- 1Department of Biotechnology, The Catholic University of Korea, Gyeonggi, South Korea
- 2Department of Systems Biology, Division of Life Sciences, and Institute for Life Science and Biotechnology, Yonsei University, Seoul, South Korea
We previously reported a Corynebacterium glutamicum JH41 strain with a 58% faster growth rate through application of adaptive laboratory evolution. To verify that the fast-reproducing strain was useful as a host for recombinant protein expression, we introduced a plasmid responsible for the secretory production of a recombinant protein. The JH41 strain harboring the plasmid indeed produced the secretory recombinant protein at a 2.7-fold greater rate than its ancestral strain. To provide the reverse engineering targets responsible for boosting recombinant protein production and cell reproduction, we compared the genome sequence of the JH41 strain with its ancestral strain. Among the 15 genomic variations, a point mutation was confirmed in the 14 bases upstream of NCgl1959 (encoding a presumed siderophore-binding protein). This mutation allowed derepression of NCgl1959, thereby increasing iron consumption and ATP generation. A point mutation in the structural gene ramA (A239G), a LuxR-type global transcription regulator involved in central metabolism, allowed an increase in glucose consumption. Therefore, mutations to increase the iron and carbon consumption were concluded as being responsible for the enhanced production of recombinant protein and cell reproduction in the evolved host.
Introduction
Many attempts through microbial metabolic engineering have been made to overproduce metabolites by (1) improving the substrate uptake rate, (2) reducing the flux to undesired by-products, (3) introducing heterologous pathways and optimizing the activity of the enzyme, and (4) balancing product formation in cells, such as secretion of product to the extracellular medium (Yadav et al., 2012). However, the metabolic manipulation of microorganisms can cause flux imbalance due to the lack of understanding of the complex regulatory mechanisms essential for operation (Yadav et al., 2012). Adaptive laboratory evolution (ALE) is a reasonable approach because it not only copes with the physical defects of strains caused by genetic manipulation but also improves the productivity of cell factories (Sandberg et al., 2019; Lee and Kim, 2020). In addition, the commercialization of next-generation sequencing (NGS) technology and the development of omics tools have enabled reverse engineering by helping us better understand mechanisms of microbial regulation (Stella et al., 2019). Besides, genetically modified organism (GMO)-related issues in the food industry have increased the demand for non-GMO strains derived from ALE.
Growth rate is a significant parameter in evaluating the compatibility of industrial strains. It is generally recognized that fast growth rates can achieve high cell densities and high productivity (Feist et al., 2010). Genomic analyses of fast-growing mutant strains have revealed previously unknown metabolic engineering targets and methods to improve production hosts. Rugbjerg et al. (2018) reported that the introduction of the rpoB mutation into Escherichia coli increased the growth rate by 13% while also increasing the production rate of mevalonate, a building block of terpenoid products such as fragrances and bioplastics, by 71%. In addition, Wang et al. reported that the introduction of gntR1 and ramA mutations into Corynebacterium glutamicum increased the growth rate by 37% and the productivity of lysine by 100% (Wang et al., 2018).
Recently, we reported a fast-reproducing C. glutamicum JH41 strain, obtained through the application of ALE (Park et al., 2020). The transcriptome pattern of the evolved JH41 strain revealed that the upregulation of genes in the tricarboxylic acid cycle (TCA) cycle and respiratory chain, and oxidative stress responses allowed the growth promotion. Since C. glutamicum is a useful host for recombinant protein expression (Lee and Kim, 2018), it was proposed that the fast-reproducing JH41 strain might be an improved host for recombinant protein production and may also provide hidden genetic targets for reverse engineering of C. glutamicum hosts. In this report, the fast-reproducing JH41 was indeed verified as an improved host for recombinant protein production. Genomic analysis of the evolved strain was therefore performed to explain the reasons for the increased growth rate and recombinant protein production.
Materials and Methods
Strains and Plasmids
Corynebacterium glutamicum PT strain (ancestor of JH41) and a fast-reproducing JH41 strain (descendant of PT) were used as hosts for recombinant protein production and genomic analysis (Park et al., 2020). DNA manipulation followed the method described by Sambrook et al. using E. coli DH10B (Invitrogen Inc., Carlsbad, CA, United States) (Sambrook and Russell, 2001). The strains and plasmids are listed in Table 1 and the primers in Supplementary Table 1. Target gene-disrupted or gene-substituted C. glutamicum strains were prepared by double crossover using the pKmobsacB-based plasmids. Homologous arms of the target genes were constructed by connecting two fragments amplified from genomic DNA using overlap extension PCR (Ho et al., 1989). Oligonucleotide synthesis and plasmid sequence confirmation were performed at a facility of Bionics Inc., (Seoul, South Korea). pfu polymerase was purchased from Solgent Inc., (Daejeon, South Korea) and restriction enzymes and T4-ligase from New England Biolabs Inc., (Ipswich, MA, United States).
Media and Batch Cultures
As for the preculture for DNA manipulations, lysogeny broth (LB) complex medium was used for E. coli, and brain heart infusion (BHI) medium was used for C. glutamicum. C. glutamicum was incubated at 30°C and 200 rpm in a shaking incubator, and E. coli was incubated at 37°C and 220 rpm. For batch cultures of C. glutamicum strains, a modified MCGC minimal medium [0.9% glucose, 6 g Na2HPO4, 4 g (NH4)2SO4, 3 g KH2PO4, 1 g sodium citrate dehydrate, 1 g NaCl, 0.1 g MgSO4–7H2O, 20 mg FeSO4–7H2O, 2 mg MnSO4–H2O, 2 mg FeCl3, 1 mg thiamine–HCl, 200 μg biotin, 35 μg CaCl2, 0.5 μg ZnSO4–7H2O, 0.2 μg Na2B4O7–10H2O, 0.2 μg CuCl2–2H2O, 0.1 μg (NH4)6Mo7O24–4H2O per liter] was used (Osten et al., 1989). A 500-ml baffled flask containing 50 ml of the modified MCGC medium was used for efficient oxygen transfer. The initial cell mass was adjusted to OD600 = 0.1. Biomass was estimated by measuring optical density (OD) at 600 nm every hour and converting into g–DCW/L unit by the coefficient of 0.25.
Secretory Production of Recombinant Protein
To confirm the recombinant protein production of the hosts, pCG-H36A-agarase encoding a strong autonomous promoter and TAT-secretion signal sequence with the agarase gene from Streptomyces coelicolor was further transformed into the PT and JH41 strains (Yim et al., 2016). After incubating for 24 and 48 h in a 500-ml baffled flask containing 50 ml of MCGC media, 50 ml of the culture broth was centrifuged (12,000 × g, 4°C, 5 min). The supernatant containing the secreted protein was concentrated 50-fold using a centrifugal filter (Amicon® Ultra-15 10K, Merck Millipore, Burlington, MA, United States), and then, the buffer was changed with 50 mM Tris–HCl (pH 7.0). The prepared protein samples were diluted to final OD600 values and then loaded on a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel. The photograph of the gel was captured using a chemical fluorescence image analyzer (Chemi Doc MP System, Bio-Rad Laboratories, Inc., Hercules, CA, United States), and the intensity of the target band was compared using the ImageJ program.
Genome Sequencing
For genome sequencing, genomic DNAs of C. glutamicum PT and JH41 were isolated using a gDNA prep kit (Solgent Inc., Daejeon, South Korea) according to the manufacturer’s instructions. The genome sequence was determined using the PacBio RS II (Pacific Biosciences Inc., Menlo Park, CA, United States) at a DNA sequencing facility (ChunLab Inc.). Raw sequences were assembled with PacBio SMRT Analysis ver. 2.0 software (Pacific Biosciences Inc., Menlo Park, CA, United States). Gene prediction was performed using Glimmer 3 (Delcher et al., 2007), and annotations were performed by a homology search against the SEED database, Universal Protein Resource (UniProt) database, and eggNOG database (Disz et al., 2010). Genome sequences of the PT and JH41 strains were compared using EzGenome (http://ezgenome.ezbiocloud.net/ezg_browse) and CLgenomics program (ChunLab). Analysis of single-nucleotide polymorphisms (SNPs) and gap was performed using the MAUVE (Darling et al., 2004) and MegaX (Kumar et al., 2018) programs.
Analyses of ATP, Iron, and Glucose Concentrations
Cells were harvested from batch cultures (1 ml) at 5 h by centrifugation (8,000 × g, 4°C, 5 min) and washed twice with sterile distilled water. The cell suspension (0.8 ml) was mixed with 0.2 g of glass beads (212–300 μm) in a screw-capped tube followed by homogenization for 30 s using a bead beater (Model 607, BioSpec Inc., Bartlesville, OK, United States). After chilling in an ice bath for 1 min, homogenization was repeated five more times. Cell debris was removed by centrifugation (8,000 × g, 4°C, 5 min), and the supernatant was used for measuring intracellular ATP.
ATP concentration was estimated using an ENLITEN ATP Assay System Bioluminescence Detection Kit (#FF2000, Promega Inc., Madison, WI, United States). Light intensity was detected by a luminometer (GloMax®-96 Microplate Luminometer, Promega Inc., Madison, WI, United States) after a luciferase-driven reaction (Na et al., 2015).
