- 1School of Agriculture and Food Systems, West Virginia University, Morgantown, WV, United States
- 2School of Agriculture and Human Ecology, Tennessee Technological University, Cookeville, TN, United States
- 3Department of Pediatrics, University of California, San Francisco, Oakland, CA, United States
We examined the effects of a blend of live Saccharomyces cerevisiae, multiple probiotic bacteria, and their fermentation products on the whole blood transcriptome of newly weaned beef steers during a 56-d receiving period. Forty newly weaned Angus crossbred steers (12-h postweaning; 217 ± 4.6 kg of body weight [BW]; 202 ± 4 d of age) from three different sources were stratified by BW and randomly assigned to one of the two treatments: 1) basal diet with no additive (CON; n = 20), and 2) the basal diet supplemented with 9 g/steer/d of a multi-strain microbial additive (PRO; n = 20). The PRO additive was a blend of S. cerevisiae and the fermentation products of Enterococcus faecium, Bacillus licheniformis, B. subtilis, Lactobacillus animalis, and Propionibacterium freudenreichii. On day 56, 10 mL of blood was collected from 10 randomly selected beef steers from each treatment group prior to morning feeding. Total RNA was isolated from the whole blood samples for the determination of gene expression profiles. Differentially expressed genes (DEGs) were identified using a false discovery rate (FDR) ≤ 0.10. A total of 41 DEGs were detected; 21 genes, including TLR10, GPR183, LGR4, and FCRL1, were upregulated in steers fed the PRO additive compared to CON, while 20 genes, such as C3, DDIT4, and ADCY8 were downregulated. Gene ontology analysis of the DEGs revealed the enrichment (FDR< 0.05) of pathways related to positive regulation of inflammatory response, regulation of cytokine secretion, positive regulation of defense response, and positive regulation of response to external stimuli in beef steers fed PRO additive. No significant differences (P > 0.05) in growth performance (BW, DMI, or ADG) were observed between CON and PRO steers. In conclusion, this study revealed that beef steers fed the PRO additive exhibited differential expression of genes related to immune function and inflammatory response, suggesting an effect on immunity and stress resilience. These findings highlight the potential of multi-strain direct-fed microbials as a nutritional strategy to support immune health, resilience to stress, and overall welfare in beef cattle during the weaning and receiving period.
Introduction
The transition period following weaning is one of the most critical and stressful phases in the life of beef cattle (Justice et al., 2024). This period is often associated with multiple stressors, including transportation, vaccination, pathogen exposure, dietary changes, and adaptation to a new environment (Lynch et al., 2019; Galyean et al., 2022). If not managed properly, these stressors can reduce dry matter intake, impair growth performance, and negatively impact cattle health. Additionally, the incidence of bovine respiratory disease (BRD) remains high during the feedlot receiving phase (de Souza et al., 2018; Beenken-Bobb et al., 2023), despite efforts to reduce stress and the implementation of immunization strategies against BRD pathogens (Snowder et al., 2006). Therefore, identifying effective strategies to mitigate these stressors is crucial for enhancing the health and productivity of beef cattle.
Nutrition plays a vital role in animal health, and one promising approach to reducing stress in calves involves the use of direct-fed microbials (DFMs). Direct-fed microbials, which include various microorganisms such as lactic acid-producing and spore-forming bacteria, yeast, and fungi have gained attention for their ability to modulate gut microbiota, enhance nutrient absorption, improve immunocompetence, and reduce morbidity in newly weaned beef cattle (Adeyemi et al., 2019; Guimaraes et al., 2024; Treon et al., 2024). Additionally, microbial fermentation by-products such as short-chain fatty acids (SCFAs) have been shown to provide further benefits by modulating the immune system, reducing inflammation, and improving gut barrier function (Prajapati et al., 2023; Rafique et al., 2023). Recent strategies have focused on combining live microorganisms with their fermentation products to enhance efficacy, although the effectiveness of these approaches can vary depending on factors such as microbial strain selection, dosage, and diet composition (AlZahal et al., 2014; Ogunade et al., 2020; Ban and Guan, 2021). Treon et al. (2024) examined the effects of adding a blend of Saccharomyces cerevisiae, multiple live probiotic bacteria, and their fermentation products on the performance and health of newly weaned beef steers during a 56-d receiving period. The results from Treon et al. (2024) demonstrated that beef steers supplemented with the additive exhibited improved growth performance and reduced stress response, as indicated by a lower white blood cell concentration during the initial days after weaning compared to non-supplemented steers. However, the exact mode of action of this additive remains unknown.
RNA sequencing provides a comprehensive method for analyzing gene expression, offering critical insights into the molecular pathways influenced by DFMs, particularly those involved in immune response, inflammation, nutrient absorption, and stress regulation (Mejia-Garcia et al., 2024; Shemery et al., 2025). Additionally, RNA-seq facilitates the detection of novel gene expression patterns and the identification of biomarkers associated with improved health and productivity in cattle (Wickramasinghe et al., 2014; Perera et al., 2022). Recent applications of RNA-seq in beef cattle have identified differentially expressed immune-related genes in response to weaning (O’Loughlin et al., 2012) and dietary interventions (Wang et al., 2023), further supporting its value for evaluating nutritional strategies. Whole blood serves as a readily accessible and minimally invasive tissue for transcriptomic analysis, providing a practical approach for identifying biomarkers related to immune function and stress responses (Bai et al., 2016). Compared to tissue biopsies, blood sampling enables real-time, on-farm monitoring of physiological changes, allowing for faster assessments of cattle health and the effectiveness of dietary interventions (Melchizedek and Maria, 2023; Zoratto et al., 2024). We hypothesized that dietary supplementation of a multispecies DFM would modulate the gene expression profile, particularly those involved in immune response, inflammation, nutrient absorption, and stress regulation of newly weaned beef steers. Therefore, the objective of this study was to evaluate the effects of dietary supplementation with a blend of live Saccharomyces cerevisiae, multiple probiotic bacteria, and their fermentation products on whole blood transcriptome in newly weaned beef steers during a 56-day receiving period.
Materials and methods
Animals, housing, and feeding
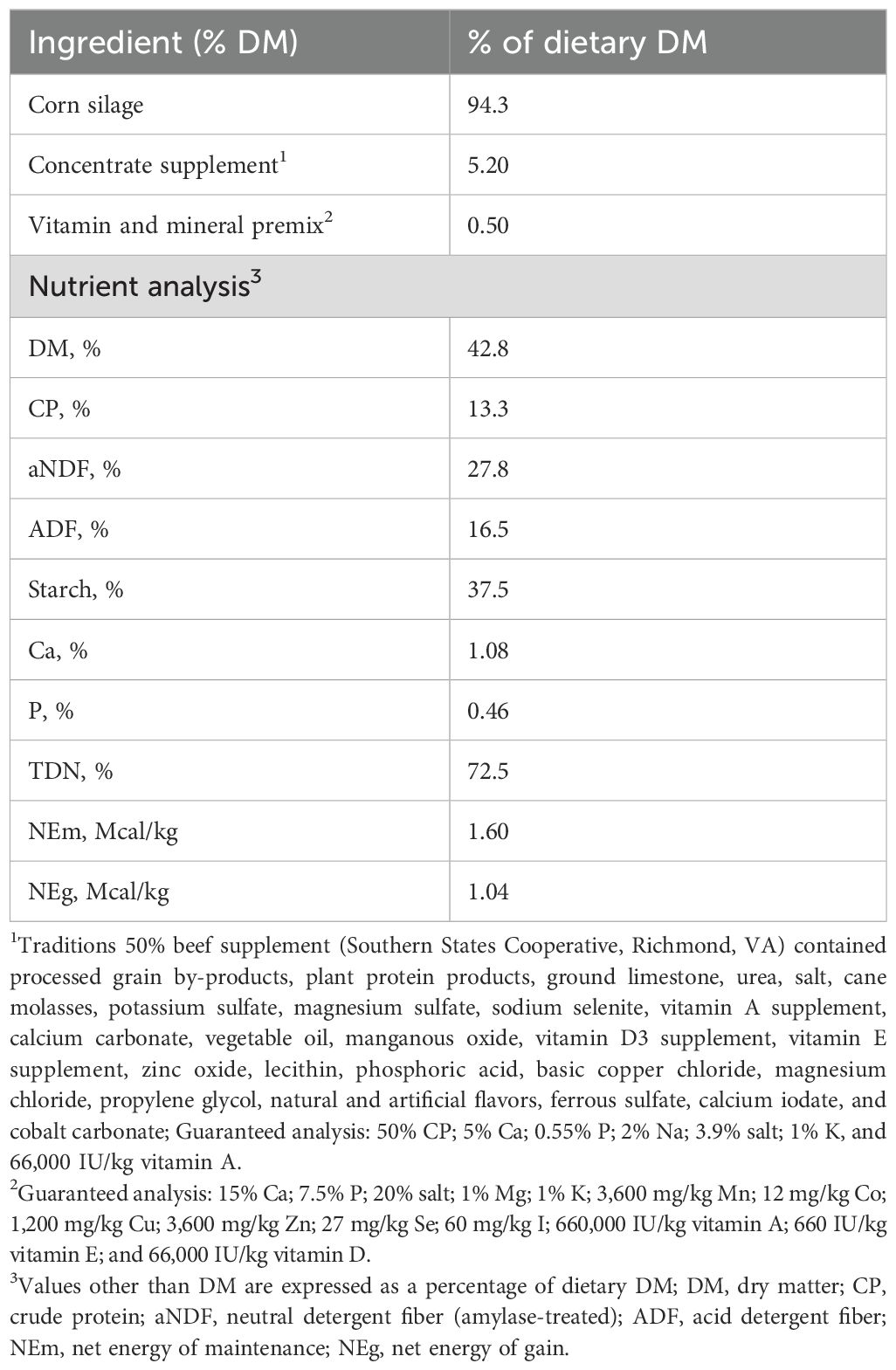
