- 1Faculty of Veterinary Science, Prince of Songkla University, Songkhla, Thailand
- 2Avian Health Research Unit, Department of Veterinary Medicine, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
Introduction: Chicken coccidiosis is a globally significant poultry disease caused by Eimeria species, which are highly pathogenic protozoa that impair growth performance and contribute to high morbidity and mortality in the poultry industry. To identify specific Eimeria species, molecular techniques have been developed in several countries as alternatives to conventional diagnostic approaches, which are labor-intensive, time-consuming, and have low accuracy in detecting mixed infections.
Methods: This study aimed to develop a SYBR Green-based real-time polymerase chain reaction assay for the identification of Eimeria species in Thailand, using DNA from five reference species (E. acervulina, E. brunetti, E. maxima, E. necatrix, and E. tenella) and 25 field samples from broiler farms in southern Thailand.
Results: The assay demonstrated high sensitivity, specificity, and reproducibility. Species-specific melting temperature profiles allowed reliable differentiation of Eimeria DNA from primer-dimers and potential contaminants. Field testing revealed a high prevalence of mixed infections, with E. tenella, E. acervulina, and E. maxima being the most common, whereas E. brunetti and E. necatrix were not detected.
Discussion: Compared with conventional gross examination, the SYBR Green-based real-time polymerase chain reaction assay proved to be a more accurate and efficient tool for diagnosing coccidiosis in commercial broiler farms, particularly in detecting subclinical and mixed-species infections.
1 Introduction
Coccidia are pathogenic intestinal protozoa that cause significant economic losses in the poultry industry worldwide, including in Thailand (Peek and Landman, 2011; Dorne et al., 2013; Del Cacho et al., 2016; Lin et al., 2017). Chicken coccidiosis is caused by Eimeria species, such as E. acervulina, E. maxima, E. necatrix, and E. tenella (Cerventes et al., 2020). Chickens are often infected with a mixture of Eimeria species, which leads to a range of clinical signs, including watery or bloody diarrhea, depression, poor growth performance, and mortality (Cerventes et al., 2020).
To prevent and control coccidiosis in broiler farms, anticoccidial drugs such as salinomycin, robenidine, and ionophores have been routinely added to feed programs for over 40 years (Peek and Landman, 2011; Kadykalo et al., 2018). However, the efficacy of these drugs has declined as a result of increasing resistance among Eimeria species (Chapman, 1984, 1986; Peek and Landman, 2003, 2006; Abbas et al., 2011; Sun et al., 2023). This resistance has led to widespread subclinical coccidiosis in broilers (Shirzad et al., 2011), which is a major concern, as it impairs growth performance and increases feed conversion ratios, resulting in financial losses for the poultry industry (Dalloul and Lillehoj, 2006; Blake et al., 2020).
Traditionally, the identification of Eimeria species has relied on clinical signs, characteristic gross intestinal lesions, and microscopic examination of oocyst morphology—methods that require highly skilled and experienced personnel (Carvalho et al., 2011b). In Thailand, coccidiosis surveillance in broiler farms is primarily conducted using these conventional approaches, especially gross lesion evaluation. This process is slow and delays the collection of epidemiological data. Moreover, subclinical lesions in medicated broiler farms often yield unreliable diagnostic information. Therefore, the application of molecular techniques for Eimeria species identification would significantly enhance the accuracy and efficiency of coccidiosis investigations.
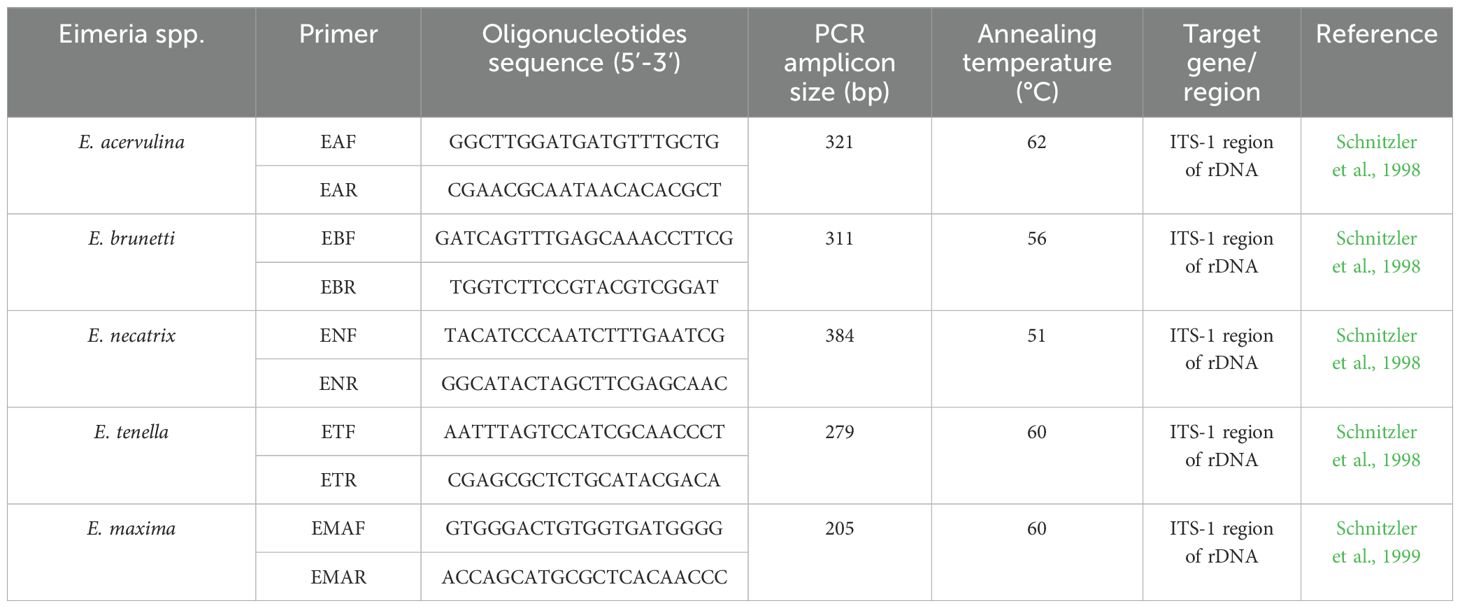
With the increasing value of the global poultry market, the development of effective diagnostic methods to monitor and control chicken coccidiosis has become a priority for the poultry industry. Several studies have developed polymerase chain reaction (PCR) and real-time PCR assays to enhance the routine diagnosis and species-level identification of Eimeria (Schnitzler et al., 1998; Haug et al., 2007; Blake et al., 2008; Kawahara et al., 2008; Morgan et al., 2009; Vrba et al., 2010; Carvalho et al., 2011a, b; Gyorke et al., 2013; Lan et al., 2017; Brown Jordan et al., 2018; Geng et al., 2021; Pajic et al., 2023).
In Thailand, monitoring of chicken coccidiosis still relies on conventional techniques, including the evaluation of specific macroscopic lesions and oocyst morphology. This study is the first to apply a SYBR Green-based real-time PCR assay for the identification of Eimeria species and to investigate chicken coccidiosis outbreaks in southern Thailand.
2 Materials and methods
2.1 Positive control sample
A positive control sample containing E. acervulina, E. maxima, E. tenella, E. brunetti, and E. necatrix was provided by the pharmaceutical company HIPRA (Bangkok, Thailand). The average concentration of each species was approximately 300 sporulated oocysts per mL.
2.2 Sample collection and preparation
Fecal samples were collected from 25 small-scale broiler farms with closed-house systems in southern Thailand between 2022 and 2024. The chickens raised on these farms were commonly of the Cobb 500, Arbor Acres, or Ross 308 breeds, managed under an all-in/all-out system. The average age of the broilers was 25–35 days, with an average weight of 1.5–2.0 kg. Feed formulation and quantity met the nutritional requirements of each breed line, and water was provided ad libitum.
The selected farms met standardized criteria, including evidence of subclinical coccidiosis—defined as moderate to poor performance without overt clinical signs such as bloody diarrhea—and the use of anticoccidial shuttle programs in all flocks. These programs included standard commercial anticoccidial drugs commonly used in poultry farming in Thailand, such as ionophores (salinomycin, narasin, and monensin), synthetic compounds (nicarbazin, diclazuril, and robenidine), and chemical-ionophore combinations (nicarbazin-narasin). Shuttle programs typically involve rotation among different classes of anticoccidial drugs to reduce resistance while maintaining control of the parasite. None of the farms reported the use of coccidiosis vaccines.
To isolate coccidian oocysts, 5–10 birds were randomly selected from each farm. The birds were weighed and humanely euthanized using carbon dioxide. Each bird underwent necropsy, and gross lesions associated with specific Eimeria species were recorded according to the criteria of Johnson and Reid (1970), to compare with real-time PCR results (Supplementary Table S1).
Intestinal contents were equally collected by scraping the mucosal surface of the duodenum, jejunum, ileum, and cecum sections using forceps, obtaining approximately 10–20 g per bird. Samples from individual birds were processed separately. Each sample was placed in 2.5% (w/v) potassium dichromate in a Ziplock plastic bag and left overnight. The next day, the samples were washed with water through a mesh sieve into a beaker, then centrifuged at 1,500 rpm for 5 min to sediment the oocysts. The supernatant was discarded, and the resuspended pellets were incubated in 2.5% (w/v) potassium dichromate at 28°C for two days to allow sporulation, then stored at 4°C (Conway and Mckenzie, 2007). The remaining suspension was examined for the presence of oocysts using a simple flotation method with saturated sodium chloride and visualized under a microscope.
