- 1Department of Orthopaedic Surgery, Thomas Jefferson University, Philadelphia, PA, United States
- 2Rothman Orthopaedic Institute, Philadelphia, PA, United States
The recent influx of machine learning centered investigations in the spine surgery literature has led to increased enthusiasm as to the prospect of using artificial intelligence to create clinical decision support tools, optimize postoperative outcomes, and improve technologies used in the operating room. However, the methodology underlying machine learning in spine research is often overlooked as the subject matter is quite novel and may be foreign to practicing spine surgeons. Improper application of machine learning is a significant bioethics challenge, given the potential consequences of over- or underestimating the results of such studies for clinical decision-making processes. Proper peer review of these publications requires a baseline familiarity of the language associated with machine learning, and how it differs from classical statistical analyses. This narrative review first introduces the overall field of machine learning and its role in artificial intelligence, and defines basic terminology. In addition, common modalities for applying machine learning, including classification and regression decision trees, support vector machines, and artificial neural networks are examined in the context of examples gathered from the spine literature. Lastly, the ethical challenges associated with adapting machine learning for research related to patient care, as well as future perspectives on the potential use of machine learning in spine surgery, are discussed specifically.
Introduction
In clinical medicine, the rise of machine learning applications represents a new era of solving healthcare problems. This is particularly true in spine surgery where algorithmic decision support tools, computer assisted navigation, and surgical robots are already being used in the clinic and operating room. While the appetite for machine learning and its role in artificial intelligence has grown amongst spine surgeons, very little discussion has revolved around how to evaluate these applications and their contributions to patient care. In 2019 alone, 82 publications (more than twice the previous year) were PubMed indexed when searching for the terms “machine,” “learning,” and “spine” together. A core component of proper peer-review requires familiarity with machine learning methodology among clinicians. Until this can be achieved, machine learning in the spine literature will either foster skepticism or flawed enthusiasm. The intricacies and real patient-safety concerns when dealing with the spine necessitates that clinicians familiarize themselves with the terminology and guiding principles of machine learning. This review will introduce the origins of the artificial intelligence field and provide an organic discussion on how to practically synthesize machine learning modalities in spine surgery. A glossary of key terms in this review can be referred to in Table 1.
A Brief History of Artificial Intelligence and Data Science
The study of artificial intelligence (AI) originated back in the summer of 1956 when Dr. John McCarthy and contemporaries gathered at Dartmouth College. They “proceeded on the basis of the conjecture that every aspect of learning or any other feature of intelligence could in principle be so precisely described that a machine could be made to simulate it (10, 11).” While this meeting of great minds was significant, progress within AI has been undulating, with great successes followed by even greater failures. Notwithstanding, the recent establishment of larger data sets (or Big Data) has enabled scientists to overcome previous obstacles. During the advent of popularized AI in the 1980's, ~1% of humankind's information was available digitally. Presently, digital information technology accounts for 99% of data, which is estimated to be 5 zettabytes (5 × 1021 bytes) (12, 13). This amount of information is greater than the sum total if one were to store genomes from every person on Earth (1 × 1019 bytes) (14). At an individual level, one can appreciate the abundance of data stored in the cloud and the expansion of stored memory on a smartphone. Over the last decade, the United States healthcare system has also benefited from the Health Information Technology for Economic and Clinical Health (HITECH) Act, which spurred the adoption of electronic medical records (15). Experts have speculated that society is rapidly approaching a point where the totality of data eclipses what can be extracted from nature itself (12). This massive amount of data has also been bolstered by large-scale commercialization of computing hardware, particularly graphics processing units or GPU (16). This increased accessibility of GPUs has allowed researchers to complete largescale machine learning tasks even at home, a feat unachievable in previous decades. Modern society is at a crossroads where we have access to inordinate amounts of data and hardware, but little guidance on how to extract meaningful information that is applicable to everyday life.
Overview of Machine Learning
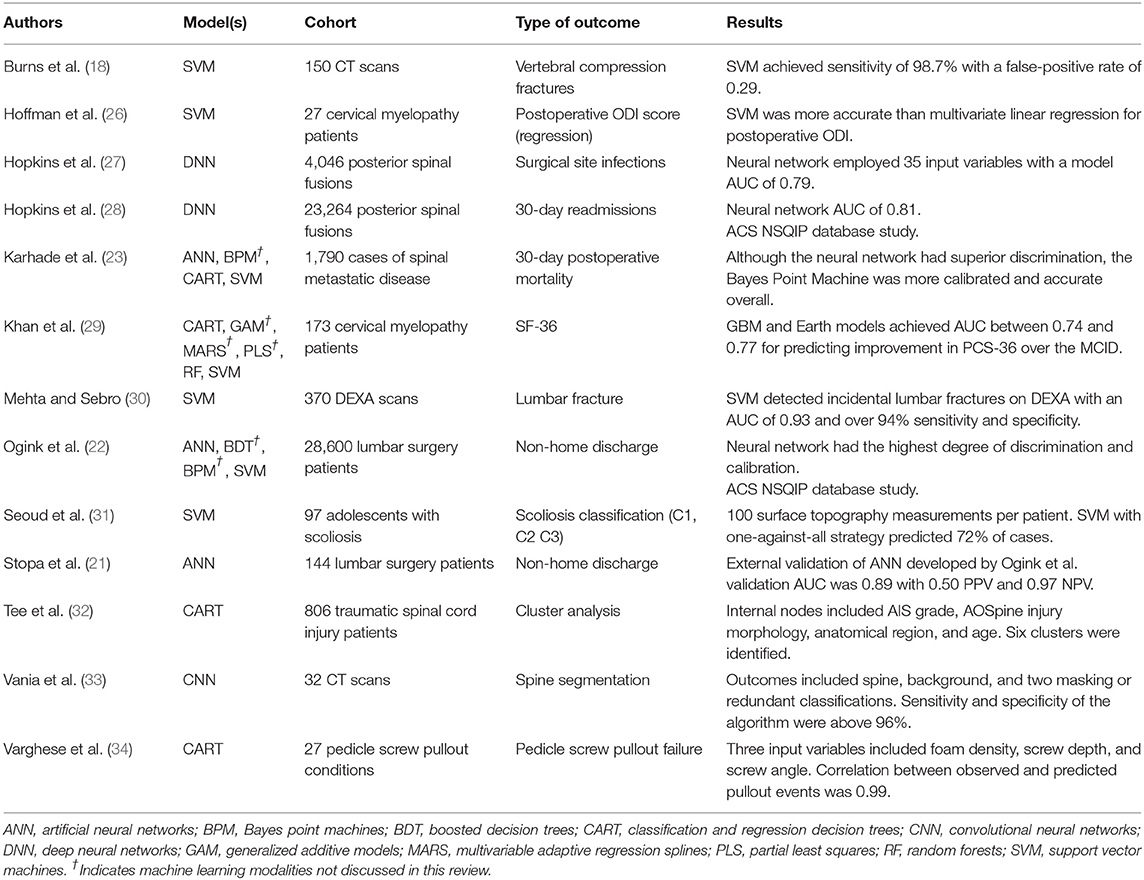
Machine Learning (ML) is a subset of AI that focuses on developing automated computer systems (learners) that predict outputs through algorithms and mathematics (7). The output represents the machine's interpretation of complex relationships that may be either linear or non-linear. Performance is graded according to its level of discrimination (probability of predicting outcomes accurately) and calibration (degree of over- or under-estimating the predicted vs. true outcome) (17). Examples of ML applications encountered by spine surgeons include image classification [i.e., automated detection of vertebral body compression fractures on CT or MRI (18–20)], preoperative risk stratification models, clinical decision support tools (21–25), among others. The purpose of this review is to define basic ML terminology, discuss the difference between ML and classical statistics, detail common ML models, and introduce examples in spine research. A summary of included references to machine learning applications in spine surgery and research are shown in Table 2.
Machine Learning Terminology
The two major forms of ML are supervised and unsupervised learning. Supervised learning entails labeled data based on a grounded truth (1, 5). For example, a database of lateral x-rays has films prelabeled as either “fracture” or “no fracture.” A portion of this data (training dataset) is analyzed to build a model that synthesizes the pattern between independent variables (i.e., pixel in an image) and dependent variables (presence or absence of pathology). Individual radiograph pixels in this example are known as feature values or vectors (1, 5). The remainder of the x-ray films (untrained dataset) are fed to the machine, which is then assessed based on its ability to accurately predict a fracture or otherwise. As such, supervised learning excels in exercises of linear classification (where the outputs are discretely defined categories) or linear regression (where the outputs are continuous values).