The use of animals in this experiment was approved by the Institutional Animal Care and Use Committees of West Virginia University (protocol number #2108046615.1). Forty newly weaned Angus crossbred beef steers (12-h postweaning; 217 ± 4.6 kg of body weight [BW]; 202 ± 4 d of age) from three different sources (West Virginia Department of Agriculture, Ben Wetzel Farm, Isaiah Smith Farm) were used. Steers were vaccinated 5 months prior to the start of the experiment and received the booster shots 2 weeks prior. The beef steers were transported approximately 160 kilometers to the research feedlot barn and immediately weighed, processed, and placed on a corn silage-based diet on the day of arrival (d 0). Processing included ear tag placement for unique radiofrequency identification and administration of appropriate vaccines, as well as an injection of dewormer. The vaccine protocol included Alpha-7/MB-1 Cattle Vaccine (Boehringer Ingelheim Animal Health, Duluth, GA), Pyramid-5 + Preresponse SQ Cattle Vaccine (Boehringer Ingelheim Animal Health), and the dewormer used was Safeguard Dewormer Suspension (Merck Animal Health, Summit, NJ). Based on d 0 BW, the steers were stratified by BW into two weight blocks. For each weight block, the beef steers were randomly assigned into one of two pens (20 steers per pen) such that each pen had a similar average BW at the beginning of the experiment. Each pen (dimensions = 11.89 × 9.75 m2) was equipped with two GrowSafe intake nodes (GrowSafe Systems Ltd., Airdrie, AB, Canada) to measure individual feed intake. Starting from d 1, the pens were randomly assigned to receive a corn silage-based diet with no additive (CON; 1 pen; n = 20) or a basal diet supplemented with 9 g per head of a PRO additive (PRO; 1 pen; n = 20) for a period of 56 d. The PRO additive (Papillon, Easton, MD) is a blend of live S. cerevisiae (1.41 billion CFU/g), multiple live bacteria (E. faecium, B. licheniformis, B. subtilis, L. animalis, and P. freudenreichii) and their fermentation products (total bacterial count = 120 million CFU/g). The basal diet was fed as a total mixed ration (TMR; Table 1), and the additive was blended into the TMR at a specific percentage, calculated based on the previous day’s average feed intake for each pen (day × intake was utilized to determine the inclusion rate for day ‘x +1’). This procedure ensured that each beef steer in every pen received the required quantity of the additive, amounting to an average of 9 g (12.7 billion CFU of S. cerevisiae and 1.08 billion CFU of total bacteria) of PRO per head per day. To prevent any risk of cross-contamination, pens for the CON and PRO groups were physically separated by barriers and the CON and PRO diets were prepared separately in dedicated feed trucks. The diets were provided ad libitum to the steers, and they had unrestricted access to water.
Body weight and intake measurement
Steers were weighed prior to morning feeding on days 0, 1, and 56. Individual feed intake was recorded using Grow-Safe intake nodes (GrowSafe Systems Ltd., Airdrie, Alberta, Canada). Average daily gain (ADG) was calculated by subtracting the initial body weight (average of days 0 and 1) from the final body weight on day 56 and dividing by the total duration of the experiment (56 days). Total mixed ration samples were collected daily, weighed, and oven-dried at 55°C for 72 hours to determine dry matter (DM) content. Dried TMR subsamples were composited within treatment, ground using a Wiley mill (Arthur H. Thomas Co., Philadelphia, PA) to pass through a 2-mm sieve, and analyzed for nutritional composition at a commercial laboratory (Dairy One Forage Laboratory, Ithaca, NY).
Blood sample collection and RNA extraction
On day 56, 10 mL of blood were collected from 10 randomly selected beef steers per treatment before morning feeding (after overnight feed withdrawal). The blood samples were taken from the jugular vein into tubes containing sodium heparin. Subsequently, subsamples of 500 µL each were immediately transferred into RNA-protect tubes (Cat. No. 76,554; Qiagen) per sample. RNA-protect tubes contained a reagent capable of lysing blood cells and stabilizing intracellular RNA. The samples were stored at -80 °C for later analyses. The use of 10 animals per treatment group for RNA-seq is consistent with established practices in livestock transcriptomics, where 6 to 10 biological replicates per group are commonly used to achieve acceptable statistical power for detecting differential gene expression, while balancing practical and economic limitations (Schurch et al., 2016; Conesa et al., 2016).
Total RNA was isolated from the whole blood samples using RNeasy Micro Kit (Cat No: 130 74,004; Qiagen, Germantown, MD), following the manufacturer’s instructions. RNA concentration was measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, 132 MA). RNA samples were screened for quality using the Agilent 2100 Bioanalyzer (Agilent Technologies; Santa Clara, CA), ensuring all samples exhibited satisfactory RNA integrity (> 8), and were used to construct sequencing libraries (n = 10 beef steers/treatment).To enhance transcriptome coverage and reduce interference from highly abundant globin transcripts, globin mRNA was depleted before library preparation using a commercial globin reduction protocol (Choi et al., 2014). Library preparation was performed by Novogene (Sacramento, CA, USA), and 150-bp pair-ended sequencing was conducted on an Illumina 1.9 HiSeq, resulting in ∼19–22 million clean reads per sample. The raw data were filtered out for low-quality reads and adapters. All samples had an effective rate greater than 93.5% (Clean reads/Raw reads × 100). Samples read mapped to their input read produced an average of 97%.
RNA-seq data analysis and quality control
All the growth performance data, including initial and final BW, DMI, and ADG were analyzed using the MIXED procedure of SAS version 9.4 (SAS Institute Inc., Cary, NC). Each animal was considered the experimental unit. The statistical model included the treatment as a fixed effect (CON vs PRO) and the random effect of animals. The initial BW was used as a covariate for the final BW. Comparisons of least square means were performed using Tukey test with significance set at P ≤ 0.05. All paired-reads FASTQ files were quality-checked using Fastqc (version 0.12.1). This step produced sequencing files with an average per base Phred score > 35, which indicates a high-quality base call. Cleaned reads were aligned to the Bos taurus genome (GCF_002263795.3_ARS-UCD2.0_genomic.fna) using STAR, Version 2.7 and the resulting sam files were converted to bam format using samtools v1.22 Reads mapping to the genes (GCF_002263795.3_ARS-UCD2.0_genomic.fna) were counted using featureCounts v1.4.6-p1 (Liao et al., 2014). Differential expression analysis was performed using PyDESeq2 in Python. Gene expression changes were visualized by Volcano plots using SRplot. Phenotypes of the differentially expressed genes identified were obtained from the Ensembl BioMart tool and filtered based on gene ID, gene name, and phenotype description. The expression with adjusted P-value ≤ 0.10 were considered to be differentially expressed. This FDR threshold was selected post hoc to balance the discovery of meaningful biological effects with statistical rigor. No additional log2 fold-change cutoffs were applied. Batch effect correction was not performed as sample processing was consistent and sequencing was conducted in a single batch. Gene ontology (GO) terms and pathways analyses of DEGs were performed using a web-based gene-ontology software (http://www.geneontology.org) as described by Ashburner et al., 2000. Significantly enriched pathways were catalogued using false discovery rate-adjusted P-values (FDR ≤ 0.05) (Benjamini and Hochberg, 1995).
Results
Growth performance
The effects of PRO supplementation on the growth performance of the beef steers are presented in Table 2. There were no differences (P > 0.05) between the two treatments for initial BW, final BW, ADG, and DMI.
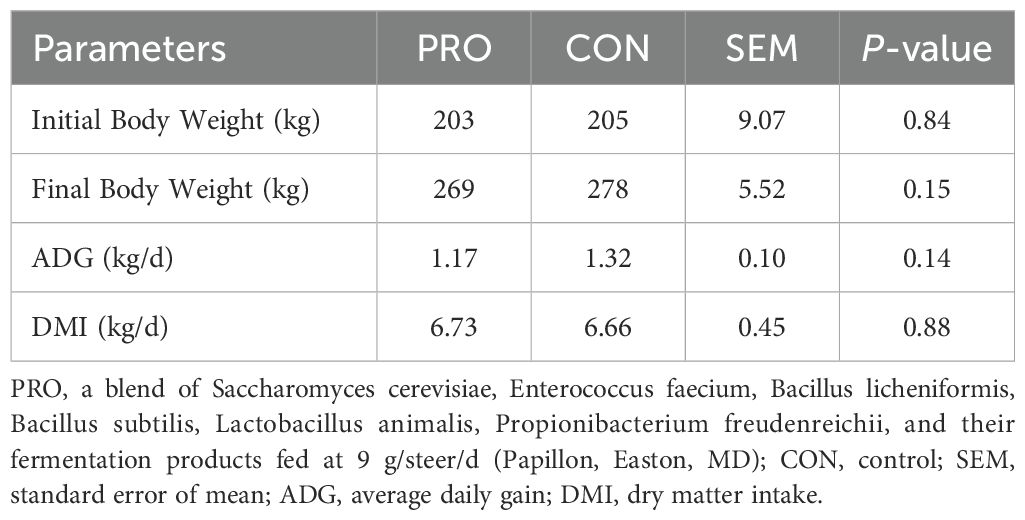
Table 2. Growth performance and dry matter intake of beef steers fed diet supplemented with a blend of Saccharomyces cerevisiae and multiple live probiotic bacteria on day 56 of the receiving period.
Sequencing coverage and read counts
After quality control and data filtering, 79,336,266 ± 5,789,255 clean reads per sample were obtained. For each sample, 95.55% to 97.83% of the reads were uniquely mapped to the Bos taurus reference genome, indicating sufficient coverage (Supplementary Table 1).
