- 1CAS Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Laboratory for Marine Fisheries Science and Food Production Processes, Pilot National Laboratory for Marine Science and Technology, Qingdao, China
- 4Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 5Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology, Qingdao, China
- 6National and Local Joint Engineering Key Laboratory of Ecological Mariculture, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
Investigating responses of organisms to stressful or new environments with selection pressure is one of the crucial problems in evolutionary biology, and it is of importance to understand the phenotypic and molecular mechanism underlying thermal tolerance under the context of the climate change. The Pacific oyster, Crassostrea gigas, inhabiting the environment with high variation in temperature, is a worldwide aquaculture species. However, summer mortality relevant to the high temperature is one of the problems challenging the oyster industry. An artificial selective breeding program was initiated to select for the thermal tolerance of oysters in an attempt to increase the summer survival rates since 2017 in our study. Parents of thermotolerance oyster selection is are based on acute thermal tolerance under controlled heat stress to strengthen the selection intensity. Furthermore, the phenotypic and the genotypic response to extreme high temperature were investigated based on the comparison of the F1 progeny of the selected and natural populations in growth, physiology, mortality rate post heat stress, genetic structure, and gene expression. The parameter of growth showed no significant change between the selected and natural populations for the 6-month-old oysters. The selected population exhibited a higher survival rate after exposure to heat stress in the laboratory, which is in line with result of the filed experience that summer mortality of selected population was significantly lower than that of natural population. Further, the respiration rate of the selected population increased at 38°C, while it increased at 35°C in the natural population. Simultaneously, metabolism-related enzymes (PK, SOD) showed higher activity levels in the selected population. Furthermore, phylogenetic analysis, population structure, and principal component analysis (PCA) suggested that the selected and natural populations exhibited genetic divergence, with eight genes (IF4A2, IF6, EIF3A, MANBA, DDX43, RECS, CAT2, and BAG4) in the selected regions showing differential expression patterns in response to heat stress in the two populations. This study suggests that artificial selection has a significant effect on phenotype and genome structure for the oyster, our study providing an alternative way to reveal the mechanism underlying thermotolerance that plays an important role to predict the potential adaptation to the climate change.
Introduction
Investigating organisms’ responses to stressful or new environments with selection pressure is one of the important problems in evolutionary biology. Moreover, it is of importance to understand the mechanism of the thermal adaptation not only because the temperature has profound effects on organisms’ physiological and biochemical reaction, but also the ability of organisms to cope with thermal stress is a significant predictor of adaptive potential to climate change. Local adaptation and phenotypic plasticity of organisms may play vital roles in forming intraspecific thermal adaptation divergence under the selection for the tolerance to suboptimal temperature (Sanford and Kelly, 2011; Yampolsky et al., 2014). Although more and more evidences appeared to support the fact that the natural selection is pervasive leading to the divergence of thermotolerance even in the marine invertebrate with long larval dispersal, it is still challenging to infer the adaptive divergence among the natural populations with high connection due to the slight genetic divergence (Li L. et al., 2018). The thermal response to acute heat stress was proved to be highly relevant to the capability to adapt to chronic stress (Yampolsky et al., 2014). The artificial selection for the thermal tolerance to the acute heat stress may provide an effective strategy to intensify the adaptive response to the high temperature under the controlled condition.
As marine molluscs distributing in the intertidal and shallow subtidal zone, oysters experience daily and seasonal variation in temperature. As such it provides an ideal model to study the thermal adaption (Przeslawski et al., 2005). The Pacific oyster Crassostrea gigas originates in East Asia and has become the primary oyster species supporting the worldwide shellfish industry, due to its rapid growth rate and high productivity (Guo, 2009). However, mass mortality is a major problem challenging the oyster industry. Significant recurrent summer losses of the C. gigas have occurred globally for over five decades and the concern over its impact on oyster aquaculture has increased in recent years (Glude, 1975; Cheney and Macdonald, 2000; Soletchnik et al., 2005; Paul-Pont et al., 2013; Alfaro et al., 2018). Elevating temperature is considered as one of the most important abiotic causing the summer mortality of oysters. It has been reported that summer mortality occurs regularly in culture farms as well as on natural oyster beds during the period of summer when temperature increased (Soletchnik et al., 2005). Elevated temperature have been proposed to increase the spread of Vibrio sp. from infected to non-infected oysters (Petton et al., 2013). Therefore, exposure to high temperatures was one of most important causes linked to summer mortality of Pacific oyster.
The artificial selection for the survival rate during summer was proved to be effective to improve the survival rate of the oyster in summer. A study on the West Coast of the United States increased survival rate among the resulting progeny. Average mortality rates of selected third generation families ranged from 13 to 23% compared with 63% of the natural group (Beattie et al., 1980; Hershberger et al., 1984). The studies in France observed a high genetic basis for survival of young C. gigas (under 1 year of age) in field trials during the summer (Samain et al., 2007). The high heritability (0.47–1.8) of this trait has also been demonstrated by divergent selection over successive years and generations (Dégremont et al., 2010). All these studies focused on the morphological traits and survival rate while the genetic and molecular mechanism underpinning the selection was missing. An artificial selective breeding program aimed at increasing the survival rate during the summer since 2017 was initiated in our study. Parents of thermotolerance oyster selection are based on acute thermal tolerance under controlled heat stress to strengthen the selection intensity. The comparison of the selected population with the natural population would provide new insight into the thermal adaptation by investigating the phenotypic and the genetic response to the artificial selection for the thermotolerance.
Most of the studies of the response to heat stress in marine mollusks focused on the physiological aspects at the organismal, cellar and molecular level (Gagnaire et al., 2006; Farcy et al., 2008; Zhang et al., 2016). Thermal stress has a pronounced impact on the physiology of many intertidal species (Yao and Somero, 2014), including reef fishes (Mora and Ospína, 2001), marine snail Chlorostoma funebralis (Gleason and Burton, 2013), geoduck clams Panopea globose (Juarez et al., 2018). Variation in temperature influences the activity of enzymes that are involved in the metabolic processes that support their growth and survival (Tang and Liu, 2005; Alter et al., 2017; Wang et al., 2018). Animals will increase metabolism to ensure adequate energy supply for survival when temperature is elevated (Mora and Maya, 2006; Sokolova et al., 2012). Beyond the thermal optimum, aerobic respiration may transition to anaerobic metabolism because of insufficient oxygen supply and decreased metabolism (Portner, 2001; Anestis et al., 2010). Transcriptomic studies revealed an extensive set of heat stress responsive genes related to energy metabolism, anti-apoptotic pathways and mRNA processing (Yang et al., 2017). There have been reports on dramatic difference in expression level of the HSP70 gene family and genes associated with oxygen binding between susceptible and thermotolerance oysters (Kim et al., 2017). Divergent transcriptional profiles of genes associated with intracellular stress mechanisms of antioxidant defense, energy metabolism and the cytoskeleton were observed in two population of Sydney rock oysters Saccostrea glomerata (one that had been selectively bred over seven generations for fast growth and disease resistance and one wild type) on the hottest pavers (McAfee et al., 2018).
The studies related genetic responses to the natural and artificial are relatively rare, especially for identification of genetic variation linked to the adaptation. A small proportion of adaptive loci were inferred based on the natural populations with slight genetic divergence (Li L. et al., 2018). Genetic analysis based on neutral marker such as microsatellite has been used to evaluate genetic differentiation of the populations of cultured oysters subject to artificial selection (Li et al., 2006) but has not produced sufficient information to understand the mechanism of selection. Next-generation sequencing technologies have promoted the generation of massive SNP markers, which may serve as a useful tool for analyzing population genetics and phylogenomics and identifying varieties (Gupta et al., 2015). Specific length amplified fragment sequencing (SLAF-seq) was developed based on high-throughput sequencing technology, and it has been widely used to elevate understanding of genetic populations in plants and animals (Li Z. et al., 2017; Ge et al., 2019). In summary, the integration of the physiological and genetic response may provide an efficient way to reveal the underpinning of the thermotolerance.
Therefore, the motivation behind this study was to further understand the mechanism in the selection for thermotolerance to acute high temperature. Specifically, we aimed to collect phenomics and genomics evidence to identify discrepant responses to elevated temperature between thermotolerance oysters after one-generation of selection and unselected oysters. The effects of thermal stress on survival rate, respiration rate, the activities of key metabolic enzymes, the expression levels of candidate genes were assessed. The genetic difference was identified by SLAF-seq approach. This study will be useful to understand the potential molecular mechanisms for heat tolerance in oysters.
Materials and Methods
Sample Collection
In October 2017, wild populations of Pacific oyster (Crassostrea gigas) were collected from the Shentanggou (36°21′ N), Qingdao, Shangdong Province, China. Oysters were cleaned to eliminate attaching organisms and acclimated in an aquarium for 14 days with sand-filtered and aerated seawater before experimentation. During the acclimation period, individuals were fed spirulina powder, and seawater was changed daily. Oysters were randomly divided into two populations (the number of individuals in each population is greater than 300). One population served as the parent of the natural population (hereinafter referred to as control population), and continued to be cultured without any treatment. The other population was subjected to heat shock at 42°C (LT50) for 1 h (Li A. et al., 2018). Specifically, oysters were submerged in preheated, aerated filtered seawater baths of 42°C for 1 h, then returned to sea water of ambient temperature for 14 days, before removing and counting dead individuals. Finally, the surviving individuals were selected as parents to produce the next generation of the thermal tolerance population (intensity of selection = 37%).
In April 2018, the progeny of selected parents and control population, were used as the experimental F1 generation population and the control population, respectively. We use 30 males and 30 females to reproduce progeny. Specifically, the eggs of the 30 females were mixed and equally divided into 30 beakers, each of which was fertilized by sperm from one of the 30 males. The larvae were reared in the hatchery and juvenile oysters were cultured in the same sea area using standard protocol (Guo et al., 2012). Six months old F1 generations were collected from the sea and acclimatized to the laboratory aquarium for 2 weeks for follow-up experiments. The treatments of experiment material and processes are shown in Figure 1.