2.3 DNA extraction
Genomic DNA from field samples was extracted from oocysts and other parasitic stages using the Quick-DNA Fecal/Soil Microbe Microprep Kit (Zymo Research, Orange, CA, USA), following the instructions of the manufacturer. Intestinal samples were directly added to ZR BashingBead™ Lysis Tubes and subjected to bead-beating using a Minilys homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) at maximum speed for 1 min to disrupt the oocyst walls. Samples then underwent four cycles of freeze-thawing. Extracted DNA was subsequently analyzed by real-time PCR using species-specific primers targeting five Eimeria species, as summarized in Table 1.
2.4 Real-time PCR assay
Real-time PCR was performed using the CFX96 Touch Detection System (Bio-Rad, CA, USA). Each 20 µL reaction mixture contained 10 µL of 2x SensiFAST™ SYBR No-ROX Mix (Meridian Bioscience, OH, USA), 0.8 µL each of forward and reverse primers (400 nM), 7.4 µL of sterile DNase-free water, and 1 µL of DNA template.
The thermal cycling conditions were as follows: initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 5 s; annealing at 51–59.5°C for 10 s (temperature depending on Eimeria species, as shown in Table 1); and extension at 72°C for 20 s. Distilled water was used as the template for the negative control. All reactions were performed in triplicate.
Fluorescence data were collected at every 0.1°C increment during the melting curve analysis. Melting temperature (Tm) was analyzed using Precision Melt Analysis Software (Bio-Rad, CA, USA) over a range of 65–95°C. Average quantification cycle (Cq) values, which reflect DNA copy number, were calculated using the Bio-Rad CFX software.
2.5 Serial DNA dilution
A 10-fold serial dilution of DNA-positive controls containing five Eimeria species was prepared and analyzed using real-time PCR. The highest genomic DNA concentration was quantified using a NanoDrop Lite spectrophotometer based on the A260/A280 absorbance ratio (Thermo Scientific, UK). The limit of detection was determined using serial dilutions, ranging from 7.5 ng/µL to 0.075 fg/µL of genomic DNA (100 to 10−8 dilution factor), along with corresponding Cq values (Bustin et al., 2009). Five dilution points (100, 10−1, 10−2, 10−3, and 10−4) were used for standard curve construction, as they consistently yielded reliable amplification within the detection range of the assay.
2.6 Statistical analysis
The prevalence of each Eimeria species was calculated as the percentage of positive samples among the total number of samples tested. Accordingly, 95% confidence intervals (CIs) were calculated using the Wilson score interval method (Wilson, 1927), which offers more accurate coverage than the standard Wald method, especially for small sample sizes and proportions near 0 or 1 (Newcombe, 1998). The Wilson score interval is computed using the following formula:
where p is the observed proportion (prevalence), n is the sample size, and z is the 1− quantile of the standard normal distribution (1.96 for a 95% CI). Fisher’s exact test was used for pairwise comparisons between species, with Bonferroni correction applied to adjust for multiple comparisons. Statistical significance was set at p < 0.05.
Cq values for each Eimeria species were determined using SYBR Green-based real-time PCR. All reactions were performed in triplicate to ensure reliability. For each species in each sample, the mean Cq value and standard deviation were calculated (Supplementary Tables S1, S2). The corresponding 95% CIs for Cq values were calculated using the following formula:
where is the mean Cq value, s is the standard deviation, n is the number of replicates (3), and is the critical t-value for n − 1 degrees of freedom at (4.303 for n = 3, α = 0.05).
One-way analysis of variance was performed to compare Cq values among the different Eimeria species, followed by Tukey’s post hoc test for pairwise comparisons. Statistical significance was set at p < 0.05.
The reproducibility of the PCR assay was evaluated by calculating the coefficient of variation (CV) for each sample across three separate testing days. A lower CV indicates greater precision and reproducibility of the real-time PCR assay. The CV was calculated using the following formula:
The standard deviation and mean were derived from the Cq values of triplicate measurements for each sample. Based on the CV, reproducibility was classified as excellent for CV values below 1%, good for values below 2%, acceptable for values below 5%, and poor for values ≥ 5% (Bustin et al., 2009).
Agreement and discrepancies between gross examination and real-time PCR results were assessed for each Eimeria species. The agreement rate was defined as the percentage of samples in which both methods produced the same result, whether positive or negative. The discrepancy rate referred to the percentage of samples for which the two methods disagreed. Discrepancies were further categorized as either Gross+/PCR− or Gross−/PCR+. McNemar’s test was used to determine whether the detection rates of the two methods differed significantly.
Prevalence estimates, 95% CIs, and reproducibility analyses were performed using Python (version 3.8) with the NumPy and Pandas libraries. All statistical analyses were conducted using R (version 4.4.3).
3 Results
3.1 Sensitivity of the real-time PCR assay and limit of detection
The sensitivity of the real-time PCR assays for the five Eimeria species was assessed using a 10-fold serial dilution of genomic DNA. The limit of detection for all five species was determined to be at the 10−4 dilution (750 fg/µL), as Cq values remained detectable up to this dilution, whereas no amplification was observed at further dilutions (10−5 to 10−8).
Standard curves were generated for each Eimeria species based on the 10-fold serial dilution. Cq values ranged from approximately 20 to 38 across the dilution series. The relationship between Cq values and the log10 of the dilution factors demonstrated strong linearity for all species, with R2 values ranging from 0.993 to 0.999, indicating excellent correlation. The slopes of the standard curves ranged from −3.465 to −4.186, corresponding to real-time PCR efficiencies between 73.3% and 94.4%. E. maxima exhibited the highest PCR efficiency (94.4%) with a slope of −3.465, whereas E. necatrix showed the lowest efficiency (73.3%) with a slope of −4.186 (Figure 1).
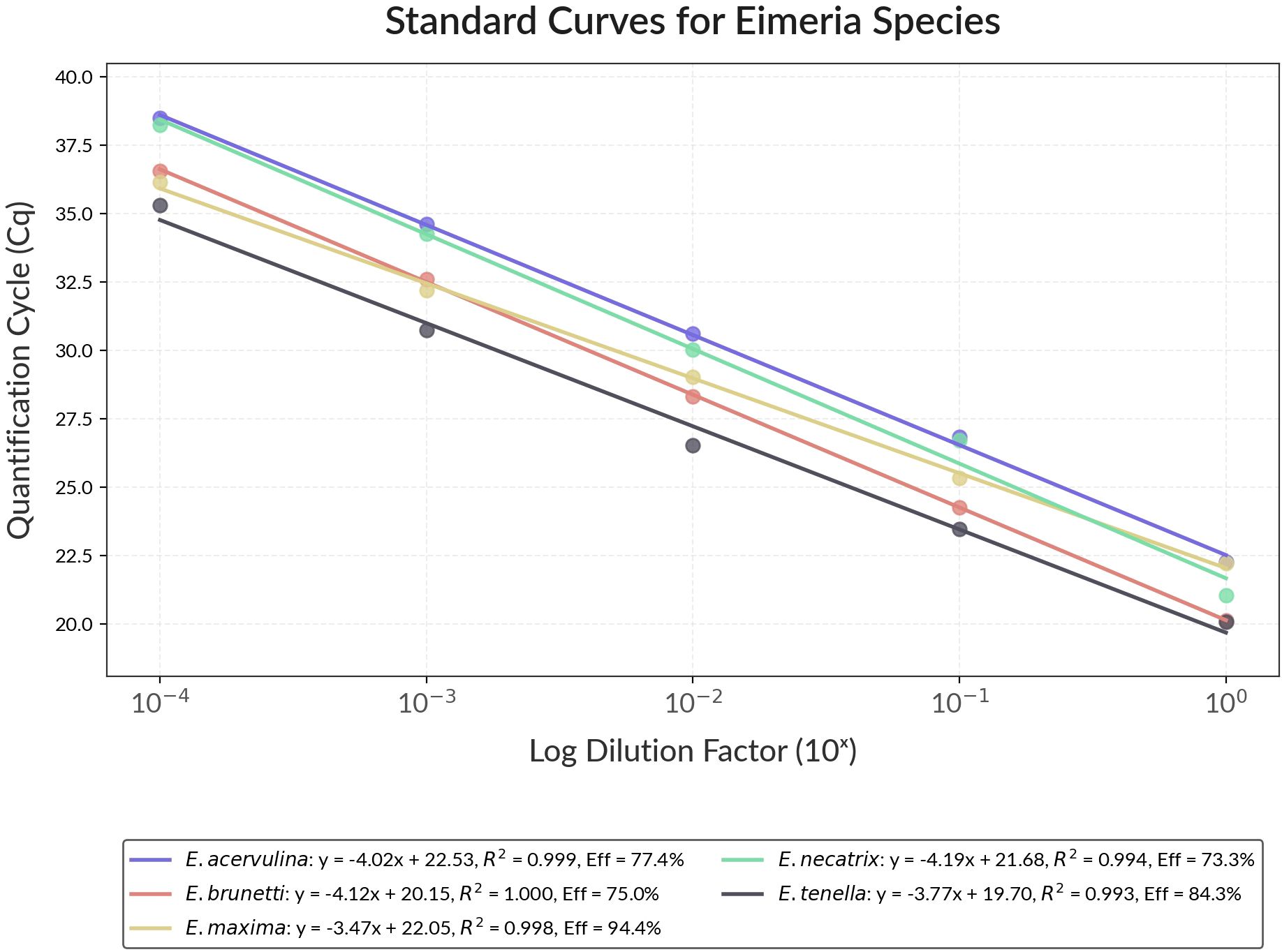
Figure 1. The graph displays standard quantification curves for five Eimeria species plotted as Quantification Cycle (Cq) versus Log Dilution Factor (10×). Each species is represented by a different colored line.
