- 1Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Clayton, VIC, Australia
- 2Melbourne Brain Centre, Florey Neuroscience Institute, Parkville, VIC, Australia
- 3ARC Centre of Excellence in Advanced Molecular Imaging, Monash University, Clayton, VIC, Australia
Background: Brinps 1–3, and Astrotactins (Astn) 1 and 2, are members of the Membrane Attack Complex/Perforin (MACPF) superfamily that are predominantly expressed in the mammalian brain during development. Genetic variation at the human BRINP2/ASTN1 and BRINP1/ASTN2 loci has been implicated in neurodevelopmental disorders. We, and others, have previously shown that Brinp1−/− mice exhibit behavior reminiscent of autism spectrum disorder (ASD) and attention deficit hyperactivity disorder (ADHD).
Method: We created Brinp2−/− mice and Brinp3−/− mice via the Cre-mediated LoxP system to investigate the effect of gene deletion on anatomy and behavior. Additionally, Brinp2−/−Brinp3−/− double knock-out mice were generated by interbreeding Brinp2−/− and Brinp3−/− mice. Genomic validation was carried out for each knock-out line, followed by histological, weight and behavioral examination. Brinp1−/−Brinp2−/−Brinp3−/− triple knock-out mice were also generated by crossing Brinp2/3 double knock-out mice with previously generated Brinp1−/− mice, and examined by weight and histological analysis.
Results: Brinp2−/− and Brinp3−/− mice differ in their behavior: Brinp2−/− mice are hyperactive, whereas Brinp3−/− mice exhibit marked changes in anxiety-response on the elevated plus maze. Brinp3−/− mice also show evidence of altered sociability. Both Brinp2−/− and Brinp3−/− mice have normal short-term memory, olfactory responses, pre-pulse inhibition, and motor learning. The double knock-out mice show behaviors of Brinp2−/− and Brinp3−/− mice, without evidence of new or exacerbated phenotypes.
Conclusion: Brinp3 is important in moderation of anxiety, with potential relevance to anxiety disorders. Brinp2 dysfunction resulting in hyperactivity may be relevant to the association of ADHD with chromosome locus 1q25.2. Brinp2−/− and Brinp3−/− genes do not compensate in the mammalian brain and likely have distinct molecular or cell-type specific functions.
Introduction
BMP/RA-inducible neural-specific protein(s) (Brinps) are a family of three highly conserved vertebrate genes that are almost exclusively expressed in neurons in the central and peripheral nervous system (Kawano et al., 2004; Terashima et al., 2010). They form part of a larger protein superfamily exemplified by the membrane attack complex/perforin (MACPF) domain. Brinp2 and Brinp3 are expressed in differentiated neurons of the neocortex, amygdala, hippocampus, and cerebellum during mammalian brain development, peaking at postnatal day 14 (Kawano et al., 2004). In the adult mouse brain, Brinp2 and Brinp3 gene expression is reduced and region-specific expression, unlike the more ubiquitous expression of Brinp1. Brinp2 and Brinp3 show a partial temporal and regional overlap in expression during development of the mammalian brain (Kawano et al., 2004). High homology between Brinp2 and Brinp3 amino acid sequence (70% identity) suggests that these two genes evolved by duplication of a common ancestor (Kawano et al., 2004; Giousoh et al., 2015), and may perform a similar, interchangeable function—to the extent that one may compensate for the other's absence. Furthermore, Brinp1 is ~50% homologous to Brinp2 and Brinp3, suggesting all three Brinps have related roles (Kawano et al., 2004).
BRINP2, BRINP3, and the MACPF superfamily member Astrotactin1 (ASTN1) are co-located in humans at chromosome 1q25.2, and in mice (Giousoh et al., 2015). Genetic variations at the 1q25.2 locus are associated with neurodevelopmental disorders, in particular attention deficit hyperactivity disorder (ADHD) (Lesch et al., 2008; Romanos et al., 2008). Two copy number variations (CNV)—one deletion and one duplication—are reported in patients with neurodevelopmental disorders (NDDs) that span the ASTN1/BRINP2 loci, suggesting that one or both of these genes is responsible for the neuropathology (Lionel et al., 2014). The patient with a CNV gain had anxiety, ASD, learning disability, and motor delay, whereas the patient with the deletion suffered developmental delay and seizures (Lionel et al., 2014). In addition, BRINP2 has been linked to substance abuse and reward dependence in two genome-wide association studies (Verweij et al., 2010; Drgon et al., 2011). Patients diagnosed with schizophrenia show altered methylation states of Brinp3 (Numata et al., 2014). Changes in BRINP3 (FAM5C) levels have also been reported to correlate with changes in the neurotransmitter norepinephrine during exercise (Karoly et al., 2012).
In non-NDD diagnoses, overexpression of BRINP2 (FAM5b) or BRINP3 (FAM5c) has been implicated in cancer: BRINP2 is amplified in oral squamous cell carcinoma (Cha et al., 2011), and BRINP3 is overexpressed in pituitary adenomas (Shorts-Cary et al., 2007). Both genes also show association with coronary heart disease (Connelly et al., 2008; Angelakopoulou et al., 2012). Presentation in similar disease suggests a common molecular function of BRINP2 and BRINP3.
Brinp1 was the first of the three Brinp genes to be studied in mice. Brinp1−/− mice generated by our group and others show hyperactivity, decreased body weight, reduced reproductive success, impaired short-term memory, altered anxiety response, and social communication impairments reminiscent of autism spectrum disorder (ASD). In the adult brain, Brinp1−/− mice exhibit increased parvalbumin-interneuron density in the neocortex and hippocampus (Kobayashi et al., 2014; Berkowicz et al., 2016).
In this study, the key aims were to determine the effect of loss of Brinp2 and/or Brinp3 on mouse anatomy and behavior, and to gain insight into the (potentially overlapping) function of these genes and their relationship with neurodevelopmental disorders. To investigate if compensation by Brinp2 or Brinp3 masks a more severe phenotype in the Brinp1−/− mice, Brinp1−/−Brinp2−/−Brinp3−/− (Brinp1/2/3) triple knock-out mice were also generated and studied.
Materials and Methods
Gene Targeting
Gene targeting methods were similar to those described for the generation of Brinp1−/− mice (Berkowicz et al., 2016). A targeting vector was constructed to alter the Brinp2 locus in mouse embryonic stem (ES) cells by homologous recombination following the general strategy outlined in Teoh et al. (2014). Separately, a targeting vector was constructed to alter the Brinp3 locus in ES cells. The vectors were built using bacterial artificial chromosome (BAC) clones RP23-97A3 and RP23-213F2 as the source of Brinp2 DNA, and BACs RP23-146N23/RP23-301J4 as the source of Brinp3 DNA. Both the Brinp2 and Brinp3 vectors comprised a neomycin transcriptional unit flanked by flippase (Flp) recognition target (FRT) elements placed in intron 3. For each vector, a loxP element was placed in the same intron immediately downstream of the neomycin cassette, while an upstream loxP element was placed in intron 2. Cre-recombinase mediated deletion of exon 3 was designed to result in a frame shift, creating a stop codon in the fourth exon. The Brinp2 and Brinp3 targeting constructs were separately electroporated into Bruce 4 C57BL/6-derived embryonic stem (ES) cells, and the targeted clone carrying the targeted allele Brinp2tm1Pib (MGI:5604614) or Brinp3tm1Pib (MGI:5604619) were identified by Southern analysis. A correctly targeted clone for each gene was injected into BALB/c blastocysts to generate chimeric mice. Chimeric Brinp2tm1Pib and chimeric Brinp3tm1Pib mice were each crossed to C57BL/6 Cre-deleter transgenic mice (Tg(CMV-cre)1Cgn) to remove exon 3 and the neomycin cassette from the targeted allele to produce animals carrying the Brinp2tm1.1Pib mutation (MGI:5604615) or Brinp3tm1.1Pib mutation (MGI:5604620). In parallel, chimeric mice were crossed to C57BL/6 Flp-deleter transgenic mice to remove the neomycin cassette only [Brinp2tm1.2Pib (MGI:5604617); Brinp3tm1.2Pib (MGI:5604621)]. “Floxed” mice heterozygous for the Brinp2tm1.1Pib mutation were inter-crossed to generate mice of all three genotypes: Brinp2+/+ (WT); Brinp2+/tm1.1Pib (het); and Brinp2tm1.1Pib/tm1.1Pib (Brinp2−/−). “Floxed” mice heterozygous for the Brinp3tm1.1Pib mutation were inter-crossed to generate mice of all three genotypes: Brinp3+/+ (WT); Brinp3+/tm1.1Pib (het); and Brinp3tm1.1Pib/tm1.1Pib (Brinp3−/−).
Genomic Analysis
Mice carrying the Brinp2−/− and Brinp3−/− floxed alleles, along with their respective WT, targeted, and Flpe mouse lines were all verified by Southern analysis. Genomic DNA isolated from the spleen was digested with AseI (Brinp2) or NdeI (Brinp3) and probed with a 500 bp 5′ homology probe. A 3′ homology arm probe was used for blotting of genomic DNA digested with BglII (Brinp2) PvuII (Brinp3). An internal probe was used to rule out random integration into the genome. Genotyping PCR confirmed absence of the neomycin cassette in floxed animals.
Breeding Brinp2/3−/− Double Knock-Out Mice
Brinp2−/− mice were mated with Brinp3−/− mice. The resultant heterozygous offspring were bred, avoiding sibling matings. As both genes are present on chromosome 1q (separated by 11.8 M bp) offspring were screened for a crossover event occurring between the Brinp2tm1.1Pib allele and the Brinp3tm1.1Pib allele, resulting in a chromosome with the mutant version of both genes.
The presence of a Brinp2tm1.1Pib Brinp3tm1.1Pib chromosome was screened for as a genotyping result showing a mouse homozygous for one floxed gene, and heterozygous for the other—a scenario only possible if a cross-over event has occurred. Mice found to have Brinp2tm1.1Pib and Brinp3tm1.1Pib on the same chromosome were bred to generate Brinp2/3−/− mice. Brinp2/3−/− mice were identified by genotyping PCR, and validated by reverse-transcriptase PCR (RT-PCR).
