- 1State Key Laboratory of Chemical Raesource Engineering, Beijing University of Chemical Technology, Beijing, China
- 2College of Engineering, University of Georgia, Athens, GA, United States
Flavonoids are a class of secondary metabolites found in plant and fungus. They have been widely used in food, pharmaceutical, and nutraceutical industries owing to their significant biological activities, such as antiaging, antioxidant, anti-inflammatory, and anticancer. However, the traditional approaches for the production of flavonoids including chemical synthesis and plant extraction involved hazardous materials and complicated processes and also suffered from low product titer and yield. Microbial synthesis of flavonoids from renewable biomass such as glucose and xylose has been considered as a sustainable and environmentally friendly method for large-scale production of flavonoids. Recently, construction of microbial cell factories for efficient biosynthesis of flavonoids has gained much attention. In this article, we summarize the recent advances in microbial synthesis of flavonoids including flavanones, flavones, isoflavones, flavonols, flavanols, and anthocyanins. We put emphasis on developing pathway construction and optimization strategies to biosynthesize flavonoids and to improve their titer and yield. Then, we discuss the current challenges and future perspectives on successful strain development for large-scale production of flavonoids in an industrial level.
Introduction
Flavonoids are a special class of naturally occurring secondary metabolites that are synthesized by plant (Pandey et al., 2016) and fungus (Correa et al., 2011). Chemically, all flavonoids possess general structure of a 15-carbon skeleton with two phenyl rings (rings A and B) connected by a heterocyclic ring (ring C). In flavonoids biosynthetic pathways, ring A is formed from malonyl-CoA, and ring B is generated from 4-coumaroyl-CoA which is synthesized from phenylalanine via the shikimate pathway (Averesch and Krömer, 2018). Based on the chemical structures, flavonoids can be divided into several different subcategories, such as chalcones, flavanones, flavones, isoflavones, flavonols, flavanonols, flavanols, and anthocyanins (Tohge et al., 2017) (Figure 1).
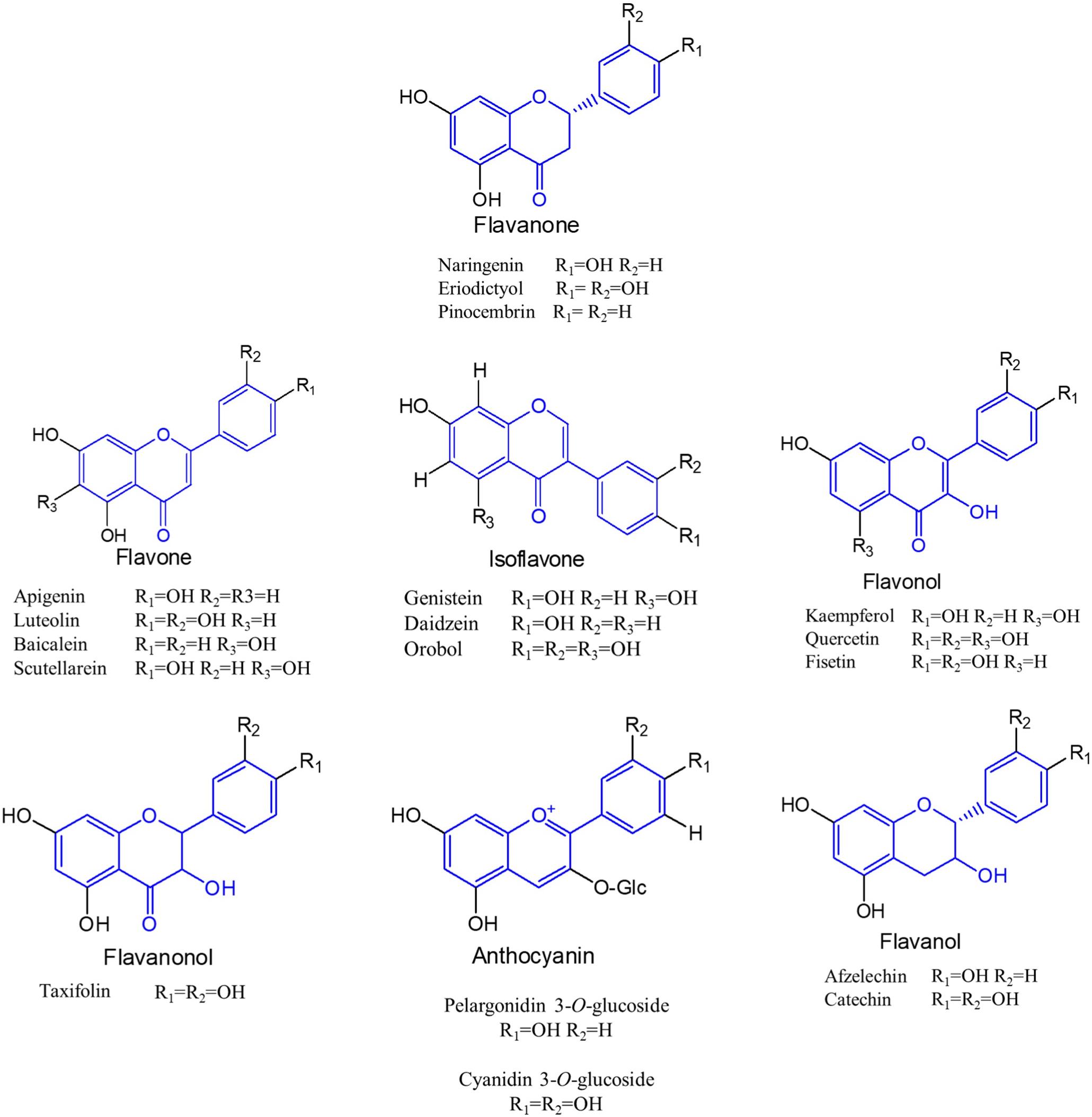
Figure 1. Molecular structures of different subclasses of flavonoids. The backbone of each subclasses is colored by blue.
Flavonoids have many important functions in plant, such as producing pigmentation for flower coloration (Nakayama et al., 2000), symbiotic nitrogen fixation (Fox et al., 2001), and UV protection (Kootstra, 1994). In addition, flavonoids demonstrate significant physiological and biochemical effects on mammals because of their unique chemical structures. Most flavonoids exhibit great antioxidant (Naderi et al., 2003), anti-inflammatory (Kim et al., 2004), antibacterial (Cushnie and Lamb, 2011), and anticancer (Ravishankar et al., 2013) activities; thus, they have been widely used in food, nutraceutical, and pharmaceutical industries (Kay et al., 2012; Georgiev et al., 2014; Hajimehdipoor et al., 2014). For example, apigenin can be used to treat high-fat and high-fructose diet–induced metabolic syndrome by reducing leptin levels and increasing adiponectin levels (Zhang et al., 2018). Epigallocatechin gallate and naringenin are able to maintain blood lipid and fat balance (Zhang et al., 2018). Quercetin, genistein, and naringenin have significant effect on reducing the expression levels of proinflammatory cytokines tumor necrosis factor α and interleukin 6 (Zhang et al., 2018). Owing to their significant bioactivities, the global market of flavonoids is rapidly growing and is expected to reach $1.05 billion in 2021 (Shah et al., 2019).
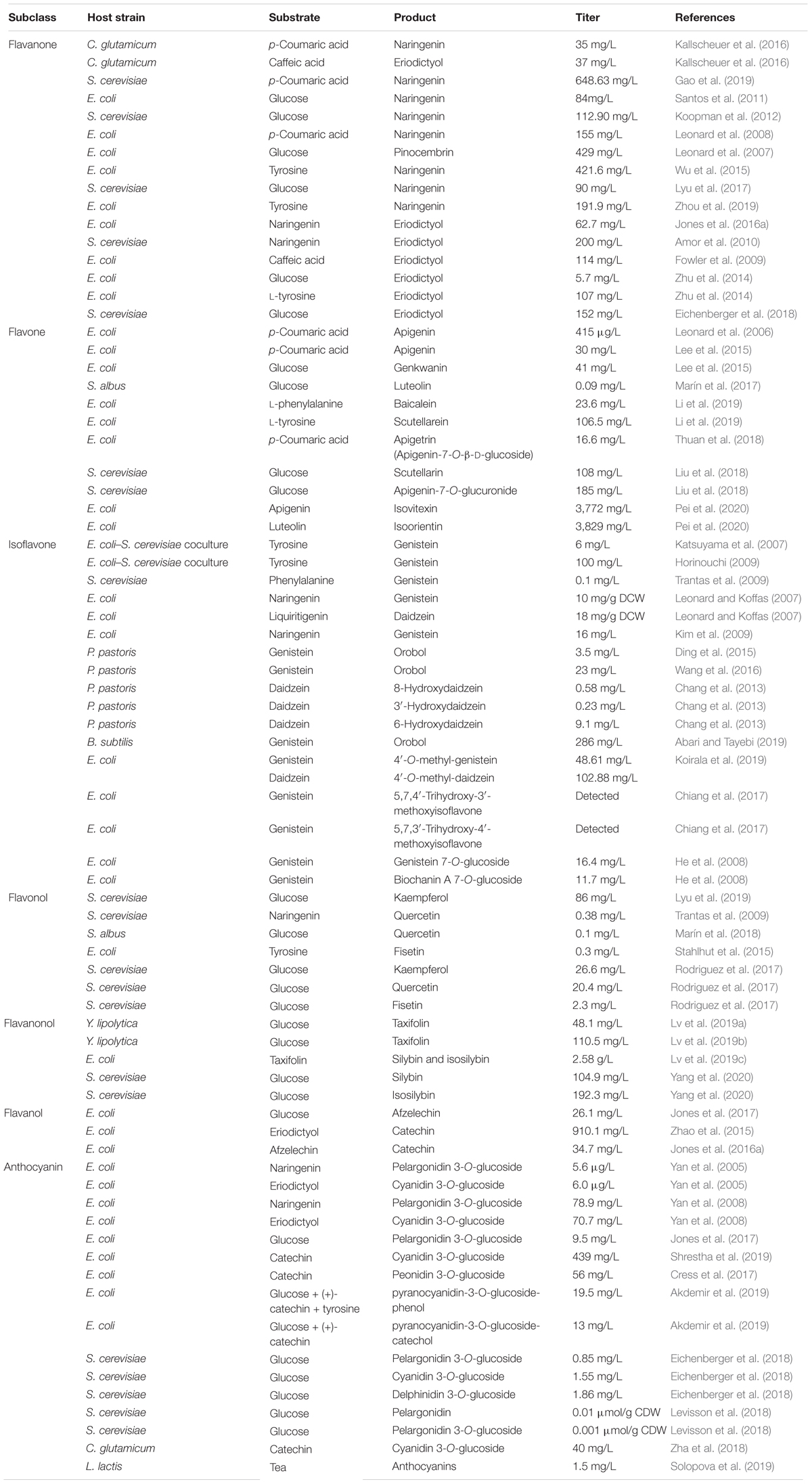
Currently, production of flavonoids predominantly relies on plant extraction, which is limited by difficulties in access and supply of the raw materials, low abundance and availability in plants, and low product yield (Jiang et al., 2005; Pyne et al., 2019). Chemical methods for production of flavonoids are also not preferred because of the complicated processes, strict reaction conditions, and poor selectivity (Pyne et al., 2019). Alternatively, introduction of the native biosynthetic pathways of flavonoids from plants into the microbial hosts through synthetic biology and metabolic engineering strategies represents a feasible approach for the production of flavonoids. In 2003, microbial synthesis of flavonoids was reported for the first time; since then, various flavonoids biosynthetic pathways have been successfully constructed in microorganisms (Hwang et al., 2003). Recently, construction of microbial cell factories for efficient production of flavonoids has made great progress (Pei et al., 2020; Wang et al., 2020; Yang et al., 2020). In this article, we recapitulate the recent and notable achievements on metabolic engineering and synthetic biology toward synthesis of flavonoids with relevant example cases (Table 1). Effects to construct flavonoids artificial biosynthetic pathways in microbes and to optimize products titer, yield, and productivity are presented.