3.2 Specificity of the real-time PCR assay
Agarose gel electrophoresis confirmed that each primer amplified only the expected target amplicon size for the corresponding Eimeria species DNA (Supplementary Figure S1). The primers used in this study were previously described (Schnitzler et al., 1998, 1999) and were designed based on the internal transcribed spacer 1 region of ribosomal DNA. This region is highly specific for Eimeria species identification because of its high interspecies variability and low intraspecies variability. The diagnostic specificity of these primers was previously validated by Schnitzler et al. (1999), who demonstrated that each primer pair specifically amplified DNA from its corresponding Eimeria species without cross-reactivity with other species or host DNA.
A single peak was observed in the Tm amplification curves for each Eimeria species when using the positive control (Figure 2), indicating specific amplification. Melting curve analysis further revealed distinct Tm profiles for all five species (Figure 3). E. acervulina exhibited the highest Tm range (87.0–87.5°C), followed by E. tenella (86.0–86.5°C), E. brunetti (81.0–81.5°C), E. maxima (79.5 ± 0.5°C), and E. necatrix (79.0°C). All species displayed characteristic peak patterns that were consistent across replicates.
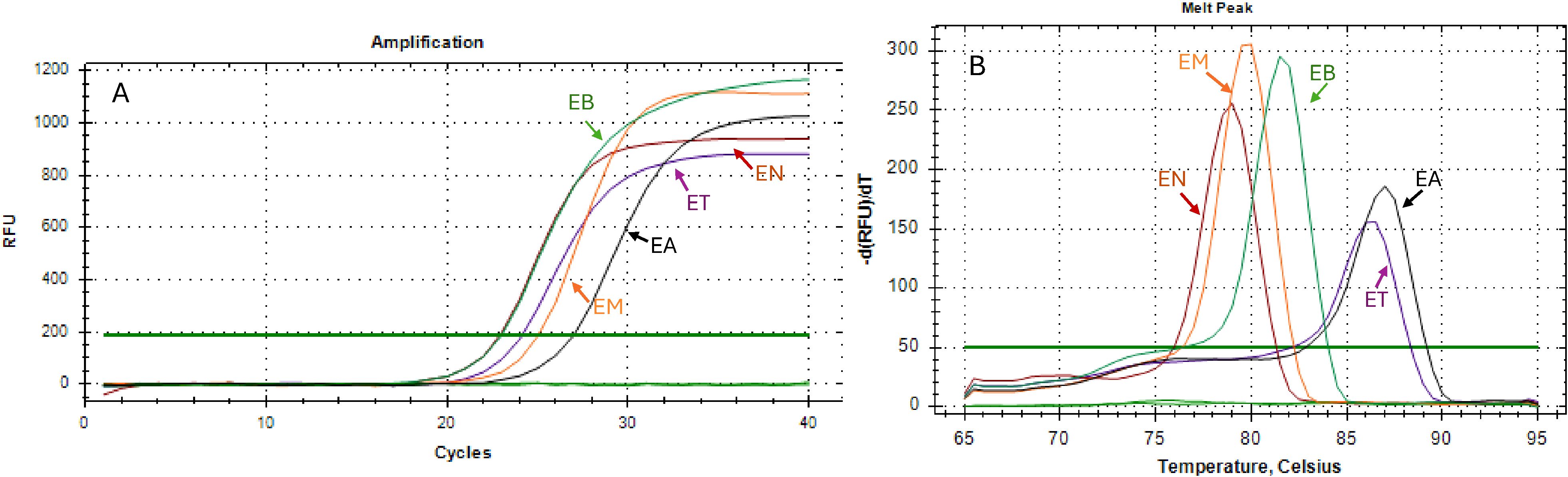
Figure 2. Real-time amplification of positive control (A) and Melting curve temperature (B). EA, Eimeria acervulina; EB, Eimeria brunetti; EM, Eimeria maxima;EN; Eimeria necatrix; and ET, Eimeria tenella.
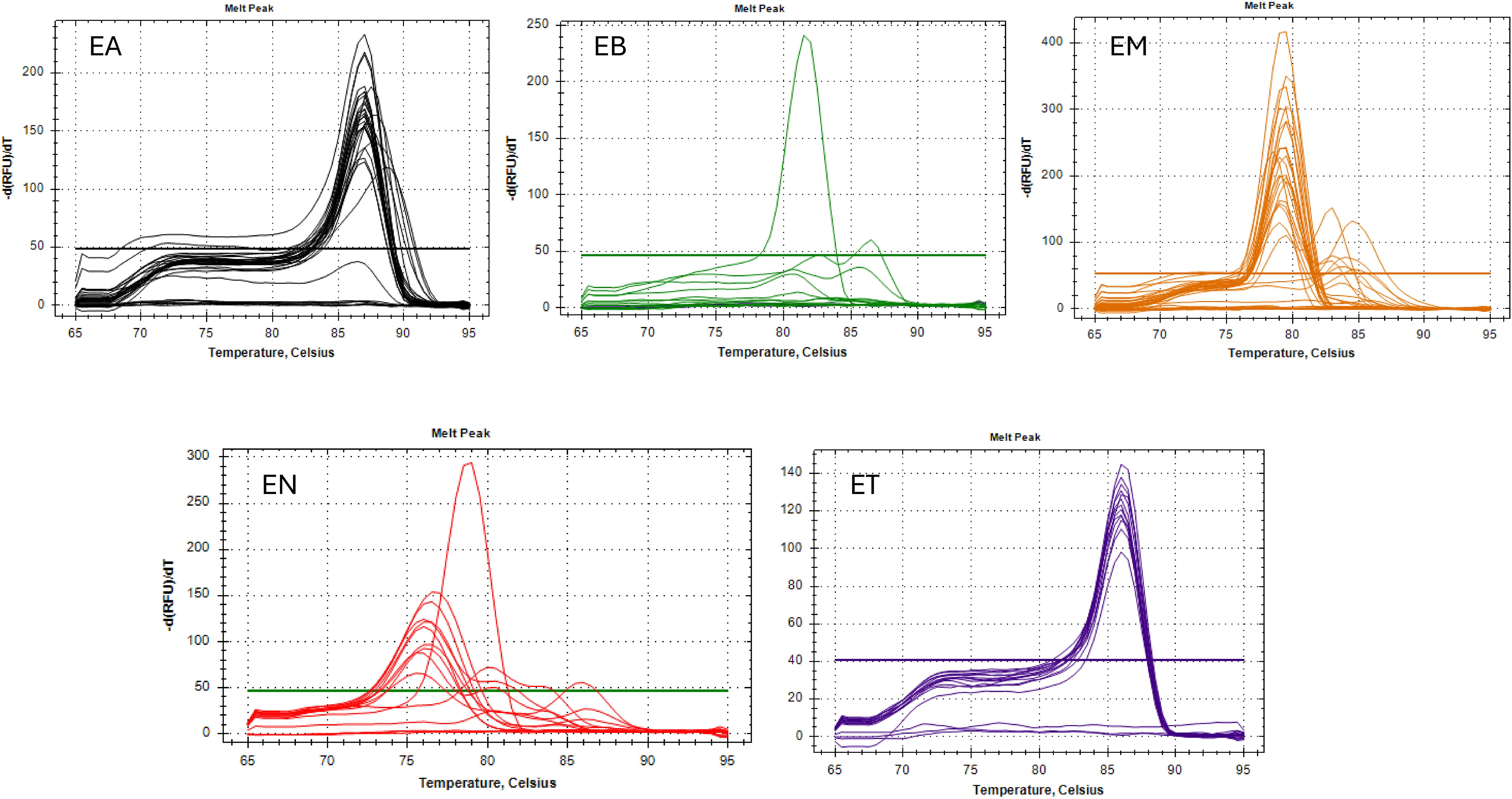
Figure 3. Melting curve analysis of 5 Eimeria spp. in field samples detected by Real-time PCR. EA, Eimeria acervulina; EB, Eimeria brunetti; EM, Eimeria maxima; EN, Eimeria necatrix; and ET, Eimeria tenella.
The melting curves for E. acervulina and E. tenella showed sharp, well-defined peaks, whereas those for E. maxima and E. necatrix occasionally exhibited secondary peaks or shoulders. The fluorescence derivative (dF/dT) values varied by species, with E. maxima showing the highest peak amplitude (approximately 400 dF/dT) and E. tenella the lowest (approximately 140 dF/dT).