Differentially expressed genes and gene ontology analysis
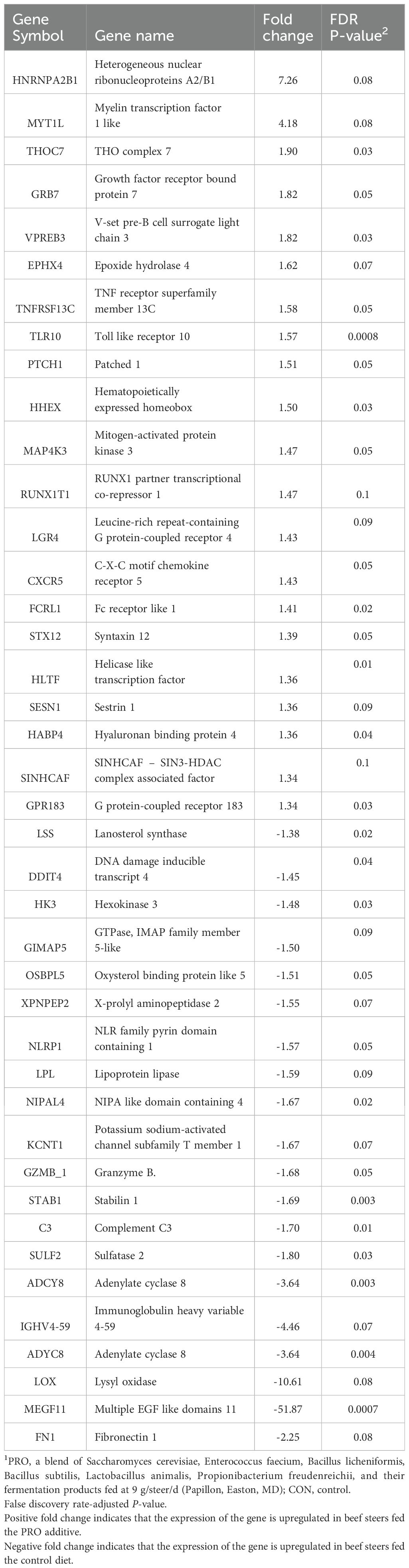
A total of 41 DEGs were detected (FDR ≤ 0.10) between the two groups (Table 3), with 20 downregulated and 21 upregulated in beef steers fed the PRO additive (Figure 1). Several downregulated genes, including C3 (FDR = 0.01; FC = -1.70), DDIT4 (FDR = 0.04; FC = -1.45), and STAB1 (FDR = 0.003; FC = -1.69), are associated with immune and inflammatory responses. In contrast, upregulated genes such as TLR10 (FDR = 0.0008; FC = 1.57), MYTIL (FDR = 0.08; FC = 4.18), GPR183 (FDR = 0.03; FC = 1.34), and LGR4 (FDR = 0.09; FC = 1.43) are involved in cytokine regulation and toll-like receptor signaling. Functional analysis and GO enrichment of these DEGs revealed significant overrepresentation of biological processes such as positive regulation of inflammatory response, cytokine secretion, response to external biotic stimuli, and lipid localization (Figure 2).
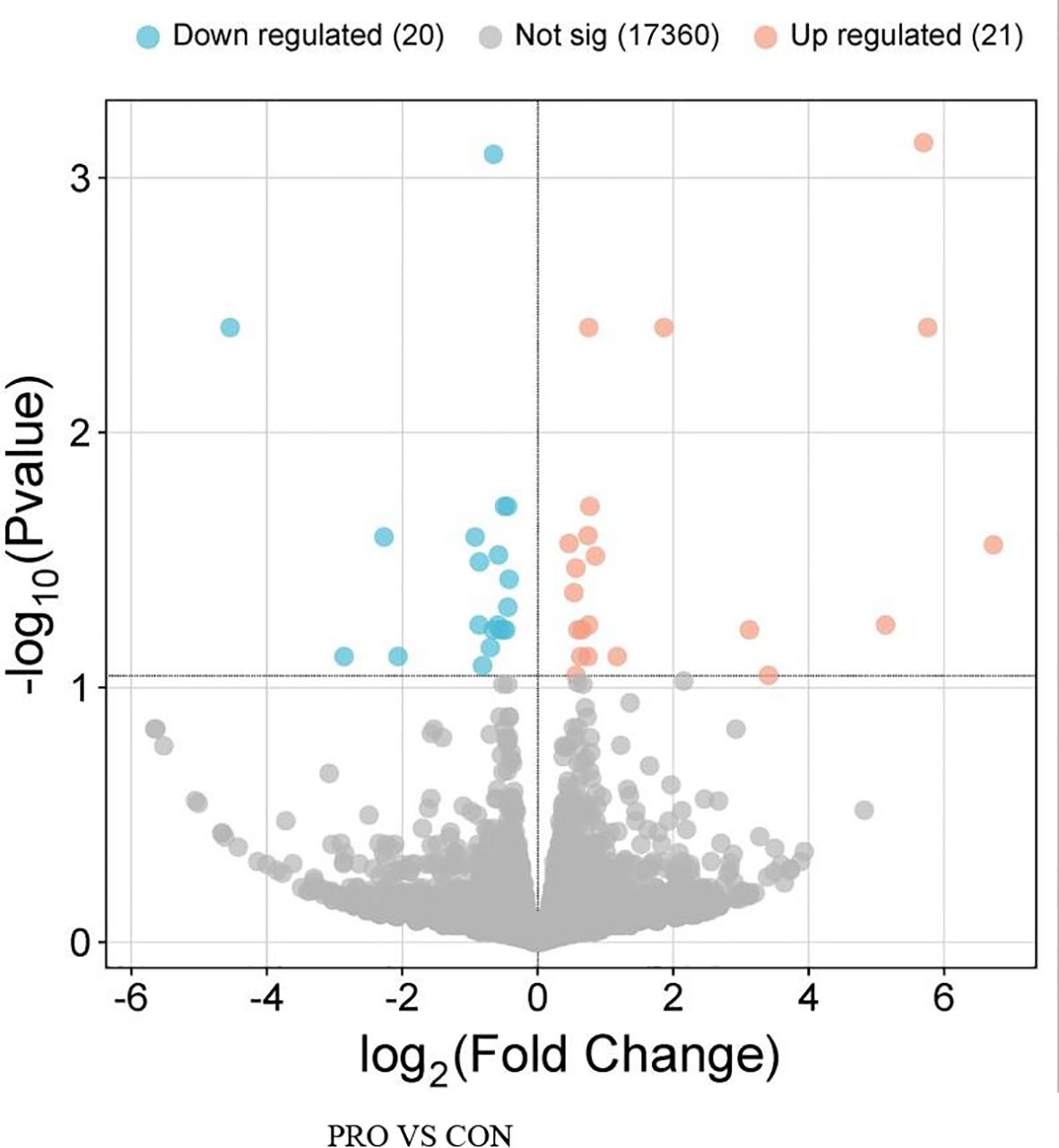
Figure 1. Volcano plot showing the number of differentially expressed genes in beef steers fed PRO, a blend of Saccharomyces cerevisiae, Enterococcus faecium, Bacillus licheniformis, Bacillus subtilis, Lactobacillus animalis, Propionibacterium freudenreichii, and their fermentation products fed at 9 g/steer/d (Papillon, Easton, MD) compared with CON, control.
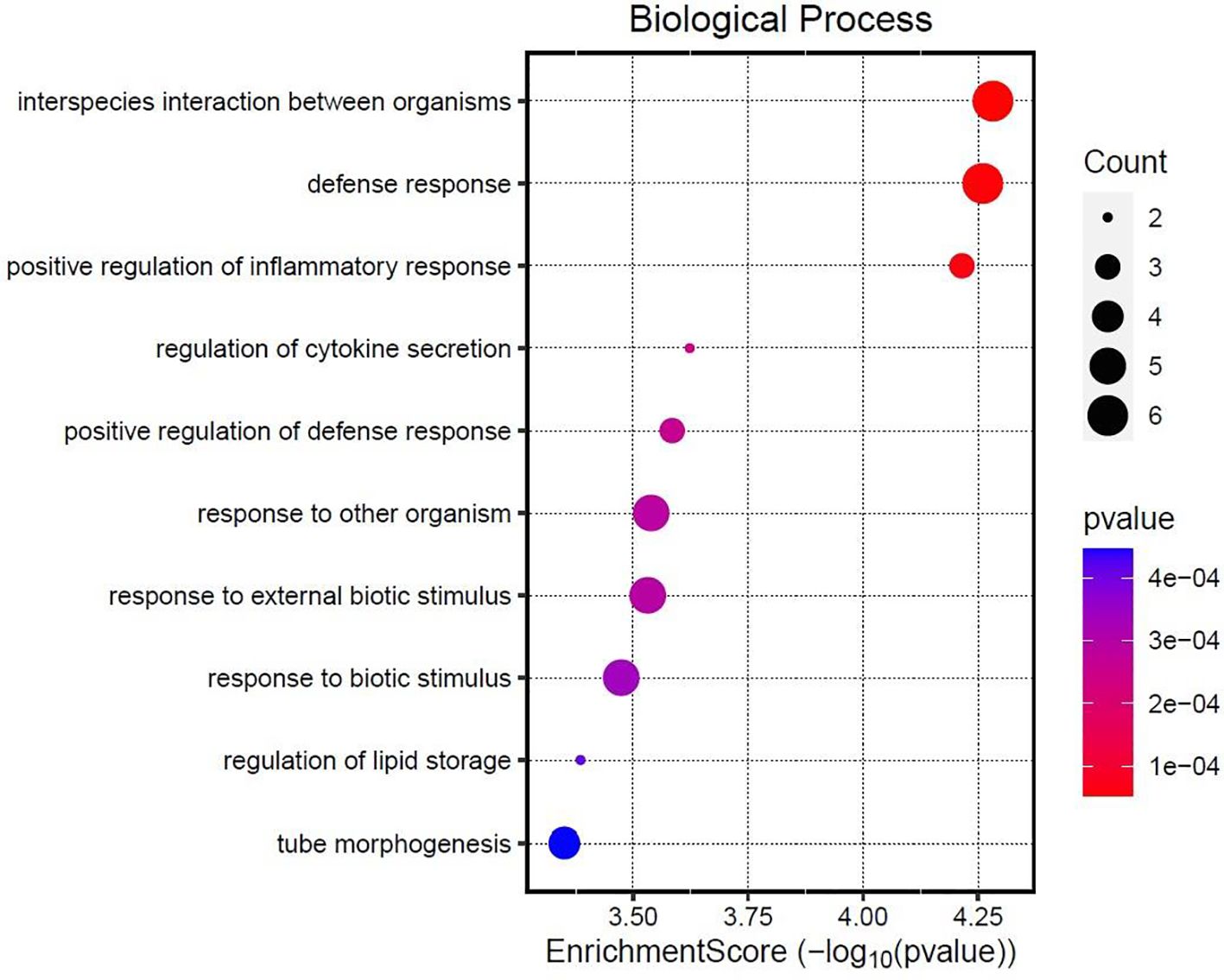
Figure 2. Enriched biological processes highlighted in the over-representation analysis of differentially expressed genes identified in PRO, a blend of Saccharomyces cerevisiae, Enterococcus faecium, Bacillus licheniformis, Bacillus subtilis, Lactobacillus animalis, Propionibacterium freudenreichii, and their fermentation products fed at 9 g/steer/d (Papillon, Easton, MD) compared with CON, control.