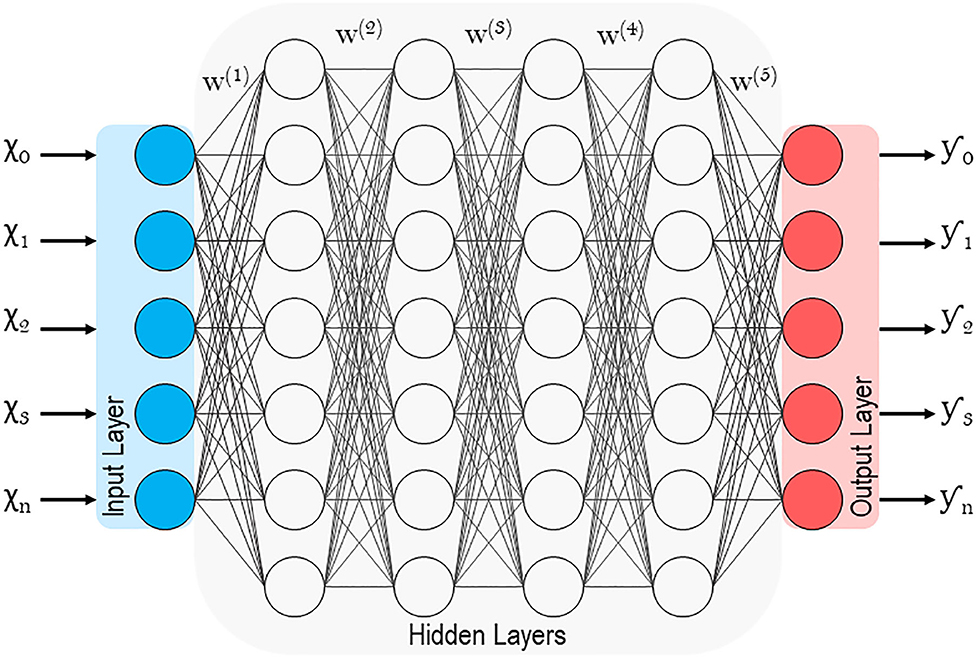
Unsupervised learning, on the other hand, involves the analysis of unlabeled datasets, and stems from neuropsychology research conducted by Dr. Donald Olding Hebb (1, 9). Hebbian theory describes the general framework (Figure 1) of neurons and their synapses, which enable humans and other animals to learn relationships and store memories (6). The proposition being that among the multitudes of neurons in the brain, it is the distinct synaptic connections between neurons and their repetitive firing that enable learning (6). Unsupervised machines (like humans) can appreciate non-linear relationships and do so without presumptions related to the data. Unsupervised learners are particularly adept at identifying clusters of related variables, detecting anomalies, and constructing artificial neural networks (detailed later) (1, 35). While unsupervised learning is thought to be the standard for the future, most current ML examples in spine surgery and clinical medicine are of the supervised variety.
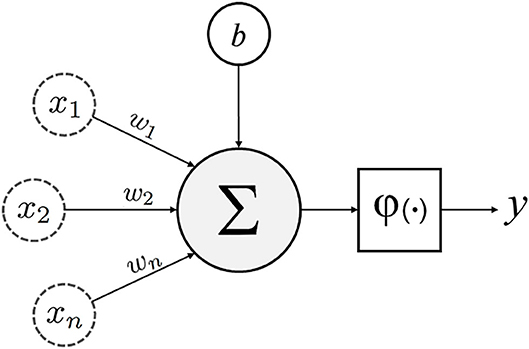
Figure 1. Schematic of an artificial neuron with inputs (x1, 2…n), weights (w1, 2…n), bias (b), transfer function (∑), activation function (φ) and output (y).
Machine Learning vs. Classical Statistics
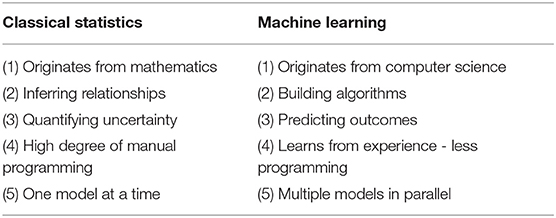
The delineation between machine learning and classical statistics is quite nebulous because learners are built upon statistical modeling (Table 3). Both modalities also rely on robust preprocessing of data that is representative of the general population. However, whereas statistics emerged from the field of mathematics, ML emerged from computer science. For purposes of simplification, the two concepts can be differentiated by the type of question needed to be answered. Classical statistics infers relationships between variables, while ML attempts to predict these relationships (36, 37). Inference (or statistics) involves testing the null vs. alternative hypothesis for an effect with a measurement of confidence. Prediction (or machine learning) involves forecasting outcomes without demanding as to why resultant relationships exist. It is also essential to highlight that while ML may appear to be more advanced than statistical analysis, neither is superior and both should be considered for predictive modeling.
To illustrate this further, a research question might ask, “What risk factors are associated with non-routine discharge after lumbar decompression and/or fusion?” In fact, multiple studies using classical statistics have already implicated that patients' age, diabetes status, cardiovascular comorbidities, functional status, among others, all contribute to non-routine discharge (38–40). And with the expertise from practicing physicians, we can reason and clarify these findings. But translating these results in a clinical setting is complex, because it is unclear how one weighs the importance of each variable when optimizing patients preoperatively. ML enables the development of tools that allow surgeons to plug-in variables and generate probabilities of a non-routine discharge. Ogink et al. recently developed learners to predict discharge to a rehabilitation or skilled nursing facility after surgery for lumbar stenosis using the American College of Surgeons National Surgical Quality Improvement Program (ACS-NSQIP) database (22). They built multiple models in parallel and ultimately arrived at a neural network that achieved high levels of discrimination and calibration with an Area Under the Curve (AUC) of 0.74 from a Receiver Operating Characteristics curve (22). This tool has since been externally validated in a smaller cohort, where 97% of patients were accurately predicted to return to home after elective lumbar surgery (21). Such algorithms warrant further independent validation, but they allow for synthesizing unwieldly large datasets in a practical way. Above all, the purpose of machine learning is performance based on indiscriminate analysis. But when practicing medicine, the ability of a learner to predict outcomes accurately must also take into consideration how and why it reaches such conclusions. This controversy of applying ML clinically is colloquially termed the black box, which will be discussed at the end of this review.
Popular Models for Machine Learning
With some basic ML terminology outlined, it is imperative that practicing physicians understand the architecture of learners encountered in peer-reviewed journals. Using examples from the spine literature, three ML modalities applicable to medicine will be discussed: (1) decision tree learning, (2) support vector machines, and (3) artificial neural networks. It is important to consider that while the following descriptions attempt to neatly categorize each model, they are flexible and can be adapted according to their needs. For example, support vector machines are often described as supervised models for linear classification, but there are many examples of them being used for unsupervised learning and non-linear classification exercises.
Decision Tree Learning
Decision tree learning, or more specifically, Classification and Regression Trees (CART) is one of the more straightforward modalities because it is better appreciated visually, rather than mathematically (3, 37, 41). By definition, a CART can analyze variables that are either categorical (classification) or continuous (regression). As shown in Figure 2, a CART is an upside-down tree with three major components (1) internal nodes, (2) branches, and (3) leaves (3, 41). Internal nodes are conditions by which the learner evaluates or measures variables. Branches are the decisions derived from each node. And leaves (or terminal nodes) represent ends of the tree where an output is finalized. The figure depicted is simplistic, and in a real-world application would only represent a branch of a much larger CART. But decision trees have a habit of becoming unnecessarily deep or involving too many layers of complexity. Trees with excessive internal nodes sub-divide data into too many small clusters, such that the outcomes are grouped in a way that are practically meaningless. A CART is a fundamentally greedy algorithm because it naturally satisfies the condition at each node, rather than optimizing conditions across the length of the tree (3). Pruning, as the name suggests, allows for incremental improvements in the tree by eliminating conditions that are less important. It is relevant here to discuss the concept of fit in both classical statistics and machine learning (42). An underfitting model has no utility because it poorly approximates potential relationships (Figure 5F). On the other hand, an overfitting model attempts to observe the smallest of associations making the model relevant only to the training dataset, and by consequence, poorly generalizable (Figure 5E) (42). In other words, overfitting learners pay too much attention to the noise in the dataset. Pruning and other adjustments are necessary to minimize overfitting and to limit the complexity of the tree, all the while optimizing accuracy.
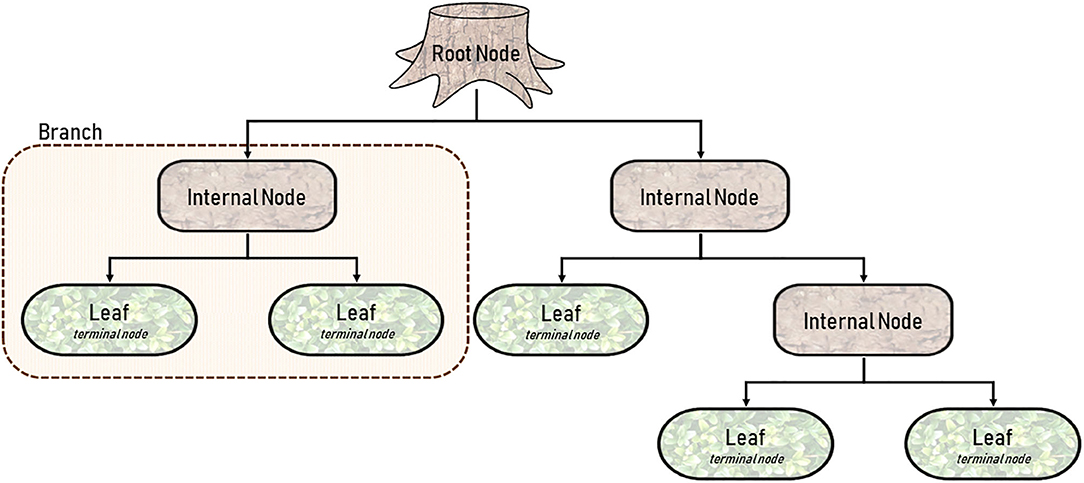
Figure 2. Diagram of a single Classification and Regression Tree (CART) with five terminal nodes or leaves.
