- 1Clean Energy Research Center, Korea Institute of Science and Technology (KIST), Seoul, South Korea
- 2Department of Biotechnology, Korea University, Seoul, South Korea
- 3Division of Energy and Environment Technology, University of Science and Technology (UST), Daejeon, South Korea
- 4Green School, Korea University, Seoul, South Korea
Yeasts are promising industrial hosts for sustainable production of fuels and chemicals. Apart from efficient bioethanol production, yeasts have recently demonstrated their potential for biodiesel production from renewable resources. The fuel-oriented product profiles of yeasts are now expanding to include non-native chemicals with the advances in synthetic biology. In this review, current challenges and opportunities in yeast engineering for sustainable production of non-native chemicals will be discussed, with a focus on the comparative evaluation of a bioethanol-producing Saccharomyces cerevisiae strain and a biodiesel-producing Yarrowia lipolytica strain. Synthetic pathways diverging from the distinctive cellular metabolism of these yeasts guide future directions for product-specific engineering strategies for the sustainable production of non-native chemicals on an industrial scale.
Introduction
Microorganisms have gained significant attention as cell factories for the sustainable production of fuels and chemicals, providing great opportunities for a bio-based economy in the post-petroleum era. Of the microbial cell factories, yeast has long been used as a proven industrial producer of fuels and chemicals. The robustness in a wide range of environmental conditions, the ease of separation from the culture medium, and no phage contamination related issues make the yeast more attractive industrial cell factory than bacteria. With recent advances in synthetic biology, the product profiles of yeasts have been expanding to include those that have never been produced naturally, such as cannabinoids, tropine, and amorphadiene (Luo et al., 2019; Ping et al., 2019; Srinivasan and Smolke, 2019; Marsafari and Xu, 2020). Given that synthetic pathways for non-native products commonly involve multiple enzymes from plants and/or other higher eukaryotes, which often require post-translational modification and are difficult to be functionally expressed in prokaryotic systems, the development of yeast cell factories is drawing more attention with its advantages as a eukaryotic system.
The model yeast Saccharomyces cerevisiae is one of the most widely developed cell factories for the production of fuels and chemicals. Industrial production of ethanol, the native product of S. cerevisiae, is in operation worldwide to supply alcoholic beverages and biofuels (Walker and Stewart, 2016; Parapouli et al., 2020). Industrial-scale production of β-farnesene by engineered S. cerevisiae strains has also been reported (Gray et al., 2014; Wehrs et al., 2019). Of the non-model yeast, Yarrowia lipolytica has recently risen as another promising industrial cell factory with a superior capacity to accumulate lipids for biodiesel production (Ma et al., 2019). Currently, taking advantage of readily accessible genetic information and highly efficient genetic tools, an increasing number of products are being added to the product profiles of the model yeast S. cerevisiae. Besides being utilized for biodiesel production, Y. lipolytica is being transformed into a cell factory platform for the production of advanced biofuels and chemicals such as isoprenoids, polyketides, drug precursors, pigments, and organic acids (Miller and Alper, 2019).
The successful use of S. cerevisiae for the industrial production of bioethanol and the superior lipid production performance of Y. lipolytica shows that these strains can provide an economically feasible method for carbohydrate-based-biofuels production (Qiao et al., 2017). As such, there is increasing hope that these strains could be used for the production of numerous non-native chemicals on an industrial scale. Their GRAS (generally recognized as safe) status, as well as their well-known genomic characteristics and widely available engineering tools, make these yeast strains attractive microbial cell factories for the production of numerous chemicals (Darvishi et al., 2017; Markham and Alper, 2018; Ekas et al., 2019; Favaro et al., 2019). Here, the recent advances in engineering S. cerevisiae and Y. lipolytica as yeast cell factories for the production of non-native chemicals will be reviewed. Non-native chemicals were classified into four large groups, (i) terpenoids, (ii) polyketides, (iii) fatty acid-derived chemicals, and (iv) others, based on the core intermediate used as a precursor, namely acetyl-CoA, malonyl-CoA, and DHAP, which are key intermediates and diverge into branched pathways in the cellular metabolism of yeasts, such as the mevalonate or shikimate pathways (Figure 1). Thus, engineering strategies to produce certain groups of chemicals could be shared and more easily expanded to transform yeasts into cell factories for specific non-native chemicals. The performance of engineered yeasts as producers of specific chemical groups was analyzed and compared to that of the intensively engineered bacterial host of E. coli to highlight the benefits of using engineered yeasts for the industrial production of non-native chemicals by maximizing their potential, which is supported by their distinctive cellular metabolism. Finally, future directions for choosing an optimal platform strain and engineering strategies will be discussed to advance yeast cell factories for the sustainable production of non-native chemicals on an industrial scale.
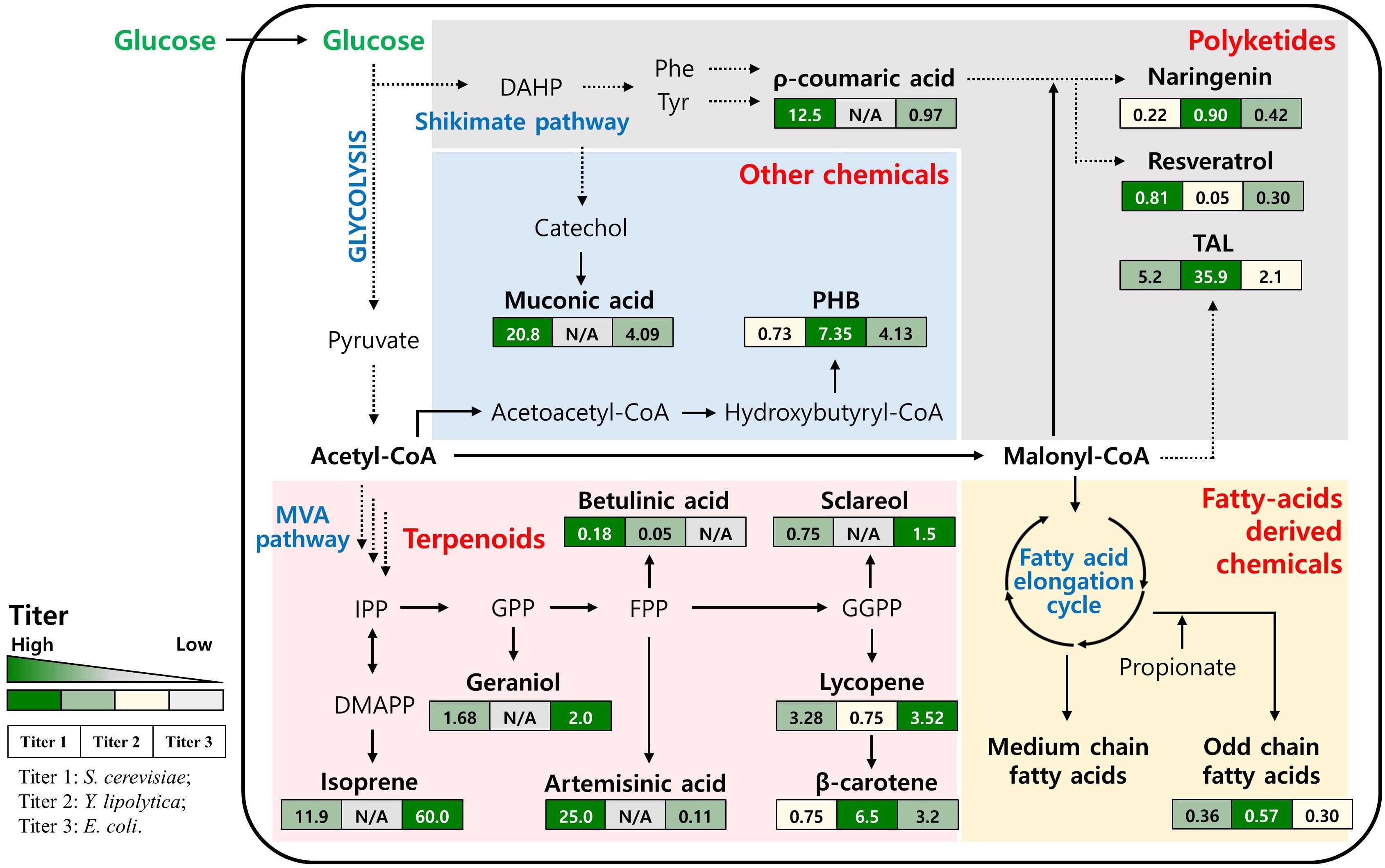
Figure 1. Simplified biosynthetic pathway and the main intermediates for production of non-native chemicals in yeasts. The values in the boxes represent the production titer obtained by engineered S. cerevisiae, Y. lipolytica and E. coli (from left to right). N/A, not available; DAHP, 3-Deoxy-D-arabinoheptulosonate 7-phosphate; Phe, Phenylalanine; Tyr, Tyrosine; TAL, Triacetic acid lactone; PHB, Polyhydroxybutyrate; IPP, Isopentenyl pyrophosphate; DMAPP, Dimethylallyl pyrophosphate; GPP, Geranyl pyrophosphate; FPP, Farnesyl pyrophosphate; GGPP, Geranylgeranyl diphosphate.
Production of Non-Native Chemicals in Yeast Cell Factories
Terpenoids
Terpenoids, also known as isoprenoids, are a large group of chemicals derived from five-carbon isoprene units with diverse applications in pharmaceuticals, flavors, colorants, and even liquid fuel alternatives (Vickers et al., 2017; Zhou, 2018). As naturally produced organic chemicals, terpenoids are conventionally extracted from natural sources such as plants, but the long cultivation time and inefficiency in extraction limit the sufficient supply of terpenoids to meet the increasing demand (Chang and Keasling, 2006; Wang et al., 2018). Thus, microbial terpenoid production has gained much attention as an alternative way to produce terpenoids. To achieve high productivity and yield in microbial hosts, strong metabolic flux through the mevalonate (MVA) pathway or methylerythritol 4-phosphate (MEP) pathway are required to sufficiently supply isopentenyl pyrophosphate (Pompon et al., 1996) and thus geranyl pyrophosphate (GPP), the main precursor of terpenoids (Zhang and Hong, 2020). In addition, functional expression of heterologous enzymes adapted from a source organism plays a critical role in achieving high titer production of terpenoids. Here, we discuss the engineering efforts put into S. cerevisiae and Y. lipolytica for the production of a few representative compounds in each class of terpenoids and compare their performance to that of E. coli, a model bacterial cell factory.