Iron consumption was estimated by measuring the remaining free iron concentration using the o-phenanthroline colorimetric method (Serra et al., 2015) after cell removal. Culture broth (1 ml) was centrifuged (12,000 × g, 4°C, 5 min), and the supernatant (100 μl) was mixed with 20 μl of bromophenol blue (0.4 g/L). After pH adjustment to 3.5 by adding 0.2% H2SO4, a further 10 μl of hydroquinone (1 g/L), 20 μl of o-phenanthroline (2.5 g/L), and distilled water (up to 250 μl) were added to the mixture. After 1 h of color development at room temperature, absorbance was measured at 510 nm. The analyses of ATP and iron concentrations were determined at least three times with three biological replicates.
To analyze the glucose consumption of JH41 and PT ramAA239G, the glucose concentration in the medium was measured by a reducing sugar quantification method using a 3,5-dinitrosalicylic acid (DNS) reagent (Deshavath et al., 2020). After cell removal, 150 μl of supernatant and 150 μl of DNS reagent were mixed and heated at 100°C for 5 min. After standing to cool for 10 min at room temperature, 200 μl of the mixture was dispensed into 96-well plates to measure absorbance at 540 nm. D-Glucose reagent was used as a standard.
Determination of DtxR Affinity on the Mutated Promoter Sequence
The affinity of negative regulator diphtheria toxin repressor (DtxR) on the mutated promoter region was determined by an electrophoresis mobility shift assay using the purified DtxR protein and the PCR-amplified DNA fragment. The dtxR gene from C. glutamicum was cloned and expressed in E. coli BL21 (DE3) with an additional 12 × His tag in the expression plasmid (pET24b-dtxR, Table 1). The protein was further purified using Ni2+ chelate affinity chromatography, as described previously (Wennerhold et al., 2005). To test the binding of DtxR to the putative target promoter DNA, the purified DtxR protein (0–350 ng, dimeric form) was dissolved with PCR-amplified DNA fragments (399 bp, 200 ng) in a 10-μl binding buffer [50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 40 mM KCl, 5% (v/v) glycerol, 1 mM dithiothreitol (DTT), and 150 μM MnCl2]. The reaction mixture was loaded onto a 15% native polyacrylamide gel containing 1 mM DTT and 150 μM MnCl2 after incubation for 30 min at room temperature. Electrophoresis was performed at room temperature and 170 V using 1 × TB (89 mM Tris base, 89 mM boric acid) supplemented with 1 mM DTT and 150 μM MnCl2 as an electrophoresis buffer. The gel was then stained with a fluorescent dye (SYBR Green I EMSA Kit, Invitrogen Inc., Carlsbad, CA, United States) according to the manufacturer’s instructions. The photograph of the gel was captured using a chemical fluorescence image analyzer (Chemi Doc MP System, Bio-Rad Laboratories, Inc., Hercules, CA, United States).
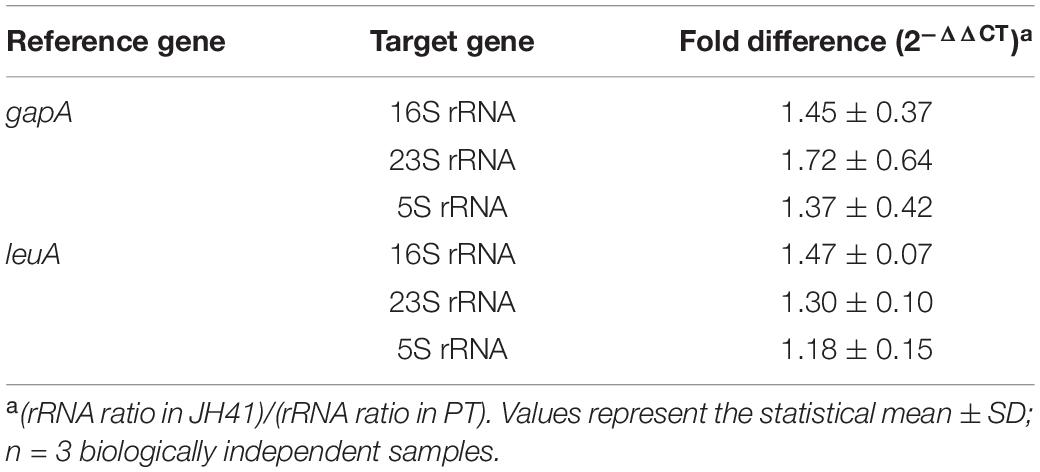
Transcription Quantification of rRNAs
The ribosomal RNAs (rRNAs) transcription level determination method was followed as described (Kwon et al., 2015). Total RNA was extracted from actively growing C. glutamicum strains using TRIzol1 reagent (Invitrogen, Carlsbad, CA, United States) and NucleoSpin1 RNA II Kit (Macherey-Nagel, Düren, Germany) after cell disruption using a Mini-Beadbeater-16 (BioSpec, Bartlesville, PA, United States) with glass beads (acid washed, 212–300 mm, Sigma-Aldrich, MO, United States). The 50 ng of total RNAs was used for complementary DNA (cDNA) synthesis using ReverTra Ace-α-1 (Toyobo, Osaka, Japan) according to the manufacturer’s instructions. THUNDERBIRD SYBR1 qPCR Mix (Toyobo, Osaka, Japan) and the Step-One Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) were used for the gene expression analysis. Step-One Plus Software ver. 2.0 (Applied Biosystems, Foster City, CA, United States) was used to quantify the relative transcription. gapA [glyceraldehyde 3-phosphate dehydrogenase (GAPDH)] and leuA (2-isopropyl malate synthase) genes were used as internal references. The relative expression of rRNAs with the reference genes was determined using the 2–Δ Δ CT (Livak) method. Primers for identifying transcription levels are shown in Supplementary Table 1.
Results
Effect of Fast-Growing JH41 Host on the Secretory Production of Recombinant Protein
Because protein is the major macromolecule in cells, the rapidly reproducing JH41 strain was supposed to be able to produce a protein at an enhanced rate. The fact that no extracellular protease activity or economical purification of secretory protein was found in C. glutamicum host (Lee and Kim, 2018) further encouraged us to hypothesize that the rapidly reproducing JH41 strain would become an improved C. glutamicum host for secretory protein production. To verify this hypothesis, a plasmid designed to secrete foreign agarase (pCG-H36A-agarase) was introduced into JH41 and PT strains. The secretory recombinant protein (agarase, 34 kDa) showed a major band in the SDS-PAGE (Figure 1A). The band intensities of the recombinant protein from the JH41 host were 7,490 AU at 24 h and 17,231 AU at 48 h, while those from the PT host were 4,780 AU at 24 h and 6,212 AU at 48 h (Figure 1B). The JH41 host produced the secretory recombinant protein (56% at 24 h and 177% at 48 h) at a greater rate than the PT host. In order to confirm the production effect of other proteins in the JH41 host, a plasmid expressing green fluorescent protein (GFP) in cytoplasm (pCG-H36A[-SS]-GFP, no signal sequence) and a plasmid secreting GFP by a TAT-signal sequence (pCG-H36A-porD-GFP) were introduced into the JH41 host (Supplementary Figure 1). The productions of cytoplasmic GFP and secreted GFP from the JH41 host were greater than those from the PT host by 5–16% and 13–116%, respectively. Therefore, the rapid-reproducing JH41 strain was an enhanced host for the production of recombinant proteins.
