- Institute of Microbiology, University of Stuttgart, Stuttgart, Germany
Aromatic amines are an important class of chemicals which are used as building blocks for the synthesis of polymers and pharmaceuticals. In this study we establish a de novo pathway for the biosynthesis of the aromatic amines para-amino-phenylethanol (PAPE) and para-amino-phenylacetic acid (4-APA) in Escherichia coli. We combined a synthetic para-amino-L-phenylalanine pathway with the fungal Ehrlich pathway. Therefore, we overexpressed the heterologous genes encoding 4-amino-4-deoxychorismate synthase (pabAB from Corynebacterium glutamicum), 4-amino-4-deoxychorismate mutase and 4-amino-4-deoxyprephenate dehydrogenase (papB and papC from Streptomyces venezuelae) and ThDP-dependent keto-acid decarboxylase (aro10 from Saccharomyces cerevisiae) in E. coli. The resulting para-amino-phenylacetaldehyde either was reduced to PAPE or oxidized to 4-APA. The wild type strain E. coli LJ110 with a plasmid carrying these four genes produced (in shake flask cultures) 11 ± 1.5 mg l−1 of PAPE from glucose (4.5 g l−1). By the additional cloning and expression of feaB (phenylacetaldehyde dehydrogenase from E. coli) 36 ± 5 mg l−1 of 4-APA were obtained from 4.5 g l−1 glucose. Competing reactions, such as the genes for aminotransferases (aspC and tyrB) or for biosynthesis of L-phenylalanine and L-tyrosine (pheA, tyrA) and for the regulator TyrR were removed. Additionally, the E. coli genes aroFBL were cloned and expressed from a second plasmid. The best producer strains of E. coli showed improved formation of PAPE and 4-APA, respectively. Plasmid-borne expression of an aldehyde reductase (yahK from E. coli) gave best values for PAPE production, whereas feaB-overexpression led to best values for 4-APA. In fed-batch cultivation, the best producer strains achieved 2.5 ± 0.15 g l−1 of PAPE from glucose (11% C mol mol-1 glucose) and 3.4 ± 0.3 g l−1 of 4-APA (17% C mol mol−1 glucose), respectively which are the highest values for recombinant strains reported so far.
Introduction
Aromatic amines (AA) are an important class of chemicals of which several are already used and characterized for different industrial applications. These chemicals can be used for the manufacturing of dyes, pesticides, drugs, plastics, semi-conductive or conductive polymers, and other industrial products (Lawrence, 2004; Vogt and Gerulis, 2005; Sousa et al., 2013; Arora, 2015; Masuo et al., 2016; Tateyama et al., 2016; Tsuge et al., 2016; Kawasaki et al., 2018). AA contain at least one benzene ring and one amino group attached directly to a benzene ring and are usually derived from petroleum-based substrates (Vogt and Gerulis, 2005); they can also be found in Nature. The AA para-aminobenzoic acid (PABA) is an intermediate of the folate biosynthesis pathway (Koma et al., 2014; Kubota et al., 2016) while ortho-aminobenzoic acid (anthranilic acid) is the first intermediate of the L-tryptophan biosynthesis pathway. The non-proteinogenic aromatic amino acid para-amino-L-phenylalanine (L-PAPA) is an important building block for biosynthesis of antibiotics like chloramphenicol (Brown et al., 1996; Chang et al., 2001; He et al., 2001) or of pristinamycin (Blanc et al., 1997). The AA para-amino-phenylethanol (PAPE) (see Figure 1), an analog of 2-phenylethanol which has a rose-like odor (Etschmann et al., 2002), is an important building block for chemical syntheses. PAPE can be used for the synthesis of aromatic polyamides (Masuo et al., 2016), polymers or copolymers (Li et al., 1999; Xu et al., 2008), and for the surface functionalization of graphene (Yadav and Cho, 2013). PAPE can also be used for the synthesis of the overactive bladder treatment drug Myrbetriq® (Sacco and Bientinesi, 2012). A classical synthesis of aromatic amines is performed by nitroreduction of aromatic compounds in the presence of acidic compounds and a reducing agent, such as solid metals (Vogt and Gerulis, 2005; Schul'tsev, 2011; Froidevaux et al., 2016).
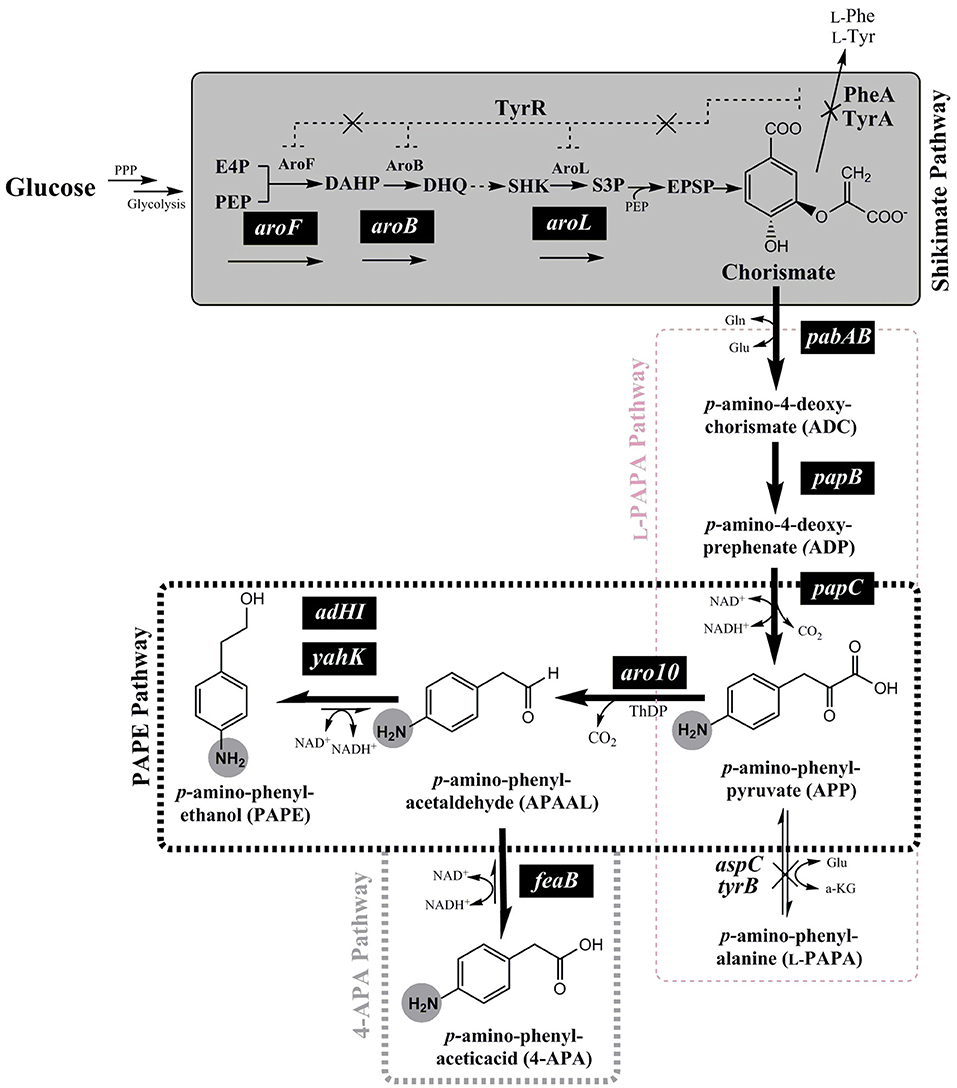
Figure 1. Schematic overview of the de-novo biosynthesis pathway of para-amino phenylethanol (PAPE) or para-amino phenylacetic acid (4-APA) via expanded shikimate pathway in E. coli strains from glucose. The genes expressed from plasmids are indicated by white letters on black backgrounds (pabAB, papB, papC, aro10, aroF, aroB, aroL, adHI, adHII, yahK, or feaB). Enzymes and molecules are abbreviated as follows: G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1.6-BP, fructose 1,6-bisphosphate; G3P, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; PPP, Pentose phosphate pathway; E4P, erythrose 4-phosphate; PEP, phosphoenolpyruvic acid; DAHP, 3-deoxy-D-arabinoheptulosonate 7-phosphate; DHQ, dehydroquinate; SHK, shikimic acid; S3P, shikimate 3-phosphate; aroF, DAHP synthase; aroB, dehydroquinate synthase; aroL, shikimate kinase I; pabAB, p-amino-4-deoxychorismate (ADC) synthase; papB, ADC mutase; papC, p-amino-4-deoxyprephenate dehydrogenase; tyrB, aromatic aminotransferase; aspC, aspartate aminotransferase; aro10, ThDP-ketoacid decarboxylase; ADHI and ADHII, alcohol dehydrogenase I and II; yahK, aldehyde reductase; feaB, phenylacetaldehyde dehydrogenase.
Due to the fast growing demand for polymers and “super-engineered” plastics (Masuo et al., 2016; Tateyama et al., 2016; Tsuge et al., 2016; Kawaguchi et al., 2017; Kawasaki et al., 2018), PAPE, p-amino-phenylacetate (4-APA) and other AAs are considered as important substrates for the synthesis of materials for novel technological applications. So they have become promising candidates for the replacement of metal or ceramic materials (Kawasaki et al., 2018). Hence, an annual global production of about 100,000 tons of AA with an expected market size of several hundred millions US dollars have been estimated (Masuo et al., 2016). Due to the high-value and industrial importance, various approaches to synthesize aromatic amines by biological routes have been started. Thus, PABA (Koma et al., 2014; Kubota et al., 2016), L-PAPA (Mehl et al., 2003; Masuo et al., 2016; Tateyama et al., 2016; Konishi et al., 2017; Mohammadi Nargesi et al., 2018), 4-aminocinnamic acid (Suvannasara et al., 2014; Konishi et al., 2016; Kawasaki et al., 2018), 4-aminohydrocinnamic acid (Tateyama et al., 2016; Kawasaki et al., 2018), have already been produced with genetically engineered Corynebacterium glutamicum or E. coli strains. More specifically for the present study, Takaya and coworkers recently described an approach to produce PAPE or 4-APA with E. coli (Masuo et al., 2016; Konishi et al., 2017). Still, the achieved titers and yields of these aromatic amines are in most cases relatively low and might not be sufficient for an intended industrial use.