Isoprene
Isoprene (2-methyl-1,3-butadiene) is a colorless and flammable compound, and the main monomer of natural rubber. In S. cerevisiae, high-titer isoprene production (11.9 g/L) has been reported through protein engineering and enhanced precursor supply (Yao et al., 2018). Specifically, the authors constructed an isoprene production pathway by introducing a mutant isoprene synthase (ISPSLN), created by saturation mutagenesis, and compartmentalized a mitochondrial MVA pathway. In addition, improving the precursor supply through co-overexpression of diphosphomevalonate decarboxylase (MVD) and isopentenyl-diphosphate δ-isomerase, encoded by MVD1 and IDI1, respectively, and redirection of the metabolic flux toward dimethylallyl pyrophosphate (DMAPP) contributed to improved metabolic flux through an isoprene synthetic pathway (Figure 2). There are no reports on the production of isoprene in Y. lipolytica, possibly due to its greater potential for the production of C30 or higher terpenoids (Larroude et al., 2018; Zhang et al., 2019). The isoprene titer reported by a bacterial counterpart of E. coli was 60 g/L. The engineered E. coli strongly expressed the heterologous MVA pathway and isoprene synthase (IspS) from Populus alba along with an enhanced pentose phosphate pathway (Figure 2). The isoprene productivity was 2 g/L/h with a yield of 850 mg/g DCW (Whited et al., 2010).
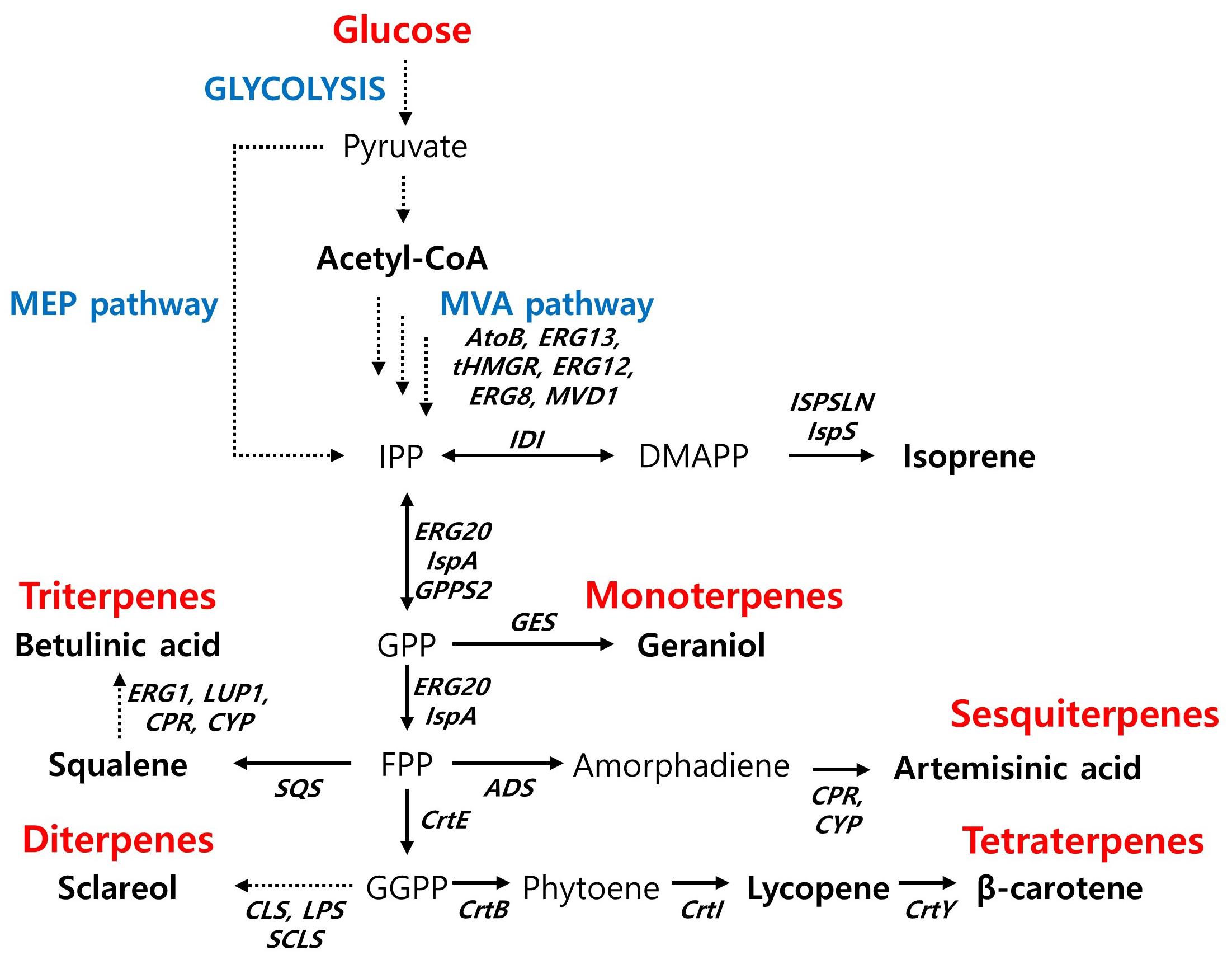
Figure 2. Biosynthetic pathway for terpenoids production and main engineering targets. AtoB, acetyl-CoA acetyltransferase; ERG13, hydroxymethylglutaryl-CoA synthase; tHMGR, truncated HMG-CoA reductase; ERG12, mevalonate kinase; ERG8, phosphomevalonate kinase; MVD1, diphosphomevalonate decarboxylase; IDI, isopentenl-diphosphate δ-isomerase; IspS, isoprene synthase; ISPSLN, mutant isoprene synthase; GPPS2, geranyl diphosphate synthase; ERG20 (IspA), farnesyl pyrophosphate synthethase; SQS, squalene synthase; ADS, amorpha-4,11-diene synthase; CPR, cytochrome P450 reductase; CYP, cytochrome P450 monooxygenase; ERG1, squalene monooxygenase; LUP1, lupeol synthase; CrtE, geranylgeranyl diphosphate synthase; CLS, 8-hydroxy copalyl diphosphate synthase; LPS, LDPP synthase; SCLS, sclareol synthase; CrtB, phytoene synthase; CrtY, lycopene beta-cyclase.
Geraniol (Monoterpene)
Geraniol (3,7-dimethylocta-trans-2,6-dien-1-ol) is one of the most widely used monoterpenes as a food additive, cosmetic ingredient, pesticide, and gasoline alternative. The highest titer of geraniol in S. cerevisiae has been reported to be 1.68 g/L through overexpression of the genes involved in the MVA pathway, tHMGR and IDI1, and the use of truncated geraniol synthase (GES) (Jiang et al., 2017; Figure 2). Of the nine candidates for heterologous GES, the GES from Catharanthus roseus (CrGES) showed the highest efficiency in geraniol production, which was further improved by truncation of a plastid targeting signal sequence required in its source organism. In addition, the researchers fused the ERG20ww (F96W/N126W), the modified FPPS solely generating GPP (Fischer et al., 2011; Ignea et al., 2014), into CrGES to enhance GPP accessibility (Figure 2). There are limited reports on monoterpene production by Y. lipolytica with a titer one order of magnitude lower than that by S. cerevisiae. Limonene and linalool have been produced at a titer of 165.3 mg/L (Cheng et al., 2019) and 6.96 mg/L (Cao et al., 2017), respectively, using minimally engineered Y. lipolytica strains. Geraniol production by Y. lipolytica has never been reported. Even with a distinctive cellular network and the lack of a native MVA pathway, the geraniol production efficiency of E. coli (Liu W. et al., 2016) has been shown to be similar to that of S. cerevisiae (2.0 vs. 1.68 g/L).
Artemisinic Acid (Sesquiterpene)
Artemisinic acid, a sesquiterpene with three isoprene units, is the precursor of artemisinin, a potent antimalarial drug (Figure 2). Based on the success by Ro et al. (2006), the highest titer of artemisinic acid (25 g/L) in S. cerevisiae has been reported through the construction of a functional biosynthetic route based on a plant-derived dehydrogenase and cytochrome P450 (Paddon et al., 2013). Possibly due to the already proven success in S. cerevisiae, no efforts have been devoted to the development of artemisinic acid-producing Y. lipolytica. Prior to the development of artemisinic acid-producing strain of S. cerevisiae, E. coli had been engineered through similar approaches involving cytochrome P450, but the titer was limited to only 105 mg/L due to the poor expression of cytochrome P450 in a prokaryotic cell factory of E. coli (Chang et al., 2007).
Sclareol (Diterpene)
Sclareol is a diterpene alcohol used as a cosmetic precursor in the fragrance industry. In S. cerevisiae, 750 mg/L of sclareol has been produced through protein engineering and deletion screening approaches (Trikka et al., 2015). To support sufficient metabolic fluxes in a sclareol synthesis pathway, a fusion protein of a modified ERG20 (F96C) and 8-hydroxycopalyl diphosphate synthase (CLS) from Cistus creticus, which catalyze the production of geranylgeranyl pyrophosphate (GGPP) and 8-hydroxycopalyl diphosphate (8-OH-CPP), respectively, were overexpressed in the strain harboring a sclareol synthase (SCLS), a class I diterpene synthase from Salvia sclarea. The engineered strain was further modified by disrupting the function of the genes selected from a heterozygous deletion screening experiments; this resulted in a 40-fold increase in the sclareol titer (750 mg/L). To our knowledge, the production of sclareol in Y. lipolytica has not been previously reported. In E. coli, a high titer (1.5 g/L) of sclareol was reportedly produced by simply introducing a heterologous pathway that converts GGPP into sclareol from S. sclarea (Schalk et al., 2012).