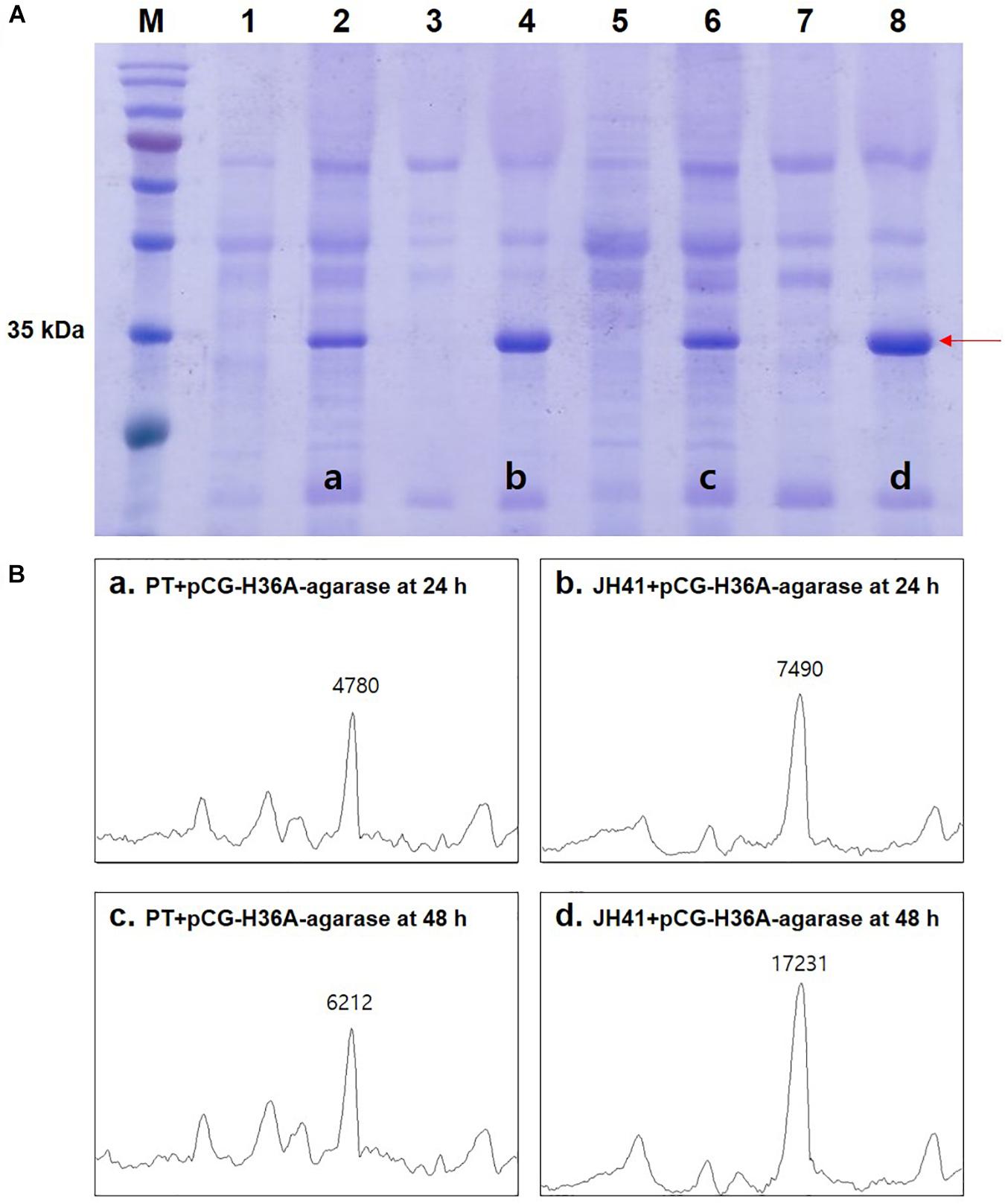
Figure 1. Effects of host on the secretory production of recombinant protein. (A) Image of sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Lane 1, PT + pCG-H36A (empty vector) at 24 h; lane 2, PT + pCG-H36A-agarase at 24 h; lane 3, JH41 + pCG-H36A (empty vector) at 24 h; lane 4, JH41 + pCG-H36A-agarase at 24 h; lane 5, PT + pCG-H36A (empty vector) at 48 h; lane 6, PT + pCG-H36A-agarase at 48 h; lane 7, JH41 + pCG-H36A (empty vector) at 48 h; lane 8, JH41 + pCG-H36A-agarase at 48 h. Arrow indicates the secreted agarase band (34 kDa). The image is from the representative experiment from three independent experiments. (B) Band density estimation of the SDS-PAGE image using ImageJ software. a: lane 2; b: lane 4; c: lane 6; d: lane 8.
Genome Analysis of the JH41 Host
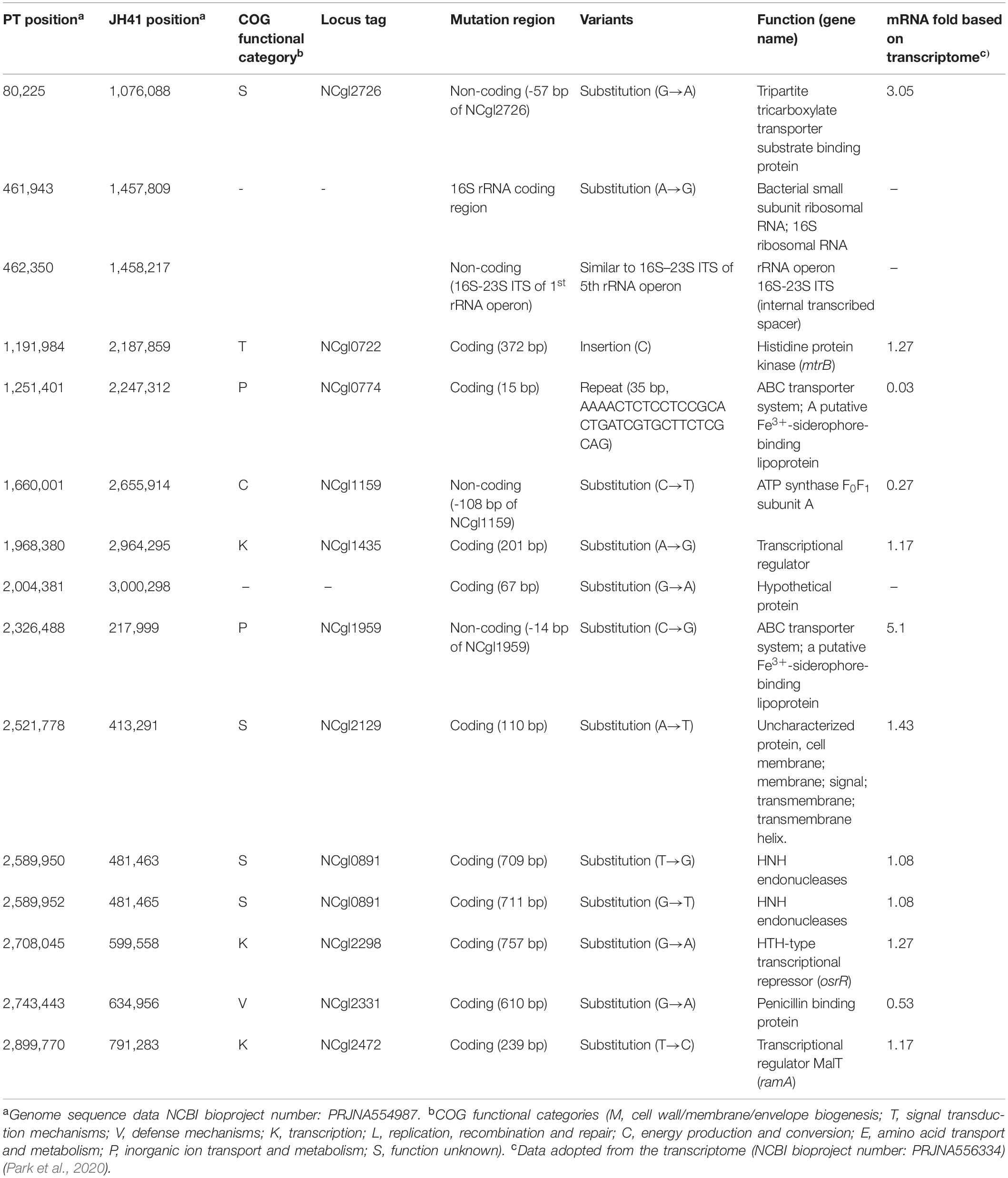
To understand the reasons for enhanced cell reproduction and to provide reverse engineering targets responsible for the improved recombinant protein production, the JH41 genome was resequenced and deposited [National Center for Biotechnology Information (NCBI) accession number PRJNA554987]. A total 15 genomic mutations were found to be accumulated in JH41 compared to its ancestral PT (Table 2). Among them, four were found in the non-coding region, including an internal transcribed spacer (ITS) mutation, and 11 in the coding region. Genomic mutations were categorized into the clusters of orthologous groups of proteins (COG) as four to the function unknown group (S), two to the transcription group (K), two to the inorganic ion transport and metabolism group (P), one to the defense mechanism (V), one to the signal transduction mechanism (T), and one to the energy production and conversion group (C). The remaining three mutations did not show protein function or were not protein-encoding regions. A 35-bp repeated mutation in the structural gene encoding a putative siderophore-binding lipoprotein A (SBP A, NCgl0774) and a substitution mutation (C→G) at the 14 bases upstream of the start codon of the gene encoding a putative siderophore-binding lipoprotein B (SBP B, NCgl1959) were identified. A condensed mutation was also found in one of the operons encoding rRNA primary transcript. One mutation was found in the 16S rRNA structural gene, and another 23-point mutations were densely observed between the 16S and 23S rRNA sequences, which are called 16S–23S internal transcribed spacer (ITS) (Osorio et al., 2005).
Mutation Effect on Iron Consumption
The mutations observed in two presumed siderophore-binding lipoproteins (NCgl0774: 35-bp repeated mutation; NCgl1959: point substitution mutation at -14 bp) suggested their possible role in the rapid reproduction of JH41. To explore the functional significance of these putative iron-importing systems (Ribeiro and Simões, 2019), the iron consumption of JH41 was profiled (Figure 2B). Iron concentration in the medium decreased more rapidly in the JH41 strain. Since iron is an essential element for bacteria and acts as a redox center for several cytochromes and iron–sulfur proteins in the electron transfer chain (Hider and Kong, 2010), it was assumed that the rapidly reproducing JH41 might harbor greater energy than its ancestor. To validate this, ATP concentrations of the fast-growing JH41 and ancestral PT were analyzed. Indeed, the intracellular ATP concentrations of JH41 were greater than those of the PT strain with a range of 19–35% during the active growing phase (Figure 2C).