Tee et al. application of decision tree learning for optimizing patient risk stratification after spinal cord injury provides a framework for understanding this modality (32). They combined different methods of assessing spinal cord function after trauma, including the American Spinal Injury Association (ASIA) Impairment Scale, total motor score (TMS) and the AOSpine classification system, to allow a decision tree to identify patient clusters that respond differently to treatment. As show in Figure 3, the cohort was first divided based on “ASIA grading (A-D)” (root node) and then evaluated at the first internal node, “AOSpine: A (compression), B (tension-band), or C (translational).” Interestingly, the learner concluded that it would be more worthwhile for the branches to keep A and B classifications together and C separate. The next internal node for each branch was binary, “cervical or thoracic injuries.” At this level in the tree, three leaves (nodes 4, 5, and 6) were finalized as these clusters were considered homogenous enough and not worth sub-dividing further. For example, node 5 represents AOSpine C injuries in the cervical region, whereas node 6 represents AOSpine C injuries in the thoracic region. Finally, the branch containing AOSpine A and B injuries in the cervical spine were passed through another internal node for “age,” generating another three leaves or clusters. The final six clusters are detailed in Table 4 (32). The results of this study provide a platform for external validation studies with other patient cohorts to compare this unique classification system with current ones. Tee et al. findings exemplify machine learning's ability to synthesize a multitude of variables that may associate non-linearly into a more easily digestible format. It is especially noteworthy that the investigators assembled a relatively large cohort of 806 patients for model building, a practice that is inconsistently applied in the spine literature.
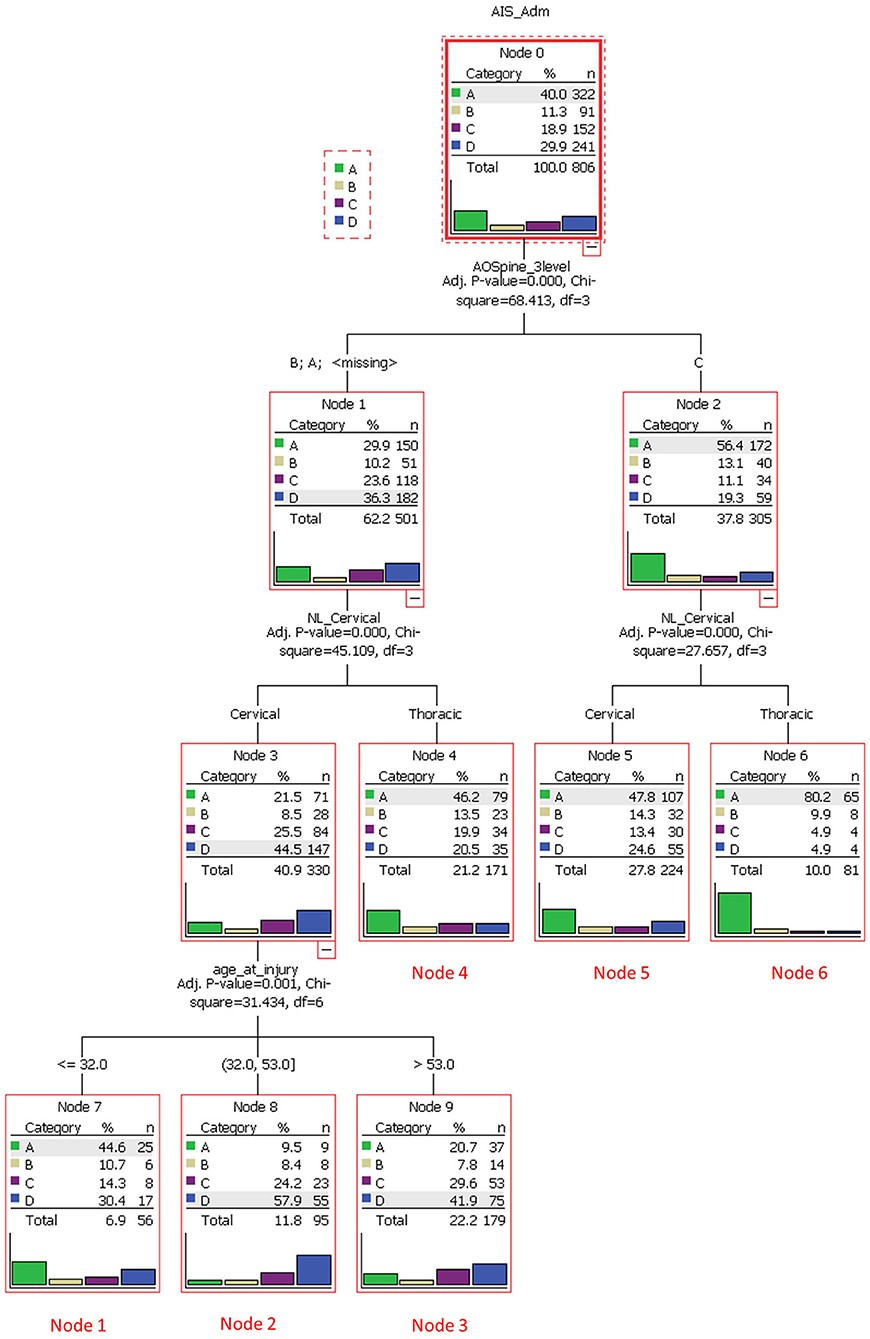
Figure 3. A decision tree analysis to stratify spinal cord injury cases and to identify clusters of homogeneous patients that would respond similarly to treatment. The root node was based on the American Spinal Injury Association Impairment Scale (AIS), which ranged from grade A through D. The subsequent internal node was based on AOSpine injury classification (class A/B or C). Each branch then underwent another node based on anatomical region (cervical or thoracic). Class A/B cervical injuries were divided further based on age. Six unique terminal nodes or clusters were identified. Reproduced with permission by Tee et al. (32).
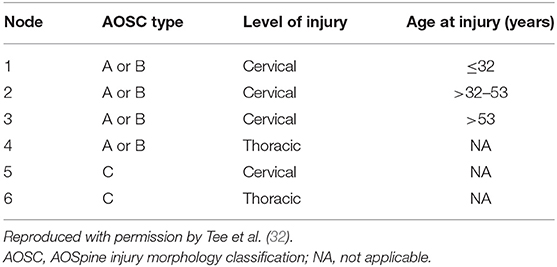
Table 4. Final cluster analysis of spinal cord injury classifications based on decision tree learning.
The need for substantial patient datasets in spine surgery is particularly noticeable when exploring ML applications for predicting patient-reported outcomes. Exploratory investigations using decision tree learning have been pursued in spine research. Khan et al. utilized seven different supervised learners to predict improvement in SF-36 (PCS/MCS) scores after surgery for degenerative cervical myelopathy (29). The architecture of their model included multiple comorbidities, physical exam findings, imaging, baseline characteristics, among others. They set the minimal clinically important difference or MCID at +4.0 points for both PCS and MCS components of the SF-36. All seven learners were similarly accurate for predicting MCS improvement postoperatively, including their CART with an AUC of 0.74. However, no learner was particularly better than logistic regression (AUC 0.71), and the performance of the PCS model was by comparison poor. Moving forward, it is likely that the spine literature will be inundated with publications running multiple statistical and ML models in parallel for comparative analysis. And while Khan et al. pilot investigation provides a framework for understanding machine learning, their sample size (130 training, 43 testing) leaves some concern as to the generalizability of the findings. The relationship between the natural history of spinal pathology, surgical interventions, and postoperative outcomes is delicate; and the proper use of ML for describing these relationships will require a multicenter and multidisciplinary effort to coalesce massive patient databases.
Lastly, decision tree learning can also help with characterizing medical device performance. Varghese and colleagues, using their own pedicle screw pullout strength protocol, showed that ML could be used to synthesize problems that have a large number of input permutations (34, 43). Their investigation involved the use of differing foam densities to mimic normal, osteoporotic, and extremely osteoporotic bone (Figure 4A). An actuator apparatus would then insert pedicle screws into the foam at three insertion angles, and three insertion depths (Figure 4B) (43). In total, 27 (33) permutations of these variables were analyzed using four separate models to determine pullout failure (<650 Newtons of force) or success (≥650 Newtons of force) (Figure 4C). Varghese et al. produced a promising model with very low error rates and an AUC of 1.00 for predicting pedicle screw failure, which was internally validated against a separate set of novel permutations (i.e., different pedicle screw insertion angles and foam densities) (Figure 6) (34). Their best learner was actually a random forest regression, which like a CART, is a subtype of decision tree analysis (41). As the name suggests, random forests sample random batches of the data, form multiple trees, and then combine the findings to construct a singular tree. Random forests minimize overfitting and other biases by employing the Law of Large Numbers, such that the average of multiple trees is more accurate than a single tree. The final decision tree constructed for pedicle screw pullout failure is shown in Figure 4E.