Metabolic Engineering of Microorganisms for Flavonoids Production
The flavonoid biosynthetic pathway in plants has been clearly illustrated that extends from the shikimate pathway (Falcone Ferreyra et al., 2012) (Figure 2). In this pathway, the native precursors phenylalanine and tyrosine are converted to p-coumaroyl-CoA. Then, one molecule of p-coumaroyl-CoA and three molecules of malonyl CoA are converted to naringenin chalcone via Claisen cyclization under the action of chalcone synthase (CHS). Sequentially, the naringenin chalcone is converted to naringenin automatically or catalyzed by the chalcone isomerase (CHI). The produced naringenin in this pathway serves as an important intermediate to synthesize other classes of flavonoids. For example, flavones are formed by expression of flavone synthase (FNS). Isoflavonoids are generated by introduction of isoflavone synthase (IFS). Coexpression of flavonoid 3-hydroxylase (F3H) and flavonol synthase (FLS) results in flavonols, whereas flavanols can be produced by coexpressing F3H, dihydroflavonol 4-reductase (DFR), and leucoanthocyanidin reductase (LAR), and further introduction of anthocyanidin synthase (ANS) into this pathway can generate anthocyanins. In recent years, with the development of metabolic engineering and synthetic biology, more and more flavonoids have been achieved biosynthesis (Shah et al., 2019; Nabavi et al., 2020).
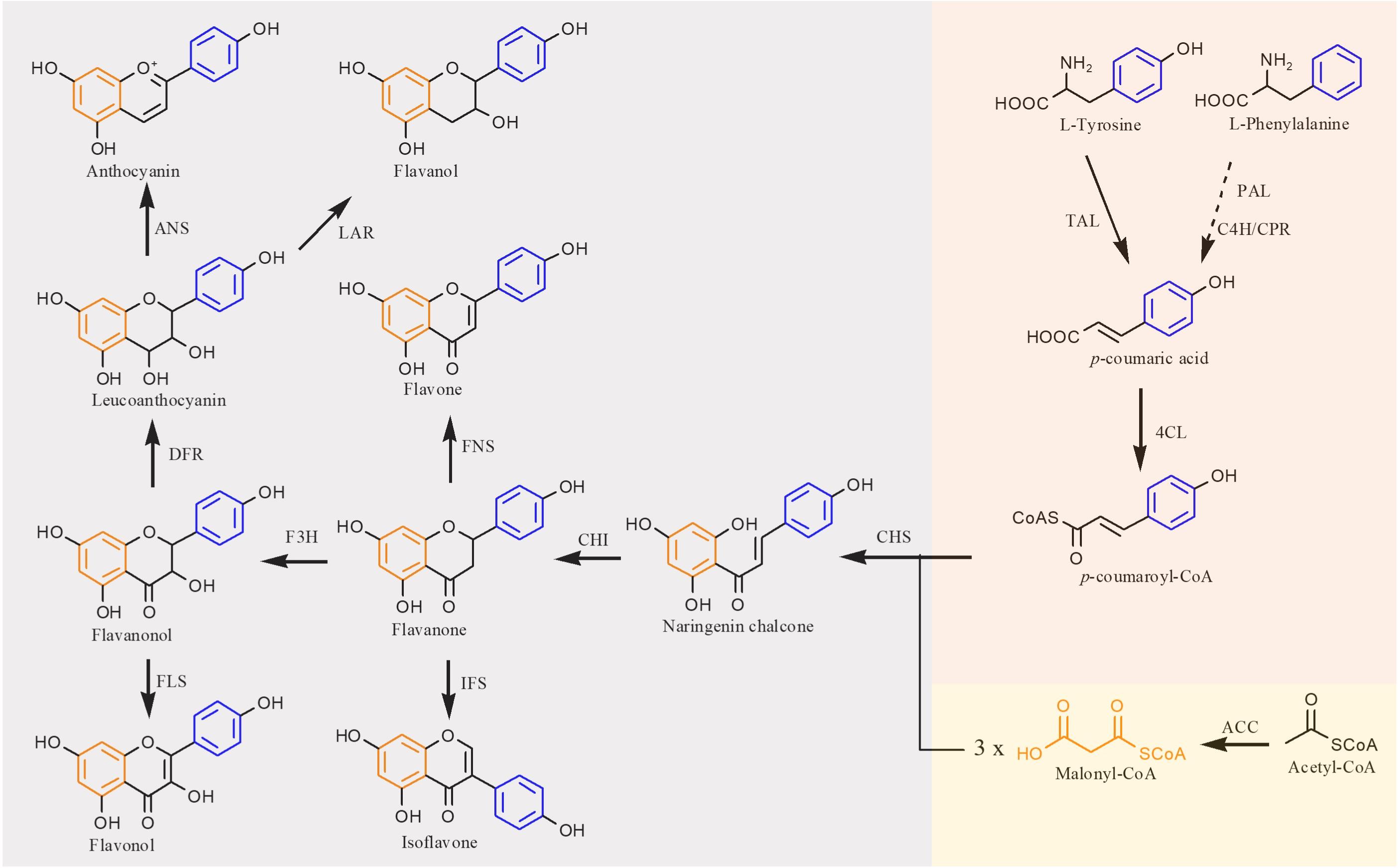
Figure 2. Biosynthetic pathways of flavonoid in plants. TAL, tyrosine ammonia lyase; PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; CPR, cytochrome P450 reductase; 4CL, 4-coumaroyl-CoA ligase; ACC, acetyl-CoA carboxylase; CHS, chalcone synthase; CHI, chalcone isomerase; FNS, flavone synthase; IFS, isoflavone synthase; F3H, flavanone 3β-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol reductase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase. Solid lines indicate a single step, and dotted lines indicate multiple steps.
Flavanones
Flavanones, including naringenin, pinocembrin and eriodictyol, are the first flavonoid products in flavonoids biosynthetic pathway. They are generally present in citrus fruits, such as grapefruit (Peterson et al., 2006), oranges (Khan et al., 2010), and lemons (Peterson et al., 2006) and have anti-inflammatory (Bano et al., 2013), antioxidant (Miranda et al., 2000), anticancer (Ketabforoosh et al., 2014), and other important pharmacological activities. The basic skeleton of flavanones is 2-phenylchromogen, which is characterized by the saturation of the C2–C3 double bond. The structure of flavanones is highly active that can be modified by hydroxylation (Britsch and Grisebach, 1986), methylation (Koirala et al., 2016), and glycosylation (Di Majo et al., 2005). Biosynthesis of naringenin from the precursor p-coumaric acid has been achieved in Corynebacterium glutamicum by overexpression of CHS and CHI from Petunia hybrida. By disrupting the competing pathways, 35 mg/L naringenin were produced in the engineered host strains (Kallscheuer et al., 2016). It was believed that the low titers were limited by the low enzyme activity of CHS. Alternatively, Gao et al. (2019) introduced the SmCHS2 from Silybum marianum into Saccharomyces cerevisiae and obtained 648.63 mg/L naringenin by exogenously feeding p-coumaric acid.
De novo biosynthesis of flavanones from simple carbon sources has also been achieved. Santos et al. introduced the genes RgTAL (encoding tyrosine ammonia lyase) from Rhodotorula glutinosa, Pc4CL (encoding 4-coumaric acid-CoA ligase) from Petroselinum crispum, PhCHS from P. hybrida, and MsCHI from Medicago sativa into a tyrosine-overproducing S. cerevisiae strain, resulting in 29 mg/L naringenin using glucose as the substrate (Santos et al., 2011). In another example, the naringenin biosynthetic pathway in Arabidopsis thaliana containing phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), cytochrome P450 reductase (CPR), 4CL, CHS, and CHI was transferred into the S. cerevisiae. The original recombinant host strain only generated 1.4 mg/L naringenin from glucose. To further improve the titer, the feedback inhibition effect was eliminated by deletion of ARO3 and introduction of the ARO4G226S mutant. As a result, the naringenin production increased to 2.8 mg/L. Furthermore, disruption of competing pathway and enhancement of the precursor p-coumaric acid supply enabled the host strain to produce 54.5 mg/L naringenin in shake flask experiments (Koopman et al., 2012). The insufficient precursor supply is still the dominant rate-limiting factors for high-level production of naringenin.
Malonyl-CoA not only serves as an important precursor for the production of flavanones but also is an essential intermediate for the synthesis of fatty acids to support cell growth in microorganisms (Takamura and Nomura, 1988). Thus, there exist a trade-off relationship between the flavanone biosynthesis and the cell growth. In order to balance the production phase and growth phase, Leonard et al. (2008) constructed a malonate assimilation pathway in a recombinant naringenin-producing Escherichia coli strain by overexpression of the malonate synthase (MatB) and malonate carrier protein (MatC) isolated from Rhizobium trifolii. Introduction of malonate assimilation pathway enables engineered E. coli strain to synthesize malonyl-CoA by exogenously feeding malonate. Finally, the recombinant E. coli strain produced 155 mg/L of naringenin, indicating a 2.7-fold improvement in titer compared with the parent strain without malonate assimilation pathway overexpression (Leonard et al., 2008). Cerulenin is a fatty acid synthase inhibitor for inhibiting the expression levels of fabB and fabF. According to the report, addition of 200 μM of cerulenin could increase the concentration of malonyl-CoA from 2 pmol/mg cell dry weight (CDW) to 105 pmol/mg CDW in 1.5 h (van Summeren-Wesenhagen and Marienhagen, 2015). By exogenously supplementing cerulenin into the cultures, the naringenin titer enhanced from 29 mg/L to 84 mg/L in engineered host strains (Santos et al., 2011). Although those strategies can effectively increase the level of malonyl-CoA in engineered host strain, thereby to enhance the titer of flavanones, the high costs of malonate and cerulenin limited their use in high-level production of flavanones. In microbes, malonyl-CoA is formed from acetyl-CoA catalyzed by acetyl-CoA carboxylase complex (ACC). Leonard et al. (2007) introduced ACC from Pseudomonas luminescens into E. coli; the titer of pinocembrin reached 196 mg/L, exhibiting a 5.8-fold increase in titer compared with that in control strains. Additionally, the availability of malonyl-CoA could also be improved by employing a clustered regularly interspaced short palindromic repeat interference (CRISPRi) system to inhibit the expression level of genes involved in essential metabolic pathways. For instance, Wu et al. (2015) utilized the CRISPRi system to down-regulate the expression level of genes fabF, fumC, fabB, sucC, and adhE in recombinant E. coli to redirect the carbon flux toward malonyl-CoA synthesis. The resultant strain generated 421.6 mg/L naringenin from tyrosine, which is the highest naringenin production reported so far (Wu et al., 2015). Recently, Lyu et al. (2017) tried to improve naringenin titer in S. cerevisiae by dividing the naringenin biosynthetic pathway into three modules: the first module containing TAL, 4CL, CHS, and CHI was applied for naringenin biosynthesis. The second module carrying acetyl-CoA synthase, ATP-citrate lyase, and ACC was used for malonyl-CoA accumulation, whereas the third module harboring the ARO4K229L mutant was employed to produce more tyrosine. Engineering and integration of those three modules into the final strain resulted in 90 mg/L of naringenin from glucose (Lyu et al., 2017). In another example, an iterative high-throughput screening method was applied to fine-tune the expression level of naringenin biosynthetic pathway genes. A promoter library containing a series of constitutive promoters with different strength was constructed. Those promoters were randomly placed upstream of naringenin biosynthetic pathway genes. After several rounds of high-throughput screening, the best performed recombinant E. coli strain produced 191.9 mg/L naringenin, representing a two-fold increase in titer compared with that in a strain optimized using traditional modular optimization strategy (Zhou et al., 2019).