Betulinic Acid (Triterpene)
Betulinic acid is a pentacyclic triterpene and is used as an antitumor, antiviral, and anti-inflammatory agent (An et al., 2020). The highest titer of betulinic acid production obtained using S. cerevisiae was reported to be 182 mg/L by combining metabolic and process engineering (Czarnotta et al., 2017). The engineered strains used in the study overexpressed heterologous lupeol synthase (LUP1) and P450 reductase (CPR) from Arabidopsis thaliana, and P450 monooxygenase (CYP) from Catharanthus roseus along with homologous squalene monooxygenase (ERG1) and tHMGR. To increase the production titer of betulinic acid, these authors performed fed-batch fermentation with excess ethanol, which supports the increased supply of acetyl-CoA and NADPH for triterpene production. When combined with nitrogen-limitation conditions, metabolic flux could be directed toward betulinic acid synthesis and thereby increased the titer and yields of betulinic acid. In Y. lipolytica, betulinic acid titer of 51.87 mg/L has been reported during flask fermentation by adapting multimodular engineering strategy (Jin et al., 2019). To maximize betulinic acid production, they divided the betulinic acid synthesis pathway into four separate modules: the CYP/CPR, mevalonate, acetyl-CoA generation, and redox cofactor supply modules. The best combinations of heterologous CYP/CPR from five different sources, namely CYP from Betula platyphylla and CPR from Medicago truncatula were overexpressed along with ERG1, ylHMG1, and squalene synthase (ERG9); this resulted in the production of betulinic acid with a titer of 51.87 mg/L. Given that this titer was obtained from flask fermentation, the production performance of Y. lipolytica could be further improved when combined with process engineering approaches applied to S. cerevisiae. There are limited reports on the betulinic acid production in E. coli possibly due to the antibacterial properties of betulinic acid (Oloyede et al., 2017).
β-Carotene (Tetraterpene)
β-carotene is an orange-colored tetraterpene pigment composed of eight isoprene units, and is found in plants, photosynthetic bacteria, and algae. It exhibits antioxidant properties and serves as a precursor of vitamin A (Arendt et al., 2016). Recent success in engineering S. cerevisiae has improved β-carotene titer up to 750 mg/L (López et al., 2019) through the combined efforts of adaptive laboratory evolution and metabolic engineering; the strain overexpressing the carotenoid biosynthetic genes crtYB, crtI, and crtE from Xanthophyllomyces dendrorhous with tHMGR were evolved for improvedβ-carotene production. Later, high-titer production of lycopene, a precursor of β-carotene with its own utility, was reported to be 3.28 g/L (Shi et al., 2019) by expressing the heterologous genes involved in the lycopene biosynthesis pathway (crtE, crtB, crtI sourced from Pantoea ananatis, Pantoea agglomerans, and Blakeslea trispora, respectively) under the control of various promoters with optimized gene expression levels (Figure 2). Given that crtB and crtI are shared in the β-carotene synthesis pathway, the production titer of β-carotene could be further improved by introducing a proper gene at the last step of the β-carotene synthesis pathway. Y. lipolytica has shown superior potential for the production of β-carotene, with the highest titer of 6.5 g/L (Larroude et al., 2018). The high metabolic flux through acetyl-CoA and its lipophilic depository of liposomes provided a favorable cellular system for the high production of β-carotene. Introduction of a heterologous β-carotene synthesis pathway encoded by carRP and carB from Mucor circinelloides with overexpression of a native MVA pathway (ylHMG1) turned a previously engineered lipid over-producing strain of Y. lipolytica into a superior β-carotene-producing strain. A comparable β-carotene titer has been achieved by E. coli (Yang and Guo, 2014). The co-expression of an optimized MEP and a hybrid MVA pathway to maximize the supply of IPP and GPP resulted in a β-carotene titer of 3.2 g/L in the strain heterologously expressing the genes involved in the β-carotene synthesis pathway from Erwinia herbicola and GPPS2 from Abies grandis.
Polyketides
Polyketides are a diverse group of secondary metabolites produced by bacteria (type I, II, III), fungi (type I, II), and plants (type III). Polyketides exhibit various bioactive properties such as anticancer, antifungal, and antiviral, and serve as important resources for pharmaceutical development (Jakočiūnas et al., 2020). Within the type III polyketides class, triacetic acid lactone, ρ-coumaric acid, naringenin, and resveratrol have been produced at high titers by fungal as well as bacterial hosts (López et al., 2019).
Triacetic Acid Lactone
Triacetic acid lactone (López et al., 2019), naturally produced by the plant Gerbera hybrid, is a simple polyketide used as a chemical building block for a range of value-added products such as sorbic acid, hexanoic acid, and acetylacetone (Chia et al., 2012). TAL is generated from two common metabolic precursors, acetyl-CoA and malonyl-CoA (Figure 3A). Recently, Markham et al. (2018) demonstrated TAL production in an engineered Y. lipolytica strain reaching a titer of 35.9 g/L, the highest amount obtained by recombinant strains during bioreactor operation (Table 1). Overexpression of ACC1, which encodes the enzyme that converts acetyl-CoA to malonyl-CoA, combined with overexpression of the pyruvate bypass pathway genes, PDC2, ALD5, and ACS1, and the β-oxidation pathway genes reinforced the already sufficient acetyl-CoA pool in Y. lipolytica leading to a high-titer TAL production (Figure 3A). In S. cerevisiae, the central carbon pathway is regulated toward TAL biosynthesis by blocking competing pathways and a protease enzyme activity resulting in 2.2 g/L of TAL production under bioreactor conditions (Cardenas and Da Silva, 2014). Moreover, TAL production in an engineered industrial strain of S. cerevisiae reached a titer of 5.2 g/L by employing fed-batch cultivation in a bioreactor with ethanol feeding (Saunders et al., 2015). In E. coli, a similar titer of TAL (2.1 g/L) was reported from the shake-flask culture of an engineered strain expressing a designed TAL reporter (Tang et al., 2013).
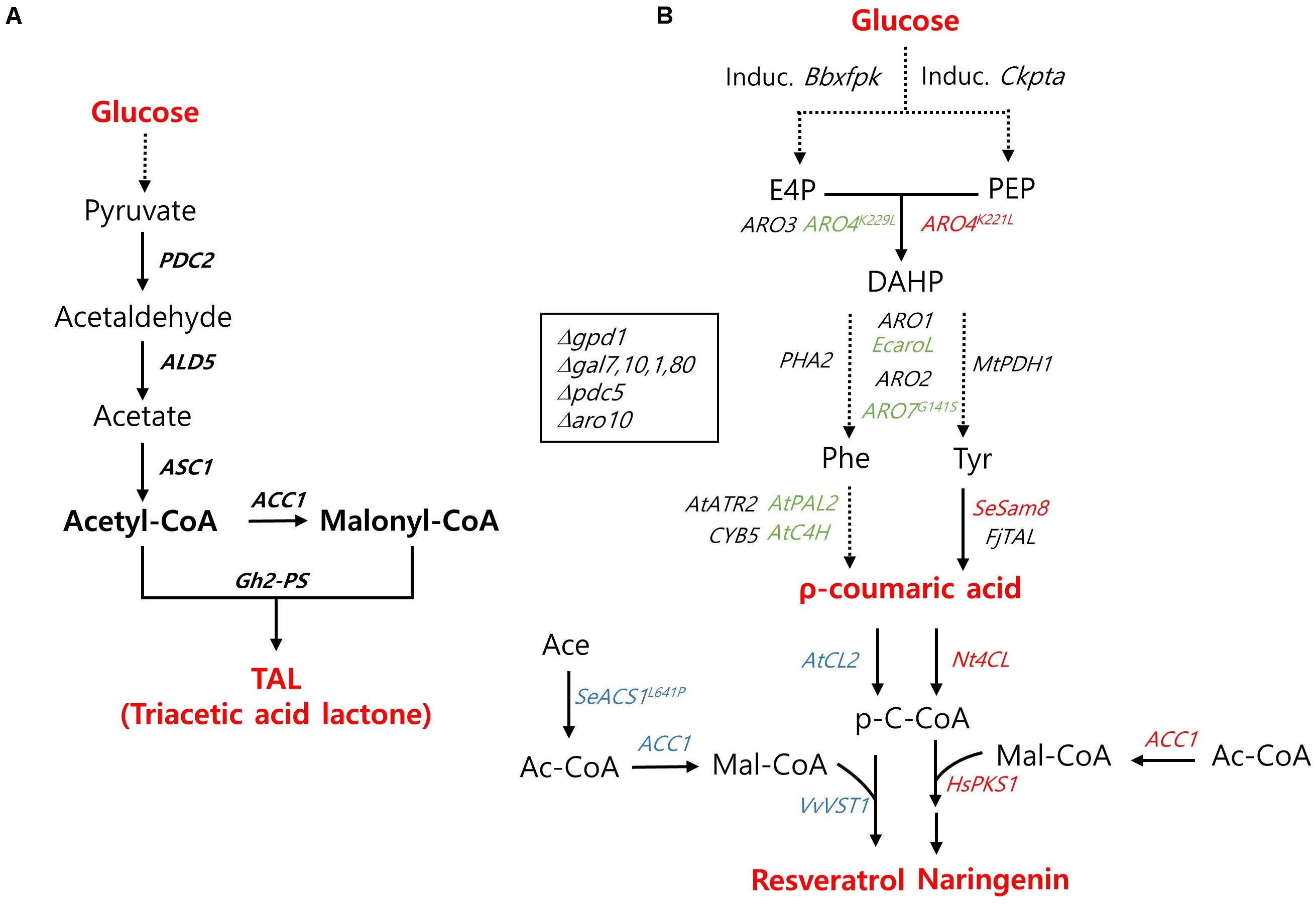
Figure 3. Biosynthetic pathway for polyketides production and main engineering targets. (A) A pathway for triacetic acid lactone production. (B) A pathway for the production of ρ-coumaric acid, resveratrol, and naringenin. The genes used specifically for the production ρ-coumaric acid, resveratrol, and naringenin are indicated with green, blue, and red colors, respectively. PDC, pyruvate decarboxylase; ALD5, aldehyde dehydrogenase 5; ACS1, acetyl-CoA synthase 1; ACC1, acetyl-CoA carboxylase; Gh2-PS, Gerbera hybrida 2-pyrone synthase; Induc. Bbxfpk, Induc. Ckpta, The inducible phosphoketolase (PHK) pathway including a Bifidobacterium breve phosphoketolase (Bbxfpk) and a Clostridium kluyveri phosphotransacetylase (Ckpta); ARO3, DAHP synthase; ARO4K229L, ARO4K221L, L-tyrosine-feedback-insensitive DAHP synthase; ARO1, pentafunctional arom protein; EcaroL, Escherichia coli shikimate kinase; ARO2, chorismate synthase; ARO7G141S, L-tyrosine-feedback-insensitive chorismate mutase; PHA2, prephenate dehydratase; MtPDH1, Medicago truncatula prephenate dehydrogenase; AtC4H, Arabidopsis thaliana cinnamic acid hydroxylase; AtPAL2, A. thaliana phenylalanine ammonia lyase; AtATR2, A. thaliana cytochrome P450 reductase; CYB5, Cytochrome b5; FjTAL, Flavobacterium johnsoniae tyrosine ammonia lyase; SeSam8, Saccharothrix espanaensis tyrosine amino lyase; AtCL2, Nt4CL, Arabidopsis thaliana, Nicotiana tabacum 4-coumaroyl-CoA ligase; VvVST1, Vitis vinifera resveratrol synthase; SeACS1L641P, Salmonella enteric acetyl-CoA synthetase; HsPK1, Huperiza serrata chalcone synthase; gpd1, Glycerol-1-phosphatase; gal7, galactose-1-phosphate uridyl transferase; gal10, UDP-glucose-4-epimerase; gal1, Galactokinase; gal80, transcriptional regulator; aro10, phenylpyruvate decarboxylase.