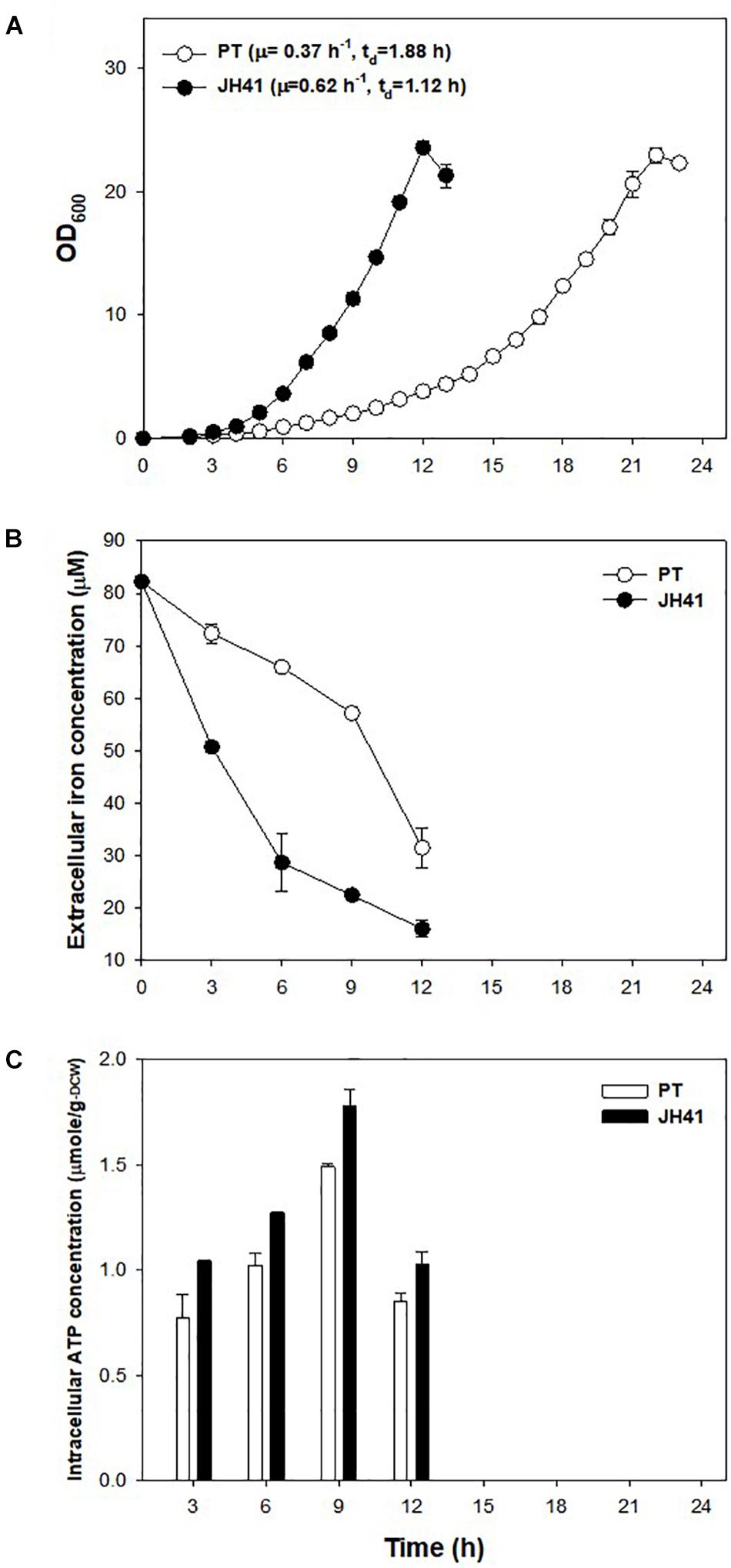
Figure 2. Profiles of growth, iron consumption, and intracellular ATP concentration of the JH41 host. (A) Batch culture growth profiles of PT and JH41 hosts. Cultures were in a 500-ml baffled flask containing 50 ml of modified MCGC medium. Each data point represents the mean ± SD; n = 3 biologically independent samples. (PT: white; JH41: black). (B) Iron consumption in PT and JH41 hosts. Samples were taken from the batch cultures (500-ml baffled flask containing 50 ml of modified MCGC medium). Iron concentration of the medium was measured by the o-phenanthroline colorimetric method. Error bars show a statistical mean ± SD; n = 3 biologically independent samples. (PT: white; JH41: black). (C) Intracellular ATP concentrations in PT and JH41 strains. Samples were taken from the batch cultures (500-ml baffled flask containing 50 ml of modified MCGC medium). Intracellular ATP was measured by the luciferin–luciferase reaction immediately after disrupting cells. Error bars show a statistical mean ± SD; n = 3 biologically independent samples (PT: white; JH41: black).
To assess which of the putative siderophore-binding lipoproteins affected iron consumption, a PT strain harboring the genomic NCgl0774 gene with the precise 35-bp repeated sequence (PT NCgl0774:35-bp) was constructed for complementation tests. The constructed PT NCgl0774:35-bp strain showed no differences compared with the PT strain in the growth profile (Supplementary Figure 2). This result indicated that the 35-bp repeat mutation on the putative siderophore-binding protein A was not a prominent factor contributing to the rapid reproduction of JH41.
Another point mutation found in the JH41 strain was located at -14 bp of the start codon of NCgl1959 encoding a putative siderophore-binding lipoprotein B, in the middle of the consensus sequence of the DtxR binding site (TTAGGCAAGGCTACCTTTTTGCCTATG; bold: DtxR binding consensus sequence; underline: mutation C→G; italic: start codon), where DtxR is a repressor of the genes responsible for iron uptake and protection against free radical damage (Wennerhold and Bott, 2006). The mutation on the DtxR binding sequence was assumed to derepress the downstream NCgl1959. To this end, we also found that the transcriptome data show a 5.1-fold upregulation of NCgl1959 in JH41 [Table 2; messenger RNA (mRNA) fold based on transcriptome column]. To verify the hypothesis, the binding affinity between the NCgl1959 regulatory region and DtxR was investigated by gel shift assay (Figure 3A). The binding affinity of the His-tag purified DtxR protein on the DNA fragment carrying the point mutation was only 40% of the affinity of the original sequence, which suggested that the mutation would have caused the derepression of NCgl1959 by reducing affinity to the transcription repressor, DtxR. Further, complementation studies were performed by constitutively expressing NCgl1959, the putative siderophore-binding lipoprotein B, on a plasmid in the PT strain. The PT strain expressing NCgl1959 (PT + pSL360-SBP B) showed more rapid reproduction (75% increase) (Figure 3B), greater iron consumption (Figure 3C), and higher ATP concentrations (Figure 3D) than the parental PT strain. Therefore, the derepression or overexpression of the putative siderophore-binding protein (NCgl1959) enabled C. glutamicum to consume more iron and further increase intracellular ATP concentration.
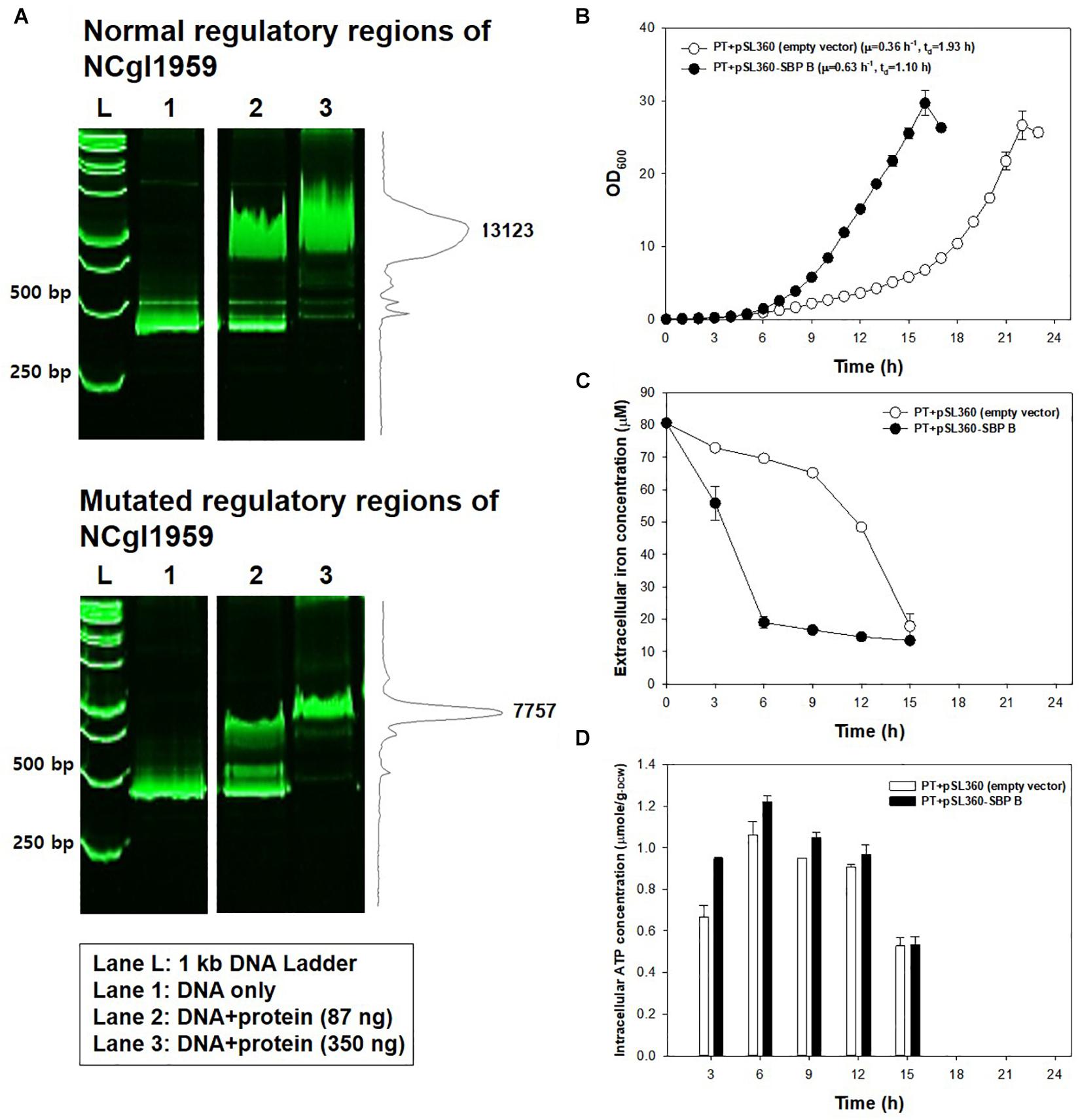
Figure 3. Effects of the mutation at 14 bases upstream of NCgl1959. (A) Native polyacrylamide gel image showing the affinity of DtxR protein to the normal DNA fragment (up) and to the mutated DNA fragment (down). L: size marker; lane 1: DNA only; lane 2: DNA + DtxR (87 ng); lane 3: DNA + DtxR (350 ng). The DNA fragments were stained with SYBR Green. The chromatogram on the right side represents the intensity of bands by ImageJ software. The image is from the representative experiment from three independent experiments. (B) Growth profile of PT strain harboring plasmid constitutively expressing NCgl1959. Each data point is the mean ± SD; n = 3 biologically independent samples. (PT + empty vector: white; PT + pSL360-SBP B: black). (C) Iron consumption in PT strain harboring plasmid constitutively expressing NCgl1959. The iron concentration in the medium was measured by the o-phenanthroline colorimetric method. All strains were cultured in a 500-ml baffled flask containing 50 ml of modified MCGC medium. Bar heights and error bars show a statistical mean ± SD; n = 3 biologically independent samples (PT + empty vector: white; PT + pSL360-SBP B: black). (D) Intracellular ATP concentrations in PT strain harboring plasmid constitutively expressing NCgl1959. The intracellular ATP luciferin–luciferase reaction was measured immediately after harvest during the actively growing phase. Point and error bars show a statistical mean ± SD; n = 3 biologically independent samples (PT + empty vector: white; PT + pSL360-SBP B: black).