On the basis of the naringenin biosynthetic pathway, eriodictyol can be produced by introducing F3′H into this pathway. A 4-hydroxyphenylacetate 3-hydroxylase (EcHpaBC) was identified and characterized from E. coli. It is able to directly transform the naringenin into eriodictyol with 62.7 mg/L of eriodictyol produced (Jones et al., 2016a). In another example, a cytochrome P450 F3′H isolated from Gerbera hybrida was functionally expressed in S. cerevisiae, resulting in 200 mg/L eriodictyol from naringenin (Amor et al., 2010). In addition, by directly feeding caffeic acid rather than p-coumaric acid as the precursor, eriodictyol can also be biosynthesized via overexpressing 4CL, CHS, and CHI. Fowler et al. (2009) introduced 4CL, CHS, and CHI into E. coli; 114 mg/L eriodictyol was obtained by exogenously supplementing caffeic acid into the cultures. De novo biosynthesis of eriodictyol from simple carbon source in E. coli was also achieved by coexpressing TAL, 4CL, CHS, and CHI with the cytochrome P450 F3′H. In this study, a fusion protein (tF3′H-tCPR) was created by fusing a truncated F3′H with a truncated CPR to improve the solubility and enzyme activity of F3′H. Finally, only 5.7 mg/L of eriodictyol was obtained using glucose as the substrate (Zhu et al., 2014). The low catalytic efficiency of F3′H is still the rate-limiting factor in this pathway. It is believed that compared with yeasts, bacterial hosts are usually difficult to express cytochrome P450 enzymes because they are unable to perform posttranslational modifications and expression of membrane proteins. When transferring the de novo eriodictyol biosynthetic pathway into S. cerevisiae, the titer of eriodictyol improved to 152 mg/L from glucose (Eichenberger et al., 2018).
Flavones
Structurally, flavones correspond to a flavonoid subgroup that characterized by a 2-phenylchromogen-4-one backbone and have a double bond between C2 and C3. Flavones are commonly found in fruits and vegetables, such as carrots, parsley, mint, and red and chili peppers (Panche et al., 2016) and exhibit great health benefits such as anti-inflammatory (Huang and Ho, 2010), anticancer (Li-Weber, 2009), antioxidant (Škerget et al., 2005), hypoglycemic, and hypolipidemic activities (Sharma et al., 2008) and prevention of heart disease (Hirvonen et al., 2001). Apigenin, a common flavone, can be synthesized from naringenin catalyzed by FNS. The first case about biosynthesis of apigenin in microorganisms was reported in 2006. Leonard et al. (2006) introduced 4CL, CHS, CHI, and an FNS from P. crispum into E. coli; 415 μg/L of apigenin was produced from p-coumaric acid, and the conversion rate was only 2.9% (Leonard et al., 2006). Since then, continuous researches have been reported to improve the apigenin production. Lee et al. (2015) constructed an apigenin biosynthetic pathway in E. coli composed of 4CL from Oryza sativa, CHS from Populus euramericana, CHI from Medicago truncatula and FNS from Parsley in E. coli, 23 mg/L of apigenin was obtained from p-coumaric acid. Then the titer was further increased to 30 mg/L by enhancing the expression levels of 4CL and CHS (Lee et al., 2015). Hydroxylation of apigenin at the 3′ position results in luteolin. This reaction usually catalyzed by F3′H. Recently, Marin et al. constructed a de novo luteolin biosynthetic pathway in Streptomyces albus by employing the F3′H from A. thaliana and generated 0.09 mg/L of luteolin (Marín et al., 2017). Very recently, baicalein (5,6,7-trihydroxyflavone) and scutellarein (4′,5,6,7-tetrahydroxyflavone) have also been synthesized in E. coli. Overexpression of PAL from Rhodotorula toruloides, 4CL and FNS from P. crispum, CHS from P. hybrida, CHI from M. sativa, F6′H (encoding flavonoid 6′-hydroxylase) from Scutellaria baicalensis, and CPR from A. thaliana, baicalein, and scutellarein can be produced from tyrosine and phenylalanine, respectively (Figure 3). To remove the rate-limiting step, a hydrophilic modification 2B1 was incorporated with an N-terminal truncated F6′H to increase its solubility in E. coli, resulting in 8.5 mg/L baicalein and 47.1 mg/L scutellarein in shake flask experiments. Further enhancement of malonyl-CoA availability greatly improved the titers to 23.6 mg/L baicalein and 106.5 mg/L scutellarein (Li et al., 2019).
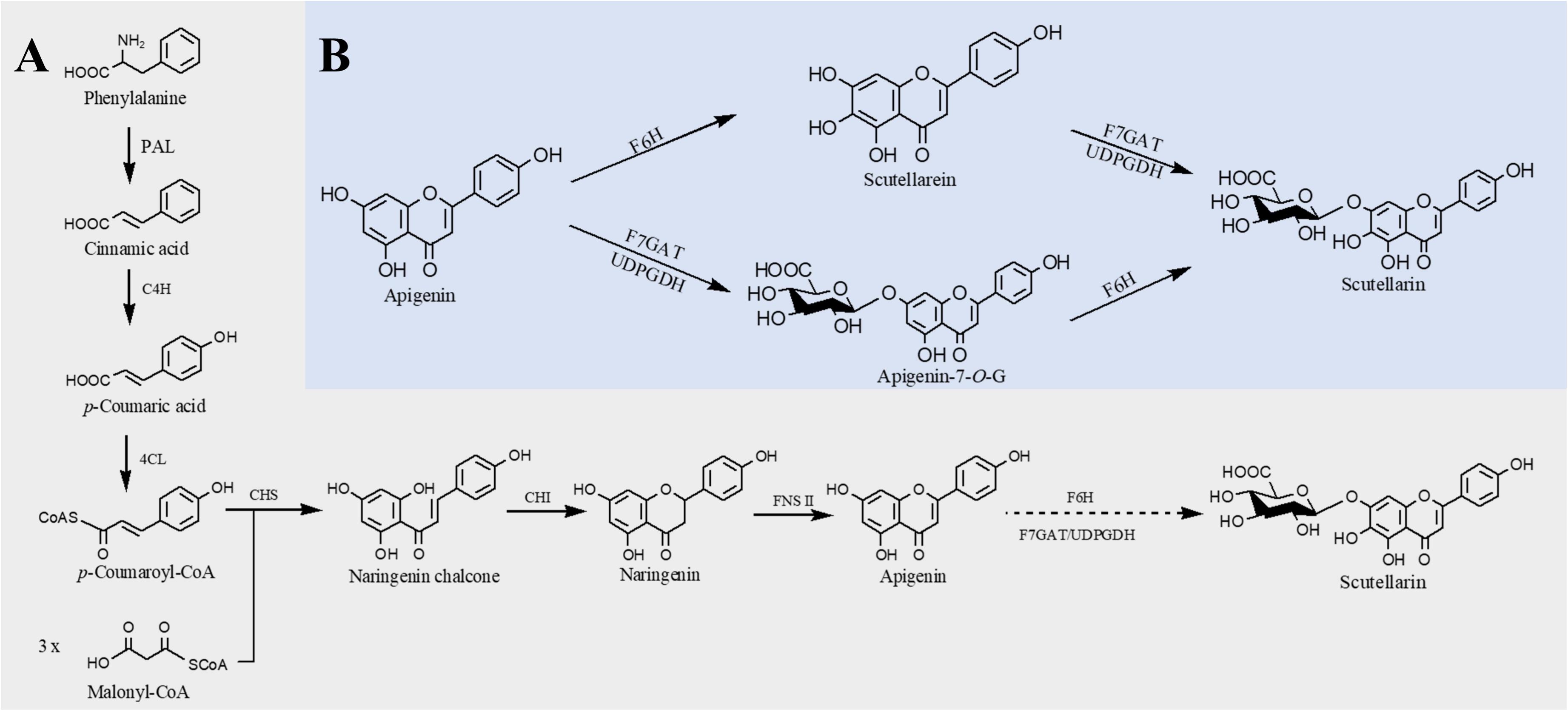
Figure 3. Microbial production of scutellarin in E. coli. (A) Biosynthesis of scutellarin from phenylalanine; (B) biosynthesis of scutellarin from apigenin. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumaroyl-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; FNSII, flavone synthase II; F6H, flavone 6-hydroxylase; F7GAT, flavonoid-7-O-glucuronosyltransferase; UDPGDH, UDP-glucose dehydrogenase. Solid lines indicate a single step, and dotted lines indicate multiple steps.
Except for hydroxylation, other modifications such as methylation and glycosylation also contribute to expand the structural diversity of flavones. For example, by introducing a methyl group at C7 site of apigenin, one compound known as genkwanin is obtained. Compared with apigenin, genkwanin exhibits many novel properties including antibacterial and radical scavenging activities. Lee et al. (2015) introduced apigenin 7-O-methyltransferase (POMT7) from Populus deltoides into an apigenin overproducing E. coli strain to construct a de novo biosynthetic pathway for genkwanin production; the titer of genkwanin reached 41 mg/L by using glucose as the carbon source. By introducing glucosyl group into the chemical structure of apigenin at the C7 site, the product apigenin-7-O-β-D-glucoside appears several additional bioactivities, such as antiproliferative and higher antioxidant activities. Tuan et al. achieved apigenin-7-O-β-D-glucoside production from p-coumaric acid in E. coli by employing a glycosyltransferase (PaGT3) from Phytolacca americana. To eliminate by-products formation, a coculture strategy was used by separating the entire pathway into two strains. The first strain contains 4CL from Nicotiana tabacum, CHS from P. hybrida, CHI from M. sativa, and FNS from Parsley to generate apigenin from p-coumaric acid, whereas the second strain carries PaGT3 and an UDP-glucose overproduction system to convert apigenin into apigenin-7-O-β-D-glucoside. By optimization of initial inoculum ratio of the two strains, the titer of apigenin-7-O-β-D-glucoside reached 16.6 mg/L, which was 2.5 times higher than monoculture producing strain (Thuan et al., 2018). Breviscapine is a crude extract of flavonoids isolated from Erigeron breviscapus. It has been used for treatment of cardiovascular and cerebrovascular diseases for a long time (Zhong et al., 2005). The main active components in breviscapine are scutellarin and apigenin-7-O-glucuronide. Liu et al. (2018) identified two key enzymes involved in breviscapine biosynthetic pathway in E. breviscapus: one is flavonoid 7-O-glucuronosyltransferase, and the other is flavone-6-hydroxylase. Introduction of those two enzymes into an apigenin overproducing S. cerevisiae strain resulted in 108 mg/L of scutellarin and 185 mg/L of apigenin-7-O-glucuronide using glucose as the carbon source (Liu et al., 2018). Very recently, a C-glycosyltransferase (Gt6CGT) from Gentiana triflora was identified and characterized. It exhibits significant enzyme activities for conversion of apigenin and luteolin into isovitexin and isoorientin, respectively. This enzyme was coupled with a sucrose synthase (GmSUS) from Glycine max to regenerate the UDP-glucose. After optimizing coupled reaction conditions, the production of isovitexin and isoorientin reached 3,772 and 3,829 mg/L with molar conversion rate of 97.1 and 94.7%, respectively (Pei et al., 2020).
