.ymthe.2016.12.010
Han, K.-Y., Tran, J. A., Chang, J.-H., Azar, D. T., and Zieske, J. D. (2017). Potential role of corneal epithelial cell-derived exosomes in corneal wound healing and neovascularization. Sci. Rep. 7:40548. doi: 10.1038/srep40548
Han, S., Xu, Y., Sun, J., Liu, Y., Zhao, Y., Tao, W., et al. (2020). Isolation and analysis of extracellular vesicles in a Morpho butterfly wing-integrated microvortex biochip. Biosens. Bioelectron. 154:112073. doi: 10.1016/j.bios.2020.112073
Harrell, C. R., Simovic Markovic, B., Fellabaum, C., Arsenijevic, A., Djonov, V., Arsenijevic, N., et al. (2018). Therapeutic potential of mesenchymal stem cell-derived exosomes in the treatment of eye diseases. Adv. Exp. Med. Biol. 1089, 47–57. doi: 10.1007/5584_2018_219
He, D., Ho, S.-L., Chan, H.-N., Wang, H., Hai, L., He, X., et al. (2019). Molecular-recognition-based DNA nanodevices for enhancing the direct visualization and quantification of single vesicles of tumor exosomes in plasma microsamples. Anal. Chem. 91, 2768–2775. doi: 10.1021/acs.analchem.8b04509
He, G.-H., Zhang, W., Ma, Y.-X., Yang, J., Chen, L., Song, J., et al. (2018). Mesenchymal stem cells-derived exosomes ameliorate blue light stimulation in retinal pigment epithelium cells and retinal laser injury by VEGF-dependent mechanism. Int. J. Ophthalmol. 11, 559–566. doi: 10.18240/ijo.2018.04.04
Heavner, W., and Pevny, L. (2012). Eye development and retinogenesis. Cold Spring Harb. Perspect. Biol. 4:a008391. doi: 10.1101/cshperspect.a008391
Hoshino, D., Kirkbride, K. C., Costello, K., Clark, E. S., Sinha, S., Grega-Larson, N., et al. (2013). Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 5, 1159–1168. doi: 10.1016/j.celrep.2013.10.050
Huang, T., and Deng, C. X. (2019). Current progresses of exosomes as cancer diagnostic and prognostic biomarkers. Int. J. Biol. Sci. 15, 1–11. doi: 10.7150/ijbs.27796
Hudry, E., Martin, C., Gandhi, S., György, B., Scheffer, D. I., Mu, D., et al. (2016). Exosome-associated AAV vector as a robust and convenient neuroscience tool. Gene Ther. 23, 380–392. doi: 10.1038/gt.2016.11
Ibrahim, A., and Marban, E. (2016). Exosomes: fundamental biology and roles in cardiovascular physiology. Annu. Rev. Physiol. 78, 67–83. doi: 10.1146/annurev-physiol-021115-104929
Jangamreddy, J. R., Haagdorens, M. K. C., Mirazul Islam, M., Lewis, P., Samanta, A., Fagerholm, P., et al. (2018). Short peptide analogs as alternatives to collagen in pro-regenerative corneal implants. Acta Biomater. 69, 120–130. doi: 10.1016/j.actbio.2018.01.011
Johnstone, R. M., Adam, M., Hammond, J. R., Orr, L., and Turbide, C. (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262, 9412–9420.