3.3 Cq values in the real-time PCR assay
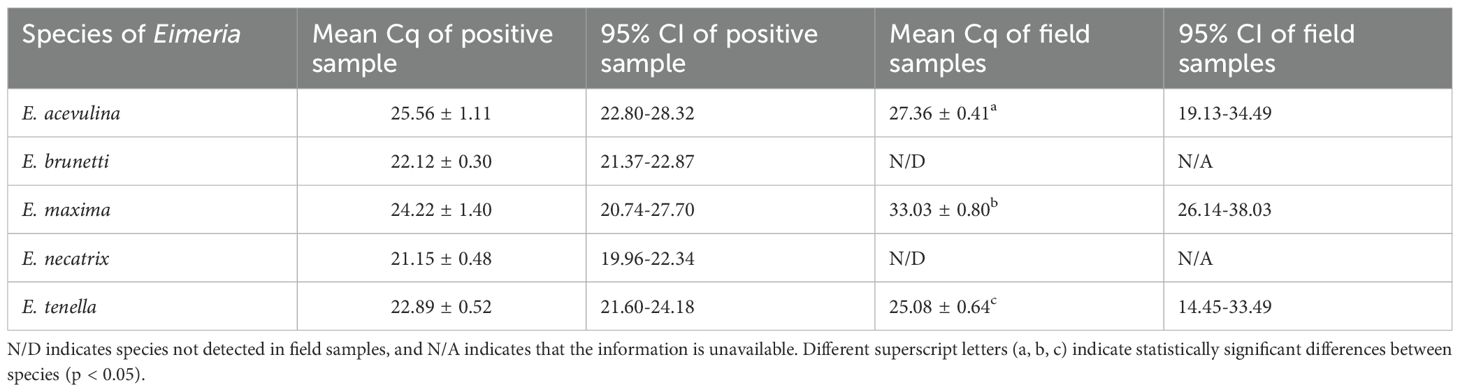
No amplification was observed in the non-template controls for any of the Eimeria species in the real-time PCR assay (Supplementary Table S2). The mean Cq values and their 95% CIs were calculated for all five Eimeria species in both the positive control and field samples (Table 2).
In the positive control, the mean Cq values were as follows: 25.56 ± 2.75 for E. acervulina, 22.12 ± 0.74 for E. brunetti, 24.22 ± 3.47 for E. maxima, 21.15 ± 1.19 for E. necatrix, and 22.89 ± 1.29 for E. tenella.
Among the field samples, E. tenella exhibited the lowest mean Cq value (23.45 ± 0.67), indicating the highest concentration of target DNA, followed by E. acervulina (27.32 ± 0.63). E. maxima had the highest mean Cq value (32.18 ± 1.19), suggesting the lowest DNA concentration among the detected species.
Statistical comparisons revealed that E. tenella had significantly lower Cq values than E. acervulina (p = 0.015), indicating a higher DNA load. In contrast, E. maxima had significantly higher Cq values than E. acervulina (p < 0.001), suggesting lower DNA levels. The most pronounced difference was observed between E. tenella and E. maxima, with E. tenella exhibiting significantly lower Cq values (p < 0.001), further confirming its higher target DNA concentration (Table 3).
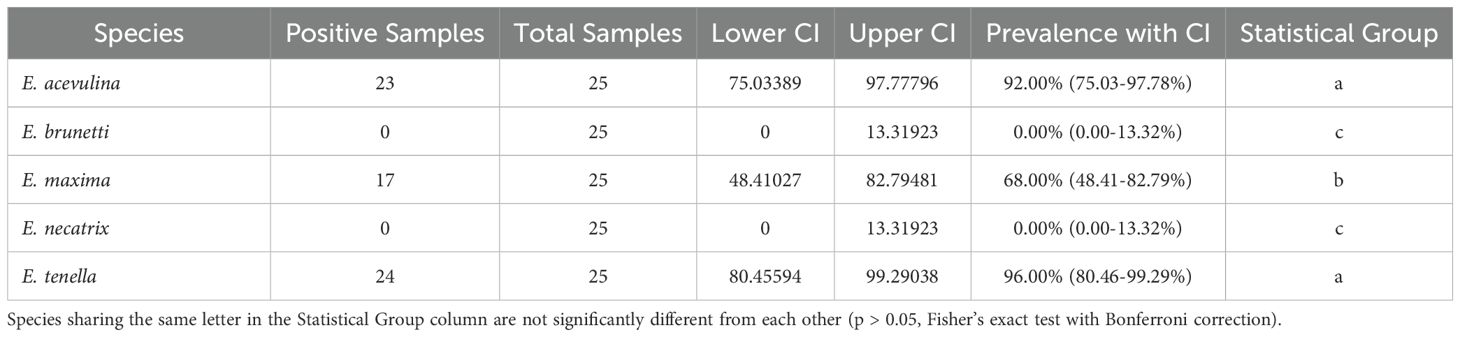
3.4 Prevalence of Eimeria species
E. tenella had the highest prevalence, detected in 96% of samples (95% CI: 80.46–99.29%), followed by E. acervulina at 92% (95% CI: 75.03–97.78%). E. maxima was detected in 68% of samples (95% CI: 48.41–82.79%), whereas E. brunetti and E. necatrix were not detected in any samples (0%; 95% CI: 0–13.32%; Table 3).
The half-width of the CI reflected the precision of the prevalence estimates. E. brunetti and E. necatrix had the narrowest intervals (6.66%), followed by E. tenella (9.42%) and E. acervulina (11.37%). In contrast, E. maxima showed the widest interval (17.19%), indicating greater uncertainty in its prevalence estimate compared with the other species.
Statistical analysis showed that E. tenella, E. acervulina, and E. maxima had significantly higher prevalence rates than E. brunetti and E. necatrix (p < 0.001). The difference in prevalence between E. maxima and E. tenella was also statistically significant (p = 0.023), whereas no significant difference was observed between E. tenella and E. acervulina (p = 1.000).
Analysis of infection patterns revealed that mixed infections were most common, as 64% of samples contained all three detected species (E. acervulina, E. maxima, and E. tenella). Dual infections involving E. acervulina and E. tenella were present in 28% of samples. Single-species infections were rare, with E. maxima and E. tenella each occurring alone in only 4% of cases (Figure 4).
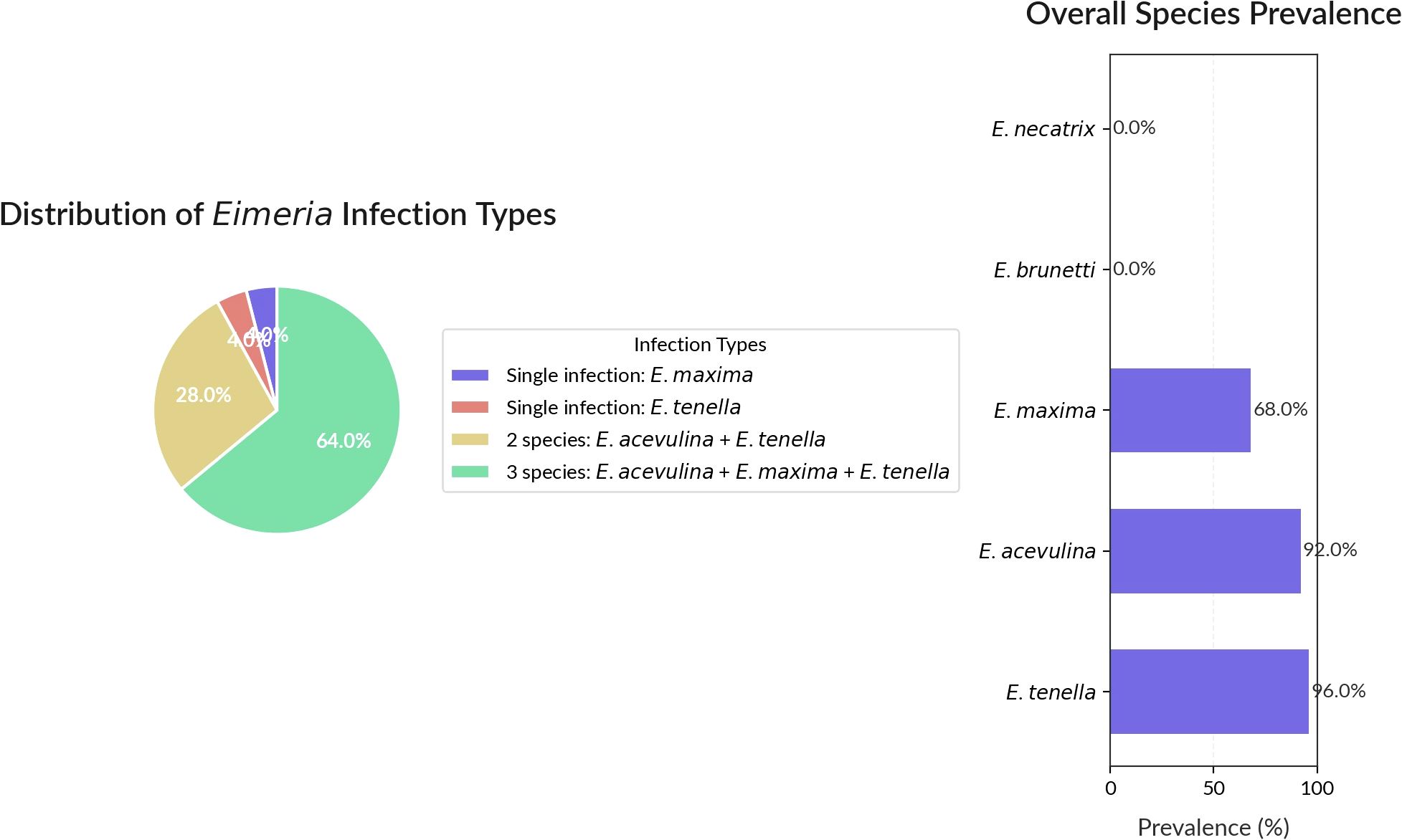
Figure 4. The pie chart demonstrates the distribution of infection types, the horizontal bar chart presents the overall prevalence of each Eimeria species, and the table provides the prevalence percentages.