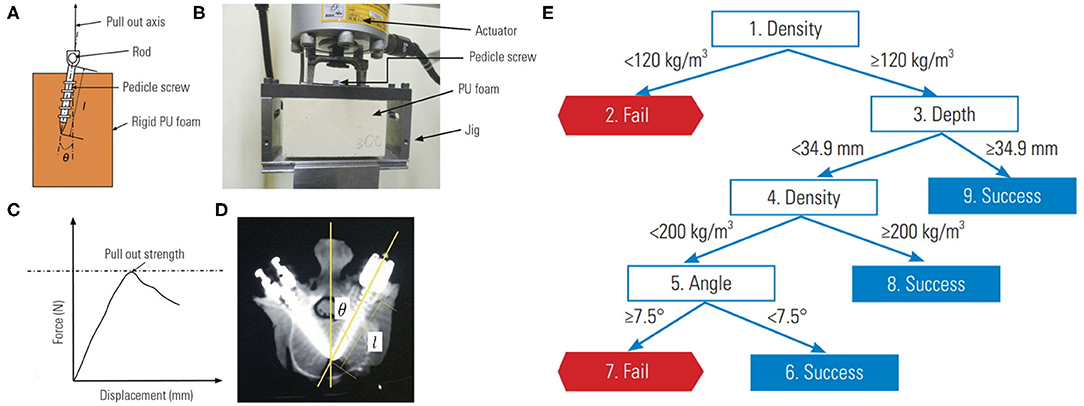
Figure 4. Biomechanical model for testing pedicle screw pullout-strength. (A) Schematic of rigid polyurethane foam to mimic normal, osteoporotic, and extremely osteoporotic bone. (B) Apparatus to test pedicle screw pull-out. (C) Force vs. displacement graph from pullout studies. (D) Anatomy of pedicle screw instrumentation. (E) Decision tree learning to predict pedicle screw pullout success vs. failure in relation to foam density, screw depth and insertion angle. Reproduced with permission by Varghese et al. (34).
Support Vector Machines
Support vector machines (SVMs) are also a commonly encountered ML modality in clinical literature. SVMs are intuitive and best appreciated graphically as shown in Figure 5. Although comparable to CARTs in exercises of linear classification or regression, SVMs accomplish such goals by constructing a hyperplane (8, 44). For a classification exercise, the hyperplane represents a line (or plane) that maximizes the distance between two categorical outcomes, which is also known as a maximum-margin hyperplane (Figure 5A). But as one can appreciate in Figure 5B, not all two-dimensional representations of data (only “x” and “y” coordinates) can be separated linearly with a hyperplane in that same dimension. Often, mathematical transformations or kernel functions are needed to transform the data into a higher dimension (44). As shown in Figure 5C, the same dataset plotted in three dimensions (3D) can be easily separated by a hyperplane. This transformation is prototypical and involves the inclusion of a “z” coordinate that equates the product of x and y, such that each outcome is plotted in 3D as (x,y,z) or (x,y,x*y). This is also known as a linear kernel. The byproduct of an SVM for an otherwise linearly inseparable dataset is shown in Figure 5D, where higher dimensional hyperplanes are represented as a circle in lower dimensions. However, like pruning, kernels can be overly extrapolated leading to overfitting and generating sub-clusters of outcomes that are incidental and practically meaningless (Figure 5E).
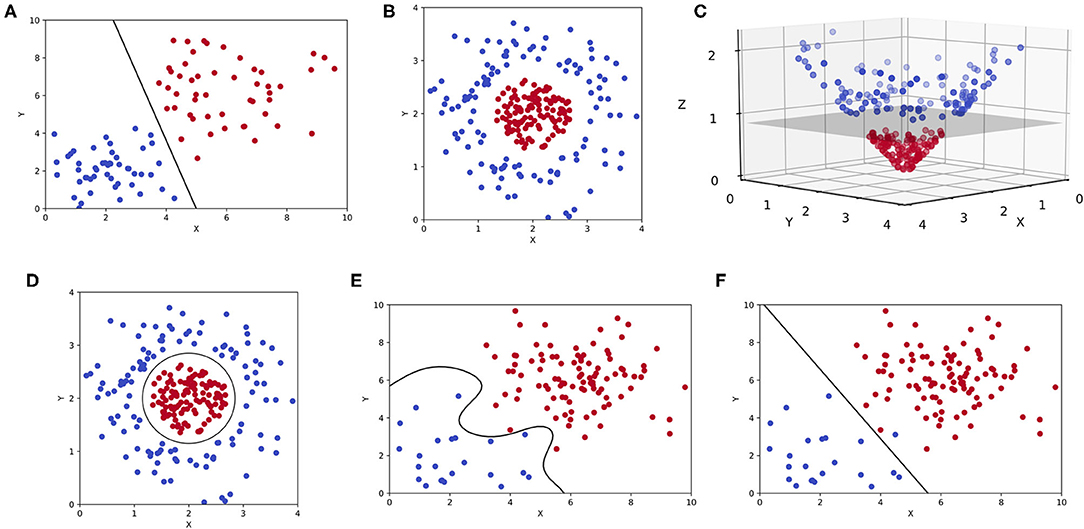
Figure 5. Support vector machines (SVMs). (A) Two-dimensional representation of a binary classification problem in an SVM represented by two features (x, y) with a maximum-margin hyperplane. (B) The same binary classification problem as before, but the outcomes are linearly inseparable in two-dimensions. (C) A linear kernel is used to transform the previous plot, allowing for a hyperplane to be constructed in three-dimensions (x, y, z). (D) A two-dimensional representation of the same hyperplane created in higher dimensions. (E) An overfitting SVM that is being influenced by outlier data or noise. (F) An underfitting SVM that fails to maximize the distance between two outcomes in a binary classification task.
Delving into the literature, SVMs are popular for classifying and detecting the presence of spine pathology on imaging. For example, a common problem when managing patients with osteoporosis arises from missed fractures on routine DEXA (Dual-Energy X-Ray Absorptiometry) scans (30). Given separate management guidelines for osteoporotic patients with and without fractures, Mehta and Sebro developed a model to detect incidental lumbar spine fractures from a large cohort of routine DEXA scans (30). The two outcomes or classifiers were “control” and “fracture.” The input variables to characterize the model included baseline demographics and ancillary data from the DEXA scan (i.e., bone mineral density, Z-scores, T-scores, among others). They conducted four SVMs in parallel, using different types of kernel functions, but ultimately arrived at a linear kernel with a high AUC of 0.93 against the training set, and an AUC of 0.90 against the test set (30). Their investigation exemplifies the potential of ML for automated detection of pathology. Such innovation can minimize missed diagnoses that are critical to quality care, especially in this case for incidental lumbar fractures on routine DEXA, where the error rate has been reported to be as high as 15.8% (45).
Another example of an image classification task achieved through SVMs was conducted by Seoud et al. The investigators attempted to determine scoliosis curve based on a modified Lenke classification system (C1, C2, or C3) for adolescents by analyzing surface topography data captured by multiple cameras (31). As a learning point, this is an example of applying SVMs with 3 outcomes (or classifications) instead of two. Seoud and colleagues addressed this problem by opting for a “one-against-all” approach, where the model compares C1 scoliosis curves to C2/C3 curves (46). And as discussed previously, the learner finds the ideal dimension where the outcomes can be linearly separated with the largest margin of distance between points. In this example, an overfitting model would be one where the SVM describes sub-clusters of scoliosis classifications that are clinically irrelevant. Seoud et al. model for classification based on topography alone accurately predicted over 72% of cases (31).
In addition to image classification tasks, SVMs have also been applied for predicting outcomes after spine surgery. Hoffman et al. prospectively evaluated patients undergoing surgery for degenerative cervical myelopathy, and attempted to predict postoperative outcomes including Oswestry Disability Index (ODI), modified Japanese Orthopedic Association scale (mJOA), and handgrip pressure (26). Their model illustrated how SVMs can also be used for regression. In contrast to classification, support vector regressions involve hyperplanes that minimize the distance between variables because the goal is to predict a continuous variable rather than a discrete one. Hoffman and colleagues also constrained the model to three input variables in order to curtail overfitting, which included preoperative ODI, symptom duration, and handgrip pressure. When compared to a traditional multiple linear regression, they achieved a higher goodness-of-fit or R2 of 0.93 via the SVM (26). While the prospective study design was a strength, the cohort was limited to only 20 patients. Herein lies the perpetual conflict between statistical power and generalizability when using ML. Models for predicting risk necessitate prospective data, but the feasibility of large datasets is limited to national databases, which are likely heterogeneous and retrospective.
Overall, SVMs are well-suited for general purpose machine learning (particularly in medicine) because tuning kernels allows for clinicians to assign appropriate weights according to their knowledge in that field (8). SVMs are also excellent tools for problems dealing with high dimensional data where the number of features far exceeds the number of observations or samples (47). Common examples of high dimensional data in clinical medicine include baseline demographics, preoperative risk factors, or gene expression levels. However, if the separation between two outcomes is unclear within a reasonable number of dimensions, SVMs struggle. And because SVMs are overly reliant on finely tuned kernels, the resultant models are only applicable to solving single problems (i.e., tools for predicting outcomes for cervical vs. lumbar surgery have to be separately and manually engineered). Counterintuitive to what has been discussed, SVMs are not proficient with very large data sets where the number of observations far exceed features (opposite of high dimensional data). As the number of points or samples increase, so does the noise, generating far too many outliers above and below the hyperplane (48).