Brinp2/3 knock-out mice were maintained as a homozygous colony to prevent recombination re-occurring in order to conserve the Brinp2/Brinp3tm1PB alleles. A WT line from the same parental origins was maintained in parallel.
Breeding Brinp1/2/3−/− Triple Knock-Out Mice
Female Brinp2/3−/− mice were bred with male Brinp1−/− mice, resulting in mice heterozygous for Brinp1tm1.1Pib, Brinp2tm1.1Pib, and Brinp3tm1.1Pib. Triple heterozygous mice were mated to generate Brinp1/2/3−/− mice at an expected frequency of 1/16, along with WT littermates. Additionally, male mice of genotype Brinp1+/−/2−/−/3−/− were mated with female triple heterozygous mice in an attempt to increase triple knock-out mice frequency. Brinp1/2/3−/− mice were identified by genotyping PCR, and validated by RT-PCR.
RT-PCR
RNA was extracted from the whole brain of WT, het and knock-out mice from each Brinp line and reverse transcribed into cDNA (SSIII First Strand Synthesis, Life Sciences). Primers used for Brinp1 primers were designed to Exon 2: 5′-CTGGGACAGACCAACATGTCTC and Exon 6: 3′-GCTCTCCGTGCTTTGCAGAAGG, to produce a 526 bp WT product or a 336 bp floxed product.
Brinp2 primers were designed to Exon 2: 5′-wordGGACTGGCTGCTCACAGACCG and Exon 4: 3′-wordGTGCTCTCTCTGTCAATGAAG, to produce a 439 bp WT product or a 247 bp floxed product.
Brinp3 primers were designed to Exon 2: 5′-wordCCCCTTCGACTGGCTCCTCTC and Exon 5: 3′-wordCCTGTCCGTGTTTCTGTCACC, to produce a 510 bp WT product or a 221 bp floxed product. PCR conditions: 95°C 60 s (95°C 30 s, 61°C 30 s, 72°C 30 s) × 35, 72°C 120 s. RT-PCR products were cut out of a 2% agarose gel and sequenced.
Animals
C57BL/6 floxed mice and wild type littermates were generated from heterozygous breeders in all studies. Mice were genotyped from tail snips collected at postnatal day 10 (P10). For each knock-out line, mice were housed with mixed genotype littermates; with a maximum of five adults per box. Mice were housed in individually ventilated, sawdust lined Thoren cages, and fed ad libitum on Barastoc rodent feed with constant access to water. Mice were maintained on a 12 h light/dark cycle (light 7 a.m.–7 p.m.) and with a controlled room temperature of 18–24°C. Cages and bedding were changed weekly. All breeding and experiments were approved by the Monash University Animal Ethics Committee.
Weighing
Brinp2−/−, Brinp3−/− Brinp2/3−/−, Brinp1/2/3−/− mice, along with their respective WT littermates were weighed weekly between 3 and 12 weeks.
Histology
Twenty-five organs per mouse were compared to their respective WT littermates from juvenile (7–8 week old) Brinp2−/−, Brinp3−/−, Brinp2/3−/−, and Brinp1/2/3−/− mice. Tissue was prepared as formalin fixed, paraffin wax embedded sections (10 μm). Hematoxylin and Eosin (H&E) staining was carried out by the Australian Phenomics Network (http://www.australianphenomics.org.au/). Histopathological assessments and clinical hematological analysis was performed on two mice per genotype and compared to WT littermates. The same 25 organ types were examined for macromorphological abnormalities as previously described (Berkowicz et al., 2016). For a list of organs examined, refer to the characterization of Brinp1−/− mice.
Reproductive Phenotyping
Three breeding pairs per genotype (WT/WT, Triple-Het/Triple-Het, and Triple-Het ♂/Brinp1−/+2−/−3−/− ♂) were set up and monitored over 5 months. Mice were first used as breeders at 7–8 weeks of age. The number of pups were recorded at birth and at weaning (postnatal day 21).
Behavioral Testing
Cohort sizes were 9–12 mice, aged 3–4 months. Where possible, a ratio of 1:1 females/males was tested. Mice were habituated to the testing facility for 1 week, then habituated to the testing room overnight. Mice were tested by experimenters blind to genotype and in random order. Tests were separated by a minimum of 1 day. WT and knock-out mice were tested in the same testing sessions. Lighting conditions were 30 lux for all behavioral tests. Testing arenas were cleaned with Equinade disinfectant (lavender scent) between trials. In all instances, mice had previously been habituated to the disinfectant whilst housed at the testing facility. Behavioral tests were performed in an identical manner to previously described for the behavioral characterization of Brinp1−/− mice (Berkowicz et al., 2016), as follows.
Visual Placing Test
Mice were lifted by the tail to a height of 15 cm and lowered onto a mesh grid within 1 s, decelerating as the grid approached. The distance of the animal's nose from the grid was measured the moment before the mouse extended its forelimbs toward it. A single trial was performed per animal.
Olfaction Test
Relative levels of sniffing behavior was investigated within an area that contained 2 squares of filter paper (4 × 4 cm), one containing 275 μl of peanut butter (at three dilutions—1:10; 1:100; 1:1000) and one containing 275 μl of water, over a 3 min period. The animals were first acclimatized to the area for a 15 min period.
Rotarod
Mice were pre-trained on the Rotarod (Ugo Basil) for two initial trials at a constant speed of 4 rpm for 5 min, followed by a third trial accelerating from 4 to 40 rpm over 5 min. Testing was carried out the following day by 4 × 5 min accelerating trials with an inter-trial interval of 30 min.
Elevated Plus Maze
Mice were placed on an elevated platform (material: Perspex, color: beige) at a height of 40 cm above the floor. The platform comprised of two open arms and two closed arms (each 4.5 cm wide, 30 cm in length), connected by a central square (6 × 6 cm). The two closed arms were protected by a 15 cm high wall. Mice were placed on the center square and video recorded whilst exploring the maze. Time and frequency of entry into each arm, and average velocity throughout the trial time of 5 min were recorded. Tracking software: Noldus Ethovision 3.0. Frequency of peering down, and entries to the far end of the open arm, were counted manually. Entries into the far ends of the open arms were defined as all feet greater than halfway along the arm.
Y-Maze
Mice were tested in two trials of a Plexiglass Y-maze (material: Perspex, color: gray) with each of the three arms having a distinctive visual cue at the end. Dimensions of each arm were 30 × 10 cm, with a triangle center zone of 10 cm equal sides. Mice received a random association between visual cues and arm location. In Trial 1, a partition blocked off the left arm of the maze. The mouse was placed at the end of the home arm, facing away from the center. The time spent in each of the two available arms over 10 min was recorded. Mice were rested for 2 h. In trial 2, testing was repeated in a second trial with the partition removed and all three arms made accessible. Time in each arm and average velocity was recorded for trial 2 for 10 min. Tracking software: Noldus Ethovision XT 5.0.
Acoustic Startle and Pre-pulse Inhibition (PPI)
Mice were placed individually inside a Perspex cylinder, closed at both ends. The cylinder was placed upon a platform sensitive to weight displacement, within a sound attenuating box with a background sound level (San Diego Instruments Startle Response System). The background white noise level was set to 70 dB. To measure acoustic startle, a strong 40 ms startle sound was played and startle response was measured by the jumping reflex (<1 s) as weight displacement on the platform. Pre-Pulse Inhibition (PPI) was measured as the percentage reduction in startle response when a non-startling 20 ms pre-pulse of (a) 4 db (b) 8 db (c) 16 db above the 70 dB background sound was played 100 ms prior to the startle sound.
Three Chamber Social Interaction Test
Identical rectangular wire cages were placed in equivalent positions in the left and right chambers of a three-chamber plexiglass box (600 × 400 × 250 mm). Mice were habituated to the empty cages in the left and right chambers (trial 1) and time interacting with each cage was recorded. In trial 2, an unfamiliar C57BL/6J WT sex-matched mouse was introduced to one of the cages, and time interacting with each cage was again recorded. Each trial lasted 10 min, with average velocity recorded for each trial. The mice serving as strangers were habituated to placement under the wire cage for 5 min prior to the test. Mice were tracked using Cleversys Tracking and Topscan software. The interaction zone was defined by the software as an unmarked perimeter zone of 2 cm around the metal cages. Interaction time was defined as nose within the interaction zone. The chambers were cleaned between trials with Equinade disinfectant (lavender scent).
Statistical Analysis
Survival rates of total numbers of mice weaned was analyzed by Chi-square test. Number of surviving mice per litter was analyzed by one-way ANOVA. Postnatal weight, olfaction test, Rotarod, Elevated Plus Maze, and Y-maze performance, and PPI analyzed by repeat measures two-way ANOVA. Vision test, startle response and EPM and Y-maze secondary data was analyzed by Student t-test. Habituation trials and social interaction test were analyzed by one-way ANOVA. Average velocity for trial 1 (habituation) and trial 2 of the social interaction test was analyzed by two-way repeat measures ANOVA. Male and female mice were analyzed separately, unless otherwise stated. Behavioral data was represented as the mean ± standard deviation. A value of p < 0.05 was used to determine significance.
Results
Targeting of Brinp2 Knock-Out Mice
A conditional Brinp2 targeted allele (Brinp2tm1/Pib) was designed to allow Cre-recombinase-mediated, tissue-specific deletion of Brinp2 (Figure 1A). Mice lacking Brinp2 in all tissues were generated by breeding animals carrying the targeted allele with animals expressing Cre-recombinase from the two-cell embryonic stage onwards (global Cre-deleter). Progeny exhibiting deletion of the selection cassette and third exon of Brinp2 (Brinp2tm1.1/Pib) were inter-bred to generate homozygous Brinp2tm1.1Pib/tm1.1Pib (Brinp2−/−) animals and WT littermates. Correct targeting and deletion of exon 3 was confirmed by Southern analysis (Figure 1B), RT-PCR (Figure 1E), and DNA sequencing of RT-PCR products (Figure 1F). The resultant mRNA lacking exon 3 reflects forced splicing between the intron 2 donor and intron 4 acceptor, fusing exons 2 and 4, and changing the reading frame to introduce a truncating stop codon (Figure 1F). The predicted mutant protein would comprise the cleavable signal peptide (33 aa) and 56 aa of the 750 aa mature BRINP2 protein, and contains no recognizable functional domains. Hence this 56 aa form is missing over 92% of the BRINP2 amino acid sequence, and would be highly unlikely to fold correctly. It is therefore likely degraded shortly after synthesis, which is the generally accepted fate of truncated or misfolded proteins (Hiller et al., 1996).
