- Department of Biology, University of Puget Sound, Tacoma, WA, United States
Fungal infection is a common source of egg loss in reptiles, a taxa that rarely expresses egg-tending behaviors or other forms of parental care. In the absence of parental care, one potential mechanism for antifungal egg protection is the vertical transfer of symbiotic bacteria from the maternal cloaca to eggshell surfaces that inhibit fungal growth. We examined the inhibition of four fungal strains isolated from the natural environment of striped plateau lizards (Sceloporus virgatus) when challenged against each of seven bacteria strains isolated from female cloacae. Twenty-three of 28 pairwise interactions showed >80% inhibition of fungal growth, with 18 pairs showing >95% inhibition, similar to Serratia marcescens D1, a lab strain known to have potent antifungal activity. Whole genome sequencing of one cloacal strain (Serratia marcescens strain 1) identified 6 genes with antifungal products that may break down or outcompete fungi, including Chitinase B and ferri- bacillibactin esterase, and further identified 11 genes that may contribute to biosynthetic pathways for antimicrobial secondary metabolites. Overall, we document strong fungal inhibition by maternal S. virgatus cloacal microbes, which are known to transfer to eggshells during oviposition. Comparative work is needed to determine whether vertically-transmitted antifungal microbes are a common source of egg-protection among reptiles, as well as other oviparous species, particularly in the absence of apparent parental care.
Introduction
Pathogenic fungi in nest environments have been widely associated with egg mortality in a variety of oviparous animals. For example, Carvalho et al. (2019) determined that Fusarium and Penicillium, along with other fungi, were responsible for killing the embryos of Tegu lizard eggs. Fusarium fungi are also responsible for egg mortality in sea turtles and freshwater turtles (Sarmiento-Ramírez et al., 2010; Sarmiento-Ramirez et al., 2014; Carranco et al., 2022). Similarly, fungal pathogens have been shown to infect eggs from Iberian rock lizards and tree frogs, increasing egg mortality, inducing early hatching, and decreasing offspring fitness (Villa, 1979, Warkentin et al., 2001; Moreira and Barata, 2005).
Fungal infection of eggs can be limited through behavioral means in species that provide parental care. Two common methods to prevent infection are nest tending, which can limit exposure to harmful microbes while vertically transmitting beneficial antibacterial microbes (Ruiz-Castellano et al., 2016; van Veelen et al., 2017), and egg smearing, in which parents coat eggs with antimicrobial secretions or beneficial microbes (Hosokawa et al., 2006; D’Alba and Shawkey, 2015; Martínez-García et al., 2015). Both of these methods are common in birds and insects, and variations on egg smearing have also been observed in squid and salamanders (Lauer et al., 2007; Kerwin et al., 2019; Suria et al., 2020).
However, many species do not provide care for eggs, meaning that any protection provided by parents must be provisioned at the time of oviposition. In sea turtles (Eretmochelys imbricata; Sarmiento-Ramirez et al., 2014) and striped plateau lizards (Sceloporus virgatus; Bunker et al., 2021), these protections appear to be sourced from antifungal microbes which reside in the maternal cloaca and are passed to eggs during oviposition – a method that may be far more widespread than currently recognized. The S. virgatus cloacal microbiome is largely dominated by gram-negative Enterobacteriaceae bacteria, including Serratia species (Bunker et al., 2022), which are known to have antifungal properties (Gutiérrez-Román et al., 2015; Dhar Purkayastha et al., 2018). Indeed, eggs that pass through the cloaca, relative to those that are removed by dissection, have reduced fungal attachment and improved hatch success and hatchling phenotype (Bunker et al., 2021).
In this study, we directly tested whether bacterial strains isolated from the cloaca of S. virgatus inhibit fungal growth. We used culture-based methods to explore the in vitro interactions between several cloacal microbes and potentially pathogenic fungi isolated from the soil of the natural habitat of S. virgatus. We aimed to identify both bacterial taxa that may have a protective function in the nest environment and fungal species that may overcome these protections. We measured fungal growth in the presence of cloacal bacteria both qualitatively and quantitatively through spore formation. Additionally, we used whole genome sequencing to identify possible pathways of antifungal action, such as hydrolytic and/or competitive compounds.
Methods
Bacterial isolates
Bacterial isolates were obtained from female Sceloporus virgatus cloacal microbes. We captured lizards near the Southwestern Research Station (SWRS) in Cochise County, AZ. This and all work with lizards was approved by the University of Puget Sound IACUC (PS18002) and Arizona Game and Fish (SP649069). Cloacal microbes were sampled using BD ESwab Collection kits; a sterile swab was inserted in the cloaca and gently rotated, then placed in the liquid Amies solution. Microbes were eluted into the solution, and half the volume was stored at -80°C in a 25% glycerol solution for later use. The remaining volume was used for separate studies (Bunker et al., 2021, 2022). Bacterial samples were grown from frozen stocks on Luria broth (LB) agar plates, and morphologically distinct colonies were restreaked in triplicate to ensure each strain was uniquely isolated. CloneID™ 1X Colony PCR Master Mix (Lucigen) was used to execute whole colony PCR on the 16S rRNA gene from each morphotype isolated, and PCR products were sequenced by Macrogen (Seoul, South Korea). Taxonomy was identified through the National Center for Biotechnology Information (NCBI) BLASTn function (Camacho et al., 2009), and sequences were uploaded to the NCBI database (see Supplementary Table S1 for accession numbers). A sample of each isolated bacterial strain was stored for future use in a 25% glycerol solution at -80°C. These stocks were utilized, as needed, for experimental trials and cultured by incubation in LB media at 30°C while shaking at 300 RPM.
Non-cloacal bacterial strains were also used, to comparatively assess the relative strength of the antifungal properties of cloacal bacteria. These were obtained from previously existing frozen glycerol stocks, and included Bacillus megaterium, Escherichia coli DH5α, and Serratia marcescens D1 (Carolina Biological Supply), the latter of which is known to have antifungal properties and served as a positive control.