3.5 Reproducibility analysis
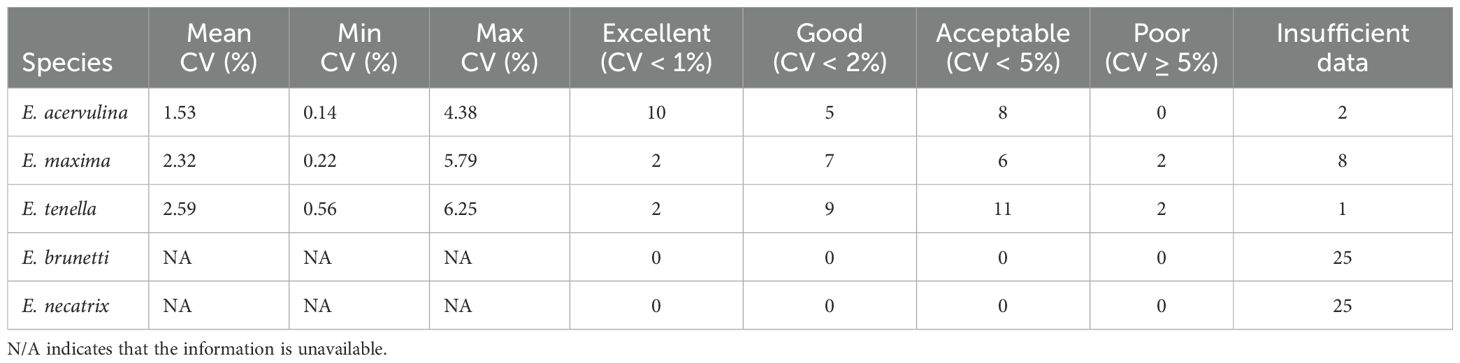
The reproducibility of the real-time PCR assay was evaluated for all five Eimeria species using 25 samples per species, with measurements taken on three separate days (Table 4). E. acervulina showed the highest reproducibility, with a mean CV of 1.53%, followed by E. maxima (2.32%) and E. tenella (2.59%). No valid data were available for E. brunetti and E. necatrix, as all Cq values for these species were negative.
Analysis of reproducibility categories revealed that E. acervulina had the highest proportion of samples with excellent reproducibility, as 43.5% of valid samples (10 out of 23) had a CV below 1%. E. maxima showed good reproducibility in 41.2% of valid samples (CV < 2%), whereas E. tenella had 45.8% of valid samples with acceptable reproducibility (CV < 5%).
3.6 Agreement and discrepancies between gross examination and real-time PCR results
The level of agreement between gross examination and real-time PCR varied considerably among the five Eimeria species (Table 5). E. acervulina showed an agreement rate of 44% and a discrepancy rate of 56%. E. maxima had a 72% agreement rate and a 28% discrepancy rate. E. necatrix and E. brunetti each demonstrated a 100% agreement rate, with no discrepancies observed. E. tenella exhibited a 56% agreement rate and a 44% discrepancy rate.
The nature of discrepancies also differed by species. For E. acervulina, all discrepancies (14 out of 25 samples) were cases where gross examination was negative, whereas real-time PCR was positive (Gross−/PCR+). For E. maxima, discrepancies included both Gross+/PCR− cases (two out of 25 samples) and Gross−/PCR+ cases (five out of 25 samples). For E. tenella, all discrepancies (11 out of 25 samples) were Gross−/PCR+ cases. No discrepancies were recorded for E. necatrix and E. brunetti, as all samples tested negative by both methods.
McNemar’s test revealed a significant difference between the two methods, with real-time PCR detecting significantly more positive samples for E. acervulina (p = 0.0005) and E. tenella (p = 0.0026). However, no significant difference was found between the methods for E. maxima (p = 0.4497; Figure 5).
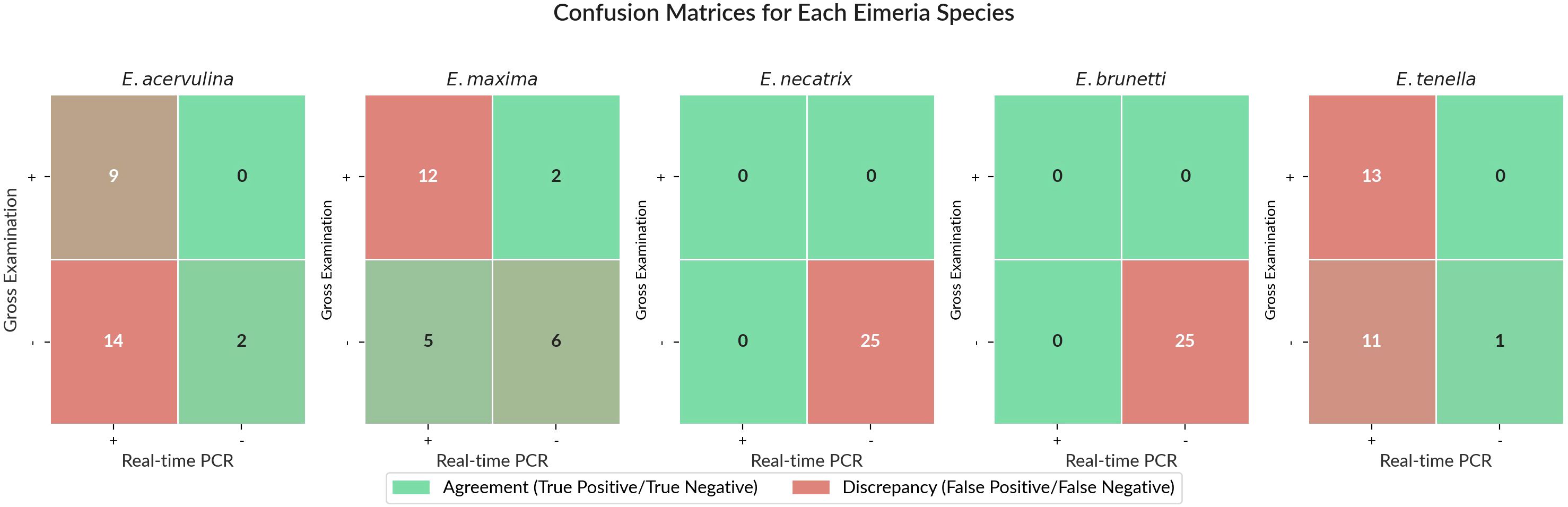
Figure 5. Confusion matrices showing the agreement and discrepancy between gross examination and real-time PCR for five Eimeria species. Each cell shows the number of samples in each category. Green cells represent agreement between the two methods (true positives in the upper left and true negatives in the lower right), while red cells represent discrepancy (false positives in the upper right and false negatives in the lower left).
4 Discussion
In this study, SYBR Green-based real-time PCR was used to identify Eimeria species. Real-time PCR is recognized for its high specificity, sensitivity, and reproducibility in DNA detection (Bustin et al., 2009). In particular, SYBR Green offers a productive and cost-effective alternative to hybridization probe-based PCR for Eimeria identification (Kawahara et al., 2008). Efficient oocyst rupture and DNA extraction are critical for detecting protozoan cysts in fecal samples, as DNA yield and quality directly influence PCR performance. Incomplete rupture may cause false negatives, whereas excessive disruption can fragment DNA (Haug et al., 2007; Kumar et al., 2014; Reginato et al., 2020).
Our optimized protocol, which incorporates bead-beating in lysis tubes, improved rupture efficiency and reduced co-extraction of PCR inhibitors from fecal samples (Schrader et al., 2012). This method is particularly suitable for Eimeria, whose oocysts resist both chemical and mechanical disruption (Guy et al., 2004). By controlling bead size, quantity, and homogenization parameters, our mechanical disruption technique effectively breaks the robust oocyst wall while minimizing DNA damage, preserving genomic integrity for accurate PCR analysis (Hachimi et al., 2024).
The limit of detection for Eimeria DNA by real-time PCR was approximately 750 fg. Given that one sporozoite contains about 75 fg of DNA according to Cornelissen et al. (1984), this corresponds to the detection of approximately 1–1.2 sporulated oocysts in a mixed DNA sample of five species, or roughly two sporozoites per species. These results align with previous studies reporting detection thresholds between 1 and 13.6 sporozoites (Haug et al., 2007; Blake et al., 2008; Kawahara et al., 2008; Oliveira et al., 2011). In contrast, conventional PCR had a detection limit of 25 oocysts in earlier work (Schnitzler et al., 1998). However, because we did not conduct a direct sensitivity comparison between real-time and conventional PCR, no definitive conclusions can be drawn from our dataset.
Standard curves generated for all five Eimeria species demonstrated excellent linearity across five orders of magnitude (R2 > 0.99), indicating strong quantification capability for diagnostic use (Bustin et al., 2009). According to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines, optimal PCR efficiency ranges from 90% to 110%, with corresponding slopes between –3.1 and –3.6. In this study, efficiencies ranged from 73.3% to 94.4%, with E. maxima achieving the highest (94.4%) and E. necatrix the lowest (73.3%). The values for the remaining species ranged from 75.0% to 84.3%. This variability may be explained by several factors. Differences in genomic structure can affect primer binding and amplification success (Reid et al., 2014). High GC content can also hinder amplification efficiency; for example, E. necatrix has a higher GC composition than E. maxima, which may contribute to its lower efficiency (Haug et al., 2007; Opel et al., 2010). Furthermore, species-specific DNA secondary structures may impede polymerase activity and reduce amplification efficiency (Bustin and Huggett, 2017). Despite the lower efficiency observed with E. necatrix, consistent use of standardized protocols, transparent reporting of efficiency values, and appropriate standard curves support reliable and reproducible quantification—even when amplification performance is suboptimal (Bustin et al., 2009).