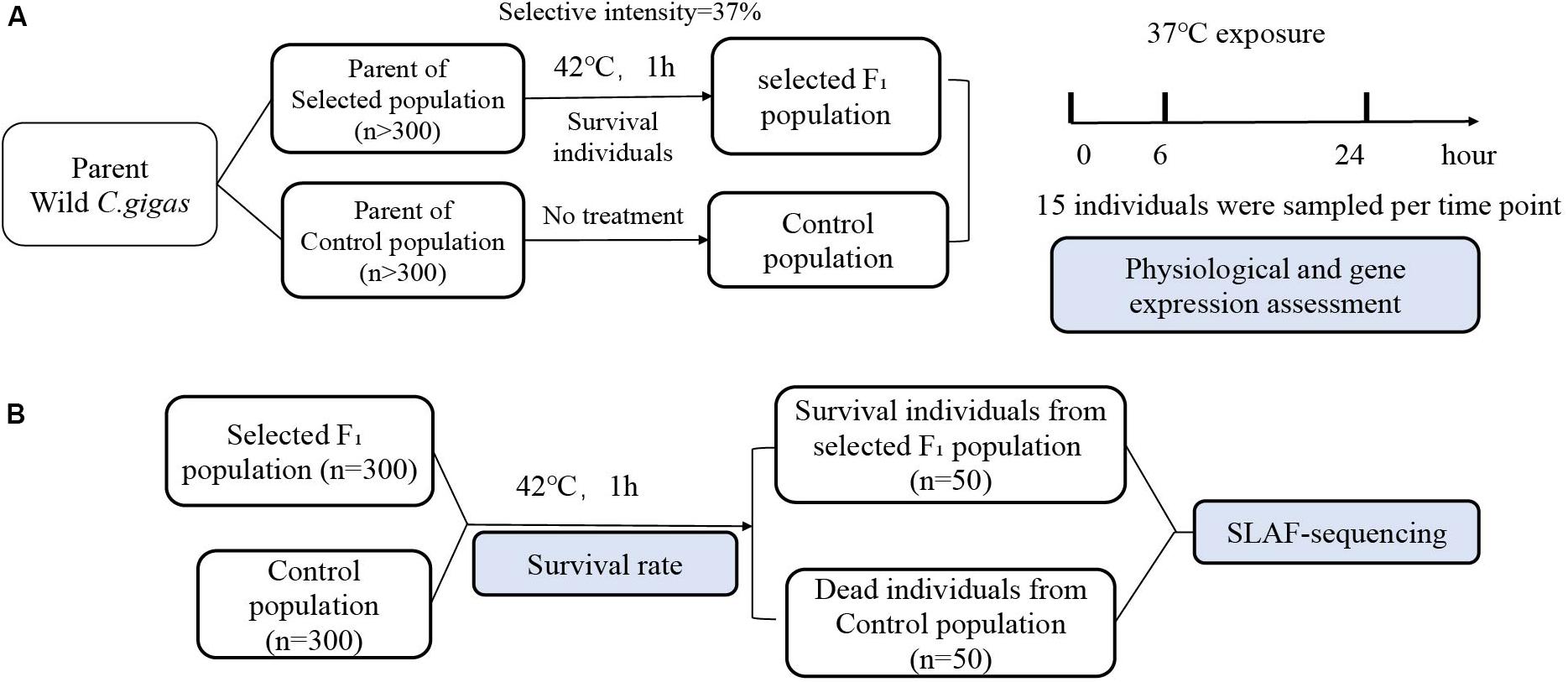
Figure 1. Flow chart of the experiment. (A) Wild C. gigas was randomly divided into two population (n > 300). One population was subjected to heat shock at 42°C for 1 h, and surviving individuals served as the parent of the next generation of selection for thermotolerance population, while the other population served as the control population without any treatment. The individuals from selected F1 population and control population were exposed at 37°C seawater. Gills from 15 individuals were sampled at each time points of 0, 6, 24 h, which used to determine physiological parameters and gene expression. (B) After heat shock at 42°C for 1 h, selected F1 population and control population were kept at ambient temperature restoration for 2-weeks recovery, and mortality was recorded. The gills of dead individuals from control population during the recovery period and that of the survival individuals from selected F1 population after 2 weeks were sampled for SLAF-sequencing (n = 50).
Growth Measurements
The individuals of the selected F1 generation were used as the selected population and the individuals of control F1 generation were used as the control population. A total of 150 oysters were collected from each population. The wet weight and shell height were measured after cleaning the epibionts.
Analysis of Thermal Tolerance
Thermotolerance of the populations was assessed by detecting the survival rate post-acute thermal treatment. Six-month-old oysters were used to examined thermal tolerance (n = 300). The oysters from each population were subjected to heat shock treatment at 42°C (LT50) for 1 h and then removed to ambient temperature. During the recovery process, oysters were fed spirulina powder, and the seawater was changed daily. Visual monitoring of oyster shell activity (open or closed) was performed. The individuals whose shell remained open even after being challenged were considered dead and immediately removed. The survival rate after heat shock for 20 days was recorded.
Respiration Rate Analysis
Six-month-old oysters of similar size (n = 6, shell height: 42.76 ± 3.01 mm [selected population], 41.33 ± 3.56 mm [control population], p = 0.215) were randomly selected to estimate respirate rate at different temperature. One oyster was measured at a time. Individuals were cleaned using seawater to avoid the influence of periphyton. The metabolic rate was measured at 20°C to simulate the natural environment and at 35, 37, 38, and 39°C to simulate acute heat stress, respectively. Individuals were placed in a 330 mL glass bottle filled with sand-filtered, air-saturated seawater at the designed temperature, which was regulated using a water bath. The seawater was mixed using a magnetic stirrer bar beneath the chamber. Needle-type fiber-optic microsensor oxygen sensor and temperature probe were connected to the oxygen transmitter that simultaneously recorded the temperature and oxygen concentration of water every 3 s for 1 h (Li A. et al., 2017). The slope of the decrease in oxygen concentration was calculated as the respiration rate (mg/mL/h).
Assessment Impact of Heat Stress
The gills of oysters after heat shock were used to examine the physiological parameters and expression level of heat responsive genes. Oysters from each population were randomly divided into three replicates and subjected to heat stress of seawater at 37°C for 24 h (shell height: 41.7 ± 5.63 mm [selected population], 40.38 ± 6.64 mm [control population], p = 0.14). The gills of five oysters were individually sampled from each replicate per population at 0, 6, 24 h (n = 15 per sampling time per population). The gills were immediately frozen in the liquid nitrogen and stored at –80°C for follow-up analysis of physiology and gene expression.
Physiological Assessment
According to the ratio of weight: volume = 1: 9, 30 ∼ 40 mg of frozen gill tissue from each sample of every sample time of each population was added to an excess volume (9X) of precooled saline and ground to homogenate in an ice bath. The supernatant was diluted to a 1% homogenate by adding ninefold saline after centrifuging for 10 min at 2500 rpm and 4°C. The 1% homogenate was then stored at –80°C until required for further analysis.
The fundamental physiological indexes, including the content of total protein and malondialdehyde (MDA), and the enzymatic activity of pyruvate kinase (PK), and superoxide dismutase (SOD), were chosen to evaluate the dynamic physiological responses to high temperature in oysters (Li A. et al., 2018). The concentration of protein was used to normalize other enzyme activity indexes. The first three indexes were measured using the corresponding kit from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). SOD levels were determined using the Total Superoxide Dismutase Assay kit with NBT (Beyotime, Shanghai, China). Absorbance values were measured using the Varioskan Flash (Thermo Fisher Scientific, Waltham, MA, United States).
Field Experiment
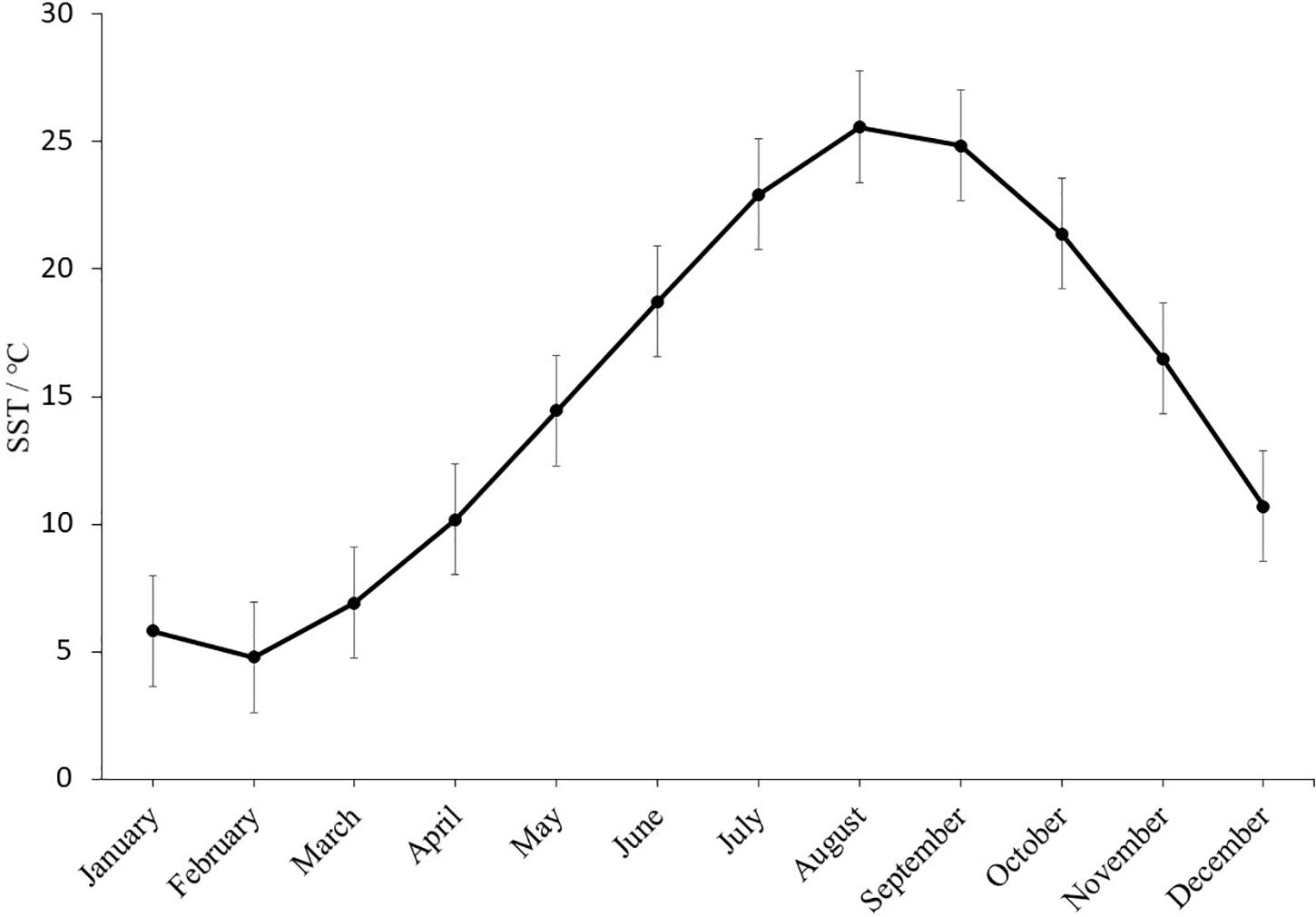
Oysters were cultured in Shentanggou, Qingdao. The monthly average sea surface temperature (SST) of Qingdao in 2019 was showed in Figure 2 which is not significantly different from the past 18 years (Ghaffari et al., 2019). The highest average sea surface temperature was observed in August is 25.57 ± 0.71°C. The salinity of seawater was 30.0 ± 0.5‰. When the F1 oysters were 19-month-old, the survival rate and shell height of the selected and control population were recorded to investigate the thermal resistance of the selected population in the field.