Discussion
Growth performance metrics such as BW, DMI, and ADG are critical indicators of how dietary interventions affect the health and productivity of beef steers. In the present study, supplementation with the PRO additive did not result in statistically significant differences in final BW, DMI, or ADG between treatment groups. These findings are consistent with several previous studies where supplementation with DFMs or yeast-based products showed limited or no effects on performance during the receiving period, particularly in lower-stress environments (Deters et al., 2018; Hall et al., 2018).
The immune system plays a critical role in maintaining health and resilience in newly weaned beef steers, especially during stressful periods like weaning, often associated with increased susceptibility to infections and immune dysregulation (Munteanu and Schwartz, 2022). Dietary interventions, such as supplementation with DFMs have shown the potential to modulate immune responses and enhance animal health by influencing key immune pathways (Ban and Guan, 2021). The microbial additive used in this study contains several microbes, including S. cerevisiae, lactic acid bacteria, and their fermentation products, known to directly or indirectly interact with components of the immune system.
Yeast (Saccharomyces cerevisiae) and its derivatives are widely recognized for their ability to modulate the immune system in farm animals (Maturana et al., 2023). Saccharomyces cerevisiae and its cell wall components, such as β-glucans and mannans, interact with immune cells via pattern recognition receptors (PRRs) like Dectin-1 and TLR2, thereby activating innate immune responses (Lesage and Bussey, 2006; Ogunade et al., 2021). These interactions stimulate the production of pro-inflammatory cytokines (e.g., IL-6, TNF-α) and enhance the phagocytic activity of macrophages and neutrophils (Levin, 2005). Additionally, yeast cell wall polysaccharides improve gut barrier integrity by promoting the production of tight junction proteins, reducing intestinal permeability, and supporting gut-associated lymphoid tissue (GALT), a central component of the immune system (Zhou et al., 2023). Studies have shown that supplementing animals with S. cerevisiae enhances immune cell proliferation and function, particularly increasing the activity of macrophages, dendritic cells, and T lymphocytes (Chou et al., 2017; Mahmoud et al., 2020). For example, dietary supplementation with a hydrolyzed mannan- and glucan-rich yeast fraction in newly received feedlot cattle improved the immune response of beef steers (Pukrop et al., 2018). Furthermore, yeast supplementation has been associated with improved gut microbiota composition, fostering a beneficial microbial environment that supports both local and systemic immune responses (Pascual et al., 2020).
Lactic acid-producing bacteria (LAB), including Lactobacillus and Bifidobacterium species, along with their fermentation products, also play a crucial role in immune modulation and inflammation control (Fijan, 2014). Lactic acid-producing bacteria produce organic acids, such as lactic acid and acetic acid, which lower the intestinal pH, creating unfavorable conditions for pathogenic bacteria while promoting the growth of beneficial microbes (de Vrese and Schrezenmeir, 2008; Fidan et al., 2022). This shift in gut microbiota composition strengthens the intestinal barrier and enhances mucosal immunity. Moreover, LAB stimulate the production of antimicrobial peptides, cytokines, and immunoglobulins (IgA) in the gut, contributing to infection defense (Hernández-González et al., 2021; Anjana and Tiwari, 2022). Fermentation products from LAB, such as short-chain fatty acids (SCFAs), further promote anti-inflammatory responses by regulating immune cell functions (Fusco et al., 2023). In particular, butyrate has been shown to reduce the production of pro-inflammatory cytokines and promote a regulatory T-cell phenotype, supporting immune homeostasis (Shin et al., 2023). Studies indicate that supplementation with LAB or their fermentation products in livestock improves immune function, reduces inflammation, and enhances resilience to stress-induced immunosuppression (García-Burgos et al., 2020; Lin et al., 2022).
The results of our study revealed the differential expression of five genes (TLR10, LGR4, GPR183, C3, and DDIT4) from critical immune pathways, including the positive regulation of inflammatory response, regulation of cytokine secretion, positive regulation of defense response, and response to external biotic stimuli in beef steers fed PRO feed additive compared with CON (Table 4). The expressions of TLR10, GPR183, and LGR4 were upregulated, while the expressions of C3 and DDIT4 were downregulated in beef steers fed the PRO feed additive compared with CON. To initiate an effective host defense against microbial pathogens, immune cells must first detect specific molecular patterns from invading microbial pathogens (Li and Wu, 2021). Toll-like receptors (TLRs) are a class of pattern-recognition receptors that play a critical role in recognizing microbial pathogens by detecting pathogen-associated molecular patterns (Janssens and Beyaert, 2003; Duan et al., 2022). Among them, TLR10 is expressed in immune cells such as neutrophils, macrophages, and dendritic cells and has been implicated in modulating innate immune responses through intracellular signaling pathways involved in immune cell recruitment (Balachandran et al., 2015). Additionally, studies have also reported that TLR10 exhibits anti-inflammatory properties (Fore et al., 2020; Mourits et al., 2020; Knez et al., 2023). For instance, Hess et al. (2017) showed that TLR10 acts as an inhibitor of MyD88-dependent and independent pathways. In their experiment, they used monoclonal Abs (activator of TLR10) and observed a reduction in the production of pro-inflammatory cytokines, IL-6, and TNF-α. Thus, the upregulation of this gene in beef steer-fed PRO additive may suggest that the additive may prepare the innate immune system for a more effective response to pathogens and promote a more regulated inflammatory environment.
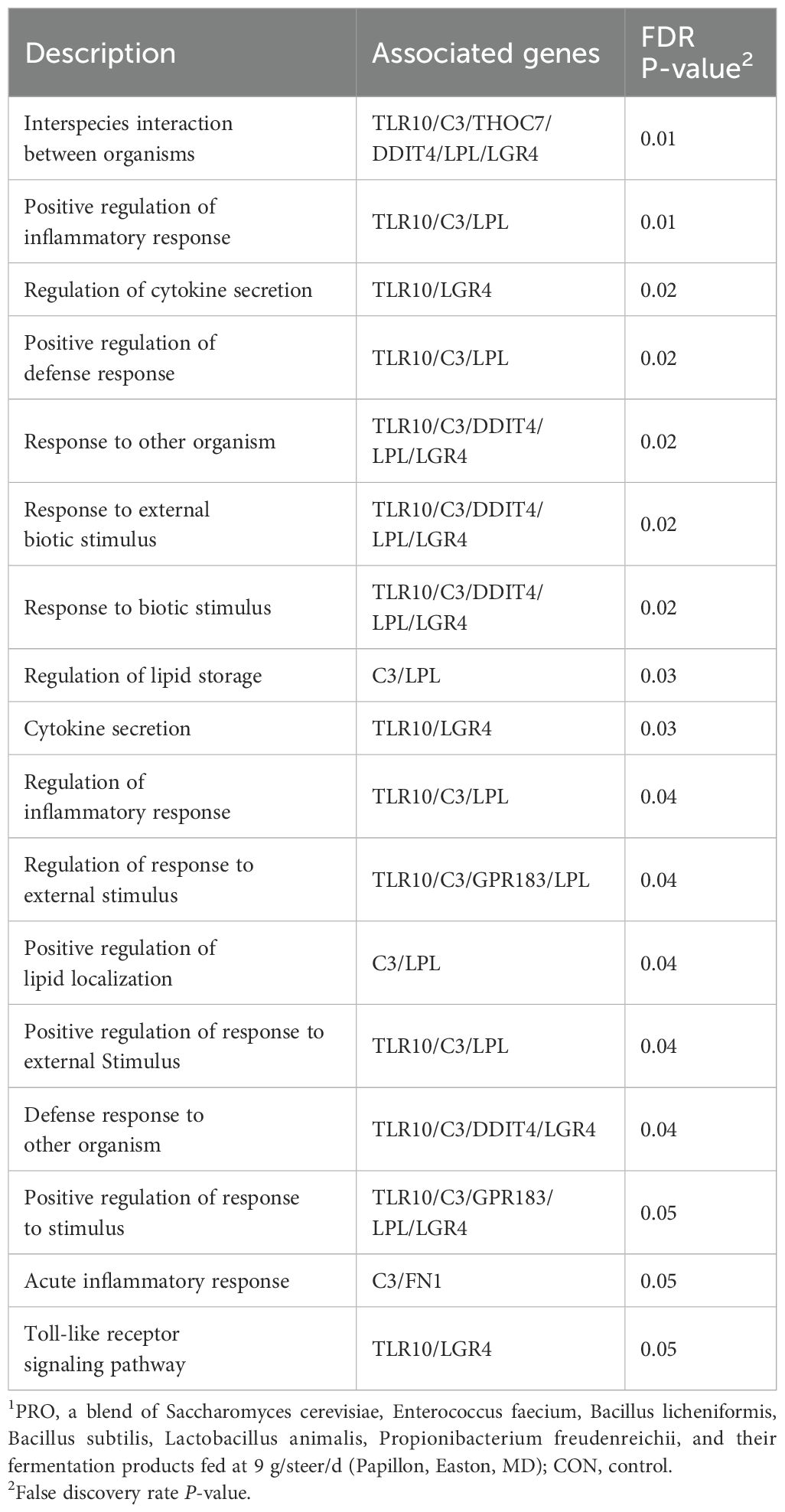
Table 4. Enriched biological processes (FDR ≤ 0.05) highlighted in the over-representation analysis of the differentially expressed genes in PRO compared to CON1.