The point mutation found at the -108 region of the NCgl1159 (subunit A of F0F1-ATP synthase) might be involved in ATP generation (Table 2). The growth profile of the PT host containing the genomic mutation at the -108 region of the NCgl1159 (PT NCgl1159C–108T), however, was not different from that of the PT host (Supplementary Figure 3).
Investigating further, known regulators related to iron metabolism were found to be disrupted in the PT genome. Along with DtxR, the repressor of the putative siderophore-binding lipoprotein B expression, RipA, which is known to be a gene-enhancing regulator for iron-consuming proteins and is repressed by DtxR (Wennerhold et al., 2005), were chosen as the disruption targets. Part of the coding sequences of DtxR (1.9→1.0 kb) and RipA (1.7→0.9 kb) were removed from the genome of PT strain using a double crossover method. The PT ΔdtxR strain showed a 34% faster growth rate and the PT ΔripA (47% slower growth rate) compared with PT strain (Figure 4). The disruption of RipA (leading downregulation of iron-consuming genes) led the cell to double more slowly, and the disruption of DtxR (leading to overexpression of iron-importing genes) resulted in the faster doubling.
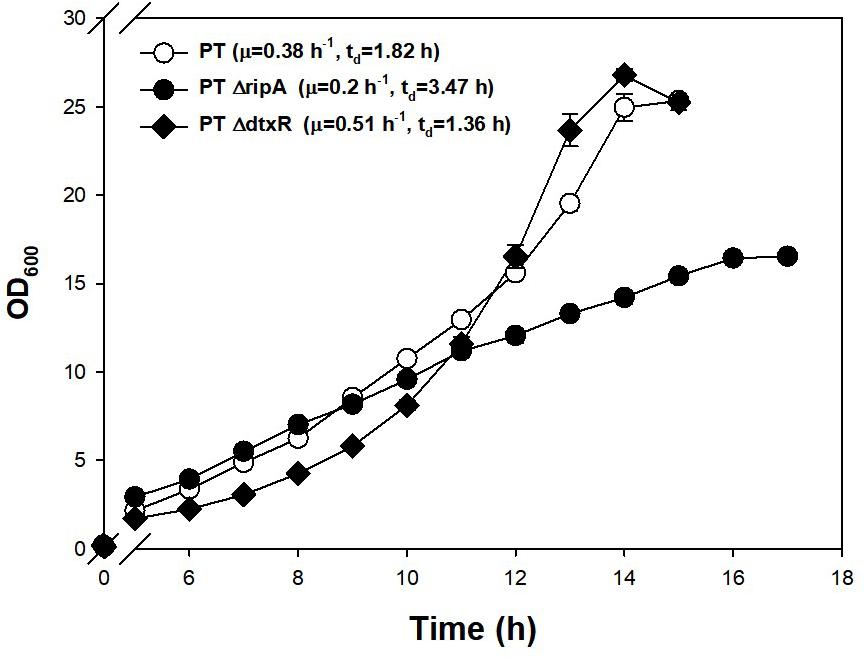
Figure 4. Effect of genomic disruptions of iron-relevant regulator genes (ripA, dtxR) on the growth of PT strain. The PT ΔripA and PT ΔdtxR strains were incubated at 30°C, 200 rpm in a shaking incubator. A 500-ml baffled flask containing 50 ml of the modified MCGC media was used for efficient oxygen transfer. The exponential growth rate (μ) in batch culture was determined by linear regression of log biomass concentrations over each process time. Each data point is the mean ± SD; n = 3 biologically independent samples (PT: white circle; PT ΔripA: black circle; PT ΔdtxR: black rhombus).
Mutation Effect on Glucose Consumption
Another notable mutation in JH41 was on the NCgl2472 gene (A239G), encoding the LuxR-type global transcription regulator RamA protein, which controls the expression of genes involved in sugar uptake, glycolysis, acetate, and many other metabolic pathways (Auchter et al., 2011). Wang et al. and Graf et al. reported a ramA mutation found in another fast-growing strain of C. glutamicum, and the mutation was described as a key mutation because it enabled substrate uptake and increased metabolic flux (Wang et al., 2018; Graf et al., 2019). To verify that the ramAA239G mutation in the JH41 strain also contributed to the enhanced cell division, the genomic point mutation was introduced to PT strain by homologous recombination to construct PT ramAA239G strain. Indeed, the PT ramAA239G strain consumed glucose at 0.89 ± 0.03 g–glucose/g–DCW⋅h, whereas PT and JH41 consumed it at 0.64 ± 0.04 and 1.11 ± 0.01 g–glucose/g–DCW⋅h, respectively (Figure 5). Therefore, the ramAA239G mutation led the PT strain to increase the rates of glucose consumption by 39% and growth by 40%. The increase in glucose uptake would have provided the increased precursor metabolites and energy to C. glutamicum cells.
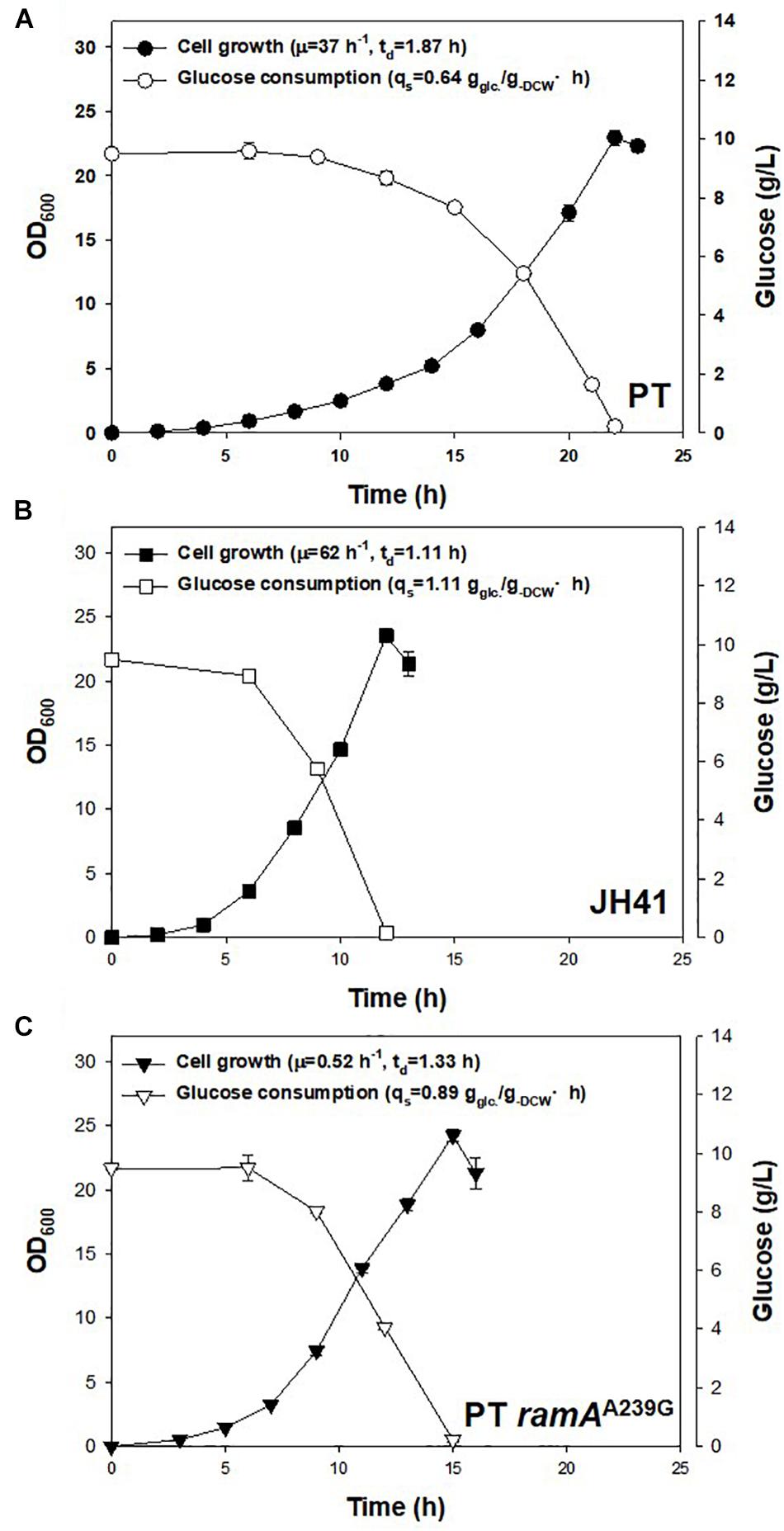
Figure 5. Effects of genomic ramAA239G mutation in PT strain on growth rate and glucose consumption. The growth rate and glucose consumption profiles of (A) PT, (B) JH41, and (C) PT ramAA239G. Batch cultures were performed in a 500-ml baffled flask containing 50 ml of 0.9% glucose modified MCGC medium. Cell densities of C. glutamicum PT, JH41, and PT ramAA239G were measured every 2 h. Glucose consumption was measured by 3,5-dinitrosalicylic acid (DNS) reduction sugar quantification method. Each data point represents mean ± SD; n = 3 biologically independent samples. (biomass: black; glucose concentration: white).