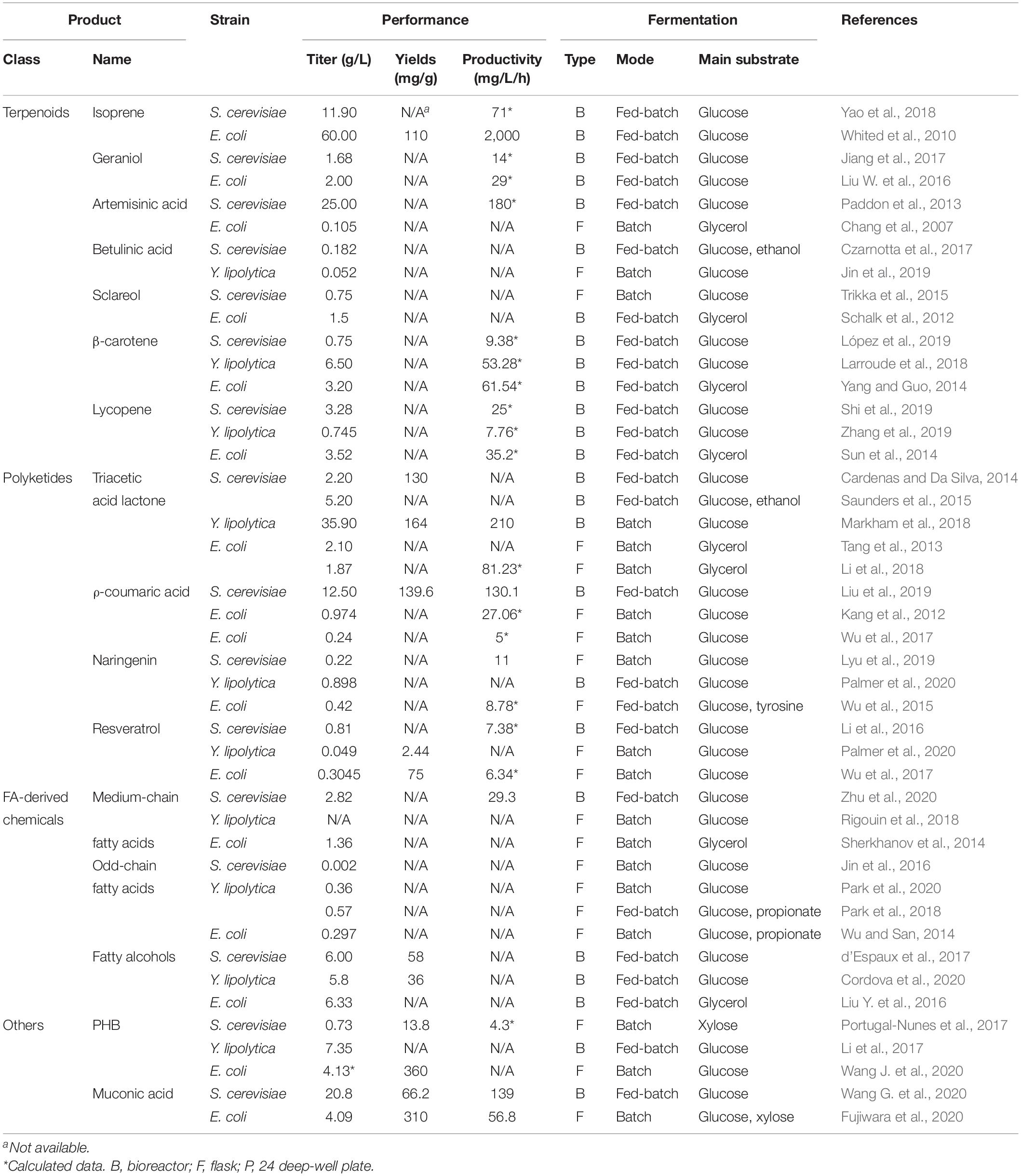
Table 1. Comparison of the production titer of non-native chemicals by engineered S. cerevisiae, Y. lipolytica and E. coli.
ρ-Coumaric Acid
ρ-Coumaric acid is an aromatic amino acid (AAA) that serves as a starting material for numerous high-value biochemicals such as flavors, fragrances, nutraceuticals, and pharmaceuticals. Recently, a recombinant diploid QL60 strain of S. cerevisiae produced 12.5 g/L of ρ-coumaric acid with a productivity of 0.13 g/L/h and a yield of 0.14 g/g sugar under glucose-limited fed-batch fermentation conditions (Liu et al., 2019). This demonstrated superior performance of S. cerevisiae in the production of ρ-coumaric acid over an E. coli strain, in which 168–974 mg/L was produced during flask culture (Kang et al., 2012; Morelli et al., 2017). This remarkable performance of ρ-coumaric acid biosynthesis in S. cerevisiae was achieved by intensive engineering; systematic engineering of the AAA pathway through debottlenecking a shikimate pathway, enhancing cytochrome P450 activity by overexpression of AtCYB5 and AtATR2; and diverting carbon flux from glycolysis to erythrose 4-phosphate (E4P) by introducing a heterologous phosphoketolase (PHK)-based pathway. Optimization of carbon flux through interconnecting points between glycolysis and the AAA pathway was also critical for high-titer ρ-coumaric acid production (Figure 3B).
Naringenin
Naringenin is a key bioactive polyketide derived from ρ-coumaric acid, from which numerous flavonoids can be synthesized. Palmer et al. (2020) demonstrated de novo production of naringenin from Y. lipolytica, with the highest titer among those using S. cerevisiae and E. coli. In this study, they introduced the tyrosine-based ρ-coumaroyl-CoA- naringenin pathway and enhanced the metabolic flux from acetyl-CoA to malonyl-CoA through the overexpression of ACC1 and PEX10 (Figure 3B). The effectiveness of the upregulation of the peroxisomal matrix protein encoded by PEX10, previously reported to strongly improve the production of TAL, increased acetyl-CoA pool via refluxing the β-oxidation pathway. Besides, the introduction of mutant and/or heterologous enzymes, ARO4K221L and a tyrosine amino lyase encoded by SeSam8 from Saccharothrix espanaensis, resolved the rate-limiting steps contributing to the higher titer (Figure 3B). The final strain of engineered Y. lipolytica produced 124.1 mg/L of naringenin in flask culture, which was further increased to 898 mg/L during bioreactor operation. Lyu et al. (2019) applied multiple approaches for enhancing the metabolic flux toward naringenin via a tyrosine-based pathway. More interestingly, they investigated the beneficial effects of the co-culture system, in which the whole pathway was divided between two strains to relieve the heavy burden of complicated biosynthesis pathways and surfactant supplementation (Tween 80) on the improved naringenin production in S. cerevisiae (Lyu et al., 2019). In a flask culture, the naringenin titer of S. cerevisiae was higher than that of Y. lipolytica (220 vs. 124.1 mg/L), implying the potential of S. cerevisiae as a production host of naringenin as demonstrated in resveratrol production (discussed in the next paragraph). In E. coli, 421.6 mg/L of naringenin was produced in flask culture condition (Wu et al., 2015).
Resveratrol
Resveratrol shares its precursor with naringenin; ρ-coumaroyl-CoA is converted either to resveratrol or naringenin by stilbene synthase (STS) or the serial reaction of chalcone synthase (CHS) and chalcone isomerase (CHI), respectively. With the introduction of a phenylalanine-based ρ-coumaroyl-CoA—route, S. cerevisiae demonstrated higher resveratrol productivity over Y. lipolytica or E. coli. Li et al. adapted the engineering strategy used for ρ-coumaric acid production; overexpression of AtATR2, ARO1/2, and ARO3 encoded by AtPAL2, EcaroL/ARO7G141S, and RO4K229L, and enhanced availability of malonyl-CoA by overexpressing SeACSL641P, ACC1S659A, S1157A encoding acetyl-CoA synthetase and acetyl-CoA carboxylase (Li et al., 2016) along with ARO10 deletion (Figure 3B; Li et al., 2016). As mentioned above, S. cerevisiae efficiently produced 812 mg/L of resveratrol during fed-batch fermentation in a bioreactor, which was similar to the naringenin production of Y. lipolytica under bioreactor conditions (Table 1). Under shake-flask conditions, Y. lipolytica produced 48.7 mg/L of resveratrol (Palmer et al., 2020). In E. coli, 304.5 mg/L of resveratrol was produced in flask culture conditions (Wu et al., 2017; Table 1). Despite the lack of post-transcriptional modification, E. coli has shown a higher-titer production of plant-based polyketides. The main engineering strategies employed in E. coli mainly focused on increasing the malonyl-CoA pool and balancing the synthetic pathway via gene expression regulation. Recent reports on the newly developed CRISPRi system, which controls gene expression through repression rather than conventional gene knock-out, applied in E. coli offered finely tuned metabolic flux effectively delivered through the newly introduced synthetic pathway (Wu et al., 2015, 2017).
Fatty Acid-Derived Chemicals
Fatty acids are essential compounds for sustaining cell membranes. With the help of synthetic biology, fatty acids are transformed into non-native chemicals with a wide range of applications. As fatty alcohols and fatty esters have been frequently discussed in previous reviews (Hu et al., 2019; Munkajohnpong et al., 2020), we focused on medium-chain and odd-chain fatty acids for which yeast engineering efforts have made noticeable progress in recent years, due to their importance as platform chemicals for the replacement of petroleum in the chemical industry.