The ability of immune cells to move to the sites of infection or inflammation is essential for effective defense and clearance of invading pathogens (Alberts et al., 2002). G Protein-Coupled Receptor 183 (GPR183), also known as EBI2, is a G-protein-coupled receptor that plays a key role in directing immune cell migration, particularly for B cells, T cells, and dendritic cells, to the sites of infection or inflammatory response (Barington et al., 2018; Emgård et al., 2018). In mice, it has been found that deficiencies of GPR183 resulted in impaired positioning of T and B cells within secondary lymphoid organs, leading to defects in T cell-dependent immune response (Hannedouche et al., 2011; Bartlett et al., 2020). Similarly, Chu et al. (2018) demonstrated that GPR183 knockout mice show increased susceptibility to Citrobacter rodentium infection, highlighting the receptor's role in intestinal immune defense. In addition to its role in immune cell migration, GPR183 has pro-inflammatory activities, particularly in the intestine (Misselwitz et al., 2021). Leucine-rich repeat-containing G-Protein-Coupled Receptor 4 (LGR4) is known to be enriched in various organs, including the liver, kidney, intestine, bone, reproductive system (ovary, testis, and mammary gland), intestinal tract, pancreas to nervous system cells (Ordaz-Ramos et al., 2021; Zhang et al., 2023). The LGR4 gene has been demonstrated to be one of the few GPCRs upregulated during macrophage M2-type polarization, indicating a potential function in modulating macrophage-mediated immunological responses (Wang et al., 2019). Taken together, the increased expression of TLR10, GPR183, and LGR4 in beef steers fed the supplemental PRO additive suggests that these animals had a better ability than CON for immune monitoring, pathogen detection, and controlled inflammatory responses, which are crucial for maintaining immune homeostasis and resilience to infections in newly weaned beef steers.
The observed downregulation of C3 and DDIT4 in the beef steers fed supplemental PRO additive compared with the CON provides further insight into the immunomodulatory effects of the supplemental diets. Complement Component 3 is a central component of the complement system, which plays a key role in innate immunity activation and inflammation (Peng et al., 2016; Bai et al., 2022). It is an integral part of the complement system and a key molecule of the three complement reaction pathways: the classical pathway (triggered by antibodies), the lectin pathway (triggered by lectin), and the alternative pathway (triggered directly on pathogen surfaces) (Charles et al., 2021). The decreased expression of C3 in beef steers fed supplemental PRO additive could indicate a more controlled inflammatory state, preventing potential tissue damage caused by overactive immune signaling. Additionally, this modulation might indicate the immunomodulatory properties of the supplemental diet, which could help the animals maintain immune homeostasis under stressful conditions. DNA Damage-Inducible Transcript 4 (DDIT4), also known as Regulated in Development and DNA Damage Response 1 (REDD1), is involved in cellular stress responses and acts as a negative regulator of the mTOR pathway, which is important for cell growth and metabolism (Tirado-Hurtado et al., 2018). The downregulation of DDIT4 in beef steers fed the PRO may suggest mitigated stressors or altered stress response pathways, reducing the need for DDIT4's regulatory functions and potentially leading to better overall immune function. By modulating the expression of DDIT4, the additive may influence pathways associated with immune cell resilience, potentially supporting their function under stress conditions commonly experienced by newly weaned calves. However, these gene expression changes are suggestive and do not confirm functional outcomes.
Beyond the genes identified through GO analysis, several additional genes, including MEGF11, STAB1, FCRL1, ADCY8, HHEX, TRAF5, and HLTF, exhibited significant expression changes in beef steers fed the PRO additive compared to CON. Fc Receptor-Like 1, HHEX, TRAF5, and HLTF were up-regulated, while MEGF11, STAB1, and ADCY8 were downregulated in beef steers fed the PRO additive compared to CON. Fc Receptor-like 1 (FCRL1) is located on chromosome 3 of cattle and plays a role in the immune system by encoding proteins that function as receptors on the surface of immune cells (Daëron, 2016; Zhao et al., 2019);. These receptors are known to interact with antibodies and other molecules involved in regulating immune activation and immune function of B cells (Yousefi et al., 2023). An in vitro study revealed that the synaptic accumulation of B cell receptors (BCRs) was significantly impaired in FCRL1-knockout primary B cells. Using total internal reflection fluorescence microscopy, the study demonstrated that FCRL1 plays a critical role in promoting the efficient clustering of BCRs at the immunological synapse during B cell activation, suggesting that FCRL1 acts as a positive regulator in this process (Zhao et al., 2019). Additionally, in mice, BCR ligation in Fcrl1-deficient primary splenic B cells demonstrated impaired proliferation, as evidenced by lower Carboxyfluoresce in succinimidyl ester dilution compared to wild-type (WT) control B cells (DeLuca et al., 2021). Therefore, the upregulation of FCRL1 in beef steers fed the PRO additive suggests that the additive may enhance immune function by promoting more efficient B cell receptor (BCR) clustering and activation. Hematopoietically expressed homeobox (Hhex) is abundantly expressed in natural killer (NK) cells, and its deletion results in significant impairment in lymphoid development, impacting NK cell maturation and functionality (Goh et al., 2020). Natural killer cells are innate lymphocytes best known for their functional efficacy against transformed and virus-infected cells (Mujal et al., 2021). Jackson et al. (2017) showed that Hhex-deficient mice lack mature B cells because of impaired IL-7 signaling, dysregulated expression of cell cycle genes, increased apoptosis, and developmental arrests of early progenitors. Consequently, the upregulation of Hhex in beef steers fed the PRO additive suggests a potential enhancement of NK cell development and functionality, which is crucial for effective innate immune responses.
TNF receptor-associated factor 5 (TRAF5) belongs to the TRAF family, which plays an important role in transducing intracellular signals via TNF receptor superfamily molecules, particularly in regulating immune and inflammatory responses (Nakano et al., 1997). The TRAF5 gene functions as an adaptor protein, facilitating the transmission of signals from TNF receptors and other immune receptors, activating key pathways such as NF-κB and AP-1, both crucial for driving inflammation, immune activation, and cytokine production (Au and Yeh, 2013; Arkee and Bishop, 2020). The upregulation of TRAF5 in beef steers fed the additive suggests an improved ability to mount stronger immune responses, indicating that the additive has a role in promoting a more efficient immune defense mechanism in these animals. Helicase-like Transcription Factor (HLTF) is a DNA helicase that plays a key role in maintaining genomic stability by participating in the repair of damaged DNA, particularly during replication stress (Chavez et al., 2018; Bai et al., 2024). Its upregulation in beef steers fed additives could indicate an enhanced ability of the cells to manage DNA damage and maintain genomic integrity, particularly under stress conditions.
The expressions of MEGF11, STAB1, and ADCY8 were downregulated in response to the dietary supplementation PRO additive as compared with the CON. Multiple EGF-Like Domains Protein 11 (MEGF11) is involved in cell adhesion and neural development, particularly influencing synaptic connectivity (Kay et al., 2012; Ray et al., 2020). Recent studies indicate that knockdown of MEGF11 significantly affects Protein kinase B, mTOR, and NF-κB signaling, as well as the decreased expression of transcription factors like NF-κB p65, CREB, and AP-1 (Chiu et al., 2020). The downregulation of MEGF11 may suggest reduced inflammation, modulated immune responses, and possibly improved overall health and resilience in beef steers fed the PRO additive. Stabilin-1 (STAB1) encodes a transmembrane receptor primarily involved in clearing extracellular molecules such as modified lipids, dead cells, and pathogens (Arias-Alpizar et al., 2021). In fact, STAB1's presence can promote an anti-inflammatory M2 macrophage phenotype, which is associated with tissue repair and resolution of inflammation (Soler Palacios et al., 2020). In the current study, the downregulation of STAB1 may serve as a mechanism to return to homeostatic STAB1 mRNA levels following a transient upregulation. Adenylate cyclase 8 (ADCY8) is a member of the adenylyl cyclase (AC) family that encodes the enzyme adenylyl cyclase 8 (AC8), which plays a critical role in converting ATP to cyclic AMP (Guo et al., 2022; Devasani and Yao, 2022). Cyclic AMP is a secondary messenger that modulates various biological processes, including cellular responses to hormones and stress signals (Yan et al., 2016). In mice, it has been found that over-expression of AC8 in cardiomyocytes increased cellular stress that cardiomyocytes experience during normal physiological aging (Kumar et al., 2024). Thus, the decreased expression of ADCY8 in beef steers fed the supplemental PRO additive, compared to the CON group, may suggest that feeding the supplemental PRO additive alleviated the stress that the CON group continued to experience.