Artificial Neural Networks
Lastly, Artificial neural networks (ANNs) are of particular interest because they are associated with deep learning, which has been traditionally unsupervised (1, 49, 50). Supervised models, as discussed previously, involve feature values that are highly discriminatory because they have been meticulously engineered with intricate knowledge of the subject matter (in this case spine surgery) (48). Deep learning circumvents this through representation learning, where the learner automatically classifies raw unlabeled data (51). With minimal human engineering, these unsupervised learners generate highly discriminatory feature extractors that characterize the input-output relationship, while ignoring irrelevant variations. Like in Figure 6, ANNs extrapolate the single neuron construct in Hebbian learning into an entire network where hidden layers or intermediate representations help refine the network of input-output synapses between artificial neurons (1). For a more technical and in-depth review of deep learning and ANNs please refer to the work by Emmert-Streib et al. (52)
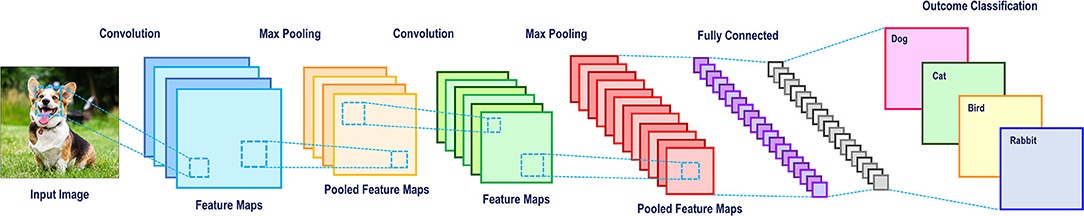
ANNs are adept in computer vision (53–56), natural language processing (51, 57), and predicting downstream effects of genetic mutations (58–60). Computer vision is of interest to spine surgeons as it may potentially increase the efficiency and accuracy of reporting patient imaging. The classical computer vision task is identifying a “dog” in a photo (Figure 9). Manually extracting features is near impossible because no two photos of dogs are the same. Practically, humans recognize dogs in photos despite variations in their pose, environment, lighting or orientation of the photo, among others. However, machines can only interpret pixels in an image, none of which are specific to a dog. Convolutional neural networks (CNNs), with the help of multiple hidden layers, are particularly adept at computer vision tasks and can be visualized graphically in Figure 7. The first hidden layer in a CNN convolves or filters the native input, extracting the “important” information and generates a feature map (a representation of the input). Subsequent max-pooling reduces complexity and minimizes overfitting by creating a more abstract form of the previous feature map and thus more applicable to generic pictures of dogs. This process can be repeated for the desired number of hidden layers. Once all feature maps have been considered, the images are flattened and the desired output (dog or otherwise) can be generated. In many ways, CNNs are more so learning to identify small arrangements or motifs that resemble dogs. This concept is known as local connectivity, meaning two neighboring pixels are considered more relevant than two distant pixels (61). Interestingly, CNNs structurally resemble the hierarchy and pathway used by the human visual cortex found in the occipital lobe (62). Multilayer neural networks like these are also essential for the development of fully automated robots and self-driving automobiles (57).
In spine surgery, computer vision technology has risen in parallel with the use of computer assisted navigation, robotic surgery, and augmented reality in the operating room, all of which require high fidelity 3D reconstructions of the spinal column from computed tomography or magnetic resonance imaging scans (33, 63–67). This is achieved through automated segmentation and detection of vertebrae via ANNs. Vania et al. recently reported the results of their CNN for automated vertebral column segmentation with a unique classification system (33). Instead of the traditional classifiers of “vertebrae” vs. “not vertebrae,” they implemented four classifications (background, spine, and two redundant classifiers) as show in Figure 8 (33). They did this in order to minimize overfitting so that the learner could consider variabilities in vertebral width and length outside of the training dataset. Their model generated a sensitivity and specificity of 0.97 and 0.99, respectively, both of which were either better or comparable to other commonly applied methods (33). In addition to spinal segmentation, significant strides have also been made in automated detection of vertebral compression and posterior element fractures, as well as the grading of lumbar stenosis (18–20, 54). The potential for successful translation for preoperative and intraoperative care is promising in spine surgery. For example, automation would allow for consistent application of sagittal deformity parameters by minimizing manual measurements and displaying associated risk factors all in one software ecosystem.
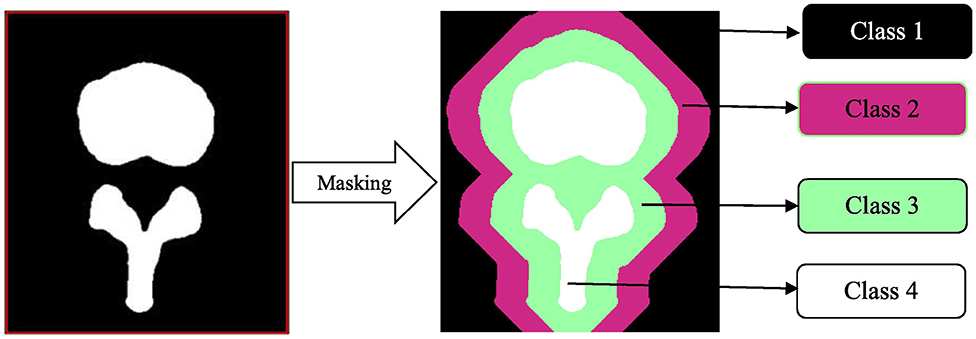
Figure 8. Masking in a convolutional neural network. This automated vertebral column segmentation model incorporates masking to generate two redundant classifiers for a traditionally binary classification task. Class 1 represents the background. Class 2 and 3 are unique redundant classifiers. Class 4 is the spine. By doing this, the machine becomes more adept at identifying varieties of vertebra, instead of the background. Reproduced with permission by Vania et al. (33).
While supervised learners, including CARTs and SVMs, have been used to predict postoperative outcomes, there is evidence to suggest that ANNs may be the preferred method for such tasks going forward (22, 23, 27, 28, 68). Kim et al. utilized an ANN to predict cardiac and wound complications, venous thromboembolism (VTE), and mortality rates after posterior lumbar fusion from an ACS-NSQIP cohort (68). Their learner was rather informative because they addressed the problem of low complication incidence by applying ADASYN (adaptive synthetic sampling approach to imbalanced learning). As shown in Figure 9, ADASYN generates multiple synthetic cohorts with positive complications that can be compared with controls, essentially creating multiple ANNs with different weights. The final ANN achieved an AUC of 0.71 for predicting cardiac complications postoperatively, which was superior to both logistic regression and American Society of Anesthesiologists (ASA) score (68). However, the regression model proved to be superior to the ANN for predicting VTE, mortality, and wound complications. In another investigation, Hopkins et al. applied an ANN with 35 input variables on over 4,000 cases of posterior spinal fusions to predict surgical site infections (27). Their model reliably predicted both infected and non-infected cases with an AUC of 0.79 across all their neural network iterations. However, the model unexpectedly demonstrated that intensive care unit admission and increasing Charlson Comorbidity Score were protective against surgical site infections, both findings of which are contradictory to the literature (27). The inability to interpret what seems like inconsistent findings is a key dilemma when applying ML in clinical medicine. Though, it is possible that such associations exist in a non-linear fashion that cannot be appreciated intuitively. And while surgeon's acumen and experience must be integrated with decision support tools, there is still significant deficits in these models before they can be safely (and without hesitancy) applied when patient lives are at stake.
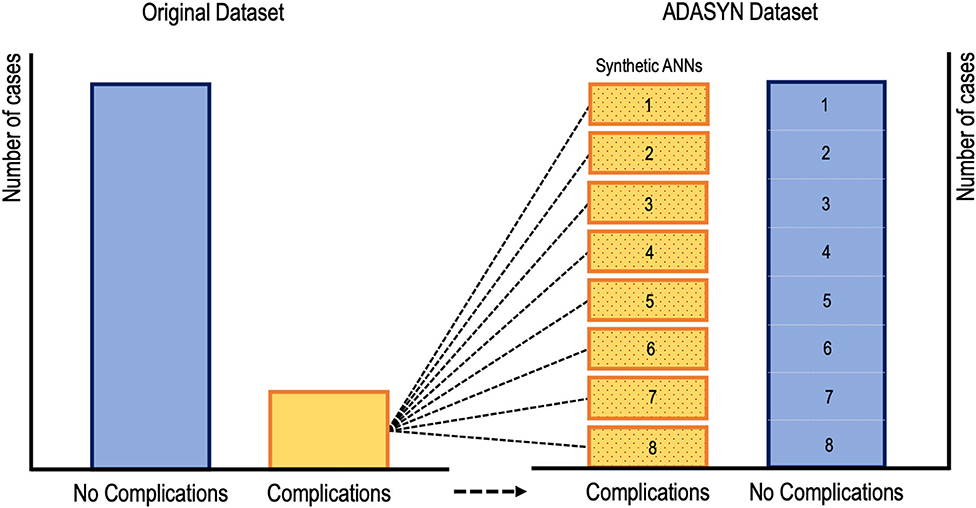
Figure 9. Visual representation of oversampling low incidence complications via adaptive synthetic sampling approach to imbalanced learning or ADASYN. Oversampled synthetic neural networks are created and then compared to subsets of the “no complication” cohort (68).
