- 1Department of Physiology, School of Medicine, Wayne State University, Detroit, MI, United States
- 2Department of Biological Sciences, Wayne State University, Detroit, MI, United States
While the rapid development of CRISPR/CAS9 technology has allowed for readily performing site-specific genomic editing in non-rodent species, an emerging challenge is to select the most suitable species to generate animal models for the study of human biology and diseases. Improving CRISPR/CAS9 methodology for more effective and precise editing in the rabbit genome to replicate human disease is an active area of biomedical research. Although rabbits are more closely related to humans than mice (based on DNA sequence analysis), our whole-genome protein database search revealed that rabbits have more missing human protein sequences than mice. Hence, precisely replicating human diseases in rabbits requires further consideration, especially in studies involving essential functions of the missing proteins. For example, rabbits lack calponin 2, an actin-associated cytoskeletal protein that is important in the pathogenesis of inflammatory arthritis, atherosclerosis, and calcific aortic valve disease. The justification of using rabbits as models for human biomedical research is based on their larger size and their closer phylogenetic distance to humans (based on sequence similarity of conserved genes), but this may be misleading. Our findings, which consider whole-genome protein profiling together with actual protein expressions, serve as a warning to the scientific community to consider overall conservation as well as the conservation of specific proteins when choosing an animal model to study a particular aspect of human biology prior to investing in genetic engineering.
Introduction
The rapid development of CRISPR/Cas9 technology for readily editing mammalian genomes has allowed for the efficient generation of genetically engineered animal models for the study of human diseases in non-rodent species (Liu et al., 2018). The rabbit, in particular, has become a commonly used animal model for the study of molecular mechanisms underlying human biology and disease (Esteves et al., 2018). Historically, rabbits have been utilized for the development of vaccines (starting with Louis Pasteur’s rabies vaccine in 1881) (Pasteur, 1885), production and characterization of antibodies (Weber et al., 2017), early molecular immunology (Esteves et al., 2018), and experimental surgery (Calasans-Maia et al., 2009), among other applications, due to advantages such as strong immune response to foreign antigens, greater availability of cells and tissues from a single animal, and ease of manipulation because of the larger size. Rabbits are considered an intermediate between broadly studied rodents and larger but costlier mammals. In recent years, using CRISPR/Cas9 technology, genetically modified rabbits have been promoted in studies of human diseases ranging from muscular dystrophy (Sui et al., 2018) and hypertrophy (Lv et al., 2016) to retinal degeneration (Kondo et al., 2009), atherosclerosis (Wang et al., 2013), and X-linked hypophosphatemia (Sui et al., 2016). Intrigued by the increasing power and efficiency to generate mutant rabbits for models of human diseases and the rich historical literature of rabbit research on immunological (Esteves et al., 2018) and cardiovascular disorders (Hasenfuss, 1998), we explored this approach for a potential expansion of our research on calponin, a family of regulators of actin cytoskeleton, that plays a role in the regulation of actin cytoskeleton-mediated cell motility (Liu and Jin, 2016a).
Three homologous genes (CNN1, CNN2, and CNN3) have evolved in vertebrates to encode three isoforms of calponin (calponin 1, 2, and 3). Each of the calponin isoform genes is composed of seven exons with a variable region at the C-terminus that differentiates the isoforms (Figure 1). Although the calponin isoforms have significantly diverged during evolution, each isoform is well conserved in the vertebrate phylum as has been shown for numerous species, including channel catfish, Western clawed frogs, brown tree snakes, black flying foxes, chickens, mice, hamsters, chimpanzees, and humans, among others (Liu and Jin, 2016a). The expression and function of each calponin isoform are distinct. Calponin 1 is specifically expressed in fully differentiated smooth muscle cells and contributes to regulating contractility (Takahashi et al., 2000; Feng et al., 2019). Calponin 2 is expressed in a broader range of cell types, including fibroblasts, macrophages, cancer cells, and others to participate in regulating cellular responses to mechanical signals, cell proliferation, and motility (Qiu et al., 2017; Plazyo et al., 2018, 2019). Calponin 3 is the least studied of the three isoforms. It is expressed in the brain with a potential contribution to neural plasticity (Ferhat et al., 2003) and in placenta where it mediates trophoblastic cell fusion (Shibukawa et al., 2010).