Although PRO additive supplementation did not affect BW, DMI, and ADG in our study, the RNAseq analysis revealed that beef steers fed PRO exhibited increased expression of immune-related genes, suggesting that DFM may enhance immune competence and resilience. While no measurable improvements in performance were observed, these findings support the role of DFM in priming the immune system, potentially better preparing animals for future stressors, such as pathogen exposure and environmental challenges. However, sampling only at day 56 may have missed the acute post-weaning window, when DFMs are typically most active. Future studies should incorporate earlier and multiple sampling points to better capture the temporal dynamics of DFM-induced effects. Additionally, while independent qPCR validation of key differentially expressed genes was not conducted, particularly given that the FDR threshold was relaxed to 0.10, the transcriptome data provide a comprehensive overview of gene expression changes in response to the PRO additive. These findings offer valuable insights into the role of multi-strain DFM supplements in immune modulation and stress resilience in newly weaned beef steers, which can inform future studies aimed at further validating these gene expression patterns.
Conclusion
Our findings demonstrate that dietary supplementation with the PRO additive was associated with modulation of immune-related gene expression in newly weaned beef cattle. The upregulation of critical genes associated with immune recognition, regulation of defense response, and inflammatory regulation indicates an improved capacity for pathogen defense and a more balanced immune response in beef steers receiving the PRO additive. Additionally, the downregulation of certain genes, such as C3, DDIT4, MEGF11, STAB1, and ADCY8, suggests a strategic modulation of inflammatory responses and stress-related pathways, potentially promoting better overall health and recovery during the challenging weaning period. This study adds to the growing body of evidence supporting the use of multi-strain DFM supplements to promote immune health and stress resilience in livestock management. Future studies involving larger cohorts of steers are warranted to confirm these effects and assess the translational potential of PRO additive supplementation in commercial beef production settings.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SAMN48310144.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of West Virginia University (IACUC Protocol Number: 2108046615.1). The study was conducted in accordance with the local legislation and institutional requirements. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AA: Data curation, Conceptualization, Writing – review & editing, Methodology, Investigation, Writing – original draft, Formal Analysis, Visualization. SJ: Investigation, Methodology, Writing – review & editing, Data curation. IM: Data curation, Writing – review & editing. YL: Data curation, Writing – review & editing. TS: Data curation, Writing – review & editing. GT: Data curation, Writing – review & editing. IO: Writing – review & editing, Funding acquisition, Visualization, Resources, Project administration, Validation, Conceptualization, Methodology, Supervision, Data curation. CA: Data curation, Validation, Conceptualization, Supervision, Project administration, Methodology, Writing – review & editing, Resources, Investigation, Formal Analysis, Visualization, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research study was funded by the U.S. Department of Agriculture hatch multi-state regional project W-3010.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2025.1623311/full#supplementary-material
References
Adeyemi J. A., Harmon D. L., Compart D. M. P., and Ogunade I. M. (2019). Effects of a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products in the diet of newly weaned beef steers: growth performance, whole-blood immune gene expression, serum biochemistry, and plasma metabolome1. J. Anim. Sci. 97, 4657–4667. doi: 10.1093/jas/skz308
Alberts B., Johnson A., Lewis J., Raff M., Roberts K., and Walter P. (2002). “Innate immunity,” in Molecular Biology of the Cell, 4th (New York, NY, USA: Garland Science). Available at: https://www.ncbi.nlm.nih.gov/books/NBK26846/.
AlZahal O., McGill H., Kleinberg A., Holliday J. I., Hindrichsen I. K., Duffield T. F., et al. (2014). Use of a direct-fed microbial product as a supplement during the transition period in dairy cattle. J. Dairy Sci. 97, 7102–7114. doi: 10.3168/jds.2014-8248
Anjana and Tiwari S. K. (2022). Bacteriocin-producing probiotic lactic acid bacteria in controlling dysbiosis of the gut microbiota. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.851140
Arias-Alpizar G., Koch B., Hamelmann N. M., Neustrup M. A., Paulusse J. M. J., Jiskoot W., et al. (2021). Stabilin-1 is required for the endothelial clearance of small anionic nanoparticles. Nanomed.: Nanotechnol. Biol. Med. 34, 102395. doi: 10.1016/j.nano.2021.102395
Arkee T. and Bishop G. A. (2020). TRAF family molecules in T cells: multiple receptors and functions. J. Leukoc. Biol. 107, 907–915. doi: 10.1002/JLB.2MR1119-397R
Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Au P. Y. B. and Yeh W. C. (2013). “Physiological roles and mechanisms of signaling by TRAF2 and TRAF5,” in Madame curie bioscience database (Austin, TX, USA: Landes Bioscience). Available at: https://www.ncbi.nlm.nih.gov/books/NBK6132/.
Bai G., Endres T., Kühbacher U., Mengoli V., Greer B. H., Peacock E. M., et al. (2024). HLTF resolves G4s and promotes G4-induced replication fork slowing to maintain genome stability. Mol. Cell. 84, 3044–3060.e11. doi: 10.1016/j.molcel.2024.07.018
Bai H., Mu L., Qiu L., Chen N., Li J., Zeng Q., et al. (2022). Complement C3 regulates inflammatory response and monocyte/macrophage phagocytosis of streptococcus agalactiae in a teleost fish. Int. J. Mol. Sci. 23, 15586. doi: 10.3390/ijms232415586
Bai X., Zheng Z., Liu B., Ji X., Bai Y., and Zhang W. (2016). Whole blood transcriptional profiling comparison between different milk yield of Chinese Holstein cows using RNA-seq data. BMC Genomics 17, 512. doi: 10.1186/s12864-016-2901-1
Balachandran Y., Knaus S., Caldwell S., and Singh B. (2015). Toll-like receptor 10 expression in chicken, cattle, pig, dog, and rat lungs. Vet. Immunol. Immunopathol. 168, 184–192. doi: 10.1016/j.vetimm.2015.09.007
Ban Y. and Guan L. L. (2021). Implication and challenges of direct-fed microbial supplementation to improve ruminant production and health. J. Anim. Sci. Biotechnol. 12, 109. doi: 10.1186/s40104-021-00630-x
Barington L., Wanke F., Niss Arfelt K., Holst P. J., Kurschus F. C., and Rosenkilde M. M. (2018). EBI2 in splenic and local immune responses and in autoimmunity. J. Leukoc. Biol. 104, 313–322. doi: 10.1002/JLB.2VMR1217-510R
Bartlett S., Gemiarto A. T., Ngo M. D., Sajiir H., Hailu S., Sinha R., et al. (2020). GPR183 regulates interferons, autophagy, and bacterial growth during mycobacterium tuberculosis infection and is associated with TB disease severity. Front. Immunol. 11. doi: 10.3389/fimmu.2020.601534
Beenken-Bobb A. M., Dornbach C. W., Deters E. L., Shike D. W., Hansen S. L., and McCann J. C. (2023). Effects of injectable vitamin C at weaning and prior to transit on growth performance of early-weaned beef steers. J. Anim. Sci. 101, skac307. doi: 10.1093/jas/skac307
Benjamini Y. and Hochberg Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Statist Soc. Ser. B. 57, 289–300. doi: 10.2307/2346101
Charles A., Janeway J., Travers P., Walport M., and Shlomchik M. J. (2001). “The complement system and innate immunity,” in Immunobiology: the immune system in health and disease, 5th Edition (New York, NY, USA: Garland Science). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK27100/ (Accessed October 21, 2024).