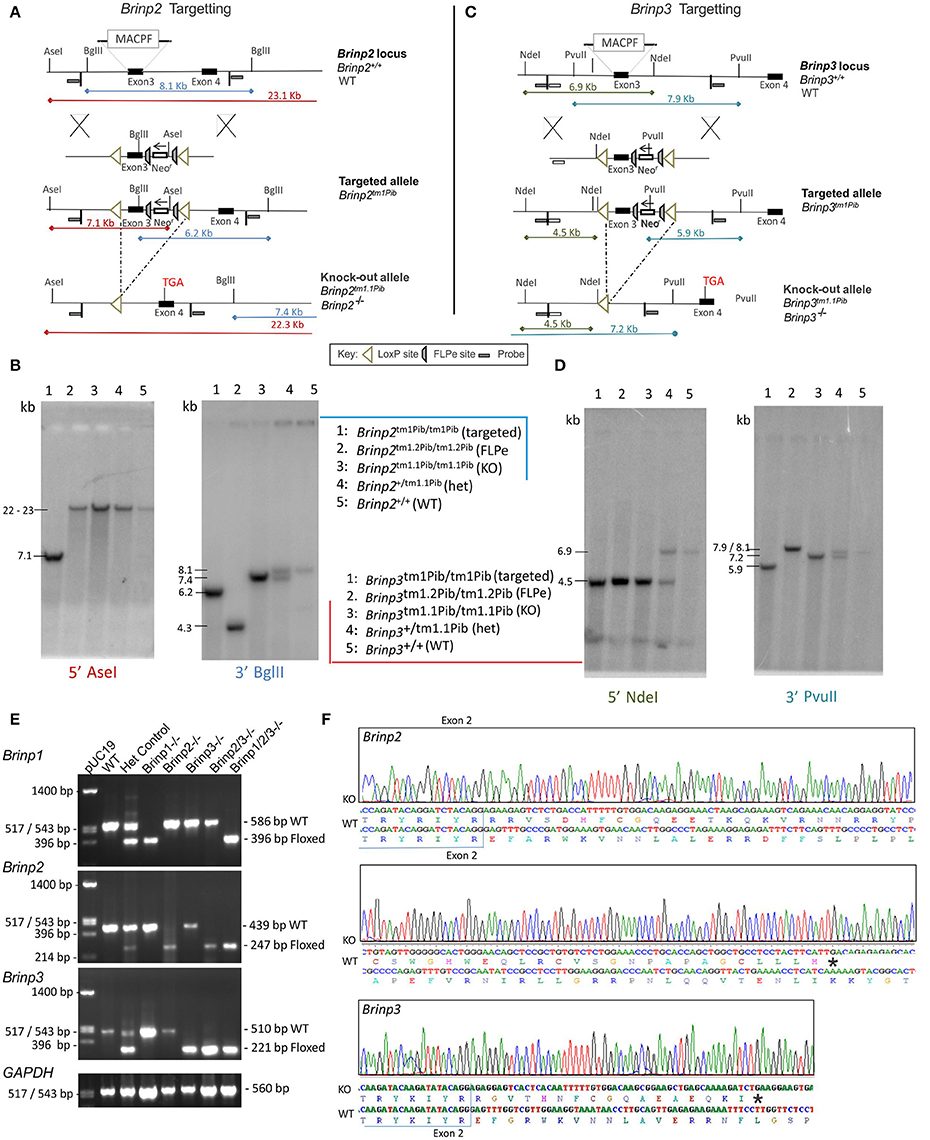
Figure 1. Brinp2 and Brinp3 Targeting. (A) The Brinp2 targeting vector was designed with a Neomycin resistance (Neo-r) cassette after exon 3, and FRT sites placed before and after the Neo-r cassette. The 192 bp 3rd exon of Brinp2 contains the start of the Membrane Attack Complex/Perforin (MACPF) domain. LoxP sites flank exon 3 and the Neo-r cassette. When crossed with a global Cre-deleter mouse line, the recombination of LoxP sites resulted in the deletion of exon 3 and the Neo-r cassette. (B) Brinp2 Southern Blot: splenic genomic DNA was cleaved with PstI and BglII and hybridized to 500 bp genomic DNA probes from the 5′ region (Asel) and 3′ region (Bgl II) of the targeting construct. In wild-type DNA, species of 23.1 kb (Asel) and 8.1 kb (Bgl II) were detected. The 8.1 kb (Bgl II) species was not present in DNA from Brinp2−/− mutants, replaced with shorter species 7.4 kb (BglII). (C) The Brinp3 targeting vector was designed with a Neo-r cassette after exon 3, and FRT sites positioned before and after the Neo-r Cassette. The 289 bp 3rd exon of Brinp3 contains the start of the MACPF domain. LoxP sites flank exon 3 and the Neo-r cassette. When crossed with a global Cre-deleter mouse line, the recombination of LoxP sites resulted in the deletion of exon 3 and the Neo-r cassette. (D) Brinp3 Southern Blot: splenic genomic DNA was cleaved with PstI and BglII and hybridized to 500 bp genomic DNA probes from the 5′ region (Ndel) and 3′ region (PvuII) of the targeting construct. In wild-type DNA, species of 6.9 kb (Ndel) and 7.9 kb (PvuII) were detected. These products were not present in DNA from Brinp3−/− mutants, replaced with shorter species of 4.5 kb (Ndel) and 7.2 kb (PvuII). (E) Validation of knock-out line using cDNA derived from brain tissue mRNA from WT, heterozygous and Brinp2−/− Brinp3−/− Brinp2/3−/− Brinp1/2/3−/− mice. For each genotype, PCR product sizes correspond to the removal of exon3 in the knock-out allele. (F) Sequencing of the Brinp2−/− and Brinp3−/− allele RT-PCR products showed the expected absence of exon 3, and that splicing fuses exons 2 and 4, resulting in a frame shift that introduces a stop codon after 50 (Brinp2−/−) and 18 (Brinp3−/−) residues. Sequencing results for Brinp1−/− allele can be found in previously published work (Berkowicz et al., 2016).
Targeting of Brinp3 Knock-Out Mice
In an identical manner to the Brinp2 targeted mouse line, a conditional Brinp3 targeted allele (Brinp3tm1/Pib) was designed to allow Cre-recombinase-mediated, tissue-specific deletion of Brinp3 (Figure 1C). Mice lacking Brinp3 in all tissues were created by breeding animals carrying the Brinp3 targeted allele with animals expressing global Cre-deleter mice. Progeny exhibiting deletion of the selection cassette and third exon of Brinp3 (Brinp3tm1.1/Pib) were inter-bred to generate homozygous Brinp3tm1.1Pib/tm1.1Pib (Brinp3−/−) animals and WT littermates. Deletion of exon 3 was confirmed by Southern analysis (Figure 1D), RT-PCR (Figure 1E), and DNA sequencing of RT-PCR products (Figure 1F). The resultant mRNA lacking exon 3 reflects forced splicing between the intron 2 donor and intron 4 acceptor, fusing exons 2 and 4, and changing the reading frame to introduce a truncating stop codon (Figure 1F). The predicted mutant protein would comprise the cleavable signal peptide (33 aa) and 44 aa of the 733 aa mature BRINP3 protein, and contains no recognizable functional domains. Hence this 44 aa form is missing over 93% of the BRINP3 amino acid sequence, and would be highly unlikely to fold correctly, and therefore again likely degraded shortly after synthesis.
Generation and Validation of Multigenic Brinp Deletion
Validation of Brinp2/3−/− and Brinp1/2/3−/− mice was performed by RT-PCR of cDNA derived from knock-out mouse brain tissue of each genotype. Only PCR products of sizes corresponding with the exon 3-deleted Brinp2tm1.1Pib and Brinp3tm1.1Pib alleles were present for Brinp2/3−/− mice, confirming homozygosity of both knock-out alleles (Figure 1E). For the Brinp1/2/3−/− mouse cDNA, only PCR products of sizes corresponding with the exon3-deleted Brinp1tm1.1Pib, Brinp2tm1.1Pib, and Brinp3tm1.1Pib alleles were present, confirming homozygosity of all three knock-out exon-3 deleted alleles (Figure 1E).
Brinp2−/− and Brinp3−/− Mice Are Viable and Appear at Mendelian Frequencies; Brinp1/2/3−/− Mice Exhibit Poor Viability
Litters bred from Brinp2+/− or Brinp3+/− heterozygous crosses were monitored for survival. Mice were genotyped at age of weaning (day 21). Genotypes were assessed for Mendelian inheritance (25% of total progeny expected to be knock-out mice) as an indicator of normal in utero and neonatal viability. Brinp2−/− and Brinp3−/− mice demonstrated close to Mendelian inheritance for the number of knock-outs surviving to age of weaning (Brinp2−/−: 9/50 = 22.5%, Brinp3−/−: 10/41 = 24%) and litter sizes from homozygous breeders were normal (Brinp2−/−: 6.4 ± 2.9 SD), Brinp3−/−: 6.9 ± 1.9 SD). Brinp2/3−/− homozygotes also produced litters of standard size (6.0 ± 1.9 SD) indicating normal viability.