Specific-Locus Amplified Fragment (SLAF) Sequencing Analysis
Sample Collection and DNA Extraction
After heat shock at 42°C for 1 h, the F1 selected and control populations were placed to restore at room temperature for 2 weeks. The gills of individuals from F1 control population that died during the recovery period, and those of the live individuals from the F1 selected population were sampled after 2 weeks, before freezing in liquid nitrogen and storing at –80°C (n = 50 per population). Thus, the oysters with the potential largest differences in thermal tolerance were used for genomic analysis to identify SNPs and candidate genes associated with thermal tolerance. Total DNA was extracted from the gill tissue using the TIANamp Marine Animals DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. DNA quality and concentration were measured by 1.0% agarose gel electrophoresis and UV spectrometry on a NanoDrop 2000 device, respectively.
SLAF Library Construction and Sequencing
Simulated restriction enzyme digestion was performed on the oyster genome in order to avoid repetitive SLAF tags and to obtain a relatively uniform distribution of restriction fragments in the genome for maximum SLAF-seq efficiency. As a result, genomic DNA was digested with RsaI and HaeIII restriction enzyme combination. To evaluate the efficiency and accuracy of the experiment procedure, Oryza sativa ssp. Japonica DNA (Ohyanagi, 2006) was served as control to evaluate the availability of the enzyme digestion. Specific-locus amplified fragment library construction was carried out as previously described (Sun et al., 2013). DNA fragments of 264–314 bp were defined as SLAF tags and diluted for pair-end sequencing on an Illumina High-seq 2500 system (Illumina, Inc., San Diego, CA, United States) at Beijing Biomarker Technologies Corporation.
SLAF-seq Data Processing and SNP Marker Development
The raw paired-end reads were mapped to the oyster reference genome (oyster_v9, GenBank accession number GCA_000297895.1) using BWA software (Li and Durbin, 2009). The position of clean reads was located by comparison with the reference genome, the sequencing depth of each sample was counted, and mutation were detected. The development of SNP markers was performed by both GATK (McKenna et al., 2010) and SAMtools (Li et al., 2009) analysis, the intersection of SNP markers obtained by the two methods as the final reliable SNPs.
Population Genetic Structure
Filtering high-quality SNPs for downstream analysis according to the following criteria: minor allele frequency ≥ 5% and the integrity of each SNP ≥ 80%. A total of 379,002 SNPs from 100 individuals were development for analysis of phylogenetic relationships and population structure. MEGA X (Kumar et al., 2018) was employed to construct phylogenetic trees of each sample with Kimura 2-parameter based on the neighbor joining (NJ) method. The bootstrap repeated 1,000 times was applied to assure the reliable of the phylogenetic trees. The population structure was analyzed using the ADMIXTURE program (Alexander et al., 2009). We conducted a K-means clustering analysis supposing the K-value ranged from 1 to 10. The clustering results were cross-validated, and the optimal group number was determined according to the valley value of the cross-validation error rate. A good K-value will exhibit a low cross-validation error compared to other K-values. Principal component analysis (PCA) was performed with EIGENSOFT program (Price et al., 2006). We can access the relationship among samples through PCA analysis which can assist evolutionary analysis.
Identification of Differentially Selected Regions and Functional Enrichment Analysis
The divergence index, F-statistics (Fst) and the index of nucleotide polymorphism (θπ) were calculated via the PopGenome package (Pfeifer et al., 2014) based on 100 – kb sliding windows in 10 – kb steps. We calculated and used the θπ ratio (θπ, control population/θπ, selected population) and Fst values between the two populations to detect genomic regions with signatures of selection. The region with significantly low and high θπ ratios (low 5% regions and top 5% regions) and significantly high Fst values (top 5% regions) was identified as selected regions with strong selection signals. Gene Ontology (GO) enrichment analysis of target genes were performed in the topGO R package. GO terms with a p-value < 0.05 were considered significantly enriched and genes on these terms were considered as candidate genes.
Expression Level of Candidate Genes
The materials we used here were described in above-mentioned assessment impact of heat stress. GO terms with a p-value < 0.05 were considered significantly enriched and the genes in these terms were identified as candidate genes. Although not enriched in the GO analysis, BAG4 was selected due to its potential role in temperature adaptation, and was also found in the region of top 5% Fst in our study. Therefore, BAG4 was considered a candidate gene.
The gills were used for total RNA extraction with the RNA prep Pure Tissue kit (Tiangen, China) following manufacturer’s protocol. All RNAs were treated with DNase I to avoid genomic contamination in the RNA. The quality and concentration of RNA were assessed using 1.2% agarose gel electrophoresis and a NanoDrop 2000 spectrophotomer. Equal amounts (1 μg) of RNA from five oysters at each sample time of each population were pooled together to reduce the effect of error (three biological replicates × three technical replicates). Reverse transcription was performed using PrimeScript RT reagent Kit (TaKaRa Bio, Shiga, Japan) on 1 μg of total RNA to synthesize cDNA following the manufacture’s protocol. Synthetic cDNA was diluted 20-fold and its expression was determined.
The 18 candidate genes enriched in GO terms with p < 0.05 and BAG4 were selected for real-time PCR analysis. The gene of elongation factor 1 alpha (EF1-α) was used as a reference gene due to its low expressional variability during heat stress (Li A. et al., 2017). Primers designed for each gene with Primer Premier 5.0. qPCR was performed using the ABI7500 Fast Real-Time Detection System (Applied Biosystems, Foster City, CA, United States). The total volume of the reaction system was 20 μL and consisted of 10 μL SYBR Green 2X Supermix (TaKaRa), 6.8 μL DEPC H2O, 0.4 μL of each primer pair and ROX DyeII, 2 μL diluted cDNA. The program started with a 30 s activation of DNA polymerase at 95°C, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. The melt curve stage was conducted as: 15 s at 95°C, 1 min at 60°C, 30 s at 95°C, and 15 s at 60°C. The expression level of gene of induced transcripts (6 and 24 h) was calculated using the Livak 2–ΔΔCT method (Livak and Schmittgen, 2001). The expression of the control population was used as a control to calculated basal expression level.
Statistical Analysis
All data were expressed as mean ± standard error of mean (SEM). Statistics analysis was performed with SPSS 22.0 statistical package (IBM, NY, United States). The differences in shell height and wet weight between the two populations were analyzed using one-way analysis of variance (ANOVA), followed by Duncan’s multiple range tests for post hoc comparison of means. The same method was used to calculate the variation in respiration rate, gene expression, and physiological parameters between the two populations. Survival time in the laboratory survival module was determined by the Kaplan–Meier analysis with long-rank test. The survival rate in field was obtained by counting the number of deaths and survival on each of attachment substance. The level of statistically significant differences was set at p ≤ 0.05.
Results
Phenotypic Variations
Growth
The shell height and wet height of selected population were 41.70 ± 5.63 mm, 8.88 ± 2.87 g respectively, and those of control population were 40.38 ± 6.64 mm, 8.71 ± 3.58 g (Figure 3A). There was no significant difference in shell height and wet weight between the selected and control populations (p > 0.05).
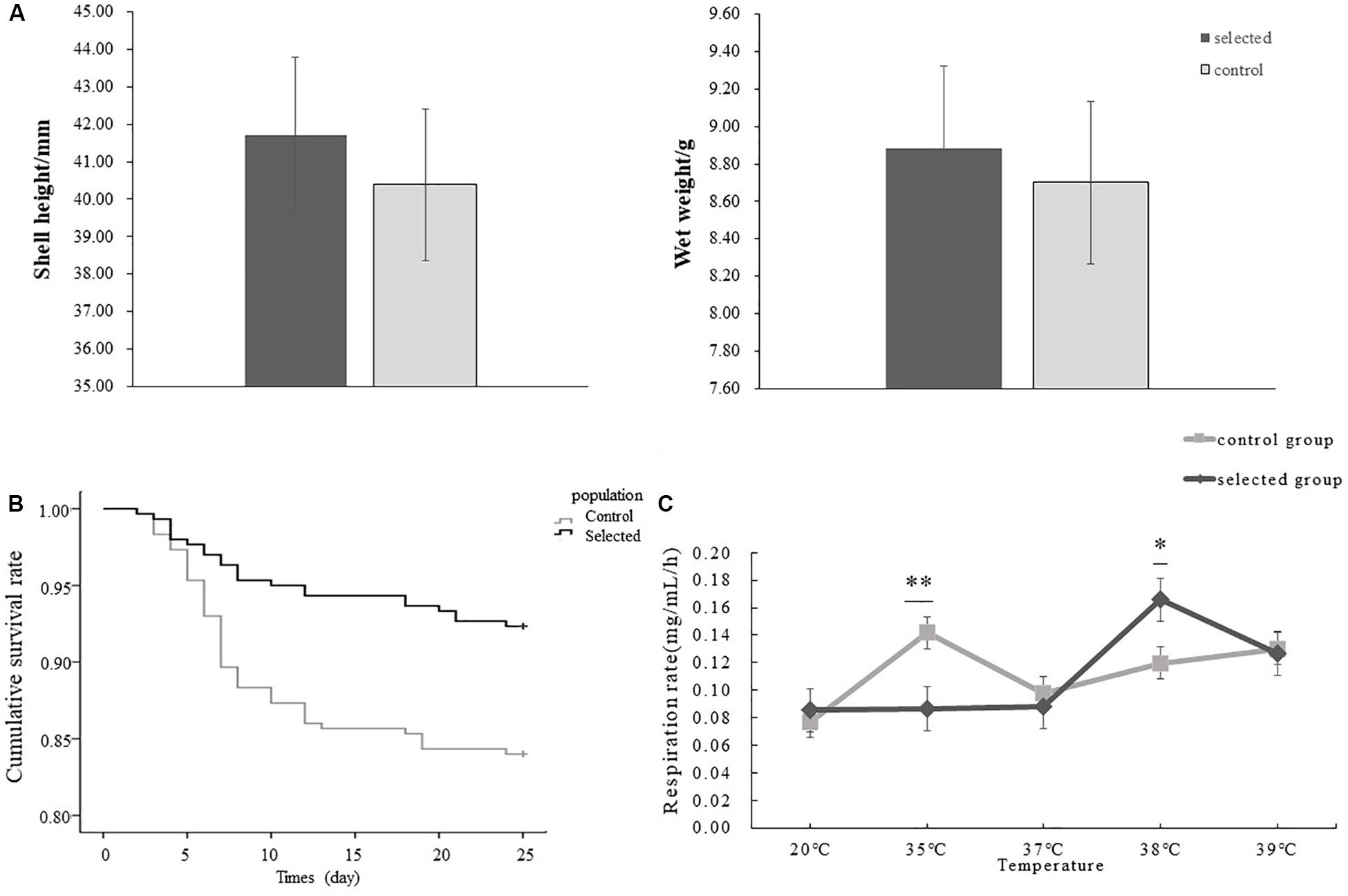
Figure 3. Comparison of growth and heat tolerance between selected and control populations, respectively, and comparison of the effect of elevating temperature on respiration between two populations. The error bar denotes the standard error of the mean. ∗indicated p < 0.05, ∗∗indicates p < 0.01. (A) The shell height and wet height of selected and control populations, respectively (n = 150 per population). (B) The survival rates of selected oysters and control oysters after heat shock at 42°C for 1 h (n = 300 per population). (C) The effect of increasing temperature on the respiration between selected oysters and control oysters (n = 6, six oysters from each population were used for each temperature).