Chavez D. A., Greer B. H., and Eichman B. F. (2018). The HIRAN domain of helicase-like transcription factor positions the DNA translocase motor to drive efficient DNA fork regression. J. Biol. Chem. 293, 8484–8494. doi: 10.1074/jbc.RA118.002905
Chiu J. H., Tseng L. M., Huang T. T., Liu C. Y., Wang J. Y., Huang C. P., et al. (2020). MEGF11 is related to tumour recurrence in triple negative breast cancer via chemokine upregulation. Sci. Rep. 10, 8060. doi: 10.1038/s41598-020-64950-0
Choi I., Bao H., Kommadath A., Hosseini A., Sun X., Meng Y., et al. (2014). Increasing gene discovery and coverage using RNA-seq of globin RNA reduced porcine blood samples. BMC Genomics 15, 954. doi: 10.1186/1471-2164-15-954
Chou W. K., Park J., Carey J. B., McIntyre D. R., and Berghman L. R. (2017). Immunomodulatory effects of saccharomyces cerevisiae fermentation product supplementation on immune gene expression and lymphocyte distribution in immune organs in broilers. Front. Vet. Sci. 4. doi: 10.3389/fvets.2017.00037
Chu C., Moriyama S., Li Z., Zhou L., Flamar A.-L., Klose C. S. N., et al. (2018). Anti-microbial functions of group 3 innate lymphoid cells in gut-associated lymphoid tissues are regulated by G-protein-coupled receptor 183. Cell Rep. 23, 3750–3758. doi: 10.1016/j.celrep.2018.05.099
Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., et al. (2016). A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13. doi: 10.1186/s13059-016-0881-8
Daëron M. (2016). Fc receptors and fc receptor-like molecules within the immunoreceptor family. Encycloped. Immunobiol. 2, 360–370. doi: 10.1016/B978-0-12-374279-7.02017-8
DeLuca J. M., Murphy M. K., Wang X., and Wilson T. J. (2021). FCRL1 regulates B cell receptor-induced ERK activation through GRB2. J. Immunol. 207, 2688–2698. doi: 10.4049/jimmunol.2100218
de Souza K. A., Cooke R. F., Schubach K. M., Brandão A. P., Schumaher T. F., Prado I. N., et al. (2018). Performance, health and physiological responses of newly weaned feedlot cattle supplemented with feed-grade antibiotics or alternative feed ingredients. Animal 12, 2521–2528. doi: 10.1017/S1751731118000551
Deters E. L., Stokes R. S., Genther-Schroeder O. N., and Hansen S. L. (2018). Effects of a Saccharomyces cerevisiae fermentation product in receiving diets of newly weaned beef steers. II. Digestibility and response to a vaccination challenge. J. Anim. Sci. 96, 3906–3915. doi: 10.1093/jas/sky247
Devasani K. and Yao Y. (2022). Expression and functions of adenylyl cyclases in the CNS. Fluids Barriers CNS. 19, 23. doi: 10.1186/s12987-022-00322-2
de Vrese M. and Schrezenmeir J. (2008). “Probiotics, prebiotics, and synbiotics,” in Food Biotechnology. Eds. Stahl U., Donalies U. E. B., and Nevoigt E. (Berlin, Heidelberg, Germany: Springer), 1–66. doi: 10.1007/10_2008_097
Duan T., Du Y., Xing C., Wang H. Y., and Wang R. F. (2022). Toll-like receptor signaling and its role in cell-mediated immunity. Front. Immunol. 13. doi: 10.3389/fimmu.2022.812774
Emgård J., Kammoun H., García-Cassani B., Chesné J., Parigi S. M., Jacob J.-M., et al. (2018). Oxysterol sensing through the receptor GPR183 promotes the lymphoid-tissue-inducing function of innate lymphoid cells and colonic inflammation. Immunity 48, 120–132.e8. doi: 10.1016/j.immuni.2017.11.020
Fidan H., Esatbeyoglu T., Simat V., Trif M., Tabanelli G., Kostka T., et al. (2022). Recent developments of lactic acid bacteria and their metabolites on foodborne pathogens and spoilage bacteria: Facts and gaps. Food Biosci. 47, 101741. doi: 10.1016/j.fbio.2022.101741
Fijan S. (2014). Microorganisms with claimed probiotic properties: an overview of recent literature. Int. J. Environ. Res. Public Health 11, 4745–4767. doi: 10.3390/ijerph110504745
Fore F., Indriputri C., Mamutse J., and Nugraha J. (2020). TLR10 and its unique anti-inflammatory properties and potential use as a target in therapeutics. Immune Netw. 20, e21. doi: 10.4110/in.2020.20.e21
Fusco W., Lorenzo M. B., Cintoni M., Porcari S., Rinninella E., Kaitsas F., et al. (2023). Short-chain fatty-acid-producing bacteria: key components of the human gut microbiota. Nutrients 15, 2211. doi: 10.3390/nu15092211
Galyean M. L., Duff G. C., and Rivera J. D. (2022). Galyean Appreciation Club Review: revisiting nutrition and health of newly received cattle—what have we learned in the last 15 years?. J. Anim. Sci. 100, skac067. doi: 10.1093/jas/skac067
García-Burgos M., Moreno-Fernández J., Alférez M. J. M., Díaz-Castro J., and López-Aliaga I. (2020). New perspectives in fermented dairy products and their health relevance. J. Funct. Foods. 72, 104059. doi: 10.1016/j.jff.2020.104059
Goh W., Scheer S., Jackson J. T., Hediyeh-Zadeh S., Delconte R. B., Schuster I. S., et al. (2020). Hhex directly represses BIM-dependent apoptosis to promote NK cell development and maintenance. Cell Rep. 33, 108285. doi: 10.1016/j.celrep.2020.108285
Guimaraes O., Preedy G., Fox J. T., Cappellozza B. I., Theurer M. E., Davis T. C., et al. (2024). A novel direct-fed microbial impacts growth performance and supports overall health of feedlot cattle. Ruminants 4, 267–279. doi: 10.3390/ruminants4020019
Guo R., Liu T., Shasaltaneh M. D., Wang X., Imani S., and Wen Q. (2022). Targeting adenylate cyclase family: new concept of targeted cancer therapy. Front. Oncol. 12. doi: 10.3389/fonc.2022.829212
Hall J. B., Laarman A. H., Reynolds M. K., and Smith W. K. (2018). Performance of backgrounding steers fed diets containing monensin or a lactobacillus fermentation product. Transl. Anim. Sci. 2, S130–S133. doi: 10.1093/tas/txy035
Hannedouche S., Zhang J., Yi T., Shen W., Nguyen D., Pereira J. P., et al. (2011). Oxysterols direct immune cell migration via EBI2. Nature 475, 524–527. doi: 10.1038/nature10280
Hernández-González J. C., Martínez-Tapia A., Lazcano-Hernández G., García-Pérez B. E., and Castrejón-Jiménez N. S. (2021). Bacteriocins from lactic acid bacteria. a powerful alternative as antimicrobials, probiotics, and immunomodulators in veterinary medicine. Anim. (Basel). 11, 979. doi: 10.3390/ani11040979
Hess N. J., Felicelli C., Grage J., and Tapping R. I. (2017). TLR10 suppresses the activation and differentiation of monocytes with effects on DC-mediated adaptive immune responses. J. Leukoc. Biol. 101, 1245–1252. doi: 10.1189/jlb.3A1116-492R
Jackson J. T., Shields B. J., Shi W., Di Rago L., Metcalf D., Nicola N. A., et al. (2017). Hhex regulates hematopoietic stem cell self-renewal and stress hematopoiesis via repression of cdkn2a. Stem Cells 35, 1948–1957. doi: 10.1002/stem.2648
Janssens S. and Beyaert R. (2003). Role of toll-like receptors in pathogen recognition. Clin. Microbiol. Rev. 16, 637–646. doi: 10.1128/CMR.16.4.637-646.2003
Justice S. M., Smith W. B., Mullenix M. K., Tigue D. A., Elmore M. F., Dillard S. L., et al. (2024). Assessment of weaning and backgrounding management practices used by Alabama beef cattle producers. Appl. Anim. Sci. 40, 81–90. doi: 10.15232/aas.2023-02467
Kay J., Chu M., and Sanes J. (2012). MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature 483, 465–469. doi: 10.1038/nature10877
Knez Š., Narat M., and Ogorevc J. (2023). Differential gene expression induced by different TLR agonists in A549 lung epithelial cells is modulated by CRISPR activation of TLR10. Biomolecules 13, 19. doi: 10.3390/biom13010019
Kumar V., Bermea K. C., Kumar D., Singh A., Verma A., Kaileh M., et al. (2024). RelA-mediated signaling connects adaptation to chronic cardiomyocyte stress with myocardial and systemic inflammation in the ADCY8 model of accelerated aging. Geroscience 46, 4243–4262. doi: 10.1007/s11357-024-01121-3
Lesage G. and Bussey H. (2006). Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 317–343. doi: 10.1128/MMBR.00038-05
Levin D. E. (2005). Cell wall integrity signaling in saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291. doi: 10.1128/MMBR.69.2.262-291.2005
Li D. and Wu M. (2021). Pattern recognition receptors in health and diseases. Sig Transduct. Target. Ther. 6, 1–24. doi: 10.1038/s41392-021-00687-0
Liao Y., Smyth G. K., and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lin A., Yan X., Wang H., Su Y., and Zhu W. (2022). Effects of lactic acid bacteria-fermented formula milk supplementation on ileal microbiota, transcriptomic profile, and mucosal immunity in weaned piglets. J. Anim. Sci. Biotechnol. 13, 113. doi: 10.1186/s40104-022-00762-8
Lynch E., McGee M., and Earley B. (2019). Weaning management of beef calves with implications for animal health and welfare. J. Appl. Anim. Res. 47, 167–175. doi: 10.1080/09712119.2019.1594825
Mahmoud A. H. A., Slate J. R., Hong S., Yoon I., and McGill J. L. (2020). Supplementing a Saccharomyces cerevisiae fermentation product modulates innate immune function and ameliorates bovine respiratory syncytial virus infection in neonatal calves. J. Anim. Sci. 98, skaa252. doi: 10.1093/jas/skaa252
Maturana M., Castillejos L., Martin-Orue S. M., Minel A., Chetty O., Felix A. P., et al. (2023). Potential benefits of yeast Saccharomyces and their derivatives in dogs and cats: a review. Front. Vet. Sci. 10. doi: 10.3389/fvets.2023.1279506
Mejia-Garcia A., Fernandez G. J., Echeverri L. F., Balcazar N., and Acin S. (2024). RNA-seq analysis reveals modulation of inflammatory pathways by an enriched-triterpene natural extract in mouse and human macrophage cell lines. Heliyon 10, e24382. doi: 10.1016/j.heliyon.2024.e24382
Melchizedek A. and Maria L. V. (2023) Intelligent wearable devices and biosensors for monitoring cattle health conditions: A review and classification - ScienceDirect. Available online at: https://www.sciencedirect.com/science/article/abs/pii/S2352648322001039 (Accessed February 4, 2025).