Fungal isolates and spore suspensions
Fungal stocks were created by isolating soil fungus that had infected S. virgatus eggs. Eggs were obtained from field-collected gravid females induced to oviposit using a 0.1 mL injection of oxytocin. Eggs were laid onto sanitized paper towels, collected with sterile forceps as they were laid, and weighed. Eggs were incubated in a 0.8 mL/g mixture of sterile vermiculite and soil slurry; the slurry was a 1:10 ratio of soil collected near SWRS mixed with sterile water. Eggs that failed to hatch due to fungal fouling were swabbed, fungi on swabs were eluted into Amies solution, and the sample was stored at -80°C in a 25% glycerol solution until plating. Fungal cultures were grown on Malt Extract Agar (MEA) until morphologically distinct mycelia appeared, and unique strains were isolated and cultured in triplicate. Fungi were vouchered as living cultures in sterile water and stored at room temperature. DNA from fungal vouchers was extracted using the Extract-N-AmpTM Tissue PCR Kit (Millipore Sigma). The ITS gene was amplified, then sequenced by Eton Biosciences (San Diego, CA).
Each fungal species (Fusarium sp., Aspergillus sp., Neocosmospora rubicola, and Purpureocillium lilacinum) was grown from vouchers on MEA plates infused with three antibiotics: kanamycin (50 μg/mL), ampicillin (100 μg/mL), and tetramycin (15 μg/mL) to discourage bacterial growth. Once fungal lawns covered the agar surface, spore suspensions were created based on Ellwood et al. (2006), as follows. Lawns were flooded with 10 mL of a sterile 5 M NaCl solution, then the plates were incubated at 30°C for approximately 30 min. After incubation, spores were gently agitated using a sterile metal spreader and collected using a micropipette. Spore suspensions were filtered through sterile cheesecloth to remove hyphal fragments. The concentration of spores in suspensions was determined using a hemocytometer and a dissecting microscope. Spore suspensions were diluted to concentrations of 106 spores/mL with the 5 M NaCl solution.
Qualitative assays
Direct bacterial–fungal challenge plates were designed based on Miller et al. (2021). Bacterial strains were incubated in LB media for 48 h at 30°C while shaking at 300 RPM, until cultures reached saturation. Cultures were then diluted to a low optical density (absorbance < 0.10 at 600 nm, determined by Genesys 50 UV-visible spectrometer) with sterile LB. 100 μL of each diluted bacteria culture was mixed with 2 mL of liquid 0.75% LB agar, then poured over a 60 mm x 15 mm solidified LB agar plate. Each plate was inoculated with 103 fungal spores using a micropipette. We included a plate with fungus only as a positive control, and a plate without bacteria nor fungus as a negative control. Plates were incubated at 30°C for 4 to 7 days, until the positive control plate showed signs of sporulation (Fusarium sp.: 4 d, Aspergillus sp.: 6 d, N. rubicola: 7 d, P. lilacinum: 7 d), and were then photographed and qualitatively assessed for relative fungal growth. Each cloacal bacterial strain was challenged once against each fungal strain, whereas the non-cloacal bacterial strains were only challenged against P. lilacinum and N. rubicola.
Quantitative assays
Fungal inhibition assays were created using a protocol modified from Miller et al. (2021). Bacteria strains were grown to their maximum optical density, then diluted (absorbance < 0.10 at 600 nm) with sterile LB. Then 103 spores of a given fungal strain (Fusarium sp., Aspergillus sp., N. rubicola, and P. lilacinum) were combined with 100 μL of each bacterial isolate, creating bacterial–fungal liquid cocultures. A liquid culture containing 100 μL of sterile LB and 103 fungal spores, as well as a LB plate inoculated with 103 fungal spores were prepared as positive controls, and 100 μL of sterile LB only was prepared as a negative control. Liquid cultures were incubated at 30°C while shaking at 300 RPM. Incubation was considered complete after the fungal control plate demonstrated signs of sporulation. At that time, the number of spores in each liquid culture was counted using a hemocytometer and a dissecting microscope. Cloacal bacterial strains were tested with all four fungal strains, and non-cloacal bacterial strains were challenged against P. lilacinum and N. rubicola. Assays were replicated 5 times per bacterial strain for P. lilacinum and N. rubicola, and 3 times per bacterial strain for Fusarium sp. and Aspergillus sp.
Data analysis
For each fungal strain, we performed one-way ANOVAs to compare the concentration of spores across bacterial treatments; tests were run separately for cloacal bacterial strains and for non-cloacal lab strains. When incubated with cloacal bacterial strains, spore counts for Fusarium and P. lilacinum were square root transformed to meet parametric assumptions, and a Dunnett’s post hoc test (Signorell, 2025) was used to compare the average concentration of spores in each bacterial treatment to the average concentration of spores in the positive control. Spore counts for P. lilacinum when incubated with non-cloacal strains were log transformed to meet assumptions and analyzed in the same way. Spore counts for Aspergillus and N. rubicola did not meet parametric assumptions following standard transformations, and were instead analyzed with Kruskal-Wallis tests followed by pairwise Dunn’s tests comparing each bacterial strain to the positive control. P-values were adjusted with a Bonferroni adjustment to account for the number of tests. All analyses were performed using R (version 4.0.2, R Core Team 2020). Data and scripts are available at zenodo.org (doi: 10.5281/zenodo.15556854).
Whole genome sequencing
A sample of cloacal Serratia marcescens strain 1, which showed strong inhibition of all four fungi, was sent to the Microbial Genome Sequencing Center (MiGS, Pittsburgh, PA) for whole genome sequencing to identify potential genes that could account for antifungal properties. They utilized Illumina and Oxford Nanopore (ONT) sequencing technology to produce raw reads for processing. MiGS assembled and annotated the reads. Quality control and adapter trimming was performed with bcl2fastq and porechop (Wick, 2018), which linked annotations directly to the associated Uniprot entry (Uniprot Consortium), for Illumina and ONT sequencing, respectively. Hybrid assembly with Illumina and ONT reads was performed with Unicycler (Wick, 2017). Assembly statistics were recorded with QUAST (Gurevich et al., 2013). Assembly annotation was performed with Prokka (Seemann 2014). The genome sequence was deposited at DDBJ/ENA/GenBank under the accession JBNRUO000000000. The version described in this paper is version JBNRUO010000000.