Mutation Effect on Ribosome Components
The ITS mutation (condensed 23 point mutations, Table 2) located in the non-coding region of rRNA primary transcript in the genome of JH41 was assumed to influence the maturation of rRNA from its pre-transcript, and this might contribute to the increased number of ribosomes. To confirm this hypothesis, concentrations of the mature 5S, 16S, and 23S rRNAs in the JH41 strain were compared with those in the PT strain by quantitative PCR (Table 3). The fast-growing JH41 strain harbored higher concentrations of mature rRNAs (45% more 16S rRNA, 72% more 23S rRNA, 37% more 5S rRNA) compared to the PT strain. Along with increases in the three rRNAs, our previous paper also reported that most of the mRNAs encoding ribosomal proteins were upregulated (Supplementary Table 2) (Park et al., 2020). These results enabled us to conclude the ITS mutation in rRNA primary transcript supported the fast growth of JH41 by increases in matured rRNAs and ribosomal proteins, and therefore, the increase in ribosome components might be a reverse engineering target to enhance the production of recombinant protein even though the genomic introduction of the multiple mutations in the one ITS region among the 6 rRNA operons would be technically difficult.
Combinatorial Effect of SBP B and RamA Mutations on Recombinant Protein
In the above results, the mutations on the SBP B upregulation (SBP BC–14G) and the glucose-uptake regulator (ramAA239G) were the clear genetic factors affecting the growth rate of JH41. To evaluate the combinatorial effect of the two mutations on the growth profile and recombinant protein production, the genome of the PT host was mutated to have either the SBP BC–14G mutation (PT SBP BC–14G host) or the ramAA239G mutation (PT ramAA239G host) or to have both mutations (PT SBP BC–14G ramAA239G, in short, PT S + R). The growth rate of the SBP B genome-edited host (PT SBP BC–14G) was 0.56 h–1 (tD = 1.23 h) and that of the RamA genome-edited host (PT ramAA239G) was 0.52 h–1 (tD = 1.33 h), which were 51 and 41% faster doubling, respectively, than that of PT host (growth rate = 0.37 h–1, tD = 1.87 h). The double mutant of ramAA239G and SBP BC–14G host (PT S + R) showed the fasted growth rate of 0.66 h–1 (tD = 1.05 h), which was 78% higher than that of PT and even faster than that of the evolved JH41 host (0.61 h–1, tD = 1.13 h) (Figure 6A).
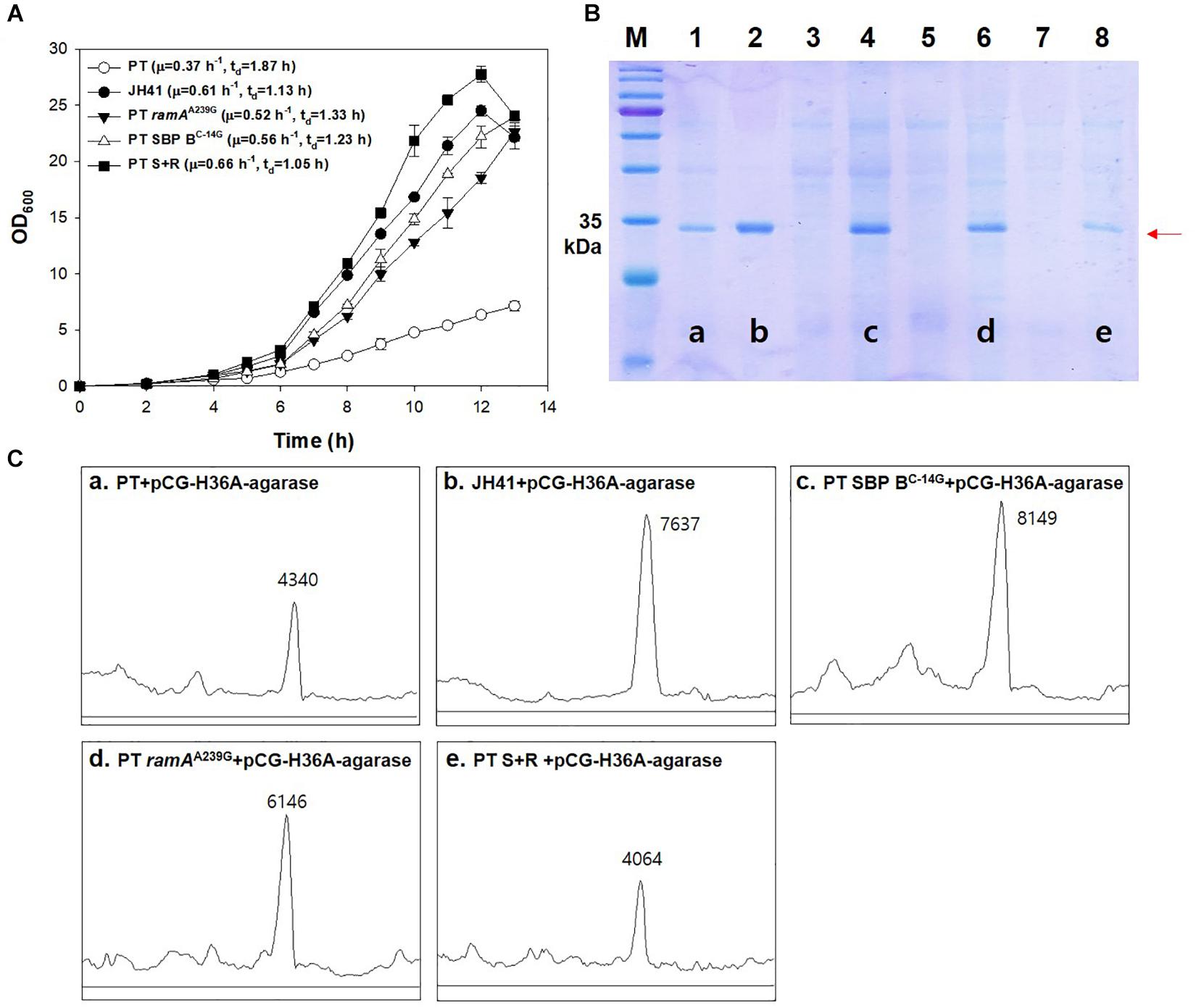
Figure 6. Effects of genomic ramAA239G and SBP BC–14G mutation in PT strain on growth rate and the secretory production of recombinant protein. (A) Batch culture growth profiles of PT and JH41 strains. The growth rate profiles of PT, JH41, PT ramAA239G, PT SBP BC–14G, and PT S + R. Batch cultures were performed in a 500-ml baffled flask containing 50 ml of 0.9% glucose modified MCGC medium. Cell densities of C. glutamicum PT, JH41, PT ramAA239G, PT SBP BC–14G, and PT S + R were measured every 1 h. Each data point represents mean ± SD; n = 3 biologically independent samples. (PT: white circle; JH41: black circle; PT ramAA239G: black triangle; PT SBP BC–14G: white triangle; PT S + R: Black square). (B) Image of sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Lane 1, PT + pCG-H36A (empty vector); lane 2, JH41 + pCG-H36A-agarase; lane 3, PT SBP BC–14G + pCG-H36A (empty vector); lane 4, PT SBP BC–14G + pCG-H36A-agarase; lane 5, PT ramAA239G + pCG-H36A (empty vector); lane 6, PT ramAA239G + pCG-H36A-agarase; lane 7, PT S + R + pCG-H36A (empty vector); lane 8, PT S + R + pCG-H36A-agarase. Agarase production was confirmed after 24 h cultivation. Arrow indicates the secreted agarase band (34 kDa). The image is from the representative experiment from three independent experiments. (C) Band density estimation of the SDS-PAGE image using ImageJ software. a: lane 1; b: lane 2; c: lane 4; d: lane 6; e: lane 8.