In this study, DNA from both positive controls and field samples was successfully amplified by SYBR Green-based real-time PCR, producing single, species-specific Tm peaks. The clear separation between high-Tm species (E. acervulina and E. tenella) and low-Tm species (E. maxima and E. necatrix) enabled reliable species differentiation without additional post-PCR processing. The higher Tm observed for E. tenella (85.5°C) and E. necatrix (84.5°C), compared with E. acervulina (83.5°C), E. maxima (82.5°C), and E. brunetti (81.5°C), reflects their higher GC content. This pattern aligns with established biophysical principles, as GC base pairs form three hydrogen bonds—more than the two in AT pairs—requiring greater thermal energy to denature (Santalucia, 1998; Ririe et al., 1997; Shchyolkina et al., 2000). Consequently, amplicons with higher GC content showed correspondingly higher Tm values, supporting their utility in species identification.
The occasional secondary peaks observed in E. maxima and E. necatrix samples may indicate sequence variation within the amplified regions. This is consistent with prior findings of substantial genetic diversity among Eimeria species, which can affect real-time PCR amplification patterns (Lew et al., 2003; Kumar et al., 2015; Prakashbabu et al., 2017). Alternatively, these peaks could result from primer-dimers or non-specific amplification (Lan et al., 2004). However, their consistent appearance at species-specific temperatures suggests that they likely represent genuine intraspecies variants. Morris and Gasser (2006) similarly showed that reproducible secondary peaks during melting curve analysis often reflect true genetic polymorphisms within the internal transcribed spacer 1 and other genomic regions, rather than technical artifacts. This phenomenon is particularly common in genetically diverse Eimeria species, such as E. maxima and E. necatrix. Although informative, these secondary peaks may introduce diagnostic ambiguity in field samples, where species identification often relies on melting temperature (Tm) profiles. Misinterpretation of secondary peaks may lead to false-positive or false-negative results, thereby compromising the accuracy of epidemiological assessments and the effectiveness of control strategies, particularly with common mixed infections (Scipioni et al., 2008).
Reproducibility analysis for E. acervulina detection yielded a CV of 1.53%, with 43.5% of samples demonstrating excellent reproducibility (CV < 1%). These values align with established precision standards for quantitative molecular diagnostics. The real-time PCR assays for E. maxima and E. tenella showed comparable reproducibility, with mean CVs of 2.32% and 2.59%, respectively—both within the acceptable threshold (CV < 5%) for reliable quantitative molecular assays (Bustin et al., 2009).
Cq values and their 95% CIs provided key insights into Eimeria species abundance and assay precision. E. tenella and E. acervulina exhibited lower Cq values in field samples, consistent with their higher prevalence and DNA concentration in infected birds. Measurement precision, reflected by CI width, was greater for E. acervulina (mean CI width: 0.86) than for E. maxima (mean CI width: 1.64), possibly owing to the same factors influencing real-time PCR efficiency (Blake et al., 2006). The wider CIs observed for E. maxima indicated greater variability in quantification, warranting cautious interpretation of infection intensity estimates.
The consistent detection of all five Eimeria species in positive controls confirmed assay specificity and sensitivity. However, the absence of E. brunetti and E. necatrix in field samples could not be evaluated for biological relevance because of their non-detection. Although Cq values generally correlated with oocyst load, real-time PCR cannot precisely quantify absolute oocyst numbers because of factors affecting DNA yield, such as the degree of sporulation, extraction efficiency, and infection stage at sampling (Haug et al., 2007; Morgan et al., 2009; Raj et al., 2013).
All field samples tested positive for chicken coccidiosis by real-time PCR. E. acervulina was detected in 23 of 25 samples (92%; 95% CI: 75.03–97.78%), E. maxima in 17 samples (68%; 95% CI: 48.41–82.79%), and E. tenella in 24 samples (96%; 95% CI: 80.46–99.29%). The wider CI for E. maxima may reflect greater variability in its detection across samples. Mixed infections involving all three detected species were the most common among broiler farms in southern Thailand (64%). Neither E. brunetti nor E. necatrix was detected in any of the samples. To date, E. brunetti infection has not been reported in broiler farms in Thailand, whereas E. necatrix is typically observed in breeder flocks or older chickens (9–14 weeks), likely owing to its limited oocyst output in younger birds and greater susceptibility to anticoccidial drugs (Cerventes et al., 2020). Species-specific prepatent periods also influence detection rates, as E. acervulina (97–98 h) and E. tenella (115–120 h) have shorter prepatent periods than E. maxima (121–140 h) (Conway and Mckenzie, 2007; Cha et al., 2018). In addition, E. acervulina and E. tenella produce significantly more oocysts per infected cell than E. maxima, increasing their likelihood of detection (Williams, 2001). The absence of mucosal content collected from the rectum may have limited the detection sensitivity for E. brunetti, which primarily infects the lower intestinal tract (Cerventes et al., 2020). During the samples collection process, the efficiency of oocyst recovery may have differed among various Eimeria species. This study did not assess the uniformity of oocyst purification across different intestinal sections which may affect the detection sensitivity among species. These biological factors likely explain why E. acervulina and E. tenella were consistently the most prevalent species across broiler farms compared with E. maxima. All farms in this study used anticoccidial drugs in their feed programs, indicating that chicken coccidiosis remains widespread and may involve drug-resistant strains in this region. Notably, no positive samples were found for E. mitis, E. precox, E. lata, E. nagambie, and E. zaria—three of which are operational taxonomic units reported in Australia, India, the United States, and Europe (Jaramillo-Ortiz et al., 2023).
The perfect agreement between gross examination and real-time PCR results for E. necatrix and E. brunetti likely resulted from the absence of these species in the sample population. In the present study, all samples were collected exclusively from broiler farms. This observation aligns with previous research suggesting that E. necatrix and E. brunetti are uncommon in modern commercial broiler operations (Williams, 2005; Brown Jordan et al., 2018; Jaramillo-Ortiz et al., 2023). For the three detected species (E. acervulina, E. maxima, and E. tenella), agreement between methods ranged from moderate to poor. Most discrepancies involved gross examination failing to detect infections identified by real-time PCR. This is consistent with earlier research showing that PCR-based methods offer higher sensitivity than traditional diagnostic techniques (Haug et al., 2008; Vrba et al., 2010). The superior sensitivity of molecular techniques has important implications for coccidiosis surveillance in commercial broiler systems, as subclinical infections may remain undetected with gross examination alone, potentially leading to underestimated disease prevalence and avoidable production losses.
This study had several limitations. First, the sample size was relatively small (25 farms), which may limit the generalizability of the findings. Second, oocyst counting and histopathological examination were not performed, which could have provided additional insights into infection intensity and improved the accuracy of species identification. Third, although the real-time PCR assay is highly sensitive, it also has several limitations when applied in field conditions for detecting Eimeria infections in chickens. Real-time PCR may detect DNA from non-viable parasites or be inhibited by PCR inhibitors from an environment, potentially leading to an overestimation of true prevalence. It requires precise knowledge of infection timing, which is often difficult to determine field isolates, it cannot distinguish between different life stages of the parasite, potentially leading to misinterpretation if sampling isn’t carefully timed. In addition, achieving standardized protocols across diverse field conditions is difficult but necessary for comparability (Nolan et al., 2015). Lastly, this study lacks performance indicators such as weight gain or feed conversion ratio (FCR), which would facilitate a more straightforward interpretation of the impact on growth performance based solely on Eimeria species detection.
Despite these limitations, our findings provide important comparative insights into the diagnostic performance of gross examination and real-time PCR for Eimeria detection. Mixed infections involving multiple Eimeria species present significant diagnostic challenges for conventional methods. They complicate lesion scoring, obscure subclinical lesions suppressed by anticoccidial drugs, and hinder the identification of distinct Eimeria subpopulations (Morgan et al., 2009; Sun et al., 2009; Vrba et al., 2010; Shirzad et al., 2011). Although newer technologies—such as probe-based quantitative PCR (qPCR), loop-mediated isothermal amplification (LAMP), and next-generation sequencing—have emerged, real-time PCR remains more economically viable in several countries (Arya et al., 2005; Pereira-Gómez et al., 2021) In the context of Thailand, the widespread use of advanced molecular diagnostics is often constrained by infrastructural, financial, and technical limitations. As such, real-time PCR serves as a feasible and scalable diagnostic option, effectively balancing diagnostic accuracy with the realities of local resource availability and laboratory capabilities.
In conclusion, this study successfully developed and validated a SYBR Green-based real-time PCR assay for the identification of Eimeria species in the poultry industry in Thailand. The assay demonstrated high sensitivity, specificity, and reproducibility. Distinct Tm profiles enabled reliable differentiation of E. acervulina, E. brunetti, E. maxima, E. necatrix, and E. tenella. Field sample analysis revealed a high prevalence of mixed Eimeria infections in commercial broiler farms, with E. tenella, E. acervulina, and E. maxima as the predominant species, whereas E. brunetti and E. necatrix were not detected. The molecular assay proved significantly more sensitive than gross examination, identifying infections that would have been missed using traditional methods. This increased sensitivity has important implications for coccidiosis surveillance, as subclinical infections—often undetected by gross examination alone—can still lead to considerable production losses. The frequent occurrence of mixed Eimeria infections in medicated broiler farms, despite the use of anticoccidial shuttle programs, highlights the ongoing challenge of effective coccidiosis control in commercial poultry operations. Although real-time PCR is effective for Eimeria spp. detection, it exhibits limitations in the precise quantification of absolute Eimeria oocyst numbers. Its precision can be hindered by small sample sizes, PCR inhibitors in fecal samples, and detection of DNA from non-viable parasites, potentially overestimating prevalence.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal studies were approved by Standard of animal research, Research and development office, Prince of Songkla University (ethic document no. MHESI 68014/1779). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
BC: Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. SP: Data curation, Supervision, Software, Validation, Writing – review & editing. KL: Investigation, Methodology, Writing – review & editing. PP: Investigation, Methodology, Writing – review & editing. AS: Investigation, Methodology, Writing – review & editing. AL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by an academic service fund from Huvepharma Thailand Ltd. to Faculty of Veterinary Science, Prince of Songkla University (MorOr117.1.7/64-02). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
We thank to the poultry companies, and the small-scale poultry farm owners for kindly providing the animal samples and sharing knowledge and information. Thanks to HIPRA company, Thailand for the positive control in this study. Thanks to Dr. Niroj Kijphakapanith for helping with sample collection. We gratefully thank Dr. Sumeth Sapchukun who greatly supported us during this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2025.1533577/full#supplementary-material
Supplementary Figure 1 | Primer annealing time and DNA band pictures.