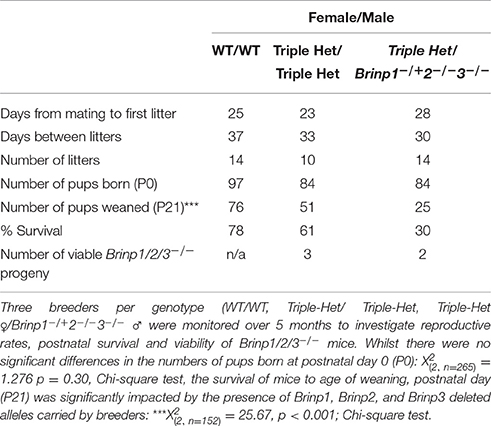
It proved difficult to generate practical numbers of viable Brinp1/2/3−/− progeny due to both the low expected frequency from triple heterozygous matings (1 in 16) and the compromised postnatal viability of pups from mothers carrying the Brinp1−/− alleles (see Berkowicz et al., 2016). Three triple-het breeders mated for a total of 5 months produced three triple KO mice at a frequency of 3/51 (5.9%), and therefore viable at close to expected frequency of 1/16 (6.3%), however reduced overall litter survival from triple het breeders meant only 3 out of a total of 84 mice pups born were viable triple knock-out mice (Table 1). To increase the likelihood of generating enough triple knock-out mice for a minimum behavioral cohort of 10 within a similar age, female triple heterozygous breeders were paired with male Brinp1+/−/2−/−/3−/− mice. These breeders produced normal litter sizes at birth, but many progeny died before weaning, similar to mice carrying Brinp1 knock-out allele. The addition of the male mice carrying the Brinp2/3−/− mutation resulted in a significant decrease in offspring survival and indicates the absence of Brinp2 and Brinp3, when combined with Brinp1 heterozygosity in the male breeder impacts litter survival (Supplementary Figure 1). These limitations meant that it was not possible to generate a triple knock-out behavioral cohort within an appropriate age range.
Brinp2−/−, Brinp3−/−, Brinp2/3−/−, and Brinp1/2/3−/− Mice Exhibit Reduced Body Mass, but Normal Gross Morphology
Brinp2−/−, Brinp3−/−, Brinp2/3−/−, and Brinp1/2/3−/− mice were weighed weekly from 3 to 12 weeks of age, alongside their respective WT littermates. Knock-out mice from all four lines showed some reduction in body mass. Female Brinp2−/− mice displayed normal weight from infant to adult, whereas male Brinp2−/− mice showed reduced body mass from 7 weeks onwards, weighing 10% less as adults at week 12 (Figure 2A). Brinp3−/− mice weighed less than their WT littermates in the first few weeks of weighing (3–5 weeks) for both males and females, before their weights recovered to that of their WT littermates (Figure 2B). Female Brinp2/3−/− mice were smaller in the first few weeks, before recovering to a weight similar to that of WT. Male Brinp2/3−/− mice weighed less than WT controls as adults (Figure 2C). Overall, Brinp2/3−/− mice show a trend in body mass reduction that reflects a combination of the Brinp2−/− and Brinp3−/− mice weight profiles.
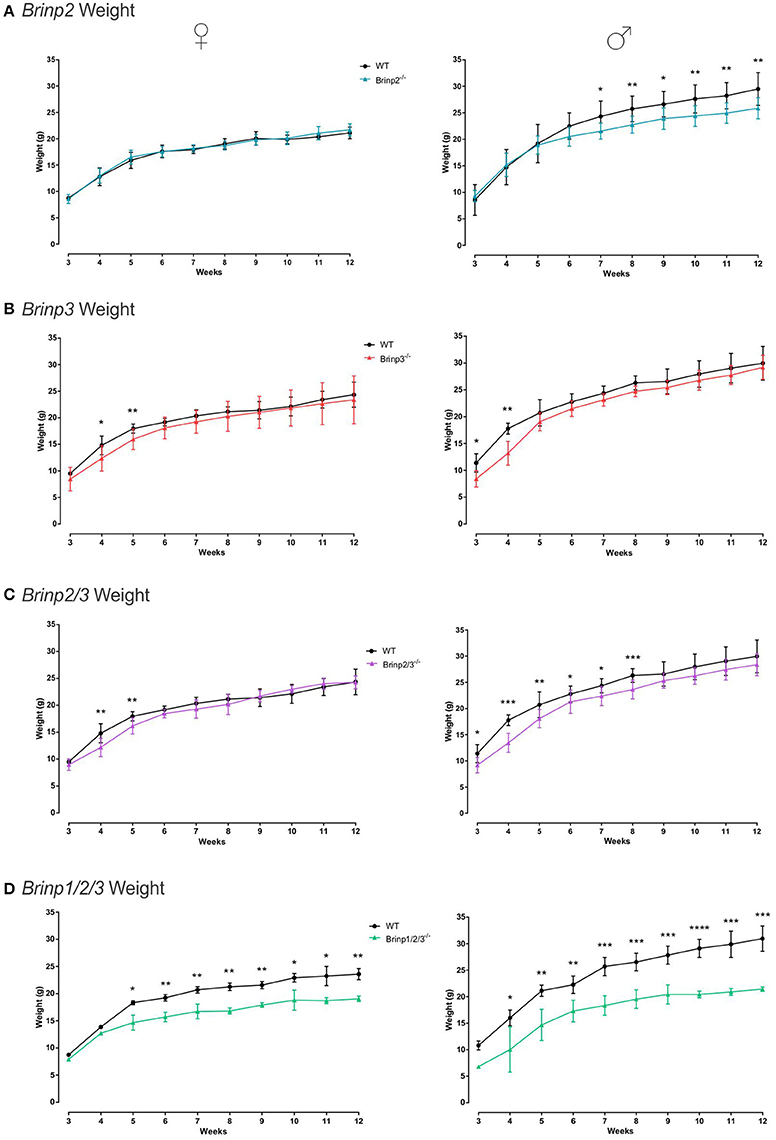
Figure 2. Brinp2, Brinp3, Brinp2/3, and Brinp1/2/3 weights. (A) Brinp2−/− mice weighed from week 3 to 12. Female Brinp2−/− mice show normal weight between 3 and 12 weeks by repeat measures two-way ANOVA: F(1, 15) = 0.155, p = 0.699. Male Brinp2−/− mice show a significant reduction in body weight from 6 weeks of age onwards. By repeat measures two-way ANOVA male Brinp2−/− mice show a significant interaction of genotype × week: F(9, 117) = 5.645, p < 0.001. N = 8 females, 8 males per genotype. (B) Brinp3−/− mice weighed from week 3 to 12. Both Male and Female Brinp3−/− mice show reduced body mass from weeks 3 to 5, before recovering to near normal weight from week 6 onwards. Female Brinp3−/− mice show a significant interaction of genotype × week: F(8, 96) = 2.115, p = 0.042, repeat measures two-way ANOVA. Male Brinp3−/− mice show a significant reduction in body weight to their littermates F(1, 12) = 8.602, p = 0.013, and a significant genotype × week interaction F(9, 108) = 3.919, p < 0.001, repeat measures two-way ANOVA. N = 7 females, 7 males per genotype. (C) Brinp2/3−/− mice weighed from week 3 to 12. Female Brinp2/3−/− mice show a significant week × genotype interaction effect: F(9, 117) = 3.065, p = 0.002. Male Brinp2/3−/− mice show a significant decrease in body weight, analyzed by repeat measures two-way ANOVA: F(1, 14) = 6.228, p = 0.026, and a significant interaction effect between week × genotype: F(9, 126) = 2.461, p = 0.013. N = 8 females, 8 males per genotype. (D) Brinp1/2/3−/− mice weighed from week 3 to 12. Female Brinp1/2/3−/− mice show a significant reduction in body weight: F(1, 3) = 47.738, p = 0.007, with a significant interaction between week × genotype: F(9, 27) = 5.736, p < 0.001, repeat measures two-way ANOVA, n = 3 WT, 2 Brinp1/2/3−/− mice. Male Brinp1/2/3−/− mice also show a significant decrease in body weight: F(1, 8) = 54.708, p < 0.001, with a significant interaction between week × genotype: F(8, 64) = 3.005, p = 0.006, repeat measures two-way ANOVA, n = 5 WT, 5 Brinp1/2/3−/− mice. Results represented as the mean ± SD.
Female and male Brinp1/2/3−/− mice weighed significantly less than WT littermates, and less than Brinp2−/−, Brinp3−/−, or Brinp2/3−/− mice (Figure 2D). Triple Brinp knock-out mice also weighed less the Brinp1−/− mice (Berkowicz et al., 2016), indicating a cumulative effect of the triple gene knock-out on body mass.
A full histological examination of juvenile mice for Brinp2−/−, Brinp3−/−, Brinp2/3−/−, and Brinp1/2/3−/− mice at 7–8 weeks of age showed normal organ development (25 organs examined) including normal structures in the brain and spinal cord. In all cases, brains appeared symmetrical, with normal myelination, and no ventricular dilation observed (data not shown).
Brinp2−/−, Brinp3−/−, and Brinp2/3−/− Mouse Behavior
To evaluate the effect of Brinp2 and Brinp3 loss on neurological function, the behavior of the single and double knock-out lines were assessed. In an initial screen, Brinp2−/− and Brinp3−/− and Brinp2/3−/− mice showed normal auditory, visual, and olfactory capabilities (Supplementary Figures 2A,B). Mice showed no impairments in motor co-ordination on the Rotarod (Supplementary Figures 3A–C), with female Brinp2−/− mice showing improved performance on this test.
Brinp3−/− Knock-Out Mice Exhibit Marked Changes in Exploratory Behavior
To examine whether anxiety was affected in the knock-out mice, animals were allowed to freely explore an elevated plus maze (EPM) for 5 min. Mice normally show a species-typical preference for the walled closed arms, and show some hesitance to enter the more exposed open arms. Mice that show a high level of reluctance to approach the open arm are interpreted as having an anxiety-like phenotype.
A pronounced phenotype was exhibited by male and female Brinp3−/− mice, which spent significantly longer on the open arms and center square, and less time in the closed arms (Figure 3A), indicating reduced anxiety levels. The velocity of Brinp3−/− mice was normal for this test, ruling out hyperactivity (Figure 3B). Brinp3−/− mice showed a reduced latency time to enter the open arm of the EPM (Figure 3C), and increased number of entries into the open arms (Figure 3D). Additionally, Brinp3−/− mice showed prolonged exploration time at the exposed ends of the open arms (Figure 3E), and exhibited peering down behavior at the edges of the open arms (Figure 3F).