Karanjia, R., Chahal, J., Ammar, M., and Sadun, A. A. (2017). Treatment of Leber’s hereditary optic neuropathy. Curr. Pharm. Des. 23, 624–628. doi: 10.2174/1381612823666170125164856
Kestens, V., Bozatzidis, V., De Temmerman, P.-J., Ramaye, Y., and Roebben, G. (2017). Validation of a particle tracking analysis method for the size determination of nano- and microparticles. J. Nanopart. Res. 19:271. doi: 10.1007/s11051-017-3966-8
Kibria, G., Ramos, E. K., Wan, Y., Gius, D. R., and Liu, H. (2018). Exosomes as a drug delivery system in cancer therapy: potential and challenges. Mol. Pharm. 15, 3625–3633. doi: 10.1021/acs.molpharmaceut.8b00277
Klingeborn, M., Dismuke, W. M., Bowes Rickman, C., and Stamer, W. D. (2017). Roles of exosomes in the normal and diseased eye. Prog. Retin. Eye Res. 59, 158–177. doi: 10.1016/j.preteyeres.2017.04.004
Klingeborn, M., Stamer, W. D., and Bowes Rickman, C. (2018). Polarized exosome release from the retinal pigmented epithelium. Adv. Exp. Med. Biol. 1074, 539–544. doi: 10.1007/978-3-319-75402-4_65
Knight, A. R., Taylor, E. L., Lukaszewski, R., Jensen, K. T., Jones, H. E., Carre, J. E., et al. (2018). A high-sensitivity electrochemiluminescence-based ELISA for the measurement of the oxidative stress biomarker, 3-nitrotyrosine, in human blood serum and cells. Free Radic. Biol. Med. 120, 246–254. doi: 10.1016/j.freeradbiomed.2018.03.026
Koritzinsky, E. H., Street, J. M., Star, R. A., and Yuen, P. S. T. (2017). Quantification of exosomes. J. Cell. Physiol. 232, 1587–1590. doi: 10.1002/jcp.25387
Kreimer, S., Belov, A. M., Ghiran, I., Murthy, S. K., Frank, D. A., and Ivanov, A. R. (2015). Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J. Proteome Res. 14, 2367–2384. doi: 10.1021/pr501279t
Kreimer, S., and Ivanov, A. R. (2017). Rapid Isolation of extracellular vesicles from blood plasma with size-exclusion chromatography followed by mass spectrometry-based proteomic profiling. Methods Mol. Biol. 1660, 295–302. doi: 10.1007/978-1-4939-7253-1_24
Langevin, S. M., Kuhnell, D., Orr-Asman, M. A., Biesiada, J., Zhang, X., Medvedovic, M., et al. (2019). Balancing yield, purity and practicality: a modified differential ultracentrifugation protocol for efficient isolation of small extracellular vesicles from human serum. RNA Biol. 16, 5–12. doi: 10.1080/15476286.2018.1564465
Lei, Y., Garrahan, N., Hermann, B., Fautsch, M. P., Johnson, D. H., Hernandez, M. R., et al. (2011). Transretinal degeneration in ageing human retina: a multiphoton microscopy analysis. Br. J. Ophthalmol. 95, 727–730. doi: 10.1136/bjo.2010.180869
Lim, J., Choi, M., Lee, H., Kim, Y. H., Han, J. Y., Lee, E. S., et al. (2019). Direct isolation and characterization of circulating exosomes from biological samples using magnetic nanowires. J. Nanobiotechnol. 17:1. doi: 10.1186/s12951-018-0433-3
Lin, B., Utleg, A. G., Yi, E. C., Xie, T., Shannon, P., White, J. T., et al. (2004). Proteomic analysis of human prostasomes. Mol. Cell. Proteomics 3:S38.
Lozano-Ramos, I., Bancu, I., Oliveira-Tercero, A., Pilar Armengol, M., Menezes-Neto, A., Del Portillo, H. A., et al. (2015). Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles 4:27369. doi: 10.3402/jev.v4.27369
Ma, M., Li, B., Zhang, M., Zhou, L., Yang, F., Ma, F., et al. (2020). Therapeutic effects of mesenchymal stem cell-derived exosomes on retinal detachment. Exp. Eye Res. 191:107899. doi: 10.1016/j.exer.2019.107899
Matheis, N., Lantz, M., Grus, F. H., Ponto, K. A., Wolters, D., Brorson, H., et al. (2015). Proteomics of orbital tissue in thyroid-associated orbitopathy. J. Clin. Endocrinol. Metab. 100, E1523–E1530. doi: 10.1210/jc.2015-2976
