- 1Faculty of Medicine, Institute of Histology and Embryology, University of Ljubljana, Ljubljana, Slovenia
- 2Department of Cardiovascular Surgery, University Medical Centre Ljubljana, Ljubljana, Slovenia
- 3Clinical Institute of Genomic Medicine, University Medical Centre Ljubljana, Ljubljana, Slovenia
MicroRNAs are highly investigated for their role in the pathogenesis of cardiovascular diseases. Nevertheless, evidence for clinical implementation is still lacking. In our systematic review, we evaluated the potential of microRNAs as pathophysiological and diagnostic biomarkers of heart failure. We identified 72 differentially expressed microRNA molecules among groups of heart failure patients and control groups by searching the PubMed database. We did not identify a substantial overlap of differentially expressed microRNAs among different studies; only five microRNAs (miR-1228, miR-122, miR-423-5p, miR-142-3p, and exosomal miR-92b-5p) were differentially expressed in more than one included study. Gene set enrichment analysis, based on the gene targets of microRNAs presented in the included studies, showed that gene targets of differentially expressed microRNAs were enriched in the MAPK, TGFβ, PI3K-Akt, and IL-2 signaling pathways, as well as apoptosis pathway, p53 activity regulation, and angiogenesis pathway. Results of our systematic review show that there is currently insufficient support for the use of any of the presented microRNAs as pathophysiological or prognostic biomarkers in the clinical setting.
Introduction
Heart failure is a major cause of morbidity and mortality worldwide. The prevalence of heart failure is on the rise, and it has been estimated that it will grow further and reach 10% of the general population in 2030 (1). It is defined as the inability of the heart to supply the peripheral tissues with a sufficient amount of blood and oxygen to meet their metabolic needs and is mainly a result of age-related cardiovascular conditions and associated changes in cardiovascular structure and function (1). Heart failure is predominantly caused by an underlying myocardial disease (especially myocardial ischemia); however, other cardiac diseases, including valve diseases, endocardial, or pericardial abnormalities and disorders in the heart rhythm, may also result in diminished cardiac function (2). Clinical and research criteria for heart failure are heterogeneous and prone to misclassification. Heart failure is usually diagnosed by physical examination, laboratory workup, cardiovascular imaging, and hemodynamic catheterization (3). One of the main challenges in diagnosing heart failure is the identification of reliable biomarkers. Natriuretic peptides are the most extensively studied and used biomarkers in heart failure (4). The serum brain natriuretic peptide is currently the only routinely used biomarker for heart failure with class 1A recommendation from both American and European guidelines (1, 5). Fibrosis markers galectin-3 and soluble suppression of tumorigenicity-2 have been included in the ACC/AHA guidelines (strength of recommendation 2b), but their clinical value is still uncertain (1, 5). In addition to proteins, circulating microRNAs gained significant interest as potential novel heart failure biomarkers.
MicroRNAs (miRNAs) are small (~22 nucleotides long) endogenous non-coding RNAs that play an important role by regulating post-transcriptional gene expression. They act to either inhibit the translation of messenger RNA or to induce the degradation of specific mRNA (6–8). MiRNAs circulating in the blood were found to be protected from degradation caused by endogenous RNases. Evidence suggests that resistance to degradation is achieved by binding of miRNA to carrier molecules, such as Argonaute 2, nucleophosmin, and lipoproteins (HDL), and by the packaging of miRNA in microparticles such as exosomes, microvesicles, or apoptotic bodies (9–11). Stability in blood, differential expression in healthy tissue vs. pathologically changed tissue, and robust laboratory methods that detect the expression of miRNAs with a high degree of sensitivity and specificity all make miRNAs good candidates as biomarkers of the disease (12).
Differentially expressed miRNA patterns were found to be associated with various pathophysiological mechanisms of heart failure, such as cardiac remodeling, hypertrophy, apoptosis, and hypoxia (13–32). The involvement of miRNAs in various pathophysiological mechanisms of heart failure, diverse subtypes of heart failure investigated in studies, different design and methods of the studies, and the difference in inclusion/exclusion criteria for patient selection, however, mean that their role in heart failure and their potential as biomarkers remains elusive (33).
In this review, we aim to provide an overview of current scientific support for the use of miRNAs as pathophysiological and diagnostic biomarkers for heart failure.
Methods
Search Strategy
The literature search was conducted in the PubMed database until August 2019, using the following terms: (“MiRNAs” OR “microRNAs”) AND (“heart failure” OR “HF”) in the title/abstract. We limited our search to articles written in English. The “AND” operator was used to create all possible combinations of selected terms. The literature search was conducted independently by two authors (AP and KP) who reached consensus on all of the research papers.
Study Selection and Data Collection
Research papers were initially retrieved as title and abstract and screened for eligibility. All selected research papers were then retrieved as complete manuscripts and checked for compliance with inclusion and exclusion criteria. We included human studies meeting the following criteria: (1) heart failure diagnosis based on clinical features and confirmed with echocardiography; (2) the patients had measured specific miRNA expression level at the time of heart failure exacerbation/follow up examination; (3) case–control study design; (4) qPCR, real-time PCR, microarray, and RNAseq are acceptable methods to evaluate the expressions of miRNAs. The exclusion criteria applied to the studies were as follows: (1) patients had received medications before blood/serum samples were collected; (2) heart failure as an immediate consequence of acute myocardial infarction; (3) heart failure studied only on subpopulations of patients, i.e., diabetics; (4) research papers that were not focused on heart failure but were focused only on the specific pathophysiological mechanism leading to heart failure (i.e., hypertension, atherosclerosis, arrhythmias); and (5) the same cohort was already studied in other research papers. Newcastle-Ottawa quality assessment scale (34) was used to assess the quality of included research papers. For each research paper included in the systematic review, the following data were extracted: authors, year of publication, study population geographical origin, number of heart failure patients and controls, selected miRNAs, sample source, and employed diagnostic criteria for heart failure. Additionally, we checked for the information on performed normalization of the differential microRNA expression.