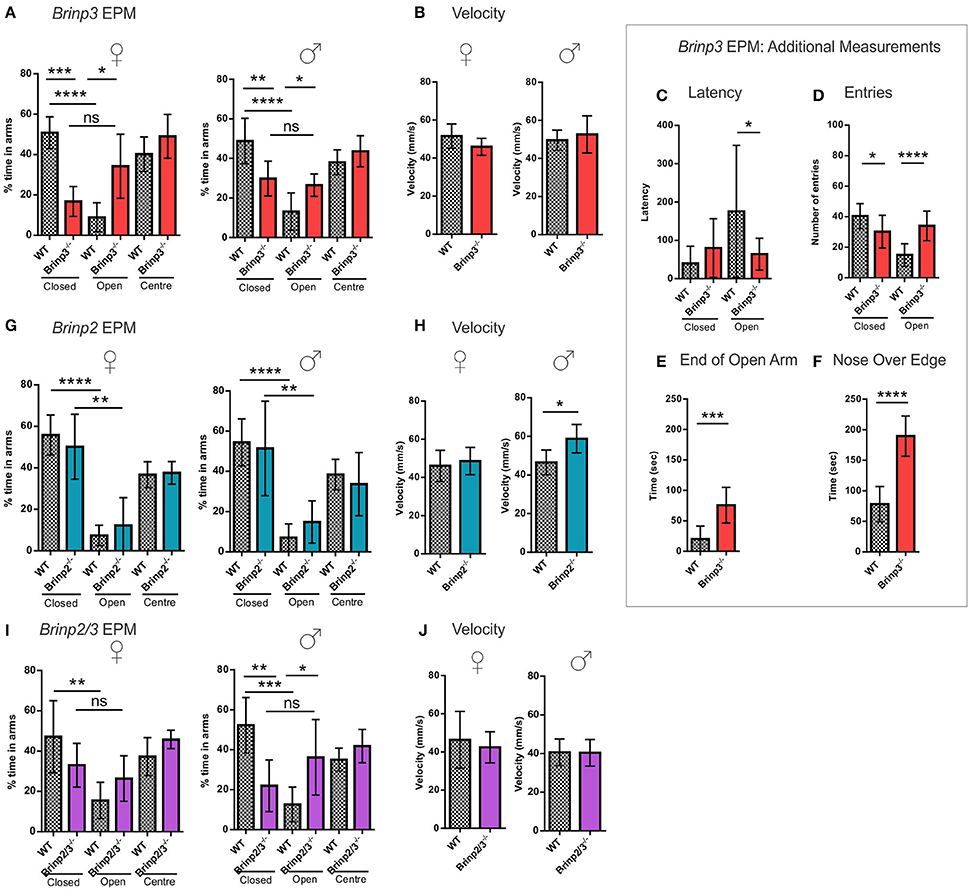
Figure 3. Brinp3−/− and Brinp2/3−/− mice show increased exploration of the Elevated Plus Maze. (A) Brinp3−/− mice spend more time in the open arms relative to the closed arms of the maze, indicating reduced anxiety. Analysis by repeat measure ANOVA shows a genotype × arm interaction: Female: F(2, 16) = 15.359, p <0.001, male: F(2, 24) = 4.407, p = 0.023. N = 5 female, 7 male mice per genotype. (B) Normal Brinp3−/− mice average velocity during EPM testing, female: t(8) = 1.629, p = 0.142, male: t(12) = 0.731, p = 0.479, Student’s t-test. (C) Brinp3−/− mice show a significantly reduced latency to enter the open arm: t(22) = 2.187, p = 0.040, unpaired student t-test. (D) Brinp3−/− show a significantly reduced number of entries into the closed arm: t(21) = 2.575, p = 0.018 and a significant increase into the open arm: t(21) = 5.334, p <0.0001, unpaired Student’s t-test. (E) Brinp3−/− mice spend significantly more time peering over the edge of the open arms compared to WT littermates, t(14) = 7.190, p <0.0001, Student’s t-test. (F) Brinp3−/− mice spend significantly more time at the end of the open arms (defined as >50% of arm length from center), t(14) = 4.346, p = 0.0007, unpaired Student’s t-test. (G) No significant effect on arm duration for Brinp2−/− mice, female: F(2, 20) = 0.552, p = 0.584, male: F(2, 20) = 0.483, p = 0.624, repeat measures two-way ANOVA. N = 6 female, 6 male mice per genotype. (H) Male Brinp2−/− mice show increased average velocity during EPM testing, female: t(10) = 0.571, p = 0.581, male: t(9) = 2.95, p = 0.016, Student’s t-test. (I) Brinp2/3−/− mice: Analysis by repeat measure ANOVA shows a genotype × arm interaction for Brinp2/3−/− male mice for percentage time in each arm: Female: F(2, 16) = 1.762, p = 0.203, male: F(2, 20) = 4.571, p = 0.023. N = 5 female, 6 male mice per genotype. (J) Normal Brinp2/3−/− mice average velocity during EPM testing, female: t(10) = 0.523, p = 0.616, male: t(10) = 0.055, p = 0.958, Student’s t-test. ns = not significant. *p <0.05, **p <0.01, ***p <0.001, ****p <0.0001. Female and male data was combined for graphs (C–F). Results represented as the mean ± SD.
In contrast to the Brinp3−/− mice, Brinp2−/− mice did not show an increased preference for the open arms of the EPM, indicating a normal response to potential danger (Figure 3G). However, male but not female mice did show increased velocity in the test, consistent with hyperactivity (Figure 3H).
An increase in open arm exploration was also apparent in the Brinp2/3−/− mice (Figures 3I,J), consistent with the absence of Brinp3 alone, suggesting that this reduced anxiety phenotype is not modified by the absence of Brinp2.
Brinp2−/−, Brinp3−/−, and Brinp2/3−/− Mice Exhibit Normal Sensory Gating and Short-Term Memory
The startle response and PPI test is used to measure sensory gating in mice, and models deficits in human subjects diagnosed with schizophrenia. Brinp2−/−, Brinp3−/−, and Brinp2/3−/− mice were able to inhibit the startle response when primed with pre-pulses of 4, 8, and 16 dB (Supplementary Figures 4A,B). The ability of all three knock-out lines to gate their startle response indicates that these mice do not model this aspect of human schizophrenia.
Brinp2−/−, Brinp3−/−, and Brinp2/3−/− mice were tested for spatial learning and memory in the Y-maze. Following a 2-h interval after exploring the home and a single arm of the maze, mice were recorded exploring the three arms of the maze. As expected, WT control mice for each line preferred to explore the novel arm in this test. All three knock-out lines tested also showed the species-typical significant increase in time spent exploring the novel arm compared to the familiar arm, indicating normal short-term memory (Figures 4A–C). Brinp2−/− mice moved at a statistically significantly increased velocity for this test (Figure 4D), consistent with hyperactivity, whereas Brinp3−/− and Brinp2/3−/− mice did not show a significant increase in locomotor activity (Figures 4E,F).
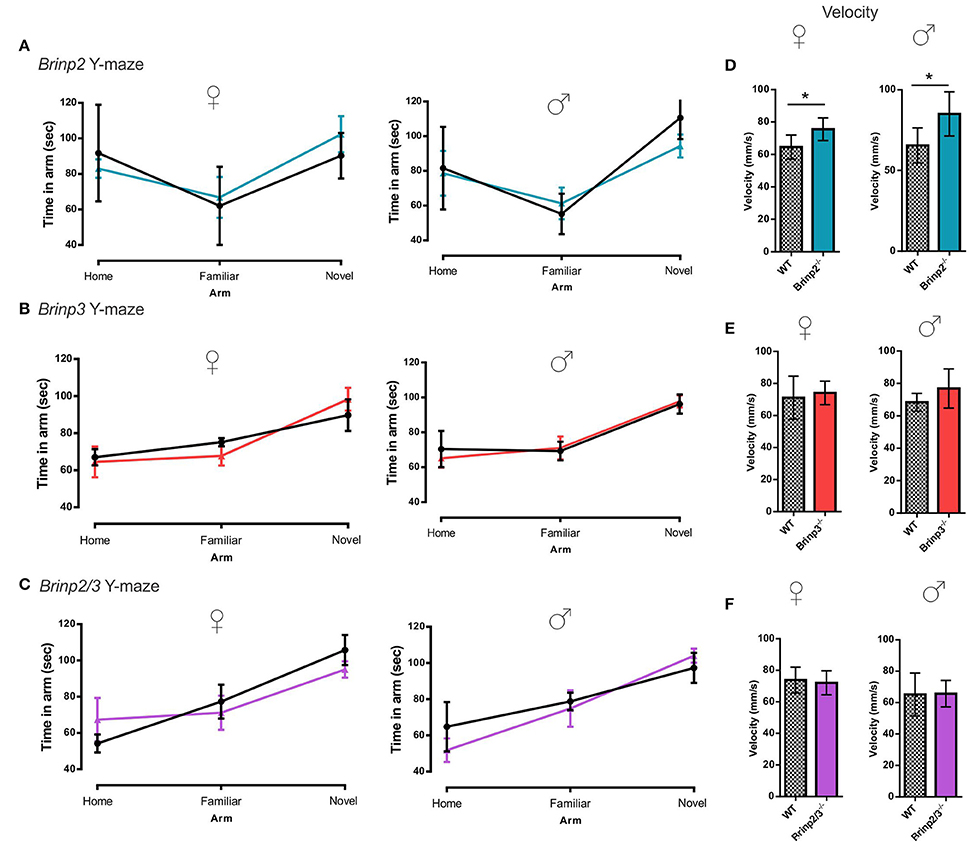
Figure 4. Y-maze: normal short-term memory. (A) Brinp2−/− Y-maze: analysis by repeat measures two-way ANOVA shows no interaction effect between genotype × arm; female: F(2, 20) = 0.842, p = 0.446, male: F(2, 20) = 1.521, p = 0.243. N = 6 female, 6 male mice per genotype. (B) Brinp3−/− Y-maze: No interaction effect between genotype × arm; female: F(2, 16) = 0.582, p = 0.574, male: F(2, 24) = 0.136, p = 0.874. N = 5 female, 7 male mice per genotype, two-way repeat measures ANOVA. (C) Brinp2/3−/− Y-maze: No interaction effect between genotype × arm; female: F(2, 20) = 0.791, p = 0.467, male: F(2, 20) = 0.864, p = 0.436. N = 6 female, 6 male mice per genotype, two-way repeat measures ANOVA. (D) Brinp2−/− mice show increased average velocity during Y-maze testing, female: t(10) = 2.637, p = 0.025, male: t(10) = 2.504, p = 0.037, Student's t-test. (E) Normal Brinp3−/− mice average velocity during Y-maze testing, female: t(8) = 0.435, p = 0.675, male: t(12) = 1.694, p = 0.116, Student's t-test. (F) Normal Brinp2/3−/− mice average velocity during Y-maze testing, female: t(10) = 0.385 p = 0.709 male: t(10) = 0.082, p = 0.937, Student's t-test.*p < 0.05, Data presented as the mean ± SD.