May, L. A., Kramarenko, I. I., Brandon, C. S., Voelkel-Johnson, C., Roy, S., Truong, K., et al. (2013). Inner ear supporting cells protect hair cells by secreting HSP70. J. Clin. Invest. 123, 3577–3587. doi: 10.1172/jci68480
Mead, B., Logan, A., Berry, M., Leadbeater, W., and Scheven, B. A. (2013). Intravitreally transplanted dental pulp stem cells promote neuroprotection and axon regeneration of retinal ganglion cells after optic nerve injury. Investig. Ophthalmol. Vis. Sci. 54, 7544–7556. doi: 10.1167/iovs.13-13045
Meldolesi, J. (2018). Exosomes and ectosomes in intercellular communication. Curr. Biol. 28, R435–R444. doi: 10.1016/j.cub.2018.01.059
Melo, S. A., Luecke, L. B., Kahlert, C., Fernandez, A. F., Gammon, S. T., Kaye, J., et al. (2015). Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182. doi: 10.1038/nature14581
Men, Y. Q., Yelick, J., Jin, S. J., Tian, Y., Chiang, M. S. R., Higashimori, H., et al. (2019). Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat. Commun. 10:4136. doi: 10.1038/s41467-019-11534-w
Milane, L., Singh, A., Mattheolabakis, G., Suresh, M., and Amiji, M. M. (2015). Exosome mediated communication within the tumor microenvironment. J. Control. Release 219, 278–294. doi: 10.1016/j.jconrel.2015.06.029
Nadal-Nicolás, F. M., Vidal-Sanz, M., and Agudo-Barriuso, M. (2018). The aging rat retina: from function to anatomy. Neurobiol. Aging 61, 146–168. doi: 10.1016/j.neurobiolaging.2017.09.021
Pisitkun, T., Shen, R. F., and Knepper, M. A. (2004). Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 101, 13368–13373. doi: 10.1073/pnas.0403453101
Pocsfalvi, G., Stanly, C., Vilasi, A., Fiume, I., Capasso, G., Turiak, L., et al. (2016). Mass spectrometry of extracellular vesicles. Mass Spectrom. Rev. 35, 3–21. doi: 10.1002/mas.21457
Poe, A. J., and Knowlton, A. A. (2018). Exosomes and cardiovascular cell-cell communication. Essays Biochem. 62, 193–204. doi: 10.1042/EBC20170081
Raghu, D., Christodoulides, J. A., Christophersen, M., Liu, J. L., Anderson, G. P., Robitaille, M., et al. (2018). Nanoplasmonic pillars engineered for single exosome detection. PLoS One 13:e0202773. doi: 10.1371/journal.pone.0202773
Rance, G., Kearns, L. S., Tan, J., Gravina, A., Rosenfeld, L., Henley, L., et al. (2012). Auditory function in individuals within Leber’s hereditary optic neuropathy pedigrees. J. Neurol. 259, 542–550. doi: 10.1007/s00415-011-6230-7
Rodrigues, G., Hoshino, A., Kenific, C. M., Matei, I. R., Steiner, L., Freitas, D., et al. (2019). Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 21, 1403–1412. doi: 10.1038/s41556-019-0404-4
Saldanha, I. J., Lindsley, K., Do, D. V., Chuck, R. S., Meyerle, C., Jones, L. S., et al. (2017). Comparison of clinical trial and systematic review outcomes for the 4 most prevalent eye diseases. JAMA Ophthalmol. 135, 933–940. doi: 10.1001/jamaophthalmol.2017.2583
Samaeekia, R., Rabiee, B., Putra, I., Shen, X., Park, Y. J., Hematti, P., et al. (2018). Effect of human corneal mesenchymal stromal cell-derived exosomes on corneal epithelial wound healing. Investig. Ophthalmol. Vis. Sci. 59, 5194–5200. doi: 10.1167/iovs.18-24803
Samulski, R. J., and Muzyczka, N. (2014). AAV-mediated gene therapy for research and therapeutic purposes. Annu. Rev. Virol. 1, 427–451. doi: 10.1146/annurev-virology-031413-085355
Seo, N., Akiyoshi, K., and Shiku, H. (2018). Exosome-mediated regulation of tumor immunology. Cancer Sci. 109, 2998–3004. doi: 10.1111/cas.13735
Sexton, R. E., Mpilla, G., Kim, S., Philip, P. A., and Azmi, A. S. (2019). Ras and exosome signaling. Semin. Cancer Biol. 54, 131–137. doi: 10.1016/j.semcancer.2019.02.004