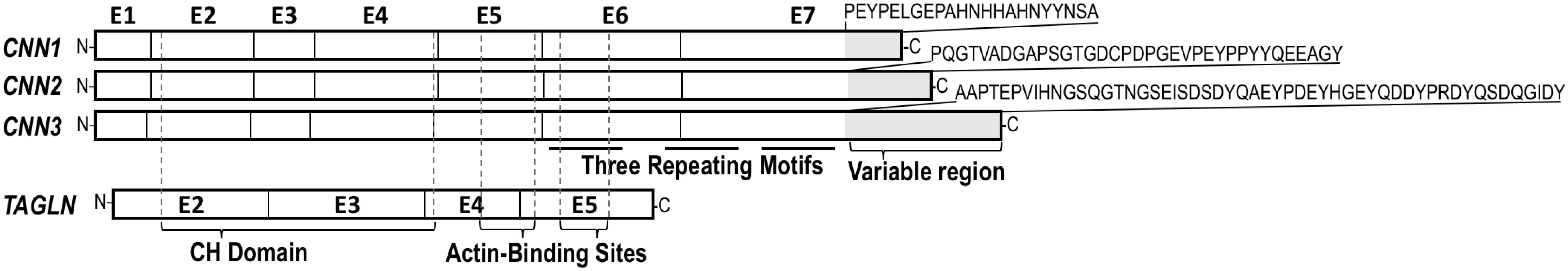
Figure 1. Human calponin isoforms and transgelin. CNN1 (GenBank NC_000019.10, region 11538775.11550323), CNN2 (GenBank NC_000019.10, region 1026608.1039065), and CNN3 (GenBank NC_000001.11, region complement 94896949.94927223) genes each consists of seven exons. The three calponin isoforms are conserved in structure except the C-terminal region that is diverged in length and amino acid sequences as shown in the figure. TAGLN (SM22alpha) (GenBank NC_000011.10, region 117199294.117207465) contains only four coding exons corresponding to the conserved core structure of calponin family. Positions of the CH (calponin homology) domain and two actin binding sites are indicated in the alignment map.
Extensive research in the past three decades demonstrates that each of the calponin isoforms have evolved with discrete functions and expression regulation as observed across a variety of vertebrate species (Liu and Jin, 2016a). The present study provides the first report that the rabbit is a unique species that lacks calponin 2, a cytoskeleton regulatory protein conserved in all other vertebrates studied to date. Using Western blotting to examine actual protein expression and whole-genome comparison to reveal the difference between humans and rabbits versus humans and mice, our data present a striking example of the loss of a gene with important functions, suggesting that caution should be used when selecting rabbits as animal models to study human biology and disease.
Results
Considering that the three calponin isoforms are conserved in all vertebrate species studied in the past, it was surprising that our initial examination failed to detect the calponin 2 protein in all of the rabbit tissues that are known to express calponin 2 in humans and mice. Using specific, previously characterized monoclonal antibodies (Hossain et al., 2006), we assessed the expression of all calponin isoforms in total protein extracts from the trachea, lungs, small intestine, large intestine, spleen, bladder, uterus, aorta, heart, brain, kidney, liver, and pancreas by Western blot. Figure 2 demonstrates that only calponin 1 and calponin 3 were detected in the rabbit, whereas the cell motility regulator, calponin 2, was not found. We verified the results by multiple immunological examinations using several monoclonal and polyclonal antibodies against calponin 2 with confirmed cross-species reactivity (Jin et al., 1996) (data not shown).