Future Perspectives on Machine Learning and Spine Surgery
Machine learning and artificial intelligence are progressively becoming more commonplace in modern society. We all in some ways either actively or passively contribute to Big Data through the use of smartphones, online shopping, wearables, among other activities even unbeknown to us. Moreover, the average physician is even more “plugged-in” to the modern technological ecosystem, given the use of electronic medical records, decision support tools, and imaging software. In spine surgery specifically, the nature of dealing with vital anatomic structures in the operating room instills an eagerness for innovations that might balance operative efficiency, patient safety, and surgical outcomes. Machine learning is at the core of AI advancement in healthcare and there are definite reasons for optimism.
As discussed previously, machine learning applications for computer vision will continue to optimize computer assisted navigation systems used by spine surgeons. AI implementation in the operating room has begun to transcend beyond what was previously possible through the use of augmented or mixed reality (69–72). Nguyen et al. in a trial of augmented reality for pedicle screw insertion with navigation, designed a virtual road map that was superimposed on the surgical site of patients undergoing spinal fusion (69). Their intention was to address the underlying obstacle of surgeons memorizing optimal screw trajectory provided by navigation, which is typically displayed away from the surgical site. By installing two overhead stereoscopic cameras, they coordinated intraoperative video with data sourced from the navigation's infrared tracking system. A representation of their innovative design is shown in Figure 10 (69). While they did not attempt to display their augmented reality system through a headset, other investigators have undertaken pilot studies as proof of concept with devices such as the Microsoft HoloLens (71–75). In the shoulder arthroplasty literature, Gregory et al. presented a proof of concept study using the Hololens to superimpose a 3D hologram of a patient's scapula in real time during a shoulder replacement (74). This application of mixed reality in the operating room was impressive because the headset did not need to be synced to a navigation system, the hologram could be adjusted in space, and the surgeon's point-of-view could be teleconferenced to others (Figure 11). Looking forward, these innovations in computer vision for the spine may also pave the way for significant improvements for surgical robots. Spine surgery robots presently appear rudimentary when compared to those utilized for minimally invasive gastrointestinal, urologic, and gynecologic surgeries. And while there is little reported on even a semi-automated robot for the spine, machine learning advancements may change this trajectory as it has for self-driving cars. However, spine surgeons (for patient safety concerns) may purposefully interact with robots in a slave-and-master paradigm in order to maintain total control over the machine. Using the five levels of autonomy described by the Society of Automotive Engineers, ranging from “no” to “full” automation, experts have postulated that clinical medicine may only ever incorporate up to “conditional” automation, where the machine both drives and monitors the circumstances, but humans are available for backup (9, 76).
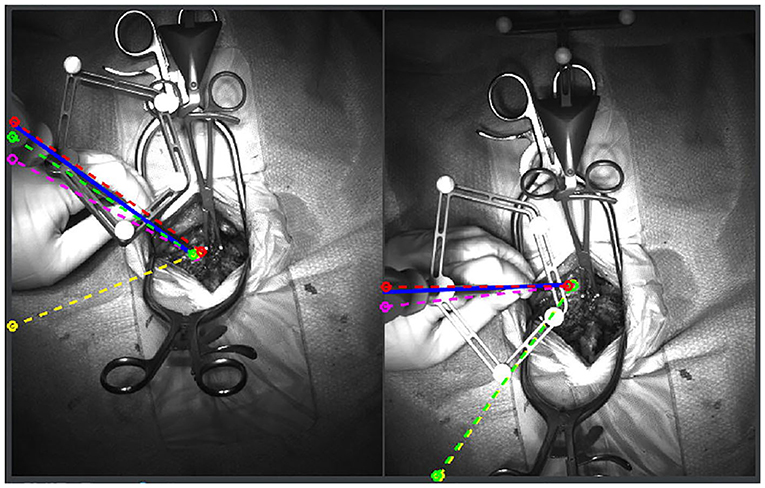
Figure 10. Augmented reality system that superimposes pedicle screw trajectories from computer assisted navigation onto the operating field. By minimizing the need to memorize trajectories from a separate screen, the surgeon is more readily able to identify safe zones. The blue, red, pink, yellow, and green lines represent correct, medial, lateral, superior and inferior breaches, respectively. Reproduced with permission by Nguyen et al. (69).
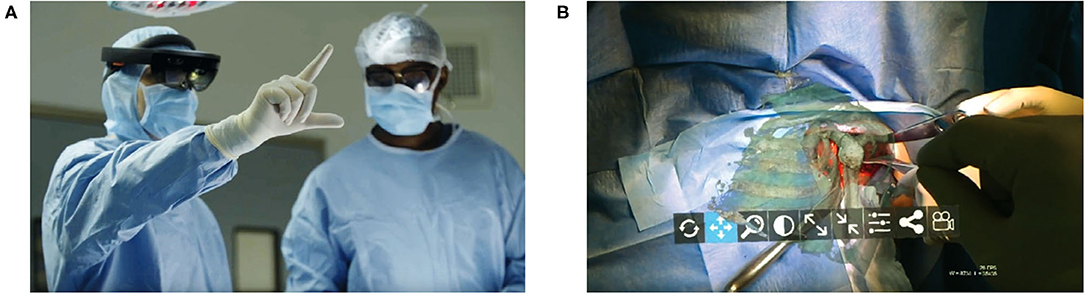
Figure 11. A proof of concept application of Microsoft Hololens for reverse total shoulder arthroplasty. (A) The surgeon is able to view in real-time and place in space a 3D hologram from a CT of the patient's scapula. (B) A 3D hologram of the patient's scapula is superimposed intraoperatively in order to fully visualize the glenoid and other relevant anatomy. Reproduced with permission by Gregory et al. (74).
Finally, as foreshadowed in the Overview of Machine Learning section, a major component of artificial intelligence research involves the ethical challenges of implementing machine learning for clinical practice (2, 4, 77–79). This has colloquially been termed the black box, which is the near impossible task of interpreting or explaining as to how a learner reaches the conclusions that it does, no matter how accurate it is (2, 4). And though the black box is typically attributed to ANNs and deep learning, it is also problematic for supervised learning. If a machine is learning non-linear associations in a manner that is hidden from both the engineer and the consumer, there will undoubtedly be apprehension toward the safety of an otherwise promising tool. As described by Dr. Alex John London, a professor of philosophy and artificial intelligence at Carnegie Mellon University, “the most powerful machine learning techniques seem woefully incomplete because they are atheoretical, associanist, and opaque.” As mentioned earlier in the study by Hopkins et al. for predicting surgical site infections, their neural network operated according to associations that oppose what spine surgeons consider grounded truths (27). To characterize this further, Caruana and colleagues published an infamous and equally informative machine learning model for predicting mortality after inpatient admission for pneumonia. While their learner was accurate, it reasoned that asthmatic patients with pneumonia should receive less aggressive care because on average they do better than non-asthmatics with pneumonia (80). This suggested course of action was in direct opposition to modern management guidelines for asthmatics, who are regularly provided the most aggressive care. However, Caruana et al. learner was not attuned to such contextual guidelines. Thus, from a prediction standpoint, asthmatics with pneumonia in an intensive care unit were observed (by the model) as experiencing better outcomes relative to the general population that is treated more conservatively. This harkens back to the point previously discussed regarding the importance of understanding exactly which question the model is being asked to answer. Beyond the black box, other ethical and logistical obstacles in machine learning in medicine include distributional shift (training datasets that may be biased toward race or socioeconomic status or simply outdated), insensitivity to impact (predictive tools that underestimate the consequences of a false positive or false negative outcome), and reward hacking (the machine learns unexpected means of achieving an outcome that cheat the system) (4).
While the challenge of explaining machine learning's method for reasoning persists, it draws some similarities to the way clinical medicine is practiced in the present. Physicians, much like deep learners, often treat patients using some component of their clinical experience or gestalt (difficult to explain) in addition to their technical knowledge (easy to explain). And the solution to this problem may involve a combination of (1) accepting the black box of machine learning, and (2) testing them rigorously against multiple patient cohorts (79). Altogether, these examples from the literature suggest the need for a healthy level of skepticism toward machine learning, and a willingness to appreciate its methodology.