Bioinformatic Analysis/Gene Set Enrichment Analysis
Using miRTarBase (release 8.0 beta), we identified all known gene targets for selected miRNAs that were differentially expressed in the research papers included in the systematic review (35). We then developed the list of all target genes related to 72 miRNAs identified in Table 1 and performed gene set enrichment analysis using Enrichr (36, 37). Results from KEGG, BioPlanet, and Panther databases were analyzed.
Results
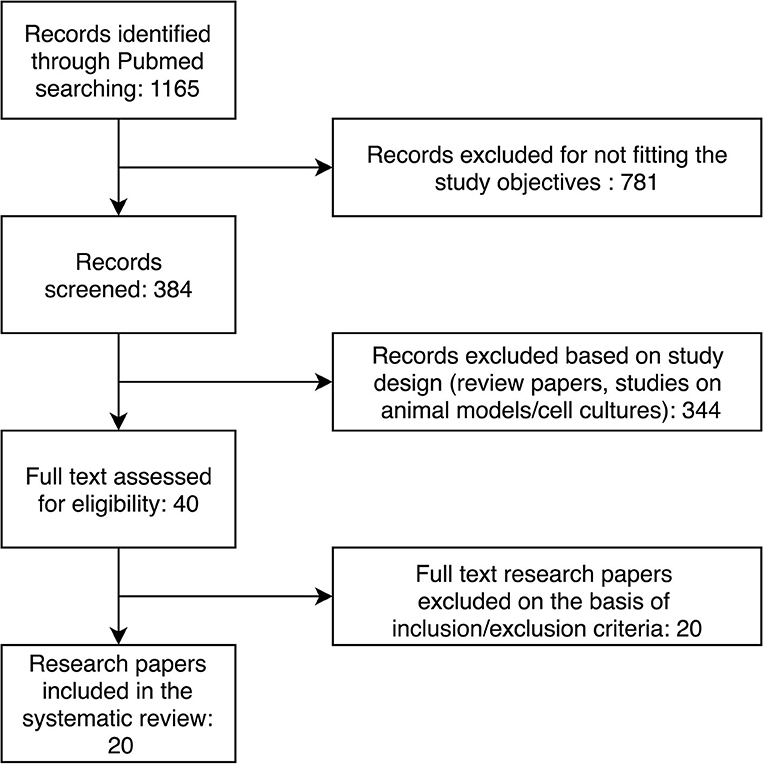
Through the initial keyword search, we retrieved 1,165 research papers. The further selection process is presented in Figure 1. After applying inclusion and exclusion criteria, we identified 20 research papers to include in our systematic review. The characteristics of the included studies are presented in Table 1. Selected studies described 72 differentially expressed miRNAs in patients with heart failure compared to controls without heart failure.
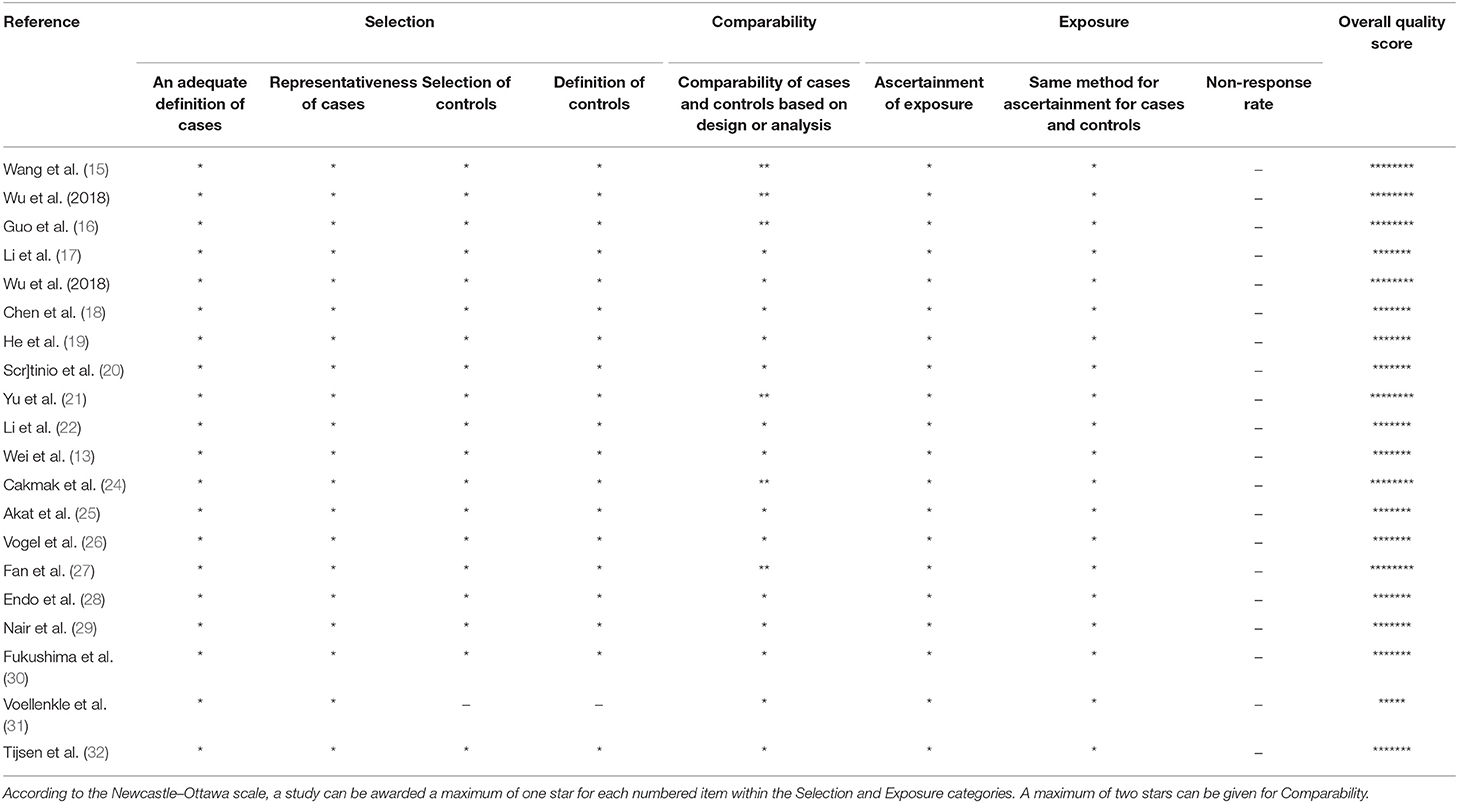
The Newcastle-Ottawa questionnaire was applied to assess the quality of the included studies (Table 2). The most common overall quality score was 7/9. Important shortcomings of studies included in the systematic review were suboptimal comparability of cases and controls as well as different definitions for cases and controls in the included studies.
The majority of included studies (17/18 RT-qPCR studies) provides information on performed normalization. However, there was a discrepancy in the normalization methods (exogenous or endogenous controls) and selected control microRNAs. The most commonly used control microRNAs were U6 snRNA, i.e., in seven studies (13–15, 19, 22, 23, 29, 32) and miR-39 in three studies (16, 20, 27).
By searching miRTarBase (35), we identified 2,052 potential gene targets for selected differentially expressed miRNAs and performed gene set enrichment analysis using Enrichr (36, 37). To reveal a pathophysiologically important set of genes, we investigated KEGG, BioPlanet, and Panther databases. The results are presented in Table 3.