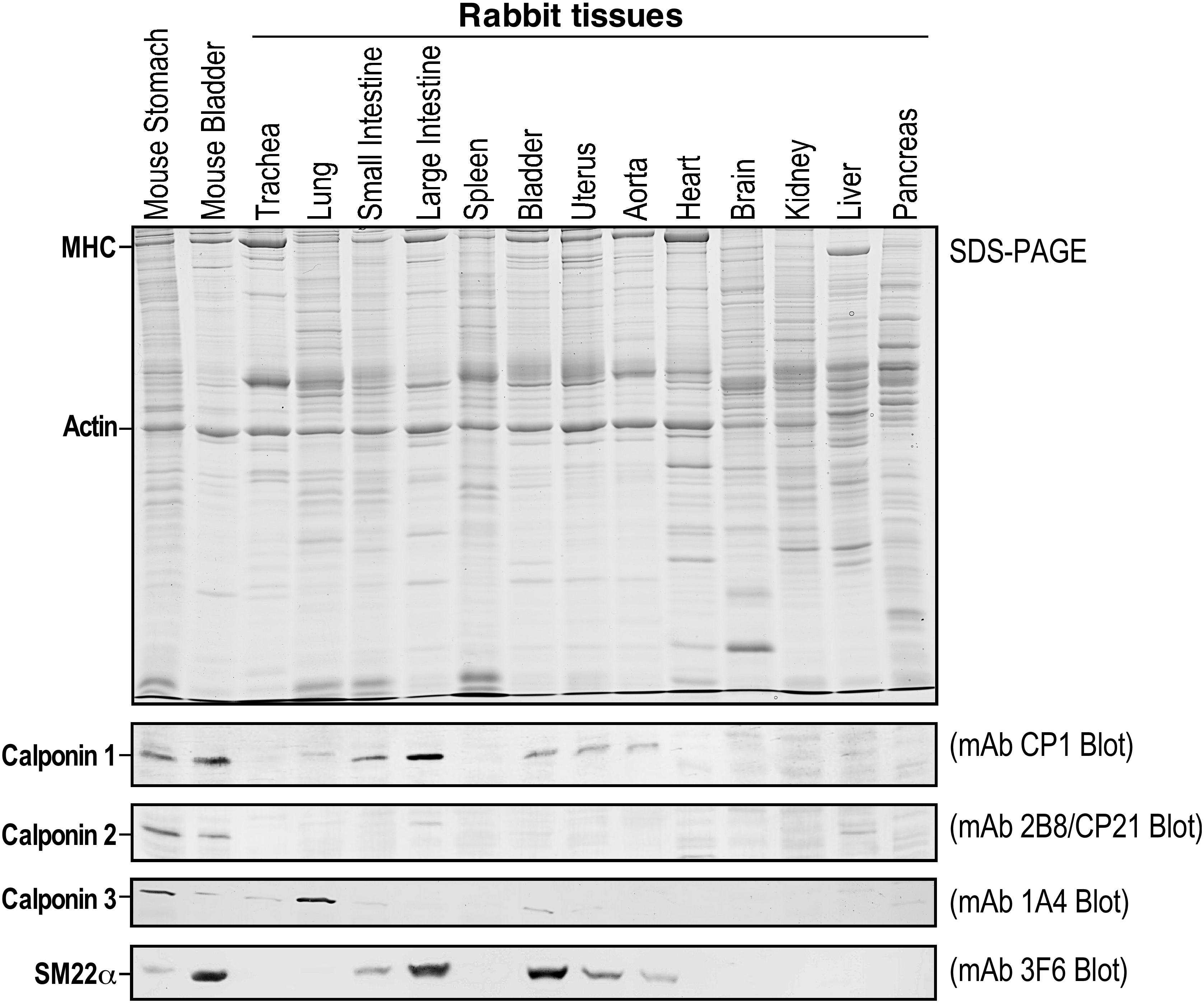
Figure 2. Calponin 2 is absent in rabbits. Rabbit tissue lysates were subjected to SDS-PAGE (top panel) and immunoblotting using monoclonal antibodies (mAbs) specific to calponin 1 (CP1) (Jin et al., 1996), calponin 2 (2B8/CP21) (Hossain et al., 2006), calponin 3 (1A4 raised against mouse calponin 3 expressed from cloned cDNA) or SM22alpha/transgelin (3F6) (Liu et al., 2017) (lower panels). While calponin 1 and calponin 2 have similar mobility in the SDS-gel used for Western blotting, calponin 3 and SM22alpha are readily distinguished from calponin 1 and calponin 2 by their molecular weights. Mouse stomach and bladder were used as positive controls. Whereas the expression of calponin 1, calponin 3, and SM22alpha was detected in specific tissue types as previously shown in mice and humans, calponin 2 is completely absent in rabbit tissues as demonstrated in the mAb 2B8/CP21 blot. MHC, myosin heavy chain.
We then searched the Ensembl and NCBI sequence databases and found no entry for rabbit calponin 1 and calponin 2, confirming the lack of calponin 2 as originally observed by Western blot. However, the calponin 1 protein is detected at significant levels in rabbit smooth muscles using Western blotting (Figure 2). To verify the initial search results, human CNN1, CNN2, and CNN3 isoforms were used as query sequences in BLAST searches against mouse and rabbit protein databases. To investigate the presence of the calponin genes in mice and rabbits, we collected proteins with a BLASTP E-value less than 10–30. Protein sequences were aligned using MUSCLE (Edgar, 2004). A maximum likelihood tree was constructed using PhyML (Guindon et al., 2010). The results found only one calponin (CNN3) in the rabbit. This was further confirmed by TBLASTN searches using human CNN1, CNN2, and CNN3 as query sequences, all of them match the same genomic region and CNN3 has the highest similarity. Therefore, the presence of the apparently functional CNN1 gene in the rabbit was not detectable by using the common sequence database searches, demonstrating the critical value of direct detections at the protein level (Figure 2). We also performed a HMMER search at https://www.ebi.ac.uk/Tools/hmmer/search/phmmer (Potter et al., 2018). When the query search is with the human CNN2 protein, then the top HMMER hit is calponin 3 in rabbit and the second best hit is to transgelin. These HMMER results confirm the BLASTP results.