The secretory protein productions in the PT SBP BC–14G and the PT ramAA239G hosts are also shown to be greater than that from the PT host (Figures 6B,C). The SDS-PAGE band intensity of the secretory recombinant proteins from the PT SBP BC–14G and the PT ramAA239G hosts were 8,149 and 6,146 AU, respectively, where those from PT host was 4,130 AU. The fastest growing PT S + R hosts, however, secreted the recombinant protein at only 4,064 AU, which is even lower than that of the PT host. Therefore, the mutations in ramAA239G and SBP BC–14G, respectively, resulted in the increased growth rates and protein production. Meanwhile, the combination of the two genes showed a remarkable increase in the growth rate but was found to have a negative effect on protein production. The iron metabolism regulator-deleted strains (PT ΔdtxR and PT ΔripA) also decreased the recombinant agarase production by 34 and 37%, respectively, than that from PT host (Supplementary Figure 4).
Discussion
The evolved C. glutamicum JH41 strain with increased growth rate was able to produce 2.7-fold greater secretory recombinant protein than PT host (Figure 1). The confirmed genetic factors that contributed to the increased growth rates and became the reverse engineering targets to enhance production of recombinant proteins were (1) SBP BC–14G that enhanced iron transport leading the increase of intracellular energy and (2) ramAA239G that enhanced glucose uptake leading to the increase in energy and metabolite pool. High levels of intracellular ATP have been known to improve target products by improving substrate uptake, cell growth, biosynthesis, and resistance to toxic compounds (Hara and Kondo, 2015). The authors also reported that the productions of recombinant proteins (green fluorescence protein and alkaline protease) were increased in the highly energized E. coli host by the overexpression of phosphoenolpyruvate carboxykinase (Pck) (Kim et al., 2012). A high level of intracellular energy can provide favorable conditions for energy-consuming reactions such as charging amino acids to transfer RNA (tRNA), translational elongation, folding, and secretion. In addition, a high intracellular energy state can upregulate the genes involved in the biosynthetic pathways for amino acid supply (Kwon et al., 2008).
The point substitution mutation in the regulatory region of NCgl1959 in JH41 strain, resulted in the increase in iron consumption by derepressing the cognate genes (Figure 3). Although the function of NCgl1959 protein in C. glutamicum has not yet been clearly identified, the NCgl1959 protein was estimated to have a similar 3D structure with the known siderophore-binding lipoproteins (E. coli FhuD and Bacillus cereus YfiY) (Clarke et al., 2000; Zawadzka et al., 2009) (Supplementary Figure 5). One might speculate that NCgl1959 would play the role of iron complex uptake. The imported iron could be used for the assembly of electron transfer chain components (i.e., iron–sulfur reaction center for NADH dehydrogenase; heme–iron for cytochrome c oxidase and cytochromes) and redox-containing proteins (i.e., heme-dependent catalase and peroxidase) (Park et al., 2020), and therefore, it is considered that the enhanced iron consumption in JH41 strain would contribute to generating more energy to the respiratory chain, along with a simultaneous increase in defense capability against oxidative stress.
In particular, RamA has been shown to act as a positive regulator of the TCA cycle genes, sdhCAB (succinate dehydrogenase operon) and acn (aconitase) (Bussmann et al., 2009; Teramoto et al., 2011). The point mutation ramAA239G (corresponding to RamAY80C) of JH41 strain was located at the GAF-2 domain (amino acid positions 8 and 146), the same domain of the reports from Graf et al. and Wang et al. (Wang et al., 2018; Graf et al., 2019), and the GAF domain has been found in the cyclic nucleotide phosphodiesterase superfamily in other species (Fawcett et al., 2000; Schultz, 2009). Therefore, we considered that the ramAA239G would have enabled the JH41 strain to increase glucose consumption by altering the cyclic nucleotide-mediated signals, thereby resulting in increased substrate uptake. The increase in carbon source in the JH41 host would have been favorable cellular conditions for the recombinant protein production by providing enhanced pools of precursor metabolites, along with more energy.
We do not understand why the combination of the two mutations of ramAA239G and SBP BC–14G showed a faster growth rate exceeding JH41 and less production of recombinant protein (Figure 6). The H36 promoter for the recombinant protein production was one of the most powerful synthetic promoter known in C. glutamicum (Yim et al., 2013), and it was able to promote the secretory production of agarase from the PT ramAA239G and the PT SBP BC–14G along with the JH41 hosts. There might be unknown factor responsible for this mismatch between fast growth and less protein production (Nordholt et al., 2017). Further basic studies on cellular physiology are required to understand this unexpected phenomenon.
The condensed 23 point mutations occurred in the ITS region of the first rRNA operon among the six rRNA operons of C. glutamicum JH41. The multimutated first ITS sequence was similar to the sequence of the ITS in the fifth rRNA operon. A tRNA gene is located between 16S rRNA and 23S rRNA in E. coli rRNA pretranscript, while C. glutamicum does not contain tRNA in this sequence region. Although the processing mechanism of the rRNA pretranscript in C. glutamicum remains unknown, the ITS mutation in JH41 might have facilitated RNase processing, as in the case of E. coli. Therefore, the processing of the polycistronic primary transcript might have been increased to form mature ribosomes by the ITS mutation, having observed the increases in rRNAs and ribosomal proteins. The increase in mature ribosome components would contribute to the enhancement of recombinant protein productions, as observed in JH41.
Additional genetic factors enhancing growth and recombinant protein production remain veiled. It is worth mentioning another complementation on the genomic mutation on the coding region of NCgl2298 (osrR, G757A = D253N), an HTH-type transcription regulator known to upregulate the tolerance genes against oxidative stress and heat stress (Hong et al., 2016). The mutation on the osrR was expected to release the stress tension that was provoked by the enhanced iron and carbon uptakes. The growth of the PT NCgl2298G757A was, however, no better than that of the PT strain (Supplementary Figure 6A). The combination of mutations (i.e., the mutations on ramA + osrR) would be required for further understanding.
Besides, a mutation in the -109 region of NCgl1159 (F0F1-ATP synthase subunit A), which was expected to affect cell growth by altering ATP generation, did not increase the growth rate (Supplementary Figure 3). The transcription levels of ATP synthase components in JH41 were only half those in our previous paper (Park et al., 2020), so the half-expressed ATP synthase might have been somehow balanced for the enhanced energy generation under the increased iron and carbon uptake conditions. Another possible mutation to consider is NCgl2331, a penicillin-binding protein involved in cell defense. Substituting A for the 610th G of the NCgl2331 gene in the PT strain resulted in marginal increase (14%) in growth rate (Supplementary Figure 6B). Penicillin-binding proteins are membrane-associated macromolecules and are expected to play a key role in cell biosynthesis and cell division (Macheboeuf et al., 2006).
In this respect, our discovery offers essential cues for the construction of an efficient cell factory for the production of recombinant proteins and expands the biological knowledge base. Studies on the reverse engineering of the other mutations including the ITS condensed mutations might lead us to further improvement on C. glutamicum host.
Data Availability Statement
The JH41 genome was resequenced and deposited in NCBI accession number: PRJNA554987.