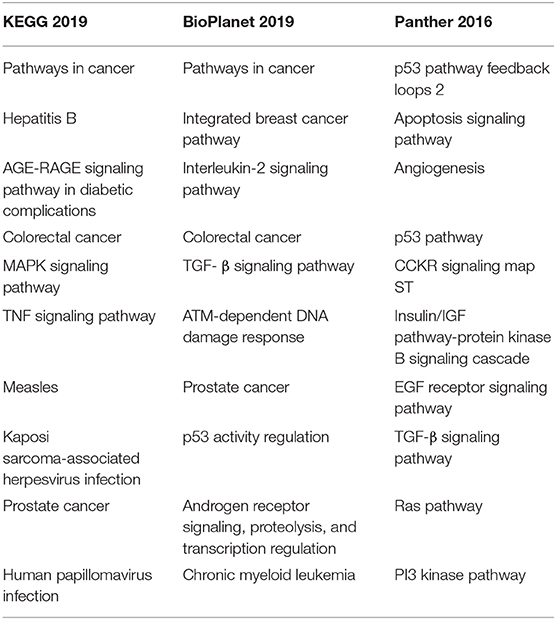
Table 3. Top enriched pathways defined by gene targets of differentially expressed miRNA in studies investigating serum and plasma samples.
Discussion
We identified 72 differentially expressed miRNAs among groups of heart failure patients and control groups with a systematic review. Among 72 differentially expressed miRNAs, only 5 miRNAs, namely, miR-1228, miR-122, miR-423-5p, miR-142-3p, and exosomal miR-92b-5p were differentially expressed in more than one included study.
Two studies found increased levels of miR-1228 and miR-122 in patients with heart failure (24, 26). Vogel et al. found high expression of miR-1228 in CD15+ granulocytes and speculated that differences in leukocyte subpopulations might influence the inflammatory processes known to play an important role in the development and progression of heart failure (26, 26, 38–41). They found miR-122 to be among the most significantly up-regulated miRNAs with good discriminative power as a single marker for systolic heart failure (26). MiR-122 was previously investigated in the porcine cardiogenic shock model, where it was significantly up-regulated in blood samples (42).
Studies by Tijsen et al. and Fan et al. found miR423-5p to be significantly increased in heart failure patients compared to healthy controls and speculated that miR423-5p could be a significant predictor of heart failure diagnosis (27, 32). They also found that miR423-5p correlated with NT-proBNP (27, 32). While Tijsen et al. found a correlation between miR423-5p and NYHA classification, Fan et al. did not find plasma levels of miR423-5p to correlate with NYHA functional class or left ventricular ejection fraction values (27). Tijsen et al. did not specify the subpopulation of heart failure patients, while Fan et al. studied the subpopulation of heart failure patients with dilated cardiomyopathy. Goren et al. reported higher plasma levels of miR423-5p in patients suffering from heart failure due to dilated cardiomyopathy (43). MiR423-5p was up-regulated in array studies of failing human myocardium (44).