To obtain a comprehensive understanding of this unexpected finding, we also performed systemic BLASTP homology searches of all human protein genes against mouse and rabbit genomes with E-values < 10–5. The BLASTP search revealed the absence of 843 distinct human protein coding genes in rabbit proteome (Table 1), among which many are critical to essential functions such as immune regulation, cell cycle progression, mitochondrial energetics, transcription/chromatin remodeling, lipid metabolism, ER/endosome trafficking, calcium signaling, neuropeptides, and spermatogenesis (Supplementary Table S1). In addition, gene ontology (GO) analysis detected 157 human genes missing in the rabbit genome (Supplementary Figure S1).
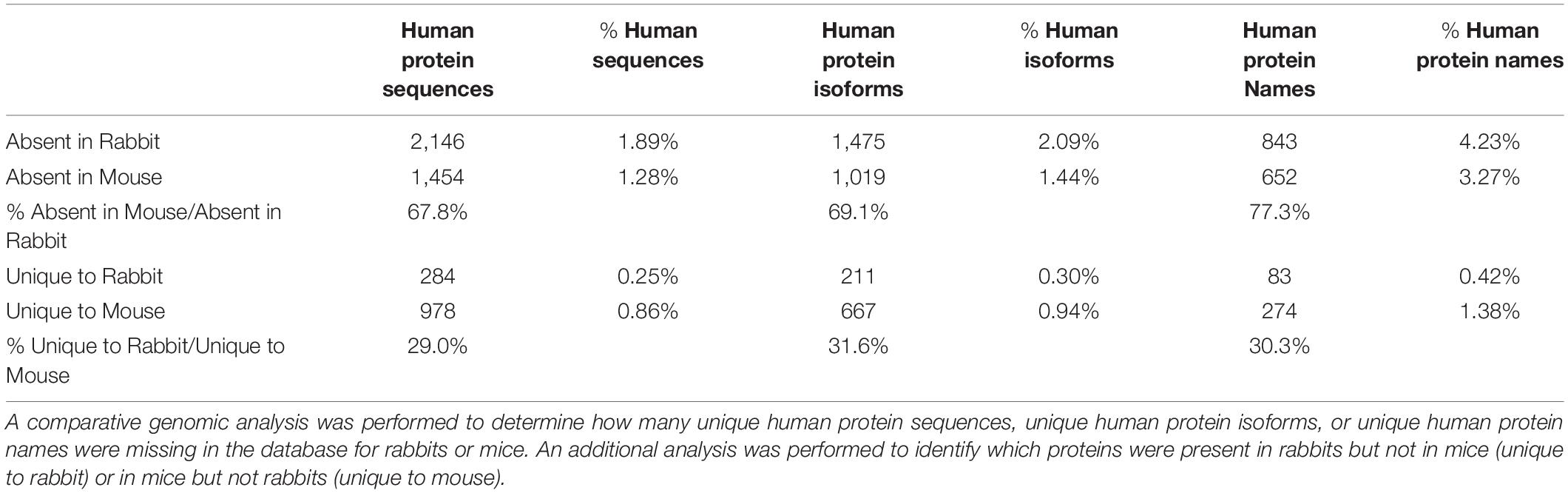
Mice have been extensively used to develop genetically modified models for the research of human diseases. Although the use of mice is continuously criticized as unideal in replicating human physiology and disease (Nature Medicine Editorial, 2013), the same comparison of human and mouse genomes found an absence of a significantly smaller number (652) of distinct human protein coding genes in mice (Table 1). Both rabbits and mice belong to the Glires clade with the mouse lineage having a longer branch length than the rabbit lineage based on the sequence similarity of conserved genes (Murphy et al., 2001). However, the sequence alignment approach to classification may be misleading, as whole-genome protein profiling appears to be more informative. While rabbits are considered phylogenetically closer to humans than mice based on DNA sequence similarities, the fact that mice have a profile of protein coding genes that are more similar to that of humans than that of rabbits to humans raises concern. To quantitatively evaluate this potentially high impact finding, genomic comparison results in Table 1 demonstrate that the proportion of unique human protein sequences, protein isoforms, or total protein entries in the databases all showed approximately 30% more missing in rabbits than that missing in mice.