Medium-Chain Fatty Acids (MCFAs)
MCFAs have gained attention as jet fuel replacements, platform chemicals, and ingredients for plastics and cosmetics. Recently, an MCFA-producing S. cerevisiae strain was developed by adapting a type I bacterial fatty acid synthesis pathway. Through the directed evolution of TPO1 transporter and laboratory adaptive evolution, the engineered strain with improved MCFA tolerances produced almost 1 g/L of MCFAs (Zhu et al., 2020; Figure 4A). In Y. lipolytica, the native FAS enzyme was simply modified to synthesize MCFAs based on its superior capacity for long-chain fatty acid (LCFA) production. Although the MCFA content (45%) in its total lipids was agreeable, the titer was insignificant, possibly because its FAS systems evolved toward a highly efficient LCFA production system (Rigouin et al., 2017). E. coli has also shown a similar range of MCFA titer (1.36 g/L) through metabolic engineering approaches along with enhancing the tolerance toward MCFAs (Sherkhanov et al., 2014).
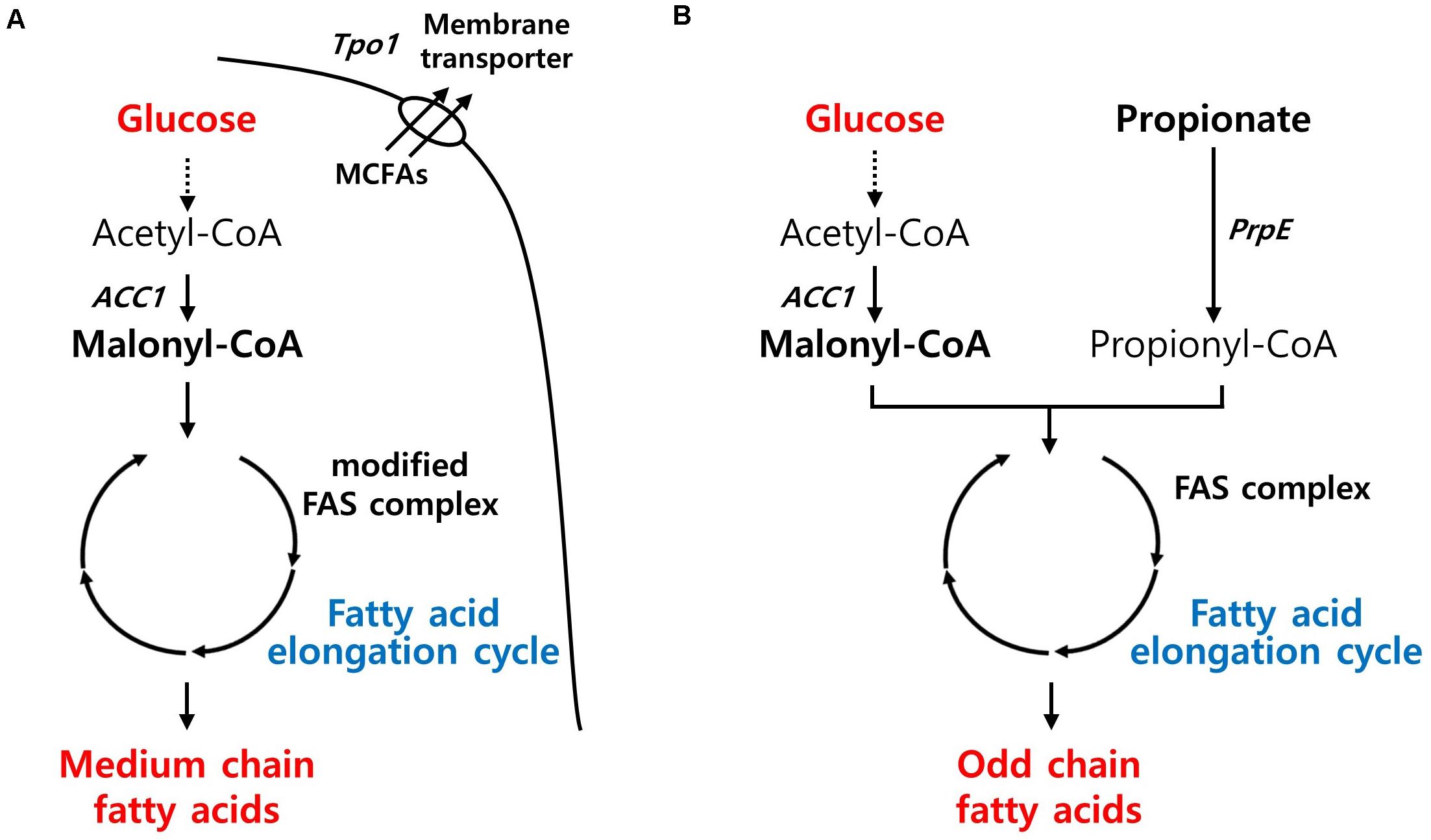
Figure 4. Biosynthetic pathway for fatty-acids derived chemicals production and main engineering targets. (A) A pathway for MCFAs production. (B) A pathway for OCFAs production. ACC1, acetyl-CoA carboxylase; Tpo1, polyamine transporter; PrpE, propionate-CoA ligase; FAS, fatty acid synthase.
Odd-Chain Fatty Acids (OCFAs)
In nature, most fatty acids derived from microorganisms contain even numbers of carbon, and rare microbes produce minute amounts of OCFAs with the exception of propionic acid. Recently, microbial production of OCFAs has also attracted considerable interest because of its potential benefits to human health, such as diabetes (Weitkunat et al., 2017) or adiposity (Aglago et al., 2017). Modifying the fatty acid synthesis pathway, in which acetyl-CoA and malonyl-CoA are used as starting materials, is not simple unless odd chain fatty acids such as propionic acid are fed as substrates. Recently, Y. lipolytica has shown promise as a cell factory for the production of OCFAs by harnessing the power of superior LCFA production capacity. When the metabolic flux toward LCFA production in an obese strain diverged toward OCFAs, Y. lipolytica produced 360 mg/L of OCFAs with a chain length of over C15. By supplying endogenous propionyl-CoA from oxaloacetate through a newly introduced synthetic pathway, the engineered strain with the genetic background for high lipid production was able to produce OCFAs without the supplementation of propionic acid (Park et al., 2020; Figure 4B). In E. coli, the overexpression of propionyl-CoA synthase from Salmonella enterica and acyl-ACP thioesterase from Ricinus communis and Umberllularia california enabled the production of 276 mg/L of OCFAs, with chain lengths of C11–C15, comparable to that in Y. lipolytica, in the presence of optimized propionate supplementation (Wu and San, 2014).
Other Chemicals
Polyhydroxybutyrate (PHB)
PHB has recently emerged as a promising candidate for the sustainable production of biodegradable thermoplastics. PHB is naturally accumulated as energy storage molecules under non-optimal conditions by several bacterial species such as Cupriavidus necator (previously known as Ralstonia eutropha) and Methylobacterium. However, these native producers still have drawbacks to be served as a production host for PHB commercialization due to their intracellular lysis of PHB, poor tolerance toward harsh environments, restriction of metabolic substrates, and unfamiliar genetic characterization (Madison and Huisman, 1999). Thus, the feasibility of PHB production in non-native hosts has been investigated. High-titer PHB production was achieved by an engineered oleaginous yeast Y. lipolytica harboring a heterologous PHB biosynthetic pathway based on PhaA, PhaB, and PhaC from a native producer of C. necator (Figure 5A; Li et al., 2017). To strengthen the availability of cytosolic acetyl-CoA, acetate, an essential precursor for PHB production, was fed as a sole carbon source, resulting in a PHB content of 10.2% with a high titer of 7.35 g/L during fed-batch fermentation in a bioreactor (Li et al., 2017). In S. cerevisiae, high PHB content was achieved during xylose fermentation (Portugal-Nunes et al., 2017). An engineered xylose-utilizing strain of S. cerevisiae with a heterologously expressed PHB pathway based on PhaA and PhaC1 from C. necator and NADH-preferred PhaB1 from Allochromatium vinosum (Figure 5A) produced 730 mg/L PHB with a content of 16.4% during xylose fermentation with additional nitrogen supply under an anaerobic shake-flask culture condition (Portugal-Nunes et al., 2017). PHB production in E. coli has been shown to be more promising than that in yeast strains. Wang et al. reported the role of truncated lipopolysaccharide (LPS) in E. coli in improving PHB production as a rebalanced carbon and nitrogen metabolism with increased precursors and energy levels. By introducing a PHB synthesis pathway based on PhaA, PhaB, and PhaC from C. necator (Figure 5A) as well as a truncated LPS, the engineered strain of E. coli produced PHB with a superior content and yield of 84.3% and 360 mg/g, respectively, under shake-flask culture conditions (Wang J. et al., 2020). In terms of PHB content and yield, the production capacity of the engineered E. coli was highly competitive with the natural PHB-producing strain of C. necator, which produced 232 g/L of PHB with a content of 80% and a yield of 380 mg/g during glucose fed-batch fermentation under phosphate limitation conditions in a bioreactor (Ryu et al., 1997).