The results regarding the differential expression of miR142-3p were conflicting (29, 31). Vollenkle et al. found miR142-3p to be significantly increased in patients with non-ischemic dilated cardiomyopathy, while Nair et al. found miR142-3p to be downregulated in patients with heart failure due to dilated cardiomyopathy. Sample sources in both studies were peripheral blood mononuclear cells (29, 31).
Wu et al. investigated the role of serum exosomal miR-92b-5p in two different subpopulations of heart failure patients (patients with heart failure with reduced ejection fraction and patients with dilated cardiomyopathy caused by acute heart failure). They found increased expression levels of exo-miR-92b in both patient subgroups compared to control groups. Both studies also found miR-92b to be positively related to the left atrium diameter, left ventricular end-diastolic dimension, and left ventricular end-systolic dimension and negatively related to left ventricular ejection fraction and left ventricular fractional shortening (14, 23). Due to sequence similarity with miR-92a, which was studied in the mouse model, they predicted]that miR-92b could be involved in angiogenesis and functional recovery of ischemic tissues (45); however, they acknowledged the need of further basic research to prove the pathophysiological role of miR-92b in heart failure.
Through a thorough assessment of previously published studies, we established that the evidence of the magnitude of effect and certainty of the evidence, contextual factors including improvement of the predictive diagnostic value of investigated miRNAs, and pathophysiological action of miRNA in heart failure are still poorly investigated at present. Similarly, analytical and clinical validity, as well as clinical utility, have not been assessed yet (46, 47). Therefore, further studies are needed, which would follow the scientific statement on the criteria for the evaluation of novel markers of cardiovascular risk (48), which emphasize the importance of research design, representative at-risk population, adequate outcome events, and inclusion of measures of both discrimination and accuracy among others.
Based on the results of this systematic review, it is evident that none of the miRNAs could be considered to be used as a biomarker in the clinical setting. Therefore, we performed gene set enrichment analysis on 2,558 genes that were defined as targets of 72 differentially expressed miRNAs to investigate if there are any common enriched pathways related to pathophysiological processes of heart failure. We found support for an association of pathway enrichment results with heart failure for MAPK signaling pathway, TGFβ signaling pathway, PI3K-Akt signaling pathway, IL-2 signaling pathway, apoptosis pathway, p53 activity regulation, and angiogenesis pathway.
Cardiac angiogenesis, especially of microvasculature, in the setting of pathological cardiac hypertrophy, is insufficient to maintain adequate perfusion. Impaired production of NO from dysfunctional endothelial cells is one of the most researched pathophysiological mechanisms that impair angiogenesis (49). Inhibition of NO production in cardiomyocytes was shown to rapidly increase the production of reactive oxygen species, to activate p38 MAP kinase and enhance TGFβ and TNFα expression (50).
Mitogen-activated protein kinase (MAPK) signaling cascades are considered to be important regulators of cardiac hypertrophic response (51–53). Numerous reports have demonstrated that p38 MAPK inhibition can reduce cardiomyocyte growth in response to hypertrophic stimuli in vitro (54–57). Furthermore, chronic activation of the p38 MAPK pathway has been associated with the induction of hypertrophic response in cultured cardiomyocytes (55–57). Myocardial ischemia was also found to induce p38 MAPK activation, while p38 MAPK inhibition has been demonstrated to attenuate apoptosis in ischemia/reperfusion-injured hearts (58).
Several studies have already linked different miRNAs with the PI3K-Akt pathway (59–61). Two miRNAs, identified in our systematic review, were implied to have a role in the regulation of the PI3K-Akt pathway, namely, miR-200a-3p and miR302s. MiR-200a-3p accelerated cardiac hypertrophy by directly modulating WDR1 and simultaneously regulating PTEN/PI3K/AKT/CREB/WDR1 pathway (60), while the miR302-367 cluster was found to impair autophagy to worsen cardiac hypertrophy through silencing PTEN and consequently activating PI3K/AKT/mTOR pathway (59). Selective activation of distinct PI3K signaling pathways was also shown in a longitudinal canine model of heart failure and cardiac regeneration (62).
The results of previously published studies on the effects of IL-2 on heart-related pathologies are inconsistent. On the one hand, plasma levels of IL-2 were shown to be elevated in patients with acute myocardial infarction, angina pectoris, and dilated cardiomyopathy, while on the other hand, there are reports that suggest a potential therapeutic effect of IL-2 in the setting of acute myocardial infarction (63, 64).
TGFβ was found to be involved in cardiac remodeling (65). Specifically, numerous studies indicated that increased TGFβ1 expression plays an important role in heart hypertrophy, cardiac fibrosis, and cardiomyocyte apoptosis (66–70). Apoptosis importantly contributes to cardiomyocyte death in acute myocardial infarction (71, 72). A high grade of apoptosis is also present in the setting of unstable angina pectoris (73) and correlates with parameters of progressive left ventricular remodeling (74, 75). Increased p53 expression levels and other components of apoptosis pathways were reported in the myocardial tissue of patients with heart pathologies and were found to progressively increase in the process of heart failure (76–78). For example, it was shown that elevated levels of p53 in the myocardium, as a consequence of hypoxic stress due to acute myocardial infarction, led to the apoptosis of cardiomyocytes (79). P53 was also shown to be up-regulated due to the cardiotoxic effects on myocytes caused by doxorubicin treatment (80).
We discovered that many of the most enriched pathways were associated with different types of cancer, especially prostate cancer, colorectal cancer, and chronic myeloid leukemia. This discovery is not all that surprising since more than 50% of the miRNA genes are located in regions associated with cancer (81).