Shao, H. L., Chung, J., Balaj, L., Charest, A., Bigner, D. D., Carter, B. S., et al. (2012). Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 18, 1835–1840. doi: 10.1038/nm.2994
Shao, H. L., Im, H., Castro, C. M., Breakefield, X., Weissleder, R., and Lee, H. H. (2018). New technologies for analysis of extracellular vesicles. Chem. Rev. 118, 1917–1950. doi: 10.1021/acs.chemrev.7b00534
Sitar, S., Kejzar, A., Pahovnik, D., Kogej, K., Tusek-Znidaric, M., Lenassi, M., et al. (2015). Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal. Chem. 87, 9225–9233. doi: 10.1021/acs.analchem.5b01636
Sokolova, V., Ludwig, A.-K., Hornung, S., Rotan, O., Horn, P. A., Epple, M., et al. (2011). Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf. BBiointerfaces 87, 146–150. doi: 10.1016/j.colsurfb.2011.05.013
Subra, C., Grand, D., Laulagnier, K., Stella, A., Lambeau, G., Paillasse, M., et al. (2010). Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 51, 2105–2120. doi: 10.1194/jlr.M003657
Sun, W., Li, Z., Zhou, X., Yang, G., and Yuan, L. (2019). Efficient exosome delivery in refractory tissues assisted by ultrasound-targeted microbubble destruction. Drug Deliv. 26, 45–50. doi: 10.1080/10717544.2018.1534898
Sun, Y., Shi, H., Yin, S., Ji, C., Zhang, X., Zhang, B., et al. (2018). Human mesenchymal stem cell derived exosomes alleviate type 2 diabetes mellitus by reversing peripheral insulin resistance and relieving β-cell destruction. ACS Nano 12, 7613–7628. doi: 10.1021/acsnano.7b07643
Sung, B. H., Ketova, T., Hoshino, D., Zijlstra, A., and Weaver, A. M. (2015). Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 6:7164. doi: 10.1038/ncomms8164
Suri, F., Yazdani, S., and Elahi, E. (2018). LTBP2 knockdown and oxidative stress affect glaucoma features including TGFβ pathways, ECM genes expression and apoptosis in trabecular meshwork cells. Gene 673, 70–81. doi: 10.1016/j.gene.2018.06.038
Tao, W. A., Chen, I. H., and Iliuk, A. (2017). Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. FASEB J. 31:1. doi: 10.1073/pnas.1618088114
Tauro, B. J., Greening, D. W., Mathias, R. A., Ji, H., Mathivanan, S., Scott, A. M., et al. (2012). Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56, 293–304. doi: 10.1016/j.ymeth.2012.01.002
Taylor, D. D., and Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21. doi: 10.1016/j.ygyno.2008.04.033
Thery, C., Amigorena, S., Raposo, G., and Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3:Unit3.22. doi: 10.1002/0471143030.cb0322s30
Tkach, M., and Thery, C. (2016). Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232. doi: 10.1016/j.cell.2016.01.043
Tofaris, G. K. (2017). A critical assessment of exosomes in the pathogenesis and stratification of Parkinson’s disease. J. Parkinsons Dis. 7, 569–576. doi: 10.3233/JPD-171176
van der Pol, E., Hoekstra, A. G., Sturk, A., Otto, C., van Leeuwen, T. G., and Nieuwland, R. (2010). Optical and non-optical methods for detection and characterization of microparticles and exosomes. J. Thromb. Haemost. 8, 2596–2607. doi: 10.1111/j.1538-7836.2010.04074.x
van Niel, G., D’Angelo, G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228. doi: 10.1038/nrm.2017.125
Vogel, R., Coumans, F. A. W., Maltesen, R. G., Boing, A. N., Bonnington, K. E., Broekman, M. L., et al. (2016). A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing. J. Extracell. Vesicles 5:31242. doi: 10.3402/jev.v5.31242