The pathway to PAPE runs in analogy to the classical Ehrlich pathway for 2-phenylethanol (2-PE) production from L-phenylalanine in yeasts (Etschmann et al., 2002; Stark et al., 2003; Hua and Xu, 2011; Kim et al., 2014a; Suastegui and Shao, 2016) with the only difference that L-PAPA is the starting amino acid. The Ehrlich pathway to 2-PE consists of a transaminase (aminotransferase) reaction on phenylalanine to deliver phenylpyruvate, which in turn is decarboxylated by a ThDP-dependent keto-acid decarboxylase (KDC) to phenylacetaldehyde. Phenylacetaldehyde can then be reduced to 2-PE by an oxidoreductase (alcohol dehydrogenase, ADH). Alternatively, phenylacetaldehyde can be oxidized to phenylacetate by an aldehyde dehydrogenase (Hazelwood et al., 2008). Whereas, the conversion of L-phenylalanine to 2-PE is a biotransformation from a more expensive starting material, a biosynthesis of 2-PE (or PAPE) would start from an inexpensive carbon source as glucose and include the aromatic biosynthesis pathway to phenylpyruvate/L-phenylalanine (APP/L-PAPA) as well. By establishing these necessary enzyme activities in recombinant E. coli cells, Takaya and coworkers reported titers for PAPE and 4-APA titers of about 0.24 and 0.19 g l−1 from glucose, respectively, with their genetically modified strains (Masuo et al., 2016).
In the present study, we followed this idea for de-novo PAPE/4-APA biosynthesis pathway in E. coli. The overall pathway contains an upstream pathway module from glucose to chorismate (naturally occurring in E. coli) and a downstream pathway module from chorismate to PAPE or 4-APA via p-amino-phenylpyruvate (APP) (see Figure 1). We also took care of improved provision of possibly limiting intermediates. Thereby we were able to improve the PAPE and 4-APA titers.
Materials and Methods
Bacterial Strains
All E. coli K-12 strains and plasmids employed in this study are listed in Table S1. All used primers are listed in Table S2. E. coli DH5α was used for cloning experiments and E. coli LJ110 (W3110) was used as wild-type strain.
Bacterial Culture Medium, Enzymes, and Reagents
Lysogeny broth (LB, 10 g l−1 tryptone, 5 g l−1 yeast extract, and 10 g l−1 NaCl) was used as complex media (Sambrook et al., 2001). The AA production was performed in minimal medium (Gerhardt et al., 1984) which contained 3 g l−1 KH2PO4, 12 g l−1 K2HPO4, 5 g l−1 (NH4)2SO4, 0.3 g l−1 MgSO4·7H2O, 0.1 g l−1 NaCl, 0.1125 g l−1 Fe(II)SO4·7H2O/Na citrate 15 ml (from the solution of 7.5 g l−1 FeSO4 and 100 g l−1 sodium citrate), 0.015 g l−1 CaCl2·2H2O, 7.5 μg l−1 thiamine, 0.02 mg ml−1 L-phenylalanine (L-Phe), 0.02 mg ml−1 L-tyrosine (L-Tyr), 3 g l−1 aspartic acid (if required) and 4.5 g l−1 glucose. The shake flask cultivation was carried out in 250 mL Erlenmeyer flasks containing 20 ml of minimal medium. The cultures were grown at 37°C on an orbital shaker with a stirring speed of 150 rpm. The gene expressions were induced with isopropyl β-D-thiogalactopyranoside (IPTG, the final concentration was 0.5 mM) at an OD600 of 0.6. If required, antibiotics were added at appropriate concentrations of kanamycin 50 μg ml−1 and ampicillin 100 μg ml−1. para-amino-phenylethanol (PAPE) was purchased from Sigma-Aldrich Chemie GmbH (Munich, Germany), para-amino phenylacetic acid (4-APA) from Merck KGaA (Darmstadt, Germany), IPTG was from PEQLAB Biotechnologie GmbH (Erlangen, Germany) and all other chemical were either from Carl Roth GmbH (Karlsruhe, Germany), Fluka (Taufkirchen, Germany), or Merck KGaA (Darmstadt, Germany) and were of the highest available purity. All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, USA). KOD Hot Start DNA Polymerase for gene amplification was from Novagen, Merck KGaA (Darmstadt, Germany), Taq DNA polymerase used for screening-PCR was from Genaxxon Bioscience GmbH (Ulm, Germany). Plasmid preparation and gel extraction kits were obtained from Macherey-Nagel (Düren, Germany) and DNA sequencing services were provided by GATC Biotech GmbH (Konstanz, Germany).
Construction of the Plasmids for PAPE or 4-APA Biosynthesis Pathway
To enable the PAPE or 4-APA biosynthesis we constructed the plasmid pC53BCA, pC53BCAY, pC53BCAHII, or pC53BCAF, respectively. These plasmids are based on pC53BC, which already carries the genes pabAB, from C. glutamicum and papBC from Streptomyces venezuelae (Mohammadi Nargesi et al., 2018). The ThDP-dependent ketoacid decarboxylase gene aro10 was amplified from genomic DNA of Saccharomyces cerevisiae W3118 (Vandenhazel et al., 1992) by using the primer pair aro10-Fw/Rw (Table S2). The aro10 carrying PCR product was first introduced by blunt-end ligation into the EcoRV-restricted cloning vector pBluescript KS to generate pBSK-aro10 and sequence-verified before continuing with the construction process. Then the aro10 containing DNA fragment was isolated by BamHI-XbaI restriction which was ligated to a BamHI/XbaI restricted plasmid DNA of pJF119EH or pC53BC subsequently to yield pJFA10 and pC53BCA, respectively. Genes encoding alcohol dehydrogenases (I and II) ADH1 and ADH2 (De Smidt et al., 2008) from Saccharomyces cerevisiae W3118, and genes for aldehyde reductase (yahK) and phenylacetaldehyde dehydrogenase (feaB) from Escherichia coli BW25113 were PCR amplified from genomic yeast or E. coli DNA, respectively. The following primer pairs were used: ADHI-Fw/-Rw, ADHII-Fw/-Rw, yahK-Fw/-Rw and feaB-Fw/-Rw (Table S2). The resulting PCR products of ADH1, ADH2, yahK or feaB were each ligated blunt end first into the EcoRV restricted pBluescript KS cloning vector to generate pBSK-ADHI, pBSK-ADHII, pBSK-yahK, or pBSK-feaB, respectively. The correct sequences were verified by sequencing (GATC, Konstanz, Germany). The fragments containing ADH1, ADH2, yahK, or feaB genes were double-digested with SphI/Sbf I restricted and ligated into a SphI/SbfI digested pC53BCA vector to generate pC53BCAHI, pC53BCAHII, pC53BCAY, or pC53BCAF, respectively. The digested fragment containing feaB was also ligated to SphI/Sbf I restricted pJFA10 expression plasmid to generate pJFA10F. Additionally, yahK or feaB fragments were digested by SmaI/KpnI and then ligated into SmaI/KpnI restricted pJNTaroFBL, yielding pJNTaroFBL-yahK or pJNTaroFBL-feaB, respectively.
Construction of aspC, tyrB, and tyrR Deletion Mutants
For chromosomal gene disruption the one-step recombineering inactivation method was used (Datsenko and Wanner, 2000). For PCR amplification of the FRT-flanked chloramphenicol resistance cassette for the aspC deletion the primer pair Del-aspC-fw/-rev was used. For the deletion of tyrB, the primer pair Del-tyrB-fw/-rev was used with pCAS30-FRT-cat-FRT as template for the chloramphenicol resistance cassette (Tables S1, S2) (Vallon et al., 2008). The obtained DNA amplificates were consecutively transformed into E. coli FUS4 (Gottlieb et al., 2014) carrying the λ-Red recombinase expression vector pKD46 (Datsenko and Wanner, 2000). Knock-in mutant colonies were selected on LB agar plates with chloramphenicol (Cm; 30 mg l−1) and were tested by colony PCR for correct location using the control primer Ko-aspC-fw/-rev or Ko-tyrB-fw/-rev (Table S2). After each successful integration the chloramphenicol resistance gene cat was removed by using the plasmid pCP20 as previously described (Cherepanov and Wackernagel, 1995). The tyrR deletion was performed as described previously (Mohammadi Nargesi et al., 2018). The chromosomal gene deletions were verified by PCR.
Verification of Aro10 Specificity Toward APP via Whole Cell Biotransformation
To verify the substrate specificity of Aro10 toward para-amino-phenylpyruvate (APP), a resting whole cell biotransformation approach was applied. Bio-L-PAPA (wherein “Bio” refers to microbial production of aromatic monomers) was produced from glucose as described previously (Mohammadi Nargesi et al., 2018). An overnight LB culture of E. coli LJ110 with pJFA10 or pJFA10F was used to inoculate 50 ml LB media to an OD600 of 0.1 in a 500 ml Erlenmeyer flask and was incubated by shaking at 30°C and 150 rpm. At an OD600 of about 0.6, IPTG was added to the culture (final concentration 0.5 mM). After 6 h incubation (OD600 ~5) the cells were collected by centrifugation (6000 g and 10 min), washed twice with potassium phosphate buffer (200 mM, pH~7.2) and resuspended in a reaction mixture (10 mL) containing ~1 g l−1 Bio-L-PAPA, 200 mM potassium phosphate buffer (pH ~ 7.2) and 0.25 mM thiamine diphosphate (ThDP) to start the biotransformation with an OD600 ~ 16–18. Resting cells were incubated in a rotary shaking incubator at 110 rpm at 30°C. L-PAPA consumption and PAPE or 4-APA production were determined by taking samples at 2 h intervals over a period of 8 h. The direct product of Aro10 action, p-aminophenylacetaldehyde (APAAL), could not be measured.
Shake Flask Batch and Fed-Batch Cultivation for PAPE or 4-APA Production With E. coli
An overnight culture growing in minimal medium was used to inoculate 20 ml of minimal medium in a 250 ml Erlenmeyer flask. The medium was supplemented with 0.02 mg ml−1 L-Phe, L-Tyr, 3 mg ml−1 L-aspartic acid (if applicable) and 4.5 g l−1 glucose. The shake flasks were cultivated at 37°C with 150 rpm. After 6 h (batch cultivation) or 24 h (fed-batch cultivation), IPTG was added to a final concentration of 0.5 mM and the temperature was reduced to 30°C. In order to start the fed-batch cultivation ~ 4.5 g l−1 glucose was fed to the culture in intervals of 12 h. In order to increase the biomass production, additional pulses of 0.01 mg ml−1 L-Phe and L-Tyr were given concomitant with IPTG addition. During the cultivation 5 g l−1, sodium bicarbonate was added to adjust the pH and 4 g l−1 ammonia sulfate was given as an ammonia source. Samples were taken every 12 h. A portion of the samples was used for optical cell density measurement, and the rest were centrifuged at 22,000 g for 10 min. The supernatants were stored at −28°C until further analysis. All the experiments in this report were conducted in three independent replicates.
PAPE and 4-APA Susceptibility Assays
PAPE or 4-APA susceptibility of E. coli strains was determined by monitoring cell growth at various PAPE or 4-APA concentrations by measuring the OD600nm (Cary 50 Bio, Varian). An overnight culture grown in minimal media with 4.5 g l−1 glucose was used to inoculate the minimal media with an initial OD600 of 0.1. Cell growth was monitored for 48 h.