Author Contributions
ML, JP, and KP performed the experiments. ML and PK designed the experiments. JK and PK raised grant and supervised. ML and PK wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the grants from the National Research Foundation of Korea (2016R1E1A1A01943552).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.588070/full#supplementary-material
References
Auchter, M., Cramer, A., Huser, A., Ruckert, C., Emer, D., Schwarz, P., et al. (2011). RamA and RamB are global transcriptional regulators in Corynebacterium glutamicum and control genes for enzymes of the central metabolism. J. Biotechnol. 154, 126–139. doi: 10.1016/j.jbiotec.2010.07.001
Bussmann, M., Emer, D., Hasenbein, S., Degraf, S., Eikmanns, B. J., and Bott, M. (2009). Transcriptional control of the succinate dehydrogenase operon sdhCAB of Corynebacterium glutamicum by the cAMP-dependent regulator GlxR and the LuxR-type regulator RamA. J. Biotechnol. 143, 173–182. doi: 10.1016/j.jbiotec.2009.06.025
Clarke, T. E., Ku, S. Y., Dougan, D. R., Vogel, H. J., and Tari, L. W. (2000). The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat. Struct. Biol. 7, 287–291. doi: 10.1038/74048
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Deshavath, N. N., Mukherjee, G., Goud, V. V., Veeranki, V. D., and Sastri, C. V. (2020). Pitfalls in the 3, 5-dinitrosalicylic acid (DNS) assay for the reducing sugars: interference of furfural and 5-hydroxymethylfurfural. Int. J. Biol. Macromol. 156, 180–185. doi: 10.1016/j.ijbiomac.2020.04.045
Disz, T., Akhter, S., Cuevas, D., Olson, R., Overbeek, R., Vonstein, V., et al. (2010). Accessing the SEED genome databases via Web services API: tools for programmers. BMC Bioinformatics 11:319. doi: 10.1186/1471-2105-11-319
Fawcett, L., Baxendale, R., Stacey, P., McGrouther, C., Harrow, I., Soderling, S., et al. (2000). Molecular cloning and characterization of a distinct human phosphodiesterase gene family: PDE11A. Proc. Natl. Acad. Sci. U.S.A. 97, 3702–3707. doi: 10.1073/pnas.050585197
Feist, A. M., Zielinski, D. C., Orth, J. D., Schellenberger, J., Herrgard, M. J., and Palsson, B. O. (2010). Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab. Eng. 12, 173–186. doi: 10.1016/j.ymben.2009.10.003
Graf, M., Haas, T., Müller, F., Buchmann, A., Harm-Bekbenbetova, J., Freund, A., et al. (2019). Continuous adaptive evolution of a fast-growing Corynebacterium glutamicum strain independent of protocatechuate. Front. Microbiol. 10:1648. doi: 10.3389/fmicb.2019.01648
Hara, K. Y., and Kondo, A. (2015). ATP regulation in bioproduction. Microb. Cell. Fact. 14:198. doi: 10.1186/s12934-015-0390-6
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657. doi: 10.1039/b906679a
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. doi: 10.1016/0378-1119(89)90358-2
Hong, E. J., Kim, P., Kim, E. S., Kim, Y., and Lee, H. S. (2016). Involvement of the osrR gene in the hydrogen peroxide-mediated stress response of Corynebacterium glutamicum. Res. Microbiol. 167, 20–28. doi: 10.1016/j.resmic.2015.09.005
Kim, H. J., Kwon, Y. D., Lee, S. Y., and Kim, P. (2012). An engineered Escherichia coli having a high intracellular level of ATP and enhanced recombinant protein production. Appl. Microbiol. Biotechnol. 94, 1079–1086. doi: 10.1007/s00253-011-3779-0
Kim, S. H., Kim, M. S., Lee, B. Y., and Lee, P. C. (2016). Generation of structurally novel short carotenoids and study of their biological activity. Sci. Rep. 6:21987. doi: 10.1038/srep21987
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwon, S. K., Kim, S. K., Lee, D. H., and Kim, J. F. (2015). Comparative genomics and experimental evolution of Escherichia coli BL21(DE3) strains reveal the landscape of toxicity escape from membrane protein overproduction. Sci. Rep. 5:16076. doi: 10.1038/srep16076
Kwon, Y. D., Lee, S. Y., and Kim, P. (2008). A physiology study of Escherichia coli overexpressing phosphoenolpyruvate carboxykinase. Biosci. Biotechnol. Biochem. 72, 1138–1141. doi: 10.1271/bbb.70831
Lee, M. J., and Kim, P. (2018). Recombinant protein expression system in Corynebacterium glutamicum and its application. Front. Microbiol. 9:2523. doi: 10.3389/fmicb.2018.02523
Lee, S., and Kim, P. (2020). Current status and applications of adaptive laboratory evolution in industrial microorganisms. J. Microbiol. Biotechnol. 30, 793–803. doi: 10.4014/jmb.2003.03072
Macheboeuf, P., Contreras-Martel, C., Job, V., Dideberg, O., and Dessen, A. (2006). Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30, 673–691. doi: 10.1111/j.1574-6976.2006.00024.x
Na, Y. A., Lee, J. Y., Bang, W. J., Lee, H. J., Choi, S. I., Kwon, S. K., et al. (2015). Growth retardation of Escherichia coli by artificial increase of intracellular ATP. J. Ind. Microbiol. Biotechnol. 42, 915–924. doi: 10.1007/s10295-015-1609-6
Nordholt, N., van Heerden, J., Kort, R., and Bruggeman, F. J. (2017). Effects of growth rate and promoter activity on single-cell protein expression. Sci. Rep. 7:6299. doi: 10.1038/s41598-017-05871-3
Osorio, C. R., Collins, M. D., Romalde, J. L., and Toranzo, A. E. (2005). Variation in 16S-23S rRNA intergenic spacer regions in Photobacterium damselae: a mosaic-like structure. Appl. Environ. Microbiol. 71, 636–645. doi: 10.1128/AEM.71.2.636-645.2005
Osten, C. H. V. D., Gioannetti, C., and Sinskey, A. J. (1989). Design of a defined medium for growth of Corynebacterium glutamicum in which citrate facilitates iron uptake. Biotechnol. Lett. 11, 11–16. doi: 10.1007/bf01026778
Park, J., Lee, S., Lee, M. J., Park, K., Lee, S., Kim, J. F., et al. (2020). Accelerated growth of Corynebacterium glutamicum by up-regulating stress-responsive genes based on transcriptome analysis of a fast-doubling evolved strain. J. Microbiol. Biotechnol. 30, 1420–1429. doi: 10.4014/jmb.2006.06035
Park, S. D., Lee, S. N., Park, I. H., Choi, J. S., and Jeong, W. K. (2004). Isolation and characterization of transcriptional elements from Corynebacterium glutamicum. Microbiol. Biotechnol. 14, 789–795.
Ribeiro, M., and Simões, M. (2019). Advances in the antimicrobial and therapeutic potential of siderophores. Environ. Chem. Lett. 17, 1485–1494. doi: 10.1007/s10311-019-00887-889
Rugbjerg, P., Feist, A. M., and Sommer, M. O. A. (2018). Enhanced metabolite productivity of Escherichia coli adapted to glucose M9 minimal medium. Front. Bioeng. Biotechnol. 6:166. doi: 10.3389/fbioe.2018.00166
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning : A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sandberg, T. E., Salazar, M. J., Weng, L. L., Palsson, B. O., and Feist, A. M. (2019). The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 56, 1–16. doi: 10.1016/j.ymben.2019.08.004
Schafer, A., Tauch, A., Jager, W., Kalinowski, J., Thierbach, G., and Puhler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Schultz, J. E. (2009). Structural and biochemical aspects of tandem GAF domains. Handb. Exp. Pharmacol. 191, 93–109. doi: 10.1007/978-3-540-68964-5_6
Serra, P., Moliner-Martínez, Y., Herráez-Hernández, R., Verdú-Andrés, J., and Campíns-Falcó, P. (2015). Simplifying iron determination with o-phenanthroline in food ashes using 2-nitrophenol as an acid-base indicator. Food Anal. Methods 9, 1150–1154. doi: 10.1007/s12161-015-0294-4
Stella, R. G., Wiechert, J., Noack, S., and Frunzke, J. (2019). Evolutionary engineering of Corynebacterium glutamicum. Biotechnol. J. 14:e1800444. doi: 10.1002/biot.201800444
Teramoto, H., Inui, M., and Yukawa, H. (2011). Transcriptional regulators of multiple genes involved in carbon metabolism in Corynebacterium glutamicum. J. Biotechnol. 154, 114–125. doi: 10.1016/j.jbiotec.2011.01.016
Wang, Z., Liu, J., Chen, L., Zeng, A. P., Solem, C., and Jensen, P. R. (2018). Alterations in the transcription factors GntR1 and RamA enhance the growth and central metabolism of Corynebacterium glutamicum. Metab. Eng. 48, 1–12. doi: 10.1016/j.ymben.2018.05.004
Wennerhold, J., and Bott, M. (2006). The DtxR regulon of Corynebacterium glutamicum. J. Bacteriol. 188, 2907–2918. doi: 10.1128/JB.188.8.2907-2918.2006
Wennerhold, J., Krug, A., and Bott, M. (2005). The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J. Biol. Chem. 280, 40500–40508. doi: 10.1074/jbc.M508693200
Yadav, V. G., De Mey, M., Lim, C. G., Ajikumar, P. K., and Stephanopoulos, G. (2012). The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab. Eng. 14, 233–241. doi: 10.1016/j.ymben.2012.02.001
Yim, S. S., An, S. J., Kang, M., Lee, J., and Jeong, K. J. (2013). Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol. Bioeng. 110, 2959–2969. doi: 10.1002/bit.24954
Yim, S. S., Choi, J. W., Lee, R. J., Lee, Y. J., Lee, S. H., Kim, S. Y., et al. (2016). Development of a new platform for secretory production of recombinant proteins in Corynebacterium glutamicum. Biotechnol. Bioeng. 113, 163–172. doi: 10.1002/bit.25692
Keywords: Corynebacterium glutamicum, host for recombinant protein, iron consumption, cellular energy, glucose consumption, ribosome
Citation: Lee MJ, Park J, Park K, Kim JF and Kim P (2020) Reverse Engineering Targets for Recombinant Protein Production in Corynebacterium glutamicum Inspired by a Fast-Growing Evolved Descendant. Front. Bioeng. Biotechnol. 8:588070. doi: 10.3389/fbioe.2020.588070
Received: 28 July 2020; Accepted: 09 November 2020;
Published: 09 December 2020.
Edited by:
Yu Wang, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, ChinaReviewed by:
Zhi-Gang Jeff Qian, Shanghai Jiao Tong University, ChinaDae-Hee Lee, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South Korea
Copyright © 2020 Lee, Park, Park, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pil Kim, a2ltcEBjYXRob2xpYy5hYy5rcg==
†Present address: Jihoon Park, Durae Corporation, Gangwon, South Korea
 Min Ju Lee1
Min Ju Lee1 Jihyun F. Kim
Jihyun F. Kim Pil Kim
Pil Kim