Wang, L., Hu, L., Zhou, X., Xiong, Z., Zhang, C., Shehada, H. M. A., et al. (2017). Exosomes secreted by human adipose mesenchymal stem cells promote scarless cutaneous repair by regulating extracellular matrix remodelling. Sci. Rep. 7:13321. doi: 10.1038/s41598-017-12919-x
Wang, Y., Grenell, A., Zhong, F., Yam, M., Hauer, A., Gregor, E., et al. (2018). Metabolic signature of the aging eye in mice. Neurobiol. Aging 71, 223–233. doi: 10.1016/j.neurobiolaging.2018.07.024
Wassmer, S. J., Carvalho, L. S., György, B., Vandenberghe, L. H., and Maguire, C. A. (2017). Exosome-associated AAV2 vector mediates robust gene delivery into the murine retina upon intravitreal injection. Sci. Rep. 7:45329. doi: 10.1038/srep45329
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442, 368–373. doi: 10.1038/nature05058
Witwer, K. W., Buzas, E. I., Bemis, L. T., Bora, A., Lasser, C., Lotvall, J., et al. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2:20360. doi: 10.3402/jev.v2i0.20360
Wong, E. H. C., Dong, Y. Y., Coray, M., Cortada, M., Levano, S., Schmidt, A., et al. (2018). Inner ear exosomes and their potential use as biomarkers. PLoS One 13:e0198029. doi: 10.1371/journal.pone.0198029
Woo, J., Sharma, S., and Gimzewski, J. (2016). The role of isolation methods on a nanoscale surface structure and its effect on the size of exosomes. J. Circ. Biomark. 5:11. doi: 10.5772/64148
Xiong, W., Grillet, N., Elledge, H. M., Wagner, T. F. J., Zhao, B., Johnson, K. R., et al. (2012). TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell 151, 1283–1295. doi: 10.1016/j.cell.2012.10.041
Żak, M., Klis, S. F., and Grolman, W. (2015). The Wnt and Notch signalling pathways in the developing cochlea: formation of hair cells and induction of regenerative potential. Int. J. Dev. Neurosci. 47(Pt B), 247–258. doi: 10.1016/j.ijdevneu.2015.09.008
Zhang, J., Li, S., Li, L., Li, M., Guo, C., Yao, J., et al. (2015). Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13, 17–24. doi: 10.1016/j.gpb.2015.02.001
Zhang, J. Y., Chen, C. Y., Hu, B., Niu, X., Liu, X. L., Zhang, G. W., et al. (2016). Exosomes derived from human endothelial progenitor cells accelerate cutaneous wound healing by promoting angiogenesis through Erk1/2 signaling. Int. J. Biol. Sci. 12, 1472–1487. doi: 10.7150/ijbs.15514
Zhang, M. D., Jin, K., Gao, L., Zhang, Z. K., Li, F., Zhou, F. F., et al. (2018). Methods and technologies for exosome isolation and characterization. Small Methods 2:1800021. doi: 10.1002/smtd.201800021
Zhang, S., Chuah, S. J., Lai, R. C., Hui, J. H. P., Lim, S. K., and Toh, W. S. (2018). MSC exosomes mediate cartilage repair by enhancing proliferation, attenuating apoptosis and modulating immune reactivity. Biomaterials 156, 16–27. doi: 10.1016/j.biomaterials.2017.11.028
Keywords: exosome, biogenesis, composition, isolation, eye, hair cell
Citation: Jiang P, Zhang S, Cheng C, Gao S, Tang M, Lu L, Yang G and Chai R (2020) The Roles of Exosomes in Visual and Auditory Systems. Front. Bioeng. Biotechnol. 8:525. doi: 10.3389/fbioe.2020.00525
Received: 15 April 2020; Accepted: 04 May 2020;
Published: 03 June 2020.
Edited by:
Mingqiang Li, Sun Yat-sen University, ChinaReviewed by:
Renjun Pei, Suzhou Institute of Nano-tech and Nano-bionics (CAS), ChinaXi Chen, Nanjing University, China
Jia Huang, Tongji University, China
Copyright © 2020 Jiang, Zhang, Cheng, Gao, Tang, Lu, Yang and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renjie Chai, cmVuamllY0BzZXUuZWR1LmNu; Guang Yang, Z3lhbmczMjFAaG90bWFpbC5jb20=; Ling Lu, ZW50bHVsaW5nNjBAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Pei Jiang1†
Pei Jiang1† Song Gao
Song Gao Mingliang Tang
Mingliang Tang Renjie Chai
Renjie Chai