Discussion
Three homologous calponin isoform genes have evolved in vertebrates (Liu and Jin, 2016a). Our finding of the lack of CNN2 gene in rabbits (Figure 2) calls for caution in using rabbit models in calponin or actin regulation-related research. While CNN1 functions in smooth muscle contractility (Yamamura et al., 2007) and CNN3 is a isoform with unclear functions (Liu and Jin, 2016a), calponin 2 functions in many cell motility-related functions. The loss of calponin 2 in myeloid immune cells in mice has been shown to modulate inflammatory responses (Plazyo et al., 2019), such as attenuating inflammatory arthritis (Huang et al., 2016) and the pathogenesis of atherosclerosis (Liu and Jin, 2016b). CNN2 knock out in mice also reduces myofibroblast differentiation and the development of calcific aortic valve disease (Plazyo et al., 2018) and slows down platelet adhesion in thrombosis (Hines et al., 2014). Reduced expression of calponin 2 is found in metastatic cancer cells (Moazzem Hossain et al., 2014). Therefore, the lack of CNN2 in rabbits is anticipated to have multiple pathophysiological effects. Our data, reported here, aims to timely inform the research community of an important precaution in order to avoid the resource-consuming construction of genetically modified rabbit models that may be less effective for many areas of research.
The three calponin isoforms have distinct expressions in different cell and tissue types (Figure 2) reflecting specific functions. If co-expressed in the same cell, calponin isoforms may potentially have complementary functions (Feng et al., 2019). Calponins also share high sequence similarities with transgelin (TAGLN), also named SM22alpha (Morgan and Gangopadhyay, 2001). Phylogenetic analysis demonstrated that TAGLN is clearly related to the calponin gene family (Figure 3). In the genomic search of the rabbit, the second best BLAST hit of each human calponin protein is a transgelin homolog. Although SM22alpha is considerably shorter than calponins 1, 2, and 3, it contains the conserved core structures of calponins (Figure 1). The mechanoregulation of SM22alpha in fibroblasts also resemble that of calponin 2 (Liu et al., 2017), suggesting potentially functional overlap. Therefore, the functional impacts of loss of CNN2 in rabbit smooth muscles and myofibroblast biology may be partially compensated by calponin 1 and/or SM22alpha (Liu et al., 2017; Feng et al., 2019).
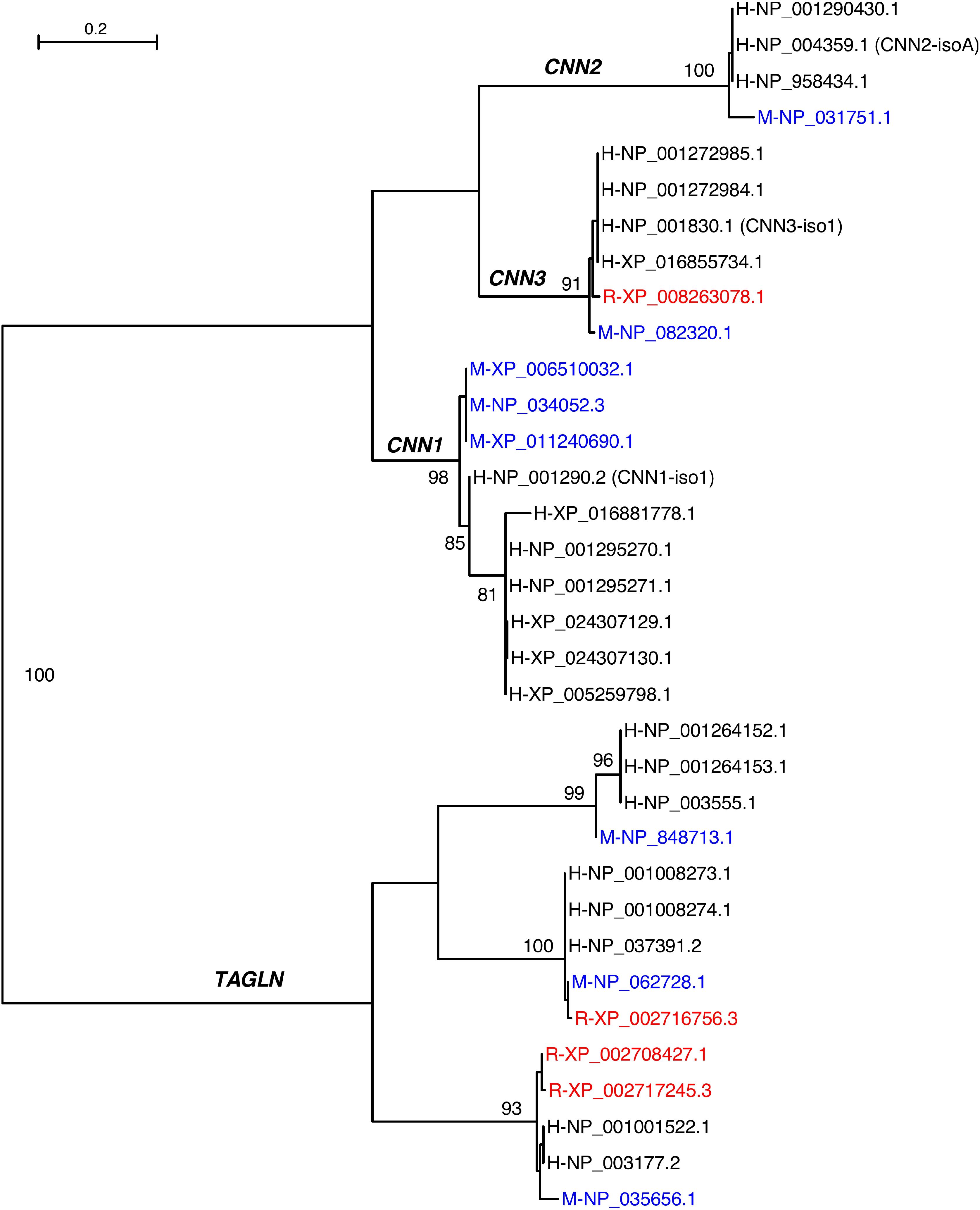
Figure 3. Maximum likelihood phylogeny of calponin and transgelin homologs from humans (H), mice (M), and rabbits (R). Mouse proteins are in blue whereas rabbit proteins are in red. The primary entries of the three human calponin isoforms are indicated in parentheses. The branches are supported by bootstrap values of which values greater than 80% are shown.
Consistent with the fact that rabbits survive well without the CNN2 gene, deletion of CNN2 in mice does not cause lethality (Huang et al., 2008). The absence of calponin 2 in rabbits presents a natural calponin 2-null model that can be employed to study the compensatory functions of calponin 1 and 3 in actin-mediated cellular processes in multiple cell types. In the meantime, comparative genomic analysis can reveal other missing human genes that are critical for the research on particular mechanisms of human diseases to guide targeted humanizations of rabbit and other animal models, for example by using the increasingly powerful CRISPR/Cas9 technology.