The systematic evaluation of diagnostic and prognostic values of miRNA presented in this study has some limitations due to different subpopulations of patients with various heart failure phenotypes across different studies, a small number of patients per study, and different technical approaches for miRNA identification and analysis (targeted sequencing or hypothesis-free approach and different methods of normalization). Furthermore, inferrals about a pathophysiological mechanism based on pathway analysis of circulating microRNA should be made with caution since heart tissue was not analyzed directly. Reporting bias may exist to some extent because we only included research papers written in English or because only positive results were published.
Conclusions
The results of our systematic review on the role of miRNAs as pathophysiological and diagnostic biomarkers of heart failure show that there is currently insufficient support for the use of any of the presented miRNAs in the clinical setting. Gene set enrichment analysis showed that gene targets of differentially expressed miRNAs were enriched in pathways playing an important role in the MAPK, TGFβ, PI3K-Akt, PDGF, and IL-2 signaling pathways, as well as, apoptosis pathway, p53 activity regulation, and angiogenesis pathway. To establish the definite value of miRNAs as pathophysiological and diagnostic markers, future experimental studies employing the same methodological design and performed on large sample sizes are needed.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
miRNA/miR, microRNA; HF, heart failure; CHF, chronic heart failure; AHF, acute heart failure; HFrEF, heart failure with reduced ejection fraction; ACC/AHA/HFSA, American College of Cardiology/American Heart Association/Heart Failure Society of America; DC, diastolic dysfunction; DCM-AHF, dilated cardiomyopathy–acute heart failure; NIDCM, non-ischemic dilated cardiomyopathy; ICM, ischemic cardiomyopathy; DCM, dilated cardiomyopathy; LVEF, left ventricular ejection fraction; NYHA, New York Heart Association classification of heart failure; HDL, high-density lipoprotein; PCR, polymerase chain reaction; snRNA, small nuclear RNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; NT-proBNP, N-terminal pro b-type natriuretic peptide; EGAPP Work Group, The Evaluation of Genomic Applications in Practice and Prevention Work Group; MAPK, mitogen-activated protein kinase; TGFβ, transforming growth factor beta; NO, nitric oxide; IL-2, interleukin 2; TA, targeted analysis; HyF, hypothesis-free; PBMC, peripheral blood mononuclear cells.
References
1. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JG, Coats AJ. 2016 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. (2016) 70:309–10. doi: 10.5603/KP.2016.0141
2. Tanai E, Frantz S. Pathophysiology of heart failure. Compr Physiol. (2016) 6:187–214. doi: 10.1002/cphy.c140055
3. Ziaeian B, Fonarow GC. Epidemiology and aetiology of heart failure. Nat Rev Cardiol. (2016) 13:368–78. doi: 10.1038/nrcardio.2016.25
4. Lin DCC, Diamandis EP, Januzzi JL, Maisel A, Jaffe AS, Clerico A. Natriuretic peptides in heart failure. Clin Chem. (2014) 60:1040–6. doi: 10.1373/clinchem.2014.223057
5. Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Mark H. 2013 ACCF/AHA guideline for the management of heart failure: a report of the american college of cardiology foundation/american heart association task force on practice guidelines. J Am Coll Cardiol. (2013) 62:e147–239. doi: 10.1161/CIR.0b013e31829e8776
6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
8. Mirzaei H, Yazdi F, Salehi R, Mirzaei HR. SiRNA and epigenetic aberrations in ovarian cancer. J Cancer Res Ther. (2016) 12:498–508. doi: 10.4103/0973-1482.153661
9. Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B. Delivery of MicroRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. (2009) 2:ra81. doi: 10.1126/scisignal.2000610
10. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF. Argonaute2 complexes carry a population of circulating micrornas independent of vesicles in human plasma. Proc Natl Acad Sci USA. (2011) 108:5003–8. doi: 10.1073/pnas.1019055108
11. Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. (2010) 38:7248–59. doi: 10.1093/nar/gkq601
12. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
13. Wei XJ, Han M, Yang FY, Wei GC, Liang ZG, Yao H, et al. Biological significance of MiR-126 expression in atrial fibrillation and heart failure. Brazilian J Med Biol Res. (2015) 48:983–9. doi: 10.1590/1414-431x20154590
14. Wu T, Chen Y, Du Y, Tao J, Zhou Z, Yang Z. Serum exosomal MiR-92b-5p as a potential biomarker for acute heart failure caused by dilated cardiomyopathy. Cell Physiol Biochem. (2018) 46:1939–50. doi: 10.1159/000489383
15. Wang G, Zheng X, Zheng Y, Rui C, Zhang M, Sun Y. Construction and analysis of the lncRNA-MiRNA-MRNA network based on competitive endogenous RNA reveals functional genes in heart failure. Mol Med Rep. (2019) 19:994–1003. doi: 10.3892/mmr.2018.9734