Analytical Methods
To analyze PAPE and 4-APA amounts, culture samples were centrifuged at 22,000 g for 10 min. The supernatants were transferred to new tubes and were frozen at −28°C until further use. The PAPE and 4-APA concentrations were measured by high-pressure liquid chromatography (HPLC; Agilent Technologies Series 1200 system, USA) using a symmetry C18 silica column (Prontosil, 250 × 4 mm, CS Chromatography Service GmbH, Langerwehe, Germany) with a pre-column. 40 mM Na2SO4 (pH adjusted ~2.7 with methanesulfonic acid) were used as the mobile phase with a flow rate of 1 ml min−1 at 40°C. Absorption was detected at 210 nm with a DAD detector (Agilent Technologies Series 1260 Infinity DAD system). For LC-MS analysis of PAPE and 4-APA, the same HPLC method was used as described above, except that the mobile phase was replaced by 0.1% formic acid solution. The mass was determined using an Agilent 1260 system with 1260 Infinity DAD detector and an Agilent 6130 mass spectrometer (Agilent Technologies Waldbronn, Germany). Glucose concentration was monitored by using Glucose Medi-Test stripes (Macherey Nagel, Düren, Germany) and by HPLC on an Organic acid column (300 × 8 mm, CS Chromatography Service GmbH, Langerwehe, Germany) at 40°C. 5 mM H2SO4 was used as mobile phase with a flow rate of 0.6 ml min−1. Absorption was detected by the refractive index (RI) detector. The compounds were quantified using standard curves of the respective commercial chemicals.
Results
Susceptibility of E. coli Cells to PAPE and 4-APA
As it is known that 2-phenylethanol (2-PE), a structural analog of PAPE, is toxic to E. coli at doses above 1–1.5 g l−1 (Masker and Eberle, 1972; Lucchini et al., 1993; Kang et al., 2012), we tested the growth behavior of E. coli toward the addition of PAPE or 4-APA. The PAPE or 4-APA susceptibility was assayed by monitoring the biomass formation of E. coli wild-type strain LJ110 in minimal medium with varying concentrations of PAPE or 4-APA. At concentrations up to 30 mM of PAPE or 4-APA, only little effects on the growth of E. coli were observed (Figure S1). Cells were still able to grow in the presence of 40 mM PAPE or 4-APA, but the growth rate (μ) was decreased to 0.27 h−1 (PAPE) or 0.38 h−1 (4-APA) compared to the absence (μ~0.67 h−1). A complete growth inhibition was observed at a PAPE or 4-APA concentration of 50 mM (Figure S1). From this observation we reasoned that PAPE and 4-APA are compounds to be produced with recombinant E. coli strains without extreme toxicity to the producer cells.
Construction of a Recombinant Route to PAPE and 4-APA in E. coli
In order to enable the production of the aromatic amines PAPE and/or 4-APA in E. coli a recombinant route has to be established. The key metabolite chorismate needs to be converted to p-amino-phenylpyruvate (APP) via 4-amino-4-deoxy-chorismate (ADC), which is also the precursor of PABA (Figure 1). For this purpose, we used the plasmid pC53BC, which carries the fused gene pabAB (ADC synthase) from Corynebacterium glutamicum (Wubbolts et al., 2005; Kozak, 2006; Stolz et al., 2007), papB (ADC mutase) and papC (p-amino-4-deoxyprephenate dehydrogenase) from Streptomyces venezuelae (Brown et al., 1996; He et al., 2001; Mohammadi Nargesi et al., 2018) (Figures 1, S3). We showed in a recent study the biosynthesis of L-PAPA in recombinant E. coli strains using pC53BC (Mohammadi Nargesi et al., 2018). In order to form PAPE and 4-APA, a decarboxylation of APP to p-amino-phenylacetaldehyde (APAAL) is necessary. In bacteria, fungi and yeasts certain ThDP-dependent enzymes are known to decarboxylate 2-keto-acids like pyruvate or phenylpyruvate to the corresponding aldehydes (Delaplaza et al., 2004; Siegert et al., 2005; Vuralhan et al., 2005; Yep et al., 2006, 2008; Küberl et al., 2011; Masuo et al., 2015). The ThDP-dependent 2-keto-acid decarboxylase Aro10 from Saccharomyces cerevisiae has been described (Vuralhan et al., 2005; Kneen et al., 2011) to have a broad substrate specificity and thus might also accept the p-amino-substituted APP. Therefore, we cloned the aro10 gene from S. cerevisiae strain W3118 and expressed it in E. coli (Figure S3). If APP would be used as substrate by Aro10, p-amino-phenylacetaldehyde (APAAL) and CO2 would be formed. Subsequently, APAAL could be further converted either reductively to the alcohol PAPE (Atsumi et al., 2010; Koma et al., 2012; Rodriguez and Atsumi, 2014) or by oxidation to the acid 4-APA (Hanlon et al., 1997) (see Figure 1). An Aro10 enzyme assay with APP, however, was not possible as this compound is not commercially available and should be rather unstable, as previously reported for 4-amino-4-deoxychorismate and 4-amino-4-deoxyprephenate (Teng et al., 1985; Wubbolts et al., 2005; Konishi et al., 2017).
We therefore decided to first perform a biotransformation assay with L-PAPA as surrogate substrate; L-PAPA had been previously purified from culture supernatants of a recombinant E. coli producer strain (“Bio”-L-PAPA) (Mohammadi Nargesi et al., 2018). We reasoned that L-PAPA can be taken up by E. coli cells and can be transaminated intracellularly to APP by the existent aminotransferases (TyrB and/or AspC). For the biotransformation approach, we used E. coli LJ110/pJF119EH as control and E. coli LJ110/pJFA10 to enable a plasmid borne expression of aro10. Furthermore, we used E. coli LJ110/pJFA10F to express additionally feaB from E. coli encoding a phenylacetaldehyde dehydrogenase (Ferrandez et al., 1997; Hanlon et al., 1997; Koma et al., 2012; Zhang et al., 2017). After 8 h of incubation, conversion of L-PAPA neither to PAPE nor to APA could be detected by E. coli LJ110/pJF119EH cells (Figure 2). L-PAPA was converted to PAPE by E. coli LJ110/pJFA10 with a conversion yield of 0.69 g g l−1 PAPE/L-PAPA and a titer of 0.69 ± 0.034 g l−1 PAPE (Figure 2A). A conversion yield of 0.80 g g l−1 4-APA/L-PAPA with E. coli LJ110/pJFA10F was detected with a titer of 0.89 ± 0.032 g l−1 (Figure 2B). Reciprocally, 0.12 ± 0.03 g l−1 4-APA were detected with E. coli LJ110/pJFA10 cells and 0.075 ± 0.005 g l−1 PAPE in E. coli LJ110/pJFA10F as by-products. This indicated that APAAL is diverted to at least two different products (Figure 2).
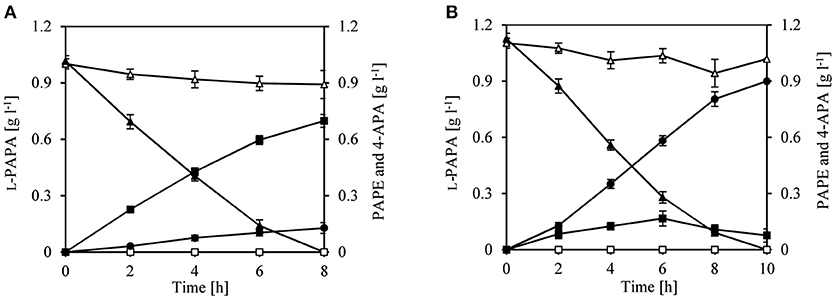
Figure 2. Whole cell biotransformation of para-amino-L-phenylalanine (L-PAPA) with resting cells of E. coli LJ110/pJFA10 and LJ110/pJFA10F. The Biotransformation was performed at 30°C on a rotary shake with 110 rpm and an initial L-PAPA concentration of 1–1.1 g l−1 (triangles). Resting cells of E. coli LJ110/pJF119EH (empty symbols), LJ110/pJFA10 (A; filled symbols) and LJ110/pJFA10F (B; filled symbols) were used. The PAPE formation (squares) and 4-APA (circles) were determined by HPLC. The data represent the mean and standard deviations from measurements of three biological replicates.
Apparently, Aro10 accepted APP as substrate and catalyzed the synthesis of the precursor of PAPE and APA, APAAL. Based upon this finding, we went on to construct the plasmids pC53BCA and pC53BCAF (Figure S3) to allow in vivo biosynthesis of either PAPE or APA from glucose as carbon source. Plasmid pC53BCA contains aro10, whereas pC53BCAF contains both aro10 and feaB (see Table S1 and Figure S3). These plasmids were introduced into wild type strain E. coli LJ110. When grown in shake flasks with glucose as sole carbon source, the resulting strains E. coli LJ110/pC53BCA produced 11 ± 1.5 mg l−1 PAPE or E. coli LJ110/pC53BCAF produced 36 ± 5 mg l−1 4-APA (Figure 3). While we took this as an initial proof that our strategy worked, it clearly left room for improvement in productivities.
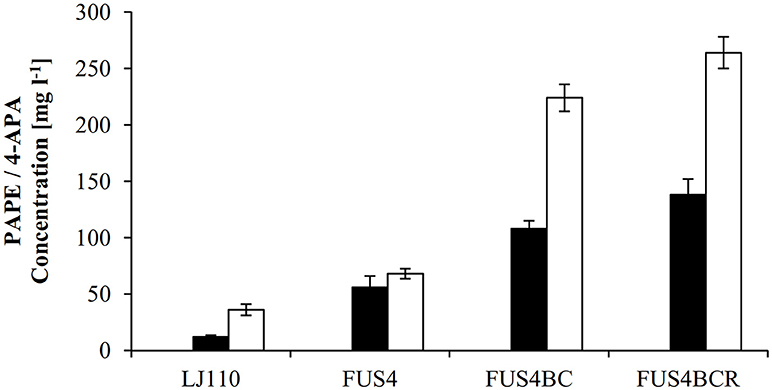
Figure 3. Overview of PAPE or 4-APA production by different E. coli strains. The E. coli strains LJ110, FUS4, FUS4BC, and FUS4BCR with either pC53BCA for PAPE production (filled columns), or with pC53BCAF for 4-APA production (empty columns) were compared. The PAPE and 4-APA titers were determined by HPLC measurements after 48h batch cultivation in minimal media with 4.5 g l−1 glucose. The data represent the mean and standard deviations from measurements of three biological replicates.