The comparative genomics data further showcases that mice actually miss fewer human protein coding genes than that rabbits (Table 1). While the mouse has long been regarded simply as a convenient animal model for advantages such as low cost of housing, relatively short reproductive cycle, large litter size, and ease of genetic manipulations among others, its evaluation should now be revisited for being a more preferable choice over the rabbit due to its higher similarity to humans in genome-wide protein profiles (Supplementary Table S1). Consistent with the substantial divergence of rabbit proteome with that of humans, numerous studies have alluded to the divergence of molecular, cellular, and physiological mechanisms between rabbits and humans.
Improvements in genome editing, such as the generation of point mutations (Liu et al., 2018), will certainly facilitate the production of rabbit models for research, but precautions are imperative to thoroughly consider their precise representation of human conditions. Our example of evaluating animal models of human biology and diseases involving calponin-mediated mechanisms shows that when selecting an animal model to study a specific physiologic mechanism or disease of humans, intuitive preference for larger sized animals may not always translate into closer genetic background that determines the relevance to the research objective, especially in integrative pathophysiological and translational studies.
Finally, it is worth pointing out that, since the differences in whole-genome protein profiles between human and various vertebrates would certainly affect the function of individual proteins in vivo and during development, one must consider these differences to understand the often complex phenotype of a particular genetic modification of interest engineered in a particular animal model of human disease. To achieve this goal, it is necessary to reliably detect missing genes in a genome by sensitive homology search analysis. In this present study, we applied multiple homology search methods along with protein level confirmation to reliably demonstrate the lack of a functional Cnn2 gene in rabbit. This two-level approach – a combination of tests for protein expression and genome search analysis – provides the power to improve our understanding of animal models and their application in biomedical studies.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Wayne State University.
Author Contributions
OP conducted the experiments, analyzed the data, drafted the manuscript, and approved the submitted version of the manuscript. WH designed and conducted the bioinformatic studies, analyzed the data, drafted the manuscript, and approved the submitted version of the manuscript. J-PJ designed the study, analyzed the data, drafted the manuscript, and approved the submitted version of the manuscript.