Survival
The survival rate after heat shock at 42°C for 1 h was used to evaluate the thermal resistance of oysters. The selected oysters exhibited a significant higher survival rate (91.67%) during the recovery period compared control oysters (83.33%) following post-acute heat stress (p < 0.05) (Figure 3B). The first mortality of oysters under post treatment was observed in both selected and control populations on day 2. The highest mortality was observed on day 4 in the selected population while on day 7 in the control population. The continuous low and average mortality was observed after highest mortality.
Respiration Rate
The effects of temperature on oxygen consumption rate in two populations were not significant at 20°C (p > 0.05). Specifically, the average oxygen consumption of the selected population was 0.085 mg/mL/h while that of control population was 0.077 mg/mL/h. However, the differences were significant when temperature increased to 35°C (p < 0.01), and the average oxygen consumption of the control population reached 0.142 mg/mL/h. In contrast oxygen consumption in the selected population was 0.087 mg/mL/h. The respiration rate of the selected oysters was significantly lower than that of unselected oysters at 35°C, but the opposite result was shown when the temperature increased from 35 to 38°C (p < 0.05), when the average oxygen consumption of the selected population reached 0.166 mg/mL/h compared with 0.120 mg/mL/h in the control population (Figure 3C).
Physiological Assessment
Significant effects of time on MDA concentration were observed in the selected population which displayed a tendency to accumulate during exposure to high temperature. Moreover, we observed significant differences at 6 h between the selected and control populations (p < 0.05). Total protein content of the selected population was significantly higher than that of the control population during heat stress (p < 0.01). Furthermore, there was significant variation in activity of enzymes between the two populations. The activity of SOD and PK in both populations showed a trend of increasing in the beginning and then decreasing during heat shock. Moreover, the enzyme activities of SOD and PK were significantly higher in the selected population than in the control population during heat stress (p < 0.05) (Figure 4).
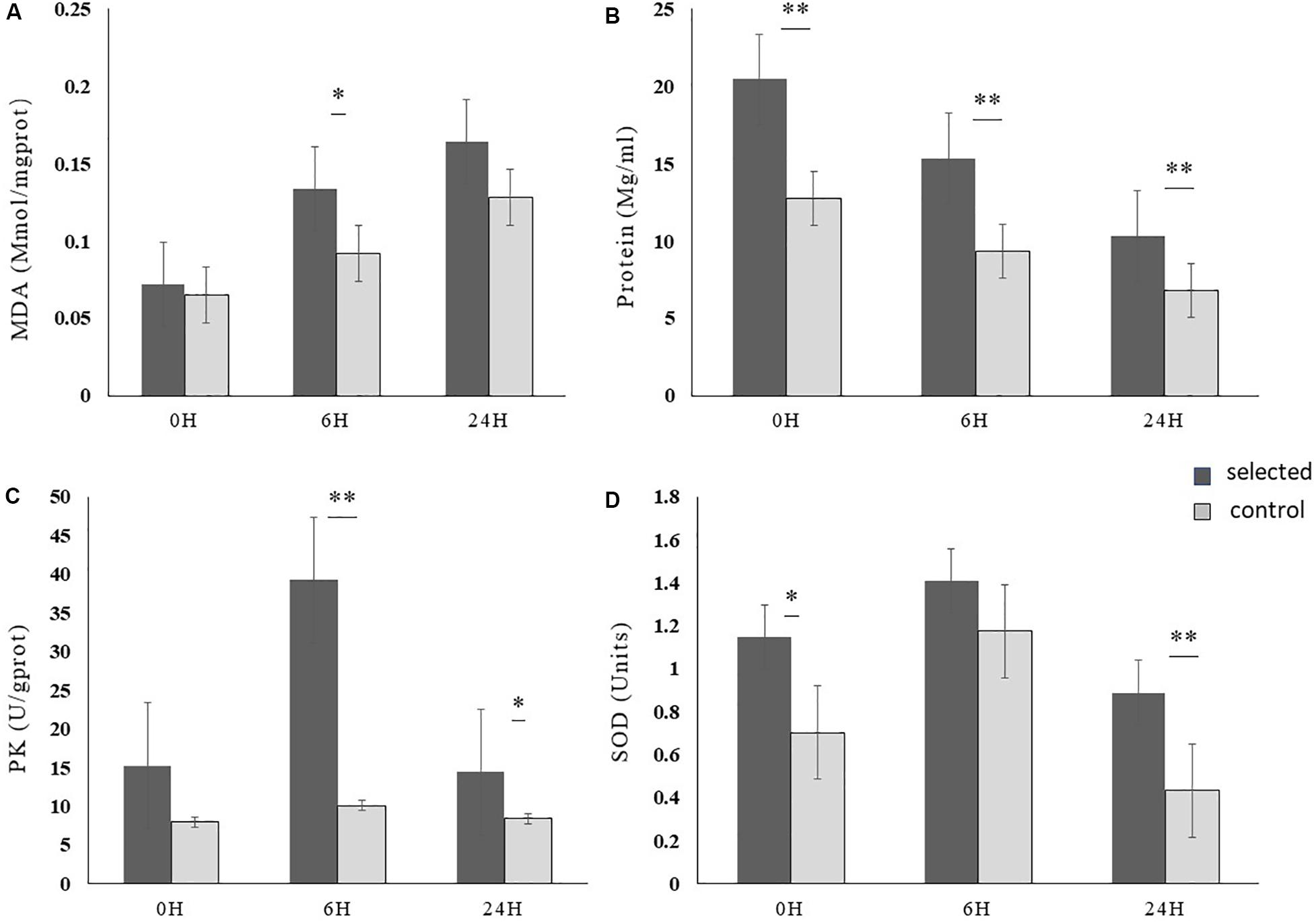
Figure 4. The physiological parameter of selected and control oysters during heat shock at 37°C for 24 h (n = 15). (A) MDA, malondialdehyde; (B) protein; (C) PK, pyruvate kinase; (D) SOD, superoxide dismutase. Bars denote standard deviation. ∗Indicates p < 0.05, ∗∗indicates p < 0.01.
Field Experiment
The mortality of the 19-month-old selected population was lower than the control population during summer under field conditions, which is consistent with our laboratory result. The survival rate of the selected population was 74.19% which is significantly higher than the value of 62.33% (p < 0.05) for control population. The average shell height of the selected and control populations was 69.53 and 73.38 mm, respectively. The shell height of the selected population was not significantly different from the control population (p > 0.05).
Sequencing Analysis
SLAF Sequencing and SNP Discovery
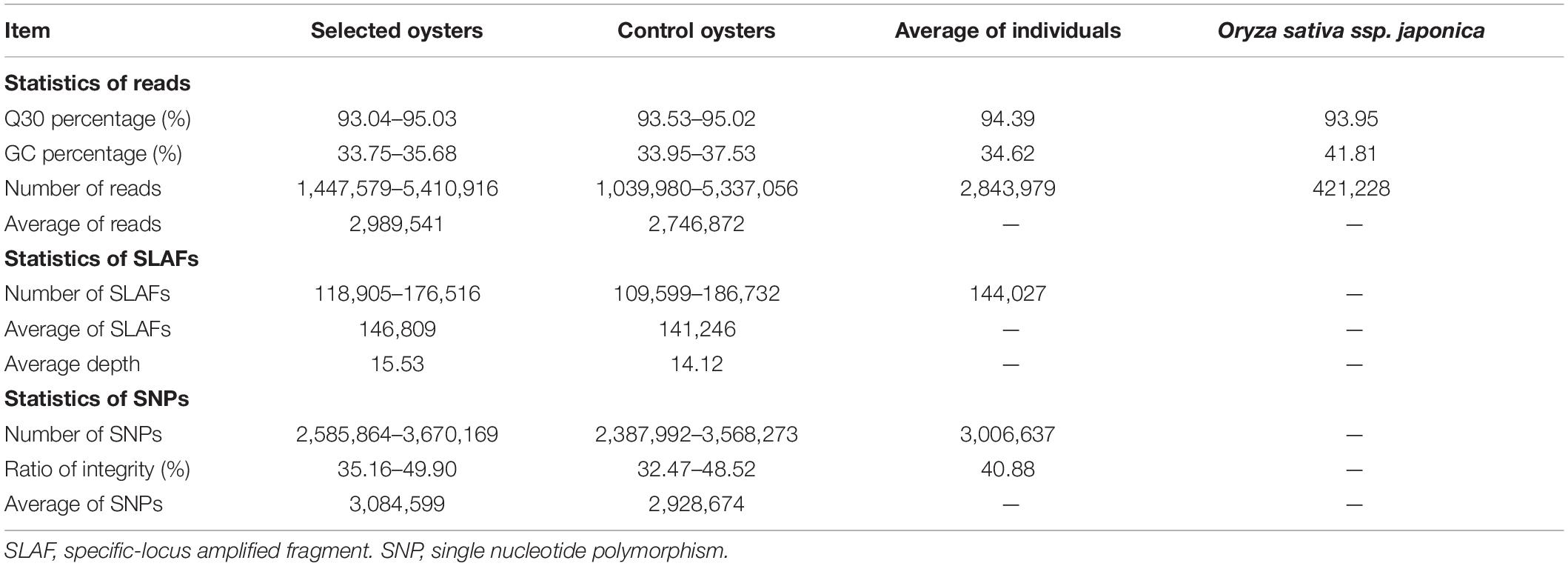
A total of 286.82 million reads were obtained from 100 individuals, which showed the average Q30 was 94.40%, and the average GC content was 34.55%, indicating the high quality of the generated data (Table 1). A total of 593,103 SLAF tags were obtained, including 454,594 polymorphic SLAF tags, with an average sequencing depth of 14.83x. In addition, Oryza sativa ssp. Japonica was used as a control in this project. Our results revealed that the percentage of paired-end mapped reads and normally digestion of control was 96.31 and 91.08%, respectively, indicating that the SLAF-seq process was normal and reliable. After genomic mapping and SNP calling with GATK and SAMtools, a total of 7,354,783 SNPs were revealed using all individuals, of which the per individual averages of 198,765 were intergenic, 300,145 were intron and 368 were exons. A total of 1,076,631 indels were identified in this program, of which 9,095 were in coding regions.