Elimination of Competing Pathways and Negative Regulation to Enhance Carbon Flux Toward PAPE or 4-APA
To augment PAPE or 4-APA production, we then turned to a more advanced E. coli LJ110-derived strain, FUS4 (Gottlieb et al., 2014) which we also had successfully used for L-PAPA formation (Mohammadi Nargesi et al., 2018). This strain carries gene deletions of pheA (chorismate mutase/prephenate dehydratase) and tyrA (chorismate mutase/prephenate dehydrogenase), as these gene products directly compete for the same precursor, chorismate. Furthermore, an extra copy of aroFBL (as a gene cassette) is chromosomally inserted (see Table S1). When grown in shake flasks with glucose as sole C source, the PAPE titer was enhanced to 56 ± 10 mg l−1 (Figure 3) with E. coli FUS4/pC53BCA. On the other hand, the 4-APA titer was also increased to 68 ± 4.5 mg l−1 by E. coli FUS4/pC53BCAF.
Next, to reduce the competing carbon flux toward L-PAPA by transamination of APP by TyrB and AspC (Hayashi et al., 1993), the strain FUS4BC was applied. FUS4BC has gene deletions of the aminotransferase genes tyrB and aspC, which leads to an aspartic acid auxotrophy (see Table S1). FUS4BC cells, when transformed with pC53BCA and pC53BCAF, showed increased PAPE or 4-APA titers of 108 ± 7.5 and 224.6 ± 14.2 mg l−1 (Figure 3), respectively. Thus, we concluded that the aminotransferases TyrB and AspC are, indeed, competitors for APP and their removal allows improved formation of both, PAPE or 4-APA.
The transcriptional regulator TyrR (gene tyrR) is involved in the tight regulation of genes involved in shikimate pathway (Pittard et al., 2005). We had already seen positive effects on L-PAPA production with tyrR-negative strains of E. coli (Mohammadi Nargesi et al., 2018). To alleviate TyrR regulation, tyrR gene was thus deleted in strain FUS4BC to yield FUS4BCR. After introduction of pC53BCA or pC53BCAF into strain FUS4BCR, we grew the strains in shake flasks with glucose as sole C source. The respective product titers increased further to 138 ± 8 mg l−1 PAPE and 264 ± 12 mg l−1 4-APA (Figure 3). This is about 2.5 and 4 times higher than FUS4/pC53BCA or FUS4/pC53BCAF, respectively (Figure 3) and significantly higher than with strain FUS4BC. During PAPE production, 23 ± 7.5 mg l−1 4-APA as by-product was observed in FUS4BCR/pC53BCA. We assume that accumulation of 4-APA by FUS4BCR/pC53BCA is due to an endogenous phenylacetaldehyde dehydrogenase (FeaB, AldB and/or AldH) activity in E. coli as described before (Ho and Weiner, 2005; Jo et al., 2008; Koma et al., 2012; Zhang et al., 2017).
Effect of Additionally Expressed Dehydrogenase/Reductase Activities on PAPE Synthesis
As described above, Aro10 apparently is able to produce APAAL from APP. This aldehyde then can either be reduced to an alcohol (PAPE) or may be oxidized to 4-APA. In E. coli, there have been several reports on various genes which encode real or predicted activities of alcohol dehydrogenases/aldehyde reductases (Atsumi et al., 2008a; Koma et al., 2012; Kunjapur et al., 2014; Rodriguez and Atsumi, 2014) or of phenylacetaldehyde dehydrogenase (Hanlon et al., 1997; Koma et al., 2012; Zhang et al., 2017). As we looked for genes which encode aldehyde reductases or alcohol dehydrogenase which might facilitate the PAPE production in our E. coli strains, we decided to clone heterologous genes from the yeast S. cerevisiae (alcohol dehydrogenase genes ADH1 and ADH2) as well as the endogenous E. coli yahK gene, which had been proposed to be most efficient for the production of aromatic alcohols (Koma et al., 2012). The three genes were cloned separately into plasmid pC53BCA to yield the new plasmids pC53BCAHII, pC53BCAHI, and pC53BCAY (Figure S3). Each plasmid was then introduced into our best producer stain so far, FUS4BCR (Table S1, Figure S3). The cell growth and the PAPE formation were observed for 48 h in shake flask batch cultures with glucose as sole C source.
First, to our surprise, FUS4BCR/pC53BCAHII did not produce PAPE at all; rather 4-APA was detected at 85 mg l−1 (Table 1). This finding could be rationalized if the recombinant ADHII enzyme in E. coli either does not reduce APAAL to PAPE, or even catalyzes the re-oxidation of PAPE thereby counteracting the action of endogenous E. coli aldehyde reductases.
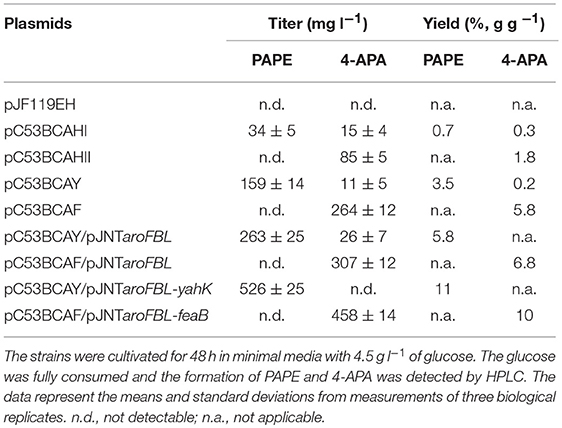
Table 1. PAPE and 4-APA production by E. coli FUS4BCR strains harboring different combinations of plasmids.
The expression of ADH1 (plasmid pC53BCAHI) resulted in a titer of about 40 mg l−1 while the overexpression of the plasmid-borne yahK led to an increased PAPE titer of 159 mg l−1 (Table 1). Thus, from the tested genes, yahK overexpression gave best results for PAPE production.
Plasmid-Borne Overexpression of Genes to Further Enhance PAPE or 4-APA Production
One common strategy to enhance the flux through the shikimate pathway is increasing the activity of major rate-limiting steps in the pathway (Báez-Viveros et al., 2007; Sprenger, 2007a; Gosset, 2009; Chávez-Béjar et al., 2013; Gottlieb et al., 2014). Hence to improve the flux toward chorismate as the main precursor for APP biosynthesis, we turned to a two-plasmid system with the additional vector pJNT-aroFBL (Mohammadi Nargesi et al., 2018) which confers kanamycin resistance. This vector is compatible with the pC53 plasmid series and was introduced into FUS4BCR/pC53BCAY and FUS4/pC53BCAF. In shake flask batch cultivations, the PAPE titer of FUS4BCR/pC53BCAY/pJNT-aroFBL increased to 263 ± 15 mg l−1 PAPE (Table 1), which was ~2-fold higher compared to the FUS4BCR/pC53BCAY (Table 1). Within 48 h, FUS4BCR/pC53BCAF/pJNT-aroFBL consumed 4.5 g l−1 glucose completely and produced 307 ± 12 mg l−1 4-APA, which is about 1.5-fold more than the concentration of 4-APA observed with the one plasmid strain FUS4BCR/pC53BCAF.
To see whether an additional copy of either yahK or feaB genes from E. coli would lead to a further increase of products, they were cloned into pJNT-aroFBL generating pJNT-aroFBL-yahK and pJNT-aroFBL-feaB, respectively (Figure S3). These additional gene overexpressions led to a final titer of PAPE (526 ± 25 mg l−1) in the case of yahK and 4-APA (458 ± 14.5 mg l−1) in the case of feaB. In both strains no reciprocal by-product (4-APA and PAPE, respectively) was detectable (Table 1). Therefore, we decided to use strain FUS4BCR/pC53BCAY/pJNT-aroFBL-yahK and FUS4BCR/pC53BCAF/pJNT-aroFBL-feaB for fed-batch cultivations for PAPE or 4-APA, respectively.
Fed-Batch Cultivations for PAPE and 4-APA Production
We changed the cultivation conditions to fed-batch to see whether product titers of PAPE and 4-APA could be further improved. After 168 h of cultivation of strain FUS4BCR/pC53BCAY/pJNT-aroFBL-yahK, 33.3 g l−1 glucose were consumed and a titer of 2.5 ± 0.15 g l−1 PAPE with a yield of 0.08 PAPE/g of glucose (11% C mol PAPE mol−1 glucose) was reached (Figure 4A). FUS4BCR/pC53BCAF/pJNT-aroFBL-feaB gave a final product concentration of 3.4 ± 0.3 g l−1 4-APA from 25.3 g l−1 glucose with a yield of 0.14 4-APA/g of glucose (17% C mol 4-APA mol−1 glucose) after 168 h (Figure 4B). To the best of our knowledge these are the highest reported PAPE and 4-APA titers with recombinant E. coli strains.
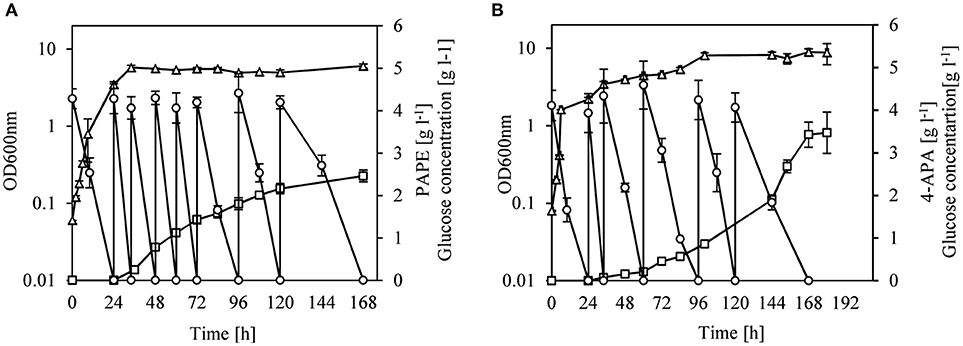
Figure 4. Fed-batch cultivation of E. coli FUS4BCR/pC53BCAY/pJNTaroFBL-yahK (A) or E. coli FUS4BCR/pC53BCAF/pJNTaroFBL-feaB (B) for PAPE or 4-APA production. Cell growth (empty triangles) was determined by measuring OD600, glucose concentration (empty circles) and the total PAPE or 4-APA production (empty squares) in the minimal media were determined by using HPLC. The cells were cultivated in 20 ml minimal media for 168 h. The data represent the mean and standard deviations from measurements of three biological replicates.
Discussion
In this study we established a de novo biosynthesis pathway for two aromatic amine products, PAPE and 4-APA in E. coli which share a common precursor, APAAL. We could demonstrate that E. coli can efficiently produce PAPE and 4-APA for the first time in gram scale from glucose. Recently, Takaya and coworkers have also studied the microbial biosynthesis of PAPE and 4-APA with recombinant E. coli strains (Masuo et al., 2016). However, to reach the high titer of 2.8 g l−1 PAPE and 2.7 g l−1 4-APA, these authors used whole cell biotransformations using the expensive L-PAPA as substrate with E. coli cells overexpressing the aro10 gene from yeast and relying on the endogenous aminotransferase activities of E. coli (Masuo et al., 2016).