Funding
This work was supported in part by grants from the National Institutes of Health (HL133162, HL127691, and HL138007) to J-PJ and from the National Science Foundation (ISO1838291) and the Wayne State University (URG2018) to WH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Xia Hou for collecting the rabbit tissues.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00042/full#supplementary-material
References
Calasans-Maia, M. D., Monteiro, M. L., Ascoli, F. O., and Granjeiro, J. M. (2009). The rabbit as an animal model for experimental surgery. Acta Circ. Bras. 24, 325–328. doi: 10.1590/s0102-86502009000400014
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Esteves, P. J., Abrantes, J., Baldauf, H. M., BenMohamed, L., Chen, Y., Christensen, N., et al. (2018). The wide utility of rabbits as models of human diseases. Exp. Mol. Med. 50:66. doi: 10.1038/s12276-018-0094-1
Feng, H. Z., Wang, H., Takahashi, K., and Jin, J. P. (2019). Double deletion of calponin 1 and calponin 2 in mice decreases systemic blood pressure with blunted length-tension response of aortic smooth muscle. J. Mol. Cell Cardiol. 129, 49–57. doi: 10.1016/j.yjmcc.2019.01.026
Ferhat, L., Esclapez, M., Represa, A., Fattoum, A., Shirao, T., and Ben-Ari, Y. (2003). Increased levels of acidic calponin during dendritic spine plasticity after pilocarpine-induced seizures. Hippocampus 13, 845–858. doi: 10.1002/hipo.10136
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hasenfuss, G. (1998). Animal models of human cardiovascular disease, heart failure and hypertrophy. Cardiovasc. Res. 39, 60–76. doi: 10.1016/s0008-6363(98)00110-2
Hines, P. C., Gao, X., White, J. C., D’Agostino, A., and Jin, J. P. (2014). A novel role of h2-calponin in regulating whole blood thrombosis and platelet adhesion during physiologic flow. Physiol. Rep. 2:e12228. doi: 10.14814/phy2.12228
Hossain, M. M., Smith, P. G., Wu, K., and Jin, J. P. (2006). Cytoskeletal tension regulates both expression and degradation of h2-calponin in lung alveolar cells. Biochemistry 45, 15670–15683. doi: 10.1021/bi061718f
Huang, Q. Q., Hossain, M. M., Sun, W., Xing, L., Pope, R. M., and Jin, J. P. (2016). Deletion of calponin 2 in macrophages attenuates the severity of inflammatory arthritis in mice. Am. J. Physiol. Cell Physiol. 311, C673–C685. doi: 10.1152/ajpcell.00331.2015
Huang, Q. Q., Hossain, M. M., Wu, K., Parai, K., Pope, R. M., and Jin, J. P. (2008). Role of H2-calponin in regulating macrophage motility and phagocytosis. J. Biol. Chem. 283, 25887–25899. doi: 10.1074/jbc.M801163200
Jin, J. P., Walsh, M. P., Resek, M. E., and McMartin, G. A. (1996). Expression and epitopic conservation of calponin in different smooth muscles and during development. Biochem. Cell. Biol. 74, 187–196. doi: 10.1139/o96-019
Kondo, M., Sakai, T., Komeima, K., Kurimoto, Y., Ueno, S., Nishizawa, Y., et al. (2009). Generation of a transgenic rabbit model of retinal degeneration. Invest. Ophthalmol. Vis. Sci. 50, 1371–1377. doi: 10.1167/iovs.08-2863
Liu, R., Hossain, M. M., Chen, X., and Jin, J. P. (2017). Mechanoregulation of SM22alpha/Transgelin. Biochemistry 56, 5526–5538. doi: 10.1021/acs.biochem.7b00794
Liu, R., and Jin, J. P. (2016a). Calponin isoforms CNN1, CNN2 and CNN3: regulators for actin cytoskeleton functions in smooth muscle and non-muscle cells. Gene 585, 143–153. doi: 10.1016/j.gene.2016.02.040
Liu, R., and Jin, J. P. (2016b). Deletion of calponin 2 in macrophages alters cytoskeleton-based functions and attenuates the development of atherosclerosis. J. Mol. Cell Cardiol. 99, 87–99. doi: 10.1016/j.yjmcc.2016.08.019
Liu, Z., Chen, M., Chen, S., Deng, J., Song, Y., Lai, L., et al. (2018). Highly efficient RNA-guided base editing in rabbit. Nat. Commun. 9:2717. doi: 10.1038/s41467-018-05232-2