Brinp3−/− and Brinp2/3−/− Mice Exhibit a Mild Reduction in Sociability; Brinp2−/− Mice Show Increased Locomotor Activity
The three chamber social interaction test is routinely used to investigate sociability in rodents (Silverman et al., 2010). In the habituation trial (trial 1) mice were allowed to explore the whole arena, including left and right chambers containing empty cages. Brinp2−/−, Brinp3−/−, and Brinp2/3−/− mice spent a normal amount of time exploring the cages, without preference for left or right chambers (Supplementary Figure 5A). Brinp2−/− mice exhibited increased velocity in this trial (Figure 5A), resulting an increased total distance traveled (Supplementary Figure 5B). Neither Brinp3−/− nor Brinp2/3−/− mice showed significant increases in velocity/distance traveled (Figure 5A and Supplementary Figure 5B).
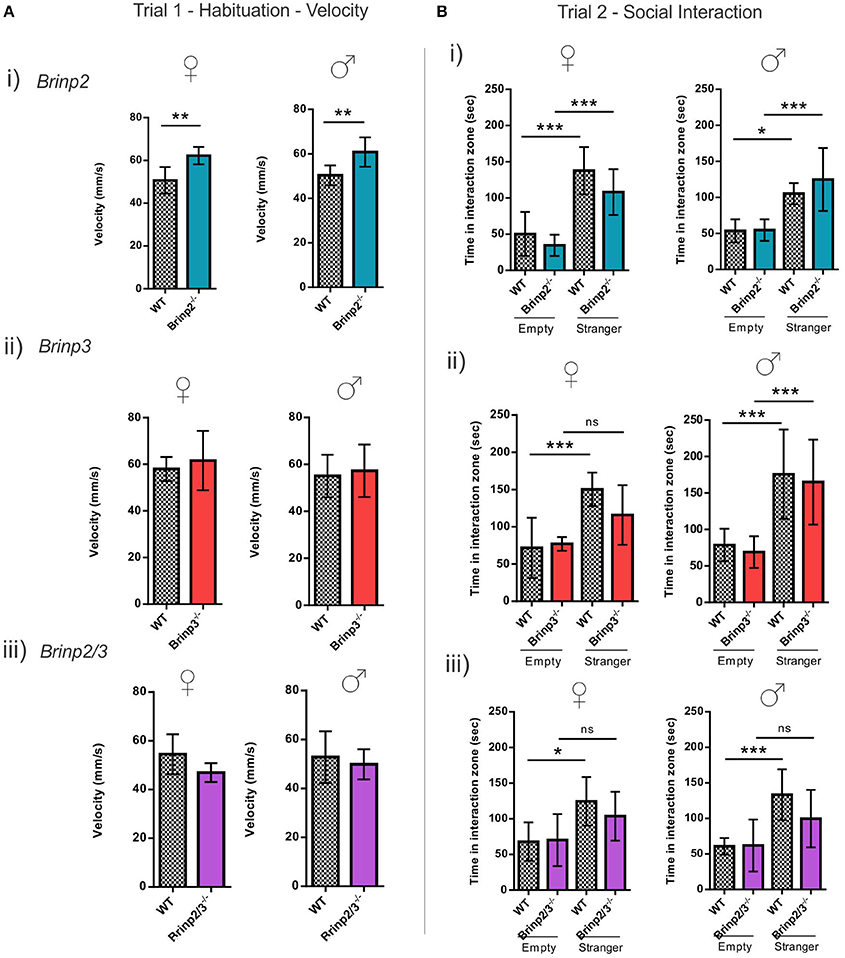
Figure 5. Social interaction of Brinp2−/−, Brinp3−/−, and Brinp2/3−/− knock-out mice. (A) Velocity Trial 1: (i) Brinp2−/− mice exhibited increased average velocity over 10 min trial interval whilst habituating to the area in trial 1, indicating hyperactivity. Female: p = 0.0035, Male: p = 0.0091, Student's t-test. (ii) and (iii) Brinp3−/− and Brinp2/3−/− showed normal activity (velocity) during the habituation trial. Brinp3−/− female: p = 0.5708 Brinp3−/− male: p = 0.6920, Brinp2/3−/− female: p = 0.0938 Brinp2/3−/− male: p = 0.5956, Student's t-test. (B) Social Interaction Trial 2: (i) Brinp2−/− mice display the expected significant increase in interaction time with the stranger mouse compared to an empty cage, indicating normal sociability: Female: F(3, 23) = 17.598, p < 0.001, male: F(3, 22) = 8.310, p < 0.001, one way ANOVA. Tukey HSD post-hoc test: Female: WT empty—WT stranger: p < 0.001, Brinp2−/− empty—Brinp2−/− stranger: p = 0.001, WT empty—Brinp2−/− empty: p = 0.772, WT stranger—Brinp2−/− stranger: p = 0.297. Male: WT empty—WT stranger: p = 0.033, Brinp2−/− empty—Brinp2−/− stranger: p = 0.001, WT empty—Brinp2−/− empty: p = 1.000, WT stranger—Brinp2−/− stranger: p = 0.582, N = 6 females, 6 males per genotype. (ii) Brinp3−/− (female only) mice do not show a significant increase in interaction time with the stranger mouse compared to an empty cage: Brinp3−/− female: F(3, 19) = 7.057, p = 0.003, male: F(3, 27) = 10.868, P < 0.001, one way ANOVA. Tukey HSD post-hoc test: Female: WT empty—WT stranger: p = 0.005, Brinp3−/− empty—Brinp3−/− stranger: p = 0.233, WT empty—Brinp3−/− empty: p = 0.993, WT stranger—Brinp3−/− stranger: p = 0.329. Male: WT empty—WT stranger: p = 0.003, Brinp3−/− empty—Brinp3−/− stranger: p = 0.003, WT empty—Brinp3−/− empty: p = 0.977, WT stranger—Brinp3−/− stranger: p = 0.969, N = 5 females, 7 males per genotype. (iii) Brinp2/3−/− mice do not show a significant increase in interaction time with the stranger mouse compared to an empty cage: Brinp2/3−/− female: F(3, 21) = 3.977, p = 0.025, male: F(3, 23) = 6.606, p = 0.003, one way ANOVA. Tukey HSD post-hoc test: Female: WT empty—WT stranger: p = 0.036, Brinp2/3−/− empty—Brinp2/3−/− stranger: p = 0.399, WT empty—Brinp2/3−/− empty: p = 0.999, WT stranger—Brinp2/3−/− stranger: p = 0.722. Male: WT empty—WT stranger: p = 0.006, Brinp2/3−/− empty—Brinp2/3−/− stranger: p = 0.229, WT empty—Brinp2/3−/− empty: p = 1.000, WT stranger—Brinp2/3−/− stranger: p = 0.321, N = 6 females, 6 males per genotype. ns = not significant. *p < 0.05, **p < 0.01, ***p < 0.001. Data presented as the mean ± SD.
Whilst hyperactive, Brinp2−/− mice exhibited normal sociability in this test. In contrast, Brinp3−/− females and Brinp2/3−/− mice of both sexes did not show the expected increase in interaction time when investigating the cage containing a stranger mouse compared to an empty cage (Figure 5B). No significant difference was detected between WT and knock-outs (Brinp3−/− and Brinp2/3−/−) in the interaction time with the stranger mouse. Overall, these finding suggest that absence of Brinp3 reduces sociability.
Discussion
In this study we have found that absence of Brinp2 or Brinp3 alone or in combination has no overt effect on mouse anatomy, reproduction, or viability, but has an age-specific negative effect on bodyweight. Knockout mice exhibit behavioral traits consistent with NDDs, including hyper-exploration, hyperactivity and reduced sociability.
Brinp3−/− Mice Exhibit Hyper-Exploratory Behavior
The reduced aversion of Brinp3−/− mice to the open arms of the Elevated Plus Maze demonstrates these mice have an altered response to potential danger: specifically, that the Brinp3−/− anxiety response is lessened. A similar but less pronounced phenotype is exhibited by Brinp1−/− mice (Kobayashi et al., 2014; Berkowicz et al., 2016). The fact that both Brinp1−/− and Brinp3−/− mice exhibit this phenotype, along with sociability changes, may indicate a role for both genes in regulating anxiety as well as sociability. Consistent with this suggestion, both Brinp1 and Brinp3 are highly expressed in the adult amygdala complex, a brain region governing fear and other emotions (Prather et al., 2001; Kalin et al., 2004).
The repetitive peering down behavior of Brinp3−/− mice has been previously reported in the dopamine transporter (DAT knock-out) mouse model for ADHD as the “cliff avoidance reaction” (Yamashita et al., 2013), which could indicate that changes in synaptic dopamine levels may contribute to this behavior in Brinp3−/− mice. Another explanation could be changes in levels of the stress-response neurotransmitter norepinephrine in the Brinp3−/− mice, as Brinp3 expression correlates with changes in norepinephrine levels (Goddard et al., 2010; Karoly et al., 2012). Future steps to investigate underlying physiology, as well as the use of fear response paradigms, such as fear avoidance and fear conditioning, may further elucidate the role of Brinp3 in relation to human anxiety disorders.
Increased Locomotor Activity of Brinp2−/− Mice
Brinp2−/− mice exhibit increased locomotor activity, a phenotype also observed in Brinp1−/− mice (Kobayashi et al., 2014; Berkowicz et al., 2016). This hyperactivity may model the reported ADHD in patients with alterations at the 1q25.2 locus (Lesch et al., 2008; Romanos et al., 2008; Lionel et al., 2014). To determine whether these mice show face validity for human ADHD, the Brinp2−/− mice would require testing for attention and impulsivity as key diagnostic criteria. The 5-choice serial reaction time test is an ideal approach, as this paradigm has been established for assessing both attention and impulse control in rodents (Higgins and Breysse, 2008; Asinof and Paine, 2014).