From the results obtained by the Takaya group and the present report with biotransformation with externally added L-PAPA as substrate it becomes apparent that the gene product of the yeast aro10 gene encodes a decarboxylase with a broader substrate range than reported before (Vuralhan et al., 2005; Kneen et al., 2011). We should emphasize however that we could not detect formation of the aldehyde APAAL; rather, we detected the follow-up products, either the reduced PAPE or the oxidized 4-APA. The approach of Takaya and coworkers differs from our report as the former group applied a synthetic gene cluster of papABC from Pseudomonas fluorescens for para-amino phenylpyruvate (APP) production in E. coli (Masuo et al., 2016). In our study, a combination of the pabAB fusion gene from C. glutamicum and of the codon-optimized genes of papB and papC from S. venezuelae was used to allow a conversion of chorismate to APP (Mohammadi Nargesi et al., 2018). We combined the APP biosynthesis pathway with the yeast Ehrlich pathway in E. coli by recruiting Aro10 to enable PAPE and 4-APA production (Figure 1) (Vuralhan et al., 2003, 2005; Vogt and Gerulis, 2005; Atsumi et al., 2008b; Kneen et al., 2011; Machas et al., 2017; Mohammadi Nargesi et al., 2018). Already the wild type strain E. coli LJ110 harboring pC53BCA or pC53BCAF was able to produce 11 ± 1.5 mg l−1 PAPE or 36 ± 5 mg l−1 4-APA; the correct mass of these products was confirmed by mass spectrometry (Figure S2). This result indicated that the de-novo PAPE or 4-APA biosynthesis pathway was successfully established and that the endogenous aldehyde reductases/dehydrogenases of E. coli (Atsumi et al., 2008b, 2010; Koma et al., 2012; Kunjapur et al., 2014; Rodriguez and Atsumi, 2014) are able to convert APAAL to PAPE or 4-APA, respectively.
In order to improve the flux toward the products, we decided to use more advanced E. coli strains. E. coli strain FUS4 (Gottlieb et al., 2014) carries gene deletions (pheA-tyrA) which remove the activities of the bifunctional enzymes and thus allow increased chorismate flux to the desired products as the formation of L-Tyr and L-Phe is disabled (Backman et al., 1990; Rüffer et al., 2004; Sprenger, 2007a,b; Sun et al., 2011; Gottlieb et al., 2014; Weiner et al., 2014). As a certain disadvantage for growth in minimal media, however, supplementation of both aromatic amino acids is necessary. Strain FUS4-when transformed with plasmids pC53BCA or pC53BCAF, respectively—yielded increased titers of PAPE or 4-APA compared to the wild type host strain. The PAPE titer was improved five times to 56 ± 10 mg l−1 while the 4-APA titer was almost doubled to 68 ± 4.5 mg l −1 (Figure 3). Thus, the removal of competing pathways resulted in an improved formation of PAPE and/or 4-APA.
In E. coli, several aminotransferases are known to catalyze terminal steps in aromatic amino acid biosynthesis (Fotheringham et al., 1986; Inoue et al., 1988; Hayashi et al., 1993; Pittard, 1996; Marienhagen et al., 2005). For the transamination of the aromatic amino acids L-phenylalanine and L-tyrosine, four aminotransferases (AspC, TyrB, IlvE, and AvtA) are involved in E. coli (Pittard, 1996; Rodriguez et al., 2014; Li et al., 2016). Previous studies had already shown that the deletions of genes aspC and tyrB led to an aspartic acid auxotrophy but also to an increased precursor supply of aromatic ketoacids like phenylpyruvate or 4-hydroxyphenylpyruvate (Liu et al., 2014, 2015; Pugh et al., 2014; Li et al., 2016). In good agreement with these former studies, the use of E. coli FUS4BC which lacks aspC and tyrB, led to an almost doubling of the titer for PAPE to 108 ± 7.5 mg l−1 and the 4-APA titer of 224.6 ± 14.2 mg l−1 was 3-fold higher (Figure 3).
Furthermore, the transcriptional repressor tyrR was deleted as its inactivation is known to enhance the expression of aromatic biosynthesis genes, like aroG, tyrB, aroP, tyrA, and aroL (Pittard, 1996; Bongaerts et al., 2001; Pittard et al., 2005; Salgado et al., 2006). The beneficial effect of a tyrR deletion had been already shown for the biosynthesis of aromatic amino acids like L-Tyr (Lutke-Eversloh and Stephanopoulos, 2007), L-Phe (Doroshenko et al., 2015), L-DOPA (Munoz et al., 2011; Das et al., 2018), and recently L-PAPA (Mohammadi Nargesi et al., 2018). The cultivation of FUS4BCR/pC53BCA or/pC53BCAF increased the PAPE or 4-APA titer up to 27 or 17% than FUS4BC/pC53BCA or/pC53BCAF, respectively (Figure 3).
The reduction or the oxidation of APAAL to PAPE and 4-APA in E. coli is accomplished by endogenous aldehyde reductases encoded by the yqhD, yjgB, yahK, or other genes (in total 13 known endogenous aldehyde reductases) (Atsumi et al., 2008a; Koma et al., 2012; Rodriguez and Atsumi, 2014) or phenylacetaldehyde dehydrogenase encoded by feaB in E. coli (Hanlon et al., 1997), respectively. Although E. coli FUS4BCR/pC53BCA produced PAPE already with a reasonable titer, a formation of the by-product 4-APA with ~23 ± 7.5 mg l−1 was detected. This indicated that the endogenous dehydrogenase gene (feaB) in the genome of E. coli, enables the conversion of APAAL to 4-APA, which limits the PAPE production (Masuo et al., 2016; Machas et al., 2017). To avoid an unwanted 4-APA accumulation we tested three genes yahK, ADH1, and ADH2 for their potential roles in accelerating PAPE formation as the role of these genes had been discussed before by others (Dickinson et al., 2003; Koma et al., 2012; Kim et al., 2014a,b). Unexpectedly however, introduction of pC53BCAHII into strain E. coli FUS4BCR resulted in a complete loss of PAPE production (Table 1). This could be explained by a preference of the recombinant yeast ADHII enzyme to catalyze the oxidation of alcohols instead of a reduction of aldehydes (Thomson et al., 2005; De Smidt et al., 2008; Kang et al., 2012). When the gene for ADHI from yeast was introduced on pC53BCAHI plasmid into strain FUS4BCR this led to lower level of PAPE than in the strain without an extra alcohol dehydrogenase FUS4BCR/pC53BCA, e.g., 34.3 ± 5.2 mg l−1 (see Figure 3 and Table 1). Also for ADHI enzyme reports pointed to its ability to oxidize primary alcohols at high concentrations (Schöpp and Aurich, 1976; De Smidt et al., 2008; Atsumi et al., 2010). So in comparison of the three alcohol dehydrogenases/aldehyde reductases which we tested, the cloned endogenous aldehyde reductase encoded by yahK (Koma et al., 2012; Rodriguez and Atsumi, 2014) on pC53BCAY led to increased PAPE production in E. coli FUS4BCR. Simultaneously this led to a reduction of the by-product 4-APA to 11 ± 5 mg l−1 (Table 1). To ensure a sufficient supply of chorismate for the biosynthesis of PAPE or 4-APA, an increased carbon flux through the shikimate pathway is needed. This can be achieved by deregulation of expression of aroF (DAHP synthase) and relieving limiting enzymatic reactions of DHQ synthase (encoded by aroB) and shikimate kinase II (encoded by aroL) (Dell and Frost, 1993; Oldiges et al., 2004; Báez-Viveros et al., 2007; Sprenger, 2007b). Therefore, we used the plasmid pJNT-aroFBL to overexpress the genes aroF, aroB, and aroL (Mohammadi Nargesi et al., 2018). Indeed, the two plasmid combination of pJNTaroFBL with pC53BCAY or pC53BCAF in FUS4BCR strains, resulted in an increased carbon flux to chorismate and eventually to the desired products. The co-expression of aroFBL led to about 1.5-fold increase of PAPE or 4-APA titer in FUS4BCR pC53BCAY/pJNTaroFBL (263 ± 15 mg l−1) or FUS4BCR pC53BCAF/pJNT-aroFBL (307 ± 12 mg l−1). The yield was increased about 40% from 3.5% PAPE/glucose (g g−1) (Table 1) to 5.8% PAPE/glucose (g g−1) and a 17% increase in yield was observed for 4-APA (Table 1). This beneficial effect of an increased flux through the shikimate pathway for PAPE and 4-APA formation was also shown for other compounds derived from the key metabolite chorismate (Sprenger, 2007b; Kang et al., 2012; Rodrigues et al., 2013; Yao et al., 2013; Gottlieb et al., 2014; Lin et al., 2014; Weiner et al., 2014; Mohammadi Nargesi et al., 2018).
As small amounts of PAPE or 4-APA were still detectable as byproducts during the production of 4-APA or PAPE (Table 1), respectively, we decided to integrate an additional gene copy of genes yahK or feaB or on the vector pJNT-aroFBL. Compared to the previous strains, the additional gene copies of yahK or feaB caused a 90% and 47% enlargement of the PAPE and 4-APA titers (Table 1). The unwanted side-reactions for PAPE or 4-APA (oxidation or reduction of APAAL, respectively) could be avoided without inactivation of dehydrogenase (feaB) or oxidoreductase (yahK, yqhD, yjgB, and the other 10 genes), respectively (Rodriguez and Atsumi, 2014; Machas et al., 2017).
Finally, to increase the PAPE and 4-APA titers, the cultivation condition was changed to a fed-batch cultivation in shake flask. A high titer of 2.5 ± 0.15 g l−1 PAPE or 3.4 ± 0.3 g l−1 4-APA could be reached which corresponds to a yield of 11 or 17% carbon mol mol−1 with glucose after 168 h (Figure 4). This titer is higher than the previously described titers of PAPE (0.24 g l−1) and 4-APA (0.19 g l−1) in minimal media with 20 g l−1 glucose (supplemented with tryptone and yeast extract) from the Takaya group (Masuo et al., 2016; Konishi et al., 2017). Even though we reached the gram scale in shake flask the cultivation was not optimal in term of oxygen supply and pH stability. A cultivation in a pH- and O2-controlled bioreactor could be advantageous and will be analyzed in future studies. Furthermore, as we observed a negative effect of PAPE and 4-APA on the growth of E. coli at higher concentration (Figure S1), a deletion of the global stress regulator rpoS may be beneficial for the aromatic amine production as it was seen for the production of the diamine putrescine (Qian et al., 2009).