Isoflavones
Isoflavones differ from flavones by the position of the phenyl group at the C3 site, which is at the C2 site in flavones. They represent a class of phytoestrogens produced by the leguminous plants (Kaufman et al., 1997; Chon, 2013) such as soybean, green bean, alfalfa sprout, and cowpea. Isoflavones act as estrogens on human body and can be used to treat various hormonal disorders such as prostate cancer (Kumar et al., 2004), breast cancer (Dong and Qin, 2011), osteoporosis (Taku et al., 2011), and cardiovascular diseases (Watanabe et al., 2002). Biosynthesis of isoflavones can be achieved from flavanones catalyzed by IFS, a membrane-bound cytochrome P450 monooxygenase. Utilization of naringenin and liquiritigenin as the substrates for IFS, resulting in the generation of genistein and daidzein, respectively. Katsuyama et al. (2007) introduced CHS, CHI, and an IFS isolated from Glycyrrhiza echinata into the yeast strain S. cerevisiae; 0.34 mg/L genistein was produced by feeding N-acetylcysteamine-attached p-coumaric acid as the precursor. To achieve biosynthesis of genistein from tyrosine, a naringenin overproducing E. coli strain, was cocultured with a recombinant S. cerevisiae carrying the IFS gene, which resulted in 6 mg/L genistein from tyrosine (Katsuyama et al., 2007). The titer was further enhanced to 100 mg/L by optimization of coculture conditions (Horinouchi, 2009). To avoid metabolite transport limitations through the cell walls of two different strains, it is beneficial to assemble entire pathway into a single strain. Trantas et al. (2009) reconstituted the entire genistein biosynthetic pathway in S. cerevisiae by coexpression of seven genes encoding PAL, CPR, C4H, 4CL, CHS, CHI, and IFS. The best engineered strain synthesized 0.1 mg/L genistein when the cultures were fed with phenylalanine as the precursor (Trantas et al., 2009). However, expression of IFS in bacteria is very difficult because of the absence of the electron transfer system. In order to functionally express IFS in E. coli, a redox partner CPR from Catharanthus roseus was fused to IFS from G. max by a glycine–serine–threonine linker sequence, which enabled it to efficiently transfer electron from NAD(P)H to substrate. Then, the N-terminus of this fusion protein was truncated and appended to a tailor-made membrane recognition signal. Expression of this construct in E. coli enabled efficient production of 10 and 18 mg/g dry cell weight of genistein and daidzein from naringenin and liquiritigenin, respectively (Leonard and Koffas, 2007). In another study, an IFS from red clover was truncated by removing the first 21 amino acids on the N-terminus, and then it was fused to a CPR from rice. The recombinant E. coli expressing this fusion protein can produce 16 mg/L genistein from naringenin (Kim et al., 2009).
The chemical structures of isoflavones also can be conjugated with hydroxyl (Tahara et al., 1991), methyl (Edwards and Dixon, 1991), and glycosyl (Ismail and Hayes, 2005) groups by hydroxylases, methyltransferases and glucosyltransferase. Overexpression of a P450 hydroxylase (CYP57B3) from Aspergillus oryzae and a CPR from S. cerevisiae in Pichia pastoris led to 3.5 mg/L of orobol (3′-hydroxygenistein) in a fed-batch culture fed with genistein as the precursor (Ding et al., 2015). Wang et al. further increased the titer of orobol to 23 mg/L by construction of a P. pastoris mutant strain via periodic hydrogen peroxide treatment (Wang et al., 2016). The CYP57B3 also has been used to hydroxylate daidzein in a recombinant P. pastoris strain. Interestingly, when using daidzein as the substrate, three orthohydroxylated daidzein derivatives were obtained, including 8-hydroxydaidzein (8-OHDe), 3′-hydroxydaidzein (3′-OHDe), and 6-hydroxydaidzein (6-OHDe), with a conversion rate of 2.4, 0.9, and 36.3%, respectively (Chang et al., 2013). In order to improve the catalytic efficiency of hydroxylase and avoid by-products formation, a tyrosinase from Bacillus megaterium was functionally displayed on Bacillus subtilis spores using CotE as an anchor protein (Hosseini-Abari et al., 2016). This spore displayed tyrosinase is able to efficiently hydroxylate 1 mM genistein into 1 mM orobol with a 100% conversion rate in 90 min (Abari and Tayebi, 2019). The diversity of isoflavones also can be expanded by methylation reactions. Koirala et al. (2019) constructed a recombinant E. coli strain for methylation of isoflavones by overexpressing native EcSAM encoding S-adenosy-L-methionine (SAM) synthase and SaOMT2 encoding an O-methyltransferase from Streptomyces avermitilis. When using genistein and daidzein as the substrates, 4′-O-methyl-genistein and 4′-O-methyl-daidzein were obtained. After optimization of substrate concentrations, incubation time, and culture conditions, 46.81 mg/L of 4′-O-methyl-genistein and 102.88 mg/L of 4′-O-methyl-daidzein were generated (Koirala et al., 2019). In another example, genistein was converted into 5,7,3′-O-trihydroxy-4′-methoxyisoflavone and 5,7,4′-trihydroxy-3′-methoxyisoflavone by two sequential bioreactions including 3′-hydroxylation catalyzed by one recombinant E. coli strain containing tyrosinase from B. megaterium and then methylation catalyzed by another recombinant E. coli strain expressing O-methyltransferase from Streptomyces peucetius (Chiang et al., 2017).
By introduction of glucosyltransferases, the isoflavone glycosides can be produced. Heterologous overexpression of an UDP-glucosyltransferase UGT71G1 from M. truncatula enabled the recombinant E. coli strain to synthesize 16.4 mg/L genistein 7-O-glucoside and 11.7 mg/L biochanin A 7-O-glucoside from genistein and biochanin A in 500 mL TB culture medium (He et al., 2008).
Flavonols and Flavanols
Flavonols can be considered as the derivatives of flavones; their structural difference is on the presence of a 3-hydroxy group on flavonols, which is lacking on flavones. Flavonols are commonly found in tea, red wines, fruits, and vegetables (Panche et al., 2016). Flavonols have a wide range of applications in pharmaceutical industries. For example, quercetin has been proven to be effectively inhibit the metastasis of melanoma cells (Caltagirone et al., 2000). Fisetin has the potential to treat neurodegenerative diseases (Fazel Nabavi et al., 2016) such as Alzheimer disease and Huntington disease. Biosynthesis of flavonols can be achieved by coexpressing F3H and FLS with flavanones as the precursor. Lyu et al. (2019) introduced F3H and FLS into a naringenin-producing S. cerevisiae strain Y-22, which rendered the recombinant strain the ability to generate kaempferol. Further optimization strategies including screening of efficient pathway enzymes, disruption of competing pathways, enhancement of the precursors PEP and E4P supply, and mitochondrial engineering of F3H and FLS significantly improved the kaempferol titer to 86 mg/L (Lyu et al., 2019). Quercetin is a 3′-hydroxylated derivative of kaempferol. Biosynthesis of quercetin was achieved by extending kaempferol biosynthetic pathway via overexpressing F3′H in S. cerevisiae. When naringenin was added into the cultures, the best engineered host strain produced 0.38 mg/L of quercetin during 70-h cultivation (Trantas et al., 2009). Recently, de novo biosynthesis of quercetin in actinomycetes was reported for the first time. In this study, an artificial pathway containing the TAL from Rhodobacter capsulatus, 4CL from S. coelicolor, CHS and CHI from G. max, naringenin 3-dioxygenase from P. crispum, and FLS and F3′H from A. thaliana was constructed and introduced into S. albus and S. coelicolor. The recombinant S. albus produced the higher quercetin titer of 0.1 mg/L (Marín et al., 2018). Fisetin has very similar chemical structure with quercetin; hydroxylation of fisetin at the C5 site results in quercetin. According to the structural similarity between fisetin and quercetin, a novel biosynthetic pathway for the production of fisetin was developed by recruiting liquiritigenin as the intermediate. Heterologous overexpression of TAL, 4CL, CHS, and chalcone reductase (CHR) from Astragalus mongholicus and CHI, F3H, FLS, flavonoid 3′-monooxygenase (FMO), and CPR in E. coli resulted in 0.3 mg/L of fisetin from tyrosine (Stahlhut et al., 2015). Recently, de novo biosynthesis of kaempferol, quercetin, and fisetin from simple carbon source in S. cerevisiae was reported. The best engineered host strain produced 26.6 mg/L of kaempferol, 20.4 mg/L of quercetin, and 2.3 mg/L of fisetin directly from glucose (Rodriguez et al., 2017).
Taxifolin, also known as dihydroquercetin, belongs to the flavanonols. Biosynthesis of taxifolin can be achieved by employment of eriodictyol as the direct precursor under the catalysis of F3H. Recently, Lv et al. (2019a) constructed a taxifolin biosynthetic pathway consisting TAL, 4CL, CHS, CHI, F3′H, CPR, and an F3H from Solanum lycopersicum. Integration of this pathway into the genome of Yarrowia lipolytica resulted in 48.1 mg/L of taxifolin (Lv et al., 2019a). Then, several metabolic engineering optimization strategies, such as improving precursor supply, enhancing copy number of the rate-limiting enzyme CHS, and screening efficient CPR, were utilized to increase the taxifolin titer. The final engineered strain generated 110.5 mg/L taxifolin using glucose as the carbon source (Lv et al., 2019b). Taxifolin can be used as the precursor to synthesize other important compounds. Very recently, Lv et al. (2019c) introduced a vanillyl alcohol oxidase (PsVAO) from Penicillium simplicissimum and an ascorbate peroxidase (APX1) from milk thistle into E. coli strain to build up an enzymatic cascade that is able to efficiently convert taxifolin and eugenol to silybin and isosilybin. Under the best culture conditions, a total amount of 2.58 g/L silybin and isosilybin was generated with a molar conversion rate of 76.7% in a 3-L fermenter (Lv et al., 2019c). On this basis, Yang et al. (2020) achieved biosynthesis of silybin and isosilybin from glucose in S. cerevisiae for the first time (Figure 4). In this study, the biosynthetic pathways for production of coniferyl alcohol and taxifolin were individually reconstructed in two different strains. Systematic optimization of those two pathways enabled the host strains to respectively synthesize 201.1 mg/L of coniferyl alcohol and 336.8 mg/L of taxifolin. Using APX1 as the catalyst, 104.9 mg/L of silybin and 192.3 mg/L of isosilybin were produced from coniferyl alcohol and taxifolin, with a yield of 62.5% (Yang et al., 2020).