16. Guo M, Luo J, Zhao J, Shang D, Lv Q, Zang P. Combined use of circulating MiR-133a and NT-ProBNP improves heart failure diagnostic accuracy in elderly patients. Med Sci Monitor. (2018) 24:8840–8. doi: 10.12659/MSM.911632
17. Li G, Song Y, Li YD, Jie LJ, Wu WY, Li JZ. Circulating miRNA-302 family members as potential biomarkers for the diagnosis of acute heart failure. Biomark Med. (2018) 12:871–80. doi: 10.2217/bmm-2018-0132
18. Chen F, Yang J, Li Y, Wang H. Circulating microRNAs as novel biomarkers for heart failure. Hellenic J Cardiol. (2018) 59:209–14. doi: 10.1016/j.hjc.2017.10.002
19. He X, Ji J, Wang T, Wang MB, Chen XL. Upregulation of circulating MiR-195-3p in heart failure. Cardiology. (2017) 138:107–14. doi: 10.1159/000476029
20. Scrutinio D, Conserva F, Passantino A, Iacoviello M, Lagioia R, Gesualdo L. Circulating MicroRNA-150-5p as a novel biomarker for advanced heart failure: a genome-wide prospective study. J Heart Lung Transpl. (2017) 36:616–24. doi: 10.1016/j.healun.2017.02.008
21. Yu M, Liang W, Xie Y, Long Q, Cheng X, Liao YH. Circulating MiR-185 might be a novel biomarker for clinical outcome in patients with dilated cardiomyopathy. Sci Rep. (2016) 6:33580. doi: 10.1038/srep33580
22. Li H, Fan J, Yin Z, Wang F, Chen C, Wang DW. Identification of cardiac-related circulating microrna profile in human chronic heart failure. Oncotarget. (2015) 7:33–45. doi: 10.18632/oncotarget.6631
23. Wu T, Chen Y, Du Y, Tao J, Li W, Zhou Z. Circulating exosomal MiR-92b-5p is a promising diagnostic biomarker of heart failure with reduced ejection fraction patients hospitalized for acute heart failure. J Thor Dis. (2018) 10:6211–20. doi: 10.21037/jtd.2018.10.52
24. Cakmak HA, Coskunpinar E, Ikitimur B, Barman HA, Karadag B, Tiryakioglu NO. The prognostic value of circulating micrornas in heart failure: preliminary results from a genome-wide expression study. J Cardiovasc Med. (2015) 16:431–7. doi: 10.2459/JCM.0000000000000233
25. Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J. Comparative RNA-sequencing analysis of myocardial and circulating small rnas in human heart failure and their utility as biomarkers. Proc Natl Acad Sci USA. (2014) 111:11151–6. doi: 10.1073/pnas.1401724111
26. Vogel BA, Keller KS, Leidinger FP, Sedaghat-Hamedani F, Kayvanpour E, Kloos W. Multivariate MiRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur Heart J. (2013) 34:2812–23. doi: 10.1093/eurheartj/eht256
27. Fan KL, Zhang HF, Shen J, Zhang Q, Li XL. Circulating microRNAs levels in chinese heart failure patients caused by dilated cardiomyopathy. Indian Heart J. (2013) 65:12–6. doi: 10.1016/j.ihj.2012.12.022
28. Endo K, Naito Y, Ji X, Nakanishi M, Noguchi T, Goto Y. MicroRNA 210 as a biomarker for congestive heart failure. Biol Pharm Bull. (2013) 36:48–54. doi: 10.1248/bpb.b12-00578
29. Nair N, Kumar S, Gongora E, Gupta S. Circulating miRNA as novel markers for diastolic dysfunction. Mol Cell Biochem. (2013) 376:33–40. doi: 10.1007/s11010-012-1546-x
30. Fukushima Y, Nakanishi M, Nonogi H, Goto Y, Iwai N. Assessment of plasma miRNAs in congestive heart failure. Circ J. (2011) 75:336–40. doi: 10.1253/circj.CJ-10-0457
31. Voellenkle C, Van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. (2010) 42:420–6. doi: 10.1152/physiolgenomics.00211.2009
32. Tijsen AJ, Creemers EE, Moerland PD, De Windt LJ, Van Der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. (2010) 106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297
33. Vegter EL, Van Der Meer P, De Windt LJ, Pinto YM, Voors AA. microRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail. (2016) 18:457–68. doi: 10.1002/ejhf.495
34. Wells GA, Shea B, O'Connell D, Peterson J. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Ottawa, ON: Ottawa Hospital Research Institute (2000).
35. Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW. MiRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. (2018) 46:D296–302. doi: 10.1093/nar/gkx1067
36. Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV. Enrichr: interactive and collaborative html5 gene list enrichment analysis tool. BMC Bioinform. (2013) 14:128. doi: 10.1186/1471-2105-14-128
37. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. (2016) 44:W90–7. doi: 10.1093/nar/gkw377
38. Marchant DJ, Boyd JH, Lin DC, Granville DJ, Garmaroudi FS, McManus BM. Inflammation in myocardial diseases. Circ Res. (2012) 110:126–44. doi: 10.1161/CIRCRESAHA.111.243170
39. Pfister R, Sharp SJ, Luben R, Wareham NJ, Khaw KT. Differential white blood cell count and incident heart failure in men and women in the EPIC-norfolk study. Eur Heart J. (2012) 33:523–30. doi: 10.1093/eurheartj/ehr457
40. Korantzopoulos P, Vlachou C, Kotsia A, Kalantzi K, Barbouti A, Galaris D. Leukocyte labile iron pool in patients with systolic heart failure. Hellenic J Cardiol. (2012) 53:95–100.