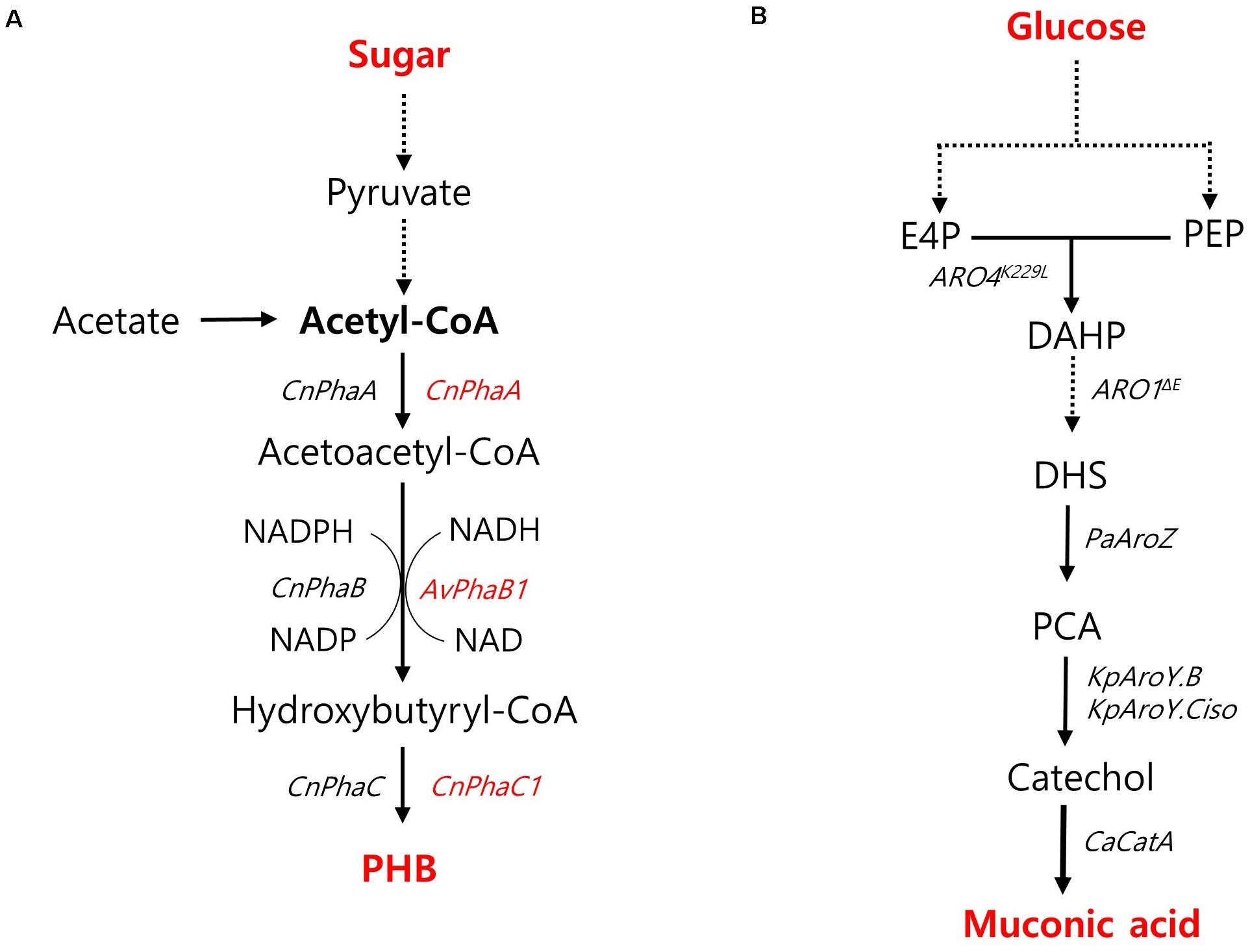
Figure 5. Biosynthetic pathway for other chemicals production and main engineering targets. (A) A pathway for PHB production. The genes specifically used for S. cerevisiae and Y. lipolytica are indicated with red and black colors, respectively. (B) A pathway for muconic acid production. Cn, Cupriavidus necator; Av, Allochromatium vinosum; PhaA, acetyl-CoA acetyl transferase; PhaB, acetoacetyl-CoA reductase; PhaC, polyhydroxyalkanoate synthase. ARO4K229L, L-tyrosine-feedback-insensitive DAHP synthase; ARO1ΔE, AROM protein without shikimate dehydrogenase domain; PaAroZ, Podospora anserine DHS dehydratase; KpAroY.B, KpAroY.Ciso, Klebsiella pneumonia PCA decarboxylase subunits; CaCatA, Candida albicans catechol 1,2-dioxygenase.
Muconic Acid
Muconic acid is another platform chemical with the potential for production of bio-plastics, such as nylon-6,6, polyurethane and polyethylene terephthalate, as well as cosmetics and pharmaceuticals (Xie et al., 2014). Wang G. et al. (2020) developed high muconic acid-producing S. cerevisiae through biosensor-aided genome engineering. From a UV-mutagenesis library, they screened out a mutant strain harboring missense mutations in several native genes, such as PWP2, EST2, ATG1, DIT1, CDC15, CTS2, and MNE1, duplicated aroZ, and catA, which are involved in the introduced muconic acid pathway (Figure 5B). The overexpression of protocatechuic acid (PCA) decarboxylase and AROM protein without the shikimate dehydrogenase domain (Aro1pΔ E) led to the high production of muconic acid, resulting in a muconic acid titer of 20.8 g/L during pH-controlled fed-batch fermentation. In E. coli, co-fermentation of glucose and xylose resulted in 4.09 g/L of muconic acid in a shake-flask condition (Fujiwara et al., 2020; Table 1).
Challenges and Opportunities in Non-Native Chemical Production
Based on success as a proven industrial cell factory for biofuel production, yeasts are transforming into a cell factory for the production of non-native chemicals. In addition to the benefit from their robustness, yeasts offer ease of reactor operation and cell separation as well as low energy consumption with lower fermentation temperature (Liu et al., 2019). By exploiting the advantages of eukaryotic systems, yeast demonstrated its potential for the biosynthesis of plant-derived metabolites by better supporting the functional expression of heterologous enzymes requiring post-translational modification or providing intracellular structures for proper enzyme activity (Siddiqui et al., 2012). For instance, membrane-bound cytochrome P450 oxidase, a crucial enzyme for the biosynthesis of plant metabolites, can be functionally expressed in yeast (Pompon et al., 1996). Due to these advantages, the two main yeast cell factories, namely S. cerevisiae and Y. lipolytica, could potentially be used for the production of plant-derived chemicals, especially terpenes with three or more isoprene units and polyketides. S. cerevisiae has been shown to produce higher titers of artemisinic and betulinic acids. Moreover, Y. lipolytica has shown comparable performance with S. cerevisiae for the production of β-carotene and lycopene (in terms of titer), even with relatively minimal engineering. The higher titers in terpene production by S. cerevisiae were often based on subcellular compartmentalization using mitochondria or peroxisomes, implying the critical role of proper intracellular structures. The potential of Y. lipolytica in triterpene production could be improved by adapting the engineering strategy of compartmentalization applied to S. cerevisiae. In the strain development for the biosynthesis of β-carotene and lycopene in S. cerevisiae and Y. lipolytica, balanced enzyme expression levels played a critical role in enhancing production titer, especially at the rate-limiting steps mediated by crtI and crtE, the two main engineering targets. Even with limited attempts, Y. lipolytica has shown greater titer in the production of β-carotene, naringenin, and TAL by taking advantage of the high accessibility of acetyl-CoA over S. cerevisiae and E. coli. As more engineering tools are developed, and thus more sophisticated tuning of gene expression would be possible, Y. lipolytica could be used as a promising industrial host for the production of tetraterpene such as β-carotene. Moreover, polycistronic gene expression systems, which allow equivalent expression and regulation of multiple genes by one promoter in bacterial hosts, have been recently investigated and successfully developed for fungal hosts such as S. cerevisiae (Souza-Moreira et al., 2018). Therefore, the construction of a non-native chemical production pathway involving numerous heterologous genes would facilitate the development of yeast cell factories. Interestingly, unconventional carbon sources other than glucose, such as xylose, acetate, and propionic acid, have been shown to better support metabolic flux through the biosynthesis pathway for non-native chemicals (Wu and San, 2014; Li et al., 2017; Portugal-Nunes et al., 2017; Wu et al., 2019). Thus, non-native chemical production by the engineered yeast strains with non-native carbon metabolism could offer better promises for industrial-scale production of non-native chemicals.
Conclusion
In this review, we discussed the current status of non-native biochemical production by model and non-model yeasts of S. cerevisiae and Y. lipolytica, respectively. The comparison of the potential of these yeasts in the production of non-native chemicals presented can serve to identify the production host of choice. By taking advantage of their distinctive cellular metabolism and characteristics as eukaryotic systems, these yeasts offer great potential for the industrial production of various non-native chemicals. Recent technological developments, such as high-throughput screening methods, are accelerating the application of synthetic biology to develop and upgrade yeast cell factories in a more efficient manner. Through target-specific engineering strategies with an expanded carbon source portfolio and optimized fermentation conditions of a single/co-culture system, yeasts could be successfully transformed into favorable industrial cell factories for the production of a wide range of non-native chemicals.
Author Contributions
JK and PH wrote and edited the manuscript. S-ML provided conception of this review and edited the manuscript. All authors contributed to the article and approved the submitted manuscript.
Funding
This work was supported by the Korea Institute of Science and Technology (KIST) Institutional Program (Grant No. 2E30170). The authors also appreciate the additional support by the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (Information & Communication Technology) (Grant No. NRF-2020M1A2A2080847).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aglago, E. K., Biessy, C., Torres-Mejia, G., Angeles-Llerenas, A., Gunter, M. J., Romieu, I., et al. (2017). Association between serum phospholipid fatty acid levels and adiposity in Mexican women. J. Lipid Res. 58, 1462–1470. doi: 10.1194/jlr.P073643
An, T., Zha, W., and Zi, J. (2020). Biotechnological production of betulinic acid and derivatives and their applications. Appl. Microbiol. Biotechnol. 104, 3339–3348. doi: 10.1007/s00253-020-10495-1
Arendt, P., Pollier, J., Callewaert, N., and Goossens, A. (2016). Synthetic biology for production of natural and new-to-nature terpenoids in photosynthetic organisms. Plant J. 87, 16–37. doi: 10.1111/tpj.13138
Cao, X., Wei, L.-J., Lin, J.-Y., and Hua, Q. (2017). Enhancing linalool production by engineering oleaginous yeast Yarrowia lipolytica. Bioresour. Technol. 245, 1641–1644. doi: 10.1016/j.biortech.2017.06.105
Cardenas, J., and Da Silva, N. A. (2014). Metabolic engineering of Saccharomyces cerevisiae for the production of triacetic acid lactone. Metab. Eng. 25, 194–203. doi: 10.1016/j.ymben.2014.07.008
Chang, M. C. Y., Eachus, R. A., Trieu, W., Ro, D.-K., and Keasling, J. D. (2007). Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat. Chem. Biol. 3, 274–277. doi: 10.1038/nchembio875
Chang, M. C. Y., and Keasling, J. D. (2006). Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2, 674–681. doi: 10.1038/nchembio836
Cheng, B.-Q., Wei, L.-J., Lv, Y.-B., Chen, J., and Hua, Q. (2019). Elevating limonene production in oleaginous Yeast Yarrowia lipolytica via genetic engineering of limonene biosynthesis pathway and optimization of medium composition. Biotechnol. Bioprocess Eng. 24, 500–506. doi: 10.1007/s12257-018-0497-9
Chia, M., Schwartz, T. J., Shanks, B. H., and Dumesic, J. A. (2012). Triacetic acid lactone as a potential biorenewable platform chemical. Green Chem. 14, 1850–1853. doi: 10.1039/C2GC35343A
Cordova, L. T., Butler, J., and Alper, H. S. (2020). Direct production of fatty alcohols from glucose using engineered strains of Yarrowia lipolytica. Metab. Eng. Commun. 10:e00105. doi: 10.1016/j.mec.2019.e00105
Czarnotta, E., Dianat, M., Korf, M., Granica, F., Merz, J., Maury, J., et al. (2017). Fermentation and purification strategies for the production of betulinic acid and its lupane-type precursors in Saccharomyces cerevisiae. Biotechnol. Bioeng. 114, 2528–2538. doi: 10.1002/bit.26377
Darvishi, F., Fathi, Z., Ariana, M., and Moradi, H. (2017). Yarrowia lipolytica as a workhorse for biofuel production. Biochem. Eng. J. 127, 87–96. doi: 10.1016/j.bej.2017.08.013
d’Espaux, L., Ghosh, A., Runguphan, W., Wehrs, M., Xu, F., Konzock, O., et al. (2017). Engineering high-level production of fatty alcohols by Saccharomyces cerevisiae from lignocellulosic feedstocks. Metab. Eng. 42, 115–125. doi: 10.1016/j.ymben.2017.06.004
Ekas, H., Deaner, M., and Alper, H. (2019). Recent advancements in fungal-derived fuel and chemical production and commercialization. Curr. Opin. Biotechnol. 57, 1–9. doi: 10.1016/j.copbio.2018.08.014
Favaro, L., Jansen, T., and van Zyl, W. H. (2019). Exploring industrial and natural Saccharomyces cerevisiae strains for the bio-based economy from biomass: the case of bioethanol. Crit. Rev. Biotechnol. 39, 800–816. doi: 10.1080/07388551.2019.1619157
Fischer, M. J. C., Meyer, S., Claudel, P., Bergdoll, M., and Karst, F. (2011). Metabolic engineering of monoterpene synthesis in yeast. Biotechnol. Bioeng. 108, 1883–1892. doi: 10.1002/bit.23129
Fujiwara, R., Noda, S., Tanaka, T., and Kondo, A. (2020). Metabolic engineering of Escherichia coli for shikimate pathway derivative production from glucose–xylose co-substrate. Nat. Commun. 11:279. doi: 10.1038/s41467-019-14024-1
Gray, D., Sato, S., Garcia, F., Eppler, R., and Cherry, J. (2014). Integrated Biorefinery Project Summary Final Report - Public Version. Emeryville, CA: Amyris, Inc.