UniProt was utilized to research putative genes identified by the annotation process. 16S rRNA sequences were matched to existing organisms using BLAST. Genes that produce secondary metabolites with antimicrobial capabilities were identified using antiSMASH (Blin et al., 2019). Strict alignment parameters were used to identify putative gene clusters.
Results
Sequencing of the 16s rRNA gene and BLASTn comparison identified the seven bacterial strains isolated from the S. virgatus cloacal microbiome as the following: Citrobacter amalonaticus, Citrobacter sp., Enterobacter ludwigii, two strains of Enterococcus faecalis, and two strains of Serratia marcescens (Supplementary Table S1). Fungal strains identified from ITS sequencing were: Aspergillus sp., Fusarium sp., Purpureocillium lilacinum, and Neocosmospora rubicola.
Viability of fungi when plated with cloacal bacteria
Aspergillus grew to similar size on the control plate and the plates with the two E. faecalis strains, but did not grow on the plates with the other five cloacal bacterial strains (Supplementary Figure S1). P. lilacinum and N. rubicola grew to similar size on the control plate and the plate with E. faecalis strain 1, and did not grow on the plates with the other six cloacal bacterial strains; note that E. faecalis strain 2 appeared to swarm when plated with these two fungi (Supplementary Figure S1). Fusarium, our fastest growing fungus, grew to similar size on the control plate and the two E. faecalis plates, grew a small colony on the C. amalonaticus plate, and did not grow when plated with the remaining four cloacal bacterial strains (Supplementary Figure S1).
When interacting with the non-cloacal bacteria, both P. lilacinum and N. rubicola grew on the control plate, the B. megaterium treatment plate, and the E. coli DH5α treatment plate, however these fungi did not grow on the S. marcescens D1 treatment plate (Supplementary Figure S2).
Fungal spore formation when interacting with cloacal bacteria
Spore formation of each fungal species was individually compared across 7 cloacal bacterial strains and a control, to determine variation in spore formation when in coculture (or control) conditions. When incubated with cloacal bacterial strains, all four fungal strains were found to vary significantly across bacterial treatments (Aspergillus: χ2 = 18.05, df = 7, p = 0.012; P. lilacinum: F = 19.09, df = 7,32, p < 0.001; Fusarium: F = 43.33, df = 7,16, p < 0.001; N. rubicola: χ2 = 18.73, df = 7 p = 