41. Uthamalingam S, Patvardhan EA, Subramanian S, Ahmed W, Martin W, Daley M. Utility of the neutrophil to lymphocyte ratio in predicting long-term outcomes in acute decompensated heart failure. Am J Cardiol. (2011) 107:433–8. doi: 10.1016/j.amjcard.2010.09.039
42. Andersson P, Gidlöf O, Braun OO, Götberg M, van der Pals J, Olde B. Plasma levels of liver-specific mir-122 is massively increased in a porcine cardiogenic shock model and attenuated by hypothermia. Shock. (2012) 37:234–8. doi: 10.1097/SHK.0b013e31823f1811
43. Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of MicroRNAs in patients with heart failure. Eur J Heart Fail. (2012) 14:147–54. doi: 10.1093/eurjhf/hfr155
44. Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, Van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. (2007) 116:258–67. doi: 10.1161/CIRCULATIONAHA.107.687947
45. Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. (2009) 324:1710–3. doi: 10.1126/science.1174381
46. Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP working group: genomic profiling to assess cardiovascular risk to improve cardiovascular health. Genet Med. (2010) 12:839–43. doi: 10.1097/GIM.0b013e3181f872c0
47. Teutsch SM, Bradley LA, Palomaki GE, Haddow JE, Piper M, Calonge N. The evaluation of genomic applications in practice and prevention (EGAPP) initiative: methods of the EGAPP working group. Genet Med. (2009) 11:3–14. doi: 10.1097/GIM.0b013e318184137c
48. Hlatky MA, Greenland P, Arnett DK, Ballantyne CM, Criqui MH, Elkind MS. Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the american heart association. Circulation. (2009) 19:2408–16. doi: 10.1161/CIRCULATIONAHA.109.192278
49. Carnicer R, Crabtree MJ, Sivakumaran V, Casadei B, Kass DA. Nitric oxide synthases in heart failure. Antiox Redox Signal. (2013) 18:1078–99. doi: 10.1089/ars.2012.4824
50. Wenzel S, Rohde C, Wingerning S, Roth J, Kojda G, Dieter Schlüter K. Lack of endothelial nitric oxide synthase-derived nitric oxide formation favors hypertrophy in adult ventricular cardiomyocytes. Hypertension. (2007) 49:193–200. doi: 10.1161/01.HYP.0000250468.02084.ce
51. Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. (1999) 11:211–8. doi: 10.1016/S0955-0674(99)80028-3
52. Rose BA, Force T, Wang M. Mitogen-activated protein kinase signaling in the heart: angels versus demons in a heart-breaking tale. Physiol Rev. (2010) 90:1507–46. doi: 10.1152/physrev.00054.2009
53. Tanaka K, Honda M, Takabatake T. Redox regulation of MAPK pathways and cardiac hypertrophy in adult rat cardiac myocyte. J Am Coll Cardiol. (2001) 37:676–85. doi: 10.1016/S0735-1097(00)01123-2
54. Liang Q, Molkentin JD. Redefining the roles of P38 and JNK signaling in cardiac hypertrophy: dichotomy between cultured myocytes and animal models. J Mol Cell Cardiol. (2003) 35:1385–94. doi: 10.1016/j.yjmcc.2003.10.001
55. Nemoto S, Sheng Z, Lin A. Opposing effects of jun kinase and P38 mitogen-activated protein kinases on cardiomyocyte hypertrophy. Mol Cell Biol. (1998) 8:3518–26. doi: 10.1128/MCB.18.6.3518
56. Wang Y, Huang S, Sah VP, Ross J Jr, Brown JH, Han J, et al. Cardiac muscle cell hypertrophy and apoptosis induced by distinct members of the P38 mitogen-activated protein kinase family. J Biol Chem. (1998) 273:2161–8. doi: 10.1074/jbc.273.4.2161
57. Zechner D, Thuerauf DJ, Hanford DS, Mcdonough PM, Glembotski CC. A role for the p38 mitogen-activated protein kinase pathway in myocardial cell growth, sarcomeric organization, and cardiac-specific gene expression. J Cell Biol. (1997) 139:115–27. doi: 10.1083/jcb.139.1.115
58. Yokota T, Wang Y. P38 MAP kinases in the heart. Gene. (2016) 575:369–76. doi: 10.1016/j.gene.2015.09.030
59. Jin L, Zhou Y, Han L, Piao J. MicroRNA302-367-PI3K-PTEN-AKT-MTORC1 pathway promotes the development of cardiac hypertrophy through controlling autophagy. In Vitro Cell Dev Biol Animal. (2019) 56:112–9. doi: 10.1007/s11626-019-00417-5
60. Yang X, Chen G, Chen Z. MicroRNA-200a-3p is a positive regulator in cardiac hypertrophy through directly targeting WDR1 as well as modulating PTEN/PI3K/AKT/CREB/WDR1 signaling. J Cardiovasc Pharmacol. (2019) 74:453–61. doi: 10.1097/FJC.0000000000000732
61. Zhang CJ, Huang Y, Lu JD, Lin J, Ge ZR, Huang H. Upregulated MicroRNA-132 rescues cardiac fibrosis and restores cardiocyte proliferation in dilated cardiomyopathy through the phosphatase and tensin homolog–mediated pi3k/akt signal transduction pathway. J Cell Biochem. (2019) 120:1232–44. doi: 10.1002/jcb.27081
62. Braz JC, Gill RM, Corbly AK, Jones BD, Jin N, Vlahos CJ. Selective activation of PI3Kα/Akt/GSK-3β signalling and cardiac compensatory hypertrophy during recovery from heart failure. Eur J Heart Fail. (2009) 11:739–48. doi: 10.1093/eurjhf/hfp094