Author Contributions
MC and NP were responsible for reviewing publications for inclusion in the review, drafting of the manuscript, and creating table and figures for the manuscript. JC and KN were responsible for critical revision of the manuscript for important intellectual content related to machine learning methodology and spine surgery research. AV was responsible for the conception of the review, supervision, and critical revision of the manuscript for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This study was funded internally by the Rothman Orthopaedic Institute and the Department of Orthopaedic Surgery at Thomas Jefferson University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
2. Watson DS, Krutzinna J, Bruce IN, Griffiths CE, McInnes IB, Barnes MR, et al. Clinical applications of machine learning algorithms: beyond the black box. BMJ Clin Res Ed. (2019) 364:l886. doi: 10.1136/bmj.l886
3. Krzywinski M, Altman N. Classification and regression trees. Nat Methods. (2017) 14:757–8. doi: 10.1038/nmeth.4370
4. Challen R, Denny J, Pitt M, Gompels L, Edwards T, Tsaneva-Atanasova K. Artificial intelligence, bias and clinical safety. BMJ Qual Saf. (2019) 28:231–7. doi: 10.1136/bmjqs-2018-008370
5. Panchmatia JR, Visenio MR, Panch T. The role of artificial intelligence in orthopaedic surgery. Brit J Hosp Med. (2018) 79:676–81. doi: 10.12968/hmed.2018.79.12.676
6. Munakata Y, Pfaffly J. Hebbian learning and development. Developmental Sci. (2004) 7:141–8. doi: 10.1111/j.1467-7687.2004.00331.x
7. Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. (2015) 349:255–60. doi: 10.1126/science.aaa8415
8. Noble WS. What is a support vector machine? Nat Biotechnol. (2006) 24:1565–7. doi: 10.1038/nbt1206-1565
9. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. (2019) 25:44–56. doi: 10.1038/s41591-018-0300-7
10. Russell SJ, Norvig P. Artificial Intelligence: A Modern Approach. 3rd ed. NJ: Pearson Education (2010).
11. McCarthy J, Minsky ML, Rochester N, Shannon CE. A proposal for the dartmouth summer research project on artificial intelligence. AI Magazine, Palo Alto, CA (2006) 27:12.
12. Gillings MR, Hilbert M, Kemp DJ. Information in the Biosphere: Biological and Digital Worlds. Trends Ecol Evol. (2016) 31:180–9. doi: 10.1016/j.tree.2015.12.013
13. Hilbert M, López P. The World's Technological Capacity to Store, Communicate, and Compute Information. Science. (2011) 332:60–5. doi: 10.1126/science.1200970
14. Landenmark HKE, Forgan DH, Cockell CS. An Estimate of the Total DNA in the Biosphere. PLoS Biol. (2015) 13:e1002168. doi: 10.1371/journal.pbio.1002168
15. Ratwani RM, Reider J, Singh H. A decade of health information technology usability challenges and the path forward. JAMA. (2019) 321:743. doi: 10.1001/jama.2019.0161
16. Mittal S, Vaishay S. A survey of techniques for optimizing deep learning on GPUs. J Syst Architect. (2019) 99:101635. doi: 10.1016/j.sysarc.2019.101635
17. Alba AC, Agoritsas T, Walsh M, Hanna S, Iorio A, Devereaux PJ, et al. Discrimination and calibration of clinical prediction models. JAMA. (2017) 318:1377. doi: 10.1001/jama.2017.12126
18. Burns JE, Yao J, Summers RM. Vertebral body compression fractures and bone density: automated detection and classification on CT images. Radiology. (2017) 284:788–97. doi: 10.1148/radiol.2017162100
19. Bar A, Wolf L, Amitai OB, Toledano E, Elnekave E. Compression fractures detection on CT. Proceeding. (2017) 10134:40–8. doi: 10.1117/12.2249635
20. Frighetto-Pereira L, Rangayyan RM, Metzner GA, Azevedo-Marques PM de, Nogueira-Barbosa MH. Shape, texture and statistical features for classification of benign and malignant vertebral compression fractures in magnetic resonance images. Comput Biol Med. (2016) 73:147–56. doi: 10.1016/j.compbiomed.2016.04.006
21. Stopa BM, Robertson FC, Karhade AV, Chua M, Broekman MLD, Schwab JH, et al. Predicting nonroutine discharge after elective spine surgery: external validation of machine learning algorithms. J Neurosurg Spine. (2019) 26:1–6. doi: 10.3171/2019.5.SPINE1987
22. Ogink PT, Karhade AV, Thio QCBS, Gormley WB, Oner FC, Verlaan JJ, et al. Predicting discharge placement after elective surgery for lumbar spinal stenosis using machine learning methods. Eur Spine J. (2019) 28:1433–40. doi: 10.1007/s00586-019-05928-z
23. Karhade AV, Thio QCBS, Ogink PT, Shah AA, Bono CM, Oh KS, et al. Development of machine learning algorithms for prediction of 30-day mortality after surgery for spinal metastasis. Neurosurgery. (2018) 85:E83–91. doi: 10.1093/neuros/nyy469
24. Thio QCBS, Karhade AV, Ogink PT, Raskin KA, Bernstein KDA, Calderon SAL, et al. Can machine-learning techniques be used for 5-year survival prediction of patients with chondrosarcoma? Clin Orthop Relat R. (2018) 476:2040–8. doi: 10.1097/CORR.0000000000000433
25. Ramkumar PN, Karnuta JM, Navarro SM, Haeberle HS, Iorio R, Mont MA, et al. Preoperative prediction of value metrics and a patient-specific payment model for primary total hip arthroplasty: development and validation of a deep learning model. J Arthroplast. (2019) 34:2228–34.e1. doi: 10.1016/j.arth.2019.04.055
26. Hoffman H, Lee SI, Garst JH, Lu DS, Li CH, Nagasawa DT, et al. Use of multivariate linear regression and support vector regression to predict functional outcome after surgery for cervical spondylotic myelopathy. J Clin Neurosci. (2015) 22:1444–9. doi: 10.1016/j.jocn.2015.04.002
27. Hopkins BS, Mazmudar A, Driscoll C, Svet M, Goergen J, Kelsten M, et al. Using artificial intelligence (AI) to predict postoperative surgical site infection: a retrospective cohort of 4046 posterior spinal fusions. Clin Neurol Neurosur. (2020) 192:105718. doi: 10.1016/j.clineuro.2020.105718
28. Hopkins BS, Yamaguchi JT, Garcia R, Kesavabhotla K, Weiss H, Hsu WK, et al. Using machine learning to predict 30-day readmissions after posterior lumbar fusion: an NSQIP study involving 23,264 patients. J Neurosurg Spine. (2019) 32:1–8. doi: 10.3171/2019.9.SPINE19860
29. Khan O, Badhiwala JH, Witiw CD, Wilson JR, Fehlings MG. Machine learning algorithms for prediction of health-related quality-of-life after surgery for mild degenerative cervical myelopathy. Spine J Official J North Am Spine Soc. (2020) 1–11. doi: 10.1016/j.spinee.2020.02.003. [Epub ahead of print].
30. Mehta SD, Sebro R. Computer-aided detection of incidental lumbar spine fractures from routine dual-energy X-ray absorptiometry (DEXA) studies using a support vector machine (SVM) classifier. J Digit Imaging. (2019) 33:1–7. doi: 10.1007/s10278-019-00224-0
31. Seoud L, Adankon MM, Labelle H, Dansereau J, Cheriet F. Prediction of scoliosis curve type based on the analysis of trunk surface topography. In: 2010 IEEE Int Symposium Biomed Imaging Nano Macro. Rotterdam (2010) p. 408–11. doi: 10.1109/ISBI.2010.5490322
32. Tee JW, Rivers CS, Fallah N, Noonan VK, Kwon BK, Fisher CG, et al. Decision tree analysis to better control treatment effects in spinal cord injury clinical research. J Neurosurg Spine. (2019) 31:1–9. doi: 10.3171/2019.3.SPINE18993
33. Vania M, Mureja D, Lee D. Automatic spine segmentation from CT images using convolutional neural network via redundant generation of class labels. J Comput Des Eng. (2019) 6:224–32. doi: 10.1016/j.jcde.2018.05.002
34. Varghese V, Krishnan V, Kumar GS. Evaluating Pedicle-Screw Instrumentation Using Decision-Tree Analysis Based on Pullout Strength. Asian Spine J. (2018) 12:611–21. doi: 10.31616/asj.2018.12.4.611
35. Galbusera F, Casaroli G, Bassani T. Artificial intelligence and machine learning in spine research. Jor Spine. (2019) 2:e1044. doi: 10.1002/jsp2.1044
36. Bzdok D, Altman N, Krzywinski M. Statistics versus machine learning. Nat Methods. (2018) 15:233–4. doi: 10.1038/nmeth.4642
37. Bzdok D, Krzywinski M, Altman N. Machine learning: a primer. Nat Methods. (2017) 14:1119–20. doi: 10.1038/nmeth.4526
38. Soffin EM, Beckman JD, Beathe JC, Girardi FP, Liguori GA, Liu J. Trends in ambulatory laminectomy in the USA and key factors associated with successful same-day discharge: a retrospective cohort study. Hss J. (2019) 16:72–80. doi: 10.1007/s11420-019-09703-0
39. Best MJ, Buller LT, Falakassa J, Vecchione D. Risk factors for nonroutine discharge in patients undergoing spinal fusion for intervertebral disc disorders. Iowa Orthop J. (2015) 35:147–55.