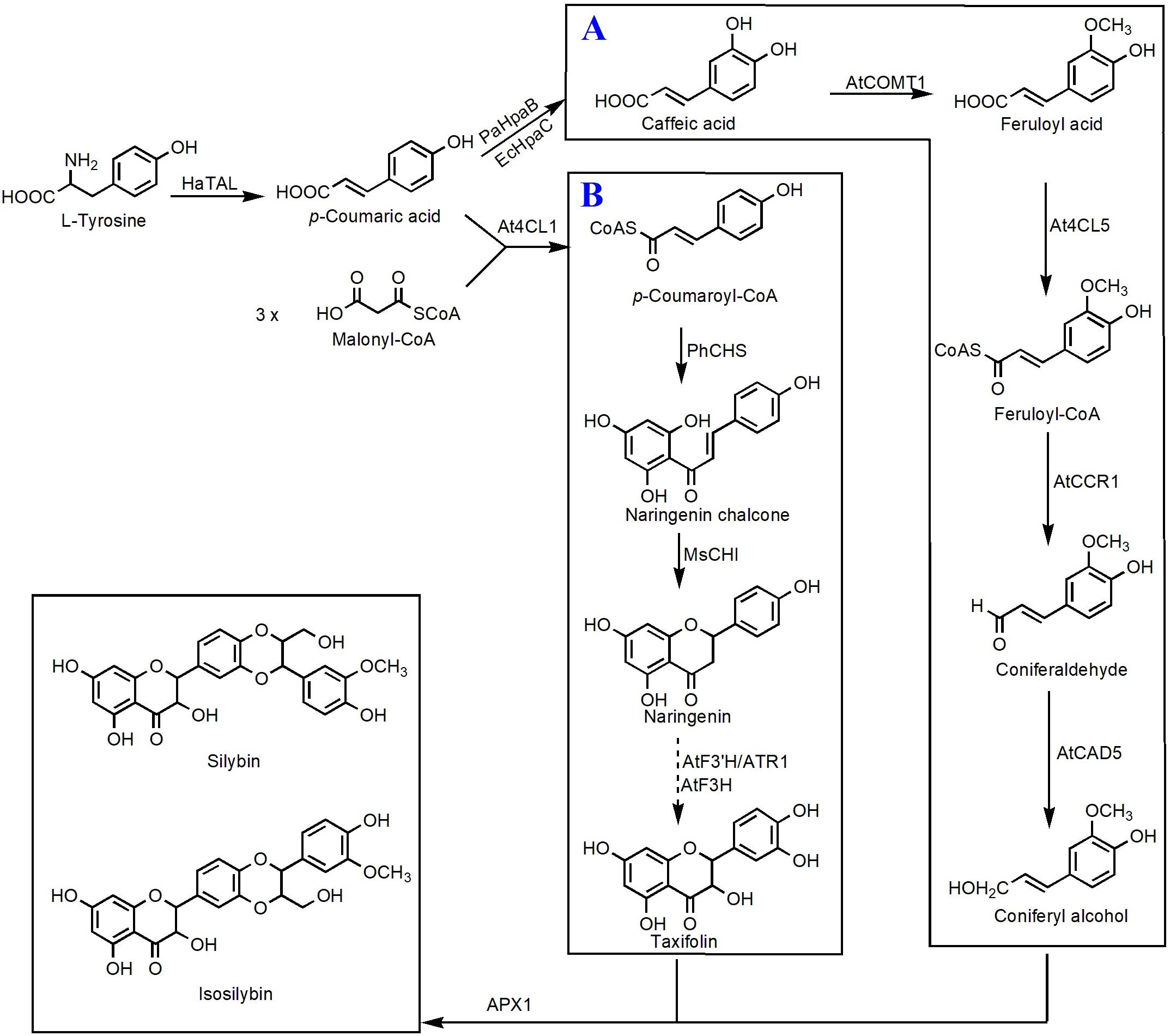
Figure 4. Microbial production of silybin and isosilybin in S. cerevisiae. (A) Biosynthesis of coniferyl alcohol from caffeic acid; (B) biosynthesis of taxifolin from p-coumaroyl-CoA. HaTAL, tyrosine ammonia lyase from H. aurantiacus; PaHpaB, 4-hydroxyphenylacetate 3-monooxygenase from P. aeruginosa; EcHpaC, flavin reductase from E. coli; AtCOMT1, O-methyltransferase 1 from A. thaliana; At4CL5, 4-coumarate-CoA 5 ligase from A. thaliana; AtCCR1, cinnamoyl-CoA reductase 1 from A. thaliana; AtCAD5, cinnamyl alcohol dehydrogenase 5 from A. thaliana; At4CL1, 4-coumarate-CoA 1 ligase from A. thaliana; PhCHS, chalcone synthase from P. hybrida; MsCHI, chalcone isomerase from M. sativa; AtF3′H, flavonoid 3′-monooxygenase from A. thaliana; ATR1, cytochrome P450 reductase 1 from A. thaliana; AtF3H, flavanone 3-hydroxylase from A. thaliana; APX1, ascorbate peroxidase 1 from S. marianum. Solid lines indicate a single step, and dotted lines indicate multiple steps.
Flavanols, also known as flavan-3-ols, are occurring abundantly in fruits such as apple, peach, and pear (Panche et al., 2016). It has been reported that quite a high level of flavanols was detected in cocoa (Neukam et al., 2007). An overwhelming body of research evidence demonstrated that regular flavanols intake can prevent hypertension and cardiovascular and cerebrovascular diseases (Schroeter et al., 2006). Biosynthesis of flavanols can be achieved by extending the flavanones biosynthetic pathway via introduction of F3H, DFR, and LAR. Afzelechin and catechin are the most important representatives of the class of flavanols. Afzelechin is a typical flavan-3-ol, which is synthesized by using naringenin as the precursor. Recently, de novo biosynthesis of afzelechin from glucose in microorganism was achieved for the first time. In this study, a polyculture system was developed by separating the entire biosynthetic pathway into three parts: glucose to p-coumaric acid, p-coumaric acid to naringenin, and naringenin to afzelechin. Those three parts were individually introduced into three different E. coli strains. The best engineered production system generated 26.1 mg/L afzelechin in shake flask experiments (Jones et al., 2017). In another example, F3H from Camellia sinensis, DFR from Fragaria ananassa, and LAR from Desmodium uncinatum were coexpressed in E. coli, thus constructing a recombinant E. coli strain that is able to synthesize catechin using eriodictyol as the precursor. Enhancement of the NADPH availability, optimization of pathway gene copy number, and employment of a series of scaffolding proteins enabled the production of 910.1 mg/L catechin from eriodictyol, with a conversion rate of 91% (Zhao et al., 2015).
Alternatively, according to the structural similarity between catechin and afzelechin, it is believed that biosynthesis of catechin can also be achieved by hydroxylation of afzelechin at the C3′ site. Jones et al. employed the non-P450 hydroxylase EcHpaBC to directly hydroxylate afzelechin with catechin titer of 34.7 mg/L (Jones et al., 2016a).
Anthocyanins
Anthocyanins are a class of flavonoids containing polyhydroxy or polymethoxy derivatives of 2-phenylbenzophyryllium in their chemical structures. They are naturally occurring pigments widespread in virous flowers, vegetables, fruits, and cereal grains (Jackman et al., 1987; Wrolstad, 2004). Anthocyanins present black, blue, purple, or red in color, depending on the pH of the environment (Heredia et al., 1998; Torskangerpoll and Andersen, 2005). Anthocyanins possess great antioxidant (Miguel, 2011), anticancer (Bowen-Forbes et al., 2010), and antidiabetic (Mojica et al., 2017) activities, thus exhibiting great health-promoting potentials. The biosynthetic pathway of anthocyanins extended from the flavanols biosynthetic pathway by overexpression of ANS and glycosyltransferases. Yan et al. (2005) constructed a four-step biosynthetic pathway for the production of anthocyanin in E. coli by introduction of F3H, DFR, an ANS from Malus domestica and a 3-O-glucosyltransferase (PGT8) from P. hybrida. As a result, 5.6 μg/L pelargonidin 3-O-glucoside and 6.0 μg/L cyanidin 3-O-glucoside were produced by exogenously adding naringenin and eriodictyol into the cultures (Yan et al., 2005). Further enhancement of the UDP-glucose supply and construction of an ANS-PGT8 fusion protein greatly improved the titers to 78.9 mg/L pelargonidin 3−O−glucoside and 70.7 mg/L cyanidin 3−O−glucoside (Yan et al., 2008). De novo biosynthesis of pelargonidin 3-O-glucoside is very difficult to achieve because the biosynthetic process is too complicated and involves too many enzymatic steps. Recently, to overcome this issue, a polyculture system was constructed to improve the titer of pelargonidin 3-O-glucoside. In this system, the entire pelargonidin 3-O-glucoside biosynthetic pathway containing 15 enzymes and transcriptional factors was distributed into four independent E. coli strains, which allowed for the distribution of metabolic burden. When using glucose as the carbon source, the best performed polyculture system produced 9.5 mg/L pelargonidin 3-O-glucoside (Jones et al., 2017). Very recently, Shrestha et al. (2019) constructed cyanidin 3-O-glucoside biosynthetic pathway in E. coli by overexpression of ANS from P. hybrida and cyanidin 3-O-glucosyltransferase (At3GT) from A. thaliana. In order to balance the expression level of ANS and At3GT, a synthetic bio-brick vector system was developed to express ANS, At3GT, and UDP-glucose biosynthetic pathway genes under different promoters. The results showed that one engineered strain carrying At3GT and ANS driven by the Ptrc promoter and UDP-glucose biosynthetic pathway genes under PT7 promoter resulted in the highest cyanidin 3-O-glucoside titer of 439 mg/L in 36-h cultivation (Shrestha et al., 2019). In another example, a methylated anthocyanin, peonidin 3-O-glucoside, was produced in engineered E. coli by extra expression of an O-methyltransferase. Screening of efficient pathway enzymes and improving the SAM availability via a CRISPRi system enhanced the peonidin 3-O-glucoside titer to 56 mg/L (Cress et al., 2017). Very recently, coculture systems were developed in E. coli strains to produce pyranoanthocyanins. In this work, 4-vinylphenol and 4-vinylcatechol producers were separately cocultured with cyanidin-3-O-glucoside producer cells to generate pyranocyanidin-3-O-glucoside-phenol and pyranocyanidin-3-O-glucoside-catechol. After optimization of the coculture conditions, up to 19.5 mg/L of pyranocyanidin-3-O-glucoside-phenol and 13 mg/L of pyranocyanidin-3-O-glucoside-catechol were synthesized from glucose (Akdemir et al., 2019).
Actually, E. coli is not a preferred host to produce anthocyanins because of too many cytochrome P450 enzymes involved in the biosynthetic pathway. Thus, a lot of efforts have been made to construct anthocyanins biosynthetic pathway in S. cerevisiae. For instance, Eichenberger et al. (2018) first reported de novo biosynthesis of pelargonidin-3-O-glucoside, cyanidin-3-O-glucoside, and delphinidin-3-O-glucoside in engineered S. cerevisiae strains. By screening of efficient glucosyltransferase, 0.85 mg/L pelargonidin 3−O−glucoside, 1.55 mg/L cyanidin 3-O-glucoside, and 1.86 mg/L delphinidin-3-O-glucoside were produced in the best engineered strains (Eichenberger et al., 2018). In another example, Levisson et al. (2018) constructed a biosynthetic pathway for the production of pelargonidin 3-O-glucoside starting from glucose in S. cerevisiae. The host strain was engineered to prevent by-product formation and to improve the precursors supply, the resultant strain generated 0.01 μmol/g CDW pelargonidin and 0.001 μmol/g CDW pelargonidin 3-O-glucoside in controlled aerobic batch conditions (Levisson et al., 2018). Except E. coli and S. cerevisiae, several other microorganisms have also demonstrated the ability to synthesize anthocyanins. Zha et al. (2018) constructed a recombinant C. glutamicum strain to produce cyanidin 3-O-glucoside from catechin by overexpression of ANS from P. hybrida and 3GT from A. thaliana. Further adjustment of the expression level of ANS and 3GT, process optimization, and improvement of UDP-glucose availability enabled the production of 40 mg/L cyanidin 3-O-glucoside (Zha et al., 2018). In another study, the Lactococcus lactis was engineered to produce some unusual anthocyanins using tea as substrate. Various red–purple cyanidin and delphinidin, orange, and yellow pyranoanthocyanidins were produced at a total titer of 1.5 mg/L by the end of 16 h (Solopova et al., 2019).