Brinp2−/− mice show a reduction in body weight as adults (males only). This reduced body mass may reflect increased energy expenditure due to hyperactivity. The enhanced Rotarod performance by female Brinp2−/− mice may also be due to increased locomotor activity, as reported for other hyperactive mice (Graham and Sidhu, 2010; Bohuslavova et al., 2016). It is intriguing that Brinp2−/− mice are hyperactive, whilst Brinp2/3−/− mice are not. Perhaps the absence of Brinp3 results in behavior that reduces overall locomotor activity. For example, frequent peering-down along the edges of the open arms of the EPM would reduce overall horizontal plane velocity.
Brinp2/3−/− Mice Resemble Brinp3−/− Mice
The phenotype of Brinp2/3−/− mice shows a high degree of similarity to that of the Brinp3−/−mice. The weight profile is almost identical, as is the hyper-exploratory phenotype detected on the elevated plus maze, normal velocity in various tests, and the changes in sociability. These results suggest that apart perhaps from locomotor activity, there is no significant phenotypic alteration or enhancement in the double knock-out mice compared to the single knock-out mice. Therefore, the genes do not compensate to mask phenotypic behaviors of the single knock-out mice.
Brinp2 and Brinp3 show partial overlap in expression profile during development, including co-expression in brain regions that include the cerebellum, neocortex, and olfactory bulb (Kawano et al., 2004). It is however notable that there are regions where Brinp2 and Brinp3 show distinct expression profiles, e.g., Brinp2 is highly expressed in the CA1, CA2, and CA3 regions of the adult hippocampus, whilst Brinp3 is predominantly expressed in the dentate gyrus. Brinp3 also shows broader expression in the cerebellum (Kawano et al., 2004). Taken with our findings, this suggests that either Brinp2/Brinp3 have distinct molecular functions, or Brinp2 and Brinp3 carry out the same role, but function in distinct neuronal subtypes.
Comparison with the Brinp1−/− Mice Phenotype
Comparing Brinp2−/− and Brinp3−/− mice phenotypes to previously reported Brinp1−/− mice (Kobayashi et al., 2014; Berkowicz et al., 2016), it is evident that there is some areas of overlap, but overall mice do not show the same breadth of severity as the Brinp1−/− mice. For instance, neither Brinp2−/− or Brinp3−/− mice exhibit reduced viability, impaired short-term memory and do not show the same degree of reduced sociability or decreased body weight. This is consistent with the observation that Brinp1 is the most highly and ubiquitously expressed of the three genes during brain development (Kawano et al., 2004). The overlap in some, but not all phenotypes between mice lacking Brinp2−/− or Brinp3−/− with the Brinp1−/− mice indicates possible shared Brinp1–Brinp2 and Brinp1–Brinp3 molecular functions.
Brinp1/2/3−/− Mice: Initial Insights into Mice Lacking All Brinps
The poor survival of litters from Brinp1/2/3 heterozygous breeders is likely related to the absence of Brinp1, as we have previously reported (Berkowicz et al., 2016). Although the additional absence of Brinp2 and Brinp3 likely exacerbates the phenotype, as appears to be the case when viability of litters drops further when male mice of genotype Brinp1−/+2−/−3−/− are bred with female triple-het mice. This drop in viability of litters above that of litters heterozygous for all three Brinp genes may be due to the cumulative effect of the absence of Brinp3 and Brinp1 in reducing sociability, and therefore affecting parental care of offspring. Altogether, a different breeding strategy, or a much larger breeding colony of triple-hets is needed for generating sufficient numbers of triple KO out mice for a behavioral testing cohort of 10 mice of a similar age.
The survival past weaning of some Brinp1/2/3−/− triple knock-out mice indicates that Brinps are not essential for embryonic or neonatal development. The absence of all three Brinps results in an ostensibly normal, but significantly smaller mouse, with normal gross morphology of all neural and non-neural tissue, implying that Brinps are not essential for organogenesis or neurogenesis. The normal gross anatomy of the Brinp1/2/3−/− mice is surprising given the high expression of Brinps in overlapping regions during murine neural development (Kawano et al., 2004). A greater level of histopathological analysis, using layer specific markers, is needed to determine whether Brinp1/2/3−/− mice have altered neural architecture.
The reduction in body weight of Brinp1/2/3−/− mice may reflect the observed phenotype of Brinp1−/− mice. Male Brinp1/2/3−/− mice especially are significantly smaller from even the Brinp1−/− mice (Berkowicz et al., 2016). This may be the additive effect of the reduced body weight of Brinp1−/− and Brinp2−/− mice, both of which weigh less than WT as adults.
Concluding Remarks
This study is a first step in understanding the effect of Brinp2 and Brinp3 genes on cognitive function. The observed changes in Brinp3−/− anxiety response, and increased Brinp2−/− locomotor activity may be relevant to the genetic variation at the 1q25.2 locus associated with patients diagnosed with ADHD, anxiety and other NDDs. Brinp2−/− and Brinp3−/− genes do not appear to compensate in the mammalian brain and are therefore likely have distinct molecular or cell specific functions.
Author Contributions
PB and JW conceived the study. SB and PB designed and co-ordinated the study. SB carried out knock-out mouse validation, mouse reproductive and weight analysis, statistical analysis, compiled the figures. SB drafted the manuscript with revision by PB. TF performed behavioral testing of mice. All authors reviewed and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. Jeanette Rientjes, Arianna Nenci, and Jose Gonzalez (Monash Gene Targeting Facility) for contract production of the Brinp2tm1/Pib mice and Brinp3tm1/Pib mice and Southern Blot validation. Behavioral testing was carried out by TF, Neuroscience Research Services. Thank you to Dr. Emma Burrows and Professor Anthony Hannan for advice on behavioral experiments. Necroscopies were performed by the Australian Phenomics Network (APN). JW and PB groups were funded by NHMRC Program grant 490900. JW is a National Health and Medical Research Council of Australia Senior Principal Research Fellow. JW also acknowledges the support of an Australian Research Council Federation Fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fnbeh.2016.00196
Supplementary Figure 1. Reduced litter survival when breeding Triple-het and Triple-het × Brinp1−/+2−/−3−/− mice. Breeders were monitored for litter size at birth and litter size at age of weaning (P21). (A) No significant differences in number of pups per litter at postnatal day 0, from WT/WT, Triple-Het/Triple-Het and Triple-Het ♀/Brinp1−/+2−/−3−/− ♂ parents, F(2, 35) = 2.702, p = 0.0811, one-way ANOVA. (B) The combined Brinp1, Brinp2, and Brinp3 deleted allele of breeders impacted the number of pups weaned at postnatal day 21, from WT/WT, Triple-Het/ Triple-Het and Triple-Het ♀/Brinp1−/+2−/−3−/− ♂ parents. F(2, 35) = 7.142, p<0.0025, one-way ANOVA. Tukey HSD multiple comparisons tests showed significant differences: WT × WT and Triple-Het × Triple-Het: p = 0.842, WT × WT and Triple-Het × Brinp1−/+2−/−3−/−: p = 0.0025, Triple-Het × Triple-Het and Triple-Het × Brinp1−/+2−/−3−/−: p = 0.027.*p < 0.05, **p < 0.001, N = 3 breeding pairs per genotype, 10–14 litters per genotype.
Supplementary Figure 2. Vision and Olfaction. (A) Normal vision for Brinp2−/−, Brinp3−/− and Brinp2/3−/− mice: (i) Brinp2−/− Vision test female: t(10) = 0.674, p = 0.516 male: t(10) = 0.277, p = 0.787, Student's t-test. (ii) Brinp3 Vision test female: t(8) = 1.265, p = 0.242 male: t(12) = 0.000, p > 0.999, Student's t-test. (iii) Brinp2/3 Vision test female: t(9) = 0.302, p = 0.770 male: t(10) = 0.568, p = 0.583, Student's t-test. (B) Normal olfaction for Brinp2−/− and Brinp3−/− mice: (i) Brinp2 Olfaction female: F(1, 10) = 1.773, p = 0.213, male: F(1, 10) = 0.081, p = 0.781, repeat measures two-way ANOVA. (ii) Brinp3 Olfaction female: F(1, 8) = 1.513, p = 0.254, male: F(1, 12) = 0.538, p = 0.477, repeat measures two-way ANOVA. Data presented as the mean ± SD.
Supplementary Figure 3. Rotarod. (A) Female Brinp2−/− mice show significant improvement in latency to fall on the Rotarod; female: F(1, 10) = 10.464, p = 0.009, male: F(1, 10) = 0.769, p = 0.401, repeat measures two-way ANOVA. N = 6 female, 6 male mice per genotype. (B) Brinp3−/− mice do not show significant motor co-ordination impairment on the Rotarod; female: F(1, 8) = 1.337, p = 0.281, male F(1, 12) = 1.483, p = 0.247, repeat measures two-way ANOVA. N = 5 female, 7 male mice per genotype. (C) Brinp2/3−/− mice do not show significant motor co-ordination impairment on the Rotarod; female: F(1, 8) = 1.560, p = 0.247, male F(1, 10) = 0.087, p = 0.774, repeat measures two-way ANOVA. N = 6 female, 6 male mice per genotype. Data presented as the mean ± SD.
Supplementary Figure 4. Startle and Pre-pulse Inhibition (PPI). (A) Normal startle response for Brinp2, Brinp3, and Brinp2/3 mice: (i) Brinp2−/− startle female: t(9) = 0.1178, p = 0.908, male: t(10) = 0.4818, p = 0.640, Student's t-test. (ii) Brinp3−/− startle female: t(8) = 0.6158, p = 0.555, male: t(12) = 0.2630, p = 0.797, Student's t-test. (iii) Brinp2/3−/− startle female: t(9) = 0.7216, p = 0.489, male: t(10) = 0.2195, p = 0.831, Student's t-test. (B) Normal Pre Pulse Inhibition (PPI) for Brinp2−/−, Brinp3−/−, and Brinp2/3−/− mice: (i) Brinp2−/− PPI female: F(1, 10) = 3.551, p = 0.089, male: F(1, 10) = 0.783, p = 0.397, repeat measures two-way ANOVA. (ii) Brinp3−/− PPI female: F(1, 8) = 0.276, p = 0.614, male: F(1, 12) = 0.056, p = 0.817, repeat measures two-way ANOVA. (iii) Brinp2/3−/− PPI female: F(1, 9) = 0.056, p = 0.818, male: F(1, 10) = 0.133, p = 0.753, repeat measures two-way ANOVA. Data presented as the mean ± SD.