40. Morcos MW, Jiang F, McIntosh G, Ahn H, Dea N, Abraham E, et al. Predictive Factors for Discharge Destination Following Posterior Lumbar Spinal Fusion: A Canadian Spine Outcome and Research Network (CSORN) Study. Global Spine J. (2018) 9:219256821879709. doi: 10.1177/2192568218797090
41. Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Nat Methods. (2017) 14:318–41. doi: 10.1201/9781315139470-12
42. Lever J, Krzywinski M, Altman N. Model selection and overfitting. Nat Methods. (2016) 13:703–4. doi: 10.1038/nmeth.3968
43. Varghese V, Kumar GS, Krishnan V. Effect of various factors on pull out strength of pedicle screw in normal and osteoporotic cancellous bone models. Med Eng Phys. (2016) 40:28–38. doi: 10.1016/j.medengphy.2016.11.012
44. Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol. (2019) 19:64. doi: 10.1186/s12874-019-0681-4
45. Bazzocchi A, Ferrari F, Diano D, Albisinni U, Battista G, Rossi C, et al. Incidental findings with dual-energy X-ray absorptiometry: spectrum of possible diagnoses. Calcified Tissue Int. (2012) 91:149–56. doi: 10.1007/s00223-012-9609-2
46. Hsu C-W, Lin C-J. A comparison of methods for multiclass support vector machines. IEE Trans Neural Netw. (2002) 13:415–25. doi: 10.1109/72.991427
47. Fu H, Archer KJ. High-dimensional variable selection for ordinal outcomes with error control. Brief Bioinform. (2020). doi: 10.1093/bib/bbaa007. [Epub ahead of print].
48. Domingos P. A few useful things to know about machine learning. Commun Acm. (2012) 55:78. doi: 10.1145/2347736.2347755
49. LeCun Y. Deep learning hardware: past, present, and future. In: 2019 International Solid-State Circuits Conference (ISSCC). San Francisco, CA (2019). p. 12–9. doi: 10.1109/ISSCC.2019.8662396
50. LeCun Y. The power and limits of deep learning. Res Technol Manage. (2018) 61:22–7. doi: 10.1080/08956308.2018.1516928
51. Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal. (2013) 35:1798–828. doi: 10.1109/TPAMI.2013.50
52. Emmert-Streib F, Yang Z, Feng H, Tripathi S, Dehmer M. An introductory review of deep learning for prediction models with big data. Front Artif Intell. (2020) 3:4. doi: 10.3389/frai.2020.00004
53. Michelson JD. CORR insights®: what are the applications and limitations of artificial intelligence for fracture detection and classification in orthopaedic trauma imaging? A systematic review. Clin Orthop Relat Res. (2019) 477:2492–4. doi: 10.1097/CORR.0000000000000912
54. Roth HR, Wang Y, Yao J, Lu L, Burns JE, Summers RM. Deep convolutional networks for automated detection of posterior-element fractures on spine CT. In: SPIE Medical Imaging. San Diego, CA (2016). doi: 10.1117/12.2217146
55. Lindsey R, Daluiski A, Chopra S, Lachapelle A, Mozer M, Sicular S, et al. Deep neural network improves fracture detection by clinicians. Proc National Acad Sci USA. (2018) 115:11591–6. doi: 10.1073/pnas.1806905115
56. Gale W, Oakden-Rayner L, Carneiro G, Bradley AP, Palmer LJ. Detecting hip fractures with radiologist-level performance using deep neural networks. arXiv [Preprint]. (2017). arXiv:1711.06504.
57. Yu K-H, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. (2018) 2:719–31. doi: 10.1038/s41551-018-0305-z
58. Quang D, Chen Y, Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. (2014) 31:761–3. doi: 10.1093/bioinformatics/btu703
59. Quang D, Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. (2016) 44:e107. doi: 10.1093/nar/gkw226
60. Kamps R, Brandão R, Bosch B, Paulussen A, Xanthoulea S, Blok M, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. (2017) 18:308. doi: 10.3390/ijms18020308
61. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Adv Neural Inform Process Syst. (2012) 25:1097–105. doi: 10.1145/3065386
62. Yazdan-Shahmorad A, Silversmith DB, Kharazia V, Sabes PN. Targeted cortical reorganization using optogenetics in non-human primates. Elife. (2018) 7:e31034. doi: 10.7554/eLife.31034
63. Lessmann N, van Ginneken B, de Jong PA, Išgum I. Iterative fully convolutional neural networks for automatic vertebra segmentation and identification. Med Image Anal. (2018) 53:142–55. doi: 10.1016/j.media.2019.02.005
64. Chen H, Dou Q, Wang X, Qin J, Cheng JCY, Heng P-A. 3D fully convolutional networks for intervertebral disc localization and segmentation. In: International Conference on Medical Imaging and Augmented Reality. Hong Kong (2016). p. 375–82. doi: 10.1007/978-3-319-43775-0_34
65. Kim YJ, Ganbold B, Kim KG. Web-based spine segmentation using deep learning in computed tomography images. Healthc Inform Res. (2020) 26:61–7. doi: 10.4258/hir.2020.26.1.61
66. Alsofy SZ, Stroop R, Fusek I, Saravia HW, Sakellaropoulou I, Yavuz M, et al. Virtual reality-based evaluation of surgical planning and outcome of monosegmental, unilateral cervical foraminal stenosis. World Neurosurg. (2019) 129:e857–65. doi: 10.1016/j.wneu.2019.06.057
67. Alsofy SZ, Nakamura M, Ewelt C, Kafchitsas K, Fortmann T, Schipmann S, et al. Comparison of stand-alone cage and cage-with-plate for monosegmental cervical fusion and impact of virtual reality in evaluating surgical results. Clin Neurol Neurosur. (2020) 191:105685. doi: 10.1016/j.clineuro.2020.105685
68. Kim JS, Merrill RK, Arvind V, Kaji D, Pasik SD, Nwachukwu CC, et al. Examining the ability of artificial neural networks machine learning models to accurately predict complications following posterior lumbar spine fusion. Spine. (2018) 43:853–60. doi: 10.1097/BRS.0000000000002442
69. Nguyen NQ, Priola SM, Ramjist JM, Guha D, Dobashi Y, Lee K, et al. Machine vision augmented reality for pedicle screw insertion during spine surgery. J Clin Neurosci. (2020) 72:350–6. doi: 10.1016/j.jocn.2019.12.067
70. Burström G, Nachabe R, Persson O, Edström E, Terander AE. Augmented and virtual reality instrument tracking for minimally invasive spine surgery: a feasibility and accuracy study. Spine. (2019) 44:1097–104. doi: 10.1097/BRS.0000000000003006
71. Gibby JT, Swenson SA, Cvetko S, Rao R, Javan R. Head-mounted display augmented reality to guide pedicle screw placement utilizing computed tomography. Int J Comput Ass Rad. (2018) 14:525–35. doi: 10.1007/s11548-018-1814-7
72. Deib G, Johnson A, Unberath M, Yu K, Andress S, Qian L, et al. Image guided percutaneous spine procedures using an optical see-through head mounted display: proof of concept and rationale. J Neurointerv Surg. (2018) 10:1187–91. doi: 10.1136/neurintsurg-2017-013649
73. Elmi-Terander A, Skulason H, Söderman M, Racadio J, Homan R, Babic D, et al. Surgical navigation technology based on augmented reality and integrated 3D intraoperative imaging. Spine. (2016) 41:E1303–11. doi: 10.1097/BRS.0000000000001830
74. Gregory TM, Gregory J, Sledge J, Allard R, Mir O. Surgery guided by mixed reality: presentation of a proof of concept. Acta Orthop. (2018) 89:480–3. doi: 10.1080/17453674.2018.1506974
75. Tepper OM, Rudy HL, Lefkowitz A, Weimer KA, Marks SM, Stern CS, et al. Mixed reality with hololens. Plast Reconstr Surg. (2017) 140:1066–70. doi: 10.1097/PRS.0000000000003802
76. Wen W, Kuroki Y, Asama H. The sense of agency in driving automation. Front Psychol. (2019) 10:2691. doi: 10.3389/fpsyg.2019.02691
77. Vayena E, Blasimme A, Cohen IG. Machine learning in medicine: addressing ethical challenges. PLoS Med. (2018) 15:e1002689. doi: 10.1371/journal.pmed.1002689
78. Char DS, Shah NH, Magnus D. Implementing machine learning in health care — addressing ethical challenges. New Engl J Med. (2018) 378:981–3. doi: 10.1056/NEJMp1714229
79. London AJ. Artificial intelligence and black-box medical decisions: accuracy versus explainability. Häst Cent Rep. (2019) 49:15–21. doi: 10.1002/hast.973
Keywords: machine learning, deep learning, artificial intelligence, spine surgery, orthopedic surgery
Citation: Chang M, Canseco JA, Nicholson KJ, Patel N and Vaccaro AR (2020) The Role of Machine Learning in Spine Surgery: The Future Is Now. Front. Surg. 7:54. doi: 10.3389/fsurg.2020.00054
Received: 14 April 2020; Accepted: 13 July 2020;
Published: 21 August 2020.
Edited by:
Vassilios S. Nikolaou, National and Kapodistrian University of Athens, GreeceReviewed by:
Alberto Di Martino, University of Bologna, ItalyKonstantinos Markatos, Biomedical Research Foundation of the Academy of Athens (BRFAA), Greece
Copyright © 2020 Chang, Canseco, Nicholson, Patel and Vaccaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jose A. Canseco, am9zZS5jYW5zZWNvQHJvdGhtYW5vcnRoby5jb20=
 Michael Chang
Michael Chang Jose A. Canseco
Jose A. Canseco Kristen J. Nicholson2
Kristen J. Nicholson2 Neil Patel
Neil Patel Alexander R. Vaccaro
Alexander R. Vaccaro