Discussion and Perspectives
For the past few years, with the increasing knowledge about the natural biosynthetic pathways of flavonoids and the rapid development of synthetic biology and metabolic engineering, more and more microbial cell factories have been constructed to produce flavonoids from their direct precursors or from renewable carbon sources (Zhu et al., 2014; Wu et al., 2016b). However, commercial and industrial scale-up synthesis of those chemicals has yet been achieved because of the low product titer, yield, and productivity. In order to greatly improve the flavonoids titer in microorganisms, several challenges need to be addressed. First, the enzymes involved in flavonoids biosynthetic pathway, especially cytochrome P450s and postmodification enzymes, are usually redox partner dependence and have lower enzyme activity and poor selectivity (Stahlhut et al., 2015; Delmulle et al., 2018), which greatly limited the production efficiency of flavonoids. Recently, a lot of strategies have been made to improve their catalytic efficiency and specificity, such as rational protein engineering (Li et al., 2019), directed evolution (Pandey et al., 2016), substrate engineering (Li et al., 2020), and redox-partner engineering (Stahlhut et al., 2015). However, those efforts showed little effects on tackling the problem. We believe that utilization of the promiscuous of some efficient bacterial non-P450 oxygenases and hydroxylases to replace the plant cytochrome P450 enzymes serves as a promising alternative. Second, the flavonoids biosynthetic pathway involves lengthy reaction steps and complicated regulations that significantly limit production yield. To overcome this obstacle, some researchers attempted to construct coculture or polyculture system to distribute metabolic burden by dividing the entire biosynthetic pathway into several independent host strains (Ganesan et al., 2017; Thuan et al., 2018; Wang et al., 2020). However, the metabolite transport limitation and the difficulty in keeping stable and reliable coculture system are the primary challenges for a wide-range application of this approach (Jones et al., 2016b, 2017). Therefore, it is quite important for further optimization of the current coculture systems or seeking novel strategies to overcome this issue. Third, the production efficiency of flavonoids biosynthetic pathway is also limited by the insufficient supply of the precursors, especially the aromatic amino acids and malonyl-CoA (Koopman et al., 2012; Delmulle et al., 2018). However, those precursors also serve as essential intermediates to support cell growth; thus, it is very important to precisely distribute the carbon flux between the central metabolic pathways and the heterologous biosynthetic pathways. Recently, construction of dynamic regulatory elements to fine-tune the carbon flux between cell growth and products synthesis has been shown as a powerful tool to improve microbial synthesis efficiency (Wu et al., 2014, 2016a; Zhou et al., 2019). In addition, oleaginous yeast strains including Yarrowia, Candida, Lipomyces, and Rhodotorula are able to accumulate a high level of malonyl-CoA (Lv et al., 2019b), which serve as the promising hosts for flavonoid biosynthesis. The rapid and continued development of advanced tools and technologies is expected to create new and exciting opportunities in achieving efficient and economical production of flavonoids.
Author Contributions
HS and JW drafted the manuscript. XSh, JW, and XSu revised the manuscript. YY and QY supervised this work. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (21908003, 21636001, and 21776008), the National Key Research and Development Program of China (2018YFA0903000 and 2018YFA0901800), and the Fundamental Research Funds for the Central Universities (buctrc201911).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Beijing University of Chemical Technology.
References
Abari, A. H., and Tayebi, M. (2019). Bioconversion of genistein to orobol by Bacillus subtilis spore displayed tyrosinase and monitoring the anticancer effects of orobol on MCF-7 Breast Cancer Cells. Biotechnol. Bioprocess Eng. 24, 507–512. doi: 10.1007/s12257-019-0067-9
Akdemir, H., Silva, A., Zha, J., Zagorevski, D. V., and Koffas, M. A. G. (2019). Production of pyranoanthocyanins using Escherichia coli co-cultures. Metab. Eng. 55, 290–298. doi: 10.1016/j.ymben.2019.05.008
Amor, I. L.-B., Hehn, A., Guedon, E., Ghedira, K., Engasser, J. M., Chekir-Ghedrira, L., et al. (2010). Biotransformation of naringenin to eriodictyol by Saccharomyces cerevisiea functionally expressing flavonoid 3′ hydroxylase. Nat. Prod. Commun. 5, 1893–1898. doi: 10.1177/1934578x1000501211
Averesch, N. J., and Krömer, J. O. (2018). Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds-present and future strain construction strategies. Front. Bioeng. Biotechnol. 6:32. doi: 10.3389/fbioe.2018.00032
Bano, S., Javed, K., Ahmad, S., Rathish, I., Singh, S., Chaitanya, M., et al. (2013). Synthesis of some novel chalcones, flavanones and flavones and evaluation of their anti-inflammatory activity. Eur. J. Med. Chem. 65, 51–59. doi: 10.1016/j.ejmech.2013.04.056
Bowen-Forbes, C. S., Zhang, Y., and Nair, M. G. (2010). Anthocyanin content, antioxidant, anti-inflammatory and anticancer properties of blackberry and raspberry fruits. J. Food Compost. Anal. 23, 554–560. doi: 10.1016/j.jfca.2009.08.012
Britsch, L., and Grisebach, H. (1986). Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur. J. Biochem. 156, 569–577. doi: 10.1111/j.1432-1033.1986.tb09616.x
Caltagirone, S., Rossi, C., Poggi, A., Ranelletti, F. O., Natali, P. G., Brunetti, M., et al. (2000). Flavonoids apigenin and quercetin inhibit melanoma growth and metastatic potential. Int. J. Cancer 87, 595–600. doi: 10.1002/1097-0215(20000815)87:4<595::aid-ijc21<3.0.co;2-5
Chang, T., Chao, S., and Chen, Y. (2013). Production of ortho-hydroxydaidzein derivatives by a recombinant strain of Pichia pastoris harboring a cytochrome P450 fusion gene. Process Biochem. 48, 426–429. doi: 10.1016/j.procbio.2013.02.014
Chiang, C., Chang, Y., Wu, J., and Chang, T. (2017). Production and anti-melanoma activity of methoxyisoflavones from the biotransformation of genistein by two recombinant Escherichia coli strains. Molecules 22:87. doi: 10.3390/molecules22010087
Chon, S. (2013). Total polyphenols and bioactivity of seeds and sprouts in several legumes. Curr. Pharm. Des. 19, 6112–6124. doi: 10.2174/1381612811319340005
Correa, M. J. C., Nunes, F. M., Bitencourt, H. R., Borges, F. C., Guilhon, G. M. S. P., Arruda, M. S. P., et al. (2011). Biotransformation of chalcones by the endophytic fungus Aspergillus flavus isolated from Paspalum maritimum trin. J. Braz. Chem. Soc. 22, 1333–1338. doi: 10.1590/S0103-50532011000700019
Cress, B. F., Leitz, Q. D., Kim, D. C., Amore, T. D., Suzuki, J. Y., Linhardt, R. J., et al. (2017). CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Fact. 16:10. doi: 10.1186/s12934-016-0623-3
Cushnie, T. T., and Lamb, A. J. (2011). Recent advances in understanding the antibacterial properties of flavonoids. Int. J. Antimicrob. Agents 38, 99–107. doi: 10.1016/j.ijantimicag.2011.02.014
Delmulle, T., De Maeseneire, S. L., and De Mey, M. (2018). Challenges in the microbial production of flavonoids. Phytochem. Rev. 17, 229–247. doi: 10.1007/s11101-017-9515-3
Di Majo, D., Giammanco, M., La Guardia, M., Tripoli, E., Giammanco, S., and Finotti, E. (2005). Flavanones in Citrus fruit: Structure-antioxidant activity relationships. Food Res. Int. 38, 1161–1166. doi: 10.1016/j.foodres.2005.05.001
Ding, H., Chiang, C., Tzeng, W., and Chang, T. (2015). Identification of 3′-hydroxygenistein as a potent melanogenesis inhibitor from biotransformation of genistein by recombinant Pichia pastoris. Process Biochem. 50, 1614–1617. doi: 10.1016/j.procbio.2015.06.007
Dong, J., and Qin, L. (2011). Soy isoflavones consumption and risk of breast cancer incidence or recurrence: a meta-analysis of prospective studies. Breast Cancer Res. Treat. 125, 315–323. doi: 10.1007/s10549-010-1270-8
Edwards, R., and Dixon, R. A. (1991). Isoflavone O-methyltransferase activities in elicitor-treated cell suspension cultures of Medicago sativa. Phytochemistry 30, 2597–2606. doi: 10.1016/0031-9422(91)85107-B
Eichenberger, M., Hansson, A., Fischer, D., Dürr, L., and Naesby, M. (2018). De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 18:11. doi: 10.1093/femsyr/foy046
Falcone Ferreyra, M. L., Rius, S., and Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3:222. doi: 10.3389/fpls.2012.00222
Fazel Nabavi, S., Braidy, N., Habtemariam, S., Sureda, A., Manayi, A., and Mohammad Nabavi, S. (2016). Neuroprotective effects of fisetin in Alzheimer’s and Parkinson’s Diseases: From chemistry to medicine. Curr. Top. Med. Chem. 16, 1910–1915. doi: 10.2174/1568026616666160204121725
Fowler, Z. L., Gikandi, W. W., and Koffas, M. A. G. (2009). Increased malonyl coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavanone production. Appl. Environ. Microbiol. 75, 5831–5839. doi: 10.1128/AEM.00270-09
Fox, J. E., Starcevic, M., Kow, K. Y., Burow, M. E., and McLachlan, J. A. (2001). Nitrogen fixation: endocrine disrupters and flavonoid signalling. Nature 413, 128–130. doi: 10.1038/35093163
Ganesan, V., Li, Z., Wang, X., and Zhang, H. (2017). Heterologous biosynthesis of natural product naringenin by co-culture engineering. Synth. Syst. Biotechnol. 2, 236–242. doi: 10.1016/j.synbio.2017.08.003
Gao, S., Lyu, Y., Zeng, W., Du, G., Zhou, J., and Chen, J. (2019). Efficient Biosynthesis of (2S)-Naringenin from p-Coumaric Acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 68, 1015–1021. doi: 10.1021/acs.jafc.9b05218
Georgiev, V., Ananga, A., and Tsolova, V. (2014). Recent advances and uses of grape flavonoids as nutraceuticals. Nutrients 6, 391–415. doi: 10.3390/nu6010391
Hajimehdipoor, H., Shahrestani, R., and Shekarchi, M. (2014). Investigating the synergistic antioxidant effects of some flavonoid and phenolic compounds. Res. J. Pharmacogn. 1, 35–40.