0.009; Figures 1a–d). Post-hoc tests indicated that fungal growth was significantly inhibited in the presence of all seven bacterial strains for Aspergillus and N. rubicola, and for six and five out of seven bacterial strains for P. lilacinum and Fusarium, respectively (excluding one or both E. faecalis strains), relative to growth seen in control cultures. Bacterial inhibition of fungal growth was extremely successful with percent reductions in fungal growth over 80%, often over 95% (for 18 of 28 combinations), and up to 99.9% (Table 1).
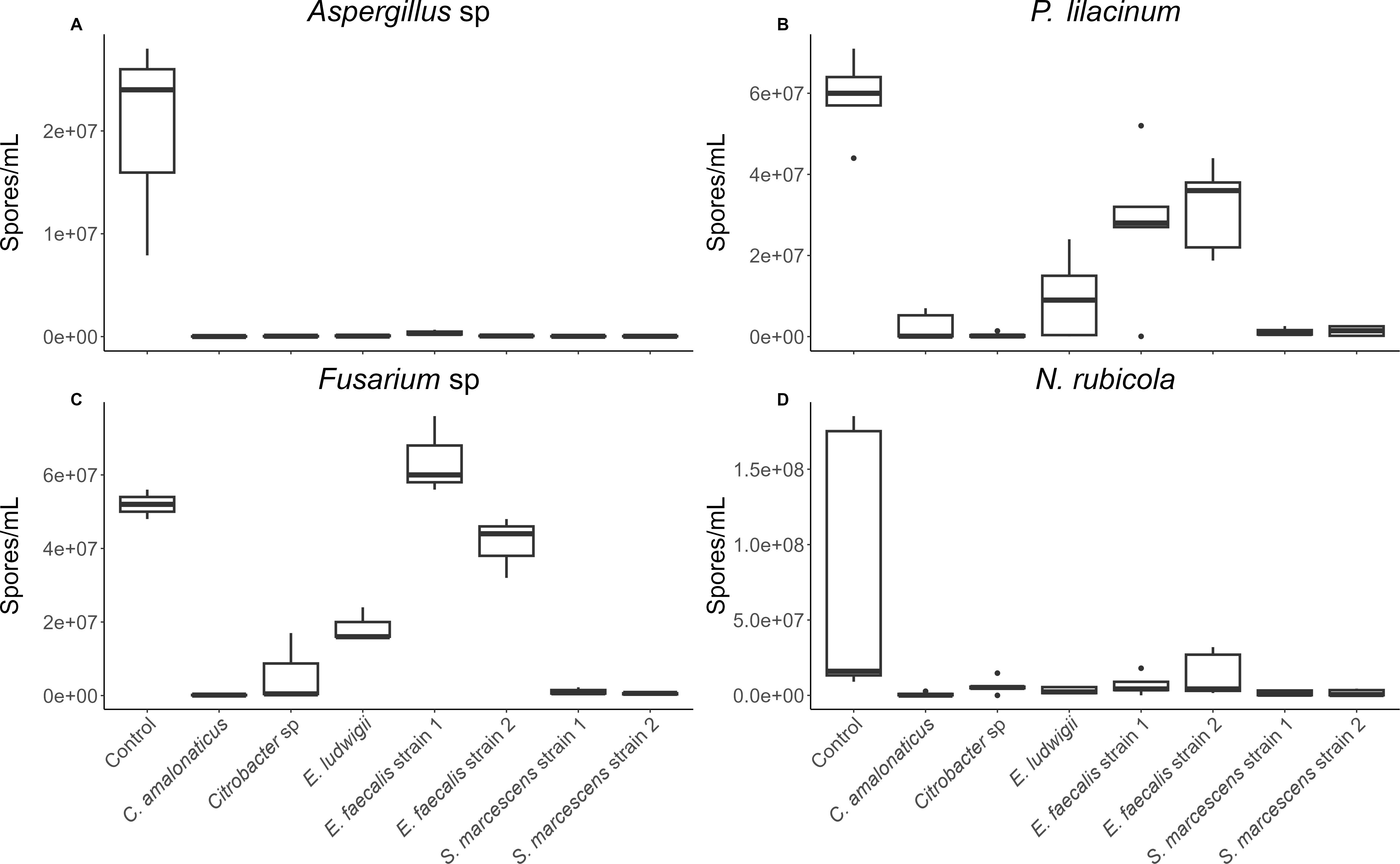
Figure 1. Number of (A) Aspergillus, (B) P. lilacinum, (C) Fusarium, and (D) N. rubicola spores/mL counted when fungus was incubated individually (control) or with several Sceloporus virgatus cloacal bacteria strains. Boxes indicate median and quartiles, and whiskers indicate 95% confidence intervals. N = 3 assays/bacterial strain for Aspergillus and Fusarium. N = 5 assays/bacterial strain for P. lilacinum and N. rubicola. Incubation conditions significantly affected growth of each fungal strain (p ≤ 0.012). All fungal-bacterial combinations successfully reduced fungal growth except P. lilacinum and Fusarium competed against E faecalis. P values from post-hoc tests are in Table 1.

Table 1. Percent reduction of fungal spores in bacterial challenges compared to the control, sample sizes, and p-values from post-hoc tests comparing each bacterial treatment to the control treatment.
P. lilacinum and N. rubicola were also compared against non-cloacal bacterial strains, in the same manner. There were significant differences between treatment types overall (P. lilacinum: F = 184.8, df = 3,16, p < 0.001; N. rubicola: χ2 = 16.29, df = 3, p = 0.001; Figures 2a, b), and between controls and bacterial treatments based on Dunnet’s tests. The scale of these changes were substantially smaller when fungi were treated with B. megatirium and E. coli DH5α, but similar to the cloacal bacteria when treated with S. marcescens D1, which is known to have antifungal properties (Table 2).
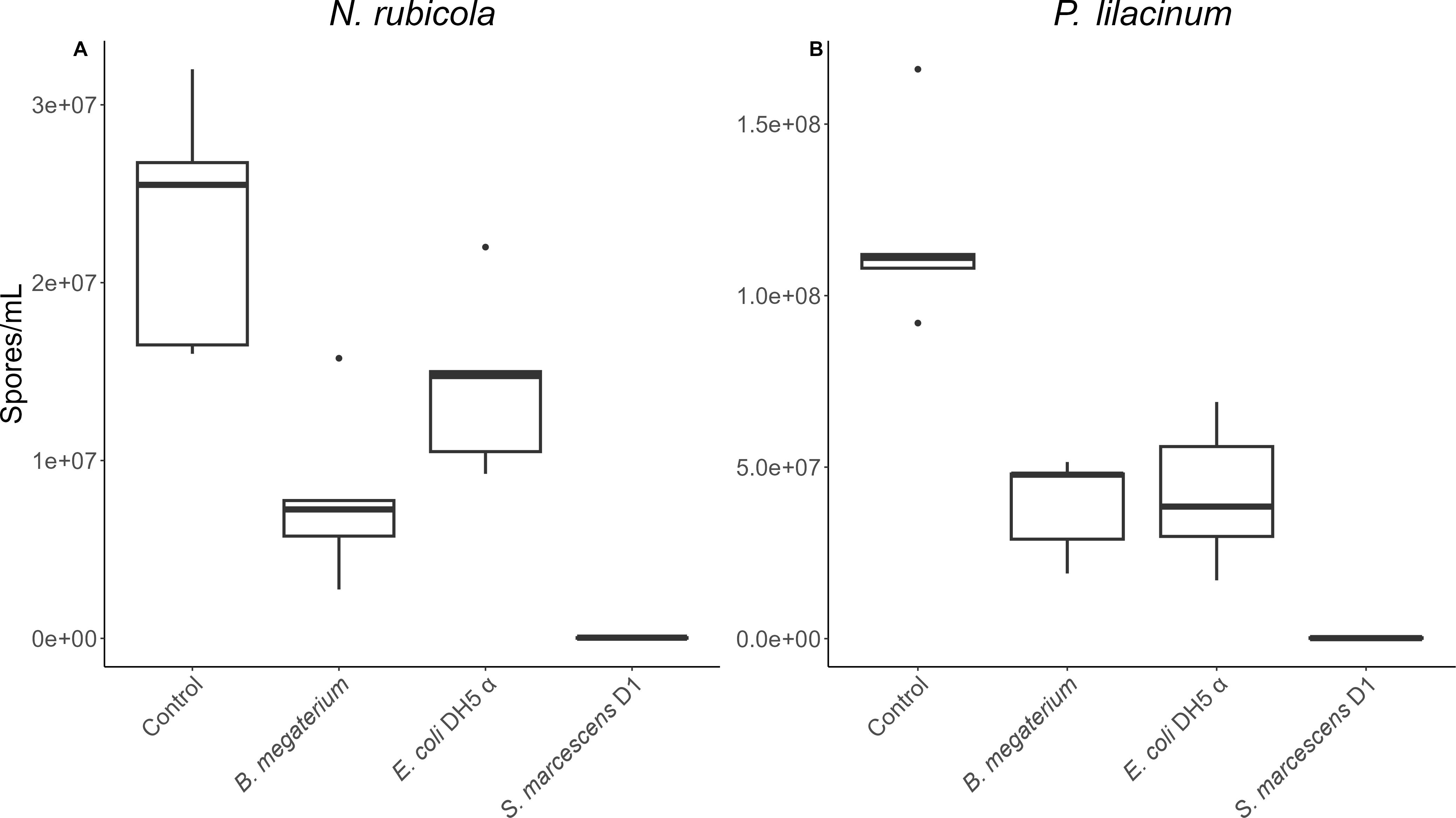
Figure 2. Number of (A) N. rubicola and (B) P. lilacinum spores/mL counted when fungus was incubated individually (control) or with several non-cloacal bacteria strains. Boxes indicate median and quartiles, and whiskers indicate 95% confidence intervals. n=5 assays/bacterial strain. Incubation conditions significantly affected fungal growth (p ≤ 0.001). P values from post-hoc tests are in Table 2.