Population Structure and Phylogenetic Relationship
A strict quality control filtering of SNPs was performed to identify 379,002 SNPs used for further analysis. Phylogenetic analysis was conducted to determine the relationships between the two populations (100 individuals). Neighbor-joining cluster analysis divided 100 individuals into three groups (Figure 5A). The result clearly showed that the clustering of most selected individuals separated from the control population, and a few control individuals mixed with the selected population, which was consistent with the assignments made by PCA. The PCA analysis separated the 100 individuals into three clusters. The first three PCs accounted for 15.7% of the population-wide SNP variation, with PC1 accounting for 7.78% (Figure 5B).

Figure 5. Genetic structure analysis of 100 individuals which include 50 individuals from selected oysters and others from control oysters based on 379,002 SNPs. (A) The neighbor-joining phylogenetic tree divided into three clusters. (B) PCA plot showed three clusters. The red color represents control oysters and blue represents selected oysters in (A,B). (C) Cross validation error rates corresponding to different K values show K = 3 is the best. (D) Population structure with a K-value of 3 and each color indicate one cluster.
The estimated membership fractions for different values of K ranged from 1 to 10, and the maximum likelihood revealed by the population structure showed an optimum value of 3 (K = 3) (Figures 5C,D), which indicated that the entire population could be categorized into three groups: group 1, group 2, and group 3. Group 1 contained 27 individuals from the control population. Group 2 contained 23 individuals, two of which were from the control population, while the remainder were from the selected population. Group 3 contained 50 individuals, 29 of which were from the selected population, and 21 of which were from the control population (Table 2).
Identification of Selected Regions and Candidate Genes
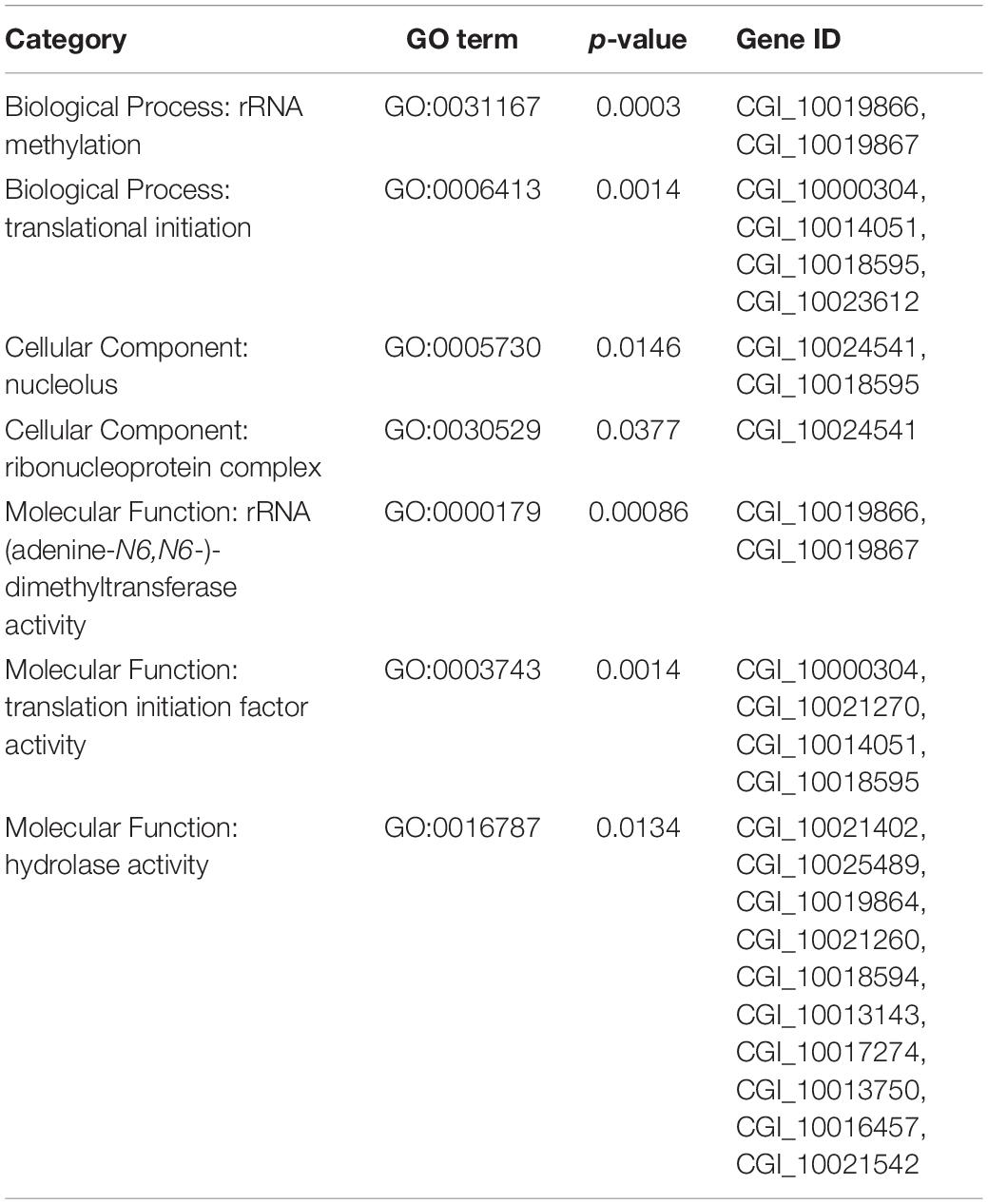
To obtain the genomic difference between the selected and control populations, a SNP-based Fst and θπ estimate was performed to identify selected regions. The value of the top 5% paired- Fst ranged from 0.0851 to 0.3253. As a result, analysis identified 472 genes from 115 genomic regions showing selection signals between the two populations. GO enrichment analysis of all 472 genes under selection showed that seven GO terms covering 18 genes were significantly over-represented (p < 0.05) (Table 3). The previously identified adaptive gene, BAG4 (Li L. et al., 2018), which is in the selected regions of top 5% Fst value was also selected as a candidate gene for subsequent gene expression investigation.
The Expression Level of Candidate Genes
The expression of the 19 genes in the selected region were further investigated by real-time PCR. The relative induced expression of eight genes (IF4A2, IF6, EIF3A, MANBA, DDX43, RECS, CAT2, and BAG4) were significantly different between the selected and control populations (p < 0.05), and the primers of each gene are shown in Table 4. The relative basal expression under ambient environment of six genes (IF6, EIF3A, DDX43, RECS, CAT2, and BAG4) in the control population exhibited significantly higher expression than in the selected population (1.42–2.64-fold change, p < 0.05) (Figure 6). In the selected population, the expression level of IF4A2, DDX43, and BAG4 showed up-regulation under heat stress, whereas induced expression of IF6, EIF3A, MANBA, RECS, and CAT2 showed down-regulation. The changed range of genes expression in selected population from 0.41 to 35.41-fold. However, most genes in the control population showed more moderate expression patterns under heat stress, the induced genes expression changed range from 0.55 to 2.27-fold (Figure 7).

Table 4. The genes with differential expression level between selected and control oysters and the primers used for q-PCR quantification.
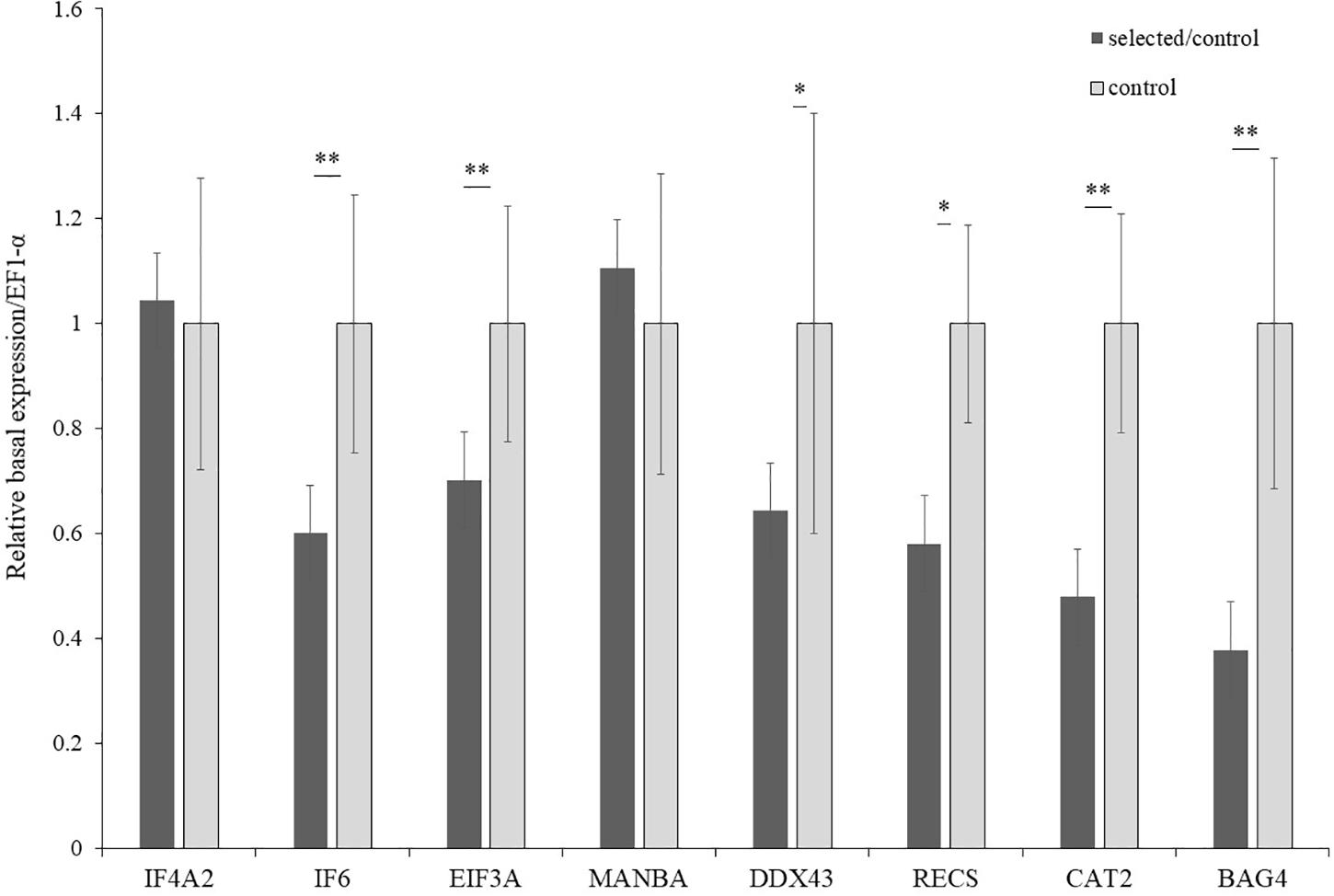
Figure 6. Basal expression of candidate genes in selected and control populations (n = 15). ∗Indicated p < 0.05; ∗∗indicated p < 0.01.