He, X., Li, W., Blount, J. W., and Dixon, R. A. (2008). Regioselective synthesis of plant (iso) flavone glycosides in Escherichia coli. Appl. Microbiol. Biotechnol. 80, 253–260. doi: 10.1007/s00253-008-1554-7
Heredia, F. J., Francia-Aricha, E. M., Rivas-Gonzalo, J. C., Vicario, I. M., and Santos-Buelga, C. (1998). Chromatic characterization of anthocyanins from red grapes-I. pH effect. Food Chem. 63, 491–498. doi: 10.1016/S0308-8146(98)00051-X
Hirvonen, T., Pietinen, P., Virtanen, M., Ovaskainen, M.-L., Häkkinen, S., Albanes, D., et al. (2001). Intake of flavonols and flavones and risk of coronary heart disease in male smokers. Epidemiology 12, 62–67. doi: 10.1097/00001648-200101000-00011
Horinouchi, S. (2009). Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Curr. Opin. Chem. Biol. 13, 197–204. doi: 10.1016/j.cbpa.2009.02.004
Hosseini-Abari, A., Kim, B.-G., Lee, S.-H., Emtiazi, G., Kim, W., and Kim, J.-H. (2016). Surface display of bacterial tyrosinase on spores of Bacillus subtilis using CotE as an anchor protein. J. Basic Microbiol. 56, 1331–1337. doi: 10.1002/jobm.201600203
Huang, Y., and Ho, S. (2010). Polymethoxy flavones are responsible for the anti-inflammatory activity of citrus fruit peel. Food Chem. 119, 868–873. doi: 10.1016/j.foodchem.2009.09.092
Hwang, E. I., Kaneko, M., Ohnishi, Y., and Horinouchi, S. (2003). Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 69, 2699–2706. doi: 10.1128/AEM.69.5.2699-2706.2003
Ismail, B., and Hayes, K. (2005). β-Glycosidase activity toward different glycosidic forms of isoflavones. J. Agric. Food Chem. 53, 4918–4924. doi: 10.1021/jf0404694
Jackman, R. L., Yada, R. Y., Tung, M. A., and Speers, R. A. (1987). Anthocyanins as food colorants-a review. J. Food Biochem. 11, 201–247. doi: 10.1111/j.1745-4514.1987.tb00123.x
Jiang, H., Wood, K. V., and Morgan, J. A. (2005). Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 2962–2969. doi: 10.1128/AEM.71.6.2962-2969.2005
Jones, J. A., Collins, S. M., Vernacchio, V. R., Lachance, D. M., and Koffas, M. A. G. (2016a). Optimization of naringenin and p-coumaric acid hydroxylation using the native E. coli hydroxylase complex. HpaBC. Biotechnol. Prog. 32, 21–25. doi: 10.1002/btpr.2185
Jones, J. A., Vernacchio, V. R., Sinkoe, A., Collins, S. M., Ibrahim, M. H. A., Lachance, D. M., et al. (2016b). Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab. Eng. 35, 55–63. doi: 10.1016/j.ymben.2016.01.006
Jones, J. A., Vernacchio, V. R., Collins, S. M., Shirke, A. N., Xiu, Y., Englaender, J. A., et al. (2017). Complete biosynthesis of anthocyanins using E. coli polycultures. mBIO 8:e00621-17. doi: 10.1128/mBio.00621-17
Kallscheuer, N., Vogt, M., Stenzel, A., Gätgens, J., Bott, M., and Marienhagen, J. (2016). Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 38, 47–55. doi: 10.1016/j.ymben.2016.06.003
Katsuyama, Y., Miyahisa, I., Funa, N., and Horinouchi, S. (2007). One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 73, 1143–1149. doi: 10.1007/s00253-006-0568-2
Kaufman, P. B., Duke, J. A., Brielmann, H., Boik, J., and Hoyt, J. E. (1997). A comparative survey of leguminous plants as sources of the isoflavones, genistein and daidzein: implications for human nutrition and health. J. Altern. Complement Med. 3, 7–12. doi: 10.1089/acm.1997.3.7
Kay, C. D., Hooper, L., Kroon, P. A., Rimm, E. B., and Cassidy, A. (2012). Relative impact of flavonoid composition, dose and structure on vascular function: a systematic review of randomised controlled trials of flavonoid-rich food products. Mol. Nutr. Food Res. 56, 1605–1616. doi: 10.1002/mnfr.201200363
Ketabforoosh, S. H., Kheirollahi, A., Safavi, M., Esmati, N., Ardestani, S. K., Emami, S., et al. (2014). Synthesis and Anti-Cancer Activity Evaluation of New Dimethoxylated Chalcone and Flavanone Analogs. Arch. Pharm. 347, 853–860. doi: 10.1002/ardp.201400215
Khan, M. K., Abert-Vian, M., Fabiano-Tixier, A.-S., Dangles, O., and Chemat, F. (2010). Ultrasound-assisted extraction of polyphenols (flavanone glycosides) from orange (Citrus sinensis L.) peel. Food Chem. 119, 851–858. doi: 10.1016/j.foodchem.2009.08.046
Kim, D. H., Kim, B.-G., Jung, N. R., and Ahn, J.-H. (2009). Production of genistein from naringenin using Escherichia coli containing isoflavone synthase-cytochrome P450 reductase fusion protein. J. Microbiol. Biotechnol. 19, 1612–1616. doi: 10.4014/jmb.0905.05043
Kim, H. P., Son, K. H., Chang, H. W., and Kang, S. S. (2004). Anti-inflammatory plant flavonoids and cellular action mechanisms. J. Pharmacol. Sci. 3, 229–245. doi: 10.1254/jphs.crj04003x
Koirala, N., Pandey, R. P., Thuan, N. H., Ghimire, G. P., Jung, H. J., Oh, T. J., et al. (2019). Metabolic engineering of Escherichia coli for the production of isoflavonoid-4′-O-methoxides and their biological activities. Biotechnol. Appl. Biochem. 66, 484–493. doi: 10.1002/bab.1452
Koirala, N., Thuan, N. H., Ghimire, G. P., Van Thang, D., and Sohng, J. K. (2016). Methylation of flavonoids: chemical structures, bioactivities, progress and perspectives for biotechnological production. Enzyme Microb. Technol. 86, 103–116. doi: 10.1016/j.enzmictec.2016.02.003
Koopman, F., Beekwilder, J., Crimi, B., van Houwelingen, A., Hall, R. D., Bosch, D., et al. (2012). De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb. Cell Fact. 11:155. doi: 10.1186/1475-2859-11-155
Kootstra, A. (1994). Protection from UV-B-induced DNA damage by flavonoids. Plant Mol. Biol. 26, 771–774. doi: 10.1007/BF00013762
Kumar, N. B., Cantor, A., Allen, K., Riccardi, D., Besterman-Dahan, K., Seigne, J., et al. (2004). The specific role of isoflavones in reducing prostate cancer risk. Prostate 59, 141–147. doi: 10.1002/pros.10362
Lee, H., Kim, B. G., Kim, M., and Ahn, J.-H. (2015). Biosynthesis of two flavones, apigenin and genkwanin in Escherichia coli. J. Microbiol. Biotechnol. 25, 1442–1448. doi: 10.4014/jmb.1503.03011
Leonard, E., Chemler, J., Lim, K. H., and Koffas, M. A. G. (2006). Expression of a soluble flavone synthase allows the biosynthesis of phytoestrogen derivatives in Escherichia coli. Appl. Microbiol. Biotechnol. 70, 85–91. doi: 10.1007/s00253-005-0059-x
Leonard, E., and Koffas, M. A. (2007). Engineering of artificial plant cytochrome P450 enzymes for synthesis of isoflavones by Escherichia coli. Appl. Environ. Microbiol. 73, 7246–7251. doi: 10.1128/AEM.01411-07
Leonard, E., Lim, K. H., Saw, P. N., and Koffas, M. A. G. (2007). Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl. Environ. Microbiol. 73, 3877–3886. doi: 10.1128/AEM.00200-07
Leonard, E., Yan, Y., Fowler, Z. L., Li, Z., Lim, C.-G., Lim, K. H., et al. (2008). Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol. Pharm. 5, 257–265. doi: 10.1021/mp7001472
Levisson, M., Patinios, C., Hein, S., de Groot, P. A., Daran, J.-M., Hall, R. D., et al. (2018). Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb. Cell Fact. 17:103. doi: 10.1186/s12934-018-0951-6
Li, J., Tian, C., Xia, Y., Mutanda, I., Wang, K., and Wang, Y. (2019). Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine. Metab. Eng. 52, 124–133. doi: 10.1016/j.ymben.2018.11.008
Li, Z., Jiang, Y., Guengerich, F. P., Ma, L., Li, S., and Zhang, W. (2020). Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 295, 833–849. doi: 10.1074/jbc.REV119.008758
Liu, X., Cheng, J., Zhang, G., Ding, W., Duan, L., Yang, J., et al. (2018). Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 9, 1–10. doi: 10.1038/s41467-018-02883-z
Li-Weber, M. (2009). New therapeutic aspects of flavones: the anticancer properties of Scutellaria and its main active constituents Wogonin, Baicalein and Baicalin. Cancer Treat. Rev. 35, 57–68. doi: 10.1016/j.ctrv.2008.09.005
Lv, Y., Edwards, H., Zhou, J., and Xu, P. (2019a). Combining 26s rDNA and the Cre-loxP system for iterative gene integration and efficient marker curation in Yarrowia lipolytica. ACS Synth. Biol. 8, 568–576. doi: 10.1021/acssynbio.8b00535
Lv, Y., Marsafari, M., Koffas, M. A. G., Zhou, J., and Xu, P. (2019b). Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis. ACS Synth. Biol. 8, 2514–2523. doi: 10.1101/614099
Lv, Y., Xu, S., Lyu, Y., Zhou, S., Du, G., Chen, J., et al. (2019c). Engineering enzymatic cascades for the efficient biotransformation of eugenol and taxifolin to silybin and isosilybin. Green Chem. 21, 1660–1667. doi: 10.1039/C8GC03728K
Lyu, X., Ng, K. R., Lee, J., Mark, R., and Chen, W. (2017). Enhancement of naringenin biosynthesis from tyrosine by metabolic engineering of Saccharomyces cerevisiae. J. Agric. Food Chem. 65, 6638–6646. doi: 10.1021/acs.jafc.7b02507
Lyu, X., Zhao, G., Ng, K. R., Mark, R., and Chen, W. (2019). Metabolic engineering of Saccharomyces cerevisiae for de novo production of kaempferol. J. Agric. Food Chem. 67, 5596–5606. doi: 10.1021/acs.jafc.9b01329
Marín, L., Gutiérrez-del-Río, I., Entrialgo-Cadierno, R., Villar, C. J., and Lombó, F. (2018). De novo biosynthesis of myricetin, kaempferol and quercetin in Streptomyces albus and Streptomyces coelicolor. PLoS One 13:e0207278. doi: 10.1371/journal.pone.0207278
Marín, L., Gutiérrez-del-Río, I., Yagüe, P., Manteca, Á, Villar, C. J., and Lombó, F. (2017). De novo biosynthesis of apigenin, luteolin, and eriodictyol in the actinomycete Streptomyces albus and production improvement by feeding and spore conditioning. Front. Microbiol. 8:921. doi: 10.3389/fmicb.2017.00921
Miguel, M. G. (2011). Anthocyanins: Antioxidant and/or anti-inflammatory activities. J. Appl. Pharm. Sci. 1, 7–15.