Supplementary Table 1 | Gross lesion and real-time results table.
Supplementary Table 2 | The Cq value and Tm of 5 Eimeria spp.
References
Abbas R. Z., Iqbal Z., Blake D., Khan M. N., and Saleemi M. K. (2011). Anticoccidial drug resistance in fowl coccidia: the state of play revisited. J. World’s Poult. Sci. 67, 337–349. doi: 10.1017/S004393391100033X
Arya M., Shergill I. S., Williamson M., Gommersall L., Arya N., and Patel H. R. (2005). Basic principles of real-time quantitative PCR. Expert Rev. Mol. Diagn. 5, 209–219. doi: 10.1586/14737159.5.2.209
Blake D. P., Hesketh P., Archer A., Shirley M. W., and Smith A. L. (2006). Eimeria maxima: the influence of host genotype on parasite reproduction as revealed by quantitative real-time PCR. Int. J. Parasitol. 36, 97–105. doi: 10.1016/j.ijpara.2005.09.011
Blake D. P., Knox J., Dehaeck B., Huntington B., Rathinam T., Ravipati V., et al. (2020). Re-calculating the cost of coccidiosis in chickens. Vet. Res. 51, 115. doi: 10.1186/s13567-020-00837-2
Blake D. P., Qin Z., Cai J., and Smith A. L. (2008). Development and validation of real-time polymerase chain reaction assays specific to four species of Eimeria. Avian Pathol. 37, 89–94. doi: 10.1080/03079450701802248
Brown Jordan A., Blake D., Beard J., Beharry A., Serrette L., Soleyn A., et al. (2018). Molecular identification of eimeria species in broiler chickens in Trinidad, West Indies. Vet. Sci. 5, 12. doi: 10.3390/vetsci5010012
Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Bustin S. and Huggett J. (2017). qPCR primer design revisited. Biomol. Detect. Quantif. 14, 19–28. doi: 10.1016/j.bdq.2017.11.001
Carvalho F. S., Wenceslau A. A., Teixeira M., and Albuquerque G. R. (2011a). Molecular diagnosis of Eimeria species affecting naturally infected Gallus gallus. Genet. Mol. Res. 10, 996–1005. doi: 10.4238/vol10-2gmr1043
Carvalho F. S., Wenceslau A. A., Teixeira M., Matos carneiro J. A., Melo A. D., and Albuquerque G. R. (2011b). Diagnosis of Eimeria species using traditional and molecular methods in field studies. Vet. Parasitol. 176, 95–100. doi: 10.1016/j.vetpar.2010.11.015
Cerventes H. M., Mcdougald L. R., and Jerkins M. C. (2020). “Cocidiosis,” in Diseases of Poultry, 14 ed. Eds. Swayne D. E., Boulianne M., Logue C. M., Mcdougald L. R., David V. N., Suarez L., De Wit S., Grimes T., Johnson D., Kromm M., Prajitno T. Y., Rubinoff I., and Zavala G. (John Wiley & Sons, Inc, NJ United states), 1193–1217.
Cha J. O., Zhao J., Yang M. S., Kim W. I., Cho H. S., Lim C. W., et al. (2018). Oocyst-shedding patterns of three eimeria species in chickens and shedding pattern variation depending on the storage period of eimeria tenella oocysts. J. Parasitol. 104, 18–22. doi: 10.1645/16-132
Chapman H. D. (1984). Drug resistance in avian coccidia (a review). Vet. Parasitol. 15, 11–27. doi: 10.1016/0304-4017(84)90106-7
Chapman H. D. (1986). Isolates of Eimeria tenella: studies on resistance to ionophorous anticoccidial drugs. Res. Vet. Sci. 41, 281–282. doi: 10.1016/S0034-5288(18)30617-9
Conway D. P. and Mckenzie M. E. (2007). Poultry coccidiosis. 3 ed (Ames (IA: Blackwell Publishing Professional).
Cornelissen A. W., Overdulve J. P., and van der Ploeg M. (1984). Determination of nuclear DNA of five eucoccidian parasites, Isospora (Toxoplasma) gondii, Sarcocystis cruzi, Eimeria tenella, E. acervulina and Plasmodium berghei, with special reference to gamontogenesis and meiosis in I. (T.) gondii. Parasitology 88, 531–553. doi: 10.1017/s0031182000054792
Dalloul R. A. and Lillehoj H. S. (2006). Poultry coccidiosis: recent advancements in control measures and vaccine development. Expert Rev. Vaccines 5, 143–163. doi: 10.1586/14760584.5.1.143
Del Cacho E., Gallego M., Lillehoj H. S., Quilez J., Lillehoj E. P., and Sanchez-acedo C. (2016). Induction of protective immunity against experimental Eimeria tenella infection using serum exosomes. Vet. Parasitol. 224, 1–6. doi: 10.1016/j.vetpar.2016.04.043
Dorne J. L., Fernandez-cruz M. L., Bertelsen U., Renshaw D. W., Peltonen K., Anadon A., et al. (2013). Risk assessment of coccidostatics during feed cross-contamination: animal and human health aspects. Toxicol. Appl. Pharmacol. 270, 196–208. doi: 10.1016/j.taap.2010.12.014
Geng T., Ye C., Lei Z., Shen B., Fang R., Hu M., et al. (2021). Prevalence of Eimeria parasites in the Hubei and Henan provinces of China. Parasitol. Res. 120, 655–663. doi: 10.1007/s00436-020-07010-w
Guy R. A., Xiao C., and Horgen P. A. (2004). Real-time PCR assay for detection and genotype differentiation of Giardia lamblia in stool specimens. J. Clin. Microbiol. 42, 3317–3320. doi: 10.1128/JCM.42.7.3317-3320.2004
Gyorke A., Pop L., and Cozma V. (2013). Prevalence and distribution of Eimeria species in broiler chicken farms of different capacities. Parasite 20, 50. doi: 10.1051/parasite/2013052
Hachimi O., Falender R., Davis G., Wafula R. V., Sutton M., Bancroft J., et al. (2024). Evaluation of molecular-based methods for the detection and quantification of Cryptosporidium spp. in wastewater. Sci. Total Environ. 947, 174219. doi: 10.1016/j.scitotenv.2024.174219
Haug A., Gjevre A. G., Thebo P., Mattsson J. G., and Kaldhusdal M. (2008). Coccidial infections in commercial broilers: epidemiological aspects and comparison of Eimeria species identification by morphometric and polymerase chain reaction techniques. Avian Pathol. 37, 161–170. doi: 10.1080/03079450801915130
Haug A., Thebo P., and Mattsson J. G. (2007). A simplified protocol for molecular identification of Eimeria species in field samples. Vet. Parasitol. 146, 35–45. doi: 10.1016/j.vetpar.2006.12.015
Jaramillo-Ortiz J. M., Burrell C., Adeyemi O., Werling D., and Blake D. P. (2023). First detection and characterisation of Eimeria zaria in European chickens. Vet. Parasitol. 324, 110068. doi: 10.1016/j.vetpar.2023.110068
Johnson J. and Reid W. M. (1970). Anticoccidial drugs: lesion scoring techniques in battery and floor-pen experiments with chickens. Exp. Parasitol. 28, 30–36. doi: 10.1016/0014-4894(70)90063-9
Kadykalo S., Roberts T., Thompson M., Wilson J., Lang M., and Espeisse O. (2018). The value of anticoccidials for sustainable global poultry production. Int. J. Antimicrob. Agents 51, 304–310. doi: 10.1016/j.ijantimicag.2017.09.004
Kawahara F., Taira K., Nagai S., Onaga H., Onuma M., and Nunoya T. (2008). Detection of five avian Eimeria species by species-specific real-time polymerase chain reaction assay. Avian Dis. 52, 652–656. doi: 10.1637/8351-050908-Reg.1
Kumar S., Garg R., Banerjee P. S., Ram H., Kundu K., Kumar S., et al. (2015). Genetic diversity within ITS-1 region of Eimeria species infecting chickens of north India. Infect. Genet. Evol. 36, 262–267. doi: 10.1016/j.meegid.2015.09.023
Kumar S., Garg R., Moftah A., Clark E. L., Macdonald S. E., Chaudhry A. S., et al. (2014). An optimised protocol for molecular identification of Eimeria from chickens. Vet. Parasitol. 199, 24–31. doi: 10.1016/j.vetpar.2013.09.026
Lan L. H., Sun B. B., Zuo B. X., Chen X. Q., and Du A. F. (2017). Prevalence and drug resistance of avian Eimeria species in broiler chicken farms of Zhejiang province, China. Poult. Sci. 96, 2104–2109. doi: 10.3382/ps/pew499
Lan Y., Xun S., Tamminga S., Williams B. A., Verstegen M. W., and Erdi G. (2004). Real-time PCR detection of lactic acid bacteria in cecal contents of eimeria tenella-lnfected broilers fed soybean oligosaccharides and soluble soybean polysaccharides. Poult. Sci. 83, 1696–1702. doi: 10.1093/ps/83.10.1696
Lew A. E., Anderson G. R., Minchin C. M., Jeston P. J., and Jorgensen W. K. (2003). Inter- and intra-strain variation and PCR detection of the internal transcribed spacer 1 (ITS-1) sequences of Australian isolates of Eimeria species from chickens. Vet. Parasitol. 112, 33–50. doi: 10.1016/s0304-4017(02)00393-x
Lin R. Q., Lillehoj H. S., Lee S. K., Oh S., Panebra A., and Lillehoj E. P. (2017). Vaccination with Eimeria tenella elongation factor-1alpha recombinant protein induces protective immunity against E. tenella and E. maxima infections. Vet. Parasitol. 243, 79–84. doi: 10.1016/j.vetpar.2017.06.003
Morgan J. A., Morris G. M., Wlodek B. M., Byrnes R., Jenner M., Constantinoiu C. C., et al. (2009). Real-time polymerase chain reaction (PCR) assays for the specific detection and quantification of seven Eimeria species that cause coccidiosis in chickens. Mol. Cell Probes 23, 83–89. doi: 10.1016/j.mcp.2008.12.005
Morris G. M. and Gasser R. B. (2006). Biotechnological advances in the diagnosis of avian coccidiosis and the analysis of genetic variation in Eimeria. Biotechnol. Adv. 24, 590–603. doi: 10.1016/j.bioteChadv.2006.06.001
Newcombe R. G. (1998). Improved confidence intervals for the difference between binomial proportions based on paired data. Stat. Med. 17, 2635-2650.