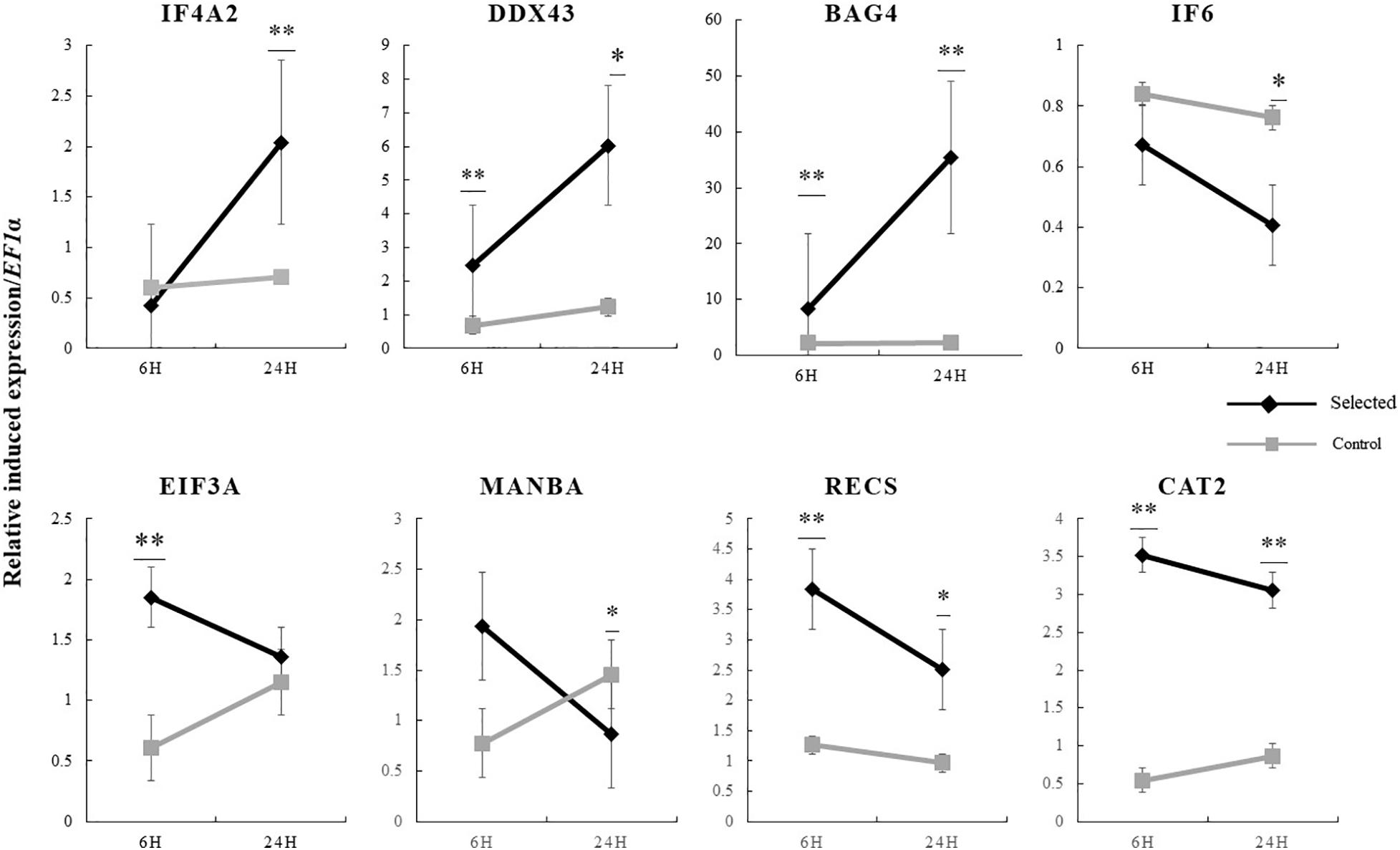
Figure 7. Induced expression of candidate genes at 6 and 24 h in response to heat stress in selected oysters and control oysters (n = 15) and significance analysis was conducted. ∗p < 0.05; ∗∗p < 0.01.
Discussion
In this study, the genetic and phenotypic effect of one-generation selection oysters targeting to improve the thermal tolerance was evaluated. The selection population showed higher survival rate, increased oxygen consumption at higher temperatures, and higher enzyme activity than control population (p < 0.05). In addition, the two populations exhibited difference in both genome structure and gene expression.
No significant differentiation of growth was observed between the selected and control populations in our study. This result is consistent with studies which have shown that selection on survival during the summer period after three generations does not have any impact on growth (Dégremont et al., 2010). Moreover, selection for growth was conducted but no effect on survival was observed in Sydney rock oyster Saccostrea glomerata (Hand et al., 2004). Our results show no significant difference of growth between the selected and control populations, which suggested that the selection for thermotolerance would not affect the growth at least at juvenile stage (<1-year-old) Pacific oysters.
We used the survival rate after acute heat stress to assess the thermotolerance of oysters. High heritability for survival during summer was observed, which suggests that selective breeding could effectively improve the thermotolerance of juvenile Pacific oysters (Dégremont et al., 2007). In this study, the survival rate of the progeny post exposure to acute heat stress at 42°C in the selected population was significantly higher than that in the control population even only after one-generation selection, suggesting the thermotolerance of selected oysters was improved. This result was in line with our expectations, which demonstrated that our methodology for thermotolerance selection of oysters was effective. Moreover, it showed that the mortality of selected population was lower than that of the control population under field conditions (p < 0.05). The selected population demonstrated a better survival rate than the control population, indicating a positive response to selection for survival. This result is consistent with the results of a previous study that significant higher survival was observed in the “high survival” selected population during summer at three different sites for F2 and F3 generations in 6-month-old Pacific oysters (Dégremont et al., 2010).
Temperature is one of the most important abiotic factors affecting the physiological response in marine organisms (Windisch et al., 2011). In this study, the oxygen consumption rate of Pacific oyster was greatly affected by temperature and showed a fluctuation with increase in temperature. The respiration rate obtained in this study increased with temperature, up to a maximum or optimum limit beyond which they rapidly decreased. This was consistent with the result of previous studies for other bivalve species, the temperature has a significant effect on oxygen consumption and respiratory energy loss of Calafia mother-of-pearl oyster, Pinctada mazatlanica, which suggested the existence of compensatory mechanisms that facilitate the organism to adapt to the temperature changes (Bayne and Newell, 1983; Saucedo et al., 2004). The individuals from the control population exposed to acute thermal stress (35°C) showed an increment in their metabolic rates compared to the condition (20°C), which is a trend also observed in different species of marine bivalves such as the flat oyster Ostrea edulis and mother-of-pearl oyster Pinctada mazatlanica (Haure, 1998; Saucedo et al., 2004). The oxygen consumption rate of selective oysters increased at higher temperature (38°C) compared to the control population, indicating the respiration of selective oysters reacted less sensitively than unselected oysters at these temperatures by increasing rapidly and the selective oysters had a higher tolerance to high temperature than the control population. Furthermore, 35 and 38°C represent different stress strength, the selected population showed higher oxygen consumption rate under higher stress strength than control population, indicating that the selected population has greater capacity of oxygen supply that decides the wider scope for metabolic adjustment, which is limited to a particular thermal tolerance window. Thus, the wider thermotolerance window of selected population decides higher survival at extreme high temperature compared with control population. This phenomenon is common in aquatic animals (Portner, 2001; Portner et al., 2006; Portner and Knust, 2007). The greater capacity of oxygen supply in selected population may be the reason that selected population is more thermotolerance. Low metabolic sensitivity of selected population to fluctuations of temperature may translate into lower long-term metabolic costs and higher heat tolerance. The metabolic rate increases as temperature increases, generating higher production of reactive oxygen species or ROS (Lushchak and Bagnyukova, 2006). Marine invertebrates are equipped with antioxidant enzymes such as SOD, CAT, and glutathione peroxidase to prevent cellular damage from ROS accumulation, such as lipid peroxidation, protein degradation and DNA breaking (Lesser, 2006). The activity of enzymes PK and SOD first exhibited an increasing pattern and then a decreasing pattern during the period of heat stress, which indicated the transition from aerobic to anaerobic metabolism (Anestis et al., 2010; Sussarellu et al., 2012). Enzyme activity and total protein of the selective population were significantly higher than those of the control population at most time points (p < 0.05). However, the concentration of MDA did not exhibit divergence between the two populations. This result was consistent with our previous study (Li A. et al., 2017), which showed southern oysters inhabiting warmer inshore locations had higher enzyme activity than northern oysters. The higher metabolic, enzymes’ activities could be explained as protective mechanism to alleviate oxidative damage from elevated amounts of ROS for organism’s cope with extremely high temperature (Abele et al., 2002).
In the present study, we used SLAF-seq to analyze the genetic structure and differentially selected regions between the selected and control populations using a reducing representation library sequencing method (Sun et al., 2013). It is clearly indicated that the surviving oysters from selected population and dead oyster from the control population exhibited divergence in genome structure after the acute stress selection for thermotolerance. This suggested the selection of heat stress is directional rather than random. Moreover, 115 differentially selected regions including 472 genes were identified, which provide a comprehensive view of the genome-wide basis for genetic difference between the two populations.
Population structure analysis identified three subgroups instead of two. The result is consistent with the phylogenetic analysis and PCA. This may be due to that fact that artificial selection is at the early stage and the restricted genetic differentiation between the oysters with one-generation selection and the unselected oysters. As the quantitative trait, thermotolerance of the oyster shows the continuous distribution. Furthermore, the third subgroup may represent oysters with moderate thermotolerance. In another oyster selection program for growth improvement, the population structure analysis showed that the selected lines and wild populations were not divided into two groups even after continuous selective breeding of six-generations (Zhong et al., 2016).
In this study, the top 5% Fst and θπ values between selected and control populations were used to determine the selected regions, in which 18 genes were enriched in p < 0.05 GO terms and were considered to be candidate for gene expression detection. In addition, BAG4 in the region with top 5% Fst also was selected because of its potential function in temperature adaptation (Li L. et al., 2018). Surprisingly, no canonical heat shock protein family were enriched in our study. This is consistent with the result that no significant difference of expression of HSP genes between selected and control population (unpublished).