63. Cao CM, Xia Q, Tu J, Chen M, Wu S, Wong TM. Cardioprotection of interleukin-2 is mediated via κ-opioid receptors. J Pharmacol Exp Ther. (2004) 309:560–7. doi: 10.1124/jpet.103.061135
64. Bouchentouf M, Williams P, Forner KA, Cuerquis J, Michaud V, Paradis P. Interleukin-2 enhances angiogenesis and preserves cardiac function following myocardial infarction. Cytokine. (2011) 56:732–38. doi: 10.1016/j.cyto.2011.09.023
65. Dobaczewski M, Chen W, Frangogiannis NG. Transforming growth factor (tgf)-β signaling in cardiac remodeling. J Mol Cell Cardiol. (2011) 51:600–6. doi: 10.1016/j.yjmcc.2010.10.033
66. Lijnen PJ, Petrov VV, Fagard RH. Induction of cardiac fibrosis by transforming growth factor-β1. Mol Genet Metabol. (2000) 71:418–35. doi: 10.1006/mgme.2000.3032
67. Hein S, Arnon E, Kostin S, Schönburg M, Elsässer A, Polyakova V. Progression from compensated hypertrophy to failure in the pressure-overloaded human: heart structural deterioration and compensatory mechanisms. Circulation. (2003) 107:984–91. doi: 10.1161/01.CIR.0000051865.66123.B7
68. Li RK, Li G, Mickle DAG, Weisel RD, Merante F, Luss H. Overexpression of transforming growth factor-β1 and insulin-like growth factor-i in patients with idiopathic hypertrophic cardiomyopathy. Circulation. (1997) 96:874–81. doi: 10.1161/01.CIR.96.3.874
69. Fielitz J, Hein S, Mitrovic V, Pregla R, Zurbrügg HR, Warnecke C. Activation of the cardiac renin-angiotensin system and increased myocardial collagen expression in human aortic valve disease. J Am Coll Cardiol. (2001) 37:1443–9. doi: 10.1016/S0735-1097(01)01170-6
70. Ghavami S, Shojaei S, Yeganeh B, Ande SR, Jangamreddy JR, Mehrpour M. Autophagy is a regulator of TGF-B1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis. (2015) 6:e1696. doi: 10.1038/cddis.2015.36
71. Saraste A, Pulkki K, Kallajoki M, Henriksen K, Parvinen M, Voipio-Pulkki LM. Apoptosis in human acute myocardial infarction. Circulation. (1997) 95:320–3. doi: 10.1161/01.CIR.95.2.320
72. Olivetti G, Quaini F, Sala R, Lagrasta C, Corradi D, Bonacina E. Acute myocardial infarction in humans is associated with activation of programmed myocyte cell death in the surviving portion of the heart. J Mol Cell Cardiol. (1996) 28:2005–16. doi: 10.1006/jmcc.1996.0193
73. Teringova E, Tousek P. Apoptosis in ischemic heart disease. J Transl Med. (2017) 15:1–7. doi: 10.1186/s12967-017-1191-y
74. Baldi A, Abbate A, Bussani R, Patti G, Melfi R, Angelini A. Apoptosis and post-infarction left ventricular remodeling. J Mol Cell Cardiol. (2002) 34:165–74. doi: 10.1006/jmcc.2001.1498
75. Abbate A, Biondi-Zoccai GG, Bussani R, Dobrina A, Camilot D, Feroce F. Increased myocardial apoptosis in patients with unfavorable left ventricular remodeling and early symptomatic post-infarction heart failure. J Am Coll Cardiol. (2003) 41:753–60. doi: 10.1016/S0735-1097(02)02959-5
76. Moorjani N, Westaby S, Narula J, Catarino PA, Brittin R, Tim J. Effects of left ventricular volume overload on mitochondrial and death-receptor-mediated apoptotic pathways in the transition to heart failure. Am J Cardiol. (2009) 103:1261–8. doi: 10.1016/j.amjcard.2009.01.013
77. Park M, Cook AR, Lim JT, Sun Y, Dickens BL. Apoptosis predominates in nonmyocytes in heart failure. Am J Physiol Heart Circ Physiol. (2009) 297:785–91. doi: 10.1152/ajpheart.00310.2009
78. Song H, Conte JV, Foster AH, Mclaughlin JS, Wei C. Increased p53 protein expression in human failing myocardium. J Heart Lung Transpl. (1999) 18:744–9. doi: 10.1016/S1053-2498(98)00039-4
79. Naito AT, Okada S, Minamino T, Iwanaga K, Liu ML, Sumida T, et al. Promotion of CHIP-mediated P53 degradation protects the heart from ischemic injury. Circ Res. (2010) 106:1692–702. doi: 10.1161/CIRCRESAHA.109.214346
80. L'Ecuyer T, Sanjeev S, Thomas R, Novak R, Das L, Campbell W. DNA damage is an early event in doxorubicin-induced cardiac myocyte death. Am J Physiol Heart Circ Physiol. (2006) 291:H1273–80. doi: 10.1152/ajpheart.00738.2005
Keywords: heart failure, biomarker (BM), epigenetics (DNA methylation, histone modifications), microRNA (miR), systematic (literature) review
Citation: Peterlin A, Počivavšek K, Petrovič D and Peterlin B (2020) The Role of microRNAs in Heart Failure: A Systematic Review. Front. Cardiovasc. Med. 7:161. doi: 10.3389/fcvm.2020.00161
Received: 13 February 2020; Accepted: 31 July 2020;
Published: 15 October 2020.
Edited by:
George W. Booz, University of Mississippi Medical Center School of Dentistry, United StatesReviewed by:
Donato Santovito, Hospital of the University of Munich, GermanyHamed Mirzaei, Kashan University of Medical Sciences, Iran
Copyright © 2020 Peterlin, Počivavšek, Petrovič and Peterlin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Peterlin, YW5hLnBldGVybGluQG1mLnVuaS1sai5zaQ==
 Ana Peterlin
Ana Peterlin Karolina Počivavšek2
Karolina Počivavšek2 Borut Peterlin
Borut Peterlin