Nolan M. J., Tomley F. M., Kaiser P., and Blake D. P. (2015). Quantitative real-time PCR (qPCR) for replication - Implications for experimental refinement and animal welfare. Parasitol. Int. 64, 464–470. doi: 10.1016/j.parint.2015.06.010
Oliveira U. C., Fraga J. S., Licois D., Pakandl M., and Gruber A. (2011). Development of molecular assays for the identification of the 11 Eimeria species of the domestic rabbit (Oryctolagus cuniculus). Vet. Parasitol. 176, 275–280. doi: 10.1016/j.vetpar.2010.10.054
Opel K. L., Chung D., and Mccord B. R. (2010). A study of PCR inhibition mechanisms using real time PCR. J. Forensic Sci. 55, 25–33. doi: 10.1111/j.1556-4029.2009.01245.x
Pajic M., Todorovic D., Knezevic S., Prunic B., Velhner M., Andric D. O., et al. (2023). Molecular investigation of eimeria species in broiler farms in the province of Vojvodina, Serbia. Life (Basel) 13, 1039. doi: 10.3390/life13041039
Peek H. W. and Landman W. J. (2003). Resistance to anticoccidial drugs of Dutch avian Eimeria spp. field isolates originating from 1996, 1999 and 2001. Avian Pathol. 32, 391–401. doi: 10.1080/0307945031000121149
Peek H. W. and Landman W. J. (2006). Higher incidence of Eimeria spp. field isolates sensitive for diclazuril and monensin associated with the use of live coccidiosis vaccination with paracox-5 in broiler farms. Avian Dis. 50, 434–439. doi: 10.1637/7486-121205R.1
Peek H. W. and Landman W. J. (2011). Coccidiosis in poultry: anticoccidial products, vaccines and other prevention strategies. Vet. Q 31, 143–161. doi: 10.1080/01652176.2011.605247
Pereira-Gómez M., Fajardo Á., Echeverría N., López-tort F., Perbolianachis P., Costábile A., et al. (2021). Evaluation of SYBR Green real time PCR for detecting SARS-CoV-2 from clinical samples. J. Virol. Methods 289, 114035. doi: 10.1016/j.jviromet.2020.114035
Prakashbabu B. C., Thenmozhi V., Limon G., Kundu K., Kumar S., Garg R., et al. (2017). Species occurrence varies between geographic regions and poultry production systems and may influence parasite genetic diversity. Vet. Parasitol. 233, 62–72. doi: 10.1016/j.vetpar.2016.12.003
Raj G. D., Aarthi S., Selvabharathi R., Raman M., Blake D. P., and Tomley F. M. (2013). Real-time PCR-based quantification of Eimeria genomes: a method to outweigh underestimation of genome numbers due to PCR inhibition. Avian Pathol. 42, 304–308. doi: 10.1080/03079457.2013.790531
Reginato C. Z., Bräunig P., Portella L. P., Mortari A. P. G., Minuzzi C. E., Sangioni L. A., et al. (2020). DNA extraction methods for molecular detection of Eimeria spp. in cattle and sheep. Pesquisa Vet. Bras. 40, 514–518. doi: 10.1590/1678-5150-Pvb-6625
Reid A. J., Blake D. P., Ansari H. R., Billington K., Browne H. P., Bryant J., et al. (2014). Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res. 24, 1676–1685. doi: 10.1101/gr.168955.113
Ririe K. M., Rasmussen R. P., and Wittwer C. T. (1997). Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245, 154–160. doi: 10.1006/abio.1996.9916
Santalucia J. Jr. (1998). A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 95, 1460–1465. doi: 10.1073/pnas.95.4.1460
Schnitzler B. E., Thebo P. L., Mattsson J. G., Tomley F. M., and Shirley M. W. (1998). Development of a diagnostic PCR assay for the detection and discrimination of four pathogenic.Eimeria species of the chicken. Avian Pathol. 27, 490–497. doi: 10.1080/03079459808419373
Schnitzler B. E., Thebo P. L., Tomley F. M., Uggla A., and Shirley M. W. (1999). PCR identification of chicken Eimeria: a simplified read-out. Avian Pathol. 28, 89–93. doi: 10.1080/03079459995091
Schrader C., Schielke A., Ellerbroek L., and Johne R. (2012). PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 113, 1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x
Scipioni A., Mauroy A., Ziant D., Saegerman C., and Thiry E. (2008). A SYBR Green RT- PCR assay in single tube to detect human and bovine noroviruses and control for inhibition. Virol. J. 5, 94. doi: 10.1186/1743-422X-5-94
Shchyolkina A. K., Borisova O. F., Livshits M. A., Pozmogova G. E., Chernov B. K., KlemenT R., et al. (2000). Parallel-stranded DNA with mixed AT/GC composition: role of trans G.C base pairs in sequence dependent helical stability. Biochemistry 39, 10034–10044. doi: 10.1021/bi9913909
Shirzad M. R., Seifi S., Gheisari H. R., Hachesoo B. A., Habibi H., and Bujmehrani H. (2011). Prevalence and risk factors for subclinical coccidiosis in broiler chicken farms in Mazandaran province, Iran. Trop. Anim. Health Prod. 43, 1601–1604. doi: 10.1007/s11250-011-9876-3
Sun X. M., Pang W., Jia T., Yan W. C., He G., Hao L. L., et al. (2009). Prevalence of Eimeria species in broilers with subclinical signs from fifty farms. Avian Dis. 53, 301–305. doi: 10.1637/8379-061708-Resnote.1
Sun H. C., Su X. Y., Fu Y., Hao L. L., Zhou W., Zhou, et al. (2023). Pathogenicity and drug resistance of the isolate from Yiwu, Zhejiang province, eastern China. Poult. Sci. 102, 102845. doi: 10.1016/j.psj.2023.102845
Vrba V., Blake D. P., and Poplstein M. (2010). Quantitative real-time PCR assays for detection and quantification of all seven Eimeria species that infect the chicken. Vet. Parasitol. 174, 183–190. doi: 10.1016/j.vetpar.2010.09.006
Williams R. B. (2001). Quantification of the crowding effect during infections with the seven Eimeria species of the domesticated fowl: its importance for experimental designs and the production of oocyst stocks. Int. J. Parasitol. 31, 1056–1069. doi: 10.1016/s0020-7519(01)00235-1
Williams R. B. (2005). Intercurrent coccidiosis and necrotic enteritis of chickens: rational, integrated disease management by maintenance of gut integrity. Avian Pathol. 34, 159–180. doi: 10.1080/03079450500112195
Keywords: broiler chicken, coccidiosis, Eimeria species, real-time polymerase chain reaction, SYBR Green, Thailand
Citation: Chanchayanon B, Pakpinyo S, Limpavithayakul K, Promvijit P, Sriprad A and Limsatanun A (2025) Identification of five Eimeria species in broiler farms in southern Thailand using SYBR Green-based real-time polymerase chain reaction. Front. Anim. Sci. 6:1533577. doi: 10.3389/fanim.2025.1533577
Received: 24 November 2024; Accepted: 09 June 2025;
Published: 10 July 2025.
Edited by:
Virginia Marugan-Hernandez, Royal Veterinary College (RVC), United KingdomReviewed by:
Mariela Lujan Tomazic, INTA-CONICET, ArgentinaKelsilandia Aguiar Martins, Royal Veterinary College (RVC), United Kingdom
Shan Randima Nawarathne, Chungnam National University, Republic of Korea
Copyright © 2025 Chanchayanon, Pakpinyo, Limpavithayakul, Promvijit, Sriprad and Limsatanun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arithat Limsatanun, YXJpdGhhdC5sQHBzdS5hYy50aA==
 Baramee Chanchayanon
Baramee Chanchayanon Somsak Pakpinyo
Somsak Pakpinyo Kriengwich Limpavithayakul
Kriengwich Limpavithayakul Promwit Promvijit1
Promwit Promvijit1 Arithat Limsatanun
Arithat Limsatanun