Of the 19 genes showing genomic differentiation between the two populations, eight genes (IF4A2, IF6, EIF3A, MANBA, DDX43, RECS, CAT2, and BAG4) also diverged in transcriptional expression response to heat stress, which suggested divergence of genomic structure may regulate the expression of those genes and the expression differentiation may mediate the superior thermotolerance in the selected oysters. IF4A2, IF6, and EIF3A are related with protein synthesis. The studies of the three genes are related with diseases in human, such as diabetes, gallbladder cancer and nasopharyngeal carcinoma. IF6 and EIF3A play regulatory roles at translation initiation process as a rate-limiting step (Cheyssac et al., 2006; Liu et al., 2011; Golob-Schwarzl et al., 2019). The proteins of DDX43 and RECS are helicase. The overexpression of DDX43 was found in human cancer (Ambrosini et al., 2014). The proteins of MANBA represent one of the most thermostable versions and probably hydrolyze extracellular galactomannans to monosaccharides as energy sources in hyperthermophilic eubacterium (Duffaud et al., 1997). CAT2 is involved in dehydrated process of the fermentation of 4-aminobutyrate to ammonia, acetate, and butyrate in anaerobe (Peter and Wolfgang, 1990). The studies of these genes are rare in invertebrates. The potential effects of these genes in responding to high temperature require further investigations for oysters. It is worth noting that expression pattern of BAG4 in this study is consistent with previous study. BAG4, in intertidal populations and southern populations whose habitat exposed to higher temperature and more fluctuating in temperature exhibited higher plasticity compared with subtidal populations and northern populations respectively under heat stress (Li A. et al., 2018; Li L. et al., 2018). This suggested BAG4 may play an important role in thermal adaptation.
The selected population exhibited higher expression flexibility in candidate genes (p < 0.05). Specifically, IF4A2, DDX43, and BAG4 showed a high extent of up-regulated in response to heat shock in the selected population. The other five genes in the selected population exhibited high extent down-regulation in response to elevated temperature, which indicated the selected population has a higher gene expression plasticity under thermal stress. This was consistent with our previous study that plasticity in gene expression positively correlated with evolved divergence, high plasticity in gene expression may be adapt and favored by organisms under thermal regime (Li L. et al., 2018). Some studies have showed that in order to adapt to fluctuating environments organisms have evolved higher phenotypic plasticity than those habited in the stable environment, which suggested that plasticity may be adaptive (Kenkel and Matz, 2016).
Conclusion
We explored divergence of phenotype and genome between the selected and control populations after one-generation selection for thermotolerance in oysters. From analyses of different levels of biological organization (e.g., growth, thermotolerance, oxygen consumption rate, enzyme activities, gene expression, and genetic structure), we can propose that selected population has evolved higher phenotypic plasticity ensuring the oysters have potential adaptation to face extreme high temperature. Furthermore, 115 differentially selected regions including 472 genes were identified indicated the divergence of phenotype between selected and natural population maybe have a genetic background. GO analysis identified and q-PCR result indicated eight genes (IF4A2, IF6, EIF3A, MANBA, DDX43, RECS, CAT2, and BAG4) may be critical in response to thermal stress for oysters. Further, the result of BAG4 was consistent with our previous studies, which suggested its vital function in thermal adaptation.
Data Availability Statement
The raw sequence reads have been submitted to the Sequence Read Archive (SRA) in the NCBI, and accession number PRJNA604406.
Ethics Statement
This study was conducted on oysters which are not included in the Animals (Scientific Procedures) Act. All experimental works was reviewed and approved by the Experimental Animal Ethics Committee, Institute of Oceanology, Chinese Academy of Sciences, China.
Author Contributions
GZ, HQ, and LL conceived the experiment. WW and RC collected oysters and performed the spawning. AL and XW contributed to the experiment in parental oysters. FD performed the experiment, collected and analyzed the data. FD, LL, AL, and GZ wrote and edited the manuscript.
Funding
This work was supported by the National Key R&D Program of China (No. 2018YFD0900304 to LL), Project funded by China Postdoctoral Science Foundation (No. 2019TQ0324 to AL), Key Deployment Project of Centre for Ocean Mega-Research of Science, Chinese Academy of Sciences (COMS2019Q06 to AL and LL), and the Technology and the Modern Agro-Industry Technology Research System (No. CARS-49 to LL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the laboratory for their technical advice and helpful discussions.
References
Abele, D., Heise, K., Portner, H. O., and Puntarulo, S. (2002). Temperature-dependence of mitochondrial function and production of reactive oxygen species in the intertidal mud clam Mya arenaria. J. Exp. Biol. 205, 1831–1841.
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Alfaro, A. C., Nguyen, T. V., and Merien, F. (2018). The complex interactions of Ostreid herpesvirus 1, Vibrio bacteria, environment and host factors in mass mortality outbreaks of Crassostrea gigas. Rev. Aquac. 11, 1148–1168. doi: 10.1111/raq.12284
Alter, K., Andrewartha, S. J., Morash, A. J., Clark, T. D., Hellicar, A. D., León, R. I., et al. (2017). Hybrid abalone are more robust to multi-stressor environments than pure parental species. Aquaculture 478, 25–34. doi: 10.1016/j.aquaculture.2017.04.035
Ambrosini, G., Khanin, R., Carvajal, R. D., and Schwartz, G. K. (2014). Overexpression of DDX43 mediates MEK inhibitor resistance through RAS Upregulation in uveal melanoma cells. Mol. Cancer Ther. 13, 2073–2080. doi: 10.1158/1535-7163.MCT-14-0095
Anestis, A., Pörtner, H. O., Karagiannis, D., Angelidis, P., Staikou, A., and Michaelidis, B. (2010). Response of Mytilus galloprovincialis (L.) to increasing seawater temperature and to marteliosis: metabolic and physiological parameters. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 156, 57–66. doi: 10.1016/j.cbpa.2009.12.018
Bayne, B. L., and Newell, R. C. (1983). Physiological energetics of marine molluscs. Mollusca 4, 407–515. doi: 10.1016/B978-0-12-751404-8.50017-7
Beattie, J. H., Chew, K. K., and Hershberger, W. K. (1980). Differential survival of selected strains of pacific oysters (Crassostrea-Gigas) during summer mortality. Proc. Natl. Shellfish. Assoc. 70, 184–189.
Cheney, D., and Macdonald, B. (2000). summer mortality of pacific oysters inifial findings on multiple environmental stressors in Puget Sound, Washington, 1998. J. Shellfish Res. 19, 353–359.
Cheyssac, C., Dina, C., Lepretre, F., Vasseur-Delannoy, V., Dechaume, A., Lobbens, S., et al. (2006). EIF4A2 is a positional candidate gene at the 3q27 locus linked to type 2 diabetes in French families. Diabetes Metab. Res. Rev. 55, 1171–1176. doi: 10.2337/diabetes.55.04.06.db05-1298
Dégremont, L., Bédier, E., and Boudry, P. (2010). Summer mortality of hatchery-produced Pacific oyster spat (Crassostrea gigas). II. Response to selection for survival and its influence on growth and yield. Aquaculture 299, 21–29. doi: 10.1016/j.aquaculture.2009.11.017
Dégremont, L., Ernande, B., Bédier, E., and Boudry, P. (2007). Summer mortality of hatchery-produced Pacific oyster spat (Crassostrea gigas). I. Estimation of genetic parameters for survival and growth. Aquaculture 262, 41–53. doi: 10.1016/j.aquaculture.2006.10.025
Duffaud, G. D., Mccutchen, C. M., and Leduc, P. (1997). Purification and characterization of extremely thermostable {beta}-mannanase, {beta}-mannosidase, and {alpha}-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl. Environ. Microbiol. 63, 169–177. doi: 10.1109/50.337485
Farcy, É., Voiseux, C., Lebel, J.-M., and Fiévet, B. (2008). Transcriptional expression levels of cell stress marker genes in the Pacific oyster Crassostrea gigas exposed to acute thermal stress. Cell Stress Chaperones 14, 371–380. doi: 10.1007/s12192-008-0091-8
Gagnaire, B., Frouin, H., Moreau, K., Thomas-Guyon, H., and Renault, T. (2006). Effects of temperature and salinity on haemocyte activities of the Pacific oyster, Crassostrea gigas (Thunberg). Fish Shellfish Immunol. 20, 536–547. doi: 10.1016/j.fsi.2005.07.003
Ge, Y., Zhang, T., Wu, B., Tan, L., Ma, F., Zou, M., et al. (2019). Genome-wide assessment of avocado germplasm determined from specific length amplified fragment sequencing and transcriptomes: population structure, genetic diversity, identification, and application of race-specific markers. Genes 10:215. doi: 10.3390/genes10030215
Ghaffari, H., Wang, W., Li, A., Zhang, G., and Li, L. (2019). Thermotolerance divergence revealed by the physiological and molecular responses in two oyster subspecies of Crassostrea gigas in China. Front. Physiol. 10:1137. doi: 10.3389/fphys.2019.01137
Gleason, L. U., and Burton, R. S. (2013). Phenotypic evidence for local adaptation to heat stress in the marine snail Chlorostoma (formerly Tegula) funebralis. J. Exp. Mar. Biol. Ecol. 448, 360–366. doi: 10.1016/j.jembe.2013.08.008
Glude, J. (1975). “A summary report of Pacific coast oyster mortality investigations,” in Proceedings of the Third US.-Japan Meeting on Aquaculture, Tokyo, 15–16.
Golob-Schwarzl, N., Wodlej, C., Kleinegger, F., Gogg-Kamerer, M., Birkl-Toeglhofer, A. M., Petzold, J., et al. (2019). Eukaryotic translation initiation factor 6 overexpression plays a major role in the translational control of gallbladder cancer. J. Cancer Res. Clin. Oncol. 145, 2699–2711. doi: 10.1007/s00432-019-03030-x
Guo, X. (2009). Use and exchange of genetic resources in molluscan aquaculture. Rev. Aquac. 1, 251–259. doi: 10.1111/j.1753-5131.2009.01014.x
Guo, X., Li, Q., Wang, Q. Z., and Kong, L. F. (2012). Genetic mapping and QTL analysis of growth-related traits in the Pacific oyster. Mar. Biotechnol. 14, 218–226. doi: 10.1007/s10126-011-9405-4
Gupta, S. K., Baek, J., Carrasquilla-Garcia, N., and Penmetsa, R. V. (2015). Genome-wide polymorphism detection in peanut using next-generation restriction-site-associated DNA (RAD) sequencing. Mol. Breed. 35:145. doi: 10.1007/s11032-015-0343-0
Hand, R. E., Nell, J. A., and Thompson, P. A. (2004). Studies on triploid oysters in Australia: XIII. Performance of diploid and triploid Sydney rock oyster, Saccostrea glomerata (Gould, 1850), progeny from a third generation breeding line. Aquaculture 233, 93–107. doi: 10.1016/j.aquaculture.2003.09.017
Haure, J. (1998). Influence of temperature on clearance and oxygen consumption rates of the flat oyster Ostrea edulis: determination of allometric coefficients. Aquaculture 169, 211–224.