To further augment the PAPE or 4-APA production performance we have to consider to improve the E4P and PEP precursor supply for future strain developments (Patnaik et al., 1995; Bongaerts et al., 2001; Chandran et al., 2003; Sprenger, 2007b; Gosset, 2009; Rodriguez et al., 2014). It is very likely that a positive effect will also be observed for PAPE and 4-APA production as we detected it for L-phenylalanine and L-PAPA production (Gottlieb et al., 2014; Mohammadi Nargesi et al., 2018; Trondle et al., 2018). In addition an even more improved flux through the shikimate pathway may increase the productivity as it was demonstrated that an increased flux through the L-lysine pathway augmented the biosynthesis of the diamine cadaverine (Qian et al., 2011). Although we already minimized the byproduct formation by overexpression of feaB or yahK, we have to consider also to eliminate the genes to suppress phenylacetic acid/4-hydroxyphenylacetic acid pathway in a further improved process as it was done previously (Satoh et al., 2012; Bai et al., 2014; Machas et al., 2017; Xue et al., 2017; Liu et al., 2018).
In conclusion, our study demonstrated that E. coli is a suitable chassis strain for both, PAPE and 4-APA production. By a combination of improved flux, avoidance of by-product formation and a change in the cultivation condition a gram scale production of PAPE and 4-APA was achieved.
Availability of Data and Materials
The dataset(s) supporting the conclusions of this article are all included within the article and additional files.
Author Contributions
BM performed the experiments. GS and J-WY provided guidance for the experimental setups. BM, GS, and J-WY wrote the final manuscript. All authors approved the final version of the manuscript.
Funding
BM was supported by a personal grant of the Ministry of Science, Research and Technology of Iran (MSRT; 91000277).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Jana Hoffmann (Institute of Industrial Genetics, University of Stuttgart) for kindly providing the yeast strain, to Dr. Bernd A. Nebel (Institute of Technical Biochemistry, University of Stuttgart) for helping with the MS measurements. We are deeply grateful to the Ministerium für Wissenschaft und Kunst (MWK), Baden-Württemberg, Germany for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2018.00201/full#supplementary-material
References
Arora, P. K. (2015). Bacterial degradation of monocyclic aromatic amines. Front. Microbiol. 6, 820–820. doi: 10.3389/fmicb.2015.00820
Atsumi, S., Cann, A. F., Connor, M. R., Shen, C. R., Smith, K. M., Brynildsen, M. P., et al. (2008a). Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10, 305–311. doi: 10.1016/j.ymben.2007.08.003
Atsumi, S., Hanai, T., and Liao, J. C. (2008b). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. doi: 10.1038/nature06450
Atsumi, S., Wu, T.-Y., Eckl, E.-M., Hawkins, S. D., Buelter, T., and Liao, J. C. (2010). Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 85, 651–657. doi: 10.1007/s00253-009-2085-6
Backman, K., O'connor, M. J., Maruya, A., Rudd, E., Mckay, D., Balakrishnan, R., et al. (1990). Genetic engineering of metabolic pathways applied to the production of phenylalanine. Ann. N. Y. Acad. Sci. 589, 16–24. doi: 10.1111/j.1749-6632.1990.tb24231.x
Báez-Viveros, J. L., Flores, N., Juárez, K., Castillo-España, P., Bolivar, F., and Gosset, G. (2007). Metabolic transcription analysis of engineered Escherichia coli strains that overproduce L-phenylalanine. Microb. Cell Fact. 6, 30–30. doi: 10.1186/1475-2859-6-30
Bai, Y., Bi, H., Zhuang, Y., Liu, C., Cai, T., Liu, X., et al. (2014). Production of salidroside in metabolically engineered Escherichia coli. Sci. Rep. 4, 6640–6640. doi: 10.1038/srep06640
Blanc, V., Gil, P., Bamas-Jacques, N., Lorenzon, S., Zagorec, M., Schleuniger, J., et al. (1997). Identification and analysis of genes from Streptomyces pristinaespiralis encoding enzymes involved in the biosynthesis of the 4-dimethylamino-L-phenylalanine precursor of pristinamycin I. Mol. Microbiol. 23, 191–202. doi: 10.1046/j.1365-2958.1997.2031574.x
Bongaerts, J., Kramer, M., Muller, U., Raeven, L., and Wubbolts, M. (2001). Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab. Eng. 3, 289–300. doi: 10.1006/mben.2001.0196
Brown, M. P., Aidoo, K. A., and Vining, L. C. (1996). A role for pabAB, a p-aminobenzoate synthase gene of Streptomyces venezuelae ISP5230, in chloramphenicol biosynthesis. Microbiology 142, 1345–1355. doi: 10.1099/13500872-142-6-1345
Chandran, S. S., Yi, J., Draths, K. M., Von Daeniken, R., Weber, W., and Frost, J. W. (2003). Phosphoenolpyruvate availability and the biosynthesis of shikimic acid. Biotechnol. Progr. 19, 808–814. doi: 10.1021/bp025769p
Chang, Z., Sun, Y., He, J., and Vining, L. C. (2001). p-Aminobenzoic acid and chloramphenicol biosynthesis in Streptomyces venezuelae: gene sets for a key enzyme, 4-amino-4-deoxychorismate synthase. Microbiology 147, 2113–2126. doi: 10.1099/00221287-147-8-2113
Chávez-Béjar, M. I., Balderas-Hernandez, V. E., Gutiérrez-Alejandre, A., Martinez, A., Bolívar, F., and Gosset, G. (2013). Metabolic engineering of Escherichia coli to optimize melanin synthesis from glucose. Microb. Cell Fact. 12, 108–108. doi: 10.1186/1475-2859-12-108
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-A
Das, A., Tyagi, N., Verma, A., Akhtar, S., and Mukherjee, K. J. (2018). Metabolic engineering of Escherichia coli W3110 strain by incorporating genome-level modifications and synthetic plasmid modules to enhance L-Dopa production from glycerol. Prep. Biochem. Biotechnol. 7, 1–12. doi: 10.1080/10826068.2018.1487851
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De Smidt, O., Du Preez, J. C., and Albertyn, J. (2008). The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Res. 8, 967–978. doi: 10.1111/j.1567-1364.2008.00387.x
Delaplaza, M., Fernandezdepalencia, P., Pelaez, C., and Requena, T. (2004). Biochemical and molecular characterization of α-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol. Lett. 238, 367–374. doi: 10.1016/j.femsle.2004.07.057
Dell, K. A., and Frost, J. W. (1993). Identification and removal of impediments to biocatalytic synthesis of aromatics from D-glucose: rate-limiting enzymes in the common pathway of aromatic amino acid biosynthesis. J. Am. Chem. Soc. 115, 11581–11589. doi: 10.1021/ja00077a065
Dickinson, J. R., Salgado, L. E. J., and Hewlins, M. J. E. (2003). The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278, 8028–8034. doi: 10.1074/jbc.M211914200
Doroshenko, V. G., Livshits, V. A., Airich, L. G., Shmagina, I. S., Savrasova, E. A., Ovsienko, M. V., et al. (2015). Metabolic engineering of Escherichia coli for the production of phenylalanine and related compounds. Appl. Biochem. Microbiol. 51, 733–750. doi: 10.1134/S0003683815070017
Etschmann, M. M. W., Bluemke, W., Sell, D., and Schrader, J. (2002). Biotechnological production of 2-phenylethanol. Appl. Microbiol. Biotechnol. 59, 1–8. doi: 10.1007/s00253-002-0992-x
Ferrandez, A., Prieto, M. A., Garcia, J. L., and Diaz, E. (1997). Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 406, 23–27. doi: 10.1016/S0014-5793(97)00228-7
Fotheringham, I. G., Dacey, S. A., Taylor, P. P., Smith, T. J., Hunter, M. G., Finlay, M. E., et al. (1986). The cloning and sequence analysis of the aspC and tyrB genes from Escherichia coli K12. Comparison of the primary structures of the aspartate aminotransferase and aromatic aminotransferase of E. coli with those of the pig aspartate aminotransferase isoenzymes. Biochem. J. 234, 593–604. doi: 10.1042/bj2340593
Froidevaux, V., Negrell, C., Caillol, S., Pascault, J.-P., and Boutevin, B. (2016). Biobased amines: from synthesis to polymers; present and future. Chem. Rev. 116, 14181–14224. doi: 10.1021/acs.chemrev.6b00486
Gerhardt, P., Murray, R. G. E., Wood, W., and Krieg, N. (1984). Methods for General and Molecular Bacteriology. Washington, DC: ASM Press.
Gosset, G. (2009). Production of aromatic compounds in bacteria. Curr. Opin. Biotechnol. 20, 651–658. doi: 10.1016/j.copbio.2009.09.012
Gottlieb, K., Albermann, C., and Sprenger, G. A. (2014). Improvement of L-phenylalanine production from glycerol by recombinant Escherichia coli strains: the role of extra copies of glpK, glpX, and tktA genes. Microb. Cell Fact. 13:96. doi: 10.1186/s12934-014-0096-1
Hanlon, S. P., Hill, T. K., Flavell, M. A., Stringfellow, J. M., and Cooper, R. A. (1997). 2-Phenylethylamine catabolism by Escherichia coli K-12: gene organization and expression. Microbiology 143, 513–518. doi: 10.1099/00221287-143-2-513
Hayashi, H., Inoue, K., Nagata, T., Kuramitsu, S., and Kagamiyama, H. (1993). Escherichia coli aromatic amino acid aminotransferase: characterization and comparison with aspartate aminotransferase. Biochemistry 32, 12229–12239. doi: 10.1021/bi00096a036
Hazelwood, L. A., Daran, J. M., Van Maris, A. J., Pronk, J. T., and Dickinson, J. R. (2008). The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266. doi: 10.1128/AEM.02625-07
He, J., Magarvey, N., Piraee, M., and Vining, L. C. (2001). The gene cluster for chloramphenicol biosynthesis in Streptomyces venezuelae ISP5230 includes novel shikimate pathway homologues and a monomodular non-ribosomal peptide synthetase gene. Microbiology 2331, 2817–2829. doi: 10.1099/00221287-147-10-2817
Ho, K. K., and Weiner, H. (2005). Isolation and characterization of an aldehyde dehydrogenase encoded by the aldB gene of Escherichia coli. J. Bacteriol. 187, 1067–1073. doi: 10.1128/JB.187.3.1067-1073.2005
Hua, D., and Xu, P. (2011). Recent advances in biotechnological production of 2-phenylethanol. Biotechnol. Adv. 29, 654–660. doi: 10.1016/j.biotechadv.2011.05.001
Inoue, K., Kuramitsu, S., Aki, K., Watanabe, Y., Takagi, T., Nishigai, M., et al. (1988). Branched-chain amino acid aminotransferase of Escherichia coli: overproduction and properties. J. Biochem. 104, 777–784. doi: 10.1093/oxfordjournals.jbchem.a122549
Jo, J. E., Raj, S. M., Rathnasingh, C., Selvakumar, E., Jung, W. C., and Park, S. (2008). Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-hydroxypropionaldehyde as a substrate. Appl. Microbiol. Biotechnol. 81, 51–60. doi: 10.1007/s00253-008-1608-x
Kang, S.-Y., Choi, O., Lee, J., Hwang, B., Uhm, T.-B., and Hong, Y.-S. (2012). Artificial biosynthesis of phenylpropanoic acids in a tyrosine overproducing Escherichia coli strain. Microb. Cell Fact. 11, 153–153. doi: 10.1186/1475-2859-11-153
Kawaguchi, H., Ogino, C., and Kondo, A. (2017). Microbial conversion of biomass into bio-based polymers. Bioresour. Technol. 245, 1664–1673. doi: 10.1016/j.biortech.2017.06.135
Kawasaki, Y., Aniruddha, N., Minakawa, H., Masuo, S., Kaneko, T., and Takaya, N. (2018). Novel polycondensed biopolyamide generated from biomass-derived 4-aminohydrocinnamic acid. Appl. Microbiol. Biotechnol. 102, 631–639. doi: 10.1007/s00253-017-8617-6
Kim, B., Cho, B. R., and Hahn, J. S. (2014a). Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol. Bioeng. 111, 115–124. doi: 10.1002/bit.24993
Kim, B., Park, H., Na, D., and Lee, S. Y. (2014b). Metabolic engineering of Escherichia coli for the production of phenol from glucose. Biotechnol. J. 9, 621–629. doi: 10.1002/biot.201300263
Kneen, M. M., Stan, R., Yep, A., Tyler, R. P., Saehuan, C., and Mcleish, M. J. (2011). Characterization of a thiamin diphosphate-dependent phenylpyruvate decarboxylase from Saccharomyces cerevisiae. FEBS J. 278, 1842–1853. doi: 10.1111/j.1742-4658.2011.08103.x
Koma, D., Yamanaka, H., Moriyoshi, K., Ohmoto, T., and Sakai, K. (2012). Production of aromatic compounds by metabolically engineered Escherichia coli with an expanded shikimate pathway. Appl. Environ. Microbiol. 78, 6203–6216. doi: 10.1128/AEM.01148-12
Koma, D., Yamanaka, H., Moriyoshi, K., Sakai, K., Masuda, T., Sato, Y., et al. (2014). Production of p-aminobenzoic acid by metabolically engineered Escherichia coli. Biosci. Biotechnol. Biochem. 78, 350–357. doi: 10.1080/09168451.2014.878222
Konishi, K., Naoki, T., Shunsuke, M., and Zhou, S. (2016). 4-Amino Cinnamic Acid Production Method Using Enzyme. US patent US20160362.