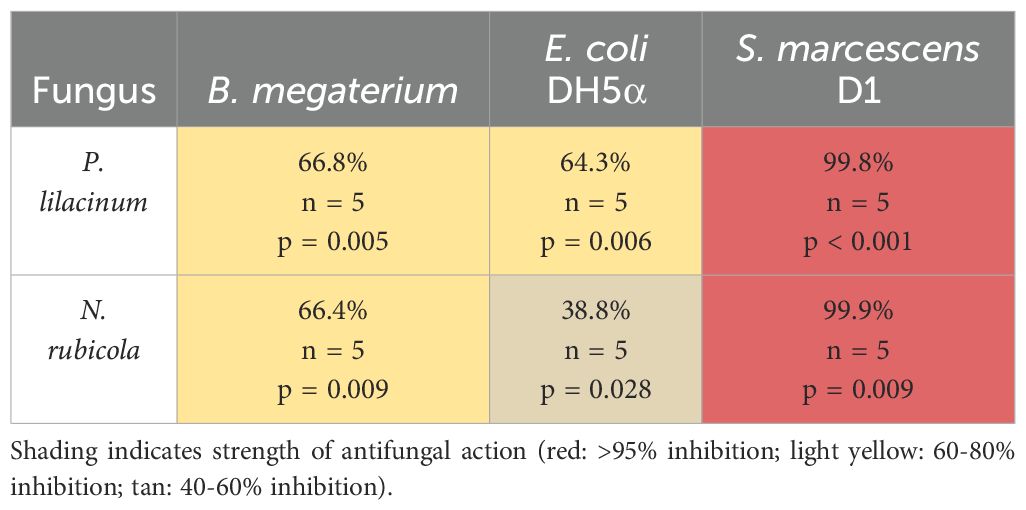
Table 2. Percent reduction in fungal spore counts in each non-cloacal bacterial treatment compared to the control, sample sizes, and p-values from post-hoc tests comparing each bacterial treatment to the control treatment.
Potential antifungal pathway
To look for gene loci associated with antifungal activity, we selected Serratia marcescens strain 1 for whole genome sequencing. Sequencing yielded 30 contigs with an N50 value of 1.55 Mbp (largest contig: 1.89 Mbp) and there were no ambiguous bases. GC content was in the expected range for bacterial genomes (59.3%), total assembly length was 5.9 Mbp, and estimated coverage was around 80x. Analysis of the Serratia marcescens strain 1 genome identified 6 genes with potential antifungal products (Table 3), including the gene pathway for Chitinase B and ferri-bacillibactin esterase. Further, we identified 11 genomic regions that bear resemblance to biosynthetic gene clusters that produce antimicrobial secondary metabolites (Supplementary Table S2). Most of the identified secondary metabolite-producing genes have products that have been characterized to have some antimicrobial properties, although not fungus-specific.

Table 3. Six genes of interest identified in the genome of Serratia marcescens strain 1, isolated from the cloaca of female Sceloporus virgatus.
Discussion
Both qualitative and quantitative results indicate that cloacal bacteria of S. virgatus females can significantly inhibit fungal growth, supporting previous evidence (Bunker et al., 2021) that these bacteria offer antifungal protection to eggs in the nest environment. The two methodological approaches provided some different patterns across the various bacterial-fungal combinations. For example, Aspergillus was more successful against the two strains of E. faecalis when growing on plates than when growing in liquid culture. In contrast, P. lilacinum was less successful against E. faecalis strain 2 when grown on plates than when grown in liquid culture, perhaps due to the swarming behavior of this strain when grown on surfaces.
Based on inhibition of spore formation, Aspergillus was the most sensitive fungi, with fungal spore formation significantly reduced (>95% inhibition) when cultured with each of the seven cloacal bacterial strains. N. rubicola was also significantly reduced when challenged with all seven cloacal bacterial strains (>80% inhibition). The most successful bacterial strains were C. amalonaticus and the two Serratia marcescens strains, as these strains consistently and strongly reduced fungal growth (>95% inhibition) across all four fungal strains. In contrast, the two E. faecalis strains only inhibited two of the four fungal strains. Although non-cloacal strains B. megatirium and E. coli DH5α (which served as controls) also reduced fungal spore growth, the scale of the reduction was generally much lower than that produced by the cloacal strains and by the positive control non-cloacal strain, Serratia marcescens D1. This indicates that, while co-culturing itself can reduce fungal growth, possibly through resource competition, there are additional antifungal mechanisms (i.e., stronger competitive abilities and/or metabolite and protein production) at work in the cloacal strains. Antifungal effects in natural nests, where multiple bacterial taxa are acting in combination, may be even stronger than those documented in our two-species interactions.