Hershberger, W. K., Perdue, J. A., and Beattie, J. H. (1984). Genetic selection and systematic breeding in Pacific oyster culture. Aquaculture 39, 237–245. doi: 10.1016/0044-8486(84)90269-2
Juarez, O. E., Lafarga-De la Cruz, F., Leyva-Valencia, I., Lopez-Landavery, E., Garcia-Esquivel, Z., Diaz, F., et al. (2018). Transcriptomic and metabolic response to chronic and acute thermal exposure of juvenile geoduck clams Panopea globosa. Mar. Genomics 42, 1–13. doi: 10.1016/j.margen.2018.09.003
Kenkel, C. D., and Matz, M. V. (2016). Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evol. 1:0014. doi: 10.1038/s41559-016-0014
Kim, B.-M., Kim, K., Choi, I.-Y., and Rhee, J.-S. (2017). Transcriptome response of the Pacific oyster, Crassostrea gigas susceptible to thermal stress: a comparison with the response of tolerant oyster. Mol. Cell. Toxicol. 13, 105–113. doi: 10.1007/s13273-017-0011-z
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lesser, P. (2006). Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278. doi: 10.1146/annurev.physiol.68.040104.110001
Li, A., Li, L., Song, K., Wang, W., and Zhang, G. (2017). Temperature, energy metabolism, and adaptive divergence in two oyster subspecies. Ecol. Evol. 7, 6151–6162. doi: 10.1002/ece3.3085
Li, A., Li, L., Wang, W., Song, K., and Zhang, G. (2018). Transcriptomics and fitness data reveal adaptive plasticity of thermal tolerance in oysters inhabiting different tidal zones. Front. Physiol. 9:825. doi: 10.3389/fphys.2018.00825
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, L., Li, A., Song, K., Meng, J., Guo, X., Li, S., et al. (2018). Divergence and plasticity shape adaptive potential of the Pacific oyster. Nat. Ecol. Evol. 2, 1751–1760. doi: 10.1038/s41559-018-0668-2
Li, Q., Yu, H., and Yu, R. (2006). Genetic variability assessed by microsatellites in cultured populations of the Pacific oyster (Crassostrea gigas) in China. Aquaculture 259, 95–102. doi: 10.1016/j.aquaculture.2006.05.030
Li, Z., Wei, S., Li, H., Wu, K., Cai, Z., Li, D., et al. (2017). Genome-wide genetic structure and differentially selected regions among Landrace, Erhualian, and Meishan pigs using specific-locus amplified fragment sequencing. Sci. Rep. 7:10063. doi: 10.1038/s41598-017-09969-6
Liu, R. Y., Dong, Z., Liu, J., Yin, J. Y., Zhou, L., Wu, X., et al. (2011). Role of eIF3a in regulating cisplatin sensitivity and in translational control of nucleotide excision repair of nasopharyngeal carcinoma. Oncogene 30, 4814–4823. doi: 10.1038/onc.2011.189
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lushchak, V. I., and Bagnyukova, T. V. (2006). Temperature increase results in oxidative stress in goldfish tissues. 1. Indices of oxidative stress. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 143, 30–35. doi: 10.1016/j.cbpc.2005.11.017
McAfee, D., Cumbo, V. R., Bishop, M. J., and Raftos, D. A. (2018). Intraspecific differences in the transcriptional stress response of two populations of Sydney rock oyster increase with rising temperatures. Mar. Ecol. Prog. Ser. 589, 115–127. doi: 10.3354/meps12455
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mora, C., and Maya, M. F. (2006). Effect of the rate of temperature increase of the dynamic method on the heat tolerance of fishes. J. Therm. Biol. 31, 337–341. doi: 10.1016/j.jtherbio.2006.01.005
Mora, C., and Ospína, A. (2001). Tolerance to high temperatures and potential impact of sea warming on reef fishes of Gorgona Island (tropical eastern Pacific). Mar. Biol. 139, 765–769. doi: 10.1007/s002270100626
Ohyanagi, H. (2006). The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 34, D741–D744. doi: 10.1093/nar/gkj094
Paul-Pont, I., Evans, O., Dhand, N. K., Rubio, A., Coad, P., and Whittington, R. J. (2013). Descriptive epidemiology of mass mortality due to Ostreid herpesvirus-1 (OsHV-1) in commercially farmed Pacific oysters (Crassostrea gigas) in the Hawkesbury River estuary, Australia. Aquaculture 42, 146–159. doi: 10.1016/j.aquaculture.2013.12.009
Peter, W., and Wolfgang, B. (1990). Assay of 4-hydroxybutyryl-CoA dehydratase from Clostridium aminobutyricum. FEMS Microbiol. Lett. 70, 187–191. doi: 10.1111/j.1574-6968.1990.tb13976.x
Petton, B., Pernet, F., Robert, R., and Boudry, P. (2013). Temperature influence on pathogen transmission and subsequent mortalities in juvenile Pacific oysters Crassostrea gigas. Aquac. Environ. Interact. 3, 257–273. doi: 10.3354/aei00070
Pfeifer, B., Wittelsburger, U., Ramos-Onsins, S. E., and Lercher, M. J. (2014). PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936. doi: 10.1093/molbev/msu136
Portner, H. O. (2001). Climate change and temperature-dependent biogeography: oxygen limitation of thermal tolerance in animals. Naturwissenschaften 88, 137–146. doi: 10.1007/s001140100216
Portner, H. O., and Knust, R. (2007). Climate change affects marine fishes through the oxygen limitation of thermal tolerance. Science 315, 95–97. doi: 10.1126/science.1135471
Portner, H. O., Peck, L. S., and Hirse, T. (2006). Hyperoxia alleviates thermal stress in the Antarctic bivalve, Laternula elliptica: evidence for oxygen limited thermal tolerance. Polar Biol. 29, 688–693. doi: 10.1007/s00300-005-0106-1
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A., and Reich, D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. doi: 10.1038/ng1847
Przeslawski, R., Davis, A. R., and Benkendorff, K. (2005). Synergistic effects associated with climate change and the development of rocky shore molluscs. Glob. Chang. Biol. 11, 515–522. doi: 10.1111/j.1365-2486.2005.00918.x
Samain, J. F., Dégremont, L., Soletchnik, P., Haure, J., Bédier, E., Ropert, M., et al. (2007). Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture 268, 227–243. doi: 10.1016/j.aquaculture.2007.04.044
Sanford, E., and Kelly, M. W. (2011). Local adaptation in marine invertebrates. Ann. Rev. Mar. Sci. 3, 509–535. doi: 10.1146/annurev-marine-120709-142756
Saucedo, P. E., Ocampo, L. A., Monteforte, M., and Bervera, H. (2004). Effect of temperature on oxygen consumption and ammonia excretion in the Calafia mother-of-pearl oyster, Pinctada mazatlanica (Hanley, 1856). Aquaculture 229, 377–387.
Sokolova, I. M., Frederich, M., Bagwe, R., Lannig, G., and Sukhotin, A. A. (2012). Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates. Mar. Environ. Res. 79, 1–15. doi: 10.1016/j.marenvres.2012.04.003
Soletchnik, P., Lambert, C., and Costil, K. (2005). Summer mortality of Crassostrea Gigas (Thunberg) in relation to environmental rearing conditions. J. Shellfish Res. 24, 197–207.
Sun, X., Liu, D., Zhang, X., Li, W., Liu, H., Hong, W., et al. (2013). SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8:e58700. doi: 10.1371/journal.pone.0058700
Sussarellu, R., Fabioux, C., Sanchez, M. C., Goïc, N. L., Lambert, C., Soudant, P., et al. (2012). Molecular and cellular response to short-term oxygen variations in the Pacific oyster Crassostrea gigas. J. Exp. Mar. Biol. Ecol. 412, 87–95. doi: 10.1016/j.jembe.2011.11.007
Tang, B., and Liu, B. (2005). Oxygen consumption and ammonia-N excretion of Meretrix meretrix in different temperature and salinity. Chin. J. Oceanol. Limnol. 23, 469–474. doi: 10.1007/BF02842693
Wang, J., Dong, B., Yu, Z. X., and Yao, C. L. (2018). The impact of acute thermal stress on green mussel Perna viridis: oxidative damage and responses. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 222, 7–15. doi: 10.1016/j.cbpa.2018.04.001
Windisch, H. S., Kathöver, R., Pörtner, H. O., Frickenhaus, S., and Lucassen, M. (2011). Thermal acclimation in Antarctic fish: transcriptomic profiling of metabolic pathways. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R1453–R1466. doi: 10.1152/ajpregu.00158.2011
Yampolsky, L. Y., Schaer, T. M. M., and Ebert, D. (2014). Adaptive phenotypic plasticity and local adaptation for temperature tolerance in freshwater zooplankton. Proc. Biol. Sci. 281:20132744. doi: 10.1098/rspb.2013.2744
Yang, C., Gao, Q., Liu, C., Wang, L., Zhou, Z., Gong, C., et al. (2017). The transcriptional response of the Pacific oyster Crassostrea gigas against acute heat stress. Fish Shellfish Immunol. 68, 132–143. doi: 10.1016/j.fsi.2017.07.016
Yao, C.-L., and Somero, G. N. (2014). The impact of ocean warming on marine organisms. Chin. Sci. Bull. 59, 468–479. doi: 10.1007/s11434-014-0113-0
Zhang, G., Li, L., Meng, J., Qi, H., Qu, T., Xu, F., et al. (2016). Molecular basis for adaptation of oysters to stressful marine intertidal environments. Annu. Rev. Anim. Biosci. 4, 357–381. doi: 10.1146/annurev-animal-022114-110903
Keywords: Pacific oyster, artificial selection, thermotolerance, genetic structure, physiological traits
Citation: Ding F, Li A, Cong R, Wang X, Wang W, Que H, Zhang G and Li L (2020) The Phenotypic and the Genetic Response to the Extreme High Temperature Provides New Insight Into Thermal Tolerance for the Pacific Oyster Crassostrea gigas. Front. Mar. Sci. 7:399. doi: 10.3389/fmars.2020.00399
Received: 29 February 2020; Accepted: 08 May 2020;
Published: 30 June 2020.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Vengatesen Thiyagarajan (Rajan), The University of Hong Kong, Hong KongLiqiang Zhao, The University of Tokyo, Japan
Yunwei Dong, Ocean University of China, China
Copyright © 2020 Ding, Li, Cong, Wang, Wang, Que, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guofan Zhang, Z2Z6aGFuZ0BxZGlvLmFjLmNu; Li Li, bGlsaUBxZGlvLmFjLmNu
 Fangfang Ding1,2
Fangfang Ding1,2 Ao Li
Ao Li Rihao Cong
Rihao Cong Xinxing Wang
Xinxing Wang Wei Wang
Wei Wang Guofan Zhang
Guofan Zhang Li Li
Li Li