Konishi, K., Takaya, N., and Masuo, S. (2017). Method for Producing Aaniline Derivative by Fermentation From Carbon Source. Patent WO2015141791A1.
Kozak, S. (2006). Mikrobielle Biosynthese von (3S,4R)-4-Amino-3-hydroxycyclohexa-1,5-diencarbonsäure mit rekombinanten Escherichia coli Zellen: Molekulargenetische und biochemische Untersuchungen. Dissertation, University of Stuttgart, Germany.
Küberl, A., Schneider, J., Thallinger, G. G., Anderl, I., Wibberg, D., Hajek, T., et al. (2011). High-quality genome sequence of Pichia pastoris CBS7435. J. Biotechnol. 154, 312–320. doi: 10.1016/j.jbiotec.2011.04.014
Kubota, T., Watanabe, A., Suda, M., Kogure, T., Hiraga, K., and Inui, M. (2016). Production of para-aminobenzoate by genetically engineered Corynebacterium glutamicum and non-biological formation of an N-glucosyl byproduct. Metab. Eng. 38, 322–330. doi: 10.1016/j.ymben.2016.07.010
Kunjapur, A. M., Tarasova, Y., and Prather, K. L. (2014). Synthesis and accumulation of aromatic aldehydes in an engineered strain of Escherichia coli. J. Am. Chem. Soc. 136, 11644–11654. doi: 10.1021/ja506664a
Lawrence, S. A. (2004). Amines: Synthesis, Properties and Applications. Cambridge: Cambridge University Press.
Li, F.-F., Zhao, Y., Li, B.-Z., Qiao, J.-J., and Zhao, G.-R. (2016). Engineering Escherichia coli for production of 4-hydroxymandelic acid using glucose–xylose mixture. Microb. Cell Fact. 15, 90–90. doi: 10.1186/s12934-016-0489-4
Li, L., Hiroshi, S., Koichiro Yonetake, A., and Ueda, M. (1999). Synthesis and characterization of ordered poly(amide–ester)s from isophthaloyl chloride and 4-(2-aminoethyl)phenol. Macromolecules 32, 3851–3858. doi: 10.1021/ma981919u
Lin, Y., Sun, X., Yuan, Q., and Yan, Y. (2014). Extending shikimate pathway for the production of muconic acid and its precursor salicylic acid in Escherichia coli. Metab. Eng. 23, 62–69. doi: 10.1016/j.ymben.2014.02.009
Liu, S. P., Liu, R. X., El-Rotail, A. A. M. M., Ding, Z. Y., Gu, Z. H., Zhang, L., et al. (2014). Heterologous pathway for the production of L-phenylglycine from glucose by E. coli. J. Biotechnol. 186, 91–97. doi: 10.1016/j.jbiotec.2014.06.033
Liu, S. P., Zhang, L., Mao, J., Ding, Z. Y., and Shi, G. Y. (2015). Metabolic engineering of Escherichia coli for the production of phenylpyruvate derivatives. Metab. Eng. 32, 55–65. doi: 10.1016/j.ymben.2015.09.007
Liu, X., Li, X.-B., Jiang, J., Liu, Z.-N., Qiao, B., Li, F.-F., et al. (2018). Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab. Eng. 47, 243–253. doi: 10.1016/j.ymben.2018.03.016
Lucchini, J. J., Bonnaveiro, N., Cremieux, A., and Legoffic, F. (1993). Mechanism of bactericidal action of phenethyl alcohol in Escherichia coli. Curr. Microbiol. 27, 295–300. doi: 10.1007/BF01575995
Lutke-Eversloh, T., and Stephanopoulos, G. (2007). L-tyrosine production by deregulated strains of Escherichia coli. Appl. Microbiol. Biotechnol. 75, 103–110. doi: 10.1007/s00253-006-0792-9
Machas, M. S., Mckenna, R., and Nielsen, D. R. (2017). Expanding upon styrene biosynthesis to engineer a novel route to 2-phenylethanol. Biotechnol. J. 12, 1700310 (1–10). doi: 10.1002/biot.201700310
Marienhagen, J., Kennerknecht, N., Sahm, H., and Eggeling, L. (2005). Functional analysis of all aminotransferase proteins inferred from the genome sequence of Corynebacterium glutamicum. J. Bacteriol. 187, 7639–7646. doi: 10.1128/JB.187.22.7639-7646.2005
Masker, W. E., and Eberle, H. (1972). Effect of phenethyl alcohol on deoxyribonucleic acid-membrane association in Escherichia coli. J. Bacteriol. 109, 1170–1174.
Masuo, S., Osada, L., Zhou, S., Fujita, T., and Takaya, N. (2015). Aspergillus oryzae pathways that convert phenylalanine into the flavor volatile 2-phenylethanol. Fungal Genet. Biol. 77, 22–30. doi: 10.1016/j.fgb.2015.03.002
Masuo, S., Zhou, S., Kaneko, T., and Takaya, N. (2016). Bacterial fermentation platform for producing artificial aromatic amines. Sci. Rep. 6, 25764. doi: 10.1038/srep25764
Mehl, R. A., Anderson, J. C., Santoro, S. W., Wang, L., Martin, A. B., King, D. S., et al. (2003). Generation of a bacterium with a 21 amino acid genetic code. J. Am. Chem. Soc. 125, 935–939. doi: 10.1021/ja0284153
Mohammadi Nargesi, B., Trachtmann, N., Sprenger, G. A., and Youn, J. W. (2018). Production of p-amino-L-phenylalanine (L-PAPA) from glycerol by metabolic grafting of Escherichia coli. Microb. Cell Fact. 17:149. doi: 10.1186/s12934-018-0996-6
Munoz, A. J., Hernandez-Chavez, G., De Anda, R., Martinez, A., Bolivar, F., and Gosset, G. (2011). Metabolic engineering of Escherichia coli for improving L-3,4-dihydroxyphenylalanine (L-DOPA) synthesis from glucose. J. Ind. Microbiol. Biotechnol. 38, 1845–1852. doi: 10.1007/s10295-011-0973-0
Oldiges, M., Kunze, M., Degenring, D., Sprenger, G. A., and Takors, R. (2004). Stimulation, monitoring, and analysis of pathway dynamics by metabolic profiling in the aromatic amino acid pathway. Biotechnol. Progr. 20, 1623–1633. doi: 10.1021/bp0498746
Patnaik, R., Spitzer, R. G., and Liao, J. C. (1995). Pathway engineering for production of aromatics in Escherichia coli: Confirmation of stoichiometric analysis by independent modulation of AroG, TktA, and Pps activities. Biotechnol. Bioeng. 46, 361–370. doi: 10.1002/bit.260460409
Pittard, J. (1996). “Biosynthesis of the aromatic amino acids,” in Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn., eds F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, et al. (Washington, DC: ASM Press), 458–484.