Hu, Y., Zhu, Z., Nielsen, J., and Siewers, V. (2019). Engineering Saccharomyces cerevisiae cells for production of fatty acid-derived biofuels and chemicals. Open Biol. 9:190049. doi: 10.1098/rsob.190049
Ignea, C., Pontini, M., Maffei, M. E., Makris, A. M., and Kampranis, S. C. (2014). Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synthetic Biol. 3, 298–306. doi: 10.1021/sb400115e
Jakočiūnas, T., Klitgaard, A. K., Kontou, E. E., Nielsen, J. B., Thomsen, E., Romero-Suarez, D., et al. (2020). Programmable polyketide biosynthesis platform for production of aromatic compounds in yeast. Synth. Syst. Biotechnol. 5, 11–18. doi: 10.1016/j.synbio.2020.01.004
Jiang, G.-Z., Yao, M.-D., Wang, Y., Zhou, L., Song, T.-Q., Liu, H., et al. (2017). Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae. Metab. Eng. 41, 57–66. doi: 10.1016/j.ymben.2017.03.005
Jin, C. C., Zhang, J. L., Song, H., and Cao, Y. X. (2019). Boosting the biosynthesis of betulinic acid and related triterpenoids in Yarrowia lipolytica via multimodular metabolic engineering. Microb. Cell Fact. 18:77. doi: 10.1186/s12934-019-1127-8
Jin, Z., Wong, A., Foo, J. L., Ng, J., Cao, Y.-X., Chang, M. W., et al. (2016). Engineering Saccharomyces cerevisiae to produce odd chain-length fatty alcohols. Biotechnol. Bioeng. 113, 842–851. doi: 10.1002/bit.25856
Kang, S.-Y., Choi, O., Lee, J. K., Hwang, B. Y., Uhm, T.-B., and Hong, Y.-S. (2012). Artificial biosynthesis of phenylpropanoic acids in a tyrosine overproducing Escherichia coli strain. Microb. Cell Fact. 11:153. doi: 10.1186/1475-2859-11-153
Larroude, M., Celinska, E., Back, A., Thomas, S., Nicaud, J.-M., and Ledesma-Amaro, R. (2018). A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol. Bioeng. 115, 464–472. doi: 10.1002/bit.26473
Li, M., Schneider, K., Kristensen, M., Borodina, I., and Nielsen, J. (2016). Engineering yeast for high-level production of stilbenoid antioxidants. Sci. Rep. 6:36827. doi: 10.1038/srep36827
Li, Y., Qian, S., Dunn, R., and Cirino, P. C. (2018). Engineering Escherichia coli to increase triacetic acid lactone (TAL) production using an optimized TAL sensor-reporter system. J. Industr. Microbiol. Biotechnol. 45, 789–793. doi: 10.1007/s10295-018-2062-0
Li, Z.-J., Qiao, K., Liu, N., and Stephanopoulos, G. (2017). Engineering Yarrowia lipolytica for poly-3-hydroxybutyrate production. J. Industr. Microbiol. Biotechnol. 44, 605–612. doi: 10.1007/s10295-016-1864-1
Liu, Q., Yu, T., Li, X., Chen, Y., Campbell, K., Nielsen, J., et al. (2019). Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 10:4976. doi: 10.1038/s41467-019-12961-5
Liu, W., Xu, X., Zhang, R., Cheng, T., Cao, Y., Li, X., et al. (2016). Engineering Escherichia coli for high-yield geraniol production with biotransformation of geranyl acetate to geraniol under fed-batch culture. Biotechnol. Biofuels 9:58. doi: 10.1186/s13068-016-0466-5
Liu, Y., Chen, S., Chen, J., Zhou, J., Wang, Y., Yang, M., et al. (2016). High production of fatty alcohols in Escherichia coli with fatty acid starvation. Microb. Cell Fact. 15:129. doi: 10.1186/s12934-016-0524-5
López, J., Cataldo, V. F., Peña, M., Saa, P. A., Saitua, F., Ibaceta, M., et al. (2019). Build Your Bioprocess on a Solid Strain—β-Carotene Production in Recombinant Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 7:171. doi: 10.3389/fbioe.2019.00171
Luo, X., Reiter, M. A., d’Espaux, L., Wong, J., Denby, C. M., Lechner, A., et al. (2019). Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 567, 123–126. doi: 10.1038/s41586-019-0978-9
Lyu, X., Zhao, G., Ng, K. R., Mark, R., and Chen, W. N. (2019). Metabolic Engineering of Saccharomyces cerevisiae for De Novo Production of Kaempferol. J. Agricult. Food Chem. 67, 5596–5606. doi: 10.1021/acs.jafc.9b01329
Ma, Y.-R., Wang, K.-F., Wang, W.-J., Ding, Y., Shi, T.-Q., Huang, H., et al. (2019). Advances in the metabolic engineering of Yarrowia lipolytica for the production of terpenoids. Bioresource Technol. 281, 449–456. doi: 10.1016/j.biortech.2019.02.116
Madison, L. L., and Huisman, G. W. (1999). Metabolic Engineering of Poly(3-Hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21. doi: 10.1128/MMBR.63.1.21-53.1999
Markham, K. A., and Alper, H. S. (2018). Synthetic biology expands the industrial potential of Yarrowia lipolytica. Trends Biotechnol. 36, 1085–1095.