Antifungal mechanisms were investigated through whole genome sequencing. We identified several genes that code for products with direct antifungal effects, including chitinase B, a 2-nitroimidazole transporter, and validamycin A dioxygenase. Most notably, chitinases are widely known to facilitate antifungal activity in Serratia by breaking down chitin in fungal cell walls (Gutiérrez-Román et al., 2015; Dhar Purkayastha et al., 2018). We have preliminary evidence that five of our seven cloacal bacteria strains (all but the two E. faecalis strains) can grow on chitin minimum media, suggesting chitinase activity (Supplementary Figure S3). The 2-nitroimidazole transporter could be beneficial both in defense against antimicrobial compounds for Serratia, as well as having an antagonistic effect on other microbes (Seki et al., 1970; Ogasawara et al., 2015). Validomycin A is part of a larger validomycin complex that has known antifungal properties (Guirao-Abad and Sánchez-Fresneda, 2013). We also identified the presence of a gene for ferri-bacillibactin esterase, which has been shown to kill fungus by starving them of environmental iron (Yu et al., 2011), suggesting competitive interactions may underlie the inhibition of fungal growth documented herein. Finally, we identified several secondary metabolites that have general antimicrobial effects, or support pathways with potential antifungal effects (Supplementary Table S1). Future work should examine which of these (and other) genes are upregulated when in the presence of fungi. For instance, Serratia strains isolated from amphibian skin are known to confer resistance to the global fungal pathogen, Batrachochytium dendrobatidis (Bd), implicated in global amphibian declines; when co-cultured with Bd these strains show upregulation of genes associated with nitrate reductases, nitrite transporters, and acyl-coenzyme A dehydrogenase supporting their involvement in mediating antifungal action (Madison et al., 2017).
Though the underlying mechanism(s) are unresolved, our findings indicate an effective antifungal function of maternal S. virgatus cloacal microbes. The five strains most successful at inhibiting fungal growth (two Citrobacter, Enterobacter, and two Serratia) are all from Enterobacteriaceae, the family that dominates the S. virgatus cloacal microbiome, comprising 77% of the microbial community during the reproductive season (Bunker et al., 2022). We have previously found that cloacal microbes transfer to eggshells and increase egg survival and hatchling fitness of S. virgatus offspring (Bunker et al., 2021; Bunker and Weiss, 2024). A similar phenomenon has been observed in sea turtles, whose eggs are often infected by Fusarium (Sarmiento-Ramirez et al., 2014). Maternal transmission of microbiota is widespread across the animal kingdom, including many species that, like the majority of squamate reptiles, are oviparous and lack parental care (Rowe et al., 2020; Nyholm, 2020). Among oviparous species, the microbial nest environment and its maternal contributions are key aspects of the “nidobiome,” which has recently been posited as an integral part of neonate development (Campos-Cerda and Bohannan, 2020).
Despite the apparent efficacy of the S. virgatus cloacal microbiome as an antifungal agent, reptile eggs of other species are often killed by fungal infections (Otaño et al., 2014; Badiane et al., 2017; Carvalho et al., 2019; Gleason et al., 2020), and interactions between fungi and cloacal microbes are likely more complex in the natural nest environment than in our in vitro cocultures. For instance, in the nest, fungus may experience a more ideal growing environment, and may be able to establish in the nest on unfertilized or failed eggs and then opportunistically transfer to neighboring eggs (Moreira and Barata, 2005). These factors could help to reconcile the very strong inhibitory effects of the maternal microbiome in our experimental system, and known rates of fungal infection and egg failure in natural reptile nests. However, anecdotally, egg failure rates for S. virgatus both in natural settings and when challenged with fungus in a lab setting appear to be low (unpublished data). The nesting ecology of this temperate species is relatively unique in that oviposition is triggered by the onset of the North American monsoon rains (Vinegar, 1975). Nesting in periods of increased moisture may have put strong selective pressure on S. virgatus for its potent antifungal microbes. Future comparative studies, across reptile taxa, are needed to assess this possibility.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: Zenodo, https://doi.org/10.5281/zenodo.15556854. 16S rRNA sequences are uploaded to the NCBI database, with accession numbers listed in Supplementary Table S1. The genome sequence was deposited at DDBJ/ENA/GenBank under the accession JBNRUO000000000.
Ethics statement
The animal study was approved by University of Puget Sound Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MLB: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. OP: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. MEB: Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft. MM: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – review & editing. SW: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Science Foundation (1755408) and the University of Puget Sound.
Acknowledgments
We thank Helena Heyer-Grey, Sarah Sanz, Temple Dees, Robert Walls, and Brenda Lundt for assistance in the lab.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/famrs.2025.1589793/full#supplementary-material
Supplementary Figure 1 | Variation in growth of fungal species when incubated with Sceloporus virgatus cloacal bacteria on LB agar. The left column shows fungal growth in the absence of bacteria. Swarming by E. faecalis strain 2 was observed when paired with P. lilacinum and N. rubicola.
Supplementary Figure 2 | Variation in growth of fungal species when incubated with non-cloacal bacteria on LB agar. The left column shows fungal growth in the absence of bacteria.
Supplementary Figure 3 | Growth of seven bacteria isolated from the Sceloporus virgatus cloaca on chitin minimum media. Presence of bacterial growth on this media indicates bacterial chitinase activity. The unnumbered section of the plate had no bacteria.
Supplementary Table 1 | Identities and accession numbers of bacterial strains isolated from the Sceloporus virgatus cloacal microbiome.
Supplementary Table 2 | Serratia marcescens strain 1’s putative genes and their antimicrobial secondary metabolites. Output is taken from antiSMASH. The genome region, product type, and base pair location (nt) are given for each putative gene. Accompanying information is for the most similar known cluster and what type of molecule it produces, as well as the molecule’s identity and gene cluster’s similarity to the sample genome. n/a is assigned to regions with structural similarity to antimicrobial-producing clusters but lack exact sequence matches to functionally characterized clusters.