Supplementary Figure 5. Habituation trial of three-chamber social interaction test (Trial 1). (A) Habituation trial of the three chamber social interaction test, showing interaction time between empty cages. No significant preference between the left/right chambers for Brinp2−/−, Brinp3−/−, or Brinp2/3−/− mice. (i) Brinp2−/− SI Test Trial 1: female: F(3, 23 = 0.371, p = 0.775, male: F(3, 23) = 0.119, p = 0.948, one-way ANOVA, N = 6 females, 6 males per genotype. (ii) Brinp3−/− SI Test Trial 1: female: F(3, 19) = 0.399, p = 0.756, male: F(3, 27) = 1.144, p = 0.352, one-way ANOVA, N = 5 females, 7 males per genotype. (iii) Brinp2/3−/− SI Test Trial 1: female: F(3, 23) = 0.621, p = 0.609, male: F(3, 21) = 0.086, p = 0.967, one-way ANOVA, N = 6 females, 6 males per genotype. (B) Distance Travelled Trial 1. (i) Brinp2−/− mice traveled a significantly increased distance over 10 min trial interval whilst habituating to the arena, indicating hyperactivity. Female: p = 0.0163, male: p = 0.0061, Student's t-test. (ii–iii) Brinp3−/− and Brinp2/3−/− showed normal activity (distance traveled) during the habituation trial. Brinp3−/− female: p = 0.5199, Brinp3−/− male: p = 0.6300, Brinp2/3−/− female: p = 0.5030, Brinp2/3−/− male: p = 0.4608, Student's t-test.
References
Angelakopoulou, A., Shah, T., Sofat, R., Shah, S., Berry, D. J., Cooper, J., et al. (2012). Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: cardiovascular biomarker genetics collaboration. Eur. Heart J. 33, 393–407. doi: 10.1093/eurheartj/ehr225
Asinof, S. K., and Paine, T. A. (2014). The 5-choice serial reaction time task: a task of attention and impulse control for rodents. J. Vis. Exp. 90:e51574. doi: 10.3791/51574
Berkowicz, S. R., Featherby, T. J., Qu, Z., Giousoh, A., Borg, N. A., Heng, J. I., et al. (2016). Brinp1−/− mice exhibit autism-like behaviour, altered memory, hyperactivity and increased parvalbumin-positive cortical interneuron density. Mol. Autism 7, 1–20. doi: 10.1186/s13229-016-0079-7
Bohuslavova, R., Dodd, N., Macova, I., Chumak, T., Horak, M., Syka, J., et al. (2016). Pax2-islet1 transgenic mice are hyperactive and have altered cerebellar foliation. Mol. Neurobiol. doi: 10.1007/s12035-016-9716-6. [Epub ahead of print].
Cha, J. D., Kim, H. J., and Cha, I. H. (2011). Genetic alterations in oral squamous cell carcinoma progression detected by combining array-based comparative genomic hybridization and multiplex ligation-dependent probe amplification. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 111, 594–607. doi: 10.1016/j.tripleo.2010.11.020
Connelly, J. J., Shah, S. H., Doss, J. F., Gadson, S., Nelson, S., Crosslin, D. R., et al. (2008). Genetic and functional association of FAM5C with myocardial infarction. BMC Med. Genet. 9:33. doi: 10.1186/1471-2350-9-33
Drgon, T., Johnson, C. A., Nino, M., Drgonova, J., Walther, D. M., and Uhl, G. R. (2011). “Replicated” genome wide association for dependence on illegal substances: genomic regions identified by overlapping clusters of nominally positive SNPs. Am. J. Med. Genet. B Neuropsychiatr. Genet. 156, 125–138. doi: 10.1002/ajmg.b.31143
Giousoh, A., Vaz, R., Bryson-Richardson, R. J., Whisstock, J. C., Verkade, H., and Bird, P. I. (2015). Bone morphogenetic protein/retinoic acid inducible neural-specific protein (brinp) expression during Danio rerio development. Gene Expr. Patterns 18, 37–43. doi: 10.1016/j.gep.2015.05.002
Goddard, A. W., Ball, S. G., Martinez, J., Robinson, M. J., Yang, C. R., Russell, J. M., et al. (2010). Current perspectives of the roles of the central norepinephrine system in anxiety and depression. Depress. Anxiety 27, 339–350. doi: 10.1002/da.20642
Graham, D. R., and Sidhu, A. (2010). Mice expressing the A53T mutant form of human alpha-synuclein exhibit hyperactivity and reduced anxiety-like behavior. J. Neurosci. Res. 88, 1777–1783. doi: 10.1002/jnr.22331
Higgins, G. A., and Breysse, N. (2008). Rodent model of attention: the 5-choice serial reaction time task. Curr. Protoc. Pharmacol. Chapter 5, Unit 5.49. doi: 10.1002/0471141755.ph0549s41
Hiller, M. M., Finger, A., Schweiger, M., and Wolf, D. H. (1996). ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725–1728.
Kalin, N. H., Shelton, S. E., and Davidson, R. J. (2004). The role of the central nucleus of the amygdala in mediating fear and anxiety in the primate. J. Neurosci. 24, 5506–5515. doi: 10.1523/JNEUROSCI.0292-04.2004
Karoly, H. C., Stevens, C. J., Magnan, R. E., Harlaar, N., Hutchison, K. E., and Bryan, A. D. (2012). Genetic influences on physiological and subjective responses to an aerobic exercise session among sedentary adults. J. Cancer Epidemiol. 2012:540563. doi: 10.1155/2012/540563
Kawano, H., Nakatani, T., Mori, T., Ueno, S., Fukaya, M., Abe, A., et al. (2004). Identification and characterization of novel developmentally regulated neural-specific proteins, BRINP family. Brain Res. Mol. Brain Res. 125, 60–75. doi: 10.1016/j.molbrainres.2004.04.001
Kobayashi, M., Nakatani, T., Koda, T., Matsumoto, K., Ozaki, R., Mochida, N., et al. (2014). Absence of BRINP1 in mice causes increase of hippocampal neurogenesis and behavioral alterations relevant to human psychiatric disorders. Mol. Brain 7:12. doi: 10.1186/1756-6606-7-12
Lesch, K. P., Timmesfeld, N., Renner, T. J., Halperin, R., Röser, C., Nguyen, T. T., et al. (2008). Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J. Neural Transm. 115, 1573–1585. doi: 10.1007/s00702-008-0119-3
Lionel, A. C., Tammimies, K., Vaags, A. K., Rosenfeld, J. A., Ahn, J. W., Merico, D., et al. (2014). Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum. Mol. Genet. 23, 2752–2768. doi: 10.1093/hmg/ddt669
Numata, S., Ye, T., Herman, M., and Lipska, B. K. (2014). DNA methylation changes in the postmortem dorsolateral prefrontal cortex of patients with schizophrenia. Front. Genet. 5:280. doi: 10.3389/fgene.2014.00280
Prather, M. D., Lavenex, P., Mauldin-Jourdain, M. L., Mason, W. A., Capitanio, J. P., Mendoza, S. P., et al. (2001). Increased social fear and decreased fear of objects in monkeys with neonatal amygdala lesions. Neuroscience 106, 653–658. doi: 10.1016/S0306-4522(01)00445-6
Romanos, M., Freitag, C., Jacob, C., Craig, D. W., Dempfle, A., Nguyen, T. T., et al. (2008). Genome-wide linkage analysis of ADHD using high-density SNP arrays: novel loci at 5q13.1 and 14q12. Mol. Psychiatry 13, 522–530. doi: 10.1038/mp.2008.12
Shorts-Cary, L., Xu, M., Ertel, J., Kleinschmidt-Demasters, B. K., Lillehei, K., Matsuoka, I., et al. (2007). Bone morphogenetic protein and retinoic acid-inducible neural specific protein-3 is expressed in gonadotrope cell pituitary adenomas and induces proliferation, migration, and invasion. Endocrinology 148, 967–975. doi: 10.1210/en.2006-0905
Silverman, J. L., Yang, M., Lord, C., and Crawley, J. N. (2010). Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 11, 490–502. doi: 10.1038/nrn2851
Teoh, S. S., Vieusseux, J., Prakash, M., Berkowicz, S., Luu, J., Bird, C. H., et al. (2014). Maspin is not required for embryonic development or tumour suppression. Nat. Commun. 5:3164. doi: 10.1038/ncomms4164
Terashima, M., Kobayashi, M., Motomiya, M., Inoue, N., Yoshida, T., Okano, H., et al. (2010). Analysis of the expression and function of BRINP family genes during neuronal differentiation in mouse embryonic stem cell-derived neural stem cells. J. Neurosci. Res. 88, 1387–1393. doi: 10.1002/jnr.22315
Verweij, K. J. H., Zietsch, B. P., Medland, S. E., Gordon, S. D., Benyamin, B., Nyholt, D. R., et al. (2010). A genome-wide association study of Cloninger's temperament scales: implications for the evolutionary genetics of personality. Biol. Psychol. 85, 306–317. doi: 10.1016/j.biopsycho.2010.07.018
Keywords: Brinp2, Brinp3, knock-out mice, ADHD, anxiety, neurodevelopmental disorders
Citation: Berkowicz SR, Featherby TJ, Whisstock JC and Bird PI (2016) Mice Lacking Brinp2 or Brinp3, or Both, Exhibit Behaviors Consistent with Neurodevelopmental Disorders. Front. Behav. Neurosci. 10:196. doi: 10.3389/fnbeh.2016.00196
Received: 26 July 2016; Accepted: 29 September 2016;
Published: 25 October 2016.
Edited by:
Allan V. Kalueff, St. Petersburg State University, RussiaReviewed by:
Gregg Stanwood, Florida State University, USAMichael Arthur Van Der Kooij, University of Mainz, Germany
Copyright © 2016 Berkowicz, Featherby, Whisstock and Bird. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susan R. Berkowicz, U3VzaWUuYmVya293aWN6QG1vbmFzaC5lZHU=
 Susan R. Berkowicz
Susan R. Berkowicz Travis J. Featherby2
Travis J. Featherby2