Miranda, C. L., Stevens, J. F., Ivanov, V., McCall, M., Frei, B., Deinzer, M. L., et al. (2000). Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J. Agric. Food Chem. 48, 3876–3884. doi: 10.1021/jf0002995
Mojica, L., Berhow, M. A., and De Mejia, E. G. (2017). Black bean anthocyanin-rich extracts as food colorants: Physicochemical stability and antidiabetes potential. Food Chem. 229, 628–639. doi: 10.1016/j.foodchem.2017.02.124
Nabavi, S. M., Šamec, D., Tomczyk, M., Milella, L., Russo, D., Habtemariam, S., et al. (2020). Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 38:107316. doi: 10.1016/j.biotechadv.2018.11.005
Naderi, G. A., Asgary, S., Sarraf-Zadegan, G. N., and Shirvany, H. (2003). “Anti-oxidant effect of flavonoids on the susceptibility of LDL oxidation,” in Vascular Biochemistry, eds G. N. Pierce, J. Wigle, and P. Zahradka (Berlin: Springer), 193–196. doi: 10.1007/978-1-4615-0298-2_27
Nakayama, T., Yonekura-Sakakibara, K., Sato, T., Kikuchi, S., Fukui, Y., Fukuchi-Mizutani, M., et al. (2000). Aureusidin synthase: a polyphenol oxidase homolog responsible for flower coloration. Science 290, 1163–1166. doi: 10.1126/science.290.5494.1163
Neukam, K., Stahl, W., Tronnier, H., Sies, H., and Heinrich, U. (2007). Consumption of flavanol-rich cocoa acutely increases microcirculation in human skin. Eur. J. Nutr. 46, 53–56. doi: 10.1007/s00394-006-0627-6
Panche, A., Diwan, A., and Chandra, S. (2016). Flavonoids: an overview. J. Nutr. Sci. 5:e47. doi: 10.1017/jns.2016.41
Pandey, R. P., Parajuli, P., Koffas, M. A. G., and Sohng, J. K. (2016). Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 34, 634–662. doi: 10.1016/j.biotechadv.2016.02.012
Pei, J., Sun, Q., Gu, N., Zhao, L., Fang, X., Tang, F., et al. (2020). Production of isoorientin and isovitexin from luteolin and apigenin using coupled catalysis of glycosyltransferase and sucrose synthase. Appl. Biochem. Biotechnol. 190, 601–615. doi: 10.1007/s12010-019-03112-z
Peterson, J. J., Beecher, G. R., Bhagwat, S. A., Dwyer, J. T., Gebhardt, S. E., Haytowitz, D. B., et al. (2006). Flavanones in grapefruit, lemons, and limes: A compilation and review of the data from the analytical literature. J. Food Compost. Anal. 19, S74–S80. doi: 10.1016/j.jfca.2005.12.009
Pyne, M. E., Narcross, L., and Martin, V. J. (2019). Engineering plant secondary metabolism in microbial systems. Plant Physiol. 179, 844–861. doi: 10.1104/pp.18.01291
Ravishankar, D., Rajora, A. K., Greco, F., and Osborn, H. M. (2013). Flavonoids as prospective compounds for anti-cancer therapy. Int. J. Biochem. Cell Biol. 45, 2821–2831. doi: 10.1016/j.biocel.2013.10.004
Rodriguez, A., Strucko, T., Stahlhut, S. G., Kristensen, M., Svenssen, D. K., Forster, J., et al. (2017). Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour. Technol. 245, 1645–1654. doi: 10.1016/j.biortech.2017.06.043
Santos, C. N. S., Koffas, M. A. G., and Stephanopoulos, G. (2011). Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab. Eng. 13, 392–400. doi: 10.1016/j.ymben.2011.02.002
Schroeter, H., Heiss, C., Balzer, J., Kleinbongard, P., Keen, C. L., Hollenberg, N. K., et al. (2006). (–)-Epicatechin mediates beneficial effects of flavanol-rich cocoa on vascular function in humans. Proc. Natl. Acad. Sci. U.S.A. 103, 1024–1029. doi: 10.1073/pnas.0510168103
Shah, F. L. A., Ramzi, A. B., Baharum, S. N., Noor, N. M., Goh, H.-H., Leow, T. C., et al. (2019). Recent advancement of engineering microbial hosts for the biotechnological production of flavonoids. Mol. Biol. Rep. 46, 6647–6659. doi: 10.1007/s11033-019-05066-1
Sharma, B., Balomajumder, C., and Roy, P. (2008). Hypoglycemic and hypolipidemic effects of flavonoid rich extract from Eugenia jambolana seeds on streptozotocin induced diabetic rats. Food Chem. Toxicol. 46, 2376–2383. doi: 10.1016/j.fct.2008.03.020
Shrestha, B., Pandey, R. P., Darsandhari, S., Parajuli, P., and Sohng, J. K. (2019). Combinatorial approach for improved cyanidin 3-O-glucoside production in Escherichia coli. Microb. Cell Fact. 18:7. doi: 10.1186/s12934-019-1056-6
Škerget, M., Kotnik, P., Hadolin, M., Hraš, A. R., Simonič, M., and Knez, Ž (2005). Phenols, proanthocyanidins, flavones and flavonols in some plant materials and their antioxidant activities. Food Chem. 89, 191–198. doi: 10.1016/j.foodchem.2004.02.025
Solopova, A., Van Tilburg, A. Y., Foito, A., Allwood, J. W., Stewart, D., Kulakauskas, S., et al. (2019). Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate. Metab. Eng. 54, 160–169. doi: 10.1016/j.ymben.2019.04.002
Stahlhut, S. G., Siedler, S., Malla, S., Harrison, S. J., Maury, J., Neves, A. R., et al. (2015). Assembly of a novel biosynthetic pathway for production of the plant flavonoid fisetin in Escherichia coli. Metab. Eng. 31, 84–93. doi: 10.1016/j.ymben.2015.07.002
Tahara, S., Ingham, J. L., Hanawa, F., and Mizutani, J. (1991). 1H NMR chemical shift value of the isoflavone 5-hydroxyl proton as a convenient indicator of 6-substitution or 2′-hydroxylation. Phytochemistry 30, 1683–1689. doi: 10.1016/0031-9422(91)84234-J
Takamura, Y., and Nomura, G. (1988). Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon and energy metabolism of Escherichia coli K12. Microbiology 134, 2249–2253. doi: 10.1099/00221287-134-8-2249
Taku, K., Melby, M. K., Nishi, N., Omori, T., and Kurzer, M. S. (2011). Soy isoflavones for osteoporosis: an evidence-based approach. Maturitas 70, 333–338. doi: 10.1016/j.maturitas.2011.09.001
Thuan, N. H., Chaudhary, A. K., Van Cuong, D., and Cuong, N. X. (2018). Engineering co-culture system for production of apigetrin in Escherichia coli. J. Ind. Microbiol. Biotechnol. 45, 175–185. doi: 10.1007/s10295-018-2012-x
Tohge, T., de Souza, L. P., and Fernie, A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68, 4013–4028. doi: 10.1093/jxb/erx177
Torskangerpoll, K., and Andersen, ØM. (2005). Colour stability of anthocyanins in aqueous solutions at various pH values. Food Chem. 89, 427–440. doi: 10.1016/j.foodchem.2004.03.002
Trantas, E., Panopoulos, N., and Ververidis, F. (2009). Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab. Eng. 11, 355–366. doi: 10.1016/j.ymben.2009.07.004
van Summeren-Wesenhagen, P. V., and Marienhagen, J. (2015). Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin. Appl. Environ. Microbiol. 81, 840–849. doi: 10.1128/AEM.02966-14
Wang, R., Zhao, S., Wang, Z., and Koffas, M. A. G. (2020). Recent advances in modular co-culture engineering for synthesis of natural products. Curr. Opin. Biotechnol. 62, 65–71. doi: 10.1016/j.copbio.2019.09.004
Wang, T., Tsai, Y., Yu, I., and Chang, T. (2016). Improving 3′-hydroxygenistein production in recombinant Pichia pastoris using periodic hydrogen peroxide-shocking strategy. J. Microbiol. Biotechnol. 26, 498–502. doi: 10.4014/jmb.1509.09013
Watanabe, S., Uesugi, S., and Kikuchi, Y. (2002). Isoflavones for prevention of cancer, cardiovascular diseases, gynecological problems and possible immune potentiation. Biomed. Pharmacother. 56, 302–312. doi: 10.1016/S0753-3322(02)00182-8
Wrolstad, R. E. (2004). Anthocyanin pigments-Bioactivity and coloring properties. J. Food Sci. 69, C419–C425. doi: 10.1111/j.1365-2621.2004.tb10709.x
Wu, J., Du, G., Chen, J., and Zhou, J. (2015). Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci. Rep. 5:13477. doi: 10.1038/srep13477
Wu, J., Yu, O., Du, G., Zhou, J., and Chen, J. (2014). Fine-Tuning of the Fatty Acid Pathway by Synthetic Antisense RNA for Enhanced (2S)-Naringenin Production from L-Tyrosine in Escherichia coli. Appl. Environ. Microbiol. 80, 7283–7292. doi: 10.1128/AEM.02411-14
Wu, J., Zhang, X., Dong, M., and Zhou, J. (2016a). Stepwise modular pathway engineering of Escherichia coli for efficient one-step production of (2S)-pinocembrin. J. Biotechnol. 231, 183–192. doi: 10.1016/j.jbiotec.2016.06.007
Wu, J., Zhang, X., Zhou, J., and Dong, M. (2016b). Efficient biosynthesis of (2S)-pinocembrin from d-glucose by integrating engineering central metabolic pathways with a pH-shift control strategy. Bioresour. Technol. 218, 999–1007. doi: 10.1016/j.biortech.2016.07.066
Yan, Y., Chemler, J., Huang, L., Martens, S., and Koffas, M. A. G. (2005). Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 71, 3617–3623. doi: 10.1128/AEM.71.7.3617-3623.2005
Yan, Y., Li, Z., and Koffas, M. A. G. (2008). High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol. Bioeng. 100, 126–140. doi: 10.1002/bit.21721
Yang, J., Liang, J., Shao, L., Liu, L., Gao, K., Zhang, J., et al. (2020). Green production of silybin and isosilybin by merging metabolic engineering approaches and enzymatic catalysis. Metab. Eng. 59, 44–52. doi: 10.1016/j.ymben.2020.01.007
Zha, J., Zang, Y., Mattozzi, M., Plassmeier, J., and Koffas, M. A. G. (2018). Metabolic engineering of Corynebacterium glutamicum for anthocyanin production. Microb. Cell Fact. 17:143. doi: 10.1186/s12934-018-0990-z
Zhang, J., Zhao, L., Cheng, Q., Ji, B., Yang, M., Sanidad, K. Z., et al. (2018). Structurally different flavonoid subclasses attenuate high-fat and high-fructose diet induced metabolic syndrome in rats. J. Agric. Food Chem. 66, 12412–12420. doi: 10.1021/acs.jafc.8b03574
Zhao, S., Jones, J. A., Lachance, D. M., Bhan, N., Khalidi, O., Venkataraman, S., et al. (2015). Improvement of catechin production in Escherichia coli through combinatorial metabolic engineering. Metab. Eng. 28, 43–53. doi: 10.1016/j.ymben.2014.12.002
Zhong, H., Deng, Y., Wang, X., and Yang, B. (2005). Multivesicular liposome formulation for the sustained delivery of breviscapine. Int. J. Pharm. 301, 15–24. doi: 10.1016/j.ijpharm.2005.04.001
Zhou, S., Lyu, Y., Li, H., Koffas, M. A. G., and Zhou, J. (2019). Fine-tuning the (2S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy. Biotechnol. Bioeng. 116, 1392–1404. doi: 10.1002/bit.26941
Keywords: metabolic engineering, natural products, flavonoids, pathway optimization, microorganism
Citation: Sheng H, Sun X, Yan Y, Yuan Q, Wang J and Shen X (2020) Metabolic Engineering of Microorganisms for the Production of Flavonoids. Front. Bioeng. Biotechnol. 8:589069. doi: 10.3389/fbioe.2020.589069
Received: 30 July 2020; Accepted: 14 September 2020;
Published: 07 October 2020.
Edited by:
Wei Luo, Jiangnan University, ChinaReviewed by:
John Andrew Jones, Miami University, United StatesWeihong Jiang, Shanghai Institutes for Biological Sciences (CAS), China
Copyright © 2020 Sheng, Sun, Yan, Yuan, Wang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Wang, d2FuZ2ppYUBtYWlsLmJ1Y3QuZWR1LmNu; Xiaolin Shen, c2hlbnhsQG1haWwuYnVjdC5lZHUuY24=
 Huakang Sheng1
Huakang Sheng1 Xinxiao Sun
Xinxiao Sun Yajun Yan
Yajun Yan Qipeng Yuan
Qipeng Yuan