References
Badiane A., Pérez i de Lanuza G., del C. García-Custodio M., Carazo P., and Font E. (2017). Colour patch size and measurement error using reflectance spectrophotometry. Methods Ecol. Evol. 8, 1585–1593. doi: 10.1111/mee3.2017.8.issue-11
Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research 47 , W81–W87. doi: 10.1093/nar/gkz310
Bunker M. E., Arnold A. E., and Weiss S. L. (2022). Wild microbiomes of striped plateau lizards vary with reproductive season, sex, and body size. Sci. Rep. 12, 1–12. doi: 10.1038/s41598-022-24518-6
Bunker M. E., Elliott G., Heyer-Gray H., Martin M. O., Arnold A. E., and Weiss S. L. (2021). Vertically transmitted microbiome protects eggs from fungal infection and egg failure. Anim. Microb. 3, 43. doi: 10.1186/s42523-021-00104-5
Bunker M. E. and Weiss S. L. (2024). The reproductive microbiome and maternal transmission of microbiota via eggs in Sceloporus virgatus. FEMS Microbiol. Ecol. 100, fiae011. doi: 10.1093/femsec/fiae011
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Campos-Cerda F. and Bohannan B. J. M. (2020). The nidobiome: A Framework for understanding microbiome assembly in neonates. Trends Ecol. Evol. 35, 573–582. doi: 10.1016/j.tree.2020.03.007
Carranco A. S., Gillingham M. A. F., Wilhelm K., Torres M. L., Sommer S., and Romo D. (2022). Transcending sea turtles: First report of hatching failure in eggs of an Amazonian freshwater turtle with symptoms of the fungal emerging disease fusariosis. Transbound. Emerg. Dis. 69, e3282–e3288. doi: 10.1111/tbed.14596
Carvalho A. M., Souza L. K. H., Ataídes F. S., and Péres Junior A. K. (2019). Eggs of tegu lizard, Salvator merianae (Duméril & Bilbron 1839) (Squamata; Teiidae) damaged by fungal infections. Braz. J. Biol. 80, 112–114. doi: 10.1590/1519-6984.193388
Chakraborty K., Kizhakkekalam V., Joy M., and Chakraborty R. (2022). Bacillibactin class of siderophore antibiotics from a marine symbiotic Bacillus as promising antibacterial agents. Appl. Microbiol. Biotechnol. 106, 329–340. doi: 10.1007/s00253-021-11632-0
D’Alba L. and Shawkey M. D. (2015). Mechanisms of antimicrobial defense in avian eggs. J. Ornithol. 156, 399–408. doi: 10.1007/s10336-015-1226-1
Dhar Purkayastha G., Mangar P., Saha A., and Saha D. (2018). Evaluation of the biocontrol efficacy of a Serratia marcescens strain indigenous to tea rhizosphere for the management of root rot disease in tea. PloS One 13, 0191761. doi: 10.1371/journal.pone.0191761
Ellwood S., Kamphuis L., Pfaff T., Oliver R., Samac D. A., Foster-Hartnett D., et al. (2006). “Inoculation and growth with foliar pathogenic fungi,” in The Medicago truncatula Handbook (The Samuel Roberts Noble Foundation, Ardmore, OK).
Gleason F. H., Allerstorfer M., and Lilje O. (2020). Newly emerging diseases of marine turtles, especially sea turtle egg fusariosis (SEFT), caused by species in the Fusarium solani complex (FSSC). Mycology 11, 184–194. doi: 10.1080/21501203.2019.1710303
Guirao-Abad J. P. and Sánchez-Fresneda R. (2013). Analysis of validamycin as a potential antifungal compound against Candida albicans. Int. Microbiol. 16, 217–225. doi: 10.2436/20.1501.01.197
Gurevich A., Saveliev V., Vyahhi N., and Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Gutiérrez-Román M. I., Holguín-Meléndez F., Dunn M. F., Guillén-Navarro K., and Huerta-Palacios G. (2015). Antifungal activity of Serratia marcescens CFFSUR-B2 purified chitinolytic enzymes and prodigiosin against Mycosphaerella Fijiensis, causal agent of black Sigatoka in banana (Musa spp.). BioControl 60, 565–572. doi: 10.1007/s10526-015-9655-6
Hosokawa T., Kikuchi Y., Nikoh N., Shimada M., and Fukatsu T. (2006). Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PloS Biol. 4, e337. doi: 10.1371/journal.pbio.0040337
Kerwin A. H., Gromek S. M., Suria A. M., Samples R. M., Deoss D. J., O’Donnell K., et al. (2019). Shielding the next generation: Symbiotic bacteria from a reproductive organ protect bobtail squid eggs from fungal fouling. mBio, 10, 10.1128/mbio.02376-19. doi: 10.1128/mBio.02376-19
Lauer A., Simon M. A., Banning J. L., André E., Duncan K., and Harris R. N. (2007). Common cutaneous bacteria from the Eastern red-backed salamander can inhibit pathogenic fungi. Copeia 2007, 630–640. doi: 10.1643/0045-8511(2007)2007[630:CCBFTE]2.0.CO;2
Madison J. D., Berg E. A., Abarca J. G., Whitfield S. M., Gorbatenko O., Pinto A., et al. (2017). Characterization of Batrachochytrium dendrobatidis inhibiting bacteria from amphibian populations in Costa Rica. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00290
Martínez-García Á., Soler J. J., Rodríguez-Ruano S. M., Martínez-Bueno M., Martín-Platero A. M., Juárez-García N., et al. (2015). Preening as a vehicle for key bacteria in hoopoes. Microbial. Ecol. 70, 1024–1033. doi: 10.1007/s00248-015-0636-1
Miethke M., Klotz O., Linne U., May J. J., Beckering C. L., and Marahiel M. A. (2006). Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 61, 1413–1427. doi: 10.1111/j.1365-2958.2006.05321.x
Miller D. L., Smith E. A., and Newton I. L. G. (2021). A bacterial symbiont protects honey bees from fungal disease. mBio 12, 10.1128/mbio.00503-21. doi: 10.1128/mbio.00503-21
Mital A. (2009). Synthetic nitroimidazoles: biological activities and mutagenicity relationships. Sci. Pharm. 77, 497–520. doi: 10.3797/scipharm.0907-14
Moreira P. L. and Barata M. (2005). Egg mortality and early embryo hatching caused by fungal infection of Iberian rock lizard (Lacerta monticola) clutches. Herpetol. J. 15, 265–272.