Lv, Q., Yuan, L., Deng, J., Chen, M., Wang, Y., Zeng, J., et al. (2016). Efficient generation of myostatin gene mutated rabbit by CRISPR/Cas9. Sci. Rep. 6:25029. doi: 10.1038/srep25029
Moazzem Hossain, M., Wang, X., Bergan, R. C., and Jin, J. P. (2014). Diminished expression of h2-calponin in prostate cancer cells promotes cell proliferation, migration and the dependence of cell adhesion on substrate stiffness. FEBS Open Biol. 4, 627–636. doi: 10.1016/j.fob.2014.06.003
Morgan, K. G., and Gangopadhyay, S. S. (2001). Invited review: cross-bridge regulation by thin filament-associated proteins. J. Appl. Physiol. (1985) 91, 953–962. doi: 10.1152/jappl.2001.91.2.953
Murphy, W. J., Eizirik, E., Johnson, W. E., Zhang, Y. P., Ryder, O. A., and O’Brien, S. J. (2001). Molecular phylogenetics and the origins of placental mammals. Nature 409, 614–618. doi: 10.1038/35054550
Plazyo, O., Liu, R., Moazzem Hossain, M., and Jin, J. P. (2018). Deletion of calponin 2 attenuates the development of calcific aortic valve disease in ApoE(-/-) mice. J. Mol. Cell Cardiol. 121, 233–241. doi: 10.1016/j.yjmcc.2018.07.249
Plazyo, O., Sheng, J. J., and Jin, J. P. (2019). Down-regulation of Calponin 2 contributes to the quiescence of lung macrophages. Am. J. Physiol. Cell Physiol. 317, C749–C761. doi: 10.1152/ajpcell.00036.2019
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Qiu, Z., Chu, Y., Xu, B., Wang, Q., Jiang, M., Li, X., et al. (2017). Increased expression of calponin 2 is a positive prognostic factor in pancreatic ductal adenocarcinoma. Oncotarget 8, 56428–56442. doi: 10.18632/oncotarget.17701
Shibukawa, Y., Yamazaki, N., Kumasawa, K., Daimon, E., Tajiri, M., Okada, Y., et al. (2010). Calponin 3 regulates actin cytoskeleton rearrangement in trophoblastic cell fusion. Mol. Biol. Cell. 21, 3973–3984. doi: 10.1091/mbc.E10-03-0261
Sui, T., Xu, L., Lau, Y. S., Liu, D., Liu, T., Gao, Y., et al. (2018). Development of muscular dystrophy in a CRISPR-engineered mutant rabbit model with frame-disrupting ANO5 mutations. Cell Death Dis. 9:609. doi: 10.1038/s41419-018-0674-y
Sui, T., Yuan, L., Liu, H., Chen, M., Deng, J., Wang, Y., et al. (2016). CRISPR/Cas9-mediated mutation of PHEX in rabbit recapitulates human X-linked hypophosphatemia (XLH). Hum. Mol. Genet. 25, 2661–2671.
Takahashi, K., Yoshimoto, R., Fuchibe, K., Fujishige, A., Mitsui-Saito, M., Hori, M., et al. (2000). Regulation of shortening velocity by calponin in intact contracting smooth muscles. Biochem. Biophys. Res. Commun. 279, 150–157. doi: 10.1006/bbrc.2000.3909
Wang, Y., Niimi, M., Nishijima, K., Waqar, A. B., Yu, Y., Koike, T., et al. (2013). Human apolipoprotein A-II protects against diet-induced atherosclerosis in transgenic rabbits. Arterioscler. Thromb. Vasc. Biol. 33, 224–231. doi: 10.1161/ATVBAHA.112.300445
Weber, J., Peng, H., and Rader, C. (2017). From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp. Mol. Med. 49:e305. doi: 10.1038/emm.2017.23
Keywords: CRISPR/Cas9 gene editing, rabbit, comparative genomics, protein profiling, calponin 2
Citation: Plazyo O, Hao W and Jin J-P (2020) The Absence of Calponin 2 in Rabbits Suggests Caution in Choosing Animal Models. Front. Bioeng. Biotechnol. 8:42. doi: 10.3389/fbioe.2020.00042
Received: 05 October 2019; Accepted: 21 January 2020;
Published: 28 February 2020.
Edited by:
Robert Friedman, University of South Carolina, United StatesReviewed by:
Duo Xu, Cornell University, United StatesJunjie Yue, Institute of Biotechnology (CAAS), China
Jia Liu, ShanghaiTech University, China
Copyright © 2020 Plazyo, Hao and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weilong Hao, aGFvd0B3YXluZS5lZHU=; Jian-Ping Jin, amppbkBtZWQud2F5bmUuZWR1
 Olesya Plazyo
Olesya Plazyo Weilong Hao
Weilong Hao Jian-Ping Jin
Jian-Ping Jin