Pittard, J., Camakaris, H., and Yang, J. (2005). The TyrR regulon. Mol. Microbiol. 55, 16–26. doi: 10.1111/j.1365-2958.2004.04385.x
Pugh, S., Mckenna, R., Osman, M., Thompson, B., and Nielsen, D. R. (2014). Rational engineering of a novel pathway for producing the aromatic compounds p-hydroxybenzoate, protocatechuate, and catechol in Escherichia coli. Process Biochem. 49, 1843–1850. doi: 10.1016/j.procbio.2014.08.011
Qian, Z. G., Xia, X. X., and Lee, S. Y. (2009). Metabolic engineering of Escherichia coli for the production of putrescine: a four carbon diamine. Biotechnol. Bioeng. 104, 651–662. doi: 10.1002/bit.22502
Qian, Z. G., Xia, X. X., and Lee, S. Y. (2011). Metabolic engineering of Escherichia coli for the production of cadaverine: a five carbon diamine. Biotechnol. Bioeng. 108, 93–103. doi: 10.1002/bit.22918
Rodrigues, A. L., Trachtmann, N., Becker, J., Lohanatha, A. F., Blotenberg, J., Bolten, C. J., et al. (2013). Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab. Eng. 20, 29–41. doi: 10.1016/j.ymben.2013.08.004
Rodriguez, A., Martinez, J. A., Flores, N., Escalante, A., Gosset, G., and Bolivar, F. (2014). Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell Fact. 13:126. doi: 10.1186/s12934-014-0126-z
Rodriguez, G. M., and Atsumi, S. (2014). Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab. Eng. 25, 227–237. doi: 10.1016/j.ymben.2014.07.012
Rüffer, N., Heidersdorf, U., Kretzers, I., Sprenger, G. A., Raeven, L., and Takors, R. (2004). Fully integrated L-phenylalanine separation and concentration using reactive-extraction with liquid-liquid centrifuges in a fed-batch process with E. coli. Bioproc. Biosyst. Eng. 26, 239–248. doi: 10.1007/s00449-004-0354-4
Sacco, E., and Bientinesi, R. (2012). Mirabegron: a review of recent data and its prospects in the management of overactive bladder. Ther. Adv. Urol. 4, 315–324. doi: 10.1177/1756287212457114
Salgado, H., Gama-Castro, S., Peralta-Gil, M., Diaz-Peredo, E., Sanchez-Solano, F., Santos-Zavaleta, A., et al. (2006). RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 34, D394–397. doi: 10.1093/nar/gkj156
Sambrook, J., Fritsch, E. F., and and, Maniatis, T. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Satoh, Y., Tajima, K., Munekata, M., Keasling, J. D., and Lee, T. S. (2012). Engineering of a tyrosol-producing pathway, utilizing simple sugar and the central metabolic tyrosine, in Escherichia coli. J. Agric. Food Chem. 60, 979–984. doi: 10.1021/jf203256f
Schöpp, W., and Aurich, H. (1976). Kinetics and reaction mechanism of yeast alcohol dehydrogenase with long-chain primary alcohols. Biochem. J. 157, 15–22. doi: 10.1042/bj1570015
Schul'tsev, A. L. (2011). Thermal dehydration of 2-(4-aminophenyl)ethanol. Russ. J. Gen. Chem. 81, 2300–2303. doi: 10.1134/S1070363211110132
Siegert, P., Mcleish, M. J., Baumann, M., Iding, H., Kneen, M. M., Kenyon, G. L., et al. (2005). Exchanging the substrate specificities of pyruvate decarboxylase from Zymomonas mobilis and benzoylformate decarboxylase from Pseudomonas putida. Protein Eng. Des. Sel. 18, 345–357. doi: 10.1093/protein/gzi035
Sousa, A. C., Martins, L. O., and Robalo, M. P. (2013). Laccase catalysed homocoupling of primary aromatic amines towards the biosynthesis of dyes. Adv. Synth. Catal. 355, 2908–2917. doi: 10.1002/adsc.201300501
Sprenger, G. A. (2007a). “Aromatic amino acids,” in Amino Acid Biosynthesis–Pathways, Regulation and Metabolic Engineering, ed V. Wendisch (Berlin; Heidelberg: Springer Berlin Heidelberg), 93–127.
Sprenger, G. A. (2007b). From scratch to value: engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl. Microbiol. Biotechnol. 75, 739–749. doi: 10.1007/s00253-007-0931-y
Stark, D., Kornmann, H., Munch, T., Sonnleitner, B., Marison, I. W., and Von Stockar, U. (2003). Novel type of in situ extraction: Use of solvent containing microcapsules for the bioconversion of 2-phenylethanol from L-phenylalanine by Saccharomyces cerevisiae. Biotechnol. Bioeng. 83, 376–385. doi: 10.1002/bit.10679
Stolz, M., Peters-Wendisch, P., Etterich, H., Gerharz, T., Faurie, R., Sahm, H., et al. (2007). Reduced folate supply as a key to enhanced L-serine production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 73, 750–755. doi: 10.1128/AEM.02208-06
Suastegui, M., and Shao, Z. (2016). Yeast factories for the production of aromatic compounds: from building blocks to plant secondary metabolites. J. Ind. Microbiol. Biotechnol. 43, 1611–1624. doi: 10.1007/s10295-016-1824-9
Sun, Z., Ning, Y., Liu, L., Liu, Y., Sun, B., Jiang, W., et al. (2011). Metabolic engineering of the L-phenylalanine pathway in Escherichia coli for the production of S- or R-mandelic acid. Microb. Cell Fact. 10, 71–71. doi: 10.1186/1475-2859-10-71
Suvannasara, P., Tateyama, S., Miyasato, A., Matsumura, K., Shimoda, T., Ito, T., et al. (2014). Biobased polyimides from 4-aminocinnamic acid photodimer. Macromolecules 47, 1586–1593. doi: 10.1021/ma402499m
Tateyama, S., Masuo, S., Suvannasara, P., Oka, Y., Miyazato, A., Yasaki, K., et al. (2016). Ultrastrong, transparent polytruxillamides derived from microbial photodimers. Macromolecules 49, 3336–3342. doi: 10.1021/acs.macromol.6b00220
Teng, C. Y. P., Ganem, B., Doktor, S. Z., Nichols, B. P., Bhatnagar, R. K., and Vining, L. C. (1985). Shikimate derived metabolites.15. Total Synthesis of (+/–)-4-amino-4-deoxychorismic acid–a key intermediate in the biosynthesis of para-aminobenzoic acid and L-(para-aminophenyl)alanine. J. Am. Chem. Soc. 107, 5008–5009. doi: 10.1021/ja00303a038
Thomson, J. M., Gaucher, E. A., Burgan, M. F., De Kee, D. W., Li, T., Aris, J. P., et al. (2005). Resurrecting ancestral alcohol dehydrogenases from yeast. Nat. Genet. 37, 630–635. doi: 10.1038/ng1553
Trondle, J., Albermann, C., Weiner, M., Sprenger, G. A., and Weuster-Botz, D. (2018). Phosphoenolpyruvate transporter enables targeted perturbation during metabolic analysis of L-phenylalanine production with Escherichia coli. Biotechnol. J. 13:e1700611. doi: 10.1002/biot.201700611
Tsuge, Y., Kawaguchi, H., Sasaki, K., and Kondo, A. (2016). Engineering cell factories for producing building block chemicals for bio-polymer synthesis. Microb. Cell Fact. 15, 19–19. doi: 10.1186/s12934-016-0411-0
Vallon, T., Ghanegaonkar, S., Vielhauer, O., Muller, A., Albermann, C., Sprenger, G., et al. (2008). Quantitative analysis of isoprenoid diphosphate intermediates in recombinant and wild-type Escherichia coli strains. Appl. Microbiol. Biotechnol. 81, 175–182. doi: 10.1007/s00253-008-1707-8
Vandenhazel, H. B., Kiellandbrandt, M. C., and Winther, J. R. (1992). Autoactivation of proteinase-A initiates activation of yeast vacuolar zymogens. Eur. J. Biochem. 207, 277–283. doi: 10.1111/j.1432-1033.1992.tb17048.x
Vogt, P. F., and Gerulis, J. J. (2005). “Amines, aromatic,” in Ullmann's Encyclopedia of Industrial Chemistry, Vol. 2 (Weinheim: Wiley-VCH), 699–719. doi: 10.1002/14356007.a02_037
Vuralhan, Z., Luttik, M. A. H., Tai, S. L., Boer, V. M., Morais, M. A., Schipper, D., et al. (2005). Physiological characterization of the ARO10 dependent, broad substrate specificity 2-oxo acid decarboxylase activity of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 3276–3284. doi: 10.1128/AEM.71.6.3276-3284.2005
Vuralhan, Z., Morais, M. A., Tai, S.-L., Piper, M. D. W., and Pronk, J. T. (2003). Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69, 4534–4541. doi: 10.1128/AEM.69.8.4534-4541.2003
Weiner, M., Albermann, C., Gottlieb, K., Sprenger, G. A., and Weuster-Botz, D. (2014). Fed-batch production of L-phenylalanine from glycerol and ammonia with recombinant Escherichia coli. Biochem. Eng. J. 83, 62–69. doi: 10.1016/j.bej.2013.12.001
Wubbolts, M. G., Bongaerts, J. J., Bovenberg, R. A., Kozak, S., Sprenger, G. A., and Müller, M. (2005). Biosynthetic production of 4-amino-4-deoxychorismate (ADC) and [3,4r]-4-amino-3-hydroxycyclohexa-1,5-diene-1-carboxylic acid (3,4-CHA). Patent EP1602730A2.
Xu, Z., Wang, S., Wang, L., Fang, Z., and Wang, X. (2008). Preparation and pyroelectric properties of poly(pentafluorostyrene)-r-poly(4-vinylaniline) copolymer films. Macromol. Symp. 261, 144–147. doi: 10.1002/masy.200850119
Xue, Y., Chen, X., Yang, C., Chang, J., Shen, W., and Fan, Y. (2017). Engineering Eschericha coli for enhanced tyrosol production. J. Agric. Food Chem. 65, 4708–4714. doi: 10.1021/acs.jafc.7b01369
Yadav, S. K., and Cho, J. W. (2013). Functionalized graphene nanoplatelets for enhanced mechanical and thermal properties of polyurethane nanocomposites. Appl. Surf. Sci. 266, 360–367. doi: 10.1016/j.apsusc.2012.12.028
Yao, Y. F., Wang, C. S., Qiao, J., and Zhao, G. R. (2013). Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway. Metab. Eng. 19, 79–87. doi: 10.1016/j.ymben.2013.06.001
Yep, A., Kenyon, G. L., and Mcleish, M. J. (2006). Determinants of substrate specificity in KdcA, a thiamin diphosphate-dependent decarboxylase. Bioorg. Chem. 34, 325–336. doi: 10.1016/j.bioorg.2006.08.005
Yep, A., Kenyon, G. L., and Mcleish, M. J. (2008). Saturation mutagenesis of putative catalytic residues of benzoylformate decarboxylase provides a challenge to the accepted mechanism. Proc. Natl. Acad. Sci. U.S.A. 105, 5733–5738. doi: 10.1073/pnas.0709657105
Keywords: Escherichia coli, aromatic amines, para-amino-phenylethanol, para-amino-phenylacetic acid, para-amino-phenylacetaldehyde, phenylpyruvate decarboxylase
Citation: Mohammadi Nargesi B, Sprenger GA and Youn J-W (2019) Metabolic Engineering of Escherichia coli for para-Amino-Phenylethanol and para-Amino-Phenylacetic Acid Biosynthesis. Front. Bioeng. Biotechnol. 6:201. doi: 10.3389/fbioe.2018.00201
Received: 28 September 2018; Accepted: 10 December 2018;
Published: 04 January 2019.
Edited by:
Thomas Bartholomäus Brück, Technische Universität München, GermanyReviewed by:
Zhi-Gang Jeff Qian, Shanghai Jiao Tong University, ChinaRobert Kourist, Graz University of Technology, Austria
Copyright © 2019 Mohammadi Nargesi, Sprenger and Youn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung-Won Youn, anVuZy13b24ueW91bkBpbWIudW5pLXN0dXR0Z2FydC5kZQ==
 Behrouz Mohammadi Nargesi
Behrouz Mohammadi Nargesi Georg A. Sprenger
Georg A. Sprenger Jung-Won Youn
Jung-Won Youn