Nyholm S. V. (2020). In the beginning: egg–microbe interactions and consequences for animal hosts. Philos. Trans. R. Soc. B: Biol. Sci. 375, 20190593. doi: 10.1098/rstb.2019.0593
Ogasawara H., Ohe S., and Ishihama A. (2015). Role of transcription factor NimR (YeaM) in sensitivity control of Escherichia coli to 2-nitroimidazole. FEMS Microbiol. Lett. 362, 1–8. doi: 10.1093/femsle/fnu013
Otaño N. B. N., Piña C. I., Bucsinsky A. M., and Arambarri A. M. (2014). Fungal diversity on broad-snouted caiman (Caiman latirostris) eggs, and their effects on hatchlings. Herpetol. J. 42, 217–222.
Plabutong N., Ekronarongchai S., Niwetbowornchai N., Edwards S. W., Virakul S., Chiewchengchol D., et al. (2020). The inhibitory effect of validamycin A on Aspergillus flavus. Int. J. Microbiol. 2020, 397415. doi: 10.1155/2020/3972415
R Core Team. (2020). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/.jo.
Rowe M., Veerus L., Trosvik P., Buckling A., and Pizzari T. (2020). The Reproductive Microbiome: An emerging driver of sexual selection, sexual conflict, mating systems, and reproductive isolation. Trends Ecol. Evol. 35, 220–234. doi: 10.1016/j.tree.2019.11.004
Ruiz-Castellano C., Tomás G., Ruiz-Rodríguez M., Martín-Gálvez D., and Soler J. J. (2016). Nest material shapes eggs bacterial environment. PloS One 11, e0148894. doi: 10.1371/journal.pone.0148894
Sarmiento-Ramírez J. M., Abella E., Martín M. P., Tellería M. T., López-Jurado L. F., Marco A., et al. (2010). Fusarium solani is responsible for mass mortalities in nests of loggerhead sea turtle, Caretta caretta, in Boavista, Cape Verde. FEMS Microbiol. Lett. 312, 192–200. doi: 10.1111/j.1574-6968.2010.02116.x
Sarmiento-Ramirez J., van der Voort M., Raaijmakers J., and Dieguez-Uribeondo J. (2014). Unraveling the microbiome of eggs of the endangered sea turtle Eretmochelys imbricata identifies bacteria with activity against the emerging pathogen Fusarium falciforme. PloS One 9, e95206. doi: 10.1371/journal.pone.0095206
Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seki Y., Nakmura T., and Okami Y. (1970). Accumulation of 2-aminoimidazole by streptomyces eurocidicus. J. Biochem. 67, 389–396. doi: 10.1093/oxfordjournals.jbchem.a129262
Signorell A. (2023). DescTools: tools for descriptive statistics. R package version 0.99.59, https://github.com/AndriSignorell/DescTools/, https://andrisignorell.github.io/DescTools/.
Suria A. M., Tan K. C., Kerwin A. H., Gitzel L., Abini-Agbomson L., Bertenshaw J. M., et al. (2020). Hawaiian bobtail squid symbionts inhibit marine bacteria via production of specialized metabolites, including new bromoalterochromides BAC-D/D’. mSphere 5, e00166–e00120. doi: 10.1128/msphere.00166-20
van Veelen H. P. J., Falcao Salles J., and Tieleman B. I. (2017). Multi-level comparisons of cloacal, skin, feather and nest-associated microbiota suggest considerable influence of horizontal acquisition on the microbiota assembly of sympatric woodlarks and skylarks. Microbiome 5, 156. doi: 10.1186/s40168-017-0371-6
Villa J. (1979). Two fungi lethal to frog eggs in Central America. Copeia 1979, 650–655. doi: 10.2307/1443873
Vinegar M. B. (1975). Demography of the striped plateau lizard, Sceloporus virgatus. Ecology 56, 172–182. doi: 10.2307/1935309
Warkentin K. M., Currie C. R., and Rehner S. A. (2001). Egg-killing fungus induces early hatching of red-eyed treefrog eggs. Ecology 82, 2860–2869. doi: 10.1890/0012-9658(2001)082[2860:EKFIEH]2.0.CO;2
Wick R. R. (2018). Porechop: Adapter trimmer for Oxford Nanopore reads. [Software]. https://github.com/rrwick/Porechop.
Wick R. R., Judd L. M., Gorrie C. L., and Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Computational Biology 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Keywords: antifungal, egg protection, Fusarium, microbes, nest, reptile, Sceloporus, Serratia
Citation: Boulet ML, Perotti ON, Bunker ME, Martin MO and Weiss SL (2025) Maternal cloacal bacteria of striped plateau lizards inhibit fungal growth. Front. Amphib. Reptile Sci. 3:1589793. doi: 10.3389/famrs.2025.1589793
Received: 08 March 2025; Accepted: 22 May 2025;
Published: 10 June 2025.
Edited by:
Douglas Woodhams, University of Massachusetts Boston, United StatesReviewed by:
Joseph Madison, Xavier University of Louisiana, United StatesCaroline Beck, University of Otago, New Zealand
Patrick Kearns, University of Massachusetts Boston, United States
Copyright © 2025 Boulet, Perotti, Bunker, Martin and Weiss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stacey L. Weiss, c3dlaXNzQHB1Z2V0c291bmQuZWR1
 McKenna L. Boulet
McKenna L. Boulet Olivia N. Perotti
Olivia N. Perotti Marie E. Bunker
Marie E. Bunker Mark O. Martin
Mark O. Martin Stacey L. Weiss
Stacey L. Weiss