Markham, K. A., Palmer, C. M., Chwatko, M., Wagner, J. M., Murray, C., Vazquez, S., et al. (2018). Rewiring Yarrowia lipolytica toward triacetic acid lactone for materials generation. Proc. Natl. Acad. Sci. U.S.A. 115:2096. doi: 10.1073/pnas.1721203115
Marsafari, M., and Xu, P. (2020). Debottlenecking mevalonate pathway for antimalarial drug precursor amorphadiene biosynthesis in Yarrowia lipolytica. Metab. Eng. Commun. 10:e00121. doi: 10.1016/j.mec.2019.e00121
Miller, K. K., and Alper, H. S. (2019). Yarrowia lipolytica: more than an oleaginous workhorse. Appl. Microbiol. Biotechnol. 103, 9251–9262. doi: 10.1007/s00253-019-10200-x
Morelli, L., Zór, K., Jendresen, C. B., Rindzevicius, T., Schmidt, M. S., Nielsen, A. T., et al. (2017). Surface Enhanced Raman Scattering for Quantification of p-Coumaric Acid Produced by Escherichia coli. Anal. Chem. 89, 3981–3987. doi: 10.1021/acs.analchem.6b04428
Munkajohnpong, P., Kesornpun, C., Buttranon, S., Jaroensuk, J., Weeranoppanant, N., and Chaiyen, P. (2020). Fatty alcohol production: an opportunity of bioprocess. Biofuels Bioprod. Biorefining 14, 986–1009. doi: 10.1002/bbb.2112
Oloyede, H. O. B., Ajiboye, H. O., Salawu, M. O., and Ajiboye, T. O. (2017). Influence of oxidative stress on the antibacterial activity of betulin, betulinic acid and ursolic acid. Microb. Pathog. 111, 338–344. doi: 10.1016/j.micpath.2017.08.012
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051
Palmer, C. M., Miller, K. K., Nguyen, A., and Alper, H. S. (2020). Engineering 4-coumaroyl-CoA derived polyketide production in Yarrowia lipolytica through a β-oxidation mediated strategy. Metab. Eng. 57, 174–181. doi: 10.1016/j.ymben.2019.11.006
Parapouli, M., Vasileiadis, A., Afendra, A. S., and Hatziloukas, E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31. doi: 10.3934/microbiol.2020001
Park, Y.-K., Dulermo, T., Ledesma-Amaro, R., and Nicaud, J.-M. (2018). Optimization of odd chain fatty acid production by Yarrowia lipolytica. Biotechnol. Biofuels 11:158. doi: 10.1186/s13068-018-1154-4
Park, Y. K., Ledesma-Amaro, R., and Nicaud, J.-M. (2020). De novo biosynthesis of odd-chain fatty Acids in Yarrowia lipolytica Enabled by Modular Pathway Engineering. Front. Bioeng. Biotechnol. 7:484. doi: 10.3389/fbioe.2019.00484
Ping, Y., Li, X., You, W., Li, G., Yang, M., Wei, W., et al. (2019). De novo production of the plant-derived tropine and pseudotropine in yeast. ACS Synthetic Biol. 8, 1257–1262. doi: 10.1021/acssynbio.9b00152
Pompon, D., Louerat, B., Bronine, A., and Urban, P. (1996). “Yeast expression of animal and plant P450s in optimized redox environments,” in Methods in Enzymology, eds E. F. Johnson and M. R. Waterman (Cambridge, MA: Academic Press), 51–64. doi: 10.1016/s0076-6879(96)72008-6
Portugal-Nunes, D. J., Pawar, S. S., Lidén, G., and Gorwa-Grauslund, M. F. (2017). Effect of nitrogen availability on the poly-3-d-hydroxybutyrate accumulation by engineered Saccharomyces cerevisiae. AMB Exp. 7:35. doi: 10.1186/s13568-017-0335-z
Qiao, K., Wasylenko, T. M., Zhou, K., Xu, P., and Stephanopoulos, G. (2017). Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat. Biotechnol. 35, 173–177. doi: 10.1038/nbt.3763
Rigouin, C., Croux, C., Borsenberger, V., Ben Khaled, M., Chardot, T., Marty, A., et al. (2018). Increasing medium chain fatty acids production in Yarrowia lipolytica by metabolic engineering. Microb. Cell Fact. 17:142. doi: 10.1186/s12934-018-0989-5
Rigouin, C., Gueroult, M., Croux, C., Dubois, G., Borsenberger, V., Barbe, S., et al. (2017). Production of medium chain fatty acids by Yarrowia lipolytica: combining molecular design and TALEN to engineer the fatty acid Synthase. ACS Synthet. Biol. 6, 1870–1879. doi: 10.1021/acssynbio.7b00034
Ro, D.-K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. doi: 10.1038/nature04640
Ryu, H. W., Hahn, S. K., Chang, Y. K., and Chang, H. N. (1997). Production of poly(3-hydroxybutyrate) by high cell density fed-batch culture of Alcaligenes eutrophus with phospate limitation. Biotechnol. Bioeng. 55, 28–32. doi: 10.1002/(sici)1097-0290(19970705)55:1<28::aid-bit4>3.0.co;2-z
Saunders, L. P., Bowman, M. J., Mertens, J. A., Da Silva, N. A., and Hector, R. E. (2015). Triacetic acid lactone production in industrial Saccharomyces yeast strains. J. Industr. Microbiol. Biotechnol. 42, 711–721. doi: 10.1007/s10295-015-1596-7
Schalk, M., Pastore, L., Mirata, M. A., Khim, S., Schouwey, M., Deguerry, F., et al. (2012). Toward a biosynthetic route to sclareol and amber odorants. J. Am. Chem. Soc. 134, 18900–18903. doi: 10.1021/ja307404u
Sherkhanov, S., Korman, T. P., and Bowie, J. U. (2014). Improving the tolerance of Escherichia coli to medium-chain fatty acid production. Metab. Eng. 25, 1–7. doi: 10.1016/j.ymben.2014.06.003
Shi, B., Ma, T., Ye, Z., Li, X., Huang, Y., Zhou, Z., et al. (2019). Systematic Metabolic Engineering of Saccharomyces cerevisiae for Lycopene Overproduction. J. Agricult. Food Chem. 67, 11148–11157. doi: 10.1021/acs.jafc.9b04519
Siddiqui, M. S., Thodey, K., Trenchard, I., and Smolke, C. D. (2012). Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 12, 144–170. doi: 10.1111/j.1567-1364.2011.00774.x
Souza-Moreira, T. M., Navarrete, C., Chen, X., Zanelli, C. F., Valentini, S. R., Furlan, M., et al. (2018). Screening of 2A peptides for polycistronic gene expression in yeast. FEMS Yeast Res. 18. doi: 10.1093/femsyr/foy036
Srinivasan, P., and Smolke, C. D. (2019). Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids. Nat. Commun. 10:3634. doi: 10.1038/s41467-019-11588-w
Sun, T., Miao, L., Li, Q., Dai, G., Lu, F., Liu, T., et al. (2014). Production of lycopene by metabolically-engineered Escherichia coli. Biotechnol. Lett. 36, 1515–1522. doi: 10.1007/s10529-014-1543-0
Tang, S.-Y., Qian, S., Akinterinwa, O., Frei, C. S., Gredell, J. A., and Cirino, P. C. (2013). Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter. J. Am. Chem. Soc. 135, 10099–10103. doi: 10.1021/ja402654z
Trikka, F. A., Nikolaidis, A., Athanasakoglou, A., Andreadelli, A., Ignea, C., Kotta, K., et al. (2015). Iterative carotenogenic screens identify combinations of yeast gene deletions that enhance sclareol production. Microb. Cell Fact. 14:60. doi: 10.1186/s12934-015-0246-0
Vickers, C. E., Williams, T. C., Peng, B., and Cherry, J. (2017). Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 40, 47–56. doi: 10.1016/j.cbpa.2017.05.017
Walker, G., and Stewart, G. (2016). Saccharomyces cerevisiae in the Production of Fermented Beverages. Beverages 2:30. doi: 10.3390/beverages2040030
Wang, C., Liwei, M., Park, J.-B., Jeong, S.-H., Wei, G., Wang, Y., et al. (2018). Microbial platform for terpenoid production: Escherichia coli and Yeast. Front. Microbiol. 9:2460. doi: 10.3389/fmicb.2018.02460
Wang, G., Øzmerih, S., Guerreiro, R., Meireles, A. C., Carolas, A., Milne, N., et al. (2020). Improvement of cis, cis-Muconic Acid Production in Saccharomyces cerevisiae through Biosensor-Aided Genome Engineering. ACS Synth. Biol. 9, 634–646. doi: 10.1021/acssynbio.9b00477
Wang, J., Ma, W., Fang, Y., Zhang, H., Liang, H., Li, Y., et al. (2020). Truncating the Structure of Lipopolysaccharide in Escherichia coli Can Effectively Improve Poly-3-hydroxybutyrate Production. ACS Synth. Biol. 9, 1201–1215. doi: 10.1021/acssynbio.0c00071
Wehrs, M., Tanjore, D., Eng, T., Lievense, J., Pray, T. R., and Mukhopadhyay, A. (2019). Engineering robust production microbes for large-scale cultivation. Trends Microbiol. 27, 524–537. doi: 10.1016/j.tim.2019.01.006
Weitkunat, K., Schumann, S., Nickel, D., Hornemann, S., Petzke, K. J., Schulze, M. B., et al. (2017). Odd-chain fatty acids as a biomarker for dietary fiber intake: a novel pathway for endogenous production from propionate. Am. J. Clin. Nutr. 105, 1544–1551. doi: 10.3945/ajcn.117.152702
Whited, G. M., Feher, F. J., Benko, D. A., Cervin, M. A., Chotani, G. K., McAuliffe, J. C., et al. (2010). TECHNOLOGY UPDATE: development of a gas-phase bioprocess for isoprene-monomer production using metabolic pathway engineering. Industr. Biotechnol. 6, 152–163. doi: 10.1089/ind.2010.6.152
Wu, H., and San, K.-Y. (2014). Engineering Escherichia coli for odd straight medium chain free fatty acid production. Appl. Microbiol. Biotechnol. 98, 8145–8154. doi: 10.1007/s00253-014-5882-5
Wu, J., Du, G., Chen, J., and Zhou, J. (2015). Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci. Rep. 5:13477. doi: 10.1038/srep13477
Wu, J., Zhou, P., Zhang, X., and Dong, M. (2017). Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J. Industr. Microbiol. Biotechnol. 44, 1083–1095. doi: 10.1007/s10295-017-1937-9
Wu, Y., Xu, S., Gao, X., Li, M., Li, D., and Lu, W. (2019). Enhanced protopanaxadiol production from xylose by engineered Yarrowia lipolytica. Microb. Cell Fact. 18:83. doi: 10.1186/s12934-019-1136-7
Xie, N.-Z., Liang, H., Huang, R.-B., and Xu, P. (2014). Biotechnological production of muconic acid: current status and future prospects. Biotechnol. Adv. 32, 615–622. doi: 10.1016/j.biotechadv.2014.04.001
Yang, J., and Guo, L. (2014). Biosynthesis of β-carotene in engineered E. coli using the MEP and MVA pathways. Microb. Cell Fact. 13:160. doi: 10.1186/s12934-014-0160-x
Yao, Z., Zhou, P., Su, B., Su, S., Ye, L., and Yu, H. (2018). Enhanced Isoprene Production by Reconstruction of Metabolic Balance between Strengthened Precursor Supply and Improved Isoprene Synthase in Saccharomyces cerevisiae. ACS Synth. Biol. 7, 2308–2316. doi: 10.1021/acssynbio.8b00289
Zhang, C., and Hong, K. (2020). Production of Terpenoids by Synthetic Biology Approaches. Front. Bioeng. Biotechnol. 8:347. doi: 10.3389/fbioe.2020.00347
Zhang, X.-K., Nie, M.-Y., Chen, J., Wei, L.-J., and Hua, Q. (2019). Multicopy integrants of crt genes and co-expression of AMP deaminase improve lycopene production in Yarrowia lipolytica. J. Biotechnol. 289, 46–54. doi: 10.1016/j.jbiotec.2018.11.009
Zhou, Y. J. (2018). Expanding the terpenoid kingdom. Nat. Chem. Biol. 14, 1069–1070. doi: 10.1038/s41589-018-0167-4
Keywords: non-native chemicals, Saccharomyces cerevisiae, Yarrowia lipolytica, yeast engineering, biorefinery
Citation: Kim J, Hoang Nguyen Tran P and Lee S-M (2020) Current Challenges and Opportunities in Non-native Chemical Production by Engineered Yeasts. Front. Bioeng. Biotechnol. 8:594061. doi: 10.3389/fbioe.2020.594061
Received: 12 August 2020; Accepted: 24 November 2020;
Published: 14 December 2020.
Edited by:
Farshad Darvishi, Alzahra University, IranReviewed by:
Jiazhang Lian, Zhejiang University, ChinaMario Andrea Marchisio, Tianjin University, China
Fu-Xing Niu, Sun Yat-sen University, China
Copyright © 2020 Kim, Hoang Nguyen Tran and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sun-Mi Lee, c21sZWVAa2lzdC5yZS5rcg==
†These authors share first authorship
 Jiwon Kim
Jiwon Kim Phuong Hoang Nguyen Tran
Phuong Hoang Nguyen Tran Sun-Mi Lee
Sun-Mi Lee