Misselwitz B., Wyss A., Raselli T., Cerovic V., Sailer A. W., Krupka N., et al. (2021). The oxysterol receptor GPR183 in inflammatory bowel diseases. Br. J. Pharmacol. 178, 3140–3156. doi: 10.1111/bph.15311
Mourits V. P., Arts R. J. W., Novakovic B., Matzaraki V., de Bree L. C. J., Koeken V. A. C. M., et al. (2020). The role of Toll-like receptor 10 in modulation of trained immunity. Immunology 159, 289–297. doi: 10.1111/imm.13145
Mujal A. M., Delconte R. B., and Sun J. C. (2021). Natural killer cells: from innate to adaptive features. Annu. Rev. Immunol. 39, 417–447. doi: 10.1146/annurev-immunol-101819-074948
Munteanu C. and Schwartz B. (2022). The relationship between nutrition and the immune system. Front. Nutr. 9. doi: 10.3389/fnut.2022.1082500
Nakano H., Shindo M., Yamada K., Yoshida M. C., Santee S. M., Ware C. F., et al. (1997). Human TNF receptor-associated factor 5 (TRAF5): cDNA cloning, expression and assignment of the TRAF5 gene to chromosome 1q32. Genomics 42, 26–32. doi: 10.1006/geno.1997.4697
O’Loughlin A., Lynn D. J., McGee M., Doyle S., McCabe M., and Earley B. (2012). Transcriptomic analysis of the stress response to weaning at housing in bovine leukocytes using RNA-seq technology. BMC Genomics 13, 250. doi: 10.1186/1471-2164-13-250
Ogunade I. M., McCoun M., Idowu M. D., and Peters S. O. (2020). Comparative effects of two multispecies direct-fed microbial products on energy status, nutrient digestibility, and ruminal fermentation, bacterial community, and metabolome of beef steers. J. Anim. Sci. 98, skaa201. doi: 10.1093/jas/skaa201
Ogunade I. M., Taiwo G., Estrada-Reyes Z. M., Yun J., Pech-Cervantes A. A., and Peters S. O. (2021). Effects of a blend of mannan and glucan on growth performance, apparent nutrient digestibility, energy status, and whole-blood immune gene expression of beef steers during a 42-d receiving period. Trans. Anim. Sci. 5, txaa226. doi: 10.1093/tas/txaa226
Ordaz-Ramos A., Rosales-Gallegos V. H., Melendez-Zajgla J., Maldonado V., and Vazquez-Santillan K. (2021). The role of LGR4 (GPR48) in normal and cancer processes. Int. J. Mol. Sci. 22, 4690. doi: 10.3390/ijms22094690
Pascual A., Pauletto M., Giantin M., Radaelli G., Ballarin C., Birolo M., et al. (2020). Effect of dietary supplementation with yeast cell wall extracts on performance and gut response in broiler chickens. J. Anim. Sci. Biotechnol. 11, 40. doi: 10.1186/s40104-020-00448-z
Peng M., Niu D., Wang F., Chen Z., and Li J. (2016). Complement C3 gene: Expression characterization and innate immune response in razor clam Sinonovacula constricta. Fish Shellfish Immunol. 55, 223–232. doi: 10.1016/j.fsi.2016.05.024
Perera T. R. W., Skerrett-Byrne D. A., Gibb Z., Nixon B., and Swegen A. (2022). The future of biomarkers in veterinary medicine: emerging approaches and associated challenges. Anim. (Basel). 12, 2194. doi: 10.3390/ani12172194
Prajapati N., Patel J., Singh S., Yadav V. K., Joshi C., Patani A., et al. (2023). Postbiotic production: harnessing the power of microbial metabolites for health applications. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1306192
Pukrop J. R., Brennan K. M., Funnell B. J., and Schoonmaker J. P. (2018). Effect of a hydrolyzed mannan- and glucan-rich yeast fraction on performance and health status of newly received feedlot cattle1. J. Anim. Sci. 96, 3955–3966. doi: 10.1093/jas/sky255
Rafique N., Jan S. Y., Dar A. H., Dash K. K., Sarkar A., Shams R., et al. (2023). Promising bioactivities of postbiotics: A comprehensive review. J. Agric. Food Res. 14, 100708. doi: 10.1016/j.jafr.2023.100708
Ray T. A., Cochran K., Kozlowski C., Wang J., Alexander G., Cady M. A., et al. (2020). Comprehensive identification of mRNA isoforms reveals the diversity of neural cell-surface molecules with roles in retinal development and disease. Nat. Commun. 11, 3328. doi: 10.1038/s41467-020-17009-7
Schurch N. J., Schofield P., Gierliński M., Cole C., Sherstnev A., Singh V., et al. (2016). How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 22, 839–851. doi: 10.1261/rna.053959.115
Shemery A., Gibson M., Gorrell E., Daniel D., Piontkivska H., and Novak C. M. (2025). RNA-sequencing reveals altered gene expression in the ventromedial hypothalamus following predator odor exposure. F1000Research 13, 648. doi: 10.12688/f1000research.152034.2
Shin Y., Han S., Kwon J., Ju S., Choi T. G., Kang I., et al. (2023). Roles of short-chain fatty acids in inflammatory bowel disease. Nutrients 15, 4466. doi: 10.3390/nu15204466
Snowder G. D., Van Vleck L. D., Cundiff L. V., and Bennett G. L. (2006). Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J. Anim. Sci. 84, 1999–2008. doi: 10.2527/jas.2006-046
Soler Palacios B., Nieto C., Fajardo P., González de la Aleja A., Andrés N., Dominguez-Soto Á., et al. (2020). Growth hormone reprograms macrophages toward an anti-inflammatory and reparative profile in an MAFB-dependent manner. J. Immunol. 205, 776–788. doi: 10.4049/jimmunol.1901330
Tirado-Hurtado I., Fajardo W., and Pinto J. A. (2018). DNA damage inducible transcript 4 gene: the switch of the metabolism as potential target in cancer. Front. Oncol. 8. doi: 10.3389/fonc.2018.00106
Treon E., Sidney T., Taiwo G., Idowu M., Leal Y., Ologunagba D., et al. (2024). Effects of dietary supplementation of a blend of Saccharomyces cerevisiae, multiple live probiotic bacteria, and their fermentation products on performance, health, and rumen bacterial community of newly weaned beef steers during a 56-d receiving period. Trans. Anim. Sci. 8, txad143. doi: 10.1093/tas/txad143
Wang J., Fan H., Li M., Zhao K., Xia S., Chen Y., et al. (2023). Integration of non-coding RNA and mRNA profiles reveals the mechanisms of rumen development induced by different types of diet in calves. Genes 14, 1093. doi: 10.3390/genes14051093
Wang X., Iyer A., Lyons A. B., Körner H., and Wei W. (2019). Emerging roles for G-protein coupled receptors in development and activation of macrophages. Front. Immunol. 10. doi: 10.3389/fimmu.2019.02031
Wickramasinghe S., Cánovas A., Rincon G., and Medrano J. (2014). RNA-Sequencing: A tool to explore new frontiers in animal genetics. Livestock Sci. 166, 206–216. doi: 10.1016/j.livsci.2014.06.015
Yan K., Gao L. N., Cui Y. L., Zhang Y., and Zhou X. (2016). The cyclic AMP signaling pathway: Exploring targets for successful drug discovery (Review). Mol. Med. Rep. 13, 3715. doi: 10.3892/mmr.2016.5005
Yousefi Z., Sharifzadeh S., Zare F., and Eskandari N. (2023). Fc receptor-like 1 (FCRL1) is a novel biomarker for prognosis and a possible therapeutic target in diffuse large B-cell lymphoma. Mol. Biol. Rep. 50, 1133–1145. doi: 10.1007/s11033-022-08104-7
Zhang N., Yuan M., and Wang J. (2023). LGR4: A new receptor member in endocrine and metabolic diseases. Endocr. Rev. 44, 647. doi: 10.1210/endrev/bnad003
Zhao X., Xie H., Zhao M., Ahsan A., Li X., Wang F., et al. (2019). Fc receptor-like 1 intrinsically recruits c-Abl to enhance B cell activation and function. Sci. Adv. 5, eaaw0315. doi: 10.1126/sciadv.aaw0315
Zhou J., Fu Y., Qi G., Dai J., Zhang H., Wang J., et al. (2023). Yeast cell-wall polysaccharides improve immunity and attenuate inflammatory response via modulating gut microbiota in LPS-challenged laying hens. Int. J. Biol. Macromol. 224, 407–421. doi: 10.1016/j.ijbiomac.2022.10.133
Keywords: beef cattle, differentially expressed genes, immune competence, inflammatory response, weaning
Citation: Ajiboye A, Johnson S, Idowu M, Leal Y, Sidney T, Taiwo G, Ogunade IM and Ashwell C (2025) Whole blood RNA-seq analysis reveals the immunomodulatory effects of a supplemental multi-strain direct-fed microbial in the diet of newly weaned beef steers1. Front. Anim. Sci. 6:1623311. doi: 10.3389/fanim.2025.1623311
Received: 05 May 2025; Accepted: 18 June 2025;
Published: 18 July 2025.
Edited by:
David L. Harmon, University of Kentucky, United StatesReviewed by:
Ravikanthreddy Poonooru, University of Missouri, United StatesYudi Adinata, Ministry of Agriculture, Indonesia
Copyright © 2025 Ajiboye, Johnson, Idowu, Leal, Sidney, Taiwo, Ogunade and Ashwell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ibukun M. Ogunade, aWJ1a3VuLm9ndW5hZGVAbWFpbC53dnUuZWR1; Christopher Ashwell, Y2hyaXN0b3BoZXIuYXNod2VsbEBtYWlsLnd2dS5lZHU=
†These authors share first authorship
 Adekunle Ajiboye
Adekunle Ajiboye Samanthia Johnson
Samanthia Johnson Modoluwamu Idowu2
Modoluwamu Idowu2 Yarahy Leal
Yarahy Leal Taylor Sidney
Taylor Sidney Ibukun M. Ogunade
Ibukun M. Ogunade Christopher Ashwell
Christopher Ashwell